- 1Food Safety and Enteric Pathogens Research Unit, National Animal Disease Center, ARS, USDA, Ames, IA, USA
- 2Agroecosystems Management Research Unit, National Laboratory for Agriculture and the Environment, ARS, USDA, Ames, IA, USA
- 3Department of Biological Sciences and Biotechnology, Hannam University, Daejeon, South Korea
- 4Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, Ames, IA, USA
Salmonella colonization of food animals is a concern for animal health and public health as a food safety risk. Various obstacles impede the effort to reduce asymptomatic Salmonella carriage in food animals, including the existence of numerous serovars and the ubiquitous nature of Salmonella. To develop an intervention strategy that is non-specific yet effective against diverse Salmonella serovars, we explored the prophylactic use of a cytokine to decrease Salmonella in swine by boosting the host’s innate immune system. Granulocyte-colony stimulating factor (G-CSF) is the major cytokine regulating the production, differentiation, function, and survival of neutrophils. Neutrophils play a critical role in the response to Salmonella; therefore, we evaluated the vectored-delivery of porcine G-CSF as a prophylactic to reduce Salmonella in pigs. Crossbred pigs, 5 weeks of age, were intramuscularly injected with a replication-defective human adenovirus (Ad5) engineered to express porcine G-CSF (Ad5-G-CSF, n = 9). Control pigs received the same Ad5 vector lacking the gene encoding G-CSF (Ad5-empty, n = 7). Four days later, all pigs (n = 16) were intranasally inoculated with 1 × 107 colony forming unit (CFU) of Salmonella enterica serovar Typhimurium UK1. At 2 and 3 days post-challenge with Salmonella, Ad5-G-CSF-treated pigs shed significantly less Salmonella (~103 CFU/g) in their feces than Ad5-empty-treated pigs (~104–105 CFU/g; P < 0.05). A significant 4-log reduction in tonsil colonization was also observed in the Ad5-G-CSF-treated pigs at 7 days post-challenge (P < 0.05). In the gastrointestinal tract, the Peyer’s patch region of the ileum exhibited a significant 0.5-log reduction in colonization in the Ad5-G-CSF-treated pigs (P < 0.05). The microbiota of all challenged pigs was assessed by sequencing and analyzing the V1–V3 region of the 16S rRNA gene from fecal DNA samples. The microbial community structure of Salmonella-challenged pigs was less disturbed post-challenge in the Ad5-G-CSF-treated pigs than the Ad5-empty-treated pigs. This suggests that Ad5-G-CSF administration mitigated changes in the microbial community structure caused by Salmonella challenge. Collectively, these data suggest that delivery of a targeted immunostimulant to enhance neutropoiesis may be a strategy to reduce Salmonella colonization, potentially during periods of immunological stress.
Introduction
Asymptomatically colonized food animals are a major reservoir of the human foodborne pathogen Salmonella (1–3). Intervention strategies are needed to not only decrease the overall prevalence of Salmonella in food animals but also reduce an animal’s susceptibility to Salmonella during times of production stress, such as farrowing, weaning, mixing, and transportation. Controlling Salmonella is challenging due to the broad host range, ubiquitous distribution, and number of Salmonella serovars (>2,500). To overcome the complexity of Salmonella, management strategies that target innate immune mechanisms warrant exploration to control the commensal-like state of this human foodborne pathogen in the gastrointestinal tract of animals contributing to our food (animal) supply.
An animal’s innate immune system offers multiple pathways that can be modulated to fight disease-causing agents without activation of the adaptive immune system, which is the primary target of vaccination strategies. Instead, bolstering an innate immune response during stressful events in animal production or periods of immune dysfunction could reduce pathogen recrudescence and infection susceptibility. One possible intervention to address this vulnerability is the prophylactic use of biotherapeutic proteins, such as cytokines [reviewed in Ref. (4)]. Granulocyte-colony stimulating factor (G-CSF) is a cytokine involved in the production, differentiation, and function of granulocytes (especially neutrophils) from bone marrow (5–7). Neutrophils are phagocytic cells of the innate immune system, and their killing mechanism provides a critical first line of defense against bacterial and viral infections (8). Recombinant human G-CSF (Neulasta, Amgen Inc.) is FDA-approved for use in humans to decrease the incidence of infection in neutropenic patients receiving myelosuppressive anti-cancer drugs (9). Recombinant bovine G-CSF (Imrestor, Elanco) has also been approved by the FDA for use in dairy cattle to restore neutrophil function and neutrophil numbers during periparturient immune suppression (10–13). Previous work by our group established that the delivery of a replication-defective human adenovirus 5 encoding porcine G-CSF increased the number of functional neutrophils in circulation (14), thus demonstrating the potential for modulating the swine immune system by targeting the G-CSF pathway.
