- 1National Veterinarian Diagnostic Center, China Animal Disease Control Center, Beijing, China
- 2Department of Inspection Technology Research, China Institute of Veterinary Drug Control, Beijing, China
- 3South China Agricultural University, Guangzhou, China
MicroRNAs (miRNAs) are a kind of small non-coding RNA molecules that could regulate multiple biological pathways at posttranscriptional level. Over 2,000 miRNAs have so far been discovered in humans, and many of them are found to be linked to various kinds of diseases. Thus, miRNAs are being considered as clinical diagnostic and therapeutic targets. With the discovery of high stability of circulating miRNAs in various kinds of mammalian body fluids, the potential of circulating miRNAs as diagnostic/prognostic biomarkers of infectious diseases aroused great interest among researchers. As far as human diseases are concerned, some biomarkers based on circulating miRNAs have been progressed to clinical application. In veterinary fields, however, this concept is only beginning to come into view. In this review, we summarize an update of preclinical studies on using circulating miRNAs as diagnostic biomarkers to combat infectious diseases that affect domestic animals.
Introduction
MicroRNAs (miRNAs) are small, endogenously expressed, non-coding RNA transcripts, about 22 nucleotides in length, with unique sequences targeting mRNAs for posttranscriptional regulation. Various researches have showed that miRNAs play important roles in various physiological and pathological processes, such as immune response, inflammatory response, and tumor occurrence (1, 2). The dysregulations of miRNAs have also been implicated in various kind of diseases, including diabetes, kidney disease, cancer, and many infectious diseases. Since 2008, the stable existence of miRNAs was identified in different type of body fluids (such as urine, serum, and plasma) (3–5). The application prospect of circulating miRNAs as non-invasive diagnostic biomarkers of disease status attracts the interest of researchers (5–7). At present, compared to the studies of circulating miRNAs in human diseases, only very limited studies are focused on miRNAs in veterinary infectious diseases and most are related to cellular miRNAs, instead of circulating miRNAs. Recently, the significance of functions of miRNAs in the host response to several veterinary virus diseases has been comprehensively reviewed (8, 9). In this review, we mainly focus on the research progress of using circulating miRNAs as diagnostic biomarkers for infectious diseases of domestic animals.
Function of miRNAs
Over 30 years ago, the first miRNA was identified in the nematode Caenorhabditis elegans with the identification of the developmental regulator lin-4 (10). In the year of 2000, another miRNA, let-7, regulating the timing of C. elegans development, was found to be conserved in a wide range of animal species, including human, and this result indicates this kind of small regulatory RNAs has a more general biological function (11, 12). In 2002, Calin and colleagues found that miR-15 and miR-16 were tumor suppressors for chronic lymphocyte leukemia, which demonstrated for the first time the relationship between dysregulation of intracellular miRNAs and disease (13). Shortly afterward, researchers found that the expression levels of let-7 were closely related to prognosis for lung cancer survival (14). More and more research literatures have highlighted that alterations in miRNA expression profile were related to a wide range of human diseases, especially in the studies of various cancers (15–17).
In terms of biological function of miRNAs, it was first reported in the researches about mouse models deficient for Dicer and DGCR8. DGCR8 plays an important role in miRNA biogenesis, while Drosha participates in other RNA metabolic pathways. Therefore, knockout of the two genes resulted in loss of most miRNAs and embryonic lethality (18, 19). However, individual miRNAs are seems to be not required for specification of individual tissues. In addition, miRNAs are usually required for maintaining tissue homeostasis, since the expression levels of many tissue-restricted miRNAs are usually downregulated in the process of illness (12).
Currently, over 2,000 miRNAs have been reported according to results of human transcriptome, and it is believed that over 60% of coding genes are regulated by miRNAs in the genome (20). These findings indicate miRNAs may participate in the vast majority of physiological pathways. Recently, the immunoregulatory roles of miRNAs during infection have been widely studied. For example, various miRNAs have been demonstrated to regulate the toll-like receptor 4 pathway in the host innate immune response (21, 22).
Circulating miRNAs as Diagnostic Biomarkers
A large number of studies have demonstrated that dysregulation of circulating miRNAs are associated with a wide variety of diseases progression and syndromes. At present, the relationships between profile changes of circulating miRNAs and human diseases (such as virus infections, cancers, and liver injury) have been widely reported (5, 23, 24). In the respect of veterinary infectious diseases, many studies have also demonstrated the correlation between expression profiles of circulating miRNAs and various bacteriosis (Paratuberculosis), virus disease [foot-and-mouse disease, bovine viral diarrhea (BVD)], and parasitosis (Echinococcosis).
