Abstract
Metastasis remains a primary cause of cancer-related mortality, with its intricate mechanisms continuing to be uncovered through advancing research. Among the various regulatory processes involved, RNA modification has emerged as a critical epitranscriptomic mechanism influencing cancer metastasis. N6-methyladenosine (m6A), recognized as one of the most prevalent and functionally significant RNA modifications, plays a central role in the regulation of RNA metabolism. In this review, we explore the multifaceted role of m6A in the different stages of cancer metastasis, including epithelial–mesenchymal transition, invasion, migration, and colonization. In addition to summarizing the current state of our understanding, we offer insights into how m6A modifications modulate key oncogenic pathways, highlighting the implications of recent discoveries for therapeutic interventions. Furthermore, we critically assess the limitations of previous studies and propose areas for future research, including the potential for targeting m6A as a novel approach in anti-metastatic therapies. Our analysis provides a comprehensive understanding of the regulatory landscape of m6A in metastasis, offering directions for continued exploration in this rapidly evolving field.
Introduction
A critical phase in cancer progression is the dissemination of malignant cells to distant tissues or organs via the bloodstream or lymphatic vessels, known as metastasis (Karras et al., 2024). This portends a poor prognosis, considering that a minimum of 60% of fatalities associated with cancer can be attributed to the presence of metastasis. Cancer metastasis is a multi-step cellular process, known as the “invasion-metastasis cascade,” which includes a series of processes including local invasion, intravasation, survival in the circulation, arrest at a distant organ site, extravasation, micro-metastasis formation, and metastatic colonization (Valastyan and Weinberg, 2011; Gerstberger et al., 2023) (Figure 1). Research shows that tumor metastasis is regulated by a series of complex networks (Shi et al., 2024), such as epitranscriptome regulation which occurs at the RNA level. The presence of modified nucleotides on RNA molecules, known as RNA modifications, has been known since the 1950s (Cohn, 1960; Littlefield and Dunn, 1958). To date, researchers have discovered more than 170 chemical modifications in various RNA classes in prokaryotes and eukaryotes (Cappannini et al., 2024). These chemical markers exert important influences on various cellular processes in eukaryotes (Chen et al., 2016; Shi et al., 2020). Among them, N6-methyladenosine (m6A) represents the most prevalent and thoroughly investigated modification found in messenger RNA (mRNA) and long noncoding RNA (lncRNA), affecting RNA metabolism and expression by regulating RNA splicing, nuclear export, decay, translation, and stability, and plays a vital role in cancer metastasis (Deng et al., 2023).
FIGURE 1
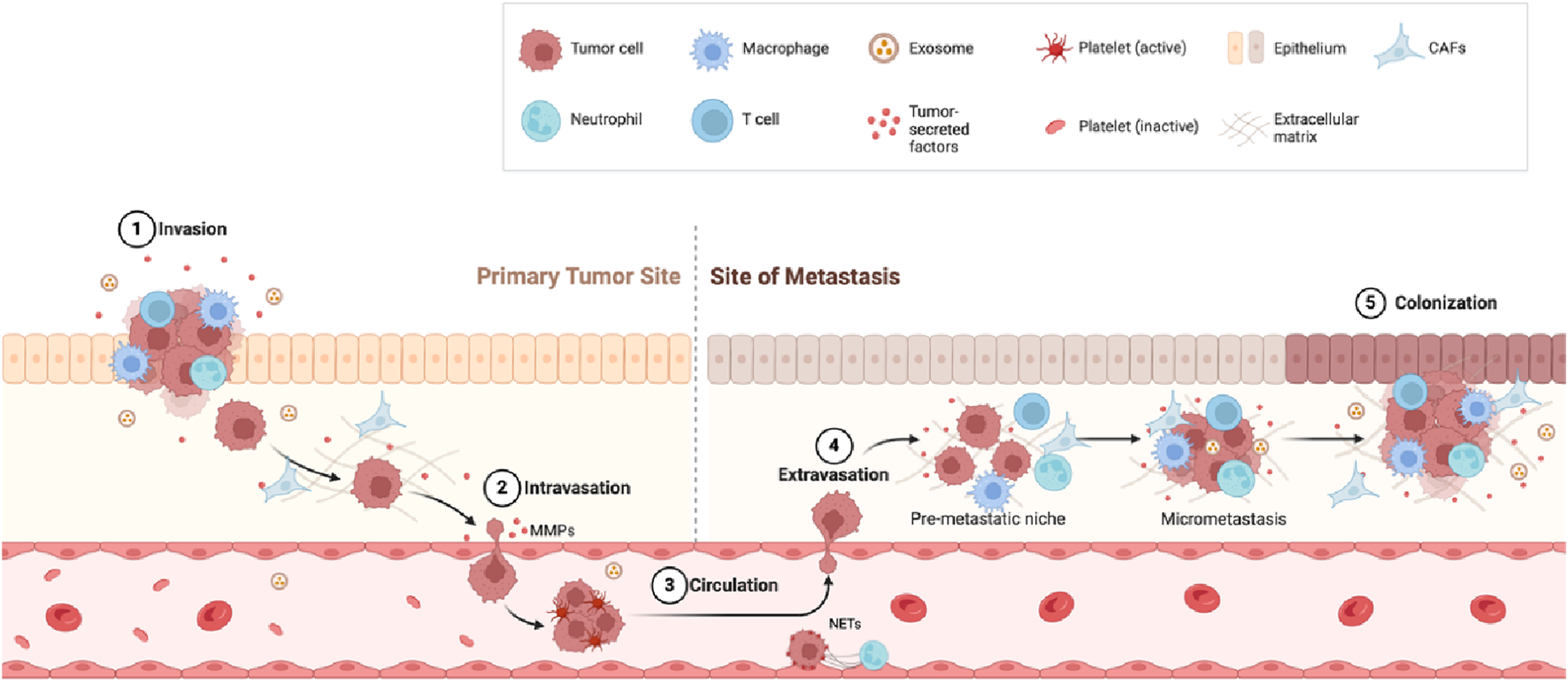
Metastasis steps. Invasion: Tumor cells break through surrounding tissue with help from immune cells such as macrophages and neutrophils. Intravasation: Tumor cells enter the bloodstream by breaking down the tissue barrier using matrix metalloproteinases (MMPs). Circulation: Tumor cells travel through the blood, protected by platelets and other factors. Extravasation: Tumor cells exit the bloodstream and settle in a new site (pre-metastatic niche). Colonization: Tumor cells grow into new tumors at the metastatic site.
Extensive research has been conducted on the tumor invasion-metastasis cascade (Valastyan and Weinberg, 2011; Shi et al., 2024; Steeg, 2016). This article focuses on summarizing the role of m6A in various steps of cancer metastasis and the related mechanisms, while also pointing out the limitations and development prospects of m6A-related research.
m6A
The m6A modification consists of the insertion of a methyl group at the 6th nitrogen atom of adenine and is the most intensively studied mRNA modification. m6A methylation is present in 20%–40% of all transcripts encoded by mammalian cells (Dubin and Taylor, 1975; Desrosiers et al., 1974). This modification is not only ubiquitous in mammalian mRNA, but also occurs in a variety of non-coding RNAs, including ribosomal (rRNA), transfer (tRNA), small nucleolar (snoRNA), long non-coding (lncRNA), small nuclear (snRNA), circular (circRNA), and microRNA (miRNA) (Sendinc and Shi, 2023). m6A is usually embedded in the sequence 5′-RRACH-3′ and exhibits diverse distribution patterns within eukaryotic transcriptomes. In mammals, m6A modification is preferentially located within the 3′-untranslated region (3′-UTR) adjacent to the stop codon and is also prevalent in long exons, suggesting a potential role in the regulation of mRNA splicing (Schwartz et al., 2014; Dominissini et al., 2012; Meyer et al., 2012). In Arabidopsis mRNA, this m6A modification is found to be enriched in both the 3′- and 5′-UTR (Luo et al., 2014). m6A modification can regulate RNA processing, maturation, splicing, degradation, stability, nuclear export, and translation, thereby regulating RNA expression and function (Sommer et al., 1978; Wang et al., 2015; Xiao et al., 2016; Roundtree et al., 2017; Wang X. et al., 2014).
The deposition of the vast majority of m6A modifications on RNA depends on the methyltransferase-like protein 3 (METTL3)/METTL14/Wilms’ tumour-associated protein (WTAP) complex; METTL3 is responsible for methylation reactions. METTL14 functions as an allosteric activator; and the guide protein WTAP facilitates the recruitment of the METTL3/METTL14 heterodimer (Liu et al., 2014; Ping et al., 2014). Recently, METTL5, METTL16, and zinc finger CCHC type 4 (ZCCHC4) were found to be novel m6A methyltransferases in eukaryotes (Pinto et al., 2020; van Tran et al., 2019; Su et al., 2022). The demethylation process is controlled by demethylases such as fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKHB5) (Jia et al., 2011; Zheng et al., 2013). There is currently no known eraser or reader for METTL5, which is responsible for the m6A modification of 18S rRNA. The elimination of m6A or m6Am by FTO depends on its subcellular distribution. In the nucleus and cytoplasm, the FTO substrate is mainly m6Am and m6A, respectively (Mauer et al., 2017). Different reader proteins can mediate RNA processing, stabilization, translation, and degradation by recognizing m6A modifications. The most famous m6A readers are the YTH domain protein family (YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2) and insulin-like growth factor 2 mRNA-binding proteins (IGF2BP) family, the heterogeneous nuclear ribonucleoprotein (HNRNP) protein family, in addition to eukaryotic translation initiation factor 3 (eIF3). YTHDF1, a member of the YTH domain protein family, can interact with m6A modifications located at the mRNA stop codon or 3′-UTR and promote mRNA translation by recruiting eIF3. While there are speculations about the involvement of eIF4G-dependent ring formation in this process, it remains unclear at present how eIF3 is recruited to the stop codon to enhance translation (Wang et al., 2015). Interestingly, a small number of m6A-modified mRNAs are located in the 5′-UTR region, and YTHDF1 directly recruits eIF3 through m6A and assembles the translation initiation complex at the 5′-UTR without the help of cap-binding proteins (Meyer et al., 2015). YTHDF2 mainly regulates the degradation of m6A-modified RNA (Wang X. et al., 2014). YTHDF3 is also thought to promote translation, and some studies have found that it is associated with RNA degradation (Chang et al., 2020; Shi et al., 2017). YTHDC1 promotes pre-mRNA splicing and export through interaction with splicing factors via the m6A modification (Xiao et al., 2016). YTHDC2 is capable of interacting with ribosomal 18S rRNA, which leads to an increase in the translation efficiency of its target genes. Concurrently, this interaction results in a reduction of the abundance of their corresponding mRNA (Hsu et al., 2017; Mao et al., 2019). Proteins of the IGF2BP family contribute to recruiting RNA stabilizers to enhance mRNA stability (Huang et al., 2018). HNRNPA2B1 was shown to selectively recognize methylated pri-microRNAs to promote microRNA maturation (Alarcón et al., 2015a; Alarcón et al., 2015b). HNRNPC is a prevalent nuclear RNA-binding protein that plays a crucial role in the processing of pre-mRNA. m6A modification changes the spatial structure of mRNA and lncRNA to promote the binding of HNRNPC, affecting the alternative splicing of target RNA, thereby affecting gene expression and maturation in the nucleus (Liu et al., 2015) (Figure 2).
