- 1School of Computer Science, South China Normal University, Guangzhou, China
- 2Grado Department of Industrial and Systems Engineering, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
Sleep apnea (SA) is a common chronic sleep breathing disorder, which would cause stroke, cognitive decline, cardiovascular disease, or even death. The SA symptoms often manifest as frequent breathing interruptions during sleep and most individuals with sleeping disorders are not aware of the SA events. Using a portable device with single-lead ECG signal is an effective way to help an individual to monitor their sleep conditions at home. However, the SA detection performance of ECG-based methods is still difficult to meet the clinical practice requirement. In this study, we propose an end-to-end spatio-temporal learning-based SA detection method, which consists of multiple spatio-temporal blocks. Each block has the identical architecture with a convolutional neural network (CNN) layer, a max-pooling layer, and a bi-gated recurrent unit (BiGRU) layer. This architecture with repeated spatio-temporal blocks can well capture the morphological spatial feature information as well as the temporal feature information from ECG signals. The proposed SA detection model was evaluated on the publicly available datasets of PhysioNet Apnea-ECG dataset (Apnea-ECG) and University College Dublin Sleep Apnea Database (UCDDB). Extensive experimental results show that our proposed SA model on both Apnea-ECG and UCDDB datasets achieves state-of-the-art results, which are obviously superior to existing ECG-based SA detection methods. It means that our proposed method has the potential to be deployed into a healthcare system to provide a sleep monitoring service, which can screen out SA population with high risk and help to take timely interventions to prevent serious consequences.
1. Introduction
Sleep apnea (SA) is a sleep disorder in which breathing is interrupted several times during sleeping. Its typical symptoms include headache, insomnia and others, and it can be potentially serious (Russell et al., 2014). Without prompt and appropriate treatment measures, patients with SA would suffer from serious complications such as stroke (King and Cuellar, 2016), cognitive decline (Vanek et al., 2020), cardiovascular disease (Lin et al., 2020), and even death. SA is considered by some researchers to be a recognized independent risk factor for stroke, such that individuals with SA have an approximately twofold greater risk of stroke compared with those without SA (Lyons and Ryan, 2015). This indicates that SA is a great threat to the global physical and mental health, with ~936 million adults (male and female) aged 30–69 years worldwide suffering from mild to severe obstructive sleep apnea, and 425 million adults aged 30–69 years suffering from moderate to severe obstructive sleep apnea (Benjafield et al., 2019). Due to the prevalence of SA, it is very vital to screen out individuals with SA and take timely interventions.
In clinical practice, polysomnography (PSG) is the gold standard test to diagnose SA. However, conducting PSG is expensive and often unavailable due to the shortage of physical therapists and sleep monitoring units (Graco et al., 2018). PSG with many biomedical sensors, including electroencephalogram (EEG), electro-oculogram (EOG), electromyogram (EMG), electrocardiogram (ECG) and pulse oximetry as well as airflow and respiratory effort, is performed as an SA test overnight in a sleep laboratory or specific unit in a hospital (Ali et al., 2019). This can be quite cumbersome and uncomfortable, so the collected signals cannot reflect the individuals' sleep conditions. In addition, a physical therapist must be available when the PSG is conducted in the hospital, which significantly restricts the screening of people with SA.
Home Sleep Test (HST) is an alternative to PSG for SA diagnosis, which is usually conducted overnight outside of the hospital or sleep laboratory (Rosen et al., 2017). Portable devices, which are simple, of low-cost and easy to operate, have been developed to enable the patients to monitor their sleep conditions in an unattended home environment. Gaiduk et al. (2020) have developed a system based on pressure sensors that can work independently and via wireless connection, which is as accurate as the current technology. However, this pressure sensor-based approach is highly sensitive to pressure, and the pressure signal can be easily contaminated with noise from the external environment. ECG is considered to be one of the most relevant physiological signals for the SA detection because patient's heart rate increases when SA occurs (Somers et al., 2008; Wang T. et al., 2019). ECG contains valuable information about the cardiorespiratory system and is therefore of great importance for SA detection (Bahrami and Forouzanfar, 2021). Over the past twenty years, various approaches have been proposed for the automated SA detection using HRV and EDR signals which can be derived from ECG (Gutiérrez-Tobal et al., 2015; Faust et al., 2016; Smruthy and Suchetha, 2017; Viswabhargav et al., 2019). In addition, ECG can be easily recorded. Therefore, ECG-based portable devices represent a better option. A wearable ECG acquisition system has been developed (Weder et al., 2015), which is designed as a chest strap that can continuously monitor ECG signals for multiple nights. As a more comfortable option, Ankhili et al. (2018) developed a reliable, washable ECG monitoring undergarment that can record and send the ECG signal wirelessly to a smartphone to analyze the ECG signal in real-time.
Using ECG signals can greatly reduce the complexity of diagnostic SA tests and allow for better monitoring of physiological changes in the patient (Bsoul et al., 2011). Several algorithms have been proposed for ECG-based SA detection. These algorithms generally include a first step of feature extraction from the original ECG signals, and then these features are used as the input and fed into a classification model (Baty et al., 2020). Sharma and Sharma (2016) extracted features from QRS complex waves using Hermite decomposition. Then, these features were combined with time series features, and least squares-support vector machine (LS-SVM) was used as a classifier for SA detection. Song et al. (2015) introduced a classifier that blends an SVM with a hidden Markov model (HMM) to take advantage of the time-dependent nature of SA segments. In recent years, deep neural networks (DNN) with end-to-end training are also applied to build SA detection models. Li et al. (2018) used HMM, ANN, and ECG signals for the identification of SA segments. Feng et al. (2020) used an unsupervised neural network to learn features, and they improved the performance of the classifier by taking into account the time-dependence and imbalance problems.
Although aforementioned models achieved promising results, there still exists a gap between their SA detection performance and the requirement of actual applications. Note that spatial patterns and temporal correlations are both important for SA detection. For example, the R-peak has salient spatial features on the ECG signals; while the RR intervals can be derived from the temporal dependencies. In reality, RR intervals are frequently utilized in SA detection (Bahrami and Forouzanfar, 2021). However, the spatio-temporal correlations are seldom utilized in the existing SA detection models (Sharan et al., 2020; Chen et al., 2021; Yang et al., 2022). Bahrami and Forouzanfar (2021) has used a hybrid CNN and LSTM network to extract spatio-temporal feature from ECG signals. However, they only use a simple combination of CNN and LSTM networks. To improve the performance of SA detection, a spatio-temporal learning based DNN model is proposed in this paper. In order to take advantage of more spatio-temporal dependencies, multiple adjacent segments are concatenated and used as the input of the proposed model. A spatio-temporal learning block is designed which is composed of a one-dimensional convolutional neural network (CNN), a max-pooling layer, and a bidirectional gated recurrent unit (BiGRU). Multiple spatio-temporal blocks are stacked in the proposed model to extract long-range spatial and temporal correlations. As a result, this model can fully utilize the multiple concatenated ECG segments. Moreover, an attention mechanism is employed to further utilize the global correlations by using the high-level forward and backward spatio-temporal features. These characteristics have made our model different with the other spatio-temporal model (Bahrami and Forouzanfar, 2021). The performance of our model has outperformed the one of Bahrami and Forouzanfar (2021). Experimental results on two public domain datasets of PhysioNet Apnea-ECG dataset (Apnea-ECG) and University College Dublin Sleep Apnea Database (UCDDB) showed that CNN-BiGRU achieved the competitive performance to the previous state-of-the-art methods. The main contributions of this study can be listed as follows:
• To fully extract spatio-temporal information from ECG signals, we proposed a spatio-temporal learning-based model called CNN-BiGRU with multiple spatio-temporal blocks, each block of which consists of a one-dimensional CNN layer, a max-pooling layer, and a BiGRU layer.
