- 1Program in Physiology and Experimental Medicine, Peter Gilgan Centre for Research and Learning, SickKids Research Institute, SickKids Hospital, Toronto, ON, Canada
- 2Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada
E26 transformation-specific (Ets) family of transcription factors are characterized by the presence of Ets-DNA binding domain and have been found to be highly involved in hematopoiesis and various tissue differentiation. ESE-1, or Elf3 in mice, is a member of epithelium-specific Ets sub-family which is most prominently expressed in epithelial tissues such as the gut, mammary gland, and lung. The role of ESE-1 during embryogenesis had long been alluded from 30% fetal lethality in homozygous knockout mice and its high expression in preimplantation mouse embryos, but there has been no in-depth of analysis of ESE-1 function in early development. With improved proteomics, gene editing tools and increasing knowledge of ESE-1 function in adult tissues, we hereby propose future research directions for the study of ESE-1 in embryogenesis, including studying its regulation at the protein level and at the protein family level, as well as better defining the developmental phase under investigation. Understanding the role of ESE-1 in early development will provide new insights into its involvement in tissue regeneration and cancer, as well as how it functions with other Ets factors as a protein family.
Introduction
E26 transformation-specific (Ets) transcription factors are characterized by the presence of conserved Ets-DNA binding domain which recognizes a core sequence of GGAA/T, consisting a protein family of at least 27 members in human and 26 in mice (Bult et al., 2008; Hollenhorst et al., 2011). Numerous studies in cell lines and gene disruption animal models have identified function of individual ETS factors as well as their evolutionary relationships, which are reviewed elsewhere (Bartel et al., 2000; Sharrocks, 2001; Oikawa and Yamada, 2003; Gutierrez-Hartmann et al., 2007). However, most studies have focused on single-protein analysis or in vitro DNA binding assays that do not reveal Ets proteins acting in vivo (Sementchenko and Watson, 2000), when global analyses of Ets factors have revealed that over 16 of Ets members are co-expressed in a given cell type and more than 8 ubiquitously so (Hollenhorst et al., 2004). The potential Ets targets can extensively overlap (Hollenhorst et al., 2007) due to the conservation of mode and specificity of the Ets-DNA binding domain (Sharrocks, 2001; Wei et al., 2010).
Ets factors have been divided into four sub-groups by difference in preferred Ets-binding motifs, which were determined by variations in amino-acids that interact with the backbone of the core recognition sequence (Wei et al., 2010). Nevertheless, there is still redundancy within sub-groups with factors binding to the same cognition signal. Class I factors, for example, include at least 11 Ets factors and mainly recognize CCGGAA/T, while Class III factors, which include ESE-1 and at least 5 others, demonstrate strong preference for GCGGAAC (Wei et al., 2010). Therefore, other determinants of specificity, such as protein-protein interactions with other transcription factors via distinct protein domains and additional regulatory sequences flanking the core Ets-binding site on target genes, are as important to recognize how Ets factors function as a family within a cell. This means that manipulation of a single Ets factor may result in a mitigated phenotype due to functional compensation from other members. This particularly applies to embryogenesis, where some Ets factors have been shown to be redundant (Kageyama et al., 2006). Therefore, more holistic approaches are required to understand the interplay of Ets factors during early development.
In this Perspective, we focus on a well-characterized member of a subfamily of Ets factors called ESE-1, or Epithelium-specific Ets transcription factor 1, to provide an overall direction of future research on ESE-1 and other Ets factors in fetal development. ESE-1 homolog in mice is known as Elf3. Epithelial-specific Ets factors, or ESEs, constitute a subfamily of Ets factors which include ESE-1/Elf3, ESE-2/Elf5, ESE-3/Ehf, and PDEF, whose expression patterns are known to be mostly restricted to epithelial tissues (Sharrocks, 2001). The role of ESE-1 is more thoroughly reviewed elsewhere (Oliver et al., 2012), and has been studied primarily in epithelium differentiation. ESE-1 therefore has received less attention regarding its role in early development, despite evidence suggesting its involvement as discussed below. With recent advances in proteomics and genomics as well as gene editing tools such as CRISPR/Cas, it is time to build more knowledge on how ESE-1 is modulated at protein level and how it functions in combination with other Ets factors during embryogenesis. Analysis of Ets intra-family interaction and regulation during embryogenesis will provide better insight into other protein families that are likewise comprised of multiple and redundant members such as HOX, GATA, and FOX proteins which share highly similar DNA-binding domains (Messina et al., 2004), as well as providing functional insight into the role of ESE-1 in cancer and tissue regeneration.
The Role of ESE-1 in Embryonic Development
The involvement of ESE-1 in early development was first identified in the diminished survival of Elf3 homozygous knockout mice, where targeted deletion of Elf3 resulted in 30% of fetal lethality at around embryonic day 11.5 (E11.5) (Ng et al., 2002). However, no gross histopathological anomalies were found to explain the cause of death (Ng et al., 2002). Similarly, disruption of Elf3 by siRNA injection reduced embryo survival by 50%, while co-suppression of three prominent Ets factors expressed in mouse preimplantation embryos Elf3, Etsrp71, and Spic blocked development to blastocysts by 70%, involving reduction of key regulatory genes eIF-1A and Oct3/4 that contain Ets-binding sites (EBS) in their promoters (Kageyama et al., 2006). Recent reports have also shown inhibitory effect of ESE-1 on stem cell transcription factor Oct4 expression by binding to EBS in the conserved region 2 (CR2) of the Oct4 promoter during retinoic acid (RA)-induced differentiation of human embryonic carcinoma cell line NCCIT (Park et al., 2014). Oct4 is essential in the maintenance of stem cell pluripotency and its loss of expression leads to cell differentiation (Pan et al., 2002). Therefore, the inverse relationship between Elf3 and Oct4 and the close involvement of ESE-1 in epithelial cell differentiation in various tissue types such as skin, small intestine and mammary gland may indicate that ESE-1 is a marker of epithelial tissue development. However, the exact gene targets or functional mechanism of ESE-1/Elf3 during early development have not been elucidated.
