- 1Department of Food Science, The Pennsylvania State University, University Park, PA, United States
- 2Sensory Evaluation Center, The Pennsylvania State University, State College, PA, United States
- 3Microbiome Center, HUCK Institute for Life Sciences, The Pennsylvania State University, State College, PA, United States
Microbial diversity present on grapes in wineries, and throughout fermentation has been associated with important metabolites for final wine quality. Although microbiome-metabolome associations have been well characterized and could be used as indicators of wine quality, the impact of regionality on the microbiome and metabolome is not well known. Additionally, studies between microbiome and metabolome have been conducted on single species grape such as Vitis vinifera instead of other species and interspecific hybrids. Although the Pennsylvania wine industry is relatively young compared to California, the industry has been experiencing rapid growth over the past decade and is expected to continue to grow in the future. Pennsylvania’s climate of cold winters and high levels of rainfall throughout the growing season favors cultivation of interspecific hybrid grapes such as Vitis ssp. Chambourcin, one of the most commonly grown hybrid varieties in the state. Chambourcin is a prime candidate for studying the impact of regionality on microbiome-metabolome interactions as interspecific hybrid varieties could shape the future of winemaking. Here, we identify for the first time the regional distribution of microbial communities and their interactions with volatile metabolome during fermentation (0–20 days) by integrating high throughput Illumina sequencing (16S and ITS) and headspace-solid phase microextraction-gas chromatography-mass spectrometry. Analyzing 88 samples from nine wineries in the Central and East Pennsylvania regions, we observed high microbial diversity during early stages of fermentation (1–4 days) where non-Saccharomyces yeasts such as Starmerella and Aureobasidium and non-Oenococcus bacteria, Sphingomonas, likely contribute to microbial terroir to the resulting wines. Furthermore, key differentiators between two regions in Pennsylvania, as identified by LEfSe analysis, include the fungal genera Cladosporium and Kazachstania and the bacterial genera Lactococcus and Microbacterium. Moreover, 29 volatile fermentation metabolites were discriminated significantly (variable importance in projection > 1) between the two regions as shown by Partial Least Squares-Discriminant Analysis. Finally, Spearman’s correlation identified regional differences of microbial-metabolite associations throughout fermentation that could be used for targeted microbiome manipulation to improve wine quality and preserve regionality. In summary, these results demonstrate the microbial signatures during fermentation and differential microorganisms and metabolites further support impact of regionality on Chambourcin wines in Pennsylvania.
Introduction
Microbial communities play critical roles in complex fermentation systems such as winemaking. Several lines of evidence suggest that changes in microbial diversity and abundance throughout fermentation and winemaking can influence the physicochemical properties of final wines, control wine spoilage, and alter wine perception (Bokulich et al., 2013). In addition, native microbial populations present on grapes, in the vineyard, in the soil, and in wine processing facilities contribute to final wine quality and characteristics relative to sensory properties (Romano et al., 2019). Previous studies have demonstrated that non-Saccharomyces genera such as Hanseniaspora, Torulaspora, Pichia, and Metschnikowia can significantly and positively influence flavor profiles of final wine (Pinto et al., 2015; Bozoudi and Tsaltas, 2016; Salvetti et al., 2016; Mezzasalma et al., 2017). For example, co-inoculation of Pichia kluyveri with Saccharomyces cerevisiae can enhance 3-mercaptohexyl acetate concentration responsible for passion/grape fruit aromas in Sauvignon Blanc wines (Carrau et al., 2020). Additionally, lactic acid bacteria (LAB) such as Lactobacillus and Pediococcus play important roles in citric acid metabolism and the synthesis of esters such as diethyl succinate esters (fruity aroma) impacting final wine flavor (Inês and Falco, 2018). Terroir is a well-known concept to winemakers. Terroir is an expression that captures unique features of a region such as environmental factors and winemaking practices that can influence final wines and shape product identity within a wine region (Marlowe and Bauman, 2019). Previous studies have shown that unique microbial populations or “microbial fingerprint” present on grape berries and throughout fermentation associated at a specific geographical location can influence distinct wine characteristics in the wine region (Bokulich et al., 2014; Gilbert et al., 2014). Thus, the contribution of microbial populations on vineyards and in winery environments could also be considered a unique feature contributing terroir and can be targeted to enhance final wine quality. Understanding how microbial terroir impacts regionality of wine including the characterization of microbial terroir related to wine fault and spoilage as well as consumer perception would allow a winemaker to consider practices that preserve and enhance microorganisms within the vineyard and in wineries. This would allow for targeted control or manipulation of microbial terroir to increase flavor complexity and preserve regionality (Capozzi et al., 2015; Bokulich et al., 2016).
Pennsylvania (PA) is traditionally known as a large juice and jelly grape producer. The Pennsylvania Wine Industry is emerging as an important economic sector in Pennsylvania and has experienced continuous growth both in terms of number of wineries as well as gallons of wine produced (Thompson et al., 2019). In 2018, sales from PA wineries contributed to approximately $ 418.3 million to the state’s economy (John Dunham and Associates, 2019). One of the major challenges of growing wine grapes in the region is climate. Pennsylvania exhibits cooler temperature with humid climates characteristic of the East Coast of the United States presenting the ideal environment for growth of hybrid grapes compared to Vitis vinifera (Reisch et al., 1993; Homich et al., 2016; Thompson et al., 2019). Chambourcin, pronounced “SHAM-bour-sin,” is a French-American hybrid (Seyve-Villard 12-417 x Chancellor) wine grape variety with a relatively dark skin and neutral flavor (Robinson et al., 2012). Compared to V. vinifera, previous research suggests that Chambourcin is more tolerant to temperature fluctuations and resistant to cold temperatures (Dombrosky and Gajanan, 2013; Gardner, 2016; Homich et al., 2016). In addition, Chambourcin grape berries are more tolerant to disease pressures such as downy mildew and powdery mildew (Barlass et al., 1987; Hartman and Beale, 2008). In Pennsylvania, this variety is the most abundant hybrid grape grown in the Central, South West, and South East regions, making Chambourcin an important grape cultivar for winemaking (Dewey, 2017). A survey of 39 PA wine and grape growers indicate that winter injury followed by disease pressure is the most relevant challenge in the region (Centinari et al., 2016). Therefore, these versatile and unique characteristics of Chambourcin grown in PA could lead to a more sustainable viticulture resulting in an economic benefit while maintaining wine quality (Santos et al., 2020). Although hybrid grapes represent an important part of many winemaking regions especially in Eastern United States, the majority of wine studies have still focused on V. vinifera varieties in warmer climate areas such as California (Homich et al., 2016; Coia and Ward, 2017). Moreover, the wide variety of environmental conditions and viticultural areas within Pennsylvania can contribute to diversity in the microbial terroir, potentially leading to distinctive organoleptic wine properties from different regions. To the best of our knowledge, few field studies on actual wineries have explored how microbial diversity and the predominance of unique taxa associates with wine volatiles from hybrid grapes (Bokulich et al., 2016; Mezzasalma et al., 2017). Previous studies of microbial and metabolic dynamics focused on laboratory scale fermentation using selected microbial strains as opposed to relevant industrial environment (Torrea and Ancín, 2002; Azzolini et al., 2013). These gaps in knowledge impedes our understanding of hybrid grape selection for winemaking that could be important when dealing with changing microclimate within wine and grape growing regions. Therefore, investigating the microbiomes associated with hybrid grapes and how this microbial ecology impacts wine aroma characteristics through direct sampling within actual wineries is a necessary first step toward achieving stable and high quality of wines produced by interspecific varieties.
To address this knowledge gap in the impact of microbial populations on hybrid grapes in winemaking, we utilize an Illumina-based next generation sequencing (NGS) approach together with untargeted volatile metabolomics in a Chambourcin model system. Here, our aims are to (1) characterize the Chambourcin fermentation microbiome, (2) determine the impact of regional differences on microbial populations and volatiles, and (3) identify associations between regionally differential microbial taxa and volatile metabolites. To achieve this, we collected 88 commercial samples from 0 to 20 days of fermentation roughly correlating with early, mid-, and late fermentation stages of winemaking from wineries in the Central and East regions of Pennsylvania. To characterize wine volatile metabolites, we used gas chromatography-mass spectrometry (GC-MS) with headspace-solid-phase microextraction (HS-SPME) for non-targeted metabolite profiling of volatile compounds in all samples. Together, our work provides important insights into distinct regional characteristics of microbiome dynamics and volatile metabolome during the Chambourcin fermentation process. Therefore, understanding regional microbial signatures and volatile metabolites would allow for future targeted microbiome manipulation to improve Pennsylvania wine characteristics and competitiveness on a national market.
Materials and Methods
Sample Collection and DNA Extraction
Samples were collected during the 2019 vintage from a total of 9 commercial wineries located in two PA regions, the East region (n = 4) and the Central region (n = 5; Supplementary Figure 1). Wineries were located within ∼ 36 km in the East region and within 73 km in the Central region, with up to 228 km between wineries from the two regions. All selected wineries and vineyards grew and processed their own Chambourcin into wine. Participating wineries were provided with sample collection and handling instructions which included a survey log for sample handling outlining fermentation stages designated S1–10 based on time. These pre-determined fermentation stages for sample collection were selected based on recommendations by head winemakers from two of the nine participating wineries taking into account practical aspects of sampling. Of particular importance in this study, wineries did not change existing winemaking procedures, and thus were not prevented from using commercial S. cerevisiae for initial fermentation and/or O. oeni to initiate malolactic fermentation (MLF), however, this information was asked for in the survey log (Supplementary Table 1). Preserving winemaking practices in this study was important to capture individual winery practices that may influence microbial communities and volatile metabolite compositions. At each of the 10 pre-determined sampling points, 50 mL of the fermenting must/wine were collected in duplicate into provided sterile centrifuge tubes (VWR, Radnor, PA, United States) over a 20-day period (Supplementary Table 2); the sampling protocol was developed together with two winemakers at two of the participating wineries. Samples were stored immediately after sampling at −20°C until pick-up by the research team, transfer on dry ice to the Penn State campus at University Park, PA within 1 day, and further storage at −80°C until microbiome and metabolome analyses. Due to uncontrollable circumstances, one sample was lost during transportation and another sample was not collected during winemaking by winery staff resulting in a total of 88 unique samples, sampled in biological duplicate (n = 88; 9 wineries × 10 fermentation stages minus 2 incomplete samples that were unable to process due to transport and handling issues).
Total genomic DNA was extracted and prepared for microbiome sequencing as previously published with minor modifications (Bokulich et al., 2016). Samples from different fermentation stages and wineries were thawed and centrifuged at 8,000 × g for 15 min and supernatants were discarded. Next, pellets were washed with ice-cold phosphate-buffered saline, PBS (pH 7.4) prepared based on the protocol (Cold Spring Harbour Laboratories, 2006) and centrifuged at 8,000 × g for 15 min. Wash and centrifugation steps were repeated three times. DNA was extracted from approximately 200 mg of washed pellets from each sample using Quick-DNATM Fecal/Soil Microbe Miniprep DNA extraction kit (Zymo Research, Irvine, CA, United States). DNA concentration obtained of each sample was quantified using NanodropOne (Thermo Scientific, Waltham, MA, United States) and quality was monitored by the 260/280 ratio. DNA samples were normalized to 3 ng/μL by dilution with nuclease-free water (Life Technologies Corporation, Carlsbad, CA, United States) and stored at −80°C until further use.
Library Preparation and Sequencing
Fungal and bacterial populations in collected samples were characterized by amplicon-based sequencing of the internal transcribed spacer 2 (ITS2) sequence and the V4 domain of 16s rRNA gene, respectively. The first round PCR amplification (25 μL reaction volume) for each sample included 12 ng of DNA template, 1 × KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, United States), 0.25 μM of each primer, nuclease-free water (Life Technologies Corporation, Carlsbad, CA, United States), and 0.05 mg/mL bovine serum albumin (BSA; Sigma-Aldrich, Saint Louis, MO, United States). The fungal ITS2 locus was amplified using the forward primer ITS9 (5′-GAA CGC AGC RAA IIG YGA-3′) and reverse primer ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′; Nordberg et al., 2014), with forward Illumina adapter overhang sequences (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG-[ITS2 sequences]-3′), and reverse Illumina adapter overhang sequences (5′- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G-[ITS2 sequences]-3′; PCR Amplicon, PCR Clean-up, and Index PCR, 2013). PCR amplification was carried out initially at 98°C for 5 min, followed by 30 cycles at 95°C for 45 s, 55°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 5 min. The V4 region of bacterial 16S rRNA genes were amplified with forward primer 515F (5′-GTG YCA GCM GCC GCG GTA A-3′; Parada et al., 2016) and reverse primer 806R (5′-GGACTACNvGGGTWTCTAAT-3′; Apprill et al., 2015), with forward Illumina adapter overhang sequences (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG-[16S rRNA v4 genes sequences]-3′), and reverse Illumina adapter overhang sequences (5′- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G-[16S rRNA v4 genes sequences]-3′). Reaction conditions consisted of 98°C for 2 min, followed by 25 cycles at 95°C for 15 s, 59°C for 15 s, and 72°C for 15 s, with a final extension at 72°C for 5 min. PCR amplicons were purified using GenEluteTM PCR Clean-Up Kit (Sigma-Aldrich, Saint Louis, MO, United States) to remove single primers and primer dimers.
Purified PCR amplicons were submitted to The Pennsylvania State University HUCK Institutes of the Life Sciences Genomics Core Facility for Illumina paired-end library preparation, cluster generation, and 250-bp paired-end sequencing. Purified fungal and bacterial PCR amplicons from every twelve samples were pooled together and analyzed using a 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, United States) to assess untargeted artifacts present in samples for quality control. The same quality control and cleanup protocol was applied to the index PCR step. Equimolar concentrations of pooled libraries containing PCR amplicons were sequenced using 250-bp paired-end sequencing on an Illumina MiSeq instrument (Illumina, San Diego, CA, United States).
Microbiome-Based Bioinformatic Data Analysis Pipeline
Raw sequences obtained from Illumina MiSeq comprising of bacterial and fungal DNA were analyzed using QIIME2 v2019.7 (Bokulich et al., 2018) and the resulting data in Casava 1.8 paired-end demultiplexed format was imported using the qiime tools import plugin.
Forward and reverse reads of bacterial 16S rRNA gene sequences were truncated at base position 196 and 204, respectively, followed by denoising with the q2-DADA2 plugin (Callahan et al., 2016). One sample (PA19_02_S10) was first removed due to low sequencing reads (reads < 1,000). Amplicon sequence variants (ASVs) from a total of 86 bacterial samples were classified using the q2-feature-classifier plugin and a pre-trained Naïve Bayes classifier (Bokulich et al., 2018) with the SILVA 128 99% OTU reference database (Quast et al., 2013) for taxonomic identification. Another sample (PA19_09_S4) was removed due to unidentified taxonomic sequences. To obtain a phylogenetic tree for diversity analyses, we used a fragment-insertion plugin based on the SEPP algorithm (Janssen et al., 2018) to phylogenetically place the ASVs into the high quality preconstructed reference SILVA v128 99% identity tree (Yilmaz et al., 2014).
Raw fungal ITS2 sequences were trimmed using the q2-ITSxpress plugin (Rivers et al., 2018) and denoised using the q2-DADA2 plugin (Callahan et al., 2016). Three samples were removed due to low sequencing reads (reads < 3,000), so a total of 85 fungal samples were used for the following procedures. The q2-feature-classifier and a Naïve Bayes classifier were used for fungal taxonomy identification with the UNITE ver8 99% OTU (UNITE Community, 2019) database, trained on the full reference sequences without any extraction. To obtain a phylogenetic tree for diversity analyses, fungal ASVs were pre-filtered if sequences were lower than 80% identity to any reference sequence and clustered against the UNITE ver8 99% OTUs reference database (UNITE Community, 2019), using the QIIME vsearch cluster-features-closed-reference plugin (Nilsson et al., 2019). Clustered sequences were then aligned with a pre-built phylogenic reference tree made by the UNITE ITS extension database and the SILVA 18S database using the q2-ghost-tree plugin to construct a reference-based fungal phylogenetic tree (Fouquier et al., 2016).
