- 1Animal Diagnostic Laboratory, Pennsylvania State University, University Park, PA, United States
- 2Center for Infectious Disease Dynamics, Pennsylvania State University, University Park, PA, United States
- 3Pennsylvania Animal Diagnostic Laboratory, New Bolton Center, University of Pennsylvania, Philadelphia, PA, United States
- 4Pennsylvania Veterinary Laboratory, Harrisburg, PA, United States
Avibacterium paragallinarum (historically called Hemophilus paragallinarum) causes infectious coryza (IC), which is an acute respiratory disease of chickens. Recently, outbreaks of IC have been reported in Pennsylvania (PA) in broilers, layer pullets, and laying hens, causing significant respiratory disease and production losses. A tentative diagnosis of IC can be made based on history, clinical signs, and characteristic gross lesions. However, isolation and identification of the organism are required for a definitive diagnosis. Major challenges with the bacteriological diagnosis of A. paragallinarum include that the organism is difficult to isolate, slow-growing, and can only be successfully isolated during the acute stage of infection and secondary bacterial infections are also common. As there were very limited whole genomes of A. paragallinarum in the public databases, we carried out whole-genome sequencing (WGS) of PA isolates and based on the WGS data analysis; we designed a novel probe-based PCR assay targeting a highly conserved sequence in the recN, the DNA repair protein gene of A. paragallinarum. The assay includes an internal control, with a limit of detection (LOD) of 3.93 genomic copies. The PCR efficiency ranged between 90 and 97%, and diagnostic sensitivity of 98.5% compared with conventional gel-based PCR. The test was highly specific, and no cross-reactivity was observed with other species of Avibacterium and a range of other common poultry respiratory viral and bacterial pathogens. Real-time PCR testing on 419 clinical samples from suspected flocks yielded 94 positives and 365 negatives in agreement with diagnostic bacterial culture-based detection. We also compared the recN PCR assay with a previous HPG-2 based real-time PCR assay which showed a PCR efficiency of 79%.
Introduction
Infectious coryza (IC) is an acute upper respiratory disease of growing broilers and layers, caused by Avibacterium paragallinarum, a gram-negative bacterium previously called Haemophilus paragallinarum (1, 2). The illness is associated with reduced egg production in layers and decreased bodyweight due to impaired food and water consumption in broilers (1). The most common clinical signs in chickens infected with A. paragallinarum include facial edema, nasal exudates, sneezing, and conjunctivitis (3). Previously, Page classified the bacterium with a slide agglutination test into serovars A, B, and C (4) whereas the modified Kume scheme based on hemagglutination test and its modifications describe 9 serovars within the species (5–7).
A tentative diagnosis of IC is often made based on history, clinical signs, and characteristic gross lesions. Isolation and identification of the organism are mostly hindered by its fastidious growth characteristics as well as the concurrent colonization of other bacteria in the same respiratory niches. Furthermore, the chronic stage of infection, prior antimicrobial treatments and any delay in sample processing have been shown to interfere with effective recovery of A. paragallinarum from diagnostic samples using conventional bacteriological methods (1). Despite the worldwide distribution of A. paragallinarum and a major cause of significant economic losses to the poultry industry; the true prevalence, incidence, and overall disease dynamics of IC in poultry flocks is not well-understood. The lack of rapid and sensitive diagnostic tools is one of the reasons for the limited understanding of the ecology and epidemiology of IC.
Recently, IC outbreaks have been reported from multiple states in the US and IC is now considered as an emerging respiratory disease of chickens in the North Eastern US. Since early 2019, several outbreaks of IC have been reported in broilers, layer pullets and laying hens, in Pennsylvania (PA) causing high morbidity with significant respiratory illness and significant production losses (8). IC continues to be reported from adjacent states like Delaware and Maryland and remains a major poultry health concern. This necessitates the development of advanced real-time PCR assays employing amplicon-specific probes, which are highly sensitive and precise for the rapid and accurate detection of pathogens from clinical samples.
