- Department of Molecular Biology, UT Southwestern Medical Center, Dallas, TX, USA
Multipotent neural stem cells (NSCs) possess the ability to self-renew and differentiate into both neurons and glia. However, the detailed mechanisms underlying NSC fate decisions are not well understood. Recent work suggests that the interaction between cell type specific transcription factors and microRNAs (miRNAs) is important as resident neural stem/progenitor cells give rise to functionally mature neurons. Recently, we demonstrated that the transcriptional repressor REST (RE1-silencing transcription factor) is essential to prevent precocious neuronal differentiation and maintain NSC self-renewal in the adult hippocampus. Here we show that REST is required for orchestrating the expression of distinct subsets of miRNAs in primary mouse NSC cultures, a physiologically relevant cell type. Using miRNA array profiling, we identified known REST-regulated miRNA genes, as well as previously uncharacterized REST-dependent miRNAs. Interestingly, in response to proliferation and differentiation stimuli, REST-regulated miRNAs formed distinct clusters and displayed variable expression dynamics. These results suggest that REST functions in a context-dependent manner through its target miRNAs for mediating neuronal production.
Introduction
The process of neuronal production from multipotent neural stem cells (NSCs) continues in the postnatal and adult mammalian brain in two discrete locations: the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus (Ming and Song, 2005; Kempermann et al., 2008). Here, NSCs self-renew and give rise to fate-restricted precursors that ultimately differentiate into mature neurons and astrocytes (Ming and Song, 2005; Kempermann et al., 2008). SGZ and SVZ neurogenesis is regulated by both extrinsic factors, such as receptor-mediated signaling pathways (Ming and Song, 2005), and intrinsic factors, such as epigenetic mechanisms (Hsieh and Gage, 2004). Understanding how the epigenome integrates extrinsic and intrinsic signals in multipotent NSCs is critical to harness their therapeutic potential in regenerative medicine.
Epigenetic mechanisms, including histone modifications and chromatin remodeling, have emerged to play critical roles in NSC self-renewal and differentiation (Hsieh and Gage, 2004). Disruptions in epigenetic regulation within the brain have been associated with neurodevelopmental and neuropsychiatric disorders (e.g., Angelman, Prader–Willi, and Rett syndrome; Amir et al., 1999; Sahoo et al., 2008; Mabb et al., 2011). For example, the transcriptional repressor REST has been the focus of research for many years, and is linked to neurological disorders such as Huntington’s disease and epilepsy (Zuccato et al., 2003; Jessberger et al., 2007). REST is a zinc finger protein with a DNA-binding domain in the middle region and two repressor domains in its N- and C-terminal regions (Ballas and Mandel, 2005). REST exerts its repressor function through a 21–23 bp DNA element known as RE1. Moreover, recent bioinformatics analyses reveal more than 1300 RE1 sites throughout the mammalian genome (Bruce et al., 2004; Johnson et al., 2007), consistent with the widespread role of REST.
We recently demonstrated that REST is required for the coordination of the neuronal gene program to ensure a balance between neuronal production and maintaining the NSC pool (Gao et al., 2011). How REST controls stage-specific neuronal gene expression is still unclear. In addition to regulating coding genes, REST has also been shown to regulate neurogenesis through the actions of microRNAs (miRNAs or miRs), such as miR-124a (Conaco et al., 2006). These authors found that REST and miR-124a are expressed in a reciprocal manner and that miR-124a is a direct target of REST. MiRNAs belong to a class of non-coding RNAs ∼22 nucleotides in length and conserved in metazoans (Bartel, 2004). By binding 3′UTRs of target messenger RNAs, miRNAs degrade their transcript and/or inhibit translation (Bartel, 2004). Since this first study, the list of miRNAs that are predicted or regulated by REST has increased (Wu and Xie, 2006; Otto et al., 2007), however none of these studies address whether these targets are dependent on the physiological role of REST per se.
Here using primary mouse NSCs derived from a floxed REST allele, we show that REST is required for the proper expression of miRNA genes, many of which are potential novel REST targets. The loss of REST results in different subsets of miRNA expression changes in NSCs grown under proliferation versus differentiation conditions. In addition, we found that the expression of REST-regulated miRNAs dynamically changes over time. These results reveal the context-dependent nature of REST regulation and suggest that loss of REST and altered expression of miRNAs may potentially contribute to the premature neuronal differentiation and exhaustion of the NSC pool observed in REST-deficient mice.
Materials and Methods
Primary NSC Culture
The derivation, differentiation, viral infection, and immunostaining of primary mouse NSCs were detailed in our previous study (Gao et al., 2011). Briefly, mouse NSCs were dissociated from dissected hippocampi and lateral ventricles from 3-week old REST loxP/loxP or WT littermate mice. Pooled hippocampal and SVZ NSCs (“neurospheres”) were cultured in DMEM:F12 supplemented with N2 and B27 (Invitrogen) in the presence of FGF2 (20 ng/ml), EGF (20 ng/ml), and heparin (5 μg/ml; proliferation conditions) on uncoated tissue culture plates. To delete REST, neurospheres were trypsinized and infected in suspension with GFP (control) or Cre-GFP adenovirus (1/10,000 of 1 × 1010 pfu/ml stock; University of Iowa) in N2/B27 medium containing FGF2/EGF/heparin. Three days after Ad-Cre infection, where over 95% of the NSCs were GFP+, the recombination rate was determined to be greater than 80–90% (Gao et al., 2011). NSCs were induced to differentiate with retinoic acid (RA; 1 nM) and forskolin (5 nM) for indicated time points. Semi-quantitative RT-PCR was carried out to confirm induction of select neuronal and/or REST target genes (Gao et al., 2011).
MiRNA Array Profiling
For miRNA array profiling, qRT-PCR and validation, total RNA samples were prepared using the miRNeasy kit (QIAGEN). Five micrograms of total RNA (from control and cKO cells) were sent to LC Sciences (Houston, TX, USA) for miRNA array analysis using Sanger miRBase Release 16.0. Multiple redundant regions were included on the chip and each region further comprised a miRNA probe region plus multiple control probes. Statistics were done with ANOVA analysis. Hierarchical clustering analyses of miRNA array profiling data were done using the software CLUSTER (Eisen et al., 1998). Bioinformatics prediction of RE1 sites was accomplished using TFbind (Tsunoda and Takagi, 1999).
To confirm the miRNA array profiling results, one microgram of total RNA was used for reverse transcription using NCode miRNA first-strand synthesis and QPCR kits (Invitrogen). QPCRs were performed using an ABI 7900HT instrument. The PCR program consisted of 2 min at 50°C and 2 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 58°C. Primer quality was analyzed by dissociation curves. MiRNA expression was normalized to the RNA input (Lossos et al., 2003). Statistics were performed using unpaired Student’s t-test. Values of p ≤ 0.05 were considered significant.
