- 1Department of Veterinary Science, Institute for Hygiene and Infectious Diseases of Animals, Justus Liebig University, Giessen, Germany
- 2Tierärztliche Klinik für Kleintiere am Kaiserberg, Duisburg, Germany
- 3Institute of Bacterial Infections and Zoonoses, Friedrich-Loeffler-Institute, Federal Research Institute for Animal Health, Jena, Germany
- 4Department of Bacteriology, Host-Pathogen Interaction and Diagnostics, Wageningen Bioveterinary Research, Lelystad, Netherlands
In 2021, a case of canine brucellosis diagnosed in a dog with orchitis was presented to a veterinary practice in Germany. Serological testing excluded Brucella (B.) canis as a causative agent, but molecular analysis revealed the presence of B. suis biovar 1. Since biovar 1 is not endemic in Europe and the dog had no history of travel to endemic areas, a comprehensive epidemiological investigation was conducted using whole genome sequence data to determine the source of infection. We describe the clinical progress of the animal and the potential infection of a veterinary clinic employee. The findings highlight the importance of considering less common Brucella species as possible causes of canine brucellosis. The data also emphasize that it is quite challenging to identify Brucella species in a routine diagnostic laboratory and to conduct epidemiological investigations to unveil possible transmission routes.
1. Introduction
Brucellosis primarily manifests in the reproductive system of various terrestrial and aquatic mammals. It has been associated with abortion and other reproductive as well as chronic disorders, thus inflicting significant economic losses and posing a threat to human health due to its zoonotic potential (1). It is caused by the genus Brucella, a coccoid, Gram-negative, aerobic to microaerophilic rod that is slow-growing and requires complex nutrients for culturing (2). Of the six classical Brucella species, namely, B. melitensis (sheep and goats), B. abortus (cattle), B. suis (swine, hares, and reindeer), B. canis (dogs), B. ovis (sheep), and B. neotomae (rodents), only the first four are well-known relevant terrestrial zoonotic pathogens (3–5). For humans, B. melitensis is considered the most pathogenic species, followed by B. suis, B. abortus, and B. canis (5–10).
Canine brucellosis is most commonly caused by B. canis and is mainly noticed by infertility and reproductive failure in dog breeding kennels (11, 12). It has a high affinity for the testes, epididymides, prostate, and uterus but also colonizes the lymphatic organs, eyes, and spinal column. Typical clinical signs in females are infertility in form of conception failure, embryo resorption, late-time abortion, and vaginal discharge after abortion or parturition; in males, epididymitis, orchitis, or prostatitis may be noticed. Uveitis, discospondylitis, and meningitis may occur as symptoms independent of the reproductive tract or as long-term effects (12–15). The main path of infection is sexual transmission (11, 16). A study concerning the incidence of B. canis in Europe between 2011 and 2016 detected 5.4% of serologically positive animals. However, only samples from dogs with suspected B. canis infections were tested. Therefore, the actual incidence of canine brucellosis due to B. canis in Europe remains unknown (17).
In addition to B. canis, the causal agents of canine brucellosis have also been sporadically identified as B. suis, B. abortus, and B. melitensis (18). Concerning the five biovars of B. suis, biovars (bv) 1–3 are the predominant cause of porcine brucellosis and manifest in the reproductive tissues, causing reproductive disorders and infertility (19). Bv 1 exhibits the highest zoonotic potential and virulence concerning humans and pigs (8, 20): In regions where B. suis bv 1 is endemic, such as Australia or the United States, infection of humans and dogs associated with hunting or feeding raw meat of feral pigs/wild hogs has been documented (21–27). While B. suis bv 1 is most prevalent in Southern Asia, North and South America, and Oceania, B. suis bv 2 is present in European brown hare populations in Europe and frequently found in wild boars and domestic pigs (28, 29). Unlike bv 1, bv 2 rarely infects humans (30).1
In Europe, there are few reports of B. suis bv 1 infections in humans (20, 31–34); in dogs, infection with B. suis bv 1 was reported in Berlin, Germany, in 1978 and in the Netherlands in 2016, both presumably caused by consumption of uncooked meat (32, 33).
The objective of this report is to provide a comprehensive account of the clinical presentation, microbiological characteristics, and molecular analysis of a case of canine brucellosis caused by B. suis bv 1 in Germany, including investigations of the source by molecular typing.
2. Methods
2.1. Case history
In July 2021, a 2-year-old male intact Rhodesian Ridgeback was presented to a primary care veterinary practice in North Rhine-Westphalia for apathy and inappetence. The dog showed lameness, a reluctance to lie down, and bilateral painful swollen testes. Suspecting an infective orchitis, the dog was treated with amoxicillin/clavulanic acid (12.5 mg/kg bid). After 2 days of antibiotic therapy, the dog was neutered because clinical signs did not improve. At surgery, purulent exudation from the testes and a thickened spermatic cord were noted. Antibiotic therapy was modified by administering amoxicillin/clavulanic acid (20 mg/kg bid) and marbofloxacin (2 mg/kg qd).
Four days after the dog was neutered, it was referred to a veterinary clinic due to fever, vomiting, and weight loss. Abdominal sonography revealed ascites and an enlarged, inhomogeneous prostate. Blood analysis resulted in leukocytosis (28/μl), neutrophilia (21.56/μl), monocytosis (4.38/μl), hypoalbuminemia (2.2 g/dl), hypochloremia (105 mg/dl), and a high level of C-reactive protein (0.85 mg/dl). Analysis of urine showed high levels of leukocytes, erythrocytes, protein (30 mg/dl), high specific gravity (1050), and presumably the presence of coccoid bacteria. The ascitic fluid was revealed to be a septic exudate with a specific gravity of 1,026, total protein of 30 g/dl, cell count of 107,200 cells/μl, the presence of phagocytosed intracellular coccoid bacteria, activated mesothelial cells, and degenerated neutrophils. During the following diagnostic laparotomy the next day, 2 L of a brownish exudate was collected, peritonitis was noted throughout the abdominal cavity, and multiple abscesses were found in the left spermatic cord. Subsequently, the left spermatic cord was resected, and abdominal lavage was performed. Following surgical intervention, a tentative diagnosis of brucellosis was established, and the dog was placed in the isolation ward. Antibiotic treatment was adjusted to trimethoprim/sulfamethoxazole (5 mg/kg qd) and doxycycline (10 mg/kg qd). Clinical symptoms improved, and the dog had a full recovery within 7 days.
