- 1School of Intelligent Medicine, China Medical University, Shenyang, Liaoning, China
- 2Center for Biomedical Engineering, School of Information Science and Technology, Fudan University, Shanghai, China
- 3Department of Medical Imaging, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning, China
- 4Public Health Supervision Technology Department, Liaoning Provincial Center for Disease Prevention and Control, Liaoning, China
- 5Department of Radiology, Shengjing Hospital of China Medical University, Shenyang, China
- 6Department of Radiology, The First Affiliated Hospital of Dalian Medical University, Dalian, China
- 7Department of Scientific Research and Academic, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, China
Objectives: Evaluating response to epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) is crucial in non-small cell lung cancer (NSCLC) patients with brain metastases (BM). To explore values of multi-sequence MRI in early assessing response to EGFR-TKIs in non-small cell lung cancer (NSCLC) patients with BM.
Approach: A primary cohort of 133 patients (January 2018 to March 2024) from center one and an external cohort of 52 patients (May 2017 to December 2022) from center two were established. Radiomics features were extracted from 4 mm brain-tumor interface (BTI) and whole BM region across T1-weighted contrast enhanced (T1CE) and T2-weighted (T2W) and T2 fluid-attenuated inversion recovery (T2-FLAIR) MRI sequences. The most relevant features were selected using the U test and least absolute shrinkage and selection operator (LASSO) method to develop the multi-sequence models based on BTI (RS-BTI-COM) and BM (RS-BM-COM). By integrating RS-BTI-COM with peritumoral edema volume (VPE), the combined model was built using logistic regression. Model performance was evaluated using the area under the ROC curve (AUC), sensitivity (SEN), specificity (SPE) and accuracy (ACC).
Main Results: The constructed RS-BTI-COM demonstrated a higher association with early response to EGFR-TKI therapy than RS-BM-COM. The combined RS-BTIplusVPE, incorporating BTI-based radiomics features and VPE, exhibited the highest AUCs (0.843–0.938), SPE (0.808–0.905) and ACC (0.712–0.875) in the training, internal validation, and external validation cohort, respectively.
Significance: The study developed a validated non-invasive model (RS-BTIplusVPE) based on integrating BTI-based radiomics features and VPE, which showed improved prediction of EGFR-TKI response in NSCLC patients with BM compared to tumor-focused models.
1 Introduction
Lung cancer has become the leading cause of cancer deaths globally, accounting for more than 25% of all cancer deaths each year (Brainard and Farver, 2019). Non-small cell lung cancer (NSCLC) is the most common subtype of lung cancer, accounting for approximately 85% of all lung cancers (Gridelli et al., 2015). Studies have shown that approximately 40% of NSCLC cases have distal metastases at the time of diagnosis (Little et al., 2007; Schuchert and Luketich, 2003). Brain or central nervous system as the most common site of metastasis in NSCLC with the incidence rate of 40%–50% (Sørensen et al., 1988; Yawn et al., 2003). The development of brain metastasis (BM) would severely lead to poor prognosis for NSCLC patient, with the median overall survival ranging between 2–9 months (Peters et al., 2016).
In recent years, epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) therapy has been shown to display superior efficacy compared to standard chemotherapy (Ettinger et al., 2017), and has profoundly impacted the therapeutic landscape of NSCLC (Yuan et al., 2019; Lynch et al., 2004). However, approximately one-third of TKI-treated NSCLC patients do not benefit from the EGFR-TKI therapy. Their tumors continue to have rapid progression despite the treatment (Kawaguchi et al., 2014; Spigel et al., 2017), which indicates that many patients may be at risk for rapid deterioration of clinical symptoms, delayed treatment, poor prognosis and even death (Yang et al., 2013).
The Response Evaluation Criteria for Solid Tumours (RECIST 1.1) provides an objective, standardized method for assessing the efficacy of EGFR-TKIs (Eisenhauer et al., 2009), and requires the visual assessment of the tumor size based on radiological imaging (Mayerhoefer et al., 2020). Magnetic Resonance Imaging (MRI) has been widely and routinely used to assess response to EGFR-TKI. However, it relies heavily on visual assessment, making it highly subjective and less accurate (Mayerhoefer et al., 2020; Chetan and Gleeson, 2021). This limitation arises from the lack of preoperative specific biomarkers in MRI imaging that can identify patients who would benefit from EGFR-TKI therapy. Previous studies (Hsiao et al., 2020; Guo et al., 2020; Li et al., 2015) have explored molecular markers for predicting EGFR-TKI efficacy, including soluble cadherin-3, genetic alterations, and COX-2 serum levels, but these markers have not been widely validated. For PET/CT imaging, recent studies (Zhu et al., 2022; Agüloğlu et al., 2022; Shao et al., 2020) have shown its potential in predicting treatment response and progression-free survival, but its role in predicting the response to EGFR-TKI therapy in BM remains unclear. Therefore, a novel method that enable early predict response to EGFR-TKI before treatment is essential for the development of an appropriately individualized treatment regimens.
Radiomics have been addressed to the field of precision medicine for making individual therapeutic decisions based on medical imaging data (Sharpton et al., 2014). Previous studies have shown that the development of radiomic can be helpful to identify valuable features associated with EGFR-TKI response, enabling non-invasive assessment of therapeutic efficacy of EGFR-TKI (Chetan and Gleeson, 2021; Sharpton et al., 2014; Zhang et al., 2023; Song et al., 2020; Mu et al., 2020; Song et al., 2018; Bera et al., 2022). Fan’s group recently conducted radiomic studies investigating the role of brain MRI on BM for assessment of response to EGFR-TKI (Fan et al., 2023a; Fan et al., 2022). However, the studies only evaluated T1-weighted contrast enhanced (T1CE) and T2-weighted (T2W) MRI, and neglected the potential value of brain T2 fluid-attenuated inversion recovery (T2-FLAIR) sequence. The T2-FLAIR sequence can effectively suppress cerebrospinal fluid signals and highlight adjacent lesions, and has been widely used in the diagnosis of central nervous system diseases (Tha et al., 2009). Moreover, the T2-FLAIR can quantify the degree of peritumoral edema and inflammation in BM (Zakaria et al., 2021). Previous reports have demonstrated that features derived from T2-FLAIR are strongly correlated with gene mutation status (Wang et al., 2021) and immune responses (Madi et al., 2022). While, the role of T2-FLAIR MRI on BM for predicting EGFR-TKI therapeutic efficacy has not yet been investigated.
Besides, the BM has a unique microenvironment (Eichler et al., 2011). The interface between brain parenchyma and tumor (brain-to-tumor interface, BTI) represents the area that metastatic tumor cells interact with endocranial brain cells and patient’s immune system (Berghoff et al., 2013). This region has garnered significant interest in recent years, with BTI-focused radiomics proven to effectively indicate the extent of brain invasion (Xiao et al., 2021b; Joo et al., 2021; Li et al., 2021) and assess tumor grading (Zhao et al., 2024). This underscores the potential utility of the BTI in computer-aided diagnosis of BM. This study aims to evaluate the value of the MRI image of BM for predicting response to EGFR-TKI therapy based on the BTI area in NSCLC patients. Our findings and developed models were further tested with an external validation set.
