- 1Department of Cellular, Computational and Integrative Biology (CiBiO), University of Trento, Trento, Italy
- 2Department of Neuroscience, Scuola Internazionale Superiore di Studi Avanzati (SISSA), Trieste, Italy
- 3Institute of Molecular Life Sciences, University of Zurich, Zürich, Switzerland
- 4Department of Medicine, NYU Langone Medical Center, New York, NY, United States
Proteinopathies are a large group of neurodegenerative diseases caused by both genetic and sporadic mutations in particular genes which can lead to alterations of the protein structure and to the formation of aggregates, especially toxic for neurons. Autophagy is a key mechanism for clearing those aggregates and its function has been strongly associated with the ubiquitin-proteasome system (UPS), hence mutations in both pathways have been associated with the onset of neurodegenerative diseases, particularly those induced by protein misfolding and accumulation of aggregates. Many crucial discoveries regarding the molecular and cellular events underlying the role of autophagy in these diseases have come from studies using Drosophila models. Indeed, despite the physiological and morphological differences between the fly and the human brain, most of the biochemical and molecular aspects regulating protein homeostasis, including autophagy, are conserved between the two species.In this review, we will provide an overview of the most common neurodegenerative proteinopathies, which include PolyQ diseases (Huntington’s disease, Spinocerebellar ataxia 1, 2, and 3), Amyotrophic Lateral Sclerosis (C9orf72, SOD1, TDP-43, FUS), Alzheimer’s disease (APP, Tau) Parkinson’s disease (a-syn, parkin and PINK1, LRRK2) and prion diseases, highlighting the studies using Drosophila that have contributed to understanding the conserved mechanisms and elucidating the role of autophagy in these diseases.
1. Introduction
Drosophila melanogaster is an established model organism for developmental studies and due to the remarkable conservation of the signaling regulating autophagy, it has been used to better understand the relationship of this catabolic pathway with the genetic conditions that in humans are responsible of a class of neuronal diseases called proteinopathies (PPs). Autophagy is a key cellular pathway that, together with the ubiquitin-proteasome system (UPS), controls protein homeostasis by degrading misfolded proteins or exhausted organelles otherwise detrimental to the cells (Kocaturk and Gozuacik, 2018). Autophagy and UPS are closely linked, in fact protein ubiquitination is a key step for the cargo recognition by the autophagic receptors and alterations in one pathway may affect the activity of the other (Waite et al., 2022). Both pathways are crucial for cell survival particularly in neurons where their perturbation causes age-associated disorders including neurodegenerative diseases (Rai et al., 2022). In this review, we will illustrate the contribution of Drosophila studies to the understanding of the role of autophagy in PPs induced by mutations in genes responsible for the most common neurodegenerative diseases (summarized in Table 1). Furthermore, we will discuss how flies could be used to further improve our understanding of the mechanisms that control these diseases, particularly those that are linked to mutations in genes that are physiologically involved in the control of the autophagic-proteostatic pathway.
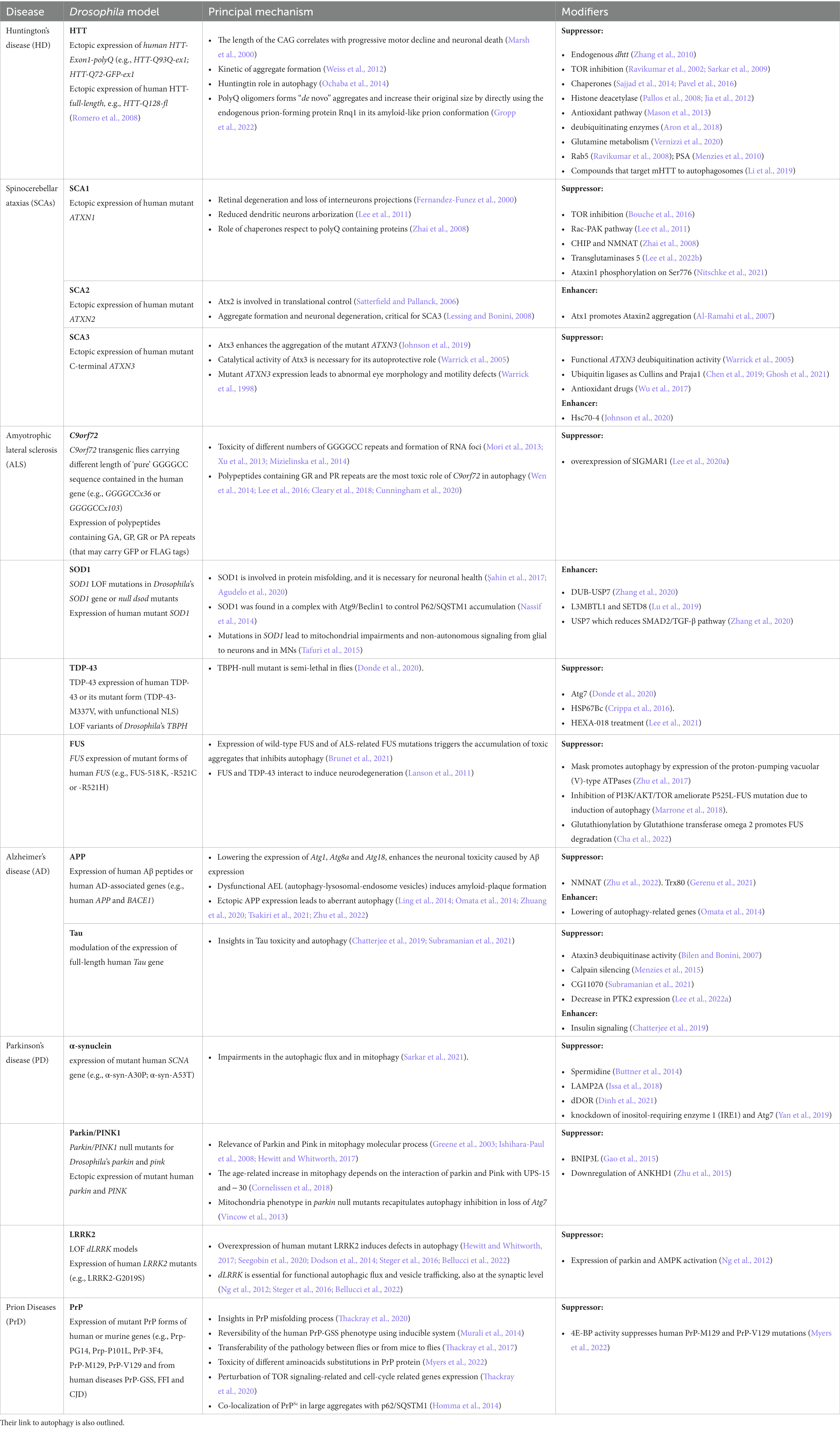
Table 1. Drosophila models of human proteinopathies discussed in this article, their principal mechanism, and components of pathways (modifiers) that can either suppress on enhance the toxic phenotype.
2. Drosophila versus human brain
Drosophila ‘s studies played a crucial role in understanding brain function and development, leading to significant advances in neuroscience. Drosophila’s brain contains about 200,000 neurons, which are quite similar to mammalian neurons in terms of electrophysiological properties, and form a complex network of interactions recently mapped at high resolution (Zheng et al., 2018), and glial cells that represents 5–10% of the total cell population within the central nervous system (CNS). The CNS is composed of multiple specific and distinct brain compartments (Ito et al., 2014) that are interconnected and synergistically cooperating to control complex behaviors, such as learning, flight control, courtship, grooming, and memory-driven behaviors. Despite the small size, the overall brain organization and regional division shows fundamental similarities to the network structure of the mammalian brain. Indeed, the neural organization underlying primitive functions, such as the perception of odors, taste, vision, sound, gravity, and the circuits regulating feeding and satiety are very similar to those in humans (Tsubouchi et al., 2017; Jayakumar and Hasan, 2018). Drosophila CNS can be divided into two histological regions: the neuronal cell cortex, where all the neurons cell bodies are located, and the neuropil, where axons and dendrites project. This represents a major difference with vertebrates, where the cell bodies are in different areas of the brain depending on the circuit that they are regulating. In addition, in the fruit fly, the lateral connections between neighboring projection-neurons of the same compartment are made not only through interneurons (Liou et al., 2018) but also through dendro-dendritic synapses, which are rare or absent in most vertebrate neuropiles. In terms of morphology, while in vertebrates most neurons are multipolar, in invertebrates there are mostly unipolar (Rolls, 2011; Shah et al., 2016; Smarandache-Wellmann, 2016). Despite these differences, both vertebrate and invertebrate neuronal circuits are plastic which means that their structure and physiology are modified in response to stimuli, both extrinsic and intrinsic, during development and in adult life (Holtmaat and Svoboda, 2009; Lin et al., 2013; Peretti et al., 2015). The electrophysiological properties of the fruit fly neurons are also very similar to those in mammals. Their firing activity depends on Na+ and K+ fluxes that affect the membrane potential, and they communicate using vesicle release of conserved neurotransmitters (i.e., acetylcholine, GABA, glutamate) and neuromodulators (i.e., biogenic amines and neuropeptides) at the synapses. In Drosophila there are several classes of glia, mostly classified based on their morphology function and association with neurons. The perineural glia (PNG), which surrounds the central nervous system, is required to filter nutrients, while the sub-perineural glia (SPG) forms the septate junctions and together form the blood–brain barrier (BBB). This is different than in humans, where the BBB is composed of astrocytes and microglia, however also in Drosophila the BBB prevents paracellular diffusion and controls the influx and efflux of soluble molecules (Hindle and Bainton, 2014; Babatz et al., 2018; Kim et al., 2020). Within the CNS there are other classes of glia cells such as the cortex glia, associated with the neuronal cell bodies, unsheathing glia, that surrounds neuropils and astrocytes that associates closely with the synaptic neuropils and support neurons, similarly to those of the mammalian glia. These classes of glia are abundant and are involved in nervous system development, circuit assembly, synaptic plasticity and neurons support (Freeman, 2015).
3. Drosophila’s tools to study proteinopathies (PPs)
The availability of a wide variety of transgenic strains, advanced genetic tools and databases have enabled the rapid development of Drosophila models for nearly 75% of human neuronal diseases including proteinopathies PPs (Ugur et al., 2016). The simplicity of fly’s brain architecture and the highly conserved function of genes involved in neuronal development, highlights another advantage for its use. Furthermore, Drosophila’s genome generally harbors only one orthologue of the human counterpart therefore, mutation of a single gene generally leads to loss-of-function phenotypes, without redundant effects due to the presence of compensatory paralogues. The availability of unique and advanced genetic techniques allows for the rapid generation of transgenes in which the manipulation of the gene of interest (GOI) is performed in a short time and with unique precision in specific tissues and organs. The most common approach used is based on the yeast derived UAS/Gal4 system, in which a line carrying a tissue specific promoter fused with the transactivating domain of Gal4, is crossed with another line carrying the GOI (overexpression or its RNAi) cloned under the control of the UAS (Upstream regulating sequence). In the progeny, the binding of Gal4 to the UAS sequence will express or reduce the GOI in the tissue of interest (Brand and Perrimon, 1993). If manipulation of the GOI induces lethality, tissue expression control can be achieved by co-expression of the temperature sensitive inhibitor GAL80ts, which acts as a transcriptional repressor of Gal4 at its permissive temperature, or by the Gene-Switch system which relies on a modified version of Gal4 which can be temporally induced by the administration of the drug RU486 (Walters et al., 2019). Variants of the UAS/Gal4 system, such as LexA/Op and QUAS, can be used to drive the expression of different transgenes concurrently with the UAS/Gal4. This is particularly useful for example to study the presence of non-autonomous signals between different organs, tissues or cells in vivo (Potter et al., 2010). Additionally, loss-of-function mutations or overexpression of genes can be easily obtained using CRISPR/Cas9 technology and commercially available sgRNA lines deposited at the Bloomington Stock Center (Ewen-Campen et al., 2017; Zirin et al., 2022). More recently, novel optogenetic techniques have been successfully developed to study protein misfolding in vivo in the brain using Drosophila models of Alzheimer’s disease (AD), Parkinson’s disease (PD), and TDP-43/ALS (Lim et al., 2021). The morphological defects induced by the expression of genes responsible for human PPs (Figures 1A,B) can be easily analyzed after their expression in Drosophila (Figure 1C) using ex vivo dissection of the entire larval or in adult-fly brains, using different techniques ranging from immunofluorescence to super resolution microscopy (SEM or TEM; Figure 1D). Furthermore, motor defects caused by neuronal degeneration can be characterized by analyzing larval motility defects or by analyzing the decline of negative geotaxis in adults, which is the natural ability of flies to climb against gravity (Figure 1E). PPs are characterized by the formation of protein aggregates, which can be visualized by immunofluorescence using specific antibodies, or with the expression of fusion proteins where the gene of interest is fused with fluorochromes such as GFP or RFP (Figure 1F). Biochemical assays, using techniques such as western blot or filter-trap analysis, can show variations in molecular weight and the presence of large insoluble aggregates (Figure 1K). Cell death or induced inflammation at sites of aggregate formation can be studied directly by immunofluorescence assays using appropriate markers (e.g., cleaved caspase, TUNEL assay, or the presence of secreted immune-modulators, like eiger/TNFα or SPARK). Finally, one of the great advantages of using fruit flies as a model for PPs is that the onset of these diseases is relatively rapid, for example the expression of human mutant forms of Huntingtin HTTQ97GFP, in neurons leads to the formation of aggregates which are already visible within 48–72 h of larval development (Figure 1F). This is extremely advantageous since it allows to perform genetic or chemical screening to identify in a short time genes or drugs that reduce the formation of the aggregates (Figures 1G,J). These inhibitors can be chemically optimized (Figure 1H) to improve their qualities and finally be tested on human cells derived from patients (Figure 1I). However, questions remain as to why flies develop aggregates so early in their development, compared to vertebrates. One hypothesis is because flies lack a more complex, adaptive immune system that could control the onset of these diseases as it does in other model systems. In summary, the possibility to target specific mutations into subpopulation of neurons or glia and the ability to rapidly observe their effect cell autonomously or across the neural networks, combined with the short life cycle and the propensity to produce many offspring confirms Drosophila as an excellent model for studying human diseases including proteinopathies.
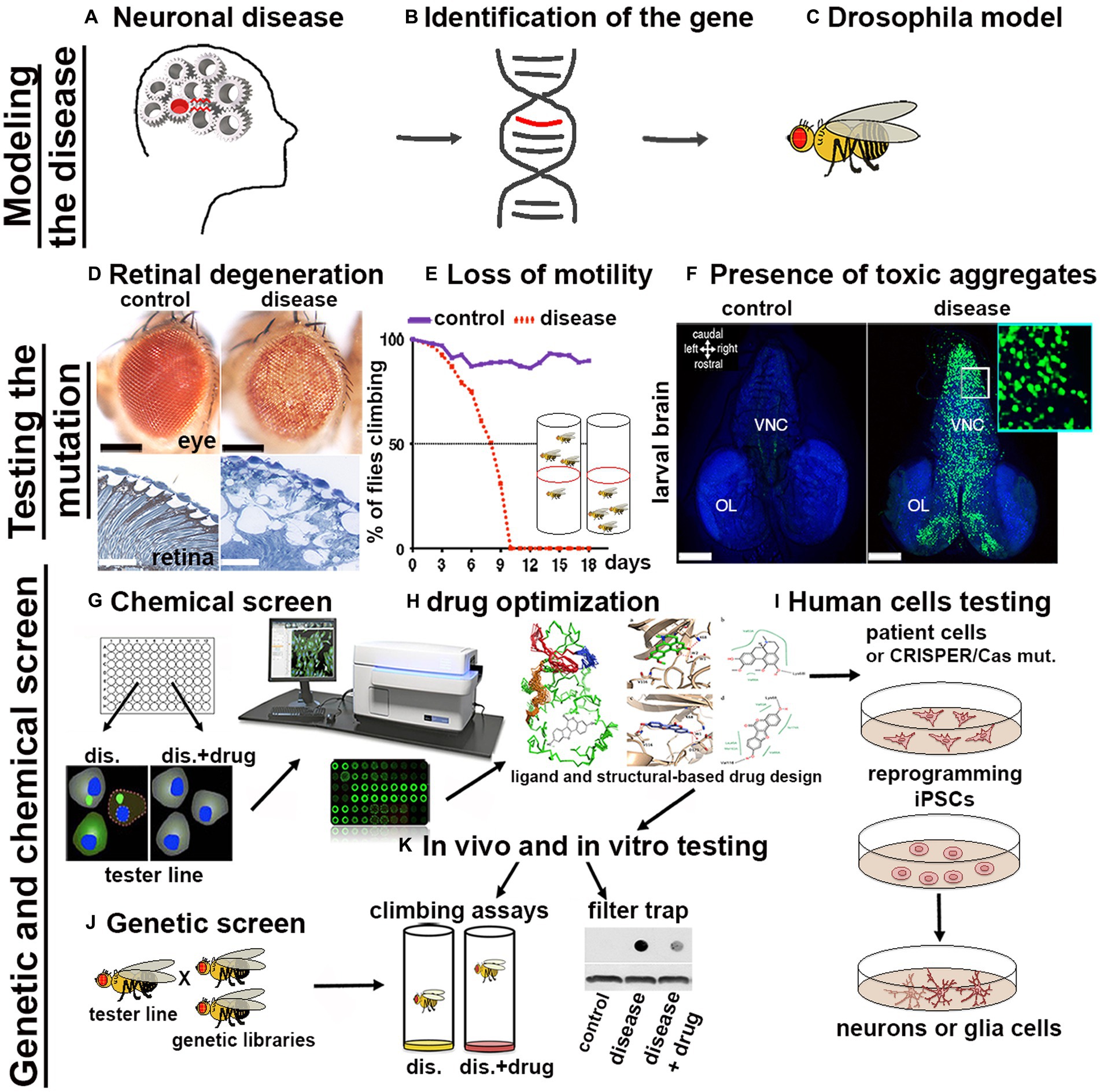
Figure 1. Scheme of a pipeline to characterize genes associated with proteinopathies and to perform High-throughput Screens with small chemical compounds, to develop new therapeutical strategies in humans. (A–C) From the identification of a gene related to a human disease to the generation of the transformants carrying the GOI. (D–F) The effect of the mutant-disease genes can be tested at cellular and behavioral levels. (D) For example, the expression of exon1 of the human mutant Huntingtin containing 93-CAGs (HTTQ93) in the retina using the GMR-Gal4 promoter leads to loss of the pigmentation in the ommatidia of the compound eye (as seen in the upper image obtained using a stereo microscope) and to retinal degeneration accompanied by defects in tissue morphology and neuronal death [outlined by the white spot of missing tissues visible by transmission electron microscopy (TEM) showed in the images below; (Vernizzi et al, 2020)]. (E) Expression of mutant HTT in neurons using ELAV-Gal4 induces neuronal defects that can be indirectly quantified by measuring the decline over time of the animal motility (negative geotaxis assay). (F) Using the Elav promoter we can express the human HTT exon-1 with 97 CAGs as a fusion protein with GFP (HTT-GFP), and show the formation of HTT-GFP aggregates in larval neurons already at 48-72 hours of age. Photo in panel F, to the right is shown a larval brain of Elav-LexOP-HTTGFP-LexA larvae at 72 hrs AEL, control is to the left. BLUE stains the nuclei (inset 63x). (G–K) Potential pipeline for a High throughput screen (HTS) to identify drugs that reduce the formation of toxic aggregates. (G) Drosophila cells, induced to express the HTTGFP construct are cultured in medium containing small chemical compounds (libraries); analysis of the changes in GFP expression can be quantified using a microplate reader (TECAN). Compounds capable of reducing GFP expression will highlight potential pathways that could be involved in the reduction of mHTT aggregates. (J) Drosophila HD models can be used to perform genetic screens to analyze in vivo the expression of components of these pathways; (K) Drosophila HD models will be fed with the small compounds identified to analyze their effect in ameliorating animal motility or in reducing the size of mHTT aggregates; analyzed directly by immunofluorescence in the brain or by filter-trap assays using either organs or from whole animals. (H) For better performance, chemical drugs can be modified using ligand and structure-based drug design to improve their characteristics and then they can be tested again in vivo in Drosophila HD models such as in K. (I) Finally, the candidate drugs will be tested using human cells differentiated to iPSCs and then to neurons of glia, or directing to neurons iNs (Hong and Do, 2019) starting from cells of patients or from cells from healthy donors in which the specific mutation is introduced using the CRISPR/CAS9 technique.
