- 1 Institute for Virus Research, Kyoto University, Kyoto, Japan
- 2 Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Kyoto, Japan
In neural stem/progenitor cells, expression of the Notch effector Hes1, a transcriptional repressor, oscillates with a period of 2–3 h by negative feedback, and Hes1 oscillations induce the oscillatory expression of the proneural gene Neurogenin2 (Ngn2) and the Notch ligand gene Delta-like1 (Dll1). Dll1 oscillation leads to the mutual activation of Notch signaling between neighboring cells, thereby maintaining a group of cells in the undifferentiated state. Not all cells express Hes1 in an oscillatory manner: cells in boundary regions such as the isthmus express Hes1 in a sustained manner, and these cells are rather dormant with regard to proliferation and differentiation. Thus, Hes1 allows cell proliferation and differentiation when its expression oscillates but induces dormancy when its expression is sustained. After Hes1 expression is repressed, Ngn2 is expressed in a sustained manner, promoting neuronal differentiation. Thus, Ngn2 leads to the maintenance of neural stem/progenitor cells by inducing Dll1 oscillation when its expression oscillates but to neuronal differentiation when its expression is sustained. These results indicate that the different dynamics of Hes1 and Ngn2 lead to different outcomes.
Introduction
Neuroepithelial cells, which constitute the wall of the neural tube, proliferate by repeating symmetric cell division, where each neuroepithelial cell divides into two neuroepithelial cells (progenitor expansion phase, Figure 1; Alvarez-Buylla et al., 2001; Fishell and Kriegstein, 2003; Fujita, 2003; Götz and Huttner, 2005; Miller and Gauthier, 2007). As the wall of the neural tube becomes thicker, neuroepithelial cells elongate and become radial glial cells, which have cell bodies in the ventricular zone, and radial fibers reaching the pial surface (Figure 1). Radial glial cells were previously thought of as specialized glial cells that guide neuronal migration. Later, it was found that radial glial cells undergo asymmetric cell division, where each radial glial cell divides into two distinct cell types, a radial glial cell and an immature neuron or a basal progenitor (neurogenic phase, Figure 1; Malatesta et al., 2000; Miyata et al., 2001; Noctor et al., 2001). Immature neurons migrate outside of the ventricular zone along radial fibers into the cortical plate, where these cells become mature neurons, whereas basal progenitors migrate into the subventricular zone (SVZ), proliferate further and give rise to more neurons. Radial glial cells give rise to many different types of neurons, initially deep layer neurons and then superficial layer neurons later, by repeating asymmetric cell division. Radial glial cells also give rise to oligodendrocytes and ependymal cells and finally differentiate into astrocytes (gliogenic phase, Figure 1). Both neuroepithelial and radial glial cells are considered neural stem/progenitor cells.
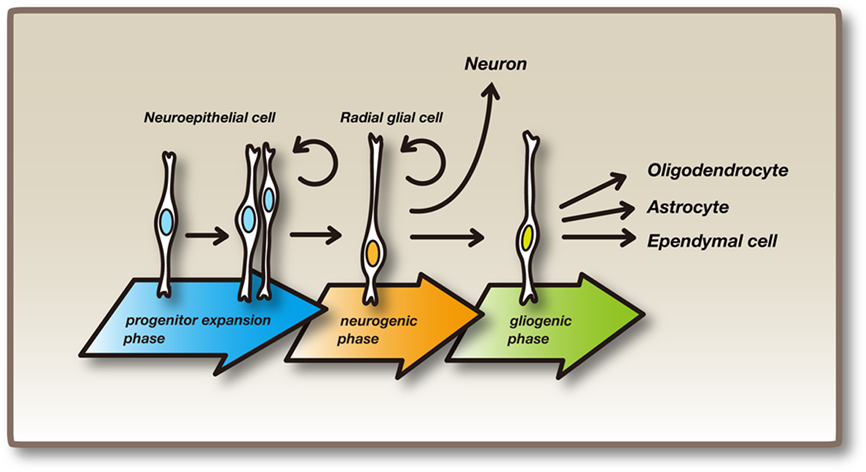
Figure 1. Neural stem/progenitor cells and their differentiation. Initially, neuroepithelial cells undergo repeated self-renewal by symmetric division (progenitor expansion phase). As development proceeds, neuroepithelial cells elongate to become radial glial cells, which have cell bodies on the inner side (called the ventricular zone) of the neural tube and radial fibers that reach the outer surface. Radial glial cells gives rise to neurons or basal progenitors (neurogenic phase). After the production of neurons, some radial glial cells give rise to oligodendrocytes and ependymal cells. Radial glial cells finally differentiate into astrocytes (gliogenic phase). Both neuroepithelial cells and radial glial cells are considered embryonic neural stem/progenitor cells.
