- 1School of Medicine, University of Connecticut Health Center, Farmington, CT, United States
- 2The Jackson Laboratory for Genomic Medicine, Farmington, CT, United States
- 3The Jackson Laboratory, Bar Harbor, ME, United States
Substance use disorders (SUDs) remain a significant public health challenge, affecting tens of millions of individuals worldwide each year. Often comorbid with other psychiatric disorders, SUD can be poly-drug and involve several different substances including cocaine, opiates, nicotine, and alcohol. SUD has a strong genetic component. Much of SUD research has focused on the neurologic and genetic facets of consumption behavior. There is now interest in the role of the gut microbiome in the pathogenesis of SUD. In this review, we summarize current animal and clinical evidence that the gut microbiome is involved in SUD, then address the underlying mechanisms by which the gut microbiome interacts with SUD through metabolomic, immune, neurological, and epigenetic mechanisms. Lastly, we discuss methods using various inbred and outbred mice models to gain an integrative understanding of the microbiome and host genetic controls in SUD.
Introduction
Substance use disorder (SUD) is a mental condition affecting the brain and is characterized in part by chronic dependence despite negative social, mental, and physical health consequences. Addiction represents the most severe form of SUD, with affected individuals often incapable of maintaining abstinence despite the will to discontinue substance use. Importantly, the effects of addiction can persist beyond cessation of substance use, suggesting that lasting physiological changes in the brain are involved (Koob et al., 2008). An expansive list of substance classes are related to SUD, ranging from legal forms, including alcohol, to illicit or controlled forms, including cocaine and opioids. Alcohol use disorder (AUD) remains the most prevalent SUD, however, poly-drug use within individuals is not uncommon (Lipari and Van Horn, 2017).
Addictive substances can affect multiple pathways controlling brain function. Alteration of the dopaminergic system is one common mechanism shared by all substances in the establishment of recurrent substance use (Nestler, 2005). Notably, substance misuse leads to increased levels of dopamine release and chronic signal imbalance in D2-like dopamine receptors in the ventral striatum, particularly in the nucleus accumbens (Koob and Le Moal, 2001; Nestler, 2005). Termed the mesolimbic pathway, substances of misuse operate to “hijack” the host reward system critical for pleasurable response and reinforcement to rewarding stimuli, as well as memory and emotional processes. Furthermore, the chronic use of these substances leads to neuroplasticity and structural reorganizations, and drug-specific changes in neurotransmitter transporters and receptors (Bliss et al., 2003; Kauer, 2004). Despite the central role of dopamine in SUD, other neurotransmitters such as serotonin and gamma-aminobutyric acid (GABA) also play an important role in SUD, depending on the dosage and frequency of substance use, as well as intrinsic biological factors such as host genetics (Prom-Wormley et al., 2017).
Previous human linkage studies have shown that children of individuals affected by SUD are at a greater risk for SUD, suggesting a genetic component to the risk of developing the disorder (Kreek et al., 2004). SUD involving cocaine confers the highest genetic risk of any substance, estimated at approximately 70% heritability (Goldman et al., 2005). Numerous genetic loci have been implicated in the heritability of risk for cocaine SUD, most notably genes involved in dopamine transport (Drd2, Drd4) and metabolism (Dbh) (Table 1). AUD also show strong genetic factors with variants in genes involved in alcohol metabolism (Adh1b, Aldh2) and diverse variants of smaller effect in dopaminergic systems (Table 1). However, discordant SUD rates found in identical twins suggest that genetics alone does not entirely explain the incidence of SUD (Agrawal and Lynskey, 2008). Aside from socio-economic factors studied extensively (Stone et al., 2012), there is intense interest in examining the role of the gut microbiome in SUD. The gut microbiota are a diverse population of microorganisms that constantly interact with the central nervous system (CNS) through complicated metabolomic, immune, neurological, and epigenetic pathways, making them potential sources of environmental influence on SUD.
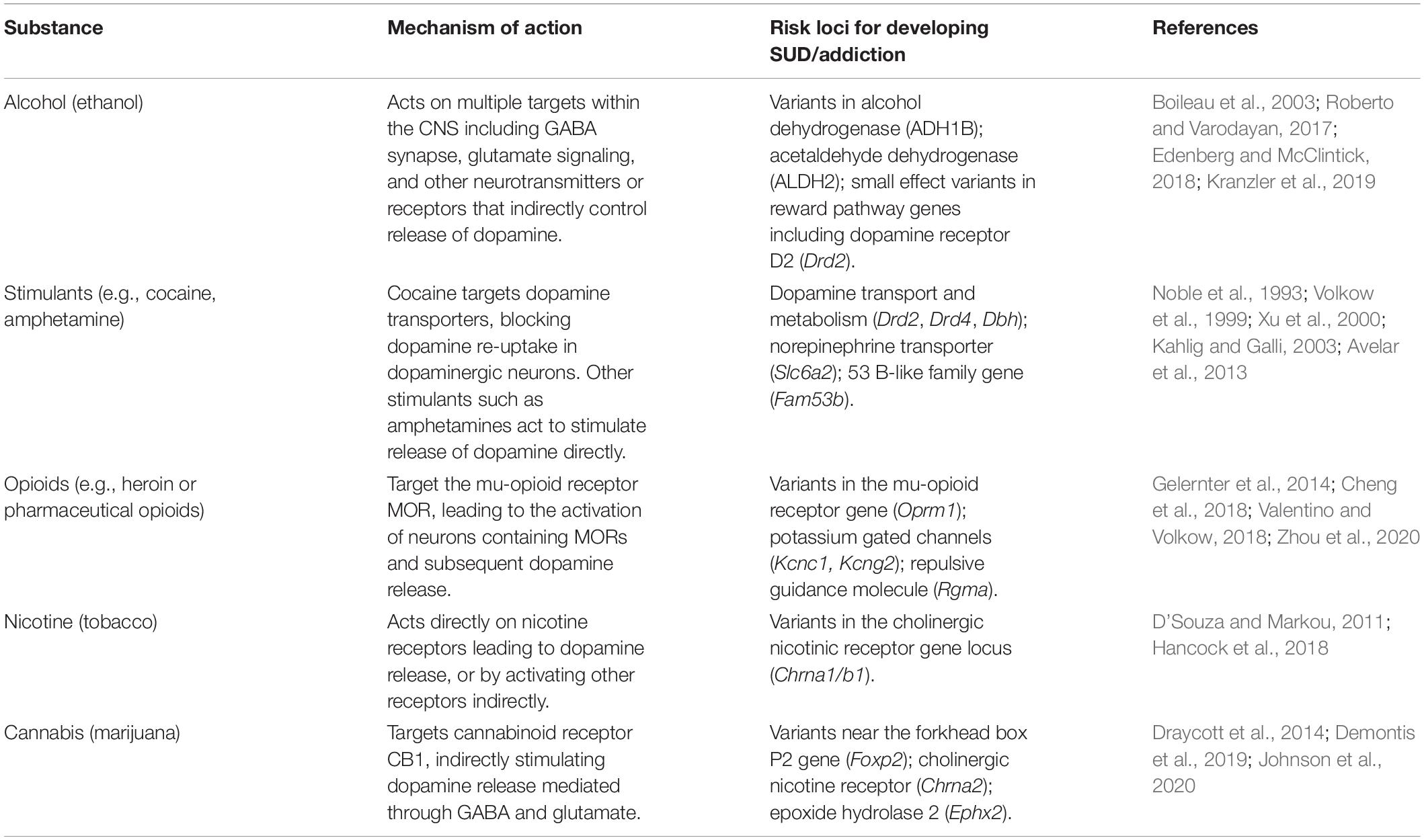
