- 1Center for Bio-based Chemistry, Korea Research Institute of Chemical Technology (KRICT), Ulsan, Republic of Korea
- 2Department of Biotechnology, Duksung Women’s University, Seoul, Republic of Korea
The oleaginous yeast Yarrowia lipolytica has emerged as a powerful chassis for the sustainable production of high-value nutraceuticals. Its innate metabolism, characterized by a high flux towards the key precursor acetyl-CoA, makes it an ideal host for synthesizing complex molecules like carotenoids, flavonoids, and specialty lipids. This review summarizes recent progress in engineering Y. lipolytica cell factories, focusing on the synergistic application of metabolic engineering and synthetic biology. Key strategies discussed include enhancing precursor supply, redirecting metabolic flux away from competing pathways, and optimizing heterologous gene expression. We highlight the use of advanced tools like organelle compartmentalization to improve reaction efficiency and biosensor-driven screening to accelerate strain development. Furthermore, systems biology approaches utilizing multi-omics data are proving crucial for identifying novel engineering targets and overcoming metabolic bottlenecks. This review consolidates these advancements and discusses future perspectives for creating robust, industrially-relevant Y. lipolytica platforms for the bio-based economy.
1 Introduction
The global nutraceutical market has undergone significant expansion, driven by growing consumer concerns about health and wellness. However, traditional production methods, relying on direct plant extraction or complex chemical synthesis, face several challenges including low yield, seasonal availability, environmental concerns, and scalability limitations. Microbial biosynthesis has emerged as a promising alternative for sustainable nutraceutical production, offering advantages such as controlled production conditions and reduced environmental impact (Yuan and Alper, 2019; Thakur et al., 2023).
Among various microbial platforms, Yarrowia lipolytica has been receiving remarkable attention as a versatile platform for nutraceutical production (Zhang T. L. et al., 2023). While traditional model microbial hosts like Escherichia coli and Saccharomyces cerevisiae are well-established for biotechnological production, Y. lipolytica offers unique advantages for nutraceutical synthesis. Unlike S. cerevisiae, the intrinsic ability of Y. lipolytica to accumulate large amounts of lipids provides a vast intracellular pool of the key metabolic precursor acetyl-CoA, which is a critical building block for a wide array of high-value nutraceuticals, including terpenoid, flavonoid, and sphingolipid (Sun et al., 2023). The Generally Recognized as Safe (GRAS) status of Y. lipolytica confers significant regulatory advantages for nutraceutical production, particularly when compared to E. coli, which lacks GRAS designation for food-related applications. Furthermore, Y. lipolytica exhibits exceptional metabolic flexibility and possesses sophisticated genetic engineering tools, including CRISPR-Cas9 gene editing platforms (Lee et al., 2024). These advances have facilitated the engineering of metabolic pathways for strain improvement (Park and Ledesma-Amaro, 2023). While previous reviews have provided valuable insights into its capacity, there is growing interest in the high-value nutraceuticals that offer therapeutic potential and commercial opportunities (Madhavan et al., 2023).
This review provides a comprehensive overview of Y. lipolytica’s remarkable capabilities in nutraceutical production. It focuses on acetyl-CoA–derived nutraceuticals such as terpenoids, flavonoids, and sphingolipids, excepting other classes such as polyunsaturated fatty acids, phenolics, and amino acid derivatives. By consolidating recent advancements and future research directions, we identify opportunities to translate academic potential of Y. lipolytica into robust industrial applications for nutraceutical production.
2 Advances in strategies for cell factory development
2.1 Metabolic engineering approach
The development of efficient Y. lipolytica-based cell factories relies on strategic metabolic engineering to redirect cellular resources toward target product biosynthesis. Central to this approach is the manipulation of key metabolic fluxes, particularly the acetyl-CoA pool, which serves as the primary building block for numerous nutraceutical compounds (Huang et al., 2018; Marella et al., 2018).
Several strategies have been employed to enhance acetyl-CoA availability, including engineering the lipolysis pathway and the acetyl-CoA biosynthetic pathway. Enhanced lipolysis is a direct approach to increase acetyl-CoA availability. Improvement of lipolysis critically depends on the upregulation of lipase expression, whether native or heterologous, along with the optimization of lipase secretion pathways (Fickers et al., 2011; Park and Nicaud, 2020). The β-oxidation pathway, converting fatty acids to acetyl-CoA units, is a prime target for metabolic engineering associated with acetyl-CoA pool. Peroxisomal β-oxidation involves a complex enzymatic machinery including six acyl-CoA oxidases, the bifunctional enzyme, and thiolase. Coordinated overexpression of these enzymes has been shown to substantially enhance β-oxidation capacity (Liu et al., 2022; Dong T. et al., 2025). Additionally, optimization of initial step of fatty acid activation can significantly enhance acetyl-CoA production from β-oxidation (Qin et al., 2025; Yang et al., 2025). Pyruvate dehydrogenase complex (Pdc), representing a major source of acetyl-CoA from glycolytic flux, serves as a critical control point for acetyl-CoA biosynthesis. Balanced expression of subunits (Pda1, Pdb1, and Lat1) can increase the overall capacity of this multienzyme complex (Zhou et al., 2012; Guo et al., 2014). Engineering of Pdc regulation to reduce feedback inhibition is also a key strategy for sustained acetyl-CoA production (Moreira et al., 2021). Optimization of NAD+ regeneration systems is crucial to ensure the continued operation of the Pdc under high-flux conditions (Liu et al., 2019).
Beyond the conventional pyruvate dehydrogenase route, alternative pathways for acetyl-CoA synthesis can be engineered to provide additional flux capacity. The citrate lyase pathway represents an attractive alternative, particularly for cytosolic acetyl-CoA production (Blazeck et al., 2014; Dulermo et al., 2015). Heterologous expression of ATP citrate lyase enables the conversion of citrate to acetyl-CoA and oxaloacetate, effectively transporting acetyl units from the mitochondria to the cytoplasm. Engineering of citrate transport systems and balancing of citrate cycle flux are also essential for this strategy (Zhang et al., 2014). Overexpression of acetyl-CoA carboxylase has been shown to increase malonyl-CoA pools, essential for fatty acid synthesis and subsequent lipid-derived compounds (Tai and Stephanopoulos, 2013; Qiao et al., 2015). In addition, introduction of heterologous acetyl-CoA synthetase variants has further enhanced acetyl-CoA availability (Xu et al., 2016; Liu et al., 2019). The acetyl-CoA synthetase can convert acetate directly to acetyl-CoA in an ATP-dependent manner. Engineering of acetate availability through controlled hydrolysis of acetyl-containing compounds and optimization of ATP supply can enhance contribution of this pathway to the acetyl-CoA pool (Ma et al., 2024).
Successful cell factory development requires precise control of metabolic flux distribution. Metabolic flux redirection is the process of converting the secured precursor into the target compound without loss. This is accomplished by finely tuning the expression levels of the enzymes that catalyze each step of the biosynthetic pathway. For instance, disruption of the β-oxidation pathway prevents fatty acid degradation, thereby preserving lipid precursors for nutraceutical production (Beopoulos et al., 2008; Ledesma-Amaro et al., 2016; Ledesma-Amaro and Nicaud, 2016). Additionally, balanced expression of pentose phosphate pathway (PPP) enzymes maintains adequate NADPH supply while preserving glycolytic flux for acetyl-CoA production (Beopoulos et al., 2009; Qiao et al., 2017; Wang et al., 2020).