Immune and non-immune cells could actively release miRNAs into extracellular environments (25, 26). In 2008, researchers found for the first time that circulating miRNAs were present in the samples of serum and plasma. Subsequently, circulating miRNAs were also found in other body fluids, such as saliva, urine as well as semen (27, 28). For a long time period, invasive tissue biopsies served as the main diagnostic methods for cancer; however, the existence of circulating miRNAs in blood samples of cancer parents undoubtedly significantly promotes the development of diagnostic techniques for cancer (4, 5, 29, 30). For some infectious diseases that lack of blood-based diagnostic methods, circulating miRNA is likely a good choice in term of establishing new diagnostic methods. For instance, human tuberculosis is an infectious disease that a number of researches have established new diagnostics methods using circulating miRNAs, and currently, dozens of circulating miRNAs have been identified related to this disease. However, the results obtained by different researchers exhibited somewhat inconsistency, which may be due to the different test samples (serum and sputum), different test technologies (miRNA-seq, microarray, and RT-qPCR), and data normalization methods (31).
In addition, circulating miRNAs are remarkably stable under harsh conditions, such as extended storage, boiling, low or high pH, as well as multiple freeze–thaw cycles (3, 5, 23). A study of detecting bovine serum samples from experimental infections with Mycobacterium avium subspecies paratuberculosis (MAP) demonstrated that the circulating miRNA profile of samples, which were stored at −20°C for 10–15 years, was remarkably similar to that of fresh serum samples (stored at −80°C for less than one year) (7). In another study, researchers evaluated the stability of miRNA levels in plasma and serum from healthy dogs after storage at room temperature for different time points, and it was found that miRNAs were sufficiently stable in serum or plasma stored at room temperature for 1 h but not for 24 h (32). The remarkable stability of circulating miRNAs in body fluids is considered to be mainly attributed to two mechanisms: (1) circulating miRNAs could form a protein–miRNA complex with argonaute proteins or high-density lipo-proteins and (2) they could incorporate into exosomes (33).
Based on all these features of circulating miRNAs mentioned above, they have attracted great attention of researchers as a kind of potential diagnostic biomarker for various diseases.
Analysis of Circulating miRNA
As mentioned previously, circulating miRNAs can be detected in many kind of biofluids, while majority of current studies about circulating miRNAs are based on serum and plasma. In addition, the abundance of circulating miRNAs varies in different type of body fluids (27, 34). Due to this reason, here we only focus on discussion of the main points of detecting circulating miRNAs in plasma or serum samples.
Previous studies about analysis of serum and plasma samples demonstrate that different sample treatment methods can affect the quality of extracted miRNAs (34, 35). For example, as heparin could inhibit downstream PCR assay, collection of peripheral blood samples should use EDTA-coated tubes (36, 37). In addition, while separation of serum or plasma, hemolysis should be strictly avoided, as intracellular miRNAs from platelets and erythrocytes can introduce significant bias in the expression profiles of circulating miRNAs (34, 35, 38). To determine whether the serum and plasma samples are contaminated by hemolysis, the relative expression levels of the specific miR-451 of erythrocyte and the stable miR-23a could be used as an indicator (35).
Currently, several miRNA extraction kits are available to extract circulating miRNAs from blood. In a recent study, Guo and colleagues compared the performance of five kits (ThermoFisher Scientific Ambion TRIzol LS Reagent, Qiagen Circulating Nucleic Acid Kit, QiaSymphony RNA extraction kit, Qiagen miRNEasy, and the Exiqon MiRCURY RNA Isolation Kit) for isolating circulating extracellular sRNAs, including miRNAs, and their variant isoforms (isomiRs), and transfer RNA-derived small RNAs (tDRs), and other miscellaneous sRNAs. In the respect of isolation of miRNAs, Ambion TRIzol LS produced the most reads for miRNAs, the repeatability of QiaSymphony RNA extraction kit was the highest, and Circulating Nucleic Acid Kit detected the greatest number of miRNAs. In addition, Circulating Nucleic Acid Kit also detected the most singleton miRNAs, which could explain why this kit was able to detect the most miRNAs (39).
As miRNAs are different from mRNAs in the respects of biochemistry and molecular structure, the techniques for miRNAs detection are not completely the same as that of mRNAs detection (40–42). Here, we mainly describe three frequently used techniques of miRNAs detection.