2 Materials and method
2.1 Patients
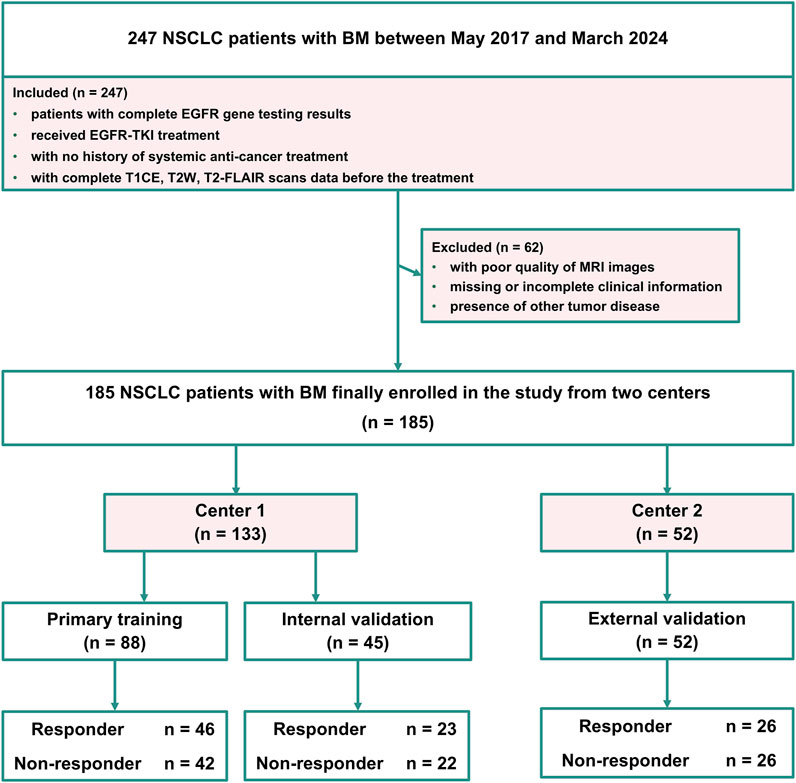
The MRI data of patients with BM included in this retrospective study were obtained with the approval of the local ethics committee (number: 20,220,659), and all patient data were anonymized to ensure confidentiality. Center one enrolled 133 patients with EGFR-mutant NSCLC with BM from January 2018 to March 2024, forming the primary cohort used to construct the training set and internal validation set. Center two included 52 patients with EGFR-mutant NSCLC with BM from May 2017 to December 2022, serving as an external validation cohort. The inclusion criteria for all the patients were as follows: (i) patients with complete EGFR gene testing results, (ii) received EGFR-TKI treatment, (iii) with no history of systemic anti-cancer treatment, and (iv) with complete T1CE, T2W and T2-FLAIR scans data before the treatment. Patients were excluded if they met any of the following criteria: (i) with poor quality of MRI images, (ii) missing or incomplete clinical information, or (iii) presence of other tumor disease. The response to EGFR-TKI treatment was evaluated using the RECIST 1.1 criteria (Therasse et al., 2000; Therasse et al., 2006). The primary cohort (Center 1) was divided into a training cohort and an internal validation cohort in a stratified 2:1 ratio. Stratified random sampling was performed using the strata function in R with the srswor method, ensuring random assignment without replacement. The external cohort (Center 2) was utilized for independent external validation of the developed radiomics models. Figure 1 shows the patient inclusion flowchart.
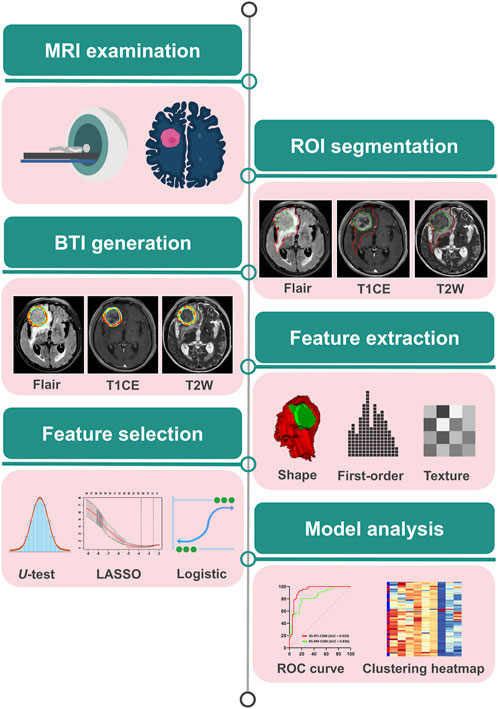
2.2 MRI protocol and tumor delineation
T1CE, T2W, and T2-FLAIR sequences were acquired on a 3.0 T scanner for image analysis. Details of the brain MRI scanning devices and parameters were listed in Supplementary Material 1. Regions of interest (ROIs) segmentation was performed by the first radiologist (Y.J.H., with 4 years’ experience), who had no knowledge of the patient’s clinicopathological information, except for the tumour location. ROIs of the BM and peritumoral edema area (PEA) in the brain were manually drawn using the open-source software ITK-SNAP (version 3.6.1, www.itksnap.org). All manual depictions were validated by the second radiologist (Z.G.Y., with 19 years’ experience).
2.3 BTI generation
We utilized a semi-automatic segmentation algorithm to delineate the BTI region. In the Python software, the manually outlined BM contour was eroded by 2 mm inwards and dilated by 2 mm outwards, resulting in a final annular region with a width of 4 mm. After obtaining the ROI of the BTI region, we calculate the entropy value of the BTI region to quantify the entropy of the pixel intensity distribution within the region, reflecting its texture complexity and information content. The formula for calculating the entropy is shown in the Supplementary Material 2. The peritumoral edema volume (VPE) was computed based on the PEA that was demarcated by using the ITK-SNAP software. In summary, the three regions used in this study are: (i) BM, manually delineated for feature extraction and BM-based model development; (ii) PEA, the edema region surrounding the BM, manually delineated to calculate the value of VPE; and (iii) BTI, an annular region derived by expanding and contracting the BM contour through a python script, containing information about the interface between the PEA and the active tumor region of BM, used for feature extraction and BTI-based model development. The representative brain MR images of three different sequences are shown in the (Supplementary Figure S1).
2.4 Feature extraction
First-order, texture, shape and filter features were extracted from the manually segmented tumor regions and the BTI region. The first-order features can provide information about the overall brightness, contrast, and distribution of an image. The texture features include 24 Gray-level co-occurrence matrix, 14 Gray-level dependence matrix, 16 Gray-level size zone matrix and 16 Gray-level run length matrix. The shape features quantitatively describe the 2D size and shape of the ROIs. To obtained filtered features, the original images were filtered by using eight filters, which include Wavelet, square, squareroot, laplacian of Gaussian, logarithm, gradient, exponential, and local binary pattern 2D/3D, then used to calculate textural and first-order features. Finally, a total of 1,967 radiomics features were generated for each MR sequence (T1CE, T2W and T2-FLAIR). Imaging preprocessing and feature calculating were performed using the Pyradiomics package (version 3.0.1) according to a previous report (van Griethuysen et al., 2017).
