- 1USDA, ARS, National Laboratory for Agriculture and the Environment, Ames, IA, USA
- 2USDA, ARS, National Animal Disease Center, Ames, IA, USA
- 3Embrapa Swine and Poultry, Concórdia, Brazil
- 4Department of Biological Sciences and Biotechnology, Hannam University, Daejeon, South Korea
Swine are often asymptomatic carriers of Salmonella spp., and interventions are needed to limit colonization of swine to enhance food safety and reduce environmental contamination. We evaluated the attenuation and potential vaccine use in pigs of a Salmonella enterica serovar Typhimurium mutant of rfaH, the gene encoding the RfaH antiterminator that prevents premature termination of long mRNA transcripts. Pigs inoculated with wild-type S. Typhimurium exhibited a significant elevation in average body temperature (fever) at 1 and 2 days post-inoculation; rfaH-inoculated pigs did not (n = 5/group). During the 7-day trial, a significant reduction of Salmonella in the feces, tonsils, and cecum were observed in the rfaH-inoculated pigs compared to wild-type inoculated pigs. To determine whether vaccination with the rfaH mutant could provide protection against wild-type S. Typhimurium challenge, two groups of pigs (n = 14/group) were intranasally inoculated with either the rfaH mutant or a PBS placebo at 6 and 8 weeks of age and challenged with the parental, wild-type S. Typhimurium at 11 weeks of age. The average body temperature was significantly elevated in the mock-vaccinated pigs at 1 and 2 days post-challenge, but not in the rfaH-vaccinated pigs. Fecal shedding at 2 and 3 days post-challenge and colonization of intestinal tract tissues at 7 days post-challenge by wild-type S. Typhimurium was significantly reduced in the rfaH-vaccinated pigs compared to mock-vaccinated pigs. Serological analysis using the IDEXX HerdChek Swine Salmonella Test Kit indicated that vaccination with the rfaH mutant did not stimulate an immune response against LPS. These results indicate that vaccination of swine with the attenuated rfaH mutant confers protection against challenge with virulent S. Typhimurium but does not interfere with herd level monitoring for Salmonella spp., thereby allowing for differentiation of infected from vaccinated animals (DIVA).
Introduction
An estimated one million cases of foodborne illness are attributed to non-typhoidal Salmonella spp. each year in the U.S. at a predicted cost of $2.3 billion (1, 2). Non-typhoidal Salmonella spp. are a leading cause of hospitalization (35%) and death (28%) in the U.S. due to foodborne disease (2). Of 14 foodborne pathogens that cause 95% of foodborne illnesses, hospitalizations, and death in the U.S., non-typhoidal Salmonella spp. are the leading cause of disease burden (27%) measured by quality-adjusted life years (QALYs) (3).
Most of the >2,400 Salmonella serovars are ubiquitous in the environment and many can colonize food producing animals and poultry as well as wild animals and birds without causing overt disease. In the U.S., ~68,000 cases of pork-associated, human salmonellosis occur each year with an estimated social cost of $218 million annually (4). Of 104 pathogen–food combinations, the annual disease burden due to S. enterica in pork is estimated as the 13th highest. The most recent study from the USDA’s National Animal Health Monitoring System, Swine 2006, reported that 52.6% of swine production sites (representing 94% of the U.S. swine inventory) were positive for Salmonella spp. (5). Swine that are Salmonella carriers are a food safety risk for consumers of pork, an animal health risk to non-colonized/uninfected pigs, an economical risk to producers due to reduced feed conversion/efficiency (6, 7), and an environmental risk due to fecal shedding of the pathogen into swine manure that is used as a soil amendment. Interventions are needed to limit the colonization of swine with Salmonella spp. and reduce the risks to public health, animal health, and the environment.
The potential use of a Salmonella enterica serovar Typhimurium rfaH mutant as a vaccine candidate has been described (8, 9). The RfaH antiterminator prevents the premature termination of long mRNA transcripts encoded by large operons. A 5′ cis-acting operon polarity suppressor (ops) element is required to enhance transcription elongation from operons in the RfaH regulon (10, 11). S. Typhimurium rfaH mutants have decreased transcription of genes encoding lipopolysaccharide (LPS) core, O-antigen, Salmonella Pathogenicity Island 4 (SPI-4), flagellum/chemotaxis, and type III secretion system 1 (T3SS-1) (12). However, the decreased transcription of some of these operons may be due indirectly to the loss of LPS. A S. Typhimurium SL1344 rfaH mutant is attenuated for virulence (both oral and intraperitoneal inoculation) in the BALB/c mouse model of systemic disease compared to wild-type SL1344 (8). Immunization of BALB/c mice with three doses of an rfaH mutant [5 × 107 colony forming units (CFU)] at 2-week intervals was protective against challenge with 5 × 107 CFU of wild-type SL1344 at 2 weeks following the final vaccine dose (0/4 vs. 4/4 deaths for vaccinated and naïve mice). Further investigation indicated that vaccination of BALB/c mice with three increasing doses (2 × 107, 2 × 108, and 2 × 109 CFU) of the SL1344 rfaH mutant at 2-week intervals resulted in significant protection against homologous (86%) and heterologous (40%) challenge with 5 × 106 CFU of either wild-type SL1344 or wild-type S. Enteritidis NCTC13349, respectively (13). These results indicate that vaccination of mice with the SL1344 rfaH mutant provides some cross-protection against other serovars in addition to S. Typhimurium.
