- 1School of Veterinary Medicine, Wollo University, Dessie, Ethiopia
- 2Shire Agricultural Technical Vocational and Education Training College, Shire, Ethiopia
- 3College of Veterinary Sciences, Mekelle University, Mekelle, Ethiopia
- 4College of Agriculture and Veterinary Medicine, Addis Ababa University, Addis Ababa, Ethiopia
Escherichia coli O157:H7 is an emerging and major zoonotic foodborne pathogen. It has an increasing concern about the spread of antimicrobial-resistant strains. This study aimed to isolate and characterize Shiga toxin-producing E. coli O157:H7 from raw milk, yogurt, and meat of bovine origin and determine their antimicrobial susceptibility pattern. A cross-sectional study was conducted from December 2014 to June 2015, and a total of 284 milk and meat samples were collected from different sources in Mekelle. The collected samples were analyzed for the presence of E. coli and Shiga toxin-producing E. coli O157:H7 and the determination of their antimicrobial susceptibility pattern following the standard bacteriological and molecular techniques and procedures and antimicrobial sensitivity test. Out of the total 284 samples, 70 (24.6%) were bacteriologically positive for E. coli and 14.3% were found to be Shiga toxin-producing E. coli O157:H7. Of note, 100% of E. coli isolates carried the pal gene and 41.7% eaeA gene (EHEC). Of these EHEC isolates, 40% and 60% were positive for stx1 and stx2, respectively. E. coli isolates showed the highest level of susceptibility to gentamycin (91.7%) but the highest level of resistance to amoxicillin (95.8%). Of the tested isolates, 18 (75%) of E. coli showed multidrug-resistant. This study revealed the occurrence of Shiga toxin-producing E. coli O157:H7 in foods of bovine origin in the study area. In conclusion, a nationwide phenotypic and molecular characterization, in-depth typing, and drug-resistant gene identification of E. coli O157:H7 should be undertaken.
Introduction
In Ethiopia, both food shortage and lack of appropriate food safety assurance systems are problems that have become obstacles to the country's economic development and public health safety (1, 2). Animal products are generally regarded as high-risk commodities in respect of pathogen contents, natural toxins, and other possible contaminants and adulterants (3). Foodborne microorganisms are major pathogens affecting food safety and cause human illness worldwide as a result of the consumption of foodstuff, mainly animal products contaminated with vegetative pathogens or their toxins. Most of these microbes have zoonotic importance, resulting in a significant impact on both public health and economic sectors (4). Moreover, the emergence of multidrug-resistant (MDR) pathogens presents a global challenge for treating and preventing disease spread through zoonotic transmission (5).
Data regarding foodborne diseases are extremely scarce at a national level, and a few studies conducted in different parts of the country showed the poor sanitary conditions of catering establishments and the presence of pathogenic organisms such as Campylobacter, Salmonella, Staphylococcus aureus, Bacillus cereus, and Escherichia coli (6–10).
Escherichia coli found in humans can be categorized on basis of genetic and clinical criteria into the following three main groups: commensal, pathogenic (enteric or diarrheagenic), and extraintestinal pathogenic E. coli (ExPEC) (11). The typical diarrheagenic strains include enterotoxigenic (ETEC), enterohemorrhagic (EHEC), enteroinvasive (EIEC), enteropathogenic (EPEC), and enteroaggregative (EAEC) E. coli (12, 13). The E. coli that cause enteric disease have been divided into pathotypes, based on their virulence factors and mechanisms by which they cause disease (11, 14). One of these pathotypes, called Shiga toxin-producing E. coli (STEC), refers to those strains of E. coli that produce at least 1 member of a class of potent cytotoxins called Shiga toxin. The STEC is also called verotoxin-producing E. coli (VTEC). The names Shiga toxin (Stx), derived from similarity to a cytotoxin produced by Shigella dysenteriae serotype 1 (15), and verotoxin (VT), based on cytotoxicity for Vero cells (16), are used interchangeably. Those STEC that cause hemorrhagic colitis and hemolytic uremic syndrome are called enterohemorrhagic E. coli (EHEC) (14, 17). The Shiga toxin-producing E. coli O157 is synonymous with E. coli O157:H7 (18, 19). Pathogenicity of E. coli O157:H7 is encoded by a variety of plasmid, bacteriophage, and chromosomal genes (20). The key virulence factor for the subset of EHEC is the Shiga toxin (Stx) family, which contains two subgroups, namely, Stx1 and Stx2 that share approximately 55% amino acid homology (11). The ability to produce Shiga toxin was acquired from a bacteriophage presumably directly or indirectly from Shigella (20).
Shiga toxin-producing E. coli is recognized as an important cause of food-borne disease in humans and causes large outbreaks worldwide (21). E. coli O157:H7 is the leading cause of hemorrhagic colitis and hemolytic-uremic syndrome (HUS), a life-threatening sequela characterized by a triad of acute renal failure, microangiopathic hemolytic anemia, and thrombocytopenia in humans (22, 23). People of all ages are susceptible to infection with STEC. However, the young and the elderly are more susceptible and are more likely to develop more serious symptoms (24). Domestic and wild animals are sources of EHEC O157:H7 but the major animal carriers are healthy domesticated ruminants, primarily cattle, and to a lesser extent, sheep, and possibly goats. Fresh meat and raw milk are, nevertheless, considered common vehicles for E. coli, particularly for the EHEC (O157:H7) strain. Contamination of meat usually occurs during animal slaughter, as a result of poor slaughter practices, abattoir hygiene, and animal handling practices (20, 25).
Foods of bovine origin are frequently implicated in human outbreaks of Shiga toxin-producing E. coli (STEC) O157 (26, 27). Shiga-toxigenic E. coli is transmitted by the fecal-oral route by either consumption of contaminated food or water, from direct contact with infected animals, or via person-to-person contact. Moreover, a series of studies on the resistance of E. coli isolated from animals and humans have strongly suggested that those bacteria that are resistant to antimicrobials used in animals would also be resistant to antimicrobials used in humans (28–30). In general, antimicrobial resistance is a global public health problem, and growing scientific evidence indicates that it is negatively impacted by both human and animal antimicrobial usage (31). Although antimicrobial therapy is not the primary tool for treating infections caused by STEC O157:H7, MDR STEC O157:H7 is a public health issue as those strains participate in a reservoir of resistance genes that could be easily exchanged between Enterobacteriaceae in the host and the environment (32). However, less is currently known about the molecular basis of MDR in STEC O157:H7 isolates of food origin (33). Therefore, it is of paramount importance to systematically investigate and characterize this recurring food-borne disease. Thus, the current study was conducted to isolate and characterize Shiga toxin-producing E. coli O157:H7 from raw milk, yogurt, and meat of bovine origin from different sources in the study area and to determine the antimicrobial susceptibility pattern of the isolates.
