- 1Department of Soil Ecology, Helmholtz Centre for Environmental Research, Halle, Germany
- 2Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 3Crop Research Unit, Institute of Agricultural and Nutritional Sciences, Martin Luther University Halle-Wittenberg (MLU), Halle, Germany
- 4German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany
Under climate change scenarios for temperate regions in Europe, prolonged droughts pose a major threat to barley production, but few studies have been conducted on stress mitigation strategies using plant-beneficial rhizobacteria. With this in mind, we isolated and screened a culture collection of drought-tolerant bacteria from the barley rhizosphere. From this collection, we assembled a 16-member consortium based on their relative abundances in the rhizosphere after drought and in vitro osmotic stress tolerance (Drought Tolerant Synthetic microbial Community/”DT-SynCom”). Members of the DT-SynCom range from Proteobacteria to Firmicutes and Actinobacteria. We used Oxford Nanopore and Illumina technologies to assemble complete genomes. Whole genome annotation revealed the presence of a number of genes associated with plant growth promoting traits such as IAA biosynthesis, ACC deaminase activity and siderophore production. In vitro assays confirmed auxin production, ACC deaminase activity, siderophore production, inorganic P solubilization, and cellulase and chitinase activities by the selected bacterial strains. The consortium members were not antagonistic to each other, and were either neutral or beneficial to barley shoot and root growth of barley when applied individually in vitro. To clarify the effect of the designed DT-SynCom on barley drought tolerance, a pot experiment was conducted under drought stress conditions. The DT-SynCom reduced the number of wilting leaves and had a positive effect on barley growth under drought. The results of the research suggest that the members of the barley DT-SynCom have beneficial plant growth promoting traits that result in improved plant growth under drought stress.
1 Introduction
The negative impacts of climate change affect the planet since the 20th century (Feulner, 2017) and recent research shows that global warming and its accompanying adverse effects, such as prolonged periods of drought, will continue to intensify in the near future (Lee et al., 2023). For instance, the 2018-2020 heat and drought events in Europe showed an intensity unprecedented in the last 250 years (Rakovec et al., 2022). Such increasing temperatures and prolonged drought periods adversely affect the food production and poses a threat to food security at a global scale (Verschuur et al., 2021; Funk et al., 2018). In recent decades, up to 10% of the world’s yield reductions in crop production have been linked to extreme weather disasters (Lesk et al., 2016). Drought caused by climate change is one of the main reasons for declining crop yields (Bodner et al., 2015; Gupta et al., 2020). With 1.5°C of global warming over the historic baseline seemingly unavoidable (Lamboll et al., 2023), developing methods to help plants survive expected future drought-intense conditions (Balting et al., 2021) is of great importance with respect to ensuring food security. One of the most promising, environmental friendly solutions to help plants withstand drought stress involves equipping them with drought-adapted plant microbiomes (Liu et al., 2022).
Apart from the physiological responses plants use to withstand drought periods, they also have their specific rhizosphere and root microbial communities (rhizosphere and root microbiome) that can contribute to drought tolerance (Rahnama et al., 2023). Water scarcity adjusts the composition of the microbial communities, and drought-adapted microbial taxa have the potential to not only withstand drought stress themselves but to also mitigate drought stress in the host plants (Liu et al., 2020; Coleman-Derr and Tringe, 2014; Azarbad et al., 2020). Bacterial traits and bacteria associated with such stress tolerance are referred to as plant growth promoting traits (PGPTs) and plant growth promoting (PGPB) bacteria. For instance, inoculation of barley with bacteria from Flavobacteriaceae and Paenibacillaceae has been shown to enhance tolerance of moderate drought stress (Xu et al., 2023). This is attributed to the production of auxins such as indole-3-acetic acid (IAA) by the bacteria which induces rhizosheath formation in the host roots (Xu et al., 2023). Similarly root bacteria capable of producing 1-aminocyclopropane-1-carboxylate (ACC) deaminase have been shown to help promote plant growth under drought stress. This is caused by reducing levels of the plant growth suppressing stress hormone ethylene (C2H4) in the host by cleaving its precursor molecule (ACC) into ammonia and α-ketobutyrate (Ojuederie and Babalola, 2023; Shahid et al., 2023). Plant growth under water scarcity can also be aided by biofilm formation. Biofilms are extracellular matrices created upon the secretion of metabolic byproducts known as exopolysaccharides (EPS) by bacteria. Bacterial biofilms have been demonstrated to reduce water loss in not only the bacteria but also the host root by helping maintain humidified microenvironment (Roberson and Firestone, 1992; Morcillo and Manzanera, 2021). However, despite these noted benefits, and their vital importance for the maintenance of agricultural yields, a direct link between the structure of plant growth promoting microbial communities and increased drought resistance of the plants has rarely been substantiated (Hartman and Tringe, 2019).
Diploid barley (Hordeum vulgare L.) is the fourth-most cultivated crop worldwide representing an important source for food and animal feed (Cai et al., 2020; Elakhdar et al., 2021). It is a cereal crop with moderate to poor drought tolerance (Ghotbi-Ravandi et al., 2014), and water scarcity inhibits its growth and limits its productivity (Cai et al., 2020). Barley represents an emerging model to study molecular host-microbiota interactions as its genome has been sequenced, mutants have been generated (Mascher et al., 2017; Mahdi et al., 2022), and it has been used as a model host plant for investigating host-microbe interactions with pathogenic and beneficial fungi (Sarkar et al., 2019; Hilbert et al., 2019). Numerous greenhouse studies with drought-tolerant and drought-sensitive barley cultivars under precisely controlled environmental conditions have demonstrated a variety of changes at the morphological, biochemical, physiological, and molecular levels to tolerate drought stress (Sallam et al., 2019). However, research on the barley microbiome has shown that certain microbial taxa are enriched in the barley rhizosphere and roots, including members of the families Comamonadaceae, Flavobacteriaceae, and Rhizobiaceae, irrespective of the tested barley genotypes and their drought tolerance (Bulgarelli et al., 2015). For this purpose, the research priority program SPP 2125 of the German Research Foundation (“Deconstruction and Reconstruction of the Plant Microbiota, DECRyPT”) has set the goal to gain a deeper understanding of the plant microbiota through systematic reductionist approaches, including the deconstruction and reconstruction of beneficial plant-associated microbial communities (Deutsche Forschungsgemeinschaft, 2023).
At present, specific studies on the application of synthetic microbial communities (SynComs) to barley under drought stress are limited, but the beneficial effects of certain bacteria on plant growth are well established, and shifts in microbial communities under water stress have been documented. For example, inoculation of maize with SynCom has been shown to reduce leaf temperature, decrease turgor loss under severe drought conditions, and accelerate recovery upon rehydration (Armanhi et al., 2021). Similarly, a stable 15-member bacterial SynCom promoted plant growth and enhanced drought resilience in Brachypodium compared to untreated controls (Yadav et al., 2024). In contrast, studies on barley under drought stress have mainly focused on single strain inoculations. For example, inoculation of barley with the plant growth promoting rhizobacterium Serratia odorifera in combination with biochar improved plant growth, biomass, and seed germination under drought conditions (Gul et al., 2023). These results highlight the potential of SynComs to improve drought tolerance in cereals and show that single strain inoculations can improve drought tolerance in barley. However, the application of SynComs specifically designed for barley under drought stress remains unexplored. Therefore, our study to assemble a drought-tolerant SynCom tailored for barley is a relevant and valuable step towards understanding and harnessing microbial communities to improve drought resilience in this important crop. To build on this, we aimed to investigate the effects of inoculating a drought tolerant synthetic microbial community (DT-SynCom) consisting of individually isolated and trait-screened microorganisms into the barley rhizosphere. As a basis for this, we examined barley rhizospheres exposed to predicted future drought conditions to identify root bacteria enriched in hosts that survived these (simulated) conditions. We then isolated pure cultures of these strains (17 in total) with the intent to construct a DT-SynCom. In this regard, we performed experiments to determine the presence of PGPTs through in vitro experiments. Additionally, to both enhance the representation of genomic resources for soil bacteria and gain insights into the genomes of these strains, we sequenced the genomes using both long read (Oxford Nanopore) and short read (Illumina) technologies and assembled these using a state-of-the-art hybrid assembly approach to produce near-complete genomes. The in vitro experiments and genome annotations revealed the presence of PGPTs such as auxin production, cellulose and chitin degradation, IAA biosynthesis, nitrogen fixation and inorganic P solubilization in one or more strains. Experiments revealed that the strains were not antagonistic towards one another and were not detrimental to barley root and shoot growth when applied individually. Application of the DT-SynCom to barley hosts experiencing drought stress in pot experiments revealed mild to moderate protective effects suggesting that refined versions of this SynCom may be beneficial for conferring drought-tolerance in barley.
2 Materials and methods
2.1 Collection of bacteria from barley rhizosphere
Sampling took place at the Global Change Experimental Facility (GCEF) in Bad Lauchstädt (51° 23’ 30N, 11° 52’ 49E) on the site of the field research station of the Helmholtz Centre for Environmental Research (https://www.ufz.de/index.php?en=42385). Specifics regarding the communities and other details regarding the GCEF sites as well as strains isolated and used for this study can be found in previously published literature (Breitkreuz et al., 2021b, 2021, 2020). In brief: the Bad Lauchstädt field site is characterized by a temperate continental climate with a mean annual precipitation of 483 mm, a mean annual temperature of 9.7°C (1993-2013) (Schädler et al., 2019), and a Haplic Chernozem soil type (Driessen, 2001; Schädler et al., 2019). The GCEF design comprises three grassland management types and two agriculture approaches (conventional (CF) and organic (OF)) that are all subjected to either ambient (current) or future climatic conditions. All strains involved in this study were exclusively isolated from CF and OF plots exposed to the aforementioned future climate. The future climate treatment here comprises a 20% reduction of rainfall in summer (Jun-Aug), while precipitation is increased by 10% in spring (March-May) and autumn (September-November), thereby simulating the expected climate in 2080 (Schädler et al., 2019). Winter barley cultivar Antonella was grown in the GCEF. The variety is characterized by moderate to good lodging resistance, above average leaf health and average winter hardiness and drought tolerance. Two dates were selected, in order to ensure sampling of rhizosphere bacteria from a rapidly growing plant development stage (stem elongation stage, BBCH 37-39) as well as a post-flowering development stage (grain filling stage, BBCH 75-77). On both dates, rhizosphere soil was collected by gently loosening the root-attached soil of individual healthy plants by hand (Breitkreuz et al., 2020). After thorough mixing, 0.25 g of soil was suspended in 25 ml of double distilled water by stirring with a magnetic stirrer for two minutes (IKA, RCT basic). An ultrasonic bath was then used to dissociate bacteria from soil particles, and samples were stirred for two minutes. Double distilled water and the soil suspension were mixed at a ratio of 1:200, and 50 µl of this new suspension was used to inoculate Pikovskaya’s agar medium containing tricalcium phosphate as the sole phosphate source and cycloheximide to suppress proliferation of fungal contaminants (Pikovskaya, 1948). After incubation at 25°C for three weeks, colonies were selected on the basis of phosphate solubilization ability as indicated by bright halos around the colonies. Colonies were also selected on the basis of phenotypic characteristics such as color and shape to collect a diverse set of bacterial strains for the construction of a Drought Tolerant Synthetic bacterial Community (DT-SynCom). The selected strains were then purified by incubating on yeast malt extract (YME) agar medium at 25°C for three days.
2.2 Drought tolerance measurements of barley rhizosphere strains
Drought tolerance measurements were conducted on polyethylene glycol (PEG) agar by comparing the growth of the strains on YME agar medium to YME agar medium surface-infiltrated with PEG 8000 following previous work (Breitkreuz et al., 2020; Bouremani et al., 2024; Wang et al., 2014). The PEG concentration was set to 500 g/L (PEG0.5) which corresponds to an osmotic potential level of around 1.1 MPa to simulate severe drought stress (Verslues et al., 2006). Colony diameters on the control (PEG0) and PEG0.5 with 5 replicates each were cross-compared in ImageJ (Schneider et al., 2012) using overhead photographs taken against a dark background. Drought stress tolerance was determined as percentage difference in colony diameters between PEG0 and PEG0.5 and statistical significance was evaluated using a t-test.
2.3 Partial 16S rDNA sequencing of barley rhizosphere isolates
DNA was extracted by addition of PEG to bacterial suspensions followed by vortexing with glass beads following prior work (Breitkreuz et al., 2020). Polymerase chain reaction (PCR) to amplify the DNA was performed using the primers 27F and 1492R (Lane, 1991) and Promega Green (Promega, Madison, WI, USA). Subsequently, partial 16S rRNA sequencing was performed with the primer BAC 341F (Muyzer et al., 1993) for Sanger sequencing. at LGC Genomics (Berlin, Germany). Sequences were trimmed manually using Sequencher v5.4.5 (Gene Codes Corporation, 2016), and then compared with type strain reference sequences of the National Center of Biotechnology Information (NCBI) to establish the identities of the bacterial strains. The primary aim of Sanger sequencing was to provide sequences of each strain that will overlap with the ASVs provided by amplicon sequencing. The 16S rDNA Sanger sequencing analysis also grouped the strains into different genera (Supplementary Table S1).
2.4 Strain selection for drought tolerant synthetic bacterial community
Amplicon sequencing variants (ASVs) that were more abundant under the simulated future climate than under ambient climate in the GCEF (Breitkreuz et al., 2021a) as well as under drought than under well-watered conditions in a greenhouse pot experiment with soil from Bad Lauchstädt (Breitkreuz et al., 2021b) were extracted from the respective datasets. The differentially abundant ASVs were cross-compared with the 16S rDNA sequences of the culture collection, and bacterial isolates with identical or highly similar 16S rDNA sequence were identified. Out of this strain pool, sixteen PEG-tolerant strains, displaying same growth rate on PEG0 and PEG0.5, were chosen for the DT-SynCom (for a comprehensive workflow of the study see Figure 1).
Figure 1. Workflow of the study, illustrating the work described in the Materials and methods section. Created with BioRender.com.