Table 1. Substances involved in SUD and addiction, their mechanism of action and genetic risk loci for development of SUD/addiction.
Although methods for investigating the microbiome in various diseases have made tremendous advancements in the last decade (Quigley and Gajula, 2020), studies of SUD in humans remain challenging because of the difficulty in controlling for complex experimental variables (for example, diet and host genotype). Model organisms such as mice provide excellent opportunities for studying the microbiome and its interaction with the host due to greater control over genetic and environmental confounders. In addition, mice of diverse but known genetic backgrounds can be harnessed to interrogate complex interactions between host genotype and the gut microbiota that may be important for risk of developing SUD (Poltorak et al., 2018). This review will provide a concise overview of recent insights on mechanisms of interaction between the gut microbiota and CNS, in addition to methodological considerations for studying the gut microbiome in SUD in animal models.
The Gut–Brain Axis and Substance Use
The human gastrointestinal tract is home to a large community of microbiota on the order of trillions of cells. Likewise, the collective gene content of the microbiome encodes a staggering amount of functional potential, with an estimated 150 times greater number of genes than that of the human or mouse genome (Grice and Segre, 2012). Despite a core microbiome, fluctuations in composition are often observed after administering antibiotics, non-antibiotic medication, and diet intervention (Caporaso et al., 2011). This deviation from an individual-defined normal or “healthy” microbiome in the context of disease is termed dysbiosis, and a significant body of evidence has linked the dysbiotic microbiome as a contributor of disease and recently mental health (Shreiner et al., 2015; Capuco et al., 2020). There are multiple potential facets of gut host–microbe interactions to explain these disease associations, requiring much interdisciplinary research focus. Emerging evidence suggests prominent roles in host–microbe immune interaction, crosstalk between the gut and brain via neurotransmitters, and a role for bacterial metabolites including short-chain fatty acids (SCFA) in the pathophysiology of SUD. A summary of representative studies of the microbiome and SUD in animal models is provided in Table 2. For a comprehensive summary see the recent review by Angoa-Pérez and Kuhn (2021).
Interactions Between the Microbiota and the Immune System in SUD
The gut microbiota produce an immense number of foreign antigens to which the host immune system must tolerate under normal homeostasis (Swiatczak and Cohen, 2015). The intestinal epithelium provides a physical barrier between the microbiota and immune cells, largely regulating the immune response to commensal microbes. Evidence suggests increased intestinal barrier permeability (leaky gut) in SUD, particularly AUD (Leclercq et al., 2014). Alcohol consumption has long been associated with developing a leaky gut through oxidative stress, which allows for increased translocation of bacterial products into the lamina propria. Here, microbial antigens can interact directly with gut-localized dendritic cells and macrophages and upregulate multiple pro-inflammatory cytokines production, leading to a heightened local and systemic inflammatory response.
Immune cytokines have modulatory effects on behavior. Levels of pro-inflammatory cytokines, including IL-1β, TNF-α, and IL-8, are increased in response to numerous substances of abuse (Meckel and Kiraly, 2019). Interestingly, this pro-inflammatory response to substance use is akin to the response to bacterial lipopolysaccharide (LPS) through signaling via toll-like receptor 4 (Northcutt et al., 2015). This pro-inflammatory response further implicates the interaction between the gut microbiota and the immune system in SUD pathology. It was observed that alcohol and opiates use leads to an enrichment of pro-inflammatory Proteobacteria in the gut. Likewise, anti-inflammatory responses are altered in instances of substance use which leads to an unbalanced neuroimmune response. Anti-inflammatory cytokines such as IL-10 have been shown to modulate and even reverse anxiety behaviors related to substance use (Patel et al., 2021). Furthermore, IL-10 could entirely diminish the escalation of alcohol intake by modulating GABA signaling in the amygdala. Thus, the link between the gut microbiota and the immune system via neuroimmune pathways are important research areas in SUD and addiction.
