- Center for Food Safety, Department of Food Science, University of Arkansas, Fayetteville, AR, United States
Poultry meat is an important source of zoonotic Salmonella infection. Oral vaccination of chickens with live attenuated Salmonella during grow-out is an attractive approach to control Salmonella colonization in the chicken gastrointestinal tract. In this study, we report the construction of methionine-dependent and growth of Salmonella Typhimurium mutant strains with methionine auxotrophy (ΔmetR and ΔΔmetRmetD) and survival in chicken feed and fecal matrices. The methionine auxotroph mutant ΔΔmetRmetD grew slowly on L-methionine but failed to grow on D-methionine, as expected, and exhibited lower affinity for methionine compared with the isogenic parent strain (ΔmetR single mutant) in whole-cell affinity experiments. Preliminary data conducted as part of a previously published bird challenge study indicated that the methionine auxotroph was less effective at protection in chickens to a challenge with virulent wild-type parent strain but generated greater Salmonella-specific serum IgG. Although the auxotroph could not sustain itself in minimal media it was able to survive when incubated in the presence of chicken and fecal material. The immune response appears promising but further work may be needed to alter low-affinity methionine transporters and methionine biosynthesis genes in combination with the knock-out of the high affinity transporter metD reported here to ensure timely clearance of the candidate vaccine strain.
Introduction
As an essential amino acid for animals, methionine is an important component of animal feeds, including poultry feed (1). Methionine is one of the nutritionally limiting components of animal feeds and is limited in plant proteins. Methionine is essential for protein synthesis and serves as a source of methyl groups for the biosynthesis of lipids, biotin, nucleic acids, and polyamines (2, 3).
Methionine synthesis and uptake have been extensively investigated in Escherichia coli and Salmonella Typhimurium (4–7). Transport of methionine into the bacterial cell is mediated by both a high-affinity transporter (Km approximately 0.1 mM) and one or more low-affinity transporters (Km approximately 20–40 mM) (6). The high-affinity transporter is referred to as metD in both E. coli and S. Typhimurium, and mutants in this transporter are unable to transport D-methionine. More recently, the metD transporter gene has been sequenced and shown to consist of an operon comprised of three genes, recently named metNIQ (8). We have tested the hypothesis that a methionine auxotrophic strain of S. Typhimurium with limited uptake and synthesis of methionine can serve as an effective vaccine for the pre-harvest control of Salmonella in poultry. Poultry is a significant source of food-borne Salmonella illness in humans (9). Thus, pre-harvest control measures such as vaccination are desirable.
One approach to poultry vaccination with live attenuated Salmonella has focused on auxotrophy by deletion of genes encoding essential regulators of metabolism. One of these is the regulation of synthesis and uptake of methionine. The metR gene encodes a transcription factor of the LysR family that regulates several genes of the methionine biosynthesis pathway. The metR controls primarily genes involved with the last steps of methionine biosynthesis: metF, metE, and metH. The metF gene product produces a methyl donor, 5-methyltetrahydrofolate, which provides the terminal methyl group for methionine. Both metE and metH encode cobalamin-independent and cobalamin-dependent enzymes, respectively, that add the terminal methyl group to homocysteine to form methionine (6, 10). The metD deletion eliminates the high-affinity methionine transporter (7). We hypothesized that use of this mutant in combination with the metR deletion might further reduce the ability of a Salmonella vaccine strain to survive in the host by limiting methionine uptake to that of the remaining (low-affinity) methionine transporters.
In this study, single (ΔmetR) and double (ΔΔmetRmetD) S. Typhimurium UK-1 mutants were constructed and characterized as potential vaccine strains for control of Salmonella colonization. Here we present preliminary data on the ΔΔmetRmetD unpublished part of the bird challenge study (11) and compare it with the previously published responses to the wild-type parent strain of S. Typhimurium, UK-1 (positive control) and a PBAD-mviN vaccine strain from our past research (11). The PBAD-mviN strain is a genetically attenuated strain that has the native promoter of the mviN gene (a gene required for cell wall synthesis) (12) removed and replaced with an arabinose-inducible promoter (PBAD) and the gene encoding the upstream activator, araC. By growing this strain in arabinose, but then washing away this medium and inoculating the washed cells orally to the chicken, the bacterium undergoes delayed lysis as cell wall synthesis shuts down (11). To assess the environmental characteristics of the methionine auxotrophs, growth kinetics in minimal medium with L-methionine as well as growth curves in D-methionine, and survival of the auxotroph strains in chicken feed and feces are presented in the current study.
