Abstract
At the moment of their union, fertilizing gametes (sperm and oocyte) are transcriptionally silent: gene expression has to be initiated within the resulting embryo, a process termed embryonic genome activation, EGA. Until recently, EGA was believed to occur at the two-cell stage (mouse) or four-to-eight-cell stage (human), but new evidence from single-cell RNA-sequencing (scRNAseq) suggests that it initiates at the one-cell stage in both species. Precise time-course scRNA-seq of mouse one-cell embryos revealed an EGA program referred to as immediate EGA, iEGA: iEGA occurred from within 4 h of fertilization, mainly from the maternal genome, with paternal genomic transcription from ∼10 h. Significant low-magnitude upregulation similarly occurred in healthy human one-cell embryos. In both species, new transcripts were canonically spliced, and expression predicted embryonic processes and regulatory transcription factors (TFs) associated with cancer, including MYC/c-Myc. Blocking their activities in mouse one-cell embryos induced acute developmental arrest and disrupted iEGA. Inhibiting c-Myc induced upregulation of hundreds of genes, implying that they are normatively repressed, a phenomenon we term embryonic genome repression, EGR. iEGA is downregulated coincidentally with a subsequent, higher-amplitude wave of gene expression (referred to as ‘major EGA’ or ‘major ZGA’) in two-cell (mouse) or 4–8-cell (human) embryos. We suggest that iEGA is continuous with gene expression previously termed ‘minor EGA’ (or ‘minor ZGA’) and that the regulation of iEGA and major EGA are distinctive. The pattern of gene upregulation in iEGA illuminates processes involved at the onset of development, with implications for epigenetic inheritance, stem cell-derived embryos and cancer.
1 Introduction
When a sperm and an oocyte (egg) combine in fertilization, they are transcriptionally silent (Balhorn, 1982; Zuccotti et al., 1995). Transcription must therefore be initiated on the newly-formed embryonic genome, a process generically referred to as embryonic genome activation, EGA. This Perspective considers how EGA is initiated in mouse and human embryos, with implicit relevance to other mammalian species. A central tenet is that mouse and human EGA begin in one-cell embryos during fertilization.
Fertilization describes the period linking sperm-oocyte fusion to chromosome mingling just prior to the first mitotic cytokinesis: the gamete-to-embryo transition (see Box 1 for a glossary of terms) (Yanagimachi, 1994). In the mouse, fertilization takes around 16 h, and in humans a little longer (Suzuki et al., 2016) (Figure 1). The product is a presumptively totipotent cell capable of engendering the full-term development of an individual (Condic, 2014) (Box 1). The emergence of totipotency during fertilization coincides with multiple integrated dynamic processes, including meiotic progression (Yanagimachi, 1994), signalling fluxes that involve calcium oscillations (Cuthbertson et al., 1981) and phospho-relays (Perry and Verlhac, 2008), transmission (to the embryo) and activation of maternal factors (including protein and RNAs carried over from the oocyte following sperm union; Wu and Dean, 2020), the onset of maternal transcript (and other maternal factor) degradation (Clegg and Piko, 1983), extensive and parent-specific chromatin remodeling including genome demethylation (Adenot et al., 1997; Liu et al., 2014; Mayer et al., 2000), pronucleus formation (Yanagimachi, 1994), and a program of intracellular force changes involving surges during chromatin remodelling and cytokinesis (Duch et al., 2020). Although little is known about them, additional changes, including organelle reconfiguration (e.g., migration and restructuring), macromolecular trafficking and phase separation (Hyman et al., 2014; Ling et al., 2022), are likely to play formative roles, not least because in cellular terms, mouse and human one-cell embryos are relatively large (≥170 pL, compared to ∼4 pL for a typical somatic cell). Studying the emergence of totipotency is made more challenging by the likelihood of functional redundancy and complementarity during fertilization. For example, phospholipase C-zeta, which is the oocyte-activating trigger for embryogenesis delivered by a fertilizing sperm, is dispensable for developmental activation (Hachem et al., 2017). Calcium ion mobilization during mammalian fertilization presumptively activates phospho-signalling but is dispensable for full-term development (Suzuki et al., 2010). Compensation may also confound the analysis of transcriptional regulation, producing multiple, occasionally incompatible, inferred mechanisms (Gassler et al., 2022; Ji et al., 2023).
BOX 1 Glossary of selected terms used.
| Term | Definition |
|---|---|
| Fertilization | Period linking gamete fusion to parental chromosome mingling (syngamy) following pronuclear membrane breakdown (Yanagimachi, 1994) |
| Minor EGA, minor ZGA | Transcription in late (defined without good temporal resolution) one-cell embryos (Hamatani et al., 2004; Xue et al., 2013): approximately, S-phase of one-cell to G1-phase of two-cell stages. |
| Major EGA, major ZGA | Transcription in two-cell (mouse) or four-to-eight-cell (human) embryos (Braude et al., 1988; Hamatani et al., 2004; Xue et al., 2013). |
| Plenipotent | Able to give rise to any embryonically-derived cell type (Condic, 2014). |
| Pluripotent | Able to give rise to any embryonically-derived cell type present in the embryo proper |
