- Department of Biology, Salem State University, Salem, MA, United States
Cilia assembly is accompanied by rapid and highly coordinated transcription of hundreds of genes. Cilia gene regulation has been studied extensively in both metazoans and unicellular model organisms. The forkhead and RFX family transcription factors regulating cilia genes in animals were first identified 25 years ago and considerable molecular details of the regulatory processes have been described since then. While many of the most important early studies of cilia gene regulation were done in unicellular organisms, additional molecular players need to be discovered for a more complete understanding in these organisms. In this concise review, written primarily for students new to the field, I present a brief history of research on cilia gene regulation, highlight some key metazoan discoveries from the last decade, and discuss gaps in our understanding of cilia gene regulation in unicellular model organisms with a focus on Chlamydomonas reinhardtii.
1 Introduction
Cilia (a.k.a. Flagella in some contexts) extend from eukaryotic cells to carry out important functions in motility and sensory perception. Cilia are complex organelles including a ciliary membrane with distinct composition surrounding a ciliary matrix and a microtubule-based axoneme (Brown and Witman, 2014; Mill et al., 2023). The requirement for nodal cilia in determining left-right asymmetry during vertebrate development (Nonaka et al., 1998) and for primary cilia in signaling pathways critical for development and homeostasis (Pazour et al., 2000) led to intense interest in cilia over the last quarter century (Brown and Witman, 2014; Mill et al., 2023). Research on cilia includes studies on the regulated expression of hundreds of genes encoding cilia proteins (hereafter ‘cilia genes’). This review focuses on the historical and recent discoveries in cilia gene regulation with emphasis on metazoan transcription factors and underexplored regulatory mechanisms in unicellular organisms, particularly Chlamydomonas reinhardtii. As the mini-review format makes it difficult to cite all relevant literature, I apologize to authors I have excluded and direct interested readers to other excellent reviews describing cilia gene expression in more depth (Choksi et al., 2014; Lefebvre and Rosenbaum, 1986; Lewis and Stracker, 2021; Thomas et al., 2010).
2 A brief history of research on cilia and transcription
Research on expression of cilia genes can be viewed as a series of convergences. Discoveries on gene expression occurred in parallel with new cilia discoveries until these paths converged and led to critical breakthroughs in cilia gene expression.
2.1 Connecting cilia and gene expression across phyla
The 1960s were an important period for research on cilia and transcription. The foundation for understanding gene expression was established when theoretical and experimental approaches to DNA structure and function (Crick, 1958; Dounce, 1953; Franklin and Gosling, 1953; Watson and Crick, 1953) and genetic approaches to gene regulation and the genetic code (Crick et al., 1961; Jacob and Monod, 1961) converged at the beginning of the decade with the discovery of messenger RNA (Brenner et al., 1961; Cobb, 2015; Gros et al., 1961; Matthaei and Nirenberg, 1961). Microscopy and biochemistry of cilia identified axonemal dynein arms as the location of the ATPase for ciliary motility (Gibbons, 1963; Gibbons and Rowe, 1965). Early studies on cilia growth were in Chlamydomonas moewusii and sea urchin (Auclair and Siegel, 1966; Lewin, 1953). Soon after gene expression principles were discovered, studies on ciliary regeneration demonstrated its value as a model system for regulated gene expression (Rosenbaum and Child, 1967). Importantly, cilia regeneration and incorporation of tritiated leucine into TCA insoluble protein were inhibited by cycloheximide, making it clear that new protein synthesis is required for assembly of full-length cilia (Rosenbaum and Child, 1967).
Bookending the decade, Rosenbaum et al. (1969) published a watershed paper establishing C. reinhardtii as a model organism for cilia gene regulation. When cycloheximide inhibited protein synthesis during deciliation, cells formed half-length cilia, indicating a preexisting pool of cilia proteins. This also suggested that a reduced concentration of one or more proteins in this pool might be a limiting factor for full-length cilia assembly (Rosenbaum et al., 1969). Pulse labeling of arginine-requiring cells with arginine-3H during cilia regeneration showed that incorporation of newly synthesized proteins into cilia peaked when the precursor pool in the cell body was nearly depleted. Colchicine inhibition of cilia growth also indicated that new protein synthesis could occur in the absence of cilia elongation (Rosenbaum et al., 1969). Later work showed that this increase in cilia protein synthesis is largely due to mRNA accumulation (Minami et al., 1981; Silflow and Rosenbaum, 1981). Additional progress in Chlamydomonas is discussed in more detail below, especially in Section 4.
Early studies on cilia regeneration in Tetrahymena and sea urchin showed that ability to regenerate cilia is conserved across phyla (Auclair and Siegel, 1966; Rosenbaum and Carlson, 1969). Unlike in Chlamydomonas, cycloheximide treatment of Tetrahymena at the time of deciliation completely blocked cilia regeneration suggesting that synthesis of some limiting protein is necessary to utilize the cilia precursor pool present at the time of deciliation (Guttman and Gorovsky, 1979; Rannestad, 1974; Rosenbaum and Carlson, 1969). Cilia protein synthesis in starved regenerating Tetrahymena cells can be detected above the protein synthesis background in starved non-deciliated cells revealing a dramatic increase in tubulin synthesis (Guttman and Gorovsky, 1979). This increase in tubulin synthesis is preceded by an increase in tubulin gene transcription resulting in increased tubulin mRNA available for translation (Bird and Zimmerman, 1980; Guttman and Gorovsky, 1979). Cilia regeneration in sea urchin embryo does occur in the presence of the protein synthesis inhibitor puromycin, indicating that, like in Chlamydomonas, a preexisting pool of cilia precursor proteins can be used in the absence of translation (Auclair and Siegel, 1966). However, new protein synthesis, including synthesis of tubulin and other less abundant cilia proteins, is induced during regeneration (Stephens, 1977). Increased tubulin mRNA abundance could be blocked by the transcription inhibitor actinomycin D (Merlino et al., 1978), and nuclear run-on assays confirmed that increased tubulin mRNA abundance during cilia regeneration is due in part to increased transcription (Gong and Brandhorst, 1987).
The dramatic transformation of Naegleria gruberi from amoeba to an elongated cell with cilia that occurs on transferring cells to nutrient-free media (Fulton and Dingle, 1967) was also developed during this period as a model to study cilia gene expression. The majority of tubulin incorporated into the growing cilia is newly synthesized during this differentiation period (Kowit and Fulton, 1974) and is specific to cilia (Kennard et al., 2025). Differentiation-specific tubulin synthesis can be inhibited by actinomycin D (Fulton and Kowit, 1975) and translatable tubulin mRNA increases during differentiation (Lai et al., 1979). Nuclear run-on assays showed that the increase can largely be attributed to increased transcription (Lee and Walsh, 1988). Application of microarrays and RNA-seq to study differentiation-specific gene induction in Naegleria revealed increased expression of multiple cilia genes including a cilia-specific network of microtubule binding proteins (Fritz-Laylin and Cande, 2010; Kennard et al., 2025). Taken together, the studies described above established non-vertebrate model organisms as important contributors to a broad understanding of cilia gene regulation.
2.2 Connecting cilia to specific transcription factors
Work in the late 1990s implicated specific transcription factors in the regulation of cilia genes. Interest in cilia expanded after mouse embryonic nodal cilia were shown to be required to establish normal left-right asymmetry (Nonaka et al., 1998) and cilia were first connected to polycystic kidney disease (Pazour et al., 2000; Qin et al., 2001). Initial characterization of transcription factors in the 1980s led to studies of tissue specific expression patterns in the 1990s (Clevidence et al., 1993; Maniatis et al., 1987). The first transcription factor shown to co-occur with cilia assembly in both space and time was mammalian HFH-4 (FOXJ1) (Blatt et al., 1999; Hackett et al., 1995) a forkhead family protein which is also required for mouse motile cilia assembly (Chen et al., 1998). Its orthologs in Drosophila and Caenorhabditis elegans regulate specialization of cilia in sensory neurons (Cachero et al., 2011; Newton et al., 2012, Brocal-Ruiz et al., 2023).