Materials and methods
Study area
This study was conducted from December 2014 to June 2015 in Mekelle City. Mekelle is the capital city of Tigray Regional State, situated approximately 783 km North of Addis Ababa at 38.5° East longitude and 13.5° North latitude at an altitude of 2,300 above sea level. The city has a total population of 215,546 (34), 308 cafeterias, 292 restaurants, 258 supermarkets, and an active urban-rural exchange of goods which has 30,000 micro and small enterprises (35). The weather condition is hot and dry. The mean annual rainfall of the area is 628.8 mm and an annual average temperature of 21°C (36).
Study design and study population
A cross-sectional study on Shiga toxin-producing E. coli O157:H7 was conducted from December 2014 to June 2015 on raw milk, yogurt, and meat samples collected from different sources and parts of Mekelle City, Tigray, Ethiopia. The study populations comprised purposively selected milking dairy cows and slaughtered cattle found in Mekelle City.
Sampling technique and sample collection
A total of 284 samples of bovine origin, comprised of raw milk (n = 145), yogurt (n = 48), and meat (n = 91), were collected using a purposive random sampling technique. These samples were collected based on the willingness of the owners until the required sample size was achieved. Raw milk samples were aseptically collected directly from the teats of lactating cows (n = 100), whole-sellers (n = 17), cafeterias (n = 28), and similarly the yogurt samples were collected from dairy farms (n = 26) and cafeterias (n = 22) using a sterile universal bottle. However, the raw meat samples were collected from abattoirs (n = 55), butchery shops (n = 16), and restaurants (n = 20) during slaughtering and selling. Then, the sections of meat samples were aseptically removed and placed in separate sterile plastic bags to prevent spilling and cross-contamination. Both samples were transported to the Veterinary Microbiology Laboratory of the College of Veterinary Sciences, Mekelle University using an icebox and stored at +4°C until the laboratory work was conducted.
Isolation and identification of E. coli and E. coli O157:H7
Microbiological procedures
Isolation of E. coli was attempted according to Quinn et al. (37) with slight modification. A part of each sample (10 ml or 10 g) was enriched in peptone water (HiMedia, Mumbai, India) (90 ml) and was incubated at 37°C for 24 h. Enriched samples were inoculated on MacConkey Agar (MCA) (HiMedia, Mumbai, India) by four flame techniques, and plates were incubated at 37°C for 24 h. Pink-colored colonies were considered presumptive of E. coli. Gram staining was performed as per procedures described by Merchant and Packer (38) to determine the size, shape, and arrangement of bacteria. The organisms revealed Gram-negative, pink-colored with rod-shaped appearance, and arranged in single or in pairs were suspected as E. coli.
A single well-isolated colony was picked from MCA and streaked on Eosin Methylene Blue Agar (EMB) (HiMedia, Mumbai, India) and incubated at 37°C for 24 h. The characteristic green metallic sheen growth of colonies is a presumptive identification for E. coli. Colony morphology and colors on MCA and EMB agar plates together with the Gram-stain procedure were used as an initial identification of E. coli colonies (39). Such colonies were taken from EMB into nutrient broth and agar for further biochemical examination. Standard biochemical tests (catalase test, indole, methyl red, Voges-Proskauer test, nitrate reduction, citrate utilization, and urease production) were used as confirmation of identification (40–46). The Triple Sugar Iron test was performed according to Vanderzant and Splittstresser (47). Carbohydrate fermentation tests of the isolates were performed according to the method of Simmons (43). Colonies that were confirmed as E. coli were further subcultured onto MacConkey Agar with Sorbitol to differentiate E. coli O157: H7 from other strains, especially lactose fermenters E. coli. A bacterial strain that was used as a quality control organism in this study was a standard strain of E. coli ATCC 25922.
PCR amplification of the pal, eaeA stx1, and sxt2 virulence genes of E. coli
Escherichia coli genomic DNA extraction and purification were performed as per the protocol given by PureLink® Genomic DNA Purification Kit, USA, for Gram-negative organisms, and the total genomic DNA was checked by running on 1.0% agarose gel.
The DNA of all presumptive isolates was subjected to multiplex PCR for analyzing the presence of the pal (48), eaeA (49), stx1, and stx2 (50) genes and further modified by Fitzmaurice (51). The amplification was performed according to the protocol reported by Parekh and Subhash (52) and Wang et al. (53) using the following specific primers: ECPAL-F-5′GGC AAT TGC GGC ATG TTC TTC C3′ and ECPAL-R-5′CCG CGT GAC CTT CTA CGG TGA3′ for pal gene of E. coli (280 bp); EHEC-F-5′TGG TAC GGG TAA TGA AAA3′ and EHEC-R-5′AAT AGC CTG GTA GTC TTG T3′ for eaeA gene of EHEC (360 bp); Stx1-F-5′ATA AAT CGC CAT TCG TTG ACT AC3′ and Stx1-R-5′AGA ACG CCC ACT GAG ATC ATC3′ for stx1 (180 bp); and Stx2-F-5′GGC ACT GTC TGA AAC TGC TCC3′ and Stx2-R-5′TCG CCA GTT ATC TGA CAT TCT G3′ for stx2 (255 bp). Each reaction mixture (50 μl) consisted of 5 μl of 10× reaction buffer (500 mM KCl, 15 mM MgCl2, 100 mM Tris HCl pH 8.3, 0.1%w/v gelatin), 5 μl of template DNA, 1 μl of each primer (the primers were used at a final concentration of 100 M), 3 μl of 10 mM dNTP mixture (at a concentration of 100 μl each), and 1 μl of Taq polymerase (3 U/l). The remaining volume of the reaction mixture was nuclease-free water. The amplification was carried out using a Tianlong PCR Thermocycler with thermal cycling conditions of an initial denaturation at 94°C for 6 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, and with a final extension at 72°C for 6 min. Finally, PCR products were separated in a horizontal equipment system by running on a 1.5% (w/v) agarose gel containing 0.5 g/ml ethidium bromide for 55 min at 110 V using 1XTAE buffer (40 mM Tris, 1 mM EDTA, and 20 mM glacial acetic acid, pH 8.0). The amplicons were visualized under UV-light gel doc, and their molecular weight was estimated by comparing them with a 100 bp DNA molecular weight marker (Solis BioDyne, Tartu, Estonia) (54).
