- 1The First School of Clinical Medicine, Lanzhou University, Lanzhou, China
- 2Department of Urology, The First People’s Hospital of Lanzhou City, Lanzhou, China
- 3West China School of Medicine, Sichuan University, Chengdu, China
- 4Department of laboratory, The First Hospital of Lanzhou University, Lanzhou, China
- 5Department of General Surgery, The First Hospital of Lanzhou University, Lanzhou, China
Cholangiocarcinoma is a highly heterogeneous malignant tumor, including intrahepatic cholangiocarcinoma, hepatoportal cholangiocarcinoma and distal cholangiocarcinoma. Its incidence is increasing worldwide and currently accounts for approximately 15% of all primary liver cancers and 3% of all gastrointestinal malignancies. There is a lack of early diagnostic methods for cholangiocarcinoma, and the overall treatment effect is poor, with a 5-year survival rate of less than 25%. New biomarkers are urgently needed in clinical practice to improve the current diagnosis and treatment status. Circulating tumor DNA (ctDNA) is DNA fragments released by tumor cells, which can show tumor-specific gene mutations (such as IDH1/2, FGFR2 fusion) and epigenetic modifications (such as abnormal methylation). With the rapid development of tumor liquid biopsy technology, ctDNA has been gradually applied in solid tumors such as lung cancer and colorectal cancer due to its high sensitivity and dynamic monitoring capabilities. This review systematically introduces ctDNA technology and its progress in early screening, early diagnosis, treatment response, and prognosis monitoring of cholangiocarcinoma. In addition, this review also summarizes the challenges and limitations of current ctDNA technology and analyzes future hot research directions.
1 Background
Cholangiocarcinoma (CCA) is a highly heterogeneous malignant tumor originating from the bile duct epithelium, and its pathological type is mostly adenocarcinoma. CCA can be divided into intrahepatic cholangiocarcinoma (iCCA) and extrahepatic cholangiocarcinoma (eCCA) according to the anatomical location. ECCA includes hilar cholangiocarcinoma (hCCA) and distal cholangiocarcinoma (dCCA). There are significant differences in the treatment strategies and prognosis of CCA in different anatomical locations (Valle et al., 2021). In recent years, the incidence of CCA has continued to increase, accounting for about 3% of gastrointestinal malignancies. Among them, iCCA is the second most common primary liver cancer after hepatocellular carcinoma, accounting for about 15% (Banales et al., 2020). There are significant regional differences in the incidence of CCA, with Asia having the highest incidence of CCA worldwide. The age-standardized incidence is highest in northeastern Thailand (85 cases/100,000), followed by Gwangju, South Korea (8.8 cases/100,000). The incidence in the West is lower than in Asia, with the highest incidence in Italy (3.4 cases/100,000) (Khan et al., 2019; Qurashi et al., 2025). The number of biliary tract cancer (BTC) cases worldwide has increased by 84.8% in the past 20 years (1990–2019), and the number of new cases in China has increased by 211% (Chen et al., 2022; Su et al., 2024). As the population ages, the incidence of BTC in China is expected to continue to rise in the next decade. In addition, CCA is highly malignant and lacks specific early detection methods. Most patients are already in the late stage when diagnosed, missing the opportunity for surgical treatment (Moss et al., 2007). The prognosis of CCA is poor, with a 5-year overall survival rate (OS) of less than 25% (Kamsa-Ard et al., 2020).
Tumor liquid biopsy, as an emerging minimally invasive sampling and detection method, focuses on detecting blood or body secretions, such as tumor cells, molecules and metabolites. Compared with traditional tissue biopsy, liquid biopsy has the advantages of simple operation and minimal invasiveness, which significantly improves patient acceptance and examination feasibility (Ma et al., 2024). As emerging liquid biopsy biomarkers, cell free DNA (cfDNA) and circulating tumor DNA (ctDNA) have shown great potential in cancer treatment and monitoring. ctDNA is a tumor-derived DNA fragment released from tumor cells through apoptosis, necrosis or active secretion, which can reflect the genomic and epigenomic characteristics of the tumor (Diehl et al., 2008). The dynamic changes of its concentration have been proven to be closely related to tumor burden and microenvironment characteristics, and it is more suitable for tumor efficacy evaluation than protein markers such as CA19-9 (Heitzer et al., 2019). With the advancement of technologies such as real-time quantitative PCR (rt-qPCR), digital droplet PCR (ddPCR), Sanger sequencing, and next-generation sequencing (NGS), it is expected that the detection of gene mutations and abnormal DNA methylation in ctDNA will replace tumor pathological biopsy and tumor marker detection (Ståhlberg et al., 2017; Hannigan et al., 2019; Mody et al., 2019; Olmedillas-López et al., 2022).
Currently, the application of ctDNA in CCA is mostly limited to small sample exploratory studies and lacks standardized testing procedures. This review will systematically review the scientific research evidence on the early diagnosis, efficacy response and prognosis detection of CCA for the first time, and explore the prospects and research hotspots of ctDNA in the clinical diagnosis and treatment of CCA.
2 The biological origin, molecular characteristics and detection methods of ctDNA
ctDNA is a circulating free DNA (cfDNA) that is released into biological fluids by cancer cells through apoptosis, necrosis, or active release (Normanno et al., 2025). cfDNA in normal humans is mainly derived from white blood cells and stromal cells and is rapidly cleared within a few minutes to 1–2 h (Sun et al., 2015; Li et al., 2023). On the one hand, compared with cfDNA, the base fragments of ctDNA from cancer patients are about 20–50 base pairs, which is significantly shorter than cfDNA from normal cells (Underhill et al., 2016). This feature makes it more stable than cfDNA, and ctDNA can still provide reliable monitoring data when tumor heterogeneity is large. On the other hand, ctDNA has a short half-life (about 15 min) and can be used as a real-time tumor marker to quickly reflect the dynamic changes of the tumor (Ma et al., 2024). It is these two characteristics of ctDNA that give it obvious advantages over traditional biopsy markers (Figure 1 Mechanism of ctDNA release).
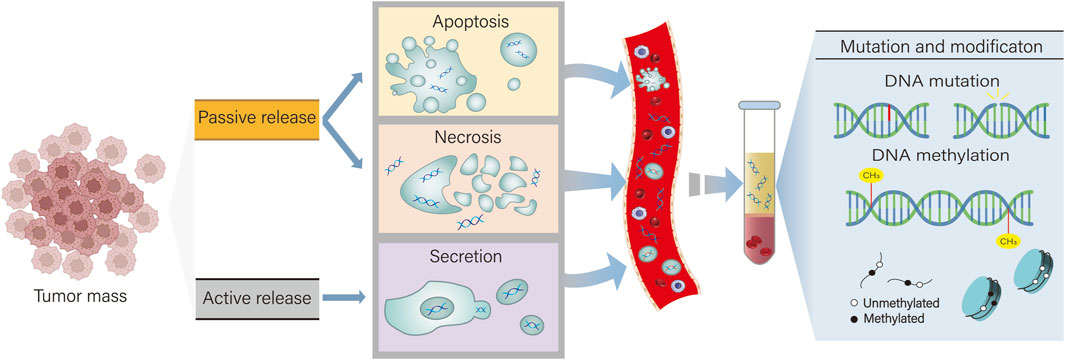
Figure 1. ctDNA has two types of release: active secretion and passive release. Active secretion refers to the secretion of ctDNA by tumor cells via exosomes or microvesicles. Passive release of ctDNA occurs when cells undergo apoptosis and necrosis. By detecting DNA mutations and abnormal methylation in ctDNA, it can assist in the diagnosis and treatment of cholangiocarcinoma.
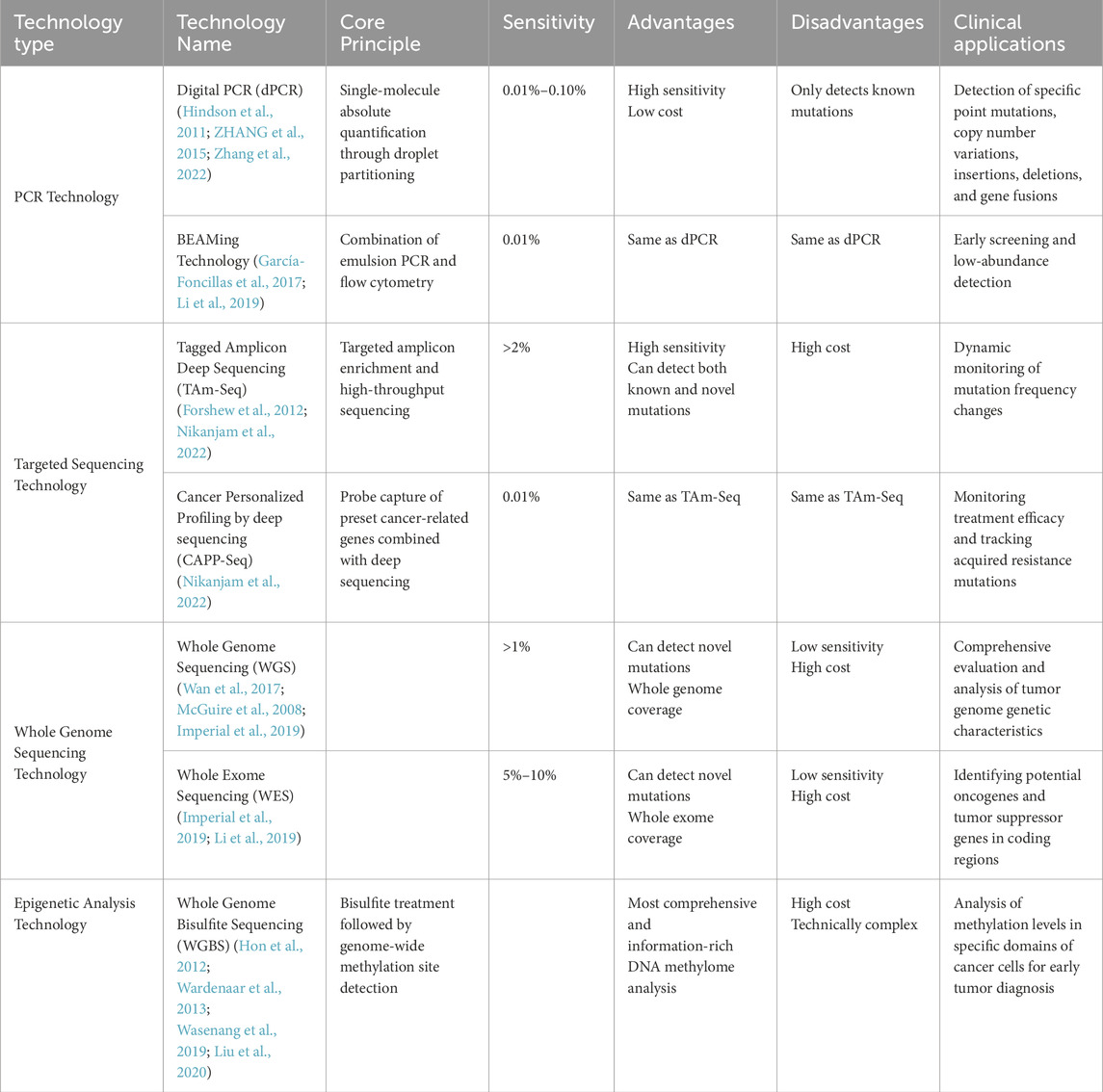
Currently, ctDNA detection methods mainly include mutation detection based on digital PCR (dPCR), next-generation sequencing (NGS), and whole genome sequencing (WGS), etc. These methods have their own advantages and disadvantages, but all have the potential to be used for early monitoring of tumors, real-time detection of dynamic changes, and monitoring of disease progression (Table 1 Comparison of methods for ctDNA detection).
