- 1National Laboratory for Agriculture and the Environment, United States Department of Agriculture (USDA), ARS, Ames, IA, United States
- 2National Animal Disease Center, United States Department of Agriculture (USDA), ARS, Ames, IA, United States
- 3Crop Production and Pest Control Research, United States Department of Agriculture (USDA), ARS, West Lafayette, IN, United States
In recent years, multidrug-resistant (MDR) Salmonella enterica serovar Heidelberg (S. Heidelberg) has been associated with numerous human foodborne illness outbreaks due to consumption of poultry. For example, in 2011, an MDR S. Heidelberg outbreak associated with ground turkey sickened 136 individuals and resulted in 1 death. In response to this outbreak, 36 million pounds of ground turkey were recalled, one of the largest meat recalls in U.S. history. To investigate colonization of turkeys with an MDR S. Heidelberg strain isolated from the ground turkey outbreak, two turkey trials were performed. In experiment 1, 3-week-old turkeys were inoculated with 108 or 1010 CFU of the MDR S. Heidelberg isolate, and fecal shedding and tissue colonization were detected following colonization for up to 14 days. Turkey gene expression in response to S. Heidelberg exposure revealed 18 genes that were differentially expressed at 2 days following inoculation compared to pre-inoculation. In a second trial, 1-day-old poults were inoculated with 104 CFU of MDR S. Heidelberg to monitor transmission of Salmonella from inoculated poults (index group) to naive penmates (sentinel group). The transmission of MDR S. Heidelberg from index to sentinel poults was efficient with cecum colonization increasing 2 Log10 CFU above the inoculum dose at 9 days post-inoculation. This differed from the 3-week-old poults inoculated with 1010 CFU of MDR S. Heidelberg in experiment 1 as Salmonella fecal shedding and tissue colonization decreased over the 14-day period compared to the inoculum dose. These data suggest that young poults are susceptible to colonization by MDR S. Heidelberg, and interventions must target turkeys when they are most vulnerable to prevent Salmonella colonization and transmission in the flock. Together, the data support the growing body of literature indicating that Salmonella establishes a commensal-like condition in livestock and poultry, contributing to the asymptomatic carrier status of the human foodborne pathogen in our animal food supply.
Introduction
Food-producing animals such as swine, cattle, and poultry are a major reservoir of the human foodborne pathogen Salmonella (1, 2). While some Salmonella serovars can cause disease in food-producing animals, most serovars colonize these animals asymptomatically, resulting in the hosts becoming carriers and intermittent shedders of Salmonella (1). Poultry (turkey and chicken) are frequent carriers of Salmonella, and poultry products represent about 58% of the salmonellosis cases associated with products regulated by the Food Safety Inspection Service (3). The prevalence of foodborne disease outbreaks caused by Salmonella enterica serovar Heidelberg (S. Heidelberg) has increased over the last decade (4). One of the largest meat recalls in U.S. history resulted from a multistate outbreak of multidrug-resistant (MDR) S. Heidelberg in 2011 that caused 136 confirmed cases of human foodborne disease (39% hospitalization rate) and the recall of 36 million pounds of ground turkey meat (5–7). Other recent outbreaks of foodborne illness involving S. Heidelberg include contact with dairy bull calves (https://www.cdc.gov/salmonella/heidelberg-11-16/index.html), an international in-flight catered meal (8), and chicken that sickened 634 case patients in 29 states and Puerto Rico (38% hospitalization rate) (9). Analysis of invasive non-typhoidal Salmonella isolated in the U.S. from 1996 to 2007 indicated that ~14% of S. Heidelberg isolates were from human bloodstream infections with resistance to one or more antimicrobial agents being associated with increased risk for invasive disease (10). S. Heidelberg is responsible for 7% of human deaths due to non-typhoidal Salmonella in the U.S. (11), the second most frequent serovar causing mortality following serovar Typhimurium. The prevalence of multidrug resistance (resistance to three or more antibiotic classes) in S. Heidelberg has increased 2.6-fold since 2004 (12), including resistance to ampicillin, gentamicin, streptomycin, tetracycline, chloramphenicol, kanamycin, and sulfisoxazole (5, 9). Based on 2013 data from the National Antimicrobial Resistance Monitoring System, ~33% of S. Heidelberg isolates that cause human foodborne disease are MDR (13).
