- 1Department Sensors and Modelling, Leibniz Institute for Agricultural Engineering and Bioeconomy e. V. (ATB), Potsdam, Germany
- 2Institute for Animal Hygiene and Environmental Health, Veterinary Centre for Resistance Research, Freie Universität Berlin (FU), Berlin, Germany
- 3Institute of Veterinary Epidemiology and Biostatistics, Veterinary Centre for Resistance Research, Freie Universität Berlin (FU), Berlin, Germany
Introduction: Antimicrobial resistance (AMR) can spread in microorganisms through the transfer of antimicrobial resistance genes (ARGs). Livestock husbandry is one of the pathways for AMR emergence and transmission. Chicken manure contains valuable nutrients for agricultural field fertilization and can be used as input material for biogas production by anaerobic digestion (AD). However, usually, chicken manure also contains quite high levels of ARGs. In this study, we investigated the presence and temporal dynamics of ARGs against different antibiotic classes in chicken manure and anaerobic digestate as a source of AMR spread.
Methods: To get an overview of the ARG profiles, we quantified the abundances of 374 ARGs by high-throughput (HT)-PCR. We studied eight selected ARGs (tetA, tetX, sul1, sul2, lnuF, emrD, aadA, and tnpA) using qPCR in chicken manure from different flocks and animal ages and in digestate from different AD time points.
Results: Chicken manure showed higher amounts of ARGs compared to digestate, which was characterized by a higher ARG diversity. We observed that the effect of chicken age differed between the flocks. ARG abundances in digestate from different time points and different treatment conditions did not exhibit major changes.
Conclusion: The flocks’ variability had no relevant effect on ARG abundances in chicken manure, likely due to similar growth conditions. However, manure ARG content increased with the age of the chickens. In our experimental batch setup, AD was more effective in reducing AMR microorganisms than reducing ARGs. Further investigations on process optimization or the application of pre-treatment methods could enhance ARG reduction. Notably, pre-mixing chicken manure with material from a biogas plant prior to processing resulted in a lower ARG load compared to untreated chicken manure.
1 Introduction
Antibiotic resistance is a global problem [World Health Organization, 2023]. The WHO estimated that 1.27 million deaths were directly attributed to antibiotic resistance and 4.95 million deaths were associated with antibiotic resistance in 2019 worldwide (World Health Organization, 2023). The reason for antibiotic resistance is antimicrobial-resistant (AMR) bacteria harboring antibiotic resistance genes (ARGs), which can spread between bacteria of animals, humans, and the environment (Zhuang et al., 2021). Ding et al. (2023); Skandalis et al. (2021), and Zhuang et al. (2021) described the pathways through which ARGs are transferred between different environments, including hospitals and farms. The connection among all these environments contributes to the One Health concept, which emphasizes the interdependence of human, animal, and environmental health. German livestock husbandry consumed 309 tons of antibiotics in 2023, and, therefore, farms are one of the sources of AMR emergence and spread (German Federal Institute for Risk Assessment, 2025). In 2023, the European Union (EU) produced 20.6 tons of pork and 13.3 tons of poultry meat (eurostat Statistics Explained, 2024), leading to huge manure production. Köninger et al. (2021) reported that annually approximately 1.4 billion tons of livestock manure were produced in the EU and the United Kingdom between 2016 and 2019. Manure is a valuable source for agricultural field fertilization as it contains different nutrients. However, it can transfer different pollutants, such as ARGs, into the environment (Peng et al., 2022). One suitable way to use manure for energy (biogas) production and pollution reduction is anaerobic digestion (AD) (Congilosi and Aga, 2021; Zhang et al., 2013, 2012). This process is suitable for the reduction of pathogens and AMR bacteria (Atanasova et al., 2025; Thomas, 2023). AD also leads to ARG reduction (Mortezaei et al., 2024; Song et al., 2017).
The following genes are examples of those that are widespread and of particular concern due to their relevance in both human and animal health. The genes emrD and tetA encode efflux pumps. The genes tetX and lnuF encode enzymes that inactivate antibiotic molecules. Resistance to aminoglycosides can be transferred by aminoglycoside nucleotidyltransferase encoded by aadA. The genes sul1 and sul2 encode an enzyme that replaces the antibiotic target in the bacterial cell, therefore, enabling resistance to sulfonamide (CARD: https://card.mcmaster.ca/analyze/rgi; Alcock et al., 2023). Together with ARGs, mobile genetic elements (MGEs) play a role in transferring antibiotic resistance. One example that is frequently encountered in the environment is the transposon tnpA, which was shown to be associated with antibiotic resistance (Partridge et al., 2018). Associations between MGEs and ARGs could indicate horizontal gene transfer (Waseem et al., 2019). Investigating ARGs and their dynamics in chicken manure and during AD helps to assess AMR spread in the environment. There are only a limited number of studies that compare the amount of frequent ARGs in chicken manure from different flocks, animal ages, and at different AD time points. Therefore, the aims of this study were 1) to identify frequently appearing ARGs in chicken manure and anaerobic digestate from chicken manure, 2) to investigate the variation in ARG abundances in chicken manure from different flocks and chickens of different ages, and 3) to determine ARG temporal dynamics during AD in the mesophilic temperature range.