Regulation of ESE-1 Expression
Promoter analysis of ESE-1 has revealed four main regulatory regions Ets, CAAT, TATA, and NFκB (Oettgen et al., 1999). However, different regions of the upstream segments of ESE-1 gene may be more critical than others during development and differentiation. For example, Hou et al. demonstrated that in retinoic acid-differentiated murine embryonal carcinoma (EC) cell line F9, there is a substantial increase in the utilization of an upstream regulator region approximately 2 kb upstream of the transcription start site that was responsible for the increase in the ESE-1 promoter site (Hou et al., 2004). While it is well-known that long-range enhancers have important role in mammalian gene regulation (Heintzman and Ren, 2009), factors which bind to the enhancer region of ESE-1 gene have not been well-defined. A conserved 30 base pair (bp) ESE-1 enhancer sequence (ESS) was also shown to be essential in regulating ESE-1 transcription in response to epithelial differentiation signals, controlled by two ESS-binding protein complexes that are of uncertain identity (Neve et al., 2006). In mouse embryo, elf3 has been shown to be expressed at low levels in mouse meiosis II (MII) oocytes, and increase at one-cell stage embryo, briefly decline at the two-cell stage, and increase again until the blastocyst stage (Kageyama et al., 2006). Therefore, more investigation is needed to elucidate the ESE-1 enhancer elements and their binding nuclear factors, as well as signals which activate ESE-1 gene transcription during embryogenesis. The most notable regulator of ESE-1 so far known is NFκB, which is required for ESE-1 induction in response to pro-inflammatory cytokines (Rudders et al., 2001; Wu et al., 2008). Whether this applies to embryogenesis, however, is unknown. Additionally, ESE-1 has been shown to downregulate its own cytokine-induced expression (Wu et al., 2008), possibly by auto-inhibition through intramolecular interaction between the N-terminal transactivation domain and the C-terminal ETS domain (Kopp et al., 2007). Other non-transcriptional mechanisms of regulation of ESE-1, such as through microRNAs (Di et al., 2014; Qin et al., 2015) has never been explored. Modern molecular techniques such as long range interactions (4C experiments) (Cai et al., 2016) or knock-down of specific transcription factors motifs on the promotor and enhancers of ESE-1 using CRISPR/Cas9 may reveal interesting information about ESE-1 regulation in the future.
Building an Overall Picture and Approaches for the Future
As mentioned above, there is dearth of information on the exact role of ESE-1 during embryogenesis, despite evidence suggesting its involvement during blastocyst formation and organogenesis. We therefore outline three main future research objectives for the study of ESE-1 function during early development:
Better Analysis of ESE-1 Function at the Protein Level
Most expression analysis on parts of mouse embryo for the expression of ESE-/elf3 and ESE-3/ehf in different tissues have so far focused on quantifying levels of mRNA, by Northern blot or RT-PCR of isolated tissues and cell lines. Since there has not been a longitudinal analysis of expression during mouse embryonic development, we assessed the mRNA levels of ESE-1/Elf3 and ESE-3/Ehf, whose Ets domain is 84% identical to ESE-1 Ets domain (Kas et al., 2000), in wild-type (WT) mice during E7.5-E18.5 by limited cycle RT-PCR as a preliminary analysis (Figure 1A). Albeit with varying degrees, we found that most of the Elf3 and Ehf expression occurred between E13.5 and E18.5 during active organogenesis and tissue differentiation and in E7.5 following gastrulation (Loebel et al., 2003; Tam and Loebel, 2007; Takaoka and Hamada, 2012). Further analysis by quantitative RT-PCR may help confirm and quantify these expression levels. However, RNA-based studies are limited in that they identify transcriptional changes that are regulated both directly and indirectly (Sementchenko and Watson, 2000), and that they do not reveal protein modulations which may be more functionally relevant, by mechanisms such as protein-protein interactions and post-translational modifications that alter protein activity or stability. It is understandable that there has been lack of high quality antibodies which could recognize Ets factors of interest in the past. However, with increased availability of better ESE-1 antibodies and proteomic tools (Sydor and Nock, 2003; Pauly et al., 2013), it is time to delve into studying the expression and function of ESE-1/Elf3 at protein level.