The RFX protein, DAF-19, was identified in a screen for mutants with defective dye filling in sensory cilia in C. elegans (Perkins et al., 1986) and was found to regulate cilia genes via a 14-bp target sequence, the X box (Swoboda et al., 2000). RFX genes have only been found in the genomes of unikont organisms. However, not all ciliated unikonts have RFX transcription factors and some non-ciliated unikonts do have RFX. Piasecki et al. (2010) concluded from these observations that RFX originated in an early unikont ancestor and only later began regulating cilia genes in an early animal or ancestor of animals and choanoflagellates. The important recent discovery that choanoflagellates also use RFX to regulate cilia genes clearly pushes this event back to a common ancestor of choanoflagellates and animals (Coyle et al., 2023). In addition, some lineages secondarily lost cilia and maintained RFX which regulates non-cilia genes. Additional details on FOXJ1 and RFX regulation are discussed below.
2.3 Cataloging cilia genes
Understanding cilia gene regulation needs to include identifying which genes are regulated. Early studies developing methods for purifying and fractionating cilia in Tetrahymena and Chlamydomonas began to reveal the complexity of the cilia proteome (Gibbons, 1963; Piperno et al., 1977; Witman et al., 1972). Genome sequencing allowed comparison of genomes of ciliated and non-ciliated organisms identifying hundreds of candidate cilia genes (Avidor-Reiss et al., 2004; Li et al., 2004; Merchant et al., 2007). A recent study using additional tools now available for phylogenetic profiling identified 152 new strong candidate cilia genes (Dobbelaere et al., 2023). Genome-wide searches and comparative genomics identified multiple new cilia genes with X box sequences in their promoters in C. elegans (Blacque et al., 2005; Chen et al., 2006; Efimenko et al., 2005) and Drosophila (Laurençon et al., 2007). Proteomic analyses of purified cilia added more cilia proteins and linked them with specific cilia subfractions (Ishikawa et al., 2012; Pazour et al., 2005; Smith et al., 2005). As a complementary approach, transcriptomics supported many of the results of the earlier studies and added additional cilia genes. Some of these studies attempted to establish a transcriptional profile during normal or experimentally induced ciliogenesis (Albee et al., 2013; Fritz-Laylin and Cande, 2010; Kennard et al., 2025; Patir et al., 2020; Quigley and Kintner, 2017; Stolc et al., 2005; Zones et al., 2015). In others, transcriptome analysis was performed after the expression of RFX or forkhead transcription factors was disrupted or induced ectopically to identify genes regulated by the transcription factors in question (Chung et al., 2014; Lemeille et al., 2020; Phirke et al., 2011; Stubbs et al., 2008). Various studies have also made ciliome databases accessible to the research community including CilDB, Syscilia Gold Standard, CiliaCarta, and CilioGenics, and the Chlamydomonas flagellar proteome database (Arnaiz et al., 2014; Elliott et al., 2023; Pazour et al., 2005; Pir et al., 2024; van Dam et al., 2013; van Dam et al., 2019).
3 Expanding understanding in metazoans
Involvement of FOXJ1 and RFX family transcription factors in the expression of animal cilia genes was discovered over 20 years ago (Choksi et al., 2014). FOXJ1 regulates the expression of proteins needed for motility including dynein arms and radial spokes (Lewis and Stracker, 2021). RFX family transcription factors are more generally involved in transcription of proteins needed for assembly of both motile and immotile cilia (Choksi et al., 2014). Considerable details have also emerged about functional specialization of cilia through the use of different combinations of transcription factors and their upstream regulatory networks.
3.1 RFX and forkhead interactions in functional specialization of cilia
Interactions between FOXJ1 and RFX family transcription factors have become increasingly clear during the development of specific ciliary functions in different contexts. Correct specification of ciliated dendrites on the chordotonal neurons in Drosophila is dependent on both RFX and the distant FOXJ1 relative, fd3F (Cachero et al., 2011; Newton et al., 2012). Similarly, C. elegans FKH-8 (a forkhead TF) and DAF-19 (an RFX TF) interact with each other in regulation of cilia genes for assembly of sensory cilia (Brocal-Ruiz et al., 2023). Surprisingly, mouse FOXJ1and Xenopus FOXN4 are able to functionally substitute for FKH-8 for C. elegans cilia gene expression, suggesting that co-regulation of cilia genes by RFX and forkhead proteins may be an ancient connection. Human RFX3 and FOXJ1 act together in human airway MCCs (Didon et al., 2013) as described in more detail below. RFX2 interacts with FOXJ1 for cilia gene expression in larval MCCs in Xenopus. In these interactions FOXJ1 may be most often bound to distal enhancers while RFX2 is bound to proximal promoter sequences and recruits FOXJ1 to the promoters by dimerization and chromatin looping (Quigley and Kintner, 2017). It is unknown whether similar interactions occur in vertebrates outside of motile ciliogenesis. A more detailed description of the specific gene regulatory network in MCC differentiation is presented below.
3.2 Multiciliogenesis
In humans and mice, cells with dozens of motile cilia (multiciliated cells, MCC) are found on the respiratory epithelium (Figure 1A), in brain ventricles, and in reproductive organs (Brooks and Wallingford, 2014). During commitment of progenitor cells to MCC differentiation, the expression of FOXJ1 and RFX2/3 is tightly regulated. Three key regulators of FOXJ1 and RFX expression are the geminin family proteins GEMC1 and MCIDAS (a.k.a. multicilin) and the transcription factor p73 (Lu et al., 2019; Marshall et al., 2016; Terré et al., 2016). Despite tissue-specific differences in these regulatory pathways (Wildung et al., 2019), a relatively clear picture has emerged for respiratory and oviduct epithelia (Figure 1B) (Lewis and Stracker, 2021). In these tissues, GEMC1 acts upstream of MCIDAS (Lu et al., 2019) and forms a complex with p73 and the transcription factor E2F5. This complex regulates the expression of downstream regulators of cilia genes including FOXJ1, RFX3, and p73, itself, such that loss of either GEMC1 or p73 led to loss of respiratory and oviduct MCCs (Lalioti et al., 2019). MCIDAS acting downstream of GEMC1 and in a complex with E2F4 or E2F5 activates FOXJ1 and other genes involved in the extensive basal body duplication needed to nucleate multiple cilia (Lu et al., 2019). The story in ciliated cells in the brain is somewhat less clear. However, loss of p73 in the choroid plexus leads to an upregulation of E2F/MCIDAS activity and upregulation of microRNA-449 which compensates for the absence of p73 (Wildung et al., 2019). Additional work will be necessary to fully describe these interactions in brain epithelial cells.
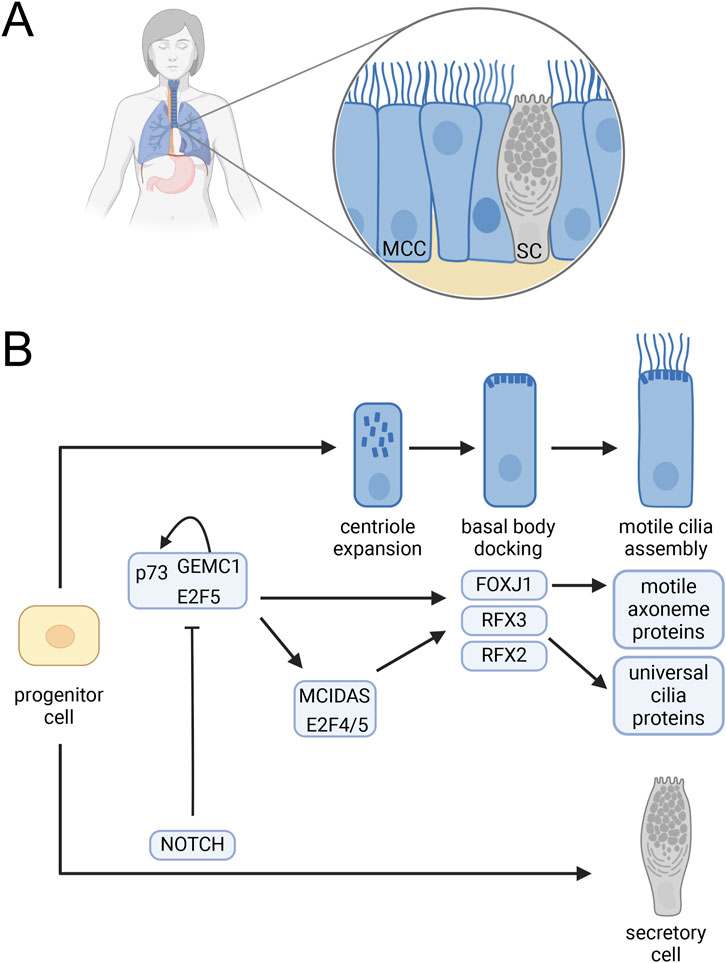
Figure 1. Multiciliated cell differentiation. (A) Mammalian respiratory epithelium including MCC interspersed with secretory cells. (B) Regulation of MCC development. A complex containing GEMC1 and the E2F5 and p73 transcription factors activates p73 as well as MCIDAS and the FOXJ1, RFX2 and RFX3 ciliogenesis transcription factors. MCIDAS in complex with E2F4 or E2F5 also activates FOXJ1, RFX2, and RFX3 and promotes centriole expansion. FOXJ1 promotes basal body docking and activates expression of axonemal proteins required for cilia motility. RFX2 and RFX3 activate the expression of many cilia genes needed for assembly of all cilia including the motile cilia on MCC. Notch signaling blocks GEMC1 complex activation leading to a secretory cell fate (Lalioti et al., 2019; Lewis and Stracker, 2021; Lu et al., 2019). Created in BioRender. Brown, (2025) https://BioRender.com/z01l282.