Materials and Methods
Bacterial Strains
A wild-type S. Typhimurium UK-1 strain was utilized to construct potential vaccine strains. A nalidixic acid (NA) resistant S. Typhimurium UK-1 derived from the wild type was used as the challenge strain. The PBAD-mviN vaccine strain discussed in the current study for comparative purposes was generated from UK-1 as described in our previous report (11).
Construction of ΔΔmetRmetD S. Typhimurium UK-1
Single and double deletion mutants affecting methionine metabolism were produced in S. Typhimurium strain UK-1. This strain and the plasmids for the Red recombinase system were obtained from Dr. Young Min Kwon, Department of Poultry Science, University of Arkansas (Fayetteville, AR, USA). The Red recombinase system was used for the targeted gene deletions as previously described (13). Briefly, disruption of the targeted genes was accomplished by first transforming strain UK-1 with plasmid pKD46. This plasmid confers ampicillin resistance and harbors the genes for phage lambda Red recombinase, which mediates the exchange of DNA between the gene of Salmonella to be deleted and the gene disruption construct. This plasmid also contains a temperature-sensitive origin of replication, which facilitates its removal following recombination. Gene disruption constructs were synthesized by amplifying a region of plasmid pKD4 (13) by polymerase chain reaction (PCR), from the P1 site (nucleotides 31–50 of pKD4) to the P2 site (nt. 1488–1507). This region consists of a central kanamycin (Kan) resistance gene (encoding aminoglycoside 3′-phosphotransferase), flanked by two FLP recognition target (FRT) sites. Genomic DNA sequences corresponding to upstream and downstream regions surrounding the appropriate target gene of strain UK-1 were subsequently introduced on either side of the FRT-KanR-FRT region by overlap extension PCR (14), using the primers indicated in Table 1.
The method for deletion of the metD transporter sequence, which is comprised of the metNIQ operon is shown in Figure 1. The resulting PCR products were gel purified, treated with restriction endonuclease Dpn1 to degrade any trace amount of template pKD4, gel purified again, and then electroporated into electroporation-competent UK-1:pKD4 cells that had been grown at 30°C in Luria-Bertani (LB) broth with 1 mM arabinose to induce the Red recombinase. After a 1 h incubation of the electroporated cells at 37°C in SOC medium, the electroporated cells were spread on LB/Kan agar plates and grown at 37°C overnight. Four tranformants were then streaked for isolation on LB/Kan, and tested on LB/ampicillin plates to confirm curing of plasmid pKD46.
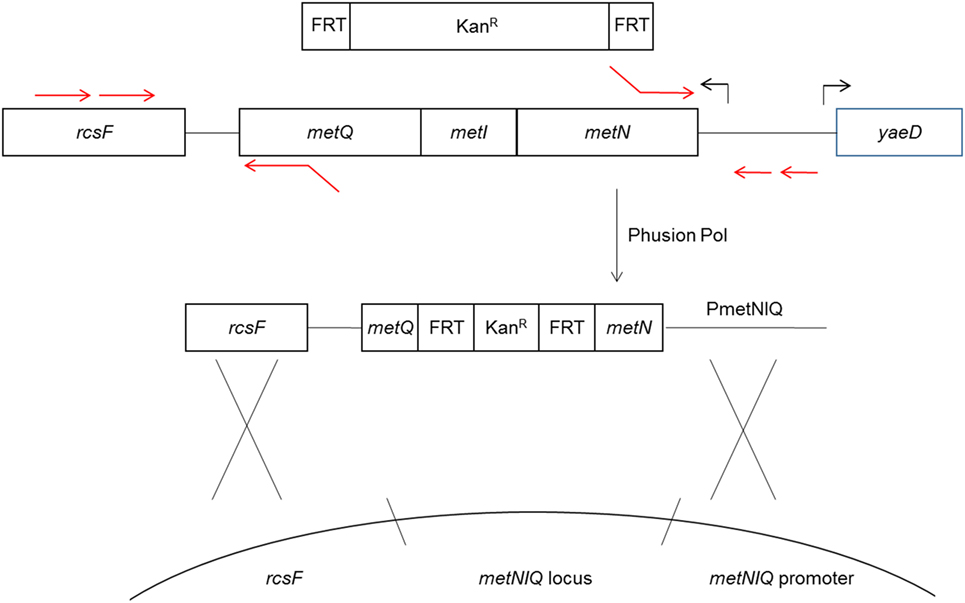
Figure 1. Diagram of metD mutant construction. The metR mutant was constructed in a similar fashion. The metD transporter consists of three subunits, encoded by the metNIQ operon. Primer sequences fusing the metN and metQ sequences to FRT-Kan expression cassette sequences (13) were used to delete the entire metI gene and part of the metN and metQ genes, replacing these with the FRT-Kan cassette. The resulting construct was introduced into the metD locus by electroporation and homologous recombination using the Red recombinase system (13).