If neural stem/progenitor cells are prematurely depleted, not only is the total number of cells reduced but also later-born cell types are lacking, indicating that the maintenance of neural stem/progenitor cells until the final stage of development is essential to produce the proper number of cells and the full diversity of cell types (Hatakeyama et al., 2004). Notch signaling has been shown to play an important role in the maintenance of neural stem/progenitor cells (Fortini, 2009; Kopan and Ilagan, 2009; Pierfelice et al., 2011). Here we review the recent progress on the mechanism and role of Notch signaling in neural development.
The Core Pathway of Notch Signaling
Proneural genes such as the basic helix-loop-helix (bHLH) transcriptional activators Mash1 and Neurogenin2 (Ngn2) induce neuronal differentiation (Bertrand et al., 2002; Ross et al., 2003). These genes also induce the expression of Notch ligands such as Delta-like1 (Dll1) and Jagged1, which activate Notch signaling in neighboring cells (Figure 2; Castro et al., 2006). The ubiquitin ligase Mindbomb is required in Dll1-expressing cells for the Dll1-induced activation of Notch signaling (Ito et al., 2003; Yoon et al., 2008). Upon activation of the transmembrane protein Notch, the Notch intracellular domain (NICD) is released from the transmembrane portion and transferred to the nucleus, where NICD forms a complex with the DNA-binding protein Rbpj and the transcriptional co-activator Maml (Figure 2). The NICD–Rbpj–Maml complex induces the expression of bHLH transcriptional repressors such as Hes1 and Hes5 (Honjo, 1996; Artavanis-Tsakonas et al., 1999; Ohtsuka et al., 1999; Kageyama et al., 2007). Hes1 and Hes5 then repress the expression of proneural genes and Dll1, thereby leading to the inhibition of neuronal differentiation and the maintenance of neural stem/progenitor cells (Figure 2). Thus, neurons inhibit their neighboring cells from differentiating into the same cell types, a process known as lateral inhibition. This regulation maintains asymmetric division into a neural stem/progenitor cell and a differentiating daughter neuron. However, this regulation also suggests that the maintenance of neural stem/progenitor cells depends on neighboring neurons expressing Dll1, raising the question as to how neural stem/progenitor cells are maintained during the early stages before neurons are generated.
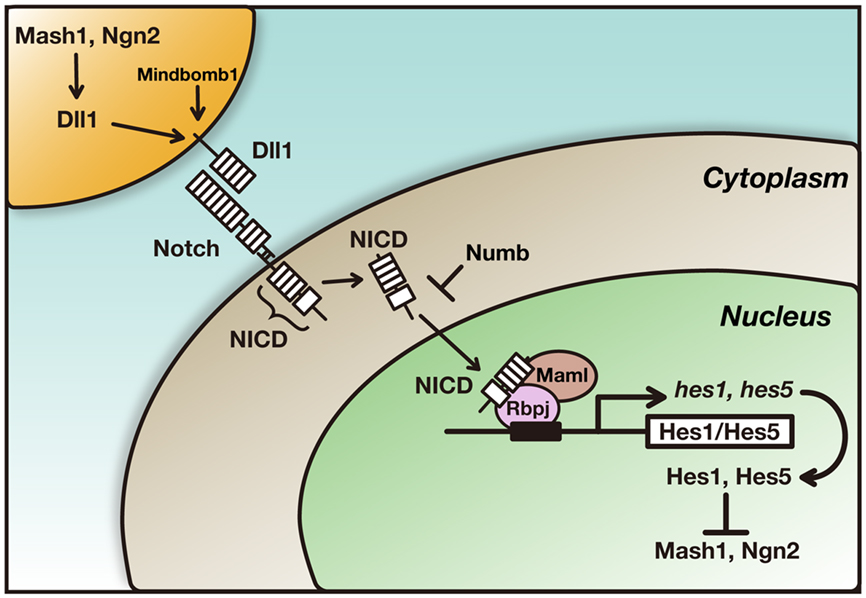
Figure 2. The core pathway of Notch signaling. Proneural genes such as Mash1 and Ngn2 promote neuronal differentiation and induce the expression of Dll1, which in turn activates Notch in neighboring cells. Upon activation of Notch, the Notch intracellular domain (NICD) is released from the transmembrane portion and transferred to the nucleus, where it forms a complex with the DNA-binding protein Rbpj and the transcriptional co-activator Maml. The NICD–Rbpj–Maml complex induces the expression of transcriptional repressor genes such as Hes1 and Hes5. Hes1 and Hes5 then repress the expression of proneural genes and Dll1, thereby leading to the maintenance of neural stem/progenitor cells. Numb inhibits Notch signaling and induces neuronal differentiation.