2.2 Synthetic biology-based approach
Synthetic biology is an innovative field that designs and assembles novel genetic parts and devices to build new biological systems with functions not found in nature. It provides a powerful toolbox that elevates the predictability and efficiency of metabolic engineering beyond traditional methods, enabling the creation of sophisticated cellular systems. In Y. lipolytica, two prominent strategies are subcellular compartmentalization and the implementation of biosensors for dynamic metabolic control (Kulagina et al., 2021; Zhang T. L. et al., 2023).
The complex cellular organelle structure of Y. lipolytica, including peroxisomes and lipid droplets, offers particular opportunities for metabolic compartmentalization (Navarro-Espíndola et al., 2020). This strategy involves targeting biosynthetic pathways to specific organelles to increase substrate and enzyme concentration, isolate metabolic intermediates, and alleviate the cytotoxicity (Gu et al., 2023; Ma et al., 2024). The highly developed peroxisomal system, in particular, has been exploited (Dusseaux et al., 2020; Kulagina et al., 2021). For instance, engineering peroxisomal import mechanisms through peroxisomal targeting signal modifications has enabled the successful compartmentalization of carotenoid biosynthetic pathways, resulting in improved yields (Chen et al., 2023; Park K. et al., 2024; Soldat et al., 2024). Similarly, mitochondrial engineering has shown great potential. Targeting the mevalonate (MVA) pathway to mitochondria has been demonstrated to enhance the availability of target precursor while maintaining cellular energy homeostasis (Zhu et al., 2022; Liu L. et al., 2024).
Furthermore, the implementation of biosensors enables not only high-throughput screening (HTS) for the rapid selection of high-efficiency strains, but also dynamic real-time control of metabolic pathways (Qiu et al., 2020; Arnesen and Borodina, 2022). Transcription factor-based biosensors that respond to key metabolites such as acetyl-CoA, malonyl-CoA, and farnesyl diphosphate have been successfully developed (Wei et al., 2017; Liu S. C. et al., 2024). These biosensors can be integrated into feedback control circuits that automatically regulate gene expression in response to the intracellular concentration of a target molecule, to optimize metabolic flux and product formation.
2.3 Systems biology-based approach
To overcome the limitations of conventional metabolic engineering, systems biology-based approaches are increasingly being adopted for the rational development of Y. lipolytica cell factories. The application of systems biology through comprehensive multi-omics analysis has provided a powerful tool for identifying the engineering targets. In particular, genome-scale metabolic models (GEMs) integrated with transcriptomic, proteomic, and metabolomic data provide unprecedented insights into cellular behavior and bottlenecks (Kavscek et al., 2015; Wei et al., 2017; Yang et al., 2024).
This approach has proven highly effective across different classes of nutraceuticals. In terpenoid production, for example, comparative transcriptomics has revealed key competing pathways, enabling targeted gene deletions that significantly boost precursor flux (Worland et al., 2020; Zhang et al., 2020). Similarly, for bioactive lipids, a multi-omics analysis identified a critical link between amino acid catabolism and product synthesis, leading to gram-scale titers (Ma et al., 2020; Gasovic et al., 2023). In polyphenol biosynthesis, systems-level optimization of heterologous gene expression has been critical for balancing complex pathways and minimizing byproduct formation (Wang et al., 2022; Mitri et al., 2024). Overall, the shift towards data-driven, systems-level engineering is accelerating the development of robust strains for high-value nutraceutical production.
3 Nutraceutical production in Y. lipolytica as cell factory
3.1 Terpenoids production
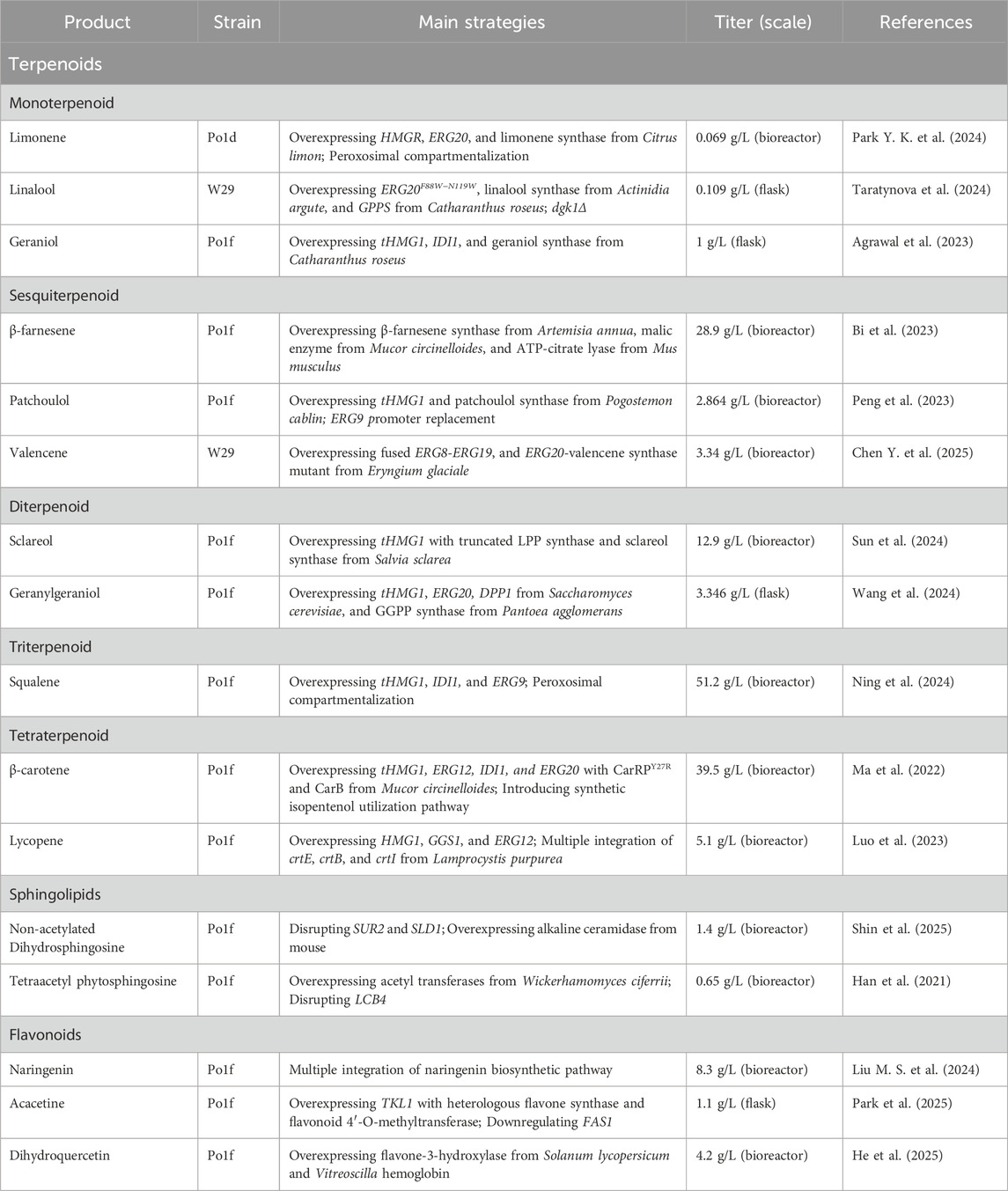
The oleaginous nature of Y. lipolytica provides intrinsic advantages for terpenoids biosynthesis, owing to its efficient lipid metabolism, which can be redirected through targeted metabolic engineering (Figure 1A). Significant progress has been made in the production of various terpenoids (Table 1).