FIGURE 2
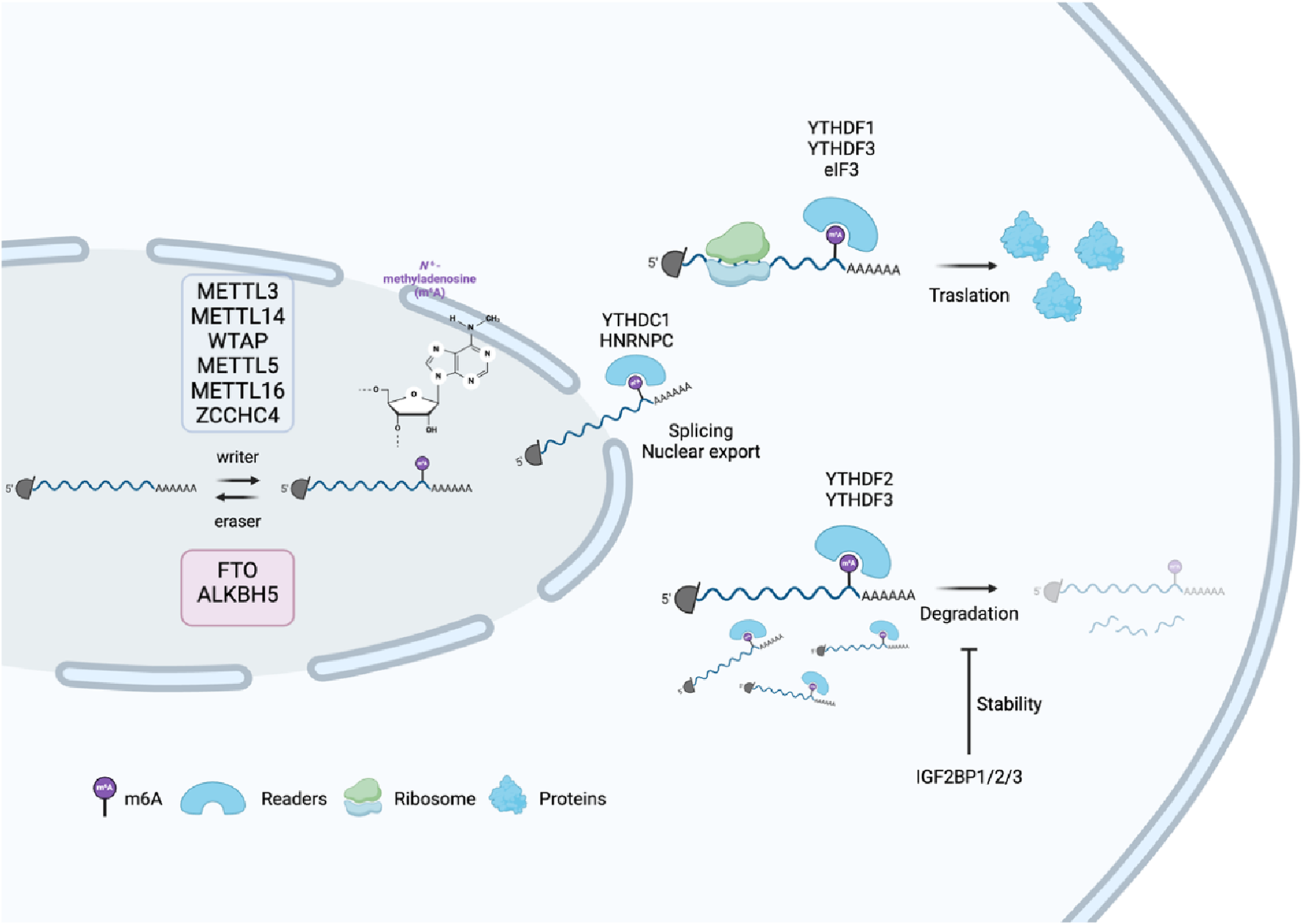
Process of m6A modification and its molecular functions. m6A modification of mRNA is added by “writers” (e.g., METTL3 and METTL14) and removed by “erasers” (FTO and ALKBH5), influencing mRNA fate. m6A-binding “readers,” such as YTHDC1, regulate splicing and export to the nucleus, whereas YTHDF1/3 promote translation in the cytoplasm. Other readers, such as YTHDF2, trigger mRNA degradation, and IGF2BP proteins stabilize mRNA.
Cancer metastasis
Tumor metastasis is formed after a series of complex cellular processes known as the “invasion-metastasis cascade.” Epithelial cells of the original tumor (Karras et al., 2024): locally invade the basement membrane (BM) and extracellular matrix (ECM) (Valastyan and Weinberg, 2011), penetrate the blood vessel (Gerstberger et al., 2023), survive from the vascular transport pressure (Shi et al., 2024), trapped at distant organ locations (Cohn, 1960) undergo extravasation into the parenchymal tissue of distant organs (Littlefield and Dunn, 1958), survive in metastatic tissues to form micro-metastases, and (Cappannini et al., 2024) restart their proliferation program at the metastatic site to give rise to macroscopic, clinically detectable tumors (Valastyan and Weinberg, 2011). The process of tumor metastasis is controlled by the intrinsic molecular signaling pathways of tumor cells, and the interaction between tumor cells and the surrounding environment also plays an essential role. The regulation of m6A modification plays a role in every stage of the process (Figure 3) (Table 1).
FIGURE 3
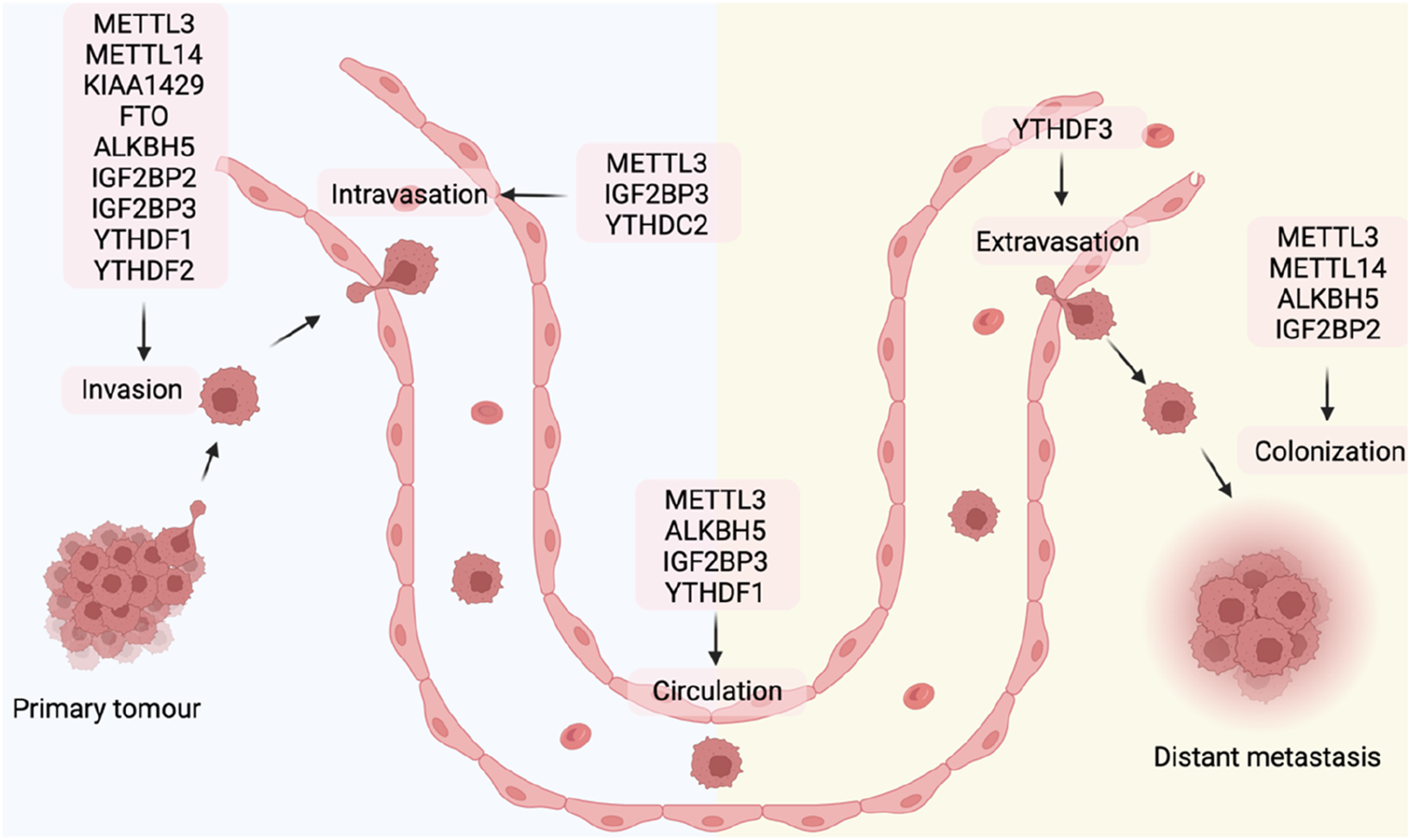
m6A regulators in different steps of tumor metastasis.
TABLE 1
| Stages of cancer metastasis | Cancer | Model | Type | Regulator | Target | M6A stie and mechanism | Axis | Function | References |
|---|---|---|---|---|---|---|---|---|---|
| Local invasion | Ovarian cancer | Tumor xenograft nude mouse, in vitro cell culture | Eraser | ALKBH5 | Snail1 | CDS/stability | FSH/CREB/ALKBH5/Snail/EMT | Promote metastasis and EMT | Xu et al. (2024) |
| Colorectal cancer | Subcutaneous tumors and lung metastases BALB/c nude mouse, in vitro cell culture | Writer | METTL3 | Snail1 | Not mentioned | METTL3/Snail/CXCL2/M2 macrophage | Promote metastasis and EMT | Ouyang et al. (2024) | |
| Head and neck squamous carcinoma | Popliteal LN metastasis nude mouse, in vitro cell culture | Reader | IGF2BP2 | Slug/Snail2 | CDS/stability | IGF2BP2/Slug/EMT | Promote metastasis and EMT | Yu et al. (2022) | |
| Breast cancer | In vitro cell culture | Eraser | FTO | ZEB1 | Not mentioned | piR-17560//FTO/ZEB1 | Promotes chemoresistance and EMT | Ou et al. (2022) | |
| NSCLC | In vitro cell culture | Reader | IGF2BP3 | TWSIT1 | 3‘UTR/stability | IGF2BP3/m 6 A/TWIST1 | Promote metastasis | Cui et al. (2022) | |
| HCC | In vitro cell culture | Writer | METTL3 | lnc-TSPAN12 | stability | METTL3/lnc-TSPAN12/EIF3I/SENP1/Wnt/β-catenin | Promote metastasis and EMT | Li et al. (2024) | |
| Cervical cancer | In vitro cell culture | Writer/Reader | METTL3/YTHDF2 | CTNNB1 | 5’UTR/instability | METTL3/YTHDF2/CTNNB1 | Promote metastasis | Li et al. (2022) | |
| Cervical cancer | In vitro cell culture | Writer/Reader | METTL3/IGF2BP3 | c-MET | Stability | METTL3/IGF2BP3/c-MET/decrease β-catenin membrane localization | Promote metastasis | Li et al. (2022) | |
| Cervical cancer/Lung cancer | In vitro cell culture | Write | METTL3 | E2F1 | Stability | METTL3/E2F1/decrease CTNNB1 transcription | Promote metastasis | Li et al. (2022) | |
| Cervical cancer/Lung cancer/Liver cancer | In vitro cell culture | Writer/Reader | METTL3/YTHDF1 | CTNNB1 | 5′UTR/inhibit translate | METTL3/YTHDF1/eIF4E1/CTNNB1 | Promote metastasis | Li et al. (2022) | |
| Cervical cancer | Zebrafish in vivo study, in vitro cell culture | Eraser | ALKBH5 | MMP2/9 | Increase mRNA and protein level | MALAT1/ALKBH5/MMP2 MMP9 | Promote metastasis | Wu et al. (2023) | |
| HCC | Lung metastasis BALB/c nude mouse, in vitro cell culture | Writer | KIAA1429 | circDLC1 | Decrease RNA level | KIAA1429/circDLC1/HuR/MMP1 | Inhibit metastasis | Liu et al. (2021) | |
| Esophageal squamous cell carcinoma | Subcutaneous xenograft mouse model, in vitro cell culture | Eraser | FTO | MMP13 | Increase mRNA and protein level | FTO/MMP13 | Promote metastasis | Liu et al. (2020) | |
| Renal cancer | In vitro cell culture | Writer | METTL14 | P2RX6 | Decrease mRNA and protein level | ATP-P2RX6-Ca2+ -p-ERK1/2-MMP9 | Inhibit metastasis | Gong et al. (2019) | |
| Melanoma | In vitro cell culture | Writer | METTL3 | MMP2 | Increase protein level | METTL3/MMP2 | Promote metastasis | Dahal et al. (2019) | |
| Intravasation | Colorectal cancer | Zebrafish model animals, in vitro cell culture | Writer | METTL3 | LINC00662/VEGFA | RNA stability | METTL3/LINC00662/VEGFA | Promote angiogenesis | Zhang et al. (2022) |
| Gastric cancer | Liver metastasis nude mouse, in vitro cell culture | Writer/Reader | METTL3/IGF2BP3 | HDGF | mRNA stability | P300/METTL3/IGF2BP3/HDGF | Promote angiogenesis | Wang et al. (2020) | |
| Bladder Cancer | Mettl3flox/flox and Mettl3 KO mice, in vitro cell culture | Writer | METTL3 | TEK/VEGFA | Increase protein level | METTL3/TEK/VEGFA | Promote angiogenesis | Wang et al. (2021) | |
| Colorectal cancer | Subcutaneous xenografts nude mice, in vitro cell culture | Reader | IGF2BP3 | VEGF | CDS/mRNA stability | IGF2BP3/VEGF | Promote angiogenesis | Yang et al. (2020) | |
| Lung cancer | Subcutaneous xenografts nude mouse, in vitro cell culture | Reader | YTHDC2 | VEGFA | 5′UTR/translation | YTHDC2/eIF4GI/VEGFA | Promote angiogenesis | Zhang et al. (2023) | |
| Survival in circulation | Malignant glioma | Orthotopic and subcutaneous transplantations C57BL/6 mice, in vitro cell culture | Reader | IGF2BP3 | CFS3 | 3′UTRmRNA stability | IGF2BP3/CSF3/NET/NETosis | Promote NET | Dai et al. (2024) |
| Melanoma | Lung metastasis Mettl3 knock out C57BL/6 genetic mouse, in vitro cell culture | Writer/Reader | METTL3/YTHDF1 | SPRED2 | CDS/translation | METTL3/YTHDF1/SPRED2/NF-kB STAT3/macrophage polarization | Decrease macrophage polarization and inhibit metastasis | Yin et al. (2021) | |
| Arrest at a distant organ site | Colorectal cancer | Subcutaneous tumors and lung metastases BALB/c nude mouse, in vitro cell culture | Writer | METTL3 | Snail1 | Not mentioned | METTL3/Snail/CXCL2/M2 macrophage | Promote lung metastasis | Ouyang et al. (2024) |
| NSCLC | Brain orthotopic transplantation BALB/c nude mice, in vitro cell culture | Eraser | ALKBH5 | Not mentioned | Not mentioned | BCAT1/ALKBH5/N-cadherin Vimentin/EMT | Promote brain metastasis | Mao et al. (2024) | |
| Extravasation | Breast Cancer | Brain metastasis and orthotopic transplantation nude mice, in vitro cell culture | Reader | YTHDF3 | ST6GALNAC5/GJA1/EGFR/VEGFA | 5′UTR/translation | YTHDF3/ST6GALNAC5/GJA1/EGFR/VEGFA | Promote blood-brain barrier extravasation, angiogenesis, and outgrow | Chang et al. (2020) |
| Micro-metastasis formation | Breast cancer | Xenograft Mouse Model, in vitro cell culture | Reader | IGF2BP2 | ZNF281 | 3′UTR/mRNA stability | lncSNHG5/IGF2BP2/ZNF281/CCL2/CCL5 | Promote premetastatic niche formation | Zeng et al. (2022) |
| Metastatic colonization | Glioblastoma | Foxn1nu/nu athymic nude mice, in vitro cell culture | Eraser | ALKBH5 | FOXM1 | 3′UTR | ALKBH5-FOXM1 | Promote cancer stem cell growth | Zhang et al. (2017) |
| Glioblastoma | Orthotopic intracranial mouse model, in vitro cell culture | Writer | METTL3 | SOX2 | 3′UTR/mRNA stability | METTL3/SOX2 | Promote cancer stem cell growth | Visvanathan et al. (2018) | |
| Leukemia | Xenograft NSG Mouse Model, in vitro cell culture | Writer | METTL3 | MYC/BCL2/PTEN | Translation | METTL3/c-MYC/BCL2/PTEN | Maintenance of the hematopoietic stem cell program | Vu et al. (2017) | |
| Bladder cancer | In vitro cell culture | Writer | METTL14 | Notch1 | mRNA instability | METTL14/Notch 1 | Inhibit tumor initiating cells self-renewal | Gu et al. (2019) |
Cancer metastasis regulated through m6A methylation in different types of cancer.
