- 1Department of Agriculture and Animal Health, College of Agriculture and Environmental Sciences, University of South Africa, Johannesburg, South Africa
- 2Section Veterinary Public Health, Department of Paraclinical Sciences, Faculty of Veterinary Science, University of Pretoria, Pretoria, South Africa
- 3Department of Biomedical and Diagnostic Sciences, College of Veterinary Medicine, University of Tennessee, Knoxville, Knoxville, TN, United States
Background: While surveillance of antimicrobial drug resistance is ongoing in human medicine in South Africa, there is no such activity being performed in veterinary medicine. As a result, there is a need to investigate antimicrobial resistance among enterococci isolated from dogs in South Africa to improve understanding of the status of antimicrobial drug resistance given its public and veterinary public health importance. This study investigated antimicrobial resistance and factors associated with resistance profiles of enterococci isolated from dogs presented for veterinary care at a veterinary teaching hospital in South Africa.
Methods: In total 102 Enterococcus isolated between 2007 and 2011 by a bacteriology laboratory at a teaching hospital were included in this study. Antimicrobial susceptibility of the isolates was determined against a panel of 18 antimicrobials using the Kirby Bauer disc diffusion technique. Univariate analysis was used to assess simple associations between year, season, breed group, age group, sex, and specimen as covariates and extensive drug resistance (XDR) as the outcome. Variables that were significant in the univariate analysis at a generous p-value ≤ 0.2 were included in the multivariable logistic models to investigate predictors of XDR.
Results: All the Enterococcus isolates were resistant to at least one antimicrobial. High proportions of isolates were resistant against lincomycin (93%), kanamycin (87%), orbifloxacin (85%), and aminogycoside-lincosamide (77%). Ninety three percent (93%), 35.3, and 8.8% of the isolates exhibited multi-drug, extensive-drug and pan-drug resistance, respectively. Only year was significantly (p = 0.019) associated with extensive-drug resistance.
Conclusion: Given the zoonotic potential of Enterococcus spp., the high antimicrobial resistance and multi-drug resistance observed in this study are a public health concern from one health perspective. The identified resistance to various antimicrobials may be useful in guiding clinicians especially in resource scarce settings where it is not always possible to perform AST when making treatment decisions.
Introduction
Enterococci are Gram-positive, non-spore forming facultative anaerobic bacteria that can survive a wide range of temperatures, pH, hyperosmolarity, and prolonged desiccation (1–3). Enterococcus faecalis, Enterococcus faecium, and Enterococcus durans, are the most commonly isolated Enterococcus species from animals. However, E. faecalis and E. faecium are the most common in dogs (4–6).
The public health significance of enterococci is based on their ability to develop resistance and horizontally transfer this resistance to other bacteria (7). This is confirmed by a number of studies that have reported enterococci as an important source of resistant genes for organisms such as Listeria spp. (8, 9) and Staphylococcus aureus (10, 11). The public health importance of enterococci is exacerbated by the fact that pets are a putative reservoir of antimicrobial-resistant bacteria for humans (12).
Available evidence suggests that enterococci tend to develop resistance to most antibiotics including: penicillin, chloramphenicol, tetracyclines, rifampin, fluoroquinolones, aminoglycosides, and vancomycin (13–15). This resistance is both intrinsic and acquired (13–17). In view of this, antibiotic resistance is a problem that complicates therapy in patients with enterococcal infection, with potential to compromise successful treatment of enterococci infection (18). Resistant enterococci have also been observed in poultry and poultry abattoir workers (19, 20). Recent reports suggest that resistance among enterococci organisms from humans is not only limited to first line antimicrobials such as the beta-lactams, but has also been observed against antimicrobials such as vancomycin that are reserved for use as the second line of defense (21, 22).
Enterococci are opportunistic pathogens that form part of the normal microbial flora in the gastrointestinal tracts of humans, animals, and birds (23–27). When found outside the gut, they are indicators of fecal contamination (3). However, over the past decade, enterococci have become the leading cause of nosocomial and community-acquired infections in human medicine (18, 28–30).
Some of the conditions caused by enterococci include endocarditis, bacteraemia, urinary tract infections, neonatal sepsis (31, 32), diarrhea, biliary tract infection, peritonitis, post-operative infection, and post-partum endomyometritis (32). Moreover, enterococci have become increasingly resistant to antimicrobial agents commonly used in veterinary hospitals (18, 19, 28–30).
There is no evidence of studies in South Africa that have investigated antimicrobial resistance profiles of Enterococcus species in dogs. Furthermore, while surveillance of antimicrobial resistance is ongoing in human medicine in South Africa, currently there is no such activity being performed in veterinary medicine. As a result, there is a need to investigate antimicrobial resistance among enterococci isolated from dogs in South Africa. This study investigated antimicrobial resistance and factors associated with resistance profiles of enterococci isolated from dogs presented for veterinary care at a veterinary teaching hospital in South Africa. This study is based on the hypothesis that presupposes that resistance profiles of clinical enterococci isolated from dogs admitted at a veterinary teaching hospital are influenced by several host factors. This study is intended to contribute to improved understanding of antimicrobial resistance profiles of enterococci from dogs, and information from this study will enable clinicians to make decisions regarding the most appropriate and effective antimicrobial therapies to treat enterococci infections in dogs.
Methods
Ethical Statement
This study was approved by the Research Ethics Committee of the University of South Africa (REC Reference Number: 2019/CAES_AREC/107). When owners of animals present their animals for treatment at the Veterinary Teaching Hospital where the data were obtained, they are requested to sign a consent form granting permission for samples and data collected for the purpose of diagnosis to also be used for research purposes. To ensure anonymity and confidentiality of patients and their owners, study findings are reported in aggregated form. As a result, no patient identifiable information is included in this manuscript.
Study Area
This retrospective data used in this study came from a laboratory associated with veterinary teaching hospital in Gauteng province, South Africa. The province spans approximately over 18,000 km2 and consists of rural, urban and peri-urban areas. Based on the 2019 mid-year population estimates, the province has a human population of 15,176,115 people and it is the most populated province in South Africa (33).
South Africa has been dubbed a dog country with a population of 9.1 million dogs. Residents of Gauteng province keep dogs for various reason that include: for personal security, as companions, and to keep feral cats away from their compounds (34, 35). Oosthuizen et al. (36) observed that 28% of the dogs in selected provinces of South Africa that included Gauteng Province, were stray dogs while 72% had owners. This notwithstanding, keeping dogs is regulated by city by-laws. These by-laws for example, prescribe the number of dogs that one can keep on their premises, and requires that all dog owners ensure that at all times the dog is confined to the premises of the owner (37). With respect to access to veterinary services, this is skewed in favor of dogs in urban and peri urban areas where veterinary services are readily available. However, this is not the case for dogs in rural and/or formerly disadvantaged areas (areas where the majority of black South Africans reside). In South Africa, while antibiotics used in cattle are available over the counter without a prescription, this is not the case with antibiotics use in companion animals such as dogs. In the later, antibiotics are only available by prescription from a qualified veterinarian.