2.2. Sampling
During neutering at the primary care veterinary practice, purulent exudate from the testes and spermatic cord was collected for microbiological examination. This sample was sent to the Institute of Hygiene and Infectious Diseases of Animals at the Justus Liebig University in Giessen, Germany (IHIT). In addition, during a posterior laparotomy performed at the veterinary clinic, ascitic fluid and abscess material were collected. These samples were sent to an external diagnostic laboratory for microbiological analysis. Blood samples taken during laparotomy were sent to the IHIT for serological testing for B. canis.
To determine the source of infection, the local veterinary department obtained samples of the dogs' feed provided by the owners, a commercially available raw meat diet consisting of beef, horse, salmon (origin unknown), and kangaroo sourced from Australian farms. Only salmon and kangaroo meat were available at the time of the investigation and were subsequently subjected to testing for Brucella species at the corresponding Chemical and Veterinary Analytical Institute.
2.3. Microbiological investigation
For initial microbiological analysis, the sample was streaked on standard nutrient agar (Oxoid, Wesel, Germany) containing 5% defibrinated sheep blood (blood agar) and on water-blue metachrome-yellow lactose agar, according to Gassner (Sifin Diagnostics GmbH, Berlin, Germany). The plates were incubated at 37°C for 48 h in ambient air. For bacterial enrichment, the sample was cultivated for 24 h at 37°C in standard I nutrient broth (E. Merck KG, Darmstadt, Germany) and streaked on 5% sheep blood agar and Gassner agar. For the detection of microaerophilic bacteria, the sample was additionally streaked on brain-heart infusion agar (Oxoid) and incubated for 5 days in 10% CO2 at 37°C. Schaedler agar (Becton Dickinson GmbH, Heidelberg, Germany) and Zeissler agar (E. Merck KG) were incubated for 72 h at 37°C under anaerobic conditions in a jar using the AnaeroGen™ gas sachets (Oxoid). After 48 h, abundant growth (>200) of small, non-hemolytic, shiny colonies was evident on blood agar. Similar colonies were also observed on brain-heart-infusion agar. Identification with matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS, Bruker Daltonics, Bremen, Germany) using the standard MBT Compass reference library (version 10.0.0.0) and the Security-Relevant (SR) Library was performed.
For further identification, the isolate was sent to the reference laboratory of the World Organization for Animal Health (WOAH) and National Reference Laboratory (NRL) for brucellosis in animals, the Institute of Bacterial Infections and Zoonosis at the Friedrich-Loeffler-Institut, Jena, Germany. Pathogen identification and biovar typing were conducted by the so-called Bruce-Ladder and New Bruce-Ladder polymerase chain reaction (PCR) (35, 36).
The dogs' feed was analyzed using cultural detection methods and real-time multiplex PCR according to the published protocols for the detection of brucellosis in cattle, pigs, sheep, and goats of the NRL (37).
2.4. Serological investigation
Serological testing for B. canis was performed with B. canis-specific antigens (in-house antigen) using the tube agglutination test (TAT) at the IHIT. In addition, serological tests [slide agglutination test (SAT), complement fixation test, and Rose Bengal Test] with B. abortus antigens (IDEXX Montpellier SAS, Montpellier, France) cross-reacting for B. abortus, B. suis, and B. melitensis were performed at the NRL for brucellosis in animals.
2.5. Genomic characterization
2.5.1. DNA isolation and whole genome sequencing
DNA was extracted from pure cultures using the High Pure PCR Template Preparation Kit (Roche Molecular Systems, Pleasanton, CA, United States). For short-read sequencing by Illumina technology, a genomic library was prepared using the NexteraXT kit (Illumina Inc., San Diego, CA, United States), which was sequenced on a MiSeq (Illumina Inc., San Diego, CA, United States) in paired-end mode. For obtaining a closed genome, the DNA was additionally sequenced by Nanopore technology on a MinION Mk1B device (Oxford Nanopore Technologies Ltd., Oxford, England). The corresponding genomic library was prepared with the Ligation Sequencing kit (SQK-LSK109) and barcoded using the EXP-NBD 104 kit (Oxford Nanopore Technologies Ltd., Oxford, England). This library was run on a R9.4.1 flow cell for 24 h.
2.5.2. De novo assembly and annotation
Reads from both Illumina and Nanopore technologies were used for combined de novo genome assembly using microPIPE (38) with basecalling in super-accuracy mode. The assembly quality statistics were assessed using QUAST version 5.0.2 (39), and the annotation was carried out using Bakta version 1.6.1 with database version 4.0 (40).
2.5.3. Genome comparison and genotyping
For determining the origin of the isolate, NCBI's Sequence Read Archive (SRA) and RefSeq genome databases were browsed (accessed in January 2023) for B. suis bv 1 sequences (Supplementary Table 1). Furthermore, MLVAbank [https://microbesgenotyping.i2bc.paris-saclay.fr/; accessed February 9, 2023 (41)] was searched for multiple locus variable number of tandem repeats analysis (MLVA) profiles with similarity to the investigated isolate displaying a maximum of three alleles difference.
The average nucleotide identity of the newly assembled genome compared to other B. suis bv 1 strains deposited in the NCBI RefSeq database was assessed using fastANI version 1.1 (42). To exclude that the isolate was identical to the B. suis bv 1 vaccine strain S2, an in silico PCR using a script by Egon A. Ozer (version 0.5.1) (https://github.com/egonozer/in_silico_pcr) was conducted with primers IclRP1 (5′-TGGCAAGAGCGGTTTCAG-3′) and IclRP2 (5′-TCCAAGGTCGGCTACGAA-3′) (43). In silico MLVA was carried out using the MISTReSS (https://github.com/Papos92/MISTReSS) as described by Sacchini et al. (44). Based on the differences in alleles, a minimum spanning tree was calculated using GrapeTree version 1.0 (45) with the implemented MSTreeV2 algorithm. In addition, core genome multilocus sequence typing (cgMLST) using Ridom Seqsphere+ version 7.7 (46) and the scheme by Abdel-Glil et al. (47) was conducted. Foreign strains for which exclusively raw reads have been deposited on NCBI were assembled using Shovill version 1.0.4 (https://github.com/tseemann/shovill) with the SPAdes assembler and the option “- - trim”. CgMLST allelic distances were used for the calculation of a minimum spanning tree as implemented in Ridom Seqsphere+. Typing based on core genome single nucleotide polymorphisms (cgSNPs) was performed by using Snippy version 4.6.0 (https://github.com/tseemann/snippy) using B. suis bv 1 strain 1330 (GCF_000223195.1) as reference. In this analysis, SRA data was included. The cgSNP alignment was used for the calculation of a phylogenetic maximum likelihood tree using RAxML version 8.2.12 (48) with the GTRGAMMA model. The tree was visualized using FigTree version 1.4.3 (http://tree.bio.ed.ac.uk/soft-ware/figtree/).
