- 1College of Tobacco Science and Engineering, Zhengzhou University of Light Industry, Zhengzhou, China
- 2Technology Research Developing Center, Shenzhen Tobacco Industrial Co., Ltd., Shenzhen, China
- 3Department of Biosciences, COMSATS University Islamabad, Islamabad, Pakistan
Over the past 20 years, researchers have used multi-omics techniques to study microbial diversity and metabolic function on tobacco leaves. The unique metabolic function of tobacco microorganisms has attracted extensive attention from researchers, which is an important research field in tobacco industry to improve the intrinsic quality of tobacco leaf with microbial agents. The microorganisms are particularly rich on the surface of tobacco leaf, and their metabolic function is closely related to the change of tobacco leaf chemical composition. Some microorganisms have important metabolic functions, such as: degrading macromolecular and harmful substances in tobacco leaves, and they have different degradation rates and pathways for the substances. At present, many functions of tobacco leaf microorganisms have not been fully verified and analyzed. In the future, more novel culture methods are needed to screen and isolate microorganisms on the surface of tobacco leaves, deeply tap their metabolic potential, explore the application value of microorganisms in the tobacco industry, and further promote the innovation and development of the industry.
1 Introduction
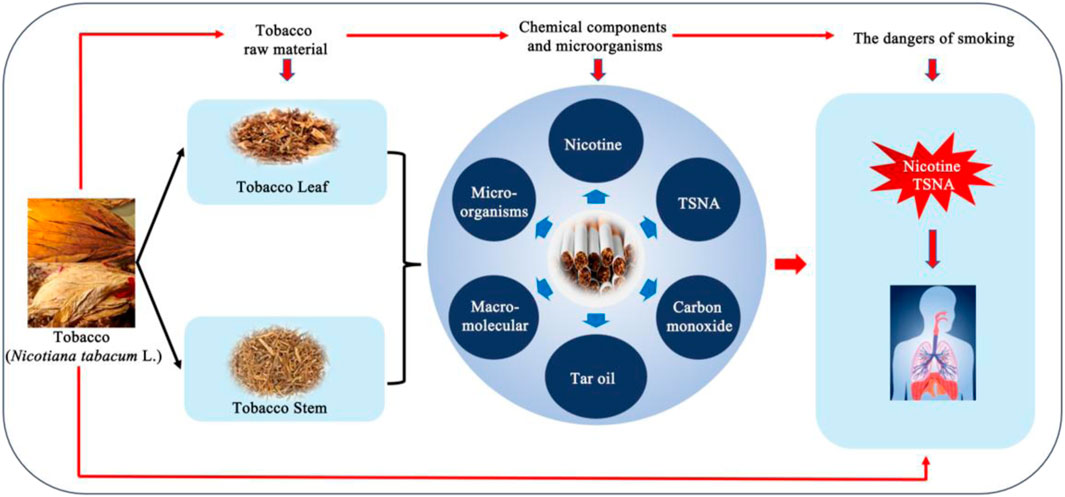
Tobacco (Nicotiana tabacum L.) is an important agricultural nonfood crop and serves as the primary raw material for tobacco products (Zhang et al., 2020). Globally, China is the largest producer of tobacco where it is extensively cultivated in regions of South China, including Hainan and Sichuan (Xing et al., 2021). Tobacco products contain more than 6,000 chemicals, such as proteins, cellulose, starch, nicotine, tobacco-specific nitrosamines (TSNA) and numerous other toxicants (Chopyk et al., 2017b). It is well established that nicotine and TSNA, the predominant nitrogenous compounds in tobacco plants, play a crucial role in driving smoking addiction (Wang et al., 2018a) and pose a substantial threat to smokers’health (Chopyk et al., 2017b) (Figure 1).
The high concentration of starch, protein and other macromolecules causes an irritating burnt odor in tobacco leaves during combustion and reduce sensory quality. These macromolecules organic substances significantly affect the release of harmful substances in the flue gas during incomplete combustion (Yuan et al., 2006). Therefore, reducing the content of macromolecules and harmful substances in cigarette products can reduce the addiction and health hazards of cigarettes to smoking consumers (Figure 1). Previous studies have reported that the specific functional microorganisms are highly effective in improving the quality (Huang et al., 2022a). These microorganisms can enhance tobacco quality by improving aroma, degrading harmful substances, such as nicotine (Liu et al., 2015; Fitzpatrick, 2018; Xia et al., 2018; Mu et al., 2020; Zhang et al., 2022), TSNA (Wang and Huang, 2006; Shan et al., 2011; Jiang et al., 2021b), macromolecules (Li et al., 2006), and optimizing other desirable characteristics for smoking products during the aging process of tobacco leaves (Wang et al., 2018b).
Thus, the specific objectives of this review are: 1) to explore the community composition of tobacco microorganisms. 2) to outline the functions of tobacco microorganisms and discuss their biodegradation 3) to recap the isolation and culture techniques for tobacco-degrading microorganisms and recommend future research avenues in tobacco microbiology. This review provides an updated overview of the detailed description of microbial community composition on tobacco and its relationship with tobacco’s chemical composition, offering a more comprehensive and detailed analysis than previous reviews.
2 The structure and determinants of the tobacco microorganism community
2.1 The community composition of tobacco microorganisms
The microbial species identified through molecular techniques are abundant on tobacco leaves, in cigarettes, and across various smokeless tobacco brands (Chopyk et al., 2017b). It has been found that the number of culturable bacteria in tobacco leaves collected from plantations ranges between 2 × 103 CFU/g and 7 × 105 CFU/g, while those from manufacturing plants range between 2 × 103 CFU/g and 8 × 103 CFU/g (Larsson et al., 2008). Most existing research concentrating on the microbial diversity during different aging periods of tobacco leaves had been reported in the previous studies (Huang et al., 2010; Chen et al., 2018; Wang et al., 2018a; Zhang et al., 2020; Hu et al., 2021; Liu F. et al., 2021). High throughput sequencing analysis has revealed that the dominant bacterial communities on the surfaces of aging flue-cured tobacco belong to 48 genera, 36 families, and 7 phyla (Wang et al., 2018b). The predominant genera on flue-cured tobacco leaves include Bacillus, Pseudomonas, Enterobacter, Sphingomonas, Pantoea and Methylobacterium (Huang et al., 2010). And, cigar tobacco also exhibits a higher relative abundance of Limnobacter, Brevundimonas, unidentified_Cyanobacteria and Pseudomonas, with most of these species classified within just two bacterial phyla: Proteobacteria and Cyanobacteria (Xing et al., 2021). The core bacterial operational taxonomic units (OTUs) identified in cigarette tobacco comprise Bacillus pumilus, Rhizobium sp., Sphingomonas sp., unidentified members of Enterobacteriaceae, Pantoea sp., Pseudomonas oryzihabitans and Pseudomonas putida (Chopyk et al., 2017b). The bacterial communities in smokeless tobacco are primarily dominated by the phyla Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes (Han et al., 2016; Tyx et al., 2016; Smyth et al., 2019). Additionally, some bacterial endophytes have been identified in the leaves of tobacco, such as Clostridium sp. (Saito et al., 2008). At present, studies on the community composition of endophytic bacteria in tobacco leaf are very limited. Future studies need to further explore the diversity of endophytic bacteria in tobacco leaf and explore its importance in the process of tobacco leaf quality improvement.
Fungi are also an important group in the microbial composition of tobacco leaves (Chen et al., 2020). Studies have reported that the number of culturable fungi in plantation-grown tobacco leaves ranges from 0.3 × 103 CFU/g to 3 × 103 CFU/g and can reach up to 8 × 103 CFU/g in the leaves collected from a manufacturing plants (Larsson et al., 2008). The diversity of fungi in the phyllosphere of tobacco leaves have been extensively studied using high throughput sequencing technologies (Lv et al., 2013). These studies revealed the existence of most abundant fungal genera including Alternaria, Phoma, Cercospora, Aspergillus and Rhizopus on tobacco leaves during curing (Chen et al., 2020). Whereas, genera such as Cladosporium, Epicoccum, Trichoderma, Nigrospora, Penicillium, Chaetomium and Fusarium are most frequently isolated and cultured from flue-cured and non-flue-cured tobacco leaves (Nagrale et al., 2016). Moreover, some fungal endophytes have also been found in the leaves, stems and roots of tobacco plants. At the phylum level, Ascomycota and Basidiomycota dominate fungal endophyte communities (Yuan et al., 2018; Jiang et al., 2021a). Genera such as Altermaria, Apiotrichum, Cladosporium, and Microdium are particularly abundant in both ordinary and “cherry-red” tobacco (Jiang et al., 2021a).