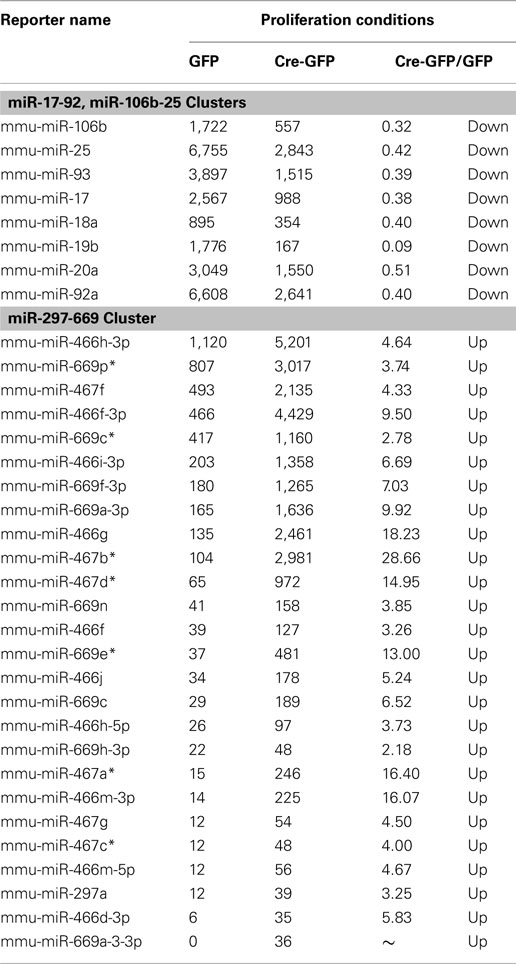
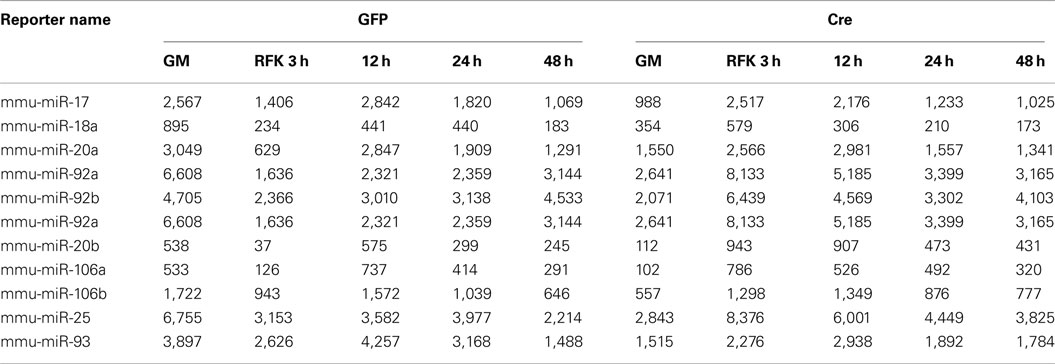
For the array profiling experiment, we conducted the following quality control steps before performing in-depth analyses. As expected, we did not detect any significant global differences in the total expression of miRNAs between the control and cKO cells (Figure A1 in Appendix). For both groups, out of the total 1040 miRNAs examined, only a quarter have significant expression (defined as having an intensity above 32 as the service provider recommends) in each experimental condition (Figure A2 in Appendix). About 8% have an expression value above 500 but account for more than 90% of the total miRNA expression (Figure A3 in Appendix). About 2% had an intensity score above 5000 but account for more than 60% of the total expression. The top expressed miRNAs are either ubiquitous (e.g., let-7 family) across lineages or neural enriched (e.g., miR-9, -21, -132, -16, -26a, -99b, and -125; Cao et al., 2006; Table A1 in Appendix). When we examined the expression of miRNA gene clusters including miR-17-92, -106b-25, -106a-363, and -297-669 loci, we observed a strong correlation among miRNAs from the same loci (Figure 2; Figure A4 and Table A2 in Appendix). In particular, from the miR-297-669 gene locus that comprised more than 70 miRNAs, all miRNAs with an intensity of 32 or above displayed similar expression dynamics throughout the experimental series (Figure A4 in Appendix).
Chromatin Immunoprecipitation and QPCR Analysis
As previously detailed (Gao et al., 2011), control and cKO cells were cultured with the desired treatments at indicated time points, crosslinked, and harvested. The chromatin immunoprecipitation (ChIP) assay was then performed following manufacturer’s instructions (Invitrogen). Antibodies used for ChIP were as follows: REST/NRSF (Upstate), mSin3A (Santa Cruz), and CoREST (Upstate). ChIP experiments were from independent chromatin preparations. QPCR and quantification of the results were performed as previously described (Gao et al., 2011). All primer sets were subjected to a dissociation curve analysis. The relative enrichment was determined by taking the absolute quantity ratios of specific IPs normalized to the input groups.
Results
Identification of REST-Dependent MiRNA Genes in NSCs under Proliferating Conditions
We recently demonstrated conditional deletion of REST led to reduced proliferation and precocious neuronal differentiation in hippocampal and SVZ mixed NSC cultures (Gao et al., 2011). Consistent with these findings, we observed dramatic upregulation of neuronal and/or REST target genes (Gao et al., 2011). To begin evaluating REST’s possible function in miRNA regulation, we performed miRNA microarray profiling using the same culture system (Gao et al., 2011). Floxed REST NSCs under proliferation conditions expressed the progenitor markers Nestin and Sox2 and generated Tuj1-positive neuronal cells and GFAP-positive astroglial cells under differentiation conditions consistent with their multipotentiality (Figure 1A). As indicated in Section “Materials and Methods,” we deleted REST by Ad-Cre-GFP infection of floxed REST NSCs to generate REST conditional knockout (cKO) NSCs. Consistent with REST deletion at a recombination efficiency of 80–90%, we confirmed robust upregulation of REST target neuronal genes NeuroD1 and Tuj1 in cKO NSCs, as well as precocious neuronal differentiation under proliferation conditions (Figure 1B; Gao et al., 2011). Upregulation of the immature neuroblast marker doublecortin (DCX) also confirmed that REST deletion precociously induced neuronal differentiation (Figure 1B; Gao et al., 2011).