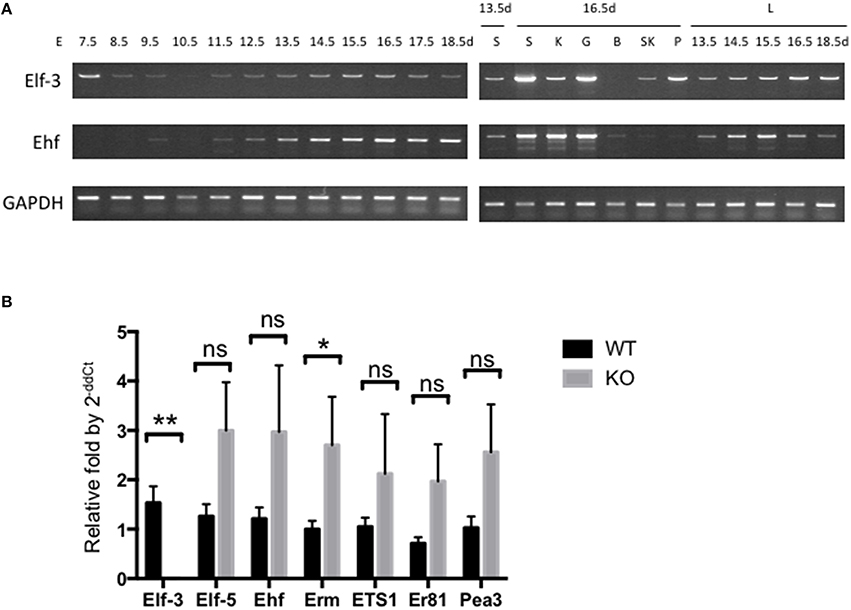
Figure 1. Expression of ESE-1/Elf3 and ESE-3/Ehf during mouse embryogenesis. (A) Limited cycle RT-PCR analysis of Elf3 and Ehf gene expression. Total mRNA from whole mouse embryo were collected at indicated time points of development (Left Panel), as well as from individual organs (Right Panel) such as lung (L), salivary gland (S), kidney (K), gut (G), brain (B), skeletal muscle (SK), and Pancreas (P). The isolated mRNA were subjected to limited cycle RT-PCR analysis across different time points ranging from Embryonic Day (E) 7.5–18.5. GAPDH gene was included as an internal control. RT-PCR products from each data point were loaded onto agarose gel to visualize their size and intensity. Six to eight embryos were collected at early data points (E7.5-E10.5), and data shows n > 3 for each data point. (B) RT-qPCR of various Ets factors in wild-type and Elf3−∕− adult mouse lung. The graph shows n = 4–6 per data point, and statistical analysis was done by two-tailed Student t-test with Welch's correction where appropriate. “ns” indicates not significant; * and ** indicates P < 0.05 and P < 0.01, respectively.
One of the important determinants of Ets specificity is tissue expression pattern (Hollenhorst et al., 2004; Wei et al., 2010). However, there has been no comprehensive study on the localization of Ets proteins during development, and known facts on ESE-1 expression have been derived from Northern blot analysis on human fetal tissues (Oettgen et al., 1997), and E19 mouse embryo tissues (Tymms et al., 1997). Therefore, immunohistochemistry of Elf3 in fixed mouse fetus over the time course of development may be a good starting point to get better understanding of ESE-1 protein expression and its tissue distribution pattern. Understanding protein expression and modulation will provide stepping stones to ascertaining ESE-1 function and regulation at the protein level. Additionally, Ets factors are known to undergo various post-translational modifications such as phosphorylation, glycosylation, ubiquitination, sumoylation, and acetylation (Tootle and Rebay, 2005). Post-translational modification can influence protein activity, protein-protein interactions, protein-DNA interactions, subcellular localization, and stability, thus significantly increasing the functional versatility of a protein (Tootle and Rebay, 2005). The most promising post-translational modification in ESE-1 is phosphorylation, where ESE-1 has previously been shown to be stabilized by serine phosphorylation, which altered its subcellular localization and protein half-life in breast cancer (Manavathi et al., 2007). NetPhos2.0 server predicts 15 potential serine phosphorylation sites on ESE-1 protein with P-score of >0.95. Thus, whether ESE-1 is post-translationally modified during embryogenesis should be further investigated, and the development of phospho-ESE-1 antibody may help in functional analysis of phosphorylated ESE-1.
ESE-1/Elf3 is also unique to the family of Ets factors that it contains two AT-hook domains which can potentially enable ESE-1 to bind in the minor groove of AT-rich DNA and interact with other proteins (Kopp et al., 2007). Protein binding partners have been shown to be critical in regulating Ets factor function (reviewed in Li et al., 2000), but there is limited literature on ESE-1 binding partners, only identified through GST-pull down assays or immunoprecipitations in cell lines such as endothelial cells and 293T cell line (Rudders et al., 2001; Longoni et al., 2013). Therefore following identification and characterization of ESE-1/Elf3 protein expression during embryogenesis, mass spectrometric analysis of embryonic proteins (Nagano et al., 2005) altered in Elf3−∕− embryonic stem cells (ES) and co-immunoprecipitated binding partners in WT mouse ES nuclear extract (Liang et al., 2008) will provide great insights into its function during early development through protein interaction.
Better Analysis of ESE-1 Function As a Member of the Ets Protein Family
Identification Ets target genes has mostly been derived from gain or loss of function studies. Single knockout animal models have been useful in revealing function of individual Ets factors by distinct phenotypes (Bartel et al., 2000). However, there has been virtually no in-depth analysis of changes in other Ets factor expression or post-translational modifications, while studies indicate that over two-thirds of Ets family members are co-expressed in most cell types (Hollenhorst et al., 2004). Notably, Ets factors are expressed temporally during differentiation, showing sharp transitions in expression that correlate with critical commitment (Anderson et al., 1999) or growth events (Bhat et al., 1987), and distinct pattern of co-expression during tumorigenesis (Galang et al., 2004). Therefore, understanding this combinatorial nature of Ets family members is essential in identifying the physiological function of Ets factors in vivo.
Ets factors are known to physically and functionally interact with each other, such as the binding between Tel and Fli-1 (Kwiatkowski et al., 1998) and between Ets2 with Ets1 and Erg (Basuyaux et al., 1997). Analysis of other Ets factors in Elf3 KO mouse lungs also revealed upregulation of other Ets factors in the absence of Elf3, notably Erm (Figure 1B). One approach to studying the interplay between other Ets factors, therefore, would be to examine changes in expression and protein activity of other Ets factors in homozygous knockout mice of a single or double Ets gene(s), thus identifying possible source of compensation from other Ets family members. High-throughput analyses of transcriptional binding specificities have already been shown to be more physiologically relevant than single-protein studies, as exemplified by comparison study of homeobox protein family (Wei et al., 2010). This similarly applies to revealing tissue-specific interactions of Ets proteins, where examples of Ets family interaction have shown tissue-dependent functional redundancy of Ets factors. For example, in Ets2 knockout mice, Ets1 along with other gene targets such as MMP3, MMP9, and uPA are reduced in skin but unchanged in mammary gland (Yamamoto et al., 1998). Protein microarrays are available for tissue-specific interactions (Poetz et al., 2005; Stoll et al., 2005; Dominguez et al., 2008), and initial analyses of tissue expression pattern would be critical to guide this investigation.
