- 1Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Provincial, College of Animal Science and Technology, Northwest A&F University, Xianyang, China
- 2Shaanxi Provincial Engineering and Technology Research Center of Cashmere Goats, Yulin University, Yulin, China
- 3Life Science Research Center, Yulin University, Yulin, China
The Booroola fecundity (FecB) gene, as the first major fecundity gene identified in Booroola sheep, has attracted careful attention. So far, previous research have uncovered the FecB mutation (Q249R) as the main mutation by virtue of which sheep exhibits multiple lambing phenomena. This mutation is now being intensively studied and widely used. However, such effect of the FecB mutation has not been applied to goats, and similar types of the FecB gene in goats still need to be studied. Thus, the current study attempted to verify potential mutations in the goat FecB gene as well as investigate their functions related to fecundity. First, FecB expression was investigated in six different goat tissues, and we found that FecB expression was highest in the mammary gland, followed by the ovary. Next, the influence of the FecB gene was analyzed from a new perspective, where five potential copy number variations (CNVs) (CNV1–5) within the FecB gene were identified for the first time, and then their effects on litter size were measured. Our results point out that CNV3 (P = 3.44E-4) and CNV5 (P = 0.034) could significantly influence the litter size of goats. Identically, the combination genotype of CNV3 and CNV5 which consisted of their dominant genotypes was also significantly associated with goat litter size (P = 7.80E-5). Hence, CNV3 and CNV5 could serve as potential DNA molecular markers applied to DNA editing and DNA microarray. Additionally, the abovementioned study has laid a theoretical foundation for the detection of potential fertility-related quantitative trait loci within the goat FecB gene.
Introduction
The Booroola fecundity (FecB) gene, also named the bone morphogenetic protein receptor 1B (BMPR1B) gene, was first recognized as a high-prolificacy major gene in Booroola sheep (1) due to the multi-lamb phenomenon contributed by the FecB mutation (Q249R) (2, 3). Moreover, as a receptor of bone morphogenetic proteins (BMPs) (4), it is involved in the regulation of follicle-stimulating hormone (FSH) levels, which regulates the ovulation rate in animals and humans (5, 6). In detail, the diminution of the BMP signal reduced by Q249R leads to an increase in follicle-stimulating hormone (FSHR) and luteinizing hormone receptor (LHR) density and also reduces apoptosis to increase ovulation (7–9). Furthermore, the attenuation of BMP signaling can maintain the reserve of primordial follicles while promoting follicular growth and ovulation, which contributes to the overall fertility of a female (10). Subsequently, a previous study showed that the Q249R mutation can regulate the expression of BMP/SMAD signaling and significantly upregulate the expression of fecundity-related genes such as STAR, BMP6, and BMP2 (11). Moreover, it can increase ovulation by suppressing the expression of SMAD6, which acts as an inhibitor of the BMP/SMAD signaling pathway (12).
To date, the FecB gene has been deeply studied in several breeds and strains of sheep such as Hu, Chinese Merino prolific meat strain, Suffolk, Dorset, and Charolais sheep breeds (13, 14), especially the Q249R locus which was successfully applied in sheep breeding as a significant DNA marker. Although only Q249R is regarded as the major mutation related to high fertility in sheep, there are also several identified mutations within the FecB gene worth studying—for instance, g.29362047T>C and g.29427689G>A were found to be significantly associated with litter size in Hu sheep (15). In addition to single-nucleotide polymorphisms (SNPs), five insertion/deletions (indels) (4, 10, 12, 17, and 23 bp) in the BMPR1B gene were also verified in Chinese Australian White sheep. However, among them, only the 10-bp indel significantly affected the sheep litter size (16). In goat, a previous study has revealed that Smad signaling, steroidogenesis, and cell viability in granulosa cells have been altered upon modulation of the FecB gene which was similar to that documented in sheep breeds carrying the FecB mutation (17). Moreover, several research were devoted to the detection of potential mutations in goat FecB gene, but no major high-fertility-related mutation was discovered in goat until now. In detail, C94T was identified to exert a significant influence on the litter size of Liaoning cashmere goats (18). Apart from that, three novel SNPs, including G773C, A775G, and G777A, have also been uncovered in Assam hill and Markhoz goats, respectively, but their specific functions still need to be studied (19, 20). Altogether, as a major fecundity-related gene, the FecB gene as well as its function has attracted careful attention (20–25), and a number of SNPs and indels within the FecB gene have also been examined to varying degrees (26–28). However, almost all studies have focused on sheep, while no major quantitative trait locus (QTL) has yet been identified in goats. Additionally, we found that all relevant research focus on SNPs and indels without paying attention to copy number variation (CNV), which is also one of the promising DNA markers.