Materials and Methods
Real-Time PCR Assay Design
Based on the comparative genome analysis of different PA isolates of A. paragallinarum that we generated using Illumina MiniSeq platform (Genbank accession #CP051642, CP051641, CP051640, CP051639, CP051638, CP051637, and CP051636) and several other sequences in GenBank, we determined that the DNA repair protein gene recN was a highly conserved gene. Previous studies also confirmed that recN gene is present in all serovars of A. paragallinarum and was considered as a potential housekeeping gene for gene expression studies (9). We designed specific primers and probe targeting a 99bp region of the recN gene of A. paragallinarum using Primer Express 3.0.1 software (Applied Biosystems, Foster City, CA).
Synthetic Gene Fragments to Establish Limit of Detection
Two hundred and forty four base pair (bp) synthetic double-stranded DNA fragment for recN gene amplicon (gBlock Gene Fragment- Integrated DNA Technologies, Coralville, IA) was used as a positive standard to determine the limit of detection (LOD) for the assay. The number of genomic copies per μL was determined using the molecular weight of the gBlock as per the manufacturer's instructions. Ten-fold serial dilutions of the standard were made starting from 103 to generate a standard curve using three technical replicates for each dilution. The synthetic DNA fragments are used to avoid the problem of copy number variation in different strains and a standardized dilution series could be thus used instead of the reference strain standards.
Bacteria and Viruses Used for Assessment of Cross-Reactivity
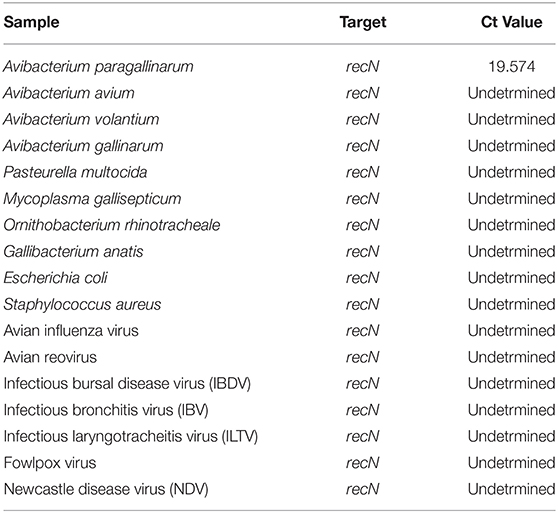
Reference strain of A. paragallinarum (ATCC 29545) along with 37 clinical isolates from commercial flocks of PA, cultured and identified as A. paragallinarum in the laboratory were used to evaluate the diagnostic sensitivity of the developed assay. The clinical A. paragallinarum isolates were identified using biochemical reactions, growth conditions, matrix- assisted laser desorption/ionization time-of-flight identification system (MALDI-TOF MS, Bruker Daltonics, Bremen, Germany) and a positive result in a conventional PCR specific for A. paragallinarum developed by Chen et al. (10). Analytical specificity was tested using various Avibacterium sp. isolates including A. avium (ATCC 29546), A. gallinarum (ATCC 13360), A. gallinarum (ATCC 13361), A. volantium (ATCC 14385), and common respiratory pathogens including infectious bursal disease virus (IBDV D-78 strain), infectious bronchitis virus (IBV- Georgia 2008 type strain), infectious laryngotracheitis virus (ILTV—Lt-IVAX vaccine strain), fowlpox virus, Newcastle disease virus (NDV- Lasota strain), avian influenza virus (AIV), avian reovirus (S-1133 strain), Mycoplasma gallisepticum (6/85 strain), and Pasteurella multocida (M-9 strain). All bacterial and virus isolates were obtained from either the American Type Culture Collection (ATCC) or from the National Veterinary Services Laboratory (NVSL), Ames, Iowa, USA.