2 Materials and methods
2.1 Study design and sampling
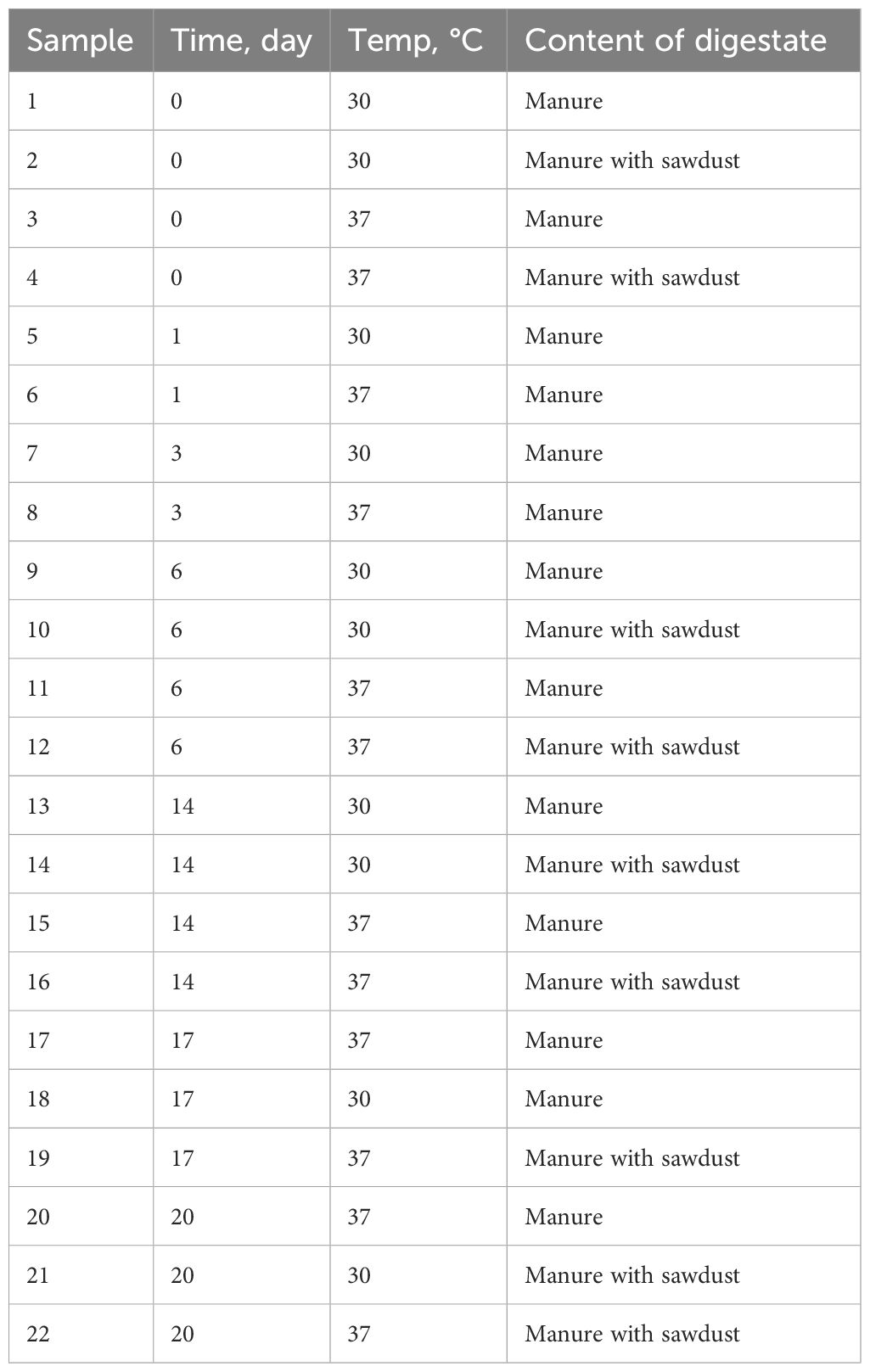
Details of the performed AD experiments were described in the study of Atanasova et al. (2025). In brief, chicken manure for AD was collected from a conventional broiler farm in Brandenburg, Germany. Inoculum for AD was collected from a biogas plant running on maize silage and cow slurry. Four different setups were used for the experiment: 1) AD with chicken manure at 30°C, 2) AD with chicken manure and sawdust addition at 30°C, 3) AD with chicken manure at 37°C, and 4) AD with chicken manure and sawdust addition at 37°C. Sawdust additions served as an additional carbon source and optimized the AD reaction to a higher biogas yield. During the AD experiment, digestate samples were collected every day from day 0 until day 9 and every second day from day 10 until day 20 and stored at -20°C. For ARG quantifications by qPCR, we selected digestate samples from days 0, 1, 3, 6, 14, 17, and 20 (Table 1). Samples from day 0 and day 1 were chosen as representatives of the starting condition at the beginning of the AD experiment. Microbiological cultivation results (Atanasova et al., 2025) showed a rapid change in E. coli survival during the first week of the experiment. Therefore, we performed qPCR analyses of samples from day 3 as a middle time point before E. coli was undetectable. On day 6, cultivated E. coli were under the detection limit at 37°C. On day 14, E. coli in digestate samples from reactors at 30°C were below the detection limit (see Atanasova et al., 2025). Day 17 was the last day of microbiological and chemical analyses and day 20 was the last day of sampling.
For monitoring selected ARGs in chicken manure, broiler litter was collected from the same conventional farm in Brandenburg, Germany a few months later. We took samples from five different flocks (4, 5, 6, 10, and 11) at three different time points (chickens that were 1 week, 3 weeks, and 5 weeks old). Additionally, the farmer provided us with information regarding the medical treatments (date, drug, dose, and duration) for each flock.
2.2 DNA isolation
DNA isolation from chicken manure was performed using a QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. For DNA isolation from the digestate samples, the samples were processed according to the manufacturer’s instructions using the Soil FastDNA™ SPIN Kit (MP Biomedicals, California, USA). DNA concentration and purity were measured using a NanoDrop (NanoPhotometer® N60/N50, IMPLEN, Munich, Germany).
2.3 HT-qPCR SmartChip
To get an overview of the ARG composition and abundance in chicken manure and digestate, we sent one mixture of chicken manure DNA and one mixture of anaerobic digestate DNA to Resistomap (Helsinki, Finland). We pooled several DNA samples from different time points together to cover a wider range of the presented ARGs. In the results, we named the samples “Chicken manure” that contained DNA of 1-week and 3-week-old chicken manure, from 2 and 3 different barns, respectively. “Digestate” samples consisted of DNA samples from AD with chicken manure at 37°C on day 3 and 14; chicken manure and sawdust at 37°C on day 6, and at 30°C on day 14. At Resistomap, a SmartChip HT-qPCR (Wafergen Inc, USA) with 384 genes (374 ARGs, 9 taxonomic genes, and 16S rRNA) was performed. The abundance of each gene was quantified in triplicate. As a result, relative abundances were calculated using the ΔC method, with 16S rRNA serving as the reference gene. The selection of ARGs was the standard setup of the ARG2.1 chip or the One Health Package. ARGs measured by the chip represented resistances against all the important antibiotic classes, some integrons, and MGEs associated with AMR. A detailed list of ARGs measured by HT-qPCR is shown in Supplementary Figures 1, 2. As output, we obtained gene abundances relative to 16S rRNA and gene copy numbers.
