- 1Department of Biomedical Sciences, Iowa State University College of Medicine, Ames, IA, United States
- 2Diamond V, Cedar Rapids, IA, United States
Prior studies revealed that yeast fermentation products, specifically XPC™ and related products (Diamond V, Cedar Rapids, IA), serve as viable food safety tools across multiple food animal species including cattle and poultry. Providing this supplement in feed leads to reduced prevalence, load, virulence, and antibiotic resistance of foodborne pathogens such as Salmonella and Escherichia coli O157:H7. These findings are worthy of further study, especially when coupled with the enhanced growth and performance observed with these products. Mechanistically, XPC appears to modulate these effects through the immune system and gut microbiome. Herein we further investigated this product and demonstrate that XPC mediates an enhancement of immunocyte killing of Salmonella in calves fed the product. Additionally, these studies reveal that XPC reduces the lymph node infiltration, invasiveness, and antibiotic resistance of Salmonella in dairy calves fed the product-consistent with findings observed in poultry and adult beef cattle. Furthermore, the reduction in invasiveness does not lead to a rebound hyperinvasive phenotype in Salmonella obtained from XPC-fed animals. In summary, these studies suggest that XPC reduces the invasion of Salmonella and may alter various phenotypic characteristics of the pathogen.
Introduction
The presence of important foodborne pathogens continues to prove a formidable challenge to overcome. Two important foodborne pathogens, Salmonella enterica and shiga toxin-producing Escherichia coli serotypes such as O157:H7 (STEC), are serious concerns in food animal production management.
Yeast fermentation products, specifically the Saccharomyces cerevisiae fermentation product designated as XPC™, appears to impact enteric pathogens across multiple livestock species (1–3). Traditionally, XPC has been used for its beneficial production effects (4, 5). However, studies into its anti-pathogenic effects began after anecdotal observations indicated that there was a decrease in environmental Salmonella in chicken laying barns as part of the National Poultry Improvement Plan environmental monitoring. Questions began to emerge as to the potential use and efficacy of XPC as a pathogen mitigation tool, as well as the mechanisms underlying its effects. Recent and ongoing studies suggest that XPC, and derivatives, is a viable intervention with uses in food animal production (1–3).
Given the anecdotal reports of XPC-mediated reduction of Salmonella in poultry, the anti-Salmonella effects of XPC were examined in dairy calves. An investigator-blinded study addressed the hypothesis that XPC plus SmartCare™, an XPC derivative that is included in milk replacer, reduces the impact of Salmonella in dairy calves on milk (1). This study indicated that the consequences of feeding calves XPC plus SmartCare are an overall improvement of gastrointestinal health and development as well as a significant reduction in Salmonella load and virulence (1).
The positive results from the dairy calf study prompted an analogous study in beef cattle using an XPC derivative specific for those animals and is designated as NaturSafe™. This investigator-blinded study examined the effects of NaturSafe on fecal shedding, lymph node colonization, virulence, and antibiotic resistance of Salmonella as well as the prevalence and load of STEC (2). The results indicated that NaturSafe mediated a decrease in Salmonella load, a decrease in Salmonella fecal shedding, and a decrease in Salmonella colonization of the lymph nodes when compared to the Control diet (2).
Another study was conducted to examine the anti-Salmonella properties of XPC in regards to the load, prevalence, virulence, and antibiotic resistance of Salmonella in a controlled experiment with broilers. These studies revealed an XPC-mediated reduction of load and prevalence of Salmonella as well as a reduction in virulence and antibiotic resistance (3).
The aforementioned studies prompted further investigations into the anti-Salmonella effects of XPC. Specifically, the studies presented herein were designed to address the following hypotheses: XPC plus SmartCare enhances the immune function of calves whereby Salmonella is cleared; the feeding of XPC plus SmartCare leads to anti-virulence and anti-antibiotic resistance effects on Salmonella present in calves; and, the XPC-mediated inhibition of virulence is sustained and does not lead to an exaggerated rebound effect once the pathogen exits an XPC-fed animal.
