- 1Department of Cell Biology, University of Texas Southwestern Medical Center, Dallas, TX, United States
- 2Department of Molecular Biology, University of Wyoming, Laramie, WY, United States
Nuclear morphology is a defining cellular feature, differing based on cell type, tissue type, and species. In healthy cells, nuclear morphology is generally tightly regulated and maintained; however, dynamic changes in nuclear morphology are observed under certain conditions, for instance in early embryos and in some immune cells. Deviations in normal nuclear morphology are linked to numerous diseases, including most cancers and premature aging syndromes. Many regulators of nuclear morphology have been identified, encompassing both intranuclear, cytoplasmic, and extracellular factors. Of note, recent studies have converged on chromatin and chromatin-associated proteins as key determinants of nuclear morphology and dynamics. In this review we discuss how the chromatin landscape regulates nuclear morphology in both normal and diseased cellular states. Additionally, we highlight emerging technologies that promise to bridge critical gaps in our understanding of nuclear morphology, including new approaches to probe nuclear structure and the use of synthetic cells.
1 Introduction
Cells vary substantially in size and morphology, ranging from yeast cells, which have a diameter of 3 μm, to nerve cells in the neck of the giraffe, which can be 3 m in length. A fundamental question is how organelle size and shape are tuned to support the structure and function of such diverse cell types. The regulation of nuclear size and shape is one area of particular interest (Marshall, 2002; Levy and Heald, 2012; Edens and Levy, 2014; Jevtic et al., 2015).
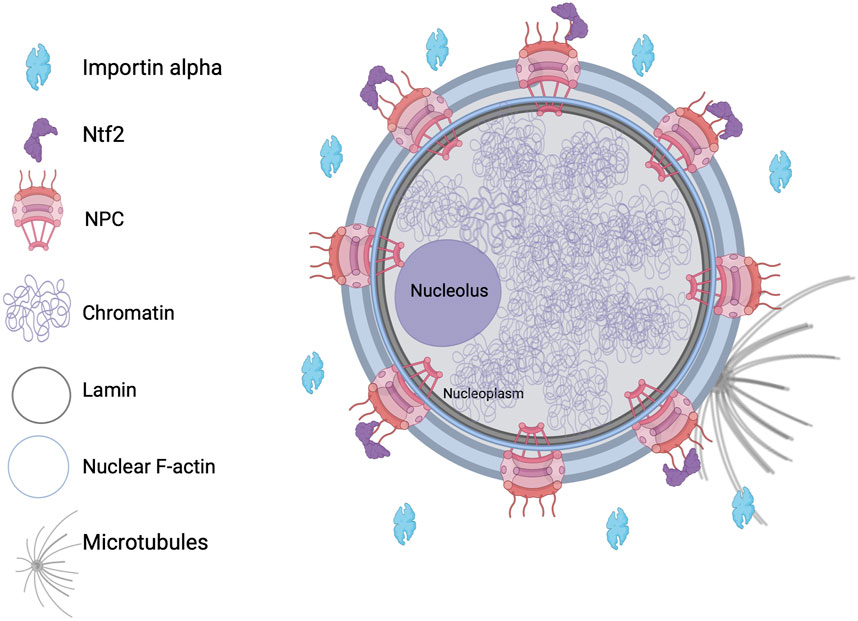
In eukaryotic cells, the nucleus is comprised of a double-membrane nuclear envelope (NE) that encloses the DNA and is often continuous with the endoplasmic reticulum. Nucleoporins are the proteins that make up nuclear pore complexes (NPCs) which span the NE, traversing through the inner and outer nuclear membranes (Alber et al., 2007; Fernandez-Martinez and Rout, 2009). Selective transport of molecules occurs through these NPCs (D'Angelo and Hetzer, 2008; Lin and Hoelz, 2019). The nuclear lamina is a meshwork that lines the nucleoplasmic face of the inner nuclear membrane and is composed of intermediate filament lamin proteins and additional interacting proteins, providing mechanical support and structural organization (Figure 1).
The maintenance of organelle shape and size is likely critical for cellular function. Under physiological conditions, nuclear sizes typically scale with cell sizeto maintain a constant nuclear-to-cytoplasmic ratio (Conklin, 1912; Gregory, 2005; Jorgensen et al., 2007; Neumann and Nurse, 2007; Hara and Kimura, 2009; Jevtic and Levy, 2015; Vukovic et al., 2016; Cantwell and Nurse, 2019; Sengupta et al., 2025). With respect to shape, nuclei are generally roughly spherical, although there are variations based on cell type, species, and differentiation status. Important questions in cell biology relate to the mechanisms responsible for the regulation of nuclear morphology.
