- 1Department of Bone and Soft Tissue Cancer, The Affiliated Cancer Hospital of Zhengzhou University and Henan Cancer Hospital, Zhengzhou, China
- 2School of Public Health, Zhengzhou University, Zhengzhou, Henan, China
- 3Department of Orthopaedic Oncology Surgery, National Center for Orthopaedics, Beijing Jishuitan Hospital, Capital Medical University, Beijing, China
- 4Musculoskeletal Tumor Center, Peking University People’s Hospital, Beijing, China
Therapeutic resistance is a formidable barrier in cancer treatment, necessitating innovative solutions to enhance drug efficacy. Exosomes, with their unparalleled biocompatibility, low immunogenicity, and robust cargo protection, have emerged as groundbreaking nanocarriers. This review unveils the transformative potential of exosomes in overcoming drug resistance - encompassing chemotherapy, targeted therapy, and immunotherapy - in a wide spectrum of tumors. Through advanced genetic and non-genetic modifications, exosomes can dramatically enhance drug targeting and cytotoxicity, offering unprecedented precision in treatment. We explore state-of-the-art exosome engineering techniques, their revolutionary applications in clinical trials, and their promise as the next Frontier in therapeutic innovation. This comprehensive review aims to capture the cutting-edge developments and future directions of exosome-based therapies, positioning them as a cornerstone of next-generation oncology.
1 Introduction
Cancer therapy resistance continues to pose a significant barrier to improving patient survival, leading to suboptimal responses to chemotherapy, targeted therapy, and immunotherapy across various malignancies (Katz and Shaked, 2015; Tilsed et al., 2022; Gotwals et al., 2017; Chang et al., 2021). Traditional strategies aimed at overcoming resistance, such as combination drug regimens and dose escalation, are often compromised by tumor heterogeneity, adaptive signaling reprogramming, and microenvironment-mediated protection. As a result, there is an urgent need to design delivery systems that can effectively bypass resistance mechanisms while improving therapeutic specificity and efficacy.
Extracellular vesicles (EVs) have emerged as a remarkable class of naturally occurring nanoparticles that play a pivotal role in intercellular communication (van Niel et al., 2018; Namee and O'Driscoll, 2018). EVs are classified into: exosomes (30–150 nm), which originate from the inward budding of endosomal multivesicular bodies and are released upon fusion with the plasma membrane; microvesicles (100–1,000 nm), which are generated through outward budding of the plasma membrane; and apoptotic bodies (500–2000 nm), which are formed during the terminal stages of programmed cell death (van der Pol et al., 2012).
Among these, exosomes demonstrate the most promising characteristics for therapeutic development. In contrast to microvesicles and apoptotic bodies, which exhibit greater heterogeneity and unpredictable immunogenicity, exosomes are distinguished by their superior biostability, inherent tropism mediated by tetraspanins and integrins, and low immunogenicity, rendering them ideal carriers for precision medicine applications (Gurung et al., 2021; Zhang Y. et al., 2020). Their endosomal origin ensures a more homogeneous composition, while their nanoscale size enhances deep tumor penetration through the enhanced permeability and retention. Additionally, their membrane structure can be readily engineered for drug encapsulation, genetic editing payloads, or ligand-based targeting strategies (Zhang et al., 2015; Sadeghi et al., 2023).
While EVs as a broad category offer significant biological and translational insights, engineered exosomes emerge as a promising next-generation tool to address cancer therapy resistance. Their ultra-low immunogenicity, exceptional capacity to cross biological barriers, and significant potential for both autologous and allogeneic applications further enhance their attractiveness for clinical translation.
In this review, we firstly examined the molecular intricacies of chemotherapy resistance, focusing on how engineered exosomes can deliver drugs with enhanced precision, evade drug efflux pumps, and disrupt intracellular resistance mechanisms (Yang E. et al., 2020; Fidelle et al., 2023). Next, we explored the conundrum of targeted therapy resistance, in which engineered exosomes present a novel paradigm. By tailoring their surface ligands and modifying their cargo, these vesicles can target aberrant signaling pathways, effectively reinstating drug sensitivity in resistant tumors (Liang et al., 2021). Precision engineering not only addresses the current limitations of kinase inhibitors and monoclonal antibodies but also opens new avenues for synergistic treatments, combining molecular therapy with biological reprogramming. Engineered exosomes offer a dual approach in immunotherapy, in which resistance often manifests through immune evasion or checkpoint upregulation (Lu et al., 2024; Yue et al., 2023). They can be designed to enhance the recruitment and activation of immune cells while simultaneously delivering molecules that inhibit immunosuppressive pathways, therefore re-establishing immune surveillance. The capacity to manipulate both the immune system and tumor cells positions exosomes a pivotal tool in reinvigorating immune-based cancer treatments.
However, alongside their remarkable potential, significant challenges remain. The scalability of exosome production, the consistency of their engineering, and their long-term biocompatibility in human systems are critical hurdles (Zhu et al., 2020; Tenchov et al., 2022). This review not only addresses these technical and translational barriers but also proposes forward-thinking strategies to refine exosome engineering, optimize their clinical application, and overcome the limitations currently facing their widespread use. By synthesizing cutting-edge discoveries and proposing novel hypotheses, this review provides a comprehensive and forward-looking examination of how engineered exosomes can catalyze the next-generation of cancer therapeutics. With their unique adaptability, these vesicles hold the promise of revolutionizing cancer treatment, offering a dynamic, multi-functional platform capable of outpacing even the most resilient forms of drug resistance.
2 Molecular complexities of chemotherapy resistance
Chemotherapy resistance constitutes a complex, multifactorial, and dynamic process. A comprehensive understanding of the molecular mechanisms underlying this resistance is critical for the development of targeted interventions. This section outlines the key molecular contributors, such as non-coding RNAs, proteins, and lipids, with an emphasis on the major signaling pathways implicated in chemoresistance across various human cancers. Elucidating the operational mechanisms and interconnections of these systems offers vital insights for the design of effective therapeutic strategies, including the engineered exosomes.
2.1 The molecular landscape of chemotherapy resistance
Chemotherapy resistance arises from a complex web of molecular interactions, in which cancer cells evolve to escape the cytotoxic effects of drugs. Central to this are multiple molecular players, including miRNAs, LncRNAs, circRNAs, and proteins (Table 1) (Desterro et al., 2020; Wang et al., 2019; Zhang X. et al., 2020; Eptaminitaki et al., 2022). Previous studies have demonstrated that these molecules interact within the cell’s intricate signaling networks to enhance drug efflux, prevent drug-induced apoptosis, or repair chemotherapy-induced DNA damage (Garofalo and Croce, 2013; Xia et al., 2021; Peng et al., 2020). Overcoming this resistance requires an in-depth understanding of how each molecule contributes to the survival of cancer cells, thus paving the way for novel therapeutic strategies that exploit these vulnerabilities.
2.2 Role of mRNAs in drug metabolism and transport
mRNA-mediated regulation plays a crucial role in chemotherapy resistance primarily through the expression of drug-metabolizing enzymes and efflux transporters (Fabbri et al., 2021; Lin and Kuang, 2024; Shao et al., 2015). Genes encoding multidrug resistance proteins (MRPs) (Mahaffey et al., 2009; Di Giacomo et al., 2019; Noma et al., 2008) and P-glycoprotein (P-gp) (Zhang L. et al., 2022; Shen et al., 2014) are tightly regulated by mRNAs, which, when overexpressed, lead to enhanced drug efflux, reduced intracellular drug concentration, and diminished cytotoxic effects of chemotherapy. The upregulation of mRNAs coding for DNA repair proteins, such as BRCA1 (Zhu et al., 2023; Xu Z. et al., 2020; Qin et al., 2017) and RAD51 (Short et al., 2011; Krumm et al., 2016; Liu et al., 2019), further exacerbates resistance by enabling cancer cells to repair the DNA damage inflicted by chemotherapy agents, such as cisplatin and doxorubicin. Such molecular overexpression forms the backbone of acquired resistance in various cancers, highlighting the necessity of therapies that target the transcriptional regulation of these key players.
2.3 The role of microRNAs as post-transcriptional regulators of chemoresistance
microRNAs (miRNAs) are small, non-coding RNAs that exert profound effects on chemotherapy resistance by post-transcriptionally modulating gene expression during cell survival, apoptosis, and drug metabolism (Rastgoo et al., 2017; Leonetti et al., 2019; Iqbal et al., 2019). For instance, miR-21 targets tumor suppressor genes such as PTEN, thereby promoting the activation of the PI3K/AKT survival pathway, which allows cancer cells to evade drug-induced apoptosis (Kim EH. et al., 2024; Fu et al., 2017; Cao et al., 2017; Chen et al., 2018). Similarly, miR-155 (Pouliot et al., 2012; Van Roosbroeck et al., 2017) and miR-221 (Xu et al., 2019; Fornari et al., 2017; Wang et al., 2016) are known to modulate key resistance pathways, including those related to the expression of apoptosis inhibitors and DNA repair enzymes. miRNAs act as critical switches in therapeutic resistance by fine-tuning the balance between pro- and anti-apoptotic signals, making them prime targets for therapeutic intervention.
2.4 Master regulators of long non-coding RNAs in chemoresistance
Long non-coding RNAs (lncRNAs) have gained increasing attention as master regulators of gene expression, acting through various mechanisms, such as chromatin modification, transcriptional control, and post-transcriptional processing (Singh et al., 2022; Gao et al., 2023; Han M. et al., 2020). lncRNAs, such as HOTAIR (Raju et al., 2023) and MALAT1 (Hou et al., 2023), are often overexpressed in resistant tumors, where they influence a wide array of resistance-associated processes. For instance, HOTAIR promotes epigenetic silencing of tumor suppressor genes through PRC2 recruitment, driving chemotherapy resistance by enhancing cellular survival (Yang E. et al., 2024; Ling et al., 2017). On the other hand, MALAT1 modulates alternative splicing events and promotes the epithelial-mesenchymal transition (EMT), an essential process linked to resistance and metastasis (Gupta et al., 2024; Li et al., 2017; Mao et al., 2021). The versatility of lncRNAs in modulating diverse signaling pathways highlights their central role in chemoresistance, making them attractive candidates for targeted therapies aimed at disrupting resistance networks.
2.5 Circular RNAs sponging miRNAs to sustain resistance
Circular RNAs (circRNAs) function as molecular sponges that sequester miRNAs and prevent them from inhibiting their target mRNAs (Xu X. et al., 2020; Cui et al., 2020; Ma et al., 2020). Overexpression of numerous circRNAs in chemoresistant cancer cells plays a crucial role in the maintenance of oncogenic pathways by inhibiting tumor-suppressive miRNAs. For instance, circHIPK3 enhances resistance by sequestering miR-637 and miR-485-3p, thus hindering its ability to downregulate anti-apoptotic proteins such as Bcl-2 (Zhang et al., 2019; Lai et al., 2020). The circRNA/miRNA/mRNA axis represents a finely tuned regulatory mechanism that cancer cells exploit to escape chemotherapy-induced cell death, offering a new layer of complexity to resistance biology.
2.6 Protein and lipid-mediated mechanisms of chemotherapy resistance
Proteins and lipids serve as essential components of the cellular response network that facilitates tumor cells in evading the cytotoxic effects of chemotherapy. These macromolecules contribute to drug resistance through a variety of interconnected processes, including detoxification, efflux, DNA damage repair, and inhibition of apoptosis.
Among the detoxification systems, glutathione S-transferases (GSTs) play a critical role by catalyzing the conjugation of electrophilic anticancer agents with glutathione. This enzymatic modification enhances drug hydrophilicity, thereby promoting their inactivation and elimination, which ultimately diminishes therapeutic efficacy (Lv et al., 2023; Pljesa-Ercegovac et al., 2018).
The Bcl-2 family of anti-apoptotic proteins, particularly Mcl-1 (Leber et al., 2018; Song et al., 2020), Bcl-2, and Bcl-xL (Lopez et al., 2022; Haselager et al., 2021), are pivotal regulators of the mitochondrial (intrinsic) apoptotic pathway. These proteins inhibit mitochondrial outer membrane permeabilization (MOMP) by sequestering pro-apoptotic members like Bax and Bak, thus preventing cytochrome c release and subsequent caspase activation. Their upregulation enables cancer cells to survive under genotoxic stress, rendering them resistant to apoptosis-inducing chemotherapeutics.
Membrane lipids, especially phosphatidylserine (PS) and sphingolipids, also contribute to chemoresistance by modulating plasma membrane composition, fluidity, and curvature. Changes in lipid raft structure influence the activity of membrane-bound proteins, including drug efflux transporters and death receptors. Notably, PS externalization has been linked to immune evasion and decreased drug uptake (Rajeev Krishnan et al., 2020; Chen Y. et al., 2024).
A major protein-mediated resistance mechanism involves the ATP-binding cassette transporter family, including ABCB1, MRP1/ABCC1, and BCRP/ABCG2 (Zhang W. et al., 2025; Mohammad et al., 2018). These transmembrane proteins harness ATP hydrolysis to actively extrude a wide spectrum of chemotherapeutic agents from the cell, thereby reducing intracellular drug concentrations and diminishing cytotoxic effects. Their expression is frequently modulated by transcriptional programs downstream of NF-κB (Walther et al., 2015; Li et al., 2019), which is commonly activated within the tumor microenvironment.
Concurrently, enhanced DNA damage repair mechanisms contribute to survival following chemotherapy-induced genotoxic stress. Tumor cells can upregulate critical DNA repair effectors such as ATM, ATR, and BRCA1/2, enabling efficient repair of DSBs and crosslinks induced by agents like platinum compounds and topoisomerase inhibitors (Brownlie et al., 2023). This elevated repair capacity compromises drug efficacy and fosters resistance, particularly in pancreas cancer (Crowley et al., 2021) and triple-negative breast cancer (Geenen et al., 2018). In addition to these classical repair pathways, translesion synthesis (TLS) represents an error-prone DNA damage tolerance mechanism (Shilkin et al., 2020a). This process is mediated by specialized low-fidelity DNA polymerases, which enable the continuation of DNA replication at sites of chemotherapy-induced damage, thereby preventing replication fork stalling, but at the expense of an elevated mutation rate (Patel et al., 2021; Shilkin et al., 2020b). By inducing genomic instability, TLS contributes to the survival of tumor cells, such as colorectal tumor cells (Das et al., 2024) and prostate tumor cells (Williams et al., 2024), under the selective pressure of drug treatment, ultimately promoting the development of chemotherapy resistance.
Lastly, evasion of apoptosis remains a defining characteristic of resistant tumors. Downregulation or functional inhibition of pro-apoptotic proteins such as Bax, Bak, and Caspase-3, coupled with persistent activation of survival signaling pathways - including PI3K/AKT/mTOR, MAPK/ERK, and NF-κB - collectively shift cellular fate toward survival rather than death (Li et al., 2025; Mok et al., 2022). These pathways also enhance the expression of effector proteins involved in metabolism and transcription, further consolidating the resistant phenotype.