Figure 1. Biosynthetic pathways of major nutraceuticals in Y. lipolytica. (A) Terpenoid biosynthesis: The pathway starts from acetyl-CoA and proceeds through the mevalonate (MVA) pathway to produce IPP and DMAPP, the universal C5 precursors. (B) Flavonoid biosynthesis: This pathway utilizes precursors from both the shikimate pathway and acetyl-CoA pool. (C) Sphingolipid biosynthesis: This pathway begins with serine and palmitoyl-CoA to form the basic ceramide structure. Metabolites: HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; E4P, erythrose 4-phosphate; PEP, 2-phosphoenolpyruvate; Ru5P, ribulose 5-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate 7-phosphate. Genes and Enzymes: ERG10, acetyl-CoA acetyltransferase; ERG13, 3-hydroxy-3-methylglutarylCoA synthase; HMG1, 3-hydroxy-3-methylglutarylCoA reductase; ERG12, mevalonate kinase; ERG8, phosphomevalonate kinase; ERG19, mevalonate diphosphate decarboxylase; IDI1, isopentenyl diphosphate isomerase; ERG20, geranyl/farnesyl diphosphate synthase; ERG9, squalene synthase; PS, α-pinene synthase; CS, 1,8-cinelool synthase; BFS, β-farnesene synthase; PD, phytoene desaturase; BCO, β-carotene 15,15′-dioxygenase; RDH, retinol dehydrogenase; CCD, carotenoid-cleaving dioxygenase; ARO1, ARO2, multifunctional enzymes; ARO7, chorismite mutase; TYR1, prephenate dehydrogenase; TAL, tyrosine ammonia lyase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; F4′OMT, flavonoid 4′-O-methyltransferase; F3′H, flavonoid 3′-hydroxylase; CPR, cytochrome P450 reductase; F3H, flavanone-3-hydroxylase; FLS, flavanol synthase; FMO, flavonoid 3′-monooxygenase; PT, prenyltransferase; OMT, O-methyltransferase; LCB1, LCB2, TSC3-1, TSC3-2, serine palmitoyltransferase; TSC10, 3-ketosphinanine reductase; LAC1, LAG1, TDA4, ceramide synthase; SUR2, c4-hydroxylase; SCS7, sphingolipid α-hydroxylase; DES1, sphingolipid Δ4-desaturase; YXC1, ceramidase; SLD1, sphingolipid Δ8-desaturase; MTS1, sphingolipid C9 methyltransferase; HSX11, Glucosylceramide synthase; PHSAT, phytosphingosine acetyl transferase; ACER, alkaline ceramidase.
Recent advancements in monoterpenoid production have primarily involved the heterologous expression of plant-derived monoterpene synthases (MS). Numerous efforts have focused on enhancing acetyl-CoA supply and optimizing the expression of key enzymes in the MVA pathway, such as HMG-CoA synthase and reductase. D-limonene production, for example, has been demonstrated through expressing a fused construct of Erg20 and Citrus limon D-limonene synthase (LS) along with HMG-CoA reductase, yielding up to 69.3 mg/L (Park Y. K. et al., 2024). Linalool has been produced via heterologous expression of (S)-linalool synthase (LIS) and geranyl pyrophosphate (GPP) synthase (Taratynova et al., 2024). Geraniol titers of approximately 1 g/L in flask fermentation were achieved through overexpressing of three copies of a plant-derived geraniol synthase (GS), along with single-copy of truncated HMG1 (tHMG1), IDI1, ERG10, and ERG13 (Agrawal et al., 2023). Additional monoterpenes, such as α-pinene and 1,8-cinelool, have been produced by expressing their respective synthases (Wei et al., 2021; Bhoir et al., 2025).
Sesquiterpenoid production has targeted high-value compounds such as β-farnesene, patchoulol, and valencene. By introducing heterologous malic enzyme and ATP-citrate lyase, coupled with fermentation process optimization, β-farnesene production titers reached 28.9 g/L (Bi et al., 2023). Patchoulol biosynthesis was enhanced by integrating a mutant patchoulol synthase (PCS) from Pogostemon cablin and optimizing farnesyl pyrophosphate (FPP) availability, resulting in a 1684-fold increase and final titers of 2.864 g/L in bioreactor fermentation (Peng et al., 2023). Likewise, valencene production has been achieved titers of up to 3.34 g/L by screening and selection of highly active valencene synthase (VS) (Chen Y. et al., 2025).
Diterpenoid production represents greater challenges due to the complexity of geranylgeranyl diphosphate (GGPP) biosynthesis and the requirement for specialized enzymes. Sclareol production has been demonstrated through GGPP accumulation, enabling by MVA pathway optimization and co-expression of (13E)-8α-hydroxylabden-15-yl diphosphate synthase (LPPS) and sclareol synthase (SCS) from Salvia sclarea (Sun et al., 2024; Chen J. et al., 2025). Geranylgeraniol, an important component in essential oils, has reached 3.346 g/L in the flask fermentation via overexpression of GGPP synthase (GGPPS) and a heterologous GGPP phosphatase (GGPPP) (Wang et al., 2024).
Triterpenoid production has focused on bioactive compounds. The production of squalene, a representative high-value acyclic triterpenoid compound, has exceeded 51.2 g/L by combining strategies including multi-copy gene integration, peroxisomal compartmentalization, and enhanced FPP availability (Liu Z. Y. et al., 2024; Ning et al., 2024). The biosynthesis of lupeol and betulinic acid has been achieved through expression of plant-derived lupeol synthases (LS) and cytochrome P450 monooxygenases (CYP) with cytochrome P450 reductase (CRP), along with engineering of the MVA pathway and lipid metabolism (Jin et al., 2019; Zhang et al., 2020; Li et al., 2025). However, these efforts are still constrained by the complex enzymatic machinery and cofactor requirements.
Among terpenoid classes, tetraterpenoids, particularly carotenoids, represents one of the most successful terpenoids produced in Y. lipolytica. β-carotene production has reached over 39.5 g/L through the reconstruction of the carotenoid biosynthesis pathway using phytoene synthase (PHS) and lycopene cyclase (LC) from Xanthophyllomyces dendrorhous (CrtI, and CrtYB) and Mucor circinelloides (CarB, and CarRP). This achievement was driven by optimized codon usage, balanced enzyme expression, and protein engineering to alleviate substrate inhibition (Liu et al., 2021; Ma et al., 2022). Lycopene production has been improved to over 5.1 g/L through multicopy integration of bacterial crtE, crtB, and crtI genes from Pantoea ananatis (Luo et al., 2023). Astaxanthin, a high-value carotenoid with pharmaceutical applications, has been produced at up to 2.8 g/L through the complete pathway implementation, including β-carotene hydroxylase (BCH) and ketolase (BCK) enzymes from algae (Abdullah et al., 2025). Additionally, production of zeaxanthin and β-cryptoxanthin has been demonstrated through pathway optimization (Zhang G. L. et al., 2023). Other C40 terpenoids such as retinol and β-ionone have also been produced through engineered biosynthetic pathways (Chen et al., 2022; Park et al., 2022; Ren et al., 2024; Shi et al., 2024).