Understanding the interactions of a foodborne pathogen with its food animal host is important for managing food safety risk; therefore, we investigated the pathogenicity, colonization and transmission potential of an MDR S. Heidelberg strain from the 2011 ground turkey outbreak in a natural poultry host—commercial turkeys. The MDR S. Heidelberg strain colonized the spleen and tissues of the digestive tract of the turkey without causing noticeable clinical symptoms. Gene expression analysis of blood from 3-week-old turkeys at two days post-inoculation (dpi) suggested only a mild response to the 1010 CFU challenge, with 18 genes identified as differentially expressed. In young poults less than one week old, MDR S. Heidelberg from inoculated poults was efficiently transmitted to naive poults; these data suggest that young poults are susceptible to colonization by MDR S. Heidelberg which may allow for the development of an asymptomatic carrier state in turkeys, thereby confirming this vulnerability as a critical control point to reduce food safety risk in poultry. Collectively, the lack of clinical symptoms and limited gene expression in 3-week-old turkeys in response to the MDR S. Heidelberg outbreak strain paired with the efficient transmission, colonization, and proliferation of the strain in newly hatched poults provide insight into potential factors that contribute to the successful colonization of turkey farms with MDR S. Heidelberg thereby leading to the recent outbreaks with this human foodborne pathogen.
Materials and Methods
Bacterial Strains and Selective Medium
An MDR S. Heidelberg strain BSX 126 (2011K-1138; CVM41579) isolated from ground turkey and associated with a 2011 ground turkey outbreak was used for this study (6). Strain BSX 126 is resistant to ampicillin, tetracycline, streptomycin, and gentamicin. In experiment 1 (described below), BSX 126 was inoculated into a turkey, isolated from the spleen at 7 days post-inoculation (dpi) and designated strain SB 395. Growth of S. Heidelberg on XLT-4 medium indicated that this serovar is a weak H2S producer. Similar to our investigation of S. Choleraesuis (14), reducing the tergitol concentration in XLT-4 (Becton, Dickinson and Co., Sparks, MD, USA) to 25% of the normal level allowed S. Heidelberg to produce H2S, resulting in the visualization of black colonies following 48 h of incubation. Therefore, the bacterial growth medium for culture of S. Heidelberg from turkeys was XLT-4 containing 25% tergitol (1.15 ml/l), tetracycline (15 µg/ml), streptomycin (50 µg/ml), and novobiocin (40 µg/ml).
Animal Trials and Sample Processing
Experiment 1
Sixteen 1-day-old tom (male) turkey poults were group housed for two weeks. Fecal samples from the group pen tested negative for Salmonella twice using qualitative bacteriology as previously described (15). Turkeys were separated in individual pens and inoculated by oral gavage with 108 (n = 8) or 1010 (n = 7) CFU of MDR S. Heidelberg strain BSX 126 at 3 weeks of age. Cloacal temperatures were measured using a Medline thermometer, model # MDS9850B (Mundelein, IL, USA) at 0, 1, 2, and 3 days post-inoculation (dpi). Salmonella levels in the feces were determined at 0, 1, 2, 3, 7, 10, and 14 dpi using quantitative and qualitative bacteriology as previously described (16). At 7 dpi, four turkeys from the 108 CFU inoculated group and four turkeys from the 1010 CFU inoculated group were euthanized, and tissues [crop, liver, spleen, small intestine (near the cecum), cecum, and cloaca] were collected for Salmonella enumeration as previously described (16). At 14 dpi, the remaining turkeys (3–4) were euthanized and evaluated as described earlier.
Experiment 2
Thirty-nine 1-day-old tom (male) turkey poults were group housed for the trial. Fecal samples obtained from the shipping crate tested negative for Salmonella using qualitative bacteriology. On the day of arrival at NADC, 20 poults (index birds) were inoculated with 2 × 104 CFU SB 395 in 0.25 ml PBS by oral gavage and 19 poults (sentinel birds) received 0.25 ml PBS. At 8 days following MDR S. Heidelberg inoculation into the index birds, 10 turkeys from each group (index and sentinel) were euthanized, and the cecum and spleen were collected for Salmonella enumeration. The remaining turkeys in both experimental groups were euthanized at 9 days following MDR S. Heidelberg inoculation of index birds and the tissues harvested for Salmonella enumeration.