Gauteng province experiences four seasons: summer (November–March), autumn (April–May), winter (June–August), and spring (September–October). Winter is the driest season, while summer (especially December and January) is the wettest season. The province experiences annual maximum temperatures of about 22°C in the south and 25°C in the north (38).
Isolation of Enterococcus spp. and Testing for Antimicrobial Susceptibility
All samples used in the study were processed by the bacteriology laboratory associated with a teaching veterinary academic hospital. The laboratory follows standardized protocols for isolation of Enterococcus spp. described by Quinn et al. (39). Furthermore, the laboratory conducts antimicrobial susceptibility testing using the Kirby Bauer disc diffusion technique following the guidelines described by the Clinical and Laboratory Standards Institute (CLSI) (40–46). The susceptibility profile of the Enterococcus isolates included in this study were determined against a panel consisting of the following 18 drugs: 30 μg-amikacin, 20/10 μg amoxicillin-clavulanic acid,100 μg carbenicillin,30 μg ceftazidime, 30 μg cephalothin,30 μg chloramphenicol, 30 μg doxycycline, 5 μg enrofloxacin,10 μg gentamicin, 30 μg imipenem, 30 μg kanamycin, 2 μg lincomycin, 100 μg lincospectin,5 μg orbifloxacin, 10 μg penicillin-G, 25 μg trimethoprim_sulfamethoxazole, 10 μg tobramycin, and 15 μg tylosin tartrate (Oxoid Ltd., Cambridge, UK).
The laboratory that provided the data also follows the CLSI protocol for isolation of enterococci organisms and uses clinical breakpoints, to determine breakpoints of each isolate (40–46). However, where there are no clinical breakpoints, it adopts epidemiological cut-off points. Furthermore, where there are no veterinary break points, the laboratory uses human breakpoints.
The laboratory reports antimicrobial susceptibility profile of the isolates as susceptible, intermediate or resistant. However, the data provided did not include isolates that were classified as intermediate.
For purpose of this study, antimicrobial resistance (AMR) was defined as resistance to at least one of the tested antimicrobials. Multidrug resistance (MDR) was defined as resistance to at least one agent in more than three antimicrobial categories, while extensive drug resistance (XDR) was defined as resistance to at least one agent in all but two or fewer antimicrobial categories. Finally, pan-drug (PDR) resistance was defined as resistance to all antimicrobial categories tested (47).
Data Source
Laboratory records of 102 clinical isolates of Enterococcus spp. from dogs presented at a veterinary teaching hospital between January 2007 and December 2011 were included in this study. The inclusion criteria included the following: an isolate had to have data on AMR and other explanatory variables to be included in the study or the analysis. Fields extracted from the records included: breed, sex, age, date sample was submitted, type of specimen submitted, organ system giving rise to the sample as well as culture and antimicrobial susceptibility test results.
Data Management, Rationale of Data Analysis Approach and Data Analysis
Data Management
To prepare the data for analysis, dog breeds were categorized based on the American Kennel Club classification into breed groups (48). The age of dog was analyzed both as a numeric and categorical variable. As a categorical variable, age was categorized as described by Chen et al. (49), and was consequently recoded into five categories (<2, 2–4, >4–7, >7–10, and >10 years old) (Table 1). Since several types of specimen were submitted to the laboratory for isolation of enterococci, this variable was re-coded into a five-category variable.
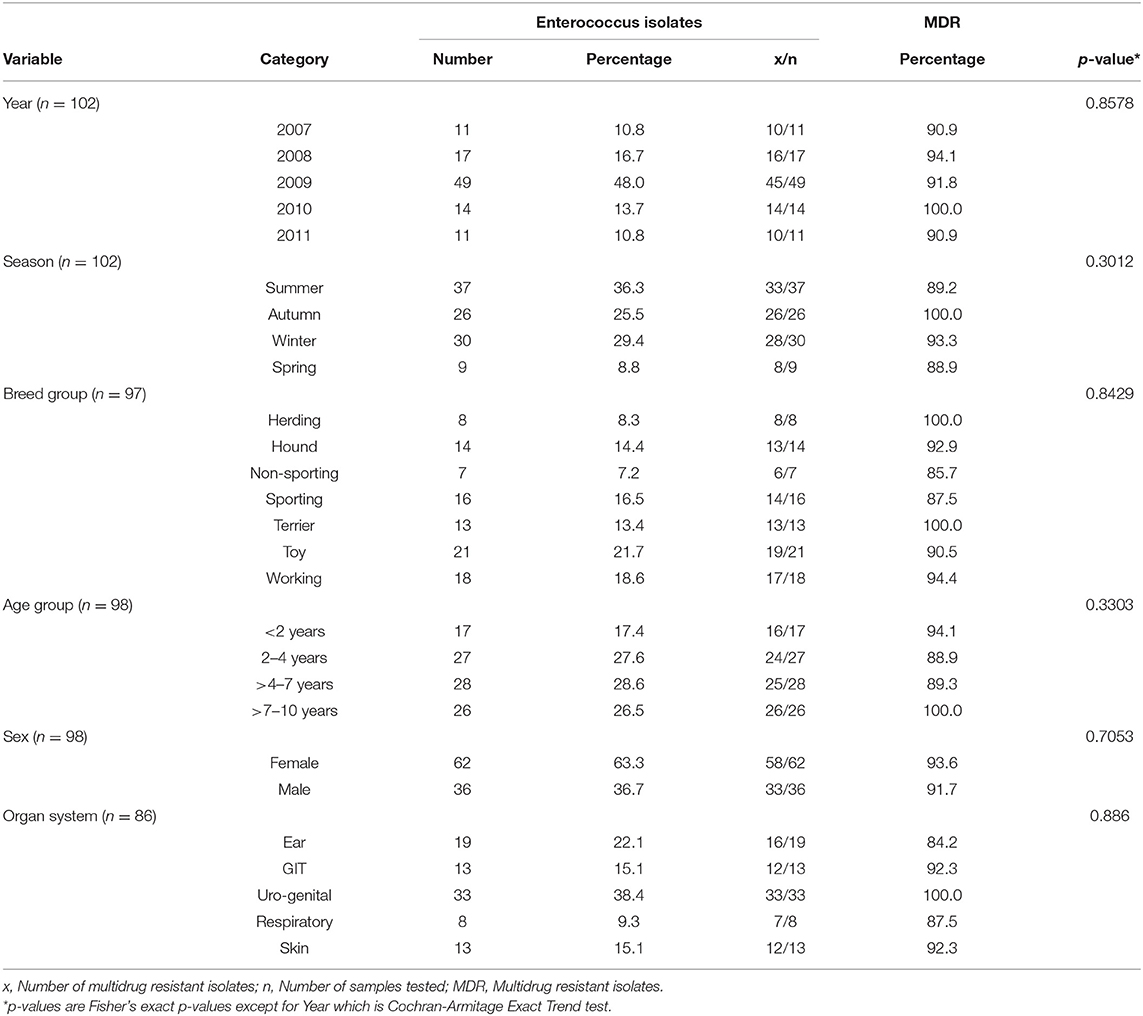
Table 1. Distribution of Enterococcus and multidrug resistant enterococcal isolates among dog specimens tested at a teaching veterinary hospital in South Africa, 2007-2011.