3.2 Sphingolipids production
Sphingolipids are essential structural components of eukaryotic plasma membranes and play a crucial role in cellular processes such as cell signaling. Sphingolipid biosynthesis in Y. lipolytica encompasses diverse bioactive compounds with significant nutraceutical potential because this microorganism possesses native C4-desaturase gene for synthesis of sphingosine-based sphingolipids (Murakami et al., 2015; Megyeri et al., 2016; Shin et al., 2025). The foundation of sphingolipid production relies on sphingoid bases, particularly sphingosine, dihydrosphingosine, and phytosphingosine (Figure 1C).
Ceramide production represents a challenging endeavor requiring coordinated expression of multiple enzymes including serine palmitoyltransferase, 3-ketodihydrosphingosine reductase, and ceramide synthase. Recent research showed a new possibility for human glucosylceremides production in Y. lipolytica via disrupting C4-hydroxylase and Δ8 desaturase. In addition, integration of heterologous alkaline ceramidase is an important strategy to enhance production levels of non-acetylated long-chain bases, including dihydrosphingosine and sphingosine (Shin et al., 2025).
Tetraacetyl-phytosphingosine, an acetylated derivative of phytosphingosine, has been produced at up to 650 mg/L via co-expression of Wickerhamomyces ciferrii derived acetyl transferases, Sli1p and Atf2p and sphingoid long-chain base kinase gene deletion, along with optimized culture conditions (Han et al., 2021).
3.3 Flavonoids production
Recent progress has established Y. lipolytica as a promising chassis for flavonoids biosynthesis, with initial efforts has focused on the efficient production of the key precursor naringenin (Figure 1B). This was achieved by enhancing shikimate flux and increasing the availability of malonyl-CoA and erythrose-4-phosphate. Major engineering strategies included the overexpression of enzymes such as Aro4K221L, Aro2, Aro7G139S, a heterologous tyrosine ammonia lyase, and transketolase, along with downregulation of fatty acid synthase and 4-hydroxyphenylpyruvate dioxygenase (Park et al., 2025). Utilizing a high-efficiency multi-copy integration system, naringenin production was increased to 8.3 g/L in fed-batch fermentation (Liu M. S. et al., 2024).
Based on naringenin-producing strain, acacetin production has been accomplished at a titer of 1.1 g/L by introducing codon-optimized flavone synthase and flavonoid 4′-O-methyltransferase. Enzyme copy number optimization and controlled fermentation conditions further enhanced the production yield (Park et al., 2025). A similar modular engineering strategy was applied to produce dihydroquercetin. By identifying and amplifying a highly active flavanone-3-hydroxylase, and improving cofactor supply and oxygen availability, dihydroquercetin production has reached 4.2 g/L in fed-batch culture (He et al., 2025). In addition, other flavonoids, including quercetin, icaritin, and hesperetin have been successfully produced in Y. lipolytica through various engineered biosynthetic pathways (Dong Y. X. et al., 2025; Sun et al., 2025; Wang et al., 2025).
4 Conclusion and future perspectives
Y. lipolytica has established as a versatile platform for nutraceutical production, demonstrating biosynthetic capabilities across diverse compounds (Table 1). The achievement of high-value products such as β-carotene and ceramides emphasizes its significant potential for sustainable nutraceutical production (Shin et al., 2025).
Despite these promising developments, several challenges remain, including the need to achieve economically viable production levels for complex nutraceuticals while managing inherent metabolic trade-offs (Lee et al., 2025; Shin et al., 2025). However, rapid progress in synthetic biology and systems biology provide possibilities for overcoming these limitations. Current metabolic engineering strategies typically focus on individual pathways without considering global metabolic networks and regulatory mechanisms. Next-generation strain development requires integrated multi-scale modeling approaches that simultaneously account for metabolic, regulatory, and evolutionary restrictions (Kim et al., 2019; Gasovic et al., 2023; Duman-Özdamar et al., 2025).
The integration of kinetic models with regulatory networks will enable more accurate prediction of engineering outcomes, while machine learning applications to large-scale omics datasets can reveal hidden regulatory relationships and guide sophisticated control strategies. The combination of rational metabolic engineering with evolutionary approaches, particularly adaptive laboratory evolution (ALE) provides synergistic opportunities for robust strain development (Chen et al., 2024; Yook et al., 2025). ALE under production-relevant conditions can identify beneficial mutations inaccessible through rational design alone, and its integration with systematic genome editing can accelerate strain optimization.
CRISPR-based technologies beyond conventional gene editing offer opportunities for precise metabolic control (Schwartz et al., 2019). CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) systems enable reversible, tunable gene regulation without permanent modifications (Schwartz et al., 2018; Misa and Schwartz, 2021). Multiplexed CRISPR platforms allow simultaneous modulation of multiple targets, enhancing the precision and complexity of metabolic engineering strategies (Gao et al., 2016; Bae et al., 2020).
Synthetic biology facilitates the construction of sophisticated regulatory circuits, including biosensors for real-time monitoring and dynamic flux control. Improving heterologous enzyme performance through directed evolution and rational protein engineering will be crucial for enhanced productivity (Zhang and Shi, 2021; Hu et al., 2024).
Compartmentalizing biosynthetic pathways within specific organelles represents a promising strategy for reducing metabolic burden and optimizing cofactor availability (Kulagina et al., 2021). Well-developed peroxisomal and mitochondrial systems of Y. lipolytica offer excellent platforms for implementing this approach.
Beyond metabolic capabilities, practical considerations for industrial-scale production are critical. The high lipid content of Y. lipolytica can complicate the extraction and purification of the target nutraceutical from the complex biomass matrix. Therefore, developing cost-effective and efficient recovery processes is a key area of ongoing research to ensure the economic viability of Y. lipolytica-based production platforms.
The integration of these advanced approaches with a systems-level understanding holds significant promise for fully realizing the potential of Y. lipolytica in sustainable nutraceutical production. Future success will depend on continued advances in systems biology, sophisticated engineering tools, and multiple optimization strategies. Ultimately, Y. lipolytica-based production systems will play an increasingly crucial role for high-quality, sustainable nutraceuticals.