2.4 qPCR
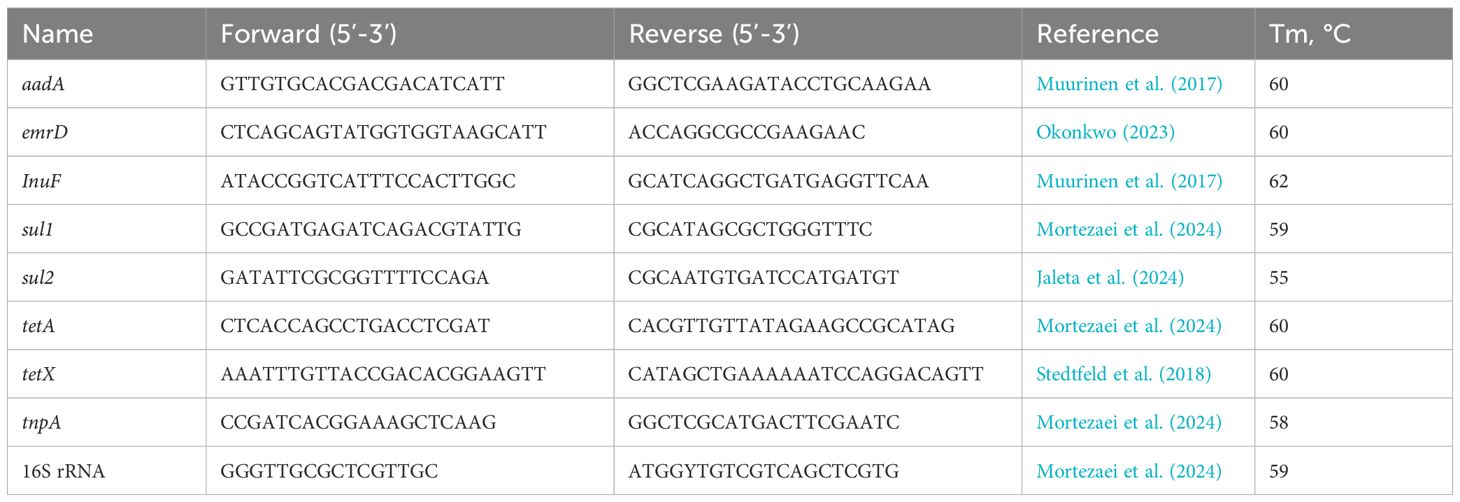
We investigated the detailed abundance and temporal dynamics of eight ARGs (aadA, emrD, lnuF, tetA, tetX, sul1, sul2, and tnpA) in chicken manure and digestate samples using qPCR. One of the main criteria for ARG selection was their high abundance detected by the SmartChip system, reflecting their widespread occurrence in the environment. Additional selection criteria for qPCR genes included the use of specific antibiotic classes on the farm during the study period, such as sulfamethoxazole and lincomycin, and the clinical relevance of these ARGs in both human and veterinary medicine. Primer (Merck Millipore, Missouri USA) information is shown in Table 2. The efficiency of all the primers was tested and it was 90%–100%. All samples were measured in triplicate. Each reaction mixture contained 20 µL, including 1 µL of DNA, 10µL SYBR Green Master I (Light Cycler 480 SYBR Green I Master, Roche), 1 µL forward primer, 1µl reverse primer (10 mM), and 7 µL MiliQ water. Samples were measured in a thermocycler (BioRad CFX384 Real-Time system, California, USA) under the following program: pre-incubation cycle for 180s at 95°C, 40 cycles of amplification for 10s at 95°C, primer annealing for 20 s at the primer-specific temperature (55°C-62°C, according to Table 2), and elongation for 20 s at 72°C. Additionally, a melting curve analysis was performed from 65°C to 95°C with a temperature increase of 0.5°C every 5s. The abundances of all the ARGs were normalized to 16S rRNA abundance. Fold change values were calculated by dividing the abundance of the measured samples by the reference sample, which was always the first one in the time series (week 1 for chicken manure and day 0 for AD).
2.5 Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics, version 25 (SPSS, Inc., Chicago, IL). We applied a linear regression analysis with the fold change of genes as the dependent variable and several independent variables [for AD: days, temperature, and substrate additive (sawdust); chicken manure: time and flock]. All two-way interactions were included, and non-significant ones were removed stepwise. Hochberg’s post-hoc tests were run for pairwise comparisons of the days (AD) or time and flock (chicken manure). Confidence intervals at 95% were calculated when applicable. A p-value <0.05 was considered significant.
3 Results
3.1 ARGs by HT-qPCR in chicken manure and digestate
Out of 374 ARGs potentially detectable on the Resistomap ARG2.1 chip, the maximum detected ARG number was 277 in digestate compared to 167 in chicken manure (Figure 1). In the digestate sample, the number of different detected ARGs was 110 (approx. 66%) more than in the chicken manure sample. This meant that the ARG diversity in the digestate was higher than in chicken manure. The ARG classes with the most detected genes were in both types of samples: MGE, multidrug resistance (MDR), aminoglycosides, and macrolide-lincosamide-streptogramin B (MLSB). In chicken manure, the two dominant ARG classes were MGE (29 detected genes) and aminoglycosides (28 detected genes), whereas for digestate, the two dominant ARG classes were MGE (39 detected genes) and MLSB (36 detected genes).
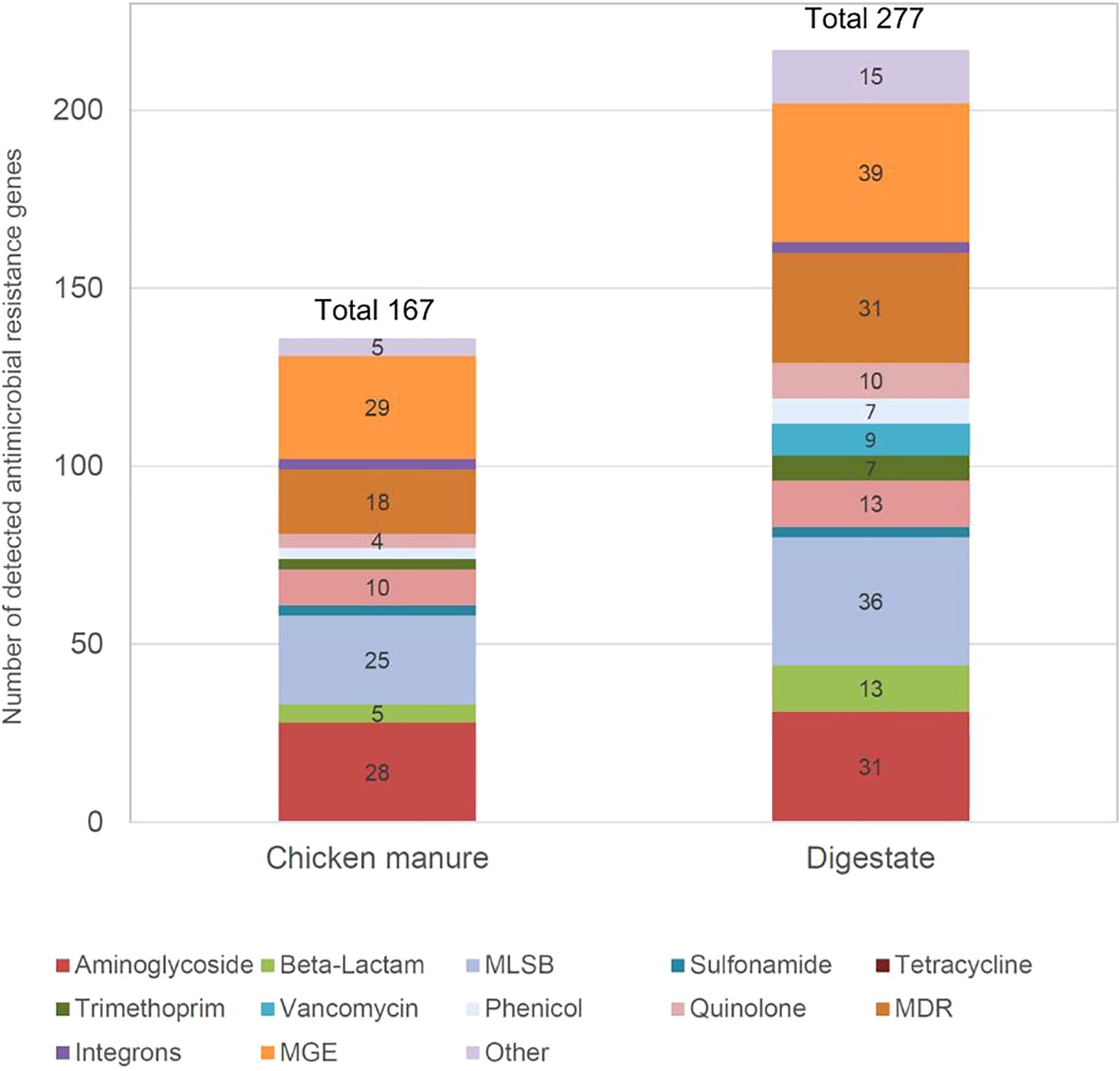
Figure 1. Number of detected antimicrobial resistance genes using a Resistomap HT-qPCR SmartChip in chicken manure and digestate. MDR, multidrug resistance; MLSB, macrolide-lincosamide-streptogramin B; MGEs, mobile genetic elements.