Materials and Methods
Assessment of Immune Clearance of Salmonella in Dairy Calves
All animal experiments were approved by the Institutional Animal Care and Use Committee at Iowa State University. Newborn dairy calves (n = 20 newborn Holstein heifers and bulls in each treatment group) were fed daily doses of XPC plus SmartCare for 2 weeks as part of a previous study (1). Prior to experimental infection with Salmonella, approximately 4 mL of whole blood was collected into an EDTA tube, of which 3 mL was transferred into a microfuge tube and subjected to density gradient centrifugation (the other 1 mL was submitted for CBC analysis). The erythrocyte fraction was then removed and 120 μL of the buffy coat interface was collected and aliquoted into six separate tubes containing RPMI media, to which 107 colony-forming units of statically grown Salmonella (approximate multiplicity of infection = 100; grown in LB broth) were added and the tubes were incubated at 37°C. After 1 h, extracellular (i.e., non-invasive) bacteria were killed by the addition of 50 μg/mL gentamicin. At 0, 1, 2, 4, 8, and 12 h post-killing, leukocytes were centrifuged and the gentamicin-containing media was removed and replaced with 50 μL of phosphate-buffered saline containing 1% Triton which lyses the leukocytes. Lysates were then plated on XLD agar that was incubated overnight at 37°C. The following day, black-centered colonies were enumerated and Salmonella survival/cell was calculated as number of recovered colonies/number of leukocytes per 20 μL blood (derived from the CBC analyses). The strains used were gentamicin-susceptible S. Dublin SGI1 (6) and S. Typhimurium LNWI (7). Studies were performed in triplicate on two occasions for each of the 40 calves used in the study (1).
These same newborn dairy calves (1) were fed daily doses of XPC plus SmartCare for 2 weeks and then experimentally infected with multiresistant Salmonella DT104, followed by continued feeding of XPC plus SmartCare for another 5 weeks. At the end of the study, calves were euthanized and Salmonella were cultured from the superficial cervical lymph nodes. The exterior surface of the lymph nodes was rinsed in ethanol, and then the lymph nodes were placed in whirl packs and smashed with a rubber mallet. An equal volume of LB broth was then added and the contents were massaged and left to sit at room temperature for 1 h. An aliquot was then plated on XLD agar and then incubated overnight at 37°C. Salmonella were then enumerated on XLD agar. Studies were performed in triplicate on two occasions for each of the 40 calves used in the study (1).
Assessment of Virulence and Antibiotic Resistance of Salmonella in Dairy Calves
Salmonella isolated from dairy calves (1) were subjected to the same virulence and antibiotic resistance assays described previously (2, 3), i.e., standard tissue culture invasion assays involving HEp-2 cells (8) and micro-broth dilution antibiogram assays using MICs and resistance breakpoint concentrations. For tissue culture invasion, Salmonella were added to and extracted from tissue culture cells in a manner similar to that described for the leukocyte assays (except that a higher concentration of antibiotic was used since the strain is gentamicin-resistant). Antibiogram assays focused on florfenicol resistance since this resistance is encoded on an integron present in the input strain. That is, the studies presented herein assessed the ability of XPC to reduce both the invasiveness and integron-mediated antibiotic resistance of Salmonella obtained from dairy calves from a prior study (1). Studies were performed in triplicate on two occasions for each of the 40 calves used in the study (1). Invasion was recorded as % invasiveness, i.e., number of bacteria recovered/number of bacteria added to cells hila expression was recorded as an inverse of the number of PCR cycles required to visualize an amplicon. Percent resistant to florfenicol was determined as the number of colonies that grew in the breakpoint concentration (32 ug/mL) of florfenicol divided by the number of colonies assayed. Percent containing SGI was derived as the number of colonies that were PCR(+) for the SGI1 integron divided by the number of colonies assessed.
