- Department of Developmental Biology, Kent Waldrep Foundation Center for Basic Neuroscience Research on Nerve Growth and Regeneration, University of Texas Southwestern Medical Center, Dallas, TX, USA
Myelination of axons by oligodendrocytes in the central nervous system is essential for normal neuronal functions. The failure of remyelination due to injury or pathological insults results in devastating demyelinating diseases. Oligodendrocytes originate in restricted regions of the embryonic ventral neural tube. After migration to populate all areas of the brain and spinal cord, oligodendrocyte precursors undergo a temporally well-defined series of molecular and structural changes, ultimately culminating in the cessation of proliferation, and the elaboration of a highly complex myelin sheath. The emergence of microRNAs (miRNAs) as potent regulators of gene expression at the posttranscriptional level has broad implications in all facets of cell biology. Recent studies have demonstrated a critical role of miRNAs in oligodendrocyte development, including cell proliferation, differentiation, and myelin formation. In this review, we will highlight and discuss the recent understanding of functional links of miRNAs to regulatory networks for central myelination, as well as perspectives on the role of miRNAs in demyelinating diseases.
Introduction
Oligodendrocytes (OLs) produce myelin sheaths that enwrap the internodal segments of axons to allow axonal insulation and saltatory conduction of action potentials in the vertebrate nervous system. During CNS development, OLs arise after successive stages of lineage progression from OL precursors (OPCs) to immature OLs and finally to mature myelinating OLs (Pfeiffer et al., 1993). This complex process initiates during embryonic development and continues into young adult life. It is regulated by extracellular signals and a network of intrinsic/intracellular factors with coordinated action in a spatially and temporally specific manner. Abnormal development and/or maintenance of myelin sheaths impairs propagation of action potential along nerve fibers, leading to nerve degeneration associated with acquired and inherited disorders including devastating multiple sclerosis (MS) and leukodystrophies (Trapp et al., 1998; Franklin, 2002; Mar and Noetzel, 2010).
During oligodendrocyte lineage progression, positive regulators are typically induced, whereas negative regulators are downregulated. This balance of positive and negative regulatory circuitry likely controls the differentiation timing of myelinating cells. A cohort of spatiotemporally specific factors that promote OL differentiation including Olig1/2, Sox10, YY1, Nkx2.2, Zfp488, MRF, and Zfp191 have been shown to (Xin et al., 2005; Wang et al., 2006; He et al., 2007; Li et al., 2007; Emery et al., 2009; Howng et al., 2010; Kuspert et al., 2011) and coordinate and balance inhibitory signaling pathways such as BMP, Wnt, and Notch signaling (Grinspan et al., 2000; Mekki-Dauriac et al., 2002; See et al., 2004; Fu et al., 2009; Emery, 2010). These inhibitory signaling pathways mediate through specific downstream effectors such as BMP/Smads, Wnt/β-catenin, Notch/Hes factors, ID2/4, Sox5/6, and NF1a/b at different developmental stages to inhibit oligodendrocyte differentiation and myelination (Grinspan et al., 2000; Mekki-Dauriac et al., 2002; See et al., 2004; Fu et al., 2009; Emery, 2010).
MicroRNAs (miRNAs) are emerging as important regulators of various aspects of CNS development by modulating, mainly downregulating, expression of extrinsic and intrinsic effectors, and activities of signaling networks in specific cell types and thereby, control fate specification, and differentiation of neural stem cells (Shi et al., 2010) and neurogenic niches (Brett et al., 2011).
In this review we summarize and discuss recent findings of miRNAs in regulating differentiation of myelinating cells in the CNS, with a focus on how specific miRNAs integrate into regulatory circuitries to control OL development in physiological and pathological conditions.
miRNA Biogenesis, Expression, and Function
MicroRNAs are short 20–25 nt non-coding RNAs processed from endogenous genomic loci that bind primarily to the 3′ untranslated region (UTR) of target mRNAs through an imperfect match to repress their translation and stability (Bartel, 2009). Like transcriptional regulators, miRNAs can cause large-scale changes in the proteome of a cell, thus regulating nearly every single process of development and disease (Bartel, 2009; Liu and Olson, 2010; Vo et al., 2010).
The biogenesis of miRNAs is tightly regulated in a spatially and temporally controlled manner and responds to environmental cues and cellular states (Figure 1). miRNAs are transcribed in most cases by RNA polymerase II to primary precursors transcripts (pri-miRNAs) that are first processed in the nucleus by Drosha and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8) into an 60–100 hairpin intermediate form (pre-miRNAs). The pre-miRNAs are then transported by the exportin 5-Ran GTPase shuttle system into the cytoplasm, where they are further cleaved by Dicer, an RNase III-like endonuclease to yield mature miRNAs (Figure 1). miRNAs are also produced by two non-canonical, Drosha- or Dicer-dependent pathways. In the first pathway, the early processing step is done by spliceosome and by a debranching enzyme that yields a short hairpin that is ready for further processing by Dicer. These non-canonical miRNAs have been termed mirtrons (Cheng et al., 2007; Ruby et al., 2007). In the second pathway, short hairpin RNAs (shRNAs) are processed by unknown nucleases into pre-microRNAs and are further processed into miRNAs by Dicer. miRNAs that are derived in this way have been termed endogenous shRNA-derived miRNAs (Babiarz et al., 2008). Additionally, recent studies describe a few miRNAs that are processed in a Drosha independent manner (Han et al., 2009), whereas other miRNAs appear to be Dicer1 independent (Cao et al., 2009; Chong et al., 2010).