Microbiota-Derived Neurotransmitters
Neurotransmitters intuitively play a critical role in the development and maintenance of all forms of SUD. Numerous bacteria have been described as capable of producing a number of neurotransmitters including GABA, serotonin, and dopamine (Yano et al., 2015; Strandwitz, 2018). A culture study showed that Bacillus species can produce dopamine. Furthermore, germ-free and antibiotic-treated mice have shown that the gut microbiota influences the production and turnover of dopamine outside the CNS (Strandwitz, 2018). Similar fluctuations in GABA levels are observed in mice treated with antibiotics (Yunes et al., 2020). Lactobacillus, Bifidobacteria, and Bacteroides species were identified to have the capability to produce GABA (Barrett et al., 2012; Strandwitz et al., 2019). Mice given specific GABA-producing Lactobacillus or Bifidobacteria have shown altered behavioral phenotypes (Yunes et al., 2020). Curiously, neurotransmitters produced by the gut microbiota may not under normal circumstances directly affect the brain, as whether these molecules cross the blood–brain barrier is still debated (Boonstra et al., 2015). However, increasing evidence has shown that the blood–brain barrier can fluctuate between states of increased permeability (Braniste et al., 2014). It may also be that the gut microbiota, rather than producing neurotransmitters themselves, affect neurotransmitter production or receptor expression indirectly via signals delivered through the vagus nerve (Bravo et al., 2011). Studies are ongoing to identify the full breadth of neurotransmitter-producing strains in the gut microbiome and the mechanisms by which microbial messages influence host behavior in the context of SUD.
Gut Microbiome Mediated Epigenetic Mechanisms
Substance use leads to changes in gene expression in the brain in critical signaling pathways in the reward circuitry (Walker et al., 2018). Regulation of gene activation and repression is controlled in part epigenetically by adding and removing histone post-translational modifications. Common modifications include acetylation and methylation, which work to activate or silence gene expression, respectively. Importantly, these modifications are reversible; they can be added or removed by dedicated enzymes for this process, based on cellular cues. Histone acetylation, which leads to gene activation by opening the chromatin structure, increases at the nucleus accumbens in response to drug use for most substances (Renthal and Nestler, 2009). Alcohol is a unique case because the byproduct of ethanol metabolism is acetate itself. Acetate derived from alcohol metabolism post-consumption is readily incorporated into the brain, and in mice is associated with spatial memory and preference for the rewarding stimulus of alcohol (Mews et al., 2019). Inhibition or deletion of genes involved in acetylation, histone acetyltransferase (HAT), and histone deacetylase (HDAC) leads to alterations in sensitivity and drug-related behaviors (Cadet, 2016). These lines of evidence implicate the epigenetics of the brain in chronic SUD and addiction, and current research is focusing on the specific genetic loci affected by epigenetic alteration in response to substance use.
The gut microbiota produces large quantities of SCFAs, predominantly acetate, propionate, and butyrate, through carbohydrate metabolism and the breakdown of dietary fiber (Dalile et al., 2019). Butyrate, a potent HDAC inhibitor, can modulate host epigenetics through similar pathways as those affected by substance use (Simon-O’Brien et al., 2015). The microbial contribution to epigenetic modification may be important not only for current SUD but also the risk of developing SUD before initial drug intake, by predisposition toward the activation of addiction and reward pathways in the brain (Meckel and Kiraly, 2019).
Studying the Microbiome in SUD Using Inbred Mice
The use of laboratory rodents to study the neurobiological aspects of SUD has been well documented, and the use of animal research to study the environmental effects of the microbiome is expanding rapidly (Fowler and Kenny, 2012; Meckel and Kiraly, 2019). Mouse models prove useful for investigating the interactions between host genetics and the gut microbiome in substance use. For example, C57BL/6 (B6) mice consume sufficient alcohol levels to mimic binge-like drinking episodes, making this strain a popular choice for modeling binge alcohol intake (Thiele et al., 2014). Further work in this model strain investigates links between alcohol consumption, the gut microbiota, host gene expression, and drinking-related phenotypes. Furthermore, rodent models of selectively inbred “high drinkers” have been proposed for the use of screening drugs aimed at reducing binge-like drinking behaviors (Crabbe et al., 2017). Using single or selectively inbred strains effectively isolates environmental variables of interest for experimentation by controlling for genetic variation in the host. Considering the complex diversity of the microbiota, isolating the microbial effects while controlling for host genetics is prudent. Despite the utility of inbred strains in isolating environmental effects, this method does not fully consider both the genetic and environmental complexity of SUD in humans, underpinning the critical issue of translatability of rodent models.
Diversity Outbred Mouse Models
Much effort has been placed on developing recombinant-inbred mouse strains to capture greater genetic diversity in strains that are more applicable to complex disorders such as SUD in humans. In 2004, a large multi-institutional initiative led to the creation of the collaborative cross (CC) strains by crossing among a set of eight mouse strains which represent >90% of the genetic diversity of Mus musculus and its subspecies (Threadgill et al., 2011). Because CC mice originate from founder strains with defined genomes, this panel provides a high degree of genetic diversity that can be accurately mapped with genotyping techniques (Srivastava et al., 2017). Expanding further on the CC mouse panel are the diversity outbred (DO) strains, which provide even greater genetic and phenotypic diversity by outbreeding, producing unique individual heterozygous mice (Churchill et al., 2012).