Assessment of Salmonella Invasion After the Pathogen Exits an XPC-Fed Host
Salmonella obtained from cattle (1, 2) and broilers (3) were subjected to serial tissue culture invasion assays. Colonies were collected en masse and then incubated with HEp-2 tissue culture cells as per Feye et al. (3). Invasion was then determined and hilA expression was then quantitated (2, 3) for successive incursions into the tissue culture cells, with the bacteria recovered from cells serving as the input strain for the next round of invasion assay. Specifically, bacteria were recovered from animals on XLD plates (i.e., zero generation) and then suspended in PBS and directly added to a fresh set of HEp-2 cells. The invasion assay was then repeated and the bacteria recovered (i.e., the first generation) were used in the subsequent invasion assay. Studies were performed in triplicate for each assay, and were performed once for the calf isolates (1), once for the beef cattle isolates (2), and once for the poultry isolates (3). Data were then pooled for all three animal-specific isolates and reported as an averaged set of data. Data are compared to the wild-type statically grown strain that was not exposed to an animal.
Statistical Analyses
For data with multiple sampling time points, a repeated measures analysis was used with the Bonferroni ad hoc test for multiple comparisons (Prism Graph Pad 7.0). For data with a single measurement, an analysis of variance (ANOVA) was conducted with the Tukey's ad hoc test for multiple comparisons.
Results
Immune Clearance of Salmonella in Dairy Calves
Our previous studies with SmartCare and XPC (1) revealed diminished Salmonella fecal shedding and intestinal carriage in calves fed the products, when compared to calves fed the Control diet. To assess the possibility that the leukocytes from XPC-fed calves are clearing the Salmonella, peripheral blood cells were examined for the ability of the leukocytes to temporally reduce the presence of Salmonella. As shown in Figure 1, the number of Salmonella was equivalent in both groups of calves (n = 5 calves/group) at the first time point but the numbers of Salmonella dropped significantly (50–75%) at each successive time point for leukocytes obtained from XPC-fed calves when compared to Control-fed calves. Importantly, this mechanism seems to be conserved across two divergent Salmonella enterica serovars- Typhimurium and Dublin.
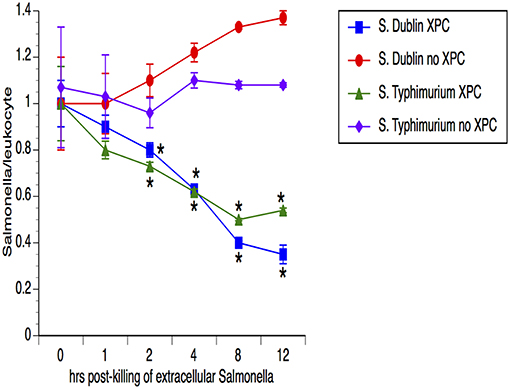
Figure 1. Assessment of Salmonella lysis by leukocytes obtained from calves fed XPC™ compared to calves that did not receive XPC. Peripheral blood cells were examined for the ability of the leukocytes to reduce the presence of Salmonella in calves fed XPC plus SmartCare. Specifically, whole blood was taken and incubated with Salmonella for 1 h. After 1 h, an enumerated portion of leukocytes were removed and exposed to gentamicin in order to kill the non-engulfed/non-invasive Salmonella. Leukocytes were then lysed with Triton and lysates were plated on XLD agar plates that were incubated overnight at 37°C for colony enumeration the next day. Salmonella/leukocyte was then calculated based on the number of Salmonella recovered and the number of leukocytes incubated with the Salmonella. This process was repeated at multiple time points in order to determine the ability of the leukocytes to temporally kill Salmonella. The strains used were S. Dublin SGI1 (6) and S. Typhimurium LNWI (7). Data presented are the mean ± sem from leukocytes obtained from 10 calves from each group on at least two separate occasions. *p < 0.05 vs. XPC for the same strain.
