Abstract
Autophagy is a crucial mechanism implicated in both aging and cardiovascular disease, which are two closely interconnected conditions. Modulation of autophagy is expected to have profound impacts on cellular aging and maintenance of cardiovascular functions under physiological or pathological conditions. Consequently, modulation of autophagy could be an effective strategy for counteracting age-induced vascular and cardiac remodelling as well as alleviating cardiovascular disease. The present review comprehensively elucidates the multifaceted impacts of autophagy on aging of the cardiovascular system. We comprehensively analyse both vascular and cardiac tissues, including vascular and cardiac malignancies, in distinct contexts. We also emphasize the significance of non-coding RNAs (ncRNAs) in the epigenetic regulation of gene expression and their roles as biomarkers of cardiovascular pathologies while maintaining clear distinctions between the vascular and cardiac tissues. Preclinical and clinical models are described herein to highlight the importance of ncRNAs in disease treatment by considering their involvement in the modulation of autophagy within the cardiocirculatory system. Finally, we conducted a comprehensive meta-analysis of transcriptomic data to underscore the paramount importance of autophagy while demonstrating it as a process that is frequently dysregulated in both cardiac and vascular cells under pathological conditions. The findings presented herein emphasize the importance of investigating novel strategies for modulating autophagy as a potential therapeutic approach to the management of age-related cardiovascular disorders.
1 Introduction
Cardiovascular disease (CVD) is a term used to indicate the range of conditions affecting the heart and blood vessels. The four main types of CVDs are coronary artery disease (CAD), strokes and transient ischemic attacks, peripheral artery disease, and aortic disease. The aorta is the primary blood vessel in the body that serves as the conduit for transporting blood from the heart to various regions of the body. One of the common problems of the aorta is an aneurysm, where the aorta becomes weakened and its wall tends to bulge outward; this region could become a site of possible rupture and cause hemorrhage that would prevent sustenance of the other parts of the body. Similar problems are known to occur in peripheral artery diseases, which are characterized by occlusion of the peripheral arteries that prevent normal blood flow. The obstruction of blood flow to specific regions of the brain can lead to cerebral infarctions, which can result in permanent brain damage or even death. Mini cerebral infarctions are defined as transient ischemic events. In both types of infarctions, the symptoms include the inability to smile, drooping of the mouth or eye, the impossibility of fitting both arms, and the inability to speak or understand communication with another person. The obstruction of oxygen-rich blood supply to the heart can also cause coronary heart diseases. Among these, it is possible to recognize angina pectoris that causes chest pain, heart attack, or heart failure. Angina is typically not life-threatening but serves as an early warning sign of the risk of a potential heart attack or stroke. In contrast, a heart attack (myocardial infarction or MI) is a serious medical emergency characterized by sudden blockage of blood supply to the heart as well as heart failure caused by the inability of the heart to pump blood to the body.
The exact causes of CVDs are not clear, but there are several risk factors that can increase the possibility of developing cardiovascular conditions. In Europe, there were approximately 1.7 million deaths in the year 2020 from diseases of the circulatory system (343 deaths per 100,000 inhabitants; Eurostat, 2024), while the number of such deaths in the United States that had been decreasing until 2019 started increasing after the COVID-19 pandemic, reaching approximately 454.5 deaths per 100,000 inhabitants in 2022 (Woodruff et al., 2024). Interestingly, the World Health Organization reported in 2021 that the three leading causes of death were ischemic heart disease (∼9 millions), COVID-19 (slightly less than the former), and stroke (∼7 millions) (World Bank Group, 2024). Excluding the impacts of COVID-19 on death, the most prevalent pathologies responsible for mortality are associated with the cardiocirculatory system. CVD is most common in people older than 50 years, and the risk of developing CVD increases as people age further. Men are more affected than women (Figure 1) and are likely to develop CVD at an earlier age. Moreover, unhealthy dietary habits can lead to high cholesterol and high blood pressure, which are two of the important risk variables for developing CVDs. Some additional factors associated with increased risk of developing CVDs are smoking that can damage and narrow blood vessels; inactivity that can cause high blood pressure, high cholesterol, and excess bodyweight; diabetes that can damage blood vessels and cause their narrowing because of high blood sugar levels; hereditary conditions; and ethnicity (black non-Hispanic persons are the most affected in the United States) (U.S. Centers for Disease Control and Prevention, 2024). Chronic infections appear to be emerging risk factors for the development of CVDs even if they are not associated with aging. Notably, infection by the hepatitis C virus is associated with elevated incidence of atherosclerosis, strokes, and ischemic attacks (Domont and Cacoub, 2016). Chronic infection by the human immunodeficiency virus is well-known to be associated with various cardiovascular conditions, including heart failure, stroke, coronary artery plaques, and atherosclerosis (Longenecker et al., 2016; So-Armah et al., 2020; Perkins et al., 2023). Recently, there is a growing body of evidence suggesting potential associations between chronic endodontic infections and CVDs (Koletsi et al., 2021). Globally, CVDs are the most common causes of death and are more prevalent than cancers (Figure 2).
FIGURE 1
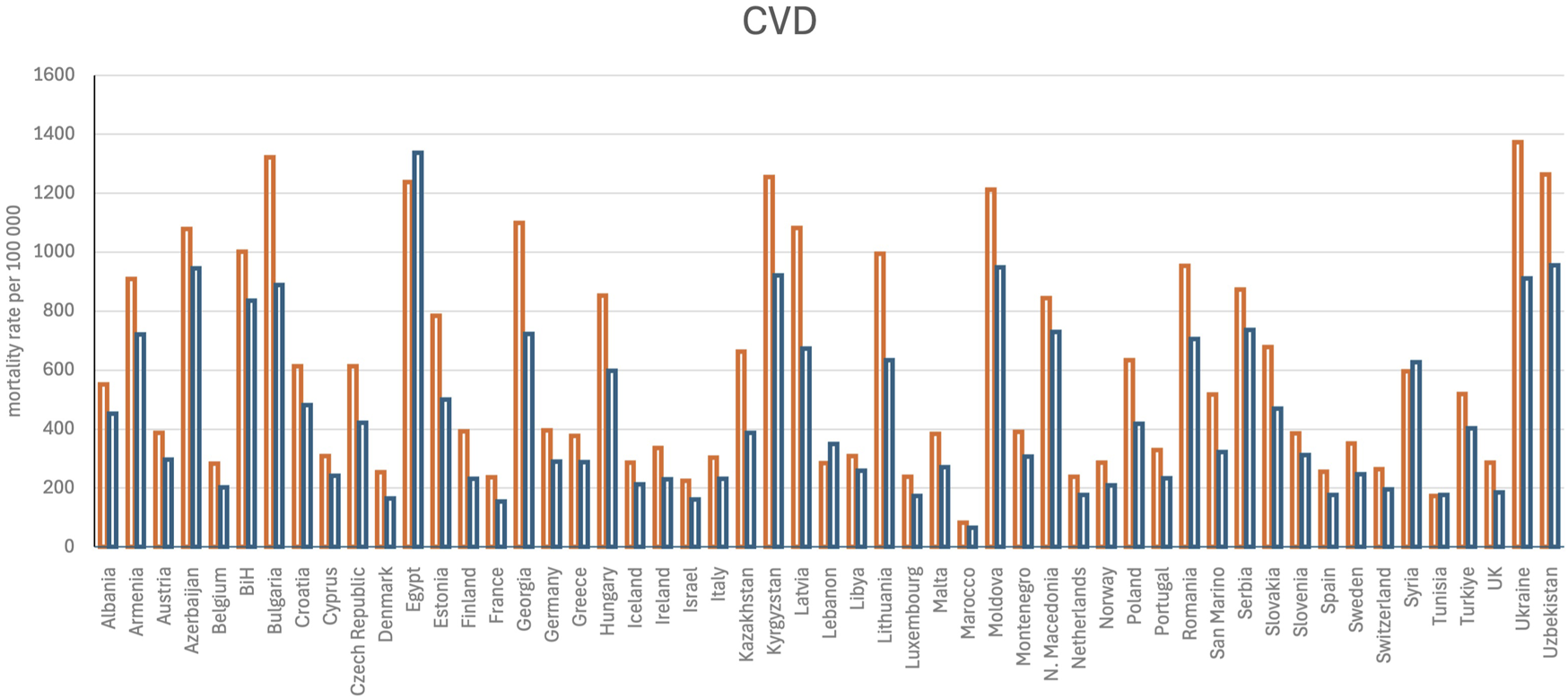
Age-standardized mortality rate per 100,000 persons for cardiovascular diseases (CVDs). The data were retrieved from Timmis et al. (2024). The orange bars represent men, and blue bars indicate women. UK, United Kingdom; BiH, Bosnia and Herzegovina.
FIGURE 2
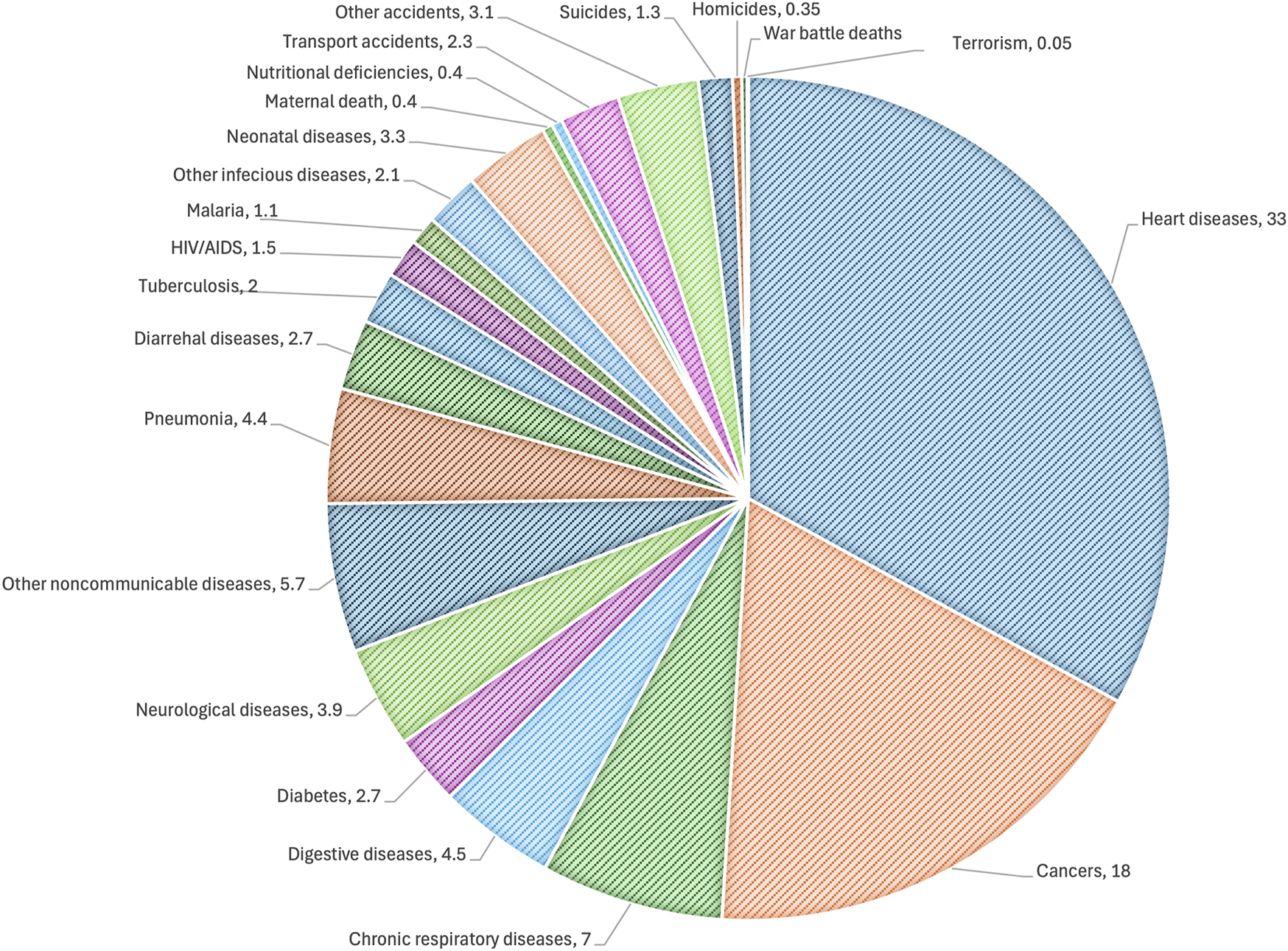
Distribution showing the global causes of death. The data were retrieved from the IHME global burden and disease and global terrorism databases. The data are representative for 2019, when the total number of deaths was 55 million.
Notably, under pathological cardiac conditions, the tissues undergo substantial remodeling, necessitating turnover of the molecules and organelles akin to developmental processes. This is based on a catabolic mechanism called autophagy. Several studies have described the importance of autophagy in CVDs and the possibilities of its modulation for therapeutic interventions (Mei et al., 2014; Sciarretta et al., 2018; Jiang et al., 2023). Autophagic activities have been found to decrease with age, likely contributing to the accumulation of damaged macromolecules and organelles from aging (Barbosa et al., 2019; Aman et al., 2021). Therefore, autophagic decline occurring during aging can contribute to the development of CVDs. Non-coding RNAs (ncRNAs) have been demonstrated to play crucial roles in diverse biological processes, including aging (Dhahbi, 2016; He et al., 2018; Sherazi et al., 2023; Wagner et al., 2024a; Ugalde et al., 2024) and CVDs (Poller et al., 2017; Kohlmaier et al., 2023; Jiang and Zhang, 2024). In this work, we emphasize the interactions between mechanisms associated with aging that are related to the development of CVDs; further, we focus on coding genes that regulate autophagy and experience expression modulations related to aging as well as CVD development. Excluding the descriptions of genes associated with autophagy, we primarily consider recent publications (from the last 5 years) on aging and CVDs in this review. Moreover, we consider transcriptomic data from different CVDs (human coronary plaques, failing human heart, ischemic cardiomyopathy, and idiopathic dilated cardiomyopathy) to demonstrate the importance of autophagy modulations in these pathologies.
2 Approaches for investigating the relationships between CVDs and autophagy-associated ncRNAs
2.1 Literature search
A comprehensive literature search was performed in PubMed to select articles published from 2019 to 2024. The search terms used included the following keywords: “cardiovascular disease,” “CVD,” “heart,”, “cardiac’,” “vasculature’,” “vascular,” “aging,” “ageing,” “non-coding RNAs,” “ncRNAs,” “microRNAs,” “miRNAs,” “long non-coding RNAs,” “lncRNAs,” “circular RNAs,” “circRNAs,” and “autophagy.” The information on autophagy was sourced from articles without any restriction on the year of publication.
2.2 Meta-analysis based on databases
Data were downloaded from the Gene Expression Omnibus (GEO) and GEO RNA-seq Experiments Interactive Navigator (GREIN) (Mahi et al., 2019) databases. Our database search encompassed the terms “cardiovascular disease” and “cardiovascular system” and prioritized studies conducted exclusively on humans. We excluded studies involving blood and blood cells, in vitro studies utilizing specific cells, or studies on modulation of specific genes. In contrast to the literature search, we utilized data published between 2014 and 2024 here as the data retrieved from the past 5 years were deemed insufficient. To avoid introducing alterations that could be associated with various normalisation approaches, we opted to utilize data that were already normalized. The differentially expressed genes (DEGs) were identified using AltAnalyze software (Emig et al., 2010; Salomonis et al., 2010; Olsson et al., 2016). Genes with normalized expressions below 0.3 (no log) were removed from the analysis, and a moderate t-test with Benjamini–Hochberg correction was performed to identify the DEGs. The normalized data are shown in Supplementary Table S1. The DEGs were then categorized by an overrepresentation approach implemented in easyGSEA (Cheng et al., 2021), and the Venn diagram was obtained using Venny 2.0 (Oliveros, 2007).
2.3 Approaches to investigate the roles of ncRNAs in autophagy within CVDs
The ncRNAs regulate gene expressions through several mechanisms that impact the approaches used to understand their functions. The first step here is to understand if their expressions are altered under different pathological conditions. In this regard, RNA sequencing is preferred to microarray nowadays because of the similar costs and greater ability to distinguish between the long non-coding and micro RNA (lncRNA and miRNA) isoforms (isomiRs) that have demonstrated more importance (Tomasello et al., 2021). For instance, van der Kwast et al. (2020) utilized 5′Dumbbell-PCR (5′DB-PCR) to support the RNA sequencing data to demonstrate that WT-miR-411 and iso-miR-411 exhibit differential expressions between the primary human umbilical arterial fibroblasts and human umbilical venous endothelial cells (HUVECs) with different target pools. The 5′DB-PCR is based on annealing of the two stem loops at the 3′ and 5′ ends of a miRNA. Although stem loops are used as sequence bases for annealing PCR primers, the gaps or overlaps in isomiRs strongly impact the efficacy of ligation as well as annealing of the TaqMan probe partially complementary to the microRNA and partially to the 3′ adapter sequences (Tomasello et al., 2021). In particular, isomiRs remain relatively unexplored and present an open problem because miRNAs and isomiRs regulate different targets; their analyses may furnish new and alternative approaches to treating autophagy induced by ischemic events. Different isomiRs can be formed as consequences of the altered activities of RNAses type III Drosha and Dicer involved in miRNA generation, RNA editing processes, and DNA mutations (single-nucleotide polymorphisms). These are described in several databases that have collected sequencing results. Two different databases have described ncRNAs that are specifically involved in CVDs, namely, CVDncR (Wu et al., 2020) and CARDIO-LNCRNAs (Jiang et al., 2019). Databases are valuable resources for studying ncRNAs (Balamurali and Stoll, 2020; Zhang et al., 2020a), especially those that describe validated interactions, such as the Encyclopedia of RNA Interactomes (Yang et al., 2011; Li et al., 2014). Indeed, identification of ncRNA interactors would enable formulation of testable hypotheses and facilitate planning of validation experiments. Notably, ncRNAs exhibit higher cell-type-specific expression patterns than mRNAs. Therefore, their identification in individual cells is crucial. This is now feasible through various approaches based on single-cell RNA sequencing (scRNA-seq) (Jovic et al., 2022). Although scRNA-seq offers significant advantages, such as the ability to capture cellular heterogeneity, it has a critical a limitation in the form of loss of histological information. Spatial transcriptomic data may address this limitation by simultaneously capturing both the transcriptomic and spatial information while preserving the information of individual cells (Williams et al., 2022). Consequently, the study of ncRNAs in CVDs would require development of specialized databases based on scRNA-seq and spatial transcriptomic data obtained from specific tissues (e.g. heart and vascular tissues). It is crucial to acknowledge that scRNA-seq has limited sensitivity and lacks the ability to reliably detect low-abundance transcripts. This inherent limitation poses a challenge to the analysis of ncRNAs exhibiting lower expression levels than miRNAs.
3 Normal autophagy
Autophagy is a highly conserved cellular process in eukaryotes that involves the sequestration of cytoplasmic components into double-membrane vesicles called autophagosomes, which are then delivered to lysosomes for degradation and recycling (Yorimitsu and Klionsky, 2005; Mizushima and Klionsky, 2007). This process plays crucial roles in cellular homeostasis, adaptation to nutrient limitations, and protein turnover (Mizushima and Klionsky, 2007). Autophagy is regulated by a complex network of proteins and can be induced by various stressors, including starvation and endoplasmic reticulum stress (Ryter et al., 2013); it can also occur through bulk degradation or selective pathways targeting specific cargoes, such as aggregated proteins or dysfunctional mitochondria (Ryter et al., 2013). The process consists of several steps, namely, initiation, sequestration, transport to lysosomes, degradation, and utilization of degraded products, each of which can potentially serve different functions (Mizushima and Klionsky, 2007). Three types of autophagy have been identified in mammals, namely, macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Tettamanti et al., 2019). Tables 1–3 collectively summarize the proteins involved in the initial stages of autophagy, membrane shaping and autophagosome formation, and fusion of autophagosomes with lysosomes, respectively. Microautophagy differs from macroautophagy because it is not based on the formation of new membranes to isolate small pieces of cytoplasm for degradation. During microautophagy, the degradable portion (cargo) is selected by invaginations or protrusions of the membranes of the endo-lysosomal compartments. The cargos are then recognized by Atg8, Nbr1, Hsc70, or other proteins. Microautophagy can also uptake cytoplasmic materials non-selectively (see Table 4 for the proteins involved). However, CMA does not use membrane structures to select the degradable material (Kaushik and Cuervo, 2018). Here, the chaperone complex recognizes KFERQ-like pentapeptides that permit translocation of unfolded proteins to the lysosomes through pores formed by the multimers of Lamp2A (Agarraberes and Dice, 2001) (see Table 5 for the proteins involved).
TABLE 1
| Macroautophagy | |||
|---|---|---|---|
| Starvation-induced initiation | |||
| Protein | Complex | Function | References |
| mTorc1 | Inhibition of Ulk1 | Fujioka et al. (2014) | |
| Ulk1 | Ulk complex | Autophagosome fusion | Fujioka et al. (2014) |
| Fip200 | Ulk1 interacting protein | Turco et al. (2019) | |
| Atg13 | Target for TOR kinase signaling | Yamamoto et al. (2016) | |
| Atg101 | Interactor of Atg13 | Fujioka et al. (2014) | |
| Atg9 | Associated with vesicles | Ren et al. (2023b) | |
| Cargo-driven assembly | |||
| In addition to proteins involved in the starvation-induced autophagy | |||
| Ndp52 | Sequestrosome | Receptor for ubiquitin-coated proteins | Vargas et al. (2019) |
| Sqstm1 | Ubiquitin binding | Turco et al. (2019) | |
| Tax1bp1 | Ubiquitin-binding adapter | Turco et al. (2021) | |
| Nbr1 | Autophagy receptor for selective autophagic degradation of peroxisomes | Turco et al. (2021) | |
| Optn | Vesicle trafficking (recruiting of Atg9 vesicles) | Yamano et al. (2020) | |
| p62 | Assembly of Ulk complex | Turco et al. (2021) | |
Autophagy initiation.
Macroautophagy induced by starvation starts with formation of the Ulk complex near the endoplasmic reticulum membrane and recruits Atg9 vesicles via interaction with the Atg13–Atg101 subcomplex. Alternatively, cargo-driven assembly commences with the intervention of adaptors such as p62, Ndp52, and Tax1bp1, which initiate assembly of the Ulk complex through interactions with FIP200; here, Atg9 vesicles are recruited by Optn.
TABLE 2
| Membrane elongation | References | ||
|---|---|---|---|
| Beclin1 | Pi3kc3 complex I | Mammalian ortholog of the yeast autophagy-related gene 6 (Atg6) | Baskaran et al. (2014) |
| Vps15 | Class 3 phosphoinositide 3-kinase (PI3K) | Baskaran et al. (2014) | |
| Vps34 | Class 3 phosphoinositide 3-kinase (PI3K) | Baskaran et al. (2014) | |
| Nrbf2 | Association with PI3K complex I (PI3KC3-C1) | Baskaran et al. (2014) | |
| Atg14 | Determines localization of the autophagy-specific PI3-kinase complex PI3KC3-C1 | Baskaran et al. (2014) | |
| Proteins involved in starvation-induced autophagy | Ulk complex | ||
| Atg5 | In combination with autophagy protein 12 (Atg12), functions as an E1-like activating enzyme in a ubiquitin-like conjugating system | Bozic et al. (2020) | |
| Atg12 | Works in combination with Atg5 | Bozic et al. (2020) | |
| Atg16L1 | Part of a large protein complex that is necessary for autophagy | Bozic et al. (2020) | |
| Wipi2 | Regulates the assembly of multiprotein complexes | Dooley et al. (2014) | |
| Ub | Ubiquitins are involved in protein labeling for degradation | Kraft et al. (2010) | |
| Atg8 | Ubiquitin protein ligase binding activity and autophagosome assembly | Bozic et al. (2020) | |
| Atg9 | Association between the endoplasmic reticulum (ER) and the cup-shaped membrane structure (known as phagophore or isolation membrane) | Autophagosome assembly | Maeda et al. (2020) |
| Wipi4 | Involved in autophagosome assembly downstream of Wipi2 | Bakula et al. (2017) | |
| Atg2 | Lipid transfer protein involved in autophagosome assembly | Osawa et al. (2019) | |
| Tmem41b | Involved in autophagosome assembly. Located in the ER and mitochondria-associated ER membranes | Ghanbarpour et al. (2021) | |
| Vmp1 | Transmembrane protein that plays a key regulatory role in autophagy | Ghanbarpour et al. (2021) | |
| Dfcp1 | Recruitment of proteins involved in membrane trafficking | Nähse et al. (2023) | |
| Membrane closure and autophagosome formation | |||
|---|---|---|---|
| Vps2 | Escrt-III | Formation of endocytic multivesicular bodies | Takahashi et al. (2018) |
| Vps20 | Formation of endocytic multivesicular bodies | Takahashi et al. (2018) | |
| Vps24 | Formation of endocytic multivesicular bodies | Takahashi et al. (2018) | |
| Snf7 | Formation of endocytic multivesicular bodies | Zhou et al. (2019) | |
| Vps60 | Formation of endocytic multivesicular bodies | Zhen et al. (2020) | |
| Did2 | Escrt-III | Formation of endocytic multivesicular bodies | Pfitzner et al. (2020) |
| Ist1 | Interacts with components of endosomal sorting complexes required for transport | Pfitzner et al. (2020) | |
| Vps4 | Associated with the endosomal compartments | Mejlvang et al. (2018) | |
Membrane shaping during autophagy.
During the membrane-elongation step, the Ulk complex recruits the class III phosphatidylinositol 3-kinase complex I (PI3KC3–C1) that produces PI(3)P and further recruits its effector proteins: Dfcp1 to omegasomes; Wipi2 and Wipi4 to phagophores. Wipi4 directs Atg2 to the phagophore membrane, which then transfers phospholipids from the ER along with Atg9, Vmp1, and Tmem41b. Wipi2 recruits the Atg12–Atg5–Atg16l1 complex to promote LC3 lipidation on the phagophore membrane. ESCRT machinery is then involved in autophagosome closing.