Culture Conditions and Genomic DNA Preparation
Avibacterium sp. were cultured on chocolate agar (CHOC; Remel, ThermoFisher Scientific, Waltham, MA) for 48 h at 37°C with 5–7% CO2. DNA was extracted from plates with purified colonies using MagMAX Pathogen RNA/ DNA Kit (Applied Biosystems).
Conventional PCR
Conventional PCR was performed on 2 μL of extracted DNA to amplify a 500 bp region as previously described (10). Briefly, 3 μL each of forward (5′-CAA GGT ATC GAT CGT CTC TCT ACT −3′) and reverse (5′-TGA GGG TAG TCT TGC ACG CGA AT-3′) primers were used at 10 μM concentrations in a total reaction volume of 50 μL containing AmpliTaq Gold® DNA Polymerase (Applied Biosystems) and 25 mM magnesium chloride (Promega) in GeneAmp assay buffer II (Applied Biosystems). Reactions were carried out using a 9,600 GeneAmp PCR system (Perkin Elmer, Waltham, MA) with the following conditions: 95°C 4 min; 40 cycles of 94°C 1 min, 63°C 1 min, 72°C 30 s, and a cycle of 94°C 1 min, 63°C 1 min, 72°C 10 min. The reaction product (20 μL) was visualized on a 1.5% agarose gel containing 0.5 mg ethidium bromide per mL of agarose solution.
Real-Time PCR
Purified primers and probe were purchased from Integrated DNA Technologies (IDT). The PCR reaction was prepared in a volume of 25 μl consisting of 5 μL of the template DNA, 12.5 μL of master mix solution (VetMAX-Plus qPCR Master; Applied Biosystems), 1 μL of each 10 μM forward (recN FWD Primer 5′- GAACAAGACCCTTATCGCTTACAAG −3′) and reverse (recN REV Primer 5′- ACTCACTAATTCTTCCGCTTTTACATT −3′) primers, 0.3 μL of 10 μM fluorogenic probe [recN Probe 5′ [FAM] CAGGCACTGCAATTAGCCCGCAA [BHQ-1]-3′], 1 μL of Xeno IPC/VIC) and 4.2 μL of DNase and RNase-free water. The reactions were performed using a 7,500 Fast Real-Time PCR System (Applied Biosystems) with the following cycling conditions: an enzyme activation cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 45 sec. All samples were run in triplicates, and the experiment was repeated thrice using nuclease free water as no target control in all reactions. With an aim to ensure accurate PCR results and to reduce the likelihood of false negatives VetMAX Xeno IPC DNA (Applied Biosystems) in a concentration of 10,000 copies/μL was introduced at the nucleic acid isolation/preparation step and carried through the qPCR workflow as an internal control.
Analytical Sensitivity-Limit of Detection
Serial 10-fold dilutions of a gBlock gene fragment of recN gene of A. paragallinarum was PCR tested in triplicates, and cycle threshold (Ct) values for each were plotted to obtain standard curves, slopes, and R2 values. The gBlock was resuspended to 10 ng/ml stock solution with approximately 1.52 × 1012 copies/ml. The LOD for A. paragallinarum PCR assay was estimated from the values of the standard curve generated through the assay. The assay included three technical replicates.
Analytical Specificity of recN PCR Assay
Pathogens most frequently encountered in clinical samples from chickens with respiratory illnesses were tested on this real-time assay to ensure specificity. The genomic content extraction from the pathogens were performed using the MagMAX Pathogen RNA/DNA kit as described previously.
Evaluation of Diagnostic Sensitivity and Specificity
Identification of Clinical Isolates
Thirty-seven clinical isolates from PA commercial flocks cultured and identified as A. paragallinarum using bacterial culture in the Pennsylvania State University Animal Diagnostic Laboratory were used. DNA was extracted from plates with purified colonies using the extraction kit, MagMAX Pathogen RNA/ DNA kit (Applied Biosystems). Although the colony count was not standardized between extraction preparations, the initial DNA concentrations yielded comparable Ct values. The isolates were also tested using the conventional PCR described above.