| Totipotent | Cell that is normatively able to give rise to an entire individual (Condic, 2014). In the mouse, only two cell types are totipotent: one-cell embryos and the blastomeres of a two-cell embryo (Katayama et al., 2010; Rossant, 1976; Tarkowski, 1959). Defining ‘totipotency’ to include cells that can give rise to all cell types does not capture additional tiers of information necessary to choreograph full development. Cells that can normatively give rise to all cell types but not offspring have been labelled plenipotent (Condic, 2014). |
| Zygote | One-cell embryo |
| Zygotic genome activation, ZGA | ZGA has been used synonymously with EGA for historical reasons, but is inappropriate when describing processes that occur in two-, four- or eight-cell embryos (e.g., major ZGA), because they do not occur in zygotes. |
FIGURE 1
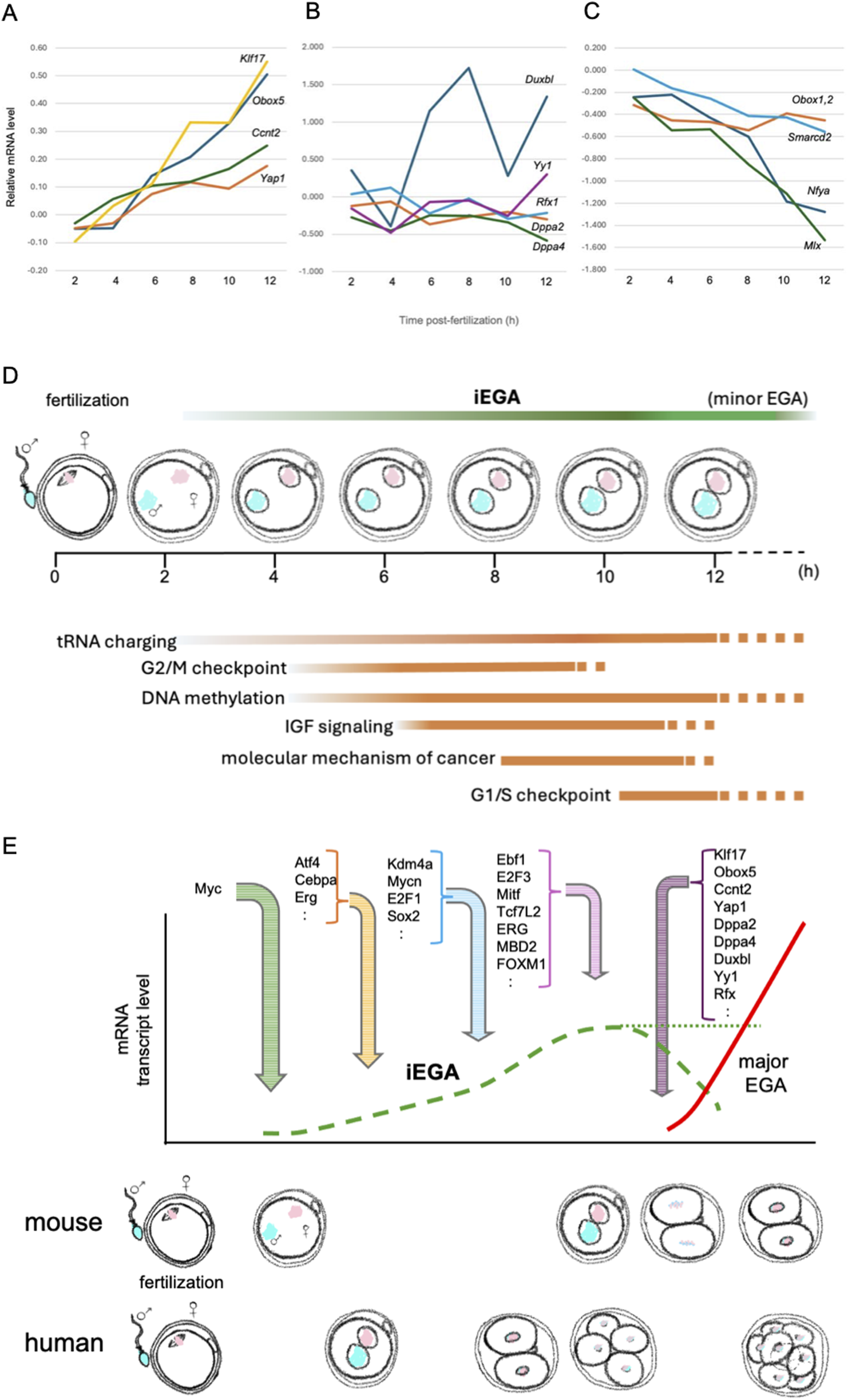
Expression profiles of candidate major EGA activating TF mRNAs during iEGA. (A)Klf17, Obox5, Ccnt2 and Yap1 are iEGA genes whose levels increase (Asami et al., 2023). Y-axis: log2FC. (B) Other predicted major EGA TF mRNA levels do not change, or (C) decrease. For all analyses, FDR<5%. (D) The succession of pathways predicted by iEGA (FDR<5%) include tRNA charging (e.g., EARS2, HARS), G2/M checkpoint-associated genes (e.g., CHEK1, CKS1B), DNA methylation-associated genes (e.g., HIST1H4A, SAP30), IGF signaling-associated genes (e.g., CSNK2A1, IGFBP4), genes associated with the molecular mechanism of cancer (e.g., AKT1, WNT4) and G1/S checkpoint-associated genes (e.g., CCNE2, TP53). (E) Predicted mouse and human gene activators in iEGA and major EGA (distinct waves of transcription). Mouse and human iEGA profiles overlap. Transcription regulators were inferred by IPA of mouse iEGA (FDR<5%) at 4, 6, 8 and 12 h, and are indicated where they are first predicted to act.
Because these processes reflect and determine the intracellular milieu during fertilization, understanding them should lead to better models of totipotency establishment, maintenance and exit. This is challenging because they are complex, occur at small scale in low numbers of transient cell types (e.g., one-cell embryos) and are integrative, thwarting reductionist approaches. Totipotency is a transitory state and totipotent stem cells do not exist. Parallels have been drawn between the two-cell embryonic state and a sub-population within pluripotent stem cell (PSC) cultures, referred to as two-cell-like cells in the mouse (Macfarlan et al., 2012); a corresponding state has been identified between eight-cell embryos and eight-cell-like PSCs in humans (Mazid et al., 2022). Such parallels may indicate overlaps between the mechanisms that regulate pluripotency and totipotency, but the embryos and stem cells in which they occur are respectively distinct transcriptionally, structurally, metabolically, morphologically and developmentally.
The onset of EGA provides a read-out of the processes controlling totipotency: it is a response to cellular events in the nascent one-cell embryo that are critical for the emergence of the totipotent state. We refer to the initiation of EGA in the first 12 h after fertilization in the mouse, as immediate EGA, iEGA, and argue that an analogous process occurs in human one-cell embryos (Asami et al., 2022; Asami et al., 2023). Gene expression has previously been reported in human and mouse late one-cell embryos, termed ‘minor EGA’ (‘minor ZGA’; Box 1) in the mouse, but it has not been defined with temporal precision and its biological role has not been acknowledged (Abe et al., 2015; Hamatani et al., 2004; Park et al., 2013; Wang et al., 2004; Xue et al., 2013; Zeng and Schultz, 2005). We argue that minor EGA is a continuation of iEGA (Asami et al., 2023). iEGA is followed by ‘major EGA’ (‘major ZGA’; Box 1) in mouse two-cell embryos and at the four-to-eight-cell stage in humans (Hamatani et al., 2004; Park et al., 2013; Wang et al., 2004; Xue et al., 2013; Zeng and Schultz, 2005) (Figure 1).
We now consider iEGA in the context of major EGA and preparation for preimplantation development. We also introduce the notion of embryonic genome repression, EGR, which corresponds to a specific profile of transcriptional repression identified in the mouse during iEGA.
2 The onset of embryonic transcription: iEGA
Profiling the onset of embryonic transcription has proven elusive. Studies have relied on embryos derived in vivo for which the time of fertilization was indeterminate, even though, in the mouse, oocytes are fertilizable for >12 h post-ovulation (Marston and Chang, 1964), the time of coitus and duration of sperm passage and fusion at the fertilization site varies (Suarez, 1987), and one-cell embryo morphology and time since fertilization are not reliably correlated (Adenot et al., 1997). Some studies (Table 1) used hundreds or thousands of embryos (Ko et al., 2000; Abe et al., 2015; Hamatani et al., 2004; Park et al., 2013; Wang et al., 2004), potentially smoothing signals (Olsen and Baryawno, 2018) and precluding the degree of inter-embryo synchrony necessary for accurate one-cell embryo transcriptome profiling. In most cases, transcripts have been isolated by poly(A) capture, but the length of poly(A) tails is controlled in early embryos as a means to regulate translation, potentially skewing outputs such that they reflect mRNA polyadenylation in addition to de novo gene expression (Blower et al., 2013; Daniels et al., 1995; Hamatani et al., 2004; Temeles and Schultz, 1997). Additionally, maternally-derived mRNA in one-cell embryos (Qiao et al., 2020) may compromise the detection of significant, low-amplitude embryonic gene expression.