RNA Isolation and Sequencing from Turkey Blood
Using the LeukoLOCK™ Fractionation & Stabilization Kit (ThermoFisher Scientific), blood was collected and fractionated from the wing vein of 3-week-old turkeys (from experiment 1) before inoculation as well as 2 dpi with 1010 CFU S. Heidelberg following NCAH SOP-ARU-0300. RNA from the leukocyte population (white blood cells) was extracted using the LeukoLOCK™ Total RNA Isolation System (ThermoFisher Scientific). RNA quality and quantity were analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). Libraries were constructed using the Illumina TruSeq RNA Sample Prep Kit v2 and were sequenced on an Illumina HiSeq 2500 in a 100-cycle paired-end sequencing run (Illumina Inc., San Diego, CA, USA) at the Iowa State University DNA core facility. Sequence data were imported, quality trimmed in CLC Genomic workbench V 9.5.2, and mapped to the Meleagris gallopavo reference assembly 5.0 (17). Expression values were calculated only using uniquely mapped reads. Empirical analysis of differential gene expression was performed using the EdgeR statistical test, implemented in CLC Genomic workbench, on the raw unique reads (18). Gene expression differences greater than 1.5-fold with false discovery rate-adjusted P-values less than 0.05 were considered significant.
Results and Discussion
Fecal Shedding, Tissue Colonization, and Transmission of an MDR S. Heidelberg Outbreak Strain in Turkeys
To evaluate the pathogenicity of an MDR S. Heidelberg outbreak strain in turkeys, 3-week-old turkey poults were inoculated with 108 or 1010 CFU and monitored for fecal shedding of Salmonella and changes in body (cloacal) temperature. Using Bonferroni’s multiple comparison test, no significant difference in average body temperatures at 1, 2, or 3 dpi compared to pre-inoculation was observed at either inoculation dose (data not shown); thus, no fever was induced in the turkeys following S. Heidelberg challenge. Fecal shedding of Salmonella was detected out to 10 dpi for the 108 CFU inoculated turkeys and to 14 dpi for the 1010 CFU inoculated turkeys (Figure 1). During the first week post-inoculation, an ~1-log difference in Salmonella shedding between the 108 and 1010 CFU inoculated birds was measured, and an ~1-log reduction, regardless of inoculation dose, occurred each day in the turkeys for the first 3 days.
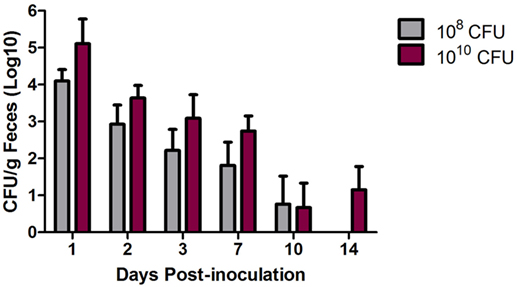
Figure 1. Multidrug-resistant (MDR) Salmonella enterica serovar Heidelberg (S. Heidelberg) fecal shedding from turkeys. At 3 weeks of age, individually housed turkeys were inoculated with 108 (n = 8) or 1010 (n = 7) CFU of MDR S. Heidelberg. Feces was collected at the indicated time points, and quantitative and qualitative bacteriology was performed to determine fecal shedding of MDR S. Heidelberg. Four turkeys from each group were euthanized at day 7 dpi resulting in smaller groups for the remaining time points. Error bars indicate SEM.
Tissue colonization of S. Heidelberg was determined at 7 and 14 dpi for the crop, liver, spleen, small intestine (near the cecum), cecum, and cloaca (Figure 2). At 7 dpi, S. Heidelberg was detected in the crop (108 dose; 2/4 birds), spleen (108 and 1010 doses; 1/4 and 4/4 birds, respectively), small intestine (1010 dose; 1/4 birds), cecum (108 and 1010 doses; 2/4 and 3/4 birds, respectively), and cloaca (108 and 1010 doses; 2/4 and 3/4 birds, respectively). At 14 dpi, Salmonella was only detected in the cecum (2/4 birds) and cloaca (1/4 birds) samples in the turkeys inoculated with S. Heidelberg at 108 CFU. S. Heidelberg was not detected in the liver at 7 dpi and was therefore not evaluated at 14 dpi.