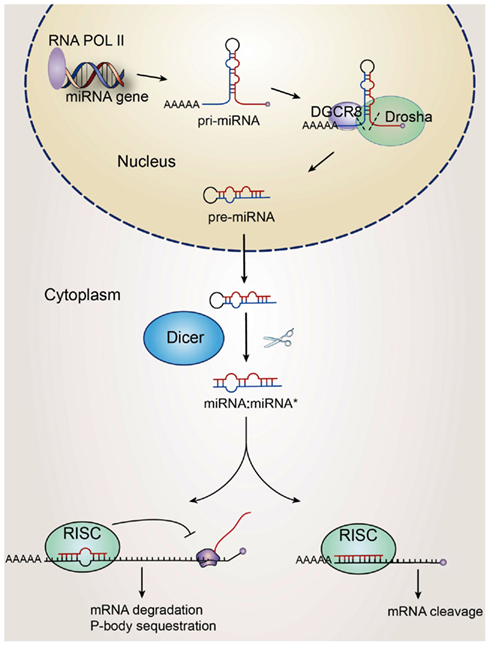
Figure 1. Biosynthesis of miRNAs. The miRNA gene is transcribed by RNA polymerase II as pri-miRNAs which are then processed by Drosha to produce long miRNA precursor (pre-miRNA). The pre-miRNAs are transported to the cytoplasm where they are further cleaved by the endoribonuclease Dicer to mature ∼22 nt long miRNA–miRNA *duplex. In this process Dicer interacts with the TRBP, PACT, and Argonaute proteins, which form the RNA-induced silencing complex (RISC). Within this complex, one strand of the miRNA duplex is removed and the single stranded miRNA, complementary to the target mRNA, remains in the complex and becomes functional. Seven to eight base pair “seed” sequence in 5′ miRNAs is partially complementary to the 3 UTR of mRNA targets, and induces posttranscriptional silencing through mechanisms such as mRNA destabilization and translational repression.
The mature miRNAs are then incorporated into the RNA-induced silencing complex (RISC), which contains an Argonaute-subfamily member, that then targets the 3′ UTR region of the target mRNAs by an imperfect match between the miRNA and the mRNA, to induce posttranscriptional gene silencing mainly through mRNA destabilization (Guo et al., 2010; Figure 1). A short “seed” sequence complementarity in 5′ miRNA (position 2–8) allows a single miRNA to pair with the 3′UTR of hundreds of mRNAs (Bartel, 2009). To add a level of complexity, miRNAs are also found to recognize seedless sites, 5′UTRs, and coding regions of mRNAs (Asirvatham et al., 2008; Tay et al., 2008; Lal et al., 2009). Although a majority of regulatory miRNAs appears to have a negative regulatory role, there are a few cases of miRNAs that can activate translation of target mRNAs (Vasudevan et al., 2007; Schwartz et al., 2008).
MicroRNAs are shown to widely participate in neurogenesis, including cell fate determination, neural patterning, synaptic plasticity, activity-dependent regulation, and neurological diseases (Kosik, 2006; Vo et al., 2010; Zhao et al., 2010), however its role in OL development is still is still in its infancy. Recently, OL development and miRNA fields have converged with the identification of OL-highly enriched miRNAs that are required for OL differentiation.
Dynamic Expression of miRNAs during Oligodendrocyte Development
MicroRNA expression profiling analysis of the oligodendrocyte lineage performed in rodents and in oligodendroglial cells derived from human ES cells (Lau et al., 2008; Letzen et al., 2010) reveals a cohort of miRNAs whose expression dynamically changes during the differentiation process. Although a few miRNAs in these two microarray screens are common, the majority of them are not overlapping at the same developmental stage, which might be due to the different cell sources or stages used (rat and human). However, the top twenty miRNAs showing highest expression in rat OPCs, such as miR-130a, miR-7, miR-16, miR-17, and miR-20a, are also highly expressed inhuman glial progenitors. Similarly, miR-17, miR-20a, miR-21, miR-16, miR-103, and miR-107 identified in rat premyelinating OL showed overlapping expression with the human OPCs (Lau et al., 2008; Letzen et al., 2010). Currently the functions of these miRNAs in OL development remain elusive.
