- 1Biotechology Research Center, Technology Innovation Institute, Abu Dhabi, United Arab Emirates
- 2Medical Biotechnology Laboratory, Dr.B.R. Ambedkar Center for Biomedical Research, University of Delhi, Delhi, India
- 3Molecular Microbiology, School of Clinical and Experimental Sciences, Faculty of Medicine, University of Southampton, Southampton, United Kingdom
Tackling antimicrobial resistance requires the development of new drugs and vaccines. Artificial intelligence (AI) assisted computational approaches offer an alternative to the traditionally empirical drug and vaccine discovery pipelines. In this mini review, we focus on the increasingly important role that AI now plays in the development of vaccines and provide the reader with the methods used to identify candidate vaccine candidates for selected multi-drug resistant bacteria.
1 Introduction
Increased life expectancy in the developed world in the 20th century can be attributed to several factors – sanitation infrastructure and clean water, increased food security, population-based healthcare systems, mass vaccination programs for the most prevalent infectious diseases of childhood and the use of antibiotics. The discovery of penicillin in 1928 by Alexander Fleming and its purification by Howard Florey and Ernst Chain in 1940 ushered in the ‘antibiotic age’ and laid the foundation for exploring the potential of a plethora of novel antimicrobials (Hutchings et al., 2019; Lima et al., 2020; Vila et al., 2020). Currently, it takes about 12 years to get a drug to the market for public use and the process is hugely expensive, with “median development costs for a new antibiotic exceeding $1 billion and sponsor costs of ~$350m to complete post-approval work and manufacture of the compound during its first 10 years on the market” (Wouters et al., 2020). However, not long after the first clinical use of penicillin, micro-organisms were observed to acquire antibiotic resistance through several different mechanisms (Christaki et al., 2020; Huemer et al., 2020; Larsen et al., 2022). The World Health Organization (WHO) has highlighted the priorities for tackling drug resistance in bacteria, viruses, parasites, and fungi, which requires a globally coordinated multi-sector approach (Tacconelli et al., 2018). Antimicrobial resistance has undeniably been aggravated by i) the extensive use of broad-spectrum antibiotics in agriculture, veterinary and medical practices, ii) in self-medication, iii) with poor diagnostics, and iv) with a heavy patient load on healthcare systems (Sulis et al., 2022). Antibiotic resistance is also spreading rapidly in the environment due to contamination of water bodies through the discharge of untreated domestic sewage, hospital wastewater, effluents from the antibiotic manufacturing units, open defecation, and mass bathing. These contaminated water bodies may lead to the massive growth of resistant bacteria and act as a pool for antibiotic resistance transfer (Davies and Davies, 2010; Perry et al., 2016; Hutchings et al., 2019; Christaki et al., 2020). Prevailing antibiotic resistance has led to a rapidly diminishing pool of bioactive compounds and plausible solutions to combat the challenges posed by pathogen resistance include i) the modification of existing antibiotic classes (repurposing), ii) exploring new structural classes, and iii) the development of prophylactic vaccines. Recent antimicrobial development regimes use resistance gene(s) screening, whole genome(s) sequencing, and correct pathogen detection approaches (Vila et al., 2020; Wang et al., 2022).
Increased antibiotic resistance in micro-organisms, specifically amongst bacteria, has stimulated the development of novel antimicrobials and vaccines using Artificial Intelligence (AI), Machine Learning (ML) and Neural Networks (NN) (Dalsass et al., 2019; Stokes et al., 2020; David et al., 2021; Sahayasheela et al., 2022). The AI-based learning techniques can be categorized into three classes: 1) supervised learning, where the prediction model learns from the previous examples of classification, 2) unsupervised learning, where the prediction model learns by exploring different patterns in the training data, and 3) reinforcement learning, where the prediction model learns by implementing a scheme of operations depending upon reward sequences and penalty. In general, development of an AI-based prediction model requires data pre-processing followed by encoding, feature extraction, training/learning from the features, and testing, validation and evaluation on unseen data with the help of different AI platforms available to the developer. Some of the widely used AI platforms include TensorFlow, Google AI, Microsoft Azure, OpenAI, amongst many. TensorFlow is an open-source ML platform that offers a broad range of AI algorithms to build and design deep learning models from training data of interest. Google AI is a division of Google, focusing on artificial intelligence through the Google Cloud AI platform that hosts several AI tools armored through generative AI (for prototyping and testing generative AI models), vertex AI (to create, train, test, monitor, tune, and deploy ML and AI models), natural language AI (derive insights from unstructured text using Google machine learning), translation AI (multilingual with fast, dynamic machine translation), vision and video AI (detect objects, understand text, videos and more), and so on. The Google AI platform is another extensive suite of open-source AI algorithms with a fast and easy implementation to build used defined prediction models. Microsoft Azure is a comprehensive set of cloud services that offers solutions for AI analytics through its AI tools and frameworks, equipped with multi-layered data protection. OpenAI is an AI research and deployment driven company focusing on benefiting humanity through artificial general intelligence, controlled through a unique capped-profit model, and it delivers very powerful models (e.g. Generative Pre-trained Transformer 4, GPT-4) that are trained to generate ‘human-like’ text and to enable multi-domain applications. ChatGPT and DALL-E2 are among the famous models developed by OpenAI. OpenAI is a potential game-changing state-of-the-art AI platform with the potential to translate languages, write articles and answer operator questions. An AI platform can enable users to develop their own deep leaning models and to utilize pre-trained models for their specific research problems. Trained prediction models can be evaluated with different evaluation matrices that include mean absolute error (MSE), root mean square error (RMSE), various statistics derived from confusion matrix, accuracy, precision, area under curve (AUC), receiver operating (ROC) curve, etc. The prediction model can be implemented for practical purposes after the desired accuracy - as defined by the different evaluation matrices - is achieved (Wang et al., 2019; Yang et al., 2019; Jiang et al., 2020).
