Abstract
Long noncoding RNAs (lncRNAs) are a class of noncoding transcripts characterized with more than 200 nucleotides of length. Unlike their names, some short open reading frames are recognized for them encoding small proteins. LncRNAs are found to play regulatory roles in essential cellular processes such as cell growth and apoptosis. Therefore, an increasing number of lncRNAs are identified with dysregulation in a wide variety of human cancers. SNHG7 is an lncRNA with upregulation in cancer cells and tissues. It is frequently reported with potency of promoting malignant cell behaviors in vitro and in vivo. Like oncogenic/tumor suppressor lncRNAs, SNHG7 is found to exert its tumorigenic functions through interaction with other biological substances. These include sponging target miRNAs (various numbers are identified), regulation of several signaling pathways, transcription factors, and effector proteins. Importantly, clinical studies demonstrate association between high SNHG7 expression and clinicopathological features in cancerous patients, worse prognosis, and enhanced chemoresistance. In this review, we summarize recent studies in three eras of cell, animal, and human experiments to bold the prognostic, diagnostic, and therapeutic potentials.
Introduction
Initially based on the central dogma of molecular biology lasting for decades, sequential flow of cell genetic information was defined through RNAs, which encoded proteins, and so messenger RNA (mRNA) were considered mediators of template DNA and downstream proteins (Crick, 1970). However, exceptions were gradually made, and RNAs that did not directly encode any protein or polypeptide were identified. Transfer RNAs (tRNA), ribosomal RNAs (rRNAs), and small nuclear and nucleolar RNAs (snRNAs and snoRNAs, respectively) were recognized as groups of non-protein-coding RNAs (ncRNAs) with functions in the translation of coding mRNAs and modification or processing of other RNAs (Hombach and Kretz, 2016). Nowadays, we know that a minority of large genomes in complex eukaryotic organisms encode protein or polypeptide strands, and a majority [for instance, 98% in humans (Elgar and Vavouri, 2008)] does not encode for amino acids. This great proportion, formerly called “junk DNA,” however, is mainly [e.g., two thirds of the mammalian genomes (Mattick, 2001)] transcribed to thousands of RNA transcripts, including various types of known ncRNAs that are demonstrated to be involved in critical cellular processes through conducting regulatory functions (Najafi et al., 2022). By employment of high-throughput technologies, such as RNA-seq, identification of novel ncRNAs is accelerated, and new members are being introduced constantly (Taheri et al., 2021). Although the role of ncRNAs is not yet clear, however, their involvement in essential life processes have caused them to be the architects of complexity in eukaryotes (Mattick, 2001). The number of functional ncRNAs are growing, and several show regulatory roles on gene expression.
The size of the transcript is the main discriminating parameter used for classification of ncRNAs. Based on a size limit, ncRNAs are divided in two short and long classes. MicroRNAs (miRNAs), rRNAs, tRNAs, and snRNAs/snoRNAs are several described subclasses of short ncRNAs with a total length shorter than 200 nucleotides (Amin et al., 2019). Among them, miRNAs are studied more broadly compared with others, an increasing number identified in mammalian cells, and also a number are reported with altered expression in various human diseases.
Long noncoding RNAs (lncRNAs) are the second class of ncRNAs with characteristic length of >200 nucleotides. Thousands of lncRNA-related genes have been identified in the human genome and corresponding transcripts reported in large quantities by a large number ranging from 10,000 to 60,000 in human cells (Guttman et al., 2009; Iyer et al., 2015). They have been identified in a wide variety of eukaryotic species, and several show conserved sequences among different organisms suggesting evolution pressure (Ramírez-Colmenero et al., 2020). A number of exclusive properties have made lncRNAs different compared with regular mRNAs. These remarkable differences include characteristic biogenesis, localization, structure, and roles (Quinn and Chang, 2016). Unlike protein-coding RNAs, lncRNAs are mainly transcribed from regulatory and noncoding sequences such as promotors, enhancers, and introns. Furthermore, they could be generated from shared sequences with other transcripts (Al-Tobasei et al., 2016) although some researchers consider lncRNAs as noises or byproducts of transcription (Gao et al., 2020). Unlike their names, some short open reading frames are recognized for them that encode for small proteins (Hartford and Lal, 2020). According to the location of transcription, lncRNAs are classified into intronic and intergenic. Structurally, lncRNAs can be found in linear and circular forms, which are mainly referred to as the former structures; however, circular RNAs also have been found with regulatory functions and roles in pathogenesis of various human cancers (Rahmati et al., 2021; Sayad et al., 2021). LncRNAs show specific expression in cell-, tissue-, and developmental stage–specific manners (Sarropoulos et al., 2019). Their biogenesis is also forced to more strict regulation relative to protein-coding transcripts that, along with their conservation among species, suggests critical regulatory functions for lncRNAs (Dahariya et al., 2019). Several strategies, including ribonuclease P cleavage, processing by ribonucleoproteins, and circularization via backsplicing, play a role in biogenesis of lncRNAs (Dahariya et al., 2019). Same as mRNAs, lncRNAs undergo post-transcriptional modifications on processing such as capping and polyadenylation at 5ˊ and 3ˊ ends, respectively, splicing and base modifications (Sarropoulos et al., 2019). They are mainly located at the nucleus exerting their epigenetic and gene expression regulatory functions via altering the histone modifications or transcription control through several mechanisms, including scaffold, signal, guide, and decoy (Zhang et al., 2019a; Dahariya et al., 2019). Through these ways, lncRNAs in interactions with DNA, proteins, and other RNAs, play a role in various biological phenomena, such as cell differentiation and reprogramming, organ development, immune responses, and cell cycle control (Statello et al., 2021).
Accordingly, a set of lncRNAs is found to be deregulated in various human disorders. An association between expression level of these transcripts and pathogenesis in major health conditions confirms critical roles of lncRNAs in essential health-affecting processes. Among an increasing number of pathogenic lncRNAs, a handful, such as XIST, MALAT1, HOTAIR, H19, ANRIL, and MEG3, are the best known and most often found transcripts. Playing a role particularly in cancer development and progression, differentially expressed lncRNAs in cancer tissues are functionally subdivided into two group of oncogenic and tumor suppressors.
Small nucleolar RNA host gene 7 (SNHG7) is among the oncogenic lncRNAs with progressive effects in multiple human cancers although a single study suggests tumor suppressor function for SNHG7 in pituitary adenoma (Xue and Ge, 2020). Its corresponding gene, located on chromosome 9q34.3, encodes a 2157-base-pair-long transcript. SNHG7 was reported for the first time in 2013 by Chaudhry in X-ray-treated lymphoblastoid cells (Chaudhry, 2013). Rather than regulation of transcription factors, translation, or stability of mRNAs involved in several diseases such as cardiac fibrosis, hepatic fibrosis, and cardiac hypertrophy in addition to helping fracture repair (Chen et al., 2019a; Jing et al., 2019; Yu et al., 2019; Wang et al., 2020a), SNHG7 is found to be overregulated in cancer tissues compared with healthy tissues in a wide variety of human malignancies, including bladder, prostate, gastric, colorectal, and pancreatic cancers (Li et al., 2018a; Zhong et al., 2018a; Cheng et al., 2019a; Han et al., 2019; Zhang et al., 2020a). This upregulation also is demonstrated to accelerate cancer progression. It is shown that SNHG7 is negatively regulated by insulin-like growth factor 1 (IGF1) signaling at the post-transcriptional level through the MAPK pathway to control cell proliferation (Boone et al., 2020). In cell and animal studies, SNHG7 is shown with oncogenic roles in accordance with clinicopathological features and also diagnostic and prognostic values in cancerous patients. In this review, we have gathered recent findings on the oncogenic roles of this lncRNA in three levels of cell, animal, and human studies with a focus on clinical results predicting SNHG7 as a novel biomarker for different types of human cancers.
Cell Line Studies
Through study of SNHG7 knockdown or overexpression in cancer cell lines, it is demonstrated that expression of this oncogenic lncRNA promotes malignant features of the cells in vitro. This universal finding, although opposite effects have been described for SNHG7 at least in two distinct experiments (Pei et al., 2020; Huang et al., 2020), is reported for a broad spectrum of cancer cell types, such as breast, colorectal, bladder, gastric, liver, etc. Proliferation and colony formation experiments have unveiled increased cell and colony numbers in cancer cells in response to SNHG7 simulated excess expression compared with baseline conditions. Accordingly, reduced apoptosis consistent with elevated tumor cell growth hypothesizes the role of this lncRNA in cancer progression. Migratory and invasive potentials of cancer cells also show enhancement in Transwell and Matrigel assays, respectively. Conferring chemoresistance or desensitization has been concluded from cellular studies in which increased sensitivity of cancer cells to conventional chemotherapy agents and/or radiotherapy is seen on SNHG7 knockdown. For example, in two distinct experiments, enhanced sensitivity of breast cancer cells to Adriamycin and Trastuzumab is shown when SNHG7 is silenced or its target sponged miRNA (miR-34a or miR-186, respectively) is overexpressed (Li et al., 2020a; Zhang et al., 2020b). Knockdown studies employing RNA silencing confirm the overexpression experiment results by reversing the SNHG7 impacts on malignant cells behaviors. Via making a network, lncRNAs are known to affect expression of a specific target miRNA. Dual luciferase reporter and RNA immunoprecipitation (RIP) assays confirm the association between SNHG7 and target miRNA consistent with bioinformatics predictions. These interactions seem to be conducted via complementary sequences as binding sites on miRNA for SNHG7. This regulatory effect is mainly repressive, and expression levels in quantitative real-time polymerase chain reaction (qRT-PCR) reveal a negative correlation between both. It is hypothesized that through downregulation of the target miRNA, SNHG7 as a competing endogenous RNA (ceRNA) or sponger exerts its regulatory impacts on downstream transcription factors playing a role in some signaling pathways (Figure 1). Activation of an oncogenic signaling pathway demonstrates why these lncRNAs are considered to have tumor promoting potentials. A handful of evidence on the acceleration of the cell cycle in response to SNHG7 overexpression or arrest in a phase under knockdown conditions suggests indirect enhancing influences of this lncRNA on cell proliferation and differentiation, which consequently, leads to cancer progression. For instance, She et al. (2018) find that SNHG7 upregulates the Fas apoptotic inhibitory molecule 2 (FAIM2) through sponging miR-193b in non-small cell lung cancer (NSCLC) cells. In silico investigations demonstrate binding sites for miR-193b on the SNHG7 sequence. FAIM2 is a membrane protein; shows antiapoptotic activity; is upregulated in several cancers; and is already known to promote tumor cell proliferation, migration, and invasion in lung cancer cells (She et al., 2016). Repression of proapoptotic proteins, such as Bax, and SIRT1-associated pyroptosis also benefits reduced tumor cell death (Xu et al., 2019; Chen et al., 2020a). Thus, it is not surprising to see repressed apoptosis frequently reported on SNHG7 overexpression, which means steady growth of cancer cells. Enhanced glycolysis through upregulation of lactate dehydrogenase A (LDHA) in the tumor microenvironment is another finding on SNHG7 overexpression, which can help the cancer cell economy (Zhang et al., 2019b; Pei et al., 2021). SNHG7 also causes arrest in the G1/G0 phase of the cell cycle (Wang et al., 2017a; Xu et al., 2018); regulates signaling pathways, such as Wnt/β-Catenin and AKT/mTOR pathways; and represses tumor suppressors, such as P15 and P16 (Wang et al., 2017b; Li et al., 2020b; Chi et al., 2020; Du et al., 2020). Furthermore, an elevated neovascularization rate following SNHG7 overexpression is consistent with tumor progression conditions (Li et al., 2018b). Playing a role in regulation of cellular processes, signaling pathways, transcription factors, and particularly via sponging miRNAs, SNHG7 is described as an oncogenic lncRNA with upregulation in various types of cancer cells (Figure 2).