Direct Detection in Clinical Samples
Clinical samples (n = 419) comprising swabs from sinus, choana, oropharynx, lung, air sac or trachea from chickens with suspected respiratory disease submitted to the Pennsylvania State University Animal Diagnostic Laboratory (ADL) were used for evaluation of diagnostic sensitivity. Swabs were vortexed and agitated well in saline and 300 μl of the broth was used to extract DNA using the MagMAX Pathogen RNA/ DNA kit (Applied Biosystems) following the manufacturer's instructions. The samples were also simultaneously cultured for bacterial isolation and identification for comparison.
Comparative Efficacy of the New recN qPCR vs. Previous HPG-2 qPCR
The efficiency of recN based PCR was compared to a previously validated assay targeting the HPG-2 (Haemophilus paragallinarum, the historical name for A. paragallinarum) region of the bacterium (11). DNA was extracted from broth cultures of A. paragallinarum (ATCC 29545) using the MagMAX Pathogen RNA/ DNA Kit. The DNA extract from each 10-fold dilution was PCR tested in triplicate, and cycle threshold (Ct) values were plotted to obtain standard curves, slopes, and R2 values.
As per the published protocol, HPG-2 PCR reaction was prepared in a volume of 25 μl consisting of 2 μL of the template DNA, 12.5 μL of master mix solution (VetMAX-Plus qPCR Master; Applied Biosystems), 1.76 μL of each 10 μM forward (HPG-2 FWD Primer 5′- GCAAAAGACTACCAGCAAGGATAAT −3′) and reverse (HPG-2 REV Primer 5′- CCTTACCCAAATATAATGTTCCACATT −3′) primers, 0.66 μL of 10 μM fluorogenic probe (Probe 5′ 6FAM-TCCTAGTTAG- CATTATTGC-MGBNFQ 3′), 1 μL of Xeno IPC/VIC) and 2.32 μL of DNase and RNase-free water. The reactions were carried out on Applied Biosystems 7,500 Fast Real time PCR machine with cycling parameters as: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, followed by 60°C for 1 min.
Statistical Analysis
Cohen's kappa was estimated to determine the agreement between the two tests. The level of statistical significance was set at 0.05. The sensitivity and specificity of the recN based PCR were calculated by comparing it with the conventional gel-based assay.
Results
Analytical Sensitivity and Specificity
The efficiency of recN based assay using genomic fragments of gBlock dilutions was between 90 and 97% with R2 0.99. The 3.93 copy numbers samples produced a mean Ct of 35.97, which resulted in a consistent LOD for the assay (Figure 1).
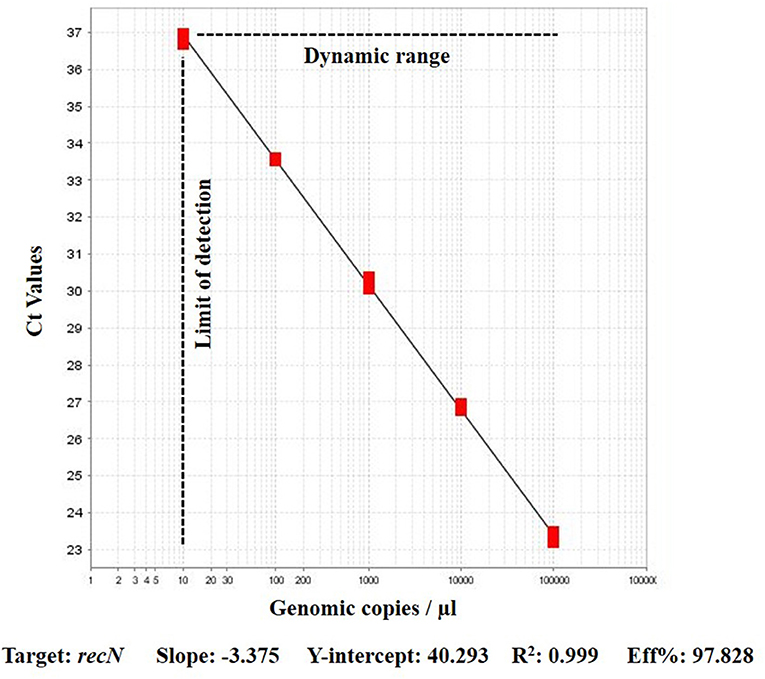
Figure 1. Performance of real-time PCR assay on serial dilutions of gBlock genomic fragments for A. paragallinarum recN target.