It is worth noting that tobacco-associated microorganisms also include potentially pathogenic species such as P. putida (Chopyk et al., 2017a; 2017b). And some fungi on tobacco leaves, such as Alternaria (Wang H. et al., 2016), Phoma Rhizopus, Epicoccum (Guo et al., 2020), are known plant pathogens. Up to now, the community composition of pathogenic microorganisms in tobacco microorganisms has not been reported, and the risk of ingestion and exposure to humans has not been analyzed and discussed in depth.
2.2 Factors influencing the community composition of tobacco microorganisms
The community composition of tobacco-associated microorganisms is dynamic, and influenced by various environmental factors, such as temperature, humidity and pH (Chopyk et al., 2017b; Chen et al., 2020). In bacterial community on the tobacco leaf of flue-curing procedure, the abundance of Pantoea and Variovorax is positively correlated with temperature and humidity, whereas the abundance of Nesterenkonia, Staphylococcus, Chryseomonas, Rhodococcus, Paracoccus, Serratia and Ralstonia shows a negative correlation with temperature and humidity (Hu et al., 2021). In the fungal community of the tobacco leaf phyllosphere during curing of leaves, the abundance of Golovinomyces is significantly affected by temperature, while the abundance of Alternaria, Phoma, Trichoderma, Leptosphaerulina, Gibellulopsis and Candida is notably impacted by relative humidity (Chen et al., 2020). One study also found that the fungal community diversity presented an obvious negative correlation with temperature and humidity during the flue-curing process (Hu et al., 2021). Furthermore, the community composition and diversity of tobacco-associated microorganisms are also influenced by fermentation duration (Liu F. et al., 2021). For example, the relative abundance of Klebsiella variicola, Klebsiella pneumoniae, Serratia, and Salmonella initially increases, peaking after 16 h before subsequently decreasing as the fermentation process continues to 24 and 36 h (Huang et al., 2024). Additionally, spatiotemporal variations affect the community composition of tobacco-associated microorganisms (Xing et al., 2021). For instance, the fungal community structure on the surface of tobacco leaves varies between different areas (Chen et al., 2018).
3 Functional roles of tobacco microorganisms
Tobacco microorganisms play crucial metabolic roles (Huang et al., 2024), which have a close connection with the chemical components of tobacco leaves (Liu F. et al., 2021). Previous study reported many attractive metabolic capacities in the aging flue-cured tobaccos microorganisms, including those involved in amino acid metabolism, carbohydrate metabolism, vitamin metabolism, the biosynthesis of flavors and fragrances, and the degradation of harmful compounds such as nicotine and nitrite (Wang et al., 2018b). During the aging process of tobacco leaves, the metabolic activities of microorganisms can consume protein and cellulose in tobacco leaves (Mou et al., 2020). At the same time, the macromolecular substances in tobacco leaf were decomposed into small molecular flavor substances such as viololanone, damalone and furfural (Mou et al., 2020). Moreover, it has been observed that the introduction of fungal agents during the aging of tobacco leaves can promote the co-regulation of chemical composition (Li et al., 2009) and facilitate the conversion of compounds within the leaves (Zheng et al., 2003).
Numerous reports have highlighted the crucial role of tobacco microorganisms in degrading macromolecular substances and harmful compounds (Chen et al., 2015; Liu F. et al., 2021; Huang et al., 2022a; 2022b), including the degradation of both types of hydrocarbons, i.e., aliphatic non-methane and aromatic compounds and other harmful substances (Liu F. et al., 2021). Several investigations have been published on the degradation of substances such as β-carotenes, starch, protein, phytosterols, as well as the harmful compounds like nicotine and TSNA (Table 1). For example, Bacillus species are known to degrade cellulose (Li et al., 2006), proteins (Chen et al., 2015), carotene (Huang et al., 2022a) and other compounds, thereby reducing irritation, bitterness, and astringency in tobacco during combustion. Consequently, several Bacillus species, including Bacillus subtilis, Bacillus coagulans, Bacillus circulans, Bacillus megaterium and Bacillus thuringiensis, have been employed to enhance the development of desirable aromas and improve the smoking qualities of tobacco (Zhao et al., 2007; Wang et al., 2018b). Sphingomonas sp. (Ma et al., 2016), P. putida HSM-C2 (Huang et al., 2022b), Agrobaterium tumefaciens sp. (Wang and Huang, 2006) have also been employed to degrade other substances, such as chlorogenic acid, coumarin, and TSNA. In addition, some fungi, including Trametes versicolor (Su et al., 2016), Trametes hirsute (Su et al., 2016), Phanerochaete chrysosporium (Su et al., 2016), Moniliales Gliocephalias sp. (Yang X. et al., 2014) have also shown effectiveness in degrading materials such as hemicellulose, and cellulose (Table 1).
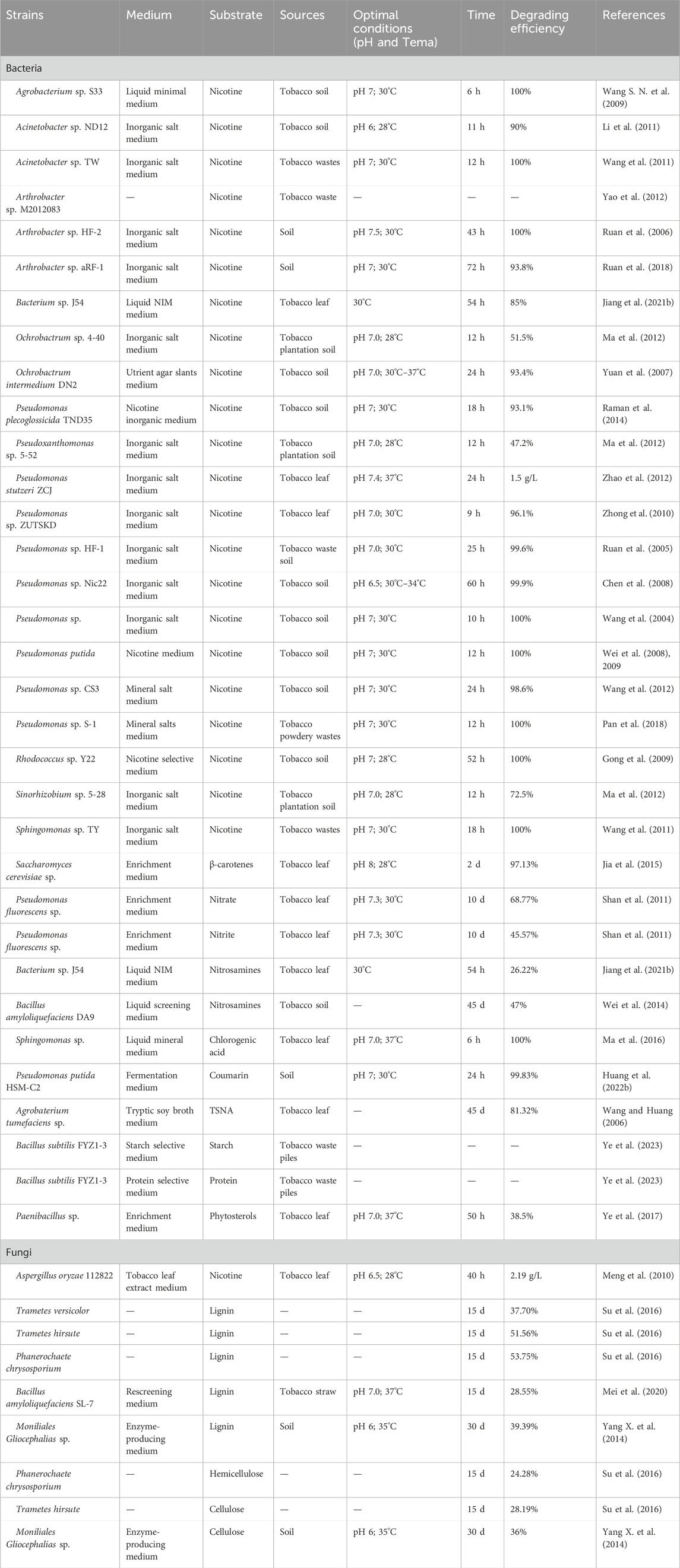
Table 1. Summary of the tobacco components degradation by bacterial isolates and their culture media.