• We employed an attention mechanism to further exploit the high-level forward and backward spatio-temporal features from the last spatio-temporal block, which was able to extract the global correlations from multiple ECG signal segments.
• Experiment results on two public domain datasets of Apnea-ECG and UCDDB showed that the proposed CNN-BiGRU achieved a state-of-the-art performance, which outperforms the previous state-of-the-art methods. It means that the proposed CNN-BiGRU can be potentially deployed into a medical system to provide the SA monitoring service.
The remainder of this paper is organized as follows. Section 2 details the composition of the CNN-BiGRU model. In Sections 3 and 4 the results are presented and discussed. The conclusion is presented in Section 5.
2. Methods
2.1. Overview
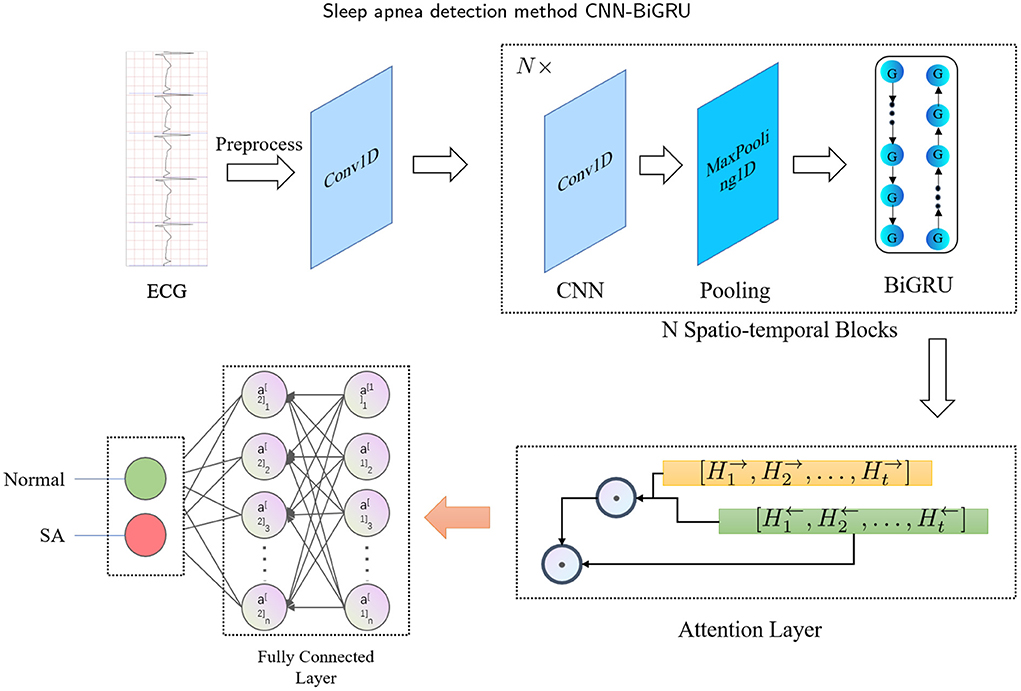
The main idea of this study is to develop a fully automated (or end-to-end) spatio-temporal learning-based SA detection method, which is illustrated in Figure 1. First, in the pre-processing phase, the RR intervals and R-peak amplitudes are extracted from combining adjacent and labeled segments. Then, the RR intervals and R-peak amplitudes are fed into the proposed CNN-BiGRU model, which recycles through multiple spatio-temporal blocks to capture high-level spatio-temporal features. The whole method can be mathematically defined as follows: Given the input X ∈ ℝD, a mapping function f : X → {0, 1} is learned, where D is the dimensionality of the input, and 0 and 1 denote normal and SA, respectively. Specifically, the spatio-temporal block consists of CNN, max-pooling and BiGRU layers. Furthermore, the attention mechanism is used to extract the most effective part of the spatio-temporal features and improve the accuracy. Finally, the fully connected layer is used to identify whether the labeled segment belongs to SA.
2.2. Preprocessing
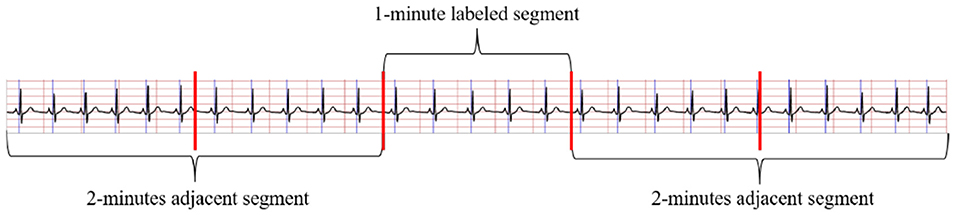
The method in Wang T. et al. (2019) to obtain the RR intervals and R-peak amplitudes were applied in this study to pre-process ECG signals. Considering adjacent segment information is useful for SA detection in each segment. As shown in Figure 2, both the labeled segment and its surrounding ECG signal ±2 segments (five segments total of 1 min) were extracted and processed. Firstly, the Hamilton algorithm (Hamilton, 2002) was used to find R-peaks and adjust the detection peak to match the local signal maximum. Then, RR intervals were calculated and the amplitudes were extracted using the locations of the R-peaks, while anomalous signals were removed. For physiologically unexplained points, median filters were chosen to solve the extracted RR intervals (Chen et al., 2015). The final problem of unequal time intervals between the RR intervals and amplitudes was tackled by cubic interpolation, which yielded 900 RR interval points and 900 amplitudes over 5 min segments.
2.3. The proposed CNN-BiGRU for SA recognition
The proposed CNN-BiGRU consists of spatio-temporal blocks of CNNs and BiGRUs, an attention layer and fully connections layers. These layers are introduced in detail as follows.
2.3.1. Convolutional neural network
Convolutional neural networks are among the most common and efficient techniques that are widely used in various signal and image processing applications (Fan et al., 2018, 2019; Wang et al., 2018). A lightweight CNN model can be trained on a mixed-scale graph in order to extract deep features for the detection of obstructive SA (Mashrur et al., 2021). In this study, we used a one-dimensional CNN to extract spatial dependencies from ECG signals, which is mathematically defined as follows:
where x, w, L are the input, filter, and filter size, respectively.
The next layer of the CNN is generally the pooling layer. The max-pooling layer can be used to reduce mean value shift errors caused by bad initialization of weights (Wang L. et al., 2019). In this paper, the max-pooling layer was used to decrease the computational effort and to mitigate the overfitting problem by selecting the maximum value of each feature.
2.3.2. Gated recurrent unit
Gated recurrent units (GRU) (Cho et al., 2014) represent a more advanced alternative to the simple recurrent neural network (RNN) and are more capable of learning long-term dependencies than vanilla RNN (Zhang et al., 2022). While both GRU and Long Short-Term Memory (LSTM) units have gating units that regulate the flow of information within the unit, GRUs do not have a separate memory unit, only update and reset gates. The j-th hidden unit of each mentioned gate is defined as follows:
Reset gates:
where σ and [·]j are the logistic sigmoid function and the j-th element of a vector, respectively; t is the time step, x denotes the input, and h〈t−1〉 represents the previous hidden state.
