- 1Department of Microbiology, College of Veterinary Medicine, King Faisal University, Al-Ahsa, Saudi Arabia
- 2Department of Biomedical Sciences, College of Veterinary Medicine, King Faisal University, Al-Hofuf, Saudi Arabia
- 3Department of Pharmacology, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafrelsheikh, Egypt
- 4Department of Virology, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafrelsheikh, Egypt
The bovine viral diarrhea virus (BVDV) consists of two species and various subspecies of closely related viruses of varying antigenicity, cytopathology, and virulence-induced pathogenesis. Despite the great ongoing efforts to control and prevent BVDV outbreaks and the emergence of new variants, outbreaks still reported throughout the world. In this review, we are focusing on the molecular biology of BVDV, its molecular pathogenesis, and the immune response of the host against the viral infection. Special attention was paid to discuss some immune evasion strategies adopted by the BVDV to hijack the host immune system to ensure the success of virus replication. Vaccination is one of the main strategies for prophylaxis and contributes to the control and eradication of many viral diseases including BVDV. We discussed the recent advances of various types of currently available classical and modern BVDV vaccines. However, with the emergence of new strains and variants of the virus, it is urgent to find some other novel targets for BVDV vaccines that may overcome the drawbacks of some of the currently used vaccines. Effective vaccination strategy mainly based on the preparation of vaccines from the homologous circulating strains. The BVDV-E2 protein plays important role in viral infection and pathogenesis. We mapped some important potential neutralizing epitopes among some BVDV genomes especially the E2 protein. These novel epitopes could be promising targets against the currently circulating strains of BVDV. More research is needed to further explore the actual roles of these epitopes as novel targets for the development of novel vaccines against BVDV. These potential vaccines may contribute to the global eradication campaign of the BVDV.
Introduction
BVDV is one of the most important viruses affecting bovine species throughout the world (1–4). It is a complex cocktail of two strains of the virus inducing various clinical syndromes of the affected animals (5). BVDV was first identified in the USA in a herd of cattle that suffered from acute gastroenteritis with high mortality rates (6). Since that time, BVDV represents one of the main viral pathogens of cattle in several regions across the globe (7–9). Some classical live attenuated and inactivated vaccines against BVDV are being in use for many decades for the immunization of the animals. Some of these vaccines usually trigger and induce the production of some levels of both humoral and cell-mediated immune responses (10). However, the emergence of new strains and variants hampered the efficacy of these conventional vaccines. Thus, the identification of some novel vaccines that match the new circulating strains in certain regions of the world would be of great impact on the contentious combating efforts for the BVDV. The eradication of the BVDV mainly depends on several factors including rigorous and continuous monitoring of the virus strains and variants, development of the most up-to-date diagnostic with high throughput ability to detect various strains and variants, and any potential new emergent strains or variants of the virus simultaneously as well as the development of homologous vaccines. We already mapped some novel neutralizing epitopes across the genome of BVDV, particularly the E2 gene. These epitopes could represent some unique targets for the production of novel vaccines against BVDV in the future. However, confirmation and validation of these potential epitopes require further studies.
Morphology, Structure, and Classification of BVDV
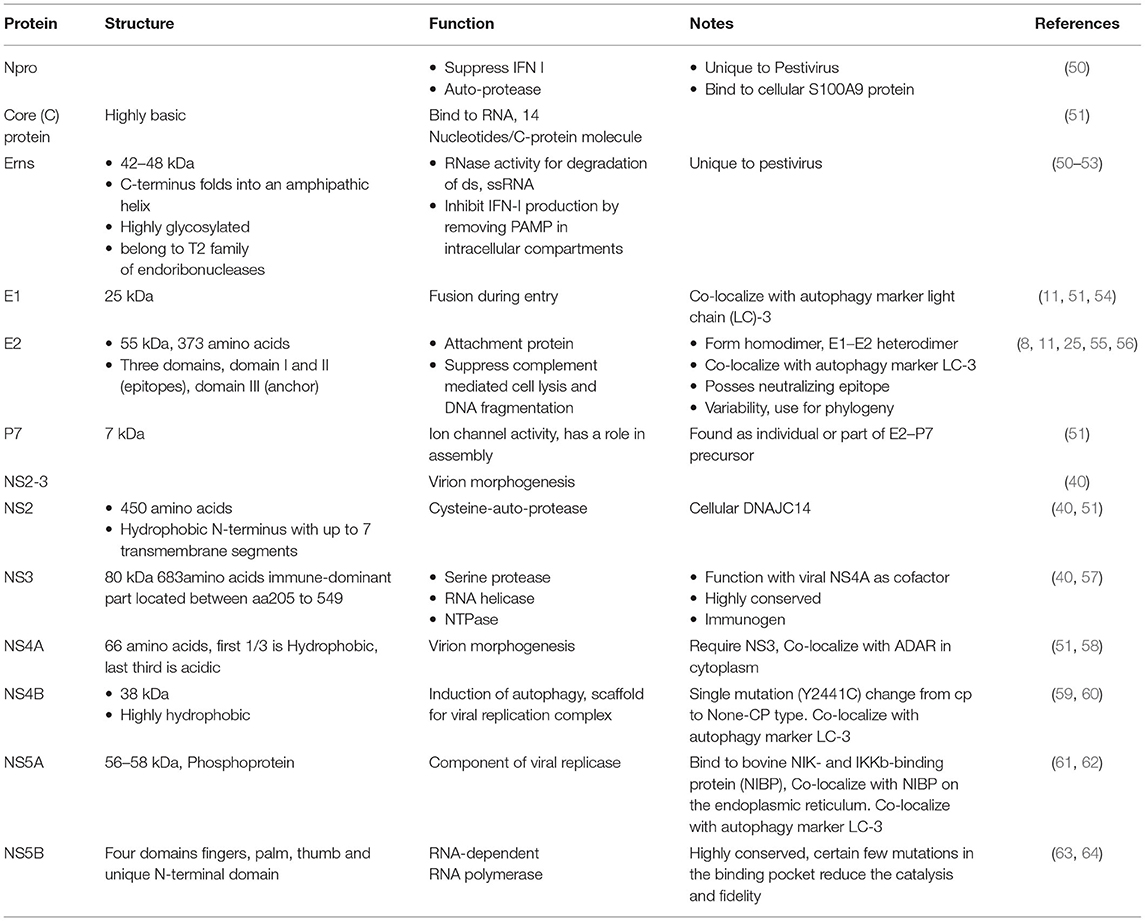
The BVDV particles are spherical to semi-spherical in shape (Figure 1). The virus particle consists of an outer bi-lipid layer envelope surrounding an electron-dense core as revealed by cryo-electron microscopy and negative staining electron microscopy (11, 12). There is some variation in the size of virus particles with a diameter of approximately 50 nm (range between 40 and 60 nm) for the majority of virus particles, but about 2% of the particles show a diameter of ~65 nm (11, 12).
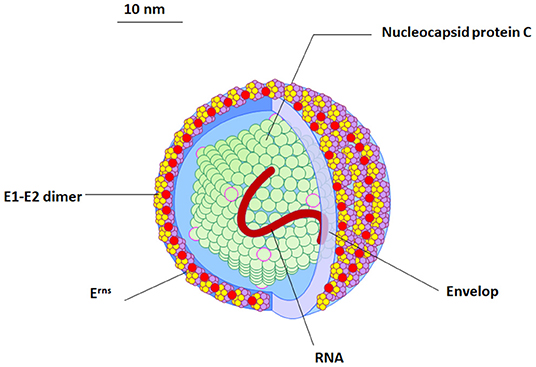
Figure 1. The morphology and structure of BVDV virus. An illustration of the BVDV particle showing the morphology and structure of the virus. Four structural proteins (Protein C, Erns, E1, and E2) are enclosing RNA (red). The outer viral protein coat contains important E1–E2 heterodimers which is required for the virus entry.
The BVDV belongs to the genus Pestivirus in the family Flaviviridae, which includes other viruses affecting sheep and swine species (border disease virus and classical swine fever virus, respectively) (13). Both genotyping and serotyping are effective tools for variation mapping and classification of BVDV (14). Two genotypes of BVDV were identified based on their cytopathology and cell culture growing abilities in cell culture (BVDV-1 and BVDV-2). Meanwhile, there are two known biotypes within each genotype of the virus. The major effect of the BVDV infection in cattle is the reproductive, respiratory, and immunosuppression while, viral infection is rarely causing diarrhea and digestive tract problems (15).
Consequently, sequence relatedness became an essential parameter of species assignment (14). The HoBi-like virus was recently identified in Europe from the fetal bovine sera imported from Brazil (14). This virus was potentially classified as one of the atypical pestiviruses or belong to the BVD-3 serotypes (14).
Genome Structure and Organization of BVDV
The BVDV genome is ~12.3 kilo-bases (kb) in size and it consists of a single open reading frame (ORF) flanked by short 5′- and 3′-untranslated regions (UTRs) (16). The 5′-UTR of BVDV contains an internal ribosomal entry site (IRES) that functions to initiate the translation of a single polyprotein. The IRES is composed of 3 helices that contain two highly variable regions (17). The 3′-UTR contains conserved stem-loops instead of the poly-A tail and has sites for binding for several host cell microRNAs (16, 18). The ORF encodes a single large polyprotein that is post-translationally processed into four structural proteins and eight non-structural ones. The genome organization is as follows (NH2–Npro/C/Erns/E1/E2/p7/NS2/NS3/NS4A/NS4B/NS5A/NS5B–COOH) (Figure 2) (19). The BVDV ribosomal frameshifting is mainly due to the missing of one nucleotide in the Npro coding region, which occurs in the SD-1 strain of BVDV resulting in a reduction in viral RNA and viral protein in infected cells (20).
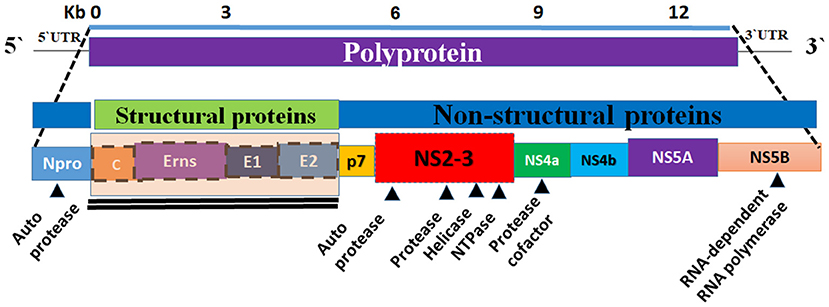
Figure 2. Graphical representation of the genome organization of BVDV. The BVDV genome is composed of a single strand of RNA (the bottom panel). The viral genome encodes a polyprotein (top panel). The central panel showing the composition of the viral genome (the structural and non-structural proteins). Eight non-structural proteins (Npro, p7, NS2, NS3, Ns4a, NS4b, and NS5b) and four structural proteins (C, Erns, E1, and E2) are encoded by polyprotein (middle and bottom panels). The non-structural proteins are encoding the viral proteases, helicase, NTPase, and RdRP.
Three regions are usually targeted for genotyping of Pestivirus the 5′UTR, which contains intra-species and inter-species conserved motifs, and the Npro region, which is a unique region to Pestiviruses (14). Those two regions out of them were frequently used for genotyping, especially 5′UTR (14, 21–23). However, the third region is the coding region of E2 protein also showed high variability and was frequently used for genotyping of Pestiviruses (8). Based on partial or complete genome sequencing, BVDV-1 has been classified into at least 22 subtypes (BVDV-1a to BVDV-1v) (24) while BVDV-2 and HoBi-like virus divided into 4 subtypes (a to d) (25).