2.5 DT-SynCom: inter-strain interaction testing using a cross-streak method
To address if any of the DT-SynCom isolates displayed in vitro inhibitory or stimulatory activity, they were cross-stroken on Plant Nutrition Medium (PNM) agar with pH adjusted to 5.7 (Egorov, 1985). Each test strain was streaked vertically in the center of its own agar plate, and 10 µl of each (remaining) DT-SynCom strain was streaked horizontally against the strain of interest. Care was taken to avoid possible interactions between the non-test strains. The plates were incubated at 25°C for 72 hours and inspected for visible signs of antagonism. Namely, the growth of each strain and the gaps between strains (i.e., the inhibition zones) were measured and gaps larger than 0.5 cm were interpreted as an indication of antagonism between the strains in question.
2.6 DT-SynCom: screening for plant growth promoting traits
2.6.1 IAA production using Salkowski reagent
Indole–3–acetic acid (IAA) produced by each strain was quantified using the Salkowski reagent (Gordon and Weber, 1951). The quantification of IAA was performed following a modified protocol described by Gang et al. (2019). Bacterial strains were first cultured overnight at 30°C using nutrient broth medium, and were then transferred into fresh nutrient broth supplemented with L-tryptophan and incubated in the dark for 96 hours. For quantification of IAA production, 1.5 ml of each culture was centrifuged and equal volumes of the supernatant and Salkowski reagent were mixed together and incubated for 30 minutes at 30°C in darkness. The presence of IAA was qualitatively assessed by checking for the development of a pink color. Quantitative measurements were made spectrophotometrically at 536 nm with caliberation against an IAA standard curve. Spectrophotometric measurements were repeated every 24 hours until no further production of IAA was detected.
2.6.2 1-aminocyclopropane-1-carboxylate (ACC) utilization
ACC utilization was characterized following the method of Penrose and Glick (2003). Fresh ACC solution and ninhydrin were prepared before the start of the experiment. ACC stock solution (0.5 M) was sterilized using a 0.2 µm membrane filter and added to autoclaved DF (Dworkin and Foster) minimal medium to acquire the final ACC concentration of 3 mM. Ninhydrin reagent was prepared and the ACC production assay performed following the protocol described previously (Li et al., 2011). A single bacterial colony was transferred from solid to liquid LB (Luria-Bertani) medium and incubated overnight at 28°C on a shaker at 200 rpm. From each culture, 2 ml was centrifuged, the pellet was washed twice with 1 ml of liquid DF medium, and suspended in 2 ml of DF minimal medium with ACC. After 24 hours at 28°C on a shaker at 200 rpm, the cells were pelleted, and 100 µl of the supernatant was diluted 1:10 with DF medium. Of the dilution, 60 µl were mixed with 120 µl of ninhydrin reagent, incubated for 30 minutes in a pre-warmed heating block at 100°C and measured at 570 nm. Bacterial strains with lower absorbance values compared with the uninoculated supernatant were considered to be ACC-utilizing.
2.6.3 Cellulose degradation
Single colonies of bacterial strains were suspended in 200 µl sterilized tap water, of which 1 µl aliquots were then plated onto cellulose agar. The plates were then cultured at 25°C for one week. Subsequently, the agar plates were stained for 15 minutes with Congo Red (1 g/L) and de-stained with 1 M NaCl (also 15 minutes). Overhead photographs of the plates taken against a dark background were evaluated using ImageJ to check for bacterial growth indicating ability to degrade cellulose. Activity was classified as 0 (no growth), 1 (growth, but no activity), 2 (growth with agar decoloration), 3 (growth with halo), with class 3 being most strongly indicative of cellulose degradation. All isolates were tested with five replicates.
2.6.4 Siderophore production
Siderophore production was evaluated using the blue agar chrome azurol S (CAS) assay following an established protocol (Schwyn and Neilands, 1987). In brief, CAS agar media were prepared following standard protocol and were inoculated with 1 µl aliquots of the strains (prepared as indicated in sub-section “Cellulose degradation” above). The plates were incubated at 25°C for 3 days and were evaluated as described in the paragraph above.
2.6.5 Chitin degradation
Chitin agar medium was prepared following previous work (Hsu and Lockwood, 1975) and was inoculated as described in Section “Cellulose degradation”. After incubation at 25°C for 2 weeks, the plates were evaluated for signs of ability to degrade chitin as indicated in sub-section “Cellulose degradation”.
2.7 DT-SynCom: genome sequencing and characterization
2.7.1 Illumina (short read) sequencing
Three day old bacterial colonies grown on YME agar medium were transferred into 1x phosphate buffered saline (PBS) buffer and gently mixed and subsequently pelleted by centrifugation. DNA extraction was performed using the QIAGEN DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) following manufacturer instructions. DNA purity and quantity were measured with a NanoDrop (ThermoFisher Scientific, Waltham, MA, USA) and DNA size by gel electrophoresis. DNA extracts were processed following the standard protocol of the Nextera XT DNA Library Preparation Kit (Illumina, 2023). Purified indexed libraries were quantified using Qubit (ThermoFisher Scientific), pooled into equal-mole sets, and DNA sequencing was carried out using an Illumina MiSeq to 2 x 300 bp nucleotide lengths.
2.7.2 Nanopore (long read) sequencing
DNA was extracted and quality controlled as described above for short read sequencing. Library preparation (DNA repair and end-prep, native barcoding, pooling and sequencing adapter ligation) and sequencing on an Oxford Nanopore instrument (GridIOn, 10.4.1 Flow Cell, 95h runtime, R10.4.1 flow cell, 200 bp minimum read length, minimum Q score 10) were performed by Eurofins Genomics (Konstanz, Germany).
2.7.3 Hybrid genome assembly and annotation
Illumina short reads were quality controlled using FastQC v0.12.1 (Andrews, 2010) and Nanopore long reads with NanoPlot v1.42.0 (Coster and Rademakers, 2023). Long read-first assemblies (supported and bridged by short read data) were produced using Hybracter v0.7.3 (Bouras et al., 2024) using default parameters for the “hybrid-single” assembly mode. Only one sample (Iso16) was assembled with additional parameters, namely, “-c 4000000 –subsample_depth 200” was supplied alongside. The parameters “–flyeModel “–nano-raw”“ was specified for all assemblies. The assemblies were subsequently annotated using Bakta v1.9.3 (Schwengers et al., 2021) under default parameters with the “–compliant” flag set. The assemblies were quality controlled by inspecting a number of different metrics. These included basic metrics produced by Hybracter itself, Bakta, and seqkit v2.8.0 (Shen et al., 2016) such as number of contigs, assembly size, N50, GC content, contig completeness, and annotation statistics. Genome completeness, contamination, and strain heterogeneity were evaluated using CheckM v1.2.2 (Parks et al., 2015). Coverage of the generated assemblies by the short and long read data sets was estimated using minimap2 v2.28-r1209 (Li, 2018, 2021) (to map reads to the assemblies; with option “-ax sr” for short reads and “-ax map-ont” for long reads), index from samtools v1.20 (Danecek et al., 2021) (to index the mappings), and mosdepth v0.3.8 (Pedersen and Quinlan, 2018) to estimate coverage (with option “–fast-mode”). One hybrid assembly (Iso19) which was deemed unsatisfactory due to unacceptable levels of strain heterogeneity as indicated by CheckM (implying possible mixing of closely related strains in this isolate) was reassembled with long read data only using Flye v2.9-b1768 (Kolmogorov et al., 2019) under default parameters.
2.7.4 In silico identification of plant-growth promoting traits
The Bakta annotations were examined for specific KEGG identifiers, gene names, and gene product descriptions to identify genes associated with plant-growth promoting traits (Supplementary Material S1_pgpt_genome_search_terms). The protein sequences predicted by Bakta were also submitted to PLaBAse v1.01 (Patz et al., 2021) to perform a second round of comprehensive annotation for plant growth-promoting traits in the genomes. Secondary metabolite biosynthetic gene clusters in the assembled genomes were additionally identified using antiSMASH v7.1.0 (Blin et al., 2023) invoked via the parallelization wrapper multiSMASH v0.3.1 (Reitz, 2024).
2.8 DT-SynCom: effects of individual members on barley germination and seedling growth
Germination and growth assessments were carried out based on earlier work (Guglielmetti et al., 2013). The Golden Promise cultivar was used for all germination and growth experiments and was chosen for two reasons. First, as it is a universally susceptible cultivar it is an ideal platform for detecting plant response to drought and SynCom inoculation. Second, this research was conducted within a research program (https://ag-zuccaro.botanik.uni-koeln.de/decrypt), where four research groups used Golden Promise, allowing for meaningful cross-comparisons and facilitating ongoing collaboration within the priority program. For the germination experiment, barley seeds were first surface sterilized by immersion in 6% hypochlorite for 1 hour and washing with sterile distilled water for 30 minutes afterwards. This washing step was repeated multiple times until the smell of chloride became imperceptible. The sterilized seeds were then inoculated with individual DT-SynCom species as follows. Bacterial cells of each DT-SynCom strain were grown overnight on a shaker at 120 rpm at 28°C in a liquid Tryptic Soy Broth medium (TSB), pH 7.3 +/- 0.2. Each bacterial culture was resuspended in TSB medium when the culture reached an optical density (OD) of 0.5 at 600 nm. The sterilized seeds were immersed in the bacterial suspensions for two hours, then placed on sterile, moistened paper towels and left to germinate in the dark at room temperature. Each sterilized seed was inoculated with a single DT-SynCom strain. Uninoculated seeds were used as controls. After four days of germination the following parameters were recorded: the number of germinated seeds, number of roots and leaves, lengths of roots and leaves and the root as well as leaf fresh and dry weights. For the barley growth tests, seed sterilization was performed as described above (but without bacterial inoculation) with the seeds being germinated again for four days. Subsequently, seedling roots were soaked in bacterial inoculum (prepared as above) for two hours at room temperature with the seedlings being transferred to 1/10 PMN agar medium and grown for five days under sterile conditions thereafter. Uninoculated seedlings were used as controls. The following parameters were recorded after the growth period: numbers of roots and leaves, root and leaf lengths, and the fresh and dry weight of leaves and roots.
2.9 DT-SynCom: barley growth and drought tolerance in a greenhouse experiment
Barley (Golden Promise) seeds were surface sterilized and germinated in the dark at room temperature four days prior the experiment. For DT-SynCom preparation each bacterial culture was resuspended in TSB medium and optical density was measured to reach OD 0.5 at 600 nm. Seedling roots were then immersed in either DT-SynCom or MgCl2 (control group) for two hours and planted in soil (potting soil, Einheits Erde, Classic Profisubstrat soil). Plants were grown for three weeks in a growth chamber with a day/night cycle of 14 hours of light, with day and night temperatures maintained at 20°C and 16°C respectively. Relative humidity was set at 65% and plants were watered at 80% of water holding capacity (WHC). Pot field capacity was determined beforehand by weighing soil into pots and oversaturating with water. The soil was then allowed to drain for two days to reach its natural water holding capacity, while being weighed twice a day. A plastic bag was taped over the top of the pot to prevent water loss by evaporation. The soil was then dried in an oven at 105°C for 24 hours and the water holding capacity was calculated by subtracting the dry weight from the weight of the soil at field capacity. Drought stress was induced on day 21 by watering the drought group at 30% WHC once every three days for a period of 21 days. A total of four treatments were observed, five plants per treatment: Control (80% WHC), Control +DT-SynCom inoculation, Drought (30% WHC) and Drought +DT-SynCom. Plants were observed and measured daily for changes in plant height, leaf number and wilting. Barley leaf wilting was used as a proxy for drought response. Leaf angle images were taken after three weeks of drought stress (30% WHC watering for the drought stressed plants vs. 80% WHC watering for the control plants). Three plants of each treatment were photographed and leaf wilting angle determined by measuring the angle between the blade of the first leaf and the stem. For the analysis ANOVA and Tukey test were used (P < 0.05).
3 Results
3.1 Isolation and drought-stress tolerance testing of a bacterial community from barley rhizosphere
A total of 245 purified bacterial strains were isolated from the rhizospheres of conventionally (CF) and organically (OF) farmed barley harvested at stem elongation (BBCH 37-39) and grain filling (BBCH 75-77) stages at the Global Change Experimental Facility in Bad Lauchstädt, Germany. Partial 16S rDNA was sequenced to group the strains (Supplementary Material S2_barley_strains). Partial 16S rDNA was determined by Sanger sequencing to group the strains and to provide a template for later comparison with amplicon sequencing data (Supplementary Material S2). Phyllobacterium (66 isolates, 27%), Streptomyces (49, 20%), and Pseudomonas (35, 14%) were dominant in the culture collection, followed by Rhizobium (29, 11%), Mesorhizobium (18, 7.3%), Priestia (11, 4.5%) and Arthrobacter (8, 3.8%). The relative contributions of Pseudomonas and Rhizobium spp. was lower and the abundance of Streptomyces higher in the grain filling than stem elongation stage, respectively (Figure 2). While Rhizobium was more common than Pseudomonas in OF, the opposite pattern occurred in CF (Figure 2). Many isolated strains were found from both growth stages, and these included Arthrobacter pascens, Mesorhizobium australicum, M. ciceri, Phyllobacterium brassicacearum, P. ifriqiyense, P. loti, Priestia aryabhattai, Pseudomonas corrugata, P. koreensis, P. lini, R. grahamii, R. indigoferae, R. viscosum, R. fascians, S. beijiangensis, S. canus, S. cyaneochromogenes and S. rishiriensis.