3.3 Other emerging regulatory mechanisms
3.3.1 Auto fatty acylation and dimerization
Using recombinant RFX3 in the presence of clickable fatty acid analogs Chen et al. (2018) found that RFX3 autoacylates at a conserved cysteine (544) in its dimerization domain. Mutation of this cysteine reduced fatty acylation and dimerization but not nuclear localization. In an RFX3 null background, expression of WT RFX3, but not C544S RFX3 drove the expression of three known RFX3 targets, suggesting that RFX3 fatty acylation is required for normal expression of cilia genes (Chen et al., 2018).
3.3.2 Regulation by corticosteroid receptors
ChIP-seq analysis in the rat hippocampus identified over 50 cilia genes bound by mineralocorticoid receptors (MRs). MRs normally respond to circadian and stress-induced changes in adrenal gland hormone release (Mifsud et al., 2021). MR bound at sites on the DNA that are also bound by RFX3, suggesting a possible RFX3/MR interaction. Supporting that hypothesis, MR agonists were required for neuronal differentiation and ciliogenesis and MR antagonists prevented differentiation and cilia growth (Mifsud et al., 2021). Together these results suggest that a response to corticosteroids through MR and RFX3 is required for ciliogenesis involved in neuronal development.
3.3.3 Mitochondrial stress
Recent studies have indicated a signaling connection between mitochondria and cilia (Bae et al., 2019). Perturbing the function of mitochondria in astrocytes led to robust activation of multiple cilia genes and FOXJ1 (Ignatenko et al., 2022). The astrocytes remained monociliated despite the role of FOXJ1 in multiciliogenesis in other contexts. However, the cilia were abnormally long and contorted due to an unidentified mechanism. It is possible that the upregulation of cilia genes only occurs as a pathological response (Ignatenko et al., 2022), but it will be important to explore the intriguing possibility that mitochondrial function might be connected to cilia gene regulation under normal circumstances.
4 Cilia and transcription in Chlamydomonas
During Chlamydomonas cilia regeneration, new protein synthesis begins within the first few minutes and peaks around the time that the pre-existing cilia precursor pool in the cell body would be depleted without translation (Lefebvre et al., 1978; Rosenbaum et al., 1969; Weeks et al., 1977). This increase in translation corresponds with a peak in cilia gene mRNA abundance as shown initially for tubulin genes (Minami et al., 1981; Silflow and Rosenbaum, 1981). The mechanisms of the increases and decreases in mRNA abundance remain largely unknown although some of the story has been revealed (Figure 2).
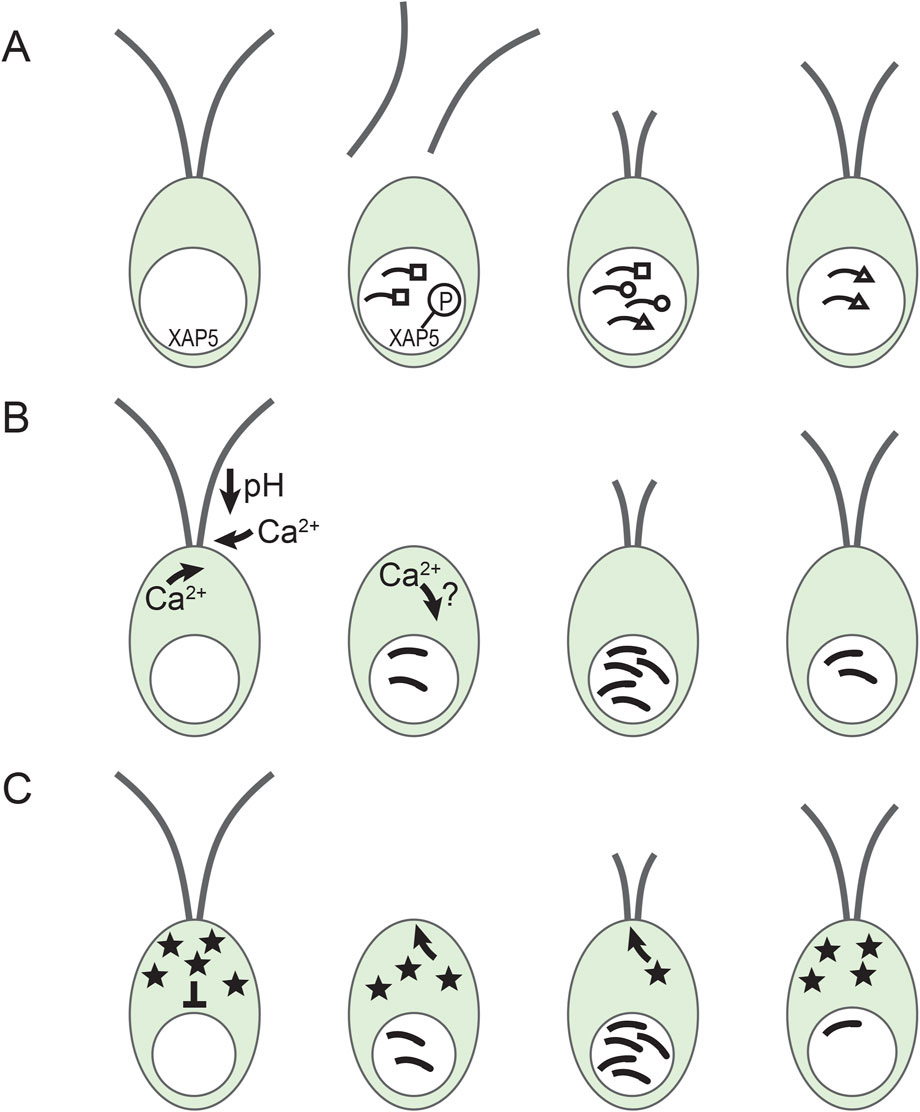
Figure 2. Gene regulation during cilia regeneration in Chlamydomonas. (A) Different temporal expression patterns and XAP5. Cilia genes are expressed in groups with different expression patterns. Depicted here are early (squares) middle (circles) and late (triangle) genes (Schloss et al., 1984). In addition, nuclear localized XAP5 is phosphorylated rapidly after deciliation by an unidentified kinase. (B) Calcium involvement in cilia gene induction. Upon pH shock, extracellular calcium enters the cell and stimulates the release of additional calcium from intracellular stores and triggers deciliation (Quarmby and Mahjoub, 2023). Calcium entry is needed for maximal gene induction concomitant with the initiation of cilia regrowth (Cheshire and Keller, 1991). How the intracellular calcium concentration changes are involved in normal gene induction is still unclear. (C) The repressor sequestration model. A constitutively produced repressor (stars) blocks cilia gene expression. After deciliation, the repressor is sequestered in rapidly growing cilia, reducing the effective concentration of the repressor in the cell body and allowing increased transcription of cilia genes. As cilia approach full length, cilia assembly slows down allowing the repressor to accumulate and cilia gene expression to slow down (Perlaza et al., 2023).