3. Results
3.1. Microbiological analysis
Initial identification of the isolate, denominated 21RB23181, with MALDI-TOF-MS revealed Brucella melitensis, yielding a score of 2.39. Although species identification must be considered questionable due to the fact that the commercial database only provides reference spectra for B. melitensis, it has to be assumed that genus identification is correct. Therefore, after differentiation as Brucella sp., the results were immediately forwarded to the local veterinary department, and the sample was handled in compliance with official protective measures (49, 50). The isolate was tested by the NRL for brucellosis in animals by PCR, which identified 21RB23181 as a B. suis bv 1 strain.
The sample of ascitic fluid and abscess material sent to an external diagnostic laboratory did not result in the detection of Brucella sp. Similarly, the remaining samples of salmon and kangaroo meat obtained by the local veterinary department tested negative for Brucella sp.
3.2. Serological analysis
Serological testing of a blood sample for B. canis using the TAT assay yielded a negative result (<40 IU/ml). However, when tested with antigens specific to the B. melitensis, B. abortus, and B. suis group, the sample showed a positive result (SAT 841 IU/ml, complement fixation test 1,189 SensE/ml, and positive in the Rose Bengal test).
3.3. Genomic analysis
3.3.1. Genome characterization and similarity
Using a combined assembly approach of Illumina and Nanopore reads, the genome of 21RB23181 could be assembled to completion at a mean coverage of 271×. The genome consisted of two circular contigs of 2,107,952 and 1,207,151 bp with an average GC content of 57.25% and 3,113 predicted coding sequences.
The highest ANI values were observed for B. suis bv 1 strains Human/AR/US/1981 (99.9958% ANI) and VBI22 (99.9949% ANI), both isolated in the United States, and vaccine strain S2 (99.9946% ANI) isolated in China. In silico PCR yielded a negative result for the S2-specific primer pair, thus ruling out the possibility that 21RB23181 was the vaccine strain.
3.3.2. Genotyping using allele-based methods
Although WGS offers the possibility to compare strains at the nucleotide level, the lack of genome sequences from reported isolates necessitates resorting to comparing MLVA profiles, despite the lower resolution. Profiles published in MLVAbank and in literature (see Supplementary Table 2) were included. Remarkably, in the resulting minimum spanning tree (Figure 1), no clustering of strains according to their origin could be observed. Especially strains from Croatia, the United States, and Argentina were scattered in the tree. With the exception of one allele, the MLVA profile of 21RB23181 matched that of a strain isolated in Denmark in 1987 (BCCN#87-85) and strain B93-0078, which originated from cattle in the United States in 1993. Despite the closer geographic location, the distance to Brucella isolates from a dog (WBVR_2016) and a hare in the Netherlands (WBVR_2017) displayed higher differences with four differing alleles each.
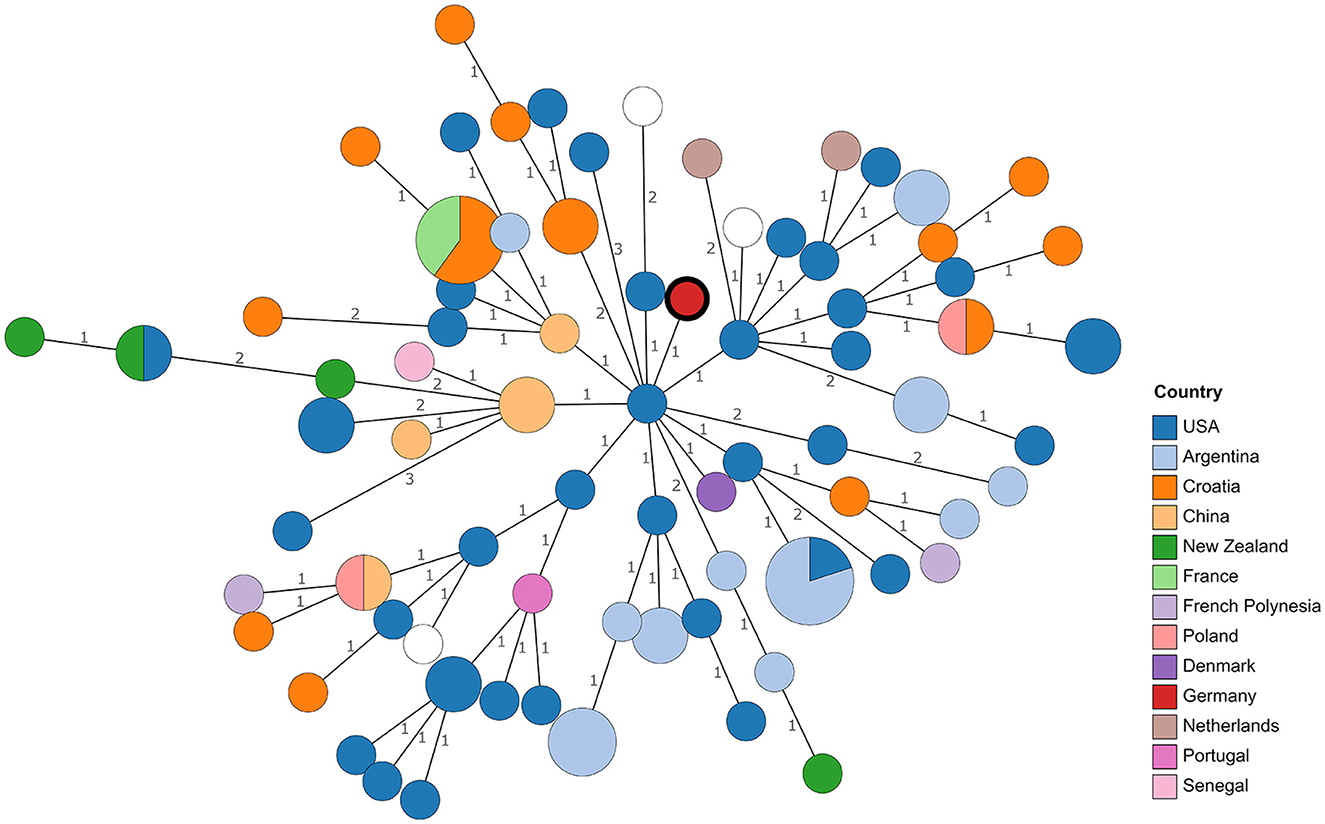
Figure 1. Minimum spanning tree based on MLVA profile differences. Numbers on branches indicate allele differences. Leaves are colored according to the origin of the strains. The leaf representing isolate 21RB23181 has a bold margin. For empty leaves, the origin is unknown.