Reverse transcription quantitative real-time PCR is the most widely used technology for detecting miRNA expression level. As the sequences of miRNAs are very short, the experimental flow is different from the conventional reverse transcription PCR of mRNAs. At present, there are two common strategies used to detect the expression of circulating miRNAs: (1) using stem-loop reverse transcription followed by TaqMan PCR analysis (43) and (2) polyadenylated and reverse transcribed by a poly(T) adapter for quantitative RT-PCR using the miRNA-specific forward primer and sequence complementary to the poly(T) adapter as the reverse primer (44). It is worth noting that RT-qPCR-based technologies are unable to identify new miRNAs and special attention is needed for the design of standardized internal controls (40, 45, 46).
Hybridization-based technologies usually depend on DNA capture probes that are immobilized on a microarray so that the fluorescent signal intensities could be quantified to evaluate expression levels of different miRNAs. At present, many commercial miRNA array products are available, such as GeneChip® miRNA Arrays (Affymetrix), miRCURY LNA™ microRNA Arrays (Exiqon), and SurePrint miRNA Microarrays (Agilent) (47). Compared to other methods, the specificity and dynamic range of hybridization-based technologies are relatively lower. Thus, the results of microarray methods are usually needed to verify by RT-qPCR (48). Recently, the NanoString Technologies, Inc. established a new hybridization-based method (nCounter® miRNA Expression Assay) that does not require a PCR amplification step or direct labeling of target miRNAs (49). This approach has several advantages: it is as sensitive as RT-PCR; it has high throughput; and it could test up to 800 distinct miRNA variant targets in the same assay (31). However, similar to RT-qPCR, these hybridization-based methods are unable to identify novel miRNAs.
Unlike the methods mentioned above, miRNA-seq methods are able to discover global expression profiling of the whole miRNA transcriptome from certain biological sample (50). Compared to microarrays, the most considerable advantage of miRNA-seq is that it does not limit to detection of known miRNAs. miRNA-seq starts with constructing a cDNA sequence library reversely transcribed from short sRNAs selected by different methods, for example, size-selected gel electrophoresis. The prepared, indexed, and pooled cDNA library is subsequently sequenced using different sequencing platforms. However, special attention should be paid that technical biases inherent to different sequencing technologies may generate unreal reads (51, 52).
Differential Expression of Circulating miRNAs in Veterinary Infectious Diseases
Foot-and-Mouth Disease (FMD)
Foot-and-mouth disease, caused by foot-and-mouth disease virus, is a highly contagious infectious disease with significant economic impact. FMD can affect livestock and wild cloven-hoofed animals worldwide. This disease is characterized by fever and blister-like sores in the mouth, on the teats, on the tongue and lips, and between the hooves.
Using miRNA PCR array plates, Stenfeldt and colleagues detected the alterations of miRNA levels in bovine serum samples collected from three different phases of FMDV infection (acute, persistent, and convalescent phases) compared to uninfected animals. A total of 169 abundant miRNAs were detected in serum samples, and 3 differentially expressed miRNAs were identified: the expression of miR-1281 was significantly reduced at both acute and persistent infection stages; bta-miR-17-5p was expressed in the highest level at acute infection stage, whereas the expression level of bta-miR-31 was the highest during FMDV persistence. In addition, as cattle that cleared infection resembled the baseline profile, the authors considered that serum miRNA profiling could be used for identification of subclinically infected FMDV carriers (53).
Bovine Viral Diarrhea
Bovine viral diarrhea virus (BVDV) is an important pathogen of cattle with a global distribution and causes major economic losses (54). Animals infected with BVDV could exhibit a variety of clinical symptoms including diarrhea, depression, and pyrexia ranging from clinically mild to severe. However, in many cases, the symptoms of BVDV infections are subclinical and difficult to detect (55–57).
To identify circulating miRNAs that could be used for biomarkers indicating the timing of BVDV exposure to cattle, Taxis and colleagues investigated miRNA profiles of BVDV infected colostrum deprived male Holstein calves at four different time points: prior to infection (day 0) and at 4, 9, and 16 days postinfection. In accordance with short duration of fever and lymphopenia, the expression levels of two circulating miRNAs (Bta-miR-423-5p and Bta-miR-151-3p) were different between BVDV challenged group and control group across time examined. However, only the expression level of Bta-miR-151-3p was significantly increased at the time point of 9 days postinfection compared to the control animals. Thus, these two miRNAs could not be considered as appropriate diagnostic biomarkers (58). It is widely known that persistently infected (PI) animals are the primary source of BVDV infection, and eliminating PI animals is the main measure of prevention and control of BVD. Thus, identification of different level of expression of circulating miRNAs in serums of PI cattle could be an idea approach for seeking diagnostic biomarkers of this disease.