For ESE subfamily, ESE-1 and ESE-3 are known to be expressed mostly in the epithelium in both fetal and adult tissues (Kas et al., 2000; Tugores et al., 2001; Silverman et al., 2002), and ESE-1 has been shown to upregulate ESE-3 during inflammation (Wu et al., 2008). To study whether ESE-1 and ESE-3 expression therefore overlap during embryogenesis, we performed in situ hybridization of the gut, lungs, and salivary glands as representative tissues for ESE-1 and ESE-3 expression. Interestingly, we found a differential pattern of expression in the two ESEs where ESE-1 was more predominantly expressed in the gut compared to ESE-3 (Figure 2A), and ESE-3 being slightly more robust in the lungs (Figure 2B), and both ESE's were present at similar levels in the salivary gland (Figure 2C). Therefore, conclusions drawn from one study using different cell type and condition may not apply to other tissue models, and specific definition of experimental context is vital to study Ets factors during early development as discussed further below.
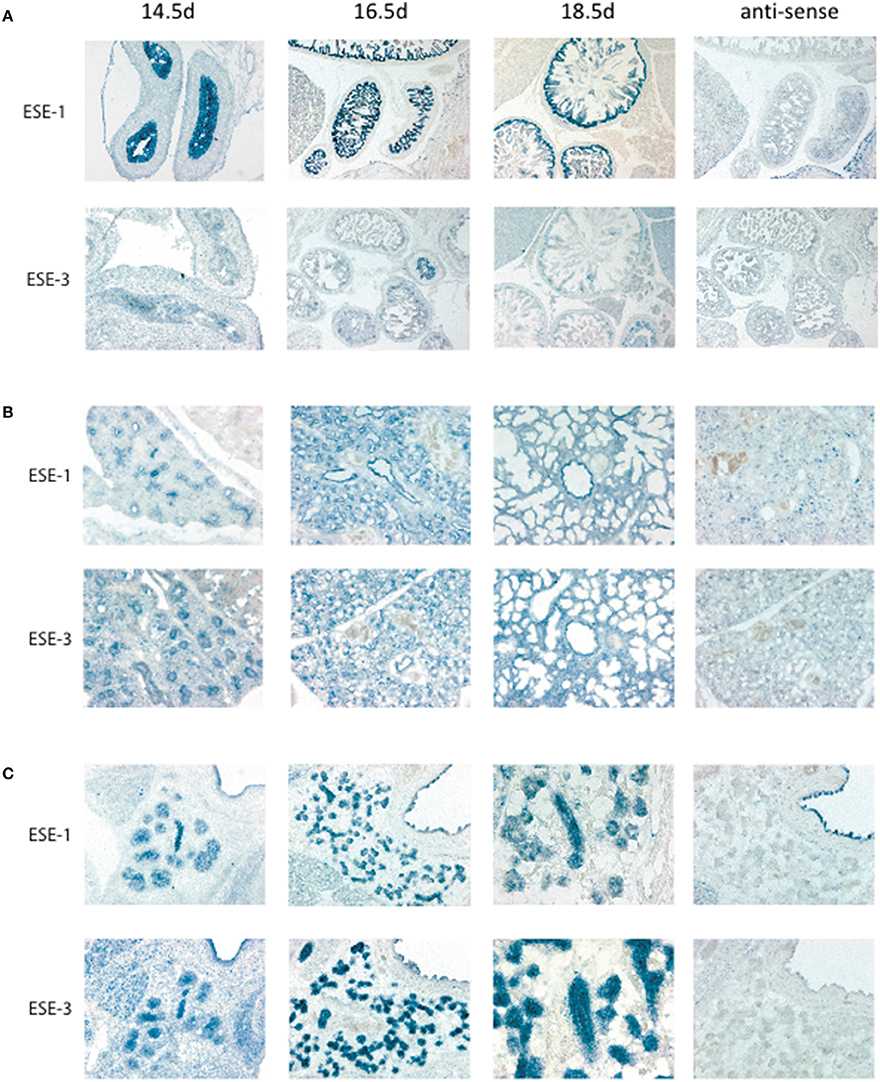
Figure 2. In situ hybridization of Elf3 and Ehf for tissue localization during development. In situ hybridization was performed as previously described in Hui and Joyner (1993). Mouse Elf3 full length cDNA plasmid was used as template for the digoxygenin-labeled RNA probe. Elf3 and Ehf were detected in (A) Gut, (B) Lung, and (C) Salivary gland with anti-sense probe as a negative control. Pictures were taken at different time points from Embryonic Day (E) 14.5–18.5. Figure shows a representative result from n > 3 mice for each data point.
Better Definition of the Physiological Context
Embryogenesis and fetal development consist of multiple steps which involve rapid changes in cellular symmetry, position, survival, and differentiation at the whole organismal level. The microenvironment of a cell, such as stem cell niche, has a critical impact on the cellular developmental potential and differentiation (Martinez-Agosto et al., 2007; Lane et al., 2014). Therefore, precise definition of the developmental phase of interest is important to more accurately represent the particular in vivo microenvironment in vitro. Determination of specific time point and tissue context during development, instead of generic differentiation models using embryonic carcinoma cell lines (Hou et al., 2004; Park et al., 2014), will thus be more informative in designing in vitro models representative of the developmental stage of interest to study the role of ESE-1 during embryogenesis, and relate the information back to the in vivo setting.
