- 1Neuroscience Institute, Department of Biology, Nagoya University, Furo-cho, Japan
- 2Department of Biological Sciences, Inha University, Incheon, South Korea
- 3Center for Hypothalamic Research, University of Texas Southwestern Medical Center, Dallas, TX, United States
Sleep and metabolism are interconnected homeostatic states; the sleep cycle can be entrained by the feeding cycle, and perturbation of the sleep often results in dysregulation in metabolism. However, the neuro-molecular mechanism by which metabolism regulates sleep is not fully understood. We investigated how metabolism and feeding regulate sleep using satiety quiescence behavior as a readout in Caenorhabditis elegans, which shares certain key aspects of postprandial sleep in mammals. From an RNA interference-based screen of two neuropeptide families, RFamide-related peptides (FLPs) and insulin-like peptides (INSs), we identified flp-11, known to regulate other types of sleep-like behaviors in C. elegans, as a gene that plays the most significant role in satiety quiescence. A mutation in flp-11 significantly reduces quiescence, whereas over-expression of the gene enhances it. A genetic analysis shows that FLP-11 acts upstream of the cGMP signaling but downstream of the TGFβ pathway, suggesting that TGFβ released from a pair of head sensory neurons (ASI) activates FLP-11 in an interneuron (RIS). Then, cGMP signaling acting in downstream of RIS neurons induces satiety quiescence. Among the 28 INSs genes screened, ins-1, known to play a significant role in starvation-associated behavior working in AIA is inhibitory to satiety quiescence. Our study suggests that specific combinations of neuropeptides are released, and their signals are integrated in order for an animal to gauge its metabolic state and to control satiety quiescence, a feeding-induced sleep-like state in C. elegans.
Introduction
From simple invertebrates to complicated humans, most animals exhibit a behavioral state of sleep. Circadian rhythm can be entrained by feeding, and perturbation of the rhythm often results in obesity. In addition, the neuropeptide orexin regulates both sleep and feeding, suggesting that the two behaviors are linked (Bass and Takahashi, 2010; Marcheva et al., 2010). However, the role of metabolism in sleep and the underlying neuronal mechanisms that connect metabolism to sleep are not fully understood.
Caenorhabditis elegans, with its powerful genetics and simple nervous system, emerges as an ideal model to study neuro-molecular mechanisms underlying sleep. Two types of sleep, developmentally timed sleep (DTS), and stress-induced sleep (SIS), share fundamental aspects of sleep in other animals (Flavell et al., 2020). Additionally, we reported that the C. elegans behavioral state satiety quiescence mimics certain aspects of post-prandial sleep in mammals (You et al., 2008). After feeding, rodents exhibit behavioral sequence of satiety: termination of meals, reduction of locomotion, and sleep (Gibbs et al., 1973; Antin et al., 1975). C. elegans also becomes quiescent after feeding. This quiescence induced by satiety is dependent on the animal’s metabolic state. If the animal is undernourished, either by being deprived of food, or by having defects in the feeding or digestion processes, the animal shows little quiescence. During satiety quiescence, an animal is inactive and exhibits a sleep-like posture. The longer an animal is starved, the more quiescent it becomes after being refed. The animal’s response to touch is reduced during satiety quiescence; the animal no longer exhibits a full escape response but instead, returns to quiescence almost immediately after being touched, suggesting that their touch perception is dampened during satiety quiescence. These observations suggest satiety quiescence contains several key components of behavioral state of sleep and thus provides us a unique readout to study how metabolism might regulate sleep or sleep-like states.
Satiety quiescence is regulated by insulin, cGMP, and TGFβ signaling. In C. elegans, the three signaling pathways regulate dauer formation during development, which is also critically dependent on the animal’s metabolic state. Lack of any of these three signals drives the animal to enter the dauer state, a dormant stage to survive unfavorable environment (Hu, 2007). The wrong decision of whether to enter the reproductive cycle or the dauer state is fatal. The animal either would not survive if it enters to reproductive cycle in a harsh environment or would be outcompeted while the other animals prosper if it becomes dauer in a nutritious environment. The fact that the same three pathways control both the critical developmental choice and satiety quiescence, indicates that these three pathways are used to ensure the animals of metabolic wellbeing.
We found that TGFβ released from the head sensory neuron pair ASI is necessary for satiety quiescence. Lack of TGFβ signal reduces quiescence and increases fat storage, again linking metabolism and sleep by a single molecular pathway (Gallagher et al., 2013b). The conserved roles of cGMP and TGFβ signaling in feeding have also been discovered in mammals (Valentino et al., 2011; Tsai et al., 2013, 2018; Folgueira et al., 2018), suggesting a similar set of molecules regulate metabolism and sleep in many animals. Satiety quiescence requires intact fat metabolism mediated by the SREBP-SCD (sterol regulatory element-binding protein – stearoyl-CoA desaturase) pathway, indicating that a communication between the nervous system and the organs of energy storage is necessary (Hyun et al., 2016).
Neuropeptides (NP) are the major signaling molecules that control feeding and energy homeostasis in most of the animals including invertebrates (Li and Kim, 2008; Nassel et al., 2019). C. elegans has a total of 121 neuropeptides divided into three families: 34 RFamide-related peptides (FLP) characterized by a C-terminal Arg-Phe-amide motif, 47 neuropeptide-like proteins (NLP), and 40 insulin-like proteins (INS) (Li and Kim, 2008). We and others have demonstrated that many of these peptides serve conserved functions in metabolism and feeding. NLP-24, a C. elegans opioid capable of binding to human μ-opioid receptors, regulates fasting responses (Cheong et al., 2015). FLP-7, a C. elegans tachykinin, centrally regulates peripheral fat storage by controlling the transcription level of a hormone-sensing lipase in periphery (Palamiuc et al., 2017). NLP-75 the C. elegans oxytocin/vasopressin regulates reproductive behavior and gustatory associative learning (Beets et al., 2012; Garrison et al., 2012). NLP-38 is also critical for the animal to form a memory to avoid salt concentration associated with starvation condition (Peymen et al., 2019).
RF-amide peptides play roles in diverse behaviors such as social interaction, reproduction, and feeding (Parhar et al., 2016; Quillet et al., 2016). In zebrafish, the RFamide neuropeptide VF (NPVF) and the npvf-expressing neurons are necessary and sufficient to promote sleep (Lee et al., 2017). In C. elegans, both FLPs and NLPs regulate sleep: FLP-18, NLP-2, NLP-22, and NLP-14 regulate DTS and FLP-13, FLP-24, and NLP-8 do SIS (Nelson et al., 2013; Nagy et al., 2014; Nath et al., 2016; Iannacone et al., 2017; Honer et al., 2020; Van der Auwera et al., 2020).
Caenorhabditis elegans has 40 insulin-like ligands all of which act on the insulin receptor DAF-2 (Murphy and Hu, 2013). The pathway has conserved components, including positive regulators of PI3K (AGE) and AKT (AKT) as well as the negative regulators of PTEN (DAF-18) and FOXO (DAF-16). The insulin pathway regulates vast arrays of physiological processes, including L1 arrest, dauer decision, fat and sugar metabolism and longevity (Murphy and Hu, 2013). The 40 ins genes redundantly function as either agonists or antagonists to the DAF-2 receptor and can regulate a specific process such as L1 arrest or fat metabolism or all of the processes (Zheng et al., 2018). Among the 40 ins genes, ins-1 is most similar to the human insulin in its sequence and antagonizes DAF-2 insulin receptor (Pierce et al., 2001). INS-1 plays roles in salt conditioning, olfactory feedback, and thermotaxis plasticity by starvation (Tomioka et al., 2006; Chalasani et al., 2010; Takeishi et al., 2020). These studies indicate that INS peptides play critical roles in conveying the animal’s metabolic states. Furthermore DAF-16, the C. elegans FOXO ortholog and the major downstream target of insulin signaling, is required for sleep homeostasis and is essential for DTS (Driver et al., 2013; Bennett et al., 2018). However, despite its conserved role in feeding and metabolism, the roles of insulin signaling in sleep has not been systemically tested in C. elegans.