Antibiotic susceptibility testing
The isolates of E. coli were screened for in vitro antimicrobial susceptibility using the agar disk diffusion method described by Bauer et al. (55). For this, the following seven different antibiotic disks (Oxoid Ltd., Basingstoke, Hampshire, England) with their concentrations given in parentheses were used in the antibiograms: amoxicillin (AMX) (2 μg), doxycycline (DOX) (30 μg), erythromycin (ERY) (15 μg), gentamicin (GEN) (10 μg), penicillin (PEN) (10 μg), trimethoprim-sulfamethoxazole (SXT) (20 μg), and tetracycline (TET) (30 μg). The selection of these antibiotics was based on the availability and frequent use of these antimicrobials in the study area both in veterinary and human medicine. Four to five well-isolated colonies from nutrient agar plates were transferred into tubes containing 5 ml of a normal saline solution until it achieved the 0.5 McFarland turbidity standards, and then a sterile cotton swab was dipped into the adjusted suspension and then spread evenly over the entire surface of the plate of Mueller-Hinton agar (Oxoid Ltd., Basingstoke, Hampshire, England) to obtain uniform inoculums. The plates were then allowed to dry for 3–5 min. Antibiotic-impregnated disks were then applied to the surface of the inoculated plates with sterile forceps. After 18–24 h of incubation at 37°C, the plates were examined, and the clear zone (inhibition zones of bacterial growth around the antibiotic disk including the disk) diameter for individual antimicrobial agents was measured using a digital caliper and then translated into susceptible (S), intermediate (I), and resistant (R) categories according to the interpretation table of the Clinical and Laboratory Standard Institute (56).
Data management and analysis
All collected data were entered into Microsoft Excel Sheet and analyzed through the SPSS version 16. Accordingly, descriptive statistics such as percentages and frequency distribution were used to determine the occurrence of the food items, and the chi-square (χ2) test was applied to assess the association.
Results
The occurrence of E. coli and E. coli O157:H7
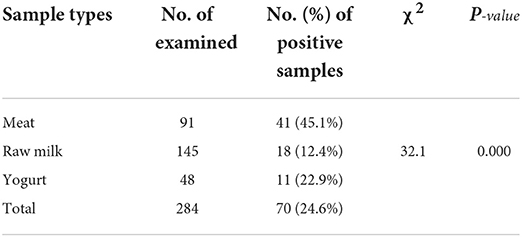
Out of the total 284 samples collected from the different sample sources, 70 (24.6%) were found to be positive for E. coli. The sample-based positivity of meat, raw milk, and yogurt samples were 41 (45.1%), 18 (12.4%), and 11 (22.9%), respectively. There was a significant difference (χ2 = 32.1; p = 0.000) among the different sample types in the occurrence of E. coli (Table 1). Of the total isolated E. coli isolates, 14.29% were found to be Shiga toxin-producing E. coli O157:H7.
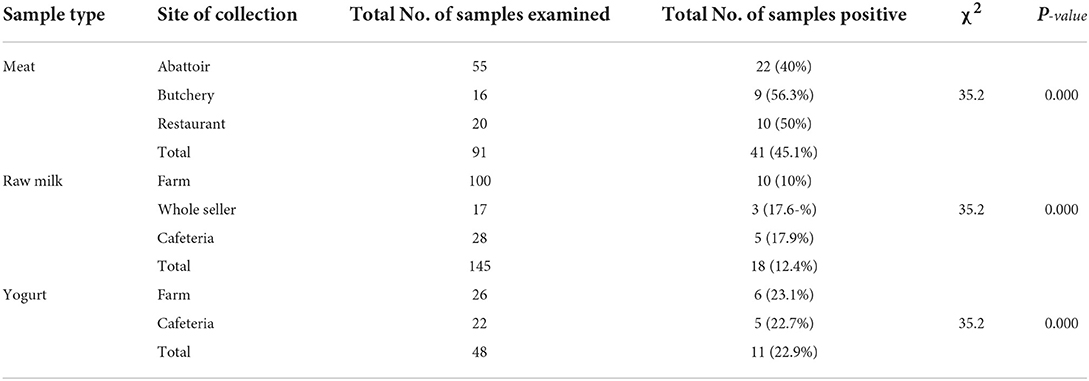
Occurrence rates of 56.3%, 40%, and 50% were recorded in meat samples collected from butchery, abattoir, and restaurant, respectively. The positivity of E. coli for raw milk samples collected from farms, whole-sellers, and cafeterias were 10, 17.6, and 17.9%, respectively, whereas yogurt samples collected from farms and cafeterias were found to be 23.1 and 22.7%, respectively. The chi-square analysis result revealed that all sample collection sources showed a significant difference (χ2= 35.2; p = 0.000) in E. coli positivity (Table 2).
PCR amplification results of the virulence genes of E. coli
The PCR result indicated that all (100%) of the tested isolates of E. coli carried the pal gene and 41.67% eaeA gene (EHEC). Among the eaeA gene-carried E. coli isolates, 40% carried the stx1 gene and 60% of them carried the stx2 gene.