TABLE 3
| Lysosome fusion and formation of autolysosome | References | ||
|---|---|---|---|
| Rab7 | RAS-related GTP-binding proteins that are important regulators of vesicular transport | Wang et al. (2016) | |
| Epg5 | Involved in autophagy | Wang et al. (2016) | |
| Plekhm1 | Acts as a multivalent adapter protein to regulate Rab7-dependent fusion events | McEwan et al. (2015) | |
| Vamp7 | SNARE complex | Involved in targeting and/or fusion of transport vesicles to their target membranes | Bas et al. (2018) |
| Vamp8 | VAMP8 is a SNARE involved in autophagy through direct control of autophagosome membrane fusion with the lysosome membrane | Bas et al. (2018) | |
| Snap29 | Snap29 is a SNARE involved in autophagy | Bas et al. (2018) | |
| Stx17 | Stx17 is a SNARE involved in autophagy | Itakura et al. (2012) | |
| Stx7 | Mediates endocytic trafficking from early to late endosomes and lysosomes | Bas et al. (2018) | |
| Ykt6 | Mediates vesicle docking and fusion to a specific acceptor cellular compartment | Matsui et al. (2018) | |
| Recycling | |||
| Stx17 | Stx17 is a SNARE involved in autophagy | Itakura et al. (2012) | |
| Atg9 | Associated with vesicles | Zhou et al. (2022) | |
| Snx17 | Critical regulator of endosomal recycling | Zhou et al. (2022) | |
| Snx4 | Involved in autophagosome assembly by regulating trafficking and recycling of phospholipid scramblase ATG9A | Zhou et al. (2022) | |
| Snx5 | Involved in several stages of intracellular trafficking | Zhou et al. (2022) | |
| Kif5b | Microtubule-dependent motor required for normal distribution of mitochondria and lysosomes | Du et al. (2016) | |
| Dnm2 | Catalyzes the hydrolysis of GTP and utilizes this energy to mediate vesicle scission | Du et al. (2016) | |
| Pip5k1b | Catalyses the phosphorylation of phosphatidylinositol 4-phosphate (PtdIns(4)P/PI4P) to form phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2/PIP2), a lipid second messenger that regulates several cellular processes like signal transduction, vesicle trafficking, actin cytoskeleton dynamics, cell adhesion, and cell motility | Rong et al. (2012) | |
Formation of autolysosomes and endosomal recycling.
Subsequent to autophagosome closing, lysosomes are tethered to the autophagosomes by Plekhm1, Epg5, and Rab7, while the two SNARE complexes Stx17–Snap29–Vamp7/8 and Ykt6–Snap29–Stx7 trigger fusion. Lysosomal membrane proteins on autolysosomes are recycled via autophagic lysosome reformation, whereas autophagosomal membrane proteins are recycled via autophagosomal component recycling.
TABLE 4
| Microautophagy | References | |
|---|---|---|
| Atg30 | Key player in the selection of peroxisomes as cargo and delivery to autophagy for pexophagy | Farré et al. (2013) |
| Atg39 | Autophagy of perinuclear ER/nucleus under nitrogen deprivation | Otto and Thumm (2021) |
| Sec62 | Intervenes during recovery from ER stress | Loi et al. (2019) |
| Hsc70 | Cargo recognition | Sahu et al. (2011) |
| cGAS | Cyclic GMP-AMP synthase: cytosolic DNA sensor | Zhao et al. (2021b) |
Microautophagy.
Microautophagy is regulated by the invagination of endosomal or lysosomal membranes to incorporate cytoplasmic material. It can be recognized from Atg8, Nbr1, Ub (proteins shared with macroautophagy as noted in the table), Atg30, Atg39, Sec62, cGAS, Hsc70, and Tsg101.
TABLE 5
| Chaperone-mediated autophagy | ||
|---|---|---|
| Protein | Function | References |
| Hsc70 (also known as Hspa8) | Cytoplasmic Hsc70: recognition of KFERQ-like motif binding to lysosome-associated membrane protein (LAMP2A) Lysosomal Hsc70: substrate translocation in the lysosome |
Kaushik and Cuervo (2018) |
| Hsp40 or Dnabj1 | Co-chaperones that participate in substrate unfolding when bound to Hsc70 and membrane | Kaushik and Cuervo (2018) |
| HOP (Hsp70–Hsp90 organizing protein) | Kaushik and Cuervo (2018) | |
| Hsp90 | Bandyopadhyay et al. (2008) | |
| HIP (Hsp70-interacting protein) | Kaushik and Cuervo (2018) | |
| Cathepsins | Protein degradation within the lysosome | Kaminskyy and Zhivotovsky (2012) |
| Lamp2A | Receptor for internalization in the lysosomes of targeted proteins | Qiao et al. (2023) |
| GFAP | Filament protein involved in modulation of stability of Lamp2A | Mehrbod et al. (2019) |
| Ef1a | Partner of GFAP | Kaushik and Cuervo (2018) |
List of proteins involved in chaperone-mediated autophagy.
The process starts with recognition of the KFERQ-like motif in the proteins that are degraded by Hsc70 and co-chaperones. This complex is guided to the Lamp2A receptor. Lamp2A is formed on the surfaces of the lysosomes through GFAP. The lysosome component of Hsc70 allows the protein to be degraded to enter the lysosome where it is degraded by cathepsins.
4 Autophagy in aging and age-related CVDs
4.1 Autophagy in aging
Aging is the most important risk factor for age-related diseases, such as neurodegenerative diseases, CVDs, metabolic diseases, musculoskeletal diseases, and diseases of the immune system (Kubben and Misteli, 2017; Guo et al., 2022). Many elderly people have multiple comorbidities with advancing age (Guo et al., 2022). López-Otín et al. (2023) have suggested twelve molecular, cellular, and systemic hallmarks of aging as follows: DNA instability, telomere attrition, epigenetic alterations, loss of proteostasis, disabled macroautophagy, deregulated nutrient-sensing mechanisms, mitochondrial dysfunction, cellular senescence, stem-cell exhaustion, altered intercellular communication, chronic inflammation, and dysbiosis. These hallmarks are interdependent, meaning that the experimental accentuation or attenuation of a specific hallmark can affect the others as well (Guo et al., 2022). Impairment of autophagy was proposed for the first time as a hallmark of aging by Aman et al. (2021). There is increasing evidence that autophagy-related gene expressions and autophagic activities decrease with age in different tissues in different species. Forced genetic impairment of autophagy has been reported to accelerate the decline of cellular functions (Guo et al., 2022), where tissue-specific knockout of the autophagy related 7 (ATG7) or 5 (ATG5) gene exhibits phenotypes similar to those found in aging (Rubinsztein et al., 2011). Conversely, increase in autophagic activity has been associated with delayed aging in animal models (Rubinsztein et al., 2011; Tabibzadeh, 2023) as the restoring the expressions of autophagy genes can counteract age-related damage and decline (Yang et al., 2010; Bjedov et al., 2020; Hosoda et al., 2023; Huang et al., 2023). Unfortunately, the mechanisms by which autophagic components or processes decrease with age remain unclear (Chung and Chung, 2019), making it difficult to implement rejuvenation interventions impacting the altered mechanisms associated with autophagy.
Growing evidence supports that autophagy induced by calorie restriction (CR) has a substantial beneficial role (Chung and Chung, 2019); CR has been proven to increase both the health and lifespan in a wide range of animal models (Hwangbo et al., 2020) as well as support healthy human aging (Belsky et al., 2017). Therefore, CR is regarded as the gold standard for many aging intervention methods. Although CR has clearly diverse effects in counteracting the aging process, the exact mechanisms are still under investigation. Modulation of autophagy has a deep impact on cellular aging because it protects cellular functionality in several ways. In physiological conditions, autophagy plays crucial roles in inhibiting premature cell death by regulating the intracellular nutrient levels and metabolite availability, facilitating the turnover of cytoplasmic organelles, alleviating cellular stress, mitigating inflammation, preserving the self-renewal potential of stem cells, and maintaining the differentiation capacity and plasticity (Guo et al., 2022; Tabibzadeh, 2023) to modulate senescence (Tabibzadeh, 2023). This protection offered by autophagy is progressively lost during aging (Tabibzadeh, 2023).
4.2 Aging of the cardiovascular system
In the cardiocirculatory system, vascular aging is a complex process that causes structural and functional changes in the blood vessels and can be considered vascular remodeling. Vascular aging is characterized by increased arterial wall thickness and decreased lumen diameter, with an overall increase in the ratio of wall thickness to lumen diameter, reduced elasticity (increased stiffness), increased collagen, and reduced elastin deposition in the extracellular matrix (ECM) (Rizzoni et al., 2019; Koutouroushis and Sarkar, 2021). Four types of cells are involved in vascular remodeling: fibroblasts in the adventitial layer, vascular smooth muscle cells (VSMCs) in the median layer, endothelial cells (ECs) in the intimal layer, and macrophages in the blood stream (Qi et al., 2019). One of the main reasons for vascular remodeling is the transition of VSMCs from the contractile and low-proliferative phenotype to highly proliferative synthetic cells characterized by high production of ECM (Qi et al., 2019; Lehners et al., 2018). Aged vasculature often presents dysfunctional ECs with diminished production of nitric oxide, which is responsible for vascular dilatation, tone regulation, and inflammation inhibition. Furthermore, the senescence of ECs can result in reduced proliferation, which in turn allows VSMC migration and ECM deposition (Qi et al., 2019; Liu et al., 2024a).
The heart is also affected by structural and functional alterations with age and age-related changes, including reduced myocardial contractile capacity, wall thickening consequent to increased cardiomyocyte size, fibrosis (pathological deposition of ECM) leading to ventricular stiffness, stenosis (inflammation and calcification of the valves) resulting in narrowing and stiffness, and impaired conduction transmission with loss of the pacemaker cells (Obas and Vasan, 2018; Tracy et al., 2020; Koutouroushis and Sarkar, 2021). All these changes mediate the decline of cardiac functions and increase heart vulnerability to stress in the elderly. As a result, the risks of CVDs such as strokes, MI, atrial fibrillation, and atherosclerosis also increase (Koutouroushis and Sarkar, 2021; Chang et al., 2020; Yan et al., 2021). However, studies on cardiovascular remodeling and functioning during the process of aging (Zhang et al., 2023a; Zhao et al., 2024) are less common than studies about CVDs (Watanabe et al., 2020; Stassen et al., 2022; Elghazaly et al., 2023; Serio et al., 2023; Sheng et al., 2024; Kinoshita et al., 2024) because normal healthy aging is rarely studied as a disease-like entity (Jusic et al., 2022).
4.3 Role of autophagy in CVDs
Autophagy can both protect cardiovascular function and promote vascular remodeling. Protective autophagy often serves to maintain cardiovascular functions under physiological conditions, but excessive or dysregulated autophagy can contribute to disease development under pathological conditions (Mei et al., 2014; Sciarretta et al., 2018; Ren et al., 2023a). Therefore, modulating autophagy can be a successful strategy for counteracting age-induced vascular and cardiac remodeling to relieve CVDs. Herein, we describe recent studies (from the last 5 years) about the beneficial and detrimental roles of autophagy in the aging of the cardiovascular system and CVDs.
4.3.1 Role of autophagy in vascular tissues
Several studies support the notion that autophagy critically regulates the proliferation, migration, and matrix secretion of the VSMCs (Qi et al., 2019; Koutouroushis and Sarkar, 2021) as well as inflammation and senescence of the ECs (Koutouroushis and Sarkar, 2021; Liu et al., 2024c). Autophagy has a beneficial role of delaying senescence in the vascular ECs. Conversely, autophagy plays a detrimental role of promoting phenotype switching in VSMCs, with a few exceptions (Fang et al., 2023; Shu et al., 2024; 2025; Jiang et al., 2025). Therefore, it is important to consider the cell type before determining if autophagy is detrimental or beneficial. Many studies highlight the importance of signaling pathway modulation to activate or inhibit autophagy. For example, downmodulation of PI3K/AKT/mTOR signaling is well known to promote autophagy (this pathway is also associated with longevity, aging, and cardiovascular health) (Koutouroushis and Sarkar, 2021). Next, we present recent results on autophagy modulation through regulation of the signaling pathways. In fact, investigations on the impact of autophagy via direct modulation of the autophagy-related proteins or studies on the impacts of autophagy on other cellular processes/pathways are scarce.
Li.et al. (2021a) associated the induction of autophagy with VSMC proliferation during hypertension. The mechanoresponsive nuclear envelope proteins are already associated with vascular remodeling in response to hypertension (Qi et al., 2016). Li et al. (2021b) demonstrated that suppression of lamina A/C and emerin can induce autophagy via the mTOR pathway, which in turn promotes VSMC proliferation and vascular remodeling. More recently, Shen et al. (2024a) studied angiotensin-II (Ang-II)-induced aortic dissection, which can be prevented by S-adenosylmethionine (SAM) through inhibition of autophagy and the cellular phenotypic switching of VSMCs; here, the activation of the PI3K/AKT/mTOR signaling pathway is the proposed molecular mechanism. Moreover, changes in transcription regulation were demonstrated to be involved in the autophagic modulation of vascular cells. During the development of aortic dissection (separation of the layers of the aortic wall), CCAAT/enhancer binding protein (C/EBPα) binding to the PIK3C2A promoter (gene encoding type II PI3Ks) can activate autophagy and phenotypic switching of the VSMCs from contractile to synthetic cells (Lu et al., 2022). The role of the C/EBPα transcription factor is still under investigation in VMSCs and has been recently reported to be associated with the regulation of vascular calcification (Chen et al., 2022a). Continuing with the transcription factors involved in the modulation of autophagy, the upregulation of NK2 homeobox 3 (NKX2-3), which is involved in tissue differentiation and organ development, has been shown to enhance autophagy through the AMPK/mTOR signaling pathway as well as modulate the proliferation and migration of VMSCs along with vascular remodeling (Zheng et al., 2021b). The forkhead box protein O (FOXO) transcription factors have multiple roles in the regulation of autophagy, as noted by Cheng (2019). FOXO3a promotes VSMC phenotype switching by enhancing autophagy in Ang-II-induced aortic aneurysms (Lu et al., 2021), while the peroxisome-proliferation-activated receptor γ (PPARγ) attenuates H2O2-induced senescence in VSMCs via the mTORC2/FOXO3a/autophagy signaling pathway (Kim et al., 2023). Furthermore, ECs have been considered in the evaluations of vascular senescence. Zhang et al. (2023a) investigated the roles of CD44, a cell surface adhesion molecule involved in angiogenesis and cardiac remodeling, in the senescence of vascular ECs. During aging, the upregulation of CD44 leads to reduced levels of PIK3R4 and PIK3C3, which are key components of the PI3K complex; this reduction in the activity of the PI3K complex results in a decline in autophagy and subsequent senescence of the ECs (Zhang et al., 2023a).
Aside from the PI3K/AKT/mTOR pathway that triggers autophagy, upregulation of heat shock protein-110 (HSP110) promotes proliferation, migration, and autophagy of pulmonary artery smooth muscle cells (PASMCs) during pulmonary hypertension. Here, HSP110 regulates the YAP/TAZ-TEAD4 pathway involved in cellular proliferation and angiogenesis. The TEA domain transcription factor 4 (TEAD4) regulates HSP110 transcription by binding to the HSP110 promoter (Liu et al., 2022). In arteriosclerosis obliterans (peripheral arterial presentation of atherosclerosis), downregulation of the Grb2-associated binder 1 (GAB1) protein has been shown to significantly increase autophagy in ECs through activation of the MAPK pathways, which in turn inhibit cell proliferation and migration (Qian et al., 2020). Interestingly, Yu et al. (2023a) recently discovered a new function of myosin 1b (MYO1B) in ECs; they showed that the expression of MYO1B increases during aging and is responsible for intracellular calcium homeostasis through its interaction with leucine-rich repeat kinase 2 (LRRK2), inducing the augmentation of intracellular calcium to impair autophagy, promote the senescence of ECs, and promote vascular aging (Yu et al., 2023a).
Although rarer than the analyses of signaling pathways and transcription factors regulating autophagy, we present some studies focusing on the direct modulation of autophagy through the regulation of autophagy proteins during aging or CVD. In Ang-II induced vascular remodeling, the overexpression of transmembrane member 16a (TMEM16a) ameliorates vascular remodeling and inhibits the proliferation of VSMCs. TMEM16a is a subunit of the calcium-activated chloride channels that can inhibit autophagy via modulation of the interactions between p62, BCL2, BECLIN1, and VPS34 while decreasing VSP34 activity (Lv et al., 2020). After coronary intervention, overexpression of methyltransferase-like 3 (METTL3) can increase autophagy by promoting the expressions of ATG5 and ATG7 proteins in VSMCs and thereby inhibiting vascular remodeling (Fang et al., 2023). Additionally, various enzymes may be involved in autophagy, such as nattokinase that possesses antioxidative and anti-inflammatory effects while suppressing the inflammation of ECs by inducing autophagy through activation of the transcription factor serum response factor (SRF) and glycoprotein thrombospondin 1 (THBS1), both of which are associated with inflammation regulation (Chiu et al., 2024).
Autophagy can also impact molecular and cellular mechanisms by degrading proteins or protein complexes to affect vascular remodeling. Yu et al. (2022) studied vascular remodeling after aortic allograft and demonstrated that autophagy activation upregulates the expression of the transcription factor sex-determining region Y box (SOX9) by degrading p27; p27 is a transcriptional corepressor that blocks the expression of SOX9 in association with p130 and E2F4; in turn, SOX9 promotes VMSCs of the synthetic phenotype (Yu et al., 2022). In conclusion, most recent studies have focused on the roles of autophagy in vascular remodeling in the context of specific pathologies. Only a few studies have investigated the contributions of autophagy to vascular aging (Yu et al., 2023a; Zhang et al., 2023b), although it is widely acknowledged that advanced age is one of the primary risk factors of CVDs.
4.3.2 Role of autophagy in cardiac tissues
As in other tissues, autophagy in the heart decreases with aging. Since autophagy is required for the maintenance of cardiac structure and functions, it has been proposed that a decline in autophagy may be associated with the aging process of the heart (Obas and Vasan, 2018; Yan et al., 2021). Over the last 5 years, several studies have investigated the action mechanisms of molecules modulating CVDs (Guo et al., 2021; Aziz et al., 2022; Liao et al., 2022; Nagarajan et al., 2023; Shen et al., 2024b; Ou et al., 2024), and only a few of these works propose new pathways and mechanisms by which autophagy could be involved in cardiac aging and diseases. In the present review, we focus on these recent studies. Liu et al. (2024a) showed that decreased nuclear cardiac troponin I (cTnI) impairs autophagy during aging by downregulating the transcription factor Fos proto-oncogene, which in turn reduces ATG5 expression. Different studies have demonstrated that activation of autophagy is a prosurvival mechanism to reduce cellular stress and remove the organelles damaged during MI; therefore, enhancing autophagy is a promising mode of treatment for heart diseases (Sciarretta et al., 2018; Huang et al., 2023). For example, knockout mice for NOD-, LRR-, and pyrin-domain-containing protein 3 (NLRP3) showed inhibition of the PI3K/AKT/mTOR pathway and enhanced autophagy, resulting in reduced cardiac damage (Marín-Aguilar et al., 2020). Alternatively, the activation of glycogen synthase kinase 3 beta (GSK-3β) promoted autophagy through the phosphorylation of Unc-51 like autophagy activating kinase 1 (ULK1) to prevent cardiac aging (Chen et al., 2021a).
4.4 Roles of ncRNAs in autophagy in vascular and cardiac cancers
Vascular and cardiac tissues are very susceptible to alterations that can lead to the development of tumors, and autophagy plays a dual role in tumors. During the early phases of tumor formation, autophagy functions as a tumor suppressor to restore homeostasis and eliminate cellular aberrations. Indeed, misfolded proteins and organelles as well as reactive oxygen species (ROS) are removed through basal autophagy, thus avoiding genomic damage that could lead to carcinogenesis. Conversely, in the later phases, autophagy may either support or facilitate tumor growth by allowing the tumor cells to adapt to a stressful environment. Vascular tumors, such as hemangiomas and hemangioendotheliomas, are formed during infancy and childhood but are usually left as is until complete involution. For a comprehensive review on the ncRNAs involved in infantile hemangiomas, see Wang et al. (2024a). Although the ncRNAs directly involved in autophagy have not been identified yet, it is interesting to note that several ncRNAs involved in infantile hemangiomas are involved in apoptosis regulation (Wang et al., 2024a). This represents a response to autophagy since excessive autophagy in the context of specific diseases could result in cell death through apoptosis (Xi et al., 2022). Unlike hemangiomas and hemangioendotheliomas, angiosarcomas can arise at any age and predominantly affect elderly persons, with poor long-term prognosis (Sepulveda and Buchanan, 2014; Ota et al., 2023; Wagner et al., 2024a). The incidence of angiosarcoma remains uncertain, ranking between 0.15 and 0.33 per 100,000 person-years (Wagner et al., 2024b). The significance of miRNAs in angiosarcoma is demonstrated by the evidence that mutations in the RNA helicase/RNase III Dicer are associated with angiosarcoma pathogenesis (Loh et al., 2023). Dicer is a crucial protein involved in the production of miRNAs; although the influences of miRNAs in angiosarcomas have been discussed recently (Modarresi Chahardehi et al., 2024), there are no established associations between ncRNAs and autophagy in angiosarcomas. Notably, the contribution of autophagy to angiosarcomas remains poorly understood. Both inhibition and a high level of autophagy have the potential to prevent tumorigenesis (Suzuki et al., 2022; Yang et al., 2023). An intriguing observation in sarcomas is that their treatment, including radiotherapy, chemotherapy, immunotherapy, and targeted therapy, induces modulation of both miRNA and lncRNA expressions; these modulations are guided by tumor resistance (Chen et al., 2022b). Consequently, ncRNAs have central roles in not only treatment but also monitoring of the treatment effects. Primary heart cancer is one of the rarest neoplastic entities, with an incidence of 0.33%. The low incidence of heart cancer may be related to the mechanical forces exerted by the contraction of cardiomyocytes that could disrupt the adhesion and survival of cancer cells, including cells that may have adhered to the intramuscular endothelium (Kump, 2024). Aside from their rarity, the prognosis of cardiac tumors is usually poor with an overall survival of 12–17 months after diagnosis. The diagnosis is usually made late in such cases because the symptoms often begin after a stroke or an ischemic attack caused by detached tumor tissue or thrombus (Geronikolou et al., 2021). For example, myxoma is the most common type of cardiac cancer that could embolize and consequentially be lethal (Campisi et al., 2022). Cardiac tumors can develop in adults between the ages of 50 and 70 years, as exemplified by myxoma and malignant mesothelioma. Alternatively, they can manifest in infancy and childhood, as in the case of rhabdomyoma, rhabdomyosarcoma, and fibroma (Campisi et al., 2022). As discussed previously, during the late phases of cancer, aberrant autophagy increases intracellular stress and leads to DNA damage, which in turn could promote cancer progression. The role of autophagy in adult cardiac cancer is not clear since the related studies are scarce. It has been reported that myxoma can upregulate autophagy (Jantuan et al., 2021; Sramek et al., 2021) and that there is close interplay between autophagy and the immune system in this cancer (Sramek et al., 2021). Inhibition of autophagy in myxoma has not yet been investigated; nevertheless, autophagy is a key resistance mechanism in rhabdomyosarcoma. Indeed, autophagy inhibitors can be beneficial in combination with cancer therapy (Zarrabi et al., 2023).
Cardiac myxoma is a significant contributor to stroke in young adults, and its diagnosis poses challenges in patients presenting with stroke owing to the absence of diagnostic biomarkers. To identify the ncRNAs involved in ischemic stroke caused by myxoma, Ma et al. (2025) compared tumor tissues between patients with cardiac myxoma-related ischemic stroke (CM-IS) and patients with cardiac myxoma (CM). Furthermore, since they were interested in tumor communication, these authors evaluated miRNA, lncRNA, and mRNA in exosomes purified from the plasma samples of patients with CM-IS and CM. In the plasma samples, they identified 74 differentially expressed miRNAs, 12 lncRNAs, and 693 mRNAs, while in the tumor-derived tissue samples, they identified 61 miRNAs, 67 lncRNAs, and 433 mRNAs (Ma et al., 2025). Notably, among the upregulated miRNAs in the CM-IS-derived plasma, miR-486 and miR-96 were also upregulated in the CM-IS tissue samples, supporting the idea that CM-IS tissues are responsible for their secretion; miR-96 promotes MI-induced apoptosis by targeting antiapoptotic genes (Wang et al., 2021), while miR-486 exhibits protective roles against cardiac ischemia-reperfusion (I/R) injury and myocardial apoptosis (Bei et al., 2022). However, the results concerning the secreted miRNAs and their potential roles in regulating apoptotic processes remain inconclusive. The identified miRNAs secreted by the CM-IS tissues may be involved in both the induction and protection of apoptosis; this discrepancy may be influenced by the absence of gender differences in the study and primarily by the heterogeneity of the patient ages. Most CM-IS patients enrolled in the study were approximately 55 years of age, with the extremes being 38-year-old and 70-year-old subjects. Notably, the older patient had the smallest tumor size. Interestingly, miR-486 has been reported to modulate cardiomyocyte cell size and inflammatory responses in heart failure (Verjans et al., 2019). The pleiotropic activity of miRNA may be another factor influencing these variable results. Hence, understanding the types of cells that express the studied miRNAs could provide insights into their functions in relation to the genes expressed by the specific cell types. Indeed, different cells expressing different genes could be associated with the functions of miRNAs. Lastly, one aspect that is not considered in the above studies is the subcellular localization of miRNAs and their compartmentalisation, which could affect their availability and consequently their activities.