The use of AI computational approaches validated fast-tracked novel drugs, drug repurposing and vaccine designs during the COVID-19 pandemic, along with developing infection diagnostics (Shrock et al., 2020; DeGrace et al., 2022; Thomas et al., 2022). AI assisted prediction of B-cell and T-cell epitopes and novel vaccine candidates is rigorously driving immuno-informatics-based approaches towards developing refined AI- and ML-based prediction servers. AI systems reason to find the microbial components that are unlikely to mutate or alter, to guarantee that a vaccine remains effective for a long duration. Computational analysis aided with ML algorithms have played a pivotal role in vaccine development. For example, AI-based approaches can provide structural and molecular insights on SARS-CoV2 viruses and predict the viral components that can trigger potentially protective immune responses and interpret experimental findings. By combining data from multiple experimental and real-world sources, AI can monitor the genetic changes (mutations) in the SARS-CoV2 genome over time to maximize future vaccine efficacy (Waltz, 2020). Different AI and ML techniques have already demonstrated their potential in diverse healthcare and biomedicine related fields, for example by accelerating the discovery of novel antimicrobials in a cost-effective manner, reducing expenses on equipment, synthesis, and human resources. Some well-known AI/ML techniques include support vector machine (SVM), logistic regression (LR), random forest (RF), and different types of neural networks, including multi-layer perceptron (MLP), recurrent neural networks (RNN), convolutional neural networks (CNN) and deep neural networks (DNN), amongst others (Wang et al., 2019; Jiang et al., 2020; Stokes et al., 2020; Vila et al., 2020; Sahayasheela et al., 2022; Thomas et al., 2022). The SVM algorithms implement a supervised statistical leaning approach for linear and non-linear data classification and regression analysis. The LR algorithms utilize standard logistic (sigmoid), and probability (log-odds) functions derived from the explanatory variables to predict the outcome variables. The RF algorithms implement an extensive multiple decision tree approach to learn from the training data and perform the prediction. These algorithms utilize bagging and feature randomness to generate a random forest of decision trees. The MLP algorithms implement a completely connected feed-forward artificial neural network with at least three hidden layers along with an input layer and an output layer. The flow of information passes from input layer to output layers through hidden layers, controlled by an activation function. The RNN algorithms are the advanced version of the MLP algorithms where the flow of information can also occur in a cyclical manner in the hidden layer to impart recurrent learning from the input layer. Likewise, the CNN algorithms are regularized MLP algorithms. The regularization nullifies the possibility of over fitting of data during training due to fully connected layers in the MLP. The CNN algorithms implement convolutions functions instead of matrix multiplication functions. The DNN algorithms broadly cover MLP, RNN, CNN, and several other neural network algorithms. The neural network that implements multiple hidden layers can be categorized as DNN. It should be noted that the list of different AI techniques mentioned above is not exhaustive and providing their fundamental details are beyond the scope of this mini-review.
It is worth mentioning also that AI and ML are intricately related (as both are supervised learning), but conceptually different. AI is a broad field of computer science focused on creating machines or systems that can perform tasks requiring human intelligence, such as problem-solving, reasoning, and understanding natural language. ML is a subset of AI that focuses on the development (training, fine tuning, testing, and deployment) of task-orientated prediction models. ML involves training machines to learn from data and make predictions or decisions without explicit programming. In essence, ML is a technique used within the broader field of AI to achieve ‘intelligent behaviour’. Hereon in, we will use the broader term AI to embrace the diverse subfields of AI and avoid any ambiguity and inconsistency. The field of AI is anticipated to flourish in coming years to become a vital tool to combat microbial infectious diseases. Considering the principles of AI based techniques, the focus of this mini-review is to provide an overview of current AI practices in the discovery and development of vaccines.
2 AI in vaccine development
The 21st century has seen unparalleled and innovative growth and development of cutting-edge technologies to benefit society (Jaffee et al., 2017; Vetrano et al., 2022). AI and ML technologies have made significant contributions to the healthcare and medicine sectors, with the market expected to expand to $45.2 billion by 2025 (Kruk et al., 2018; Ahmed et al., 2020). Recent advances in high throughput experimental techniques have generated a considerable amount of ‘big data’ in the healthcare and biomedical sectors. These data sets fuel AI to deliver highly accurate projections and predictions in the fields of vaccine development and drug discovery. Notably, ~40% of companies engaged in drug discovery are using various AI techniques to identify drug targets and for novel drug design (Lee et al., 2022; Zeng et al., 2022; Blanco-González et al., 2023).
The emergence of new infectious pathogens and increased antimicrobial resistance amongst existing pathogens has accelerated AI-assisted vaccine development in recent years (Thomas et al., 2022). The availability of high throughput genomic and proteomic data garnered from various infectious diseases could serve as a catalyst for developing reliable AI driven predictive models. Vaccine design benefits from an AI-assisted better understanding of the pathogen infection cycle at the genetic, molecular, and cellular levels (Ong and He, 2022; Goodswen et al., 2023).
Identification of potential antigen(s) is the important step for vaccine development. Conventional experimental approaches for antigen detection were laborious and time intensive and the arrival of Reverse Technology (RV) has revolutionized the field of vaccine development. RV is a computationally assisted genome-based approach of vaccine design that circumvents the necessity of developing bacterial cultures to prioritize vaccine targets. The analysis of pathogen genomes with computational bioinformatics has benefitted the screening of potential vaccine candidates significantly. Recent advances in RV techniques are assisted by the incorporation of modern AI techniques at various stages of vaccine development, and these AI based models are beginning to expedite the discovery and optimization of potential vaccine candidates. The contribution of AI computational tools to the development of SARS-CoV-2 vaccines has demonstrated the potential of AI for vaccine development against different microbial pathogens. One of the major challenges for AI is to accurately identify the potential antigens that can trigger host immune responses from amongst the thousands of pathogen components. In the following section we discuss the relevance of AI in vaccine development, its checkpoints, and potential solutions.
