- 1 Department of Genetics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA
- 2 Case Comprehensive Cancer Center, School of Medicine, Case Western Reserve University, Cleveland, OH, USA
- 3 Center for RNA Molecular Biology, School of Medicine, Case Western Reserve University, Cleveland, OH, USA
Alternative splicing in eukaryotes plays an important role in regulating gene expression by selectively including alternative exons. A wealth of information has been accumulated that explains how alternative exons are selected in a developmental stage- or tissue-specific fashion. However, our knowledge of how cells respond to environmental changes to alter alternative splicing is very limited. For example, although a number of alternative exons have been shown to be regulated by calcium level alterations, the underlying mechanisms are not well understood. As calcium signaling in neurons plays a crucial role in essential neuronal functions such as learning and memory formation, it is important to understand how this process is regulated at every level in gene expression. The significance of the dynamic control of alternative splicing in response to changes of calcium levels has been largely unappreciated. In this communication, we will summarize the recent advances in calcium signaling-mediated alternative splicing that have provided some insights into the important regulatory mechanisms. In addition to describing the cis-acting RNA elements on the pre-mRNA molecules that respond to changes of intracellular calcium levels, we will summarize how splicing regulators change and affect alternative splicing in this process. We will also discuss a novel mode of calcium-mediated splicing regulation at the level of chromatin structure and transcription.
Calcium signaling plays a pivotal role in many functions of the cell at the molecular level. It is particularly important in neurons for expression of genes essential for learning and memory formation, and disruption of calcium signaling can cause neuronal cell death leading to loss of memory (reviewed in West et al., 2001; Berridge, 2009). In neurons, changes in intracellular calcium levels cause changes in membrane potential that control neuronal excitability (reviewed in Berridge, 1998). Many studies have demonstrated that calcium signaling can regulate gene expression at the transcription level (reviewed in Zhang et al., 2009; Alonso and Garcia-Sancho, 2011). However, a number of recent studies are revealing an intriguing new post-transcriptional level of regulation by calcium signaling: at the level of alternative pre-mRNA splicing. It has been observed that an alteration of the intracellular calcium level can lead to differential expression of numerous alternative exons (reviewed in Xie, 2008). These observations have very significant implications in terms of the dynamic control of alternative splicing.
More than 95% of the protein-coding genes in humans undergo alternative splicing to provide a varied proteome (Pan et al., 2008; Wang et al., 2008). Studies carried out during the last three decades have generated a wealth of information that contributes to our understanding of cell type- and developmental stage-specific regulation of alternative splicing. However, we know surprisingly little about how cells respond to stimuli to modulate alternative splicing. Only a handful of studies have explored the underlying mechanisms by which calcium signaling regulates alternative splicing. A review published in 2008 provided a comprehensive summary of calcium-mediated splicing control (Xie, 2008). In this review, the author compiled a list of nearly 20 alternative exons that were demonstrated to undergo calcium-mediated regulation. Next, he discussed the potential mechanisms of calcium-mediated splicing control using three well-studied examples. Finally, he speculated how calcium signaling influences neuronal functions and disease development. Since 2008, several more studies have been published, one of which demonstrated an exciting new level of regulation. The current review is aimed at providing an in-depth discussion of the molecular mechanisms of the dynamic control of alternative splicing regulated by calcium signaling. An important goal is to draw attention to this under-explored area of research. Building on information gained from a limited number of studies, we will point out important future directions.
Regulation of Alternative Splicing
Production of protein-coding mRNAs in mammalian cells involves two tightly coupled processes: transcription of the pre-mRNA from the gene and pre-mRNA processing, which consists of 5′ end capping, removal of introns, and polyadenylation of the 3′ end of the pre-mRNA. RNA processing factors are loaded onto the pre-mRNA co-transcriptionally, some through the C terminal domain (CTD) of the largest subunit of RNA polymerase II (RNA Pol II) (Misteli and Spector, 1999; Hirose and Manley, 2000; Lewis and Tollervey, 2000; Phatnani et al., 2004; Rosonina and Blencowe, 2004; Bentley, 2005; Egloff and Murphy, 2008). The classical view of alternative splicing regulation involves short conserved RNA sequences (enhancers and silencers) found in exons and introns of the pre-mRNA molecule, where splicing regulators such as serine/arginine proteins (SR proteins) and heterogeneous ribonucleoproteins (hnRNPs) bind (reviewed in Black, 2003; Jurica and Moore, 2003). Upon binding to an enhancer or silencer sequence, the splicing regulators either recruit the spliceosome, the basal splicing machinery, to or prevent the spliceosome from interacting with a particular splice site to regulate the associated alternative exon. Many splicing regulators are tissue-specific and hence are thought to be important in providing developmental stage-specific and functional isoforms of proteins for a particular tissue (Black, 2003; Grosso et al., 2008).