The Kan resistance marker was subsequently removed from the genomic insertion sites, leaving a gene deletion, by introducing a second plasmid, pCP20, which expresses a second recombinase, the S. cerevisiae FLP recombinase and confers ampicillin resistance (15). The FLP mediates the removal of the antibiotic resistance marker by recombining the flanking FRT sites. The FLP was induced and pCP20 removed by shifting the temperature from 30 to 42°C as described previously (13). Removal of the KanR genomic insertion and pCP20 were confirmed by failure to grow on LB/Kan and LB/ampicillin, respectively, with appropriate growth on LB in parallel.
Whole-Cell Affinity Measurements
Whole-cell affinity measurements were conducted as described previously (16). Briefly, cultures of the ΔmetR single mutant and ΔΔmetRmetD double mutant were grown in M9 minimal medium + 10 μM L-methionine at 37°C for 16 h and then diluted to an OD600 of 0.05 in M9 minimal medium + L-methionine at 3, 7, 10, 13, 15, and 17 μM in culture tubes containing 4 ml each of minimal medium at the L-methionine concentrations indicated, and three technical replicate culture tubes were prepared at each methionine concentration. Cultures were grown at 37°C in a shaking water bath at 220 RPM, and the OD600 was measured every 15 min for a total of 6 h. The replicate data were then averaged and transformed for Lineweaver–Burk plots using Microsoft Excel software.
Vaccination
Details of the vaccination challenge trial have been described elsewhere (11). Briefly male Cobb 500 broiler chicks (Siloam Springs, AR, USA) were obtained on day of hatch and randomly assigned to four pre-sterilized Horsfall units. A University of Arkansas Institutional Animal Care and Use Committee-approved protocol was used to ensure humane treatment of the chickens. Chicks vaccinated with ΔmetRΔmetD double mutant Salmonella was one treatment group (designated Group 2) of the four treatment groups (Group 1: unvaccinated, challenged; Group 3: vaccinated with the PBAD-mviN vaccine strain Salmonella, challenged), and Group 4: vaccinated with the wild-type parent strain UK-1, challenged. The vaccine and control inocula were grown for 16 h in LB broth at 37oC, followed by three washes in phosphate-buffered saline (PBS) and adjustment of the cell density by dilution in PBS to 5 × 108 cells/ml. Chicks were orally inoculated with 1 × 108 CFU Salmonella cells via sterile gavage needle on Day 2 post-hatch and again on Day 7 post-hatch. The unvaccinated chicks received an equal volume (0.2 ml) of sterile PBS via sterile gavage needle on Days 2 and 7 post-hatch. The challenge strain, which had been passaged repeatedly through chicks to increase its virulence followed by cryopreservation, was grown for 16 h in LB + 20 μg/ml NA. The challenge strain was then passaged twice for 8 h each passage to ensure a log phase culture. The resulting cells were then diluted to 5 × 108 cells/ml with PBS and orally inoculated via sterile gavage needle to chickens at 2 weeks post-hatch with 1 × 108 cells (0.2 ml).