Interestingly, some miRNAs, such as miR-205, exhibit a high level of expression in human embryonic stem cells and early neural progenitor stages, while they are downregulated during early OPCs transition. However, their expression level becomes upregulated again during mid to late OPC differentiation (Letzen et al., 2010). This dynamic expression of miRNAs reflects the complexity of miRNAs during differentiation, suggesting that miRNAs may have unrelated roles at various stages of lineage development. Specifically, miR-205 contains a conserved 8 mer complementary site within the Cldn11 3’-UTR, encoding for a transmembrane protein Claudin-11 critical for tight junction formation in the myelin sheath (Gow et al., 1999; Bronstein et al., 2000).
Another miRNA, miR-199a-5p increases in early OPCs derived from human ES cells and displays a strong target bias toward C11Orf9 (Figure 2). miR-199a-5p has three sites with evolutionarily conserved 8 mer seed matches to the 3’ UTR of C11Orf9, suggesting a high probability for miRNA–mRNA interactions. This gene, C11Orf, is speculated to be the human ortholog to the mouse MRF, a protein that has been shown to be critical for oligodendrocyte maturation and myelin production (Emery et al., 2009). Thus, it is possible that the downregulation in miR-199a-5p expression may be concomitant with the initiation of MBP expression.
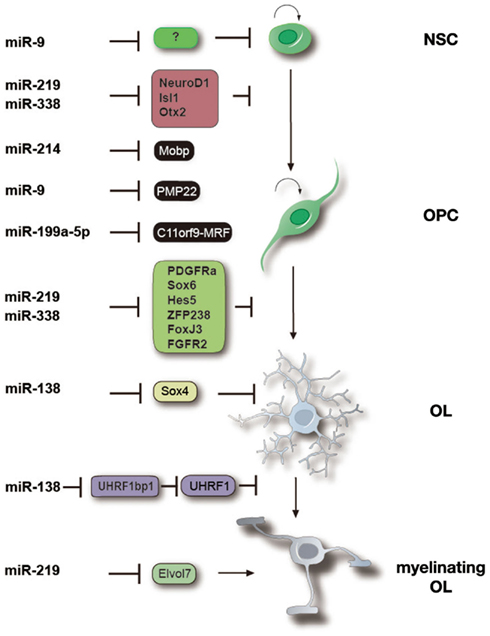
Figure 2. miRNAs function at multiple stages to control oligodendrocyte differentiation and myelination. Specific miRNAs involved in regulating the neural precursor to OPC transition, OPC differentiation, OL maturation and myelination as well as the documented and predicted targets of those miRNAs are shown. miR-9, miR-214, and miR-199a-5p are highly expressed in OPCs and prevent premature differentiation by blocking expression of myelin components including Mobp, PMP22 and maturation promoting factors such as MRF/C11orf9. miR-138, -219 and -338 are more highly expressed in OLs and promote OL differentiation by inhibiting expression of differentiation inhibitors (e.g., Hes5, Sox6, and Sox4) and proliferation signaling molecules such as PDGFRα and FGFR, as well as neuronal differentiation factors (e.g., Zfp238 and FoxJ3). miR-219 may also regulate OL terminal maturation and myelin maintenance by targeting fatty acid elongases such as Elvol7. The expression pattern of miRNAs is likely correlated to their functions at different stages of OL lineage progression.
miR-214 is highly expressed in rat OPCs but undetectable in OLs. It has a strong evolutionarily conserved 8 mer target site to Mobp, which is important for providing proper structural properties of myelin (Letzen et al., 2010; Figure 2), suggesting that miR-214 might block myelin sheath formation.
Other miRNAs may function to maintain myelin structure. miR-23 is a negative regulator of lamin B1 (Lin and Fu, 2009). Excessive Lamin B1 expression has been shown to cause severe CNS myelin loss in adult-onset autosomal dominant leukodystrophy patients by repressing production of myelin proteins such as MBP, PLP, and MOG. This suggests that miRNAs such as miR-23 could play an important role in myelin maintenance (Lin and Fu, 2009).
In general, mRNAs exhibit a high degree of stage and tissue specificity. The expression profile of miRNAs strictly depends on the differentiation program occurring. As consequence, miRNAs in the control of OL fate specification are still largely unknown at present and warrant further investigation.
miRNAs in Neural and Glial Specification
Deletion of miRNA-processing enzyme Dicer in neural progenitor cells was found to control the switch of neurogenesis to gliogenesis (Kawase-Koga et al., 2009; Zheng et al., 2010). A series of evidence indicate that miRNAs play critical roles in neuronal and astroglial cell fate decision.
One of the most highly expressing miRNAs in the CNS that regulates neural/glial specification is miR-124. During neurogenesis, miR-124 is undetectable or expressed at low levels in progenitor cells and is upregulated in differentiating and mature neurons (Deo et al., 2006). Overexpression of the neurogenic miR-124 in HeLa cells represses expression of many non-neuronal genes, and activates a gene expression program more similar to that of neuronal cells (Lim et al., 2005). Although inhibiting miR-124 did not affect neuronal differentiation, at least in chick neural tube (Cao et al., 2007), more recently, miR-124 was shown to promote neuronal differentiation and cell cycle exit of neural stem cells in the subventricular zone, one of the neurogenic niches of the adult mammalian brain (Cheng et al., 2009).