Rationale of Data Analysis and Data Analysis
Rationale of data analysis
Since the outcome (XDR status; Yes = 1/No = 0) was a binary variable, a binary logistic regression model was fitted to the data to investigate the association between the explanatory variables and the outcome (XDR).
Data Analysis
All the statistical analyses were performed in SAS 9.4 statistical package (SAS Institute Inc., Cary, NC, USA).
Descriptive Statistics
The age of the dogs from which samples were obtained was not normally distributed (Shapiro-Wilk W = 0.958; p = 0.0035), and therefore the median and interquartile range (IQR) were calculated. Percentages or proportions of categorical variables were calculated and presented as tables.
Inferential Statistics
Models to identify predictors of XDR
Since all isolates in the study dataset were resistant to at least one antimicrobial (AMR) and almost all isolates were multidrug resistant (MDR), the associations between predictor variables; year, season, breed group, age group, sex, and organ system and AMR or MDR were not assessed. Therefore, only the association between the predictor variables listed above and one outcome variable; XDR status (Yes = 1/No = 0) was assessed using a logistic regression model.
The model building
Building of the models was done in two steps: the 1st step involved building univariable logistic regression models to identify potential predictors associated with the outcome at a relaxed α ≤ 0.20. Variables with p ≤ 0.20 in the univariable model were selected for inclusion in the multi-variable model. The 2nd step involved fitting a multivariable logistic regression model using manual backwards selection method with the level of significance set at α ≤ 0.05.
Confounding was assessed by comparing the change in model coefficients with and without the suspected confounders. If the removal of a suspected confounding variable resulted in a ≥20% change in the coefficient of any variable in the model, the variable that had been removed was considered a confounder and was thus retained in the model regardless of whether it was significantly associated with the outcome variable or not.
Odds ratios (ORs) and their 95% confidence intervals were computed for variables included in the final model. Hosmer-Lemeshow goodness-of-fit test was used to assess model fit.
Results
The Number of Enterococcus Isolates
The proportions of Enterococcus species isolated from dog clinical samples submitted to the bacteriology laboratory between 2007 and 2011 are presented in Table 1. A total of 102 Enterococcus spp. were isolated over the study period. The highest percentage were isolated in 2009 (48%) and the lowest numbers were isolated in both 2007 and 2011.
Stratified by season of isolation, most of the 102 Enterococcus isolates, were isolated in summer (36.3%). In terms of breeds, 21.7% of the isolates came from toy breeds, while the working breeds contributed the least number of isolates.
Dogs aged 5–7 and 2–4 years old yielded the highest number of isolates (28.6 and 27.6%, respectively). The lowest number of isolates were from dogs aged <2 years.
Stratified by sex, majority of the 102 isolates, were recovered from female dogs (63.3%). When the study population was stratified by sample type, the uro-genital specimens contributed 38.4% of isolates, which was the highest, followed by ear specimens that contributed 22.1% of the isolates. The respiratory system contributed the least number of isolates.
Multi-Drug Resistant Isolates
More than 80% of the Enterococcus isolates included in this study were MDR. As shown in Table 1, the number of isolates that were MDR did not vary significantly across years or seasons. Likewise, there was no significant variations in MDR across breed groups, age groups, sex, and specimen types.
Antimicrobial Drug Resistance (AMR)
All the Enterococcus isolates included in the study were resistant to at least one antimicrobial agent, and a very high proportion of these isolates exhibited resistance against the following antimicrobial groups: Lincosamide (94.1%), aminoglycosides (92.2%), fluoroquinolones (85.3%), and β-lactams (78.4) (Table 2).
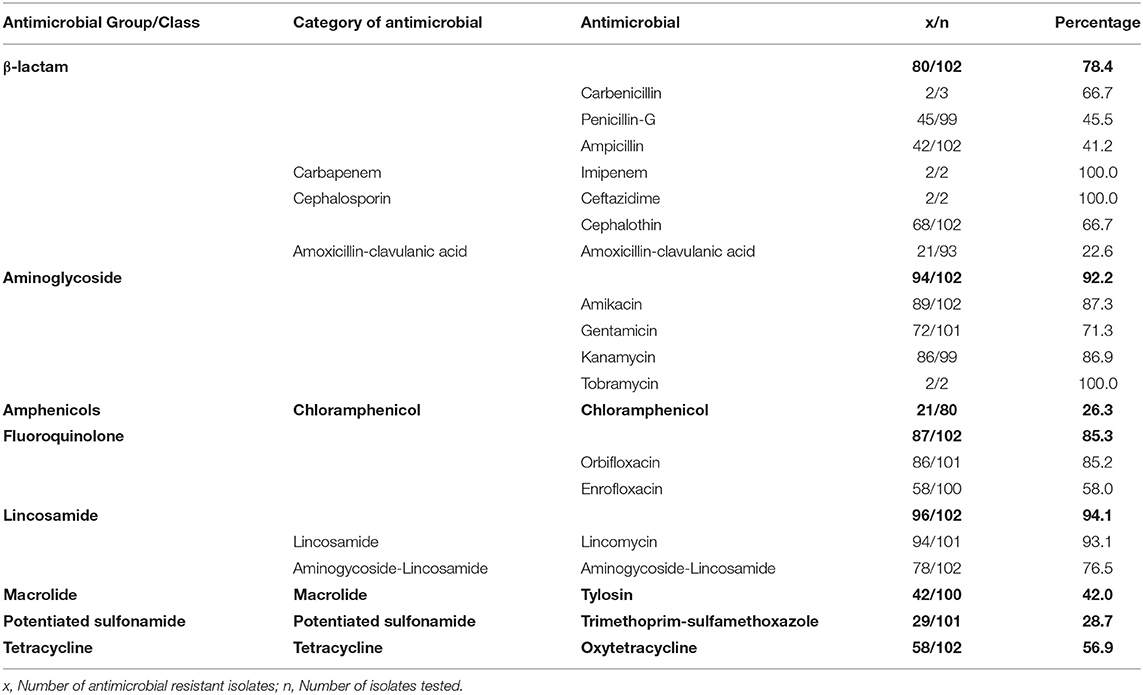
Table 2. Antimicrobial resistance profile of Enterococcus isolates from dogs tested at a teaching veterinary hospital in South Africa, 2007–2012.
Over half of the isolates were resistant to each of the following individual drugs: carbenicillin (66.7%), cephalothin (66.7%), amikacin (87.3%), gentamicin (71.3%), kanamycin (96.9%), tobramycin (100%), orbifloxacin (85.2%), enrofloxacin (58.0%), lincomycin (93.1%), aminogycoside-lincosamide (76.5%), and doxycycline (56.9%) (Table 2). On the contrary, relatively low proportions of isolates exhibited resistance against amphenicols (26.3%), Amoxicillin-clavulanic acid (22.6%) and potentiated sulfonamides (28.7%) (Table 2).