At the time of chicken necropsy reported previously (11), ceca and ilea organs were collected aseptically and transferred to sterile sample bags, subsequently removed and transferred to 10 ml tetrathionate (TT) broth for enrichment. The TT broth was incubated for 24 h at 37°C, followed by streaking of a 10 μl loopful of the TT broth for isolation on Brilliant Green (BG) agar supplemented with 20 μg/ml NA and another 10 μl loopful on BG agar supplemented with 50 μg/ml Kan + 1 mM arabinose. Agar plates were incubated for 24 h at 37°C, examined for Salmonella colony appearance to enumerate the NA—resistant S. Typhimurium challenge strain per gram cecal content, and live vaccine strains (Kan—resistant Salmonella per gram of cecal content). A 0.1 g portion of cecal content was aseptically added to a sterile microtube, weighed, and then combined with nine volumes of sterile PBS to obtain a 1:10 dilution, followed by vortexing. After serial dilution in sterile PBS, mixed cecal contents were spread-plated aseptically onto selective media (BG + 20 μg/ml NA) for enumeration of the respective treatment groups. Since S. Typhimurium is naturally resistant to novobiocin (NO), the wild-type control NA-sensitive strain was distinguished from the NA-resistant challenge strain by direct plating of cecal contents from the wild-type-inoculated chickens on both BG + NA and BG + NO plates, and subtracting the number of colonies on BG + NA (challenge strain) from the total colonies on BG + NO (challenge strain + wild-type control strain).
Enzyme-Linked Immunosorbent Assay (ELISA)
As described previously (11), indirect ELISA reactions were performed by placing 1 μg of Salmonella protein from sonicated UK-1 cells in each well of 96-well microlon medium binding microtiter plates (Grainer, Frickenhausen, Germany) after diluting to 100 μl in carbonate buffer, pH 9.6. After allowing proteins to bind for 2 h 37°C, the plates were allowed to air-dry overnight at 23°C then blocked with Superblock (Thermo Scientific, Rockford, IL, USA) 2 h, 37°C. Chicken sera from unvaccinated and vaccinated chickens from all treatment groups were serially diluted in Superblock and the plates incubated at 37°C, 2 h followed by washing four times with ELISA plate wash buffer (50 mM Tris pH 8, 140 mM NaCl, 0.05% Tween-20). Anti-chicken IgG-HRP conjugate was diluted 1:20,000 with Superblock and plates incubated 37°C, 1 h, followed by washing four times in wash buffer. The TMB substrate (KPL, Gaithersburg, MD, USA) was incubated 10 min in each well, stopped with 1 N HCl and absorbance measured in a Tecan Infinite M200 plate reader at 450 nm.
Growth and Survival of Mutants in M9, Feed, and Fecal Broth
Wild-type parent strain and mutants ΔmetR and ΔmetRΔmetD were grown in M9 minimal medium supplemented with 10 μM L- or D-methionine as indicated for 16 h and subsequently washed three times in PBS. Five grams of chicken feed were blended with 100 ml dionized water at high speed for 3 min. The same was done for 5 g of chicken feces, separately. These blends were then filtered through cheese cloth and autoclaved. These were subsequently aliquoted into sterile culture tubes and the washed Salmonella was added to a final density of 1 × 106 cells/ml. The cultures were incubated at 37°C, 200 RPM and growth of the cultures was monitored by spread-plating and colony counting of appropriate dilutions on LB medium at 0, 2, 5, 24, 48, 96, 264, and 504 h.
Statistical Analysis
The enumeration of the challenge (NA-resistant marker) strain in ceca of unvaccinated and vaccinated chickens was compared by a two-tailed Student’s t-test, using the Microsoft Excel program.
Results and Discussion
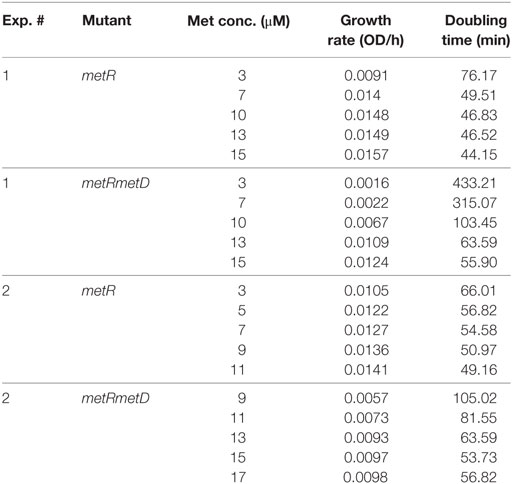
Growth curves of single ΔmetR and ΔΔmetRmetD in M9 minimal medium supplemented with L- or D-methionine are shown in Figure 2. Both mutants exhibited reduced growth when compared with the control strain (S. Typhimurium UK-1) in both media with either L- or D-methionine. The double mutant exhibited little or no growth in D-methionine, as expected. The growth rates and doubling times of the mutants in M9 minimal medium are shown in Table 2. The growth rates were 10–20-fold lower than that reported by Froelich et al. for an E. coli methionine auxotroph (16). The whole-cell affinities of the single and double mutants for methionine are shown in Figure 3. Whole-cell affinity measurements indicated that the ΔΔmetRmetD double mutant had a consistently lower affinity (Ks) for L-methionine compared with the ΔmetR single mutant (Figure 3). The Ks of the metRmetD double mutant was 5.90 and 251.6 μM in Experiments 1 and 2, respectively, while the corresponding values for the metR single mutant were 3.38 and 1.54 μM. Except for the outlier value of 251.6 μM, these values are similar to the Ks values of 6.41 and 7.00 reported by Froelich et al. for an E. coli methionine auxotroph, ATCC 23798 (16). The discrepancy between the much lower growth rate of the Salmonella auxotroph described in the present paper compared with the E. coli ATCC 23798 could be due to genetic differences. The genotype of ATCC 23798 is not known, but the parent strain is described as having been mutagenized with N-methyl-N-nitrosoguanidine (17).