2.5 Feature selection and model construction
To select highly correlated and minimally redundant features, we employed the following feature selection strategy: Firstly, we applied the Mann-Whitney U test to the features, where features with a p-value <0.05 were considered significantly different and retained for further screening. Secondly, the most predictive features were determined using the least absolute shrinkage and selection operator (LASSO) with ten-fold cross-validation (Tibshirani, 1997). Thirdly, we identified the most predictive features using logistic regression with stepwise selection based on Akaike Information Criterion (AIC) minimization, derived from the BTI and BM regions (Pan, 2001). Finally, we evaluated these most predictive features using intra-class correlation coefficient (ICC) (Leijenaar et al., 2013). We randomly selected thirty patients to assess the reproducibility of the selected features. Features with an ICC >0.80 were considered to have better reproducibility and were retained for further construction. More detailed information on the ICC calculation can be found in the Supplementary Material 1.
In the subsequent model construction process, we predicted the response to EGFR-TKI therapy using a logistic regression classifier, a widely accepted and effective machine learning classifier, implemented using the glmnet package in R (Bera et al., 2022). Specifically, the selected most predictivefeatures were used to develop radiomics models based on the BTI region, the combined radiomics signatures (RSs) named RS-BTI-COM was constructed by integrating all the features from T1CE, T2W and T2-FLAIR sequences to predict response to EGFR-TKI treatment. Similarly, the RS-BM-COM were established based on BM. Finally, the VPE was incorporated into the RS-BTI-COM as a clinical model, resulting in a new model that integrates both radiomics features and VPE, referred to as RS-BTIplusVPE.
2.6 Statistical analysis
Clinical factors between responders and non-responders were statistically analyzed using SPSS (version 27.0) and R language (version 4.2.2). The Mann-Whitney U test and chi-square test were used for continuous and categorical variables, respectively. A p-value of less than 0.05 was considered statistically significant. Receiver Operating Characteristic (ROC) curves and Area Under Curve (AUC) were used to assess the ability of features to predict response to EGFR-TKIs. Delong’s test (DeLong et al., 1988) was used to compare differences in AUC values of the models. Figure 2 shows the study design. This study was approved by the ethics committee on 23 August 2022.
3 Results
3.1 Patient characteristics
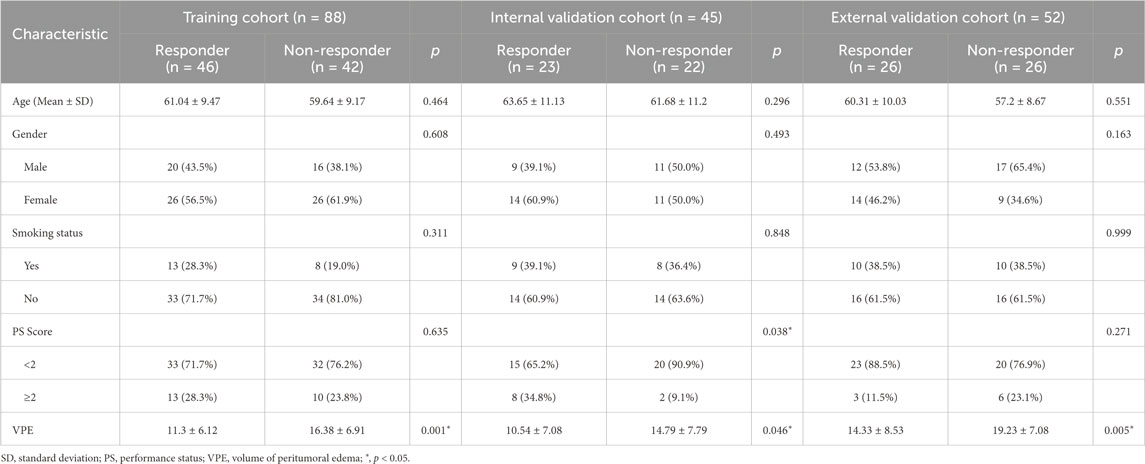
Table 1 listed the demographic and clinical characteristics of all patients with BM from NSCLC in both centers. There was no statistical significance in age, gender, smoking status and performance status (PS) of the patients. While, for predicting response to EGFR-TKI, a significant difference (p < 0.05) in VPE was found between the responder and non-responder groups across all three cohorts, which suggests that VPE may be correlated with patients’ responses to EGFR-TKI treatment (Fan et al., 2023b).
3.2 Prediction performance of the multi-sequence radiomics signatures
A total of nine radiomics features were selected from the BTI region as the most predictive features with LASSO logistic regression. Supplementary Material 2 shows the process of LASSO-based selection of the features (Supplementary Figure S2). From T1CE, T2W and T2-FLAIR MRI, there were four, one and four features were identified as the most predictive features and used to establish the combined radiomics models (RS-BTI-COM). Formulas of the developed radiomics models were listed in Supplementary Material 1.
Table 2 compares predictive performance of the constructed RSs based on the BTI and BM region. The multi-sequence fused RS-BTI-COM achieves higher AUC, SEN, SPE and ACC compared with RS-BM-COM. A possible explanation is that the invasion of metastatic brain tumor cells involves interactions with the brain microenvironment, including changes in astrocytes and vascular structures. The invasive process may lead to dynamic alterations of the BTI, which could be associated with the response to EGFR-TKI (Lorger and Felding-Habermann, 2009). Figure 3 depicted ROC curves of the established RS-BTI-COM and RS-BM-COM.
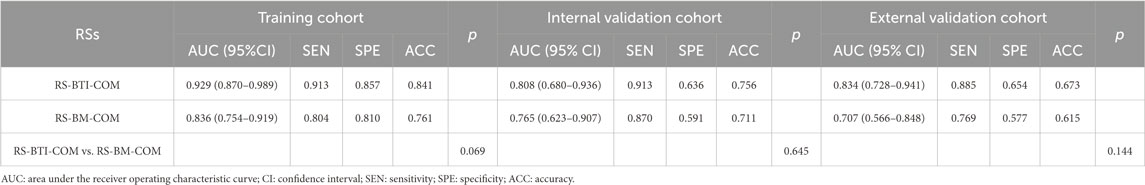
Table 2. Performance of the developed models based on BM and BTI for determining response to EGFR-TKI.