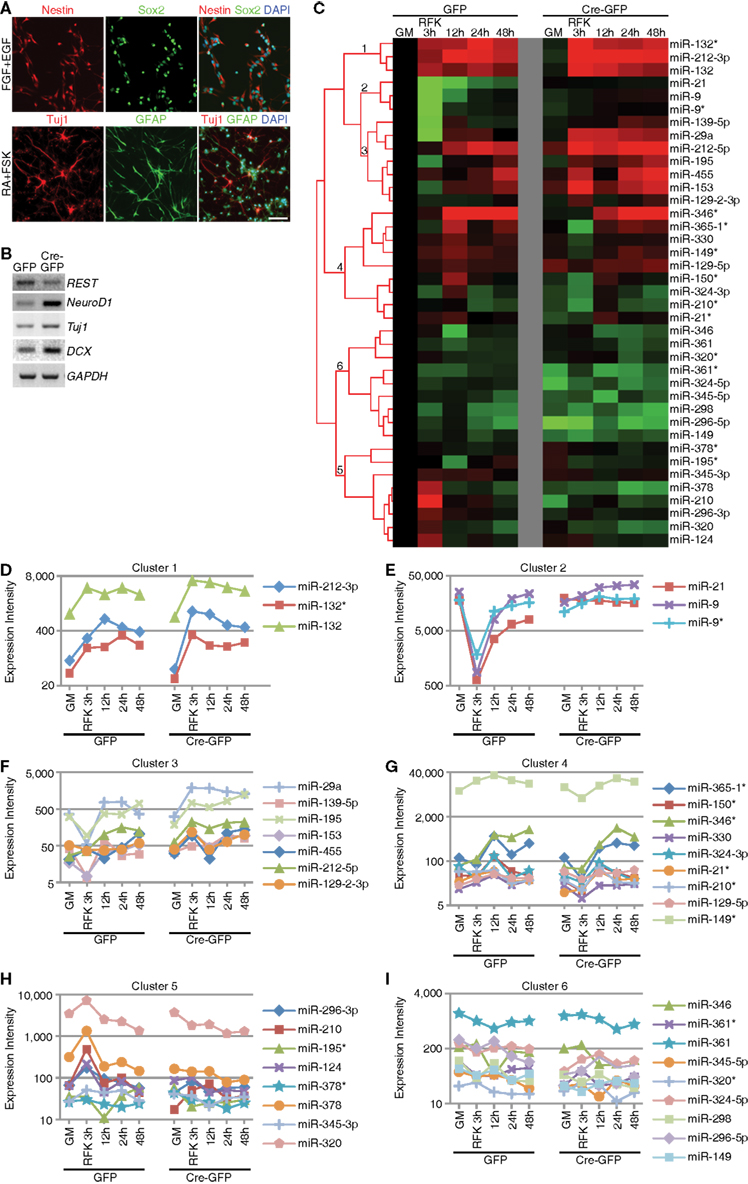
Figure 1. Context-dependent regulation of REST miRNA targets in mouse NSCs. (A) Successful derivation of multipotent REST cKO NSCs was confirmed using NSC markers (Nestin and Sox2), neuronal (Tuj1) and astrocytic markers (GFAP). Scale bar: 40 μm. (B) REST deletion by Ad-Cre-GFP viral infection in vitro resulted in upregulation of its target genes and neuronal genes, as well as precocious neuronal differentiation (not shown). (C–I) Hierarchical clustering analysis showed that known REST miRNA targets could be categorized in six different clusters (labeled 1–6) according to their expression dynamics during differentiation and responsiveness to REST ablation.
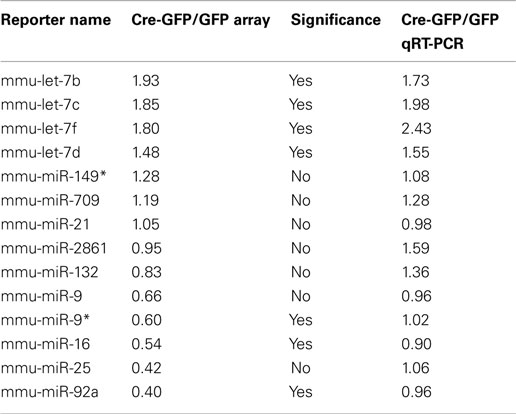
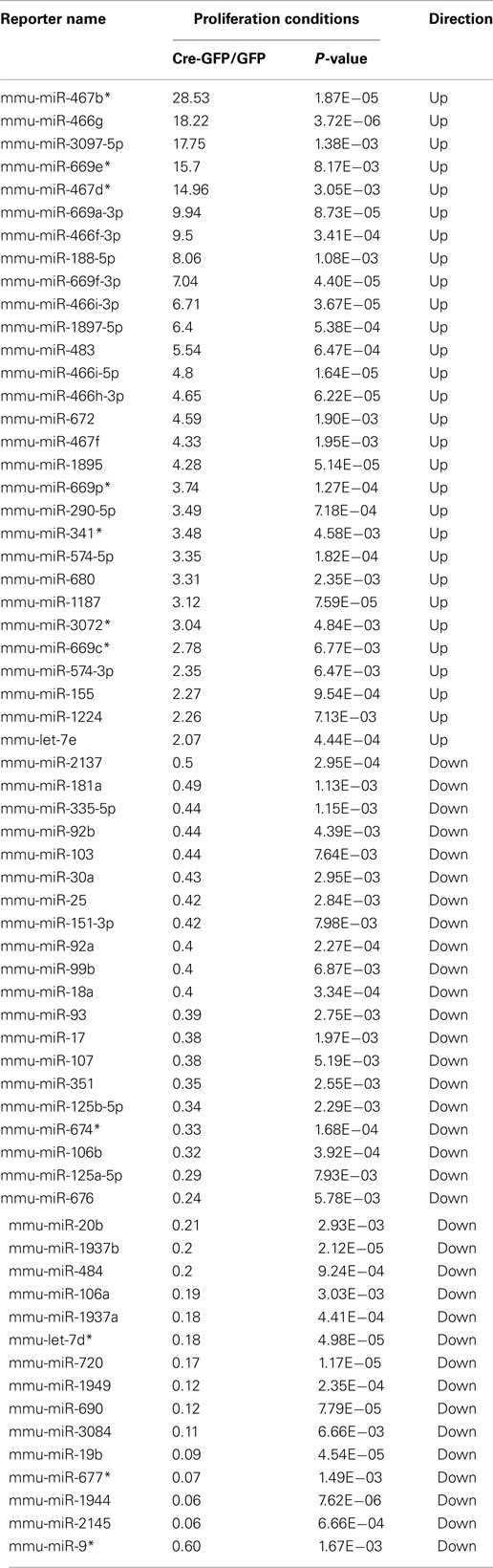
First, we validated the microarray profiling analysis by performing quality control steps (see Materials and Methods; Figures A1–A4 in Appendix and Tables A1– A3 in Appendix). We then performed qRT-PCR analysis to confirm a subset of miRNAs (either with the highest expression and/or largest fold change) from the array data and confirmed ∼70% of the array results (Table A3 in Appendix). We identified a subset of miRNAs that are significantly changed (p < 0.01) in cKO NSCs compared to control cells (Table A4 in Appendix). Among them, there were 29 miRNAs with over twofold expression increase and 35 miRNAs with over twofold expression decrease in cKO NSCs (Table A4 in Appendix). The only two known REST targets that were significantly changed in cKO NSCs compared to control cells were miR-30a (Cre-GFP/GFP = 0.43; p-value 2.95E−03) and miR-9* (Cre-GFP/GFP = 0.6; p-value 1.67E−03). However, instead of being upregulated, they were downregulated by 2.3 and 1.6 fold, respectively, in cKO NSCs compared to control cells. Since there are currently over 40 known REST miRNA targets (Wu and Xie, 2006; Otto et al., 2007), it is surprising that we did not observe expression changes of additional REST miRNA targets, at least under proliferation conditions.