2.1 Reverse vaccinology and newer AI-based methods
In general, most vaccines in use today induce antibody-mediated immunity, and they include live-attenuated vaccines, inactivated vaccines, messenger RNA vaccines, toxoid vaccines, recombinant and conjugate vaccines, viral vector vaccines, etc. Identification of the protein/peptide antigen that stimulates immunity is the first step in vaccine design (Schubert-Unkmeir and Christodoulides, 2013; Guimaraes et al., 2015; Hardt et al., 2016; Vetter et al., 2018) and AI methods can deliver reliable antigen identification in a time efficient manner and with improved accuracy. Vaxijen was one of the first AI driven prediction methods for antigen identification, and it assumed that antigenicity was inherently encoded in the protein sequence and could be captured directly through the chemical properties of the amino acid residues. The prediction model was trained on the chemical properties of known bacterial antigens to deliver highly reliable and robust results for antigen identification and quantification (Doytchinova and Flower, 2007).
Affordable and fast sequencing technologies have made microbial whole genome sequences available widely and publicly. The first successful implementation of RV resulted in the eventual development of the meningitis vaccine Bexsero/4CMenB, which includes the antigens Neisseria Heparin Binding Antigen (NHBA, previously known as Genome-derived Neisseria Antigen (GNA) 2132), factor H binding protein, (fHBP or lipoprotein (LP)2086, previously known as GNA1870) and Neisserial adhesin A (NadA, previously known as GNA1994). In addition, GNA1030 and GNA2091 were selected because they also induced protective immunity and these were fused to NHBA and fHBP, respectively, which enhanced immune responses to the individual antigens (Pizza et al., 2000; Masignani et al., 2019; Deghmane and Taha, 2022). Bexsero is currently the only RV-developed bacterial vaccine that has been introduced into the routine immunization schedules for children. It should be stressed that the development of Bexsero vaccine did not include direct assistance of modern-day AI techniques but laid the foundation of incorporating various in silico tools into a RV based vaccine development pipeline. The development of Bexsero is a noteworthy achievement in vaccinology, independent from contemporary AI technologies.
Computational analysis and screening of several different pathogen genomes and proteomes is being used broadly to identify potential vaccine candidates using RV (Ong and He, 2022; Thomas et al., 2022). Exploring host-pathogen interactions at the molecular level and implementation of genomic and proteomics approaches should provide novel insights into the mechanisms of acquiring protective immunity and assist in developing next-generation vaccines. Several computational methods/pipelines have been developed since the introduction of RV and these use various AI techniques (Hardt et al., 2016; Vetter et al., 2018; Bradley et al., 2019; Yang et al., 2019; Ong and He, 2022; Thomas et al., 2022) (Table 1).
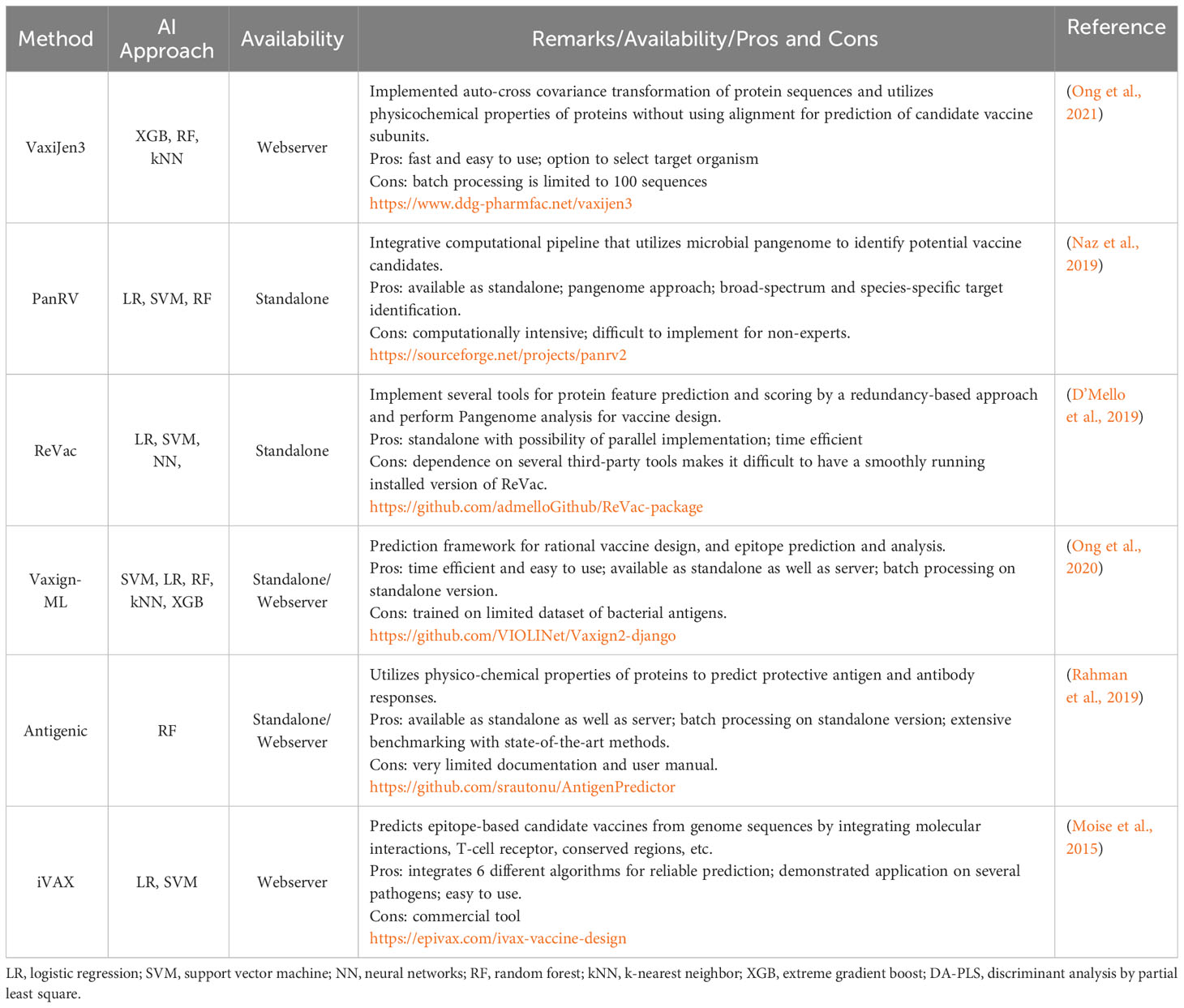
Table 1 A summary of AI-based state of the art methods/pipelines for identifying potential vaccine candidates.
