- 1Institute of Biology, Humboldt Universität zu Berlin, Berlin, Germany
- 2Institute of Chemistry, Humboldt Universität zu Berlin, Berlin, Germany
The non-conventional yeast Pichia pastoris (syn. Komagataella phaffii) has become a powerful eukaryotic expression platform for biopharmaceutical and biotechnological applications on both laboratory and industrial scales. Despite the fundamental role that artificial transcription factors (ATFs) play in the orthogonal control of gene expression in synthetic biology, a limited number of ATFs are available for P. pastoris. To establish orthogonal regulators for use in P. pastoris, we characterized ATFs derived from Arabidopsis TFs. The plant-derived ATFs contain the binding domain of TFs from the plant Arabidopsis thaliana, in combination with the activation domains of yeast GAL4 and plant EDLL and a synthetic promoter harboring the cognate cis-regulatory motifs. Chromosomally integrated ATFs and their binding sites (ATF/BSs) resulted in a wide spectrum of inducible transcriptional outputs in P. pastoris, ranging from as low as 1- to as high as ∼63-fold induction with only small growth defects. We demonstrated the application of ATF/BSs by generating P. pastoris cells that produce β-carotene. Notably, the productivity of β-carotene in P. pastoris was ∼4.8-fold higher than that in S. cerevisiae, reaching ∼59% of the β-carotene productivity obtained in a S. cerevisiae strain optimized for the production of the β–carotene precursor, farnesyl diphosphate, by rewiring the endogenous metabolic pathways using plant-derived ATF/BSs. Our data suggest that plant-derived regulators have a high degree of transferability from S. cerevisiae to P. pastoris. The plant-derived ATFs, together with their cognate binding sites, powerfully increase the repertoire of transcriptional regulatory modules for the tuning of protein expression levels required in metabolic engineering or synthetic biology in P. pastoris.
Introduction
Owing to the high yield of recombinant protein production, high similarity of the glycosylation pattern to that in mammalian cells (Balamurugan et al., 2007; Gao et al., 2021), and appropriate folding and secretion of recombinant proteins to the extracellular environment (Yang et al., 2013), yeast Pichia pastoris (syn. Komagataella phaffii) is a broadly used cell factory for the production of various biopharmaceuticals such as insulin (Kjeldsen et al., 1999), hepatitis B surface antigen (Ana Vassileva et al., 2001), human serum albumin (Mallem et al., 2014), and many industrial chemicals including phytase (Tang et al., 2009), lipase (Su et al., 2010), and mannanase (Chen et al., 2020). In contrast to its well−known function as a protein expression system, a limited set of characterized, species-specific synthetic biological tools have been established for P. pastoris (Sanjana et al., 2012; Fischer and Glieder, 2019). Therefore, more efforts should be devoted to developing novel synthetic biology tools to establish P. pastoris as a robust chassis for the recombinant protein production of biosynthesis of natural products (Gao et al., 2021).
Transcriptional regulators play an important role in heterologous protein production (Hartner et al., 2008; Sanjana et al., 2012). P. pastoris, like other eukaryotic organisms, requires a distinct promoter (and terminator) to control the expression of each gene (Tang et al., 2020). Although diverse regulators have been developed in recent years for synthetic biology applications in prokaryotic and eukaryotic cells (Hartner et al., 2008; Bruckner et al., 2015; Naseri et al., 2017; Ji et al., 2019), a limited number of artificial regulators are available for P. pastoris. To express heterologous protein and multi-enzymes of a heterologous pathway in P. pastoris, its native promoters are commonly used (Massahi and Çalık, 2018; Xu et al., 2018; Prattipati et al., 2020). For example, promoter libraries created by the deletion and duplication of putative transcription factor binding sites within the methanol inducible alcohol oxidase 1 (AOX1) promoter (Hartner et al., 2008) or the single point mutation of the constitutive glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter (Waterham et al., 1997) are being used in P. pastoris. However, employing native regulatory elements for heterologous pathway engineering may cause metabolic burden on the cell as the formation of the product competes with cell proliferation and growth (Wu et al., 2016; Naseri et al., 2017) and be undesirable when an orthogonal (minimal interfere with the native cellular processes) and controllable (allowing expression at desired time) system is needed (Naseri and Koffas, 2020). Additionally, the repetitious use of the same regulator in multiple expression cassettes results in genetic instability (Peng et al., 2015). To address the aforementioned challenges, orthogonal transcription factors (TFs) allowing separation of biomass accumulation from the subsequent target molecule production phase, tight control, and tuneable expression of the heterologous gene is desirable (Naseri and Koffas, 2020).
Natural TFs typically contain two functional domains, namely DNA-binding domain (DBD) that target its binding site (BS) within the gene promoter, and activation domain (AD) that activates transcription by interacting with the basal transcription machinery of the cell. Additionally, TFs require nuclear localization signal (NLS) to import to the nucleus of the eucaryotic cell. Recently, pairs of artificial transcription factors (ATFs) and synthetic target promoters [minimal promoter fused to binding site(s) of the ATF were developed as orthogonal regulatory modules. ATFs are based on DBD of various heterologous TF or synthetic DBDs fused to various ADs and NLS. Expression of ATFs is usually controlled by either exogenous chemical inducers [e.g., isopropyl-β-D-thiogalactopyranoside (IPTG)] (Li et al., 2020) or by the light of specific wavelengths (Hartmann et al., 2020), allowing control of the timing of ATF expression and subsequent binding to the synthetic promoter driving expression of a target gene. Until now, inducible ATFs, based on synthetic transcription activator-like effector proteins (TALE) and dead CRISPR-associated protein (dCas9) (Sanjana et al., 2012; Gaj et al., 2013; Machens et al., 2017) have been established for yeast Saccharomyces cerevisiae (Naseri et al., 2019). However, little attention has been paid to the implementation of TALE- and dCas9-derived regulators for metabolic engineering mainly due to their relatively large size (> 5.5 kb), the repetitive structure of TALE-based ATFs and low transcriptional activation by dCas9-derived ATFs (Gao et al., 2014). Surprisingly, the small size ATFs based on heterologous DBDs of plant TFs (∼ 2.5–∼ 3 kb) have recently been developed as strong regulators for S. cerevisiae (10-fold stronger than the yeast constitutive and strong TDH3 promoter).
Plant TFs are grouped into diverse families according to conserved motifs that define their DBD. Over 2000 TFs belonging to different families are identified in higher plants, and approximately half of them are plant-specific. Such TFs are potentially well-suited for establishing orthogonal regulators in yeast (or other heterologous) systems. Plant-derived regulators are based on DBD or full-length (consists of DBD and native AD) of various plant-specific TFs, N- or C-terminally fused to various ADs from yeast GAL4 or plant EDLL and NLS and synthetic promoters harboring one to multiple copies of the cognate cis-regulatory motifs upstream of a yeast CYC1 minimal promoter driving a target gene expression (Naseri et al., 2017). The expression of ATF is under the control of an IPTG-inducible yeast GAL1 promoter. Upon binding ATF to its BS, the downstream gene will be expressed (Naseri et al., 2017). By employing only nine (three weak, three medium, and three strong) plant-derived ATFs and their corresponding BSs, we previously established a combinatorial library of S. cerevisiae variants that allow a wide range of gene expression fine-tuning (Naseri et al., 2019). Considering the similarity between S. cerevisiae and P. pastoris, functional regulators of S. cerevisiae are expected to be transferable to P. pastoris (Ito et al., 2020). A recent study showed that among the 72 terminators in S. cerevisiae, 41 had a high degree of transferability to P. pastoris (Ito et al., 2020).
Here, we focused on characterizing plant-derived ATFs for orthogonal gene expression in P. pastoris. We integrated a collection of ATF/BSs, covering low to high transcriptional strength, into the GUT1 (glycerol utilization 1) locus (Vogl et al., 2018) of P. pastoris. The data show that plant-derived ATF/BSs had a high degree of transferability from S. cerevisiae to P. pastoris. Moreover, 30% of the plant-derived regulators of our collection resulted in higher transcriptional output than the constitutive, strong GAPDH promoter (Hans R. Waterham et al., 1997) of P. pastoris. Additionally, we compared the carotenoid production ability of P. pastoris and S. cerevisiae capable of fabricating high levels of the β-carotene precursor, farnesyl diphosphate (FPP). Accumulation of FPP in S. cerevisiae was achieved by upregulating the expression of ERG20 (FPP synthase), GDH2 (glutamate dehydrogenase), and HMG1 (3-hydroxy-3-methyl-glutaryl-CoA reductase) (Verwaal et al., 2007; Paradise et al., 2008) using inducible, strong plant-derived ATF/BSs, and by deleting LPP1 (lipid phosphate phosphatase), DPP1 (diacylglycerol diphosphate phosphatase), and GDH1 (NADP + glutamate dehydrogenase) (Scalcinati et al., 2012; Trikka et al., 2015). Upon expressing β-carotene pathway genes under the control of plant-derived ATF/BSs, P. pastoris produced 70% of the β-carotene obtained from S. cerevisiae with optimized background. In conclusion, our data confirm the high transcriptional capacity of plant-derived regulators for bioengineering applications in P. pastoris, a suitable host for production of biopharmaceuticals.
Materials and Methods
General
Plasmids were constructed using NEBuilder HiFi DNA assembly (New England Biolabs, Frankfurt am Main, Germany) and SLiCE cloning (Zhang et al., 2012). Plasmid and primer sequences are given in Supplementary Tables 1, 2, respectively. PCR amplification of DNA fragments was performed using high-fidelity polymerases: Phusion Polymerase (Thermo Fisher Scientific), Q5 DNA Polymerase (New England Biolabs, Frankfurt am Main, Germany), or PrimeSTAR GXL DNA Polymerase (Takara Bio, Saint-Germain-en-Laye, France) according to the manufacturer’s recommendations. All restriction enzymes were purchased from New England Biolabs (Frankfurt am Main, Germany). Amplified DNA fragments were gel-purified prior to further use. The primers were ordered from Biomers.net (Ulm, Germany). Escherichia coli ER2925, NEB 5α, or NEB 10β cells (New England Biolabs) were transformed with the plasmids. Strains were grown in Luria-Bertani medium containing an appropriate selection marker at 37°C (Ampicillin, 100 μg/mL or Kanamycin, 50 μg/mL). The plasmid constructs were confirmed by sequencing (Microsynth Seqlab, Goettingen, Germany).
Genetic transformation of the plasmids or linearized DNA fragments from S. cerevisiae was performed using the LiAc/SS carrier DNA/PEG method (Gietz and Schiestl, 2007). The adopted LiAc/SS carrier DNA/PEG method was used to transform P. pastoris with plasmids or linearized DNA fragments (Ito et al., 2018). The yeast strains were grown at 30°C in yeast extract peptone dextrose adenine (YPDA)-rich medium [1% (w/v) bacto yeast extract, 2% (w/v) peptone, 2% (w/v) glucose, 0.04% (w/v) adenine hemisulfate, and 2% (w/v) agar for solid medium] or in an appropriate synthetic complete (SC) media [0.67% (w/v) yeast nitrogen base with ammonium sulfate, 2% (w/v) glucose, and 2% (w/v) agar for solid medium] lacking amino acids to allow selection for transformed cells. When required, glucose was replaced by 2% (w/v) glucose. Single copy integration of each linearized plasmid into the GUT1 site of the P. pastoris genome was verified by colony PCR using primers GNPP001 and GNPP002.