Furthermore, investigations into tobacco microorganism isolates have confirmed that certain bacteria exhibit a high degradation efficiency (>99%) for nicotine (Table 1). Notable examples include Agrobacterium sp. S33 (Wang S. N. et al., 2009), Acinetobacter sp. TW (Wang et al., 2011), Pseudomonas sp. Nic22 (Chen et al., 2008), Pseudomonas sp. (Wang et al., 2004), P. putida (Wei et al., 2008; 2009), Rhodococcus sp. Y22 (Gong et al., 2009), and Sphingomonas sp. TY (Wang et al., 2011). Therefore, it is of great scientific significance and practical value to search for functional strains with high degradation ability to nicotine to improve tobacco quality. However, some bacteria have low degradation efficiency, such as Pseudoxanthomonas sp. 5-52 (47.2%), Ochrobactrum sp. 4-40 (51.5%) and Sinorhizobium sp. 5-28 (72.5%) (Ma et al., 2012). Additionally, fungi also play an important role in the degradation of tobacco macromolecular substances, such as lignin and so on (Table 1), but the degradation efficiency of lignin is relatively low, such as T. versicolor (37.70%) (Su et al., 2016), T. hirsute (51.56%) (Su et al., 2016) and Bacillus amyloliquefaciens SL-7 (28.55%) (Mei et al., 2020). The different degradation efficiency may be related to the degradation characteristics of strains and culture conditions (Ruan et al., 2005; Gaekwad and Vinchurkar, 2018; Huang et al., 2022b). For instance, the degradation efficiency of nicotine was increased within the pH range (5.5–7.5) and decreased within the pH range (7.5–9.5) (Ruan et al., 2005). Ruan et al. found that the rising of temperature from 29°C to 41°C, could lead into profound decrease in the nicotine degradation (Ruan et al., 2005). While, Huang et al. revealed that variations in carbon or nitrogen source type, and ammonium nitrate contents cause a significant impact on the degradation rate of coumarin (Huang et al., 2022b).
The role of microorganisms in the fermentation process of tobacco leaves is closely related to the enzymatic reaction, as microorganisms secrete various enzymes (e.g., alpha-amylase (Ullah et al., 2021), protease (Yogesh and Halami, 2015), cellulase (Araújo et al., 2021) and so on) to the exocytosomes during their growth and development. These enzymes released into the extracellular environment catalyze the decomposition or synthesis of certain substances in tobacco leaves. Study reported that the neutral aroma-enhancing compound was positively correlated with the carbohydrate-active enzymes, such as glycoside hydrolase, glycosyltransferase, polysaccharide lyase, carbohydrate esterase, and auxiliary active enzyme (Huang et al., 2024). Based on the enzyme-producing characteristics of different microorganisms, some hybrid strains have also been utilized to optimize tobacco fermentation. For example, co-cultivation of Bacillus amyloliquefaciens LB with high alpha-amylase activity and Bacillus kochii SC with high neutral protease activity has been used to improve sensory quality of flue-cured tobacco (Wu et al., 2021). And the complementary culture of Erwinia carotovora could effectively degrade pectin and cellulose by producing pectin- and cellulose-degrading enzymes and then be used for the production of the neutral aroma-enhancing compound (Huang et al., 2024). Microorganisms on the surface of tobacco leaves produce not only xylanase, cellulase, pectinase, protease and amylase (Fan et al., 2013), but also nicotine-degrading enzymes (Table 2).
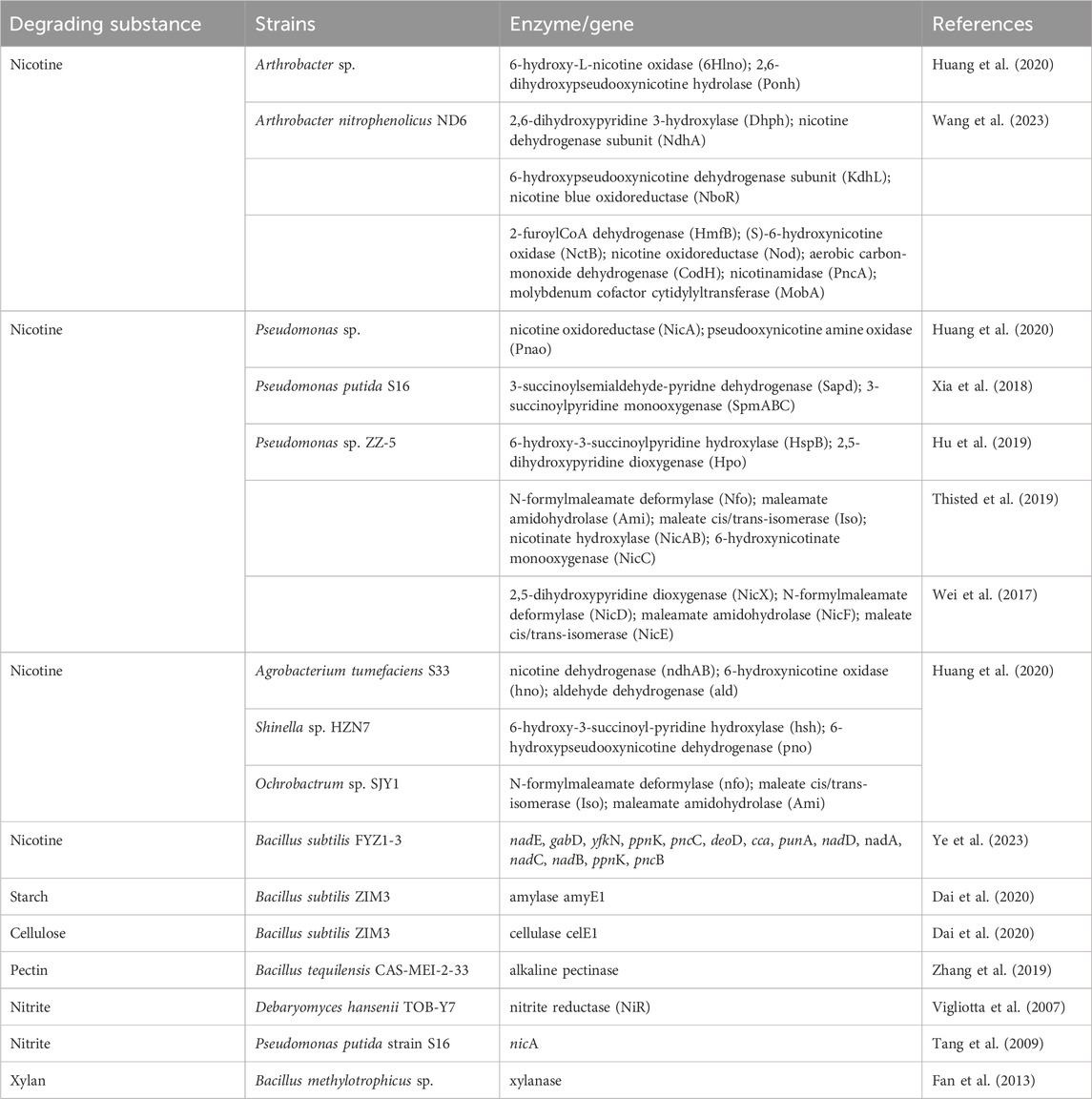
Table 2. The key enzymes and genes in the microorganisms for degradation of chemical components of tobacco.
In recent years, the metabolism of harmful substances in prokaryotes, including the catabolic pathways for its degradation and the enzymes involved during the pathways, has been systemically investigated (Meng et al., 2010; Fitzpatrick, 2018; Huang et al., 2020), and nicotine metabolism is one of the most extensively studied pathways. (Fitzpatrick, 2018; Huang et al., 2020; Mu et al., 2020). In bacteria, three nicotine degradation pathways have been reported: the pyridine pathway, the pyrrolidine pathway and the hybrid Pathway (a combination of pyridine and pyrrolidine pathways, known as the VPP pathway) (Meng et al., 2010; Fitzpatrick, 2018; Huang et al., 2020). In fungi, such as Aspergillus oryzae, the demethylation pathway has been reported to be employed, while the pathways in the eukaryote remain unclear (Meng et al., 2010) (Figure 2). Key enzymes, such as Pnao, Pno and Ponh, have been identified as the representative enzymes in the pyrrolidine, pyridine, and VPP pathways, respectively (Table 2) (Mu et al., 2020). The methylamine from pseudooxynicotine and 6-hydroxypseudooxynicotine were removed by Pnao in the pyrrolidine pathway and Pno in the VPP pathway, while 2,6-dihydroxypseudooxynicotine was hydrolyzed to 2,6-dihydroxypyridine and 4-methylaminobutyrate by Ponh in the pyridine pathway (Mu et al., 2020). Additionally, amylase amyE1 and cellulase celE1 can be produced by Bacillus subtilis ZIM3, which can simultaneously degrade both starch and cellulose (Table 2) (Dai et al., 2020). Compared to nicotine, the metabolism of TSNA and macromolecule (such as starch, lignin and protein) has not been fully resolved in the strains (Table 1). Therefore, this situation limits the in-depth analysis to metabolism pathways and functions of TSNA and macromolecule in the environment.