In patients who obtain tissue specimens, the combined use of multiple detection methods to perform tumor-informed ctDNA testing can further improve the ctDNA detection rate. Tumor-informed ctDNA testing refers to the development of a custom assay panel based on the patient’s tumor tissue sequencing results (such as WES/WGS) to perform more in-depth sequencing of known mutations in the tumor. Research results in non-small cell lung cancer, pancreatic cancer, colorectal cancer, and breast cancer show that tumor-informed ctDNA detection technology can significantly increase the chances of capturing and identifying ctDNA fragments carrying targeted mutations, achieving high-sensitivity detection at low ctDNA concentration levels, and is particularly suitable for minimal residual disease (MRD) monitoring and recurrence warning (Watanabe et al., 2022; Chen et al., 2023; Santonja et al., 2023; Nesic et al., 2024; Sasaki et al., 2025). Currently, there are few reports on this technology in CCA.
Tumor-naive ctDNA testing does not rely on tissue specimens and is used for preliminary screening or in scenarios where tissue is unavailable. It has the advantages of being non-invasive, capable of dynamic monitoring, and having a wide detection range. The results of the study showed that tumor-naive ctDNA testing could assist in the early diagnosis of CCA and guide custom treatment selection (such as initial screening of targeted drugs), but the sensitivity depends on the ctDNA concentration, and negative results are more likely to occur when the tumor burden is low (Miura et al., 2024).
3 Characteristics of ctDNA in CCA
In CCA, due to severe tumor interstitial fibrosis and sparse vascular distribution, the efficiency of ctDNA release is significantly lower than that of other solid tumors (such as colorectal cancer). However, its fragment characteristics (such as short fragmentation, and terminal oxidative damage) and epigenetic abnormalities are still highly heterogeneous (Nakamura et al., 2015; Cristiano et al., 2019). The results of ctDNA testing are highly consistent with those of tissue biopsies, with a sensitivity of up to 84.8% in terminal cancer patients (Hwang et al., 2024). Through non-invasive liquid biopsy technology, ctDNA can comprehensively reflect the genomic characteristics of CCA, including high-frequency driver gene mutations, signal pathway abnormalities, and pathological classification information, providing a basis for personalized treatment.
3.1 Common ctDNA mutations and their clinical significance
3.1.1 KRAS
KRAS mutations were more frequently detected in eCCA, with G12D (37.0%), G12V (24.0%), and Q61H (8.2%) being the main sites. KRAS G12/13 mutation is an important prognostic marker for CCA and is closely associated with shortened OS and recurrence-free survival (RFS) (sensitivity 80%, specificity 93%). Meanwhile, ddPCR analysis of plasma ctDNA showed that patients with higher KRAS G12/G13 mutant allele frequency (MAF) and CA19-9 levels (MAF >0.174% and CA19-9 >49.99 U/mL) had a significantly worse prognosis (15.8 vs. 39.0 months; p = 0.046) (Thongyoo et al., 2025). It can be found that ctDNA detection found that KRAS is associated with poor survival in CCA. This approach offers a less invasive diagnostic alternative to traditional biopsy methods and provides critical insights into the potential of cfDNA analysis to act as a predictive tool for patient survival. Although targeted therapies for the KRAS G12C mutation, such as sotolacizumab, have been approved by the FDA, the development of inhibitors targeting other KRAS mutation subtypes is still in its early stages. In addition, studies have shown that inhibition of casein kinase 2 (CK2) may affect metabolic pathways in KRAS mutant CCA, providing a new research direction for targeting KRAS-driven tumor metabolism and is expected to provide a potential new approach for the treatment of CCA (Lee D. S. et al., 2024). In addition, KRAS mutations induce high expression of PD-L1/CTLA-4 by activating the MAPK pathway, forming an immunosuppressive microenvironment, thereby leading to resistance to immunotherapy (Job et al., 2020).
3.1.2 TP53
Homologous recombination repair (HR) pathway gene mutations (such as TP53, BRCA2 and RAD51D) have also been shown to be associated with a poor prognosis in CCA. TP53 mutation is the most common mutation type in CCA, with a detection rate of up to 38.1% (Silverman et al., 2019). Sanger sequencing of bile cfDNA revealed that patients with HR pathway mutations had significantly shorter survival than those without mutations (P = 0.0049), which may be related to the accelerated tumor progression caused by genomic instability caused by DNA damage repair defects (Yin et al., 2024). In addition, the study also found that patients with HR pathway mutations may be more sensitive to PARP inhibitors (such as olaparib), suggesting that it can be used as a biomarker for targeted therapy to improve patient prognosis. However, there are still few clinical trials of PARP inhibitors for CCA, and their efficacy needs to be further verified.
3.1.3 IDH1
It has been reported that approximately 13%–25% of iCCA patients have isocitrate dehydrogenase 1 (IDH1) mutations (Rizzo et al., 2021). Berchuck et al. performed NGS analysis on ctDNA in the blood of 1,671 patients with advanced BTC, and the results showed that 9.1% of patients had IDH1 mutations. Among patients with IDH1 mutations detected in tissues, 87% (41/47) of patients also had these mutations detected in ctDNA. This shows that IDH1 mutations have high tissue consistency and can be used for targeted therapy (Berchuck et al., 2022). Ivosidenib, an oral, selective mutant IDH1 inhibitor, has shown promising results in a Phase III trial, significantly improving progression-free survival (PFS) and OS in advanced CCA (Abou-Alfa et al., 2020a). Resistance is particularly evident in IDH1-targeted therapies and may be related to acquired resistance mutations in IDH1 or IDH2 that prevent ivosidenib from binding to its target site (Cleary et al., 2022).
3.1.4 FGFR
The incidence of rearrangement or fusion changes (3.5%) was higher than that of amplification (2.6%), and mutation events were rare (0.9%) (Helsten et al., 2016). Alberto et al. conducted a retrospective study of 18 iCCA patients who were found to have FGFR2 fusions. Plasma samples were analyzed using a custom hybrid capture gene panel with NGS (VHIO-iCCA panel). The results showed that patients in the high ctDNA group had a worse prognosis than those in the low ctDNA group (median PFS 6.53 months vs. 13.3 months, P = 0.0018; median OS 10.6 months vs. 21.2 months, P = 0.0198). In addition, 16 patients (88.9%) were found to have positive FGFR2 fusion events in plasma ctDNA, proving that VHIO-iCCA can accurately detect FGFR2 fusion in plasma ctDNA, thereby quickly screening patients who benefit better from targeted therapy. (González-Medina et al., 2024). A variety of FGFR receptor inhibitors have been developed, such as infigratinib, futibatinib, pemigatinib, erdafitinib and derazantinib, etc., and phase I and phase II trials of these drugs have shown good results (Javle et al., 2018; Goyal et al., 2019; Abou-Alfa et al., 2020b; Bahleda et al., 2019; Mazzaferro et al., 2019). Goyal et al. reported evidence of acquired resistance to FGFR inhibitors in CCA. PCR and Sanger sequencing of plasma ctDNA were performed in three patients receiving FGFR inhibitors, and the results showed that the gatekeeper mutation FGFR2 V564F was found in all patients, suggesting that secondary mutations in the FGFR2 kinase domain are a potential resistance mechanism (Goyal et al., 2017).
4 Clinical application of ctDNA in CCA
CCA is a malignant tumor originating from the bile duct epithelium. Due to its lack of specific clinical symptoms, most patients are diagnosed at an advanced stage (Moss et al., 2007). Advanced CCA has poor treatment efficacy, so early diagnosis can significantly reduce the mortality of CCA and is crucial to improving patient prognosis. In recent years, with the continuous development of biomarkers and detection technologies, early diagnosis methods for CCA have continued to improve (Figure 2 Tumor liquid biopsy for CCA).
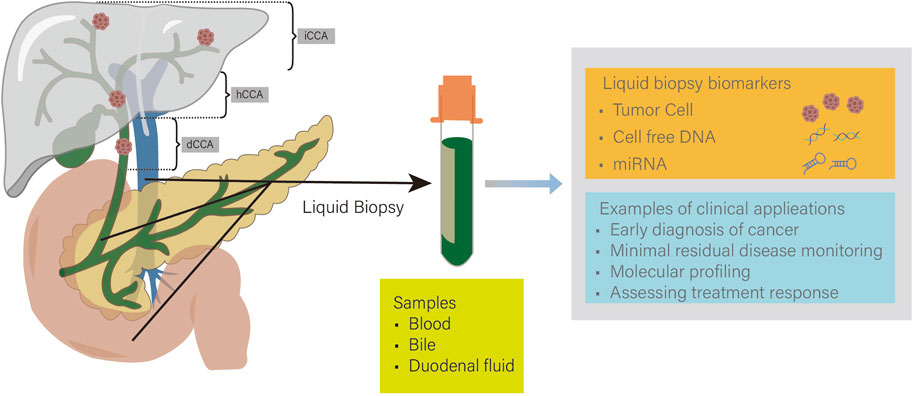
Figure 2. iCCA, intrahepatic cholangiocarcinoma; hCCA, hilar cholangiocarcinoma; dCCA, distal cholangiocarcinoma. Tumor liquid biopsy techniques are widely used in cholangiocarcinoma (CCA). Common samples include blood, bile, and duodenal fluid. Detection of tumor cells, cell free DNA (cfDNA), and microRNA (miRNA) can enable early diagnosis and personalized treatment of CCA.
4.1 Early diagnosis and screening of CCA
4.1.1 Traditional diagnostic methods
Imaging examinations (ultrasound/CT/MRI) are still the first-line tools for CCA screening. Common manifestations include bile duct obstruction, dilatation, and masses. They can evaluate tumor morphology (such as local invasion, vascular encapsulation) and metastasis status (Khan et al., 2012; Rushbrook et al., 2024). However, imaging methods have low sensitivity for small tumors and early lesions, and can only provide structural information of the tumor, but cannot reveal the molecular biological characteristics of the tumor and pathological changes at the cellular level (European Association for the Study of the Liver, 2023). Endoscopic examination techniques such as endoscopic ultrasound (EUS), magnetic resonance cholangiopancreatography (MRCP) and endoscopic retrograde cholangiopancreatography (ERCP) can directly observe bile duct lesions, detect early lesions and tiny tumors, and can obtain pathological information when combined with tissue biopsy. For example, ERCP combined with brush cytology can obtain tissue or cells, but its sensitivity is low (45%), so negative cytology does not exclude the diagnosis of CCA (Navaneethan et al., 2015). However, endoscopic examination is greatly affected by the quality of tissue samples and the sampling location, and has low sensitivity. In addition, the entry of the endoscope into the bile duct may lead to related complications, such as pancreatitis and bleeding (Tamada et al., 2011). Pathological biopsy is still the gold standard for diagnosing bile duct cancer. Pathological diagnosis can be performed directly through brush cytology, fine needle puncture or percutaneous methods. However, tissue specimens are difficult to obtain and can only reflect information about the sampling site, which is greatly affected by sample heterogeneity. Moreover, as an invasive examination, it is easy to cause harm to patients and is not convenient for continuous monitoring of disease progression (Vaidyanathan et al., 2018).
Compared with the above traditional methods, liquid biopsy is a minimally invasive sample collection method, especially suitable for patients or high-risk groups who cannot obtain tissue samples (Heitzer et al., 2019). Carbohydrate antigen 19–9 (CA19-9) is currently the most commonly used serum tumor marker for CCA diagnosis. Its false positive rate in patients with biliary obstruction exceeds 50%, and Lewis antigen-negative individuals (about 10% of the population) cannot secrete CA19-9, which seriously limits its universality (Ballehaninna and Chamberlain, 2012; Liang et al., 2015; Luo et al., 2016). The American Association for the Study of Liver Diseases and the American College of Gastroenterology both pointed out that the clinical application of CA19-9 faces limitations in CCA (European Association for the Study of the Liver, 2022; Boberg et al., 2006; Levy et al., 2005; Sinakos et al., 2011; Lindor et al., 2015; Bowlus et al., 2023).