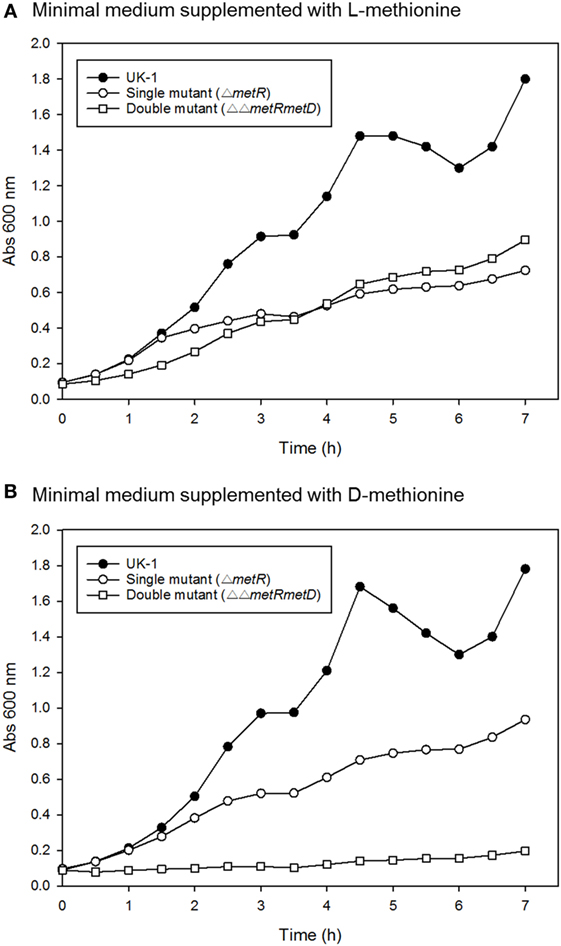
Figure 2. Growth curves of S. Typhimurium wild-type (UK-1), single (ΔmetR), and double mutant (ΔΔmetRmetD) mutant in minimal medium supplemented with (A) L-methionine or (B) D-methionine.