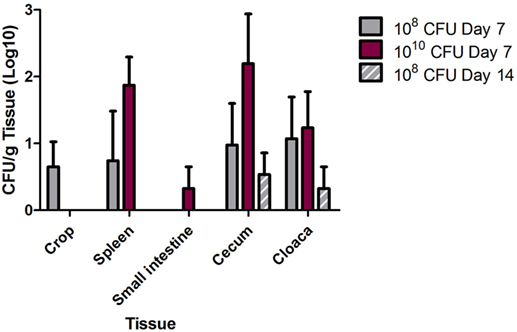
Figure 2. Multidrug-resistant (MDR) Salmonella enterica serovar Heidelberg (S. Heidelberg) tissue colonization in turkeys. At 3 weeks of age, individually housed turkeys were inoculated with 108 (n = 8) or 1010 (n = 7) CFU of MDR S. Heidelberg. At day 7 following Salmonella inoculation, four turkeys from each group were euthanized, and the remaining turkeys were euthanized at 14 dpi. Tissues (crop, spleen, small intestine, cecum, and cloaca) were harvested following euthanasia for quantitative and qualitative bacteriology to determine tissue colonization by MDR S. Heidelberg. Error bars indicate SEM.
In a separate study, 1-day-old poults (n = 20) were inoculated with 2 × 104 CFU of the MDR S. Heidelberg outbreak strain and group housed with mock-inoculated poults (n = 19). At 8 and 9 dpi, all poults were euthanized, and spleen and cecal colonization was determined. The spleens from five index and seven sentinel poults were positive for MDR S. Heidelberg whereas the cecum in all poults were colonized by Salmonella (Figure 3). For the index and sentinel poults, Salmonella levels were 5.6 and 6.3 Log10 CFU/g cecum tissue, respectively (Figure 3). Thus, exposure to MDR S. Heidelberg earlier in life (1-day-old poults) resulted in higher cecal colonization rates in poults compared to challenge at a later time in life (3 weeks, Figure 2), even with a lower dose of ~104 CFU (compared to the 1010 dose at 3 weeks of age). Interestingly, at 8/9 days following oral inoculation of the 1-day-old turkey poults with MDR S. Heidelberg, cecum colonization was increased ~2 Log10 CFUs compared to the inoculum dose regardless of whether birds were directly inoculated (index) or following transmission (sentinel) from inoculated poults. In our experience with challenging swine or turkeys with various Salmonella serovars, we typically observe a considerable decrease in Salmonella CFUs for fecal shedding and tissue colonization by 7 dpi compared to the initial inoculation dose, not an increase as measured in this study. However, our previous experiments were performed with pigs or turkeys that were 3 weeks of age or older. This suggests that the development and maturation of host factors such as immunity and/or the intestinal microbiota play an important role in limiting Salmonella colonization in older swine and poultry. Our results with MDR S. Heidelberg in turkeys are consistent with an experiment by Menconi et al. who demonstrated that day-of-hatch turkey poults inoculated with ~106 CFU S. Heidelberg were colonized with 7.04 and 6.05 Log10 CFU/g cecal contents at 24 and 72 h following inoculation, respectively, with Salmonella present in the cecal tonsils of all poults (20/20) at both time points (19). These results indicate that the level of S. Heidelberg in the cecal contents of poults at 72 h did not decrease from the inoculum level. The findings of Menconi et al. in turkeys were not replicated in two trials in which day-of-hatch broiler chicks were inoculated with either 105 or 106 CFU of serovar Heidelberg (19); at 72 h following inoculation of chicks with Salmonella, the cecal contents were colonized with either 1.08 or 2.96 Log10 CFU S. Heidelberg/g tissue. Thus, whereas in turkeys the inoculum dose and the level of cecal content colonization at 72 h were similar, in broiler chicks the colonization level decreased compared to the inoculation dose of S. Heidelberg. The authors specifically noted this difference indicating that turkey poults were more susceptible to serovar Heidelberg colonization compared to broiler chicks (19). S. Heidelberg colonization of the turkey cecal tonsils and cecal contents could be reduced by inoculating poults with a mixed culture of lactic acid bacteria (LAB) 1 h after Salmonella inoculation. The inoculation of poults with LAB reduced S. Heidelberg cecal content colonization by ~4 Log10 CFU/g content and cecal tonsil colonization by 55% at 72 h following Salmonella inoculation in comparison to inoculation with S. Heidelberg alone (19). This supports a role of the turkey intestinal microbiota in limiting colonization of S. Heidelberg either due to direct inoculation (e.g., probiotic administration) or maturation of the microbial community with age. The quantity of S. Heidelberg in the cecum or cecal tonsils in both our experiment and the trial by Menconi et al. indicates similar levels of colonization (106 CFU), potentially suggesting a threshold for niche colonization in the turkey cecum. Our results further extend the findings of Menconi et al. by demonstrating that in newly hatched turkey poults, S. Heidelberg can efficiently colonize the cecum through transmission of the pathogen within the flock. Efficient colonization of young turkey poults may contribute to lifelong colonization of turkeys with S. Heidelberg and a human foodborne risk for consumption of turkey meat.