4.1.2 The potential of ctDNA as a non-invasive early diagnostic tool
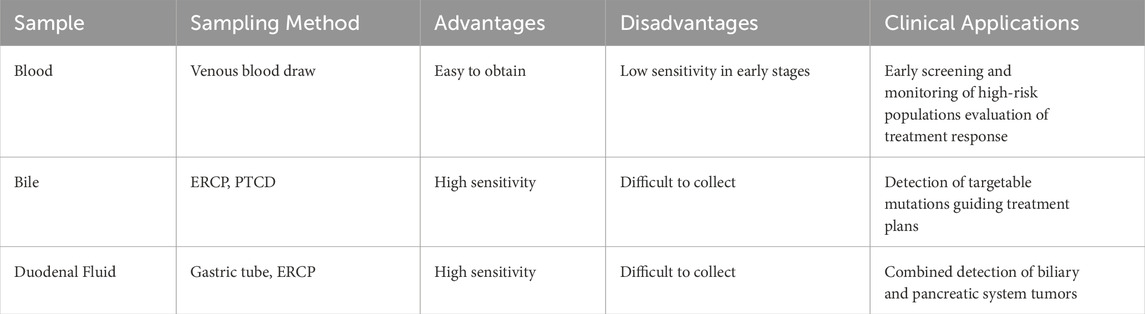
In recent years, ctDNA has demonstrated significant advantages in the early diagnosis, disease monitoring and prognosis assessment of CCA due to its non-invasive, real-time monitoring, and dynamic change monitoring. Genetic and epigenetic changes in ctDNA are closely related to changes in tumor tissue, which lay the foundation for the application of ctDNA in CCA (Ettrich et al., 2019). Currently, cfDNA detection of CCA has been achieved through multiple sample sources such as blood, bile, and duodenal fluid (Table 2 Comparison of liquid biopsy specimens for CCA).
Blood ctDNA: Multiple studies have confirmed that blood ctDNA is as effective as traditional tissue genomic analysis and can be used as a marker for the diagnosis of CCA (Wasenang et al., 2019; Yang J. D. et al., 2021; Awosika et al., 2024). Hwang S et al. showed that the ctDNA in the blood of patients with BTC was consistent with the gene profile of tumor tissue, with a sensitivity of 84.8% and a positive predictive value (PPV) of 79.4% (Hwang et al., 2024).
Bile cfDNA: As the environment for the growth of bile duct tumor cells, bile is rich in tumor markers released by tumor cells through paracrine or autocrine pathways, making the detection of bile tumor markers an important means for early diagnosis of bile duct cancer and monitoring of disease progression (Shu et al., 2024; Liu et al., 2023). Multiple studies have reported that the mutation spectrum of bile cfDNA (such as TP53, KRAS, IDH1) is highly consistent with that in tumor tissue (P < 0.001) (Shen et al., 2019; Arechederra et al., 2022; Yin et al., 2024). By combining machine learning to optimize variant screening methods, the sensitivity of bile cfDNA detection can be increased by 2 times (Ito et al., 2024). Moreover, the content of bile cfDNA is much higher than that of plasma ctDNA, reaching 68.2 times that of plasma. The detection rate of driver mutations (bile 54% vs. plasma 17%) and sensitivity (96.2% vs. 31.6%) are significantly higher than those in plasma (Driescher et al., 2020; Arrichiello et al., 2022; Ito et al., 2024; Yin et al., 2024). Although bile sample acquisition is more technically demanding than blood sampling, biliary drainage constitutes an integral part of the initial therapeutic protocol for CCA patients with obstructive jaundice. Consequently, bile cfDNA analysis is particularly suitable for patients with biliary tract obstruction (Kearney et al., 2023; Shu et al., 2024).
Duodenal fluid ctDNA: Duodenal fluid (DF) contains components from the bile duct, pancreatic duct, gastric juice, and intestinal juice, and is mainly affected by pancreatic juice and bile. A study on DF analysis found that the cfDNA concentration of DF was significantly higher than that of plasma cfDNA, and it was more advantageous than plasma cfDNA in detecting low-abundance variants, and could reflect the overall microenvironment of the pancreatic and biliary system (González-Medina et al., 2024; Tavano et al., 2024). Compared with the precise detection of CCA using bile cfDNA, DF is more suitable for the combined detection of biliary and pancreatic system tumors.
In addition to detecting specific mutations in ctDNA, cancer type-specific methyl groups can also be found (Khosla et al., 2024). Wasenang et al. used a qPCR-based methylation-sensitive high-resolution melting (MS-HRM) method and found that hypermethylation of OPCML, HOXA9, and HOXD9 genes in serum ctDNA can effectively distinguish CCA from benign biliary diseases (such as gallstones), with a sensitivity and specificity of 62.5% and 100%, respectively (Wasenang et al., 2019). The NGS-based targeted methylation detection system developed by the Circulating Cell-free Genome Atlas (CCGA; NCT02889978) Consortium has validated the diagnostic value of cfDNA methylation patterns in a multi-cancer cohort study covering CCA (Liu et al., 2020; 2018). This technology can simultaneously analyze hundreds of methylation sites, significantly improving the detection throughput. Yang et al. found that the sensitivity of reduced-representation bisulfite sequencing (RRBS) technology in the early detection of ctDNA methylation combination markers can reach 76% and the specificity is 94% (Yang J. D. et al., 2021). Moreover, among patients with low levels of the traditional marker serum CA19-9 (≤100 U/mL), the test was able to successfully identify 64% of patients who were suitable for transplantation or surgical resection. The above research results show that ctDNA methylation markers can be effectively used as an early screening and identification tool for CCA. In the future, its clinical application can be further broadened through combined detection with other tumor biopsy markers.
In summary, ctDNA and bile cfDNA have great clinical potential for the accurate detection of CCA-related gene mutations. They can not only make up for the shortcomings of tissue biopsy, but provide a more sensitive and accurate detection method for early diagnosis, disease progression monitoring and targeted therapy of CCA.
In the future, the integration of liquid biopsy with imaging genomics and artificial intelligence is expected to achieve full-cycle management of CCA: “Non-invasive early-stage cancer detection, precise classification, and dynamic monitoring”.
4.2 Clinical prognostic assessment of CCA
4.2.1 Current clinical prognosis of CCA
Early-stage CCA lacks specific clinical and diagnostic methods and has a poor prognosis, with a 5-year OS rate of only 3%–24% (Tawarungruang et al., 2021). After receiving the standard treatment of surgery combined with adjuvant therapy, 60%–70% of patients will still experience recurrence, and the 5-year survival rate is usually less than 25% (Brandi et al., 2012; Banales et al., 2016; Blechacz, 2017). Monitoring tools related to CCA prognosis are relatively lacking, and expanding prognostic monitoring methods is crucial to improving the clinical management of CCA.
4.2.2 Clinical value of ctDNA prognostic assessment
Although CA19-9 and carcinoembryonic antigen (CEA) are widely used in the diagnosis and disease monitoring of CCA in clinical practice, their sensitivity and specificity are still low, and their role as independent prognostic markers is limited. In recent years, ctDNA has shown potential as a powerful prognostic biomarker for postoperative MRD in multiple cancer types (Huffman et al., 2022; Kotani et al., 2023; Hou et al., 2022). ctDNA can be used to detect tiny residual lesions early after surgery, thereby providing more accurate disease monitoring and prognosis assessment, and providing a basis for personalized medicine.
ctDNA positivity can serve as a strong predictor of decreased disease-free survival (DFS). Many studies in the CRC field (such as the GALAXY, BESPOKE, and DYNAMIC trials) have demonstrated that the level of ctDNA is closely related to the patient’s prognosis. Patients with persistently positive ctDNA or ctDNA conversion from negative to positive after surgery have a poor prognosis, while patients with ctDNA conversion from positive to negative have a significant survival benefits (Yu et al., 2025). The ctDNA mutation levels of 60 plasma samples were evaluated using NGS, and the results showed that patients with low ctDNA levels had a progression-free survival (PFS) of 12.3 months and an overall survival (OS) of 22.6 months, which were significantly longer than those of patients with high ctDNA levels (PFS = 5.8 months, OS = 9.4 months), suggesting that ctDNA can serve as a predictor of disease burden and treatment response (González-Medina et al., 2024).
The variant allele frequency (VAF) can reflect the degree of tumor clonal advantage. Hwang S et al. confirmed that the median OS of patients with VAF>3.9% in the NGS results of blood ctDNA was only 4.9 months, which was significantly lower than that of patients with VAF≤0.9% (16.4 months, p = 3.8 × 10−7) (Hwang et al., 2024). Allele frequency variance (AFV), as a measure of the uniformity of VAF distribution, can reflect the genomic instability (GI) of tumors. High AFV values are usually associated with greater tumor clonal heterogeneity, high chromosomal instability, and subclonal evolution, features that usually predict a poor prognosis. Liu R et al. proposed that AFV can be used as a new method to evaluate the prognosis of BTC. Patients with higher preoperative AFV levels had poorer OS (p = 0.004), and preoperative AFV showed better predictive value compared with postoperative AFV (Liu et al., 2024).
NGS is a commonly used ctNDA detection method in clinical practice, which can detect low-frequency mutations below 0.1%. At the same time, different panels of NGS can analyze the full spectrum of mutations such as point mutations, insertions and deletions (Indel), copy number variations (CNV), and fusion genes, reducing technical costs. Currently, NGS is widely used in MRD monitoring, prognosis assessment, and drug resistance mechanism research of CCA. However, recent studies have also showed that liver function status may affect NGS plasma ctDNA concentration determination (González-Medina et al., 2024). Studies have found that the NGS plasma ctDNA level in patients with liver dysfunction is 2.1 times higher than that in patients with normal liver function, which may be due to decreased liver metabolic capacity, resulting in reduced ctDNA clearance. Therefore, future studies should consider the impact of liver function on ctDNA load to avoid misjudging disease progression due to liver dysfunction.
4.3 Dynamic monitoring of CCA treatment
4.3.1 Post-operative monitoring
Although radical resection can remove most of the tumor tissue, MRD may still exist, which cannot be detected by conventional imaging examinations. As a sensitive prognostic marker, ctDNA can effectively detect MRD and thus assess the risk of relapse in patients. Postoperative ctDNA detection can be used for early monitoring of CCA recurrence and provide a basis for adjuvant therapy decisions.
Compared with traditional markers and imaging examinations, ctDNA has higher sensitivity in predicting recurrence. In a study of 56 patients with curatively resected stage I-III BTC, blood ctDNA abnormalities were detected in 93.8% of recurrent patients at an average of 3.7 months in advance, and ctDNA had a stronger predictive ability for recurrence than traditional CA19-9 levels (HR = 1.17 [95% CI, 0.24 to 5.71]; P = 0.844) (Yu et al., 2025). A study by Reinert T et al. showed that ctDNA can detect recurrence several months earlier (174–222 days) than imaging examinations in monitoring tumor recurrence (Reinert et al., 2019).
A CCA-based subanalysis of the STAMP Phase II trial showed that ctDNA levels were positively correlated with tumor burden. During postoperative MRD monitoring, plasma ctDNA was evaluated by 16-plex PCR next-generation sequencing, and ctDNA positivity indicated that residual lesions were not completely eliminated and was significantly associated with worse DFS (HR = 1.8; 95% CI 1.06–3.07; p = 0.029) (Yoo Changhoon et al., 2024). During adjuvant chemotherapy, persistently positive ctDNA indicates that the patient will relapse 100% after surgery and their RFS is significantly shortened (Yoo C. et al., 2024).
These results show that ctDNA has a higher sensitivity in predicting recurrence than traditional biomarkers and imaging examinations. With the advancement of larger-scale prospective studies, tumor-specific ctDNA detection is expected to become an important recurrence monitoring biomarker in clinical practice and promote the establishment of a precise postoperative monitoring system.
