- 1Department of Neurology, Zhoushan Hospital, Wenzhou Medical University, Zhoushan, Zhejiang, China
- 2Centre for Intelligent Healthcare, Coventry University, Coventry, United Kingdom
- 3National Medical Research Association, Leicester, United Kingdom
- 4Department of Neurology, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 5Department of Medical Genetics and Center for Rare Diseases, Second Affiliated Hospital, Zhejiang University School of Medicine and Zhejiang Key Laboratory of Rare Diseases for Precision Medicine and Clinical Translation, Hangzhou, Zhejiang, China
Editorial on the Research Topic
Genetic insights and diagnostic innovations in cerebrovascular and cerebrospinal fluid disorders
Genetics-centered precision neurology is reshaping both research and clinical practice in cerebrovascular disease and cerebrospinal fluid-related disorders. Technological advancements in multimodal neuroimaging, standardized laboratory testing, next-generation sequencing, transcriptomics, and extracellular-vesicle analytics are collectively accelerating the acquisition, management, processing, and interpretation of novel biomarkers for clinical practice. The impact is evident in earlier etiologic clarification, finer-grained risk stratification, and biomarker-informed monitoring that spans the acute phase through rehabilitation. Within this Research Topic, Genetic Insights and Diagnostic Innovations in Cerebrovascular and Cerebrospinal Fluid Disorders, we assembled 13 contributions—reviews, bioinformatics analyses, clinical cohort studies, neuroimaging investigations, and preclinical experimentation—that collectively illustrate how genetics and novel biomarkers are being applied to cerebrovascular and cerebrospinal fluid-related diseases. Taken together, these studies provide up-to-date examples of how molecular insights can be translated into implementable diagnostic tools and mechanism-informed therapeutic strategies. The details are summarized in Supplementary Table 1.
We observed that multiple low-cost and readily obtainable blood biomarkers show promise for risk stratification and outcome prediction. Building on a randomized study, Mitra et al. showed that Short Message Service–guided exercise improved post-stroke six-minute walk test (6MWT) performance and attenuated the decline in brain-derived neurotrophic factor (BDNF). Changes in choline acetyltransferase (ChAT) activity and in the ChAT/butyrylcholinesterase (ChAT/BChE) index correlated with the 6MWT outcomes, with stronger signals observed in women for ChAT activity. This synchronized acquisition of biomarker and behavioral endpoints highlights the potential of peripheral markers as tools for monitoring treatment response.
In an observational analysis of 1,470 older adults with acute ischemic stroke (AIS), Huang et al. identified a non-linear inverse association between a lower hemoglobin-to-red blood cell distribution width ratio (HRR) and a higher risk of unfavorable 3-month outcomes. Restricted cubic spline modeling revealed an optimal inflection point at 10.70, with an area under the curve of approximately 0.64. Lower HRR values signaled greater risk. As a zero-additional-cost index derived from routine hematology tests, the HRR may offer practical value for bedside risk stratification.
In a cohort of AIS patients treated with intravenous thrombolysis, Li et al. evaluated the etiology-dependent prognostic value of lymphocyte-related ratios. The neutrophil-to-lymphocyte ratio (NLR) consistently predicted 90-day outcomes across the Trial of Org 10,172 in Acute Stroke Treatment (TOAST) subtypes with subtype-specific cutoffs and was associated with adverse outcomes in each category, whereas the lymphocyte-to-monocyte ratio (LMR) showed predictive value primarily in the large-artery atherosclerosis subtype.
Using a large United States adult cohort and multivariable logistic regression, Ye et al. examined the neutrophil percentage-to-albumin ratio (NPAR) in relation to stroke prevalence and found that a higher NPAR was associated with greater prevalence. These findings provide new insights for primary prevention and support the NPAR as a practical tool for estimating stroke likelihood.
Drawing on a clinical database of critically ill patients with acute brain injury, Wang J. et al. assessed the stress hyperglycemia ratio (SHR) and showed that it independently predicts both short- and long-term mortality. When combined with the Glasgow Coma Scale (GCS) and ventilation status, the SHR further improved risk stratification, supporting its use as a practical and feasible quantitative metric in intensive care settings.
In a large cross-sectional analysis of the National Health and Nutrition Examination Survey (NHANES), Xu H. et al. used weighted multivariable models with stratified interaction testing and provided the first population-level evidence of an independent inverse association between the serum klotho and stroke risk. The association was consistent across most subgroups. These results suggest that anti-aging endocrine pathways may modulate cerebrovascular risk, indicating the possibility of developing hormone biomarker panels for risk stratification.