Therefore, in this work, we aimed to investigate potential CNVs within the BMPR1B gene and, for the first time, measure their effects on goat litter size, thereby trying to provide information for DNA editing and microarray as well as laying the theoretical foundation for improving goat fecundity.
Materials and Methods
Animals, Total RNA, and Genomic DNA Isolation
In order to obtain RNA samples for further experiments, a total of six different tissues (corpus luteum, large follicle, skeletal muscle, uterus, ovary, and mammary gland) of the goat were collected (n = 3). Total RNA extraction was carried out using TRIzol total RNA extraction reagent (Takara, Dalian, China), following the instructions of the manufacturer. Then, 1% agarose gel electrophoresis in 6 × loading buffer was used to evaluate the integrity of total RNA. RNA was then quantified and qualitatively analyzed using a Nanodrop 2000 Spectrophotometer. Qualified RNA samples were stored at −80°C. To accomplish the further experiment, first-strand cDNA was synthesized by Prime Script™ RT Reagent Kit (Takara, Dalian, China). After that, these cDNAs were conserved at −20°C for subsequent experiments.
Shaanbei white cashmere (SBWC) female goats (2–3 years) that were healthy and under the same feeding and management conditions were used as the DNA sample in this work. All genomic DNAs were isolated from the ear tissue of 312 female goats according to the protocol of our previous study (29) and then stored at −40°C. In addition, all litter size and growth trait records of these goats were provided by the staff from farm records.
RNA Experiment: mRNA Expression
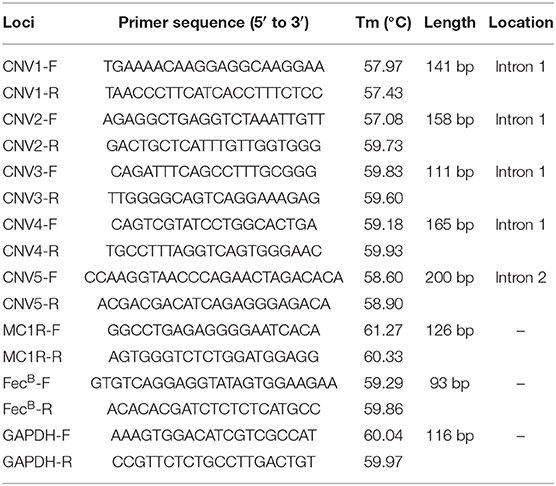
A pair of primers for qPCR (FecB-F: F′-GTGTCAGGAGGTATAGTGGAAGAA-R′; FecB-R: F′- ACACACGATCTCTCTCATGCC-R′) was designed according to the mRNA sequence (accession number: NM_001285575.1) of the goat FecB gene in the NCBI database (https://www.ncbi.nlm.nih.gov/gene) via primer premier 6.0 (Table 1). It has covered all exons of the goat FecB gene to ensure cDNA amplification. The qPCR reaction system (10 μl) comprised of 5 μl of 2 × SYBR Premix Ex Taq (Takara Biotech, Dalian, China), 1 μl of cDNA, 0.5 μl of each primer, and 3 μl of ddH2O, using the same protocol as in our previous study (30). GAPDH was used as the reference gene (31). The results were then analyzed by 2−ΔΔCt method (32, 33).
DNA Experiment: Genotyping of CNV Within the FecB Gene
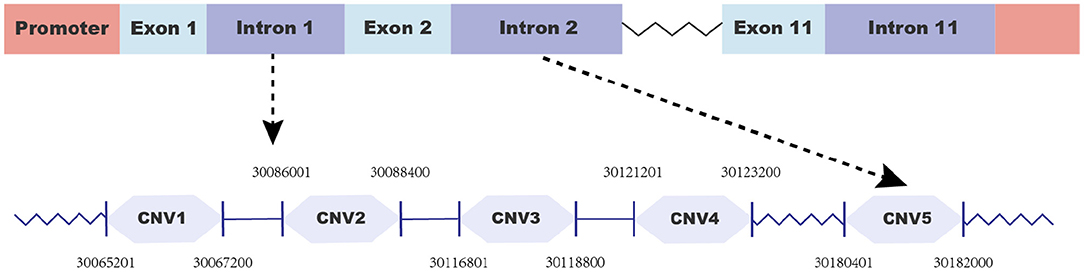
To identify copy number variations (CNVs), we first searched for potential CNVs in the goat FecB gene in the Animal Omics database (http://animal.nwsuaf.edu.cn) and found five CNV loci. Additionally, the figure of the goat FecB gene was drawn using Adobe Illustrator 2021 (Adobe, USA) as shown in Figure 1. When we analyzed these CNVs in our detected population, five pairs of primers (Table 1) were designed by NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast), and qPCR was executed to further validate the CNVs. The qPCR reaction system (10 μl) contained 5 μl of 2 × SYBR Premix Ex Taq (Takara Biotech), 0.5 μl of goat genomic DNA, 0.5 μl of each primer, and 3.5 μl of ddH2O, using the program in our previous study (30). Then, the copy number of the goat FecB gene was calculated using 2*2−ΔCt method, where ΔCt = Ct target gene - Ct reference gene, and MC1R gene was the reference gene (28). Eventually, based on the value of 2*2−ΔCt, the CNVs were divided into three types: “loss” (2*2−ΔCt <2), “median” (2*2−ΔCt = 2), and “gain” (2*2−ΔCt > 2).