Human granulocytes exhibit multi-lobed nuclei, connected by short channels of nucleoplasm (Skinner and Johnson, 2017). Spindly-shaped or fusiform nuclei are commonly seen in human fibrocytes and syncytial endosperm of flowering plants like Arabidopsis thaliana (Skinner and Johnson, 2017). Pathological conditions, such as cancers and laminopathies, are associated with significant alterations in nuclear morphology. Mutations in lamin A are known to cause Hutchinson-Gilford progeria syndrome, which is characterized by dysmorphic nuclei (Scaffidi and Misteli, 2006). Enlarged nuclei are observed in various forms of cancer. Furthermore, lobulated nuclei are characteristic of adenocarcinomas, and nuclei with grooves and clefts are seen in thyroid cancers (Zink et al., 2004). These anomalies in nuclear morphologies are often used by clinicians for diagnostic purposes. Nuclear inclusions and abnormal nuclei are also associated with neurodegenerative disease (Woulfe, 2008). Importantly, biological processes, including gene expression and cell migration, can be influenced by nuclear morphology. For example, preventing dynamic changes in nuclear morphology impedes the transition of cells to S phase (Aureille et al., 2019). Cells with altered nuclear elasticity due to changes in lamin A/C levels exhibit altered migration capabilities (Bell and Lammerding, 2016).
In eukaryotes, genomic DNA is wrapped around histone octamers, comprising two copies each of the core histones H2A, H2B, H3, and H4, along with the linker histone H1. This facilitates the formation of highly condensed chromatin which, along with RNAs and other proteins, enables the packaging of DNA within the nucleus (Luger et al., 2012). Chromatin structure can be broadly divided into two types: heterochromatin which is more condensed and inhibitory for DNA metabolic processes and euchromatin which is more open and accessible to DNA-binding factors. The dynamism between euchromatin and heterochromatin is regulated by ATP-dependent chromatin remodelers, post-translational histone modifications like methylation, acetylation, and phosphorylation, and direct DNA methylation (Suganuma and Workman, 2011; Nodelman and Bowman, 2021; Mattei et al., 2022) (Figure 2). Of note, aberrations in these processes are the etiology of several diseases and disorders. Altered promoter methylation leads to aberrant gene expression in neurodevelopmental disorders like Parkinson’s, Alzheimer’s, and Huntington’s disease (Lu et al., 2013). Gain and loss of function mutations in histone modifiers are observed in various forms of cancer, such as lung squamous cell carcinoma (Brennan et al., 2017; Papillon-Cavanagh et al., 2017; Farhangdoost et al., 2021), clear cell renal carcinoma (Abuzeid et al., 1987), myeloid neoplasms, breast and prostate cancer (Varambally et al., 2002), and others (Janssen and Lorincz, 2022). Chromatin is the major occupant of the nucleus and, as such, it is perhaps not surprising that the chromatin landscape can modulate nuclear morphology. In this review, we first provide a broad overview of known regulators of nuclear morphology and then focus on more recent studies that highlight the roles of chromatin structure and factors in regulating nuclear morphology and, in turn, cellular and nuclear function.
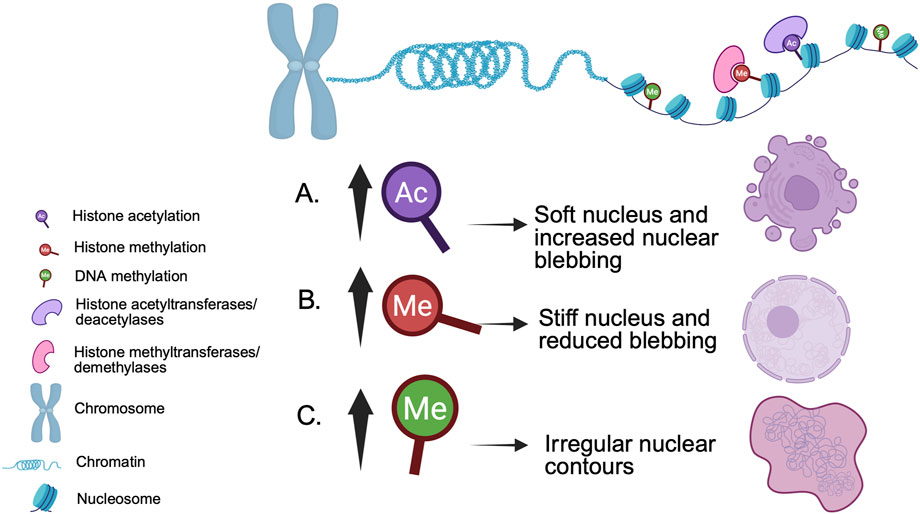
Figure 2. Example epigenetic modifications and their impact on nuclear morphology. (A) Nuclear softening and blebs caused by increased histone acetylation are reversed by (B) increased histone methylation (Stephens et al., 2018). (C) Increased DNA methylation is associated with irregularly shaped nuclei (de Capoa et al., 1996; Rougier et al., 1998).