The pace of development in vaccine-informatics requires that emerging novel technologies are integrated with the existing state-of-the-art methods/pipelines for improved accuracy in antigen selection and optimization. However, this integration is ongoing and yet to be achieved.
2.2 Success stories of AI assisted vaccine development against multidrug resistant bacteria
Recent advances of AI techniques, especially several variants of ML with an ability to rationally learn from the features derived from large volumes of data, have resulted in several applications in medicine and healthcare (Yang et al., 2019; Stokes et al., 2020; Melo et al., 2021; Sahayasheela et al., 2022; Thomas et al., 2022). We are potentially entering a technologically advanced, efficient, and productive era of vaccine development, supported with generous global finance. The fast-tracked development of various COVID-19 mRNA vaccines is irrefutable proof-of-concept for AI-assisted vaccine development against different pathogens (Polack et al., 2020; Feikin et al., 2022; Zheng et al., 2022). Several studies have used the AI driven methods described in Table 1 to try and develop bacterial vaccines. For example, in silico identification of 22 membrane proteins as potential antigens in the Helicobacter pylori proteome was done using AI approaches (Rahman et al., 2020). Likewise, Acinetobacter baumannii was studied with the help of AI based methods, and an outer membrane protein named FilF (a putative pilus assembly protein), was proposed and experimentally validated as a potential vaccine candidate (Singh et al., 2016). In a separate study of 33 A. baumannii genomes, AI driven RV methods identified candidates to develop vaccines to protect against infection with antibiotic resistance strains (Chiang et al., 2015). Computational identification and characterization of T-cell epitopes in Mycobacterium spp has also been reported. The immunoinformatics of Mycobacterium tuberculosis (Mtb) aided by several AI based methods/tools, identified immunogenic epitopes with the potential for inclusion in candidate vaccines for testing in follow up in vitro studies (Panigada et al., 2002; Hossain et al., 2017; Das et al., 2021). In Table 2, we summarize studies that have used AI-RV platforms to develop vaccines against major multidrug resistant bacteria.
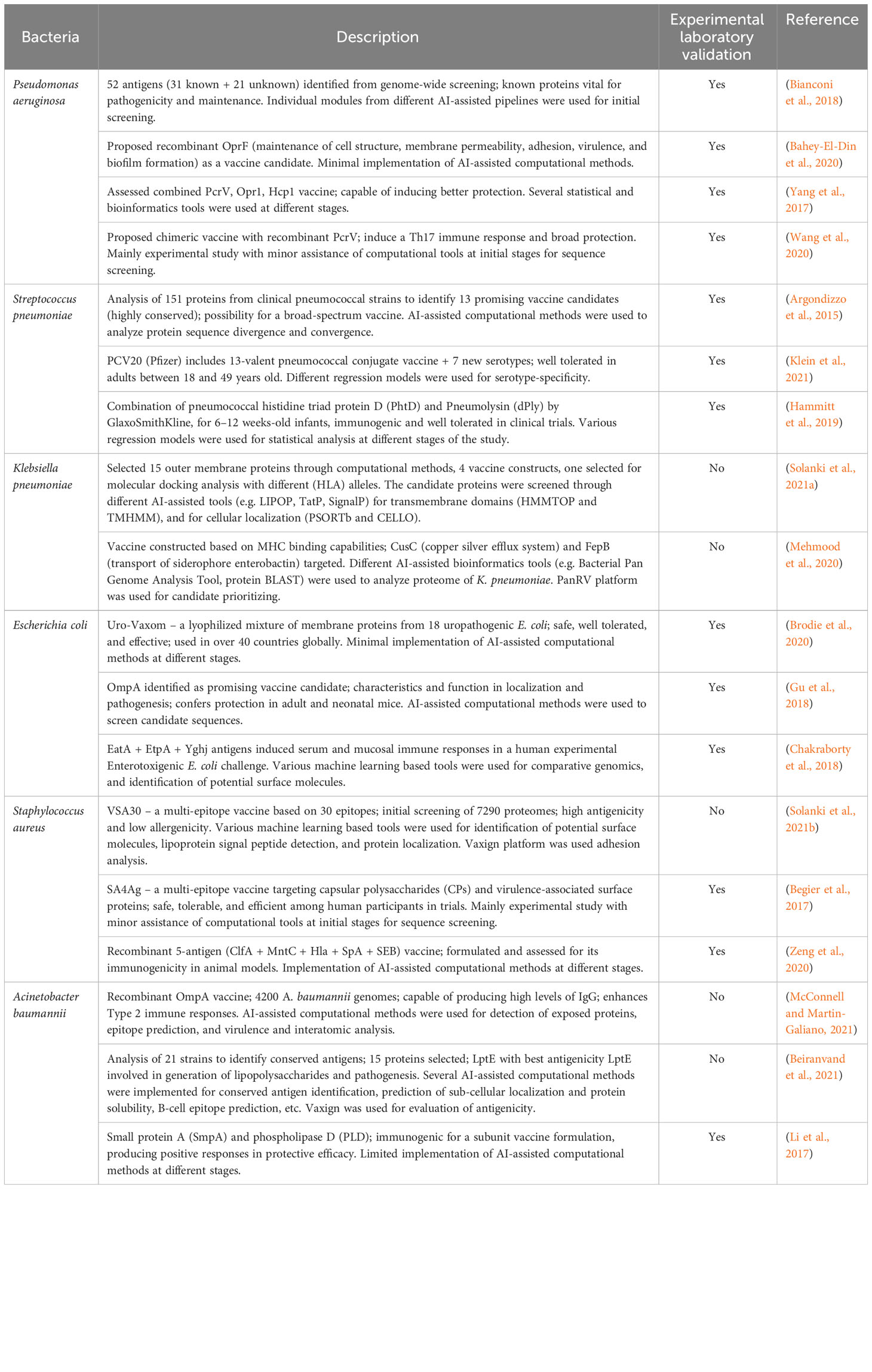
Table 2 A summary of AI powered RV platforms to develop vaccines against multidrug resistant bacteria.