Statistical Analyses
The genotype frequencies of the five CNVs in the tested population was calculated, and the haplotypes were constructed via SHEsis online program (http://analysis.bio-x.cn). After that, the genotype distributions of these CNVs in all single lamb and multi-lamb individuals were analyzed using chi-square test (χ2).
Besides that, a linear model was used to estimate the effect of fixed factors on goat litter size (28): Yijk = μ + ai + ßj + eijk, where Yijk is the litter size, μ is the overall mean value for each trait, ai is the fixed-factor age, ßj is the fixed-factor genotype, and eijk is the random error. For CNVs that displayed more than two genotypes, one-way ANOVA was used to assess their relationship with litter size; for <2 group, t-test was performed.
Results
mRNA Expression
mRNA expression was determined in the mammary gland, corpus luteum, large follicle, skeletal muscle, ovary, and uterus tissues. Our results demonstrated that FecB expression was highest in the mammary gland and was 10-fold higher than that in other tissues. To add, the relative expression of the goat FecB gene in the skeletal muscle, uterus, and ovary varied from 10- to 50-fold changes compared to those in the corpus luteum and large follicle which were used as control (Figure 2).
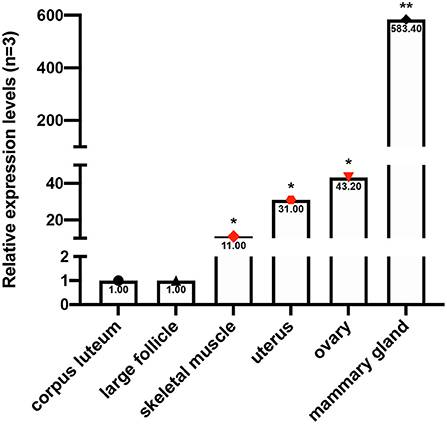
Figure 2. The mRNA expression profile of goat FecB gene, using the corpus luteum and large follicle as references (selecting three individuals for each tissue with three parallel repeats). *P < 0.05; **P < 0.01.
CNV Detection: Genotype Frequency, LD Analysis, and Haplotype Construction
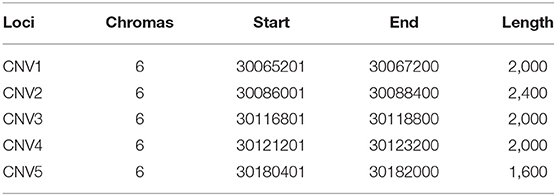
To make the detected CNVs easier to describe, all of them were named according to their location at the goat chromosome as CNV1–5 (Table 2). Moreover, CNV2–5 displayed three genotypes (“loss,” “median,” and “gain”), while CNV1 only manifested two genotypes (“median” and “gain”) (Table 3). Furthermore, “gain” was the dominant genotype among all of them.
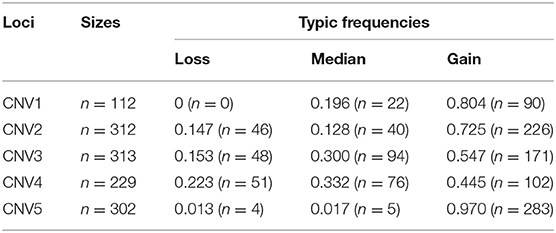
Table 3. Typical frequencies of copy number variations within FecB gene in Shaanbei white cashmere goats.