In the cgMLST analysis, 21RB23181 exhibited at least 31 allele differences compared to other B. suis bv 1 strains (Figure 2). The strain VBI22 isolated from cattle in the United States displayed the highest degree of similarity. In this analysis, a B. suis bv 1 isolate from a human brucellosis case in Germany in 2018 (Bw_180660) was also included. However, it displayed 45 allele differences from the German dog isolate, so no connection between these two cases could be inferred.
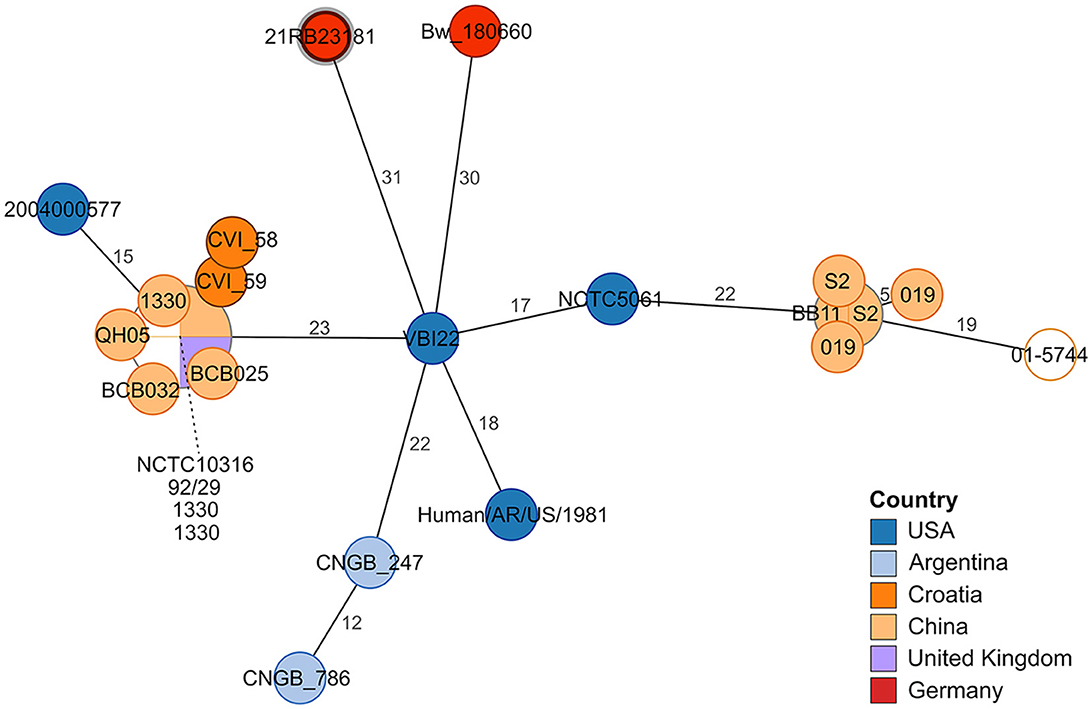
Figure 2. Minimum spanning tree based on allelic distances determined by cgMLST analysis. The numbers on the branches indicate allele distances. The leaves are colored according to the strain's origin. The leaf representing isolate 21RB23181 has a bold margin. For better readability, the names of clustering leaves are connected to the leaves by a dashed line. For empty leaves, the origin is unknown.
3.3.3. Genotyping using SNPs
In a cgSNP approach (Figure 3), 21RB23181 was compared to B. suis bv 1 isolates for which raw read data were available (Supplementary Table 1). In total, 476 core genome SNPs were called. The isolate 21RB23181 was most similar to strains originating from Tonga, with the highest SNP identity to a strain isolated in 1979 from a human in Tonga (69 SNPs). However, differences between strains from the United States and Argentina were only slightly higher, ranging between 77 and 96 SNPs. Based on this result, the genomes of the strains from Tonga were assembled using the raw sequencing data, and cgMLST analysis was repeated (Supplementary Figure 1). The allelic profiles of two strains from Tonga exhibited higher concordance with 21RB23181 than the US American strain VBI22, differing in 27 alleles, but still, this difference is not much smaller than that of the US American strain. These strains from Tonga are partially also represented in the MLVA tree; however, in the MLVAbank database, their origin is given as New Zealand (B13-0234, B13-0236, B13-0237, and B13-0239). In the MLVA, 21RB23181 differs in three alleles from the strains originating from Tonga.