2 Regulators of nuclear morphology
An important determinant of nuclear morphology is the nuclear lamina (Figure 1). The nuclear lamina contributes to the ability of the nucleus to respond to mechanical forces and to withstand intracellular and extracellular forces to avoid catastrophe (Swift et al., 2013; Swift and Discher, 2014; Morival et al., 2025). Lamin A/C restricts nuclear deformation while facilitating movement through narrow channels with smooth surfaces. Experiments with fibroblasts cultured on arrays of fibronectin-coated micropost barriers that mimic collagen fiber bundles showed that lamin A/C facilitates nuclear passage between slender obstacles. Nuclei containing lamin A/C preserve their oval shape despite local indentations. Conversely, nuclei deficient in lamin A/C experience severe distortion and become entangled around obstacles, impeding movement. This supports a model where the nucleus deforms like a droplet, with lamin A/C providing surface tension that allows for local invaginations that enable forward movement while maintaining overall shape (Katiyar et al., 2022).
Nuclei formed in Xenopus laevis egg extracts in the presence of dynamic F-actin exhibit a bilobed shape, with distinct membrane compositions in each lobe and F-actin concentrated at the inner nuclear envelope (Figure 1). Adding lamin A, not present in Xenopus eggs, results in more spherical nuclei. This indicates that a balance of forces exerted by nuclear F-actin and lamin A influences nuclear shape. Nuclear F-actin filaments, nucleated by formins, are thought to exert outward forces that alter nuclear morphology unless counterbalanced by lamin A (Mishra and Levy, 2022a). Microtubules can also influence nuclear morphology, for instance short term proteasome inhibition causes microtubule-mediated NE deformation independently of nuclear import (Sengupta et al., 2025). Nuclear lamin concentration can influence nuclear growth and size, with low and high levels leading to increased and decreased nuclear size, respectively, regardless of the type of lamin expressed (Jevtic et al., 2015). The phosphorylation of lamin B3 by protein kinase C plays a role in controlling nuclear size during early X. laevis development and in mammalian cells, indicating that this mechanism of nuclear size regulation is conserved (Edens and Levy, 2014; Edens et al., 2017).
Nucleocytoplasmic transport is crucial for nuclear size regulation, as increased nuclear influx and inhibited efflux are associated with nuclear volume expansion and blebbing. In fission yeast, excessive accumulation of mRNA and protein within the nucleus results in enlarged nuclear size (Kume et al., 2017). Factors that regulate nuclear import, such as Importin alpha and Nuclear Transport Factor 2 (NTF2), play a more significant role in determining nuclear size than the amount of DNA (Figure 1), perhaps by regulating the amount of nuclear lamins that are imported (Levy and Heald, 2010). Radial growth phase primary melanoma cells exhibit larger nuclei than normal melanocytes, in particular when NTF2 levels are reduced. In patient-derived melanoma cells, increasing NTF2 expression leads to reduced nuclear size, decreased cell motility and proliferation, and enhanced apoptosis, implicating NTF2 as a melanoma tumor suppressor (Vukovic et al., 2021).
The nucleoporin ELYS plays a vital role in the assembly of the NPC following mitosis. In mammalian cells, a reduction in ELYS levels results in fewer NPCs, impaired nuclear import, diminished localization of nuclear lamin B2, and smaller nuclei. Some of these effects can be rescued by enhancing nuclear import through the overexpression of Importin alpha. Conversely, ELYS overexpression leads to a higher density of NPCs, increased import of nuclear lamin B2, and larger nuclei (Jevtic et al., 2019). In other work, mutations that lead to the clustering and/or misplacement of NPCs result in changes to the shape of the nucleus (Cohen et al., 2003; Tamura and Hara-Nishimura, 2011; Joseph-Strauss et al., 2012). While these experiments show a correlation between NPC number/distribution and nuclear size, other studies suggest that NPC assembly and nuclear expansion are independently regulated (Ryan et al., 2003; Doucet et al., 2010; Titus et al., 2010; McCloskey et al., 2018).