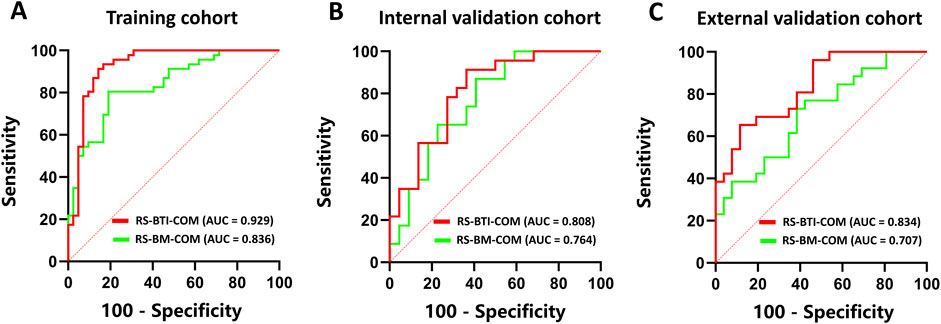
Figure 3. ROC curves of RS-BTI-COM and RS-BM-COM illustrating the performance of radiomics models for predicting response to EGFR-TKI in the training (A), internal validation (B), and external validation (C) cohorts.
3.3 Prediction performance of the combined model
The RS-BTI-COM was then integrated with VPE to establish the RS-BTIplusVPE. Table 3 compared performances of the VPE, RS-BTI-COM and RS-BTIplusVPE. When the VPE was used alone to predict response to EGFR-TKI, the AUCs yielded were ranged from 0.674 to 0.725 in primary and external cohorts. The RS-BTIplusVPE exhibited the highest AUCs (0.843–0.938), SPE (0.808–0.905) and ACC (0.712–0.875). Delong’s test indicates a significant difference (p < 0.05) between VPE and RS-BTIplusVPE in both the training and internal validation cohorts. These results demonstrated that VPE may serve as a clinical indicator, providing complementary information for BTI-based radiomic models. Figure 4 depicted ROC curves of the VPE and RS-BTIplusVPE.
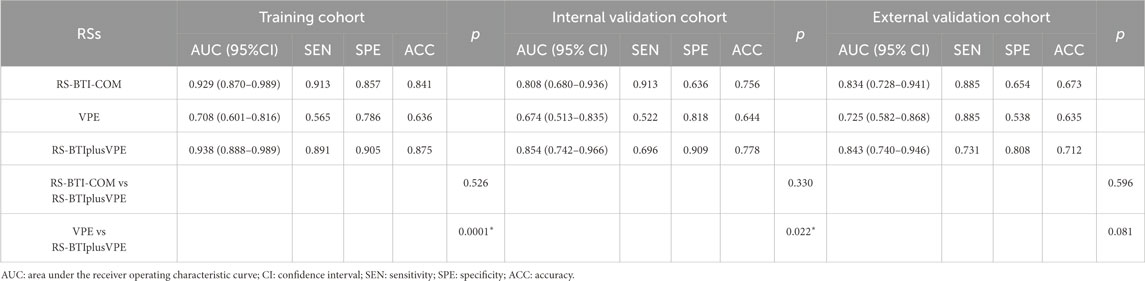
Table 3. Performance of VPE, RS-BTI-COM and combined models for determining the response to EGFR-TKI.
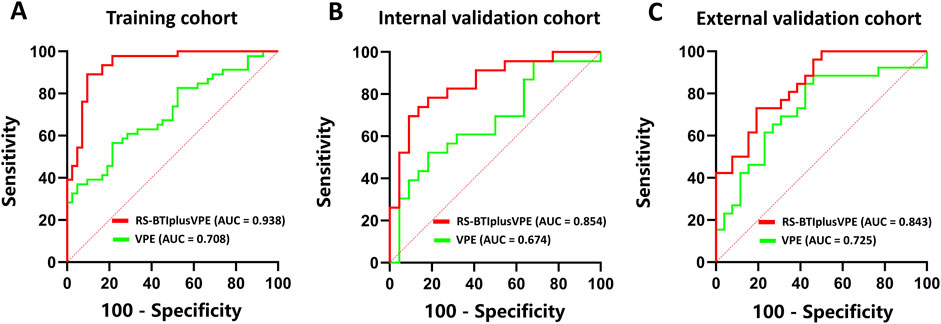
Figure 4. ROC curves of VPE and RS-BTIplusVPE in the training (A), internal validation (B) and external validation (C) cohorts.
3.4 Radiomics features analysis
A total of nine radiomics features were identified as key predictors for determining the response to EGFR-TKI treatment. Among these, eight features were classified under the textural category, reflecting complex patterns within the image data, while only the remaining one features belonged to the first-order category, capturing essential statistical information from the voxel intensity distributions. These selected features play a crucial role in differentiating between responders and non-responders to EGFR-TKI therapy. Detailed performance metrics of the identified features are summarized in Table 4. The detailed meanings of the identified features are provided in Supplementary Material 1. Supplementary Material 2 shows cluster analyses of the selected radiomics features (Supplementary Figure S3).
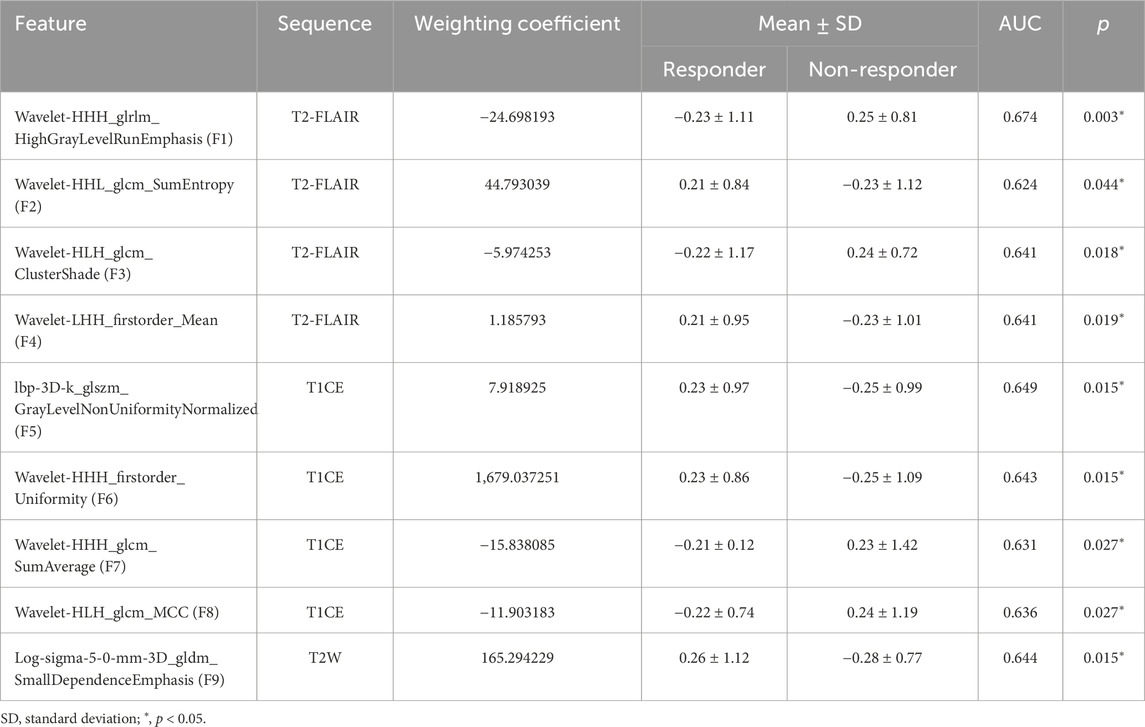
Table 4. Performance metrics of the selected features for predicting EGFR-TKI response in NSCLC patients with BM in the training cohort.
