- 1Department of Pathogen Biology and Biosecurity, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China
- 2Institute of Human Virology, Guangzhou, China
- 3Faculty of Forensic Medicine, Guangdong Province Translational Forensic Medicine Engineering Technology Research Center, Guangzhou, China
Electrophilic compounds from natural products (NPs) and metabolites can covalently modify the cysteines of target proteins to induce biological activities. To facilitate the discovery of novel NPs and metabolites, chemical probes with various thiol groups—mimicking the reactivity of cysteine—have been developed. These probes are designed to react with electrophilic groups of NPs and metabolites in an electrophilic addition mechanism, with the resulting adducts having molecular masses which equal to the sum of the probe and the target compound. This principle has been fundamental to analyzing mass spectrometry (MS) data and calculating the exact molecular weights of the target compound. In this study, we report a phenol thiol probe initially designed to mimic cysteine reacts with Mollugin and other structurally related NPs in an electrophilic free radical addition mechanism, and thus leads to the incorporation of not only the thiol probe but also a hydroxyl group in the adducts. Our results demonstrate that the phenol thiol group of the probe cannot always represent the thiol in cysteine to discover novel NPs or metabolites that can covalently modify cysteines.
1 Introduction
Natural product (NP) or metabolite compounds containing electrophilic groups can covalently modify the cysteines of target proteins to induce biological activities (Berdan et al., 2019; Gehrtz and London, 2021; Yang et al., 2012; Hoch et al., 2020; Qin et al., 2020; Byun et al., 2023). Given the scarcity and high chemical diversity of such compounds, a variety of chemical probes have been developed to facilitate the discovery and identification of novel electrophilic NPs or metabolites from extracts of cells or body fluids (Hughes, 2021; Wesche et al., 2017; Reimer and Hughes, 2017; Castro-Falcón et al., 2016; Jiang et al., 2020; Weerapana et al., 2010). The key part of these probes is the “war head”, which is designed to react with the electrophilic groups of the target compounds. We previously reported a nucleophilic chemical probe (Probe 1) with a phenol thiol group as the “war head” for the discovery of novel electrophilic NPs as cysteine modulators (Figure 1) (Gao et al., 2022). This chemical probe can afford accurate mass information of NPs, by assuming that the phenol thiol group is added to the NP and the molecular weight of the product is the sum of the molecular weights of the probe and NP. This mechanism also has been successfully applied to other chemical probes that employ alky thiol or cysteine as the “war head”.
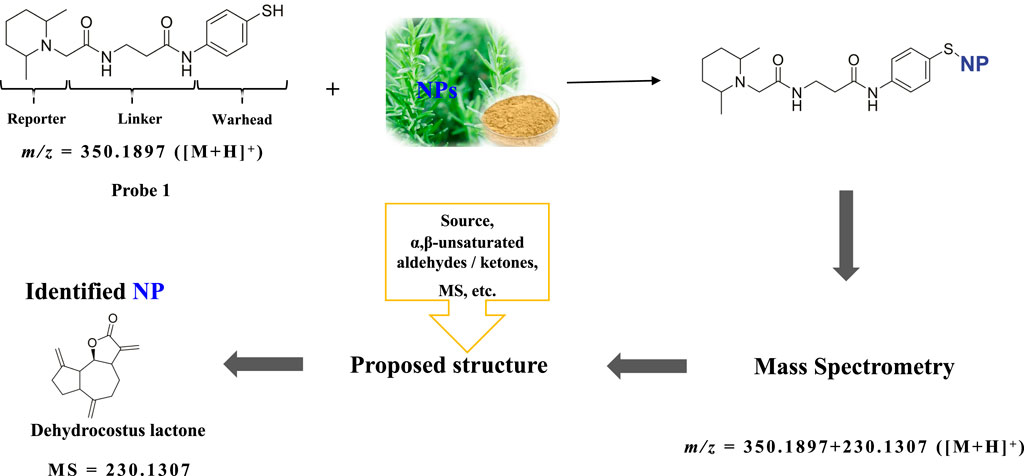
Figure 1. Working mechanism of Probe 1 and rationale of the deduction of the molecular weight of the NP. Probe 1 reacts with the electrophilic NP in the NP extract. The accurate molecular mass of the product can be detected from high resolution MS1, from which the molecular weight and molecular formula can be calculated. Dehydrocostus lactone is employed as an example.
We applied Probe 1 to discover new electrophilic NPs from various source of NPs such as the fruit stalk of Pedicellus melo, the fruit body of Ganoderma lucidum, gamboge (resin from Garcinia hanburyi), the flower of Chrysanthemum morifolium Ramat, cocklebur, and the leaf of Ginkgo biloba. We identified some electrophilic NPs that have been reported such as cucurbitacin B, gambogic acid A and xanthatin. We also successfully discovered a novel NP BG-1,then clarified its structure and identified potential target proteins (Gao et al., 2022). These successful results showed the efficacy of the assumption on the calculation of target NP’s accurate molecular weight. However, in our recent effort to identify electrophilic NPs from Rubia cordifolia L. (madder), we found a new type of addition reaction of Probe 1 with some NPs that did not follow the assumption above. More importantly, we discovered that Probe 1 did not always capture the NPs that can react with cysteine. These “off-target” effects showed that it is necessary to carefully verify the accuracy of “war heads” of our and other chemical probes.