The relative abundances of ARG classes in chicken manure were much higher compared to digestate (Figure 2), except for beta-lactams, where both were similar (relative abundance in chicken manure was 0.093 and in digestate was 0.101). The highest relative abundance was observed for integrons class, consisting of four different genes (Supplementary Figures 1, 2), with 0.978 for chicken manure and 0.036 in digestate. The ARGs with the highest relative abundance in chicken manure were the aminoglycoside, MLSB, and tetracycline classes. Similarly, digestate showed the highest abundances of the aminoglycoside, MLSB, and beta-lactam ARG classes.
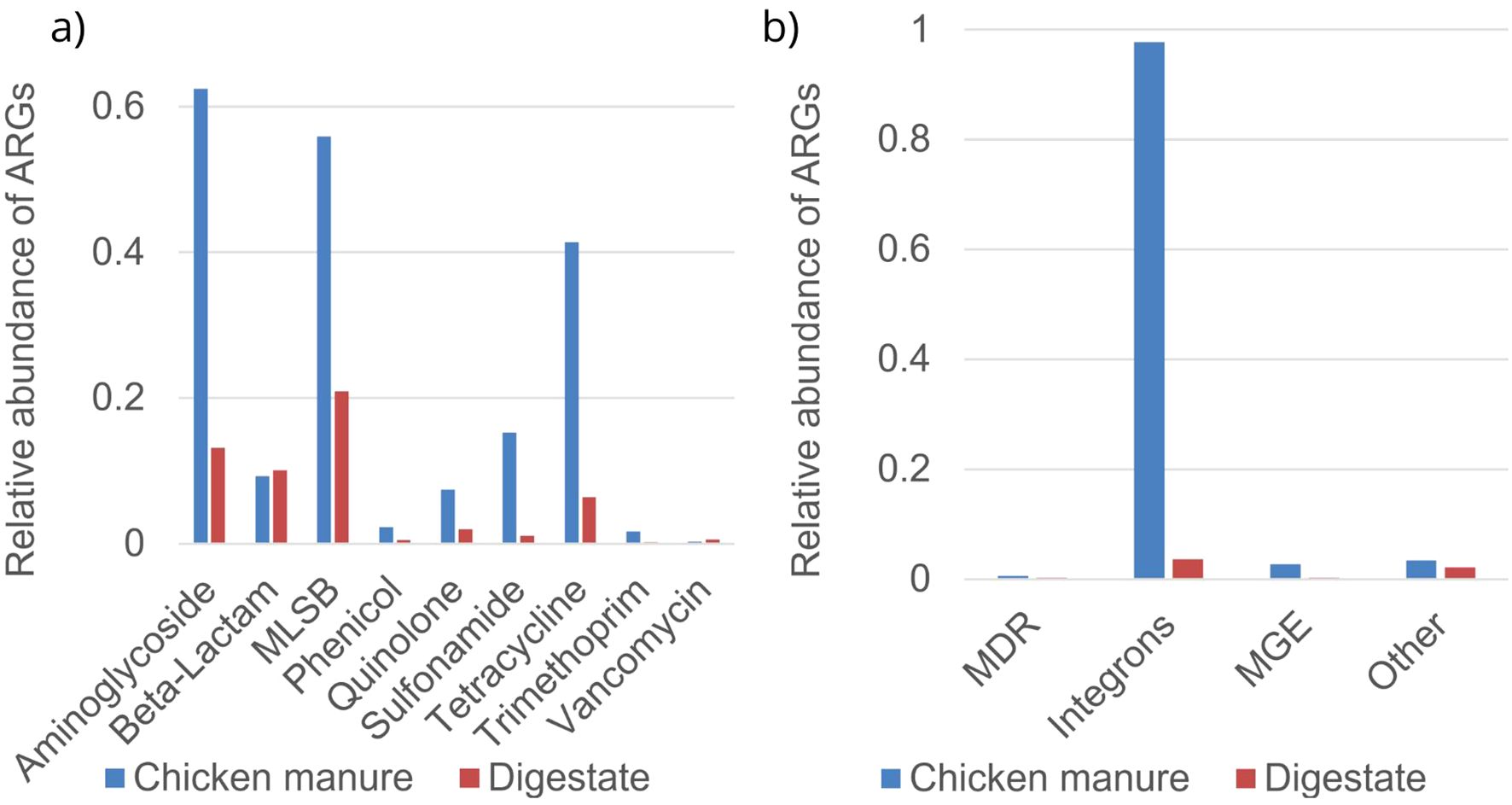
Figure 2. Average relative abundance normalized to 16S rRNA of antibiotic resistance gene (ARG) classes using a Resistomap HT-qPCR SmartChip in chicken manure and digestate. (a): according to antibiotic class, (b) according to multidrug resistance (MDR), integrons, mobile genetic elements (MGEs), and others.
The analyses of the relative abundances of single ARGs (Supplementary Figures 1, 2) in the aminoglycoside group revealed that the genes aadA_1 (0.079), aadA1_2 (0.071), and aadA2_3 (0.069) showed high detection frequencies in chicken manure, while aadA7 (0.028) was predominantly in digestate. In chicken manure, the MLSB class gene lnuC (0.018) was the most abundant, while in digestate, the dominant genes were lnuB (0.037) and ermF_1 (0.035). Chicken manure contained high levels of sul1 (0.004) and sul2 (0.012) from the sulfonamide group, as well as tet39 (0.246), tetX (0.098), and tetW (0.079) from the tetracycline class. Tetracycline resistance genes tetM (0.011) and tetD (0.008) were mainly present in the digestate. In the integrons class, intI1_1 (1.73) and intI2_2 (0.083) were highly abundant in chicken manure and intI3 (0.032) was more frequent in digestate. Chicken manure showed a high abundance of the MGE, tnpA_1 (0.120), while digestate had high levels of trbC (0.012).
3.2 ARGs in chicken manure by qPCR
After getting an overview of the ARG diversity and abundances in chicken manure and the anaerobic digestate by the SmartChip HT-qPCR, we focused on a more detailed investigation of eight selected ARGs using qPCR. All the selected genes (tetA, tetX, sul1, sul2, tnpA, emrD, InuF, and aadA) were detectable in chicken manure from animals of different ages and from different flocks.
Age and flock significantly influenced all the genes, but the effect of age differed between the flocks (Supplementary Table 1). The abundance of the tetA gene increased between week 1 and week 3, except for flock 4, where it decreased and had a maximum on week 5. Flock 6 had significantly lower values (p <0.001) than all other flocks (Figure 3). Concerning tetX, the values increased significantly in flock 6 (p <0.001), but remained relatively stable in all other flocks. In flocks 4 and 6, tnpA had the highest values on week 3, while flock 11 had its lowest values at that time point. Concerning sul1 and sul2, flock 11 again had the lowest values on week 3, with all other flocks peaking on week 3. The abundance of the emrD gene had increasing trends in flocks 6, 10, and 11, but decreased in flocks 4 and 5. The values of lnuF peaked on week 3 in flocks 4 and 6, while flock 11 showed the highest values on week 5. Flocks 4 and 6 had very similar abundances of tnpA, sul1, and lnuF (Figure 3).
