- 1Yunnan Provincial Key Laboratory of Animal Nutrition and Feed Science, Faculty of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
- 2School of Agriculture and Life Sciences, Kunming University, Kunming, China
Introduction: The misuse of antibiotics threatens animal health and food safety and limits the development of livestock and poultry farming. In animal husbandry, probiotics can be used as an alternative to antibiotics because of their environmental protection and high efficiency. Parabacteroides goldsteinii (PG) has positive anti-inflammatory effects in mice; however, its role in other animals remains unknown.
Methods: The results showed that cell viability with the PG supernatant was highest at a 20-fold dilution ratio. Exposure of IPEC-J2 cells to LPS (1 μg/mL) significantly increased IL-6 and IL-8 concentrations, which triggered an inflammatory response. PG attenuated the LPS-induced inflammatory response by inhibiting the release of pro-inflammatory cytokines (IL-6) and promoting the release of anti-inflammatory cytokines (IL-10). Transcriptome analysis showed that the average number of clean reads in the LPS and LPS-PG groups was 39,311,061 and 38,085,237, respectively. In total, 2,126 differentially expressed genes (DEGs) were identified between the LPS and LPS-PG groups, including 36 up-regulated and 2090 down-regulated genes. Some DEGs, such as IL-6R and NF-кB, are related to inflammation. GO analysis was used to annotate the functions of the DEGs, and the results showed that biological regulation, cellular anatomical entity, and binding were dominant. KEGG enrichment analysis showed that the DEGs were significantly enriched in the PI3K-AKT signaling pathway, protein export pathway, and antigen processing and presentation pathway.
Discussion: These results indicate that P. goldsteinii is a promising probiotic for maintaining and improving intestinal health in piglets.
1 Introduction
With the rising global demand for animal-derived products, intensive livestock farming has become essential for maximizing production efficiency (1). However, this has led to increased disease risks, prompting the widespread use of antibiotics in animal husbandry. However, long-term abuse of antibiotics reduces disease resistance in animals, increases resistance among pathogenic bacteria, and leads to drug residues in animal products (2). These factors endanger food safety and limit the development of livestock and poultry farming. As several antibiotics are forbidden in livestock feed, it is important to identify feed additives that can replace antibiotics. Therefore, developing environmentally friendly and safe antibiotic alternatives is a priority in livestock farming.
Probiotics are considered green, safe, and can effectively maintain the microbial balance of the host gut and improve immune function (3). They are considered viable antibiotic alternatives because they are non-toxic, leave no residues, do not contribute to antibiotic resistance, are cost-effective, promote animal health, and are environmentally sustainable (4). Furthermore, probiotic metabolites can have various beneficial effects on the host. For example, short-chain fatty acids (5), indoles (6), vitamins (7), and polyphenols (8) play protective roles in intestinal barrier function. Studies have shown that probiotics such as Bacillus subtilis (9), Clostridium butyricum (10), and Lactobacillus spp. (11) effectively improve growth performance, digestive enzyme activity, and the intestinal morphology of livestock. Previous studies on probiotics have primarily focused on aerobic and facultative anaerobic bacteria. In contrast, anaerobic bacteria such as Parabacteroides goldsteinii (PG) have not been widely investigated and are thus not commonly used in livestock and poultry farming. PG helps to reduce inflammation in mice (12), ameliorates chronic obstructive pulmonary disease (COPD) induced by cigarette smoke in mice, reduces intestinal inflammation, and enhances mitochondrial and ribosome activity in colon cells (43). Thus, PG could be a promising next-generation probiotic.
In this study, we established an LPS-induced porcine small intestinal epithelial cell line (IPEC-J2) to explore the effects of PG extracts by comparing cytokine levels and analyzing its mechanism of action in IPEC-J2 cells. These results help to establish a theoretical basis for the use of PG in the pig farming industry.
2 Methods
2.1 Culture and preparation of PG supernatant
PG was obtained from the Microbe Division, Institute of Physical and Chemical Research (Japan) and cultured in Gifu Anaerobic Medium (GAM) using an LAI-3 T-N anaerobic incubator (Shanghai Longyue Instrument Equipment Co., Ltd., China). The conditions and operation steps of the Institute of Physical and Chemical Research (Japan) were strictly followed. When the OD600 reached 0.591 and there were 7.9 × 107 CFUs, the cultivation was terminated. DNA extraction was performed immediately using 1 mL of the bacterial solution. The supernatant was collected via centrifugation at 5,000 × g for 20 min and stored at −80°C.
2.2 IPEC-J2 cell culture
IPEC-J2 cells were cultured in high-glucose DMEM containing 10% FBS, 10,000 U/mL penicillin, and 10,000 μg/mL streptomycin in a 37°C and 5% CO2 incubator (13). The medium was changed every 2 days. When a confluence of ~80% was reached, cells were trypsinized and seeded in six-well cell culture plates (3 × 105 cells/well). After overnight culture, cells were starved for 6 h in Arg-free DMEM-F12. The PG-derived supernatant was diluted 20, 100, 500, 2,500, and 12,500-fold and added to the culture medium for probiotic functional analysis.
2.3 Cell viability assay
After overnight culture in 96-well plates (1 × 105 cells/well, 100 μL medium/well), cells were treated with different concentrations of LPS and dilutions of the PG bacterial solution supernatant for 24 h. Thiazolyl blue tetrazolium bromide (MTT; 10 μL) was added to each well, and the cells were incubated at 37°C and 5% CO2 for 4 h. Dimethyl sulfoxide (DMSO; 100 μL) was added, and the mixture was incubated at 37°C for 10 min. Finally, the absorbance was evaluated at 540 nm after shaking for 10 min (14).
2.4 Construction of the experimental model
IPEC-J2 cells were cultured as described above, and when 80% confluence was reached, they were treated with LPS (0.2, 1, 5, 25, and 125 μg/mL) derived from Escherichia coli for 24 h.
After the inflammation model were established, IPEC-J2 cells were administered LPS (1 μg/mL), and co-incubated with PG supernatant (0- or 20-fold dilution). After 24 h, cells were harvested via centrifugation.
2.5 RNA-seq
Total RNA of IPEC-J2 cell was extracted from the supernatant of logarithmically growing IPEC-J2 cells using TRIzol (Invitrogen, United States) according its introduction. The concentrations of RNA were detected by NanoDrop 2,000 spectrophotometer (Thermo Scientific, United States), and its integrity was determined by the RNA Nano 6,000 Assay Kit (Agilent Technologies, United States) in the Agilent Bioanalyzer 2,100 system (Agilent Technologies, United States). A cDNA library was constructed using the Illumina VAHTS Universal V10 RNA-seq Library Prep Kit (MGI Tech, China) following manufacturer’s protocols. Poly-A tailed mRNA was enriched using oligo (dT)-coated magnetic beads, and the mRNA was fragmented via ultrasound. The fragmented mRNA was used as a template, and random oligonucleotides were used as primers to synthesize the first strand of cDNA using the M-MuLV reverse transcriptase system. The RNA strand was degraded using RNase H, and the second cDNA strand was synthesized using DNA polymerase I and dNTPs. The purified double-stranded cDNA was end-repaired and A-tailed, and the sequencing joints were connected. cDNA (~200 bp) was obtained using AMPure XP beads, amplified using, and the products were then purified using AMPure XP beads. Double-ended sequencing was performed using an Illumina NovaSeq 6000 sequencing system in Lingen Biotechnology Ltd., Shanghai, China.