Another method based on the secondary structure of the 5′UTR palindromic region has been adopted for typing of Pestivirus. A software for palindromic nucleotide substitution (PNS) typing of Pestivirus was developed and used to type 543 sequences into 9 species in the genus Pestivirus (26). Similarly, PNS typing of 281 strains of BVDV-1 showed that it segregated into 15 genotypes (BVDV-1a to−1o) with 4 common PANs in the variable loci, V1, V2, and V3, of the 5′UTR that characterize BVDV-1 (27). However, PNS typing of 536 Pestivirus strains showed that 32 strains, that were isolated from a small ruminant with a clinical picture of border disease, were assigned to BVDV-1, BVDV-2, CSFV, and tentative BDV-2 (28). Homologous recombination has been reported to naturally occurs in members of Pestivirus, including BVDV-1 and−2, emphasizing the need to build genotyping on the sequence of multiple regions (29, 30). The results of genotyping usually agree with serotyping (14). The phylogenetic analysis was based on the 5′UTR and the E2 sequences of 30 Argentinean isolates of the BVDV. About 76% of these isolates were belonging to the BVDV1b however, the BVDV (1a, 2a, and 2b) were also detected in this study (31). Species of the Pestivirus usually showed some degree of antigenic relatedness, and the titer of neutralizing antibody in sera from infected/vaccinated animals against viruses belonging to the same species are several-fold higher than the titer against viruses from other Pestivirus species (14). Based on the virus neutralization test, there is some antigenic variability within the HoBi-like virus, and higher antigenic variability between the HoBi-like virus and BVDV-2, and even higher antigenic variability with BVDV-1 (23, 32, 33). A serosurveillance study on Hobi virus was conducted in Argentina (34). This study reported the detection of antibodies in sera of 12 large animals. The same study reported no or very mild antibody titers of other BVDV strains (BVD1a, BVDV1b, and BVD2) (34).
On the other hand, serotyping is not always constant with speciation based on host origin and clinical picture. Two Pestivirus isolates from sheep and goats with signs of border disease showed genetic and antigenic characteristics suggestive of a new species closer to the classic swine fever virus (35). Similarly, strains isolated from beef cattle and genotyped as BVDV-2a have shown a high ability to react with both anti-BVDV-1 and anti-BVDV-2 (36).
As the virus neutralization test (VNT) solely was not always sufficient for serotyping. To improve the performance of this serotyping approach, some monoclonal antibodies were also for differentiation of various BVDV serotypes and considered as diagnostic markers (14, 37). The level of cross-reactivity varies according to the targeted protein was reported using various monoclonal antibodies (38). For instance, cross-reactivity between BVDV-1/-2 and HoBi-like virus using anti-Erns or anti-NS2/3 monoclonal antibodies was higher than that using an anti-E2 monoclonal antibody (33). Genotyping by sequencing some key regions in the BVDV genome such as the 5′UTR is one of the most useful tools for virus identification (39). One of the best examples is the study conducted in China and identified that BVDV1b and BVDV1c are the predominant subgenotypes in some cattle populations in this region (39). Additional typing method based on the cytopathic effects of the virus growing on the infected cell culture. Accordingly, Pestiviruses were classified into two biotypes, cytopathic (CP), and non-cytopathic (NCP) strains (40). NCP-BVDV was further divided according to the exaltation of Newcastle disease virus (END) in certain cell cultures, for e.g., RK13, into two types END+ and END− (41–43).
One of the limitations of antigenic characterization of BVDV is the antigenic diversity and cross-neutralization among isolates (44). A recent study employed a multivariate analysis for visualization of virus neutralization results to analyze the antigenic relationships between vaccine strains and some field isolates. Based on the demonstrated clustering patterns between the isolates the study concluded that BVDV-1 and BVDV-2 had the greatest antigenic differences (44).
Until recently, only four species were grouped in the genus Pestivirus including BVDV, CSFV, BDV, and unassigned Pestiviruses (26, 45, 46). Recently, Smith et al. proposed host-independent names for these species (Pestivirus A to D) and the addition of 7 new species (Pestivirus E to K) based on their sequence relatedness to Pestiviruses (Table 1) (13). According to ICVT, no change was introduced on the taxon of Pestivirus up to the time of writing this review (48). However, six viruses were reported from bat and rodents in China and proposed to form 6 new species based on their phylogenetic divergence (47). The sequence divergence of more than 25% with other Pestiviruses based on complete genome sequence was suggested as a criterion to assign separate species, as with the Brazilian strains of the HoBi-like virus that showed sequence similarity of 66.3 to 68.1% with representatives of Pestivirus species (49).
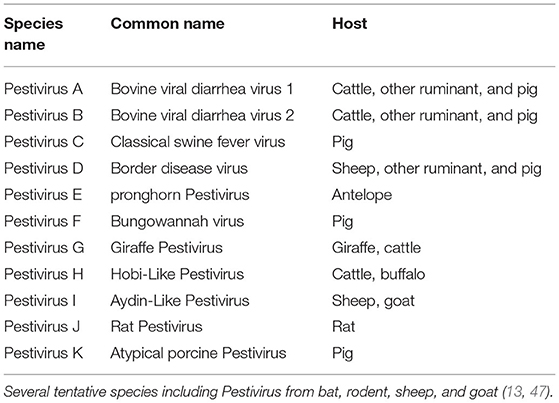
Table 1. Member of the genus Pestivirus, adapted from Smith et al. (13).
Processing and Maturation of the BVDV Polyproteins
BVDV encodes a single polyprotein that is post-translationally cleaved into four structural proteins (C, Erns, E1, and E2) (Figure 2) and 8 nonstructural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B). Table 2 summarizes the main aspects of BVDV proteins. The structural proteins can be classified as three envelope proteins (Erns, E1, and E2) and one capsid protein (nucleocapsid protein C). Among the structural proteins, C protein is the most abundant protein, followed by Erns, while E1 and E2 showed limited presence. However, on the surface of the virion, E2 is the most abundant surface protein and induces an immune response against BVDV, followed by Erns. All envelop proteins are produced as precursor protein Erns/E1E2, which passes two-step cleavage reactions to produce the free envelope proteins Erns, E1, and E2. Both E1 and E2 contain transmembrane domains, while Erns is only anchored to the membranes, which also allows for the secretion of Erns (65, 66). Compared with MDBK cells, lipids of BVDV contain more cholesterol, sphingomyelin, and hexosyl-ceramide and fewer glycerol-phospholipids with unknown mechanisms of lipid sorting. Cholesterol and sphingomyelin were shown to be important for BVDV entry (11).
The Erns protein is one of the unique proteins in the Pestiviruses' genome which can bind with nucleotide substrates and with evidence of ribonuclease activity (Figure 3). This protein was found to be highly similar in structure to T2 ribonucleases from plants and fungi (67). The majority of Erns protein is secreted into the infected cell culture media (68). Erns is believed to make heterodimer very early during the virus replication which suggests its roles in the virus attachment to the target cells (68).
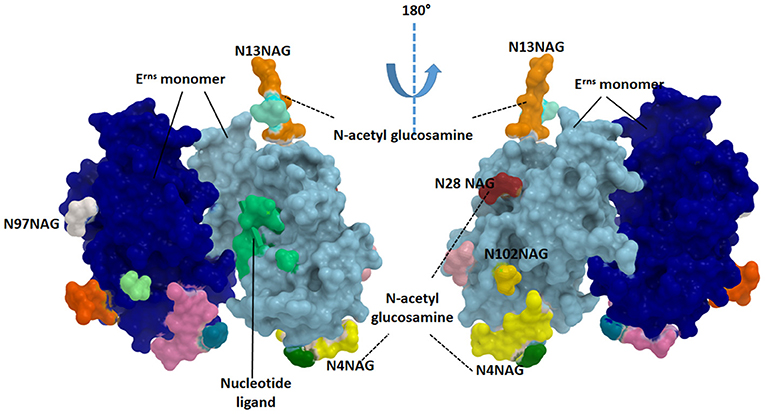
Figure 3. Structure of the catalytic domain of envelope Erns glycoprotein. Each monomer is showing in blue and cyan colors. The glycosylated sites are provided in green (N4NAG), orange (N13NAG), red (N28NAG), white (N97NAG), and yellow (N102NAG). The figure was derived from the structure deposited in the protein data bank (PDB ID 4dvn).
Erns released from the infected cells interfere with immune response by the degradation of the circulating nucleic acids (69). Erns shows RNase activity in the intracellular compartment, thus preventing IFN production by degrading RNA and removing the resistant pathogen-associated molecular pattern (PAMP), thus maintaining the appropriate milieu for persistent infection (52). As an RNAse protein, Erns is a glycosylated protein carrying several N-acetyl glucosamine molecules (Figure 3). Erns activity is not confined to bovine cells. Extracellularly added Erns was shown to be uptaken into bovine turbinate cells, probably by clathrin-dependent endocytosis, and to remain active for a long time after been engulfed. Degradation of viral RNA takes place in endosomal compartments before reaching cytosol (70). The Erns protein belongs to the T2 family of endoribonucleases that preferably cleave ssRNAs. However, monomeric Erns showed the ability to cleave dsRNA and RNA in DNA/RNA, methylated RNA/RNA hybrid (53).
The surface of the BVDV mainly consists of the E2–E2 homodimers and the E1–E2 heterodimers (11, 71). This heterodimer is the most important protein for virus fusion with the host cells (72). However, due to the absence of hydrophobic core and lacking fusogenic sequences, the E2 function in virus fusion was not proposed and the functionally uncharacterized E1 might carry out this role (73). The main function of the E2 glycoprotein (55 kDa) is the attachment to the host cell by forming the homodimers beside the mentioned heterodimerization with E1 (11). The E2 ectodomain consists of three domains with a total span of 140A with no known fusion motifs (Figure 4). Domain I and II are Ig-like domains with 90 and 78 amino acid residues, respectively. Domain III consists of 175 amino acids that form three β-sheet modules (IIIa-IIIc) (55). The c-terminus contain a single-span transmembrane anchor that retained the E2 glycoprotein in ER (74). Post-translationally processed E2 contains four glycans and eight disulfide linkages. A ninth disulfide link is used to form an end-to-end homodimer, a linkage that explains the need to low pH activation to initiate fusion (55).
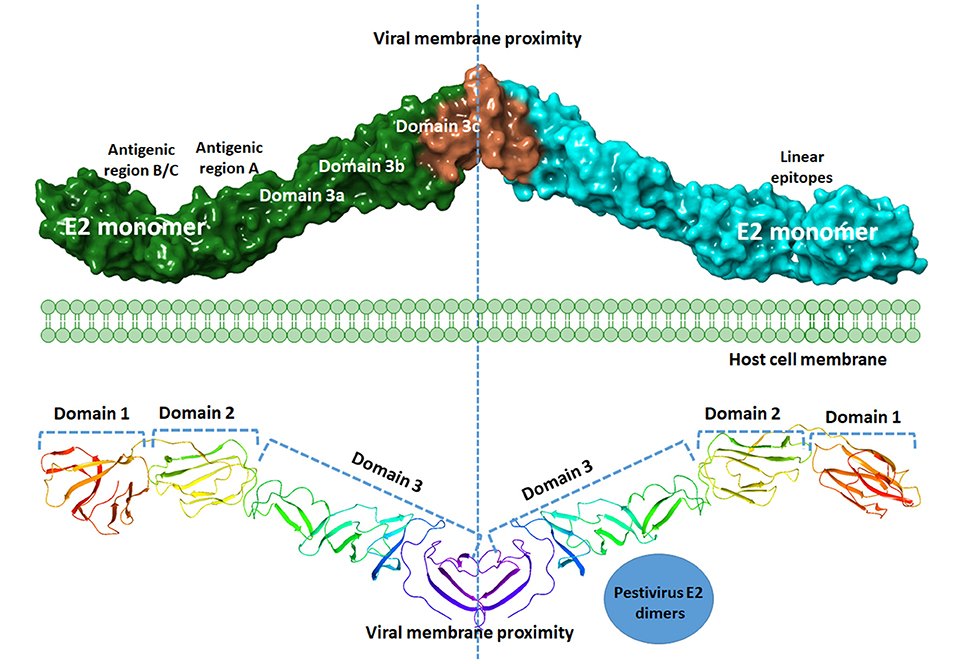
Figure 4. Graphical representation of BVDV E2 dimers. A model of the graphical representation of the E2 dimers. Cartoon (upper panel) or surface representation (lower panel) are provided. The domain contents of each monomer, antigenic sites, and the subdomains are highlighted. Each E2 monomer contains three domains, domain 1, 2, and 3. Domains can be divided into subdomains 3a, 3b, and 3c. Three regions of linear epitopes comprising antigenic regions A, B, and C. E2 glycoprotein is thought to share in the membrane fusion process during virus entry, yet the exact mechanism is to be investigated. The figure was derived from the structure deposited in the protein data bank (PDB ID 4jnt).
