Abstract
Hepatocellular carcinoma (HCC) is the most frequent and deadly type of liver cancer. While the underlying molecular mechanisms are poorly understood, it is documented that lncRNAs may play key roles. Many HCC-associated lncRNAs have been linked to HBV and HCV infection, mediating gene expression, cell growth, development, and death. Studying the regulatory mechanisms and biological functions of HCC-related lncRNAs will assist our understanding of HCC pathogenesis as well as its diagnosis and management. Here, we address the potential of dysregulated lncRNAs in HCC as diagnostic and therapeutic biomarkers, and we evaluate the oncogenic or tumor-suppressive properties of these lncRNAs.
Introduction
Primary liver cancer is more common in males than in women and is associated with a high fatality rate worldwide (Sung et al., 2021). Viral infection (HBV, HCV), aflatoxin contamination of food, heavy alcohol use, being overweight or obese, smoking, and having type 2 diabetes have all been associated with the development of HCC (Marrero et al., 2005; Singal and El-Serag, 2015; McGlynn et al., 2021). Currently, effective treatments for HCC include targeted therapy, surgical resection, chemotherapy, radiotherapy, interventional therapy, and immunotherapy (Heimbach et al., 2018). However, since HCC is often diagnosed at a late stage, overall survival (OS) remains inadequate (Singal et al., 2022). Thus, it is crucial to improve diagnostic and therapeutic strategies to prolong survival in HCC patients and reduce the global cancer burden.
LncRNAs are RNA molecules that are 200 nucleotides or longer but do not code for proteins. Most are nuclear, with some located in the cytoplasm. The involvement of lncRNAs in tumorigenesis, specifically, as they modulate both gene expression and signal transduction, is well documented (Carlevaro-Fita and Johnson, 2019; Maass et al., 2019; Morf et al., 2020). Evidence suggests that LncRNAs have a role in every stage of HCC progression, from initial cell proliferation and differentiation through invasion, infiltration, and metastasis (Huo et al., 2017; Lim et al., 2019; Huang Z. et al., 2020). Understanding the pathophysiology of HCC and discovering important tumor markers to increase the sensitivity and specificity of HCC diagnosis and produce HCC-specific medicines may be achieved via research into the biological regulatory mechanisms and activities of lncRNAs. Here, we review the functions and regulatory activities of HCC-related lncRNAs and outline their involvement in the promotion or inhibition of HCC tumorigenesis and, based on this, analyze their potential for diagnosing and treating HCC (Table 1).
TABLE 1
| LncRNAs | Roles in HCC | Molecular mechanism | Functions | References |
|---|---|---|---|---|
| LEU2 | Oncogene | LncRNA DLEU2/HBx/EZH2/PRC2 | The transcription and replication of cccDNA in HBV-related HCC | Salerno et al. (2020) |
| PCNAP1 | Oncogene | LncRNA PCNAP1/miR-154/PCNA | HBV replication and HBV cccDNA accumulation | Feng et al. (2019) |
| LINC01149 | Oncogene | LINC01149/miR128-3p/MICA | Promoting HBV remission but increases the risk of HCC | Zhong et al. (2020) |
| IFI6 | Oncogene | LncRNA-IFI6/IFI6 | Increasing HCV replication | Liu et al. (2019) |
| Linc-Pint | Tumor suppressor | Linc-Pint/SRPK2 | Inhibiting HCV infection | Khatun et al. (2021) |
| SNHG6 | Oncogene | LncSNHG6/MTORC1 | Accelerating the progression of NAFLD to HCC | Liu et al. (2022) |
| H19 | Oncogene | AFB1/E2F1/lncRNA H19 | Promoting HCC growth and invasion | Lv et al. (2014) |
| AY927503 | Oncogene | LncRNA AY/H1FX | Promoting HCC metastasis | Kang et al. (2019) |
| LncMER52A | Oncogene | LncMER52A/YY1 | Promoting HCC cells migration, invasion, EMT and metastasis | Wu et al. (2020) |
| ID2-AS1 | Tumor suppressor | LncRNA-ID2-AS1/HDAC8/ID2 | Inhibiting HCC cells migration, invasion, and metastasis | Zhou et al. (2020) |
| TCAM1P-004 | Tumor suppressor | EZH2/lncRNA TCAM1P-004 | Inhibiting HCC cells proliferation | Xu et al. (2019) |
| RP11-598D14.1 | Tumor suppressor | EZH2/RP11-598D14.1/STAU1/PFKFB4 | Inhibiting HCC cells proliferation | Xu et al. (2019) |
| LINC00662 | Oncogene | LINC00662/MAT1A/AHCY | Promoting HCC cells proliferation, invasion, and metastasis | Guo et al. (2020) |
| SNHG6 | Oncogene | LncRNA SNHG6/miR-1297/MAT2ALncRNA SNHG6/miR-1297/FUS/MAT1A | Promoting hepatocarcinogenesis | Guo et al. (2018) |
| GATA3-AS | Oncogene | LncRNA GATA3-AS/KIAA1429/GATA3 | Enhancing HCC growth and metastasis | Lan et al. (2019a) |
| H19 | Oncogene | NSUN2/lncRNA H19/G3BP1 | Promoting HCC poor differentiation | Sun Z. et al. (2020) |
| LncMER52A | Oncogene | LncMER52A/p120-catenin/Rac1/Cdc42 | Promoting HCC cells invasion and metastasis | Wu et al. (2020) |
| miR503HG | Tumor suppressor | LncRNA miR503HG/HNRNPA2B1/p52/p65 | Inhibiting HCC invasion and metastasis | Wang et al. (2018) |
| LINC01138 | Oncogene | IGF2BP1/IGF2BP3/LINC01138/PRMT5 | Promoting HCC proliferation, tumorigenicity, invasion and metastasis | Li et al. (2018) |
| CASC2 | Tumor suppressor | LncRNA CASC2/miR-367/FBXW7 | Promoting HCC cells EMT | Wang Y. et al. (2017) |
| MUF | Oncogene | LncRNA-MUF/ANXA2/miR-34a/Snail1 | Promoting HCC cells EMT | Yan et al. (2017) |
| MCM3AP-AS1 | Oncogene | LncRNA MCM3AP-AS1/miR-194-5p/FOXA1 | Promting HCC proliferation, cell cycle progression and growth | Wang et al. (2019a) |
| lnc-Ip53 | Oncogene | LncRNA lnc-Ip53/HDAC1/p53/p300 | Promoting HCC growth and chemoresistance | Zhang et al. (2020b) |
| PSTAR | Tumor suppressor | LncRNA PSTAR/HNRNPK/p53 | Inhibiting HCC cells proliferation and tumorigenicity | Qin et al. (2020) |
| TSLNC8 | Tumor suppressor | LncRNA TSLNC8/TKT/IL-6/STAT3 | Inhibiting HCC invasion and metastasis | Zhang J. et al. (2018) |
| HOXD-AS1 | Oncogene | STAT3/LncRNA HOXD-AS1/miR-130a-3p/SOX4/EZH2/MMP2 | Promoting HCC metastasis | Wang H. et al. (2017) |
| CSMD1-1 | Oncogene | LncRNA CSMD1-1/MYC | Promoting HCC cells proliferation, migration, invasion, tumor growth and metastasis | Liu J. et al. (2020) |
| PXN-AS1-IR3 | Oncogene | lncRNA PXN-AS1-IR3/MYC | Promoting HCC metastasis | Zhou H. et al. (2022) |
| H19 | Oncogene | TGF-β/Tgfbr2/Sox2/lncRNA H19 | Promoting TIC proliferation, survival and progenitor capacity | Zhang J. et al. (2019) |
| UTGF | Oncogene | TGF-β/SMAD/lnc-UTGF | Promoting HCC metastasis | Wu et al. (2021) |
| PTTG3P | Oncogene | LncRNA PTTG3P/PI3K/AKT/PTTG1 | Promoting HCC cells proliferation, migration, invasion, tumorigenesis and metastasis | Huang et al. (2018) |
| CASC11 | Oncogene | LncRNA CASC11/PI3K/AKT/mTOR | Promoting HCC proliferation, cell mobility, apoptosis, and cellular metabolism | Song et al. (2020) |
| T-UCR uc.158- | Oncogene | Wnt/β-catenin/lncRNA T-UCR uc.158- | Promoting HCC growth | Carotenuto et al. (2017) |
| Lnc-UCID | Oncogene | miR-148a/lnc-UCID/DHX9/CDK6 | Promoting HCC cells G1/S transition and growth | Wang T. et al. (2019) |
| Lnc-APUE | Oncogene | Lnc-APUE/HNF4α/miR-20b/E2F1 | Promoting HCC cells G1/S transition and growth | Li et al. (2021) |
| PINT87aa | Tumor suppressor | LncRNA PINT87aa/FOXM1/PHB2 | Inducing HCC cells senescence | Xiang et al. (2021) |
| HULC | Oncogene | LncRNA HULC/miR15a/P62 | Promoting HCC cells growth and autophagy | Xin et al. (2018) |
| MALAT1 | Oncogene | BA/lncRNA MALAT1/miR-22-3p/IAP | Inhibiting HCC growth, induceing HCC cells death and apoptosis | Chen F. et al. (2020) |
| NEAT1 | Tumor suppressor | p53/lncRNA NEAT1/miR-362-3p/MIOX | Increasing the anti-tumor activity of erastin and RSL3 | Zhang Y. et al. (2022) |
| NEAT1_2 | Tumor suppressor | MTORC1/lncRNA NEAT1_2/NONO/SFPQ | Restraining AKT-mTORC1-mediated aerobic glycolysis and liver tumor development | Zhang H. et al. (2022) |
| MALAT1 | Oncogene | mTORC1/lncRNA MALAT1/TCF7L2 | Regulating HCC glucose metabolism, enhancing glycolysis, and inhibiting gluconeogenesis | Malakar et al. (2019) |
| WFDC21P | Tumor suppressor | Nur77/lncRNA WFDC21P/PFKP/PKM2 | Inhibiting HCC proliferation, growth, metastasis | Guan et al. (2020) |
| LINC01554 | Tumor suppressor | LINC01554/PKM2/Akt/mTOR | Impairing HCC cells aerobic glycolysis | Zheng et al. (2019) |
| HNF1A-AS1 | Tumor suppressor | HNF1α/lncRNA HNF1A-AS1/SHP-1 | Inhibiting HCC tumorigenesis and metastasis | Ding et al. (2018) |
| NEAT1 | Oncogene | LncRNA NEAT1/miR-124-3p/ATGL/DAG/FFA | Promoting HCC cells growth, disrupting the lipolysis of HCC cells | Liu et al. (2018) |
| PRLH1 | Oncogene | p53/lncRNA PRLH1/RNF169 | Promoting HCC cells proliferation and the HR-mediated DNA repair | Deng et al. (2019) |
| NIHCOLE | Oncogene | LncRNA NIHCOLE | Promoting HCC cells ligation efficiency of DNA double-strand breaks | Unfried et al. (2021) |
| HAND2-AS1 | Oncogene | LncRNA HAND-AS1/INO80/BMPR1A | Promoting HCC stem cells self-renewal | Wang et al. (2019b) |
| lncRNA-MUF | Oncogene | LncRNA-MUF/ANXA2/Wnt/β-catenin | Promoting HCC EMT | Yan et al. (2017) |
| LINC01133 | Oncogene | LINC01133/ANXA2/STAT3/cyclinD1 | Promoting HCC aggressive and EMT | Yin et al. (2021) |
| SNHG10 | Oncogene | LncRNA SNHG10/miR-150-5p/c-Myb/RPL4/SOX9 | Promoting HCC cells proliferation, invasion, migration, cell cycle and EMT | Lan et al. (2019b) |
| HCCL5 | Oncogene | TGF-β1/lncRNA HCCL5/ZEB1 | Promoting HCC cells viability, migration, and EMT | Peng et al. (2019) |
| HOXD-AS1 | Oncogene | LncRNA HOXD-AS1/miR19a/ARHGAP11A/RGS3/MEK/ERK | Promoting HCC metastasis and growth, Inhibiting Dox-induced apoptosis | Lu et al. (2017) |
| LINC01278 | Oncogene | Wnt/β-catenin/LINC01278/TCF-4/miR-1258/Smad2/3 | Promoting HCC cells migration, invasion and metastasis | Huang W. J. et al. (2020) |
Some dysregulated lncRNAs in HCC and their roles.
The relationship between lncRNAs and risk factors of hepatocellular carcinoma
Hepatitis B virus infection
The hepatitis B virus (HBV) infection is linked to an elevated risk of developing HCC (Hepatitis, 2018). HBV can be identified as a possible cause of HCC if the replication of viral DNA is detected. The viral HBV X protein (HBx) has been shown to induce host gene expression and promote HCC progression (Kim et al., 1991; Hepatitis, 2018). Patients with HBV infection remain in a chronic condition because HBx regulates the transcription of the HBV gene template covalently closed circular DNA (cccDNA), which in turn modifies the chromatin structure of the virus (Protzer, 2015; Wei and Ploss, 2020; Wei and Ploss, 2021).
LncRNAs have been linked to HCC caused by HBV infection (Zhang et al., 2014; Wang et al., 2021). The lncRNA DLEU2 is induced to be transcribed by HBx in HBV-infected cells, leading to an increase in DLEU2 levels in infected hepatocytes. In HBV-related HCC, the transcription and replication of cccDNA, together with the transcriptional activation of genes downstream of Enhancer of Zeste Homolog 2/Polycomb Repressive Complex 2 (EZH2/PRC2), leads to HCC via a cascade of events. This suggests that the lncRNA DLEU2-HBx interaction may be useful as a target for treating HBV-related HCC (Salerno et al., 2020). PCNAP1, another lncRNA, also induces HBV replication and resultant increases in cccDNA. This occurs because PCNAP1 sponges miR-154, promoting expression of hepatic Proliferating cell nuclear antigen (PCNA), which is required for cccDNA formation and is involved in DNA replication, repair, RNA modification, and chromatin remodeling. PCNA then binds to HBc, enabling anchorage to the cccDNA mini-chromosome and subsequent vital replication and cccDNA accumulation (Kelman, 1997; Feng et al., 2019; Wei and Ploss, 2020; Wei and Ploss, 2021) (Figure 1). A lncRNA variation was shown to increase HCC risk but aid in HBV remission via a genome-wide association analysis. The authors identified an rs2844512G>C variant in LINC01149, which enabled LINC01149 to sponge miR128-3p and lower its expression, leading to increased levels of MHC class-I chain-related gene A (MICA). On the one hand, MICA promotes HBVSR (hepatitis B virus spontaneous recovery), which benefits from the efficient activation of NK cell recognition and cytotoxicity by the interaction of MICA with NKG2D on NK cells, resulting in the lysis of infected hepatocytes. On the other hand, MICA increases the risk of HCC, as LINC01149 contributes to increased soluble MICA (sMICA) levels by targeting specific alleles. Studies have shown that higher levels of sMICA can promote NK cell depletion and tumor immune escape, which may be related to the fact that up-regulated MICA can support persistent HBV infection, leading to increased susceptibility to HCC (Groh et al., 2002; El-Gazzar et al., 2013; Tong et al., 2013; Zhong et al., 2020).