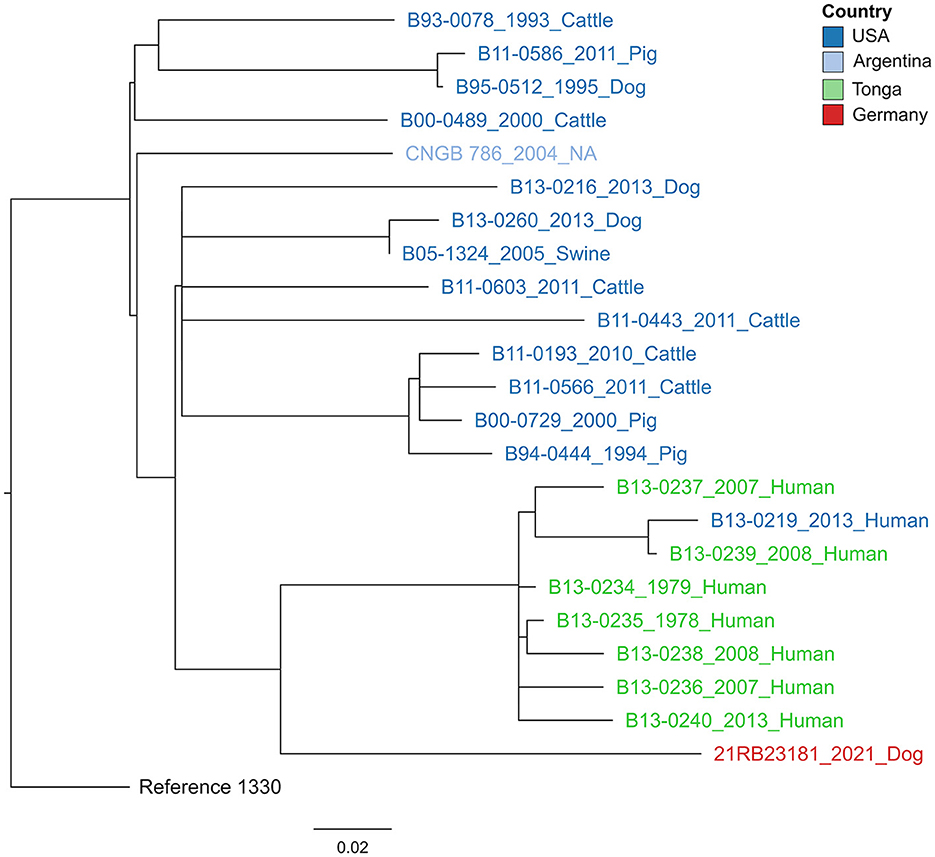
Figure 3. Maximum likelihood tree based on core genome SNP alignment. The tree is rooted in the reference strain B. suis bv 1 strain 1330. The colors of the labels give the origin of the isolates. The year of isolation and host are given after the strain names. The bar indicates the number of base substitutions per site.
3.4. Feasible measures concerning possible transmission
Immediately after the detection of Brucella sp., the zoonotic risk for the involved veterinary practices and the microbiological laboratory was assessed. The employees with increased risk due to close contact with the animal or handling the microbiological sample and the pure culture before identification of the isolate were informed and subjected to serological control. One staff member of the primary care veterinary practice who had been in close contact with the animal showed a slightly increased IgM antibody value of 21 U/ml (reference range, <15 U/ml) using Brucella IgM and IgG antibody enzyme-linked immunosorbent assay (ELISA) (Virion/Serion, Würzburg, Germany) ~14 days after the first contact with the dog. IgG antibody and immune-capture-agglutination test values (bestbion dx GmbH, Cologne, Germany) remained within the normal range. The affected employee also reported symptoms like fatigue and night sweats lasting for about a week. Symptoms improved rapidly while receiving doxycycline (100 mg bid for 3 months). The follow-up serology 6 weeks later still showed slightly elevated IgM values of 19 U/ml but no increase in IgG antibody values. The Brucella immune-capture-agglutination test resulted negative again. However, no attempts were made to isolate the bacterium by direct culture. To date, 1.5 years later, the employee continues to be symptom-free.
4. Discussion
This report illustrates the first notified B. suis infection in a dog in Germany since 1978. The infected individual was an intact male Rhodesian Ridgeback suffering from orchitis with symptoms including fever, testicular swelling and pain, abscessed spermatic cord, free abdominal fluid due to peritonitis, anorexia, and weakness. Microbiological analysis of an abscessed testis after neutering revealed the presence of Brucella sp., identified by PCR as B. suis bv 1.
In areas where B. suis bv 1 is endemic in wildlife, canine infection with the pathogen is not uncommon. The incidence of infection in dogs has been reported to increase in certain regions of the United States and Australia (51–53). In most of these cases, exposure to feral pigs through hunting or the consumption of raw feral pig meat was associated with the infection. In Europe, only two cases of B. suis infection in dogs have been reported to date: In 1978, B. suis bv 1 was detected in two male dogs in Berlin, Germany, showing fever and orchitis/epididymitis. Since they were kept only in urban surroundings and bv 1 is not endemic in Germany, the dog feed (raw meat from Eastern European countries) was suspected as the source of infection (33). The second case reported was a dog infected with B. suis bv 1 in the Netherlands in 2016. It had no history of hunting or travel to B. suis bv 1 endemic countries but was fed uncooked hare meat imported from Argentina. Via PCR, B. suis bv 1 was detected in these hare carcasses and characterized in silico by MLVA and MLST. Both samples of the dog and the meat showed high similarity in MLVA (32).
Unfortunately, in the case presented here, it was not possible to determine the source of infection. The patient was neither a hunting nor a breeding dog, although contact with infected vaginal fluid or urine from bitches could not be excluded. The dog, raised in Germany, had a history of travel to the Netherlands but had never been to B. suis bv 1 endemic regions. Since cases of B. suis bv 1 infection were associated with consumption of raw meat, this route of transmission is probable but uncertain, especially since Brucella sp. could not be detected in the analyzed meat. This result is limited by the inability to test all types of meat and the uncertainty regarding whether the tested meat was from the same batch consumed during the time of infection. Given the variable incubation period of Brucella infection, which can range from 2 weeks to several months, some time may have elapsed from infection to the onset of symptoms (11).
Transmission of B. suis from dogs to humans has not yet been clearly demonstrated. A single report from the United States suggests a B. suis infection in a woman as a result of handling aborted canine fetuses without gloves (54). However, in the case reported here, the serological results of the staff member do not ultimately prove infection. An increase in IgM antibodies alone may indicate a non-specific reaction and not necessarily an infection. The symptoms presented by the affected employee were characteristic of a Brucella infection but were not specific and therefore inconclusive. Unfortunately, no cultural or direct evidence by PCR has been obtained, leaving the possibility of human infection as speculative but noteworthy, especially since a case of B. suis bv 1 infection in a human without any serological response has also been reported (55). In Europe, only two human cases of infection with B. suis bv 1 were described to the authors' knowledge: A Spanish medical waste treatment plant worker was infected with B. suis bv 1 in 2014, probably after an accidental puncture with a contaminated needle (34). Presumably as a consequence of private meat processing, a German became infected with B. suis bv 1 in 2018 (20).