Construction of Plant-Derived ATF and Promoter Pair Clones
To integrate the plasmid into the GUT1 locus of the P. pastoris genome, 1000-bp LHA (primers GNPP003/GNPP004) and 1,000-bp RHA (primers GNPP005/GNPP006) were PCR-amplified from the genomic DNA of the GS115 P. pastoris strain. A PmeI site was added to the 5′-end of GNPH004 and GNPH005. The expression plasmids, listed in Supplementary Table 3 (Naseri et al., 2017), were digested with NotI-HF and PmeI and assembled with the LHA and RHA fragments, resulting in the plasmids pGNPP1 to pGNPP7 listed in Supplementary Table 4. Subsequently, the pGNPP1 to pGNPP7 plasmids were digested with AatII. A synthetic promoter containing a plant TF binding site and a downstream GFP fragment (primers GNPP007/GNPP008) were PCR-amplified from the reporter plasmids listed in Supplementary Table 5 (Naseri et al., 2017). The PCR-amplified fragments were assembled in the digested plasmids to generate integration the plasmids pGNPP008 to pGNPP0024 listed in Supplementary Table 6. To construct a positive control plasmid, pGN005B was digested with NotI-HF and BamHI and subsequently gel-purified to remove a 650-bp fragment. The remaining 6,500-bp fragment was assembled with the PCR-amplified GUT1 integration locus (primers GNPP004/GNPP009, on pGNPP001) and the GAPDH1 promoter (primers GNPP010/GNPP011, on yeast GS115 DNA). The generated integration plasmid was named pGNPP025. pGNPP integration plasmids (see section “Results”) were linearized with PmeI (present within the GU1 homology buffer) and used to transform the JC308 P. pastoris strain (Lin Cereghinoa et al., 2001). Integration takes place at the GUT1 locus of the yeast genome. Positive clones were selected on SC-URA3 medium. The generated P. pastoris strains are listed in Table 1. Subsequently, expression of the chimeric TFs was controlled by an IPTG-inducible promoter, and their transactivation capacity was tested against one, two, or four TF binding sites placed upstream of the CYC1 minimal promoter that controls GFP expression.
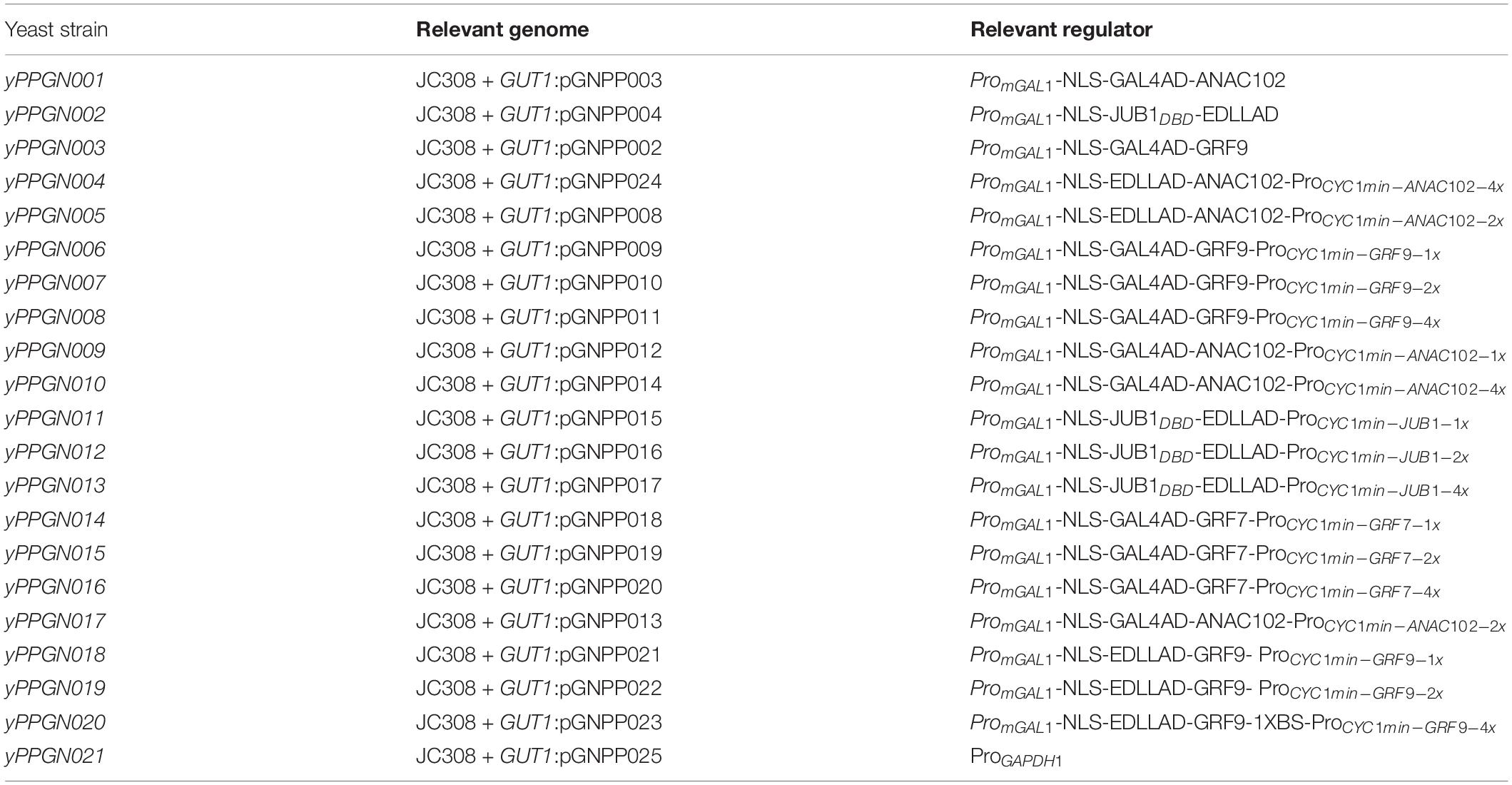
Table 1. List of P. pastoris strains for characterization of plant-derived ATF/BSs generated in JC308 background (Lin Cereghinoa et al., 2001).
Construction of Plasmids and Donors for the Upregulation of Genes in S. cerevisiae
Plasmid pL0A_0-1 (Hochrein et al., 2017) was digested with PmeI and SbfI and assembled with: (i) PCR-amplified ATAF1-derived ATF [primers GNPP012/GNPP013, on Entry vector X_mGAL1_NLS_GAL4 AD_ATAF1_4X (Naseri et al., 2019)] and ERG20 fused to its native terminator (primers GNPP014/GNPP015, on yeast BY4741 DNA) to generate pGNSC001; (ii) PCR-amplified JUB1-derived ATF [primers GNPP012/GNPP016, on Entry vector X_mGAL1_NLS_ GAL4 AD_ JUB1_2X (Naseri et al., 2019)] and GDH2 fused to its native terminator (primers GNPP017_1/GNPP017_2, on yeast YPH500 DNA) to generate pGNSC002; and (iii) PCR-amplified ANAC102-derived ATF [primers GNPP012/GNPP018, on Entry vector X_mGAL1_NLS_GAL4 AD_ANAC102_2X (Naseri et al., 2019)] and tHMG1 fused to its native terminator (primers GNPP019_1/GNPP019_2, on yeast YPH500 DNA) to generate pGNSC003. Donors containing plant-derived ATF-fused GDH2-, tHMG1-, and ERG20-encoding DNA (see section “Results”) were amplified from pGNSC001, pGNSC002, and pGNSC003 using primers GNSC020/GNSC021, GNSC022/GNSC023, and GNSC024/GNSC025, respectively. Transformation of strain Gen 0.2 with the three donors and plasmid pTAJAK105 (Bao et al., 2015; Table 1) allowed integration of the GDH2, tHMG1, and ERG20 donors into the ADE2.a (Bao et al., 2015), his3D1 (Bao et al., 2015), and ura3-52 (Bao et al., 2015) sites of the genome, respectively. Selection for integration was carried out on a medium lacking yeast auxotrophic marker LEU (SC-LEU), since the LEU2 gene is encoded on pTAJAK105. When colonies appeared, the transformation plates were replicated on non-selective plates (YPDA media) to remove the pTAJAK105 plasmid.
Construction of Plasmids and Donors for the Inactivation of Genes
The PCR-amplified SNR52 promoter (primers GNPP026/GNPP027, on pCRCT) and PDC1 terminator (primers GNPP028/GNPP029, on yeast Gen 0.1 DNA; Naseri et al., 2019) were assembled in XhoI/AgeI-digested pCRCT (Bao et al., 2015). A sequence containing gRNA targeting the DPP1 gene (392-bp downstream of the start codon), tracrRNA, and the SUP4 terminator was introduced after the SNR52 promoter through the primer sequences. The generated plasmid was named pGNSC004.
The PCR-amplified CYC1 terminator (primers GNPP030/GNPP031, on p426-SNR52p-gRNA.CAN1.Y-SUP4t; Addgene #43803) was assembled in SmaI/SalI-digested pGNSC004, introducing a gRNA targeting LPP1 (429-bp downstream of the start codon) and tracrRNA after the SNR52 promoter. The resulting plasmid was named pGNSC005.
SalI/AscI-digested pGNSC004 was assembled with double-stranded oligonucleotides (primers GNPP032/GNPP033), introducing a gRNA targeting the GDH1 gene (658-bp downstream of the start codon). The generated plasmid was named pGNSC006.
Subsequently, NotI-digested pGNSC004 was assembled with the PCR-amplified LPP1 gRNA encoding cassette (primers GNPP034/GNPP035, on pGNSC005). The generated plasmid was called pGNSC007.
Subsequently, NotI/PacI-digested pGNSC007 was assembled with the PCR-amplified GDH1-gRNA encoding cassette (primers GNPP036/GNPP037, on pGNSC006). The generated plasmid was called pGNSC008.
Moreover, three donor DNAs, allowing a frameshift mutation in DPP1, LPP1, and GDH1, were constructed by annealing single-strand oligonucleotides (primers GNPP038/GNPP039, GNPP040/GNPP041, and GNPP042/GNPP043, respectively). Each donor contains a 50-bp homology arm in the up- and downstream targeting sequence of the gene. Three donors, together with pGNSC008 (expressing gRNAs, tracer RNAs, and iCas9 protein), were used to transform yeast cells. Selection for integration was carried out on a medium lacking the yeast auxotrophic marker URA (SC-URA), since the URA3 gene is encoded on pGNSC008. When colonies appeared, the transformation plates were replicated on non-selective plates (YPDA media) to remove the pGNSC008 plasmid.
FPP-Producing S. cerevisiae Strains
We combined the previously reported HI-CRISPR system (Bao et al., 2015) and three plant-derived ATF/BSs, namely NLS-GAL4AD-JUB1-2X (Naseri et al., 2019), NLS-GAL4AD-ANAC102-2X (Naseri et al., 2019), and NLS-GAL4AD-ATAF1-4X (Naseri et al., 2019), to overexpress yeast genes, GDH2, tHMG1, and ERG20, respectively. To construct a strain overexpressing GDH2, tHMG1, and ERG20, previously characterized integration sites, shown to exhibit a high integration efficiency, were targeted (Bao et al., 2015). The GDH2, tHMG1, and ERG20 donors were integrated, respectively, at genomic ADE2.a, his3D1, and ura3-52 loci of yeast Gen 0.2, where each donor is integrated into a single locus (Figure 5A). Each donor contains an IPTG-inducible ATF/BS upstream of a CDS and ends with a yeast terminator. The 50-bp overhang sequences up- and downstream of each donor allow its integration into pre-designed genomic loci.