FIGURE 1
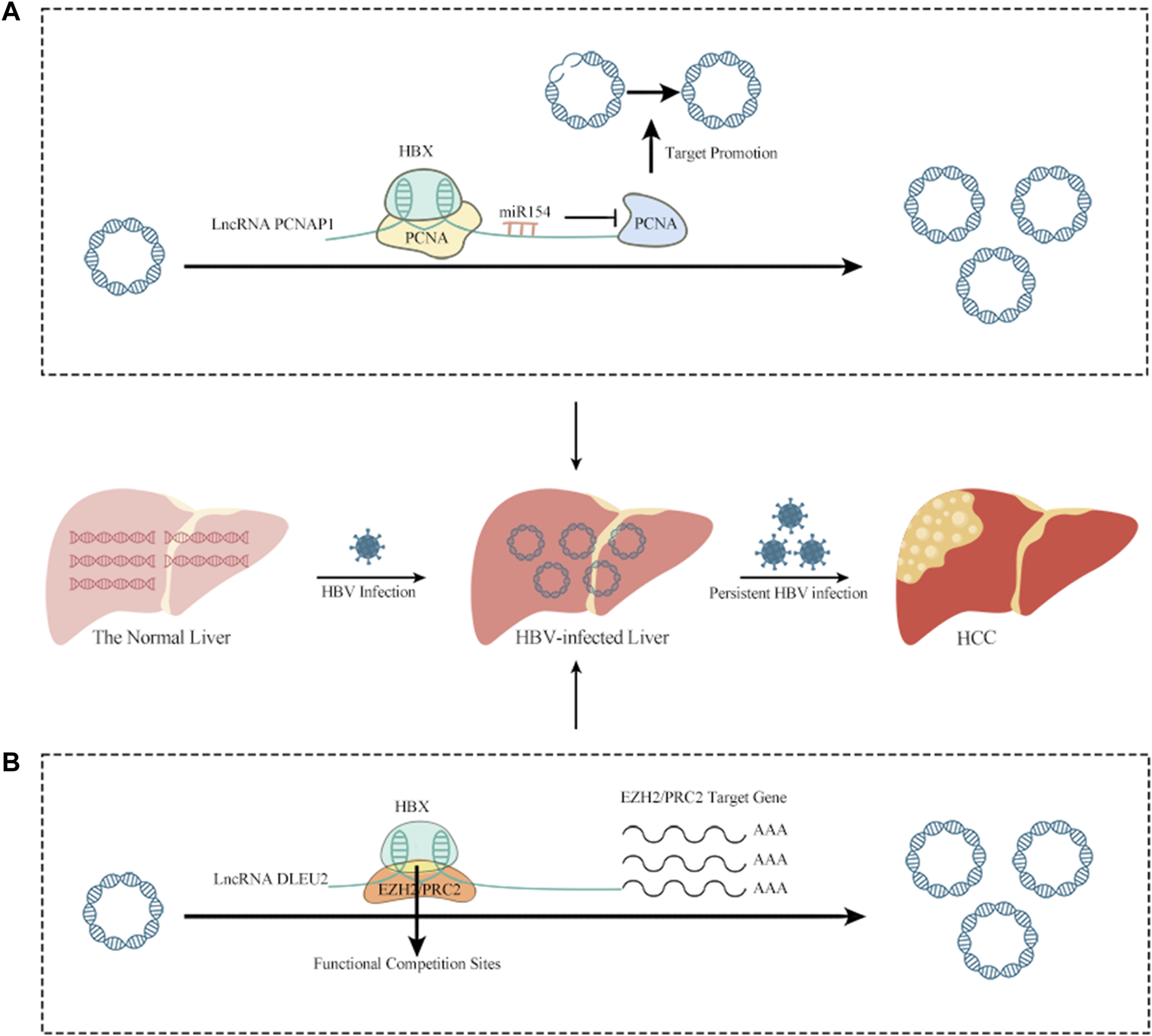
The relationship between lncRNAs and HBV infection of HCC. (A) PCNAP1 sponges miR-154, promoting expression of hepatic PCNA, which is required for cccDNA formation and is involved in DNA replication, repair, RNA modification, and chromatin remodeling. PCNA then binds to HBc, enabling anchorage to the cccDNA mini-chromosome and subsequent vital replication and cccDNA accumulation. (B) In HBV-infected cells, HBx binds to the lncRNA DLEU2 promoter, promoting DLEU2 transcription and its subsequent accumulation in infected hepatocytes. Blocking the repressive function of EZH2/PRC2 and activating transcription leads to the transcription and replication of cccDNA in HBV-related HCC, together with the transcriptional activation of genes downstream of EZH2/PRC2 and, ultimately leading to HCC.
Hepatitis C virus infection
Chronic hepatitis C (CHC) is the result of an hepatitis C virus (HCV) infection. Obesity, insulin resistance, and hepatic steatosis can further drive the progression of CHC to HCC, influenced by the host’s metabolic state. However, in late CHC, liver problems such as ageing, inflammation, and post-sustained viral response (SVR) metabolic dysfunction are risk factors for HCC, although direct-acting antiviral (DAA) has dramatically increased the cure rate of CHC (Reig et al., 2016; Innes et al., 2018; Nishibatake Kinoshita et al., 2019; Leslie et al., 2022).
HCV disrupts gene expression in hepatocytes, seen in hypomethylation of whole-genome DNA and the dysregulation of enhancer expression. Even if HCV is cured, these abnormal epigenetic and transcriptional changes are not restored, resulting in a persistently elevated risk of HCV-induced HCC (Hlady et al., 2022). LncRNAs also regulate gene expression in HCV-infected hepatocytes. For instance, lncRNA-IFI6 mediated modification of histones stimulates HCV replication by preventing IFI6 transcription (Liu et al., 2019) and the levels of lncRNA Linc-Pint are reduced in hepatocytes infected with HCV. Linc-Pint binds to Serine-arginine protein kinase 2 (SRPK2), reducing the expression of the latter and inhibiting de novo lipogenesis, thus blocking HCV infection (Khatun et al., 2021).
Other risk factors
Many studies have shown that not only obesity, smoking, and diabetes mellitus type 2 (Dhanasekaran and Felsher, 2019; Fujii et al., 2020; Premkumar and Anand, 2021; Qi et al., 2021) but also nutritional factors are risk important factors for HCC (Ruiz-Margáin et al., 2021). Non-alcoholic fatty liver disease (NAFLD) is a potential outcome of malnutrition. Some studies have shown that the lncRNA SNHG6 may promote the transition from NAFLD to HCC, possibly through activation of cholesterol-induced mechanistic target of rapamycin complex 1(MTORC1) (Campisano et al., 2019; Liu et al., 2022). However, nutritional imbalance, including high fructose or high sugar intake, can also stimulate hepatic steatosis as well as induce hepatic fibrosis through the gut-liver axis, potentially leading to HCC (Febbraio and Karin, 2021). However, some plant foods, such as potatoes, carrots, apples, and papaya, are reported to prevent HCC (Zhang D. et al., 2020; Saeed et al., 2022).
Excess alcohol consumption has been reported to disrupt the gut flora and translocate into the bloodstream, contributing to liver damage (Meroni et al., 2019; Moreno-Gonzalez and Beraza, 2021). Moreover, alcohol can synergize with HBV/HCV infection to promote the metabolic reprogramming of hepatocytes/liver tumor initiating cells (TICs), initiating their self-renewal and potentially leading to HCC (Machida, 2020). Dysregulation of lncRNAs has been observed in alcohol-induced HCC (Zheng et al., 2018) although the mechanism by which they cooperate with alcohol-induced HCC remains to be studied.
It is well documented that aflatoxin B1 (AFB1) causes HCC. AFB1 interacts synergistically with HBV/HCV to promote HCC development through epigenetic modifications (Ferreira et al., 2019; McCullough and Lloyd, 2019; Rushing and Selim, 2019). Studies have found that AFB1 exposure can exert immunosuppressive properties through lncRNAs (Ferreira et al., 2019; Chao et al., 2022). AFB1 interacts with E2 promoter binding factor 1 (E2F1) and upregulates lncRNA H19 levels, promoting HCC growth and metastasis (Lv et al., 2014). Further, it was found that AFB1 may induce the up-regulation of anti-apoptotic lncRNA expression to promote increased AFB1 carcinogenesis (Shi et al., 2016; Liu X. et al., 2020).
LncRNAs and gene expression in hepatocellular carcinoma
The functional structure of chromatin is the basis of epigenetics. Chromatin modifications, including histone modifications and DNA methylation, are important “languages” for regulating gene expression, thus potentially modulating the entire structure and functions of the cell. However, numerous studies have shown that dysregulation of these chromatin modification languages can cause aberrant gene expression, which is one of the key causes of many malignancies (Zhao et al., 2021).
Chromatin modification
LncRNAs are also implicated in epigenetic modifications of chromatin in HCC (Marrero et al., 2005). LncRNA AY927503 (AY) is strongly expressed in HCC, where it promotes metastasis through regulation of integrin subunit alpha-V gene (ITGAV) expression. The underlying mechanism is that AY interacts with histone 1 FX (H1FX), leading to the recruitment of epigenetically modified histone H3 lysine 4 trimethylation (H3K4me3) and histone H3-lysine 9-lysine 14 acetylation (acH3K9/14) to the promoter region of integrin αV (ITGAV), displacing tri-methylation at lysine 27 of histone H3 (H3K27me3) and H1FX. This, in turn, leads to chromatin remodeling through the actions of RNA polymerase II to promote cancer progression (Kang et al., 2019). LncMER52A is only expressed in HCC cells, and its activation is regulated both by chromatin modification and the transcription factor yin-yang 1 (YY1). High levels of lncMER52A are associated with significantly increases in H3K4me3 and H3K27ac. Furthermore, YY1 binds the lncMER52A promoter to activate lncMER52A transcription. These epigenetic mechanisms promote HCC tumorigenesis (Wu et al., 2020). LncRNA-ID2-AS1, on the other hand, acts as a tumor suppressor, preventing HCC migration and metastasis. Mechanistically, by controlling the binding of H3K27ac to the ID2 promoter and RNA polymerase II (RNAPII) to the ID2 enhancer domain, ID2-AS1 acts as a chromatin modifier to regulate ID2 transcription. This results in epigenetic modification and activation of the enhancer and modulation of ID2 expression which, in turn, regulates Epithelial-Mesenchymal Transition (EMT) progression to suppress HCC metastasis (Zhou et al., 2020).
Two lncRNAs, TCAM1P-004 and RP11-598D14.1, were discovered as tumor suppressors in HCC by a genome-wide screen and functional analysis. EZH2, a major component of the Polycomb repressive complex 2 (PRC2), acts as a histone methyltransferase and hence silences the expression of tumor suppressor genes. Transcriptional activity for TCAM1P-004 and RP11-598D14.1 is suppressed as a result of EZH2 catalyzing the production of H3K27me3. Additionally, the study authors discovered that RP11-598D14.1 interacts with the RNA-binding protein Staufen1 (STAU1) to suppress the expression of the oncogene phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4). RP11-598D14.1 expression is low in HCC, potentially leading to increased levels of PFKFB4 which, in turn, may lead to HCC (Xu et al., 2019).
Methylation modification of genes
The hypomethylation status of the genome has been linked to HCC (Meunier et al., 2021). High levels of LINC00662 are seen in HCC, associated with downregulation of methionine adenosyltransferase 1A (MAT1A) and S-adenosylhomocysteine hydrolase (AHCY). This reduces the levels of S-adenosylmethionine (SAM) and increases those of S-adenosylhomocysteine (SAH) levels, altering methylation of the LINC00662 promoter and, in turn, leading to hypomethylation of the genome. Upregulation of MAT1A and AHCY contributes to antitumor effects (Guo et al., 2020). The lncRNA SNHG6 acts as a “molecular switch” to modulate intracellular S-adenosylmethionine (SAMe) concentrations through two coupled positive feedback loops. First, SNHG6 acts as a ceRNA to inhibit miR-1297 expression, thus increasing methionine adenosyltransferase 2A (MAT2A) levels. Second, SNHG6 stabilizes fused in sarcoma (FUS) mRNA by reducing its miR-1297-induced decay. Then, FUS binds to MAT1A mRNA and hinders nuclear export of MAT1A mRNA, thereby reducing MAT1A protein synthesis. It can be concluded that the reduction of MAT1A and the increase of MAT2A, two key enzymes of SAMe, can suppress SAMe levels which, in turn, leads to genome-wide hypomethylation. Further, it has been found that exogenous SAMe can inhibit the effect of SNHG6 on genome-wide hypomethylation, which provides new evidence for tumor methylation therapy (Guo et al., 2018).
RNA modification
More than 100 chemical modifications of RNA have been shown over the past 50 years using a variety of methods. These highly modified forms of covalent binding have been extensively characterized, with the best documented including N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methyladenosine (m1A), internal 7-methylguanosine (m7G)), RNA cap methylations, pseudouridine, and uridylation (Barbieri and Kouzarides, 2020). RNA modification plays a significant part in cancer development, and proteins associated with the process can serve as potential anticancer targets (Delaunay and Frye, 2019; Boriack-Sjodin et al., 2022).
The lncRNA GATA3-AS serves as a guide RNA to bind KIAA1429 to GATA binding protein 3 (GATA3) pre-mRNA, and KIAA1429 is a component of the m6A methyltransferase complex. Increased m6A methylation of the 3′UTR of GATA3 pre-mRNA is induced in HCC cells after KIAA1429 targeting. As a result, GATA3 expression is downregulated, cancer and metastasis are promoted, and normal hepatocytes lose their stability as a result of the HuR-GATA3 interaction (Shi et al., 2020). Catalyzing m5C RNA modifications, NSUN2 is an RNA methyltransferase. Many studies have shown that lncRNA H19 is carcinogenic, and it has been suggested that this is related to its RNA modification. Sun et al. demonstrated that in HCC cells, NSUN2 increased the methylation level of H19, which was found to be positively correlated with poor HCC differentiation. Moreover, the oncoprotein GTPase-activating protein SH3 domain-binding protein 1 (G3BP1) can specifically bind to the m5C-modified lncRNA H19, a further indication of the part played by the m5C modification of H19 lncRNA in HCC malignancy (Sun Z. et al., 2020). Other related RNA modifications of lncRNAs in HCC remain to be studied (Shi et al., 2020; Luo et al., 2022).