4.1 Message abundance changes
4.1.1 Increased transcription and changes in message stability
The increase in mRNA abundance is due to increases in both transcription and mRNA stability. Run-on transcription of tubulin RNAs increased in nuclei isolated from cilia regenerating cells compared with nuclei from non-regenerating cells. This was the first direct demonstration that new transcription contributes to tubulin mRNA accumulation during regeneration (Keller et al., 1984). Baker et al. (1984) confirmed this result with in vivo 32P pulse labeling and found that tubulin mRNA stability is doubled in deciliated cells compared with non-deciliated cells. Cis DNA sequence elements have been identified that regulate increased expression during cilia regeneration of the TUB2 β-tubulin gene, the TUA1 α-tubulin gene, and the DIC2 (a.k.a. ODA6) gene encoding flagellar outer arm dynein intermediate chain 2 (a.k.a. IC70) (Bandziulis and Rosenbaum, 1988; Davies et al., 1992; Davies and Grossman, 1994; Kang and Mitchell, 1998; Periz and Keller, 1997). After cilia regenerate, the stabilization of tubulin mRNAs switches to rapid degradation, a translation-dependent process that is independent of the normal deadenylation-dependent pathway operating on the same mRNAs prior to deciliation (Baker et al., 1986; 1984; Baker and Liggit, 1993; Gera and Baker, 1998). Thus, increases in transcription and mRNA stability both contribute to the accumulation of cilia gene mRNAs and the decrease in message abundance following regeneration involves a novel pathway that has not yet been characterized.
4.1.2 Complex patterns of mRNA abundance change
Although expression of most cilia genes increases minutes after deciliation, timing is different for different groups of genes (Albee et al., 2013; Lefebvre et al., 1978; Remillard and Witman, 1982; Schloss et al., 1984). Lefebvre et al. (1978) found that tubulin synthesis remained high for hours while synthesis of other proteins peaked in under an hour. Remillard and Witman (1982) used 2-D gels to connect proteins to specific ciliary structures (e.g., radial spokes) and showed that proteins found together in a certain structure were produced with similar kinetics. Subsequently, dot blot hybridization identified three classes of RNAs that were increased at early, middle, and late times during regeneration (Figure 2A) (Schloss et al., 1984). Albee et al. (2013) using RNA-seq identified 16 different expression profiles. Again, genes encoding proteins that work together often had a similar expression pattern (Albee et al., 2013). It is likely that these complex expression patterns involve several parallel regulatory pathways that have yet to be discovered.
It is unclear whether the mechanisms regulating cilia gene expression during the cell cycle are the same as those during cilia regeneration. Chlamydomonas cells in synchronized cultures resorb their cilia around the G1/S transition and regrow them with most cells being ciliated by the transition to the post-mitotic phase. Levels of mRNAs encoding intraflagellar transport (IFT) proteins were strictly regulated with peak accumulation occurring during cilia assembly in S/M (Wood et al., 2012). RNA-seq analysis during the diurnal cycle showed that many cilia genes are coordinately regulated during cilia assembly and exhibit different expression clusters (Zones et al., 2015). However, since these studies do not have the same temporal resolution as cilia regeneration studies (Albee et al., 2013) it is unknown whether the regeneration expression clusters are relevant to cilia growth during the cell cycle. Interestingly, some mutants induce cilia genes in cycling vegetative cells but not in non-cycling gametes, suggesting distinct cilia gene regulation during the cell cycle (Lefebvre et al., 1988).
4.2 Transcription factors
Importantly, the FOXJ1 and RFX transcription factors are not found in Chlamydomonas (Chu et al., 2010; Piasecki et al., 2010). Recently, the first transcription factor, XAP5, regulating Chlamydomonas cilia genes was identified (Li et al., 2018). Consistent with the hypothesis that multiple pathways control cilia genes, xap5 mutant cells downregulated expression of some cilia genes whereas others were unaffected. Nuclear localization of XAP5 was needed for expression of XAP5-dependent genes (Figure 2A). Sequence-specific binding of XAP5 to cilia gene promoters assisted in the recruitment of RNA polymerase II to those promoters indicating that XAP5 is a transcription factor (Li et al., 2018). Although XAP5 is required for basal expression of several cilia genes and for upregulation of some of those genes following pH shock, it is currently unclear whether XAP5 is required for the normal induction of cilia genes during regrowth since the xap5 mutant lacks cilia.
While XAP5 orthologs are found broadly in eukaryotes, their presence in a genome does not predict presence of cilia and so far, XAP5 has only been shown to regulate cilia genes in Chlamydomonas and mice. In mice, XAP5 and its paralog XAP5L act antagonistically during spermatogenesis with XAP5 promoting expression of multiple cilia genes including FOXJ1 and RFX transcription factors, whereas XAP5L represses many of those same genes (Wang et al., 2025). The Naegleria gruberi and Tetrahymena thermophila genomes both lack XAP5 (Eisen et al., 2006; Fritz-Laylin et al., 2010; Tegenfeldt et al., 2025). Emphasizing that XAP5 proteins have other functions outside of regulating cilia genes, Schizosaccharomyces pombe which lacks cilia, does have an XAP5 orthologous gene in its genome (Tegenfeldt et al., 2025; Wood et al., 2002).
Since XAP5 only regulates some cilia genes, additional transcription factors likely regulate the XAP5-independent Chlamydomonas cilia genes. Three other predicted transcription factors have been suggested as good candidates due to their increased expression following deciliation and during cell cycle regulated cilia growth (Albee et al., 2013; Sale and Dutcher, 2023; Zones et al., 2015). These are Cre02. g103450, encoding a protein with a domain 50%–60% similar over 100 amino acids to MYB domains in plants and animals, Cre03. g201250 encoding a protein with zinc finger and G-patch nucleic acid binding domains, and Cre04. g228400, encoding a WRKY family plant and algae specific transcription factor (Craig et al., 2023; Sale and Dutcher, 2023). Interestingly, although Cre03. g201250 and Cre04. g22840 mRNA levels peaked around the time that most cells are ciliated, Cre02. g103450 level remained elevated over the next 10 h of day in light-synchronized cultures (Zones et al., 2015). This difference suggests possible functional differences between Cre02. g103450 and the other two genes. It will be important to analyze cilia gene expression during regeneration and the cell cycle in mutants lacking these proteins.
4.3 Signaling pathways
Upregulation of cilia genes in Chlamydomonas is likely to involve initiating events associated with cilia that stimulate signaling to the nucleus activating one or more transcription factors. The initiating signal and molecular details of the signaling pathway(s) are still not known, but some clues have emerged and are discussed below (Quarmby and Mahjoub, 2023; Sale and Dutcher, 2023).
4.3.1 Calcium
Calcium signaling regulates cilia motility, pH shock deciliation, cilia growth and maintenance, and cilia gene expression (Brown et al., 2012; Cheshire and Keller, 1991; Quader et al., 1978; Quarmby and Mahjoub, 2023). Under normal conditions, pH shock or mechanical shearing lead to deciliation, selective accumulation of mRNAs from cilia genes, and regrowth of cilia. Manipulating calcium concentration uncoupled these events (Cheshire and Keller, 1991). Cells deciliated in 10−7 M calcium did not regenerate cilia or accumulate cilia mRNAs until calcium was added, after which submaximal accumulation of cilia mRNAs occurred. When calcium was present during deciliation but lowered immediately after, a submaximal cilia mRNA accumulation occurred after deciliation that was uncoupled from cilia growth. If these cells are maintained in ∼10−7 M calcium, they begin to regrow cilia around 135 min after deciliation. This regrowth was accompanied by a small peak of mRNA abundance (Cheshire and Keller, 1991). In addition, wild-type cells accumulate cilia mRNAs in response to experimentally stimulated cilia resorption and mutants lacking cilia or unable to deciliate and cells treated to block calcium influx accumulate cilia mRNAs submaximally (Cheshire et al., 1994; Evans and Keller, 1997; Lefebvre, 1980; Lefebvre et al., 1978). Together these results indicate that maximal cilia gene induction is likely to result from a combination of responses from overlapping signaling pathways, some of which are calcium dependent (Figure 2B) (Cheshire and Keller, 1991).
4.3.2 Kinases
Many signaling pathways involve kinases that add phosphate groups to modify the activity of target proteins. Several Chlamydomonas cilia associated kinases have been identified (Mahjoub et al., 2004; Pazour et al., 2005; Wang et al., 2019; Wilson and Lefebvre, 2004). However, no kinases have yet been definitively connected with cilia gene induction. Interestingly, nuclear localized XAP5 transcription factor was phosphorylated within 1 min after pH shock deciliation (Figure 2A) and mutant XAP5 that could not be phosphorylated did not support cilia growth (Li et al., 2018). Identifying the XAP5 phosphorylating kinase could be an important step in identifying signaling pathways for cilia gene induction during regeneration.