2 Materials and methods
2.1 Materials and reagents
We used commercially available reagents and solvents without further purification. Preparative thin-layer chromatography (TLC) silica gel plates (HSGF254) were purchased from Yantai Jiangyou Silica Gel Development Co., Ltd., with a coating thickness of 0.4–0.5 mm and a pH range of 6.2–6.8. TLC visualization was performed under UV light (254 nm or 365 nm) or by phosphomolybdic acid staining. Flash chromatography was carried out using 300–400 mesh silica gel. The following reagents were used: ethanol, ascorbic acid, methanol (HPLC grade), dimethyl sulfoxide (DMSO), dichloromethane (DCM), 3,5-dimethylphenol thiol (DPT), tetrahydrofuran (THF), N,N-dimethylformamide (DMF), tert-butyl hydroperoxide (TBHP), 2,2,6,6-tetramethylpiperidin-1-oxyl (TEMPO), Glabridin, and various phenol thiol and small thiol molecules for chemical synthesis. All reagents were purchased from commercial suppliers including Shanghai Macklin Biochemical Co., Ltd., Anhui Zesheng Technology Co., Ltd., Shanghai Titan Scientific Co., Ltd., and Shanghai TCI Development Co., Ltd., and used as received. Rubia cordifolia L. (Rubiaceae) was purchased from Anguo Medicine Source Trading Co., Ltd. (MoYuan Medicinal Materials) and authenticated by Dr. Peng Guangtian of the Department of Medicinal Botany, College of Chinese Materia Medica, Guangzhou University of Chinese Medicine. The R. cordifolia extract was prepared in-house. Mollugin was obtained from Baoji Herbest Bio-Tech Co., Ltd. (batch number HR4014W5, purity ≥98%). HPLC analysis by Herbest Bio-Tech confirmed a retention time (RT = 7.44 min) consistent with the reference standard and a purity of 99.6%, meeting enterprise specifications.
2.2 The general methods of the reaction of probe 1 with NPs or natural extract
The NP (natural extract) (10 mg), ascorbic acid (2 mg) and Probe 1 (3 mg) was dissolved in certain solvent (2 mL, methanol, DCM or DMF depending on the solubility of NP or natural extract) in a flask. The reaction was monitored with TLC. After TLC showed that the NP spot disappeared (or after reacting with the natural extract for 24 h), the reaction solvent was removed with vacuum and the residue was stored in −40°C as analysis samples. Right before the analysis by liquid chromatograph mass spectrometer (LC-MSn), the sample was dissolved with methanol to afford a 20 μg/mL (20 ppm) solution in the total weight of NP, probe 1 and ascorbic acid.
2.3 The general methods of chromatography and mass spectrometry
LC-MS/MS analysis was performed with a Q Exactive mass spectrometer (Thermofisher Scientific, United States), equipped with electrospray sources and Dionex UltiMate 3000 UHPLC (Thermofisher Scientific, United States). Aqueous mobile phase (A) was 0.1% formic acid in water and organic mobile phase (B) was acetonitrile. Samples (2 µL) were analyzed with a Hypersil GOLD-C18 HPLC column (2.1 × 100 mm, 1.9 µm) (Thermofisher Scientific, United States) with mobile phase in flow rate of 0.3 mL/min. The details of the HPLC gradient and the settings of mass spectrometry were described in support information.
2.4 Preparation method of Rubia cordifolia L. Extract
Rubia cordifolia L. raw material (500 g) was macerated with 1 L of 95% (v/v) ethanol at room temperature for 72 h. The resulting ethanol extract was concentrated to a semi-solid state using a rotary evaporator, and residual ethanol was completely removed by vacuum drying. The obtained extract was aliquoted into airtight amber vials and stored at −40°C until use.
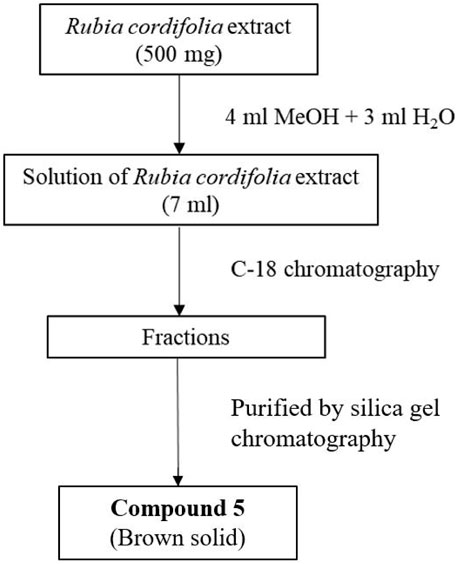
2.5 Isolation and purification of compound 5 from extract of Rubia cordifolia L
Ethanol extract of R. cordifolia L. (500 mg) was dispensed in 60% methanol water solution (7 mL) and the solution was loaded to a C-18 column (40 g) and purified with gradient (12 mL/min) described in Supplementary Table S1. The eluted fractions were tested with high resolution mass spectrometry to identify the fractions containing the targeted molecular weight (m/z = 285.1117 [M + H]+). These fractions were collected and the solvent was removed to afford light brown solid. The obtained solid further purified with silica gel column to afford pure compound 5 (3.2 mg).
2.6 Method for reacting Mollugin or Glabridin with various phenol thiols or thiols
Mollugin or Glabridin (1 equivalent) was dissolved in 3–5 mL of methanol/dichloromethane/tetrahydrofuran/DMF together with phenol thiol or thiol compound (1.2 equivalent), and the mixture was stirred at room temperature. The reaction progress was monitored by TLC. Upon completion of the reaction, the solvent was removed by rotary evaporation under reduced pressure, and the residue was purified by silica gel column chromatography. The purified product was structurally confirmed by LC-MS, NMR, or MS.
3 Results
3.1 Probe 1 reacts with Mollugin through an unexpected mechanism
In this study, we utilized Probe 1 to discover cysteine-targeted NPs from R. cordifolia L. (madder), a traditional medicinal plant widely used in Asia (Wen et al., 2022). The reaction was monitored by LC-MS as previously described (Supplementary Tables S1, S2). A qualified adduct signal with m/z = 650.2894 was detected (Figures 2A,B) and we deduce the expected exact mass of the corresponding NP with the established principle. However, this mass did not match with any known NPs previously reported from R. cordifolia L. Intriguingly, the observed mass could be rationalized if an additional oxygen atom was incorporated into the product formed from the reaction of Probe 1 with Mollugin, a well-documented bioactive compound from R. cordifolia L. (Wen et al., 2022; Zhu et al., 2013). To validate this hypothesis, we reacted a Mollugin standard with Probe 1 and observed an identical LC-MS signal to that obtained from the R. cordifolia L. extract (Figures 2A–D). Subsequent isolation of the NP (compound 5) from the extract of R. cordifolia L. was performed (Scheme 1), and it was confirmed as Mollugin via MSn and NMR (Nuclear Magnetic Resonance) (Supplementary Figures S1–S6). We also confirmed the structure of the adduct of Mollugin and Probe-1 with MS2 (Supplementary Figure S7). We proposed potential structures for the reaction product as compounds 1–4 (Figure 2E). These results demonstrated that the phenol thiol group of Probe 1 reacts with Mollugin through a mechanism distinct from the previously established one.