Based on the conserved roles of neuropeptides in regulation of homeostasis, such as in energy balance and in sleep, we performed an RNA interference-based screen of two neuropeptide families, FLPs and INSs. Among the 28 tested flp genes, RNA interference (RNAi) of 10 FLPs altered satiety quiescence, while flp-11, known to regulate other types of sleep-like behaviors in C. elegans plays the most important role. flp-11 mutants show reduction in satiety quiescence, whereas over-expression lines show enhancement. FLP-11 acts in the interneuron RIS; a mutant that carries a mutation in the gene aptf-1, a transcription factor that functions in RIS, is also defective in satiety quiescence. From a genetic analysis, we found that FLP-11 acts potentially upstream of the cGMP signaling and downstream of the TGFβ pathway, suggesting a potential neural circuit. Among the 28 tested ins genes, ins-1, known to play a significant role in starvation-associated behavior working in AIA interneurons and receiving input from ASI neurons (Tomioka et al., 2006), is inhibitory to satiety quiescence. Our results could suggest a neural circuit where internal nutrient status is integrated to generate an appropriate behavioral output such as satiety quiescence.
Materials and Methods
Strains
The wild-type strain was C. elegans variant Bristol, strain N2. Mutant strains were, GR1396 eri-1 IV; lin-15b X, VC2324 flp-6(ok3056) V, RB1990 flp-7(ok2625) X, PT501 flp-8(pk360) X, RB2067 flp-9(ok2730) IV, RB1989 flp-10(ok2624) IV, VC1669 aptf-1(gk794) II, HBR507 flp-11(tm2706) X, RB1863 flp-12(ok2409) X. These strains are available from the Caenorhabditis Genetics Center (CGC). YJ233 flp-6(ok3056) V, YJ234 flp-7(ok2625) X, YJ235 flp-9(ok2730) IV and YJ236 flp-10(ok2624) IV were outcrossed 2–4 times from the original strain. The flp-11 overexpression (OE) lines, YJ258 and YJ259, were generated by injecting the reporter Psur-5::mCherry with the target plasmid (pPD95.77, Pflp-11::flp-11::GFP) carrying the flp-11 gene into N2 strain animals (Sunny Biotech). The double mutant YJ262 flp-11(tm2706) X; egl-4(ks62) IV was generated by crossing HBR507 flp-11(tm2706) X with FK234 egl-4(ks62) IV. YJ 263 flp-11(tm2706) X; egl-4(ad450sd) IV was generated by crossing HBR507 flp-11(tm2706) X with DA521 egl-4(ad450sd) IV. The double mutant of flp-11 OE; daf-7 was generated by crossing YJ258 with CB1372 daf-7(e1372) III. All animals were maintained at 20°C on Escherichia coli strain HB101 unless indicated otherwise.
RNAi Screening
Among 34 flp genes and 40 ins genes, 28 available clones of flp genes and ins genes from Ahringer feeding library were tested by bacteria-mediated feeding RNAi (Fraser et al., 2000). The plates containing NGM agar with 1 mM IPTG and 50 μg/ml carbenicillin were inoculated with bacterial cultures grown 16–18 h for each target gene. HT115 bacteria, an RNase III-deficient E. coli was used. The strain GR1396 (eri-1 IV; lin-15B X) was used to enhance RNAi sensitivity (Ruvkun G., personal communication). Three L4 stage animals were transferred to the plates and 36 h later the adults were removed. Another 36 h later, the progeny L4 animals were picked to perform satiety quiescence assay. For each test, 7–9 concurrent control animals treated with empty vector (L4440) containing RNAi bacteria and 15–18 animals treated with an RNAi containing bacteria were used. Except for the RNAi clones whose treatment resulted in no significant difference compared to the control (gray bars in Figures 1, 3), the experiment was repeated at least twice.
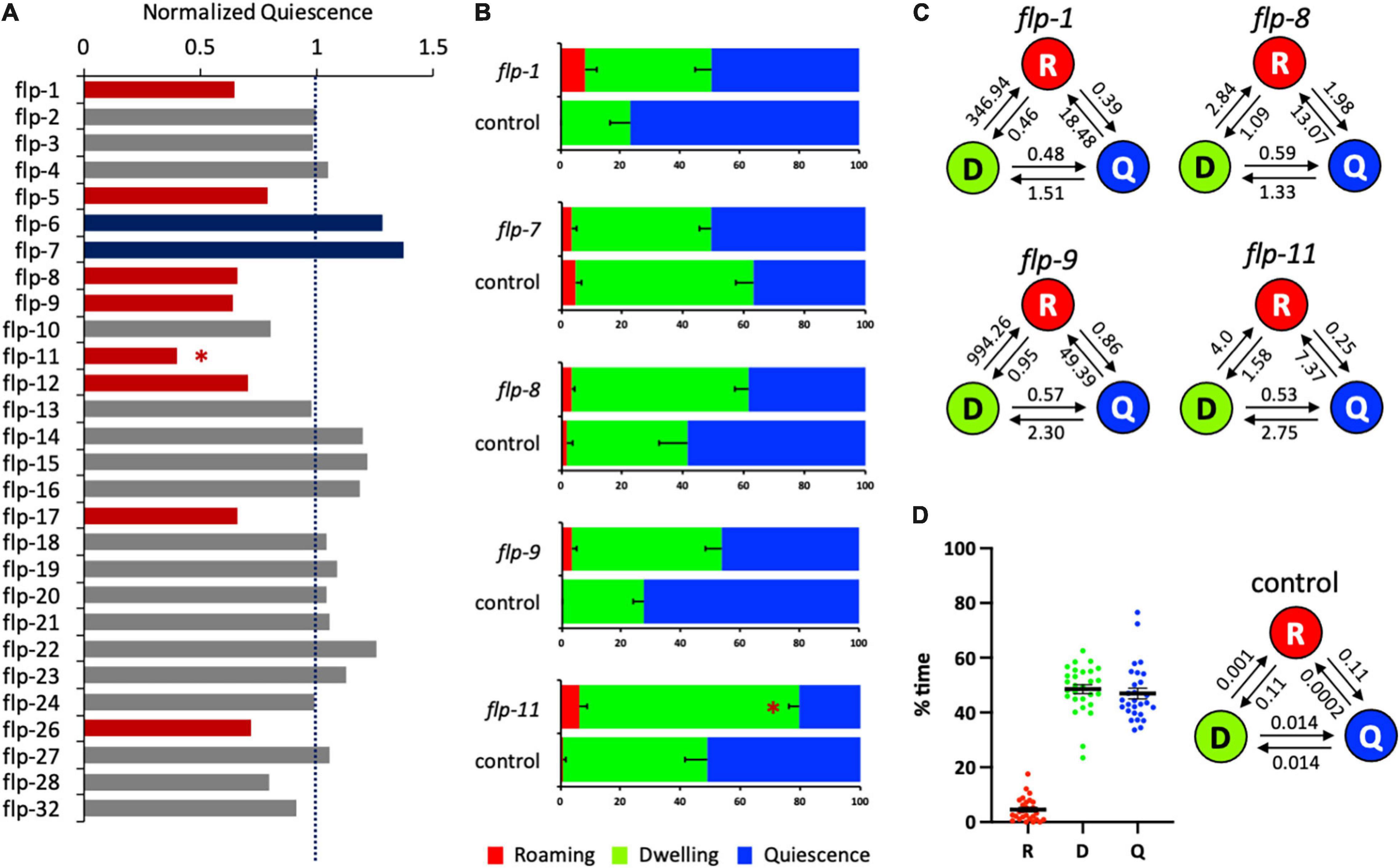
Figure 1. Several flp genes regulate satiety quiescence. (A) The normalized time in quiescence after knockdown of individual flp genes by RNA interference. The percent time in quiescence of each flp is normalized to the percent time in quiescence of the concurrent control (RNAi vector only). Each RNAi was performed with the sample number of 15–18 of tested animals with 7–9 control animals. Red bars indicate that RNAi of the gene reduced satiety quiescence, whereas blue bars indicate that RNAi of the gene enhanced satiety quiescence with an approximate cut-off by ±20% of the normalized value (see Table 1). (B) The individual data of the flp genes including whose RNAi showed significant changes in satiety quiescence. After two generations of feeding RNAi, the age-synchronized 1-day-old adults were tested for their locomotive activity by measuring the percent time they spent in roaming, dwelling and quiescence. (C) The transition rates of the animals treated with RNAi of flp-1, flp-8, flp-9, and flp-11. The circles of R, D, and Q represent each state of roaming, dwelling, and quiescence, respectively. The rates are shown above the arrow that indicates the direction of transition. (D) Distribution of the values of each state (left) and average transition rates (right) of the 28 independently tested RNAi control groups. (∗p < 0.009 by Student’s t-test after the Bonferroni correction. The error bars indicate SEM).