Together, the protein- and lipid-mediated mechanisms do not operate independently but instead form an interdependent cooperative resistance network that supports tumor survival under therapeutic pressure. Consequently, therapeutic strategies aimed at reversing resistance must target this multifactorial framework, either by employing combinatorial regimens or engineered exosomes delivery platforms.
2.7 Epigenetic modulation in chemotherapy resistance
Epigenetic alterations - heritable changes in gene expression that occur without modifications to the DNA sequence - play a pivotal role in the development and maintenance of chemotherapy resistance across a wide range of malignancies. These epigenetic modifications, which encompass DNA methylation, histone tail modifications, and chromatin remodeling, dynamically regulate gene accessibility and transcriptional activity, thereby influencing tumor cell phenotype, survival, and drug responsiveness.
Aberrant DNA methylation is one of the most prevalent epigenetic mechanisms implicated in resistance. Hypermethylation of CpG islands within the promoter regions of tumor suppressor genes such as PTEN (Maeda et al., 2015), MLH1 (Geissler et al., 2024), and CDKN2A (Wang et al., 2024) leads to their transcriptional silencing, thereby promoting cell survival, impairing DNA damage response, and facilitating evasion of apoptosis (Ogino et al., 2007; Hiraki et al., 2010).
Histone modifications, including acetylation, methylation, phosphorylation, and ubiquitination of histone H3 and H4 tails, further modulate chromatin architecture and transcriptional activation. Histone deacetylation induces chromatin condensation and gene silencing, which can downregulate DNA repair inhibitors or pro-apoptotic mediators, supporting a resistant phenotype (Yang WB. et al., 2020; Bhaskara, 2015). Conversely, aberrant histone methylation, as H3K27me3 catalyzed by EZH2, is associated with transcriptional activation, a process linked to multidrug resistance (Marsolier et al., 2022; Yamagishi et al., 2024; Xiong et al., 2022).
Overall, the plasticity of the epigenome enables cancer cells to adapt to therapeutic stress and develop drug-tolerant states without necessitating genetic mutations. Consequently, targeting epigenetic regulators represents a compelling strategy for overcoming resistance and achieving sustained responses when combined with chemotherapy or advanced engineered exosomes delivery systems.
3 Unique biological features of exosomes in tumor therapy
3.1 Mimicking natural communication of exosome surface signatures
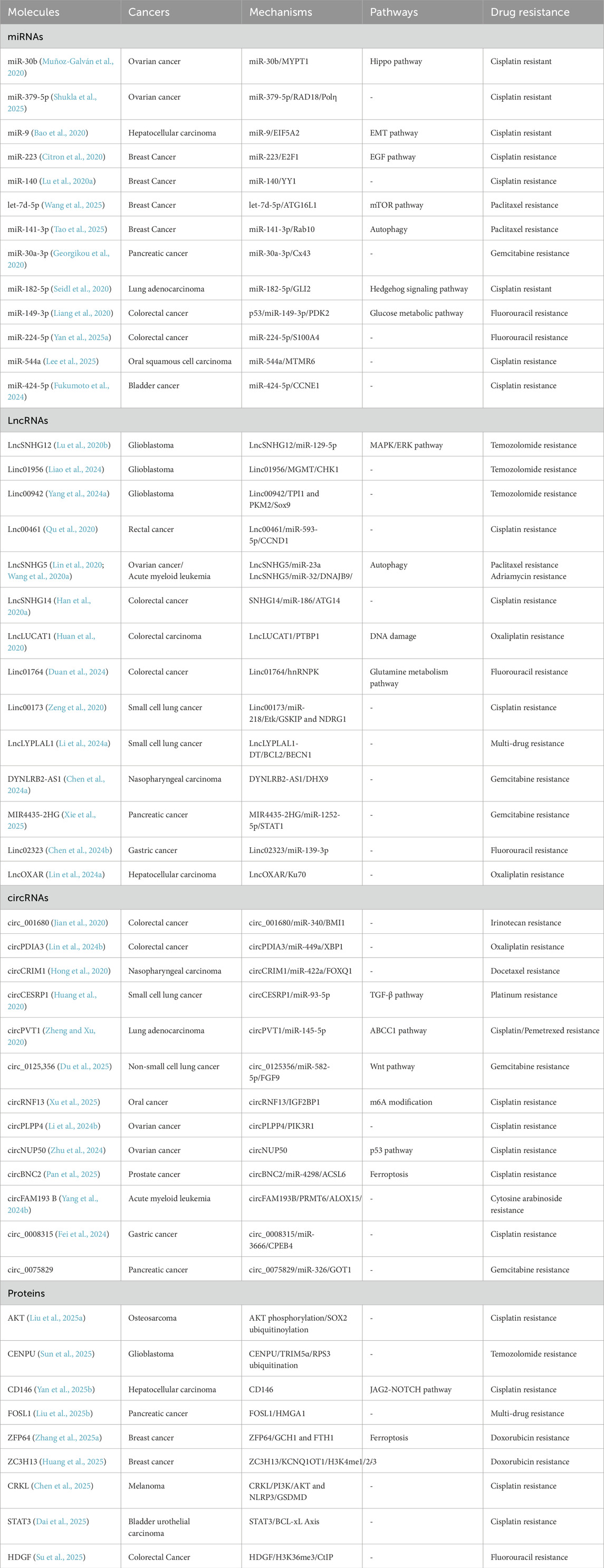
Unlike synthetic nanoparticles, exosomes are naturally derived vesicles that retain the surface signatures of their parent cells (Lai et al., 2022). This includes a wide array of surface proteins, such as tetraspanins (CD63, CD81), integrins, and major histocompatibility complex (MHC) molecules, which facilitate their interaction with target cells (Figure 1) (Salunkhe et al., 2020).
Figure 1. Biological characteristics of exosomes. Key surface proteins, including CD9, CD63, CD81, and CD82, are illustrated for their roles in maintaining membrane protein stability, facilitating oligomerization, and modulating immune responses. Intra-vesicular proteins such as ALIX, TSG101, HSP90, and various molecular chaperones are involved in cargo sorting and the biogenesis of exosomes. The exosomal cargo, which encompasses a diverse array of proteins, phospholipids, and nucleic acids (DNA and RNA), plays a pivotal role in mediating intercellular communication. The illustration further delineates the biological functions of exosomes, such as immune modulation, regulation of membrane proteins, mediation of cargo transfer, and their contributions to tumor microenvironment heterogeneity and exosome trafficking.
The presence of these proteins allows exosomes to engage in natural cellular communication processes, including receptor-mediated uptake and fusion with target cells (Kalluri and LeBleu, 2020; Pathania et al., 2021). This biocompatibility and ability to mimic natural cell-to-cell interactions confer exosomes with a distinct advantage in drug delivery, as they can evade immune detection and circulate for extended periods, ensuring more efficient delivery of therapeutic cargo (Figure 2) (Fu et al., 2021; Farooqi et al., 2018).

Figure 2. Biological properties of exosomes in tumor therapy. The endosomal pathway of exosome biogenesis begins with endocytosis, progresses through early and late endosomal stages, and culminates in the formation of multivesicular bodies (MVBs). These MVBs subsequently fuse with the plasma membrane, thereby releasing exosomes—nanoscale vesicles ranging from 30 to 150 nm in diameter—into the extracellular environment. This process is closely associated with several key features of exosomes that are pertinent to tumor biology, including their low immunogenicity, capacity to infiltrate hypoxic tumor microenvironments, and inherent targeting abilities. Additionally, the figure illustrates surface protein markers such as CD63, CD81, TSG101, integrins, and MHC, which play crucial roles in mediating intercellular communication and immune evasion. Moreover, functional domains involved in tumor microenvironment adaptation, drug delivery, and surface recognition are emphasized, underscoring the multifaceted utility of exosomes in both physiological and therapeutic contexts.
3.2 Tumor navigation and microenvironment penetration
The ability of exosomes to navigate complex tumor microenvironments is another critical feature that sets them apart from conventional nanoparticles. Tumors are characterized by dense extracellular matrices and hypoxic regions that create physical barriers to drug delivery (Yang M. et al., 2021). Due to their small size and flexible lipid bilayer, exosomes can penetrate these biological barriers (He et al., 2022; Li and Nabet, 2019). Furthermore, exosomes can be engineered to display peptides or proteins that enhance their ability to navigate these hostile environments, such as integrin-mediated adhesion molecules that enable them to home in on specific tumor sites (Qiao et al., 2020). In a study using exosomes to deliver small interfering RNAs (siRNAs) targeting KRAS in pancreatic cancer, researchers found that the exosomes penetrated deeply into the tumor core, significantly suppressing tumor growth (Kamerkar et al., 2017). This natural ability to traverse challenging tumor environments enhances the therapeutic potential of exosome-based delivery systems.
3.3 Enhancing drug delivery efficiency through natural targeting mechanisms
Exosomes also possess intrinsic targeting capabilities that can be harnessed to improve the efficiency of drug delivery (Meng et al., 2020). They contain specific receptors that facilitate the selective uptake of exosomes, enabling precise cargo delivery. For instance, exosomes derived from dendritic cells or macrophages can be engineered to express ligands that target receptors overexpressed on tumor cells, such as LFA-1 or ICAM-1, further improving the specificity of drug delivery (Guo and Jiang, 2020; Zhang W. et al., 2022). Compared with synthetic nanoparticles, which often rely on passive targeting mechanisms, exosomes can actively engage with specific cellular receptors, improving drug delivery efficiency while minimizing off-target effects (Liang et al., 2023).
4 Engineered exosomes for precision drug delivery and mechanism disruption
4.1 Engineering exosomes for targeted delivery
The specificity of engineered exosomes lies in their ability to be precisely tailored for targeted drug delivery (Tran et al., 2020; Gao et al., 2018). By functionalizing the exosome surface with ligands that bind to receptors overexpressed on cancer cells, such as EGFR, HER2, or PSMA, these vesicles can home in on tumor sites with remarkable accuracy (Jiang et al., 2024; Kim R. et al., 2024; Cheng et al., 2023). This targeted approach significantly reduces off-target effects and enhances the therapeutic index of the chemotherapeutic agents encapsulated within exosomes. Previous studies have demonstrated the efficacy of engineered exosomes engineered to carry doxorubicin directly to tumor cells, bypassing drug efflux pumps and delivering their cytotoxic payload (Liu et al., 2023; Zhu S. et al., 2022). These precision delivery systems have immense potential for overcoming the specific challenges of conventional nanoparticles.
4.2 Evading drug efflux pumps
One of the primary mechanisms by which cancer cells acquire multidrug resistance is through the overexpression of ATP-binding cassette (ABC) transporters, such as P-glycoprotein (P-gp/ABCB1), multidrug resistance-associated proteins (MRPs), and breast cancer resistance protein (BCRP/ABCG2) (Li et al., 2015; To et al., 2024). These transmembrane efflux pumps utilize ATP hydrolysis to actively expel chemotherapeutic agents from the intracellular milieu into the extracellular environment, thereby reducing intracellular drug concentrations below cytotoxic thresholds and compromising therapeutic efficacy. Notably, P-gp is highly expressed in various resistant tumor types and exhibits broad substrate specificity, encompassing taxanes, anthracyclines, and vinca alkaloids (Guo et al., 2024; Das et al., 2021).
Engineered exosomes possess several strategic advantages for evading recognition and export by these efflux systems (Dad et al., 2021; Chen et al., 2024d). First, the phospholipid bilayer of exosomes encapsulates their cargo - whether small molecule drugs, nucleic acids, or proteins - thereby providing a steric and structural barrier that prevents direct interaction between the therapeutic agent and membrane-bound drug efflux pumps during early cellular uptake. This cloaking mechanism ensures that the drug remains unrecognized as a substrate by efflux proteins at the initial stages of cell entry.
Second, engineered exosomes can be designed to encapsulate functional siRNAs, shRNAs, or miRNAs (Zhan et al., 2022; Shan et al., 2022) that specifically downregulate the expression of key efflux pump genes such as ABCB1, ABCC1, and ABCG2. For instance, the exosome-mediated delivery of anti-ABCB1 siRNA has been demonstrated to effectively re-sensitize drug-resistant tumor cells to standard chemotherapy regimens (Wang C. et al., 2020). This gene-silencing approach targets the fundamental cause of resistance by inhibiting the biosynthesis of efflux transporters. In resistant ovarian cancer models, exosomes engineered to deliver both paclitaxel and anti-MDR1 siRNA successfully increased drug retention and cytotoxicity, providing proof-of-concept for this dual-delivery approach (Wang C. et al., 2020).
Furthermore, exosomes exhibit intrinsic targeting capabilities through the expression of tetraspanins, integrins, and engineered surface ligands, enabling precise delivery of their cargo to specific subcellular compartments, including the perinuclear region, endoplasmic reticulum, or lysosomes (Zhu et al., 2019). These microenvironments are typically inaccessible to plasma membrane-localized drug pumps, thus providing an additional mechanism for spatial avoidance and enhancing the accumulation of cytotoxic payloads in regions where they exert maximal efficacy.
Collectively, these evasion strategies empower engineered exosomes to circumvent or functionally neutralize one of the most significant barriers to chemotherapy success: the drug efflux pump system. By integrating physical shielding, intracellular trafficking, gene silencing, and spatial targeting, exosomes serve as both stealth carriers and active modulators of drug resistance networks, offering a robust platform for restoring chemosensitivity in refractory malignancies.
4.3 Disrupting intracellular resistance mechanisms
The intracellular landscape of resistant cancer cells is fraught with molecular defenses that are designed to withstand chemotherapy-induced stress. Engineered exosomes can be harnessed to disrupt these defenses by delivering molecules that target the key resistance pathways. For instance, exosomes loaded with Bcl-2 inhibitors can re-sensitize cancer cells to apoptosis by negating the anti-apoptotic effects of Bcl-2 proteins (Tao et al., 2020). Similarly, exosomes can be engineered to deliver PARP inhibitors, which disrupt DNA repair pathways in cells deficient in homologous recombination, thereby inducing synthetic lethality in resistant tumors (Jung et al., 2018). Furthermore, the delivery of CRISPR/Cas9 systems via exosomes allows for precise gene editing, enabling the knockout of resistance-associated genes such as PARP1 in drug-resistant cancers (Kim et al., 2017). These strategies not only enhance the cytotoxicity of chemotherapy but also address the root molecular causes of resistance, offering a multi-pronged approach to treatment.
The molecular complexities of chemotherapy resistance, driven by the intricate interplay of coding and non-coding RNAs, proteins, and lipids, present significant challenges for effective cancer treatment. Engineered exosomes offer a promising solution by providing precision drug delivery, evading drug efflux pumps, and disrupting key intracellular resistance mechanisms (Figure 3). Their unique biological features, including their natural surface signatures and ability to navigate tumor microenvironments, further enhance their therapeutic potential. As research into exosome engineering continues to advance, these vesicles hold great promise for overcoming chemotherapy resistance and improving cancer therapeutic outcomes.