TABLE 1
| Species | Data type | References | Accession no(s) | Embryo preparation | Embryo stages | Library preparation method | Cell numbers | Cut-off |
|---|---|---|---|---|---|---|---|---|
| Mouse | 3′-EST sequences | Ko et al. (2000) | C75935-C81630, C85044-C88357, AU014577-AU024803, AU040095-AU046300 | Natural mating | mII to blastocyst | Total RNA-derived PCR-based cDNA library construction | >1,528 x mIl, 1,137 × 1C, 397 × 2C, 32 × 4C, 230 × 8C, 42 × 16C, 40 x blastocyst | na |
| Microarray | Hamatani et al. (2004) | GSE936 | Natural mating | mII to blastocyst | Quickprep micro poly-A RNA Extraction Kit | 500 x mII, 500 × 1C, 500 × 2C, 500 × 4C, 500 × 8C, 500 x morula, 500 x blastocyst | FDR <1% | |
| Microarray | Wang et al. (2004) | (Deposited Arrayexpress) | Natural mating (defined by phCG) | GV to blastocyst | Total RNA derived cDNA synthesis | <60 x GV, <55 x mII, <70 × 1C, <134 × 2C, <131 × 4C, <95 × 8C, <42 × 8C, <70 x blastocyst | na | |
| Affymetrix MOE430 microarray | Zeng and Schultz (2005) | Not given | Natural mating | mII, 1C and 2C | cRNA preparation according to the Affymetrix Small Sample Prep Technical Bulletin | Pools of ∼325 eggs; 335 1C embryos; 380 2-cell embryos | FDR <5% | |
| RNA-sequencing | Park et al. (2013) | DRA001066 | IVF | mII to 4C | Total RNA-seq libraries: SOLiD Total RNA-seq kit | 10,000 cells from each stage | FDR <5% | |
| Single-cell RNA-sequencing | Xue et al. (2013) | GSE44183 | Natural mating (defined by hCG) | mII to morula | Tang et al., 2010 (Illumina) | Single cell | FDR <5% | |
| RNA-sequencing | Deng et al., 2014 | GSE45719 | Natural mating (defined by hCG) | mII to blastocyst | Poly(A) RNA-seq libraries, Smart-seq2 (Takara Clontech) | Single cell | na | |
| RNA-sequencing | Abe et al. (2015) | not given | IVF | mII to blastocyst | Total RNA RNA-Seq libraries, mRNA-seq Sample Preparation Kit (Illumina) | 3,000 x mII, 3,000 × 1C, 4,500 × 2C, 2,800 x 4C, 1,400 x morula, and 700 x blastocysts |
na | |
| Single-cell RNA-sequencing | Fan et al. (2015) | GSE53386 | Natural mating | mII to blastocyst | SUPeR-seq | Single cell | p-value <0.05 | |
| RNA-sequencing | Abe et al. (2018) | DRA006557 | IVF | mII and 2C | Total RNA RNA-Seq libraries, mRNA-Seq Sample Preparation Kit (Illumina) | 4,500 embryos | na | |
| Smart-seq2 long-read RNA-sequencing | Qiao et al. (2020) | GSE138760 | Natural mating | mII to blastocyst | Total RNA cDNA amplified via the Smart-seq2 protocol | Pools of 150 oocytes; 150 × 1C; 100 × 2C; 50 × 4C; 25 × 8C; 20 × 32-64C blastocyst | unknown | |
| RNA-sequencing | Zhang et al. (2022) | GSE169632 | IVF | mII to blastocyst | Total RNA-seq libraries: SMART-Seq Stranded Kit (Takara Clontech) Poly(A) RNA-seq libraries: SMARTer ultralow input RNA cDNA preparation kit (Takara Clontech) | 100 to 250 oocytes or embryos | FDR <1% | |
| Single-cell RNA-sequence and DNA microarray | Asami et al. (2023) | GSE222130, GSE64648, GSE64649 and GSE64650 | ICSI | mII and 1C (2-, 4-, 6-, 8-, 10-, 12-hpf) | SMARTer Stranded Total RNA-Seq Kit v1 and 2 – Pico Input Mammalian (Takara Clontech) | Single cell | FDR <5% | |
| Human | Microarray | Vassena et al. (2011) | GSE29397 | Not stated | mII to blastocyst | Affymetrix Human Gene 1.0 ST array | Not stated | p-value <0.05 |
| Single-cell RNA-sequencing | Yan et al. (2013) | GSE36552 | IVF | mII to blastocyst | Step-by-step single-cell RNA-seq TrueSeq DNA library preparation kit (Illumina) | Single cell | p-value <0.01 | |
| Single-cell RNA-sequencing | Xue et al. (2013) | GSE44183 | ICSI | mII to morula | Tang et al., 2010 (Illumina) | Single cell | FDR <5% | |
| Single-cell RNA-sequencing | Leng et al. (2019) | GSE133856 | IVF | mII to morula | Total RNA cDNA amplified via the Smart-seq2 protocol | Single cell | p-value <0.05 | |
| Single-cell RNA-sequencing | Asami et al. (2022) | GSE157834 | ICSI | mII and 1C | Clontech SMARTer Total RNA-Seq Kit Pico Input (V2) system (Takara Clontech) | Single cell | FDR <5% |
Summary of mouse and human datasets containing one- and two-cell embryo transcriptomes.
Recent high-resolution, polyadenylation-independent scRNA-seq time-course profiling of precisely-staged mouse one-cell embryos has addressed several of these caveats (Asami et al., 2023). Embryo synchrony within 5 min per time-point was achieved by coordinated microinjection and, following scRNA-seq of embryos at different points on the resulting time-course, revealed a program of embryonic gene expression initiating within 4 h of fertilization (Asami et al., 2023). Using a false discovery rate (FDR) of <5%, 1,777 genes were found to be upregulated in iEGA (i.e., within the first 12 h of sperm-egg union) compared to mature, fertilizable metaphase II (mII) oocytes. Of these, ∼90% were predicted to be RNA polymerase II- (PolII-) generated transcripts that were canonically-spliced, with transcription predominantly from the maternal genome (Asami et al., 2023). Analogous time-course profiling in human embryos is impracticable, but scRNA-seq of imprecisely-staged, apparently healthy (e.g., bipronuclear; 2PN) human one-cell embryos also revealed significant (FDR <5%) transcriptional upregulation. At 12.7% (FDR<5%), the overlap between mouse iEGA and presumptive iEGA in human one-cell embryos (henceforth referred to as iEGA) was modest, consistent with human embryo genetic heterogeneity (e.g., they were from different ethnicities), lack of synchrony, and the precise time of fertilization being unknown, any of which might conceal human-mouse similarities. However, pathways predicted for mouse and human iEGA overlapped, with the human dataset most closely corresponding to an early (4 h) timepoint in the mouse time-course (Asami et al., 2022). Mouse iEGA pathway terms included cell cycle regulation (e.g., the meiotic-to-mitotic cell cycle transition), metabolism (e.g., IGF signalling) and DNA methylation (e.g., transcriptional repression signalling) (Mayer et al., 2000; Wossidlo et al., 2011; Yoshida et al., 2007). Many analogous pathway terms are represented in human iEGA (Asami et al., 2022) (Figure 1). Consistent with a previous report of murine endogenous retrovirus (MuERV) expression in one-cell embryos (Kigami et al., 2003), LTR, Pol and Gag genes (Rowe and Trono, 2011) were upregulated at hundreds of loci from 8 h, showing that MuERV gene activation is a feature of iEGA (Asami et al., 2023). Upregulated genes in human one-cell embryos also included 63 endogenous retrovirus (hERV) loci (Asami et al., 2022).
3 Regulation of iEGA
What might we infer from iEGA about upstream and downstream transcription regulators in one-cell embryos? The putative EGA regulator gene, Dux (Hendrickson et al., 2017; De Iaco et al., 2017), is upregulated in mouse iEGA (Asami et al., 2023). Dux has been shown to recruit p300/CBP to regulatory regions in minor EGA genes including Obox4, Zscan4s and Usp17s, independently of p300/CBP catalytic acetylation (Xiao et al., 2025); recruitment of p300/CBP may also be primed by H3.3S31ph (Zhang et al., 2025). This may help facilitate PolII localization and the transition from minor to major EGA (Xiao et al., 2025). However, although genetic deletion experiments show that Dux promotes early development in the mouse (possibly by regulating the cell cycle), not all Dux target genes are dysregulated in Dux-null embryos, some of which complete full-term development (De Iaco et al., 2020). Moreover, Dux-responsive genes were not upregulated in iEGA, suggesting that iEGA is independent of Dux and its paralogs (Asami et al., 2023). In humans, DUX4 (the mouse Dux ortholog) (Hendrickson et al., 2017) and other TFs postulated to drive human cleavage-stage (major) EGA, including OCT4 (Gao et al., 2018) and LEUTX (Jouhilahti et al., 2016), were absent from iEGA (Asami et al., 2022).