The relative abundance of non-Saccharomyces or non-Oenococcus taxa at the genus level was normalized using the values determined by the reads per taxon divided by the number of summing reads for each sample (reads of S. cerevisiae or O. oeni were excluded). The relative abundance of S. cerevisiae and O. oeni was determined by the reads of the taxon at the species level divided by the number of summing reads for each sample. Alpha-diversity (within-sample) was measured using Faith’s phylogenetic diversity (Faith’s PD; Faith, 1992) and Pielou’s Evenness (Pielou, 1966) within the q2-diversity plugin using rarefied counts (i.e., normalized to the same reads across samples; normalized to 1,669 for the 16S dataset, and 16,306 for the filtered ITS2 dataset). Boxplots for alpha diversity were created using the R package ggplot2 (Wickham, 2016) in R version 3.6.3 (R Core Team, 2020) via RStudio version 1.2.13351. Pairwise comparisons between fermentation stages relative to stage 1 were performed using the Kruskal–Wallis rank-based approach for non-parametric data. A false discovery rate (FDR) adjusted p-value (q-value) was used to indicate statistical significance (q-value < 0.05; Nahm, 2016). Beta diversity was performed for quantitative measures of microbial community dissimilarity using weighted UniFrac distance metrics (Lozupone et al., 2007). Distance metrics were exported from QIIME2 and imported into R to be visualized in a Principal Coordinate Analysis (PCoA) plot using the R package qiime2R (Bisanz, 2018). Pairwise comparisons of beta diversity were tested using permutational multivariate analysis of variance (PERMANOVA) with 999 permutations, a non-parametric approach of multivariate analysis of dissimilarity based on pairwise distances (McArdle and Anderson, 2001; Supplementary Table 3).
Differentiation of microbial communities between Central and East regions at the different taxonomic levels was analyzed by the Linear discriminant analysis (LDA) Effect Size (LEfSe). Any fungal or bacterial taxa representing less than 0.01% of the total bacterial or fungal reads was filtered to avoid the influence of erroneous reads. LEfSe supports multidimensional groups comparisons and enables identification of differences between groups by coupling standard tests for statistical significance; here, a non-parametric factorial Kruskal–Wallis (KW) sum-rank test and LDA scores were used to estimate the effect size of differentially abundant taxa (Segata et al., 2011). Microbial communities were considered significantly different if their differentiation between two regions had a p-value < 0.05 and a log10 transformed LDA score > 3.
Volatile Compounds Analysis
Volatile compounds present in the 88 samples obtained at each of the 10 fermentation stages were analyzed by headspace solid phase microextraction coupled to gas chromatography-mass spectrometry (HS-SPME-GC-MS; Agilent Technologies, Santa Clara, CA, United States), using a Robotic autosampler (Gerstel, Linthicum Heights, MD, United States). A 2 cm 50/30 μm divinylbenzene- carboxen-polydimethysiloxane (DVB/CAR/PDMS) SPME fiber (Sigma-Aldrich, Inc., Saint Louis, MO, United States) was chosen for extracting volatile compounds, based on previous literature (Hopfer et al., 2013). Each sample vial contained 2 mL of sample, 3 g of NaCl (DotScientific, Burton, MI, United States), 0.5 g D-gluconic acid lactone (Sigma-Aldrich) as an inhibitor of grape β-glucosidase activity (Pedneault et al., 2013) and 10 μL of an internal standard (IS; 2-octanol 13.7 mg/L and d8-naphthalene 9.9 mg/L in Methanol; Sigma-Aldrich) for the normalization of volatile compounds from each sample. Each vial was incubated at 30°C for 5 min with shaking at 250 rpm after which the SPME fiber was exposed to the headspace for 30 min at 30°C. Extracted volatiles where thermally desorbed for 10 min in the hot (250°C) inlet equipped with a SPME inlet liner (Sigma-Aldrich) and separated in constant flow mode (1 mL/min ultrapure Helium, Praxair, State College, PA, United States) on a Rtx-WAX capillary column (30 m × 0.25 mm ID, 0.25 μm film thickness; Restek, Bellefonte, PA, United States) with an oven program starting at 30°C for 1 min, followed by a 10°C/min temperature ramp to 250°C, with a final hold of 5 min. Volatiles were identified in scan mode (33–350 amu; 8 scans/s) in electron ionization (EI) at 70 eV with the MS interface, ion source and quadrupole temperatures held at 250, 230, and 150°C. An alkane standard (C8-C20; Sigma-Aldrich) was analyzed alongside the samples to calculate retention indices (RIs) for each metabolite. Data from a total of 83 samples were further processed described below as 5 samples (PA19_05 S1-S5) were lost during laboratory preparation.
For GCMS data processing, common contaminating ions (147, 148, and 149, 207, 221, 267, and 281 m/z) were removed, followed by the Savizky-Golay filter in OpenChrom version 1.3.0 (Wenig and Odermatt, 2010). The PaRAllel FACtor analysis 2 (PARAFAC2)-based Deconvolution and Identification System (PARADISe) version 3.9 (Johnsen et al., 2017) was used to deconvolute overlapping signals, lower the signal-to-noise (S/N) ratio of chromatographic peaks, and address retention time shifts. The settings used were non-negativity and performance of 5,000 iterations for manual set retention time intervals. One to seven components calculated from the model were determined by the user to differentiate the underlying co-eluting metabolites and baseline. Deconvoluted mass spectra were then identified using the National Institute of Standards and Technology (NIST14) mass spectral library version 2.2 (Mikaia et al., 2014). A spectral library match of at least 70%, a verification of calculated RIs with literature values, and the compound being present at the last fermentation stage (S10) in all nine wineries were set as the selective cutoff for identified metabolites (n = 64; Supplementary Table 4). The rationale for selection of these core Chambourcin metabolites was that metabolites identified in the final fermentation stage (S10; day 20) more closely mimic final Chambourcin wine composition and thus represent important Chambourcin wine metabolites. For statistical analysis, volatile metabolites were log2-transformed to correct for data skewness and Pareto-scaled to reduce the effect of highly abundant metabolites, using MetaboAnalyst version 4.0 (Chong et al., 2019), prior to partial least squares-discriminant analysis (PLS-DA).
Partial least squares-discriminant analysis was performed in MetaboAnalyst to identify volatiles that differed between the two regions throughout fermentation. Volatiles with variable importance in projection (VIP) values of greater 1 were deemed important for the differentiation of regional volatile profiles. Loading values from principle component 1 and 2 were listed for the importance of volatile compounds in the PLS-DA model (Supplementary Table 5). The quality of the PLS-DA model was estimated by the cumulative R2, representing the coefficient of determination (goodness of fit), and the cumulative Q2, representing the coefficient of prediction (goodness of prediction), calculated by Leave-one-out cross-validation (LOOCV; Supplementary Figure 2). To test the effectiveness of the PLS-DA discrimination model, a permutation test with 1,000 permutations was conducted based on the ratio of between group sum of squares and within group sum of squares (B/W-ratio; Supplementary Figure 3).
Correlation Analyses of Microbiome and Metabolomes
A list of significantly different fungal and bacterial populations from LEfSe analysis and VIP scores of volatile metabolites from PLS-DA were extracted from the Central and East regions. To visualize associations between fungal or bacterial taxa and volatile metabolites, a Spearman’s rank correlation was performed using the ggcorrplot R package where correlation coefficients with FDR-adjusted p-value of less than 0.05 were retained in the correlation heatmaps (Alboukadel Kassambara, 2019).
Results
Microbial Diversity During Early Stages of Chambourcin Fermentation Provides Insights Into Pennsylvania Regional Identity of Microbiome
In this study, our aim was to characterize the impact of hybrid grape microbial populations throughout fermentation in commercial winemaking. It is important to note that we did not ask different winemaking practices such as the addition of commercial S. cerevisiae and Oenococcus oeni and the use of sulfur dioxide during fermentation to be changed for our study in order to preserve commercial winery and vineyard practices in Pennsylvania (Supplementary Table 1). To obtain a clearer picture of the taxonomic distribution throughout the 10 fermentation stages sequence reads obtained from S. cerevisiae and O. oeni were excluded and displayed separately (Figure 1).
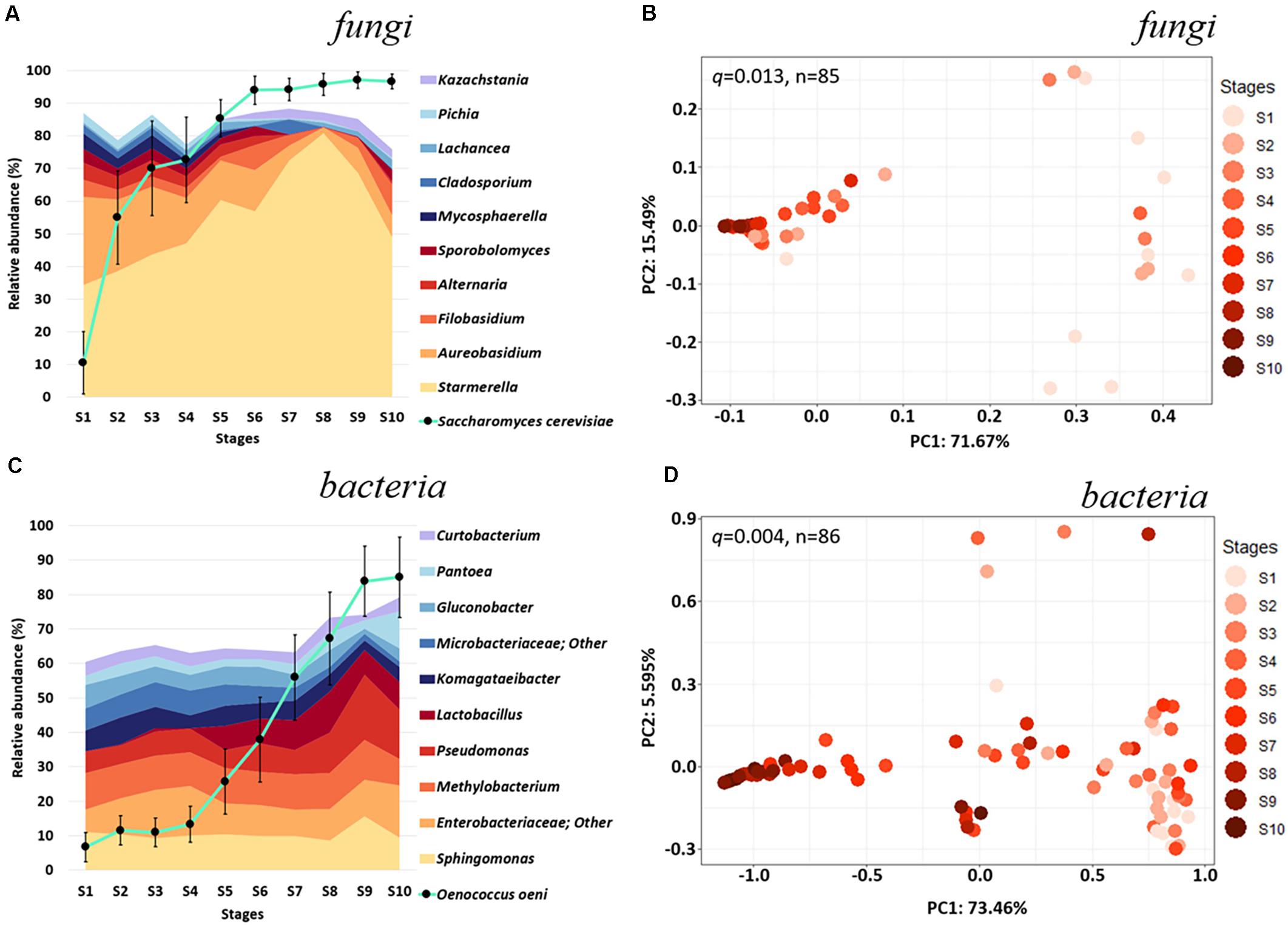
Figure 1. Microbial diversity across fermentation stages of Chambourcin red wine. Relative taxonomic distribution at the genus level displaying the top 10 (A) fungal and (C) bacterial community abundance throughout fermentation minus Saccharomyces cerevisiae and Oenococcus oeni (∙ connected by turquoise line). Taxonomic distributions for each timepoint represent the average abundance of microbial taxa detected in all samples from each fermentation stage. Weighted UniFrac PCoA for (B) fungal (n = 85) and (D) bacteria (n = 86) communities categorized by fermentation stages. Relative abundances of S. cerevisiae and O. oeni were obtained from their reads normalized to total sample reads. Error bars in (A,C) denote variance (Standard Error of the Mean, SEM) of S. cerevisiae and O. oeni abundances. Statistical significance for was determined by PERMANOVA (n = 999; Figures 1B,D and Supplementary Table 3).
Analysis of fungal taxonomy highlight a few non-Saccharomyces yeasts were predominant throughout fermentation (Figure 1A). Starmerella was the most abundant non-Saccharomyces yeast across all stages (34.31% in S1 and 48.88% in S10), followed by Aureobasidium (26.97% in S1 and 6.69% in S10), Filobasidium (5.19% in S1 and 9.51% in S10), and Alternaria (5.27% in S1 and 0.98% in S10). Together, these non-Saccharomyces yeast account for over 71.74% and 66.06% of the non-Saccharomyces fungal populations in our fermentation stage 1 and 10, respectively. Interestingly, while the abundance of most non-Saccharomyces yeast decreased in relative abundance throughout fermentation, Kazachstania increased in relative abundance toward the end of fermentation from below detection level in S1 to 2.50% in stage 10 (Figure 1A). S. cerevisiae accounted for 18.04% of the total fungal community in stage 1 and eventually becomes the dominant species by the end of fermentation (94.42%, S10; Figure 1A).
Based on beta-diversity, fermentation of Chambourcin which included the addition of commercial S. cerevisiae and O. oeni strongly influenced the structure of the fungal community with significant dissimilarity (q = 0.013, pseudo-F = 18.58) between the initial (S1) and the end of fermentation (S10; Figure 1B). In the later stages of the fermentation, the compositions of the fungal communities from 9 wineries seem to converge, possibly due to the selective pressures of S. cerevisiae driving fermentation (Supplementary Table 3).
Analyses of bacterial communities indicated that among the non-Oenococcus taxa, Sphingomonas (11.02% in S1 and 9.40% in stage 10), an unidentified Enterobacteriaceae genus (6.61% in S1 and 15.23% in S10), and Methylobacterium (10.58% in S1 and 7.55% in S10) were the most abundant throughout the 10 Chambourcin fermentation stages (Figure 1C). Additionally, Pseudomonas (6.24 in S1 and 14.50% in S10) and Lactobacillus (0.12% in S1 and 7.95% in S10) showed relatively higher abundance among non-Oenococcus bacteria in the middle to later fermentation stages (S4–S10). Two acetic acid bacteria (AAB) genera, Komagataeibacter and Gluconobacter, were more abundant during early fermentation (5.98% and 6.85% in S1 and 7.86% and 5.42% in S2), compared to middle and late fermentation (4.30% and 3.70% in S10; Figure 1C). Relative abundances of the majority of non-Oenococcus bacteria seemed to fluctuate less throughout fermentation compared to the fungi (Figures 1A–C). Similar to S. cerevisiae, O. oeni accounted for only 6.67% of the total bacterial community in S1 but dominated the bacterial population by the end of fermentation (85.07%, S10; Figure 1C). This increase was not as rapid as for the commercial yeast until stage S5 where the amount of O. oeni grew more rapidly and took place after S7 (relative abundance > 50% of total bacteria populations), which support the survey that participating wineries added O. oeni during later stages of fermentation (S5–S9; Supplementary Table 1). Interestingly, beta-diversity analysis (Figure 1D) for the bacterial communities showed a high dissimilarity between the 9 different wineries, which did not converge to the same extent as for the fungal communities in the later stages of fermentation. These fermentations did, however, undergo a significant change in the composition of the bacterial community between the beginning (S1) and end (S10) of fermentation (q = 0.004, pseudo-F = 49.34; Figure 1D and Supplementary Table 3).