With respect to genetics and large-vessel structural phenotypes, Wang J.-W. et al. examined the association between the apolipoprotein E (APOE) genotype and extracranial carotid artery (ECA) tortuosity in a Chinese cohort and found that the ε2 allele may be associated with the increased tortuosity of the ECA, whereas the ε4 allele might be a protective factor. These observations suggest that lipid-metabolism genotypes may influence the geometry of cerebral arteries.
By integrating peripheral blood transcriptomes from multiple Gene Expression Omnibus cohorts, Wang and Liu further identified ABCA1, CLEC4E, and IRS2 as potential key biomarkers and therapeutic targets for cardioembolic stroke and ischemic stroke, serving as shared feature genes. Their expression correlates closely with neutrophil infiltration and autophagy activation, and a nomogram based on these markers demonstrates potential clinical applicability. This progression from single-gene signals to systems-level networks suggests translatable diagnostic and therapeutic targets.
In symptomatic intracranial atherosclerotic stenosis (sICAS), Xu X. et al. linked cerebral perfusion patterns to infarct topography and early neurological outcomes. Specific perfusion abnormality profiles were associated with cortical and subcortical infarct distributions, as well as short-term clinical trajectories, underscoring the central role of hemodynamic compromise in the risk stratification of sICAS and identifying the candidates for intensified hemodynamic management. The penumbra-infarct core mismatch volume in CT perfusion, with a Tmax of >4s defining the penumbra, was associated with early neurological outcomes in patients with sICAS.
On the translational front, using a murine transient middle cerebral artery occlusion model, Wang L.-P. et al. evaluated the anti-inflammatory and blood–brain barrier (BBB)–protective effects of oligodendrocyte precursor cell (OPC) transplantation. OPCs reduced neuroinflammation, preserved BBB integrity, decreased infarct volume, and improved neurobehavioral outcomes, with benefits associated with Wnt/β-catenin signaling. These findings indicate a promising therapeutic strategy for ischemic stroke.
From a phytochemistry perspective, Yu et al. reviewed natural compounds derived from traditional Chinese medicine (TCM) that regulate microglial polarization to achieve neuroprotection after ischemic stroke. Multiple classes of compounds inhibit pro-inflammatory polarization and/or promote protective polarization, thereby exerting neuroprotective effects within a multitarget network. The review systematically catalogs candidate molecules and pathways, summarizes delivery innovations, and emphasizes the need for standardized pharmacology, pharmacokinetics, and quality control to advance standardized and personalized TCM treatment and management of ischemic stroke.
Liang et al. presented a comprehensive synthesis of blood-based biomarkers in ischemic stroke, covering coagulation and fibrinolysis pathways, endothelial dysfunction markers, inflammatory mediators, neuronal and axonal injury markers, and extracellular vesicles with their circular RNAs. The review also surveys contemporary detection platforms and assay methodologies, providing critical guidance for clinical implementation. Across these categories, many candidates show promise for etiologic subtyping, early neurological deterioration, and prognostic assessment, thereby bridging molecular mechanisms with deployable diagnostic assays.
Using bibliometric methods, Ding et al. comprehensively appraised exosome research in ischemic stroke, focusing on endogenous and therapeutic exosomes, engineered cargo, and delivery across the BBB. This data-driven landscape provides valuable references and resources to guide further exploration of exosome-based diagnostics and therapeutics.
This Research Topic delineates some research hotspots of genetics in cerebral circulation and relevant diseases. The Research Topic spans bibliometric analysis and methodological reviews (Liang et al.; Ding et al.), low-cost hematologic ratios for risk stratification and prognosis (Huang et al.; Li et al.; Ye et al.; Wang J. et al.), endocrine and rehabilitation-related markers (Mitra et al.; Xu H. et al.), genetic and molecular biomarkers (Wang J.-W. et al.; Wang and Liu; Liang et al.; Ding et al.), and perfusion-based imaging phenotypes linked to early clinical outcomes (Xu X. et al.). The cell-based and natural-product interventions establish a foundation for mechanism-guided therapies (Wang L.-P. et al.; Yu et al.). These contributions also advance clinical decision support by integrating inexpensive hematologic indexes with imaging and transcriptomic information, aiming to enhance diagnostic precision for acute management, etiologic classification, prognostic stratification, and rehabilitation follow-up.