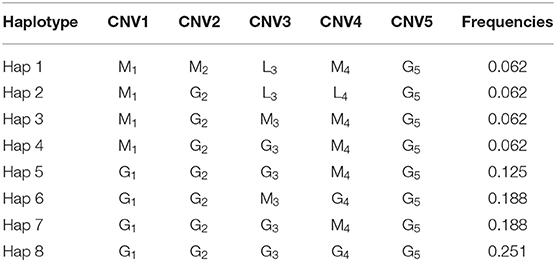
Hereafter, eight haplotypes were constructed among these five CNVs. In detail, G1G2G3G4G5, of which the frequency was 0.251, accounted for the most, while the frequencies of G1G2G3M4G5, G1G2M3G4G5, G1G2G3M4G5, M1G2G3M4G5, M1G2M3M4G5, M1G2L3L4G5, and M1M2L3M4G5 were 0.125, 0.188, 0.188, 0.125, 0.062, 0.062, 0.062, and 0.062, respectively (Table 4).
Association Analysis
To test whether these five CNVs exerted a significant effect on the litter size of goats, we analyzed the association between them and the litter. Our results point out that CNV3 (P = 3.44E-4) and CNV5 (P = 0.034) were significantly associated with goat litter size (Figure 3; Table 5). The dominant genotype for CNV3 was “median,” while the dominant genotype for CNV5 was “gain.” Interestingly, the dominant combination genotype M3G5 could also have a significant influence on litter size (P = 7.80E-5) (Figure 4; Table 6). Additionally, the distribution of different genotypes in different litter-type populations significantly differed from each other (Table 7).
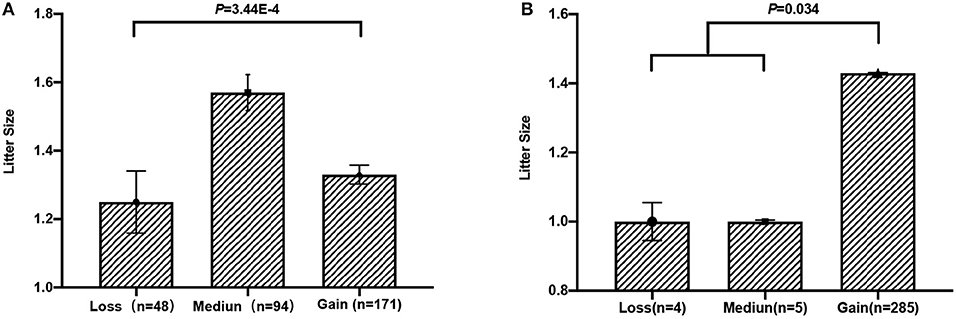
Figure 3. Association analysis between litter size and copy number variations (CNVs) in Shaanbei white cashmere goats. (A) Association between CNV3 and litter size. (B) Association between CNV5 and litter size.
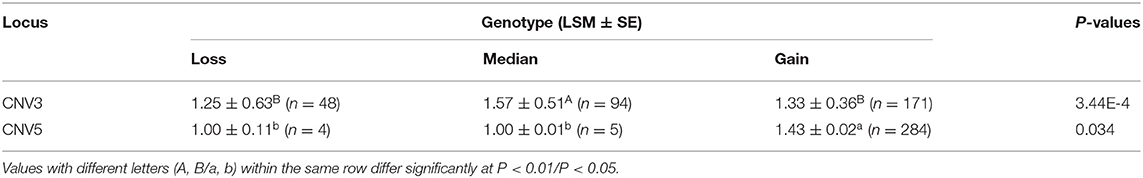
Table 5. Association analyses between litter size and copy number variation types in Shaanbei white cashmere goats.
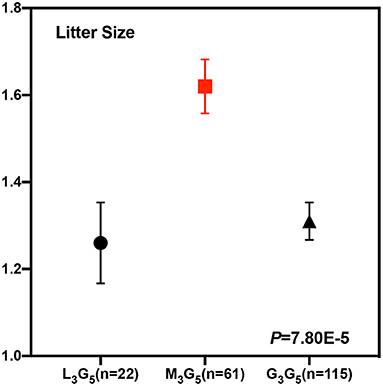
Figure 4. Least squares mean and standard error for litter size of the different combination genotypes of CNV3 and CNV5 within the FecB gene in Shaanbei white cashmere goats. Combination genotypes with a sample size of less than five were not calculated.
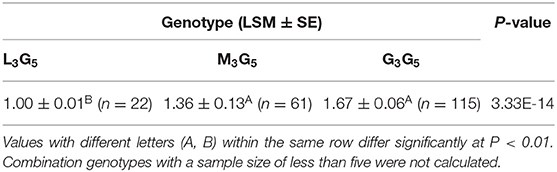
Table 6. Least squares mean and standard error for litter size of different combination genotypes of CNV3 and CNV5 within the FecB gene in Shaanbei white cashmere goats.