Many transcription activators predicted by both mouse and human iEGA (FDR<5%) were oncogenes, including (with corresponding mouse species orthologs), MYC, MYCN, RABL6, FYN and E2F4 (Asami et al., 2022; 2023). In the mouse, trans-activation by c-Myc, Mycn, Erg and Atf4, whose human counterparts have well-documented roles in cancer (Baluapuri et al., 2020; Papas et al., 1990; Shen-Li et al., 2000; Wortel et al., 2017), was predicted to have occurred within 8 h of fertilization (Asami et al., 2023). Each of these proteins was present in mouse mII oocytes and one-cell embryos, although corresponding mRNAs were often undetectable: c-Myc was present in immature oocytes and localized to spindles in mII oocytes (Alexandrova et al., 1995; Asami et al., 2023; González-Prieto et al., 2015). RNA-seq following high-sensitivity assay for transposase-accessible chromatin (ATAC-seq) of human one-cell embryos (albeit abnormal, 3PN) revealed regions of open chromatin that were distal to major EGA regulatory regions, enriched for transcription factor-binding sites and overlapping with DNA hypomethylated domains (Wu et al., 2018). Many of these distal regions become inaccessible after major EGA in a transcription-dependent manner. Such chromatin dynamics are conserved in mice.
Both c-Myc, and its canonical heterodimeric co-activating partner, Max, were present in oocytes and one-, two- and four-cell embryos (Asami et al., 2023). The c-Myc cleavage product, Myc-nick (Conacci-Sorrell et al., 2010) was present in one-cell embryos, and other isoforms became readily detectable in cleavage-stage embryos, indicative of dynamic c-Myc regulation during and after iEGA. Treating mouse one-cell embryos with structurally distinct inhibitors of c-Myc-Max heterodimerization (MYCi975 and 10058-F4) that block c-Myc gene-regulatory activity (Han et al., 2019; Huang et al., 2006) induced embryo developmental arrest at one- or two-cell stages (Asami et al., 2023). scRNA-seq following 10058-F4 treatment identified 577 genes expressed at reduced levels compared to untreated controls (FDR<5%), including 95.4% of the differentially expressed genes that overlapped with iEGA genes. The list contained known c-Myc targets and predicted involvement (p < 0.01) in G2/M DNA damage regulation, cell-cycle control of chromosome replication and nucleotide excision repair. Perhaps c-Myc potentiates iEGA by poised transcription complex formation (Rahl et al., 2010) in mII oocytes and modulates transcription akin to its amplification of gene expression in cancer (Lin et al., 2012). Inhibiting the predicted iEGA TF, Mycn, disrupted embryo morphology, impeded cytokinesis, blocked early development and disrupted iEGA gene upregulation. Similarly, blocking the cancer-associated TFs, Erg or Atf4, which are also present in one-cell embryos and predicted to have iEGA-regulatory roles, also impeded preimplantation development (Asami et al., 2023). Thus, iEGA is a predictor of TFs that contribute to the onset of embryonic transcription, and which in many cases are regulatory oncogenes.
The number of iEGA genes (1,777 in mouse [FDR<5%], 1,322 in human [FDR<10%]) is clearly more than a handful (Ji et al., 2023), but in both mouse and human one-cell embryos, the amplitude of expression upregulation in iEGA was typically <2-fold and the mean log2 fold-change in mouse was 0.77 ± 0.03 (FDR<5%) (Asami et al., 2022; 2023). Increases were not population effects attributable to high-expressing outliers, as the analyses were on single cells (scRNA-seq). However, the induction of target genes by MYC is typically less than twofold (Baluapuri et al., 2020) and genes specific to ES cells undergo only modest increases during pluripotency induction (Chronis et al., 2017). The polycomb repressor is a pluripotency modulator that responds to low levels of transcription (Berrozpe et al., 2017), and a pleiotropic activator-repressor with a key gene-regulatory role in ES cells, CTCF, also exerts only a modest (less than 2-fold) effect on transcripts when CTCF and RAD21 are depleted (Narita et al., 2025). The nucleosome remodeling and deacetylation (NuRD) is essential for pluripotency, but eliminating its activity in ES cells results in gene expression changes of mostly less than twofold (Bornelöv et al., 2018; Miller et al., 2016). Modest gene expression level changes of less than twofold may thus be a hallmark of cellular potency regulation and transitions.
One-cell embryos represent the only obligate developmental node through which gamete-derived chromatin passes. Thus, iEGA may provide a unique read-out of chromatin marks transmitted from parents via their gametes with the potential to mediate epigenetic inheritance of acquired parental traits (Fitz-James and Cavalli, 2022; Asami et al., 2023).
4 Embryonic genome repression, EGR, and an iEGA ‘off’ switch
In addition to blocking iEGA, 10058-F4 treatment of mouse one-cell embryos caused upregulation of 923 genes (i.e., 61.5% of genes that were differentially expressed; FDR<5%) (Asami et al., 2023). This suggests that c-Myc either directly or indirectly represses gene expression in one-cell embryos (Perry et al., 2023). We refer to targeted suppression of transcription in the first 12 h after sperm-egg union as embryonic genome repression, EGR. It is possible that the disruption of EGR by 10058-F4 treatment contributes, at least partly, to acute developmental attenuation.
Some EGR genes become upregulated during preimplantation development (50 in mouse major EGA), indicating that for some, repression is transient. EGR pathways reflect downstream transitions from one-cell-stage to blastocyst development: lipid biosynthesis for the ∼26% plasma membrane area increase attending the first cell division (Duch et al., 2020; Pratt, 1990), maternal factor catabolism (Wu and Dean, 2020), and the downstream metabolic transition from oxidative phosphorylation to glycolysis (Redel et al., 2012).
It is established that c-Myc can behave as a transcriptional repressor (Grandori et al., 2000; Wanzel et al., 2003) and may thus act as a poised transcriptional (co)repressor that switches to or from (co)activator-mode, raising the possibility that c-Myc is differentially regulated to perform each function simultaneously at different loci in one-cell embryos. Like c-Myc, other transcriptional regulators predicted by iEGA have documented activator and repressor functions, including Atf4, Erg, Mycn, E2F1, Mitf, c-Rel and Foxm1 (Asami et al., 2023), but it is unknown whether they contribute to EGR. Our understanding about these activator-repressors largely derives from adult disease, but their principal normative physiological roles may include regulating early development.
Expression of most (61.6%) mouse iEGA genes had markedly declined by the two-cell stage (Asami et al., 2023). In the mouse, this coincides with major EGA, in which there is a relatively high-amplitude transcriptional upregulation of a larger set of genes (see below) (Hamatani et al., 2004). This situation is mirrored by human embryos when allowance is made for the later occurrence of human major EGA at the eight-cell stage: human one-cell embryo (iEGA) transcript levels remained elevated until around the eight-cell stage, when they declined (Asami et al., 2022). It thus appears iEGA is switched off as major EGA is initiated in human and mouse embryos.
5 Waves of early embryonic transcription
The dynamics of mouse EGA include iEGA (which segues to minor EGA) in one-cell embryos, followed by major EGA at the two-cell stage (Hamatani et al., 2004; Park et al., 2013; Wang et al., 2004; Xue et al., 2013; Zeng and Schultz, 2005). Mouse iEGA is refractory to the Pol II inhibitor, α-amanitin (Asami et al., 2023), which is a feature shared with minor EGA (Hamatani et al., 2004; Zeng and Schultz, 2005). In minor EGA, as in genome activation in one-cell embryos following nuclear transfer, the carboxy-terminal domain of the PolII large catalytic subunit, Rpb1, is hypophosphorylated (Bellier et al., 1997; Miyamoto et al., 2018). Rpb1 is canonically phosphorylated in active PolII and indeed becomes phosphorylated in major EGA (Bellier et al., 1997), so iEGA seems to be mediated by a distinctive PolII mechanism. These findings are consistent with iEGA and minor EGA employing related and idiosyncratic transcriptional mechanisms, and support the idea that they describe the same transcriptional phase.