In the current study, an rfaH knockout mutant of Salmonella enterica serovar Typhimurium was constructed to determine whether: (1) the strain was attenuated in pigs, (2) would protect swine against virulent S. Typhimurium challenge, while (3) allowing for DIVA using the IDEXX HerdChek Swine Salmonella Test Kit. This is the first report of an rfaH mutant evaluation in swine and our results indicate that rfaH-vaccinated pigs have reduced disease severity, pathogen fecal shedding, and intestinal colonization due to virulent S. Typhimurium challenge, but swine vaccination did not interfere with the herd level monitoring system for Salmonella spp.
Materials and Methods
Strain Construction and Growth Conditions
The S. Typhimurium BBS 202 (rfaH:neo) strain was constructed by recombineering as previously described (14). Briefly, oligos oBBI 189 (atgcaatcctggtatttactgtactgcaaacgcgggcaacttca gcatagagcagtgacgtagtcgc) and oBBI 190 (ctaaatcttgcgaaaaccggtgttttttacgctctgcttcactt cgatagctgaatgagtgacgtgc) were used to PCR amplify the oBBI 92/93-neo template for synthesis of a linear knockout fragment. The 5′ end (bold) of oBBI 189 and oBBI 190 have homology to the 5′ and 3′ rfaH target sequences (respectively) whereas the 3′ end (underlined) of the oligos have homology to oBBI 92/93-neo encoding an antibiotic resistance gene flanked by universal sequences that truncate potential translation of the target gene. Following PCR amplification, gel electrophoresis was performed using a 1% agarose gel in TBE and the oBBI 189/190-neo knockout fragment was gel purified using a BioRad Freeze ‘N Squeeze column (Hercules, CA, USA). The linear knockout fragment containing the neo gene was electroporated into arabinose-induced competent cells of S. Typhimurium BSX 7 (14) containing the plasmid pKD46 encoding Lambda exo, bet, and gam to facilitate recombination of the DNA fragment (15). Kanamycin-resistant transformants were screened by PCR to confirm the replacement of rfaH with the neo gene and this strain was stocked as BBS 195. Due to the rfaH mutant being a poor recipient for P22 binding, the rfaH gene was cloned into a plasmid to complement the rfaH mutation using primers oBBI 193 (caggaagacgcgttaaatcg) and oBBI 194 (gaacgatcgctaaatcttgc). The oBBI 193 primer binds within yigC (STM3978), ~625 bp upstream of the codon for the rfaH start methionine. The binding site for the oBBI 194 primer overlaps with the rfaH stop codon. Primers oBBI 193 and oBBI 194 were used to amplify the rfaH region from the S. Typhimurium χ4232 genome, and the PCR product was cloned using the pBAD TOPO TA Expression Kit (Invitrogen, Carlsbad, CA, USA) followed by transformation into One Shot TOP10 competent E. coli. The pBAD-oBBI193/194 plasmid was extracted from the TOP10 strain and transformed into BBS 195 (rfaH:neo), single colony isolated, and stocked as BBS 201. Using a P22 high-transducing phage lysate grown on BBS 201, the rfaH:neo mutation was transferred to wild-type S. Typhimurium χ4232, creating BBS 202.
The S. Typhimurium strains χ4232 and BBS 202 were grown in LB in the presence of nalidixic acid without and with kanamycin, respectively. Antibiotics were used at the following concentrations: ampicillin (100 μg/ml) for pKD46, kanamycin (50 μg/ml), and nalidixic acid (30 μg/ml).
Ethics Statement
All experimental procedures involving animals were in compliance with the recommended principles described in the Guide for the Care and Use of Laboratory Animals by the National Research Council of the National Academies and were approved by the USDA-ARS, National Animal Disease Center, Animal Care, and Use Committee.
Animal Trial #1
Ten crossbred, conventionally raised piglets from five Salmonella-fecal-negative sows were weaned at 12 days of age and shipped to the National Animal Disease Center, Ames, IA, USA. Siblings from each litter were divided and raised in two isolation rooms (n = 5/room). Piglets tested fecal-negative for Salmonella spp. thrice over a 4-week period using bacteriological culture with selective enrichment (16). At 6 weeks of age (day zero), pigs received an intranasal inoculation of 1 ml PBS containing either 1 × 109 CFU of BBS 202 (rfaH:neo; n = 5) or S. Typhimurium χ4232 (wild-type; n = 5). Clinical symptoms and pathogen fecal shedding were monitored at 0, 1, 2, 3, 5, and 7 days post-inoculation (dpi) (see below).