4.5 Non-coding RNAs in aging and age-related diseases
The Encyclopedia of DNA Elements project has unveiled that less than 3% of the human genome codes for proteins (ENCODE Project Consortium, 2012) but it is pervasively transcribed into RNA molecules known as ncRNAs (Jensen et al., 2013). Among these, miRNAs, lncRNAs, and circular RNAs (circRNAs) have been shown to be involved in the regulation of gene expression to control cellular processes (Panni et al., 2020). Herein, we review the most recent studies (from the last 5 years) that highlight the involvement of ncRNAs in the modulation of autophagy with the aim of identifying those that can serve as novel therapeutic targets for CVDs.
4.6 MicroRNAs modulating autophagy in CVDs
The miRNAs are often short ncRNAs of approximately 19–22 nucleotides length that regulate gene expressions by repressing translation or inducing mRNA degradation of the target transcripts. This regulation is typically achieved through sequence-specific binding to the 3′ untranslated region (3′UTR) of the target mRNA (Bushati and Cohen, 2007). Studies have recognized miRNAs as important regulators of aging processes (Kinser and Pincus, 2020) and pathogenesis of CVDs (Guo et al., 2022). The following studies underscore the significance of miRNAs in both direct and indirect regulation of autophagy. Notably, miRNAs exert an indirect influence on autophagy by targeting mRNAs encoding proteins involved in the signaling pathways governing autophagy (i.e. mTOR pathway).
4.6.1 MicroRNAs modulating autophagy in vascular tissues
Aberrant autophagy can promote vascular remodeling and development of CVDs. The expression of myocardin is essential for maintaining the contractile phenotype of VSMCs as it inhibits autophagy by the miR30a/BECLIN1 axis (Shi et al., 2022). Similarly, miR-130a inhibits autophagy by targeting ATG2B in the VSMCs (Zheng et al., 2021b). MiR-125b-1-3p ameliorates atherosclerosis in mice by enhancing autophagy in the VMSCs via the RRAGD/mTOR/ULK1 axis. Ras-related GTP binding D (RRAGD) is a member of the Rag GTPase family that mediates mTOR signaling in autophagy regulation (Chen et al., 2024a). Moreover, miR-145-5p promotes autophagy via the AMPK/mTOR/ULK1 pathway by targeting calcium-/calmodulin-dependent protein kinase II delta (CaMKIIδ) in atherosclerotic VSMCs; CaMKIIδ is a serine/threonine protein kinase that can activate AMPK (Zhang et al., 2022a). MiR-874-5p targets SIRT3 to induce autophagy in the PASMCs under hypoxic pulmonary hypertension; SIRT3 has recently been reported to be involved in the regulation of autophagy (Zhang et al., 2020b). However, miR-92a expression is associated with CVDs and inhibits autophagy by targeting FOXO3 in the ECs (Cao et al., 2024). Furthermore, miR-483-5p targeting TIMP metallopeptidase inhibitor 2 (TIMP2) promotes atherosclerosis development and endothelial dysfunction by inhibiting autophagy. In fact, TIMP2 downregulation is associated with progression of CVDs (Zhu et al., 2023). For comprehensive reviews on the ncRNAs involved atherosclerosis, the readers are referred to other works (Yuan et al., 2021; Singh et al., 2022). Figure 3A presents a summary of the miRNAs influencing autophagy in CVDs, while Table 6 provides a comprehensive overview of the studies reported herein, including the pathological conditions and models utilized.
FIGURE 3
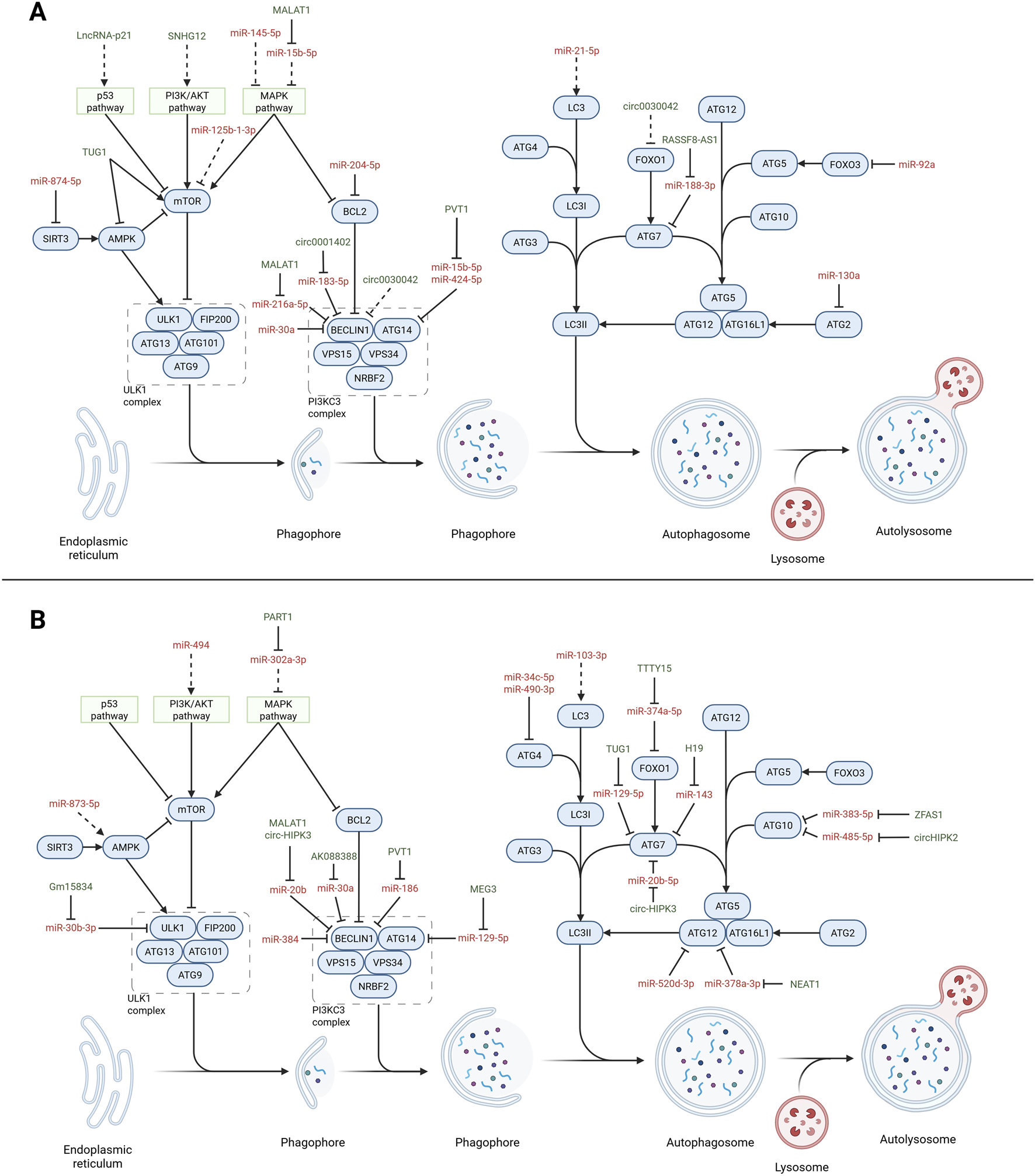
Non-coding RNAs (ncRNAs) affecting autophagy in CVDs. Overview of the autophagy pathway (blue) highlighting the interactions with microRNAs (red) and long non-coding RNAs (lncRNAs; green) in regulating the autophagy process in CVDs. The dotted lines indicate indirect interactions. (A) Non-coding RNAs involved with vascular tissues. MicroRNAs: miR-874-5p (Zhang et al., 2020b), miR-204-5p (Tian et al., 2024), miR-183-5p (Lin et al., 2024), miR-30a (Shi et al., 2022), miR-15b-5p (Zhang et al., 2023a), miR-424-5p (Zhang et al., 2023b), miR-188-3p (Song et al., 2023), miR-92a (Cao et al., 2024), miR-130a (Zheng et al., 2021a), miR-125b-1-3p (Chen et al., 2024a), miR-145-5p (Zhang et al., 2022a), miR-21-5p (Ke et al., 2022), and miR-216a-5p (Wang et al., 2019b). Long non-coding and circular RNAs: SNHG12 (Li et al., 2019b), MALAT1 (Wang et al., 2019c; Zhu et al., 2019), PVT1 (Zhang et al., 2023c), RASSF8-AS1 (Song et al., 2023), circ0001402 (Lin et al., 2024), and circ00300442 (Yu et al., 2021). (B) Non-coding RNAs involved with cardiac tissues. MicroRNAs: miR-30a (Wang et al., 2019a), miR-873-5p (Zhu et al., 2024), miR-129-5p (Mi et al., 2023), miR-34c-5p (Zhang et al., 2022b), miR-374a-5p (Chen et al., 2021c), miR-520d-3p (Wu et al., 2021c), miR-383-5p (Liu et al., 2024a), miR-485-5p (Zhou et al., 2020), miR-103-3p (Xue et al., 2023), miR-384-5p (Zhang et al., 2019), miR-490-3p (Wu et al., 2021b), miR-494 (Ning et al., 2020), miR-143 (Lv et al., 2021), miR-20b (Wang et al., 2019c; Qiu et al., 2021), miR-378a-3p (Zhao et al., 2020), miR-302a-3p (Zeng et al., 2024), and miR-186 (Ouyang et al., 2020). Long non-coding and circular RNAs: AK088388 (Wang et al., 2019a), TUG1 (You et al., 2020; Tan et al., 2023), H19 (Lv et al., 2021), MALAT1 (Wang et al., 2019c), NEAT1 (Zhao et al., 2020), PART1 (Zeng et al., 2024), MEG3 (Mi et al., 2023), TTTY15 (Chen et al., 2021c), ZFAS1 (Liu et al., 2024b), circHIPK2 (Zhou et al., 2020), and circ-HIPK3 (Qiu et al., 2021).Created with Biorender.com.
TABLE 6
| MicroRNAs | Condition/Disease | Models | Non-coding RNA roles | References |
|---|---|---|---|---|
| miR-103-3p | HF | HL-1 CMs treated with Ang-II | Promotes autophagy by targeting HLF transcription factor and FYCO1 that interact with LC3 | Xue et al. (2023) |
| miR-125b-1-3p | AS | Apoe −/− mouse and MOVAS cells (mouse VMSCs) | Promotes autophagy by targeting RRAGD, which is a mediator of the mTOR signaling pathway | Chen et al. (2024a) |
| miR-130 | AS | Human VSMCs | Inhibits autophagy by targeting ATG2B | Zheng et al. (2021b) |
| miR-145-5p | AS | Human aortic VSMCs | Promotes autophagy via the AMPK/mTOR/ULK1 pathway by targeting CaMKIIδ | Zhang et al. (2022b) |
| miR-204-5p | AS | ApoE −/− mouse, human umbilical vein ECs, and human aortic SMCs | Targets BCL2 in VSMCs | Tian et al. (2024) |
| Targets RUNX2 in ECs | ||||
| miR-21-5p | AS | Endothelial colony-forming cells | Promotes autophagy by targeting SIPA1L2 that interacts with LC3 | Ke et al. (2022) |
| miR-30a | Vascular proliferative diseases | Human aortic VSMCs | Inhibits autophagy by targeting BECLIN1 | Shi et al. (2022) |
| miR-34c-5p | Cardiac hypertrophy | Isoprenaline-treated mice and rat CMs | Inhibits autophagy by targeting ATG4B | Zhang et al. (2022a) |
| miR-384-5p | I/R-induced myocardial injury | H/R treated H9C2 CMs | Inhibits autophagy by targeting BECLIN1 | Zhang et al. (2019) |
| miR-483-5p | AS | Human umbilical vein ECs treated with ox-LDL | Inhibits autophagy by targeting TIMP2 | Zhu et al. (2023) |
| miR-490-3p | I/R-induced myocardial injury | I/R mouse model | Inhibits autophagy by targeting ATG4 | Wu et al. (2021c) |
| miR-494 | I/R-induced myocardial injury | H/R treated H9C2 CMs | Inhibits autophagy by targeting SIRT1 | Ning et al. (2020) |
| miR-520d-3p | H/R-induced myocardial injury | I/R injury rat model and H/R-treated human CMs | Inhibits autophagy by targeting ATG12 | Wu et al. (2021b) |
| miR-873-5p | Myocardial infarction | MSCs | Promotes autophagy by regulating the AMPK signaling pathway | Zhu et al. (2024) |
| miR-874-5p | Hypoxic pulmonary hypertension | Rats exposed to chronic hypoxia and PASMCs | Promotes autophagy by targeting SIRT3 | Zhang et al. (2020b) |
| miR-92a | CVDs | EA.hy926 cells (ECs) | Inhibits autophagy by targeting FOXO3 | Cao et al. (2024) |
MicroRNAs regulating autophagy in cardiovascular diseases (CVDs).
ECs: endothelial cells, CMs: cardiomyocytes, MSCs: mesenchymal stem cells, SMCs: smooth muscle cells, VSMCs: vascular smooth muscle cells, PASMCs: pulmonary artery smooth muscle cells, ox-LDL: oxidized low-density lipoprotein, Ang-II: angiotensin-II, H/R: hypoxia/reoxygenation, I/R: ischemia/reperfusion, AS: atherosclerosis, HF: heart failure.
4.6.2 MicroRNAs modulating autophagy in cardiac tissues
The roles of miRNAs in I/R-induced myocardial injuries have been widely investigated; miR-494 (Ning et al., 2020), miR-384-5p (Zhang et al., 2019), and miR-490-3p (Wu et al., 2021a) have been shown to alleviate myocardial injury via autophagy inhibition by targeting sirtuin 1 (SIRT1), BECLIN1, and autophagy related 4A cysteine peptidase (ATG4), respectively. Moreover, autophagy inhibition promotes hypertrophy via the miR-34c-5p/ATG4B axis (Zhang et al., 2022b). The overexpression of miRNA-520d-3p inhibits the expression of autophagy related 12 (ATG12) to attenuate apoptosis in myocardial cells after hypoxia/reoxygenation (H/R) (Wu et al., 2021a). In the cell model for heart failure progression, miR-103-3p has been shown to promote autophagy by targeting the hepatic leukemia factor (HLF) transcription factor as well as FYVE and coiled-coil domain autophagy adaptor 1 (FYCO1) that directly interact with LC3 to increase autophagic flux (Xue et al., 2023). Finally, mesenchymal stem cells (MSCs) may have cardioprotective properties through the promotion of angiogenesis following infarction. MiR-873-5p suppression in the MSCs enhances autophagy by regulating the AMPK signaling pathway (Zhu et al., 2024). The miRNAs regulating autophagy in CVDs are shown in Figure 3B and Table 6, including the pathological conditions and models used.
4.6.3 Circulating ncRNAs
The potential roles of circulating miRNAs in cell–tissue communication are strongly supported by their stability related to their abilities to associate with lipoproteins and proteins or to remain within vesicles that allow miRNAs to be exported or imported from the cells through mechanisms involving vesicle and protein vector trafficking (Mori et al., 2019). Extracellular vesicles secrete not only miRNAs but also mRNAs, lncRNAs, and circRNAs (Kim et al., 2017). Endothelial colony-forming cells are ECs that mediate vascular repair and secrete exosomes containing miR-21-5p in atherosclerosis; these exosomes deliver miR-21-5p to target signal induced proliferation associated 1 like 2 (SIPA1L2) that interacts with LC3 and rescues autophagy in the ECs (Ke et al., 2022). Recently, Tian et al. (2024) investigated the communication between VSMCs and ECs. During atherosclerosis, activation of autophagy in the ECs induces packing and secretion of miR-204-5p; this targets BCL2 when absorbed by the ECs and runt-related transcription factor 2 (RUNX2) when absorbed by the VSMCs to alleviate smooth muscle cell calcification (Tian et al., 2024). In fact, RUNX2 has been reported to have a detrimental role in the cardiovascular system (Chen et al., 2021b). Notably, several lncRNAs were identified in exosomes released from MSCs treated with atorvastatin (Huang et al., 2020); these findings hold significant importance because atorvastatin is commonly prescribed to prevent CVD in individuals with abnormal lipid levels. Furthermore, exosomes are crucial for mediating cardioprotective mechanisms (Wei et al., 2021). Consequently, statins not only modulate HMG-CoA reductase (HMGCR) but also influence the secretion of lncRNAs that could synergistically contribute to physiological processes. This discovery presents compelling evidence of a positive therapeutic response to a pharmacological intervention that is not specifically targeted to lncRNAs but rather modulates their activity, thereby underscoring the physiological relevance. A summary of the studies reported herein is presented in Table 6.
4.7 Long ncRNAs modulating autophagy in CVDs
The lncRNAs are a heterogeneous class of regulatory ncRNAs that are poorly conserved among species; lncRNAs are subdivided into other categories according to their genomic context or function (Alessio et al., 2020). They regulate gene expressions at all levels of genome activity (transcriptional, RNA processing, translation, and post-translation) by interacting with DNA, RNA, or proteins (Mattick et al., 2023). LncRNAs are the basis of important pathological processes in aging and age-related diseases (He et al., 2018). To date, the proposed mechanisms primarily focus on lncRNAs functioning as competitive endogenous RNAs (ceRNAs) that compete with targets for miRNA binding; ceRNAs can contact miRNAs to modulate their availability within cells and ability to bind to the mRNA targets (Giza et al., 2014). Hence, ceRNAs are also called “sponges.” LncRNAs can modulate autophagy-related protein expressions or impact the signaling pathways to regulate autophagy by sponging miRNAs. The following sections highlight the importance of ncRNAs in the modulation of cellular processes and autophagy as well as provide a guidance for studies that on the use of the same RNAs as therapeutic agents. Indeed, unlike miRNAs that have multiple targets, lncRNAs offer more specificity at the expense of less conservation of their primary structures across species.
4.7.1 Long ncRNAs modulating autophagy in vascular tissues
In the human aortic VSMC model of atherosclerosis, the lncRNA RASSF8 antisense RNA 1 (RASSF8-AS1) sponges miR-188-3p to elevate autophagy-related 7 (ATG7) expression and induce autophagy (Song et al., 2023). Similarly, the lncRNA plasmacytoma variant translocation 1 (PVT1) enhances autophagy to alleviate hypoxia-induced apoptosis of ECs by binding to miR-15b-5p and miR-424-5p, thereby avoiding the miRNAs targeting autophagy-related 14 (ATG14) (Zhang et al., 2023c). In several cases, dysfunction of ECs can be caused by inflammation; the potential of lncRNAs in modulating inflammation and subsequent autophagy has been demonstrated. LINC00346 acts as a miRNA-637 sponge to positively regulate the expression of NLRP1, a member of the NLR family (Ge et al., 2023) of proteins that are involved in the immune system to help regulate the process of inflammation. Recently, the serum level of NLRP1 was shown to be positively related to unstable angina (Zong et al., 2023). Contrarily, downregulation of the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) impacts the level of miR-19b-3p, increasing its availability and permitting the inhibition of hypoxia-inducible factor 1α (HIF-1α) (Liu et al., 2020). HIF-1α is known to be involved in apoptosis, autophagy, and inflammation; therefore, the modulation of MALAT1 may be an alternative method of reducing apoptosis, autophagy, and inflammation. Wang et al. (2019a) demonstrated that MALAT1 has a direct impact on autophagy; in the HUVEC model of atherosclerosis, this lncRNA sponges miR-216a-5p to positively regulate BECLIN1 and promote autophagy (Wang et al., 2019b). Moreover, MALAT1 contributes to atherosclerosis by inhibiting autophagy in the endothelial progenitor cells via miR-15b-5p/MAPK1. Mitogen-activated protein kinase 1 (MAPK1), whose expression is associated with atherosclerosis, can activate the mTOR signaling pathway to inhibit autophagy (Zhu et al., 2019).
The previously reported results describe the function of lncRNAs as miRNA sponges, indicating their localization within the cytoplasm of cells. However, this location is not exclusive for lncRNAs as they may also be present in the nucleus, where they interact with the chromatin and transcription machinery to regulate gene expressions. For example, the overexpression of lncRNA-p21 enhances autophagy and attenuates senescence in Ang-II-induced damage to the ECs. The lncRNA-p21 activates the SESN2/AMPK/TSC2 pathway by promoting the transcriptional activity of p53 (Li et al., 2021a); p53 is an important modulator of p21 that regulates cell cycle and senescence. Both lncRNA-p21 and the lncRNA taurine upregulated gene 1 (TUG1) are modulated in Ang-II-induced damage to the ECs; TUG1 downregulation reduces vascular damage by permitting the action of miR-9-5p, which represses the concentration of the transcript for the C-X-C motif chemokine receptor 4 (CXCR4) (Shi et al., 2024). CXCR4 is a CXC chemokine receptor involved in different signalling transductions of ECs. Moreover, TUG1 knockdown in the EC model of atherosclerosis was reported to promote autophagy via the AMPK/mTOR pathway (You et al., 2020). Comprehensive reviews on the ncRNAs involved in atherosclerosis are available elsewhere (Yuan et al., 2021; Singh et al., 2022). Figure 3A summarizes the impacts of lncRNAs on the autophagy pathway in CVDs. Tables 7, 8 report the pathological conditions and models used for the descriptions of the lncRNA functions in vascular tissues and cardiomyocytes, respectively.
TABLE 7
| Long non-coding RNAs | Condition/Disease | Models | Non-coding RNA roles | References |
|---|---|---|---|---|
| AC136007.2 | I/R injury | I/R injury rat model; oxygen-glucose deprivation and reoxygenation-treated SH-SY5Y cells | Inhibits autophagy via the PI3K/AKT/mTOR signaling pathway | Liu et al. (2021) |
| LINC00346 | AS | Human umbilical vein ECs treated with ox-LDL | Regulates inflammation by sponging miR-637 to modulate NLRP1 expression | Ge et al. (2023) |
| LncRNA-p21 | Hypertension | Human endothelial progenitor cells | Activates the SESN2/AMPK/TSC2 pathway by promoting transcriptional activity of p53 | Li et al. (2021c) |
| MALAT 1 | AS | Human umbilical vein ECs treated with ox-LDL | Promotes autophagy by sponging miR-216a-5p to induce BECLIN1 | Wang et al. (2019b) |
| MALAT1 | Hypoxia | Human umbilical vein ECs in hypoxic conditions | Regulates inflammation by sponging miR-19b-3p to modulate HIF-1α expression | Liu et al. (2020) |
| MALAT1 | AS | Endothelial progenitor cells | Inhibits autophagy by sponging miR-15b-5p to induce MAPK1 | Zhu et al. (2019) |
| RASSF8-AS1 | AS | Human aortic VSMCs | Promotes autophagy by sponging miR-188-3p to induce ATG7 | Song et al. (2023) |
| SNHG12 | I/R injury | Rat model of middle cerebral artery occlusion, I/R treated MSCs, and rat brain microvascular ECs | Inhibits autophagy via the PI3K/AKT/mTOR signaling pathway | Li et al. (2019b) |
| TUG1 | Hypertension | Ang-II-treated human umbilical vein ECs and spontaneously hypertensive rat | Sponges miR-9-5p to induce CXCR4 expression | Shi et al. (2024) |
| TUG1 | AS | Human umbilical vein ECs | Promotes autophagy via the AMPK/mTOR pathway | You et al. (2020) |
Long non-coding RNAs regulating autophagy in endothelial and vascular muscle cells.
ECs: endothelial cells, MSCs: mesenchymal stem cells, SMCs: smooth muscle cells, VSMCs: vascular smooth muscle cells, ox-LDL: oxidized low-density lipoprotein, Ang-II: angiotensin-II, I/R: ischemia/reperfusion, AS: atherosclerosis.
TABLE 8
| Long non-coding RNAs | Condition/Disease | Models | Non-coding RNA roles | References |
|---|---|---|---|---|
| AK088388 | Myocardial I/R injury | H/R treated HL-1 CMs | Promotes autophagy by sponging miR-30a to induce BECLIN1 | Wang et al. (2019a) |
| DCRF | Diabetic cardiomyopathy | Diabetic rat models and primary rat CMs | Promotes autophagy via the miR-551b-5p/PCDH17 axis | Feng et al. (2019) |
| FOXD3-AS1 | Myocardial I/R injury | Oxygen-glucose deprivation and reoxygenation-treated H9C2 CMs | Promotes autophagy by activating the NFκB/COX2/iNOS signaling pathway | Tong et al. (2019) |
| H19 | Myocardial I/R injury | I/R injury mouse model and H/R-treated HL-1 CMs | Promotes autophagy by sponging miR-143 to induce ATG7 | Lv et al. (2021) |
| LncRNA 2810403D21Rik/Mirf | Myocardial infarction | Neonatal mice CMs and myocardial infarction mouse model | Inhibits autophagy by sponging miR-26a to induce USP15 | Liang et al. (2020) |
| LncRNA Gm15834 | Myocardial hypertrophy | Ang-II treated HL-1 and AC16 CMs; transverse aortic constriction mouse model | Promotes autophagy by sponging miR-30b-3p to induce ULK1 | Song et al. (2021) |
| MALAT1 | Myocardial I/R injury | Oxygen-glucose deprivation and reoxygenation-treated H9C2 CMs | Promotes autophagy by sponging miR-20b to induce BECLIN1 | Wang et al. (2019c) |
| MEG3 | HF | Isoprenaline-treated mouse and H9C2 CMs treated with H2O2 | Promotes autophagy by sponging miR-129-5p to induce ATG14 | Mi et al. (2023) |
| NEAT1 | Myocardial I/R injury | Primary rat CMs in hypoxia condition | Promotes autophagy by sponging miR-378a-3p to induce ATG12 | Zhao et al. (2020) |
| PART1 | Myocardial I/R injury | H/R-treated AC16 CMs | Promotes autophagy by sponging miR-302a-3p to induce TFAP2C | Zeng et al. (2024) |
| PVT1 | Myocardial I/R injury | H/R-treated AC1 CMs | Promotes autophagy by sponging miR-186 to induce BECLIN1 | Ouyang et al. (2020) |
| TTTY15 | Myocardial I/R injury | I/R injury mouse model and H/R-treated H9C2 CMs | Promotes autophagy by sponging miR-374a-5p to induce FOXO1 | Chen et al. (2021c) |
| TUG1 | HF | AC16 cells CMs treated with H2O2 | Promotes autophagy by sponging miR-129-5p to induce ATG7 | Tan et al. (2023) |
| XIST | Myocardial I/R injury | I/R injury mouse model and H/R-treated H9C2 CMs | Inhibits autophagy via the miR-133a/SOCS2 axis | Li et al. (2019b) |
| ZFAS1 | Hypoxia-induced myocardial injury | H9C2 CMs in hypoxic conditions | Promotes autophagy by sponging miR-383-5p to induce ATG10 | Liu et al. (2024c) |
Long non-coding RNAs regulating autophagy in cardiomyocytes.