Ubiquitination-associated degradation
Ubiquitination-deubiquitination is a post-translational modification associated with protein degradation via the ubiquitin-proteasomal pathway. Dysregulation of ubiquitination can lead to cancer development and there have been numerous investigations into the targeting of ubiquitin-proteasomal pathway for cancer treatment. However, these efforts have not proved entirely successful due to drug resistance and research is now being directed into the identification of suitable biomarkers to expand the treatment strategy. LncRNAs are involved in the cellular ubiquitination degradation pathway in HCC, and may represent feasible therapeutic targets (Narayanan et al., 2020; Sun T. et al., 2020; Cockram et al., 2021; Dang et al., 2021).
High expression of lncMER52A is related to poor HCC prognosis. LncMER52A promotes HCC tumorigenesis through modulation of p120-catenin/Rac1/Cdc42 signaling. LncMER52A inhibits the ubiquitin/proteasome-dependent degradation of p120-catenin, thus promoting its stability, as well as regulating the activity of downstream GTPases leading to its oncogenic effect (Wu et al., 2020). LncRNA miR503HG levels are reduced in HCC and have been linked to poor prognosis, suggesting its potential as a biomarker. Interacting with heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) and so inhibiting NF-kB signaling has been demonstrated to prevent HCC metastasis. Inhibition of the NF-kB signaling pathway and suppression of metastasis are the results of LncRNA miR503HG’s role in lowering HNRNPA2B1 protein levels through increased ubiquitination and degradation of the protein, as well as via the degradation of the p52 and p65 mRNAs that influence NF-kB signaling (Wang et al., 2018). The LncRNA LINC01138 was shown to be upregulated in HCC, and studies proved its oncogene status in this disease. IGF2BP1/IGF2BP3 stabilizes LINC01138 and, in HCC, LINC01138 binds specifically to protein arginine methyltransferase 5 (PRMT5), preventing its ubiquitination and degradation and enabling its oncogenic function. Several PRMT5 inhibitors have been used in cancer therapy, indicating that the study of the carcinogenic mechanisms of lncRNAs will further our understanding of HCC, allowing the development of new drugs targeting its pathways and effective prediction of patient prognosis (Li et al., 2018).
Competing endogenous RNA mechanism
It is been shown that lncRNAs may play a role in carcinogenesis as ceRNAs (competing endogenous RNAs) (Figure 2). Successful biomarkers may now be investigated with the use of ceRNAs, due to the advancement of ceRNA network research (Chan and Tay, 2018; Qi et al., 2020).
FIGURE 2
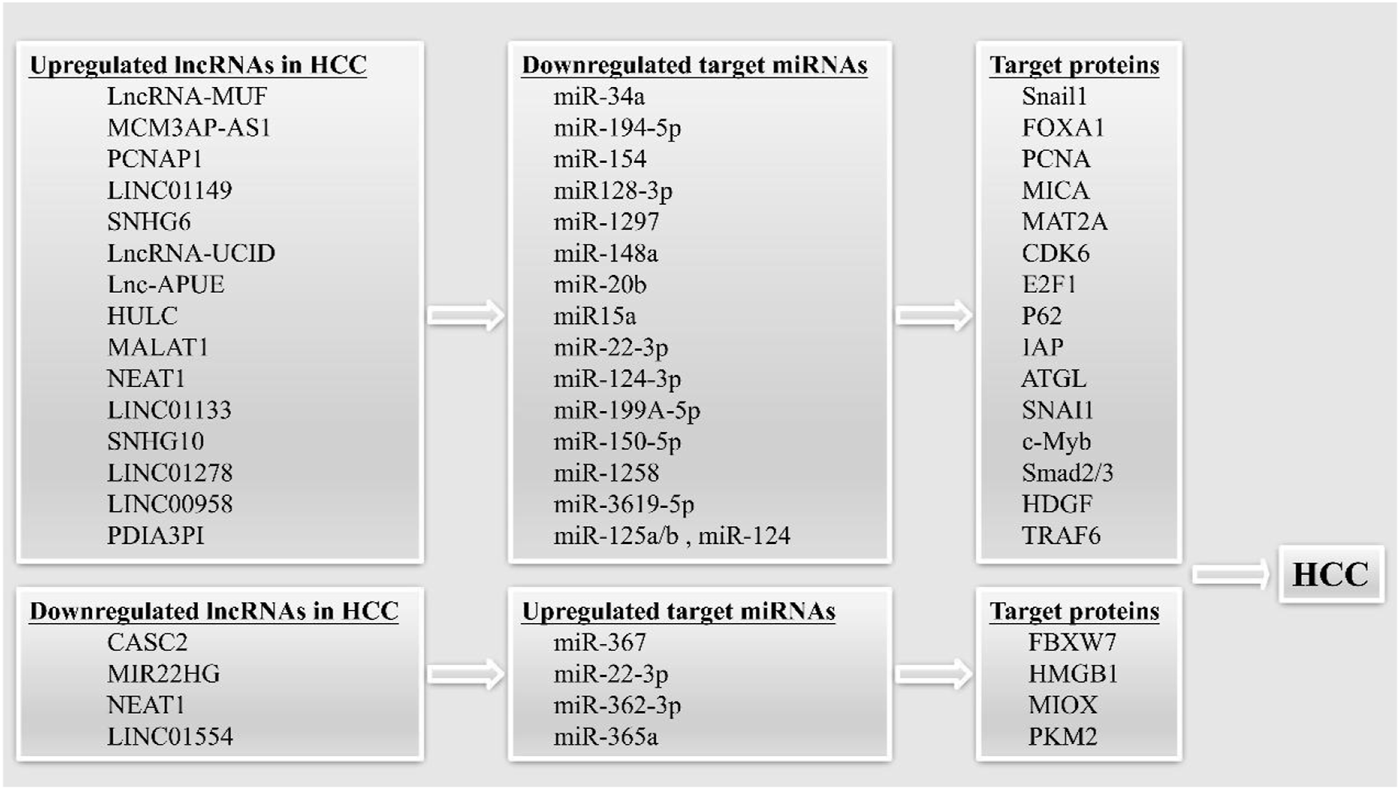
LncRNAs can act as competing endogenous RNAs (ceRNAs) in HCC. The upregulated and downregulated lncRNAs can sponging target miRNAs, leading to increased or decreased expression of target proteins, thereby promoting the occurrence of HCC.
The tumor suppressor lncRNA CASC2 functions as a ceRNA, decreasing miR-367 levels and causing an increase in F-box with 7 tandem WD40 (FBXW7) levels, which in turn inhibits EMT and invasion and migration in HCC cells (Wang Y. et al., 2017). LncRNA-MUF has been shown to have high expression in HCC and to be associated with a dismal prognosis in this disease. By acting as a ceRNA, the LncRNA-MUF suppresses the tumor-suppressing function of miR-34A, which in turn increases Snail1 expression and promotes EMT (Yan et al., 2017). Researchers have also identified a tumor suppressor lncRNA, MIR22HG, which is downregulated in HCC. On the one hand, MIR22HG inhibited the expression of HMGB1 through miR-22-3p, inhibiting high mobility group box-1 (HMGB1) signaling to reduce HCC cell proliferation and metastasis. On the other hand, MIR22HG was found to compete with human antigen R (HuR)-targeted oncogenes for binding to HuR, thus destabilizing the oncogenes and reducing HCC progression (Zhang D. Y. et al., 2018). MCM3AP-AS1 was identified by Wang et al. as a lncRNA that served as a ceRNA and bound miR-194-5p to induce forkhead box protein A1 (FOXA1) overexpression and HCC (Wang et al., 2019b).
The relationships between lncRNAs and signaling pathways in hepatocellular carcinoma
The involvement of lncRNAs in cancer-associated signaling pathways is well-documented. It is, thus, important to investigate these pathways to identify potential targets for drug development (Peng et al., 2017; Lin and Yang, 2018).
p53 signaling
The acetylation of p53 is crucial for its function as a tumor suppressor (Wang et al., 2016). When p53 is activated, it binds directly to the p53-responsive element (p53RE) in the lncRNA lnc-Ip53 promoter, inducing lnc-Ip53 transcription. As a result, Lnc-Ip53 interacts with histone deacetylase 1 gene (HDAC1) to inhibit its ubiquitination, which in turn increases HDAC1 levels and consequently p53 deacetylation. To prevent the acetylation of p53 by p300, Lnc-Ip53 also competes with p53 for binding to p300. These findings indicate that p53/lnc-Ip53 forms a negative feedback loop that inhibits p53 acetylation/activity and thus promotes both HCC development and chemoresistance (Zhang et al., 2020b). Increased p53 signaling and HCC suppression are the results of lncRNA PSTAR’s regulation of p53 activity by preventing de-SUMOylation of heterogeneous nuclear ribonucleoprotein K (Qin et al., 2020).
STAT3 signaling
Reduced expression of the tumor suppressor LncRNA TSLNC8 is associated with a poor prognosis in HCC. Several kinds of tumors show persistent activation and high levels of signal transducer and activator of transcription 3 (STAT3) (Yu et al., 2009; He and Karin, 2011; Zou et al., 2020). Interactions between TSLNC8 and transketolase (TKT) and STAT3 suppress phosphorylation and transcription of STAT3, hence inactivating the IL-6/STAT3 pathway and lowering tumorigenesis. TSLNC8 inhibits phosphorylation of STAT3 at tyrosine 705, or Y705, while increasing phosphorylation of serine 727, or S727. This results in increased levels of phosphorylated STAT3-S727 and decreased levels of phosphorylated STAT3-Y705 in the nucleus. Because of this change, STAT3’s phosphorylation state and nuclear localization are both modified, which in turn affects the protein’s biological function. The authors further found that TKT and STAT3 phosphorylation at either Tyr705 or Ser727 can independently predict HCC prognosis. Interestingly, TSLNC8 may prevent the interaction of TKT and STAT3. Overexpression of TSLNC8 blocks interactions between TKT and STAT3, suggesting competition between TSLNC8 and TKT or STAT3, affecting STAT3 phosphorylation. IL-6 is known to activate STAT3, enhancing phosphorylation at Tyr705 and reducing phosphorylation at Ser727. In addition, as a result of its effect on the IL-6-STAT3 pathway, TSLNC8 is also able to inhibit carcinogenesis in HCC. Therefore, the TSLNC8-TKT/IL-6-STAT3 axis shows promise as a therapeutic target for HCC (Zhang J. et al., 2018). LncRNA HOXD-AS1, which is controlled by STAT3 and increased in HCC, was discovered to increase SRY-related high-mobility-group box 4 (SOX4) expression by binding competitively to miR-130a-3p and activating two direct SOX4 targets, EZH2 and matrix metallopeptidase 2 (MMP2). This resulted in the promotion of HCC metastasis (Wang H. et al., 2017).
MYC signaling
LncRNA CSMD1-1 is upregulated in HCC, where it bind specifically to MYC to inhibit its ubiquitination and degradation, thereby enhancing MYC signaling to promote HCC progression (Liu J. et al., 2020). LncRNA PXN-AS1-IR3 is recently identified abnormally spliced isoform produced by DDX17-induced intron retention in the third intron of lncRNA-PXN-AS1. DDX17 is upregulated in HCC where it induces MYC transcriptional activation by inducing PXN-AS1-IR3 production which, in turn, promotes HCC metastasis (Zhou H. et al., 2022).
TGF-β signaling
Transforming growth factor-β (TGF-β) has both pro- and anticancer effects in HCC pathogenesis (Majumdar et al., 2012). An “early TGF-β signature” has been described (Coulouarn et al., 2008), characterized by lower tumor stage, reduced serum AFP, and improved patient survival, indicative of tumor suppression by TGF-β. TGFBR2 incactivation in tumor-initiating hepatocytes (TICs) was found to promote HCC progression, also suggesting a tumor-suppression role for TGF-β in TICs. Notably, deletion of transforming growth factor-β receptors 2 (TGFBR2) elevated levels of lncRNA H19, similar to the effects of SRY-related high-mobility-group box 2 (SOX2) overexpression. As SOX2 functions as a transcription factor in promoting the cancer stem cell phenotype, this suggests that the tumor-suppressing effects of TGF-β are associated with SOX2 inhibition of lncRNA H19 transcription in TICs (Zhang J. et al., 2019). Furthermore, a positive feedback loop involving TGF-β/SMAD/lnc-UTGF has been discovered, which stimulates HCC metastasis. When TGF-β is present, it promotes SMAD2/3 binding to the lnc-UTGF promoter, which in turn stimulates transcription of lnc-UTGF, which then interacts with and stabilizes SMAD2/4 mRNA. HCC metastasis is facilitated by the loop because of its enhancement of TGF-β signaling (Wu et al., 2021).
PI3K/AKT signaling
LncRNA PTTG3P levels are raised in HCC in association with pituitary tumor-transforming gene 1 (PTTG1) expression. Pituitary tumour-transforming 3, pseudogene (PTTG3P) activates the PI3K/AKT pathway, and elevated levels of PTTG1 and PI3K/AKT pathway activation induce the G1/S phase transition in HCC cells, increasing proliferation and promoting the EMT (Huang et al., 2018). YY1 is reported to target the lncRNA CASC11 promoter, leading to increased expression of the lncRNA in HCC. CASC11 also upregulates E2F1 expression through recruitment of eukaryotic translation initiation factor 4A3 (EIF4A3), activating the NF-κB and PI3K/AKT/mTOR pathways to promote HCC progression. Programmed cell death-Ligand 1 (PD-L1) levels were also reduced after CASC11 inhibition, suggesting that CASC11-mediated HCC progression is linked to PD-L1 induction of immune escape (Song et al., 2020).
Wnt signaling
Investigation of the T-UCR downstream of the Wnt/β-catenin pathway in HCC identified a lncRNA T-UCR uc.158-. T-UCR activation through Wnt signaling may promote HCC development and progression. It was shown that uc.158- was exclusively turned on in cancer cells, indicating a specialized involvement in Wnt signaling and a possible therapeutic target (Carotenuto et al., 2017).
Relationships between lncRNAs and hepatocellular carcinoma cell cycle regulation
Malignant proliferation is a hallmark of malignant tumors. Cell cycle dysregulation contributes to this prolonged proliferation (Matthews et al., 2022). Several proteins, such as cyclin-dependent kinases (CDK4/6) are responsible for controlling the G1/S phase transition, which is crucial in determining cell growth or arrest (Choi and Anders, 2014). In cancer cells, the normal regulation of the cell cycle is dysregulated, leading to uncontrolled cell division (Matthews et al., 2022). Cancer development and progression are mediated by regulatory networks associated with lncRNAs, as pointed out by Guiducci and Stojic (2021).