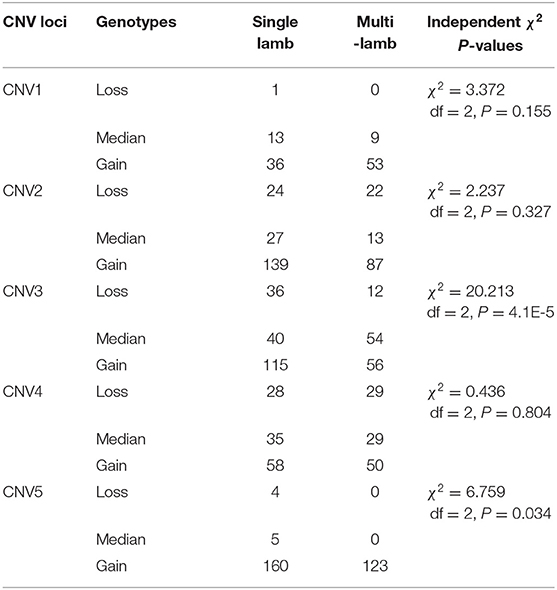
Table 7. Type distribution between mothers of single lamb and multi-lamb in Shaanbei white cashmere goats.
Discussion
The FecB gene, which serves as a major gene for high fecundity, has been shown to contribute to the multi-lambing phenomena in sheep. Currently, it is gradually becoming clear how the FecB gene regulates sheep fecundity, while the specific mechanism of the goat FecB gene is still poorly understood and the first major QTL still needs to be disclosed. Here we identified the expression of the FecB gene in six different goat tissues and strikingly found that it had the highest expression in the mammary gland, indicating that the FecB gene might also function as a lactation regulator of goats. According to a previous study, an increase in litter size stimulates the expression of RFamide-related peptide mRNA, which might contribute to lactational anestrus in rats (34). Additionally, as the litter size increases, milk yield increases (35–38), and so does the uptake of net daily amino acid by the mammary gland, which is the basis for milk yield and lactoprotein production (39). In short, the above results indicate that the lactation would be mediated by an increase in litter size. Thus, the FecB gene, which significantly affects litter size, also presumably has a function related to lactation in goats. This might be the reason why the FecB gene had the highest expression in the mammary gland in goats. To our best knowledge, this is the first time that the FecB gene has shown such a high expression in the mammary gland, which leads to an indication that it is worth investigating whether the FecB gene has a lactation-related function, especially in populations with multiple lambs. Then, following the mammary gland, high expression in the ovary indicated that the FecB gene might be involved in ovarian activity, thereby influencing goat fertility. However, whether the FecB gene could impact goat fecundity has not yet been sufficiently studied (17).
Until now, reported studies have found a series of polymorphisms associated with fecundity within the FecB gene in sheep. However, related research analyzing the association between mutations in the FecB gene and fertility in goat are relatively scanty—for instance, G773C was identified to be unique in Assam hill goat, while the association between goat fecundity still needed to be studied (19). T242C was uncovered in Barbari, Beetal, Black Bengal, Ganjam, Jhakana, Osmanabadi as well as Sangamneri goat breeds, but the effect of genotypes was non-significant on litter size (40). Moreover, C94T within the goat FecB gene was revealed to be significantly associated with litter size in Liaoning cashmere goats (18). Interestingly, a related work investigated the FecB mutation in goat since its significant influence on multiple lambing in sheep. However, the results pointed out that the FecB mutation did not exert the same effect in goat, which led to a hint that novel significant mutations still need discovering (20). Many reports altogether revealed lots of polymorphisms within the FecB gene associated with fecundity in sheep, while potential mutations still need discovery in goats. No related research also paid attention on the CNV within this gene. In this study, we first identified five potential CNVs in the FecB gene and measured their effects on litter size in goats. Our results pointed out that the “gain” genotype, rather than another two genotypes, displayed the highest frequency in all five CNVs. Identically, the G1G2G3G4G5 haplotype was also the dominant haplotype with the highest frequency. In addition to the distribution of the five CNVs, we further analyzed their effect on goat litter size, which was the main purpose of this work. The results based on a large experimental population showed that CNV3 and CNV5 exerted significant effects on litter size in goats (P < 0.05), with “median” and “Gain” displaying a superior phenotype, respectively. In addition, the combination genotypes of CNV3 and CNV5 could also have the same effects on goat litter size. Notably, the combined genotype of “median” (CNV3) and “gain” (CNV5) performed superior litter size, which is consistent with the abovementioned results. Thus, CNV3 and CNV5 could serve as effective DNA markers applied to marker-assisted selection breeding, and individuals, especially with combined genotype “M3G5” should be selected to improve the litter size of goats.