4.3.2 Chemotherapy and targeted therapy
Most CCA patients are already in the metastatic or locally advanced stage at the time of diagnosis, and chemotherapy (such as gemcitabine/cisplatin regimen) has become the standard treatment for unresectable or recurrent CCA. ctDNA plays an important role in this process. By monitoring the changes in specific gene mutations in ctDNA and the fluctuations in tumor load, it can reflect the patient’s response to treatment in real time. This can help patients optimize follow-up treatment strategies and provide timely personalized and precise treatment. Specifically, a decrease in ctDNA levels after chemotherapy indicates a reduction in tumor burden, indicating that the treatment is effective. If ctDNA turns negative after chemotherapy, it may indicate complete remission (CR) and the patient has a better treatment response. However, patients whose ctDNA levels do not decrease or continue to increase may indicate chemotherapy resistance or disease progression, with a higher risk of recurrence and significantly lower DFS than those whose ctDNA levels decrease. In addition, the study by Hwang S et al. showed that high variant allele frequency in ctDNA was closely associated with poor prognosis after gemcitabine/cisplatin chemotherapy, specifically manifested as a significant shortening of OS and PFS (p = 6.9 × 10−6 and p = 3.8 × 10−7) (Hwang et al., 2024). This study further demonstrated the important role of ctDNA in evaluating chemotherapy effects and prognosis.
Molecular targeted therapy and immunotherapy have gradually become the core of personalized treatment for CCA, especially in patients with specific gene mutations (such as FGFR2, IDH1, MSI-H, TMB-H) or ERBB2 gene amplification (Hwang et al., 2024). Currently, targeted therapy is widely used in patients with CCA, and dynamic monitoring of ctDNA can help predict the effect of targeted therapy. If the targeted mutation in ctDNA disappears or decreases, it usually means that the targeted therapy is effective and the patient is sensitive to the drug; if the targeted mutation does not decrease, it may indicate the presence of primary drug resistance, and the targeted therapy is not effective. However, patients may develop acquired resistance mutations during treatment, and ctDNA can be used to monitor the emergence of these resistance mutations, thereby helping to dynamically adjust treatment options (Goyal et al., 2017). Plasma NGS analysis results showed that approximately 34.3% of patients carried mutations that could be targeted for treatment, such as FGFR2, IDH1, MSI-H, and ERBB2, which makes ctDNA play an important role in personalized treatment decisions (Hwang et al., 2024). Individualized precision treatment is particularly critical for patients with different subtypes (Chen et al., 2024).
Currently, FGFR2 fusion mutations (approximately 10%–20%) and IDH1 gene mutations (approximately 15%–20%) are the most common mutations in CCA and have been widely used in clinical practice with FDA-approved targeted therapies (Abou-Alfa et al., 2020a; Uson Junior and Borad, 2022). Various FGFR inhibitors (such as infigratinib (Abou-Alfa et al., 2020a), Pemigatinib (Abou-Alfa et al., 2020b), Futibatinib (Goyal et al., 2023), and the recently observed Tasurgratinib (Morizane et al., 2025), etc.) have shown good therapeutic effects in CCA patients. However, patients with FGFR2 fusion may develop mutations such as FGFR2 V564F and N550K after receiving FGFR inhibitors, leading to drug resistance. Traditional ctDNA detection methods have a low sensitivity for FGFR2 fusion mutations, at only 18% (Vaidyanathan et al., 2018). However, there have been advances in ctDNA profiling techniques in recent years. Hwang S et al.'s study showed that ctDNA profiling can efficiently detect FGFR2 fusion mutations and comprehensively capture tumor genetic changes (Hwang et al., 2024). The optimized ctDNA method specifically targeting FGFR2 fusions (FGFR-Dx) developed by Julie W et al. has for the first time verified the feasibility of FGFR2 fusion detection at low VAF (0.5%) levels in BTC patients (Reeser et al., 2024). This method significantly improves the sensitivity of FGFR2 detection (92.9%, compared with the original 18%), and is lower in cost. It is expected to help more patients adjust their treatment plans in time when drug-resistant mutations appear early. The results of a randomized, double-blind, placebo-controlled phase 3 study (ClarIDHy) showed that ivosidenib can significantly improve the median PFS of patients with advanced IDH1 mutation CCA (2.7 months vs. 1.4 months, P < 0.0001), and has a good benefit in OS, but its objective response rate (tumor size reduction >30%) is only 2% (Abou-Alfa et al., 2020a). Patients receiving ivosidenib may develop new IDH1 R132C mutations or activating IDH2 mutations (R172V), resulting in drug failure (Harding et al., 2018; Lowery et al., 2019). ctDNA testing can identify these mutations in a timely manner, providing evidence for treatment adjustments.
4.3.3 ctDNA monitoring of drug resistance
Drug resistance is one of the main reasons for CCA treatment failure. Drug resistance can be divided into primary resistance (no initial response to treatment) and acquired resistance (disease progression during treatment). Its potential mechanisms include secondary mutations of driver genes (such as EGFR, PI3K/Akt, Erk and NF-κB), epigenetic remodeling (such as abnormal DNA methylation) and cell apoptosis escape (Varghese et al., 2021). These resistance mechanisms not only affect the efficacy of chemotherapy and targeted therapy, but may also accelerate tumor metastasis and recurrence. Therefore, real-time tracking of drug resistance is crucial for developing individualized treatment plans and evaluating patient prognosis. ctDNA can break through the limitations of a single tissue biopsy and simultaneously monitor multiple drug resistance mechanisms, providing a basis for dynamically adjusting treatment plans. A study of 42 patients with advanced gastrointestinal tumors showed that among 23 resistant patients, the positive rate of detecting blood ctDNA drug-resistant mutations using NGS and ddPCR reached 87% (20/23), significantly higher than the 48% (11/23) of tissue biopsy. The proportion of ctDNA detection of multiple drug resistance mechanisms (40%) was 4.4 times higher than that of tissue biopsy (9%) (Parikh et al., 2019). Nevertheless, the clinical significance of ctDNA in monitoring drug resistance in CCA has not yet been fully confirmed, and more prospective studies are still needed to verify its application value.
4.4 CCA metastasis detection
CCA is highly invasive and metastatic, with common metastatic sites including the liver, peritoneum, lymph nodes, and lungs. Metastatic disease is usually associated with progressive disease and a poor prognosis. Therefore, early detection of metastatic lesions is crucial to improve the prognosis of CAA. Traditional imaging methods (such as CT and MRI) are difficult to detect small and occult metastatic lesions in time. The level of ctDNA is closely related to the degree of tumor differentiation. Poorly differentiated tumors are usually more metastatic. However, it has no significant correlation with the number of metastatic sites (Pietrasz et al., 2017). The concordance between ctDNA and metastatic tissue was significantly higher than that between primary tumors (Kruger et al., 2018). In patients with CCA, distant metastases such as the liver, lung, and peritoneum are often accompanied by increased plasma ctDNA abundance. Different metastatic sites are often accompanied by different types of ctDNA mutations: TP53 mutations are more common in intrahepatic metastasis; KRAS mutations are common in lung metastasis, indicating enhanced tumor invasiveness; peritoneal dissemination is associated with inactivation mutations of CTNNB1 and BAP1, suggesting that drug resistance may increase (Amato et al., 2019). Furthermore, ctDNA metastasis did not differ significantly with patient gender, age or metastatic site, further enhancing its broad applicability as a metastasis monitoring tool (Del Re et al., 2017).
5 Challenges and limitations of ctDNA testing
As an emerging liquid biopsy technology, ctDNA detection has shown great potential in early diagnosis of tumors, efficacy evaluation, and recurrence monitoring. However, it still faces many challenges and limitations in practical applications, mainly including the following aspects.
5.1 Technical sensitivity and specificity issues
The abundance of ctDNA in the blood is low in the early stages of tumors or in the remission stage after treatment, and may be as low as 0.1% (Crowley et al., 2013; Anagnostou and Velculescu, 2024; Zhang et al., 2024). Targeted methods for detecting ctDNA have reached their current biological limits. Although existing detection technologies (such as dPCR, NGS, etc.) continue to improve, it is still difficult to achieve high-sensitivity detection in low-abundance ctDNA (Kim et al., 2023; Normanno et al., 2025). Clonal hematopoiesis (CH) is a condition in which somatic mutations (such as mutations in genes such as DNMT3A, TET2, and ASXL1) exist in healthy individuals. cfDNA produced by clonal hematopoietic cells may interfere with ctDNA detection, leading to false positives (Imai et al., 2022). Studies have shown that clonal hematopoietic (CH) mutations were detected in the plasma of 29.8% of patients with gastric cancer, 10% of prostate cancer, and 30% of patients with metastatic renal cell carcinoma, with a high risk of misdiagnosis (Bacon et al., 2020; Jensen et al., 2021; Lee K. S. et al., 2024). Currently, there is a lack of mature methods to exclude clonal hematopoietic (CH) variants. Inclusion of clonal hematopoietic (CH) variants in commercial ctDNA targeted panels is expected to help distinguish true ctDNA from other somatic expansions in vivo (Bacon et al., 2020).
5.2 Sample collection and standardization problems
Although blood-based ctDNA analysis shows great promise in molecular diagnosis of various cancer types, the ratio and stability of ctDNA are low, and various stages such as collection and analysis greatly affect the results (Lampignano et al., 2020; Pascual et al., 2022; Shin et al., 2022). Paul et al. divided the key parts of ctDNA analysis workflow into two parts: pre-analysis and analysis. The key parts of pre-analysis include: sample collection, storage, transportation, elution and extraction of ctDNA, etc.; the key parts of analysis workflow include ctDNA quantification, analysis input and identification (van der Leest and Schuuring, 2024). Although the current international guidelines (the “Use of Circulating Tumor DNA for Curative-Intent Solid Tumor Drug Development Guidance for Industry” issued by the FDA) have put forward framework recommendations, the ctDNA enrichment and detection schemes for different cancer types (such as lung cancer vs. bile duct cancer) still need to be refined. Establishing a full-process quality control system from sample collection to reporting will be the key to clinical promotion.
5.3 Technical cost and multi-center verification issues
The technology of ctDNA detection is complex and involves multiple high-investment processes. The high technical cost is one of the important reasons that limits its large-scale clinical application. Deep sequencing and personalized sequencing technologies are expensive, and body fluids such as pancreatic juice and bile require special extraction reagents, which also increase costs. A retrospective observational cohort study from Italy showed that tumor profiling using comprehensive NGS panels improved patients’ eligibility to personalized therapies (De Micheli et al., 2025). However, the diagnostic cost per patient ranges from $5500 to $8400 (NSCLC: $8400; CCA: $5500; PC: $6600).
Many different ctDNA tests claim to have specificity and sensitivity, but there are few independent or cross-platform validation studies. One report showed that a very poor ctDNA correlation was found between liquid biopsy platforms Guardant360 and PlasmaSELECT when examining 42 genes that overlapped between the two platforms (Torga and Pienta, 2018). Although this was a small study involving 40 metastatic prostate cancer patients, 25 of 40 had alterations in overlapping genes, only three had complete congruence, six had partial congruence, and sixteen had no congruence. Differences in detection methods, differences in instrument sensitivity, and lack of standardization in threshold setting make multi-center validation of ctDNA still challenging. Unfortunately, there is currently a lack of research in this area (Cherney et al., 2025).
5.4 Tumor heterogeneity and dynamic changes
Different stages and anatomical locations of CCA may lead to differences in genetic information between ctDNA, and this heterogeneity increases the complexity of ctDNA detection. In AJCC stage II and distal CCA, the sensitivity of ctDNA in detecting copy number variations (CNVs) is significantly reduced (Shen et al., 2019). The concordance between ctDNA and tissue results was high, 74% overall, and 92% for iCCA, but was lower for eCCA (55%) (Ettrich et al., 2019). Oliver et al.'s study showed that the diagnostic accuracy and sensitivity of ctDNA detection in advanced CCA were as high as 97.7% and 92.3%, and the specificity was 100%. However, relevant clinical research is still needed in patients with early CCA (Zill et al., 2015). In conclusion, tumor heterogeneity, along with variations in CCA stages and types, compromises ctDNA detection performance, thereby restricting its clinical applicability.
ctDNA levels are affected by multiple factors such as tumor burden and therapeutic intervention, resulting in significant dynamic changes. In a small sample clinical study conducted by Kyung et al., ctDNA samples from 16 patients with CCA were analyzed after surgery, and new mutations were found in ctDNA of eight patients (50%) after surgery (Kim et al., 2023). Postoperative plasma mutations had a sensitivity of 44% and a specificity of 45% for detecting clinical relapses. Ettrich et al. performed ctDNA testing in 11 patients who received first-line palliative chemotherapy. Compared with baseline, 36% (4/11) of these patients had a change in the ctDNA mutational landscape. Three of them had TP53 mutations, which were not detected after treatment. The fourth patient had a PBRM1 mutation, which no longer appeared at the “progression” time point (Ettrich et al., 2019). Both studies demonstrated the potential of ctDNA testing in monitoring CCA treatment response and elucidating drug resistance mechanisms; however, their clinical generalizability is limited by small sample sizes and a single-institution study design.