CMs: cardiomyocytes, H/R: hypoxia/reoxygenation, I/R: ischemia/reperfusion, HF: heart failure.
4.7.2 Long ncRNAs modulating autophagy in cardiac tissues
Previously, we discussed the involvement of lncRNAs in the modulation of autophagy in endothelial and vascular cells; now, we extend this discussion to myocardial cells. The lncRNA maternally expressed 3 (MEG3) interacts with miR-129-5p, permitting upregulation of ATG14 in H2O2-treated cardiomyocytes (Mi et al., 2023). Therefore, MEG3 downregulation increases autophagy and reduces apoptosis. It has also been demonstrated that the transcription factor ETS proto-oncogene 2 (ETS2) promotes expression of TUG1 in cardiomyocytes under oxidative stress conditions (Tan et al., 2023); in this work, the stress condition was used as the cell model for heart failure to demonstrate that TUG1 interacts with miR-129-5p to increase the expression of ATG7. Here, the authors showed that TUG1 overexpression reverses ETS2 knockdown-mediated inhibition of cardiomyocyte apoptosis and autophagy. Indeed, ETS2 is a transcription factor involved in development and apoptosis; therefore, they suggested that the modulation of ETS2 expression may be a new therapeutic approach for the treatment of heart failure. One of the common risks factors for CVDs is diabetes, and diabetic cardiomyopathy (DCM) is a major complication of this metabolic disease. The lncRNA DCM-related factor (DCRF) is associated with the development of DCM and regulates autophagy by acting as the ceRNA of miR-551b-5p; here, DCFR binds to miR-551b-5p to upregulate protocadherin-17 (PCDH17) that increases autophagy and has been reported to aggravate heart diseases (Feng et al., 2019). Song et al. (2021) described the direct impact of the lncRNA Gm15834 on autophagy; in the cardiac hypertrophic mouse model, knockdown of the lncRNA Gm15834 was found to inhibit autophagy by downregulating ULK1 through the release of miR-133a from Gm15834.
During ischemic attacks or strokes, the heart undergoes H/R stress that can induce myocardial injury; in this condition, autophagy has a detrimental role and induces the death of cardiomyocytes (Ma et al., 2015). During myocardial injury induced by hypoxia, the lncRNA ZNFX1 antisense RNA 1 (ZFAS1) promotes autophagy to positively regulate ATG10 by sponging miR-383-5p (Liu et al., 2024c); here, the authors proposed the possibility of using ZFAS1 as a therapeutic target for myocardial injury. The H/R cardiomyocyte model was also used by Ouyang et al. (2020) to investigate the role of the lncRNA PVT1 after myocardial I/R injury, where PVT1 downregulation was shown to alleviate H/R injury by inhibiting autophagy via the miR-186/BECLIN1 axis; the expression of BECLIN1 can be modulated by MALAT1/miR-20b in cardiomyocytes (Wang et al., 2019c). In addition, H19 (Lv et al., 2021) and nuclear enriched abundant transcript 1 (NEAT1) (Zhao et al., 2020) were shown to promote autophagy in I/R-induced myocardial injury models, upregulating ATG7 by targeting miR-143 and ATG12 by targeting miR-378a-3p. Conversely, the lncRNA X-inactive specific transcript (XIST) inhibits autophagy via the miR-133a/SOCS2 axis; however, the interactions between autophagy and the suppressor of cytokine signaling 2 (SOCS2) need to be elucidated (Li et al., 2019a). Liang et al. (2020) described the involvement of the lncRNA 2810403D21Rik/AK007586/Mirf (myocardial infarction regulatory factor) in macroautophagy/autophagy inhibition through modulation of miR-26a that targets the ubiquitin-specific peptidase 15 (USP15) transcript to alleviate ischemic-stress-induced cardiac injury. Therefore, the lncRNA Mirf could be a therapeutic target for counteracting the detrimental effects on autophagy after its transcriptional activation.
Similar to miRNAs, lncRNAs impact different signaling pathways to regulate autophagy in cardiac diseases. The lncRNA testis expressed transcript Y-linked 15 (TTTY15) acts as a ceRNA for miR-374a-5p to negatively regulate forkhead box O1 (FOXO1); FOXO1 is necessary for autophagy induction to alleviate myocardial injury, so the induction of TTTY15 during myocardial I/R injury is detrimental for cardiac recovery (Chen et al., 2021c). Here, another lncRNA is overexpressed in myocardial I/R injury, namely, prostate androgen regulated transcript 1 (PART1), whose function was described by Zeng et al. (2024). These authors showed that PART1 interacts with miR-302a-3p to allow upregulation of the transcription factor activating enhancer binding protein 2C (TFAP2C), which is a member of the AP2 family of transcription factors. TFAP2C regulates dual specificity phosphatase 5 (DUSP5), which is the phosphatase of ERK1/2 that suppresses I/R-stimulated autophagy and apoptosis (Zeng et al., 2024). Both studies provide evidence regarding the importance of lncRNAs for alleviating cardiomyocyte apoptosis and autophagy induced by I/R injury (Chen et al., 2021c; Zeng et al., 2024). Similarly, the lncRNA AK088388 regulates autophagy through miR-30a (Wang et al., 2019a); here, the authors showed that miR-30a expression is downregulated, while the expressions of AK088388, BECLIN1, and LC3-II are upregulated in the H/R cardiomyocytes; moreover, miR-30a targets both AK088388 and BECLIN1. The authors demonstrated that targeting AK088388 using siRNAs and upregulating miR-30a expression using miRNA mimics can enhance the viability of the H/R cardiomyocytes. These findings support the application of ncRNAs for potentially modulating cardiomyocyte viability through autophagy regulation. The lncRNA FOXD3 antisense RNA 1 (FOXD3-AS1) promotes autophagy during myocardial I/R injury (Tong et al., 2019); its upregulation enhances the expressions of LC3 II, BECLIN1, and ATG5 along with downregulation of p62 expression in H9C2 (embryonic rat heart tissue) cells. Although this finding was only based on in vitro experiments, it supports the idea that autophagy may also be modulated by downregulation of FOXD3-AS1. For a comprehensive review on the lncRNAs involved cardiac hypertrophy and heart failure, we refer readers to Mably and Wang (2024). Figure 3B summarizes the direct impacts of lncRNAs on the autophagy pathway in CVDs, while Tables 7, 8 present the pathological conditions and models employed to delineate the functions of lncRNAs in vascular cells and cardiomyocytes, respectively.
4.8 Circular RNAs modulating autophagy in vascular and cardiac tissues
CircRNAs are lncRNAs that are covalently closed to form a loop without the 5′ and 3′ polarities (Hombach and Kretz, 2016). This structure makes them more stable than linear RNAs (Chen and Lu, 2021). The functions of circRNAs include sponging for miRNAs and proteins, acting as scaffolds for RNA binding proteins, and regulating splicing and translation (Lu and Thum, 2019). This class of ncRNAs was discovered most recently, and its involvement in CVDs is a rapidly emerging and expanding research area (Altesha et al., 2019; Xie et al., 2024). For example, hsa_circ_0001402 has been found to bind miR-183-5p, which in turn promotes proliferation and migration of VSMCs by targeting FKBP prolyl isomerase like (FKBPL) while inhibiting autophagy targeting BECLIN1. Therefore, overexpression of hsa_circ_0001402 alleviates phenotypic switching in VSMCs (Lin et al., 2024) even as circ_0002331 promotes proliferation while repressing apoptosis, autophagy, and inflammation in dysfunctional ECs. Mechanistically, circ_0002331 positively regulates cyclin D2 (CCND2) mRNA stability by interacting with ELAV like RNA binding protein 1 (ELAVL1) (Chen and Yu, 2024). CCND is a protein belonging to the cyclin family that promotes cell proliferation and reduces apoptosis in HUVECs (Wu et al., 2016). Another circRNA that regulates autophagy is hsa_circ_0030042, which is downregulated in coronary heart disease; hsa_circ_0030042 obstructs eIF4A3 recruitment to BECLIN1 and FOXO1 mRNAs, thereby inhibiting abnormal autophagy in HUVECs. Moreover, upregulation of hsa_circ_0030042 in ApoE−/− mice fed a high-fat diet was found to reduce autophagy (Yu et al., 2021). In primary mouse neonatal cardiomyocytes (MNCs) treated with H2O2, circ-HIPK2 impacts autophagy by sponging miR-485-5p; this interaction blocks miRNA activity and permits upregulation of ATG10, with consequent enhancement of autophagy, suppression of apoptosis, and acceleration of MNC proliferation (Zhou et al., 2020). Another circRNA involved in autophagy regulation is circHIPK3; using cardiomyocytes, Qiu et al. (2021) demonstrated that it acts as an endogenous miR-20b-5p sponge that permits expression of ATG7 during myocardial and H/R injuries. Figure 3 summarizes the impacts of circRNAs on the autophagy pathway in CVDs. Table 9 summarizes the studies reported herein, including the pathological conditions and models used.
TABLE 9
| Circular RNAs | Condition/Disease | Models | Non-coding RNA roles | References |
|---|---|---|---|---|
| circ_0002331 | AS | Human umbilical vein ECs treated with ox-LDL | Inhibits autophagy through stabilization of CCND2 mRNA | Chen and Yu (2024) |
| circ-HIPK2 | Myocardial injury | Primary mouse neonatal CMs treated with H2O2 | Promotes autophagy by sponging miR-485-5p to induce ATG10 | Zhou et al. (2020) |
| circ-HIPK3 | Myocardial I/R injury | I/R injury mouse model and H/R-treated neonatal mouse ventricular CMs | Promotes autophagy by sponging miR-20b-5p to induce ATG7 | Qiu et al. (2021) |
| Has_circ_0030042 | AS/coronary heart disease | Human umbilical vein ECs treated with ox-LDL and high-fat-diet-fed ApoE−/− mouse | Sponges eIF4A3 and blocks its recruitment to BECLIN1 and FOXO1 mRNAs | Yu et al. (2021) |
| hsa_circ_0001402 | Neointimal hyperplasia | Neointimal hyperplasia mouse model and human aortic SMCs | Promotes autophagy by sponging miR-183-5p to induce BECLIN1 | Lin et al. (2024) |
Circular RNAs regulating autophagy in CVDs.
ECs: endothelial cells, CMs: cardiomyocytes, SMCs: smooth muscle cells, ox-LDL: oxidized low-density lipoprotein, H/R: hypoxia/reoxygenation, I/R: ischemia/reperfusion, AS: atherosclerosis.
4.9 Preclinical models demonstrate the significance of ncRNAs as drugs for treating CVDs
RNA-based therapeutics are classified into various types as small interference RNA (siRNA)-based therapeutics, miRNA-based therapeutics, antisense oligonucleotides (ASOs), RNA aptamers, ribozymes, and mRNA-based therapeutics (Chatterjee et al., 2023). Most RNA-based drugs belong to the siRNA and ASO classes (Chatterjee et al., 2023). SiRNAs are dsRNA molecules having lengths of approximately 18–25 nucleotides and structures that can be directly recognized by and loaded onto the RISC complex. Therefore, siRNAs use cellular machinery to downmodulate specific RNA molecules (Zhu et al., 2022; Chatterjee et al., 2023; Zhang and Zhang, 2023). The newest class of miRNA-based therapeutics is currently in phase II or III of clinical trials (Nappi, 2024) and includes miRNA mimics and inhibitors. MiRNAs mimics are synthetic RNAs that can mimic the upregulation of specific miRNAs to simultaneously act on different targets, similar to endogenous miRNAs. However, miRNA inhibitors (also known as anti-miRs or antagomiRs) are a class of single-stranded RNA molecules that can bind endogenous miRNAs simulating the activities of ceRNAs to prevent miRNA activity (Chatterjee et al., 2023). ASOs are short single-stranded DNA or RNA sequences with lengths of 12–24 nucleotides that are designed to target sequence-specific RNAs. ASOs can degrade a target mRNA or lncRNA through cleavage of RNA by RNase H; moreover, ASOs can block proteins that bind RNAs to inhibit mRNA translation or can enter the nucleus to modulate splicing (Zhu et al., 2022; Zhang and Zhang, 2023). ASOs also act as anti-miRs to target and inactivate mature miRNA forms (Chatterjee et al., 2023). Before developing a drug, it is imperative to comprehend its molecular activities and test its efficacy. This necessitates the use of preclinical models, which can be divided into in vitro models comprising two- or three-dimensional cell cultures and single cell assays, ex vivo models, in vivo models, and bioinformatics-based models.
Bioinformatics-based models. With the advent of artificial intelligence (AI), models based on bioinformatics appear promising but need large amounts of data obtained from real experiments for training. Therefore, these are often based on in vitro, ex vivo, and in vivo models. Bioinformatics approaches have recently been employed to simulate cardiomyocyte calcium handling, which is crucial for the development of cardiac arrhythmias (Sutanto et al., 2020). Potassium current is known to be involved in cardiac ventricular repolarisation; thus, Meier et al. (2023) developed an in silico model considering Kv 11.1 ion-channel trafficking and gating to reveal a complex temporal regulation of cardiac electrophysiology based on temperature, hypokalemia, and dofetilide. These efforts are merely a few examples as several other works published within the last 5 years have demonstrated the significance of bioinformatics approaches in investigating the effects of electromechanical coupling and pharmacological actions on human ventricular electrophysiology (Margara et al., 2021). Bioinformatics analyses have also been conducted to comprehend thrombus formation through modelling of fluid dynamics in the left atrium (Boyle et al., 2021) and to enhance valve design for optimising structural performance (Whiting et al., 2022). With regard to RNAs, bioinformatics and AI in particular could enhance our understanding of RNA–RNA or RNA–protein interactions by elucidating the molecular structures of these molecules. The 3D structure of RNA is particularly crucial for lncRNAs as it enables their specific interactions with proteins (e.g. the lncRNA growth-arrest-specific 5 regulates cell survival through distinct structural modules) (Frank et al., 2020). Similarly, the 3′-UTR shape of the coding genes permits or prevents (if the structure masks the binding site) miRNA binding (Yang et al., 2020).
In vitro models. In vitro models are cheaper, more accessible, and simpler than ex vivo and in vivo models; hence, they can also be used in high-throughput analyses. In vitro analyses show the contributions of single-cell types but do not account for the interactions between cells. To overcome this problem, cell cultures are performed with different cells but deducing the cause–effect relationships could become complicated as the outcomes could be influenced by more than one element. One solution to this problem that can also be applied to tissues is single-cell analysis based on newly developed transcriptomic techniques (Alessio et al., 2020). These techniques are widely used to understand the composition of heart tissue and to evaluate changes in cell compositions or single-cell gene expressions in response to different pathological states (Yamada and Nomura, 2020; Wang et al., 2022; Miranda et al., 2023). The development of induced pluripotent stem cells (iPSCs) has enabled studies on the various aspects of CVDs. In fact, they allow the generation of virtually all cell types harbouring pathological mutations, thereby eliminating the need for continuous tissue sampling to investigate pathological conditions. Alternatively, they may serve as the foundation for therapeutic interventions, such as myocyte substitution in MI. Notably, cellular reprogramming, pluripotency, cardiac differentiation, and maturation are regulated by ncRNAs (Hunkler et al., 2022). Therefore, a comprehensive understanding of the biological functions of ncRNAs associated with these processes can significantly enhance the quality and quantity of stem cells as well as their respective derivatives, paving the path for safer and more efficient cell therapy approaches in heart failure. In vitro models provide more realistic representations if the cells are cultured in 3D structures. Several approaches have been employed to generate 3D human cardiac constructs from iPSCs by using scaffolds composed of hydrogels (Williams et al., 2021), bioprinters comprising cell suspensions as part of their inks (Lippi et al., 2020), cells seeded onto decellularized scaffolds (Tenreiro et al., 2021), or organoid production (Zhao et al., 2021a). Thus, different cells and even iPSCs can be integrated on a chip to reduce the sample volumes required for drug tests (Skardal et al., 2020). With regard to ncRNAs, the stiffness of the 3D structure is particularly crucial in such experiments; in fact, ncRNAs play an important role in modulating the conformation of the ECM, thereby influencing the phenotypes of the interacting cells (Akbari Dilmaghnai et al., 2021). For example, fibrosis characterized by excessive collagen production and ECM accumulation is associated with heart failure, valve tinkering, and other cardiovascular conditions. Recently, the lncRNA Airn has been found to have antifibrotic activity; it prevents ubiquitination-dependent degradation of insulin-like growth factor 2 mRNA-binding protein 2 (IMP2) and protects p53 mRNA from degradation, leading to cardiac fibroblast cell-cycle arrest (Peng et al., 2022). Consequently, variations in the outcomes obtained for various experimental conditions of in vitro models may be attributed to alterations in the stiffness of the ECM. For instance, the stiffnesses of plastics used in two-dimensional cultures are significantly different from those of hydrogels suitable for three-dimensional cultures.
Ex vivo models. In contrast to in vitro models, ex vivo preclinical models, particularly those used in perfusion experiments involving the heart, are not well-suited for analyzing ncRNAs. These models are based on living tissues maintained in an artificial environment outside the body. This artificial setting can cause alterations in RNA stability, potentially resulting in degradation or structural changes in response to the altered physiological conditions. As elucidated previously, these structural changes are crucial for the activities of the lncRNAs. Otherwise, artificial conditions can induce non-canonical ncRNA expressions that can lead to misinterpretations or misguided development of therapeutic approaches associated with RNAs. However, it is indisputable that ex vivo models are particularly useful for fluid dynamic studies, analyses on contractile functions associated with ischemia and hypoxia, and studying rhythm disorders (van Doorn et al., 2024).
Animal models. Small animals (Drosophila, zebrafish, Xenopus, mice, and rats), medium-sized animals (guinea pigs, rabbits, and cats), and large animals (dogs, pigs, sheep, and non-human primates) represent one of the cornerstones of preclinical research efforts. However, animal models have specific limitations. They possess distinct genetic backgrounds compared to humans, and the induction of various pathologies often results in incomplete recapitulation of the pathological progression observed in humans. For instance, the aging process in mice is faster than in humans. Consequently, the accumulation of age-related events that transpire in humans may not be concomitant to other alterations in mice. As a general principle, larger animals have greater biological resemblance to humans, with better translational applicability at increased costs. Recently, several lncRNAs were associated with the development of CVDs using animal models. Kcnq1ot1 is a type of lncRNA whose expression is increased in the myocardium infracted zones of rats whose left anterior descending coronary arteries (LAD) were ligated to induce MI. It was demonstrated that Kcnq1ot1 inhibits the interaction of miR-466-5p with the transcription factor TEA domain family member 1 (TEAD1) to trigger cardiomyocyte injury (Liao et al., 2020). Previously, we have underscored the importance of potassium channels in ventricular repolarization. From a genomic standpoint, it is particularly interesting that an ncRNA involved in cardiomyocyte injury is synthesized from the reverse strand of the potassium voltage-gated channel subfamily Q member 1 coding gene, where mutations in this gene are associated with inherited arrhythmias. The same experimental setup was employed to demonstrate the activity of the lncRNA MIRT2 on miR-764, where it mitigates the inhibitory effect of miR-764 on 3-phosphoinositide-dependent kinase 1 (PDK1). PDK1 is situated within the mitochondrial matrix and plays a crucial role in cellular metabolism and apoptosis (De Rosa et al., 2021). LAD ligation in rats is a widely employed technique to investigate the functions of ncRNAs in MI and has also been used to demonstrate the ability of the lncRNA FGF9-associated factor (FAF) to sponge for miR-185-5p (Gu et al., 2022). P21 activated kinase 2 (PAK2) has a cardioprotective role in several CVDs (Binder et al., 2022; Xu et al., 2022); therefore, avoiding its downmodulation through sponging of miR-185-5p that can target PAK2 has beneficial effects on hypoxia/ischemia-induced pyroptosis (Gu et al., 2022).
Instead of relying solely on rats, it is feasible to replicate the previously described methodology for rats to simulate MI in mice. This approach was used to demonstrate that the lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) binds to miR-450b-5p (Yu et al., 2024). It is not surprising that NEAT1 interacts with miRNAs because it is involved in several RNA processes, such as mRNA polyadenylation, retention in the nucleus, or pri-miRNA processing (Knutsen et al., 2022). We previously discussed the role of the lncRNA 2810403D21Rik/AK007586/Mirf in regulating autophagy by modulating the activity of miR-26a. It is widely recognized that miRNAs do not have single targets. Su et al. (2020) demonstrated that miR-26a may also be involved in the modulation of cardiac apoptosis. Notably, miR-26a regulates BCL2 antagonist/killer 1 (BAK1), and its sequestration by Mirf stimulates apoptosis through the release of mitochondrial cytochrome C. The lncRNA FAF interacts with miR-185-5p in hypoxia/ischemia-induced cardiomyocytes. This interaction allows upregulation of PAK2 to protect the heart (Gu et al., 2022). Another lncRNA that sponges miRNAs and is associated with I/R myocardial injury is AK020546; it avoids the action of miR-350-3p on Erb-B2 receptor tyrosine kinase 3 that activates the RAC (Rho family)-alpha serine/threonine-protein kinase (AKT) to inhibit BCL2 associated agonist of cell death/BCL2 (Bad/Bcl2) (Zhang et al., 2021c). Therefore, activation of the lncRNA AK020546 can protect against myocardial apoptosis.
Additional intriguing evidence on the interactions between lncRNAs and miRNAs has been proposed by Niu et al. (2020). These authors demonstrated that Opa-interacting protein 5-antisense transcript 1 (OIP5-AS1) interacts with miR-29a to decrease the apoptosis induced by myocardial I/R injury (Niu et al., 2020). Bai et al. (2021) conducted research on the functions of the lncRNA MIAT in the mouse model of MI; their findings reveal that MIAT silencing induces reductions of the translocation protein (TSPO) at both the mRNA and protein levels in MI. Indeed, such silencing prevents mitochondrial permeability transition pore opening, thereby safeguarding the integrity of the mitochondrial membrane from hypoxic damage (Bai et al., 2021).
These studies emphasize the importance of ncRNAs in intracellular communications, particularly those between the nucleus and mitochondria. These organelles are involved in not only metabolic cell regulation but also the regulation of cell survival and gene expression modulation through epigenetic mechanisms. Notably, acetyl-CoA is synthesized in the mitochondria by metabolizing fatty acids and plays a pivotal role in the epigenetic modifications of histones (He et al., 2023). Metabolic alterations in mice with MI induced by LAD ligation can be modulated through another lncRNA called the cardiomyocyte pyroptosis-associated (CPAL). Instead of using a mechanism based on miRNA interaction, CPAL directly interacts with nuclear factor kappa B (NF-κB) to induce its activation through phosphorylation (Li et al., 2023). NF-κB plays important roles in CVDs, atherosclerosis, and diabetes; it is responsible for the regulation of genes involved in inflammation, immune responses, and fibrosis. Several therapeutic agents used to treat CVDs and diabetes, such as pimobendan and sodium–glucose cotransporter 2 inhibitors, inhibit NF-κB activation. Therefore, the possibility of using an siRNA to downmodulate the expression of the lncRNA CPAL (an NF-κB activator) is a viable alternative for modulating several detrimental aspects of CVDs.
Mitochondrial shapes and metabolic states are strictly related (Chemello et al., 2019), where mitochondrial shaping is regulated by fusion or fission proteins. Interestingly, dynamin-related protein 1 (DRP1) promotes mitochondrial fission (Gao et al., 2020) that can be modulated by the interactions between DRP1 and OIP5-AS1. These interactions prevent cellular apoptosis during MI (Niu et al., 2024). Several other lncRNAs have been demonstrated to interact with proteins and modulate different phenomena associated with MI. For instance, the lncRNA cardiac conduction regulatory RNA (CCRR) binds to the toll-like receptors 2 and 4 (TLR2 and TLR4) to inhibit inflammation during MI (Wang et al., 2024b). Additionally, the lncRNA small nucleolar RNA host gene 1 (SNHG1) interacts with phosphatase and tensin homolog (PTEN) to induce its degradation (Li et al., 2021b). The upregulation of SNHG1 has beneficial effects as PTEN degradation activates the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway to promote cardiomyocyte proliferation. Furthermore, the lncRNA DACH1 directly binds to protein phosphatase 1 catalytic subunit alpha (PP1A) to limit its dephosphorylation activity. This process is crucial for cardiac repair and regeneration after MI since the interaction of PP1A with lncRNA DACH1 permits phosphorylation of yes-associated protein 1 (YAP1), limiting its nuclear translocation (Cai et al., 2020). This exemplifies a sophisticated mechanism of cell-cycle regulation through modulation of the Hippo/YAP1 pathway by lncRNAs. The lncRNA cardiac fibroblast-associated transcript (Cfast) also interacts with a protein to modulate cardiac fibrosis. Here, Cfast interacts with coactosin-like 1 (COTL1) (Zhang et al., 2021a), which then interacts with TNF receptor associated protein 1 (TRAP1) to promote formation of the SMAD2/4 complex leading to fibrosis (Wang et al., 2015). Therefore, blocking this interaction through COTL1 sequestration on Cfast can abrogate fibrosis. Interestingly, TRAP1 is also involved in several other cellular activities, including modulation of cellular metabolism (Amoroso et al., 2014). Therefore, Cfast may be another example of an RNA involved in communication between the nucleus and mitochondria. The associations between lncRNAs and proteins can impact the assembly of a “transcriptosome complex” to favor or avoid gene transcription; here, the latter is exemplified by the interactions between the lncRNA scaffold attachment factor B interacting (SAIL) and SAFB (Luo et al., 2021). SAIL is downregulated in fibrotic cardiac tissues and ameliorates fibrosis when upregulated. Luo et al. (2021) demonstrated that SAIL interacts with SAFB and limits its availability to form complexes to permit transcription of profibrotic genes. The cardiac ischemia reperfusion associated Ku70 interacting lncRNA (CIRKIL) is another example that modulates the effects of myocardial I/R through interaction with a specific protein; CIRKIL is upregulated in the I/R myocardium and leads to a decrease in the nuclear translocation of Ku70 (Xiao et al., 2022), a DNA repair subunit protein that binds to DNA double-strand breaks and facilitates repair via the non-homologous end-joining repair pathway. During I/R, there is nuclear translocation of the BCL2-associated transcription factor 1 (Bclaf1) by the cardiac injury-related Bclaf1-inhibiting lncRNA (CIRBIL); once translocated into the nucleus, Bclaf1 promotes transcription of TP53 (p53) to suppress the expression of CIRBIL and activate apoptosis in the cardiomyocytes (Zhang et al., 2021b).