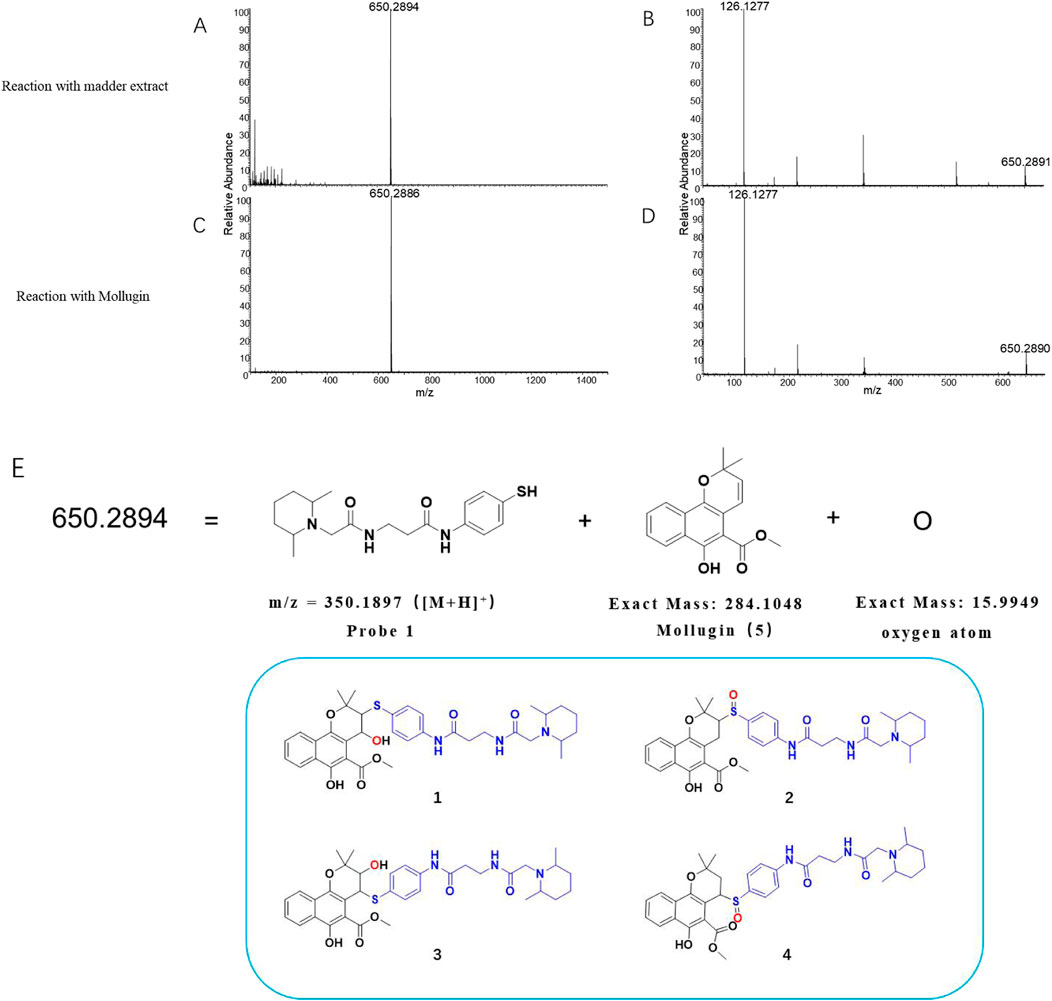
Figure 2. Identification of the effective signal at m/z = 650.2894. (A) Madder extract reacted with Probe 1, showing the parent ion with m/z = 650.2894 in MS1. (B) Madder extract reacted with Probe 1, showing the parent ion with m/z = 650.2894 in MS2. (C) Mollugin standard reacted with Probe 1, showing the parent ion with m/z = 650.2886 in MS1. (D) Mollugin standard reacted with Probe 1, showing the parent ion with m/z = 650.2886 in MS2. (E) Possible structures of the compounds containing Probe 1 (blue), Mollugin (black), and an oxygen atom (red), corresponding to the parent ion with m/z = 650.2894.