2.6 Analysis of interleukin (IL)-6, IL-8, and IL-10 levels
IL-6, IL-8, and IL-8 concentrations were measured using the Interleukin-6, Interleukin-8, and Interleukin-10 ELISA Assay Kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.7 RNA-seq data analysis
The raw sequencing reads were filtered for low-quality sequences (such as adapter sequences, N > 10% and junction sequences) by Trimmomatic (SLIDINGWINDOW:4:15 MINLEN:75) (version 0.36)1 to ensure the reliability of the sequencing results (15). The base composition and quality of the clean reads were then analyzed. Then HISAT22 software was used to align clean reads to the reference genome. The overall quality of the RNA-seq data was evaluated based on the coverage, ribosomal reads, and comparison of the regional statistics. The Q20 (the proportion of bases with a phred score greater than 20), Q30 The Q20 (the proportion of bases with a phred score greater than30) and GC content were used to determine the RNA-Seq quality.
To account for the effects of sequencing coverage and gene length, the number of read counts was normalized using the fragments per kilobase of exon model per million mapped reads (FPKM) method and used as the input for subsequent analysis (16). Principal component analysis (PCA) was used to evaluate the reproducibility of the samples and exclude outliers. Differentially expressed genes (DEGs) were analyzed using edgeR. Genes with a log2 fold change (|log2FC|) ≥ 1 and a false-discovery rate (FDR)-adjusted p-value (padj) < 0.05 between the LPS and LPS-PG groups were considered differentially expressed.
The Gene Ontology (GO) database3 was used to investigate the differences in biological functions of the DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database4 was used to investigate signaling and metabolic pathways associated with the DEGs (17).
2.8 DEG verification using qPCR
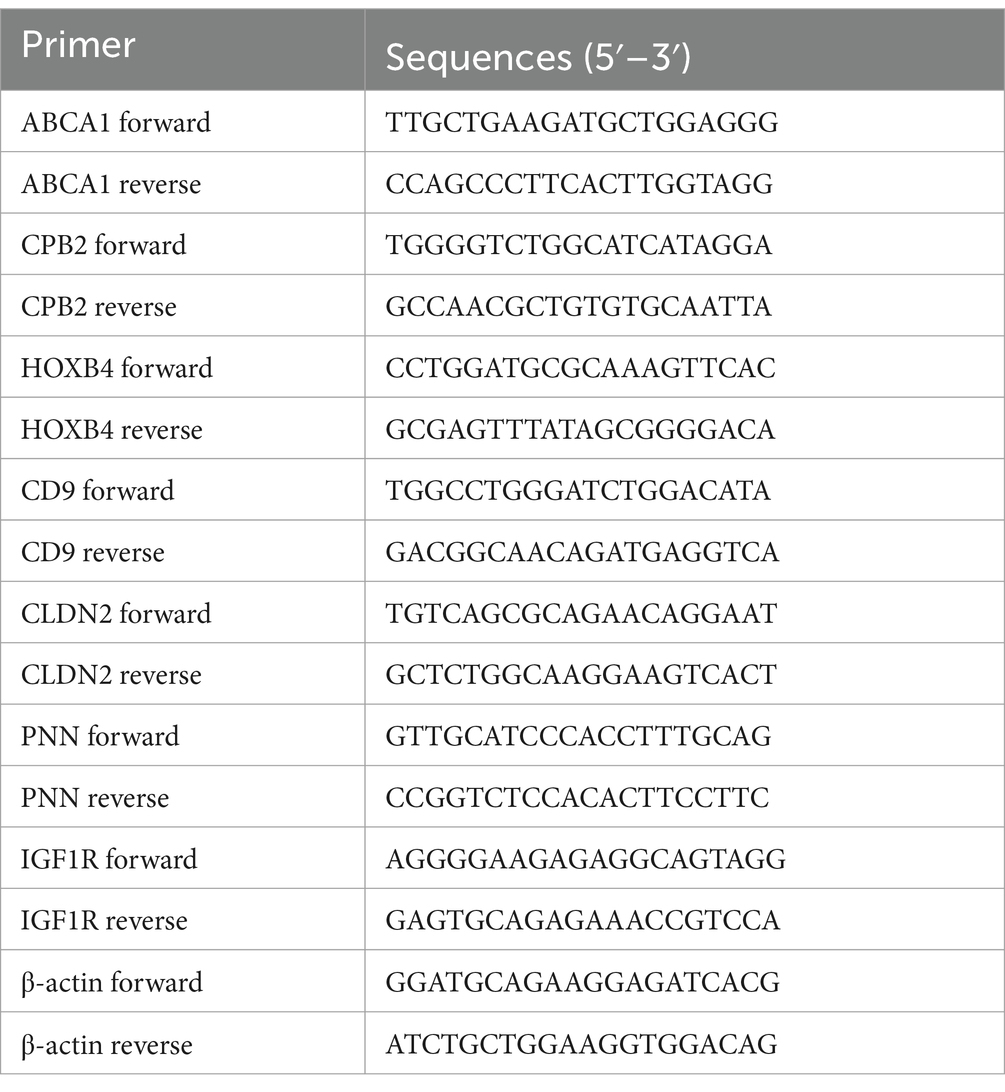
To validate the RNA-seq results, eight DEGs were randomly selected and qPCR was performed using the SYBR Green Supermix kit (Bio-Rad, United States) to verify the expression of DEGs in the LPS and LPS-PG groups. Primers were designed using the NCBI Primer BLAST tool (Table 1).5 cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The PCR reaction condition was as follows: 95°C for 30 s, s, 40 cycles of 95°C for 5 s, 56°C for 30 s, and 72°C for 1 min. The β-actin gene was used as an internal reference, and the target mRAN expression was calculated using the 2–ΔΔCT method.
2.9 Statistical analysis
Data are expressed as the mean ± SEM. SPSS 27.0 was used to conduct one-way ANOVA. Duncan’s test was used to determine significance at p < 0.05. Twelve cell samples were used for RNA-Seq. FPKM was used to detect DEGs between the LPS and LPS-PG groups.
3 Results
3.1 Effect of the PG supernatant on IPEC-J2 cell viability
Cell viability was significantly higher (p < 0.05) with 500-, 100-, and 20-fold dilutions of PG supernatant than in the control group (Figure 1). Overall, cell-viability was highest with the 20-fold PG dilution, and consequently, this was used for subsequent experiments.
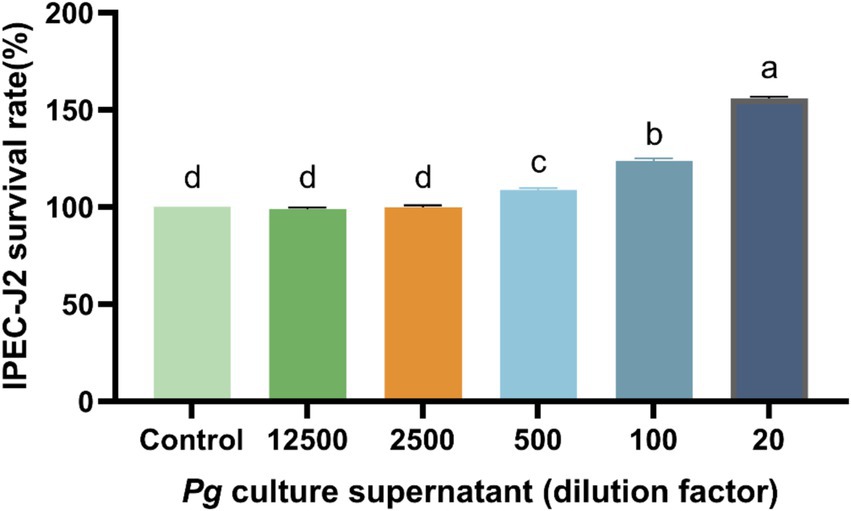
Figure 1. Effect of PG culture supernatant on IPEC-J2 cell viability. IPEC-J2 cells were cultured in 96-well plates and treated with various dilution factor of PG culture supernatant (ranging from 20 to 12,500) for 24 h. The results, presented as the mean ± standard error of the mean (SEM). Values with different lowercase letters indicate significant differences.