Previous findings on ESE-1 provide hints to the most promising time points and tissue settings to focus on. Hierarchical clustering of the transcriptome over the time course of embryonic development, for example, revealed that E11.5-E12.0, during which Elf3−∕− embryos show partial fetal lethality, is associated with dramatic induction of genes involved in adult hemoglobin production and erythrocyte differentiation (Wagner et al., 2005). The transcription of the beta hemoglobin chain starts at E12.0, including other genes involved in hemoglobin biosynthesis such as erythroid associated differentiation factor (ERAF), and heme binding protein 1 (HEBP1) (Wagner et al., 2005). Also, E11.5-E12.0 is the critical time point where definitive hematopoiesis begins in the liver (Wagner et al., 2005). Therefore, defects in hemoglobin synthesis may have been partially responsible for embryonic lethality in Elf3−∕− mice, and studying the effect of ESE-1 deletion by genome editing (Peters et al., 2008; Musunuru, 2013) in in vitro models of erythropoiesis such as human inducible pluripotent stem cells (iPSCs) (Malik et al., 1998; Lapillonne et al., 2010; Kobari et al., 2012) may help to identify contribution of ESE-1 in embryonic hemoglobin synthesis.
The involvement of ESE-1 in lung development may similarly provide a good model, given distinct pattern of expression of ESE-1 in the human fetal lung vs. the adult tissue, the availability of in vitro models in 3D cultures to study branching morphogenesis from embryonic lung explants (Warburton et al., 2005; Del Moral and Warburton, 2010), and well-characterization of signaling pathways involved in lung development such as FGF10 and SHH signaling (Iber and Menshykau, 2013), which are the two most important regulatory diffusible proteins that facilitate outgrowth of lung buds and branching. One way to test whether Erm, an Ets factor which was highly expressed in Elf3−∕− lung (Figure 1B), functionally compensate for the loss of Elf3 would be to generate Elf3−∕−Erm−∕− double knockout and investigate the fetal lung development. However, while mice have been used as the primary model for the study of mammalian development given their genetic similarity to humans, short generation time, and the ease of gene manipulation, the human and mouse airways are anatomically very different. For example, basal cells which are limited to trachea in mice are present throughout the human airways (Rock et al., 2009; Rock and Hogan, 2011), and Clara cells which are confined mostly to distal bronchiole airways in humans are found throughout the murine airways (Boers et al., 1999; Rawlins et al., 2009). Therefore, another significant advantage of studying lung morphogenesis would be the availability of human cell models of 3D lung morphogenesis using immortalized human bronchial epithelial cells (Franzdottir et al., 2010; Kaisani et al., 2014), which have been shown to be multipotent (Delgado et al., 2011), to complement the animal studies.
Concluding Remarks
The role of ESE-1 during embryogenesis had long been implicated in previous discoveries, but there has been no in-depth analysis of ESE-1 function in early development. With improved proteomics and gene editing tools that are now available, therefore, future studies which elucidate the role of ESE-1 during embryonic development will provide not only better insights into ESE-1 function in specific cell differentiation pathways, but also its interactions with other Ets family members. In addition, understanding how epithelium-specific Ets factors contribute to normal embryonic development by, for example, global gene expression analysis in systemic knockdown study (Atabakhsh et al., 2012), will help us to better understand how they take part in disorders and abnormalities which share signaling pathways and gene clusters involved in embryogenesis, such as cancer (Monk and Holding, 2001; Kim and Orkin, 2011; Atabakhsh et al., 2012; Smith and Sturmey, 2013) and tissue regeneration (Poss, 2010). This may open new opportunities to tackle aberrant cell differentiation or repair mechanisms that pose barriers to effective treatment, while building on our current body of knowledge on how multi-member protein families function collectively.
Author Contributions
CL and JW contributed equally to the manuscript. CL critically evaluated the data and wrote the manuscript. WJ designed the experiments and performed embryo analysis. YX performed data analysis and finalized the figures. JH supervised the project and critically reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by research grants to JH from the Canadian Institutes of Health Research (MOP 125882), Cystic Fibrosis Canada (Grant ID #3023), Cystic Fibrosis Foundation Therapeutics (HU15XX0). CL is a recipient of Queen Elizabeth II/Canadian Arthritis Network Scholarships in Science and Technology from University of Toronto and SickKids Restracomp Studentship from SickKids Hospital. All animal experiments were conducted in accordance with humane animal use protocol (AUP) approved by Toronto Center for Phenogenomics (TCP) in Toronto, Canada.
References
Anderson, M. K., Hernandez-Hoyos, G., Diamond, R. A., and Rothenberg, E. V. (1999). Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126, 3131–3148.
Atabakhsh, E., Wang, J. H., Wang, X., Carter, D. E., and Schild-Poulter, C. (2012). RanBPM expression regulates transcriptional pathways involved in development and tumorigenesis. Am. J. Cancer Res. 2, 549–565.