Results in Table 3 show that very high levels of resistance were consistently observed against the following drugs for the duration of the study: amikacin, cephalothin, kanamycin, lincomycin, aminoglycoside-lincosamide, and orbifloxacin. However, the number of isolates that were resistant to other antimicrobials fluctuated over the study period. Additionally, the proportion of isolates resistant to antimicrobials such as amikacin, ampicillin, and penicillin G, seemed to increase until 2010 and then dropped either drastically or slightly in 2011.
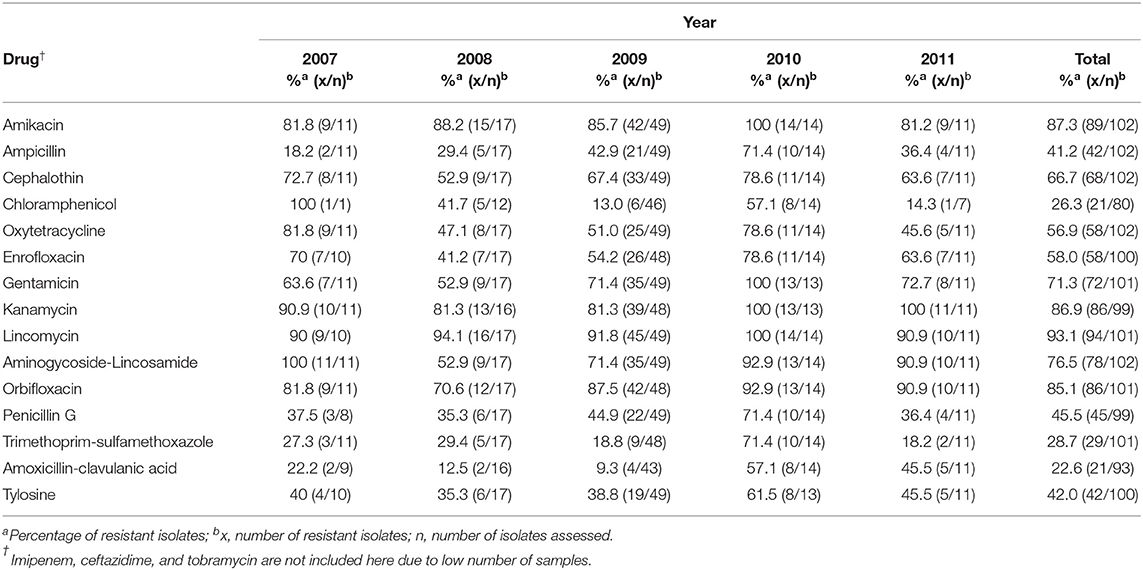
Table 3. Distribution of AMR Enterococcus isolates from dogs presented at a teaching veterinary hospital by year for the period 2007-2011.
Pan-Drug Resistance (PDR)
Eight percent (7.84%; n = 8/102) of the isolates in this study exhibited PDR, and were isolated from uro-genital (n = 4), ear (n = 2), and GIT (n = 1) samples. But as shown in Table 4, two (n = 2) of the eight (n = 8) PDR isolates had missing organ information.
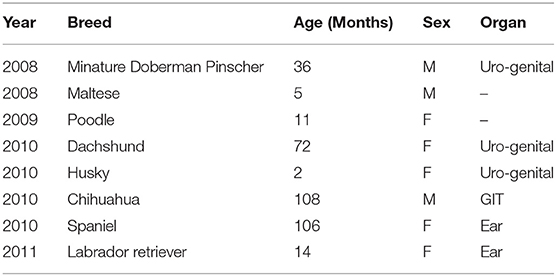
Table 4. The year of isolation and characteristics of dogs with PDR Enterococcus species infections whose samples were processed at the veterinary academic hospital, 2007-2012.
As was the case with XDR isolates, most the PDR isolates (62.5%; n = 5/8) came from female dogs, and from the uro-genital tract (37.5%; n = 3/8). The median age of dogs with PDR infections was 36 months (3 years). However, overall, the range of the age of dogs included in this study was 2 to 108 months (Table 4).
Predictors of Extensive-Drug Resistance (XDR)
Overall, 35.3% (n = 36/102) of the Enterococcus isolates tested, exhibited XDR. Only year had a statistically significant association with XDR (p = 0.0019) and hence it was the only variable included in the final XDR model. The odds of isolates being XDR were lower for isolates obtained in 2008 (OR: 0.109, p = 0.012), 2009 (OR: 0.101, p = 0.002), and 2010 (OR: 0.078, p = 0.014) compared to those obtained 2011 (Table 5).
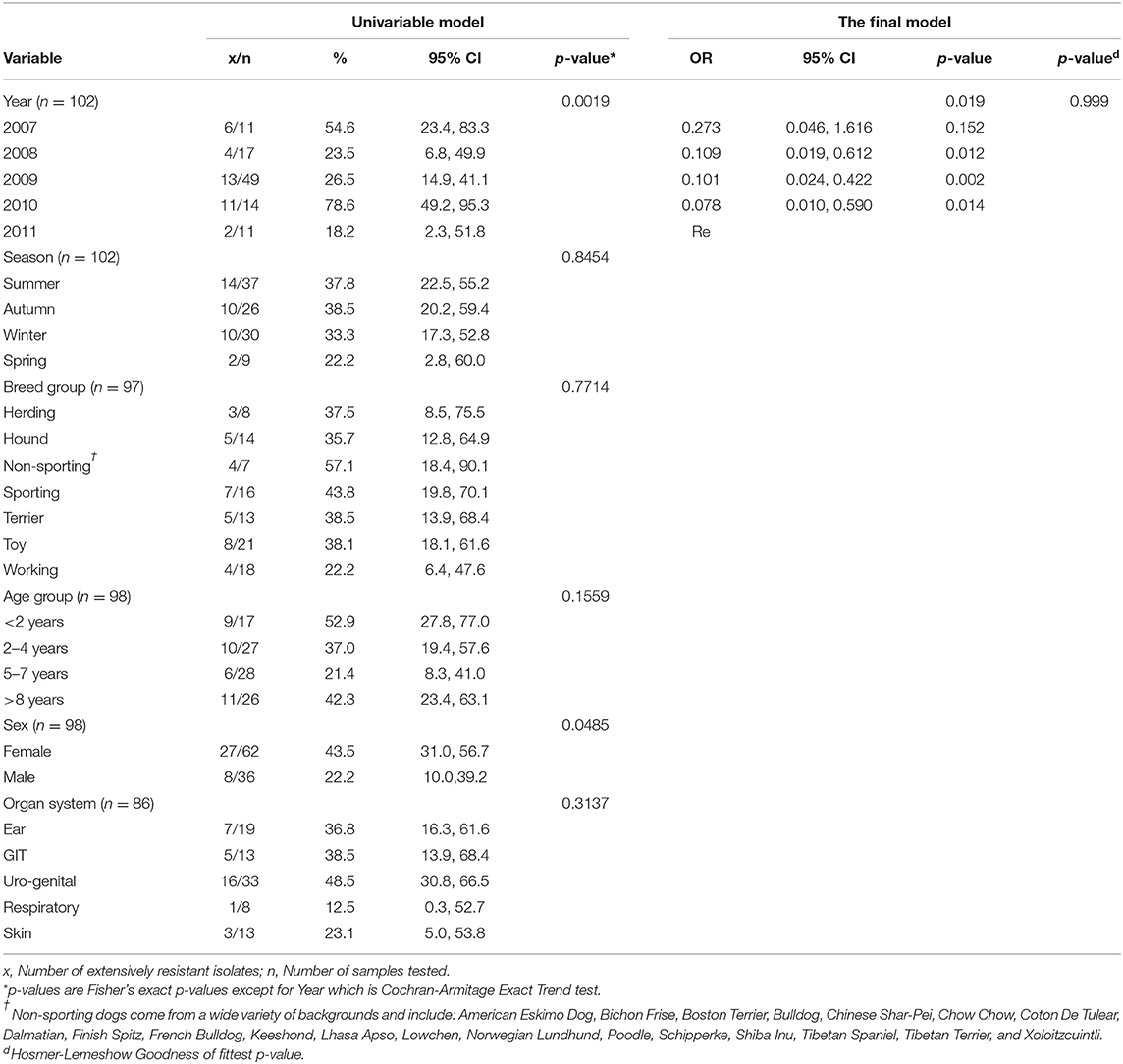
Table 5. Distribution of XDR enterococci from dog specimens submitted by the teaching hospital in South Africa between 2007 and 2011, and the association between predictor variables and the outcome (XDR).