To address the possibility that these calves were more efficient at translocating the Salmonella to lymph nodes, peripheral lymph nodes (superficial cervical) were excised at euthanasia and subjected to selective culture for Salmonella. As shown in Figure 2, Salmonella was significantly less abundant (~80% reduced) in lymph nodes from XPC-fed calves when compared to Control-fed calves.
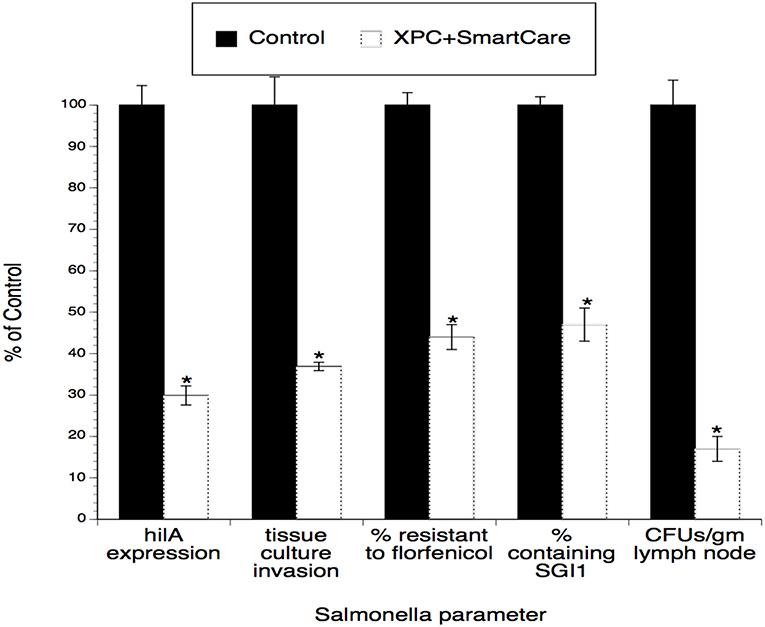
Figure 2. Assessment of Salmonella obtained from lymph nodes of calves fed XPC™ plus SmartCare™, when compared to calves that did not receive the products. Newborn dairy calves were fed daily doses of XPC plus SmartCare for two weeks and then experimentally infected with multiresistant Salmonella DT104, followed by continued feeding of XPC plus SmartCare for another 5 weeks (1). At the end of the study, calves were euthanized and Salmonella were cultured from the superficial cervical lymph nodes. Salmonella were enumerated and subjected to the virulence and antibiotic resistance assays described in other studies (2, 3). Data presented are the mean ± sem from data obtained from 10 calves from each group. *p < 0.05 vs. Control.
Diminished Virulence and Antibiotic Resistance of Salmonella in Dairy Calves
Since the input strain used in the poultry studies (2) was also used in the calf studies (1), similar virulence and antibiotic resistance assays were performed using Salmonella recovered from the calves. As shown in Figure 3, tissue culture invasiveness, hilA expression, antibiotic resistance, and prevalence of the antibiotic resistance element (SGI1) were all significantly diminished (~60–70% reductions) in Salmonella recovered from XPC-fed calves.