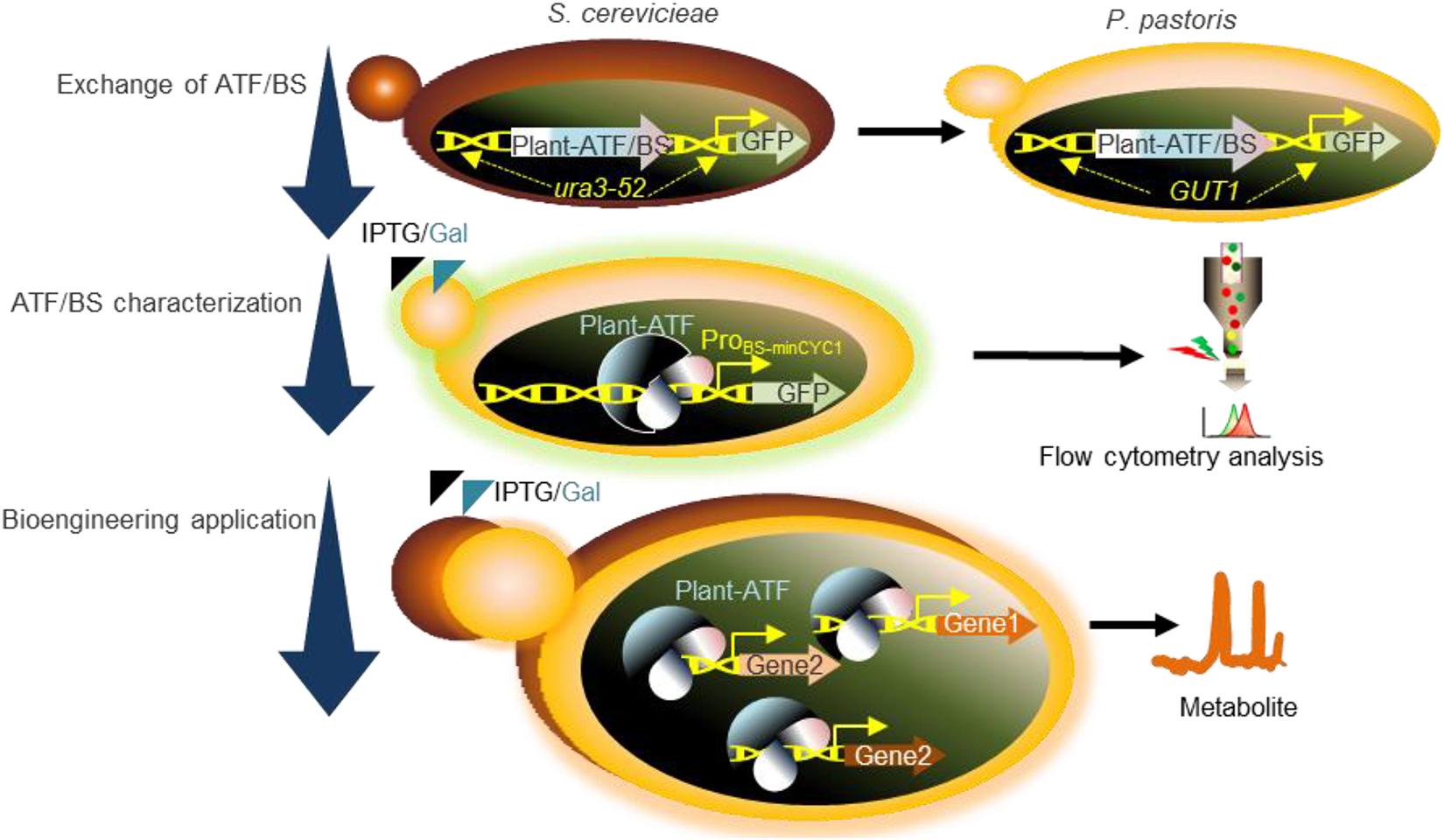
Figure 1. Schematic overview of the workflow used in the present study. Exchange of ATF/BSs: our plant-derived GFP-ATF/BSs deriving GFP expression, previously characterized following integration into the ura3-52 locus of S. cerevisiae (Naseri et al., 2017), were inserted into the GUT1 locus of P. pastoris. ATF/BS characterization: following the addition of IPTG and galactose (inducers), GFP expression was determined by flow cytometry. Bioengineering application: characterized ATF/BSs were employed to tune the protein expression levels required for metabolic engineering in P. pastoris. ATF/BS, artificial transcription factor and its corresponding binding site; GUT1, glycerol utilization 1; ProBS–minCYC1, synthetic promoter containing the binding sites of plant TFs upstream of the minimal CYC1 promoter.
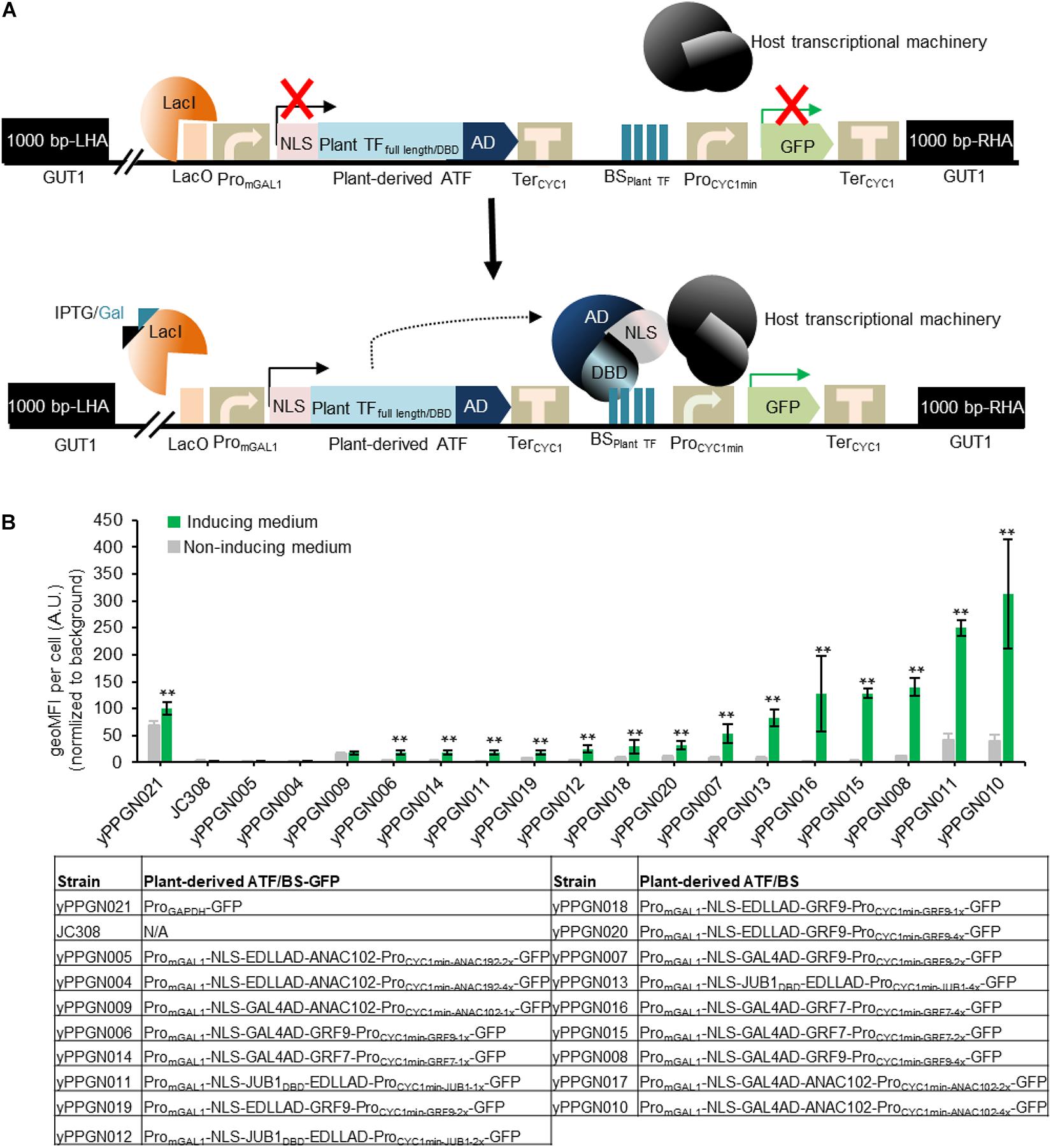
Figure 2. Library of genome-integrated, plant-based ATF/BSs in P. pastoris. (A) Schematic overview of plant-derived ATFs and BSs used in the present study. The ATF cassette contains an mGAL1 promoter with a lacO site at the 5′ end (induced by IPTG and galactose), NLS, and full-length plant TFs or their DBDs fused in different combinations with GAL4AD or EDLLAD and TerCYC1. The transactivation capacity of the ATFs was tested against the TF BSs inserted as one (1x), two (2x), or (4x) copies upstream of the CYC1 minimal promoter driving GFP reporter expression. The 1,000-bp LHA and RHA allow integration of the regulator cassette into the GUT1 sites of the genome. The expression of ATFs was controlled by an IPTG-inducible GAL1 promoter. Constitutive expression of the repressor (LacI) inhibits the expression of plant-derived ATFs, while the addition of inducers (IPTG and galactose) results in ATF expression. Binding of the ATF to its cognate BS within the CYC1 minimal promoter drives GFP expression. Fluorescence output is measured in the absence and presence of inducer. AD, activation domain; ATF, artificial transcription factor; BSplant TF, binding site of the plant transcription factor; BS, binding site; DBD, DNA binding domain; GFP, green fluorescent protein; IPTG, isopropyl-β-D-thiogalactopyranoside; GUT1, glycerol utilization 1; LHA, left homology arm; NLS, nucleus localization signal; ProminCYC1, minimal CYC1 promoter; PromGAL1, modified GAL1 promoter carry LacO site; LacI, Lac repressor; TerCYC1, CYC1 terminator; RHA, right homology arm. (B) Transcriptional output of plant-derived ATF/BSs in P. pastoris. The GFP output was tested in the absence and presence of IPTG and galactose. The types of ATF used to generate yPPGN004—yPPGN020 strains are listed in the table. P. pastoris strain yPPGN021 was used as a positive control, where the GFP expression is controlled by GAPDH promoter allowing constitutive expression of GFP in both inducing- and non-inducing medium. EDLLAD, activation domain from plant; GAL4, GAL4 activation domain of yeast; GRF7 and GRF9, Growth-Regulating Factor 7 and 9; JUB1, NAC TF JUNGBRUNNEN1; ANAC102, A NAC TF 102; VP64AD, viral activation domain. Gray, non-induction medium; green, induction medium. Data are expressed as the geometric mean ± SD of the fluorescence intensity obtained from three cultures, each derived from an independent yeast colony and determined in three technical replicates. Data are normalized to the average geoMFI of the background fluorescence obtained from the wild-type JC308 strain (Lin Cereghinoa et al., 2001). Asterisks indicate a statistically significant difference from the non-induction medium (Student’s t-test; **p < 0.01). AU, arbitrary units; geoMFI, geometric mean fluorescence. The full data are shown in Supplementary Data 1.