Recently, Yoo et al. (2009) showed that miR-124 and miR-9 facilitate the conversion of human fibroblasts into neurons by modifying the subunit composition of the BAF chromatin-remodeling complex, thereby transforming it into a neuronal specific chromatin remodeler. miR-9is highly expressed in mouse OPCs and targets the 3′UTR of an mRNA encoding peripheral myelin protein Pmp22 (Figure 2). Together with the inhibition of astrocytic GFAP, miR-9 may repress expression of non-lineage related proteins in oligodendrocytes and function as a guardian to maintain oligodendroglial identity (Krichevsky et al., 2006; Figure 2). It will be interesting to investigate whether overexpression of miR-9 in cooperation with OL-specific transcription factors, might also convert fibroblast into oligodendrocyte-like cells.
Control of Oligodendrocyte Proliferation by miRNAs
As in many other cell types, the initiation of OL differentiation is very tightly linked to the cessation of OPC proliferation and cell cycle withdrawal (Temple and Raff, 1985; Barres and Raff, 1994; Raff et al., 1998). However, targeted disruption of cell cycle components that results in proliferation arrest is often insufficient to promote the program of OL differentiation (Casaccia-Bonnefil and Liu, 2003). Deletion of Dicer1 gene in oligodendrocyte lineage cells induced OPC proliferation, and a drastic reduction in myelination, suggesting that miRNAs are required for normal OPC cell cycle exit and differentiation (Dugas et al., 2010; Zhao et al., 2010). Those two studies, identified that miR-219, miR-138, and miR-338 may function by targeting OPC-expressed genes whose repression is necessary to promote the rapid cessation of proliferation coupled to OL differentiation.
Like miR-138, miR-219, and miR-338, miR-17–92 miRNA cluster, also known as Oncomir-1, is highly enriched in oligodendrocytes (Lau et al., 2008; Budde et al., 2010). Targeted inactivation of miR-17–92 cluster leads to a reduction in the number of oligodendrocytes, in vivo (Budde et al., 2010). Indeed, expression of the Oncomir-1 in primary cultures of OPCs promotes cell proliferation by influencing Akt signaling (Budde et al., 2010). Consistent with the role of this miRNA in promoting cell survival (Ventura et al., 2008), miR-17–92 cluster plays an important role in OPC survival and proliferation.
On the other hand, miR-192, a miRNAs induced during OL differentiation, has been directly implicated in arresting the cell cycle by coordinately repressing several G1-S and G2-M cell cycle checkpoint control genes (Georges et al., 2008). Another OL-induced miRNA, miR-181a, functions as a tumor suppressor in gliomas (Shi et al., 2008).
Recently, by using an elegant mosaic mouse model, Liu et al. (2011) demonstrated that OPCs are a major cell of origin of glioma, which might link the role of miRNAs in promoting OL differentiation by inhibiting OPC proliferation-promoting genes. Specifically, if OL-expressed miRNAs inhibit cellular proliferation, the loss of these miRNAs could create a permissive environment for tumorigenesis. In fact, OL-enriched miR-219, miR-138, and miR-192 are downregulated in brain tumor samples relative to normal tissue control (Ferretti et al., 2009).
Collectively, these results suggest that miRNAs are essential in determining oligodendroglial cell number by promoting OPC cell cycle exit and survival in a temporally specific fashion. Indeed, miRNAs may offer novel avenues for future therapeutic development in CNS tumors.
Role of Specific miRNAs in Oligodendrocyte Differentiation
Several groups have performed miRNA microarray profiling on the CNS tissues of wild-type and myelin deficient Dicer1 knockout mice and identified a cohort of miRNAs that are downregulated in myelin deficient Dicer1 mutants (Shin et al., 2009; Zhao et al., 2010). Among them, miR-219 and miR-338 are substantially increased at the onset of oligodendrocyte myelination, and in mature oligodendrocytes and play a positive role in promoting OPC differentiation. Expression of stable miRNA mimics for either miR-219 or miR-338-5p and miR-338-3p together can promote OPC differentiation and partially rescue the differentiation deficit of Dicer1-deleted OPCs in vitro (Dugas et al., 2010; Zhao et al., 2010). Overexpression of miR-219 or miR-338 miRNA is sufficient to promote precocious expression of oligodendrocyte lineage markers in the developing chick and mouse CNS (Zhao et al., 2010). Consistently, knock-down of these two miRNAs in cultured OPCs (Dugas et al., 2010; Zhao et al., 2010) or miR-219 in zebrafish embryos (note that miR-338 is not detectable in the CNS of zebrafish embryos; Zhao et al., 2010) inhibits OPC differentiation suggesting that miR-219 is both necessary and sufficient for oligodendrocyte differentiation.