3.2 Effect of LPS on IL-6 and IL-8 secretion from IPEC-J2 cells
Compared with the control group, LPS (0.2, 1, 5, 25, or 125 μg/mL) induced a significant increase (p < 0.05) in IL-6 and IL-8 levels (Figure 2). Specifically, 1 μg/mL of LPS induced the highest IL-6 and IL-8 levels, and consequently, this LPS concentration was used to construct the IPEC-J2 cellular inflammatory model.
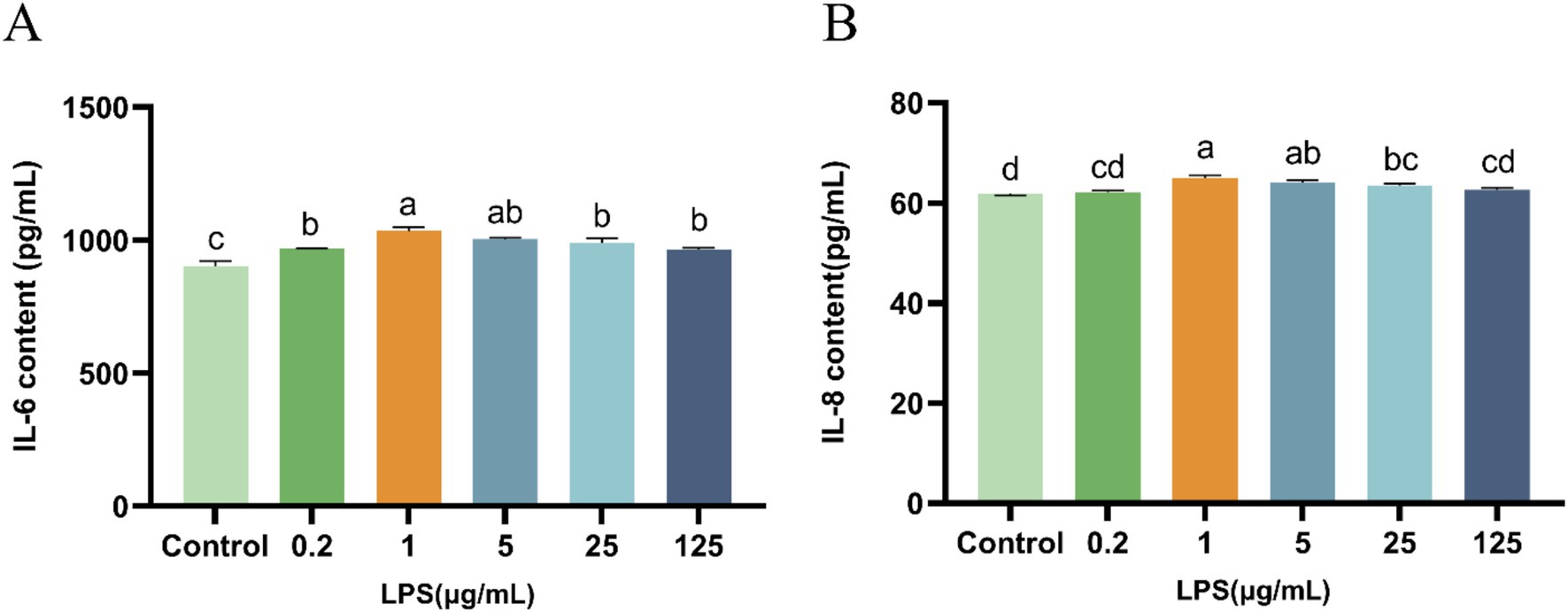
Figure 2. Effect of LPS on IL-6 and IL-8 secretion from IPEC-J2 cells. (A) IL-6. (B) IL-8. IPEC-J2 cells were cultured 96-well plates and treated with LPS (0.2, 1, 5, 25, and 125 μg/mL). The results, presented as the mean ± standard error of the mean (SEM). Values with different lowercase letters indicate significant differences.
3.3 Effect of PG supernatant on IL-6, IL-8, and IL-10 levels in LPS-induced IPEC-J2 cells
LPS induced a significant increase (p < 0.05) in IL-6 and IL-8 levels and a decrease (p < 0.05) in IL-10 levels in IPEC-J2 cells (Table 2). However, IL-6 and IL-8 levels were significantly decreased in the presence of PG supernatant, whereas IL-10 levels were significantly increased (p < 0.05) in LPS-induced IPEC-J2 cells (Table 2).
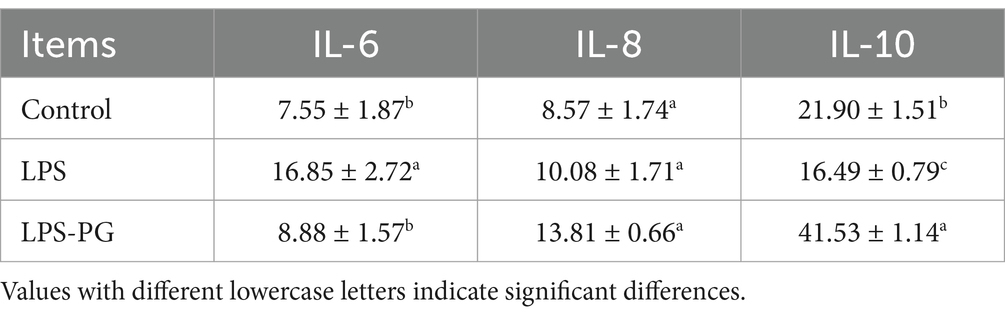
Table 2. The effect of PG culture supernatant on LPS induction of IPEC-J2 cell levels of IL-6, IL-8, and IL-10.
3.4 RNA-seq analysis
To investigate the molecular mechanism by which the PG supernatant alleviates LPS-induced inflammation in IPEC-J2 cells, RNA-Seq of the LPS and LPS-PG groups was conducted.
The LPS and LPS-PG groups had an average of 39,311,061 and 38,085,237 clean reads, respectively. The sequencing quality of the Q20 (error rate < 1%) and Q30 (error rate< 0.1%) samples was 98.28–99.23% and 94.26–97.19%, respectively. The sequencing data from the samples were aligned to the pig reference genome, with an average of 93.18% of the reads mapped to the reference genome (Table 3).
3.5 DEG analysis
The LPS and LPS PG groups were analyzed, and 2,126 significantly differentially expressed genes (DEGs) were identified, including 36 upregulated genes and 2090 down-regulated genes (Figure 3). The DEGs included IL-6R, NF-κB1, IGF1R, and ABCA1, which are associated with cellular inflammation.