Animal Trial #2
Twenty-eight crossbred, conventionally raised piglets from four Salmonella-fecal-negative sows were weaned at 12 days of age and shipped to the National Animal Disease Center, Ames, IA, USA. Equally divided from each litter, the pigs were raised in two isolation rooms (n = 14/room) and tested fecal-negative for Salmonella spp. twice over a 4-week period using bacteriological culture with selective enrichment. At 6 weeks of age, pigs received a 1 ml intranasal inoculation of either 1 × 109 BBS 202 or a placebo (PBS); a booster inoculation occurred 2 weeks later at 8 weeks of age. At 11 weeks of age, all pigs were intranasally challenged with PBS containing 1 × 108 CFU of wild-type S. Typhimurium (χ4232) and monitored for clinical disease symptoms, Salmonella-fecal shedding and intestinal colonization over a 7-day period (see below).
Sample Collection and Processing
Swine body temperatures were determined using a rectal thermometer, and blood samples were obtained via the jugular vein at each inoculation time point as well as 7 days post-challenge. Fecal samples were obtained for quantitative and qualitative Salmonella culture analysis as previously described (16) with the addition of XLT-4 medium containing the appropriate antibiotics to select for each strain (nalidixic acid for χ4232; nalidixic acid and kanamycin for BBS 202). At 7 dpi, all pigs were euthanized and necropsies performed to obtain tissue samples from the tonsil and the intestinal tract (ileal Peyer’s Patches, ileocecal lymph nodes, and cecum) for quantitative and qualitative Salmonella culture analysis as previously described with slight modification (17). One hundred microliters of each homogenized tissue (1 g of tissue combined with 2 ml of PBS) was aliquoted onto BGS and XLT-4 media containing kanamycin and/or nalidixic acid (see below).
To differentiate between the wild-type and rfaH mutant strains in the bacteriological culture assays, fecal and tissues samples were plated on medium containing nalidixic acid alone and medium containing both nalidixic acid and kanamycin. As both χ4232 and BBS 202 are resistant to nalidixic acid, the medium with nalidixic acid and kanamycin was used to identify BBS 202. Subtraction of the number of colonies present on the kanamycin–nalidixic acid medium from the colonies present on the nalidixic acid only medium determined the number of χ4232 colonies.
All statistical analyses were performed in GraphPad Prism 5.01 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analysis of body temperatures for each treatment group was performed using repeated measures ANOVA followed by Bonferroni’s Multiple Comparison Test. Statistical analysis of Salmonella present (Log10 colony forming units per gram) in fecal samples was analyzed by two-way ANOVA followed by Bonferroni post-tests. Statistical analysis of Salmonella present (Log10 colony forming units per gram) in tissues at 7 dpi was analyzed using the Two Sample t-test for the Means.
Fecal Score Analysis
Each fecal sample in animal trial #2 was assigned a diarrhea score (18) by the same four evaluators at the time of collection (prior to information concerning shedding status) using a scale of 1–5 (1 = dry feces, 2 = moist feces, 3 = mild diarrhea, 4 = severe diarrhea, and 5 = watery diarrhea). Statistical analysis of diarrhea scores was analyzed by two-way ANOVA followed by Bonferroni post-tests.
IDEXX HerdChek Swine Salmonella Antibody Assay
Serum antibody analysis to LPS antigen derived from Salmonella serogroups B, C1, and D was performed using the IDEXX HerdChek Swine Salmonella Test Kit (IDEXX Europe B.V., Hoofddorp, Netherlands). The assay and interpretation were performed per the manufacturer’s instructions. Briefly, the 30-min incubation was performed and the sample to positive (S/P) ratio was calculated for each sample using S/P = (sample mean − negative control mean)/positive control mean − negative control mean). A sample’s S/P ratio of ≥0.25 was considered positive and <0.25 was considered negative. Analysis of piglet serum was performed in-house whereas sow screening was performed by Boehringer Ingelheim, Vetmedica, Inc. (Ames, IA, USA).
Results and Discussion
rfaH mutant of S. Typhimurium is attenuated in swine
An rfaH mutant of S. Typhimurium was constructed and tested for attenuation in vivo. Following a 1 × 109 CFU inoculation, the average body temperature (fever) of pigs inoculated with the parental, wild-type S. Typhimurium χ4232 was significantly higher at day 1 (39.9°C) and day 2 (39.3°C) post-inoculation compared to day 0 (38.2°C) for the wild-type strain (Figure 1A; P < 0.05). Also, the average body temperature of pigs inoculated with wild-type S. Typhimurium was significantly higher at 1 dpi compared to pigs inoculated with the rfaH mutant (Figure 1A; 39.9°C vs. 38.5°C; P < 0.05). In fact, the average body temperature of the pigs inoculated with the rfaH mutant did not significantly increase throughout the 7-day trial. Swine fecal shedding of the S. Typhimurium rfaH mutant was significantly reduced (up to 2.5 logs) at days 3, 5, and 7 dpi compared to wild-type S. Typhimurium (Figure 1B; P < 0.05). Furthermore, tissue colonization was significantly lower in the tonsil (>2 logs) and cecum (2.5 logs) in the pigs inoculated with the rfaH mutant compared to wild-type S. Typhimurium (Figure 1C; P < 0.05), although no difference in colonization of the ileal Peyer’s Patches or ileocecal lymph nodes occurred. Thus, the rfaH mutant is attenuated for clinical symptoms (fever) with reduced fecal shedding and tissue colonization compared to the parental, wild-type S. Typhimurium.