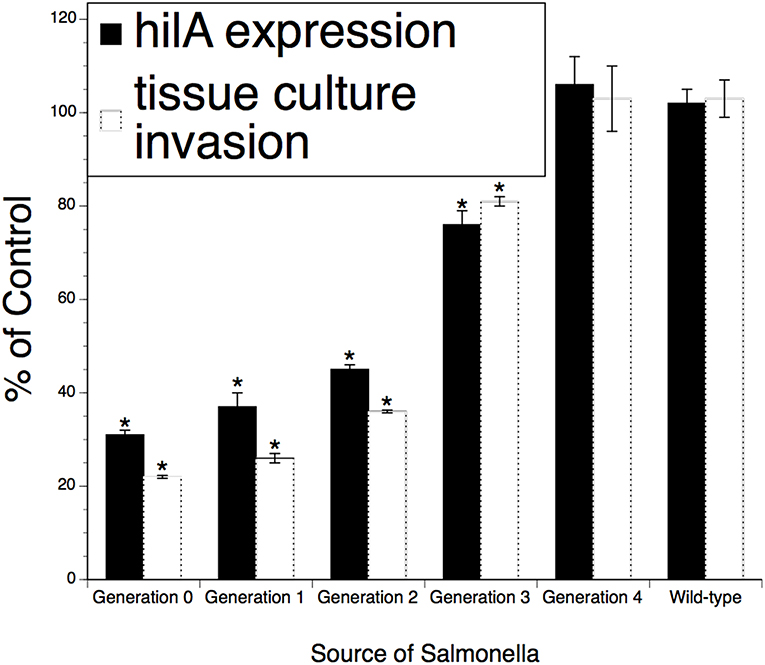
Figure 3. Persistence of the effect of XPC™ on the hi/A expression and invasiveness of Salmonella recovered from feces of naturally infected animals fed XPC (or a Control diet) for >28 days. Salmonella were recovered from treatment-specific animals (cattle and broilers) and then serially subjected to the invasion assays examining cell penetration and virulence gene expression. Generations 1–4 represent Salmonella serially recovered from tissue culture studies while Generation 0 represents Salmonella directly recovered from animals. *p < 0.05 vs. wild-type.
Restoration of Salmonella Invasion After the Pathogen Exits an XPC-Fed Host
To investigate the possibility that XPC mediates a partially sustainable hypo-invasive state in Salmonella and the microbe will return to the fully invasive state or even become hyperinvasive after exiting an XPC-fed animal, a series of temporal invasion assays were performed on isolates recovered from XPC-fed poultry and cattle. As shown in Figure 3, invasiveness and hila expression were ~70–80% reduced after recovery from animals (“Generation)” and then returned to baseline after three successive incursions (designated as “Generations”) into tissue culture cells. Neither the invasiveness nor the hila expression went above the baseline observed for Salmonella recovered from Control-fed animals or from the wild-type strain. Thus, it appears that, after exiting an XPC-fed animal, the Salmonella need to invade three additional and subsequent hosts before returning to the fully virulent state as per the wild-type strain. These data include Salmonella obtained from the experimentally infected dairy calves (1), naturally infected beef cattle (2), and from poultry that were experimentally infected (3). Thus, the data represent a variety of naturally occurring Salmonella serotypes from a variety of hosts.
Discussion
Herein we demonstrate that XPC mediates an enhancement of immunocyte killing of Salmonella in calves fed the product. Additionally, these studies reveal that XPC reduces the lymph node infiltration, invasiveness, and antibiotic resistance of Salmonella in dairy calves fed the product- consistent with findings observed in poultry (3) and adult beef cattle (2). Furthermore, the reduction in invasiveness does not lead to an immediate rebound hyperinvasive phenotype in Salmonella obtained from XPC-fed animals.
Through the implementation of XPC as a feed additive, there is an apparent advantage for reducing important foodborne illnesses attributed to food animal production. Evidence continues to show that XPC leads to a reduction in Salmonella prevalence, GI colonization, shedding, virulence, and antibiotic resistance across multiple species. Reducing the former three attributes will coordinately reduce the number of Salmonella entering the food supply. The reduced virulence in Salmonella, which is supported by a marked decrease in hilA expression and tissue culture invasiveness, has implications for both cattle and chickens where the microbe is an opportunist and a commensal, respectively. The net result is that Treated animals harbor Salmonella that are less efficient at causing disease in the current host or in a downstream mammalian host, thus requiring a significantly higher infectious dose for eliciting salmonellosis. XPC also directly decreases antimicrobial resistance and the benefit of this effect is 2-fold. The first benefit is a restoration of susceptibility to medically important antibiotics that occasionally need to be used for treating salmonellosis. The second benefit is a reduction of the presence of antibiotic resistance-encoding genetic elements, like plasmids and integrons, that allow for the global transfer of genetic information from commensal to pathogen and from pathogen to pathogen. Beyond transferring antibiotic resistance genotypes, these elements have been implicated in the transfer of information that improves the fitness (9) and virulence (10) of pathogenic bacteria. The mechanism for this effect is unknown, but may involve a dampening of antibiotic resistance gene expression or an elimination of the elements (plasmids or integrons).