In addition to regulation by splicing factors, alternative splicing can be controlled by the rate of transcription as was first shown by cell transfection experiments. A change in the rate of RNA synthesis provides a change in the secondary structure of the pre-mRNA, which in turn affects the splicing event (Eperon et al., 1988). This was confirmed when different levels of alternative exon inclusion were observed for the same gene using promoters of different strengths to drive transcription. To explain the effects on alternative splicing by transcription, two models have been proposed in the literature: the recruitment model and the kinetic coupling model. The recruitment model suggests that splicing regulators are recruited to pre-mRNA through their association with the transcription machinery, in particular the CTD of Pol II, to regulate splicing choices, whereas the kinetic model suggests that splicing is controlled by the rate of elongation of Pol II. The slower rate of transcription elongation allows more time for the usage of weak splice sites (reviewed in Kornblihtt, 2005; Allo et al., 2011; Luco et al., 2011).
The rate of elongation of transcription is thought to be controlled by chromatin structure and the level of compaction of the intragenic regions. The following recent studies support an emerging idea that chromatin structure and histone modifications are major contributors in regulation of alternative splicing events (Allo et al., 2011; Luco et al., 2011). First, nucleosomes were found to be enriched on exons as compared to introns by analysis using the deep sequencing technique in combination with computational predictions (reviewed in Kaplan, 2009; Kornblihtt et al., 2009; Schor et al., 2010). The general hypothesis is that the nucleosomes provide a “roadblock or speed bump” for RNA Pol II elongation, which in turn provides more time for the splicing machinery to recognize alternative exons (Allo et al., 2011; Luco et al., 2011). In addition to the exon definition concept (Berget, 1995), where the splicing machinery recognizes intron–exon boundaries by physically interacting with the splice sites, this is thought to provide another mode of control at the DNA level for splice site recognition. Support for this hypothesis came from the observation that there is an enrichment of RNA Pol II at alternatively spliced exons in comparison to constitutive exons, thereby providing the splicing machinery more time to access weak splice sites present at the alternative splice sites (Brodsky et al., 2005).
Secondly, chromatin immunoprecipitation (ChIP) studies indicate that exons are differentially marked with specific histone modifications such as trimethylation of lysine 36 of histone 3 (Kolasinska-Zwierz et al., 2009; Spies et al., 2009). Thirdly, studies have demonstrated that specific histone marks can recruit splicing factors to the transcription site. For example, H3K4me3 recruits the U2 snRNP, a component of the spliceosome, via the CHD1 protein that interacts with the histone mark (Sims et al., 2007). In addition, H3K9me3 recruits hnRNPs through the HP1 proteins in Drosophila (Piacentini et al., 2009). These studies indicate that histone modifications can have a direct impact on splicing factor recruitment. Lastly, a very recent study indicates that Hu proteins, upon binding to their target sequences on the pre-mRNA, influence the acetylation status of histones H3 and H4 leading to a localized change in transcription elongation rate that further impacts exon skipping of at least two alternative exons (Zhou et al., 2011).
Calcium and Splicing
Several studies have shown that disturbance of the physiological balance of calcium can contribute to alterations of alternative splicing events, especially in neuronal cells which have the most diverse RNA population (Xie and Black, 2001; Black and Grabowski, 2003; An and Grabowski, 2007; Lee et al., 2007; Schor et al., 2009). In each of these studies, depolarization was induced in cells with introduction of high extracellular potassium, which triggers calcium signaling by opening voltage-dependent calcium channels (Figure 1). An increase in internal calcium level causes skipping of exon 5 and exon 21 of NMDA receptor type 1 (NMDAR1) in hippocampal neurons (Han et al., 2005; An and Grabowski, 2007; Lee et al., 2007) as well as the STREX exon of the BK channel in GH3 pituitary cells (Xie and Black, 2001). These two splicing events, which are regulated by calcium signaling, have been studied in much detail and the results have provided important insights into how splicing regulation by calcium signaling can influence neuronal functions and diseases (Xie, 2008). In the case of the STREX exon of the BK channel, the regulated inclusion of the exon is thought to fine-tune the electrical activity of neurons (Xie, 2008). It has been shown that inclusion of the exon increases the sensitivity of the BK channels to voltage and calcium, providing a feedback loop regulatory mechanism (Li et al., 2007, 2009; Xie, 2008; and references therein). In the case of NMDAR1, the inclusion or exclusion of exons 5 and 21 affects the localization and membrane trafficking of the NMDA receptor. This localization and trafficking in turn regulates the synaptic strength to stabilize the neuronal firing rate (homeostatic plasticity in neurons) (Lee et al., 2007; Li et al., 2007; and references therein).
