- 1Department of Molecular Biology, Russian State Center for Quality and Standardization of Veterinary Drugs and Feed (VGNKI), Moscow, Russia
- 2Laboratory of Cell Biology, Lopukhin Federal Research and Clinical Center of Physical-Chemical Medicine of FMBA of Russia (Lopukhin FRCC PCM), Moscow, Russia
Introduction
Polymerase chain reaction (PCR) as a diagnostic method has gained enormous notability during the coronavirus pandemic (1). Indeed, PCR is an indispensable tool for rapidly assessing the spread of a disease spreading during viral infection outbreaks in the human population, animal herds and poultry. More generally, PCR is one of the primary methods for diagnostics of viral infections in clinical and veterinary medicine.
Regarding bacterial pathogens, PCR-based diagnostics play a more modest role since laboratories rely mostly on culture-based methods (2). Nevertheless, the World Organization for Animal Health (WOAH, founded as OIE) recommends the use of PCR assay as a supplement to traditional diagnostic methods for Mycoplasma spp., Mycobacterium tuberculosis, Pasteurella multocida and Chlamydia spp. For example, recommendations to employ genetic tests are present in the WOAH guidelines for the diagnostics of mycoplasmosis (3), tuberculosis (4), fowl cholera (5), and chlamydiosis (6). However, no WOAH diagnostics guidelines have yet been developed for contagious diseases, such as infectious coryza, bordetellosis, ornitobacteriosis, serositis, etc.
Some bacterial pathogens that cause the above diseases are difficult to culture in vitro. For instance, Ornithobacterium rhinotracheale, which causes bird ornitobacteriosis, and Avibacterium paragallinarum, which causes infectious coryza are very demanding to culture conditions (7, 8). In addition, frequent mixed infections can seriously hamper the identification of pathogens (9). For such diseases, the PCR assay can become a primary diagnostic method. Furthermore, the PCR-based identification of pathogens circulating in an animal population can also be helpful in the choosing a vaccination strategy (10). Indeed, papers on the development and validation of novel PCR-based diagnostic assays for bacterial pathogens are regularly published in veterinary journals.
When designing PCR assays, developers follow a standard workflow consisting of three main stages: in silico stage, the evaluation of analytical performance of PCR, and evaluation of its diagnostic performance (11, 12). An in silico bioinformatics analysis involves the use of computational tools and databases to check the quality of designed PCR primers. By performing the in silico bioinformatics analysis, potential problems such as primer-dimer formation, hairpin formation, primer mispriming, and false negative or false positive results can be avoided. Here, we briefly review the in silico stage of the recently published PCR assays, focusing on how the authors account for genetic polymorphism that can cause false negative results in a PCR assay. For this mini-review, we compared nearly three dozen real-time PCR diagnostic assays developed in the last 5 years to identify bacteria causing upper respiratory tract diseases in animals (Table 1). We considered only those PCR assays that assessed the diagnostic performance of PCR tests. In the end, we formulate some suggestions for improving the presentation of in silico analysis in publications.
Common practice for in silico analysis of genetic polymorphism to develop PCR assays
Intra-species genetic variation can compromise the diagnostic sensitivity of PCR, increasing the likelihood of false negative results. For instance, genetic polymorphism can seriously impair primer annealing, especially if a variable nucleotide is at the 3′ end of the primer. Moreover, the impact of genetic polymorphism on PCR efficiency can be even more significant if the polymorphic position is at the annealing site of the PCR probe. Guidelines for creating diagnostic PCR tests recommend taking genetic polymorphism into account during the assay development (20), but this recommendation is not specific enough. For example, there are no quantitative endpoints for considering genetic polymorphisms. There are also no guidelines for presenting the results of the in silico analysis of genetic polymorphisms.