Figure 3. Mechanisms of engineered exosome targeting to overcome chemotherapy resistance. Subfigure (A) illustrates the targeted delivery of therapeutic agents via ligand-modified exosomes. Subfigure (B) demonstrates the capacity of exosomes to bypass drug efflux pumps, thereby enhancing intracellular drug accumulation. Subfigure (C) showcases exosomes loaded with a variety of therapeutic cargoes—including Bcl-2 inhibitors, PARP inhibitors, CRISPR/Cas9 systems, miRNA, circular RNA, and long non-coding RNA, which work synergistically to modulate drug resistance mechanisms. The central figure synthesizes these elements, depicting how engineered exosomes can overcome both intrinsic and acquired drug resistance by targeting key pathways and molecules associated with resistance within tumor cells.
5 Engineered exosomes-a breakthrough in overcoming targeted therapy resistance
Targeted therapies designed to inhibit specific oncogenic pathways, such as RTKs or the PI3K/AKT/mTOR axis, often encounter resistance due to downstream signaling components mutations or compensatory activation of alternative pathways. Engineered exosomes, with their inherent biological compatibility and ability to be tailored for precise therapeutic intervention, provide a novel solution for these resistance mechanisms. This section explores how exogenous surface ligand modifications and endogenous cargo alterations allow for the reinstatement of drug sensitivity in resistant tumors, opening new avenues for synergistic treatments while addressing current therapeutic bottlenecks.
5.1 Surface ligand modifications of exogenous exosomes for precision targeting of resistant tumors
Targeted therapies, such as monoclonal antibodies or tyrosine kinase inhibitors (TKIs), are often undermined by developing resistance mechanisms such as secondary mutations in RTKs (EGFR and HER2) or increased expression of alternative receptors such as MET and AXL (Kim et al., 2017; Fu et al., 2022). Engineered exosomes can be precisely modified on their surface to carry ligands that selectively bind to receptors overexpressed on resistant cancer cells. This ensures that the therapeutic payloads are delivered with high specificity and efficacy. For instance, anti-EGFR antibodies can be conjugated to exosome surfaces to target EGFR-mutated cancers that have developed resistance to TKIs (Toh et al., 2024). These exosomes, carrying both targeting antibodies and therapeutic cargo, can bypass the resistance mechanism by directly delivering their payload to resistant cancer cells. Similarly, HER2-targeting aptamers have been used to modify exosomes for enhanced delivery to HER2-positive breast cancer cells, overcoming resistance to HER2 inhibitors such as trastuzumab (Tao et al., 2023).
Moreover, using cell-penetrating peptides such as TAT or iRGD on exosome surfaces significantly improves their ability to penetrate deeply into solid tumors. These peptides facilitate exosome uptake by resistant cancer cells, thereby further enhancing the precision of drug delivery (Zhu Z. et al., 2022; Gan et al., 2024). For instance, iRGD-modified exosomes have demonstrated superior targeting of metastatic colon cancer, where traditional therapies have failed (Lin et al., 2021). Precision targeting through surface ligand modification not only re-sensitizes resistant tumors but also minimizes off-target effects, which are often a limiting factor in conventional targeted therapies.
5.2 Disrupting resistance pathways of endogenous cargo modifications
While surface modifications enable exosomes to selectively target resistant cancer cells, the actual therapeutic potential lies in their ability to carry and deliver diverse types of cargo that disrupt the underlying resistance mechanisms. By loading exosomes with siRNAs, miRNAs, or CRISPR/Cas9 gene-editing tools, cancer cells can be reprogrammed at the molecular level, restoring their sensitivity to targeted therapies. One prominent example is the use of exosomes loaded with siRNAs targeting MEK or ERK. In cancers where mutations in the MAPK pathway confer resistance to BRAF inhibitors, such as melanoma, these exosome-encapsulated siRNAs can silence MEK or ERK, thereby inhibiting compensatory survival signaling and restoring the efficacy of BRAF-targeted therapies (Vella et al., 2017). Moreover, exosomes loaded with miRNAs that downregulate the PI3K/AKT/mTOR pathway, a common escape route in numerous resistant cancers, have been exhibited to reverse resistance to PI3K inhibitors (Zhao et al., 2020; Yang C. et al., 2021; Xue et al., 2024).
Another powerful application of engineered exosomes is their ability to deliver CRISPR/Cas9 systems, which can edit genes responsible for resistance. For instance, in glioblastoma, exosomes delivering CRISPR/Cas9 tools targeting mutant KRAS G12C have demonstrated the ability to knock out the mutant allele, reinstating the tumor’s temozolomide resistance (Zhao et al., 2024). This cargo flexibility offers a significant advantage over traditional drug delivery methods, which are often constrained by poor intracellular uptake or rapid drug degradation. In contrast, exosomes can deliver their cargo directly into the cytoplasm, bypassing endosomal degradation and ensuring efficient gene silencing or protein inhibition. The precise delivery of customized cargo enables engineered exosomes to tackle molecular circuits that enable cancer cells to evade targeted therapies, offering a robust solution to multidimensional resistance mechanisms.
While exogenous engineering enables direct and multifunctional functionalization of purified exosomes, endogenous engineering provides superior biocompatibility and preserves vesicle integrity. However, the latter is often constrained by lower cargo loading efficiency and cell type dependence. Future applications could potentially leverage a hybrid strategy that integrates the strengths of both methodologies.
5.3 Exosome-mimicking nanoplatforms - a comparative evaluation
While natural exosomes possess distinct advantages in terms of biocompatibility, biological targeting, and functional cargo delivery, challenges such as scalability and loading consistency have driven the development of EV mimetics. These synthetic or semi-synthetic vesicles are designed to replicate the structural and functional features of natural exosomes while offering enhanced control over composition and manufacturing processes.
EV mimetics encompass platforms such as liposomes, polymeric nanovesicles, cell membrane-coated nanoparticles, and extruded nanovesicles (Du et al., 2022). While some mimetics, particularly membrane-coated particles, can preserve surface proteins for homing capability, others primarily serve as physicochemical models that lack the endogenous signaling capacity of exosomes (Gangadaran and Ahn, 2020). For instance, lipid-based mimetics like PEGylated liposomes may exhibit improved circulatory stability but often fail to fully recapitulate the complex surface architecture and cargo fidelity of native exosomes (Wei et al., 2023; Kooijmans et al., 2016).
Compared with engineered exosomes, EV mimetics present several limitations (Kim et al., 2020). First, they generally lack the complete repertoire of membrane proteins, adhesion molecules, and endogenous ligands required for natural biodistribution and cell-specific uptake. Second, mimetics do not reproduce the functional RNA and protein signatures of native exosomes, restricting their role in dynamic cell signaling and therapeutic reprogramming. Additionally, certain EV mimetic systems have demonstrated higher immunogenicity and reduced in vivo stability, raising concerns regarding long-term safety and clinical translation. Although their production is more standardized, this standardization comes at the expense of diminished biological function. In contrast, engineered exosomes retain the complete spectrum of native exosomal functions, such as precise intercellular communication, tissue-specific tropism, and immunologic tolerance. Their capacity to incorporate functional RNAs, signaling proteins, and membrane ligands renders them inherently advantageous as carriers for bioactive therapeutics. With advancements in both endogenous and exogenous engineering methodologies, exosomes can now be tailored for enhanced targeting specificity, cargo enrichment, and immune modulation while preserving their intrinsic biological advantages.
Consequently, although EV mimetics may serve as temporary alternatives in contexts where standardization is paramount, their applications remain largely complementary. Naturally engineered exosomes provide a more extensive and promising application scope, especially within complex biological systems requiring native bio-functionality and therapeutic adaptability. Ongoing innovations in exosome bioengineering are expected to establish them as leading candidates for next-generation targeted therapy platforms.
5.4 Translational challenges and opportunities of engineered exosomes
Although engineered exosomes offer substantial promise in overcoming resistance to targeted therapy, several challenges must be addressed before their full potential can be realized in clinical settings. One of the bottlenecks is their inability to address tumor heterogeneity, the presence of diverse cell populations within a tumor that may exhibit different resistance mechanisms (Labrie et al., 2022). Engineered exosomes have the potential to overcome this hurdle by delivering multiple therapeutic agents simultaneously, enabling a multi-pronged attack on resistant tumors. For example, exosomes loaded with both KRAS G12D siRNAs and TP53 mRNA, can simultaneously suppress multiple resistance pathways in cancers such as pancreatic cancer (Chiang et al., 2023). This versatility allows exosomes to address not only primary resistance mechanisms but also compensatory pathways that may arise following treatment.
Another major obstacle is the scalability of exosome production (Rehman et al., 2023). Current methods of exosome isolation, such as ultracentrifugation or size-exclusion chromatography, are labor-intensive and not easily scalable for clinical-grade production (Kim et al., 2022). Automated microfluidic systems that allow for the continuous production and isolation of exosomes are being developed as a solution to this bottleneck (Zhao et al., 2022; Zhao et al., 2019).
Additionally, variability in exosome content depending on the cell source and isolation technique raises concerns regarding consistency and standardization. Another challenge is the potential immunogenicity of repeated exosome administrations (Herrmann et al., 2021). Although exosomes occur naturally, their engineered variants may elicit immune responses over time. To mitigate this, researchers have investigated the use of immunologically inert exosomes derived from hypoimmunogenic cells or artificial lipid vesicles that mimic exosomal structures. Advances in synthetic biology have also offered the possibility of creating “immune-quiet” exosomes that can circulate in the bloodstream without triggering an immune response (Tan et al., 2024; Lin et al., 2022).
In conclusion, the combination of surface ligands and cargo modifications in engineered exosomes presents a powerful new approach to overcoming the limitations of targeted therapy. While challenges remain, ongoing innovations in exosome engineering, production scalability, and safety optimization have paved the way for their clinical translation. By employing the unique properties of exosomes, such as their precise targeting, deep tumor penetration, and cargo versatility, future cancer therapies could see a paradigm shift, with engineered exosomes at the forefront of personalized and effective cancer treatment strategies.
6 Revolutionizing immune surveillance of engineered exosomes in immunotherapy resistance
Immunotherapy offers the potential to harness the immune system of the body to combat cancer. However, numerous tumors develop resistance to immunotherapy through immune evasion mechanisms and the upregulation of immune checkpoints, such as PD-L1 or CTLA-4, which hinder the immune system’s ability to recognize and destroy tumor cells (Peng et al., 2019; Menzies et al., 2022; Pardoll, 2012). This challenge necessitates innovative solutions, and engineered exosomes have emerged as a promising platform. These nanovesicles offer a dual approach by enhancing the recruitment and activation of immune cells while simultaneously delivering molecules to inhibit immunosuppressive pathways, re-establishing immune surveillance. In this section, we investigate the potential of engineered exosomes to reinvigorate immune-based cancer treatments. We explored their role in enhancing immune cell recruitment and activation, delivering immunosuppressive pathway inhibitors, and manipulating both immune and tumor cells to overcome resistance while addressing the challenges and solutions in their clinical application (Figure 4).
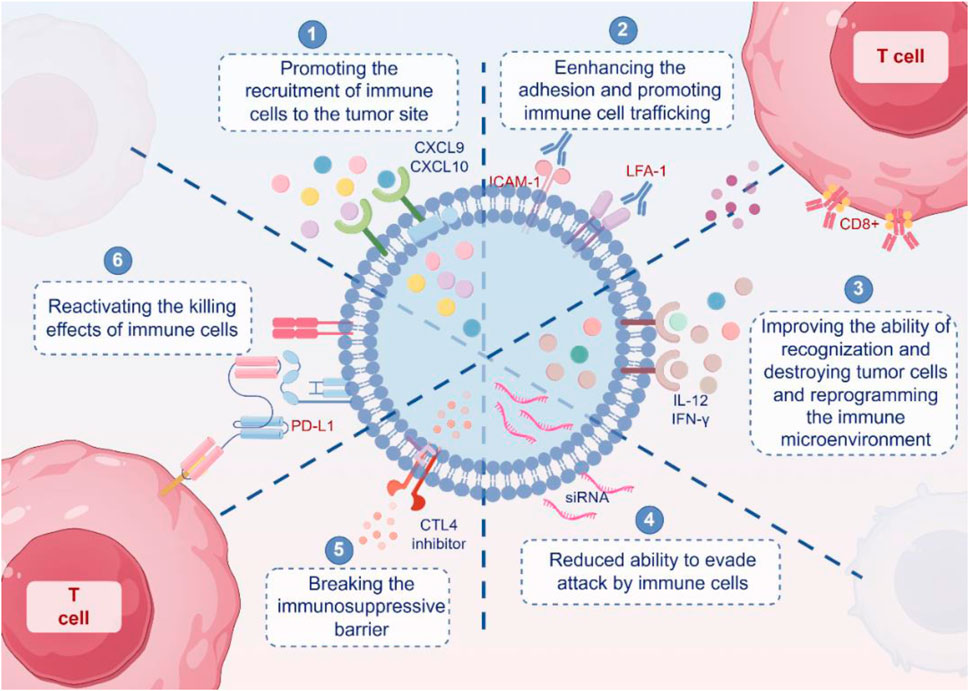
Figure 4. Mechanism of engineered exosomes reshaping immunotherapy. Engineered exosomes enhance anti-tumor immunity through multiple mechanisms. First, exosomes carrying chemokines such as CXCL9 and CXCL10 promote the recruitment of immune cells to tumor sites. Second, exosomes expressing adhesion molecules such as ICAM-1 and LFA-1 facilitate the migration and infiltration of immune cells into the tumor microenvironment. Third, exosomes containing cytokines such as IL-12 and IFN-γ enhance immune cell activation and recognition, while also modulating the tumor microenvironment. Fourth, engineered exosomes can reduce tumor immune escape by inhibiting immunosuppressive signaling pathways. Fifth, exosomes can serve as delivery vehicles for checkpoint inhibitors, such as CTLA-4 and PD-L1 siRNA, thereby helping to overcome immune tolerance. Sixth, immune-stimulatory exosomes can reactivate cytotoxic T lymphocytes and restore their tumor-killing capacity. Collectively, these findings illustrate how exosome engineering can reshape anti-tumor immune responses and counteract immune evasion mechanisms.
6.1 Roles of engineered exosomes in enhancing immune cell recruitment and activation
A key aspect of immunotherapy involves enhancing the recruitment of immune cells, specifically T cells, dendritic cells, and NK cells, to the tumor microenvironment (Kubli et al., 2021; Galon and Bruni, 2019). Tumor resistance to immunotherapy often results from immune cell exclusion, in which tumors create an immunosuppressive microenvironment that impairs immune cell infiltration and activation (O'Donnell et al., 2019; Shimasaki et al., 2020). Engineered exosomes offer an innovative method for circumventing these barriers by being tailored to actively recruit and stimulate immune cells.
The design of exosomes that express surface ligands or chemokines that attract immune cells is a crucial breakthrough. Exosomes can be engineered to display ligands, such as ICAM-1 or LFA-1, which facilitate the recruitment of T cells by enhancing the adhesion and promoting immune cell trafficking to tumor sites (Nolte-'t Hoen et al., 2009; Segura et al., 2005). Additionally, exosomes can carry CXCL9 and CXCL10 chemokines, which are known for their role in attracting cytotoxic T cells, thereby amplifying the immune response against tumors (Zhang LS. et al., 2024). This targeted approach not only improves immune cell presence in the tumor microenvironment but also ensures that activated T cells reach and infiltrate the tumor mass, overcoming immune exclusion.