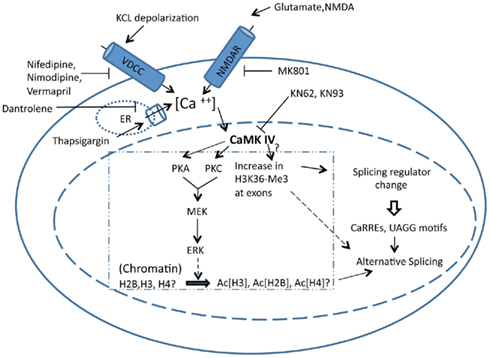
Figure 1. Stimulation of a cell using various drugs can increase or decrease intracellular calcium by affecting various channels and pumps associated with calcium influx. Increase in intracellular calcium activates calcium/calmodulin-dependent kinases, which influence splicing and/or localization of several splicing factors. These splicing factors can in turn regulate splicing by binding to CaRREs or UAGG motifs in the pre-mRNA. In addition, CaMK can phosphorylate protein kinases A and C, which feed into the downstream MAPK pathway, eventually phosphorylating ERK. Phosphorylated ERK plays a role in hyperacetylation of histones H2B, H3, and H4, which affects splicing by controlling the rate of transcription. CaMK also increases trimethylation of histone H3K36 at specific exons, although the mechanism by which CaMK does it is not known. The increase in trimethylation of histone H3K36 can directly affect splicing by recruiting splicing factors.
Treatment of neurons by various drugs increases intracellular calcium levels by affecting calcium channels and pumps. Agonists of NMDA receptors, such as NMDA, and glutamate can increase calcium influx through the NMDA receptors, whereas antagonists such as MK801 can inhibit the influx of calcium through NMDA receptors. Opening of L-type calcium channels can be inhibited by use of pharmacological agents such as nifedipine, nimodipine, and verapamil (Xie, 2008; Yoneyama et al., 2011; Figure 1). Thapsigargin increases intracellular calcium by inhibiting the sarco/endoplasmic reticulum calcium pump, whereas use of dantrolene can inhibit the release of the calcium from the sarco/endoplasmic reticulum (Yoneyama et al., 2011) (Figure 1). In addition to these two genes, it has been shown that increasing calcium in the cell by using any of these pharmacological agents affects the changes in splice site selection for many genes (for a complete list see Table 1 in the review by Xie, 2008). Also, an exon-array performed on human neuroblastoma IMR-32 cells after depolarization with potassium chloride at different time points or after treatment with thapsigargin identified a number of genes that showed changes in splicing and transcript levels. Splicing changes were observed in mRNAs of genes such as potassium channels, neuroendocrine secretory proteins and metabolic enzymes (McKee et al., 2007).
Calcium and RNA Elements
It is thought that just like the RNA elements in exons and introns where specific splicing regulators bind to regulate the inclusion or skipping of the exon, there are specific short conserved RNA sequences under the regulation of calcium. The current working theory is that increased intracellular calcium levels result in the activation of calcium/calmodulin-dependent kinase IV (CaMK IV), which regulates splice site selection through specific RNA elements named CaMK IV responsive RNA elements (CaRREs) (Ares, 2007; Lee et al., 2007; Xie, 2008). Two such CaRRE sequences, CACAUNRUUAU (N: any nucleotide, R: purine) and GUGGUAGA, have been identified (Xie et al., 2005; Lee et al., 2007; Xie, 2008). Importantly, when these CaRREs were inserted into a mini-gene reporter, the mini-gene alternative splicing was regulated by CaMK IV activity (Xie and Black, 2001; Xie et al., 2005; Lee et al., 2007). Other than the CaRREs, a distinct UAGG motif in the pre-mRNA has been shown to be required for depolarization-mediated splicing regulation. The UAGG motif-mediated splicing regulation can be inhibited by KN93, a calcium/calmodulin-dependent kinase inhibitor (An and Grabowski, 2007). In addition, KN93 and nifedipine, an L-type calcium channel blocker, were shown to block the exclusion of the STREX exon (thought to be under CaRRE control) caused by potassium chloride-mediated depolarization (Xie and Black, 2001; Xie et al., 2005). Lastly, cerebella of CaMK IV heterozygous mice showed increased STREX exon inclusion (Xie et al., 2005).