4. Mechanism of autophagy
Autophagy is a conserved mechanism with different specific catabolic functions in different tissues and cells. For example, in conditions of nutrient deficiency it represents a survival mechanism that generates amino acids and bioenergetic substrates to allow cell survival. However, autophagy is also used to eliminate toxic debris and exhausted organelles to maintain cellular clearance, particularly relevant in aging neurons, where physiological reduction in autophagy can cause their premature loss. Autophagy can occur in three different forms: microautophagy that is mediated by small cargo-containing vesicles on the lysosomal membrane; chaperone-mediated autophagy (CMA), in which the chaperone Hsc70, a member of the Hsp70 heat shock protein family, recognizes cargo proteins containing KFERQ-like motifs and delivers them to the lysosomes via the receptor lysosome-associated membrane protein 2A (LAMP2A); and the most studied macroautophagy herein referred as autophagy, mediated by a subset of ATG proteins encoded by the ATG gene-family, originally identified in yeast in response to nutrient starvation (Fleming et al., 2022). The product of the ATGs genes is responsible for the initial formation of the phagophore that envelops cytoplasmic cargoes in a double membrane (autophagosomes) which subsequently fuses with the lysosomes for the degradation of the cargo by hydrolases. Due to the specificity of different cargoes, autophagy is now studied and classified into more specific pathways (selective autophagy) such as: lipophagy (lipid droplets), ERphagy (endoplasmic reticulum), mitophagy (mitochondria), pexophagy (peroxisomes), aggrephagy (protein aggregates) and xenophagy (bacteria and viruses; Galluzzi et al., 2017). An important step in selective autophagy is the interaction of the ubiquitinated cargo-proteins with autophagic receptors, such as p62/SQSTM1 (Ref2(P) in flies) that by binding LC3 (ATG8a in flies) via the LIRs (LC3-interacting regions) sequences, recruits cargoes into the autophagosome for degradation. The amino acids produced in the lysosome by hydrolysis of the cargo can directly re-activate the TORC1 complex, located on the lysosomal membrane, thus blocking the autophagic flux (Abu-Remaileh et al., 2017; Wyant et al., 2017). Indeed, TOR kinase, which is part of the TORC1 complex, acts as negative regulator of autophagy by phosphorylating specific sites of the Ser/Tre kinase ULK1/2 (ATG1 in flies) thus destabilizing the initiator complex of the autophagy process that is composed by ATG13, ATG101, FIP200 (Figure 2). This is an essential step for the phagophore assembly since this complex activates local phosphatidylinositol-3-phosphate (PI3P) production at membrane structures called omegasome, that recruits the WIPI2 (WD repeat domain phosphoinositide-interacting proteins) and DFCP1 (zinc-finger FYVE domain-containing protein 1). These proteins are important for the recruitment of the ATG6L1-ATG5-ATG12 complex, responsible for the ATG3-mediated conjugation of LC3 proteins to membrane phosphatidylethanolamines (PEs) to form the membrane double bond. In this process, LC3-I is cleaved at the membrane and converted into LC3-II, process that is also used as a signature of autophagic flux efficiency and typically visualized in western blot assays (Figure 2A). Ectopic expression of LC3 fused to a Fluorescent Proteins is used as a marker of the autophagosome and to quantify the autophagic vesicles (in Figure 2B is shown Atg8a-mCherry expressed in neurons of the calyx together with GFP using the Elav-Gal4 promoter). Autophagy is also regulated by AMPK, a kinase that is activated by cellular stress and low levels of ATP. AMPK phosphorylates ULK1/2 (ATG1 in flies) at sites different than those targets of TORC1. This promotes the formation of the initiator complex of autophagy (Figure 2; Kim et al., 2011). Therefore, molecules capable of inducing basal autophagy, such as rapamycin (a potent TORC1 inhibitor), or metformin (which activates AMPK) represent important therapeutic targets in proteinopathies. The use of simple animal models such as Drosophila was particularly relevant to the discovery and initial characterization of such molecules.
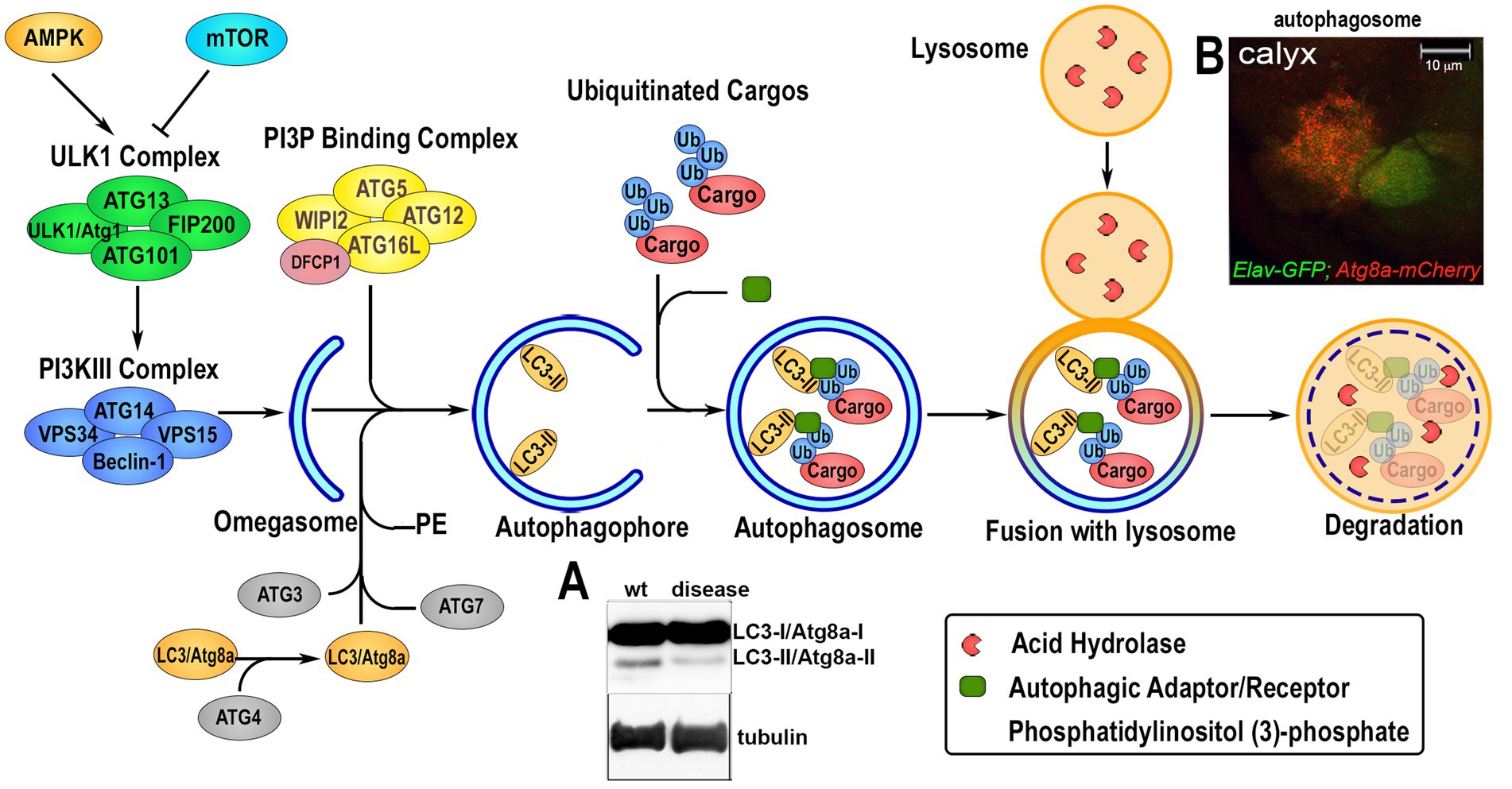
Figure 2. Overview of the autophagic flux. The induction of the autophagic process is regulated upstream by 5†’-AMP-activated Protein Kinase (AMPK) and Target of Rapamycin (TOR), that modulate the activation of the Unc51-like Kinase 1 (ULK1/Atg1) via phosphorylation and the formation of the initiator the ULK1-Complex composed by ULK1, Autophagy-related protein 13 (ATG13), ATG101 and FIP200. Upon activation, ULK1/Atg1 Complex in turn activates via phosphorylation the phosphatidylinositol-3-kinase III (PI3KIII) Complex, which comprises vacuolar protein sorting 34 (VPS34), VPS15, ATG14 and Beclin-1. This allows its translocation on the ER membrane and the production of an isolation membrane enriched in phosphatidylinositol (3)-phosphate (PI3P). PI3P induces the recruitment of the PI3P Binding Complex, consisting of ATG5, ATG12, ATG16L and WD Repeat Domain, Phosphoinositide Interacting 2 protein (WIPI2), on the growing autophagophore (omegasome) and the Double FYVE-containing protein 1 (DFCP1). This process enhances the ATG3-mediated binding of Microtubule-associated protein 1A/1B-light chain 3 (LC3) to phosphatidylethanolamine (PE) on the autophagosomal membrane where is lipidated. This leads to its cleavage from LC3-I/ATG8a-I to LC3-II/ATG8-II, which is considered a feature of an active autophagic flux and can be visualized and quantified by western blot analysis (panel A). Ubiquitin-tagged proteins are recognized by specific autophagic adaptors/receptors, such as p62/SQSTM1/Ref2(P), with a mechanism that is selective for each different organelles or cellular structure called selective autophagy. The cargo receptors bind LC3/ATG8a and transport the cargo into the autophagophore where its content is hydrolyzed upon fusion with lysosome. Autophagosome formation can be visualized in vivo by ectopic expression of LC3/Atg8a fused to GFP or mCherry fluorescent proteins that form small “puncta” on the autophagosome membrane, and fluorescence can be quantified using imaging processing programs. Panel B shows the UAS-Atg8a-mCherry staining pattern under physiological conditions analyzed in the calyx region of Drosophila larval brain, where neuronal cells are visualized by expressing UAS-GFP using the ELAV-Gal4 promoter. Co-localization of mCherry puntae within GFP is used to quantify the presence of the autophagosome in neurons.
4.1. Drosophila’s autophagy a link to neuronal degeneration
Most of the steps in the autophagy pathway were first identified and characterized in the Drosophila fat body, where there is a physiological induction of autophagy during metamorphosis to allow the animal to survive starvation (Rusten et al., 2004; Scott et al., 2004). Furthermore, because of the big size of the fat cells (50 μm), the fat body was used to better visualize the autophagosome maturation using ATG8a conjugated with mCherry or GFP (Figure 1B). Subsequently, genetic studies have delineated the important control of autophagy in longevity (Maruzs et al., 2019). First, Atg7d77 mutants showed increased oxidative stress and levels of ubiquitinated proteins, and presented defects in climbing and shortening of lifespan (Juhasz et al., 2007). Similar data were obtained with ATG8a mutants, while its overexpression was shown to promote neuronal survival by controlling oxidative stress and to induce longevity (Simonsen et al., 2008). Work in mice confirmed the role of ATG5 in promoting the survival of Purkinje cells and of ATG7 in preventing axonal degeneration (Komatsu et al., 2007; Nishiyama et al., 2007). Recently, GWAS studies in mice and human cells confirmed ATG5, ATG7 and identified ATG101 and ATG16L1 as crucial for maintaining neuronal survival in particular, ATG7 was associated with complex neurodevelopmental disorders in patients, confirming the role of these autophagic genes in the control of neuronal homeostasis (Wertz et al., 2020; Collier et al., 2021).
5. Neurodegenerative proteinopathies or (PPs)
Neurodegenerative Proteinopathies (PPs) are a large family of diseases that share common pathogenic features such as misfolded protein aggregations, neuroinflammation, oxidative stress and mitochondrial dysfunction that contribute to cellular degeneration of the neural tissue (Fleming et al., 2022). Either induced by genetic alteration or by an age-related or stochastic event, alterations of the natural conformation of proteins leads to the formation of oligomers or aggregates with consequent loss of the physiological function of the protein (Rai et al., 2022). Accumulation of aggregates in PPs is commonly due to different mechanisms that include: (i) irreversible formation of aggregates with new strong intermolecular interactions; (ii) inefficacious cell clearance; (iii) seeding and spread of pathological aggregates between cells, evident for prion-like diseases, but identified also in other proteinopathies where the presence of old aggregates function as a seed for the new one (seeds effect; Soto and Pritzkow, 2018). Thus, the central pathogenic role of protein aggregation in these diseases underlines the importance of endogenous mechanisms that control the proteostasis networks, such as the unfolded protein response (UPR), autophagy-lysosome pathway and chaperone activity (Klaips et al., 2018). In fact, age-related disruption of these proteostasis networks or their genetic alterations, contribute to the early onset of these diseases (Fleming et al., 2022; Rai et al., 2022). Deregulation of autophagy has been linked to the pathogenesis on many neuronal diseases thanks to the use of several animal models. In the current review, we will discuss only the most conserved and frequently mutated genes that are the cause of PPs (summarized in Table 1) whose characterization in flies has shown their role in protein homeostasis and autophagy, and those include HD, SCAs, ALS, AD, PD, and prion-like diseases (Pr-D).
5.1. Huntington’s disease
Huntington’s disease (HD) is an inherited neurodegenerative disorder caused by a dominant mutation in the first exon of the huntingtin (htt) gene that leads to an expansion of the CAG trinucleotide sequence (longer than 35 repeats), resulting in a protein with a long polyglutamine-Q stretch. The clinical onset of the disease is generally at middle age, however the length of the mutation is inversely proportional to the onset of the disease in fact, patients with long CAG show signs already at a young age (Gusella and MacDonald, 1995). The human htt gene encodes for a protein of 350 KDa that contains four HEAT (Huntingtin, elongation factor 3, protein phosphatase 2A and TOR kinase) domains, structurally related to the ARM (armadillo) repeats, and few PEST (peptide sequence rich in proline, glutamic acid, serine, and threonine) domains, that act as substrates for proteolytic enzymes, including caspases or calpain that cleave at amino acids 552 and 586 to produce an N-terminal fragment containing the polyQ domain (Ehrnhoefer et al., 2011). Post translational modifications (PTMs) regulates these proteolytic events and modification of the N-terminus region, such as arginine methylation (Migazzi et al., 2021; Ratovitski et al., 2022), neddylation (Ghosh and Ranjan, 2022), acetylation (Gottlieb et al., 2021), palmitoylation (Lemarie et al., 2021) and phosphorylation at Ser13 and Ser15, are relevant to modulate HTT localization and aggregate formation (Arbez et al., 2017; Chatterjee et al., 2021). These proteolytic cleavages occur also in the mutant HTT (mHTT) resulting in the formation of N-terminus truncated fragments that compete with endogenous HTT and favor the formation of toxic aggregates (Graham et al., 2006; Barbaro et al., 2015; Koyuncu et al., 2017; Ast et al., 2018). The presence of mutant HTT protein induces cellular defects leading to cell death, particularly of neurons in the striatum and cerebral cortex, leading to motor defects and rapid cognitive decline (Bates et al., 2015).
5.1.1. Huntingtin and autophagy
The physical interaction between endogenous HTT and p62/SQSTM1 was shown to facilitates the binding of HTT to ULK1/Atg1 releasing its negative regulation by mTOR, thus promoting autophagy (Rui et al., 2015). The fact that HTT protein contains sequences with structural homology to Atg23, vacuole protein 8 (Vac8ar) and Atg11, containing LC3-interacting repeats (LIRs) sequences (WxxL) present only in cargo receptors suggested a role for endogenous HTT to control autophagy (Ochaba et al., 2014; Martin et al., 2015). Further data showed that myristylation at its N-terminus directed the truncated HTT to the endoplasmic reticulum to initiate autophagosome-formation (Martin et al., 2014). Conversely, the inefficient myristoylation of the mutant HTT promoted the formation of large aggregates that bind p62/SQSTM1 but were not transported to the cargo resulting in an empty autophagosome (Martinez-Vicente et al., 2010; Martin et al., 2015). The human huntingtin gene (htt) is conserved across evolution and its homologue in flies, dhtt, encodes for a protein that shares an overall 24% identity with the human counterpart but contains only one CAG triplet positioned at its N-terminus (Li et al., 1999). While in vertebrates the function of htt is essential for development and is required for neuronal survival (Nguyen et al., 2013), flies dhtt is not necessary for development. However, dhtt knockout animals exhibit a reduced Mushroom body, learning impairment, age-related impaired of motility and shortened lifespan (Zhang et al., 2009). The role of endogenous HTT in autophagy was first outlined in dhtt mutant animals that showed defective developmental autophagy and increased ubiquitination of p62/Ref(2)P (Drosophila’s p62/SQSTM1; Ochaba et al., 2014) and like its orthologue in vertebrates, Ref(2)P accumulated in brains of aged animals (Nezis et al., 2008; Bartlett et al., 2011). Furthermore, endogenous dhtt was shown to counteract the toxic effect of ectopic expression of the mutant human htt-exon-1-Q93 (Zhang et al., 2009), corroborating the protective role of endogenous HTT evidenced also in mice (Van Raamsdonk et al., 2005). More recently, loss of function mutation in the dhtt gene was shown to enhance defects in axon outgrowth in the Mushroom body due to decreased function of the App gene, the Drosophila orthologue of the human amyloid beta-precursor protein (APP) gene, responsible of Alzheimer’s disease, suggesting an interaction between those genes that may control the onset of AD (Marquilly et al., 2021).
5.1.2. Drosophila models of HD
A few models of Drosophila HD have been generated over the years (Lewis and Smith, 2016). They are mostly based on the expression of the N-terminal fragments of the human htt gene or on the use of htt-exon-1 mutated to carry different lengths of the CAG tract; while few models have been generated using the full-length mutant human gene due to the high toxicity of its expression when carrying the polyQ expansion tract (Romero et al., 2008). Because of their viability, flies carrying mutations in the htt-exon-1 (mHTT) were widely used to study the function of the polyQ, and to demonstrated that the length of the CAG tract is proportional to a progressive neuronal degeneration and decline in animal motility (Jackson et al., 1998; Marsh et al., 2000; Krench and Littleton, 2017). Furthermore, expression of mHTT in neurons showed a proportional increase in the number and size of aggregates that were already visible at 48–72 h of development (Figure 1F), indicating that Drosophila could be an efficient model to study the kinetic and inhibitors of aggregates formation in vivo. Indeed, genetic screens for aggregate-phenotype modifiers, identified several pathways including TOR signaling (Ravikumar et al., 2004; Sarkar et al., 2007), the chaperons CCT (Sajjad et al., 2014; Pavel et al., 2016), histone deacetylase (Pallos et al., 2008; Jia et al., 2012), antioxidant pathways (Mason et al., 2013), deubiquitinating enzymes (Aron et al., 2018), enzymes involved in glutamine metabolism (Vernizzi et al., 2020), early endosomal protein Rab5 (Ravikumar et al., 2008), Puromycin-sensitive aminopeptidase (PSA; Menzies et al., 2010) and very recently the novel chemical compounds that function as linkers between mHTT and LC3 able to target only the mutant form of HTT to the autophagosome leaving intact the wild-type allele in a selective manner (Li et al., 2019).
5.2. Spinocerebellar ataxias
Spinocerebellar ataxias (SCAs) are a heterogeneous group of inherited disorders that affects the spinal cord and the cerebellum and are characterized by loss of Purkinje cells and cerebellar atrophy. SCAs present mutations in 40 different genes many of which carry an expansion of the polyglutamine tract such as SCA1, SCA2, SCA3/MJD (Machado-Joseph disease), SCA6, SCA7, SCA17 and DRPLA (Dentatorubral-pallidoluysian atrophy; Klockgether et al., 2019). Here we will focus only on the functional mechanisms characterized in Drosophila models for SCA1-3 (Warrick et al., 2005), since models for CACNA1A responsible for SCA6 (Tsou et al., 2016; Sujkowski et al., 2022), SCA7 (Jackson et al., 2005), SCA17 (Ren et al., 2011) and DRPLA (Charroux and Fanto, 2010) only describe motility defect in the adult flies.
5.2.1. SCA1
SCA1 is an autosomal dominant disease caused by an expansion of the CAG triplet in the coding region of the Ataxin1 (ATXN1) gene. This gene encodes for a protein with RNA binding capacity that associate to transcriptional regulators at promoter regions (Yue et al., 2001). Patients affected by SCA1 represent 6% of the individuals affected by cerebellar ataxias, and their mutant ATXN1 carries a tract of more than 39 CAG as compared to approximately 20 in the wild-type gene. The mutation leads to the production of a protein with a long polyQ that forms insoluble aggregates, visible in the nuclei of the Purkinje cells (Stoyas and La Spada, 2018). Furthermore, ATXN1 has been shown to favor the toxicity of human pathogenic ATXN2 in mice-models; on the contrary loss of function of ATXN1 increased the stability of BACE1-mRNA, enhancing amyloidogenic cleavage of APP in a mouse model of AD (see Section 5.4.1) and outlining a function of ATXN1 also in AD (Suh et al., 2019).
5.2.1.1. Ataxin1 and autophagy
A specific function for Ataxin1 in controlling the autophagic pathway in flies has not been described, however TOR inhibitors in flies were shown to ameliorate the toxic effect of mutant Ataxin1 through the release of the inhibitory role or TORC1 on the lysosomal transcription factor Mitf (Drosophila homologue of human TFEB; Bouche et al., 2016). Drosophila harbors a gene encoding for Ataxin1 (dAtx) that lacks the polyQ domain, nevertheless its overexpression led to phenotypes like those obtained by human ATXN1 overexpression, but different from those observed upon overexpression of the polyQ alone. Genetic experiments showed that Atx1 interacts through its conserved AXH domain with the zinc-finger transcription factor Senseless (Sens). This mechanism is conserved for its mammal’s homolog, the growth factor independence-1 (Gfi-1) and proposed as a potential pathogenic mechanism for SCA1 (Tsuda et al., 2005).
5.2.1.2. Drosophila models of SCA1
Expression of the human mutant ATXN1 gene in Drosophila’s neurons leads to retinal degeneration and loss of axonal projections of the interneurons in the ventral nerve cord of the brains (Fernandez-Funez et al., 2000), expression of the pathogenic form of SCA1 or SCA3 in Drosophila larval dendritic neurons reduced their arborization with disruption of F-actin dendritic structures, defect that is partially rescued by Rac-PAK signaling activation (Lee et al., 2011). In vivo screens using the adult eye helped define the role of chaperones to proteins with an expanded polyQ. In particular, the chaperone-dependent E3 ubiquitin (Ub) ligase CHIP (carboxyl terminus of Hsp70-interacting protein) and the NAD synthase nicotinamide mononucleotide adenylyl transferase (NMNAT) together with Hsp70 mediate activation of the proteosome pathway (Zhai et al., 2008). Genetic experiments have delineated the role of transglutaminases (TGs), a class of enzymes that catalyzes the formation of cross-links between glutamine residues within proteins thus increasing their stiffness and insolubility (Muma, 2007). The relevance of these enzymes in proteotoxicity and autophagy was first demonstrated using a Drosophila model for PD and later also in HD, where TG2 phosphorylation by PINK blocked its degradation favoring proteotoxicity and mitochondria degradation. Further experiments, both in Drosophila and in human cells, showed that TG2 reduction ameliorates the defects in both PD and HD (Karpuj et al., 2002; McConoughey et al., 2010; Min et al., 2015). Similarly, a role for TG5 in enhancing the toxicity of mutant ATXN1 was shown in Drosophila models and in cells from SCA1-patients where TG5 was shown to colocalize with Ataxin1 (Lee et al., 2022b) further supporting a functional role for this enzymes in the disease. Genetic screen and chemical inhibitors revealed the RAS-MAPK–MSK1 as an important axis in the control of Ataxin1 aggregate formation (Park et al., 2013). Using small inhibitors of the MSK1/MAPK complex it has been demonstrated in mice and flies that the phosphorylation of Ataxin1 on Ser776 is relevant for its stability, while mutation of this residue in the wt protein reduced the level of Ataxin1-82Q in mice suggesting that Ser776 could represent a novel substrate for both drug and allele-specific therapies (Nitschke et al., 2021).