3.2 Elucidation of the structure of the reaction product and that Mollugin does not react with cysteine
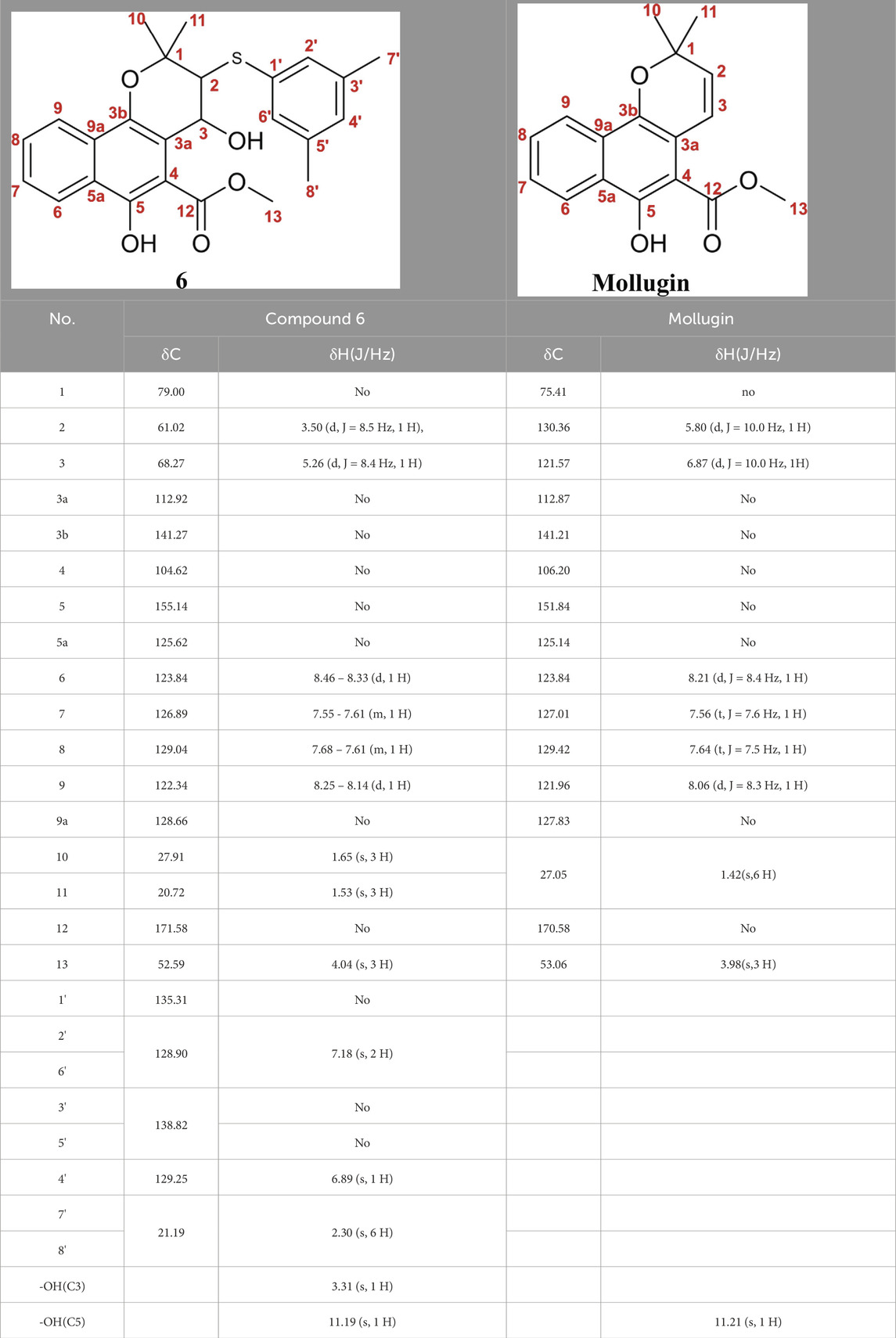
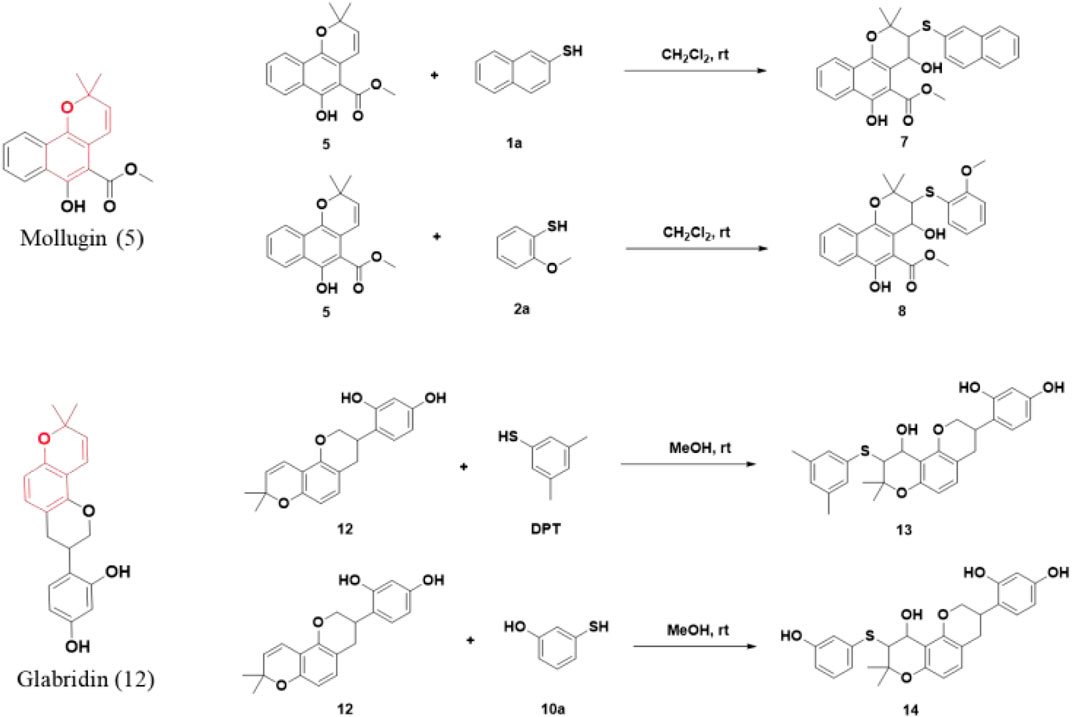
Subsequently, Mollugin was reacted with 3,5-dimethyl phenol thiol (DPT), as a surrogate for Probe 1, and the structure of the product was elucidated (Figure 3). Comprehensive analytical characterization of the product was conducted (Supplementary Figures S8–S15). These data enabled detailed assignment of the proton (H) and carbon (C) signals for compound 6, which were systematically compared with those of Mollugin (Table 1). Analysis of the H-H and C-H correlation spectra from 2D NMR revealed the addition of a phenol thiol group to the C3 carbon of the alkene moiety in compound 6 (Figure 3B). In addition to DPT, other phenol thiols reacted with Mollugin under mild conditions (25°C, catalyst-free), yielding the corresponding products (Figure 4A). Notably, Mollugin exhibited no reactivity toward alkyl thiols, including cysteine, under the same conditions (Figure 4B). The formation of compounds 7–10 was confirmed by LC-MS, with observed molecular weights consistent with the expected structures (Figures 4C,D). This distinct selectivity toward phenol thiols demonstrated that Mollugin could not modulate cysteine as initially expected, highlighting its unique reactivity profile.