On the other hand, miR-138 exhibits transient expression in early postmitotic oligodendrocytes. Expression of miR-138 stimulates early differentiation, but inhibits oligodendrocyte terminal differentiation (Dugas et al., 2010).
Fine-Tuning Oligodendrocyte Differentiation through Temporal Negative Feedback Loops
miR-219 and miR-338 target a number of genes that have previously been shown to be involved in inhibiting OPC differentiation and maintaining OPCs in the proliferative state, including PDGFRα, Hes5, and Sox6 (Dugas et al., 2010; Zhao et al., 2010; Figure 2). Interestingly, miR-219 may also target other transcription factors such as Zfp238, FoxJ3, NeuroD1, Isl1, and Otx2 that are potentially involved in promoting neurogenesis (Dugas et al., 2010; Zhao et al., 2010; Figure 2). Since overexpression of these pro-neural factors is able to inhibit OPC differentiation, miRNA-mediated inhibition of the factors may restrict differentiation of neural progenitors to the oligodendrocyte lineage. Together, a network of miRNAs individually or cooperatively regulates distinct stages of oligodendrocyte lineage progression, as well as safeguards against the expression of neuronal and other cell lineage genes.
Besides regulating the differentiation program, miR-219 also regulates a lipid metabolism such as by targeting fatty acid elongase ELOVL7, which synthesizes very long chain of fatty acid (Figure 2). Accumulation of fatty acids were observed in PLP-CreERT Dicer-floxed mice (Shin et al., 2009), suggesting miRNAs such as miR-219 also play a role in myelin membrane homeostasis. Collectively, miRNAs function at multiple stages during oligodendrocyte lineage development to direct and fine-tune oligodendrocyte differentiation processes (Figure 2).
miRNAs in Demyelinating Diseases
The progressive loss of CNS myelin in patients with MS results from the combined effects of damage to oligodendrocytes and remyelination failure. A common feature of the demyelinated lesions is the presence of OPCs at the premyelinating stage. However, the mechanistic basis for the impaired myelin repair is not completely understood. Since MS is heterogeneous with respect to clinical manifestations and responses to therapy, the identification of biomarkers appears desirable for an improved diagnosis of MS as well as for monitoring of disease activity and treatment response.
MicroRNA profiles have been established from active and inactive demyelinating lesions (Junker et al., 2009) and peripheral blood cells (Du et al., 2009; Keller et al., 2009; Otaegui et al., 2009) from MS patients. Those studies documented differences in miRNA expression patterns in MS patients compared to healthy controls as well as in relapse versus remission of the disease. Due to substantial gliosis and immune cell infiltration in demyelinating lesions, strongly upregulated miRNAs in tissue lesions are mainly assigned to astrocytes, T cells, and monocytes (Junker et al., 2009). Indeed, miRNA profiling in isolated cells by laser capture microdissection from active and inactive MS lesions shows that the upregulated miRNAs in active MS lesions, such as miR-155, miR-650, miR-34a, and miR-326, are mainly assigned to astrocytes and infiltrating immune cells (Junker et al., 2009). Consistently, Dicer1 ablation in mature OLs results in demyelination and inflammatory neuronal degeneration similar to that of MS (Dugas et al., 2010), suggesting that miRNAs are not only key players during development but also during myelin maintenance or disease formation.
miR-219 and miR-338 are highly expressed in mature oligodendrocytes and their expression is lost when Dicer1 function is specifically ablated in mature oligodendrocytes (Shin et al., 2009). Strikingly, mature OL-enriched miRNAs such as miR-219 and miR-338 are the least detectable miRNAs in chronic MS lesions (Junker et al., 2009), suggesting that these miRNAs may also function in human OL maturation and myelin repair. On the other hand, since miR-219 and miR-338 are highly expressed in OLs, the decrease of those miRNAs expression might reflect the mature myelinating OL loss in the MS plaques. Further studies will be necessary to elucidate the role of those miRNAs in MS pathogenesis and whether those miRNAs can promote remyelination process.
Other miRNAs such as miRNA-23 may contribute to demyelinating diseases such as adult-onset autosomal dominant leukodystrophy by targeting lamin B1 (Lin and Fu, 2009). Elevated level of LaminB1 by gene duplications preferentially leads to myelin loss in the CNS.
As miRNAs are more stable than mRNAs, they are good candidates as disease biomarkers. Moreover, expression of miRNAs can be detected and quantified by sensitive luminescence-based, fluorescence-based and nanotechnology-based methods (Kong et al., 2009). The use of miRNAs as biomarkers has gained growing interest in the last few years. Blood serum and plasma are important sample types for investigating miRNAs as biomarkers. For example, Keller et al. (2009) investigated the expression profile of human miRNAs in whole blood cells of MS patients and found 165 miRNAs significantly up- or down-regulated in patients with relapsing/remitting MS as compared to healthy controls. The best single miRNA marker, miR-145, allows discriminating MS patients from controls with a specificity of 89.5%, suggesting that single miRNA, or a cohort of miRNAs, may have the potential to serve as diagnostic biomarkers for relapsing/remitting MS.