Alpha-diversity indices, namely, Faith’s PD and Pielou’s evenness, were used to measure microbial richness and evenness within each fermentation stage, followed by a Kruskal–Wallis non-parametric test to detect significant changes between fermentation stages relative to S1 (Figure 2). We observed the highest richness (Faith’s PD = 10.59) and evenness (Pielou’s evenness = 0.40) within the fungal communities for S1, followed by a significant decline in both richness and evenness (q < 0.05) of the fungal communities during S3 through S5 (Figures 2A,B). This decline was characterized by a dramatic decrease in the number of different taxa followed by a succession of remaining taxa in the community (Figures 2A,B).
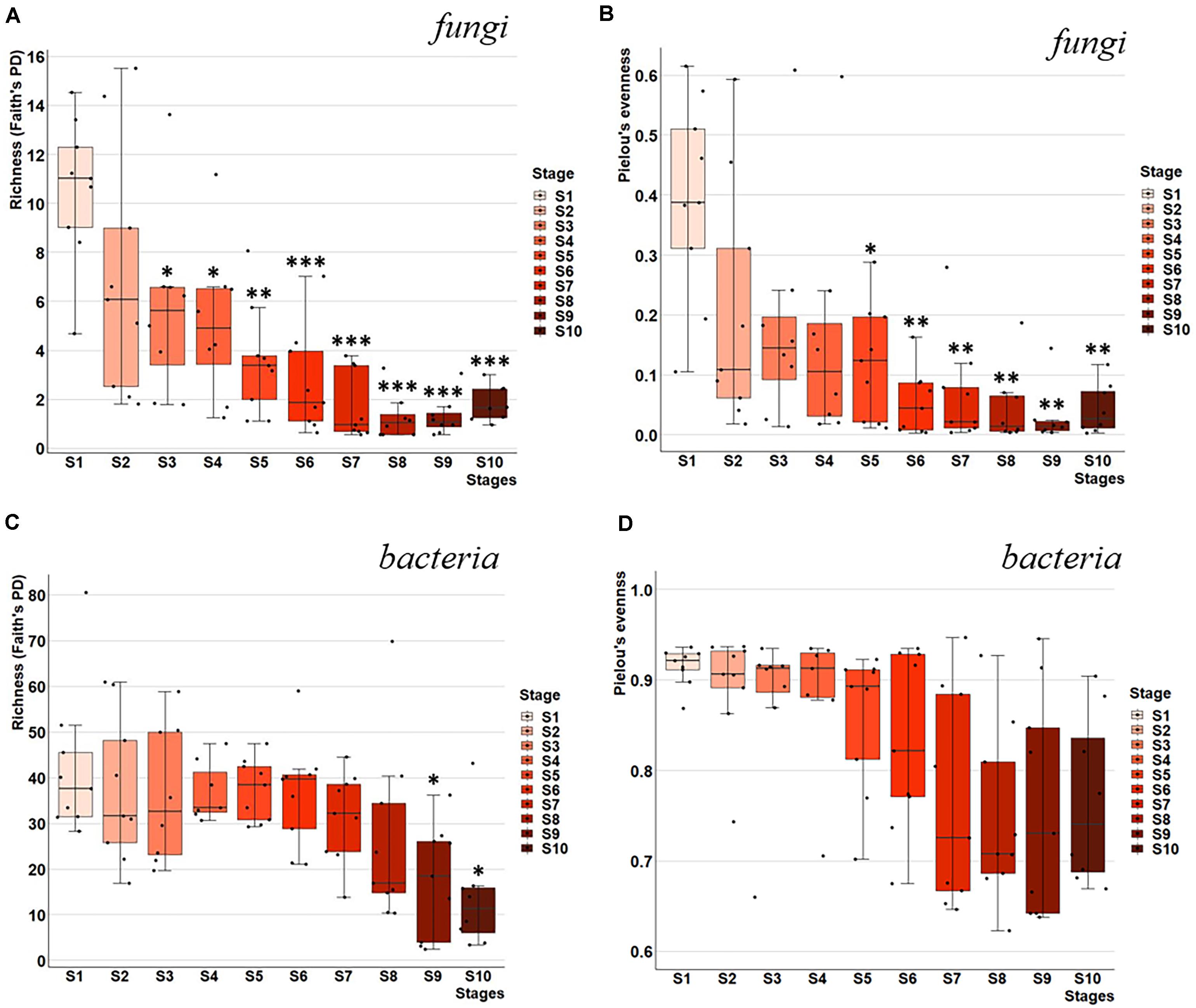
Figure 2. Microbial richness and evenness throughout fermentation stages of Chambourcin red wine. Rarefied alpha diversity distribution of microbial richness and evenness throughout fermentation stages for samples collected across nine wineries and ten fermentation stages. Faith’s phylogenetic diversity (PD) of richness for (A) fungal and (C) bacterial communities, and Pielou’s evenness for (B) fungal and (D) bacterial communities. Box plots represent the 1.5*IQR (Inter quartile range) divided by the square root of n, which corresponds to the 95% confidence interval. Significant differences between fermentation stages relative to stage 1 were determined using Kruskal–Wallis test with FDR-adjusted p-value (q-value). *q-value < 0.05, **q-value < 0.01, and ***q-value < 0.005.
In contrast, bacterial communities demonstrated much higher microbial richness compared to fungal community in S1 (Faith’s PD of 42.18 vs. 10.59; Figures 2A,C). Bacterial richness and evenness did not decrease significantly in the early stages of fermentation. While both richness and evenness decreased throughout fermentation, only richness showing a significant decrease in the last two fermentation stages S9 and S10 compared to S1 (q < 0.05; Figures 2C,D).
Differential Microbial Signatures Between Pennsylvania Central and East Regions
Besides changes in fungal and bacterial diversity obtained from ten predetermined sampling stages throughout Chambourcin fermentation (see section “Materials and Methods”), samples collected from 9 different wineries located in two different regions in PA also allowed comparison of fermentation microbiome between Central and East regions. This comparison is motivated by the research question “What is the impact of regional differences on microbial populations throughout fermentation stages in Chambourcin?”. We found that S. cerevisiae dramatically increased in abundance during early timepoints of fermentation (S1–S3) in samples from both regions, however, non-Saccharomyces yeast population showed a different pattern (Figures 3A,B). For both regions, the highest non-Saccharomyces yeast abundance was observed in earlier stages of fermentation; S1 and S2 for samples from the Central region and S1 for the East region. For both regions, the most abundant non-Saccharomyces yeast and filamentous fungi in S1 were Starmerella and Aureobasidium. However, their relative abundances and rank order differed between these two regions: Starmerella was the most dominant non-Saccharomyces yeast in the Central region (46.46% vs. 19.13% in East) while Aureobasidium was the most dominant species in the East region (30.69% vs. 23.99% in Central; Figures 3A,B). We further observed that samples from the East region showed higher abundances of Mycosphaerella and Cladosporium in the early stages S1 to S5 and Lachancea throughout the fermentation (S1–S10), while Alternaria was more abundant in samples from the Central region from S1 to S6. Furthermore, Kazachstania was only detected in samples from the Central region during middle and later fermentation stages (S5–S10; Figures 3A,B).
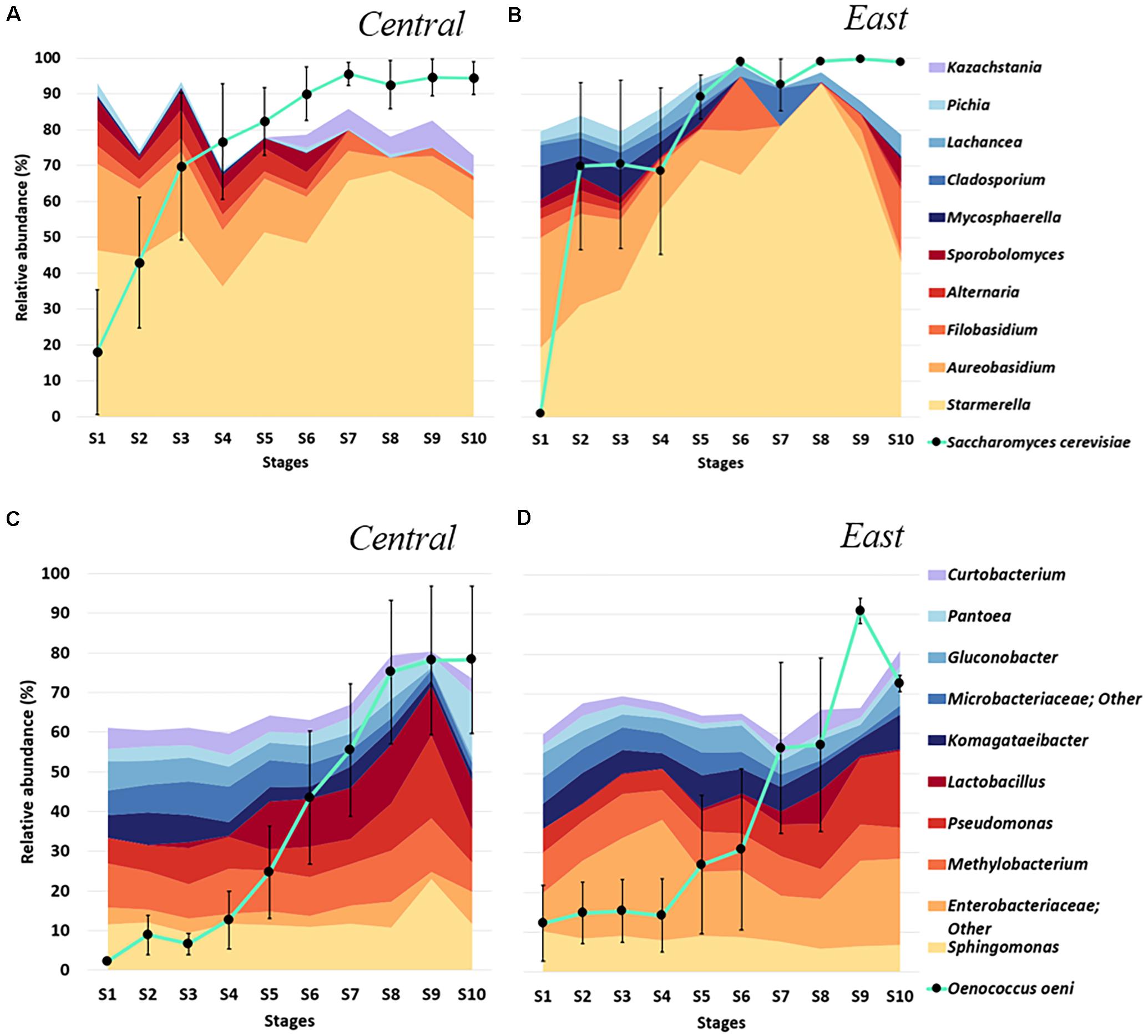
Figure 3. Microbial diversity across fermentation stages of Chambourcin winemaking in two Pennsylvania regions, (A,C) Central and (B,D) East. Relative taxonomic distribution at the genus level for the top 10 (A) fungal and (C) bacteria communities throughout fermentation minus S. cerevisiae and O. oeni (∙ connected by turquoise line). The taxonomic distribution at each timepoint represents the average abundance of microbial taxa detected in all samples from one region at each fermentation stage. Relative abundances of S. cerevisiae and O. oeni were obtained from their reads normalized to total sample reads. Error bars denote variance (Standard Error of the Mean, SEM) of S. cerevisiae and O. oeni abundances.
Among the bacteria, O. oeni which was the predominant species in samples from both regions beginning at the mid- fermentation stages (S7, >50% of whole bacteria populations), mirroring the impact of added commercial S. cerevisiae (Figures 3C,D). Similar to changes in fungal communities, the non-Oenococcus bacteria showed regional differences in samples collected from wineries in the Central vs. East regions of PA. Samples from the East region showed a higher abundance of Enterobacteriaceae related genera while Sphingomonas was more abundant in samples from the Central region throughout fermentation whereas Lactobacillus was more abundant observed in samples obtained from S5–S10 from the Central region. Interestingly, we also observed that the relative abundance of the genus Methylobacterium was relatively stable throughout fermentation in samples from both regions (Figures 3C,D). Taken together, these preliminary taxonomic distributions suggested distinct microbial fingerprints in Chambourcin samples throughout fermentation between the East and Central regions of Pennsylvania.
We employed LDA with effect size (LEfSe) to identify differences in abundance in fungal and bacterial communities at different taxonomic levels throughout fermentation stages of Chambourcin winemaking between the East and Central regions. Using an LDA score of larger than 3 and p < 0.05 as a cut-off, a total of 7 fungal species and 7 bacterial genera were identified as being significantly different in all samples from both regions (Figure 4). Analysis of the fungal taxa demonstrate that the abundance of Cladosporium tenuissimum was significantly higher in samples from the East vs. Central region across all fermentation stages, followed by Botryosphaeria agaves, Neofusicoccum parvum, Lachancea fermentati, Lachancea thermotolerans, and Pichia terricola (Figure 4A). In addition, within the Saccharomycetales order, the fungal species Kazachstania humills was identified as significantly more abundant in samples from the Central region (Figure 4A).