4 Discussion
NSCLC is the most common subtype of lung cancer, therefore, early assessment of the response to targeted therapy can significantly benefit the personalized treatment of NSCLC patients (Ai et al., 2018). Most previous studies have focused on predicting the efficacy of EGFR-TKI therapy only based on the primary lung tumor (Wang et al., 2019; Zhao et al., 2017; Chen et al., 2022; Xu et al., 2019). Fan et al. (Fan et al., 2022; Fan et al., 2023b; Fan et al., 2023a) have revealed that important information for predicting the response to EGFR-TKI therapy also exists within BM. However, these studies have primarily concentrated on T1-weighted and T2W MRI sequences. The potential of T2-FLAIR in reflecting the microenvironment and medical mechanisms of brain metastases remains unexplored. In this study, the RS-BTIplusVPE model was developed and validated for the early prediction of the response to EGFR-TKI therapy in NSCLC patients with BM, which integrating BTI radiomics features from T1CE, T2W and T2-FLAIR MRI sequences and VPE. The RS-BTIplusVPE model outperformed BTI-based model, demonstrating that VPE is significantly associated with the response to EGFR-TKI therapy (p < 0.05). Additionally,our research confirmed that the BTI region exhibits greater heterogeneity than whole BM area, containing more information that plays a positive role in predicting EGFR-TKI treatment response. Furthermore, we found that incorporating the T2-FLAIR MRI sequence enables the extraction of valuable features from the BTI region more effectively.
BTI is a biologically active zone where tumor cells invade and spread into adjacent brain tissue while interacting with the surrounding brain microenvironment, including immune components (Tabassum et al., 2023). At this interface, tumor cells can also evade immune surveillance by expressing immunosuppressive molecules, such as PD-L1, thereby promoting tumor growth and dissemination (Xiao et al., 2021b). Studies have shown (Li et al., 2023) that the activity of immune cells (e.g., macrophages and T cells) at the tumor margins within the BTI microenvironment can directly impact the patient’s therapeutic prognosis. This interaction between the BTI microenvironment and the host immune system may provide valuable insights into tumor behavior under EGFR-TKI treatment, suggesting that the BTI region holds more predictive value than features derived solely from the tumor mass of BM. Additionally, peritumoral edema is another common feature in the BTI, often associated with pro-inflammatory and angiogenic factors (e.g., VEGF) released by tumor cells, which compromise vascular integrity and lead to fluid leakage into the brain parenchyma (Chen et al., 2024). MRI-based studies (Joo et al., 2021) have indicated that radiomic features of the BTI, such as tumor margin blurriness, are closely related to tumor invasiveness, providing critical information for assessing tumor progression and predicting response to targeted therapies. Our findings also suggest that the BTI in BMs likely contains important information relevant to therapeutic response, further supporting the potential of BTI-based radiomics in improving clinical decision-making for targeted treatments. Further research is needed to deepen our understanding of the biological mechanisms underlying these findings.
The incorporation of the T2-FLAIR sequence in brain MRI is crucial for extracting high-value features from the BTI region. T2-FLAIR is particularly effective at suppressing cerebrospinal fluid signals, thereby enhancing the visibility of peritumoral edema and other abnormal brain tissues (Zhang et al., 2021). Given that peritumoral edema is a common feature in the BTI region, T2-FLAIR allows for the extraction of valuable radiomic features that are difficult to capture using other sequences like T1 or T2-weighted images. Studies have shown that combining T2-FLAIR with radiomic analysis significantly improves the accuracy of predicting tumor invasiveness and treatment response, especially in gliomas and BMs (Dvořák, 2015; Rajkumar and Kavitha, 2011). The enhanced contrast between abnormal and normal brain tissues provided by T2-FLAIR facilitates more precise segmentation and feature extraction in the BTI, which is critical for evaluating the biological behavior of the tumor (Xiao et al., 2021a). Moreover, T2-FLAIR-based analysis of the BTI has been associated with more accurate predictions of therapeutic outcomes. By capturing subtle changes at the tumor-brain interface, T2-FLAIR-derived features offer deeper insights into tumor invasiveness and progression, supporting the development of more personalized treatment strategies (Aboian et al., 2022).
In feature analysis of this study, we found that eight texture features and only one first-order extracted from the BTI region showed significant differences between patients with and without a response to EGFR-TKI therapy (p < 0.05). According to the texture feature analysis, we found that four texture features extracted from the gray-level co-occurrence matrix (GLCM) in the BTI region showed significant differences between patients with and without a response to EGFR-TKI therapy (p < 0.05). These GLCM-based features reflect the disorder or randomness of intensity values, which are typically associated with varying cellular densities or the presence of different tissue types (e.g., tumor infiltration) (Qin et al., 2017). This finding suggests that the heterogeneity within the BTI region is closely related to the efficacy of targeted therapy observed in this study. Moreover, the only first-order feature extracted from the T2-FLAIR sequence within the BTI region, which reflects the average intensity, effectively reveals the pattern of edema spread and its predictive value for response to EGFR-TKI (Liu et al., 2021). This study also found a correlation between the value of VPE and treatment response (p < 0.05) across all three cohorts, suggesting that features from this region could serve as potential predictive markers of therapeutic efficacy.
We assessed the value of VPE in predicting the response to EGFR-TKI therapy in NSCLC patients with brain metastases. While VPE has been recognized as an important imaging marker in differentiating between primary brain tumors and BMs (Baris et al., 2016), its role in predicting therapeutic outcomes remains uncertain. Our analysis showed that although VPE contributed to model performance, its predictive ability was relatively limited compared to other radiomic features (such as those derived from the BTI region). VPE reflects the extent of vasogenic edema surrounding the tumor, which is associated with factors like tumor-induced disruption of the blood-brain barrier and local inflammation (Esquenazi et al., 2017). However, it does not fully capture the biological complexity related to EGFR-TKI response. This suggests that while VPE is a useful imaging marker for assessing tumor burden and edema, its contribution to predicting EGFR-TKI response is limited, and further research is needed to explore its underlying biological mechanisms.
This study has several limitations. First, the retrospective nature of the study and the limited sample size necessitate further validation of the model using prospective data. Second, although the BTI region were derived from MR images, pathological confirmation was not performed due to the study’s retrospective design. Third, while BTI region segmentation was automated, tumor and peritumoral edema segmentation was done manually, introducing potential subjective bias. Fourth, future work will focus on exploring the correlations between MRI-based morphological variations and histological microstructure to enhance the intratumoral partitioning algorithm. Fifth, although this study suggests a potential correlation between the 4 mm BTI and the efficacy of EGFR-TKI treatment, it lacks the determination of the optimal expansion distance for generating the BTI ROI. Lastly, future studies should further investigate the relationship between tumor heterogeneity in brain tissue and EGFR-TKI treatment response, aiming to explore the underlying biological mechanisms and enhance the interpretability of radiomics models.