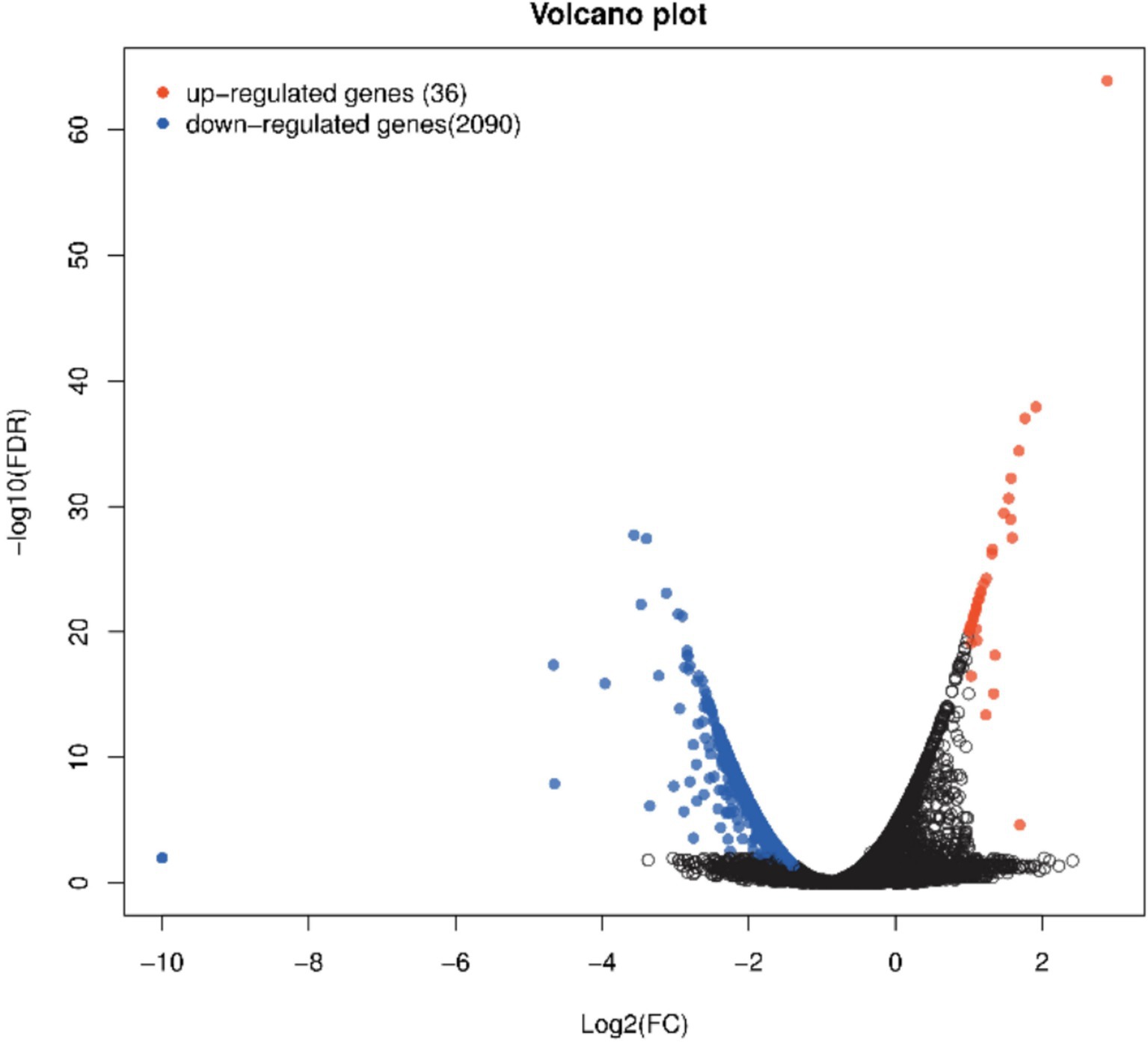
Figure 3. Volcano plot of significant genes from DEseq analysis of n = 6 LPS vs. n = 6 LPS-PG. Red dots show significant genes defined by |logFC| ≥ 1, FDR ≤ 0.05. Blue dots show significantly downregulated genes defined by |logFC| ≥ 1, FDR ≤ 0.05.
3.6 GO enrichment analysis
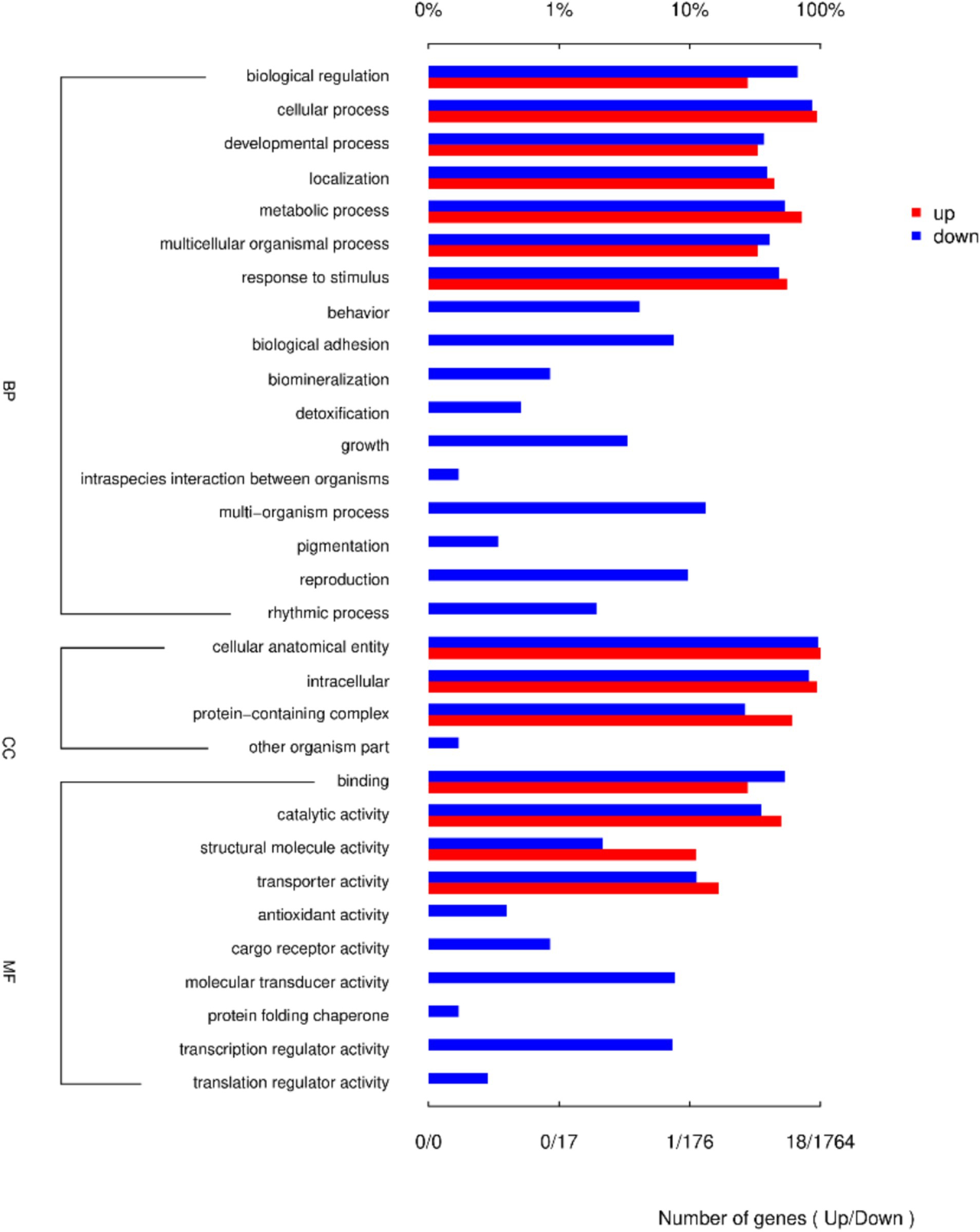
GO enrichment analysis identified biological processes (BP), cellular components (CC), and molecular functions (MF). Biological regulation, cellular process, and developmental process were the most enriched BP terms; cellular anatomical entity, intracellular, and protein-containing complexes were the most enriched CC terms; and binding, catalytic activity, and structural molecule activity were the most enriched MF terms (Figure 4).