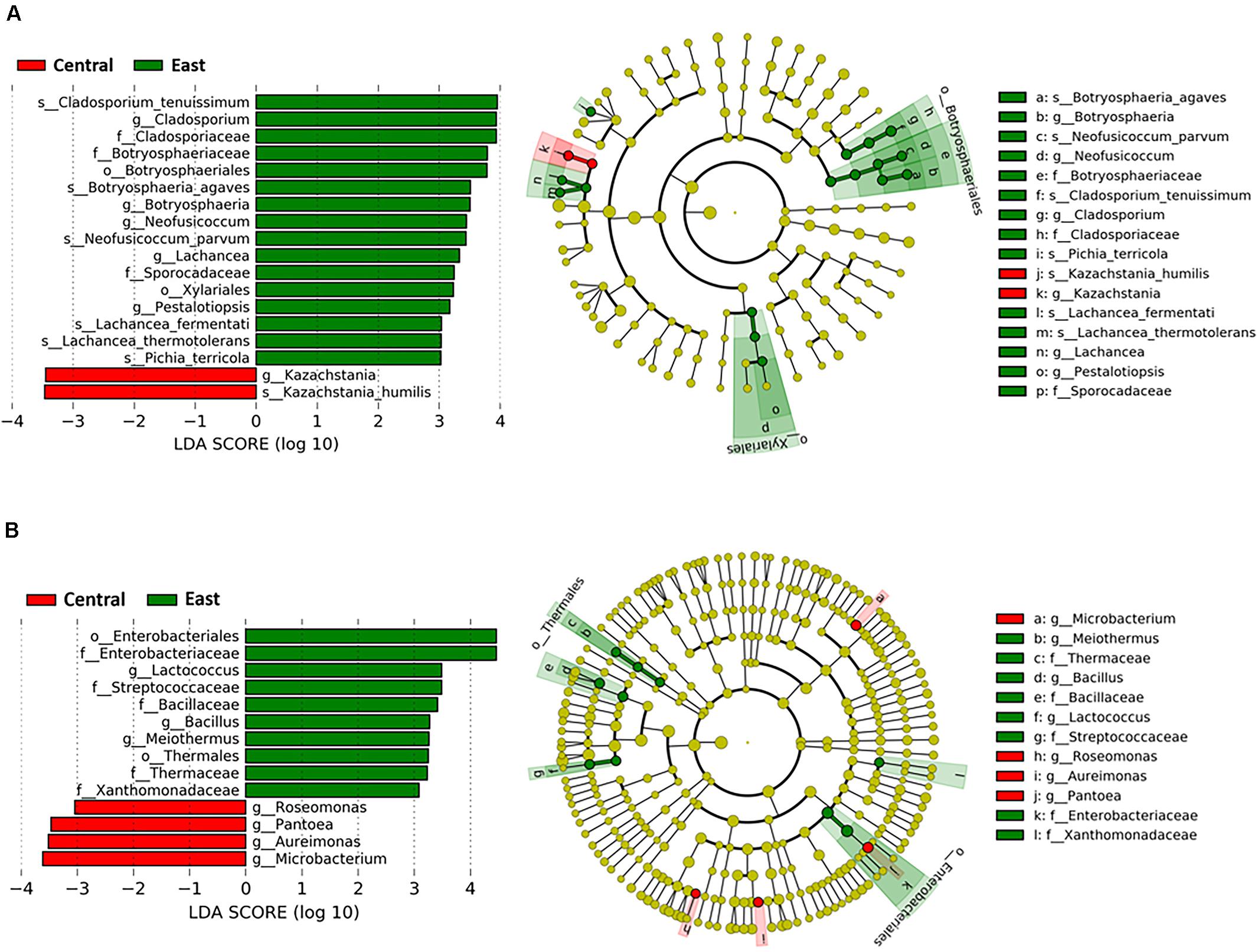
Figure 4. Taxa that discriminated between samples from the Central and East regions throughout Chambourcin fermentation. Linear discriminant analysis with effect size (LEfSe) analyses were performed using microbial relative abundance data. Data shown are the log10 linear discrimination analysis (LDA) scores following LEfSe analysis and the hierarchy of differential taxa visualized as cladograms for different taxonomic level comparisons between two regions for the (A) fungal, and (B) bacterial communities. A cut-off criterion of LDA scores >3 and p-value < 0.05 was used.
Based on our LDA score cut off, some notable differences in bacterial genera were evident between samples from the East and Central regions. In the East region, LEfSe identified Lactococcus as the most differential genus followed by Bacillus and Meiothermus. On the other hand, Microbacterium was the most significantly discriminating for the Central region, followed by Aureimonas, Pantoea, and Roseomonas (LDA > 3, p < 0.05; Figure 4B). Therefore, LEfSe analysis enabled identification of 14 differential fungal species and bacterial genera between the East and Central regions that were selected for further downstream analyses.
Differences in Volatile Metabolome Throughout Fermentation Can Help Explain Differences in Chambourcin Wine Characteristics From Central and East Regions
Volatile metabolites emitted from samples collected across all fermentation stages from each of the nine wineries in the East and Central regions were analyzed by HS-SPME-GC-MS. To identify Chambourcin-associated volatile metabolites, we identified 64 core volatile compounds which were detected across all wineries in the last fermentation stage (S10) as target compounds for downstream analyses (Supplementary Tables 4–7).
To explore potential regional differences in volatile profiles, we investigated the distribution of individual volatiles throughout fermentation in samples from the Central and East regions. The top 10 most abundant volatiles showed a similar pattern for both regions, with concentration increases in 1-Butanol, 3- methyl-, followed by Octanoic acid, ethyl ester and Phenylethyl Alcohol after mid-fermentation (S5–S10; Figure 5). In general, samples from the Central region were characterized by higher concentrations of Octanoic acid, ethyl ester while samples from the East region showed higher Phenylethyl Alcohol levels. Importantly, between fermentation stages S4-S6, volatile profiles began to differentiate by region. In samples from the Central region, a dramatic increase of volatile metabolites was observed moving from S4 to S5, including the ethyl esters Hexanoic acid, ethyl ester, and Decanoic acid, ethyl ester (Figure 5A). In contrast, in samples from the East region, most volatiles dramatically increased in between stage S5 to S6 and then decreased again in concentration, including alcohols, acetate and ethyl esters such as 1-Butanol, 3- methyl-, Octanoic acid, ethyl ester, 1-Butanol, 3- methyl-, acetate, Hexanoic acid, ethyl ester, and Decanoic acid, ethyl ester (Figure 5B). In contrast to most other volatiles which showed a concentration increase throughout fermentation, 1-Hexanol decreased in concentration across the fermentation stages in a similar fashion for both regions (Figures 5A,B).
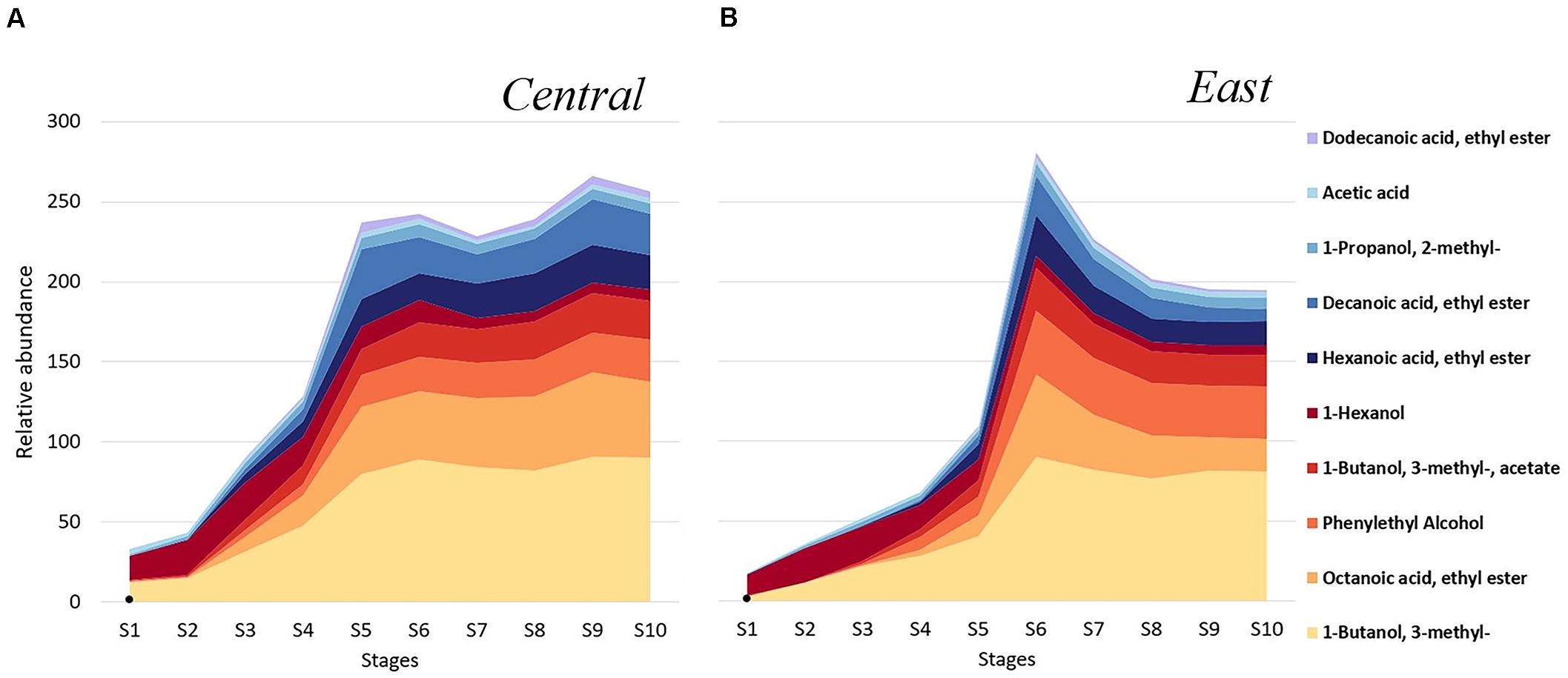
Figure 5. Chambourcin volatile metabolites distribution in wines from the Central (n = 5) and East (n = 4) regions. Shown are the top 10 most abundant volatile compounds in the samples throughout fermentation from the (A) Central, and the (B) East region. Relative abundance of each volatile metabolite was obtained from calculating metabolite peak area divided by IS peak areas which was acquired from each sample throughout each fermentation stage.
It is important to note that volatiles are produced continuously during Chambourcin wine fermentation. Regional microbial communities present on grape berries and in wineries have been shown to be an important contributor to sensory wine characteristics. To better identify volatile metabolites generated during fermentation that could be associated with specific wine-producing regions, a targeted PLS-DA model was created using the 64 core Chambourcin volatile metabolites measured from 83 fermentation samples obtained from the Central and East regions (Figure 6). A validated PLS-DA was obtained for our model based on permutation tests (p < 0.001), however, the goodness of fit and goodness of prediction indicated moderate predictive accuracy (2 components, R2 = 0.39, Q2 = 0.18). Using a score plot to visualize the separation of samples from driven by two regions, the first two model components captured 39.9% and 19.6% of the total variance, respectively, (Figure 6A and Supplementary Figures 2, 3). Our interpretation of the data suggested separation of features in the two regions to some degree: fermentation samples collected from the East region showed less variation compared to the Central region, as indicated by the size of confidence ellipses. The loadings plot in Figure 6B shows that most esters were located in the top left quadrant demonstrating high positive correlation to samples obtained from later stages of fermentation (S7–S10) from both regions. Separation along the first axis was mainly driven by differences in Hexanal (C51), (E)-2-Hexenoic acid (C49), Butanoic acid, methyl ester (C1), and Heptanal (C52) concentrations on the positive PC1 axis and Decanoic acid, ethyl ester (C18), Butanedioic acid, diethyl ester (C20), and Ethyl (S)-(-)-lactate (C10) on the negative PC 1 dimension. Furthermore, the negative PC2 axis was mainly driven by higher concentrations of 3-Buten-2-ol, 2-methyl- (C24), C1, 2-Hexen-1-ol, (Z)- (C32), and 3-Hexen-1-ol, (Z)- (C31; Supplementary Table 5). Finally, using VIP values of over 1 on either of the first two PCs as a cut-off level for metabolite discrimination between regions, the most differential volatiles (VIP > 2) were (E)-2-Hexenoic acid (C49), Heptanal (C52), and Heptanoic acid (C48), which all showed higher concentrations in samples from the East region (Figure 6C). Additional metabolites with VIP scores of more than 1.5 included Decanoic acid, ethyl ester (C18), Octanoic acid, methyl ester (C13), and Ethyl (S)-(-)-lactate (C10), which were all found in higher levels in samples from the Central region (Figures 6C,D). Using a regional PLS-DA model and VIP scores of larger than 1 as cut-off, 29 volatile metabolites from the 83 fermentation samples were selected for downstream correlation analysis.
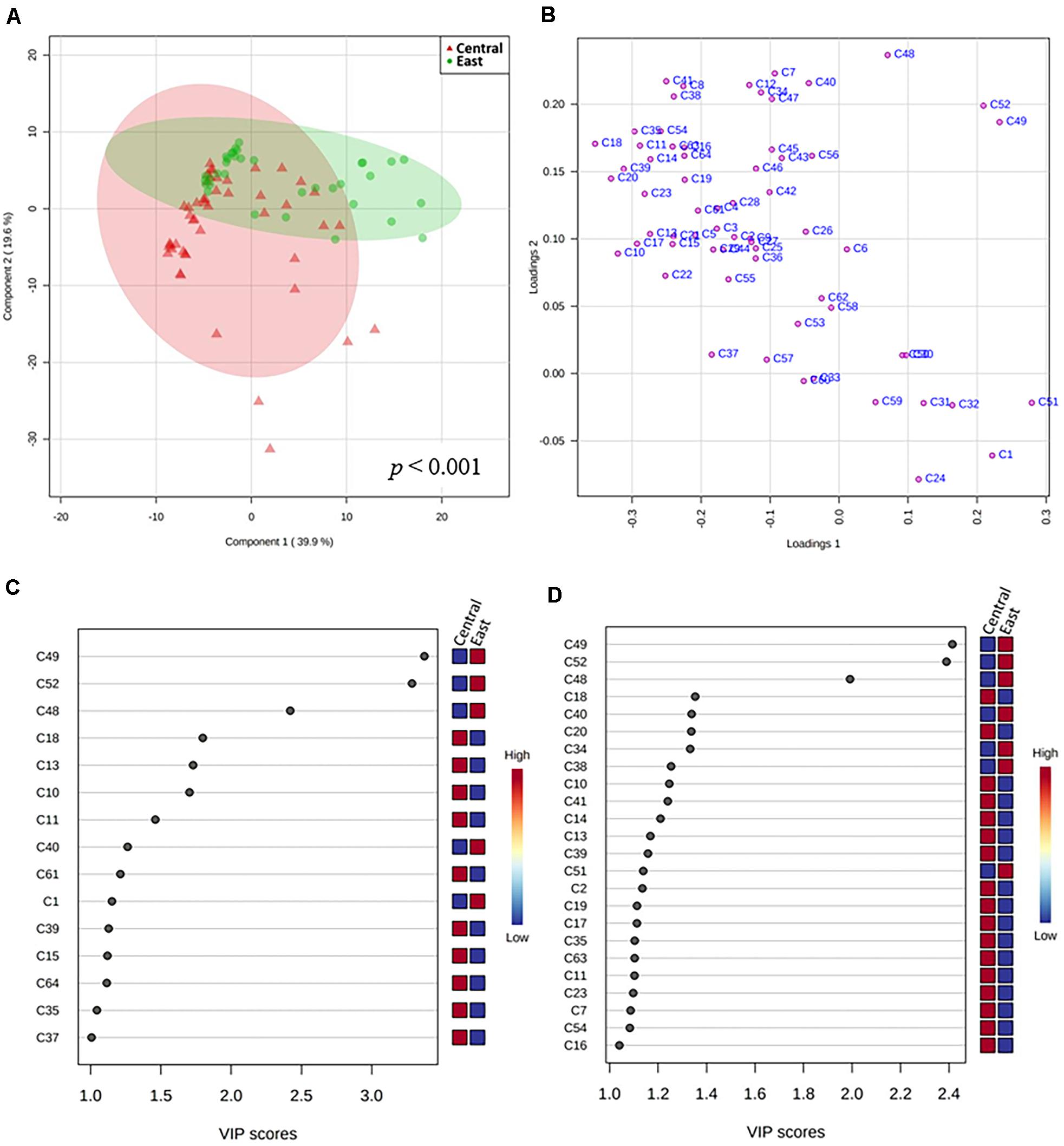
Figure 6. PLS-DA of volatile metabolites from fermentation samples collected from five Central and four East wineries. (A) Score plot showing individual samples from both regions with 95% confidence ellipses for the regions with permutation-obtained p-value of p < 0.001. (B) Loadings plot showing volatile metabolites; Important features in (C) PC1 and (D) PC2 based on variable importance in projection (VIP) values. The colored boxes on the right of the VIP scores indicate the relative concentrations of the corresponding metabolite in each region under study (VIP > 1). Note: Volatile metabolite codes are provided in Supplementary Table 5.