5.2.2. SCA2
Mutations in the human Ataxin2 (ATXN2) gene lead to spinocerebellar ataxia type 2 (SCA2) which accounts for 13% of spinocerebellar ataxias (Klockgether et al., 2019). Mutations in ATXN2 with an expansion greater than 32 CAG repeats forms insoluble cytoplasmatic aggregates visible in Purkinje and granule cells, leading to gliosis and neuronal death (Huynh et al., 2000). Additionally, mutations in ATXN2 were found to be a risk also for ALS due to their effect on increasing TP-43 toxicity (van den Heuvel et al., 2014).
5.2.2.1. Ataxin2 and autophagy
Not many studies ruled out the function of autophagy in SCA2 until recently, when it was shown that autophagy ameliorates mitochondrial dysfunction and oxidative stress in mice models for SCA2 and in cells from patients (Wardman et al., 2020). In addition, it was shown that cordycepin, a drug that activates AMPK and autophagy, reduces the abnormal accumulation of p62/SQSTM1 and of LC3 observed in cells form SCA2 patients, indicating a dysfunction in the autophagic flux in the disease (Marcelo et al., 2021). Drosophila possess one orthologue of the ATXN2 gene (Atx2) which encodes for a protein Atx2 that shares 23% of identity and 36% of similarity with the human Ataxin2 protein, but differently Drosophila Atx2 does not contain a long polyQ stretch but three short separate segments of glutamines. Structurally human Ataxin2 contains the Lilke-Sm (Lsm) and Lsm-AD domains found in RNA binding proteins (Satterfield and Pallanck, 2006), and further experiments in human and flies confirm that both Ataxin2 and Atx2 physically bind with the polyribosome suggesting that Ataxin2 may be involved in the control of translation (Satterfield et al., 2002).
5.2.2.2. Drosophila models of SCA2
Expression of human mutant ATXN2 in neurons resulted in the formation of aggregates leading to neuronal degeneration that (Lessing and Bonini, 2008). Further work demonstrated that overexpression of Drosophila Atx1 promotes the accumulation of human mutant Ataxin2 aggregates, indicating that the interaction between the two proteins could contribute to the pathogenesis of SCA1 and SCA2. This hypothesis is in line with the idea of a cooperative mechanism between Ataxin1 and Ataxin2, also suggested by the presence of Ataxin2-enriched aggregates in postmortem neurons of patients with SCA1 (Al-Ramahi et al., 2007). Further studies should be conducted using Drosophila’s model of SCA2 to better understand the physiological role of Ataxin2/Atx2 in the control of autophagy and how it could be perturbed in SCA2, to reveal novel mechanisms of intervention.
5.2.3. SCA3
Also known as Machado-Joseph disease, SCA3 is the most common type of spinocerebellar ataxia. It is characterized by a progressive neurodegeneration caused by an expansion of CAG at the 3′ end of the coding region of the ATXN3 gene, which from 12 to 40 repeat in the wild-type reaches more than 55 CAGs in the mutant form (Klockgether et al., 2019). The gene encodes for a deubiquitinase (DUB), a class of enzymes that counteracts the action of ubiquitin and important for the control protein stability of TP-43 and HTT (Doss-Pepe et al., 2003; van Well et al., 2019; Tran and Lee, 2022), responsible when mutated of ALS and HD, making DUBs potential targets for proteinopathies.
5.2.3.1. Ataxin3 and autophagy
Autophagy is compromised by mutation of ATXN3 since the expression of the autophagic markers p62/SQSTM1 and LC3 was found abnormal in the brain of patients with SCA3. Furthermore, transgenic mice expressing ATXN3-71Q exhibit large aggregates that are reduced in the presence of the autophagy-promoting gene Beclin1 (Nascimento-Ferreira et al., 2011). Puromycin-sensitive aminopeptidase (PSA), a cytosolic enzyme able to digest polyQ sequences (Menzies et al., 2010) was shown to ameliorate also Ataxin3 phenotypes both in flies and mice and to induce autophagy (Menzies et al., 2010). Drosophila, like mammals has two isoforms of Atx3 proteins that contain either two or three ubiquitin interacting motifs (2UIM) with an additional at its C-terminus (3UIM; Johnson et al., 2019). Studies in mammals and flies highlighted the different ability of these isoforms to form aggregates when carrying the polyQ expansion. In particular, 2UIM is more prone to form aggregates than 3UIM, that is rapidly degraded mainly through the proteasome pathway (Harris et al., 2010). Further studies in flies showed that the UIMs motifs of Atx3 interact with the heat shock protein cognate-4 (Hsc70-4) to enhance Ataxin3 mutant aggregation and toxicity (Johnson et al., 2021). This data complement those showing that the auto-protective function of Drosophila endogenous Atx3 depends on its deubiquitinating catalytic activity that indirectly reduce the folding of the toxic polyQ present in mutant Ataxin3 proteins as in others polyQ proteins (Warrick et al., 2005), rather than on the activation of the proteasomal degradation pathway (Tsou et al., 2015).
5.2.3.2. Drosophila models of SCA3
The most widely used models for SCA3 express the C-terminal fragment of the human ATXN3 gene with 78CAGs or 82CAGs repeats, that in the retinal neurons cause abnormal eye morphology due to neuronal and tissues degeneration and motility defects (Warrick et al., 1998, 2005). Genetic screens for modifiers of the Ataxin3 mutant phenotypes led to the identification of several genes of the ubiquitin pathways, members of Hsp70/chaperone proteins, and potential regulators of RAN translation, a Repeat-associated non-AUG (RAN) translation that generates toxic dipeptide repeat proteins (DPRs) from pathological repeat RNA expansions hat do not contain the classical methionine start codon (Cleary et al., 2018), see also for C9orf72-ALS (Section 5.3.1). Genetic screen in flies, using the eye, identified members of the ubiquitin ligase family, such as Cullins and of Praja1, that protect against the effect of mutant Ataxin3 strongly supporting the relevance of the ubiquitin and of the DUBs in the disease (Chen et al., 2019; Ghosh et al., 2021). This screens also identified selective modifiers of Ataxin3 able to rescue also Tau-R406W-AD mutation (Section 5.4.2) supporting the existence of common pathways that converge and contribute to neuronal degeneration, controlled by the Ataxin3 DUB-activity (Bilen and Bonini, 2007). In addition, Ataxin3 DUB-activity controls parkin’s ubiquitination in the modulation of “physiological” mitophagy (Durcan and Fon, 2011), and ubiquitination of K63 in the formation of SOD1 aggregates (Wang et al., 2012). Treatment with antioxidant drugs such as the Nrf2-inducers caffeic acid and resveratrol have also been shown to indirectly reduce apoptosis and to induce autophagy, both in human cells and in Drosophila models of SCA3 (Wu et al., 2017).
5.3. Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is a rare disease, with an incidence of 2–3 per 100,000 and a variable progression rate. ALS patients are diagnosed based on the degeneration of motor neurons, however, the pathophysiological heterogeneity of the disease is accompanied by a variety of other factors that are not completely understood. ALS is often associated with frontotemporal dementia (FTD), a disease that presents common mutant genes with ALS resulting in mental disability without motoneurons degeneration or movement impairment. Genetics ALS accounts for only 10–15% of all ALS cases, while the remaining are of sporadic, however up to now, more than 50 genes have been implicated in ALS and the pool of loci associated with this disease is expanding due to GWAS and whole-exome/genome sequencing. Among new pathways identified there are members of sphingolipid signaling and actin polymerization, identified in the vesicles transport network (Brenner and Freischmidt, 2022) suggesting how complex is the physiopathology of this disease. Autophagy in ALS. Deregulation of autophagy has been linked to pathogenesis of ALS thanks to the use of several animal models. ALS patients present accumulation of autophagosomes in the cytoplasm of spinal cord neurons and harbor mutations in genes involved in the autophagy machinery. Many genes have been associated to ALS and here, we will focus on C9orf72, SOD1, TD-P43, FUS since their characterization in flies has been of crucial importance to show their role in protein homeostasis and autophagy.
5.3.1. C9orf72
Mutations in the C9orf72 gene are characterized by an expansion of the hexanucleotide sequence GGGGCC (G4C2) in the first intron of the gene, which can reach up to thousands of repeats in the most affected patients (DeJesus-Hernandez et al., 2011; Renton et al., 2011). The gene encodes for a guanine nucleotide exchange involved in cellular processes such as vesicular trafficking, autophagy, lysosomal function and in the control of the immune system and recently described as part of the lysosomal membrane-complex that binds RABs for a correct lysosomal function (Taylor et al., 2016; Root et al., 2021). C9orf72 mutation causes toxicity and alteration of the autophagy-lysosomal pathway contributing to disease pathology (Beckers et al., 2021). Hypothesis regarding the consequences of C9orf72 mutation range from loss of gene function due to epigenetic transcriptional silencing of the locus, to RNA-mediated toxicity caused by the formation of RNA foci that trap both RNA transcripts and RNA binding proteins, and to the formation of toxic dipeptides (DPR) produced by non-AUG translation (RAN) associated with intronic hexanucleotide repeat expansion (Kwon et al., 2014; Wen et al., 2014; Freibaum and Taylor, 2017). The presence of DPRs also compromises the nucleocytoplasmic transport, resulting in the accumulation of toxic debris in the cytoplasm leading to neuronal death (Freibaum et al., 2015; Zhang et al., 2015). Here we have outlined few of these events and discussed their relationship with autophagy.
5.3.1.1. C9orf72 and autophagy
The relationship between C9orf72 and autophagy is somewhat controversial, since some data showed that mutation in C9orf72 cause toxicity and alteration of the autophagy-lysosomal pathway (Beckers et al., 2021), others showed that C9orf72 acts as a negative regulator of autophagy as mice mutant for the C9orf72 gene showed reduction TOR activity with nuclear translocation of TFEB and activation of the autophagy flux (Ugolino et al., 2016). Moreover, DPRs induce dysfunctions of the autophagic-lysosomal pathway by synergizing with other mechanisms of toxicity increasing the pathogenesis of the disease (Beckers et al., 2021). Growth factors and insulin/IGF influences neurodegenerative diseases including AD, PD and ALS. indeed C9orf72-G4C2 mutation has been shown to downregulate insulin/IGF signaling in both fly and human cells while activating insulin/IGF signaling enhances the toxicity of poly-GR repeats (Atilano et al., 2021). IGFs also act as neurotrophic factors for the survival of motor neurons and have therapeutic effects in a mouse model for SOD1-ALS, and in the murine motoneurons NSC34 cells expressing mutant C9orf72-G4C2 (Kaspar et al., 2003; Stopford et al., 2017). Using high-throughput screen to identify chemical modulators of DPRs, we also found that cells expressing mutant C9orf72-G4C2 have increased cAMP levels resulting in protein kinase A (PKA) activation, event that was partially rescued by pharmacological or genetic inhibition of PKA (Licata et al., 2022). Since PKA is activated by growth factors, this observation raises the question of a connection between growth pathways and C9orf72 in ALS and, although the mechanisms are not yet fully understood, we presume that they may converge to the control of cellular proteostasis or autophagy.
5.3.1.2. Drosophila models of C9orf72
Although Drosophila does not have a C9orf72 orthologue, transgenic models have been established and mimic the toxicity in humans due to its gain of function effect in flies (Mori et al., 2013; Xu et al., 2013; Mizielinska et al., 2014). The insertion of repeating sequences of GGGGCCs at different lengths was used to determine their different toxicity and ability to form structures called foci that include RNA-binding proteins and RNA (Mori et al., 2013; Mizielinska et al., 2014). RNA of the DPRs obtained by RAN translation was optimized in vitro to express as specific peptides (poly GA, GP, GR, PA, alone or fused with GFP or FLAG tags; Cleary et al., 2018). The difference in toxicity for each DPR is related to their different chemical composition and atomic charge. Indeed, highly arginine-rich GR and PR have been shown to alter the phase separation of LCD-containing proteins by changing the charge and structure of membrane organelles and their dynamics and functions (Wen et al., 2014; Lee et al., 2016). Works in flies evidenced a direct role of C9orf72-G4C2 in autophagy that led to the identification that its expression induced the accumulation of Ref(2)P and of poly-ubiquitinated proteins both in motor neurons and in whole larvae, due to defects in cargo protein degradation. These flies showed defects in lysosome formation, accompanied by reduced Mitf/TFEB nuclear localization (Cunningham et al., 2020). Recent work has demonstrated that nuclear transport of TFEB is mediated by nucleoporins (nuclear import proteins) and their expression is based on the molecular chaperone SIGMAR1/Sigma-1 (sigma-1 intracellular non-opioid receptor) mutated in ALS (Wang et al., 2022). Overexpression of SIGMAR1 in flies restores proper cellular transport, outlining the relevance of this chaperonin in nuclear import in this disease (Lee et al., 2020a).
5.3.2. Superoxide dismutase
Superoxide dismutase (SOD1) is the second most frequently mutated gene in ALS, representing approximately 20% of familial ALS. This gene encodes for SOD1 a ubiquitously cytosolic Cu/Zn-binding enzyme that homodimerizes and catalyzes the dismutation of superoxide radicals to hydrogen peroxide counteracting the toxic effect of free reactive oxygen species (ROS). SOD1 modulates protein quality control, autophagy, mitochondrial function, and axonal transport. SOD1 mutations may lead to both loss or gain of function phenotypes, making difficult to interpret the mechanisms through which its mutation leads to ALS (Brenner and Freischmidt, 2022).
5.3.2.1. Superoxide dismutase-ALS and autophagy
wt SOD1 was found in a complex with Atg9/Beclin1 to control P62/SQSTM1 expression, indeed mice mutant for SOD1-ALS show reduced Atg9/Beclin1 and increased level of P62/SQSTM1 and of ubiquitinated proteins (Nassif et al., 2014). Work in Drosophila as well as in C. elegans, identified a role for lethal(2) malignant brain tumor (L3MBTL1) a histone methyl-lysine protein, and SET domain-containing protein-8 (SETD8) in the control protein misfolding and degradation as their reduction alleviate the phenotypes induced by SOD1. This mechanism is conserved also in mammals underlining the role of chromatin modification in the control of protein quality control (Lu et al., 2019). As we know the interplay of ubiquitin-mediated autophagy is controlled also by deubiquitinases (Clague et al., 2019) and studies in Drosophila, as in other invertebrate models, outlined the negative role of the DUB-USP7 that by reverting the ubiquitination of the E3-ligase NEDD4L, reduced autophagy and the clearance of misfolded SOD1 and TDP-43 proteins by reducing the SMAD2/TGF-beta pathway (Zhang et al., 2020).
5.3.2.2. Drosophila models of SOD1
Drosophila endogenous dSOD1 is necessary for neuronal health as its null mutation results in neuronal loss. Co-expression of a mutant dSOD1 allele, carrying specific structural LOF mutations, results in formation of unfunctional wt-SOD1/dSOD1 heterodimers indicating that functional dimers are necessary for the activity of the enzyme (Phillips et al., 1995). Studies in Drosophila and in mice demonstrated that expression of human SOD1 in motor neurons reduced ROS activity, leading to longevity (Parkes et al., 1998; Missirlis et al., 2001). Further studies in flies demonstrated that the enzymatic activity of human SOD1 is not required for its toxicity: In fact, homozygous inactive SOD1 mutants (G85R, H48R, and H71Y) expression in dSOD1 null flies results in neurodegeneration, motor defects, and shortened life span, suggesting that these phenotypes are associated to SOD1-mutations rather than SOD1 activity (Şahin et al., 2017; Agudelo et al., 2020). Expression of mutants SOD-1 results in deposition of protein aggregates visible in neurons, glia and in skeletal muscles in both mice-of ALS and cells from ALS patients (Dirren et al., 2015). Mutations in SOD1 lead to mitochondrial impairment, a defect that also occurs in Drosophila, in fact mutant SOD1 accumulates in the intermembrane space of the mitochondrial membranes modifying their structure, reducing the production of ATP and causing metabolic dysfunctions particularly relevant for the activity of motor neurons (Tafuri et al., 2015; Gallart-Palau et al., 2016). A distinctive characteristic of SOD1 mutations is the presence of non-autonomous signals from glia that induce lethality in neighboring cells. This effect was demonstrated in vivo in Drosophila and mice models for SOD1-ALS (Boillee et al., 2006; Watson et al., 2008) and supported by data showing that ablation of the glia ameliorate the disease in mice mutants for SOD1-ALS (Guttenplan et al., 2020). The importance of non-autonomous signaling has been strengthened by recent work demonstrating that motor neurons from SOD1-G93A mutation-bearing mice exhibit metabolic changes that affect surrounding myocytes (Peggion et al., 2022), and likewise between glia and neurons to induce survival of neurons (Chua et al., 2022). Understanding the nature of the non-autonomous signals is relevant to better define the mechanisms in the pathogenesis of ALS and to identify new components and modalities of therapeutic intervention. Thus, the design of Drosophila ALS models using the combination of the two binaries, LexA-LexOp with UAS-Gal4, may allow for the expression of SOD1 mutations in a specific tissue, for example motor neurons, and the concomitant manipulation of glia, using the UAS-Gal4 where many lines are available at stock centers. This would allow to rapidly identify genes that can interfere/suppress SOD1 neuron lethality similar to the approach used for identify mHTT non-autonomous interactors (Bason et al., 2019).
5.3.3. TAR DNA-binding protein 43
TAR DNA-binding protein 43 (TDP-43) is a DNA/RNA-binding protein that belongs to the heterogeneous nuclear ribonucleoprotein (hnRNP) family with both nuclear and cytoplasmic functions. Functionally, TDP-43 is involved in modulation of several aspects of RNA life such as transcription, splicing, stability and turnover, degradation, alternative polyadenylation, transport, translation, and microRNA biogenesis [165]. TDP-43 is considered a key protein in ALS for two main reasons: firstly, TDP-43 is the major component of the ubiquitin-positive cytoplasmic inclusions found in spinal motor neurons of ALS patients, secondly, mutations in its gene (TARDBP) occur in around 0.9% of ALS patients.
5.3.3.1. TAR DNA-binding protein 43 and autophagy
Autophagy plays an important role in the clearance of the cytoplasmic inclusions in fact, it has been shown that the TDP43 aggregates colocalize with markers of autophagy and inhibition of autophagy increases their aggregates formation (Brady et al., 2011). Moreover, TDP-43 itself modulates autophagy, therefore creating a complex scenario whose perturbation contributes to ALS (Huang et al., 2020). Despite the recognized importance of TDP-43 in ALS, it is still unclear whether the pathogenesis of ALS is related to its reduced physiological function, since wild-type TDP-43 is sequestered in inclusion bodies and unable to interact with its physiological targets, or if it is due to the formation of toxic aggregates containing TDP-43 (Scotter et al., 2015). Loss of TARDBP in mammals downregulates histone deacetylase 6 (HDAC6), an enzyme that controls ubiquitinated protein and autophagy (Fiesel et al., 2010). Controversial results have been obtained in TARDBP knockout cells, where although the lysosomal transcription factor TFEB is upregulated, accumulation of immature autophagic vesicles and reduced expression of Atg7 are observed, suggesting that reduction of TARDBP affects other signals/genes necessary for a functional autophagy (Xia et al., 2016).
5.3.3.2. Drosophila models of TDP-43
Drosophila harbors a TARDBP orthologue, namely TBPH, and several groups have investigated its function to gain insights into ALS pathogenesis by performing loss of function studies. Interestingly, Atg7 overexpression suppresses the semi-lethality of TBPH null flies, ameliorating motility, lifespan and reducing the accumulation of Ref2(P)/P62 inclusions at the NMJ (Donde et al., 2020). Other studies in flies addressed the consequences of overexpressing wild-type or disease forms of TDP-43. Overexpression of a cytoplasmic wt-TDP-43 or of TDP-43-M337V mutant in the fly’s retina results in neurodegeneration that can be rescued by co-expression the heat shock protein HSP67Bc that promotes autophagy (Crippa et al., 2016). More recently, HEXA-018, a novel chemical compound that induces autophagy in a TOR-independent manner and ameliorates climbing activity and lifespan was identified as a suppressor of TDP-43 overexpression in flies (Lee et al., 2021).
5.3.4. Fused in sarcoma
Fused in sarcoma (FUS) is a DNA/RNA-binding protein involved in DNA repair and RNA processing, often found in stress granules (SGs) dense structures present in the cytosol composed of RNA and RNA binding proteins (Wolozin and Ivanov, 2019). Over 70 mutations in the FUS gene have been identified in patients with familial and sporadic ALS, the vast majority of which are heterozygous mutations with autosomal dominant inheritance. Considering that most mutations influence the nuclear localization signal (NLS) of the protein, recent studies have delineated the presence of intrinsically disordered regions often enriched with arginine and glycine repeats that may be prone to promote protein aggregation, when mutated in ALS perturbating SGs dynamics that may be at the heart of ALS (Vance et al., 2009; Rhine et al., 2020). RNA binding proteins have multivalent domains important for their behavior; indeed proteins like TDP-43 and FUS, that exhibit spontaneous liquid–liquid demixing upon interaction with specific targets during phase separation, have been found to interact genetically when mutated, further accelerating neurodegeneration (Lanson et al., 2011). WT and mutant FUS are incorporated into a variety of RNA granules and they accumulate in de novo paraspeckles described in spinal motor neurons of ALS-patients (An et al., 2019).