Not with standing this progress, several challenges remain: insufficient availability of high-quality specimens and multicenter external validation (Mitra et al.; Huang et al.; Li et al.; Ye et al.; Wang J. et al.; Xu H. et al.; Wang J.-W. et al.; Xu X. et al.), lack of standardization in pre-analytical workflows and analytical platforms (Wang and Liu; Liang et al.; Ding et al.), limited interpretability and portability of multimodal models (Wang and Liu; Ding et al.), constraints in clinical integration and turnaround time (Liang et al.; Ding et al.), incomplete translational and regulatory pathways (Wang L.-P. et al.; Yu et al.), and ethics concerns regarding equity and accessibility (Ye et al.; Xu H. et al.). Figure 1 provides an overview of the selected contributions and maps current limitations and future research directions. Future research priorities include establishing multicenter prospective cohorts and biobanks, refining standardized preprocessing and quality-control frameworks, and developing computational models based on multimodal big data. Current efforts could be coupled with advanced computational modeling—such as computational fluid dynamics (CFD), fluid–structure interaction (FSI), and multiphysics simulation—to reconstruct cerebral and cerebrospinal fluid dynamics (1–4). Genomics and multi-omics can delineate risk loci and pathways for pathological analysis (5–8). Furthermore, interpretable artificial intelligence (AI) approaches that integrate neuroimaging, biochemical, genetic, and hemodynamic features, along with large-scale multimodal clinical data, can improve diagnostic accuracy and prognostic performance (9–13). In parallel, mechanism-anchored early-phase translation should be accelerated to support the development and evaluation of targeted interventions. Taken together, these computational and data-driven approaches will enable mechanistic elucidation, early diagnosis, and the optimization of interventions toward individualized therapy and precise medicine.
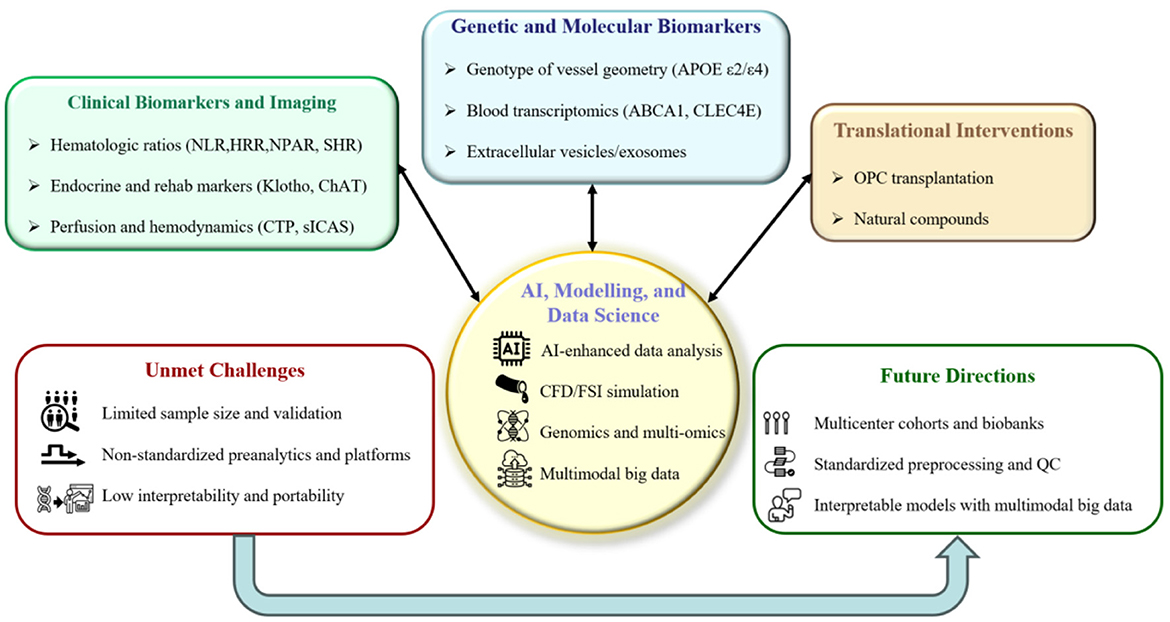
Figure 1. Overview of this Research Topic. The top colored panels summarize the Research Topics represented by the selected contributions: (i) clinical biomarkers and imaging; (ii) genetic and molecular biomarkers; and (iii) translational interventions. The central hub depicts AI, modeling, and data science, which are also prioritized as future directions. The bottom outline panels synthesize the limitations of the current state of the art and the corresponding priorities for future research.
Author contributions
LLi: Methodology, Visualization, Conceptualization, Writing – original draft, Investigation, Writing – review & editing. HL: Writing – original draft, Writing – review & editing, Investigation, Validation, Visualization, Supervision, Conceptualization. LLa: Validation, Writing – review & editing, Formal analysis. HY: Visualization, Formal analysis, Writing – original draft, Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82201513).