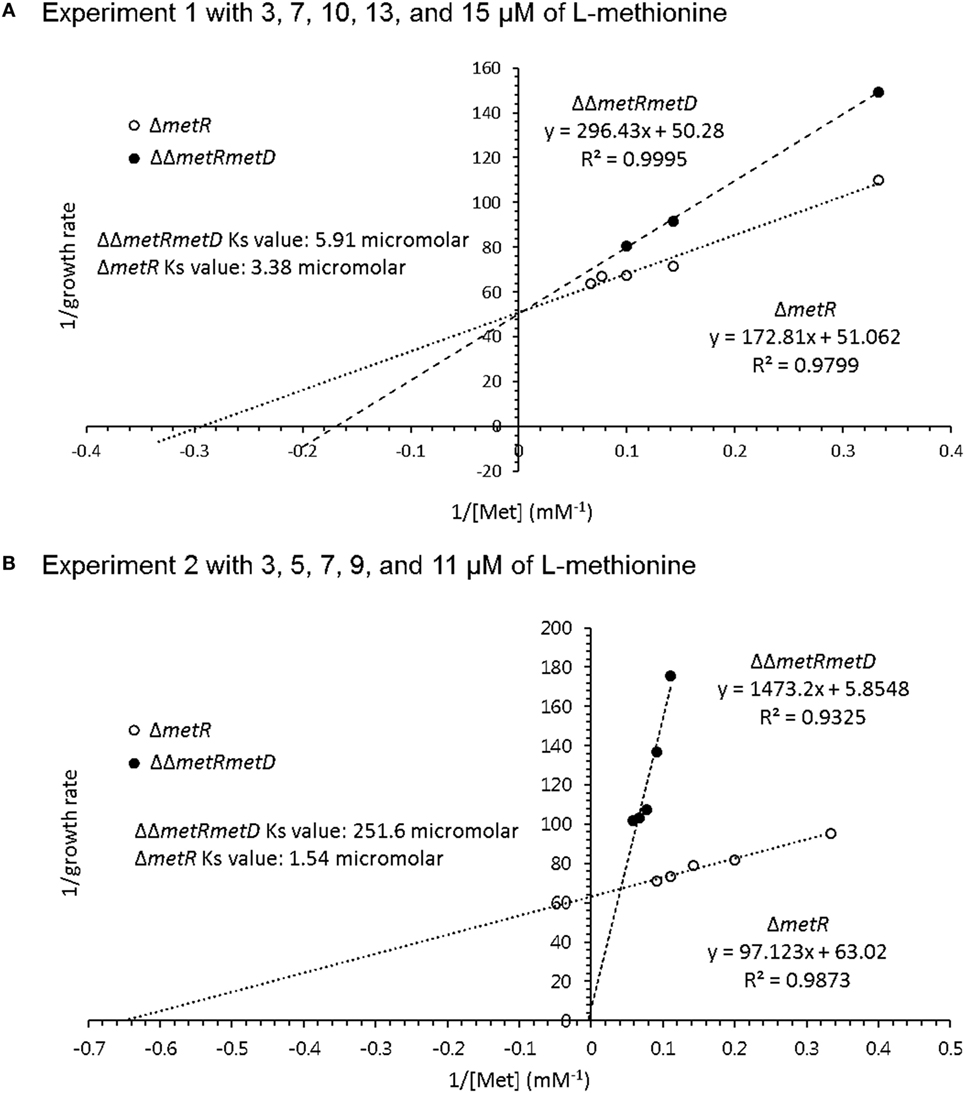
Figure 3. Whole-cell affinity comparing growth of ΔmetR and ΔΔmetRmetD mutant in minimal medium with added L-methionine. (A) Experiment 1 with 3, 7, 10, 13, and 15 μM of L-methionine; (B) Experiment 2 with 3, 5, 7, 9, and 11 μM of L-methionine.
Cecal prevalence of the Salmonella challenge strain was evaluated, and 100% were positive for the challenge strain in the unvaccinated and metRmetD vaccinated groups at the end of the trial, whereas 75 and 40% were positive for the challenge strain in the PBAD-mviN and wild-type vaccinated groups as reported previously, respectively (11). Colonization of ceca was also measured by enumeration of challenge strain colonies on selective agar plates containing NA. Both the metRmetD and PBAD-mviN vaccine strains significantly reduced (P < 0.01) the number of challenge strain Salmonella in the cecal contents when compared with the unvaccinated control group (the means ± SD were 4.71 ± 1.41 log CFU/g for metRmetD, 2.62 ± 0.8 log CFU/g for PBAD-mviN, and 6.49 ± 0.61 log CFU/g for the unvaccinated group, partially reported in our previous study of the PBAD-mviN vaccine) (11).
This suggests the vaccine strains partially protected against challenge strain colonization but based on the greater level of prevalence, the metRmetD vaccine candidate strain was not as well cleared by the birds. This is supported by two independent lines of evidence. For one, the survival curves of the methionine mutants in chicken feed and fecal material indicated a high degree of survival in these matrices versus incubation in minimal M9 medium (Figure 4). This may also be reflective of the fact that chickens vaccinated orally with the ΔΔmetRmetD mutant exhibited elevated levels of serum IgG binding specifically to Salmonella proteins in ELISA relative to an attenuated mutant strain, PBAD-mviN and the unvaccinated group. The metRmetD mutant had a mean titer of 7840 ± 1711, while the PBAD-mviN strain had a mean titer of 4520 ± 1544, and the unvaccinated group mean titer was 1700 ± 352.5 (11).