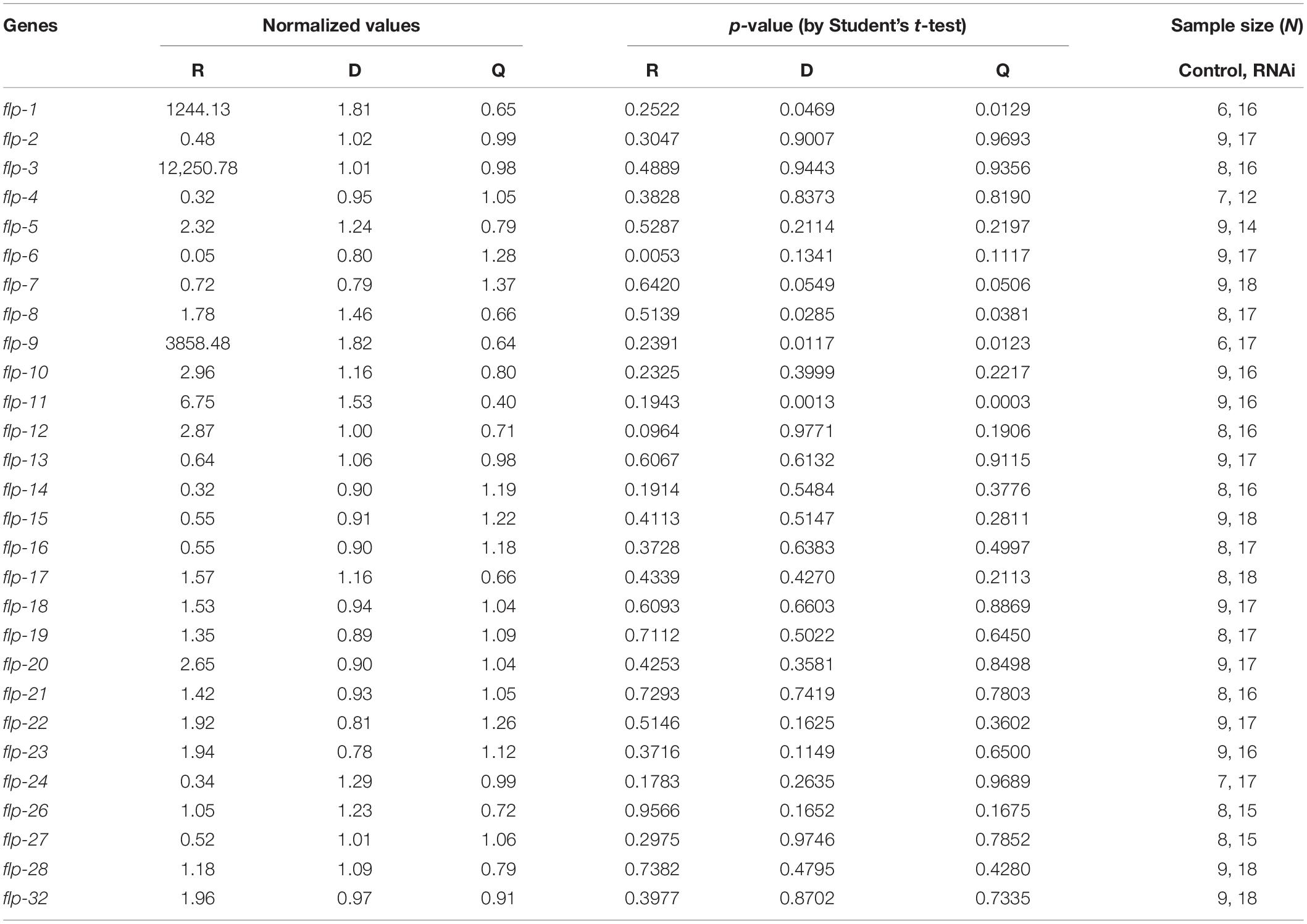
Table 1. Normalized values of each state of roaming, dwelling and quiescence, the sample size, and statistical significance of 28 flp genes.
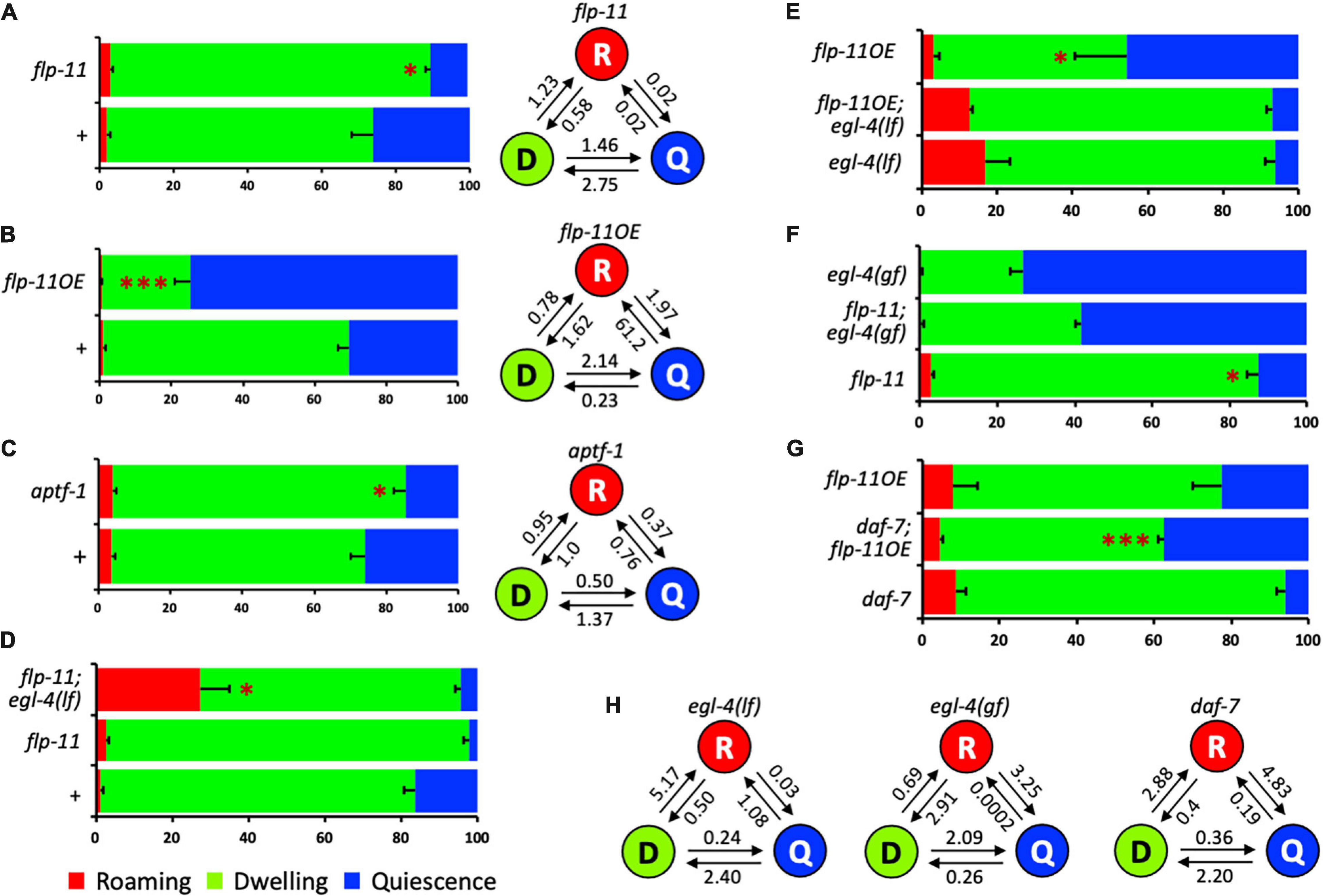
Figure 2. The potential mechanisms of flp-11 in satiety quiescence. (A) flp-11 mutants show reduced satiety quiescence, confirming the RNAi result. The percent time and the normalized transition rates among the three states are shown. (B) An overexpression line of flp-11 shows enhanced satiety quiescence. The data is a representative of two overexpression lines. The percent time and the normalized transition rates among the three states are shown. (C) aptf-1, a transcription factor acting in RIS that promotes other types of sleep behavior in C. elegans, is also required for satiety quiescence. The percent time and the normalized transition rates among the three states are shown. (D) Both flp-11 and flp-11; egl-4 loss of function mutants are defective in satiety quiescence [for comparison to egl-4 single mutant, see (E)]. Considering the roaming behavior of the flp-11; egl-4 mutants shown in the red bar, egl-4 could be epistatic to flp-11. (E) Overexpression of flp-11 does not suppress the defect in the egl-4 loss of function mutants. (F) A gain-of-function allele of egl-4(gf) rescued flp-11 loss of function, suggesting that egl-4 could act downstream of flp-11. (G) flp-11 overexpression suppresses the defect in quiescence of daf-7 mutant. (H) The transition rates of egl-4(if), egl-4(gf), and daf-7 mutants. (∗p < 0.05, ∗∗∗p < 0.001 by Student’s t-test. The error bars indicate SEM).