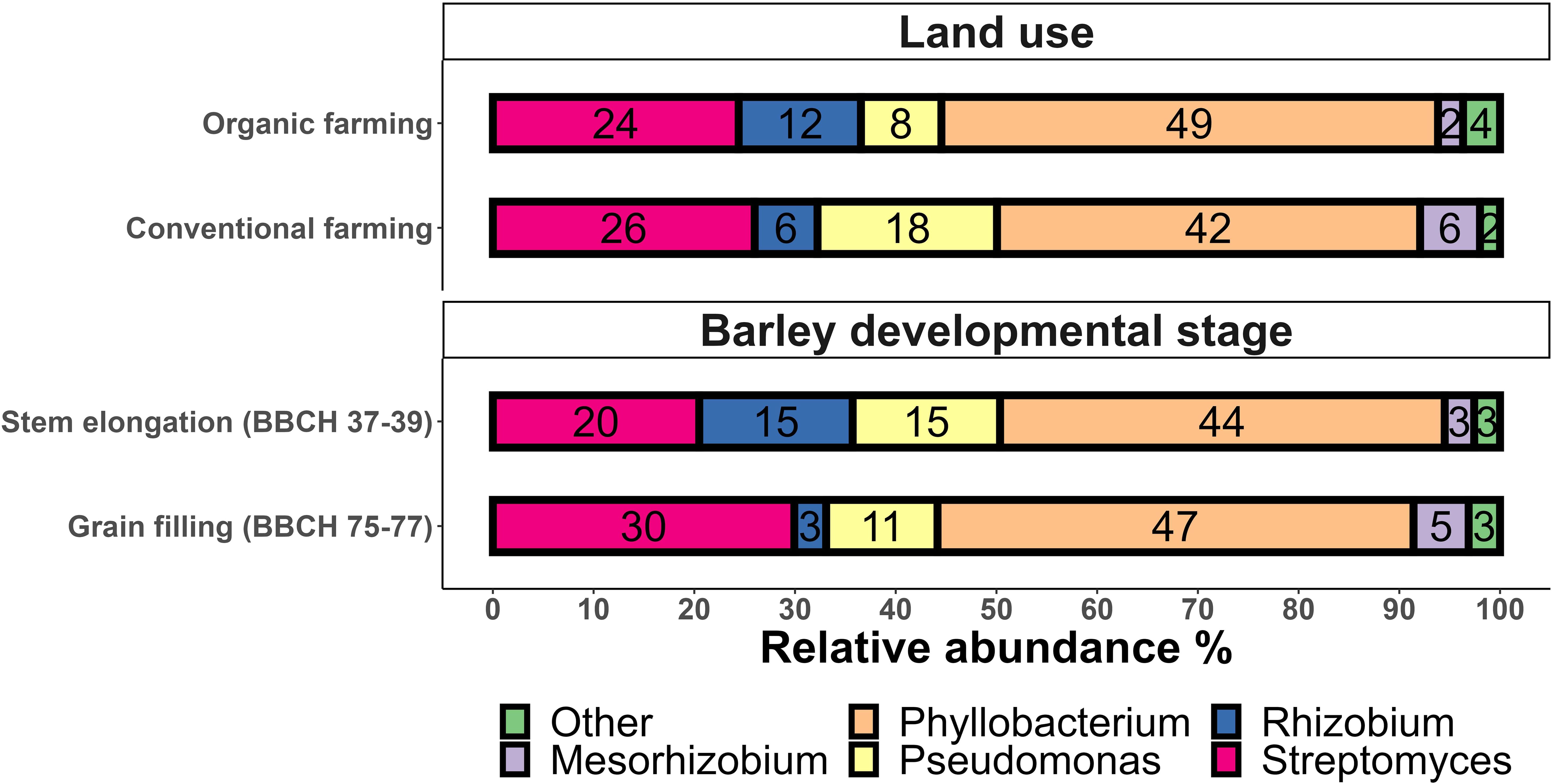
Figure 2. Relative abundances of cultivated bacteria from the barley rhizosphere at the level of most abundant genera. Samples were taken at two host developmental stages (stem elongation BBCH 37-39, grain filling BBCH 75-77) and in two management types (organic farming, conventional farming). The partial 16S rDNA gene was used to group the bacterial strains. Taxa with < 2% relative abundance were grouped together as “Other”. Numbers in bars represent rounded percentage relative abundances of each genus.
Drought stress tolerances of the strains were estimated as percentage difference in colony diameter on polyethylene glycol medium compared to control medium. Of the genera with most isolated strains, the highest values were observed for Rhizobium and Phyllobacterium, and the lowest values for Pseudomonas and Mesorhizobium (Table 1). Drought stress tolerances of Pseudomonas strains were higher for those isolates obtained from grain filling than for those obtained from stem elongation stage, and drought stress tolerances of Pseudomonas and Mesorhizobium isolates were higher for the strains isolated from OF than for those isolated from CF. Several members of other genera in the bacterial culture collection with lower abundance also demonstrated drought stress tolerance (Supplementary Material S3_drought_tolerance_peg). For example, Arthrobacter strains displayed a mean drought tolerance of 62.8% ± 19% (n = 9), and Priestia strains showed a mean drought tolerance of 64.6% ± 22% (n = 12).
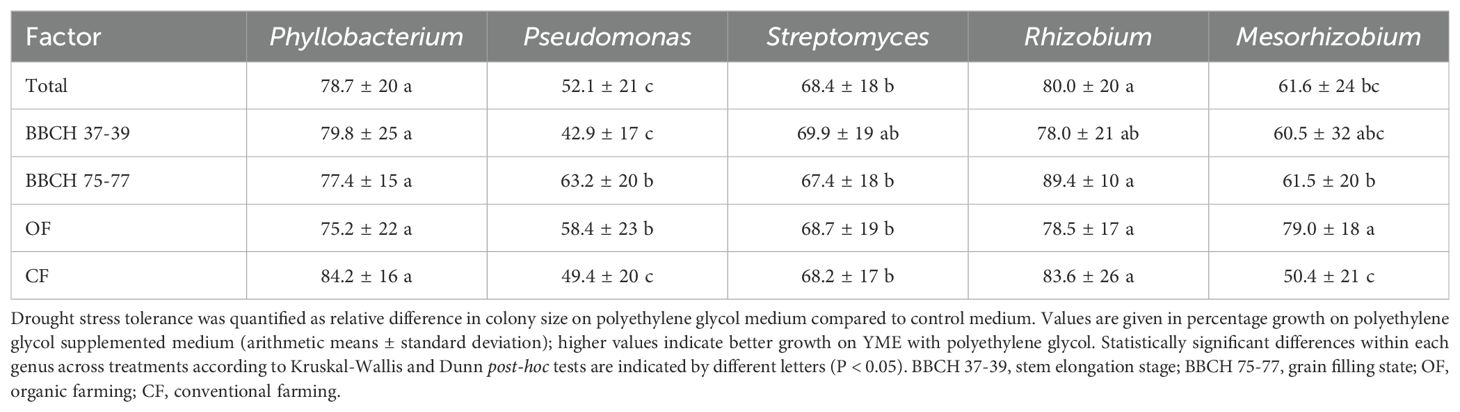
Table 1. Drought stress tolerance of the members of the dominant genera within the bacterial culture collection sorted by barley developmental stage and land use type.
3.2 Strain selection for the drought tolerant synthetic community
Potential strains for the DT-SynCom were selected from the aforementioned culture collection based on 16S rDNA metabarcoding analysis of barley rhizosphere DNA obtained from the Global Change Experimental Facility and from a greenhouse experiment conducted with soil from the GCEF site. Both datasets were used to identify bacterial ASVs that were higher in abundance in the GCEF future climate treatment and under drought stress in the greenhouse experiment than under corresponding control conditions (Breitkreuz et al., 2021a, 2021). Subsequently, the bacterial culture collection was compared with the list of enriched ASVs. The bacterial strains that were enriched under drought and additionally demonstrated a high drought stress tolerance in the polyethylene glycol assay (Supplementary Material S3_drought_tolerance_peg) were selected for inclusion in the DT-SynCom. The 16 members of the DT-SynCom comprise strains from Proteobacteria, Firmicutes, and Actinobacteria (Figure 3A), including four strains of Streptomyces, two strains each of Priestia, Phyllobacterium, and Rhizobium, and one strain each of Rhodococcus, Mycolicibacterium, Arthrobacter, Aeromicrobium, Mesorhizobium, and Pseudomonas. To confirm that the strains do not inhibit one another, they were tested for antagonism with paired growth tests on agar. None of the strains showed signs of antagonism towards each other (Figure 3B).
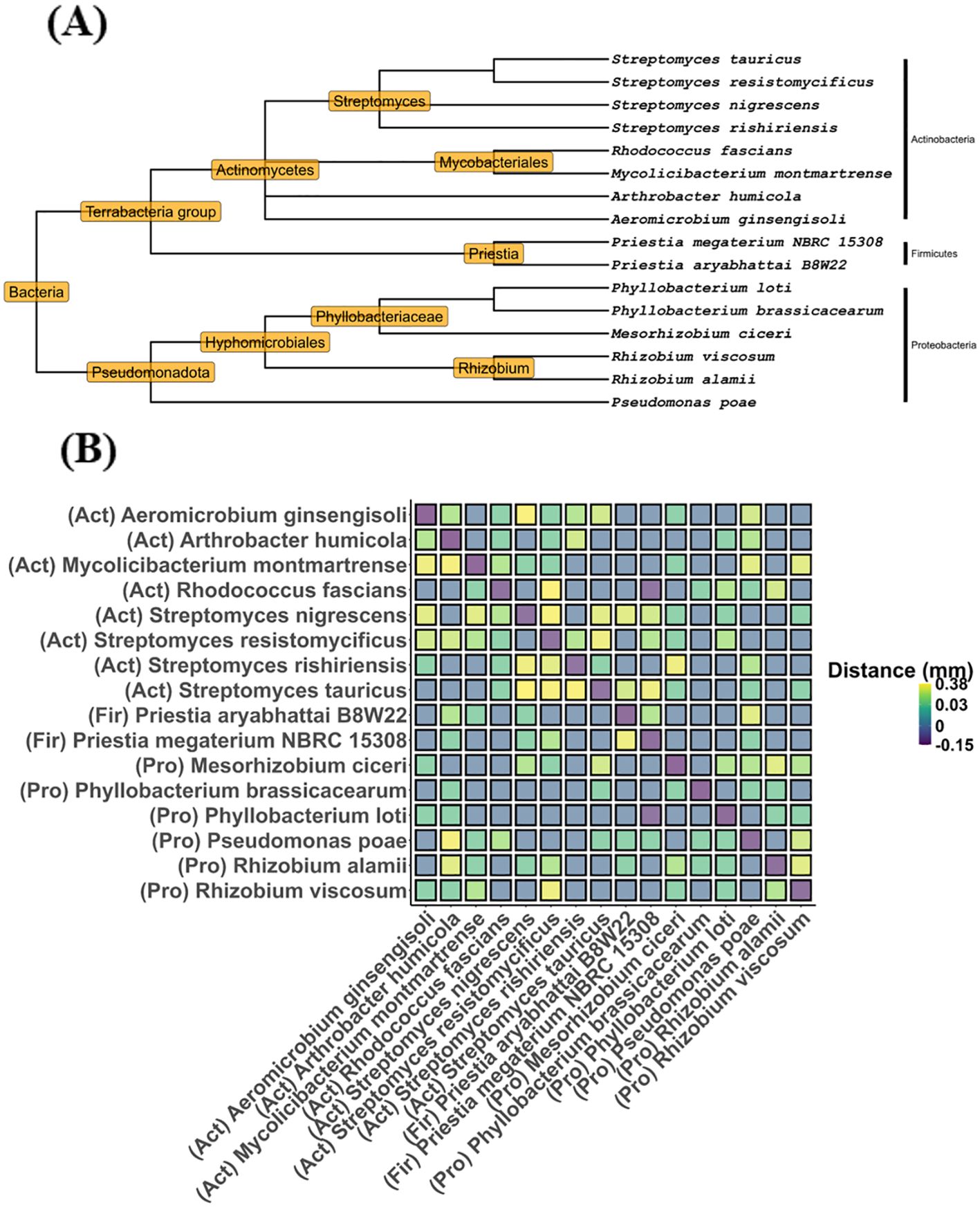
Figure 3. (A) Taxonomic tree of the bacterial strains selected for the Drought-Tolerant Synthetic bacterial Community (DT-SynCom) for barley. Species identifiers are NCBI taxonomic IDs of closest known strains based on partial 16S rDNA homology. (B) Heatmap showing mean distances (in mm) as measured in a pairwise growth challenge assay on solid agar media for all possible pairs of strains. Negative distances indicate pairs growing into one another.
3.3 DT-SynCom genome assemblies and annotations
To obtain genomes, bacterial DNA from DT-SynCom members was extracted and subjected to Illumina paired-end and Nanopore long-read sequencing yielding deeply sequenced genomes with at least 20x coverage from short read (Illumina) data and at least 50x coverage from long read (Nanopore) data respectively (Table 2). Most bacterial genomes were successfully assembled into complete or near complete genomes using a state-of-the-art “long read-first” hybrid assembly method (Table 2, Supplementary Material S4_genome_assembly_annotation_stats, refer Materials and Methods). Ten bacterial genomes featured a circularized contigs indicative of a completely assembled chromosome(s) (Table 2; Supplementary Material S4_genome_assembly_annotation_stats), and in 7 genomes contigs representing other elements such as plasmids were also assembled. According to CheckM, the DT-SynCom genomes were close to complete and contamination-free (> 99% completion in all cases; Table 2; Supplementary Material S4_genome_assembly_annotation_stats). The assembled genomes ranged in size from 4.1 Mbp to 10.9 Mbp and in GC content from 38% to 71% with 6540 ± 1595 protein coding genes (Table 2). Among the top 10 most abundant KEGG ontology categories in theDT-SynCom as a whole were transporters, ribosomal components, two-component systems, transcription factors, and genes with unknown functions (Supplementary Figure 1).