Some recombinant E2 based baculovirus vaccines were produced commercially and showed promising trends in bosting the immune-response against BVDV in combination with the inactivated BVDV in the goat model (75). Some monoclonal antibodies against E2 of the CSFV fulfilled the DIVA concept which is an important approach to differentiate between the infected and vaccinated animals (76).
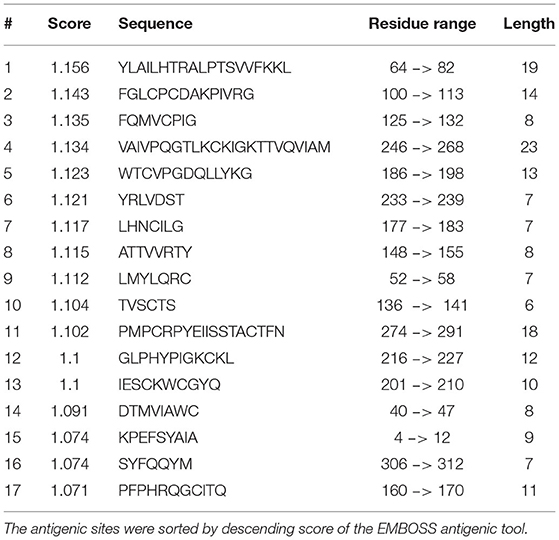
Mapping the antigenic epitopes within the E2 of BVDV-1 and -2 showed the presence of type-specific epitopes in a context of comparable antigenic structure (77). It consists of four epitopes, two in each of domain I and domain II (Figure 4). While, domain III showed no epitopes, with IIIc as the most conserved part of the E2. Most of the escaping mutants were mapped to one face of the E2, suggesting that this is the exposed face (55). For further investigation of BVDV E2 glycoprotein epitopes, the sequence of E2 protein was mapped by EMBOSS antigenic predictor tool (78). About 17 antigenic sites were predicted with six or more amino acids count (Table 3). Close inspection of the delivered sites of predicted intensity reveals the potential inclusion of domain III (Figure 5). Previous reports showed that domains I and II are the only parts of E2 that are exposed to the immune reaction (55). However, based on the EMBOSS antigenic prediction tool results, the potential finding of antigenic sites in E2 domain III gives new insights into the structure and function of E2 protein. It was predicted that only domains I and II were exposed to the immune reactions owing to their potential presence on the surface of the virus, a property that is lacking in the 3rd domain. By the presence of epitopes in domain III, the whole structure of E2 might be then exposed to the virus surface and might contribute to the immune response against the virus as well as other functions e.g., participation in membrane fusion and virus entry process.
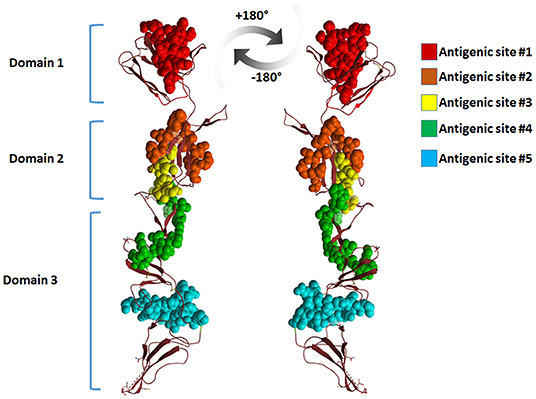
Figure 5. Cartoon representation of BVDV-E2 glycoprotein monomer showing the antigenic sites predicted by EMBOSS antigenic prediction tool. The top five antigenic sites were given in spheres with different colors. The figure was derived from the structure deposited in the protein data bank (PDB ID 4jnt).
Persistently infected animals (PI) usually developed during embryonic developments during the first 125 days of gestation (9). When these calves are born, they become a major source of infection and virus spread by secreting a large number of viruses in their body secretions and excretions (79).
Comparing the genome sequences of the BVDVs from severe acute infections and PI showed that the mutation rate was higher in viruses from PI (80). Similarly, an increase in mutation rate was reported after serial transmission in pregnant bovine, ovine (81), caprine (82), and swine (83). Certain amino acid substitutions appear after transmission from species to species (81–83). Mutation tends to concentrate in the coding regions of the structural proteins, especially the E2 protein (80–84). Several studies suggest the occurrence of natural recombination between BVDV genomes. It is usually associated with PI and concomitant immunotolerance, a situation in which mixed infection and consequent recombination may occur (30, 85). Similarly, recombination of BVDV genome with cellular RNAs in PI animals was incriminated as a mechanism that gives rise to mutant BVDV that induces the lethal mucosal disease (86). BVDV recombination was reported to occur even in the absence of a translation of viral proteins or even active viral RNA-dependent-RNA-polymerase (86, 87).
BVDV Replication Cycle and Pathogenesis
The outcomes of the BVDV infection in susceptible animals are mainly dependent on the breeding status of the animals. BVDV infection in pregnant cattle may result in several syndromes, including early embryonic death, teratogenic effects on the fetus, and the development of persistently infected animals (PI) (5). The PI plays an important role in the sustainability of the BVDV in the environment and the transmission of the virus from animal to animal as well as from one herd to another. The PI animal continues to shed the BVDV in its body secretions and excretions throughout its life, posing a major risk to other animals within the herd and in close contact herds as well (6, 19, 88). Another concern about the PI animals is they cannot be identified easily throughout the animal's life, especially immediately after birth due to the over masking effect of the antibodies from the colostrum, as described below in more details (5).
The susceptibility of cattle to the BVDV infections is mainly associated with some loci on cattle chromosome 2 (BTA2) and 26 (BTA26) (89). These regions showed their substantial contribution to the animal susceptibility to PI-BVDV infections (90). This genetic predisposition may relate to individual cattle rather than to a specific breed. This could be supported by the significant difference in the replication of BVDV in cell culture derived from various breeds and individuals animals within-herd (91).
Cross-species transmission is a highly important phenomenon that may lead to the establishment of a new reservoir for the virus which makes the control and eradication is too difficult in its natural reservoir. The transmission from PI cattle to goats was reported up to two generations of goats (80, 82, 83).
The CD46 molecule act as a BVDV receptor during virus entry. Some studies showed that the NADL-BVDV strain was able to spread from infected to susceptible cells via a CD46-independent mechanism (92). Despite the frequent association, BVDV-E2 binding to the CD46 receptor was not required for BVDV uptake, suggesting the involvement of other cellular proteins (93). Upon uptake into ovine cells (SFT-R), the NCP-BVDV enters the eclipse phase of replication that takes 8–9-h. The positive and the negative strands of the viral RNAs first appear at 4-h post-infection (hpi). In those ovine cells, the complete replication cycle takes 10–12-h, and infectious BVDV particles first appear intracellularly and extracellularly at 8 and 10 hpi, respectively. Virus titer and positive-strand RNA reach a peak at 16 hpi. NS3 and E2 proteins appear at 6 and 7 hpi, respectively, while NS2-3 gradually accumulated thereafter (94).
Replication of the NCP-BVDV is regulated via the mechanism of cleavage of NS2-3, which is performed by NS2 protease activity that requires DNAJC14, a limiting cellular cofactor. Cleavage of NS2-3 led to the generation of the NS3, an essential part of the replicate (40). During replication, the 5' UTR of BVDV stalls the 5'−3' exoribonuclease (XRN1) enzyme and repress its activity resulting in a significant increase in the half-life time of many normally short-lived cellular mRNAs (95).
The gene expression and genome protection against transposon and viruses are among many functions played by non-coding RNA, such as long ncRNA (lncRNA), short ncRNA (sncRNA), microRNA (miRNA), and transfer RNA halves (ts-RNA) (96, 97). Studies on the expression of lncRNA in BVDV infected MDBK cells revealed the enrichment of several pathways particularly related to immune response such as the T-cell receptor, TNF, Jak-STAT, apoptosis, Ras, NOD-like receptor, NF-κB, ErbB, and fatty acid biosynthesis (98, 99). Similarly, analysis of the expression of circular RNA (circ-RNAs) in BVDV infected MDBK cells suggest their involvement in the regulation of cell proliferation and apoptosis (100). An additional mechanism used by BVDV to indirectly control cellular transcriptome is the sequestration of small non-coding RNA, such as miR-17 and let-7, that is required for BVDV replication, as shown in SK-6 cells (18). The expression levels of other microRNAs, like bta-miR-423-5P and bta-miR-151-3p and some transfer RNA halves (ts-RNAs) in calves sera, differ significantly as a function of BVDV infection and time post-inoculation (97, 101). RNA interference by short interfering RNA (siRNA) that targets the 5'UTR and the envelop glycoproteins Erns, E1, and E2 coding regions induce a moderate reduction in viral titer, antigen, or RNA copy numbers in MDBK cells infected with BVDV-1 (102).
BVDV-Induced Immune Response and Immune Dysfunction
The persistent infection and dysregulated immune response are two major consequences of BVDV infections (103, 104). Protective immunity after natural infection with BVDVs is characterized by the activation of both virus-specific humoral and cellular immune responses (105). Due to their central role in guiding humoral and cell-mediated immune responses, CD4+ T helper cells, mainly targeting the NS3 and E2 proteins, are key players in the development of protective immunity against the virus (106). In opposite to the depletion of CD8+ T cells, depletion of CD4+ T helper cells was associated with higher blood viral load, prolonged viremia, and virus secretion via the nasal route (107). After B cell activation, neutralizing antibodies are detectable, starting on day 14 of the infection (105). The most effective neutralizing antibodies mainly target the surface protein E2, whereas antibodies specific to the Erns possess less neutralizing activity (108). While some antibodies target the E1, the main structural protein of BVDV (C) does not induce B cell activation and antibody production. Also, the non-structural protein NS2-3 induces a strong antibody response (108). Several studies investigated the impact of the virus biotype on the course of the immune response against BVDVs. Comparative analysis of the immune response to the NCP and CP biotypes of BVDV suggests a higher potential for NCP-BVDV to induce humoral immunity, while infection with CP-BVDV resulted in a better cell-mediated immunity (88, 109, 110).
Several studies have been recently conducted to identify the mechanisms behind BVDV-induced immune dysregulation (110–114). BVDV-induced immunosuppression has been identified in naturally infected animals, with both transient or persistent infection, as well as after experimental BVDV infection (103). Immunosuppressive effects of BVDV include changes in the immune cell composition, and altered immunophenotype of leukocytes, and several defects in immune cell function, resulting in increased disease severity of secondary infections with other pathogens. In experimentally infected cattle, BVDV infection was associated with significant changes in the bovine leukogram with reduced numbers of total leukocytes, neutrophils, lymphocytes, and platelets (115–117). The functions of both myeloid and lymphoid cells are affected by BVDV. Functional analysis of neutrophils of the PI animals revealed reduced phagocytosis capacity and decreased reactive oxygen (ROS) production, compared to neutrophils from healthy animals (118). The inhibitory effect of BVDV on neutrophils has been found strain-specific (119). Although all BVDV strains (including CP and NCP-BVDV) induced a significant decrease in the expression of the cell adhesion molecules CD18 and L-selectin on neutrophils and impaired their in vitro ROS and neutrophil extracellular traps (NET) activity, only CP-BVDV reduced the phagocytosis function. In contrast, only NCP-BVDV enhanced CD14 expression on neutrophils and improved their chemotactic activity (119). A recent study evaluated the impact of supernatant collected from macrophages infected with different BVDV strains on macrophage inflammatory response and lymphocyte apoptosis. The results of this study revealed a role for macrophages-secreted mediators in the immune dysfunction associated with highly virulent NCP-BVDV (62). An essential role of danger-associated molecular patterns (DAMPs) and danger-sensing protein complexes in the dysregulated immune response to BVDV infection has been recently reported (120). A recent experimental infection study of bovine macrophages indicated the ability of CP-BVDV-1 to activate the danger-sensing multi-protein complex, the inflammasome, in a caspase-1 dependent manner, resulting in IL-1β secretion with increased viral replication (120). The role of the DAMP S100 protein A9 (S100A9), which induces its effect via toll-like receptor (TLR)-4 / MyD88 signaling, in the BVDV-induced immunosuppression, has been recently reported (50). The results of this study revealed that in vitro BVDV replication was enhanced by inhibiting S100A9 protein expression in BVDV-infected cells using siRNA, while overexpression of S100A9 enhanced the virus-induced type-I IFN production. The strong interaction between S100A9 in infected cells and the Npro protein of BVDV suggests a role of this interaction in reducing the type-I IFN response by reducing the availability of S100A9 protein (50).