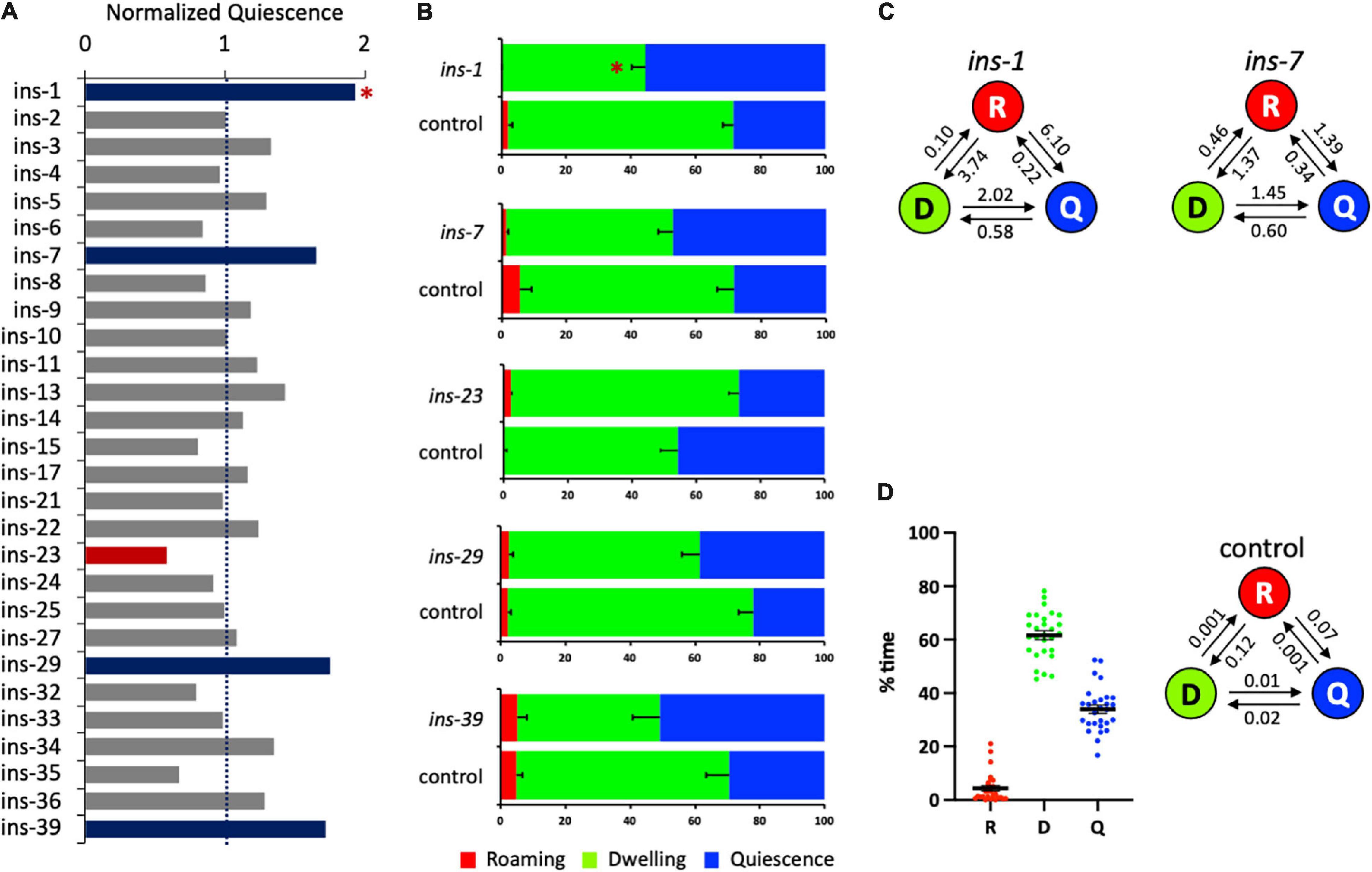
Figure 3. Several ins genes regulate satiety quiescence. (A) The normalized time in quiescence after knockdown of individual ins gene by RNA interference. The percent time in quiescence of each ins is normalized to the percent time in quiescence of the concurrent control (RNAi vector only). Each RNAi was performed with the sample number of 15–18 of tested animals with 7–9 control animals. The experiment was repeated at least three times. Red bars indicate that RNAi of the gene reduced satiety quiescence, whereas blue bars indicate that RNAi of the gene enhanced satiety quiescence. (B) The individual data of the ins genes whose RNAi resulted in changes in satiety quiescence. After two generations of feeding RNAi, the age-synchronized 1-day-old adults were tested for their locomotive activity by measuring the percent time they spent in roaming, dwelling, and quiescence. (C) The transition rates of the animals treated with RNAi of ins-1 and ins-7. (D) Distribution of the values of each state (left) and average transition rates (right) of the 28 independently tested RNAi control groups. (∗p < 0.009 by Student’s t-test after the Bonferroni correction. The error bars indicate SEM).
Satiety Quiescence Assay
Satiety quiescence was measured with an automated method using a nine-video camera monitoring system, as previously described (Gallagher et al., 2013a). For the RNAi screening, on day 1, approximately 20 L4s of the F1 generation were picked and placed on the new RNAi plates with 100 μl of the RNAi bacteria and grown for 24 h. The next day, the RNAi bacteria culture was centrifuged at 4000 RPM for 3 min and the supernatant was removed. The pellet was mixed three 1:1 dilutions of bacteria with M9 buffer. After that, 5 μl of the diluted bacteria was added to each of a 35 mm plate and dried completely, and then a single animal was transferred to each plate. One plate was placed under each video camera of the nine-camera system we built (Gallagher et al., 2013a). Measuring satiety quiescence of mutant strains was performed the same way, except instead GR1396 strain, the outcrossed mutant line was tested while N2 was used as control, and instead of the RNAi bacteria, E. coli HB101 containing mCherry was used for food. Once all nine animals were in focus, the LED light was turned off for 30 min to give the animals time to recover from the transfer. After the 30 min, the LED light was turned on and the image was captured at the rate of 1 frame/second for 30 min using the Point Grey’s FlyCap2 software. The centroid position of each worm was identified in each image using custom written software and the change in centroid position between frames was used to calculate worm movement. The movement data was analyzed using a custom written Hidden Markov Model based program (Gallagher et al., 2013a) and the percent time for each of three behavioral states, roaming, dwelling and quiescence, and the transition rates were calculated.
Statistics
The Bonferroni correction (0.05/56 = 0.0009) was applied to correct for multiple comparisons for two independent comparisons for each RNAi experiment (28 × 2 for two states) and assigned the significance accordingly.
Results
An RNAi Screen of 28 flp Genes Identified FLP-11 in RIS as a Major Regulator of Satiety Quiescence
To investigate the roles of flp genes in satiety quiescence, we performed feeding RNAi and individually knocked down 28 genes whose clones were available to us from the Ahringer library (Fraser et al., 2000). Satiety quiescence is detected under two conditions: non-fasted but fed with high quality food and fasted then refed. The animals show consistent quiescence in both settings, although it is higher in fasted and refed animals compared to non-fasted (You et al., 2008). The non-fasted condition allowed us to rapidly perform the RNAi screen for satiety quiescence in a reliable manner (Hyun et al., 2016). The animals were fed with the RNAi clone of each neuropeptide gene from L4s (the 1st generation). The L4s of the 2nd generation were picked and tested next day as young adults. Throughout the assay, the animals were continuously fed with the RNAi bacteria (see section “Materials and Methods”).