3 Chromatin and epigenetics as regulators of nuclear morphology
In the following sections, we discuss how chromatin structure and epigenetic determinants affect nuclear morphology and, if known, cell and organismal function.
3.1 Epigenetic factors, histones, and nuclear lamins
Epigenetic regulators affect nuclear dimensions and are linked to abnormal nuclear shape (Imbalzano et al., 2013; Furusawa et al., 2015; Schreiner et al., 2015; Stephens et al., 2017; Senigagliesi et al., 2019; Stephens et al., 2019a). For instance, overexpression of the histone acetyltransferase BRD4 leads to enlarged nuclei in HeLa cells (Devaiah et al., 2016), and various chromatin components, including core histones, impact nuclear shape in MCF10A cells (Tamashunas et al., 2020). Depletion of the linker histone H1.0 was found to alter nuclear shape as measured using the elliptical Fourier coefficient ratio (Tamashunas et al., 2020). In hTERT-immortalized fibroblasts, expression of disease-relevant histone H3.3 mutations (e.g., K9M, K27M, K36M) caused nuclear shape abnormalities, including reduced nuclear size, decreased nuclear circularity, and a general increase in nuclear morphology variability across cells. These observed effects were not due to changes in cell number, toxicity, or lamin A localization (Schibler et al., 2023).
Interactions between chromatin and lamins further shape nuclear architecture (Karoutas et al., 2019; Stephens et al., 2019a). Studies of nuclear mechanics showed that chromatin mediates responses to minor deformations whereas lamin A/C responds to greater forces (Stephens et al., 2017). Modifying the levels of euchromatin and heterochromatin influences nuclear structure and stiffness (Stephens et al., 2018; Stephens et al., 2019a), with increased heterochromatin reducing nuclear blebbing in compromised nuclei (Stephens et al., 2019a; Stephens et al., 2019b). The absence of the acetyltransferase MOF or its binding partners alters nuclear mechanics, associated with reduced lamin acetylation and epigenetic changes (Karoutas et al., 2019). These observations highlight that nuclear size and shape are the result of intricate interactions between chromatin and nuclear structural proteins.
3.2 Lamin-independent effects
Chromatin is critical for the ability of the nucleus to withstand and respond to mechanical force (Reddy et al., 2008; Stephens et al., 2019a). Histone modifications are known to influence chromatin structure and nuclear morphology (Figure 2). Treating mammalian cells with histone deacetylase inhibitors to enhance euchromatin softens the nucleus, increasing blebbing (Stephens et al., 2018; Kalinin et al., 2021). Conversely, histone demethylase inhibitors that increase the amount of heterochromatin stiffen the nucleus, reducing blebbing (Stephens et al., 2018). In both scenarios, nuclear morphological changes occur independently of lamin alterations.
Although lamin disruptions are typically associated with nuclear blebbing (i.e., protrusions larger than 1 µm), changes in chromatin alone can lead to bleb formation without altered lamin levels. Mouse embryonic fibroblasts (MEFs) treated with a histone deacetylase inhibitor showed increased euchromatic H3K9ac and nuclear blebbing while lamin B1 and A/C levels remained unchanged (Stephens et al., 2017). Thus, chromatin decompaction alone can trigger nuclear blebbing without lamin depletion (Stephens et al., 2018). Contrary to studies linking nuclear blebbing with lamin B absence (Shimi et al., 2008), blebs induced with valproic acid (VPA) retained lamin B1 and A/C in 50% of treated MEF cells and 30% of treated HT1080 cells. Similar lamin B1 retention was observed in nuclear blebs when cells were treated with trichostatin A or 3-Deazaneplanocin A (DZNep) (Miranda et al., 2009; Stephens et al., 2017; Stephens et al., 2018; Stephens et al., 2019b; Esmaeili et al., 2020), suggesting nuclear blebbing can occur as a result of chromatin-mediated reductions in nuclear rigidity without nuclear lamina disruptions (Stephens et al., 2018).