BVDV also showed inhibitory effects on several adaptive immune cell functions. Polyclonal mitogenic stimulation of bovine lymphocytes from infected animals induced a reduced proliferative response, as compared to cells from healthy animals (118, 121). A recent in vitro infection model reported the immunosuppressive capacity of BVDV on bovine peripheral blood mononuclear cells (PBMCs) (114). Bovine PBMCs from BVDV-naturally infected animals were more susceptible to BVDV infection with a higher apoptosis response, compared to cells from naive animals. In the same study, in vitro infection with NCP-BVDV failed to induce the expression of cell surface markers related to antigen presentation function (114). This biotype-specific immunosuppressive effect on the adaptive immune response has also been reported in a recent study that investigated the effect of infection with CP, or NCP-BVDV strains on levels of total serum IgG, IgG1, IgG2, BVDV neutralizing antibodies, and total white blood cell count (113). While the infection with CP-BVDV resulted in an early (d7 pi) decreased levels of neutralizing antibodies and leukocyte numbers, the infection with NCP-BVDV induced polarization of the immune response toward the Th1 response with the production of more antibodies of the IgG2 isotype (113).
Some earlier studies suggested a role for the higher suppressive effect of NCP-BVDV on interferon response, compared to CP-BVDV, in the establishment of persistent infection with NCP but not CP-BVDV (122). However, recent studies identified the induction of a significant IFN-alpha (IFN-α), IFN-β, and IFN-γ response in the PI fetuses after in utero infection with NCP-BVDV (123, 124). This is also supported by another study on naturally infected cattle, showing that IFN signaling is not completely inhibited in PI cattle (125). Similarly, the higher frequency of cells expressing MHC class I and II molecules in liver tissues from the PI fetuses, compared to control fetuses, indicates the induction of an immune response to NCP-BVDV infection (126). The reason behind the failure of the immune response to clear the virus and the mechanism through which BVDV-PI is established, therefore, still to be elucidated. A recent study identified different immune responses of fetal lymphoid organs to transient and persistent BVDV infections (127). The PI fetuses showed reduced expression of several genes involved in the innate immune response and antigen presentation to adaptive immune cells; transiently infected fetuses upregulated several innate immune response genes in their thymuses. Also, several adaptive immune response genes were downregulated in PI fetuses. The study suggested a role for the suppressed innate and adaptive immune responses in the developing lymphoid organs in the persistence of the BVDV in the PI animals and, on the other hand, a role for the upregulation of the innate immune response genes in transiently infected fetuses in virus clearance from these animals (127). Bovine embryonic cells showed the ability to take the function of immune cells by recognizing and responding to BVDV infection through the upregulation of genes encoding for INFα and TLR7, which are involved in inflammatory and immune responses (128).
BVDV Immune Evasion Strategies
Many viruses including BVDV use several unique immune evasion strategies to hijack the host immune response to ensure successful viral replication and spreading from one host to another. These strategies include the adaptation of several viral survival strategies, the “hit & run” approach, and viral persistence (122). These strategies favor the virus replication and spreading. On the other hand, these strategies reduce the values of the currently used diagnostic assays to identify the newly emerged strains of the virus. Furthermore, these evasion strategies will favor the emergence of new variants and strains of the virus, which may have negative impacts on the currently used BVDV vaccines in the protection of the animals at risk. The milestone of the BVDV infection among a certain population of animals is the establishment of innate immune tolerance in the pregnant animals to produce an immune tolerant and PI animal after birth (129).
Although the viral RNAs induce the IFN production and synthesis, the viral-Erns suppress the IFN production pathways triggered earlier by the viral RNAs (122, 130). The Npro induces degradation of the IRF3 (essential IFN activation factor) during the virus replication in cell culture (130). Both the Npro and the Erns act as an IFN antagonist in a non-redundant manner in the cell culture of the CP-BVDV and the NCP-BVDV strains (130). Although the NCP-BVDV strains efficiently cause evasion of the innate immune response in the affected animals (124), they can induce IFN-γ production during the acute phase of infection, especially in the PI animals (124). This indicates, that despite the marked inhibitory effects of BVDV infection on the IFN production in the affected host, it does not alter or suppress their actions (130). Another unique BVDV immune evasion strategy is the “self” and “non-self” alteration to the IFNα/β pathways (131). This selective phenomenon enables the BVDV to establish the persistence of infection in the affected animals, which maintains the circulation of the virus among a certain population of animals (131).
Recent Advances on BVDV Vaccination and Immunotherapeutic Strategies
Due to the significance of PI animals for spreading the infection, the target of vaccination strategies against BVDV mainly focuses on fetal protection, in addition to preventing clinical disease and virus-induced immune dysregulation. Control vaccination programs against BVDV involved using different types of live attenuated, killed, and recombinant vaccines (132–136). Although several scientific reports suggested their ability to prevent the clinical BVDV manifestations in cattle, live attenuated and killed BVDV vaccines differ in their safety for the vaccination of different cattle populations. Different live attenuated vaccines have been developed and widely used against BVDV, usually resulting in strong humoral and cell-mediated immune responses with solid fetal protection (133, 137). Due to its higher safety, being not able to infect the fetus and establish a persistent infection, CP-BVDV was used for the development of the most recent attenuated vaccines. On the opposite, as they can cross the placenta and infect the fetus, live attenuated vaccines based on NCP-BVDV are generally not recommended for the vaccination of pregnant animals. However, recent attempts to overcome the safety problem of NCP-BVDV involved the development of a mutant virus after deleting the Npro gene and inactivating the endoribonuclease activity of Erns (112, 137). Although some reports indicated that the mutated virus can induce a strong immune response without crossing the placenta (112, 113, 137), a recent work, however, reported the ability of the vaccine virus to cross the placenta (138). This indicates the need for further studies for evaluating the safety of this type of vaccine.
The different adverse effects associated with live attenuated vaccines, including the intrauterine infection of pregnant animals and immunosuppressive effects of the vaccine virus, have prompted the need for safe killed vaccines that can be given at any age and stage of pregnancy (139–141). Although they are safer than live attenuated vaccines, killed vaccines have lower immunogenicity and need therefore to be injected several times with slowly developing immunity. In a recent report, vaccination with different killed BVDV vaccines failed to induce cross-protective antibody response against all used virus strains (142, 143). Although the application of killed vaccines is associated with high antibody titers, the effectivity of the induced cell-mediated immunity is variable (144, 145). Fetal protection after vaccination with killed vaccines varies from incomplete to satisfactory (141, 146, 147).
To enhance the reduced immunogenicity of killed vaccines, several different powerful adjuvants have been used to improve the immune response after vaccination. A recent work evaluated the effectiveness of using immunomodulatory adjuvants in the BVDV vaccine design. The administration of a subunit vaccine formulated of the BVDV type-2 E2 protein with a novel adjuvant containing a mixture of a TLR 3 agonist, poly (I:C); an innate defense regulatory peptide; and water-soluble polymer, poly[di(sodium carboxyl atoethyl phenoxy)]-phosphazene (PCEP) induced the development of a robust immune response. In addition to inducing a strong humoral immune response, the vaccine resulted in cross-presentation with the development of both virus-specific CD4+ and CD8+ T-cell-mediated immune responses (148). For achieving a balance between vaccine safety and immunogenicity, combined administration of killed and live attenuated BVD vaccines has been recently suggested to induce reproductive protection in cows (149). The potential advantage of a DNAprime–protein boost vaccination approach has been recently demonstrated in mice primed with a plasmid encoding the E2 protein and boosted with adjuvant recombinant E2 protein. The combination of a DNA prime with protein boost vaccination was effective in eliciting high neutralizing antibody titers and cellular helper and cytotoxic immune responses against the virus (150). An earlier study showed the cross-protection between the type-1 and type-2 BVDVs (151). Vaccination of some female cattle with the bivalent MLV vaccines from both the BVDV and the BoHV-1 was linked to some undesired reproductive problems, especially abortion (133, 152). An interesting study recently developed a quadrivalent recombinant vaccine to overcome the side effects of the bivalent vaccines (BoHV-1 and the BVDV) (153). This study showed superior results of the newly developed quadrivalent vaccine with much more potent neutralizing activities against both viruses. It also showed the cross-protection among the two types of the BVDV with minimal reproductive failure compared to the other commercially available vaccines (153). A further recent work reported the development and efficacy of the first targeted subunit vaccine against BVDV (154). The vaccine was based on the fusion of the BVDV structural protein E2 with a single-chain antibody for targeting the E2 antigen to the major histocompatibility complex (MHC) class II molecules on antigen-presenting cells. The developed subunit vaccine induced a rapid and sustained BVDV-specific neutralizing antibody response in cattle (154). The immunogenicity of a recombinant Lactobacillus vaccine (using Lactobacillus casei strain W56 as antigen carrier) constitutively expressing BVDV E2 protein fused with cholera toxin B subunit as an adjuvant has been recently evaluated in a mice model (136). The study reported the induction of protective mucosal, humoral, and cellular immune responses in vaccinated mice and suggested employing the used strategy for vaccine development against BVDV (136).
Several studies have been conducted aiming at the development of novel anti-BVDV therapeutic strategies. Based on their advantageous small size and stable chemical structure, a recent approach investigated the therapeutic potential of newly developed single-domain antibodies (Nanobodies) against the nonstructural protein 5 (NS5B) of BVDV, which play an essential role in viral replication (155). In vitro analysis confirmed the interaction between the NS5B-nanobody and the BVDV NS5B protein, resulting in a marked suppressive effect on BVDV replication (155). A further therapeutic approach against BVDV infection proved the anti-BVDV activity of a biologically active recombinant bovine IFN-lambda (IFN-λ) (156). Systemic administration of IFN-λ to cattle experimentally infected with BVDV induced a systemic type-I IFN response, prevented BVDV replication and the development of the clinical disease, and enhanced the humoral immune response against the virus (156). Although these novel therapeutic strategies may contribute to BVDV control in individual animals, they are difficult to be implemented in commercial animal herds.
Maternal Immunity and Vaccination of Newborn Animals Against BVDV
The colostrum uptake supplies newborn calves with maternal antibodies, which prevent BVDV infection during the first weeks after birth (157). Maternal antibodies-mediated protection may, however, last for up to 9 months. The duration of protection in the newborn animal depends on the decrease in virus-specific antibodies in calf serum, which is mainly affected by the initial titer of colostrum's maternal antibodies (157). Although maternal antibodies are important for passive immunity and early protection, they may interfere with mounting active immune responses in vaccinated newborn calves (132, 158–162). Upon the vaccination of seven weeks-aged calves against BVDV, a T-cell mediated immune response developed despite the presence of circulating maternal antibodies (163). A recent BVDV vaccination study suggested a significant impact of virus biotype; method of attenuation, presentation, and use of adjuvant on the immune response of colostrum deprived calves and recommended the consideration of these variables when vaccinating newborn calves (111). The combination between a modified-live BVDV vaccine and the administration of some injectable trace minerals resulted in an enhanced immune response (titer of neutralizing antibodies) and improved health status of beef calves challenged with BVDV2 (164).