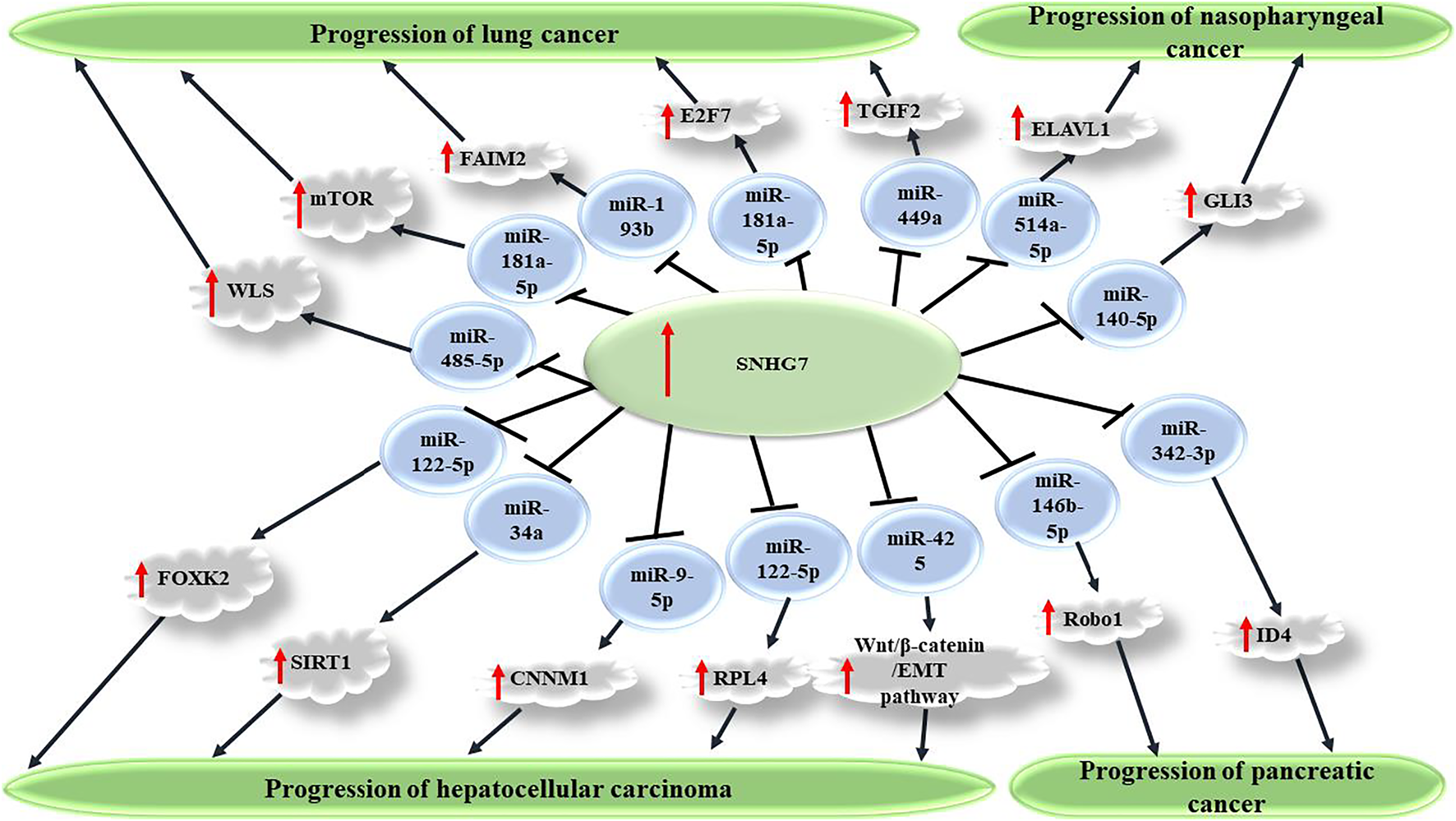
FIGURE 1
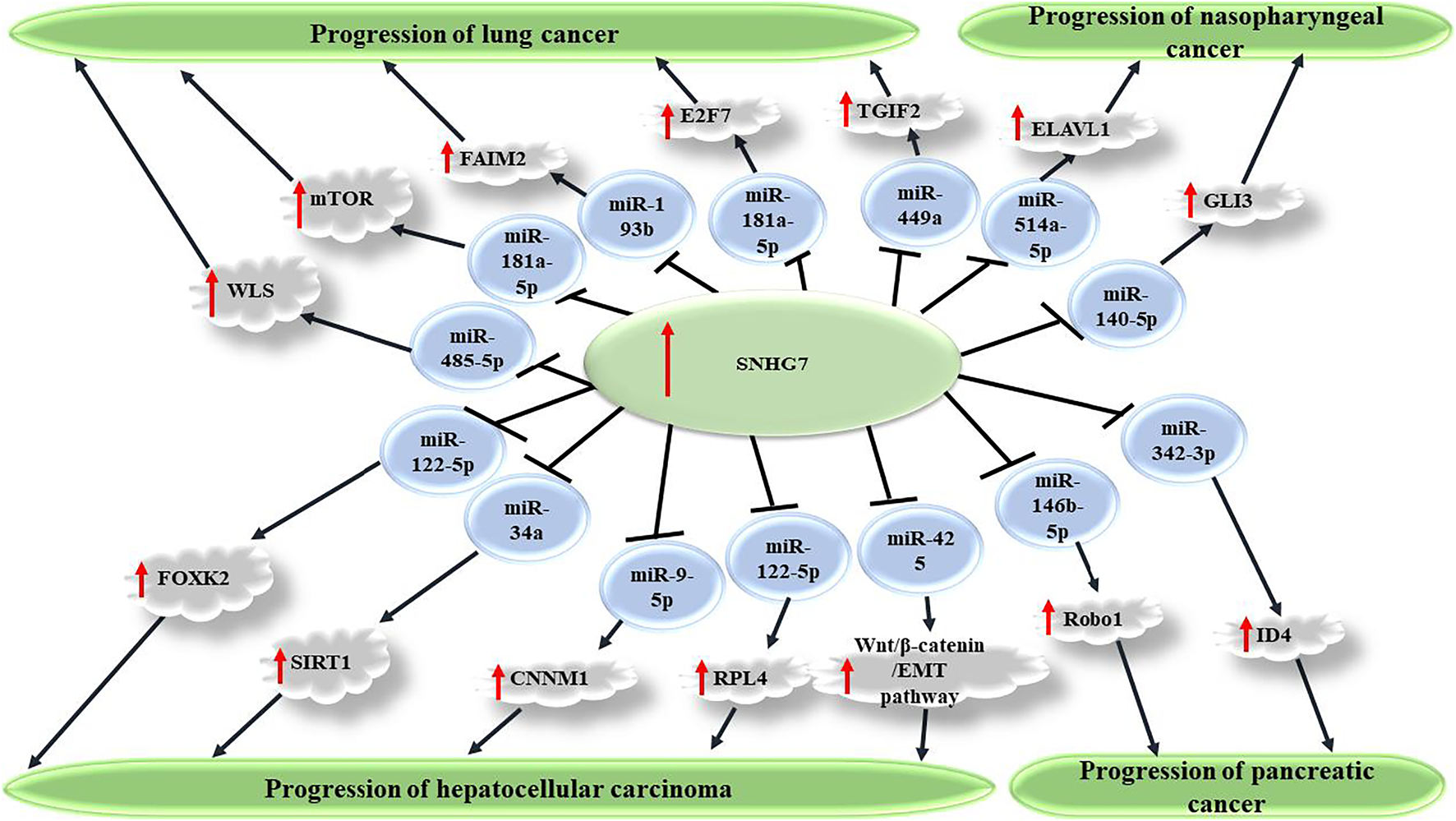
SNHG7 promotes carcinogenesis via sponging miRNAs and consequently upregulating several transcription factors. Through repression of target miRNAs, including miR-449a, miR-181a-5p, miR-193b, and miR-485-5p, SNHG7 causes increased expression of TGIF2, mTOR, FAIM2, and WLS factors, which consequently promote malignant features of cancer cells and enhanced carcinogenesis in non-small cell lung carcinoma. SNHG7 also plays a role in progression of several other cancers, including nasopharyngeal, pancreatic, and hepatocellular carcinoma.
FIGURE 2
Signaling pathways and cellular processes are affected by SNHG7 to enhance tumor progression in cancer cells. Through sponging miR-34a, SNHG7 activates several signaling pathways, such as Notch-1 and PI3K/Akt/mTOR in breast and colorectal cancer, respectively. Other pathways, such as Wnt/b-catenin and K-ras/ERK/cyclinD1, are also affected via other target miRNAs. The effect of SNHG7 on enhancement of glycolysis through upregulation of lactate dehydrogenase A (LDHA) has beneficial effects for cancer cells metabolism.
Animal Studies
Xenograft animal experiments with inoculation of cancer cells into nude mice try to simulate the cancer conditions in an animal model. BALB/C nude mice are used to evaluate the effect of lncRNA upregulation and/or downregulation in vivo. Cancer cells transfected with a vector expressing small heterogenous RNA (shRNA) for overexpression or small interfering RNA for knockdown of the lncRNA along with a vector expressing a control scrambled sequence are injected into the flank of nude mice to establish the xenograft mouse model. Size and volume of the tumor created in the mice is then calculated to compare the tumor growth after sacrificing the animals. Using immunohistochemistry for detection of Ki-67 as a proliferation marker in excised tumor tissues, it is feasible to assess the implanted tumor cell proliferation. Xenograft animal experiments in a number of studies demonstrate that SNHG7 knockdown suppresses tumor growth via decreasing tumor size in vivo, whereas faster tumor growth is reported for SNHG7-overexpressing implanted cells compared with control animals. This effect is reported for SNHG7 silencing in various cancer models (Table 1). Decreased tumor metastasis or repression of some carcinogenic signaling pathways, such as the Notch pathway, is also reported in other animal studies (Sun et al., 2019). For instance, several studies assess the role of SNHG7 knockdown on hepatocellular carcinoma (HCC) growth in xenograft mice (Yang et al., 2019; Yao et al., 2019; Xie et al., 2020; Zhao et al., 2021). Yang et al. (2019) demonstrate lower tumor volume and percentage of Ki-67-stained HCCLM3 cells and less lung metastasis of HCCLM3 cells in an SNHG7 knockdown mice group compared with the control group. Additionally, Yao et al. (2019) show that the expression of the metastasis-associated protein matrix metalloproteinase-9 (MMP-9) is increased in SNHG7-overexpressing HepG2 implanted cells, suggesting a mechanism for enhancing the effect of SNHG7 on tumor metastasis. Collectively, promoted tumor growth on SNHG7 overexpression and/or suppressed tumor proliferation on SNHG7 knockdown is reported in a body of studies. These results, along with cellular findings, confirm the oncogenic role of SNHG7, and the knockdown achievements may suggest therapeutic potentials for anticancer therapies.