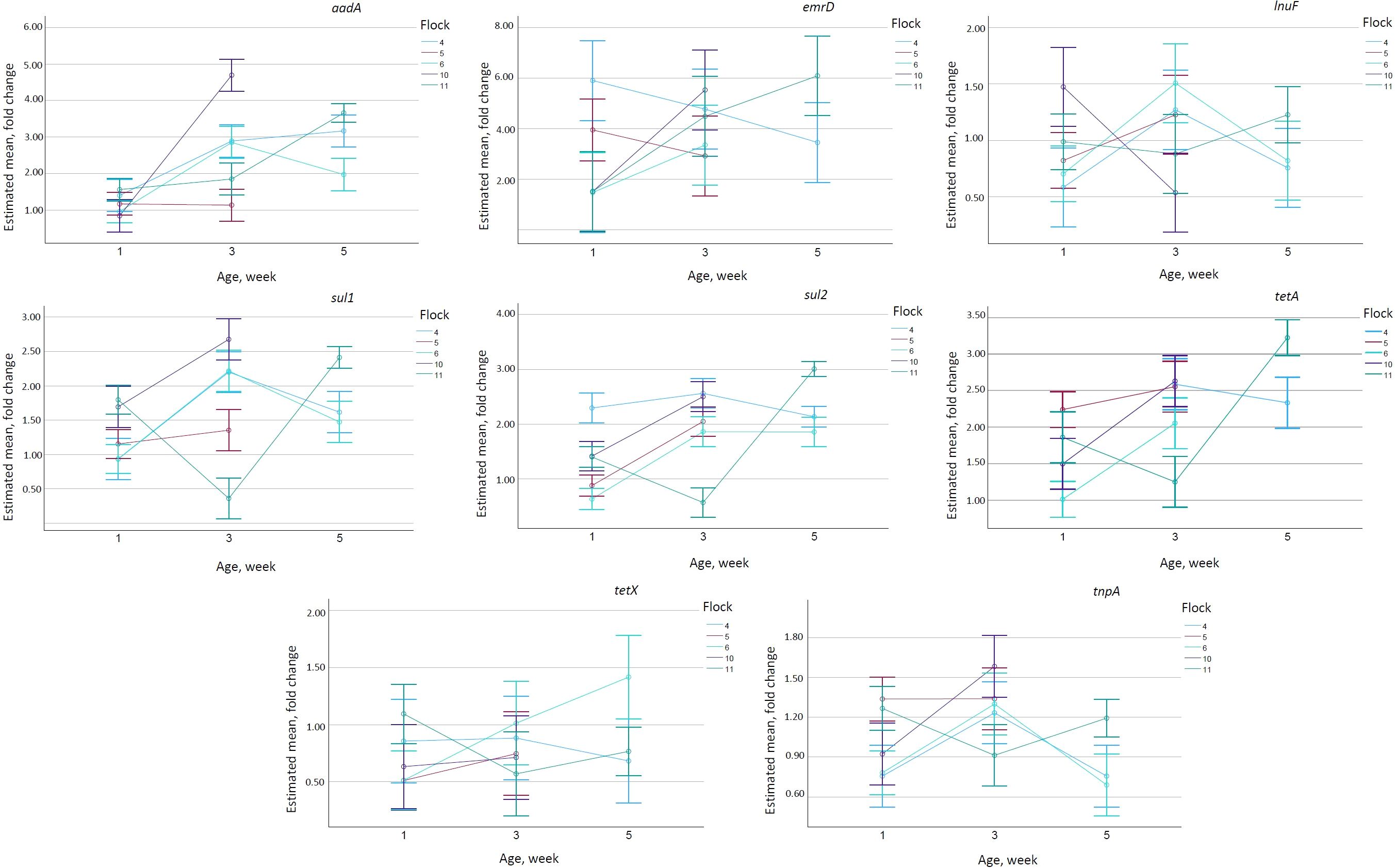
Figure 3. Estimated mean from UNIANOVA model for all of selected genes (aadA, emrD, lnuF, sul1, sul2, tetA, tetX, and tnpA). Non-estimated means are not plotted. Error bars show 95% confidence interval.
3.3 ARGs in digestate by qPCR
The results of the regression models for the digestate samples are shown in Supplementary Table 2. For all genes, except for emrD and tetX, sawdust had no significant impact, while temperature had a significant impact on the fold change of all genes. The genes aadA, tetA, sul1, sul2, and tetX had significantly lower fold changes at 30°C compared to 37°C, while emrD had higher values at 30°C. Regarding the substrates, significant interactions between the substrate and the temperature were observed for tetX. For the emrD gene, the effect of substrate also differed between the days, with higher values in the sawdust group at days 6, 14, and 20, but not at day 0. Interactions between day and temperature occurred for both emrD and tetX. These genes had interactions between the independent parameters day-sawdust (p = 0.016, resp., 0.015) and day-temperature (p <0.001, resp., 0.005), meaning temperature and sawdust were significant depending on the day.
4 Discussion
4.1 ARGs by HT-qPCR
In our study, ARGs from the aminoglycoside, MLSB, and tetracycline classes were dominant in chicken manure. Similar findings were reported by Peng et al. (2022); Błażejewska et al. (2022), and Sun et al. (2024), who investigated ARGs in manure from broiler farms and confirmed our results. Chicken manure is used for biogas production. Notably, Germany is the largest biomethane producer in Europe (European Biogas Association, 2020). Already in 2018, 9,000 biogas plants were performing anaerobic digestion (Appunn, 2016). The AD process starts with hydrolytic and fermentative bacteria, followed by acetogens and methanogens (Yangin-Gomec et al., 2020), which come from biogas plants, mainly consisting of maize silage and cattle slurry, and finally, chicken manure is mixed with this inoculum. In our digestate (chicken manure and inoculum), the majority of fresh matter by weight (approx. 99%) came from maize silage and cattle slurry. However, this is only 6% of the dry matter weight because the highest proportion of digestate is water. We compared raw material (manure) to digestate (manure with inoculum) to estimate the impact of the inoculum on chicken manure. Due to the fact that our digestate contained a mixture of biogas plant inoculum and chicken manure, it had higher ARG diversity and lower ARG relative abundance (Figure 2). Peng et al. (2022) compared the ARG content in different types of manure. Cattle slurry showed the lowest abundance of ARGs compared to poultry and pig manure, which is in agreement with our study. The differences in ARG content can be attributed to variations in the microbiome. Several studies have investigated this question. Peng et al. (2022) and Wang et al. (2019) related ARG abundance to different microbiome compositions in chicken manure and cattle slurry. Yue et al. (2022) and Gurmessa et al. (2021) considered Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes to be phyla associated with ARG abundance. Gurmessa et al. (2021) reported Firmicutes and Bacteroidetes were the dominant phyla in cattle slurry, while Firmicutes and Actinobacteria were the prevalent phyla in poultry manure. Additionally, the microbiome changed during the AD process, which could contribute to ARG changes as well. Riaz et al. (2020) reported a slight increase in the prevalence of the phyla Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes after anaerobic digestion. This can lead to no decrease or even an increase in ARG abundance. Yangin-Gomec et al. (2020) reported differences between inoculum and digested chicken manure microbiomes. The inoculum, which initially contained a diverse range of phyla without a high prevalence of any specific one, became a digestate with a dominant prevalence of Firmicutes and Actinobacteria, which are associated with the abundance of ARGs.