Standard serological tests for suspected canine brucellosis will only detect B. canis, as there is no serological cross-reaction between B. canis and B. suis since they have different antigenic characteristics. B. canis and B. ovis carry a rough lipopolysaccharide (LPS) without the most external antigen, the O-polysaccharide, while all other Brucella species are smooth Brucella strains with the O-polysaccharide present in the LPS (2, 56). This can lead to false-negative results for infection with Brucella species other than B. canis, so direct culture is preferred. Still, accurate identification of isolates using the standard method of MALDI-TOF MS is challenging, as only reference spectra for B. melitensis are available in the database. Given that this organism is highly clonal and demands methods with high discriminatory potential, whole genome sequencing is widely employed for the identification and differentiation of Brucella sp. (47, 57). Three years before the presented case, brucellosis was also diagnosed in a dog in the Netherlands, and B. suis bv 1 was isolated (32). However, based on the MLVA results, no epidemiological connection could be drawn between both cases. Likewise, the isolate from a human brucellosis case in Germany in 2018 (20) markedly differed from 21RB23181. Determining the geographic origin of the infection source in the presented case is hampered by the lack of comprehensive sequencing data. Regarding Europe, only a limited number of B. suis bv 1 WGS data is available, of which 21RB23181 did not show a notable similarity. With regard to the known distribution of B. suis bv 1 strains, it can be expected that the strain is not of endemic origin but imported, maybe from the Pacific region (e.g., Polynesia), as SNP typing revealed a higher similarity to strains from Tonga. The fact that MLVA would put 21RB23181 closer to US American isolates as the allelic distance to Tonga strains was comparably high can be disregarded. It was shown for B. melitensis that MLVA results can lead to false conclusions regarding strain origin and that WGS-based methods are more appropriate (57).
Despite its recovery, the dog was euthanized after being diagnosed with B. suis bv 1, given the high risk of zoonotic transmission. Brucella disseminates through direct contact, ingestion, or aerosolization of body fluids. It may be intermittently shed for up to 60 weeks and remains persistent for at least 2 years after inoculation (12). Due to its intracellular nature and periodic bacteremia, antibiotic treatment is often unsuccessful. The most promising therapy involves a combination of tetracyclines (high-dose doxycycline or minocycline for 1–2 months) and aminoglycosides (streptomycin or gentamicin for the first 2 weeks). However, relapses can occur shortly after the discontinuation of the antibiotic. Given the high risk of zoonotic transmission, prolonged shedding, and poor treatment options, euthanasia of affected dogs is recommended (12).
Herewith, we report the detection of the third case of canine brucellosis caused by B. suis bv 1 in Europe. Whole genome sequencing was used to determine the phylogenetic relationship of the isolate to other strains, with the aim of tracing the origin of the infection and identifying possible transmission routes. However, due to the limited availability of relevant sequence data, it was not possible to clearly determine the origin of the isolates, but connections to other European cases could be excluded. Although transmission from dogs to humans has not been clearly demonstrated, B. suis bv 1 is a highly virulent lineage that frequently infects humans with mild to severe symptoms, posing a threat to dog owners and veterinary personnel. Laboratories should be aware of the difficulties in culturing and serological testing that can result in underdiagnosis of this disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB60627.
Author contributions
EP-B, CE, and FM supervised the project. EP-B, CE, and SA conducted microbiological analyses. HB performed genome sequencing and wrote sections of the manuscript. AK contributed B. suis genome data. HB and FM analyzed sequencing data. JS and SA drafted the first version of the manuscript. All authors contributed to the manuscript revision, read, and approved the submitted version.
Acknowledgments
We would like to thank the dog owners for detailed information about the dog's history; the staff from the veterinary practice for detailed information about the dog's clinical progress and for providing data from the serological testing; Dr. Thomas Mönig, head of the local veterinary department, and the Chemical and Veterinary Analytical Institute of Westphalia for the cooperation concerning the investigations of the dog feed.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1233118/full#supplementary-material
Footnotes
References
1. Figueiredo P de, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. Pathogenesis and immune-biology of brucellosis: review of Brucella-host interactions. Am J Pathol. (2015) 185:1505–17. doi: 10.1016/j.ajpath.2015.03.003
2. Corbel MJ, Menachem B. Genus I. Brucella Meyer and Shaw 1920. In: Garrity GM, Bell JA, Lilburn T, editors. Class I. Alphaproteobacteria class. nov: In: Brenner, D.J., Krieg, N.R., Staley, J.T. (eds) Bergey's Manual® of Systematic Bacteriology. 2nd ed. Boston, MA: Springer (2005). p. 370–86.
3. Glynn MK, Lynn TV. Brucellosis. J Am Vet Med Assoc. (2008) 233:900–8. doi: 10.2460/javma.233.6.900
4. Yagupsky P, Morata P, Colmenero JD. Laboratory diagnosis of human brucellosis. Clin Microbiol Rev. (2019) 33:3. doi: 10.1128/CMR.00073-19
5. Young EJ, Corbel MJ. Brucellosis: Clinical and Laboratory Aspects. Boca Raton, FL: Taylor & Francis Group (1989). Available online at: https://ebookcentral.proquest.com/lib/kxp/detail.action?docID=6269747 (accessed April 14, 2023).
6. Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet Microbiol. (2010) 140:392–8. doi: 10.1016/j.vetmic.2009.06.021
7. Dentinger CM, Jacob K, Lee LV, Mendez HA, Chotikanatis K, McDonough PL, et al. Human Brucella canis infection and subsequent laboratory exposures associated with a Puppy, New York City, 2012. Zoonoses Public Health. (2015) 62:407–14. doi: 10.1111/zph.12163
8. Lucero NE, Ayala SM, Escobar GI, Jacob NR. Brucella isolated in humans and animals in Latin America from 1968 to 2006. Epidemiol Infect. (2008) 136:496–503. doi: 10.1017/S0950268807008795
9. Lucero NE, Corazza R, Almuzara MN, Reynes E, Escobar GI, Boeri E, et al. Human Brucella canis outbreak linked to infection in dogs. Epidemiol Infect. (2010) 138:280–5. doi: 10.1017/S0950268809990525
10. Lucero NE, Escobar GI, Ayala SM, Jacob N. Diagnosis of human brucellosis caused by Brucella canis. J Med Microbiol. (2005) 54 (Pt 5):457–61. doi: 10.1099/jmm.0.45927-0
11. Hollett RB. Canine brucellosis: outbreaks and compliance. Theriogenology. (2006) 66:575–87. doi: 10.1016/j.theriogenology.2006.04.011
12. Davidson PA, Skyes JE. Canine Brucellosis. In: Sykes JE, editor. Greene's Infectious Diseases of the Dog and Cat. 5th ed. St. Louis: Elsevier Saunders (2022). p. 876–92.
13. Makloski CL. Canine brucellosis management. Vet Clin N Am Small Anim Pract. (2011) 41:1209–19. doi: 10.1016/j.cvsm.2011.08.001
14. Wanke MM. Canine brucellosis. Anim Reprod Sci. (2004) 82–3:195–207. doi: 10.1016/j.anireprosci.2004.05.005
15. Hohmann M. Canine Brucellose - Ein Globalisierungsproblem? Deutsches Tierärzteblatt (2012). p. 1066–70. Available online at: https://www.bund estieraerztekammer.de/btk/dtbl/archiv/2012/artikel/DTBl_08_2012_Brucellose.pdf (accessed April 25, 2023).
16. Holst BS, Löfqvist K, Ernholm L, Eld K, Cedersmyg M, Hallgren G. The first case of Brucella canis in Sweden: background, case report and recommendations from a northern European perspective. Acta Vet Scand. (2012) 54:18. doi: 10.1186/1751-0147-54-18
17. Buhmann G, Paul F, Herbst W, Melzer F, Wolf G, Hartmann K, et al. Canine brucellosis: insights into the epidemiologic situation in Europe. Front Vet Sci. (2019) 6:151. doi: 10.3389/fvets.2019.00151
18. Woldemeskel M. Zoonosis due to Brucella suis with special reference to infection in dogs (carnivores): a brief review. OJVM. (2013) 3:213–21. doi: 10.4236/ojvm.2013.33034
19. Olsen SC, Boggiatto P, Nol P, Samartino L. Brucellosis. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Jianqiang Z, editors. Diseases of Swine. 11th ed. Hoboken, NJ: Wiley-Blackwell/American Association of Swine Veterinarians (2019). p. 778–91.
20. Zange S, Schneider K, Georgi E, Scholz HC, Antwerpen MH, Walter MC, et al. A headache with surprising outcome: first case of brucellosis caused by Brucella suis biovar 1 in Germany. Infection. (2019) 47:863–8. doi: 10.1007/s15010-019-01312-7
21. Irwin MJ, Massey PD, Walker B, Durrheim DN. Feral pig hunting: a risk factor for human brucellosis in north-west NSW? N S W Public Health Bull. (2009) 20:192–4. doi: 10.1071/NB09023
22. Eales KM, Norton RE, Ketheesan N. Brucellosis in northern Australia. Am J Trop Med Hyg. (2010) 83:876–8. doi: 10.4269/ajtmh.2010.10-0237
23. Centers for Disease Control and Prevention. Brucella suis infection associated with feral swine hunting in three states, 2007-2008. Morbid Mortal Wkly Rep. (2009) 58:618–21.
24. Mason RJ, Fleming PJ. Serological survey for Brucella antibodies in feral pigs from eastern Australia. Aust Vet J. (1999) 77:331–2. doi: 10.1111/j.1751-0813.1999.tb10276.x
25. Crichton R, Medveczky NE. The identity, distribution and epizootiological significance of Brucella isolates in Australia, 1981 to 1985. Aust Vet J. (1987) 64:48–52. doi: 10.1111/j.1751-0813.1987.tb16128.x
26. Norton TH, Thomas AD. Letter: Brucella suis in feral pigs. Aust Vet J. (1976) 52:293–4. doi: 10.1111/j.1751-0813.1976.tb00122.x
27. Aldrick SJ. Typing of Brucella strains from Australia and Papua-New Guinea received by the regional W.H.O. Brucellosis Centre. Aust Vet J. (1968) 44:130–3. doi: 10.1111/j.1751-0813.1968.tb09054.x
28. Massis F de, Zilli K, Di Donato G, Nuvoloni R, Pelini S, Sacchini L, et al. Distribution of Brucella field strains isolated from livestock, wildlife populations, and humans in Italy from 2007 to 2015. PLoS ONE. (2019) 14:e0213689. doi: 10.1371/journal.pone.0213689
29. Grégoire F, Mousset B, Hanrez D, Michaux C, Walravens K, Linden A, et al. serological and bacteriological survey of brucellosis in wild boar (Sus scrofa) in Belgium. BMC Vet Res. (2012) 8:80. doi: 10.1186/1746-6148-8-80
30. EFSA. Porcine brucellosis (Brucella suis). EFSA J. (2009) 7:1–112. doi: 10.2903/j.efsa.2009.1144
31. Kutlu M, Cevahir N, Erdenlig-Gürbilek S, Akalin S, Uçar M, Sayin-Kutlu S. The first report of Brucella suis biovar 1 isolation in human in Turkey. J Infect Public Health. (2016) 9:675–8. doi: 10.1016/j.jiph.2016.01.011
32. van Dijk MAM, Engelsma MY, Visser VXN, Spierenburg MAH, Holtslag ME, Willemsen PTJ, et al. Brucella suis infection in dog fed raw meat, the Netherlands. Emerg Infect Dis. (2018) 24:1127–9. doi: 10.3201/eid2406.171887