CIRBIL levels are reduced in I/R hearts; consequently, therapeutic approaches to reverse cardiomyocyte death based on modulation of CIRBIL expression must be based on upregulation of CIRBIL. However, this approach may pose challenges as cells require transfection with a plasmid that exceeds the dimensions of conventional siRNAs used for gene silencing. Furthermore, transcription of lncRNAs should be precisely regulated to avoid toxicity associated with excess concentrations, which can lead to binding with non-canonical proteins. In contrast to CIRBIL, cardiac ischemia reperfusion associated p53 interacting lncRNA (CIRPIL) inhibits apoptosis of cardiomyocytes exposed to anoxia. Jiang et al. (2022) demonstrated that CIRPIL operates in the direction opposite to the lncRNA CIRBIL; notably, CIRPIL sequesters p53 in the cytoplasm and prevents its translocation into the nucleus. This mechanism inhibits the activation of apoptotic genes and prevents cardiomyocyte death. P53 is also central to the function of lncRNA-6395 and binds to it to avoid p53 ubiquitination and degradation. Therefore, upregulation of lncRNA-6395 during I/R injury promotes cardiomyocyte apoptosis through stabilisation of p53 that permits its nuclear translocation (Zhan et al., 2022). The lncRNA HOX transcript antisense RNA (Hotair) is upregulated in response to I/R stimuli; Hotair exerts protective effects on oxidative stress and cardiac myocyte apoptosis by binding to the enhancer of zeste homolog 2 (EZH2) to modulate the expression of miR-451 involved in the regulation of myocyte apoptosis and oxidative stress (Meng et al., 2020). The action mechanism of Hotair is unique and seems to be a combination of two mechanisms described earlier, namely, lncRNA–protein and lncRNA–miRNA interactions. Hotair interacts with a protein, but this interaction modulates the expression of a miRNA, thereby regulating the expression of another protein. Yu et al. (2023b) showed the importance of the lncRNA myeloid RNA regulator of Bim-induced death (Morrbid) to protect the heart from acute MI; the authors showed a correlation with serpin family E member 1 (SERPINE1) without demonstrating a direct interaction with the lncRNA.
By leveraging a unique cardiac characteristic, Fu et al. (2022) discovered genes associated with cardiac regeneration and suggested the potential for activating this process, which is diminished in the adult heart. Notably, mammalian cardiomyocytes are capable of regeneration for a limited period following birth. Apical resection (AR by exposure of the left ventricular apex and surgical amputation) in 1-day-old mice was shown to trigger a regenerative response that restored the damaged heart to its normal anatomical and functional state (Porrello et al., 2011). Fu et al. (2022) leveraged AR to identify the lncRNA natriuretic peptide A antisense RNA 1 (NAPPA-AS1) that exhibited inhibitory effects on cardiomyocyte proliferation. NAPPA-AS1 expression is downregulated during cardiomyocyte regeneration following AR activation, which enables regeneration. NAPPA-AS1 interacts with the protein splicing factor SFPQ that is involved in DNA repair; consequently, the absence of this lncRNA during cardiomyocyte proliferation allows SFPQ functions, i.e. DNA damage prevention and subsequent cardiomyocyte proliferation. A unique study conducted by Aghagolzadeh et al. (2023) on mouse heart aimed to evaluate gene expressions at the single-cell level during MI; this study is notable for its innovative approach that enables the identification of cell composition changes during the development of pathologies. Furthermore, since lncRNAs exhibit more cell-type-specific expressions compared to coding RNAs, it is crucial to ascertain which cells express specific lncRNAs. Notably, Aghagolzadeh et al. (2023) demonstrated that cardiac cell identity is solely defined by the expressions of lncRNAs in single-cell experiments. Additionally, they identified a novel lncRNA named fibrogenic LOX-locus enhancer RNA (FIXER) that interacts with the protein chromobox 4 (CBX4) to guide gene expression regulators to the promoter of the RUNX family transcription factor 1 (RUNX1). This interaction induces the expression of RUNX1, which positively regulates the expressions of fibrogenic genes. From a therapeutic perspective, downregulation of FIXER has been shown to limit fibrosis and improve heart function.
In MI, the fibroblasts and cells involved in vascular system development must also be targeted for replacement in addition to cardiomyocytes. Atherosclerosis and thrombosis affect the functionality of tissues supplied by arteries and veins, respectively, and can impair vascular function. Angiogenesis is crucial for vascular development, and neovascularization is the bodily response to ischemia. Identifying lncRNAs that stimulate neovascularization holds potential for the treatment of MI. Wu et al. (2021b) addressed this challenge by comparing the cardioprotective effects of embryonic stem-cell-derived MSCs (ES-MSCs) with human bone-marrow-derived MSCs (BM-MSCs); they identified lncRNAs upregulated in ES-MSCs compared to BM-MSCs and found that the stem-cell-derived angiogenic lncRNA (SCDAL) induces the expression of growth differentiation factor 6 (GDF6), which promotes endothelial angiogenesis. Notably, the activation of GDF6 is mediated by recruitment of the SWI/SNF chromatin-remodeling protein SNF5 to the GDF6 promoter through SCDAL (Wu et al., 2021b). MSCs can be modified to inhibit the expression of the lncRNA small nucleolar host gene 12 (SNHG12), which significantly enhances their effects in activating the PI3K/AKT/mTOR signaling pathway both in vitro and in vivo (Li et al., 2019a). Therefore, modified MSCs may ameliorate I/R injuries via the PI3K/AKT/mTOR signaling pathway. Another lncRNA that modulates the AMPK/mTOR pathway during ischemic stroke is AC136007.2. Liu et al. (2021) demonstrated that intraventricular lncRNA AC136007.2 administration reduced cerebral infarction and brain edema in a rat model of ischemic stroke. Several other lncRNAs are involved in atherosclerosis, heart failure, arrhythmia, cardiomyopathies, and diabetic cardiomyopathy, as described by Xie et al. (2025).
Large animal models. Compared to rodents, pigs have many advantages as animal models given that their anatomy, physiology, metabolism, and immune system are more similar to those of humans; thus, pigs were used to demonstrate that the lncRNA rhabdomyosarcoma 2-associated transcript (RMST) acts as a competitive endogenous RNA of miR-24-3p. Ma et al. (2023) evaluated the possibility of modulating RMST to alleviate cardiac fibrosis and improve cardiac function in a porcine model of MI; they demonstrated that the downmodulation of RMST improves fibrosis and cardiac functions after MI.
4.10 Application of ncRNAs in the treatment of CVDs
Although ncRNAs present promising opportunities in medicine, there are concerns regarding potential adverse effects and undesirable outcomes caused by off-targeting. Indeed, single-stranded or double-stranded RNAs utilized for therapeutic purposes may be recognized by the immune system as viruses, potentially triggering undesirable immune responses. In the case of siRNAs, immune stimulation reflects special siRNA sequences, siRNA delivery vehicle types, and RNAi-directed RNA cleavage products. Therefore, siRNA sequence modulation can be used to modulate immune responses. It is known that avoiding immunostimulatory motifs, such as 5′-UGU-3′, 5′-UGUGU-3′ (Judge et al., 2005), and 5′-GUCCUUCAA-3′ (Hornung et al., 2005), can limit immunostimulation. Additionally, nucleotide chemical modifications, such as 2′-O-methyl purines, 2′-fluoropyrimidines, 2-thiouracil or pseudouracil, PS linkage modifications, 5-methyl-C, and N6-methyl-A pseudouridine, can prevent immune activation (Meng and Lu, 2017). RNA-based therapies still have challenges related to delivery. These are simpler in the cases of cells that can be purified from patients before being in vitro cultivated, treated, and reimplanted in patients (e.g. blood cells), but reaching specific tissues like the heart or vessels that are broadly distributed in the body is more complicated. In fact, treatment through the blood system is often affected by the filtration ability of the liver. These topics have been recently reviewed by Androsavich (2024) and Nappi (2024); therefore, in this review, we focus on RNA-based therapeutics to treat CVDs. As discussed in the previous sections, ncRNAs are involved in several processes of CVDs, and the modulation of autophagy by ncRNAs is a valid alternative to classical approaches. Current RNA-based drug candidates aim to modulate aspects associated with the development of hypertension and vascular permeability. For example, they aim to correct dyslipidemia to limit atherosclerosis (Burvill et al., 2024; National Library of Medicine, 2024; Nissen et al., 2024a, 2024b) or prevent angiotensin II synthesis to modulate hypertension (Bakris et al., 2024; National Library of Medicine, 2024). It is known that overexpression of miR-26a attenuates ischemic-stress-induced cell death by activating autophagy through targeting Usp15 (Liang et al., 2020). Consequently, upregulation of miR-26a via miRNA mimics may confer beneficial effects on ischemic stress. However, miR-26a and its corresponding miRNA mimic can be captured by lncRNAs that are upregulated in MI mice. As discussed previously, the lncRNA 2810403D21Rik/AK007586/Mirf acts as a sponge for miR-26a, thereby inhibiting its beneficial effects. From a therapeutic standpoint, the previously described interaction underscores the importance of comprehending the expressions of lncRNAs or circRNAs that can influence the activities of therapeutic miRNAs.
The translation of basic discoveries into treatments for human diseases (via the “bench-to-bedside” approach) is a lengthy and risky process. It has been estimated that discovery to approval of a new drug could take more than 13 years and could fail in 99.9% of cases. Only 0.1% of new drug candidates from preclinical studies are approved by regulatory authorities. This phenomenon is known as the valley of death (Seyhan, 2019; Everts and Drew, 2024). Currently, the only RNA-based therapy approved by the United States Food and Drug Administration (USFDA) or European Medicines Agency (EMA) for treating CVDs is Leqvio®, which is also known as inclisiran or KJX839 (Novartis Pharmaceuticals). Leqvio is indicated for lowering the concentration of low-density lipoprotein cholesterol (LDL-C) in adults affected by heterozygous familial hypercholesterolemia or clinical atherosclerotic CVD (EMA/FDA 2021). Inclisiran is a double-stranded siRNA targeting proprotein convertase subtilisin kexin type 9 (PCSK9) mRNA; PCSK9 downregulation increases LDL-C receptor recycling and expression on the hepatocyte cell surface. This increases LDL-C uptake to lower blood LDL levels (U.S. Food and Drug Administration, 2024). There are several ongoing clinical trials in phase 3 or 4 to extend the indications and usage of Leqvio. Olpasiran (Amgen), SLN360 (Silence Therapeutics), and Pelacarsen (Novartis Pharmaceuticals) are some of the new drug candidates that are now in clinical experimentation for the treatment of atherosclerosis by targeting the mRNA of apolipoprotein (a) to lower lipoprotein(a). Lowering the amount of lipoprotein(a) could reduce the risk of CVD. Furthermore, Zilebesiran (Alnylam Pharmaceuticals) and ADX-850 (ADARx pharmaceutical, Inc.) have been developed to act on the angiotensin system to treat hypertension (National Library of Medicine, 2024). Currently, there is only one RNA-based drug candidate targeting a ncRNA; it is an ASO named CDR132L that was designed to bind miR-132-3p; it was developed to treat heart failure because blocking the pathological overexpression of miR-132-3p reduces heart remodeling and improves cardiac functions in heart-failure patients (National Library of Medicine, 2024). It has been initially assessed for antimiR-132 therapeutic efficacy, pharmacokinetic efficacy, and safety in a large animal (pig) model of heart failure (Foinquinos et al., 2020). Subsequently, CDR132L has been assessed in phase 1 (Täubel et al., 2021) and then phase 2 (Bauersachs et al., 2024) trials. Table 10 presents a summary of the ongoing clinical trials aimed at developing effective treatments for CVDs.
TABLE 10
| Candidate | Sponsor | Type | Target | Condition | Phase | ID NCT | Study starting year | References |
|---|---|---|---|---|---|---|---|---|
| Olpasiran AMG 890 |
Amgen | siRNA | Apolipoprotein (a) | ASCVD | 3 | NCT05581303 | 2022 | Burvill et al. (2024) |
| CDR132L | Cardior Pharmaceuticals GmbH | ASO | miR-132-3p | Heart failure | 2 | NCT05953831 | 2024 | |
| Heart failure, myocardial infarction | 2 | NCT05350969 | 2022 | Bauersachs et al. (2024) | ||||
| Zerlasiran SLN360 | Silence Therapeutics PLC | siRNA | Apolipoprotein (a) | ASCVD | 2 | NCT05537571 | 2023 | Nissen et al. (2024a), Nissen et al. (2024b) |
| ADX-850 | ADARx Pharmaceutical, Inc. | siRNA | Hypoxanthine phosphoribosyltransferase | Hypertension | 1 | NCT06205628 | 2024 | |
| Zilebesiran ALN-AGT01 | Alnylam Pharmaceuticals | siRNA | Angiotensinogen | Hypertension | 2 | NCT04936035 | 2021 | Bakris et al. (2024) |
| 2 | NCT05103332 | 2021 | ||||||
| 2 | NCT06272487 | 2024 | ||||||
| 2 | NCT06423352 | 2024 | ||||||
| Pelacarsen TQJ230 | Novartis Pharmaceuticals | ASO | Apolipoprotein (a) | ASCVD | 3 | NCT04023552 | 2019 | Burvill et al. (2024) |
| 3 | NCT06267560 | 2024 | ||||||
| 3 | NCT05900141 | 2023 | ||||||
| Inclisiran Leqvio® KJX839 |
Novartis Pharmaceuticals | siRNA | Proprotein convertase subtilisin/kexin type 9 (PCSK9) | ASCVD | 4 | NCT06501443 | 2024 | |
| Icahn School of Medicine at Mount Sinai | ASCVD | 3 | NCT06494501 | 2024 | ||||
| Novartis Pharmaceuticals | ASCVD | 4 | NCT06431763 | 2024 | ||||
| Jose Seijas Amigo | Ischemic heart disease, acute coronary syndrome | 4 | NCT06421363 | 2024 | ||||
| Novartis Pharmaceuticals | Atherosclerosis, myocardial infarction | 4 | NCT06372925 | 2024 | ||||
| University of Louisville | Atherosclerosis, coronary artery disease | 4 | NCT06280976 | 2024 | ||||
| Novartis Pharmaceuticals | ASCVD | 3 | NCT05030428 | 2021 |
Ongoing clinical trials for CVDs.
ASCVD: Atherosclerotic cardiovascular disease. Source: https://clinicaltrials.gov/
5 Meta-analysis: identification of genes implicated in CVDs
Gene expression data from coronary plaques (GSE236610; Widlansky et al., 2023) as well as left ventricles of patients with pediatric idiopathic dilated cardiomyopathy (GSE99321; Tatman et al., 2017) and heart failure (GSE46224; Yang et al., 2014) were retrieved using the parameters described in the methods section. A total of 773 differentially expressed genes (DEGs) were found by comparing samples from stable CAD with those from acute coronary syndrome (Supplementary Table S2), 274 DEGs were obtained by comparing controls and patients with idiopathic dilated cardiomyopathy (Supplementary Table S2), and 555 DEGs were noted by comparing controls and heart failure (Supplementary Table S2). Interestingly, if we simply compare these lists of DEGs, two genes were common among all studies (Figure 4A): Ena/vasodilator-stimulated phosphoprotein-like (EVL) and GABA type A receptor-associated protein like 2 (GABARAPL2). Both genes are important for autophagy. EVL is a member of the Ena/VASP protein family that functions as a highly efficient actin elongation factor; it was demonstrated that EVL is important for autophagosome formation and trafficking and that it colocalizes with MAP1LC3/LC3 so that mammalian ATG9A forms a ring-like structure around EVL-LC3 (Li et al., 2024). GABARAPL2 is a member of the ATG8 family that is involved in autophagy and is associated with autophagosomes (Jacquet et al., 2021; Chen et al., 2024b). Considering the functions of the genes shared by at least two datasets, we showed that these genes are involved in pathways that are directly related to autophagy (Figure 4B). The genes associated with the pathways represented in Figure 4B are as follows: PEX12, UBB, UBE2H, UBE2Q2, and RNF40 for protein ubiquitination; TUBA3E, TUBA3D, KIF13A, USP11, and RGS11 for protein folding; TUBA3E, GABARAPL2, TUBA3D, HBB, and HSP90AB1 for autophagy; UBB, UBH, and UBE2Q2 for synthesis of active ubiquitin; UBB, UBH, UBE2Q2, and SKP1 for ubiquitin-mediated proteolysis; TUBA3E, TBA3D, and HSP90AB1 for HSP90 chaperone cycle. Interestingly, proteins coded from genes associated with previously cited pathways are strictly related; they essentially form two groups of proteins, namely those associated with ubiquitination and those associated with autophagy and chaperone-mediated protein folding (Figure 5).
FIGURE 4
Representation of differentially expressed genes (DEGs). (A) Venn diagram of the DEGs reported in different studies. (B) Pathway enrichment analysis of the DEGs common to at least two studies. RA, Reactome database; KEGG, Kyoto encyclopedia of genes and genomes pathway database.
FIGURE 5
Network of DEGs shared by at least two studies and related to autophagy. The DEGs shared by at least two studies code for proteins related with autophagy as well as proteins that interact with each other to form a network described by two big clusters—one associated with ubiquitination (blue nodes) and another associated with autophagy and chaperone-mediated protein folding (red and green nodes, respectively)—according to the Reactome database. The light-green edges represent interactions retrieved by text mining, violet ones represent experimentally identified interactions, and light-blue ones represent interactions sourced from curated databases.
6 Conclusions and future perspectives
The present review considers recently published works demonstrating the importance of autophagy in aging and CVDs. Cardiovascular aging contributes significantly to the pathogenesis of age-related CVDs. Oxidative stress, mitochondrial dysfunction, and inflammation are some of the hallmarks of aging, and their involvement in CVDs underscores their crucial roles in the development of these conditions. Oxidative stress is regulated by reactive oxygen species and reactive nitrogen species, which are among the primary intracellular signal transducers that sustain autophagy. Conversely, dysfunctional mitochondria are eliminated through mitophagy. Notably, as the myocardium is a highly oxidative metabolic tissue, mitochondria play central roles in maintaining optimal cardiac functions. Alterations to or impaired elimination of non-functional mitochondria may pose significant challenges to myocardial functions. Furthermore, autophagy exerts significant effects on the induction and modulation of inflammatory responses (Pang et al., 2022). Consequently, processes that are directly associated with aging and the development of CVDs are either modulated by or modulate autophagy. However, the initial processes associated with aging that initiate the development of CVDs remain poorly understood.
Autophagy modulation is regarded as a promising mechanism of programmed cell death that has the potential to prevent and treat a wide range of disorders and diseases, including CVDs. The pivotal step in developing an effective therapeutic strategy lies in comprehending the precise and accurate causes of diseases. Furthermore, it is imperative to determine whether autophagy serves as a cytoprotective mechanism or as a cytotoxic/cytostatic agent in the progression and prevention of diseases. To this end, it is crucial to recall that although numerous models have been developed to study the end stages of CVD, including human models, the normal healthy aging process is seldom investigated as a disease-like entity. Studies investigating cardiovascular remodeling and functions throughout the aging process as well as circulating biomarkers indicative of cardiovascular functions and risk of CVDs in aging individuals are limited, hindering understanding of the underlying causes of CVD initiation and development associated with aging. Currently, the most feasible approach is to identify the genes that undergo alterations during aging and assess whether these genes may also be involved in phenotype modulations in CVD models. To the best of our knowledge, there are no studies that systematically follow aging and monitor the state of the cardiovascular system in conjunction with omics analyses to evaluate the responses using a systems biology approach. Herein, we furnish several tables to summarize the principal knowledge and genes associated with autophagy while avoiding an extensive explanation of this mechanism since several reviews have already discussed this aspect. To sustain the central role of autophagy in CVDs, we performed a meta-analysis of different transcriptomic studies demonstrating that shared DEGs are involved in the regulation of autophagy. Considering the pervasive transcription of mammalian genomes and the importance of ncRNAs, this review describes these RNAs by also considering their potential utilization as therapeutic agents. The pleiotropic nature of miRNAs makes them difficult to use in therapy because it is important to avoid off targets in such cases. This aspect can be mitigated by using tissue-specific promoters to express miRNAs or peculiar delivery systems to target the desired cells. Another approach is to modulate genes that are ubiquitously expressed, such as in the case of Leqvio.
Most of the recently discussed ncRNAs are lncRNAs. They are important for cell functions because they can modulate gene expressions by modulating the actions of miRNAs or by interacting with DNA or transcription factors. Unfortunately, their poor conservation, which is contrary to that of miRNAs, limits the possibility of studies using different organisms and hence the knowledge of their functions and applications in therapy. Despite the challenges to the therapeutic applications of ncRNAs, they offer novel, valid, and accessible alternatives to fine-tuning autophagy. Several nucleic-acid-based therapies have been approved by the USFDA or EMA (Scalabrin and Cagnin, 2025) that are primarily based on ASOs or siRNAs. As in cancer, autophagy has a dual effect in the cardiovascular system. Enhanced autophagy in the heart can confer cardioprotective effects, but its excessive activation can be detrimental and lead to excessive degradation of intracellular components with subsequent cardiomyocyte death. In the future, ncRNAs that regulate autophagy can be utilized as substitutes for conventional drugs. Alternatively, by targeting specific ncRNAs, the sensitivity to routine drugs can be enhanced substantially, potentially enabling the reduction of their concentration and the consequent alleviation of side effects. RNAs have multifaceted applications beyond therapeutic purposes. Their expressions are altered prior to protein expressions, making them valuable diagnostic tools. NcRNAs can be secreted from cells to facilitate intercellular communication. This enables their identification through blood sampling, allowing not only diagnosis but also monitoring of pathology progression and assessing the efficacies of therapeutic interventions. In the context of substitution therapy, such as enzyme replacement therapy approved for Pompe disease (Colella, 2024), future knowledge on the mechanisms inducing reduced expressions of specific ncRNAs that are predisposed to or induce CVDs during aging may be used to undo the alterations and restore ncRNA expressions in the appropriate cells. RNAs have the advantage that they do not integrate into the genome to prevent potential tumor alterations and are stable for a certain period before being degraded; the development of therapeutic RNAs is a faster process than the development of therapeutic proteins, and these RNAs are not immunogenic if modified properly.