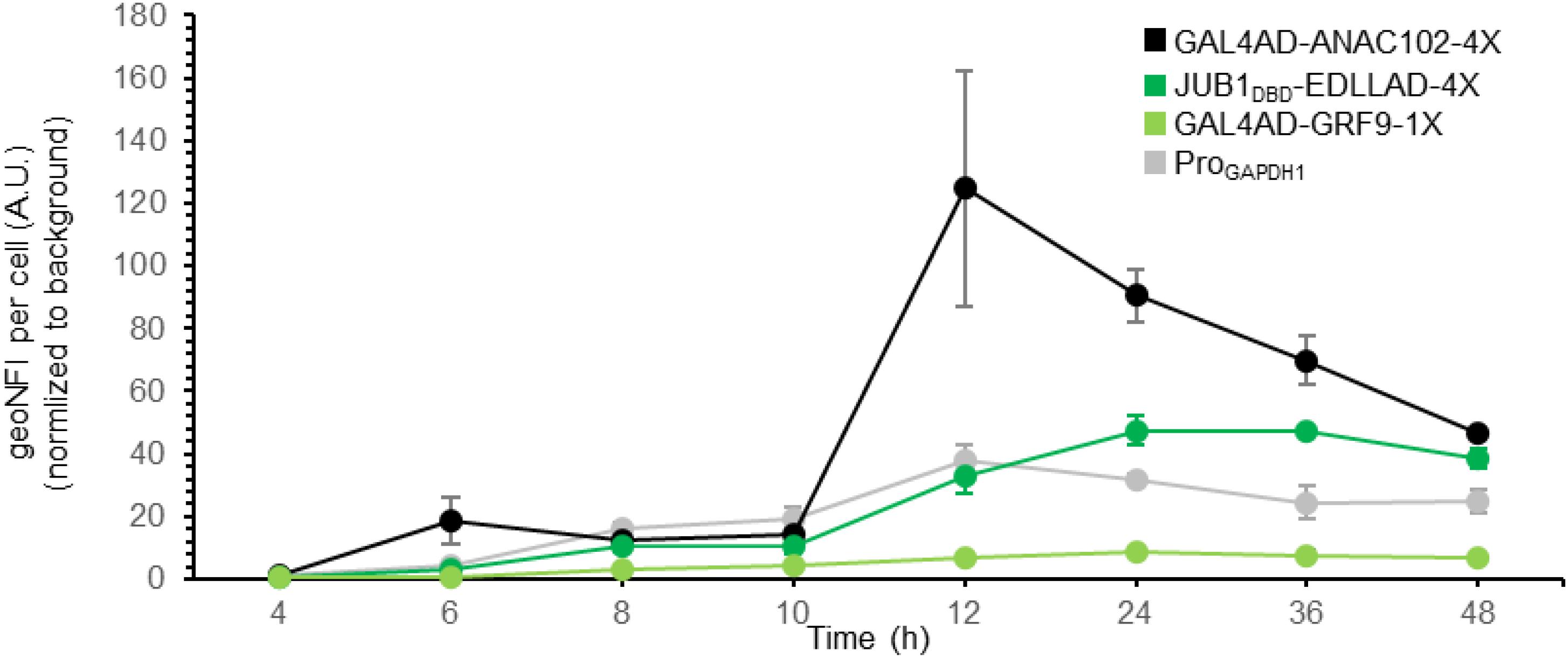
Figure 3. Time course of plant-derived ATF/BS-dependent reporter gene induction. Time course of reporter gene induction, resulting from plant-derived ATFs consisting of GAL4AD and ANAC102 (black), the DBD of JUB1 and EDLLAD (green), or GAL4AD and GRF9 (light green) in combination with “4,” “4,” or “1” copies of their BS, respectively, was compared with the fluorescent output of the constitutive GAPDH promoter (gray). The output of GFP was measured by flow cytometry after 4, 6, 8, 10, 12, 24, 36, and 48 h. The samples were harvested at each time point and analyzed for GFP expression. At each time point, the medium was replaced with the fresh induction medium. EDLLAD, activation domain from plant; GAL4AD, GAL4 activation domain of yeast; GRF9, Growth-Regulating Factor 9; JUB1, NAC TF JUNGBRUNNEN1; ANAC102, A NAC TF 102; DBD, DNA binding site; Pro, promoter. Data are expressed as the geometric mean ± SD of the fluorescence intensity obtained from three cultures, each derived from an independent yeast colony and determined in two technical replicates. Data are normalized to the average geoMFI of the background fluorescence obtained from wild-type JC308. AU, arbitrary units; geoMFI, geometric mean fluorescence. The full data are shown in Supplementary Data 2.
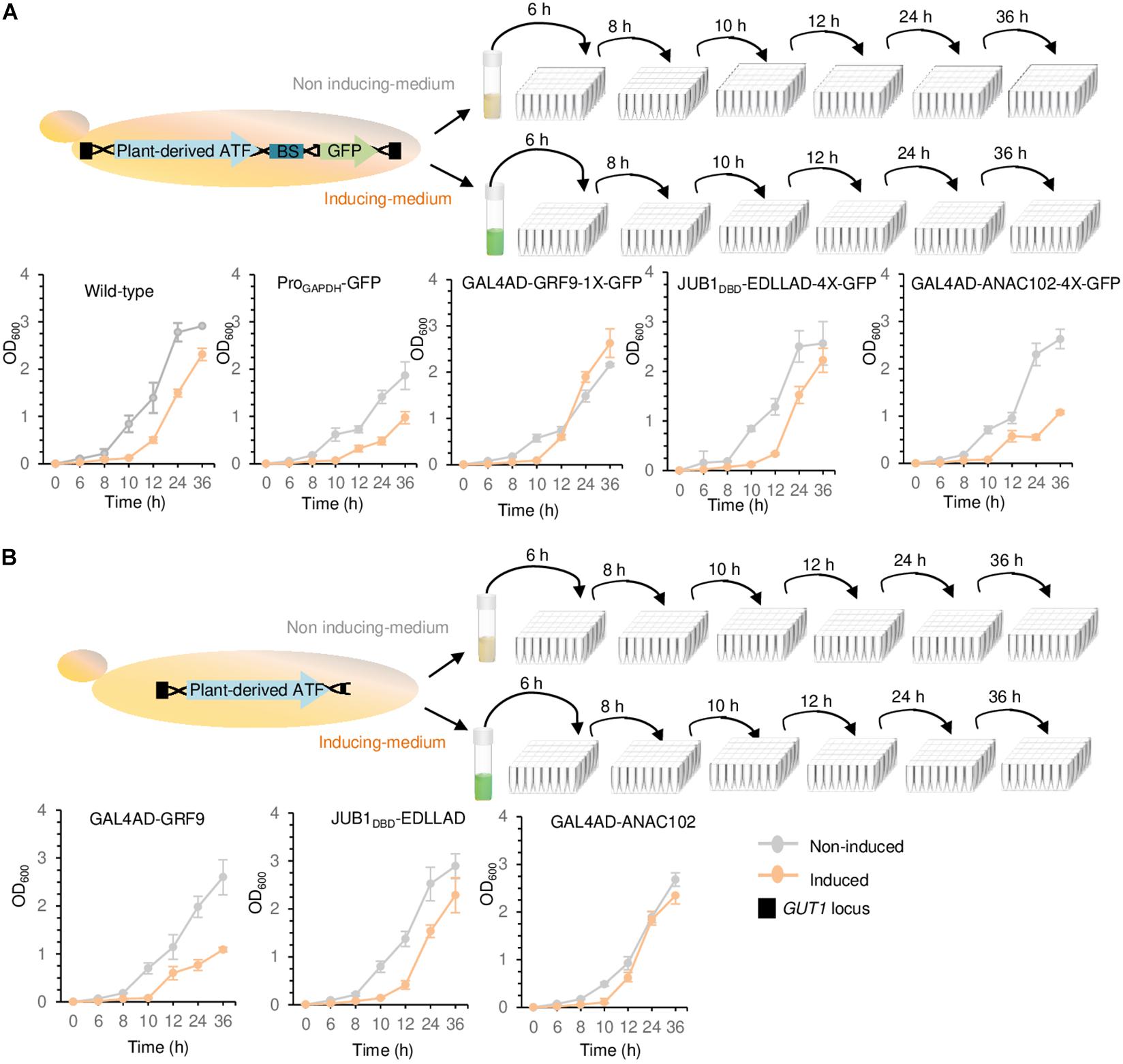
Figure 4. Growth assays. (A) Growth assay of yeast cells containing chromosomally integrated driver and reporter modules. (B) Growth assay of yeast cells containing chromosomally integrated driver modules. The strains containing either the integrated driver/reporter or driver modules were inoculated in induction (orange line) and non-induction (gray line) medium, followed by OD600 measurement at 0, 6, 8, 10, 12, 24, and 36 h. Wild-type JC308 was used as a control. “1X” and “4X” indicate one or four copies of the TF binding site, respectively. To simplify the figure, the modified GAL1 promoter and CYC1 terminator located upstream and downstream of plant-derived ATFs, NLS, and the minimal CYC1 promoter located upstream of GFP are not shown. EDLLAD, activation domain from plant; GAL4AD, GAL4 activation domain of yeast; GRF9, Growth-Regulating Factor 9; JUB1, NAC TF JUNGBRUNNEN1; ANAC102, A NAC TF 102; DBD, DNA binding site; Pro, promoter. Data are expressed as the mean OD600 values ± SD obtained from three cultures, each derived from an independent yeast colony and determined in three technical replicates. OD600, optical density at 600 nm. The full data are shown in Supplementary Data 3.