5.3.4.1. Autophagy and FUS
Similarly, to TP-43, autophagy seems to play a crucial positive role in the elimination of toxic aggregates also in FUS-related ALS. Expression of wild-type FUS and ALS-related FUS mutations triggers mechanisms that lead the accumulation of toxic cytoplasmic aggregates and to inhibition of autophagy (Brunet et al., 2021). Furthermore, work in flies and human iPSCs demonstrated that the P525L-FUS mutation alters stress granule dynamics resulting in inhibition of PI3K/AKT/TOR signaling which indirectly increases autophagy by a yet unknown mechanism (Marrone et al., 2018).
5.3.4.2 Drosophila models of FUS
Genetic screens using flies identified Mask, an Ankyrin-repeat containing protein, that promotes the expression of the proton-pumping vacuolar (V)-type ATPases, favoring the elimination of FUS aggregates via lysosomal autophagy (Zhu et al., 2017). Recently, attention has been placed on post translational modifications that interfere with the protein phase separation of FUS, indeed the enzyme glutathione transferase omega 2 (GstO2) was shown to reduce cytoplasmic FUS level and the formation of FUS aggregates both in Drosophila and in mouse neuronal cells overexpressing FUS. Glutathionylation of FUS promotes its separation into the liquid phase suggesting how an accurate analysis of the mechanism driving protein phase separation might be a promising target for novel therapeutic strategies (Cha et al., 2022).
5.4. Alzheimer’s disease
Alzheimer’s disease (AD) is the most common form of dementia in elders (Gonzales et al., 2022), characterized by progressive neurodegeneration and cognitive impairment. This pathology is considered a proteinopathy because it is characterized by the accumulation of extracellular amyloid plaques, enriched in amyloid-β peptide (Aβ42), intracellular neurofibrillary tangles, composed of hyperphosphorylated Tau, and reactive gliosis. Most cases of AD are sporadic and only a small percentage shows clear familial autosomal dominant inheritance: the familial cases are predominantly early-onset forms associated with fully penetrant mutations in the Amyloid precursor protein (APP), and the γ-secretases Presenilin 1 and 2 (PSEN1 and 2; Karch et al., 2014). APP encodes for a type I transmembrane protein involved in several neuronal functions, many of which have been discovered and studied in Drosophila (Gunawardena and Goldstein, 2001). APP can be processed following two different pathways: the so-called amyloidogenic pathway, consisting in a subsequent action of β- and γ-secretases (BACE and PSENs) that leads to the formation of several fragments including the Aβ, and the non-amyloidogenic pathway, where the α- and γ-secretases lead to the production of different APP fragments (Vassar et al., 2009; Zhang et al., 2014). The fruit fly represents a powerful model to study AD in fact, the genes associated with AD are evolutionary conserved and models obtained by overexpression of human Aβ peptide, Tau or of APP carrying pathological mutations mimic neurodegeneration and deficit in memory/cognitive abilities.
5.4.1. Amyloid precursor protein and amyloidopathies
Mutations in APP, PS1, and PS2 genes, favor the production of the Aβ42 peptides and this, together with the presence of the amyloid plaques, shaped the “Amyloid Hypothesis” that proposes the imbalance between production and clearance of Aβ42 as the main cause of AD development (Selkoe and Hardy, 2016). To date, therapies for AD are still missing, underlying the need to better understand APP function/processing and the consequences of Aβ42 and tau aggregation.
5.4.1.1 Autophagy in APP and amyloidopathies
Autophagy plays a complex and dual role in AD since it regulates both Aβ eneration and clearance (Mputhia et al., 2019). Several studies support a protective role: Atg5, Beclin1, and ULK1 were shown to be involved in Aβ degradation (Tian et al., 2011) and the adaptor protein AP2, a member of a complex responsible for the internalization of cargos in clathrin-mediated endocytosis, was shown to induce the degradation of the APP-βCTF (C-terminal fragment) via autophagy, affecting Aβ production (Tian et al., 2013). Moreover, autophagic degradation of APP-CTF and Aβ is promoted by NRBF2, a member of the class of III phosphatidylinositol 3-kinase (PtdIns3K) complex that regulates autophagosome maturation (Cai et al., 2021). On the other hand, activation of autophagy might play a detrimental role in AD depending on the context. For example, activation of TFEB, plays opposite effects on Aβ production in neurons depending on the expression level of APP or of the βCTF (Yamamoto et al., 2019; Song et al., 2020).
5.4.1.2 Drosophila models of APP and amyloidopathies
Drosophila harbors an orthologue of APP, namely APPL and since the Aβ sequence is not conserved Drosophila’ models of AD that contributed to gain insight into the role of autophagy were mostly obtained by overexpression of either human Aβ peptide or of human AD-associated genes. The protective action of autophagy has been proven by modulating the levels of autophagic activity in flies expressing human Aβ peptide. For example, lowering the expression of autophagy-related genes, such as Atg1, Atg8a and Atg18, strongly enhances the neuronal toxicity caused by Aβ expression (Omata et al., 2014). In line with this, Atg8a overexpression in an Alzheimer Drosophila model obtained by overexpression of human amyloid precursor (hAPP) and beta-secretase 1 (hBACE1), enhances stress-resistance, slows down neurodegeneration and improves lifespan (Tsakiri et al., 2021). In a similar AD model, promotion of autophagy by the Nicotinamide mononucleotide adenylyltransferase (NMNAT) reduced amyloid plaques formation (Zhu et al., 2022). In flies overexpressing Aβ42, the protective role of autophagy was demonstrated by the expression of Thioredoxin-80 (Trx80), which prevents the accumulation of the toxic peptides in the brain and rescues lifespan by inducing autophagosome formation (Gerenu et al., 2021). On the other hand, Ling and colleagues showed that dysfunctional AEL (autophagy-endosomal-lysosomal) plays a crucial role in Aβ42 accumulation. In fact, the abnormal accumulation of Aβ42 within AEL vesicles was reduced when the functional autophagy was decreased (modulation of Atg5 or Atg12 levels), suggesting that dysfunctional autophagy–endosomal–lysosomal may cause Amyloid-like plaque formation (Ling et al., 2014). Moreover, it was recently shown that ectopic APP expression is sufficient to trigger aberrant autophagy through its interaction with the carboxyl-terminus of Hsc70-interacting protein (CHIP), a U-box type chaperone associated E3 ubiquitin ligase via transcriptional upregulation of BACE1 and PSEN inducing an AD-like neurodegeneration (Zhuang et al., 2020).
5.4.2. Tau and tauopathies
Tau is a group of six microtubule-associated proteins all derived from alternative splicing of the MAP/Tau gene, they are abundant in the CNS, where they are necessary for the stabilization of the microtubules important for axonal transport. In the presence of specific mutations or during aging, Tau becomes hyperphosphorylated and changes its structure, decreases its ability to bind microtubules, leading to the formation of insoluble filaments that accumulate as neurofibrillary tangles resulting in tauopathies, a hallmark of AD (Goedert et al., 2017; Hernandez et al., 2022). Recent work pointed to a role of Tau within the nucleus where it associates with nucleolar proteins, suggesting the presence of other mechanisms activated by mutation of Tau (Anton-Fernandez et al., 2022).
5.4.2.1. Autophagy in tauopathies
Autophagy plays a protective role in tauopathies, as it favors the clearance of the soluble and the insoluble Tau form the last present in the aggregates. Furthermore, autophagy helps the movement of Tau through vesicles involved in its transport into neurons.
5.4.2.2. Drosophila models of tauopathies
Drosophila possesses a tau gene with 46% identity and 66% similarity to the corresponding human Tau. Flies have been successfully used to pin down numerous pathways that control Tau toxicity via autophagy. For example, insulin signaling can induce abnormal Tau phosphorylation and accumulation, inhibiting its autophagic clearance and worsening the disease (Chatterjee et al., 2019). Silencing calpain, a family of proteases that inhibit autophagy, is protective against Tau toxicity in the Drosophila-eye, an effect that requires a functional autophagy since it was lost in Atg8a mutant flies (Menzies et al., 2015). A microRNAs screen aimed at identifying suppressors of Tau-mediated phenotypes, clarified that CG11070, a target of miR-9a mediates Tau ubiquitination and degradation primarily via the autophagy-lysosome system. Interestingly, this function is conserved for the human orthologue UBE4B in cells of neuroblastoma (Subramanian et al., 2021). In AD patients Tau protein is often hyper-phosphorylated and one of the kinases responsible is PTK2/FAK (focal adhesion kinase; Lee et al., 2022a). PTK2 plays a critical role in proteinopathies as it favors abnormal phosphorylation of proteins, including TDP-43 and p62/SQSTM1 leading to the formation of ubiquitinated aggregates and neurotoxicity induced by the dysregulation of the ubiquitin–proteasome system (UPS; Lee et al., 2020b). This mechanism is conserved for Tau (wt or mutant P301L) and also present in Drosophila where it was shown that attenuation of PTK2 expression reduces the phosphorylation of Ref2(P)/p62 ameliorating the motility of Tau expressing flies (Lee et al., 2022a). This also suggests the presence of a feed-back loop between PTK2 and p62 dysregulated in tauopathies.
5.5. Parkinson’s disease
Parkinson’s disease (PD) is the second most common neurodegenerative disorder after Alzheimer’s disease and another example of tauopathy (Poewe et al., 2017). PD symptoms include slow movement, postural imbalances, resting tremor, and muscle rigidity. Non-motor symptoms are also common and include memory loss, psychiatric symptoms, sleep problems, and pain. These symptoms stem from dopaminergic (DA) neuron loss in a part of the brain known as the substantia nigra pars compacta (SN), which leads to a lack of dopamine in other regions of the brain including the striatum. Both the affected regions are involved in movement and muscle control (Maiti et al., 2017). Treatment with the dopamine precursor levodopa or drugs such as dopamine agonists can temporarily improve many of the motor symptoms of PD however, no treatment currently exist able to prevent the neurodegenerative processes, underlying the importance to study the mechanisms responsible for DA neuron loss (Poewe et al., 2017). PD is characterized by the presence of Lewy Bodies, cytoplasmic inclusions consisting of a variety of misfolded proteins including α-synuclein and phosphorylated tau, in the SN and throughout the brain. As for Alzheimer’s Disease, most cases of PD are sporadic, thought to be triggered by environmental factors. However inherited forms of PD, linked to mutations in several different genes including α-synuclein, LRRK2, GBA, Parkin, PINK1, and Vps35 do exist, and their pathology and progression is very similar or indistinguishable from the sporadic one.
5.5.1 Autophagy in AD
The misfolded proteins accumulated in the Lewy Bodies are thought to be toxic, triggering neuroinflammation and cell death (Lashuel et al., 2013), and even though the exact mechanisms leading to their accumulation and toxicity are not completely understood, it seems that they include mitochondrial dysfunction, oxidative stress, and disruption of autophagy-mediated protein clearance (Karabiyik et al., 2017; Maiti et al., 2017). Interestingly, most of the genes associated with PD, such as Parkin, PINK1, and LRRK2, control mitochondria activity and are involved in autophagy-mediated degradation/ubiquitination of proteins to control the turnover of damaged mitochondria (mitophagy; Winklhofer, 2014; Rahman and Morrison, 2019). Most of the PD-associated genes are expressed and functionally conserved in Drosophila, therefore both loss of function of endogenous genes or models harboring different human mutations were generated (Hewitt and Whitworth, 2017). Work performed in the fruit fly has largely contributed to clarifying the role of these genes in autophagy/mitophagy. In the following paragraphs, we will discuss the most common Drosophila models of PD used to study autophagy modulation in this disease.
5.5.2. Alpha-synuclein (α-syn)
The SNCA gene encodes for α-synuclein (α-syn), a protein that is abundantly expressed in the nervous system where it is found in membranes and is a major components of the Lewy Bodies. Mutations in α-syn have been identified in both familial and sporadic cases of PD (Lesage and Brice, 2009). While some carry single-amino acid substitutions that increased α-synuclein secretion (Guan et al., 2020), most mutations promote α-syn aggregation and fibrils formation (i.e., E46K, H50Q, A53T), while others (i.e., G51D and A53E) decelerate α-syn aggregation (Flagmeier et al., 2016; Guan et al., 2020). This suggests that the effects of PD-associated pathogenic mutations on α-syn behavior are quite complex and probably modulated by different signaling.
5.5.2.1. Drosophila models of α-synuclein
Flies do not possess an orthologue of α-syn therefore models have been produced by ectopically expressing human mutants of α-syn. A large part of studies conducted in flies point to a dysfunction of autophagy in presence of α-syn mutation and to a positive role of autophagy in α-syn induced degeneration: ectopic expression of human α-syn leads to F-actin-mediated impairment in autophagic flux, accumulation of abnormal autophagosomes, impairment in mitophagy, leading to defects in cellular bioenergetics (Sarkar et al., 2021). Treatment of flies overexpressing α-syn with spermidine, a naturally occurring polyamine known to prolong lifespan by inducing autophagy, protects against α-syn induced neurotoxicity (Buttner et al., 2014). Overexpression of the human lysosomal membrane protein LAMP2A fully prevents the behavioral defects induced by neuronal expression of a PD-associated mutant form of α-syn (SNCA/A30P) as well as reduces the α-syn accumulation in older flies. Moreover, LAMP2A expression upregulates Atg5 that stimulates macroautophagy (Issa et al., 2018). Similarly, the progressive locomotor decline and the loss of dopaminergic (DA) neurons caused by the human α-syn-A30P variant is reduced by pan-neuronal overexpression of dDOR (orthologue of the human tumor protein 53-induced nuclear protein 1 (TP53INP1), able to activate autophagy and basal mitophagy (Dinh et al., 2021). Another group demonstrated that knockdown of inositol-requiring enzyme 1 (IRE1) or Atg7 reverses α-syn-A30P-induced neurodegeneration in terms of lifespan, locomotor activity, and DA neuron loss (Yan et al., 2019). This suggests that IRE1 and Atg7 may play different roles in the classical autophagy mechanism of survival and their loss activates protective mechanisms thereby explaining the reduction of toxic proteins and neuron survival in PD.
5.5.3. Parkin/Pink1
One of the molecular characteristics of PD are defects in mitophagy, the process that allows the elimination of damaged mitochondria, and this leads to accelerated neurodegeneration.
5.5.3.1. Drosophila models of Parkin/Pink1
Parkin and Pink1 are involved in mitochondria quality control and turnover (Shimura et al., 2000), the recruitment of the E3 ubiquitin ligase Parkin on the damaged mitochondrial membrane by the kinase Pink1, controls mitochondrial turnover by inducing a physiological level of mitophagy (Truban et al., 2017). Drosophila harbors only one parkin and PINK1 orthologue (Clark et al., 2006) and their function is highly conserved. Several models have been created both using endogenous parkin and PINK1 null mutants and by overexpressing human genes harboring pathogenic mutations, that closely recapitulate many PD features including DA loss, decreased lifespan, and motor defects (Ishihara-Paul et al., 2008; Hewitt and Whitworth, 2017). In particular, these models have been fundamental to understand the molecular mechanisms linking parkin and Pink1 to mitophagy. Studies using flies’ flight muscles identified their relevance in the control physiological mitophagy (Greene et al., 2003), furthermore, new studies outline how the increase in mitophagy observed in aging depends on the parkin/Pink1 interaction and on the activity of two deubiquitinases USP15 and 30 (Cornelissen et al., 2018). The mitochondria phenotype observed in parkin null mutants recapitulates general autophagy inhibition obtained by loss of Atg7, supporting the physiological role of parkin in the activation of mitophagy as clearance pathway (Vincow et al., 2013). Many proteins have been identified that may affect parkin/Pink1 modulation of mitophagy: Fbxo7, whose gene PARK15 is mutated in early-onset autosomal recessive forms of Parkinson, directly interacts with parkin to control its translocation to the mitochondrial membrane and functionally interacts with Pink1 to regulate parkin-dependent mitophagy (Burchell et al., 2013). The mitochondrial protein BNIP3L, a BH3 protein member of the Bcl2 family, interacts with Pink1 to induce mitophagy, and work in flies’ muscles helped to understand the role of human BNIP3 as suppressor of Pink1-induced-mitophagy (Zhang et al., 2016). BNIP3L was identified as a substrate for PARKIN2, and this ubiquitination recruits the autophagic cargo receptor NBR1 to the mitochondria to induce PARKIN2-mediated mitophagy. Even if PARKIN2 is present only in mammals, it was demonstrated using Drosophila that ectopic expression of BNIP3L rescues the mitochondrial defects of PINK1 mutant but not the effect of parkin mutant, outlining that PARKIN2 is the principal substrate for BNIP3L and it is necessary for BNIP3L clearance of mitochondria (Gao et al., 2015). Using flies, it was also demonstrated how the reduction of the human orthologue of Mask (ANKHD1), a scaffolding protein that inhibits mitophagy, rescues parkin/PINK1 mutant defects, suggesting that the human ANKHD1 might represent an interesting novel therapeutic target for treating PD associated to parkin/Pink1 mutations (Zhu et al., 2015). A novel mechanistic link between mitophagy and translation lies in the evolutionarily conserved function of EFTU, a mitochondrial translation elongation factor, that acts as substrate and interacts with Pink1 independently of parkin. Pink1 regulates mitophagy by restraining EFTU in the cytosol after phosphorylation on Ser222, inhibiting mitophagy by interacting with the Atg5-Atg12 complex formation (Lin et al., 2020). Despite several studies strongly linking parkin/Pink1 to mitophagy, Lee et al. (2018) demonstrated that basal mitophagy is not strongly reduced in the muscles of parkin/PINK1 mutant animals, and similar results were obtained using mouse models for PD under conditions of high metabolic demand. A possible explanation could be that the metabolic state of cells could influence mitophagy, particularly in animals carrying parkin/PINK1 mutations (McWilliams et al., 2018).
5.5.4. Leucine rich repeat kinase-2
Leucine rich repeat kinase-2 (LRRK2) is a leucine-rich repeat cytoplasmic kinase found also on the mitochondrial outer membrane, which contains a central core with a ROC-GTPase and other protein–protein interaction domains, suggesting multi-functional activity. LRRK2 is involved in several signaling pathways including vesicle trafficking, mitochondrial function, autophagic and lysosomal pathways, protein translation, neurite outgrowth, and cytoskeletal arrangement (Cookson, 2010). Mutations in LRRK2 are the most common genetic cause of both familial (41%) and sporadic (1–3%) PD (Jia et al., 2022; Oun et al., 2022) and among the function of LRRK2 that have been already clarified, it seems that its role in endomembrane trafficking may play a role in the pathogenesis of PD (Roosen and Cookson, 2016). Notably, recent experiments in human cells, also demonstrated that LRRK2 reduction is protective against the formation of α-syn aggregates suggesting a common signaling between these proteins. These studies pointed to glial cell function, in which the LRRK2 mutation interferes with the ability of glia to clear neuronal α-syn aggregates with a mechanism that is still unclear. The relevance of astrocytes in PD and proteinopathies is an important topic that needs attention (see Section 6) as more and more data delineate the presence/relevance of non-autonomous signals between astrocytes and neurons critical for neuronal clearance and survival (Streubel-Gallasch et al., 2021).
5.5.4.1. Drosophila models of LRRK2
Drosophila harbors a single LRRK2 orthologue gene (dLRKK) therefore it has been used to better understand the physiological functions of LRRK2 using loss of function models or overexpression of human LRRK2 variants responsible for PD (Hewitt and Whitworth, 2017; Seegobin et al., 2020). Overexpression of pathogenic mutant forms of LRRK2 causes defects in endolysosomal and autophagic assembly, and LRRK2-targeted therapies in PD are based on this molecular event. However, several studies in Drosophila have shown that dLRKK reduction affects lysosomal compartments and leads to the accumulation of dysfunctional autophagosomes, suggesting that proper expression of LRRK2 is required for functional signaling of autophagy and vesicle trafficking (Dodson et al., 2014; Seegobin et al., 2020). LRRK2 controls the phosphorylation of several small GTPases of the Ras-associated binding protein (Rab) family responsible for vesicle trafficking at the synapse level (Steger et al., 2016; Bellucci et al., 2022). Another relevant observation is the interaction of LRRK2 with other proteins leading to PD. Indeed, Ng and colleagues demonstrated that overexpression of the disease-associated LRRK2/G2019S mutant in flies flight muscles induces mitochondrial pathology and impairs locomotion by phenocopying the behavior of parkin-null flies, whereas in DA neurons it leads to significantly enlarged mitochondria. In both cases, the toxic effects of LRKK2 mutants were rescued by co-expression of parkin or activation of AMPK, known to reduce TOR signaling and to activate autophagy (Ng et al., 2012), suggesting a cooperative function in mitochondria between LRRK2 and parkin.
5.6. Prion diseases
Prion diseases (PrD) are a group of disorders characterized by the presence of aberrantly shaped proteins, called prions (proteinaceous infectious particles) encoded by the PRNP gene, that cause the accumulation of misfolded aggregated proteins in the central nervous system (Geschwind, 2015). Some forms occur sporadically, others are inherited or acquired through and include Creutzfeldt-Jacob disease (CJD), Gerstmann-Straussler-Scheinker syndrome (GSS), Fatal Familial Insomnia (FFI), and Kuru disease. Their characteristic is to induce transmissible spongiform encephalopathies (TSE) which lead to inflammation and neuronal death, with symptoms and severity depending on the type of prion and the animal species (Geschwind, 2015).