Cell cycle
The 850–1030 nt domain of Lnc-UCID has been found to have a role in cell cycle progression by binding competitively to DExH-Box helicase 9 (DHX9) and lowering DHX9’s binding to the 3′UTR of CDK6 mRNA. Proliferation was stimulated because CDK6 levels were raised, the G1/S transition was favored, and post-transcriptional regulation of CDK6 expression was blocked. In addition, miR-148A could bind directly to lnc-UCID resulting in its downregulation, suggesting that increased levels of lnc-UCID in HCC may be the consequence of either its amplification or reduced expression of miR-148a (Wang et al., 2019a). Furthermore, lnc-APUE, which is elevated in HCC and associated with patients’ risk of recurrence-free survival (RFS), has been shown to have a role in the cell cycle. The G1/S transition and tumor development are also stimulated by Lnc-APUE. Hepatocyte nuclear factor 4α (HNF4α) binds the lnc-APUE promoter to reduce lnc-APUE transcription, which in turn decreases E2F1 expression and cell proliferation, consistent with reports that E2F1 regulates the cell cycle. In HCC, lnc-APUE can sponge miR-20b, thereby preventing miR-20b-mediated repression of E2F1, leading to increased levels of E2F1 expression and levels and augmented G1/S transition (Li et al., 2021).
Cellular senescence and death
Excessive stress can result in an arrest of cell proliferation, a phenomenon known as cellular senescence. Harnessing cellular senescence has emerged as a promising modality for antitumor therapy (Wang et al., 2022). The induction of cancer cell senescence is a key step in an anti-tumor program that leads to cell cycle arrest, which prevents cancer progression by limiting cancer cell proliferation. However, senescent cells do not lose their functionality permanently or completely. According to the available data, senescent cancer cells may re-enter the cell cycle and reestablish cancer stemness, which can result in tumor recurrence. Therefore, anti-aging therapy, or the elimination of senescent cancer cells, may boost the efficacy of cancer ageing treatment and reduce the likelihood of tumor recurrence. The challenge facing current cancer aging treatment is mainly a lack of biomarkers for accurate detection of the aging status of the cancer cells. Using drugs to trigger senescence in different cancer cell lines, Jochems et al. established a SENCAN classifier aimed at identifying senescence-specific genes. Several genes with lncRNA signatures were included in the SENCAN classifier, indicating their involvement in cancer cell senescence (Muñoz-Espín and Serrano, 2014; Jochems et al., 2021).
HCC senescent cells produce the LncRNA PINT87aa, which has been demonstrated to promote senescence and suppress proliferation in HCC cells. PINT87aa binds to forkhead box M1 (FOXM1) to inhibit prohibitin 2 (PHB2) transcription, further inhibiting PHB2-mediated mitophagy, blocking the cell cycle, and inducing cellular senescence (Xiang et al., 2021).
Autophagy is important in tumorigenesis (White et al., 2015), and lncRNA HULC has been shown to be involved in autophagy in HCC cells. However, lncRNA HULC is highly elevated in HCC, and its promotion of autophagy is reflected in increased expression of Beclin 1 and the autophagy marker LC3II. By encouraging autophagy, phosphatase and tensin homolog (PTEN) is more effectively ubiquitinated and degraded. However, by decreasing miR15a levels, HULC increases the production of p62, which is essential for PTEN’s autophagic breakdown. This suggests that HULC reduces PTEN through the autophagy-p62-mediated ubiquitin-proteasomal system, thereby promoting the development of HCC (Xin et al., 2018). Betulinic acid (BA) has been shown in several studies to induce cell death in various cancers. In HCC, BA was shown to block the carcinogenic lncRNA MALAT1 by lowering its levels. This in turn promoted apoptosis of HCC cells by targeting inhibitor of apoptosis (IAP) through miR-22-3p, therefore preventing HCC from progressing. BA was also found to induce autophagic flux in HCC cells, resulting, ultimately, in cell death, with IAPs likely to be key to this regulatory process (Chen F. et al., 2020).
Programmed cell death known as ferroptosis has just lately been identified (Mou et al., 2019). According to the literature, the lncRNA NEAT1 induces ferroptosis in HCC cells. After subjecting cells to ferroptosis inducers erastin and RAS-selective lethal (RSL3), the scientists discovered p53 upregulated NEAT1 levels. Ferroptosis elicited by erastin and RSL3 is facilitated by NEAT1 sponging miR-362-3p to increase myo-Inositol oxygenase (MIOX) levels. There is evidence that ferroptosis inducers enhance cellular sensitivity to chemotherapy (Liang et al., 2019). Since lncRNAs seem to play important roles in HCC ferroptosis, NEAT1 may be a viable therapeutic target when used in conjunction with treatments that specifically target ferroptosis (Zhang Y. et al., 2022).
Cellular metabolism
Malignancy is also characterized by the resetting of cellular metabolism. Cancer cells promote tumor growth via the use of internal mechanisms that drive metabolic alterations and interactions with cytokines in the tumor microenvironment (TME) (Dey et al., 2021; Martínez-Reyes and Chandel, 2021).
Cancer cells typically use glycolysis to maintain their metabolic energy and promote tumor growth, termed the “Warburg effect” (Warburg, 1925). The “Warburg effect” is characteristic of cancer and involves the promotion of cancer growth by switching glucose production from oxidative phosphorylation to aerobic glycolysis through the MTORC1 pathway (Szwed et al., 2021). It is documented that HCC is closely related to aerobic glycolysis. MTORC1 inhibits lncRNA NEAT1_2 activation in HCC, and NEAT1_2 is essential for paraspeckles, constituting its “architectural backbone.” Additionally, it can sequester NONO and SFPQ, both RNA-binding proteins (Yamazaki et al., 2018). Reductions in NEAT1_2 levels lead to the release of NONO/SFPQ, allowing NONO to bind to specific motifs in key glycolytic enzymes. This regulates pre-mRNA splicing of these enzymes leading to the promotion of aerobic glycolysis for supplying energy. This binding can be inhibited by rapamycin. Rapamycin inhibits HCC cell growth under high-glucose conditions, and NEAT1_2 knockdown attenuates this inhibition, further suggesting the effects of rapamycin on HCC glucose metabolism-related processes, possibly through inhibition of the mTORC1 -NEAT1_2 axis (Zhang H. et al., 2022). MALAT1, a lnRcNA, is also linked to the Warburg effect. Because of mTORC1’s stimulation of cap-dependent translation, MALAT1 stimulates the translation of the transcription factor TCF7L2, which in turn increases the expression of glycolytic genes and negatively regulates gluconeogenesis to preserve the oncogenic features of HCC cells. Cancer glucose metabolism may benefit from a novel approach including the suppression of TCFL2 or downregulation of the lncRNA MALAT1 (Malakar et al., 2019). LncRNA WFDC21P inhibits HCC tumorigenesis in both cell lines and in vivo through Nur77 enhancement of WFDC21P expression. The interaction of WFDC21P with two key glycolytic enzymes, phosphofructokinase-P (PFKP) and pyruvate kinase M2 (PKM2), is enhanced by WFDC21P overexpression. Inhibiting PFKP oligomerization and enzymatic activity, as well as preventing nuclear translocation of PKM2 and its role as a transcriptional coactivator leads to decreased glycolysis and impacts HCC cell proliferation and metastasis (Yu and Li, 2017; Guan et al., 2020). LINC01554 mediates glycolysis, and its downregulation in HCC suppresses tumorigenesis. LINC01554 also promotes the ubiquitination-mediated degradation of the glycolytic enzyme PKM2 and inhibits Akt/mTOR signaling to reduce glycolysis in HCC cells. Increased levels of miR-365a have also been found to reduce LINC01554 expression. Most importantly, the results of one of these studies showed that a nude mouse model of tumors formed by LINC01554-transfected cells were reduced by the Akt inhibitor MK2206. Based on these findings, LINC01554 has the potential as a prognostic marker in Akt inhibitor-treated patients with HCC (Zheng et al., 2019).
Fat metabolism is also important in HCC progression. By controlling the expression of Src homology 2 domain-containing protein tyrosine phosphatase 1 (SHP-1), a protein involved in glucose and lipid metabolism in the liver, hepatocyte nuclear factor 1α (HNF1α) inhibits HCC. To slow the development of HCC, sorafenib boosts SHP-1 activity and inhibits EMT produced by TGF-β (Shih et al., 2001; Dubois et al., 2006; Lopez-Ruiz et al., 2011; Tai et al., 2012; Xu et al., 2014; Fan et al., 2015). The lncRNA HNF1A-AS1 has been identified to have a role in HNF1’s anti-HCC actions. It has been shown that hepatic nuclear factor 1 (HNF1) binds to the HNF1A-AS1 promoter, which in turn stimulates the production of HNF1A-AS1. To suppress HCC, HNF1A-AS1 acts as a phospho-activator, connecting directly with SHP-1 to boost its phosphatase activity (Ding et al., 2018). As shown by Liu et al. lncRNA NEAT1 is involved in abnormal lipolysis. NEAT1 levels are increased in HCC where they suggest poor prognosis. NEAT1 competes with miR-124-3p for binding and then promotes increased expression of adipose triglyceride lipase (ATGL), which disrupts lipolysis in HCC cells, and it exerts pro-tumor effects by promoting diacylglycerol (DAG) and free fatty acid (FFA) production. ATGL can also promote the upregulation of the oncogene PPARα in HCC and promotes proliferation in HCC cells (Liu et al., 2018).
Cell repair
Double-strand breaks (DSBs) represent the most serious form of DNA damage. Repair of DSBs occurs through homologous recombination (HR) and non-homologous end joining (NHEJ). In these two pathways, many oncogenes have been found to be involved in DSB repair signaling and may mediate malignancy (Khanna and Jackson, 2001). These authors identified a novel HR-promoting factor, the lncRNA PRLH1, which was upregulated in p53-mutated HCC samples and repressed by p53. PRLH1 can specifically bind to the DNA repair protein RNF169, forming an HR-repair functional complex, and promote the recruitment and retention of RNF169 at DSB sites, thereby promoting HCC HR repair and increasing proliferation in HCC cells (Deng et al., 2019). LncRNA NIHCOLE (lncRNA LINC02163) can target the NHEJ repair mechanism by binding to effector proteins promote the ligation efficiency of DSBs, thereby maintaining proliferation (Unfried et al., 2021).
Relationships between lncRNAs and the tumor microenvironment in hepatocellular carcinoma
Stem cells
Tumor cells that may self-replicate and differentiate in several ways are called cancer stem cells (CSCs) (Nassar and Blanpain, 2016). Evidence suggests that CSCs are important drivers of HCC growth and represent one of the reasons for cancer recurrence and drug resistance after chemotherapy/radiotherapy (Wang and Jacob, 2011). Markers of liver CSCs have been linked to poor patient prognosis. Lee et al. investigated the research status of related CSCs markers and related stem cell regulatory factors. These tumor markers identify distinct stem cell lineages that promote HCC development and characterize important mechanisms by which cancer stem cell-intrinsic regulators contribute to the tumor-initiating potential of HCC (Lee et al., 2022). There are many abnormally expressed lncRNAs in HCC, which not only play important roles in tumorigenesis but also maintain the characteristics of HCC stem cells (Huo et al., 2017).
A recently identified lncRNA, HAND2-AS1, is strongly expressed in hepatic CSCs. HAND2-AS1 has been found to promote self-renewal in liver CSCs and HCC development through the bone morphogenetic protein (BMP) signaling pathway. HAND2-AS1 binds the INO80 complex, recruiting it to the type IA bone morphogenetic protein receptor (BMPR1A) promoter, thereby inducing BMPR1A expression and activating BMP signaling (Wang et al., 2019c). Liu W. et al. (2020), using lipopolysaccharide induction of hepatic progenitor cells to fibroblasts, observed an increased risk of hepatic progenitor cell (HPC) carcinogenesis. Lipopolysaccharides/Toll-like receptor 4 (LPS/TLR4) signaling mediates HPC development into myofibroblasts that show high levels of IL-6 and TNF-α, potentially promoting upregulation of EGFR and downregulation of PTEN through lncRNA regulation, leading to abnormal Ras and p53 signaling and, ultimately, inducing the malignant transformation of HPCs, potentially leading to HCC development.
EMT/metastasis
The EMT (Epithelial to Mesenchymal Transition) has been lined with various aspects of malignancy, including metastasis and drug resistance (Pastushenko and Blanpain, 2019). It is particularly associated with tumor metastasis (Bakir et al., 2020).
Increased lncRNA-MUF levels are associated with a dismal outlook in patients with HCC. This EMT is stimulated by LncRNA-MUF binding to annexin A2 (ANXA2), which in turn activates Wnt/β-catenin. Elevated expression of lncRNA-MUF has been linked to EMT in HCC-associated mesenchymal stem cells (HCC-MSCs) (Yan et al., 2017). LINC01133 is upregulated in HCC, interacting with annexin A2 (ANXA2) and activating ANXA2/STAT3/cyclinD1 signaling which, in turn, promotes HCC progression. LINC01133 also sponges miR-199A-5p to upregulate SNAI1, thereby promoting the EMT (Yin et al., 2021). HCC and lung metastases showed upregulation of the lncRNAs SNHG10 and its homolog SCARNA13. SNHG10 was discovered to boost c-Myb activity via stabilizing ribosomal protein L4 (RPL4) mRNA and promoting RPL4 expression, both of which were previously thought to occur independently of one another. Therefore, SNHG10, SCARNA13, and their downstream effector SRY-related high-mobility-group box 9 (SOX9) were all upregulated in HCC as a result of this. Cancer cells overexpressing SOX9 are known to exhibit increased cell proliferation and EMT (Lan et al., 2019b). Increased expression of lncRNA HCCL5 was reported by Peng et al., a functional lncRNA, in a TGF-β1-induced EMT model. When Zinc-finger E-box binding protein 1 (ZEB1) reached the HCCL5 promoter and super-enhancer, it immediately bonded to them and began driving transcription of that gene. HCCL5 promotes EMT progression in HCC cells by regulating EMT-related transcription factors (Peng et al., 2019). LncRNA HOXD-AS1 levels are significantly raised in metastatic HCC, where it appears to upregulate Rho GTPase-activating protein 11A (ARHGAP11A) through competitive binding to miR19a. HOXD-AS1 overexpression also downregulated regulator of G-protein signaling 3 (RGS3), leading to reduced doxorubicin (Dox)-induced apoptosis. Interestingly, RGS3 has been suggested to negatively regulate MEK/ERK signaling, and HOXD-AS1 is known to reduce RGS3 expression, which can block HCC cell apoptosis and promote proliferation through activation of MEK/ERK signaling (Lu et al., 2017). LncRNA LINC01278 levels are elevated in HCC where it negatively modulates the expression of miR-1258, thereby upregulating Smad2/3 and promoting HCC metastasis. Wnt/β-catenin signaling was shown to be involved in the regulation of LINC01278 by these authors as well. Elevated levels of β-catenin expression in HCC cells cause T cell factor 4 (TCF-4) to bind to the LINC01278 promoter, resulting in the overexpression of this gene. As with TGF-β/Smad signaling, miR-1258 has been shown to target Smad2/3 and suppress it. Based on these findings, LINC01278 may play a role in HCC metastasis by mediating the Wnt/β-catenin and TGF-β/Smad pathways (Huang W. J. et al., 2020). To facilitate -mediated m6A methylation of the GATA3 pre-mRNA 3′UTR, Lan et al. discovered a lncRNA, GATA3-AS, which interacts with KIAA1429 and the GATA3 pre-mRNA. HuR acted as a negative regulator of GATA3 expression by binding to GATA3 pre-mRNA, which decreased GATA3 expression and promoted HCC (Lan et al., 2019a). LncRNA 34a is also upregulated in HCC and has been associated with bone metastasis. While miR-34a targets Smad4 through the TGF- pathway to suppress HCC malignancy and bone metastasis, LncRNA 34a epigenetically represses miR-34a production by inducing methylation of the miR-34a promoter and histone deacetylation in HCC cells. This inhibitory effect of miR-34a was prevented by lncRNA 34a, which in turn mediates HCC bone metastasis (Zhang L. et al., 2019).