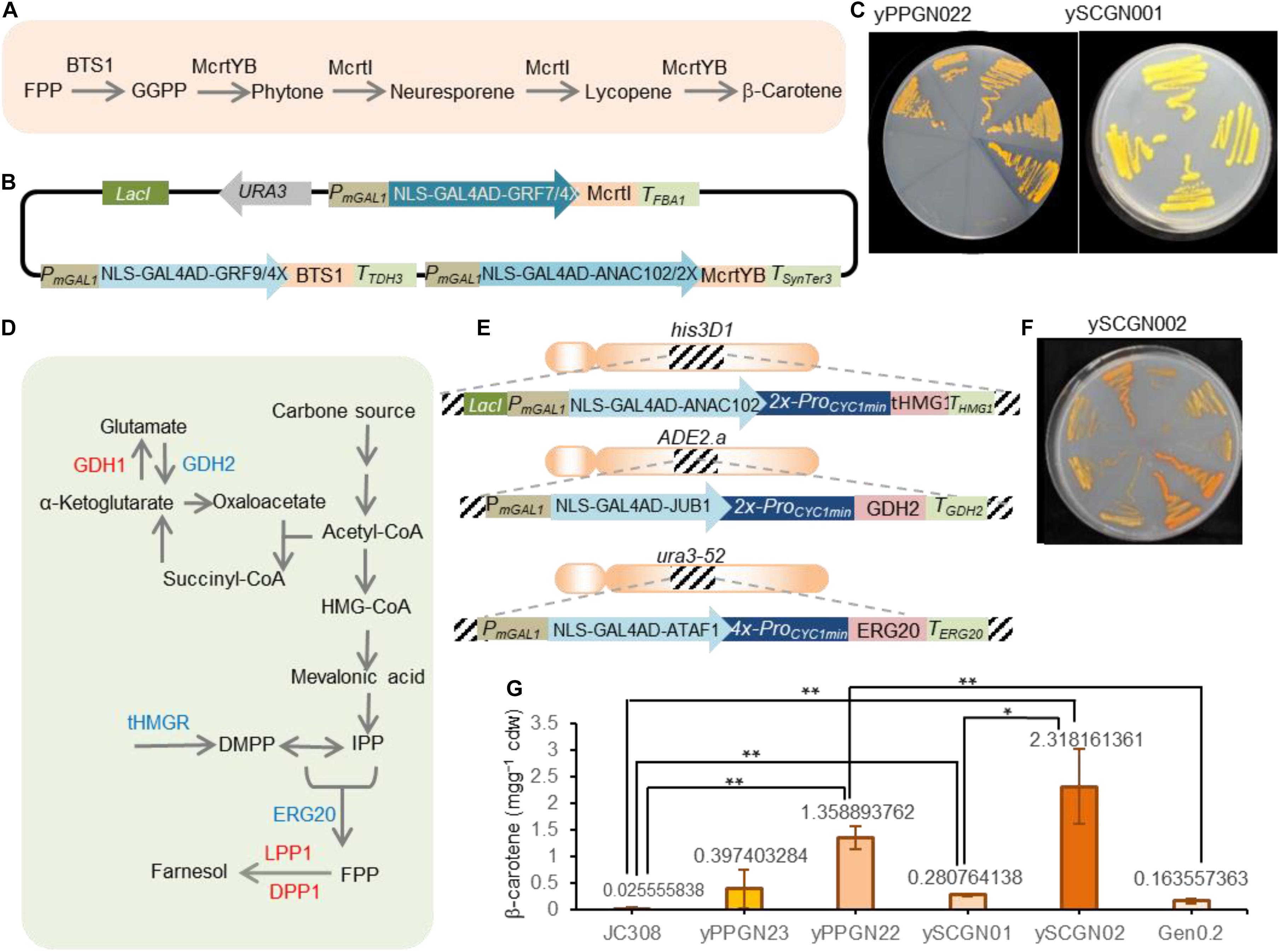
Figure 5. Production of β-carotene in yeast strains. (A) Scheme showing the β-carotene production from FPP precursor. BTS1, geranylgeranyl diphosphate synthase; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; McrtYB, optimized phytoene synthase/lycopene cyclase; McrtI, phytoene desaturase. (B) Scheme showing the β-carotene-encoding plasmid. The pCAROTENE plasmid was constructed as described by Naseri et al. (2019). McrtI, McrtYB, and BTS1 genes are under the control of ATF/BSs NLS-GAL4AD-GRF9-4X, NLS-GAL4AD-GRF7-4X, and NLS-GAL4AD-ANAC102-4X. The ATF/BS-McrtI, -McrtYB-, and -BTS1 modules are flanked by the IPTG inducible modified GAL1 promoter. Additionally, the ATF/BS-McrtI module encodes the LacI repressor. Selection on SC-URA media allows screening for successful plasmid integration. To simplify the figure, the CYC1 terminator located downstream of ATF is not shown. AD, activation domain; ATF, artificial transcription factor; BSplant TF, binding site of the plant TF; DBD, DNA binding domain; IPTG, isopropyl-β-D-thiogalactopyranoside; NLS, nucleus localization signal; ProminCYC1, CYC1 minimal promoter; Ter, terminator. (C) Representative plate of the constructed β-carotene producing strains. The pCAROTENE plasmid was transformed to P. pastoris and S. cerevisiae to generate the yPPGN022 and ySCGN01, respectively. Upon adding IPTG, plant-derived ATFs were expressed and β-carotene was produced. Colors of carotenoid-producing yeast strains in YPDA media for the yPPGN022 and ySCGN01 strains. (D) Scheme illustration of FPP precursor production in yeast. To redirect yeast metabolisms toward the FPP production, GDH2, ERG20, and tHMG1 overexpression (blue), deleted or inactive DPP1, LPP1 and GDH1 (red) are needed. ERG20, FPP synthase; DPP1 and LPP1, lipid phosphate phosphatases; tHMG1, truncated HMG-CoA reductase, GDH1, NADP + -glutamate dehydrogenase; GDH2, NAD + -dependent glutamate dehydrogenase; HMG-CoA, 3-hydroxy-3methylglutaryl-CoA; IPP, isopentenyl diphosphate; DMAPP: dimethylallyl diphosphate. (E) Scheme representing overexpressing donors for the enhanced production of FPP. ATFs NLS-GAL4AD-ANAC102-2X, NLS- GAL4AD-JUB1-2X, and NLS-GAL4AD-ATAF1-4X are fused to tHMG1-THMG1, GDH2-TGDH2, and ERG20-TERG20. The tHMG1, GDH2, and ERG20 donors are flanked by 50-bp homology arms to integrate into the his3D1, ADE2.a, and ura3-52 loci, respectively. In each donor, a modified GAL1 promoter is located upstream of the plant-derived ATF. Additionally, the tHMG1 donor encodes LacI. The BS of ATFs is placed upstream of the CYC1 minimal promoter to drive gene expression. NLS, nuclear localization signal; JUB1, plant JUNGBRUNNEN1 transcription factor; GAL4AD, yeast GAL4 activation domain; ANAC102, NAC domain containing protein 102; ATAF1, Arabidopsis thaliana Activating Factor 1; “2X”, and “4X” indicate two, or four copies of the plant TF binding site, respectively. (F) Colors of the carotenoid-producing ySCGN02 S. cerevisiae strain (optimized for FPP production) in YPDA media. (G) SFC analysis of the carotenoid content of yeast strains. Values represent the mean ± SD of three independent colonies after 3 days of cultivation. P. pastoris strains yPPGN023 containing pCAROTENE-PGAP plasmid and J308 were used as a positive and negative controls, while Gen 0.2 was used as a negative S. cerevisiae control. Asterisks indicate a statistically significant difference (Student’s t-test; *p < 0.05; **p < 0.01). Cdw, cell dry weight. Full data are shown in Supplementary Data 4.
For inactivation of the GDH1, DPP1, and LPP1 genes, three donors, together with pCRCT-GDH1-DPP1-LPP1 (expressing gRNAs, tracer RNAs, and iCas9 protein), were used to transform yeast cells, where each donor is integrated into a single locus. Each donor contains the 50-bp overhang sequences up- and downstream of its integration into target genomic sites, and incorporates an 8-bp deletion included within the target gene, thus introducing a frameshift mutation. The ySCGN0.1 or ySCGN0.2 strains were obtained by integrating overexpression or inactivation donors into the yeast genome, respectively. The ySCGN1.2 strain harbors inactivated genes, lpp1, dpp1, and gdh1, in addition to overexpressed GDH2, tHMG1, and ERG20.
Induction Experiments, Flow Cytometry, and Data Analysis
To determine the effect of plant-derived ATF/BS on GFP fluorescence output, single colonies of yeast reporter strains were inoculated into 250 μl SC-URA medium containing 2% glucose in 96-well deep-well plates. Plates were incubated for 16 h at 30°C in a rotary shaker at 230 rpm. The precultures were used to inoculate main cultures in 250 μl YPDA containing 2% glucose (non-induction medium) or YPDA containing 2% galactose and 20 mM IPTG (induction medium) to an OD600 ∼0.1. Each strain was inoculated in three technical replicates per experiment. Cells were grown at 30°C for 12 h in a rotary shaker at 230 rpm. Samples were harvested after 4, 6, 8, 10, 12, 24, 36, and 48 h (see section “Results”) and analyzed using a BD FACSCalibur Flow Cytometer (BD Biosciences). At each time point, the medium was replaced with the fresh induction medium. GFP fluorescence values were obtained from a minimum of 10,000 cells in each sample. The geometric mean of the GFP fluorescence per cell was calculated using FlowJo Software.
Growth Assays
Growth assays were done similarly to induction experiments, except that experimental cultures were inoculated to an OD600 of ∼0.05 and grown at 30°C and 230 rpm in a rotary shaker. OD600 was measured after 6, 8, 10, 12, 24, and 36 h. At each time point, the medium was removed and the fresh induction medium was added to each well.
Positive Control for β-Carotene Production
The β-carotene pathway genes McrtI-, McrtYB-, and BTS1-CDSs under the control of inducible ATFs NLS-GAL4AD-GRF7/4X, NLS-GAL4AD-ANAC102/2X, and NLS-GAL4AD-GRF9/4X were previously assembled in Destination vector I to generate plasmid pCAROTENE (see COMPASS protocol; Naseri et al., 2019). Destination vector I was digested with EcoRI/SalI and used in a two-way assembly reaction using PCR-amplified ProGAPDH sequence (PROGAPMCRTI_for/PROGAPMCRTI_rev, on genomic DNA of the GS115 P. pastoris) and McrtI CDS fussed to TerFBA1 (MCRTIPOS_for/MCRTIPOS_rev, on Entry vector X-McrtI; Naseri et al., 2019) to generate pMCRTI_PGAP. Next, NotI-digested pCOMPASS32 was used in an assembly reaction using PCR-amplified ProGAPDH sequence (PROGAPMCRTI_for/PROGAPMCRTI_rev, on genomic DNA of the GS115 P. pastoris) and BTS1 CDS fussed to TerTDH3 (BTSPOS_for/BTSPOS_rev, on Entry vector X-BTS1; Naseri et al., 2019). NotI-digested pCOMPASS33 was used in an assembly reaction using PCR-amplified ProGAPDH sequence (PROGAPMCRTI_for/PROGAPMCRTI_rev, on genomic DNA of the GS115 P. pastoris) and McrtYB CDS fussed to TerSYN3 (MCRTYBPOS_for/MCRTYBPOS_rev, on Entry vector X-McrtYB; Naseri et al., 2019) to generate pMCRTYB_PGAP. I-CeuI- digested pMCRTI_PGAP was used in an assembly reaction using PCR-amplified ProGAPDH-BTS1-TerTDH3 (PGAP_BTS1_for/PGAP_BTS1_rev, on pBTS1-PGAP). The resulting plasmid was called pMCRTI-BTS1_PGAP. Then, FseI/I-SceI-digested pMCRTI-BTS1_PGAP was used in an assembly reaction using PCR-amplified ProGAPDH–McrtYB-TerSYN3 (PGAP_MCRTYB_for/PGAP_MCRTYB_rev, on pMCRTYB-PGAP). The resulting plasmid was called pCAROTENE-PGAP.
β-Carotene Production
The yeast strains transformed with pCAROTENE plasmid (Naseri et al., 2019) were plated on SC-URA [supplemented with 2% (w/v) glucose]. Cells were grown at 30°C for 3–4 days. The cells were then plated on induction SC-URA medium plates containing 20 mM IPTG and 2% (w/v) galactose and 1 M sorbitol. Cells were grown at 30°C for 3–4 days. Subsequently, colony PCR was performed, followed by sequencing. Three independent colonies were chosen for β-carotene analysis by SFC. Yeast colonies were inoculated into 4 mL non-induction medium SC-URA [supplemented with 2% (w/v) glucose] and grown for 18–24 h at 30°C and 230 rpm in a rotary shaker. Subsequently, the pre-cultures were used to inoculate main cultures (50 mL) in induction medium [YPDA with the 20 mM IPTG, 2% (w/v) galactose, and 1 M sorbitol]. All shake-flask cultures were inoculated from pre-cultures to an initial OD600 of 0.1. Cells were then grown in a 500 mL shake flask for 3 days at 30°C and 230 rpm to saturation.
Supercritical Fluid Chromatography (SFC)
Carotenoid extraction from saturated culture was carried out according to the acetone extraction method with some modifications (Lian et al., 2017). The stationary phase yeast cells were collected by centrifugation at 13,000 × g for 1 min and cell precipitates were resuspended in 1 mL of 3N HCl, boiled for 5 min, and then cooled in an ice-bath for 5 min. The lysed cells were washed with ddH2O and resuspended in 400 μL acetone to extract β-carotene. The cell debris was removed by centrifugation. The acetone supernatant was collected in 2-mL microcentrifuge tubes. This extraction procedure was repeated until the cell pellet was white.
SFC analyses were carried out in triplicates. Carotenoids were separated by SFC WatersTM in 5 min using packed Silica column 4.6 × 150 mm (5 μm) with carbon dioxide modified with 2% (v/v) Tetrahydrofuran (THF) as a mobile phase. The column temperature was set to 40°C and the backpressure was 100 bar, at a flow rate of 1 mL/min. The results were detected using a photodiode array (PDA) detector at 510 nm and analyzed with ChromScope Software.