Another innovative approach is the delivery of immune-stimulating cytokines via exosomes. For instance, exosomes containing IL-12 or IFN-γ can facilitate the activation and proliferation of CD8+ cytotoxic T cells, thereby improving their capacity to identify and eliminate tumor cells (Zhang et al., 2010). These cytokine-loaded exosomes can induce a shift from an immunosuppressive to an immunostimulatory tumor microenvironment, reprogramming the immune landscape in favor of effective tumor eradication. Furthermore, exosomes carrying GM-CSF (granulocyte-macrophage colony-stimulating factor) can enhance dendritic cell activation, improving antigen presentation and boosting T-cell activation (Dai et al., 2008; Yaddanapudi et al., 2019).
This capacity of engineered exosomes to manipulate immune cell recruitment and activation forms the foundation of their role in overcoming immune resistance mechanisms. Exosomes re-establish an immune-permissive environment that supports long-term anti-tumor immunity by actively recruiting immune cells and ensuring their sustained activation.
6.2 Functions of engineered exosomes in rebuilding immune surveillance and inhibiting immune-suppressive pathways
While enhancing immune cell recruitment is crucial, the reactivation of immune surveillance within the tumor microenvironment depends heavily on inhibiting the immunosuppressive pathways that tumors use to escape detection. Immune checkpoints such as PD-1/PD-L1 and CTLA-4 serve as critical regulators of immune tolerance, and their upregulation in tumors leads to T-cell exhaustion and impaired immune responses (Dammeijer et al., 2020; Li Y. et al., 2024). Engineered exosomes provide a unique platform for delivering checkpoint inhibitors directly into the tumor microenvironment, thus overcoming these immune evasion mechanisms. Exosomes can be loaded with anti-PD-L1 antibodies, enabling the blockade of PD-L1 on tumor cells and the reactivation of T cell function (Poggio et al., 2019). Besides, exosomes engineered to carry CTLA-4 inhibitors can enhance the activity of regulatory T cells, shifting the balance from immune suppression to immune activation (Shi et al., 2020; Zocchi et al., 2020). Furthermore, exosomes can be designed to carry siRNAs that target immunosuppressive pathways at the gene expression level. For instance, exosomes loaded with siRNAs targeting PD-L1 mRNA can reduce PD-L1 expression on the surface of tumor cells, thereby diminishing their ability to evade T cell attack (Zhong et al., 2023).
Restoring immune surveillance through exosomal delivery of checkpoint inhibitors and gene silencing marks a significant advancement in overcoming the limitations of current immunotherapy methods. By specifically targeting the mechanisms involved in immune evasion, engineered exosomes not only increase the effectiveness of existing immune checkpoint therapies but also minimize systemic toxicity and enhance immune system activation within the tumor microenvironment.
6.3 Dual manipulation of immune and tumor cells
The true power of engineered exosomes lies in their ability to simultaneously manipulate both the immune system and tumor cells, creating a synergistic effect that reactivates immune-based cancer treatments. This dual approach not only enhances immune cell function but also directly influences tumor cell behavior, allowing for a comprehensive therapeutic strategy that tackles both sides of the immune evasion coin.
On the immune side, exosomes can be engineered to carry T cell-activating ligands such as OX40 L or 4-1BBL, which boost the proliferation and activation of effector T cells (Xie et al., 2013). This strategy enhances the immune response by providing additional co-stimulatory signals that overcome the exhaustion typically observed in tumor-infiltrating lymphocytes. The combination of T-cell activation and checkpoint inhibition creates a robust immune assault on the tumor, making it difficult for cancer cells to develop further resistance.
Simultaneously, engineered exosomes can deliver tumor-targeting molecules, such as TNF-related apoptosis-inducing ligand (TRAIL), which induces apoptosis in cancer cells without affecting healthy tissue (Zhang X. et al., 2024). This selective killing of tumor cells helps reduce the tumor burden while maintaining immune pressure on the remaining cancer cells. The delivery of apoptotic agents through exosomes is particularly advantageous because exosomes can evade endosomal degradation, ensuring efficient delivery to the cytoplasm of the target cells.
Despite these promising advantages, several challenges must be addressed to fully realize the potential of exosomes in immunotherapy. One major hurdle is the complexity of the tumor microenvironment, which varies widely between patients and even within different regions of the same tumor. Tumors can evolve mechanisms to resist exosome-mediated therapies, such as altering the expression of receptors required for exosome uptake. To overcome this, future exosome-based therapies may require personalized engineering, where exosomes are designed based on a patient’s specific tumor biology, ensuring that they target the most relevant immune-suppressive pathways. Scaling up the production of engineered exosomes for clinical use remains technically challenging. Although exosomes are highly biocompatible, the production process is labor-intensive and requires advanced technologies such as ultrafiltration or tangential flow filtration. Innovations in biomanufacturing, including synthetic exosomes and automated exosome production platforms, are being explored to streamline this process and make large-scale exosome therapies feasible.
In conclusion, engineered exosomes represent a groundbreaking tool for reinvigorating immune-based cancer treatment. By enhancing immune cell recruitment, inhibiting immunosuppressive pathways, and manipulating both immune and tumor cells, exosomes offer a comprehensive solution to overcome immunotherapy resistance. While challenges remain, ongoing advancements in exosome engineering, production, and personalized therapy design hold significant promise for the future of cancer immunotherapy.
7 Clinical applications and translational advances of exosomes
The clinical translation of extracellular vesicles, particularly exosomes, has gained significant momentum in recent years. Due to their biocompatibility, low immunogenicity, and intrinsic targeting capacity, exosomes have been extensively investigated in various therapeutic and diagnostic applications across oncology, neurology, and inflammatory diseases.
According to ClinicalTrials.gov, over 200 clinical trials have been registered to evaluate the safety, efficacy, and feasibility of exosome-based strategies (Table 2). For instance, exosomes derived from mesenchymal stem cells (MSCs) are being studied and have been tested in conditions ranging from glioblastoma (e.g., NCT03608631) to pancreatic cancer (NCT02530047). Additionally, tumor-derived exosomes are being evaluated as biomarkers for liquid biopsy and as platforms for cancer vaccines.
In the context of cancer drug resistance, the delivery of small interfering RNAs (siRNAs) and chemotherapeutic agents via exosomes is being explored to overcome resistance pathways. For example, exosome-encapsulated formulations of paclitaxel and doxorubicin have demonstrated enhanced tumor accumulation and efficacy in preclinical models, and early-phase clinical trials are ongoing to assess their translational potential in resistant breast and ovarian cancers.
Despite these advances, several challenges remain unresolved. These include the lack of standardized GMP-grade production protocols, batch-to-batch variability, and regulatory ambiguity regarding exosome classification - as biologics, drugs, or combination products. In contrast, while EV mimetics are easier to manufacture at scale, they lack the complex surface signaling and immune interactions necessary for full biological mimicry and have yet to demonstrate comparable clinical potential.
Overall, engineered exosomes provide a unique combination of therapeutic versatility and clinical relevance that distinguishes them from other nanocarriers. The expanding body of clinical trial data highlights their potential; however, further advancements in isolation standardization, long-term safety evaluation, and regulatory harmonization will be critical for achieving widespread clinical adoption.
8 Conclusion
Engineered exosomes have emerged as transformative agents in combating therapy resistance in cancer, offering a multifaceted approach to enhance the effectiveness of chemotherapy, targeted therapy, and immunotherapy. By leveraging the inherent biocompatibility and versatile engineering potential of exosomes, we can precisely target and disrupt the molecular foundations of drug resistance, effectively sensitizing tumors to conventional treatments. This review has emphasized the crucial role of exosomes in delivering therapeutic agents with unparalleled specificity, modulating both tumor and immune cells, and restoring immune surveillance. Despite promising advancements, challenges such as scalable production and personalized design need to be addressed to fully exploit their clinical potential. As we continue refining exosome-based strategies and integrating cutting-edge technologies, these biological nanocarriers are poised to redefine the landscape of cancer treatment by ushering in a new era of personalized and resilient therapeutic modalities.
Author contributions
PZ: Conceptualization, Data curation, Writing – review and editing. KC: Data curation, Methodology, Writing – review and editing. WL: Validation, Writing – review and editing, Investigation. XN: Writing – review and editing, Formal Analysis, Methodology. XW: Resources, Writing – review and editing, Formal Analysis. JW: Formal Analysis, Writing – review and editing. WY: Writing – review and editing, Conceptualization, Validation. XT: Validation, Investigation, Writing – review and editing. WT: Visualization, Funding acquisition, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by National Natural Science Foundation of China (82303914), Henan Medical Science and Technology Research and Development Plan Key Projects Jointly Constructed by the Provincial and Ministerial Departments in 2023 (SBGJ202302021), Henan Medical Science and Technology Research and Development Plan Youth Projects Jointly Constructed by the Provincial and Ministerial Departments (SBGJ202303023), and Science and technology in Henan Province (242102310351).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor YZ declared a shared affiliation with the author WL at the time of review.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bao, Y., Zhang, Y., Lu, Y., Guo, H., Dong, Z., Chen, Q., et al. (2020). Overexpression of microRNA-9 enhances cisplatin sensitivity in hepatocellular carcinoma by regulating EIF5A2-mediated epithelial-mesenchymal transition. Int. J. Biol. Sci. 16 (5), 827–37. doi:10.7150/ijbs.32460
Bhaskara, S. (2015). Histone deacetylases 1 and 2 regulate DNA replication and DNA repair: potential targets for genome stability-mechanism-based therapeutics for a subset of cancers. Cell cycleGeorget. Tex 14 (12), 1779–85. doi:10.1080/15384101.2015.1042634
Brownlie, J., Kulkarni, S., Algethami, M., Jeyapalan, J. N., Mongan, N. P., Rakha, E. A., et al. (2023). Targeting DNA damage repair precision medicine strategies in cancer. Curr. Opin. Pharmacol. 70, 102381. doi:10.1016/j.coph.2023.102381
Cao, L., Chen, J., Ou, B., Liu, C., Zou, Y., and Chen, Q. (2017). GAS5 knockdown reduces the chemo-sensitivity of non-small cell lung cancer (NSCLC) cell to cisplatin (DDP) through regulating miR-21/PTEN axis. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 93, 570–9. doi:10.1016/j.biopha.2017.06.089
Chang, L., Ruiz, P., Ito, T., and Sellers, W. R. (2021). Targeting pan-essential genes in cancer: challenges and opportunities. Cancer cell 39 (4), 466–79. doi:10.1016/j.ccell.2020.12.008
Chen, J., Zhou, C., Li, J., Xiang, X., Zhang, L., Deng, J., et al. (2018). miR-21-5p confers doxorubicin resistance in gastric cancer cells by targeting PTEN and TIMP3. Int. J. Mol. Med. 41 (4), 1855–66. doi:10.3892/ijmm.2018.3405
Chen, K. L., Huang, S. W., Yao, J. J., He, S. W., Gong, S., Tan, X. R., et al. (2024a). LncRNA DYNLRB2-AS1 promotes gemcitabine resistance of nasopharyngeal carcinoma by inhibiting the ubiquitination degradation of DHX9 protein. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 76, 101111. doi:10.1016/j.drup.2024.101111
Chen, Z., Zhang, X., Li, Z., Zhang, H., and Wang, Z. (2024b). lncRNA LINC02323 predicts adverse neoadjuvant chemotherapy outcomes of gastric cancer patients and regulates cell sensitivity to 5-fluorouracil by negatively modulating miR-139-3p. Ann. Med. 56 (1), 2424513. doi:10.1080/07853890.2024.2424513
Chen, Y., Chen, C. Y., Huang, H., Luo, Z., Mu, Y., Li, S., et al. (2024c). Knocking Down of Xkr8 enhances chemotherapy efficacy through modulating tumor immune microenvironment. J. Control. release official J. Control. Release Soc. 370, 479–89. doi:10.1016/j.jconrel.2024.04.041