The journey of vaccine development using traditional approaches has been very successful against several viral and bacterial pathogens. However, efficacious vaccines are yet to be developed for several infectious diseases, owing to the high rate of mutagenesis and sequence variability, antigenic complexity, and pathogen persistence. The lack of efficient vaccines against tuberculosis, and several viral pathogens including herpes simplex virus, respiratory syncytial virus, and human immunodeficiency virus, present some serious challenges to AI-ML models applied to vaccine development. Implementation of novel AI techniques along with the advances in nucleic acid and viral vectors could revolutionize vaccine development programs.
3 Conclusions and prospectus
The WHO asserts that antimicrobial resistance (AMR) is one of the top 10 global threats to human health and development. In 2022, at least 1.27 million deaths per year are directly attributable to AMR (Murray et al., 2022) and by 2050, 10 million deaths are estimated per annum from infections caused by AMR bacteria, with economic healthcare-associated costs in the trillions of dollars (USD). Vaccines represent a prophylactic strategy to combat AMR, although it is worth mentioning that vaccine development against certain pathogens is hampered by a lack of understanding of the mechanisms of infection and of how immunity develops. The accelerated development of COVID-19 vaccines has shown how state-of-the-art biomedical technologies can serve society during pandemic crises. It is inevitable that new and developing AI and ML techniques will be used to develop vaccines for infections caused by many viruses, bacteria and parasites that have proved particularly intractable. Implementation of advanced AI techniques in research dates to the start of 21st century. Undoubtedly, the development of AlphaFold (Higgins, 2021; Jumper et al., 2021; Binder et al., 2022; Varadi et al., 2022), a deep ML-based method for protein structure prediction, and the discovery of halicin, a potent antibiotic compound identified with a deep learning approach (Stokes et al., 2020) has imparted a belief amongst scientific communities of the potential of AI driven methods and technologies.
In summary, the integration of AI in vaccine development represents a transformative leap forward, expediting the process, enhancing precision, and broadening our understanding of infectious diseases. Whilst AI will significantly streamline the development of vaccines, we emphasize that these technologies are not a panacea. The integration of AI should always complement and not replace, rigorous ‘wet’ laboratory experimentation and animal and human trials, which remain indispensable steps in ensuring vaccine safety, efficacy, and are essential for regulatory approval. Nevertheless, as we continue to harness the power of AI-driven tools, the future holds great promise for the rapid creation of effective vaccines, particularly for emerging biological threats. We predict that AI assisted research and innovation in antimicrobial and vaccine development programs will ultimately improve global human health and increase life expectancy.
Author contributions
RahulK: Writing – original draft, Writing – review & editing. RK: Writing – review & editing. MC: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
RahulK is thankful to Biotechnology Research Center, Technology Innovation Institute, Abu Dhabi, UAE for its support and resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author MC declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed Z., Mohamed K., Zeeshan S., Dong X. (2020). Artificial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine. Database 2020. doi: 10.1093/database/baaa010
Argondizzo A. P., Da Mota F. F., Pestana C. P., Reis J. N., De Miranda A. B., Galler R., et al. (2015). Identification of proteins in Streptococcus pneumoniae by reverse vaccinology and genetic diversity of these proteins in clinical isolates. Appl. Biochem. Biotechnol. 175 (4), 2124–2165. doi: 10.1007/s12010-014-1375-3
Bahey-El-Din M., Mohamed S. A., Sheweita S. A., Haroun M., Zaghloul T. I. (2020). Recombinant N-terminal outer membrane porin (OprF) of Pseudomonas aeruginosa is a promising vaccine candidate against both P. aeruginosa and some strains of Acinetobacter baumannii. Int. J. Med. Microbiol. 310 (3), 151415. doi: 10.1016/j.ijmm.2020.151415
Begier E., Seiden D. J., Patton M., Zito E., Severs J., Cooper D., et al. (2017). SA4Ag, a 4-antigen Staphylococcus aureus vaccine, rapidly induces high levels of bacteria-killing antibodies. Vaccine 35 (8), 1132–1139. doi: 10.1016/j.vaccine.2017.01.024
Beiranvand S., Doosti A., Mirzaei S. A. (2021). Putative novel B-cell vaccine candidates identified by reverse vaccinology and genomics approaches to control Acinetobacter baumannii serotypes. Infect. Genet. Evol. 96, 105138. doi: 10.1016/j.meegid.2021.105138
Bianconi I., Alcala-Franco B., Scarselli M., Dalsass M., Buccato S., Colaprico A., et al. (2018). Genome-based approach delivers vaccine candidates against Pseudomonas aeruginosa. Front. Immunol. 9. doi: 10.3389/fimmu.2018.03021
Binder J. L., Berendzen J., Stevens A. O., He Y., Wang J., Dokholyan N. V., et al. (2022). AlphaFold illuminates half of the dark human proteins. Curr. Opin. Struct. Biol. 74, 102372. doi: 10.1016/j.sbi.2022.102372