Results
Plant-Derived ATF/BS Units
A central aim of our work was to establish inducible, heterologous regulators for future use in synthetic biology dealing with P. pastoris. To this end, 17 ATF/BS combinations from two Arabidopsis TF families, namely: Growth-Regulating Factor 7 (GRF7) (Kim et al., 2012), GRF9 (Kim et al., 2003), the NAC TFs JUNGBRUNNEN1 (JUB1) (Wu et al., 2012), and ANAC102 (Christianson et al., 2009), were selected from our library of plant-derived ATF/BSs developed for S. cerevisiae (Naseri et al., 2017). The selected ATF/BS combinations resulted in weak (NLS-EDLLAD-GRF9/1x, NLS-JUB1DBD-EDLLAD/1x, NLS-J UB1DBD-EDLLAD/2x, NLS-GAL4AD-GRF7/1x, NLS-EDLLAD-GRF9/2x, NLS-GAL4AD-GRF9/1x, NLS-GAL4AD-GRF9/2x; 0–400 AU), medium (NLS-GAL4AD-GRF9/4x, NLS-GAL4AD-ANAC102/1x, NLS-EDLLAD-GRF9/4x, NLS-GAL4AD-GR F7/2x, NLS-JUB1DBD-EDLLAD/1x, NLS-GAL4AD-GRF7/4x; 4 00–1,200 AU), and strong (NLS-GAL4AD-ANAC102/2x, N LS-GAL4AD-ANAC102/4x, NLS-EDLLAD-ANAC102/2x, NLS-EDLL-GAL4AD-ANAC102/4x, NLS-EDLLAD-ANAC102/2x, NLS-EDLLAD-ANAC102/4x; 1,200–8,000 AU) transcriptional outputs and low basal expression, upon integration into the ura3-52 locus of the genome for transcriptional level characterization (Naseri et al., 2017). The selected ATF/BSs spanned a transcriptional activity (determined by GFP expression levels) ranging from ∼0.1- to ∼9-fold as compared with the strong TDH3 promoter in S. cerevisiae (Supplementary Figure 1; Naseri et al., 2017). To ensure a fair comparison of the ATF/BSs in P. pastoris, each of the regulator cassettes was inserted into the GUT1 locus, which leads to high specific integration rates by native homologous recombination and low random genomic integration frequency (Schwarzhans et al., 2016; Vogl et al., 2018). Following selection of a single GFP-transgene, the transcriptional output of plant-derived ATF/BSs was assessed using flow cytometry. The characterized ATF/BSs were subsequently used for metabolic engineering applications by producing β-carotene in P. pastoris (Figure 1).
A Wide Range of Transcriptional Outputs for Plant-Derived ATF/BSs in P. pastoris
Plant-derived ATFs, consisting of different combinations of full-length plant TFs or their DBDs, heterologous ADs, and different numbers of the respective cis-regulatory motifs located upstream of the yeast CYC1 minimal promoter, were integrated into the GUT1 locus in the P. pastoris genome. GUT1 is an essential enzyme for glycerol metabolism. Strains with deleted gut1 (due to integration of ATF/BS cassettes within GUT1 locus) cannot grow on glycerol (Vogl et al., 2018), while growth on glucose or galactose is not reduced. Following the addition of inducer (20 mM IPTG and 2% galactose), ATFs are expressed and target their BSs within the synthetic promoter (upstream of the yeast CYC1 minimal promoter), resulting in GFP expression (Figure 2A). As shown in Figure 2B, we achieved a wide spectrum of transcriptional outputs for plant-derived ATF/BSs. Of note, five of the ATFs (30%) resulted in up to a ∼3.13-fold higher GFP reporter output than the yPPGN021 control strain expressing the native GAPDH promoter (Figure 2B, strains yPPGN016, yPPGN008, yPPGN011, and yPPGN010). We observed the strongest transcriptional activation capacity for NAC TF-derived ATF NLS- GAL4AD-ANAC102 in combination with 2 or 4 copies of its BS (Figure 2B, yPPGN005 and yPPGN004). Surprisingly, the EDLLAD-ANAC102 combination resulted in a low transcriptional output in P. pastoris despite being categorized as a strong regulator in S. cerevisiae (Naseri et al., 2017). A combination of EDLLAD and plant TFs typically results in a high expression level of the reporter gene in S. cerevisiae (Naseri et al., 2017); however, EDLLAD-derived ATFs (yPPGN04, yPPGN04, yPPGN18-20) resulted in low-to-medium expression of the reporter gene in all our tested drivers/reporters in P. pastoris.
Characterization of Plant-Derived ATF/BSs in P. pastoris
We previously showed that by employing only nine inducible plant-derived ATF/BSs, a library of S. cerevisiae variants with a million members was established, allowing the fine-tuning of naringenin and β-carotene over a wide range (Naseri et al., 2019). Hence, here we aimed to further prove the transcriptional activation capacity of our plant-derived ATF/BSs for metabolic engineering purposes in P. pastoris, rather than extending the size of our library. For full exploitation of the plant-derived ATF/BSs, we further characterized the transcriptional output of a weak GRF9-, medium JUB1DBD-, and strong ANAC102-derived ATF/BS (yPPGN06, yPPGN13, and yPPGN10, respectively) in comparison with the constitutive GAPDH promoter (Waterham et al., 1997) in induction medium over time (Figure 3). Cells were grown in 20 mL induction medium in 100-mL flasks. The output of GFP was measured by flow cytometry over a 48-h time-course.
JUB1DBD- and ANAC102-derived ATF/BS allow tuning GFP expression from low to high responses, compared the GAPDH promoter of P. pastoris. The GFP fluorescent output of the JUB1DBD-derived ATF/BS (Figure 3, green line) reached to the level of GFP was expressed from the GAPDH promoter (Figure 3, gray line) 12 h post-induction and was stable for at least 24 h. In contrast, the strong ANAC102-derived ATF/BS (Figure 3, black line) mediated a much higher expression output 12 h post-induction as compared with the GAPDH promoter. However, the reporter expression decreased dramatically and reached the level derived from the GAPDH promoter after 48 h. Growing P. pastoris to a high density is of great benefit when producing a heterologous protein (Karbalaei et al., 2020); however, a high level of oxygen is required to achieve a high yield, which demands a culture vessel to deliver a sufficient amount of oxygen to the yeast (Eck et al., 2018). Due to the inadequate essential nutrients (amino acids, carbohydrates, minerals, vitamins), hormones, growth factors, and change of the physicochemical environment (pH, osmotic pressure), as well as limited oxygen availability (Krause et al., 2016), the reporter expression derived from plant-derived ATFs could be negatively affected in our study. This effect might be more severe in the case of strong ANAC102-derived ATF In contrast, we observed that the weak GRF9-derived ATF/BS (Figure 3, light green line) resulted in a stable expression level of the reporter over time.
Growth Effects of Plant-Derived ATF and Promoter Pairs in P. pastoris
We studied the effect of a weak GRF9-, medium JUB1DBD-, and strong ANAC102-derived driver/reporter (yPPGN06, yPPGN13, and yPPGN10, respectively) on cell growth. Following integration of the ATF/BS-GFP modules into the GUT1 locus of the genome, cell growth in induction (Figure 4A, solid orange line) and non-induction (Figure 4A, solid gray line) medium was assessed over 36 h. Cells expressing chromosomally integrated GFP under the control of the constitutive GAPDH promoter were used as a positive control. The growth curves of wild-type JC308 cells (not containing ATF) in induction (dashed orange line) and non-induction (dashed gray line) medium are included in all growth plots. The growth of wild-type JC308 was reduced in the induction medium as compared with that in the non-induction medium, demonstrating a negative effect of galactose on cell growth. The strain expressing the strong driver/reporter GAL4AD-ANAC102-2X-GFP (in induction medium) showed similar growth defects to the positive control strain (constitutively expressing the GFP reporter under the control of the GAPDH promoter). In contrast, the strains expressing the medium JUB1DBD-EDLLAD-4X-GFP and weak GAL4AD-GRF9-1X-GFP drivers/reporters displayed only minor growth reduction as compared with the wild-type control strain. Moreover, our data reveal that when the ATFs are present within the genome but are not expressed (non-induction medium), yeast growth was only slightly affected. To assess whether the growth penalty is related to the high level of GFP production, strains expressing only the driver modules GAL4AD-ANAC102. JUB1DBD-EDLLAD, or GAL4AD-GRF9 (yPPGN01, yPPGN02, and yPPGN03 strains, respectively) (no reporter module) were generated (Figure 4B). JUB1DBD-EDLLAD strain showed similar growth defects to the JUB1DBD-EDLLAD-4X-GFP expressing both the driver and reporter, suggesting that cell growth was slightly affected by the expression of fluorescent reporter protein derived from the binding sites of medium JUB1-derived ATF/BS. We observed that the expression of the strong ANAC102-derived ATF driver slightly affects the cell growth (Figure 4B, orange line), while the presence of the ANAC102 driver/reporter module resulted in a growth defect similar to constitutive ProGAPDH (Figure 4A, orange line). Moreover, the cells expressing ATFs showed a minor growth defect in the non-induction medium, compared to the cells expressing ProGAPDH (Figure 4B, gray line), emphasizing that the use of inducible promoters for the expression of ATFs may minimize the consumption of energy and nutrient resources and maximize biomass production capacity prior to the production phase.
Plant-Derived ATF/BS Capacity for β-Carotene Production in P. pastoris and S. cerevisiae
We sought to confirm the transcriptional ability of plant-derived ATF/BSs for metabolic engineering applications in P. pastoris by testing a well-known phenotype. Although P. pastoris is a non-carotenogenic yeast, it has been used for the production of carotenoids (Araya-Garay et al., 2012). To convert FPP precursor to β-carotene, geranylgeranyl diphosphate synthase (BTS1), phytoene synthase/lycopene cyclase (McrtYB), and phytoene desaturaseare (McrtI) enzymes are needed (Figure 5A). Here, we transformed yeast P. pastoris cells (wild-type JC308 background) with the episomal pCAROTENE plasmid (Naseri et al., 2019; Figure 5B) encoding the McrtI-, McrtYB-, and BTS1-CDS of the β-carotene pathway under the control of the inducible ATF/BSs, NLS-GAL4AD-GRF9-4X, NLS-GAL4AD-GRF7-4X, and NLS-GAL4AD-ANAC102-4X (Naseri et al., 2017, ∼1.27—∼3.13-fold stronger than ProGAPDH) to generate the yPPGN022 strain (Table 2). Selection for the assembly was carried out on a medium lacking the yeast auxotrophic marker, URA3 (the selection marker gene is encoded on the plasmid carrying β-carotene pathway genes). Following induction of ATF expression, orange colonies were observed on the yeast plate (Figure 5C for yPPGN022). After transformation with the pCAROTENE plasmid, we observed stronger β-carotene accumulation in the yPPGN022 P. pastoris strain than that in the S. cerevisiae strain (Table 1 and Figure 5C for ySCGN01) [ySCGN01 has wild-type Gen 0.2 S. cerevisiae background (Naseri et al., 2019)] in induction medium (production phase). To further compare the β-carotene productivity of the two hosts, P. pastoris and S. cerevisiae, we constructed an optimized S. cerevisiae cell factory for isoprenoid production by redirecting carbon flux toward the production of the isoprenoid precursor, FPP (Figure 5D; Scalcinati et al., 2012; Lopez et al., 2015). Using CRISPR/Cas9-mediated one-step double-strand breaks (Bao et al., 2015): (i) three modules expressing IPTG-inducible super-strong plant-derived ATF/BSs were integrated into the genome to upregulate the expression of GDH2, tHMG1, and ERG20 (Figure 5E and Table 2 for ySCGN0.1); and (ii) DPP1, LPP1, and GDH1 were inactivated (Table 2 for ySCGN0.2). Transformation of the ySCGN1.2 strain, carrying both upregulation and inactivation groups (Table 2), with the pCAROTENE plasmid produced yeast colonies with a more intense color (Figure 5F and Table 2 for ySCGN02) as compared with the P. pastoris yPPGN022 strain. We selected three colonies from each strain for the quantitative determination of β-carotene content by supercritical fluid chromatography (SFC).