Chen, Z., Xiong, M., Tian, J., Song, D., Duan, S., and Zhang, L. (2024d). Encapsulation and assessment of therapeutic cargo in engineered exosomes: a systematic review. J. Nanobiotechnology 22 (1), 18. doi:10.1186/s12951-023-02259-6
Chen, J., Xu, M., Wu, F., Wu, N., Li, J., Xie, Y., et al. (2025). CRKL silencing inhibits melanoma growth and enhances its chemotherapy sensitivity through the PI3K/AKT and NLRP3/GSDMD pathways. Biochem. Pharmacol. 235, 116840. doi:10.1016/j.bcp.2025.116840
Cheng, W., Sun, Y., Zhao, G., Khan, A., Zhang, J., Zhang, Z., et al. (2023). A novel peptide-templated AgNPs nanoprobe for theranostics of prostate cancer. Biosens. and Bioelectron. 223, 114978. doi:10.1016/j.bios.2022.114978
Chiang, C. L., Ma, Y., Hou, Y. C., Pan, J., Chen, S. Y., Chien, M. H., et al. (2023). Dual targeted extracellular vesicles regulate oncogenic genes in advanced pancreatic cancer. Nat. Commun. 14 (1), 6692. doi:10.1038/s41467-023-42402-3
Citron, F., Segatto, I., Vinciguerra, G. L. R., Musco, L., Russo, F., Mungo, G., et al. (2020). Downregulation of miR-223 expression is an early event during mammary transformation and confers resistance to CDK4/6 inhibitors in luminal breast cancer. Cancer Res. 80 (5), 1064–77. doi:10.1158/0008-5472.CAN-19-1793
Crowley, F., Park, W., and O'Reilly, E. M. (2021). Targeting DNA damage repair pathways in pancreas cancer. Cancer metastasis Rev. 40 (3), 891–908. doi:10.1007/s10555-021-09983-1
Cui, C., Yang, J., Li, X., Liu, D., Fu, L., and Wang, X. (2020). Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol. cancer 19 (1), 58. doi:10.1186/s12943-020-01180-y
Dad, H. A., Gu, T. W., Zhu, A. Q., Huang, L. Q., and Peng, L. H. (2021). Plant exosome-like nanovesicles: emerging therapeutics and drug delivery nanoplatforms. Mol. Ther. J. Am. Soc. Gene Ther. 29 (1), 13–31. doi:10.1016/j.ymthe.2020.11.030
Dai, S., Wei, D., Wu, Z., Zhou, X., Wei, X., Huang, H., et al. (2008). Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther. J. Am. Soc. Gene Ther. 16 (4), 782–90. doi:10.1038/mt.2008.1
Dai, W. C., Chen, T. H., Peng, T. C., He, Y. C., Hsu, C. Y., and Chang, C. C. (2025). Blockade of the STAT3/BCL-xL axis leads to the cytotoxic and cisplatin-sensitizing effects of fucoxanthin, a marine-derived carotenoid, on human bladder urothelial carcinoma cells. Mar. drugs 23 (2), 54. doi:10.3390/md23020054
Dammeijer, F., van Gulijk, M., Mulder, E. E., Lukkes, M., Klaase, L., van den Bosch, T., et al. (2020). The PD-1/PD-L1-Checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer cell 38 (5), 685–700.e8. doi:10.1016/j.ccell.2020.09.001
Das, T., Anand, U., Pandey, S. K., Ashby, C. R., Assaraf, Y. G., Chen, Z. S., et al. (2021). Therapeutic strategies to overcome taxane resistance in cancer. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 55, 100754. doi:10.1016/j.drup.2021.100754
Das, A., Gkoutos, G. V., and Acharjee, A. (2024). Analysis of translesion polymerases in colorectal cancer cells following cetuximab treatment: a network perspective. Cancer Med. 13 (1), e6945. doi:10.1002/cam4.6945
Desterro, J., Bak-Gordon, P., and Carmo-Fonseca, M. (2020). Targeting mRNA processing as an anticancer strategy. Nat. Rev. Drug Discov. 19 (2), 112–29. doi:10.1038/s41573-019-0042-3
Di Giacomo, S., Briz, O., Monte, M. J., Sanchez-Vicente, L., Abete, L., Lozano, E., et al. (2019). Chemosensitization of hepatocellular carcinoma cells to sorafenib by β-caryophyllene oxide-induced inhibition of ABC export pumps. Archives Toxicol. 93 (3), 623–34. doi:10.1007/s00204-019-02395-9
Du, Y., Wang, H., Yang, Y., Zhang, J., Huang, Y., Fan, S., et al. (2022). Extracellular vesicle mimetics: preparation from top-down approaches and biological functions. Adv. Healthc. Mater. 11 (19), e2200142. doi:10.1002/adhm.202200142
Du, X., Luo, W., Li, H., Gu, Q., Huang, P., Wang, C., et al. (2025). Hsa_circ_0125356 promotes gemcitabine resistance by modulating WNT canonical and non-canonical pathways via miR-582-5p/FGF9 axis in non-small cell lung cancer. Mol. cancer 24 (1), 59. doi:10.1186/s12943-025-02259-0
Duan, R., Zhai, Y., Wang, Q., Zhao, L., Wang, Y., Yu, N., et al. (2024). LINC01764 promotes colorectal cancer cells proliferation, metastasis, and 5-fluorouracil resistance by regulating glucose and glutamine metabolism via promoting c-MYC translation. MedComm 5 (11), e70003. doi:10.1002/mco2.70003
Eptaminitaki, G. C., Stellas, D., Bonavida, B., and Baritaki, S. (2022). Long non-coding RNAs (lncRNAs) signaling in cancer chemoresistance: from prediction to druggability. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 65, 100866. doi:10.1016/j.drup.2022.100866
Fabbri, L., Chakraborty, A., Robert, C., and Vagner, S. (2021). The plasticity of mRNA translation during cancer progression and therapy resistance. Nat. Rev. Cancer 21 (9), 558–77. doi:10.1038/s41568-021-00380-y
Farooqi, A. A., Desai, N. N., Qureshi, M. Z., Librelotto, D. R. N., Gasparri, M. L., Bishayee, A., et al. (2018). Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 36 (1), 328–34. doi:10.1016/j.biotechadv.2017.12.010
Fei, Y., Cao, D., Li, Y., Wang, Z., Dong, R., Zhu, M., et al. (2024). Circ_0008315 promotes tumorigenesis and cisplatin resistance and acts as a nanotherapeutic target in gastric cancer. J. Nanobiotechnology 22 (1), 519. doi:10.1186/s12951-024-02760-6
Fidelle, M., Rauber, C., Alves Costa Silva, C., Tian, A. L., Lahmar, I., de La Varende, A. M., et al. (2023). A microbiota-modulated checkpoint directs immunosuppressive intestinal T cells into cancers. Sci. (New York, NY) 380 (6649), eabo2296. doi:10.1126/science.abo2296
Fornari, F., Pollutri, D., Patrizi, C., La Bella, T., Marinelli, S., Casadei Gardini, A., et al. (2017). In hepatocellular carcinoma miR-221 modulates sorafenib resistance through inhibition of Caspase-3-Mediated apoptosis. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 23 (14), 3953–65. doi:10.1158/1078-0432.CCR-16-1464
Fu, X., He, Y., Wang, X., Peng, D., Chen, X., Li, X., et al. (2017). Overexpression of miR-21 in stem cells improves ovarian structure and function in rats with chemotherapy-induced ovarian damage by targeting PDCD4 and PTEN to inhibit granulosa cell apoptosis. Stem cell Res. and Ther. 8 (1), 187. doi:10.1186/s13287-017-0641-z
Fu, P., Zhang, J., Li, H., Mak, M., Xu, W., and Tao, Z. (2021). Extracellular vesicles as delivery systems at nano-/micro-scale. Adv. drug Deliv. Rev. 179, 113910. doi:10.1016/j.addr.2021.113910
Fu, K., Xie, F., Wang, F., and Fu, L. (2022). Therapeutic strategies for EGFR-Mutated non-small cell lung cancer patients with osimertinib resistance. J. Hematol. and Oncol. 15 (1), 173. doi:10.1186/s13045-022-01391-4
Fukumoto, W., Okamura, S., Tamai, M., Arima, J., Kawahara, I., Fukuda, I., et al. (2024). Development of a novel treatment based on PKMYT1 inhibition for cisplatin-resistant bladder cancer with miR-424-5p-dependent cyclin E1 amplification. BMC cancer 24 (1), 1333. doi:10.1186/s12885-024-13109-5
Galon, J., and Bruni, D. (2019). Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 18 (3), 197–218. doi:10.1038/s41573-018-0007-y
Gan, Y., Hao, Q., Han, T., Tong, J., Yan, Q., Zhong, H., et al. (2024). Targeting BRIX1 via engineered exosomes induces nucleolar stress to suppress cancer progression. Adv. Sci. Weinheim, Baden-Wurttemberg, Ger. 11. doi:10.1002/advs.202407370
Gangadaran, P., and Ahn, B. C. (2020). Extracellular Vesicle- and extracellular vesicle mimetics-based drug delivery systems: new perspectives, challenges, and clinical developments. Pharmaceutics 12 (5), 442. doi:10.3390/pharmaceutics12050442
Gao, X., Ran, N., Dong, X., Zuo, B., Yang, R., Zhou, Q., et al. (2018). Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 10 (444), eaat0195. doi:10.1126/scitranslmed.aat0195
Gao, Y., Tong, M., Wong, T. L., Ng, K. Y., Xie, Y. N., Wang, Z., et al. (2023). Long noncoding RNA URB1-Antisense RNA 1 (AS1) suppresses Sorafenib-Induced ferroptosis in hepatocellular carcinoma by driving ferritin phase separation. ACS nano 17 (22), 22240–58. doi:10.1021/acsnano.3c01199
Garofalo, M., and Croce, C. M. (2013). MicroRNAs as therapeutic targets in chemoresistance. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 16 (3-5), 47–59. doi:10.1016/j.drup.2013.05.001
Geenen, J. J. J., Linn, S. C., Beijnen, J. H., and Schellens, J. H. M. (2018). PARP inhibitors in the treatment of triple-negative breast cancer. Clin. Pharmacokinet. 57 (4), 427–37. doi:10.1007/s40262-017-0587-4
Geissler, F., Nesic, K., Kondrashova, O., Dobrovic, A., Swisher, E. M., Scott, C. L., et al. (2024). The role of aberrant DNA methylation in cancer initiation and clinical impacts. Ther. Adv. Med. Oncol. 16, 17588359231220511. doi:10.1177/17588359231220511
Georgikou, C., Yin, L., Gladkich, J., Xiao, X., Sticht, C., Torre, C., et al. (2020). Inhibition of miR30a-3p by sulforaphane enhances gap junction intercellular communication in pancreatic cancer. Cancer Lett. 469, 238–45. doi:10.1016/j.canlet.2019.10.042
Gotwals, P., Cameron, S., Cipolletta, D., Cremasco, V., Crystal, A., Hewes, B., et al. (2017). Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 17 (5), 286–301. doi:10.1038/nrc.2017.17
Guo, Q., and Jiang, C. (2020). Delivery strategies for macromolecular drugs in cancer therapy. Acta Pharm. Sin. B 10 (6), 979–86. doi:10.1016/j.apsb.2020.01.009
Guo, Y., Ashrafizadeh, M., Tambuwala, M. M., Ren, J., Orive, G., and Yu, G. (2024). P-glycoprotein (P-gp)-driven cancer drug resistance: biological profile, non-coding RNAs, drugs and nanomodulators. Drug Discov. today 29 (11), 104161. doi:10.1016/j.drudis.2024.104161
Gupta, S., Silveira, D. A., Piedade, G. P. S., Ostrowski, M. P., Mombach, J. C. M., and Hashimoto, R. F. (2024). A dynamic Boolean network reveals that the BMI1 and MALAT1 axis is associated with drug resistance by limiting miR-145-5p in non-small cell lung cancer. Non-coding RNA Res. 9 (1), 185–93. doi:10.1016/j.ncrna.2023.10.008
Gurung, S., Perocheau, D., Touramanidou, L., and Baruteau, J. (2021). The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal. CCS 19 (1), 47. doi:10.1186/s12964-021-00730-1
Han, Y., Zhou, S., Wang, X., Mao, E., and Huang, L. (2020a). SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 121, 109580. doi:10.1016/j.biopha.2019.109580
Han, M., Qian, X., Cao, H., Wang, F., Li, X., Han, N., et al. (2020b). lncRNA ZNF649-AS1 induces trastuzumab resistance by promoting ATG5 expression and autophagy. Mol. Ther. J. Am. Soc. Gene Ther. 28 (11), 2488–502. doi:10.1016/j.ymthe.2020.07.019
Haselager, M., Thijssen, R., West, C., Young, L., Van Kampen, R., Willmore, E., et al. (2021). Regulation of Bcl-XL by non-canonical NF-κB in the context of CD40-induced drug resistance in CLL. Cell death Differ. 28 (5), 1658–68. doi:10.1038/s41418-020-00692-w
He, G., Peng, X., Wei, S., Yang, S., Li, X., Huang, M., et al. (2022). Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol. cancer 21 (1), 19. doi:10.1186/s12943-021-01440-5
Herrmann, I. K., Wood, M. J. A., and Fuhrmann, G. (2021). Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 16 (7), 748–59. doi:10.1038/s41565-021-00931-2
Hiraki, M., Kitajima, Y., Nakafusa, Y., Nakamura, J., Hashiguchi, K., Sumi, K., et al. (2010). CpG island methylation of BNIP3 predicts resistance against S-1/CPT-11 combined therapy in colorectal cancer patients. Oncol. Rep. 23 (1), 191–7. doi:10.3892/or_00000622
Hong, X., Liu, N., Liang, Y., He, Q., Yang, X., Lei, Y., et al. (2020). Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol. cancer 19 (1), 33. doi:10.1186/s12943-020-01149-x
Hou, J., Zhang, G., Wang, X., Wang, Y., and Wang, K. (2023). Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy resistance. Biomark. Res. 11 (1), 23. doi:10.1186/s40364-023-00467-8
Huan, L., Guo, T., Wu, Y., Xu, L., Huang, S., Xu, Y., et al. (2020). Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol. cancer 19 (1), 11. doi:10.1186/s12943-019-1122-z
Huang, W., Yang, Y., Wu, J., Niu, Y., Yao, Y., Zhang, J., et al. (2020). Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell death Differ. 27 (5), 1709–27. doi:10.1038/s41418-019-0455-x
Huang, L., Han, L., Liang, S., and Han, G. (2025). Molecular mechanism of ZC3H13 -mediated ferroptosis in doxorubicin resistance of triple negative breast cancer. Cell Biol. Toxicol. 41 (1), 52. doi:10.1007/s10565-024-09980-4
Iqbal, M. A., Arora, S., Prakasam, G., Calin, G. A., and Syed, M. A. (2019). MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol. aspects Med. 70, 3–20. doi:10.1016/j.mam.2018.07.003
Jian, X., He, H., Zhu, J., Zhang, Q., Zheng, Z., Liang, X., et al. (2020). Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. cancer 19 (1), 20. doi:10.1186/s12943-020-1134-8
Jiang, J., Lu, Y., Chu, J., Zhang, X., Xu, C., Liu, S., et al. (2024). Anti-EGFR ScFv functionalized exosomes delivering LPCAT1 specific siRNAs for inhibition of lung cancer brain metastases. J. nanobiotechnology 22 (1), 159. doi:10.1186/s12951-024-02414-7
Jung, K. O., Jo, H., Yu, J. H., Gambhir, S. S., and Pratx, G. (2018). Development and MPI tracking of novel hypoxia-targeted theranostic exosomes. Biomaterials 177, 139–48. doi:10.1016/j.biomaterials.2018.05.048
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Sci. (New York, NY) 367 (6478). doi:10.1126/science.aau6977
Kamerkar, S., LeBleu, V. S., Sugimoto, H., Yang, S., Ruivo, C. F., Melo, S. A., et al. (2017). Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546 (7659), 498–503. doi:10.1038/nature22341