Fungal Taxa Strongly Associate With Volatile Metabolites in the East Region While Volatile Metabolome in the Central Region Associates With Bacterial Taxa
In this section, we aim to identify associations between microbial communities and volatile metabolites that differentiate between regions by performing a quantitative Spearman’s correlation analysis. Using all significant correlation coefficients (q < 0.05) between seven fungal species and seven bacterial genera and 29 volatile metabolites, we created a correlation heatmap to help us explain microbiome-metabolome differences of associations that are characteristic of Central and East regions (Figure 7 and Supplementary Figure 4). Most significant associations between microbial communities and volatile metabolites were negatively correlated (Fungi, 85% in the Central and 88% in the East; Bacteria, 83% in the Central and 67% in the East; Figure 7). Among those associations, we could observe that East region was characterized by strong associations between metabolites and the fungal population, while the Central was more characterized by bacterial associations. Nevertheless, it should be noted that microorganisms did not necessarily produce the correlated compound themselves, but rather may influence the overall production or consumption by the community.
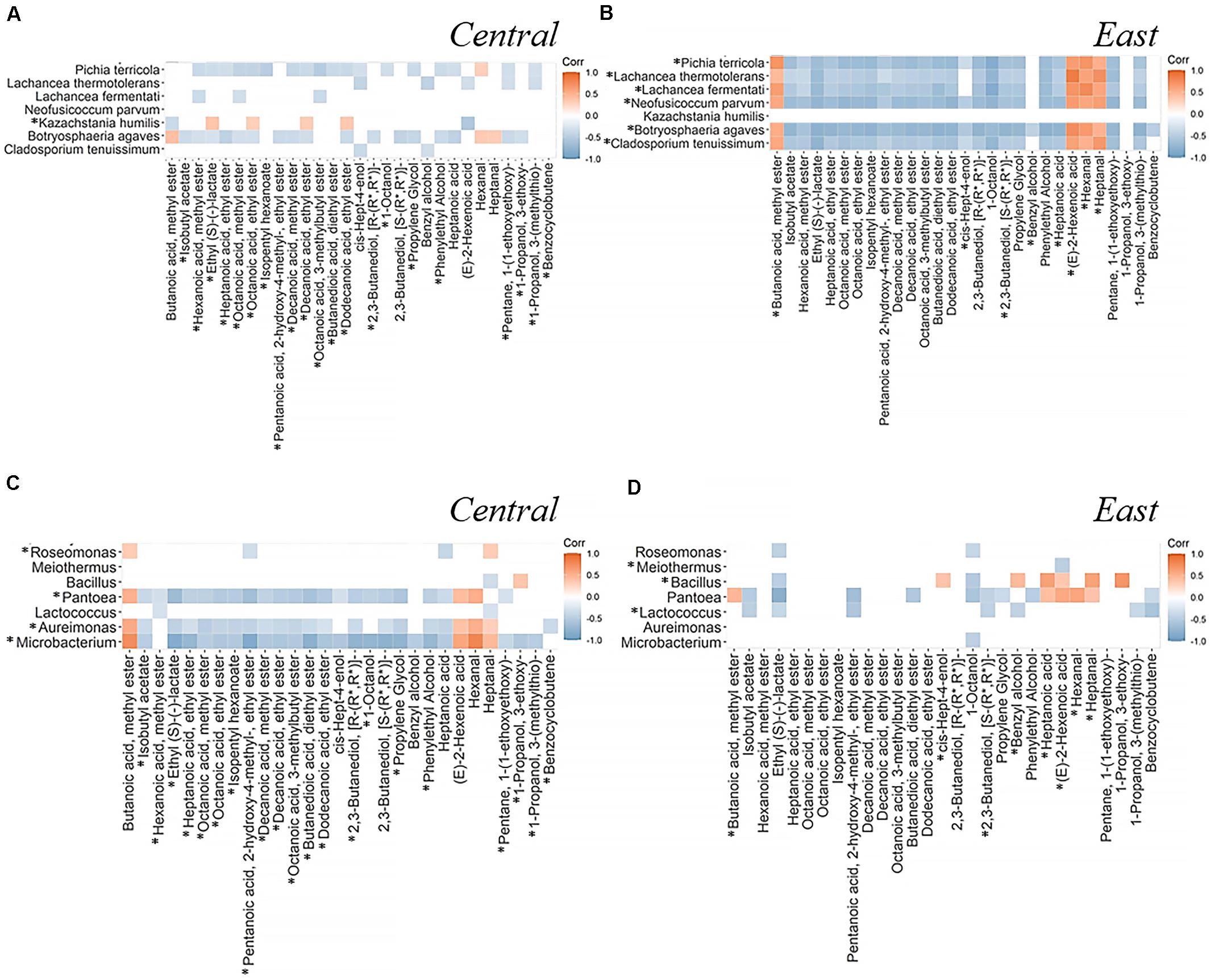
Figure 7. Spearman’s correlation analysis between significantly different microorganisms identified by LEfSe and volatile metabolites identified by PLS-DA. Data shown here are the correlation coefficients between seven fungi (A,B) or seven bacteria (C,D) and 29 volatile metabolites across fermentation stages in the Central (A,C) and East (B,D) regions. Positive correlations on the heatmap are represented by orange and negative correlation are in gray. Spearman’s correlation significance indicated by coefficient with q-value < 0.05 (coefficients with q > 0.05 are shown in white block). The comprehensive correlation heatmaps of microbial communities and volatile metabolites are provided in the Supplementary Figure 4. *highlighted the features with higher abundance for the indicated region.
As for the fungal community, Neofusicoccum parvum did not demonstrate a significant correlation in the Central region but showed a strong microbiome-metabolome correlation in the East region (q-value < 0.05). On the other hand, Lachancea thermotolerans was negatively correlated with cis-Hept-4-enol and Benzyl alcohol in the Central region but there were no significant correlations of these two metabolites in the East region. Notably, (E)-2-Hexenoic acid showed a positive correlation with L. thermotolerans in the East, which was opposite from the Central. Interestingly, Kazachstania humilis was only detected in the Central region and demonstrated opposing patterns of correlations compared with other six differential fungi. For instance, it demonstrated positive correlations with Ethyl (S)-(-)-lactate, Octanoic acid, ethyl ester and Decanoic acid, ethyl ester while negative correlations were shown in other six fungal species (Figures 7A,B).
Analysis of microbiome-metabolome interactions focused on bacterial communities highlight a total of seven different bacterial genera with distinct patterns of associations with 29 volatile metabolites from the East and Central region. For instance, genera Pantoea, Aureimonas, and Microbacterium were key features in the Central region where as Bacillus and Lactococcus were key features in the East (Figures 7C,D). Meanwhile, there were no significant correlations shown between Meiothermus and metabolites in the Central region, instead, this genus showed negative association with (E)-2-Hexenoic acid in the East region. Interestingly, Pantoea was negatively correlated with Heptanoic acid in the Central region but positively correlated with this compound in the East. In the Central region, Bacillus negatively correlated with Heptanal but the opposite correlation was found in the East. Unlike the fungal community where most species exhibited similar correlation patterns, it seemed to be more common that bacterial taxa exhibit negative associations with metabolites within a region, for example, Heptanal and 1-Propanol, 3-ethoxy- in the Central region and Benzyl alcohol and (E)-2-Hexenoic acid in the East region.
Discussion
Changes in Microbial Communities Throughout Chambourcin Fermentation Provide Insights Into the Microbiome of Interspecific Grape Varieties
Recent studies have demonstrated that microbial populations present on grape berries, in wineries, and throughout fermentation can impact final wine characteristics and contribute to terroir (Bozoudi and Tsaltas, 2016). While most studies on wine microbiome focus on V. vinifera grape varieties, the focus of this study is on Chambourcin, an interspecific hybrid grape variety widely grown in Pennsylvania and the Midwest and Eastern regions of the United States. Hybrid grapes such as Chambourcin are of great interest to the wine industry due to their resistance to diseases and adaptability to changing climates (González-Centeno et al., 2019; Santos et al., 2020). The ability to understand how microbial populations on hybrid grapes impact specific wine characteristics could help grape growers and winemakers monitor spoilage microorganisms on berries, prevent potential wine faults, and control final wine quality. In our study, alpha and beta diversity analyses of Chambourcin microbiome demonstrate decrease diversity of native microbial populations throughout fermentation. It is important to note that all participating wineries added commercial S. cerevisiae and O. oeni as oppose to spontaneous fermentation which is one factor that can impact microbial diversity.
The use of commercial S. cerevisiae and O. oeni during winemaking is a common practice in winemaking. Commercial S. cerevisiae is added relatively early during fermentation to ensure a complete fermentation and reduce the risk of stuck or sluggish fermentation. However, this practice has been shown to significantly decrease microbial diversity during fermentation (Bokulich et al., 2016; Dimitrios et al., 2019) resulting in lower complexity of wine important aroma compounds (Steensels and Verstrepen, 2014; Bozoudi and Tsaltas, 2016). Previous literature on wine microbiology emphasizes the role of fungal community, especially yeasts in winemaking. However, diversity analysis in our study demonstrate that non-Oenococcus bacterial populations maintain a certain level of population evenness without a significant dominant taxon throughout fermentation. Our results deviate from prior literature that reported the increased abundance of Gluconobacter, Komagataeibacter, or Enterobacteriaceae during fermentation. The difference in distribution patterns of bacterial communities and other wine-related studies gives a distinct bacterial signature of Pennsylvania Chambourcin fermentation (Bokulich et al., 2016; Marzano et al., 2016; Jiang et al., 2020).
Native fungal populations present on grape berries are important for winemaking and for final wine quality (Jolly et al., 2014). Despite the common practice of adding commercial S. cerevisiae to fermenting must, native fungal populations present during early stages of fermentation have been reported to establish a microbial fingerprint that influences production of important wine volatile compounds (Steensels and Verstrepen, 2014; Andorrà et al., 2019). For example, fungal population in Cabernet Sauvignon must from four different California regions were dominated by filamentous fungi, such as Cladosporium spp., Botryotinia cinerea, and Penicillium spp. (Bokulich et al., 2014). For Cannonau grape must samples collected from four localities in Sardinia, Italy the fungal community was dominated primarily by the genera, Aureobasidium, Alternaria, and Hanseniaspora (Mezzasalma et al., 2017). In our study on interspecific Chambourcin grape fermentation, Starmerella, Aureobasidium, Filobasidium, and Alternaria dominated fungal population during early fermentation (first 4 days), after which S. cerevisiae became the dominant species.
The abundance of native fungal populations on PA Chambourcin could be a characteristic differentiator of final wines. One specific example is observation of high levels of Starmerella bacillaris (synonym Candida zemplinina, abbreviated Starm. bacillaris; Sipiczki, 2003) throughout Chambourcin fermentation (except S. cerevisiae) in wine grapes must and this could be an early indicator of wine quality. S. bacillaris is frequently found on overripe grape berries because of its fructophilic character indicative of high sugar content in grapes during harvest (Englezos et al., 2017; Horváth et al., 2020). In addition, an interesting application for Starm. bacillaris is as a biocontrol fungus for preventing Botrytis cinerea infection which is often associated with overripe grapes and is a challenge grapevine in temperate climates (Lemos et al., 2016). The presence of high levels of Starm. bacillaris in our fermentation microbiome analysis could explain why we did not detect B. cinerea but only small amount of Botrytis caroliniana in the grape must (data not shown). On the other hand, adding high level of Starm. bacillaris followed by S. cerevisiae in the beginning of fermentation through sequential inoculation of wine grapes with increased maturity (high sugar content) has been highlighted as a strategy to reduce ethanol levels (Englezos et al., 2017; Goold et al., 2017). Furthermore, this inoculation practice can increase desirable compounds such as glycerol. In addition, co-inoculation of Starm. bacillaris and S. cerevisiae resulted in production of target volatile metabolite such as 2-phenylethanol, 1-hexanol, 2-methyl 1-propanol, and acetic acid that contributed to the sensory properties of Montepulciano red wines, which could be one of factors explaining our volatile compound compositions during early fermentation (Tofalo et al., 2016). In support of this, high levels of Starm. bacillaris observed in our study during early stages of fermentation likely contributes to microbial terroir and wine quality.
Next, Aureobasidium (A. pullulans) was the second most abundant fungus during early fermentation in our study. A. pullulans is well-known for the production of amylase and β-glucosidase enzymes that aid in the release of glycosylated aroma volatiles which has been shown to improve aroma perception of red wines (Baffi et al., 2013; Englezos et al., 2018). Our volatile analyses of Chambourcin samples throughout fermentation detected higher levels of phenylethyl alcohol, 3-methyl-1-butanol, and octanoic acid, ethyl ester which could be explained by previous studies that characterize the role of A. pullulans on production of typical flavor compounds of red wine (Verginer et al., 2010; Bozoudi and Tsaltas, 2018). From a spoilage standpoint, the antimicrobial activity of A. pullulans against spoilage fungi such as B. cinerea, or the bacterial pathogen Staphylococcus aureus could offer some protection during winemaking (Cho et al., 2015; Bozoudi and Tsaltas, 2018). On the other hand, considering the regionality of colonization, A. pullulans has been found in high abundance at harvest in Italy, Spain, Australia, South Africa and Canada (Wang et al., 2015; Bozoudi and Tsaltas, 2016) while it was not detected at harvest of Merlot, Cabernet Sauvignon and Cabernet Franc grapes in the Bordeaux area, France (Renouf et al., 2005). Therefore, it might indicate that the Central and East PA regions are also one of the areas suitable for A. pullulans taking its role during wine fermentation.
An interesting observation in our study is the persistence of Filobasidium (F. magnum) throughout fermentation (S1–10). Filobasidium magnum was found to be present on V. vinifera grape berries but not in the unfiltered wine (Kačániová et al., 2020) indicating that this species could play a role before alcoholic fermentation begins. In addition, F. magnum has also been isolated from apples and pears (Glushakova and Kachalkin, 2017). While previous studies have demonstrated that F. magnum is a ubiquitous in vineyards and on grape berries, the impact of F. magnum on production of wine volatile metabolites is not known. We hypothesize that its presence on different fruits and in high sugar environments would make this genera good candidates to study for potential use as starters in winemaking (Lee et al., 2011).
During wine fermentation, LAB and AAB are two main bacterial groups that are known to impact final wine characteristics (Bozoudi and Tsaltas, 2016). In this study, Sphingomonas, Enterobacteriaceae, Methylobacterium, Pseudomonas, Lactobacillus, and Komagataeibacter were the most abundant bacterial genera detected during Chambourcin fermentation. Previous studies demonstrate that Sphingomonas and Methylobacterium constitute 6–13% of the total bacterial population present on different V. vinifera grapes (Bokulich et al., 2013; Kántor et al., 2017) while Sphingomonas was shown to positively correlate with fermentation rate (Brix hr1; Bokulich et al., 2016). Likewise, Enterobacteriaceae was found to be abundant in different V. vinifera grape musts undergoing either spontaneous or inoculated fermentations showing its universal patterns in the red wine system (Pinto et al., 2015; Bokulich et al., 2016).