The SFC data (Figure 5G) demonstrate a ∼4.8-fold higher β-carotene level in the yPPGN022 P. pastoris strain than that in the ySCGN01 S. cerevisiae strain. As a further control, the episomal pCAROTENE-PGAP plasmid expressing the three genes from the GAPDH promoter was transformed into J308 P. pastoris strain to generate yPPGN023 strain. The SFC data (Figure 5G) demonstrate that the strain with ATF/BS control modules produced ∼3.4-fold more β-carotene than the strain with GAPDH promoters. We observed a relatively huge variation of β-carotene production levels in different replicates in the case of yPPGN23 strain. A common issue often encountered with the episomal expression systems is an unstable product output because of the limited control of copy number, and segregational instability even in selective medium (Futcher and Carbon, 1986). In the case of pCAROTENE-PGAP, the metabolic burden raised from the constitutive expression of the pathway enzymes may cause this effect to be more severe. In contrast, presence of the inducible promoter upstream of plant-derived ATFs allows β-carotene production after adding IPTG and galactose (in the production phase). Therefore, a considerable production of β-carotene was achieved in the case of pCAROTENE, compared to pCAROTENE-PGAP (Figure 5G). Although the production of β-carotene in the ySCGN02 S. cerevisiae strain was markedly improved as compared with that in the ySCGN01 wild-type strain (by ∼8.2-fold), it was only ∼1.7-fold higher than the level of β-carotene accumulation in the yPPGN022 strain, confirming the high transcriptional capability of plant-derived ATF/BSs for metabolic engineering applications in P. pastoris.
Discussion
Historically, S. cerevisiae is the most often used synthetic biology workhorse for heterologous protein production. Recently, other yeasts such as K. phaffii, Kluyveromyces lactis, and Yarrowia lipolytica have emerged as advantageous cell factories. Among these, K. phaffii, commonly known as P. pastoris, has gained attention for marketing novel recombinant therapeutics (Chen et al., 2020; Karbalaei et al., 2020; Spice et al., 2020; Zhang et al., 2020). To uncover and navigate the huge potential of P. pastoris in bioengineering, we need to establish straightforward strategies to assemble multi-component DNA constructs harboring genes and other essential regulatory elements. This situation decidedly relies on the availability of regulator elements, such as TFs and their cis-regulatory elements, which can be combined to establish the synthetic regulatory systems. For example, GoldenPiCS is a modular Golden Gate-derived P. pastoris cloning system that allows the creation of complex genetic constructs in a relatively short period of time (Prielhofer et al., 2017). However, genetic tools allowing the orthogonal regulation of cellular activities at diverse levels (DNA replication, transcription, translation, and others) are still not sufficiently established in P. pastoris (Kang et al., 2017).
In the past, few native promoters of P. pastoris have been used for the controlled expression of heterologous coding sequences in yeast (Waterham et al., 1997; Qin et al., 2011). Although these regulatory sequences allow gene expression over a wide range of activities, control over them is accomplished by endogenous yeast gene regulatory networks ruled by native TFs, which may not be attractive since minimal interference with the host endogenous regulatory network is highly preferred in synthetic biology. Additionally, the most efficient designs ideally incorporate a biomass production phase followed by a target production phase. By establishing a collection of inducible regulators, the expression of regulators, and therefore any potential negative interactions with the host genome, metabolites, and proteins that impose fitness costs, can be postponed until a desired time. We previously established a library of ATF/BSs using several TF families from the higher plant, Arabidopsis thaliana (that are absent from non-plant pro- and eukaryotes) to control gene expression in S. cerevisiae (Naseri et al., 2017). We hypothesized that such TFs may be principally well-suited for establishing orthologous gene regulatory elements in P. pastoris.
Here, we tested 17 different combinations of plant-derived ATFs and their BSs following chromosomal integration and observed a wide spectrum of inducible transcriptional outputs (Figure 2). A low transcriptional output and minimum growth defects observed for plant-derived ATF/BSs in non-induction medium (Figures 2, 4) render them suitable regulators, allowing separation of biomass accumulation from the target molecule production phase. With the exception of EDLLAD-ANAC102-derived ATFs that resulted in a low transcriptional output in P. pastoris, we observed a high degree of regulator transferability from S. cerevisiae to P. pastoris. In other words, the strongest regulators led to the highest mean expression of reporter protein, regardless of the host of choice. To demonstrate a useful application of plant-derived ATF/BSs, we studied β-carotene productivity in P. pastoris, where plant-derived ATF/BSs were used to control gene expression. P. pastoris was able to produce ∼1.35 mg β-carotene/g (cdw), which is ∼4.8-fold greater than that produced by S. cerevisiae. By implementing the CRISPR/Cas9-mediated one-step multigene modification system (Bao et al., 2015) and inducible super-strong plant-derived ATF/BSs (up to 10-fold stronger than the S. cerevisiae constitutive TDH3 promoter) (Naseri et al., 2017), we increased the endogenous supply by S. cerevisiae through the redirection of its endogenous metabolic flux toward FPP, a key precursor of a non-native β-carotene product. We proved plant-derived ATFs are a promising tool for fine-tuning the gene expression, either via controlling the heterologous protein production or reviewing the metabolisms of the microbial chassis (Figure 5). This strain can be used for maximized production of other isoprenoid products including squalene (Paradise et al., 2008; Ding et al., 2014). Ito et al. (2020) showed tuneable gene expression in P. pastoris using endogenous terminators of S. cerevisiae (Ito et al., 2020). Here, we also report the successful transferability of terminators and promoters, such as the CYC1 minimal promoter, from S. cerevisiae to P. pastoris (Figure 5A), in addition to plant-derived ATF/BSs.
IPTG is expensive and induction is irreversible. Moreover, galactose (required for IPTG-inducible system) negatively affects yeast cell growth (Figure 5). Therefore, IPTG/galactose responsive GAL1 promoter is not the best choice for industrial-scale production. The plant-derived ATFs are compatible with other chemically inducible promoters and light-controlled molecular switches that may be more promising alternatives for industrial application (Losi et al., 2018). We tested the functionality of the plant-derived cis-regulatory elements only within the frame of the yeast CYC1 minimal promoter. Another aspect for further refinement is the evaluation of other minimal promoters (Schlabach et al., 2010) from either yeast or plants, which can be established and modified by altering the number of ATF binding sites. This will allow establishment of a vast spectrum of controllable transcriptional regulatory systems, strongly supporting future applications in synthetic biology.
P. pastoris is a promising cell factory for the high-level expression of heterologous proteins for industrial and pharmaceutical applications, owing to the growth to high cell densities in mineral medium in batch-fed cultivation (Krause et al., 2010), the possibility of targeting proteins for secretion, and the low amount of secreted endogenous proteins (Barrero et al., 2018). However, engineering microorganisms for industrial-scale production remains a challenge, even if we succeed in establishing cutting-edge synthetic biology tools and assembly methods. In other words, the use of multiple circuits to build functional complex systems is a demanding task, and a large number of diverse synthetic constructs need to be created. This situation strongly demands parallel and smart methods, referred to as combinatorial optimization methods, allowing the simultaneous assembly of large numbers (up to thousands or even millions) of variants (Naseri and Koffas, 2020). The plant-derived ATF/BSs could be employed in the frame of combinatorial optimization methods, such as our COMPASS (Naseri et al., 2019; Naseri and Mueller-Roeber, 2020), for pathway optimization in P. pastoris. Our ATF/cis-regulatory library therefore represents an excellent starting point for further improvements to orthogonal regulatory systems in yeast.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
GN conceived the project, designed, performed the experiments, analyzed the data, and wrote the manuscript. CA designed SFC experiment. KP, HH, and CA performed and analyzed SFC experiment. All authors read and approved the manuscript.
Funding
We acknowledge support by the German Research Foundation (DFG) and the Open Access Publication Fund of Humboldt-Universität zu Berlin.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful Yansheng Zhang (Chinese Academy of Sciences, Beijing, China) for plasmid encoding McrtI and McrtYB; Michael K. Jensen (Technical University of Denmark, Kongens Lyngby, Denmark) for pTAJAK-105; Geoffrey Lin-Cereghino (University of the Pacific, United States) for yeast strain JC308 strain, Sascha Jung for yeast strain with GS115 background (Technische University of Berlin, Berlin, Germany). We thank to Marc Werten (Wageningen University and Research Centre, Wageningen, Netherlands) for his constructive feedback.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.676900/full#supplementary-material
References
Araya-Garay, J. M., Vallejo, J., Veiga-Crespo, P., Sánchez-Pérez, A., and Villa, T. G. (2012). Construction of a novel Pichia pastoris strain for production of xanthophylls. AMB Express 2:24. doi: 10.1186/2191-0855-2-24
Balamurugan, V., Reddy, G. R., and Suryanarayana, V. V. S. (2007). Pichia pastoris: a notable heterologous expression system for the production of foreign proteins—Vaccines. Indian J. Biotechnol. 6, 175–186.
Bao, Z., Xiao, H., Liang, J., Zhang, L., Xiong, X., Sun, N., et al. (2015). Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 4, 585–594. doi: 10.1021/sb500255k
Barrero, J. J., Casler, J. C., Valero, F., Ferrer, P., and Glick, B. S. (2018). An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb. Cell Fact. 17:161.
Bruckner, K., Schafer, P., Weber, E., Grutzner, R., Marillonnet, S., and Tissier, A. (2015). A library of synthetic transcription activator-like effector-activated promoters for coordinated orthogonal gene expression in plants. Plant J. 82, 707–716. doi: 10.1111/tpj.12843
Chen, M., Wang, J., Lin, L., Wei, W., Shen, Y., and Wei, D. (2020). High-level expression of a beta-mannanase (manB) in Pichia pastoris GS115 for mannose production with Penicillium brevicompactum fermentation pretreatment of soybean meal. Bioprocess Biosyst. Eng. 44, 549–561. doi: 10.1007/s00449-020-02467-6
Christianson, J. A., Wilson, I. W., Llewellyn, D. J., and Dennis, E. S. (2009). The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149, 1724–1738. doi: 10.1104/pp.108.131912
Ding, M. Z., Yan, H. F., Li, L. F., Zhai, F., Shang, L. Q., Yin, Z., et al. (2014). Biosynthesis of Taxadiene in Saccharomyces cerevisiae: selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy. PLoS One 9:e109348. doi: 10.1371/journal.pone.0109348
Eck, A., Schmidt, M., Hamer, S., Ruff, A. J., Forster, J., Schwaneberg, U., et al. (2018). Improved microscale cultivation of Pichia pastoris for clonal screening. Fungal Biol. Biotechnol. 5:8.
Fischer, J. E., and Glieder, A. (2019). Current advances in engineering tools for Pichia pastoris. Curr. Opin. Biotechnol. 59, 175–181. doi: 10.1016/j.copbio.2019.06.002
Futcher, B., and Carbon, J. (1986). Toxic effects of excess cloned centromeres. Mol. Cell. Biol. 6, 2213–2222. doi: 10.1128/mcb.6.6.2213-2222.1986
Gaj, T., Gersbach, C. A., and Barbas, C. F. III (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Gao, J., Jiang, L., and Lian, J. (2021). Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products. Synth. Syst. Biotechnol. 6, 110–119. doi: 10.1016/j.synbio.2021.04.005
Gao, X., Tsang, J. C., Gaba, F., Wu, D., Lu, L., and Liu, P. (2014). Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 42:e155. doi: 10.1093/nar/gku836
Gietz, R. D., and Schiestl, R. H. (2007). Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 1–4. doi: 10.1038/nprot.2007.17
Hartmann, D., Smith, J. M., Mazzotti, G., Chowdhry, R., and Booth, M. J. (2020). Controlling gene expression with light: a multidisciplinary endeavour. Biochem. Soc. Trans. 48, 1645–1659. doi: 10.1042/bst20200014
Hartner, F. S., Ruth, C., Langenegger, D., Johnson, S. N., Hyka, P., Lin-Cereghino, G. P., et al. (2008). Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 36:e76. doi: 10.1093/nar/gkn369
Hochrein, L., Machens, F., Gremmels, J., Schulz, K., Messerschmidt, K., and Mueller-Roeber, B. (2017). AssemblX: a user-friendly toolkit for rapid and reliable multi-gene assemblies. Nucleic Acids Res. 45:e80.