Using single-tube extraction for A. paragallinarum ATCC 29545 with Xeno DNA on serial dilutions, yielded positive detection on conventional endpoint gel-based PCR up to 10−4 dilutions. However, the recN based real-time PCR detected A. paragallinarum ATCC 29545 DNA in all dilutions up to 10−6 dilution. No cross reactivity was found with other tested respiratory pathogens (Ct value > 36; Table 1).
Diagnostic Sensitivity and Specificity
Overall agreement between the real time recN based PCR and conventional PCR testing performed at the laboratory using the A. paragallinarum isolates was 98.5%, including 36 of 37 real time PCR-positive and 30 of 30 negative cultures producing matching results. The Ct values for positive cultures ranged between 23 and 36. The internal positive control (XENO—IPC) from those isolated using the MagMAX kit amplified with an average Ct of 30.82.
Real-time PCR testing on 419 clinical samples from suspected flocks yielded 100% (94 positives and 365 negatives) agreement with diagnostic bacterial culture followed by identification by MALDI-TOF analyzer.
Comparison PCR
A strong agreement was found among the recN based assay results and the previously validated HPG-2 based PCR. The HPG-2 based PCR detected A. paragallinarum ATCC 29545 DNA in dilutions up to the 10−4 dilution. The efficiency of HPG-2 based assay evaluated from the standard curve generated using the dilutions was 79% with R2 0.996 (Figure 2). The positive amplification control included in the assay amplified with a Ct value of 16.614 in recN based assay whereas that using HPG-2 as the target had the amplification only at a Ct of 18.6415 (Table 2). A strong agreement was found between the outcomes of the new test and HPG-2 PCR (kappa = 0.96; p < 0.05).
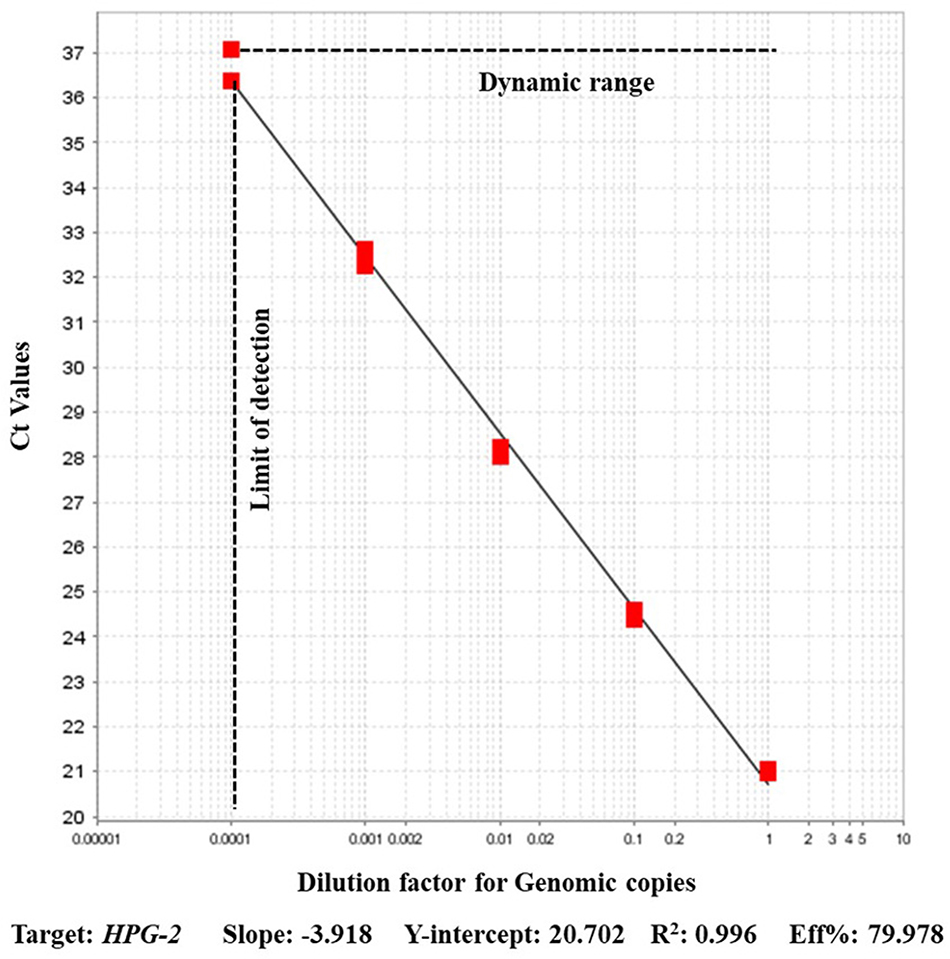
Figure 2. Performance of real-time PCR assay on serial dilutions of Avibacterium paragallinarum (ATCC 29545) for the detection of HPG-2 target.
Discussion
Infectious coryza continues to be a global threat to the poultry industry and has recently been recognized as an emerging infectious disease of poultry in the eastern US. A. paragallinarum bacterial respiratory disease of poultry, which requires robust methods for accurate diagnosis. Bacteriological diagnosis of A. paragallinarum is often challenging due to its fastidious nature. It is well-known that real-time PCR (rtPCR) assays employing amplicon-specific probes are highly sensitive and precise for the rapid and accurate detection of pathogens from clinical samples (12–14).