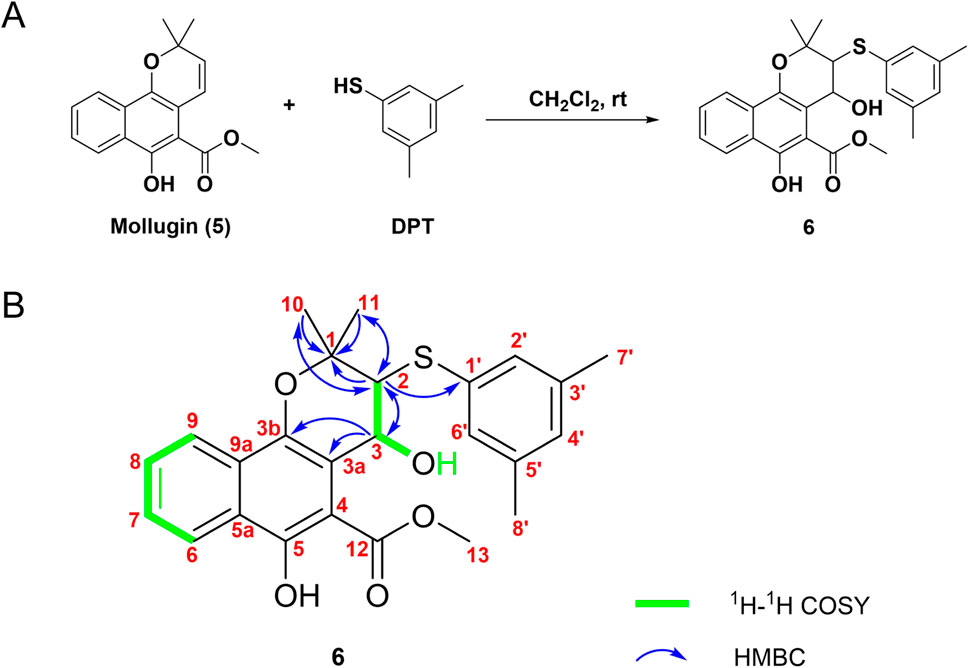
Figure 3. Confirmation of the structure of compound 6. (A) Mollugin reacts with DPT. (B) Structure of compound 6 and correlation of with 1H and 13C NMR data.
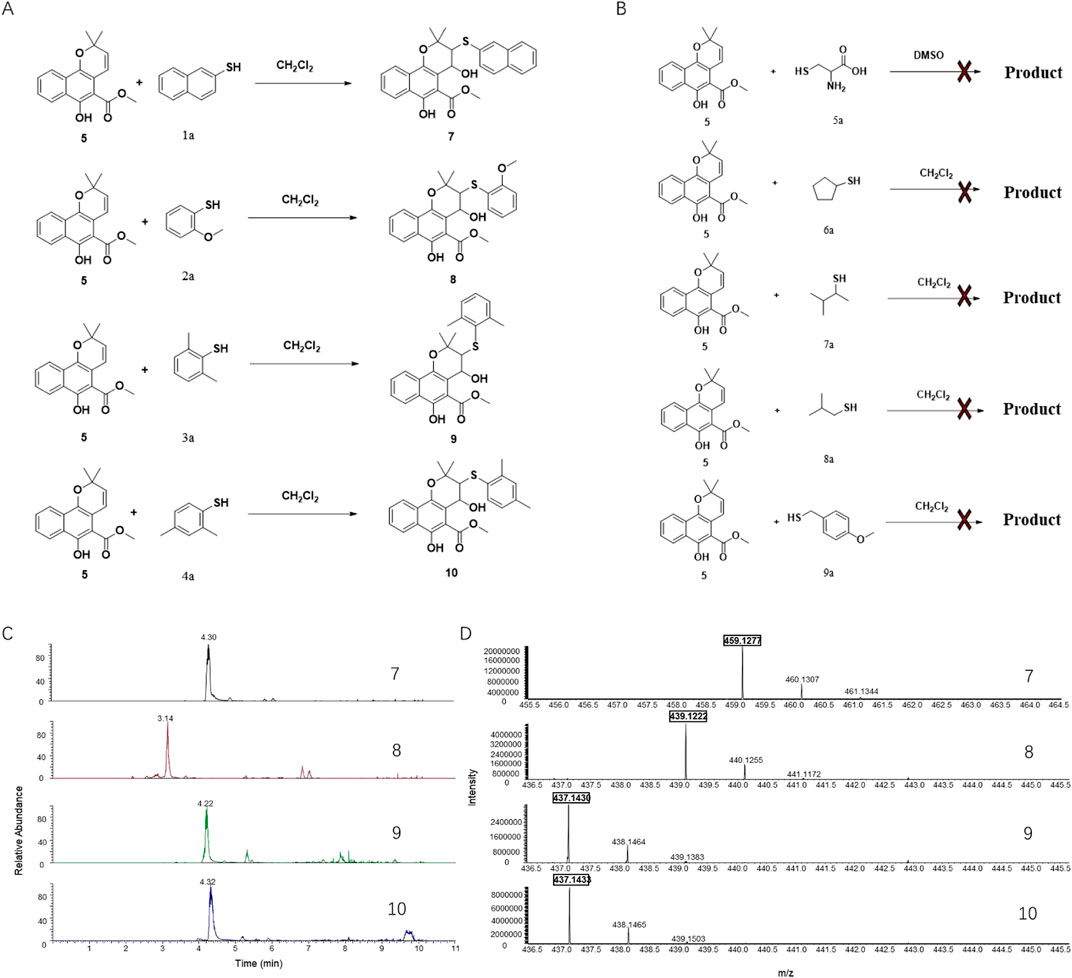
Figure 4. Mollugin does not react with various alkyl thiols but reacts with various phenol thiols, and the products demonstrate the addition of a hydroxyl group. (A) Mollugin can react with various phenol thiols. (B) Mollugin does not react with various alkyl thiols. (C) Chromatograms of the products (compounds 7–10) derived from the reactions of Mollugin with phenol thiols. (D) MS spectra of the products (compounds 7–10) derived from the reactions of Mollugin with phenol thiols.