4.3.3 Repressors
An early response leading to cilia gene induction could be activation of an activator protein or inactivation of a repressor protein. Perlaza et al. (2023) recently proposed and tested two repressor-based models. Their repressor sequestration model postulated that a repressor, or a protein that activates a repressor, is continually produced and is preferentially sequestered in rapidly growing cilia (Figure 2C). This model reproduced the essential features of cilia length control and cilia gene expression dynamics. It also correctly predicted that mutants with impaired IFT transport have lower accumulation of cilia mRNAs. If alleviation of repression is a fundamental mode of upregulation for the pulse of cilia genes, identifying the postulated repressor will be a critical next step.
5 Discussion
Sixty years ago, cilia regeneration was established as a model for gene induction (Rosenbaum and Child, 1967). Recently, much has been learned about the transcription factors FOXJ1, RFX2, and RFX3, including upstream signaling pathways, interactions with other transcription factors, and tissue-specific differences in regulation (Lewis and Stracker, 2021). However, many tissue-specific details need clarification. For instance, how microRNAs and p73 coordinate MCC differentiation in the choroid plexus is still not completely understood. Autoacylation of RFX3 also raises the question of what other post-translational modifications are important.
Cilia gene regulation in unicellular organisms is primed for exploration (Marshall, 2024; Quarmby and Mahjoub, 2023; Rosenbaum, 2009; Sale and Dutcher, 2023). FOXJ1 and RFX-family proteins are absent in Chlamydomonas, Tetrahymena, and Naegleria. (Piasecki et al., 2010). XAP5 orthologs are found broadly in eukaryotes, but not in all ciliated organisms. For instance, the Naegleria and Tetrahymena genomes lack XAP5 (Tegenfeldt et al., 2025). Other transcription factors, yet to be connected with cilia, must regulate many cilia genes in these organisms. Calcium is involved in Chlamydomonas cilia gene regulation via unknown pathways (Cheshire and Keller, 1991). In addition, it will be important to identify the recently proposed repressor (Perlaza et al., 2023). Powerful molecular (Picariello et al., 2020) and omics tools now available in Chlamydomonas make this an excellent time to revisit previous approaches that identified mutants defective in cilia gene regulation (Lefebvre et al., 1988). Targeted disruption of candidate genes and screens for mutants unable to induce cilia genes during cilia regeneration or with constitutive expression of cilia genes in the presence of full-length cilia are likely to uncover many signaling proteins and additional transcription factors in the coming years.
Author contributions
JB: Conceptualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funding for the publication charge was provided by the SSU College of Arts and Sciences, the SSU Department of Biology, the SSU Massachusetts State College Association Fund for Continuing Scholarship, and a Scholarship Support Grant to JMB from the SSU Center for Research and Creative Activities.
Acknowledgments
I would like to thank Drs. Laura Laranjo and Ryan Fisher for critical reading of the manuscript and the SSU College of Arts and Sciences, Department of Biology, and Center for Research and Creative Activities for assistance with publication charges.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
IFT, intraflagellar transport; MCC, multiciliated cell; MR, mineralocorticoid receptor.
References
Albee, A. J., Kwan, A. L., Lin, H., Granas, D., Stormo, G. D., and Dutcher, S. K. (2013). Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in chlamydomonas reinhardtti. G3 GenesGenomesGenetics 3, 979–991. doi:10.1534/g3.113.006338
Arnaiz, O., Cohen, J., Tassin, A.-M., and Koll, F. (2014). Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 3, 9. doi:10.1186/2046-2530-3-9
Auclair, W., and Siegel, B. W. (1966). Cilia regeneration in the sea urchin embryo: evidence for a pool of ciliary proteins. Science 154, 913–915. doi:10.1126/science.154.3751.913
Avidor-Reiss, T., Maer, A., Koundakjian, E., Polyanovsky, A., Keil, T. A., Subramaniam, S., et al. (2004). Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117, 527–539. doi:10.1016/s0092-8674(04)00412-x
Bae, J.-E., Kang, G. M., Min, S. H., Jo, D. S., Jung, Y.-K., Kim, K., et al. (2019). Primary cilia mediate mitochondrial stress responses to promote dopamine neuron survival in a Parkinson’s disease model. Cell. Death Dis. 10, 952. doi:10.1038/s41419-019-2184-y
Baker, E. J., Keller, L. R., Schloss, J. A., and Rosenbaum, J. L. (1986). Protein synthesis is required for rapid degradation of tubulin mRNA and other deflagellation-induced RNAs in chlamydomonas reinhardi. Mol. Cell. Biol. 6, 54–61. doi:10.1128/mcb.6.1.54
Baker, E. J., and Liggit, P. (1993). Accelerated poly(A) loss and mRNA stabilization are independent effects of protein synthesis inhibition on a-tubulin mRNA in Chlamydomonas.
Baker, E. J., Schloss, J. A., and Rosenbaum, J. L. (1984). Rapid changes in tubulin RNA synthesis and stability induced by deflagellation in chlamydomonas. J. Cell. Biol. 99, 2074–2081. doi:10.1083/jcb.99.6.2074
Bandziulis, R. J., and Rosenbaum, J. L. (1988). Novel control elements in the alpha-1 tubulin gene promoter from Chlamydomonas reinhardii. Mol. Gen. Genet. MGG 214, 204–212. doi:10.1007/BF00337712
Bird, R. C., and Zimmerman, A. M. (1980). Induction of tubulin synthesis during cilia regeneration in growing Tetrahymena. Exp. Cell. Res. 128, 199–205. doi:10.1016/0014-4827(80)90403-6
Blacque, O. E., Perens, E. A., Boroevich, K. A., Inglis, P. N., Li, C., Warner, A., et al. (2005). Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15, 935–941. doi:10.1016/j.cub.2005.04.059
Blatt, E. N., Yan, X. H., Wuerffel, M. K., Hamilos, D. L., and Brody, S. L. (1999). Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am. J. Respir. Cell. Mol. Biol. 21, 168–176. doi:10.1165/ajrcmb.21.2.3691
Brenner, S., Jacob, F., and Meselson, M. (1961). An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190, 576–581. doi:10.1038/190576a0
Brocal-Ruiz, R., Esteve-Serrano, A., Mora-Martínez, C., Franco-Rivadeneira, M. L., Swoboda, P., Tena, J. J., et al. (2023). Forkhead transcription factor FKH-8 cooperates with RFX in the direct regulation of sensory cilia in Caenorhabditis elegans. eLife 12, e89702. doi:10.7554/eLife.89702
Brooks, E. R., and Wallingford, J. B. (2014). Multiciliated cells. Curr. Biol. 24, R973–R982. doi:10.1016/j.cub.2014.08.047
Brown, J. M., Dipetrillo, C. G., Smith, E. F., and Witman, G. B. (2012). A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell. Sci. 125, 3904–3913. doi:10.1242/jcs.107151
Brown, J. M., and Witman, G. B. (2014). Cilia and diseases. Bioscience 64, 1126–1137. doi:10.1093/biosci/biu174
Cachero, S., Simpson, T. I., Zur Lage, P. I., Ma, L., Newton, F. G., Holohan, E. E., et al. (2011). The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 9, e1000568. doi:10.1371/journal.pbio.1000568
Chen, B., Niu, J., Kreuzer, J., Zheng, B., Jarugumilli, G. K., Haas, W., et al. (2018). Auto-fatty acylation of transcription factor RFX3 regulates ciliogenesis. Proc. Natl. Acad. Sci. 115, E8403-E8412–E8412. doi:10.1073/pnas.1800949115
Chen, J., Knowles, H. J., Hebert, J. L., and Hackett, B. P. (1998). Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Invest. 102, 1077–1082. doi:10.1172/JCI4786
Chen, N., Mah, A., Blacque, O. E., Chu, J., Phgora, K., Bakhoum, M. W., et al. (2006). Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 7, R126. doi:10.1186/gb-2006-7-12-r126
Cheshire, J. L., Evans, J. H., and Keller, L. R. (1994). Ca2+ Signaling in the Chlamydomonas flagellar regeneration system: cellular and molecular responses. J. Cell. Sci. 107, 2491–2498. doi:10.1242/jcs.107.9.2491
Cheshire, J. L., and Keller, L. R. (1991). Uncoupling of chlamydomonas flagellar gene expression and outgrowth from flagellar excision by manipulation of Ca2+. J. Cell. Biol. 115, 1651–1659. doi:10.1083/jcb.115.6.1651