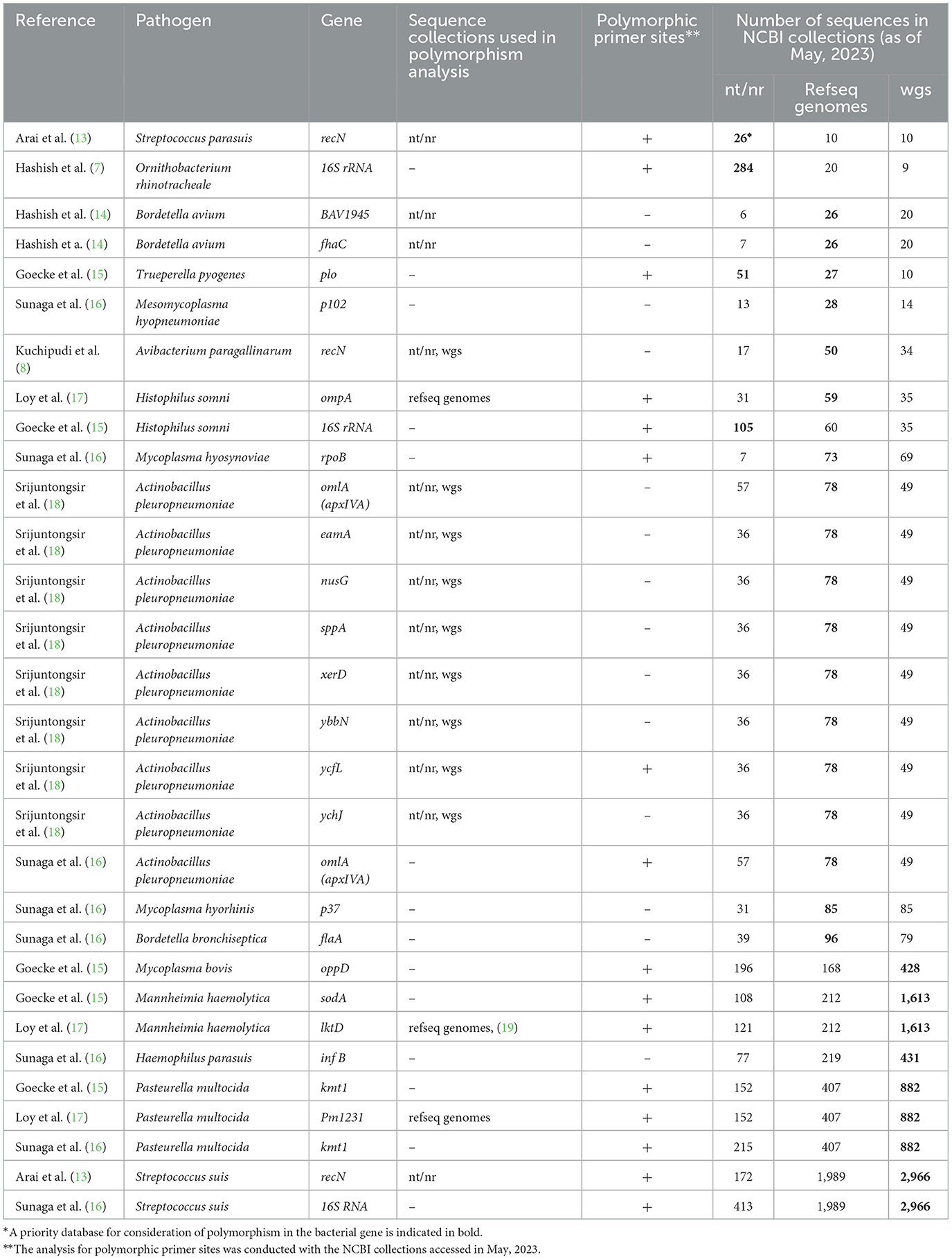
Not surprisingly, the authors describe the in silico analysis with varying levels of detail and depth (Table 1). For example, all authors used the BLAST tool (21) to check how the sequence variation within a bacterial species could affect the PCR sensitivity. However, in many cases they did not specify which NCBI database was searched: the nucleotide collection (nt/nr), the RefSeq genome database (refseq genomes), or whole-genome shotgun database (wgs) (22, 23). Thus, the BLAST search coverage was not clear in many cases.
Moreover, the authors were choosing different NCBI collections for analysis of genetic polymorphisms (Table 1). Most likely, the choice depended on personal preferences. However, the choice of the database may have depended on the target gene and the number of genomes for a bacterial species in the NCBI collections. In our sample, nearly 40 percent of the PCR target gene sequences were submitted to the NCBI sequence collections as a result of whole genome sequencing. However, other target genes, including phylogenetic genetic markers (16S rRNA, recN), virulence genes (kmt1, oppD, omlA), and multilocus sequence typing gene (infA) were submitted to the nt/nr database as result of Sanger sequencing. Therefore, for these genes, intra-species polymorphism may be more widely represented in the nt/nr database compared to the wgs database. For instance, the Streptococcus parasuis recN gene used as a target gene by Arai et al. (13) is a phylogenetic marker with 26 sequences in the nt/nr database and only 10 sequences in the wgs database. On the other hand, the wgs database is the most representative for bacterial species, the whole genome of which has been sequenced more than 150 times. The refseq genomes database is optimal for bacterial species with intermediate number of genomes sequenced (Table 1). It should be emphasized that NCBI Genbank collections may not accurately reflect true genetic polymorphism due to uneven representation of different regions, annotation inaccuracies, or technical sequencing errors. So, in silico analysis should always be accompanied by verification of PCR sensitivity and specificity on real samples.