The mechanism by which CaMK IV influences specific RNA elements still remains an open question. One theory is that an increase in the calcium level activates CaMK IV, which eventually regulates splicing through these specific RNA elements in the pre-mRNA (CaRREs and UAGG). This is supported by the fact that the observed changes in splicing after increases in the intracellular calcium level can be inhibited in most of these cases by the use of either CaMK inhibitors or by other inhibitors that affect calcium levels in the cell [for a complete list see Table 1, Xie, 2008)]. Although the detailed molecular mechanisms remain largely unknown on how CaMK levels influence splicing through the CaRRE RNA elements, a hint came from the studies of the UAGG motifs. In this case, hnRNP A1 was found to bind to these specific motifs in pre-mRNA and eventually influence splice site selection in neurons (An and Grabowski, 2007). Subsequent studies demonstrated that hnRNP L is the splicing factor that binds to a CaRRE RNA element influencing splice site selection (Yu et al., 2009). This regulation of hnRNP L on the CaRRE1 RNA elements was further shown to be dependent on phosphorylation of hnRNP L, which can be induced by CaMK IV as shown in in vitro assays using hnRNP L and CaMK IV purified from mammalian cells (Yu et al., 2009). A SELEX analysis indicated that a “CA” repeat motif is sufficient for the CaMK IV-mediated regulation of alternative splicing through hnRNP L (Li et al., 2009).
Interestingly, the CaMK IV-hnRNP L cascade may also modulate alternative splicing of a non-neuronal gene, CD45 (PTPRC). Alternative splicing of the CD45 transcript is tightly regulated during B cell development. In early peripheral lymphocytes, exons 4, 5, and 6 are included, while at later stages of B cell development, none of the three exons are included (Ogimoto et al., 1993). Several studies demonstrated that splicing regulation of exons 4 and 6 is mediated by hnRNP L, polypyrimidine tract-binding protein (PTB)-associated splicing factor (PSF), and hnRNP LL (Melton et al., 2007; Topp et al., 2008). Treatment of early stages of B cells with ionomycin, an ionophore which increases intracellular calcium, led to skipping of all of these alternative exons (Ogimoto et al., 1993). It remains to be determined whether there exist CaRRE elements where hnRNP L could potentially bind and regulate splicing of these exons. However, an exonic splicing silencer (ESS) that has been identified to be bound by hnRNP L and PSF is indeed CA-rich, similar to the CaRRE elements (Melton et al., 2007).
It is highly likely that many additional calcium responsive RNA elements remain to be identified. The recently developed high-throughput technologies will undoubtedly be very helpful in this regard. Presumably, these elements are under the influence of calcium signaling and can be bound by different splicing regulators in regulation of the expression of the alternative exons with which they are associated (Figure 1).
Calcium and Splicing Factors
How does calcium signaling influence the expression of splicing regulators themselves? Recent studies appear to reveal a theme for this regulation. Changes in calcium levels regulate cellular localization of a number of RNA-binding proteins (RBPs). For example, cortical neurons depolarized by treatment with potassium chloride were shown to have an increased localization of the splicing factor hnRNPA1 in the nucleus (An and Grabowski, 2007). Interestingly, for a number of RBPs, the change in cellular localization is the consequence of regulated alternative splicing. A clear example is Fox-1/A2BP1, where exon 19 inclusion is repressed when neuronal cells are depolarized by potassium chloride (Lee et al., 2009). The skipping of exon 19 causes a frame shift in the protein, which leads to a more nuclear localization of Fox-1/A2BP1 protein (Lee et al., 2009). Recently, a genome-wide analysis of mice with a central nervous system-specific gene depletion of Rbfox1 (A2BP1) revealed splicing changes in multiple transcripts that encode proteins important for synaptic functions and neuronal excitation (Gehman et al., 2011). Another example is exon III of the Tra2beta1 transcript, which appears to be a more complicated story. When rats were treated with pilocarpine, a drug that functions as an agonist of muscarinic acetylcholine receptor and elevates calcium levels in the cell, complex results that are suggestive of a cell-specific differential response to calcium level changes were obtained. The treatment led to a reduced inclusion of exon 3 of Tra2beta1 in the hippocampus and increased inclusion in the brain cortex (Daoud et al., 1999). In a subsequent study from the same group, it was shown that treatment of primary cortical neurons with thapsigargin as well as pilocarpine increased accumulation of the Tra2beta1 protein in the cytoplasm (Daoud et al., 2002). Furthermore, translocation of Tra2beta1 to the cytoplasm was also observed in ischemic induction, which increases the intracellular calcium levels. Interestingly, these treatments led to splicing changes of a downstream target (Daoud et al., 2002). Taken together, these studies strongly suggest that alterations in intracellular calcium levels may alter the localization of many of the spliceosomal components in neurons. Such changes can lead to alterations in splice site selection of downstream target genes that are regulated by these splicing factors.