Identification of REST-Dependent MiRNA Genes in NSCs under Differentiation Conditions
We hypothesized that the failure to observe significant upregulation of a large number of REST target miRNAs may be due to the lack of differentiation stimuli, which would be consistent with our previous findings that REST deletion results in enhanced neuronal differentiation when primed with RA and forskolin (FSK; Gao et al., 2011). Thus, we treated cKO and control NSCs with RA and FSK and performed miRNA profiling. We performed hierarchical cluster analysis and identified expression changes of known REST miRNA target genes, which were grouped into 6 clusters (Figures 1C–I). Cluster 1 contained miRNAs that were upregulated during differentiation and further enhanced upon REST deletion (Figure 1D). Clusters 2 and 3 contained miRNAs whose expression was initially downregulated and eventually restored during differentiation (Figures 1E–F). Loss of REST blocked this transient downregulation by augmenting their expression throughout the differentiation process (Figures 1E–F). The difference between cluster 2 and 3 was that cluster 2 contained highly expressed miRNAs. Cluster 4 contained miRNAs that were upregulated upon differentiation induction, which was blocked by REST ablation (Figure 1G). Cluster 5 contained miRNAs whose expression either persisted or slightly decreased upon RA and FSK treatment, which was further reduced with REST deletion (Figure 1H). Finally, cluster 6 contained miRNAs which did not display altered expression, regardless of REST ablation, either in proliferation or differentiation conditions, suggesting that REST may not be required for their expression in this context (Figure 1I). These results confirm that a subset of known REST target miRNAs were indeed subjected to REST regulation in mouse NSCs under differentiation, but not proliferation conditions. These results are also consistent with previous findings where downregulation of REST failed to globally activate or de-repress REST target gene expression (Chen et al., 1998; Ballas et al., 2005; Conaco et al., 2006), highlighting the context-dependent and dynamic nature of REST-dependent miRNA changes.
Dynamic Regulation of REST-Dependent MiRNA Genes during Differentiation
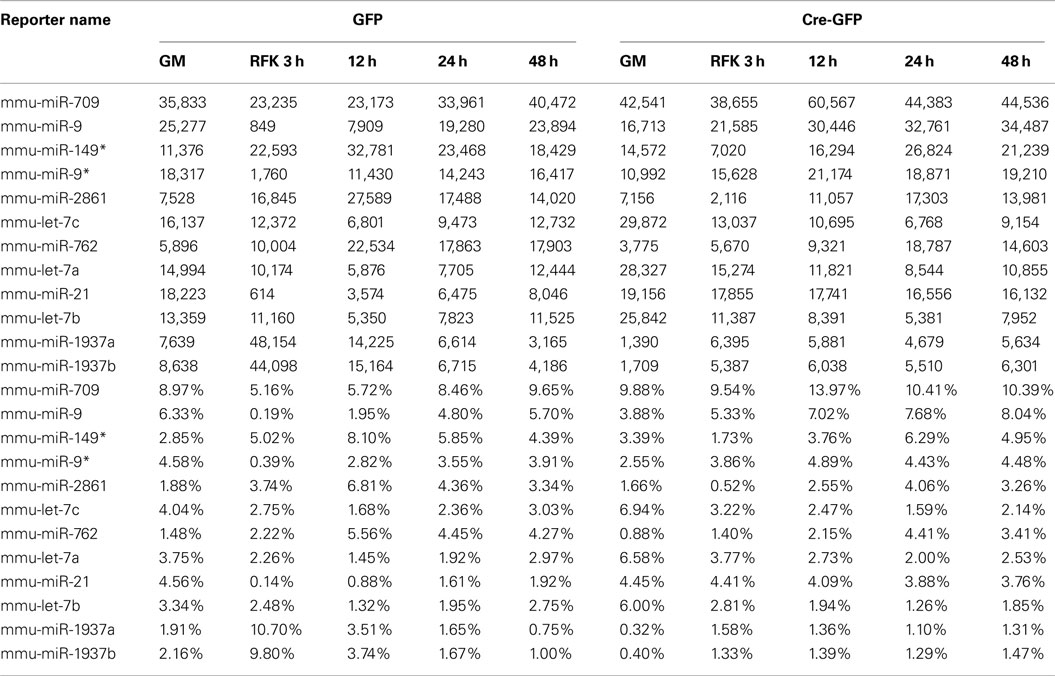
Among the miRNAs whose expression were dynamically altered during differentiation induction, several groups are worth mentioning. The first class represented “high-expressers” (Figure 2A and Table A5 in Appendix). MiR-709, previously implicated in tumorigenesis and the DNA damage response (Li et al., 2011; Tamminga et al., 2008), was the overall highest expressed single miRNA accounting for 5–14% of the total miRNAs expressed. The let-7 family of miRNAs was the highest expressed class and together comprised 9–32% of the total miRNA expression. Other high-expressers included miR-9, -9*, and -21 which are previously described REST targets that function in neural differentiation (Gao, 2010). We also observed robust upregulation of miR-1937a and -1937b, small miRNA-like RNAs of tRNA origin, immediately after RA and FSK treatment. They were the two highest expressed miRNA species, together accounting for 21% of the total miRNA expression in control cells 3 h after RA and FSK treatment, a time point when NSCs transiently increased their proliferation as they differentiate. Interestingly, this dramatic upregulation was abolished by REST ablation, which simultaneously prevented the transient increase in proliferation (Gao et al., 2011). This observation is in line with the finding that increased tRNA synthesis promotes proliferation and oncogenesis (Berns, 2008; Marshall et al., 2008), perhaps through an antagonistic mechanism to miRNA and siRNA silencing (Haussecker et al., 2010). Besides high-expressing miRNAs, another noteworthy class of miRNA changes is the miR-17-92 class and its two paralogous clusters, highly conserved between species and recently identified as oncomiRs (He et al., 2005; Mendell, 2008; Figure 2B and Table A6 in Appendix). The expression of the miR-17-92 family was downregulated immediately after RA and FSK treatment but subsequently restored with extended time points (Figure 2B). Interestingly, REST ablation prevented their initial downregulation, but instead led to their upregulation in the presence of differentiation stimuli.