Previously, a gel-based PCR assay published in 1996 has been extensively used for the detection of A. paragallinarum from diagnostic samples (10). Based on this HPG gel based PCR assay, a 5′ Taq nuclease assay targeting a smaller region of the gene called HPG-2 of A. paragallinarum was developed in 2008 (15) and the workflow was recently validated in 2019 (11). However, these assays are designed based on the study published in 1996, when the whole genome sequence information of Avibacterium paragallinarum was not available. Furthermore, recently a lateral flow test has been developed for the rapid detection of A. paragallinarum (16). While this assay offered a rapid diagnosis, it suffered from lack of specificity and did not offer ability to distinguish the commensal species of Avibacterium such as A. avium, A. endocarditis, A. gallinarum, and A. volantium.
We generated whole genome sequences (WGS) of 18 A. paragallinarum isolates from recent outbreaks in Pennsylvania (January through April 2019). Based on the analysis of WGS of PA isolates and the ATCC reference strain, we developed a novel probe based real-time PCR assay. The recN gene in A. paragallinarum is a DNA-dependent repair protein (17). Previous reports on recN based phylogenetic comparison showed a very narrow species relationship between strains of A. volantium, A. avium, A. endocarditidis, and other Avibacterium species (17). A high recN gene divergence was also reported between the A. paragallinarum strains and groups of other isolates (17). To predict the whole-genome similarity, G-C content and phylogeny of selected taxa within the Pasteurellaceae, recN was also used as a candidate gene for multi-locus sequencing (18). Furthermore, our whole genome sequence analysis of PA isolates and other sequences in GenBank, identified recN as the highly conserved gene across all the three serovars of A. paragallinarum. Therefore, the newly developed recN PCR is an invaluable tool to diagnose low levels of A. paragallinarum rapidly and accurately from clinical samples. To date 419 clinical samples from suspected flocks were tested using the recN based PCR yielded 100% agreement with diagnostic bacterial culture-based detection.