Splicing change as a result of altered calcium level was also observed with transcripts of other splicing factors. However, in these cases, it is not clear if altered splicing leads to localization changes of the resulting proteins. Intron 6 in the pre-mRNA encoding Ania-6 (cyclin L1) was shown to be retained when calcium levels were increased with the use of glutamate in striatal neurons. Paradoxically, potassium chloride depolarization that also increases calcium levels led to less intron retention (Berke et al., 2001; Sgambato et al., 2003). Furthermore, exon 3 of hnRNP H3 and exon 2 of RNP-S1 were shown to be skipped when cortical neurons were depolarized by potassium chloride (An and Grabowski, 2007).
Calcium Levels Affect Splicing by Regulating Chromatin Modifiers
Recently, various studies have indicated that calcium signaling can influence chromatin modification, which in turn influences splice site selection. For example, an increase in intracellular calcium by depolarization using potassium chloride induces acetylation of H2B in the hippocampus of rats (Maharana et al., 2010). The elevated level of acetylation is reversed by addition of KN93, a CaMK inhibitor, suggesting that the acetylation of H2B requires CaMK (Maharana et al., 2010). The acetylation of H2B is also blocked by U0126, a specific inhibitor targeting ERK phosphorylation, suggesting that the increased acetylation is dependent on ERK phosphorylation. ERK is a known target for phosphorylation by CaMK; however, the mechanism by which ERK acetylates H2B remains unknown (Maharana et al., 2010). Furthermore, the treatment of rat hippocampal neurons with NMDA increases calcium influx, and activates the protein kinase A (PKA) or protein kinase C (PKC) pathway, which enhances ERK-dependent acetylation of Histone H3 (Levenson et al., 2004; Chandramohan et al., 2008). These data suggest that an increase in intracellular calcium levels can influence the acetylation levels of histones (Figure 1).
Changes of acetylation levels of histones by intracellular calcium signaling may lead to splicing pattern changes. It has been shown that cells treated with trichostatin A (TSA), a potent inhibitor of histone deacetylation, exhibit changes in mini-gene splicing (Nogues et al., 2002). In addition, studies carried out using yeast indicate that hyperacetylation of H3 by histone deacetylase (HDAC) depletion can delay the release of U2 snRNP, and perturb the assembly of spliceosome components (Gunderson et al., 2011). Recently an exon-array study showed that in HeLa cells treated with sodium butyrate (NaB), a potent HDAC inhibitor, 683 genes showed changes in splice site selection, including Tau, which is expressed in the central nervous system and associated with fronto-temporal dementia and Parkinson’s disease (Hnilicova et al., 2011).
Depolarization by potassium chloride induces hyperacetylation of histones H3 and H4 globally in N2A cells and this hyperacetylation is reversed when cells are treated with normal media suggesting that global hyperacetylation is a direct consequence of increased intracellular calcium (Schor et al., 2009). This depolarization and recovery coincide with the alternative splicing of NCAM exon 18, which is skipped when cells are depolarized and included after recovery (Schor et al., 2009). ChIP studies showed an increase in H3K9ac in the intragenic chromatin region between exons 17 and 19 in response to depolarization (Schor et al., 2009). These data suggest that hyperacetylation of histones in response to increases in calcium affects the splice site selection of exon 18. This idea was further supported by the use of TSA, which mimicked the results of depolarization (Schor et al., 2009).