The conserved mechanism for the anti-Salmonella effects of XPC is largely unknown. Research has been initiated to elucidate this interesting mechanism, though the literature is highly suggestive of two mechanisms: augmenting gut immunophysiology and enhancing microbial communities. Sustained inflammation resulting from any number of sources ultimately leads to a reduction in gut-barrier integrity, increases circulating endogenous endotoxin, and is linked to decreased feed intake and susceptibility to disease in food animals (11–13). The utilization of fermented products specifically addresses a number of these inflammatory issues by enhancing gut physiology resulting in reduced pathogenesis of important foodborne pathogens across multiple species (1, 3, 14).
XPC appears to directly enhance commensal microbial populations, leading to an improvement of bacterial competition, gastrointestinal physiology, and nutrient utilization. The underlying mechanisms governing the success of XPC may also be pertinent to the reduction of other pathogens that extend beyond the foodborne pathogens discussed herein. Therefore, XPC has the potential to be a significant and economically viable solution for curtailing foodborne pathogens and further studies will continue to enhance our understanding of postbiotics on physiology.
Ethics Statement
This study was carried out in accordance with the recommendations of the Iowa State University Institutional Animal Care and Use Committee. The protocol was approved by the committee.
Author Contributions
All university investigators performed experiments for the 261 study, with KF, KA and SC performing the animal experiments. The manuscript was initially authored by KF and then revised by SC. The other investigators (KA, JC, JW, GS-M, DM, and HP) then provided editorial comments that culminated in the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
These studies were funded by Diamond V, Cedar Rapids, IA, USA. The university investigators (KF, JC, KA, JW, GS-M, and SC) were blinded as to the treatments provided by the sponsors (DM and HP). A third party revealed the treatments after the investigators disclosed the results. Part of these findings first appeared in an author's thesis (KF), which can be accessed online at https://lib.dr.iastate.edu/cgi/viewcontent.cgi?article=7134&context=etd (15).
References
1. Brewer M, Anderson K, Yoon I, Scott M, Carlson S. Amelioration of salmonellosis in pre-weaned dairy calves fed Saccharomyces cerevisiae fermentation products in feed and milk replacer. Veter Microbiol. (2014) 172:248–55. doi: 10.1016/j.vetmic.2014.05.026
2. Feye K, Anderson K, Scott M, Henry D, Dorton K, Depenbusch B, et al. Abrogation of Salmonella and E. coli O157:H7 in feedlot cattle fed a proprietary Saccharomyces cerevisiae fermentation prototype. J Veter Sci Technol. (2016) 7:350. doi: 10.4172/2157-7579.1000350
3. Feye K, Anderson K, Scott M, McIntyre D, Carlson S. Inhibition of the virulence, antibiotic resistance, and fecal shedding of multiple antibiotic-resistant Salmonella Typhimurium in broilers fed Original XPC. Poult Sci. (2016) 95:2902–10. doi: 10.3382/ps/pew254
4. Price KL, Totty HR, Lee HB, Utt MD, Fitzner GE, Yoon I, et al. Use of Saccharomyces cerevisiae fermentation product on growth performance and microbiota of weaned pigs during Salmonella infection. J Anim Sci. (2010) 88:3896–908. doi: 10.2527/jas.2009-2728