3.3 The proposed radical reaction mechanism and its validation
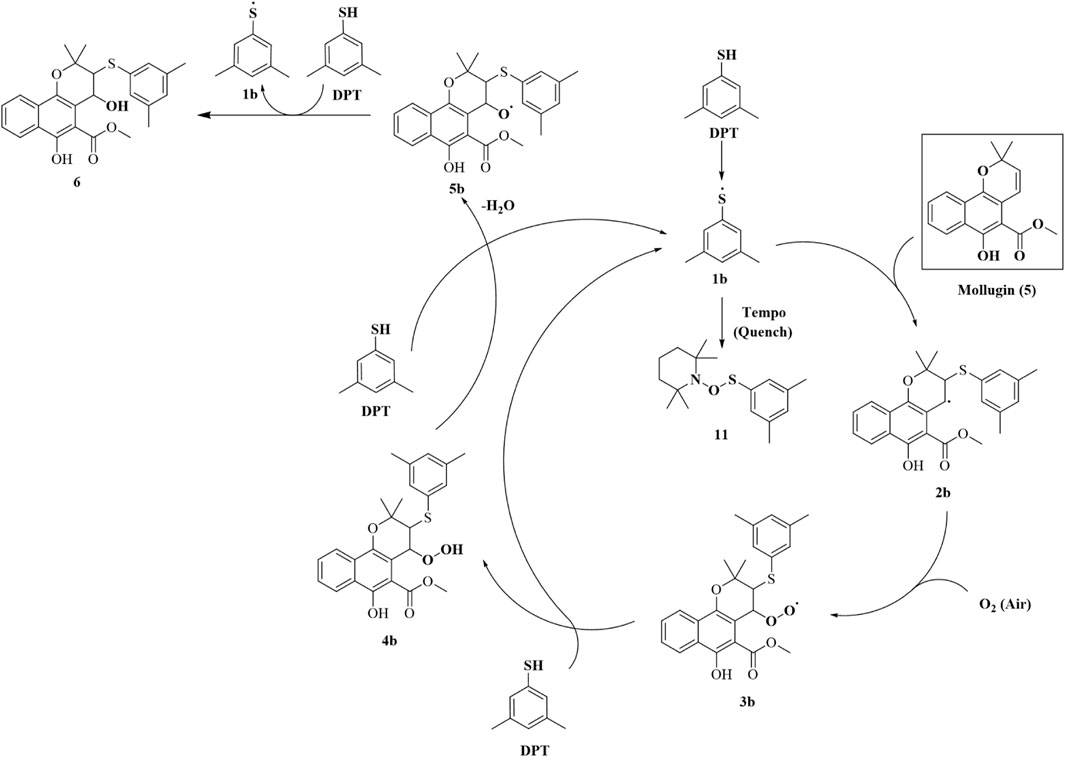
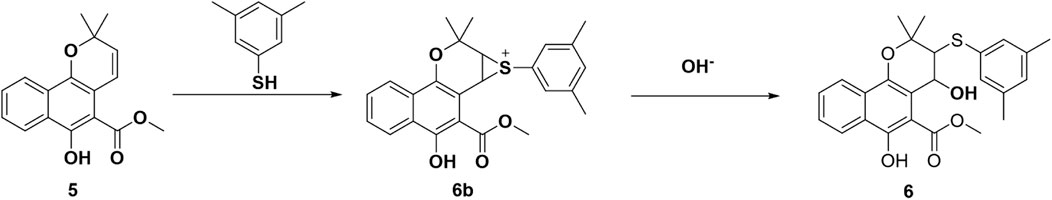
The addition of both a phenol thiol group and a hydroxyl group to the alkene bond of Mollugin is not a common electrophilic reaction in which typically only the nucleophilic molecule is added to the electrophilic NP without concomitant hydroxylation. This observation led us to propose a free radical addition mechanism, which has been reported to result in hydroxyl group addition (Scheme 2) (Zhou et al., 2015; Choudhuri et al., 2018; Wang et al., 2016). First, the clear selectivity toward phenol thiols over alky thiols aligns with the known propensity of phenol thiols to generate radicals under mild conditions, even without of strong radical inducers, a reactivity not typically observed with alkyl thiols. Second, free radical scavenger TEMPO and initiator TBHP can effectively abolish or facilitate this reaction (Figure 5; Supplementary Figures S16, 17) and the adduct of TEMPO and DPT was also isolated (Supplementary Figures S19–S21). Third, we ruled out a sulfonium-based mechanism (Taniguchi, 2006), in which the phenol thiol would form a sulfonium intermediate with the alkene bond of Mollugin, followed by nucleophilic attack by a hydroxyl group to yield the final product (Scheme 3). The other nucleophilic bromide anion did not afford any expected addition product, showing that the sulfonium is unlikely to be involved in this reaction (Supplementary Figure S18). Finally, we demonstrated that Glabridin, a NP containing the same reactive moiety as Mollugin, undergoes a similar reaction with phenol thiol (Figure 6). The structure of the corresponding synthetic product has been confirmed by NMR and MS (Supplementary Figures S22–S33). This observation suggests that the reaction mechanism is not unique to Mollugin but is likely applicable to a broader range of NPs with diverse structures.