By contrast, major EGA constitutes a second transcriptional wave, rather than a simple continuation of iEGA (Asami et al., 2023). In this model, iEGA is initiated by maternal factors immediately after fertilization, but largely yields to a new program of higher-amplitude expression driven by distinctive TFs in major EGA (Figure 1). Proposed modulators of major EGA include Ccnt2 (Zhang et al., 2022), Dppa2 (Eckersley-Maslin et al., 2019), Dppa4 (Guo et al., 2024), DUXBL (Vega-Sendino et al., 2024), Kdm1a (Ancelin et al., 2016), Klf17 (Hu et al., 2024), MLX (Wang et al., 2022), NAT10 (Cui et al., 2025), Nr5a2 (Gassler et al., 2022), Nuclear transcription factor Y subunit-α, NFYA (Lu et al., 2016), Obox (Ji et al., 2023), PRDM10 (Seah et al., 2025), Rfx1 (Wang et al., 2022), Smarcd2 (Zhang et al., 2022), Tprx1 (Zou et al., 2022), Yap1 (Yu et al., 2016) and YY1 (Wang et al., 2024). Of these, Obox5 (FDR, 2.08E-26), Yap1 (FDR, 1.65E-04), Ccnt2 (FDR, 3.42E-03) and Klf17 (FDR, 6.69E-22) are upregulated in iEGA (Figure 1). Treatment of mouse one-cell embryos with the protein synthesis inhibitor, cycloheximide, disrupts major EGA (i.e., at the two-cell stage) and development (Hu et al., 2024), suggesting that the translation of canonically-spliced transcripts produced during iEGA contributes to the transcriptional circuitry required for major EGA. Indeed, genes for several putative major EGA regulators (including orphan receptor genes, but not Nr5a2) are themselves upregulated in iEGA, suggesting that iEGA primes major EGA (Figure 1). We now describe selected examples of putative major EGA regulators.
Obox. The PRD-like homeobox domain transcription factor family, Obox, apparently regulates major EGA, as mice deficient for maternally-transcribed Obox1-5 and Obox7 expressed at the one-cell stage underwent impaired transcription and two-to-four-cell arrest (Ji et al., 2023). Activation by Obox family members is thought to exhibit redundancy and involves depositing H3K27ac at GC-poor promoter and enhancer regions, opening chromatin and pre-configuring PolII (Liu et al., 2020). This redundancy may involve the related TF, Tprx1 (Zou et al., 2022).
Nr5a2. The orphan nuclear receptor, Nr5a2, has been shown to upregulate major EGA genes in mouse two-cell embryos and to be required for progression beyond the two-cell stage (Gassler et al., 2022). Nr5a2 promotes chromatin accessibility and binds to motifs within short interspersed nuclear element (SINE) B1 (B1-elements) in the mouse, and in human counterparts, Alu retrotransposons, present in cis-regulatory regions of major EGA genes (Gassler et al., 2022). Chemical inhibition of Nr5a2 resulted in significant (FDR<5%) under-expression of 535 genes compared to controls (Gassler et al., 2022). However, genetic removal of maternal Nr5a2 produced the conclusion that Nr5a2 is dispensable for major EGA and that Nr5a2 may cooperate with other developmental TFs, such as Krüppel-like factors (below), to establish robust chromatin accessibility (Festuccia et al., 2024). In addition, Smart-seq analysis has suggested that Nr5a2 is activated after major EGA (Oomen et al., 2025). The evolutionary processes that gave rise to B1/Alu-mediated major EGA also require explanation. B1-elements and Alu-repeats evolved from 7SL RNA and are mostly found in introns and upstream gene regulatory elements (Tsirigos and Rigoutsos, 2009): open chromatin at major EGA genes may thus not be the result of retroviral integration, but its cause. Integration at hundreds or thousands of loci associated with major EGA presumably occurred independently in mouse and human genomes to yield a selective advantage (or at least, not a disadvantage), but transcriptional activation must clearly have been successful before retroposition took place. The roles of Nr5a2 and retrotransposons in major EGA are therefore unclear. Genes encoding nuclear receptor family members Nr3c1, Nr2e1 and Nr6a1 are upregulated in iEGA, whereas Nr5a2 is neither upregulated in, nor a predicted regulator of iEGA (Asami et al., 2023). There is little evidence that Nr5a2 functions as a pioneer factor for iEGA.
Klf17. Krüppel-like factor 17 (Klf17) involvement in mouse and human major EGA has recently been inferred from genetic and proteomic analyses, and it may mediate PolII pre-configuration at the early two-cell stage (Taubenschmid-Stowers et al., 2022; Hu et al., 2024).
Yap1. Yes-associated protein 1 (Yap1) is highly expressed in mouse and human oocytes and early embryos. Maternally-derived Yap1 is necessary for major EGA, and Yap1 gradually translocates from the cytoplasm to the nucleus during early development (Yu et al., 2016). The Yap1 gene is expressed in mouse iEGA 6 h post-fertilization, and it is a predicted upstream regulator of iEGA genes including Rrm2, Cdc25a and Pdcl whose expression increases after 10 h (Asami et al., 2023).
6 Relationship of EGA/EGR to embryoids
The genesis in vitro of embryoids (e.g., blastoids) from naïve PSCs skips multiple embryonic processes that follow fertilization (Boiani, 2024; Kagawa et al., 2022; Rivron et al., 2018). Among these processes are iEGA, major EGA and the downregulation of iEGA transcripts (EGR) that more-or-less coincides with major EGA (Asami et al., 2022; 2023; Perry et al., 2023). The expression profiles of genes undergoing regulation during EGA and EGR may be important for key down-stream developmental events, but they are manifest before the establishment of naïve pluripotency in blastocyst-stage embryos. Thus, even though the intracellular history of iEGA expression may be critical to mapping the developmental trajectory of embryoids, it may be difficult to trace back from later developmental stage embryos (e.g., blastocysts) or other entities from which naïve PSCs are derived. This concept potentially undermines confidence about the downstream - perhaps far-downstream - developmental potential of systems derived from naïve PSCs.
7 Concluding comments
We suggest that two waves of embryonic transcription follow fertilization in early preimplantation development: iEGA and major EGA. Both share conserved features with their respective counterparts in mouse and human. iEGA reflects the initiation of transcription in one-cell embryos, and minor EGA is a continuation of it. The second gene expression wave (major EGA) involves both a boost in transcriptional amplitude and qualitative differences compared to iEGA. Blocking iEGA can precipitate acute developmental arrest (Asami et al., 2023), suggesting that it is of critical functional importance. Cell cycle pathways feature in mouse and human iEGA, and may play roles in the intricate and anomalous choreography between cell volume and cell cycle length in early embryos. Moreover, iEGA may prime major EGA, as genes for several candidate regulators of major EGA are upregulated in iEGA. This has mechanistic implications and suggests that iEGA holds clues to the identities of major EGA regulators. Similarly, iEGA may illuminate chromatin alterations in mouse and human gametes that reflect parentally-acquired traits (e.g., obesogenic traits): epigenetic inheritance (Fitz-James and Cavalli, 2022). Thus, iEGA may be both of functional importance in its own right, and report additional key steps in the early embryo.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries may be directed to the authors.
Ethics statement
This work was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MA: Formal Analysis, Visualization, Writing – original draft, Data curation. ACFP: Supervision, Conceptualization, Funding acquisition, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors received grant support for the research, authorship and publication of this article from the UK Medical Research Council (MR/W024845/1).