TABLE 1
| Cancer type | Animal models | Function | References (s) |
|---|---|---|---|
| Pancreatic cancer | Nude mice | Δ SNHG7: ↓ tumor growth | Jian and Fan, (2021) |
| Female BALB/C nude mice | Δ SNHG7: ↓ tumor growth | Cheng et al. (2019b) | |
| Breast cancer | BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Zhang et al. (2020b) |
| BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Li et al. (2020d) | |
| BALB/c athymic nude mice | Δ SNHG7: ↓ tumor growth, ↓EMT, and ↓Notch-1 pathway | Sun et al. (2019) | |
| Colorectal cancer | Nude mice | Δ SNHG7: ↓ tumor growth | Li et al. (2018b) |
| Lung cancer (non-small cell lung cancer; NSCLC) | Nude mice | Δ SNHG7: ↓ tumor growth | Li et al. (2020b) |
| Athymic nude mice | Δ SNHG7: ↓ tumor growth | Wang et al. (2020b) | |
| Nude mice | Δ SNHG7: ↓ tumor growth | She et al. (2018) | |
| Liver cancer (hepatocellular carcinoma; HCC) | BALB/c male nude mice | Δ SNHG7: ↓ tumor growth | Zhao et al. (2021) |
| BALB/c male nude mice | Δ SNHG7: ↓ tumor growth | Xie et al. (2020) | |
| BALB/c nude mice | Δ SNHG7: ↓ tumor growth, and ↓metastasis | Yang et al. (2019) | |
| BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Yao et al. (2019) | |
| Gastric cancer | BALB/c mice | Δ SNHG7: ↓ tumor growth | Wang et al. (2017b) |
| Bladder cancer | Male nude mice | Δ SNHG7: ↓ tumor growth | Wang et al. (2020c) |
| Pituitary adenocarcinoma | Nude mice | Δ SNHG7: ↓ tumor growth | Yue et al. (2021) |
| Neuroblastoma | BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Jia et al. (2020) |
| Glioma | BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Du et al. (2020) |
| Thyroid | BALB/c nude mice | Δ SNHG7: ↓tumor cell proliferation, and ↓131I resistance | Chen et al. (2021) |
| Glioblastoma | BALB/c nude mice | Δ SNHG7: ↓ tumor growth, and ↓metastasis | Ren et al. (2018) |
| Ovarian cancer | BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Bai et al. (2020) |
| Cervical cancer | BALB/c nude mice | Δ SNHG7: ↓ tumor growth | Zhao et al. (2020) |
| Prostate cancer | BALB/c nude mice | Δ SNHG7: ↓ tumor growth, and ↑cell cycle arrest | Qi et al. (2018) |
Effects of SNHG7 on tumor growth and metastasis in animal studies.
Human Studies
Consistent with cellular findings, enormous expression assessments using qRT-PCR analysis demonstrate elevated SNHG7 expression in tissues retrieved from cancerous patients compared with healthy adjacent tissues. Increased SNHG7 tissue expression is frequently found to be associated with worse clinicopathological features, which are used in clinical classification and staging of human malignancies. Importantly, patients with more advanced clinicopathological characteristics are predicted to have worse prognosis and severe outcomes. These include larger tumor size, more advanced clinical stage, poor histologic grade, deeper tumor invasion, and lymph node metastasis in accordance with high SNHG7 expression in the affected patients (Zeng et al., 2019; Pang et al., 2020; Zhu et al., 2021). This value is also shown in malignancies with broad and different features and in meta-analyses pooling data of tens of studies (Yu et al., 2021a; Yi et al., 2021; Yu et al., 2021b). For example, in acute myeloid leukemia (AML), an association between SNHG7 and SNHG12 lncRNAs and specific clinical/molecular features, including white blood cell (WBC) counts and mutations in IDH1, RUNX1, and NPM1 genes, shows high value of SNHG7 in correlation with extensive features (Shi et al., 2020). These demonstrations suggest that elevated SNHG7 expression predicts poor clinicopathological characteristics. In other words, high SNHG7 expression can predict worse outcomes following poor clinicopathological determinants. In accordance with clinicopathological findings, SNHG7 also shows correlation with prognostic parameters. Survival analysis using a Kaplan–Meier curve indicates shorter survival time in overall survival (OS) and disease-free survival (DFS) for patients with high SNHG7 expression relative to those with low levels. This finding is reported for various human cancers, for which survival analysis is conducted (Table 2). For example, in three distinct studies that reported survival analyses in HCC patients, among a total of 150 patients, poorer OS time was reported separately for the patients with elevated tissue SNHG7 expression in comparison to those with low levels (Yang et al., 2019; Zhao et al., 2021; Yao et al., 2019). Additionally, recurrence is predicted to happen in shorter durations and higher rates among patients with high SNHG7 expression (Zhang et al., 2020c). Interestingly, Cox regression analyses confirm the predictive value of SNHG7 as an independent prognostic factor among cancerous patients. This is particularly reported in several district experiments on human malignancies such as gastric cancer, cervical cancer, HCC, and liver metastasis following hepatectomy in CRC patients (Table 2) (Zeng et al., 2019; Zhang et al., 2020a; Zhang et al., 2020c; Shen et al., 2020). As for diagnostic values, an area under curve (AUC) of 0.84 in the receiver operating characteristic (ROC) curve is reported for SNHG7 in CRC patients (Hu et al., 2019). Importantly, SNHG7 is demonstrated as a potential therapeutical target as it is identified in several studies to lead to enhanced chemoresistance to several anticancer agents such as Cisplatin, Trastuzumab, and Folfirinox in the cancer cells (Chen et al., 2019d; Li et al., 2020a; Zhang et al., 2020b; Dai et al., 2020; Cheng et al., 2021; Pei et al., 2021). Also, metformin with anticancer properties is found to exert its effects in sensitization to Paclitaxel via regulation of SNHG7/miR-3127-5p-mediated autophagy in ovarian cancer cells (Yu et al., 2020). In another study, metformin is demonstrated to suppress growth of hypopharyngeal cancer cells through epigenetic silencing of SNHG7 (Wu et al., 2019). Taken together, human studies suggest SNHG7 lncRNA with promising diagnostic, prognostic, and therapeutic potentials in various types of cancer.
TABLE 2
| Cancer type | Clinical samples | Expression change in tumor tissues compared to normal tissues | Associated clinical features | Kaplan–Meier analysis | Multivariate cox regression | References (s) |
|---|---|---|---|---|---|---|
| Lung cancer | 36 cancerous patient tissues and matched NATs | Upregulated | — | Patients with elevated expression levels of SNHG7 demonstrated decreased OS rate compared to those with lower levels | — | Li et al. (2020b) |
| 30 cancerous patient tissues and matched NATs | Upregulated | — | — | — | Wang et al. (2020b) | |
| Esophageal cancer | 40 cancerous patient tissues and matched NATs | Upregulated | — | — | — | Wang et al. (2021) |
| Liver (hepatocellular carcinoma; HCC) | 30 cancerous patient tissues and matched NATs | Upregulated | Tumor size, TNM grade, and Distant metastasis | Log-rank test demonstrated that patients with high SNHG7 expression had poorer OS. | — | Zhao et al. (2021) |
| 25 cancerous patient tissues and matched NATs | Upregulated | — | — | — | Chen et al. (2020a) | |
| 80 cancerous patient tissues and matched NATs | Upregulated | Tumor stages, tumor grade, and vascular invasion | Patients with high SNHG7 expression levels had poor OS. | — | Yang et al. (2019) | |
| 40 cancerous patient tissues and matched NATs | Upregulated | TNM stage, and tumor metastasis | Elevated SNHG7 expression was markedly associated with poor OS in hepatic carcinoma patients | — | Yao et al. (2019) | |
| 100 cancerous patient tissues and matched NATs | Upregulated | Tumor number, lymph node metastasis, and clinical stage | Patients with high SNHG7 expression demonstrated worse OS and PFS relative to those with low levels | SNHG7 expression acts as an independent prognostic factor in HCC patients | Shen et al. (2020) | |
| Synchronous colorectal liver metastasis (SCLM) | 96 SCLM patients | Upregulated | Differentiation of primary tumor, invasion depth of primary focus, lymph node metastases, number of liver metastases, and liver metastasis grade | Patients with high SNHG7 expression levels had poor OS. | SNHG7 expression acts as an independent prognostic factor for OS and occurrence in SCLM patients | Zhang et al. (2020c) |
| Pancreatic cancer | 50 cancerous patient tissues and matched NATs | Upregulated | tumor size, TNM stage, lymph node metastasis, and distant metastasis | Patients with elevated expression levels of SNHG7 demonstrated decreased survival rate relative to those with lower levels | — | Jian and Fan, (2021) |
| 40 cancerous patient tissues and matched NATs | Upregulated | — | Patients with high SNHG7 expression levels had poor OS. | — | Cheng et al. (2019b) | |
| Breast cancer | 43 cancerous patient tissues | Upregulated | Tumor size, TNM stage, and Ki-67 index | Patients with high SNHG7 levels had lower DFS compared to those with lower levels | — | Li et al. (2020a) |
| 50 cancerous patient tissues and matched NATs | Upregulated | Pathological stage, and lymph node metastasis | — | — | Li et al. (2020d) | |
| 837 cancerous patient tissues and matched NATs | Upregulated | — | High SNHG7 was associated with decreased survival in breast cancer patients | — | Zhang et al. (2019b) | |
| 72 cancerous patient tissues and matched NATs | Upregulated | Clinical Stage, lymph node and distant metastasis | High SNHG7 was correlated with shorter survival time in breast cancer patients | — | Luo et al. (2018) | |
| Gastric cancer | 30 cancerous patient tissues and 30 healthy tissues | Upregulated | — | — | — | Pei et al. (2021) |
| 36 cancerous patient tissues and matched NATs | Upregulated | — | — | — | Zhao and Liu, (2021) | |
| 162 cancerous patient tissues and matched NATs | Upregulated | TNM stage, depth of invasion, lymph node and distant metastasis | Patients with high SNHG7 levels showed lower OS compared to those with high SNHG7 expression | SNHG7 acts as an independent factor for poor OS in patients with gastric cancer | Zhang et al. (2020a) | |
| Bladder cancer | 60 cancerous patient tissues and matched NATs | Upregulated | Clinical stage | Patients with high SNHG7 levels showed unfavorable prognosis | — | Wang et al. (2020c) |
| 92 cancerous patient tissues and matched NATs | Upregulated | Tumor range, lymph nodes, and pathological stage | Patients with high SNHG7 levels had poor OS compared to those with low levels | — | Chen et al. (2019b) | |
| Pituitary adenocarcinoma | 30 cancerous patient tissues and matched NATs | Upregulated | — | Patients with high SNHG7 levels showed unfavorable prognosis compared to those with low levels | — | Yue et al. (2021) |
| Glioma | 30 cancerous patient tissues and matched NATs | Upregulated | — | — | — | Cheng et al. (2020) |
| 20 and 33 cancerous patient tissues and matched NATs | Upregulated | Tumor grade | — | — | (Du et al., 2020; Deng et al., 2021) | |
| Glioblastoma | 53 cancerous patient tissues and matched NATs | Upregulated | WHO Grade | -- | -- | Chen et al. (2020b) |
| 53 cancerous patient tissues and matched NATs | Upregulated | — | Patients with high SNHG7 levels had poor survival rates compared to those with low levels | — | Ren et al. (2018) | |
| Neuroblastoma | 45 cancerous patient tissues and matched NATs | Upregulated | Clinical stage | Patients with low SNHG7 levels demonstrated longer OS compared to those with high levels | — | Jia et al. (2020) |
| 92 cancerous patient tissues and matched NATs | Upregulated | Lymph node metastasis, INSS stage, and optic nerve invasion | Patients with high SNHG7 levels had poorer prognosis compared to those with high levels | — | Chi et al. (2019) | |
| Thyroid cancer | 56 normal samples and 578 tumor samples | Upregulated | Pathology stage | Patients with high SNHG7 levels shorter DFS times compared with those with low levels | — | Chen et al. (2019c) |
| Cervical cancer | 45 cancerous patient tissues and matched NATs | Upregulated | Tumor Size, FIGO Stage, and lymph-Node Metastasis | Patients with high SNHG7 levels demonstrated poorer OS compared with those with low levels | — | Zhao et al. (2020) |
| 60 cancerous patient tissues and matched NATs | Upregulated | TNM stage, lymph node metastasis, and depth of tumor invasion | Patients with high SNHG7 levels demonstrated poorer OS compared with those with low levels | SNHG7 acts as an independent factor for poor OS in patients with gastric cancer | Zeng et al. (2019) | |
| Colorectal cancer | 48 cancerous patient tissues and matched NATs | Upregulated | Clinical stage, lymph node and distant metastasis | High SNHG7 expression was correlated with poor survival | — | Shan et al. (2018) |
| 198 cancerous patient tissues and matched NATs | Upregulated | Invasion depth | High SNHG7 expression was correlated with poor OS. | SNHG7 expression is an independent prognostic risk factor for OS in CRC patients | Hu et al. (2019) | |
| Prostate cancer | 499 cancerous patient tissues and matched NATs | Upregulated | — | — | — | Han et al. (2019) |
| 42 cancerous patient tissues and matched NATs | Upregulated | Gleason score, and tumor stage | Patients with high SNHG7 expression had poor OS compared to those with low expression | — | Qi et al. (2018) | |
| 127 cancerous patient tissues and matched NATs | Upregulated | TNM stage, Gleason score, bone, and pelvic lymph node metastasis | Patients with high SNHG7 expression had poor prognosis compared to those with low expression | SNHG7 acts as an independent factor for poor prognosis in patients with prostate cancer | Xia et al. (2020) | |
| Osteosarcoma | 30 cancerous patient tissues and matched NATs | Upregulated | Tumor size, high Enneking staging, and distant metastasis | Patients with high SNHG7 levels had shorter survival time compared with those with low levels | — | Deng et al. (2018) |
| Chromophobe renal cell carcinoma | Tissue expression of 59 patients retrieved from the TCGA database and 23 NATs | Upregulated | — | SNHG7 level was associated with OS | — | He et al. (2016) |
Clinical prognostic importance of SNHG7 in human cancers.