6 Future research directions
6.1 Multiplex detection of ctDNA combined with other liquid biopsy markers
6.1.1 ctDNA-CA19-9 combined detection
In addition to traditional tumor markers, recent studies have also found that the level of syncytin-1 in bile cfDNA can be used as a new biomarker, especially with potential in the early diagnosis of CCA. The study by He JD et al. showed that syncytin-1 performed better than CA19-9, CEA and AFP in the diagnosis of bile cfDNA, with an AUC of 0.805 (95% CI: 0.719–0.890, p < 0.001), a specificity of 90.2% and a sensitivity of 75%. Combining syncytin-1 with CA19-9 increased the AUC to 0.927 (0.901 95% CI: 0.877–0.978, p < 0.001) (He et al., 2025). The results show that the combination of the two can significantly improve the efficiency and accuracy of early diagnosis, providing an important basis for the early diagnosis of CCA.
6.1.2 ctDNA-cf-miRNA combined detection
In recent years, cell-free miRNA (cf-miRNA), like ctDNA, has shown promise in diagnosing a variety of diseases, including heart disease, infection, and various cancers (Biró et al., 2020; Song et al., 2020; Hadipour et al., 2023). Studies have shown that there is a significant positive correlation between ctDNA and cf-miRNA (Prasopdee et al., 2024). The combination of plasma cf-miRNA and ctDNA can significantly improve the sensitivity and specificity of differential diagnosis of CCA compared with ctDNA detection alone (83.33%, 100% vs. 75.00%, 95.83%). This means that the combination of the two can serve as a potential diagnostic tool to distinguish CCA from other diseases and further improve the accuracy of CCA diagnosis.
6.1.3 ctDNA-CTC combined detection
Circulating tumor cells (CTCs) spread in the bloodstream through epithelial-mesenchymal transition and are closely related to tumor metastasis. They have been actively studied as prognostic biomarkers (Han et al., 2021). A study conducted by Sung et al. demonstrated that the combined detection of CTC and ctDNA improved the accuracy of predicting recurrence in BTC patients after standard treatment (surgery + adjuvant chemotherapy) (AUC = 0.92, sensitivity 93.8%, specificity 87.5%), which was much higher than CA 19–9 (AUC = 0.75, sensitivity 75.0%, specificity 75.0%) (Park et al., 2024).
The multiplex detection scheme of ctDNA combined with other liquid biopsy markers is still in its infancy, and the data integration standards of different detection platforms have not yet been unified. In the future, with the help of artificial intelligence (AI) algorithms and the development of multicenter cohorts, the clinical operability of combined markers can be further verified.
6.2 Application of ctDNA in immunotherapy
6.2.1 Predicting response to immunotherapy
ctDNA detection can analyze the dynamics of tumor clonal evolution in real time, overcome the limitations of traditional imaging in evaluating immunotherapy response (such as pseudoprogression, delayed response, etc.), and provide a non-invasive means to monitor tumor dynamics and immunotherapy response (Weber et al., 2024).
Li et al. developed a patient-specific ctDNA fingerprint based on NGS (including eight hotspot genes: BRAF, EGFR, ERBB2, KIT, KRAS, MET, NRAS, and PIK3CA), by monitoring ctDNA content fraction (CCF) levels and CCF fold changes. This method has improved the specificity and sensitivity of monitoring the response to immunotherapy in CCA and can identify tumor recurrence earlier than imaging (Li et al., 2020).
Xu et al. constructed a CNV risk scoring model based on copy number variation (CNV) in plasma ctDNA to predict the clinical outcomes of immune checkpoint inhibitor (ICI) treatment in advanced hepatobiliary cancer. The results showed that the low CNV risk group had better clinical outcomes than the high CNV risk group (median PFS: 6.17 months vs. 2.60 months, HR = 0.045; median OS: not reached vs. 6.5 months, HR = 0.39) (Yang X. et al., 2021).
6.2.2 Guidance on perioperative immunotherapy strategies
Yu et al. used ctDNA analysis to guide the adjuvant treatment of a patient with stage III CCA after resection and achieved good results. The patient started standard capecitabine adjuvant therapy on the 50th day after surgery. ctDNA was positive for two consecutive times and CA19-9 continued to rise. Before obtaining evidence of radiological recurrence, the patient was given capecitabine combined with pembrolizumab. After 112 days, ctDNA was negative and CA19-9 returned to normal after 155 days (Yu et al., 2024). This case report reveals the guiding significance of ctDNA testing for perioperative adjuvant treatment of CCA, but due to the limited sample size, the results are less credible.
Currently, there are few studies on ctDNA and CCA immunotherapy. In the future, larger-scale prospective studies are needed to demonstrate the role and clinical application value of ctDNA in CCA immunotherapy.
7 Conclusion
As an emerging tumor liquid biopsy technology, ctDNA detection has broad application prospects in CCA. ctDNA has the advantages of high tissue consistency and high sensitivity, and has shown its effectiveness in early screening and diagnosis. In the future, the combined application of ctDNA and other tumor liquid biopsy markers is expected to achieve more efficient and accurate early diagnosis of CCA. Moreover, due to the easy availability and short half-life of ctDNA samples, real-time treatment response monitoring and prognosis assessment can be provided for patients receiving treatment. In terms of treatment, by monitoring the levels of various targeted mutations such as FGFR2 and IDH1 in patients, drug resistance can be detected in a timely manner, providing patients with personalized treatment plans. In the future, there is still an urgent need to improve the ctDNA detection standards and standardize each link of sample collection, analysis and detection. As a ray of hope in CCA treatment, immunotherapy is increasingly widely used in clinical practice. ctDNA testing is expected to further broaden the use scenarios of immunotherapy, but currently there are few studies in this field and the sample size is small. In the future, there is an urgent need to conduct more clinical studies with larger sample sizes.
Author contributions
YW: Methodology, Writing – review and editing, Writing – original draft. YiL: Investigation, Writing – original draft. ZL: Writing – original draft, Data curation. YZ: Methodology, Writing – review and editing. TL: Writing – review and editing, Methodology. CT: Data curation, Writing – review and editing. JZ: Writing – review and editing, Visualization. BJ: Writing – review and editing, Visualization. JC: Supervision, Writing – review and editing. YaL: Writing – review and editing, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Gansu Provincial Natural Science Foundation (No. 24JRRA318), Gansu Provincial Health Commission Scientific Research Program (No. GSWSKY2020-11), the First Hospital of Lanzhou University Intramural Research Fund (No. LDYYYN2023-118).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abou-Alfa, G. K., Macarulla, T., Javle, M. M., Kelley, R. K., Lubner, S. J., Adeva, J., et al. (2020a). Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 21, 796–807. doi:10.1016/S1470-2045(20)30157-1
Abou-Alfa, G. K., Sahai, V., Hollebecque, A., Vaccaro, G., Melisi, D., Al-Rajabi, R., et al. (2020b). Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 21, 671–684. doi:10.1016/S1470-2045(20)30109-1
Amato, E., Mafficini, A., Hirabayashi, K., Lawlor, R. T., Fassan, M., Vicentini, C., et al. (2019). Molecular alterations associated with metastases of solid pseudopapillary neoplasms of the pancreas. J. Pathol. 247, 123–134. doi:10.1002/path.5180
Anagnostou, V., and Velculescu, V. E. (2024). Pushing the boundaries of liquid biopsies for early precision intervention. Cancer Discov. 14, 615–619. doi:10.1158/2159-8290.CD-24-0037
Arechederra, M., Rullán, M., Amat, I., Oyon, D., Zabalza, L., Elizalde, M., et al. (2022). Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut 71, 1141–1151. doi:10.1136/gutjnl-2021-325178
Arrichiello, G., Nacca, V., Paragliola, F., and Giunta, E. F. (2022). Liquid biopsy in biliary tract cancer from blood and bile samples: current knowledge and future perspectives. Explor. Target. Anti-Tumor Ther. 3, 362–374. doi:10.37349/etat.2022.00087
Awosika, J. A., Monge, C., and Greten, T. F. (2024). Integration of circulating tumor DNA in biliary tract cancer: the emerging landscape. Hepatic Oncol. 11, 2403334. doi:10.1080/20450923.2024.2403334
Bacon, J. V. W., Annala, M., Soleimani, M., Lavoie, J.-M., So, A., Gleave, M. E., et al. (2020). Plasma circulating tumor DNA and clonal hematopoiesis in metastatic renal cell carcinoma. Clin. Genitourin. Cancer 18, 322–331.e2. doi:10.1016/j.clgc.2019.12.018
Bahleda, R., Italiano, A., Hierro, C., Mita, A., Cervantes, A., Chan, N., et al. (2019). Multicenter phase I study of erdafitinib (JNJ-42756493), oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced or refractory solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 25, 4888–4897. doi:10.1158/1078-0432.CCR-18-3334
Ballehaninna, U. K., and Chamberlain, R. S. (2012). The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J. Gastrointest. Oncol. 3, 105–119. doi:10.3978/j.issn.2078-6891.2011.021
Banales, J. M., Cardinale, V., Carpino, G., Marzioni, M., Andersen, J. B., Invernizzi, P., et al. (2016). Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the european network for the study of cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 13, 261–280. doi:10.1038/nrgastro.2016.51
Banales, J. M., Marin, J. J. G., Lamarca, A., Rodrigues, P. M., Khan, S. A., Roberts, L. R., et al. (2020). Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 17, 557–588. doi:10.1038/s41575-020-0310-z
Berchuck, J. E., Facchinetti, F., DiToro, D. F., Baiev, I., Majeed, U., Reyes, S., et al. (2022). The clinical landscape of cell-free DNA alterations in 1671 patients with advanced biliary tract cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 33, 1269–1283. doi:10.1016/j.annonc.2022.09.150
Biró, O., Rigó, J., and Nagy, B. (2020). Noninvasive prenatal testing for congenital heart disease - cell-free nucleic acid and protein biomarkers in maternal blood. J. Matern.-Fetal Neonatal Med. Off. J. Eur. Assoc. Perinat. Med. Fed. Asia Ocean. Perinat. Soc. Int. Soc. Perinat. Obstet. 33, 1044–1050. doi:10.1080/14767058.2018.1508437