Ito, Y., Terai, G., Ishigami, M., Hashiba, N., Nakamura, Y., Bamba, T., et al. (2020). Exchange of endogenous and heterogeneous yeast terminators in Pichia pastoris to tune mRNA stability and gene expression. Nucleic Acids Res. 48, 13000–13012. doi: 10.1093/nar/gkaa1066
Ito, Y., Watanabe, T., Aikawa, S., Nishi, T., Nishiyama, T., Nakamura, Y., et al. (2018). Deletion of DNA ligase IV homolog confers higher gene targeting efficiency on homologous recombination in Komagataella phaffii. FEMS Yeast Res. 18:foy074.
Ji, C. H., Kim, H., and Kang, H. S. (2019). Synthetic inducible regulatory systems optimized for the modulation of secondary metabolite production in Streptomyces. ACS Synth. Biol. 8, 577–586. doi: 10.1021/acssynbio.9b00001
Kang, Z., Huang, H., Zhang, Y., Du, G., and Chen, J. (2017). Recent advances of molecular toolbox construction expand Pichia pastoris in synthetic biology applications. World J. Microbiol. Biotechnol. 33:19.
Karbalaei, M., Rezaee, S. A., and Farsiani, H. (2020). Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 235, 5867–5881. doi: 10.1002/jcp.29583
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313x.2003.01862.x
Kim, J. S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kjeldsen, T., Pettersson, A. F., and Hach, M. (1999). Secretory expression and characterization of insulin in Pichia pastoris. Biotechnol. Appl. Biochem. 29, 79–86.
Krause, M., Neubauer, A., and Neubauer, P. (2016). The fed-batch principle for the molecular biology lab: controlled nutrient diets in ready-made media improve production of recombinant proteins in Escherichia coli. Microb. Cell Fact. 15:110.
Krause, M., Ukkonen, K., Haataja, T., Ruottinen, M., Glumoff, T., Neubauer, A., et al. (2010). A novel fed-batch based cultivation method provides high cell-density and improves yield of soluble recombinant proteins in shaken cultures. Microb. Cell Fact. 9:11. doi: 10.1186/1475-2859-9-11
Li, X., Zhang, C., Xu, X., Miao, J., Yao, J., Liu, R., et al. (2020). A single-component light sensor system allows highly tunable and direct activation of gene expression in bacterial cells. Nucleic Acids Res. 48:e33. doi: 10.1093/nar/gkaa044
Lian, J., Hamedirad, M., Hu, S., and Zhao, H. (2017). Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 8:1688.
Lin Cereghinoa, G. P., Jay Sungaa, A., Johnsona, M. A., Lima, M., Gleesonb, M. A. G., and Cregg, J. M. (2001). New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene 263, 159–169. doi: 10.1016/s0378-1119(00)00576-x
Lopez, J., Essus, K., Kim, I. K., Pereira, R., Herzog, J., Siewers, V., et al. (2015). Production of β-ionone by combined expression of carotenogenic and plant CCD1 genes in Saccharomyces cerevisiae. Microb. Cell Fact. 14:84. doi: 10.1186/s12934-015-0273-x
Losi, A., Gardner, K. H., and Moglich, A. (2018). Blue-light receptors for optogenetics. Chem. Rev. 118, 10659–10709. doi: 10.1021/acs.chemrev.8b00163
Machens, F., Balazadeh, S., Mueller-Roeber, B., and Messerschmidt, K. (2017). Synthetic promoters and transcription factors for heterologous protein expression in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 5:63. doi: 10.3389/fbioe.2017.00063
Mallem, M., Warburton, S., Li, F., Shandil, I., Nylen, A., Kim, S., et al. (2014). Maximizing recombinant human serum albumin production in a Mut(s) Pichia pastoris strain. Biotechnol. Prog. 30, 1488–1496.
Massahi, A., and Çalık, P. (2018). Naturally occurring novel promoters around pyruvate branch-point for recombinant protein production in Pichia pastoris (Komagataella phaffii): Pyruvate decarboxylase- and pyruvate kinase- promoters. Biochem. Eng. J. 138, 111–120. doi: 10.1016/j.bej.2018.07.012
Naseri, G., and Koffas, M. A. G. (2020). Application of combinatorial optimization strategies in synthetic biology. Nat. Commun. 11:2446.
Naseri, G., and Mueller-Roeber, B. (2020). A Step-by-step protocol for COMPASS, a synthetic biology tool for combinatorial gene assembly. Methods Mol. Biol. 2205, 277–303. doi: 10.1007/978-1-0716-0908-8_16
Naseri, G., Balazadeh, S., Machens, F., Kamranfar, I., Messerschmidt, K., and Mueller-Roeber, B. (2017). Plant-derived transcription factors for orthologous regulation of gene expression in the yeast Saccharomyces cerevisiae. ACS Synth. Biol. 6, 1742–1756. doi: 10.1021/acssynbio.7b00094
Naseri, G., Behrend, J., Rieper, L., and Mueller-Roeber, B. (2019). COMPASS for rapid combinatorial optimization of biochemical pathways based on artificial transcription factors. Nat. Commun. 10:2615.
Paradise, E. M., Kirby, J., Chan, R., and Keasling, J. D. (2008). Redirection of flux through the FPP branch-point in Saccharomyces cerevisiae by down-regulating squalene synthase. Biotechnol. Bioeng. 100, 371–378. doi: 10.1002/bit.21766
Peng, B., Williams, T. C., Henry, M., Nielsen, L. K., and Vickers, C. E. (2015). Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb. Cell Fact. 14:91.
Prattipati, M., Ramakrishnan, K., and Sankaranarayanan, M. (2020). Pichia pastoris protein disulfide isomerase (PDI1) promoter for heterologous protein production and its sequence characterization. Enzyme Microb. Technol. 140:109633. doi: 10.1016/j.enzmictec.2020.109633
Prielhofer, R., Barrero, J. J., Steuer, S., Gassler, T., Zahrl, R., Baumann, K., et al. (2017). GoldenPiCS: a Golden Gate-derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris. BMC Syst. Biol. 11:123. doi: 10.1186/s12918-017-0492-3
Qin, X., Qian, J., Yao, G., Zhuang, Y., Zhang, S., and Chu, J. (2011). GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl. Environ. Microbiol. 77, 3600–3608. doi: 10.1128/aem.02843-10
Sanjana, N. E., Cong, L., Zhou, Y., Cunniff, M. M., Feng, G., and Zhang, F. (2012). A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171–192. doi: 10.1038/nprot.2011.431
Scalcinati, G., Partow, S., Siewers, V., Schalk, M., Daviet, L., and Nielsen, J. (2012). Combined metabolic engineering of precursor and co-factor supply to increase alpha-santalene production by Saccharomyces cerevisiae. Microb. Cell Fact. 11:117. doi: 10.1186/1475-2859-11-117
Schlabach, M. R., Hu, J. K., Li, M., and Elledge, S. J. (2010). Synthetic design of strong promoters. Proc. Natl. Acad. Sci. U.S.A. 107, 2538–2543. doi: 10.1073/pnas.0914803107
Schwarzhans, J. P., Wibberg, D., Winkler, A., Luttermann, T., Kalinowski, J., and Friehs, K. (2016). Integration event induced changes in recombinant protein productivity in Pichia pastoris discovered by whole genome sequencing and derived vector optimization. Microb. Cell Fact. 15:84.
Spice, A. J., Aw, R., Bracewell, D. G., and Polizzi, K. M. (2020). Synthesis and assembly of hepatitis B Virus-like particles in a Pichia pastoris cell-free system. Front. Bioeng. Biotechnol. 8:72. doi: 10.1007/s00253-009-2382-0
Su, G. D., Huang, D. F., Han, S. Y., Zheng, S. P., and Lin, Y. (2010). Display of candida antarctica lipase B on Pichia pastoris and its application to flavor ester synthesis. Appl. Microbiol. Biotechnol. 86, 1493–1501.
Tang, H., Wu, Y., Deng, J., Chen, N., Zheng, Z., Wei, Y., et al. (2020). Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 10:320. doi: 10.3390/metabo10080320
Tang, S., Boehme, L., Lam, H., and Zhang, Z. (2009). Pichia pastoris fermentation for phytase production using crude glycerol from biodiesel production as the sole carbon source. Biochem. Eng. J. 43, 157–162. doi: 10.1016/j.bej.2008.09.020
Trikka, F. A., Nikolaidis, A., Athanasakoglou, A., Andreadelli, A., Ignea, C., Kotta, K., et al. (2015). Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microb. Cell Fact. 14:60.
Vassileva, A., Chugh, D. A., Swaminathan, S., and Khanna, N. (2001). Expression of hepatitis B surface antigen in the methylotrophic yeast Pichia pastoris using the GAP promoter. J. Biotechnol. 88, 21–35. doi: 10.1016/s0168-1656(01)00254-1
Verwaal, R., Wang, J., Meijnen, J. P., Visser, H., Sandmann, G., Van Den Berg, J. A., et al. (2007). High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 73, 4342–4350. doi: 10.1128/aem.02759-06
Vogl, T., Gebbie, L., Palfreyman, R. W., and Speight, R. (2018). Effect of plasmid design and type of integration event on recombinant protein expression in Pichia pastoris. Appl. Environ. Microbiol. 84:e02712-17.
Waterham, H. R., Digan, M. E., Koutz, P. J., Lair, S. V., and Cregg, J. M. (1997). Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186, 37–44. doi: 10.1016/s0378-1119(96)00675-0
Wu, A., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506. doi: 10.1105/tpc.111.090894
Wu, G., Yan, Q., Jones, J. A., Tang, Y. J., Fong, S. S., and Koffas, M. A. G. (2016). Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 34, 652–664. doi: 10.1016/j.tibtech.2016.02.010
Xu, N., Zhu, J., Zhu, Q., Xing, Y., Cai, M., Jiang, T., et al. (2018). Identification and characterization of novel promoters for recombinant protein production in yeast Pichia pastoris. Yeast 35, 379–385. doi: 10.1002/yea.3301
Yang, S., Kuang, Y., Li, H., Liu, Y., Hui, X., Li, P., et al. (2013). Enhanced production of recombinant secretory proteins in Pichia pastoris by optimizing Kex2 P1’ site. PLoS One 8:e75347. doi: 10.1371/journal.pone.0075347
Zhang, X., Wang, D., Duan, Y., Zheng, X., Lin, Y., and Liang, S. (2020). Production of lycopene by metabolically engineered Pichia pastoris. Biosci. Biotechnol. Biochem. 84, 463–470. doi: 10.1080/09168451.2019.1693250
Keywords: artificial transcription factor, metabolic engineering, Pichia pastoris, Saccharomyces cerevisiae, synthetic biology
Citation: Naseri G, Prause K, Hamdo HH and Arenz C (2021) Artificial Transcription Factors for Tuneable Gene Expression in Pichia pastoris. Front. Bioeng. Biotechnol. 9:676900. doi: 10.3389/fbioe.2021.676900
Received: 06 March 2021; Accepted: 30 June 2021;
Published: 09 August 2021.
Edited by:
Dong-Yup Lee, Sungkyunkwan University, South KoreaReviewed by:
Lars M. Blank, RWTH Aachen University, GermanySang Woo Seo, Seoul National University, South Korea
Copyright © 2021 Naseri, Prause, Hamdo and Arenz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gita Naseri, Z2l0YS5uYXNlcmlAaHUtYmVybGluLmRl
†ORCID: Gita Naseri, orcid.org/0000-0002-4630-3141; Christoph Arenz, orcid.org/0000-0001-7613-9437
 Gita Naseri
Gita Naseri Kevin Prause2
Kevin Prause2 Housam Haj Hamdo
Housam Haj Hamdo