Oscillatory Expression of Notch Signaling Genes
Proneural genes and Dll1 are expressed in a salt-and-pepper pattern by neural stem/progenitor cells as early as E9.0 (progenitor expansion phase) in the developing mouse dorsal telencephalon (our unpublished observation), where neuronal formation starts around E10.5–E11.0. In addition, Hes1 is expressed at various levels by neural stem/progenitor cells (Shimojo et al., 2008). It is likely that proneural genes induce the expression of Dll1, which up-regulates Hes1 expression in neighboring cells, suggesting that Notch signaling is active in neural stem/progenitor cells before neurons are formed. This observation raises the question of why neurons are not formed during the progenitor expansion phase even though proneural genes are expressed.
Time-lapse imaging analysis revealed that Hes1 expression oscillates in neural stem/progenitor cells (Masamizu et al., 2006; Shimojo et al., 2008). Hes1 represses its own expression by directly binding to its promoter. This negative feedback leads to the disappearance of Hes1 mRNA and protein because they are extremely unstable, but the disappearance of Hes1 protein allows the next round of its expression. In this way, Hes1 expression oscillates with a period of ∼2–3 h in neural stem/progenitor cells (Figure 3; Hirata et al., 2002). Hes1 protein expression exhibits an inverse correlation with Ngn2 protein and Dll1 mRNA expression in neural stem/progenitor cells, suggesting that Hes1 oscillation induces the oscillatory expression of Ngn2 and Dll1 by periodic repression (Shimojo et al., 2008). Indeed, time-lapse imaging analysis revealed that Ngn2 and Dll1 expression oscillates in neural stem/progenitor cells, where Hes1 expression oscillates, whereas their expression is sustained in differentiating neurons, where Hes1 expression is repressed (Figure 4; Shimojo et al., 2008). It is likely that Ngn2 cannot induce neuronal differentiation when its expression oscillates, probably because many downstream genes respond rather slowly to Ngn2 expression. It seems that Ngn2 induces neuronal differentiation only when its expression becomes sustained, and when its expression oscillates, only quickly responding genes such as Dll1 are selectively induced. Dll1 oscillations may lead to the mutual activation of Notch signaling and the maintenance of neural stem/progenitor cells. Thus, depending on its expression dynamics, Ngn2 may lead to two opposite outcomes: when its expression oscillates, Ngn2 induces the maintenance of neural stem/progenitor cells, but when its expression is sustained, Ngn2 induces neuronal differentiation.
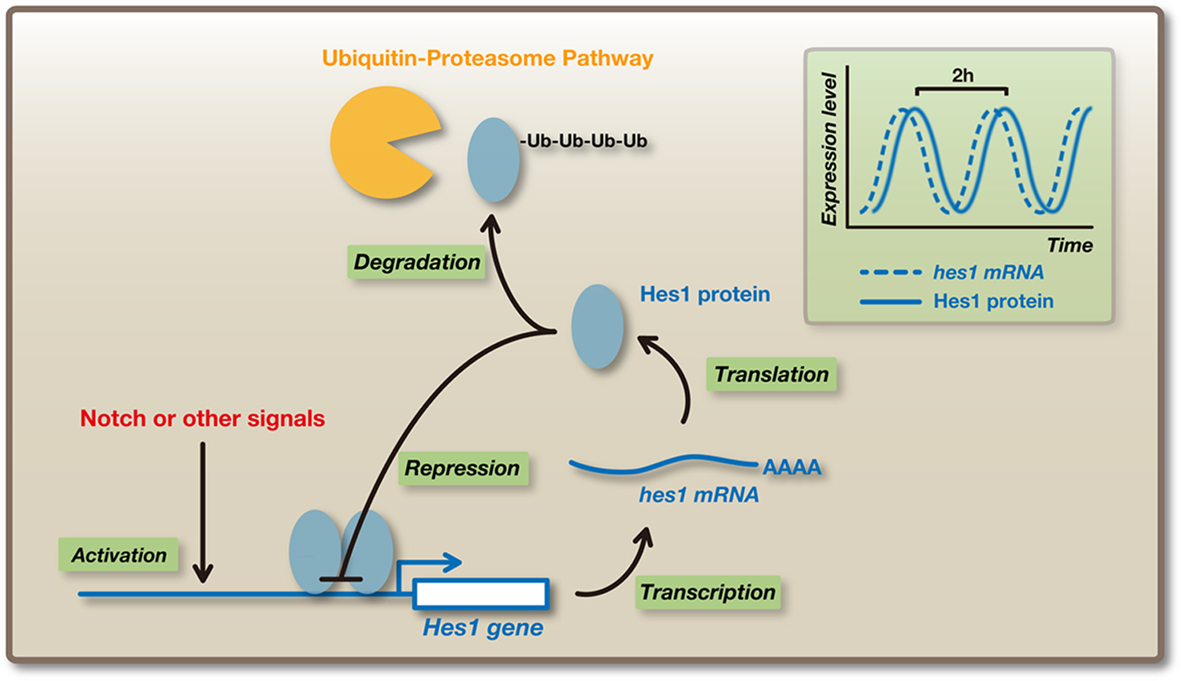
Figure 3. Oscillatory expression of Hes1. Hes1 expression oscillates with a period of ∼2 h in many cell types such as neural stem/progenitor cells and fibroblasts. Hes1 represses its own expression by directly binding to its promoter. This negative feedback leads to the disappearance of Hes1 mRNA and protein, because they are extremely unstable, allowing the next round of its expression. In this way, Hes1 autonomously starts an oscillatory expression pattern.