Discussion
In this study, we investigated using data from samples submitted to the bacteriology laboratory for isolation of enterococci and assessing antimicrobial susceptibility of enterococci isolates. This investigation revealed that most of the enterococci isolates were from urino-genital samples. We also observed that enterococci were isolated from various organ systems. The study also demonstrated that a high number of isolates were resistant to at least one antimicrobial and that a high number of isolates were also MDR and XDR. However, pan-drug resistance was not as widespread. Year was the only explanatory variable that was associated with XDR. Of noteworthy, is that results reported here showed that resistance to antimicrobials recommended for the treatment of enterococci such as amoxicillin-clavulinic Acid combination (23%), ampicillin (41%), and penicillin-G (46%), exhibited relatively lower levels of resistance compared to other antimicrobials tested in this study.
Dogs are commonly colonized by antimicrobial drug resistant enterococci and thus act as a reservoir for resistant enterococci for humans. Therefore, continued and increasing use of antimicrobials in dogs increases the risk of commensal bacteria such as enterococci developing resistance and transferring the resistance to humans. Yet emphasis has focused on food animals as source of resistance transfer to humans, with little attention given to the role of pets or companion animals as a possible source of resistant organisms and/or genes that mediate resistance (25, 50, 51).
Enterococci Isolates
Out of a total of 102 enterococci isolated from the clinical samples from dogs, the largest proportion (38.4%) were from urogenital samples. According to Weese et al. (52) and Kajihara et al. (53) enterococci are common causative pathogens in complicated urinary tract infections. Since the samples are from dogs attending a referral hospital it is possible that many of these cases had been referred to the hospital with urinary tract infection. Unfortunately, the authors were not able to verify this due to lack of data documenting why the dogs were presented for veterinary care at the teaching hospital in question.
However, Jackson et al. (50) have noted that certain enteroccocal species tend to be predominantly isolated from the rectum as opposed to other organ systems. There are several explanations for the observed difference between our findings and those reported by Jackson et al. (50). The first being that, the present study was not designed to establish the prevalence of enterococci by origin, and secondly unlike the study by Jackson et al. (50), the present study considered enterococci as a genus and did not focus on specific species such as E. faecalis and E. faecium. Thirdly, it is known that both E. faecalis and E. faecium have a primary intestinal reservoir in vertebrate animals. The other possible explanations for the difference between our findings and those of Jackson et al. (50), is that the later did not collect samples from the urogenital organs, and furthermore, while we used clinical samples, the study by Jackson and colleagues used samples from healthy dogs.
Isolation of enterococci from several organ systems, is consistent with the observation that enterococci do not only inhabit several habitats, but also inhabit various parts of the body of dogs (50). Therefore, results reported here confirm that enterococci can infect several animal organ systems and cause several infections such as urinary tract infections, hepatobiliary sepsis, endocarditis, surgical wound infections, bacteraemia, and neonatal sepsis (25, 53, 54).
Antimicrobial Drug Resistance Among Enterococci Isolates
In the study by Ossiprandi and Zerbini (51), a very high level of resistance was observed in various Enterococci spp. against antimicrobials such as erythromycin, enrofloxacin, and tetracycline. Similarly in a study conducted in Japan (25), the authors also observed that the most active resistance among enterococci from dogs was against erythromycin (44.2%) and tetracycline (44.2%), while considerable resistance was observed against lincomycin (41.6%), kanamycin (32.2%), and gentamycin (32.2%). This is consistent with our findings that show that all isolates were resistant to at least one antimicrobial agent, with very high prevalence observed against antimicrobials such as lincomycin (93.1%), aminoglycosides-lincosamide (92.2%), oxytetracycline (56.9%), orbifloxacin (85.2%), and carbenicillin (66.7%). According to Kataoka et al. (25) such high prevalence of resistance should be expected because dogs treated with antimicrobials are commonly colonized with antimicrobial resistant enterococci.
Moreover, since the isolates used in this study were from a teaching hospital which, also doubles as a referral hospital, it is possible that most of the cases from which the samples were drawn, had already been exposed to antimicrobial treatment by the time they were presented at the hospital. Furthermore, such a high occurrence of resistance among enterococci should be expected given that enterococci relative to streptococci are known to be intrinsically resistant to most commonly used antimicrobials agents and that enterococci have genetic capability of acquiring, conserving and disseminating genetic traits including genetic resistance determinants among themselves (16, 55). According to Watson (54) there has been an increase in drug-resistant E. faecalis strains, and that many antibiotics are no longer effective against infections caused by these bacteria.
The high prevalence of resistance (94.1%) observed against lincosamide, was expected. This is because enterococci are known to be intrinsically resistant to drugs such as clindamycin that inhibit protein synthesis at the chain elongation step by interfering with the transpeptidation of the 50S ribosomal subunit. Enterococci are also able to exhibit native resistance to clinically achievable concentrations of aminoglycosides, a feature which precludes their use as single agents. Therefore, the very high levels of resistance against aminoglycoside (92.2%) observed in this study suggest that aminoglycosides may be of very limited therapeutic importance in clinical practice at the hospital under study (16).