Rapid neutrophil influx into the intestines is the hallmark of a Salmonella infection (15, 16). In our previous work, cytokines involved in neutrophil production and recruitment were upregulated in swine following Salmonella challenge (17, 18). Furthermore, van Diemen et al. demonstrated higher numbers of circulating neutrophils with greater polymorphonuclear neutrophil (PMN) function in pigs bred for resistance to Salmonella enterica serovar Choleraesuis (19). Thus, we hypothesized that elevating the abundance of circulating neutrophils in pigs prior to Salmonella exposure may assist in controlling Salmonella colonization and shedding. The results demonstrate the beneficial effects of Ad5-G-CSF-induced neutrophilia on the reduction of S. enterica serovar Typhimurium (S. Typhimurium) colonization and shedding in swine, as well as decreased Salmonella-induced disturbance of the gastrointestinal microbiota, suggesting prophylactic use of porcine Ad5-G-CSF may serve as a biotherapeutic approach to reduce Salmonella in pigs.
Materials and Methods
Swine Experiment
Sixteen crossbred, conventionally reared piglets from three Salmonella-fecal-negative sows were weaned at 12 days of age and shipped to the National Animal Disease Center, Ames, IA, USA. Siblings from each litter were divided and raised in two isolation rooms. Piglets tested fecal-negative for Salmonella spp. twice over a 2-week period using bacteriological culture with selective enrichment (20). At 5 weeks of age, piglets received an intramuscular injection of 1010 TCID50/pig of a replication-defective human adenovirus (Ad5) engineered to express porcine G-CSF (Ad5-G-CSF, n = 9) (14). As previously described, Ad5-G-CSF was derived by directionally cloning G-CSF cDNA into the AdEasyTM XL System (Stratagene, La Jolla, CA, USA) and propagated in specialized AD-HEK-293 cells. Control pigs received the same Ad5 vector lacking the gene encoding G-CSF (Ad5-empty, n = 7). Four days later, all pigs (n = 16) were intranasally inoculated with 1 × 107 colony forming unit (CFU) of a nalidixic acid-resistant derivative of S. enterica serovar Typhimurium UK1 (21) that had been passaged in swine and isolated from the ileocecal lymph node of a pig (strain name: SB 377). Fecal samples were collected at 0, 1, 2, 3, and 7 days post-inoculation (d.p.i.) for microbiota analysis as well as quantitative and qualitative Salmonella culture analyses (see below). Blood samples were collected from the jugular vein at −4, −2, 0, 1, 2, 3, and 7 d.p.i. for enumeration of circulating blood cells by flow cytometry (see below). At 7 d.p.i., all pigs were euthanized and necropsied to obtain tissue samples from the tonsil and the intestinal tract (ileal Peyer’s patches, ileocecal lymph nodes, and cecum) for quantitative and qualitative Salmonella culture analysis (see below). Procedures involving animals followed humane protocols as approved by the USDA, ARS, NADC Animal Care and Use Committee in strict accordance with the recommendations in the Guide for the Care, and Use of Laboratory Animals of the National Institutes of Health.
Bacteriology
For quantitative bacteriology, 1 g of pig feces was combined with 5 ml PBS, vortexed, and 0.1 ml directly plated to XLT-4 medium (Beckton, Dickinson and Co., Sparks, MD, USA) containing 30 μg/ml of nalidixic acid. For tissue samples, 1 g of each tissue was combined with 2 ml of PBS in a whirlpak bag, pounded with a mallet, and homogenized in a Stomacher (Seward, Westbury, NY, USA) for 1 min. One hundred microliters of the resulting solution was aliquoted onto XLT-4 medium containing nalidixic acid. One hundred microliters of a 10-fold dilution of each fecal and tissue sample was also plated, and additional dilutions were performed when CFU reached >300/plate. Following 48 h of incubation at 37°C, black colonies were enumerated and a single colony from each plate was confirmed to be Salmonella by serogroup antiserum agglutination (Beckton, Dickinson and Co., Sparks, MD, USA). The total number of CFU for each quantitative tissue or fecal sample was calculated per gram of sample by obtaining the number of Salmonella per plate and multiplying by the dilution factor.
Qualitative bacteriology of Salmonella was performed as follows: 1 g (fecal) or 0.1 ml (homogenized tissue) samples were inoculated in 10 ml tetrathionate broth (TET; VWR, Rutherford, NJ, USA) for 48 h of growth at 37°C. Following incubation, 0.1 ml of each culture was transferred to 10 ml Rappaport–Vassiliadis medium (RV; Difco) and incubated at 37°C for 18–20 h. Cultures were streaked on XLT-4 medium containing nalidixic acid. Colonies suspicious for Salmonella were confirmed by serogroup antiserum agglutination.
Statistical analysis for Salmonella shedding in feces (CFU/g) was Log10-transformed and analyzed using a mixed linear model for repeated measures (Proc Mixed in SAS for Windows, version 9.2; SAS Institute Inc., Cary, NC, USA). Covariance structures within pigs across time were tested and modeled using the REPEATED statement to determine the optimal covariance structure. Linear combinations of the least-squares mean estimates were used in a priori contrasts after testing for a significant (P < 0.05) treatment group effect. Comparisons were made between each group at each time point, using a 5% level of significance (P < 0.05) to assess statistical differences. The endpoint data for bacterial colonization (CFU/g) of tissues collected at necropsy were Log10-transformed and analyzed by analysis of variance using a general linear model for unbalanced data. A 5% level of significance (P < 0.05) was used to assess statistical differences.