Author contributions
SL: Visualization, Writing – original draft. JL: Visualization, Writing – original draft. HP: Writing – original draft, Writing – review and editing. S-HB: Writing – original draft, Writing – review and editing, Conceptualization, Funding acquisition, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Development of an Integrated Process to Produce Lignocellulosic Biomass–derived Fermentable Sugars for Next Generation Biorefinery project (NRF-2022M3J5A1056173) from the National Research Foundation supported by the Korean Ministry of Science and ICT. This work was also supported by the Technology Innovation Program Production of 100% Bio-Diols through Highly-efficient Engineering Technology for Microorganisms (RS-2024-00488503) funded by the Ministry of Trade, Industry and Energy (MOTIE, Korea) and the Korea Research Institute of Chemical Technology through the core program (KS2542-10).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdullah, C. N., Liu, M. S., Chen, Q. H., Gao, S., Zhang, C. T., Liu, S. K., et al. (2025). Efficient production of astaxanthin in Yarrowia lipolytica through metabolic and enzyme engineering. Synth. Syst. Biotechnol. 10 (3), 737–750. doi:10.1016/j.synbio.2025.02.014
Agrawal, A., Yang, Z., and Blenner, M. (2023). Engineering Yarrowia lipolytica for the biosynthesis of geraniol. Metab. Eng. Commun. 17, e00228. doi:10.1016/j.mec.2023.e00228
Arnesen, J. A., and Borodina, I. (2022). Engineering of Yarrowia lipolytica for terpenoid production. Metab. Eng. Commun. 15, e00213. doi:10.1016/j.mec.2022.e00213
Bae, S. J., Park, B. G., Kim, B. G., and Hahn, J. S. (2020). Multiplex gene disruption by targeted base editing of Yarrowia lipolytica genome using cytidine deaminase combined with the CRISPR/Cas9 system. Biotechnol. J. 15 (1), e1900238. doi:10.1002/biot.201900238
Beopoulos, A., Mrozova, Z., Thevenieau, F., Le Dall, M. T., Hapala, I., Papanikolaou, S., et al. (2008). Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 74 (24), 7779–7789. doi:10.1128/Aem.01412-08
Beopoulos, A., Cescut, J., Haddouche, R., Uribelarrea, J. L., Molina-Jouve, C., and Nicaud, J. M. (2009). Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48 (6), 375–387. doi:10.1016/j.plipres.2009.08.005
Bhoir, K., Prakash, G., and Odaneth, A. (2025). Genetic engineering of Yarrowia lipolytica for 1,8-cineole production: a sustainable approach. Enzyme Microb. Technol. 189. doi:10.1016/j.enzmictec.2025.110659
Bi, H. R., Xu, C. C., Bao, Y. F., Zhang, C. W., Wang, K., Zhang, Y., et al. (2023). Enhancing precursor supply and modulating metabolism to achieve high-level production of ß-farnesene in Yarrowia lipolytica. Bioresour. Technol. 382. doi:10.1016/j.biortech.2023.129171
Blazeck, J., Hill, A., Liu, L. Q., Knight, R., Miller, J., Pan, A., et al. (2014). Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 5, 3131. doi:10.1038/ncomms4131
Chen, S. Y., Lu, Y. P., Wang, W., Hu, Y. Z., Wang, J. F., Tang, S. X., et al. (2022). Efficient production of the β-ionone aroma compound from organic waste hydrolysates using an engineered strain. Front. Microbiol. 13. doi:10.3389/fmicb.2022.960558
Chen, L., Xiao, W., Yao, M., Wang, Y., and Yuan, Y. (2023). Compartmentalization engineering of yeasts to overcome precursor limitations and cytotoxicity in terpenoid production. Front. Bioeng. Biotechnol. 11, 1132244. doi:10.3389/fbioe.2023.1132244
Chen, C., Li, Y. W., Chen, X. Y., Wang, Y. T., Ye, C., and Shi, T. Q. (2024). Application of adaptive laboratory evolution for Yarrowia lipolytica: a comprehensive review. Bioresour. Technol. 391. doi:10.1016/j.biortech.2023.129893
Chen, J., Huang, L. Z., Ye, B. C., and Zhou, Y. (2025). Combinatorial metabolic engineering of Yarrowia lipolytica for high-level production of the plant-derived diterpenoid sclareol. Microb. Cell. Fact. 24 (1). doi:10.1186/s12934-025-02744-7
Chen Y., Y., Su, L., Liu, Q., Zhang, G., Chen, H., Wang, Q., et al. (2025). Triune engineering approach for (+)-valencene overproduction in Yarrowia lipolytica. Biotechnol. J. 20 (1), e202400669. doi:10.1002/biot.202400669
Dong, T., Shu, Y., Wang, Y., Yao, M., and Xiao, W. (2025). An engineered Yarrowia lipolytica with rapid growth and efficient lipid utilization. Synth. Syst. Biotechnol. 10 (2), 495–503. doi:10.1016/j.synbio.2025.01.007
Dong Y. X., Y. X., Wei, W. P., Li, M. F., Qian, T., Xu, J. Y., Chu, X. H., et al. (2025). De novo biosynthesis of quercetin in Yarrowia lipolytica through systematic metabolic engineering for enhanced yield. Bioresour. Bioprocess. 12 (1). doi:10.1186/s40643-024-00825-w)
Dulermo, T., Lazar, Z., Dulermo, R., Rakicka, M., Haddouche, R., and Nicaud, J. M. (2015). Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1851 (9), 1107–1117. doi:10.1016/j.bbalip.2015.04.007
Duman-Özdamar, Z. E., Julsing, M. K., dos Santos, V. A. P. M., Hugenholtz, J., and Suarez-Diez, M. (2025). Model-driven engineering of Yarrowia lipolytica for improved microbial oil production. Microb. Biotechnol. 18 (3). doi:10.1111/1751-7915.70089
Dusseaux, S., Wajn, W. T., Liu, Y., Ignea, C., and Kampranis, S. C. (2020). Transforming yeast peroxisomes into microfactories for the efficient production of high-value isoprenoids. Proc. Natl. Acad. Sci. U. S. A. 117 (50), 31789–31799. doi:10.1073/pnas.2013968117
Fickers, P., Marty, A., and Nicaud, J. M. (2011). The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol. Adv. 29 (6), 632–644. doi:10.1016/j.biotechadv.2011.04.005
Gao, S., Tong, Y., Wen, Z., Zhu, L., Ge, M., Chen, D., et al. (2016). Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J. Ind. Microbiol. Biotechnol. 43 (8), 1085–1093. doi:10.1007/s10295-016-1789-8
Gasovic, S. J., Dietrich, D., Gläser, L., Cao, P., Kohlstedt, M., and Wittmann, C. (2023). Multi-omics view of recombinant Yarrowia lipolytica: enhanced ketogenic amino acid catabolism increases polyketide-synthase-driven docosahexaenoic production to high selectivity at the gram scale. Metab. Eng. 80, 45–65. doi:10.1016/j.ymben.2023.09.003