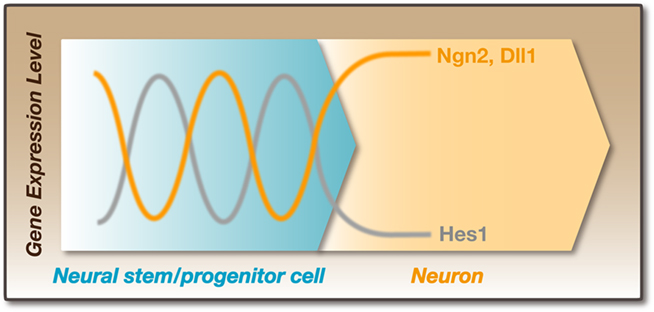
Figure 4. Dynamic expression in neural stem/progenitor cells. Hes1 expression oscillates with a period of ∼2–3 h in neural progenitor cells. In these cells, Ngn2 and Dll1 expression also oscillates. Hes1 protein expression exhibits an inverse correlation with Ngn2 protein and Dll1 mRNA expression. In contrast, the expression of Ngn2 and Dll1 is sustained in post-mitotic neurons, which lose Hes1 expression. It is likely that Dll1 oscillation periodically activates Notch signaling between neighboring cells. This reciprocal regulation is particularly important for the maintenance of neural stem/progenitor cells before the formation of neurons.
The expression dynamics of Ngn2 (oscillatory versus sustained) are regulated by Hes1 (oscillatory versus no expression). However, the precise mechanism by which Hes1 oscillations stop is not known. One possible mechanism is Numb-mediated inhibition of Notch signaling (Figure 2). Numb is asymmetrically distributed into a prospective neuron during asymmetric cell division (Shen et al., 2002; Ohtsuka et al., 2006). Numb-expressing cells lose Hes1 expression and differentiate into neurons, while Numb-negative cells maintain Hes1 expression (Ohtsuka et al., 2006).
The above observations raise the possibility that Ngn2 and Dll1 oscillations enable the maintenance of neural stem/progenitor cells without the aid of neurons. When Hes1 expression is low in a subset of cells (Cell 1 in the left panel of Figure 5), Ngn2 and Dll1 expression becomes high, leading to the activation of Notch signaling and the up-regulation of Hes1 in neighboring cells (Cell 2 in the left panel of Figure 5). In the latter cells, high levels of Hes1 repress Ngn2 and Dll1 expression, but due to oscillations, Hes1 expression becomes low after ∼1 h, while Ngn2 and Dll1 expression becomes high (Cell 2 in the right panel of Figure 5), leading to the activation of Notch signaling in the former cells (Cell 1 in the right panel of Figure 5). In this way, Dll1 oscillations lead to the mutual activation of Notch signaling between neural stem/progenitor cells (Shimojo et al., 2008). Thus, oscillatory expression is advantageous for maintaining a group of undifferentiated cells. These observations also suggest that Notch signaling is not a one-way mechanism (neuron to neural stem/progenitor), but functions by reciprocal transmission (neural stem/progenitor to neural stem/progenitor). At later stages (neurogenic phase), many differentiating neurons express Dll1 in a sustained manner, thereby activating Notch signaling in neural stem/progenitor cells. At these stages, Ngn2 and Dll1 expression mostly occurs in neurons but not in neural stem/progenitor cells, and therefore their expression is sustained, although Hes1 expression in neural stem/progenitor cells oscillates even at these later stages.