Local invasion
Local invasion describes the mechanism by which cancer cells penetrate the BM and invade the adjacent tumor-associated stroma. The writers, erasers, and readers of m6A modification play important roles in this process (Figure 4). The junctions between epithelial cells are an important obstacle to the spread of cancer cells, especially the intercellular junctions mediated by E-cadherin (Berx and van Roy, 2009; Vleminckx et al., 1991). These junctions hold epithelial cells together and prevent detachment of individual epithelial cells from neighboring cells. To surmount this challenge, cancer cells may initiate a key cell biological process called epithelial–mesenchymal transition (EMT). This biological process causes polarized cells to acquire mesenchymal characteristics, with alterations in the expression of cellular adhesion molecules and the cytoskeleton, promoting migration and invasion, increasing tumor stemness, and enhancing resistance to chemotherapy and immunotherapy (Nieto et al., 2016; Yilmaz and Christofori, 2009). The opposite procedure is called mesenchymal–epithelial transition (MET). It is worth noting that both EMT and MET are essential for the metastatic process. EMT enables primary tumor cells to acquire the ability to migrate, whereas MET halts the cellular migration phase, thereby promoting better colonization of tumor cells at distant metastatic sites. In living organisms, EMT plays a role in several physiological and pathological processes such as inflammation, wound repair, tissue fibrosis, cancer progression, and embryonic development (Grande et al., 2015; Gros and Tabin, 2014). Cells exist in a continuum of states with inherent plasticity, progressively acquiring various transformation intermediates, rather than being confined to two polarized cell identities. EMT represents a fluid and progressive transition from the epithelial to the mesenchymal cell phenotype. Tumor cells in the intermediate phases of the EMT process display a combination of epithelial and mesenchymal features, enhancing their ability to survive, spread, and colonize distant metastatic sites. EMT is tightly controlled by a complex regulatory network orchestrated by numerous intrinsic and extrinsic factors, such as various transcription factors (TFs), epigenetic changes, post-translational modifications, and non-coding RNA-mediated mechanisms. These factors drive the mesenchymal shift by downregulating epithelial markers and upregulating mesenchymal-associated markers (Huang et al., 2022; Stemmler et al., 2019).
FIGURE 4
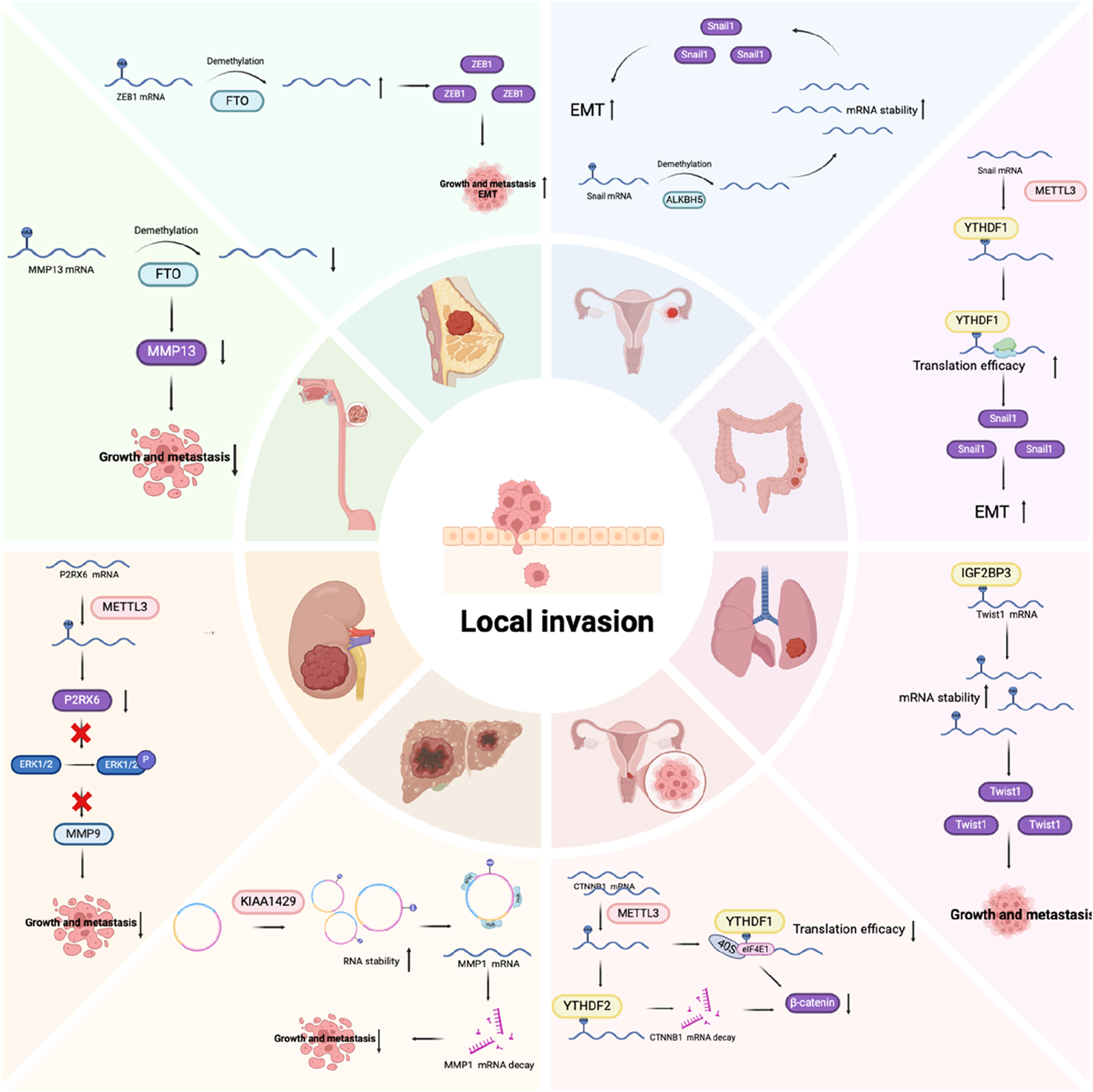
The role of m6A regulators in the invasion stage of tumor metastasis.
The major EMT-related TFs (EMT-TFs) are snail family transcriptional repressor 1 (SNAIL1) and SNAIL2/Slug, zinc finger E-box binding homeobox 1/2 (ZEB1/2), and twist family BHLH transcription factor 1/2 (TWIST1/2) (Lamouille et al., 2014). SNAIL1 directly inhibits the formation of E-cadherin and tight junction proteins, while upregulating mesenchymal phenotype markers like fibronectin and vimentin (Dong et al., 2012; Tong et al., 2012). SNAIL2/Slug facilitates the disruption of cell adhesion and polarity, while enhancing migratory and invasive abilities (Casas et al., 2011). ZEB1/2 belong to the human ZEB family. They are zinc finger TFs that attach to regulatory gene sequences at the E-box, either promoting or inhibiting transcription. Notably, ZEB1 and ZEB2 exert contrasting effects on various cell lines, suggesting that their function is highly dependent on the environment. For example, in a melanoma mouse model, ZEB2 inhibits cancer metastasis, whereas ZEB1 promotes tumor initiation and progression (Caramel et al., 2013). In clinical research, reduced ZEB2 levels correlated with lower survival rates in melanoma patients, whereas elevated ZEB1 expression was linked to poorer clinical prognoses (Denecker et al., 2014). TWIST1/2 belong to the basic helix-loop-helix (BHLH) TF family. Like SNAIL, TWIST1 can suppress E-cadherin expression while upregulating N-cadherin, leading to a reduction in cell adhesion and an enhancement in cell motility. This shift plays a crucial role in facilitating cellular migration and promoting metastatic potential in cancer (Yang et al., 2008). m6A modifications have been confirmed to directly affect the expression of these EMT-TFs, thereby affecting the EMT process. For example, Snail1 mRNA itself will be modified by m6A, and its regulation depends on the writing protein METTL3 or the erasing protein ALKBH5. Its final expression is determined by the reading protein YTHDF1 or 2, and thus regulates the expression levels of E- and N-cadherin, promoting the EMT process (Xu et al., 2024; Ouyang et al., 2024). In head and neck squamous cell carcinoma, SNAIL2 is also modified by m6A. IGF2BP2 has the capacity to recognize and attach to m6A-modified sites, and promote its mRNA stability, thereby regulating the EMT process and promoting lymph node metastasis (Yu et al., 2022). Similarly, ZEB1/2 and Twist can also be regulated in an m6A-dependent manner, thereby affecting the EMT process of tumor cells (Ou et al., 2022; Cui et al., 2022; Cai et al., 2024). Research has demonstrated that noncoding RNA are of essence in EMT regulation. lncRNA often acts as a molecular sponge to interact with miRNA, inhibiting the degradation of EMT-TF to regulate EMT (Yuan et al., 2014; Grelet et al., 2017). The m6A modification can indirectly affect EMT by affecting the expression of non-coding RNA. For example, in hepatocellular carcinoma, it was found that the upregulation of METTL3 promoted the deposition of m6A modification on LncRNA Tetraspanin 12 (TSPAN12), enhanced the stability of LncRNA, and upregulated the expression of lncRNA TSPAN12, acting as a scaffold, promoting the interaction of EIF3I and SUMO specific peptidase 1 (SENP1), thus activating the Wnt/β-catenin signaling pathway to promote EMT (Li et al., 2024). Wnt, transforming growth factor-β (TGF-β), PI3K-AKT, Notch, and other signaling pathways are likewise implicated in the EMT regulatory framework. The key molecules in these pathways are also often regulated by m6A (Tan et al., 2022; Liu et al., 2019; Li et al., 2020). In the common Wnt/β-catenin pathway, Wnt protein activates β-catenin to regulate EMT. Catenin beta 1 (CTNNB1) (encoding β-catenin) mRNA itself can be modified by m6A and is associated with METTL3 expression. METTL3 promotes increased YTHDF2-dependent mRNA degradation and YTHDF1-dependent translation inhibition through m6A modification at the 5ʹ-UTR of the CTNNB1 mRNA. METLL3 can also reduce the cell membrane localization of β-catenin through m6A/IGF2BP3/c-MET, thereby reducing E-cadherin. In summary, METTL3 modulates EMT by regulating β-catenin expression and its subcellular localization (Li et al., 2022). It can be seen that the EMT process is regulated by a complex network, and m6A modification plays a critical role in it. Interestingly, we found that the role of m6A modification may be completely different in different tumor types or cells, which shows that the complexity of RNA modification may be determined by different environmental factors, and ultimately the different expressions of target molecules mediated by different readers ultimately determine the direction of EMT.