Antimicrobial susceptibility profile of E. coli isolates
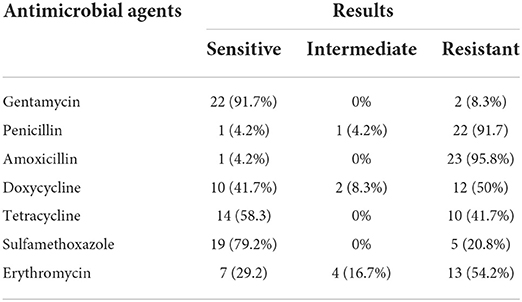
The E. coli strains isolated from meat, raw milk, and yogurt showed the highest level of susceptibility to gentamycin (91.7%). However, the highest level of resistance was observed against amoxicillin (95.8%) (Table 3). MDR was detected in 75% of the isolates.
Discussion
This study revealed an overall positivity rate of 24.6% for E. coli. This finding was in line with the reports of Bedasa et al. (57) (20%), Sebsibe and Asfaw (58) (20.2%), Messele et al. (59) (21.6%), Tadese et al. (60) (23.4%), Abebe et al. (61) (23.7%), Haileselassie et al. (10) (27.3%), and Haftu et al. (62) (27.3%). It was higher than the reports of Kumar et al. (63) (8.15%), Yakubu et al. (64) (9.23%), Atnafie et al. (65) (12.38%), Mohammed et al. (66) (15.89%), and Ababu et al. (67) (19%). However, it was lower than the findings of Disassa et al. (68) (33.9%), Thaker et al. (69) (38%), Negesse et al. (70) (46.26%), Balcha et al. (71) (61.7%), and Ali et al. (72) (63%). This study also revealed an overall positivity of 14.3% for Shiga toxin-producing E. coli O157:H7 (EHEC). This finding was almost in agreement with the reports of Bekele et al. (73) (10.2%), Abebe et al. (61) (10.4%), Mekuria and Beyene (74) (10.4%), Akomoneh et al. (75) (10.9%), Beyi et al. (76) (11.43%), Mohammadi et al. (77) (17.47%), and Balcha et al. (71) (18%). However, it was higher than the findings of Abdissa et al. (78) (0.54%), Yakubu et al. (64) (1.92%), Geresu and Regassa (79) (2.1%), Mohammed et al. (65) (2.4%), Carney et al. (80) (2.4%), Disassa et al. (68) (2.9%), Ahmed and Shimamoto (81) (3.38%), Bedasa et al. (57) (3.5%), Hiko et al. (82) (4.2%), Ababu et al. (67) (5.2%), Sebsibe and Asfaw (58) (5.4%), Gutema et al. (83) (6.3%), Vanitha et al. (84) (8.8%), Dutta et al. (85) (9%), Tadese et al. (60) (9.1%), and Haile et al. (86) (9.3%). However, it was lower than the finding of Garbaj et al. (87) (25%), Llorente et al. (88) (36.1%), Nehoya et al. (89) (41.66%), and Islam et al. (90) (69%). The variation might be due to differences in sample types of food of bovine origin, sources of the samples, methodological approaches, diagnostic techniques, calculation/interpretation, geographical locations, hygienic conditions, handling and transportation of samples, and contamination rates from utensils and personnel. Generally, in the present findings, the positivity of E. coli was higher in the samples collected from butchery, restaurants, cafeterias, and whole sellers other than the other sources. This might be due to food establishments in the study area being found to have poor sanitation and were not maintained well, specifically, poor repair condition of kitchens, improperly managed toilet facilities, inappropriate solid waste receptacles, lack of standard dishwashing compartments, and lack of handwashing lavatories in a large number of establishments (8).
The antimicrobial susceptibility test results of E. coli strains isolated from meat, raw milk, and yogurt showed the highest level of susceptibility for gentamycin (91.7%). It was almost in agreement with the reports of Ababu et al. (67) (100%), Bedasa et al. (57) (82.5%), and Hailu (91) (81.82%). The 79.2% isolates were susceptible to trimethoprim-sulfamethoxazole, which was in agreement with the reports of Hailu (91) (78.79%). However, the highest level of resistance was observed against amoxicillin (95.8%), which was in line with the reports of Tadese et al. (60) (100%) and Abdissa et al. (78) (100%). In addition, 41.7% of the isolates were resistant to tetracycline, which was in agreement with the reports of Messele et al. (59) (47.6%) but was higher than the findings of Carvalho et al. (92) (4%), Bekele et al. (73) (5.1%), Mohammadi et al. (77) (23.08%), Tadese et al. (60) (27.7%), and Mora et al. (93) (32%). However, it was lower than the findings of Ahmed and Shimamoto (81) (80.6%), Disassa et al. (68) (81.8%), Ababu et al. (67) (63.63%), and Hailu (91) (60.61%). This study also revealed that 20.8% of the isolates showed resistance against Sulfamethoxazole, which was lower than the findings of Mora et al. (93) (36%). In general, in this study, 75% of the isolates developed MDR, which was higher than the reports of Messele et al. (59) (46.0%) and Tadese et al. (60) (66.3%) but was lower than the report of Mekuria and Beyene (74) (93.2%). Antimicrobial resistance may arise either spontaneously by selective pressure or due to antimicrobial misuse by humans or overuse in the feeding or treatment of animals by farmers (94).
Conclusion
This study revealed a relatively high occurrence of E. coli as well as Shiga toxin-producing E. coli O157:H7 in food of bovine origin in Mekelle City and isolates developed a high level of MDR (a bacterium that is resistant to more than one antibiotic) to the antimicrobials tested. Hence, foods of bovine origin can serve as a potential vehicle for transmitting E. coli, in particular, Shiga toxin-producing E. coli O157:H7, and its presence indicates a serious public health hazard and gives a warning signal for the possible occurrence of a foodborne outbreak in humans through the consumption of raw or undercooked food items. As a result, both its presence and development of MDR should receive particular attention and is an alert for the concerned bodies. Therefore, a coordinated effort is needed to reduce or eliminate the risk posed by these pathogens at various stages in the food chains and on the appropriate use of antimicrobials both in veterinary and human treatment regimes. Moreover, awareness creation should be made on foodborne diseases caused by Shiga toxin-producing E. coli O157:H7 with due consideration to the safe handling and consumption of food of animal origin and selection and safe use of antimicrobials. In conclusion, a nationwide phenotypic and molecular characterization, in-depth typing, and drug-resistant gene identification of E. coli O157:H7 should be undertaken.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Institutional Ethical and Environmental Considerations Review Committee, College of Veterinary Sciences, Mekelle University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
GG, MW, YT, and NeA participated in conception and proposal development, sample collection, laboratory analysis, supervision, validation, data analysis, and write-up of the original draft and reviewing and editing of the final version of the manuscript. AK, MA, NiA, HT, and AB were involved in the conception and proposal development, data analysis, validation, write-up of the original draft, and reviewing and editing of the final version of the manuscript. All authors contributed to the final version of the manuscript and approved the submission.