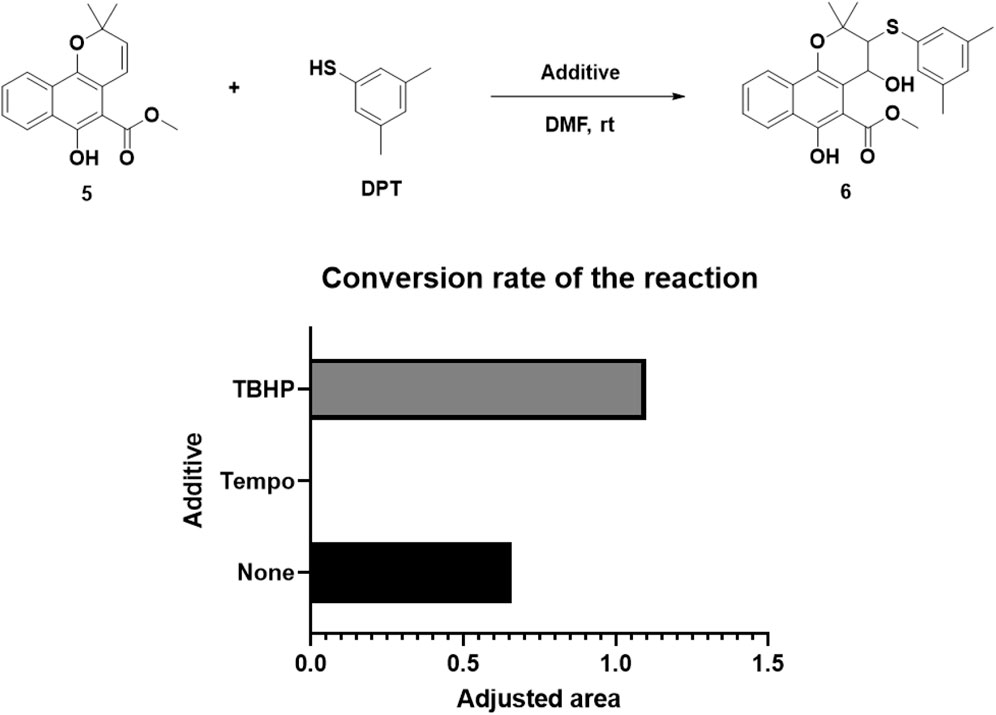
Figure 5. The influence of various additives on the reaction conversion rate was quantified by analyzing the chromatographic peak area of the target product. The adjusted area is defined as the peak area ratio of the reaction product to the internal standard.
4 Discussion
Chemical probes capable of reacting with specific functional groups have been employed as powerful tools in the discovery of novel compounds from NPs and metabolites. The reaction between the “war head” of these probes and the functional groups in target compounds is vital for obtaining the key information, such as the accurate molecular weight that can be used to deduce the molecular formula of the target compound. However, the chemical properties of the “war head” can only partially represent those of the desired molecules. In this study, we utilized a phenol thiol group as the “war head” in Probe 1, based on the assumption that it could mimic the reactivity of the thiol group in cysteine. Our unpublished studies of Probe 1 showed that its phenol thiol group exhibits higher nucleophilic reactivity than the alky thiol group toward electrophilic groups such as unsaturated carbonyls. Interestingly, here we observed that the phenol thiol “war head” reacted with Mollugin, whereas cysteine was unreactive under the same conditions. This discrepancy suggests that Probe 1 may yield false-positive results in the discovery of cysteine-targeting NPs. Furthermore, Mollugin afforded an unexpected product incorporating Probe 1 together with a hydroxyl group. This result emphasized the necessity of carefully interpreting raw MS data to avoid erroneous conclusions regarding the molecular masses of target compounds.
Mollugin has been reported as one of the bioactive compounds in R. cordifolia L., exerting its effects through mechanisms related with antioxidant pathways such as the Keap1–Nrf2 axis (Liu et al., 2023). This has led to the hypothesis that Mollugin may interact with key cysteine residues in Keap1. However, our findings demonstrate that Mollugin does not react with cysteine, suggesting that its mechanism of action is unlikely to be direct modification of cysteine residues of target proteins. These results indicated that the compounds discovered by Probe-1 may not conjugated with cysteine residuals. Instead, the high selectivity of Mollugin toward phenol thiols implies that it can be developed as a selective “war head” for targeting biologically relevant phenol thiols (Deaton et al., 2008; Hoyle et al., 2010), while avoiding the interference from alkyl thiols such as from cysteine or glutathione.