Acknowledgments
We appreciate the contributors to this Research Topic.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2025.1691759/full#supplementary-material
Abbreviations
AI, artificial intelligence; APOE, apolipoprotein E; CFD, computational fluid dynamics; ChAT, choline acetyltransferase; CTP, computed tomography perfusion; FSI, fluid–structure interaction; HRR, hemoglobin-to-red blood cell distribution width ratio; QC, quality control; NLR, neutrophil-to-lymphocyte ratio; NPAR, neutrophil percentage-to-albumin ratio; SHR, stress hyperglycemia ratio; sICAS, symptomatic intracranial atherosclerotic stenosis; OPC, oligodendrocyte precursor cell.
References
1. Liu Y, Li S, Liu H, Tian X, Liu Y, Li Z, et al. Clinical implications of haemodynamics in symptomatic intracranial atherosclerotic stenosis by computational fluid dynamics modelling: a systematic review. Stroke Vasc Neurol. (2025) 10:16–24. doi: 10.1136/svn-2024-003202
2. Liu H, Zheng D, Wang D, Shi L, Leng X, Wong LKS, et al. State-of-the-art computational models of circle of willis with physiological applications: a review. IEEE Access. (2020) 8:156261–73. doi: 10.1109/ACCESS.2020.3007737
3. Fillingham P, Levendovszky SR, Andre J, Bindschadler M, Friedman S, Kurt M, et al. Noninvasive, patient-specific computational fluid dynamics simulations of dural venous sinus pressures in idiopathic intracranial hypertension. Brain Multiphys. (2023) 5:100081. doi: 10.1016/j.brain.2023.100081
4. Liu H, Pan F, Lei X, Hui J, Gong R, Feng J, et al. Effect of intracranial pressure on photoplethysmographic waveform in different cerebral perfusion territories: a computational study. Front Physiol. (2023) 14:1085871. doi: 10.3389/fphys.2023.1085871
5. Li W, Shao C, Zhou H, Du H, Chen H, Wan H, et al. Multi-omics research strategies in ischemic stroke: a multidimensional perspective. Ageing Res Rev. (2022) 81:101730. doi: 10.1016/j.arr.2022.101730
6. Montaner J, Ramiro L, Simats A, Tiedt S, Makris K, Jickling GC, et al. Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat Rev Neurol. (2020) 16:247–64. doi: 10.1038/s41582-020-0350-6
7. Debette S, Paré G. Stroke genetics, genomics, and precision medicine. Stroke. (2024) 55:2163–8. doi: 10.1161/STROKEAHA.123.044212
8. Bordes C, Sargurupremraj M, Mishra A, Debette S. Genetics of common cerebral small vessel disease. Nat Rev Neurol. (2022) 18:84–101. doi: 10.1038/s41582-021-00592-8
9. Bonkhoff AK, Grefkes C. Precision medicine in stroke: towards personalized outcome predictions using artificial intelligence. Brain. (2022) 145:457–75. doi: 10.1093/brain/awab439
10. Czap AL, Sheth SA. Overview of imaging modalities in stroke. Neurology. (2021) 97(20 Suppl 2):S42–51. doi: 10.1212/WNL.0000000000012794
11. Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine learning-based model for prediction of outcomes in acute stroke. Stroke. (2019) 50:1263–5. doi: 10.1161/STROKEAHA.118.024293
12. Akay EMZ, Hilbert A, Carlisle BG, Madai VI, Mutke MA, Frey D. Artificial intelligence for clinical decision support in acute ischemic stroke: a systematic review. Stroke. (2023) 54:1505–16. doi: 10.1161/STROKEAHA.122.041442
Keywords: cerebrovascular diseases (CVDs), neuroimaging, multi-omics data analysis, molecular genetics, artificial intelligence
Citation: Li L, Liu H, Lan L and Yu H (2025) Editorial: Genetic insights and diagnostic innovations in cerebrovascular and cerebrospinal fluid disorders. Front. Neurol. 16:1691759. doi: 10.3389/fneur.2025.1691759
Received: 24 August 2025; Accepted: 22 September 2025;
Published: 21 October 2025.
Edited and reviewed by: Huifang Shang, Sichuan University, China
Copyright © 2025 Li, Liu, Lan and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Yu, aGFveXV6anVAemp1LmVkdS5jbg==
†ORCID: Haipeng Liu orcid.org/0000-0002-4212-2503
 Ling Li
Ling Li Haipeng Liu
Haipeng Liu Linfang Lan
Linfang Lan Hao Yu
Hao Yu