Blechacz, B. (2017). Cholangiocarcinoma: current knowledge and new developments. Gut Liver 11, 13–26. doi:10.5009/gnl15568
Boberg, K. M., Jebsen, P., Clausen, O. P., Foss, A., Aabakken, L., and Schrumpf, E. (2006). Diagnostic benefit of biliary brush cytology in cholangiocarcinoma in primary sclerosing cholangitis. J. Hepatol. 45, 568–574. doi:10.1016/j.jhep.2006.05.010
Bowlus, C. L., Arrivé, L., Bergquist, A., Deneau, M., Forman, L., Ilyas, S. I., et al. (2023). AASLD practice guidance on primary sclerosing cholangitis and cholangiocarcinoma. Hepatol. Balt. Md 77, 659–702. doi:10.1002/hep.32771
Brandi, G., Tavolari, S., and Biasco, G. (2012). Genomic and genetic characterization of cholangiocarcinoma identifies therapeutics targets for tyrosine kinase inhibitors. Gastroenterology 143, e20–e21. doi:10.1053/j.gastro.2012.07.120
Chen, K., Yang, F., Shen, H., Wang, C., Li, X., Chervova, O., et al. (2023). Individualized tumor-informed circulating tumor DNA analysis for postoperative monitoring of non-small cell lung cancer. Cancer Cell 41, 1749–1762.e6. doi:10.1016/j.ccell.2023.08.010
Chen, S., Han, K., Song, Y., Liu, S., Li, X., Wang, S., et al. (2022). Current status, trends, and predictions in the burden of gallbladder and biliary tract cancer in China from 1990 to 2019. Chin. Med. J. (Engl.) 135, 1697–1706. doi:10.1097/CM9.0000000000002258
Chen, Z., Gao, J., Li, Z., Ma, D., Wang, Y., Cheng, Q., et al. (2024). Integrative analysis reveals different feature of intrahepatic cholangiocarcinoma subtypes. Liver Int. Off. J. Int. Assoc. Study Liver 44, 2477–2493. doi:10.1111/liv.16015
Cherney, B., Diaz, A., Chavis, C., Ghattas, C., Evans, D., Arambula, D., et al. (2025). The next-generation sequencing quality initiative and challenges in clinical and public health laboratories. Emerg. Infect. Dis. 31, 14–17. doi:10.3201/eid3113.241175
Cleary, J. M., Rouaisnel, B., Daina, A., Raghavan, S., Roller, L. A., Huffman, B. M., et al. (2022). Secondary IDH1 resistance mutations and oncogenic IDH2 mutations cause acquired resistance to ivosidenib in cholangiocarcinoma. NPJ Precis. Oncol. 6, 61. doi:10.1038/s41698-022-00304-5
Cristiano, S., Leal, A., Phallen, J., Fiksel, J., Adleff, V., Bruhm, D. C., et al. (2019). Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389. doi:10.1038/s41586-019-1272-6
Crowley, E., Di Nicolantonio, F., Loupakis, F., and Bardelli, A. (2013). Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 10, 472–484. doi:10.1038/nrclinonc.2013.110
Del Re, M., Vivaldi, C., Rofi, E., Vasile, E., Miccoli, M., Caparello, C., et al. (2017). Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci. Rep. 7, 7931. doi:10.1038/s41598-017-08297-z
De Micheli, V., Agnelli, L., Conca, E., Rabsiun Aramburu, V. L., Baggi, A., Vingiani, A., et al. (2025). Impact of comprehensive genomic profiling and molecular tumour board on costs and access to tailored therapies: real-world observational study. BMJ Open 15, e099134. doi:10.1136/bmjopen-2025-099134
Diehl, F., Schmidt, K., Choti, M. A., Romans, K., Goodman, S., Li, M., et al. (2008). Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990. doi:10.1038/nm.1789
Driescher, C., Fuchs, K., Haeberle, L., Goering, W., Frohn, L., Opitz, F. V., et al. (2020). Bile-based cell-free DNA analysis is a reliable diagnostic tool in pancreatobiliary cancer. Cancers 13, 39. doi:10.3390/cancers13010039
Ettrich, T. J., Schwerdel, D., Dolnik, A., Beuter, F., Blätte, T. J., Schmidt, S. A., et al. (2019). Genotyping of circulating tumor DNA in cholangiocarcinoma reveals diagnostic and prognostic information. Sci. Rep. 9, 13261. doi:10.1038/s41598-019-49860-0
European Association for the Study of the Liver (2022). EASL clinical practice guidelines on sclerosing cholangitis. J. Hepatol. 77, 761–806. doi:10.1016/j.jhep.2022.05.011
European Association for the Study of the Liver (2023). EASL-ILCA clinical practice guidelines on the management of intrahepatic cholangiocarcinoma. J. Hepatol. 79, 181–208. doi:10.1016/j.jhep.2023.03.010
Forshew, T., Murtaza, M., Parkinson, C., Gale, D., Tsui, D. W. Y., Kaper, F., et al. (2012). Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 4, 136ra68. doi:10.1126/scitranslmed.3003726
García-Foncillas, J., Alba, E., Aranda, E., Díaz-Rubio, E., López-López, R., Tabernero, J., et al. (2017). Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann. Oncol. 28, 2943–2949. doi:10.1093/annonc/mdx501
González-Medina, A., Vila-Casadesús, M., Gomez-Rey, M., Fabregat-Franco, C., Sierra, A., Tian, T. V., et al. (2024). Clinical value of liquid biopsy in patients with FGFR2 fusion-positive cholangiocarcinoma during targeted therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 30, 4491–4504. doi:10.1158/1078-0432.CCR-23-3780
Goyal, L., Meric-Bernstam, F., Hollebecque, A., Valle, J. W., Morizane, C., Karasic, T. B., et al. (2023). Futibatinib for FGFR2-Rearranged intrahepatic cholangiocarcinoma. N. Engl. J. Med. 388, 228–239. doi:10.1056/NEJMoa2206834
Goyal, L., Saha, S. K., Liu, L. Y., Siravegna, G., Leshchiner, I., Ahronian, L. G., et al. (2017). Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 7, 252–263. doi:10.1158/2159-8290.CD-16-1000
Goyal, L., Shi, L., Liu, L. Y., Fece de la Cruz, F., Lennerz, J. K., Raghavan, S., et al. (2019). TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov. 9, 1064–1079. doi:10.1158/2159-8290.CD-19-0182
Hadipour, M., Fasihi Harandi, M., Mirhendi, H., and Yousofi Darani, H. (2023). Diagnosis of echinococcosis by detecting circulating cell-free DNA and miRNA. Expert Rev. Mol. diagn. 23, 133–142. doi:10.1080/14737159.2023.2178903
Han, S. Y., Park, S. H., Ko, H. S., Jang, A., Seo, H. I., Lee, S. J., et al. (2021). Vimentin-positive circulating tumor cells as diagnostic and prognostic biomarkers in patients with biliary tract cancer. J. Clin. Med. 10, 4435. doi:10.3390/jcm10194435
Hannigan, B., Ye, W., Mehrotra, M., Lam, V., Bolivar, A., Zalles, S., et al. (2019). Liquid biopsy assay for lung carcinoma using centrifuged supernatants from fine-needle aspiration specimens. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 30, 963–969. doi:10.1093/annonc/mdz102
Harding, J. J., Lowery, M. A., Shih, A. H., Schvartzman, J. M., Hou, S., Famulare, C., et al. (2018). Isoform switching as a mechanism of acquired resistance to mutant isocitrate dehydrogenase inhibition. Cancer Discov. 8, 1540–1547. doi:10.1158/2159-8290.CD-18-0877
He, J., Qian, J., Li, X., Zhao, X., Meng, W., and Zhuang, X. (2025). Bile-derived cfDNA of Syncytin-1 and SLC7A11 as a potential molecular marker for early diagnosis of cholangiocarcinoma. J. Gastrointest. Cancer 56, 55. doi:10.1007/s12029-025-01180-5
Heitzer, E., Haque, I. S., Roberts, C. E. S., and Speicher, M. R. (2019). Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88. doi:10.1038/s41576-018-0071-5
Helsten, T., Elkin, S., Arthur, E., Tomson, B. N., Carter, J., and Kurzrock, R. (2016). The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 22, 259–267. doi:10.1158/1078-0432.CCR-14-3212
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J., et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610. doi:10.1021/ac202028g
Hon, G. C., Hawkins, R. D., Caballero, O. L., Lo, C., Lister, R., Pelizzola, M., et al. (2012). Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258. doi:10.1101/gr.125872.111
Hou, J. Y., Chapman, J. S., Kalashnikova, E., Pierson, W., Smith-McCune, K., Pineda, G., et al. (2022). Circulating tumor DNA monitoring for early recurrence detection in epithelial ovarian cancer. Gynecol. Oncol. 167, 334–341. doi:10.1016/j.ygyno.2022.09.004
Huffman, B. M., Aushev, V. N., Budde, G. L., Chao, J., Dayyani, F., Hanna, D., et al. (2022). Analysis of circulating tumor DNA to predict risk of recurrence in patients with esophageal and gastric cancers. JCO Precis. Oncol. 6, e2200420. doi:10.1200/PO.22.00420
Hwang, S., Woo, S., Kang, B., Kang, H., Kim, J. S., Lee, S. H., et al. (2024). Concordance of ctDNA and tissue genomic profiling in advanced biliary tract cancer. J. Hepatol. 82, 649–657. doi:10.1016/j.jhep.2024.10.020
Imai, M., Nakamura, Y., Sunami, K., Kage, H., Komine, K., Koyama, T., et al. (2022). Expert panel consensus recommendations on the use of circulating tumor DNA assays for patients with advanced solid tumors. Cancer Sci. 113, 3646–3656. doi:10.1111/cas.15504
Imperial, R., Nazer, M., Ahmed, Z., Kam, A. E., Pluard, T. J., Bahaj, W., et al. (2019). Matched whole-genome sequencing (WGS) and whole-exome sequencing (WES) of tumor tissue with circulating tumor DNA (ctDNA) analysis: complementary modalities in clinical practice. Cancers 11, 1399. doi:10.3390/cancers11091399
Ito, S., Ando, M., Aoki, S., Soma, S., Zhang, J., Hirano, N., et al. (2024). Usefulness of multigene liquid biopsy of bile for identifying driver genes of biliary duct cancers. Cancer Sci. 115, 4054–4063. doi:10.1111/cas.16365
Javle, M., Lowery, M., Shroff, R. T., Weiss, K. H., Springfeld, C., Borad, M. J., et al. (2018). Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 36, 276–282. doi:10.1200/JCO.2017.75.5009
Jensen, K., Konnick, E. Q., Schweizer, M. T., Sokolova, A. O., Grivas, P., Cheng, H. H., et al. (2021). Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference. JAMA Oncol. 7, 107–110. doi:10.1001/jamaoncol.2020.5161
Job, S., Rapoud, D., Dos Santos, A., Gonzalez, P., Desterke, C., Pascal, G., et al. (2020). Identification of four immune subtypes characterized by distinct composition and functions of tumor microenvironment in intrahepatic cholangiocarcinoma. Hepatol. Balt. Md 72, 965–981. doi:10.1002/hep.31092
Kamsa-Ard, S., Luvira, V., Suwanrungruang, K., Kamsa-Ard, S., Luvira, V., Santong, C., et al. (2020). Cholangiocarcinoma trends, incidence, and relative survival in Khon Kaen, Thailand from 1989 through 2013: a population-based cancer registry study. 29 (5), 108–109. doi:10.2188/jea.JE20190251
Kearney, J. F., Weighill, D., and Yeh, J. J. (2023). Promise of bile circulating tumor DNA in biliary tract cancers. Cancer 129, 1643–1645. doi:10.1002/cncr.34715
Khan, S. A., Davidson, B. R., Goldin, R. D., Heaton, N., Karani, J., Pereira, S. P., et al. (2012). Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 61, 1657–1669. doi:10.1136/gutjnl-2011-301748
Khan, S. A., Tavolari, S., and Brandi, G. (2019). Cholangiocarcinoma: epidemiology and risk factors. Liver Int. Off. J. Int. Assoc. Study Liver 39 (1), 19–31. doi:10.1111/liv.14095