5.6.1 Autophagy in PrD
PrPC, responsible for this class of diseases, is an abundant protein inserted into the cell membrane via a GPI domain located at its C-terminus while the N-terminal tail contains functional glycosylation sites (Scheckel and Aguzzi, 2018). PrPC undergoes conformational changes transforming into an infectious misfolded isoform, named scrapie-associated prion protein (PrPSc) responsible for the toxic effect of the infection (Sandberg et al., 2011). PrD and autophagy. Proteostasis and autophagy play a central role in protecting neurons from the toxic effect of PrPSc (Thellung et al., 2022). Several studies have outlined a direct role of PrPC in the control of autophagy: in fact, the reduction of PrPC impairs the cellular response to oxidative stress with consequent dysregulation of the autophagic flux. Conversely, ectopic expression of PrPC appears to have a protective role by inducing autophagy under stressful conditions (Jeong and Park, 2015). Inducing autophagy is beneficial for the disease indeed, treating mice expressing the infective prions with rapamycin prolong their survival, while in yeast the induction of autophagy reduces the formation of prion protein PrPSc (Speldewinde et al., 2015). Similarly, transcriptomic analysis using the Drosophila model of PrPSc revealed a perturbation in genes component of TOR signaling suggesting a role in the control of translation by mutant PrPSc (Thackray et al., 2020). Furthermore, in the brains of prion-infected rodents the level of p62/SQSTM1 increases and co-localizes with PrPSc in large aggregates surrounding the perinuclear region of the cells (Homma et al., 2014). This mechanism resembles that observed in HD, where p62/SQSTM1 accumulates in the perinuclear area due to the formation of large ubiquitinated aggregates thus blocking its ability to transport cargo proteins into the autophagosome. However, the ubiquitin-binding protein p62/SQSTM1 not only mediates autophagy but also controls the activation of the Nuclear factor erythroid 2 (Nrf2), a key regulator of the cellular antioxidant response (Bellezza et al., 2018; Gureev et al., 2020). Thus inactivation of p62/SQSTM1 in prion diseases may contribute to the higher oxidative stress present in the cells (Shah et al., 2018). Recently, p62/SQSTM1 was found in cytoplasmic inclusions together with PrPC and the E3-ligase/TRAF6. Moreover, its activity was required for proper redistribution of PrPC into the insoluble cell fraction suggesting a novel physiological interaction between p62/SQSTM1 and PrPC (Masperone et al., 2022). Recent evidence suggested that neurodegeneration in prion diseases may be the consequence of defective glia as astrocytes have been shown to deposit mutant PrP aggregates and propagate prions to neurons and other cells (seed effect; Tahir et al., 2022). To this end we would like to suggest that Drosophila could be an optimal model to start exploring the contribution of glia to neuronal survival/death during prion pathogenesis using the combination of binary systems (see Sections 2 and 6).
5.6.2 Drosophila models of PrD
Drosophila models of PrD. Drosophila genome does not contain a PrP orthologue, making flies ideal for identifying the gain-of-function mechanisms associated with PrP misfolding (Fernandez-Funez et al., 2017). Common PrP mutations were first created in Drosophila using a mutation found in the CJD-Prp-PG14 family with nine additional repeats (Deleault et al., 2003). Prp-PG14 was expressed in neurons of the brain, in photoreceptors and pigmented cells of the eye but surprisingly, Prp-PG14 did not accumulate in the neurons and flies showed no behavioral or neuropathological abnormalities. In contrast, the expression of Prp-P101L (the murine orthologue of human PrP-P102L) resulted in locomotor defects and accumulation of PrP-P101L aggregates and vacuolation similar to what observed in neurons of GSS patients (Gavin et al., 2006). Surprisingly, using an inducible system it was shown that PrP-GSS phenotypes could be reverted and its level decreased when its expression was blocked, representing the first demonstration of reversibility of a phenotype reported in a genetic model for prion disease (Murali et al., 2014). Drosophila is also susceptible to exogenous sources of prions, indeed, it has been shown that PrP toxic phenotype can be transferred from fly to fly and from mice to fly. In these experiments, Drosophila larvae expressing variants of the PrPWT or carrying the toxic PrP3F4 mutant epitope were exposed to a homogenate of mouse or Drosophila brains expressing amino acid substitutions associated with the human prion diseases FFI and CJD (Thackray et al., 2017). These experiments demonstrated that both Drosophila adult-animals PrPWT or PrP3F4 exposed to prions show a significant decline in locomotor ability and of survival, indicating that the cellular and molecular components for prion replication and toxicity, were present and functional also in flies (Thackray et al., 2017). Furthermore, transcriptomic analysis using a Drosophila model of mutant PrP revealed a perturbation in cell cycle genes, regulator of protein synthesis and mitochondrial function, revealing data very similar to those in mammalian hosts undergoing to prion disease, further supporting the idea that flies are a well-established animal model to study mammalian prion biology (Thackray et al., 2020). More recently, new transgenic flies expressing different mutations of PrP in neurons highlight the function of PERK (EIF2AK3 in humans) and of the activating transcription factor 4 (ATF4), members of the UPR response in mediating PrP toxicity. A partially protective activity was shown for 4E-BP (EIF4EBP2 in humans), in the disease induced by human PrP-M129 and PrP-V129 mutations, on the contrary mutations in human PrP-(N159D, D167S, N174S) revealed a partial reduction of the toxic activity induced by co-expression of N159 mutation (Myers et al., 2022), highlighting how Drosophila can be used to study important amino acid substitutions to link PrP structural propensities to its toxicity.
6. Why using Drosophila to study PPs
6.1. To study non autonomous glial-neuron signaling in vivo
Cell-autonomous degeneration of neurons was considered the main outcome of neurodegenerative proteinopathies until several studies highlighted the causal role of glia and the relevance of non-cell autonomous signals that may influence neuronal health (Hickman et al., 2018; Yamanaka and Komine, 2018; Acioglu et al., 2021). Glial cells play important physiological functions in the CNS trough non-autonomous signals such as release of small molecules necessary for neuronal survival or inducing toxicity causing their death (Acioglu et al., 2021; Goodman and Bellen, 2022). The exploitation of cell type-specific expression systems, such as UAS-Gal4 and LexA-LexOp, would make fruit flies a suitable model to study the non-autonomous interactions between cells (see section 2). Indeed, it has been demonstrated that non-cell autonomous mechanisms play a key role in mutation for SOD1 (ALS), LKRR2 (PD) and in HD (Van Harten et al., 2021), with mechanisms not totally clear. Phagocytic glia may be activated by the dying neurons promoting their clearance but this may also worsen the disease as glia releases inflammatory neurotoxic cytokines toxic for the cells as shown for HD (Crotti et al., 2014; Creus-Muncunill and Ehrlich, 2019). A non-autonomous mechanisms is activated by the expression in glia of the chaperon DNAJB6, ortholog of the human HSP-DNAJ, that protects neuronal degeneration and extend lifespan (Bason et al., 2019), A new component is the Triggering Receptor Expressed on Myeloid cells (TREM2), a microglial phagocytic transmembrane receptor, conserved also in flies, for which genetic variations have been associated with senile dementia and increased risk of AD (Filipello et al., 2022). Data in mammals showed that its N-terminal cleavage blocks amyloid-β oligomerization in neurons exerting a protective role in AD. TREM2 also enhances glia metabolism and its function in clearance (Filipello et al., 2022); we may speculate that in AD TREM soluble-extracellular domain may act non-autonomously to activate its protecting signals on Aβ oligomerization.
6.2. To study protein-aggregate formation in vivo
Aggregates can be visualized ex vivo in organs (brain, muscles) either by immunofluorescence or by using the gene expressed as fusion protein with fluorescent proteins. The size and number of aggregates can be easily quantified with imaging techniques and applied to visualize changes upon performing: edible screens using chemical libraries; genetic screens for interactors or to identify signals, i.e., between glia and neurons that modify aggregates formation and dimensions. Glia-mediated phagocytosis might also favor prion-like seeding mechanisms characteristic of some PPs that drive aggregate formation (Pearce et al., 2015; Soto and Pritzkow, 2018; Donnelly et al., 2022). The seed-mechanism occurs also in flies and it was demonstrated in a model for HD using FRET/FRASE where the migration of mHTT aggregates was monitored in vivo in fly brains, and it has been proposed that glia phagocytosis might be involved in the rapid grow of mHTT aggregates in vivo (Ast et al., 2018). This observation is corroborated by a new mechanism in yeast showing that polyQ oligomers forms “de novo” aggregates and increase their original size by directly using the endogenous prion-forming protein Rnq1 in its amyloid-like prion conformation (Gropp et al., 2022).
6.3. To study the cross-talks of genes acting in the same proteinopathies
As we have seen, mutations in some PP can affect other genes suggesting common pathways to be analyzed. For example, Dewan and colleagues identified pathogenic HTT repeat expansions in patients diagnosed with FTD/ALS neurodegenerative disorders with mutations in TDRPH (Dewan et al., 2021), while polyQ expansions in the ATXN2 gene, that are normally associated with the onset of SCA2, have been observed in some forms of ALS (van den Heuvel et al., 2014). In addition, increasing evidence points to prion-like transmission as a mechanism underlying the development of many proteinopathies, driven by α-syn, HTT, SOD1, Tau, TDP-43, which can be uptake by the cytoplasm of acceptor cells from the brain (i.e., phagocytic microglia; Bayer, 2015; Pearce et al., 2015; Jo et al., 2020; Holec et al., 2022; Trist et al., 2022). Again Drosophila models can be easily engineered to co-express different mutations and their cross-talks with specific pathways such as autophagy or cell death, or their response to specific chemical drugs, can be easily analyzed in vivo.
6.4. To identify biomarkers
Nowadays much attention is paid to the identification of biomarkers for an early diagnosis/detection of proteinopathies (Doroszkiewicz et al., 2022). To analyze the direct secretion of non-autonomous signals (see section 6.1), or conveyed by Extracellular Vesicles (EV; Quiroz-Baez et al., 2020; Picca et al., 2022) or by exosomes (Beatriz et al., 2021; Zhang et al., 2021; Kumari and Anji, 2022) could identify molecules that may have a pathological role in the diseases (Jackson et al., 2022). In Drosophila, markers are available to identify EVs or exosomes, this, together with the advantage of generating a large progeny and the use of advanced biochemical tools, could accelerate this area of intervention.
In conclusion, the field of proteinopathies is in continuous evolution and there is still much to uncover about the misfolding process that is at the basis of the protein accumulation and disease propagation. Drosophila represents a powerful tool to dissect the many aspects that we still need to understand and represent an important alternative to the use of other model organisms. For example, the short time frame needed for the formation of the toxic aggregates in flies is an incredible advantage when studying the dynamic of aggregates formation upon modulation of signaling pathways or with chemical compounds. These data can be complemented with those obtained directly from drug screen using 3Dculture of patients-derived organoids rather than using 1D tissue culture lines. Drosophila can also by-pass mouse models for proteinopathies in the initial phase of characterization of novel human genes whose function is still unknown, using genetic screens. However, there are also some pitfalls to consider since the lack of an immune system and of proper microglia limits a deeper understanding of the physiology of the PPs. Nevertheless, we would like to underline the relevance of implementing Drosophila studies of the function of glia in PPs since it is more targetable for therapies than neurons, and because many data suggest the presence of non-autonomous cellular mechanisms, that are difficult to address in the available mammalian models.
Author contributions
SS, CL, CC, LT, and LV wrote and revised the review. PB and AS organized the topics and wrote and revised the review. All authors contributed to the article and approved the submitted version.
Funding
5 × 1000 tax campaigned for the research on neurodegenerative diseases (University of Trento).
Acknowledgments
We thank all members of Bellosta’s and Soldano’s laboratories for their comments on the review. We apologize to colleagues whose work was not cited in this manuscript due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu-Remaileh, M., Wyant, G. A., Kim, C., Laqtom, N. N., Abbasi, M., Chan, S. H., et al. (2017). Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. doi: 10.1126/science.aan6298
Acioglu, C., Li, L., and Elkabes, S. (2021). Contribution of astrocytes to neuropathology of neurodegenerative diseases. Brain Res. 1758:147291. doi: 10.1016/j.brainres.2021.147291
Agudelo, A., St. Amand, V., Grissom, L., Lafond, D., Achilli, T., Sahin, A., et al. (2020). Age-dependent degeneration of an identified adult leg motor neuron in aDrosophilaSOD1 model of ALS. Biology Open 9, 1–12. doi: 10.1242/bio.049692
Al-Ramahi, I., Perez, A. M., Lim, J., Zhang, M., Sorensen, R., De Haro, M., et al. (2007). dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 3:e234. doi: 10.1371/journal.pgen.0030234
An, H., Skelt, L., Notaro, A., Highley, J. R., Fox, A. H., La Bella, V., et al. (2019). ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol. Commun. 7:7. doi: 10.1186/s40478-019-0658-x
Anton-Fernandez, A., Valles-Saiz, L., Avila, J., and Hernandez, F. (2022). Neuronal nuclear tau and neurodegeneration. Neuroscience, 518, 178–184. doi: 10.1016/j.neuroscience.2022.07.015
Arbez, N., Ratovitski, T., Roby, E., Chighladze, E., Stewart, J. C., Ren, M., et al. (2017). Post-translational modifications clustering within proteolytic domains decrease mutant huntingtin toxicity. J. Biol. Chem. 292, 19238–19249. doi: 10.1074/jbc.M117.782300
Aron, R., Pellegrini, P., Green, E. W., Maddison, D. C., Opoku-Nsiah, K., Oliveira, A. O., et al. (2018). Deubiquitinase Usp12 functions noncatalytically to induce autophagy and confer neuroprotection in models of Huntington's disease. Nat. Commun. 9:3191. doi: 10.1038/s41467-018-05653-z
Ast, A., Buntru, A., Schindler, F., Hasenkopf, R., Schulz, A., Brusendorf, L., et al. (2018). mHTT seeding activity: a marker of disease progression and neurotoxicity in models of Huntington's disease. Mol. Cell 71:e676, 675–688.e6. doi: 10.1016/j.molcel.2018.07.032
Atilano, M. L., Gronke, S., Niccoli, T., Kempthorne, L., Hahn, O., Moron-Oset, J., et al. (2021). Enhanced insulin signalling ameliorates C9orf72 hexanucleotide repeat expansion toxicity in Drosophila. elife 10, 1–21. doi: 10.7554/eLife.58565
Babatz, F., Naffin, E., and Klambt, C. (2018). The Drosophila blood-brain barrier adapts to cell growth by unfolding of pre-existing Septate junctions. Dev. Cell 47:e693, 697–710.e3. doi: 10.1016/j.devcel.2018.10.002
Barbaro, B. A., Lukacsovich, T., Agrawal, N., Burke, J., Bornemann, D. J., Purcell, J. M., et al. (2015). Comparative study of naturally occurring huntingtin fragments in Drosophila points to exon 1 as the most pathogenic species in Huntington's disease. Hum. Mol. Genet. 24, 913–925. doi: 10.1093/hmg/ddu504
Bartlett, B. J., Isakson, P., Lewerenz, J., Sanchez, H., Kotzebue, R. W., Cumming, R. C., et al. (2011). p62, ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 7, 572–583. doi: 10.4161/auto.7.6.14943
Bason, M., Meister-Broekema, M., Alberts, N., Dijkers, P., Bergink, S., Sibon, O. C. M., et al. (2019). Astrocytic expression of the chaperone DNAJB6 results in non-cell autonomous protection in Huntington's disease. Neurobiol. Dis. 124, 108–117. doi: 10.1016/j.nbd.2018.10.017
Bates, G. P., Dorsey, R., Gusella, J. F., Hayden, M. R., Kay, C., Leavitt, B. R., et al. (2015). Huntington disease. Nat. Rev. Dis. Primers. 1:15005. doi: 10.1038/nrdp.2015.5
Bayer, T. A. (2015). Proteinopathies, a core concept for understanding and ultimately treating degenerative disorders? Eur. Neuropsychopharmacol. 25, 713–724. doi: 10.1016/j.euroneuro.2013.03.007
Beatriz, M., Vilaca, R., and Lopes, C. (2021). Exosomes: innocent bystanders or critical culprits in neurodegenerative diseases. Front. Cell Dev. Biol. 9:635104. doi: 10.3389/fcell.2021.635104
Beckers, J., Tharkeshwar, A. K., and Van Damme, P. (2021). C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels. Autophagy 17, 3306–3322. doi: 10.1080/15548627.2021.1872189
Bellezza, I., Giambanco, I., Minelli, A., and Donato, R. (2018). Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 1865, 721–733. doi: 10.1016/j.bbamcr.2018.02.010
Bellucci, A., Longhena, F., and Spillantini, M. G. (2022). The role of Rab proteins in Parkinson's disease Synaptopathy. Biomedicine 10, 1–17. doi: 10.3390/biomedicines10081941
Bilen, J., and Bonini, N. M. (2007). Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 3, 1950–1964. doi: 10.1371/journal.pgen.0030177
Boillee, S., Vande Velde, C., and Cleveland, D. W. (2006). ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron 52, 39–59. doi: 10.1016/j.neuron.2006.09.018
Bouche, V., Espinosa, A. P., Leone, L., Sardiello, M., Ballabio, A., and Botas, J. (2016). DrosophilaMitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 12, 484–498. doi: 10.1080/15548627.2015.1134081
Brady, O. A., Meng, P., Zheng, Y., Mao, Y., and Hu, F. (2011). Regulation of TDP-43 aggregation by phosphorylation andp62/SQSTM1. J. Neurochem. 116, 248–259. doi: 10.1111/j.1471-4159.2010.07098.x
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. doi: 10.1242/dev.118.2.401
Brenner, D., and Freischmidt, A. (2022). Update on genetics of amyotrophic lateral sclerosis. Curr. Opin. Neurol. 35, 672–677. doi: 10.1097/WCO.0000000000001093
Brunet, M. A., Jacques, J. F., Nassari, S., Tyzack, G. E., Mcgoldrick, P., Zinman, L., et al. (2021). TheFUSgene is dual‐coding with both proteins contributing toFUS‐mediated toxicity. EMBO Rep. 22:e50640. doi: 10.15252/embr.202050640
Burchell, V. S., Nelson, D. E., Sanchez-Martinez, A., Delgado-Camprubi, M., Ivatt, R. M., Pogson, J. H., et al. (2013). The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 16, 1257–1265. doi: 10.1038/nn.3489
Buttner, S., Broeskamp, F., Sommer, C., Markaki, M., Habernig, L., Alavian-Ghavanini, A., et al. (2014). Spermidine protects against α-synuclein neurotoxicity. Cell Cycle 13, 3903–3908. doi: 10.4161/15384101.2014.973309
Cai, C. Z., Yang, C., Zhuang, X. X., Yuan, N. N., Wu, M. Y., Tan, J. Q., et al. (2021). NRBF2 is a RAB7 effector required for autophagosome maturation and mediates the association of APP-CTFs with active form of RAB7 for degradation. Autophagy 17, 1112–1130. doi: 10.1080/15548627.2020.1760623