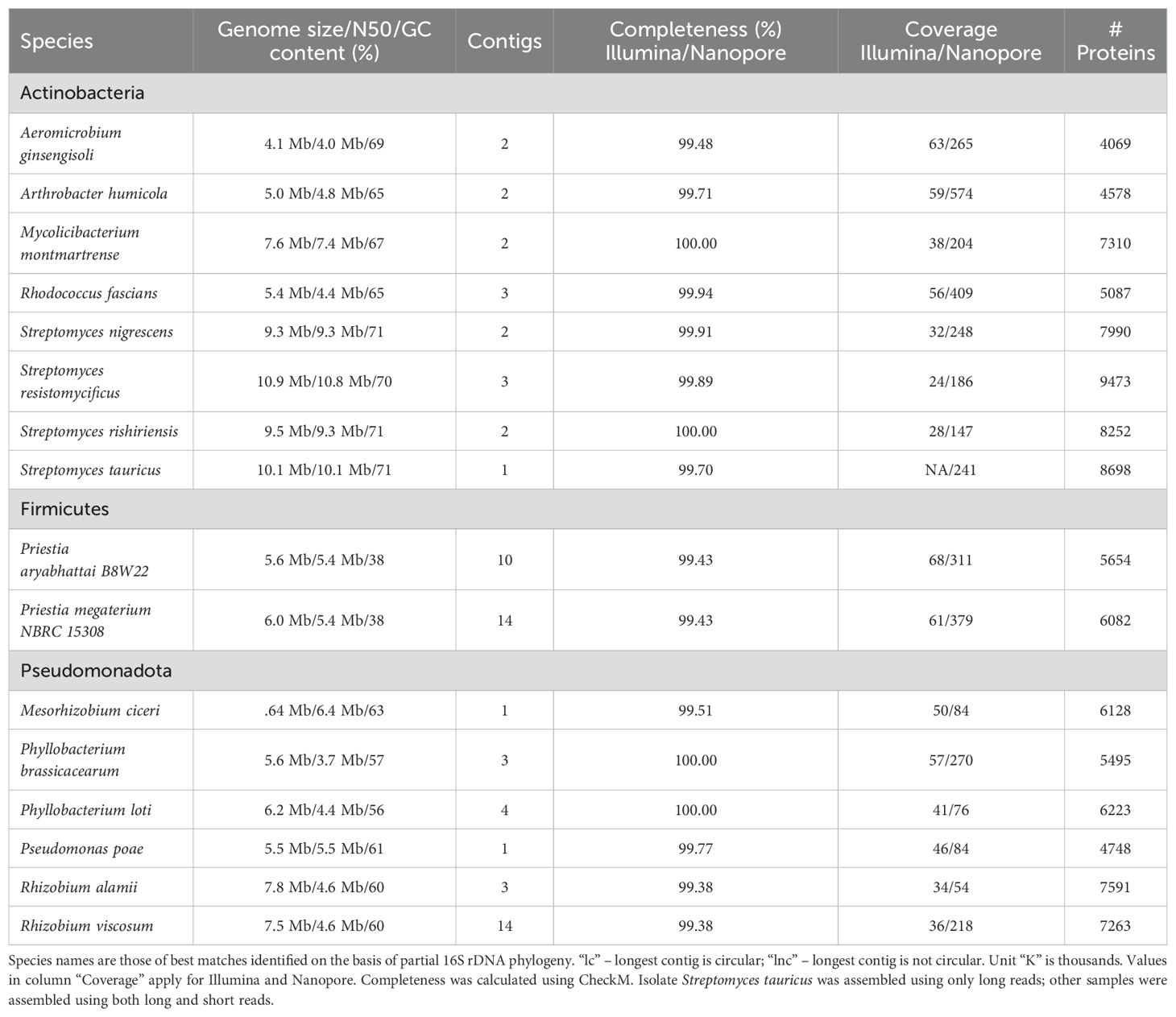
Table 2. Bacterial genome assembly and annotation metrics of 16 barley rhizosphere isolates composing the DT-SynCom.
3.4 Genomic and in vitro evidence for plant-growth promoting (PGP) traits in the DT-SynCom
To establish genomic evidence for PGP traits, multiple sets of PGP genes were identified via examination of the genome annotations (Figure 4; Supplementary Figure 2A, Supplementary Material S5_bakta_pgpg_results.csv). Common to all species were the PGP genes related to phosphate solubilization and transport, nitrogen fixation, siderophore biosynthesis and transporter and indole acetic acid biosynthesis (Supplementary Figure 2A). Part of the genomes contained genes for ACC deaminase (8 genomes), chitinase (12 genomes) and cellulose (8 genomes). No associations were observed between phylogenetic associations of taxa and the presence/absence of PGP genes, with the exception of the two Firmicutes species both lacking genes for ACC deaminase, cellulose degradation and chitin degradation. Similarly no strong association were observed between the location of the PGP genes, i.e., on contigs corresponding to the nuclear chromosome versus other contigs, in any other the species (Supplementary Figure 3, Supplementary Material S5_bakta_pgpg_results.csv). antiSMASH was used to determine the presence of secondary metabolite clusters (SMGCs) in the genomes of the DT-SynCom strains (Supplementary Material S6_antismash_region_counts_annotated). The highest numbers of SMGCs was found in Streptomyces resistomycificus (42 SMGCs), followed by Streptomyces nigrescens (35 SMGCs), and Streptomyces tauricus (34 SMGCs), while the strains with lowest number of SMGCs had 5 SMGCs per genome respectively (Aeromicrobium ginsengisoli, Arthrobacter humicola, Phyllobacterium brassicacearum, Phyllobacterium loti). For strains belonging to the same genus, the number of SMGCs per strain was similar, as expected. The most frequent SMGCs encoded terpene biosynthesis clusters and these were found in 13 of the 16 strains (Supplementary Material S6_antismash_region_counts_annotated). A redox cofactor SMGC was detected in 8 genomes as the next most frequent one. Ectoine SMGC was the only one directly associated with bacterial drought tolerance. This is due to the function of ectoine as an osmoprotectant. Ectoine SMGCs were found in the S. nigrescens, S. resistomycificus, S. rishiriensis, S. tauricus and R. fascians genomes. We also found secondary metabolite clusters that can contribute to drought stress resistance indirectly, for example, through nutritive, protective and signaling roles. These include SMGCs encoding siderophore biosynthesis, detected in genomes of A. humicola, both Priestia strains (P. aryabhattai, P. megaterium), and all four Streptomyces strains (S. nigrescens, S. resistomycificus, S. rishiriensis, S. tauricus). SMGCs associated with arylpolyene biosynthesis were found in the genomes of the proteobacteria P. poae, R. alamii, R. viscosum and the actinobacteria R. fascians. SMGCs associated with melanin production were found only in the genomes of S. resistomycificus and S. tauricus.
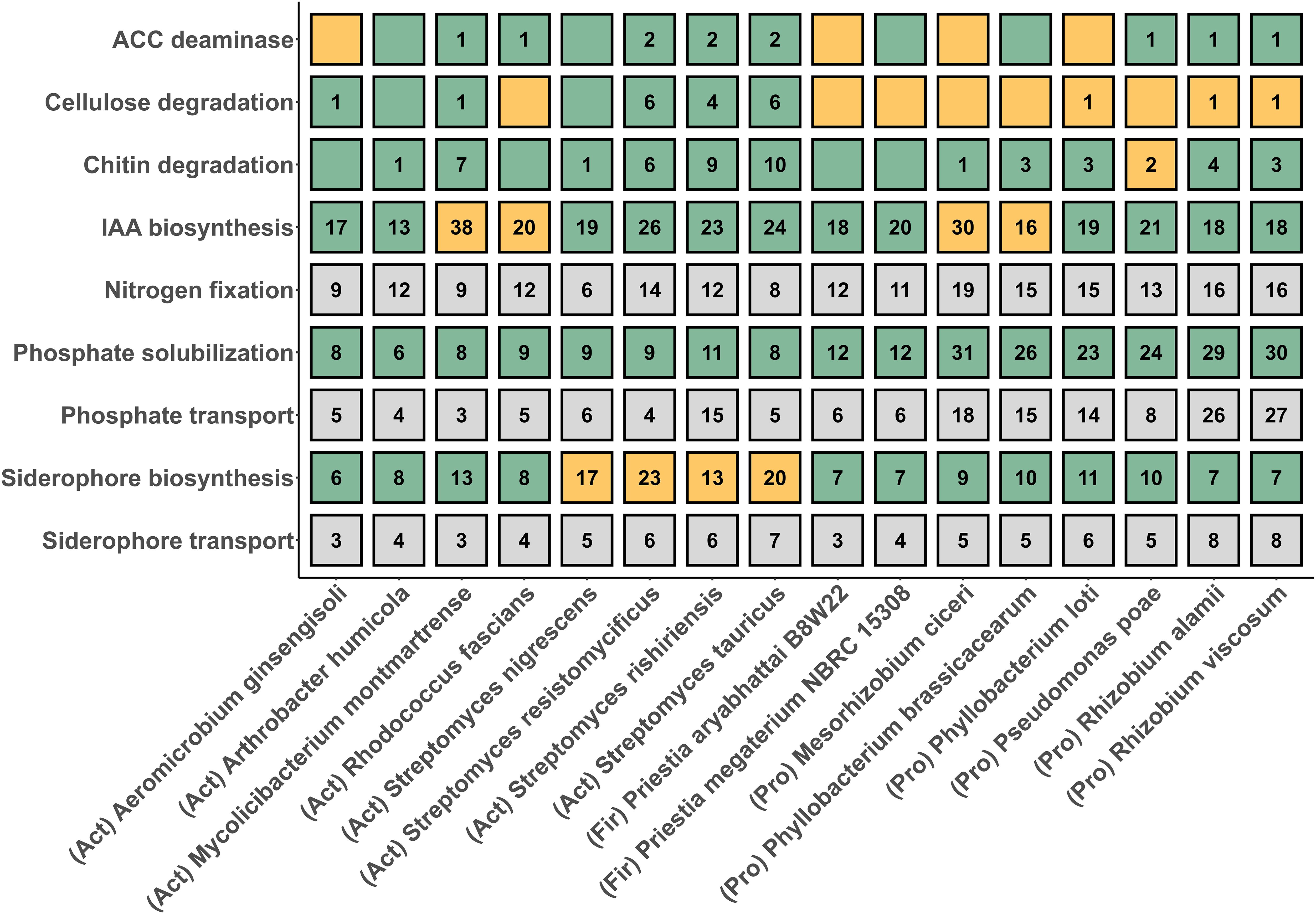
Figure 4. Potential plant growth promoting traits (PGPTs) detected by barley DT-SynCom genome annotation and observed by in vitro activity assays. Numbers indicated number of genes associated with that particular PGPT. Colored boxes indicate whether PGPT activity was measurable by in vitro assay. Grey - no in vitro assay performed; green - activity detected; orange - no activity detected. Phyla are indicated in brackets before species names: Act - Actinobacteria, Fir - Firmicutes, Pro - Proteobacteria.
To collect in vitro evidence for PGP traits, members of the DT-SynCom were tested for different traits (Figure 4; Supplementary Figure 2B, Supplementary Material S7_pgpt_experimental_data). Most Actinobacteria strains showed cellulose degrading activity with the highest activities for S. tauricus, S. resistomycifus and S. rishiriensis. In contrast, none of the Firmicutes, Proteobacteria or Rhodococcus fascians (belonging to Actinobacteria) showed cellulose degrading activity (Figure 4; Supplementary Figure 2B). All DT-SynCom members except Pseudomonas poae showed chitinase activity. Highest activity was displayed by S. nigrescens. All DT-SynCom species solubilized phosphate in vitro (Figure 4; Supplementary Figure 2B). A majority of species from phylum Proteobacteria were strong phosphate solubilizers (with halo formation on selective media) as were three Actinobacteria species: Streptomyces nigrescens, Streptomyces resistomycifus, and Streptomyces rishiriensis (Figure 4; Supplementary Figure 2B). Rhizobium spp. appear to be weak phosphate solubilizers while both Firmicutes taxa (Priestia spp.) were moderate phosphate solubilizers. Weak (Arthrobacter humicola) to moderate siderophore biosynthesis activity was detected in all other Actinobacteria species except for Streptomyces spp. (Figure 4; Supplementary Figure 2B). Both species from genus Priestia (phylum Firmicutes) as well as a majority of the species from phylum Proteobacteria also displayed weak siderophore biosynthetic activity. Phyllobacterium loti and Pseuodomonas poae both showed moderate siderophore biosynthetic activity as indicated by their ability to grow on selective media with accompanying surface discoloration (Figure 4; Supplementary Figure 2B). Besides four species (Aeromicrobium ginsengisoli, Priestia aryabhattai B8W22, Mesorhizobium ciceri, Phyllobacterium loti) all consortium members showed ACC deaminase activity in vitro (Figure 4; Supplementary Figure 2B). Rhodococcus fascians, Streptomyces rishiriensis, Streptomyces tauricus, and Rhizobium alamii showed strong ACC deaminase activity, with the rest showing moderate activity. IAA biosynthesis was found in all species except Mycolicibacterium montmartrense (Actinobacteria), Rhodococcus fascians (Actinobacteria), Mesorhizobium ciceri (Proteobacteria), Phyllobacterium brassicacearum (Proteobacteria).
3.5 In vitro effects of DT-SynCom member application on seed germination and early growth
To test the effects on barley seed germination and seedling growth in vitro, seeds were treated with individual DT-SynCom strains and grown in vitro in a growth chamber for four days. None of the strains inhibited the germination of barley seeds (Figure 5). Treatment with Aeromicrobium ginsengisoli increased root biomass in seedlings (Figure 5A) while decreasing leaf moisture content (Figure 5F). Inoculation with Pseudomonas poae decreased leaf biomass (Figure 5B), root length (Figure 5C), and leaf length (Figure 5D). Inoculation with Rhodococcus fascians decreased leaf length (Figure 5D) and increased leaf moisture content (Figure 5F). Application of Rhizobium viscosum decreased leaf length (Figure 5D). None of the strains affected root moisture content in a statistically significant manner (Figure 5E).