Katz, O. B., and Shaked, Y. (2015). Host effects contributing to cancer therapy resistance. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 19, 33–42. doi:10.1016/j.drup.2014.12.002
Kim, S. M., Yang, Y., Oh, S. J., Hong, Y., Seo, M., and Jang, M. (2017). Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. release official J. Control. Release Soc. 266, 8–16. doi:10.1016/j.jconrel.2017.09.013
Kim, H., Kim, D., Nam, H., Moon, S., Kwon, Y. J., and Lee, J. B. (2020). Engineered extracellular vesicles and their mimetics for clinical translation. Methods San Diego, Calif. 177, 80–94. doi:10.1016/j.ymeth.2019.10.005
Kim, J., Li, S., Zhang, S., and Wang, J. (2022). Plant-derived exosome-like nanoparticles and their therapeutic activities. Asian J. Pharm. Sci. 17 (1), 53–69. doi:10.1016/j.ajps.2021.05.006
Kim, E. H., Ryu, Y., Choi, J., Park, D., Lee, J. W., Chi, S. G., et al. (2024a). Targeting miR-21 to overcome P-glycoprotein drug efflux in doxorubicin-resistant 4T1 breast cancer. Biomaterials Res. 28, 0095. doi:10.34133/bmr.0095
Kim, R., Mun, B., Lim, S., Park, C., Kim, J., Lim, J., et al. (2024b). Colorimetric detection of HER2-Overexpressing-Cancer-Derived exosomes in mouse urine using magnetic-polydiacetylene nanoparticles. Small Weinheim der Bergstrasse, Ger. 20 (13), e2307262. doi:10.1002/smll.202307262
Kooijmans, S. A. A., Fliervoet, L. A. L., van der Meel, R., Fens, M., Heijnen, H. F. G., van Bergen En Henegouwen, P. M. P., et al. (2016). PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J. Control. release official J. Control. Release Soc. 224, 77–85. doi:10.1016/j.jconrel.2016.01.009
Krumm, A., Barckhausen, C., Kücük, P., Tomaszowski, K. H., Loquai, C., Fahrer, J., et al. (2016). Enhanced histone deacetylase activity in malignant melanoma provokes RAD51 and FANCD2-Triggered drug resistance. Cancer Res. 76 (10), 3067–77. doi:10.1158/0008-5472.CAN-15-2680
Kubli, S. P., Berger, T., Araujo, D. V., Siu, L. L., and Mak, T. W. (2021). Beyond immune checkpoint blockade: emerging immunological strategies. Nat. Rev. Drug Discov. 20 (12), 899–919. doi:10.1038/s41573-021-00155-y
Labrie, M., Brugge, J. S., Mills, G. B., and Zervantonakis, I. K. (2022). Therapy resistance: opportunities created by adaptive responses to targeted therapies in cancer. Nat. Rev. Cancer 22 (6), 323–39. doi:10.1038/s41568-022-00454-5
Lai, J., Xin, J., Fu, C., and Zhang, W. (2020). CircHIPK3 promotes proliferation and metastasis and inhibits apoptosis of renal cancer cells by inhibiting MiR-485-3p. Cancer cell Int. 20, 248. doi:10.1186/s12935-020-01319-3
Lai, J. J., Chau, Z. L., Chen, S. Y., Hill, J. J., Korpany, K. V., Liang, N. W., et al. (2022). Exosome processing and characterization approaches for research and technology development. Adv. Sci. Weinheim, Baden-Wurttemberg, Ger. 9 (15), e2103222. doi:10.1002/advs.202103222
Leber, B., Kale, J., and Andrews, D. W. (2018). Unleashing blocked apoptosis in cancer cells: new MCL1 inhibitors find their groove. Cancer Discov. 8 (12), 1511–4. doi:10.1158/2159-8290.CD-18-1167
Lee, K. Y., Oh, S. Y., Lee, H. J., Kwon, T. G., Kim, J. W., Shin, C. G., et al. (2025). MTMR6 downregulation contributes to cisplatin resistance in oral squamous cell carcinoma. Cancer cell Int. 25 (1), 30. doi:10.1186/s12935-025-03654-9
Leonetti, A., Assaraf, Y. G., Veltsista, P. D., El Hassouni, B., Tiseo, M., and Giovannetti, E. (2019). MicroRNAs as a drug resistance mechanism to targeted therapies in EGFR-Mutated NSCLC: current implications and future directions. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 42, 1–11. doi:10.1016/j.drup.2018.11.002
Li, I., and Nabet, B. Y. (2019). Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. cancer 18 (1), 32. doi:10.1186/s12943-019-0975-5
Li, S., Zhang, W., Yin, X., Xing, S., Xie, H. Q., Cao, Z., et al. (2015). Mouse ATP-binding cassette (ABC) transporters conferring multi-drug resistance. Anti-cancer agents Med. Chem. 15 (4), 423–32. doi:10.2174/1871520615666150129212723
Li, P., Zhang, X., Wang, H., Wang, L., Liu, T., Du, L., et al. (2017). MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol. cancer Ther. 16 (4), 739–51. doi:10.1158/1535-7163.MCT-16-0591
Li, Y., He, L. R., Gao, Y., Zhou, N. N., Liu, Y., Zhou, X. K., et al. (2019). CHD1L contributes to cisplatin resistance by upregulating the ABCB1-NF-κB axis in human non-small-cell lung cancer. Cell death and Dis. 10 (2), 99. doi:10.1038/s41419-019-1371-1
Li, S., Lv, J., Li, Z., Zhang, Q., Lu, J., Huo, X., et al. (2024a). Overcoming multi-drug resistance in SCLC: a synergistic approach with venetoclax and hydroxychloroquine targeting the lncRNA LYPLAL1-DT/BCL2/BECN1 pathway. Mol. cancer 23 (1), 243. doi:10.1186/s12943-024-02145-1
Li, H., Lin, R., Zhang, Y., Zhu, Y., Huang, S., Lan, J., et al. (2024b). N6-methyladenosine-modified circPLPP4 sustains cisplatin resistance in ovarian cancer cells via PIK3R1 upregulation. Mol. cancer 23 (1), 5. doi:10.1186/s12943-023-01917-5
Li, Y., Sharma, A., and Schmidt-Wolf, I. G. H. (2024c). Evolving insights into the improvement of adoptive T-cell immunotherapy through PD-1/PD-L1 blockade in the clinical spectrum of lung cancer. Mol. cancer 23 (1), 80. doi:10.1186/s12943-023-01926-4
Li, K., Yap, Y. Q., Moujalled, D. M., Sumardy, F., Khakham, Y., Georgiou, A., et al. (2025). Differential regulation of BAX and BAK apoptotic activity revealed by small molecules. Sci. Adv. 11 (10), eadr8146. doi:10.1126/sciadv.adr8146
Liang, Y., Hou, L., Li, L., Li, L., Zhu, L., Wang, Y., et al. (2020). Dichloroacetate restores colorectal cancer chemosensitivity through the p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene 39 (2), 469–85. doi:10.1038/s41388-019-1035-8
Liang, Y., Duan, L., Lu, J., and Xia, J. (2021). Engineering exosomes for targeted drug delivery. Theranostics 11 (7), 3183–95. doi:10.7150/thno.52570
Liang, Y., Iqbal, Z., Lu, J., Wang, J., Zhang, H., Chen, X., et al. (2023). Cell-derived nanovesicle-mediated drug delivery to the brain: principles and strategies for vesicle engineering. Mol. Ther. J. Am. Soc. Gene Ther. 31 (5), 1207–24. doi:10.1016/j.ymthe.2022.10.008
Liao, X., Zhang, S., Li, X., Qian, W., Li, M., Chen, S., et al. (2024). Dynamic structural remodeling of LINC01956 enhances temozolomide resistance in MGMT-Methylated glioblastoma. Sci. Transl. Med. 16 (767), eado1573. doi:10.1126/scitranslmed.ado1573
Lin, S., and Kuang, M. (2024). RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives. Nat. Rev. Gastroenterology and hepatology 21 (4), 267–81. doi:10.1038/s41575-023-00884-y
Lin, H., Shen, L., Lin, Q., Dong, C., Maswela, B., Illahi, G. S., et al. (2020). SNHG5 enhances paclitaxel sensitivity of ovarian cancer cells through sponging miR-23a. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 123, 109711. doi:10.1016/j.biopha.2019.109711
Lin, D., Zhang, H., Liu, R., Deng, T., Ning, T., Bai, M., et al. (2021). iRGD-modified exosomes effectively deliver CPT1A siRNA to Colon cancer cells, reversing oxaliplatin resistance by regulating fatty acid oxidation. Mol. Oncol. 15 (12), 3430–46. doi:10.1002/1878-0261.13052
Lin, Z., Wu, Y., Xu, Y., Li, G., Li, Z., and Liu, T. (2022). Mesenchymal stem cell-derived exosomes in cancer therapy resistance: recent advances and therapeutic potential. Mol. cancer 21 (1), 179. doi:10.1186/s12943-022-01650-5
Lin, Z., Huang, Z., Qiu, J., Shi, Y., Zuo, D., Qiu, Z., et al. (2024a). m(6)A-mediated lnc-OXAR promotes oxaliplatin resistance by enhancing Ku70 stability in non-alcoholic steatohepatitis-related hepatocellular carcinoma. J. Exp. and Clin. cancer Res. CR 43 (1), 206. doi:10.1186/s13046-024-03134-4
Lin, J., Lyu, Z., Feng, H., Xie, H., Peng, J., Zhang, W., et al. (2024b). CircPDIA3/miR-449a/XBP1 feedback loop curbs pyroptosis by inhibiting palmitoylation of the GSDME-C domain to induce chemoresistance of colorectal cancer. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 76, 101097. doi:10.1016/j.drup.2024.101097
Ling, Z., Wang, X., Tao, T., Zhang, L., Guan, H., You, Z., et al. (2017). Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J. Exp. and Clin. cancer Res. CR 36 (1), 159. doi:10.1186/s13046-017-0629-7
Liu, G., Yu, M., Wu, B., Guo, S., Huang, X., Zhou, F., et al. (2019). Jab1/Cops5 contributes to chemoresistance in breast cancer by regulating Rad51. Cell. Signal. 53, 39–48. doi:10.1016/j.cellsig.2018.09.010
Liu, J., Sun, Y., Zeng, X., Liu, Y., Liu, C., Zhou, Y., et al. (2023). Engineering and characterization of an artificial drug-carrying vesicles nanoplatform for enhanced specifically targeted therapy of glioblastoma. Adv. Mater. Deerf. Beach, Fla 35 (41), e2303660. doi:10.1002/adma.202303660
Liu, Y., Kang, L., Luo, J., Yang, M., Wang, D., Ye, J., et al. (2025a). Targeting AKT as a promising strategy for SOX2-positive, chemoresistant osteosarcoma. Bone Res. 13 (1), 25. doi:10.1038/s41413-024-00395-9
Liu, X., Zhang, X., Zeng, T., Chen, Y., Ye, L., Wang, S., et al. (2025b). FOSL1 drives the malignant progression of pancreatic cancer cells by regulating cell stemness, metastasis and multidrug efflux system. J. Transl. Med. 23 (1), 268. doi:10.1186/s12967-025-06304-w
Lopez, A., Reyna, D. E., Gitego, N., Kopp, F., Zhou, H., Miranda-Roman, M. A., et al. (2022). Co-targeting of BAX and BCL-XL proteins broadly overcomes resistance to apoptosis in cancer. Nat. Commun. 13 (1), 1199. doi:10.1038/s41467-022-28741-7
Lu, X., Liu, R., Wang, M., Kumar, A. K., Pan, F., He, L., et al. (2020a). MicroRNA-140 impedes DNA repair by targeting FEN1 and enhances chemotherapeutic response in breast cancer. Oncogene 39 (1), 234–47. doi:10.1038/s41388-019-0986-0
Lu, C., Wei, Y., Wang, X., Zhang, Z., Yin, J., Li, W., et al. (2020b). DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol. cancer 19 (1), 28. doi:10.1186/s12943-020-1137-5
Lu, Q., Kou, D., Lou, S., Ashrafizadeh, M., Aref, A. R., Canadas, I., et al. (2024). Nanoparticles in tumor microenvironment remodeling and cancer immunotherapy. J. Hematol. and Oncol. 17 (1), 16. doi:10.1186/s13045-024-01535-8
Lv, N., Huang, C., Huang, H., Dong, Z., Chen, X., Lu, C., et al. (2023). Overexpression of glutathione S-Transferases in human diseases: drug targets and therapeutic implications. Antioxidants Basel, Switz. 12 (11), 1970. doi:10.3390/antiox12111970
Ma, S., Kong, S., Wang, F., and Ju, S. (2020). CircRNAs: biogenesis, functions, and role in drug-resistant tumours. Mol. cancer 19 (1), 119. doi:10.1186/s12943-020-01231-4
Maeda, M., Murakami, Y., Watari, K., Kuwano, M., Izumi, H., and Ono, M. (2015). CpG hypermethylation contributes to decreased expression of PTEN during acquired resistance to gefitinib in human lung cancer cell lines. Lung cancer Amsterdam, Neth. 87 (3), 265–71. doi:10.1016/j.lungcan.2015.01.009
Mahaffey, C. M., Zhang, H., Rinna, A., Holland, W., Mack, P. C., and Forman, H. J. (2009). Multidrug-resistant protein-3 gene regulation by the transcription factor Nrf2 in human bronchial epithelial and non-small-cell lung carcinoma. Free Radic. Biol. and Med. 46 (12), 1650–7. doi:10.1016/j.freeradbiomed.2009.03.023
Mao, T. L., Fan, M. H., Dlamini, N., and Liu, C. L. (2021). LncRNA MALAT1 facilitates ovarian cancer progression through promoting chemoresistance and invasiveness in the tumor microenvironment. Int. J. Mol. Sci. 22 (19), 10201. doi:10.3390/ijms221910201
Marsolier, J., Prompsy, P., Durand, A., Lyne, A. M., Landragin, C., Trouchet, A., et al. (2022). H3K27me3 conditions chemotolerance in triple-negative breast cancer. Nat. Genet. 54 (4), 459–68. doi:10.1038/s41588-022-01047-6
Meng, W., He, C., Hao, Y., Wang, L., Li, L., and Zhu, G. (2020). Prospects and challenges of extracellular vesicle-based drug delivery system: considering cell source. Drug Deliv. 27 (1), 585–98. doi:10.1080/10717544.2020.1748758
Menzies, A. M., Pires da Silva, I., Trojaniello, C., Vieu, E., Amaria, R. N., Zimmer, L., et al. (2022). CTLA-4 blockade resistance after relatlimab and nivolumab. N. Engl. J. Med. 386 (17), 1668–9. doi:10.1056/NEJMc2119768
Mohammad, I. S., He, W., and Yin, L. (2018). Understanding of human ATP binding cassette superfamily and novel multidrug resistance modulators to overcome MDR. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 100, 335–48. doi:10.1016/j.biopha.2018.02.038
Mok, E. H. K., Leung, C. O. N., Zhou, L., Lei, M. M. L., Leung, H. W., Tong, M., et al. (2022). Caspase-3-Induced activation of SREBP2 drives drug resistance via promotion of cholesterol biosynthesis in hepatocellular carcinoma. Cancer Res. 82 (17), 3102–15. doi:10.1158/0008-5472.CAN-21-2934
Muñoz-Galván, S., Felipe-Abrio, B., Verdugo-Sivianes, E. M., Perez, M., Jiménez-García, M. P., Suarez-Martinez, E., et al. (2020). Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the hippo pathway and increasing the stemness. Mol. cancer 19 (1), 7. doi:10.1186/s12943-020-1130-z
Namee, N. M., and O'Driscoll, L. (2018). Extracellular vesicles and anti-cancer drug resistance. Biochimica biophysica acta Rev. cancer 1870 (2), 123–36. doi:10.1016/j.bbcan.2018.07.003
Nolte-'t Hoen, E. N., Buschow, S. I., Anderton, S. M., Stoorvogel, W., and Wauben, M. H. (2009). Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 113 (9), 1977–81. doi:10.1182/blood-2008-08-174094