In this study a strong agreement was found among the recN based assay results and the HPG-2 based PCR. Although, the previous study reported 89–111% efficiency with HPG-2 detection (11), we were only able to replicate the HPG-2 based assay with 79% efficiency. The new recN based qPCR assay has consistently been performed with a higher efficiency of 90–97%. PCR efficiency is a significant factor for the quantification of the target DNA in unknown samples (13). Consequently, PCR efficiency affects the detection and quantification limits of a PCR assay. Clinical samples for bacteriological diagnosis can have varying levels of the bacterial load depending on the type of sample, collection method, and stage of the disease. Sequence-verified, dsDNA gBlocks Gene Fragments, used to generate standard curve in this study are a great alternative to culture -based methods as it reduces the chances for pipetting error, and saves time diluting and plating multiple bacterial isolates.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
SK conceived and designed the study. MB and SK analyzed the WGS data and designed the PCR assay. MY, JW, RB, and RN carried out the PCR assay development and validation. PD, EW-P, SD, DK, and SL carried out the necropsies and collection of clinical samples. TP, TM, and DB carried out the bacterial isolation and identification. DT, BJ, SK, MS, and RN analyzed the data and interpretation. MS, MY, and SK drafted the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This study was funded by grant support from the Pennsylvania Soybean Board (OSP # 213542), 252 Pennsylvania Department of Agriculture (OSP#189021), and the United States Department of 253 Agriculture (OSP#197702).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the technical staff in the avian pathology sections of all the three labs in the Pennsylvania Animal Diagnostic Laboratory System (PADLS) for their assistance in sample collection for the test evaluation.
References
1. Blackall P. Infectious coryza: overview of the disease and new diagnostic options. Clin Microbiol Rev. (1999) 12:627–32. doi: 10.1128/CMR.12.4.627
2. Blackall PJ, Christensen H, Beckenham T, Blackall LL, Bisgaard M. Reclassification of Pasteurella gallinarum,[Haemophilus] paragallinarum, Pasteurella avium and Pasteurella volantium as Avibacterium gallinarum gen. nov, comb nov, Avibacterium paragallinarum comb nov, Avibacterium avium comb nov and Avibacterium volantium comb nov. Int J Syst Evol Microbiol. (2005) 55:353–62. doi: 10.1099/ijs.0.63357-0
3. Blackall PJ, Soriano-Vargas E. Infectious coryza and related bacterial infections. Dis Poult. (2020) 1:890–906. doi: 10.1002/9781119371199.ch20
4. La P. Haemophilus infections in chickens. I. Characteristics of 12 Haemophilus isolates recovered from diseased chickens. Am J Vet Res. (1962) 23:85–95.
5. Kume K, Sawata A, Nakai T, Matsumoto M. Serological classification of Haemophilus paragallinarum with a hemagglutinin system. J Clin Microbiol. (1983) 17:958–64. doi: 10.1128/JCM.17.6.958-964.1983
6. Blackall P, Eaves L, Aus G. Serotyping of Haemophilus paragallinarum by the Page scheme: comparison of the use of agglutination and hemagglutination-inhibition tests. Avian Dis. (1990) 643–5. doi: 10.2307/1591258
7. Blackall PJ, Eaves LE, Rogers DG. Proposal of a new serovar and altered nomenclature for Haemophilus paragallinarum in the Kume hemagglutinin scheme. J Clin Microbiol. (1990) 28:1185–7. doi: 10.1128/JCM.28.6.1185-1187.1990