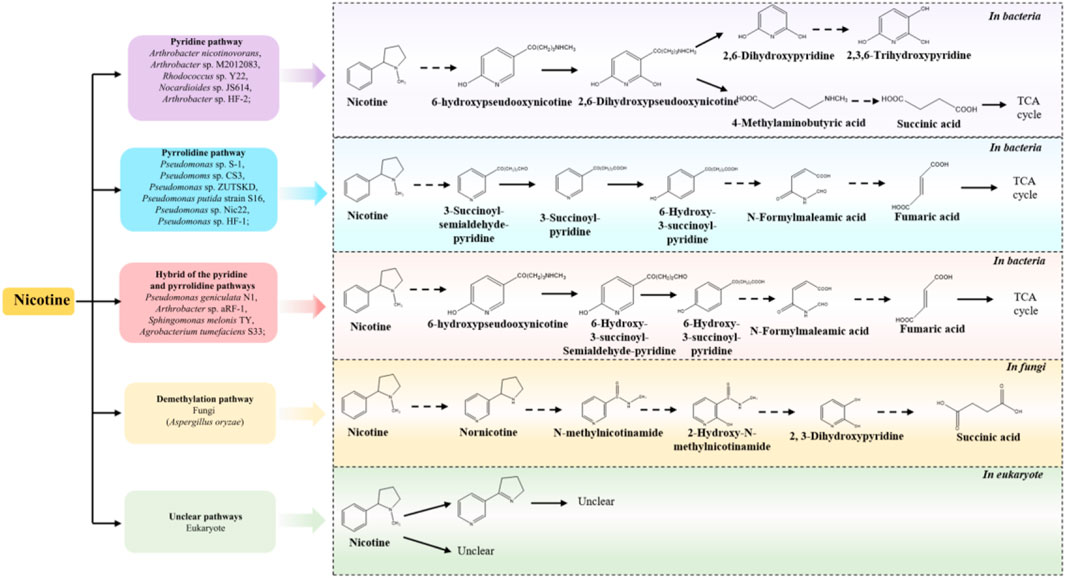
Figure 2. The degradation pathways of nicotine by bacteria, fungi and eukaryotes (adapted from Meng et al. (2010); Huang et al. (2020); Ganas et al. (2008); Ruan et al. (2006); Pan et al. (2018); Wang et al. (2012); Zhong et al. (2010); Tang et al. (2008); Chen et al. (2008); Ruan et al. (2006); Liu R. et al. (2021); Ruan et al. (2018); Wang et al. (2011); Wang S. N. et al. (2009)).
Studies indicate that many bacteria possess all the essential genetic elements for nicotine catabolism (Mu et al., 2020). Nicotine-degrading genes annotated from the metagenome data, include ndhA, nctB, kdhL, nboR, and dhponh are found to be actively involved in the pyridine pathway, which play an important roles in whole process of nicotine metabolism (Wang et al., 2023). Interestingly, ndhA and nctB are also the critical genes in the VPP pathway (Wang et al., 2023). Study reported that the ndhA gene is annotated to encode the isoquinoline 1-oxidoreductase alpha subunits, which can catalyze the hydroxylation of isoquinoline to 1-oxo-1,2-dihydroisoquinoline (Table 2) (Li et al., 2016). Additionally, genomic analysis of Bacillus subtilis FYZ1-3 revealed 14 functional genes associated with nicotine metabolism, and primarily located on the distinct genomic island of Bacillus subtilis FYZ1-3 (Ye et al., 2023). The homologous genes involved in nicotine catabolism, such as cup, ponh, kdhL, TR2, kdhM, kdhS, nit, ndhL, ndhS, ndhM, coxG, dhph, pkc, mox, TR1, 6hlno have been identified in Nocardioides sp. JS614 and Arthrobacter nicotinovorans (Ganas et al., 2008). Furthermore, the genome of Bacillus. subtilis FYZ1-3 has been shown to harbor multiple metabolic pathways and numerous genes related to the degradation of carbohydrate and proteins. These include pathways for starch and sucrose metabolism (47 genes), glycolysis/gluconeogenesis (39 genes), amino sugar and nucleotide sugar metabolism (43 genes), biosynthesis of amino acids (122 genes), and alanine, aspartate and glutamate metabolism (33 genes) (Ye et al., 2023). In recent years, some organisms have been well utilized in the process of tobacco aging and fermentation, but most of the functions of microorganisms have not been developed and utilized. Therefore, developing more tobacco microbial resources and understanding their ecological functions have important guiding significance for the improvement of tobacco quality.
4 The application of functional microorganisms
Previous studies have reported that the addition of functional microorganisms can significantly improve the sensory quality of tobacco, such as Arthrobacter sp. (Xu et al., 2021), Aureobasidium pullulans, Stenotrophomonas maltophilia, H3-1. For example, the addition of functional strains (Bacillus amyloliquefaciens LB, Bacillus kochii SC and Bacillus subtilis subsp.) could promote an increase in aroma, softness and a decrease in irritation (Wu et al., 2021; Huang et al., 2022a). Klebsiella variicola H8 has the functions that increase neutral aroma-enhancing compound production, decrease the nicotine level and the water-soluble total sugar content in the reconstituted tobacco leaf concentrate solution (Huang et al., 2024). And the combination of aroma-producing yeast, Lactobacillus debrueckii, and Rhizopus had the most significant improvement in aroma, taste and smoke. Furthermore, some enzyme produced by microorganisms can significantly improve the quality of tobacco leaves, such as protease, amylozyme, pectinase, cellulase, which can catalyze the hydrolysis of corresponding substrates to produce flavoring substances and the precursors, so the corresponding catalytic hydrolysis mechanism has become one of the research hotspots in the tobacco industry.
5 Isolation and culturing strategies of tobacco microorganisms
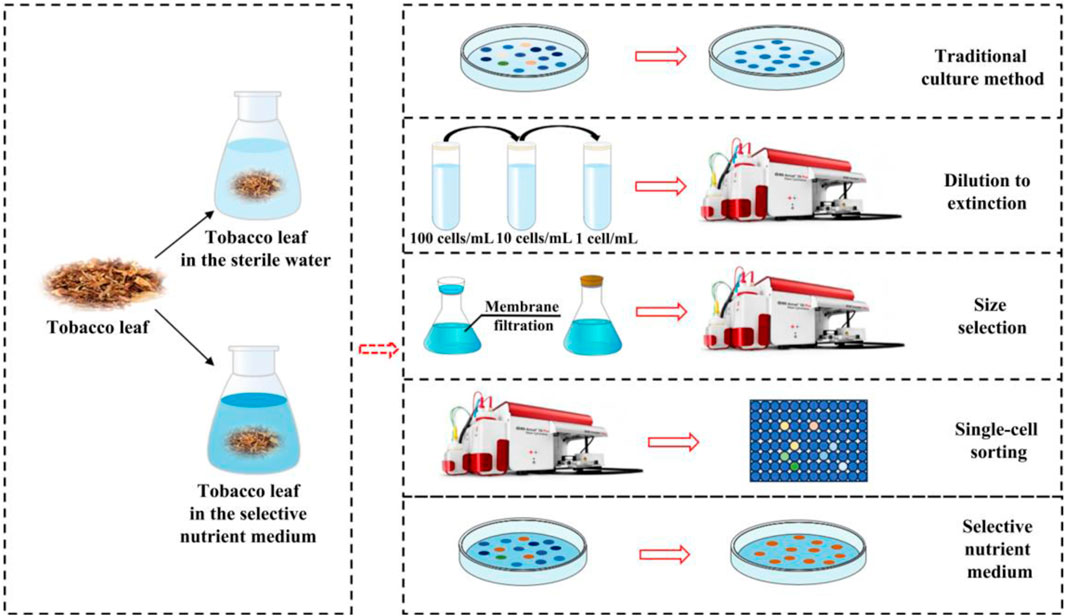
Traditional microbiological methods have played a crucial role in successfully isolating numerous microorganisms of interest and continue to be invaluable tools for cultivation (Lewis et al., 2021). During the isolation and cultivation of tobacco microorganisms, a variety of traditional techniques are employed (Figure 3). Common media, such as LB medium and inorganic salt medium, are typically used for this purpose (Chen et al., 2008; Mei et al., 2020). Additionally, selective nutrient media containing specific substrates, such as proteins, starch, and nicotine, are used to enrich specific microbial taxa (Raman et al., 2014; Ye et al., 2023). It is worth noting that successful isolation of strains using these approaches requires considerable time and patience, as well as meticulous optimization of media compositions and different physicochemical conditions (Lewis et al., 2021). Despite these refined efforts, the vast majority (>99%) of the microorganisms in the natural environment remained uncultured under laboratory conditions (Wang et al., 2021; Hu et al., 2022). To overcome the limitations of traditional culture methods, several innovative techniques have been developed to enhance microbial isolation and cultivation. For instance, single-cell sorting Via flow cytometry have been employed to isolate a greater diversity of strains from the tobacco microbial community, and two functional strains, Bacillus amyloliquefaciens LB (with high alpha-amylase activity) and Bacillus kochii SC (with high neutral protease activity) were successfully cultured (Wu et al., 2021). While other advanced techniques such as size selection, and dilution-to-extinction, have yet to demonstrate their universal applicability across different species and environments, they have already shown promise in culturing the marine bacteria (Figure 3) (Hu et al., 2022). For example, size selection, also referred as filtration, has been combined with flow cytometry to culture the small-sized bacteria (Hu et al., 2022). Similarly, dilution-to-extinction has been used to culture the marine bacterium, Candidatus Fonsibacter ubiquis LSUCC0530) in the previous study (Henson et al., 2018). In addition, bacteria with low nucleic acid content have been successfully cultured by using a combination of size selection, dilution-to-extinction and flow cytometry (Wang Y. et al., 2009), highlighting the potential and advantages of these key techniques in the isolation and culture of previously uncultured microorganisms (Wang Y. et al., 2009; Henson et al., 2018; Hu et al., 2022). In future studies, these innovative methods can be applied to the isolation and cultivation of uncultured tobacco microorganisms, paving the way for new discoveries and advancements in microbial research.