OS: overall survival, DFS: disease-free survival, PFS: progression-free survival.
Discussion
LncRNAs are a group of ncRNA transcripts defined with a length of >200 nucleotides. Although not elucidated, however, a number of regulatory functions are described for lncRNAs. They are involved in controlling several biological processes, such as cell cycle and proliferation. Accordingly, dysregulation of lncRNAs is identified in a number of human malignancies, suggesting diagnostic and therapeutic potentials. SNHG7 is an lncRNA that has been studied as an oncogenic transcript in a handful of cellular and animal experiments. It is upregulated in cancer cells and tissues retrieved from cancerous patients. SNHG7 is shown to be predominantly localized in the cytoplasm, where it serves as a ceRNA to sponge miRNAs and control expression of downstream targets (Hu et al., 2020). In vitro experiments frequently demonstrate a promoted malignant phenotype of cancer cells on SNHG7 overexpression, whereas its knockdown reverses tumor cell proliferation, migration, and invasion and enhances apoptosis. These regulatory effects are thought to be conducted through an axis of action affecting translation and stability of several transcription factors and signaling pathways mediated by sponging target miRNAs. Not a single one, but plenty of miRNAs are identified to be sponged by SNHG7 (see Table 3). Xenograft animal studies confirm the oncogenic role of SNHG7 as tumor growth and metastasis of grafted cancer cells are promoted, whereas SNHG7 knockdown represses them (see Table 1). For a reported association between upregulated SNHG7 expression and worse clinicopathological characteristics in cancerous patients, clinical studies support oncogenic features of SNHG7. Eventually, Kaplan–Meier survival and Cox univariate and multivariate analyses suggest SNHG7 as a potential prognostic and diagnostic biomarker for human malignancies. Importantly, knockdown experiences and also the contributing role of SNHG7 in chemoresistance suggest it as a potential therapeutic target, which can benefit the anticancer therapies.
TABLE 3
| Cancer type | Targets/Regulators and signaling pathways | Assessed cell lines | Function | References(s) |
|---|---|---|---|---|
| Lung cancer | miR-485-5p/WLS axis | H1650, H1975, A549 and H1299 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Li et al. (2020c) |
| miR-181a-5p/AKT/mTOR axis | A549, and NCI-H1299 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion and ↑apoptosis | Li et al. (2020b) | |
| miR-193b/FAIM2 axis | Beas‐2B, H125, 95D, and A549 | ↑↑ SNHG7: ↑↑FAIM2: ↑tumor cell proliferation, ↑migration, and ↑invasion | She et al. (2018) | |
| miR-181a-5p/E2F7 Axis | NCI-H520, SPC-A1, H-23, and BEAS-2B | Δ SNHG7: ↓tumor cell viability, ↓colony formation, ↓migration, ↓invasion and ↑apoptosis | Wang et al. (2020b) | |
| miR-449a/TGIF2 axis | BEAS-2B, A549, and H1299 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↓EMT | Pang et al. (2020) | |
| FAIM2 | BEAS-2B, H125, 95D, and A594 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion and ↑apoptosis | She et al. (2016) | |
| miR-34a-5p | NSCLC cells | ↑↑ SNHG7: ↑tumor cell proliferation | Chai et al. (2021) | |
| Esophageal cancer | miR-625/SNHG7 axis | TE1, EC109, TE13, and YES2 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Wang et al. (2021) |
| — | HEEC, Eca109, EC9706, TE-10, and TE-11 | Δ SNHG7: ↓tumor cell proliferation, ↑cell cycle arrest, and ↑apoptosis | Xu et al. (2018) | |
| Nasopharyngeal cancer | miR-514a-5p/ELAVL1 axis | NP69, CNE1, CNE2, C666–1 and HNE1 | ↑↑ SNHG7: ↑tumor cell proliferation, and ↑colony formation | Hu et al. (2020) |
| miR-140-5p/GLI3 axis | CNE1, HONE1, C666-1, and CNE2 | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, ↓drug resistance, and ↑apoptosis | Dai et al. (2020) | |
| Liver cancer (hepatocellular carcinoma; HCC) | miR-122-5p/FOXK2 axis | SNU449, Hep3B, and THLE-2 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↓EMT | Zhao et al. (2021) |
| miR-34a/SIRT1 axis | THLE-3, HEK-293, HepG2, and SK-hep-1 | Δ SNHG7: ↑NLRP3-dependent pyroptosis | Chen et al. (2020a) | |
| miR-9-5p/CNNM1 axis | THLE-3, BEL-7404, HCCLM3, Hep3B and HepG2 | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, and ↑apoptosis | Xie et al. (2020) | |
| miR-122-5p/RPL4 axis | Hhu7, Hep3B, HCCLM3, and MHCC97H | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Yang et al. (2019) | |
| miR‐425/Wnt/β‐catenin/EMT pathway | HepG2, and HCC-LM3 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Yao et al. (2019) | |
| Pancreatic cancer | miR-146b-5p/Robo1 axis | PANC-1, SW 1990, BxPC-3 and AsPC-1 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion and ↑apoptosis | Jian and Fan, (2021) |
| miR-342-3p/ID4 axis | HPDE6-C7, HEK293T, AsPC-1, BxPC-3, SW 1990, PANC-1, and PaCa-2 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Cheng et al. (2019b) | |
| Notch1/Jagged1/Hes-1 Signaling Pathway | PANC-1, and AsPC-1 | ↑↑ SNHG7: ↑stemness, and ↓ apoptosis SNHG7 regulates Folfirinox resistance in pancreatic cancer cells | Cheng et al. (2021) | |
| Breast cancer | miR-15a | MCF7, and T47D | Δ SNHG7: ↓tumor cell proliferation, and ↓invasion | Li et al. (2020d) |
| miR-34a | MCF-7, and MDA-MB-231 | Δ SNHG7: ↑chemosensitivity of cancer cells to Adriamycin | Li et al. (2020a) | |
| miR-186 | SK-BR-3, and AU565 | Δ SNHG7: ↓ tumor cell proliferation, ↓migration and ↓EMT, and ↑apoptosis in chemoresistant cancer cells Δ SNHG7: ↑Trastuzumab sensitivity | Zhang et al. (2020b) | |
| miR-34a-5p/LDHA (Glycolysis) axis | MCF10A, MDA-MMB-436, HS578T, SKBR3, MDA-MB-231, and MCF-7 | Δ SNHG7: ↓ tumor cell proliferation, and ↓glycolysis | Zhang et al. (2019b) | |
| miR-381 | MCF-10A, ZR-75–1, HCC-1973, MDA-MB-231, and MDA-MB-468 | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, and ↓invasion | Gao and Zhou, (2019) | |
| miR-34a/Notch-1 pathway | MCF-10A, MCF-7, MDA-MB-231, MDA-MB-157, and MDA-MB-435 | Δ SNHG7: ↓tumor cell proliferation, and ↓invasion | Sun et al. (2019) | |
| miR-186 | MCF-10A, MCF-7, MDA-MB-231 and SKBR3 | Δ SNHG7: ↓tumor cell proliferation, and ↓invasion | Luo et al. (2018) | |
| Colorectal cancer | miR-23a-3p/CXCL12 axis | SW480, LoVo, RKO, and HCT116 | Δ SNHG7: ↓tumor cell viability, ↓proliferation, and ↓migration | Liu et al. (2020) |
| miR-193b/K-ras/ERK/cyclinD1 axis | — | Δ SNHG7: ↓tumor cell proliferation, and ↑apoptosis | Liu et al. (2019) | |
| miR-34a/GALNT7/PI3K/Akt/mTOR pathway | FHC, caco2, SW480, SW620, Hct116, and LoVo | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, ↓vasculogenic mimicry, ↓cell cycle progression, and ↑apoptosis | Li et al. (2018b) | |
| miR-216b/GALNT1 axis | FHC, SW480, SW620, LOVO, and HCT-116 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion and ↑apoptosis | Shan et al. (2018) | |
| Gastric cancer | miR-34a/LDHA (Glycolysis) axis | HGC27, and AGS | Δ SNHG7: ↓ tumor cell viability and ↑chemosensitivity of cancer cells to cisplatin | Pei et al. (2021) |
| miR-485-5p | HS746 T, HGC-27, SNU-1, AGS, and GES-1 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Zhao and Liu, (2021) | |
| miR-34a/Snail/EMT axis | GES-1, MKN-45, SGC-7901, and N87 | Δ SNHG7: ↓tumor cell migration, and ↓invasion | Zhang et al. (2020a) | |
| P15 and P16 | GES-1, BGC823, MGC803, SGC7901, N87, and AGS | Δ SNHG7: ↓tumor cell migration, ↓colony formation, ↑apoptosis, and ↑cell cycle arrest | Wang et al. (2017b) | |
| Bladder cancer | miR-2682-5p/ELK1/Src/FAK signaling pathway | T24, SW780, J82, UM-UC-3, 5637, and SE780 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↑apoptosis | Wang et al. (2020c) |
| Bax, p21, and E-cadherin | SW780, T24, UMUC, and 5637 | Δ SNHG7: ↓tumor cell proliferation, ↓invasion, ↑apoptosis, and ↑expression of Bax, p21 and E-cadherin proteins | Xu et al. (2019) | |
| Wnt/β-catenin pathway | SV-HUC-1, T24, 5637, 253 J, TCC, J82, and EJ | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, ↓migration, and ↑cell cycle arrest | Chen et al. (2019b) | |
| — | SV-HUC-1, T24, J82, and SW780 | Δ SNHG7: ↓tumor cell proliferation, ↓invasion, ↓EMT, and ↑apoptosis | Zhong et al. (2018b) | |