Exosomes
Exosomes have been studied extensively and shown to have a role in invasion, migration, and EMT in a variety of malignancies (Bebelman et al., 2018; McAndrews and Kalluri, 2019; Dai et al., 2020).
The exosome secretion process begins when multivesicular bodies (MVB) fuses with the plasma membrane, releasing intraluminal membrane vesicle (ILV). The SNARE proteins are the mediators of MVB-membrane fusion. LncRNA HOTAIR is upregulated in HCC where it promotes exosome secretion. According to the work of Yang et al. (2019), HOTAIR promotes co-localization of vesicle-associated membrane protein 3 (VAMP3) and synaptosome associated protein 23 (SNAP23) to affect the development of the SNARE complex, leading to MVB fusion with the plasma membrane. Additionally, phosphorylation of SNAP23 by HOTAIR stimulates exosomal release.
Application of lncRNAs in the clinical diagnosis and treatment of hepatocellular carcinoma
Recent years have seen several breakthroughs in the ability to detect and diagnose HCC (Ye et al., 2019; Ahn et al., 2021; Nault and Villanueva, 2021; Zhang et al., 2021). lncRNAs are suitable choices for biomarkers. Liquid biopsy is an emerging testing modality that is being developed. Compared with traditional tissue biopsy, liquid biopsy can repeatedly obtain tumor samples and dynamically monitor the overall tumor status, reducing the bias caused by tumor heterogeneity (Heitzer et al., 2019; Ye et al., 2019; Moldogazieva et al., 2021). Saliva testing is an important aspect of liquid biopsy and has the advantage of being non-invasive and easy to collect. Research has found that salivary lncRNA lnc-PCDH9-13:1 was found to show more powerful diagnostic sensitivity and specificity than serum AFP and could be used as a specific biomarker for early and AFP-negative HCC (Xie et al., 2018). Extracellular vesicles are now also thought to mediate the process of cancer development. An optimized human plasma exLR-sequencing (exLR-seq) method has recently been proposed for the detection of long RNAs, including lncRNAs, in extracellular vesicles (EVs). Blood exLR heterogeneity may serve as biomarkers for HCC diagnosis and the development of an exLR profile based on exLR liquid biopsy may differentiate cancer patients from healthy persons (Li et al., 2019). HCC is characterized by elevated levels of the lncRNA NONHSAT122051, which is favorably connected with clinicopathological characteristics (such as poorly differentiated tumor, PVTT, and AFP levels) (Yao et al., 2017). In addition, PXN-AS1-IR3, a lncRNA, was shown to have increased levels in both HCC tissues and the serum of extrahepatic metastases (Zhou H. et al., 2022). This suggests that in the future, we can use liquid biopsy to identify these abnormal lncRNAs and determine the malignancy and clinical features of HCC based on this.
Drug resistance is a significant challenge to cancer chemotherapy, and the acquisition of tumor drug resistance is associated with tumor heterogeneity interacting with the surrounding microenvironment (Bhattacharya et al., 2021). Regarding anticancer drug resistance, Nussinov et al. (2021) described the research status and molecular mechanisms in detail, showing that lncRNAs were also involved in HCC drug resistance.
Progression and a poor prognosis in HCC are directly connected to resistance to chemotherapeutic treatments. A lncRNA, PDIA3P1, which regulates the chemoresistance of tumor cells, shows upregulation in HCC and is also highly expressed after antitumor treatment with DNA-damaging agents. Since PDIA3P1 acts as a ceRNA by binding to miR-125a/b and miR-124 to upregulate TRAF6 expression and enhance NF-kB signaling to suppress cancer cell apoptosis, its overexpression confers chemoresistance to HCC cells. Additionally, exosomes were found to mediate the degradation of PDIA3P1. The increase in PDIA3P1 levels induced by the DNA-damaging agent Dox was reduced when hMTR4, an important cofactor of exosomes, was overexpressed. These results provide additional evidence that targeting PDIA3P1 is critical for increasing the sensitivity of HCC to chemotherapeutics, and they show that exosomes play a role in the lncRNA PDIA3P1-mediated response to DNA damage (Xie et al., 2020). LncRNA ZFAS1 was observed to be significantly upregulated in HCC and recurrent tumors. Treatment with sorafenib leads to ZFAS1-induced promotion of stemness and normal EMT functioning in HCC, and it is possible that sorafenib-resistant stem-like cells may be present and may, after prolonged exposure to the drug, multiply and lead to overall drug resistance. Therefore, the elimination of quiescent CSC-like cells or silencing ZFAS1 could prevent sorafenib resistance and prolong survival in HCC patients (Zhou K. et al., 2022).
Many lncRNAs have been shown to be associated with increased tumor burden and poor prognosis based on their differential expression between normal and malignant tissues, and these lncRNAs are preferentially expressed in healthy testes or brain, where they are predicted to be oncogenes. Perhaps, in targeting these up-regulated lncRNAs with oncogenic potential, it is possible to comprehensively consider tissues that preferentially express these targets, and develop more comprehensive targeted therapy regimens to reduce the occurrence or progression of cancer (Unfried et al., 2019). Based on an RNA-binding protein (RBP)-centered approach, these authors identified a novel lncRNA, LINC00326, which is minimally expressed under normal physiological conditions and is speculated to act as an inhibitor to prevent the transformation of normal hepatocytes to malignant cells (Søndergaard et al., 2022). Zuo et al. (2020) identified a lncRNA, LINC00958, with elevated levels in HCC. The authors exploited tumor promotion by the LNC00958/miR-3619-5p/HDGF axis in HCC to design a novel nanoplatform for systemic therapy of HCC in which LINC00958 siRNA was encapsulated. This nanosystem significantly reduces tumor burden and prolongs survival, indicating that lncRNAs have great potential in using nanotechnology to treat malignant tumors.
The use of nanoplatforms to treat diseases is widely studied, and nanocomplexes targeting lncRNAs hold a wide range of promise (van der Meel et al., 2019). siRNA tumor-targeting nanocomplexes carrying the lncRNA MALAT1 not only inhibit the malignant behavior of glioblastoma multiforme (GBM) cells, but when combined with temozolomide (TMZ), they also increase the chemosensitivity of TMZ and improve the prognosis of GBM patients (Kim et al., 2018). A study shows that targeted delivery of ASO to the brain using glucose-encapsulated polymeric nanocarriers can reduce target lncRNA expression in the brain and treat central nervous system disorders (Min et al., 2020). Precise delivery of lncRNA lncAFAP1-AS1 siRNA using a nanoparticle (NP) platform improves the effectiveness of radiation therapy for triple-negative breast cancer (TNBC), which provides a rationale for combining targeted lncRNA therapy with radiotherapy (Bi et al., 2020). A combination therapy that co-writes nanoparticles, small interfering RNA/antisense oligonucleotide (siRNA/ASO) and lncRNA into transplanted neural stem cells (NSC), which targets the lncRNA Pnky and prevents Pnky-mediated differentiation of NSC into astrocytes and loss of therapeutic effect. This shows that nanomedicines targeting lncRNAs can enhance the great potential of stem cell-based treatments for stroke (Lin et al., 2021). Zuo et al. (2020) identified a lncRNA, LINC00958, with elevated levels in HCC. The authors exploited tumor promotion by the LNC00958/miR-3619-5p/HDGF axis in HCC to design a novel nanoplatform for systemic therapy of HCC in which LINC00958 siRNA was encapsulated. This nanosystem significantly reduces tumor burden and prolongs survival. Summarize the above, lncRNAs have great potential in using nanotechnology to treat malignant tumors.
Targeting risk factors for HCC and treating patients with HBV infection is also an important treatment pathway. Positive results from phase IIb clinical trials of the drug bepirovirsen, which is being developed against HBV. Significant reductions in HBsAg and HBV DNA were seen in treated CHB (Chronic Hepatitis B) patients, which contributed to improved clinical outcomes. Bepirovirsen is an ASO that specifically recognizes HBV mRNA and recruits the liver’s own enzymes to eliminate HBV-derived RNA, HBV DNA and viral proteins, achieving a functional cure for CHB patients, which helps avoid HCC (Yuen et al., 2021).
As one of the important types of solid tumors, the abnormal construction of extracellular matrix (ECM) is closely related to its development in HCC, where collagen, a major component of ECM, mediates the carcinogenic effects of HCC (Feng et al., 2017). One study found that aspirin (ASA), an anti-inflammatory drug, targets P4HA2, a key enzyme in collagen formation, to block collagen deposition. Further, lncRNA LMCD1-AS1 was found to target miRNA let-7g, which induced upregulation of target gene P4HA2 expression at post-transcriptional level, and P4HA2 induced liver fibrosis. Since the development of HCC is often associated with fibrosis, targeting the fibrotic process in HCC can be a good therapeutic strategy (Wang T. et al., 2019). A 26-year clinical follow-up study also provides support that regular, long-term aspirin use reduces the risk of HCC (Simon et al., 2018).
Conclusion
Advances in science and technology have led to reductions in both the incidence and mortality of HCC. Nevertheless, HCC remains one of the most deadly cancers in the world. Therefore, improving the survival rate of HCC patients is an urgent need. For early-stage HCC, surgical resection or local therapy is usually recommended, while for intermediate-advanced HCC, transarterial chemo- and radioembolization (TACE/TARE) or systemic therapy is recommended (Heimbach et al., 2018). Therefore, for early-stage patients, we need to effectively exclude risk factors and improve HCC detection, leading to early diagnosis and early treatment. For patients with advanced disease, we should try our best to prolong the patient’s life and improve their quality of life. Currently, the most commonly used imaging method for monitoring HCC is CT/MRI, and the most effective biological method is measurement of blood AFP levels. However, this is far from enough, and new methods for the diagnosis and treatment of HCC are needed (Kanwal and Singal, 2019).
In the past, lncRNAs were considered transcriptional “noise” as they do not encode proteins, and were thought to have no real biological function (Louro et al., 2009; Rosenbloom et al., 2012). However, with the discovery of the first lncRNA H19 (Brannan et al., 1990; Gabory et al., 2009), the subsequent identification of lncRNA XIST mediation of X-chromosome inactivation (Brown et al., 1991), and advances in high-throughput sequencing, a large number of lncRNAs have been discovered (Iyer et al., 2015). These lncRNAs not only act as regulators to participate in the whole process of gene expression regulation (Zhang Y. et al., 2018; Cao et al., 2019; Qian et al., 2019; Gao et al., 2021), but also have the potential to encode functional peptides (Choi et al., 2019). In both the malignant tumor itself and its tumor microenvironment, and even in cell-to-cell communication, lncRNAs act as bridges to transmit information, and can guide cancer development and progression, mediating cancer cell proliferation, invasion, metastasis, and chemoresistance (Zhang et al., 2020a; Wang et al., 2020; Liu et al., 2021; Xu et al., 2022). In short, lncRNAs have great research prospects. Whether functioning as oncogenes or tumor suppressors, tumor biomarkers can be studied to identify associated targets in signal transduction pathways regulated by these lncRNAs. In conclusion, the lncRNA research on HCC is promising.
Regarding the early detection of HCC, liquid biopsy method can be used to detect circulating tumor cells (CTCs) and cell-free DNA (cfDNA). This helps with monitoring HCC (Chen V. L. et al., 2020) and has implications for the study of lncRNAs in HCC. Systemic therapy for patients with advanced HCC, including chemotherapy, targeted therapy, and immunotherapy, has also developed rapidly in the past few years (Llovet et al., 2018). Of course, this is inseparable from the study of cancer signaling pathways. However, due to the high morbidity and mortality of HCC and its drug resistance, we urgently need a new drug that overturns the traditional treatment to improve or even cure HCC patients. lncRNA drugs are the trend of future research. To date, due to the large number of LncRNAs, the variety of regulatory modalities, and the difficulties in targeting delivery and related immune responses, no clinical translation has been completed, and no HCC drugs or clinical trials have yet been conducted using lncRNAs as targets. It is promising that with the continuous development and intelligent transformation of nanotechnology, siRNA drugs, antisense nucleotide drugs, miRNA drugs, etc. can be developed by targeting lncRNAs in the future and delivering drugs to target lesions stably and precisely through nanoplatforms, which not only has great potential but also needs to face great challenges (de Lázaro and Mooney, 2021; Kumar et al., 2021; Mast et al., 2021).
Statements
Author contributions
YH conceived the conception and wrote the manuscript. HoC and Q-QY provided a lot of valuable advice and contributed to the revision. YZ, HZ, and HaC provided many constructive suggestions and discussions. All authors read and approved the final manuscript.