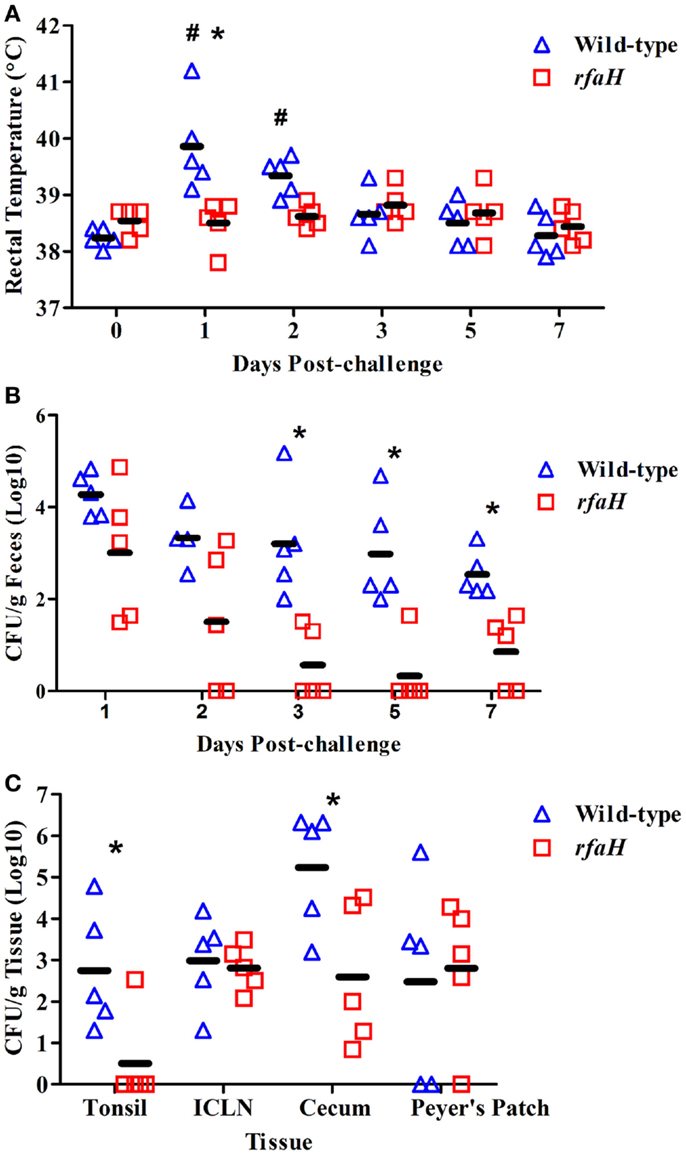
Figure 1. Average rectal temperature, fecal shedding, and tissue colonization were significantly reduced in pigs inoculated with the rfaH mutant compared to the parental wild-type S. Typhimurium. At 6 weeks of age, swine were inoculated with 1 ml of PBS containing either 1 × 109 colony forming units (CFU) of BBS 202 (rfaH:neo; n = 5) or S. Typhimurium χ4232 (wild-type; n = 5). (A) Swine rectal temperature at 0, 1, 2, 3, 5, and 7 dpi. (B) Salmonella bacteriological analysis (colony forming units per gram) of swine fecal samples obtained at 1, 2, 3, 5, and 7 dpi. (C) Salmonella bacteriological analysis (colony forming units per gram) of the tonsil, ileocecal lymph nodes (ICLN), Peyer’s Patches, and cecum obtained during necropsy at 7 dpi. *Significant difference (P < 0.05) between swine inoculated with wild-type S. Typhimurium compared to the rfaH mutant at the indicated time point. #Significant difference (P < 0.05) in wild-type inoculated pigs at the indicated time point compared to day 0.