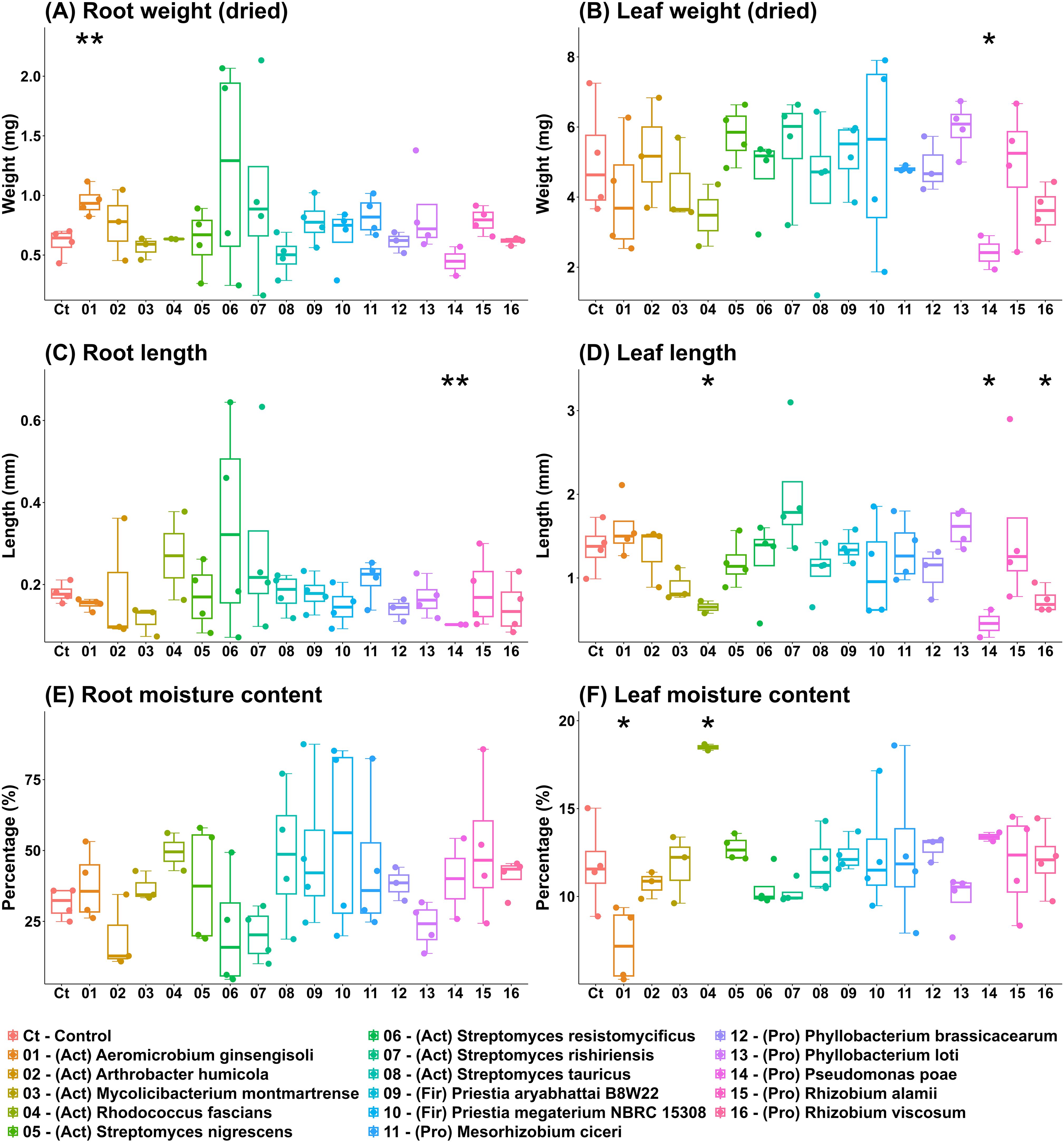
Figure 5. In vitro germination effects of DT-SynCom members on barley seeds. Individual DT-SynCom members were applied to barley seeds, and the seedlings were grown in vitro for four days. Root and leaf biomass (A, B), root and leaf lengths (C, D) and moisture contents (E, F) were estimated at harvest. Asterisks indicate difference to control (no inoculation) value according to ANOVA, with * P < 0.05 and ** P < 0.01.
Similarly to germination study, seedling inoculation with individual strains did not result in seedling growth inhibition (Figure 6). Inoculation with Streptomyces resistomycificus increased leaf moisture content while treatment with Mesorhizobium ciceri decreased it (Figure 6F). Root moisture content was increased after the treatment with Rhodococcus fascians, Streptomyces resistomycificus, Streptomyces tauricus, Phyllobacterium brassicacearum, Rhizobium alamii (Figure 6E). None of the strains affected root and leaf biomass, or root and leaf length in a statistically significant manner (Figures 6A–D).
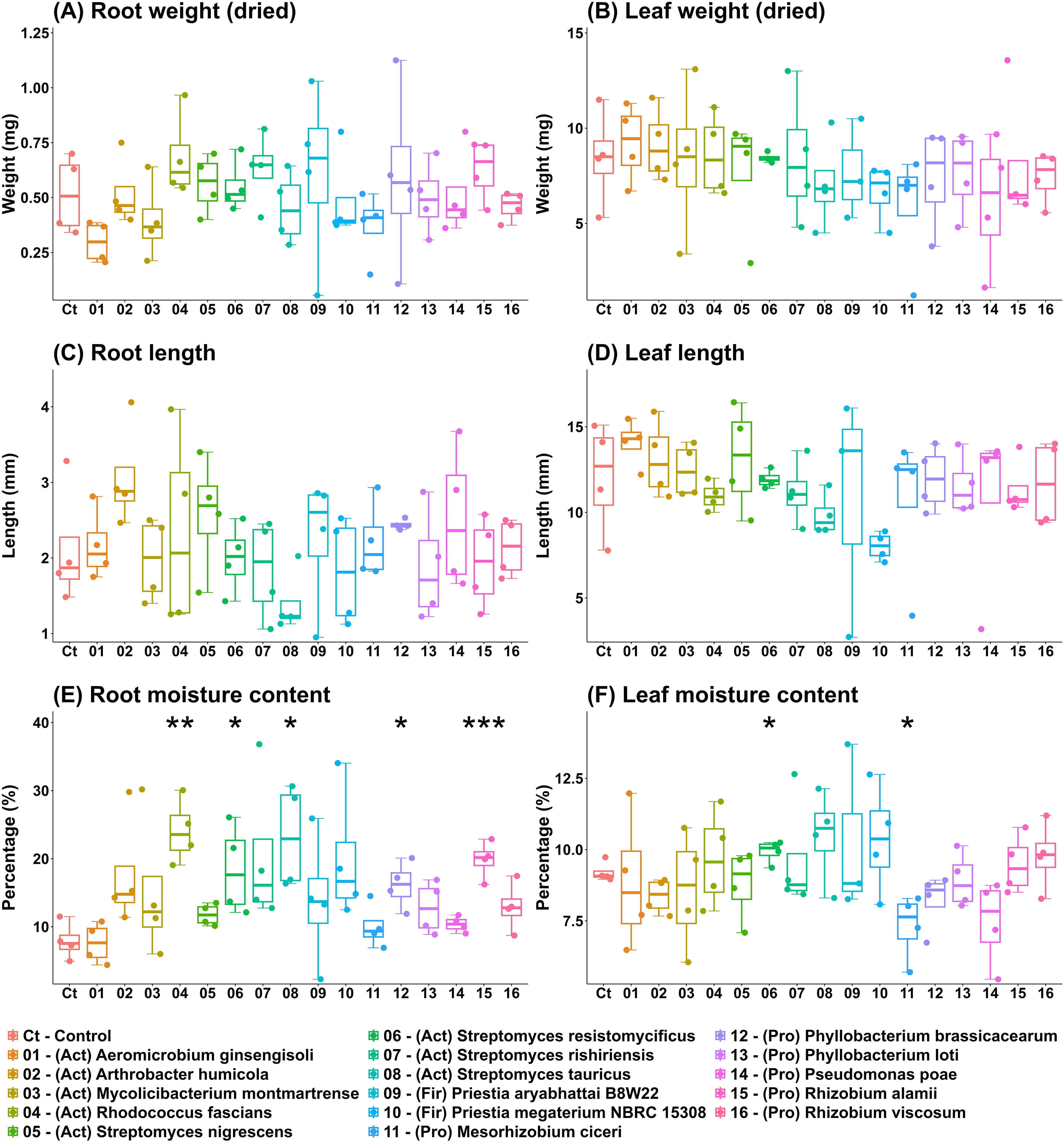
Figure 6. In vitro growth effects of DT-SynCom members on barley seedlings. Individual DT-SynCom members were applied to five days old barley seedlings, and the seedlings were grown in vitro for five days. Root and leaf biomass (A, B), root and leaf lengths (C, D) and moisture contents (E, F) were estimated at harvest. Asterisks indicate difference to control (no inoculation) value according to ANOVA, with * P < 0.05 and ** P < 0.01.
3.6 Greenhouse trial for the evaluation of barley growth and drought tolerance after DT-SynCom application
Severe wilting of barley under drought as measured by differences in first leaf angle was attenuated by treatment with DT-SynCom (Figure 7). Effects of DT-SynCom on barley when watered sufficiently (control) or exposed to 2 weeks drought (drought) was tested subsequently (Figure 8). Leaf length increased but leaf dry weight decreased in the DT-SynCom treatment in the control conditions (Figures 8C, F). Under drought, DT-SynCom application tended to increase leaf length, but the difference was not significant. Additionally, DT-SynCom application contributed to an increase in leaf dry weight during drought stress (Figures 8C, F); this appears to be statistically insignificant potentially due to the presence of an outlier. In general, drought stress caused a strong reduction of leaf biomass with SynCom application attenuating this reduction but not in a statistically significant manner (Figure 8F). Application of SynCom did not show significant differences in lead count (Figure 8A), number of yellow leaves (Figure 8B), plant height (Figure 8D) or root dry weight (Figure 8E).
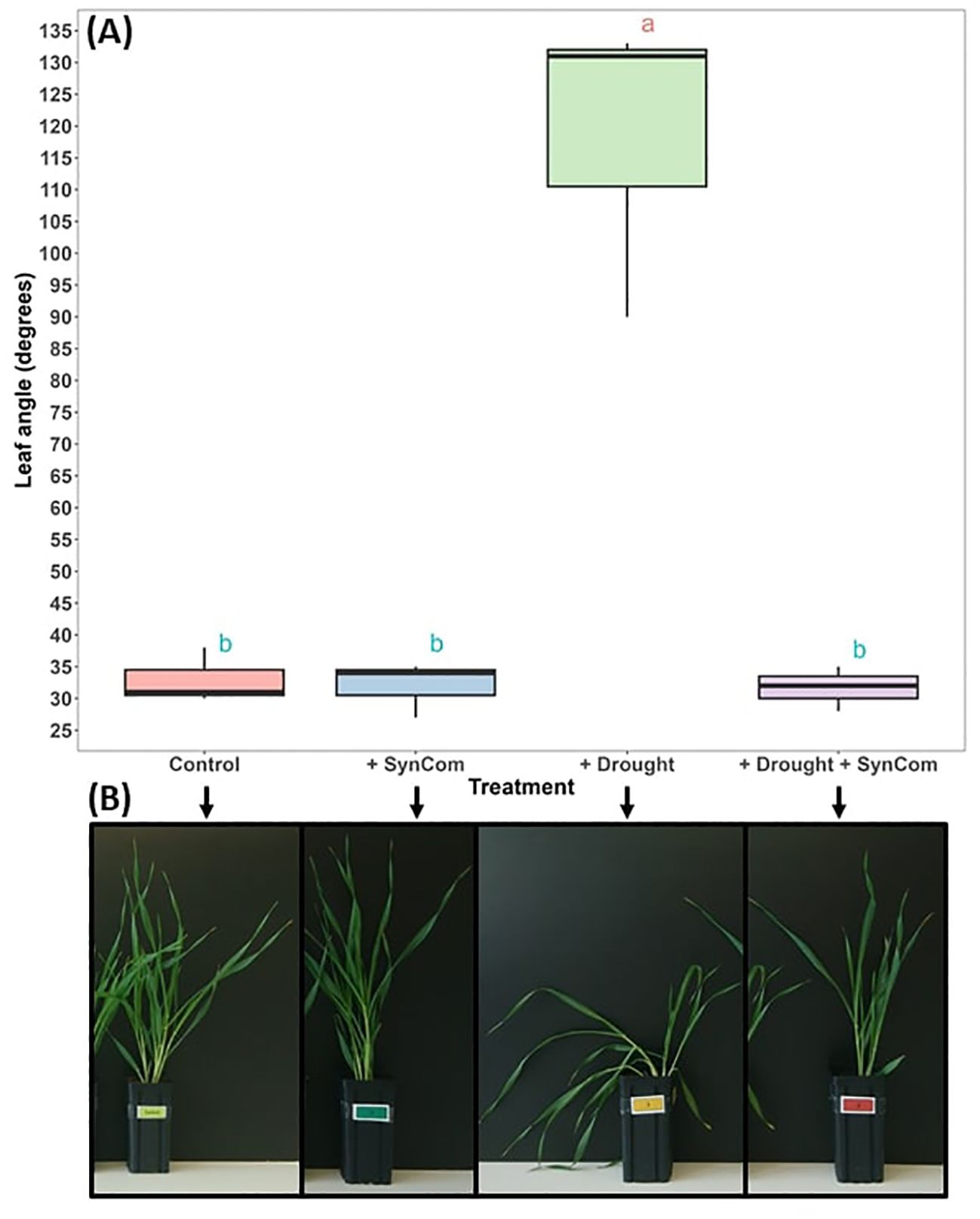
Figure 7. Synthetic community application reduces wilting in barley. (A) Average leaf angle of the first three leaves of barley seedlings. Different letters indicate statistically significant differences between treatments (p < 0.05, ANOVA and Tukey HSD test); a corresponds to more wilted leaves, while b corresponds to less wilted leaves. (B) Example of corresponding wilting/wilted phenotype in the drought treatment without and with DT-SynCom.