Statements
Author contributions
SS: Writing – original draft. SC: Conceptualization, Funding acquisition, Supervision, Writing – original draft, and Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by grant no. PRIN 2022NBFJNT to SC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1520850/full#supplementary-material
References
1
Agarraberes F. A. Dice J. F. (2001). A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci.114, 2491–2499. 10.1242/jcs.114.13.2491
2
Aghagolzadeh P. Plaisance I. Bernasconi R. Treibel T. A. Pulido Quetglas C. Wyss T. et al (2023). Assessment of the cardiac noncoding transcriptome by single-cell RNA sequencing identifies FIXER, a conserved profibrogenic long noncoding RNA. Circulation148, 778–797. 10.1161/CIRCULATIONAHA.122.062601
3
Akbari Dilmaghnai N. Shoorei H. Sharifi G. Mohaqiq M. Majidpoor J. Dinger M. E. et al (2021). Non-coding RNAs modulate function of extracellular matrix proteins. Biomed. and Pharmacother.136, 111240. 10.1016/j.biopha.2021.111240
4
Alessio E. Bonadio R. S. Buson L. Chemello F. Cagnin S. (2020). A single cell but many different transcripts: a journey into the world of long non-coding RNAs. Int. J. Mol. Sci.21, 302. 10.3390/ijms21010302
5
Altesha M.-A. Ni T. Khan A. Liu K. Zheng X. (2019). Circular RNA in cardiovascular disease. J. Cell Physiol.234, 5588–5600. 10.1002/jcp.27384
6
Aman Y. Schmauck-Medina T. Hansen M. Morimoto R. I. Simon A. K. Bjedov I. et al (2021). Autophagy in healthy aging and disease. Nat. Aging1, 634–650. 10.1038/s43587-021-00098-4
7
Amoroso M. R. Matassa D. S. Sisinni L. Lettini G. Landriscina M. Esposito F. (2014). TRAP1 revisited: novel localizations and functions of a “next-generation” biomarker (review). Int. J. Oncol.45, 969–977. 10.3892/ijo.2014.2530
8
Androsavich J. R. (2024). Frameworks for transformational breakthroughs in RNA-Based medicines. Nat. Rev. Drug Discov.23, 421–444. 10.1038/s41573-024-00943-2
9
Aziz S. G.-G. Pourheydar B. Chodari L. Hamidifar F. (2022). Effect of exercise and curcumin on cardiomyocyte molecular mediators associated with oxidative stress and autophagy in aged Male rats. Microvasc. Res.143, 104380. 10.1016/j.mvr.2022.104380
10
Bai X. Yang C. Jiao L. Diao H. Meng Z. Wang L. et al (2021). LncRNA MIAT impairs cardiac contractile function by acting on mitochondrial translocator protein TSPO in a mouse model of myocardial infarction. Signal Transduct. Target Ther.6, 172. 10.1038/s41392-021-00538-y
11
Bakris G. L. Saxena M. Gupta A. Chalhoub F. Lee J. Stiglitz D. et al (2024). RNA interference with zilebesiran for mild to moderate hypertension: the KARDIA-1 randomized clinical trial. JAMA331, 740–749. 10.1001/jama.2024.0728
12
Bakula D. Müller A. J. Zuleger T. Takacs Z. Franz-Wachtel M. Thost A.-K. et al (2017). WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun.8, 15637. 10.1038/ncomms15637
13
Balamurali D. Stoll M. (2020). Non-coding RNA databases in cardiovascular research. Non-Coding RNA6, 35. 10.3390/ncrna6030035
14
Bandyopadhyay U. Kaushik S. Varticovski L. Cuervo A. M. (2008). The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol.28, 5747–5763. 10.1128/MCB.02070-07
15
Barbosa M. C. Grosso R. A. Fader C. M. (2019). Hallmarks of aging: an autophagic perspective. Front. Endocrinol.9, 790. 10.3389/fendo.2018.00790
16
Bas L. Papinski D. Licheva M. Torggler R. Rohringer S. Schuschnig M. et al (2018). Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J. Cell Biol.217, 3656–3669. 10.1083/jcb.201804028
17
Baskaran S. Carlson L.-A. Stjepanovic G. Young L. N. Kim D. J. Grob P. et al (2014). Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife3, e05115. 10.7554/eLife.05115
18
Bauersachs J. Solomon S. D. Anker S. D. Antorrena-Miranda I. Batkai S. Viereck J. et al (2024). Efficacy and safety of CDR132L in patients with reduced left ventricular ejection fraction after myocardial infarction: rationale and design of the HF-REVERT trial. Eur. J. Heart Fail26, 674–682. 10.1002/ejhf.3139
19
Bei Y. Lu D. Bär C. Chatterjee S. Costa A. Riedel I. et al (2022). miR-486 attenuates cardiac ischemia/reperfusion injury and mediates the beneficial effect of exercise for myocardial protection. Mol. Ther.30, 1675–1691. 10.1016/j.ymthe.2022.01.031
20
Belsky D. W. Huffman K. M. Pieper C. F. Shalev I. Kraus W. E. (2017). Change in the rate of biological aging in response to caloric restriction: CALERIE biobank analysis. J. Gerontol. A Biol. Sci. Med. Sci.73, 4–10. 10.1093/gerona/glx096
21
Binder P. Nguyen B. Collins L. Zi M. Liu W. Christou F. et al (2022). Pak2 regulation of Nrf2 serves as a novel signaling nexus linking ER stress response and oxidative stress in the heart. Front. Cardiovasc Med.9, 851419. 10.3389/fcvm.2022.851419
22
Bjedov I. Cochemé H. M. Foley A. Wieser D. Woodling N. S. Castillo-Quan J. I. et al (2020). Fine-tuning autophagy maximises lifespan and is associated with changes in mitochondrial gene expression in drosophila. PLoS Genet.16, e1009083. 10.1371/journal.pgen.1009083
23
Boyle P. M. Del Álamo J. C. Akoum N. (2021). Fibrosis, atrial fibrillation and stroke: clinical updates and emerging mechanistic models. Heart107, 99–105. 10.1136/heartjnl-2020-317455
24
Bozic M. van den Bekerom L. Milne B. A. Goodman N. Roberston L. Prescott A. R. et al (2020). A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep.21, e48412. 10.15252/embr.201948412
25
Burvill A. Watts G. F. Norman R. Ademi Z. (2024). Early health technology assessment of gene silencing therapies for lowering lipoprotein(a) in the secondary prevention of coronary heart disease. J. Clin. Lipidol.18, e946–e956. 10.1016/j.jacl.2024.08.012
26
Bushati N. Cohen S. M. (2007). microRNA functions. Annu. Rev. Cell Dev. Biol.23, 175–205. 10.1146/annurev.cellbio.23.090506.123406
27
Cai B. Ma W. Wang X. Sukhareva N. Hua B. Zhang L. et al (2020). Targeting LncDACH1 promotes cardiac repair and regeneration after myocardium infarction. Cell Death Differ.27, 2158–2175. 10.1038/s41418-020-0492-5
28
Campisi A. Ciarrocchi A. P. Asadi N. Dell’Amore A. (2022). Primary and secondary cardiac tumors: clinical presentation, diagnosis, surgical treatment, and results. Gen. Thorac. Cardiovasc Surg.70, 107–115. 10.1007/s11748-021-01754-7
29
Cao W. Zhao B. Gui L. Sun X. Zhang Z. Huang L. (2024). The role and mechanism of action of miR-92a in endothelial cell autophagy. Mol. Med. Rep.30, 172. 10.3892/mmr.2024.13296
30
Chang K. Kang P. Liu Y. Huang K. Miao T. Sagona A. P. et al (2020). TGFB-INHB/activin signaling regulates age-dependent autophagy and cardiac health through inhibition of MTORC2. Autophagy16, 1807–1822. 10.1080/15548627.2019.1704117
31
Chatterjee S. Bhattacharya M. Lee S.-S. Chakraborty C. (2023). An insight of different classes of RNA-Based therapeutic, nanodelivery and clinical status: current landscape. Curr. Res. Biotechnol.6, 100150. 10.1016/j.crbiot.2023.100150
32
Chemello F. Grespi F. Zulian A. Cancellara P. Hebert-Chatelain E. Martini P. et al (2019). Transcriptomic analysis of single isolated myofibers identifies miR-27a-3p and miR-142-3p as regulators of metabolism in skeletal muscle. Cell Rep.26, 3784–3797. 10.1016/j.celrep.2019.02.105
33
Chen X. Lu Y. (2021). Circular RNA: biosynthesis in vitro. Front. Bioeng. Biotechnol.9, 787881. 10.3389/fbioe.2021.787881
34
Chen F. Yu X. (2024). Circ_0002331 interacts with ELAVL1 to improve ox-LDL-Induced vascular endothelial cell dysfunction via regulating CCND2 mRNA stability. Cardiovasc Toxicol.24, 625–636. 10.1007/s12012-024-09865-2
35
Chen Y. Maejima Y. Shirakabe A. Yamamoto T. Ikeda Y. Sadoshima J. et al (2021a). Ser9 phosphorylation of GSK-3β promotes aging in the heart through suppression of autophagy. J. Cardiovasc Aging1, 9. 10.20517/jca.2021.13
36
Chen Y. Zhao X. Wu H. (2021b). Transcriptional programming in arteriosclerotic disease: a multifaceted function of the Runx2 (Runt-Related transcription factor 2). Arterioscler. Thromb. Vasc. Biol.41, 20–34. 10.1161/ATVBAHA.120.313791
37
Chen Y.-Q. Yang X. Xu W. Yan Y. Chen X.-M. Huang Z.-Q. (2021c). Knockdown of lncRNA TTTY15 alleviates myocardial ischemia-reperfusion injury through the miR-374a-5p/FOXO1 axis. IUBMB Life73, 273–285. 10.1002/iub.2428
38
Chen H.-H. Zhang T.-N. Zhang F.-Y. Zhang T. (2022a). Non-coding RNAs in drug and radiation resistance of bone and soft-tissue sarcoma: a systematic review. eLife11, e79655. 10.7554/eLife.79655
39
Chen P. Hong W. Chen Z. Gordillo-Martinez F. Wang S. Fan H. et al (2022b). CCAAT/Enhancer-Binding protein alpha is a novel regulator of vascular smooth muscle cell osteochondrogenic transition and vascular calcification. Front. Physiol.13, 755371. 10.3389/fphys.2022.755371
40
Chen J. Zhao H. Liu M. Chen L. (2024a). A new perspective on the autophagic and non-autophagic functions of the GABARAP protein family: a potential therapeutic target for human diseases. Mol. Cell Biochem.479, 1415–1441. 10.1007/s11010-023-04800-5
41
Chen X. Cao Y. Guo Y. Liu J. Ye X. Li H. et al (2024b). microRNA-125b-1-3p mediates autophagy via the RRAGD/mTOR/ULK1 signaling pathway and mitigates atherosclerosis progression. Cell Signal118, 111136. 10.1016/j.cellsig.2024.111136
42
Cheng X. Yan J. Liu Y. Wang J. Taubert S. (2021). eVITTA: a web-based visualization and inference toolbox for transcriptome analysis. Nucleic Acids Res.49, W207–W215. 10.1093/nar/gkab366
43
Cheng Z. (2019). The FoxO-Autophagy axis in health and disease. Trends Endocrinol. Metab.30, 658–671. 10.1016/j.tem.2019.07.009
44
Chiu H.-W. Chou C.-L. Lee K.-T. Shih C.-C. Huang T.-H. Sung L.-C. (2024). Nattokinase attenuates endothelial inflammation through the activation of SRF and THBS1. Int. J. Biol. Macromol.268, 131779. 10.1016/j.ijbiomac.2024.131779
45
Chung K. W. Chung H. Y. (2019). The effects of calorie restriction on autophagy: role on aging intervention. Nutrients11, 2923. 10.3390/nu11122923
46
Colella P. (2024). Advances in pompe disease treatment: from enzyme replacement to gene therapy. Mol. Diagn Ther.28, 703–719. 10.1007/s40291-024-00733-x
47
De Rosa V. Iommelli F. Terlizzi C. Leggiero E. Camerlingo R. Altobelli G. G. et al (2021). Non-canonical role of PDK1 as a negative regulator of apoptosis through macromolecular complexes assembly at the ER–Mitochondria interface in oncogene-driven NSCLC. Cancers13, 4133. 10.3390/cancers13164133
48
Dhahbi J. M. (2016). “Small noncoding RNAs in senescence and aging,” in Cellular ageing and replicative senescence. Editors RattanS. I. S.HayflickL. (Cham: Springer International Publishing), 287–312. 10.1007/978-3-319-26239-0_15
49
Domont F. Cacoub P. (2016). Chronic hepatitis C virus infection, a new cardiovascular risk factor?Liver Int.36, 621–627. 10.1111/liv.13064
50
Dooley H. C. Razi M. Polson H. E. J. Girardin S. E. Wilson M. I. Tooze S. A. (2014). WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell55, 238–252. 10.1016/j.molcel.2014.05.021
51
Du W. Su Q. P. Chen Y. Zhu Y. Jiang D. Rong Y. et al (2016). Kinesin 1 drives autolysosome tubulation. Dev. Cell37, 326–336. 10.1016/j.devcel.2016.04.014
52
Elghazaly H. McCracken C. Szabo L. Malcolmson J. Manisty C. H. Davies A. H. et al (2023). Characterizing the hypertensive cardiovascular phenotype in the UK biobank. Eur. Heart J. Cardiovasc Imaging24, 1352–1360. 10.1093/ehjci/jead123
53
Emig D. Salomonis N. Baumbach J. Lengauer T. Conklin B. R. Albrecht M. (2010). AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res.38, W755–W762. 10.1093/nar/gkq405
54
ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74. 10.1038/nature11247
55
Eurostat (2024). Cardiovascular diseases statistics. Available online at: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Cardiovascular_diseases_statistics (Accessed October 26, 2024).
56
Everts M. Drew M. (2024). Successfully navigating The Valley of death: the importance of accelerators to support academic drug discovery and development. Expert Opin. Drug Discov.19, 253–258. 10.1080/17460441.2023.2284824
57
Fang Z.-M. Zhang S.-M. Luo H. Jiang D.-S. Huo B. Zhong X. et al (2023). Methyltransferase-like 3 suppresses phenotypic switching of vascular smooth muscle cells by activating autophagosome formation. Cell Prolif.56, e13386. 10.1111/cpr.13386
58
Farré J.-C. Burkenroad A. Burnett S. F. Subramani S. (2013). Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep.14, 441–449. 10.1038/embor.2013.40
59
Feng Y. Xu W. Zhang W. Wang W. Liu T. Zhou X. (2019). LncRNA DCRF regulates cardiomyocyte autophagy by targeting miR-551b-5p in diabetic cardiomyopathy. Theranostics9, 4558–4566. 10.7150/thno.31052
60
Foinquinos A. Batkai S. Genschel C. Viereck J. Rump S. Gyöngyösi M. et al (2020). Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat. Commun.11, 633. 10.1038/s41467-020-14349-2
61
Frank F. Kavousi N. Bountali A. Dammer E. B. Mourtada-Maarabouni M. Ortlund E. A. (2020). The lncRNA growth arrest specific 5 regulates cell survival via distinct structural modules with independent functions. Cell Rep.32, 107933. 10.1016/j.celrep.2020.107933
62
Fu W. Ren H. Shou J. Liao Q. Li L. Shi Y. et al (2022). Loss of NPPA-AS1 promotes heart regeneration by stabilizing SFPQ-NONO heteromer-induced DNA repair. Basic Res. Cardiol.117, 10. 10.1007/s00395-022-00921-y
63
Fujioka Y. Suzuki S. W. Yamamoto H. Kondo-Kakuta C. Kimura Y. Hirano H. et al (2014). Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol.21, 513–521. 10.1038/nsmb.2822
64
Gao F. Reynolds M. B. Passalacqua K. D. Sexton J. Z. Abuaita B. H. O’Riordan M. X. D. (2020). The mitochondrial fission regulator DRP1 controls post-transcriptional regulation of TNF-α. Front. Cell Infect. Microbiol.10, 593805. 10.3389/fcimb.2020.593805
65
Ge J.-Y. Yan X.-J. Yang J. Jin H. Sun Z.-K. Guo J.-L. et al (2023). LINC00346 regulates NLRP1-mediated pyroptosis and autophagy via binding to microRNA-637 in vascular endothelium injury. Cell Signal109, 110740. 10.1016/j.cellsig.2023.110740
66
Geronikolou S. A. Pavlopoulou A. Chrousos G. P. Cokkinos D. V. (2021). Interactions networks for primary heart sarcomas. Cancers (Basel)13, 3882. 10.3390/cancers13153882
67
Ghanbarpour A. Valverde D. P. Melia T. J. Reinisch K. M. (2021). A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. U. S. A.118, e2101562118. 10.1073/pnas.2101562118
68
Giza D. E. Vasilescu C. Calin G. A. (2014). MicroRNAs and ceRNAs: therapeutic implications of RNA networks. Expert Opin. Biol. Ther.14, 1285–1293. 10.1517/14712598.2014.920812
69
Gu J. Shi J.-Z. Wang Y.-X. Liu L. Wang S.-B. Sun J.-T. et al (2022). LncRNA FAF attenuates hypoxia/ischaemia-induced pyroptosis via the miR-185-5p/PAK2 axis in cardiomyocytes. J. Cell Mol. Med.26, 2895–2907. 10.1111/jcmm.17304
70
Guo J. Chen W. Bao B. Zhang D. Pan J. Zhang M. (2021). Protective effect of berberine against LPS-Induced endothelial cell injury via the JNK signaling pathway and autophagic mechanisms. Bioengineered12, 1324–1337. 10.1080/21655979.2021.1915671
71
Guo J. Huang X. Dou L. Yan M. Shen T. Tang W. et al (2022). Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal Transduct. Target Ther.7, 391. 10.1038/s41392-022-01251-0
72
He J. Tu C. Liu Y. (2018). Role of lncRNAs in aging and age-related diseases. Aging Med. Milt.1, 158–175. 10.1002/agm2.12030
73
He W. Li Q. Li X. (2023). Acetyl-CoA regulates lipid metabolism and histone acetylation modification in cancer. Biochimica Biophysica Acta (BBA) - Rev. Cancer1878, 188837. 10.1016/j.bbcan.2022.188837
74
Hombach S. Kretz M. (2016). Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol.937, 3–17. 10.1007/978-3-319-42059-2_1
75
Hornung V. Guenthner-Biller M. Bourquin C. Ablasser A. Schlee M. Uematsu S. et al (2005). Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med.11, 263–270. 10.1038/nm1191
76
Hosoda R. Nakashima R. Yano M. Iwahara N. Asakura S. Nojima I. et al (2023). Resveratrol, a SIRT1 activator, attenuates aging-associated alterations in skeletal muscle and heart in mice. J. Pharmacol. Sci.152, 112–122. 10.1016/j.jphs.2023.04.001
77
Huang P. Wang L. Li Q. Tian X. Xu J. Xu J. et al (2020). Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc Res.116, 353–367. 10.1093/cvr/cvz139
78
Huang H. Wang T. Wang L. Huang Y. Li W. Wang J. et al (2023). Saponins of Panax japonicus ameliorates cardiac aging phenotype in aging rats by enhancing basal autophagy through AMPK/mTOR/ULK1 pathway. Exp. Gerontol.182, 112305. 10.1016/j.exger.2023.112305
79
Hunkler H. J. Groß S. Thum T. Bär C. (2022). Non-coding RNAs: key regulators of reprogramming, pluripotency, and cardiac cell specification with therapeutic perspective for heart regeneration. Cardiovasc. Res.118, 3071–3084. 10.1093/cvr/cvab335
80
Hwangbo D.-S. Lee H.-Y. Abozaid L. S. Min K.-J. (2020). Mechanisms of lifespan regulation by calorie restriction and intermittent fasting in model organisms. Nutrients12, 1194. 10.3390/nu12041194
81
Itakura E. Kishi-Itakura C. Mizushima N. (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell151, 1256–1269. 10.1016/j.cell.2012.11.001
82
Jacquet M. Guittaut M. Fraichard A. Despouy G. (2021). The functions of Atg8-family proteins in autophagy and cancer: linked or unrelated?Autophagy17, 599–611. 10.1080/15548627.2020.1749367
83
Jantuan E. Chiu B. Chiu B. Shen F. Oudit G. Y. Sergi C. (2021). The tumor microenvironment May trigger lymphoproliferation in cardiac myxoma. Transl. Oncol.14, 100911. 10.1016/j.tranon.2020.100911
84
Jensen T. H. Jacquier A. Libri D. (2013). Dealing with pervasive transcription. Mol. Cell52, 473–484. 10.1016/j.molcel.2013.10.032
85
Jiang X. Zhang M. (2024). The roles of long noncoding RNA NEAT1 in cardiovascular diseases. Hypertens. Res.47, 735–746. 10.1038/s41440-023-01551-0
86
Jiang C. Ding N. Li J. Jin X. Li L. Pan T. et al (2019). Landscape of the long non-coding RNA transcriptome in human heart. Brief. Bioinform20, 1812–1825. 10.1093/bib/bby052
87
Jiang Y. Yang Y. Zhang Y. Yang J. Zhang M.-M. Li S. et al (2022). Cytoplasmic sequestration of p53 by lncRNA-CIRPILalleviates myocardial ischemia/reperfusion injury. Commun. Biol.5, 716. 10.1038/s42003-022-03651-y
88
Jiang B. Zhou X. Yang T. Wang L. Feng L. Wang Z. et al (2023). The role of autophagy in cardiovascular disease: cross-Interference of signaling pathways and underlying therapeutic targets. Front. Cardiovasc. Med.10, 1088575. 10.3389/fcvm.2023.1088575
89
Jiang Y. Wei Z.-Y. Song Z.-F. Yu M. Huang J. Qian H.-Y. (2025). Platelet membrane-modified exosomes targeting plaques to activate autophagy in vascular smooth muscle cells for atherosclerotic therapy. Drug Deliv. Transl. Res. 10.1007/s13346-025-01792-1
90
Jovic D. Liang X. Zeng H. Lin L. Xu F. Luo Y. (2022). Single-cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med.12, e694. 10.1002/ctm2.694
91
Judge A. D. Sood V. Shaw J. R. Fang D. McClintock K. MacLachlan I. (2005). Sequence-dependent stimulation of the Mammalian innate immune response by synthetic siRNA. Nat. Biotechnol.23, 457–462. 10.1038/nbt1081
92
Jusic A. Thomas P. B. Wettinger S. B. Dogan S. Farrugia R. Gaetano C. et al (2022). Noncoding RNAs in age-related cardiovascular diseases. Ageing Res. Rev.77, 101610. 10.1016/j.arr.2022.101610
93
Kaminskyy V. Zhivotovsky B. (2012). Proteases in autophagy. Biochim. Biophys. Acta1824, 44–50. 10.1016/j.bbapap.2011.05.013
94
Kaushik S. Cuervo A. M. (2018). The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol.19, 365–381. 10.1038/s41580-018-0001-6
95
Ke X. Liao Z. Luo X. Chen J.-Q. Deng M. Huang Y. et al (2022). Endothelial colony-forming cell-derived exosomal miR-21-5p regulates autophagic flux to promote vascular endothelial repair by inhibiting SIPL1A2 in atherosclerosis. Cell Commun. Signal20, 30. 10.1186/s12964-022-00828-0
96
Kim K. M. Abdelmohsen K. Mustapic M. Kapogiannis D. Gorospe M. (2017). RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA8. 10.1002/wrna.1413
97
Kim S. G. Sung J. Y. Kang Y. J. Choi H. C. (2023). PPARγ activation by fisetin mitigates vascular smooth muscle cell senescence via the mTORC2-FoxO3a-autophagy signaling pathway. Biochem. Pharmacol.218, 115892. 10.1016/j.bcp.2023.115892
98
Kinoshita D. Suzuki K. Yuki H. Niida T. Fujimoto D. Minami Y. et al (2024). Coronary plaque phenotype associated with positive remodeling. J. Cardiovasc Comput. Tomogr.18, 401–407. 10.1016/j.jcct.2024.04.009
99
Kinser H. E. Pincus Z. (2020). MicroRNAs as modulators of longevity and the aging process. Hum. Genet.139, 291–308. 10.1007/s00439-019-02046-0
100
Knutsen E. Harris A. L. Perander M. (2022). Expression and functions of long non-coding RNA NEAT1 and isoforms in breast cancer. Br. J. Cancer126, 551–561. 10.1038/s41416-021-01588-3
101
Kohlmaier A. Holdt L. M. Teupser D. (2023). Long noncoding RNAs in cardiovascular disease. Curr. Opin. Cardiol.38, 179–192. 10.1097/HCO.0000000000001041
102
Koletsi D. Iliadi A. Tzanetakis G. N. Vavuranakis M. Eliades T. (2021). Cardiovascular disease and chronic endodontic infection. Is there an association? A systematic review and meta-analysis. Int. J. Environ. Res. Public Health18, 9111. 10.3390/ijerph18179111
103
Koutouroushis C. Sarkar O. (2021). Role of autophagy in cardiovascular disease and aging. Cureus13, e20042. 10.7759/cureus.20042
104
Kraft C. Peter M. Hofmann K. (2010). Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol.12, 836–841. 10.1038/ncb0910-836
105
Kubben N. Misteli T. (2017). Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol.18, 595–609. 10.1038/nrm.2017.68
106
Kump D. S. (2024). Mechanisms underlying the rarity of skeletal muscle cancers. Int. J. Mol. Sci.25, 6480. 10.3390/ijms25126480
107
Lehners M. Dobrowinski H. Feil S. Feil R. (2018). cGMP signaling and vascular smooth muscle cell plasticity. J. Cardiovasc Dev. Dis.5, 20. 10.3390/jcdd5020020
108
Li J.-H. Liu S. Zhou H. Qu L.-H. Yang J.-H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res.42, D92–D97. 10.1093/nar/gkt1248
109
Li Y. Guo S. Liu W. Jin T. Li X. He X. et al (2019a). Silencing of SNHG12 enhanced the effectiveness of MSCs in alleviating ischemia/reperfusion injuries via the PI3K/AKT/mTOR signaling pathway. Front. Neurosci.13, 645. 10.3389/fnins.2019.00645
110
Li Z. Zhang Y. Ding N. Zhao Y. Ye Z. Shen L. et al (2019b). Inhibition of lncRNA XIST improves myocardial I/R injury by targeting miR-133a through inhibition of autophagy and regulation of SOCS2. Mol. Ther. Nucleic Acids18, 764–773. 10.1016/j.omtn.2019.10.004
111
Li C. Lin L. Zhang L. Xu R. Chen X. Ji J. et al (2021a). Long noncoding RNA p21 enhances autophagy to alleviate endothelial progenitor cells damage and promote endothelial repair in hypertension through SESN2/AMPK/TSC2 pathway. Pharmacol. Res.173, 105920. 10.1016/j.phrs.2021.105920
112
Li H.-P. Liu J.-T. Chen Y.-X. Wang W.-B. Han Y. Yao Q.-P. et al (2021b). Suppressed nuclear envelope proteins activate autophagy of vascular smooth muscle cells during cyclic stretch application. Biochim. Biophys. Acta Mol. Cell Res.1868, 118855. 10.1016/j.bbamcr.2020.118855
113
Li M. Zheng H. Han Y. Chen Y. Li B. Chen G. et al (2021c). LncRNA Snhg1-driven self-reinforcing regulatory network promoted cardiac regeneration and repair after myocardial infarction. Theranostics11, 9397–9414. 10.7150/thno.57037
114
Li J. Xue H. Xu N. Gong L. Li M. Li S. et al (2023). CPAL, as a new mediator of cardiomyocyte metabolic alterations and pyroptosis, regulates myocardial infarction injury in mice. Engineering20, 49–62. 10.1016/j.eng.2022.08.012
115
Li Y. Zhang Y. Wang M. Su J. Dong X. Yang Y. et al (2024). The Mammalian actin elongation factor ENAH/MENA contributes to autophagosome formation via its actin regulatory function. Autophagy20, 1798–1814. 10.1080/15548627.2024.2347105
116