When satiety quiescence was measured using an automated system (Gallagher et al., 2013a), the knockdown of 10 flp genes showed altered satiety quiescence (Figure 1A and Table 1). Previous studies defined several behavioral states based on locomotive activities (Fujiwara et al., 2002; Gray et al., 2005; Ben Arous et al., 2009; Gallagher et al., 2013a; Flavell et al., 2020). Roaming is a state when an animal moves straight in a relatively high speed to explore, dwelling is a state when an animal moves back and forth in a low speed to exploit, and quiescence is a state when an animal is not moving. When we examined individual locomotive activity regarding the percent time the animal spent in each of three behavioral states, RNAi of five flp genes (flp-1, 7, 8, 9, and 11) reduced the percent time in quiescence, although the only one that reached statistical significance after the correction was flp-11 (Figure 1B). RNAi of flp-7, a C. elegans tachykinin implicated in nutrient sensing and lipid metabolism (Palamiuc et al., 2017), shows a tendency to enhance satiety quiescence. We also examined the normalized transition rates between each state. Transition rates are analogous to rate constants of first-order chemical reactions, and in this case reflect the tendency of the animal to switch from one state to another (Gallagher et al., 2013a). Due to the very low percent time that animals spend in roaming, there are extremely small values of transition rates entering roaming (for example, Figure 1D, 0.001 for dwelling to roaming, 0.0002 for quiescence to roaming), and so the normalized transition rates between roaming and the other two states are often not meaningful. However, the transition rates between dwelling and quiescence could imply the duration of each quiescence bout because more frequent switching could result in a short duration. When normalized to the transition rates of concurrent controls, RNAi of the five flps reduced the transition rates from dwelling to quiescence but increased the rates from quiescence to dwelling, suggesting their quiescence bouts are likely reduced (Figure 1C).
Satiety quiescence is extremely variable especially when the animals are tested under the non-fasted condition. Therefore, we always run concurrent controls which were picked as L4s among the progeny from the same mother, cultivated with the control RNAi bacteria under the exact same condition as the group treated with the testing RNAi. When we perform the experiment, we provide as consistent conditions as possible for the temperature, humidity, and the time of the day to ensure that the errors of the control are minimum. To assess the variation, we combined the data from all 28 control groups from each independent experiment (where each control group consists of 6–9 animals) and analyzed the distribution of the percent time of roaming (R), dwelling (D), and quiescence (Q) and found most of the values are within 20% of variation with a few outliers (Figure 1D). We also calculated the average values of the transition rates of the 28 control groups (Figure 1D). The data show that the animals in the control groups have an equal tendency to switch from dwelling to quiescence and from quiescence to dwelling. The animals spent most of their time dwelling or quiescent as the transition rates of either dwelling or quiescence to enter roaming are extremely low.
Among the five, flp-11 RNAi reduced satiety quiescence most. flp-11 is known to regulate two types of sleep, DTS and SIS, in C. elegans. When we tested flp-11 mutants, they also showed reduction in satiety quiescence, confirming the RNAi result (Figure 2A). In contrast to the flp-11 loss-of-function mutant, a flp-11 overexpression (flp-11OE) under its own promoter enhanced satiety quiescence (Figure 2B). flp-11 mutants increase transition rates from quiescence to dwelling, whereas flp-11OE decreases it, suggesting flp-11 is required for long bouts of satiety quiescence. flp-11 regulates DTS and SIS through its action in an interneuron RIS. To examine whether satiety quiescence is also regulated by the flp-11 action in RIS, we tested a mutant of aptf-1, which is necessary for RIS function (Turek et al., 2016). The mutant showed reduced quiescence, as the flp-11 mutant did, suggesting that flp-11 regulates satiety quiescence through its action in RIS (Figure 2C).
ELG-4 Likely Acts Downstream of FLP-11 to Regulate Satiety Quiescence
Satiety quiescence requires function of egl-4, a cGMP-dependent protein kinase (You et al., 2008). To investigate whether flp-11 genetically interacts with egl-4, we generated three different double mutants: (1) flp-11 loss-of-function with egl-4 loss-of-function, (2) flp-11OE with egl-4 loss-of-function, and (3) flp-11 loss-of-function with egl-4 gain-of-function. The double mutant of flp-11 loss-of-function with egl-4 loss-of-function shows a similar phenotype to that of egl-4 loss-of-function single mutants – reduced percent of quiescence and enhanced of roaming (Figures 2D,E and Table 2). In addition, overexpression of FLP-11 does not rescue the loss-of-function mutant of egl-4. Finally, egl-4 gain-of-function restored satiety quiescence to flp-11 mutants (Figure 2F). Taken together, these results suggest that EGL-4 can act downstream of FLP-11. When we examined the normalized transition rates of egl-4 loss-of-function, gain-of-function and daf-7 mutants, they also showed the changes in the transition rates between dwelling and quiescence, consistent with their changes in percent time the animals spent in the two states (Figure 2H).
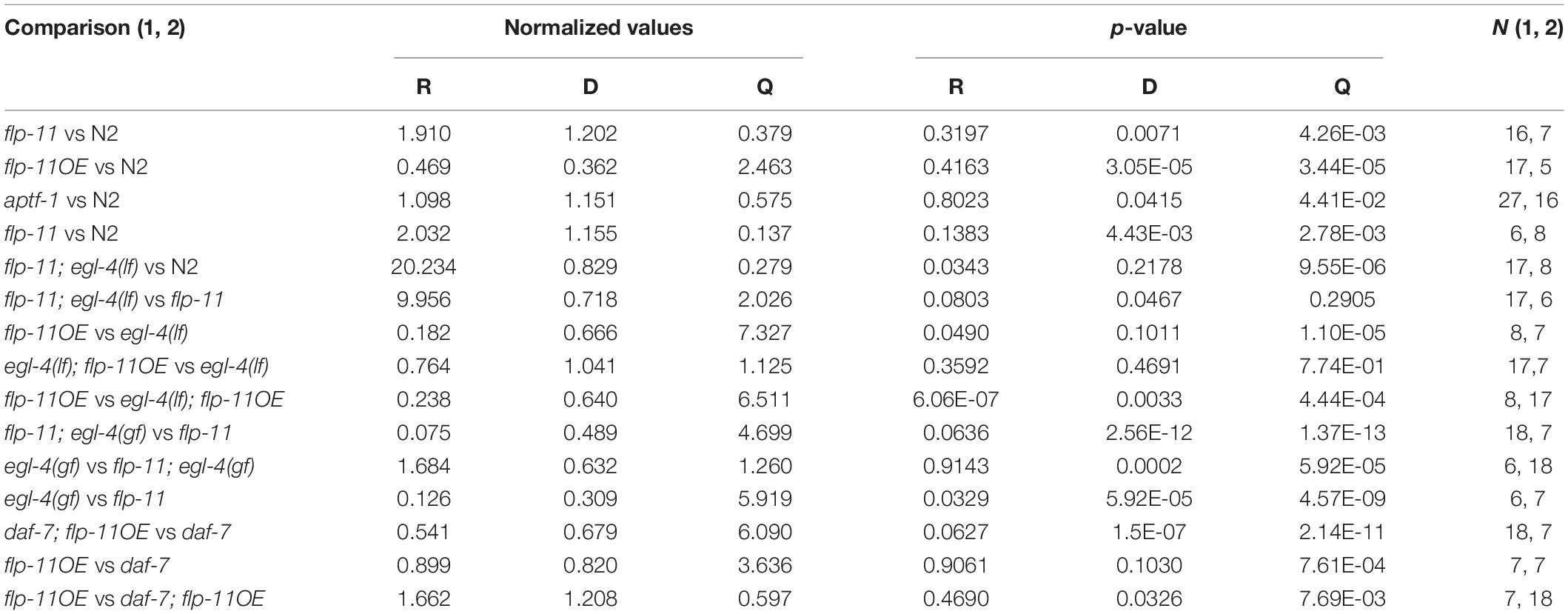
Table 2. Normalized values of each state of roaming, dwelling and quiescence, the sample size, and statistical significance of flp-11, flp-11OE, egl-4(lf), egl-4(gf), daf-7, and the double mutants.
FLP-11 in RIS Acts Downstream of DAF-7 in ASI
In C. elegans, out of a total of 302 neurons, the ASI neuron is the key neuron to relay the animal’s metabolic state and is therefore an important regulator of developmental progression, feeding behavior, and aging processes. Under adverse conditions such as starvation, C. elegans larvae enter a developmental diapause known as the dauer larva (Hu, 2007; Baugh and Hu, 2020). In the presence of food, ASI prevents the animal from becoming dauer and thus promotes reproductive growth (Bargmann and Horvitz, 1991). Calorie restriction extends life span in an ASI dependent manner (Bishop and Guarente, 2007). ASI is the sole source of TGFβ under normal growth conditions, and upregulation of TGFβ in ASI when the animal is sated is required for satiety (Gallagher et al., 2013b).