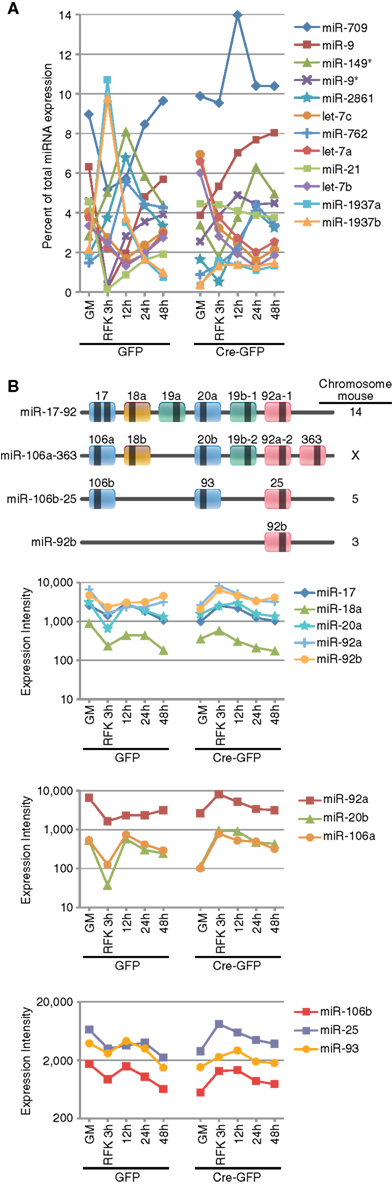
Figure 2. REST ablation modulates the expression of numerous miRNAs during differentiation. (A) The top expressed miRNAs included the let-7 family, miR-709, -9, -21, and -1937a/b. Most of the top expressed miRNAs are either ubiquitously expressed or neural specific/enriched species and many undergo dynamic changes over the course of neural differentiation or response to REST ablation. (B) miRNAs from the oncomiR miR-17-92 and its two paralogous clusters are dynamically regulated during RA and FSK induced NSC differentiation. Loss of REST prevents the initial downregulation mediated by RA and FSK.
Identification of Potential Novel REST miRNA Targets
Since concordantly regulated genes will cluster together with genome-wide profiling (Eisen et al., 1998), we reasoned there may be potential novel REST target miRNAs residing in the same cluster with known REST miRNA targets. To examine this, we used the CLUSTER software (Eisen et al., 1998) and performed unsupervised clustering analysis with all the miRNAs that have an expression value above 32 in the majority of the ten samples. We first validated the clustering strategy using the miR-297-669 cluster that contained dozens of miRNA genes under the same transcriptional regulation (Calabrese et al., 2007). As expected, regardless of the clustering algorithm used, miRNAs from this cluster readily grouped together (Figure A5 in Appendix), confirming the validity of our approach.
Next, we focused our analysis on clusters containing multiple known REST miRNA target genes. As shown in Figures 3A,B and Figure A6 in Appendix, two neighboring clusters contained at least nine known REST miRNA target genes together with dozens of other miRNAs. We used the TFBind software (Tsunoda and Takagi, 1999) to search for putative consensus RE1 site(s) flanking these miRNA genes. Together with a REST ChIP-sequencing (seq) study in our laboratory (unpublished results), we identified many potential REST target miRNAs within these two clusters (Figure 3C). In total, we have identified over 80 potential novel REST miRNA target genes through bioinformatics analysis and genome-wide profiling. The expanded list is shown in Table A7 in Appendix.
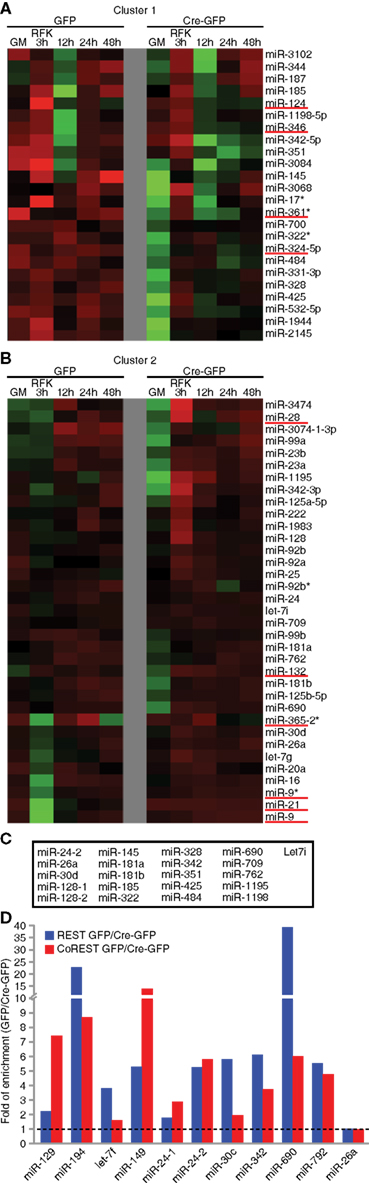
Figure 3. Identification of novel REST miRNA targets. (A,B) In unsupervised clustering analyses, known REST miRNA targets formed clusters that also contained other miRNAs. Examples of clusters containing known REST miRNA targets (underlined in red) (C) Combination of bioinformatic analysis and REST ChIP-seq identified novel REST miRNA target genes in Cluster 1 and 2. (D) REST ChIP-QPCR analysis confirmed the binding of REST and CoREST to novel miRNA target genes (seven out of nine randomly chosen), except miR-24-1 and -26a that fell below the twofold cut-off. MiR-129 and -149 were included as known miRNA target controls.
Finally, we randomly selected nine putative novel REST miRNA targets and performed ChIP-QPCR of cKO and control NSCs under proliferation conditions. Notably, for all of them except two, we were able to detect 2 fold or more enrichment in control cells compared to cKO NSCs (Figure 3C). For comparison, miR-129 and -149 were included as known miRNA target controls. In addition to REST, its corepressors CoREST and Sin3a are recruited to target promoter regions in a REST-dependent manner (Gao et al., 2011). We performed CoREST and Sin3a ChIP-QPCR analysis and found decreased recruitment of both CoREST and Sin3a (data not shown) in cKO NSCs (Figure 3D). Taken together, miRNA profiling expression data and ChIP-QPCR demonstrated we can identify known as well as novel REST-dependent miRNA targets in NSCs.
Discussion
Central to the understanding of brain development and pathogenesis is how genetic information in the genome is differentially and dynamically retrieved to achieve distinct neural cell fates. As guardian of the neuronal genome, REST is a master transcriptional repressor critical for brain development and homeostasis (Ballas and Mandel, 2005; Gao et al., 2011). MiRNAs are abundant and diverse in the brain, thought to be particularly important for the robustness and fine-tuning of gene regulation (Flynt and Lai, 2008; Gao, 2010). Hence, the identification of REST-dependent miRNAs in a physiologically relevant NSC system is beneficial for probing cell fate decisions in the brain.
In the present study, we performed genome-wide profiling and identified a collection of miRNAs that are dynamically regulated in NSCs during differentiation. At least some of these miRNAs are expected to possess stage-specific roles and underlie the ability of multipotent NSCs to self-renew and differentiate. In light of REST’s role in neural development, it is interesting that a subset of REST-dependent miRNAs also exhibit dynamic modes of expression during differentiation. While some miRNAs (e.g., miR-9, -21, and -132) have been shown to have roles in specific stages of either neuronal or glial differentiation (Gao, 2010), the functional significance of many others remain to be elucidated.