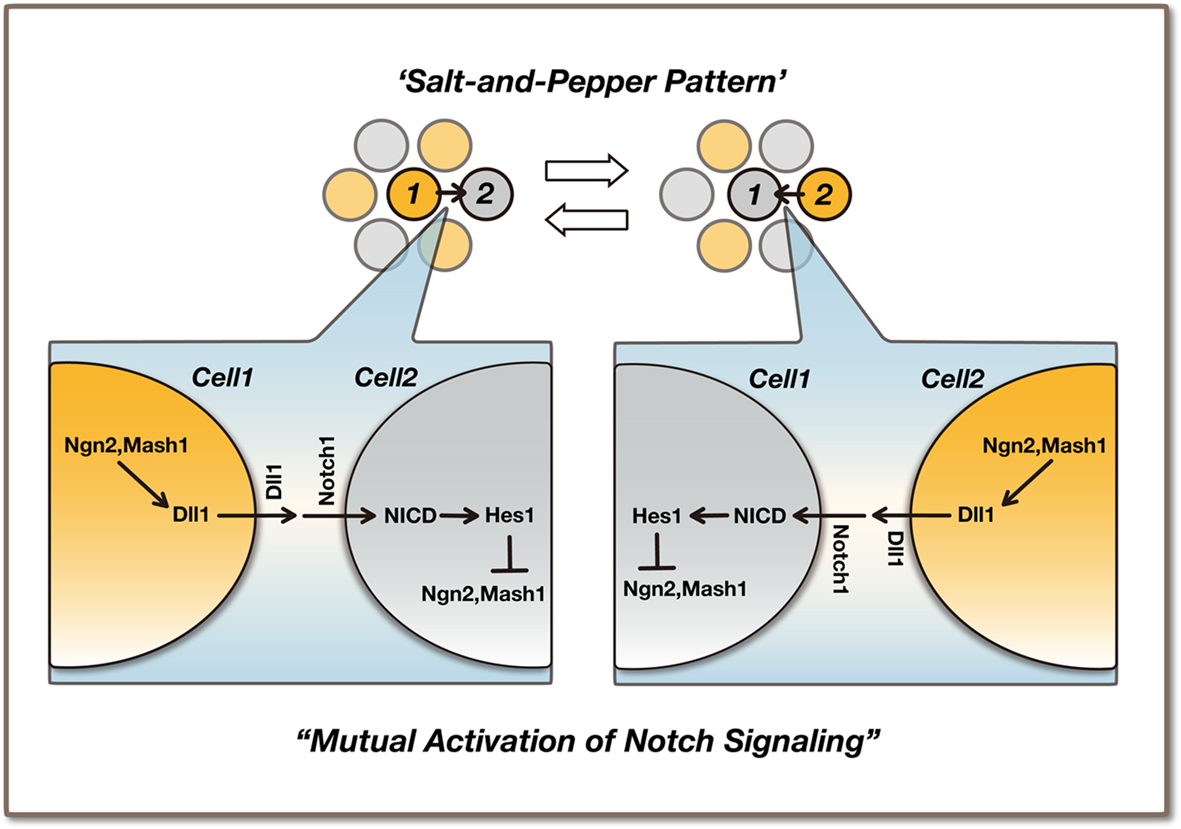
Figure 5. Maintenance of neural stem/progenitor cells by the mutual activation of Notch signaling. Ngn2 and Dll1 oscillations are regulated by Hes1 oscillation in neural stem/progenitor cells. Ngn2 oscillation may be advantageous for the maintenance/proliferation of neural stem/progenitor cells during the early stages of development, because it induces Dll1 expression and activates Notch signaling without promoting neuronal differentiation. A salt-and-pepper pattern induced by lateral inhibition is just a snapshot of oscillatory expression. The precise interactive regulation among Dll1-positive and -negative cells remains to be determined.
It has been thought that lateral inhibition amplifies stochastic variation, creates a salt-and-pepper pattern in gene expression and selects subsets of cells for neuronal differentiation: Ngn2-positive or Dll1-positive neural stem/progenitor cells are initially selected to become neurons when the neurogenic phase starts, while negative cells remain neural stem/progenitor cells (Artavanis-Tsakonas et al., 1999). However, time-lapse imaging analysis indicated that a salt-and-pepper pattern is just a snapshot of oscillatory expression; therefore, neither Ngn2-positive nor Dll1-positive cells are initially selected to become neurons. Thus, the traditional view of salt-and-pepper pattern formation induced by lateral inhibition should be revised (Kageyama et al., 2008).
Basal Progenitors and Outer Subventricular Zone Progenitors
Basal progenitors in the SVZ, which are derived from radial glial cells by Tbr2, retract their apical and basal processes and generally divide only once to generate two neurons (Sessa et al., 2008). Thus, basal progenitors have a limited proliferation ability. In these cells, Hes1 and Hes5 expression is down-regulated, suggesting that the Notch-Hes1/5 pathway is not active in basal progenitors (Mizutani et al., 2007; Kawaguchi et al., 2008). It was recently shown that the developing human neocortex has an expanded outer region in the SVZ (OSVZ), and that progenitors in this region (OSVZ progenitors) divide multiple times and generate a large number of neurons (Fietz et al., 2010; Hansen et al., 2010). These cells seem to play a major role in the expansion of the cortex. OSVZ progenitors have radial glia-like morphology and extend radial fibers to the pial surface. However, they lack apical processes and therefore are not in contact with the ventricular surface. Like radial glia, OSVZ progenitors express Hes1, and inhibition of Notch signaling by treatment with a γ-secretase inhibitor induces OSVZ progenitors to differentiate into neurons or Tbr2+ basal progenitors (Hansen et al., 2010), suggesting that the Notch-Hes1 pathway is required for maintenance of OSVZ progenitors (Figure 6). It is possible that the cells that migrate into the SVZ may become OSVZ progenitors when Notch signaling is active, whereas they may become basal progenitors when Notch signaling is inactive. It remains to be determined how Notch signaling is regulated in the SVZ and whether Hes1 expression oscillates in OSVZ progenitors, as observed in radial glia.