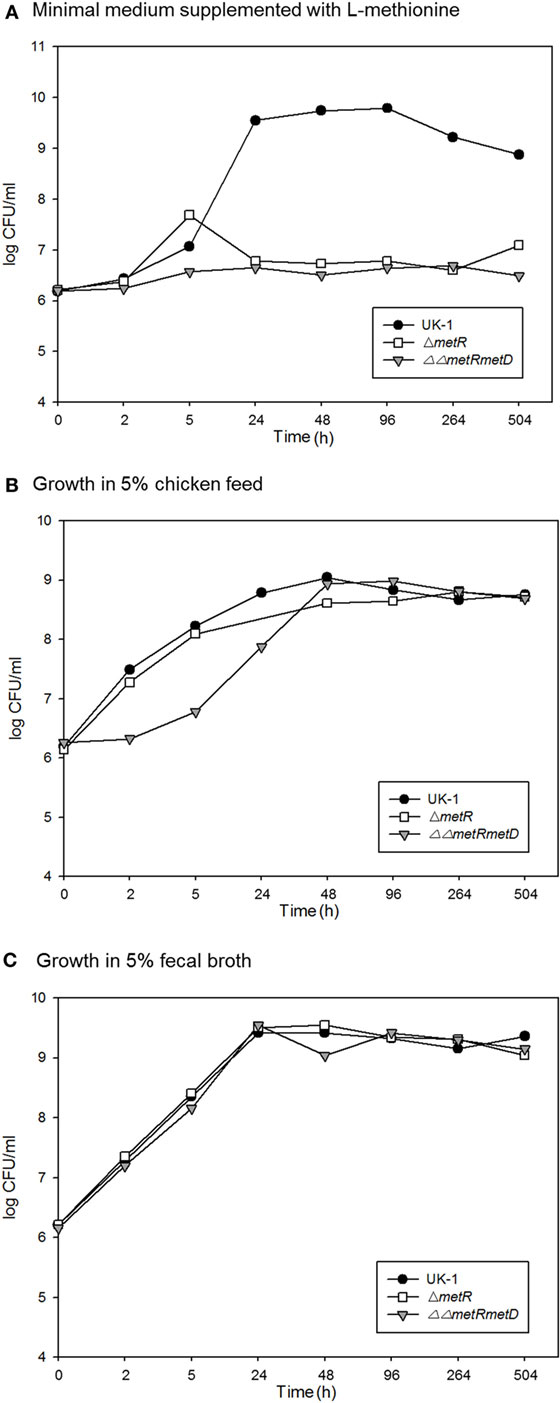
Figure 4. Survival over time of the wild-type parent strain (S. Typhimurium UK-1) and methionine auxotrophic mutants in (A) M9 minimal medium; (B) 5% chicken feed; (C) 5% chicken feces.
Given the superior immune response of the metRmetD mutant, this strain may warrant further research as a vaccine strain. However, this mutant does not appear to be easily cleared out by the inoculated birds and this could be problematic from an environmental contamination standpoint. There are possible remedies for this. To eliminate this problem and further reduce intracellular survival, further investigations may be required that eliminate some of the low-affinity methionine transporters in combination with the knock-out of the high affinity transporter metD reported here. Finally, different genes involved in methionine biosynthesis could also be targeted in addition to the transport genes. For example, S. Gallinarum metC mutants have been shown to possess diminished virulence capabilities, lowered levels in reticuloendothelial organs and competitiveness defects in challenged birds (18).
In conclusion, new vaccine strains (ΔmetR single mutant and ΔΔmetRmetD) were constructed in this study. The methionine auxotroph ΔΔmetRmetD generated a greater Salmonella-specific serum IgG level and reduced the level of Salmonella in cecal contents of approximately 100-fold relative to the unvaccinated control group. Particular combinations of methionine biosynthesis and transport mutants could result in optimal vaccine candidates that can be retained sufficiently to stimulate an optimal immune response but yet easily cleared via dietary manipulation.
Author Contributions
PR performed experiments, drafted the manuscript, collected test data, and analyzed the data. SK drafted and revised the manuscript. PR, SP and SR designed the study and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was funded by USDA-SBIR grant award 2012-33610-19529.