Gu, Y., Lu, X. Y., Liu, T. K., Song, Y. J., Sang, E., Ding, S. R., et al. (2023). Engineering the oleaginous yeast Yarrowia lipolytica to produce nutraceuticals: from metabolic design to industrial applications. Food Bioeng. 2 (3), 187–199. doi:10.1002/fbe2.12062
Guo, H. W., Madzak, C., Du, G. C., Zhou, J. W., and Chen, J. (2014). Effects of pyruvate dehydrogenase subunits overexpression on the α-ketoglutarate production in Yarrowia lipolytica WSH-Z06. Appl. Microbiol. Biotechnol. 98 (16), 7003–7012. doi:10.1007/s00253-014-5745-0
Han, C., Jang, M., Kim, M. J., Han, M. H., Lee, K. R., Hahn, J. S., et al. (2021). Engineering Yarrowia lipolytica for de novo production of tetraacetyl phytosphingosine. J. Appl. Microbiol. 130 (6), 1981–1992. doi:10.1111/jam.14931
He, Q., Yue, M. Y., Liu, M. S., Ren, X. F., Wang, H. J., Chen, J. B., et al. (2025). De novo biosynthesis of dihydroquercetin in an engineered Yarrowia lipolytica with enhanced (2S)-eriodictyol. Food Biosci. 64. doi:10.1016/j.fbio.2025.105912
Hu, M., Ge, J., Jiang, Y., Sun, X., Guo, D., and Gu, Y. (2024). Advances and perspectives in genetic expression and operation for the oleaginous yeast Yarrowia lipolytica. Synth. Syst. Biotechnol. 9 (4), 618–626. doi:10.1016/j.synbio.2024.05.003
Huang, Y. Y., Jian, X. X., Lv, Y. B., Nian, K. Q., Gao, Q., Chen, J., et al. (2018). Enhanced squalene biosynthesis in Yarrowia lipolytica based on metabolically engineered acetyl-CoA metabolism. J. Biotechnol. 281, 106–114. doi:10.1016/j.jbiotec.2018.07.001
Jin, C. C., Zhang, J. L., Song, H., and Cao, Y. X. (2019). Boosting the biosynthesis of betulinic acid and related triterpenoids in Yarrowia lipolytica via multimodular metabolic engineering. Microb. Cell. Fact. 18, 77. doi:10.1186/s12934-019-1127-8
Kavscek, M., Bhutada, G., Madl, T., and Natter, K. (2015). Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst. Biol. 9, 72. doi:10.1186/s12918-015-0217-4
Kim, M., Park, B. G., Kim, E. J., Kim, J., and Kim, B. G. (2019). In silico identification of metabolic engineering strategies for improved lipid production in Yarrowia lipolytica by genome-scale metabolic modeling. Biotechnol. Biofuels 12, 187. doi:10.1186/s13068-019-1518-4
Kulagina, N., Besseau, S., Papon, N., and Courdavault, V. (2021). Peroxisomes: a new hub for metabolic engineering in yeast. Front. Bioeng. Biotechnol. 9, 659431. doi:10.3389/fbioe.2021.659431
Ledesma-Amaro, R., and Nicaud, J. M. (2016). Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 61, 40–50. doi:10.1016/j.plipres.2015.12.001
Ledesma-Amaro, R., Lazar, Z., Rakicka, M., Guo, Z., Fouchard, F., Coq, A. C., et al. (2016). Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 38, 115–124. doi:10.1016/j.ymben.2016.07.001
Lee, S. M., Lee, J. Y., Hahn, J. S., and Baek, S. H. (2024). Engineering of Yarrowia lipolytica as a platform strain for producing adipic acid from renewable resource. Bioresour. Technol. 391. doi:10.1016/j.biortech.2023.129920
Lee, H., Song, J., and Seo, S. W. (2025). Engineering Yarrowia lipolytica for the production of beta-carotene by carbon and redox rebalancing. J. Biol. Eng. 19 (1), 6. doi:10.1186/s13036-025-00476-1
Li, X. Y., Jiao, L. C., Zhao, G. W., Li, Y. C., Yan, Y. J., and Yan, J. Y. (2025). Multidimensional metabolic engineering of Yarrowia lipolytica for highly efficient biosynthesis of betulinic acid. Green Chem. 27 (23), 6855–6868. doi:10.1039/d5gc01276g
Liu, H., Marsafari, M., Wang, F., Deng, L., and Xu, P. (2019). Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab. Eng. 56, 60–68. doi:10.1016/j.ymben.2019.08.017
Liu, M. M., Zhang, J., Ye, J. R., Qi, Q. S., and Hou, J. (2021). Morphological and metabolic engineering of Yarrowia lipolytica to increase β-carotene production. ACS Synth. Biol. 10 (12), 3551–3560. doi:10.1021/acssynbio.1c00480
Liu, Y., Zhang, J., Li, Q., Wang, Z., Cui, Z., Su, T., et al. (2022). Engineering Yarrowia lipolytica for the sustainable production of beta-farnesene from waste oil feedstock. Biotechnol. Biofuels Bioprod. 15 (1), 101. doi:10.1186/s13068-022-02201-2
Liu, L., Zhao, K., and Liu, Z. (2024). Construction and regulation of the abscisic acid biosynthesis pathway in Yarrowia lipolytica. J. Agric. Food. Chem. 72 (13), 7299–7307. doi:10.1021/acs.jafc.4c00223
Liu, M. S., Wu, J. J., Yue, M. Y., Ning, Y., Guan, X., Gao, S., et al. (2024). YaliCMulti and YaliHMulti: stable, efficient multi-copy integration tools for engineering Yarrowia lipolytica. Metab. Eng. 82, 29–40. doi:10.1016/j.ymben.2024.01.003
Liu, S. C., Xu, L., Sun, Y., Yuan, L., Xu, H., Song, X., et al. (2024). Progress in the metabolic engineering of Yarrowia lipolytica for the synthesis of terpenes. Biodes. Res. 6, 0051. doi:10.34133/bdr.0051
Liu, Z. Y., Huang, M. K., Chen, H., Lu, X. Y., Tian, Y., Hu, P. C., et al. (2024). Metabolic engineering of Yarrowia lipolytica for high-level production of squalene. Bioresour. Technol. 394. doi:10.1016/j.biortech.2023.130233
Luo, Z., Shi, J. T., Chen, X. L., Chen, J., Liu, F., Wei, L. J., et al. (2023). Iterative gene integration mediated by 26S rDNA and non-homologous end joining for the efficient production of lycopene in Yarrowia lipolytica. Bioresour. Bioprocess. 10 (1). doi:10.1186/s40643-023-00697-6
Ma, J., Gu, Y., Marsafari, M., and Xu, P. (2020). Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J. Ind. Microbiol. Biotechnol. 47 (9-10), 845–862. doi:10.1007/s10295-020-02290-8
Ma, Y. S., Liu, N., Greisen, P., Li, J. B., Qiao, K. J., Huang, S. W., et al. (2022). Removal of lycopene substrate inhibition enables high carotenoid productivity in Yarrowia lipolytica. Nat. Commun. 13 (1). doi:10.1038/s41467-022-28277-w
Ma, Y., Shang, Y., and Stephanopoulos, G. (2024). Engineering peroxisomal biosynthetic pathways for maximization of triterpene production in Yarrowia lipolytica. Proc. Natl. Acad. Sci. U. S. A. 121 (5), e2314798121. doi:10.1073/pnas.2314798121
Madhavan, A., Arun, K. B., Alex, D., Anoopkumar, A. N., Emmanual, S., Chaturvedi, P., et al. (2023). Microbial production of nutraceuticals: metabolic engineering interventions in phenolic compounds, poly unsaturated fatty acids and carotenoids synthesis. J. Food. Sci. Technol. 60 (8), 2092–2104. doi:10.1007/s13197-022-05482-5