5 Conclusion
In conclusion, our study developed a validated non-invasive model (RS-BTIplusVPE) by integrating multi-sequence radiomic model and VPE, which showed improved prediction of EGFR-TKI response in NSCLC patients with brain metastases compared to tumor-focused models. Our findings were validated with clinically obtained data from two centers, which may indicate good potential of our model for assisting in clinical decision-making.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: The datasets generated and/or analysed during the current study are not publicly available due to restrictions but are available from the corresponding authors on reasonable request. Requests to access these datasets should be directed to WJ, eGlhb3lhODM5MjFAMTYzLmNvbQ==.
Ethics statement
The studies involving humans were approved by Cancer Hospital of China Medical University (20220806YG). The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’; legal guardians/next of kin because this retrospective study was approved by the Medical Ethics Committee of Cancer Hospital of China Medical University (20220806YG), and informed consent was waived.
Author contributions
CY: Conceptualization, Methodology, Writing – original draft. YS: Validation, Writing – original draft. MJ: Software, Writing – original draft. YF: Validation, Writing – review and editing. YH: Resources, Writing – review and editing. QZ: Resources, Writing – review and editing. YZ: Data curation, Writing – review and editing. YW: Writing – review and editing. XJ: Conceptualization, Writing – review and editing. ZW: Resources, Writing – review and editing. ZY: Investigation, Writing – review and editing. BS: Investigation, Writing – review and editing. WJ: Data curation, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Science and Technology Joint Program Fund Project of Liaoning (2023JH2/101700175) and General Program from Department of Education of Liaoning Province (JYTMS20230132).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1525989/full#supplementary-material
References
Aboian, M., Bousabarah, K., Kazarian, E., Zeevi, T., Holler, W., Merkaj, S., et al. (2022). Clinical implementation of artificial intelligence in neuroradiology with development of a novel workflow-efficient picture archiving and communication system-based automated brain tumor segmentation and radiomic feature extraction. Front. Neurosci. 16, 860208. doi:10.3389/fnins.2022.860208
Agüloğlu, N., Akyol, M., Kömek, H., and Katgi̇, N. (2022). The prognostic value of 18F-FDG PET/CT metabolic parameters in predicting treatment response before EGFR TKI treatment in patients with advanced lung adenocarcinoma. Mol. Imaging Radionucl. Ther. 31 (2), 104–113. doi:10.4274/mirt.galenos.2022.24650
Ai, X., Guo, X., Wang, J., Stancu, A. L., Joslin, P. M. N., Zhang, D., et al. (2018). Targeted therapies for advanced non-small cell lung cancer. Oncotarget 9, 37589–37607. doi:10.18632/oncotarget.26428
Baris, M. M., Celik, A. O., Gezer, N. S., and Ada, E. (2016). Role of mass effect, tumor volume and peritumoral edema volume in the differential diagnosis of primary brain tumor and metastasis. Clin. Neurol. Neurosurg. 148, 67–71. doi:10.1016/j.clineuro.2016.07.008
Bera, K., Braman, N., Gupta, A., Velcheti, V., and Madabhushi, A. (2022). Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat. Rev. Clin. Oncol. 19, 132–146. doi:10.1038/s41571-021-00560-7
Berghoff, A. S., Rajky, O., Winkler, F., Bartsch, R., Furtner, J., Hainfellner, J. A., et al. (2013). Invasion patterns in brain metastases of solid cancers. Neuro Oncol. 15, 1664–1672. doi:10.1093/neuonc/not112
Brainard, J., and Farver, C. (2019). The diagnosis of non-small cell lung cancer in the molecular era. Mod. Pathol. 32, 16–26. doi:10.1038/s41379-018-0156-x
Chen, N., Li, R., Jiang, M., Guo, Y., Chen, J., Sun, D., et al. (2022). Progression-free survival prediction in small cell lung cancer based on radiomics analysis of contrast-enhanced CT. Front. Med. (Lausanne) 9, 833283. doi:10.3389/fmed.2022.833283
Chen, Y., Lin, H., Sun, J., Pu, R., Zhou, Y., and Sun, B. J. A. R. (2024). Texture feature differentiation of glioblastoma and solitary brain metastases based on tumor and tumor-brain interface. Acad. Radiol. 32, 400–410. doi:10.1016/j.acra.2024.08.025
Chetan, M. R., and Gleeson, F. V. (2021). Radiomics in predicting treatment response in non-small-cell lung cancer: current status, challenges and future perspectives. Eur. Radiol. 31, 1049–1058. doi:10.1007/s00330-020-07141-9
Delong, E. R., Delong, D. M., and Clarke-Pearson, D. L. (1988). Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845. doi:10.2307/2531595
DvořáK, P. (2015). “Brain tumor detection and segmentation in multisequence MRI,”. Ph. D. Thesis (Brno: Electrical Engineering and Communication).