33. Hellmann E, Sprenger HU. Brucella suis infections in the dog. Berl Munch Tierarztl Wochenschr. (1978) 91:385–7.
34. Compés Dea C, Guimbao Bescós J, Alonso Pérez de Ágreda JP, Muñoz Álvaro PM, Blasco Martínez JM, Villuendas Usón MC. Epidemiological investigation of the first human brucellosis case in Spain due to Brucella suis biovar 1 strain 1330. Enferm Infecc Microbiol Clin. (2017) 35:179–81. doi: 10.1016/j.eimc.2016.06.005
35. García-Yoldi D, Marín CM, Miguel MJ de, Muñoz PM, Vizmanos JL, López-Goñi I. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin Chem. (2006) 52:779–81. doi: 10.1373/clinchem.2005.062596
36. López-Goñi I, García-Yoldi D, Marín CM, Miguel MJ de, Barquero-Calvo E, Guzmán-Verri C, et al. New Bruce-ladder multiplex PCR assay for the biovar typing of Brucella suis and the discrimination of Brucella suis and Brucella canis. Vet Microbiol. (2011) 154:152–5. doi: 10.1016/j.vetmic.2011.06.035
37. Friedrich-Loeffler-Institut. Brucellose der Rinder, Schweine, Schafe und Ziegen: Amtliche Methode und Falldefinition: Amtliche Methode und Falldefinition. Friedrich-Loeffler-Institut & Friedrich-Loeffler-Institut, Amtliche Methodensammlung und Falldefinitionen: Anzeigepflichtige Tierseuchen, Greifswald - Insel Riems, Germany. (2021).
38. Murigneux V, Roberts LW, Forde BM, Phan M-D, Nhu NTK, Irwin AD, et al. MicroPIPE: validating an end-to-end workflow for high-quality complete bacterial genome construction. BMC Genom. (2021) 22:474. doi: 10.1186/s12864-021-07767-z
39. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST quality assessment tool for genome assemblies. Bioinformatics. (2013) 29:1072–5. doi: 10.1093/bioinformatics/btt086
40. Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom. (2021) 7. doi: 10.1099/mgen.0.000685
41. Grissa I, Bouchon P, Pourcel C, Vergnaud G. Online resources for bacterial microevolution studies using MLVA or CRISPR typing. Biochimie. (2008) 90:660–8. doi: 10.1016/j.biochi.2007.07.014
42. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. (2018) 9:5114. doi: 10.1038/s41467-018-07641-9
43. Nan W, Tan P, Wang Y, Xu Z, Mao K, Peng D, et al. Duplex PCR for differentiation of the vaccine strain Brucella suis S2 and B. suis biovar 1 from other strains of Brucella spp. Vet J. (2014) 201:427–8. doi: 10.1016/j.tvjl.2014.05.033
44. Sacchini L, Wahab T, Di Giannatale E, Zilli K, Abass A, Garofolo G, et al. Whole genome sequencing for tracing geographical origin of imported cases of human brucellosis in Sweden. Microorganisms. (2019) 7:3. doi: 10.3390/microorganisms7100398
45. Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. (2018) 28:1395–404. doi: 10.1101/gr.232397.117
46. Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, et al. Updating benchtop sequencing performance comparison. Nat Biotechnol. (2013) 31:294–6. doi: 10.1038/nbt.2522
47. Abdel-Glil MY, Thomas P, Brandt C, Melzer F, Subbaiyan A, Chaudhuri P, et al. Core genome multilocus sequence typing scheme for improved characterization and epidemiological surveillance of pathogenic Brucella. J Clin Microbiol. (2022) 60:e0031122. doi: 10.1128/jcm.00311-22
48. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
49. TRBA 466 Classification of Prokaryotes (Bacteria and Archaea) Into Risk Groups: Technical Rule for Biological Agents. GMBI (2010). p. 1428–667.
50. TRBA 100 Protective Measures for Activities Involving Biological Agents in Laboratories: Technical Rule for Biological Agents. GMBI (2013). p. 1010–42.
51. Landis M, Rogovskyy AS. Closing the brief case: Brucella suis infection in a household of dogs. J Clin Microbiol. (2022) 60:e0098521. doi: 10.1128/jcm.00985-21
52. Mor SM, Wiethoelter AK, Lee A, Moloney B, James DR, Malik R. Emergence of Brucella suis in dogs in New South Wales, Australia: clinical findings and implications for zoonotic transmission. BMC Vet Res. (2016) 12:199. doi: 10.1186/s12917-016-0835-0
53. Ramamoorthy S, Woldemeskel M, Ligett A, Snider R, Cobb R, Rajeev S. Brucella suis infection in dogs, Georgia, USA. Emerg Infect Dis. (2011) 17:2386–7. doi: 10.3201/eid1712.111127
54. Nicoletti PL, Quinn BR, Minor PW. Canine to human transmission of brucellosis. N Y State J Med. (1967) 67:2886–7.
55. Naha K, Dasari S, Pandit V, Seshadri S. A rare case of seronegative culture–proven infection with Brucella suis. Aust Med J. (2012) 5:340–3. doi: 10.4066/AMJ.2012.1177
56. Cosford KL. Brucella canis: an update on research and clinical management. Can Vet J. (2018) 59:74–81.
Keywords: canine brucellosis, zoonosis, Brucella suis biovar 1, epididymitis, reproductive disease, whole genome sequencing, genotyping, raw meat diet
Citation: Aurich S, Schneider J, Brangsch H, Koets A, Melzer F, Ewers C and Prenger-Berninghoff E (2023) Brucella suis biovar 1 infection in a dog with orchitis in Germany. Front. Vet. Sci. 10:1233118. doi: 10.3389/fvets.2023.1233118
Received: 01 June 2023; Accepted: 10 July 2023;
Published: 03 August 2023.
Edited by:
Yogesh Chander, Varigen Biosciences Corporation, United StatesReviewed by:
Giuseppe Marruchella, University of Teramo, ItalyHosny El-Adawy, Friedrich Loeffler Institut, Germany
Copyright © 2023 Aurich, Schneider, Brangsch, Koets, Melzer, Ewers and Prenger-Berninghoff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie Aurich, U29waGllLkF1cmljaEB2ZXRtZWQudW5pLWdpZXNzZW4uZGU=
†These authors share first authorship
 Sophie Aurich
Sophie Aurich Juliane Schneider2†
Juliane Schneider2† Hanka Brangsch
Hanka Brangsch Ad Koets
Ad Koets Falk Melzer
Falk Melzer Christa Ewers
Christa Ewers Ellen Prenger-Berninghoff
Ellen Prenger-Berninghoff