There are numerous advantages to utilizing DO mice for mapping phenotypic traits to host genotype. Because they originate from the same founder strains as the CC panel, their genomes are well defined and suited for fine-detail trait mapping (Svenson et al., 2012). Furthermore, DO mice traits can be recovered from inbred CC lines to test for allelic effects. Research into benzene toxicity in DO mice revealed that harnessing this genetic heterogeneity in the mouse better reflected the diverse reactions to benzene exposure seen in humans (French et al., 2015). Efforts using heterogeneous stock (HS) mice, with similar genetic backgrounds as DO mice, have correlated genes with sex-specific ethanol preference and chronic consumption phenotypes, and similar results have been corroborated in DO mice (Hitzemann et al., 2020). In the context of SUD, DO mice may be used to study the interaction between host genes, the environment (i.e., microbiome), and phenotypic differences between strains that reflect similar genetic diversity in humans.
Reproducibility Through a Controlled Environment
Despite considerable effort, reproducibility in microbiome research using animal models remains challenging. Careful considerations must be made when studying the relationship between genetics, phenotype, and the microbiome in an animal. Microbiome differences in mice can be seen based on external factors such as vendor, environmental and facility conditions, and co-housing with other mice (Justice and Dhillon, 2016). When controlling for genetics by using inbred strains, mouse-to-mouse variation in microbiome composition is observed. A consistent methodology plays a crucial role in the reproducibility of microbiome research in animals, as circadian rhythm can also have confounding effects for sampling at different times of the day (Thaiss et al., 2014). In order to reduce confounding individual variation of the microbiota, strategies for the rotation of mice and mixing of bedding may be used so that all mice in the experiment are normalized to the same common microbiota. Furthermore, greater reproducibility can be achieved by defining a standard microbiota composition for a given model or experiment (Witjes et al., 2020). Thus, experimental replication is more likely to be achieved by knowing the target microbiota important for obtaining similar outcomes.
Though defining a normal microbiota for an experiment or model organism may lead to a greater likelihood of reproducibility, it raises the question of translatability since a standardized microbiome is not realistic in humans. Here the concept of increased reproducibility is at odds with the fact that human disorders such as SUD are not simply defined systems. Just as genetically diverse DO mice better reflect the natural heterogeneity seen in humans, so too is there value in assessing natural variation in the microbiome. The advent of “wildlings” by implanting lab-strain mouse embryos into wild mice provides a useful tool for studying a more naturally derived microbiome in an effort to enhance translatability (Rosshart et al., 2019). Ultimately, the choice to standardize and how will depend on the experimental question; whether there is a necessity to study the full breadth of genetic and microbial heterogeneity such as that in humans, to discover interactions between host and microbe, or to isolate as best as possible specific genetic and environmental factors for studying direct effects.
Discussion
Addiction and substance use research has seen renewed enthusiasm with a focus on the innate gut microbiota. Numerous studies have proposed a connection between the gut microbiota and the CNS via the gut–brain axis, providing a mechanism by which these microbes influence the host. Often drug use is associated with dysbiosis of the gut microbiome, and these changes may be critical to the establishment and maintenance of addiction by altering signals between the gut and the brain. Thus, a probiotic intervention has garnered much interest as a novel form of therapy for SUD. Recent evidence shows promising effects of probiotics in treating other mental disorders, including anxiety and depression, both of which are often co-morbid with SUD (Abildgaard et al., 2017; Hadizadeh et al., 2019; Kim and Kim, 2019; Liu et al., 2020). Fecal microbial transplant is also an area of intense research. Studies in mice have shown that transplantation of fecal microbiota of individuals diagnosed with AUD leads to altered social and anxiety behaviors and increased preference toward alcohol (Zhao et al., 2020). Reciprocally, transplantation of microbiota from healthy donors reduced anxiety and depressive behaviors in mice exposed to alcohol (Xu et al., 2018). More research is needed to validate the effects of FMT for other addictive substances and whether dysbiosis perpetuates the chronic nature of addiction. Future work will focus on untangling the mechanisms of gut microbiota modulation on the host addiction behaviors to identify target pathways and functions for potential probiotic therapeutics, such as those focused on SCFA production.
Gut microbiome dysbiosis is evident after chronic and acute substance use. However, more research is needed into whether the gut microbiome may also serve as a source of environmental risk for the development of SUD and addiction or be used as a predictor of future SUD. Furthermore, interactions between host genetics and gut microbiota deserve greater attention. Recent evidence has suggested that host genetics can have strong effects on the composition and function of the gut microbiome (Goodrich et al., 2014; Abildgaard et al., 2017; Kim and Kim, 2019; Korach-Rechtman et al., 2019; Liu et al., 2020). Using animal models from diverse genetic backgrounds will pave the way toward understanding complex interactions between genetic traits and the microbiota, especially given that genetically diverse mice respond differently to probiotic interventions (McVey Neufeld et al., 2018). Lastly, researchers must consider the balance between the reproducibility of using inbred mouse strains to test specific hypotheses versus the translatability of genetically diverse mice, as both methods will be critical to not only bolstering our understanding of the microbiome in SUD and addiction but also its applicability to a diverse human population.
Author Contributions
JR drafted the initial version of the manuscript. YZ, GW, and JB contributed to and edited the final manuscript. All authors approved the final manuscript.