3.3 Chromatin structure and DNA amount
An emerging viewpoint is that mechanical properties of the nucleus can influence nuclear morphology (Dahl et al., 2008; Stephens et al., 2019b). Alterations in chromatin modifications and spatial arrangement are linked to changes in nuclear shape (Stephens et al., 2018; Stephens et al., 2019a; Heijo et al., 2020; Flores et al., 2021). Elevating heterochromatin levels with histone demethylase inhibitors leads to increased chromatin stiffness that can rectify abnormal nuclear shapes (Stephens et al., 2018). As the nucleus expands, condensing chromatin takes up a smaller fraction of the nuclear space and increasing nuclear histone levels through the histone chaperone Npm2 further encourages nuclear enlargement (Chen et al., 2019). Conversely, supplementing Xenopus egg extract with the histone methyltransferase Set9 (Batista and Helguero, 2018) or DNA methyltransferase inhibitor Zebularine (Yoo et al., 2004) increased the proportion of nuclear space filled by chromatin, with Set9 elevating H3K4me1 and Zebularine reducing 5-methylcytosine levels. These nuclei showed diminished growth and smaller final sizes, further highlighting how chromatin structure can affect nuclear size (Chen et al., 2024).
Nuclei formed in Xenopus egg extract almost completely stopped growing when treated with DNA degrading benzonase, regardless of the nuclear size when treatment was initiated (Chen et al., 2024). Nuclear F-actin plays a role in the expansion of the nucleus (Baarlink et al., 2017; Huang et al., 2022) but benzonase-treated nuclei formed in extracts with intact actin still failed to expand. These findings suggest that DNA is crucial for nuclear growth, even in the presence of F-actin (Chen et al., 2024). Massive increases in DNA content can affect nuclear size as nuclei assembled in X. laevis egg extract using axolotl sperm chromatin, which contains 20-fold more DNA than Xenopus, exhibited a doubling in nuclear cross-sectional area (Chen et al., 2024). This indicates that nuclear size can be responsive to DNA quantity (Levy and Heald, 2010; Heijo et al., 2020). When transcriptionally inert Xenopus egg extracts were treated with VPA to increase H3K9ac or DZNep to decrease H3K27me3, nuclei were smaller without any change in nuclear import rate. Conversely, increasing H3K27me3 with the histone demethylase inhibitor Methylstat or increasing 5-methylcytosine levels with the DNA methylator NDMA led to significant nuclear enlargement. This study showed that even with similar nuclear import rates, altered chromatin structure can reduce or increase nuclear growth in a transcription-independent manner (Chen et al., 2024).
3.4 Cellular mechanics and mechanosensation
The intermediate filament protein Keratin 17 can be found within the nucleus, interacting with proteins that play a role in organizing chromatin. Keratin 17 knockout in tumor-derived HeLa and A431 cell lines leads to a decrease in histone methylation and acetylation with a concomitant increase in nuclear size (Jacob et al., 2020). Fibroblasts, the predominant cells within connective tissues, are subjected to substantial compressive forces from the surrounding extracellular matrix and fluid during activities like walking, sitting, and sleeping. Applying a static compressive force to cultured mouse fibroblasts increases nuclear levels of histone deacetylase 3, which promotes heterochromatin formation. Upon removal of the compressive force, the cells revert to their original chromatin condensation state (Damodaran et al., 2018). Whether nuclear morphology is affected under these conditions is an interesting area for future study.
Cell stretching activates mechanosensitive channels, triggering transient calcium influx (Kim et al., 2015) and increased chromatin compaction (Heo et al., 2015; Heo et al., 2016; Le et al., 2016). Mechano-transduction mediated by these channels can protect against abnormal nuclear morphology. For example, in VPA-treated MEFs that exhibit chromatin decompaction, increasing the extracellular concentration of magnesium chloride induced heterochromatin formation through histone methyltransferases, leading to a reduction in nuclear blebbing. These cells also showed increased short-extension nuclear spring constants with no change in long-extension stiffness (Stephens et al., 2019b). Interestingly, MEF cells co-treated with VPA and the transcription inhibitor alpha-amanitin also exhibited reduced nuclear blebbing (Berg et al., 2023). Treating SKOV3 cells with trichostatin A to increase histone acetylation led to chromatin decompaction and a reduction in extracellular vesicle production upon cellular compression (Toth et al., 2004).