Future Directions
Although BVDV reported for more than 6 decades, it is still one of the main common viral threats to the bovine industry. Despite the great research ongoing progress on BVDV from different aspects including molecular biology, pathogenesis, immune response, and control measures, many aspects need further studies. The continuous emergence of new variants and strains of the BVDV may hamper the efficacy of the currently available diagnostic assays and vaccines. Exploring the functional activities of the mapped neutralizing epitopes within the BVDV-E2 gene may help in the development of effective vaccines against viral infection shortly. Searching for new potential reservoirs for the BVDV is one of the understudied research lines. Identification of new BVDV reservoir/s may help in minimizing the spillover of the virus from these unknown hosts to the bovine and ovine species. Further substantial efforts are needed to combat the emergence and spread of BVDV to eradicate such an important virus from various regions across the world.
Author Contributions
AA-K, JH, MK, MH, and AA-M prepared the original draft and revised the manuscript. AA-M acquired funding. All authors have read and agreed to the published version of the manuscript.
Funding
The authors wish to thank King Abdul-Aziz City for Science and Technology for their generous funding through the Strategic Technologies program, Grant No. 12-BIO3152-06.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Fray MD, Paton DJ, Alenius S. The effects of bovine viral diarrhoea virus on cattle reproduction in relation to disease control. Anim Reprod Sci. (2000) 60–1, 615–27. doi: 10.1016/S0378-4320(00)00082-8
2. Chang L, Qi Y, Liu D, Du Q, Zhao X, Tong D. Molecular detection and genotyping of bovine viral diarrhea virus in Western China. BMC Vet Res. (2021) 17:66. doi: 10.1186/s12917-021-02747-7
3. Scharnbock B, Roch FF, Richter V, Funke C, Firth CL, Obritzhauser W, et al. A meta-analysis of bovine viral diarrhoea virus (BVDV) prevalences in the global cattle population. Sci Rep. (2018) 8:14420. doi: 10.1038/s41598-018-32831-2
4. Hou P, Zhao G, Wang H, He H. Prevalence of bovine viral diarrhea virus in dairy cattle herds in eastern China. Trop Anim Health Prod. (2019) 51:791–8. doi: 10.1007/s11250-018-1751-z
5. Lanyon SR, Hill FI, Reichel MP, Brownlie J. Bovine viral diarrhoea: pathogenesis and diagnosis. Vet J. (2014) 199:201–9. doi: 10.1016/j.tvjl.2013.07.024
6. Olafson P, Mac CA, Fox FH. An apparently new transmissible disease of cattle. Cornell Vet. (1946) 36:205–13.
7. Al-Khaliyfa MA, Abuelzein EM, Gameel AA. Identification of cattle persistently infected with BVDV by ear-notch testing in Saudi Arabia. Vet Rec. (2010) 167:660–1. doi: 10.1136/vr.c5270
8. Mirosław P, Polak MP. Variability of E2 protein-coding sequences of bovine viral diarrhea virus in polish cattle. Virus Genes. (2020) 56:515–21. doi: 10.1007/s11262-020-01756-2
9. Grooms DL. Reproductive consequences of infection with bovine viral diarrhea virus. Vet Clin North Am Food Anim Pract. (2004) 20:5–19. doi: 10.1016/j.cvfa.2003.11.006
10. Newcomer BW, Chamorro MF, Walz PH. Vaccination of cattle against bovine viral diarrhea virus. Vet Microbiol. (2017) 206:78–83. doi: 10.1016/j.vetmic.2017.04.003
11. Callens N, Brügger B, Bonnafous P, Drobecq H, Gerl MJ, Krey T, et al. Morphology and molecular composition of purified bovine viral diarrhea virus envelope. PLOS Pathog. (2016) 12:e1005476. doi: 10.1371/journal.ppat.1005476
12. Wegelt A, Reimann I, Granzow H, Beer M. Characterization and purification of recombinant bovine viral diarrhea virus particles with epitope-tagged envelope proteins. J Gen Virol. (2011) 92:1352–7. doi: 10.1099/vir.0.029330-0
13. Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Scott Muerhoff A, et al. Proposed revision to the taxonomy of the genus pestivirus, family Flaviviridae. J Gen Virol. (2017) 98:2106–12. doi: 10.1099/jgv.0.000873
14. Bauermann FV, Ridpath JF, Weiblen R, Flores EF. HoBi-like viruses: an emerging group of pestiviruses. J Vet Diagn Invest. (2013) 25:6–15. doi: 10.1177/1040638712473103
15. Yamane D, Kato K, Tohya Y, Akashi H. The relationship between the viral RNA level and upregulation of innate immunity in spleen of cattle persistently infected with bovine viral diarrhea virus. Vet Microbiol. (2008) 129:69–79. doi: 10.1016/j.vetmic.2007.11.004
16. Kokkonos KG, Fossat N, Nielsen L, Holm C, Hepkema WM, Bukh J, et al. Evolutionary selection of pestivirus variants with altered or no microRNA dependency. Nucleic Acids Res. (2020) 48:5555–71. doi: 10.1093/nar/gkaa300
17. Burks J, Zwieb C, Muller F, Wower I, Wower J. Comparative structural studies of bovine viral diarrhea virus IRES RNA. Virus Res. (2011) 160:136–42. doi: 10.1016/j.virusres.2011.06.002
18. Scheel TKH, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, et al. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. (2016) 19:409–23. doi: 10.1016/j.chom.2016.02.007
19. Neill J. Molecular biology of bovine viral diarrhea virus. Biologicals. (2013) 41:2–7. doi: 10.1016/j.biologicals.2012.07.002
20. Fan ZC, Bird RC. An alternative−1/+2 open reading frame exists within viral N(pro)(1-19) region of bovine viral diarrhea virus SD-1. Virus Res. (2012) 163:341–51. doi: 10.1016/j.virusres.2011.10.022
21. Bazzucchi M, Bertolotti L, Ceglie L, Giammarioli M, Rossi E, Rosati S, et al. Complete nucleotide sequence of a novel bovine viral diarrhea virus subtype 1 isolate from Italy. Arch Virol. (2017) 162:3545–8. doi: 10.1007/s00705-017-3486-y
22. Giangaspero M, Yesilbag K, Apicella C. Who's who in the bovine viral diarrhea virus type 1 species: genotypes L and R. Virus Res. (2018) 256:50–75. doi: 10.1016/j.virusres.2018.07.009
23. Mishra N, Rajukumar K, Pateriya A, Kumar M, Dubey P, Behera SP, et al. Identification and molecular characterization of novel and divergent hobi-like pestiviruses from naturally infected cattle n India. Vet Microbiol. (2014) 174:239–46. doi: 10.1016/j.vetmic.2014.09.017
24. Oguzoglu TÇ, Koç BT, Coşkun N, Dogan F, Duran-Yelken S. Endless variety for bovine virus diarrhea viruses: new members of a novel subgroup into pestivirus A from Turkey. Trop Anim Health Prod. (2019) 51:1083–7. doi: 10.1007/s11250-018-01787-w
25. Mishra N, Kalaiyarasu S. Bovine viral diarrhea virus. In: Malik YS, Singh RK, Yadav MP, editors. Recent Advances in Animal Virology. Bhopal: Indian Council of Agricultural Research-National Institute of High Security Animal Diseases (2019). doi: 10.1007/978-981-13-9073-9_14
26. Giangaspero M, Apicella C, Harasawa R. Numerical taxonomy of the genus pestivirus: new software for genotyping based on the palindromic nucleotide substitutions method. J Virol Methods. (2013) 192:59–67. doi: 10.1016/j.jviromet.2013.04.023
27. Giangaspero M, Harasawa R. Characterization of genotypes among bovine viral diarrhea virus type 1 strains according to palindromic nucleotide substitutions in the genomic 5'-untranslated region. J Virol Methods. (2014) 194:34–53. doi: 10.1016/j.jviromet.2013.10.003
28. Giangaspero M. Genetic variation of border disease virus species strains. Vet Ital. (2011) 47:415–35.
29. Weber MN, Streck AF, Silveira S, Mósena ACS, Silva MSD, Canal CW. Homologous recombination in pestiviruses: identification of three putative novel events between different subtypes/genogroups. Infect Genet Evol. (2015) 30:219–24. doi: 10.1016/j.meegid.2014.12.032
30. Irianingsih SH, Poermadjaja B, Wuryastuti H, Wasito R. Genetic recombination of bovine viral diarrhea virus subgenotype−1a and−1c in persistently infected dairy cattle. Indones J Biotechnol. (2020) 25:120-6. doi: 10.22146/ijbiotech.54111
31. Pecora A, Malacari DA, Ridpath JF, Perez Aguirreburualde MS, Combessies G, Odeon AC, et al. First finding of genetic and antigenic diversity in 1b-BVDV isolates from Argentina. Res Vet Sci. (2014) 96:204–12. doi: 10.1016/j.rvsc.2013.11.004
32. Dias RK, Cargnelutti JF, Weber MN, Canal CW, Bauermann FV, Ridpath JF, et al. Antigenic diversity of Brazilian isolates of HoBi-like pestiviruses. Vet Microbiol. (2017) 203:221–8. doi: 10.1016/j.vetmic.2017.03.021
33. Bauermann FV, Flores EF, Ridpath JF. Antigenic relationships between bovine viral diarrhea virus 1 and 2 and HoBi virus: possible impacts on diagnosis and control. J Vet Diagn Invest. (2012) 24:253–61. doi: 10.1177/1040638711435144
34. Pecora A, Perez Aguirreburualde MS, Malacari DA, Zabal O, Sala JM, Konrad JL, et al. Serologic evidence of HoBi-like virus circulation in Argentinean water buffalo. J Vet Diagn Invest. (2017) 29:926–9. doi: 10.1177/1040638717720246
35. Postel A, Schmeiser S, Oguzoglu TC, Indenbirken D, Alawi M, Fischer N, et al. Close relationship of ruminant pestiviruses and classical swine fever virus. Emerge Infect Dis. (2015) 21:4. doi: 10.3201/eid2104.141441
36. Yan L, Pace LW, Baughman B, Wilson FD, Zhang S, Zhang MZ. Failed detection of bovine viral diarrhea virus 2 subgenotype a (BVDV-2a) by direct fluorescent antibody test on tissue samples due to reduced reactivity of field isolates to raw anti-BVDV antibody. J Vet Diagn Invest. (2016) 28:150–7. doi: 10.1177/1040638715626483
37. Deregt D, Van Rijn PA, Wiens TY, Van Den Hurk J. Monoclonal antibodies to the E2 protein of a new genotype (type 2) of bovine viral diarrhea virus define three antigenic domains involved in neutralization. Virus Res. (1998) 57:171–81. doi: 10.1016/S0168-1702(98)00095-1
38. Deregt D, Masri SA, Cho HJ, Bielefeldt Ohmann H. Monoclonal antibodies to the p80/125 gp53 proteins of bovine viral diarrhea virus: their potential use as diagnostic reagents. Can J Vet Res. (1990) 54:343–8.