Ebola
The disease of Ebola caused by Ebola virus (EBOV) is a hemorrhagic infectious disease. This disease usually causes a severe hemorrhagic fever in humans and non-human primates. Early diagnosis of EBOV is of significant importance for implementation of effective interventions and prevention of the spread of infection. However, EBOV-infected patients usually do not exhibit typical symptom at the early stage for diagnosis; therefore, current diagnostic methods, such as detecting viral RNA or antigen in suspected patients, are only effective at the late stage (59).
Chen and colleagues identified a miRNA-like fragment (miR-VP-3p) generated by EBOV in the serum of Ebola patients, which was detectable before EBOV genomic RNA in the serum. Thus, this miRNA-like fragment may sever as a biomarker for early diagnosis of this disease (59). In another study, Duy and colleagues tested the circulating miRNA profiles in both rhesus macaques and humans. In total, eight circulating miRNAs (hsa-miR-146a-5p, hsa-miR-18b-5p, hsa-miR-21-3p, hsa-miR-22-3p, hsa-miR-29a-3p, hsa-miR-432-5p, hsa-miR-511-5p, and hsa-miR-596) were selected as an EBOV classifier. Further studies indicated that this classifier could correctly categorized infection status in 86% human and rhesus macaque samples (64/74). More importantly, 50% (6/12) presymptomatic rhesus macaques could also be diagnosed using this classifier (60). Using circulating miRNAs (or miRNA-like fragment), the two early diagnosis methods for Ebola will significantly promote controlling of fulminating infectious disease in the world. In addition, the finding of miR-VP-3p was a powerful evidence that RNA viruses were able to express miRNA-like small RNAs.
Paratuberculosis
Paratuberculosis (also called Johne’s disease) is a chronic disease caused by MAP. This disease can cause substantial economic losses to cattle industry due to increased premature culling, replacement costs, decreased milk yield, reduced feed conversion efficiency, fertility problems, reduced slaughter values, and increased susceptibility to other disease or conditions. However, the lack of accurate and reliable diagnostic tests is the main challenge faced in the control of paratuberculosis in cattle (61).
To identify novel diagnostic and prognostic miRNA biomarkers, Farrell and colleagues applied RNA-seq approaches to detect the circulating miRNA profiles in serum from an experimental JD infection model (six experimental MAP-challenged calves and six age-matched controls). However, the miRNA expression levels were almost the same when comparing serum collected from MAP-challenged animals to the control group at 6 months postinfection (62). In another study, the circulating miRNA expression profiles of seropositive cattle (n = 5) and seronegative cattle (n = 7) were also detected at both early and late stages of infection, while no significant differentially expressed circulating miRNAs were detected (7). Recently, Malvisi and colleagues investigated the miRNAs related to MAP infection, and seven miRNAs (bta-mir-19b, bta-mir-19b-2, bta-mir-1271, bta-mir-100, bta-mir-301a, bta-mir-32, and Novel:14_7917) were reduced and two (bta-mir-6517 and bta-mir-7857) increased in positive animals vs. unexposed animals (63). However, it is worth noting that Malvisi and colleagues studied the miRNA profiles of whole blood, rather than circulating miRNAs of serum or plasma. As is known to all, many components in the whole blood are rich in miRNA; thus, there is no comparison between this study and the two previous ones.
Staphylococcus aureus Infection
Staphylococcus aureus is one of the most prevalent pathogens causing chronic intramammary infection in cattle, responsible for substantial milk, quality, and economic loss in dairy farming worldwide. Sun and colleagues investigated the miRNA expression profiles of milk exosomes from four Holstein cows, whose mammary gland was infected with S. aureus, during mid-lactation prior to and after infection (48 h). Two miRNAs (bta-miR-142-5p and bta-miR-223) were identified as the potential indicators for early diagnosis of bacterial infection of the mammary gland (64).
Echinococcosis
Echinococcosis, caused by Echinococcus granulosus sensu lato (cystic echinococcosis) or Echinococcus multilocularis (alveolar echinococcosis), has become a major population health problem and economic importance worldwide. About 466 million people living in Asian region, over half exposed inhabitants (in particular herdsman and farmers), are at high risk of infection with E. granulosus and E. multilocularis (65).
As antibodies of this pathogen are unable to be detected at the early stages of infection (66), effective biomarkers for early and specific diagnosis of E. multilocularis are urgently needed. To find potential diagnostic targets for echinococcosis, Guo and colleagues compared the circulating miRNA expression profiles in the serum collected from the E. multilocularis-infected and uninfected mice at 4 weeks post-infection, using RNA sequencing method. In total, the expression levels of 58 circulating miRNAs were different, of which 21 were upregulated and 37 were significantly reduced.