MicroRNA profiling in different demyelinating diseases to identify miRNAs that reflect the fundamental feature of demyelinating pathology may point to new biomarkers for demyelinating diseases such as MS. Further functional analyses of these miRNAs in regulating expression of target mRNAs are critical to understand their impact on pathogenesis and progress of the disease.
Conclusion
The discovery of miRNAs enables a deeper insight into complexity of gene regulatory networks during the myelination process. It is clear that understanding OL development in general requires a deeper appreciation for epigenetic mechanisms that modulate OL identity, including their interplay with genetic and other cell-intrinsic factors and cell-extrinsic and cell–cell cues, such as those derived from the microenvironment. Efforts are underway to collect and analyze the individual components of the entire miRNA network, and their functional interplay with regulatory circuitry that controls the myelination program during development and myelin regeneration after injury.
The functional significance of some of these miRNAs is beginning to emerge. miRNAs play multiple roles at various stages of OL development, including in the initial production of fate-specified OPCs, in the differentiation of mature OLs and generation of compact CNS myelin during development, and in the maintenance of functional myelin sheaths in adult animals.
Understanding miRNA signatures during development and in disease states, will be valuable both for diagnostic and therapeutic purposes. miRNAs have an advantage over mRNAs as they are more stable and do not undergo a significant decay during the tissue sample processing (Jung et al., 2010). The robustness of profiling data suggests that miRNAs are potential biomarkers for diagnosis or monitoring demyelinating diseases. Further uncovering key functions for miRNAs will illuminate important aspects in regulating myelination processes. New knowledge about these small regulatory molecules will offer novel therapeutic interventions by which disease-related miRNAs could be antagonized or functionally restored for myelin repair. The future challenge will be to translate this knowledge into improved outcome for patients with demyelinating diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Yang Yu for figure drawing. This study was funded in part by grants from the US National Institutes of Health (NS072427) and the National Multiple Sclerosis Society (RG3978) to Q. Richard Lu.
References
Asirvatham, A. J., Gregorie, C. J., Hu, Z., Magner, W. J., and Tomasi, T. B. (2008). MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol. Immunol. 45, 1995–2006.
Babiarz, J. E., Ruby, J. G., Wang, Y., Bartel, D. P., and Blelloch, R. (2008). Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785.
Barres, B. A., and Raff, M. C. (1994). Control of oligodendrocyte number in the developing rat optic nerve. Neuron 12, 935–942.
Brett, J. O., Renault, V. M., Rafalski, V. A., Webb, A. E., and Brunet, A. (2011). The microRNA cluster miR-106b∼25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany N. Y.) 3, 108–124.
Bronstein, J. M., Tiwari-Woodruff, S., Buznikov, A. G., and Stevens, D. B. (2000). Involvement of OSP/claudin-11 in oligodendrocyte membrane interactions: role in biology and disease. J. Neurosci. Res. 59, 706–711.
Budde, H., Schmitt, S., Fitzner, D., Opitz, L., Salinas-Riester, G., and Simons, M. (2010). Control of oligodendroglial cell number by the miR-17-92 cluster. Development 137, 2127–2132.
Cao, F., Li, X., Hiew, S., Brady, H., Liu, Y., and Dou, Y. (2009). Dicer independent small RNAs associate with telomeric heterochromatin. RNA 15, 1274–1281.
Cao, X., Pfaff, S. L., and Gage, F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536.
Casaccia-Bonnefil, P., and Liu, A. (2003). Relationship between cell cycle molecules and onset of oligodendrocyte differentiation. J. Neurosci. Res. 72, 1–11.
Cheng, H. Y., Papp, J. W., Varlamova, O., Dziema, H., Russell, B., Curfman, J. P., Nakazawa, T., Shimizu, K., Okamura, H., Impey, S., and Obrietan, K. (2007). microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829.
Cheng, L. C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Chong, M. M., Zhang, G., Cheloufi, S., Neubert, T. A., Hannon, G. J., and Littman, D. R. (2010). Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 24, 1951–1960.
Deo, M., Yu, J. Y., Chung, K. H., Tippens, M., and Turner, D. L. (2006). Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235, 2538–2548.
Du, C., Liu, C., Kang, J., Zhao, G., Ye, Z., Huang, S., Li, Z., Wu, Z., and Pei, G. (2009). MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10, 1252–1259.
Dugas, J. C., Cuellar, T. L., Scholze, A., Ason, B., Ibrahim, A., Emery, B., Zamanian, J. L., Foo, L. C., McManus, M. T., and Barres, B. A. (2010). Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611.