39. Zhong F, Li N, Huang X, Guo Y, Chen H, Wang X, et al. Genetic typing and epidemiologic observation of bovine viral diarrhea virus in Western China. Virus Genes. (2011) 42:204–7. doi: 10.1007/s11262-010-0558-4
40. Isken O, Postel A, Bruhn B, Lattwein E, Becher P, Tautz N. CRISPR/Cas9-Mediated knockout of DNAJC14 verifies this chaperone as a pivotal host factor for RNA replication of pestiviruses. J Virol. (2019) 93:e01714–8. doi: 10.1128/JVI.01714-18
41. Muhsen M, Aoki H, Ikeda H, Fukusho A. Biological properties of bovine viral diarrhea virus quasispecies detected in the RK13 cell line. Arch Virol. (2013) 158:753–63. doi: 10.1007/s00705-012-1538-x
42. Shiokawa M, Omatsu T, Katayama Y, Nishine K, Fujimoto Y, Uchiyama S, et al. END-phenomenon negative bovine viral diarrhea virus that induces the host's innate immune response supports propagation of BVDVs with different immunological properties. Virology. (2019) 538:97–110. doi: 10.1016/j.virol.2019.09.016
43. Kozasa T, Abe Y, Mitsuhashi K, Tamura T, Aoki H, Ishimaru M, et al. Analysis of a pair of END+ and END- viruses derived from the same bovine viral diarrhea virus stock reveals the amino acid determinants in Npro responsible for inhibition of type I interferon production. J Vet Med Sci. (2015) 77:511–08. doi: 10.1292/jvms.14-0420
44. Mosena ACS, Falkenberg SM, Ma H, Casas E, Dassanayake RP, Walz PH, et al. Multivariate analysis as a method to evaluate antigenic relationships between BVDV vaccine and field strains. Vaccine. (2020) 38:5764–72. doi: 10.1016/j.vaccine.2020.07.010
45. Schweizer M, Peterhans E. Pestiviruses. Annu Rev Anim Biosci. (2014) 2:141–63. doi: 10.1146/annurev-animal-022513-114209
46. Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, et al. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol. (2017) 98:2–3. doi: 10.1099/jgv.0.000672
47. Wu Z, Liu B, Du J, Zhang J, Lu L, Zhu G, et al. Discovery of diverse rodent and bat pestiviruses with distinct genomic and phylogenetic characteristics in several Chinese provinces. Front Microbiol. (2018) 9:2562. doi: 10.3389/fmicb.2018.02562
48. Ictv ICOTOV. Genus: Pestivirus. (2020). Available online at: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/flaviviridae/361/genus-pestivirus (accessed Decemper 16, 2020).
49. Mósena ACS, Cibulski SP, Weber MN, Silveira S, Silva MS, Mayer FQ, et al. Genomic and antigenic relationships between two 'HoBi'-like strains and other members of the pestivirus genus. Arch Virol. (2017) 162:3025–34. doi: 10.1007/s00705-017-3465-3
50. Darweesh MF, Rajput MKS, Braun LJ, Rohila JS, Chase CCL. BVDV Npro protein mediates the BVDV induced immunosuppression through interaction with cellular S100A9 protein. Microb Pathog. (2018) 121:341–9. doi: 10.1016/j.micpath.2018.05.047
51. Tautz N, Tews BA, Meyers G. The molecular biology of pestiviruses. Adv Virus Res. (2015) 93:47–160. doi: 10.1016/bs.aivir.2015.03.002
52. Zürcher C, Sauter KS, Schweizer M. Pestiviral erns blocks TLR-3-dependent IFN synthesis by LL37 complexed RNA. Vet Microbiol. (2014) 174:399–408. doi: 10.1016/j.vetmic.2014.09.028
53. Lussi C, Sauter KS, Schweizer M. Homodimerisation-independent cleavage of dsRNA by a pestiviral nicking endoribonuclease. Sic Rep. (2018) 8:8226. doi: 10.1038/s41598-018-26557-4
54. Rajput MKS, Abdelsalam K, Darweesh MF, Braun LJ, Kerkvliet J, Hoppe AD, et al. Both cytopathic and non-cytopathic bovine viral diarrhea virus (BVDV) induced autophagy at a similar rate. Vet Immunol Immunopathol. (2017) 193:1–9. doi: 10.1016/j.vetimm.2017.09.006
55. Li Y, Wang J, Kanai R, Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. PNAS. (2013) 110:6805–10. doi: 10.1073/pnas.1300524110
56. Ostachuk AN. Bovine viral diarrhea virus structural protein E2 as a complement regulatory protein. Arch Virol. (2016) 161:1769–82. doi: 10.1007/s00705-016-2835-6
57. Mahmoodi P, Shapouri MRSA, Ghorbanpour M, Ekhtelat M, Hajikolaei MRH, et al. Epitope mapping of bovine viral diarrhea virus nonstructural protein 3. Vet Immunol Immunopathol. (2014) 161:232–9. doi: 10.1016/j.vetimm.2014.08.012
58. Mohamed Y, Bangphoomi N, Yamane D, Suda Y, Kato K, Horimoto T, et al. Physical interaction between bovine viral diarrhea virus nonstructural protein 4A and adenosine deaminase acting on RNA (ADAR). Arch Virol. (2014) 159: 11735–41. doi: 10.1007/s00705-014-1997-3
59. Suda Y, Murakami S, Horimoto T. Bovine viral diarrhea virus non?structural protein NS4B induces autophagosomes in bovine kidney cells. Arch Virol. (2019) 164:255–60. doi: 10.1007/s00705-018-4045-x
60. Bashir S, Kossarev A, Martin VC, Paeshuyse J. Deciphering the Role of Bovine Viral Diarrhea Virus Non-Structural NS4B Protein in Viral Pathogenesis. Vet Sci. (2020) 7:169. doi: 10.3390/vetsci7040169
61. Zahoor MA, Yamane D, Yassir Mahgoub Mohamed, Suda Y, Kobayashi K, Kato K, et al. Bovine viral diarrhea virus non-structural protein 5A interacts with NIK- and IKKb-binding protein. J Gen Virol. (2010) 91:1939–48. doi: 10.1099/vir.0.020990-0
62. Abdelsalam K, Rajput M, Elmowalid G, Sobraske J, Thakur N, Abdallah H, et al. The effect of bovine viral diarrhea virus (BVDV) strains and the corresponding infected-macrophages' supernatant on macrophage inflammatory function and lymphocyte apoptosis. Viruses. (2020) 12:701. doi: 10.3390/v12070701
63. Curti E, Jaeger J. Residues Arg283, Arg285, and Ile287 in the nucleotide binding pocket of bovine viral diarrhea virus NS5B RNA polymerase affect catalysis and fidelity. J Virol. (2013) 87:199–207. doi: 10.1128/JVI.06968-11
64. Newcomer B, Neill J, Marley M, Ridpath J, Givens M. Mutations induced in the NS5B gene of bovine viral diarrhea virus by antiviral treatment convey resistance to the compound. Virus Res. (2013) 174:5–100. doi: 10.1016/j.virusres.2013.03.007
65. Rümenapf T, Unger G, Strauss JH, Thiel HJ. Processing of the envelope glycoproteins of pestiviruses. J Virol. (1993) 67:3288–94. doi: 10.1128/JVI.67.6.3288-3294.1993
66. Bintintan I, Meyers G. A new type of signal peptidase cleavage site identified in an RNA virus polyprotein. J Biol Chem. (2010) 285:8572–84. doi: 10.1074/jbc.M109.083394
67. Krey T, Bontems F, Vonrhein C, Vaney MC, Bricogne G, Rümenapf T, et al. Crystal structure of the pestivirus envelope glycoprotein erns and mechanistic analysis of its ribonuclease activity. Structure. (2012) 20:862–73. doi: 10.1016/j.str.2012.03.018
68. Lazar C, Zitzmann N, Dwek RA, Branza-Nichita N. The pestivirus E(rns) glycoprotein interacts with E2 in both infected cells and mature virions. Virology. (2003) 314:696–705. doi: 10.1016/S0042-6822(03)00510-5
69. Mätzener P, Magkouras I, Rümenapf T, Peterhans E, Schweizer M. The viral RNase Erns prevents IFN type-I triggering by pestiviral single-and double-stranded RNAs. Virus Res. (2009) 140:15–23. doi: 10.1016/j.virusres.2008.10.015
70. Zürcher C, Sauter KS, Mathys V, Wyss F, Schweizer M. Prolonged activity of the pestiviral RNase Erns as an interferon antagonist after uptake by clatherin-mediated endocytosis. J Virol. (2014) 88:7235–43. doi: 10.1128/JVI.00672-14
71. Weiland E, Stark R, Haas B, Rümenapf T, Meyers G, Thiel J, et al. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J Virol. (1990) 64:3563–9. doi: 10.1128/JVI.64.8.3563-3569.1990
72. Wang Z, Nie Y, Wang P, Ding M, Deng H. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology. (2004) 330:332–41. doi: 10.1016/j.virol.2004.09.023
73. El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. (2013) 3:30–5. doi: 10.1016/j.celrep.2012.12.001
74. Radtke C, Tews B. Retention and topology of the bovine viral diarrhea virus glycoprotein E2. J Gen Virol. (2017) 98:2482–94. doi: 10.1099/jgv.0.000912
75. Chung YC, Cheng LT, Zhang JY, Wu YJ, Liu SS, Chu CY. Recombinant E2 protein enhances protective efficacy of inactivated bovine viral diarrhea virus 2 vaccine in a goat model. BMC Vet Res. (2018) 14:194. doi: 10.1186/s12917-018-1520-2
76. Van Rijn PA. A common neutralizing epitope on envelope glycoprotein E2 of different pestiviruses: implications for improvement of vaccines and diagnostics for classical swine fever (CSF)? Vet Microbiol. (2007) 125:150–6. doi: 10.1016/j.vetmic.2007.05.001
77. Jelsma H, Loeffen W, Van Beuningen A, Van Rijn P. Preliminary mapping of non-conserved epitopes on envelope glycoprotein E2 of Bovine viral diarrhea virus type 1 and 2. Vet Microbiol. (2013) 166:195–9. doi: 10.1016/j.vetmic.2013.06.008
78. Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. (2000) 16:276–77. doi: 10.1016/S0168-9525(00)02024-2
79. Loneragan GH, Thomson DU, Montgomery DL, Mason GL, Larson RL. Prevalence, outcome, and health consequences associated with persistent infection with bovine viral diarrhea virus in feedlot cattle. J Am Vet Med Assoc. (2005) 226:595–601. doi: 10.2460/javma.2005.226.595
80. Neill J, Newcomer B, Marley S, Ridpath J, Givens M. Genetic change in the open reading frame of bovine viral diarrhea virus is introduced more rapidly during the establishment of a single persistent infection than from multiple acute infections. Virus Res. (2011) 158:140–5. doi: 10.1016/j.virusres.2011.03.024
81. Kuca T, Passler T, Newcomer B, Neill J, Galik P, Riddell K, et al. Identification of conserved amino acid substitutions during serial infection of pregnant cattle and sheep with bovine viral diarrhea virus. Front Microbiol. (2018) 9:1109. doi: 10.3389/fmicb.2018.01109
82. Bachofen C, Vogt H, Stalder H, Mathys T, Zanoni R, Hilbe M, et al. Persistent infections after natural transmission of bovine viral diarrhoea virus from cattle to goats and among goats. Vet Res. (2013) 44:32. doi: 10.1186/1297-9716-44-32