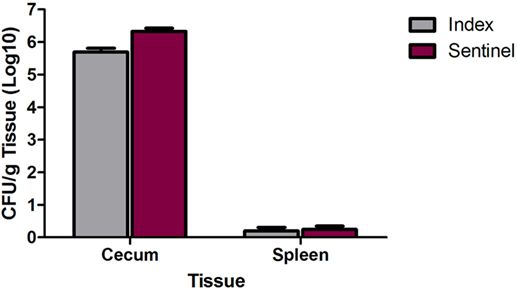
Figure 3. Cecum and spleen colonization by multidrug-resistant (MDR) Salmonella enterica serovar Heidelberg (S. Heidelberg) in index and sentinel poults. At 1 day of age, 20 poults (index) were directly inoculated with 2 × 104 CFU of MDR S. Heidelberg and group housed with 19 poults (sentinel). At days 8 and 9 following index inoculation, 10 poults from each group were euthanized the first day, and the remaining birds were euthanized the second day to harvest tissues. Quantitative and qualitative bacteriology was performed on the cecum and spleen to determine MDR S. Heidelberg tissue colonization and Salmonella transmission from index to sentinel poults. Error bars indicate SEM.
Transcriptional Response of Commercial Turkeys to an MDR S. Heidelberg Outbreak Strain
Gene expression analysis of 3-week-old turkeys in response to an MDR S. Heidelberg outbreak strain was conducted by RNA-Seq using total RNA isolated from peripheral blood before and 2 days after inoculation (day 2/day 0). Eighteen genes were differentially expressed at 2 dpi compared to pre-inoculation (Table 1). The expression of three genes was significantly upregulated (RUFY3, LOC104911311, and SERPINB10). The gene ontology biological process annotation of RUFY3 suggests a role in positive regulation of cell migration; overexpression of RUFY3 caused the formation of F-actin-protrusive structures (invadopodia) and the induction of migration and invasion in human gastric cancer cell line SGC-7901 (20). The predicted gene description for LOC104911311 is “cytokine receptor common subunit beta-like.” SERPINB10 (a.k.a. PI10, bomapin) is a member of the superfamily of serine proteinase inhibitors (serpins) that are key regulators in biological processes ranging from complement activation, coagulation, cellular differentiation, tumor suppression, apoptosis, and cell motility (21). A study by Schleef and Chuang described a role for PI10 in the inhibition of tumor necrosis factor alpha-induced cell death (22).
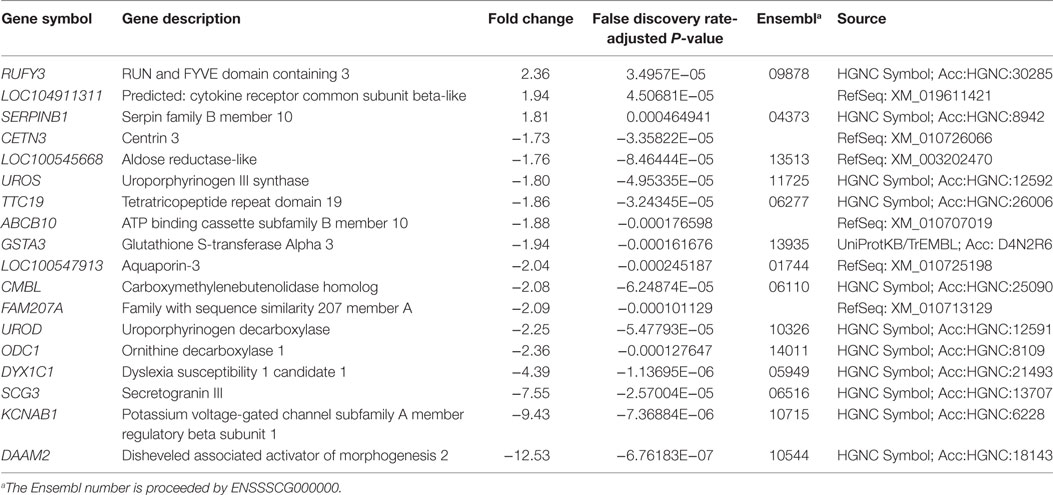
Table 1. Differentially expressed turkey genes in response to multidrug-resistant Salmonella enterica serovar Heidelberg (2 dpi/0 dpi).