Choksi, S. P., Lauter, G., Swoboda, P., and Roy, S. (2014). Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141, 1427–1441. doi:10.1242/dev.074666
Chu, J. S. C., Baillie, D. L., and Chen, N. (2010). Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol. Biol. 10, 130. doi:10.1186/1471-2148-10-130
Chung, M.-I., Kwon, T., Tu, F., Brooks, E. R., Gupta, R., Meyer, M., et al. (2014). Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife 3, e01439. doi:10.7554/eLife.01439
Clevidence, D. E., Overdier, D. G., Tao, W., Qian, X., Pani, L., Lai, E., et al. (1993). Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc. Natl. Acad. Sci. 90, 3948–3952. doi:10.1073/pnas.90.9.3948
Cobb, M. (2015). Who discovered messenger RNA? Curr. Biol. 25, R526–R532. doi:10.1016/j.cub.2015.05.032
Coyle, M. C., Tajima, A. M., Leon, F., Choksi, S. P., Yang, A., Espinoza, S., et al. (2023). An RFX transcription factor regulates ciliogenesis in the closest living relatives of animals. Curr. Biol. 33, 3747–3758.e9. doi:10.1016/j.cub.2023.07.022
Craig, R. J., Gallaher, S. D., Shu, S., Salomé, P. A., Jenkins, J. W., Blaby-Haas, C. E., et al. (2023). The Chlamydomonas Genome Project, version 6: reference assemblies for mating-type plus and minus strains reveal extensive structural mutation in the laboratory. Plant Cell. 35, 644–672. doi:10.1093/plcell/koac347
Crick, F. H. C., Barnett, L., Brenner, S., and Watts-Tobin, R. J. (1961). General nature of the genetic code for proteins. Nature 192, 1227–1232. doi:10.1038/1921227a0
Davies, J. P., and Grossman, A. R. (1994). Sequences controlling transcription of the Chlamydomonas reinhardtii beta 2-tubulin gene after deflagellation and during the cell cycle. Mol. Cell. Biol. 14, 5165–5174. doi:10.1128/mcb.14.8.5165
Davies, J. P., Weeks, D. P., and Grossman, A. R. (1992). Expression of the arylsulfatase gene from the β2 -tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 20, 2959–2965. doi:10.1093/nar/20.12.2959
Didon, L., Zwick, R. K., Chao, I. W., Walters, M. S., Wang, R., Hackett, N. R., et al. (2013). RFX3 modulation of FOXJ1 regulation of cilia genes in the human airway epithelium. Respir. Res. 14, 70. doi:10.1186/1465-9921-14-70
Dobbelaere, J., Su, T. Y., Erdi, B., Schleiffer, A., and Dammermann, A. (2023). A phylogenetic profiling approach identifies novel ciliogenesis genes in Drosophila and C. elegans. EMBO J. 42, e113616. doi:10.15252/embj.2023113616
Efimenko, E., Bubb, K., Mak, H. Y., Holzman, T., Leroux, M. R., Ruvkun, G., et al. (2005). Analysis of xbx genes in C. elegans. Development 132, 1923–1934. doi:10.1242/dev.01775
Eisen, J. A., Coyne, R. S., Wu, M., Wu, D., Thiagarajan, M., Wortman, J. R., et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286. doi:10.1371/journal.pbio.0040286
Elliott, K. H., Balchand, S. K., Bonatto Paese, C. L., Chang, C.-F., Yang, Y., Brown, K. M., et al. (2023). Identification of a heterogeneous and dynamic ciliome during embryonic development and cell differentiation. Development 150, dev201237. doi:10.1242/dev.201237
Evans, J. H., and Keller, L. R. (1997). Calcium influx signals normal flagellar RNA induction following acid shock of Chlamydomonas reinhardtii. Plant Mol. Biol. 33, 467–481. doi:10.1023/A:1005727806897
Franklin, R. E., and Gosling, R. G. (1953). Molecular configuration in sodium thymonucleate. Nature 171, 740–741. doi:10.1038/171740a0
Fritz-Laylin, L. K., and Cande, W. Z. (2010). Ancestral centriole and flagella proteins identified by analysis of Naegleria differentiation. J. Cell. Sci. 123, 4024–4031. doi:10.1242/jcs.077453
Fritz-Laylin, L. K., Prochnik, S. E., Ginger, M. L., Dacks, J. B., Carpenter, M. L., Field, M. C., et al. (2010). The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 140, 631–642. doi:10.1016/j.cell.2010.01.032
Fulton, C., and Dingle, A. D. (1967). Appearance of the flagellate phenotype in populations of Naegleria amebae. Dev. Biol. 15, 165–191. doi:10.1016/0012-1606(67)90012-7
Fulton, C., and Kowit, J. D. (1975). Programmed synthesis of flagellar tubulin during cell differentiation in Naegleria. Ann. N. Y. Acad. Sci. 253, 318–332. doi:10.1111/j.1749-6632.1975.tb19210.x
Gera, J. F., and Baker, E. J. (1998). Deadenylation-dependent and -independent decay pathways for alpha1-tubulin mRNA in Chlamydomonas reinhardtii. Mol. Cell. Biol. 18, 1498–1505. doi:10.1128/mcb.18.3.1498
Gibbons, I. R. (1963). Studies on the protein components of cilia from tetrahymena pyriformis. Proc. Natl. Acad. Sci. 50, 1002–1010. doi:10.1073/pnas.50.5.1002
Gibbons, I. R., and Rowe, A. J. (1965). Dynein: a protein with adenosine triphosphatase activity from cilia. Science 149, 424–426. doi:10.1126/science.149.3682.424
Gong, Z. Y., and Brandhorst, B. P. (1987). Stimulation of tubulin gene transcription by deciliation of sea urchin embryos. Mol. Cell. Biol. 7, 4238–4246. doi:10.1128/mcb.7.12.4238
Gros, F., Hiatt, H., Gilbert, W., Kurland, C. G., Risebrough, R. W., and Watson, J. D. (1961). Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature 190, 581–585. doi:10.1038/190581a0
Guttman, S. D., and Gorovsky, M. A. (1979). Cilia regeneration in starved tetrahymena: an inducible system for studying gene expression and organelle biogenesis. Cell. 17, 307–317. doi:10.1016/0092-8674(79)90156-9
Hackett, B. P., Brody, S. L., Liang, M., Zeitz, I. D., Bruns, L. A., and Gitlin, J. D. (1995). Primary structure of hepatocyte nuclear factor/forkhead homologue 4 and characterization of gene expression in the developing respiratory and reproductive epithelium. Proc. Natl. Acad. Sci. 92, 4249–4253. doi:10.1073/pnas.92.10.4249
Ignatenko, O., Malinen, S., Rybas, S., Vihinen, H., Nikkanen, J., Kononov, A., et al. (2022). Mitochondrial dysfunction compromises ciliary homeostasis in astrocytes. J. Cell. Biol. 222, e202203019. doi:10.1083/jcb.202203019
Ishikawa, H., Thompson, J., Yates, J. R., and Marshall, W. F. (2012). Proteomic analysis of mammalian primary cilia. Curr. Biol. 22, 414–419. doi:10.1016/j.cub.2012.01.031
Jacob, F., and Monod, J. (1961). Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356. doi:10.1016/s0022-2836(61)80072-7
Kang, Y., and Mitchell, D. R. (1998). An intronic enhancer is required for deflagellation-induced transcriptional regulation of a chlamydomonas reinhardtii dynein gene. Mol. Biol. Cell. 9, 3085–3094. doi:10.1091/mbc.9.11.3085
Keller, L. R., Schloss, J. A., Silflow, C. D., and Rosenbaum, J. L. (1984). Transcription of alpha- and beta-tubulin genes in vitro in isolated Chlamydomonas reinhardi nuclei. J. Cell. Biol. 98, 1138–1143. doi:10.1083/jcb.98.3.1138
Kennard, A. S., Velle, K. B., Ranjan, R., Schulz, D., and Fritz-Laylin, L. K. (2025). Tubulin sequence divergence is associated with the use of distinct microtubule regulators. Curr. Biol. 35, 233–248.e8. doi:10.1016/j.cub.2024.11.022
Kowit, J. D., and Fulton, C. (1974). Programmed synthesis of tubulin for the flagella that develop during cell differentiation in Naegleria gruberi. Proc. Natl. Acad. Sci. U. S. A. 71, 2877–2881. doi:10.1073/pnas.71.7.2877
Lai, E. Y., Walsh, C., Wardell, D., and Fulton, C. (1979). Programmed appearance of translatable flagellar tubulin mRNA during cell differentiation in Naegleria. Cell. 17, 867–878. doi:10.1016/0092-8674(79)90327-1
Lalioti, M.-E., Arbi, M., Loukas, I., Kaplani, K., Kalogeropoulou, A., Lokka, G., et al. (2019). GemC1 governs multiciliogenesis through direct interaction with and transcriptional regulation of p73. J. Cell. Sci. 132, jcs228684. doi:10.1242/jcs.228684
Laurençon, A., Dubruille, R., Efimenko, E., Grenier, G., Bissett, R., Cortier, E., et al. (2007). Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 8, R195. doi:10.1186/gb-2007-8-9-r195
Lee, J. H., and Walsh, C. J. (1988). Transcriptional regulation of coordinate changes in flagellar mRNAs during differentiation of Naegleria gruberi amebae into flagellates. Mol. Cell. Biol. 8, 2280–2287. doi:10.1128/mcb.8.6.2280