5. Lensing M, van der Klis J, Yoon I, Moore D. Efficacy of Saccharomyces cerevisiae fermentation product on intestinal health and productivity of coccidian-challenged laying hens. Poult Sci. (2012) 91:1590–7. doi: 10.3382/ps.2011-01508
6. Xiong N, Brewer M, Day T, Kimber M, Barnhill A, Carlson S. Evaluation of the pathogenicity and virulence of three strains of Salmonella organisms in calves and pigs. Am J Veter Res. (2010) 71:1170–7. doi: 10.2460/ajvr.71.10.1170
7. Wu MT, Carlson SA, Meyerholz DK. Cytopathic effects observed upon expression of a repressed collagenase gene present in Salmonella and related pathogens: mimicry of a cytotoxin from multiple antibiotic-resistant Salmonella enterica serotype Typhimurium phagetype DT104. Microb Pathogen. (2002) 32:1–9. doi: 10.1006/mpat.2002.0535
8. Gianella RA, Washington O, Gemski P, Formal SB. Invasion of HeLa cells by Salmonella typhimurium: a model for study of invasiveness of Salmonella. J Infect Dis. (1973) 128:69–75. doi: 10.1093/infdis/128.1.69
9. Kempf A, Hulsebus H, Akbar S. Multiple plasmids contribute to antibiotic resistance and macrophage survival in vitro in cmy2-bearing Salmonella enterica. Foodborne Pathog Dis. (2016) 13:398–404. doi: 10.1089/fpd.2015.2067
10. Carlson S, Sharma V, McCuddin Z, Rasmussen M, Franklin S. Involvement of a Salmonella genomic island 1 gene in the rumen protozoan-mediated enhancement of invasion for multiple-antibiotic-resistant Salmonella enterica serovar Typhimurium. Infect Immunity. (2007) 75:792–800. doi: 10.1128/IAI.00679-06
11. Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. (2009) 9:799–809. doi: 10.1038/nri2653
12. Gaggìa F, Mattarelli P, Biavati B. Probiotics and prebiotics in animal feeding for safe food production. Int J Food Microbiol. (2010) 141:15–28. doi: 10.1016/j.ijfoodmicro.2010.02.031
13. Mani V, Weber TE, Baumgard LH, Gabler NK. Growth and development symposium: endotoxin, inflammation, and intestinal function in livestock. J Anim Sci. (2012) 90:1452–65. doi: 10.2527/jas.2011-4627
14. Vieira AT, Fukumori C, Ferreira CM. New insights into therapeutic strategies for gut microbiota modulation in inflammatory diseases. Clin Trans Immunol. (2016) 5:e87. doi: 10.1038/cti.2016.38
Keywords: Salmonella, antibiotic resistance, virulence, yeast fermentation products, food safety
Citation: Feye KM, Carroll JP, Anderson KL, Whittaker JH, Schmidt-McCormack GR, McIntyre DR, Pavlidis HO and Carlson SA (2019) Saccharomyces cerevisiae Fermentation Products That Mitigate Foodborne Salmonella in Cattle and Poultry. Front. Vet. Sci. 6:107. doi: 10.3389/fvets.2019.00107
Received: 20 November 2018; Accepted: 22 March 2019;
Published: 10 April 2019.
Edited by:
Wageha Awad, University of Veterinary Medicine Vienna, AustriaReviewed by:
Lisa Bielke, The Ohio State University, United StatesYves Millemann, École Nationale Vétérinaire d'Alfort, France
Copyright © 2019 Feye, Carroll, Anderson, Whittaker, Schmidt-McCormack, McIntyre, Pavlidis and Carlson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steve A. Carlson, c3RldmVjQGlhc3RhdGUuZWR1
 Kristina M. Feye1
Kristina M. Feye1 John H. Whittaker
John H. Whittaker Hilary O. Pavlidis
Hilary O. Pavlidis Steve A. Carlson
Steve A. Carlson