83. Kuca T, Passler T, Newcomer B, Neill J, Galik P, Riddell K, et al. Changes introduced in the open reading frame of bovine viral diarrhea virus during serial infection of pregnant swine. Front Microbiol. (2020) 11:1138. doi: 10.3389/fmicb.2020.01138
84. Neill J, Dubovi E, Ridpath J. Identification of amino acid changes in the envelope glycoproteins of bovine viral diarrhea viruses isolated from alpaca that may be involved in host adaptation. Vet Microbiol. (2015) 179:299–03. doi: 10.1016/j.vetmic.2015.06.007
85. Kovago C, Hornyak Á, Kekesi V, Rusvai M. Demonstration of homologous recombination events in the evolution of bovine viral diarrhoea virus by in silico investigations. Acta Vet Hung. (2016) 64:401–14. doi: 10.1556/004.2016.038
86. Becher P, Tautz N. RNA recombination in pestiviruses: cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol. (2011) 8:216–24. doi: 10.4161/rna.8.2.14514
87. Büning MK, Meyer D, Austermann-Busch S, Roman-Sosa G, Rümenapf T, Becher P. Nonreplicative RNA recombination of an animal plus-strand RNA virus in the absence of efficient translation of viral proteins. Genome Biol Evol. (2017) 9:817–29. doi: 10.1093/gbe/evx046
88. Wilhelmsen CL, Bolin SR, Ridpath JF, Cheville NF, Kluge JP. Experimental primary postnatal bovine viral diarrhea viral infections in six-month-old calves. Vet Pathol. (1990) 27:235–43. doi: 10.1177/030098589002700404
89. Neibergs H, Zanella R, Casas E, Snowder G, Wenz J, Neibergs J, et al. Loci on Bos taurus chromosome 2 and Bos taurus chromosome 26 are linked with bovine respiratory disease and associated with persistent infection of bovine viral diarrhea virus. J Anim Sci. (2011) 89:907–15. doi: 10.2527/jas.2010-3330
90. Zanella R, Casas E, Snowder G, Neibergs H. Fine mapping of loci on BTA2 and BTA26 associated with bovine viral diarrhea persistent infection and linked with bovine respiratory disease in cattle. Front Genet. (2011). 2:82. doi: 10.3389/fgene.2011.00082
91. Weber MN, Bauermann FV, Gómez-Romero N, Herring AD, Canal CW, Neill JD, et al. Variation in pestivirus growth in testicle primary cell culture is more dependent on the individual cell donor than cattle breed. Vet Res Commun. (2016) 41:1–7. doi: 10.1007/s11259-016-9666-5
92. Maurer K, Krey T, Moennig V, Thiel HJ, Rumenapf T. CD46 is a cellular receptor for bovine viral diarrhea virus. J Virol. (2004) 78:1792–9. doi: 10.1128/JVI.78.4.1792-1799.2004
93. Riedel C, Chen H, Reichart U, Lamp B, Laketa V, Rumenapf T. Real time analysis of bovine viral diarrhea virus (BVDV) infection and its dependence on bovine CD46. Viruses. (2020) 12:116. doi: 10.3390/v12010116
94. Mishra N, Mathapati BS, Rajukumar K, Nema RK, Behera SP, Dubey SC. Molecular characterization of RNA and protein synthesis during a one-step growth curve of bovine viral diarrhoea virus in ovine (SFT-R) cells. Res Vet Sci. (2010) 89:130–2. doi: 10.1016/j.rvsc.2010.01.003
95. Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJT, Keene JD, et al. XRN1 stalling in the 5' UTR of hepatitis C virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLOS Pathog. (2015) 11:e1004708. doi: 10.1371/journal.ppat.1004708
96. Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. (2013) 14:16010–39. doi: 10.3390/ijms140816010
97. Taxis T, Bauermann F, Ridpath J, Casas E. Analysis of tRNA halves (tsRNAs) in serum from cattle challenged with bovine viral diarrhea virus. Genet Mol Biol. (2019) 42:374–9. doi: 10.1590/1678-4685-gmb-2018-0019
98. Ma Q, Li L, Tang Y, Fu Q, Liu S, Hu S, et al. Analyses of long non-coding RNAs and mRNA profiling through RNA sequencing of MDBK cells at different stages of bovine viral diarrhea virus infection. Res Vet Sci. (2017) 115:508–16. doi: 10.1016/j.rvsc.2017.09.020
99. Gao X, Niu C, Wang Z, Jia S, Han M, Ma Y, et al. Comprehensive analysis of IncRNA expression profiles in cytopathic biotype BVDV-infected MDBK cells provides an insight into biological contexts of host-BVDV interactions. Virulence. (2020) 12:20–34. doi: 10.1080/21505594.2020.1857572
100. Li C, Li X, Hou X, Ni W, Zhang M, Li H, et al. Comprehensive analysis of circRNAs expression profiles in different periods of MDBK cells infected with bovine viral diarrhea virus. Res Vet Sci. (2019) 125:52–60. doi: 10.1016/j.rvsc.2019.05.005
101. Taxis T, Bauermann F, Ridpath J, Casas E. Circulating MicroRNAs in serum from cattle challenged with bovine viral diarrhea virus. Front Genet. (2017) 8:91. doi: 10.3389/fgene.2017.00091
102. Mishra N, Rajukumar K, Kalaiyarasu S, Behera S, Nema R, Dubey S. Small interfering RNAs targeting viral structural envelope protein genes and the 5′-UTR inhibit replication of bovine viral diarrhea virus in MDBK cells. Acta Virol. (2011) 55:279–82. doi: 10.4149/av_2011_03_279
103. Walz PH, Chamorro MF, S MF, Passler T, Van Der Meer F A, et al. Bovine viral diarrhea virus: an updated American college of veterinary internal medicine consensus statement with focus on virus biology, hosts, immunosuppression, and vaccination. J Vet Intern Med. (2020) 34:1690–706. doi: 10.1111/jvim.15816
104. Chase CC. The impact of BVDV infection on adaptive immunity. Biologicals. (2013) 41:52–60. doi: 10.1016/j.biologicals.2012.09.009
105. Collen T, Morrison WI. CD4(+) T-cell responses to bovine viral diarrhoea virus in cattle. Virus Res. (2000) 67:67–80. doi: 10.1016/S0168-1702(00)00131-3
106. Collen T, Carr V, Parsons K, Charleston B, Morrison WI. Analysis of the repertoire of cattle CD4(+) T cells reactive with bovine viral diarrhoea virus. Vet Immunol Immunopathol. (2002) 87:235–8. doi: 10.1016/S0165-2427(02)00088-0
107. Howard CJ, Clarke MC, Sopp P, Brownlie J. Immunity to bovine virus diarrhoea virus in calves: the role of different T-cell subpopulations analysed by specific depletion in vivo with monoclonal antibodies. Vet Immunol Immunopathol. (1992) 32:303–14. doi: 10.1016/0165-2427(92)90052-R
108. Donis RO. Molecular biology of bovine viral diarrhea virus and its interactions with the host. Vet Clin North Am Food Anim Pract. (1995) 11:393–423. doi: 10.1016/S0749-0720(15)30459-X
109. Lambot M, Douart A, Joris E, Letesson JJ, Pastoret PP. Characterization of the immune response of cattle against non-cytopathic and cytopathic biotypes of bovine viral diarrhoea virus. J Gen Virol. (1997) 78 (Pt. 5):1041–7. doi: 10.1099/0022-1317-78-5-1041
110. Maldonado N, Fredericksen F, Espineira C, Toledo C, Oltra J, La Barra V, et al. BVDV-1 induces interferon-beta gene expression through a pathway involving IRF1, IRF7, NF-B kappa activation. Mol Immunol. (2020) 128:33–40. doi: 10.1016/j.molimm.2020.09.018
111. Falkenberg SM, Dassanayake RP, Palmer MV, Silveira S, Roth JA, Gauger E, et al. Changes in circulating lymphocytes and lymphoid tissue associated with vaccination of colostrum deprived calves. Vaccine. (2020) 38:7268–77. doi: 10.1016/j.vaccine.2020.09.046
112. Meyers G, Ege A, Fetzer C, Von Freyburg M, Elbers K, Carr V, et al. Bovine viral diarrhea virus: prevention of persistent fetal infection by a combination of two mutations affecting Erns RNase and Npro protease. J Virol. (2007) 81:3327–38. doi: 10.1128/JVI.02372-06
113. Rajput MKS, Darweesh MF, Braun LJ, Mansour SMG, Chase CCL. Comparative humoral immune response against cytopathic or non-cytopathic bovine viral diarrhea virus infection. Res Vet Sci. (2020) 129:109–16. doi: 10.1016/j.rvsc.2020.01.012
114. Risalde MA, Romero-Palomo F, Lecchi C, Ceciliani F, Bazzocchi C, Comazzi S, et al. BVDV permissiveness and lack of expression of co-stimulatory molecules on PBMCs from calves pre-infected with BVDV. Comp Immunol Microbiol Infect Dis. (2020) 68:101388. doi: 10.1016/j.cimid.2019.101388
115. Burciaga-Robles LO, Step DL, Krehbiel CR, Holland BP, Richards CJ, Montelongo MA, et al. Effects of exposure to calves persistently infected with bovine viral diarrhea virus type 1b and subsequent infection with mannheima haemolytica on clinical signs and immune variables: model for bovine respiratory disease via viral and bacterial interaction. J Anim Sci. (2010) 88:2166–78. doi: 10.2527/jas.2009-2005
116. Carlos-Valdez L, Wilson BK, Burciaga-Robles LO, Step DL, Holland BP, Richards CJ, et al. Effect of timing of challenge following short-term natural exposure to bovine viral diarrhea virus type 1b on animal performance and immune response in beef steers. J Anim Sci. (2016) 94:4799–808. doi: 10.2527/jas.2016-0712
117. Kelling CL, Steffen DJ, Topliff CL, Eskridge KM, Donis RO, Higuchi DS. Comparative virulence of isolates of bovine viral diarrhea virus type II in experimentally inoculated six- to nine-month-old calves. Am J Vet Res. (2002) 63:1379–84. doi: 10.2460/ajvr.2002.63.1379
118. Brown GB, Bolin SR, Frank DE, Roth JA. Defective function of leukocytes from cattle persistently infected with bovine viral diarrhea virus, and the influence of recombinant cytokines. Am J Vet Res. (1991) 52:381–7.
119. Thakur N, Evans H, Abdelsalam K, Farr A, Rajput MKS, Young AJ, et al. Bovine viral diarrhea virus compromises neutrophil's functions in strain dependent manner. Microb Pathog. (2020) 149:104515. doi: 10.1016/j.micpath.2020.104515
120. Morales-Aguilar A, Lopez-Reyes Y, Regalado-Huitron M, Sarmiento-Silva RE, Arriaga-Pizano L, Benitez-Guzman A. The nadl strain of bovine viral diarrhea virus induces the secretion of Il-1beta through caspase 1 in bovine macrophages. Res Vet Sci. (2020) 131:131–6. doi: 10.1016/j.rvsc.2020.04.014
121. Johnson DW, Muscoplat CC. Immunologic abnormalities in calves with chronic bovine viral diarrhea. Am J Vet Res. (1973) 34:1139–41.