Update gates:
Output:
The weight matrices Wr, Wz, Wx, Ur, Uz, and Ux are learned during training. The candidate hidden state is computed as follows:
The GRU can be heavily dependent on the dataset and the corresponding task, and the Apnea-ECG dataset works better with GRU than with LSTM, since it has fewer parameters and faster training (Chung et al., 2014). In this work, a bidirectional GRU was used to capture richer temporal information. By recursively computing the hidden states Ht in the forward and backward directions, the forward sequence F and the backward sequence B were obtained, respectively. This can be mathematically defined as follows:
where n denotes the number of GRUs and s represents the total number of the time step.
2.3.3. Attention mechanism
Dot-product attention (Luong et al., 2015) was used to extract the global correlation information from the input multiple ECG segments. Specifically, dot-production attention is applied on the forward sequence F and the backward sequence B of the BiGRU unit within the last spatio-temporal block. The attention score is calculated as follows:
Using the attention mechanism allows the model to pay more attention to specific high-level spatial-temporal dependency information, improving the accuracy of the model. Note that dot-product attention is fast and spatially efficient because it enables a highly optimized code for matrix multiplication (Vaswani et al., 2017).
2.3.4. Proposed CNN-BiGRU model
To better extract the spatio-temporal features of ECG signals, we have specially designed a SA detection model, named CNN-BiGRU. The proposed CNN-BiGRU model is mainly composed of a CNN layer, multiple stacked spatio-temporal learning blocks, an attention layer, and fully connected layers. First, a convolutional layer was used to extract the base features before using spatio-temporal blocks. A spatio-temporal learning block consists of a one-dimensional CNN, a max-pooling layer, and a BiGRU unit. The use of multiple spatio-temporal blocks enables the CNN-BiGRU model to extract high-level spatio-temporal features from the ECG signal. Specifically, our model is able to extract local spatial features of the R-peaks, as well as global temporal features of the heartbeat intervals. Then, the attention score of the fused high-level forward and backward features from the spatio-temporal blocks was calculated. This attention mechanism is able to further utilize the global spatio-temporal correlations from the multiple ECG segments. Finally, three dense layers were used for classification. Additionally, some of the layers were immediately followed by a dropout layer to mitigate the effects of overfitting. The mathematical expression of the whole model computation process is as follows: For the CNN input X, the output C is:
where g denotes the ReLU activation function g(x) = max(0, x) and W denotes the convolution kernel.
As previously mentioned, this study uses BiGRU with the matrices Wr, Ur, Wz, Uz, Wx, and Ux as the parameters to be learned. After reducing the size of the feature map through the max-pooling layer, the output C of the CNN was fed into the BiGRU. The output of the spatio-temporal block can be mathematically represented as follows:
where F and B are the stacked hidden states in the forward and backward directions, respectively. If the next layer of the BiGRU was a CNN, then F and B were concatenated along the channel dimension, otherwise the attention score a was calculated as follows:
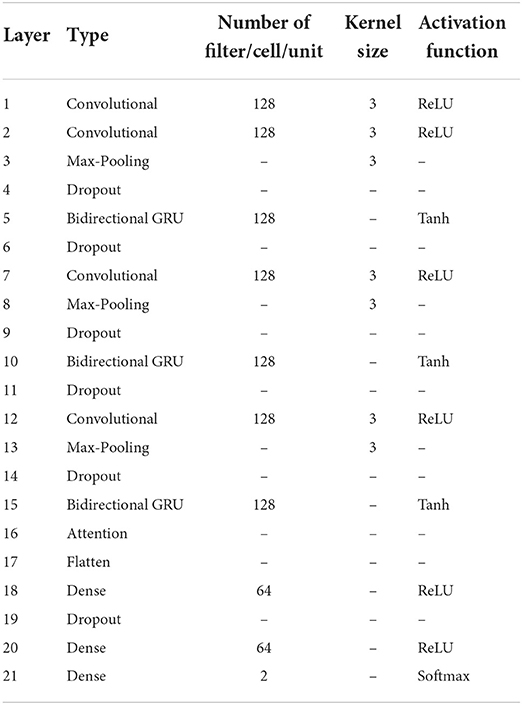
Finally, the attention score was entered into the fully connected layer for classification, and the labeled segments were classified to be SA or normal. Table 1 has listed the architecture details of the proposed CNN-BiGRU model. Specifically, the architecture contains three spatio-temporal blocks (see the layers of 2–5, 7–10, and 12–15, respectively in Table 1). And the dropout ratios in Table 1 have all been set to 0.2.
2.4. Experimental settings
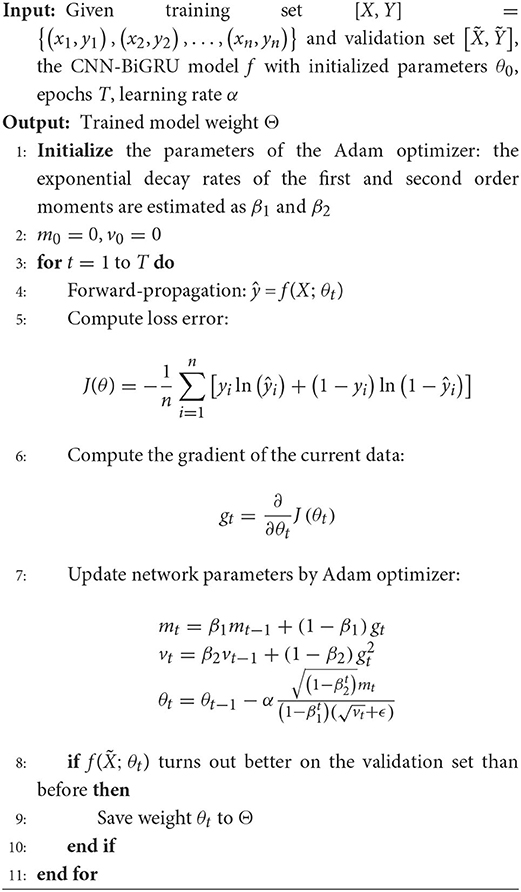
In order to enable an enhanced performance of the CNN-BiGRU model, the number of spatio-temporal blocks was tuned from 1 to 5. Adam optimizer (Kingma and Ba, 2017) and binary cross-entropy loss function were applied for parameter optimization. The learning rate and batch size were set to 0.001 and 128, respectively. The proposed model was trained for 40 epochs. In each training epoch, the model parameters were evaluated using the validation set and the best model parameters were saved to perform classification on the test set. Detailed training methods are described in Algorithm 1. Our model was implemented using the Tensorflow framework with a Tesla P100-PCIE-16GB graphics card.
Various performance metrics, such as precision, specificity, F1 score, sensitivity, and accuracy, were used to assess the performance of the proposed model. These metrics are defined as follows:
where FP, TP, FN, and TN stand for “false positive,” “true positive,” “false negative,” and “true negative,” respectively. The SA class is the positive class in this study, while the normal class is the negative class.
This model was also evaluated using the receiver operating characteristic (ROC) and the related area under the curve (AUC).