Funding
JR, YZ, and JB were funded by R21 AA027858 and YZ, GW, and JB were funded by U01DA043809. JR was also funded by Ruth L. Kirschtein T32 NRSA award 5T32AA007290-38.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abildgaard, A., Elfving, B., Hokland, M., Wegener, G., and Lund, S. (2017). Probiotic treatment reduces depressive-like behaviour in rats independently of diet. Psychoneuroendocrinology 79, 40–48. doi: 10.1016/j.psyneuen.2017.02.014
Agrawal, A., and Lynskey, M. T. (2008). Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction 103, 1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x
Angoa-Pérez, M., and Kuhn, D. M. (2021). Evidence for Modulation of Substance Use Disorders by the Gut Microbiome: hidden in Plain Sight. Pharmacol. Rev. 73, 571–596. doi: 10.1124/pharmrev.120.000144
Avelar, A. J., Juliano, S. A., and Garris, P. A. (2013). AMPHETAMINE AUGMENTS VESICULAR DOPAMINE RELEASE IN THE DORSAL AND VENTRAL STRIATUM THROUGH DIFFERENT MECHANISMS. J. Neurochem. 125, 373–385. doi: 10.1111/jnc.12197
Barrett, E., Ross, R. P., O’Toole, P. W., Fitzgerald, G. F., and Stanton, C. (2012). γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 113, 411–417. doi: 10.1111/j.1365-2672.2012.05344.x
Bergeson, S. E., Nipper, M. A., Jensen, J., Helms, M. L., and Finn, D. A. (2016). Tigecycline Reduces Ethanol Intake in Dependent and Non-Dependent Male and Female C57BL/6J Mice. Alcohol. Clin. Exp. Res. 40, 2491–2498. doi: 10.1111/acer.13251
Bliss, T. V. P., Collingridge, G. L., Morris, R. G. M., Thomas, M. J., and Malenka, R. C. (2003). Synaptic plasticity in the mesolimbic dopamine system. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 358, 815–819. doi: 10.1098/rstb.2002.1236
Boileau, I., Assaad, J.-M., Pihl, R. O., Benkelfat, C., Leyton, M., Diksic, M., et al. (2003). Alcohol promotes dopamine release in the human nucleus accumbens. Synapse 49, 226–231. doi: 10.1002/syn.10226
Boonstra, E., de Kleijn, R., Colzato, L. S., Alkemade, A., Forstmann, B. U., and Nieuwenhuis, S. (2015). Neurotransmitters as food supplements: the effects of GABA on brain and behavior. Front. Psychol. 6:1520. doi: 10.3389/fpsyg.2015.01520
Braniste, V., Al-Asmakh, M., Kowal, C., Anuar, F., Abbaspour, A., Tóth, M., et al. (2014). The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6:263ra158. doi: 10.1126/scitranslmed.3009759
Bravo, J. A., Forsythe, P., Chew, M. V., Escaravage, E., Savignac, H. M., Dinan, T. G., et al. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. PNAS 108, 16050–16055. doi: 10.1073/pnas.1102999108
Cadet, J. L. (2016). Dysregulation of Acetylation Enzymes in Animal Models of Psychostimulant use Disorders: evolving Stories. Curr. Neuropharmacol. 14, 10–16. doi: 10.2174/1570159X13666150121230133
Caporaso, J. G., Lauber, C. L., Costello, E. K., Berg-Lyons, D., Gonzalez, A., Stombaugh, J., et al. (2011). Moving pictures of the human microbiome. Genome Biol. 12:R50. doi: 10.1186/gb-2011-12-5-r50
Capuco, A., Urits, I., Hasoon, J., Chun, R., Gerald, B., Wang, J. K., et al. (2020). Gut Microbiome Dysbiosis and Depression: a Comprehensive Review. Curr. Pain Headache Rep. 24:36. doi: 10.1007/s11916-020-00871-x
Cheng, Z., Zhou, H., Sherva, R., Farrer, L., Kranzler, H. R., and Gelernter, J. (2018). Genome-wide association study identifies a regulatory variant of RGMA associated with opioid dependence in European Americans. Biol. Psychiatry 84, 762–770. doi: 10.1016/j.biopsych.2017.12.016
Chi, L., Mahbub, R., Gao, B., Bian, X., Tu, P., Ru, H., et al. (2017). Nicotine Alters the Gut Microbiome and Metabolites of Gut–Brain Interactions in a Sex-Specific Manner. Chem. Res. Toxicol. 30, 2110–2119. doi: 10.1021/acs.chemrestox.7b00162
Churchill, G. A., Gatti, D. M., Munger, S. C., and Svenson, K. L. (2012). The diversity outbred mouse population. Mamm. Genome 23, 713–718. doi: 10.1007/s00335-012-9414-2
Crabbe, J. C., Ozburn, A. R., Metten, P., Barkley-Levenson, A., Schlumbohm, J. P., Spence, S. E., et al. (2017). High Drinking in the Dark (HDID) mice are sensitive to the effects of some clinically relevant drugs to reduce binge-like drinking. Pharmacol. Biochem. Behav. 160, 55–62. doi: 10.1016/j.pbb.2017.08.002
Dalile, B., Van Oudenhove, L., Vervliet, B., and Verbeke, K. (2019). The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478. doi: 10.1038/s41575-019-0157-3
Demontis, D., Rajagopal, V. M., Thorgeirsson, T. E., Als, T. D., Grove, J., Leppälä, K., et al. (2019). Genome-wide association study implicates CHRNA2 in cannabis use disorder. Nat. Neurosci. 22, 1066–1074. doi: 10.1038/s41593-019-0416-1
Draycott, B., Loureiro, M., Ahmad, T., Tan, H., Zunder, J., and Laviolette, S. R. (2014). Cannabinoid Transmission in the Prefrontal Cortex Bi-Phasically Controls Emotional Memory Formation via Functional Interactions with the Ventral Tegmental Area. J. Neurosci. 34, 13096–13109. doi: 10.1523/JNEUROSCI.1297-14.2014
D’Souza, M. S., and Markou, A. (2011). Neuronal Mechanisms Underlying Development of Nicotine Dependence: implications for Novel Smoking-Cessation Treatments. Addict. Sci. Clin. Pract. 6, 4–16.