6 Challenges and future perspectives
Based on the above review and discussion, several challenges and perspectives should be considered in future research:
Although some tobacco microorganisms have been cultured, the number of strains capable of degrading large molecular substances and harmful compounds remains very limited. To date, only a few strains with a high efficiency in nicotine degradation have been reported. However, it is crucial to isolate or discover more functional strains, particularly those capable of degrading macromolecular substances and the harmful compounds. This will definitely develop a more precise understanding to accurately determine microbial characteristics including their metabolism and physiology as well as their ecological roles.
Although the previous studies have reported the intermediate degradation products and metabolic mechanisms of nicotine in the different strains, the comprehensive pathways, metabolic mechanisms and associated genes involved in the degradation of TSNA, β-carotenes, starch, protein and other compounds in tobacco microorganisms remain to be elucidated. In tobacco leaves, microorganisms are in a state of coexistence, displaying a competitive or symbiotic relationship with respect to substance utilization. Therefore, it is essential to conduct in-depth studies on the dynamic and long-term interactions between different microbial communities and their impact on improving tobacco quality.
Currently, the genetic mechanisms underlying the degradation of many tobacco-associated substances are not well understood. The integration of genomics, metagenomics, proteomics and systems biology represents a powerful approach to uncover the bacterial degradation mechanisms and provide valuable insights for further development of functional enzymes and genes. Additionally, advancing molecular biology techniques for the isolation and cultivation of functional tobacco microorganisms would be a crucial area for future research.
7 Conclusion
Tobacco microorganisms play a crucial role in enhancing tobacco quality, which represent a diverse group, primarily consisting of bacteria and fungi with marked metabolic capabilities, including amino acid metabolism, carbohydrate metabolism, vitamin metabolism, and the biosynthesis of flavors and fragrances. Additionally, these microorganisms secrete a variety of enzymes into the exocytosomes that can catalyze the decomposition or synthesis of certain substances in tobacco leaves. Tobacco microorganisms display remarkable degradation functions on the substances such as nicotine, TSNA, β-carotenes, starch, protein, and phytosterols, and contribute an important role in the enhancement of tobacco quality. Up to now, some organisms have been well utilized in the process of tobacco aging and fermentation, but most of the functions of microorganisms have not been explored and elucidated. Therefore, it is necessary to develop more microbial separation and culture methods for tobacco leaves, further explore the influence of microorganisms on the chemical composition of tobacco leaves, reveal the specific mechanism of their regulation and improvement of tobacco leaf quality, and provide a new scientific perspective and potential application path for the high-quality production of tobacco industry in the future.
Author contributions
WH: Writing – original draft, Writing – review and editing, Software. JaY: Software, Investigation, Writing – original draft. JF: Software, Writing – original draft. KI: Validation, Visualization, Writing – original draft. PY: Conceptualization, Investigation, Writing – original draft. SH: Supervision, Validation, Writing – original draft. DM: Funding acquisition, Writing – review and editing. JnY: Formal Analysis, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by Key Research and Development Project of China National Tobacco Corporation (110202202006) and Major Science and Technology Project of the China National Tobacco Corporation (110202201053).
Conflict of interest
Author JF was employed by Shenzhen Tobacco Industrial Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Araújo, E. A., Dias, A. H. S., Kadowaki, M. A. S., Piyadov, V., Pellegrini, V. O. A., Urio, M. B., et al. (2021). Impact of cellulose properties on enzymatic degradation by bacterial GH48 enzymes: structural and mechanistic insights from processive Bacillus licheniformis Cel48B cellulase. Carbohydr. Polym. 264, 118059. doi:10.1016/j.carbpol.2021.118059
Chen, C., Li, X., Yang, J., Gong, X., Li, B., and Zhang, K. (2008). Isolation of nicotine-degrading bacterium pseudomonas sp. Nic22, and its potential application in tobacco processing. Int. Biodeterior. Biodegrad. 62, 226–231. doi:10.1016/j.ibiod.2008.01.012
Chen, Q., Cai, L., Wang, H., Cai, L., Goodwin, P., Ma, J., et al. (2020). Fungal composition and diversity of the tobacco leaf phyllosphere during curing of leaves. Front. Microbiol. 11, 554051. doi:10.3389/fmicb.2020.554051
Chen, S., Lin, J., Bao, K., Fan, J., Zhang, R., Chen, Y., et al. (2018). High-throughput sequencing fungal community structures in aging tobacco strips from different growing areas and stalk positions. Tob. Sci. Technol. 51, 12–19. doi:10.16135/j.issn1002-0861.2017.0270
Chen, X., Wang, X., Zeng, X., Dang, L., Yang, Y., Ling, J., et al. (2015). Screening of protein- degrading bacterial strains and their effects on tobacco. Acta Agric. Jiangxi 27, 90–91. doi:10.19386/j.cnki.jxnyxb.2015.06.022
Chopyk, J., Chattopadhyay, S., Kulkarni, P., Claye, E., Babik, K. R., Reid, M. C., et al. (2017a). Mentholation affects the cigarette microbiota by selecting for bacteria resistant to harsh environmental conditions and selecting against potential bacterial pathogens. Microbiome 5, 22. doi:10.1186/s40168-017-0235-0
Chopyk, J., Chattopadhyay, S., Kulkarni, P., Smyth, E. M., Hittle, L. E., Paulson, J. N., et al. (2017b). Temporal variations in cigarette tobacco bacterial community composition and tobacco-specific nitrosamine content are influenced by brand and storage conditions. Front. Microbiol. 8, 358. doi:10.3389/fmicb.2017.00358
Dai, J., Dong, A., Xiong, G., Liu, Y., Hossain, Md. S., Liu, S., et al. (2020). Production of highly active extracellular amylase and cellulase from Bacillus subtilis ZIM3 and a recombinant strain with a potential application in tobacco fermentation. Front. Microbiol. 11, 1539. doi:10.3389/fmicb.2020.01539
Fan, Y., Lin, M., Li, X., Li, Z., Chen, Y., and Han, S. (2013). Screening and identification of a xylanase-producing strain during tobacco fermentation. Mod. Food Sci. Technol. 29, 2658–2662. doi:10.13982/j.mfst.1673-9078.2013.11.029
Fitzpatrick, P. F. (2018). The enzymes of microbial nicotine metabolism. Beilstein J. Org. Chem. 14, 2295–2307. doi:10.3762/bjoc.14.204