Funding
This study was funded by the College of Veterinary Sciences, Mekelle University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
DNA, deoxyribonucleic acid; E. coli O157, Escherichia coli O157; E. coli, Escherichia coli; EAEC, Enteroaggregative E. coli; EHEC, Enterohemorrhagic E. coli; EIEC, Enteroinvasive E. coli; EPEC, Enteropathogenic E. coli; ETEC, Enterotoxigenic E. coli; ExPEC, Extraintestinal pathogenic E. coli; HUS, hemolytic-uremic syndrome; MDR, multidrug-resistant; PCR, polymerase chain reaction; STEC, Shiga toxin-producing E. coli; Stx, Shiga toxin; TTP, thrombotic thrombocytopenic purpura; VT, verotoxin; VTEC, verotoxin-producing E. coli.
References
1. FAO/WHO. Joint FAO/WHO Food Standards Programmed FAO/WHO Coordinating Committee for Africa, Seventeenth Session. Rabat: FAO/WHO (2007).
2. Ayalew H, Birhanu A, Asrade, B. Review on food safety system: Ethiopian perspective. Afr J Food Sci. . (2013) 7:431–40. doi: 10.5897/AJFS2013.1064
3. Yousuf AH M, Ahmed MK, Yeasmin S, Ahsan N, Rahman MM, Islam MM. Prevalence of microbial load in shrimp, Penaeus Monodon and Prawn, Macrobrachium Rosenbergii from Bangladesh. World J Agricultural Sci. (2008) 4:852–5.
4. Abebe E, Gugsa G, Ahmed M. Review on major food-borne zoonotic bacterial pathogens. J Trop Med. (2020) 2020:1–19. doi: 10.1155/2020/4674235
5. Sheikh SW, Ali A, Ahsan A, Shakoor S, Shang F, Xue T. Insights into emergence of antibiotic resistance in acid-adapted Enterohaemorrhagic Escherichia coli. Antibiotics. (2021) 10:522. doi: 10.3390/antibiotics10050522
6. Molla B, Alemayehu D, Salah W. Sources and distribution of Salmonella serotypes isolated from food animals, slaughterhouse personnel and retail meat products in Ethiopia: 1997-2002. Ethiop J Health Dev. (2003) 17:63–70. doi: 10.4314/ejhd.v17i1.9782
7. Kumie A, Mezene A, Amsalu A, Tizazu A, Bikila B. The sanitary condition of food and drink establishments in Awash-Sebat Kilo Town, Afar Region. Ethiop J Health Dev. (2006) 20:201–3. doi: 10.4314/ejhd.v20i3.46856
8. Zeru K, Kumie A. Sanitary conditions of food establishments in Mekelle Town, Tigray, North Ethiopia. Ethiop J Health Dev. (2007) 21:1–9. doi: 10.4314/ejhd.v21i1.10025
9. Woldemariam T, Asrat D, Zewde G. Prevalence of thermophilic Campylobacter species in carcasses from sheep and goats in an Abattoir in Debre-Zeit area, Ethiopia. Ethiopian J Health Dev. (2009) 23:229–33. doi: 10.4314/ejhd.v23i3.53245
10. Haileselassie M, Taddele H, Adhana K, Kalayou S. Food safety knowledge and practices of abattoir and butchery shops and the microbial profile of meat in Mekelle City, Ethiopia. Asian Pac J Trop Biomed. (2013) 3:407–12. doi: 10.1016/S2221-1691(13)60085-4
11. Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. (2004) 2:123–40. doi: 10.1038/nrmicro818
12. DiRita V. Enteric bacteria: secretory diarrhea. In:Engleberg, NC, DiRita V, Dermody TS, , editors. Mechanisms of Microbial Disease 4th ed. Baltimore, MD: Lippincott Williams and Wilkins (2007). p.190–9.
13. DiRita V. Invasive and tissue-damaging enteric bacterial pathogens: bloody diahhhea and dysentery. In:Engleberg, NC, DiRita V, Dermody TS, , ed. Mechanisms of Microbial Disease 4th ed. Baltimore, MD: Lippincott Williams & Wilkins (2007). p. 200–4.
14. Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. (1998) 11:142–201. doi: 10.1128/CMR.11.1.142
15. O'Brien AD, LaVeck GD, Thompson MR, Formal SB. Production of Shigella dysenteriae Type 1-Like Cytotoxin by Escherichia coli. J Infect Dis. (1982) 146:763–9. doi: 10.1093/infdis/146.6.763
16. Konowalchuk J, Speirs JI, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. (1977) 18:775–9. doi: 10.1128/iai.18.3.775-779.1977
17. Levine MM, Xu JG, Kaper JB, Lior H, Prado V, Tall B, et al. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. (1987) 156:175–82. doi: 10.1093/infdis/156.1.175
18. Constantiniu S. Emerging Shiga cytotoxin-producing E. coli. I. Enterohaemorrhagic E. coli O157: H7. Prev Med. (2002) 10:62–78.
19. Effler P, Isaacson M, Arntzen L, Heenan R, Canter P, Barrett T, et al. Factors contributing to the 50 emergence of E. coli O157 in Africa. Emerg Infect Dis. (2001) 7:812–9. doi: 10.3201/eid0705.017507
20. Kiranmayi CB, Krishnaiah N, Mallika EN. Escherichia coli O157:H7-an emerging pathogen in foods of animal origin. Vet World. (2010) 3:382–89. doi: 10.5455/vetworld.2010.382-389
21. Schmidt MA. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol. (2010) 12:1544–52. doi: 10.1111/j.1462-5822.2010.01518.x
22. Cobbaut K, Houf K, Buvens G, Habib I, De, ZL. Occurrence of non-sorbitol fermenting verocytotoxin-lacking Escherichia coli O157 on cattle farms. Vet Microbiol. (2009) 138:174–8. doi: 10.1016/j.vetmic.2009.02.008
23. Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior, et al. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. (1985) 151:775–82. doi: 10.1093/infdis/151.5.775
24. FDA. Bad Bug Book: Foodborne Pathogenic Microorganisms and Natural Toxins Handbook. 2nd ed. Silver Spring, MD: US Food and Drug Administration (2012).