Based on the role of ASI neuron in satiety quiescence, next we examined whether and how FLP-11 interacts with DAF-7 and ASI by testing a double mutant of daf-7 mutant and flp-11OE. Overexpression of FLP-11 rescues the defect of daf-7 mutants in satiety quiescence suggesting FLP-11 acting downstream of DAF-7 and ASI (Figure 2G).
An RNAi Screen of 28 ins Genes Identified ins-1 as a Negative Regulator for Satiety Quiescence
Next, we investigated the roles of ins genes in satiety quiescence, performing the same strategy of feeding RNAi combined with the Hidden Markov Model based analysis (Figure 3A and Table 3). RNAi of five ins genes altered satiety quiescence (Figure 3B): 1, 7, 23, 29, and 39. We arbitrarily set 0.6 and 1.7 as cutoffs for reduction and enhancement, respectively. While knockdown of ins-1, 7, 29, and 39 enhances satiety quiescence (ins-1 reached statistical significance), knockdown of ins-23 reduces satiety quiescence. The transition rates of ins-1 and ins-7 are shown in Figure 3C. The distribution of the values of each state of 28 independent control RNAi groups and the average values of transition rates show that the variation for quiescence duration is bigger and the animals have shorter quiescence bout compared to the flp RNAi screen. Nonetheless, the trend is consistent: the most groups are within 20% of the mean (with a few outliers), and although the animals are less quiescent, they spent most of their time either dwelling or quiescent (Figure 3D). Because INS-1 antagonizes the DAF-2 insulin receptor (Pierce et al., 2001), and because DAF-2 function is necessary for satiety quiescence (You et al., 2008), an increase of satiety quiescence by ins-1 RNAi suggests that it inhibits satiety quiescence by suppressing DAF-2 activity.
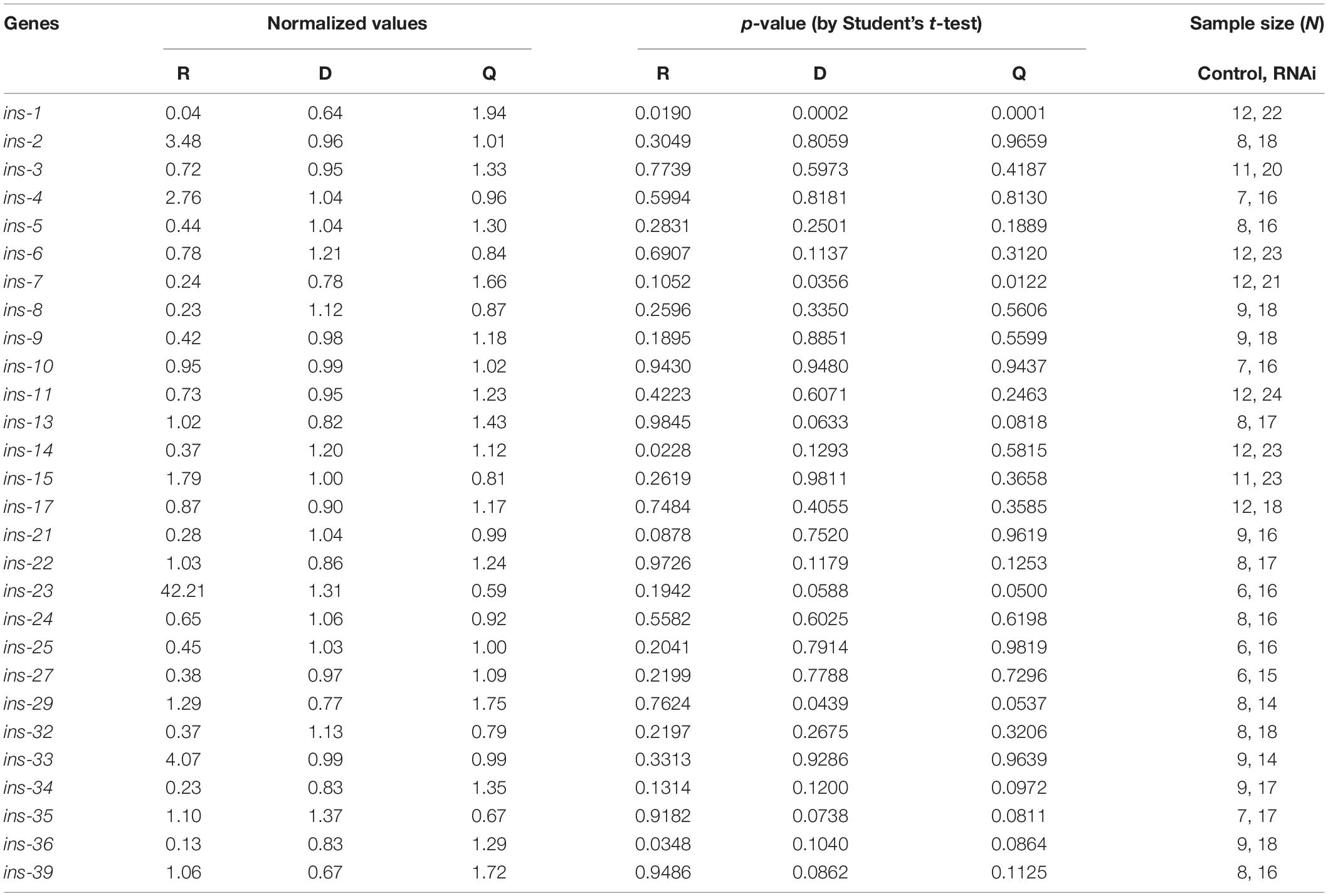
Table 3. Normalized values of each state of roaming, dwelling and quiescence, the sample size, and statistical significance of 28 ins genes.
Discussion
Among 28 tested flp genes, we found 10 FLPs that potentially regulate satiety quiescence either positively or negatively when the gene was knocked down by RNAi. Among them, flp-11, known to regulate other types of sleep-like behavior in C. elegans plays the most significant role in satiety quiescence. The flp-11 mutation reduces satiety quiescence, whereas over-expression enhances it. FLP-11 controls satiety quiescence by acting in the RIS neurons; a mutant that carries a mutation in a gene aptf-1, a transcription factor that functions in RIS, is also defective in satiety quiescence. Genetic analyses suggest that FLP-11 acts upstream of the cGMP signaling and downstream of the TGFβ pathway.
From the RNAi screen of 28 ins genes, we identified ins-1 as an antagonist for DAF-2 in satiety quiescence. ins-1 is expressed multiple neurons in the nervous system, intestine and vulval muscles (Li and Kim, 2008). It is, however, noted to mediate starvation-induced salt learning by being released from AIA. Considering that ASI is the major neuron for satiety quiescence and that AIA neurons are the main target of ASI, we suggest that INS-1 serves as a negative feedback signal to reset ASI activity when the animal feeds continuously in an abundance of food. INS-7, the 2nd best candidate, is expressed in the intestine and plays a role in aging by propagating FOXO signaling to other tissues (Murphy et al., 2007). Its expression in neurons is induced by Pseudomonas virulence (Kawli and Tan, 2008). Its downregulation is necessary for intermittent fasting-induced longevity (Honjoh et al., 2009). These findings suggest that INS-7 might mediate stress signals. The overexpression study of individual ins genes (Zheng et al., 2018) suggests ins-7 could act as either an agonist or antagonist depending on the phenotype. Our results suggest, ins-7 seems to act as an antagonist, as do ins-29 and ins-39 for satiety quiescence. In contrast, ins-23 has been suggested to be an antagonist (Matsunaga et al., 2018) or neutral (Zheng et al., 2018), but it may act as an agonist for satiety quiescence. INS-7 is expressed in URX neurons and antagonizes INS-6 released from ASI neurons by converging on RIA interneurons which play an important role in head movement and turning by synapsing on SMD or RMD head motor neurons (Gray et al., 2005). This suggests that INS-7 might regulate satiety quiescence via controlling ASI function negatively as INS-1 does.
Although satiety quiescence has not been thoroughly examined for its homeostatic properties to be defined as sleep (Raizen et al., 2008), it contains a few key components of behavioral state of sleep, such as a distinct period of inactivity and reduction of sensory perception (You et al., 2008). In addition, it is, to our knowledge, the only sleep-like behavioral state regulated mainly by metabolic state without being associated with development progresses or harsh stresses; satiety quiescence can be induced without prior starvation or stress. Therefore, our RNAi screens using this behavior as a readout would provide an insight how neuropeptides might convey the information of the metabolic state of the body to induce a sleep-like state.