| Pituitary adenocarcinoma | miR-449a | GH1, RC-4B/C, GH3 and MMQ | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↓invasion | Yue et al. (2021) |
| Glioma | miR-342-3p/AKT2 axis | A172, U87, U251, and SHG44 | ↑↑ SNHG7: ↑tumor cell proliferation, ↑migration, and ↑invasion | Cheng et al. (2020) |
| miR-506-3p/CTNNB1 axis | NHA, U87, U251, SHG44, and A172 | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, and ↑apoptosis | Du et al. (2020) | |
| miR-138-5p/EZH2 axis | LN229, A172, U251, and U87 | Δ SNHG7: ↓tumor cell proliferation | Deng et al. (2021) | |
| Glioblastoma (GBM) | miR-449b-5p/MYCN axis | NHA, T98G, U87, U251, and LN229 | Δ SNHG7: ↓GBM cell viability, ↓migration, and ↓invasion | Chen et al. (2020b) |
| miR-5095/Wnt/b-catenin pathway | HEB, A172, U87, T98G, and SHG44 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↑apoptosis | Ren et al. (2018) | |
| Neuroblastoma | miR-323a-5p and miR-342-5p/CCND1 axis | SH-SY5Y, SK-N-SH, NB-1, SK-N-AS, and HUVEC | Δ SNHG7: ↓tumor cell migration, ↓invasion, and ↓glycolysis | Jia et al. (2020) |
| miR-653‐5p/STAT2 axis | SK‐N‐AS, SK‐N‐SH, SH‐SY5Y, IMR‐32, and SK‐N‐BE Hombach and Kretz (2016) ‐C | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, ↓EMT, ↑cell cycle arrest, and ↑apoptosis | Chi et al. (2019) | |
| Ovarian cancer | EZH2/KLF2 axis | OC A2780, OCC1, H8710 and SK-OV3 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↓EMT | Bai et al. (2020) |
| Melanoma | six human UM cell lines | EZH2 | Δ SNHG7: ↓tumor cell proliferation, ↑cell cycle arrest, and ↑apoptosis | Huang et al. (2020) |
| Cervical cancer | DKK1/Wnt/β-catenin axis | H8, C-33A, CaSki, SiHa, and HeLa | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, and ↑apoptosis | Chi et al. (2020) |
| miR-485-5p/JUND axis | Ect1/E6E7, HEK-293T, Hela, SIHA, C-33A and HT-3 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↓EMT | Zhao et al. (2020) | |
| — | HeLa, and C-33A | Δ SNHG7: ↓tumor cell proliferation, and ↓invasion | Zeng et al. (2019) | |
| Thyroid cancer | miR‐449a/ACSL1 axis | Nthy‐ori‐3–1, FTC133, TPC1, BCPAP, and 8505C | Δ SNHG7: ↓tumor cell proliferation, ↓migration, and ↑apoptosis | Guo et al. (2020) |
| — | CAL62, and SW579 | Δ SNHG7: ↓tumor cell proliferation, and ↓cell cycle | Chen et al. (2019c) | |
| BDNF | K1, TPC-1, SW579, and Nthy-ori 3–1 | Δ SNHG7: ↓tumor cell proliferation, ↓colony formation, and ↑apoptosis | Wang et al. (2019) | |
| miR-9-5p/DPP4 axis | TPC-1, and B-CPAP | Δ SNHG7: ↓tumor cell proliferation, and ↓131I resistance | Chen et al. (2021) | |
| Prostate cancer | miR-324-3p/WNT2B axis | RWPE, LNCaP, PC-3, and Du-145 | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↓EMT | Han et al. (2019) |
| miR-503/cyclin D1 axis | WPMY1, LNCaP, VCaP, 22RV1, DU145, and PC3 | Δ SNHG7: ↓tumor cell proliferation, and ↓colony formation | Qi et al. (2018) | |
| Osteosarcoma | p53/DNMT1 axis | U2OS, HOS, MG-63, and Saos-2 | Δ SNHG7: ↓tumor cell proliferation, ↑cell cycle arrest, and ↑apoptosis | Zhang et al. (2019c) |
| miR-34a | hFOB1.19, MG63, SaOS2, HOS, and 143B | Δ SNHG7: ↓tumor cell proliferation, ↓migration, ↓invasion, and ↓EMT | Deng et al. (2018) | |
| miR-34a-5p/RAD9A axis | GSE70415 dataset | in situ evaluations showed that SNHG7 may enhance cell proliferation and metastasis | Wang et al. (2020d) |
An overview to the oncogenic influences of SNHG7 in cell studies of different types of cancer.
Δ: knockdown or silencing, ↓: decrease or repression, ↑: increase or induction, ↑↑: overexpression, EMT: epithelial-to-mesenchymal transition.
Exosomal lncRNAs show high stability and concentrations and, thus, can be detected in body fluids (Tellez-Gabriel and Heymann, 2019). Regarding changes in expression levels of lncRNAs and their high diagnostic values, this makes them appropriate candidates for diagnosis and prediction of prognosis in human cancers (Qian et al., 2020). Several methodologies, including ultracentrifugation, are used to isolate exosomes and then detect the RNAs within. Although, due to low costs and higher accessibility, qRT-PCR is routinely used, high-throughput technologies such as next generation sequencing (NGS) and microarrays have facilitated detection of lncRNAs (Yamada et al., 2018). LncRNAs show acceptable values as diagnostic and prognostic biomarkers for several human cancers (Qian et al., 2020). In this review, we outline the cellular, animal, and clinical studies indicating that this lncRNA is almost universally upregulated in cancer tissues, promotes malignant features of cancer cells, and has prognostic value in various malignancies; however, it seems that SNHG7 diagnostic accuracy in discrimination of human malignancies requires further investigation. Additionally, major limitations of detection methods, such as the impossibility of detecting the amplicon size, limit the number of lncRNAs that can be simultaneously detected, and nonspecific binding, which restricts the clinical application of commonly used qRT-PCR, requires more time to take the lncRNAs into the clinical setting (Jensen, 2012). Finally, there is no CRISPR-based genome editing or siRNA-based method approved or tested for suppression of SNHG7.
In conclusion, regarding a considerable number of studies that reveal oncogenic role of SNHG7 in human cancers and its prognostic value, SNHG7 is suggested as a potential cancer biomarker for human malignancies. Further investigations and more time are required for SNHG7 clinical applications in detection, prediction of prognosis, and treatment of human malignancies.
Statements
Author contributions
SG-F and SN wrote the draft and revised it. MT designed and supervised the study. BH, MH, and HJ collected the data and designed the figures and tables. All the authors read and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Al-Tobasei R. Paneru B. Salem M. (2016). Genome-wide Discovery of Long Non-coding RNAs in Rainbow trout. PLoS One11 (2), e0148940. 10.1371/journal.pone.0148940
2
Amin N. McGrath A. Chen Y.-P. P. (2019). Evaluation of Deep Learning in Non-coding RNA Classification. Nat. Mach Intell.1 (5), 246–256. 10.1038/s42256-019-0051-2
3
Bai Z. Wu Y. Bai S. Yan Y. Kang H. Ma W. et al (2020). Long Non‐coding RNA SNGH7 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with EZH2 in Ovarian Cancer. J. Cel Mol Med.24 (13), 7479–7489. PubMed PMID: 32420685. Epub 05/18. eng. 10.1111/jcmm.15373
4
Boone D. N. Warburton A. Som S. Lee A. V. (2020). SNHG7 Is a lncRNA Oncogene Controlled by Insulin-like Growth Factor Signaling through a Negative Feedback Loop to Tightly Regulate Proliferation. Sci. Rep.10 (1), 8583. PubMed PMID: 32444795. Pubmed Central PMCID: PMC7244715. Epub 2020/05/24. eng. 10.1038/s41598-020-65109-7
5
Chai R. Xu C. Lu L. Liu X. Ma Z. (2021). Quercetin Inhibits Proliferation of and Induces Apoptosis in Non-small-cell Lung Carcinoma via the lncRNA SNHG7/miR-34a-5p Pathway. Immunopharmacology and immunotoxicology43 (6), 693–703. PubMed PMID: 34448661. Epub 2021/08/28. eng. 10.1080/08923973.2021.1966032
6
Chaudhry M. (2013). Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-coding RNA in X-Rays-Treated Lymphoblastoid Cells. Ijms14 (5), 9099–9110. PubMed PMID: 23698766. eng. 10.3390/ijms14059099
7
Chen K. Abuduwufuer A. Zhang H. Luo L. Suotesiyali M. Zou Y. (2019). SNHG7 Mediates Cisplatin-Resistance in Non-small Cell Lung Cancer by Activating PI3K/AKT Pathway. Eur. Rev. Med. Pharmacol. Sci.23 (16), 6935–6943. PubMed PMID: 31486493. Epub 2019/09/06. eng. 10.26355/eurrev_201908_18733
8
Chen L. Zhu J. Zhang L. J. (2019). Long Non-coding RNA Small Nucleolar RNA Host Gene 7 Is Upregulated and Promotes Cell Proliferation in Thyroid Cancer. Oncol. Lett.18 (5), 4726–4734. PubMed PMID: 31611982. Epub 08/27. eng. 10.3892/ol.2019.10782
9
Chen W. Yu J. Xie R. Zhou T. Xiong C. Zhang S. et al (2021). Roles of the SNHG7/microRNA-9-5p/DPP4 ceRNA N-etwork in the G-rowth and 131I R-esistance of T-hyroid C-arcinoma C-ells through PI3K/Akt A-ctivation. Oncol. Rep.45 (4), 3. PubMed PMID: 33649840. Epub 03/02. eng. 10.3892/or.2021.7954
10
Chen Y. Yuan S. Ning T. Xu H. Guan B. (2020). SNHG7 Facilitates Glioblastoma Progression by Functioning as a Molecular Sponge for MicroRNA-449b-5p and Thereby Increasing MYCN Expression. Technology in Cancer Research & Treatment.