Our study showed that Lactobacillus was more abundant in later stages of fermentation (after S5) which could be due to higher tolerance to alcohol concentrations (Gold et al., 1992). This genus is one of the most relevant LAB in winemaking known for the production of volatile compounds that influence wine sensory attributes such as 2,3-Butanedione (Diacetyl) with a buttery, creamy aroma, geranium-smelling 2-Ethoxy-3,5-hexadiene, and vinegar-smelling acetic acid (Inês and Falco, 2018). On the contrary, AAB are commonly considered to be wine spoilage bacteria due to the production of acetaldehyde and acetic acid (Mamlouk and Gullo, 2013). In our study, two genera, Komagataeibacter and Gluconobacter, within the family of AAB were abundant from the first to the middle stage (S5), though they existed throughout fermentation. Komagataeibacter has been shown to decrease wine quality due to its ability to oxidize sugars and sugar alcohols (D-glucose, glycerol, and ethanol), excrete exopolysaccharides, and tolerate high acetic acid concentrations, leading to a high persistence of this bacteria in fermentation environments (Bokulich et al., 2013; Zhang et al., 2017). Likewise, Gluconobacter was reported to influence wine quality by oxidizing glucose and ethanol to acids (Drysdale and Fleet, 1988). According to our survey, four wineries (three in the Central and one in the East region) added sulfur dioxide during early fermentation stages (S1–S3; Supplementary Table 1). However, the abundances of these two AAB genera seem to be less affected by sulfur dioxide. This observation could be due to the concentration of active sulfites or the existence of higher sulfur dioxide-tolerating strains (Drysdale and Fleet, 1988; Andorrà et al., 2008).
In summary, high-throughput sequencing and biodiversity analysis demonstrate several dominant genera throughout fermentation of Chambourcin grapes that could contribute to the unique characteristics of Pennsylvania Chambourcin red wines. This highlights potential factors that can affect condition of grapes at harvest such as overripening or presence of Botrytis infection. It would be of interest in the future to compare microbial communities on V. vinifera grapes to other varieties within the same vineyard within the same harvest.
Wine Fermentation Microbiome and Dominant Taxa Identified by LEfSe Suggest Regional Differences Between Chambourcin From the Central and East Regions
Regional characteristics of wines could be influenced by microbial terroir, i.e., the overall fungal and bacterial distribution patterns present on grapes and throughout fermentation (Gilbert et al., 2014; Capozzi et al., 2015; Bokulich et al., 2016; Liu et al., 2019). Here, LEfSe analysis identified specific bacterial and fungal species that differed between samples collected from two different wine-growing regions in Pennsylvania. Cladosporium (C. tenuissimum) was found to be the most abundant fungi in samples from the East, followed by Botryosphaeria (B. agaves), and Neofusicoccum (N. parvum). The higher abundance of these three genera on PA Chambourcin could be explained by differences in climate, as prior studies reported that wet weathers or free water can lead to germination of fungal conidia (Niekerk et al., 2006; Espinoza et al., 2009; Wang et al., 2015). Although the presence of genetic material (DNA) does not indicate the presence of grape vine infection (Taylor et al., 2014), these filamentous fungi are considered plant pathogens which could result in poor grape quality and spoilage influencing final wine quality (Latorre et al., 2011; Pitt et al., 2012; Lorenzini et al., 2015).
Among the yeast community, Lachancea (L. fermentati), L. thermotolerans, and Pichia (P. terricola) also showed regional differences and were more abundant in samples from the East. First, L. fermentati and L. thermotolerans are two species most frequently isolated from grape must and wine fermentation processes (Porter et al., 2019b). Studies reported that L. fermentati showed a high SO2 tolerance (20 mg/L total SO2) and high fermentation activity in monoculture. In addition, the presence of L. fermentati is frequently associated with higher levels of Isobutanol and Isobutyric acid in Muscat wines and mixed-fermentations of L. fermentati with L. thermotolerans enhances production of monotepenes such as linalool and geraniol leading to perceivable aroma contribution in wine (Porter et al., 2019a,b). Furthermore, L. thermotolerans when co-cultured with S. cerevisiae was shown to contribute to the reduction in acetic acid and increase in Phenylethyl Alcohol and glycerol levels, which may help explain the higher Phenylethyl Alcohol content in samples from the East region (Figure 5; Ciani et al., 2006; Kapsopoulou et al., 2007; Comitini et al., 2011; Gobbi et al., 2013). Other than the direct production of volatile metabolites, P. terricola was reported to produce the extracellular enzyme, beta-glucosidase altering the sensory perception of Muscat wine wines (González-Pombo et al., 2011). Although yeasts of the genus Pichia has been previously shown to not persist past early stages of fermentation (Fleet et al., 1984), our study showed the persistence of Pichia throughout middle stages of fermentation (S1–5) suggesting a possible role in the ecology of wine fermentation in the East region. In samples from the Central region, Kazachstania (K. humilis) was the most discriminative non-Saccharomyces yeast. Previous studies mentioned that K. humilis is able to produce ethyl acetate, acetaldehyde, and ethanol during kaoliang and sourdough fermentation (DiCagno et al., 2014; Lai et al., 2019). Although not shown in grape fermentations, K. humilis appears to be able to influence the bacterial population and produce several volatile metabolites during different food fermentation, giving the potential role on wine characteristics.
Among the bacterial community, Lactococcus was the most differentially abundant genus in the East region followed by Bacillus. A member of the LAB Lactococcus has been reported to produce high-level diacetyl responsible for buttery flavor during dairy fermentation and has high association with carbonyl compounds in rice wine (Hugenholtz et al., 2000; Liu et al., 2015). Furthermore, it has been reported to carry out MLF and produce lactic acid and esters during white wine fermentation (Kurane and Ghosh, 2012). As for Bacillus, this genus has been found to have a positive correlation with pyrazines which are associated with herbal and vegetal aromas (Ren et al., 2019). In the Central region, Microbacterium was the most differentially abundant genus followed by Aureimonas, Pantoea, and Roseomonas. Although these grape epiphytic bacteria Microbacterium, Aureimonas, and Pantoae has been found on grapevine, leaves and grape must, their contribution to aroma attributes in winemaking is not known (Madhaiyan et al., 2013; Godálová et al., 2016; Salvetti et al., 2016; Liu et al., 2020). Additionally, Roseomonas species are reported to produce bacteriochlorophyll-a, a bacterial photosynthetic pigment, which might have negative impact on wine color (Hyeon and Jeon, 2017). Collectively, these wine-associated bacteria are strong candidates playing a key role in shaping microbial terroir of two Chambourcin growing regions of PA. As the use of Chambourcin and other hybrid varieties develops, monitoring microbial signatures may be important to maintaining regional qualities of final wines.
Differences of Volatile Metabolome Suggest Regionality of Chambourcin Wines and Key Metabolite Provided by PLS-DA Highlight Regional Characteristics of Chambourcin Fermentation Processes
Volatile aroma compounds contribute to the sensory properties and perception of wine (Vilanova et al., 2010). In our study, fermentation-derived volatile metabolites increased in concentration after stage 2 (=48 h) and reached a plateau around stage 6 (=7 days). Our results on accumulation of volatile alcohols and esters agree with previous literature reporting esters and higher alcohols as common wine fermentation metabolites. In Cabernet Sauvignon, volatile alcohols and esters increased in concentration in the first 24 to 36 h of fermentation reaching an exponential phase (72–84 h), after which they either decrease slightly or remain constant (Callejón et al., 2012). In addition, 1-Hexanol levels were previously reported to increase after 24 h and then decreased after 48 h, similar to our findings (Callejón et al., 2012); 1-Hexanol has been previously reported to correlate with green vegetable aroma (Zhang et al., 2015).
Following individual volatile metabolites throughout fermentation provides a baseline for aroma compounds in Chambourcin red wine which to date has not been documented. For example, 1-Butanol, 3-methyl- (Isoamyl alcohol) was the most abundant compound in samples from both regions after S6 (mid fermentation; 7 days after crush). Wine samples (S10) from the Central region were more abundant in Octanoic acid, ethyl ester (Ethyl Octanoate) whereas samples from the East region showed higher concentration of Phenylethyl Alcohol. Wine volatile profiles with abundant esters and higher alcohols are common to red wine fermentations (Morakul et al., 2013), and moreover, Isoamyl alcohol and Phenylethyl alcohol are reported as inherent alcohols for red wines produced from Cabernet Franc, Cabernet Sauvignon, Meritage, Merlot, Pinot noir, and Syrah, indicating similarity between the wines made from interspecific Chambourcin and V. vinifera grapes (Bejaei et al., 2019). At higher concentrations at 430 mg/L and more, isoamyl alcohol could impact wine quality negatively while Phenylethyl alcohol may contribute to the floral character in wines, however, a clear contribution of individual compounds to wine aroma is difficult due to the complexity of the volatile wine matrix (Morakul et al., 2013; De-La-Fuente-Blanco et al., 2016). Furthermore, we identified a number of acetate and ethyl esters present at high concentrations such as 1-Butanol, 3- methyl-, acetate, Decanoic acid, ethyl ester, and Hexanoic acid, ethyl ester, all commonly reported in V. vinifera red wines (Bejaei et al., 2019). Interestingly, Octanoic acid, ethyl ester and Decanoic acid, ethyl ester as the top abundant esters were reported as aroma enhancer compounds in Cabernet Sauvignon and Cabernet Gernischt wines (Welke et al., 2014); a similar positive contribution to wine aroma could be suggested for the Chambourcin red wines in our study. Acetic acid was the most abundant acid in Chambourcin red wine, however, well below any potential legal limits of 1.4 g/L (The standards of identity, 2016) and similar to red wines in general.
To further understand the difference of volatile profiles between regions, we use a PLS-DA modeling to discriminate differential volatile metabolites throughout fermentation. Although the power of predictive classification was low, our results suggest that the volatile profiles of wines from the two regions were distinct. Acids and aldehydes were found to be key features for regional differentiation. Higher abundance of volatile acids and aldehyde in wines seem to be caused by the activity of spoilage bacteria (Bartowsky, 2009). Future work will focus on these metabolites as potential features of wine quality Interestingly, comparing with a Spanish study of V. vinifera red wines that found the most differential volatile metabolites between regions to be higher alcohols (López-Rituerto et al., 2012), acids and aldehydes which were discriminated between Pennsylvania regions in our study could be a sign of terroir.
Regionality Demonstrated Different Patterns of Associations Between Microbial Taxa and Volatile Metabolites Throughout Fermentation Processes
Do changes in microbial communities and volatile metabolites reflect terroir? Volatile metabolites’ secretion from the grape matrix or conversion from other precursors by different enzymatic activities of microorganisms lead to fluctuations in their relative abundances (López et al., 2015; Dimitrios et al., 2019; Noecker et al., 2019). Therefore, using Spearman’s correlation and focusing on the relative abundances of key differential features in microorganisms and volatile metabolites characteristic of each region, we can begin to understand how microbial terroir influences regional Chambourcin wine volatile profiles (Li et al., 2018). Although, the interpretation of correlation is challenging in a complex fermentation system such as winemaking, we propose the associations of microbiome and metabolome as a “fingerprint” within Pennsylvania Chambourcin red wine (Steuer et al., 2003). Thus, microbiome-metabolome correlation heatmaps (Figure 7 and Supplementary Figure 4) could reveal previously unknown associations to help guide downstream analysis.
Our results support previous studies that highlight the importance of microbial terroir on regional identities of red wines (Bokulich et al., 2014; Knight et al., 2015). Specifically, microorganisms with a higher degree of correlation or a different correlated pattern between regions can be suggested to have a more important role on the regional aroma profile of Chambourcin fermentations in the certain region. Analysis of fungal community suggest that K. humilis is strongly correlations with specific ester metabolites in the Central than the East showing its influence during fermentation in the Central region. Furthermore, a study has also mentioned its ability to produce Octanoic acid, ethyl ester (Ethyl octanoate) and Decanoic acid, ethyl ester (Ethyl decanoate; Xu et al., 2019).
On the other hand, regional differences could be attributed to microbial interactions. It has been mentioned that regionality of soil microbiota and grapevine’s epiphytes and endophytes could modulate the abundances of other microorganisms as well as grape itself and eventually influence the quality of final wine products (Gilbert et al., 2014). In other words, the compositions of regional fungal and bacterial communities with their differential relative abundances could affect wine phenotypes through synergistic interactions (Roullier-Gall et al., 2020). For instance, L. thermotolerans has been reported to emulate MLF or pH reduction by co-inoculation with different yeasts or LAB (Morata et al., 2018). However, the interactions between other microorganisms and the ability to produce aroma compounds remain to be evaluated. Moreover, the biosynthesis of volatile metabolites have been studied to be microbial strains or species level-related, including acetaldehyde, acetoin, and acetic acid (Swiegers et al., 2005; Capece et al., 2013). Therefore, the differences of correlation patterns can be due to the underlying regional fungal strains and bacterial species which were limited identified based on our sequencing analyses. In both regions, our observation shows that certain fungi and bacteria correlated with specific volatile compounds displaying similar correlation patterns (Figure 7B). We hypothesize that this could be caused by exogenous factors such as environmental stress on hybrid grape integrity, procedural differences in winemaking, and hybrid grape ecology (vs. V. vinifera), especially for red wines which has been shown to be more easily influenced by these factors compared to white wines (Bubeck et al., 2020). Accordingly, it is possible that these exogenous factors other than fermentation drives changes in microbial composition and volatile metabolite production.
To our best knowledge, this is the first study to characterize the microbiome and volatile metabolome throughout fermentation in commercial wineries located in Pennsylvania using Chambourcin hybrid grapes as a research model. Characterization of microbial communities and the volatile metabolome could provide winemakers with data-based knowledge and expand our understanding of hybrid grape varieties and their final wine characteristics, wine spoilage control, and sustainable viticulture (Bozoudi and Tsaltas, 2016; Coia and Ward, 2017; Santos et al., 2020). Additionally, regional wine typicality or wine terroir together with higher wine quality typically results in increased consumer acceptance and appreciation (Belda et al., 2017). Therefore, the extent to which individual microorganisms and microbial terroir and the stability of communities preserve the regionality of wine aroma profiles over time still needs to be evaluated. Thus, through targeted microbial manipulation coupled with culture-dependent approaches as well as human and instrumental sensory analyses, it is possible to provide a comprehensive and robust approach to improve wine’s quality (Kontkanen et al., 2005; Swiegers et al., 2005; Cadot et al., 2012).
Data Availability Statement
All raw sequence data related to this study are available in the Sequence Read Archive (SRA) 877 under (The National Center for Biotechnology Information, NCBI) BioProject (Accession No. 878 PRJNA655761).
Author Contributions
HW and JW designed the experiment. HW prepared and analyzed the samples. HW produced the figures, tables, and wrote the manuscript. JW and DC advised on the microbiome analyses. HH advised on the volatile metabolite analyses. JW, HH, and DC advised on the figure and table production as well as the manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This research conducted at Penn State University Park is supported by the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project #PEN04699 and Accession #1019351, Pennsylvania Department of Agriculture, Wine Marketing and Research Board Project #44187022, and the Crouch Endowment for Viticulture, Enology, and Pomology Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully thank Pennsylvania wineries (Happy Valley Vineyards & Winery, Shade Mountain Winery & Vineyards, Benigna’s Creek Winery Inc, Cassel Vineyards of Hershey, Nissley Vineyards & Winery Estate, Galen Glen Vineyard & Winery, Black River Farms Inc, Tolino Vineyards, and Franklin Hill Vineyards) and Jacob Masser for their cooperation and for their essential help in collecting samples. The guidance on microbiome analyses for this study was possible thanks to help from Craig Praul (Genomics Core Facility), Xue Du, Chun Tang Feng, and Taejung Chung. The guidance on chemical analyses from Andrew Poveromo and Stephanie Keller. The completion of this study would not have been possible without their contributions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.614278/full#supplementary-material
Footnotes
References
Alboukadel Kassambara (2019). Visualization of a Correlation Matrix using “ggplot2.”. Available online at: http://www.sthda.com/english/wiki/ggcorrplot (accessed May 6, 2020).