Based on our results, we propose a model (Figure 4) where neuropeptides regulate satiety quiescence. In C. elegans, out of a total of 302 neurons, the ASI neuron is the key neuron to relay the animal’s metabolic state and is therefore an important regulator of developmental progression, feeding behavior and aging processes (Bargmann and Horvitz, 1991; Bishop and Guarente, 2007; Hu, 2007; Baugh and Hu, 2020). ASI is the sole source of TGFβ DAF-7 under normal growth conditions. Upregulation of DAF-7 in ASI when the animal is sated is required for satiety quiescence. The receptor DAF-1 functions in RIM and RIC to promote satiety quiescence, connecting ASI to RIM and RIC (Gallagher et al., 2013b). RIC and RIM release octopamine and tyramine (Alkema et al., 2005) and therefore potentially regulate wakefulness. In fact, RIM synapses on RIS and activate or inhibit RIS depending on RIM’s activity level (Maluck et al., 2020). Optogenetic activation of RIS results in inhibition of locomotion and pharyngeal pumping, implicating that RIS is required for executing and maintaining sleep (Steuer Costa et al., 2019). Our study suggests that ASI provides another input to RIS through DAF-7 and DAF-1 acting on RIM and RIC and convey the animal’s nutritional status to promote a sleep-like state induced by satiety. Taken together, our study provides an insight into understanding how neuropeptides regulate sleep-like behavior by unveiling the conserved molecular mechanisms and the underlying neural circuit.
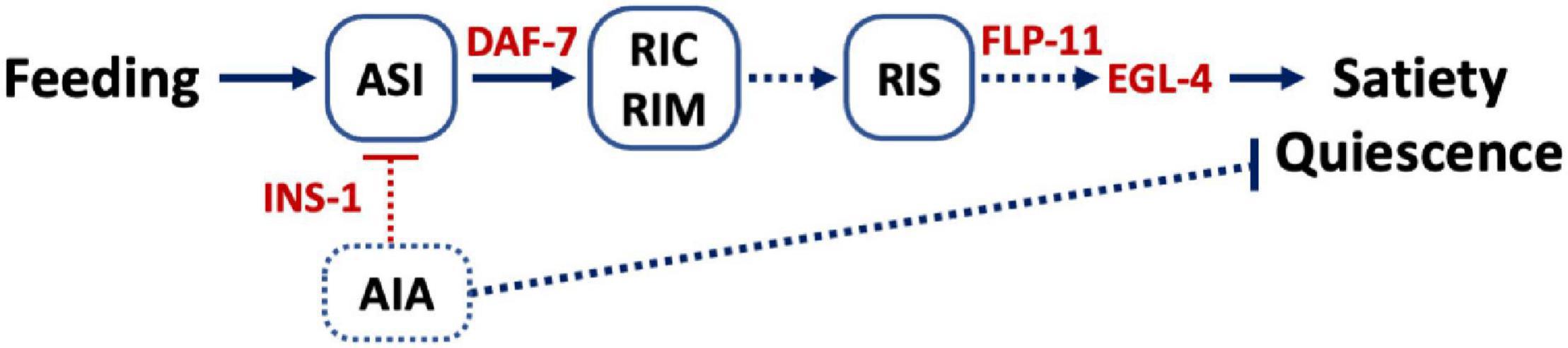
Figure 4. Model. Feeding activates ASI and promotes ASI to release DAF-7, the C. elegans TGFβ, which in turn activates the TGFβ receptors on RIM and RIC (Gallagher et al., 2013b). This activation leads to activation of RIS and release of FLP-11 to induce satiety quiescence. The cGMP signal initiated from ASI is also required for satiety quiescence. Although the exact sites of cGMP signal are unknown, our data suggest EGL-4 could act downstream of FLP-11. INS-1 presumably released from AIA could serve as a negative feedback signal to ASI and inhibit satiety quiescence. Lines indicate signaling that has been previously reported whereas dotted lines are hypothetical.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
Y-JY conceived of the study and drafted the manuscript. MM and JK helped to design experiments. MM, EU, RS, JK, and Y-JY performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Neuroscience Institute, Nagoya University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Caenorhabditis Genome Center (supported by the National Institutes of Health – Office of Research Infrastructure Programs (P40 OD010440)) and Dr. Hayashi for the strains, SunyBiotech for technical assistant, Avery for consultation regarding statistics, and Gallagher, Wyler, Yip for critical reading.
References
Alkema, M. J., Hunter-Ensor, M., Ringstad, N., and Horvitz, H. R. (2005). Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260. doi: 10.1016/j.neuron.2005.02.024
Antin, J., Gibbs, J., Holt, J., Young, R. C., and Smith, G. P. (1975). Cholecystokinin elicits the complete behavioral sequence of satiety in rats. J. Comp. Physiol. Psychol. 89, 784–790. doi: 10.1037/h0077040
Bargmann, C. I., and Horvitz, H. R. (1991). Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243–1246. doi: 10.1126/science.2006412
Bass, J., and Takahashi, J. S. (2010). Circadian integration of metabolism and energetics. Science 330, 1349–1354. doi: 10.1126/science.1195027
Baugh, L. R., and Hu, P. J. (2020). Starvation responses throughout the Caenorhabditis elegans life cycle. Genetics 216, 837–878. doi: 10.1534/genetics.120.303565
Beets, I., Janssen, T., Meelkop, E., Temmerman, L., Suetens, N., Rademakers, S., et al. (2012). Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science 338, 543–545. doi: 10.1126/science.1226860
Ben Arous, J., Laffont, S., and Chatenay, D. (2009). Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS One 4:e7584. doi: 10.1371/journal.pone.0007584
Bennett, H. L., Khoruzhik, Y., Hayden, D., Huang, H., Sanders, J., Walsh, M. B., et al. (2018). Normal sleep bouts are not essential for C. elegans survival and FoxO is important for compensatory changes in sleep. BMC Neurosci. 19:10. doi: 10.1186/s12868-018-0408-1
Bishop, N. A., and Guarente, L. (2007). Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549. doi: 10.1038/nature05904
Chalasani, S. H., Kato, S., Albrecht, D. R., Nakagawa, T., Abbott, L. F., and Bargmann, C. I. (2010). Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat. Neurosci. 13, 615–621. doi: 10.1038/nn.2526
Cheong, M. C., Artyukhin, A. B., You, Y. J., and Avery, L. (2015). An opioid-like system regulating feeding behavior in C. elegans. eLife 4:e06683. doi: 10.7554/eLife.06683
Driver, R. J., Lamb, A. L., Wyner, A. J., and Raizen, D. M. (2013). DAF-16/FOXO regulates homeostasis of essential sleep-like behavior during larval transitions in C. elegans. Curr. Biol. 23, 501–506. doi: 10.1016/j.cub.2013.02.009
Flavell, S. W., Raizen, D. M., and You, Y. J. (2020). Behavioral states. Genetics 216, 315–332. doi: 10.1534/genetics.120.303539
Folgueira, C., Barja-Fernandez, S., Gonzalez-Saenz, P., Pena-Leon, V., Castelao, C., Ruiz-Pinon, M., et al. (2018). Uroguanylin: a new actor in the energy balance movie. J. Mol. Endocrinol. 60, R31–R38. doi: 10.1530/JME-17-0263
Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. doi: 10.1038/35042517
Fujiwara, M., Sengupta, P., and McIntire, S. L. (2002). Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36, 1091–1102. doi: 10.1016/s0896-6273(02)01093-0
Gallagher, T., Bjorness, T., Greene, R., You, Y. J., and Avery, L. (2013a). The geometry of locomotive behavioral states in C. elegans. PLoS One 8:e59865. doi: 10.1371/journal.pone.0059865
Gallagher, T., Kim, J., Oldenbroek, M., Kerr, R., and You, Y. J. (2013b). ASI regulates satiety quiescence in C. elegans. J. Neurosci. 33, 9716–9724. doi: 10.1523/JNEUROSCI.4493-12.2013
Garrison, J. L., Macosko, E. Z., Bernstein, S., Pokala, N., Albrecht, D. R., and Bargmann, C. I. (2012). Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science 338, 540–543. doi: 10.1126/science.1226201
Gibbs, J., Young, R. C., and Smith, G. P. (1973). Cholecystokinin elicits satiety in rats with open gastric fistulas. Nature 245, 323–325. doi: 10.1038/245323a0
Gray, J. M., Hill, J. J., and Bargmann, C. I. (2005). A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 102, 3184–3191. doi: 10.1073/pnas.0409009101
Honer, M., Buscemi, K., Barrett, N., Riazati, N., Orlando, G., and Nelson, M. D. (2020). Orcokinin neuropeptides regulate sleep in Caenorhabditis elegans. J. Neurogenet. 34, 440–452. doi: 10.1080/01677063.2020.1830084