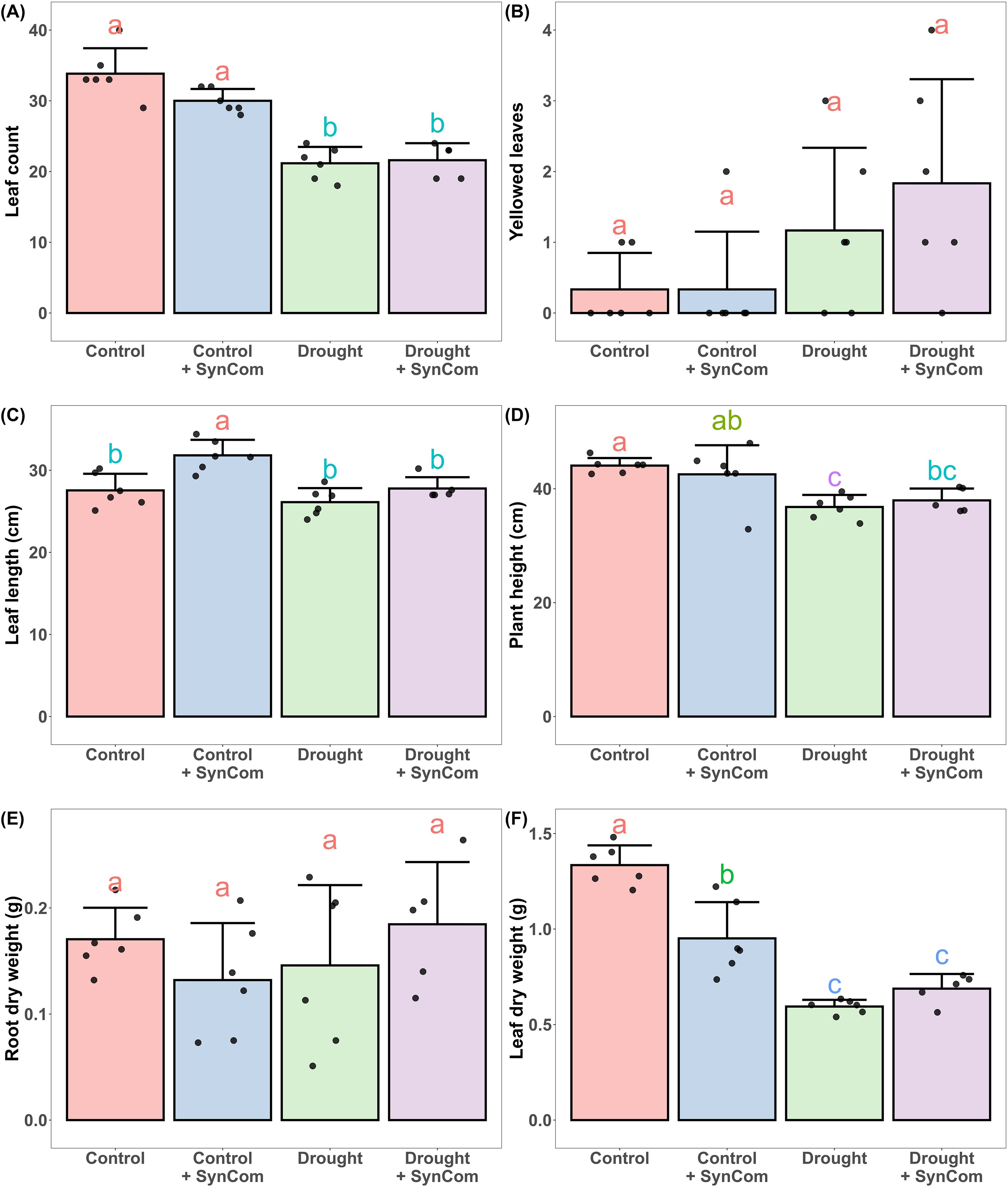
Figure 8. The effects of drought stress (Drought) and addition of the DT-SynCom (SynCom) on barley. Barley was grown with and without DT-SynCom in potting soil for 3 weeks, followed by 2 weeks of reduced watering for half of the pots. (A) Leaf count, (B) number of yellowed leaves, (C) leaf length, (D) plant height, (E) root dry weight, and (F) leaf dry weight were measured at harvest thereafter. Different indicate differences according to ANOVA and Tukey HSD test (P < 0.05). Bar heights and whiskers represent means and standard deviations respectively. Treatments that share a letter are not significantly different from each other.
4 Discussion
In this study, we assembled a drought-tolerant synthetic bacterial community (DT SynCom) for barley. It comprises sixteen strains isolated from barley rhizospheres that, according to genome sequencing, in vitro characterization and inoculation tests, complement each other in their potential to support the drought tolerance of barley.
4.1 Advantages and challenges of constructing a drought-tolerant synthetic community
The synthetic microbial communities (SynCom) is an emerging approach, designed to study plant microbe interaction in a way that more closely mimics plant microbiome (under controlled conditions). Compared to single strain inoculations, using multiple strains has shown to significantly improve the effect on quantitative plant traits (Yang et al., 2021), which might be explained by synergistic effect on plant growth promotion (Azarbad and Junker, 2024). In the context of drought stress, using multiple strains can offer a range of drought-mitigating mechanisms and may be more resilient than relying on a single strain, ultimately enhancing plant drought tolerance and expanding research possibilities.
One of the most important decisions when designing a SynCom is the choice of its size, which mainly depends on the aim of the study to be performed. Larger SynComs attempt to build a microbial community which represents the whole microbiome. They usually consist of a few hundred strains that are representative of the original phylogenetic diversity (Bai et al., 2015), thus increasing functional diversity, however the large size impairs the reproducibility of the SynCom (Xu et al., 2025). In a low-complexity SynCom, such as the DT-SynCom, representativeness is reduced to most commonly two to 30 strains per consortium (Xu et al., 2025). Using a lower number of strains may increase experimental reproducibility, causality, and allow for a more accurate analysis of microbe-plant interactions (Vorholt et al., 2017). For example, using a low complexity SynCom enables the monitoring of individual strain abundancies and their activities (Jorrin et al., 2024). The results so far indicate that DT-SynCom induces drought tolerance in barley.
When designing a SynCom for drought stress tolerance, strain compatibility is an important factor to exclude possible antagonism or strain competition for resources. To ensure that none of the bacterial strains, such as the members of Streptomyces (Schrey et al., 2012) were strong producers of antimicrobials, interbacterial antagonism was tested and found to be negligible, at least in vitro. Although reporting on pairwise strain compatibility in SynComs is rare (Marín et al., 2021), the near total absence of antagonism observed between strains in this study (Figure 3B) appears to be consistent with similar behavior reported in strains from wheat (Ijaz et al., 2019). Given that the DT-SynCom strains were selected from the field where it is very likely that their habitats in the rhizosphere overlap, it is also therefore likely that antagonistic pairs would have already been selected out of the rhizobial community. This observation is also consistent with the reporting that, in SynComs, negative interactions were more predominant between species from different kingdoms and positive (or non-negative interactions) more predominant in interactions between species from within the same kingdom (Durán et al., 2018).
4.2 Effects of DT-SynCom on early barley growth and drought tolerance
Our in vitro tests on early seedling growth (Figure 6) showed that individual strains had either no effect, or a positive effect, on plant growth. This raises the question of the effect of the individual strains upon barley hosts under drought. Previous reports present a very mixed picture regarding the effects of applying single strains from consortia individually upon the same host species. For example Yin et al. (2022), showed that while seven different SynComs reduced the wheat rot disease caused by Rhizoctonia solani AG8, inoculation with SynComs were not significantly better in this regard in comparison to inoculation with individual strains from these SynComs. Similarly, Minchev et al. (2021) reported that while the application of a SynCom reduced tomato root and foliar diseases, it was not more effective than inoculation with the most successful single strain from the consortium. Contrarily, Berendsen et al. (2018) tested the ability of single strains or their mixtures to induce stress response against foliar Hpa infection in Arabidopsis thaliana hosts and showed that while none of the single strains significantly reduced Hpa spore production, a three-strain consortium exhibited a stronger protective effect by significantly lowering spore numbers compared to non-colonized control plants. Likewise Fujiwara et al. (2016), demonstrated that single strain inoculations were not effective against the fungal phytopathogen Fusarium oxysporum, whereas paired strains were. It has also been argued by Jansson et al. (2023) that since multi-species communities have been studied for a much shorter time than single-strain inoculants, a direct comparison of the two approaches may not be instructive due to irreconcilable biases present in existing data and studies. In the context of drought tolerance, it appears that the effectiveness of single strain vs. SynCom application may be further confounded by the level of drought stress, as evidenced by the findings of Gao et al. (2024) who reported that, in pot experiments with Neopallasia pectinate, while single strains were better than SynComs at promoting plant growth under no water stress, the opposite was true for moderate water stress (35% WHC). We were unfortunately unable to examine this issue in our study due to logistical and resource constraints, as therefore posit that a pattern of effectiveness of SynCom application (as compared to single strain application) as observed by Gao et al. (2024) is to be expected with our SynCom also.
To investigate whether and to what extent each bacterial strain of the DT-SynCom contributes to plant drought tolerance in a community context, drop-out experiments can be performed (Carlström et al., 2019). For this purpose, each bacterium should be dropped out individually from the initial inoculum. Such studies have been performed in the Arabidopsis system with root and leaf bacteria and have proven to be valuable. For example, they have shown how the assembly of the leaf bacterial community is subject to priority effects and how, for example, missing strains can invade an already established microbiota (Carlström et al., 2019). In the context of plant phosphate nutrition, they have shown how, in the absence of a particular taxon of the SynCom, plant shoots accumulate higher levels of orthophosphate (Pi) than shoots colonized with the full SynCom (Finkel et al., 2019). In short, an important direction to explore is the testing of different SynCom compositions.
The main indicator of the effectiveness of this SynCom in mitigating drought stress in this study is reduced leaf wilting in the hosts (Figure 7). How DT-SynCom achieves reduced wilting in barley is not yet clear, although the trait spectrum and gene repertoire of the bacteria provides material for hypotheses to be tested. The effects on plant gene expression by wilting are complex, involving hundreds to thousands of genes (Devi et al., 2015), and future work should use RNA sequencing to compare the wilted and DT-SynCom leaves. For a better understanding of the system, this should be accompanied by light microscopic analysis of the leaves and transpiration measurements.
Wilting is essentially a passive motion that reduces exposed leaf area to reduce water consumption and is triggered by loss of turgor pressure in the cells (Fang and Xiong, 2015). This suggests that application of the SynCom potentially enhanced water availability in the hosts. In general, bacteria can improve plant drought tolerance through various traits that enhance water retention, root growth and stress resistance (Ojuederie et al., 2019; Xu et al., 2025). Among the relevant PGPTs, ACC deaminase likely plays an important role in mitigating leaf wilting as it can prevent excessive ethylene accumulation, which is a plant phytohormone well known for hastening leaf senescence and, in the process, leaf wilting (Lim et al., 2007) and plant growth promoting bacteria possessing ACC deaminase have been demonstrated to delay wilting in tomato hosts (Taiwo and Akintokun, 2025). However, ethylene is also an important signaling molecule in plant stress responses, playing a role in adaptive mechanisms such as root growth modulation and stomatal regulation. Therefore, rather than completely inhibiting ethylene production, bacterial ACC deaminase here may help fine-tune ethylene levels, preventing premature senescence while still allowing necessary stress signaling. This balance can be crucial to increase plant drought tolerance. Likewise, strains carrying genes for IAA biosynthesis were also of relevance as this trait would help enhance water transport to and through the roots (and thereby water availability in the leaves) by affecting auxin dynamics in the host (Roychoudhry and Kepinski, 2022). Finally, phosphate solubilization and transport are also relevant to leaf wilting as phosphate availability affects virtually every facet of plant health (López-Bucio et al., 2002). These PGPTs were found in the following 8 SynCom strains: M. montmartrense (Actinobacteria), R. fascians (Actinobacteria), S. resistomycificus (Actinobacteria), S. rishiriensis (Actinobacteria), S. tauricus (Actinobacteria), P. poae (Pseudomonadata), R. alamii (Pseudomonadata), and R. viscosum (Pseudomonadata). As none of these strains are Firmicutes, it is presumably possible that Firmicutes bacteria do not predominate in the mechanisms that prevent leaf wilt under drought stress. Our recent work also suggests that general stress resistance may be induced by DT-SynCom. It primes plant defense systems and improves resistance to powdery mildew infection (Corina Vlot, Anna Sommer, L.R., unpublished).
4.3 Selection of DT-SynCom members and their genomic traits relevant to drought stress
As suggested by Jing et al. (2024), the assembly and reconstitution of the barley DT-SynCom was based not only on broad taxonomy, but also on the final properties of the strains, namely the abundance patterns in amplicon sequencing analysis, related taxonomic identities and the functional traits of the bacteria. The DT-SynCom members were not only associated with ASVs, which were more abundant under the simulated future climate than under ambient climate in the GCEF (Breitkreuz et al., 2021a) and more abundant under low soil moisture in a greenhouse pot experiment with soil from Bad Lauchstädt (Breitkreuz et al., 2021b), but they also expressed osmotic stress tolerance and potential plant growth promoting activities in vitro.
We expected that a similar bacterial growth rate is important to achieve similar numbers and resource use by bacterial cells of each isolate in the consortium. For this reason, some potentially interesting isolates, such as Leclercia adecarboxylata, Pseudomonas donghuensis, and Streptomyces canus were excluded due to very low growth rates. In general, the DT-SynCom from this study is largely consistent with the “reductionist”-style SynComs reported in literature (Liu et al., 2019; Qi et al., 2022; Liu et al., 2019; Qi et al., 2022) with both its approach in terms of construction (e.g., choice of culturing media), composition (16 strains in comparison to the median 13.5 strains), and diversity (mainly comprising Proteobacteria, Actinobacteria, and Firmicutes) (Marín et al., 2021) Although SynComs reported in literature appear to be enriched for genus Pseudomonas (Marín et al., 2021) the DT-SynCom of this study is enriched for genus Streptomyces.
In our drought tolerance SynCom we identified the following four major functional groups that each contribute to plant resilience through different mechanisms: 1) stress mitigation and hormonal regulation (ACC deaminase, IAA biosynthesis), 2) Nutrient acquisition and osmotic regulation (phosphate solubilization and transport), 3) resource mobilization (cellulose and chitin degradation, 4) iron acquisition and oxidative stress protection (siderophore solubilization and transport) (Figure 4). These groups highlight the potential of our SynCom to improve drought tolerance in barley by helping to regulate plant hormones, improve nutrient uptake, promote microbial interactions and enhance stress resistance. The genes for IAA biosynthesis were found in all 16 genomes (Figure 4), suggesting an increased auxin production capacity of the SynCom. The ability of IAA-producing rhizobacteria to promote root growth, increase lateral root formation, enhance water and nutrient uptake, and contribute to coleoptile elongation has been well studied (Park et al., 2021; Metoui Ben Mahmoud et al., 2020; Spaepen et al., 2007; Ahumada et al., 2022), supporting the role of IAA-producing bacteria in plant drought resistance (Etesami and Glick, 2024). Phosphate solubilization and transport gene groups were another two important groups identified in all 16 genomes. Phosphorus is the second most limiting macronutrient for crop growth after nitrogen, and low levels of soluble phosphate can lead to plant phosphorus deficiency even in soils with adequate total phosphate (Gyaneshwar et al., 2002). Drought stress has been shown to reduce plant phosphorus levels in a variety of soils and plant species (Nagatoshi et al., 2023), highlighting the potential of DT-SynCom to alleviate plant drought stress in a variety of ways. Additionally, genes for siderophore biosynthesis and transport were also present in all 16 genomes. Although this trait does not directly enhance drought tolerance, it can still support plant growth by alleviating iron deficiency and supporting physiological and biochemical processes in plants under stressed soil conditions (Xu et al., 2021).