Vaccination of Pigs with the rfaH Mutant Significantly Reduces the Effects of Subsequent Challenge with Wild-Type S. Typhimurium
To determine whether vaccination with the rfaH mutant could provide protection against virulent S. Typhimurium challenge in swine, pigs were inoculated with the potential vaccine strain at 6 weeks of age and boosted 2 weeks later. As similarly reported in the attenuation trial above, no significant increase in average body temperature was observed following vaccination or booster vaccination of the pigs, thereby corroborating the data presented in Figure 1A for the rfaH mutant (data not shown). Mock-vaccinated pigs (n = 14) and rfaH-vaccinated pigs (n = 14) were challenged with 1 × 108 CFU of the parental, wild-type S. Typhimurium at 11 weeks of age. The swine rectal temperature increased significantly at days 1 (40.3°C) and 2 (40.1°C) post-challenge compared to day 0 (39.1°C) in the mock-vaccinated pigs (Figure 2A; P < 0.05). Moreover, the swine rectal temperature was significantly increased in mock-vaccinated pigs compared to rfaH-vaccinated pigs at days 1 (40.3°C vs. 39.4°C) and 2 (40.1°C vs. 39.3°C) post-challenge (Figure 2A; P < 0.05). Following challenge with wild-type S. Typhimurium, the average body temperature of the pigs vaccinated with the rfaH mutant did not significantly increase throughout the 7-day trial. The average diarrhea score, a subjective measurement of moisture content, was significantly higher in mock-vaccinated pigs compared to rfaH-vaccinated pigs at 2 dpi (2.7 vs. 2.1, respectively; P < 0.05). Swine fecal shedding of S. Typhimurium was significantly increased (~1.5 logs) at days 2 and 3 post-challenge in mock-vaccinated pigs compared to rfaH-vaccinated pigs (Figure 2B; P < 0.05). Furthermore, S. Typhimurium colonization was significantly decreased in the ileocecal lymph nodes, ileal Peyer’s Patches, and cecum (~10-fold) at 7 days post-challenge in rfaH-vaccinated pigs compared to mock-vaccinated pigs (Figure 2C; P < 0.05). Hence, vaccination with the rfaH mutant reduced clinical symptoms (fever and diarrhea), Salmonella-fecal shedding and intestinal colonization upon challenge with virulent S. Typhimurium.
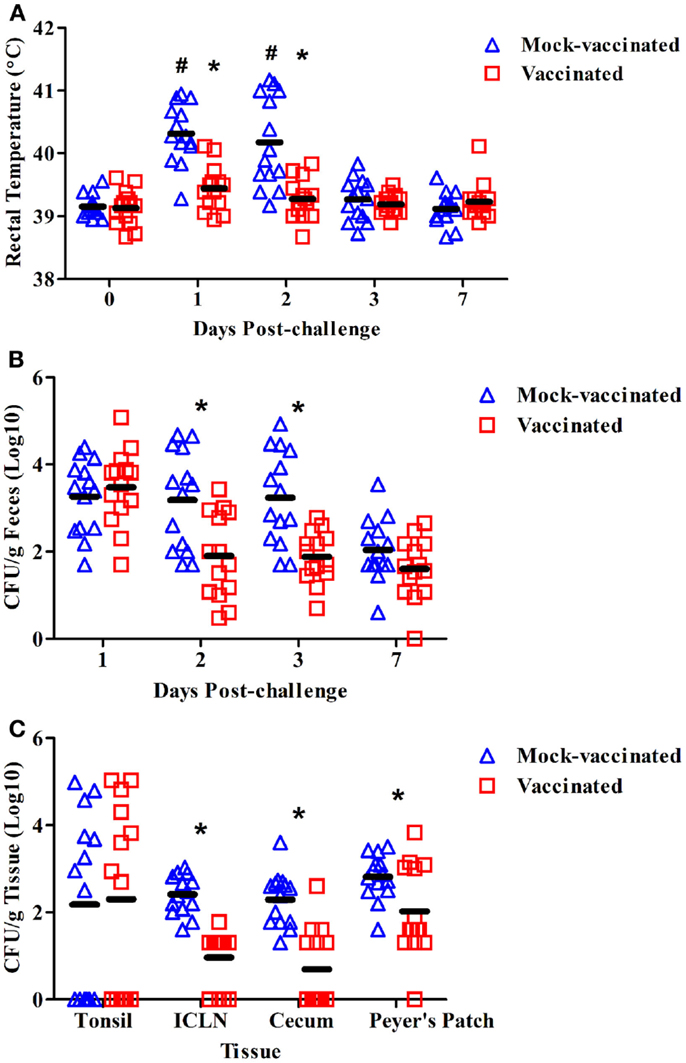
Figure 2. The rfaH-vaccinated pigs had significantly lower average rectal temperature, fecal shedding, and tissue colonization compared to the mock-vaccinated pigs following challenge with wild-type S. Typhimurium. At 6 and 8 weeks of age, pigs were inoculated with the potential rfaH vaccine strain (BBS 202) or mock–vaccinated with PBS (n = 14/group). At 11 weeks of age, all 28 pigs were challenged with 1 × 108 CFU of the parental, wild-type S. Typhimurium χ4232. (A) Swine rectal temperature at 0, 1, 2, 3, and 7 days post-challenge. (B) Salmonella bacteriological analysis (colony forming units per gram) of swine fecal samples obtained at 1, 2, 3, and 7 days post-challenge. (C) Salmonella bacteriological analysis (colony forming units per gram) of the tonsil, ileocecal lymph nodes (ICLN), Peyer’s Patches, and cecum obtained during necropsy at 7 dpi. *Significant difference (P < 0.05) comparing rfaH-vaccinated to mock-vaccinated pigs at the same time point. #Significant difference (P < 0.05) in mock-vaccinated pigs at the indicated time point compared to day 0.