Bartel, F. O., Higuchi, T., and Spyropoulos, D. D. (2000). Mouse models in the study of the Ets family of transcription factors. Oncogene 19, 6443–6454. doi: 10.1038/sj.onc.1204038
Basuyaux, J. P., Ferreira, E., Stehelin, D., and Buttice, G. (1997). The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 272, 26188–26195. doi: 10.1074/jbc.272.42.26188
Bhat, N. K., Fisher, R. J., Fujiwara, S., Ascione, R., and Papas, T. S. (1987). Temporal and tissue-specific expression of mouse ets genes. Proc. Natl. Acad. Sci. U.S.A. 84, 3161–3165. doi: 10.1073/pnas.84.10.3161
Boers, J. E., Ambergen, A. W., and Thunnissen, F. B. (1999). Number and proliferation of clara cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 159, 1585–1591. doi: 10.1164/ajrccm.159.5.9806044
Bult, C. J., Eppig, J. T., Kadin, J. A., Richardson, J. E., Blake, J. A., and Mouse Genome Database Group (2008). The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36, D724–D728. doi: 10.1093/nar/gkm961
Cai, M., Kim, S., Wang, K., Farnham, P. J., Coetzee, G. A., and Lu, W. (2016). 4C-seq revealed long-range interactions of a functional enhancer at the 8q24 prostate cancer risk locus. Sci. Rep. 6, 1–14. doi: 10.1038/srep22462
Delgado, O., Kaisani, A. A., Spinola, M., Xie, X. J., Batten, K. G., Minna, J. D., et al. (2011). Multipotent capacity of immortalized human bronchial epithelial cells. PLoS ONE 6:e22023. doi: 10.1371/journal.pone.0022023
Del Moral, P. M., and Warburton, D. (2010). Explant culture of mouse embryonic whole lung, isolated epithelium, or mesenchyme under chemically defined conditions as a system to evaluate the molecular mechanism of branching morphogenesis and cellular differentiation. Methods Mol. Biol. 633, 71–79. doi: 10.1007/978-1-59745-019-5_5
Di, J., Jiang, L., Zhou, Y., Cao, H., Fang, L., Wen, P., et al. (2014). Ets-1 targeted by microrna-221 regulates angiotensin II-induced renal fibroblast activation and fibrosis. Cell. Physiol. Biochem. 34, 1063–1074. doi: 10.1159/000366321
Dominguez, F., Gadea, B., Esteban, F. J., Horcajadas, J. A., Pellicer, A., and Simon, C. (2008). Comparative protein-profile analysis of implanted versus non-implanted human blastocysts. Hum. Reprod. 23, 1993–2000. doi: 10.1093/humrep/den205
Franzdottir, S. R., Axelsson, I. T., Arason, A. J., Baldursson, O., Gudjonsson, T., and Magnusson, M. K. (2010). Airway branching morphogenesis in three dimensional culture. Respir. Res. 11:162. doi: 10.1186/1465-9921-11-162
Galang, C. K., Muller, W. J., Foos, G., Oshima, R. G., and Hauser, C. A. (2004). Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J. Biol. Chem. 279, 11281–11292. doi: 10.1074/jbc.M311887200
Gutierrez-Hartmann, A., Duval, D. L., and Bradford, A. P. (2007). ETS transcription factors in endocrine systems. Trends Endocrinol. Metab. 18, 150–158. doi: 10.1016/j.tem.2007.03.002
Heintzman, N. D., and Ren, B. (2009). Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19, 541–549. doi: 10.1016/j.gde.2009.09.006
Hollenhorst, P. C., Jones, D. A., and Graves, B. J. (2004). Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 32, 5693–5702. doi: 10.1093/nar/gkh906
Hollenhorst, P. C., McIntosh, L. P., and Graves, B. J. (2011). Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437–471. doi: 10.1146/annurev.biochem.79.081507.103945
Hollenhorst, P. C., Shah, A. A., Hopkins, C., and Graves, B. J. (2007). Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21, 1882–1894. doi: 10.1101/gad.1561707
Hou, J., Wilder, P. J., Bernadt, C. T., Boer, B., Neve, R. M., and Rizzino, A. (2004). Transcriptional regulation of the murine Elf3 gene in embryonal carcinoma cells and their differentiated counterparts: requirement for a novel upstream regulatory region. Gene 340, 123–131. doi: 10.1016/j.gene.2004.06.007
Hui, C. C., and Joyner, A. L. (1993). A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 3, 241–246. doi: 10.1038/ng0393-241
Iber, D., and Menshykau, D. (2013). The control of branching morphogenesis. Open Biol. 3:130088. doi: 10.1098/rsob.130088
Kageyama, S., Liu, H., Nagata, M., and Aoki, F. (2006). The role of ETS transcription factors in transcription and development of mouse preimplantation embryos. Biochem. Biophys. Res. Commun. 344, 675–679. doi: 10.1016/j.bbrc.2006.03.192
Kaisani, A., Delgado, O., Fasciani, G., Kim, S. B., Wright, W. E., Minna, J. D., et al. (2014). Branching morphogenesis of immortalized human bronchial epithelial cells in three-dimensional culture. Differentiation 87, 119–126. doi: 10.1016/j.diff.2014.02.003
Kas, K., Finger, E., Grall, F., Gu, X., Akbarali, Y., Boltax, J., et al. (2000). ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J. Biol. Chem. 275, 2986–2998. doi: 10.1074/jbc.275.4.2986
Kim, J., and Orkin, S. H. (2011). Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 3, 75. doi: 10.1186/gm291
Kobari, L., Yates, F., Oudrhiri, N., Francina, A., Kiger, L., Mazurier, C., et al. (2012). Human induced pluripotent stem cells can reach complete terminal maturation: in vivo and in vitro evidence in the erythropoietic differentiation model. Haematologica 97, 1795–1803. doi: 10.3324/haematol.2011.055566