Blanco-González A., Cabezón A., Seco-González A., Conde-Torres D., Antelo-Riveiro P., Piñeiro Á., et al. (2023). The role of AI in drug discovery: challenges, opportunities, and strategies. Pharmaceuticals 16 (6), 891. doi: 10.3390/ph16060891
Bradley P., Den Bakker H. C., Rocha E. P. C., Mcvean G., Iqbal Z. (2019). Ultrafast search of all deposited bacterial and viral genomic data. Nat. Biotechnol. 37 (2), 152–159. doi: 10.1038/s41587-018-0010-1
Brodie A., El-Taji O., Jour I., Foley C., Hanbury D. (2020). A retrospective study of immunotherapy treatment with uro-vaxom (OM-89(R)) for prophylaxis of recurrent urinary tract infections. Curr. Urol 14 (3), 130–134. doi: 10.1159/000499248
Chakraborty S., Randall A., Vickers T. J., Molina D., Harro C. D., Denearing B., et al. (2018). Human experimental challenge with enterotoxigenic escherichia coli elicits immune responses to canonical and novel antigens relevant to vaccine development. J. Infect. Dis. 218 (9), 1436–1446. doi: 10.1093/infdis/jiy312
Chiang M. H., Sung W. C., Lien S. P., Chen Y. Z., Lo A. F., Huang J. H., et al. (2015). Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum. Vaccin Immunother. 11 (4), 1065–1073. doi: 10.1080/21645515.2015.1010910
Christaki E., Marcou M., Tofarides A. (2020). Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J. Mol. Evol. 88 (1), 26–40. doi: 10.1007/s00239-019-09914-3
Dalsass M., Brozzi A., Medini D., Rappuoli R. (2019). Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00113
Das R., Eniyan K., Bajpai U. (2021). Computational identification and characterization of antigenic properties of Rv3899c of Mycobacterium tuberculosis and its interaction with human leukocyte antigen (HLA). Immunogenetics 73 (5), 357–368. doi: 10.1007/s00251-021-01220-x
David L., Brata A. M., Mogosan C., Pop C., Czako Z., Muresan L., et al. (2021). Artificial intelligence and antibiotic discovery. Antibiotics (Basel) 10 (11):1376. doi: 10.3390/antibiotics10111376
Davies J., Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74 (3), 417–433. doi: 10.1128/MMBR.00016-10
Deghmane A. E., Taha M. K. (2022). Product review on the IMD serogroup B vaccine Bexsero(R). Hum. Vaccin Immunother. 18 (1), 2020043. doi: 10.1080/21645515.2021.2020043
DeGrace M. M., Ghedin E., Frieman M. B., Krammer F., Grifoni A., Alisoltani A., et al. (2022). Defining the risk of SARS-CoV-2 variants on immune protection. Nature 605 (7911), 640–652. doi: 10.1038/s41586-022-04690-5
D’Mello A., Ahearn C. P., Murphy T. F., Tettelin H. (2019). ReVac: a reverse vaccinology computational pipeline for prioritization of prokaryotic protein vaccine candidates. BMC Genomics 20 (1), 981. doi: 10.1186/s12864-019-6195-y
Doytchinova I. A., Flower D. R. (2007). VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 8, 4. doi: 10.1186/1471-2105-8-4
Feikin D. R., Higdon M. M., Abu-Raddad L. J., Andrews N., Araos R., Goldberg Y., et al. (2022). Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet 399 (10328), 924–944. doi: 10.1016/S0140-6736(22)00152-0
Goodswen S. J., Kennedy P. J., Ellis J. T. (2023). A guide to current methodology and usage of reverse vaccinology towards in silico vaccine discovery. FEMS Microbiol. Rev. 47 (2). doi: 10.1093/femsre/fuad004
Gu H., Liao Y., Zhang J., Wang Y., Liu Z., Cheng P., et al. (2018). Rational design and evaluation of an artificial escherichia coli K1 protein vaccine candidate based on the structure of OmpA. Front. Cell Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00172
Guimaraes L. E., Baker B., Perricone C., Shoenfeld Y. (2015). Vaccines, adjuvants and autoimmunity. Pharmacol. Res. 100, 190–209. doi: 10.1016/j.phrs.2015.08.003
Hammitt L. L., Campbell J. C., Borys D., Weatherholtz R. C., Reid R., Goklish N., et al. (2019). Efficacy, safety and immunogenicity of a pneumococcal protein-based vaccine co-administered with 13-valent pneumococcal conjugate vaccine against acute otitis media in young children: A phase IIb randomized study. Vaccine 37 (51), 7482–7492. doi: 10.1016/j.vaccine.2019.09.076
Hardt K., Bonanni P., King S., Santos J. I., El-Hodhod M., Zimet G. D., et al. (2016). Vaccine strategies: Optimising outcomes. Vaccine 34 (52), 6691–6699. doi: 10.1016/j.vaccine.2016.10.078
Higgins M. K. (2021). Can we alphaFold our way out of the next pandemic? J. Mol. Biol. 433 (20), 167093. doi: 10.1016/j.jmb.2021.167093
Hossain M. S., Azad A. K., Chowdhury P. A., Wakayama M. (2017). Computational identification and characterization of a promiscuous T-cell epitope on the extracellular protein 85B of mycobacterium spp. for peptide-based subunit vaccine design. BioMed. Res. Int. 2017, 4826030. doi: 10.1155/2017/4826030
Huemer M., Mairpady Shambat S., Brugger S. D., Zinkernagel A. S. (2020). Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 21 (12), e51034. doi: 10.15252/embr.202051034
Hutchings M. I., Truman A. W., Wilkinson B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80. doi: 10.1016/j.mib.2019.10.008
Jaffee E. M., Dang C. V., Agus D. B., Alexander B. M., Anderson K. C., Ashworth A., et al. (2017). Future cancer research priorities in the USA: a Lancet Oncology Commission. Lancet Oncol. 18 (11), e653–e706. doi: 10.1016/S1470-2045(17)30698-8