11
Chen Y. Peng Y. Xu Z. Ge B. Xiang X. Zhang T. et al (2019). Knockdown of lncRNA SNHG7 Inhibited Cell Proliferation and Migration in Bladder Cancer through Activating Wnt/β-Catenin Pathway. Pathol. - Res. Pract.215 (2), 302–307. PubMed PMID: 30527358. Epub 2018/12/12. eng. 10.1016/j.prp.2018.11.015
12
Chen Z. Liu Z. Shen L. Jiang H. (2019). Long Non-coding RNA SNHG7 Promotes the Fracture Repair through Negative Modulation of miR-9. Am. J. Transl Res.11 (2), 974–982. PubMed PMID: 30899396. eng.
13
Chen Z. He M. Chen J. Li C. Zhang Q. (2020). Long Non-coding RNA SNHG7 I-nhibits NLRP3-dependent P-yroptosis by T-argeting the miR-34a/SIRT1 axis in L-iver C-ancer. Oncol. Lett.20 (1), 893–901. PubMed PMID: 32566017. Epub 05/18. eng. 10.3892/ol.2020.11635
14
Cheng D. Fan J. Ma Y. Zhou Y. Qin K. Shi M. et al (2019). LncRNA SNHG7 Promotes Pancreatic Cancer Proliferation through ID4 by Sponging miR-342-3p. Cell Biosci.9 (1), 28–11. 10.1186/s13578-019-0290-2
15
Cheng D. Fan J. Qin K. Zhou Y. Yang J. Ma Y. et al (2021). LncRNA SNHG7 Regulates Mesenchymal Stem Cell through the Notch1/Jagged1/Hes-1 Signaling Pathway and Influences Folfirinox Resistance in Pancreatic Cancer. Front. Oncol.11 (3235). 10.3389/fonc.2021.719855
16
Cheng D. Fan J. Ma Y. Zhou Y. Qin K. Shi M. et al (2019). LncRNA SNHG7 Promotes Pancreatic Cancer Proliferation through ID4 by Sponging miR-342-3p. Cel Biosci.9, 28. PubMed PMID: 30949340. Pubmed Central PMCID: PMC6431029. Epub 2019/04/06. eng. 10.1186/s13578-019-0290-2
17
Cheng G. Zheng J. Wang L. (2020). LncRNA SNHG7 Promotes Glioma Cells Viability, Migration and Invasion by Regulating miR-342-3p/AKT2 axis. Int. J. Neurosci., 1–13. PubMed PMID: 32628059. Epub 2020/07/07. eng. 10.1080/00207454.2020.1790556
18
Chi C. Li M. Hou W. Chen Y. Zhang Y. Chen J. (2020). Long Noncoding RNA SNHG7 Activates Wnt/β-Catenin Signaling Pathway in Cervical Cancer Cells by Epigenetically Silencing DKK1. Cancer Biother. Radiopharm.35 (5), 329–337. PubMed PMID: 32275170. Epub 2020/04/11. eng. 10.1089/cbr.2019.3004
19
Chi R. Chen X. Liu M. Zhang H. Li F. Fan X. et al (2019). Role of SNHG7‐miR‐653‐5p‐STAT2 Feedback Loop in Regulating Neuroblastoma Progression. J. Cel Physiol.234 (8), 13403–13412. PubMed PMID: 30623419. Epub 2019/01/10. eng. 10.1002/jcp.28017
20
Crick F. (1970). Central Dogma of Molecular Biology. Nature227 (5258), 561–563. 10.1038/227561a0
21
Dahariya S. Paddibhatla I. Kumar S. Raghuwanshi S. Pallepati A. Gutti R. K. (2019). Long Non-coding RNA: Classification, Biogenesis and Functions in Blood Cells. Mol. Immunol.112, 82–92. 10.1016/j.molimm.2019.04.011
22
Dai Y. Zhang X. Xing H. Zhang Y. Cao H. Sang J. et al (2020). Downregulated Long Non-coding RNA SNHG7 Restricts Proliferation and Boosts Apoptosis of Nasopharyngeal Carcinoma Cells by Elevating microRNA-140-5p to Suppress GLI3 Expression. Cell Cycle19 (4), 448–463. PubMed PMID: 31944163. Epub 01/16. eng. 10.1080/15384101.2020.1712033
23
Deng Y. Cheng L. Lv Z. Zhu H. Meng X. (2021). lncRNA SNHG7 Promotes Cell Proliferation in Glioma by Acting as a Competing Endogenous RNA and Sponging miR-138-5p to R-egulate EZH2 E-xpression. Oncol. Lett.22 (1), 565. PubMed PMID: 34113393. Pubmed Central PMCID: PMC8185700. Epub 2021/06/12. eng. 10.3892/ol.2021.12826
24
Deng Y. Zhao F. Zhang Z. Sun F. Wang M. (2018). Long Noncoding RNA SNHG7 Promotes the Tumor Growth and Epithelial-To-Mesenchymal Transition via Regulation of miR-34a Signals in Osteosarcoma. Cancer Biother. Radiopharm.33 (9), 365–372. PubMed PMID: 29989838. Epub 2018/07/11. eng. 10.1089/cbr.2018.2503
25
Du L. Xu Z. Wang X. Liu F. (2020). Integrated Bioinformatics Analysis Identifies microRNA-376a-3p as a New microRNA Biomarker in Patient with Coronary Artery Disease. Am. J. Transl Res.12 (2), 633–648.
26
Elgar G. Vavouri T. (2008). Tuning in to the Signals: Noncoding Sequence Conservation in Vertebrate Genomes. Trends Genet.24 (7), 344–352. PubMed PMID: 18514361. Epub 2008/06/03. eng. 10.1016/j.tig.2008.04.005
27
Gao F. Cai Y. Kapranov P. Xu D. (2020). Reverse-genetics Studies of lncRNAs-What We Have Learnt and Paths Forward. Genome Biol.21 (1), 93. 10.1186/s13059-020-01994-5
28
Gao Y. T. Zhou Y. C. (2019). Long Non-coding RNA (lncRNA) Small Nucleolar RNA Host Gene 7 (SNHG7) Promotes Breast Cancer Progression by Sponging miRNA-381. Eur. Rev. Med. Pharmacol. Sci.23 (15), 6588–6595. PubMed PMID: 31378900. Epub 2019/08/06. eng. 10.26355/eurrev_201908_18545
29
Guo L. Lu J. Gao J. Li M. Wang H. Zhan X. (2020). The Function of SNHG7/miR‐449a/ACSL1 axis in Thyroid Cancer. J. Cel Biochem.121 (10), 4034–4042. PubMed PMID: 31961004. Epub 2020/01/22. eng. 10.1002/jcb.29569
30
Guttman M. Amit I. Garber M. French C. Lin M. F. Feldser D. et al (2009). Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-coding RNAs in Mammals. Nature458 (7235), 223–227. PubMed PMID: 19182780. Pubmed Central PMCID: PMC2754849. Epub 2009/02/03. eng. 10.1038/nature07672
31
Han Y. Hu H. Zhou J. (2019). Knockdown of LncRNA SNHG7 Inhibited Epithelial-Mesenchymal Transition in Prostate Cancer Though miR-324-3p/WNT2B axis In Vitro. Pathol. Res. Pract.215 (10), 152537. 10.1016/j.prp.2019.152537
32
Hartford C. C. R. Lal A. (2020). When Long Noncoding Becomes Protein Coding. Mol. Cel Biol.40 (6), e00528–19. 10.1128/MCB.00528-19
33
He H. Xu M. Kuang Y. Han X. Wang M. Yang Q. (2016). Biomarker and Competing Endogenous RNA Potential of Tumor-specific Long Noncoding RNA in Chromophobe Renal Cell Carcinoma. Ott9, 6399–6406. PubMed PMID: 27799788. eng. 10.2147/ott.s116392
34
Hombach S. Kretz M. (2016). Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol.937, 3–17. PubMed PMID: 27573892. Epub 2016/08/31. eng. 10.1007/978-3-319-42059-2_1
35
Hu W. Li H. Wang S. (2020). LncRNA SNHG7 Promotes the Proliferation of Nasopharyngeal Carcinoma by miR-514a-5p/ELAVL1 axis. BMC cancer20 (1), 376. PubMed PMID: 32370736. eng. 10.1186/s12885-020-06775-8
36
Hu Y. Wang L. Li Z. Wan Z. Shao M. Wu S. et al (2019). Potential Prognostic and Diagnostic Values of CDC6, CDC45, ORC6 and SNHG7 in Colorectal Cancer. Ott12, 11609–11621. PubMed PMID: 32021241. eng. 10.2147/ott.s231941
37
Huang W. Wu X. Xue Y. Zhou Y. Xiang H. Yang W. et al (2020). MicroRNA-3614 Regulates Inflammatory Response via Targeting TRAF6-Mediated MAPKs and NF-Κb Signaling in the Epicardial Adipose Tissue with Coronary Artery Disease. Int. J. Cardiol.
38
Iyer M. K. Niknafs Y. S. Malik R. Singhal U. Sahu A. Hosono Y. et al (2015). The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet.47 (3), 199–208. 10.1038/ng.3192
39
Jensen E. C. (2012). Real-time Reverse Transcription Polymerase Chain Reaction to Measure mRNA: Use, Limitations, and Presentation of Results. Anat. Rec.295 (1), 1–3. PubMed PMID: 22095866. Epub 2011/11/19. eng. 10.1002/ar.21487
40
Jia J. Zhang D. Zhang J. Yang L. Zhao G. Yang H. et al (2020). Long Non-coding RNA SNHG7 Promotes Neuroblastoma Progression through Sponging miR-323a-5p and miR-342-5p. Biomed. Pharmacother.128, 110293. 10.1016/j.biopha.2020.110293
41
Jian Y. Fan Q. (2021). Long Non-coding RNA SNHG7 F-acilitates P-ancreatic C-ancer P-rogression by R-egulating the miR-146b-5p/Robo1 axis. Exp. Ther. Med.21 (4), 398. PubMed PMID: 33680120. Epub 02/24. eng. 10.3892/etm.2021.9829
42
Jing L. Li S. Wang J. Zhang G. (2019). Long Non‐coding RNA Small Nucleolar RNA Host Gene 7 Facilitates Cardiac Hypertrophy via Stabilization of SDA1 Domain Containing 1 mRNA. J. Cel Biochem.120 (9), 15089–15097. PubMed PMID: 31026094. Epub 2019/04/27. eng. 10.1002/jcb.28770
43
Li L. Ye D. Liu L. Li X. Liu J. Su S. et al (2020). Long Noncoding RNA SNHG7 Accelerates Proliferation, Migration and Invasion of Non-small Cell Lung Cancer Cells by Suppressing miR-181a-5p through AKT/mTOR Signaling Pathway. Cmar12, 8303–8312. PubMed PMID: 32982425. eng. 10.2147/cmar.s258487
44
Li W. Zheng Y. Mao B. Wang F. Zhong Y. Cheng D. (2020). SNHG17 Upregulates WLS Expression to Accelerate Lung Adenocarcinoma Progression by Sponging miR-485-5p. Biochem. Biophysical Res. Commun.533 (4), 1435–1441. PubMed PMID: 33109341. Epub 2020/10/29. eng. 10.1016/j.bbrc.2020.09.130
45
Li Y. Guo X. Wei Y. (2020). LncRNA SNHG7 Inhibits Proliferation and Invasion of Breast Cancer Cells by Regulating miR-15a Expression. J. Buon25 (4), 1792–1798. PubMed PMID: 33099915. Epub 2020/10/26. eng.