3. Experimental results
3.1. Datasets
3.1.1. PhysioNet Apnea-ECG dataset
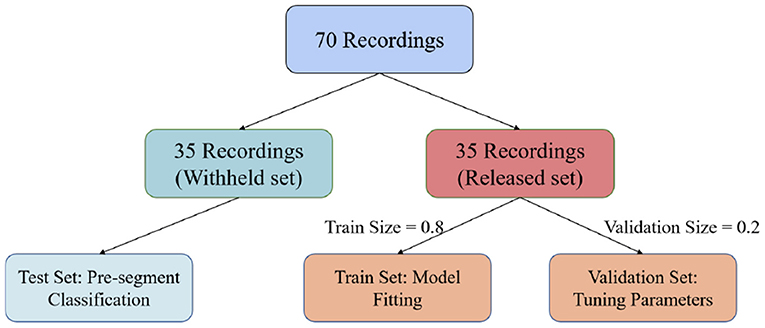
In this paper, we used the PhysioNet Apnea-ECG dataset provided by Philipps University (Penzel et al., 2000) for model evaluation. The Apnea-ECG dataset has 70 recordings, including 35 records in the released dataset (a01–a20, b01–b05, and c01–c10) and 35 records in the withheld dataset (x01–x35). And the release set is used to train the model with the withheld set used to test the model. Regarding the released set, 20% of the 35 released data were used to validate the model and tune its hyperparameters (Figure 3). ECG recordings for this dataset were obtained from subjects with an AHI (apnea hypoventilation index) between 0 and 83. And these subjects ranged in age from 27 to 63 years and their body mass indices varied between 19.2 and 45.33kg/m2. The ECG signal was sampled at 100 Hz over a range of 401 to 587 min. The experts labeled each 1 min recording segments as SA or normal. According to our pre-proccessing method, the release and withheld set contained 17,045 and 17,302 segments, respectively. The experimental results show that CNN-BiGRU achieves an outstanding performance in SA detection.
3.1.2. University college Dublin sleep Apnea database (UCDDB)
UCDDB was used as a second dataset to validate the performance of CNN-BiGRU. This database contains 25 full overnight polysomnograms from adult subjects suspected sleep-disordered breathing. ECG recordings of this dataset have been collected by a modified lead V2. We used ECG signals sampled at 128 Hz and with the durations ranging from 355 to 462 min. Following previous studies (Mostafa et al., 2017, 2020), we labeled a 1 min segment as SA if the segment contains more than 5 s of SA events. Considering the class imbalance problem of UCDDB, the data of patients without SA events (ucddb008, ucddb011, ucddb013, and ucddb018) are not used (John et al., 2021).
3.2. Classification performance on Apnea-ECG dataset
The SA detection involves two stages. The first stage is to detect whether a 1 min segment is SA. In the second stage, each patient is assessed for sleep quality overnight based on the results of the first stage.
3.2.1. Per-segment classification on Apnea-ECG dataset
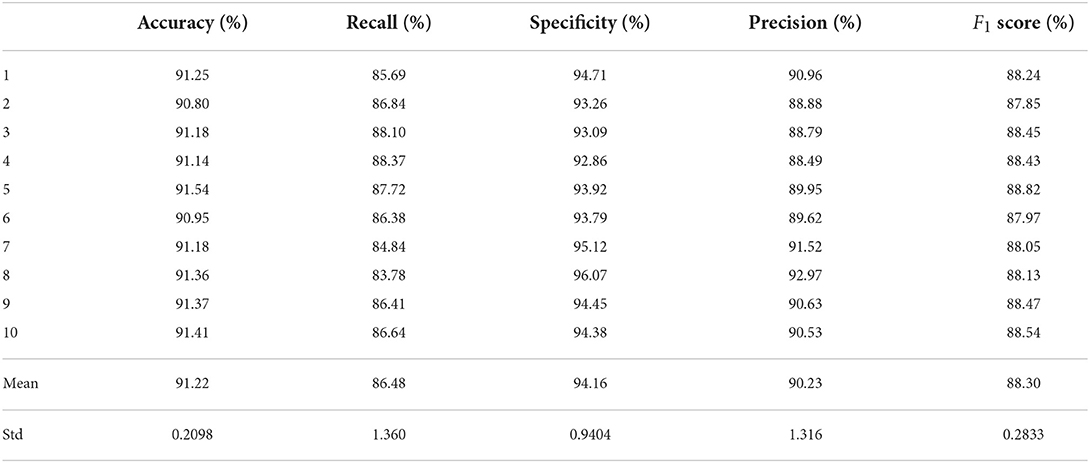
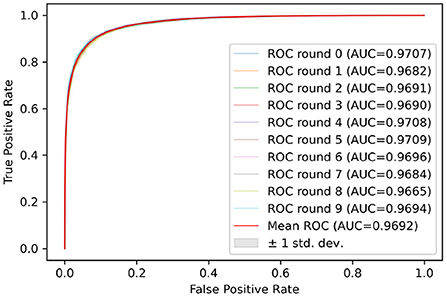
Test sets were used to evaluate the effectiveness of the proposed model. First, the pre-processed 5 min ECG segments were fed into the CNN-BiGRU network to automatically extract the features. Then, the extracted features were fed into the fully connected layers, and the ECG signal of the middle segment was classified. The results of the CNN-BiGRU model with three spatio-temporal blocks for ten runs are listed in the Table 2. It is worth noting that the 10th experiment exceeded the average on all evaluation metrics. In addition, the 5th experiment reached the highest values of 91.54 and 88.82% for the accuracy and F1 score, respectively. To evaluate the classifier more comprehensively, Figure 4 shows the ROC curve and AUC. It can be seen that the model proposed in this study is stable, with an AUC value of 0.9692±0.0013.
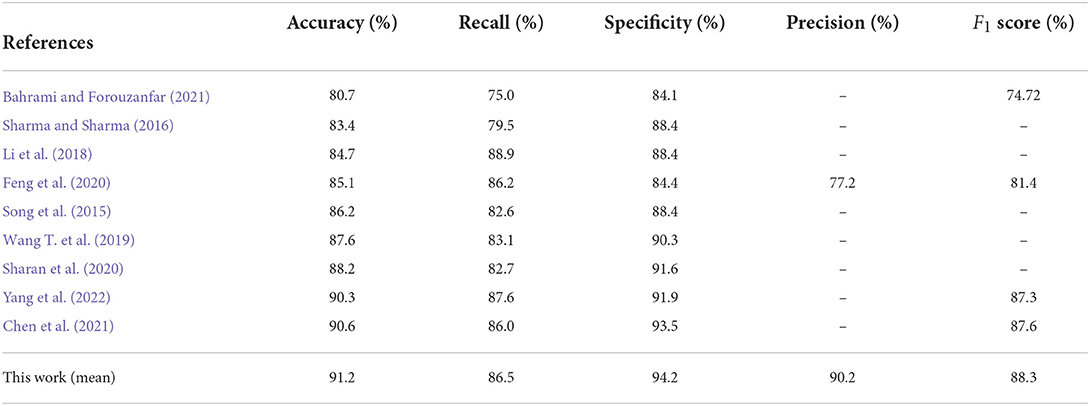
Table 3 lists the comparison results between the CNN-BiGRU model and previous state-of-the-art works on the detection of per-minute ECG signals. Notice that the compared methods and the proposed models listed in Tables 3, 4 were all trained on the release set and evaluated on the withheld set. The results show that the average performance of CNN-BiGRU outperforms the previous optimal model in terms of accuracy, specificity, and F1 metrics. It is worth noting that CNN-BiGRU only underperforms the approach of literature (Li et al., 2018; Yang et al., 2022) in terms of the recall metric. However, our model achieved the F1 score of 88.3%, which is better than that of literature (Yang et al., 2022), while literature (Li et al., 2018) did not give F1 score.