In addition, the expression levels of six circulating miRNAs, which were verified to be differently expressed at 4 weeks postinfection, were further analyzed at different time points postinfection. The expression levels of mmu-miR-107-3p, mmu-miR-103-3p, mmu-miR-146a-5p, and mmu-miR-21a-3p were found to be significantly increased compared with the early stage of infection. The expression level of mmu-miR-339-5p was significantly increased at the early stage of infection, while no difference was observed after 4-week infection. On the contrary, mmu-miR-222-3p was significantly down regulated in the course of E. multilocularis infection (67).
Porcine Whipworm
The porcine whipworm, Trichuris suis, which is epidemic all over the world, lives in the large intestine of pigs. The pigs infected with this pathogen are usually subclinical, but worm loads are usually related to reduced feed efficiency, reduced growth rates, hemorrhagic diarrhea, and death (68).
To find circulating miRNAs of importance in T. suis infections in pigs, Hansen and colleagues investigated the expression levels of 16 preselected circulating miRNAs in serum samples collected from infected and uninfected pigs at different time points postinfection. The expression level of one circulating miRNA, ssc-let-7d-3p, was significantly increased in infected pigs at 8 weeks postinfection. As the pre-patent period of T. suis is 6–7 weeks, this miRNA operating at 8 weeks cannot be detected pre-patent infection with T. suis. Thus, its usefulness as a biomarker for early diagnosis may be limited (69).
Circulating Parasite-Derived miRNAs
Currently, more and more miRNAs have been identified in parasitic helminths, and many of them can be detected in serum and plasma samples. Thus, these worm-derived miRNAs could be used as potential biomarkers for the early detection of particular helminth infections (70).
Guo and colleagues compared the expression levels of circulating miRNAs in the serum from the E. multilocularis-infected and uninfected mice, and 2 E. multilocularis-derived circulating miRNAs (emu-miR-10 and emu-miR-277) were identified in all 15 E. multilocularis-infected serum (67).
To find schistosome-specific miRNAs as potential biomarkers for the diagnosis of Schistosomiasis, Cheng and colleagues detected the miRNAs expression profiles in the plasma from rabbits infected with Schistosoma japonicum by a deep sequencing method. They identified five schistosome-specific miRNAs (Bantam, miR-3479, miR-10, miR-3096, and sja-miR-8185) in the plasma of rabbits infected with S. japonicum, and four of the five schistosome-specific miRNAs were also detected in the plasma of S. japonicum-infected mice. Among these schistosome-specific miRNAs mentioned above, miR-10 showed diagnostic potential for S. japonicum infection, while this result is still needed to be further confirmed in other animal models (71).
Table 1 provides summary information on circulating miRNA biomarker studies for diagnosis of veterinary infectious diseases.
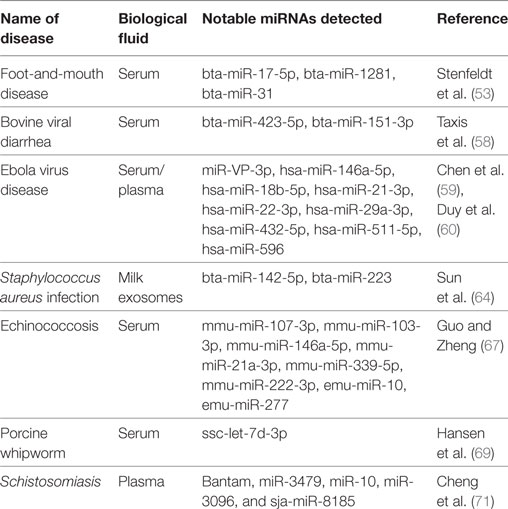
Table 1. Circulating microRNAs (miRNAs) profiled in selected studies of veterinary infectious diseases.
Conclusion
In this review, recent works of circulating miRNAs involved in diagnosis of veterinary infectious diseases were introduced. Compared to thousands of published articles about utilization of circulating miRNAs as diagnostic biomarkers in human diseases, researches focusing on circulating miRNAs in animals of veterinary relevance are still deficient. This may be mainly due to that the cost of circulating miRNAs detection is much more expensive than traditional diagnostic methods in domestic animal diseases. However, the greatest advantage of circulating miRNAs is that it could be used as markers for the early detection. Upon outbreak of infection diseases, this method could quickly identify infected animals so as to prevent the spread of the epidemic and reduce economic losses, particularly for animals of high economic value, such as breeding stocks and dairy animals. In the future, with the advances in molecular biology, it is possible that the circulating miRNAs detection method will be moved from the molecular biology lab to the clinic.