Funding
This work was supported by the grant of the Medicine and Health Care Science and Technology Development Plan Projects Foundation of Shandong Province (No. 202003100622).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Ahn J. C. Teng P. Chen P. Posadas E. Tseng H. Lu S. C. et al (2021). Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology73, 422–436. 10.1002/hep.31165
2
Bakir B. Chiarella A. M. Pitarresi J. R. Rustgi A. K. (2020). EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol.30, 764–776. 10.1016/j.tcb.2020.07.003
3
Barbieri I. Kouzarides T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer20, 303–322. 10.1038/s41568-020-0253-2
4
Bebelman M. P. Smit M. J. Pegtel D. M. Baglio S. R. (2018). Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther.188, 1–11. 10.1016/j.pharmthera.2018.02.013
5
Bhattacharya S. Mohanty A. Achuthan S. Kotnala S. Jolly M. K. Kulkarni P. et al (2021). Group behavior and emergence of cancer drug resistance. Trends Cancer7, 323–334. 10.1016/j.trecan.2021.01.009
6
Bi Z. Li Q. Dinglin X. Xu Y. You K. Hong H. et al (2020). Nanoparticles (NPs)-Meditated LncRNA AFAP1-AS1 silencing to block wnt/β-catenin signaling pathway for synergistic reversal of radioresistance and effective cancer radiotherapy. Adv. Sci.7, 2000915. 10.1002/advs.202000915
7
Boriack-Sjodin P. A. Ribich S. Copeland R. A. (2022). RNA-modifying proteins as anticancer drug targets. Nat. Rev. Drug Discov.15, 435–453. 10.1038/nrd.2018.71
8
Brannan C. I. Dees E. C. Ingram R. S. Tilghman S. M. (1990). The product of the H19 gene may function as an RNA. Mol. Cell. Biol.10, 28–36. 10.1128/mcb.10.1.28
9
Brown C. J. Lafreniere R. G. Powers V. E. Sebastio G. Ballabio A. Pettigrew A. L. et al (1991). Localization of the X inactivation centre on the human X chromosome in Xq13. Nature349, 82–84. 10.1038/349082a0
10
Campisano S. La Colla A. Echarte S. M. Chisari A. N. (2019). Interplay between early-life malnutrition, epigenetic modulation of the immune function and liver diseases. Nutr. Res. Rev.32, 128–145. 10.1017/S0954422418000239
11
Cao M. Zhao J. Hu G. (2019). Genome-wide methods for investigating long noncoding RNAs. Biomed. Pharmacother.111, 395–401. 10.1016/j.biopha.2018.12.078
12
Carlevaro-Fita J. Johnson R. (2019). Global positioning system: Understanding long noncoding RNAs through subcellular localization. Mol. Cell73, 869–883. 10.1016/j.molcel.2019.02.008
13
Carotenuto P. Fassan M. Pandolfo R. Lampis A. Vicentini C. Cascione L. et al (2017). Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut66, 1268–1277. 10.1136/gutjnl-2016-312278
14
Chan J. Tay Y. (2018). Noncoding RNA:RNA regulatory networks in cancer. Int. J. Mol. Sci.19, 1310. 10.3390/ijms19051310
15
Chao H. Ma H. Sun J. Yuan S. Dong P. Zhao A. et al (2022). Whole-Transcriptome analysis of non-coding RNA alteration in porcine alveolar macrophage exposed to aflatoxin B1. Toxins14, 373. 10.3390/toxins14060373
16
Chen F. Zhong Z. Tan H. Y. Guo W. Zhang C. Cheng C. et al (2020). Suppression of lncRNA MALAT1 by betulinic acid inhibits hepatocellular carcinoma progression by targeting IAPs via miR-22-3p. Clin. Transl. Med.10, e190. 10.1002/ctm2.190
17
Chen V. L. Xu D. Wicha M. S. Lok A. S. Parikh N. D. (2020). Utility of liquid biopsy analysis in detection of hepatocellular carcinoma, determination of prognosis, and disease monitoring: A systematic review. Clin. Gastroenterol. Hepatol.18, 2879–2902. e9. 10.1016/j.cgh.2020.04.019
18
Choi S.-W. Kim H.-W. Nam J.-W. (2019). The small peptide world in long noncoding RNAs. Brief. Bioinform.20, 1853–1864. 10.1093/bib/bby055
19
Choi Y. J. Anders L. (2014). Signaling through cyclin D-dependent kinases. Oncogene33, 1890–1903. 10.1038/onc.2013.137
20
Cockram P. E. Kist M. Prakash S. Chen S.-H. Wertz I. E. Vucic D. (2021). Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ.28, 591–605. 10.1038/s41418-020-00708-5
21
Coulouarn C. Factor V. M. Thorgeirsson S. S. (2008). Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology47, 2059–2067. 10.1002/hep.22283
22
Dai J. Su Y. Zhong S. Cong L. Liu B. Yang J. et al (2020). Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther.5, 145. 10.1038/s41392-020-00261-0
23
Dang F. Nie L. Wei W. (2021). Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ.28, 427–438. 10.1038/s41418-020-00648-0
24
de Lázaro I. Mooney D. J. (2021). Obstacles and opportunities in a forward vision for cancer nanomedicine. Nat. Mat.20, 1469–1479. 10.1038/s41563-021-01047-7
25
Delaunay S. Frye M. (2019). RNA modifications regulating cell fate in cancer. Nat. Cell Biol.21, 552–559. 10.1038/s41556-019-0319-0
26
Deng B. Xu W. Wang Z. Liu C. Lin P. Li B. et al (2019). An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination. EMBO Rep.20, e47650. 10.15252/embr.201847650
27
Dey P. Kimmelman A. C. DePinho R. A. (2021). Metabolic codependencies in the tumor microenvironment. Cancer Discov.11, 1067–1081. 10.1158/2159-8290.CD-20-1211
28
Dhanasekaran R. Felsher D. W. (2019). A tale of two complications of obesity: NASH and hepatocellular carcinoma. Hepatology70, 1056–1058. 10.1002/hep.30649
29
Ding C.-H. Yin C. Chen S.-J. Wen L.-Z. Ding K. Lei S.-J. et al (2018). The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer17, 63. 10.1186/s12943-018-0813-1
30
Dubois M.-J. Bergeron S. Kim H.-J. Dombrowski L. Perreault M. Fournès B. et al (2006). The SHP-1 protein tyrosine phosphatase negatively modulates glucose homeostasis. Nat. Med.12, 549–556. 10.1038/nm1397
31
El-Gazzar A. Groh V. Spies T. (2013). Immunobiology and conflicting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J. Immunol.191, 1509–1515. 10.4049/jimmunol.1301071
32
Fan L.-C. Shiau C.-W. Tai W.-T. Hung M.-H. Chu P.-Y. Hsieh F.-S. et al (2015). SHP-1 is a negative regulator of epithelial–mesenchymal transition in hepatocellular carcinoma. Oncogene34, 5252–5263. 10.1038/onc.2014.445
33
Febbraio M. A. Karin M. (2021). Sweet death”: Fructose as a metabolic toxin that targets the gut-liver axis. Cell Metab.33, 2316–2328. 10.1016/j.cmet.2021.09.004
34
Feng G. X. Li J. Yang Z. Zhang S. Q. Liu Y. X. Zhang W. Y. et al (2017). Hepatitis B virus X protein promotes the development of liver fibrosis and hepatoma through downregulation of miR-30e targeting P4HA2 mRNA. Oncogene36, 6895–6905. 10.1038/onc.2017.291
35
Feng J. Yang G. Liu Y. Gao Y. Zhao M. Bu Y. et al (2019). LncRNA PCNAP1 modulates Hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics9, 5227–5245. 10.7150/thno.34273
36
Ferreira R. G. Cardoso M. V. de Souza Furtado K. M. Espíndola K. M. M. Amorim R. P. Monteiro M. C. (2019). Epigenetic alterations caused by aflatoxin b1: A public health risk in the induction of hepatocellular carcinoma. Transl. Res.204, 51–71. 10.1016/j.trsl.2018.09.001
37
Fujii H. Kawada N. Japan Study Group of NAFLD (JSG-NAFLD) Japan Study Group of NAFLD (JSG-NAFLD) (2020). The role of insulin resistance and diabetes in nonalcoholic fatty liver disease. Int. J. Mol. Sci.21, 3863. 10.3390/ijms21113863
38
Gabory A. Ripoche M.-A. Le Digarcher A. Watrin F. Ziyyat A. Forné T. et al (2009). H19acts as a trans regulator of the imprinted gene network controlling growth in mice. Development136, 3413–3421. 10.1242/dev.036061
39
Gao Y. Shang S. Guo S. Li X. Zhou H. Liu H. et al (2021). Lnc2Cancer 3.0: An updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res.49, D1251–D1258. 10.1093/nar/gkaa1006
40
Groh V. Wu J. Yee C. Spies T. (2002). Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature419, 734–738. 10.1038/nature01112
41
Guan Y. Huang Q. Ai Y. Chen Q. Zhao W. Wang X. et al (2020). Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene39, 2408–2423. 10.1038/s41388-020-1158-y
42
Guiducci G. Stojic L. (2021). Long noncoding RNAs at the crossroads of cell cycle and genome integrity. Trends Genet.37, 528–546. 10.1016/j.tig.2021.01.006
43
Guo T. Gong C. Wu P. Battaglia-Hsu S.-F. Feng J. Liu P. et al (2020). LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ.27, 2191–2205. 10.1038/s41418-020-0494-3
44
Guo T. Wang H. Liu P. Xiao Y. Wu P. Wang Y. et al (2018). SNHG6 acts as a genome-wide hypomethylation trigger via coupling of miR-1297–mediated S-Adenosylmethionine–Dependent positive feedback loops. Cancer Res.78, 3849–3864. 10.1158/0008-5472.CAN-17-3833
45
He G. Karin M. (2011). NF-κB and STAT3 – key players in liver inflammation and cancer. Cell Res.21, 159–168. 10.1038/cr.2010.183
46
Heimbach J. K. Kulik L. M. Finn R. S. Sirlin C. B. Abecassis M. M. Roberts L. R. et al (2018). AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology67, 358–380. 10.1002/hep.29086
47
Heitzer E. Haque I. S. Roberts C. E. S. Speicher M. R. (2019). Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet.20, 71–88. 10.1038/s41576-018-0071-5
48
Hepatitis (2018). Hepatitis B virus infection. Nat. Rev. Dis. Prim.4, 18036. 10.1038/nrdp.2018.36
49
Hlady R. A. Zhao X. El Khoury L. Y. Luna A. Pham K. Wu Q. et al (2022). Interferon drives HCV scarring of the epigenome and creates targetable vulnerabilities following viral clearance. Hepatology75, 983–996. 10.1002/hep.32111
50
Huang J. Cao S. Ou Q. Yang B. Zheng S. Tang J. et al (2018). The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol. Cancer17, 93. 10.1186/s12943-018-0841-x
51
Huang W. J. Tian X. P. Bi S. X. Zhang S. R. He T. S. Song L. Y. et al (2020). The β-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene39, 4538–4550. 10.1038/s41388-020-1307-3
52
Huang Z. Zhou J.-K. Peng Y. He W. Huang C. (2020). The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer19, 77. 10.1186/s12943-020-01188-4
53
Huo X. Han S. Wu G. Latchoumanin O. Zhou G. Hebbard L. et al (2017). Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: Implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol. Cancer16, 165. 10.1186/s12943-017-0734-4
54
Innes H. Barclay S. T. Hayes P. C. Fraser A. Dillon J. F. Stanley A. et al (2018). The risk of hepatocellular carcinoma in cirrhotic patients with hepatitis C and sustained viral response: Role of the treatment regimen. J. Hepatol.68, 646–654. 10.1016/j.jhep.2017.10.033
55
Iyer M. K. Niknafs Y. S. Malik R. Singhal U. Sahu A. Hosono Y. et al (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet.47, 199–208. 10.1038/ng.3192
56
Jochems F. Thijssen B. De Conti G. Jansen R. Pogacar Z. Groot K. et al (2021). The cancer SENESCopedia: A delineation of cancer cell senescence. Cell Rep.36, 109441. 10.1016/j.celrep.2021.109441
57
Kang C. L. Qi B. Cai Q. Q. Fu L. S. Yang Y. Tang C. et al (2019). LncRNA AY promotes hepatocellular carcinoma metastasis by stimulating ITGAV transcription. Theranostics9, 4421–4436. 10.7150/thno.32854
58
Kanwal F. Singal A. G. (2019). Surveillance for hepatocellular carcinoma: Current best practice and future direction. Gastroenterology157, 54–64. 10.1053/j.gastro.2019.02.049
59
Kelman Z. (1997). PCNA: Structure, functions and interactions. Oncogene14, 629–640. 10.1038/sj.onc.1200886
60
Khanna K. K. Jackson S. P. (2001). DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet.27, 247–254. 10.1038/85798
61
Khatun M. Sur S. Steele R. Ray R. Ray R. B. (2021). Inhibition of long noncoding RNA linc-pint by hepatitis C virus in infected hepatocytes enhances lipogenesis. Hepatology74, 41–54. 10.1002/hep.31656
62
Kim C.-M. Koike K. Saito I. Miyamura T. Jay G. (1991). HBx gene of Hepatitis B virus induces liver cancer in transgenic mice. Nature351, 317–320. 10.1038/351317a0
63
Kim S.-S. Harford J. B. Moghe M. Rait A. Pirollo K. F. Chang E. H. (2018). Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res.46, 1424–1440. 10.1093/nar/gkx1221
64
Kumar V. Rahman M. Gahtori P. Al-Abbasi F. Anwar F. Kim H. S. (2021). Current status and future directions of hepatocellular carcinoma-targeted nanoparticles and nanomedicine. Expert Opin. Drug Deliv.18, 673–694. 10.1080/17425247.2021.1860939
65
Lan T. Li H. Zhang D. Xu L. Liu H. Hao X. et al (2019a). KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer18, 186. 10.1186/s12943-019-1106-z
66
Lan T. Yuan K. Yan X. Xu L. Liao H. Hao X. et al (2019b). LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res.79, 3220–3234. 10.1158/0008-5472.CAN-18-4044
67
Lee T. K.-W. Guan X.-Y. Ma S. (2022). Cancer stem cells in hepatocellular carcinoma — From origin to clinical implications. Nat. Rev. Gastroenterol. Hepatol.19, 26–44. 10.1038/s41575-021-00508-3
68
Leslie J. Geh D. Elsharkawy A. M. Mann D. A. Vacca M. (2022). Metabolic dysfunction and cancer in HCV: Shared pathways and mutual interactions. J. Hepatol.77, 219–236. 10.1016/j.jhep.2022.01.029
69
Li S. Zhu Y. Li R. Huang J. You K. Yuan Y. et al (2021). LncRNA lnc-APUE is repressed by HNF4 α and promotes G1/S phase transition and tumor growth by regulating MiR-20b/E2F1 Axis. Adv. Sci.8, 2003094. 10.1002/advs.202003094