The chemical characteristics of poultry manure and cattle slurry contribute to different microbiomes, leading to different ARG abundances, as indicated by Gurmessa et al. (2021). One noticeable difference is the moisture content in cattle slurry (∼ 86%) compared to chicken manure ( ∼ 24%), which is likely to cause microbiome differences. Peng et al. (2022) described a correlation between the chemical characteristics and ARG abundances of cow and sheep manure, which have low ARGs abundance and a poor nutritional content compared to poultry and pig manure, which have a higher amount of ARGs and a richer nutritional content. This suggests that there may be significant differences between raw chicken manure and digestate, which are the result of natural changes in chemical characteristics and microbiome composition during the anaerobic digestion process. Lower ARG relative abundances in digestate might be a sign of AMR reduction, but higher diversity in the future under certain conditions could lead to an increase in AMR.
WHO reported that aminoglycosides are critically important antimicrobials and tetracycline, lincomicide and sulfonamide are highly important antimicrobials. These antibiotics are frequently used in veterinary and human medicine (World Health Organization, 2024). Genes aadA (aminoglycosides), tetA, tetX (Tetracycline), sul1, sul2 (sulfonamides), and lnuF (lincomicides) were detected in our study by HT-qPCR. Gene tetA encodes an efflux pump for tetracycline residues. Another example of an efflux pump encoding is emrD. This pumps efflux of amphipathic molecules (Yin et al., 2006) and belongs to MDR ARGs. Gene tnpA is associated with ARGs because it transfers ARGs. Li et al. (2020) reported genes tetX and sul2 on the same plasmid with tnpA, and Tada et al. (2016) reported sul1. Allmeier et al. (1992) reported transposon Tn1721, including tnpA sequence, with gene tetA.
4.2 ARGs in chicken manure by qPCR
Chicken manure, as a raw material for anaerobic digestion, was investigated, aiming to identify ARG developmental trends in the farms. First, we focused on ARG diversity in chicken manure from different flocks. We analyzed the association of the fold change observations with the flocks’ diseases and treatments. At the beginning of the fattening period, flocks 10 and 11 had colibacillosis and Enterococcus infection, while flocks 4, 5, and 6 had colibacillosis and dysbacteriosis. All flocks received the same treatment of lincomycin, sulfamethoxazole and thrimethoprim, corresponding to the genes lnuF, sul1, sul2 (selected genes), dfrA, and dfrK, which had relative abundances similar to tetA, tetR, and tetM (Supplementary Figure 1). Burow et al. (2020) correlated AMR with antibiotic usage, including aminoglycosides, sulfamethoxazole, and trimethoprim. The flocks in our study were significantly different from each other, but we did not observe any association or difference between single flocks. Peng et al. (2022) compared the relative abundances of ARGs in broiler manures from different farm sizes and other types of manure. In chicken manure, single genes, such as tetX, cfr, and aadA1, varied between the farms (up to 104), while for cfxA, tet(32), and tetO_02, the differences in relative abundances were much lower (approximately 102). Peng et al. (2022) reported a significant difference between broilers raised under traditional and intensive conditions. ARG levels were higher in those from intensive farms due to high density and intensive feeding. However, no association between ARG levels and farm type for other livestock species was found. Another factor we considered in our study was age, because manure characteristics are impacted by chicken age (Prado et al., 2022). There was a general trend for all investigated ARG abundances as they all increased with chicken age. During the fattening period, the temperature in the barn is approximately 25°C, and chicken’s body temperature is approximately 40°C. This is a suitable temperature range for the spread of human pathogens (temperature 35°C–37°C) (Gutierrez et al., 2018). An increase in bacteria could cause a spread of AGRs by mobile genetic elements.
4.3 ARGs in digestate by qPCR
To sum up the results, even though there have been observed some significant differences between individual days and different AD setups (Supplementary Figure 3), the differences were marginal (lower than two-fold) and therefore negligible. Zahedi et al. (2022) reported a significant decrease only of qnrS genes, while other genes, including sul1 do not show changes after AD. Additionally, an increased temperature (55°C compared to 35°C) did not result in a higher ARG reduction. Similarly, Riaz et al. (2020) reported no change in the relative abundance of ARGs such as sul1, aadA, and tetX during the first 5 days of semi-continuous chicken manure AD and mixing with sawdust. Zhang et al. (2019) observed changes in the relative abundance of tetX mostly after day 20. In our study, the experiment was stopped after 20 days, the ARG changes that were analyzed during the first 20 days showed no changes in tetX fold change. A stable ARG level during the AD process was observed for tetM, emrF, mefA, and sul2 by Zhang et al. (2019), and in our study as well, except for emrD, which slightly increased during AD. ARGs can be transferred across various environments, making them difficult to track. Although several models have been developed to predict their spread, one of the most recent approaches involves the use of machine learning (Li et al., 2018). This complexity contributes to the challenge of accurately predicting and quantifying ARGs that pose a significant risk to human health. Zhang et al. (2019) reported changes in the microbial community during anaerobic digestion. On day 0, the dominant phyla were Firmicutes and Proteobacteria, and their relative abundances remained stable over 20 days. In contrast, the abundance of Bacteroidetes increased by approximately 20%, while Actinobacteria remained steady at approximately 5%, making it the fourth-most prevalent phylum. All of these phyla are known to be associated with ARG abundance. These findings suggest that the dominant phyla remained largely unchanged during the digestion process, indicating that ARGs likely had stable microbial hosts to persist and proliferate.
Anjum et al. (2017) showed that after thermophilic AD of poultry manure, selected ARGs were no longer detectable, except for sul1 and sul2. Song et al. (2017) investigated ARGs in swine manure and observed an increase in the abundance of tetX, sul2, and tetQ during the AD process. Song et al. (2017) tested the influence of different biomasses (swine manure mixed with different amounts of wheat straw). The experimental setup with the highest manure content showed maximum abundance of ARGs on day 3, while samples with less manure had their maximum on day 25. Additionally, the experimental setup with a higher proportion of manure to wheat (7:3) showed a decrease in total ARG abundances. Furthermore, Gao et al. (2022) reported a decrease in the abundance of sul1 and emrC in pig manure during AD in a high-solid anaerobic system at a mesophilic temperature, but the abundance of genes sul2 and aadA did not show a strong decrease. In our study, sawdust addition had no relevant impact on ARG loads, similar to the findings of the microbiological analysis (Atanasova et al., 2025), where authors reported no significant differences between E. coli concentration with and without sawdust. Based on the studies by Gao et al. (2022) and Song et al. (2017), anaerobic digestion is more suitable for ARG reduction in AD of swine manure compared to poultry manure. AD showed good results in reducing pathogens and AMR bacteria (Atanasova et al., 2025; Thomas, 2023). Furthermore, ARG reduction was achieved through AD of sewage sludge (Mortezaei et al., 2024) and pig manure (Anjum et al., 2017; Song et al., 2017). In our research, AD did not show a reduction in the abundance of ARGs, which remained stable in our setups with chicken manure. However, AMR results obtained by microbiological cultivation (Atanasova et al., 2025) showed a rapid decrease in the number of viable E. coli during AD. The selective cultivation method for AMR analyses is labor-intensive, selective of only a limited number of bacterial species, and restricted to viable bacteria. This means that selective cultivation alone is not able to cover the abundance and distribution of ARGs within a sample. qPCR enables the selection of more bacterial species and viability-independent, sensitive, and precise quantification of selected ARG abundances. Therefore, AD chicken manure is a suitable process for the reduction of AMR E. coli, while we did not observe a reduction of ARGs in our laboratory batch setups.