Andorrà, I., Landi, S., Mas, A., Guillamón, J. M., and Esteve-Zarzoso, B. (2008). Effect of oenological practices on microbial populations using culture-independent techniques. Food Microbiol. 25, 849–856. doi: 10.1016/j.fm.2008.05.005
Andorrà, I., Miró, G., Espligares, N., Maria Mislata, A., Puxeu, M., and Ferrer-Gallego, R. (2019). “Wild yeast and lactic acid bacteria of wine,” in Yeasts in Biotechnology, ed. T. P. Basso (London: IntechOpen), doi: 10.5772/intechopen.84128
Apprill, A., Mcnally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Azzolini, M., Tosi, E., Faccio, S., Lorenzini, M., Torriani, S., and Zapparoli, G. (2013). Selection of Botrytis cinerea and Saccharomyces cerevisiae strains for the improvement and valorization of Italian passito style wines. FEMS Yeast Res. 13, 540–552. doi: 10.1111/1567-1364.12054
Baffi, M. A., Tobal, T., Lago, J. H. G., Boscolo, M., Gomes, E., and Da-Silva, R. (2013). Wine aroma improvement using a β-glucosidase preparation from aureobasidium pullulans. Appl. Biochem. Biotechnol. 169, 493–501. doi: 10.1007/s12010-012-9991-2
Barlass, M., Miller, R. M., and Douglas, T. J. (1987). Development of methods for screening grapevines for resistance to infection by downy mildew. II. Resveratrol Production. Am. J. Enol. Vitic. 38, 65–68.
Bartowsky, E. J. (2009). Bacterial spoilage of wine and approaches to minimize it. Lett. Appl. Microbiol. 48, 149–156. doi: 10.1111/j.1472-765X.2008.02505.x
Bejaei, M., Cliff, M. A., Madilao, L. L., and vanVuuren, H. J. J. (2019). Modelling changes in volatile compounds in british columbian varietal wines that were bottle aged for up to 120 months. Beverages 5:57. doi: 10.3390/beverages5030057
Belda, I., Zarraonaindia, I., Perisin, M., Palacios, A., and Acedo, A. (2017). From vineyard soil to wine fermentation: microbiome approximations to explain the “terroir”, Concept. Front. Microbiol. 8:821. doi: 10.3389/fmicb.2017.00821
Bisanz, J. E. (2018). qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. Available online at: https://github.com/jbisanz/qiime2R (accessed April 27, 2020).
Bokulich, N. A., Collins, T. S., Masarweh, C., Allen, G., Heymann, H., Ebeler, S. E., et al. (2016). Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7:e00631-16. doi: 10.1128/mBio.00631-16
Bokulich, N. A., Dillon, M. R., Zhang, Y., Rideout, J. R., Bolyen, E., Li, H., et al. (2018). q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems 3:e00219-18. doi: 10.1128/msystems.00219-18
Bokulich, N. A., Ohta, M., Richardson, P. M., and Mills, D. A. (2013). Monitoring seasonal changes in winery-resident microbiota. PLoS One 8:e66437. doi: 10.1371/journal.pone.0066437
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. doi: 10.1073/pnas.1317377110
Bozoudi, D., and Tsaltas, D. (2016). “Grape microbiome: potential and opportunities as a source of starter cultures,” in Grape and Wine Biotechnology, eds A. Morata and I. Loira (London: IntechOpen). doi: 10.5772/64806
Bozoudi, D., and Tsaltas, D. (2018). The multiple and versatile roles of Aureobasidium pullulans in the vitivinicultural sector. Fermentation 4:85. doi: 10.3390/fermentation4040085
Bubeck, A. M., Preiss, L., Jung, A., Dörner, E., Podlesny, D., Kulis, M., et al. (2020). Bacterial microbiota diversity and composition in red and white wines correlate with plant-derived DNA contributions and botrytis infection. Sci. Rep. 10:13828. doi: 10.1038/s41598-020-70535-8
Cadot, Y., Caillé, S., Thiollet-Scholtus, M., Samson, A., Barbeau, G., and Cheynier, V. (2012). Characterisation of typicality for wines related to terroir by conceptual and by perceptual representations. An application to red wines from the Loire Valley. Food Qual. Prefer. 24, 48–58. doi: 10.1016/j.foodqual.2011.08.012
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callejón, R. M., Margulies, B., Hirson, G. D., and Ebeler, S. E. (2012). Dynamic changes in volatile compounds during fermentation of Cabernet Sauvignon grapes with and without skins. Am. J. Enol. Vitic. 63, 301–312. doi: 10.5344/ajev.2012.12009
Capece, A., Siesto, G., Poeta, C., Pietrafesa, R., and Romano, P. (2013). Indigenous yeast population from Georgian aged wines produced by traditional “Kakhetian” method. Food Microbiol. 36, 447–455. doi: 10.1016/j.fm.2013.07.008
Capozzi, V., Garofalo, C., Chiriatti, M. A., Grieco, F., and Spano, G. (2015). Microbial terroir and food innovation: the case of yeast biodiversity in wine. Microbiol. Res. 181, 75–83. doi: 10.1016/j.micres.2015.10.005
Carrau, F., Boido, E., and Ramey, D. (2020). Yeasts for low input winemaking: microbial terroir and flavor differentiation. Adv. Appl. Microbiol. 111, 89–121. doi: 10.1016/bs.aambs.2020.02.001
Centinari, M., Kelley, K. M., Hed, B., Miller, A., and Patel-Campillo, A. (2016). Assessing growers’ challenges and needs to improve wine grape production in Pennsylvania. J. Ext. 54, 1–8.
Cho, H. S., Lee, J. H., Cho, M. H., and Lee, J. (2015). Red wines and flavonoids diminish Staphylococcus aureus virulence with anti-biofilm and anti-hemolytic activities. Biofouling 31, 1–11. doi: 10.1080/08927014.2014.991319
Chong, J., Wishart, D. S., and Xia, J. (2019). Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinforma 68:e86. doi: 10.1002/cpbi.86
Ciani, M., Beco, L., and Comitini, F. (2006). Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 108, 239–245. doi: 10.1016/j.ijfoodmicro.2005.11.012
Coia, L. R., and Ward, D. L. (2017). The hybrid grape chambourcin has a role in quality red V. vinifera blends in a New World grape growing region. J. Wine Res. 28, 326–331. doi: 10.1080/09571264.2017.1392292
Cold Spring Harbour Laboratories (2006). Phosphate-buffered saline (PBS). Cold Spring Harb. Protoc. 2006:pdb.rec8247. doi: 10.1101/pdb.rec8247.
Comitini, F., Gobbi, M., Domizio, P., Romani, C., Lencioni, L., Mannazzu, I., et al. (2011). Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28, 873–882. doi: 10.1016/j.fm.2010.12.001
UNITE Community (2019). UNITE QIIME release for Fungi. Version 18.11. 2018. UNITE Community. doi: 10.15156/BIO/786334
De-La-Fuente-Blanco, A., Sáenz-Navajas, M. P., and Ferreira, V. (2016). On the effects of higher alcohols on red wine aroma. Food Chem. 210, 107–114. doi: 10.1016/j.foodchem.2016.04.021
Dewey, R. (2017). Get to Know Pennsylvania’s Native Grapes. Pennsylvania Winer. Assoc. Available online at: https://pennsylvaniawine.com/2017/06/13/ultimate-guide-pennsylvania-grapes/ (accessed January9, 2020).
DiCagno, R., Pontonio, E., Buchin, S., DeAngelis, M., Lattanzi, A., Valerio, F., et al. (2014). Diversity of the lactic acid bacterium and yeast microbiota in the switch from firm- to liquid-sourdough fermentation. Appl. Environ. Microbiol. 80, 3161–3172. doi: 10.1128/AEM.00309-14
Dimitrios, A. A., Kamilari, E., and Tsaltas, D. (2019). “Contribution of the microbiome as a tool for estimating wine’s fermentation output and authentication,” in Advances in Grape and Wine Biotechnology, eds A. Morata and I. Loira (London: IntechOpen). doi: 10.5772/intechopen.85692
Dombrosky, J., and Gajanan, S. (2013). Pennsylvania Wine Industry-An Assessment. Cent. Rural Pennsylvania. Available online at: https://www.rural.palegislature.us/documents/reports/pa_wine_industry_2013.pdf (accessed January 4, 2020).
Drysdale, G. S., and Fleet, G. H. (1988). Acetic acid bacteria in winemaking: a review. Am. J. Enol. Vitic. 39, 143–154.
Englezos, V., Giacosa, S., Rantsiou, K., Rolle, L., and Cocolin, L. (2017). Starmerella bacillaris in winemaking: opportunities and risks. Curr. Opin. Food Sci. 17, 30–35. doi: 10.1016/j.cofs.2017.08.007
Englezos, V., Rantsiou, K., Cravero, F., Torchio, F., Giacosa, S., Ortiz-Julien, A., et al. (2018). Volatile profiles and chromatic characteristics of red wines produced with Starmerella bacillaris and Saccharomyces cerevisiae. Food Res. Int. 109, 298–309. doi: 10.1016/j.foodres.2018.04.027
Espinoza, J. G., Briceño, E. X., Chávez, E. R., Úrbez-Torres, J. R., and Latorre, B. A. (2009). Neofusicoccum spp. associated with stem canker and dieback of blueberry in Chile. Plant Dis. 93, 1187–1194. doi: 10.1094/PDIS-93-11-1187
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Fleet, G. H., Lafon-Lafourcade, S., and Ribereau-Gayon, P. (1984). Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl. Environ. Microbiol. 48, 1034–1038. doi: 10.1128/aem.48.5.1034-1038.1984
Fouquier, J., Rideout, J. R., Bolyen, E., Chase, J., Shiffer, A., McDonald, D., et al. (2016). Ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses. Microbiome 4:11. doi: 10.1186/s40168-016-0153-6
Gardner, D. M. (2016). Tasting Chambourcin: Part I. Penn State Extension Wine & Grapes U. Available online at: https://psuwineandgrapes.wordpress.com/2016/07/15/tasting-chambourcin-part-i/ (accessed January9, 2020).
Gilbert, J. A., Van DerLelie, D., and Zarraonaindia, I. (2014). Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. U.S.A. 111, 5–6. doi: 10.1073/pnas.1320471110
Glushakova, A. M., and Kachalkin, A. V. (2017). Endophytic yeasts in Malus domestica and Pyrus communis fruits under anthropogenic impact. Microbiology 86, 128–135. doi: 10.1134/S0026261716060102
Gobbi, M., Comitini, F., Domizio, P., Romani, C., Lencioni, L., Mannazzu, I., et al. (2013). Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: a strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 33, 271–281. doi: 10.1016/j.fm.2012.10.004
Godálová, Z., Kraková, L., Puškárová, A., Bučková, M., Kuchta, T., and Piknová, L’, et al. (2016). Bacterial consortia at different wine fermentation phases of two typical Central European grape varieties: Blaufränkisch (Frankovka modrá) and Grüner Veltliner (Veltlínske zelené). Int. J. Food Microbiol. 217, 110–116. doi: 10.1016/j.ijfoodmicro.2015.10.015
Gold, R. S., Meagher, M. M., Hutkins, R., and Conway, T. (1992). Ethanol tolerance and carbohydrate metabolism in lactobacilli. J. Ind. Microbiol. 10, 45–54. doi: 10.1007/BF01583633
Goold, H. D., Kroukamp, H., Williams, T. C., Paulsen, I. T., Varela, C., and Pretorius, I. S. (2017). Yeast’s balancing act between ethanol and glycerol production in low-alcohol wines. Microb. Biotechnol. 10, 264–278. doi: 10.1111/1751-7915.12488
González-Centeno, M. R., Chira, K., Miramont, C., Escudier, J. L., Samson, A., Salmon, J. M., et al. (2019). Disease resistant bouquet vine varieties: assessment of the phenolic, aromatic, and sensory potential of their wines. Biomolecules 9:793. doi: 10.3390/biom9120793
González-Pombo, P., Fariña, L., Carrau, F., Batista-Viera, F., and Brena, B. M. (2011). A novel extracellular β-glucosidase from Issatchenkia terricola: isolation, immobilization and application for aroma enhancement of white Muscat wine. Process Biochem. 46, 385–389. doi: 10.1016/j.procbio.2010.07.016
Hartman, J., and Beale, J. (2008). Powdery Mildew of Grape. Available online at: http://www.ca.uky.edu/agc/pubs/id/id21/ (accessed January14, 2020).
Homich, L. J., Scheinberg, J. A., Elias, R. J., and Gardner, D. M. (2016). Effects of co-inoculation on wine-quality attributes of the high-acid, red hybrid variety chambourcin. Am. J. Enol. Vitic. 67, 245–250. doi: 10.5344/ajev.2015.15084
Hopfer, H., Buffon, P. A., Ebeler, S. E., and Heymann, H. (2013). The combined effects of storage temperature and packaging on the sensory, chemical, and physical properties of a cabernet sauvignon wine. J. Agric. Food Chem. 61, 3320–3334. doi: 10.1021/jf3051736
Horváth, B. O., Sárdy, D. N., Kellner, N., and Magyar, I. (2020). Effects of high sugar content on fermentation dynamics and some metabolites of wine-related yeast species Saccharomyces cerevisiae, S. uvarum and Starmerella bacillaris. Food Technol. Biotechnol. 58, 76–83. doi: 10.17113/ftb.58.01.20.6461
Hugenholtz, J., Kleerebezem, M., Starrenburg, M., Delcour, J., DeVos, W., and Hols, P. (2000). Lactococcus lactis as a cell factory for high-level diacetyl production. Appl. Environ. Microbiol. 66, 4112–4114. doi: 10.1128/AEM.66.9.4112-4114.2000
Hyeon, J. W., and Jeon, C. O. (2017). Roseomonas aerofrigidensis sp. nov., isolated from an air conditioner. Int. J. Syst. Evol. Microbiol. 67, 4039–4044. doi: 10.1099/ijsem.0.002246
Inês, A., and Falco, V. (2018). “Lactic acid bacteria contribution to wine quality and safety,” in Generation of Aromas and Flavours, ed. A. Vilela (London: IntechOpen). doi: 10.5772/intechopen.81168
Janssen, S., McDonald, D., Gonzalez, A., Navas-Molina, J. A., Jiang, L., Xu, Z. Z., et al. (2018). Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 3:e00021-18. doi: 10.1128/msystems.00021-18
Jiang, L., Su, W., Mu, Y., and Mu, Y. (2020). Major metabolites and microbial community of fermented black glutinous rice wine with different starters. Front. Microbiol. 11:593. doi: 10.3389/fmicb.2020.00593
John Dunham and Associates (2019). 2018 Economic Impact Study of the Pennsylvania Wine & Grape Industries. Available online at: https://pennsylvaniawine.com/wp-content/uploads/2019/09/PAWines_EcoImpact_FULL_2018.pdf (accessed January3, 2020).
Johnsen, L. G., Skou, P. B., Khakimov, B., and Bro, R. (2017). Gas chromatography – mass spectrometry data processing made easy. J. Chromatogr. A 1503, 57–64. doi: 10.1016/j.chroma.2017.04.052
Jolly, N. P., Varela, C., and Pretorius, I. S. (2014). Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Kačániová, M., Kunová, S., Sabo, J., Ivanišová, E., Žiarovská, J., Felsöciová, S., et al. (2020). Identification of yeasts with mass spectrometry during wine production. Fermentation 6:5. doi: 10.3390/fermentation6010005
Kántor, A., Mareček, J., Ivanišová, E., Terentjeva, M., and Kačániová, M. (2017). Microorganisms of grape berries. Proc. Latv. Acad. Sci. Sect. B Nat. Exact Appl. Sci. 71, 502–508. doi: 10.1515/prolas-2017-0087
Kapsopoulou, K., Mourtzini, A., Anthoulas, M., and Nerantzis, E. (2007). Biological acidification during grape must fermentation using mixed cultures of Kluyveromyces thermotolerans and Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 23, 735–739. doi: 10.1007/s11274-006-9283-5
Knight, S., Klaere, S., Fedrizzi, B., and Goddard, M. R. (2015). Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 5:14233. doi: 10.1038/srep14233
Kontkanen, D., Reynolds, A. G., Cliff, M. A., and King, M. (2005). Canadian terroir: sensory characterization of Bordeaux-style red wine varieties in the Niagara Peninsula. Food Res. Int. 38, 417–425. doi: 10.1016/j.foodres.2004.10.010
Kurane, A. B., and Ghosh, J. S. (2012). Malolactic fermentation of Grape Wine by Lactococcus lactis var cremoris NCIM 2402. Int. Food Res. J. 19, 1767–1770.