Fifteen genes were significantly downregulated in response to MDR S. Heidelberg challenge. A range of predicted functions for these genes includes ABC transporter (ABCB10), heme biosynthetic pathway (UROD, UROS), glutathione transferase (GSTA3), polyamine biosynthesis pathway (ODC1), calcium binding (CETN3), respiratory chain (TTC19), and voltage-gated potassium channel activity (KCNAB1). LOC100547913 is predicted to encode aquaporin-3 (AQP3). Aquaporins are involved in transepithelial fluid transport and have been implicated in cell migration by a mechanism that facilitates water transport in lamellipodia of migrating cells (23). In this regard, AQP3 has been associated with macrophage immune function via a cellular mechanism involving water and glycerol transport that results in subsequent phagocytic and migration activity (24). AQP3−/− mice had an impaired mucosal innate immune response to Citrobacter rodentium, as demonstrated by reduced crypt hyperplasia, decreased epithelial expression of IL-6 and TNF-α, and diminished bacterial clearance (25). If LOC100547913 is AQP3, downregulation of the gene may contribute to the limited immune response observed in the turkeys. The gene with the greatest reduction in expression was DAAM2, predicted to encode a key effector in the canonical Wnt signal transduction pathway involved in gene expression regulation during embryonic development and regenerative myelination (26).
Similar to our results, a study of the chicken response to S. enterica serovar Enteritidis identified SERPIN B as upregulated and AQP8 (an aquaporin) as downregulated (27). However, the number of differentially expressed genes in the MDR S. Heidelberg-challenged turkeys seemed minimal compared to other gene expression studies of poultry in response to Salmonella (28). Moreover, a recent study by our group profiling the transcriptome of 3-week-old commercial turkeys in response to S. Typhimurium challenge identified over 1,000 differentially regulated genes (manuscript in preparation). In a comparison of the gene expression changes in response to the two Salmonella serovars, 17 of the 18 genes differentially expressed in turkeys following S. Heidelberg challenge were similarly differentially expressed in response to S. Typhimurium challenge (data not shown). Taken together, this MDR S. Heidelberg outbreak strain appears capable of colonizing turkeys without inducing a strong host response (transcriptionally or clinically), conceivably due to the commensal-like state of this human foodborne pathogen in turkeys.
In summary, S. Heidelberg has been isolated from most food-producing animals, shown increased resistance to antimicrobial agents, and is among the top 5 serovars associated with human foodborne illness (29). Understanding the commensal state established by S. Heidelberg (and other Salmonella serovars) in livestock and poultry requires investigating the complex interactions of the virulence mechanisms of the particular Salmonella serovar and the host’s response to not only initial colonization but also the subsequent establishment of a persistently colonized condition. Numerous host factors play a role in this response including animal genetics, age of exposure, health and immune status, farm husbandry practices, and the microbial composition of the gastrointestinal tract. Our colonization data provide insight into the ability of this serovar to effectively evade host response systems, because the 3-week-old turkeys appeared unaffected by the inoculation with 10 billion Salmonella Heidelberg, both clinically and transcriptionally. Furthermore, the efficient transmission, colonization, and proliferation of MDR S. Heidelberg from index to sentinel poults during the first week of life suggests that limiting the introduction of Salmonella into turkey flocks during the establishment of the intestinal microbiota is critical for control of this human foodborne pathogen. Efficient colonization of turkeys at a young age by serovar Heidelberg may help explain the prevalence of this serovar in human foodborne disease including outbreaks.