Edenberg, H. J., and McClintick, J. N. (2018). Alcohol dehydrogenases, aldehyde dehydrogenases and alcohol use disorders: a critical review. Alcohol. Clin. Exp. Res. 42, 2281–2297. doi: 10.1111/acer.13904
Ezquer, F., Quintanilla, M. E., Moya-Flores, F., Morales, P., Munita, J. M., Olivares, B., et al. (2021). Innate gut microbiota predisposes to high alcohol consumption. Addict. Biol. 26:e13018. doi: 10.1111/adb.13018
Fowler, C. D., and Kenny, P. J. (2012). Utility of genetically modified mice for understanding the neurobiology of substance use disorders. Hum. Genet. 131, 941–957. doi: 10.1007/s00439-011-1129-z
French, J. E., Gatti, D. M., Morgan, D. L., Kissling, G. E., Shockley, K. R., Knudsen, G. A., et al. (2015). Diversity Outbred Mice Identify Population-Based Exposure Thresholds and Genetic Factors that Influence Benzene-Induced Genotoxicity. Environ. Health Perspect. 123, 237–245. doi: 10.1289/ehp.1408202
Gelernter, J., Kranzler, H. R., Sherva, R., Koesterer, R., Almasy, L., Zhao, H., et al. (2014). Genome-Wide Association Study of Opioid Dependence: multiple Associations Mapped to Calcium and Potassium Pathways. Biol. Psychiatry 76, 66–74. doi: 10.1016/j.biopsych.2013.08.034
Goldman, D., Oroszi, G., and Ducci, F. (2005). The genetics of addictions: uncovering the genes. Nat. Rev. Genet. 6, 521–532. doi: 10.1038/nrg1635
Goodrich, J. K., Waters, J. L., Poole, A. C., Sutter, J. L., Koren, O., Blekhman, R., et al. (2014). Human genetics shape the gut microbiome. Cell 159, 789–799. doi: 10.1016/j.cell.2014.09.053
Grice, E. A., and Segre, J. A. (2012). The Human Microbiome: our Second Genome. Annu. Rev. Genom. Hum. Genet. 13, 151–170. doi: 10.1146/annurev-genom-090711-163814
Hadizadeh, M., Hamidi, G. A., and Salami, M. (2019). Probiotic supplementation improves the cognitive function and the anxiety-like behaviors in the stressed rats. Iran. J. Basic Med. Sci. 22, 506–514. doi: 10.22038/ijbms.2019.33956.8078
Hancock, D. B., Markunas, C. A., Bierut, L. J., and Johnson, E. O. (2018). Human Genetics of Addiction: new Insights and Future Directions. Curr. Psychiatry Rep. 20:8. doi: 10.1007/s11920-018-0873-3
Hitzemann, R., Phillips, T. J., Lockwood, D. R., Darakjian, P., and Searles, R. P. (2020). Phenotypic and gene expression features associated with variation in chronic ethanol consumption in heterogeneous stock collaborative cross mice. Genomics 112, 4516–4524. doi: 10.1016/j.ygeno.2020.08.004
Johnson, E. C., Demontis, D., Thorgeirsson, T. E., Walters, R. K., Polimanti, R., Hatoum, A. S., et al. (2020). A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045. doi: 10.1016/S2215-0366(20)30339-4
Justice, M. J., and Dhillon, P. (2016). Using the mouse to model human disease: increasing validity and reproducibility. Dis. Model Mech. 9, 101–103. doi: 10.1242/dmm.024547
Kahlig, K. M., and Galli, A. (2003). Regulation of dopamine transporter function and plasma membrane expression by dopamine, amphetamine, and cocaine. Eur. J. Pharmacol. 479, 153–158. doi: 10.1016/j.ejphar.2003.08.065
Kauer, J. A. (2004). Learning Mechanisms in Addiction: synaptic Plasticity in the Ventral Tegmental Area as a Result of Exposure to Drugs of Abuse. Annu. Rev. Physiol. 66, 447–475. doi: 10.1146/annurev.physiol.66.032102.112534
Kim, S.-K., and Kim, D. H. (2019). Lactobacillus mucosae and Bifidobacterium longum Synergistically Alleviate Immobilization Stress-Induced Anxiety/Depression in Mice by Suppressing Gut Dysbiosis. 29, 1369–1374. doi: 10.4014/jmb.1907.07044
Kiraly, D. D., Walker, D. M., Calipari, E. S., Labonte, B., Issler, O., Pena, C. J., et al. (2016). Alterations of the Host Microbiome Affect Behavioral Responses to Cocaine. Sci. Rep. 6:35455. doi: 10.1038/srep35455
Koob, G. F., Kandel, D., and Volkow, N. D. (2008). “Pathophysiology of Addiction” in Psychiatry. eds T. Allan, K. Jerald, J. A. Lieberman, M. B. First, and M. Mario (United States: John Wiley & Sons, Ltd). 354–378. doi: 10.1002/9780470515167.ch22
Koob, G. F., and Le Moal, M. (2001). Drug Addiction, Dysregulation of Reward, and Allostasis. Neuropsychopharmacol 24, 97–129. doi: 10.1016/S0893-133X(00)00195-0
Korach-Rechtman, H., Freilich, S., Gerassy-Vainberg, S., Buhnik-Rosenblau, K., Danin-Poleg, Y., Bar, H., et al. (2019). Murine Genetic Background Has a Stronger Impact on the Composition of the Gut Microbiota than Maternal Inoculation or Exposure to Unlike Exogenous Microbiota. Appl. Environ. Microbiol. 85, e00826–19. doi: 10.1128/AEM.00826-19
Kranzler, H. R., Zhou, H., Kember, R. L., Vickers Smith, R., Justice, A. C., Damrauer, S., et al. (2019). Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 10:1499. doi: 10.1038/s41467-019-09480-8
Kreek, M. J., Nielsen, D. A., and LaForge, K. S. (2004). Genes associated with addiction. Neuromol. Med. 5, 85–108. doi: 10.1385/NMM:5:1:085
Leclercq, S., Matamoros, S., Cani, P. D., Neyrinck, A. M., Jamar, F., Stärkel, P., et al. (2014). Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. U. S. A. 111, E4485–E4493. doi: 10.1073/pnas.1415174111
Lee, K., Vuong, H. E., Nusbaum, D. J., Hsiao, E. Y., Evans, C. J., and Taylor, A. M. W. (2018). The gut microbiota mediates reward and sensory responses associated with regimen-selective morphine dependence. Neuropsychopharmacology 43, 2606–2614. doi: 10.1038/s41386-018-0211-9
Lipari, R. N., and Van Horn, S. L. (2017). “Trends in substance use disorders among adults aged 18 or older” The CBHSQ Report. Rockville (MD): substance Abuse and Mental Health Services Administration. (United States: Substance Abuse and Mental Health Services Administration).