122. Peterhans E, Jungi TW, Schweizer M. BVDV and innate immunity. Biologicals. (2003) 31:107–12. doi: 10.1016/S1045-1056(03)00024-1
123. Smirnova NP, Webb BT, Bielefeldt-Ohmann H, Van Campen H, Antoniazzi AQ, Morarie SE, et al. Development of fetal and placental innate immune responses during establishment of persistent infection with bovine viral diarrhea virus. Virus Res. (2012) 167:329–36. doi: 10.1016/j.virusres.2012.05.018
124. Smirnova NP, Webb BT, Mcgill JL, Schaut RG, Bielefeldt-Ohmann H, Van Campen H, et al. Induction of interferon-gamma and downstream pathways during establishment of fetal persistent infection with bovine viral diarrhea virus. Virus Res. (2014) 183:95–106. doi: 10.1016/j.virusres.2014.02.002
125. Nilson SM, Workman AM, Sjeklocha D, Brodersen B, Grotelueschen DM, Petersen JL. Upregulation of the type I interferon pathway in feedlot cattle persistently infected with bovine viral diarrhea virus. Virus Res. (2020) 278:197862. doi: 10.1016/j.virusres.2020.197862
126. Morarie-Kane SE, Smirnova NP, Hansen TR, Mediger J, Braun L, Chase C. Fetal hepatic response to bovine viral diarrhea virus infection in utero. Pathogens. (2018) 7:54. doi: 10.3390/pathogens7020054
127. Knapek KJ, Georges HM, Van Campen H, Bishop JV, Bielefeldt-Ohmann H, Smirnova NP, et al. Fetal lymphoid organ immune responses to transient and persistent infection with bovine viral diarrhea virus. Viruses. (2020) 12:816. doi: 10.3390/v12080816
128. Gonzalez Altamiranda EA, Arias ME, Kaiser GG, Mucci NC, Odeon AC, Felmer RN. Upregulation of interferon-alpha gene in bovine embryos produced in vitro in response to experimental infection with noncytophatic bovine-viral-diarrhea virus. Mol Biol Rep. (2020) 47:9959–65. doi: 10.1007/s11033-020-05958-7
129. Lussi C, Schweizer M. What can pestiviral endonucleases teach us about innate immunotolerance? Cytokine Growth Factor Rev. (2016) 29:53–62. doi: 10.1016/j.cytogfr.2016.03.003
130. Peterhans E, Schweizer M. Bvdv: a pestivirus inducing tolerance of the innate immune response. Biologicals. (2013) 41:39–51. doi: 10.1016/j.biologicals.2012.07.006
131. Schweizer M, Matzener P, Pfaffen G, Stalder H, Peterhans E. “Self” and “nonself” manipulation of interferon defense during persistent infection: bovine viral diarrhea virus resists alpha/beta interferon without blocking antiviral activity against unrelated viruses replicating in its host cells. J Virol. (2006) 80:6926–35. doi: 10.1128/JVI.02443-05
132. Moennig V, Becher P. Control of bovine viral diarrhea. Pathogens. (2018) 7:29. doi: 10.3390/pathogens7010029
133. Fulton RW, Cook BJ, Payton ME, Burge LJ, Step DL. Immune response to bovine viral diarrhea virus (BVDV) vaccines detecting antibodies to BVDV subtypes 1a, 1b, 2a, and 2c. Vaccine. (2020) 38:4032–7. doi: 10.1016/j.vaccine.2020.03.058
134. Sangewar N, Hassan W, Lokhandwala S, Bray J, Reith R, Markland M, et al. Mosaic bovine viral diarrhea virus antigens elicit cross-protective immunity in calves. Front Immunol. (2020) 11:589537. doi: 10.3389/fimmu.2020.589537
135. Koethe S, Konig P, Wernike K, Pfaff F, Schulz J, Reimann I, et al. A synthetic modified live chimeric marker vaccine against BVDV-1 and BVDV-2. Vaccines. (2020) 8:577. doi: 10.3390/vaccines8040577
136. Jia S, Huang X, Li H, Zheng D, Wang L, Qiao X, et al. Immunogenicity evaluation of recombinant Lactobacillus casei W56 expressing bovine viral diarrhea virus E2 protein in conjunction with cholera toxin B subunit as an adjuvant. Microb Cell Fact. (2020) 19:186. doi: 10.1186/s12934-020-01449-3
137. Platt R, Kesl L, Guidarini C, Wang C, Roth JA. Comparison of humoral and T-cell-mediated immune responses to a single dose of Bovela((R)) live double deleted BVDV vaccine or to a field BVDV strain. Vet Immunol Immunopathol. (2017) 187:20–7. doi: 10.1016/j.vetimm.2017.03.003
138. Wernike K, Michelitsch A, Aebischer A, Schaarschmidt U, Konrath A, Nieper H, et al. The occurrence of a commercial N(pro) and E(rns) double mutant BVDV-1 live-vaccine strain in newborn calves. Viruses. (2018) 10:274. doi: 10.3390/v10050274
139. Bolin SR, Matthews PJ, Ridpath JF. Methods for detection and frequency of contamination of fetal calf serum with bovine viral diarrhea virus and antibodies against bovine viral diarrhea virus. J Vet Diagn Invest. (1991) 3:199–203. doi: 10.1177/104063879100300302
140. Makoschey B, Van Gelder PT, Keijsers V, Goovaerts D. Bovine viral diarrhoea virus antigen in foetal calf serum batches and consequences of such contamination for vaccine production. Biologicals. (2003) 31:203–8. doi: 10.1016/S1045-1056(03)00058-7
141. Newcomer BW, Walz PH, Givens MD, Wilson AE. Efficacy of bovine viral diarrhea virus vaccination to prevent reproductive disease: a meta-analysis. Theriogenology. (2015) 83:360–5.e1. doi: 10.1016/j.theriogenology.2014.09.028
142. Sozzi E, Righi C, Boldini M, Bazzucchi M, Pezzoni G, Gradassi M, et al. Cross-reactivity antibody response after vaccination with modified live and killed bovine viral diarrhoea virus (BVD) vaccines. Vaccines. (2020) 8:374. doi: 10.3390/vaccines8030374
143. Walz PH, Riddell KP, Newcomer BW, Neill JD, Falkenberg SM, Cortese VS, et al. Comparison of reproductive protection against bovine viral diarrhea virus provided by multivalent viral vaccines containing inactivated fractions of bovine viral diarrhea virus 1 and 2. Vaccine. (2018) 36:3853–60. doi: 10.1016/j.vaccine.2018.04.005
144. Platt R, Coutu C, Meinert T, Roth JA. Humoral and T cell-mediated immune responses to bivalent killed bovine viral diarrhea virus vaccine in beef cattle. Vet Immunol Immunopathol. (2008) 122:8–15. doi: 10.1016/j.vetimm.2007.11.009
145. Stevens ET, Zimmerman AD, Butterbaugh RE, Barling K, Scholz D, Rhoades J, et al. The induction of a cell-mediated immune response to bovine viral diarrhea virus with an adjuvanted inactivated vaccine. Vet Ther. (2009) 10:E1–8. doi: 10.1016/j.vetimm.2020.110024
146. Zimmer GM, Wentink GH, Bruschke C, Westenbrink FJ, Brinkhof J, De Goey I. Failure of foetal protection after vaccination against an experimental infection with bovine virus diarrhea virus. Vet Microbiol. (2002) 89:255–65. doi: 10.1016/S0378-1135(02)00203-1
147. Rodning SP, Marley MS, Zhang Y, Eason AB, Nunley CL, Walz PH, et al. Comparison of three commercial vaccines for preventing persistent infection with bovine viral diarrhea virus. Theriogenology. (2010) 73:1154–63. doi: 10.1016/j.theriogenology.2010.01.017
148. Sadat SMA, Snider M, Garg R, Brownlie R, Van Drunen Littel-Van Den Hurk S. Local innate responses and protective immunity after intradermal immunization with bovine viral diarrhea virus E2 protein formulated with a combination adjuvant in cattle. Vaccine. (2017) 35:3466–73. doi: 10.1016/j.vaccine.2017.05.029
149. Walz PH, Givens MD, Rodning SP, Riddell KP, Brodersen BW, Scruggs D, et al. Evaluation of reproductive protection against bovine viral diarrhea virus and bovine herpesvirus-1 afforded by annual revaccination with modified-live viral or combination modified-live/killed viral vaccines after primary vaccination with modified-live viral vaccine. Vaccine. (2017) 35:1046–54. doi: 10.1016/j.vaccine.2017.01.006
150. Cai D, Song Q, Duan C, Wang S, Wang J, Zhu Y. Enhanced immune responses to E2 protein and DNA formulated with ISA 61 VG administered as a DNA prime-protein boost regimen against bovine viral diarrhea virus. Vaccine. (2018) 36:5591–9. doi: 10.1016/j.vaccine.2018.07.054
151. Dean HJ, Leyh R. Cross-protective efficacy of a bovine viral diarrhea virus (BVDV) type 1 vaccine against BVDV type 2 challenge. Vaccine. (1999) 17:1117–24. doi: 10.1016/S0264-410X(98)00329-6
152. Fulton RW, D'offay JM, Eberle R, Moeller RB, Campen HV, O'toole D, et al. Bovine herpesvirus-1: evaluation of genetic diversity of subtypes derived from field strains of varied clinical syndromes and their relationship to vaccine strains. Vaccine. (2015) 33:549–58. doi: 10.1016/j.vaccine.2014.11.033
153. Chowdhury SI, Pannhorst K, Sangewar N, Pavulraj S, Wen X, Stout RW, et al. BoHV-1-vectored BVDV-2 subunit vaccine induces BVDV cross-reactive cellular immune responses and protects against BVDV-2 challenge. Vaccines. (2021) 9:46. doi: 10.3390/vaccines9010046
154. Bellido D, Baztarrica J, Rocha L, Pecora A, Acosta M, Escribano JM, et al. A novel MHC-II targeted BVDV subunit vaccine induces a neutralizing immunological response in guinea pigs and cattle. Transbound Emerg Dis. (2020). doi: 10.1111/tbed.13952. [Epub ahead of print].
155. Duan H, Ma Z, Xu L, Zhang A, Li Z, Xiao S. A novel intracellularly expressed NS5B-specific nanobody suppresses bovine viral diarrhea virus replication. Vet Microbiol. (2020) 240:108449. doi: 10.1016/j.vetmic.2019.108449
156. Quintana ME, Cardoso NP, Pereyra R, Barone LJ, Barrionuevo FM, Mansilla FC, et al. Interferon lambda protects cattle against bovine viral diarrhea virus infection. Vet Immunol Immunopathol. (2020) 230:110145. doi: 10.1016/j.vetimm.2020.110145
157. Munoz-Zanzi CA, Thurmond MC, Johnson WO, Hietala SK. Predicted ages of dairy calves when colostrum-derived bovine viral diarrhea virus antibodies would no longer offer protection against disease or interfere with vaccination. J Am Vet Med Assoc. (2002) 221:678–85. doi: 10.2460/javma.2002.221.678
158. Platt R, Widel PW, Kesl LD, Roth JA. Comparison of humoral and cellular immune responses to a pentavalent modified live virus vaccine in three age groups of calves with maternal antibodies, before and after BVDV type 2 challenge. Vaccine. (2009) 27:4508–19. doi: 10.1016/j.vaccine.2009.05.012
159. Ellis JA, Hassard LE, Cortese VS, Morley PS. Effects of perinatal vaccination on humoral and cellular immune responses in cows and young calves. J Am Vet Med Assoc. (1996) 208:393–400.
160. Zimmerman AD, Boots RE, Valli JL, Chase CC. Evaluation of protection against virulent bovine viral diarrhea virus type 2 in calves that had maternal antibodies and were vaccinated with a modified-live vaccine. J Am Vet Med Assoc. (2006) 228:1757–61. doi: 10.2460/javma.228.11.1757
161. Zimmerman AD, Buterbaugh RE, Schnackel JA, Chase CC. Efficacy of a modified-live virus vaccine administered to calves with maternal antibodies and challenged seven months later with a virulent bovine viral diarrhea type 2 virus. Bovine Practit. (2009) 43:35–43. doi: 10.21423/bovine-vol43no1p35-43
162. Chamorro MF, Walz PH, Passler T, van Santen E, Gard J, Rodning SP, et al. Efficacy of multivalent, modified- live virus (MLV) vaccines administered to early weaned beef calves subsequently challenged with virulent Bovine viral diarrhea virus type 2. BMC Vet Res. (2015) 11:29. doi: 10.1186/s12917-015-0342-8
163. Endsley JJ, Roth JA, Ridpath J, Neill J. Maternal antibody blocks humoral but not T cell responses to BVDV. Biologicals. (2003) 31:123–5. doi: 10.1016/S1045-1056(03)00027-7
164. Bittar JHJ, Palomares RA, Hurley DJ, Hoyos-Jaramillo A, Rodriguez A, Stoskute A, et al. Immune response and onset of protection from bovine viral diarrhea virus 2 infection induced by modified-live virus vaccination concurrent with injectable trace minerals administration in newly received beef calves. Vet Immunol Immunopathol. (2020) 225:110055. doi: 10.1016/j.vetimm.2020.110055
Keywords: BVDV, genome, pathogenesis, mapping epitopes, immune evasion, immunotherapeutic, vaccines
Citation: Al-Kubati AAG, Hussen J, Kandeel M, Al-Mubarak AIA and Hemida MG (2021) Recent Advances on the Bovine Viral Diarrhea Virus Molecular Pathogenesis, Immune Response, and Vaccines Development. Front. Vet. Sci. 8:665128. doi: 10.3389/fvets.2021.665128
Received: 07 February 2021; Accepted: 16 April 2021;
Published: 14 May 2021.
Edited by:
Fuxiao Liu, Qingdao Agricultural University, ChinaReviewed by:
Demian Bellido, Vetanco S.A., ArgentinaChris Chase, South Dakota State University, United States
Copyright © 2021 Al-Kubati, Hussen, Kandeel, Al-Mubarak and Hemida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maged Gomaa Hemida, bWhlbWlkYUBrZnUuZXUuc2E=; Abdullah I. A. Al-Mubarak, YWlhbG11YmFya0BrZnUuZWR1LnNh
†These authors have contributed equally to this work
 Anwar A. G. Al-Kubati
Anwar A. G. Al-Kubati Jamal Hussen
Jamal Hussen Mahmoud Kandeel
Mahmoud Kandeel Abdullah I. A. Al-Mubarak
Abdullah I. A. Al-Mubarak Maged Gomaa Hemida
Maged Gomaa Hemida