Noma, B., Sasaki, T., Fujimoto, Y., Serikawa, M., Kobayashi, K., Inoue, M., et al. (2008). Expression of multidrug resistance-associated protein 2 is involved in chemotherapy resistance in human pancreatic cancer. Int. J. Oncol. 33 (6), 1187–94. doi:10.3892/ijo_00000108
O'Donnell, J. S., Teng, M. W. L., and Smyth, M. J. (2019). Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 16 (3), 151–67. doi:10.1038/s41571-018-0142-8
Ogino, S., Meyerhardt, J. A., Kawasaki, T., Clark, J. W., Ryan, D. P., Kulke, M. H., et al. (2007). CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Archiv Int. J. pathology 450 (5), 529–37. doi:10.1007/s00428-007-0398-3
Pan, X., Chen, K., Gao, W., Xu, M., Meng, F., Wu, M., et al. (2025). Circular RNA circBNC2 inhibits tumorigenesis by modulating ferroptosis and acts as a nanotherapeutic target in prostate cancer. Mol. cancer 24 (1), 29. doi:10.1186/s12943-025-02234-9
Pardoll, D. M. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12 (4), 252–64. doi:10.1038/nrc3239
Patel, S. M., Dash, R. C., and Hadden, M. K. (2021). Translesion synthesis inhibitors as a new class of cancer chemotherapeutics. Expert Opin. investigational drugs 30 (1), 13–24. doi:10.1080/13543784.2021.1850692
Pathania, A. S., Prathipati, P., and Challagundla, K. B. (2021). New insights into exosome mediated tumor-immune escape: clinical perspectives and therapeutic strategies. Biochimica biophysica acta Rev. cancer 1876 (2), 188624. doi:10.1016/j.bbcan.2021.188624
Peng, S., Wang, R., Zhang, X., Ma, Y., Zhong, L., Li, K., et al. (2019). EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol. cancer 18 (1), 165. doi:10.1186/s12943-019-1073-4
Peng, Y., Tang, D., Zhao, M., Kajiyama, H., Kikkawa, F., and Kondo, Y. (2020). Long non-coding RNA: a recently accentuated molecule in chemoresistance in cancer. Cancer metastasis Rev. 39 (3), 825–35. doi:10.1007/s10555-020-09910-w
Pljesa-Ercegovac, M., Savic-Radojevic, A., Matic, M., Coric, V., Djukic, T., Radic, T., et al. (2018). Glutathione transferases: potential targets to overcome chemoresistance in solid tumors. Int. J. Mol. Sci. 19 (12), 3785. doi:10.3390/ijms19123785
Poggio, M., Hu, T., Pai, C. C., Chu, B., Belair, C. D., Chang, A., et al. (2019). Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell 177 (2), 414–27.e13. doi:10.1016/j.cell.2019.02.016
Pouliot, L. M., Chen, Y. C., Bai, J., Guha, R., Martin, S. E., Gottesman, M. M., et al. (2012). Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 72 (22), 5945–55. doi:10.1158/0008-5472.CAN-12-1400
Qiao, L., Hu, S., Huang, K., Su, T., Li, Z., Vandergriff, A., et al. (2020). Tumor cell-derived exosomes home to their cells of origin and can be used as trojan horses to deliver cancer drugs. Theranostics 10 (8), 3474–87. doi:10.7150/thno.39434
Qin, T., Huang, G., Chi, L., Sui, S., Song, C., Li, N., et al. (2017). Exceptionally high UBE2C expression is a unique phenomenon in basal-like type breast cancer and is regulated by BRCA1. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 95, 649–55. doi:10.1016/j.biopha.2017.08.095
Qu, W., Huang, W., Yang, F., Ju, H., and Zhu, G. (2020). Long noncoding RNA LINC00461 mediates cisplatin resistance of rectal cancer via miR-593-5p/CCND1 axis. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 124, 109740. doi:10.1016/j.biopha.2019.109740
Rajeev Krishnan, S., De Rubis, G., Suen, H., Joshua, D., Lam Kwan, Y., and Bebawy, M. (2020). A liquid biopsy to detect multidrug resistance and disease burden in multiple myeloma. Blood cancer J. 10 (3), 37. doi:10.1038/s41408-020-0304-7
Raju, G. S. R., Pavitra, E., Bandaru, S. S., Varaprasad, G. L., Nagaraju, G. P., Malla, R. R., et al. (2023). HOTAIR: a potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol. cancer 22 (1), 65. doi:10.1186/s12943-023-01765-3
Rastgoo, N., Abdi, J., Hou, J., and Chang, H. (2017). Role of epigenetics-microRNA axis in drug resistance of multiple myeloma. J. Hematol. and Oncol. 10 (1), 121. doi:10.1186/s13045-017-0492-1
Rehman, F. U., Liu, Y., Zheng, M., and Shi, B. (2023). Exosomes based strategies for brain drug delivery. Biomaterials 293, 121949. doi:10.1016/j.biomaterials.2022.121949
Sadeghi, S., Tehrani, F. R., Tahmasebi, S., Shafiee, A., and Hashemi, S. M. (2023). Exosome engineering in cell therapy and drug delivery. Inflammopharmacology 31 (1), 145–69. doi:10.1007/s10787-022-01115-7
Salunkhe, S., Dheeraj, B. M., Chitkara, D., and Mittal, A. (2020). Surface functionalization of exosomes for target-specific delivery and in vivo imaging and tracking: strategies and significance. J. Control. release official J. Control. Release Soc. 326, 599–614. doi:10.1016/j.jconrel.2020.07.042
Segura, E., Nicco, C., Lombard, B., Véron, P., Raposo, G., Batteux, F., et al. (2005). ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 106 (1), 216–23. doi:10.1182/blood-2005-01-0220
Seidl, C., Panzitt, K., Bertsch, A., Brcic, L., Schein, S., Mack, M., et al. (2020). MicroRNA-182-5p regulates hedgehog signaling pathway and chemosensitivity of cisplatin-resistant lung adenocarcinoma cells via targeting GLI2. Cancer Lett. 469, 266–76. doi:10.1016/j.canlet.2019.10.044
Shan, S., Chen, J., Sun, Y., Wang, Y., Xia, B., Tan, H., et al. (2022). Functionalized macrophage exosomes with panobinostat and PPM1D-siRNA for diffuse intrinsic pontine gliomas therapy. Adv. Sci. Weinheim, Baden-Wurttemberg, Ger. 9 (21), e2200353. doi:10.1002/advs.202200353
Shao, H., Chung, J., Lee, K., Balaj, L., Min, C., Carter, B. S., et al. (2015). Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 6, 6999. doi:10.1038/ncomms7999
Shen, J., Wang, Q., Hu, Q., Li, Y., Tang, G., and Chu, P. K. (2014). Restoration of chemosensitivity by multifunctional micelles mediated by P-gp siRNA to reverse MDR. Biomaterials 35 (30), 8621–34. doi:10.1016/j.biomaterials.2014.06.035
Shi, Y., Zhang, J., Mao, Z., Jiang, H., Liu, W., Shi, H., et al. (2020). Extracellular vesicles from gastric cancer cells induce PD-L1 expression on neutrophils to suppress T-Cell immunity. Front. Oncol. 10, 629. doi:10.3389/fonc.2020.00629
Shilkin, E. S., Boldinova, E. O., Stolyarenko, A. D., Goncharova, R. I., Chuprov-Netochin, R. N., Khairullin, R. F., et al. (2020a). Translesion DNA synthesis and carcinogenesis. Biochem. Biokhimiia 85 (4), 425–35. doi:10.1134/S0006297920040033
Shilkin, E. S., Boldinova, E. O., Stolyarenko, A. D., Goncharova, R. I., Chuprov-Netochin, R. N., Smal, M. P., et al. (2020b). Translesion DNA synthesis and reinitiation of DNA synthesis in chemotherapy resistance. Biochem. Biokhimiia 85 (8), 869–82. doi:10.1134/S0006297920080039
Shimasaki, N., Jain, A., and Campana, D. (2020). NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19 (3), 200–18. doi:10.1038/s41573-019-0052-1
Short, S. C., Giampieri, S., Worku, M., Alcaide-German, M., Sioftanos, G., Bourne, S., et al. (2011). Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro-oncology 13 (5), 487–99. doi:10.1093/neuonc/nor010
Shukla, D., Mishra, S., Mandal, T., Charan, M., Verma, A. K., Khan, M. M. A., et al. (2025). MicroRNA-379-5p attenuates cancer stem cells and reduces cisplatin resistance in ovarian cancer by regulating RAD18/Polη axis. Cell death and Dis. 16 (1), 140. doi:10.1038/s41419-025-07430-5
Singh, D., Assaraf, Y. G., and Gacche, R. N. (2022). Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 63, 100851. doi:10.1016/j.drup.2022.100851
Song, X., Shen, L., Tong, J., Kuang, C., Zeng, S., Schoen, R. E., et al. (2020). Mcl-1 inhibition overcomes intrinsic and acquired regorafenib resistance in colorectal cancer. Theranostics 10 (18), 8098–110. doi:10.7150/thno.45363
Su, R., Wang, Q., Hu, Q., Wendurige, , Li, K., Wang, C., et al. (2025). HDGF knockout suppresses colorectal cancer progression and drug resistance by modulating the DNA damage response. Biomolecules 15 (2), 282. doi:10.3390/biom15020282
Sun, J., Zhao, W., Zhang, L., Wu, S., Xue, S., Cao, H., et al. (2025). Centromere protein U mediates the ubiquitination and degradation of RPS3 to facilitate temozolomide resistance in glioblastoma. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 80, 101214. doi:10.1016/j.drup.2025.101214
Tan, F., Li, X., Wang, Z., Li, J., Shahzad, K., and Zheng, J. (2024). Clinical applications of stem cell-derived exosomes. Signal Transduct. Target. Ther. 9 (1), 17. doi:10.1038/s41392-023-01704-0
Tao, H., Xu, H., Zuo, L., Li, C., Qiao, G., Guo, M., et al. (2020). Exosomes-coated bcl-2 siRNA inhibits the growth of digestive system tumors both in vitro and in vivo. Int. J. Biol. Macromol. 161, 470–80. doi:10.1016/j.ijbiomac.2020.06.052
Tao, B., Du, R., Zhang, X., Jia, B., Gao, Y., Zhao, Y., et al. (2023). Engineering CAR-NK cell derived exosome disguised nano-bombs for enhanced HER2 positive breast cancer brain metastasis therapy. J. Control. release official J. Control. Release Soc. 363, 692–706. doi:10.1016/j.jconrel.2023.10.007
Tao, S., Ji, Y., Li, R., Xiao, Y., Wu, H., Ye, R., et al. (2025). Layered double hydroxide LDH-loaded miR-141-3p targets RAB10 suppressing cellular autophagy to reverse paclitaxel resistance in breast cancer. ACS omega 10 (6), 5886–99. doi:10.1021/acsomega.4c09755
Tenchov, R., Sasso, J. M., Wang, X., Liaw, W. S., Chen, C. A., and Zhou, Q. A. (2022). Exosomes─Nature's lipid nanoparticles, a rising star in drug delivery and diagnostics. ACS nano 16 (11), 17802–46. doi:10.1021/acsnano.2c08774
Tilsed, C. M., Fisher, S. A., Nowak, A. K., Lake, R. A., and Lesterhuis, W. J. (2022). Cancer chemotherapy: insights into cellular and tumor microenvironmental mechanisms of action. Front. Oncol. 12, 960317. doi:10.3389/fonc.2022.960317
To, K. K. W., Huang, Z., Zhang, H., Ashby, C. R., and Fu, L. (2024). Utilizing non-coding RNA-Mediated regulation of ATP binding cassette (ABC) transporters to overcome multidrug resistance to cancer chemotherapy. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 73, 101058. doi:10.1016/j.drup.2024.101058
Toh, S. Y., Leong, H. S., Chong, F. T., Rodrigues-Junior, D. M., Ren, M. J., Kwang, X. L., et al. (2024). Therapeutic application of extracellular vesicular EGFR isoform D as a co-drug to target squamous cell cancers with tyrosine kinase inhibitors. Dev. cell 59 (16), 2189–202.e8. doi:10.1016/j.devcel.2024.07.003
Tran, P. H. L., Xiang, D., Tran, T. T. D., Yin, W., Zhang, Y., Kong, L., et al. (2020). Exosomes and nanoengineering: a match made for precision therapeutics. Adv. Mater. Deerf. Beach, Fla 32 (18), e1904040. doi:10.1002/adma.201904040
van der Pol, E., Böing, A. N., Harrison, P., Sturk, A., and Nieuwland, R. (2012). Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 64 (3), 676–705. doi:10.1124/pr.112.005983
van Niel, G., D'Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. cell Biol. 19 (4), 213–28. doi:10.1038/nrm.2017.125
Van Roosbroeck, K., Fanini, F., Setoyama, T., Ivan, C., Rodriguez-Aguayo, C., Fuentes-Mattei, E., et al. (2017). Combining Anti-Mir-155 with chemotherapy for the treatment of lung cancers. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 23 (11), 2891–904. doi:10.1158/1078-0432.CCR-16-1025
Vella, L. J., Behren, A., Coleman, B., Greening, D. W., Hill, A. F., and Cebon, J. (2017). “Intercellular resistance to BRAF inhibition can be mediated by extracellular vesicle-associated PDGFRβ,”, 19. New York, NY, 932–40. doi:10.1016/j.neo.2017.07.002
Walther, W., Kobelt, D., Bauer, L., Aumann, J., and Stein, U. (2015). Chemosensitization by diverging modulation by short-term and long-term TNF-α action on ABCB1 expression and NF-κB signaling in Colon cancer. Int. J. Oncol. 47 (6), 2276–85. doi:10.3892/ijo.2015.3189
Wang, Y., Zhao, Y., Herbst, A., Kalinski, T., Qin, J., Wang, X., et al. (2016). miR-221 mediates chemoresistance of esophageal adenocarcinoma by direct targeting of DKK2 expression. Ann. Surg. 264 (5), 804–14. doi:10.1097/SLA.0000000000001928
Wang, W. T., Han, C., Sun, Y. M., Chen, T. Q., and Chen, Y. Q. (2019). Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. and Oncol. 12 (1), 55. doi:10.1186/s13045-019-0748-z
Wang, D., Zeng, T., Lin, Z., Yan, L., Wang, F., Tang, L., et al. (2020a). Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 123, 109802. doi:10.1016/j.biopha.2019.109802
Wang, C., Guan, W., Peng, J., Chen, Y., Xu, G., and Dou, H. (2020b). Gene/Paclitaxel co-delivering nanocarriers prepared by framework-induced self-assembly for the inhibition of highly drug-resistant tumors. Acta biomater. 103, 247–58. doi:10.1016/j.actbio.2019.12.015
Wang, X., Wang, Y., Xie, M., Ma, S., Zhang, Y., Wang, L., et al. (2024). Hypermethylation of CDKN2A CpG island drives resistance to PRC2 inhibitors in SWI/SNF loss-of-function tumors. Cell death and Dis. 15 (11), 794. doi:10.1038/s41419-024-07109-3
Wang, Y., Pei, W., Yang, Y., Xia, C., Zhang, Q., Geng, Z., et al. (2025). Inhibition of XIST restrains paclitaxel resistance in breast cancer cells by targeting hsa-let-7d-5p/ATG16L1 through regulation of autophagy. Cell. Signal. 127, 111534. doi:10.1016/j.cellsig.2024.111534
Wei, X., Liu, S., Cao, Y., Wang, Z., and Chen, S. (2023). Polymers in engineering extracellular vesicle mimetics: current status and prospective. Pharmaceutics 15 (5), 1496. doi:10.3390/pharmaceutics15051496
Williams, C. S., Li, X., Jang, H., Anand, J. R., Lim, W. Y., Lee, H., et al. (2024). “Inhibition of androgen receptor exposes replication stress vulnerability in prostate cancer,” in bioRxiv: the preprint server for biology.