References
1. Bunchasak C. Role of dietary methionine in poultry production. J Poult Sci (2009) 46:169–79. doi:10.2141/jpsa.46.169
2. Brosnan JT, Brosnan ME. The sulfur-containing amino acids: an overview. J Nutr (2006) 136:1636S–40S.
3. Chalova VI, Froelich C, Ricke SC. Potential for development of an Escherichia coli-based biosensor for assessing bioavailable methionine: a review. Sensors (Basel) (2010) 10:3562–84. doi:10.3390/s100403562
4. Ayling PD, Bridgeland ES. Methionine transport in wild-type and transport-defective mutants of Salmonella Typhimurium. J Gen Microbiol (1972) 73:127–41. doi:10.1099/00221287-73-1-127
5. Ayling PD, Mojica-a T, Klopotoswski T. Methionine transport in Salmonella Typhimurium: evidence for at least one low-affinity transport system. J Gen Microbiol (1979) 114:227–46. doi:10.1099/00221287-114-2-227
6. Greene RC. Biosynthesis of methionine. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Washington, DC: ASM Press (1996). p. 542–60.
7. Merlin C, Gardiner G, Durand S, Masters M. The Escherichia coli metD locus encodes an ABC transporter which includes abc (metN), yaeE (metI), and yaeC (metQ). J Bacteriol (2002) 184:5513–7. doi:10.1128/jb.184.19.5513-5517.2002
8. Gal J, Szvetnik A, Schnell R, Kalman M. The metD D-methionine transporter locus of Escherichia coli is an ABC transporter gene cluster. J Bacteriol (2002) 184:4930–2. doi:10.1128/JB.184.17.4930-4932.2002
9. Tauxe RV. Transmission of human bacterial pathogens through poultry. In: Blankenship LC, editor. Colonization Control of Human Bacterial Enteropathogens in Poultry. NY: Academic Press (1991). p. xv–xxiii.
10. Augustus AM, Spicer LD. The MetJ regulon in gammaproteobacteria determined by comparative genomics methods. BMC Genomics (2011) 12:558. doi:10.1186/1471-2164-12-558
11. Rubinelli PM, Lee SI, Roto SM, Park SH, Ricke SC. Regulated expression of virulence gene mviN provides protective immunity and colonization control of Salmonella in poultry. Vaccine (2015) 33:5365–70. doi:10.1016/j.vaccine.2015.08.074
12. Inoue A, Murata Y, Takahashi H, Tsuji N, Fujisaki S, Kato J-i. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J Bacteriol (2008) 190:7298–301. doi:10.1128/JB.00551-08
13. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Eschericia coli K-12 using PCR products. Proc Natl Acad Sci USA (2000) 97:6640–5. doi:10.1073/pnas.120163297
14. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene (1989) 77:51–9. doi:10.1016/0378-1119(89)90358-2
15. Cherepanov PP, Wackernagel W. Gene disruption in Eschericia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene (1995) 158:9–14. doi:10.1016/0378-1119(95)00193-A
16. Froelich CA, DÍAz IBZ, Ricke SC. Construction and growth kinetics of a bioluminescent methionine auxotroph Escherichia coli strain for potential use in a methionine bioassay. J Rapid Methods Autom Microbiol (2002) 10:69–82. doi:10.1111/j.1745-4581.2002.tb00013.x
17. Adelberg EA, Mandel M, Chen GCC. Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem Biophys Res Commun (1965) 18:788–95. doi:10.1016/0006-291X(65)90855-7
Keywords: Salmonella Typhimurium, vaccine, methionine auxotrophy, poultry, ΔmetR, ΔΔmetRmetD
Citation: Rubinelli P, Kim SA, Park SH, Baker CA and Ricke SC (2017) Growth Characterization of Single and Double Salmonella Methionine Auxotroph Strains for Potential Vaccine Use in Poultry. Front. Vet. Sci. 4:103. doi: 10.3389/fvets.2017.00103
Received: 05 April 2017; Accepted: 13 June 2017;
Published: 29 June 2017
Edited by:
Subhash Verma, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaReviewed by:
Sherry Layton, Vetanco, ArgentinaPaulo Henrique Verardi, University of Connecticut, United States
Copyright: © 2017 Rubinelli, Kim, Park, Baker and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven C. Ricke, c3JpY2tlQHVhcmsuZWR1
†Present address: C. Adam Baker, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL, United States
 Peter Rubinelli
Peter Rubinelli Sun Ae Kim
Sun Ae Kim Si Hong Park
Si Hong Park Steven C. Ricke
Steven C. Ricke