Kopp, J. L., Wilder, P. J., Desler, M., Kinarsky, L., and Rizzino, A. (2007). Different domains of the transcription factor ELF3 are required in a promoter-specific manner and multiple domains control its binding to DNA. J. Biol. Chem. 282, 3027–3041. doi: 10.1074/jbc.M609907200
Kwiatkowski, B. A., Bastian, L. S., Bauer, T. R. Jr., Tsai, S., Zielinska-Kwiatkowska, A. G., and Hickstein, D. D. (1998). The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J. Biol. Chem. 273, 17525–17530. doi: 10.1074/jbc.273.28.17525
Lane, S. W., Williams, D. A., and Watt, F. M. (2014). Modulating the stem cell niche for tissue regeneration. Nat. Biotechnol. 32, 795–803. doi: 10.1038/nbt.2978
Lapillonne, H., Kobari, L., Mazurier, C., Tropel, P., Giarratana, M. C., Zanella-Cleon, I., et al. (2010). Red blood cell generation from human induced pluripotent stem cells: perspectives for transfusion medicine. Haematologica 95, 1651–1659. doi: 10.3324/haematol.2010.023556
Li, R., Pei, H., and Watson, D. K. (2000). Regulation of Ets function by protein - protein interactions. Oncogene 19, 6514–6523. doi: 10.1038/sj.onc.1204035
Liang, J., Wan, M., Zhang, Y., Gu, P., Xin, H., Jung, S. Y., et al. (2008). Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739. doi: 10.1038/ncb1736
Loebel, D. A., Watson, C. M., De Young, R. A., and Tam, P. P. (2003). Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev. Biol. 264, 1–14. doi: 10.1016/S0012-1606(03)00390-7
Longoni, N., Sarti, M., Albino, D., Civenni, G., Malek, A., Ortelli, E., et al. (2013). ETS transcription factor ESE1/ELF3 orchestrates a positive feedback loop that constitutively activates NF-kappaB and drives prostate cancer progression. Cancer Res. 73, 4533–4547. doi: 10.1158/0008-5472.CAN-12-4537
Malik, P., Barsky, L. L., Zeng, L., Hiti, A. L., and Fisher, T. C. (1998). An in vitro model of human erythropoiesis for the study of hemoglobinopathies. Ann. N.Y. Acad. Sci. 850, 382–385. doi: 10.1111/j.1749-6632.1998.tb10498.x
Manavathi, B., Rayala, S. K., and Kumar, R. (2007). Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. J. Biol. Chem. 282, 19820–19830. doi: 10.1074/jbc.M702309200
Martinez-Agosto, J. A., Mikkola, H. K., Hartenstein, V., and Banerjee, U. (2007). The hematopoietic stem cell and its niche: a comparative view. Genes Dev. 21, 3044–3060. doi: 10.1101/gad.1602607
Messina, D. N., Glasscock, J., Gish, W., and Lovett, M. (2004). An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 14, 2041–2047. doi: 10.1101/gr.2584104
Monk, M., and Holding, C. (2001). Human embryonic genes re-expressed in cancer cells. Oncogene 20, 8085–8091. doi: 10.1038/sj.onc.1205088
Musunuru, K. (2013). Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis. Model. Mech. 6, 896–904. doi: 10.1242/dmm.012054
Nagano, K., Taoka, M., Yamauchi, Y., Itagaki, C., Shinkawa, T., Nunomura, K., et al. (2005). Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics 5, 1346–1361. doi: 10.1002/pmic.200400990
Neve, R. M., Parmar, H., Amend, C., Chen, C., Rizzino, A., and Benz, C. C. (2006). Identification of an epithelial-specific enhancer regulating ESX expression. Gene 367, 118–125. doi: 10.1016/j.gene.2005.10.001
Ng, A. Y., Waring, P., Ristevski, S., Wang, C., Wilson, T., Pritchard, M., et al. (2002). Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122, 1455–1466. doi: 10.1053/gast.2002.32990
Oettgen, P., Alani, R. M., Barcinski, M. A., Brown, L., Akbarali, Y., Boltax, J., et al. (1997). Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol. Cell. Biol. 17, 4419–4433. doi: 10.1128/MCB.17.8.4419
Oettgen, P., Barcinski, M., Boltax, J., Stolt, P., Akbarali, Y., and Libermann, T. A. (1999). Genomic organization of the human ELF3 (ESE-1/ESX) gene, a member of the Ets transcription factor family, and identification of a functional promoter. Genomics 55, 358–362. doi: 10.1006/geno.1998.5681
Oikawa, T., and Yamada, T. (2003). Molecular biology of the Ets family of transcription factors. Gene 303, 11–34. doi: 10.1016/S0378-1119(02)01156-3
Oliver, J. R., Kushwah, R., and Hu, J. (2012). Multiple roles of the epithelium-specific ETS transcription factor, ESE-1, in development and disease. Lab. Invest. 92, 320–330. doi: 10.1038/labinvest.2011.186
Pan, G. J., Chang, Z. Y., Scholer, H. R., and Pei, D. (2002). Stem cell pluripotency and transcription factor Oct4. Cell Res. 12, 321–329. doi: 10.1038/sj.cr.7290134
Park, S. W., Do, H. J., Ha, W. T., Han, M. H., Yang, H. M., Lee, S. H., et al. (2014). Transcriptional regulation of OCT4 by the ETS transcription factor ESE-1 in NCCIT human embryonic carcinoma cells. Biochem. Biophys. Res. Commun. 450, 984–990. doi: 10.1016/j.bbrc.2014.06.079
Pauly, F., Dexlin-Mellby, L., Ek, S., Ohlin, M., Olsson, N., Jirstrom, K., et al. (2013). Protein expression profiling of formalin-fixed paraffin-embedded tissue using recombinant antibody microarrays. J. Proteome Res. 12, 5943–5953. doi: 10.1021/pr4003245
Peters, D. T., Cowan, C. A., and Musunuru, K. (2008). Genome Editing in Human Pluripotent Stem Cells. Cambridge, MA: StemBook.