Loss of the BM means that cancer cells can directly invade the matrix, a process usually achieved by the breakdown of the BM by matrix metalloproteinases (MMPs). In healthy tissues, MMP activity is tightly regulated through transcriptional processes and post-translational modifications. However, tumor cells can subvert this strict control in a variety of ways, ultimately leading to increased MMP function (Kessenbrock et al., 2015; Egeblad and Werb, 2002). In addition to degrading BM and ECM, MMPs can also induce the release of various growth factors in ECM and promote tumor growth (Kessenbrock et al., 2010). Studies have shown that MMPs can also be directly or indirectly regulated by m6A modification, thereby affecting the invasiveness of tumor cells (Wu et al., 2023; Liu et al., 2021; Liu et al., 2020; Gong et al., 2019; Dahal et al., 2019).
Tumor cells penetrate the BM and invade the ECM, creating numerous opportunities for them to access the circulation and disseminate to distant locations.
Intravasation
During metastasis, tumor cells enter the blood circulation through the circulatory or lymphatic system, a process called intravasation. The signaling molecule TGF-β has the ability to amplify the permeability of breast cancer cells (Giampieri et al., 2009). In addition, tumor cell-induced angiogenesis can also promote tumor cells to enter the blood circulation better. Compared with vasculature in healthy tissues, the new blood vessels induced by tumor cells are winding, leaky, and engaged in constant reconstruction, which makes it easier for tumor cells to enter the blood circulation (Carmeliet and Jain, 2011). m6A modification plays a key role in regulating tumor angiogenesis (Figure 5). Researchers have found that METTL3 is upregulated in a variety of tumors, and increased m6A deposition is associated with increased tumor angiogenesis (Zhang et al., 2022; Wang et al., 2020; Wang et al., 2021). In gastric cancer, upregulation of METTL3 is correlated with a poor prognosis for patients. Studies have found that METTL3 upregulates the m6A level of heparin binding growth factor (HDGF) mRNA, and then enhances its stability and promotes its expression through IGF2BP3, thereby increasing tumor angiogenesis and promoting gastric cancer metastasis (Wang et al., 2020). Similarly, in colon cancer, IGF2BP3 has been identified as a regulator of vascular endothelial growth factor (VEGF) mRNA stability via m6A modification, facilitating tumor angiogenesis (Yang et al., 2020). m6A modification in the 5′-UTR region of VEGFA in lung cancer triggers cap-independent translation initiation by recruiting the YTHDC2/eIF4GI complex, increasing its expression and thereby stimulating angiogenesis (Zhang et al., 2023). Once tumor cells successfully enter the blood, they will face more challenges.
FIGURE 5
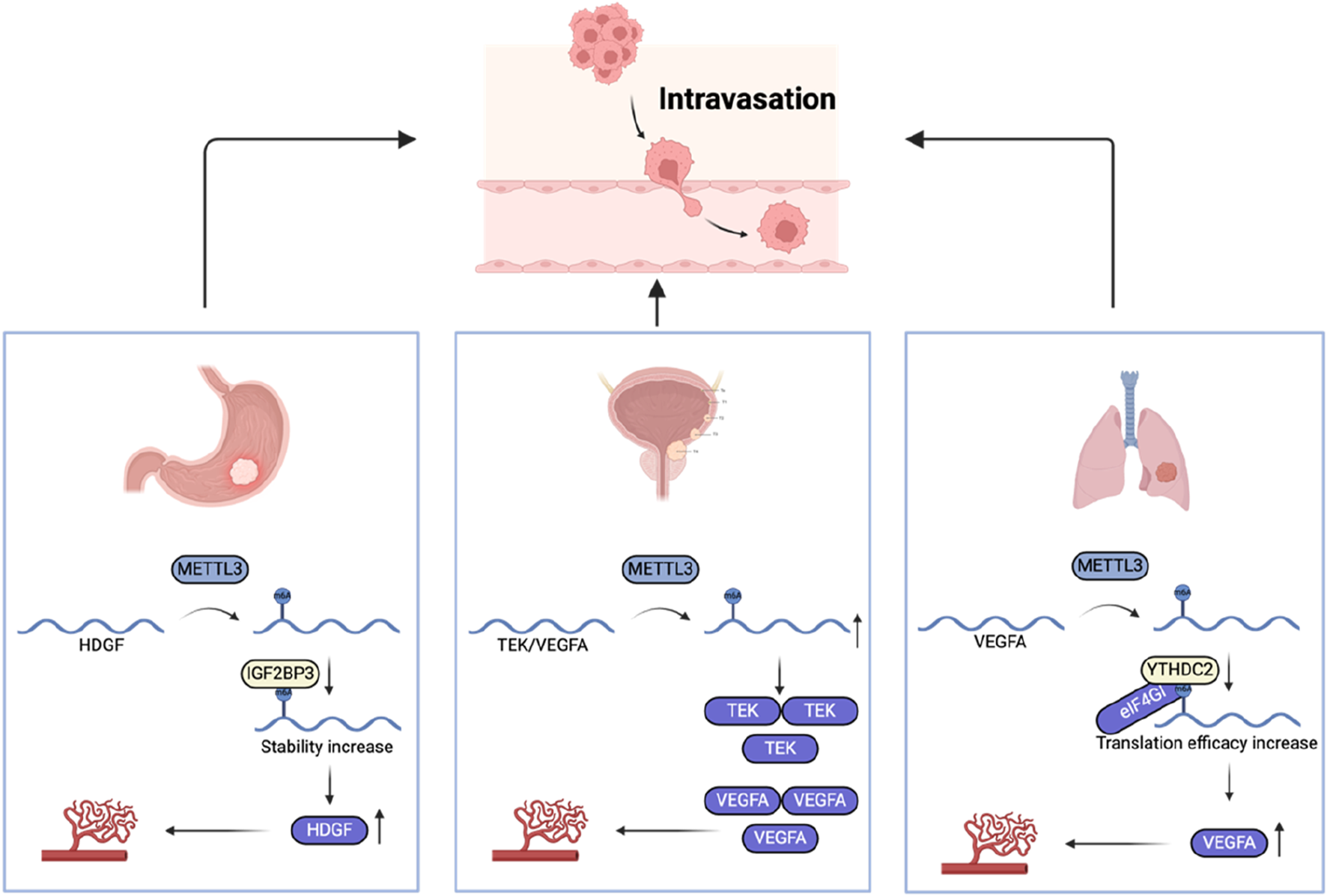
The role of m6A regulators in intravasation.
Survival in circulation
Once tumor cells successfully infiltrate the vascular cavity, they can disseminate extensively through the bloodstream. These cells are referred to as circulating tumor cells (CTCs), and they must withstand various pressures to reach distant sites (Headley et al., 2016). Firstly, to detach from the BM, CTCs often undergo a loss of integrin-dependent adhesion, which is usually required for epithelial cell survival. In the absence of attachment to the ECM, epithelial cells often undergo anoikis. Tumor cells can dynamically adapt and resist cell death by constantly sensing and responding to biochemical and biomechanical changes in their microenvironment (Mak et al., 2017; Alanko et al., 2015). Study shows that tumor matrix stiffening can activate mammalian target of rapamycin 1 (mTORC1) and achieve cell death resistance under matrix detachment through the integrin-glycogen synthase kinase 3 beta (GSK3β)-FTO-mTOR axis (Zhang et al., 2024). Secondly, CTCs must also overcome damage caused by the blood flow shear forces and immune cell-mediated attacks. A small number of CTCs have close interactions with platelets, macrophages, neutrophils, cancer-associated fibroblasts (CAFs) or myeloid-derived suppressor cells, which helps them evade the attack of the immune system in the blood and promotes their survival (Rejniak, 2016; Garrido-Navas et al., 2019). Neutrophils are the predominant white blood cells present in the bloodstream and studies suggest they can promote cancer progression (Shaul and Fridlender, 2019). The binding of CTCs to neutrophils is mediated by cell–cell junctions, anchored to the vascular endothelium to resist shear stress extravasation, a process facilitated by a range of cell adhesion molecules, including integrins, cadherins, and surface glycoproteins (Szczerba et al., 2019; Strell et al., 2007; Huh et al., 2010; Rowson-Hodel et al., 2018).
Neutrophil extracellular traps (NETs) are web-like structures composed of histones, DNA, and cytotoxic proteins originating from granules. Tumor cell-induced NETs can promote the survival and metastasis of tumor cells (Adrover et al., 2023). A study found that IGF2BP3 induced NET formation in malignant gliomas and significantly enhanced glioma cell survival (Dai et al., 2024). In liver cancer, METTL5 was found to promote the generation and release of NETs, which may further accelerate the progression of hepatocellular carcinoma. However, the precise mechanism requires further clarification (Wang et al., 2024). Tumor-associated macrophages (TAMs) can also promote the dissemination and extravasation of CTCs (Doak et al., 2018). TAMs actively communicate with CTCs, promoting their survival and aiding in their mesenchymal transformation. They secrete specific cytokines and growth factors, such as TGF-β and interleukin-6, which enhance the EMT process in CTCs. This crosstalk is essential for the mesenchymal CTCs to maintain their invasive potential, allowing them to metastasize to distant organs, such as the liver and lungs (Wei et al., 2019). Moreover, TAMs facilitate the acquisition of enhanced mechanical adhesion properties and increased cellular resilience in CTCs, thereby promoting the formation of protective cell clusters. These clusters provide a survival advantage by conferring resistance to shear stress experienced during circulation in the bloodstream, ultimately supporting their metastatic potential (Osmulski et al., 2021). This study found that TAM can promote lung metastasis of melanoma by downregulating METTL3. Mechanistic studies have demonstrated that the loss of METTL3 results in decreased sprouty related EVH1 domain containing 2 (SPRED2) translation, mediated by YTHDF1. This reduction promotes the activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and signal transducer and activator of transcription 3 (STAT3) signaling pathways, ultimately promoting enhanced tumor growth and metastasis (Yin et al., 2021).
CAFs are one of the most prevalent components of the tumor microenvironment and are crucial in facilitating tumor initiation, promoting angiogenesis, driving metastasis, and contributing to drug resistance (Chen and Song, 2019). In a mouse lung cancer metastasis model, CTCs can carry CAFs from the initial tumor site to the metastatic site (Duda et al., 2010). The study found that m6A and its writer protein METTL3, eraser protein FTO, and reader protein YTHDF2 play an essential role in CAFs promoting tumor cell metastasis and angiogenesis (Liao et al., 2024; Chen et al., 2023). However, its role in CTC survival and immune escape is still unclear. Circulating platelets can bind to CTCs in the bloodstream and form aggregates. CTCs further enhance this aggregate formation by releasing prothrombotic and procoagulant particles or through the expression of tissue factor, which promotes coagulation and may facilitate tumor metastasis (Thomas et al., 2015; Stark et al., 2018). Mediators released by platelets, such as TGF-β, have been shown to induce EMT in CTCs, thereby enhancing their invasive capabilities and promoting metastasis (Labelle et al., 2011). Platelets are thought to protect CTCs from shear stress and induce resistance to apoptosis, facilitating CTC evasion of natural killer (NK) cell attacks through various mechanisms (Egan et al., 2014; Haemmerle et al., 2017; Nieswandt et al., 1999). In general, current studies have revealed that m6A modification is partially involved in CTC circulation survival and immune escape (Figure 6), but further in-depth exploration is still needed.