One surprising observation is that miR-124 is expressed at very low levels throughout our study, even when REST is deleted. This result seems inconsistent with an earlier study (Cheng et al., 2009) but not others (Cao et al., 2007; Sanuki et al., 2011). Since miR-124 is predominantly expressed in mature neurons in vivo (Cao et al., 2007; Cheng et al., 2009; Gao, 2010; Sanuki et al., 2011), perhaps our results are not that surprising. Although we cannot exclude the possibility that the failure to observe an upregulation of miR-124 (and many other known REST miRNA targets) in NSCs under proliferation conditions was due to incomplete recombination, our recombination efficiency was always greater than 80–90%, thus we do not anticipate this to be the major reason. Importantly, we observed dramatic alterations in both protein coding and miRNA genes under the same conditions. A more plausible scenario would be that there may be contextual differences between in vitro and in vivo neuronal differentiation, and REST-dependent miRNA changes could depend on the state of maturation. Interestingly, recent studies suggest miR-9 and -124 might have complementary expression patterns, raising the possibility that they adopt overlapping and distinct roles in neurogenesis and in mature neurons (Gao, 2010). One remaining question is how different REST miRNA targets (e.g., miR-9 and -124) are regulated in distinct cell types where there are high levels of REST (Gao et al., 2011).
Despite the presence of neural developmental defects in REST knockout mice, genome-wide changes in coding or non-coding genes are not typically observed (Chen et al., 1998; Ballas et al., 2005; Conaco et al., 2006). Our results are consistent with the lack of widespread miRNA de-repression in REST cKO NSCs. Specifically, known REST miRNA target genes failed to upregulate in NSCs under proliferation conditions. However, in differentiation conditions, the expression of known REST miRNA target genes was altered and formed six clusters; each manifested distinct expression dynamics over the course of differentiation and in response to REST deletion. Furthermore, it is worth noting that the expression of some of the newly identified REST target miRNAs was also altered under proliferation conditions. Taken together, these results reveal that REST-dependent miRNA changes can be widespread, but is strictly dependent on the local gene context influenced by the proliferation or differentiation status of NSCs.
What mechanism(s) may underlie the context-dependent nature of REST miRNA target regulation? One possibility is that nuclear REST levels dictate recruitment to select RE1 sites with different binding affinities. The repression of target genes in theory could be released in a stage-wise manner based on the affinity of REST to its binding sites, as REST levels gradually decrease during differentiation. Indeed, there is evidence to suggest that RE1 sites with weaker affinity might be less repressive due to the limited recruitment of cofactors (Zheng et al., 2009; Yu et al., 2011). A non-exclusive possibility is that REST binding and de-repression of target genes involves the recruitment of additional transcriptional activators and removal of REST alone is not sufficient to upregulate target gene expression. In support of this, we previously observed that overexpression of a REST activator fusion construct (REST-VP16), but not REST knockdown/deletion alone, was able to induce robust neuronal differentiation and upregulate REST target genes, even under proliferating conditions (Gao et al., 2011). Moreover, the local chromatin environment or epigenetic histone modifications surrounding specific REST targets may affect REST-mediated gene expression changes (Yu et al., 2011). Together, any of these possibilities, or a combination of mechanisms, may explain the lack of genome-wide depression of REST miRNA targets.
In this study, we also demonstrate that combined use of clustering and bioinformatics analysis is a powerful approach to identify a number of novel REST-dependent miRNA target genes. Recently, several miRNAs were suggested as potential REST targets (Singh et al., 2008; Smith et al., 2010), further confirming the validity of our microarray profiling and bioinformatics analysis. It would be interesting in future experiments to investigate whether REST miRNA target genes within the same cluster share the same combinatorial regulation and the roles of these individual miRNAs in NSC self-renewal and differentiation.
In summary, our study identifies a collection of known and novel REST target miRNAs that are dynamically regulated during neuronal differentiation. These studies could provide valuable insights into the context-dependent nature of REST regulation as well as how stage-specific progression of neuronal differentiation is achieved. Moreover, we demonstrate it is feasible to combine cluster analysis with miRNA array profiling data to uncover novel REST miRNA targets when a well-defined consensus DNA-binding sequence is known. Future studies of REST and its miRNA targets will be important for understanding how the balance between neuronal production and maintaining the NSC pool is controlled for proper brain development and tissue homeostasis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ling Zhang for technical assistance and Jose Cabrera for graphics support. This work was supported by the following granting agencies: National Institutes of Health (R01 AG032383, R01 NS076775, and R21 MH09471501), the Cancer Prevention and Research Institute of Texas (RP100674), and the Welch Foundation (I-1660; to Jenny Hsieh). Zhengliang Gao was supported by a postdoctoral fellowship from the American Heart Association.
References
Amir, R. E., Van den Veyver, I. B., Wan, M., Tran, C. Q., Francke, U., and Zoghbi, H. Y. (1999). Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188.
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657.
Ballas, N., and Mandel, G. (2005). The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 15, 500–506.
Bruce, A. W., Donaldson, I. J., Wood, I. C., Yerbury, S. A., Sadowski, M. I., Chapman, M., Göttgens, B., and Buckley, N. J. (2004). Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. U.S.A. 101, 10458–10463.
Calabrese, J. M., Seila, A. C., Yeo, G. W., and Sharp, P. A. (2007). RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 18097–18102.
Cao, X., Pfaff, S. L., and Gage, F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536.
Cao, X., Yeo, G., Muotri, A. R., Kuwabara, T., and Gage, F. H. (2006). Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 29, 77–110.
Chen, Z. F., Paquette, A. J., and Anderson, D. J. (1998). NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142.
Cheng, L. C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Conaco, C., Otto, S., Han, J. J., and Mandel, G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 103, 2422–2427.
Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868.
Flynt, A. S., and Lai, E. C. (2008). Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9, 831–842.
Gao, F. B. (2010). Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 5, 25.
Gao, Z., Ure, K., Ding, P., Nashaat, M., Yuan, L., Ma, J., Hammer, R. E., and Hsieh, J. (2011). The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786.
Haussecker, D., Huang, Y., Lau, A., Parameswaran, P., Fire, A. Z., and Kay, M. A. (2010). Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695.
He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S. W., Hannon, G. J., and Hammond, S. M. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828–833.
Hsieh, J., and Gage, F. H. (2004). Epigenetic control of neural stem cell fate. Curr. Opin. Genet. Dev. 14, 461–469.
Jessberger, S., Nakashima, K., Clemenson, G. D. Jr., Mejia, E., Mathews, E., Ure, K., Ogawa, S., Sinton, C. M., Gage, F. H., and Hsieh, J. (2007). Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. J. Neurosci. 27, 5967–5975.
Johnson, D. S., Mortazavi, A., Myers, R. M., and Wold, B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502.
Kempermann, G., Song, H., and Gage, F. (2008). Adult Neurogenesis. New York: Cold Spring Harbor Laboratory Press.
Li, X., Sanda, T., Look, A. T., Novina, C. D., and von Boehmer, H. (2011). Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 208, 663–675.