Given that previous investigations revealing intronic variations could influence the interaction between transcription factors and host genes, we hypothesized that CNV3 and CNV5 might influence the combination ability of DNA sequence with transcription factors to indirectly influence the expression of the FecB gene (41, 42). To add, with BMPs acting as the key intraovarian factors regulating ovarian function (43–45), their biological effects will be mediated after binding to membrane-bound receptors. Mutations might influence the process BMPs combing with DNA sequence, thereby changing the related functions. However, how the two CNVs within the FecB gene influence the goat litter size still needs to be studied.
In conclusion, we have identified five CNVs within the FecB gene in SBWC goat population and found that CNV3 and CNV5 significantly influenced the litter size of goats. Moreover, this is the first report on the effect of CNVs within the FecB gene on litter size. Despite this, the specific mechanisms require further investigation.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU; protocol number NWAFAC1008).
Author Contributions
YB, XL, and CP came up with idea and revised the manuscript. YB wrote the manuscript and performed the experiments. WF, YK, KW, and YY collected the goat samples and isolated the genomic DNA. YB, YK, and LQ analyzed the data. All authors approved the final version of the manuscript for submission and contributed to the article and approved the submitted version.
Funding
This work was supported by the National Scientific and Technological Innovation Project of Undergraduate of Northwest A&F University (No. 202106001) and the National Science Foundation of China (No. 32060734).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the team of LQ at Yulin College for allowing us to collect the samples. We would also like to thank the Life Science Research Core Services of Northwest A&F University (Northern Campus) and the Agricultural Technical Station of Veterinary and Animal Husbandry of Yulin City, Shaanxi Province, PR China, for providing us with the platform.
References
1. Piper LR, Bindon BM. The Booroola Merino and the performance of medium non-Peppin crosses at Armidale. Wool Tech Sheep Bree. (1983) 31:14.
2. Davis GH. Fecundity genes in sheep. Anim Reprod Sci. (2004) 82:247–53. doi: 10.1016/j.anireprosci.2004.04.001
3. Qi MY, Xu LQ, Zhang JN, Li MO, Lu MH, Yao YC. Effect of the Booroola fecundity (FecB) gene on the reproductive performance of ewes under assisted reproduction. Theriogenology. (2020) 142:246–50. doi: 10.1016/j.theriogenology.2019.10.038
4. Otsuka F. Multiple endocrine regulation by bone morphogenetic protein system. Endocr J. (2010) 57:3–14. doi: 10.1507/endocrj.K09E-310
5. McNatty KP, Henderson KM. Gonadotrophins, fecundity genes and ovarian follicular function. J Steroid Biochem. (1987) 27:365–73. doi: 10.1016/0022-4731(87)90329-3
6. Mihm M, Evans ACO. Mechanisms for dominant follicle selection in monovulatory species: a comparison of morphological, endocrine and intraovarian events in cows, mares and women. Reprod Domest Anim. (2008) 43:48–56. doi: 10.1111/j.1439-0531.2008.01142.x
7. Xia Y, O'Shea T, Murison R, McFarlane JR. Concentrations of progesterone, follistatin, and follicle-stimulating hormone in peripheral plasma across the estrous cycle and pregnancy in merino ewes that are homozygous or noncarriers of the Booroola gene. Biol Reprod. (2003) 69:1079–84. doi: 10.1095/biolreprod.102.005512
8. Ruoss C, Tadros A, O'Shea T, McFarlane J, Almahbobi G. Ovarian follicle development in Booroola sheep exhibiting impaired bone morphogenetic protein signalling pathway. Reproduction. (2009) 138:689–96. doi: 10.1530/REP-09-0190
9. Regan SLP, McFarlane JR, O'Shea T, Andronicos N, Arfuso F, Dharmarajan A, et al. Flow cytometric analysis of FSHR, BMRR1B, LHR and apoptosis in granulosa cells and ovulation rate in merino sheep. Reproduction. (2015) 150:151–63. doi: 10.1530/REP-14-0581
10. AI-Samerria S, AI-Ali I, McFarlane JR, Almahbobi G. The impact of passive immunisation against BMPRIB and BMP4 on follicle development and ovulation in mice. Reproduction. (2015) 149:403–11. doi: 10.1530/REP-14-0451
11. Kumar S, Rajput PK, Bahire SV, Jyotsana B, Kumar V, Kumar D. Differential expression of BMP/SMAD signaling and ovarian-associated genes in the granulosa cells of FecB introgressed GMM sheep. Syst Biol Reprod Med. (2020) 66:185–201. doi: 10.1080/19396368.2019.1695977
12. Bahire SV, Rajput PK, Kumar V, Kumar D, Kataria M, Kumar S. Quantitative expression of mRNA encoding BMP/SMAD signalling genes in the ovaries of Booroola carrier and non-carrier GMM sheep. Reprod Domest Anim. (2019) 54:1375–83. doi: 10.1111/rda.13535
13. Guan F, Liu SR, Shi GQ, Yang LG. Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development. Anim Reprod Sci. (2007) 99:44–52. doi: 10.1016/j.anireprosci.2006.04.048
14. Roy J, Polley S, De S, Mukherjee A, Batabyal S, Pan S, et al. Polymorphism of fecundity genes (FecB, FecX, and FecG) in the Indian Bonpala sheep. Anim Biotechnol. (2011) 22:151–62. doi: 10.1080/10495398.2011.589239
15. Yang Z, Yang X, Liu G, Deng M, Sun B, Guo Y, et al. Polymorphisms in BMPR-IB gene and their association with litter size trait in Chinese Hu sheep. Anim Biotechnol. (2020). doi: 10.1080/10495398.2020.1789158. [Epub ahead of print].