rfaH Mutant is a DIVA Vaccine
To determine if the pigs vaccinated with the rfaH mutant were producing detectable antibodies to Salmonella LPS antigen, sera from all 28 pigs (14 mock-vaccinated and 14 rfaH-vaccinated) were assayed using the IDEXX HerdChek Swine Salmonella Test Kit at multiple time points including 11-weeks of age (5 weeks following initial inoculation with the rfaH mutant and the day of wild-type S. Typhimurium challenge). All 28 pigs were negative for antibodies to Salmonella LPS antigen by ELISA at 11-weeks of age, indicating that vaccination with the S. Typhimurium rfaH mutant did not stimulate the production of antibodies against LPS (Figure 3). Therefore, the rfaH vaccine strain could be used in conjunction with Salmonella surveillance programs in swine for DIVA. At 12-weeks of age (7-days post-challenge with wild-type S. Typhimurium), the sera from 85% of rfaH-vaccinated and 78% of mock-vaccinated pigs were positive in the LPS ELISA assay, signifying that seroconversion to Salmonella LPS antigen following wild-type S. Typhimurium challenge had occurred. These results indicate that vaccination of swine with the rfaH DIVA vaccine does not interfere with the detection of a subsequent exposure to Salmonella.
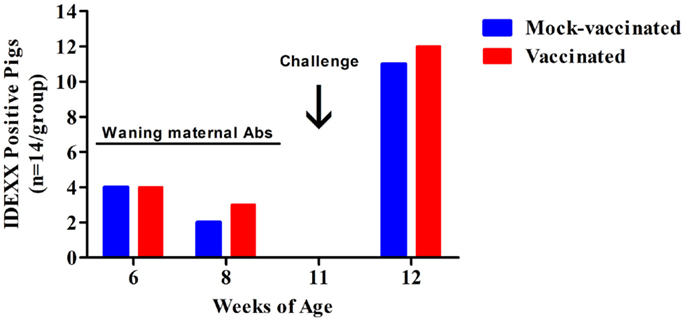
Figure 3. Serum from rfaH-vaccinated swine is negative in the IDEXX HerdChek Swine Salmonella Test Kit 5-weeks after initial vaccination. The number of pigs with serum containing antibodies to Salmonella LPS at 6-weeks (initial vaccination), 8-weeks (booster vaccination), 11-weeks (challenge), and 12-weeks (7 days post-challenge) of age. The number of positive pigs at each time point are shown. All pigs were challenged with wild-type S. Typhimurium at 11-weeks of age. A sample’s S/P ratio of ≥0.25 was considered positive and <0.25 was considered negative.
At 6-weeks of age, 8 of 28 piglets (4 pigs in each of the two treatment groups) were positive for antibodies to Salmonella LPS antigen by ELISA (Figure 3). These eight piglets were siblings from the same sow and their antibodies waned as the age of the piglets increased. Taken together, these data indicate that the antibodies to Salmonella LPS in these piglets were maternal antibodies. Salmonella surveillance data including Swine 2006 suggests that Salmonella colonization in swine herds is not unusual. From 2011 to 2013, we screened 112 sows from 4 commercial swine facilities for the presence of Salmonella spp. in feces and antibodies to Salmonella LPS in serum. Salmonella was isolated from the feces of 24 sows (21%) and antibodies to Salmonella LPS were detected in the serum of 107 sows (95%). This indicates that although not all are actively shedding Salmonella, 95% of sows had a previous exposure to Salmonella spp. Therefore, our finding that a subset of the piglets in this vaccine trial had maternally acquired antibodies to Salmonella LPS is not surprising. Furthermore, since 95% of the sows tested were Salmonella antibody positive, exposure of pigs to Salmonella on-farm is probable, indicating a need for improved interventions including vaccination.
Previous investigations have described potential DIVA vaccines for swine utilizing different Salmonella gene targets (19–21). A limitation of some currently licensed Salmonella vaccines is that vaccination produces Salmonella antibodies in the host that cannot be differentiated from antibodies produced during an active infection, and therefore could interfere with surveillance programs that monitor Salmonella in swine production facilities. To eliminate potential interference, a licensed vaccine (Salmoporc, IDT Biologika GmbH) was modified (ΔompD) and a new ELISA was developed to distinguish between infected and vaccinated animals (19). The modified vaccine did not stimulate an immune response that was detected in their anti-OmpD ELISA. However, current surveillance programs (ongoing since the early 1990s) utilize a Salmonella LPS ELISA, and switching to another ELISA method would need further validation prior to implementation in swine production. The use of S. Typhimurium LPS mutants for DIVA vaccine development has also been explored (20). Specifically, S. Typhimurium ΔrfaJ and ΔrfaL mutants when inoculated into swine did not stimulate an anti-Salmonella LPS response in contrast to the parental wild-type S. Typhimurium. However, further experiments in pigs were not performed utilizing these specific S. Typhimurium (ΔrfaJ and ΔrfaL) mutants, although the Salmoporc vaccine strain (above) was modified to incorporate the ΔrfaJ mutation (21). Inoculation of pigs with the Salmoporc-ΔrfaJ strain did not induce Salmonella-specific antibodies to LPS when serum was analyzed using the IDEXX HerdChek Swine Salmonella Test. In a S. Typhimurium transmission experiment whereby vaccinated (Salmoporc-ΔrfaJ), non-challenged pigs were exposed to S. Typhimurium-challenged “seeder” pigs, vaccination did not significantly decrease the adjusted transmission ratio compared to non-vaccinated pigs. As we performed a challenge experiment and not a transmission experiment in this study, we cannot directly compare our results to those of De Ridder et al.