3.7 KEGG pathway analysis
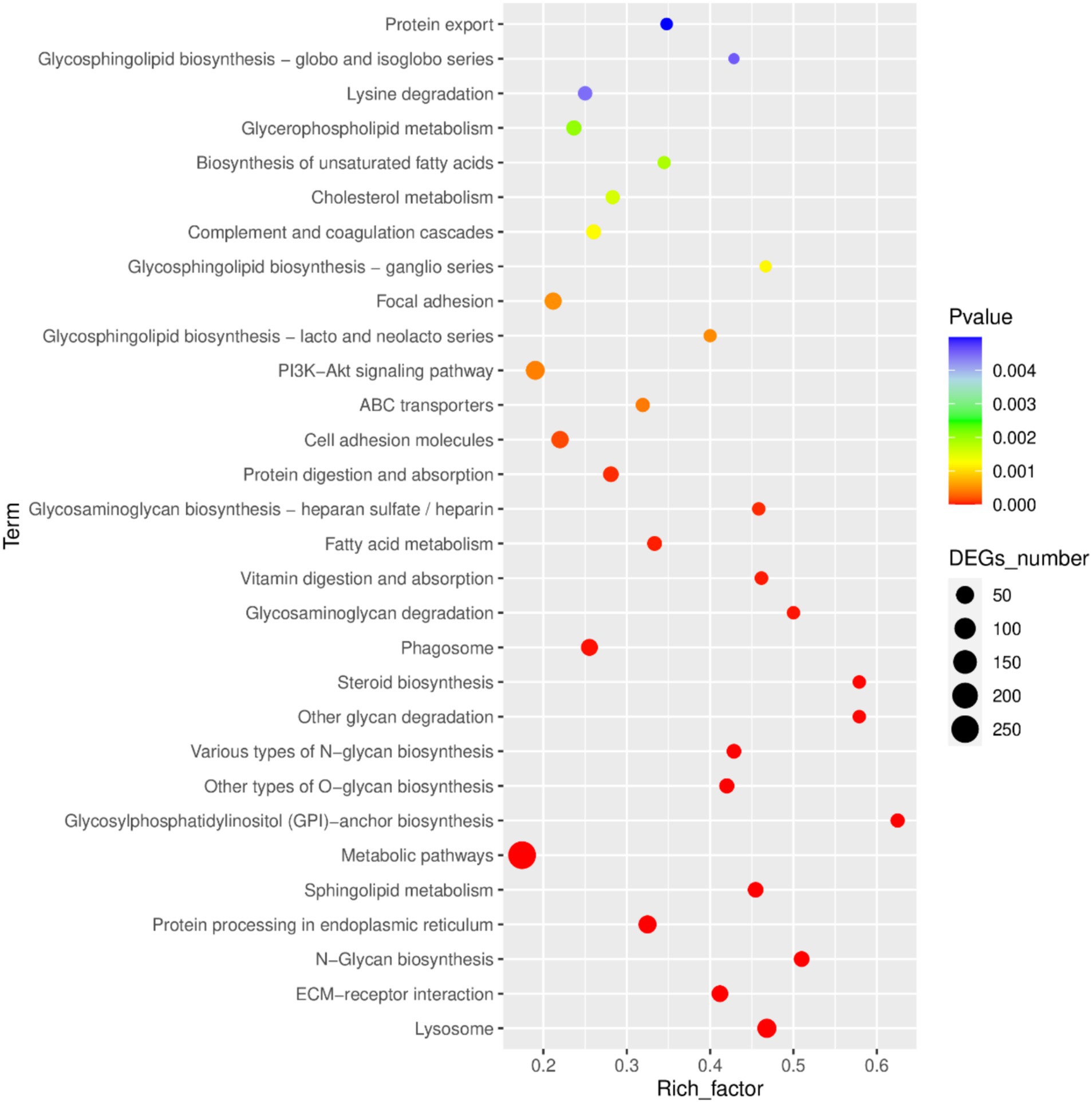
Kegg pathway analysis identified the top 30 most significantly enriched pathways, which included the PI3K-Akt signaling pathway, protein export, antigen processing and presentation, lysosome, ECM-receptor interaction, N-glycan biosynthesis, protein processing in endoplasmic reticulum, sphingolipid metabolism, metabolic pathways, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, other types of O-glycan biosynthesis, various types of N-glycan biosynthesis, other glycan degradation, steroid biosynthesis, phagosome, glycosaminoglycan degradation, vitamin digestion and absorption, fatty acid metabolism, glycosaminoglycan biosynthesis—heparan sulfate/heparin, protein digestion and absorption, cell adhesion molecules, ABC transporters, glycosphingolipid biosynthesis-lacto and neolacto series, focal adhesion, glycosphingolipid biosynthesis-ganglio series, complement and coagulation cascades, cholesterol metabolism, biosynthesis of unsaturated fatty acids, glycerophospholipid metabolism, lysine degradation, and glycosphingolipid biosynthesis-globo and isoglobo series (Figure 5).
3.8 qRT-PCR validation
Eight DEGs were selected for qRT-PCR validation of RNA-Seq results. The expression levels of the seven genes were consistent with the sequencing data (Figure 6). The results indicate that RNA-seq was highly reliable.