16. Li HX, Xu HW, Akhatayeva Z, Liu HF, Lin CJ, Han XF, et al. Novel indel variations of the sheep FecB gene and their effects on litter size. Gene. (2021) 767:145176. doi: 10.1016/j.gene.2020.145176
17. Kumar S, Punetha M, Jose B, Bharati J, Khanna S, Sonwane A, et al. Modulation of granulosa cell function via CRISPR-Cas fuelled editing of BMPR-IB gene in goats (Capra hircus). Sci Rep. (2020) 10:20446. doi: 10.1038/s41598-020-77596-9
18. Yue C, Bai WL, Zheng YY, Hui TY, Sun JM, Guo D, et al. Correlation analysis of candidate gene SNP for high-yield in Liaoning cashmere goats with litter size and cashmere performance. Anim Biotechnol. (2021) 32:43–50. doi: 10.1080/10495398.2019.1652188
19. Dutta R, Laskar S, Borah P, Kalita D, Das B, Zaman G, et al. Polymorphism and nucleotide sequencing of BMPR1B gene in prolific Assam hill goat. Mol Biol Rep. (2014) 46:3677–81. doi: 10.1007/s11033-014-3232-4
20. Dogaheh SP, Mirhoseini SZ, Tufarelli V, Hossein-Zadeh NG, Badbarin S, Colonna MA, et al. Investigating the polymorphism of bone morphogenetic protein receptor-1B (BMPR1B) gene in Markhoz goat breed. Animals. (2020) 10:1582. doi: 10.3390/ani10091582
21. Davis GH, Galloway SM, Ross IK, Gregan SM, Ward J, Nimbkar BV, et al. DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol Reprod. (2002) 66:1869–74. doi: 10.1095/biolreprod66.6.1869
22. Davis GH. Major genes affecting ovulation rate in sheep. Genet Sel Evol. (2005) 37:S11–23. doi: 10.1186/1297-9686-37-S1-S11
23. Davis GH, Balakrishnan L, Ross IK, Wilson T, Galloway SM, Lumsden BM, et al. Investigation of the Booroola (FecB) and Inverdale (FecX(I)) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim Reprod Sci. (2006) 92:87–96. doi: 10.1016/j.anireprosci.2005.06.001
24. Chu MX, Liu ZH, Jiao CL, He QY, Fang L, Ye SC, et al. Mutations in BMPR-IB and BMP15 genes are associated with litter size in Small tailed Han sheep (Ovis aries). J Anim Sci. (2007) 85:598–603. doi: 10.2527/jas.2006-324
25. Ahlawat S, Sharma R, Roy M, Mandakmale S, Prakash V, Tantia MS. Genotyping of novel SNPs in BMPR1B, BMP15, and GDF9 genes for association with prolificacy in seven Indian goat breeds. Anim Biotechnol. (2016) 27:199–207. doi: 10.1080/10495398.2016.1167706
26. Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. PNAS. (2001) 98:5104–9. doi: 10.1073/pnas.091577598
27. Souza CJ, MacDougal C, Campbell BK, McNeilly AS, Baird DT. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1B (BMPR-1B) gene. J Endocrinol. (2001) 169:R1–R6. doi: 10.1677/joe.0.169r001
28. Gootwine E, Rozov A, Bor A, Reicher S. Carrying the FecB (Booroola) mutation is associated with lower birth weight and slower post-weaning growth rate for lambs, as well as a lighter mature bodyweight for ewes. Reprod Fert Develop. (2006) 18:433–7. doi: 10.1071/RD05134
29. Zhang XL, Zhang SH, Tang Q, Jiang EH, Wang K, Lan XY, et al. Goat sperm associated antigen 17 protein gene (SPAG17): small and large fragment genetic variation detection, association analysis, and mRNA expression in gonads. Genomics. (2020) 112:5115–21. doi: 10.1016/j.ygeno.2020.09.029
30. Hui YQ, Zhang HY, Wang K, Pan CY, Chen H, Qu L, et al. Goat DNMT3B: an indel mutation detection, association analysis with litter size and mRNA expression in gonads. Theriogenology. (2020) 147:108–15. doi: 10.1016/j.theriogenology.2020.02.025