5 Conclusion
This study has confirmed that ARGs are present in high amounts in chicken manure. The types and levels of ARGs can vary depending on factors such as chicken feeding, keeping, health, treatment, animal age, and microbiome composition. In this study, we demonstrated that the trend of ARG abundances increased with the chickens’ age, while the ARG profiles between different flocks varied only marginally. Chicken manure was mixed with inoculum from a biogas plant that contained cattle slurry and maize silage for anaerobic digestion, which decreased the total ARG levels but increased ARG diversity. While AD is a common and suitable process for pathogen and AMR bacteria reduction, we did not observe a reduction of selected ARGs during the AD process in our laboratory batch experiments. This means that the risk of ARG spread to the environment by manure field fertilization may not be reduced by AD. Even if pathogens and AMR bacteria are reduced after AD, ARGs could be integrated into bacteria from the environment, and hence, acquire and spread antibiotic resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
AA: Visualization, Data curation, Validation, Investigation, Writing – review & editing, Methodology, Writing – original draft, Conceptualization. TA: Writing – review & editing, Supervision, Conceptualization, Funding acquisition. UR: Software, Resources, Funding acquisition, Methodology, Supervision, Writing – review & editing. AF: Methodology, Supervision, Writing – review & editing, Project administration, Funding acquisition, Resources. RM: Funding acquisition, Software, Visualization, Formal analysis, Project administration, Validation, Writing – review & editing, Supervision. TK: Resources, Visualization, Writing – original draft, Project administration, Data curation, Investigation, Writing – review & editing, Funding acquisition, Validation, Conceptualization, Supervision, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR-ACTION (ENVIRE project)), grant number 963864, and the Federal Ministry of Education and Research (BMBF), grant number 01KI2202B. The APC was funded by the Leibniz Institute for Agricultural Engineering and Bioeconomy e. V. (ATB).
Acknowledgments
We are grateful for the intellectual project support and exchange with the Leibniz Research Alliance “INFECTIONS”. We thank Heike Rose, Susann Sellenthin, Sabrina Hansen, and Michael Kühl (all FU, Berlin, Germany) for excellent technical advice and assistance in the laboratory. We also thank the cooperating broiler farm for providing us with chicken manure and disease treatment records.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frabi.2025.1612886/full#supplementary-material
References
Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920
Allmeier H., Cresnar B., Greck M., and Schmitt R. (1992). Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111, 11–20. doi: 10.1016/0378-1119(92)90597-I
Anjum R., Grohmann E., and Krakat N. (2017). Anaerobic digestion of nitrogen rich poultry manure: Impact of thermophilic biogas process on metal release and microbial resistances. Chemosphere 168, 1637–1647. doi: 10.1016/j.chemosphere.2016.11.132
Appunn K. (2016). Bioenergy - the troubled pillar of the Energiewende, Biogas in Germany’s energy transition.
Atanasova A., Amon T., Friese A., Rösler U., Merle R., Herrmann C., et al. (2025). Effects of carbon–to–nitrogen ratio and temperature on the survival of antibiotic-resistant and non-resistant escherichia coli during chicken manure anaerobic digestion. Poultry 4. doi: 10.3390/poultry4010009
Błażejewska A., Zalewska M., Grudniak A., and Popowska M. (2022). A comprehensive study of the microbiome, resistome, and physical and chemical characteristics of chicken waste from intensive farms. Biomolecules 12. doi: 10.3390/biom12081132
Burow E., Grobbel M., Tenhagen B.-A., Simoneit C., Szabó I., Wendt D., et al. (2020). Antibiotic resistance in escherichia coli from broiler chickens after amoxicillin treatment in an experimental environment. Microb. Drug Resist. 26, 1098–1107. doi: 10.1089/mdr.2019.0442
Congilosi J. L. and Aga D. S. (2021). Review on the fate of antimicrobials, antimicrobial resistance genes, and other micropollutants in manure during enhanced anaerobic digestion and composting. J. Hazardous Materials 405, 123634. doi: 10.1016/j.jhazmat.2020.123634
Ding D., Wang B., Zhang X., Zhang J., Zhang H., Liu X., et al. (2023). The spread of antibiotic resistance to humans and potential protection strategies. Ecotoxicology Environ. Saf. 254, 114734. doi: 10.1016/j.ecoenv.2023.114734
European Biogas Association (2020). The ‘European Biomethane Map 2020’ shows a 51% increase of biomethane plants in Europe in two years.
Gao W., Li A., Ding G., Zhang K., and Zhi S. (2022). Investigating changes in the characteristics of antibiotic resistance genes at different reaction stages of high solid anaerobic digestion with pig manure. Environ. pollut. 312, 120032. doi: 10.1016/j.envpol.2022.120032
German Federal Institute for Risk Assessment (2025). Antibiotic use in fattening animals: First annual report provides more precise data (No. 16/2023).
Gurmessa B., Ashworth A. J., Yang Y., Savin M., Moore P. A., Ricke S. C., et al. (2021). Variations in bacterial community structure and antimicrobial resistance gene abundance in cattle manure and poultry litter. Environ. Res. 197, 111011. doi: 10.1016/j.envres.2021.111011
Gutierrez C., Somoskovi A., Natarajan K., and Bell D. (2018). Need for better adherence to optimal incubation temperature for quality laboratory diagnostics and antibiotic resistance monitoring. Afr J. Lab. Med. 7, 789. doi: 10.4102/ajlm.v7i2.789
Jaleta M., Junker V., Kolte B., Börger M., Werner D., Dolsdorf C., et al. (2024). Improvements of weaned pigs barn hygiene to reduce the spread of antimicrobial resistance. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1393923
Köninger J., Lugato E., Panagos P., Kochupillai M., Orgiazzi A., and Briones M. J. I. (2021). Manure management and soil biodiversity: Towards more sustainable food systems in the EU. Agric. Syst. 194, 103251. doi: 10.1016/j.agsy.2021.103251
Li Y., Wang Q., Peng K., Liu Y., Li R., and Wang Z. (2020). Emergence of carbapenem- and tigecycline-resistant proteus cibarius of animal origin. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01940
Li L.-G., Yin X., and Zhang T. (2018). Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome 6, 93. doi: 10.1186/s40168-018-0480-x
Mortezaei Y., Williams M. R., and Demirer G. N. (2024). The fate of antibiotic resistance genes during anaerobic digestion of sewage sludge with ultrasonic pretreatment. Environ. Sci. pollut. Res. 31, 5513–5525. doi: 10.1007/s11356-023-31558-6
Muurinen J., Stedtfeld R., Karkman A., Pärnänen K., Tiedje J., and Virta M. (2017). Influence of manure application on the environmental resistome under finnish agricultural practice with restricted antibiotic use. Environ. Sci. Technol. 51, 5989–5999. doi: 10.1021/acs.est.7b00551
Okonkwo V. O. (2023). Antimicrobial resistance: molecular approaches to track antimicrobial resistance gene spread from decentralised septic tank wastewater (Scotland: University of Glasgow).