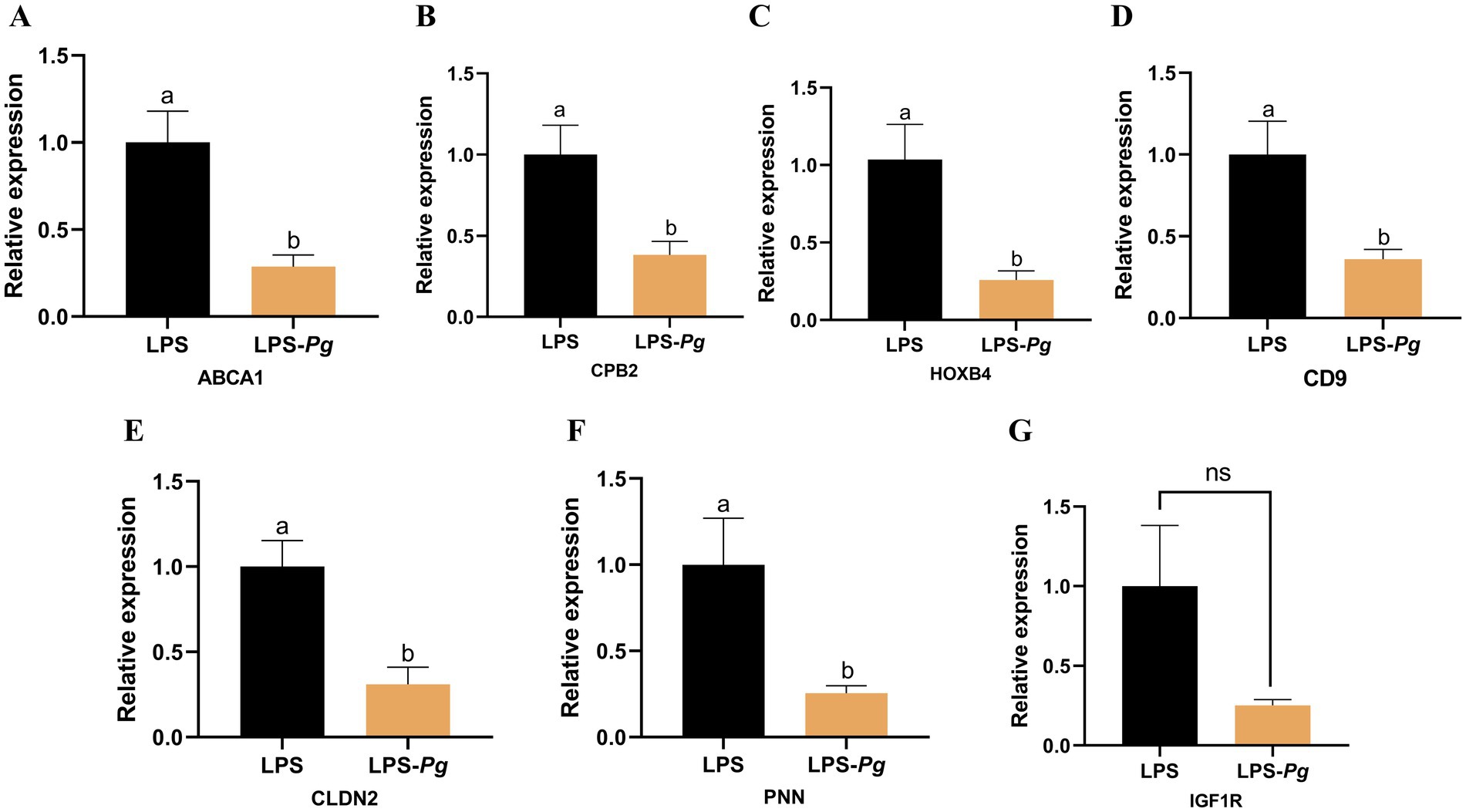
Figure 6. The DEGs were detected by qRT-PCR. (A) ABCA1. (B) CPB2. (C) HOXB4. (D) CD9. (E) CLDN2. (F) PNN. (G) IGF1R. Results are presented as mean ± SEM, significant differences (p < 0.05) are denoted by the distinct letters (a, b); unsignificant differences (p > 0.05) are denoted by the ns.
4 Discussion
Parabacteroides are a newly identified class of probiotic bacteria, which are found in the intestinal tracts of animals. Their metabolites include acetic, propionic, and butyric acids, which are produced from fermented carbohydrates and are beneficial to host health (18). PG, a member of the Parabacteroides, exerts an anti-inflammatory effect in cells. Cell viability is an important indicator used to evaluate drug efficacy (19). In this study, the administration of the PG supernatant significantly increased the viability of IPEC-J2 cells. Tong et al. previously showed that the activity and growth parameters of IPEC-J2 cells gradually improved with increasing concentrations (ranging between 2 and 10 mg/mL) of Bacillus amyloliquefaciens J and Lactiplantibacillus plantarum SN4 using solid-state fermentation (20).
LPS, commonly referred to as an endotoxin, is an integral part of the outer cell membrane of Gram-negative bacteria (21). In the context of IPEC-J2 cells, LPS primarily acts as an inducer of inflammation and apoptosis and compromises the integrity of the damaged intestinal epithelium (22). Cytokines are pivotal mediators of the intestinal mucosal immune response during acute and chronic intestinal inflammation (23). In particular, interleukins (ILs) play crucial roles in cell-to-cell transmission, activation, and immunomodulation (24). Cytokines also mediate the activation, proliferation, and differentiation of T and B cells, and are closely associated with inflammatory responses. Existing research has demonstrated that upon stimulation by either IL-1β or LPS, the level of IL-6 secreted by enteric glial cells in the intestines of adult mice increases (25). LPS induces pro-inflammatory properties and increases the cellular IL-6 and IL-8 levels. Our study shows that in comparison with the control group, 1 μg/mL LPS can effectively increase the IL-6 and IL-8 levels in IPEC-J2 cells, indicating the induction of inflammatory damage and establishment of the inflammatory model.
Inflammation is a defense mechanism of the body that involves the migration of white blood cells to damaged tissues and the production of various cytokines, including IL-6, IL-1β, and IL-10. IL-6 is a pleiotropic proinflammatory cytokine that exhibits a wide range of functions (26, 27) and is rapidly produced due to body stress, tumorigenesis, and acute inflammatory responses. Thus, IL-6 plays a role in regulating immune responses and the acute-phase reaction. IL-8 is a chemokine that is produced by various cells in response to inflammatory stimuli. The recruitment and activation of neutrophils enables them to infiltrate the inflamed site (28), thereby exacerbating the inflammatory response, and thus, they are key to many inflammatory diseases such as rheumatoid arthritis (29) and inflammatory bowel disease (30). IL-10 can maintain the integrity of the tissue epithelium and promote the healing of damaged tissues (31). It can inhibit proinflammatory responses, reduce tissue damage, and exert anti-inflammatory effects. IL-10 is a major cytokine involved in intestinal regulation. The results of this study showed that treatment of IPEC-J2 cells with the culture supernatant from PG cells decreased the levels of the pro-inflammatory factor IL-6 and increased the levels of the anti-inflammatory factor IL-10. This indicates that the PG culture supernatant has a protective effect on cells induced for inflammation using LPS and plays a key role in the repair of damaged cells involved in cellular immune regulation. Similarly, Lai et al. found that PG antagonizes the activation of the TLR4/MD-2 signaling pathway, which participates in LPS-induced inflammation in gastric epithelial cells in the Helicobacter pylori infection model. As an emerging next-generation probiotic, PG can reduce the levels of pro-inflammatory cytokines IL-1β and TNF-α in mice with colitis, alleviate inflammatory damage, and maintain the integrity of the intestinal epithelium (43).
RNA-Seq was conducted to analyze the inflammatory mechanism underlying the effect of the PG culture supernatant on LPS-induced IPEC-J2 cells. The results showed that 36 genes were upregulated in the LPS-PG group and 2090 genes were downregulated. Specifically, genes related to inflammation and immunity, such as IL-6, IL-6R, and nuclear factor-kappa B1 (NF-κB1), were significantly downregulated in the inflammatory model group with the addition of the PG culture supernatant. IL-6 and IL-6R play crucial roles in the physiological and pathological processes of the body (32). Whereas NF-κB is a protein complex that plays a crucial role in biological processes (33). Cytokine production regulates the expression of target genes, prompting the cells to synthesize and release various cytokines. These cytokines play important roles in processes such as immune regulation and inflammatory responses. Furthermore, the unregulated gene Cytochrome b is a core component of the mitochondrial respiratory chain and plays a pivotal role in the cellular energy metabolic network, providing a sustained energy supply for the efficient operation of immune cells. This energy support mechanism not only maintains the basic functions of immune cells but also significantly enhances the body’s immune defense efficiency by strengthening the phagocytic activity, signal transduction, and cytokine secretion capacity. Thus, the PG supernatant can ameliorate the inflammatory response in IPEC-J2 cells via immune-related gene regulation.