Lossos, I. S., Czerwinski, D. K., Wechser, M. A., and Levy, R. (2003). Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia 17, 789–795.
Mabb, A. M., Judson, M. C., Zylka, M. J., and Philpot, B. D. (2011). Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 34, 293–303.
Marshall, L., Kenneth, N. S., and White, R. J. (2008). Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 133, 78–89.
Mendell, J. T. (2008). miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–222.
Ming, G. L., and Song, H. (2005). Adult neurogenesis in the mammalian central nervous system. Annu. Rev. Neurosci. 28, 223–250.
Otto, S. J., McCorkle, S. R., Hover, J., Conaco, C., Han, J. J., Impey, S., Yochum, G. S., Dunn, J. J., Goodman, R. H., and Mandel, G. (2007). A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729–6739.
Sahoo, T., del Gaudio, D., German, J. R., Shinawi, M., Peters, S. U., Person, R. E., Garnica, A., Cheung, S. W., and Beaudet, A. L. (2008). Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 40, 719–721.
Sanuki, R., Onishi, A., Koike, C., Muramatsu, R., Watanabe, S., Muranishi, Y., Irie, S., Uneo, S., Koyasu, T., Matsui, R., Chérasse, Y., Urade, Y., Watanabe, D., Kondo, M., Yamashita, T., and Furukawa, T. (2011). miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 14, 1125–1134.
Singh, S. K., Kagalwala, M. N., Parker-Thornburg, J., Adams, H., and Majumder, S. (2008). REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227.
Smith, B., Treadwell, J., Zhang, D., Ly, D., McKinnell, I., Walker, P. R., and Sikorska, M. (2010). Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS ONE 5, e11109. doi:10.1371/journal.pone.0011109
Tamminga, J., Kathiria, P., Koturbash, I., and Kovalchuk, O. (2008). DNA damage-induced upregulation of miR-709 in the germline downregulates BORIS to counteract aberrant DNA hypomethylation. Cell Cycle 7, 3731–3736.
Tsunoda, T., and Takagi, T. (1999). Estimating transcription factor bindability on DNA. Bioinformatics 15, 622–630.
Wu, J., and Xie, X. (2006). Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 7, R85.
Yu, H. B., Johnson, R., Kunarso, G., and Stanton, L. W. (2011). Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 21, 1284–1293.
Zheng, D., Zhao, K., and Mehler, M. F. (2009). Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 10, R9.
Zuccato, C., Tartari, M., Crotti, A., Goffredo, D., Valenza, M., Conti, L., Cataudella, T., Leavitt, B. R., Hayden, M. R., Timmusk, T., Rigamonti, D., and Cattaneo, E. (2003). Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 35, 76–83.
Appendix
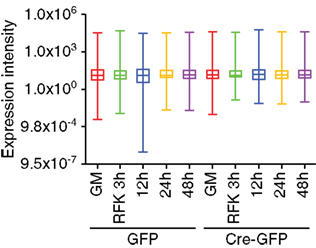
Figure A1. Array profiling of miRNAs in REST cKO NSCs. Whisker box plot analysis demonstrated there was no significant global expression differences among the GFP and Cre-GFP groups regardless of proliferation versus differentiation conditions.
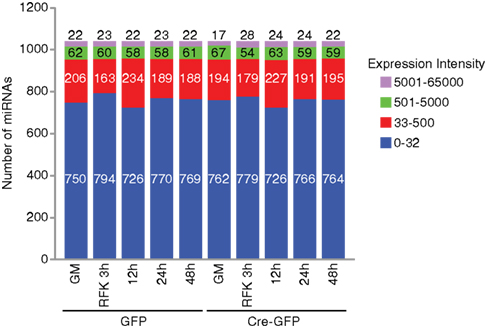
Figure A2. Distribution of the number of miRNAs examined in the array analysis. In NSCs cultured under proliferation and differentiation conditions, only a quarter of the 1040 miRNAs examined are expressed at a significant level (with an intensity above 32). Out of these significantly expressed groups, about 60 miRNAs were expressed at levels between 500 and 5000 and about 20 miRNAs were above 5000.
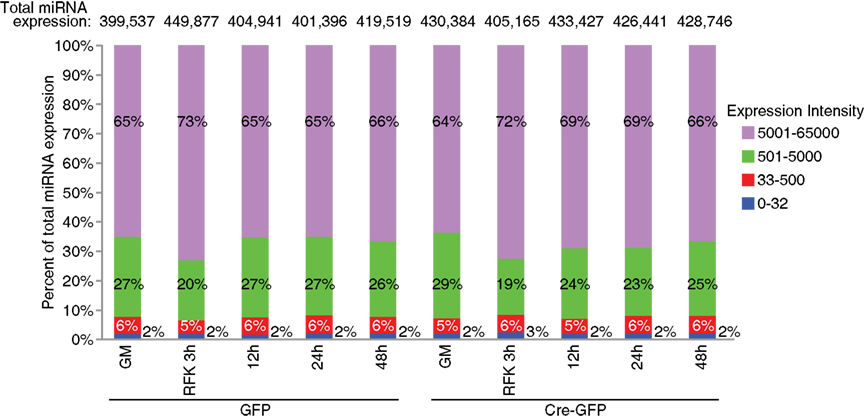
Figure A3. Expression distribution of miRNAs examined in the array analysis. The top expressed miRNAs account for the majority of the total miRNA expression: about 6% with an expression value between 500 and 5000 count for more than 20–30% of the total miRNA expression and about 2% with an intensity score above 5000 comprised 60–70% of the total expression.
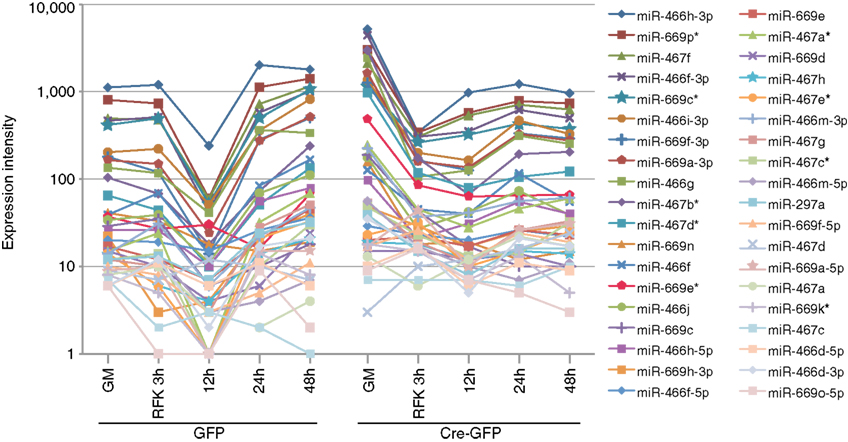
Figure A4. Validation of the array profiling by examining specific miRNA clusters. All miRNAs from the miR-297-669 loci displayed similar expression dynamics demonstrating the validity of the array analysis. Similar expression dynamics were also observed for other miRNA clusters (Figure 2 and Table A2 in Appendix).