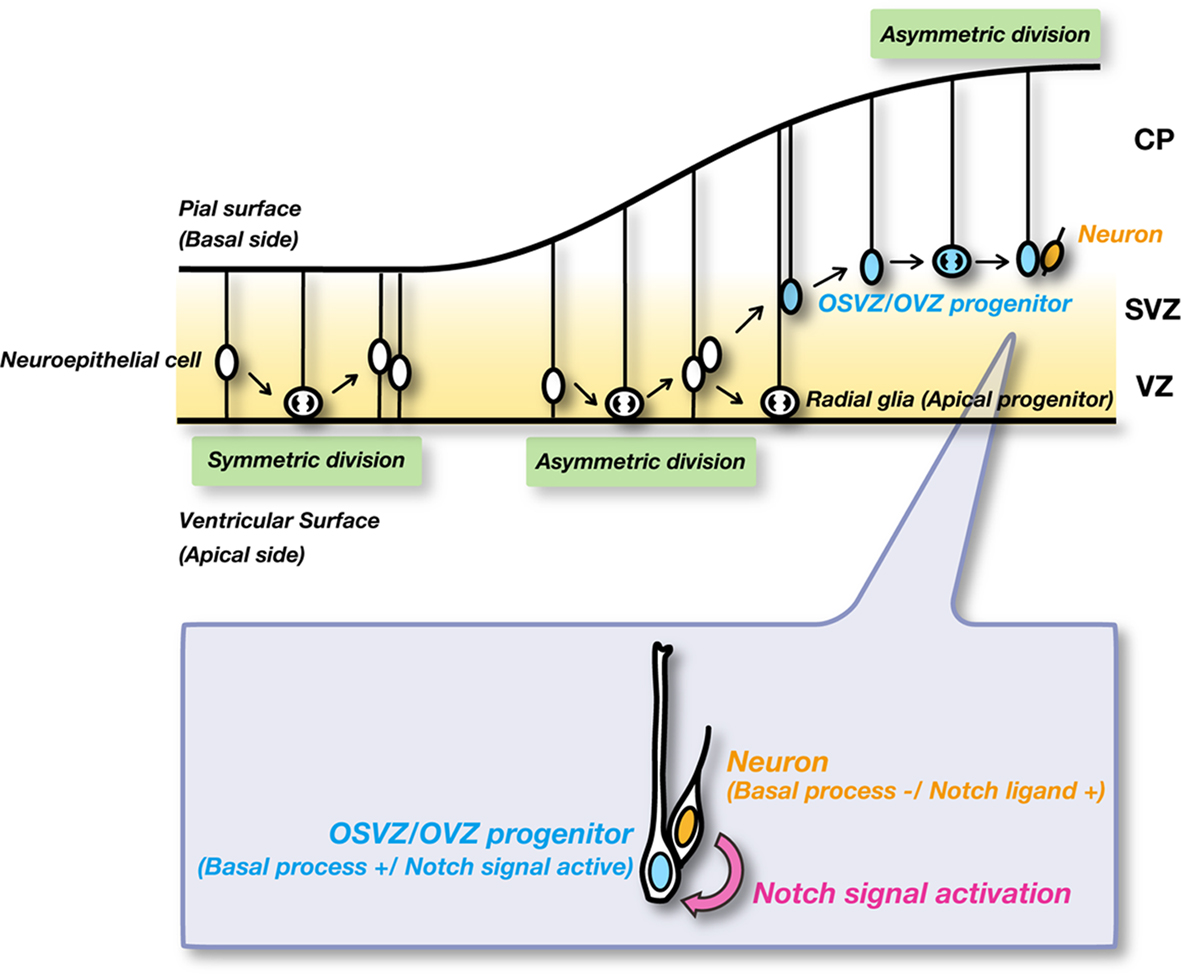
Figure 6. Notch signaling in OSVZ/OVZ progenitors. OSVZ/OVZ progenitors have radial glia-like morphology and extend radial fibers to the pial surface, but lack apical processes. These cells undergo asymmetric cell division multiple times and generate a lot of neurons. These neurons seem to express Notch ligands and activate Notch signaling in their sibling OSVZ/OVZ progenitors.