Liu, Q. F., Kim, H.-M., Lim, S., Chung, M.-J., Lim, C.-Y., Koo, B.-S., et al. (2020). Effect of probiotic administration on gut microbiota and depressive behaviors in mice. Daru 28, 181–189. doi: 10.1007/s40199-020-00329-w
McVey Neufeld, K.-A., Kay, S., and Bienenstock, J. (2018). Mouse Strain Affects Behavioral and Neuroendocrine Stress Responses Following Administration of Probiotic Lactobacillus rhamnosus JB-1 or Traditional Antidepressant Fluoxetine. Front. Neurosci. 12:294. doi: 10.3389/fnins.2018.00294
Meckel, K. R., and Kiraly, D. D. A. (2019). potential role for the gut microbiome in substance use disorders. Psychopharmacology 236, 1513–1530. doi: 10.1007/s00213-019-05232-0
Mews, P., Egervari, G., Nativio, R., Sidoli, S., Donahue, G., Lombroso, S. I., et al. (2019). Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721. doi: 10.1038/s41586-019-1700-7
Muccioli, G. G., Naslain, D., Bäckhed, F., Reigstad, C. S., Lambert, D. M., Delzenne, N. M., et al. (2010). The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 6:392. doi: 10.1038/msb.2010.46
Nestler, E. J. (2005). Is there a common molecular pathway for addiction? Nat. Neurosci. 8, 1445–1449. doi: 10.1038/nn1578
Noble, E. P., Blum, K., Khalsa, M. E., Ritchie, T., Montgomery, A., Wood, R. C., et al. (1993). Allelic association of the D2 dopamine receptor gene with cocaine dependence. Drug Alcohol Depend 33, 271–285. doi: 10.1016/0376-8716(93)90113-5
Northcutt, A. L., Hutchinson, M. R., Wang, X., Baratta, M. V., Hiranita, T., Cochran, T. A., et al. (2015). DAT isn’t all that: cocaine reward and reinforcement requires Toll Like Receptor 4 signaling. Mol. Psychiatry 20, 1525–1537. doi: 10.1038/mp.2014.177
Patel, R. R., Wolfe, S. A., Bajo, M., Abeynaike, S., Pahng, A., Borgonetti, V., et al. (2021). IL-10 normalizes aberrant amygdala GABA transmission and reverses anxiety-like behavior and dependence-induced escalation of alcohol intake. Prog. Neurobiol. 199:101952. doi: 10.1016/j.pneurobio.2020.101952
Poltorak, A., Apalko, S., and Sherbak, S. (2018). Wild-derived mice: from genetic diversity to variation in immune responses. Mamm Genome 29, 577–584. doi: 10.1007/s00335-018-9766-3
Prom-Wormley, E. C., Ebejer, J., Dick, D. M., and Bowers, M. S. (2017). The genetic epidemiology of substance use disorder: a review. Drug Alcohol Depend. 180, 241–259. doi: 10.1016/j.drugalcdep.2017.06.040
Quigley, E. M. M., and Gajula, P. (2020). Recent advances in modulating the microbiome. F1000Res. 9, F1000 Faculty Rev-46. doi: 10.12688/f1000research.20204.1
Renthal, W., and Nestler, E. J. (2009). Histone Acetylation in Drug Addiction. Semin. Cell. Dev. Biol. 20, 387–394. doi: 10.1016/j.semcdb.2009.01.005
Roberto, M., and Varodayan, F. (2017). Synaptic Targets: chronic Alcohol Actions. Neuropharmacology 122, 85–99. doi: 10.1016/j.neuropharm.2017.01.013
Rosshart, S. P., Herz, J., Vassallo, B. G., Hunter, A., Wall, M. K., Badger, J. H., et al. (2019). Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365:eaaw4361. doi: 10.1126/science.aaw4361
Shreiner, A. B., Kao, J. Y., and Young, V. B. (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75. doi: 10.1097/MOG.0000000000000139
Simon-O’Brien, E., Alaux-Cantin, S., Warnault, V., Buttolo, R., Naassila, M., and Vilpoux, C. (2015). The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict. Biol. 20, 676–689. doi: 10.1111/adb.12161
Srivastava, A., Morgan, A. P., Najarian, M. L., Sarsani, V. K., Sigmon, J. S., Shorter, J. R., et al. (2017). Genomes of the Mouse Collaborative Cross. Genetics 206, 537–556. doi: 10.1534/genetics.116.198838
Stone, A. L., Becker, L. G., Huber, A. M., and Catalano, R. F. (2012). Review of risk and protective factors of substance use and problem use in emerging adulthood. Addict. Behav. 37, 747–775. doi: 10.1016/j.addbeh.2012.02.014
Strandwitz, P. (2018). Neurotransmitter modulation by the gut microbiota. Brain Res. 1693, 128–133. doi: 10.1016/j.brainres.2018.03.015
Strandwitz, P., Kim, K. H., Terekhova, D., Liu, J. K., Sharma, A., Levering, J., et al. (2019). GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 4, 396–403. doi: 10.1038/s41564-018-0307-3