FIGURE 6
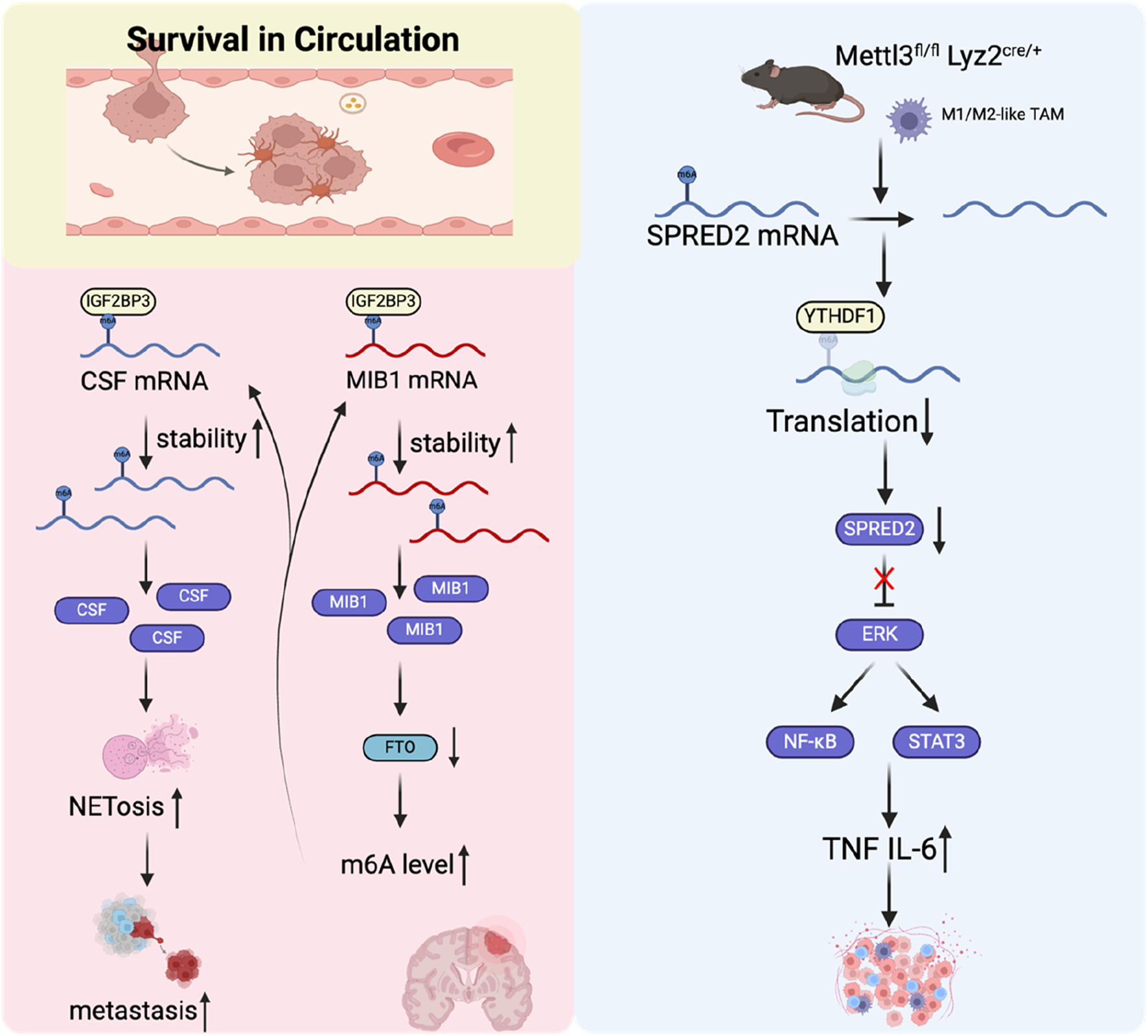
The role of m6A regulators in tumor cell survival in the circulation.
Arrest at a distant organ site
Although tumor cells theoretically have the ability to spread to various organs throughout the body, it has long been noted in clinical practice that different types of tumor cells have different preferences for metastatic target organs (Fidler, 2003). One hypothesis is that the capillary diameter of some organs is not large enough for CTCs to pass through, causing them to stagnate and then metastasize to that organ (Ewing, 1928). Another hypothesis is that CTCs have preferences and can stay in certain tissues by forming specific molecular adhesion interactions with specific tissues (Liu et al., 2017; Fidler and Poste, 2008). In the above process, CTCs as seeds have acquired many metastasis-related abilities, including EMT, tumor stem cells, and tumor-derived or induced cytokines. The microenvironment that is conducive to the survival of CTC metastasis, and its shaping also plays a key role (Liu et al., 2017). Microenvironment remodeling is driven by complex interactions between tumor cells and the ECM, as well as with various cellular components, including TAMs, mesenchymal stem cells, endothelial cells, and tumor-associated fibroblasts. Additionally, the hypoxic conditions within the tumor microenvironment and associated metabolic changes play critical roles in this remodeling process, further supporting tumor progression and metastasis (Joyce and Pollard, 2009). This study revealed that METTL3 and YTHDF1 are highly expressed in tumors from patients with colorectal cancer lung metastasis. METTL3 facilitates the EMT in colorectal cancer by adding m6A modifications to SNAIL mRNA, which enhances its stability. This, in turn, promotes the secretion of CXC motif chemokine ligand 2 (CXCL2) via activation of the NF-κB signaling pathway, contributing to tumor progression and metastasis. Further animal in vivo experiments found that secreted CXCL2 promoted the lung metastasis of colon cancer cells by recruiting M2 macrophages (Ouyang et al., 2024). Patients with brain metastases originating from non-small cell lung cancer (NSCLC), accompanied by high levels of branched chain amino acid transaminase 1 (BCAT1) protein were found to have a poor prognosis. Moreover, NSCLC cells with brain metastases had increased branched-chain amino acid (BCAA) catabolism, leading to α-Ketoglutarate (α-KG) depletion. This process resulted in the downregulation of both the expression and activity of the m6A demethylase ALKBH5, thereby impairing its ability to remove m6A modifications from target mRNAs, resulting in increased m6A deposition and elevated expression of mesenchymal-related genes. Consequently, this promoted the EMT in NSCLC cells and facilitated the proliferation of NSCLC cells within the brain, contributing to metastasis and tumor progression (Mao et al., 2024). In summary, RNA modifications play a crucial role in the process of specific tissue transfer, whether in seeds or soil changes (Figure 7).
FIGURE 7
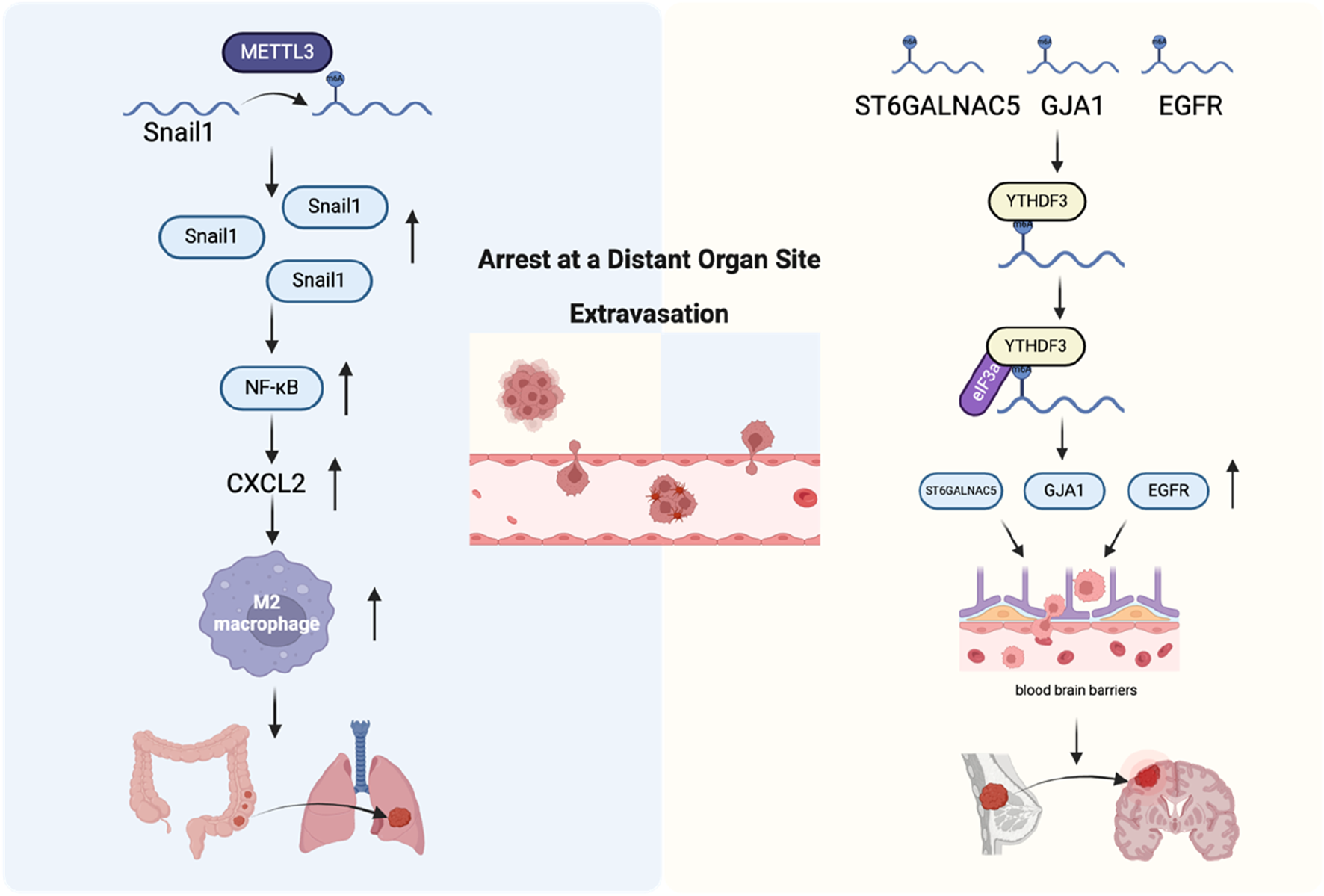
The role of m6A regulators in arrest at a distant organ site and extravasation. Left: Mechanism of colorectal cancer cell metastasis to the lungs. Right: Mechanism of breast cancer cell penetration through the blood-brain barrier and colonization in the brain.
Extravasation
During metastasis, tumor cells disseminate to distant organs and tissues through the bloodstream. They then extravasate into the tissue parenchyma by attaching to the endothelial cells of blood vessels and passing through the vessel walls to the metastatic site. This process is called extravasation (Reymond et al., 2013). Vascular extravasation and intravasation are not just opposite but also two mechanistically distinct states. The same types of cells may exist in the tumor microenvironment at the primary tumor site and the secondary metastatic site, but their functions may be completely different (Qian and Pollard, 2010). The permeability of newly formed blood vessels in the primary tumor site is very strong, which provides favorable conditions for tumor cell infiltration. However, in normal tissues at distant metastatic sites, vascular tissues are normalized, which greatly hinders the extravasation of tumor cells. Therefore, tumor cells destroy the normal connection of vascular endothelium by secreting protein angiopoietin-like 4 (Angptl4) and various cytokines, such as cyclooxygenase-2 (COX-2), epiregulin (EREG), and MMP-1/2, thereby promoting the extravasation of tumor cells (Padua et al., 2008; Gupta et al., 2007a). The study found that YTHDF3 expression increased in patients with brain metastases from breast cancer, whereas its depletion impaired the formation of brain metastases and prolonged the survival of mice. YTHDF3 can promote breast cancer cell adhesion and extravasation across the brain endothelium by enhancing interactions between cancer cells and astrocytes, as well as promoting angiogenesis in the context of breast cancer brain metastasis. This process contributes to the establishment and progression of metastatic tumors in the brain. Further studies found that YTHDF3 promoted the translation of epidermal growth factor receptor (EGFR), gap junction protein alpha 1 (GJA1), and ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ST6GALNAC5) in an m6A-dependent manner, which ultimately led to an increase in breast cancer brain metastasis (Chang et al., 2020) (Figure 7).