GO enrichment analysis showed that the function for BP enrichment included cellular processes, the function with the most genes enriched in the CC category was the cellular anatomical entity, and the function with the most genes in the molecular function category was binding. Among them, the “Antioxidant activity” pathway is important for immune responses. A close and complex relationship exists between the antioxidant activity levels of the body and the immune system as they influence and regulate each other to jointly maintain the health of the body (34, 35). KEGG analysis showed that the PI3K-AKT pathway included 62 DEGs that are significantly enriched. The PI3K/Akt signaling pathway is crucial for immune regulation and the inflammatory response (36, 37). Most of the genes were related to immune function, among which IL-6R, NF-ĸB1, VEGFA, the integrin family, laminin family, collagen family, FN1, fibroblast growth factors and their receptor, EPHA2, and ERBB2 played major roles. In the development pathways associated with immune cells, PI3K-AKT is involved in the differentiation of T cells in the thymus and maturation of B cells in the bone marrow, laying the foundation for the construction of a sound immune system (38–40). In macrophages, after recognition of pathogen-associated molecular patterns through pattern-recognition receptors, the PI3K-AKT pathway is activated, enhancing the phagocytic ability and antigen-presenting efficiency and effectively initiating an immune response (41, 42). In addition, the PI3K-AKT pathway is involved in regulating the metabolism of immune cells to provide sufficient energy for their activation and function (39). These results suggested that the PG improve the inflammatory response of IPEC-induced IPEC-J2 cell by activated the PI3K-AKT pathway.
5 Conclusion
PG reduced the secretion of the pro-inflammatory cytokine IL-6 in LPS-induced IPEC-J2 cells and increased the secretion of the anti-inflammatory cytokine IL-10, thereby exerting anti-inflammatory effects in vitro. Transcriptomic analysis suggested that PG may mitigate LPS-induced immune damage in IPEC-J2 cells by modulating the PI3K-AKT pathway. Thus, P. goldsteinii is a potential probiotic for piglets. However, several key factors, such as interactions with the gut microbiota, host immune system responses, probiotic survival in the digestive tract, and long-term effects, are not captured in this model. In the future work, the Dextran Sulfate Sodium Salt -induced piglets will used to investigate the effect of P. goldsteini in vivo study.
Data availability statement
The raw sequencing data have been deposited in the China National GeneBank Sequence Archive (CNSA) of the China National GeneBank DataBase (CNGBdb) with the accession number CNP0005800.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
JZ: Data curation, Formal analysis, Writing – original draft, Conceptualization, Methodology, Resources, Software. AL: Data curation, Formal analysis, Writing – original draft, Conceptualization, Visualization. YH: Methodology, Visualization, Data curation, Investigation, Writing – original draft. TG: Supervision, Conceptualization, Writing – original draft. YG: Supervision, Conceptualization, Writing – original draft. HP: Funding acquisition, Writing – review & editing, Conceptualization, Project administration, Resources. CD: Writing – review & editing, Project administration, Resources, Software, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Nature Science Foundation of China (32160795 and U1802234), the Industrial Innovation Talent Project of the “Xing Dian Talent Support Program” of Yunnan Province in 2022 (XDYC-CYCX-2022-0029), the Young Talent Project of the “Xing Dian Talent Support Program” of Yunnan Province in 2023 (XDYC-QNRC-2023-0403) and Yunnan Swine Industry Technology System Program (2023KJTX016).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^http://www.usadellab.org/cms/index.php?page=trimmomatic
2. ^https://ccb.jhu.edu/software/hisat2/index.shtml
References
1. Qian, Y, Song, K, Hu, T, and Ying, T. Environmental status of livestock and poultry sectors in China under current transformation stage. Sci Total Environ. (2018) 622-623:702–9. doi: 10.1016/j.scitotenv.2017.12.045
2. Wang, L, and Chai, B. Fate of antibiotic resistance genes and changes in bacterial community with increasing breeding scale of layer manure. Front Microbiol. (2022) 13:857046. doi: 10.3389/fmicb.2022.857046
3. Al-Habsi, N, Al-Khalili, M, Haque, SA, Elias, M, Olqi, NA, and Al Uraimi, T. Health benefits of prebiotics, probiotics, Synbiotics, and Postbiotics. Nutrients. (2024) 16:3955. doi: 10.3390/nu16223955
4. Krysiak, K, Konkol, D, and Korczyński, M. Overview of the use of probiotics in poultry production. Animals. (2021) 11:1620. doi: 10.3390/ani11061620
5. Burger-van Paassen, N, Vincent, A, Puiman, PJ, van der Sluis, M, Bouma, J, Boehm, G, et al. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. (2009) 420:211–9. doi: 10.1042/BJ20082222
6. Bansal, T, Englert, D, Lee, J, Hegde, M, Wood, TK, and Jayaraman, A. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun. (2007) 75:4597–607. doi: 10.1128/IAI.00630-07
7. Fathima, S, Shanmugasundaram, R, Adams, D, and Selvaraj, RK. Gastrointestinal microbiota and their manipulation for improved growth and performance in chickens. Foods. (2022) 11:1401. doi: 10.3390/foods11101401
8. Kostelac, D, Gerić, M, Gajski, G, and Frece, J. Probiotic bacteria isolated from fermented meat displays high antioxidant and anti-inflammatory potential. Mutagenesis. (2023) 38:58–63. doi: 10.1093/mutage/geac023
9. Jiao, S, Zheng, Z, Zhuang, Y, Tang, C, and Zhang, N. Dietary medium-chain fatty acid and Bacillus in combination alleviate weaning stress of piglets by regulating intestinal microbiota and barrier function. J Anim Sci. (2023) 101:skac414. doi: 10.1093/jas/skac414
10. Wang, K, Chen, G, Cao, G, Xu, Y, Wang, Y, and Yang, C. Effects of clostridium butyricum and Enterococcus faecalis on growth performance, intestinal structure, and inflammation in lipopolysaccharide-challenged weaned piglets. J Anim Sci. (2019) 97:4140–51. doi: 10.1093/jas/skz235
11. He, Y, Liang, J, Liu, Y, Zhou, X, Peng, C, Long, C, et al. Combined supplementation with Lactobacillus sp. and Bifidobacterium thermacidophilum isolated from Tibetan pigs improves growth performance, immunity, and microbiota composition in weaned piglets. J Anim Sci. (2023) 101:skad220. doi: 10.1093/jas/skad220
12. Cui, Y, Zhang, L, Wang, X, Yi, Y, Shan, Y, Liu, B, et al. Roles of intestinal Parabacteroides in human health and diseases. FEMS Microbiol Lett. (2022) 369:fnac072. doi: 10.1093/femsle/fnac072
13. Chen, L, Shen, X, Chen, G, Cao, X, and Yang, J. Effect of three-spot seahorse petroleum ether extract on lipopolysaccharide induced macrophage RAW264.7 inflammatory cytokine nitric oxide and composition analysis. J Oleo Sci. (2015) 64:933–42. doi: 10.5650/jos.ess15092