Whole Blood Cell Differential
White blood cell counts were performed via flow cytometry as previously described (14). Briefly, a 50-μl aliquot of anti-coagulated (EDTA) whole blood was added to a tube containing monoclonal antibody to porcine granulocytes (6D10, Serotech, USA) with appropriate secondary fluorochrome-labeled antibody. After a 20-min incubation, cells were fixed and red blood cells lysed with the addition of 1 ml FACS lyse (BD Biosciences, USA). Microbeads (Spherotech, USA) were added to the tube immediately prior to data acquisition on a flow cytometer (BD LSR II, Becton Dickinson, USA). A gate was drawn around the beads and events were collected on each parameter (neutrophil gate was based on forward and side scatter properties and antibody labeling) until the bead event number was 500. A ratio of total counts to bead counts was used to determine the number of neutrophils per microliter of blood. Calculation of statistical significance of neutrophils per microliter of blood by treatment group was performed using a one-way ANOVA with a Dunnett’s Multiple Comparison Test. A 5% level of significance (P < 0.05) was used to assess statistical differences.
16S rRNA Gene Sequencing and Analysis
Amplicon libraries of the 16S rRNA gene were generated and sequenced according to Kozich et al. (22), with our primers and procedures described previously (23). Briefly, PCRs contained the following: 17 μl AccuPrime Pfx SuperMix (Life Technologies, Grand Island, NY, USA), 5.0 μM each of the primers i5 + V3 and i7 + V1, and 25 ng of fecal DNA. The following PCR conditions were used: 2 min at 95°C, 22 cycles of (20 s at 95°C, 15 s at 55°C, 5 min at 72°C), 72°C for 10 min. Libraries were normalized using the SequalPrep Normalization Plate Kit (LifeTechnologies) and quantified using both Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and Kapa SYBR Fast qPCR (Kapa Biosystems, Wilmington, MA, USA). Normalized pools were sequenced using version 3 (300 × 2) chemistry on the MiSeq instrument (Illumina, San Diego, CA, USA) according to manufacturer’s instructions.
Contig assembly, sequence alignment, chimera removal, and non-bacterial sequence removal were performed in the program mothur (version 1.33.3) (24). Sequences that only occurred once or twice across all samples were removed as potentially spurious. Sequences were rarified to 3,000 sequences, clustered into operational taxonomic units (OTUs) at 97% similarity, and analyzed for community metrics, including richness (25), evenness, and diversity. Analysis of similarity (ANOSIM) and non-metric multidimensional scaling (NMDS) analyses were conducted in PAST (26). Additionally, the OTUs were assigned to bacterial taxonomy using mothur’s implementation of the SILVA database (27). One sample from a pig in the Ad5-G-CSF group at day 7 yielded insufficient sequences to be analyzed. The 16S rRNA gene sequences associated with this study were deposited in Genbank under Bioproject PRJNA339155.
Results
Both Salmonella Typhimurium Challenge and Porcine Ad5-G-CSF Administration Increased Circulating Neutrophils in Pigs
The effects of Ad5-G-CSF administration and S. Typhimurium challenge on circulating neutrophils were determined by enumerating neutrophils in the blood at various days after Ad5-G-CSF administration and S. Typhimurium challenge. S. Typhimurium challenge alone induced a significant approximately threefold increase in circulating neutrophil counts, as values post-challenge were greater when compared to values on the day of challenge (day 0) (Figure 1A). Circulating neutrophils were also enumerated in pigs that were intramuscularly injected with 1010 TCID50 Ad5-G-CSF prior to Salmonella exposure. As expected on the day of S. Typhimurium challenge (day 0), which was 4 days after Ad5-G-CSF administration, a significant neutrophilia occurred compared to pre-Ad5-G-CSF numbers (day −4, Figure 1B) or compared to Ad5-empty-treated controls on day 0 (day 0, Figure 1A). Following Salmonella challenge of the Ad5-G-CSF group, an additional significant increase in circulating neutrophils was observed at 3 and 7 d.p.i. compared to day 0 (day of Salmonella challenge). Collectively, Ad5-G-CSF administration induced a significant and sustained ~10-fold increase in the number of circulating neutrophils, and Salmonella challenge also induced significant increases in circulating neutrophils.