Ethics Statement
Procedures involving animals followed humane protocols as approved by the USDA, ARS, National Animal Disease Center Animal Care and Use Committee in strict accordance with the recommendations in the Guide for the Care, and Use of Laboratory Animals by the National Research Council of the National Academies.
Author Contributions
BB and SB designed the experiments; BB, SB, and DS were involved in acquisition of the experimental data. BB, SB, TL, GC, and DS performed data analysis and interpretation. The manuscript was drafted and revised for important intellectual content by BB, SB, TL, GC, and DS, as well as, final approval of the version to be published with agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclaimer
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HH declared a shared affiliation, with no collaboration, with the authors to the handling editor.
Acknowledgments
The authors are greatly appreciative of the outstanding technical support of Kellie Winter, Jennifer Jones, and Margaret Walker. The authors thank David Alt for assistance with the DNA sequencing facility. The MDR S. Heidelberg strain 2011K-1138 (CVM41579) was kindly provided by Patrick McDermott at the FDA Center for Veterinary Medicine, Laurel, MD, USA.
Funding
This research was supported by USDA, ARS CRIS funds.
References
1. Stevens MP, Humphrey TJ, Maskell DJ. Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci (2009) 364:2709–23. doi:10.1098/rstb.2009.0094
2. Rajan K, Shi Z, Ricke SC. Current aspects of Salmonella contamination in the US poultry production chain and the potential application of risk strategies in understanding emerging hazards. Crit Rev Microbiol (2017) 43:370–92. doi:10.1080/1040841X.2016.1223600
3. Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, et al. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis (2013) 19:407–15. doi:10.3201/eid1903.111866
4. Doyle ME. Multidrug-resistant pathogens in the food supply. Foodborne Pathog Dis (2015) 12:261–79. doi:10.1089/fpd.2014.1865
5. Folster JP, Pecic G, Rickert R, Taylor J, Zhao S, Fedorka-Cray PJ, et al. Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg from a ground turkey-associated outbreak in the United States in 2011. Antimicrob Agents Chemother (2012) 56:3465–6. doi:10.1128/AAC.00201-12
6. Hoffmann M, Zhao S, Luo Y, Li C, Folster JP, Whichard J, et al. Genome sequences of five Salmonella enterica serovar Heidelberg isolates associated with a 2011 multistate outbreak in the United States. J Bacteriol (2012) 194:3274–5. doi:10.1128/JB.00419-12
7. Routh JA, Pringle J, Mohr M, Bidol S, Arends K, Adams-Cameron M, et al. Nationwide outbreak of multidrug-resistant Salmonella Heidelberg infections associated with ground turkey: United States, 2011. Epidemiol Infect (2015) 143:3227–34. doi:10.1017/S0950268815000497
8. Rebolledo J, Garvey P, Ryan A, O’Donnell J, Cormican M, Jackson S, et al. International outbreak investigation of Salmonella Heidelberg associated with in-flight catering. Epidemiol Infect (2014) 142:833–42. doi:10.1017/S0950268813001714
9. Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, et al. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One (2016) 11:e0162369. doi:10.1371/journal.pone.0162369
10. Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, et al. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother (2011) 55:1148–54. doi:10.1128/AAC.01333-10
11. Kennedy M, Villar R, Vugia DJ, Rabatsky-Ehr T, Farley MM, Pass M, et al. Hospitalizations and deaths due to Salmonella infections, FoodNet, 1996–1999. Clin Infect Dis (2004) 38(Suppl 3):S142–8. doi:10.1086/381580
12. Brunelle BW, Bearson BL, Allen HK. Prevalence, evolution, and dissemination of antibiotic resistance in Salmonella. In: Singh OV, editor. Foodborne Pathogens and Antibiotic Resistance. Hoboken, NJ: John Wiley & Sons, Inc (2016). p. 331–48.
13. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). Human Isolates Final Report, 2013. Atlanta, GA: U.S. Department of Health and Human Services, CDC (2015).