25. Rahimi E, Kazemeini HR, Salajegheh M. E. coli O157:H7/NM prevalence in raw beef, camel, sheep, goat, and water buffalo meat in Fars and Khuzestan Provinces, Iran. Vet Res Forum. (2012) 3:13–7.
26. Bell BP, Goldoft M, Griffin PM, Davis MA, Gordon D. C., Tarr PI, et al. A multistate outbreak of E. coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA. (1994) 272:1349–53. doi: 10.1001/jama.272.17.1349
27. Dodson K, LeJeune J. E. coli O157:H7, Campylobacter jejuni, and Salmonella prevalence in cull dairy cows marketed in northeastern Ohio. J Food Prot. (2005) 68:927–31. doi: 10.4315/0362-028X-68.5.927
28. VSPA (World Society for the Protection of Animals). Resistant bacteria in people and farm animals around the world (2006).
29. Miles TD, McLaughlin W, Brown PD. Antimicrobial resistance of E. coli isolates from broiler chickens and humans. BMC Vet Res. (2006) 2:7. doi: 10.1186/1746-6148-2-7
30. Umolu PI, Ohenhen ER, Okwu IG, Ogiehor IS. Multiple antibiotic resistant index and plasmid of E. coli in Beef in Ekpoma. Am J Sci. (2006) 2:16–18.
31. Guardabassi l, Jensen LB, Kruse H. editors. Guide to Antimicrobial Use in Animals. Ames, IA: Blackwell Publishing (2008).
32. Rolain JM. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. (2013) 4:173. doi: 10.3389/fmicb.2013.00173
33. Zhao S, White DG, Ge B, Ayers S, Friedman S, English L, et al. Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl Environ Microbiol. (2001) 67:1558–64. doi: 10.1128/AEM.67.4.1558-1564.2001
34. Central Statistics Agency (CSA). Summary and statistical report of the 2007 population and housing census: population size by age and sex. Addis Ababa: Federal Democratic republic of Ethiopia population census commission. (2008).
37. Quinn PJ, Markey BK, Carter ME, Donnelly WJC, Leonard FC, Maguire D. Veterinary Microbiology and Microbial Disease. 1st ed. Oxford: Blackwell Science Ltd. (2002).
38. Merchant IA, Packer RA. Veterinary Bacteriology and Virology. 7th ed. Ames: IA: The Iowa State University Press (1969).
39. Eaton AD, Clesceri LS, Greenberg AE. Standard methods for the Examination of Water and Wastewater. 19th ed. Washington, DC: American Public Health Association (1995).
40. Jarvis CJ, Kellerman GE, Van Rensburg WJJ, Whitehead CJ. The Bacteriology Manual. 2nd ed. (1994).
41. Brenner DJ, Krieg NR, Staley JT, Garrity G. Bergey's Manual of Systematic Bacteriology. 2nd ed. New York, NY: Springer (2005).
42. MacFaddin JF. Biochemical Tests for Identification of Medical Bacteria. 3rd ed. Philadelphia, PA: Lippincott Williams and Wilkins (2000).
44. Snell EE, Wright LD. A microbiological method for the determination of nicotinic acid. J Biol Chem. (1941) 139:675–86. doi: 10.1016/S0021-9258(18)72940-2
45. Simmons J. Standard Methods for the Examination of Water and Wastewater. 11th ed. New York, NY: APHA Inc. (1960).
46. Chakraborty SP, Mahapatra SK, Roy S. Biochemical characters and antibiotic susceptibility of S. aureus isolates. Asian Pac J Trop Biomed. (2011) 1:212–6. doi: 10.1016/S2221-1691(11)60029-4
47. Vanderzant C, Splittstresser DF. Compendium of Methods for the Microbiological Examination of Foods. 3rd ed. Washington, DC: American Public Health Association (1992).
48. Kuhnert P, Nocolet J, Frey J. Rapid and accurate identification of E. coli K-12 strains. Appl Environ Microbiol. (1995) 61:4135–9. doi: 10.1128/aem.61.11.4135-4139.1995
49. Ellingson JLE, Kozickkpwski JJ, Anderson JF, Carison SA, Sharma VK. Rapid PCR detection of enterohaemorrhagic E. coli (EHEC) in bovine food products and feces. Mol Cell Probes. (2005) 19:213–7. doi: 10.1016/j.mcp.2005.01.004
50. Paton AW, Paton JC. Detection and characterization of Shiga toxigenic E. coli by using multiplex PCR assays for stx1, stx2, eaeA, Enterohemorrhagic E. coli htyA, rfbO111, and rfbO157. J Clin Microbiol. (1998) 36:598–602. doi: 10.1128/JCM.36.2.598-602.1998
51. Fitzmaurice J. Molecular Diagnostic Assay for E. coli O157:H7. (Ph.D. thesis). Department of Microbiology, National University of Ireland, University College Galway: Ireland (2003).
52. Parekh TS, Subhash R. Molecular and bacteriological examination of milk from different milk animals with special reference to coliforms. Curr Res Biotech. (2008) 1:56–63. doi: 10.3923/crb.2008.56.63
53. Wang G, Clark CG, Rodgers FG. Detection in E. coli of the genes encoding the major virulen-ce factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. (2002) 40:3613–19. doi: 10.1128/JCM.40.10.3613-3619.2002
54. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. New York, NY: Cold Spring Harbour Laboratory Press (1989).
55. Bauer AW, Kirby WMM, Sherries JC, Turck M. Antibiotic susceptibility testing by a standard single disc method. Am J Clin Pathol. (1966) 45:493–6. doi: 10.1093/ajcp/45.4_ts.493
56. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. Wayne, PA: CLSI document M100-S24 (2014).