Cha, S. J., Lee, S., Choi, H. J., Han, Y. J., Jeon, Y. M., Jo, M., et al. (2022). Therapeutic modulation of GSTO activity rescues FUS-associated neurotoxicity via deglutathionylation in ALS disease models. Dev. Cell 57:e788, 783–798.e8. doi: 10.1016/j.devcel.2022.02.022
Charroux, B., and Fanto, M. (2010). The fine line between waste disposal and recycling: DRPLA fly models illustrate the importance of completing the autophagy cycle for rescuing neurodegeneration. Autophagy 6, 667–669. doi: 10.4161/auto.6.5.12433
Chatterjee, M., Steffan, J. S., Lukacsovich, T., Marsh, J. L., and Agrawal, N. (2021). Serine residues 13 and 16 are key modulators of mutant huntingtin induced toxicity in Drosophila. Exp. Neurol. 338:113463. doi: 10.1016/j.expneurol.2020.113463
Chatterjee, S., Ambegaokar, S. S., Jackson, G. R., and Mudher, A. (2019). Insulin-mediated changes in tau hyperphosphorylation and autophagy in a Drosophila model of Tauopathy and Neuroblastoma cells. Front. Neurosci. 13:801. doi: 10.3389/fnins.2019.00801
Chen, Z. S., Wong, A. K. Y., Cheng, T. C., Koon, A. C., and Chan, H. Y. E. (2019). FipoQ/FBXO33, a Cullin-1-based ubiquitin ligase complex component modulates ubiquitination and solubility of polyglutamine disease protein. J. Neurochem. 149, 781–798. doi: 10.1111/jnc.14669
Chua, J. P., De Calbiac, H., Kabashi, E., and Barmada, S. J. (2022). Autophagy and ALS: mechanistic insights and therapeutic implications. Autophagy 18, 254–282. doi: 10.1080/15548627.2021.1926656
Clague, M. J., Urbe, S., and Komander, D. (2019). Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352. doi: 10.1038/s41580-019-0099-1
Clark, I. E., Dodson, M. W., Jiang, C., Cao, J. H., Huh, J. R., Seol, J. H., et al. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166. doi: 10.1038/nature04779
Cleary, J. D., Pattamatta, A., and Ranum, L. P. W. (2018). Repeat-associated non-ATG (RAN) translation. J. Biol. Chem. 293, 16127–16141. doi: 10.1074/jbc.R118.003237
Collier, J. J., Guissart, C., Olahova, M., Sasorith, S., Piron-Prunier, F., Suomi, F., et al. (2021). Developmental consequences of defective ATG7-mediated autophagy in humans. N. Engl. J. Med. 384, 2406–2417. doi: 10.1056/NEJMoa1915722
Cookson, M. R. (2010). The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat. Rev. Neurosci. 11, 791–797. doi: 10.1038/nrn2935
Cornelissen, T., Vilain, S., Vints, K., Gounko, N., Verstreken, P., and Vandenberghe, W. (2018). Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. elife 7, 1–14. doi: 10.7554/eLife.35878
Creus-Muncunill, J., and Ehrlich, M. E. (2019). Cell-autonomous and non-cell-autonomous pathogenic mechanisms in Huntington's disease: insights from in vitro and in vivo models. Neurotherapeutics 16, 957–978. doi: 10.1007/s13311-019-00782-9
Crippa, V., Cicardi, M. E., Ramesh, N., Seguin, S. J., Ganassi, M., Bigi, I., et al. (2016). The chaperone HSPB8 reduces the accumulation of truncated TDP-43 species in cells and protects against TDP-43-mediated toxicity. Hum. Mol. Genet. 25, 3908–3924. doi: 10.1093/hmg/ddw232
Crotti, A., Benner, C., Kerman, B. E., Gosselin, D., Lagier-Tourenne, C., Zuccato, C., et al. (2014). Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat. Neurosci. 17, 513–521. doi: 10.1038/nn.3668
Cunningham, K. M., Maulding, K., Ruan, K., Senturk, M., Grima, J. C., Sung, H., et al. (2020). TFEB/Mitf links impaired nuclear import to autophagolysosomal dysfunction in C9-ALS. elife 9, 1–35. doi: 10.7554/eLife.59419
DeJesus-Hernandez, M., Mackenzie, I. R., Boeve, B. F., Boxer, A. L., Baker, M., Rutherford, N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. doi: 10.1016/j.neuron.2011.09.011
Deleault, N. R., Dolph, P. J., Feany, M. B., Cook, M. E., Nishina, K., Harris, D. A., et al. (2003). Post-transcriptional suppression of pathogenic prion protein expression in Drosophila neurons. J. Neurochem. 85, 1614–1623. doi: 10.1046/j.1471-4159.2003.01819.x
Dewan, R., Chia, R., Ding, J., Hickman, R. A., Stein, T. D., Abramzon, Y., et al. (2021). Pathogenic Huntingtin repeat expansions in patients with Frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 109:e444, 448–460.e4. doi: 10.1016/j.neuron.2020.11.005
Dinh, E., Rival, T., Carrier, A., Asfogo, N., Corti, O., Melon, C., et al. (2021). TP53INP1 exerts neuroprotection under ageing and Parkinson's disease-related stress condition. Cell Death Dis. 12:460. doi: 10.1038/s41419-021-03742-4
Dirren, E., Aebischer, J., Rochat, C., Towne, C., Schneider, B. L., and Aebischer, P. (2015). SOD1 silencing in motoneurons or glia rescues neuromuscular function in ALS mice. Ann. Clin. Transl. Neurol. 2, 167–184. doi: 10.1002/acn3.162
Dodson, M. W., Leung, L. K., Lone, M., Lizzio, M. A., and Guo, M. (2014). Novel ethyl methanesulfonate (EMS)-induced null alleles of the Drosophila homolog of LRRK2 reveal a crucial role in endolysosomal functions and autophagy in vivo. Dis. Model. Mech. 7, 1351–1363. doi: 10.1242/dmm.017020
Donde, A., Sun, M., Jeong, Y. H., Wen, X., Ling, J., Lin, S., et al. (2020). Upregulation of ATG7 attenuates motor neuron dysfunction associated with depletion of TARDBP/TDP-43. Autophagy 16, 672–682. doi: 10.1080/15548627.2019.1635379
Donnelly, K. M., Coleman, C. M., Fuller, M. L., Reed, V. L., Smerina, D., Tomlinson, D. S., et al. (2022). Hunting for the cause: evidence for prion-like mechanisms in Huntington's disease. Front. Neurosci. 16:946822. doi: 10.3389/fnins.2022.946822
Doroszkiewicz, J., Groblewska, M., and Mroczko, B. (2022). Molecular biomarkers and their implications for the early diagnosis of selected neurodegenerative diseases. Int. J. Mol. Sci. 23, 1–22. doi: 10.3390/ijms23094610
Doss-Pepe, E. W., Stenroos, E. S., Johnson, W. G., and Madura, K. (2003). Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 23, 6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003
Durcan, T. M., and Fon, E. A. (2011). Mutant ataxin-3 promotes the autophagic degradation of parkin. Autophagy 7, 233–234. doi: 10.4161/auto.7.2.14224
Ehrnhoefer, D. E., Sutton, L., and Hayden, M. R. (2011). Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist 17, 475–492. doi: 10.1177/1073858410390378
Ewen-Campen, B., Yang-Zhou, D., Fernandes, V. R., Gonzalez, D. P., Liu, L. P., Tao, R., et al. (2017). Optimized strategy for in vivo Cas9-activation inDrosophila. Proc. Natl. Acad. Sci. U. S. A. 114, 9409–9414. doi: 10.1073/pnas.1707635114
Fernandez-Funez, P., Nino-Rosales, M. L., De Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., et al. (2000). Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408, 101–106. doi: 10.1038/35040584
Fernandez-Funez, P., Sanchez-Garcia, J., and Rincon-Limas, D. E. (2017). Drosophila models of prionopathies: insight into prion protein function, transmission, and neurotoxicity. Curr. Opin. Genet. Dev. 44, 141–148. doi: 10.1016/j.gde.2017.03.013
Fiesel, F. C., Voigt, A., Weber, S. S., Van Den Haute, C., Waldenmaier, A., Gorner, K., et al. (2010). Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221. doi: 10.1038/emboj.2009.324
Filipello, F., Goldsbury, C., You, S. F., Locca, A., Karch, C. M., and Piccio, L. (2022). Soluble TREM2: innocent bystander or active player in neurological diseases? Neurobiol. Dis. 165:105630. doi: 10.1016/j.nbd.2022.105630
Flagmeier, P., Meisl, G., Vendruscolo, M., Knowles, T. P., Dobson, C. M., Buell, A. K., et al. (2016). Mutations associated with familial Parkinson’s disease alter the initiation and amplification steps of α-synuclein aggregation. Proc. Natl. Acad. Sci. U. S. A. 113, 10328–10333. doi: 10.1073/pnas.1604645113
Fleming, A., Bourdenx, M., Fujimaki, M., Karabiyik, C., Krause, G. J., Lopez, A., et al. (2022). The different autophagy degradation pathways and neurodegeneration. Neuron 110, 935–966. doi: 10.1016/j.neuron.2022.01.017
Freeman, M. R. (2015). Drosophila central nervous system glia. Cold Spring Harb Perspect Biol 7, 1–15. doi: 10.1101/cshperspect.a020552
Freibaum, B. D., Lu, Y., Lopez-Gonzalez, R., Kim, N. C., Almeida, S., Lee, K. H., et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. doi: 10.1038/nature14974
Freibaum, B. D., and Taylor, J. P. (2017). The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 10:35. doi: 10.3389/fnmol.2017.00035
Gallart-Palau, X., Ng, C. H., Ribera, J., Sze, S. K., and Lim, K. L. (2016). Drosophila expressing human SOD1 successfully recapitulates mitochondrial phenotypic features of familial amyotrophic lateral sclerosis. Neurosci. Lett. 624, 47–52. doi: 10.1016/j.neulet.2016.05.006
Galluzzi, L., Baehrecke, E. H., Ballabio, A., Boya, P., Bravo-San Pedro, J. M., Cecconi, F., et al. (2017). Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836. doi: 10.15252/embj.201796697
Gao, F., Chen, D., Si, J., Hu, Q., Qin, Z., Fang, M., et al. (2015). The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum. Mol. Genet. 24, 2528–2538. doi: 10.1093/hmg/ddv017
Gavin, B. A., Dolph, M. J., Deleault, N. R., Geoghegan, J. C., Khurana, V., Feany, M. B., et al. (2006). Accelerated accumulation of Misfolded prion protein and spongiform degeneration in aDrosophilaModel of Gerstmann–Sträussler–Scheinker syndrome. J. Neurosci. 26, 12408–12414. doi: 10.1523/JNEUROSCI.3372-06.2006
Gerenu, G., Persson, T., Goikolea, J., Calvo-Garrido, J., Loera-Valencia, R., Pottmeier, P., et al. (2021). Thioredoxin-80 protects against amyloid-beta pathology through autophagic-lysosomal pathway regulation. Mol. Psychiatry 26, 1410–1423. doi: 10.1038/s41380-019-0521-2
Geschwind, M. D. (2015). Prion diseases. CONTINUUM Lifelong Learn. Neurol. 21, 1612–1638. doi: 10.1212/CON.0000000000000251
Ghosh, B., Karmakar, S., Prasad, M., and Mandal, A. K. (2021). Praja1 ubiquitin ligase facilitates degradation of polyglutamine proteins and suppresses polyglutamine-mediated toxicity. Mol. Biol. Cell 32, 1579–1593. doi: 10.1091/mbc.E20-11-0747
Ghosh, D. K., and Ranjan, A. (2022). HYPK coordinates degradation of polyneddylated proteins by autophagy. Autophagy 18, 1763–1784. doi: 10.1080/15548627.2021.1997053
Goedert, M., Eisenberg, D. S., and Crowther, R. A. (2017). Propagation of tau aggregates and Neurodegeneration. Annu. Rev. Neurosci. 40, 189–210. doi: 10.1146/annurev-neuro-072116-031153
Gonzales, M. M., Garbarino, V. R., Pollet, E., Palavicini, J. P., Kellogg, D. L. Jr., Kraig, E., et al. (2022). Biological aging processes underlying cognitive decline and neurodegenerative disease. J. Clin. Invest. 132, 1–10. doi: 10.1172/JCI158453
Goodman, L. D., and Bellen, H. J. (2022). Recent insights into the role of glia and oxidative stress in Alzheimer's disease gained from Drosophila. Curr. Opin. Neurobiol. 72, 32–38. doi: 10.1016/j.conb.2021.07.012
Gottlieb, L., Guo, L., Shorter, J., and Marmorstein, R. (2021). N-alpha-acetylation of Huntingtin protein increases its propensity to aggregate. J. Biol. Chem. 297:101363. doi: 10.1016/j.jbc.2021.101363
Graham, R. K., Deng, Y., Slow, E. J., Haigh, B., Bissada, N., Lu, G., et al. (2006). Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cells 125, 1179–1191. doi: 10.1016/j.cell.2006.04.026
Greene, J. C., Whitworth, A. J., Kuo, I., Andrews, L. A., Feany, M. B., and Pallanck, L. J. (2003). Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkinmutants. Proc. Natl. Acad. Sci. U. S. A. 100, 4078–4083. doi: 10.1073/pnas.0737556100
Gropp, M. H. M., Klaips, C. L., and Hartl, F. U. (2022). Formation of toxic oligomers of polyQ-expanded Huntingtin by prion-mediated cross-seeding. Mol. Cell 82:e4211, 4290–4306.e11. doi: 10.1016/j.molcel.2022.09.031
Guan, Y., Zhao, X., Liu, F., Yan, S., Wang, Y., Du, C., et al. (2020). Pathogenic mutations differentially regulate cell-to-cell transmission of α-Synuclein. Front. Cell. Neurosci. 14:159. doi: 10.3389/fncel.2020.00159
Gunawardena, S., and Goldstein, L. S. (2001). Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 32, 389–401. doi: 10.1016/S0896-6273(01)00496-2
Gureev, A. P., Sadovnikova, I. S., Starkov, N. N., Starkov, A. A., and Popov, V. N. (2020). p62-Nrf2-p62 Mitophagy regulatory loop as a target for preventive therapy of neurodegenerative diseases. Brain Sci. 10, 1–14. doi: 10.3390/brainsci10110847
Gusella, J. F., and Macdonald, M. E. (1995). Huntington's disease. Semin. Cell Biol. 6, 21–28. doi: 10.1016/1043-4682(95)90011-X
Guttenplan, K. A., Weigel, M. K., Adler, D. I., Couthouis, J., Liddelow, S. A., Gitler, A. D., et al. (2020). Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nat. Commun. 11:3753. doi: 10.1038/s41467-020-17514-9
Harris, G. M., Dodelzon, K., Gong, L., Gonzalez-Alegre, P., and Paulson, H. L. (2010). Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS One 5:e13695. doi: 10.1371/journal.pone.0013695
Hernandez, F., Ferrer, I., Perez, M., Zabala, J. C., Del Rio, J. A., and Avila, J. (2022). Tau Aggregation. Neuroscience, 518, 64–69. doi: 10.1016/j.neuroscience.2022.04.024
Hewitt, V. L., and Whitworth, A. J. (2017). Mechanisms of Parkinson's disease. Curr. Top. Dev. Biol. 121, 173–200. doi: 10.1016/bs.ctdb.2016.07.005
Hickman, S., Izzy, S., Sen, P., Morsett, L., and El Khoury, J. (2018). Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369. doi: 10.1038/s41593-018-0242-x
Hindle, S. J., and Bainton, R. J. (2014). Barrier mechanisms in the Drosophila blood-brain barrier. Front. Neurosci. 8:414. doi: 10.3389/fnins.2014.00414
Holec, S. A. M., Lee, J., Oehler, A., Ooi, F. K., Mordes, D. A., Olson, S. H., et al. (2022). Multiple system atrophy prions transmit neurological disease to mice expressing wild-type human α-synuclein. Acta Neuropathol. 144, 677–690. doi: 10.1007/s00401-022-02476-7
Holtmaat, A., and Svoboda, K. (2009). Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10, 647–658. doi: 10.1038/nrn2699
Homma, T., Ishibashi, D., Nakagaki, T., Satoh, K., Sano, K., Atarashi, R., et al. (2014). Increased expression of p62/SQSTM1 in prion diseases and its association with pathogenic prion protein. Sci. Rep. 4:4504. doi: 10.1038/srep04504
Hong, Y. J., and Do, J. T. (2019). Neural lineage differentiation from pluripotent stem cells to mimic human brain tissues. Front. Bioeng. Biotechnol. 7:400. doi: 10.3389/fbioe.2019.00400
Huang, C., Yan, S., and Zhang, Z. (2020). Maintaining the balance of TDP-43, mitochondria, and autophagy: a promising therapeutic strategy for neurodegenerative diseases. Transl. Neurodegener. 9:40. doi: 10.1186/s40035-020-00219-w
Huynh, D. P., Figueroa, K., Hoang, N., and Pulst, S. M. (2000). Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 26, 44–50. doi: 10.1038/79162
Ishihara-Paul, L., Hulihan, M. M., Kachergus, J., Upmanyu, R., Warren, L., Amouri, R., et al. (2008). PINK1 mutations and parkinsonism. Neurology 71, 896–902. doi: 10.1212/01.wnl.0000323812.40708.1f
Issa, A. R., Sun, J., Petitgas, C., Mesquita, A., Dulac, A., Robin, M., et al. (2018). The lysosomal membrane protein LAMP2A promotes autophagic flux and prevents SNCA-induced Parkinson disease-like symptoms in the Drosophilabrain. Autophagy 14, 1898–1910. doi: 10.1080/15548627.2018.1491489
Ito, K., Shinomiya, K., Ito, M., Armstrong, J. D., Boyan, G., Hartenstein, V., et al. (2014). A systematic nomenclature for the insect brain. Neuron 81, 755–765. doi: 10.1016/j.neuron.2013.12.017
Jackson, G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., et al. (1998). Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21, 633–642. doi: 10.1016/S0896-6273(00)80573-5
Jackson, N. A., Guerrero-Munoz, M. J., and Castillo-Carranza, D. L. (2022). The prion-like transmission of tau oligomers via exosomes. Front. Aging Neurosci. 14:974414. doi: 10.3389/fnagi.2022.974414
Jackson, S. M., Whitworth, A. J., Greene, J. C., Libby, R. T., Baccam, S. L., Pallanck, L. J., et al. (2005). A SCA7 CAG/CTG repeat expansion is stable in Drosophila melanogaster despite modulation of genomic context and gene dosage. Gene 347, 35–41. doi: 10.1016/j.gene.2004.12.008
Jayakumar, S., and Hasan, G. (2018). Neuronal calcium signaling in metabolic regulation and adaptation to nutrient stress. Front. Neural. Circuits 12:25. doi: 10.3389/fncir.2018.00025
Jeong, J. K., and Park, S. Y. (2015). Neuroprotective effect of cellular prion protein (PrPC) is related with activation of alpha7 nicotinic acetylcholine receptor (α7nAchR)-mediated autophagy flux. Oncotarget 6, 24660–24674. doi: 10.18632/oncotarget.4953
Jia, F., Fellner, A., and Kumar, K. R. (2022). Monogenic Parkinson's disease: genotype, phenotype, pathophysiology, and genetic testing. Genes (Basel) 13, 1–25. doi: 10.3390/genes13030471
Jia, H., Pallos, J., Jacques, V., Lau, A., Tang, B., Cooper, A., et al. (2012). Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease. Neurobiol. Dis. 46, 351–361. doi: 10.1016/j.nbd.2012.01.016
Jo, M., Lee, S., Jeon, Y. M., Kim, S., Kwon, Y., and Kim, H. J. (2020). The role of TDP-43 propagation in neurodegenerative diseases: integrating insights from clinical and experimental studies. Exp. Mol. Med. 52, 1652–1662. doi: 10.1038/s12276-020-00513-7
Johnson, S. L., Blount, J. R., Libohova, K., Ranxhi, B., Paulson, H. L., Tsou, W. L., et al. (2019). Differential toxicity of ataxin-3 isoforms in Drosophila models of Spinocerebellar Ataxia type 3. Neurobiol. Dis. 132:104535. doi: 10.1016/j.nbd.2019.104535
Johnson, S. L., Libohova, K., Blount, J. R., Sujkowski, A. L., Prifti, M. V., Tsou, W. L., et al. (2021). Targeting the VCP-binding motif of ataxin-3 improves phenotypes in Drosophila models of Spinocerebellar Ataxia type 3. Neurobiol. Dis. 160:105516. doi: 10.1016/j.nbd.2021.105516
Johnson, S. L., Ranxhi, B., Libohova, K., Tsou, W. L., and Todi, S. V. (2020). Ubiquitin-interacting motifs of ataxin-3 regulate its polyglutamine toxicity through Hsc70-4-dependent aggregation. Elife 91. doi: 10.7554/eLife.60742
Juhasz, G., Erdi, B., Sass, M., and Neufeld, T. P. (2007). Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21, 3061–3066. doi: 10.1101/gad.1600707
Karabiyik, C., Lee, M. J., and Rubinsztein, D. C. (2017). Autophagy impairment in Parkinson's disease. Essays Biochem. 61, 711–720. doi: 10.1042/EBC20170023
Karch, C. M., Cruchaga, C., and Goate, A. M. (2014). Alzheimer's disease genetics: from the bench to the clinic. Neuron 83, 11–26. doi: 10.1016/j.neuron.2014.05.041
Karpuj, M. V., Becher, M. W., Springer, J. E., Chabas, D., Youssef, S., Pedotti, R., et al. (2002). Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat. Med. 8, 143–149. doi: 10.1038/nm0202-143
Kaspar, B. K., Lladó, J., Sherkat, N., Rothstein, J. D., and Gage, F. H. (2003). Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science 301, 839–842. doi: 10.1126/science.1086137
Kim, J., Kundu, M., Viollet, B., and Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. doi: 10.1038/ncb2152
Kim, T., Song, B., and Lee, I. S. (2020). Drosophila glia: models for human neurodevelopmental and neurodegenerative disorders. Int. J. Mol. Sci. 21, 1–38. doi: 10.3390/ijms21144859
Klaips, C. L., Jayaraj, G. G., and Hartl, F. U. (2018). Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51–63. doi: 10.1083/jcb.201709072
Klockgether, T., Mariotti, C., and Paulson, H. L. (2019). Spinocerebellar ataxia. Nat. Rev. Dis. Primers. 5:24. doi: 10.1038/s41572-019-0074-3
Kocaturk, N. M., and Gozuacik, D. (2018). Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front. Cell Dev. Biol. 6:128. doi: 10.3389/fcell.2018.00128
Komatsu, M., Wang, Q. J., Holstein, G. R., Friedrich, V. L. Jr., Iwata, J., Kominami, E., et al. (2007). Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U. S. A. 104, 14489–14494. doi: 10.1073/pnas.0701311104
Koyuncu, S., Fatima, A., Gutierrez-Garcia, R., and Vilchez, D. (2017). Proteostasis of Huntingtin in health and disease. Int. J. Mol. Sci. 18, 1–18. doi: 10.3390/ijms18071568
Krench, M., and Littleton, J. T. (2017). Neurotoxicity pathways in Drosophila models of the Polyglutamine disorders. Curr. Top. Dev. Biol. 121, 201–223. doi: 10.1016/bs.ctdb.2016.07.006