Honjoh, S., Yamamoto, T., Uno, M., and Nishida, E. (2009). Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457, 726–730. doi: 10.1038/nature07583
Hyun, M., Davis, K., Lee, I., Kim, J., Dumur, C., and You, Y. J. (2016). Fat metabolism regulates satiety behavior in C. elegans. Sci. Rep. 6:24841. doi: 10.1038/srep24841
Iannacone, M. J., Beets, I., Lopes, L. E., Churgin, M. A., Fang-Yen, C., Nelson, M. D., et al. (2017). The RFamide receptor DMSR-1 regulates stress-induced sleep in C. elegans. eLife 6:e19837. doi: 10.7554/eLife.19837
Kawli, T., and Tan, M. W. (2008). Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat. Immunol. 9, 1415–1424. doi: 10.1038/ni.1672
Lee, D. A., Andreev, A., Truong, T. V., Chen, A., Hill, A. J., Oikonomou, G., et al. (2017). Genetic and neuronal regulation of sleep by neuropeptide VF. eLife 6:e25727. doi: 10.7554/eLife.25727
Maluck, E., Busack, I., Besseling, J., Masurat, F., Turek, M., Busch, K. E., et al. (2020). A wake-active locomotion circuit depolarizes a sleep-active neuron to switch on sleep. PLoS Biol. 18:e3000361. doi: 10.1371/journal.pbio.3000361
Marcheva, B., Ramsey, K. M., Buhr, E. D., Kobayashi, Y., Su, H., Ko, C. H., et al. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631. doi: 10.1038/nature09253
Matsunaga, Y., Matsukawa, T., Iwasaki, T., Nagata, K., and Kawano, T. (2018). Comparison of physiological functions of antagonistic insulin-like peptides, INS-23 and INS-18, in Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 82, 90–96. doi: 10.1080/09168451.2017.1415749
Murphy, C. T., and Hu, P. J. (2013). Insulin/insulin-like growth factor signaling in C. elegans. WormBook 1–43. doi: 10.1895/wormbook.1.164.1
Murphy, C. T., Lee, S. J., and Kenyon, C. (2007). Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 19046–19050. doi: 10.1073/pnas.0709613104
Nagy, S., Tramm, N., Sanders, J., Iwanir, S., Shirley, I. A., Levine, E., et al. (2014). Homeostasis in C. elegans sleep is characterized by two behaviorally and genetically distinct mechanisms. eLife 3:e04380. doi: 10.7554/eLife.04380
Nassel, D. R., Pauls, D., and Huetteroth, W. (2019). Neuropeptides in modulation of Drosophila behavior: how to get a grip on their pleiotropic actions. Curr. Opin. Insect. Sci. 36, 1–8. doi: 10.1016/j.cois.2019.03.002
Nath, R. D., Chow, E. S., Wang, H., Schwarz, E. M., and Sternberg, P. W. (2016). C. elegans stress-induced sleep emerges from the collective action of multiple neuropeptides. Curr. Biol. 26, 2446–2455. doi: 10.1016/j.cub.2016.07.048
Nelson, M. D., Trojanowski, N. F., George-Raizen, J. B., Smith, C. J., Yu, C. C., Fang-Yen, C., et al. (2013). The neuropeptide NLP-22 regulates a sleep-like state in Caenorhabditis elegans. Nat. Commun. 4:2846. doi: 10.1038/ncomms3846
Palamiuc, L., Noble, T., Witham, E., Ratanpal, H., Vaughan, M., and Srinivasan, S. (2017). A tachykinin-like neuroendocrine signalling axis couples central serotonin action and nutrient sensing with peripheral lipid metabolism. Nat. Commun. 8:14237. doi: 10.1038/ncomms14237
Parhar, I. S., Ogawa, S., and Ubuka, T. (2016). Reproductive neuroendocrine pathways of social behavior. Front. Endocrinol. 7:28. doi: 10.3389/fendo.2016.00028
Peymen, K., Watteyne, J., Borghgraef, C., Van Sinay, E., Beets, I., and Schoofs, L. (2019). Myoinhibitory peptide signaling modulates aversive gustatory learning in Caenorhabditis elegans. PLoS Genet. 15:e1007945. doi: 10.1371/journal.pgen.1007945
Pierce, S. B., Costa, M., Wisotzkey, R., Devadhar, S., Homburger, S. A., Buchman, A. R., et al. (2001). Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15, 672–686. doi: 10.1101/gad.867301
Quillet, R., Ayachi, S., Bihel, F., Elhabazi, K., Ilien, B., and Simonin, F. (2016). RF-amide neuropeptides and their receptors in mammals: pharmacological properties, drug development and main physiological functions. Pharmacol. Ther. 160, 84–132. doi: 10.1016/j.pharmthera.2016.02.005
Raizen, D. M., Zimmerman, J. E., Maycock, M. H., Ta, U. D., You, Y. J., Sundaram, M. V., et al. (2008). Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572. doi: 10.1038/nature06535
Steuer Costa, W., Van der Auwera, P., Glock, C., Liewald, J. F., Bach, M., Schuler, C., et al. (2019). A GABAergic and peptidergic sleep neuron as a locomotion stop neuron with compartmentalized Ca2+ dynamics. Nat. Commun. 10:4095. doi: 10.1038/s41467-019-12098-5
Takeishi, A., Yeon, J., Harris, N., Yang, W., and Sengupta, P. (2020). Feeding state functionally reconfigures a sensory circuit to drive thermosensory behavioral plasticity. eLife 9:e61167. doi: 10.7554/eLife.61167
Tomioka, M., Adachi, T., Suzuki, H., Kunitomo, H., Schafer, W. R., and Iino, Y. (2006). The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51, 613–625. doi: 10.1016/j.neuron.2006.07.024
Tsai, V. W., Macia, L., Johnen, H., Kuffner, T., Manadhar, R., Jorgensen, S. B., et al. (2013). TGF-b superfamily cytokine MIC-1/GDF15 is a physiological appetite and body weight regulator. PLoS One 8:e55174. doi: 10.1371/journal.pone.0055174
Tsai, V. W. W., Husaini, Y., Sainsbury, A., Brown, D. A., and Breit, S. N. (2018). The MIC-1/GDF15-GFRAL pathway in energy homeostasis: implications for obesity, cachexia, and other associated diseases. Cell Metab. 28, 353–368. doi: 10.1016/j.cmet.2018.07.018
Turek, M., Besseling, J., Spies, J. P., Konig, S., and Bringmann, H. (2016). Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. eLife 5:e12499. doi: 10.7554/eLife.12499
Valentino, M. A., Lin, J. E., Snook, A. E., Li, P., Kim, G. W., Marszalowicz, G., et al. (2011). A uroguanylin-GUCY2C endocrine axis regulates feeding in mice. J. Clin. Invest. 121, 3578–3588. doi: 10.1172/JCI57925
Van der Auwera, P., Frooninckx, L., Buscemi, K., Vance, R. T., Watteyne, J., Mirabeau, O., et al. (2020). RPamide neuropeptides NLP-22 and NLP-2 act through GnRH-like receptors to promote sleep and wakefulness in C. elegans. Sci. Rep. 10:9929. doi: 10.1038/s41598-020-66536-2
You, Y. J., Kim, J., Raizen, D. M., and Avery, L. (2008). Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 7, 249–257. doi: 10.1016/j.cmet.2008.01.005
Keywords: satiety, metabolism, quiescence, TGFβ, neural circuit, cyclic GMP
Citation: Makino M, Ulzii E, Shirasaki R, Kim J and You Y-J (2021) Regulation of Satiety Quiescence by Neuropeptide Signaling in Caenorhabditis elegans. Front. Neurosci. 15:678590. doi: 10.3389/fnins.2021.678590
Received: 10 March 2021; Accepted: 11 June 2021;
Published: 15 July 2021.
Edited by:
Masayuki Ikeda, University of Toyama, JapanReviewed by:
Jeffrey Donlea, University of California, Los Angeles, United StatesYu Hayashi, University of Tsukuba, Japan
Copyright © 2021 Makino, Ulzii, Shirasaki, Kim and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Young-Jai You, WW91bmctSmFpLllvdUBVVFNvdXRod2VzdGVybi5lZHU=
 Mei Makino1
Mei Makino1 Jeongho Kim
Jeongho Kim Young-Jai You
Young-Jai You