Similar cells called outer ventricular zone (OVZ) progenitors are found in the developing mouse cortex (Shitamukai et al., 2011). OVZ progenitors are located outside of the ventricular zone and retain radial fibers reaching the pial surface (basal processes). These cells undergo asymmetric cell division, where each OVZ progenitor divides into a daughter cell that inherits the radial fiber (OVZ progenitor) and the other that does not. The one that inherits the radial fiber seems to repeat asymmetric cell division multiple times, while the other differentiates into post-mitotic neurons. The former cells (OVZ progenitors) express Hes1, suggesting that Notch signaling is activated in these cells (Figure 6). Interestingly, these daughter cells (OVZ progenitor and neuron) maintain contact with each other for several hours, and neurons seem to express Notch ligands and activate Notch signaling in their sibling OVZ progenitors (Figure 6; Shitamukai et al., 2011). These observations suggest that asymmetric cell division is required to activate Notch signaling in OSVZ or OVZ progenitors by their sibling neurons.
Sustained Hes1 Expression in Boundary Cells
Not all cells express Hes1 in an oscillatory manner: cells in the roof plate, floor plate, and boundary regions, such as the isthmus, express Hes1 in a sustained manner (Baek et al., 2006). These cells are quiescent or proliferate very slowly. Furthermore, cells in these regions usually do not give rise to any neurons, probably because sustained Hes1 expression constitutively represses the expression of proneural genes and cell cycle regulators. The introduction of sustained Hes1 expression into neural stem/progenitor cells inhibits their proliferation and neuronal differentiation (Baek et al., 2006). In contrast, when Hes genes are inactivated, the cells in the roof plate, floor plate, and boundary regions can differentiate into neurons (Baek et al., 2006). These results suggest that cells with sustained Hes1 expression are rather dormant with regard to proliferation and differentiation. This feature is similar to the one observed in fibroblasts, where sustained Hes1 over expression leads to reversible quiescence (Sang et al., 2008).
The mechanism by which oscillatory versus sustained Hes1 expression is regxulated remains to be determined. In fibroblasts, Jak–Stat signaling is involved in Hes1 oscillations. Jak2 activates Stat3 by phosphorylation, and phosphorylated Stat3 (pStat3) induces Socs3 expression, which in turn inhibits Jak2. Due to this negative feedback, pStat3 and Socs3 levels oscillate in fibroblasts (Yoshiura et al., 2007). Interestingly, blockade of this pathway with a Jak inhibitor inhibits Hes1 oscillations by stabilizing the Hes1 protein, and Hes1 expression becomes steady (Yoshiura et al., 2007). Similarly, treatment with a Jak inhibitor inhibits Hes1 oscillations in neural stem/progenitor cells, suggesting that Jak–Stat signaling is also involved in the regulation of Hes1 oscillations in these cells (Shimojo et al., 2008). Another possible mechanism is the Id-mediated regulation of Hes1. Id proteins, HLH factors without a basic region, form heterodimers with Hes1 through their HLH domains and inhibit Hes1 from binding to the N box in the Hes1 promoter (Bai et al., 2007), suggesting that Id factors prevent Hes1 from negative autoregulation. However, it remains to be determined whether Id factors lead to steady Hes1 expression.
Conclusion
Oscillatory versus sustained Hes1 expression leads to different outcomes in neural stem/progenitor cells. When its expression oscillates, neural stem/progenitor cells proliferate actively and differentiate into mature cells. By contrast, when its expression is sustained, neural stem/progenitor cells become dormant. Similarly, oscillatory versus sustained Ngn2 expression leads to different outcomes. When its expression is sustained, neural stem/progenitor cells differentiate into neurons. By contrast, when its expression oscillates, neural stem/progenitor cells remain undifferentiated. Thus, not just the expression but also the dynamics of these genes are very important for the outcomes. The mechanism by which oscillatory versus sustained Hes1 expression is regulated remains to be determined. In addition, the dynamics of downstream genes for Hes1 and Ngn2 oscillations are mostly unknown. Such analysis will be required to understand the complex regulatory mechanism of neural development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alvarez-Buylla, A., Garcia-Verdugo, J. M., and Tramontin, A. D. (2001). A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2, 287–293.
Artavanis-Tsakonas, S., Rand, M.D., and Lake, R.J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770–776.
Baek, J. H., Hatakeyama, J., Sakamoto, S., Ohtsuka, T., and Kageyama, R. (2006). Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 133, 2467–2476.
Bai, G., Sheng, N., Xie, Z., Bian, W., Yokota, Y., Benezra, R., Kageyama, R., Guillemot, F., and Jing, N. (2007). Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell 13, 283–297.
Bertrand, N., Castro, D. S., and Guillemot, F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530.
Castro, D. S., Skowronska-Krawczyk, D., Armant, O., Donaldson, I. J., Parras, C., Hunt, C., Critchley, J. A., Nguyen, L., Gossler, A., Göttgens, B., Matter, J.-M., and Guillemot, F. (2006). Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell 11, 831–844.