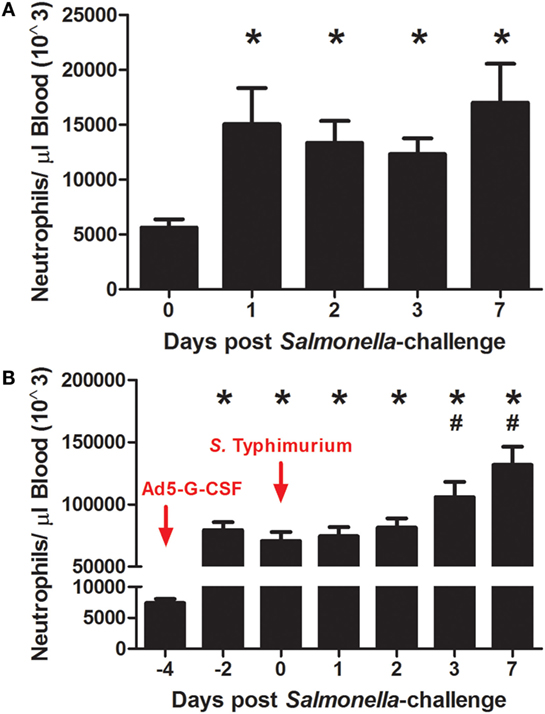
Figure 1. The number of circulating neutrophils following S. Typhimurium challenge and Ad5-G-CSF administration. Blood samples were collected at the noted day relative to Salmonella challenge for neutrophil enumeration by flow cytometry as described in Section “Materials and Methods.” Data are reported as the mean + SEM. (A) Pigs were intranasally challenged with 1 × 107 CFU of virulent S. Typhimurium UK1 strain (n = 7). Significant difference (P < 0.05) in circulating neutrophils relative to day 0 (*). (B) Pigs received a single intramuscular injection of Ad5-G-CSF followed by S. Typhimurium challenge with 1 × 107 CFU of S. Typhimurium UK1 (n = 9). Significant difference (P < 0.05) in circulating neutrophils relative to day −4 (*) or day 0 (#).
Ad5-G-CSF Treatment Reduced Salmonella Fecal Shedding and Tissue Colonization
Salmonella shedding and tissue colonization were compared between Ad5-G-CSF and Ad5-empty-treated pigs. Ad5-G-CSF-treated pigs shed significantly less Salmonella (103 CFU/g feces) when compared to the Ad5-empy-treated pigs at 2 and 3 d.p.i. (104–5 CFU/g feces) (Figure 2). This 1- to 2-log difference between the treatment groups dissipated by 7 d.p.i. as Salmonella shedding in the feces of Ad5-empty-treated pigs declined to the level of the Ad5-G-CSF-treated pigs. Typical for swine, a transient fever was observed in the S. Typhimurium-challenged pigs, peaking at 2 days post-challenge; no significant difference was observed in the elevated body temperatures between treatment groups (data not shown). Gastrointestinal tissues (ileocecal lymph nodes, Peyer’s patch region of the ileum, and cecum) were analyzed at 7 d.p.i., and all tissues were Salmonella positive in both Ad5-G-CSF-treated and Ad5-empty-treated pigs. Of these three tissues, the Peyer’s patch region of the ileum exhibited a significant 0.5-log reduction in Salmonella colonization in the Ad5-G-CSF-treated pigs compared to the Ad5-empty-treated group (Figure 3A). A striking difference in tonsil colonization was observed between treatment groups (Figure 3B). Eight of the nine Ad5-G-CSF-treated pigs harbored no detectable Salmonella in the tonsil, with only one pig being qualitatively positive for Salmonella in the tonsils (i.e., by enrichment). By contrast, all seven Ad5-empty-treated pigs harbored Salmonella in the tonsils at an average of ~10,000 CFU/g. These data suggest that prophylactic administration of Ad5-G-CSF can reduce Salmonella colonization and subsequent fecal shedding, including the tonsils that have been implicated in the carrier-status of Salmonella (28–31).
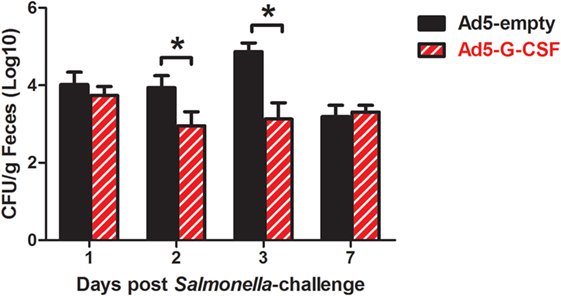
Figure 2. Fecal shedding from Salmonella-challenged pigs, with or without prior Ad5-G-CSF administration. On day 4 following Ad5-G-CSF or Ad5-empty administration, all 16 pigs were challenged with 1 × 107 CFU of S. Typhimurium UK1. Salmonella fecal shedding was monitored via bacteriological analysis of fecal samples collected at 1, 2, 3, and 7 d.p.i. *At each timepoint, significant difference (P < 0.05) in Salmonella CFU/g feces comparing pigs administered Ad5-G-CSF versus Ad5-empty.