Marella, E. R., Holkenbrink, C., Siewers, V., and Borodina, I. (2018). Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr. Opin. Biotechnol. 50, 39–46. doi:10.1016/j.copbio.2017.10.002
Megyeri, M., Riezman, H., Schuldiner, M., and Futerman, A. H. (2016). Making sense of the yeast sphingolipid pathway. J. Mol. Biol. 428 (24), 4765–4775. doi:10.1016/j.jmb.2016.09.010
Misa, J., and Schwartz, C. (2021). CRISPR interference and activation to modulate transcription in Yarrowia lipolytica. Methods Mol. Biol. 2307, 95–109. doi:10.1007/978-1-0716-1414-3_6
Mitri, S., Louka, N., Rossignol, T., Maroun, R. G., and Koubaa, M. (2024). Bioproduction of 2-phenylethanol by Yarrowia lipolytica on sugar beet molasses as a low-cost substrate. Fermentation 10 (6). doi:10.3390/fermentation10060290
Moreira, J. D., Jolicoeur, M., Schwartz, L., and Peres, S. (2021). Fine-tuning mitochondrial activity in Yarrowia lipolytica for citrate overproduction. Sci. Rep. 11 (1). doi:10.1038/s41598-020-79577-4
Murakami, S., Shimamoto, T., Nagano, H., Tsuruno, M., Okuhara, H., Hatanaka, H., et al. (2015). Producing human ceramide-NS by metabolic engineering using yeast Saccharomyces cerevisiae. Sci. Rep. 5, 16319. doi:10.1038/srep16319
Navarro-Espíndola, R., Suaste-Olmos, F., and Peraza-Reyes, L. (2020). Dynamic regulation of peroxisomes and mitochondria during fungal development. J. Fungi 6 (4), ARTN 302. doi:10.3390/jof6040302
Ning, Y., Liu, M. S., Ru, Z. Y., Zeng, W. Z., Liu, S., and Zhou, J. W. (2024). Efficient synthesis of squalene by cytoplasmic-peroxisomal engineering and regulating lipid metabolism in. Bioresour. Technol. 395. doi:10.1016/j.biortech.2024.130379
Park, Y. K., and Ledesma-Amaro, R. (2023). What makes Yarrowia lipolytica well suited for industry? Trends Biotechnol. 41 (2), 242–254. doi:10.1016/j.tibtech.2022.07.006
Park, Y. K., and Nicaud, J. M. (2020). Metabolic engineering for unusual lipid production in Yarrowia lipolytica. Microorganisms 8 (12). doi:10.3390/microorganisms8121937
Park, H., Lee, D., Kim, J. E., Park, S., Park, J. H., Ha, C. W., et al. (2022). Efficient production of retinol in Yarrowia lipolytica by increasing stability using antioxidant and detergent extraction. Metab. Eng. 73, 26–37. doi:10.1016/j.ymben.2022.06.001
Park K., K., Kim, G., Cha, S., Ham, Y., and Hahn, J. S. (2024). Efficient production of the colorless carotenoid phytoene in Yarrowia lipolytica through metabolic engineering. J. Agric. Food. Chem. 72 (48), 26786–26795. doi:10.1021/acs.jafc.4c07735
Park, Y. K., Vidal, L. S., Bell, D., Zabret, J., Soldat, M., Kavscek, M., et al. (2024). Efficient synthesis of limonene production in Yarrowia lipolytica by combinatorial engineering strategies. Biotechnol. Biofuels Bioprod. 17 (1). doi:10.1186/s13068-024-02535-z
Park, N., Ham, Y., Cha, S., Jang, B., Kim, G., Baek, S. H., et al. (2025). Metabolic engineering of Yarrowia lipolytica for enhanced production of naringenin-derived flavonoids: Apigenin and acacetin. J. Agric. Food. Chem. 73 (23), 14420–14431. doi:10.1021/acs.jafc.5c02857
Peng, Q. Q., Guo, Q., Chen, C., Song, P., Wang, Y. T., Ji, X. J., et al. (2023). High-level production of patchoulol in Yarrowia lipolytica via systematic engineering strategies. J. Agric. Food. Chem. 71 (11), 4638–4645. doi:10.1021/acs.jafc.3c00222
Qiao, K., Imam Abidi, S. H., Liu, H., Zhang, H., Chakraborty, S., Watson, N., et al. (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 29, 56–65. doi:10.1016/j.ymben.2015.02.005
Qiao, K. J., Wasylenko, T. M., Zhou, K., Xu, P., and Stephanopoulos, G. (2017). Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 35 (2), 173–177. doi:10.1038/nbt.3763
Qin, J., Liu, N., Abid, U., Coleman, S. M., Wang, Y., Fu, Q., et al. (2025). Metabolic engineering of Yarrowia lipolytica for conversion of waste cooking oil into omega-3 eicosapentaenoic acid. ACS Eng. Au 5 (2), 128–139. doi:10.1021/acsengineeringau.4c00053
Qiu, X. L., Xu, P., Zhao, X. R., Du, G. C., Zhang, J., and Li, J. H. (2020). Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab. Eng. 60, 66–76. doi:10.1016/j.ymben.2020.03.006
Ren, X. F., Liu, M. S., Yue, M. Y., Zeng, W. Z., Zhou, S. H., Zhou, J. W., et al. (2024). Metabolic pathway coupled with fermentation process optimization for high-level production of retinol in Yarrowia lipolytica. J. Agric. Food. Chem. 72 (15), 8664–8673. doi:10.1021/acs.jafc.4c00377
Schwartz, C., Curtis, N., Lobs, A. K., and Wheeldon, I. (2018). Multiplexed CRISPR activation of cryptic sugar metabolism enables Yarrowia lipolytica growth on cellobiose. Biotechnol. J. 13 (9), e1700584. doi:10.1002/biot.201700584
Schwartz, C., Cheng, J. F., Evans, R., Schwartz, C. A., Wagner, J. M., Anglin, S., et al. (2019). Validating genome-wide CRISPR-Cas9 function improves screening in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 55, 102–110. doi:10.1016/j.ymben.2019.06.007
Shi, J., Wu, Y. Y., Sun, R. Z., Hua, Q., and Wei, L. J. (2024). Synthesis of β-ionone from xylose and lignocellulosic hydrolysate in genetically engineered oleaginous yeast. Biotechnol. Lett. 46 (6), 1219–1236. doi:10.1007/s10529-024-03534-8
Shin, S. H., Moon, H. Y., Park, H. E., Nam, G. J., Baek, J. H., Jeon, C. O., et al. (2025). Elucidation and engineering of sphingolipid biosynthesis pathway in Yarrowia lipolytica for enhanced production of human-type sphingoid bases and glucosylceramides. Metab. Eng. 87, 68–85. doi:10.1016/j.ymben.2024.11.013
Soldat, M., Markus, T., Magdevska, V., Kavscek, M., Kruis, A. J., Horvat, J., et al. (2024). Screening of novel β-carotene hydroxylases for the production of β-cryptoxanthin and zeaxanthin and the impact of enzyme localization and crowding on their production in Yarrowia lipolytica. Microb. Cell. Fact. 23 (1), 298. doi:10.1186/s12934-024-02569-w
Sun, M. L., Gao, X. X., Lin, L., Yang, J., Ledesma-Amaro, R., and Ji, X. J. (2023). Building Yarrowia lipolytica cell factories for advanced biomanufacturing: challenges and solutions. J. Agric. Food. Chem. 72 (1), 94–107. doi:10.1021/acs.jafc.3c07889