Acknowledgments
We thank B. Lam and B. Hendrich for helpful suggestions during manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abe K. Yamamoto R. Franke V. Cao M. Suzuki Y. Suzuki M. G. et al (2015). The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3' processing. EMBO J.34, 1523–1537. 10.15252/embj.201490648
2
Abe K. I. Funaya S. Tsukioka D. Kawamura M. Suzuki Y. Suzuki M. G. et al (2018). Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. U. S. A.115, E6780–E6788. 10.1073/pnas.1804309115
3
Adenot P. G. Mercier Y. Renard J. P. Thompson E. M. (1997). Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development124, 4615–4625. 10.1242/dev.124.22.4615
4
Alexandrova N. Niklinski J. Bliskovsky V. Otterson G. A. Blake M. Kaye F. J. et al (1995). The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol. Cell Biol.15, 5188–5195. 10.1128/MCB.15.9.5188
5
Ancelin K. Syx L. Borensztein M. Ranisavljevic N. Vassilev I. Briseño-Roa L. et al (2016). Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. Elife5, e08851. 10.7554/eLife.08851
6
Asami M. Lam B. Y. H. Hoffmann M. Suzuki T. Lu X. Yoshida N. et al (2023). A program of successive gene expression in mouse one-cell embryos. Cell Rep.42, 112023. 10.1016/j.celrep.2023.112023
7
Asami M. Lam B. Y. H. Ma M. K. Rainbow K. Braun S. VerMilyea M. D. et al (2022). Human embryonic genome activation initiates at the one-cell stage. Cell Stem Cell29, 209–216.e4. 10.1016/j.stem.2021.11.012
8
Balhorn R. (1982). A model for the structure of chromatin in Mammalian sperm. J. Cell Biol.93, 298–305. 10.1083/jcb.93.2.298
9
Baluapuri A. Wolf E. Eilers M. (2020). Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol.21, 255–267. 10.1038/s41580-020-0215-2
10
Bellier S. Chastant S. Adenot P. Vincent M. Renard J. P. Bensaude O. (1997). Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in Mammalian embryos. EMBO J.16, 6250–6262. 10.1093/emboj/16.20.6250
11
Berrozpe G. Bryant G. O. Warpinski K. Spagna D. Narayan S. Shah S. et al (2017). Polycomb responds to low levels of transcription. Cell Rep.20, 785–793. 10.1016/j.celrep.2017.06.076
12
Blower M. D. Jambhekar A. Schwarz D. S. Toombs J. A. (2013). Combining different mRNA capture methods to analyze the transcriptome: analysis of the Xenopus laevis transcriptome. PLoS One8, e77700. 10.1371/journal.pone.0077700
13
Boiani M. MHR-ISSCR guidelines working group (2024). The future of embryoids from a reproductive science perspective. Mol. Hum. Reprod.30, gaae009. 10.1093/molehr/gaae009
14
Bornelöv S. Reynolds N. Xenophontos M. Gharbi S. Johnstone E. Floyd R. et al (2018). The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell71, 56–72.e4. 10.1016/j.molcel.2018.06.003
15
Braude P. Bolton V. Moore S. (1988). Human gene expression first occurs between the four-and eight-cell stages of preimplantation development. Nature332, 459–461. 10.1038/332459a0
16
Chronis C. Fiziev P. Papp B. Butz S. Bonora G. Sabri S. et al (2017). Cooperative binding of transcription factors orchestrates reprogramming. Cell168, 442–459.e20. 10.1016/j.cell.2016.12.016
17
Clegg K. B. Piko L. (1983). Poly (A) length, cytoplasmic adenylation and synthesis of poly(A)+ RNA in early mouse embryos. Dev. Biol.95, 331–341. 10.1016/0012-1606(83)90034-9
18
Conacci-Sorrell M. Ngouenet C. Eisenman R. N. (2010). Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell142, 480–493. 10.1016/j.cell.2010.06.037
19
Condic M. L. (2014). Totipotency: what it is and what it is not. Stem Cells Dev.23, 796–812. 10.1089/scd.2013.0364
20
Cui Z. Fu C. Ai D. Zhu H. Wang F. An X. et al (2025). NAT10 regulates zygotic genome activation and the morula-to-blastocyst transition. FASEB J.39, e70444. 10.1096/fj.202402751R
21
Cuthbertson K. S. Whittingham D. G. Cobbold P. H. (1981). Free Ca2+ increases in exponential phases during mouse oocyte activation. Nature294, 754–757. 10.1038/294754a0
22
Daniels R. Kinis T. Serhal P. Monk M. (1995). Expression of the myotonin protein kinase gene in preimplantation human embryos. Hum. Mol. Genet.4, 389–393. 10.1093/hmg/4.3.389
23
De Iaco A. Planet E. Coluccio A. Verp S. Duc J. Trono D. (2017). DUX-Family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet.49, 941–945. 10.1038/ng.3858
24
De Iaco A. Verp S. Offner S. Grun D. Trono D. (2020). DUX is a non-essential synchronizer of zygotic genome activation. Development147, dev177725. 10.1242/dev.177725
25
Duch M. Torras N. Asami M. Suzuki T. Arjona M. I. Gómez-Martínez R. et al (2020). Tracking intracellular forces and mechanical property changes in mouse one-cell embryo development. Nat. Mater.19, 1114–1123. 10.1038/s41563-020-0685-9
26
Eckersley-Maslin M. Alda-Catalinas C. Blotenburg M. Kreibich E. Krueger C. Reik W. (2019). Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev.33, 194–208. 10.1101/gad.321174.118
27
Festuccia N. Vandormael-Pournin S. Chervova A. Geiselmann A. Langa-Vives F. Coux R. X. et al (2024). Nr5a2 is dispensable for zygotic genome activation but essential for morula development. Science386, eadg7325. 10.1126/science.adg7325
28
Fitz-James M. H. Cavalli G. (2022). Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet.23, 325–341. 10.1038/s41576-021-00438-5
29
Gao L. Wu K. Liu Z. Yao X. Yuan S. Tao W. et al (2018). Chromatin accessibility landscape in human early embryos and its association with evolution. Cell173, 248–259.e15. 10.1016/j.cell.2018.02.028
30
Gassler J. Kobayashi W. Gáspár I. Ruangroengkulrith S. Mohanan A. Gómez Hernández L. et al (2022). Zygotic genome activation by the totipotency pioneer factor Nr5a2. Science378, 1305–1315. 10.1126/science.abn7478
31
González-Prieto R. Cuijpers S. A. Kumar R. Hendriks I. A. Vertegaal A. C. (2015). c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle14, 1859–1872. 10.1080/15384101.2015.1040965
32
Grandori C. Cowley S. M. James L. P. Eisenman R. N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Ann. Rev. Cell Dev. Biol.16, 653–699. 10.1146/annurev.cellbio.16.1.653
33
Guo Y. Kitano T. Inoue K. Murano K. Hirose M. Li T. D. et al (2024). Obox4 promotes zygotic genome activation upon loss of Dux. Elife13, e95856. 10.7554/eLife.95856
34
Hachem A. Godwin J. Ruas M. Lee H. C. Ferrer Buitrago M. Ardestani G. et al (2017). PLCζ is the physiological trigger of the Ca2+ oscillations that induce embryogenesis in mammals but conception can occur in its absence. Development144, 2914–2924. 10.1242/dev.150227
35
Hamatani T. Carter M. G. Sharov A. A. Ko M. S. (2004). Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell6, 117–131. 10.1016/s1534-5807(03)00373-3
36
Han H. Jain A. D. Truica M. I. Izquierdo-Ferrer J. Anker J. F. Lysy B. et al (2019). Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell36, 483–497.e15. 10.1016/j.ccell.2019.10.001