H3K36-Me3 marks have been shown to differ at constitutive and alternative exons. H3K36-Me3 marks are found at the 5′ ends of the exons and enrichment of these methyl marks correlates with increases in alternative exon inclusion (Hon et al., 2009; Kolasinska-Zwierz et al., 2009). H3K36-Me3 can recruit PTB, a splicing regulator, to the pre-mRNA through the chromatin binding protein MRG15 (Luco et al., 2010). Recruitment of splicing machinery at a weak alternative splice site assists in the inclusion of the weak exon (Allo et al., 2011; Luco et al., 2011). Calcium signaling could also affect methylation of this histone mark, as it was shown that depolarization by potassium chloride also increases H3K36-Me3 at distal exons of the NCAM gene in hippocampal neurons, but the mechanism by which calcium signaling achieves this is not clear (Schor et al., 2009).
Data obtained from these studies indicate that an increase in calcium signaling could affect chromatin modifications, which may influence alternative splicing. It is tempting to speculate that in addition to the direct effects on RBPs, this might be another mechanism by which calcium regulates alternative splicing in neurons indirectly (Figure 1).
An intriguing new study demonstrates that exon 5 splicing of CD45 pre-mRNA is modulated during development as a consequence of chromatin modification (Shukla et al., 2011). This study shows that CTCF, a zinc-finger DNA binding protein, binds to the exon 5 region of the CD45 gene, slowing down transcription by providing a pausing of RNA Pol II. This allows for exon 5 inclusion at the early stages of lymphocyte development. During maturation of lymphocytes, CTCF binding is perturbed due to increased methylation on DNA in a region where CTCF binds to exon 5, leading to exon 5 skipping (Shukla et al., 2011). Given that expression of exon 5 of CD45 was shown to respond to cellular calcium level changes, it will be interesting to investigate whether calcium signaling affects the methylation status of DNA at exon 5, thereby modulating the splicing of exon 5 of the CD45 pre-mRNA.
Perspective
All of the studies discussed above point in the direction that changes in calcium signaling can play a major role in alternative splicing in neurons and other cell types. This could possibly be a molecular mechanism by which cells control the dynamic expression of exons that are important for their development and function. The calcium levels in neurons are under “homeostatic control” and disruption of the normal calcium level in neurons leads to disruption of neuronal functions and disease development (reviewed in Marambaud et al., 2009). Genome-wide exon arrays are now available and could be a useful tool in identifying the groups of genes whose alternative splicing is regulated by calcium signaling. With the advent of newer techniques like deep sequencing and genome-wide siRNA libraries with the augmentation of bioinformatic tools, it is time to study the effect of calcium-regulated alternative splicing and its role in physiological development and disease states at the genome-wide level. It is conceivable that expression of many alternative exons is subject to calcium signaling and that the dynamic regulation of these exons involves numerous distinct RNA elements and RBPs that interact with these RNA elements. Rules of calcium signaling-regulated alternative splicing will emerge from genome-wide studies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jiuyong Xie (University of Manitoba) for critical reading of the manuscript. We thank Melissa N. Hinman and Victoria A. Fleming for helpful discussion and editing. Hua Lou is supported by a NIH grant (NS-049103).
References
Allo, M., Schor, I. E., Munoz, M. J., de la Mata, M., Agirre, E., Valcarcel, J., Eyras, E., and Kornblihtt, A. R. (2011). Chromatin and alternative splicing. Cold Spring Harb. Symp. Quant. Biol. 75, 103–111.
An, P., and Grabowski, P. J. (2007). Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 5, e36. doi: 10.1371/journal.pbio.0050036
Ares, M. Jr. (2007). Sing the genome electric: excited cells adjust their splicing. PLoS Biol. 5, e55. doi: 10.1371/journal.pbio.0050055
Bentley, D. L. (2005). Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17, 251–256.
Berke, J. D., Sgambato, V., Zhu, P. P., Lavoie, B., Vincent, M., Krause, M., and Hyman, S. E. (2001). Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32, 277–287.
Black, D. L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336.
Black, D. L., and Grabowski, P. J. (2003). Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 31, 187–216.
Brodsky, A. S., Meyer, C. A., Swinburne, I. A., Hall, G., Keenan, B. J., Liu, X. S., Fox, E. A., and Silver, P. A. (2005). Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 6, R64.
Chandramohan, Y., Droste, S. K., Arthur, J. S., and Reul, J. M. (2008). The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D-aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur. J. Neurosci. 27, 2701–2713.
Daoud, R., Da Penha Berzaghi, M., Siedler, F., Hubener, M., and Stamm, S. (1999). Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur. J. Neurosci. 11, 788–802.