Worth noting, was the observation of resistance albeit lower levels (in comparison to other drugs presented in this study) of resistance against drugs such as amoxicillin/clavulanic acid combination, ampicillin, tylosin, and penicillin G, that ordinarily constitute the empirical and definitive antimicrobial treatment for enterococci (16, 55). This relatively low resistance level against these antimicrobials is nonetheless to be expected because all enterococci organisms tend to display low-affinity penicillin-binding proteins, which leads to decreased susceptibility and hence the observed low levels of resistance against these drugs (16).
Resistance levels of 41% against ampicillin is concerning given that it is the drug of choice for empirical treatment of enteroccocal infections. According to Damborg et al. (23) ampicillin resistance is a marker for hospital—associated E. faecium. Considering this, it is possible that the isolates under study were predominantly E. faecium. The fact that the isolates were from cases treated at a referral hospital further gives credence to the suggestion that the isolates under study were predominantly E. faecium. The suggestion that most isolates recovered in this study could have been E. faecium, is also supported by Kristich et al. (16), who are of the view that ampicillin resistance tends to be rare in E. faecalis, but widespread among E. faecium. However, this could not be confirmed since the laboratory did not carry out speciation of the isolates.
Despite the observed 41% resistance against ampicillin, it cannot be precluded from the treatment of enteroccocal infections among patients presented at the teaching hospital under study (16). Which means that ampicillin can thus still be considered the drug of choice for the treatment of enterococci. However, this is only true if the organisms do not have other mechanisms for high level resistance.
Enterococci are known to be tolerant to the bactericidal activity of cell-wall active agents such as β-lactams, meaning that while enterococci can be inhibited by clinically achievable concentrations of the antibiotic, only concentrations far in excess of the inhibitory concentrations are able to kill the bacteria. For this reason, β-lactams are normally used in combination with aminoglycosides to enhance the synergistic bactericidal activity of—β-lactams (16, 55). Due to the inaccessibility of bacteria to the mammalian immune system because of its location within the cardiac vegetations, the synergism between β-lactams and aminoglycosides is important in the treatment of several conditions including endocarditis that require enhanced synergistic bactericidal activity (16). However, the high levels of resistance observed against aminoglycosides in this study, undermines the practice of taking advantage of the synergism between cell-wall active agents and aminoglycosides in the treatment of serious enterococci infections caused by enterococci.
According to Kristichet al. (16) and the CLSI (56), while trimethoprim-sulfamethoxazole, cephalosporins, aminoglycosides, and clindamycin, may appear to be active in vitro against enterococci, they have not proved to be successful in animal models. In view of this, even though only 28.7% of the isolates in this study were resistant to sulfamethoxazole-trimethoprim, this should not be interpreted to mean that this drug combination could be effective clinically. In fact, the CLSI recommends that enterococci isolates should not be reported as being susceptible even if found to have been sensitive to sulfamethoxazole-trimethoprim (46).
While high levels of resistance were consistently observed against drugs such as amikacin, cephalothin, kanamycin, lincomycin, aminoglycoside-lincosamide, and orbifloxacin for the duration of the study, the number of isolates that were resistant to other antimicrobials tended to fluctuate. This could be a reflection of antimicrobial prescription practices at the hospital. The authors postulate that clinicians could be using antimicrobials for a certain period, leading to a build up of resistance, and then withdraw the antimicrobial in question when resistance against the antimicrobial is picked frequently. During this time, the antimicrobial, which has been withdrawn, is replaced with antimicrobials from other classes with different activity spectrum or mechanisms of action. However, due to lack of data on treatment practices and patterns at the hospital, it is not possible to confirm this hypothesis.
Multidrug, Extensive, and Pan-Drug Resistance
The level of XDR (35.5%) and MDR (93.1%) observe in this study, could be explained by the fact that enterococci in addition to being intrinsically resistant and tolerant, are ordinarily capable of rapidly acquiring resistance to virtually any antimicrobial agent put into clinical use. For example, following the introduction of chloramphenicol, erythromycin, and tetracyclines, resistance to these drugs quickly emerged to the extent that prevalences of resistance attained against these drugs precluded their empirical use (16).
All XDR isolates exhibited resistance against ampicillin or other related drugs such as amoxicillin, drugs of choice in the treatment of these infections. This was expected given that β-lactams are the usually the first line of defense in the treatment against enterococci. This being a referral hospital, it is highly likely that all the dogs included in this study had previously been treated with (unknown) antibiotics.
Factors That Predicted XDR Among Enterococci Isolates
Year was the only explanatory factor that was associated with XDR. Except for isolates obtained in 2007, isolates from other earlier years (2008-2010), had significantly lower odds of being resistant compare to isolates obtained in 2011. Which suggests that the level of resistance among enterococci isolates was more likely to be higher in 2011 as compared to earlier years. A similar trend was observed among Staphylococcus aureus isolates in a study by the same authors in which increasing resistance to certain antimicrobials over the same period (2007-2011) was observed. Furthermore, a similar trend (increase in the number of isolates that were resistant to certain antimicrobials) was observed in Staphylococcus pseudintermedius over the same period (57). The reason for the observed lower odds of resistance in earlier years compared to 2011 are not clear. However, the authors are of the view that this could reflect the changing patterns in the use of antimicrobials at the hospital with a higher selection pressure exerted in 2011 compared to previous years. In view of this, data is needed to investigate prescription practices and patterns at the teaching hospital in question, so that measures can be implemented to reverse the trend and safeguard the efficacy of antimicrobials.
Limitations of the Study
This being a retrospective study that used secondary data, only variables available in the database could be investigated. This therefore, limited the variables that could be investigated in the logistic models. Furthermore, since there was no species differentiation, it was not possible to investigate differences in resistance between the two main Enterococcus species commonly isolated from dogs; E. faecalis and faecium. Moreover, the laboratory does not routinely test for vancomycin and therefore resistance status of this glycopeptide (useful for treatment of drug resistant human enteroccocal infections) could not be assessed. The laboratory uses a standard panel of antimicrobials for all susceptibility tests and hence the panel included a couple of drugs against which enterococci exhibit intrinsic resistance, which has the potential to skew the results. Lastly, the relatively low sample size implies that study findings should be interpreted with caution. These limitations notwithstanding, the findings of this study are useful in resource poor settings where it is not possible to implement regular AST to guide selection of antimicrobials for use where first line antimicrobials have failed.
Conclusion
The high levels of AMR, MDR, and XDR observed in this study are of serious public health concern from a one-health perspective. Therefore, efforts to decrease the prevalence of resistance among these organisms with zoonotic potential and the ability to spread elements mediating resistance to other organisms are urgently needed. The identified resistance levels against the different antimicrobial groups may be useful in guiding clinicians in making treatment decisions especially in resource scarce settings where it is not always possible to base treatment on AST.
Data Availability Statement
The datasets presented in this article are not readily available because they belong to a third party (the Bacteriology Laboratory of the Veterinary Academic Hospital, University of Pretoria). The authors do not have legal rights to share the data. Requests to access the datasets should be directed to Prof. Vinny Naidoo at dmlubnkubmFpZG9vQHVwLmFjLnph.