Gaekwad, I., and Vinchurkar, A. S. (2018). Isolation and identification of nicotine utilizing bacterial species from tobacco leaves. Int. Res. J. Pharm. 9, 103–106. doi:10.7897/2230-8407.09115
Ganas, P., Sachelaru, P., Mihasan, M., Igloi, G. L., and Brandsch, R. (2008). Two closely related pathways of nicotine catabolism in arthrobacter nicotinovorans and nocardioides sp. strain JS614. Archives Microbiol. 189, 511–517. doi:10.1007/s00203-007-0340-8
Gong, X., Yang, J., Duan, Y., Dong, J., Zhe, W., Wang, L., et al. (2009). Isolation and characterization of rhodococcus sp. Y22 and its potential application to tobacco processing. Res. Microbiol. 160, 200–204. doi:10.1016/j.resmic.2009.02.004
Guo, Z., Xie, H., Wang, H., Huang, Y., Chen, Q., Xiang, L., et al. (2020). Leaf spot caused by didymella segeticola on tobacco in China. Plant Dis. 104, 1559. doi:10.1094/PDIS-11-19-2398-PDN
Han, J., Sanad, Y. M., Deck, J., Sutherland, J. B., Li, Z., Walters, M. J., et al. (2016). Bacterial populations associated with smokeless tobacco products. Appl. Environ. Microbiol. 82, 6273–6283. doi:10.1128/AEM.01612-16
Henson, M. W., Lanclos, V. C., Faircloth, B. C., and Thrash, J. C. (2018). Cultivation and genomics of the first freshwater SAR11 (LD12) isolate. ISME J. 12, 1846–1860. doi:10.1038/s41396-018-0092-2
Hu, B., Gu, K., Gong, J., Zhang, K., Chen, D., He, X., et al. (2021). The effect of flue-curing procedure on the dynamic change of microbial diversity of tobaccos. Sci. Rep. 11, 5354. doi:10.1038/s41598-021-84875-6
Hu, H., Wang, L., Wang, W., Wu, G., Tao, F., Xu, P., et al. (2019). Regulatory mechanism of nicotine degradation in pseudomonas putida. Mbio 10, e00602-19. doi:10.1128/mBio.00602-19
Hu, W., Zhang, H., Lin, X., Liu, R., Bartlam, M., and Wang, Y. (2022). Characteristics, biodiversity, and cultivation strategy of low nucleic acid content bacteria. Front. Microbiol. 13, 900669. doi:10.3389/fmicb.2022.900669
Huang, H., Shang, J., and Wang, S. (2020). Physiology of a hybrid pathway for nicotine catabolism in bacteria. Front. Microbiol. 11, 598207. doi:10.3389/fmicb.2020.598207
Huang, J., Yang, J., Duan, Y., Gu, W., Gong, X., Zhe, W., et al. (2010). Bacterial diversities on unaged and aging flue-cured tobacco leaves estimated by 16S rRNA sequence analysis. Appl. Microbiol. Biotechnol. 88, 553–562. doi:10.1007/s00253-010-2763-4
Huang, S., Liu, D., Chen, M., Xi, G., Yang, P., Jia, C., et al. (2022a). Effects of bacillus subtilis subsp. on the microbial community and aroma components of flue-cured tobacco leaves based on metagenome analysis. Arch. Microbiol. 204, 726. doi:10.1007/s00203-022-03347-1
Huang, S., Wang, M., Mao, D., Rasool, A., Jia, C., Yang, P., et al. (2022b). Isolation, identification and characterization of growth parameters of pseudomonas putida HSM-C2 with coumarin-degrading bacteria. Molecules 27, 6007. doi:10.3390/molecules27186007
Huang, S., Xia, F., Wei, T., Jia, C., Qian, Y., and Mao, D. (2017). Isolation and identification of a cembratriene-4,6-diol degradation and aroma producing strain from tobacco leaf. J. Light Industry 32 (6), 73–80.
Huang, S., Zhu, L., Wang, K., Zhang, X., Mao, D., and Rasool, A. (2024). Unravel the supremacy of klebsiella variicola over native microbial strains for aroma-enhancing compound production in reconstituted tobacco concentrate through metagenomic analysis. Metabolites 14, 158. doi:10.3390/metabo14030158
Jia, B., Wei, T., Huang, S., Jia, X., Yang, J., Zhang, G., et al. (2015). Isolation and identification of a α-carotenes-degradating strain and optimization of its fermentation conditions. Food Ferment. Industries 41, 34–39. doi:10.13995/j.cnki.11-1802/ts.201501007
Jiang, Y., Chen, X., Zhao, G., Liu, J., Xie, Y., Li, Y., et al. (2021a). Endophytic fungal community of tobacco leaves and their potential role in the formation of “cherry-red” tobacco. Front. Microbiol. 12, 658116. doi:10.3389/fmicb.2021.658116
Jiang, Y., Gong, J., Chen, Y., Hu, B., Sun, J., Zhu, Y., et al. (2021b). Biodegradation of nicotine and TSNAs by bacterium sp. Strain J54. Iran. J. Biotechnol. 19, e2812. doi:10.30498/ijb.2021.240460.2812
Larsson, L., Szponar, B., Ridha, B., Pehrson, C., Dutkiewicz, J., Krysinska-Traczyk, E., et al. (2008). Identification of bacterial and fungal components in tobacco and tobacco smoke. Tob. Induc. Dis. 4, 4. doi:10.1186/1617-9625-4-4
Lei, X., Zehao, Z., Zhigang, G., Ying, W., Bin, G., Wei, C., et al. (2019). Screening and application of aroma-enhancing bacteria for tobacco. Chin. Tob. Sci. 40 (5), 60–67.
Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z., and Ettema, T. J. G. (2021). Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 19, 225–240. doi:10.1038/s41579-020-00458-8
Li, H., Duan, Y., Ma, G., Lei, L., Zhang, K., and Yang, J. (2011). Isolation and characterization of acinetobacter sp. ND12 capable of degrading nicotine. Afr. J. Microbiol. Res. 5, 1335–1341. doi:10.5897/AJMR11.210
Li, H., Xie, K., Yu, W., Hu, L., Huang, H., Xie, H., et al. (2016). Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in agrobacterium tumefaciens S33. Appl. Environ. Microbiol. 82, 1745–1755. doi:10.1128/AEM.03909-15
Li, N., Zeng, D., Dai, Y., Li, D., Wang, C., Lei, J., et al. (2009). Isolation and identification on cultivable microorganisms from cigar leaf surface. J. Anhui Agric. Sci. 37, 11857–11858. doi:10.13989/j.cnki.0517-6611.2009.25.004
Li, Y., Ding, M., Wang, J., Xu, G., and Zhao, F. (2006). A novel thermoacidophilic endoglucanase, Ba-EGA, from a new cellulose-degrading bacterium, bacillus sp.AC-1. Appl. Microbiol. Biotechnol. 70, 430–436. doi:10.1007/s00253-005-0075-x
Liu, F., Wu, Z., Zhang, X., Xi, G., Zhao, Z., Lai, M., et al. (2021). Microbial community and metabolic function analysis of cigar tobacco leaves during fermentation. MicrobiologyOpen 10, e1171. doi:10.1002/mbo3.1171
Liu, J., Ma, G., Chen, T., Hou, Y., Yang, S., Zhang, K., et al. (2015). Nicotine-degrading microorganisms and their potential applications. Appl. Microbiol. Biotechnol. 99, 3775–3785. doi:10.1007/s00253-015-6525-1
Liu, R., Bao, Z., Zhao, P., and Li, G. (2021). Advances in the study of metabolomics and metabolites in some species interactions. Molecules 26, 3311. doi:10.3390/molecules26113311
Lv, D., Ma, A., Tang, X., Bai, Z., Qi, H., and Zhuang, G. (2013). Profile of the culturable microbiome capable of producing acyl-homoserine lactone in the tobacco phyllosphere. J. Environ. Sci. 25, 357–366. doi:10.1016/S1001-0742(12)60027-8
Ma, G., Lei, L., Xia, Z., Gong, X., Zhou, W., and Yang, J. (2012). Diversity and phylogenetic analyses of nicotine-degrading bacteria isolated from tobacco plantation soils. Afr. J. Microbiol. Res. 6. doi:10.5897/AJMR12.994
Ma, Y., Wang, X., Nie, X., Zhang, Z., Yang, Z., Nie, C., et al. (2016). Microbial degradation of chlorogenic acid by a Sphingomonas sp. strain. Appl. Biochem. Biotechnol. 179, 1381–1392. doi:10.1007/s12010-016-2071-2