Liang H. Su X. Wu Q. Shan H. Lv L. Yu T. et al (2020). LncRNA 2810403D21Rik/Mirf promotes ischemic myocardial injury by regulating autophagy through targeting Mir26a. Autophagy16, 1077–1091. 10.1080/15548627.2019.1659610
117
Liao B. Dong S. Xu Z. Gao F. Zhang S. Liang R. (2020). LncRNA Kcnq1ot1 renders cardiomyocytes apoptosis in acute myocardial infarction model by up-regulating Tead1. Life Sci.256, 117811. 10.1016/j.lfs.2020.117811
118
Liao M. Xie Q. Zhao Y. Yang C. Lin C. Wang G. et al (2022). Main active components of si-miao-yong-an decoction (SMYAD) attenuate autophagy and apoptosis via the PDE5A-AKT and TLR4-NOX4 pathways in isoproterenol (ISO)-Induced heart failure models. Pharmacol. Res.176, 106077. 10.1016/j.phrs.2022.106077
119
Lin J.-J. Chen R. Yang L.-Y. Gong M. Du M.-Y. Mu S.-Q. et al (2024). Hsa_circ_0001402 alleviates vascular neointimal hyperplasia through a miR-183-5p-dependent regulation of vascular smooth muscle cell proliferation, migration, and autophagy. J. Adv. Res.60, 93–110. 10.1016/j.jare.2023.07.010
120
Lippi M. Stadiotti I. Pompilio G. Sommariva E. (2020). Human cell modeling for cardiovascular diseases. Int. J. Mol. Sci.21, 6388. 10.3390/ijms21176388
121
Liu H. Shi C. Deng Y. (2020). MALAT1 affects hypoxia-induced vascular endothelial cell injury and autophagy by regulating miR-19b-3p/HIF-1α axis. Mol. Cell Biochem.466, 25–34. 10.1007/s11010-020-03684-z
122
Liu N. Peng A. Sun H. Zhuang Y. Yu M. Wang Q. et al (2021). LncRNA AC136007.2 alleviates cerebral ischemic-reperfusion injury by suppressing autophagy. Aging (Albany NY)13, 19587–19597. 10.18632/aging.203369
123
Liu H. Zhang S. Liu Y. Ma J. Chen W. Yin T. et al (2022). Knockdown of HSP110 attenuates hypoxia-induced pulmonary hypertension in mice through suppression of YAP/TAZ-TEAD4 pathway. Respir. Res.23, 209. 10.1186/s12931-022-02124-4
124
Liu L. Zhao B. Yu Y. Gao W. Liu W. Chen L. et al (2024a). Vascular aging in ischemic stroke. J. Am. Heart Assoc.13, e033341. 10.1161/JAHA.123.033341
125
Liu R. M. Huang S. Hu D. Liu L. Sun H. C. Tian J. et al (2024b). Decreased intranuclear cardiac troponin I impairs cardiac autophagy through FOS/ATG5 in ageing hearts. J. Cell Mol. Med.28, e18357. 10.1111/jcmm.18357
126
Liu M. Zhang Y. Li Y. Shi T. Yan Y. (2024c). LncRNA Zfas1 boosts cell apoptosis and autophagy in myocardial injury induced by hypoxia via miR-383-5p/ATG10 axis. Heliyon10, e24578. 10.1016/j.heliyon.2024.e24578
127
Loh J. W. Lee J. Y. Lim A. H. Guan P. Lim B. Y. Kannan B. et al (2023). Spatial transcriptomics reveal topological immune landscapes of Asian head and neck angiosarcoma. Commun. Biol.6, 461–469. 10.1038/s42003-023-04856-5
128
Loi M. Raimondi A. Morone D. Molinari M. (2019). ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat. Commun.10, 5058. 10.1038/s41467-019-12991-z
129
Longenecker C. T. Sullivan C. Baker J. V. (2016). Immune activation and cardiovascular disease in chronic HIV infection. Curr. Opin. HIV AIDS11, 216–225. 10.1097/COH.0000000000000227
130
López-Otín C. Blasco M. A. Partridge L. Serrano M. Kroemer G. (2023). Hallmarks of aging: an expanding universe. Cell186, 243–278. 10.1016/j.cell.2022.11.001
131
Lu D. Thum T. (2019). RNA-Based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol.16, 661–674. 10.1038/s41569-019-0218-x
132
Lu W. Zhou Y. Zeng S. Zhong L. Zhou S. Song H. et al (2021). Loss of FoxO3a prevents aortic aneurysm formation through maintenance of VSMC homeostasis. Cell Death Dis.12, 378. 10.1038/s41419-021-03659-y
133
Lu H. Chen Y. Chen Y. Huang L. Chen L. (2022). C/EBPα-Mediated transcriptional activation of PIK3C2A regulates autophagy, matrix metalloproteinase expression, and phenotypic of vascular smooth muscle cells in aortic dissection. J. Immunol. Res.2022, 7465353. 10.1155/2022/7465353
134
Luo S. Zhang M. Wu H. Ding X. Li D. Dong X. et al (2021). SAIL: a new conserved anti-fibrotic lncRNA in the heart. Basic Res. Cardiol.116, 15. 10.1007/s00395-021-00854-y
135
Lv X.-F. Zhang Y.-J. Liu X. Zheng H.-Q. Liu C.-Z. Zeng X.-L. et al (2020). TMEM16A ameliorates vascular remodeling by suppressing autophagy via inhibiting Bcl-2-p62 complex formation. Theranostics10, 3980–3993. 10.7150/thno.41028
136
Lv X.-W. Wang M.-J. Qin Q.-Y. Lu P. Qin G.-W. (2021). 6-Gingerol relieves myocardial ischaemia/reperfusion injury by regulating lncRNA H19/miR-143/ATG7 signaling axis-mediated autophagy. Lab. Invest101, 865–877. 10.1038/s41374-021-00575-9
137
Ma S. Wang Y. Chen Y. Cao F. (2015). The role of the autophagy in myocardial ischemia/reperfusion injury. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis.1852, 271–276. 10.1016/j.bbadis.2014.05.010
138
Ma T. Qiu F. Gong Y. Cao H. Dai G. Sun D. et al (2023). Therapeutic silencing of lncRNA RMST alleviates cardiac fibrosis and improves heart function after myocardial infarction in mice and swine. Theranostics13, 3826–3843. 10.7150/thno.82543
139
Ma L. Liu Y.-H. Liu C. Wang S.-Q. Ma J. Li X.-Q. et al (2025). lncRNA, miRNA, and mRNA of plasma and tumor-derived exosomes of cardiac myxoma-related ischaemic stroke. Sci. Data12, 91. 10.1038/s41597-025-04410-4
140
Mably J. D. Wang D.-Z. (2024). Long non-coding RNAs in cardiac hypertrophy and heart failure: functions, mechanisms and clinical prospects. Nat. Rev. Cardiol.21, 326–345. 10.1038/s41569-023-00952-5
141
Maeda S. Yamamoto H. Kinch L. N. Garza C. M. Takahashi S. Otomo C. et al (2020). Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol.27, 1194–1201. 10.1038/s41594-020-00520-2
142
Mahi N. A. Najafabadi M. F. Pilarczyk M. Kouril M. Medvedovic M. (2019). GREIN: an interactive web platform for Re-analyzing GEO RNA-Seq data. Sci. Rep.9, 7580. 10.1038/s41598-019-43935-8
143
Margara F. Wang Z. J. Levrero-Florencio F. Santiago A. Vázquez M. Bueno-Orovio A. et al (2021). In-silico human electro-mechanical ventricular modelling and simulation for drug-induced pro-arrhythmia and inotropic risk assessment. Prog. Biophys. Mol. Biol.159, 58–74. 10.1016/j.pbiomolbio.2020.06.007
144
Marín-Aguilar F. Lechuga-Vieco A. V. Alcocer-Gómez E. Castejón-Vega B. Lucas J. Garrido C. et al (2020). NLRP3 inflammasome suppression improves longevity and prevents cardiac aging in Male mice. Aging Cell19, e13050. 10.1111/acel.13050
145
Matsui T. Jiang P. Nakano S. Sakamaki Y. Yamamoto H. Mizushima N. (2018). Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol.217, 2633–2645. 10.1083/jcb.201712058
146
Mattick J. S. Amaral P. P. Carninci P. Carpenter S. Chang H. Y. Chen L.-L. et al (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol.24, 430–447. 10.1038/s41580-022-00566-8
147
McEwan D. G. Popovic D. Gubas A. Terawaki S. Suzuki H. Stadel D. et al (2015). PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell57, 39–54. 10.1016/j.molcel.2014.11.006
148
Mehrbod P. Ande S. R. Alizadeh J. Rahimizadeh S. Shariati A. Malek H. et al (2019). The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence10, 376–413. 10.1080/21505594.2019.1605803
149
Mei Y. Thompson M. D. Cohen R. A. Tong X. (2014). Autophagy and oxidative stress in cardiovascular diseases. Biochimica biophysica acta1852, 243–251. 10.1016/j.bbadis.2014.05.005
150
Meier S. Grundland A. Dobrev D. Volders P. G. A. Heijman J. (2023). In silico analysis of the dynamic regulation of cardiac electrophysiology by Kv 11.1 ion-channel trafficking. J. Physiol.601, 2711–2731. 10.1113/JP283976
151
Mejlvang J. Olsvik H. Svenning S. Bruun J.-A. Abudu Y. P. Larsen K. B. et al (2018). Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol.217, 3640–3655. 10.1083/jcb.201711002
152
Meng Z. Lu M. (2017). RNA interference-induced innate immunity, off-target effect, or immune adjuvant?Front. Immunol.8, 331. 10.3389/fimmu.2017.00331
153
Meng K. Jiao J. Zhu R.-R. Wang B.-Y. Mao X.-B. Zhong Y.-C. et al (2020). The long noncoding RNA hotair regulates oxidative stress and cardiac myocyte apoptosis during ischemia-reperfusion injury. Oxid. Med. Cell Longev.2020, 1645249. 10.1155/2020/1645249
154
Mi S. Huang F. Jiao M. Qian Z. Han M. Miao Z. et al (2023). Inhibition of MEG3 ameliorates cardiomyocyte apoptosis and autophagy by regulating the expression of miRNA-129-5p in a mouse model of heart failure. Redox Rep.28, 2224607. 10.1080/13510002.2023.2224607
155
Miranda A. M. A. Janbandhu V. Maatz H. Kanemaru K. Cranley J. Teichmann S. A. et al (2023). Single-cell transcriptomics for the assessment of cardiac disease. Nat. Rev. Cardiol.20, 289–308. 10.1038/s41569-022-00805-7
156
Mizushima N. Klionsky D. J. (2007). Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr.27, 19–40. 10.1146/annurev.nutr.27.061406.093749
157
Modarresi Chahardehi A. Afrooghe A. Emtiazi N. Rafiei S. Rezaei N. J. Dahmardeh S. et al (2024). MicroRNAs and angiosarcoma: are there promising reports?Front. Oncol.14, 1385632. 10.3389/fonc.2024.1385632
158
Mori M. A. Ludwig R. G. Garcia-Martin R. Brandão B. B. Kahn C. R. (2019). Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab.30, 656–673. 10.1016/j.cmet.2019.07.011
159
Nagarajan N. Oka S.-I. Nah J. Wu C. Zhai P. Mukai R. et al (2023). Thioredoxin 1 promotes autophagy through transnitrosylation of Atg7 during myocardial ischemia. J. Clin. Invest133, e162326. 10.1172/JCI162326
160
Nähse V. Raiborg C. Tan K. W. Mørk S. Torgersen M. L. Wenzel E. M. et al (2023). ATPase activity of DFCP1 controls selective autophagy. Nat. Commun.14, 4051. 10.1038/s41467-023-39641-9
161
Nappi F. (2024). Non-coding RNA-targeted therapy: a state-of-the-art review. Int. J. Mol. Sci.25, 3630. 10.3390/ijms25073630
162
National Library of Medicine (2024). Clinical trials. Available online at: https://clinicaltrials.gov/(Accessed October 28, 2024).
163
Ning S. Li Z. Ji Z. Fan D. Wang K. Wang Q. et al (2020). MicroRNA-494 suppresses hypoxia/reoxygenation-induced cardiomyocyte apoptosis and autophagy via the PI3K/AKT/mTOR signaling pathway by targeting SIRT1. Mol. Med. Rep.22, 5231–5242. 10.3892/mmr.2020.11636
164
Nissen S. E. Wang Q. Nicholls S. J. Navar A. M. Ray K. K. Schwartz G. G. et al (2024a). Zerlasiran-A small-interfering RNA targeting lipoprotein(a): a phase 2 randomized clinical trial. JAMA332, 1992–2002. 10.1001/jama.2024.21957
165
Nissen S. E. Wolski K. Watts G. F. Koren M. J. Fok H. Nicholls S. J. et al (2024b). Single ascending and multiple-dose trial of zerlasiran, a short interfering RNA targeting lipoprotein(a): a randomized clinical trial. JAMA331, 1534–1543. 10.1001/jama.2024.4504
166
Niu X. Pu S. Ling C. Xu J. Wang J. Sun S. et al (2020). lncRNA Oip5-as1 attenuates myocardial ischaemia/reperfusion injury by sponging miR-29a to activate the SIRT1/AMPK/PGC1α pathway. Cell Prolif.53, e12818. 10.1111/cpr.12818
167
Niu X. Zhang J. Hu S. Dang W. Wang K. Bai M. (2024). lncRNA Oip5-as1 inhibits excessive mitochondrial fission in myocardial ischemia/reperfusion injury by modulating DRP1 phosphorylation. Cell Mol. Biol. Lett.29, 72. 10.1186/s11658-024-00588-4
168
Obas V. Vasan R. S. (2018). The aging heart. Clin. Sci. (Lond)132, 1367–1382. 10.1042/CS20171156
169
Oliveros J. C. (2007). Venny. An interactive tool for comparing lists with Venn's diagrams. Available online at: https://bioinfogp.cnb.csic.es/tools/venny/index.html (Accessed October 31, 2024).
170
Olsson A. Venkatasubramanian M. Chaudhri V. K. Aronow B. J. Salomonis N. Singh H. et al (2016). Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature537, 698–702. 10.1038/nature19348
171
Osawa T. Kotani T. Kawaoka T. Hirata E. Suzuki K. Nakatogawa H. et al (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol.26, 281–288. 10.1038/s41594-019-0203-4
172
Ota Y. Lee E. Sella E. Agarwal P. (2023). Vascular malformations and tumors: a review of classification and imaging features for cardiothoracic radiologists. Radiol. Cardiothorac. Imaging5, e220328. 10.1148/ryct.220328
173
Otto F. B. Thumm M. (2021). Mechanistic dissection of macro- and micronucleophagy. Autophagy17, 626–639. 10.1080/15548627.2020.1725402
174
Ou W. Liu H. Chen C. Yang C. Zhao X. Zhang Y. et al (2024). Spexin inhibits excessive autophagy-induced ferroptosis to alleviate doxorubicin-induced cardiotoxicity by upregulating beclin 1. Br. J. Pharmacol.181, 4195–4213. 10.1111/bph.16484
175
Ouyang M. Lu J. Ding Q. Qin T. Peng C. Guo Q. (2020). Knockdown of long non-coding RNA PVT1 protects human AC16 cardiomyocytes from hypoxia/reoxygenation-induced apoptosis and autophagy by regulating miR-186/Beclin-1 axis. Gene754, 144775. 10.1016/j.gene.2020.144775
176
Pang Y. Wu L. Tang C. Wang H. Wei Y. (2022). Autophagy-inflammation interplay during infection: balancing pathogen clearance and host inflammation. Front. Pharmacol.13, 832750. 10.3389/fphar.2022.832750
177
Panni S. Lovering R. C. Porras P. Orchard S. (2020). Non-coding RNA regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech.1863, 194417. 10.1016/j.bbagrm.2019.194417
178
Peng T. Liu M. Hu L. Guo D. Wang D. Qi B. et al (2022). LncRNA airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis. Biol. Direct17, 32. 10.1186/s13062-022-00346-6
179
Perkins M. V. Joseph S. B. Dittmer D. P. Mackman N. (2023). Cardiovascular disease and thrombosis in HIV infection. Arterioscler. Thromb. Vasc. Biol.43, 175–191. 10.1161/ATVBAHA.122.318232
180
Pfitzner A.-K. Mercier V. Jiang X. Moser von Filseck J. Baum B. Šarić A. et al (2020). An ESCRT-III polymerization sequence drives membrane deformation and fission. Cell182, 1140–1155. 10.1016/j.cell.2020.07.021
181
Poller W. Dimmeler S. Heymans S. Zeller T. Haas J. Karakas M. et al (2017). Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur. Heart J.39, 2704–2716. 10.1093/eurheartj/ehx165
182
Porrello E. R. Mahmoud A. I. Simpson E. Hill J. A. Richardson J. A. Olson E. N. et al (2011). Transient regenerative potential of the neonatal mouse heart. Science331, 1078–1080. 10.1126/science.1200708
183
Qi Y.-X. Yao Q.-P. Huang K. Shi Q. Zhang P. Wang G.-L. et al (2016). Nuclear envelope proteins modulate proliferation of vascular smooth muscle cells during cyclic stretch application. Proc. Natl. Acad. Sci. U. S. A.113, 5293–5298. 10.1073/pnas.1604569113
184
Qi Y. Dai F. Gu J. Yao W. (2019). Biomarkers in VSMC phenotypic modulation and vascular remodeling. Pharmazie74, 711–714. 10.1691/ph.2019.9743
185
Qian X. Wang H. Wang Y. Chen J. Guo X. Deng H. (2020). Enhanced autophagy in GAB1-Deficient vascular endothelial cells is responsible for atherosclerosis progression. Front. Physiol.11, 559396. 10.3389/fphys.2020.559396
186
Qiao L. Hu J. Qiu X. Wang C. Peng J. Zhang C. et al (2023). LAMP2A, LAMP2B and LAMP2C: similar structures, divergent roles. Autophagy19, 2837–2852. 10.1080/15548627.2023.2235196
187
Qiu Z. Wang Y. Liu W. Li C. Zhao R. Long X. et al (2021). CircHIPK3 regulates the autophagy and apoptosis of hypoxia/reoxygenation-stimulated cardiomyocytes via the miR-20b-5p/ATG7 axis. Cell Death Discov.7, 64. 10.1038/s41420-021-00448-6
188
Ren H. Dai R. Nik Nabil W. N. Xi Z. Wang F. Xu H. (2023a). Unveiling the dual role of autophagy in vascular remodelling and its related diseases. Biomed. Pharmacother.168, 115643. 10.1016/j.biopha.2023.115643
189
Ren X. Nguyen T. N. Lam W. K. Buffalo C. Z. Lazarou M. Yokom A. L. et al (2023b). Structural basis for ATG9A recruitment to the ULK1 complex in mitophagy initiation. Sci. Adv.9, eadg2997. 10.1126/sciadv.adg2997
190
Rizzoni D. Rizzoni M. Nardin M. Chiarini G. Agabiti-Rosei C. Aggiusti C. et al (2019). Vascular aging and disease of the small vessels. High. Blood Press Cardiovasc Prev.26, 183–189. 10.1007/s40292-019-00320-w
191
Rong Y. Liu M. Ma L. Du W. Zhang H. Tian Y. et al (2012). Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol.14, 924–934. 10.1038/ncb2557
192
Rubinsztein D. C. Mariño G. Kroemer G. (2011). Autophagy and aging. Cell146, 682–695. 10.1016/j.cell.2011.07.030
193
Ryter S. W. Cloonan S. M. Choi A. M. K. (2013). Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells36, 7–16. 10.1007/s10059-013-0140-8
194
Sahu R. Kaushik S. Clement C. C. Cannizzo E. S. Scharf B. Follenzi A. et al (2011). Microautophagy of cytosolic proteins by late endosomes. Dev. Cell20, 131–139. 10.1016/j.devcel.2010.12.003
195
Salomonis N. Schlieve C. R. Pereira L. Wahlquist C. Colas A. Zambon A. C. et al (2010). Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl. Acad. Sci. U. S. A.107, 10514–10519. 10.1073/pnas.0912260107
196
Scalabrin S. Cagnin S. (2025). Nucleic acid delivery for pathology treatment: RNA tissue delivery. Mol. Ther. Nucleic Acids36, 102459. 10.1016/j.omtn.2025.102459
197
Sciarretta S. Maejima Y. Zablocki D. Sadoshima J. (2018). The role of autophagy in the heart. Annu. Rev. Physiol.80, 1–26. 10.1146/annurev-physiol-021317-121427
198
Sepulveda A. Buchanan E. P. (2014). Vascular tumors. Semin. Plast. Surg.28, 49–57. 10.1055/s-0034-1376260
199
Serio S. Pagiatakis C. Musolino E. Felicetta A. Carullo P. Laura Frances J. et al (2023). Cardiac aging is promoted by pseudohypoxia increasing p300-Induced glycolysis. Circ. Res.133, 687–703. 10.1161/CIRCRESAHA.123.322676
200
Seyhan A. A. (2019). Lost in translation: the Valley of death across preclinical and clinical divide – identification of problems and overcoming obstacles. Transl. Med. Commun.4, 18. 10.1186/s41231-019-0050-7
201
Shen B. Wen Y. Li S. Zhou Y. Chen J. Yang J. et al (2024a). Paeonol ameliorates hyperlipidemia and autophagy in mice by regulating Nrf2 and AMPK/mTOR pathways. Phytomedicine132, 155839. 10.1016/j.phymed.2024.155839
202
Shen X. Xie X. Wu Q. Shi F. Chen Y. Yuan S. et al (2024b). S-adenosylmethionine attenuates angiotensin II-induced aortic dissection formation by inhibiting vascular smooth muscle cell phenotypic switch and autophagy. Biochem. Pharmacol.219, 115967. 10.1016/j.bcp.2023.115967
203
Sheng Y. Wang Y.-Y. Chang Y. Ye D. Wu L. Kang H. et al (2024). Deciphering mechanisms of cardiomyocytes and non-cardiomyocyte transformation in myocardial remodeling of permanent atrial fibrillation. J. Adv. Res.61, 101–117. 10.1016/j.jare.2023.09.012
204
Sherazi S. A. M. Abbasi A. Jamil A. Uzair M. Ikram A. Qamar S. et al (2023). Molecular hallmarks of long non-coding RNAs in aging and its significant effect on aging-associated diseases. Neural Regen. Res.18, 959–968. 10.4103/1673-5374.355751
205
Shi D. Ding J. Xie S. Huang L. Zhang H. Chen X. et al (2022). Myocardin/microRNA-30a/Beclin1 signaling controls the phenotypic modulation of vascular smooth muscle cells by regulating autophagy. Cell Death Dis.13, 121. 10.1038/s41419-022-04588-0
206
Shi L. Li H. Sun L. Tian C. Li H. (2024). Alleviation of angiotensin II-Induced vascular endothelial cell injury through long non-coding RNA TUG1 inhibition. Comb. Chem. High. Throughput Screen27, 1523–1532. 10.2174/0113862073265220231004071645
207
Shu Z. Li X. Zhang W. Huyan Z. Cheng D. Xie S. et al (2024). MG-132 activates sodium palmitate-induced autophagy in human vascular smooth muscle cells and inhibits senescence via the PI3K/AKT/mTOR axis. Lipids Health Dis.23, 282. 10.1186/s12944-024-02268-w
208
Shu Z. Zhang W. Sun M. Huyan Z. Xie S. Cheng H. et al (2025). Dehydrodiisoeugenol alleviates sodium palmitate-induced mitochondrial dysfunction and activates autophagy in VSMCs via the SIRT1/Nrf2 axis. Cell Biochem. Funct.43, e70074. 10.1002/cbf.70074
209
Singh D. Rai V. Agrawal D. K. (2022). Non-coding RNAs in regulating plaque progression and remodeling of extracellular matrix in atherosclerosis. Int. J. Mol. Sci.23, 13731. 10.3390/ijms232213731
210
Skardal A. Aleman J. Forsythe S. Rajan S. Murphy S. Devarasetty M. et al (2020). Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication12, 025017. 10.1088/1758-5090/ab6d36
211
So-Armah K. Benjamin L. A. Bloomfield G. S. Feinstein M. J. Hsue P. Njuguna B. et al (2020). HIV and cardiovascular disease. Lancet HIV7, e279–e293. 10.1016/S2352-3018(20)30036-9
212
Song C. Qi H. Liu Y. Chen Y. Shi P. Zhang S. et al (2021). Inhibition of lncRNA Gm15834 attenuates autophagy-mediated myocardial hypertrophy via the miR-30b-3p/ULK1 axis in mice. Mol. Ther.29, 1120–1137. 10.1016/j.ymthe.2020.10.024
213
Song Z. Han Q. Wen Z. Lv Q. Pan C. Pan Y. (2023). LncRNA RASSF8-AS1 knockdown displayed antiproliferative and proapoptotic effects through miR-188-3p/ATG7 pathway in ox-LDL-treated vascular smooth muscle cells. Ann. Transl. Med.11, 143. 10.21037/atm-22-6457
214
Sramek V. Dridi M. Papoudou-Bai A. Dumollard J. M. Péoc’h M. Karpathiou G. (2021). Autophagy in cardiac myxoma: an important puzzle piece in understanding its inflammatory environment. Pathol. Res. Pract.226, 153609. 10.1016/j.prp.2021.153609
215
Stassen J. Ewe S. H. Hirasawa K. Butcher S. C. Singh G. K. Amanullah M. R. et al (2022). Left ventricular remodelling patterns in patients with moderate aortic stenosis. Eur. Heart J. Cardiovasc Imaging23, 1326–1335. 10.1093/ehjci/jeac018
216
Su X. Lv L. Li Y. Fang R. Yang R. Li C. et al (2020). lncRNA MIRF promotes cardiac apoptosis through the miR-26a-Bak1 axis. Mol. Ther. Nucleic Acids20, 841–850. 10.1016/j.omtn.2020.05.002
217
Sutanto H. Lyon A. Lumens J. Schotten U. Dobrev D. Heijman J. (2020). Cardiomyocyte calcium handling in health and disease: insights from in vitro and in silico studies. Prog. Biophys. Mol. Biol.157, 54–75. 10.1016/j.pbiomolbio.2020.02.008
218
Suzuki T. Aoshima K. Yamazaki J. Kobayashi A. Kimura T. (2022). Manipulating histone acetylation leads to antitumor effects in hemangiosarcoma cells. Vet. Comp. Oncol.20, 805–816. 10.1111/vco.12840
219
Tabibzadeh S. (2023). Role of autophagy in aging: the good, the bad, and the ugly. Aging Cell22, e13753. 10.1111/acel.13753
220
Takahashi Y. He H. Tang Z. Hattori T. Liu Y. Young M. M. et al (2018). An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun.9, 2855. 10.1038/s41467-018-05254-w
221
Tan L. Xiong D. Zhang H. Xiao S. Yi R. Wu J. (2023). ETS2 promotes cardiomyocyte apoptosis and autophagy in heart failure by regulating lncRNA TUG1/miR-129-5p/ATG7 axis. FASEB J.37, e22937. 10.1096/fj.202202148RR
222
Tatman P. D. Woulfe K. C. Karimpour-Fard A. Jeffrey D. A. Jaggers J. Cleveland J. C. et al (2017). Pediatric dilated cardiomyopathy hearts display a unique gene expression profile. JCI Insight2, 94249. 10.1172/jci.insight.94249
223
Täubel J. Hauke W. Rump S. Viereck J. Batkai S. Poetzsch J. et al (2021). Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human phase 1b randomized, double-blind, placebo-controlled study. Eur. Heart J.42, 178–188. 10.1093/eurheartj/ehaa898
224
Tenreiro M. F. Almeida H. V. Calmeiro T. Fortunato E. Ferreira L. Alves P. M. et al (2021). Interindividual heterogeneity affects the outcome of human cardiac tissue decellularization. Sci. Rep.11, 20834. 10.1038/s41598-021-00226-5
225
Tettamanti G. Carata E. Montali A. Dini L. Fimia G. M. (2019). Autophagy in development and regeneration: role in tissue remodelling and cell survival. Eur. Zoological J.86, 113–131. 10.1080/24750263.2019.1601271
226
Tian Z. Ning H. Wang X. Wang Y. Han T. Sun C. (2024). Endothelial autophagy promotes atheroprotective communication between endothelial and smooth muscle cells via exosome-mediated delivery of miR-204-5p. Arterioscler. Thromb. Vasc. Biol.44, 1813–1832. 10.1161/ATVBAHA.123.319993
227
Timmis A. Aboyans V. Vardas P. Townsend N. Torbica A. Kavousi M. et al (2024). European society of cardiology: the 2023 atlas of cardiovascular disease statistics. Eur. Heart J.45, 4019–4062. 10.1093/eurheartj/ehae466
228
Tomasello L. Distefano R. Nigita G. Croce C. M. (2021). The MicroRNA family gets wider: the IsomiRs classification and role. Front. Cell Dev. Biol.9, 668648. 10.3389/fcell.2021.668648
229
Tong G. Wang Y. Xu C. Xu Y. Ye X. Zhou L. et al (2019). Long non-coding RNA FOXD3-AS1 aggravates ischemia/reperfusion injury of cardiomyocytes through promoting autophagy. Am. J. Transl. Res.11, 5634–5644.