8. Byukusenge M, Nissly RH, Li L, Pierre T, Mathews T, Wallner-Pendleton E, et al. Complete genome sequences of seven Avibacterium paragallinarum isolates from poultry farms in Pennsylvania, USA. Microbiol Resour Announc. (2020) 9:e00654–20. doi: 10.1128/MRA.00654-20
9. Wen S, Chen X, Xu F, Sun H. Validation of reference genes for real-time quantitative PCR (qPCR) analysis of Avibacterium paragallinarum. PLoS ONE. (2016) 11:e0167736. doi: 10.1371/journal.pone.0167736
10. Chen X, Miflin J, Zhang P, Blackall P. Development and application of DNA probes and PCR tests for Haemophilus paragallinarum. Avian Dis. (1996) 398–407. doi: 10.2307/1592238
11. Clothier KA, Stoute S, Torain A, Crossley B. Validation of a real-time PCR assay for high-throughput detection of Avibacterium paragallinarum in chicken respiratory sites. J Vet Diagn Invest. (2019) 31:714–8. doi: 10.1177/1040638719866484
12. Wittwer CT, Herrmann MG, Gundry CN, Elenitoba-Johnson KS. Real-time multiplex PCR assays. Methods. (2001) 25:430–42. doi: 10.1006/meth.2001.1265
13. Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol. (2017) 8:108. doi: 10.3389/fmicb.2017.00108
14. Hulley EN, Tharmalingam S, Zarnke A, Boreham DR. Development and validation of probe-based multiplex real-time PCR assays for the rapid and accurate detection of freshwater fish species. PLoS ONE. (2019) 14:e0210165. doi: 10.1371/journal.pone.0210165
15. Corney BG Diallo IS Wright L Hewitson G De Jong A Tolosa X . Rapid and sensitive detection of Avibacterium paragallinarum in the presence of other bacteria using a 5′ Taq nuclease assay: a new tool for diagnosing infectious coryza. Avian Pathol. (2008) 37:599–604. doi: 10.1080/03079450802449139
16. Morales Ruiz S, Bendezu J, Choque Guevara R, Montesinos R, Requena D, Choque Moreau L, et al. Development of a lateral flow test for the rapid detection of Avibacterium paragallinarum in chickens suspected of having infectious coryza. BMC Vet Res. (2018) 14:411. doi: 10.1186/s12917-018-1729-0
17. Bisgaard M, Nørskov-Lauritsen N, De Wit SJ, Hess C, Christensen H. Multilocus sequence phylogenetic analysis of Avibacterium. Microbiology. (2012) 158:993–1004. doi: 10.1099/mic.0.054429-0
Keywords: infectious coryza, respiratory disease, recN gene, A. paragallinarum, real-time PCR
Citation: Kuchipudi SV, Yon M, Surendran Nair M, Byukusenge M, Barry RM, Nissly RH, Williams J, Pierre T, Mathews T, Walner-Pendleton E, Dunn P, Barnhart D, Loughrey S, Davison S, Kelly DJ, Tewari D and Jayarao BM (2021) A Highly Sensitive and Specific Probe-Based Real-Time PCR for the Detection of Avibacterium paragallinarum in Clinical Samples From Poultry. Front. Vet. Sci. 8:609126. doi: 10.3389/fvets.2021.609126
Received: 22 September 2020; Accepted: 10 March 2021;
Published: 12 April 2021.
Edited by:
Armanda Bastos, University of Pretoria, South AfricaReviewed by:
Muhammad Zubair Shabbir, University of Veterinary and Animal Sciences, PakistanAnneke Feberwee, Royal GD, Netherlands
Marco Romito, Agricultural Research Council of South Africa (ARC-SA), South Africa
Copyright © 2021 Kuchipudi, Yon, Surendran Nair, Byukusenge, Barry, Nissly, Williams, Pierre, Mathews, Walner-Pendleton, Dunn, Barnhart, Loughrey, Davison, Kelly, Tewari and Jayarao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suresh V. Kuchipudi, c2t1Y2hpcHVkaUBwc3UuZWR1
 Suresh V. Kuchipudi
Suresh V. Kuchipudi Michele Yon
Michele Yon Meera Surendran Nair
Meera Surendran Nair Maurice Byukusenge
Maurice Byukusenge Rhiannon M. Barry1
Rhiannon M. Barry1 Ruth H. Nissly
Ruth H. Nissly Dona J. Kelly
Dona J. Kelly Deepanker Tewari
Deepanker Tewari Bhushan M. Jayarao
Bhushan M. Jayarao