Eichler, A. F., Chung, E., Kodack, D. P., Loeffler, J. S., Fukumura, D., and Jain, R. K. (2011). The biology of brain metastases-translation to new therapies. Nat. Rev. Clin. Oncol. 8, 344–356. doi:10.1038/nrclinonc.2011.58
Eisenhauer, E. A., Therasse, P., Bogaerts, J., Schwartz, L. H., Sargent, D., Ford, R., et al. (2009). New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247. doi:10.1016/j.ejca.2008.10.026
Esquenazi, Y., Lo, V. P., and Lee, K. (2017). Critical care management of cerebral edema in brain tumors. J. Intensive Care Med. 32, 15–24. doi:10.1177/0885066615619618
Ettinger, D. S., Wood, D. E., Aisner, D. L., Akerley, W., Bauman, J., Chirieac, L. R., et al. (2017). Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 15, 504–535. doi:10.6004/jnccn.2017.0050
Fan, Y., Wang, X., Dong, Y., Cui, E., Wang, H., Sun, X., et al. (2023a). Multiregional radiomics of brain metastasis can predict response to EGFR-TKI in metastatic NSCLC. Eur. Radiol. 33, 7902–7912. doi:10.1007/s00330-023-09709-7
Fan, Y., Wang, X., Yang, C., Chen, H., Wang, H., Wang, X., et al. (2023b). Brain-tumor interface-based MRI radiomics models to determine EGFR mutation, response to EGFR-TKI and T790M resistance mutation in non-small cell lung carcinoma brain metastasis. J. Magn. Reson Imaging 58, 1838–1847. doi:10.1002/jmri.28751
Fan, Y., Zhao, Z., Wang, X., Ai, H., Yang, C., Luo, Y., et al. (2022). Radiomics for prediction of response to EGFR-TKI based on metastasis/brain parenchyma (M/BP)-interface. Radiol. Med. 127, 1342–1354. doi:10.1007/s11547-022-01569-3
Gridelli, C., Rossi, A., Carbone, D. P., Guarize, J., Karachaliou, N., Mok, T., et al. (2015). Non-small-cell lung cancer. Nat. Rev. Dis. Prim. 1, 15009. doi:10.1038/nrdp.2015.9
Guo, Y., Song, J., Wang, Y., Huang, L., Sun, L., Zhao, J., et al. (2020). Concurrent genetic alterations and other biomarkers predict treatment efficacy of EGFR-TKIs in EGFR-mutant non-small cell lung cancer: a review. Front. Oncol. 10, 610923. doi:10.3389/fonc.2020.610923
Hsiao, T. F., Wang, C. L., Wu, Y. C., Feng, H. P., Chiu, Y. C., Lin, H. Y., et al. (2020). Discovery of survival predictor and early monitoring biomarkers for EGFR tyrosine kinase inhibitor therapy in lung adenocarcinoma by integrative omics analysis. FASEB J. 34 (S1), 1. doi:10.1096/fasebj.2020.34.s1.08868
Joo, L., Park, J. E., Park, S. Y., Nam, S. J., Kim, Y. H., Kim, J. H., et al. (2021). Extensive peritumoral edema and brain-to-tumor interface MRI features enable prediction of brain invasion in meningioma: development and validation. Neuro Oncol. 23, 324–333. doi:10.1093/neuonc/noaa190
Kawaguchi, T., Ando, M., Asami, K., Okano, Y., Fukuda, M., Nakagawa, H., et al. (2014). Randomized phase III trial of erlotinib versus docetaxel as second- or third-line therapy in patients with advanced non-small-cell lung cancer: docetaxel and Erlotinib Lung Cancer Trial (DELTA). J. Clin. Oncol. 32, 1902–1908. doi:10.1200/JCO.2013.52.4694
Leijenaar, R. T., Carvalho, S., Velazquez, E. R., Van Elmpt, W. J., Parmar, C., Hoekstra, O. S., et al. (2013). Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 52, 1391–1397. doi:10.3109/0284186X.2013.812798
Li, H., Wang, Y., Su, F., Li, J., and Gong, P. (2015). Monitoring of cyclooxygenase-2 levels can predict EGFR mutations and the efficacy of EGFR-TKI in patients with lung adenocarcinoma. Int. J. Clin. Exp. Pathology 8 (5), 5577–5583.
Li, N., Mo, Y., Huang, C., Han, K., He, M., Wang, X., et al. (2021). A clinical semantic and radiomics nomogram for predicting brain invasion in WHO grade II meningioma based on tumor and tumor-to-brain interface features. Front. Oncol. 11, 752158. doi:10.3389/fonc.2021.752158
Li, S., Zhang, B., Zhang, P., Xue, C., Deng, J., Liu, X., et al. (2023). Postoperative progression of intracranial grade II–III solitary fibrous tumor/hemangiopericytoma: predictive value of preoperative magnetic resonance imaging semantic features. Acta Radiol. 64, 301–310. doi:10.1177/02841851211066757
Little, A. G., Gay, E. G., Gaspar, L. E., and Stewart, A. K. (2007). National survey of non-small cell lung cancer in the United States: epidemiology, pathology and patterns of care. Lung Cancer 57, 253–260. doi:10.1016/j.lungcan.2007.03.012
Liu, J., Han, H., Xu, Y., Jin, Y., Ma, F., Mu, J., et al. (2021). A comparison of the multimodal magnetic resonance imaging features of brain metastases vs. high-grade gliomas. Am. J. Transl. Res. 13, 3543–3548.
Lorger, M., and Felding-Habermann, B. J. C. R. (2009). Capturing changes in the brain microenvironment during initial tumor cell invasion. Cancer Res. 69, 4162. doi:10.1158/0008-5472.sabcs-09-4162
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., et al. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139. doi:10.1056/NEJMoa040938
Madi, S., Xie, F., Farhangi, K., Hsu, C. Y., Cheng, S. H., Aweda, T., et al. (2022). MRI/PET multimodal imaging of the innate immune response in skeletal muscle and draining lymph node post vaccination in rats. Front. Immunol. 13, 1081156. doi:10.3389/fimmu.2022.1081156
Mayerhoefer, M. E., Materka, A., Langs, G., HäGGSTRöM, I., Szczypiński, P., Gibbs, P., et al. (2020). Introduction to radiomics. J. Nucl. Med. 61, 488–495. doi:10.2967/jnumed.118.222893
Mu, W., Jiang, L., Zhang, J., Shi, Y., Gray, J. E., Tunali, I., et al. (2020). Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 11, 5228. doi:10.1038/s41467-020-19116-x
Pan, W. (2001). Akaike's information criterion in generalized estimating equations. Biometrics 57, 120–125. doi:10.1111/j.0006-341x.2001.00120.x
Peters, S., Bexelius, C., Munk, V., and Leighl, N. (2016). The impact of brain metastasis on quality of life, resource utilization and survival in patients with non-small-cell lung cancer. Cancer Treat. Rev. 45, 139–162. doi:10.1016/j.ctrv.2016.03.009
Qin, J. B., Liu, Z., Zhang, H., Shen, C., Wang, X. C., Tan, Y., et al. (2017). Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences. Med. Sci. Monit. 23, 2168–2178. doi:10.12659/msm.901270
Rajkumar, S., and Kavitha, S. (2011). Quantitative analysis for the identification of brain tumor in CT and MRI medical images. Int. J. Adv. Res. Comput. Sci., 2, 159–165.