4.4 Impact of barley growth stage on bacterial isolates of the culture collection
In the original barley rhizosphere culture collection, several 16S rDNA affinities were found from both growth stages of barley, suggesting that they may be able to colonize the rhizosphere throughout plant development. Such potentially rhizosphere-competent and resilient bacteria could make an important contribution to the synthetic consortium (Amaya-Gómez et al., 2020; Karimi et al., 2022). Indeed, the DT-SynCom includes 7 isolates Mesorhizobium ciceri, Phyllobacterium brassicacearum, P. loti, Priestia aryabhattai, Rhizobium viscosum, Rhodocccus fascians and Streptomyces rishiriensis, that were plated on agar from both growth stages.
4.5 The barley rhizosphere microbiome in the context of wheat-associated microbial diversity
The barley rhizosphere bacterial culture collection was obtained from the same field experiment and in the same manner, on Pikovskaya agar with tricalcium phosphate, as that described for wheat (Breitkreuz et al., 2020), and not surprisingly, the 16S genera of the strains in the barley collection were similar to that earlier report. Strains of Phyllobacterium, Streptomyces, and Pseudomonas are abundant in both culture collections. However, while Pseudomonas isolates were more abundant in the wheat rhizosphere particularly in the stem elongation stage, Phyllobacterium spp. were more abundant at this stage in barley rhizosphere. Phenotypic differences between isolates, host developmental stage and land use type related differences in osmotic stress tolerance of the bacterial strains themselves were also observed (Table 1). The earlier 16S rDNA amplicon sequencing analysis of the same rhizosphere samples in the field (Breitkreuz et al., 2021a) also identified host developmental stage and land use as strong drivers of barley rhizosphere community composition, supporting the representation patterns of the strains in the barley and wheat (Breitkreuz et al., 2020) culture collections.
4.6 Future perspectives and limitations of the current study
Detection and isolation of several drought-enriched taxa from barley rhizosphere may provide plants with drought-tolerant traits that can improve barley drought resilience and our findings showed its effectiveness in reduced leaf wilting in the hosts (Figure 7), however while our experiments were conducted in a reproducible manner, controlled conditions and sterile seeds were used for the growth assays and pot experiments, thereby limiting the ability to direct our findings to real soil environments. We have shown here the production of indole-3-acetic acid, which stimulates root growth and increases water uptake. With the DT-SynCom application we found no changes in root biomass and no apparent changes in root architecture, including lateral root formation, but future work in the greenhouse and field should include analysis of root architecture to investigate this in detail. The consortium was made up of strains isolated from the rhizosphere of the cultivar Antonella, but the barley cultivar used for the experiments was Golden Promise. Golden Promise is a susceptible cultivar suitable for plant drought response research, but the study would have benefited from testing several barley cultivars, including the original host of the isolated rhizobacteria, to gain a deeper understanding of the effect of the drought-tolerant bacterial consortium on barley. Further experiments should be conducted in non-sterile soil, preferably under field conditions, to further explore the potential of DT-SynCom to improve drought tolerance in barley. Additionally, testing different SynCom compositions could have been an interesting direction to explore. For example, a study by Niu et al. (2017) examined the role of individual strains in a seven-strain SynCom assembled from maize roots by removing a single strain at a time. Similarly, Yin et al. (2022) created ten different SynComs from 14 bacterial strains isolated from the wheat rhizosphere and tested their efficiency to protect wheat form infection by Rhizoctonia solani AG8. In our study, strains carrying genes for ACC deaminase and IAA biosynthesis appear to be the most beneficial for plant drought tolerance. Testing the SynCom with and without these particular strains could have helped highlight the importance of individual members within the SynCom.
5 Conclusions
The potential of plant growth promoting bacteria in sustainable agricultural production has been recognized, mostly for reducing fertilization or improving plant health, but less frequently for improving plant drought tolerance. In this study, we provide evidence that colonization by a drought-tolerant bacterial SynCom promotes drought tolerance in barley. The introduced bacteria were obtained from the rhizosphere of barley, cultivated under future drought conditions and whole genome annotation revealed the presence of genes that translate into plant support. Screening for plant growth promoting traits (such as ACC utilization, IAA production, cellulose and chitin degradation and siderophore production) confirmed the expression of plant-beneficial activities. Strain-by-strain testing on barley showed no inhibition of barley growth or germination, while greenhouse experiments suggested SynCom’s ability to reduce barley leaf wilting under drought conditions. Further experiments are now being carried out to investigate and optimize the interactions between barley and DT-SynCom. Key remaining questions include how many members of the DT-SynCom are present during evoked barley drought tolerance and what their molecular phenotypes are (Jorrin et al., 2024). As the DT-SynCom contains both mineral phosphate solubilizing and siderophore-producing strains, as well as strains capable of degrading chitin and cellulose, their potential involvement in rhizosphere carbon and nutrient cycling shall also be investigated.
Data availability statement
The original contributions presented in the study are publicly available. The partial 16S rRNA sequences reported in this study are available from NCBI Genbank under sequential accessions PV299627 through PV299871 respectively. The raw sequencing short and long reads used for hybrid genome assembly and genome assemblies themselves are available from NCBI under Bioproject PRJNA1219633. Genome annotations and assembly statistics are available via Zenodo (https://doi.org/10.5281/zenodo.14816549).
Author contributions
LR: Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. AH: Conceptualization, Data curation, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing – review & editing. TR: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing – review & editing. MT: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
TThe author(s) declare that financial support was received for the research and/or publication of this article. This study was performed within the framework of the priority program 2125 “Deconstruction and Reconstruction of the Plant Microbiota, DECRyPT”, funded by the German Research Foundation (DFG, project number 466312020; https://gepris.dfg.de/gepris/projekt/466312020).
Acknowledgments
The authors would like to thank the Helmholtz Association and the German Science Foundation (DFG). The authors also want to thank UFZ/iDiv High-Performance Computing (HPC) cluster EVE administration for the maintenance of the EVE Cluster on which the bioinformatics analyses were performed. Assistance from Kerstin Hommel in the lab and Beatrix Schnabel for performing the short-read sequencing, as well as inputs from Dr. Luis Daniel Prada Salcedo and Dr. Qicheng Bei regarding bioinformatics are greatly appreciated. The authors would also like to thank Alga Zuccaro and Corina Vlot-Schuster for fruitful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2025.1572294/full#supplementary-material
References
Ahumada G. D., Gómez-Álvarez E. M., Dell’Acqua M., Bertani I., Venturi V., Perata P., et al. (2022). Bacterial endophytes contribute to rice seedling establishment under submergence. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.908349
Amaya-Gómez C. V., Porcel M., Mesa-Garriga L., Gómez-Álvarez M. I. (2020). A framework for the selection of plant growth-promoting rhizobacteria based on bacterial competence mechanisms. Appl. Environ. Microbiol. 86. doi: 10.1128/aem.00760-20
Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Version 0.12.1 (Barbraham Bioinformatics). Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed October 14, 2024).
Armanhi J. S. L., Souza R. S. C., Biazotti B. B., Yassitepe J.E.D.C.T., Arruda P. (2021). Modulating drought stress response of maize by a synthetic bacterial community. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.747541
Azarbad H., Junker R. R. (2024). Biological and experimental factors that define the effectiveness of microbial inoculation on plant traits: a meta-analysis. ISME Commun. 4, ycae122. doi: 10.1093/ismeco/ycae122
Azarbad H., Tremblay J., Giard-Laliberté C., Bainard L. D., Yergeau E. (2020). Four decades of soil water stress history together with host genotype constrain the response of the wheat microbiome to soil moisture. FEMS Microbiol. Ecol. 96. doi: 10.1093/femsec/fiaa098
Bai Y., Müller D. B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Balting D. F., AghaKouchak A., Lohmann G., Ionita M. (2021). Northern Hemisphere drought risk in a warming climate. NPJ Clim. Atmos. Sci. 4. doi: 10.1038/s41612-021-00218-2
Berendsen R. L., Vismans G., Yu K., Song Y., Jonge R. D., Burgman W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Blin K., Shaw S., Augustijn H. E., Reitz Z. L., Biermann F., Alanjary M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50. doi: 10.1093/nar/gkad344
Bodner G., Nakhforoosh A., Kaul H.-P. (2015). Management of crop water under drought: a review. Agron. Sustain. Dev. 35, 401–442. doi: 10.1007/s13593-015-0283-4
Bouras G., Houtak G., Wick R. R., Mallawaarachchi V., Roach M. J., Papudeshi B., et al. (2024). Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microb. Genomics 10. doi: 10.1099/mgen.0.001244
Bouremani N., Cherif-Silini H., Silini A., Rabhi N. E. H., Bouket A. C., Belbahri L. (2024). Osmotolerant plant growth promoting bacteria mitigate adverse effects of drought stress on wheat growth. AIMS Microbiol. 10, 507–541. doi: 10.3934/microbiol.2024025
Breitkreuz C., Buscot F., Tarkka M., Reitz T. (2020). Shifts between and among populations of wheat rhizosphere pseudomonas, streptomyces and phyllobacterium suggest consistent phosphate mobilization at different wheat growth stages under abiotic stress. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.03109
Breitkreuz C., Heintz-Buschart A., Buscot F., Wahdan S. F. M., Tarkka M., Reitz T. (2021a). Can we estimate functionality of soil microbial communities from structure-derived predictions? A reality test in agricultural soils. Microbiol. Spectr. 9, e0027821. doi: 10.1128/spectrum.00278-21
Breitkreuz C., Herzig L., Buscot F., Reitz T., Tarkka M. (2021b). Interactions between soil properties, agricultural management and cultivar type drive structural and functional adaptations of the wheat rhizosphere microbiome to drought. Environ. Microbiol. 23, 5866–5882. doi: 10.1111/1462-2920.15607
Bulgarelli D., Garrido-Oter R., Münch P. C., Weiman A., Dröge J., Pan Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403. doi: 10.1016/j.chom.2015.01.011
Cai K., Chen X., Han Z., Wu X., Zhang S., Qi L., et al. (2020). Screening of worldwide barley collection for drought tolerance: the assessment of various physiological measures as the selection criteria. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01159
Carlström C. I., Field C. M., Bortfeld-Miller M., Müller B., Sunagawa S., Vorholt J. A. (2019). Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 3, 1445–1454. doi: 10.1038/s41559-019-0994-z
Coleman-Derr D., Tringe S. G. (2014). Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00283
Coster W. D., Rademakers R. (2023). NanoPack2: population-scale evaluation of long-read sequencing data. Bioinf. (Oxford England) 39. doi: 10.1093/bioinformatics/btad311
Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10. doi: 10.1093/gigascience/giab008
Deutsche Forschungsgemeinschaft (2023). Priority Programme “Deconstruction and Reconstruction of the Plant Microbiota: DECRyPT&rd. Available online at: https://www.dfg.de/foerderung/info_wissenschaft/2020/info_wissenschaft_20_66/index.html.
Devi M. J., Sinclair T. R., Taliercio E. (2015). Comparisons of the effects of elevated vapor pressure deficit on gene expression in leaves among two fast-wilting and a slow-wilting soybean. PloS One 10, e0139134. doi: 10.1371/journal.pone.0139134
Driessen P. M. (2001). Lecture notes on the major soils of the world (Rome: Food and Agriculture Organization of the United Nations (World soil resources reports), 94.
Durán P., Thiergart T., Garrido-Oter R., Agler M., Kemen E., Schulze-Lefert P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e14. doi: 10.1016/j.cell.2018.10.020
Egorov N. S. (1985). Antibiotics a Scientific Approach. Antibiotic properties of microorganism cultivated in the Laboratory (Moscow: Mir Publishers).
Elakhdar A., Solanki S., Kubo T., Abed A., Elakhdar I., Khedr R., et al. (2021). Barley with improved drought tolerance: Challenges and perspectives. Environ. Exp. Bot. 201. doi: 10.1016/j.envexpbot.2022.104965
Etesami H., Glick B. R. (2024). Bacterial indole-3-acetic acid: A key regulator for plant growth, plant-microbe interactions, and agricultural adaptive resilience. Microbiol. Res. 281, 127602. doi: 10.1016/j.micres.2024.127602
Fang Y., Xiong L. (2015). General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci.: CMLS 72, 673–689. doi: 10.1007/s00018-014-1767-0
Feulner G. (2017). Global challenges: climate change. Global Challenges (Hoboken NJ) 1, 5–6. doi: 10.1002/gch2.1003
Finkel O. M., Salas-González I., Castrillo G., Spaepen S., Law T. F., Teixeira P. J. P. L., et al. (2019). The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PloS Biol. 17, e3000534. doi: 10.1371/journal.pbio.3000534
Fujiwara K., Iida Y., Someya N., Takano M., Ohnishi J., Terami F., et al. (2016). Emergence of antagonism against the pathogenic fungus Fusarium oxysporum by interplay among non-antagonistic bacteria in a hydroponics using multiple parallel mineralization. J. Phytopathol. 164, 853–862. doi: 10.1111/jph.12504
Funk C., Davenport F., Harrison L., Magadzire T., Galu G., Artan G. A., et al. (2018). Anthropogenic enhancement of moderate-to-strong el niño events likely contributed to drought and poor harvests in Southern Africa during 2016. Bull. Am. Meteorol. Soc. 99, S91–S96. doi: 10.1175/BAMS-D-17-0112.1
Gang S., Sharma S., Saraf M., Buck M., Schumacher J. (2019). Analysis of indole-3-acetic acid (IAA) production in Klebsiellaby LC-MS/MS and the Salkowski method. Bio-protocol 9, e3230. doi: 10.21769/BioProtoc.3230
Gao H., Huang Z., Chen W., Xing A., Zhao S., Wan W., et al. (2024). Mild to moderate drought stress reinforces the role of functional microbiome in promoting growth of a dominant forage species (Neopallasia pectinata) in desert steppe. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1371208
Gene Codes Corporation (2016). Sequencher® version 5.4.5 DNA sequence analysis software. Version 5.4.5 (Ann Arbor, MI, USA: Gene Codes Corporation). Available at: http://www.genecodes.com/ (Accessed October 8, 2024).