70
Li Y. Zhao J. Yu S. Wang Z. He X. Su Y. et al (2019). Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem.65, 798–808. 10.1373/clinchem.2018.301291
71
Li Z. Zhang J. Liu X. Li S. Wang Q. Chen D. et al (2018). The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun.9, 1572. 10.1038/s41467-018-04006-0
72
Liang C. Zhang X. Yang M. Dong X. (2019). Recent progress in ferroptosis inducers for cancer therapy. Adv. Mat.31, 1904197. 10.1002/adma.201904197
73
Lim L. J. Wong S. Y. S. Huang F. Lim S. Chong S. S. Ooi L. L. et al (2019). Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res.79, 5131–5139. 10.1158/0008-5472.CAN-19-0255
74
Lin B. Lu L. Wang Y. Zhang Q. Wang Z. Cheng G. et al (2021). Nanomedicine directs neuronal differentiation of neural stem cells via silencing long noncoding RNA for stroke therapy. Nano Lett.21, 806–815. 10.1021/acs.nanolett.0c04560
75
Lin C. Yang L. (2018). Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol.28, 287–301. 10.1016/j.tcb.2017.11.008
76
Liu F. Tian T. Zhang Z. Xie S. Yang J. Zhu L. et al (2022). Long non-coding RNA SNHG6 couples cholesterol sensing with mTORC1 activation in hepatocellular carcinoma. Nat. Metab.4, 1022–1040. 10.1038/s42255-022-00616-7
77
Liu J. Xu R. Mai S.-J. Ma Y.-S. Zhang M.-Y. Cao P.-S. et al (2020). LncRNA CSMD1-1 promotes the progression of Hepatocellular Carcinoma by activating MYC signaling. Theranostics10, 7527–7544. 10.7150/thno.45989
78
Liu S. J. Dang H. X. Lim D. A. Feng F. Y. Maher C. A. (2021). Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer21, 446–460. 10.1038/s41568-021-00353-1
79
Liu W. Jing Y. Gao L. Li R. Yang X. Pan X. et al (2020). Lipopolysaccharide induces the differentiation of hepatic progenitor cells into myofibroblasts constitutes the hepatocarcinogenesis-associated microenvironment. Cell Death Differ.27, 85–101. 10.1038/s41418-019-0340-7
80
Liu X. Duan X. Holmes J. A. Li W. Lee S. H. Tu Z. et al (2019). A long noncoding RNA regulates hepatitis C virus infection through interferon alpha-inducible protein 6. Hepatology69, 1004–1019. 10.1002/hep.30266
81
Liu X. Kumar Mishra S. Wang T. Xu Z. Zhao X. Wang Y. et al (2020). AFB1 induced transcriptional regulation related to apoptosis and lipid metabolism in liver of chicken. Toxins12, 290. 10.3390/toxins12050290
82
Liu X. Liang Y. Song R. Yang G. Han J. Lan Y. et al (2018). Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol. Cancer17, 90. 10.1186/s12943-018-0838-5
83
Llovet J. M. Montal R. Sia D. Finn R. S. (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol.15, 599–616. 10.1038/s41571-018-0073-4
84
Lopez-Ruiz P. Rodriguez-Ubreva J. Ernesto Cariaga A. Alicia Cortes M. Colas B. (2011). SHP-1 in cell-cycle regulation. Anticancer. Agents Med. Chem.11, 89–98. 10.2174/187152011794941154
85
Louro R. Smirnova A. S. Verjovski-Almeida S. (2009). Long intronic noncoding RNA transcription: Expression noise or expression choice?Genomics93, 291–298. 10.1016/j.ygeno.2008.11.009
86
Lu S. Zhou J. Sun Y. Li N. Miao M. Jiao B. et al (2017). The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol. Cancer16, 125. 10.1186/s12943-017-0676-x
87
Luo Y. Yao Y. Wu P. Zi X. Sun N. He J. (2022). The potential role of N7-methylguanosine (m7G) in cancer. J. Hematol. Oncol.15, 63. 10.1186/s13045-022-01285-5
88
Lv J. Yu Y.-Q. Li S.-Q. Luo L. Wang Q. (2014). Aflatoxin B1 promotes cell growth and invasion in hepatocellular carcinoma HepG2 cells through H19 and E2F1. Asian pac. J. Cancer Prev.15, 2565–2570. 10.7314/apjcp.2014.15.6.2565
89
Maass P. G. Barutcu A. R. Rinn J. L. (2019). Interchromosomal interactions: A genomic love story of kissing chromosomes. J. Cell Biol.218, 27–38. 10.1083/jcb.201806052
90
Machida K. (2020). Cell fate, metabolic reprogramming and lncRNA of tumor-initiating stem-like cells induced by alcohol. Chem. Biol. Interact.323, 109055. 10.1016/j.cbi.2020.109055
91
Majumdar A. Curley S. A. Wu X. Brown P. Hwang J. P. Shetty K. et al (2012). Hepatic stem cells and transforming growth factor β in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol.9, 530–538. 10.1038/nrgastro.2012.114
92
Malakar P. Stein I. Saragovi A. Winkler R. Stern-Ginossar N. Berger M. et al (2019). Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Res.79, 2480–2493. 10.1158/0008-5472.CAN-18-1432
93
Marrero J. A. Fontana R. J. Fu S. Conjeevaram H. S. Su G. L. Lok A. S. (2005). Alcohol, tobacco and obesity are synergistic risk factors for hepatocellular carcinoma. J. Hepatol.42, 218–224. 10.1016/j.jhep.2004.10.005
94
Martínez-Reyes I. Chandel N. S. (2021). Cancer metabolism: Looking forward. Nat. Rev. Cancer21, 669–680. 10.1038/s41568-021-00378-6
95
Mast M.-P. Modh H. Champanhac C. Wang J.-W. Storm G. Krämer J. et al (2021). Nanomedicine at the crossroads – a quick guide for IVIVC. Adv. Drug Deliv. Rev.179, 113829. 10.1016/j.addr.2021.113829
96
Matthews H. K. Bertoli C. de Bruin R. A. M. (2022). Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol.23, 74–88. 10.1038/s41580-021-00404-3
97
McAndrews K. M. Kalluri R. (2019). Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer18, 52. 10.1186/s12943-019-0963-9
98
McCullough A. K. Lloyd R. S. (2019). Mechanisms underlying aflatoxin-associated mutagenesis – implications in carcinogenesis. DNA Repair77, 76–86. 10.1016/j.dnarep.2019.03.004
99
McGlynn K. A. Petrick J. L. El-Serag H. B. (2021). Epidemiology of hepatocellular carcinoma. Hepatology73, 4–13. 10.1002/hep.31288
100
Meroni M. Longo M. Dongiovanni P. (2019). Alcohol or gut microbiota: Who is the guilty?Int. J. Mol. Sci.20, 4568. 10.3390/ijms20184568
101
Meunier L. Hirsch T. Z. Caruso S. Imbeaud S. Bayard Q. Roehrig A. et al (2021). DNA methylation signatures reveal the diversity of processes remodeling hepatocellular carcinoma methylomes. Hepatology74, 816–834. 10.1002/hep.31796
102
Min H. S. Kim H. J. Naito M. Ogura S. Toh K. Hayashi K. et al (2020). Systemic brain delivery of antisense oligonucleotides across the blood-brain barrier with a glucose-coated polymeric nanocarrier. Angew. Chem. Int. Ed. Engl.59, 8173–8180. 10.1002/anie.201914751
103
Moldogazieva N. T. Zavadskiy S. P. Terentiev A. A. (2021). Genomic landscape of liquid biopsy for hepatocellular carcinoma personalized medicine. Cancer Genomics Proteomics18, 369–383. 10.21873/cgp.20266
104
Moreno-Gonzalez M. Beraza N. (2021). The role of the microbiome in liver cancer. Cancers13, 2330. 10.3390/cancers13102330
105
Morf J. Basu S. Amaral P. P. (2020). RNA, genome output and input. Front. Genet.11, 589413. 10.3389/fgene.2020.589413
106
Mou Y. Wang J. Wu J. He D. Zhang C. Duan C. et al (2019). Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol.12, 34. 10.1186/s13045-019-0720-y
107
Muñoz-Espín D. Serrano M. (2014). Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol.15, 482–496. 10.1038/nrm3823
108
Narayanan S. Cai C.-Y. Assaraf Y. G. Guo H.-Q. Cui Q. Wei L. et al (2020). Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug resist. updat.48, 100663. 10.1016/j.drup.2019.100663
109
Nassar D. Blanpain C. (2016). Cancer stem cells: Basic concepts and therapeutic implications. Annu. Rev. Pathol.11, 47–76. 10.1146/annurev-pathol-012615-044438
110
Nault J. Villanueva A. (2021). Biomarkers for hepatobiliary cancers. Hepatology73, 115–127. 10.1002/hep.31175
111
Nishibatake Kinoshita M. Minami T. Tateishi R. Wake T. Nakagomi R. Fujiwara N. et al (2019). Impact of direct-acting antivirals on early recurrence of HCV-related HCC: Comparison with interferon-based therapy. J. Hepatol.70, 78–86. 10.1016/j.jhep.2018.09.029
112
Nussinov R. Tsai C.-J. Jang H. (2021). Anticancer drug resistance: An update and perspective. Drug resist. updat.59, 100796. 10.1016/j.drup.2021.100796
113
Pastushenko I. Blanpain C. (2019). EMT transition states during tumor progression and metastasis. Trends Cell Biol.29, 212–226. 10.1016/j.tcb.2018.12.001
114
Peng L. Jiang B. Yuan X. Qiu Y. Peng J. Huang Y. et al (2019). Super-enhancer–associated long noncoding RNA HCCL5 is activated by ZEB1 and promotes the malignancy of hepatocellular carcinoma. Cancer Res.79, 572–584. 10.1158/0008-5472.CAN-18-0367
115
Peng W.-X. Koirala P. Mo Y.-Y. (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene36, 5661–5667. 10.1038/onc.2017.184
116
Premkumar M. Anand A. C. (2021). Tobacco, cigarettes, and the liver: The smoking gun. J. Clin. Exp. Hepatol.11, 700–712. 10.1016/j.jceh.2021.07.016
117
Protzer U. (2015). Epigenetic control of HBV by HBx protein—Releasing the break?Nat. Rev. Gastroenterol. Hepatol.12, 558–559. 10.1038/nrgastro.2015.152
118
Qi X. Lin Y. Chen J. Shen B. (2020). Decoding competing endogenous RNA networks for cancer biomarker discovery. Brief. Bioinform.21, 441–457. 10.1093/bib/bbz006
119
Qi Y. Fan L. Ran D. Xu J. Wang Y. Wu J. et al (2021). Main risk factors of type 2 diabetes mellitus with nonalcoholic fatty liver disease and hepatocellular carcinoma. J. Oncol.2021, 7764817. 10.1155/2021/7764817
120
Qian X. Zhao J. Yeung P. Y. Zhang Q. C. Kwok C. K. (2019). Revealing lncRNA structures and interactions by sequencing-based approaches. Trends biochem. Sci.44, 33–52. 10.1016/j.tibs.2018.09.012
121
Qin G. Tu X. Li H. Cao P. Chen X. Song J. et al (2020). Long noncoding RNA p53-stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology71, 112–129. 10.1002/hep.30793
122
Reig M. Mariño Z. Perelló C. Iñarrairaegui M. Ribeiro A. Lens S. et al (2016). Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J. Hepatol.65, 719–726. 10.1016/j.jhep.2016.04.008
123
Rosenbloom K. R. Dreszer T. R. Long J. C. Malladi V. S. Sloan C. A. Raney B. J. et al (2012). ENCODE whole-genome data in the UCSC genome browser: Update 2012. Nucleic Acids Res.40, D912–D917. 10.1093/nar/gkr1012
124
Ruiz-Margáin A. Román-Calleja B. M. Moreno-Guillén P. González-Regueiro J. A. Kúsulas-Delint D. Campos-Murguía A. et al (2021). Nutritional therapy for hepatocellular carcinoma. World J. Gastrointest. Oncol.13, 1440–1452. 10.4251/wjgo.v13.i10.1440
125
Rushing B. R. Selim M. I. (2019). Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol.124, 81–100. 10.1016/j.fct.2018.11.047
126
Saeed R. A. Maqsood M. Saeed R. A. Muzammil H. S. Khan M. I. Asghar L. et al (2022). Plant-based foods and hepatocellular carcinoma: A review on mechanistic understanding. Crit. Rev. Food Sci. Nutr.62 (30), 1–34. 10.1080/10408398.2022.2095974
127
Salerno D. Chiodo L. Alfano V. Floriot O. Cottone G. Paturel A. et al (2020). Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut69, 2016–2024. 10.1136/gutjnl-2019-319637
128
Shi J. He J. Lin J. Sun X. Sun F. Ou C. et al (2016). Distinct response of the hepatic transcriptome to Aflatoxin B1 induced hepatocellular carcinogenesis and resistance in rats. Sci. Rep.6, 31898. 10.1038/srep31898
129
Shi Q. Xue C. Yuan X. He Y. Yu Z. (2020). Gene signatures and prognostic values of m1A-related regulatory genes in hepatocellular carcinoma. Sci. Rep.10, 15083. 10.1038/s41598-020-72178-1
130
Shih D. Q. Bussen M. Sehayek E. Ananthanarayanan M. Shneider B. L. Suchy F. J. et al (2001). Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet.27, 375–382. 10.1038/86871
131
Simon T. G. Ma Y. Ludvigsson J. F. Chong D. Q. Giovannucci E. L. Fuchs C. S. et al (2018). Association between aspirin use and risk of hepatocellular carcinoma. JAMA Oncol.4, 1683–1690. 10.1001/jamaoncol.2018.4154
132
Singal A. G. El-Serag H. B. (2015). Hepatocellular carcinoma from epidemiology to prevention: Translating knowledge into practice. Clin. Gastroenterol. Hepatol.13, 2140–2151. 10.1016/j.cgh.2015.08.014
133
Singal A. G. Lok A. S. Feng Z. Kanwal F. Parikh N. D. (2022). Conceptual model for the hepatocellular carcinoma screening continuum: Current status and research agenda. Clin. Gastroenterol. Hepatol.20, 9–18. 10.1016/j.cgh.2020.09.036
134
Søndergaard J. N. Sommerauer C. Atanasoai I. Hinte L. C. Geng K. Guiducci G. et al (2022). CCT3- LINC00326 axis regulates hepatocarcinogenic lipid metabolism. Gut71, 2081–2092. 10.1136/gutjnl-2021-325109
135
Song H. Liu Y. Li X. Chen S. Xie R. Chen D. et al (2020). Long noncoding RNA CASC11 promotes hepatocarcinogenesis and HCC progression through EIF4A3-mediated E2F1 activation. Clin. Transl. Med.10, e220. 10.1002/ctm2.220
136
Sun T. Liu Z. Yang Q. (2020). The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer19, 146. 10.1186/s12943-020-01262-x
137
Sun Z. Xue S. Zhang M. Xu H. Hu X. Chen S. et al (2020). Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene39, 6906–6919. 10.1038/s41388-020-01475-w
138