46
Li Y. Zeng C. Hu J. Pan Y. Shan Y. Liu B. et al (2018). Long Non-coding RNA-SNHG7 Acts as a Target of miR-34a to Increase GALNT7 Level and Regulate PI3K/Akt/mTOR Pathway in Colorectal Cancer Progression. J. Hematol. Oncol.11 (1), 89–17. 10.1186/s13045-018-0632-2
47
Li Y. Zeng C. Hu J. Pan Y. Shan Y. Liu B. et al (2018). Long Non-coding RNA-SNHG7 Acts as a Target of miR-34a to Increase GALNT7 Level and Regulate PI3K/Akt/mTOR Pathway in Colorectal Cancer Progression. J. Hematol. Oncol.11 (1), 89. PubMed PMID: 29970122. eng. 10.1186/s13045-018-0632-2
48
Li Z.-h. Yu N.-s. Deng Q. Zhang Y. Hu Y.-y. Liu G. et al (2020). LncRNA SNHG7 Mediates the Chemoresistance and Stemness of Breast Cancer by Sponging miR-34a. Front. Oncol.10, 592757. PubMed PMID: 33330080. eng. 10.3389/fonc.2020.592757
49
Liu K. L. Wu J. Li W. K. Li N. S. Li Q. Lao Y. Q. (2019). LncRNA SNHG7 Is an Oncogenic Biomarker Interacting with MicroRNA-193b in Colon Carcinogenesis. Clin. Lab.65 (11). 10.7754/Clin.Lab.2019.190501PubMed PMID: 31721543. Epub 2019/11/14. eng
50
Liu Y. Li Q. Tang D. Li M. Zhao P. Yang W. et al (2020). SNHG17 Promotes the Proliferation and Migration of Colorectal Adenocarcinoma Cells by Modulating CXCL12-Mediated Angiogenesis. Cancer Cel Int.20 (1), 566. PubMed PMID: 33292246. eng. 10.1186/s12935-020-01621-0
51
Luo X. Song Y. Tang L. Sun D. H. Ji D. G. (2018). LncRNA SNHG7 Promotes Development of Breast Cancer by Regulating microRNA-186. Eur. Rev. Med. Pharmacol. Sci.22 (22), 7788–7797. PubMed PMID: 30536320. Epub 2018/12/12. eng. 10.26355/eurrev_201811_16403
52
Mattick J. S. (2001). Non‐coding RNAs: the Architects of Eukaryotic Complexity. EMBO Rep.2 (11), 986–991. PubMed PMID: 11713189. eng. 10.1093/embo-reports/kve230
53
Najafi S. Tan S. C. Raee P. Rahmati Y. Asemani Y. Lee E. H. C. et al (2022). Gene Regulation by Antisense Transcription: A Focus on Neurological and Cancer Diseases. Biomed. Pharmacother.145, 112265. 10.1016/j.biopha.2021.112265
54
Pang L. Cheng Y. Zou S. Song J. (2020). Long Noncoding RNA SNHG7 Contributes to Cell Proliferation, Migration, Invasion and Epithelial to Mesenchymal Transition in Non‐small Cell Lung Cancer by Regulating miR‐449a/TGIF2 axis. Thorac. Cancer11 (2), 264–276. PubMed PMID: 31793741. Epub 12/03. eng. 10.1111/1759-7714.13245
55
Pei L.-J. Sun P.-J. Ma K. Guo Y.-Y. Wang L.-Y. Liu F.-D. (2021). LncRNA-SNHG7 Interferes with miR-34a to De-sensitize Gastric Cancer Cells to Cisplatin. Cbm30, 127–137. 10.3233/cbm-201621
56
Pei Y.-f. He Y. Hu L.-z. Zhou B. Xu H.-y. Liu X.-q. (2020). The Crosstalk between lncRNA-SNHG7/miRNA-181/cbx7 Modulates Malignant Character in Lung Adenocarcinoma. Am. J. Pathol.190 (6), 1343–1354. PubMed PMID: 32201260. Epub 2020/03/24. eng. 10.1016/j.ajpath.2020.02.011
57
Qi H. Wen B. Wu Q. Cheng W. Lou J. Wei J. et al (2018). Long Noncoding RNA SNHG7 Accelerates Prostate Cancer Proliferation and Cycle Progression through Cyclin D1 by Sponging miR-503. Biomed. Pharmacother.102, 326–332. PubMed PMID: 29571017. Epub 2018/03/24. eng. 10.1016/j.biopha.2018.03.011
58
Qian Y. Shi L. Luo Z. (2020). Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med.30, 7. 10.3389/fmed.2020.612393
59
Quinn J. J. Chang H. Y. (2016). Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet.17 (1), 47–62. 10.1038/nrg.2015.10
60
Rahmati Y. Asemani Y. Aghamiri S. Ezzatifar F. Najafi S. (2021). CiRS-7/CDR1as; an Oncogenic Circular RNA as a Potential Cancer Biomarker. Pathol. - Res. Pract.227, 153639. 10.1016/j.prp.2021.153639
61
Ramírez-Colmenero A. Oktaba K. Fernandez-Valverde S. L. (2020). Evolution of Genome-Organizing Long Non-coding RNAs in Metazoans. Front. Genet.11 (1512).
62
Ren J. Yang Y. Xue J. Xi Z. Hu L. Pan S.-J. et al (2018). Long Noncoding RNA SNHG7 Promotes the Progression and Growth of Glioblastoma via Inhibition of miR-5095. Biochem. Biophysical Res. Commun.496 (2), 712–718. PubMed PMID: 29360452. Epub 2018/01/24. eng. 10.1016/j.bbrc.2018.01.109
63
Sarropoulos I. Marin R. Cardoso-Moreira M. Kaessmann H. (2019). Developmental Dynamics of lncRNAs across Mammalian Organs and Species. Nature571 (7766), 510–514. PubMed PMID: 31243368. Epub 06/26. eng. 10.1038/s41586-019-1341-x
64
Sayad A. Najafi S. Kashi A. H. Hosseini S. J. Akrami S. M. Taheri M. et al (2021). Circular RNAs in Renal Cell Carcinoma: Functions in Tumorigenesis and Diagnostic and Prognostic Potentials. Pathol. - Res. Pract., 153720. 10.1016/j.prp.2021.153720
65
Shan Y. Ma J. Pan Y. Hu J. Liu B. Jia L. (2018). LncRNA SNHG7 Sponges miR-216b to Promote Proliferation and Liver Metastasis of Colorectal Cancer through Upregulating GALNT1. Cell Death Dis.9 (7), 722. PubMed PMID: 29915311. eng. 10.1038/s41419-018-0759-7
66
She K. Yan H. Huang J. Zhou H. He J. (2018). miR-193b Availability Is Antagonized by LncRNA-SNHG7 for FAIM2-Induced Tumour Progression in Non-small Cell Lung Cancer. Cell Prolif51 (1), e12406. PubMed PMID: 29131440. Epub 11/12. eng. 10.1111/cpr.12406
67
She K. Huang J. Zhou H. Huang T. Chen G. He J. (2016). lncRNA-SNHG7 Promotes the Proliferation, Migration and Invasion and Inhibits Apoptosis of Lung Cancer Cells by Enhancing the FAIM2 Expression. Oncol. Rep.36 (5), 2673–2680. 10.3892/or.2016.5105
68
Shen A. Ma J. Hu X. Cui X. (2020). High Expression of lncRNA-SNHG7 Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Oncol. Lett.19 (6), 3959–3963. PubMed PMID: 32382340. Pubmed Central PMCID: PMC7202315. Epub 2020/05/10. eng. 10.3892/ol.2020.11490
69
Shi J. Ding W. Lu H. (2020). Identification of Long Non-coding RNA SNHG Family as Promising Prognostic Biomarkers in Acute Myeloid Leukemia. Ott13, 8441–8450. PubMed PMID: 32922034. eng. 10.2147/ott.s265853
70
Statello L. Guo C.-J. Chen L.-L. Huarte M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cel Biol.22 (2), 96–118. 10.1038/s41580-020-00315-9
71
Sun X. Huang T. Liu Z. Sun M. Luo S. (2019). LncRNA SNHG7 Contributes to Tumorigenesis and Progression in Breast Cancer by Interacting with miR-34a through EMT Initiation and the Notch-1 Pathway. Eur. J. Pharmacol.856, 172407. 10.1016/j.ejphar.2019.172407
72
Taheri M. Najafi S. Basiri A. Hussen B. M. Baniahmad A. Jamali E. et al (2021). The Role and Clinical Potentials of Circular RNAs in Prostate Cancer. Front. Oncol.11, 781414. PubMed PMID: 34804984. eng. 10.3389/fonc.2021.781414
73
Tellez-Gabriel M. Heymann D. (2019). Exosomal lncRNAs: the Newest Promising Liquid Biopsy. Cancer Drug Resist.2 (4), 1002–1017. 10.20517/cdr.2019.69
74
Wang J. Zhang S. Li X. Gong M. (2020). LncRNA SNHG7 Promotes Cardiac Remodeling by Upregulating ROCK1 via Sponging miR-34-5p. Aging12 (11), 10441–10456. PubMed PMID: 32507765. Pubmed Central PMCID: PMC7346013. Epub 2020/06/09. eng. 10.18632/aging.103269
75
Wang L. Zhang L. Wang L. (2020). SNHG7 Contributes to the Progression of Non-small-cell Lung Cancer via the SNHG7/miR-181a-5p/E2F7 Axis. Cmar12, 3211–3222. PubMed PMID: 32440218. eng. 10.2147/cmar.s240964
76
Wang M. W. Liu J. Liu Q. Xu Q. H. Li T. F. Jin S. et al (2017). LncRNA SNHG7 Promotes the Proliferation and Inhibits Apoptosis of Gastric Cancer Cells by Repressing the P15 and P16 Expression. Eur. Rev. Med. Pharmacol. Sci.21 (20), 4613–4622. PubMed PMID: 29131253. Epub 2017/11/14. eng.