Kumari, M., and Anji, A. (2022). Biology, Novel intercellular messengers in neurodegeneration. Biology (Basel) 11:413. doi: 10.3390/biology11030413
Kwon, I., Xiang, S., Kato, M., Wu, L., Theodoropoulos, P., Wang, T., et al. (2014). Poly-dipeptides encoded by theC9orf72repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145. doi: 10.1126/science.1254917
Lanson, N. A. Jr., Maltare, A., King, H., Smith, R., Kim, J. H., Taylor, J. P., et al. (2011). A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 20, 2510–2523. doi: 10.1093/hmg/ddr150
Lashuel, H. A., Overk, C. R., Oueslati, A., and Masliah, E. (2013). The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14, 38–48. doi: 10.1038/nrn3406
Lee, J. J., Sanchez-Martinez, A., Zarate, A. M., Benincá, C., Mayor, U., Clague, M. J., et al. (2018). Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 217, 1613–1622. doi: 10.1083/jcb.201801044
Lee, K. H., Zhang, P., Kim, H. J., Mitrea, D. M., Sarkar, M., Freibaum, B. D., et al. (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cells 167:e717, 774–788.e17. doi: 10.1016/j.cell.2016.10.002
Lee, P. T., Lievens, J. C., Wang, S. M., Chuang, J. Y., Khalil, B., Wu, H. E., et al. (2020a). Sigma-1 receptor chaperones rescue nucleocytoplasmic transport deficit seen in cellular and Drosophila ALS/FTD models. Nat. Commun. 11:5580. doi: 10.1038/s41467-020-19396-3
Lee, S., Jeon, Y. M., Cha, S. J., Kim, S., Kwon, Y., Jo, M., et al. (2020b). PTK2/FAK regulates UPS impairment via SQSTM1/p62 phosphorylation in TARDBP/TDP-43 proteinopathies. Autophagy 16, 1396–1412. doi: 10.1080/15548627.2019.1686729
Lee, S., Jo, M., Kwon, Y., Jeon, Y. M., Kim, S., Lee, K. J., et al. (2022a). PTK2 regulates tau-induced neurotoxicity via phosphorylation of p62 at Ser403. J. Neurogenet., 1–10. doi: 10.1080/01677063.2022.2114471
Lee, S., Jo, M., Lee, H. E., Jeon, Y. M., Kim, S., Kwon, Y., et al. (2021). HEXA-018, a novel inducer of autophagy, rescues TDP-43 toxicity in neuronal cells. Front. Pharmacol. 12:747975. doi: 10.3389/fphar.2021.747975
Lee, S. B., Bagley, J. A., Lee, H. Y., Jan, L. Y., and Jan, Y. N. (2011). Pathogenic polyglutamine proteins cause dendrite defects associated with specific actin cytoskeletal alterations inDrosophila. Proc. Natl. Acad. Sci. U. S. A. 108, 16795–16800. doi: 10.1073/pnas.1113573108
Lee, W. S., Al-Ramahi, I., Jeong, H. H., Jang, Y., Lin, T., Adamski, C. J., et al. (2022b). Cross-species genetic screens identify transglutaminase 5 as a regulator of polyglutamine-expanded ataxin-1. J. Clin. Invest. 132. doi: 10.1172/JCI156616
Lemarie, F. L., Caron, N. S., Sanders, S. S., Schmidt, M. E., Nguyen, Y. T. N., Ko, S., et al. (2021). Rescue of aberrant huntingtin palmitoylation ameliorates mutant huntingtin-induced toxicity. Neurobiol. Dis. 158:105479. doi: 10.1016/j.nbd.2021.105479
Lesage, S., and Brice, A. (2009). Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 18, R48–R59. doi: 10.1093/hmg/ddp012
Lessing, D., and Bonini, N. M. (2008). Polyglutamine genes interact to modulate the severity and progression of neurodegeneration in Drosophila. PLoS Biol. 6:e29. doi: 10.1371/journal.pbio.0060029
Lewis, E. A., and Smith, G. A. (2016). Using Drosophila models of Huntington's disease as a translatable tool. J. Neurosci. Methods 265, 89–98. doi: 10.1016/j.jneumeth.2015.07.026
Li, Z., Karlovich, C. A., Fish, M. P., Scott, M. P., and Myers, R. M. (1999). A putative Drosophila homolog of the Huntington's disease gene. Hum. Mol. Genet. 8, 1807–1815. doi: 10.1093/hmg/8.9.1807
Li, Z., Wang, C., Wang, Z., Zhu, C., Li, J., Sha, T., et al. (2019). Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 575, 203–209. doi: 10.1038/s41586-019-1722-1
Licata, N. V., Cristofani, R., Salomonsson, S., Wilson, K. M., Kempthorne, L., Vaizoglu, D., et al. (2022). C9orf72 ALS/FTD dipeptide repeat protein levels are reduced by small molecules that inhibit PKA or enhance protein degradation. EMBO J. 41:e105026. doi: 10.15252/embj.2020105026
Lim, W. K., Kaur, P., Huang, H., Jo, R. S., Ramamoorthy, A., Ng, L. F., et al. (2021). Optogenetic approaches for understanding homeostatic and degenerative processes in Drosophila. Cell. Mol. Life Sci. 78, 5865–5880. doi: 10.1007/s00018-021-03836-4
Lin, J., Chen, K., Chen, W., Yao, Y., Ni, S., Ye, M., et al. (2020). Paradoxical Mitophagy regulation by PINK1 and TUFm. Mol. Cell 80:e612, 607–620.e12. doi: 10.1016/j.molcel.2020.10.007
Lin, S., Marin, E. C., Yang, C. P., Kao, C. F., Apenteng, B. A., Huang, Y., et al. (2013). Extremes of lineage plasticity in the Drosophila brain. Curr. Biol. 23, 1908–1913. doi: 10.1016/j.cub.2013.07.074
Ling, D., Magallanes, M., and Salvaterra, P. M. (2014). Accumulation of amyloid-like Aβ1–42in AEL (autophagy–Endosomal–Lysosomal) vesicles: potential implications for plaque biogenesis. ASN Neuro 6:AN20130044. doi: 10.1042/AN20130044
Liou, N. F., Lin, S. H., Chen, Y. J., Tsai, K. T., Yang, C. J., Lin, T. Y., et al. (2018). Diverse populations of local interneurons integrate into the Drosophila adult olfactory circuit. Nat. Commun. 9:2232. doi: 10.1038/s41467-018-04675-x
Lu, J., Periz, G., Lu, Y. N., Tang, Q., Liu, Y., Zhang, T., et al. (2019). L3MBTL1 regulates ALS/FTD-associated proteotoxicity and quality control. Nat. Neurosci. 22, 875–886. doi: 10.1038/s41593-019-0384-5
Maiti, P., Manna, J., and Dunbar, G. L. (2017). Current understanding of the molecular mechanisms in Parkinson's disease: targets for potential treatments. Transl. Neurodegener. 6:28. doi: 10.1186/s40035-017-0099-z
Marcelo, A., Afonso, I. T., Afonso-Reis, R., Brito, D. V. C., Costa, R. G., Rosa, A., et al. (2021). Autophagy in Spinocerebellar ataxia type 2, a dysregulated pathway, and a target for therapy. Cell Death Dis. 12:1117. doi: 10.1038/s41419-021-04404-1
Marquilly, C., Busto, G. U., Leger, B. S., Boulanger, A., Giniger, E., Walker, J. A., et al. (2021). Htt is a repressor of Abl activity required for APP-induced axonal growth. PLoS Genet. 17:e1009287. doi: 10.1371/journal.pgen.1009287
Marrone, L., Poser, I., Casci, I., Japtok, J., Reinhardt, P., Janosch, A., et al. (2018). Isogenic FUS-eGFP iPSC reporter lines enable quantification of FUS stress granule pathology that is rescued by drugs inducing autophagy. Stem Cell Reports 10, 375–389. doi: 10.1016/j.stemcr.2017.12.018
Marsh, J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J., et al. (2000). Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 9, 13–25. doi: 10.1093/hmg/9.1.13
Martin, D. D., Heit, R. J., Yap, M. C., Davidson, M. W., Hayden, M. R., and Berthiaume, L. G. (2014). Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Hum. Mol. Genet. 23, 3166–3179. doi: 10.1093/hmg/ddu027
Martin, D. D., Ladha, S., Ehrnhoefer, D. E., and Hayden, M. R. (2015). Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 38, 26–35. doi: 10.1016/j.tins.2014.09.003
Martinez-Vicente, M., Talloczy, Z., Wong, E., Tang, G., Koga, H., Kaushik, S., et al. (2010). Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 13, 567–576. doi: 10.1038/nn.2528
Maruzs, T., Simon-Vecsei, Z., Kiss, V., Csizmadia, T., and Juhasz, G. (2019). On the Fly: recent Progress on autophagy and aging in Drosophila. Front. Cell Dev. Biol. 7:140. doi: 10.3389/fcell.2019.00140
Mason, R. P., Casu, M., Butler, N., Breda, C., Campesan, S., Clapp, J., et al. (2013). Glutathione peroxidase activity is neuroprotective in models of Huntington's disease. Nat. Genet. 45, 1249–1254. doi: 10.1038/ng.2732
Masperone, L., Codrich, M., Persichetti, F., Gustincich, S., Zucchelli, S., and Legname, G. (2022). The E3 ubiquitin ligase TRAF6 interacts with the cellular prion protein and modulates its solubility and recruitment to cytoplasmic p62/SQSTM1-positive Aggresome-like structures. Mol. Neurobiol. 59, 1577–1588. doi: 10.1007/s12035-021-02666-6
McConoughey, S. J., Basso, M., Niatsetskaya, Z. V., Sleiman, S. F., Smirnova, N. A., Langley, B. C., et al. (2010). Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol. Med. 2, 349–370. doi: 10.1002/emmm.201000084
Mcwilliams, T. G., Prescott, A. R., Montava-Garriga, L., Ball, G., Singh, F., Barini, E., et al. (2018). Basal Mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27:e435, 439–449.e5. doi: 10.1016/j.cmet.2017.12.008
Menzies, F. M., Garcia-Arencibia, M., Imarisio, S., O'Sullivan, N. C., Ricketts, T., Kent, B. A., et al. (2015). Calpain inhibition mediates autophagy-dependent protection against polyglutamine toxicity. Cell Death Differ. 22, 433–444. doi: 10.1038/cdd.2014.151
Menzies, F. M., Hourez, R., Imarisio, S., Raspe, M., Sadiq, O., Chandraratna, D., et al. (2010). Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum. Mol. Genet. 19, 4573–4586. doi: 10.1093/hmg/ddq385
Migazzi, A., Scaramuzzino, C., Anderson, E. N., Tripathy, D., Hernandez, I. H., Grant, R. A., et al. (2021). Huntingtin-mediated axonal transport requires arginine methylation by PRMT6. Cell Rep. 35:108980. doi: 10.1016/j.celrep.2021.108980
Min, B., Kwon, Y. C., Choe, K. M., and Chung, K. C. (2015). PINK1 phosphorylates transglutaminase 2 and blocks its proteasomal degradation. J. Neurosci. Res. 93, 722–735. doi: 10.1002/jnr.23535
Missirlis, F., Phillips, J. P., and Jackle, H. (2001). Cooperative action of antioxidant defense systems in Drosophila. Curr. Biol. 11, 1272–1277. doi: 10.1016/S0960-9822(01)00393-1
Mizielinska, S., Grönke, S., Niccoli, T., Ridler, C. E., Clayton, E. L., Devoy, A., et al. (2014). C9orf72repeat expansions cause neurodegeneration inDrosophilathrough arginine-rich proteins. Science 345, 1192–1194. doi: 10.1126/science.1256800
Mori, K., Weng, S. M., Arzberger, T., May, S., Rentzsch, K., Kremmer, E., et al. (2013). TheC9orf72GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. doi: 10.1126/science.1232927
Mputhia, Z., Hone, E., Tripathi, T., Sargeant, T., Martins, R., and Bharadwaj, P. (2019). Autophagy modulation as a treatment of amyloid diseases. Molecules 24, 1–20. doi: 10.3390/molecules24183372
Muma, N. A. (2007). Transglutaminase is linked to neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 66, 258–263. doi: 10.1097/nen.0b013e31803d3b02
Murali, A., Maue, R. A., and Dolph, P. J. (2014). Reversible symptoms and clearance of mutant prion protein in an inducible model of a genetic prion disease in Drosophila melanogaster. Neurobiol. Dis. 67, 71–78. doi: 10.1016/j.nbd.2014.03.013
Myers, R. R., Sanchez-Garcia, J., Leving, D. C., Melvin, R. G., and Fernandez-Funez, P. (2022). NewDrosophilamodels to uncover the intrinsic and extrinsic factors that mediate the toxicity of the human prion protein. Dis. Model. Mech. 15, 1–15. doi: 10.1242/dmm.049184
Nascimento-Ferreira, I., Santos-Ferreira, T., Sousa-Ferreira, L., Auregan, G., Onofre, I., Alves, S., et al. (2011). Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain 134, 1400–1415. doi: 10.1093/brain/awr047
Nassif, M., Valenzuela, V., Rojas-Rivera, D., Vidal, R., Matus, S., Castillo, K., et al. (2014). Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy 10, 1256–1271. doi: 10.4161/auto.28784
Nezis, I. P., Simonsen, A., Sagona, A. P., Finley, K., Gaumer, S., Contamine, D., et al. (2008). Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180, 1065–1071. doi: 10.1083/jcb.200711108
Ng, C. H., Guan, M. S., Koh, C., Ouyang, X., Yu, F., Tan, E. K., et al. (2012). AMP kinase activation mitigates dopaminergic dysfunction and mitochondrial abnormalities inDrosophilaModels of Parkinson's disease. J. Neurosci. 32, 14311–14317. doi: 10.1523/JNEUROSCI.0499-12.2012
Nguyen, G. D., Gokhan, S., Molero, A. E., and Mehler, M. F. (2013). Selective roles of normal and mutant huntingtin in neural induction and early neurogenesis. PLoS One 8:e64368. doi: 10.1371/journal.pone.0064368
Nishiyama, J., Miura, E., Mizushima, N., Watanabe, M., and Yuzaki, M. (2007). Aberrant membranes and Double-membrane structures accumulate in the axons ofAtg5-null Purkinje cells before neuronal death. Autophagy 3, 591–596. doi: 10.4161/auto.4964
Nitschke, L., Coffin, S. L., Xhako, E., El-Najjar, D. B., Orengo, J. P., Alcala, E., et al. (2021). Modulation of ATXN1 S776 phosphorylation reveals the importance of allele-specific targeting in SCA1. Insight 6, 1–15. doi: 10.1172/jci.insight.144955
Ochaba, J., Lukacsovich, T., Csikos, G., Zheng, S., Margulis, J., Salazar, L., et al. (2014). Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U. S. A. 111, 16889–16894. doi: 10.1073/pnas.1420103111
Omata, Y., Lim, Y. M., Akao, Y., and Tsuda, L. (2014). Age-induced reduction of autophagy-related gene expression is associated with onset of Alzheimer's disease. Am. J. Neurodegener. Dis. 3, 134–142.
Oun, A., Sabogal-Guaqueta, A. M., Galuh, S., Alexander, A., Kortholt, A., and Dolga, A. M. (2022). The multifaceted role of LRRK2 in Parkinson's disease: from human iPSC to organoids. Neurobiol. Dis. 173:105837. doi: 10.1016/j.nbd.2022.105837
Pallos, J., Bodai, L., Lukacsovich, T., Purcell, J. M., Steffan, J. S., Thompson, L. M., et al. (2008). Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum. Mol. Genet. 17, 3767–3775. doi: 10.1093/hmg/ddn273
Park, J., Al-Ramahi, I., Tan, Q., Mollema, N., Diaz-Garcia, J. R., Gallego-Flores, T., et al. (2013). RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature 498, 325–331. doi: 10.1038/nature12204
Parkes, T. L., Elia, A. J., Dickinson, D., Hilliker, A. J., Phillips, J. P., and Boulianne, G. L. (1998). Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat. Genet. 19, 171–174. doi: 10.1038/534
Pavel, M., Imarisio, S., Menzies, F. M., Jimenez-Sanchez, M., Siddiqi, F. H., Wu, X., et al. (2016). CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat. Commun. 7:13821. doi: 10.1038/ncomms13821
Pearce, M. M. P., Spartz, E. J., Hong, W., Luo, L., and Kopito, R. R. (2015). Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat. Commun. 6:6768. doi: 10.1038/ncomms7768
Peggion, C., Scalcon, V., Massimino, M. L., Nies, K., Lopreiato, R., Rigobello, M. P., et al. (2022). SOD1 in ALS: taking stock in pathogenic mechanisms and the role of glial and muscle cells. Antioxidants 11, 1–24. doi: 10.3390/antiox11040614
Peretti, D., Bastide, A., Radford, H., Verity, N., Molloy, C., Martin, M. G., et al. (2015). RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature 518, 236–239. doi: 10.1038/nature14142
Phillips, J. P., Tainer, J. A., Getzoff, E. D., Boulianne, G. L., Kirby, K., and Hilliker, A. J. (1995). Subunit-destabilizing mutations in Drosophila copper/zinc superoxide dismutase: neuropathology and a model of dimer dysequilibrium. Proc. Natl. Acad. Sci. U. S. A. 92, 8574–8578. doi: 10.1073/pnas.92.19.8574
Picca, A., Guerra, F., Calvani, R., Coelho-Junior, H. J., Bucci, C., and Marzetti, E. (2022). Circulating extracellular vesicles: friends and foes in neurodegeneration. Neural Regen. Res. 17, 534–542. doi: 10.4103/1673-5374.320972
Poewe, W., Seppi, K., Tanner, C. M., Halliday, G. M., Brundin, P., Volkmann, J., et al. (2017). Parkinson disease. Nat. Rev. Dis. Primers. 3:17013. doi: 10.1038/nrdp.2017.13
Potter, C. J., Tasic, B., Russler, E. V., Liang, L., and Luo, L. (2010). The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cells 141, 536–548. doi: 10.1016/j.cell.2010.02.025
Quiroz-Baez, R., Hernandez-Ortega, K., and Martinez-Martinez, E. (2020). Insights into the proteomic profiling of extracellular vesicles for the identification of early biomarkers of Neurodegeneration. Front. Neurol. 11:580030. doi: 10.3389/fneur.2020.580030
Rahman, A. A., and Morrison, B. E. (2019). Contributions of VPS35 mutations to Parkinson's disease. Neuroscience 401, 1–10. doi: 10.1016/j.neuroscience.2019.01.006
Rai, M., Curley, M., Coleman, Z., and Demontis, F. (2022). Contribution of proteases to the hallmarks of aging and to age-related neurodegeneration. Aging Cell 21:e13603. doi: 10.1111/acel.13603
Ratovitski, T., Jiang, M., O'Meally, R. N., Rauniyar, P., Chighladze, E., Faragó, A., et al. (2022). Interaction of huntingtin with PRMTs and its subsequent arginine methylation affects HTT solubility, phase transition behavior and neuronal toxicity. Hum. Mol. Genet. 31, 1651–1672. doi: 10.1093/hmg/ddab351
Ravikumar, B., Duden, R., and Rubinsztein, D. C. (2002). Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet, 11, 1107–1117.
Ravikumar, B., Imarisio, S., Sarkar, S., O'Kane, C. J., and Rubinsztein, D. C. (2008). Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 121, 1649–1660. doi: 10.1242/jcs.025726
Ravikumar, B., Vacher, C., Berger, Z., Davies, J. E., Luo, S., Oroz, L. G., et al. (2004). Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595. doi: 10.1038/ng1362
Ren, J., Jegga, A. G., Zhang, M., Deng, J., Liu, J., Gordon, C. B., et al. (2011). A Drosophila model of the neurodegenerative disease SCA17 reveals a role of RBP-J/Su(H) in modulating the pathological outcome. Hum. Mol. Genet. 20, 3424–3436. doi: 10.1093/hmg/ddr251
Renton, A. E., Majounie, E., Waite, A., Simón-Sánchez, J., Rollinson, S., Gibbs, J. R., et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. doi: 10.1016/j.neuron.2011.09.010
Rhine, K., Makurath, M. A., Liu, J., Skanchy, S., Lopez, C., Catalan, K. F., et al. (2020). ALS/FTLD-linked mutations in FUS Glycine residues cause accelerated gelation and reduced interactions with Wild-type FUS. Mol. Cell 80:e668, 666–681.e8. doi: 10.1016/j.molcel.2020.10.014
Rolls, M. M. (2011). Neuronal polarity in Drosophila: sorting out axons and dendrites. Dev. Neurobiol. 71, 419–429. doi: 10.1002/dneu.20836
Romero, E., Cha, G. H., Verstreken, P., Ly, C. V., Hughes, R. E., Bellen, H. J., et al. (2008). Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length huntingtin accumulating in the cytoplasm. Neuron 57, 27–40. doi: 10.1016/j.neuron.2007.11.025
Roosen, D. A., and Cookson, M. R. (2016). LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol. Neurodegener. 11:73. doi: 10.1186/s13024-016-0140-1
Root, J., Merino, P., Nuckols, A., Johnson, M., and Kukar, T. (2021). Lysosome dysfunction as a cause of neurodegenerative diseases: lessons from frontotemporal dementia and amyotrophic lateral sclerosis. Neurobiol. Dis. 154:105360. doi: 10.1016/j.nbd.2021.105360
Rui, Y. N., Xu, Z., Patel, B., Chen, Z., Chen, D., Tito, A., et al. (2015). Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 17, 262–275. doi: 10.1038/ncb3101
Rusten, T. E., Lindmo, K., Juhasz, G., Sass, M., Seglen, P. O., Brech, A., et al. (2004). Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7, 179–192. doi: 10.1016/j.devcel.2004.07.005
Şahin, A., Held, A., Bredvik, K., Major, P., Achilli, T. M., Kerson, A. G., et al. (2017). Human SOD1 ALS mutations in aDrosophilaKnock-in model cause severe phenotypes and reveal dosage-sensitive gain- and loss-of-function components. Genetics 205, 707–723. doi: 10.1534/genetics.116.190850
Sajjad, M. U., Green, E. W., Miller-Fleming, L., Hands, S., Herrera, F., Campesan, S., et al. (2014). DJ-1 modulates aggregation and pathogenesis in models of Huntington's disease. Hum. Mol. Genet. 23, 755–766. doi: 10.1093/hmg/ddt466
Sandberg, M. K., Al-Doujaily, H., Sharps, B., Clarke, A. R., and Collinge, J. (2011). Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature 470, 540–542. doi: 10.1038/nature09768
Sarkar, S., Olsen, A. L., Sygnecka, K., Lohr, K. M., and Feany, M. B. (2021). α-Synuclein impairs autophagosome maturation through abnormal actin stabilization. PLoS Genet. 17:e1009359. doi: 10.1371/journal.pgen.1009359
Sarkar, S., Perlstein, E. O., Imarisio, S., Pineau, S., Cordenier, A., Maglathlin, R. L., et al. (2007). Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 3, 331–338. doi: 10.1038/nchembio883
Sarkar, S., Ravikumar, B., Floto, R. A., and Rubinsztein, D. C. (2009). Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ 16, 46–56.