Jiang Y., Yang M., Wang S., Li X., Sun Y. (2020). Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. (Lond) 40 (4), 154–166. doi: 10.1002/cac2.12012
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi: 10.1038/s41586-021-03819-2
Klein N. P., Peyrani P., Yacisin K., Caldwell N., Xu X., Scully I. L., et al. (2021). A phase 3, randomized, double-blind study to evaluate the immunogenicity and safety of 3 lots of 20-valent pneumococcal conjugate vaccine in pneumococcal vaccine-naive adults 18 through 49 years of age. Vaccine 39 (38), 5428–5435. doi: 10.1016/j.vaccine.2021.07.004
Kruk M. E., Gage A. D., Arsenault C., Jordan K., Leslie H. H., Roder-Dewan S., et al. (2018). High-quality health systems in the Sustainable Development Goals era: time for a revolution. Lancet Global Health 6 (11), e1196-e1252. doi: 10.1016/S2214-109X(18)30386-3
Larsen J., Raisen C. L., Ba X., Sadgrove N. J., Padilla-Gonzalez G. F., Simmonds M. S. J., et al. (2022). Emergence of methicillin resistance predates the clinical use of antibiotics. Nature 602 (7895), 135–141. doi: 10.1038/s41586-021-04265-w
Lee J. W., Maria-Solano M. A., Vu T. N. L., Yoon S., Choi S. (2022). Big data and artificial intelligence (AI) methodologies for computer-aided drug design (CADD). Biochem. Soc. Trans. 50 (1), 241–252. doi: 10.1042/bst20211240
Li H., Tan H., Hu Y., Pan P., Su X., Hu C. (2017). Small protein A and phospholipase D immunization serves a protective role in a mouse pneumonia model of Acinetobacter baumannii infection. Mol. Med. Rep. 16 (2), 1071–1078. doi: 10.3892/mmr.2017.6688
Lima L. M., Silva B., Barbosa G., Barreiro E. J. (2020). beta-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 208, 112829. doi: 10.1016/j.ejmech.2020.112829
Masignani V., Pizza M., Moxon E. R. (2019). The development of a vaccine against meningococcus B using reverse vaccinology. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00751
McConnell M. J., Martin-Galiano A. J. (2021). Designing multi-antigen vaccines against acinetobacter baumannii using systemic approaches. Front. Immunol. 12. doi: 10.3389/fimmu.2021.666742
Mehmood A., Naseer S., Ali A., Fatimah H., Rehman S., Kiani A. K. (2020). Identification of novel vaccine candidates against carbapenem resistant Klebsiella pneumoniae: A systematic reverse proteomic approach. Comput. Biol. Chem. 89, 107380. doi: 10.1016/j.compbiolchem.2020.107380
Melo M. C. R., Maasch J., de la Fuente-Nunez C. (2021). Accelerating antibiotic discovery through artificial intelligence. Commun. Biol. 4 (1), 1050. doi: 10.1038/s42003-021-02586-0
Moise L., Gutierrez A., Kibria F., Martin R., Tassone R., Liu R., et al. (2015). iVAX: An integrated toolkit for the selection and optimization of antigens and the design of epitope-driven vaccines. Hum. Vaccin Immunother. 11 (9), 2312–2321. doi: 10.1080/21645515.2015.1061159
Murray C. J. L., Ikuta K. S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 (10325), 629–655. doi: 10.1016/S0140-6736(21)02724-0
Naz K., Naz A., Ashraf S. T., Rizwan M., Ahmad J., Baumbach J., et al. (2019). PanRV: Pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinf. 20 (1), 123. doi: 10.1186/s12859-019-2713-9
Ong E., Cooke M. F., Huffman A., Xiang Z., Wong M. U., Wang H., et al. (2021). Vaxign2: the second generation of the first Web-based vaccine design program using reverse vaccinology and machine learning. Nucleic Acids Res. 49 (W1), W671–W678. doi: 10.1093/nar/gkab279
Ong E., He Y. (2022). ). Vaccine design by reverse vaccinology and machine learning. Methods Mol. Biol. 2414, 1–16. doi: 10.1007/978-1-0716-1900-1_1
Ong E., Wang H., Wong M. U., Seetharaman M., Valdez N., He Y. (2020). Vaxign-ML: supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics 36 (10), 3185–3191. doi: 10.1093/bioinformatics/btaa119
Panigada M., Sturniolo T., Besozzi G., Boccieri M. G., Sinigaglia F., Grassi G. G., et al. (2002). Identification of a promiscuous T-cell epitope in Mycobacterium tuberculosis Mce proteins. Infect. Immun. 70 (1), 79–85. doi: 10.1128/IAI.70.1.79-85.2002
Perry J., Waglechner N., Wright G. (2016). The prehistory of antibiotic resistance. Cold Spring Harb. Perspect. Med. 6 (6). doi: 10.1101/cshperspect.a025197
Pizza M., Scarlato V., Masignani V., Giuliani M. M., Arico B., Comanducci M., et al. (2000). Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287 (5459), 1816–1820. doi: 10.1126/science.287.5459.1809
Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. (2020). Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl. J. Med. 383 (27), 2603–2615. doi: 10.1056/NEJMoa2034577
Rahman N., Ajmal A., Ali F., Rastrelli L. (2020). Core proteome mediated therapeutic target mining and multi-epitope vaccine design for Helicobacter pylori. Genomics 112 (5), 3473–3483. doi: 10.1016/j.ygeno.2020.06.026
Rahman M. S., Rahman M. K., Saha S., Kaykobad M., Rahman M. S. (2019). Antigenic: An improved prediction model of protective antigens. Artif. Intell. Med. 94, 28–41. doi: 10.1016/j.artmed.2018.12.010
Sahayasheela V. J., Lankadasari M. B., Dan V. M., Dastager S. G., Pandian G. N., Sugiyama H. (2022). Artificial intelligence in microbial natural product drug discovery: current and emerging role. Nat. Prod Rep. 39 (12), 2215–2230. doi: 10.1039/d2np00035k