31. Kang ZH, Zhang SH, Zhu HJ, Wang Z, Yan HL, Huang YZ, et al. A 14-bp functional deletion within the CMTM2 gene is significantly associated with litter size in goat. Theriogenology. (2019) 139:49e57. doi: 10.1016/j.theriogenology.2019.07.026
32. Livak KJ, Thomas DS. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
33. Wang Z, Zhao HY, Xu YY, Zhao JN, Song Z, Bi Y, et al. Early-life lead exposure induces long-term toxicity in the central nervous system: from zebrafish larvae to juveniles and adults. Sci Total Environ. (2021) 804:150185. doi: 10.1016/j.scitotenv.2021.150185
34. Noroozi A, Jafarzadeh Shirazi MR, Tamadon A, Moghadam A, Niazi A. Increased litter size and suckling intensity stimulate mRNA of RFamide-related peptide in rats. Int J Fertil Steril. (2015) 9:380–6. doi: 10.22074/ijfs.2015.4554
35. Eisen EJ, Nagai J, Bakker H, Hayes JF. Effect of litter size at birth on lactation in mice. J Anim Sci. (1980) 50:680–8. doi: 10.2527/jas1980.504680x
36. Romero T, Beltrán MC, Rodríguez M, Olives AMD, Molina MP. Short communication: goat colostrum quality: litter size and lactation number effects. J Dairy Sci. (2013) 96:7526–31. doi: 10.3168/jds.2013-6900
37. Lv SJ, Yang Y, Li FK. Parity and litter size effects on maternal behavior of small tail Han sheep in China. Anim Sci J. (2016) 87:361–9. doi: 10.1111/asj.12441
38. Zamuner F, DiGiacomo K, Cameron AWN, Leury BJ. Effects of month of kidding, parity number, and litter size on milk yield of commercial dairy goats in Australia. J Dairy Sci. (2020) 103:954–64. doi: 10.3168/jds.2019-17051
39. Nielsen TT, Trottier NL, Stein HH, Bellaver C, Easter RA. The effect of litter size and day of lactation on amino acid uptake by the porcine mammary glands. J Anim Sci. (2002) 80:2402–11. doi: 10.2527/2002.8092402x
40. Chu MX, Mu YL, Fang L, Ye SC, Sun SH. Prolactin receptor as a candidate gene for prolificacy of Small Tail Han sheep. Anim Biotechnol. (2007) 18:65–73. doi: 10.1080/10495390601090950
41. Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. (2016) 533:95–9. doi: 10.1038/nature17939
42. Cui Y, Yan H, Wang K, Xu H, Zhang X, Zhu H, et al. Insertion/deletion within the KDM6A gene is significantly associated with litter size in goat. Front Genet. (2018) 9:91. doi: 10.3389/fgene.2018.00091
43. Chen DI, Zhao M, Mundy GR. Bone morphogenetic protein. Growth Factors. (2004) 22:233–41. doi: 10.1080/08977190412331279890
44. Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell Signal. (2004) 16:291–9. doi: 10.1016/j.cellsig.2003.08.011
Keywords: goat, FecB gene, mRNA expression, CNV, litter size
Citation: Bi Y, Feng W, Kang Y, Wang K, Yang Y, Qu L, Chen H, Lan X and Pan C (2021) Detection of mRNA Expression and Copy Number Variations Within the Goat FecB Gene Associated With Litter Size. Front. Vet. Sci. 8:758705. doi: 10.3389/fvets.2021.758705
Received: 14 August 2021; Accepted: 13 September 2021;
Published: 18 October 2021.
Edited by:
Kangfeng Jiang, Yunnan Agricultural University, ChinaReviewed by:
Mingxing Chu, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (CAAS), ChinaGuangxin E, Southwest University, China
Copyright © 2021 Bi, Feng, Kang, Wang, Yang, Qu, Chen, Lan and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianyong Lan, bGFueGlhbnlvbmc3OUAxMjYuY29t; Chuanying Pan, Y2h1YW55aW5ncGFuQDEyNi5jb20=
 Yi Bi1
Yi Bi1 Ke Wang
Ke Wang Hong Chen
Hong Chen Xianyong Lan
Xianyong Lan Chuanying Pan
Chuanying Pan