In addition, in term of human disease studies, most miRNA biomarkers reported in the literature have failed to enter clinical practice because of inconsistent and irreproducible findings. One of the important reasons is that the characteristics of clinical cases (such as age, gender, ethnicity, and medical history) are usually uncontrollable. Unlike human studies, the use of experimental animals in veterinary studies can be standardized according to the needs of experimental design, which might make the veterinary related research results more repeatable and more accurate. Thus, circulating miRNAs will have a broad application prospects in the field of veterinary diseases diagnosis.
Author Contributions
HD drafted the initial manuscript. All authors have read and approved the version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31602055 and No. 31602017).
References
1. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol (2008) 9(8):839–45. doi:10.1038/ni.f.209
2. Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med (2009) 13(1):24–38. doi:10.1111/j.1582-4934.2008.00534.x
3. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res (2008) 18(10):997–1006. doi:10.1038/cr.2008.282
4. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol (2008) 141(5):672–5. doi:10.1111/j.1365-2141.2008.07077.x
5. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A (2008) 105(30):10513–8. doi:10.1073/pnas.0804549105
6. Koberle V, Pleli T, Schmithals C, Augusto AE, Haupenthal J, Bonig H, et al. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One (2013) 8(9):e75184. doi:10.1371/journal.pone.0075184
7. Shaughnessy RG, Farrell D, Riepema K, Bakker D, Gordon SV. Analysis of biobanked serum from a Mycobacterium avium subsp paratuberculosis bovine infection model confirms the remarkable stability of circulating miRNA profiles and defines a bovine serum miRNA repertoire. PLoS One (2015) 10(12):e0145089. doi:10.1371/journal.pone.0145089
8. Samir M, Vaas LA, Pessler F. MicroRNAs in the host response to viral infections of veterinary importance. Front Vet Sci (2016) 3:86. doi:10.3389/fvets.2016.00086
9. Samir M, Pessler F. Small non-coding RNAs associated with viral infectious diseases of veterinary importance: potential clinical applications. Front Vet Sci (2016) 3:22. doi:10.3389/fvets.2016.00022
10. Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics (1980) 96(2):435–54.
11. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature (2000) 408(6808):86–9. doi:10.1038/35040556
12. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev (2015) 87:3–14. doi:10.1016/j.addr.2015.05.001
13. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A (2002) 99(24):15524–9. doi:10.1073/pnas.242606799
14. Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res (2004) 64(11):3753–6. doi:10.1158/0008-5472.CAN-04-0637
15. Bianchi F, Nicassio F, Marzi M, Belloni E, Dall’Olio V, Bernard L, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med (2011) 3(8):495–503. doi:10.1002/emmm.201100154
16. Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A (2011) 108(9):3713–8. doi:10.1073/pnas.1100048108
17. Ciesla M, Skrzypek K, Kozakowska M, Loboda A, Jozkowicz A, Dulak J. MicroRNAs as biomarkers of disease onset. Anal Bioanal Chem (2011) 401(7):2051–61. doi:10.1007/s00216-011-5001-8
18. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet (2003) 35(3):215–7. doi:10.1038/ng1253
19. Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet (2007) 39(3):380–5. doi:10.1038/ng1969
20. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi:10.1038/nrm3838
21. Yang L, Seki E. Toll-like receptors in liver fibrosis: cellular crosstalk and mechanisms. Front Physiol (2012) 3:138. doi:10.3389/fphys.2012.00138
22. He X, Jing Z, Cheng G. MicroRNAs: new regulators of toll-like receptor signalling pathways. Biomed Res Int (2014) 2014:945169. doi:10.1155/2014/945169
23. Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer (2010) 9:306. doi:10.1186/1476-4598-9-306
24. Shwetha S, Gouthamchandra K, Chandra M, Ravishankar B, Khaja MN, Das S. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep (2013) 3:1555. doi:10.1038/srep01555
25. de Candia P, De Rosa V, Casiraghi M, Matarese G. Extracellular RNAs: a secret arm of immune system regulation. J Biol Chem (2016) 291(14):7221–8. doi:10.1074/jbc.R115.708842