37
Hendrickson P. G. Dorais J. A. Grow E. J. Whiddon J. L. Lim J. W. Wike C. L. et al (2017). Conserved roles of mouse dux and human Dux4 in activating cleavage-stage genes and MERVL/HERVL retro-transposons. Nat. Genet.49, 925–934. 10.1038/ng.3844
38
Hu Y. Wang Y. He Y. Ye M. Yuan J. Ren C. et al (2024). Maternal KLF17 controls zygotic genome activation by acting as a messenger for RNA Pol II recruitment in mouse embryos. Dev. Cell59, 613–626.e6. 10.1016/j.devcel.2024.01.013
39
Huang M. J. Cheng Y. C. Liu C. R. Lin S. Liu H. E. (2006). A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol.34, 1480–1489. 10.1016/j.exphem.2006.06.019
40
Hyman A. A. Weber C. A. Jülicher F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol.30, 39–58. 10.1146/annurev-cellbio-100913-013325
41
Ji S. Chen F. Stein P. Wang J. Zhou Z. Wang L. et al (2023). OBOX regulates mouse zygotic genome activation and early development. Nature620, 1047–1053. 10.1038/s41586-023-06428-3
42
Jouhilahti E. M. Madissoon E. Vesterlund L. Töhönen V. Krjutškov K. Plaza Reyes A. et al (2016). The human PRD-like homeobox gene LEUTX has a central role in embryo genome activation. Development143, 3459–3469. 10.1242/dev.134510
43
Kagawa H. Javali A. Khoei H. H. Sommer T. M. Sestini G. Novatchkova M. et al (2022). Human blastoids model blastocyst development and implantation. Nature601, 600–605. 10.1038/s41586-021-04267-8
44
Katayama M. Ellersieck M. R. Roberts M. (2010). Development of monozygotic twin mouse embryos from the time of blastomere separation at the two-cell stage to blastocyst. Biol. Reprod.82, 1237–1247. 10.1095/biolreprod.109.082982
45
Kigami D. Minami N. Takayama H. Imai H. (2003). MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod.68, 651–654. 10.1095/biolreprod.102.007906
46
Ko M. S. Kitchen J. R. Wang X. Threat T. A. Wang X. Hasegawa A. et al (2000). Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development127, 1737–1749. 10.1242/dev.127.8.1737
47
Leng L. Sun J. Huang J. Gong F. Yang L. Zhang S. et al (2019). Single-cell transcriptome analysis of uniparental embryos reveals parent-of-origin effects on human preimplantation development. Cell Stem Cell25, 697–712.e6. 10.1016/j.stem.2019.09.004
48
Lin C. Y. Lovén J. Rahl P. B. Paranal R. M. Burge C. B. Bradner J. E. et al (2012). Transcriptional amplification in tumor cells with elevated c-Myc. Cell151, 56–67. 10.1016/j.cell.2012.08.026
49
Ling X. Liu X. Jiang S. Fan L. Ding J. (2022). The dynamics of three-dimensional chromatin organization and phase separation in cell fate transitions and diseases. Cell Regen.11, 42. 10.1186/s13619-022-00145-4
50
Liu B. Xu Q. Wang Q. Feng S. Lai F. Wang P. et al (2020). The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature587, 139–144. 10.1038/s41586-020-2847-y
51
Liu W. Yin J. Kou X. Jiang Y. Gao H. Zhao Y. et al (2014). Asymmetric reprogramming capacity of parental pronuclei in mouse zygotes. Cell Rep.6, 1008–1016. 10.1016/j.celrep.2014.02.018
52
Lu F. Liu Y. Inoue A. Suzuki T. Zhao K. Zhang Y. (2016). Establishing chromatin regulatory landscape during mouse preimplantation development. Cell165, 1375–1388. 10.1016/j.cell.2016.05.050
53
Macfarlan T. S. Gifford W. D. Driscoll S. Lettieri K. Rowe H. M. Bonanomi D. et al (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature487, 57–63. 10.1038/nature11244
54
Marston J. H. Chang M. C. (1964). The fertilizable life of ova and their morphology following delayed insemination in mature and immature mice. J. Exp. Zool.155, 237–251. 10.1002/jez.1401550211
55
Mayer W. Niveleau A. Walter J. Fundele R. Haaf T. (2000). Demethylation of the zygotic paternal genome. Nature403, 501–502. 10.1038/35000656
56
Mazid M. A. Ward C. Luo Z. Liu C. Li Y. Lai Y. et al (2022). Rolling back human pluripotent stem cells to an eight-cell embryo-like stage. Nature605, 315–324. 10.1038/s41586-022-04625-0
57
Miller A. Ralser M. Kloet S. L. Loos R. Nishinakamura R. Bertone P. et al (2016). Sall4 controls differentiation of plurip-otent cells independently of the nucleosome remodelling and deacetylation (NuRD) complex. Development143, 3074–3084. 10.1242/dev.139113
58
Miyamoto K. Nguyen K. T. Allen G. E. Jullien J. Kumar D. Otani T. et al (2018). Chromatin accessibility impacts transcriptional reprogramming in oocytes. Cell Rep.24, 304–311. 10.1016/j.celrep.2018.06.030
59
Narita T. Kilic S. Higashijima Y. Pappas G. Maskey E. Choudhary C. (2025). A unified model of gene expression control by cohesin and CTCF. bioRxiv Prepr. 10.1101/2024.11.29.625887
60
Olsen T. K. Baryawno N. (2018). Introduction to single-cell RNA sequencing. Curr. Protoc. Mol. Biol.122, e57. 10.1002/cpmb.57
61
Oomen M. E. Rodriguez-Terrones D. Kurome M. Zakhartchenko V. Mottes L. Simmet K. et al (2025). An atlas of transcription initiation reveals regulatory principles of gene and transposable element expression in early Mammalian development. Cell188, 1156–1174.e20. 10.1016/j.cell.2024.12.013
62
Papas T. S. Watson D. K. Sacchi N. Fujiwara S. Seth A. K. Fisher R. J. et al (1990). ETS family of genes in leukemia and Down syndrome. Am. J. Med. Genet. Suppl.7 (7), 251–261. 10.1002/ajmg.1320370751
63
Park S. J. Komata M. Inoue F. Yamada K. Nakai K. Ohsugi M. et al (2013). Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev.27, 2736–2748. 10.1101/gad.227926.113
64
Perry A. C. F. Asami M. Lam B. Y. H. Yeo G. S. H. (2023). The initiation of Mammalian embryonic transcription: to begin at the beginning. Trends Cell Biol.33, 365–373. 10.1016/j.tcb.2022.08.008
65
Perry A. C. F. Verlhac M.-H. (2008). Second meiotic arrest and exit in frogs and mice. EMBO Rep.9, 246–251. 10.1038/embor.2008.22
66
Pratt H. P. (1990). Phospholipid synthesis in the preimplantation mouse embryo. J. Reprod. Fertil.58, 237–248. 10.1530/jrf.0.0580237
67
Qiao Y. Ren C. Huang S. Yuan J. Liu X. Fan J. et al (2020). Author correction: high-Resolution annotation of the mouse preimplantation embryo transcriptome using long-read sequencing. Nat. Commun.12, 1767. 10.1038/s41467-021-22148-6
68
Rahl P. B. Lin C. Y. Seila A. C. Flynn R. A. McCuine S. Burge C. B. et al (2010). c-Myc regulates transcriptional pause release. Cell141, 432–445. 10.1016/j.cell.2010.03.030
69
Redel B. K. Brown A. N. Spate L. D. Whitworth K. M. Green J. A. Prather R. S. (2012). Glycolysis in preimplantation development is partially controlled by the Warburg effect. Mol. Reprod. Dev.79, 262–271. 10.1002/mrd.22017
70
Rivron N. C. Frias-Aldeguer J. Vrij E. J. Boisset J. C. Korving J. Vivié J. et al (2018). Blastocyst-like structures generated solely from stem cells. Nature557, 106–111. 10.1038/s41586-018-0051-0
71
Rossant J. (1976). Postimplantation development of blastomeres isolated from 4- and 8- cell mouse eggs. J. Embryol. Exp. Morphol.36, 283–290. 10.1242/dev.36.2.283