Satterfield, T. F., Jackson, S. M., and Pallanck, L. J. (2002). A Drosophila homolog of the Polyglutamine disease GeneSCA2Is a dosage-sensitive regulator of actin filament formation. Genetics 162, 1687–1702. doi: 10.1093/genetics/162.4.1687
Satterfield, T. F., and Pallanck, L. J. (2006). Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 15, 2523–2532. doi: 10.1093/hmg/ddl173
Scheckel, C., and Aguzzi, A. (2018). Prions, prionoids and protein misfolding disorders. Nat. Rev. Genet. 19, 405–418. doi: 10.1038/s41576-018-0011-4
Scott, R. C., Schuldiner, O., and Neufeld, T. P. (2004). Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178. doi: 10.1016/j.devcel.2004.07.009
Scotter, E. L., Chen, H. J., and Shaw, C. E. (2015). TDP-43 Proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics 12, 352–363. doi: 10.1007/s13311-015-0338-x
Seegobin, S. P., Heaton, G. R., Liang, D., Choi, I., Blanca Ramirez, M., Tang, B., et al. (2020). Progress in LRRK2-associated Parkinson's disease animal models. Front. Neurosci. 14:674. doi: 10.3389/fnins.2020.00674
Selkoe, D. J., and Hardy, J. (2016). The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 8, 595–608. doi: 10.15252/emmm.201606210
Shah, B., Lutter, D., Bochenek, M. L., Kato, K., Tsytsyura, Y., Glyvuk, N., et al. (2016). C3G/Rapgef1 is required in multipolar neurons for the transition to a bipolar morphology during cortical development. PLoS One 11:e0154174. doi: 10.1371/journal.pone.0154174
Shah, S. Z. A., Zhao, D., Hussain, T., Sabir, N., Mangi, M. H., and Yang, L. (2018). p62-Keap1-NRF2-ARE pathway: a contentious player for selective targeting of autophagy, oxidative stress and mitochondrial dysfunction in prion diseases. Front. Mol. Neurosci. 11:310. doi: 10.3389/fnmol.2018.00310
Shimura, H., Hattori, N., Kubo, S., Mizuno, Y., Asakawa, S., Minoshima, S., et al. (2000). Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305. doi: 10.1038/77060
Simonsen, A., Cumming, R. C., Brech, A., Isakson, P., Schubert, D. R., and Finley, K. D. (2008). Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. doi: 10.4161/auto.5269
Smarandache-Wellmann, C. R. (2016). Arthropod neurons and nervous system. Curr. Biol. 26, R960–R965. doi: 10.1016/j.cub.2016.07.063
Song, J. X., Malampati, S., Zeng, Y., Durairajan, S. S. K., Yang, C. B., Tong, B. C., et al. (2020). A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and tau pathology in Alzheimer's disease models. Aging Cell 19:e13069. doi: 10.1111/acel.13069
Soto, C., and Pritzkow, S. (2018). Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 21, 1332–1340. doi: 10.1038/s41593-018-0235-9
Speldewinde, S. H., Doronina, V. A., and Grant, C. M. (2015). Autophagy protects against de novo formation of the [PSI+] prion in yeast. Mol. Biol. Cell 26, 4541–4551. doi: 10.1091/mbc.E15-08-0548
Steger, M., Tonelli, F., Ito, G., Davies, P., Trost, M., Vetter, M., et al. (2016). Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. elife 5, 1–28. doi: 10.7554/eLife.12813
Stopford, M. J., Higginbottom, A., Hautbergue, G. M., Cooper-Knock, J., Mulcahy, P. J., De Vos, K. J., et al. (2017). C9ORF72 hexanucleotide repeat exerts toxicity in a stable, inducible motor neuronal cell model, which is rescued by partial depletion of Pten. Hum. Mol. Genet. 26, 1133–1145. doi: 10.1093/hmg/ddx022
Stoyas, C. A., and La Spada, A. R. (2018). The CAG-polyglutamine repeat diseases: a clinical, molecular, genetic, and pathophysiologic nosology. Handb. Clin. Neurol. 147, 143–170. doi: 10.1016/B978-0-444-63233-3.00011-7
Streubel-Gallasch, L., Giusti, V., Sandre, M., Tessari, I., Plotegher, N., Giusto, E., et al. (2021). Parkinson's disease-associated LRRK2 interferes with astrocyte-mediated alpha-Synuclein clearance. Mol. Neurobiol. 58, 3119–3140. doi: 10.1007/s12035-021-02327-8
Subramanian, M., Hyeon, S. J., Das, T., Suh, Y. S., Kim, Y. K., Lee, J. S., et al. (2021). UBE4B, a microRNA-9 target gene, promotes autophagy-mediated tau degradation. Nat. Commun. 12:3291. doi: 10.1038/s41467-021-23597-9
Suh, J., Romano, D. M., Nitschke, L., Herrick, S. P., Dimarzio, B. A., Dzhala, V., et al. (2019). Loss of Ataxin-1 potentiates Alzheimer's pathogenesis by elevating cerebral BACE1 transcription. Cells 178:e1117. doi: 10.1016/j.cell.2019.07.043
Sujkowski, A., Richardson, K., Prifti, M. V., Wessells, R. J., and Todi, S. V. (2022). Endurance exercise ameliorates phenotypes in Drosophila models of spinocerebellar ataxias. elife 11, 1–19. doi: 10.7554/eLife.75389
Tafuri, F., Ronchi, D., Magri, F., Comi, G. P., and Corti, S. (2015). SOD1 misplacing and mitochondrial dysfunction in amyotrophic lateral sclerosis pathogenesis. Front. Cell. Neurosci. 9:336. doi: 10.3389/fncel.2015.00336
Tahir, W., Thapa, S., and Schatzl, H. M. (2022). Astrocyte in prion disease: a double-edged sword. Neural Regen. Res. 17, 1659–1665. doi: 10.4103/1673-5374.332202
Taylor, J. P., Brown, R. H. Jr., and Cleveland, D. W. (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. doi: 10.1038/nature20413
Thackray, A. M., Cardova, A., Wolf, H., Pradl, L., Vorberg, I., Jackson, W. S., et al. (2017). Genetic human prion disease modelled in PrP transgenicDrosophila. Biochem. J. 474, 3253–3267. doi: 10.1042/BCJ20170462
Thackray, A. M., Lam, B., Shahira Binti Ab Razak, A., Yeo, G., and Bujdoso, R. (2020). Transcriptional signature of prion-induced neurotoxicity in aDrosophilamodel of transmissible mammalian prion disease. Biochem. J. 477, 833–852. doi: 10.1042/BCJ20190872
Thellung, S., Corsaro, A., Dellacasagrande, I., Nizzari, M., Zambito, M., and Florio, T. (2022). Proteostasis unbalance in prion diseases: mechanisms of neurodegeneration and therapeutic targets. Front. Neurosci. 16:966019. doi: 10.3389/fnins.2022.966019
Tian, Y., Bustos, V., Flajolet, M., and Greengard, P. (2011). A small‐molecule enhancer of autophagy decreases levels of Aβ and APP‐CTFviaAtg5‐dependent autophagy pathway. FASEB J. 25, 1934–1942. doi: 10.1096/fj.10-175158
Tian, Y., Chang, J. C., Fan, E. Y., Flajolet, M., and Greengard, P. (2013). Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc. Natl. Acad. Sci. U. S. A. 110, 17071–17076. doi: 10.1073/pnas.1315110110
Tran, N. N., and Lee, B. H. (2022). Functional implication of ubiquitinating and deubiquitinating mechanisms in TDP-43 proteinopathies. Front. Cell Dev. Biol. 10:931968. doi: 10.3389/fcell.2022.931968
Trist, B. G., Fifita, J. A., Hogan, A., Grima, N., Smith, B., Troakes, C., et al. (2022). Co-deposition of SOD1, TDP-43 and p62 proteinopathies in ALS: evidence for multifaceted pathways underlying neurodegeneration. Acta Neuropathol. Commun. 10:122. doi: 10.1186/s40478-022-01421-9
Truban, D., Hou, X., Caulfield, T. R., Fiesel, F. C., and Springer, W. (2017). PINK1, Parkin, and mitochondrial quality control: what can we learn about Parkinson's disease pathobiology? J. Parkinsons Dis. 7, 13–29. doi: 10.3233/JPD-160989
Tsakiri, E. N., Gumeni, S., Manola, M. S., and Trougakos, I. P. (2021). Amyloid toxicity in a Drosophila Alzheimer's model is ameliorated by autophagy activation. Neurobiol. Aging 105, 137–147. doi: 10.1016/j.neurobiolaging.2021.04.017
Tsou, W. L., Ouyang, M., Hosking, R. R., Sutton, J. R., Blount, J. R., Burr, A. A., et al. (2015). The deubiquitinase ataxin-3 requires Rad23 and DnaJ-1 for its neuroprotective role in Drosophila melanogaster. Neurobiol. Dis. 82, 12–21. doi: 10.1016/j.nbd.2015.05.010
Tsou, W. L., Qiblawi, S. H., Hosking, R. R., Gomez, C. M., and Todi, S. V. (2016). Polyglutamine length-dependent toxicity from alpha1ACT in Drosophila models of spinocerebellar ataxia type 6. Biol. Open 5, 1770–1775. doi: 10.1242/bio.021667
Tsubouchi, A., Yano, T., Yokoyama, T. K., Murtin, C., Otsuna, H., and Ito, K. (2017). Topological and modality-specific representation of somatosensory information in the fly brain. Science 358, 615–623. doi: 10.1126/science.aan4428
Tsuda, H., Jafar-Nejad, H., Patel, A. J., Sun, Y., Chen, H. K., Rose, M. F., et al. (2005). The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/senseless proteins. Cells 122, 633–644. doi: 10.1016/j.cell.2005.06.012
Ugolino, J., Ji, Y. J., Conchina, K., Chu, J., Nirujogi, R. S., Pandey, A., et al. (2016). Loss of C9orf72 enhances Autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 12:e1006443. doi: 10.1371/journal.pgen.1006443
Ugur, B., Chen, K., and Bellen, H. J. (2016). Drosophilatools and assays for the study of human diseases. Dis. Model. Mech. 9, 235–244. doi: 10.1242/dmm.023762
van den Heuvel, D. M. A., Harschnitz, O., van den Berg, L. H., and Pasterkamp, R. J. (2014). Taking a risk: a therapeutic focus on ataxin-2 in amyotrophic lateral sclerosis? Trends Mol. Med. 20, 25–35. doi: 10.1016/j.molmed.2013.09.001
Van Harten, A. C. M., Phatnani, H., and Przedborski, S. (2021). Non-cell-autonomous pathogenic mechanisms in amyotrophic lateral sclerosis. Trends Neurosci. 44, 658–668. doi: 10.1016/j.tins.2021.04.008
Van Raamsdonk, J. M., Pearson, J., Rogers, D. A., Bissada, N., Vogl, A. W., Hayden, M. R., et al. (2005). Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 14, 1379–1392. doi: 10.1093/hmg/ddi147
Van Well, E. M., Bader, V., Patra, M., Sánchez-Vicente, A., Meschede, J., Furthmann, N., et al. (2019). A protein quality control pathway regulated by linear ubiquitination. EMBO J. 38, 1–21. doi: 10.15252/embj.2018100730
Vance, C., Rogelj, B., Hortobagyi, T., De Vos, K. J., Nishimura, A. L., Sreedharan, J., et al. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211. doi: 10.1126/science.1165942
Vassar, R., Kovacs, D. M., Yan, R., and Wong, P. C. (2009). The β-Secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009
Vernizzi, L., Paiardi, C., Licata, G., Vitali, T., Santarelli, S., Raneli, M., et al. (2020). Cells, ameliorating motility in a Drosophila Model for Huntington's Disease. Cells 9, 1–23. doi: 10.3390/cells9010196
Vincow, E. S., Merrihew, G., Thomas, R. E., Shulman, N. J., Beyer, R. P., Maccoss, M. J., et al. (2013). The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl. Acad. Sci. U. S. A. 110, 6400–6405. doi: 10.1073/pnas.1221132110
Waite, K. A., Burris, A., Vontz, G., Lang, A., and Roelofs, J. (2022). Proteaphagy is specifically regulated and requires factors dispensable for general autophagy. J. Biol. Chem. 298:101494. doi: 10.1016/j.jbc.2021.101494
Walters, R., Manion, J., and Neely, G. G. (2019). Dissecting motor neuron disease with Drosophila melanogaster. Front. Neurosci. 13:331. doi: 10.3389/fnins.2019.00331
Wang, H., Ying, Z., and Wang, G. (2012). Ataxin-3 regulates aggresome formation of copper-zinc superoxide dismutase (SOD1) by editing K63-linked polyubiquitin chains. J. Biol. Chem. 287, 28576–28585. doi: 10.1074/jbc.M111.299990
Wang, S. M., Wu, H. E., Yasui, Y., Geva, M., Hayden, M., Maurice, T., et al. (2022). Nucleoporin POM121 signals TFEB-mediated autophagy via activation of SIGMAR1/sigma-1 receptor chaperone by pridopidine. Autophagy, 1–26.
Wardman, J. H., Henriksen, E. E., Marthaler, A. G., Nielsen, J. E., and Nielsen, T. T. (2020). Enhancement of autophagy and Solubilization of Ataxin-2 alleviate apoptosis in Spinocerebellar Ataxia type 2 patient cells. Cerebellum 19, 165–181. doi: 10.1007/s12311-019-01092-8
Warrick, J. M., Morabito, L. M., Bilen, J., Gordesky-Gold, B., Faust, L. Z., Paulson, H. L., et al. (2005). Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell 18, 37–48. doi: 10.1016/j.molcel.2005.02.030
Warrick, J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N., et al. (1998). Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cells 93, 939–949. doi: 10.1016/S0092-8674(00)81200-3
Watson, M. R., Lagow, R. D., Xu, K., Zhang, B., and Bonini, N. M. (2008). A drosophila model for amyotrophic lateral sclerosis reveals motor neuron damage by human SOD1. J. Biol. Chem. 283, 24972–24981. doi: 10.1074/jbc.M804817200
Weiss, K. R., Kimura, Y., Lee, W. C., and Littleton, J. T. (2012). Huntingtin aggregation kinetics and their pathological role in a Drosophila Huntington’s disease model. Genetics 190, 581–600.
Wen, X., Tan, W., Westergard, T., Krishnamurthy, K., Markandaiah, S. S., Shi, Y., et al. (2014). Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225. doi: 10.1016/j.neuron.2014.12.010
Wertz, M. H., Mitchem, M. R., Pineda, S. S., Hachigian, L. J., Lee, H., Lau, V., et al. (2020). Genome-wide in vivo CNS screening identifies genes that modify CNS neuronal survival and mHTT toxicity. Neuron 106:e78. doi: 10.1016/j.neuron.2020.01.004
Winklhofer, K. F. (2014). Parkin and mitochondrial quality control: toward assembling the puzzle. Trends Cell Biol. 24, 332–341. doi: 10.1016/j.tcb.2014.01.001
Wolozin, B., and Ivanov, P. (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20, 649–666. doi: 10.1038/s41583-019-0222-5
Wu, Y. L., Chang, J. C., Lin, W. Y., Li, C. C., Hsieh, M., Chen, H. W., et al. (2017). Treatment with Caffeic acid and resveratrol alleviates oxidative stress induced neurotoxicity in cell and Drosophila models of Spinocerebellar Ataxia Type3. Sci. Rep. 7:11641. doi: 10.1038/s41598-017-11839-0
Wyant, G. A., Abu-Remaileh, M., Wolfson, R. L., Chen, W. W., Freinkman, E., Danai, L. V., et al. (2017). mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cells 171:e612, 642–654.e12. doi: 10.1016/j.cell.2017.09.046
Xia, Q., Wang, H., Hao, Z., Fu, C., Hu, Q., Gao, F., et al. (2016). TDP‐43 loss of function increasesTFEBactivity and blocks autophagosome–lysosome fusion. EMBO J. 35, 121–142. doi: 10.15252/embj.201591998
Xu, Z., Poidevin, M., Li, X., Li, Y., Shu, L., Nelson, D. L., et al. (2013). Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 110, 7778–7783. doi: 10.1073/pnas.1219643110
Yamamoto, F., Taniguchi, K., Mamada, N., Tamaoka, A., Kametani, F., Lakshmana, M. K., et al. (2019). TFEB-mediated enhancement of the autophagy-lysosomal pathway dually modulates the process of amyloid beta-protein generation in neurons. Neuroscience 402, 11–22. doi: 10.1016/j.neuroscience.2019.01.010
Yamanaka, K., and Komine, O. (2018). The multi-dimensional roles of astrocytes in ALS. Neurosci. Res. 126, 31–38. doi: 10.1016/j.neures.2017.09.011
Yan, C., Liu, J., Gao, J., Sun, Y., Zhang, L., Song, H., et al. (2019). IRE1 promotes neurodegeneration through autophagy-dependent neuron death in the Drosophila model of Parkinson's disease. Cell Death Dis. 10:800. doi: 10.1038/s41419-019-2039-6
Yue, S., Serra, H. G., Zoghbi, H. Y., and Orr, H. T. (2001). The spinocerebellar ataxia type 1 protein, ataxin-1, has RNA-binding activity that is inversely affected by the length of its polyglutamine tract. Hum. Mol. Genet. 10, 25–30. doi: 10.1093/hmg/10.1.25
Zhai, R. G., Zhang, F., Hiesinger, P. R., Cao, Y., Haueter, C. M., and Bellen, H. J. (2008). NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature 452, 887–891. doi: 10.1038/nature06721
Zhang, S., Binari, R., Zhou, R., and Perrimon, N.. (2010). A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics 184:1165–1179.
Zhang, K., Donnelly, C. J., Haeusler, A. R., Grima, J. C., Machamer, J. B., Steinwald, P., et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. doi: 10.1038/nature14973
Zhang, N., He, F., Li, T., Chen, J., Jiang, L., Ouyang, X. P., et al. (2021). Role of Exosomes in brain diseases. Front. Cell. Neurosci. 15:743353. doi: 10.3389/fncel.2021.743353
Zhang, S., Feany, M. B., Saraswati, S., Littleton, J. T., and Perrimon, N. (2009). Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington's disease model. Dis. Model. Mech. 2, 247–266. doi: 10.1242/dmm.000653
Zhang, T., Periz, G., Lu, Y. N., and Wang, J. (2020). USP7 regulates ALS-associated proteotoxicity and quality control through the NEDD4L-SMAD pathway. Proc. Natl. Acad. Sci. U. S. A. 117, 28114–28125. doi: 10.1073/pnas.2014349117
Zhang, T., Xue, L., Li, L., Tang, C., Wan, Z., Wang, R., et al. (2016). BNIP3 protein suppresses PINK1 kinase Proteolytic cleavage to promote Mitophagy. J. Biol. Chem. 291, 21616–21629. doi: 10.1074/jbc.M116.733410
Zhang, X., Li, Y., Xu, H., and Zhang, Y. W. (2014). The gamma-secretase complex: from structure to function. Front. Cell. Neurosci. 8:427. doi: 10.3389/fncel.2014.00427
Zheng, Z., Lauritzen, J. S., Perlman, E., Robinson, C. G., Nichols, M., Milkie, D., et al. (2018). A complete Electron microscopy volume of the brain of adult Drosophila melanogaster. Cells 174:e722, 730–743.e22. doi: 10.1016/j.cell.2018.06.019
Zhu, M., Li, X., Tian, X., and Wu, C. (2015). Mask loss-of-function rescues mitochondrial impairment and muscle degeneration of Drosophila pink1 and parkin mutants. Hum. Mol. Genet. 24, 3272–3285. doi: 10.1093/hmg/ddv081
Zhu, M., Zhang, S., Tian, X., and Wu, C. (2017). Mask mitigates MAPT- and FUS-induced degeneration by enhancing autophagy through lysosomal acidification. Autophagy 13, 1924–1938. doi: 10.1080/15548627.2017.1362524
Zhu, Y., Lobato, A. G., Zhai, R. G., and Pinto, M. (2022). Human Nmnat1 promotes Autophagic clearance of amyloid plaques in a Drosophila model of Alzheimer's disease. Front. Aging Neurosci. 14:852972. doi: 10.3389/fnagi.2022.852972
Zhuang, L., Peng, F., Huang, Y., Li, W., Huang, J., Chu, Y., et al. (2020). CHIPmodulates APP‐induced autophagy‐dependent pathological symptoms inDrosophila. Aging Cell 19:e13070. doi: 10.1111/acel.13070
Keywords: protein-aggregate, protein-misfolding, autophagy, neurodegeneration, proteinopathies, non-autonomous signaling, animal model, Drosophila melanogaster
Citation: Santarelli S, Londero C, Soldano A, Candelaresi C, Todeschini L, Vernizzi L and Bellosta P (2023) Drosophila melanogaster as a model to study autophagy in neurodegenerative diseases induced by proteinopathies. Front. Neurosci. 17:1082047. doi: 10.3389/fnins.2023.1082047
Edited by:
Filomena Anna Digilio, National Research Council (CNR), ItalyReviewed by:
Alyssa N. Coyne, Johns Hopkins University, United StatesFrancesco Liguori, Santa Lucia Foundation (IRCCS), Italy
Copyright © 2023 Santarelli, Londero, Soldano, Candelaresi, Todeschini, Vernizzi and Bellosta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola Bellosta, cGFvbGEuYmVsbG9zdGFAdW5pdG4uaXQ=
†These authors have contributed equally to this work
 Stefania Santarelli
Stefania Santarelli Chiara Londero1†
Chiara Londero1† Alessia Soldano
Alessia Soldano Leonardo Todeschini
Leonardo Todeschini Luisa Vernizzi
Luisa Vernizzi Paola Bellosta
Paola Bellosta