Svenson, K. L., Gatti, D. M., Valdar, W., Welsh, C. E., Cheng, R., Chesler, E. J., et al. (2012). High-Resolution Genetic Mapping Using the Mouse Diversity Outbred Population. Genetics 190, 437–447. doi: 10.1534/genetics.111.132597
Swiatczak, B., and Cohen, I. R. (2015). Gut feelings of safety: tolerance to the microbiota mediated by innate immune receptors. Microbiol. Immunol. 59, 573–585. doi: 10.1111/1348-0421.12318
Thaiss, C. A., Zeevi, D., Levy, M., Zilberman-Schapira, G., Suez, J., Tengeler, A. C., et al. (2014). Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 159, 514–529. doi: 10.1016/j.cell.2014.09.048
Thiele, T. E., Crabbe, J. C., and Boehm, S. L. (2014). Drinking in the Dark” (DID): a Simple Mouse Model of Binge-Like Alcohol Intake. Current Protocols in Neuroscience 68, 9.49.1–12. doi: 10.1002/0471142301.ns0949s68
Threadgill, D. W., Miller, D. R., Churchill, G. A., and de Villena, F. P.-M. (2011). The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J. 52, 24–31. doi: 10.1093/ilar.52.1.24
Valentino, R. J., and Volkow, N. D. (2018). Untangling the complexity of opioid receptor function. Neuropsychopharmacology 43, 2514–2520. doi: 10.1038/s41386-018-0225-3
Volkow, N. D., Wang, G. J., Fowler, J. S., Logan, J., Gatley, S. J., Wong, C., et al. (1999). Reinforcing effects of psychostimulants in humans are associated with increases in brain dopamine and occupancy of D(2) receptors. J. Pharmacol. Exp. Ther. 291, 409–415.
Walker, D. M., Cates, H. M., Loh, Y.-H. E., Purushothaman, I., Ramakrishnan, A., Cahill, K. M., et al. (2018). Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biol. Psychiatry 84, 867–880. doi: 10.1016/j.biopsych.2018.04.009
Witjes, V. M., Boleij, A., and Halffman, W. (2020). Reducing versus Embracing Variation as Strategies for Reproducibility: the Microbiome of Laboratory Mice. Animal 10:2415. doi: 10.3390/ani10122415
Xiao, H.-W., Ge, C., Feng, G.-X., Li, Y., Luo, D., Dong, J.-L., et al. (2018). Gut microbiota modulates alcohol withdrawal-induced anxiety in mice. Toxicol. Lett. 287, 23–30. doi: 10.1016/j.toxlet.2018.01.021
Xu, F., Gainetdinov, R. R., Wetsel, W. C., Jones, S. R., Bohn, L. M., Miller, G. W., et al. (2000). Mice lacking the norepinephrine transporter are supersensitive to psychostimulants. Nat. Neurosci. 3, 465–471. doi: 10.1038/74839
Xu, Z., Liu, Z., Dong, X., Hu, T., Wang, L., Li, J., et al. (2018). Fecal Microbiota Transplantation from Healthy Donors Reduced Alcohol-induced Anxiety and Depression in an Animal Model of Chronic Alcohol Exposure. Chin. J. Physiol. 61, 360–371. doi: 10.4077/CJP.2018.BAH633
Yano, J. M., Yu, K., Donaldson, G. P., Shastri, G. G., Ann, P., Ma, L., et al. (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276. doi: 10.1016/j.cell.2015.02.047
Yunes, R. A., Poluektova, E. U., Vasileva, E. V., Odorskaya, M. V., Marsova, M. V., Kovalev, G. I., et al. (2020). A Multi-strain Potential Probiotic Formulation of GABA-Producing Lactobacillus plantarum 90sk and Bifidobacterium adolescentis 150 with Antidepressant Effects. Probiotics Antimicrob. Proteins 12, 973–979. doi: 10.1007/s12602-019-09601-1
Zhao, W., Hu, Y., Li, C., Li, N., Zhu, S., Tan, X., et al. (2020). Transplantation of fecal microbiota from patients with alcoholism induces anxiety/depression behaviors and decreases brain mGluR1/PKC ε levels in mouse. BioFactors 46, 38–54. doi: 10.1002/biof.1567
Keywords: substance use disorder, addiction, microbiome, animal models, gut–brain axis
Citation: Russell JT, Zhou Y, Weinstock GM and Bubier JA (2021) The Gut Microbiome and Substance Use Disorder. Front. Neurosci. 15:725500. doi: 10.3389/fnins.2021.725500
Received: 15 June 2021; Accepted: 12 August 2021;
Published: 31 August 2021.
Edited by:
Gary T. Hardiman, Queen’s University Belfast, United KingdomReviewed by:
Drew D. Kiraly, Icahn School of Medicine at Mount Sinai, United StatesDonald Michael Kuhn, Wayne State University, United States
Copyright © 2021 Russell, Zhou, Weinstock and Bubier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason A. Bubier, SmFzb24uQnViaWVyQGpheC5vcmc=
 Jordan T. Russell
Jordan T. Russell Yanjiao Zhou
Yanjiao Zhou George M. Weinstock
George M. Weinstock Jason A. Bubier
Jason A. Bubier