Investigations in BALB/c mice have previously demonstrated that a S. Typhimurium SL1344 rfaH mutant is attenuated and protective against challenge with wild-type S. Typhimurium SL1344 (8). Mutations in rfaH have also been constructed in S. Typhimurium UK-1 and S. Gallinarum 287/91, and virulence attenuation has been demonstrated for both of these rfaH mutants in BALB/c mice and Rhode Island Red chicks, respectively (9, 22). Furthermore, inoculation of Rhode Island Red chickens with two doses of the S. Gallinarum 287/91 rfaH mutant (χ11571) provided significant protection against challenge with S. Gallinarum χ4173. However, due to concerns by these investigators that rfaH mutants may not adequately stimulate a robust immune response (due to potentially poor colonization of the intestinal tract and reduced invasion of epithelial cells in the intestine), strains of S. Typhimurium UK-1 and S. Gallinarum 287/91 were also constructed with rfaH expression under the control of the arabinose promoter (PBAD), resulting in regulated delayed expression of rfaH (9, 22). The S. Typhimurium UK-1 ΔPrfaH178 mutant (χ9735) with regulated delayed expression of rfaH was attenuated ~100-fold compared to wild-type S. Typhimurium UK-1 but was not as attenuated as the rfaH knockout mutant (χ9445 ΔrfaH49) at >10,000-fold. In three-day old Rhode Island Red chicks, the S. Gallinarum 287/91 strain χ11386 (ΔPrfaH178) with regulated delayed expression of rfaH retained full virulence compared to the wild-type S. Gallinarum 287/91 parent. In contrast, the S. Gallinarum rfaH knockout mutant (χ11571) was attenuated ~1,000-fold. Although the mouse and chick experiments were performed with two different Salmonella serovars, the results using the regulated delayed expression of rfaH may highlight differences between animal models that may impact the efficiency of this delivery method. Clearly, the expression of rfaH following arabinose induction does not wane quick enough to attenuate the virulence of S. Gallinarum χ11386 (ΔPrfaH178) in Rhode Island Red chicks. Results from the S. Gallinarum rfaH (χ11571) vaccine trial in Rhode Island Red chickens and our S. Typhimurium rfaH vaccine trial in swine indicate that the initial concern that rfaH knockout mutants may be excessively attenuated and therefore not stimulate a sufficient immune response to provide protection against challenge may be unwarranted. Colonization of swine intestinal tissues at 7 dpi with the BBS 202 (rfaH) strain indicates that the strain is present in multiple pigs for immune stimulation.
Although the results of the vaccination trial presented in this study are encouraging, additional investigations may be proposed to further evaluate the use of an rfaH mutant as a potential vaccine candidate in swine. For example, in our pig trials, we typically euthanize the animals at 7 days post-challenge in order to quantitatively evaluate tissue colonization by Salmonella (after 7 d.p.i., quantitation in tissues steadily decreases resulting in only a qualitative assessment (±), especially with a 108 inoculum). Therefore, persistence of the rfaH mutant or of the challenge strain in the vaccinated vs. mock-vaccinated pigs over an extended period of time was not determined. Other parameters to be evaluated in future studies include inoculation route, inoculum dose, and age for vaccination (based on efficacy and industry practices), vaccination protection against Salmonella transmission from a Salmonella-shedding pig to a naïve pig, as well as evaluation of the vaccine in a larger population of pigs. Despite the need for further investigation, this initial evaluation of an rfaH mutant in swine as a potential vaccine candidate addressed three objectives: attenuation, protection, and interference with Salmonella surveillance. Vaccination of swine with the attenuated S. Typhimurium rfaH vaccine strain (BBS 202) reduced S. Typhimurium intestinal colonization and fecal shedding but did not stimulate an anti-Salmonella LPS immune response that would compromise Salmonella surveillance programs for swine herds. Thus, vaccination with the S. Typhimurium rfaH vaccine strain permits the DIVA.
Conflict of Interest Statement
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Acknowledgments
We are greatly appreciative of the outstanding technical support of Kellie Winter, Jennifer Jones, Stephanie Jones, and Ann Hoffman.
References
1. Frenzen PD, Riggs TL, Buzby JC, Breuer T, Roberts T, Voetsch D, et al. Salmonella cost estimate updated using foodnet data. Food Rev (1999) 22:10–5.
2. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States – major pathogens. Emerg Infect Dis (2011) 17:7–15. doi: 10.3201/eid1701.091101p1
3. Batz M, Hoffmann S, Morris JG Jr. Disease-outcome trees, eq-5d scores, and estimated annual losses of quality-adjusted life years (QALYs) for 14 foodborne pathogens in the United States. Foodborne Pathog Dis (2014) 11:395–402. doi:10.1089/fpd.2013.1658
4. Batz MB, Hoffmann S, Morris JG Jr. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot (2012) 75:1278–91. doi:10.4315/0362-028X.JFP-11-418
5. NAHMS. Salmonella on U.S. Swine Sites: Prevalence and Antimicrobial Susceptibility. Fort Collins, CO: U.S. Dept. of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services, Centers for Epidemiology and Animal Health (2009).