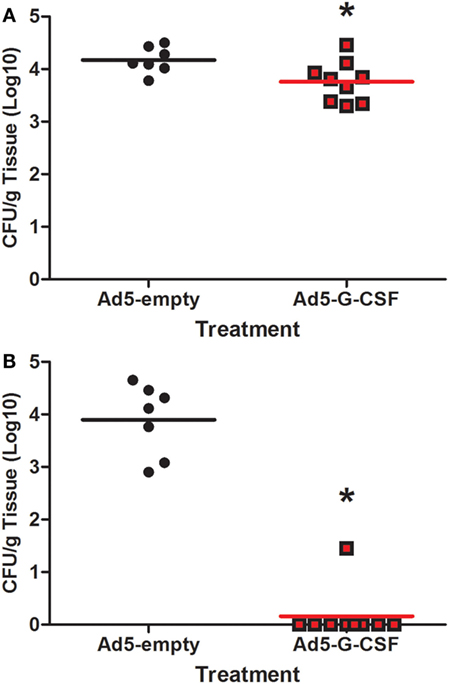
Figure 3. Tissue colonization from Salmonella-challenged pigs, with or without prior Ad5-G-CSF administration. At 7 d.p.i., Salmonella bacteriological analysis (CFU/g) of the (A) Peyer’s patch region of the ileum and (B) tonsils obtained during necropsy. *Significant difference (P < 0.05) comparing Ad5-G-CSF-treated to Ad5-empty-treated pigs at the same time point.
The Gastrointestinal Microbiota of Salmonella-Challenged Pigs Was More Stable in the Ad5-G-CSF-Treated Pigs
Fecal 16S rRNA gene sequence data were used to compare the gastrointestinal bacterial communities of the Ad5-G-CSF and Ad5-empty treatment groups following Salmonella challenge. No significant differences in indices for diversity, evenness, or richness were detected among treatments or timepoints. OTU-based analysis of bacterial community structure showed that at 7 days post-Salmonella challenge, the microbiota of pigs that had received Ad5-G-CSF was not significantly different from that of day 2 or 3 (ANOSIM, p > 0.05; R < 0.1), but the microbiota of pigs that received Ad5-empty treatment was significantly different at day 7 compared to all previous time points (ANOSIM, p < 0.05; R > 0.25). However, the difference between the Ad5-G-CSF-treated and Ad5-empty-treated groups at day 7 was insignificant. The dissimilarity of the microbiotas between days 3 and 7 was visualized via an NMDS plot, which showed the disturbed microbiota at day 7 in the Ad5-empty-treated animals compared to Ad5-G-CSF-treated animals (Figure 4). These results demonstrate that Ad5-G-CSF administration slightly decreases the beta-diversity changes in the microbiota that are caused by Salmonella challenge, suggesting that Ad5-G-CSF mitigated the disturbance to the gut microbiota that was caused by Salmonella.
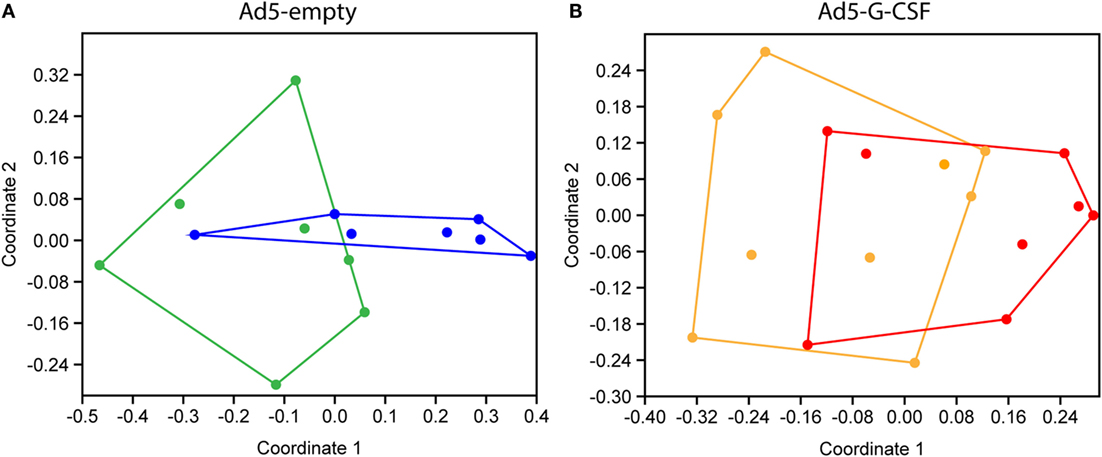
Figure 4. Non-metric multidimensional scaling (NMDS) analyses of 16S rRNA gene OTUs from Salmonella-challenged pigs with prior administration of Ad5-empty (A) or Ad5-G-CSF (B). Shown are the fecal microbiotas from samples taken at 3 days [(A), green; (B), orange] and 7 days [(A), blue; (B), red] post-Salmonella challenge. OTU cutoff of 97% similarity was used. Stress = (A), 0.1445; (B), 0.1453.