Ghotbi-Ravandi A. A., Shahbazi M., Shariati M., Mulo P. (2014). Effects of mild and severe drought stress on photosynthetic efficiency in tolerant and susceptible barley (Hordeum vulgare L.) genotypes. J. Agro. Crop Sci. 200, 403–415. doi: 10.1111/jac.12062
Gordon S. A., Weber R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant Physiol. 26, 192–195. doi: 10.1104/pp.26.1.192
Guglielmetti S., Basilico R., Taverniti V., Arioli S., Piagnani C., Bernacchi A. (2013). Luteibacter rhizovicinus MIMR1 promotes root development in barley (Hordeum vulgare L.) under laboratory conditions. World J. Microbiol. Biotechnol. 29, 2025–2032. doi: 10.1007/s11274-013-1365-6
Gul F., Khan I. U., Rutherford S., Dai Z.-C., Li G., Du D.-L. (2023). Plant growth promoting rhizobacteria and biochar production from Parthenium hysterophorus enhance seed germination and productivity in barley under drought stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1175097
Gupta A., Rico-Medina A., Caño-Delgado A. I. (2020). The physiology of plant responses to drought. Sci. (New York N.Y.) 368, 266–269. doi: 10.1126/science.aaz7614
Gyaneshwar P., Naresh Kumar G., Parekh L. J., Poole P. S. (2002). Role of soil microorganisms in improving P nutrition of plants. Plant Soil 245, 83–93. doi: 10.1023/A:1020663916259
Hartman K., Tringe S. G. (2019). Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 476, 2705–2724. doi: 10.1042/BCJ20180615
Hilbert M., Novero M., Rovenich H., Mari S., Grimm C., Bonfante P., et al. (2019). MLO differentially regulates barley root colonization by beneficial endophytic and mycorrhizal fungi. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01678
Hsu S. C., Lockwood J. L. (1975). Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl. Microbiol. 29, 422–426. doi: 10.1128/am.29.3.422-426.1975
Ijaz M., Tahir M., Shahid M., Ul-Allah S., Sattar A., Sher A., et al. (2019). Combined application of biochar and PGPR consortia for sustainable production of wheat under semiarid conditions with a reduced dose of synthetic fertilizer. Braz. J. Microbiol. 50, 449–458. doi: 10.1007/s42770-019-00043-z
Illumina (2023). Nextera XT DNA Library Prep Kit Reference Guide, (15031942). Available online at: https://support-docs.illumina.com/LP/NexteraXTRef/15031942_07_nextera-xt-product-documentation.pdf.
Jansson J. K., McClure R., Egbert R. G. (2023). Soil microbiome engineering for sustainability in a changing environment. Nat. Biotechnol. 41, 1716–1728. doi: 10.1038/s41587-023-01932-3
Jing J., Garbeva P., Raaijmakers J. M., Medema M. H. (2024). Strategies for tailoring functional microbial synthetic communities. ISME J. 18. doi: 10.1093/ismejo/wrae049
Jorrin B., Haskett T. L., Knights H. E., Martyn A., Underwood T. J., Dolliver J., et al. (2024). Stable, fluorescent markers for tracking synthetic communities and assembly dynamics. Microbiome 12, 81. doi: 10.1186/s40168-024-01792-2
Karimi E., Aliasgharzad N., Esfandiari E., Hassanpouraghdam M. B., Neu T. R., Buscot F., et al. (2022). Biofilm forming rhizobacteria affect the physiological and biochemical responses of wheat to drought. AMB Express 12, 93. doi: 10.1186/s13568-022-01432-8
Kolmogorov M., Yuan J., Lin Y., Pevzner P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Lamboll R. D., Nicholls Z. R. J., Smith C. J., Kikstra J. S., Byers E., Rogelj J. (2023). Assessing the size and uncertainty of remaining carbon budgets. Nat. Clim. Change 13, 1360–1367. doi: 10.1038/s41558-023-01848-5
Lane D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics (Chichester, UK: John Wiley and Sons). Available at: https://cir.nii.ac.jp/crid/1570291225428119552 (Accessed August 23, 2024).
Lee H., Calvin K., Dasgupta D., Krinner G., Mukherji A., Thorne P. W., et al. (2023). IPCC 2023: Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Eds. Lee H., Romero J. (Geneva, Switzerland: IPCC).
Lesk C., Rowhani P., Ramankutty N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Li H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinf. (Oxford England) 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li H. (2021). New strategies to improve minimap2 alignment accuracy. Bioinf. (Oxford England) 37, 4572–4574. doi: 10.1093/bioinformatics/btab705
Li Z., Chang S., Lin L., Li Y., An Q. (2011). A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett. Appl. Microbiol. 53, 178–185. doi: 10.1111/j.1472-765X.2011.03088.x
Lim P. O., Kim H. J., Nam H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Liu H., Brettell L. E., Qiu Z., Singh B. K. (2020). Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. doi: 10.1016/j.tplants.2020.03.014
Liu Y.-X., Qin Y., Bai Y. (2019). Reductionist synthetic community approaches in root microbiome research. Curr. Opin. Microbiol. 49, 97–102. doi: 10.1016/j.mib.2019.10.010
Liu H., Qiu Z., Ye J., Verma J. P., Li J., Singh B. K. (2022). Effective colonisation by a bacterial synthetic community promotes plant growth and alters soil microbial community. J. Sust. Agri. Env. 1, 30–42. doi: 10.1002/sae2.12008
López-Bucio J., Hernández-Abreu E., Sánchez-Calderón L., Nieto-Jacobo M. F., Simpson J., Herrera-Estrella L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–256. doi: 10.1104/pp.010934
Mahdi L. K., Miyauchi S., Uhlmann C., Garrido-Oter R., Langen G., Wawra S., et al. (2022). The fungal root endophyte Serendipita vermifera displays inter-kingdom synergistic beneficial effects with the microbiota in Arabidopsis thaliana and barley. ISME J. 16, 876–889. doi: 10.1038/s41396-021-01138-y
Marín O., González B., Poupin M. J. (2021). From microbial dynamics to functionality in the rhizosphere: A systematic review of the opportunities with synthetic microbial communities. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.650609
Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S. O., Wicker T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Metoui Ben Mahmoud O., Hidri R., Talbi-Zribi O., Taamalli W., Abdelly C., Djébali N. (2020). Auxin and proline producing rhizobacteria mitigate salt-induced growth inhibition of barley plants by enhancing water and nutrient status. South Afr. J. Bot. 128, 209–217. doi: 10.1016/j.sajb.2019.10.023
Minchev Z., Kostenko O., Soler R., Pozo M. J. (2021). Microbial consortia for effective biocontrol of root and foliar diseases in tomato. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.756368
Morcillo R. J. L., Manzanera M. (2021). The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 11 (6), 337. doi: 10.3390/metabo11060337
Muyzer G., Waal E. C. D., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Nagatoshi Y., Ikazaki K., Kobayashi Y., Mizuno N., Sugita R., Takebayashi Y., et al. (2023). Phosphate starvation response precedes abscisic acid response under progressive mild drought in plants. Nat. Commun. 14, 5047. doi: 10.1038/s41467-023-40773-1
Niu B., Paulson J. N., Zheng X., Kolter R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. United States America 114, E2450–E2459. doi: 10.1073/pnas.1616148114
Ojuederie O. B., Babalola O. O. (2023). Growth enhancement and extenuation of drought stress in maize inoculated with multifaceted ACC deaminase producing rhizobacteria. Front. Sustain. Food Syst. 6. doi: 10.3389/fsufs.2022.1076844
Ojuederie O., Olanrewaju O., Babalola O. (2019). Plant growth promoting rhizobacterial mitigation of drought stress in crop plants: implications for sustainable agriculture. Agronomy 9, 712. doi: 10.3390/agronomy9110712
Park S., Kim A.-L., Hong Y.-K., Shin J.-H., Joo S.-H. (2021). A highly efficient auxin-producing bacterial strain and its effect on plant growth. J. Genet. Eng. Biotechnol. 19, 179. doi: 10.1186/s43141-021-00252-w
Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Patz S., Gautam A., Becker M., Ruppel S., Rodríguez-Palenzuela P., Huson D. H. (2021). PLaBAse: A comprehensive web resource for analyzing the plant growth-promoting potential of plant-associated bacteria.
Pedersen B. S., Quinlan A. R. (2018). Mosdepth: quick coverage calculation for genomes and exomes. Bioinf. (Oxford England) 34, 867–868. doi: 10.1093/bioinformatics/btx699
Penrose D. M., Glick B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 118, 10–15. doi: 10.1034/j.1399-3054.2003.00086.x
Pikovskaya R. I. (1948). Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 17, 362–370.
Qi M., Berry J. C., Veley K. M., O’Connor L., Finkel O. M., Salas-González I., et al. (2022). Identification of beneficial and detrimental bacteria impacting sorghum responses to drought using multi-scale and multi-system microbiome comparisons. ISME J. 16, 1957–1969. doi: 10.1038/s41396-022-01245-4
Rahnama S., Ghehsareh Ardestani E., Ebrahimi A., Nikookhah F. (2023). Seed priming with plant growth-promoting bacteria (PGPB) improves growth and water stress tolerance of Secalemontanum. Heliyon 9, e15498. doi: 10.1016/j.heliyon.2023.e15498
Rakovec O., Samaniego L., Hari V., Markonis Y., Moravec V., Thober S., et al. (2022). The 2018–2020 multi-year drought sets a new benchmark in Europe. Earth’s Future 10, e2021EF002394. doi: 10.1029/2021EF002394
Roberson E. B., Firestone M. K. (1992). Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl. Environ. Microbiol. 58, 1284–1291. doi: 10.1128/aem.58.4.1284-1291.1992
Roychoudhry S., Kepinski S. (2022). Auxin in root development. Cold Spring Harbor Perspect. Biol. 14. doi: 10.1101/cshperspect.a039933
Sallam A., Alqudah A. M., Dawood M. F. A., Baenziger P. S., Börner A. (2019). Drought stress tolerance in wheat and barley: advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 20 (3), 3137. doi: 10.3390/ijms20133137
Sarkar D., Rovenich H., Jeena G., Nizam S., Tissier A., Balcke G. U., et al. (2019). The inconspicuous gatekeeper: endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytol. 224, 886–901. doi: 10.1111/nph.15904
Schädler M., Buscot F., Klotz S., Reitz T., Durka W., Bumberger J., et al. (2019). Investigating the consequences of climate change under different land-use regimes: a novel experimental infrastructure. Ecosphere 10, Article e02635. doi: 10.1002/ecs2.2635
Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schrey S. D., Erkenbrack E., Früh E., Fengler S., Hommel K., Horlacher N., et al. (2012). Production of fungal and bacterial growth modulating secondary metabolites is widespread among mycorrhiza-associated streptomycetes. BMC Microbiol. 12, 164. doi: 10.1186/1471-2180-12-164
Schwengers O., Jelonek L., Dieckmann M. A., Beyvers S., Blom J., Goesmann A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genomics 7. doi: 10.1099/mgen.0.000685
Schwyn B., Neilands J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shahid M., Singh U. B., Khan M. S., Singh P., Kumar R., Singh R. N., et al. (2023). Bacterial ACC deaminase: Insights into enzymology, biochemistry, genetics, and potential role in amelioration of environmental stress in crop plants. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1132770
Shen W., Le S., Li Y., Hu F. (2016). SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PloS One 11, e0163962. doi: 10.1371/journal.pone.0163962
Spaepen S., Vanderleyden J., Remans R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Taiwo M. O., Akintokun A. K. (2025). Plant growth-promoting bacteria delayed wilting and improved tomato yield when grown under water stress condition. J. Plant Nutr., 1–19. doi: 10.1080/01904167.2025.2461279
Verschuur J., Li S., Wolski P., Otto F. E. L. (2021). Climate change as a driver of food insecurity in the 2007 Lesotho-South Africa drought. Sci. Rep. 11, 3852. doi: 10.1038/s41598-021-83375-x
Verslues P. E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.-K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J.: Cell Mol. Biol. 45, 523–539. doi: 10.1111/j.1365-313X.2005.02593.x
Vorholt J. A., Vogel C., Carlström C. I., Müller D. B. (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155. doi: 10.1016/j.chom.2017.07.004
Wang S., Ouyang L., Ju X., Zhang L., Zhang Q., Li Y. (2014). Survey of plant drought-resistance promoting bacteria from Populus euphratica tree living in arid area. Indian J. Microbiol. 54, 419–426. doi: 10.1007/s12088-014-0479-3
Xu X., Dinesen C., Pioppi A., Kovács Á. T., Lozano-Andrade C. N. (2025). Composing a microbial symphony: synthetic communities for promoting plant growth. Trends Microbiol. doi: 10.1016/j.tim.2025.01.006
Xu L., Dong Z., Chiniquy D., Pierroz G., Deng S., Gao C., et al. (2021). Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 12, 3209. doi: 10.1038/s41467-021-23553-7
Xu F., Liao H., Yang J., Zhang Y., Yu P., Cao Y., et al. (2023). Auxin-producing bacteria promote barley rhizosheath formation. Nat. Commun. 14, 5800. doi: 10.1038/s41467-023-40916-4
Yadav A., Chen M., Acharya S. M., Yang Y., Zhao T. Z., Chakraborty R. (2024). A stable 15-member bacterial SynCom promotes Brachypodium growth under drought stress. doi: 10.1101/2024.09.10.612297
Yang N., Nesme J., Røder H. L., Li X., Zuo Z., Petersen M., et al. (2021). Emergent bacterial community properties induce enhanced drought tolerance in Arabidopsis. NPJ Biofilms Microbiomes 7, 82. doi: 10.1038/s41522-021-00253-0
Keywords: PGPR-induced stress resistance, microorganisms, plant beneficials, streptomyces, cereal
Citation: Rigerte L, Heintz-Buschart A, Reitz T and Tarkka MT (2025) Assembly and application of a synthetic bacterial community for enhancing barley tolerance to drought. Front. Bacteriol. 4:1572294. doi: 10.3389/fbrio.2025.1572294
Received: 06 February 2025; Accepted: 31 March 2025;
Published: 25 April 2025.
Edited by:
Anil K H Raghavendra, New South Wales Department of Primary Industries, AustraliaReviewed by:
Padhmanand Sudhakar, Kumaraguru College of Technology, IndiaEva Maria Gomez Alvarez, Sant’Anna School of Advanced Studies, Italy
Mohamed Ferioun, Sidi Mohamed Ben Abdellah University, Morocco
Copyright © 2025 Rigerte, Heintz-Buschart, Reitz and Tarkka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda Rigerte, bGluZGEucmlnZXJ0ZUB1ZnouZGU=
 Linda Rigerte
Linda Rigerte Anna Heintz-Buschart
Anna Heintz-Buschart Thomas Reitz
Thomas Reitz Mika T. Tarkka
Mika T. Tarkka