77
Wang W. Chen S. Song X. Gui J. Li Y. Li M. (20202020). ELK1/lncRNA-SNHG7/miR-2682-5p Feedback Loop Enhances Bladder Cancer Cell Growth. Life Sci.262, 118386. 10.1016/j.lfs.2020.118386
78
Wang Y. Gao Y. Guo S. Chen Z. (2020). Integrated Analysis of lncRNA-Associated ceRNA Network Identified Potential Regulatory Interactions in Osteosarcoma. Genet. Mol. Biol.43 (2), e20190090. 10.1590/1678-4685-GMB-2019-0090
79
Wang Y. Bao D. Wan L. Zhang C. Hui S. Guo H. (2021). Long Non-coding RNA Small Nucleolar RNA Host Gene 7 Facilitates the Proliferation, Migration, and Invasion of Esophageal Cancer Cells by Regulating microRNA-625. J. Gastrointest. Oncol.12 (2), 423–432. PubMed PMID: 34012636. eng. 10.21037/jgo-21-147
80
Wang Y. H. Huo B. L. Li C. Ma G. Cao W. (2019). Knockdown of Long Noncoding RNA SNHG7 Inhibits the Proliferation and Promotes Apoptosis of Thyroid Cancer Cells by Downregulating BDNF. Eur. Rev. Med. Pharmacol. Sci.23 (11), 4815–4821. PubMed PMID: 31210313. Epub 2019/06/19. eng. 10.26355/eurrev_201906_18067
81
Wang Y. Wang X. Li Z. Chen L. Zhou L. Li C. et al (2017). Two Single Nucleotide Polymorphisms (Rs2431697 and Rs2910164) of miR-146a Are Associated with Risk of Coronary Artery Disease. Ijerph14 (5), 514. 10.3390/ijerph14050514
82
Wu P. Tang Y. Fang X. Xie C. Zeng J. Wang W. et al (2019). Metformin Suppresses Hypopharyngeal Cancer Growth by Epigenetically Silencing Long Non-coding RNA SNHG7 in FaDu Cells. Front. Pharmacol.10, 143. 10.3389/fphar.2019.00143
83
Xia Q. Li J. Yang Z. Zhang D. Tian J. Gu B. (2020). Long Non-coding RNA Small Nucleolar RNA Host Gene 7 Expression Level in Prostate Cancer Tissues Predicts the Prognosis of Patients with Prostate Cancer. Medicine (Baltimore)99 (7), e18993. e. PubMed PMID: 32049793. eng. 10.1097/md.0000000000018993
84
Xie Y. Wang Y. Gong R. Lin J. Li X. Ma J. et al (2020). SNHG7 Facilitates Hepatocellular Carcinoma Occurrence by Sequestering miR-9-5p to Upregulate CNNM1 Expression. Cancer Biother. Radiopharm.35 (10), 731–740. PubMed PMID: 32397799. Epub 2020/05/14. eng. 10.1089/cbr.2019.2996
85
Xu C. Zhou J. Wang Y. Wang A. Su L. Liu S. et al (2019). Inhibition of Malignant Human Bladder Cancer Phenotypes through the Down-Regulation of the Long Non-coding RNA SNHG7. J. Cancer10 (2), 539–546. PubMed PMID: 30719150. Pubmed Central PMCID: PMC6360294. Epub 2019/02/06. eng. 10.7150/jca.25507
86
Xu L. J. Yu X. J. Wei B. Hui H. X. Sun Y. Dai J. et al (2018). LncRNA SNHG7 Promotes the Proliferation of Esophageal Cancer Cells and Inhibits its Apoptosis. Eur. Rev. Med. Pharmacol. Sci.22 (9), 2653–2661. PubMed PMID: 29771415. Epub 2018/05/18. eng. 10.26355/eurrev_201805_14961
87
Xue Y. H. Ge Y. Q. (2020). Construction of lncRNA Regulatory Networks Reveal the Key lncRNAs Associated with Pituitary Adenomas Progression. Math. Biosci. Eng.17 (3), 2138–2149. PubMed PMID: 32233527. Epub 2020/04/03. eng. 10.3934/mbe.2020113
88
Yamada A. Yu P. Lin W. Okugawa Y. Boland C. R. Goel A. (2018). A RNA-Sequencing Approach for the Identification of Novel Long Non-coding RNA Biomarkers in Colorectal Cancer. Sci. Rep.8 (1), 575. PubMed PMID: 29330370. eng. 10.1038/s41598-017-18407-6
89
Yang X. Sun L. Wang L. Yao B. Mo H. Yang W. (2019). LncRNA SNHG7 Accelerates the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cells via Regulating miR-122-5p and RPL4. Biomed. Pharmacother.118, 109386. 10.1016/j.biopha.2019.109386
90
Yao X. Liu C. Liu C. Xi W. Sun S. Gao Z. (2019). lncRNA SNHG7 Sponges miR‐425 to Promote Proliferation, Migration, and Invasion of Hepatic Carcinoma Cells via Wnt/β‐catenin/EMT Signalling Pathway. Cell Biochem Funct37 (7), 525–533. PubMed PMID: 31478234. Epub 09/02. eng. 10.1002/cbf.3429
91
Yi P. Zhang W. Yang M. Lu Q. Wu H. (2021). LncRNA SNHG7 Serves as a Potential Biomarker on the Prognosis of Human Solid Tumors: A Meta-Analysis. Cpb22 (11), 1501–1510. PubMed PMID: 33397233. Epub 2021/01/06. eng. 10.2174/1389201022666210104121207
92
Yu F. Dong P. Mao Y. Zhao B. Huang Z. Zheng J. (2019). Loss of lncRNA-SNHG7 Promotes the Suppression of Hepatic Stellate Cell Activation via miR-378a-3p and DVL2. Mol. Ther. - Nucleic Acids17, 235–244. PubMed PMID: 31272073. Epub 06/07. eng. 10.1016/j.omtn.2019.05.026
93
Yu K. Yuan W. Huang C. Xiao L. Xiao R. Zeng P. et al (2021). The Prognostic Value of Long Non-coding RNA SNHG7 in Human Cancer: A Meta-Analysis. Curr. Pharm. Biotechnol.22, 1–13. 10.2174/1389201022666210810100607
94
Yu K. Yuan W. Huang C. Xiao L. Xiao R. Zeng P. et al (2021). The Prognostic Value of Long Non-coding RNA SNHG7 in Human Cancer: A Meta-Analysis. Curr. Pharm. Biotechnol.PubMed PMID: 34375186. Epub 2021/08/11. eng. 10.2174/1389201022666210810100607
95
Yu Z. Wang Y. Wang B. Zhai J. (2020). Metformin Affects Paclitaxel Sensitivity of Ovarian Cancer Cells through Autophagy Mediated by Long Noncoding RNASNHG7/miR-3127-5p Axis. Cancer Biother. Radiopharm. PubMed PMID: 32522016. Epub 2020/06/12. eng.
96
Yue X. Dong C. Ye Z. Zhu L. Zhang X. Wang X. et al (2021). LncRNA SNHG7 Sponges miR-449a to Promote Pituitary Adenomas Progression. Metab. Brain Dis.36 (1), 123–132. PubMed PMID: 32880813. Epub 2020/09/04. eng. 10.1007/s11011-020-00611-5
97
Zeng J. Ma Y. X. Liu Z. H. Zeng Y. L. (2019). LncRNA SNHG7 Contributes to Cell Proliferation, Invasion and Prognosis of Cervical Cancer. Eur. Rev. Med. Pharmacol. Sci.23 (21), 9277–9285. PubMed PMID: 31773679. Epub 2019/11/28. eng. 10.26355/eurrev_201911_19420
98
Zhang G. D. Gai P. Z. Liao G. Y. Li Y. (2019). LncRNA SNHG7 Participates in Osteosarcoma Progression by Down-Regulating P53 via Binding to DNMT1. Eur. Rev. Med. Pharmacol. Sci.23 (9), 3602–3610. PubMed PMID: 31114984. Epub 2019/05/23. eng. 10.26355/eurrev_201905_17782
99
Zhang H. Zhang X.-Y. Kang X.-N. Jin L.-J. Wang Z.-Y. (2020). LncRNA-SNHG7 Enhances Chemotherapy Resistance and Cell Viability of Breast Cancer Cells by Regulating miR-186. Cmar12, 10163–10172. PubMed PMID: 33116871. eng. 10.2147/cmar.s270328
100
Zhang L. Fu Y. Guo H. (2019). c-Myc-Induced Long Non-coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer. J. Breast Cancer22 (4), 533–547. PubMed PMID: 31897328. eng. 10.4048/jbc.2019.22.e54
101
Zhang P. Shi L. Song L. Long Y. Yuan K. Ding W. et al (2020). LncRNA CRNDE and lncRNA SNHG7 Are Promising Biomarkers for Prognosis in Synchronous Colorectal Liver Metastasis Following Hepatectomy. Cmar12, 1681–1692. PubMed PMID: 32210611. eng. 10.2147/cmar.s233147
102
Zhang X. Wang W. Zhu W. Dong J. Cheng Y. Yin Z. et al (2019). Mechanisms and Functions of Long Non-coding RNAs at Multiple Regulatory Levels. Ijms20 (22), 5573. PubMed PMID: 31717266. eng. 10.3390/ijms20225573
103
Zhang Y. Yuan Y. Zhang Y. Cheng L. Zhou X. Chen K. (2020). SNHG7 Accelerates Cell Migration and Invasion through Regulating miR-34a-Snail-EMT axis in Gastric Cancer. Cell Cycle19 (1), 142–152. 10.1080/15384101.2019.1699753
104
Zhao D. Zhang H. Long J. Li M. (2020). LncRNA SNHG7 Functions as an Oncogene in Cervical Cancer by Sponging miR-485-5p to Modulate JUND Expression. Ott13, 1677–1689. PubMed PMID: 32161467. eng. 10.2147/ott.s237802
105
Zhao Z. Gao J. Huang S. (2021). LncRNA SNHG7 Promotes the HCC Progression through miR-122-5p/FOXK2 Axis. Dig. Dis. Sci. 10.1007/s10620-021-06918-2
106
Zhao Z. Liu X. (2021). LncRNA SNHG7 Regulates Gastric Cancer Progression by miR-485-5p. J. Oncol.2021, 6147962. PubMed PMID: 34512753. eng. 10.1155/2021/6147962
107
Zhong X. Long Z. Wu S. Xiao M. Hu W. (2018). LncRNA-SNHG7 Regulates Proliferation, Apoptosis and Invasion of Bladder Cancer Cells Assurance Guidelines. J. BUON23 (3), 776–781.
108
Zhong X. Long Z. Wu S. Xiao M. Hu W. (2018). LncRNA-SNHG7 Regulates Proliferation, Apoptosis and Invasion of Bladder Cancer Cells Assurance Guidelines. J. Buon23 (3), 776–781. PubMed PMID: 30003751. Epub 2018/07/14. eng.
109
Zhu Y. Qian X. H. Ji G. Z. Yang L. H. (2021). Clinicopathological and Prognostic Value of Long Noncoding RNA SNHG7 in Cancer Patients: a Meta-Analysis. Eur. Rev. Med. Pharmacol. Sci.25 (7), 2916–2926. PubMed PMID: 33877655. Epub 2021/04/21. eng. 10.26355/eurrev_202104_25545
Summary
Keywords
SNHG7, non-coding RNA, lncRNA, cancer, biomarker
Citation
Najafi S, Ghafouri-Fard S, Hussen BM, Jamal HH, Taheri M and Hallajnejad M (2022) Oncogenic Roles of Small Nucleolar RNA Host Gene 7 (SNHG7) Long Noncoding RNA in Human Cancers and Potentials. Front. Cell Dev. Biol. 9:809345. doi: 10.3389/fcell.2021.809345
Received
04 November 2021
Accepted
10 December 2021
Published
17 January 2022
Volume
9 - 2021
Edited by
Shiv K. Gupta, Mayo Clinic, United States
Reviewed by
Atefe Abak, Tabriz University of Medical Sciences, Iran
Macrina Beatriz Silva Cázares, Autonomous University of San Luis Potosí, Mexico
Updates
Copyright
© 2022 Najafi, Ghafouri-Fard, Hussen, Jamal, Taheri and Hallajnejad.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, mohammad_823@yahoo.com; Mohammad Hallajnejad, hallajnejad@gmail.com
This article was submitted to Molecular and Cellular Oncology, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.