Discussion
Granulocyte-colony stimulating factor is a cytokine that influences the proliferation, differentiation, maturation, function, and survival of neutrophils (32). Neutrophils are a critical mediator of antimicrobial defense during the initial stages of infection and have effects on a number of microbial targets (8). Circulating neutrophil numbers in swine have been correlated with resistance to salmonellosis, with pigs most resistant to Salmonella exhibiting higher numbers of circulating neutrophils and enhanced neutrophil function (19). In the current study, treatment of swine with Ad5-G-CSF increased the number of circulating neutrophils by 10-fold, decreased Salmonella-induced disturbance of the gastrointestinal microbiota, and reduced Salmonella fecal shedding 1–2 logs during the acute stage of infection. Thus, prophylactic use of G-CSF as an immunostimulant may be an effective strategy to reduce Salmonella in swine herds. A farm-to-consumption quantitative microbiological risk assessment (QMRA) for Salmonella in pigs in the European Union concluded that interventions should focus on decreasing the level of Salmonella in the feces of infected swine because the vast majority of human risk is derived from a subset of pigs with a high concentration of Salmonella in their feces (≥104 CFU/g) (33). In our study, the Ad5-empty-treated pigs shed Salmonella at 104–5 CFU/g, and Ad5-G-CSF treatment reduced the level of Salmonella fecal shedding to 103 CFU/g, further supporting G-CSF administration as a possible risk mitigation strategy.
The dramatic reduction in Salmonella colonization of the tonsils in the Ad5-G-CSF-treated pigs also highlights prophylactic treatment with G-CSF as a potential control strategy for persistently infected pigs. Salmonella can reside in lymph nodes and especially the tonsils (28–31). In this carrier-state, a stressful event (farrowing, weaning, or transport) can trigger Salmonella to re-emerge and reseed the gastrointestinal tract, resulting in shedding recrudescence (34, 35). Reduction of tonsil colonization, as observed in the Ad5-G-CSF-treated pigs, may reduce the reseeding process during stress. An intriguing follow-up study would be to evaluate the recrudescence of Salmonella in colonized pigs that are given Ad5-G-CSF prior to an applied stress (mixing, transportation, etc.). While additional investigations of the efficacy and safety of Ad5-G-CSF administration in swine are warranted, our data suggest that increasing the number of circulating neutrophils via Ad5-G-CSF administration may offer a non-specific yet effective method for reducing Salmonella colonization in swine.
Inflammation-associated intestinal dysbiosis can result in pathogen expansion, especially for microorganisms, such as Salmonella, that are capable of taking advantage of an inflamed environment (36). We have previously shown that Salmonella colonization of the porcine gastrointestinal tract causes a disturbance within the gut microbial community (37) and triggers an inflammatory response from the host (17, 18). Intervention strategies that target Salmonella during the initial stages of colonization could reduce overall gut inflammation and subsequently prevent the development of a “nutrient-niche” that can be selectively used by Salmonella (38). In the current study, prophylactic Ad5-G-CSF administration was beneficial in reducing the Salmonella-induced microbiota disturbance. Nevertheless, neutrophils are a primary player in the inflammatory response, and their contribution to the inflammatory response that provides an optimal environment for Salmonella expansion needs to be considered with an intervention strategy that increases neutrophils in circulation. It may be important to establish an optimal neutrophilia for the greatest Salmonella reduction with minimal neutrophil-stimulated tissue damage that, in itself, could encourage S. Typhimurium virulence factor-induced inflammation (36).
As regulatory and public scrutiny necessitates the judicious use of antibiotics in food animals (39, 40), the need for antibiotic alternatives in animal production intensifies. Naturally occurring biotherapeutics engineered for pharmaceutical application offer an alternative to antibiotic usage, especially for prophylactic or possibly metaphylactic administration during periods of anticipated stress and host susceptibility. Through the general activation of innate immune defenses, immunostimulants may provide effective pathogen reduction or elimination with broad application against bacteria and viruses that pose a food safety threat or that negatively impact animal health. Our results suggest that prophylactic use of Ad5-G-CSF in swine could decrease subclinical or clinical disease by microorganisms that are targeted by neutrophils.
Author Contributions
SB, BB, CL, HA, DM, and MK conceived and designed experiments. SB, BB, CL, HA, IL, and DM performed the experiments. SB, BB, CL, HA, and MK wrote and edited the manuscript.
Conflict of Interest Statement
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful of the superior technical support of Kellie Winter, Jennifer Jones, David Michael, Stephanie Jones, and Zahra Olson. This research was supported by USDA, ARS CRIS funds.