Sung H. Ferlay J. Siegel R. L. Laversanne M. Soerjomataram I. Jemal A. et al (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin.71, 209–249. 10.3322/caac.21660
139
Szwed A. Kim E. Jacinto E. (2021). Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev.101, 1371–1426. 10.1152/physrev.00026.2020
140
Tai W.-T. Cheng A.-L. Shiau C.-W. Liu C.-Y. Ko C.-H. Lin M.-W. et al (2012). Dovitinib induces apoptosis and overcomes sorafenib resistance in hepatocellular carcinoma through SHP-1–Mediated inhibition of STAT3. Mol. Cancer Ther.11, 452–463. 10.1158/1535-7163.MCT-11-0412
141
Tong H. V. Toan N. L. Song L. H. Bock C.-T. Kremsner P. G. Velavan T. P. (2013). Hepatitis B virus-induced hepatocellular carcinoma: Functional roles of MICA variants. J. Viral Hepat.20, 687–698. 10.1111/jvh.12089
142
Unfried J. P. Marín-Baquero M. Rivera-Calzada Á. Razquin N. Martín-Cuevas E. M. de Bragança S. et al (2021). Long noncoding RNA NIHCOLE promotes ligation efficiency of DNA double-strand breaks in hepatocellular carcinoma. Cancer Res.81, 4910–4925. 10.1158/0008-5472.CAN-21-0463
143
Unfried J. P. Serrano G. Suárez B. Sangro P. Ferretti V. Prior C. et al (2019). Identification of coding and long noncoding RNAs differentially expressed in tumors and preferentially expressed in healthy tissues. Cancer Res.79, 5167–5180. 10.1158/0008-5472.CAN-19-0400
144
van der Meel R. Sulheim E. Shi Y. Kiessling F. Mulder W. J. M. Lammers T. (2019). Smart cancer nanomedicine. Nat. Nanotechnol.14, 1007–1017. 10.1038/s41565-019-0567-y
145
Wang B. Jacob S. T. (2011). Role of cancer stem cells in hepatocarcinogenesis. Genome Med.3, 11. 10.1186/gm225
146
Wang H. Huo X. Yang X.-R. He J. Cheng L. Wang N. et al (2017). STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer16, 136. 10.1186/s12943-017-0680-1
147
Wang H. Liang L. Dong Q. Huan L. He J. Li B. et al (2018). Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics8, 2814–2829. 10.7150/thno.23012
148
Wang L. Lankhorst L. Bernards R. (2022). Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer22, 340–355. 10.1038/s41568-022-00450-9
149
Wang S.-J. Li D. Ou Y. Jiang L. Chen Y. Zhao Y. et al (2016). Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep.17, 366–373. 10.1016/j.celrep.2016.09.022
150
Wang T. Fu X. Jin T. Zhang L. Liu B. Wu Y. et al (2019). Aspirin targets P4HA2 through inhibiting NF-κB and LMCD1-AS1/let-7g to inhibit tumour growth and collagen deposition in hepatocellular carcinoma. EBioMedicine45, 168–180. 10.1016/j.ebiom.2019.06.048
151
Wang W. Han Y. Jo H. A. Lee J. Song Y. S. (2020). Non-coding RNAs shuttled via exosomes reshape the hypoxic tumor microenvironment. J. Hematol. Oncol.13, 67. 10.1186/s13045-020-00893-3
152
Wang X. Kang M. Liu C. Lin T. Han X. Jiang X. (2021). Current state and progress of research on the role of lncRNA in HBV-related liver cancer. Front. Cell. Infect. Microbiol.11, 714895. 10.3389/fcimb.2021.714895
153
Wang Y. Liu J. Yang J. Yu X. Chen Z. Chen Y. et al (2019a). Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology70, 259–275. 10.1002/hep.30613
154
Wang Y. Liu Z. Yao B. Li Q. Wang L. Wang C. et al (2017). Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer16, 123. 10.1186/s12943-017-0702-z
155
Wang Y. Yang L. Chen T. Liu X. Guo Y. Zhu Q. et al (2019b). A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol. Cancer18, 28. 10.1186/s12943-019-0957-7
156
Wang Y. Zhu P. Luo J. Wang J. Liu Z. Wu W. et al (2019c). LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J.38, e101110. 10.15252/embj.2018101110
157
Warburg O. (1925). über den Stoffwechsel der Carcinomzelle. Klin. Wochenschr.4, 534–536. 10.1007/BF01726151
158
Wei L. Ploss A. (2020). Core components of DNA lagging strand synthesis machinery are essential for Hepatitis B virus cccDNA formation. Nat. Microbiol.5, 715–726. 10.1038/s41564-020-0678-0
159
Wei L. Ploss A. (2021). Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat. Commun.12, 1591. 10.1038/s41467-021-21850-9
160
White E. Mehnert J. M. Chan C. S. (2015). Autophagy, metabolism, and cancer. Clin. Cancer Res.21, 5037–5046. 10.1158/1078-0432.CCR-15-0490
161
Wu M.-Z. Yuan Y. Huang B.-Y. Chen J.-X. Li B.-K. Fang J.-H. et al (2021). Identification of a TGF-β/SMAD/lnc-UTGF positive feedback loop and its role in hepatoma metastasis. Signal Transduct. Target. Ther.6, 395. 10.1038/s41392-021-00781-3
162
Wu Y. Zhao Y. Huan L. Zhao J. Zhou Y. Xu L. et al (2020). An LTR retrotransposon-derived long noncoding RNA lncMER52A promotes hepatocellular carcinoma progression by binding p120-catenin. Cancer Res.80, 976–987. 10.1158/0008-5472.CAN-19-2115
163
Xiang X. Fu Y. Zhao K. Miao R. Zhang X. Ma X. et al (2021). Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics11, 4929–4944. 10.7150/thno.55672
164
Xie C. Zhang L. Chen Z. Zhong W. Fang J. Zhu Y. et al (2020). A hMTR4-PDIA3P1-miR-125/124-TRAF6 regulatory Axis and its function in NF kappa B signaling and chemoresistance. Hepatology71, 1660–1677. 10.1002/hep.30931
165
Xie Z. Zhou F. Yang Y. Li L. Lei Y. Lin X. et al (2018). Lnc-PCDH9-13:1 is a hypersensitive and specific biomarker for early hepatocellular carcinoma. EBioMedicine33, 57–67. 10.1016/j.ebiom.2018.06.026
166
Xin X. Wu M. Meng Q. Wang C. Lu Y. Yang Y. et al (2018). Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer17, 94. 10.1186/s12943-018-0843-8
167
Xu E. Forest M.-P. Schwab M. Avramoglu R. K. St-Amand E. Caron A. Z. et al (2014). Hepatocyte-specificPtpn6deletion promotes hepatic lipid accretion, but reduces NAFLD in diet-induced obesity: Potential role of PPARγ. Hepatology59, 1803–1815. 10.1002/hep.26957
168
Xu F. Li C. H. Wong C. H. Chen G. G. Lai P. B. S. Shao S. et al (2019). Genome-wide screening and functional analysis identifies tumor suppressor long noncoding RNAs epigenetically silenced in hepatocellular carcinoma. Cancer Res.79, 1305–1317. 10.1158/0008-5472.CAN-18-1659
169
Xu Z. Chen Y. Ma L. Chen Y. Liu J. Guo Y. et al (2022). Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol. Ther.30, 3133–3154. 10.1016/j.ymthe.2022.01.046
170
Yamazaki T. Souquere S. Chujo T. Kobelke S. Chong Y. S. Fox A. H. et al (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell70, 1038–1053. e7. 10.1016/j.molcel.2018.05.019
171
Yan X. Zhang D. Wu W. Wu S. Qian J. Hao Y. et al (2017). Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA–MUF interaction with ANXA2 and miR-34a. Cancer Res.77, 6704–6716. 10.1158/0008-5472.CAN-17-1915
172
Yang L. Peng X. Li Y. Zhang X. Ma Y. Wu C. et al (2019). Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer18, 78. 10.1186/s12943-019-0990-6
173
Yao J. Wu L. Meng X. Yang H. Ni S. Wang Q. et al (2017). Profiling, clinicopathological correlation and functional validation of specific long non-coding RNAs for hepatocellular carcinoma. Mol. Cancer16, 164. 10.1186/s12943-017-0733-5
174
Ye Q. Ling S. Zheng S. Xu X. (2019). Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol. Cancer18, 114. 10.1186/s12943-019-1043-x
175
Yin D. Hu Z. Luo C. Wang X. Xin H. Sun R. et al (2021). LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin. Transl. Med.11, e409. 10.1002/ctm2.409
176
Yu H. Pardoll D. Jove R. (2009). STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer9, 798–809. 10.1038/nrc2734
177
Yu X. Li S. (2017). Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene36, 2629–2636. 10.1038/onc.2016.410
178
Yuen M.-F. Heo J. Jang J.-W. Yoon J.-H. Kweon Y.-O. Park S.-J. et al (2021). Safety, tolerability and antiviral activity of the antisense oligonucleotide bepirovirsen in patients with chronic Hepatitis B: A phase 2 randomized controlled trial. Nat. Med.27, 1725–1734. 10.1038/s41591-021-01513-4
179
Zhang D. Luo Y. Yishake D. Liu Z. He T. Luo Y. et al (2020). Prediagnostic dietary intakes of vitamin A and β-carotene are associated with hepatocellular-carcinoma survival. Food Funct.11, 759–767. 10.1039/C9FO02468A
180
Zhang D. Y. Zou X.-J. Cao C.-H. Zhang T. Lei L. Qi X.-L. et al (2018). Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics8, 3751–3765. 10.7150/thno.22493
181
Zhang H. Su X. Burley S. K. Zheng X. F. S. (2022). mTOR regulates aerobic glycolysis through NEAT1 and nuclear paraspeckle-mediated mechanism in hepatocellular carcinoma. Theranostics12, 3518–3533. 10.7150/thno.72581
182
Zhang J. Han C. Ungerleider N. Chen W. Song K. Wang Y. et al (2019). A transforming growth factor-β and H19 signaling Axis in tumor-initiating hepatocytes that regulates hepatic carcinogenesis. Hepatology69, 1549–1563. 10.1002/hep.30153
183
Zhang J. Li Z. Liu L. Wang Q. Li S. Chen D. et al (2018). Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology67, 171–187. 10.1002/hep.29405
184
Zhang L. Niu H. Ma J. Yuan B.-Y. Chen Y.-H. Zhuang Y. et al (2019). The molecular mechanism of LncRNA34a-mediated regulation of bone metastasis in hepatocellular carcinoma. Mol. Cancer18, 120. 10.1186/s12943-019-1044-9
185
Zhang L. Xu X. Su X. (2020a). Noncoding RNAs in cancer immunity: Functions, regulatory mechanisms, and clinical application. Mol. Cancer19, 48. 10.1186/s12943-020-01154-0
186
Zhang L. Yang J. Luo Y. Liu F. Yuan Y. Zhuang S. (2020b). A p53/lnc-Ip53 negative feedback loop regulates tumor growth and chemoresistance. Adv. Sci.7, 2001364. 10.1002/advs.202001364
187
Zhang S. Zhou Y. Wang Y. Wang Z. Xiao Q. Zhang Y. et al (2021). The mechanistic, diagnostic and therapeutic novel nucleic acids for hepatocellular carcinoma emerging in past score years. Brief. Bioinform.22, 1860–1883. 10.1093/bib/bbaa023
188
Zhang X.-D. Wang Y. Ye L.-H. (2014). Hepatitis B virus X protein accelerates the development of hepatoma. Int. J. Oncol.11, 182–190. 10.7497/j.issn.2095-3941.2014.03.004
189
Zhang Y. Luo M. Cui X. O’Connell D. Yang Y. (2022). Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ.29, 1850–1863. 10.1038/s41418-022-00970-9
190
Zhang Y. Tao Y. Liao Q. (2018). Long noncoding RNA: A crosslink in biological regulatory network. Brief. Bioinform.19, 930–945. 10.1093/bib/bbx042
191
Zhao S. Allis C. D. Wang G. G. (2021). The language of chromatin modification in human cancers. Nat. Rev. Cancer21, 413–430. 10.1038/s41568-021-00357-x
192
Zheng H. Li P. Kwok J. G. Korrapati A. Li W. T. Qu Y. et al (2018). Alcohol and hepatitis virus-dysregulated lncRNAs as potential biomarkers for hepatocellular carcinoma. Oncotarget9, 224–235. 10.18632/oncotarget.22921
193
Zheng Y.-L. Li L. Jia Y.-X. Zhang B.-Z. Li J.-C. Zhu Y.-H. et al (2019). LINC01554-Mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting akt/mTOR signaling pathway. Theranostics9, 796–810. 10.7150/thno.28992
194
Zhong R. Tian J. Fu M. Ma S. Liu L. Li J. et al (2020). LINC01149 variant modulates MICA expression that facilitates Hepatitis B virus spontaneous recovery but increases hepatocellular carcinoma risk. Oncogene39, 1944–1956. 10.1038/s41388-019-1117-7
195
Zhou H. Li F. Cheng S. Xu Y. Deng H. Gu D. et al (2022). DDX17-regulated alternative splicing that produced an oncogenic isoform of PXN-AS1 to promote HCC metastasis. Hepatology75, 847–865. 10.1002/hep.32195
196
Zhou K. Nguyen R. Qiao L. George J. (2022). Single cell RNA-seq analysis identifies a noncoding RNA mediating resistance to sorafenib treatment in HCC. Mol. Cancer21, 6. 10.1186/s12943-021-01473-w
197
Zhou Y. Huan L. Wu Y. Bao C. Chen B. Wang L. et al (2020). LncRNA ID2-AS1 suppresses tumor metastasis by activating the HDAC8/ID2 pathway in hepatocellular carcinoma. Cancer Lett.469, 399–409. 10.1016/j.canlet.2019.11.007
198
Zou S. Tong Q. Liu B. Huang W. Tian Y. Fu X. (2020). Targeting STAT3 in cancer immunotherapy. Mol. Cancer19, 145. 10.1186/s12943-020-01258-7
199
Zuo X. Chen Z. Gao W. Zhang Y. Wang J. Wang J. et al (2020). M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol.13, 5. 10.1186/s13045-019-0839-x
Summary
Keywords
hepatocellular carcinoma, long noncoding RNA, gene expression regulation, biomarker, diagnosis and therapy
Citation
Hong Y, Zhang Y, Zhao H, Chen H, Yu Q-Q and Cui H (2022) The roles of lncRNA functions and regulatory mechanisms in the diagnosis and treatment of hepatocellular carcinoma. Front. Cell Dev. Biol. 10:1051306. doi: 10.3389/fcell.2022.1051306
Received
22 September 2022
Accepted
07 November 2022
Published
18 November 2022
Volume
10 - 2022
Edited by
Xiqing Li, Henan Provincial People’s Hospital, China
Reviewed by
Jing Du, Binzhou Medical University Hospital, China
Yating Xiao, University of Chinese Academy of Sciences, China
Updates
Copyright
© 2022 Hong, Zhang, Zhao, Chen, Yu and Cui.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Qing Yu, yuqingqing_lucky@163.com; Hongxia Cui, chxjnyy@sina.com
†These authors have contributed equally to this work
This article was submitted to Cancer Cell Biology, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.