In summary, some previous works (Song et al., 2015; Sharma and Sharma, 2016) were based on feature engineering techniques that attempt to improve the performance by mapping high-dimensional training data to a low-dimensional feature space. However, this is time-consuming and ineffective. Deep learning methods can extract important features from ECG signals, and the DL-based methods (Chen et al., 2021; Yang et al., 2022) mentioned in Table 3 have all achieved good results, but their performance was inferior to that of the CNN-BiGRU model proposed in this paper. Our model uses spatio-temporal blocks, which can extract spatio-temporal features more effectively from ECG signals and provide better performance on SA classification.
3.2.2. Per-recording classification on Apnea-ECG dataset
In order to further assess the quality of the subjects' sleep, an overall SA diagnosis of the subjects' recordings was performed. Each of the subjects' recordings consisted of multiple 1 min segments. The AHI is commonly used clinically as an indicator of whether a subject is suffering from SA. An individual is considered to have SA if the subject's AHI is >5 (Ruehland et al., 2009). The formula for calculating the AHI is as follows:
where T is the number of 1 min segments and N indicates the number of 1 min SA segments.
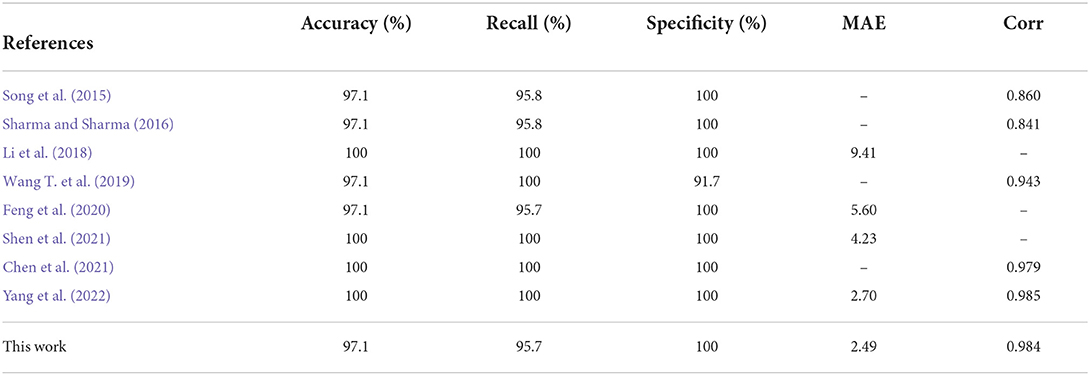
In the per-recording detection, the accuracy, sensitivity, specificity, and AUC of the CNN-BiGRU model were calculated on the retention set as 97.1, 95.7, 100, and 0.996%, respectively. The accuracy was 97.1% because the model misclassified one from the total 35 patients. More specifically, one subject (x12) with SA had an AHI of 6.75, whereas the proposed model calculated an AHI of 4.00, thus classifying the patient as normal. It is worth noting that the low precision per-segment approach may show better per-recording performance because of the relatively small amount of data in the test set (Wang T. et al., 2019). Therefore, according to the previous literature (Sharma and Sharma, 2016; Wu et al., 2021; Yang et al., 2022), the Pearson correlation coefficient (Corr) and mean absolute error (MAE) were also used as new evaluation indicators to ensure the reliability of the comparison. These metrics are defined as follows:
where N is the number of recordings, and and represent the predicted and true AHI values of the i-th recording, respectively.
Table 4 lists the comparison of the per-recording classification performance between the CNN-BiGRU model and state-of-the-art works in recent years. As mentioned above, traditional evaluation metrics do not provide a comprehensive and accurate assessment of model performance, and a better approach is to use MAE and Corr metrics. As listed in Table 4, our model achieved 2.49 and 0.984 for the MAE and Corr metrics, respectively. Our model achieved the best performance in terms of MAE metrics. On the Corr metric, literature (Yang et al., 2022) achieved the best value of 0.985, while our model achieved 0.984, which is a comparable result. Overall, our proposed model provides more competitive performance than those of the works presented in Table 4.
3.3. Classification performance on UCDDB dataset
Usually, UCDDB is utilized to evaluate the robustness of the SA detection models (Wang T. et al., 2019; Mashrur et al., 2021; Yang et al., 2022). Similarly, we evaluated our CNN-BiGRU model on UCDDB to demonstrate the model's robustness. Different from the Apnea-ECG dataset, UCDDB was not divided into the training set and test set by the original publishers. As a result, the previous works (Wang T. et al., 2019; Mashrur et al., 2021; Yang et al., 2022) had used their own splitting of training and testing sets in evaluations. In this paper, we used the same preprocessing method for the UCDDB as mentioned in Section 2.2. The difference is that the UCDDB is divided into a training set, a validation set and a test set with a proportion of 8:1:1. Due to the relatively small number of patients with SA at the UCDDB, the training set was balanced by oversampling the minority class (SA events). Meanwhile, per-recording testing is not performed for the same reason.
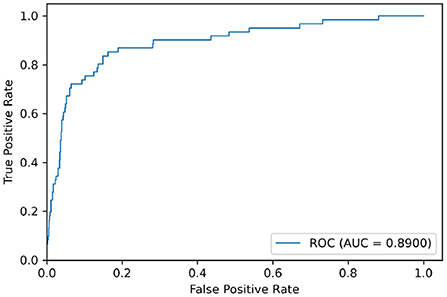
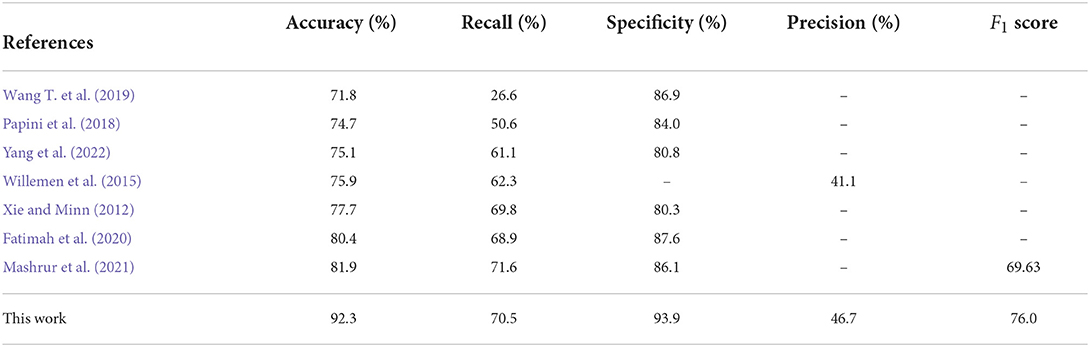
We used the model trained on the Apnea-ECG dataset to continue training on the UCDDB training set, with the experimental settings mentioned in Section 2.4. On the UCDDB test set, the performance of the CNN-BiGRU model on the accuracy, recall, specificity, and AUC metrics reached 92.3, 70.5, 93.9, and 0.890%, respectively. Figure 5 shows the ROC curves and AUC of the proposed model for per-segment detection. Table 5 lists the results of CNN-BiGRU on the test set and compares them with other detection algorithms in the literature. The results show that the CNN-BiGRU model is far superior to the previous models, with an accuracy and specificity of 92.3 and 93.9%, respectively. In regard to recall metrics, we obtained a comparative result to the works (Mashrur et al., 2021). Compared to the Apnea-ECG dataset, our model has a significant decrease in the recall metric on the UCDDB. A major reason for this is that the ratio of pre-processed SA segments to all segments is about 1%, indicating that the class imbalance is intensified. Note that it is a rough comparison in Table 5, as there is no uniform data partitioning of training set and test set for UCDDB. In summary, our CNN-BiGRU model is useful for SA detection.