Mei, J., Shen, X., Gang, L., Xu, H., Wu, F., and Sheng, L. (2020). A novel lignin degradation bacteria-Bacillus amyloliquefaciens SL-7 used to degrade straw lignin efficiently. Bioresour. Technol. 310, 123445. doi:10.1016/j.biortech.2020.123445
Meng, X., Lu, L., Gu, G., and Xiao, M. (2010). A novel pathway for nicotine degradation by aspergillus oryzae 112822 isolated from tobacco leaves. Res. Microbiol. 161, 626–633. doi:10.1016/j.resmic.2010.05.017
Mou, D., Zhou, J., Zhang, X., Zhang, X., Zhang, Q., and Zhou, X. (2020). Changes in chemical composition drive changes in the structural composition of microbial communities. Microbiol. China 47, 362–369. doi:10.13344/j.microbiol.china.190425
Mu, Y., Chen, Q., Parales, R. E., Lu, Z., Hong, Q., He, J., et al. (2020). Bacterial catabolism of nicotine: catabolic strains, pathways and modules. Environ. Res. 183, 109258. doi:10.1016/j.envres.2020.109258
Nagrale, D. T., Sharma, L., Kumar, S., and Gawande, S. P. (2016). “Recent diagnostics and detection tools: implications for plant pathogenic alternaria and their disease management,” in Current Trends in Plant Disease Diagnostics and Management Practices, 111–163. doi:10.1007/978-3-319-27312-9_5
Pan, D., Sun, M., Wang, Y., Lv, P., Wu, X., Li, Q. X., et al. (2018). Characterization of nicotine catabolism through a novel pyrrolidine pathway in pseudomonas sp. S-1. J. Agric. Food Chem. 66, 7393–7401. doi:10.1021/acs.jafc.8b01868
Raman, G., Mohan, K., Manohar, V., and Sakthivel, N. (2014). Biodegradation of nicotine by a novel nicotine-degrading bacterium, Pseudomonas plecoglossicida TND35 and its new biotransformation intermediates. Biodegradation 25, 95–107. doi:10.1007/s10532-013-9643-4
Ruan, A., Gao, Y., Fang, C., and Xu, Y. (2018). Isolation and characterization of a novel nicotinophilic bacterium, arthrobacter sp. aRF-1 and its metabolic pathway. Biotechnol. Appl. Biochem. 65, 848–856. doi:10.1002/bab.1682
Ruan, A., Min, H., Peng, X., and Huang, Z. (2005). Isolation and characterization of pseudomonas sp. strain HF-1, capable of degrading nicotine. Res. Microbiol. 156, 700–706. doi:10.1016/j.resmic.2005.02.010
Ruan, A., Min, H., and Zhu, W. (2006). Studies on biodegradation of nicotine by arthrobacter sp. Strain HF-2. J. Environ. Sci. Health 41, 1159–1170. doi:10.1080/03601230600856934
Saito, A., Kawahara, M., Ikeda, S., Ishimine, M., Akao, S., and Minamisawa, K. (2008). Broad distribution and phylogeny of anaerobic endophytes of Cluster XIVa clostridia in plant species Including Crops. Microbes Environ. 23, 73–80. doi:10.1264/jsme2.23.73
Shan, H., Chen, D., Li, J., Chen, T., Hu, H., Guo, Z., et al. (2011). Identification and primary application of TSNA degrading bacterial strain AS97 isolated from aging tobacco leaves. Acta Microbiol. Sin. 51, 1326–1333. doi:10.13343/j.cnki.wsxb.2011.10.004
Smyth, E. M., Chattopadhyay, S., Babik, K., Reid, M., Chopyk, J., Malayil, L., et al. (2019). The bacterial communities of little cigars and cigarillos are dynamic over time and varying storage conditions. Front. Microbiol. 10, 2371. doi:10.3389/fmicb.2019.02371
Su, Y., Xian, H., Shi, S., Zhang, C., Manik, S. M. N., Mao, J., et al. (2016). Biodegradation of lignin and nicotine with white rot fungi for the delignification and detoxification of tobacco stalk. BMC Biotechnol. 16, 81. doi:10.1186/s12896-016-0311-8
Tang, H., Wang, L., Meng, X., Ma, L., Wang, S., He, X., et al. (2009). Novel nicotine oxidoreductase-encoding gene involved in nicotine degradation by pseudomonas putida strain S16. Appl. Environ. Microbiol. 75, 772–778. doi:10.1128/AEM.02300-08
Tang, H., Wang, S., Ma, L., Meng, X., Deng, Z., Zhang, D., et al. (2008). A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by pseudomonas putida strain S16. Appl. Environ. Microbiol. 74, 1567–1574. doi:10.1128/AEM.02529-07
Thisted, T., Biesova, Z., Walmacq, C., Stone, E., Rodnick-Smith, M., Ahmed, S. S., et al. (2019). Optimization of a nicotine degrading enzyme for potential use in treatment of nicotine addiction. BMC Biotechnol. 19, 56. doi:10.1186/s12896-019-0551-5
Tyx, R. E., Stanfill, S. B., Keong, L. M., Rivera, A. J., Satten, G. A., Watson, C. H., et al. (2016). Characterization of bacterial communities in selected smokeless tobacco products using 16S rDNA analysis. PLoS One 11, e0146939. doi:10.1371/journal.pone.0146939
Ullah, I., Khan, M. S., Khan, S. S., Ahmad, W., Zheng, L., Shah, S. U. A., et al. (2021). Identification and characterization of thermophilic amylase producing bacterial isolates from the brick kiln soil. Saudi J. Biol. Sci. 28, 970–979. doi:10.1016/j.sjbs.2020.11.017
Vigliotta, G., Di Giacomo, M., Carata, E., Massardo, D. R., Tredici, S. M., Silvestro, D., et al. (2007). Nitrite metabolism in Debaryomyces hansenii TOB-Y7, a yeast strain involved in tobacco fermentation. Appl. Microbiol. Biotechnol. 75, 633–645. doi:10.1007/s00253-007-0867-2
Wang, A., and Huang, Q. (2006). Studies on isolation, identification and characteristics of reducing TSNA contents of burley tobacco endophytic bacteria. Acta Sci. Circumstantiae, 1914–1920. doi:10.13671/j.hjkxxb.2006.11.028
Wang, F., Li, M., Huang, L., and Zhang, X. (2021). Cultivation of uncultured marine microorganisms. Mar. Life Sci. Technol. 3, 117–120. doi:10.1007/s42995-021-00093-z
Wang, F., Men, X., Zhang, G., Liang, K., Xin, Y., Wang, J., et al. (2018a). Assessment of 16S rRNA gene primers for studying bacterial community structure and function of aging flue-cured tobaccos. AMB Express 8, 182. doi:10.1186/s13568-018-0713-1
Wang, F., Zhao, H., Xiang, H., Wu, L., Men, X., Qi, C., et al. (2018b). Species diversity and functional prediction of surface bacterial communities on aging flue-cured tobaccos. Curr. Microbiol. 75, 1306–1315. doi:10.1007/s00284-018-1525-x
Wang, H., Huang, Y., Wang, J., Chen, X., Wei, K., Wang, M., et al. (2016). Activities of azoxystrobin and difenoconazole against alternaria alternata and their control efficacy. Crop Prot. 90, 54–58. doi:10.1016/j.cropro.2016.08.022
Wang, H. H., Yin, B., Peng, X. X., Wang, J. Y., Xie, Z. H., Gao, J., et al. (2012). Biodegradation of nicotine by newly isolated pseudomonas sp. CS3 and its metabolites: a newly isolated nicotine-utilizing pseudomonas strain. J. Appl. Microbiol. 112, 258–268. doi:10.1111/j.1365-2672.2011.05208.x
Wang, M., Yang, G., Wang, X., Yao, Y., Min, H., and Lu, Z. (2011). Nicotine degradation by two novel bacterial isolates of acinetobacter sp. TW and sphingomonas sp. TY and their responses in the presence of neonicotinoid insecticides. World J. Microbiol. Biotechnol. 27, 1633–1640. doi:10.1007/s11274-010-0617-y
Wang, S. N., Liu, Z., and Xu, P. (2009). Biodegradation of nicotine by a newly isolated Agrobacterium sp. strain S33. J. Appl. Microbiol. 107, 838–847. doi:10.1111/j.1365-2672.2009.04259.x