Daoud, R., Mies, G., Smialowska, A., Olah, L., Hossmann, K. A., and Stamm, S. (2002). Ischemia induces a translocation of the splicing factor tra2-beta 1 and changes alternative splicing patterns in the brain. J. Neurosci. 22, 5889–5899.
Egloff, S., and Murphy, S. (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24, 280–288.
Eperon, L. P., Graham, I. R., Griffiths, A. D., and Eperon, I. C. (1988). Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell 54, 393–401.
Gehman, L. T., Stoilov, P., Maguire, J., Damianov, A., Lin, C. H., Shiue, L., Ares, M. Jr., Mody, I., and Black, D. L. (2011). The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 43, 706–711.
Grosso, A. R., Gomes, A. Q., Barbosa-Morais, N. L., Caldeira, S., Thorne, N. P., Grech, G., von Lindern, M., and Carmo-Fonseca, M. (2008). Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 36, 4823–4832.
Gunderson, F. Q., Merkhofer, E. C., and Johnson, T. L. (2011). Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc. Natl. Acad. Sci. U.S.A. 108, 2004–2009.
Han, K., Yeo, G., An, P., Burge, C. B., and Grabowski, P. J. (2005). A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 3, e158. doi: 10.1371/journal.pbio.0030158
Hirose, Y., and Manley, J. L. (2000). RNA polymerase II and the integration of nuclear events. Genes Dev. 14, 1415–1429.
Hnilicova, J., Hozeifi, S., Duskova, E., Icha, J., Tomankova, T., and Stanek, D. (2011). Histone deacetylase activity modulates alternative splicing. PLoS ONE 6, e16727. doi: 10.1371/journal.pone.0016727
Hon, G., Wang, W., and Ren, B. (2009). Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput. Biol. 5, e1000566. doi: 10.1371/journal.pcbi.1000566
Jurica, M. S., and Moore, M. J. (2003). Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5–14.
Kaplan, C. D. (2009). Revealing the hidden relationship between nucleosomes and splicing. Cell Cycle 8, 3633–3634.
Kolasinska-Zwierz, P., Down, T., Latorre, I., Liu, T., Liu, X. S., and Ahringer, J. (2009). Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381.
Kornblihtt, A. R. (2005). Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 17, 262–268.
Kornblihtt, A. R., Schor, I. E., Allo, M., and Blencowe, B. J. (2009). When chromatin meets splicing. Nat. Struct. Mol. Biol. 16, 902–903.
Lee, J. A., Tang, Z. Z., and Black, D. L. (2009). An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 23, 2284–2293.
Lee, J. A., Xing, Y., Nguyen, D., Xie, J., Lee, C. J., and Black, D. L. (2007). Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 5, e40. doi: 10.1371/journal.pbio.0050040
Levenson, J. M., O’Riordan, K. J., Brown, K. D., Trinh, M. A., Molfese, D. L., and Sweatt, J. D. (2004). Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 279, 40545–40559.
Lewis, J. D., and Tollervey, D. (2000). Like attracts like: getting RNA processing together in the nucleus. Science 288, 1385–1389.
Li, H., Liu, G., Yu, J., Cao, W., Lobo, V. G., and Xie, J. (2009). In vivo selection of kinase-responsive RNA elements controlling alternative splicing. J. Biol. Chem. 284, 16191–16201.
Li, Q., Lee, J. A., and Black, D. L. (2007). Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8, 819–831.
Luco, R. F., Allo, M., Schor, I. E., Kornblihtt, A. R., and Misteli, T. (2011). Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26.
Luco, R. F., Pan, Q., Tominaga, K., Blencowe, B. J., Pereira-Smith, O. M., and Misteli, T. (2010). Regulation of alternative splicing by histone modifications. Science 327, 996–1000.
Maharana, C., Sharma, K. P., and Sharma, S. K. (2010). Depolarization induces acetylation of histone H2B in the hippocampus. Neuroscience 167, 354–360.
Marambaud, P., Dreses-Werringloer, U., and Vingtdeux, V. (2009). Calcium signaling in neurodegeneration. Mol. Neurodegener. 4, 20.
McKee, A. E., Neretti, N., Carvalho, L. E., Meyer, C. A., Fox, E. A., Brodsky, A. S., and Silver, P. A. (2007). Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome Biol. 8, R159.
Melton, A. A., Jackson, J., Wang, J., and Lynch, K. W. (2007). Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol. Cell. Biol. 27, 6972–6984.
Misteli, T., and Spector, D. L. (1999). RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3, 697–705.
Nogues, G., Kadener, S., Cramer, P., Bentley, D., and Kornblihtt, A. R. (2002). Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 277, 43110–43114.