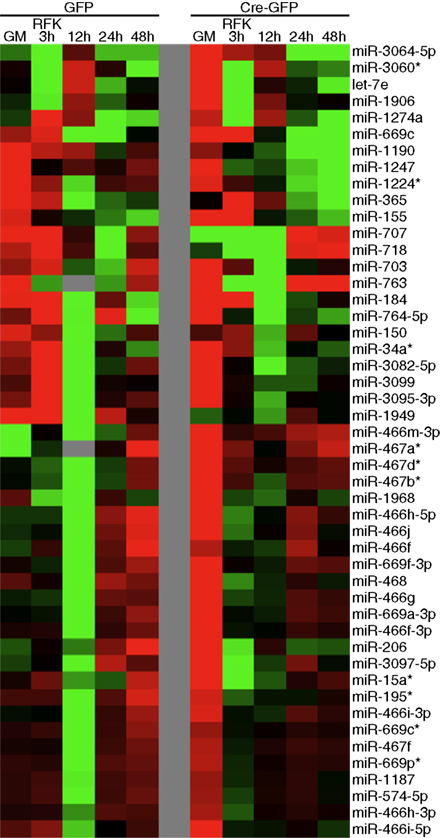
Figure A5. Validation of the clustering approach to identify potential novel REST miRNA targets. Regardless of the clustering algorithm, miRNAs from miR-297-669 locus readily form clusters, confirming the validity of our clustering approaches.
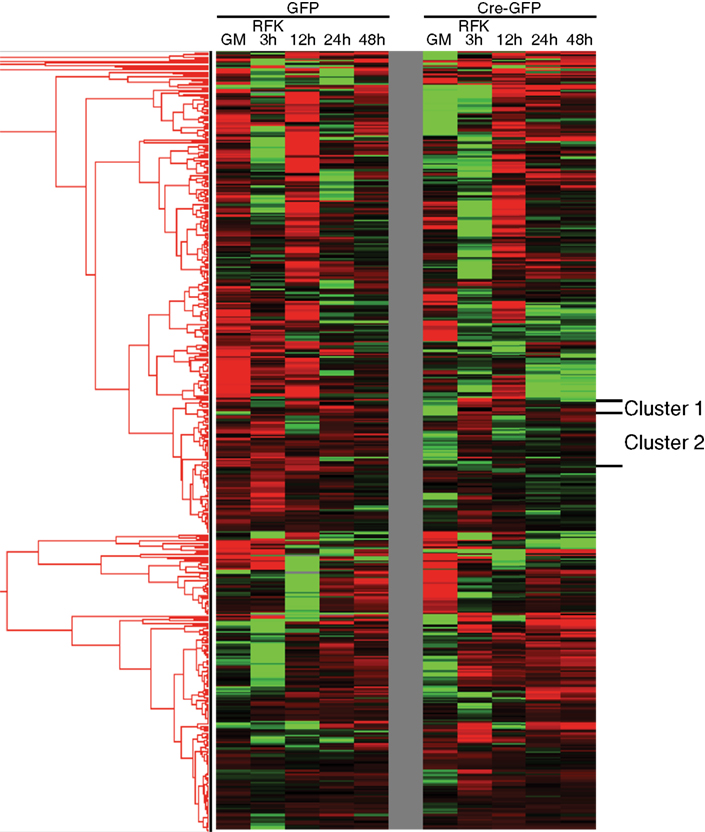
Figure A6. Clustering approach to identify potential novel REST miRNA targets. In unsupervised clustering analyses, known REST miRNA targets formed clusters that also contained other miRNAs. Cluster 1 and 2 were chosen as representative examples and presented in Figure 3. Through bioinformatic and ChIP-seq analysis, we identified numerous potential novel REST miRNA targets residing within the same clusters as known REST miRNA targets (refer to Table A7 in Appendix).
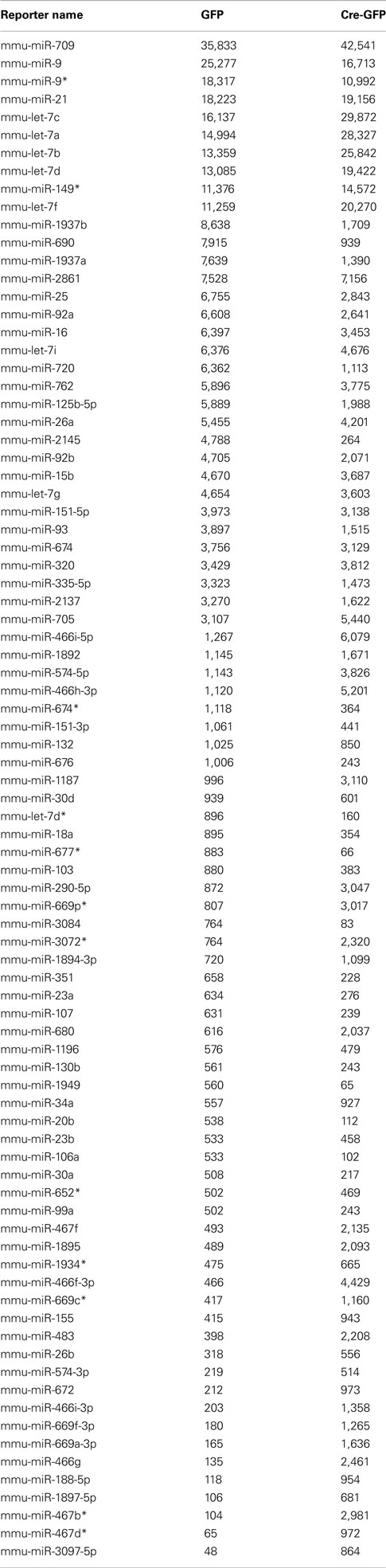
Table A1. Highly expressed miRNAs under proliferation condition in GFP (control) and Cre-GFP (experimental) groups.
Keywords: microRNAs, adult neurogenesis, transcription, repressor, epigenetic, neural stem cell
Citation: Gao Z, Ding P and Hsieh J (2012) Profiling of REST-dependent microRNAs reveals dynamic modes of expression. Front. Neurosci. 6:67. doi: 10.3389/fnins.2012.00067
Received: 06 February 2012; Accepted: 19 April 2012;
Published online: 10 May 2012.
Edited by:
Yanhong Shi, City of Hope, USAReviewed by:
Benedikt Berninger, Ludwig-Maximilians-Universität München, GermanyPeng Jin, Emory University School of Medicine, USA
Copyright: © 2012 Gao, Ding and Hsieh. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jenny Hsieh, Department of Molecular Biology, UT Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390, USA. e-mail:amVubnkuaHNpZWhAdXRzb3V0aHdlc3Rlcm4uZWR1.
†Zhengliang Gao and Peiguo Ding have contributed equally to this work.

 Peiguo Ding†
Peiguo Ding†