Author Contributions
JO was involved in study design, data analysis, interpretation of results, writing of manuscript as well as extensive editing of the manuscript. DQ was involved in study design and data management and statistical analyses and interpretation as well as reviewing of the manuscript draft. AO was involved in study design, data analysis and interpretation as well as editing of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to extend their appreciation to the Department of Tropical Diseases and Companion Animal Clinical Studies for providing access to the records used in this study. The authors also wish to extend their gratitude to both the Department of Research, Innovation and Commercialization, University of South Africa, and Carnegie African Diaspora Fellowship Program (CADFP) that jointly sponsored Dr. Agricola Odoi's trip to South Africa, which enabled him to participate in this project. We also gratefully acknowledge the reviewers for their constructive comments that helped improve the article.
Abbreviations
AKC, American Kennel Club; AMR, Antimicrobial resistance; CADFP, Carnegie African Diaspora Fellowship Program; CI, Confidence Interval; CLSI, Clinical and Laboratory Standards Institute; MDR, Multidrug resistance; PDR, Pan-drug resistance; UK, United Kingdom; US, United States; USA, United States of America; XDR, Extensive drug resistance.
References
1. Moraes PM, Perin LM, Todorov SD, Silva A, Franco BDGM, Nero LA. Bacteriocinogenic and virulence potential of Enterococcus isolates obtained from raw milk and cheese. J Appl Microbiol. (2012) 113:318–28. doi: 10.1111/j.1365-2672.2012.05341.x
2. Ali SA, Hasan KA, Bin Asif H, Abbasi A. Environmental enterococci: I prevalence of virulence, antibiotic resistance and species distribution in poultry and its related environment in Karachi, Pakistan. Lett Appl Microbiol. (2014) 58:423–32. doi: 10.1111/lam.12208
3. Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston, MA: Massachusetts Eye and Ear Infirmary (2014).
4. Ferede ZT, Tullu KD, Derese SG, Yeshanew AG. Prevalence and antimicrobial susceptibility pattern of Enterococcus species isolated from different clinical samples at black lion specialized teaching hospital, Addis Ababa, Ethiopia. BMC Res Notes. (2018) 11:793. doi: 10.1186/s13104-018-3898-0
5. Anbumani N, Menon T, Kalyani J, Mallika M. Isolation, distribution and prevalence of various species of enterococci isolated from clinical specimens in a tertiary care hospital. Indian J Pathol Microbiol. (2005) 48:534–7.
6. Bell JA, Kopper JJ, Turnbull JA, Barbu NI, Murphy AJ, Mansfield LS. Ecological characterization of the colonic microbiota of normal and diarrheic dogs. Interdiscip Perspect Infect Dis. (2008) 2008:149694. doi: 10.1155/2008/149694
7. Coburn PS, Baghdayan AS, Dolan G, Shankar N. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Mol Microbiol. (2007) 63:530–44. doi: 10.1111/j.1365-2958.2006.05520.x
8. Leclercq R, Derlot E, Weber M, Duval J, Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. (1989) 33:10–5. doi: 10.1128/aac.33.1.10
9. Niederhausern S, Sabia C, Messi P, Guerrieri E, Manicardi G, Bondi M. Glycopeptide-resistance transferability from vancomycin-resistant enterococci of human and animal source to Listeria spp. Lett Appl Microbiol. (2004) 39:483–89. doi: 10.1111/j.1472-765X.2004.01598.x
10. Noble W. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. (1992) 93:195–8. doi: 10.1016/0378-1097(92)90528-V
11. de Niederhäusern S, Bondi M, Messi P, Iseppi R, Sabia C, Manicardi G, et al. Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr Microbiol. (2011) 62:1363–7. doi: 10.1007/s00284-011-9868-6
12. Ghosh A, Dowd SE, Zurek L. Dogs leaving the ICU carry a very large multi-drug resistant Enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS ONE. (2011) 6:e22451. doi: 10.1371/journal.pone.0022451
13. Faron ML, Ledeboer NA, Buchan BW. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J Clin Microbiol. (2016) 54:2436–47. doi: 10.1128/JCM.00211-16
14. Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence. (2012) 3:421–33. doi: 10.4161/viru.21282
15. Osman KM, Badr J, Orabi A, Elbehiry A, Saad A, Ibrahim MDS, et al. Poultry as a vector for emerging multidrug resistant Enterococcus spp.: first report of vancomycin (van) and the chloramphenicol–florfenicol (cat-fex-cfr) resistance genes from pigeon and duck faeces. Microb Pathog. (2019) 128:195–205. doi: 10.1016/j.micpath.2019.01.006
16. Kristich CJ, Rice LB, Arias CA. Enterococcal infection—treatment and antibiotic resistance. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston, MA: Massachusetts Eye and Ear Infirmary. (2014).
17. Marothi YA, Agnihotri H, Dubey D. Enterococcal resistance-an overview. Indian J Med Microbiol. (2005) 23:214–9.
18. Olawale KO, Fadiora SO, Taiwo SS. Prevalence of hospital-acquired enterococci infections in two primary-care hospitals in Osogbo, southwestern Nigeria. African J Infect Dis. (2011) 5:40–6. doi: 10.4314/ajid.v5i2.66513
19. van den Bogaard AE, Willems R, London N, Top J, Stobberingh EE. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J Antimicrob Chemother. (2002) 49:497–505. doi: 10.1093/jac/49.3.497
20. Oguttu JW, Picard J, Thompson PN. Antimicrobial drug resistance among enterococci from broilers and poultry abattoir workers. J Anim Vet Adv. (2012) 11:2267. doi: 10.3923/javaa.2012.2261-2267
21. Lochan H, Moodley C, Rip D, Bamford C, Hendricks M, Davidson A, et al. Emergence of vancomycin-resistant Enterococcus at a tertiary paediatric hospital in South Africa. S Afr Med J. (2016) 106:39–43. doi: 10.7196/SAMJ.2016.v106i6.10858
22. Mahabeer Y, Lowman W, Govind CN, Swe-Swe-Han K, Mlisana KP. First outbreak of vancomycin-resistant Enterococcus in a haematology unit in Durban, South Africa. South African J Infect Dis. (2016) 31:20–4. doi: 10.1080/23120053.2015.1118819
23. Damborg P, Top J, Hendrickx APAA, Dawson S, Willems RJLL, Guardabassi L. Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections?. Appl Environ Microbiol. (2009) 75:2360–5. doi: 10.1128/AEM.02035-08
24. Wurster JI, Saavedra JT, Gilmore MS. Impact of antibiotic use on the evolution of Enterococcus faecium. J Infect Dis. (2016) 213:1862–5. doi: 10.1093/infdis/jiv598
25. Kataoka Y, Umino Y, Ochi H, Harada K, Sawada T. Antimicrobial susceptibility of enterococcal species isolated from antibiotic-treated dogs and cats. J Vet Med Sci. (2014) 76:1399–402. doi: 10.1292/jvms.13-0576
26. Hammerum AM. Enterococci of animal origin and their significance for public health. Clin Microbiol Infect. (2012) 18:619–25. doi: 10.1111/j.1469-0691.2012.03829.x
27. Santagati M, Campanile F, Stefani S. Genomic diversification of enterococci in hosts: the role of the mobilome. Front Microbiol. (2012) 3:95. doi: 10.3389/fmicb.2012.00095
28. Guzman Prieto AM, van Schaik W, Rogers MRC, Coque TM, Baquero F, Corander J, et al. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front Microbiol. (2016) 7:788. doi: 10.3389/fmicb.2016.00788
29. Huycke M. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. (1998) 4:239–49. doi: 10.3201/eid0402.980211
30. Khan HA, Ahmad A, Mehboob R. Nosocomial infections and their control strategies. Asian Pac J Trop Biomed. (2015) 5:509–14. doi: 10.1016/J.APJTB.2015.05.001
31. Agudelo Higuita NI, Huycke MM. Enterococcal disease, epidemiology, and implications for treatment. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston, MA: Massachusetts Eye and Ear Infirmary. (2014).