Sun, M. L., Han, Y. T., Yu, X., Wang, K. F., Lin, L., Ledesma-Amaro, R., et al. (2024). Constructing a green oleaginous yeast cell factory for sustainable production of the plant-derived diterpenoid sclareol. Green Chem. 26 (9). doi:10.1039/d3gc04949c
Sun, W. Z., Wang, X., Fu, M. Y., Liu, L. F., Zhang, P., Yin, B. C., et al. (2025). Metabolic engineering of Yarrowia lipolytica for enhanced de novo biosynthesis of Icaritin. ACS Synth. Biol. 14 (4), 1142–1151. doi:10.1021/acssynbio.4c00754
Tai, M., and Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15, 1–9. doi:10.1016/j.ymben.2012.08.007
Taratynova, M. O., Tikhonova, E. E., Fedyaeva, I. M., Dementev, D. A., Yuzbashev, T. V., Solovyev, A. I., et al. (2024). Boosting geranyl diphosphate synthesis for linalool production in engineered Yarrowia lipolytica. Appl. Biochem. Biotechnol. 196 (3), 1304–1315. doi:10.1007/s12010-023-04581-z
Thakur, B., Kaur, S., Rani, N., Kaur, R., Upadhyay, S. K., and Tripathi, M. (2023). Exploring microbial contributions to nutraceutical production: from ntural to dsigned foods. Mol. Biotechnol. 67. doi:10.1007/s12033-023-00937-2
Wang, J. P., Ledesma-Amaro, R., Wei, Y. J., Ji, B. Y., and Ji, X. J. (2020). Metabolic engineering for increased lipid accumulation in yarrowia lipolytica-A review. Bioresour. Technol. 313, 123707. doi:10.1016/j.biortech.2020.123707
Wang, Y. A., Liu, X. A., Chen, B. H., Liu, W., Guo, Z. K., Liu, X. Y., et al. (2022). Metabolic engineering of Yarrowia lipolytica for scutellarin production. Synth. Syst. Biotechnol. 7 (3), 958–964. doi:10.1016/j.synbio.2022.05.009
Wang, K. F., Yin, M. X., Sun, M. L., Zhao, Q. Y., Ledesma-Amaro, R., Ji, X. J., et al. (2024). Engineering Yarrowia lipolytica for efficient synthesis of geranylgeraniol. J. Agric. Food. Chem. 72 (37), 20568–20581. doi:10.1021/acs.jafc.4c06749
Wang, Y. Y., Huang, R. Q., Gao, S., Yue, M. Y., Zhang, X., Zeng, W. Z., et al. (2025). Identification of two new flavone 4′-O-methyltransferases and their application in de novo biosynthesis of (2S)-hesperetin in Yarrowia lipolytica. Synth. Syst. Biotechnol. 10 (3), 728–736. doi:10.1016/j.synbio.2025.03.003
Wei, S. S., Jian, X. X., Chen, J., Zhang, C., and Hua, Q. (2017). Reconstruction of genome-scale metabolic model of Yarrowia lipolytica and its application in overproduction of triacylglycerol. Bioresour. Bioprocess. 4. doi:10.1186/s40643-017-0180-6
Wei, L. J., Zhong, Y. T., Nie, M. Y., Liu, S. C., and Hua, Q. (2021). Biosynthesis of alpha-Pinene by genetically engineered Yarrowia lipolytica from low-cost renewable feedstocks. J. Agric. Food. Chem. 69 (1), 275–285. doi:10.1021/acs.jafc.0c06504
Worland, A. M., Czajka, J. J., Xing, Y., Harper, W. F., Moore, A., Xiao, Z. Y., et al. (2020). Analysis of Yarrowia lipolytica growth, catabolism, and terpenoid biosynthesis during utilization of lipid-derived feedstock. Metab. Eng. Commun. 11, e00130. doi:10.1016/j.mec.2020.e00130
Xu, P., Qiao, K. J., Ahn, W. S., and Stephanopoulos, G. (2016). Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. U. S. A. 113 (39), 10848–10853. doi:10.1073/pnas.1607295113
Yang, S. L., Pan, X. W., You, J. J., Guo, B. M., Liu, Z. Y., Cao, Y., et al. (2024). Systematic metabolic engineering of Yarrowia lipolytica for the enhanced production of erythritol. Bioresour. Technol. 391. doi:10.1016/j.biortech.2023.129918
Yang, Q., Tian, M., Dong, P., Zhao, Y., and Deng, Y. (2025). Engineering Yarrowia lipolytica to enhance the production of malonic acid via malonyl-CoA pathway at high titer. Adv. Sci. 12 (12), e2411665. doi:10.1002/advs.202411665
Yook, S., Deewan, A., Ziolkowski, L., Lane, S., Tohidifar, P., Cheng, M. H., et al. (2025). Engineering and evolution of Yarrowia lipolytica for producing lipids from lignocellulosic hydrolysates. Bioresour. Technol. 416. doi:10.1016/j.biortech.2024.131806
Yuan, S. F., and Alper, H. S. (2019). Metabolic engineering of microbial cell factories for production of nutraceuticals. Microb. Cell. Fact. 18 (1), 46. doi:10.1186/s12934-019-1096-y
Zhang, Y., and Shi, S. (2021). Transcription factor-based biosensor for dynamic control in yeast for natural product synthesis. Front. Bioeng. Biotechnol. 9, 635265. doi:10.3389/fbioe.2021.635265
Zhang, H. Y., Zhang, L. N., Chen, H. Q., Chen, Y. Q., Chen, W., Song, Y. D., et al. (2014). Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP:citrate lyase from Mus musculus. J. Biotechnol. 192, 78–84. doi:10.1016/j.jbiotec.2014.10.004
Zhang, J. L., Bai, Q. Y., Peng, Y. Z., Fan, J., Jin, C. C., Cao, Y. X., et al. (2020). High production of triterpenoids in Yarrowia lipolytica through manipulation of lipid components. Biotechnol. Biofuels 13, 133. doi:10.1186/s13068-020-01773-1
Zhang, G. L., Chen, J., Wang, Y. Z., Liu, Z., and Mao, X. Z. (2023). Metabolic engineering of Yarrowia lipolytica for zeaxanthin production. J. Agric. Food. Chem. 71 (37), 13828–13837. doi:10.1021/acs.jafc.3c01772
Zhang T. L., T. L., Yu, H. W., and Ye, L. D. (2023). Metabolic engineering of Yarrowia lipolytica for terpenoid production: tools and strategies. ACS Synth. Biol. 12 (3), 639–656. doi:10.1021/acssynbio.2c00569
Zhou, J. W., Yin, X. X., Madzak, C., Du, G. C., and Chen, J. (2012). Enhanced α-ketoglutarate production in Yarrowia lipolytica WSH-Z06 by alteration of the acetyl-CoA metabolism. J. Biotechnol. 161 (3), 257–264. doi:10.1016/j.jbiotec.2012.05.025
Keywords: nutraceutical, yarrowia lipolytica, microbial cell factory, sphingolipid, terpenoid, flavonoid
Citation: Lee S, Lee JH, Park HJ and Baek S-H (2025) Yarrowia lipolytica as a promising cell factory for microbial production of value-added nutraceuticals . Front. Bioeng. Biotechnol. 13:1673169. doi: 10.3389/fbioe.2025.1673169
Received: 25 July 2025; Accepted: 18 August 2025;
Published: 28 August 2025.
Edited by:
Sungmin Hwang, Korea Maritime and Ocean University, Republic of KoreaReviewed by:
Eui-Sang Cho, Graduate School of Incheon National University, Republic of KoreaKiran Kumar, United Arab Emirates University, United Arab Emirates
Copyright © 2025 Lee, Lee, Park and Baek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seung-Ho Baek, YmFla3NoQGtyaWN0LnJlLmty
†These authors share first authorship
 Soseon Lee1†
Soseon Lee1† Seung-Ho Baek
Seung-Ho Baek