Emery, B. (2010). Transcriptional and post-transcriptional control of CNS myelination. Curr. Opin. Neurobiol. 20, 601–607.
Emery, B., Agalliu, D., Cahoy, J. D., Watkins, T. A., Dugas, J. C., Mulinyawe, S. B., Ibrahim, A., Ligon, K. L., Rowitch, D. H., and Barres, B. A. (2009). Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172–185.
Ferretti, E., De Smaele, E., Po, A., Di Marcotullio, L., Tosi, E., Espinola, M. S., Di Rocco, C., Riccardi, R., Giangaspero, F., Farcomeni, A., Nofroni, I., Laneve, P., Gioia, U., Caffarelli, E., Bozzoni, I., Screpanti, I., and Gulino, A. (2009). MicroRNA profiling in human medulloblastoma. Int. J. Cancer 124, 568–577.
Franklin, R. J. (2002). Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 3, 705–714.
Fu, H., Cai, J., Clevers, H., Fast, E., Gray, S., Greenberg, R., Jain, M. K., Ma, Q., Qiu, M., Rowitch, D. H., Taylor, C. M., and Stiles, C. D. (2009). A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J. Neurosci. 29, 11399–11408.
Georges, S. A., Biery, M. C., Kim, S. Y., Schelter, J. M., Guo, J., Chang, A. N., Jackson, A. L., Carleton, M. O., Linsley, P. S., Cleary, M. A., and Chau, B. N. (2008). Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 68, 10105–10112.
Gow, A., Southwood, C. M., Li, J. S., Pariali, M., Riordan, G. P., Brodie, S. E., Danias, J., Bronstein, J. M., Kachar, B., and Lazzarini, R. A. (1999). CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99, 649–659.
Grinspan, J. B., Edell, E., Carpio, D. F., Beesley, J. S., Lavy, L., Pleasure, D., and Golden, J. A. (2000). Stage-specific effects of bone morphogenetic proteins on the oligodendrocyte lineage. J. Neurobiol. 43, 1–17.
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840.
Han, J., Pedersen, J. S., Kwon, S. C., Belair, C. D., Kim, Y. K., Yeom, K. H., Yang, W. Y., Haussler, D., Blelloch, R., and Kim, V. N. (2009). Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136, 75–84.
He, Y., Dupree, J., Wang, J., Sandoval, J., Li, J., Liu, H., Shi, Y., Nave, K. A., and Casaccia-Bonnefil, P. (2007). The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron 55, 217–230.
Howng, S. Y., Avila, R. L., Emery, B., Traka, M., Lin, W., Watkins, T., Cook, S., Bronson, R., Davisson, M., Barres, B. A., and Popko, B. (2010). ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 24, 301–311.
Jung, M., Schaefer, A., Steiner, I., Kempkensteffen, C., Stephan, C., Erbersdobler, A., and Jung, K. (2010). Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 56, 998–1006.
Junker, A., Krumbholz, M., Eisele, S., Mohan, H., Augstein, F., Bittner, R., Lassmann, H., Wekerle, H., Hohlfeld, R., and Meinl, E. (2009). MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132(Pt 12), 3342–3352.
Kawase-Koga, Y., Otaegi, G., and Sun, T. (2009). Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 238, 2800–2812.
Keller, A., Leidinger, P., Lange, J., Borries, A., Schroers, H., Scheffler, M., Lenhof, H. P., Ruprecht, K., and Meese, E. (2009). Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE 4, e7440. doi:10.1371/journal.pone.0007440
Kong, W., Zhao, J. J., He, L., and Cheng, J. Q. (2009). Strategies for profiling microRNA expression. J. Cell. Physiol. 218, 22–25.
Krichevsky, A. M., Sonntag, K. C., Isacson, O., and Kosik, K. S. (2006). Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24, 857–864.
Kuspert, M., Hammer, A., Bosl, M. R., and Wegner, M. (2011). Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 39, 1280–1293.
Lal, A., Navarro, F., Maher, C. A., Maliszewski, L. E., Yan, N., O’Day, E., Chowdhury, D., Dykxhoorn, D. M., Tsai, P., Hofmann, O., Becker, K. G., Gorospe, M., Hide, W., and Lieberman, J. (2009). miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell 35, 610–625.
Lau, P., Verrier, J. D., Nielsen, J. A., Johnson, K. R., Notterpek, L., and Hudson, L. D. (2008). Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J. Neurosci. 28, 11720–11730.
Letzen, B. S., Liu, C., Thakor, N. V., Gearhart, J. D., All, A. H., and Kerr, C. L. (2010). MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS ONE 5, e10480. doi:10.1371/journal.pone.0010480
Li, H., Lu, Y., Smith, H. K., and Richardson, W. D. (2007). Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J. Neurosci. 27, 14375–14382.
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S., and Johnson, J. M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773.
Lin, S. T., and Fu, Y. H. (2009). miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model. Mech. 2, 178–188.