4. Discussion
4.1. Hyperparameter tuning
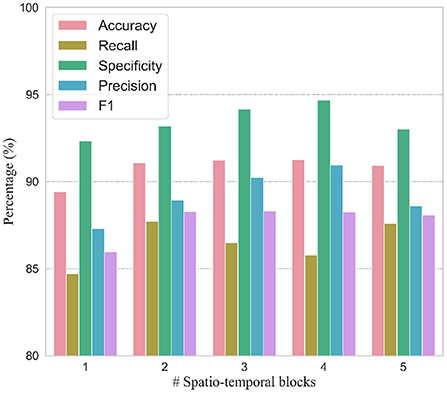
In order to verify the efficacy of spatio-temporal blocks, the number of spatio-temporal blocks was tuned from 1 to 5 on PhysioNet Apnea-ECG dataset. As shown in Figure 6 and Table 6, one spatio-temporal block model cannot effectively extract high-level spatio-temporal information. Meanwhile, too many spatio-temporal blocks also fail to learn high-level feature information due to the overfitting problem. Considering that Apnea-ECG dataset suffers from class imbalance, the F1 score became the main metric we considered. And the CNN-BiGRU model using three spatio-temporal blocks reached the highest values of 88.30% for F1 score. Therefore, we set the number of spatio-temporal blocks for CNN-BiGRU to 3 in this study.
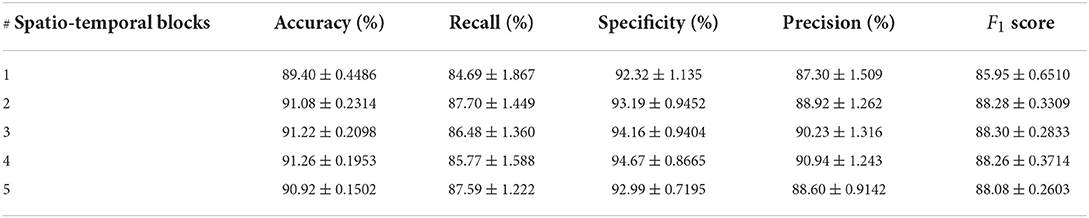
Table 6. Comparison of per-segment detection results using different numbers of spatio-temporal blocks.
4.2. Ablation study
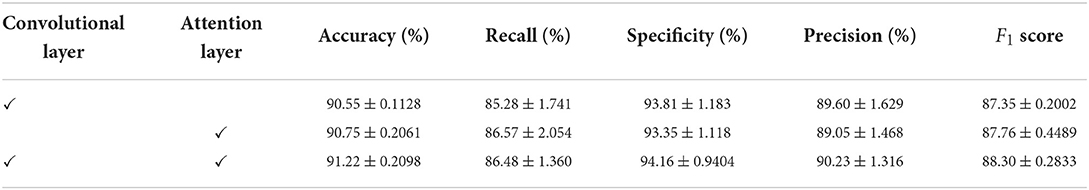
We conducted an ablation study on the Apnea-ECG dataset, considering that there was sufficient data in the Apnea-ECG dataset to fully evaluate the performance of the model. As shown previously, the CNN-BiGRU model uses a convolutional layer, spatio-temporal blocks, and an attention layer to extract features. Therefore, the results of the ablation experiments with the convolutional layer and the attention layer removed separately are listed in Table 7. It was found that removing either the convolutional layer or the attention layer will make the classification performance degrade. Specifically, the accuracy of the models with the convolutional layer removed and the attention layer removed is decreased by 0.47 and 0.67%, respectively. Overall, using the convolutional layer and attention layer improved the classification performance of the CNN-BiGRU model.
4.3. Cross-dataset evaluation
Cross-dataset evaluations are performed to demonstrate the general performance of our proposed model, using the Apnea-ECG and UCDDB datasets. Specifically, the model is trained on one dataset and evaluated directly on another dataset. When CNN-BiGRU was trained on Apnea-ECG and tested on UCDDB, an accuracy of 85.3% and an F1-score of 50.5% were achieved. Similarly, when it was trained on UCDDB and evaluated on Apnea-ECG, the accuracy was 53.8% and the F1-score was 36.3%. It is found that the performances of cross-dataset evaluation are not satisfactory. To comprehensively understand the evaluation, we performed the same cross-dataset evaluation using a previous state-of-the-art model (Chen et al., 2021) listed in Table 3. The accuracy achieved was 85.9% and the F1-score was 51.1% using the UCDDB as the testing set and Apnea-ECG as the training set. They are very slightly better than those of our model (85.9 vs. 85.3%; 51.1 vs. 50.5%). However, when it was trained with UCDDB and tested on Apnea-ECG, the accuracy and the F1-score were 45.5 and 29.2%, respectively. Obviously, our model has outperformed this previous model (53.8 vs. 45.5%; 36.3 vs. 29.2%). In general, CNN-BiGRU is superior to the compared model (Chen et al., 2021), in terms of cross-dataset evaluation.
Finally, we attribute the low performance of cross-dataset evaluation to the following reasons: (1) the populations of datasets are different. For example, subjects with central apnea and periodic respiratory episodes are included in UCDDB; (2) the different sampling rates may impact the performance (the ECG signals are sampled at 100 Hz on Apnea-ECG while 128 Hz on UCDDB); (3) UCDDB has a severe class imbalance problem. In other words, the distributions of normal and SA are quite different between the two datasets.
5. Conclusion
In this study, a novel spatio-temporal learning-based model named CNN-BiGRU was explored to classify SA events from ECG signals. Specifically, the proposed CNN-BiGRU is an end-to-end deep learning model, which consists of multiple spatio-temporal blocks. Each block has the identical architecture with a CNN layer, a max-pooling layer, and a BiGRU layer. This architecture with repeated spatio-temporal blocks can well capture the morphological spatial feature information as well as the temporal feature information from ECG signals. Experiment results on the apnea-ECG dataset showed that the proposed CNN-BiGRU achieved an accuracy of 91.22 and 97.10% for per-minute classification and per-recording classification, respectively. And the accuracy on the UCDDB dataset reached 91.24%. In contrast to the previous state-of-the-art methods, our proposed CNN-BiGRU has an obvious advantage with a big margin. It means that the CNN-BiGRU can be potentially deployed into a medical system to help physicians to screen out SA patients to avoid malignant events. In future work, we will further apply the proposed model to real healthcare systems.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at: PhysioNet Apnea-ECG: https://physionet.org/content/apnea-ecg/1.0.0/; UCDDB: https://archive.physionet.org/physiobank/database/ucddb/.
Author contributions
Conceptualization: WZ and WM. Methodology: JC and MS. Software: JC. Writing: WZ and JC. Project administration: WZ. All authors contributed to the article and approved the submitted version.