14. Bearson BL, Bearson SM, Kich JD. A DIVA vaccine for cross-protection against Salmonella. Vaccine (2016) 34:1241–6. doi:10.1016/j.vaccine.2016.01.036
15. Bearson BL, Bearson SM, Lee IS, Brunelle BW. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb Pathog (2010) 48:214–9. doi:10.1016/j.micpath.2010.03.005
16. Bearson SM, Bearson BL, Loving CL, Allen HK, Lee I, Madson D, et al. Prophylactic administration of vector-encoded porcine granulocyte-colony stimulating factor reduces Salmonella shedding, tonsil colonization, and microbiota alterations of the gastrointestinal tract in Salmonella-challenged swine. Front Vet Sci (2016) 3:66. doi:10.3389/fvets.2016.00066
17. Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg Le A, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol (2010) 8(9):e1000475. doi:10.1371/journal.pbio.1000475
18. Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26:139–40. doi:10.1093/bioinformatics/btp616
19. Menconi A, Wolfenden AD, Shivaramaiah S, Terraes JC, Urbano T, Kuttel J, et al. Effect of lactic acid bacteria probiotic culture for the treatment of Salmonella enterica serovar Heidelberg in neonatal broiler chickens and turkey poults. Poult Sci (2011) 90:561–5. doi:10.3382/ps.2010-01220
20. Wang G, Zhang Q, Song Y, Wang X, Guo Q, Zhang J, et al. PAK1 regulates RUFY3-mediated gastric cancer cell migration and invasion. Cell Death Dis (2015) 6:e1682. doi:10.1038/cddis.2015.50
21. Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem (1994) 269:15957–60.
22. Schleef RR, Chuang TL. Protease inhibitor 10 inhibits tumor necrosis factor alpha-induced cell death. Evidence for the formation of intracellular high M(r) protease inhibitor 10-containing complexes. J Biol Chem (2000) 275:26385–9. doi:10.1074/jbc.C000389200
23. Agre P, King LS, Yasui M, Guggino WB, Ottersen OP, Fujiyoshi Y, et al. Aquaporin water channels – from atomic structure to clinical medicine. J Physiol (2002) 542:3–16. doi:10.1113/jphysiol.2002.020818
24. Zhu N, Feng X, He C, Gao H, Yang L, Ma Q, et al. Defective macrophage function in aquaporin-3 deficiency. FASEB J (2011) 25:4233–9. doi:10.1096/fj.11-182808
25. Thiagarajah JR, Chang J, Goettel JA, Verkman AS, Lencer WI. Aquaporin-3 mediates hydrogen peroxide-dependent responses to environmental stress in colonic epithelia. Proc Natl Acad Sci U S A (2017) 114:568–73. doi:10.1073/pnas.1612921114
26. Lee HK, Chaboub LS, Zhu W, Zollinger D, Rasband MN, Fancy SP, et al. Daam2-PIP5K is a regulatory pathway for Wnt signaling and therapeutic target for remyelination in the CNS. Neuron (2015) 85:1227–43. doi:10.1016/j.neuron.2015.02.024
27. Matulova M, Varmuzova K, Sisak F, Havlickova H, Babak V, Stejskal K, et al. Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Vet Res (2013) 44:37. doi:10.1186/1297-9716-44-37
28. Te Pas MF, Hulsegge I, Schokker D, Smits MA, Fife M, Zoorob R, et al. Meta-analysis of chicken – Salmonella infection experiments. BMC Genomics (2012) 13:146. doi:10.1186/1471-2164-13-146
Keywords: Salmonella enterica serovar Heidelberg, multidrug-resistant, foodborne outbreak, turkey, colonization, transmission, gene expression
Citation: Bearson BL, Bearson SMD, Looft T, Cai G and Shippy DC (2017) Characterization of a Multidrug-Resistant Salmonella enterica Serovar Heidelberg Outbreak Strain in Commercial Turkeys: Colonization, Transmission, and Host Transcriptional Response. Front. Vet. Sci. 4:156. doi: 10.3389/fvets.2017.00156
Received: 29 June 2017; Accepted: 07 September 2017;
Published: 25 September 2017
Edited by:
Paul Wigley, University of Liverpool, United KingdomReviewed by:
Haiqi He, United States Department of Agriculture (USDA), United StatesGuillermo Tellez, University of Arkansas, United States
Copyright: © 2017 Bearson, Bearson, Looft, Cai and Shippy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bradley L. Bearson, YnJhZC5iZWFyc29uQGFycy51c2RhLmdvdg==
 Bradley L. Bearson
Bradley L. Bearson Shawn M. D. Bearson
Shawn M. D. Bearson Torey Looft
Torey Looft Guohong Cai
Guohong Cai Daniel C. Shippy
Daniel C. Shippy