Loy et al. (17) provided an excellent example of how to take genetic polymorphism into account. In their study, degenerate nucleotides were placed at polymorphic positions in the primer and multiple alignments were used to illustrate variable positions. However, most authors did not explicitly state whether polymorphic nucleotides were present at the primer annealing sites. As a result, we examined the primer annealing sites for polymorphism in all PCR tests listed in Table 1. In May 2023, we accessed the NCBI sequence collections, the number of sequences of which has increased significantly since the publication of the analyzed primers. We found polymorphic positions in 50% of the cases, which was common among bacterial species with the highest numbers of genomes in wgs database. However, the sodA (15) and lktD (17) genes were found to be very conservative with single-variant sequences among 1,613 Mannheimia haemolytica genomes in the wgs database. The frequency of sodA and lktD sequences with a polymorphic nucleotide in the primer was only 0.06%. In other words, the primers and probes perfectly matched the target sequence in 99.94% of the Mannheimia haemolytica genomes. It should also be noted that a BLAST search in the wgs database allows detection if a target PCR sequence is not ubiquitous for the genome sequenced of a given bacterial species. For instance, the wgs database comprises 1,613 genomes of Mannheimia haemolytica and the target sequence for PCR in the sodA gene (15) is present in 1,611 cases.
Conclusion
The NCBI sequence collections now contain thousands of genomes for a variety of pathogenic bacteria, as shown in Table 1. The abundance of data allows for comprehensive testing of primers for many sequences. On the other hand, the growth of the NCBI sequence collections will inevitably make it increasingly difficult for researchers to find perfectly conservative sites for primer design. This highlights the need for establishing pipelines that take into account genetic polymorphism when developing PCR assays for diagnostics of bacterial pathogens.
We believe that developers should clearly state the sequence database used with the BLAST tool and provide the number of bacterial gene sequences available for analysis. The presence or absence of polymorphism at the annealing sites of primers and samples should be clearly indicated. If polymorphic sites are present, it is appropriate to demonstrate the polymorphism using a multiple sequence alignment, as shown by Loy et al. (17) in the supplemental material to their publication. Moreover, a quantitative assessment may be included, such as “…the primers and probe perfectly match 99% of the genomic sequences of the target species deposited in the NCBI reference genome database…” When using previously published primers, it is important to recheck them for the presence of polymorphic sites at the annealing sites of primers and probe in current sequence databases. In addition, the authors should disclose if the PCR target sequence is absent in some bacterial genomic sequences deposited to the NCBI collections.
Author contributions
AB conceptualized the study and wrote the manuscript. EK and IS conducted literature analysis and searched the NCBI database. OP analyzed the manuals of diagnostic tests for terrestrial animals. OI secured funding for the work. All authors contributed to the article and approved the final version for submission.
Funding
This work was supported by the grant # 075-15-2021-1054 from the Ministry of Science and Higher Education of the Russian Federation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jarrom D, Elston L, Washington J, Prettyjohns M, Cann K, Myles S, et al. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ Evidence-Based Med. (2022) 27:33–45. doi: 10.1136/bmjebm-2020-111511
2. Váradi L, Luo JL, Hibbs DE, Perry JD, Anderson RJ, Orenga S, et al. Methods for the detection and identification of pathogenic bacteria: past, present, and future. Chem Soc Rev. (2017) 46:4818–32. doi: 10.1039/C6CS00693K
3. The World Organisation for Animal Health. Avian mycoplasmosis. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th Edn. The World Organisation for Animal Health (2023).
4. The World Organisation for Animal Health. Avian tuberculosis. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th Edn. The World Organisation for Animal Health (2023).
5. The World Organisation for Animal Health. Fowl Cholera. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th Edn. The World Organisation for Animal Health (2023).
6. The World Organisation for Animal Health. Avian chlamydiosis. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th Edn. The World Organisation for Animal Health (2023).
7. Hashish A, Sinha A, Sato Y, Macedo NR, El-Gazzar M. Development and validation of a new TaqMan real-time PCR for the detection of Ornithobacterium rhinotracheale. Microorganisms. (2022) 10:341. doi: 10.3390/microorganisms10020341
8. Kuchipudi SV, Yon M, Surendran Nair M, Byukusenge M, Barry RM, Nissly RH, et al. A highly sensitive and specific probe-based real-time PCR for the detection of Avibacterium paragallinarum in clinical samples from poultry. Front Vet Sci. (2021) 8:609126. doi: 10.3389/fvets.2021.609126
9. Astrup LB, Pedersen K, Farre M. Microbiological diagnoses on clinical mastitis—Comparison between diagnoses made in veterinary clinics vs. in laboratory applying MALDI-TOF MS. Antibiotics. (2022) 11:271. doi: 10.3390/antibiotics11020271
10. Kurmanov B, Zincke D, Su W, Hadfield TL, Aikimbayev A, Karibayev T, et al. Assays for identification and differentiation of Brucella species: a review. Microorganisms. (2022) 10:1584. doi: 10.3390/microorganisms10081584
11. The World Organisation for Animal Health. Principles and methods of validation of diagnostic assays for infectious disease. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th Edn. The World Organisation for Animal Health (2023).