14. Liu, QM, Xu, SS, Li, L, Pan, TM, Shi, CL, Liu, H, et al. In vitro and in vivo immunomodulatory activity of sulfated polysaccharide from Porphyra haitanensis. Carbohydr Polym. (2017) 165:189–96. doi: 10.1016/j.carbpol.2017.02.032
15. Bolger, AM, Lohse, M, and Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
16. Trapnell, C, Williams, BA, Pertea, G, Mortazavi, A, Kwan, G, van Baren, MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. (2010) 28:511–5. doi: 10.1038/nbt.1621
17. Liao, Y, Wang, J, Jaehnig, EJ, Shi, Z, and Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. (2019) 47:W199–205. doi: 10.1093/nar/gkz401
18. Fu, X, Liu, Z, Zhu, C, Mou, H, and Kong, Q. Nondigestible carbohydrates, butyrate, and butyrate-producing bacteria. Crit Rev Food Sci Nutr. (2019) 59:S130–52. doi: 10.1080/10408398.2018.1542587
19. Adan, A, Kiraz, Y, and Baran, Y. Cell proliferation and cytotoxicity assays. Curr Pharm Biotechnol. (2016) 17:1213–21. doi: 10.2174/1389201017666160808160513
20. Tong, Y, Guo, H, Abbas, Z, Zhang, J, Wang, J, Cheng, Q, et al. Optimizing postbiotic production through solid-state fermentation with Bacillus amyloliquefaciens J and Lactiplantibacillus plantarum SN4 enhances antibacterial, antioxidant, and anti-inflammatory activities. Front Microbiol. (2023) 14:1229952. doi: 10.3389/fmicb.2023.1229952
21. Zhang, X, Tian, X, Wang, Y, Yan, Y, Wang, Y, Su, M, et al. Application of lipopolysaccharide in establishing inflammatory models. Int J Biol Macromol. (2024) 279:135371. doi: 10.1016/j.ijbiomac.2024.135371
22. Yang, F, Wang, A, Zeng, X, Hou, C, Liu, H, and Qiao, S. Lactobacillus reuteri I5007 modulates tight junction protein expression in IPEC-J2 cells with LPS stimulation and in newborn piglets under normal conditions. BMC Microbiol. (2015) 15:32. doi: 10.1186/s12866-015-0372-1
23. Pizarro, TT, Dinarello, CA, and Cominelli, F. Editorial: cytokines and intestinal mucosal immunity. Front Immunol. (2021) 12:698693. doi: 10.3389/fimmu.2021.698693
24. Khan, AW, Farooq, M, Hwang, MJ, Haseeb, M, and Choi, S. Autoimmune Neuroinflammatory diseases: role of interleukins. Int J Mol Sci. (2023) 24:7960. doi: 10.3390/ijms24097960
25. Rühl, A, Franzke, S, Collins, SM, and Stremmel, W. Interleukin-6 expression and regulation in rat enteric glial cells. Am J Physiol Gastrointest Liver Physiol. (2001) 280:G1163–71. doi: 10.1152/ajpgi.2001.280.6.G1163
26. Liu, W, Han, Y, An, J, Yu, S, Zhang, M, Li, L, et al. Alternation in sequence features and their influence on the anti-inflammatory activity of soy peptides during digestion and absorption in different enzymatic hydrolysis conditions. Food Chem. (2025) 471:142824. doi: 10.1016/j.foodchem.2025.142824
27. Rose-John, S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. (2012) 8:1237–47. doi: 10.7150/ijbs.4989
28. Hammond, ME, Lapointe, GR, Feucht, PH, Hilt, S, Gallegos, CA, Gordon, CA, et al. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol. (1995) 155:1428–33. doi: 10.4049/jimmunol.155.3.1428
29. Keren, Z, Braun-Moscovici, Y, Markovits, D, Rozin, A, Nahir, M, Balbir-Gurman, A, et al. Depletion of B lymphocytes in rheumatoid arthritis patients modifies IL-8-anti-IL-8 autoantibody network. Clin Immunol. (2009) 133:108–16. doi: 10.1016/j.clim.2009.07.001
30. Jones, SC, Evans, SW, Lobo, AJ, Ceska, M, Axon, AT, and Whicher, JT. Serum interleukin-8 in inflammatory bowel disease. J Gastroenterol Hepatol. (1993) 8:508–12. doi: 10.1111/j.1440-1746.1993.tb01643.x
31. Ip, WKE, Hoshi, N, Shouval, DS, Snapper, S, and Medzhitov, R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. (2017) 356:513–9. doi: 10.1126/science.aal3535
32. Ito, H. Anti-interleukin-6 therapy for Crohn's disease. Curr Pharm Des. (2003) 9:295–305. doi: 10.2174/1381612033391900
33. Gaptulbarova, KA, Tsyganov, MM, Pevzner, AM, Ibragimova, MK, and Litviakov, NV. NF-kB as a potential prognostic marker and a candidate for targeted therapy of cancer. Exp Oncol. (2020) 42:263–9. doi: 10.32471/exp-oncology.2312-8852.vol-42-no-4.15414
34. Magrone, T, Candore, G, Caruso, C, Jirillo, E, and Covelli, V. Polyphenols from red wine modulate immune responsiveness: biological and clinical significance. Curr Pharm Des. (2008) 14:2733–48. doi: 10.2174/138161208786264098
35. Rawdin, BJ, Mellon, SH, Dhabhar, FS, Epel, ES, Puterman, E, Su, Y, et al. Dysregulated relationship of inflammation and oxidative stress in major depression. Brain Behav Immun. (2013) 31:143–52. doi: 10.1016/j.bbi.2012.11.011
36. Chu, H, Du, C, Yang, Y, Feng, X, Zhu, L, Chen, J, et al. MC-LR aggravates liver lipid metabolism disorders in obese mice fed a high-fat diet via PI3K/AKT/mTOR/SREBP1 signaling pathway. Toxins. (2022) 14:833. doi: 10.3390/toxins14120833
37. Dai, B, Wu, Q, Zeng, C, Zhang, J, Cao, L, Xiao, Z, et al. The effect of Liuwei Dihuang decoction on PI3K/Akt signaling pathway in liver of type 2 diabetes mellitus (T2DM) rats with insulin resistance. J Ethnopharmacol. (2016) 192:382–9. doi: 10.1016/j.jep.2016.07.024
38. Abdullah, L, Hills, LB, Winter, EB, and Huang, YH. Diverse roles of Akt in T cells. Immunometabolism. (2021) 3:e210007. doi: 10.20900/immunometab20210007
39. Caforio, M, de Billy, E, De Angelis, B, Iacovelli, S, Quintarelli, C, Paganelli, V, et al. PI3K/Akt pathway: the indestructible role of a vintage target as a support to the Most recent immunotherapeutic approaches. Cancers. (2021) 13:4040. doi: 10.3390/cancers13164040
40. Juntilla, MM, and Koretzky, GA. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol Lett. (2008) 116:104–10. doi: 10.1016/j.imlet.2007.12.008
41. Cevey, ÁC, Mascolo, PD, Penas, FN, Pieralisi, AV, Sequeyra, AS, Mirkin, GA, et al. Benznidazole anti-inflammatory effects in murine cardiomyocytes and macrophages are mediated by class I PI3Kδ. Front Immunol. (2021) 12:782891. doi: 10.3389/fimmu.2021.782891
42. Glaviano, A, Foo, ASC, Lam, HY, Yap, KCH, Jacot, W, Jones, RH, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. (2023) 22:138. doi: 10.1186/s12943-023-01827-6
Keywords: Parabacteroides goldsteinii, IPEC-J2 cell, lipopolysaccharide, anti-inflammatory mechanism, PI3K-Akt pathway
Citation: Zhang J, Li A, He Y, Guo T, Ge Y, Pan H and Dong C (2025) Parabacteroides goldsteinii mitigates the lipopolysaccharide-induced inflammatory response in IPEC-J2 cells. Front. Vet. Sci. 12:1577126. doi: 10.3389/fvets.2025.1577126
Edited by:
Gulbeena Saleem, University of Veterinary and Animal Sciences, PakistanReviewed by:
Baseer Ahmad, Muhammad Nawaz Shareef University of Agriculture, PakistanRavikanthreddy Poonooru, University of Missouri, United States
Copyright © 2025 Zhang, Li, He, Guo, Ge, Pan and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuilian Dong, a214eXd5Y0AxNjMuY29t; Hongbin Pan, eW5zZHl6QDE2My5jb20=
†These authors have contributed equally to this work
 Junjie Zhang
Junjie Zhang Anjian Li
Anjian Li Yang He1
Yang He1 Hongbin Pan
Hongbin Pan