Micrometastasis formation
After tumor cells pass through blood vessels, to form micrometastasis, they need to survive in the unfamiliar microenvironment of the distant metastatic site. The microenvironment at metastatic sites typically exhibits significant distinctions compared to that of the primary tumor. These differences encompass variations in stromal cell populations, ECM composition, the presence of growth factors and cytokines, and even the tissue structural organization. Tumor cells can use complex mechanisms to change the pre-metastatic niches of distant metastases before metastasis occurs, so that they can survive and form small micrometastasis after metastasis is established (Peinado et al., 2017). Tumor cells can affect the distant metastatic microenvironment in advance by secreting cytokines and exosomes. Pre-metastatic niches formation first changes the vascular permeability, followed by changes in fibroblasts in the ECM, or recruiting bone marrow-derived cells to distant metastatic sites, remodeling the ECM, and ultimately attracting CTCs to reside (Joyce and Pollard, 2009; Psaila and Lyden, 2009; Kaplan et al., 2005). In breast cancer, long noncoding RNA small nucleolar host RNA 5 (lncSNHG5) is markedly overexpressed in CAFs. It stabilizes zinc finger protein 281 (ZNF281) mRNA by interacting with the m6A-binding protein IGF2BP2, resulting in the upregulation of CC motif chemokine ligand 2 (CCL2) and CCL5 through ZNF281. This activation triggers the p38 mitogen-activated protein kinases (P38 MAPK) pathway in endothelial cells, facilitating neovascularization and increasing vascular leakage, thereby contributing to the establishment of the pre-metastatic niche (Zeng et al., 2022) (Figure 8).
FIGURE 8
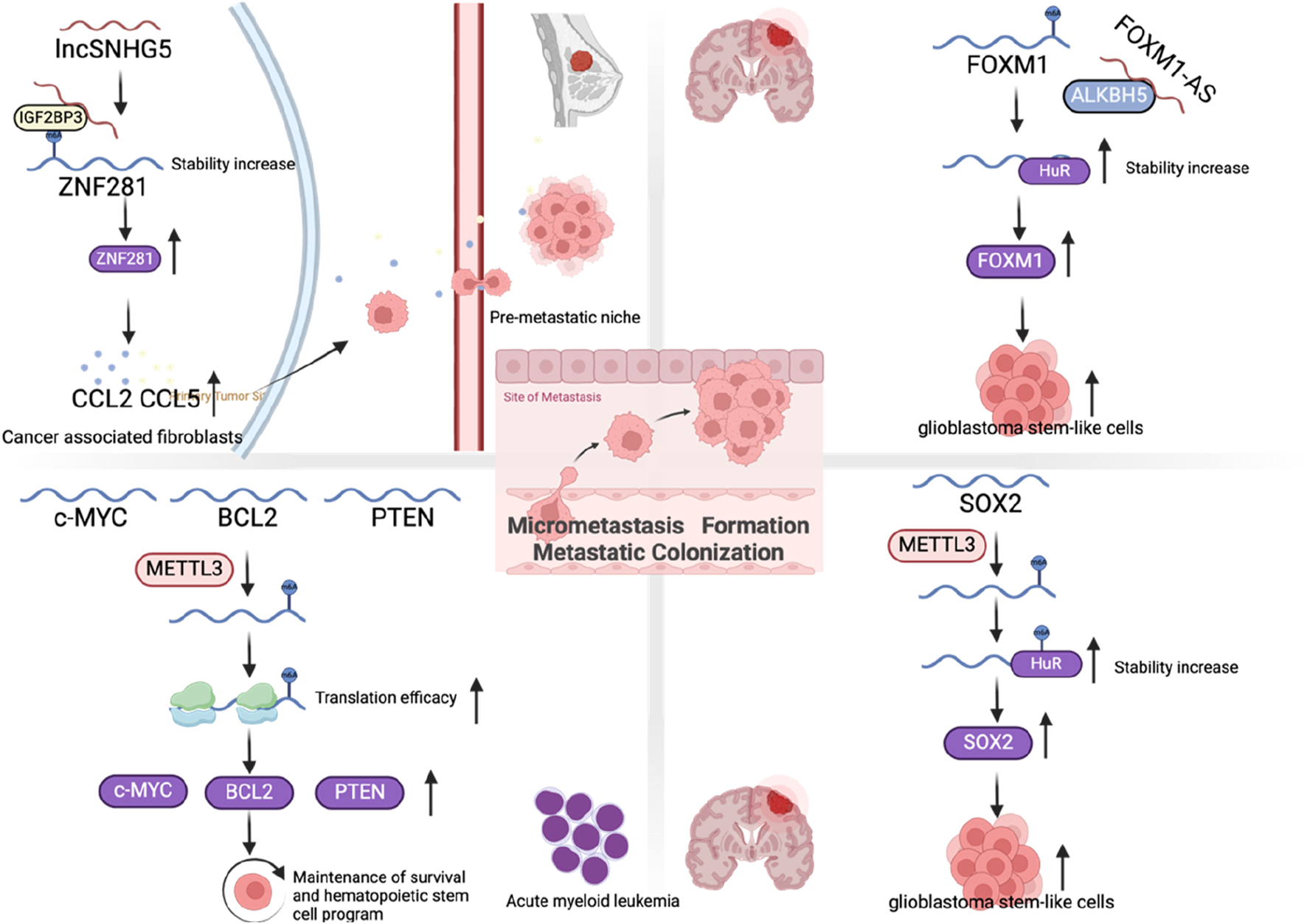
The role of m6A regulators in micrometastasis formation and metastatic colonization.
Metastatic colonization
If disseminated tumor cells successfully survive from their first encounter with the unfamiliar microenvironment of the metastatic site, this does not necessarily mean that they are guaranteed to proliferate and form macroscopic metastases. The overwhelming majority of disseminated cells are either slowly exhausted over a period of weeks to months or persist in the form of microcolonies in an apparently long-term dormant state (Gomis and Gawrzak, 2017; Sosa et al., 2014). Another key factor for successful metastatic colonization is the self-renewal ability of tumor cells. Studies suggest that there is a group of cells with high self-renewal ability inside tumor cells, called tumor-initiating (TICs) or tumor stem cells (Shackleton et al., 2009; Clevers, 2011). Previous studies have found that EMT can promote the transformation of tumor cells into TICs (Ye et al., 2015). Furthermore, the TFs from the inhibitor of differentiation (ID) family, such as ID1 and ID3, along with the homeobox TF NK2 homeobox 1 (NKX2-1), are capable of modulating the TIC phenotype in cancer cells (Ullmann et al., 2018; Winslow et al., 2011; Gupta et al., 2007b). A study revealed that m6A modification plays a critical role in regulating embryonic stem cells to maintain their self-renewal capacity (Figure 8). In mouse embryonic stem cells, the knockdown of METTL3 and METTL14 results in a loss of this self-renewal capability, highlighting the importance of these m6A-related enzymes in stem cell maintenance (Wang Y. et al., 2014). Interestingly, another study suggested that mouse embryonic stem cells that lose the m6A modification cannot completely terminate their primitive pluripotency and their differentiation potential is also limited, resulting in early embryonic death (Geula et al., 2015). In cancer stem cells, studies have shown that the depletion of FTO and ALKBH5 leads to increased m6A modification, which can significantly reduce the self-renewal ability of cancer stem cells (Zhang et al., 2017; Su et al., 2020). However, in glioma stem-like cells, it was found that METTL3 expression increased, promoting sex determining region Y-box 2 (SOX2) mRNA m6A modification immersion and increased stability, thereby maintaining its stem cell characteristics (Visvanathan et al., 2018). Similarly, in acute myeloid leukemia, depletion of METTL3 promotes cell differentiation while reducing cell proliferation (Vu et al., 2017). In contrast, in bladder cancer, it was reported that METTL14 can inhibit bladder TIC’s self-renewal ability and tumorigenesis through m6A modification of Notch1 (Gu et al., 2019). Overall, these results suggest that changes in m6A levels can significantly affect cell fate and differentiation state, but these effects may depend on the cellular context as well as the differentiation state of the cells themselves.
Conclusion and future outlook
In this review article, we mainly focus on the regulatory role of m6A modification in various stages of tumor metastasis. Tumor metastasis is a complex and inefficient process. Abnormal RNA m6A modification is closely related to tumor metastasis, which provides us with additional means and methods to interfere with tumor metastasis. In different processes of metastasis or types of tumors, m6A modification may have completely different effects. This shows that the type of effect that m6A modification exhibits is environment- and time-dependent. Writing proteins and erasing proteins are regulated by different mechanisms, and the ultimate role of m6A modification is also determined by different reading proteins. While existing studies have made significant progress in elucidating the cellular and molecular processes that propel tumor metastasis and new insights and mechanisms continue to emerge, our understanding of this process is still incomplete. The underlying mechanism of metastasis coordination by this modification remains to be further clarified. The discovery of these mechanisms provides significant potential for clinical application.
Statements
Author contributions
YZ: Conceptualization, Writing – original draft, Writing – review and editing. PC: Investigation, Writing – original draft, Writing – review and editing. ZQ: Funding acquisition, Supervision, Validation, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the 1.3.5 project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC21042).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Adrover J. M. McDowell S. A. C. He X.-Y. Quail D. F. Egeblad M. (2023). NETworking with cancer: the bidirectional interplay between cancer and neutrophil extracellular traps. Cancer Cell41, 505–526. 10.1016/j.ccell.2023.02.001
2
Alanko J. Mai A. Jacquemet G. Schauer K. Kaukonen R. Saari M. et al (2015). Integrin endosomal signalling suppresses anoikis. Nat. Cell Biol.17, 1412–1421. 10.1038/ncb3250
3
Alarcón C. R. Goodarzi H. Lee H. Liu X. Tavazoie S. Tavazoie S. F. (2015a). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell162, 1299–1308. 10.1016/j.cell.2015.08.011
4
Alarcón C. R. Lee H. Goodarzi H. Halberg N. Tavazoie S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature519, 482–485. 10.1038/nature14281
5
Berx G. van Roy F. (2009). Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb. Perspect. Biol.1, a003129. 10.1101/cshperspect.a003129
6
Cai Y. Zhou J. Xu A. Huang J. Zhang H. Xie G. et al (2024). N6-methyladenosine triggers renal fibrosis via enhancing translation and stability of ZEB2 mRNA. J. Biol. Chem.300, 107598. 10.1016/j.jbc.2024.107598
7
Cappannini A. Ray A. Purta E. Mukherjee S. Boccaletto P. Moafinejad S. N. et al (2024). MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res.52, D239–D244. 10.1093/nar/gkad1083
8
Caramel J. Papadogeorgakis E. Hill L. Browne G. J. Richard G. Wierinckx A. et al (2013). A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell24, 466–480. 10.1016/j.ccr.2013.08.018
9
Carmeliet P. Jain R. K. (2011). Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat. Rev. Drug Discov.10, 417–427. 10.1038/nrd3455
10
Casas E. Kim J. Bendesky A. Ohno-Machado L. Wolfe C. J. Yang J. (2011). Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res.71, 245–254. 10.1158/0008-5472.CAN-10-2330
11
Chang G. Shi L. Ye Y. Shi H. Zeng L. Tiwary S. et al (2020). YTHDF3 induces the translation of m6A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell38, 857–871.e7. 10.1016/j.ccell.2020.10.004
12
Chen K. Zhao B. S. He C. (2016). Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol.23, 74–85. 10.1016/j.chembiol.2015.11.007
13
Chen M. Zhang Q. Zheng S. Guo X. Cao L. Ren Y. et al (2023). Cancer-associated fibroblasts promote migration and invasion of non-small cell lung cancer cells via METTL3-mediated RAC3 m6A modification. Int. J. Biol. Sci.19, 1616–1632. 10.7150/ijbs.79467
14
Chen X. Song E. (2019). Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov.18, 99–115. 10.1038/s41573-018-0004-1
15
Clevers H. (2011). The cancer stem cell: premises, promises and challenges. Nat. Med.17, 313–319. 10.1038/nm.2304
16
Cohn W. E. (1960). Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J. Biol. Chem.235, 1488–1498. 10.1016/s0021-9258(18)69432-3
17
Cui Y. Wu Y. Wang C. Wang Z. Li Y. Jiang Z. et al (2022). Isoliquiritigenin inhibits non-small cell lung cancer progression via m6A/IGF2BP3-dependent TWIST1 mRNA stabilization. Phytomed. Int. J. Phytother. Phytopharm.104, 154299. 10.1016/j.phymed.2022.154299
18
Dahal U. Le K. Gupta M. (2019). RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res.29, 382–389. 10.1097/CMR.0000000000000580
19
Dai W. Tian R. Yu L. Bian S. Chen Y. Yin B. et al (2024). Overcoming therapeutic resistance in oncolytic herpes virotherapy by targeting IGF2BP3-induced NETosis in malignant glioma. Nat. Commun.15, 131. 10.1038/s41467-023-44576-2
20
Denecker G. Vandamme N. Akay O. Koludrovic D. Taminau J. Lemeire K. et al (2014). Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ.21, 1250–1261. 10.1038/cdd.2014.44
21
Deng X. Qing Y. Horne D. Huang H. Chen J. (2023). The roles and implications of RNA m6A modification in cancer. Nat. Rev. Clin. Oncol.20, 507–526. 10.1038/s41571-023-00774-x
22
Desrosiers R. Friderici K. Rottman F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A.71, 3971–3975. 10.1073/pnas.71.10.3971
23
Doak G. R. Schwertfeger K. L. Wood D. K. (2018). Distant relations: macrophage functions in the metastatic niche. Trends Cancer4, 445–459. 10.1016/j.trecan.2018.03.011
24
Dominissini D. Moshitch-Moshkovitz S. Schwartz S. Salmon-Divon M. Ungar L. Osenberg S. et al (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature485, 201–206. 10.1038/nature11112
25
Dong C. Wu Y. Yao J. Wang Y. Yu Y. Rychahou P. G. et al (2012). G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest.122, 1469–1486. 10.1172/JCI57349
26
Dubin D. T. Taylor R. H. (1975). The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res.2, 1653–1668. 10.1093/nar/2.10.1653
27
Duda D. G. Duyverman A. M. M. J. Kohno M. Snuderl M. Steller E. J. A. Fukumura D. et al (2010). Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl. Acad. Sci. U. S. A.107, 21677–21682. 10.1073/pnas.1016234107
28
Egan K. Cooke N. Kenny D. (2014). Living in shear: platelets protect cancer cells from shear induced damage. Clin. and Exp. Metastasis31, 697–704. 10.1007/s10585-014-9660-7
29
Egeblad M. Werb Z. (2002). New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer2, 161–174. 10.1038/nrc745
30
Ewing J. (1928). Neoplastic diseases: a treatise on tumors. Philadelphia and London: W. B. Saunders Co. Ltd.