26. Fernandez-Messina L, Gutierrez-Vazquez C, Rivas-Garcia E, Sanchez-Madrid F, de la Fuente H. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol Cell (2015) 107(3):61–77. doi:10.1111/boc.201400081
27. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem (2010) 56(11):1733–41. doi:10.1373/clinchem.2010.147405
28. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res (2011) 39(16):7223–33. doi:10.1093/nar/gkr254
29. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol (2014) 11(3):145–56. doi:10.1038/nrclinonc.2014.5
30. Ono S, Lam S, Nagahara M, Hoon DS. Circulating microRNA biomarkers as liquid biopsy for cancer patients: pros and cons of current assays. J Clin Med (2015) 4(10):1890–907. doi:10.3390/jcm4101890
31. Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, et al. Circulating microRNAs as potential biomarkers of infectious disease. Front Immunol (2017) 8:118. doi:10.3389/fimmu.2017.00118
32. Enelund L, Nielsen LN, Cirera S. Evaluation of microRNA stability in plasma and serum from healthy dogs. MicroRNA (2017) 6(1):42–52. doi:10.2174/2211536606666170113124114
33. Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol (2013) 10(9):542–52. doi:10.1038/nrgastro.2013.87
34. McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem (2011) 57(6):833–40. doi:10.1373/clinchem.2010.157198
35. Blondal T, Jensby NS, Baker A, Andreasen D, Mouritzen P, Wrang TM, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods (2013) 59(1):S1–6. doi:10.1016/j.ymeth.2012.09.015
36. Kim DJ, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JY, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn (2012) 14(1):71–80. doi:10.1016/j.jmoldx.2011.09.002
37. Li S, Chen H, Song J, Lee C, Geng Q. Avoiding heparin inhibition in circulating microRNAs amplification. Int J Cardiol (2016) 207:92–3. doi:10.1016/j.ijcard.2016.01.129
38. Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) (2012) 5(3):492–7. doi:10.1158/1940-6207.CAPR-11-0370
39. Guo Y, Vickers K, Xiong Y, Zhao S, Sheng Q, Zhang P, et al. Comprehensive evaluation of extracellular small RNA isolation methods from serum in high throughput sequencing. BMC Genomics (2017) 18(1):50. doi:10.1186/s12864-016-3470-z
40. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet (2012) 13(5):358–69. doi:10.1038/nrg3198
41. Baker M. MicroRNA profiling: separating signal from noise. Nat Methods (2010) 7(9):687–92. doi:10.1038/nmeth0910-687
42. Kolbert CP, Feddersen RM, Rakhshan F, Grill DE, Simon G, Middha S, et al. Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS One (2013) 8(1):e52517. doi:10.1371/journal.pone.0052517
43. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res (2005) 33(20):e179. doi:10.1093/nar/gni178
44. Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques (2005) 39(4):519–25. doi:10.2144/000112010
45. Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto-Sietsma SJ. Normalization panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J (2015) 29(9):3853–62. doi:10.1096/fj.15-271312
46. Marabita F, de Candia P, Torri A, Tegner J, Abrignani S, Rossi RL. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform (2016) 17(2):204–12. doi:10.1093/bib/bbv056
47. Wu D, Hu Y, Tong S, Williams BR, Smyth GK, Gantier MP. The use of miRNA microarrays for the analysis of cancer samples with global miRNA decrease. RNA (2013) 19(7):876–88. doi:10.1261/rna.035055.112
48. McLoughlin KE, Nalpas NC, Rue-Albrecht K, Browne JA, Magee DA, Killick KE, et al. RNA-seq transcriptional profiling of peripheral blood leukocytes from cattle infected with Mycobacterium bovis. Front Immunol (2014) 5:396. doi:10.3389/fimmu.2014.00396
49. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol (2008) 26(3):317–25. doi:10.1038/nbt1385
50. Keller A, Leidinger P, Bauer A, Elsharawy A, Haas J, Backes C, et al. Toward the blood-borne miRNome of human diseases. Nat Methods (2011) 8(10):841–3. doi:10.1038/nmeth.1682
51. Buschmann D, Haberberger A, Kirchner B, Spornraft M, Riedmaier I, Schelling G, et al. Toward reliable biomarker signatures in the age of liquid biopsies – how to standardize the small RNA-seq workflow. Nucleic Acids Res (2016) 44(13):5995–6018. doi:10.1093/nar/gkw545
52. Raabe CA, Tang TH, Brosius J, Rozhdestvensky TS. Biases in small RNA deep sequencing data. Nucleic Acids Res (2014) 42(3):1414–26. doi:10.1093/nar/gkt1021