12. The World Organisation for Animal Health. Development and optimisation of nucleic acid detection assays. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th Edn. The World Organisation for Animal Health (2023).
13. Arai S, Kim H, Watanabe T, Tohya M, Suzuki E, Ishida-Kuroki K, et al. Assessment of pig saliva as a Streptococcus suis reservoir and potential source of infection on farms by use of a novel quantitative polymerase chain reaction assay. Am J Vet Res. (2018) 79:941–8. doi: 10.2460/ajvr.79.9.941
14. Hashish A, Sinha A, Mekky A, Sato Y, Macedo NR, et al. Development and validation of two diagnostic real-time PCR (TaqMan) assays for the detection of Bordetella avium from clinical samples and comparison to the currently available real-time TaqMan PCR assay. Microorganisms. (2021) 9:2232. doi: 10.3390/microorganisms9112232
15. Goecke NB, Nielsen BH, Petersen MB, Larsen LE. Design of a high-throughput real-time PCR system for detection of bovine respiratory and enteric pathogens. Frontiers in veterinary science. (2021) 8. doi: 10.3389/fvets.2021.677993
16. Sunaga F, Tsuchiaka S, Kishimoto M, Aoki H, Kakinoki M, Kure K, et al. Development of a one-run real-time PCR detection system for pathogens associated with porcine respiratory diseases. J Vet Med Sci. (2020) 82:217–23. doi: 10.1292/jvms.19-0063
17. Loy JD, Leger L, Workman AM, Clawson ML, Bulut E, et al. Development of a multiplex real-time PCR assay using two thermocycling platforms for detection of major bacterial pathogens associated with bovine respiratory disease complex from clinical samples. J Vet Diagn Investigat. (2018) 30:837–47. doi: 10.1177/1040638718800170
18. Srijuntongsiri G, Mhoowai A, Samngamnim S, Assavacheep P, Bossé JT, Langford PR, et al. Novel DNA markers for identification of Actinobacillus pleuropneumoniae. Microbiology spectrum. (2022) 10:e01311–21. doi: 10.1128/spectrum.01311-21
19. Clawson ML, Murray RW, Sweeney MT, Apley MD, DeDonder KD, Capik SF, et al. Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genomics. (2016) 17:1–14. doi: 10.1186/s12864-016-3316-8
20. Toohey-Kurth K, Reising MM, Tallmadge RL, Goodman LB, Bai J, Bolin SR, et al. Suggested guidelines for validation of real-time PCR assays in veterinary diagnostic laboratories. J Vet Diagn Investigat. (2020) 32:802–14. doi: 10.1177/1040638720960829
21. Madden T. The BLAST sequence analysis tool. In: The NCBI Handbook, 2nd Edn. Bethesda, MD: National Center for Biotechnology Information (2013).
22. O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. (2016) 44:D733–45. doi: 10.1093/nar/gkv1189
Keywords: PCR assay, bacterial pathogens, nucleotide collection database, in silico analysis, assay development
Citation: Bogomazova A, Krylova E, Soltynskaya I, Prasolova O and Ivanova O (2023) In silico analysis to develop PCR assays for identification of bacterial pathogens in animals: what can we improve? Front. Vet. Sci. 10:1235837. doi: 10.3389/fvets.2023.1235837
Received: 06 June 2023; Accepted: 31 July 2023;
Published: 14 August 2023.
Edited by:
Yasser Mahmmod, Higher Colleges of Technology, United Arab EmiratesReviewed by:
Deepanker Tewari, Pennsylvania Veterinary Laboratory, United StatesAmro Hashish, Iowa State University, United States
Copyright © 2023 Bogomazova, Krylova, Soltynskaya, Prasolova and Ivanova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ekaterina Krylova, ZS5rcnlsb3ZhQHZnbmtpLnJ1
 Alexandra Bogomazova
Alexandra Bogomazova Ekaterina Krylova
Ekaterina Krylova Irina Soltynskaya
Irina Soltynskaya Olga Prasolova
Olga Prasolova Olga Ivanova1
Olga Ivanova1