57. Bedasa S, Shiferaw D, Abraha A, Moges T. Occurrence and antimicrobial susceptibility profile of Escherichia coli O157:H7 from food of animal origin in Bishoftu town, Central Ethiopia. Int J Food Contam. (2018) 5:2. doi.org/10.1186/s40550-018-0064-3. doi: 10.1186/s40550-018-0064-3
58. Sebsibe MA, Asfaw ET. Occurrence of multi-drug resistant Escherichia coli and Escherichia coli O157:H7 in meat and swab samples of various contact surfaces at abattoir and butcher shops in Jimma Town, Southwest District of Ethiopia. Infect Drug Resist. (2020) 13:3853–62. doi: 10.2147/IDR.S277890
59. Messele YE, Abdi RD, Yalew ST, Tegegne DT, Emeru BA, Werid GM. Molecular determination of antimicrobial resistance in Escherichia coli isolated from raw meat in Addis Ababa and Bishoftu, Ethiopia. Ann Clin Microbiol Antimicrob. (2017) 16:55. doi: 10.1186/s12941-017-0233-x
60. Tadese ND, Gebremedhi EZ, Moge, F, Borana BM, Marami LM, et al. Occurrence and antibiogram of Escherichia coli O157:H7 in raw beef and hygienic practices in abattoir and retailer shops in Ambo Town, Ethiopia. Vet Med Int. (2021) 2021:8846592. doi: 10.1155/2021/8846592
61. Abebe M, Hailelule A, Abrha B, Nigus A, Birhanu M, Adane, et al. Antibiogram of E. coli strains isolated from food of bovine origin in selected Woredas of Tigray Ethiopia. J Bacteriol Res. (2014) 6:17–22. doi: 10.5897/JBR2014.0126
62. Haftu R, Taddele H, Gugsa G, Kalayou S. Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large scale dairy farms of Northern Ethiopia. Trop Anim Health Pro. (2012) 44:1765–71. doi: 10.1007/s11250-012-0135-z
63. Kumar R, Prasad A. Detection of E. coli and Staphylococcus in milk and milk products in and around Pantnagar. Vet World. (2010) 3:495–6. doi: 10.5455/vetworld.2010.495-496
64. Yakubu Y, Shuaibu AB, Ibrahim AM, Hassan UL, Nwachukwu RJ. Risk of Shiga toxigenic Escherichia coli O157:H7 infection from raw and fermented milk in Sokoto Metropolis, Nigeria. J Pathol. (2018) 2018:1–5. doi.org/10.1155/2018/8938597. doi: 10.1155/2018/8938597
65. Atnafie B, Paulos D, Abera M, Tefera G, Hailu, D, et al. Occurrence of Escherichia coli O157:H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol. (2017) 17:24. doi: 10.1186/s12866-017-0938-1
66. Mohammed O, Shimelis D, Admasu P, Feyera T. Prevalence and antimicrobial susceptibility pattern of E. coli isolates from raw meat samples obtained from abattoirs in Dire Dawa City, Eastern Ethiopia. Int J Microbiol Res. (2014) 5:35–9. doi: 10.5829/idosi.ijmr.2014.5.1.82306
67. Ababu A, Endashaw D, Fesseha H. Isolation and antimicrobial susceptibility profile of Escherichia coli O157:H7 from raw milk of dairy cattle in Holeta District, Central Ethiopia. Int J Microbiol. (2020) 2020:1–8. doi: 10.1155/2020/6626488
68. Disassa N, Sibhat B, Mengistu S, Muktar Y, Belina D. Prevalence and antimicrobial susceptibility pattern of E. coli O157:H7 isolated from traditionally marketed raw cow milk in and around Asosa Town, Western Ethiopia. Vet Med Int. (2017) 2017:1–7. doi: 10.1155/2017/7581531
69. Thaker HC, Brahmbhatt MN, Nayak JB. Study on occurrence and antibiogram pattern of Escherichia coli from raw milk samples in Anand, Gujarat, India. Vet World. (2012) 5:556–9. doi: 10.5455/vetworld.2012.556-559
70. Negesse W, Fufa A, Bihonegn W. Isolation, identification and antimicrobial susceptibility profiles of E. coli O157:H7 from raw cow milk in and around Modjo Town, Ethiopia. J A Sci. (2020) 16:62–9.
71. Balcha E, Kumar A, Tassew H. Evaluation of safety of beef sold in and around Mekelle with special reference to Enterohemorrhagic Escherichia coli O157:H7. Glob Vet. (2014) 12:569–72. doi: 10.5829/idosi.gv.2014.12.04.8375
72. Ali AA, Abdelgadir WS. Incidence of Escherichia coli in raw cow's milk in Khartoum State. Br J Dairy Sci. (2011) 2:23–6.
73. Bekele T, Zewde G, Tefera G, Feleke A, Zerom K. Escherichia coli O157:H7 in raw meat in Addis Ababa, Ethiopia: Prevalence at an abattoir and retailers and antimicrobial susceptibility. Int J Food Contam. (2014) 1:4. doi: 10.1186/s40550-014-0004-9
74. Mekuria A, Beyene T. Zoonotic bacterial pathogens isolated from food of bovine in selected Woredas of Tigray, Ethiopia. World Appl Sci J. (2014) 31:1864–68.
75. Akomoneh EA, Esemu SN, Kfusi JA, Ndip RN, Ndip LM. Prevalence and virulence gene profiles of Escherichia coli O157 from cattle slaughtered in Buea, Cameroon. PLoS ONE. (2020) 15:e0235583. doi: 10.1371/journal.pone.0235583
76. Beyi AF, Fite AT, Tora E, Tafese A, Genu T, Kaba T, et al. Prevalence and antimicrobial susceptibility of Escherichia coli O157 in beef at butcher shops and restaurants in central Ethiopia. BMC Microbiol. (2017) 17:49. doi: 10.1186/s12866-017-0964-z
77. Mohammadi P, Abiri R, Rezaei M, Salmanzadeh-Ahrabi S. Isolation of Shiga toxin-producing Escherichia coli from raw milk in Kermanshah, Iran. Iran J Microbiol. (2013) 5:233–8.