230
Tracy E. Rowe G. LeBlanc A. J. (2020). Cardiac tissue remodeling in healthy aging: the road to pathology. Am. J. Physiol. Cell Physiol.319, C166-C182–C182. 10.1152/ajpcell.00021.2020
231
Turco E. Witt M. Abert C. Bock-Bierbaum T. Su M.-Y. Trapannone R. et al (2019). FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell74, 330–346. 10.1016/j.molcel.2019.01.035
232
Turco E. Savova A. Gere F. Ferrari L. Romanov J. Schuschnig M. et al (2021). Reconstitution defines the roles of p62, NBR1 and TAX1BP1 in ubiquitin condensate formation and autophagy initiation. Nat. Commun.12, 5212. 10.1038/s41467-021-25572-w
233
U.S. Centers for Disease Control and Prevention (2024). Heart Dis. Facts. Available online at: https://www.cdc.gov/heart-disease/data-research/facts-stats/index.html (Accessed October 26, 2024).
234
U.S. Food and Drug Administration (2024). Commissioner, O. Of the, 2024. Available online at: https://www.fda.gov/(Accessed October 28, 2024).
235
Ugalde A. P. Roiz-Valle D. Moledo-Nodar L. Caravia X. M. Freije J. M. P. López-Otín C. (2024). Noncoding RNA contribution to aging and lifespan. J. Gerontol. A Biol. Sci. Med. Sci.79, glae058. 10.1093/gerona/glae058
236
van der Kwast R. V. C. T. Woudenberg T. Quax P. H. A. Nossent A. Y. (2020). MicroRNA-411 and its 5’-IsomiR have distinct targets and functions and are differentially regulated in the vasculature under ischemia. Mol. Ther.28, 157–170. 10.1016/j.ymthe.2019.10.002
237
van Doorn E. C. H. Amesz J. H. Sadeghi A. H. de Groot N. M. S. Manintveld O. C. Taverne Y. J. H. J. (2024). Preclinical models of cardiac disease: a comprehensive overview for clinical scientists. Cardiovasc Eng. Technol.15, 232–249. 10.1007/s13239-023-00707-w
238
Vargas J. N. S. Wang C. Bunker E. Hao L. Maric D. Schiavo G. et al (2019). Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell74, 347–362. 10.1016/j.molcel.2019.02.010
239
Verjans R. Derks W. J. A. Korn K. Sönnichsen B. van Leeuwen R. E. W. Schroen B. et al (2019). Functional screening identifies MicroRNAs as multi-cellular regulators of heart failure. Sci. Rep.9, 6055. 10.1038/s41598-019-41491-9
240
Wagner M. J. Ravi V. Schaub S. K. Kim E. Y. Sharib J. Mogal H. et al (2024a). Incidence and presenting characteristics of angiosarcoma in the US, 2001-2020. JAMA Netw. Open7, e246235. 10.1001/jamanetworkopen.2024.6235
241
Wagner V. Kern F. Hahn O. Schaum N. Ludwig N. Fehlmann T. et al (2024b). Characterizing expression changes in noncoding RNAs during aging and heterochronic parabiosis across mouse tissues. Nat. Biotechnol.42, 109–118. 10.1038/s41587-023-01751-6
242
Wang X. Chu J. Wen C. J. Fu S. B. Qian Y. L. Wo Y. et al (2015). Functional characterization of TRAP1-like protein involved in modulating fibrotic processes mediated by TGF-β/Smad signaling in hypertrophic scar fibroblasts. Exp. Cell Res.332, 202–211. 10.1016/j.yexcr.2015.01.015
243
Wang Z. Miao G. Xue X. Guo X. Yuan C. Wang Z. et al (2016). The vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol. Cell63, 781–795. 10.1016/j.molcel.2016.08.021
244
Wang J. Bie Z. Sun C. (2019a). Long noncoding RNA AK088388 regulates autophagy through miR-30a to affect cardiomyocyte injury. J. Cell. Biochem.120, 10155–10163. 10.1002/jcb.28300
245
Wang K. Yang C. Shi J. Gao T. (2019b). Ox-LDL-induced lncRNA MALAT1 promotes autophagy in human umbilical vein endothelial cells by sponging miR-216a-5p and regulating Beclin-1 expression. Eur. J. Pharmacol.858, 172338. 10.1016/j.ejphar.2019.04.019
246
Wang S. Yao T. Deng F. Yu W. Song Y. Chen J. et al (2019c). LncRNA MALAT1 promotes oxygen-glucose deprivation and reoxygenation induced cardiomyocytes injury through sponging miR-20b to enhance beclin1-Mediated autophagy. Cardiovasc Drugs Ther.33, 675–686. 10.1007/s10557-019-06902-z
247
Wang J. Dong G. Chi W. Nie Y. (2021). MiR-96 promotes myocardial infarction-induced apoptosis by targeting XIAP. Biomed. and Pharmacother.138, 111208. 10.1016/j.biopha.2020.111208
248
Wang L. Hu S. Zhou B. (2022). Deciphering cardiac biology and disease by single-cell transcriptomic profiling. Biomolecules12, 566. 10.3390/biom12040566
249
Wang Q. Zhao C. Du Q. Cao Z. Pan J. (2024a). Non-coding RNA in infantile hemangioma. Pediatr. Res.96, 1594–1602. 10.1038/s41390-024-03250-z
250
Wang S. Xuan L. Hu X. Sun F. Li S. Li X. et al (2024b). LncRNA CCRR attenuates postmyocardial infarction inflammatory response by inhibiting the TLR signalling pathway. Can. J. Cardiol.40, 710–725. 10.1016/j.cjca.2023.12.003
251
Watanabe K. Narumi T. Watanabe T. Otaki Y. Takahashi T. Aono T. et al (2020). The association between microRNA-21 and hypertension-induced cardiac remodeling. PLoS One15, e0226053. 10.1371/journal.pone.0226053
252
Wei J. Hollabaugh C. Miller J. Geiger P. C. Flynn B. C. (2021). Molecular cardioprotection and the role of exosomes: the future is not far away. J. Cardiothorac. Vasc. Anesth.35, 780–785. 10.1053/j.jvca.2020.05.033
253
Whiting R. Sander E. Conway C. Vaughan T. J. (2022). In silico modelling of aortic valve implants - predicting in vitro performance using finite element analysis. J. Med. Eng. Technol.46, 220–230. 10.1080/03091902.2022.2026506
254
Widlansky M. E. Liu Y. Tumusiime S. Hofeld B. Khan N. Aljadah M. et al (2023). Coronary plaque sampling reveals molecular insights into coronary artery disease. Circ. Res.133, 532–534. 10.1161/CIRCRESAHA.123.323022
255
Williams K. Liang T. Massé S. Khan S. Hatkar R. Keller G. et al (2021). A 3-D human model of complex cardiac arrhythmias. Acta Biomater.132, 149–161. 10.1016/j.actbio.2021.03.004
256
Williams C. G. Lee H. J. Asatsuma T. Vento-Tormo R. Haque A. (2022). An introduction to spatial transcriptomics for biomedical research. Genome Med.14, 68. 10.1186/s13073-022-01075-1
257
Woodruff R. C. Tong X. Khan S. S. Shah N. S. Jackson S. L. Loustalot F. et al (2024). Trends in cardiovascular disease mortality rates and excess deaths, 2010-2022. Am. J. Prev. Med.66, 582–589. 10.1016/j.amepre.2023.11.009
258
World Bank Group (2024). World Bank Country and Lending Groups. Available online at: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups (Accessed October 26, 2024).
259
Wu R. Tang S. Wang M. Xu X. Yao C. Wang S. (2016). MicroRNA-497 induces apoptosis and suppresses proliferation via the Bcl-2/Bax-Caspase9-Caspase3 pathway and cyclin D2 protein in HUVECs. PLOS ONE11, e0167052. 10.1371/journal.pone.0167052
260
Wu R. Lin Y. Liu X. Zhan C. He H. Shi M. et al (2020). Phenotype-genotype network construction and characterization: a case study of cardiovascular diseases and associated non-coding RNAs. Database (Oxford)2020, baz147. 10.1093/database/baz147
261
Wu K. Chen Y. Wang D. He K. (2021a). MicroRNA-520d-3p alleviates hypoxia/reoxygenation-induced damage in human cardiomyocytes by targeting ATG-12. J. Thromb. Thrombolysis52, 429–439. 10.1007/s11239-020-02352-9
262
Wu R. Hu W. Chen H. Wang Y. Li Q. Xiao C. et al (2021b). A novel human long noncoding RNA SCDAL promotes angiogenesis through SNF5-Mediated GDF6 expression. Adv. Sci. (Weinh)8, e2004629. 10.1002/advs.202004629
263
Wu Y. Mao Q. Liang X. (2021c). Targeting the MicroRNA-490-3p-ATG4B-Autophagy axis relieves myocardial injury in ischemia reperfusion. J. Cardiovasc Transl. Res.14, 173–183. 10.1007/s12265-020-09972-9
264
Xi H. Wang S. Wang B. Hong X. Liu X. Li M. et al (2022). The role of interaction between autophagy and apoptosis in tumorigenesis (review). Oncol. Rep.48, 208. 10.3892/or.2022.8423
265
Xiao H. Zhang M. Wu H. Wu J. Hu X. Pei X. et al (2022). CIRKIL exacerbates cardiac ischemia/reperfusion injury by interacting with Ku70. Circ. Res.130, e3–e17. 10.1161/CIRCRESAHA.121.318992
266
Xie Q. Ma Y. Ren Z. Gu T. Jiang Z. (2024). Circular RNA: a new expectation for cardiovascular diseases. J. Cell Biochem.125, e30512. 10.1002/jcb.30512
267
Xie X. Huang M. Ma S. Xin Q. Wang Y. Hu L. et al (2025). The role of long non-coding RNAs in cardiovascular diseases: a comprehensive review. Noncoding RNA Res.11, 158–187. 10.1016/j.ncrna.2024.12.009
268
Xu H. Wang D. Ramponi C. Wang X. Zhang H. (2022). The P21-Activated kinase 1 and 2 as potential therapeutic targets for the management of cardiovascular disease. Int. J. Drug Discov. Pharm.5, 5. 10.53941/ijddp.v1i1.179
269
Xue P. Liu Y. Wang H. Huang J. Luo M. (2023). miRNA-103-3p-Hlf regulates apoptosis and autophagy by targeting hepatic leukaemia factor in heart failure. Esc. Heart Fail10, 3038–3045. 10.1002/ehf2.14493
270
Yamada S. Nomura S. (2020). Review of single-cell RNA sequencing in the heart. Int. J. Mol. Sci.21, 8345. 10.3390/ijms21218345
271
Yamamoto H. Fujioka Y. Suzuki S. W. Noshiro D. Suzuki H. Kondo-Kakuta C. et al (2016). The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev. Cell38, 86–99. 10.1016/j.devcel.2016.06.015
272
Yamano K. Kikuchi R. Kojima W. Hayashida R. Koyano F. Kawawaki J. et al (2020). Critical role of mitochondrial ubiquitination and the OPTN-ATG9A axis in mitophagy. J. Cell Biol.219, e201912144. 10.1083/jcb.201912144
273
Yan M. Sun S. Xu K. Huang X. Dou L. Pang J. et al (2021). Cardiac aging: from basic research to therapeutics. Oxid. Med. Cell Longev.2021, 9570325. 10.1155/2021/9570325
274
Yang L. Li P. Fu S. Calay E. S. Hotamisligil G. S. (2010). Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab.11, 467–478. 10.1016/j.cmet.2010.04.005
275
Yang J.-H. Li J.-H. Shao P. Zhou H. Chen Y.-Q. Qu L.-H. (2011). starBase: a database for exploring microRNA-mRNA interaction maps from argonaute CLIP-seq and degradome-seq data. Nucleic Acids Res.39, D202–D209. 10.1093/nar/gkq1056
276
Yang K.-C. Yamada K. A. Patel A. Y. Topkara V. K. George I. Cheema F. H. et al (2014). Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation129, 1009–1021. 10.1161/CIRCULATIONAHA.113.003863
277
Yang M. Woolfenden H. C. Zhang Y. Fang X. Liu Q. Vigh M. L. et al (2020). Intact RNA structurome reveals mRNA structure-mediated regulation of miRNA cleavage in vivo. Nucleic Acids Res.48, 8767–8781. 10.1093/nar/gkaa577
278
Yang F. Kalantari S. Ruan B. Sun S. Bian Z. Guan J.-L. (2023). Autophagy inhibition prevents lymphatic malformation progression to lymphangiosarcoma by decreasing osteopontin and Stat3 signaling. Nat. Commun.14, 978. 10.1038/s41467-023-36562-5
279
Yorimitsu T. Klionsky D. J. (2005). Autophagy: molecular machinery for self-eating. Cell Death Differ.12, 1542–1552. 10.1038/sj.cdd.4401765
280
You G. Long X. Song F. Huang J. Tian M. Xiao Y. et al (2020). Metformin activates the AMPK-mTOR pathway by modulating lncRNA TUG1 to induce autophagy and inhibit atherosclerosis. Drug Des. Devel Ther.14, 457–468. 10.2147/DDDT.S233932
281
Yu F. Zhang Y. Wang Z. Gong W. Zhang C. (2021). Hsa_circ_0030042 regulates abnormal autophagy and protects atherosclerotic plaque stability by targeting eIF4A3. Theranostics11, 5404–5417. 10.7150/thno.48389
282
Yu Q. Liu J.-X. Zheng X. Yan X. Zhao P. Yin C. et al (2022). Sox9 mediates autophagy-dependent vascular smooth muscle cell phenotypic modulation and transplant arteriosclerosis. iScience25, 105161. 10.1016/j.isci.2022.105161
283
Yu Y. Ren Y. Li Z. Li Y. Li Y. Zhang Y. et al (2023a). Myo1b promotes premature endothelial senescence and dysfunction via suppressing autophagy: implications for vascular aging. Oxid. Med. Cell Longev.2023, 4654083. 10.1155/2023/4654083
284
Yu Y. Yang H. Li Q. Ding N. Gao J. Qiao G. et al (2023b). Stress-enhanced cardiac lncRNA morrbid protects hearts from acute myocardial infarction. JCI Insight8, e165568. 10.1172/jci.insight.165568
285
Yu Q. Li Y. Zhang N. Lu J. Gan X. Chen L. et al (2024). Silencing of lncRNA NEAT1 alleviates acute myocardial infarction by suppressing miR-450-5p/ACSL4-mediated ferroptosis. Exp. Cell Res.442, 114217. 10.1016/j.yexcr.2024.114217
286
Yuan Y. Xu L. Geng Z. Liu J. Zhang L. Wu Y. et al (2021). The role of non-coding RNA network in atherosclerosis. Life Sci.265, 118756. 10.1016/j.lfs.2020.118756
287
Zarrabi A. Perrin D. Kavoosi M. Sommer M. Sezen S. Mehrbod P. et al (2023). Rhabdomyosarcoma: current therapy, challenges, and future approaches to treatment strategies. Cancers (Basel)15, 5269. 10.3390/cancers15215269
288
Zeng M. Wei X. Zhou J. Luo S. (2024). LncRNA PART1 attenuates myocardial ischemia-reperfusion injury by regulating TFAP2C/DUSP5 axis via miR-302a-3p. Korean Circ. J.54, 233–252. 10.4070/kcj.2023.0131
289
Zhan L.-F. Zhang Q. Zhao L. Dong X. Pei X.-Y. Peng L.-L. et al (2022). LncRNA-6395 promotes myocardial ischemia-reperfusion injury in mice through increasing p53 pathway. Acta Pharmacol. Sin.43, 1383–1394. 10.1038/s41401-021-00767-5
290
Zhang C. Zhang B. (2023). RNA therapeutics: updates and future potential. Sci. China Life Sci.66, 12–30. 10.1007/s11427-022-2171-2
291
Zhang C. Liang R. Gan X. Yang X. Chen L. Jian J. (2019). MicroRNA-384-5p/Beclin-1 as potential indicators for epigallocatechin gallate against cardiomyocytes ischemia reperfusion injury by inhibiting autophagy via PI3K/Akt pathway. Drug Des. Devel Ther.13, 3607–3623. 10.2147/DDDT.S219074
292
Zhang L. Ma C. Wang X. Bai J. He S. Zhang J. et al (2020a). MicroRNA-874-5p regulates autophagy and proliferation in pulmonary artery smooth muscle cells by targeting sirtuin 3. Eur. J. Pharmacol.888, 173485. 10.1016/j.ejphar.2020.173485
293
Zhang W. Yao G. Wang J. Yang M. Wang J. Zhang H. et al (2020b). ncRPheno: a comprehensive database platform for identification and validation of disease related noncoding RNAs. RNA Biol.17, 943–955. 10.1080/15476286.2020.1737441
294
Zhang F. Fu X. Kataoka M. Liu N. Wang Y. Gao F. et al (2021a). Long noncoding RNA cfast regulates cardiac fibrosis. Mol. Ther. Nucleic Acids23, 377–392. 10.1016/j.omtn.2020.11.013
295
Zhang M. Cheng K. Chen H. Tu J. Shen Y. Pang L. et al (2021b). LncRNA AK020546 protects against cardiac ischemia-reperfusion injury by sponging miR-350-3p. Aging (Albany NY)13, 14219–14233. 10.18632/aging.203038
296
Zhang Y. Zhang X. Cai B. Li Y. Jiang Y. Fu X. et al (2021c). The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia-reperfusion injury. Nat. Commun.12, 522. 10.1038/s41467-020-20844-3
297
Zhang X. Zai L. Tao Z. Wu D. Lin M. Wan J. (2022a). miR-145-5p affects autophagy by targeting CaMKIIδ in atherosclerosis. Int. J. Cardiol.360, 68–75. 10.1016/j.ijcard.2022.05.039
298
Zhang Y. Ding Y. Li M. Yuan J. Yu Y. Bi X. et al (2022b). MicroRNA-34c-5p provokes isoprenaline-induced cardiac hypertrophy by modulating autophagy via targeting ATG4B. Acta Pharm. Sin. B12, 2374–2390. 10.1016/j.apsb.2021.09.020
299
Zhang L. Yang P. Chen J. Chen Z. Liu Z. Feng G. et al (2023a). CD44 connects autophagy decline and ageing in the vascular endothelium. Nat. Commun.14, 5524. 10.1038/s41467-023-41346-y
300
Zhang P. Gong S. Li S. Yuan Z. (2023b). PVT1 alleviates hypoxia-induced endothelial apoptosis by enhancing autophagy via the miR-15b-5p/ATG14 and miR-424-5p/ATG14 axis. Biochem. Biophys. Res. Commun.671, 1–9. 10.1016/j.bbrc.2023.06.001
301
Zhang Y. Wang X. Li X.-K. Lv S.-J. Wang H.-P. Liu Y. et al (2023c). Sirtuin 2 deficiency aggravates ageing-induced vascular remodelling in humans and mice. Eur. Heart J.44, 2746–2759. 10.1093/eurheartj/ehad381
302
Zhao J. Chen F. Ma W. Zhang P. (2020). Suppression of long noncoding RNA NEAT1 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-378a-3p. Gene731, 144324. 10.1016/j.gene.2019.144324
303
Zhao D. Lei W. Hu S. (2021a). Cardiac organoid - a promising perspective of preclinical model. Stem Cell Res. Ther.12, 272. 10.1186/s13287-021-02340-7
304
Zhao M. Wang F. Wu J. Cheng Y. Cao Y. Wu X. et al (2021b). CGAS is a micronucleophagy receptor for the clearance of micronuclei. Autophagy17, 3976–3991. 10.1080/15548627.2021.1899440
305
Zhao L. Tang P. Lin Y. Du M. Li H. Jiang L. et al (2024). MiR-203 improves cardiac dysfunction by targeting PARP1-NAD+ axis in aging murine. Aging Cell23, e14063. 10.1111/acel.14063
306
Zhen Y. Spangenberg H. Munson M. J. Brech A. Schink K. O. Tan K.-W. et al (2020). ESCRT-Mediated phagophore sealing during mitophagy. Autophagy16, 826–841. 10.1080/15548627.2019.1639301
307
Zheng H. Zhai W. Zhong C. Hong Q. Li H. Rui B. et al (2021a). Nkx2-3 induces autophagy inhibiting proliferation and migration of vascular smooth muscle cells via AMPK/mTOR signaling pathway. J. Cell Physiol.236, 7342–7355. 10.1002/jcp.30400
308
Zheng L. Wang Z. Li Z. Wang M. Wang W. Chang G. (2021b). MicroRNA-130a inhibits proliferation of vascular smooth muscle cells by suppressing autophagy via ATG2B. J. Cell Mol. Med.25, 3829–3839. 10.1111/jcmm.16305
309
Zhou F. Wu Z. Zhao M. Murtazina R. Cai J. Zhang A. et al (2019). Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol.218, 1908–1927. 10.1083/jcb.201811173
310
Zhou J. Li L. Hu H. Wu J. Chen H. Feng K. et al (2020). Circ-HIPK2 accelerates cell apoptosis and autophagy in myocardial oxidative injury by sponging miR-485-5p and targeting ATG101. J. Cardiovasc Pharmacol.76, 427–436. 10.1097/FJC.0000000000000879
311
Zhou C. Wu Z. Du W. Que H. Wang Y. Ouyang Q. et al (2022). Recycling of autophagosomal components from autolysosomes by the recycler complex. Nat. Cell Biol.24, 497–512. 10.1038/s41556-022-00861-8
312
Zhu Y. Yang T. Duan J. Mu N. Zhang T. (2019). MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging (Albany NY)11, 1089–1109. 10.18632/aging.101766
313
Zhu Y. Zhu L. Wang X. Jin H. (2022). RNA-Based therapeutics: an overview and prospectus. Cell Death Dis.13, 644. 10.1038/s41419-022-05075-2
314
Zhu H. Liang H. Gao Z. Zhang X. He Q. He C. et al (2023). MiR-483-5p downregulation alleviates ox-LDL induced endothelial cell injury in atherosclerosis. BMC Cardiovasc Disord.23, 521. 10.1186/s12872-023-03496-1
315
Zhu W. Du W. Duan R. Liu Y. Zong B. Jin X. et al (2024). miR-873-5p suppression reinvigorates aging mesenchymal stem cells and improves cardiac repair after myocardial infarction. ACS Pharmacol. Transl. Sci.7, 743–756. 10.1021/acsptsci.3c00293
316
Zong J. Wang Y. Pan S. Yang Y. Peng J. Li F. et al (2023). The relationship between the serum NLRP1 level and coronary lesions in patients with coronary artery disease. Int. J. Clin. Pract.2023, 2250055. 10.1155/2023/2250055
Summary
Keywords
cardiovascular disease, autophagy, non-coding RNAs, aging, meta-analysis
Citation
Scalabrin S and Cagnin S (2025) Cardiovascular diseases in the elderly: possibilities for modulating autophagy using non-coding RNAs. Front. Cell Dev. Biol. 13:1520850. doi: 10.3389/fcell.2025.1520850
Received
31 October 2024
Accepted
09 June 2025
Published
31 July 2025
Volume
13 - 2025
Edited by
Xuehong Xu, Shaanxi Normal University, China
Reviewed by
Guifang Yan, Johns Hopkins Medicine, United States
Yi Xu, Washington University in St. Louis, United States
Updates
Copyright
© 2025 Scalabrin and Cagnin.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Cagnin, stefano.cagnin@unipd.it
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.