6. Funk J, Gebreyes WA. Risk factors associated with Salmonella prevelence on swine farms. J Swine Health Manage (2004) 5:246–51.
7. Farzan A, Friendship RM. A clinical field trial to evaluate the efficacy of vaccination in controlling Salmonella infection and the association of Salmonella-shedding and weight gain in pigs. Can J Vet Res (2010) 74:258–63.
8. Nagy G, Dobrindt U, Hacker J, Emody L. Oral immunization with an rfaH mutant elicits protection against salmonellosis in mice. Infect Immun (2004) 72:4297–301. doi:10.1128/IAI.72.7.4297-4301.2004
9. Kong Q, Liu Q, Roland KL, Curtiss R III. Regulated delayed expression of rfaH in an attenuated Salmonella enterica serovar Typhimurium vaccine enhances immunogenicity of outer membrane proteins and a heterologous antigen. Infect Immun (2009) 77:5572–82. doi:10.1128/IAI.00831-09
10. Bailey MJ, Hughes C, Koronakis V. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol Microbiol (1996) 22:729–37. doi:10.1046/j.1365-2958.1996.d01-1726.x
11. Nieto JM, Bailey MJ, Hughes C, Koronakis V. Suppression of transcription polarity in the Escherichia coli haemolysin operon by a short upstream element shared by polysaccharide and DNA transfer determinants. Mol Microbiol (1996) 19:705–13. doi:10.1046/j.1365-2958.1996.446951.x
12. Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emody L, et al. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun (2006) 74:5914–25. doi:10.1128/IAI.00619-06
13. Nagy G, Palkovics T, Otto A, Kusch H, Kocsis B, Dobrindt U, et al. “Gently rough”: the vaccine potential of a Salmonella enterica regulatory lipopolysaccharide mutant. J Infect Dis (2008) 198:1699–706. doi:10.1086/593069
14. Bearson BL, Bearson SM, Uthe JJ, Dowd SE, Houghton JO, Lee I, et al. Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect (2008) 10:807–16. doi:10.1016/j.micinf.2008.04.011
15. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A (2000) 97:6640–5. doi:10.1073/pnas.120163297
16. Bearson BL, Bearson SM, Lee IS, Brunelle BW. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb Pathog (2010) 48:214–9. doi:10.1016/j.micpath.2010.03.005
17. Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog (2008) 44:271–8. doi:10.1016/j.micpath.2007.10.001
18. Bearson SM, Allen HK, Bearson BL, Looft T, Brunelle BW, Kich JD, et al. Profiling the gastrointestinal microbiota in response to Salmonella: low versus high Salmonella shedding in the natural porcine host. Infect Genet Evol (2013) 16:330–40. doi:10.1016/j.meegid.2013.03.022
19. Selke M, Meens J, Springer S, Frank R, Gerlach GF. Immunization of pigs to prevent disease in humans: construction and protective efficacy of a Salmonella enterica serovar Typhimurium live negative-marker vaccine. Infect Immun (2007) 75:2476–83. doi:10.1128/IAI.01908-06
20. Leyman B, Boyen F, Van Parys A, Verbrugghe E, Haesebrouck F, Pasmans F. Salmonella Typhimurium LPS mutations for use in vaccines allowing differentiation of infected and vaccinated pigs. Vaccine (2011) 29:3679–85. doi:10.1016/j.vaccine.2011.03.004
21. De Ridder L, Maes D, Dewulf J, Pasmans F, Boyen F, Haesebrouck F, et al. Effect of a DIVA vaccine with and without in-feed use of coated calcium-butyrate on transmission of Salmonella typhimurium in pigs. BMC Vet Res (2013) 9:243. doi:10.1186/1746-6148-9-243
Keywords: Salmonella, vaccine, DIVA, rfaH, swine
Citation: Bearson BL, Bearson SMD, Kich JD and Lee IS (2014) An rfaH mutant of Salmonella enterica serovar Typhimurium is attenuated in swine and reduces intestinal colonization, fecal shedding, and disease severity due to virulent Salmonella Typhimurium. Front. Vet. Sci. 1:9. doi: 10.3389/fvets.2014.00009
Received: 05 June 2014; Accepted: 19 September 2014;
Published online: 09 October 2014.
Edited by:
Paul Wigley, University of Liverpool, UKCopyright: © 2014 Bearson, Bearson, Kich and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bradley L. Bearson, Agroecosystems Management Research Unit, National Laboratory for Agriculture and the Environment, USDA, ARS, 2110 University Drive, NSRIC-2103, Ames, IA 50011, USA e-mail:YnJhZC5iZWFyc29uQGFycy51c2RhLmdvdg==
 Bradley L. Bearson
Bradley L. Bearson Shawn M. D. Bearson
Shawn M. D. Bearson Jalusa D. Kich
Jalusa D. Kich In Soo Lee
In Soo Lee