Schuchert, M. J., and Luketich, J. D. (2003). Solitary sites of metastatic disease in non-small cell lung cancer. Curr. Treat. Options Oncol. 4, 65–79. doi:10.1007/s11864-003-0033-8
Shao, D., Cheng, Y., Yuan, Z. S., Jiang, B. Y., and Wang, S. X. (2020). Value of interim 18F-FDG PET/CT for predicting progression-free survival in stage ⅢB/IV EGFR-mutant non-small-cell lung cancer patients with EGFR-TKI therapy. Lung Cancer 149, 137–143. doi:10.1016/j.lungcan.2020.09.020
Sharpton, S. R., Oermann, E. K., Moore, D. T., Schreiber, E., Hoffman, R., Morris, D. E., et al. (2014). The volumetric response of brain metastases after stereotactic radiosurgery and its post-treatment implications. Neurosurgery 74, 9–16. doi:10.1227/NEU.0000000000000190
Song, J., Shi, J., Dong, D., Fang, M., Zhong, W., Wang, K., et al. (2018). A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin. Cancer Res. 24, 3583–3592. doi:10.1158/1078-0432.CCR-17-2507
Song, J., Wang, L., Ng, N. N., Zhao, M., Shi, J., Wu, N., et al. (2020). Development and validation of a machine learning model to explore tyrosine kinase inhibitor response in patients with stage IV EGFR variant-positive non-small cell lung cancer. JAMA Netw. Open 3, e2030442. doi:10.1001/jamanetworkopen.2020.30442
SøRENSEN, J. B., Hansen, H. H., Hansen, M., and Dombernowsky, P. (1988). Brain metastases in adenocarcinoma of the lung: frequency, risk groups, and prognosis. J. Clin. Oncol. 6, 1474–1480. doi:10.1200/JCO.1988.6.9.1474
Spigel, D. R., Edelman, M. J., O'Byrne, K., Paz-Ares, L., Mocci, S., Phan, S., et al. (2017). Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non-small-cell lung cancer: METLung. J. Clin. Oncol. 35, 412–420. doi:10.1200/JCO.2016.69.2160
Tabassum, M., Suman, A., Suero Molina, E., Pan, E., Di Ieva, A., and Liu, S. (2023). Radiomics and machine learning in brain tumors and their habitat: a systematic review. Cancers 15, 3845. doi:10.3390/cancers15153845
Tha, K. K., Terae, S., Kudo, K., and Miyasaka, K. (2009). Differential diagnosis of hyperintense cerebrospinal fluid on fluid-attenuated inversion recovery images of the brain. Part II: non-pathological conditions. Br. J. Radiol. 82, 610–614. doi:10.1259/bjr/29238647
Therasse, P., Arbuck, S. G., Eisenhauer, E. A., Wanders, J., Kaplan, R. S., Rubinstein, L., et al. (2000). New guidelines to evaluate the response to treatment in solid tumors. European organization for research and treatment of cancer, national cancer institute of the United States, national cancer institute of Canada. J. Natl. Cancer Inst. 92, 205–216. doi:10.1093/jnci/92.3.205
Therasse, P., Eisenhauer, E. A., and Verweij, J. (2006). RECIST revisited: a review of validation studies on tumour assessment. Eur. J. Cancer 42, 1031–1039. doi:10.1016/j.ejca.2006.01.026
Tibshirani, R. (1997). The lasso method for variable selection in the Cox model. Stat. Med. 16, 385–395. doi:10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3
Van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., et al. (2017). Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107. doi:10.1158/0008-5472.CAN-17-0339
Wang, G., Wang, B., Wang, Z., Li, W., Xiu, J., Liu, Z., et al. (2021). Radiomics signature of brain metastasis: prediction of EGFR mutation status. Eur. Radiol. 31, 4538–4547. doi:10.1007/s00330-020-07614-x
Wang, S., Shi, J., Ye, Z., Dong, D., Yu, D., Zhou, M., et al. (2019). Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 53, 1800986. doi:10.1183/13993003.00986-2018
Xiao, D., Wang, J., Wang, X., Fu, P., Zhao, H., Yan, P., et al. (2021a). Distinguishing brain abscess from necrotic glioblastoma using MRI-based intranodular radiomic features and peritumoral edema/tumor volume ratio, J. Integr. Neurosci. 20, 623–634. doi:10.31083/j.jin2003066
Xiao, D., Zhao, Z., Liu, J., Wang, X., Fu, P., Le Grange, J. M., et al. (2021b). Diagnosis of invasive meningioma based on brain-tumor interface radiomics features on brain MR images: a multicenter study. Front. Oncol. 11, 708040. doi:10.3389/fonc.2021.708040
Xu, Y., Hosny, A., Zeleznik, R., Parmar, C., Coroller, T., Franco, I., et al. (2019). Deep learning predicts lung cancer treatment response from serial medical imaging. Clin. Cancer Res. 25, 3266–3275. doi:10.1158/1078-0432.CCR-18-2495
Yang, J. J., Chen, H. J., Yan, H. H., Zhang, X. C., Zhou, Q., Su, J., et al. (2013). Clinical modes of EGFR tyrosine kinase inhibitor failure and subsequent management in advanced non-small cell lung cancer. Lung Cancer 79, 33–39. doi:10.1016/j.lungcan.2012.09.016
Yawn, B. P., Wollan, P. C., Schroeder, C., Gazzuola, L., and Mehta, M. (2003). Temporal and gender-related trends in brain metastases from lung and breast cancer. Minn Med. 86, 32–37.
Yuan, M., Huang, L. L., Chen, J. H., Wu, J., and Xu, Q. (2019). The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target Ther. 4, 61. doi:10.1038/s41392-019-0099-9
Zakaria, R., Radon, M., Mills, S., Mitchell, D., Palmieri, C., Chung, C., et al. (2021). The role of the immune response in brain metastases: novel imaging biomarkers for immunotherapy. Front. Oncol. 11, 711405. doi:10.3389/fonc.2021.711405
Zhang, L., Zhu, Y., Qi, Y., Wan, L., Ren, L., Zhu, Y., et al. (2021). T(2)-Weighted whole-brain intracranial vessel wall imaging at 3 tesla with cerebrospinal fluid suppression. Front. Neurosci. 15, 665076. doi:10.3389/fnins.2021.665076
Zhang, X., Lu, B., Yang, X., Lan, D., Lin, S., Zhou, Z., et al. (2023). Prognostic analysis and risk stratification of lung adenocarcinoma undergoing EGFR-TKI therapy with time-serial CT-based radiomics signature. Eur. Radiol. 33, 825–835. doi:10.1007/s00330-022-09123-5
Zhao, D., Chen, X., Qin, N., Su, D., Zhou, L., Zhang, Q., et al. (2017). The prognostic role of EGFR-TKIs for patients with advanced non-small cell lung cancer. Sci. Rep. 7, 40374. doi:10.1038/srep40374
Zhao, Z., Nie, C., Zhao, L., Xiao, D., Zheng, J., Zhang, H., et al. (2024). Multi-parametric MRI-based machine learning model for prediction of WHO grading in patients with meningiomas. Eur. Radiol. 34, 2468–2479. doi:10.1007/s00330-023-10252-8
Keywords: T2-FLAIR, brain metastasis, TKI therapy, MRI, radiomics
Citation: Yang C, Sun Y, Jiang M, Fan Y, Hu Y, Zhang Q, Zhang Y, Wang Y, Jiang X, Wang Z, Yang Z, Sun B and Jiang W (2025) FLAIR-based radiomics signature from brain-tumor interface for early prediction of response to EGFR-TKI therapy in NSCLC patients with brain metastasis. Front. Cell Dev. Biol. 13:1525989. doi: 10.3389/fcell.2025.1525989
Received: 11 November 2024; Accepted: 18 April 2025;
Published: 14 May 2025.
Edited by:
Manisha Singh, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
David Wasilewski, Charité University Medicine Berlin, GermanyStathis Hadjidemetriou, University of Limassol, Cyprus
Copyright © 2025 Yang, Sun, Jiang, Fan, Hu, Zhang, Zhang, Wang, Jiang, Wang, Yang, Sun and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenyan Jiang, eGlhb3lhODM5MjFAMTYzLmNvbQ==; Zhiguang Yang, eWFuZ3pnQHNqLWhvc3BpdGFsLm9yZw==; Bo Sun, c3VuYm95Y211QDE2My5jb20=
†These authors share first authorship
 Chunna Yang1†
Chunna Yang1† Ying Fan
Ying Fan Xiran Jiang
Xiran Jiang Zhiguang Yang
Zhiguang Yang Bo Sun
Bo Sun Wenyan Jiang
Wenyan Jiang