Wang, S. N., Xu, P., Tang, H. Z., Meng, J., Liu, X. L., Huang, J., et al. (2004). Biodegradation and detoxification of nicotine in tobacco solid waste by a pseudomonas sp. Biotechnol. Lett. 26, 1493–1496. doi:10.1023/B:BILE.0000044450.16235.65
Wang, Y., Hammes, F., Boon, N., Chami, M., and Egli, T. (2009). Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 3, 889–902. doi:10.1038/ismej.2009.46
Wang, Y., Luo, X., Chu, P., Shi, H., Wang, R., Li, J., et al. (2023). Cultivation and application of nicotine-degrading bacteria and environmental functioning in tobacco planting soil. Bioresour. Bioprocess. 10, 10. doi:10.1186/s40643-023-00630-x
Wei, H., Lei, L., Liu, S., Xia, Z., Liu, X., and Liu, P. (2009). PanB is involved in nicotine metabolism in pseudomonas putida. Int. Biodeterior. Biodegrad. 63, 988–992. doi:10.1016/j.ibiod.2009.02.010
Wei, H., Lei, L., Xia, Z., Liu, S., Liu, P., and Liu, X. (2008). Characterisation of a novel aerobic nicotine-biodegrading strain of Pseudomonas putida. Ann. Microbiol. 58, 41–45. doi:10.1007/BF03179443
Wei, T., Zang, J., Zheng, Y., Tang, H., Huang, S., and Mao, D. (2017). Characterization of a novel nicotine hydroxylase from pseudomonas sp. ZZ-5 that catalyzes the conversion of 6-hydroxy-3-succinoylpyridine into 2,5-dihydroxypyridine. Catalysts 7, 257. doi:10.3390/catal7090257
Wei, X., Deng, X., Cai, D., Ji, Z., Wang, C., Yu, J., et al. (2014). Decreased tobacco-specific nitrosamines by microbial treatment with bacillus amyloliquefaciens DA9 during the air-curing process of burley tobacco. J. Agric. Food Chem. 62, 12701–12706. doi:10.1021/jf504084z
Wu, X., Zhu, P., Li, D., Zheng, T., Cai, W., Li, J., et al. (2021). Bioaugmentation of Bacillus amyloliquefaciens–Bacillus kochii co-cultivation to improve sensory quality of flue-cured tobacco. Archives Microbiol. 203, 5723–5733. doi:10.1007/s00203-021-02556-4
Xia, Z., Lei, L., Zhang, H., and Wei, H. (2018). Characterization of the ModABC molybdate transport system of pseudomonas putida in nicotine degradation. Front. Microbiol. 9, 3030. doi:10.3389/fmicb.2018.03030
Xing, L., Yang, J., Jia, Y., Hu, X., Liu, Y., Xu, H., et al. (2021). Effects of ecological environment and host genotype on the phyllosphere bacterial communities of cigar tobacco (Nicotiana tabacum L.). Ecol. Evol. 11, 10892–10903. doi:10.1002/ece3.7861
Xu, Q., Li, S., Huang, S., and Mao, D. (2021). Review on tobacco-derived microorganisms and its application. J. Light Industry 36 (5), 42–50.
Yang, X., Liu, Y., Liu, Z., Lai, Y., and Chen, Y. (2014). High yield laccase producing strains screened and the study of tobacco stem degradation affected by the strains. Chin. Agric. Sci. Bull. 30, 52–57.
Yang, Z., Wu, Y., Nie, X., Zhou, P., Song, Q., Shang, H., et al. (2014). Research on growth-promoting effect of 3 tobacco-endophytic bacterial strains on crops. Acta Agric. Jiangxi 26 (11), 58–60.
Yao, Y., Tang, H., Ren, H., Yu, H., Wang, L., and Xu, P. (2012). Genome sequence of a nicotine-degrading strain of arthrobacter. J. Bacteriol. 194, 5714–5715. doi:10.1128/JB.01370-12
Ye, C., Liu, D., Huang, K., Li, D., Ma, X., Jin, Y., et al. (2023). Isolation of starch and protein degrading strain bacillus subtilis FYZ1-3 from tobacco waste and genomic analysis of its tolerance to nicotine and inhibition of fungal growth. Front. Microbiol. 14, 1260149. doi:10.3389/fmicb.2023.1260149
Ye, J., Zhang, Z., Yan, J., Hao, H., Liu, X., Yang, Z., et al. (2017). Degradation of phytosterols in tobacco waste extract by a novel Paenibacillus sp. Biotechnol. Appl. Biochem. 64, 843–850. doi:10.1002/bab.1547
Yogesh, D., and Halami, P. M. (2015). A fibrin degrading serine metallo protease of Bacillus circulans with α-chain specificity. Food Biosci. 11, 72–78. doi:10.1016/j.fbio.2015.04.007
Yuan, X.-L., Cao, M., Liu, X.-M., Du, Y.-M., Shen, G.-M., Zhang, Z.-F., et al. (2018). Composition and genetic diversity of the nicotiana tabacum microbiome in different topographic areas and growth periods. Int. J. Mol. Sci. 19, 3421. doi:10.3390/ijms19113421
Yuan, Y. J., Lu, Z. X., Huang, L. J., Bie, X. M., Lu, F. X., and Li, Y. (2006). Optimization of a medium for enhancing nicotine biodegradation by ochrobactrum intermedium DN2. J. Appl. Microbiol. 101, 691–697. doi:10.1111/j.1365-2672.2006.02929.x
Yuan, Y. J., Lu, Z. X., Huang, L. J., Li, Y., Lu, F. X., Bie, X. M., et al. (2007). Biodegradation of nicotine from tobacco waste extract by Ochrobactrum intermedium DN2. J. Industrial Microbiol. Biotechnol. 34, 567–570. doi:10.1007/s10295-007-0212-x
Zhang, G., Li, S., Xu, Y., Wang, J., Wang, F., Xin, Y., et al. (2019). Production of alkaline pectinase: a case study investigating the use of tobacco stalk with the newly isolated strain Bacillus tequilensis CAS-MEI-2-33. BMC Biotechnol. 19, 45. doi:10.1186/s12896-019-0526-6
Zhang, Q., Geng, Z., Li, D., and Ding, Z. (2020). Characterization and discrimination of microbial community and co-occurrence patterns in fresh and strong flavor style flue-cured tobacco leaves. MicrobiologyOpen 9, e965. doi:10.1002/mbo3.965
Zhang, Z., Mei, X., He, Z., Xie, X., Yang, Y., Mei, C., et al. (2022). Nicotine metabolism pathway in bacteria: mechanism, modification, and application. Appl. Microbiol. Biotechnol. 106, 889–904. doi:10.1007/s00253-022-11763-y
Zhao, L., Zhu, C., Gao, Y., Wang, C., Li, X., Shu, M., et al. (2012). Nicotine degradation enhancement by Pseudomonas stutzeri ZCJ during aging process of tobacco leaves. World J. Microbiol. Biotechnol. 28, 2077–2086. doi:10.1007/s11274-012-1010-9
Zhao, M., Wang, B., Li, F., Qiu, L., Li, F., Wang, S., et al. (2007). Analysis of bacterial communities on aging flue-cured tobacco leaves by 16S rDNA PCR-DGGE technology. Appl. Microbiol. Biotechnol. 73, 1435–1440. doi:10.1007/s00253-006-0625-x
Zheng, X., Zhang, X., Zhang, T., and Wu, X. (2003). Improving tobacco quality by fungal leavens. Microbiol. China, 10–13. doi:10.13344/j.microbiol.china.2003.06.003
Keywords: tobacco, microorganism, community composition, functions, isolation and cultivation strategies
Citation: Hu W, Yuan J, Fei J, Imdad K, Yang P, Huang S, Mao D and Yang J (2025) Shaping the future of tobacco through microbial insights: a review of advances and applications. Front. Bioeng. Biotechnol. 13:1548323. doi: 10.3389/fbioe.2025.1548323
Received: 19 December 2024; Accepted: 28 April 2025;
Published: 12 May 2025.
Edited by:
Tatjana Ilic-Tomic, University of Belgrade, SerbiaReviewed by:
Debasis Mitra, Graphic Era University, IndiaWankui Jiang, Nanjing Tech University, China
Copyright © 2025 Hu, Yuan, Fei, Imdad, Yang, Huang, Mao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Duobin Mao, bWFvZHVvYmluQDEyNi5jb20=; Jing Yang, MjAwNTA3NkB6enVsaS5lZHUuY24=
 Wei Hu
Wei Hu Jiaxing Yuan1
Jiaxing Yuan1 Jiaxiang Fei
Jiaxiang Fei Kaleem Imdad
Kaleem Imdad