Partridge S. R., Kwong S. M., Firth N., and Jensen S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31. doi: 10.1128/CMR.00088-17
Peng S., Zhang H., Song D., Chen H., Lin X., Wang Y., et al. (2022). Distribution of antibiotic, heavy metals and antibiotic resistance genes in livestock and poultry feces from different scale of farms in Ningxia, China. J. Hazardous Materials 440, 129719. doi: 10.1016/j.jhazmat.2022.129719
Prado J., Ribeiro H., Alvarenga P., and Fangueiro D. (2022). A step towards the production of manure-based fertilizers: Disclosing the effects of animal species and slurry treatment on their nutrients content and availability. J. Cleaner Production 337, 130369. doi: 10.1016/j.jclepro.2022.130369
Riaz L., Wang Q., Yang Q., Li X., and Yuan W. (2020). Potential of industrial composting and anaerobic digestion for the removal of antibiotics, antibiotic resistance genes and heavy metals from chicken manure. Sci. Total Environ. 718, 137414. doi: 10.1016/j.scitotenv.2020.137414
Skandalis N., Maeusli M., Papafotis D., Miller S., Lee B., Theologidis I., et al. (2021). Environmental spread of antibiotic resistance. Antibiotics (Basel) 10. doi: 10.3390/antibiotics10060640
Song W., Wang X., Gu J., Zhang S., Yin Y., Li Y., et al. (2017). Effects of different swine manure to wheat straw ratios on antibiotic resistance genes and the microbial community structure during anaerobic digestion. Bioresource Technol. 231, 1–8. doi: 10.1016/j.biortech.2017.01.054
Stedtfeld R. D., Guo X., Stedtfeld T. M., Sheng H., Williams M. R., Hauschild K., et al. (2018). Primer set 2.0 for highly parallel qPCR array targeting antibiotic resistance genes and mobile genetic elements. FEMS Microbiol. Ecol. 94. doi: 10.1093/femsec/fiy130
Sun B., Bai Z., Li R., Song M., Zhang J., Wang J., et al. (2024). Efficient elimination of antibiotic resistome in livestock manure by semi-permeable membrane covered hyperthermophilic composting. Bioresource Technol. 407, 131134. doi: 10.1016/j.biortech.2024.131134
Tada T., Miyoshi-Akiyama T., Shimada K., Shiroma A., Nakano K., Teruya K., et al. (2016). A Carbapenem-Resistant Pseudomonas aeruginosa Isolate Harboring Two Copies of blaIMP-34 Encoding a Metallo-β-Lactamase. PloS One 11, e0149385. doi: 10.1371/journal.pone.0149385
Thomas C. (2023). Manure management measures to reduce the risk of spreading ESBL-/AmpCproducing Escherichia coli from chicken manure into the food chain (Leibniz-Institut für Agrartechnik und Bioökonomie: Freie Universität Berlin).
Wang L., Wang J., Wang J., Zhu L., Yang L., and Yang R. (2019). Distribution characteristics of antibiotic resistant bacteria and genes in fresh and composted manures of livestock farms. Sci. Total Environ. 695, 133781. doi: 10.1016/j.scitotenv.2019.133781
Waseem H., Jameel S., Ali J., Saleem Ur Rehman H., Tauseef I., Farooq U., et al. (2019). Contributions and challenges of high throughput qPCR for determining antimicrobial resistance in the environment: A critical review. Molecules 24. doi: 10.3390/molecules24010163
World Health Organization (2024). “A risk management tool for mitigating antimicrobial resistance due to non-human use (No. ISBN 978-92-4-008461-2),” in WHO List of Medically Important Antimicrobials: a risk management tool for mitigating antimicrobial resistance due to non-human use. (Geneva: World Health Organization)
Yangin-Gomec C., Sapmaz T., and Aydin S. (2020). Impact of inoculum acclimation on energy recovery and investigation of microbial community changes during anaerobic digestion of the chicken manure. Environ. Technol. 41, 49–58. doi: 10.1080/09593330.2018.1551434
Yin Y., He X., Szewczyk P., Nguyen T., and Chang G. (2006). Structure of the multidrug transporter EmrD from Escherichia coli. Science 312, 741–744. doi: 10.1126/science.1125629
Yue Z., Zhang J., Zhou Z., Ding C., Zhang T., Wan L., et al. (2022). Antibiotic degradation dominates the removal of antibiotic resistance genes during composting. Bioresource Technol. 344, 126229. doi: 10.1016/j.biortech.2021.126229
Zahedi S., Gros M., Casabella O., Petrovic M., Balcazar J. L., and Pijuan M. (2022). Occurrence of veterinary drugs and resistance genes during anaerobic digestion of poultry and cattle manures. Sci. Total Environ. 822, 153477. doi: 10.1016/j.scitotenv.2022.153477
Zhang Y., Banks C. J., and Heaven S. (2012). Co-digestion of source segregated domestic food waste to improve process stability. Bioresource Technol. 114, 168–178. doi: 10.1016/j.biortech.2012.03.040
Zhang J., Lu T., Shen P., Sui Q., Zhong H., Liu J., et al. (2019). The role of substrate types and substrate microbial community on the fate of antibiotic resistance genes during anaerobic digestion. Chemosphere 229, 461–470. doi: 10.1016/j.chemosphere.2019.05.036
Zhang C., Xiao G., Peng L., Su H., and Tan T. (2013). The anaerobic co-digestion of food waste and cattle manure. Bioresource Technol. 129, 170–176. doi: 10.1016/j.biortech.2012.10.138
Keywords: antibiotics, anaerobic digestion fermentation, antimicrobial resistance gene (ARG), poultry, broiler, litter, high throughput qPCR (HT-qPCR)
Citation: Atanasova A, Amon T, Roesler U, Friese A, Merle R and Kabelitz T (2025) Temporal dynamics of antimicrobial resistance gene abundances in chicken manure and anaerobic digestate. Front. Antibiot. 4:1612886. doi: 10.3389/frabi.2025.1612886
Received: 16 April 2025; Accepted: 09 June 2025;
Published: 27 June 2025.
Edited by:
Kebede Amenu, International Livestock Research Institute, EthiopiaReviewed by:
Santosh Dulal, Management Sciences for Health, United StatesAchenef Beyene, University of Gondar, Ethiopia
Copyright © 2025 Atanasova, Amon, Roesler, Friese, Merle and Kabelitz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksandra Atanasova, YWF0YW5hc292YUBhdGItcG90c2RhbS5kZQ==
 Aleksandra Atanasova
Aleksandra Atanasova Thomas Amon1,2
Thomas Amon1,2 Uwe Roesler
Uwe Roesler Anika Friese
Anika Friese Roswitha Merle
Roswitha Merle Tina Kabelitz
Tina Kabelitz