Fietz, S.A., Kelava, I., Vogt, J., Wilsch-Bräuninger, M., Stenzel, D., Fish, J.L., Corbeil, D., Riehn, A., Distler, W., Nitsch, R., and Huttner, W.B. (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699.
Fishell, G., and Kriegstein, A. R. (2003). Neurons from radial glia: the consequences of asymmetric inheritance. Curr. Opin. Neurobiol. 13, 34–41.
Fortini, M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647.
Fujita, S. (2003). The discovery of the matrix cell, the identification of the multipotent neural stem cell and the development of the central nervous system. Cell Struct. Funct. 28, 205–228.
Götz, M., and Huttner, W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788.
Hansen, D.V., Lui, J.H., Parker, P.R.L., and Kriegstein, A.R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561.
Hatakeyama, J., Bessho, Y., Katoh, K., Ookawara, S., Fujioka, M., Guillemot, F., and Kageyama, R. (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539–5550.
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., and Kageyama, R. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843.
Honjo, T. (1996). The shortest path from the surface to the nucleus: RBP-J k/Su(H) transcription factor. Genes Cells 1, 1–9.
Ito, M., Kim, C.-H., Palardy, G., Oda, T., Jiang, Y.-J., Maust, D., Yeo, S.-Y., Lorick, K., Wright, G. J., Ariza-McNaughton, L., Weissman, A. M., Lewis, J., Chandrasekharappa, S. C., and Chitnis, A. B. (2003). Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67–82.
Kageyama, R., Ohtsuka, T., and Kobayashi, T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251.
Kageyama, R., Ohtsuka, T., Shimojo, H., and Imayoshi, I. (2008). Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247–1251.
Kawaguchi, A., Ikawa, T., Kasukawa, T., Ueda, H. R., Kurimoto, K., Saitou, M., and Matsuzaki, F. (2008). Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135, 3113–3124.
Kopan, R., and Ilagan, M. X. G. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233.
Malatesta, P., Hartfuss, E., and Götz, M. (2000). Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263.
Masamizu, Y., Ohtsuka, T., Takashima, Y., Nagahara, H., Takenaka, Y., Yoshikawa, K., Okamura, H., and Kageyama, R. (2006). Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl. Acad. Sci. U.S.A. 103, 1313–1318.
Miller, F. D., and Gauthier, A. S. (2007). Timing is everything: making neurons versus glia in the developing cortex. Neuron 54, 357–369.
Miyata, T., Kawaguchi, A., Okano, H., and Ogawa, M. (2001). Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron 31, 727–741.
Mizutani, K., Yoon, K., Dang, L., Tokunaga, A., and Gaiano, N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355.
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S., and Kriegstein, A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720.
Ohtsuka, T., Imayoshi, I., Shimojo, H., Nishi, E., Kageyama, R., and McConnell, S. K. (2006). Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol. Cell. Neurosci. 31, 109–122.
Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F., and Kageyama, R. (1999). Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207.
Pierfelice, T., Alberi, L., and Gaiano, N. (2011). Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855.
Ross, S. E., Greenberg, M. E., and Stiles, C. D. (2003). Basic helix-loop-helix factors in cortical development. Neuron 39, 13–25.
Sang, L., Coller, H. A., and Roberts, J. M. (2008). Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321, 1095–1100.
Sessa, A., Mao, C., Hadjantonakis, A. K., Klein, W. H., and Broccoli, V. (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56–69.
Shen, Q., Zhong, W., Jan, Y. N., and Temple, S. (2002). Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129, 4843–4853.
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in Notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64.
Shitamukai, A., Konno, D., and Matsuzaki, F. (2011). Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci. 31, 3683–3695.
Yoon, K.-J., Koo, B.-K., Im, S.-K., Jeong, H.-W., Ghim, J., Kwon, M., Moon, J.-S., Miyata, T., and Kong, Y.-Y. (2008). Mind bomb 1-expressing intermediate progenitors generate Notch signaling to maintain radial glial cells. Neuron 58, 519–531.
Keywords: basal progenitor, neuroepithelial cell, oscillatory expression, OSVZ progenitor, proneural gene, radial glia
Citation: Shimojo H, Ohtsuka T and Kageyama R (2011) Dynamic expression of Notch signaling genes in neural stem/progenitor cells. Front. Neurosci. 5:78. doi: 10.3389/fnins.2011.00078
Received: 01 April 2011;
Accepted: 03 June 2011;
Published online: 14 June 2011.
Edited by:
Nicholas Gaiano, Johns Hopkins School of Medicine, USACopyright: © 2011 Shimojo, Ohtsuka and Kageyama. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Ryoichiro Kageyama, Institute for Virus Research, Kyoto University, Shogoin-Kawahara, Sakyo-ku, Kyoto 606-8507, Japan. e-mail:cmthZ2V5YW1AdmlydXMua3lvdG8tdS5hYy5qcA==
 Hiromi Shimojo1,2
Hiromi Shimojo1,2