Lefebvre, P., Silflow, C. D., Wieben, E. D., and Rosenbaum, J. L. (1980). Increased levels of mRNAs for tubulin and other flagellar proteins after amputation or shortening of Chlamydomonas Flagella. Cell. 20, 469–477. doi:10.1016/0092-8674(80)90633-9
Lefebvre, P. A., Barsel, S.-E., and Wexler, D. E. (1988). Isolation and characterization of chlamydomonas reinhardtii mutants with defects in the induction of flagellar protein synthesis after deflagellation. J. Protozool. 35, 559–564. doi:10.1111/j.1550-7408.1988.tb04152.x
Lefebvre, P. A., Nordstrom, S. A., Moulder, J. E., and Rosenbaum, J. L. (1978). Flagellar elongation and shortening in chlamydomonas: IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J. Cell. Biol. 78, 8–27. doi:10.1083/jcb.78.1.8
Lefebvre, P. A., and Rosenbaum, J. L. (1986). Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Ann. Rev. Cell. Biol. 2, 517–546. doi:10.1146/annurev.cb.02.110186.002505
Lemeille, S., Paschaki, M., Baas, D., Morlé, L., Duteyrat, J.-L., Ait-Lounis, A., et al. (2020). Interplay of RFX transcription factors 1, 2 and 3 in motile ciliogenesis. Nucleic Acids Res. 48, 9019–9036. doi:10.1093/nar/gkaa625
Lewin, R. A. (1953). Studies on the flagella of algae. Il. Formation of flagella by Chlamydomonas in light and darkness. Ann. N. Y. Acad. Sci. 56, 1091–1093. doi:10.1111/j.1749-6632.1953.tb30293.x
Lewis, M., and Stracker, T. H. (2021). Transcriptional regulation of multiciliated cell differentiation. Semin. Cell. Dev. Biol. 110, 51–60. doi:10.1016/j.semcdb.2020.04.007
Li, J. B., Gerdes, J. M., Haycraft, C. J., Fan, Y., Teslovich, T. M., May-Simera, H., et al. (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 117, 541–552. doi:10.1016/S0092-8674(04)00450-7
Li, L., Guangmei, T., Peng, H., Meng, D., Wang, L., Hu, X., et al. (2018). New class of transcription factors controls flagellar assembly by recruiting RNA polymerase II in Chlamydomonas. Proc. Natl. Acad. Sci. 115, 4435–4440. doi:10.1073/pnas.1719206115
Lu, H., Anujan, P., Zhou, F., Zhang, Y., Chong, Y. L., Bingle, C. D., et al. (2019). Mcidas mutant mice reveal a two-step process for the specification and differentiation of multiciliated cells in mammals. Development 146, dev172643. doi:10.1242/dev.172643
Mahjoub, M. R., Qasim Rasi, M., and Quarmby, L. M. (2004). A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of chlamydomonas and mouse kidney cells. Mol. Biol. Cell. 15, 5172–5186. doi:10.1091/mbc.e04-07-0571
Maniatis, T., Goodbourn, S., and Fischer, J. A. (1987). Regulation of inducible and tissue-specific gene expression. Science 236, 1237–1245. doi:10.1126/science.3296191
Marshall, C. B., Mays, D. J., Beeler, J. S., Rosenbluth, J. M., Boyd, K. L., Santos Guasch, G. L., et al. (2016). p73 is required for multiciliogenesis and regulates the foxj1-associated gene network. Cell. Rep. 14, 2289–2300. doi:10.1016/j.celrep.2016.02.035
Marshall, W. F. (2024). Chlamydomonas as a model system to study cilia and flagella using genetics, biochemistry, and microscopy. Front. Cell. Dev. Biol. 12, 1412641. doi:10.3389/fcell.2024.1412641
Matthaei, H., and Nirenberg, M. W. (1961). The dependence of cell-free protein synthesis in E. coli upon RNA prepared from ribosomes. Biochem. Biophys. Res. Commun. 4, 404–408. doi:10.1016/0006-291X(61)90298-4
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi:10.1126/science.1143609
Merlino, G. T., Chamberlain, J. P., and Kleinsmith, L. J. (1978). Effects of deciliation of tubulin messenger RNA activity in sea urchin embryos. J. Biol. Chem. 253, 7078–7085. doi:10.1016/S0021-9258(17)38031-6
Mifsud, K. R., Kennedy, C. L. M., Salatino, S., Sharma, E., Price, E. M., Haque, S. N., et al. (2021). Distinct regulation of hippocampal neuroplasticity and ciliary genes by corticosteroid receptors. Nat. Commun. 12, 4737. doi:10.1038/s41467-021-24967-z
Mill, P., Christensen, S. T., and Pedersen, L. B. (2023). Primary cilia as dynamic and diverse signalling hubs in development and disease. Nat. Rev. Genet. 24, 421–441. doi:10.1038/s41576-023-00587-9
Minami, S. A., Collis, P. S., Young, E. E., and Weeks, D. P. (1981). Tubulin induction in C. reinhardii: requirement for tubulin mRNA synthesis. Cell. 24, 89–95. doi:10.1016/0092-8674(81)90504-3
Newton, F. G., zur Lage, P. I., Karak, S., Moore, D. J., Göpfert, M. C., and Jarman, A. P. (2012). Forkhead transcription factor Fd3F cooperates with Rfx to regulate a gene expression program for mechanosensory cilia specialization. Dev. Cell. 22, 1221–1233. doi:10.1016/j.devcel.2012.05.010
Nonaka, S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y., et al. (1998). Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 95, 829–837. doi:10.1016/S0092-8674(00)81705-5
Patir, A., Fraser, A. M., Barnett, M. W., McTeir, L., Rainger, J., Davey, M. G., et al. (2020). The transcriptional signature associated with human motile cilia. Sci. Rep. 10, 10814. doi:10.1038/s41598-020-66453-4
Pazour, G. J., Agrin, N., Leszyk, J., and Witman, G. B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell. Biol. 170, 103–113. doi:10.1083/jcb.200504008
Pazour, G. J., Dickert, B. L., Vucica, Y., Seeley, E. S., Rosenbaum, J. L., Witman, G. B., et al. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J. Cell. Biol. 151, 709–718. doi:10.1083/jcb.151.3.709
Periz, G., and Keller, L. R. (1997). DNA elements regulating alpha1-tubulin gene induction during regeneration of eukaryotic flagella. Mol. Cell. Biol. 17, 3858–3866. doi:10.1128/mcb.17.7.3858
Perkins, L. A., Hedgecock, E. M., Thomson, J. N., and Culotti, J. G. (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487. doi:10.1016/0012-1606(86)90314-3
Perlaza, K., Zamora, I., and Marshall, W. F. (2023). Role of intraflagellar transport in transcriptional control during flagellar regeneration in Chlamydomonas. Mol. Biol. Cell. 34, ar52. doi:10.1091/mbc.E22-09-0444
Phirke, P., Efimenko, E., Mohan, S., Burghoorn, J., Crona, F., Bakhoum, M. W., et al. (2011). Transcriptional profiling of C. elegans DAF-19 uncovers a ciliary base-associated protein and a CDK/CCRK/LF2p-related kinase required for intraflagellar transport. Dev. Biol. 357, 235–247. doi:10.1016/j.ydbio.2011.06.028
Piasecki, B. P., Burghoorn, J., and Swoboda, P. (2010). Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc. Natl. Acad. Sci. 107, 12969–12974. doi:10.1073/pnas.0914241107
Picariello, T., Hou, Y., Kubo, T., McNeill, N. A., Yanagisawa, H., Oda, T., et al. (2020). TIM, a targeted insertional mutagenesis method utilizing CRISPR/Cas9 in Chlamydomonas reinhardtii. PLOS ONE 15, e0232594. doi:10.1371/journal.pone.0232594
Piperno, G., Huang, B., and Luck, D. J. (1977). Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 74, 1600–1604. doi:10.1073/pnas.74.4.1600
Pir, M. S., Begar, E., Yenisert, F., Demirci, H. C., Korkmaz, M. E., Karaman, A., et al. (2024). CilioGenics: an integrated method and database for predicting novel ciliary genes. Nucleic Acids Res. 52, 8127–8145. doi:10.1093/nar/gkae554
Qin, H., Rosenbaum, J. L., and Barr, M. M. (2001). An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11, 457–461. doi:10.1016/S0960-9822(01)00122-1
Quader, H., Cherniack, J., and Filner, P. (1978). Participation of calcium in flagellar shortening and regeneration in Chlamydomonas reinhardii. Exp. Cell. Res. 113, 295–301. doi:10.1016/0014-4827(78)90369-5
Quarmby, L. M., and Mahjoub, M. R. (2023). “Deciliation,” in The Chlamydomonas sourcebook (Elsevier), 373–389. doi:10.1016/B978-0-12-822508-0.00010-1
Quigley, I. K., and Kintner, C. (2017). Rfx2 stabilizes Foxj1 binding at chromatin loops to enable multiciliated cell gene expression. PLOS Genet. 13, e1006538. doi:10.1371/journal.pgen.1006538
Rannestad, J. (1974). The regeneration of cilia in partially deciliated tetrahymena. J. Cell. Biol. 63, 1009–1017. doi:10.1083/jcb.63.3.1009
Remillard, S. P., and Witman, G. B. (1982). Synthesis, transport, and utilization of specific flagellar proteins during flagellar regeneration in Chlamydomonas. J. Cell. Biol. 93, 615–631. doi:10.1083/jcb.93.3.615
Rosenbaum, J. (2009). “A stroll through time with chlamydomonas,” in The Chlamydomonas sourcebook (Elsevier), 1–14.