78. Abdissa R, Haile W, Fite AT, Beyi AF, Agga GE, Edao BM, et al. Prevalence of Escherichia coli O157:H7 in beef cattle at slaughter and beef carcasses at retail shops in Ethiopia. BMC Infect Dis. (2017) 17:277. doi: 10.1186/s12879-017-2372-2
79. Geresu MA, Regassa S. Escherichia coli O157:H7 from food of animal origin in Arsi: Occurrence at catering establishments and antimicrobial susceptibility profile. Sci World J. (2021) 2021:1–10. 6631860. doi: 10.1155/2021/6631860
80. Carney E, O'Brien SB, Sheridan JJ, McDowell DA, Blair IS, Duffy G. Prevalence and level of Escherichia coli O157 on beef trimmings, carcasses and boned head meat at a beef slaughter plant. Food Microbiol. (2006) 23:52–9. doi: 10.1016/j.fm.2004.12.001
81. Ahmed AM, Shimamoto T. Molecular analysis of multidrug resistance in Shiga toxin-producing Escherichia coli O157:H7 isolated from meat and dairy products. Int J Food Microbiol. (2015) 193: 68–73. doi: 10.1016/j.ijfoodmicro.2014.10.014
82. Hiko A, Asrat D, Zewde G. Occurrence of Escherichia coli O157:H7 in retail raw meat products in Ethiopia. J Infect Dev Ctries. (2008) 2:389–93. doi: 10.3855/jidc.203
83. Gutema FD, Rasschaert G, Agg, GE, Jufare A, Duguma AB, et al. Occurrence, molecular characteristics, and antimicrobial resistance of Escherichia coli O157 in cattle, beef, and humans in Bishoftu Town, Central Ethiopia. Foodborne Pathog Dis. (2021) 18:2830. doi: 10.1089/fpd.2020.2830
84. Vanitha HD, Sethulekshmi C, Latha C. An epidemiological investigation on occurrence of enterohemorrhagic Escherichia coli in raw milk. Vet World. (2018) 11:1164–70. doi: 10.14202/vetworld.2018.1164-1170
85. Dutta S, Deb A, Chattopadhyay UK, Tsukamoto T. Isolation of Shiga toxin-producing Escherichia coli including O157:H7 strains from dairy cattle and beef samples marketed in Calcutta, India. J Med Microbiol. (2000) 49:765–7. doi: 10.1099/0022-1317-49-8-765
86. Haile AF, Kebede D, Wubshe AK. Prevalence and antibiogram of Escherichia coli O157 isolated from bovine in Jimma, Ethiopia: Abattoir based survey. Ethiop Vet J. (2017) 21:109–20. doi: 10.4314/evj.v21i2.8
87. Garbaj AM, Awad EM, Azwai SM, Abolghait SK, Naas HT, Moawad AA, et al. Enterohemorrhagic Escherichia coli O157 in milk and dairy products from Libya: isolation and molecular identification by partial sequencing of 16S rDNA. Vet World. (2016) 9:1184–9. doi: 10.14202/vetworld.2016.1184-1189
88. Llorente P, Barnech L, Irino K, Rumi MV, Bentancor A. Characterization of Shiga toxin-producing Escherichia coli isolated from ground beef collected in different socioeconomic strata markets in Buenos Aires, Argentina. Biomed Res Int. (2014) 2014:795104. doi: 10.1155/2014/795104
89. Nehoya KN, Hamatui N, Shilangale RP, Onywera H, Kennedy J, Mwapagha LM. Characterization of Shiga toxin-producing Escherichia coli in raw beef from informal and commercial abattoirs. PLoS One. (2020) 15:e0243828. doi: 10.1371/journal.pone.0243828
90. Islam MA, Mondol AS, Azmi IJ, de Boer E, Beumer RR, Zwietering MH, et al. Occurrence and characterization of Shiga toxin-producing Escherichia coli in raw meat, raw milk, and street vended juices in Bangladesh. Foodborne Pathog Dis. (2010) 7:1381–5. doi: 10.1089/fpd.2010.0569
91. Hailu S. Isolation, identification and antibiotic susceptibility of E. coli from diarrheic calves in and around Holeta Town, Central Ethiopia. J Vet Med Res. (2020) 7:1197. doi: 10.47739/2378-931X/1197
92. Carvalho D, Kunert-Filho HC, Simoni C, de Moraes LB, Furian TQ, Borges KA, et al. Antimicrobial susceptibility and detection of virulence-associated genes of Escherichia coli and Salmonella spp. isolated from domestic pigeons (Columba livia) in Brazil. Folia Microbiol. (2020) 65:735–45. doi: 10.1007/s12223-020-00781-w
93. Mora A, Blanco JE, Blanco M, Alonso MP, Dhabi G, Echeita A, et al. Antimicrobial resistance of Shiga toxin (verotoxin)-producing Escherichia coli O157:H7 and non-O157 strains isolated from humans, cattle, sheep and food in Spain. Res Microbiol. (2005) 156:793–806. doi: 10.1016/j.resmic.2005.03.006
Keywords: antimicrobial, bacteriological, bovine, E. coli O157:H7, food, Mekelle, molecular
Citation: Gugsa G, Weldeselassie M, Tsegaye Y, Awol N, Kumar A, Ahmed M, Abebe N, Taddele H and Bsrat A (2022) Isolation, characterization, and antimicrobial susceptibility pattern of Escherichia coli O157:H7 from foods of bovine origin in Mekelle, Tigray, Ethiopia. Front. Vet. Sci. 9:924736. doi: 10.3389/fvets.2022.924736
Received: 20 April 2022; Accepted: 26 October 2022;
Published: 17 November 2022.
Edited by:
Gabriel Arriagada, Universidad de O'Higgins, ChileReviewed by:
Kálmán Imre, Banat University of Agricultural Sciences and Veterinary Medicine, RomaniaFarhad Safarpoor Dehkordi, University of Tehran, Iran
Copyright © 2022 Gugsa, Weldeselassie, Tsegaye, Awol, Kumar, Ahmed, Abebe, Taddele and Bsrat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Getachew Gugsa, Z3Vnc2FnQHlhaG9vLmNvbQ==
 Getachew Gugsa
Getachew Gugsa Million Weldeselassie2
Million Weldeselassie2