References
1. NAHMS. Salmonella on U.S. Swine Sites – Prevalence and Antimicrobial Susceptibility. National Animal Health Monitoring System Swine 2006 N536.0109 (2009). Available from: http://www.aphis.usda.gov/animal_health/nahms/swine/downloads/swine2006/Swine2006_is_salmonella.pdf
2. Stevens MP, Humphrey TJ, Maskell DJ. Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci (2009) 364(1530):2709–23. doi:10.1098/rstb.2009.0094
3. Pires AF, Funk JA, Bolin CA. Longitudinal study of Salmonella shedding in naturally infected finishing pigs. Epidemiol Infect (2013) 141(9):1928–36. doi:10.1017/S0950268812002464
5. Freund M, Kleine HD. The role of GM-CSF in infection. Infection (1992) 20(Suppl 2):S84–92. doi:10.1007/BF01705024
6. Dale DC, Liles WC, Llewellyn C, Price TH. Effects of granulocyte-macrophage colony-stimulating factor (GM-CSF) on neutrophil kinetics and function in normal human volunteers. Am J Hematol (1998) 57(1):7–15. doi:10.1002/(SICI)1096-8652(199801)57:1<7::AID-AJH2>3.0.CO;2-0
7. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol (2013) 13(3):159–75. doi:10.1038/nri3399
8. Fournier BM, Parkos CA. The role of neutrophils during intestinal inflammation. Mucosal Immunol (2012) 5(4):354–66. doi:10.1038/mi.2012.24
9. Pfeil AM, Allcott K, Pettengell R, von Minckwitz G, Schwenkglenks M, Szabo Z. Efficacy, effectiveness and safety of long-acting granulocyte colony-stimulating factors for prophylaxis of chemotherapy-induced neutropenia in patients with cancer: a systematic review. Support Care Cancer (2015) 23(2):525–45. doi:10.1007/s00520-014-2457-z
10. Nickerson SC, Owens WE, Watts JL. Effects of recombinant granulocyte colony-stimulating factor on Staphylococcus aureus mastitis in lactating dairy cows. J Dairy Sci (1989) 72(12):3286–94. doi:10.3168/jds.S0022-0302(89)79293-6
11. Cullor JS, Fairley N, Smith WL, Wood SL, Dellinger JD, Inokuma MS, et al. Hemogram changes in lactating dairy cows given human recombinant granulocyte colony stimulating factor (r-MethuG-CSF). Vet Pathol (1990) 27(5):311–6. doi:10.1177/030098589002700502
12. Cullor JS, Smith W, Fairley N, Wood SL, Dellinger JD, Souza L. Effects of human recombinant granulocyte colony stimulating factor (HR-GCSF) on the hemogram of lactating dairy cattle. Vet Clin Pathol (1990) 19(1):9–12. doi:10.1111/j.1939-165X.1990.tb00534.x
13. Kehrli ME Jr, Cullor JS, Nickerson SC. Immunobiology of hematopoietic colony-stimulating factors: potential application to disease prevention in the bovine. J Dairy Sci (1991) 74(12):4399–412. doi:10.3168/jds.S0022-0302(91)78420-8
14. Loving CL, Kehrli ME Jr, Brockmeier SL, Bayles DO, Michael DD, Schlink SN, et al. Porcine granulocyte-colony stimulating factor (G-CSF) delivered via replication-defective adenovirus induces a sustained increase in circulating peripheral blood neutrophils. Biologicals (2013) 41(6):368–76. doi:10.1016/j.biologicals.2013.07.001
15. Tukel C, Raffatellu M, Chessa D, Wilson RP, Akcelik M, Baumler AJ. Neutrophil influx during non-typhoidal salmonellosis: who is in the driver’s seat? FEMS Immunol Med Microbiol (2006) 46(3):320–9. doi:10.1111/j.1574-695X.2006.00051.x
16. Boyen F, Pasmans F, Van Immerseel F, Donne E, Morgan E, Ducatelle R, et al. Porcine in vitro and in vivo models to assess the virulence of Salmonella enterica serovar Typhimurium for pigs. Lab Anim (2009) 43(1):46–52. doi:10.1258/la.2007.007084
17. Huang TH, Uthe JJ, Bearson SM, Demirkale CY, Nettleton D, Knetter S, et al. Distinct peripheral blood RNA responses to Salmonella in pigs differing in Salmonella shedding levels: intersection of IFNG, TLR and miRNA pathways. PLoS One (2011) 6(12):e28768. doi:10.1371/journal.pone.0028768
18. Knetter SM, Bearson SM, Huang TH, Kurkiewicz D, Schroyen M, Nettleton D, et al. Salmonella enterica serovar Typhimurium-infected pigs with different shedding levels exhibit distinct clinical, peripheral cytokine and transcriptomic immune response phenotypes. Innate Immun (2015) 21(3):227–41. doi:10.1177/1753425914525812
19. van Diemen PM, Kreukniet MB, Galina L, Bumstead N, Wallis TS. Characterisation of a resource population of pigs screened for resistance to salmonellosis. Vet Immunol Immunopathol (2002) 88(3–4):183–96. doi:10.1016/S0165-2427(02)00165-4
20. Bearson BL, Bearson SM, Lee IS, Brunelle BW. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb Pathog (2010) 48(6):214–9. doi:10.1016/j.micpath.2010.03.005
21. Bearson SM, Bearson BL, Rasmussen MA. Identification of Salmonella enterica serovar Typhimurium genes important for survival in the swine gastric environment. Appl Environ Microbiol (2006) 72(4):2829–36. doi:10.1128/AEM.72.4.2829-2836.2006
22. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl Environ Microbiol (2013) 79(17):5112–20. doi:10.1128/AEM.01043-13
23. Allen HK, Bayles DO, Looft T, Trachsel J, Bass BE, Alt DP, et al. Pipeline for amplifying and analyzing amplicons of the V1 -V3 region of the 16S rRNA gene. BMC Res Notes (2016) 9(1):1. doi:10.1186/s13104-016-2172-6
24. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol (2009) 75(23):7537–41. doi:10.1128/AEM.01541-09
25. Bunge J, Woodard L, Bohning D, Foster JA, Connolly S, Allen HK. Estimating population diversity with CatchAll. Bioinformatics (2012) 28(7):1045–7. doi:10.1093/bioinformatics/bts075
26. Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electronica (2001) 4:9.
27. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res (2013) 41(Database issue):D590–6. doi:10.1093/nar/gks1219
28. Boyen F, Haesebrouck F, Maes D, Van Immerseel F, Ducatelle R, Pasmans F. Non-typhoidal Salmonella infections in pigs: a closer look at epidemiology, pathogenesis and control. Vet Microbiol (2008) 130(1–2):1–19. doi:10.1016/j.vetmic.2007.12.017
29. Boyen F, Pasmans F, Van Immerseel F, Morgan E, Botteldoorn N, Heyndrickx M, et al. A limited role for SsrA/B in persistent Salmonella Typhimurium infections in pigs. Vet Microbiol (2008) 128(3–4):364–73. doi:10.1016/j.vetmic.2007.10.031
30. Van Parys A, Boyen F, Volf J, Verbrugghe E, Leyman B, Rychlik I, et al. Salmonella Typhimurium resides largely as an extracellular pathogen in porcine tonsils, independently of biofilm-associated genes csgA, csgD and adrA. Vet Microbiol (2010) 144(1–2):93–9. doi:10.1016/j.vetmic.2009.12.021
31. Volf J, Stepanova H, Matiasovic J, Kyrova K, Sisak F, Havlickova H, et al. Salmonella enterica serovar Typhimurium and Enteritidis infection of pigs and cytokine signalling in palatine tonsils. Vet Microbiol (2012) 156(1–2):127–35. doi:10.1016/j.vetmic.2011.10.004
32. Roberts AW. G-CSF: a key regulator of neutrophil production, but that’s not all! Growth Factors (2005) 23(1):33–41. doi:10.1080/08977190500055836
33. Snary EL, Swart AN, Simons RR, Domingues AR, Vigre H, Evers EG, et al. A quantitative microbiological risk assessment for Salmonella in pigs for the European Union. Risk Anal (2016) 36(3):437–49. doi:10.1111/risa.12586
34. Callaway TR, Morrow JL, Edrington TS, Genovese KJ, Dowd S, Carroll J, et al. Social stress increases fecal shedding of Salmonella Typhimurium by early weaned piglets. Curr Issues Intest Microbiol (2006) 7(2):65–71.
35. Verbrugghe E, Boyen F, Van Parys A, Van Deun K, Croubels S, Thompson A, et al. Stress induced Salmonella Typhimurium recrudescence in pigs coincides with cortisol induced increased intracellular proliferation in macrophages. Vet Res (2011) 42:118. doi:10.1186/1297-9716-42-118
36. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature (2010) 467(7314):426–9. doi:10.1038/nature09415
37. Bearson SM, Allen HK, Bearson BL, Looft T, Brunelle BW, Kich JD, et al. Profiling the gastrointestinal microbiota in response to Salmonella: low versus high Salmonella shedding in the natural porcine host. Infect Genet Evol (2013) 16:330–40. doi:10.1016/j.meegid.2013.03.022
38. Rivera-Chavez F, Baumler AJ. The pyromaniac inside you: Salmonella metabolism in the host gut. Annu Rev Microbiol (2015) 69:31–48. doi:10.1146/annurev-micro-091014-104108
39. FDA. The Judicious Use of Medically Important Antimicrobial Drugs in Food-Producing Animals: Guidance for Industry #209. (2012). Available from: http://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM216936.pdf
40. FDA. New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food-Producing Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI #209: Guidance for Industry #213. (2013). Available from: http://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM299624.pdf
Keywords: Salmonella, granulocyte-colony stimulating factor, swine, immune stimulation, alternatives to antibiotics
Citation: Bearson SMD, Bearson BL, Loving CL, Allen HK, Lee I, Madson D and Kehrli ME Jr. (2016) Prophylactic Administration of Vector-Encoded Porcine Granulocyte-Colony Stimulating Factor Reduces Salmonella Shedding, Tonsil Colonization, and Microbiota Alterations of the Gastrointestinal Tract in Salmonella-Challenged Swine. Front. Vet. Sci. 3:66. doi: 10.3389/fvets.2016.00066
Received: 30 June 2016; Accepted: 08 August 2016;
Published: 25 August 2016
Edited by:
Guillermo Tellez, University of Arkansas, USAReviewed by:
Jason Kindrachuk, National Institutes of Health, USASherry Layton, Vetanco S.A., Argentina
Copyright: © 2016 Bearson, Bearson, Loving, Allen, Lee, Madson and Kehrli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shawn M. D. Bearson, c2hhd24uYmVhcnNvbkBhcnMudXNkYS5nb3Y=
 Shawn M. D. Bearson
Shawn M. D. Bearson Bradley L. Bearson
Bradley L. Bearson Crystal L. Loving
Crystal L. Loving Heather K. Allen
Heather K. Allen InSoo Lee
InSoo Lee Darin Madson
Darin Madson Marcus E. Kehrli Jr.
Marcus E. Kehrli Jr.