Khosla, D., Misra, S., Chu, P. L., Guan, P., Nada, R., Gupta, R., et al. (2024). Cholangiocarcinoma: recent advances in molecular pathobiology and therapeutic approaches. Cancers 16, 801. doi:10.3390/cancers16040801
Kim, K.-H., Yi, H.-S., Lee, H., Bae, G.-E., and Yeo, M.-K. (2023). Targeting the sequences of circulating tumor DNA of cholangiocarcinomas and its applications and limitations in clinical practice. Int. J. Mol. Sci. 24, 7512. doi:10.3390/ijms24087512
Kotani, D., Oki, E., Nakamura, Y., Yukami, H., Mishima, S., Bando, H., et al. (2023). Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 29, 127–134. doi:10.1038/s41591-022-02115-4
Kruger, S., Heinemann, V., Ross, C., Diehl, F., Nagel, D., Ormanns, S., et al. (2018). Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 29, 2348–2355. doi:10.1093/annonc/mdy417
Lampignano, R., Neumann, M. H. D., Weber, S., Kloten, V., Herdean, A., Voss, T., et al. (2020). Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (pre)analytical work flows. Clin. Chem. 66, 149–160. doi:10.1373/clinchem.2019.306837
Lee, D. S., Han, M. W., Kang, Y., Kim, C., Lee, S., Kim, K.-P., et al. (2024a). CX-4945 (silmitasertib) induces cell death by impairing lysosomal utilization in KRAS mutant cholangiocarcinoma cell lines. Anticancer Res. 44, 1939–1946. doi:10.21873/anticanres.16996
Lee, K. S., Lee, C.-K., Kwon, S. S., Kwon, W. S., Park, S., Lee, S.-T., et al. (2024b). Clinical relevance of clonal hematopoiesis and its interference in cell-free DNA profiling of patients with gastric cancer. Clin. Chem. Lab. Med. 62, 178–186. doi:10.1515/cclm-2023-0261
Levy, C., Lymp, J., Angulo, P., Gores, G. J., Larusso, N., and Lindor, K. D. (2005). The value of serum CA 19-9 in predicting cholangiocarcinomas in patients with primary sclerosing cholangitis. Dig. Dis. Sci. 50, 1734–1740. doi:10.1007/s10620-005-2927-8
Li, H., Jing, C., Wu, J., Ni, J., Sha, H., Xu, X., et al. (2019). Circulating tumor DNA detection: a potential tool for colorectal cancer management. Oncol. Lett. 17, 1409–1416. doi:10.3892/ol.2018.9794
Li, J., Jiang, W., Wei, J., Zhang, J., Cai, L., Luo, M., et al. (2020). Patient specific circulating tumor DNA fingerprints to monitor treatment response across multiple tumors. J. Transl. Med. 18, 293. doi:10.1186/s12967-020-02449-y
Li, Y.-Z., Kong, S.-N., Liu, Y.-P., Yang, Y., and Zhang, H.-M. (2023). Can liquid biopsy based on ctDNA/cfDNA replace tissue biopsy for the precision treatment of EGFR-mutated NSCLC? J. Clin. Med. 12, 1438. doi:10.3390/jcm12041438
Liang, B., Zhong, L., He, Q., Wang, S., Pan, Z., Wang, T., et al. (2015). Diagnostic accuracy of serum CA19-9 in patients with cholangiocarcinoma: a systematic review and meta-analysis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 21, 3555–3563. doi:10.12659/msm.895040
Lindor, K. D., Kowdley, K. V., and Harrison, M. E.American College of Gastroenterology (2015). ACG clinical guideline: primary sclerosing cholangitis. Am. J. Gastroenterol. 110, 646–660. doi:10.1038/ajg.2015.112
Liu, F., Hao, X., Liu, B., Liu, S., and Yuan, Y. (2023). Bile liquid biopsy in biliary tract cancer. Clin. Chim. Acta Int. J. Clin. Chem. 551, 117593. doi:10.1016/j.cca.2023.117593
Liu, L., Toung, J. M., Jassowicz, A. F., Vijayaraghavan, R., Kang, H., Zhang, R., et al. (2018). Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 29, 1445–1453. doi:10.1093/annonc/mdy119
Liu, M. C., Oxnard, G. R., Klein, E. A., Swanton, C., and Seiden, M. V.CCGA Consortium (2020). Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 31, 745–759. doi:10.1016/j.annonc.2020.02.011
Liu, R., Song, Y., Hua, R., Ahmed, S., Xie, Y., Lai, C., et al. (2024). Cell-free DNA in plasma reveals genomic similarity between biliary tract inflammatory lesion and biliary tract cancer. Phenomics Cham Switz. 4, 339–351. doi:10.1007/s43657-024-00160-2
Lowery, M. A., Burris, H. A., Janku, F., Shroff, R. T., Cleary, J. M., Azad, N. S., et al. (2019). Safety and activity of ivosidenib in patients with IDH1-mutant advanced cholangiocarcinoma: a phase 1 study. Lancet Gastroenterol. Hepatol. 4, 711–720. doi:10.1016/S2468-1253(19)30189-X
Luo, G., Guo, M., Jin, K., Liu, Z., Liu, C., Cheng, H., et al. (2016). Optimize CA19-9 in detecting pancreatic cancer by lewis and secretor genotyping. Pancreatol. Off. J. Int. Assoc. Pancreatol. IAP Al 16, 1057–1062. doi:10.1016/j.pan.2016.09.013
Ma, L., Guo, H., Zhao, Y., Liu, Z., Wang, C., Bu, J., et al. (2024). Liquid biopsy in cancer: current status, challenges and future prospects. Signal Transduct. Target. Ther. 9, 336. doi:10.1038/s41392-024-02021-w
Mazzaferro, V., El-Rayes, B. F., Droz Dit Busset, M., Cotsoglou, C., Harris, W. P., Damjanov, N., et al. (2019). Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. Br. J. Cancer 120, 165–171. doi:10.1038/s41416-018-0334-0
McGuire, A. L., Caulfield, T., and Cho, M. K. (2008). Research ethics and the challenge of whole-genome sequencing. Nat. Rev. Genet. 9, 152–156. doi:10.1038/nrg2302
Miura, Y., Ohyama, H., Mikata, R., Hirotsu, Y., Amemiya, K., Mochizuki, H., et al. (2024). The efficacy of bile liquid biopsy in the diagnosis and treatment of biliary tract cancer. J. Hepato-Biliary-Pancreat. Sci. 31, 329–338. doi:10.1002/jhbp.1432
Mody, K., Kasi, P. M., Yang, J., Surapaneni, P. K., Bekaii-Saab, T., Ahn, D. H., et al. (2019). Circulating tumor DNA profiling of advanced biliary tract cancers. JCO Precis. Oncol. 3, 1–9. doi:10.1200/PO.18.00324
Morizane, C., Ueno, M., Ioka, T., Tajika, M., Ikeda, M., Yamaguchi, K., et al. (2025). Tasurgratinib in patients with cholangiocarcinoma or gastric cancer: expansion part of the first-in-human phase I study. Cancer Sci. 116, 192–203. doi:10.1111/cas.16354
Moss, A. C., Morris, E., Leyden, J., and MacMathuna, P. (2007). Malignant distal biliary obstruction: a systematic review and meta-analysis of endoscopic and surgical bypass results. Cancer Treat. Rev. 33, 213–221. doi:10.1016/j.ctrv.2006.10.006
Nakamura, H., Arai, Y., Totoki, Y., Shirota, T., Elzawahry, A., Kato, M., et al. (2015). Genomic spectra of biliary tract cancer. Nat. Genet. 47, 1003–1010. doi:10.1038/ng.3375
Navaneethan, U., Njei, B., Lourdusamy, V., Konjeti, R., Vargo, J. J., and Parsi, M. A. (2015). Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: a systematic review and meta-analysis. Gastrointest. Endosc. 81, 168–176. doi:10.1016/j.gie.2014.09.017
Nesic, M., Rasmussen, M. H., Henriksen, T. V., Demuth, C., Frydendahl, A., Nordentoft, I., et al. (2024). Beyond basics: key mutation selection features for successful tumor-informed ctDNA detection. Int. J. Cancer 155, 925–933. doi:10.1002/ijc.34964
Nikanjam, M., Kato, S., and Kurzrock, R. (2022). Liquid biopsy: current technology and clinical applications. J. Hematol. Oncol.J Hematol. Oncol. 15, 131. doi:10.1186/s13045-022-01351-y
Normanno, N., Morabito, A., Rachiglio, A. M., Sforza, V., Landi, L., Bria, E., et al. (2025). Circulating tumour DNA in early stage and locally advanced NSCLC: ready for clinical implementation? Nat. Rev. Clin. Oncol. 22, 215–231. doi:10.1038/s41571-024-00985-w
Olmedillas-López, S., Olivera-Salazar, R., García-Arranz, M., and García-Olmo, D. (2022). Current and emerging applications of droplet digital PCR in oncology: an updated review. Mol. Diagn. Ther. 26, 61–87. doi:10.1007/s40291-021-00562-2
Parikh, A. R., Leshchiner, I., Elagina, L., Goyal, L., Levovitz, C., Siravegna, G., et al. (2019). Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421. doi:10.1038/s41591-019-0561-9
Park, S. H., Lee, H. J., Kim, T. I., Lee, J., Han, S. Y., Seo, H. I., et al. (2024). Ultrashort cell-free DNA fragments and vimentin-positive circulating tumor cells for predicting early recurrence in patients with biliary tract cancer. Diagn. Basel Switz. 14, 2462. doi:10.3390/diagnostics14212462
Pascual, J., Attard, G., Bidard, F.-C., Curigliano, G., De Mattos-Arruda, L., Diehn, M., et al. (2022). ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO precision medicine working group. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 33, 750–768. doi:10.1016/j.annonc.2022.05.520
Pietrasz, D., Pécuchet, N., Garlan, F., Didelot, A., Dubreuil, O., Doat, S., et al. (2017). Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 23, 116–123. doi:10.1158/1078-0432.CCR-16-0806
Prasopdee, S., Pholhelm, M., Yusuk, S., Tangphatsornruang, S., Butthongkomvong, K., Kunjantarachot, A., et al. (2024). Investigation of plasma cell-free DNA and MiRNA in cholangiocarcinoma and opisthorchiasis viverrini patients. Asian pac. J. Cancer Prev. APJCP 25, 739–746. doi:10.31557/APJCP.2024.25.3.739
Qurashi, M., Vithayathil, M., and Khan, S. A. (2025). Epidemiology of cholangiocarcinoma. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 51, 107064. doi:10.1016/j.ejso.2023.107064
Reeser, J. W., Wing, M. R., Samorodnitsky, E., Dao, T., Smith, A., Stein, L., et al. (2024). Analytic validation of an FGFR -focused cell-free DNA liquid biopsy assay (FGFR-Dx). medRxiv., 2024.09.01.24312783. doi:10.1101/2024.09.01.24312783
Reinert, T., Henriksen, T. V., Christensen, E., Sharma, S., Salari, R., Sethi, H., et al. (2019). Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131. doi:10.1001/jamaoncol.2019.0528
Rizzo, A., Ricci, A. D., and Brandi, G. (2021). IDH inhibitors in advanced cholangiocarcinoma: another arrow in the quiver? Cancer Treat. Res. Commun. 27, 100356. doi:10.1016/j.ctarc.2021.100356
Rushbrook, S. M., Kendall, T. J., Zen, Y., Albazaz, R., Manoharan, P., Pereira, S. P., et al. (2024). British society of gastroenterology guidelines for the diagnosis and management of cholangiocarcinoma. Gut 73, 16–46. doi:10.1136/gutjnl-2023-330029
Santonja, A., Cooper, W. N., Eldridge, M. D., Edwards, P. A. W., Morris, J. A., Edwards, A. R., et al. (2023). Comparison of tumor-informed and tumor-naïve sequencing assays for ctDNA detection in breast cancer. EMBO Mol. Med. 15, e16505. doi:10.15252/emmm.202216505
Sasaki, T., Hiraki, H., Yashima-Abo, A., Nagashima, H., Endo, F., Yaegashi, M., et al. (2025). Comprehensive genome profiling-initiated tumor-informed circulating tumor DNA monitoring for patients with advanced cancer. Cancer Sci. 116, 764–774. doi:10.1111/cas.16446
Shen, N., Zhang, D., Yin, L., Qiu, Y., Liu, J., Yu, W., et al. (2019). Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep. 42, 549–560. doi:10.3892/or.2019.7177