31
Fidler I. J. (2003). The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer3, 453–458. 10.1038/nrc1098
32
Fidler I. J. Poste G. (2008). The “seed and soil” hypothesis revisited. Lancet. Oncol.9, 808. 10.1016/S1470-2045(08)70201-8
33
Garrido-Navas C. de Miguel-Perez D. Exposito-Hernandez J. Bayarri C. Amezcua V. Ortigosa A. et al (2019). Cooperative and escaping mechanisms between circulating tumor cells and blood constituents. Cells8, 1382. 10.3390/cells8111382
34
Gerstberger S. Jiang Q. Ganesh K. (2023). Metastasis. Cell186, 1564–1579. 10.1016/j.cell.2023.03.003
35
Geula S. Moshitch-Moshkovitz S. Dominissini D. Mansour A. A. Kol N. Salmon-Divon M. et al (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science347, 1002–1006. 10.1126/science.1261417
36
Giampieri S. Manning C. Hooper S. Jones L. Hill C. S. Sahai E. (2009). Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol.11, 1287–1296. 10.1038/ncb1973
37
Gomis R. R. Gawrzak S. (2017). Tumor cell dormancy. Mol. Oncol.11, 62–78. 10.1016/j.molonc.2016.09.009
38
Gong D. Zhang J. Chen Y. Xu Y. Ma J. Hu G. et al (2019). The m6A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca2+ influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J. Exp. Clin. Cancer Res.38, 233. 10.1186/s13046-019-1223-y
39
Grande M. T. Sánchez-Laorden B. López-Blau C. De Frutos C. A. Boutet A. Arévalo M. et al (2015). Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med.21, 989–997. 10.1038/nm.3901
40
Grelet S. Link L. A. Howley B. Obellianne C. Palanisamy V. Gangaraju V. K. et al (2017). A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol.19, 1105–1115. 10.1038/ncb3595
41
Gros J. Tabin C. J. (2014). Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science343, 1253–1256. 10.1126/science.1248228
42
Gu C. Wang Z. Zhou N. Li G. Kou Y. Luo Y. et al (2019). Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N6-methyladenosine of Notch1. Mol. Cancer18, 168. 10.1186/s12943-019-1084-1
43
Gupta G. P. Nguyen D. X. Chiang A. C. Bos P. D. Kim J. Y. Nadal C. et al (2007a). Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature446, 765–770. 10.1038/nature05760
44
Gupta G. P. Perk J. Acharyya S. de Candia P. Mittal V. Todorova-Manova K. et al (2007b). ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc. Natl. Acad. Sci. U. S. A.104, 19506–19511. 10.1073/pnas.0709185104
45
Haemmerle M. Taylor M. L. Gutschner T. Pradeep S. Cho M. S. Sheng J. et al (2017). Platelets reduce anoikis and promote metastasis by activating YAP1 signaling. Nat. Commun.8, 310. 10.1038/s41467-017-00411-z
46
Headley M. B. Bins A. Nip A. Roberts E. W. Looney M. R. Gerard A. et al (2016). Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature531, 513–517. 10.1038/nature16985
47
Hsu P. J. Zhu Y. Ma H. Guo Y. Shi X. Liu Y. et al (2017). Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res.27, 1115–1127. 10.1038/cr.2017.99
48
Huang H. Weng H. Sun W. Qin X. Shi H. Wu H. et al (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol.20, 285–295. 10.1038/s41556-018-0045-z
49
Huang Y. Hong W. Wei X. (2022). The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol.15, 129. 10.1186/s13045-022-01347-8
50
Huh S. J. Liang S. Sharma A. Dong C. Robertson G. P. (2010). Transiently entrapped circulating tumor cells interact with neutrophils to facilitate lung metastasis development. Cancer Res.70, 6071–6082. 10.1158/0008-5472.CAN-09-4442
51
Jia G. Fu Y. Zhao X. Dai Q. Zheng G. Yang Y. et al (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol.7, 885–887. 10.1038/nchembio.687
52
Joyce J. A. Pollard J. W. (2009). Microenvironmental regulation of metastasis. Nat. Rev. Cancer9, 239–252. 10.1038/nrc2618
53
Kaplan R. N. Riba R. D. Zacharoulis S. Bramley A. H. Vincent L. Costa C. et al (2005). VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature438, 820–827. 10.1038/nature04186
54
Karras P. Black J. R. M. McGranahan N. Marine J.-C. (2024). Decoding the interplay between genetic and non-genetic drivers of metastasis. Nature629, 543–554. 10.1038/s41586-024-07302-6
55
Kessenbrock K. Plaks V. Werb Z. (2010). Matrix metalloproteinases: regulators of the tumor microenvironment. Cell141, 52–67. 10.1016/j.cell.2010.03.015
56
Kessenbrock K. Wang C.-Y. Werb Z. (2015). Matrix metalloproteinases in stem cell regulation and cancer. Matrix Biol. J. Int. Soc. Matrix Biol.44-46, 184–190. 10.1016/j.matbio.2015.01.022
57
Labelle M. Begum S. Hynes R. O. (2011). Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell20, 576–590. 10.1016/j.ccr.2011.09.009
58
Lamouille S. Xu J. Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol.15, 178–196. 10.1038/nrm3758
59
Li B. Xiong X. Xu J. Peng D. Nie G. Wen N. et al (2024). METTL3-mediated m6A modification of lncRNA TSPAN12 promotes metastasis of hepatocellular carcinoma through SENP1-depentent deSUMOylation of EIF3I. Oncogene43, 1050–1062. 10.1038/s41388-024-02970-0
60
Li J. Chen F. Peng Y. Lv Z. Lin X. Chen Z. et al (2020). N6-Methyladenosine regulates the expression and secretion of TGFβ1 to affect the epithelial-mesenchymal transition of cancer cells. Cells9, 296. 10.3390/cells9020296
61
Li J. Xie G. Tian Y. Li W. Wu Y. Chen F. et al (2022). RNA m6A methylation regulates dissemination of cancer cells by modulating expression and membrane localization of β-catenin. Mol. Ther.30, 1578–1596. 10.1016/j.ymthe.2022.01.019
62
Liao Q. Shi H. Yang J. Ge S. Jia R. Song X. et al (2024). FTO elicits tumor neovascularization in cancer-associated fibroblasts through eliminating m6A modifications of multiple pro-angiogenic factors. Cancer Lett.592, 216911. 10.1016/j.canlet.2024.216911
63
Littlefield J. W. Dunn D. B. (1958). Natural occurrence of thymine and three methylated adenine bases in several ribonucleic acids. Nature181, 254–255. 10.1038/181254a0
64
Liu H. Lan T. Li H. Xu L. Chen X. Liao H. et al (2021). Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics11, 1396–1411. 10.7150/thno.53227
65
Liu J. Yue Y. Han D. Wang X. Fu Y. Zhang L. et al (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol.10, 93–95. 10.1038/nchembio.1432
66
Liu L. Wang J. Sun G. Wu Q. Ma J. Zhang X. et al (2019). m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol. Cancer18, 188. 10.1186/s12943-019-1119-7
67
Liu N. Dai Q. Zheng G. Parisien M. Pan T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature518, 560–564. 10.1038/nature14234
68
Liu Q. Zhang H. Jiang X. Qian C. Liu Z. Luo D. (2017). Factors involved in cancer metastasis: a better understanding to “seed and soil” hypothesis. Mol. Cancer16, 176. 10.1186/s12943-017-0742-4
69
Liu S. Huang M. Chen Z. Chen J. Chao Q. Yin X. et al (2020). FTO promotes cell proliferation and migration in esophageal squamous cell carcinoma through up-regulation of MMP13. Exp. Cell Res.389, 111894. 10.1016/j.yexcr.2020.111894
70
Luo G.-Z. MacQueen A. Zheng G. Duan H. Dore L. C. et al (2014). Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun.5, 5630. 10.1038/ncomms6630
71
Mak C. S. L. Yung M. M. H. Hui L. M. N. Leung L. L. Liang R. Chen K. et al (2017). MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol. Cancer16, 11. 10.1186/s12943-017-0582-2
72
Mao L. Wang L. Lyu Y. Zhuang Q. Li Z. Zhang J. et al (2024). Branch chain amino acid metabolism promotes brain metastasis of NSCLC through EMT occurrence by regulating ALKBH5 activity. Int. J. Biol. Sci.20, 3285–3301. 10.7150/ijbs.85672
73
Mao Y. Dong L. Liu X. M. Guo J. Ma H. Shen B. et al (2019). m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun.10, 5332. 10.1038/s41467-019-13317-9
74
Mauer J. Luo X. Blanjoie A. Jiao X. Grozhik A. V. Patil D. P. et al (2017). Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature541, 371–375. 10.1038/nature21022
75
Meyer K. D. Patil D. P. Zhou J. Zinoviev A. Skabkin M. A. Elemento O. et al (2015). 5' UTR m(6)A promotes cap-independent translation. Cell163, 999–1010. 10.1016/j.cell.2015.10.012
76
Meyer K. D. Saletore Y. Zumbo P. Elemento O. Mason C. E. Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell149, 1635–1646. 10.1016/j.cell.2012.05.003
77
Nieswandt B. Hafner M. Echtenacher B. Männel D. N. (1999). Lysis of tumor cells by natural killer cells in mice is impeded by platelets. Cancer Res.59, 1295–1300.