53. Stenfeldt C, Arzt J, Smoliga G, LaRocco M, Gutkoska J, Lawrence P. Proof-of-concept study: profile of circulating microRNAs in bovine serum harvested during acute and persistent FMDV infection. Virol J (2017) 14(1):71. doi:10.1186/s12985-017-0743-3
54. Houe H. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet Microbiol (1999) 64(2–3):89–107. doi:10.1016/S0378-1135(98)00262-4
55. Houe H. Economic impact of BVDV infection in dairies. Biologicals (2003) 31(2):137–43. doi:10.1016/S1045-1056(03)00030-7
56. Walz PH, Grooms DL, Passler T, Ridpath JF, Tremblay R, Step DL, et al. Control of bovine viral diarrhea virus in ruminants. J Vet Intern Med (2010) 24(3):476–86. doi:10.1111/j.1939-1676.2010.0502.x
57. Ridpath JF, Fulton RW. Knowledge gaps impacting the development of bovine viral diarrhea virus control programs in the United States. J Am Vet Med Assoc (2009) 235(10):1171–9. doi:10.2460/javma.235.10.1171
58. Taxis TM, Bauermann FV, Ridpath JF, Casas E. Circulating microRNAs in serum from cattle challenged with bovine viral diarrhea virus. Front Genet (2017) 8:91. doi:10.3389/fgene.2017.00091
59. Chen ZL, Liang HW, Chen X, Ke YH, Zhou Z, Yang MJ, et al. An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res (2016) 26(3):380–3. doi:10.1038/cr.2016.21
60. Duy J, Koehler JW, Honko AN, Schoepp RJ, Wauquier N, Gonzalez JP, et al. Circulating microRNA profiles of Ebola virus infection. Sci Rep (2016) 6:24496. doi:10.1038/srep24496
61. Garcia AB, Shalloo L. Invited review: the economic impact and control of paratuberculosis in cattle. J Dairy Sci (2015) 98(8):5019–39. doi:10.3168/jds.2014-9241
62. Farrell D, Shaughnessy RG, Britton L, MacHugh DE, Markey B, Gordon SV. The identification of circulating miRNA in bovine serum and their potential as novel biomarkers of early Mycobacterium avium subsp paratuberculosis infection. PLoS One (2015) 10(7):e0134310. doi:10.1371/journal.pone.0134310
63. Malvisi M, Palazzo F, Morandi N, Lazzari B, Williams JL, Pagnacco G, et al. Responses of bovine innate immunity to Mycobacterium avium subsp. paratuberculosis infection revealed by changes in gene expression and levels of microRNA. PLoS One (2016) 11(10):e0164461. doi:10.1371/journal.pone.0164461
64. Sun J, Aswath K, Schroeder SG, Lippolis JD, Reinhardt TA, Sonstegard TS. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genomics (2015) 16:806. doi:10.1186/s12864-015-2044-9
65. Zhang W, Zhang Z, Wu W, Shi B, Li J, Zhou X, et al. Epidemiology and control of echinococcosis in central Asia, with particular reference to the People’s Republic of China. Acta Trop (2015) 141(Pt B):235–43. doi:10.1016/j.actatropica.2014.03.014
66. Yamano K, Miyoshi M, Goto A, Kawase S. Time course of the antibody response in humans compared with rats experimentally infected with hepatic alveolar echinococcosis. J Helminthol (2014) 88(1):24–31. doi:10.1017/S0022149X12000685
67. Guo X, Zheng Y. Expression profiling of circulating miRNAs in mouse serum in response to Echinococcus multilocularis infection. Parasitology (2017) 144(8):1079–87. doi:10.1017/S0031182017000300
68. Roepstorff A, Mejer H, Nejsum P, Thamsborg SM. Helminth parasites in pigs: new challenges in pig production and current research highlights. Vet Parasitol (2011) 180(1–2):72–81. doi:10.1016/j.vetpar.2011.05.029
69. Hansen EP, Kringel H, Thamsborg SM, Jex A, Nejsum P. Profiling circulating miRNAs in serum from pigs infected with the porcine whipworm, Trichuris suis. Vet Parasitol (2016) 223:30–3. doi:10.1016/j.vetpar.2016.03.025
70. Cai P, Gobert GN, McManus DP. MicroRNAs in parasitic helminthiases: current status and future perspectives. Trends Parasitol (2016) 32(1):71–86. doi:10.1016/j.pt.2015.09.003
Keywords: circulating microRNA, biomarkers, infectious diseases, veterinary, diagnosis
Citation: Dong H, Gao Q, Peng X, Sun Y, Han T, Zhao B, Liu Y, Wang C, Song X, Wu J and Yang L (2017) Circulating MicroRNAs As Potential Biomarkers for Veterinary Infectious Diseases. Front. Vet. Sci. 4:186. doi: 10.3389/fvets.2017.00186
Received: 22 August 2017; Accepted: 17 October 2017;
Published: 01 November 2017
Edited by:
Jiabo Ding, China Institute of Veterinary Drug Control, ChinaReviewed by:
Yuehua Ke, Institute of Disease Control and Prevention, ChinaWen-juan Liu, Shandong University, China
Copyright: © 2017 Dong, Gao, Peng, Sun, Han, Zhao, Liu, Wang, Song, Wu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Yang, MzAyMjE1MTUzOEBxcS5jb20=
 Hao Dong
Hao Dong Qiang Gao2
Qiang Gao2