Shin, S., Woo, H. I., Kim, J.-W., Kim, Y., and Lee, K.-A. (2022). Clinical practice guidelines for pre-analytical procedures of plasma epidermal growth factor receptor variant testing. Ann. Lab. Med. 42, 141–149. doi:10.3343/alm.2022.42.2.141
Shu, L., Li, X., Liu, Z., Li, K., Shi, A., Tang, Y., et al. (2024). Bile exosomal miR-182/183-5p increases cholangiocarcinoma stemness and progression by targeting HPGD and increasing PGE2 generation. Hepatol. Balt. Md 79, 307–322. doi:10.1097/HEP.0000000000000437
Silverman, I. M., Murugesan, K., Lihou, C. F., Féliz, L., Frampton, G. M., Newton, R. C., et al. (2019). Comprehensive genomic profiling in FIGHT-202 reveals the landscape of actionable alterations in advanced cholangiocarcinoma. J. Clin. Oncol. 37, 4080. doi:10.1200/JCO.2019.37.15_suppl.4080
Sinakos, E., Saenger, A. K., Keach, J., Kim, W. R., and Lindor, K. D. (2011). Many patients with primary sclerosing cholangitis and increased serum levels of carbohydrate antigen 19-9 do not have cholangiocarcinoma. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 9, 434–9.e1. doi:10.1016/j.cgh.2011.02.007
Song, Q., Zhang, Y., Liu, H., and Du, Y. (2020). Potential of using cell-free DNA and miRNA in breast milk to screen early breast cancer. Biomed. Res. Int. 2020, 8126176. doi:10.1155/2020/8126176
Ståhlberg, A., Krzyzanowski, P. M., Egyud, M., Filges, S., Stein, L., and Godfrey, T. E. (2017). Simple multiplexed PCR-Based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat. Protoc. 12, 664–682. doi:10.1038/nprot.2017.006
Su, J., Liang, Y., and He, X. (2024). Global, regional, and national burden and trends analysis of gallbladder and biliary tract cancer from 1990 to 2019 and predictions to 2030: a systematic analysis for the global burden of disease study 2019. Front. Med. 11, 1384314. doi:10.3389/fmed.2024.1384314
Sun, K., Jiang, P., Chan, K. C. A., Wong, J., Cheng, Y. K. Y., Liang, R. H. S., et al. (2015). Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U. S. A. 112, 5503–5512. doi:10.1073/pnas.1508736112
Tamada, K., Ushio, J., and Sugano, K. (2011). Endoscopic diagnosis of extrahepatic bile duct carcinoma: advances and current limitations. World J. Clin. Oncol. 2, 203–216. doi:10.5306/wjco.v2.i5.203
Tavano, F., Latiano, A., Palmieri, O., Gioffreda, D., Latiano, T., Gentile, A., et al. (2024). Duodenal fluid analysis as a rewarding approach to detect low-abundance mutations in biliopancreatic cancers. Int. J. Mol. Sci. 25, 8436. doi:10.3390/ijms25158436
Tawarungruang, C., Khuntikeo, N., Chamadol, N., Laopaiboon, V., Thuanman, J., Thinkhamrop, K., et al. (2021). Survival after surgery among patients with cholangiocarcinoma in northeast Thailand according to anatomical and morphological classification. BMC Cancer 21, 497. doi:10.1186/s12885-021-08247-z
Thongyoo, P., Chindaprasirt, J., Aphivatanasiri, C., Intarawichian, P., Kunprom, W., Kongpetch, S., et al. (2025). KRAS mutations in cholangiocarcinoma: prevalence, prognostic value, and KRAS G12/G13 detection in cell-free DNA. Cancer Genomics Proteomics 22, 112–126. doi:10.21873/cgp.20492
Torga, G., and Pienta, K. J. (2018). Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 4, 868–870. doi:10.1001/jamaoncol.2017.4027
Underhill, H. R., Kitzman, J. O., Hellwig, S., Welker, N. C., Daza, R., Baker, D. N., et al. (2016). Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162. doi:10.1371/journal.pgen.1006162
Uson Junior, P. L. S., and Borad, M. J. (2022). Precision approaches for cholangiocarcinoma: progress in clinical trials and beyond. Expert Opin. Investig. Drugs 31, 125–131. doi:10.1080/13543784.2022.2017882
Vaidyanathan, R., Soon, R. H., Zhang, P., Jiang, K., and Lim, C. T. (2018). Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab. Chip 19, 11–34. doi:10.1039/c8lc00684a
Valle, J. W., Kelley, R. K., Nervi, B., Oh, D.-Y., and Zhu, A. X. (2021). Biliary tract cancer. Lancet lond. Engl. 397, 428–444. doi:10.1016/S0140-6736(21)00153-7
van der Leest, P., and Schuuring, E. (2024). Critical factors in the analytical work flow of circulating tumor DNA-based molecular profiling. Clin. Chem. 70, 220–233. doi:10.1093/clinchem/hvad194
Varghese, A. M., Patel, J., Janjigian, Y. Y., Meng, F., Selcuklu, S. D., Iyer, G., et al. (2021). Noninvasive detection of polyclonal acquired resistance to FGFR inhibition in patients with cholangiocarcinoma harboring FGFR2 alterations. JCO Precis. Oncol. 5, 44–50. doi:10.1200/PO.20.00178
Wan, J. C. M., Massie, C., Garcia-Corbacho, J., Mouliere, F., Brenton, J. D., Caldas, C., et al. (2017). Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238. doi:10.1038/nrc.2017.7
Wardenaar, R., Liu, H., Colot, V., Colomé-Tatché, M., and Johannes, F. (2013). Evaluation of MeDIP-chip in the context of whole-genome bisulfite sequencing (WGBS-seq) in arabidopsis. Methods Mol. Biol. Clifton N. J. 1067, 203–224. doi:10.1007/978-1-62703-607-8_13
Wasenang, W., Chaiyarit, P., Proungvitaya, S., and Limpaiboon, T. (2019). Serum cell-free DNA methylation of OPCML and HOXD9 as a biomarker that May aid in differential diagnosis between cholangiocarcinoma and other biliary diseases. Clin. Epigenetics 11, 39. doi:10.1186/s13148-019-0634-0
Watanabe, K., Nakamura, T., Kimura, Y., Motoya, M., Kojima, S., Kuraya, T., et al. (2022). Tumor-informed approach improved ctDNA detection rate in resected pancreatic cancer. Int. J. Mol. Sci. 23, 11521. doi:10.3390/ijms231911521
Weber, J. S., Carlino, M. S., Khattak, A., Meniawy, T., Ansstas, G., Taylor, M. H., et al. (2024). Individualised neoantigen therapy mRNA-4157 (V940) plus pembrolizumab versus pembrolizumab monotherapy in resected melanoma (KEYNOTE-942): a randomised, phase 2b study. Lancet lond. Engl. 403, 632–644. doi:10.1016/S0140-6736(23)02268-7
Yang, J. D., Ghoz, H., Aboelsoud, M. M., Taylor, W. R., Yab, T. C., Berger, C. K., et al. (2021a). DNA methylation markers for detection of cholangiocarcinoma: discovery, validation, and clinical testing in biliary brushings and plasma. Hepatol. Commun. 5, 1448–1459. doi:10.1002/hep4.1730
Yang, X., Hu, Y., Yang, K., Wang, D., Lin, J., Long, J., et al. (2021b). Cell-free DNA copy number variations predict efficacy of immune checkpoint inhibitor-based therapy in hepatobiliary cancers. J. Immunother. Cancer 9, e001942. doi:10.1136/jitc-2020-001942
Yin, L., Duan, A., Zhang, W., Li, B., Zhao, T., Xu, X., et al. (2024). Identification of whole-genome mutations and structural variations of bile cell-free DNA in cholangiocarcinoma. Genomics 116, 110916. doi:10.1016/j.ygeno.2024.110916
Yoo, C., George, L., Jeong, H., Ho Jeong, J., Kim, K.-P., and Lee, S. (2024a). Utility of circulating tumor DNA (ctDNA) as a predictive biomarker for disease monitoring in patients (pts) with cholangiocarcinoma (CCA) before and during adjuvant chemotherapy (ACT): sub-Analysis of the randomized phase 2 STAMP trial. | request PDF. J. Clin. Oncol. 41, 4123. doi:10.1200/JCO.2023.41.16_suppl.4123
Yoo, C., Jeong, H., Jeong, J. H., Kim, K.-P., Lee, S., Ryoo, B.-Y., et al. (2024b). Circulating tumor DNA status and dynamics predict recurrence in patients with resected extrahepatic cholangiocarcinoma. J. Hepatol. S0168-8278 (24), 861–870. doi:10.1016/j.jhep.2024.10.043
Yu, J., Avriett, T. A., Ray, C. M., and Kim, R. D. (2024). Circulating tumor DNA analysis guiding adjuvant treatment in resected stage III cholangiocarcinoma: a case report. J. Gastrointest. Oncol. 15, 485–490. doi:10.21037/jgo-23-815
Yu, J., He, A. R., Ouf, M., Mehta, R., Anaya, D. A., Denbo, J., et al. (2025). Detecting early recurrence with circulating tumor DNA in stage I-III biliary tract cancer after curative resection. JCO Precis. Oncol. 9, e2400443. doi:10.1200/PO-24-00443
Zhang, K., Fu, R., Liu, R., and Su, Z. (2024). Circulating cell-free DNA-Based multi-cancer early detection. Trends Cancer 10, 161–174. doi:10.1016/j.trecan.2023.08.010
Zhang, L., Parvin, R., Fan, Q., and Ye, F. (2022). Emerging digital PCR technology in precision medicine. Biosens. Bioelectron. 211, 114344. doi:10.1016/j.bios.2022.114344
Zhang, B., Xu, C.-W., Shao, Y., Wang, H.-T., Wu, Y.-F., Song, Y.-Y., et al. (2015). Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. Exp. Ther. Med. 9, 1383–1388. doi:10.3892/etm.2015.2221
Zill, O. A., Greene, C., Sebisanovic, D., Siew, L., Leng, J., Vu, M., et al. (2015). Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 5, 1040–1048. doi:10.1158/2159-8290.CD-15-0274
Glossary
ctDNA Circulating tumor DNA
CCA Cholangiocarcinoma
iCCA intrahepatic cholangiocarcinoma
eCCA extrahepatic cholangiocarcinoma
dCCA distal cholangiocarcinoma
OS Overall survival
cfDNA Cell free DNA
rt-qPCR real-time quantitative PCR
ddPCR digital droplet PCR
NGS Next-generation sequencing
TAm-Seq Tagged amplicon deep sequencing
CAPP-Seq Cancer personalized profiling by deep sequencing
WGS Whole genome sequencing
WES Whole exome sequencing
WGBS Whole genome bisulfite sequencing
MRD Minimal residual disease
PFS Progression-free survival
TME Tumor microenvironment
BTC Biliary tract cancer
EUS Endoscopic ultrasound
MRCP Magnetic resonance cholangiopancreatography
ERCP Endoscopic retrograde cholangiopancreatography
CA19-9 Carbohydrate antigen 19–9
DF Duodenal fluid
PPV Positive predictive value
MS-HRM Methylation-sensitive high-resolution melting
RRBS Reduced-representation bisulfite sequencing
ICI Immune checkpoint inhibitor
CEA Carcinoembryonic antigen
MRD Molecular residual disease
DFS Disease-free survival
AFV Allele frequency variance
RFS Recurrence-free survival
MAF Mutant allele frequency
HR Homologous recombination repair
CH Clonal hematopoiesis
CNVs Copy number variations
CTCs Circulating tumor cells
CCF ctDNA content fraction
NSCLC Non-small Cell Lung Cancer
PC Pancreatic Cancer
Keywords: cholangiocarcinoma, circulating tumor DNA, liquid biopsy, prognosis monitoring, tumor-informed ctDNA
Citation: Wang Y, Li Y, Liang Z, Zhang Y, Li T, Tian C, Zhao J, Jin B, Cao J and Lin Y (2025) Circulating tumor DNA in cholangiocarcinoma: current clinical applications and future perspectives. Front. Cell Dev. Biol. 13:1616064. doi: 10.3389/fcell.2025.1616064
Received: 22 April 2025; Accepted: 14 June 2025;
Published: 02 July 2025.
Edited by:
Masahide Takahashi, Fujita Health University, JapanCopyright © 2025 Wang, Li, Liang, Zhang, Li, Tian, Zhao, Jin, Cao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Lin, bGR5eV9saW55eUBsenUuZWR1LmNu
 Yi Wang
Yi Wang Yike Li1
Yike Li1 Yanyan Lin
Yanyan Lin