5 Conclusion
In conclusion, we discovered that Mollugin reacted with Probe 1 through free radical mechanism, affording the addition product in which the hydroxyl and thiol groups of Probe 1 were added to the alkene group of Mollugin. The chemical selectivity of Mollugin toward phenol thiols over cysteine showed that carefully considering subtle chemical differences between the “war head” group and its intended biological target is vital to design both selective and sensitive chemical probes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
TL: Writing – review and editing, Writing – original draft. RQ: Writing – original draft. WF: Writing – original draft. ZL: Writing – original draft. ZZ: Writing – original draft. CB: Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (21572281, 21877132, 22077145) and the Guangdong Basic and Applied Basic Research Foundation (2024A1515010326) to CB.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1629762/full#supplementary-material
References
Berdan, C. A., Ho, R., Lehtola, H. S., To, M., Hu, X. R., Huffman, T. R., et al. (2019). Parthenolide covalently targets and inhibits focal adhesion kinase in breast cancer cells. Cell Chem. Biol. 26 (7), 1027–1035.e22. doi:10.1016/j.chembiol.2019.03.016
Byun, D. P., Ritchie, J., Jung, Y. J., Holewinski, R., Kim, H. R., Tagirasa, R., et al. (2023). Covalent inhibition by a natural product-inspired latent electrophile. J. Am. Chem. Soc. 145 (20), 11097–11109. doi:10.1021/jacs.3c00598
Castro-Falcón, G., Hahn, D., Reimer, D., and Hughes, C. C. (2016). Thiol probes to detect Electrophilic natural products based on their mechanism of action. Acs Chem. Biol. 11 (8), 2328–2336. doi:10.1021/acschembio.5b00924
Choudhuri, K., Mandal, A., and Mal, P. (2018). Aerial dioxygen activation vs. thiol–ene click reaction within a system. Chem. Commun. 54 (30), 3759–3762. doi:10.1039/c8cc01359d
Deaton, D. N., Gao, E. N., Graham, K. P., Gross, J. W., Miller, A. B., and Strelow, J. M. (2008). Thiol-based angiotensin-converting enzyme 2 inhibitors:: P1 modifications for the exploration of the S1 subsite. Bioorg Med. Chem. Lett. 18 (2), 1681–1687. doi:10.1016/j.bmcl.2008.01.046
Gao, Y. Y., Li, K. L., Zhang, L. J., Chen, C., and Bai, C. (2022). A nucleophilic chemical probe targeting Electrophilic functional groups in an untargeted way to explore cysteine modulators in natural products. Acs Chem. Biol. 17 (7), 1685–1690. doi:10.1021/acschembio.2c00385
Gehrtz, P., and London, N. (2021). Electrophilic natural products as drug discovery tools. Trends Pharmacol. Sci. 42 (6), 434–447. doi:10.1016/j.tips.2021.03.008
Hoch, D. G., Abegg, D., Hannich, J. T., Pechalrieu, D., Shuster, A., Dwyer, B. G., et al. (2020). Combined omics approach identifies gambogic acid and related xanthones as covalent inhibitors of the serine palmitoyltransferase complex. Cell Chem. Biol. 27 (5), 586–597.e12. doi:10.1016/j.chembiol.2020.03.008
Hoyle, C. E., Lowe, A. B., and Bowman, C. N. (2010). Thiol-click chemistry: a multifaceted toolbox for small molecule and polymer synthesis. Chem. Soc. Rev. 39 (4), 1355–1387. doi:10.1039/b901979k
Hughes, C. C. (2021). Chemical labeling strategies for small molecule natural product detection and isolation. Nat. Prod. Rep. 38 (9), 1684–1705. doi:10.1039/d0np00034e
Jiang, Q. L., Zhu, Z. H., Shou, P. T., Teng, F., Zhu, Y., Zhao, H. J., et al. (2020). Targeting pharmacophore with probe-reactivity-guided fractionation to precisely identify Electrophilic sesquiterpenes and its activity of anti-TNBC. Phytochem. Anal. 31 (3), 322–332. doi:10.1002/pca.2898
Liu, X. J., Shen, X. F., Wang, H., Wang, J. Y., Rena, Y., Zhang, M., et al. (2023). Mollugin prevents CLP-Induced sepsis in mice by inhibiting TAK1-NF-κB/MAPKs pathways and activating Keap1-Nrf2 pathway in macrophages. Int. Immunopharmacol. 125, 111079. doi:10.1016/j.intimp.2023.111079
Qin, W., Yang, F., and Wang, C. (2020). Chemoproteomic profiling of protein-metabolite interactions. Curr. Opin. Chem. Biol. 54, 28–36. doi:10.1016/j.cbpa.2019.11.003
Reimer, D., and Hughes, C. C. (2017). Thiol-based probe for electrophilic natural products reveals that Most of the ammosamides are artifacts. J. Nat. Prod. 80 (1), 126–133. doi:10.1021/acs.jnatprod.6b00773
Taniguchi, N. (2006). Copper-catalyzed 1,2-hydroxysulfenylation of alkene using disulfide via cleavage of the S-S bond. J. Org. Chem. 71 (20), 7874–7876. doi:10.1021/jo060834l
Weerapana, E., Wang, C., Simon, G. M., Richter, F., Khare, S., Dillon, M. B. D., et al. (2010). Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468 (7325), 790–795. doi:10.1038/nature09472
Wen, M., Chen, Q., Chen, W., Yang, J., Zhou, X. G., Zhang, C. X., et al. (2022). A comprehensive review of Rubia L.: Traditional uses, phytochemistry, pharmacological activities, and clinical applications. Front. Pharmacol. 13.
Wesche, F., He, Y., and Bode, H. B. (2017). Solid-phase enrichment and analysis of electrophilic natural products. Beilstein J. Org. Chem. 13, 405–409. doi:10.3762/bjoc.13.43
Yang, J., Li, C. L., Ding, L., Guo, Q. L., You, Q. D., and Jin, S. H. (2012). Gambogic acid deactivates cytosolic and mitochondrial thioredoxins by covalent binding to the functional domain. J. Nat. Prod. 75 (6), 1108–1116. doi:10.1021/np300118c
Zhou, S. F., Pan, X. Q., Zhou, Z. H., Shoberu, A., and Zou, J. P. (2015). Air oxidative radical hydroxysulfurization of styrenes leading to β-Hydroxysulfides. J. Org. Chem. 80 (7), 3682–3687. doi:10.1021/acs.joc.5b00123
Zhu, Z. G., Jin, H., Yu, P. J., Tian, Y. X., Zhang, J. J., and Wu, S. G. (2013). Mollugin inhibits the inflammatory response in lipopolysaccharide-stimulated RAW264.7 macrophages by blocking the janus kinase-signal transducers and activators of transcription signaling pathway. Biol. Pharm. Bull. 36 (3), 399–406.
Keywords: nucleophilic chemical probe, Mollugin, cysteine, natural products, covalent modification
Citation: Liu T, Qi R, Feng W, Li Z, Zhong Z and Bai C (2025) Mollugin reacts with phenol thiol but not produce modification on cysteine discovered by a phenol thiol probe. Front. Cell Dev. Biol. 13:1629762. doi: 10.3389/fcell.2025.1629762
Received: 16 May 2025; Accepted: 17 July 2025;
Published: 20 August 2025.
Edited by:
Linhui Zhai, Tongji University, ChinaReviewed by:
Xixin He, Guangzhou University of Chinese Medicine, ChinaGang Chen, Guangdong Pharmaceutical University, China
Copyright © 2025 Liu, Qi, Feng, Li, Zhong and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan Bai, YmFpY2h1YW5AbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Tao Liu
Tao Liu Ruibing Qi1,2†
Ruibing Qi1,2†