72
Rowe H. M. Trono D. (2011). Dynamic control of endogenous retroviruses during development. Virology411, 273–287. 10.1016/j.virol.2010.12.007
73
Seah M. K. Y. Han B. Y. Huang Y. Rasmussen L. J. H. Stäubli A. J. Bello-Rodríguez J. et al (2025). Maternal PRDM10 activates essential genes for oocyte-to-embryo transition. Nat. Commun.16, 1939. 10.1038/s41467-025-56991-8
74
Shen-Li H. O'Hagan R. C. Hou H. Jr. Horner J. W. Lee H. W. DePinho R. A. (2000). Essential role for max in early embryonic growth and development. Genes Dev.14, 17–22. 10.1101/gad.14.1.17
75
Suarez S. S. (1987). Sperm transport and motility in the mouse oviduct: observations in situ. Biol. Reprod.36, 203–210. 10.1095/biolreprod36.1.203
76
Suzuki T. Asami M. Hoffmann M. Lu X. Gužvić M. Klein C. A. et al (2016). Mice produced by mitotic reprogramming of sperm injected into haploid parthenogenotes. Nat. Comm.7, 12676. 10.1038/ncomms12676
77
Suzuki T. Yoshida N. Suzuki E. Okuda E. Perry A. C. F. (2010). Full-term mouse development by abolishing Zn2+-dependent metaphase II arrest without Ca2+ release. Development137, 2659–2669. 10.1242/dev.049791
78
Tarkowski A. K. (1959). Experiments on the development of isolated blastomers of mouse eggs. Nature184, 1286–1287. 10.1038/1841286a0
79
Taubenschmid-Stowers J. Rostovskaya M. Santos F. Ljung S. Argelaguet R. Krueger F. et al (2022). 8C-like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell29, 449–459.e6. 10.1016/j.stem.2022.01.014
80
Temeles G. L. Schultz R. M. (1997). Transient polyadenylation of a maternal mRNA following fertilization of mouse eggs. J. Reprod. Fertil.109, 223–228. 10.1530/jrf.0.1090223
81
Tsirigos A. Rigoutsos I. (2009). Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Comput. Biol.5, e1000610. 10.1371/journal.pcbi.1000610
82
Vassena R. Boué S. González-Roca E. Aran B. Auer H. Veiga A. et al (2011). Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development138, 3699–3709. 10.1242/dev.064741
83
Vega-Sendino M. Lüttmann F. F. Olbrich T. Chen Y. Kuenne C. Stein P. et al (2024). The homeobox transcription factor DUXBL controls exit from totipotency. Nat. Genet.56, 697–709. 10.1038/s41588-024-01692-z
84
Wang C. Chen C. Liu X. Li C. Wu Q. Chen X. et al (2022). Dynamic nucleosome organization after fertilization reveals regulatory factors for mouse zygotic genome activation. Cell Res.32, 801–813. 10.1038/s41422-022-00652-8
85
Wang Q. T. Piotrowska K. Ciemerych M. A. Milenkovic L. Scott M. P. Davis R. W. et al (2004). A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell6, 133–144. 10.1016/s1534-5807(03)00404-0
86
Wang T. Peng J. Fan J. Tang N. Hua R. Zhou X. et al (2024). Single-cell multi-omics profiling of human preimplantation embryos identifies cytoskeletal defects during embryonic arrest. Nat. Cell Biol.26, 263–277. 10.1038/s41556-023-01328-0
87
Wanzel M. Herold S. Eilers M. (2003). Transcriptional repression by Myc. Trends Cell Biol.13, 146–150. 10.1016/s0962-8924(03)00003-5
88
Wortel I. M. N. van der Meer L. T. Kilberg M. S. van Leeuwen F. N. (2017). Surviving stress: modulation of ATF4-Mediated stress responses in normal and malignant cells. Trends Endocrinol. Metab.28, 794–806. 10.1016/j.tem.2017.07.003
89
Wossidlo M. Nakamura T. Lepikhov K. Marques C. J. Zakhartchenko V. Boiani M. et al (2011). 5-Hydroxymethylcytosine in the Mammalian zygote is linked with epigenetic reprogramming. Nat. Commun.2, 241. 10.1038/ncomms1240
90
Wu D. Dean J. (2020). Maternal factors regulating preimplantation development in mice. Curr. Top. Dev. Biol.140, 317–340. 10.1016/bs.ctdb.2019.10.006
91
Wu J. Xu J. Liu B. Yao G. Wang P. Lin Z. et al (2018). Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature557, 256–260. 10.1038/s41586-018-0080-8
92
Xiao L. Jin H. Dang Y. Zhao P. Li S. Wang S. et al (2025). DUX-mediated configuration of p300/CBP drives minor zygotic genome activation independent of its catalytic activity. Cell Rep.44, 115544. 10.1016/j.celrep.2025.115544
93
Xue Z. Huang K. Cai C. Cai L. Jiang C. Y. Feng Y. et al (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature500, 593–597. 10.1038/nature12364
94
Yan L. Yang M. Guo H. Yang L. Wu J. Li R. et al (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol.20, 1131–1139. 10.1038/nsmb.2660
95
Yanagimachi R. (1994). “Mammalian fertilization,” The physiology of reproduction. Editors KnobilE.NeillJ. D. (New York: Raven Press), 1, 189–317.
96
Yoshida N. Brahmajosyula M. Shoji S. Amanai M. Perry A. C. F. (2007). Epigenetic discrimination by mouse metaphase II oocytes mediates asymmetric chromatin remodeling independently of meiotic exit. Dev. Biol.301, 464–477. 10.1016/j.ydbio.2006.08.006
97
Yu C. Ji S. Y. Dang Y. J. Sha Q. Q. Yuan Y. F. Zhou J. J. et al (2016). Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res.26, 275–287. 10.1038/cr.2016.20
98
Zeng F. Schultz R. M. (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol.283, 40–57. 10.1016/j.ydbio.2005.03.038
99
Zhang C. Wang M. Li Y. Zhang Y. (2022). Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci. Adv.8, eabj3967. 10.1126/sciadv.abj3967
100
Zhang J. Li X. Zhao Q. Liu X. Wen D. Kong Q. et al (2025). Acetylation at lysine 27 on maternal H3.3 regulates minor zygotic genome activation. Cell Rep.44, 115148. 10.1016/j.celrep.2024.115148
101
Zou Z. Zhang C. Wang Q. Hou Z. Xiong Z. Kong F. et al (2022). Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science378, abo7923. 10.1126/science.abo7923
102
Zuccotti M. Piccinelli A. Giorgi Rossi P. Garagna S. Redi C. A. (1995). Chromatin organization during mouse oocyte growth. Mol. Reprod. Dev.41, 479–485. 10.1002/mrd.1080410410
Summary
Keywords
transcription, fertilization, one-cell embryo, embryonic genome activation (EGA), immediate EGA, zygotic genome activation (ZGA), embryonic genome repression (EGR), single-cell RNA-sequencing
Citation
Asami M and Perry ACF (2025) Mouse and human embryonic genome activation initiate at the one-cell stage. Front. Cell Dev. Biol. 13:1594995. doi: 10.3389/fcell.2025.1594995
Received
17 March 2025
Accepted
14 July 2025
Published
30 July 2025
Volume
13 - 2025
Edited by
Aimin Liu, The Pennsylvania State University (PSU), United States
Reviewed by
Alice Jouneau, l’alimentation et l’environnement (INRAE), France
Mingxiang Zhang, Colorado Center for Reproductive Medicine, United States
Updates
Copyright
© 2025 Asami and Perry.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maki Asami, m.asami@bath.ac.uk; Anthony C. F. Perry, perry135@aol.com
ORCID: Maki Asami, https://orcid.org/0000-0003-3523-0984; Anthony C. F. Perry, https://orcid.org/0000-0003-3136-5355
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.