Funding
This work was partly supported by the National Key R&D Program of China under Grant 2019YFB1804003.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ali, S. Q., Khalid, S., and Brahim Belhaouari, S. (2019). A novel technique to diagnose sleep apnea in suspected patients using their ECG data. IEEE Access 7, 35184–35194. doi: 10.1109/ACCESS.2019.2904601
Ankhili, A., Tao, X., Cochrane, C., Coulon, D., and Koncar, V. (2018). Washable and reliable textile electrodes embedded into underwear fabric for electrocardiography (ECG) monitoring. Materials 11:256. doi: 10.3390/ma11020256
Bahrami, M., and Forouzanfar, M. (2021). “Detection of sleep apnea from single-lead ECG: comparison of deep learning algorithms,” in 2021 IEEE International Symposium on Medical Measurements and Applications (Lausanne: IEE), 1–5. doi: 10.1109/MeMeA52024.2021.9478745
Baty, F., Boesch, M., Widmer, S., Annaheim, S., Fontana, P., Camenzind, M., et al. (2020). Classification of sleep apnea severity by electrocardiogram monitoring using a novel wearable device. Sensors 20:286. doi: 10.3390/s20010286
Benjafield, A. V., Ayas, N. T., Eastwood, P. R., Heinzer, R., Ip, M. S., Morrell, M. J., et al. (2019). Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir. Med. 7, 687–698. doi: 10.1016/S2213-2600(19)30198-5
Bsoul, M., Minn, H., and Tamil, L. (2011). Apnea medassist: real-time sleep apnea monitor using single-lead ECG. IEEE Trans. Inform. Technol. Biomed. 15, 416–427. doi: 10.1109/TITB.2010.2087386
Chen, L., Zhang, X., and Song, C. (2015). An automatic screening approach for obstructive sleep apnea diagnosis based on single-lead electrocardiogram. IEEE Trans. Autom. Sci. Eng. 12, 106–115. doi: 10.1109/TASE.2014.2345667
Chen, X., Chen, Y., Ma, W., Fan, X., and Li, Y. (2021). “SE-MSCNN: a lightweight multi-scaled fusion network for sleep apnea detection using single-lead ECG signals,” in 2021 IEEE International Conference on Bioinformatics and Biomedicine (Houston, TX: IEEE), 1276–1280. doi: 10.1109/BIBM52615.2021.9669358
Cho, K., van Merriënboer, B., Gulcehre, C., Bahdanau, D., Bougares, F., Schwenk, H., et al. (2014). “Learning phrase representations using RNN encoder-decoder for statistical machine translation,” in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) (Doha: Association for Computational Linguistics), 1724–1734. doi: 10.3115/v1/D14-1179
Chung, J., Çaglar Gülçehre, Cho, K., and Bengio, Y. (2014). Empirical evaluation of gated recurrent neural networks on sequence modeling. ArXiv: abs/1412.3555. doi: 10.48550/arXiv.1412.3555
Fan, X., Wang, H., Xu, F., Zhao, Y., and Tsui, K.-L. (2019). Homecare-oriented intelligent long-term monitoring of blood pressure using electrocardiogram signals. IEEE Trans. Indus. Inform. 16, 7150–7158. doi: 10.1109/TII.2019.2962546
Fan, X., Yao, Q., Cai, Y., Miao, F., Sun, F., and Li, Y. (2018). Multiscaled fusion of deep convolutional neural networks for screening atrial fibrillation from single lead short ECG recordings. IEEE J. Biomed. Health Inform. 22, 1744–1753. doi: 10.1109/JBHI.2018.2858789
Fatimah, B., Singh, P., Singhal, A., and Pachori, R. B. (2020). Detection of apnea events from ECG segments using Fourier decomposition method. Biomed. Signal Process. Control 61:102005. doi: 10.1016/j.bspc.2020.102005
Faust, O., Acharya, U. R., Ng, E., and Fujita, H. (2016). A review of ECG-based diagnosis support systems for obstructive sleep apnea. J. Mech. Med. Biol. 16:1640004. doi: 10.1142/S0219519416400042
Feng, K., Qin, H., Wu, S., Pan, W., and Liu, G. (2020). A sleep apnea detection method based on unsupervised feature learning and single-lead electrocardiogram. IEEE Trans. Instrumen. Measure. 70, 1–12. doi: 10.1109/TIM.2020.3017246
Gaiduk, M., Orcioni, S., Conti, M., Seepold, R., Penzel, T., Madrid, N. M., et al. (2020). “Embedded system for non-obtrusive sleep apnea detection*,” in 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (Montreal, QC: IEEE), 2776–2779. doi: 10.1109/EMBC44109.2020.9176075
Graco, M., Schembri, R., Cross, S., Thiyagarajan, C., Shafazand, S., Ayas, N. T., et al. (2018). Diagnostic accuracy of a two-stage model for detecting obstructive sleep apnoea in chronic tetraplegia. Thorax 73, 864–871. doi: 10.1136/thoraxjnl-2017-211131
Gutiérrez-Tobal, G. C., Álvarez, D., Gomez-Pilar, J., Campo, F. d., and Hornero, R. (2015). Assessment of time and frequency domain entropies to detect sleep apnoea in heart rate variability recordings from men and women. Entropy 17, 123–141. doi: 10.3390/e17010123
Hamilton, P. (2002). “Open source ECG analysis,” in Computers in Cardiology, (Memphis, TN: IEEE), 101–104. doi: 10.1109/CIC.2002.1166717
John, A., Cardiff, B., and John, D. (2021). “A 1D-CNN based deep learning technique for sleep apnea detection in IoT sensors,” in 2021 IEEE International Symposium on Circuits and Systems (Daegu: IEEE), 1–5. doi: 10.1109/ISCAS51556.2021.9401300
King, S., and Cuellar, N. (2016). Obstructive sleep apnea as an independent stroke risk factor: a review of the evidence, stroke prevention guidelines, and implications for neuroscience nursing practice. J. Neurosci. Nurs. 48, 133–142. doi: 10.1097/JNN.0000000000000196
Kingma, D. P., and Ba, J. (2017). Adam: a method for stochastic optimization. arXiv [Preprint]. arXiv: 1412.6980. doi: 10.48550/arXiv.1412.6980
Li, K., Pan, W., Li, Y., Jiang, Q., and Liu, G. (2018). A method to detect sleep apnea based on deep neural network and hidden Markov model using single-lead ECG signal. Neurocomputing 294, 94–101. doi: 10.1016/j.neucom.2018.03.011
Lin, C.-H., Lurie, R. C., and Lyons, O. D. (2020). Sleep apnea and chronic kidney disease: a state-of-the-art review. Chest 157, 673–685. doi: 10.1016/j.chest.2019.09.004
Luong, T., Pham, H., and Manning, C. D. (2015). “Effective approaches to attention-based neural machine translation,” in Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing (Lisbon: Association for Computational Linguistics), 1412–1421. doi: 10.18653/v1/D15-1166
Lyons, O. D., and Ryan, C. M. (2015). Sleep apnea and stroke. Can. J. Cardiol. 31, 918–927. doi: 10.1016/j.cjca.2015.03.014
Mashrur, F. R., Islam, M. S., Saha, D. K., Islam, S. R., and Moni, M. A. (2021). SCNN: Scalogram-based convolutional neural network to detect obstructive sleep apnea using single-lead electrocardiogram signals. Comput. Biol. Med. 134:104532. doi: 10.1016/j.compbiomed.2021.104532
Mostafa, S. S., Mendonça, F., Morgado-Dias, F., and Ravelo-García, A. (2017). “Spo2 based sleep apnea detection using deep learning,” in 2017 IEEE 21st International Conference on Intelligent Engineering Systems (Larnaca: IEEE), 91–96. doi: 10.1109/INES.2017.8118534