Rosenbaum, J. L., and Carlson, K. (1969). Cilia regeneration in Tetrahymena and its inhibition by colchicine. J. Cell. Biol. 40, 415–425. doi:10.1083/jcb.40.2.415
Rosenbaum, J. L., and Child, F. M. (1967). Flagellar regeneration in protozoan flagellates. J. Cell. Biol. 34, 345–364. doi:10.1083/jcb.34.1.345
Rosenbaum, J. L., Moulder, J. E., and Ringo, D. L. (1969). Flagellar elongation and shortening in chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell. Biol. 41, 600–619. doi:10.1083/jcb.41.2.600
Sale, W. S., and Dutcher, S. K. (2023). “Landmark contributions of Chlamydomonas to understanding cilia,” in The Chlamydomonas sourcebook (Elsevier), 1–34. doi:10.1016/B978-0-12-822508-0.00014-9
Schloss, J. A., Silflow, C. D., and Rosenbaum, J. L. (1984). mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol. Cell. Biol. 4, 424–434. doi:10.1128/mcb.4.3.424
Silflow, C. D., and Rosenbaum, J. L. (1981). Multiple α- and β-tubulin genes in chlamydomonas and regulation of tubulin mRNA levels after deflagellation. Cell. 24, 81–88. doi:10.1016/0092-8674(81)90503-1
Smith, J. C., Northey, J. G. B., Garg, J., Pearlman, R. E., and Siu, K. W. M. (2005). Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 4, 909–919. doi:10.1021/pr050013h
Stephens, R. E. (1977). Differential protein synthesis and utilization during cilia formation in sea urchin embryos. Dev. Biol. 61, 311–329. doi:10.1016/0012-1606(77)90301-3
Stolc, V., Samanta, M. P., Tongprasit, W., and Marshall, W. F. (2005). Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. 102, 3703–3707. doi:10.1073/pnas.0408358102
Stubbs, J. L., Oishi, I., Izpisúa Belmonte, J. C., and Kintner, C. (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 40, 1454–1460. doi:10.1038/ng.267
Swoboda, P., Adler, H. T., and Thomas, J. H. (2000). The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 5, 411–421. doi:10.1016/S1097-2765(00)80436-0
Tegenfeldt, F., Kuznetsov, D., Manni, M., Berkeley, M., Zdobnov, E. M., and Kriventseva, E. V. (2025). OrthoDB and BUSCO update: annotation of orthologs with wider sampling of genomes. Nucleic Acids Res. 53, D516–D522. doi:10.1093/nar/gkae987
Terré, B., Piergiovanni, G., Segura-Bayona, S., Gil-Gómez, G., Youssef, S. A., Attolini, C. S., et al. (2016). GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 35, 942–960. doi:10.15252/embj.201592821
Thomas, J., Morlé, L., Soulavie, F., Laurençon, A., Sagnol, S., and Durand, B. (2010). Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol. Cell. 102, 499–513. doi:10.1042/BC20100035
van Dam, T. J., Wheway, G., Slaats, G. G., Huynen, M. A., and Giles, R. H.SYSCILIA Study Group (2013). The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia 2, 7. doi:10.1186/2046-2530-2-7
van Dam, T. J. P., Kennedy, J., Lee, R., Vrieze, E. de, Wunderlich, K. A., Rix, S., et al. (2019). CiliaCarta: an integrated and validated compendium of ciliary genes. PLOS ONE 14, e0216705. doi:10.1371/journal.pone.0216705
Wang, W., Xing, J., Zhang, X., Liu, H., Liu, X., Jiang, H., et al. (2025). Control of ciliary transcriptional programs during spermatogenesis by antagonistic transcription factors. eLife 13, RP94754. doi:10.7554/eLife.94754
Wang, Y., Ren, Y., and Pan, J. (2019). Regulation of flagellar assembly and length in Chlamydomonas by LF4, a MAPK-related kinase. FASEB J. 33, 6431–6441. doi:10.1096/fj.201802375RR
Watson, J. D., and Crick, F. H. C. (1953). Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171, 737–738. doi:10.1038/171737a0
Weeks, D. P., Collis, P. S., and Gealt, M. A. (1977). Control of induction of tubulin synthesis in Chlamydomonas reinhardi. Nature 268, 667–668. doi:10.1038/268667a0
Wildung, M., Esser, T. U., Grausam, K. B., Wiedwald, C., Volceanov-Hahn, L., Riedel, D., et al. (2019). Transcription factor TAp73 and microRNA-449 complement each other to support multiciliogenesis. Cell. Death Differ. 26, 2740–2757. doi:10.1038/s41418-019-0332-7
Wilson, N. F., and Lefebvre, P. A. (2004). Regulation of flagellar assembly by glycogen synthase kinase 3 in chlamydomonas reinhardtii. Eukaryot. Cell. 3, 1307–1319. doi:10.1128/ec.3.5.1307-1319.2004
Witman, G. B., Carlson, K., Berliner, J., and Rosenbaum, J. L. (1972). CHLAMYDOMONAS FLAGELLA I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell. Biol. 54, 507–539. doi:10.1083/jcb.54.3.507
Wood, C. R., Wang, Z., Diener, D., Zones, J. M., Rosenbaum, J., and Umen, J. G. (2012). IFT proteins accumulate during cell division and localize to the cleavage furrow in chlamydomonas. PLOS ONE 7, e30729. doi:10.1371/journal.pone.0030729
Wood, V., Gwilliam, R., Rajandream, M.-A., Lyne, M., Lyne, R., Stewart, A., et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880. doi:10.1038/nature724
Keywords: cilia, flagella, transcription, Chlamydomonas, FoxJ1, RFX, XAP5, gene regulation
Citation: Brown JM (2025) Cilia and transcription: a mini review. Front. Cell Dev. Biol. 13:1582796. doi: 10.3389/fcell.2025.1582796
Received: 25 February 2025; Accepted: 30 May 2025;
Published: 09 June 2025.
Edited by:
Maureen Wirschell, McKendree University, United StatesReviewed by:
Jacinta Seraphina D’Souza, UM-DAE Centre for Excellence in Basic Sciences, IndiaCopyright © 2025 Brown. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason M. Brown, amJyb3duMkBzYWxlbXN0YXRlLmVkdQ==
 Jason M. Brown
Jason M. Brown