Poetz, O., Schwenk, J. M., Kramer, S., Stoll, D., Templin, M. F., and Joos, T. O. (2005). Protein microarrays: catching the proteome. Mech. Ageing Dev. 126, 161–170. doi: 10.1016/j.mad.2004.09.030
Poss, K. D. (2010). Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genetics 11, 710–722. doi: 10.1038/nrg2879
Qin, B., Cao, Y., Yang, H., Xiao, B., and Lu, Z. (2015). MicroRNA-221/222 regulate ox-LDL-induced endothelial apoptosis via Ets-1/p21 inhibition. Mol. Cell. Biochem. 405, 115–124. doi: 10.1007/s11010-015-2403-5
Rawlins, E. L., Okubo, T., Xue, Y., Brass, D. M., Auten, R. L., Hasegawa, H., et al. (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534. doi: 10.1016/j.stem.2009.04.002
Rock, J. R., and Hogan, B. L. (2011). Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27, 493–512. doi: 10.1146/annurev-cellbio-100109-104040
Rock, J. R., Onaitis, M. W., Rawlins, E. L., Lu, Y., Clark, C. P., Xue, Y., et al. (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 106, 12771–12775. doi: 10.1073/pnas.0906850106
Rudders, S., Gaspar, J., Madore, R., Voland, C., Grall, F., Patel, A., et al. (2001). ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-kappa B to regulate the inducible nitric-oxide synthase gene. J. Biol. Chem. 276, 3302–3309. doi: 10.1074/jbc.M006507200
Sementchenko, V. I., and Watson, D. K. (2000). Ets target genes: past, present and future. Oncogene 19, 6533–6548. doi: 10.1038/sj.onc.1204034
Sharrocks, A. D. (2001). The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827–837. doi: 10.1038/35099076
Silverman, E. S., Baron, R. M., Palmer, L. J., Le, L., Hallock, A., Subramaniam, V., et al. (2002). Constitutive and cytokine-induced expression of the ETS transcription factor ESE-3 in the lung. Am. J. Respir. Cell Mol. Biol. 27, 697–704. doi: 10.1165/rcmb.2002-0011OC
Smith, D. G., and Sturmey, R. G. (2013). Parallels between embryo and cancer cell metabolism. Biochem. Soc. Trans. 41, 664–669. doi: 10.1042/BST20120352
Stoll, D., Templin, M. F., Bachmann, J., and Joos, T. O. (2005). Protein microarrays: applications and future challenges. Curr. Opin. Drug Discov. Devel. 8, 239–252.
Sydor, J. R., and Nock, S. (2003). Protein expression profiling arrays: tools for the multiplexed high-throughput analysis of proteins. Proteome Sci. 1:3. doi: 10.1186/1477-5956-1-3
Takaoka, K., and Hamada, H. (2012). Cell fate decisions and axis determination in the early mouse embryo. Development 139, 3–14. doi: 10.1242/dev.060095
Tam, P. P., and Loebel, D. A. (2007). Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genetics 8, 368–381. doi: 10.1038/nrg2084
Tootle, T. L., and Rebay, I. (2005). Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays 27, 285–298. doi: 10.1002/bies.20198
Tugores, A., Le, J., Sorokina, I., Snijders, A. J., Duyao, M., Reddy, P. S., et al. (2001). The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. J. Biol. Chem. 276, 20397–20406. doi: 10.1074/jbc.M010930200
Tymms, M. J., Ng, A. Y., Thomas, R. S., Schutte, B. C., Zhou, J., Eyre, H. J., et al. (1997). A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene 15, 2449–2462. doi: 10.1038/sj.onc.1201427
Wagner, R. A., Tabibiazar, R., Liao, A., and Quertermous, T. (2005). Genome-wide expression dynamics during mouse embryonic development reveal similarities to Drosophila development. Dev. Biol. 288, 595–611. doi: 10.1016/j.ydbio.2005.09.036
Warburton, D., Bellusci, S., De Langhe, S., Del Moral, P. M., Fleury, V., Mailleux, A., et al. (2005). Molecular mechanisms of early lung specification and branching morphogenesis. Pediatric Res. 57, 26R–37R. doi: 10.1203/01.PDR.0000159570.01327.ED
Wei, G. H., Badis, G., Berger, M. F., Kivioja, T., Palin, K., Enge, M., et al. (2010). Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160. doi: 10.1038/emboj.2010.106
Wu, J., Duan, R., Cao, H., Field, D., Newnham, C. M., Koehler, D. R., et al. (2008). Regulation of epithelium-specific Ets-like factors ESE-1 and ESE-3 in airway epithelial cells: potential roles in airway inflammation. Cell Res. 18, 649–663. doi: 10.1038/cr.2008.57
Keywords: Ets transcription factors, ESE-1, embryogenesis, tissue differentiation
Citation: Lee CM, Wu J, Xia Y and Hu J (2016) ESE-1 in Early Development: Approaches for the Future. Front. Cell Dev. Biol. 4:73. doi: 10.3389/fcell.2016.00073
Received: 15 April 2016; Accepted: 17 June 2016;
Published: 28 June 2016.
Edited by:
Eiman Aleem, University of Arizona College of Medicine-Phoenix, USAReviewed by:
Eirini Trompouki, Max Planck Institute for Immunobiology and Epigenetics, GermanySuk-Won Jin, Yale University, USA and Gwangju Institute of Science and Technology, South Korea
Copyright © 2016 Lee, Wu, Xia and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jim Hu, amltLmh1QHV0b3JvbnRvLmNh
†These authors have contributed equally to this work.
 Chan Mi Lee
Chan Mi Lee Jing Wu
Jing Wu Yi Xia1,2
Yi Xia1,2 Jim Hu
Jim Hu