Mostafa, S. S., Morgado-Dias, F., and Ravelo-García, A. G. (2020). Comparison of SFS and MRMR for oximetry feature selection in obstructive sleep apnea detection. Neural Comput. Appl. 32, 15711–15731. doi: 10.1007/s00521-018-3455-8
Papini, G. B., Fonseca, P., Margarito, J., van Gilst, M. M., Overeem, S., Bergmans, J. W., et al. (2018). “On the generalizability of ECG-based obstructive sleep apnea monitoring: merits and limitations of the apnea-ECG database,” in 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (Honolulu, HI: IEEE), 6022–6025. doi: 10.1109/EMBC.2018.8513660
Penzel, T., Moody, G. B., Mark, R. G., Goldberger, A. L., and Peter, J. H. (2000). “The apnea-ECG database,” in Computers in Cardiology 2000. Vol. 27 (Cambridge, MA: IEEE), 255–258. doi: 10.1109/CIC.2000.898505
Rosen, I. M., Kirsch, D. B., Chervin, R. D., Carden, K. A., Ramar, K., Aurora, R. N., et al. (2017). Clinical use of a home sleep apnea test: an American academy of sleep medicine position statement. J. Clin. Sleep Med. 13, 1205–1207. doi: 10.5664/jcsm.6774
Ruehland, W. R., Rochford, P. D., O'Donoghue, F. J., Pierce, R. J., Singh, P., and Thornton, A. T. (2009). The new AASM criteria for scoring hypopneas: impact on the apnea hypopnea index. Sleep 32, 150–157. doi: 10.1093/sleep/32.2.150
Russell, M. B., Kristiansen, H. A., and Kværner, K. J. (2014). Headache in sleep apnea syndrome: epidemiology and pathophysiology. Cephalalgia 34, 752–755. doi: 10.1177/0333102414538551
Sharan, R. V., Berkovsky, S., Xiong, H., and Coiera, E. (2020). “ECG-derived heart rate variability interpolation and 1-d convolutional neural networks for detecting sleep apnea,” in 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (Montreal, QC: IEEE), 637–640. doi: 10.1109/EMBC44109.2020.9175998
Sharma, H., and Sharma, K. (2016). An algorithm for sleep apnea detection from single-lead ECG using hermite basis functions. Comput. Biol. Med. 77, 116–124. doi: 10.1016/j.compbiomed.2016.08.012
Shen, Q., Qin, H., Wei, K., and Liu, G. (2021). Multiscale deep neural network for obstructive sleep apnea detection using RR interval from single-lead ECG signal. IEEE Trans. Instrumen. Measure. 70, 1–13. doi: 10.1109/TIM.2021.3062414
Smruthy, A., and Suchetha, M. (2017). Real-time classification of healthy and apnea subjects using ECG signals with variational mode decomposition. IEEE Sensors J. 17, 3092–3099. doi: 10.1109/JSEN.2017.2690805
Somers, V. K., White, D. P., Amin, R., Abraham, W. T., Costa, F., Culebras, A., et al. (2008). Sleep apnea and cardiovascular disease: an American heart association/American college of cardiology foundation scientific statement from the American heart association council for high blood pressure research professional education committee, council on clinical cardiology, stroke council, and council on cardiovascular nursing in collaboration with the national heart, lung, and blood institute national center on sleep disorders research (national institutes of health). J. Am. Coll. Cardiol. 52, 686–717. doi: 10.1016/j.jacc.2008.05.002
Song, C., Liu, K., Zhang, X., Chen, L., and Xian, X. (2015). An obstructive sleep apnea detection approach using a discriminative hidden Markov model from ECG signals. IEEE Trans. Biomed. Eng. 63, 1532–1542. doi: 10.1109/TBME.2015.2498199
Vanek, J., Prasko, J., Genzor, S., Ociskova, M., Kantor, K., Holubova, M., et al. (2020). Obstructive sleep apnea, depression and cognitive impairment. Sleep Med. 72, 50–58. doi: 10.1016/j.sleep.2020.03.017
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., et al. (2017). “Attention is all you need,” in Proceedings of the 31st International Conference on Neural Information Processing Systems, NIPS'17 (Red Hook, NY: Curran Associates Inc.), 6000–6010.
Viswabhargav, C. S., Tripathy, R., and Acharya, U. R. (2019). Automated detection of sleep apnea using sparse residual entropy features with various dictionaries extracted from heart rate and EDR signals. Comput. Biol. Med. 108, 20–30. doi: 10.1016/j.compbiomed.2019.03.016
Wang, L., Lin, Y., and Wang, J. (2019). A RR interval based automated apnea detection approach using residual network. Comput. Methods Prog. Biomed. 176, 93–104. doi: 10.1016/j.cmpb.2019.05.002
Wang, T., Lu, C., Shen, G., and Hong, F. (2019). Sleep apnea detection from a single-lead ECG signal with automatic feature-extraction through a modified lenet-5 convolutional neural network. PeerJ 7:e7731. doi: 10.7717/peerj.7731
Wang, Z., Wang, X., and Wang, G. (2018). Learning fine-grained features via a CNN tree for large-scale classification. Neurocomputing 275, 1231–1240. doi: 10.1016/j.neucom.2017.09.061
Weder, M., Hegemann, D., Amberg, M., Hess, M., Boesel, L. F., Abächerli, R., et al. (2015). Embroidered electrode with silver/titanium coating for long-term ECG monitoring. Sensors 15, 1750–1759. doi: 10.3390/s150101750
Willemen, T., Varon, C., Dorado, A. C., Haex, B., Vander Sloten, J., and Van Huffel, S. (2015). Probabilistic cardiac and respiratory based classification of sleep and apneic events in subjects with sleep apnea. Physiol. Measure. 36:2103. doi: 10.1088/0967-3334/36/10/2103
Wu, Y., Pang, X., Zhao, G., Yue, H., Lei, W., and Wang, Y. (2021). A novel approach to diagnose sleep apnea using enhanced frequency extraction network. Comput. Methods Prog. Biomed. 206:106119. doi: 10.1016/j.cmpb.2021.106119
Xie, B., and Minn, H. (2012). Real-time sleep apnea detection by classifier combination. IEEE Trans. Inform. Technol. Biomed. 16, 469–477. doi: 10.1109/TITB.2012.2188299
Yang, Q., Zou, L., Wei, K., and Liu, G. (2022). Obstructive sleep apnea detection from single-lead electrocardiogram signals using one-dimensional squeeze-and-excitation residual group network. Comput. Biol. Med. 140:105124. doi: 10.1016/j.compbiomed.2021.105124
Keywords: sleep apnea, ECG signals, spatio-temporal learning, BiGRU, attention
Citation: Chen J, Shen M, Ma W and Zheng W (2022) A spatio-temporal learning-based model for sleep apnea detection using single-lead ECG signals. Front. Neurosci. 16:972581. doi: 10.3389/fnins.2022.972581
Received: 18 June 2022; Accepted: 18 July 2022;
Published: 05 August 2022.
Edited by:
Hui Zhou, Nanjing University of Science and Technology, ChinaReviewed by:
Shixiong Chen, Shenzhen Institutes of Advanced Technology (CAS), ChinaZiliang Xu, Fourth Military Medical University, China
Copyright © 2022 Chen, Shen, Ma and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiping Zheng, emhlbmd3ZWlwaW5nQHNjbnUuZWR1LmNu
 Junyang Chen
Junyang Chen Mengqi Shen
Mengqi Shen Wenjun Ma
Wenjun Ma Weiping Zheng
Weiping Zheng