Lai, Y.-T., Cheng, K.-C., Lai, C.-N., and Lai, Y.-J. (2019). Isolation and identification of aroma producing strain with esterification capacity from yellow water. PLoS One 14:e0211356. doi: 10.1371/journal.pone.0211356
Latorre, B. A., Briceño, E. X., and Torres, R. (2011). Increase in Cladosporium spp. populations and rot of wine grapes associated with leaf removal. Crop Prot. 30, 52–56. doi: 10.1016/j.cropro.2010.08.022
Lee, Y. J., Choi, Y. R., Lee, S. Y., Park, J. T., Shim, J. H., Park, K. H., et al. (2011). Screening wild yeast strains for alcohol fermentation from various fruits. Mycobiology 39, 33–39. doi: 10.4489/MYCO.2011.39.1.033
Lemos, W. J., Bovo, B., Nadai, C., Crosato, G., Carlot, M., Favaron, F., et al. (2016). Biocontrol ability and action mechanism of Starmerella bacillaris (synonym Candida zemplinina) isolated from wine musts against gray mold disease agent Botrytis cinerea on grape and their effects on alcoholic fermentation. Front. Microbiol. 7:1249. doi: 10.3389/fmicb.2016.01249
Li, Z., Zhang, Y., Hu, T., Likhodii, S., Sun, G., Zhai, G., et al. (2018). Differential metabolomics analysis allows characterization of diversity of metabolite networks between males and females. PLoS One 13:e0207775. doi: 10.1371/journal.pone.0207775
Liu, D., Zhang, P., Chen, D., and Howell, K. (2019). From the vineyard to the winery: how microbial ecology drives regional distinctiveness of wine. Front. Microbiol. 10:2679. doi: 10.3389/fmicb.2019.02679
Liu, S. P., Mao, J., Liu, Y. Y., Meng, X. Y., Ji, Z. W., Zhou, Z. L., et al. (2015). Bacterial succession and the dynamics of volatile compounds during the fermentation of Chinese rice wine from Shaoxing region. World J. Microbiol. Biotechnol. 31, 1907–1921. doi: 10.1007/s11274-015-1931-1
Liu, W., Li, H., Jiang, D., Zhang, Y., Zhang, S., and Sun, S. (2020). Effect of Saccharomyces cerevisiae, Torulaspora delbrueckii and malolactic fermentation on fermentation kinetics and sensory property of black raspberry wines. Food Microbiol. 91:103551. doi: 10.1016/j.fm.2020.103551
López, M. C., Mateo, J. J., and Maicas, S. (2015). Screening of β-Glucosidase and β-Xylosidase activities in four non- Saccharomyces yeast isolates. J. Food Sci. 80, C1696–C1704. doi: 10.1111/1750-3841.12954
López-Rituerto, E., Savorani, F., Avenoza, A., Busto, J. H., Peregrina, J. M., and Engelsen, S. B. (2012). Investigations of la Rioja terroir for wine production using 1H NMR metabolomics. J. Agric. Food Chem. 60, 3452–3461. doi: 10.1021/jf204361d
Lorenzini, M., Cappello, M. S., and Zapparoli, G. (2015). Isolation of Neofusicoccum parvum from withered grapes: strain characterization, pathogenicity and its detrimental effects on passito wine aroma. J. Appl. Microbiol. 119, 1335–1344. doi: 10.1111/jam.12931
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Madhaiyan, M., Hu, C. J., Roy, J. J., Kim, S.-J., Weon, H.-Y., Kwon, S.-W., et al. (2013). IUMS printed in great Britain. Int. J. Syst. Evol. Microbiol. 63, 1702–1708. doi: 10.1099/ijs.0.041020-0
Mamlouk, D., and Gullo, M. (2013). Acetic Acid Bacteria: physiology and carbon sources oxidation. Indian J. Microbiol. 53, 377–384. doi: 10.1007/s12088-013-0414-z
Marlowe, B., and Bauman, M. (2019). Terroir tourism: experiences in organic vineyards. Beverages 5:30. doi: 10.3390/beverages5020030
Marzano, M., Fosso, B., Manzari, C., Grieco, F., Intranuovo, M., Cozzi, G., et al. (2016). Complexity and dynamics of the winemaking bacterial communities in berries, musts, and wines from Apulian grape cultivars through time and space. PLoS One 11:e0157383. doi: 10.1371/journal.pone.0157383
McArdle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082[0290:fmmtcd]2.0.co;2
Mezzasalma, V., Sandionigi, A., Bruni, I., Bruno, A., Lovicu, G., Casiraghi, M., et al. (2017). Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS One 12:e0184615. doi: 10.1371/journal.pone.0184615
Mikaia, A., White, V. E., Zaikin, V., Zhu, D., Sparkman, O. D., Neta, P., et al. (2014). NIST 14 MS Database and MS Search Program v.2.2. Gaithersburg, MD: NIST.
Morakul, S., Mouret, J. R., Nicolle, P., Aguera, E., Sablayrolles, J. M., and Athès, V. (2013). A dynamic analysis of higher alcohol and ester release during winemaking fermentations. Food Bioprocess Technol. 6, 818–827. doi: 10.1007/s11947-012-0827-4
Morata, A., Loira, I., Tesfaye, W., Bañuelos, M., González, C., and Suárez Lepe, J. (2018). Lachancea thermotolerans applications in wine technology. Fermentation 4:53. doi: 10.3390/fermentation4030053
Nahm, F. S. (2016). Nonparametric statistical tests for the continuous data: the basic concept and the practical use. Korean J. Anesthesiol. 69, 8–14. doi: 10.4097/kjae.2016.69.1.8
Niekerk, J. M., van, Fourie, P. H., Halleen, F., and Crous, P. W. (2006). Botryosphaeria spp. as grapevine trunk disease pathogens. Phytopathol. Mediterr. 45, S43–S54. doi: 10.14601/Phytopathol_Mediterr-1843
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Noecker, C., Chiu, H.-C., McNally, C. P., and Borenstein, E. (2019). Defining and evaluating microbial contributions to metabolite variation in microbiome-metabolome association studies. mSystems 4:e00579-19. doi: 10.1128/msystems.00579-19
Nordberg, H., Cantor, M., Dusheyko, S., Hua, S., Poliakov, A., Shabalov, I., et al. (2014). The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 42, D26–D31. doi: 10.1093/nar/gkt1069
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Pedneault, K., Dorais, M., and Angers, P. (2013). Flavor of cold-hardy grapes: impact of berry maturity and environmental conditions. J. Agric. Food Chem. 61, 10418–10438. doi: 10.1021/jf402473u
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Pinto, C., Pinho, D., Cardoso, R., Custódio, V., Fernandes, J., Sousa, S., et al. (2015). Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front. Microbiol. 6:905. doi: 10.3389/fmicb.2015.00905
Pitt, W. M., Sosnowski, M. R., Huang, R., Qiu, Y., Steel, C. C., and Savocchia, S. (2012). Evaluation of fungicides for the management of Botryosphaeria canker of grapevines. Plant Dis. 96, 1303–1308. doi: 10.1094/PDIS-11-11-0998-RE
Porter, T. J., Divol, B., and Setati, M. E. (2019a). Investigating the biochemical and fermentation attributes of Lachancea species and strains: deciphering the potential contribution to wine chemical composition. Int. J. Food Microbiol. 290, 273–287. doi: 10.1016/j.ijfoodmicro.2018.10.025
Porter, T. J., Divol, B., and Setati, M. E. (2019b). Lachancea yeast species: origin, biochemical characteristics and oenological significance. Food Res. Int. 119, 378–389. doi: 10.1016/j.foodres.2019.02.003
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Found. Stat. Comput.
Reisch, B., Pool, R., Peterson, D., Martens, M.-H., and Henick-Kling, T. (1993). Wine and Juice Grape Varieties for Cool Climates. 1–16. Available online at: https://ecommons.cornell.edu/handle/1813/17814 (accessed January 6, 2020)
Ren, Q., Sun, L., Wu, H., Wang, Y., Wang, Z., Zheng, F., et al. (2019). The changes of microbial community and flavor compound in the fermentation process of Chinese rice wine using Fagopyrum tataricum grain as feedstock. Sci. Rep. 9:3365. doi: 10.1038/s41598-019-40337-8
Renouf, V., Claisse, O., and Lonvaud-Funel, A. (2005). Understanding the microbial ecosystem on the grape berry surface through numeration and identification of yeast and bacteria. Aust. J. Grape Wine Res. 11, 316–327. doi: 10.1111/j.1755-0238.2005.tb00031.x
Rivers, A. R., Weber, K. C., Gardner, T. G., Liu, S., and Armstrong, S. D. (2018). ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Res. 7:1418. doi: 10.12688/f1000research.15704.1
Robinson, J., Harding, J., and Vouillamoz, J. (2012). “Chambourcin,” in Wine Grapes – A Complete Guide to 1,368 Vine Varieties, Including Their Origins and Flavours (New York, NY), 218–219.
Romano, P., Ciani, M., and Fleet, G. H. (eds) (2019). Yeasts in the Production of Wine. New York, NY: Springer. doi: 10.1007/978-1-4939-9782-4
Roullier-Gall, C., David, V., Hemmler, D., Schmitt-Kopplin, P., and Alexandre, H. (2020). Exploring yeast interactions through metabolic profiling. Sci. Rep. 10:6073. doi: 10.1038/s41598-020-63182-6
Salvetti, E., Campanaro, S., Campedelli, I., Fracchetti, F., Gobbi, A., Tornielli, G. B., et al. (2016). Whole-metagenome-sequencing-based community profiles of Vitis vinifera L. cv. Corvina berries withered in two post-harvest conditions. Front. Microbiol. 7:937. doi: 10.3389/fmicb.2016.00937
Santos, J. A., Fraga, H., Malheiro, A. C., Moutinho-Pereira, J., Dinis, L.-T., Correia, C., et al. (2020). A review of the potential climate change impacts and adaptation options for European viticulture. Appl. Sci. 10:3092. doi: 10.3390/app10093092
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sipiczki, M. (2003). Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int. J. Syst. Evol. Microbiol. 53, 2079–2083. doi: 10.1099/ijs.0.02649-0
Steensels, J., and Verstrepen, K. J. (2014). Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu. Rev. Microbiol. 68, 61–80. doi: 10.1146/annurev-micro-091213-113025
Steuer, R., Kurths, J., Fiehn, O., and Weckwerth, W. (2003). Observing and interpreting correlations in metabolomic networks. Bioinformatics 19, 1019–1026. doi: 10.1093/bioinformatics/btg120
Swiegers, J. H., Bartowsky, E. J., Henschke, P. A., and Pretorius, I. S. (2005). Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11, 139–173. doi: 10.1111/j.1755-0238.2005.tb00285.x
Taylor, M. W., Tsai, P., Anfang, N., Ross, H. A., and Goddard, M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. doi: 10.1111/1462-2920.12456
The standards of identity (2016). 27 C.F.R. §4.21. GovRegs. Available online at: https://www.govregs.com/regulations/expand/title27_chapterI_part4_subpartC_section4.21#title27_chapterI_part4_subpartC_section4.21 (accessed September 22, 2020).
Thompson, L., DeVito, C., and Vigna, P. (2019). Pennsylvania Wine: Fifty Years of Progress. No. 74. Available online at: https://www.i-winereview.com/reports/Pennsylvania-i-WineReview-R74-intro.pdf (accessed June 1, 2020)
Tofalo, R., Patrignani, F., Lanciotti, R., Perpetuini, G., Schirone, M., DiGianvito, P., et al. (2016). Aroma Profile of Montepulciano d’Abruzzo wine fermented by single and co-culture starters of autochthonous Saccharomyces and Non-saccharomyces Yeasts. Front. Microbiol. 7:610. doi: 10.3389/fmicb.2016.00610
Torrea, D., and Ancín, C. (2002). Content of biogenic amines in a chardonnay wine obtained through spontaneous and inoculated fermentations. J. Agric. Food Chem. 50, 4895–4899. doi: 10.1021/jf011653h
van, J. M., Fourie, P. H., Halleen, F., and Crous, P. W. (2006). Botryosphaeria spp. as grapevine trunk disease pathogens. Phytopathol. Mediterr. 45, S43–S54. doi: 10.1094/pd-90-1490
Verginer, M., Leitner, E., and Berg, G. (2010). Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 58, 8344–8350. doi: 10.1021/jf100393w
Vilanova, M., Genisheva, Z., Masa, A., and Oliveira, J. M. (2010). Correlation between volatile composition and sensory properties in Spanish Albariño wines. Microchem. J. 95, 240–246. doi: 10.1016/j.microc.2009.12.007
Wang, C., García-Fernández, D., Mas, A., and Esteve-Zarzoso, B. (2015). Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front. Microbiol. 6:1156. doi: 10.3389/fmicb.2015.01156
Welke, J. E., Zanus, M., Lazzarotto, M., and Alcaraz Zini, C. (2014). Quantitative analysis of headspace volatile compounds using comprehensive two-dimensional gas chromatography and their contribution to the aroma of Chardonnay wine. Food Res. Int. 59, 85–99. doi: 10.1016/j.foodres.2014.02.002
Wenig, P., and Odermatt, J. (2010). OpenChrom: a cross-platform open source software for the mass spectrometric analysis of chromatographic data. BMC Bioinformatics 11:405. doi: 10.1186/1471-2105-11-405
Xu, D., Zhang, Y., Tang, K., Hu, Y., Xu, X., and Gänzle, M. G. (2019). Effect of mixed cultures of yeast and lactobacilli on the quality of wheat sourdough bread. Front. Microbiol. 10:2113. doi: 10.3389/fmicb.2019.02113
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “‘All-species Living Tree Project (LTP)”’ taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zhang, H., Xu, X., Chen, X., Yuan, F., Sun, B., Xu, Y., et al. (2017). Complete genome sequence of the cellulose-producing strain Komagataeibacter nataicola RZS01. Sci. Rep. 7:4431. doi: 10.1038/s41598-017-04589-6
Keywords: wine, hybrid grapes, fermentation, microbiome, metabolome
Citation: Wang HL, Hopfer H, Cockburn DW and Wee J (2021) Characterization of Microbial Dynamics and Volatile Metabolome Changes During Fermentation of Chambourcin Hybrid Grapes From Two Pennsylvania Regions. Front. Microbiol. 11:614278. doi: 10.3389/fmicb.2020.614278
Received: 05 October 2020; Accepted: 14 December 2020;
Published: 11 January 2021.
Edited by:
Kate Howell, The University of Melbourne, AustraliaReviewed by:
Vasileios Englezos, University of Turin, ItalyTiziana Nardi, Council for Agricultural and Economics Research (CREA), Italy
Liliana Carmen Semorile, National University of Quilmes, Argentina
Copyright © 2021 Wang, Hopfer, Cockburn and Wee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Josephine Wee, am13OTcwQHBzdS5lZHU=
 Hung Li Wang
Hung Li Wang Helene Hopfer
Helene Hopfer Darrell W. Cockburn
Darrell W. Cockburn Josephine Wee
Josephine Wee