Liu, C., Sage, J. C., Miller, M. R., Verhaak, R. G., Hippenmeyer, S., Vogel, H., Foreman, O., Bronson, R. T., Nishiyama, A., Luo, L., and Zong, H. (2011). Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146, 209–221.
Liu, N., and Olson, E. N. (2010). MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510–525.
Mekki-Dauriac, S., Agius, E., Kan, P., and Cochard, P. (2002). Bone morphogenetic proteins negatively control oligodendrocyte precursor specification in the chick spinal cord. Development 129, 5117–5130.
Otaegui, D., Baranzini, S. E., Armananzas, R., Calvo, B., Munoz-Culla, M., Khankhanian, P., Inza, I., Lozano, J. A., Castillo-Trivino, T., Asensio, A., Olaskoaga, J., and Lopez de Munain, A. (2009). Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE 4, e6309. doi:10.1371/journal.pone.0006309
Pfeiffer, S. E., Warrington, A. E., and Bansal, R. (1993). The oligodendrocyte and its many cellular processes. Trends Cell Biol. 3, 191–197.
Raff, M. C., Durand, B., and Gao, F. B. (1998). Cell number control and timing in animal development: the oligodendrocyte cell lineage. Int. J. Dev. Biol. 42, 263–267.
Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86.
Schwartz, J. C., Younger, S. T., Nguyen, N. B., Hardy, D. B., Monia, B. P., Corey, D. R., and Janowski, B. A. (2008). Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 15, 842–848.
See, J., Zhang, X., Eraydin, N., Mun, S. B., Mamontov, P., Golden, J. A., and Grinspan, J. B. (2004). Oligodendrocyte maturation is inhibited by bone morphogenetic protein. Mol. Cell. Neurosci. 26, 481–492.
Shi, L., Cheng, Z., Zhang, J., Li, R., Zhao, P., Fu, Z., and You, Y. (2008). hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 1236, 185–193.
Shi, Y., Zhao, X., Hsieh, J., Wichterle, H., Impey, S., Banerjee, S., Neveu, P., and Kosik, K. S. (2010). MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 30, 14931–14936.
Shin, D., Shin, J. Y., McManus, M. T., Ptacek, L. J., and Fu, Y. H. (2009). Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol. 66, 843–857.
Tay, Y., Zhang, J., Thomson, A. M., Lim, B., and Rigoutsos, I. (2008). MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128.
Temple, S., and Raff, M. C. (1985). Differentiation of a bipotential glial progenitor cell in a single cell microculture. Nature 313, 223–225.
Trapp, B. D., Peterson, J., Ransohoff, R. M., Rudick, R., Mork, S., and Bo, L. (1998). Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med. 338, 278–285.
Vasudevan, S., Tong, Y., and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934.
Ventura, A., Young, A. G., Winslow, M. M., Lintault, L., Meissner, A., Erkeland, S. J., Newman, J., Bronson, R. T., Crowley, D., Stone, J. R., Jaenisch, R., Sharp, P. A., and Jacks, T. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886.
Vo, N. K., Cambronne, X. A., and Goodman, R. H. (2010). MicroRNA pathways in neural development and plasticity. Curr. Opin. Neurobiol. 20, 457–465.
Wang, S. Z., Dulin, J., Wu, H., Hurlock, E., Lee, S. E., Jansson, K., and Lu, Q. R. (2006). An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development 133, 3389–3398.
Xin, M., Yue, T., Ma, Z., Wu, F. F., Gow, A., and Lu, Q. R. (2005). Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J. Neurosci. 25, 1354–1365.
Yoo, A. S., Staahl, B. T., Chen, L., and Crabtree, G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646.
Zhao, X., He, X., Han, X., Yu, Y., Ye, F., Chen, Y., Hoang, T., Xu, X., Mi, Q. S., Xin, M., Wang, F., Appel, B., and Lu, Q. R. (2010). MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65, 612–626.
Keywords: miRNAs, oligodendrocyte, myelination, feed-back regulation, neural cell fate, transcriptional control
Citation: Barca-Mayo O and Lu QR (2012) Fine-Tuning Oligodendrocyte Development by Micrornas. Front. Neurosci. 6:13. doi: 10.3389/fnins.2012.00013
Received: 11 October 2011;
Paper pending published: 06 November 2011;
Accepted: 19 January 2012;
Published online: 06 February 2012.
Edited by:
Yanhong Shi, City of Hope, USAReviewed by:
Ashok K. Shetty, Texas A&M Health Science Center College of Medicine at Scott & White, USAEdwin S. Monuki, University of California Irvine, USA
Copyright: © 2012 Barca-Mayo and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Q. Richard Lu, Department of Developmental Biology, UT Southwestern Medical Center, 6000 Harry Hines Blvd. Dallas, TX 75390-9133, USA. e-mail:cXJpY2hhcmQubHVAdXRzb3V0aHdlc3Rlcm4uZWR1