Schubert-Unkmeir A., Christodoulides M. (2013). Genome-based bacterial vaccines: current state and future outlook. BioDrugs 27 (5), 419–430. doi: 10.1007/s40259-013-0034-5
Shrock E., Fujimura E., Kula T., Timms R. T., Lee I. H., Leng Y., et al. (2020). Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 370 (6520). doi: 10.1126/science.abd4250
Singh R., Garg N., Shukla G., Capalash N., Sharma P. (2016). Immunoprotective Efficacy of Acinetobacter baumannii Outer Membrane Protein, FilF, Predicted In silico as a Potential Vaccine Candidate. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00158
Solanki V., Sharma S., Tiwari V. (2021a). Subtractive proteomics and reverse vaccinology strategies for designing a multiepitope vaccine targeting membrane proteins of Klebsiella pneumoniae. Int. J. Pept. Res. Ther. 27 (2), 1177–1195. doi: 10.1007/s10989-021-10159-2
Solanki V., Tiwari M., Tiwari V. (2021b). Subtractive proteomic analysis of antigenic extracellular proteins and design a multi-epitope vaccine against Staphylococcus aureus. Microbiol. Immunol. 65 (8), 302–316. doi: 10.1111/1348-0421.12870
Stokes J. M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N. M., et al. (2020). A deep learning approach to antibiotic discovery. Cell 180 (4), 688–702.e613. doi: 10.1016/j.cell.2020.01.021
Sulis G., Sayood S., Katukoori S., Bollam N., George I., Yaeger L. H., et al. (2022). Exposure to World Health Organization’s AWaRe antibiotics and isolation of multidrug resistant bacteria: a systematic review and meta-analysis. Clin. Microbiol. Infect. 28 (9), 1193–1202. doi: 10.1016/j.cmi.2022.03.014
Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18 (3), 318–327. doi: 10.1016/S1473-3099(17)30753-3
Thomas S., Abraham A., Baldwin J., Piplani S., Petrovsky N. (2022). Artificial intelligence in vaccine and drug design. Methods Mol. Biol. 2410, 131–146. doi: 10.1007/978-1-0716-1884-4_6
Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50 (D1), D439–D444. doi: 10.1093/nar/gkab1061
Vetrano S., Bouma G., Benschop R. J., Birngruber T., Costanzo A., D’haens G. R. A. M., et al. (2022). ImmUniverse Consortium: Multi-omics integrative approach in personalized medicine for immune-mediated inflammatory diseases. Front. Immunol. 13. doi: 10.3389/fimmu.2022.1002629
Vetter V., Denizer G., Friedland L. R., Krishnan J., Shapiro M. (2018). Understanding modern-day vaccines: what you need to know. Ann. Med. 50 (2), 110–120. doi: 10.1080/07853890.2017.1407035
Vila J., Moreno-Morales J., Balleste-Delpierre C. (2020). Current landscape in the discovery of novel antibacterial agents. Clin. Microbiol. Infect. 26 (5), 596–603. doi: 10.1016/j.cmi.2019.09.015
Waltz E. (2020). AI takes its best shot: What AI can—and can’t—do in the race for a coronavirus vaccine - [Vaccine]. IEEE Spectr. 57 (10), 24–67. doi: 10.1109/MSPEC.2020.9205545
Wang Y., Cheng X., Wan C., Wei J., Gao C., Zhang Y., et al. (2020). Development of a chimeric vaccine against pseudomonas aeruginosa based on the th17-stimulating epitopes of PcrV and AmpC. Front. Immunol. 11. doi: 10.3389/fimmu.2020.601601
Wang Z., Koirala B., Hernandez Y., Zimmerman M., Park S., Perlin D. S., et al. (2022). A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature 601 (7894), 606–611. doi: 10.1038/s41586-021-04264-x
Wang R., Pan W., Jin L., Li Y., Geng Y., Gao C., et al. (2019). Artificial intelligence in reproductive medicine. Reproduction 158 (4), R139–R154. doi: 10.1530/REP-18-0523
Wouters O. J., Mckee M., Luyten J. (2020). Estimated research and development investment needed to bring a new medicine to market 2009-2018. JAMA 323 (9), 844–853. doi: 10.1001/jama.2020.1166
Yang F., Gu J., Yang L., Gao C., Jing H., Wang Y., et al. (2017). Protective efficacy of the trivalent pseudomonas aeruginosa vaccine candidate PcrV-OprI-Hcp1 in murine pneumonia and burn models. Sci. Rep. 7 (1), 3957. doi: 10.1038/s41598-017-04029-5
Yang X., Wang Y., Byrne R., Schneider G., Yang S. (2019). Concepts of artificial intelligence for computer-assisted drug discovery. Chem. Rev. 119 (18), 10520–10594. doi: 10.1021/acs.chemrev.8b00728
Zeng X., Wang F., Luo Y., Kang S.-G., Tang J., Lightstone F. C., et al. (2022). Deep generative molecular design reshapes drug discovery. Cell Rep. Med. 3 (12), 100794. doi: 10.1016/j.xcrm.2022.100794
Zeng H., Yang F., Feng Q., Zhang J., Gu J., Jing H., et al. (2020). Rapid and broad immune efficacy of a recombinant five-antigen vaccine against staphylococcus aureus infection in animal models. Vaccines (Basel) 8 (1). doi: 10.3390/vaccines8010134
Keywords: artificial intelligence, machine learning, vaccine, bacteria, antimicrobial resistance
Citation: Kaushik R, Kant R and Christodoulides M (2023) Artificial intelligence in accelerating vaccine development - current and future perspectives. Front. Bacteriol. 2:1258159. doi: 10.3389/fbrio.2023.1258159
Received: 13 July 2023; Accepted: 25 September 2023;
Published: 09 October 2023.
Edited by:
Sandra Sousa, Universidade do Porto, PortugalReviewed by:
Daniel Alford Powell, University of Arizona, United StatesCopyright © 2023 Kaushik, Kant and Christodoulides. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Myron Christodoulides, bWM0QHNvdG9uLmFjLnVr
 Rahul Kaushik1
Rahul Kaushik1 Myron Christodoulides
Myron Christodoulides