Ogimoto, M., Katagiri, T., Hasegawa, K., Mizuno, K., and Yakura, H. (1993). Induction of CD45 isoform switch in murine B cells by antigen receptor stimulation and by phorbol myristate acetate and ionomycin. Cell. Immunol. 151, 97–109.
Pan, Q., Shai, O., Lee, L. J., Frey, B. J., and Blencowe, B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415.
Phatnani, H. P., Jones, J. C., and Greenleaf, A. L. (2004). Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43, 15702–15719.
Piacentini, L., Fanti, L., Negri, R., Del Vescovo, V., Fatica, A., Altieri, F., and Pimpinelli, S. (2009). Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 5, e1000670. doi: 10.1371/journal.pgen.1000670
Rosonina, E., and Blencowe, B. J. (2004). Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3’-end cleavage. RNA 10, 581–589.
Schor, I. E., Allo, M., and Kornblihtt, A. R. (2010). Intragenic chromatin modifications: a new layer in alternative splicing regulation. Epigenetics 5, 174–179.
Schor, I. E., Rascovan, N., Pelisch, F., Allo, M., and Kornblihtt, A. R. (2009). Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 106, 4325–4330.
Sgambato, V., Minassian, R., Nairn, A. C., and Hyman, S. E. (2003). Regulation of ania-6 splice variants by distinct signaling pathways in striatal neurons. J. Neurochem. 86, 153–164.
Shukla, S., Kavak, E., Gregory, M., Imashimizu, M., Shutinoski, B., Kashlev, M., Oberdoerffer, P., Sandberg, R., and Oberdoerffer, S. (2011). CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79.
Sims, R. J. III, Millhouse, S., Chen, C. F., Lewis, B. A., Erdjument-Bromage, H., Tempst, P., Manley, J. L., and Reinberg, D. (2007). Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28, 665–676.
Spies, N., Nielsen, C. B., Padgett, R. A., and Burge, C. B. (2009). Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 36, 245–254.
Topp, J. D., Jackson, J., Melton, A. A., and Lynch, K. W. (2008). A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14, 2038–2049.
Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., Kingsmore, S. F., Schroth, G. P., and Burge, C. B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476.
West, A. E., Chen, W. G., Dalva, M. B., Dolmetsch, R. E., Kornhauser, J. M., Shaywitz, A. J., Takasu, M. A., Tao, X., and Greenberg, M. E. (2001). Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. U.S.A. 98, 11024–11031.
Xie, J. (2008). Control of alternative pre-mRNA splicing by Ca(++) signals. Biochim. Biophys. Acta 1779, 438–452.
Xie, J., and Black, D. L. (2001). A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature 410, 936–939.
Xie, J., Jan, C., Stoilov, P., Park, J., and Black, D. L. (2005). A consensus CaMK IV-responsive RNA sequence mediates regulation of alternative exons in neurons. RNA 11, 1825–1834.
Yoneyama, M., Kawada, K., Shiba, T., and Ogita, K. (2011). Endogenous nitric oxide generation linked to ryanodine receptors activates cyclic GMP/protein kinase G pathway for cell proliferation of neural stem/progenitor cells derived from embryonic hippocampus. J. Pharmacol. Sci. 115, 182–195.
Yu, J., Hai, Y., Liu, G., Fang, T., Kung, S. K., and Xie, J. (2009). The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J. Biol. Chem. 284, 1505–1513.
Zhang, S. J., Zou, M., Lu, L., Lau, D., Ditzel, D. A., Delucinge-Vivier, C., Aso, Y., Descombes, P., and Bading, H. (2009). Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5, e1000604. doi: 10.1371/journal.pgen.1000604
Keywords: alternative splicing, depolarization, RNA-binding proteins, chromatin, histone modification, transcriptional elongation
Citation: Sharma A and Lou H (2011) Depolarization-mediated regulation of alternative splicing. Front. Neurosci. 5:141. doi: 10.3389/fnins.2011.00141
Received: 03 November 2011; Paper pending published: 23 November 2011;
Accepted: 06 December 2011; Published online: 27 December 2011.
Edited by:
Mark Reimers, Virginia Commonwealth University, USAReviewed by:
Leonard Rabinow, Université Paris Sud, FranceXi Huang, University of California San Francisco, USA
Copyright: © 2011 Sharma and Lou. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hua Lou, Department of Genetics, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA. e-mail:aHhsNDdAY2FzZS5lZHU=