32. Lewis CM, Zervos MJ. Clinical manifestations of enterococcal infection. Eur J Clin Microbiol Infect Dis. (1990) 9:111–7.
33. Department of Statistics South Africa. Statistical Release P0302 2000. (2019). Available online at: www.statssa.gov.za,aW5mb0BzdGF0c3NhLmdvdi56YQ==,Tel+27123108911 (accessed April 15, 2020).
34. McCrindle CME, Gallant J, Cornelius ST, Schoeman HS. Changing roles of dogs in urban African society: a South African perspective. Anthrozoos. (1999) 12:157–61. doi: 10.2752/089279399787000228
35. Roberto AFCI. South Africa is dog Country. Washint Post. (2014) Available online at: https://www.iol.co.za/lifestyle/family/pets/south-africa-is-dog-country-1727561 (accessed September 19, 2020).
36. Oosthuizen J, Oguttu JW, Etsebeth C, Gouws WF, Fasina FO. Risk factors associated with the occurrence of Brucella canis seropositivity in dogs within selected provinces of South Africa. J S Afr Vet Assoc. (2019) 90:1019–128. doi: 10.4102/jsava.v90i0.1956
37. City of Johannesburg Municipality Metropolitan. By-Laws Relating to Dogs and Cats for Promulgation City of Johannesburg Metropolitan Municipality By-Laws Relating to Dogs and Cats (Published under Notice No 1334 In Gauteng Provincial Gazette No 135 Dated 10 April 2006) (2006).
38. World Weather Online. Weather Averages | Monthly Average High and Low Temperature | Average Precipitation and Rainfall Days | Web Page (2019)
39. Quinn PJ, Carter ME, Markey B, Carter GR. Clinical Veterinary Microbiology. Edinburgh: Mosby Wolfe (1994).
40. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard – Fourth Edition. CLSI document VET01-A (2013).
41. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Seventeenth Informational Supplement. CLSI Document M100-S17. Clin Lab Stand Inst (2007).
42. Name P, Date R. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute (2008).
43. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptebility Testing: Nineteenth Informational Supplement M100-S19. Clinical and Laboratory Standards Institute (2009).
44. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement M100-S20. Clinical and Laboratory Standards Institute (2010).
45. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute. Twenty-first informational supplement. Approved Sta (2011).
46. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptebility Testing: Twenty-second Informational Supplement M100-S22. Clinical and Laboratory Standards Institute (2012).
47. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
49. Chen T, Legendre AM, Bass C, Mays SE, Odoi A. A case-control study of sporadic canine blastomycosis in Tennessee, USA. Med Mycol. (2008) 46:843–52. doi: 10.1080/13693780802140915
50. Jackson CRR, Fedorka-Cray PJJ, Davis JAA, Barrett JBB, Frye JGG. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J Appl Microbiol. (2009) 107:1269–78. doi: 10.1111/j.1365-2672.2009.04310.x
51. Ossiprandi MC, Zerbini L. Antimicrobial susceptibility of enterococcal species isolated from Italian dogs. In: Ossiprandi MC, editor. Antimicrobial Resistance - An Open Challenge. IntechOpen (2015). Available online at: https://www.intechopen.com/books/antimicrobial-resistance-an-open-challenge/antimicrobial-susceptibility-of-enterococcal-species-isolated-from-italian-dogs
52. Weese JS, Blondeau JM, Boothe D, Breitschwerdt EB, Guardabassi L, Hillier A, et al. Antimicrobial use guidelines for treatment of urinary tract disease in dogs and cats: antimicrobial guidelines working group of the international society for companion animal infectious diseases. Vet Med Int. (2011) 2011:263768. doi: 10.4061/2011/263768
53. Kajihara T, Nakamura S, Iwanaga N, Oshima K, Takazono T, Miyazaki T, et al. Clinical characteristics and risk factors of enterococcal infections in Nagasaki, Japan: a retrospective study. BMC Infect Dis. (2015) 15:426. doi: 10.1186/s12879-015-1175-6
54. Watson S. Enterococcus Faecalis: Causes, Symptoms, and Treatments. Heal Media (2017). Available online at: https://www.healthline.com/health/Enterococcus-faecalis (accessed December 29, 2019).
55. Werner G, Coque TM, Franz CMAP, Grohmann E, Hegstad K, Jensen L, et al. Antibiotic resistant enterococci—Tales of a drug resistance gene trafficker. Int J Med Microbiol. (2013) 303:360–79. doi: 10.1016/j.ijmm.2013.03.001
56. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement This Document Provides Updated Tables for the Clinical and Laboratory Standards Institute Antimicrobial Susceptibility Testing Standards M02-A11 and M07. (2012). Available online at: www.clsi.org (accessed June 27, 2019).
Keywords: Enterococcus species, enterococci, antimicrobial resistance, canine, dogs, multi-drug resistance, pan-drug resistance, extensive-drug resistance
Citation: Oguttu JW, Qekwana DN and Odoi A (2021) Prevalence and Predictors of Antimicrobial Resistance Among Enterococcus spp. From Dogs Presented at a Veterinary Teaching Hospital, South Africa. Front. Vet. Sci. 7:589439. doi: 10.3389/fvets.2020.589439
Received: 30 July 2020; Accepted: 07 December 2020;
Published: 07 January 2021.
Edited by:
Roswitha Merle, Freie Universität Berlin, GermanyReviewed by:
Clair L. Firth, University of Veterinary Medicine Vienna, AustriaSonja Hartnack, University of Zurich, Switzerland
Copyright © 2021 Oguttu, Qekwana and Odoi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James Wabwire Oguttu, am9ndXR0dUB1bmlzYS5hYy56YQ==
 James Wabwire Oguttu
James Wabwire Oguttu Daniel Nenene Qekwana
Daniel Nenene Qekwana Agricola Odoi
Agricola Odoi