Xia, M., Zu, X., Chen, Z., Wen, G., and Zhong, J. (2021). Noncoding RNAs in triple negative breast cancer: mechanisms for chemoresistance. Cancer Lett. 523, 100–10. doi:10.1016/j.canlet.2021.09.038
Xie, Y., Wang, L., Freywald, A., Qureshi, M., Chen, Y., and Xiang, J. (2013). A novel T cell-based vaccine capable of stimulating long-term functional CTL memory against B16 melanoma via CD40L signaling. Cell. and Mol. Immunol. 10 (1), 72–7. doi:10.1038/cmi.2012.37
Xie, B., Wu, P., Liu, H., Yang, X., and Huang, L. (2025). Long non-coding RNA MIR4435-2HG modulates pancreatic cancer stem cells and chemosensitivity to gemcitabine by targeting the miR-1252-5p/STAT1. J. Transl. Med. 23 (1), 165. doi:10.1186/s12967-025-06128-8
Xiong, X., Lai, X., Zhang, J., Meng, Q., Wang, P., Qin, S., et al. (2022). FBP1 knockdown decreases ovarian cancer formation and cisplatin resistance through EZH2-mediated H3K27me3. Biosci. Rep. 42 (9). doi:10.1042/BSR20221002
Xu, J., Su, Y., Xu, A., Fan, F., Mu, S., Chen, L., et al. (2019). miR-221/222-Mediated inhibition of autophagy promotes dexamethasone resistance in multiple myeloma. Mol. Ther. J. Am. Soc. Gene Ther. 27 (3), 559–70. doi:10.1016/j.ymthe.2019.01.012
Xu, Z., Li, X., Li, H., Nie, C., Liu, W., Li, S., et al. (2020a). Suppression of DDX39B sensitizes ovarian cancer cells to DNA-Damaging chemotherapeutic agents via destabilizing BRCA1 mRNA. Oncogene 39 (47), 7051–62. doi:10.1038/s41388-020-01482-x
Xu, X., Zhang, J., Tian, Y., Gao, Y., Dong, X., Chen, W., et al. (2020b). CircRNA inhibits DNA damage repair by interacting with host gene. Mol. cancer 19 (1), 128. doi:10.1186/s12943-020-01246-x
Xu, X., Peng, Q., Ren, Z., Han, Y., Jiang, X., Wu, Z., et al. (2025). CircRNF13 enhances IGF2BP1 phase separation-mediated ITGB1 mRNA stabilization in an m6A-dependent manner to promote oral cancer cisplatin chemoresistance. Mol. cancer 24 (1), 36. doi:10.1186/s12943-025-02239-4
Xue, Z., Liu, J., Xing, W., Mu, F., Wu, Y., Zhao, J., et al. (2024). Hypoxic glioma-derived exosomal miR-25-3p promotes macrophage M2 polarization by activating the PI3K-AKT-mTOR signaling pathway. J. nanobiotechnology 22 (1), 628. doi:10.1186/s12951-024-02888-5
Yaddanapudi, K., Meng, S., Whitt, A. G., Al Rayyan, N., Richie, J., Tu, A., et al. (2019). Exosomes from GM-CSF expressing embryonic stem cells are an effective prophylactic vaccine for cancer prevention. Oncoimmunology 8 (3), 1561119. doi:10.1080/2162402X.2018.1561119
Yamagishi, M., Kuze, Y., Kobayashi, S., Nakashima, M., Morishima, S., Kawamata, T., et al. (2024). Mechanisms of action and resistance in histone methylation-targeted therapy. Nature 627 (8002), 221–8. doi:10.1038/s41586-024-07103-x
Yan, Y. Y., Deng, Z. F., Wu, X. T., Lu, Y., Zhu, Z. Y., Wen, Q., et al. (2025a). Low miR-224-5p in exosomes confers colorectal cancer 5-FU resistance by upregulating S100A4. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 79, 101211. doi:10.1016/j.drup.2025.101211
Yan, B., Lu, Q., Gao, T., Xiao, K., Zong, Q., Lv, H., et al. (2025b). CD146 regulates the stemness and chemoresistance of hepatocellular carcinoma via JAG2-NOTCH signaling. Cell death and Dis. 16 (1), 150. doi:10.1038/s41419-025-07470-x
Yang, E., Wang, X., Gong, Z., Yu, M., Wu, H., and Zhang, D. (2020a). Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 5 (1), 242. doi:10.1038/s41392-020-00359-5
Yang, W. B., Hsu, C. C., Hsu, T. I., Liou, J. P., Chang, K. Y., Chen, P. Y., et al. (2020b). Increased activation of HDAC1/2/6 and Sp1 underlies therapeutic resistance and tumor growth in glioblastoma. Neuro-oncology 22 (10), 1439–51. doi:10.1093/neuonc/noaa103
Yang, M., Li, J., Gu, P., and Fan, X. (2021a). The application of nanoparticles in cancer immunotherapy: targeting tumor microenvironment. Bioact. Mater. 6 (7), 1973–87. doi:10.1016/j.bioactmat.2020.12.010
Yang, C., Dou, R., Wei, C., Liu, K., Shi, D., Zhang, C., et al. (2021b). Tumor-derived exosomal microRNA-106b-5p activates EMT-Cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. J. Am. Soc. Gene Ther. 29 (6), 2088–107. doi:10.1016/j.ymthe.2021.02.006
Yang, C., Li, Z., Tian, K., Meng, X., Wang, X., Song, D., et al. (2024a). LncRNA-Mediated TPI1 and PKM2 promote self-renewal and chemoresistance in GBM. Adv. Sci. Weinheim, Baden-Wurttemberg, Ger. 11 (44), e2402600. doi:10.1002/advs.202402600
Yang, X., Liu, J., Liu, W., Wu, H., Wei, Y., Guo, X., et al. (2024b). circFAM193B interaction with PRMT6 regulates AML leukemia stem cells chemoresistance through altering the oxidative metabolism and lipid peroxidation. Leukemia 38 (5), 1057–71. doi:10.1038/s41375-024-02189-8
Yang, E., Hong, B., Wang, Y., Wang, Q., Zhao, J., Cui, X., et al. (2024c). EPIC-0628 abrogates HOTAIR/EZH2 interaction and enhances the temozolomide efficacy via promoting ATF3 expression and inhibiting DNA damage repair in glioblastoma. Cancer Lett. 588, 216812. doi:10.1016/j.canlet.2024.216812
Yue, M., Hu, S., Sun, H., Tuo, B., Jia, B., Chen, C., et al. (2023). Extracellular vesicles remodel tumor environment for cancer immunotherapy. Mol. cancer 22 (1), 203. doi:10.1186/s12943-023-01898-5
Zeng, F., Wang, Q., Wang, S., Liang, S., Huang, W., Guo, Y., et al. (2020). Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate etk expression. Oncogene 39 (2), 293–307. doi:10.1038/s41388-019-0984-2
Zhan, Q., Yi, K., Cui, X., Li, X., Yang, S., Wang, Q., et al. (2022). Blood exosomes-based targeted delivery of cPLA2 siRNA and metformin to modulate glioblastoma energy metabolism for tailoring personalized therapy. Neuro-oncology 24 (11), 1871–83. doi:10.1093/neuonc/noac071
Zhang, Y., Wu, X. H., Luo, C. L., Zhang, J. M., He, B. C., and Chen, G. (2010). Interleukin-12-anchored exosomes increase cytotoxicity of T lymphocytes by reversing the JAK/STAT pathway impaired by tumor-derived exosomes. Int. J. Mol. Med. 25 (5), 695–700. doi:10.3892/ijmm_00000393
Zhang, J., Li, S., Li, L., Li, M., Guo, C., Yao, J., et al. (2015). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics, proteomics and Bioinforma. 13 (1), 17–24. doi:10.1016/j.gpb.2015.02.001
Zhang, Y., Li, C., Liu, X., Wang, Y., Zhao, R., Yang, Y., et al. (2019). circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine 48, 277–88. doi:10.1016/j.ebiom.2019.09.051
Zhang, Y., Bi, J., Huang, J., Tang, Y., Du, S., and Li, P. (2020a). Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int. J. nanomedicine 15, 6917–34. doi:10.2147/IJN.S264498
Zhang, X., Xie, K., Zhou, H., Wu, Y., Li, C., Liu, Y., et al. (2020b). Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. cancer 19 (1), 47. doi:10.1186/s12943-020-01171-z
Zhang, L., Li, Y., Hu, C., Chen, Y., Chen, Z., Chen, Z. S., et al. (2022a). CDK6-PI3K signaling axis is an efficient target for attenuating ABCB1/P-gp mediated multi-drug resistance (MDR) in cancer cells. Mol. cancer 21 (1), 103. doi:10.1186/s12943-022-01524-w
Zhang, W., Zhong, W., Wang, B., Yang, J., Yang, J., Yu, Z., et al. (2022b). ICAM-1-mediated adhesion is a prerequisite for exosome-induced T cell suppression. Dev. cell 57 (3), 329–43.e7. doi:10.1016/j.devcel.2022.01.002
Zhang, L. S., Chen, Q. C., Zong, H. T., and Xia, Q. (2024a). Exosome miRNA-203 promotes M1 macrophage polarization and inhibits prostate cancer tumor progression. Mol. Cell. Biochem. 479 (9), 2459–70. doi:10.1007/s11010-023-04854-5
Zhang, X., Taylor, H., Valdivia, A., Dasari, R., Buckley, A., Bonacquisti, E., et al. (2024b). Auto-loaded TRAIL-Exosomes derived from induced neural stem cells for brain cancer therapy. J. Control. release official J. Control. Release Soc. 372, 433–45. doi:10.1016/j.jconrel.2024.06.048
Zhang, K., Guo, L., Li, X., Hu, Y., and Luo, N. (2025a). Cancer-associated fibroblasts promote doxorubicin resistance in triple-negative breast cancer through enhancing ZFP64 histone lactylation to regulate ferroptosis. J. Transl. Med. 23 (1), 247. doi:10.1186/s12967-025-06246-3
Zhang, W., Zhu, Z., and Liu, Y. (2025b). The impact of the ATP-Binding cassette (ABC) transporter family on multidrug resistance in head and neck tumors. Mol. Biol. Rep. 52 (1), 256. doi:10.1007/s11033-025-10321-9
Zhao, Z., McGill, J., Gamero-Kubota, P., and He, M. (2019). Microfluidic on-demand engineering of exosomes towards cancer immunotherapy. Lab a chip 19 (10), 1877–86. doi:10.1039/c8lc01279b
Zhao, S., Mi, Y., Guan, B., Zheng, B., Wei, P., Gu, Y., et al. (2020). Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. and Oncol. 13 (1), 156. doi:10.1186/s13045-020-00991-2
Zhao, L., Wang, H., Fu, J., Wu, X., Liang, X. Y., Liu, X. Y., et al. (2022). Microfluidic-based exosome isolation and highly sensitive aptamer exosome membrane protein detection for lung cancer diagnosis. Biosens. and Bioelectron. 214, 114487. doi:10.1016/j.bios.2022.114487
Zhao, J., Cui, X., Zhan, Q., Zhang, K., Su, D., Yang, S., et al. (2024). CRISPR-Cas9 library screening combined with an exosome-targeted delivery system addresses tumorigenesis/TMZ resistance in the mesenchymal subtype of glioblastoma. Theranostics 14 (7), 2835–55. doi:10.7150/thno.92703
Zheng, F., and Xu, R. (2020). CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145-5p/ABCC1 axis. Biomed. and Pharmacother. = Biomedecine and Pharmacother. 124, 109828. doi:10.1016/j.biopha.2020.109828
Zhong, W., Lu, Y., Han, X., Yang, J., Qin, Z., Zhang, W., et al. (2023). Upregulation of exosome secretion from tumor-associated macrophages plays a key role in the suppression of anti-tumor immunity. Cell Rep. 42 (10), 113224. doi:10.1016/j.celrep.2023.113224
Zhu, Q., Ling, X., Yang, Y., Zhang, J., Li, Q., Niu, X., et al. (2019). Embryonic stem cells-derived exosomes endowed with targeting properties as chemotherapeutics delivery vehicles for glioblastoma therapy. Adv. Sci. Weinheim, Baden-Wurttemberg, Ger. 6 (6), 1801899. doi:10.1002/advs.201801899
Zhu, L., Sun, H. T., Wang, S., Huang, S. L., Zheng, Y., Wang, C. Q., et al. (2020). Isolation and characterization of exosomes for cancer research. J. Hematol. and Oncol. 13 (1), 152. doi:10.1186/s13045-020-00987-y
Zhu, S., Huang, H., Liu, D., Wen, S., Shen, L., and Lin, Q. (2022a). Augmented cellular uptake and homologous targeting of exosome-based drug loaded IOL for posterior capsular opacification prevention and biosafety improvement. Bioact. Mater. 15, 469–81. doi:10.1016/j.bioactmat.2022.02.019
Zhu, Z., Zhai, Y., Hao, Y., Wang, Q., Han, F., Zheng, W., et al. (2022b). Specific anti-glioma targeted-delivery strategy of engineered small extracellular vesicles dual-functionalised by Angiopep-2 and TAT peptides. J. Extracell. vesicles 11 (8), e12255. doi:10.1002/jev2.12255
Zhu, C., Xie, Y., Li, Q., Zhang, Z., Chen, J., Zhang, K., et al. (2023). CPSF6-mediated XBP1 3'UTR shortening attenuates cisplatin-induced ER stress and elevates chemo-resistance in lung adenocarcinoma. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 68, 100933. doi:10.1016/j.drup.2023.100933
Zhu, Y., Liang, L., Zhao, Y., Li, J., Zeng, J., Yuan, Y., et al. (2024). CircNUP50 is a novel therapeutic target that promotes cisplatin resistance in ovarian cancer by modulating p53 ubiquitination. J. Nanobiotechnology 22 (1), 35. doi:10.1186/s12951-024-02295-w
Zocchi, M. R., Tosetti, F., Benelli, R., and Poggi, A. (2020). Cancer nanomedicine special issue review anticancer drug delivery with nanoparticles: extracellular vesicles or synthetic nanobeads as therapeutic tools for conventional treatment or immunotherapy. Cancers 12 (7), 1886. doi:10.3390/cancers12071886
Keywords: engineered exosome, immunotherapy resistance, chemotherapy resistance, targeted therapy resistance, precise treatment
Citation: Zhang P, Chen K, Liu W, Niu X, Wang X, Wang J, Yao W, Tang X and Tian W (2025) Engineered exosomes: a promising design platform for overcoming cancer therapy resistance. Front. Cell Dev. Biol. 13:1608480. doi: 10.3389/fcell.2025.1608480
Received: 09 April 2025; Accepted: 21 July 2025;
Published: 06 August 2025.
Edited by:
Yu Zhao, Xuanwu Hospital, Capital Medical University, ChinaReviewed by:
Giulia Grisendi, University of Modena and Reggio Emilia, ItalyKanayo Ikeh, University of Vermont, United States
Copyright © 2025 Zhang, Chen, Liu, Niu, Wang, Wang, Yao, Tang and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Tang, dGFuZzE1ODc3QDE2My5jb20=; Wen Tian, dGlhbndlbjQ3NDRAenp1LmVkdS5jbg==
 Peng Zhang1
Peng Zhang1 Jiaqiang Wang
Jiaqiang Wang Wen Tian
Wen Tian