78
Nieto M. A. Huang R. Y.-J. Jackson R. A. Thiery J. P. (2016). EMT: 2016. Cell166, 21–45. 10.1016/j.cell.2016.06.028
79
Osmulski P. A. Cunsolo A. Chen M. Qian Y. Lin C. L. Hung C. N. et al (2021). Contacts with macrophages promote an aggressive nanomechanical phenotype of circulating tumor cells in prostate cancer. Cancer Res.81, 4110–4123. 10.1158/0008-5472.CAN-20-3595
80
Ou B. Liu Y. Gao Z. Xu J. Yan Y. Li Y. et al (2022). Senescent neutrophils-derived exosomal piRNA-17560 promotes chemoresistance and EMT of breast cancer via FTO-mediated m6A demethylation. Cell Death Dis.13, 905. 10.1038/s41419-022-05317-3
81
Ouyang P. Li K. Xu W. Chen C. Shi Y. Tian Y. et al (2024). METTL3 recruiting M2-type immunosuppressed macrophages by targeting m6A-SNAIL-CXCL2 axis to promote colorectal cancer pulmonary metastasis. J. Exp. Clin. Cancer Res.43, 111. 10.1186/s13046-024-03035-6
82
Padua D. Zhang X. H. F. Wang Q. Nadal C. Gerald W. L. Gomis R. R. et al (2008). TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell133, 66–77. 10.1016/j.cell.2008.01.046
83
Peinado H. Zhang H. Matei I. R. Costa-Silva B. Hoshino A. Rodrigues G. et al (2017). Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer17, 302–317. 10.1038/nrc.2017.6
84
Ping X.-L. Sun B. F. Wang L. Xiao W. Yang X. Wang W. J. et al (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res.24, 177–189. 10.1038/cr.2014.3
85
Pinto R. Vågbø C. B. Jakobsson M. E. Kim Y. Baltissen M. P. O'Donohue M. F. et al (2020). The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res.48, 830–846. 10.1093/nar/gkz1147
86
Psaila B. Lyden D. (2009). The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer9, 285–293. 10.1038/nrc2621
87
Qian B.-Z. Pollard J. W. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell141, 39–51. 10.1016/j.cell.2010.03.014
88
Rejniak K. A. (2016). Circulating tumor cells: when a solid tumor meets a fluid microenvironment. Adv. Exp. Med. Biol.936, 93–106. 10.1007/978-3-319-42023-3_5
89
Reymond N. d'Água B. B. Ridley A. J. (2013). Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer13, 858–870. 10.1038/nrc3628
90
Roundtree I. A. Luo G. Z. Zhang Z. Wang X. Zhou T. Cui Y. et al (2017). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. ELife6, e31311. 10.7554/eLife.31311
91
Rowson-Hodel A. R. Wald J. H. Hatakeyama J. O'Neal W. K. Stonebraker J. R. VanderVorst K. et al (2018). Membrane Mucin Muc4 promotes blood cell association with tumor cells and mediates efficient metastasis in a mouse model of breast cancer. Oncogene37, 197–207. 10.1038/onc.2017.327
92
Schwartz S. Mumbach M. R. Jovanovic M. Wang T. Maciag K. Bushkin G. G. et al (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep.8, 284–296. 10.1016/j.celrep.2014.05.048
93
Sendinc E. Shi Y. (2023). RNA m6A methylation across the transcriptome. Mol. Cell83, 428–441. 10.1016/j.molcel.2023.01.006
94
Shackleton M. Quintana E. Fearon E. R. Morrison S. J. (2009). Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell138, 822–829. 10.1016/j.cell.2009.08.017
95
Shaul M. E. Fridlender Z. G. (2019). Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol.16, 601–620. 10.1038/s41571-019-0222-4
96
Shi H. Chai P. Jia R. Fan X. (2020). Novel insight into the regulatory roles of diverse RNA modifications: re-defining the bridge between transcription and translation. Mol. Cancer19, 78. 10.1186/s12943-020-01194-6
97
Shi H. Wang X. Lu Z. Zhao B. S. Ma H. Hsu P. J. et al (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res.27, 315–328. 10.1038/cr.2017.15
98
Shi X. Wang X. Yao W. Shi D. Shao X. Lu Z. et al (2024). Mechanism insights and therapeutic intervention of tumor metastasis: latest developments and perspectives. Signal Transduct. Target Ther.9, 192. 10.1038/s41392-024-01885-2
99
Sommer S. Lavi U. Darnell J. E. (1978). The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol.124, 487–499. 10.1016/0022-2836(78)90183-3
100
Sosa M. S. Bragado P. Aguirre-Ghiso J. A. (2014). Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer14, 611–622. 10.1038/nrc3793
101
Stark K. Schubert I. Joshi U. Kilani B. Hoseinpour P. Thakur M. et al (2018). Distinct pathogenesis of pancreatic cancer microvesicle-associated venous thrombosis identifies new antithrombotic targets in vivo. Arteriosclerosis Thrombosis Vasc. Biol.38, 772–786. 10.1161/ATVBAHA.117.310262
102
Steeg P. S. (2016). Targeting metastasis. Nat. Rev. Cancer16, 201–218. 10.1038/nrc.2016.25
103
Stemmler M. P. Eccles R. L. Brabletz S. Brabletz T. (2019). Non-redundant functions of EMT transcription factors. Nat. Cell Biol.21, 102–112. 10.1038/s41556-018-0196-y
104
Strell C. Lang K. Niggemann B. Zaenker K. S. Entschladen F. (2007). Surface molecules regulating rolling and adhesion to endothelium of neutrophil granulocytes and MDA-MB-468 breast carcinoma cells and their interaction. Cell. Mol. Life Sci. CMLS64, 3306–3316. 10.1007/s00018-007-7402-6
105
Su R. Dong L. Li Y. Gao M. Han L. Wunderlich M. et al (2020). Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell38, 79–96. 10.1016/j.ccell.2020.04.017
106
Su R. Dong L. Li Y. Gao M. He P. C. Liu W. et al (2022). METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol.24, 205–216. 10.1038/s41556-021-00835-2
107
Szczerba B. M. Castro-Giner F. Vetter M. Krol I. Gkountela S. Landin J. et al (2019). Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature566, 553–557. 10.1038/s41586-019-0915-y
108
Tan B. Zhou K. Liu W. Prince E. Qing Y. Li Y. et al (2022). RNA 6 -methyladenosine reader YTHDC1 is essential for TGF-beta-mediated metastasis of triple negative breast cancer. Theranostics12, 5727–5743. 10.7150/thno.71872
109
Thomas G. M. Brill A. Mezouar S. Crescence L. Gallant M. Dubois C. et al (2015). Tissue factor expressed by circulating cancer cell-derived microparticles drastically increases the incidence of deep vein thrombosis in mice. J. Thromb. Haemost.13, 1310–1319. 10.1111/jth.13002
110
Tong Z. T. Cai M. Y. Wang X. G. Kong L. L. Mai S. J. Liu Y. H. et al (2012). EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene31, 583–594. 10.1038/onc.2011.254
111
Ullmann P. Rodriguez F. Schmitz M. Meurer S. K. Qureshi-Baig K. Felten P. et al (2018). The miR-371∼373 cluster represses colon cancer initiation and metastatic colonization by inhibiting the TGFBR2/ID1 signaling axis. Cancer Res.78, 3793–3808. 10.1158/0008-5472.CAN-17-3003
112
Valastyan S. Weinberg R. A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell147, 275–292. 10.1016/j.cell.2011.09.024
113
van Tran N. Ernst F. G. M. Hawley B. R. Zorbas C. Ulryck N. Hackert P. et al (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res.47, 7719–7733. 10.1093/nar/gkz619
114
Visvanathan A. Patil V. Arora A. Hegde A. S. Arivazhagan A. Santosh V. et al (2018). Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene37, 522–533. 10.1038/onc.2017.351
115
Vleminckx K. Vakaet L. Mareel M. Fiers W. van Roy F. (1991). Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell66, 107–119. 10.1016/0092-8674(91)90143-m
116
Vu L. P. Pickering B. F. Cheng Y. Zaccara S. Nguyen D. Minuesa G. et al (2017). The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med.23, 1369–1376. 10.1038/nm.4416
117
Wang G. Dai Y. Li K. Cheng M. Xiong G. Wang X. et al (2021). Deficiency of Mettl3 in bladder cancer stem cells inhibits bladder cancer progression and angiogenesis. Front. Cell Dev. Biol.9, 627706. 10.3389/fcell.2021.627706
118
Wang Q. Chen C. Ding Q. Zhao Y. Wang Z. Chen J. et al (2020). METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut69, 1193–1205. 10.1136/gutjnl-2019-319639
119
Wang Q. Huang Y. Zhu Y. Zhang W. Wang B. Du X. et al (2024). The m6A methyltransferase METTL5 promotes neutrophil extracellular trap network release to regulate hepatocellular carcinoma progression. Cancer Med.13, e7165. 10.1002/cam4.7165
120
Wang X. Lu Z. Gomez A. Hon G. C. Yue Y. Han D. et al (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature505, 117–120. 10.1038/nature12730
121
Wang X. Zhao B. S. Roundtree I. A. Lu Z. Han D. Ma H. et al (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell161, 1388–1399. 10.1016/j.cell.2015.05.014
122
Wang Y. Li Y. Toth J. I. Petroski M. D. Zhang Z. Zhao J. C. (2014b). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol.16, 191–198. 10.1038/ncb2902
123
Wei C. Yang C. Wang S. Shi D. Zhang C. Lin X. et al (2019). Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer18, 64. 10.1186/s12943-019-0976-4
124
Winslow M. M. Dayton T. L. Verhaak R. G. W. Kim-Kiselak C. Snyder E. L. Feldser D. M. et al (2011). Suppression of lung adenocarcinoma progression by Nkx2-1. Nature473, 101–104. 10.1038/nature09881
125
Wu S. Liu L. Xu H. Zhu Q. Tan M. (2023). The involvement of MALAT1-ALKBH5 signaling axis into proliferation and metastasis of human papillomavirus-positive cervical cancer. Cancer Biol. and Ther.24, 2249174. 10.1080/15384047.2023.2249174
126
Xiao W. Adhikari S. Dahal U. Chen Y. S. Hao Y. J. Sun B. F. et al (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell61, 507–519. 10.1016/j.molcel.2016.01.012
127
Xu X. Zhuang X. Yu H. Li P. Li X. Lin H. et al (2024). FSH induces EMT in ovarian cancer via ALKBH5-regulated Snail m6A demethylation. Theranostics14, 2151–2166. 10.7150/thno.94161
128
Yang M.-H. Wu M. Z. Chiou S. H. Chen P. M. Chang S. Y. Liu C. J. et al (2008). Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol.10, 295–305. 10.1038/ncb1691
129
Yang Z. Wang T. Wu D. Min Z. Tan J. Yu B. (2020). RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J. Exp. Clin. Cancer Res.39, 203. 10.1186/s13046-020-01714-8
130
Ye X. Tam W. L. Shibue T. Kaygusuz Y. Reinhardt F. Ng Eaton E. et al (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature525, 256–260. 10.1038/nature14897
131
Yilmaz M. Christofori G. (2009). EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.28, 15–33. 10.1007/s10555-008-9169-0
132
Yin H. Zhang X. Yang P. Zhang X. Peng Y. Li D. et al (2021). RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun.12, 1394. 10.1038/s41467-021-21514-8
133
Yu D. Pan M. Li Y. Lu T. Wang Z. Liu C. et al (2022). RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner. J. Exp. Clin. Cancer Res.41, 6. 10.1186/s13046-021-02212-1
134
Yuan J.-h. Yang F. Wang F. Ma J. z. Guo Y. j. Tao Q. f. et al (2014). A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell25, 666–681. 10.1016/j.ccr.2014.03.010
135
Zeng H. Hou Y. Zhou X. Lang L. Luo H. Sun Y. et al (2022). Cancer-associated fibroblasts facilitate premetastatic niche formation through lncRNA SNHG5-mediated angiogenesis and vascular permeability in breast cancer. Theranostics12, 7351–7370. 10.7150/thno.74753
136
Zhang C. Wang Y. Zhen Z. Li J. Su J. Wu C. (2024). mTORC1 mediates biphasic mechano-response to orchestrate adhesion-dependent cell growth and anoikis resistance. Adv. Sci. (Weinh)11, e2307206. 10.1002/advs.202307206
137
Zhang G. Wang T. Huang Z. Chen Y. Sun L. Xia X. et al (2022). METTL3 dual regulation of the stability of LINC00662 and VEGFA RNAs promotes colorectal cancer angiogenesis. Discov. Oncol.13, 89. 10.1007/s12672-022-00557-3
138
Zhang H. Zhou J. Li J. Wang Z. Chen Z. Lv Z. et al (2023). N6-Methyladenosine promotes translation of VEGFA to accelerate angiogenesis in lung cancer. Cancer Res.83, 2208–2225. 10.1158/0008-5472.CAN-22-2449
139
Zhang S. Zhao B. S. Zhou A. Lin K. Zheng S. Lu Z. et al (2017). m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell31, 591–606. 10.1016/j.ccell.2017.02.013
140
Zheng G. Dahl J. A. Niu Y. Fedorcsak P. Huang C. M. Li C. J. et al (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell49, 18–29. 10.1016/j.molcel.2012.10.015
Summary
Keywords
M6A, RNA, methylation, metastasis, cancer
Citation
Zhou Y, Cao P and Zhu Q (2025) The regulatory role of m6A in cancer metastasis. Front. Cell Dev. Biol. 13:1539678. doi: 10.3389/fcell.2025.1539678
Received
04 December 2024
Accepted
16 April 2025
Published
28 April 2025
Volume
13 - 2025
Edited by
Vincenza Barresi, University of Catania, Italy
Reviewed by
Udayan Bhattacharya, NewYork-Presbyterian, United States
Omprakash Shriwas, National Cancer Institute at Frederick (NIH), United States
Updates
Copyright
© 2025 Zhou, Cao and Zhu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Zhu, newzhuqing1972@163.com
‡These authors have contributed equally to this work
ORCID: Qing Zhu, orcid.org/0000-0003-2182-5709
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.