3.5 DNA damage and nuclear blebs
One of the hallmarks of many human diseases is DNA damage. MEFs and HT1080 cells treated with the DNA damaging agents cisplatin and bleomycin exhibited increased nuclear blebbing independently of passage through mitosis. Furthermore, DNA damage-induced nuclear blebs proceeded to rupture at a >90% frequency (Stephens et al., 2019b; Eskndir et al., 2025). Disrupting DNA damage response pathways, including p53, Rb, and BRCA2, can also increase the frequency of nuclear rupture (Yang et al., 2017; Kovacs et al., 2023). Micromanipulation force measurements of isolated vimentin-null MEF nuclei revealed that DNA damage significantly reduces chromatin-based nuclear stiffness (Stephens et al., 2017; Dos Santos et al., 2021; Currey et al., 2022). Heterochromatin was reduced in response to DNA damage through activation of Ataxia-telangiectasia Mutated kinase (ATM), leading to chromatin softening, reduced nuclear rigidity, and increased nuclear blebbing and rupture (Ziv et al., 2006; Ayrapetov et al., 2014; Eskndir et al., 2025). Inhibiting ATM during DNA damage treatment rescued heterochromatin levels and restored nuclear mechanics, shape, and integrity (Eskndir et al., 2025). Defects in nuclear morphology and integrity caused by DNA damage exacerbate a cycle of nuclear dysfunction in both confined and unconfined cells (Chen et al., 2018; Xia et al., 2018; Stephens et al., 2019b; Pho et al., 2024). Furthermore, inhibition of actin contractions can rescue DNA damage-induced changes in nuclear morphology (Pho et al., 2024).
Nuclear blebs often exhibit a unique chromatin signature. In MEFs, HT1080 fibrosarcoma, and PC3 prostate cancer cell lines it was recently discovered that nuclear blebs tend to exhibit reduced DNA density but that bleb formation does not correlate with lamin B1 levels (Bunner et al., 2025). Interestingly, increased DNA damage in such blebbed nuclei was independent of rupture (Chu et al., 2025). Disrupted nuclear morphology, including blebs, is associated with multiple disorders, and chromatin structure within nuclear blebs is often altered (Pujadas Liwag et al., 2025). There is direct evidence that the chromatin methylation status affects nuclear morphology. Loss of H3K9me3 increases nuclear blebbing and rupture due to decreased nuclear rigidity, while loss of H3K9me2 decreases nuclear blebbing and rupture accompanied by increased nuclear rigidity and more compact chromocenters (Manning et al., 2025). Biological processes like transcription can also regulate nuclear morphology, for instance chemical inhibition of transcription can suppress nuclear bleb formation and rupture (Berg et al., 2023). Taken together, DNA damage and chromatin status are important determinants of nuclear blebbing and rupture.
3.6 Cell migration, differentiation, and senescence
Alterations in chromatin structure and nuclear shape contribute to cell migration. In tenocytes, mechanical stretch caused reduced levels of H3K27me3 and increased levels of H3K9ac and H3K27ac, which in turn led to changes in nuclear morphology independently of lamin A/C, promoting tenocyte migration (Xu et al., 2023). Additionally, inhibiting histone methyltransferases with small molecule inhibitors affected nuclear morphology and integrity, diminished wound closure efficiency, and impeded cellular migration (Forman et al., 2024). In chicken primordial germ cells, disruption of the transcription factor Oct4 led to a reduction in H3K27ac modifications on active chromatin regions and altered cell migration (Meng et al., 2022).
The human immune system depends on a variety of cell types to establish and maintain its surveillance capabilities. Nuclear lobulation, a characteristic of many human immune cells, is a result of increased NE deformability and increased chromatin-NE interactions (Olins et al., 2001). Neutrophils adopt malleable polymorphonuclear structures by halting chromatin loop extrusion, thus enabling them to migrate through narrow interstitial spaces (Patta et al., 2024). Short range genome reorganization favors neutrophil migration such that the nucleus can alter its shape without causing chromatin damage (Jacobson et al., 2018).
Cell differentiation is often associated with changes in nuclear stiffness and morphology. For example, nuclei in embryonic stem cells are deformable and become six times stiffer in their terminal stages of differentiation (Pajerowski et al., 2007). Furthermore, increases in cell size have been shown to contribute to cellular senescence through genome dilution (Neurohr et al., 2019; Lanz et al., 2022; Lanz et al., 2024). Given that nuclear size typically scales with cell size, an interesting area for future study is whether increased nuclear size might also contribute to the onset of senescence.
3.7 Epigenetics and disease
Epigenetic modifications profoundly affect gene expression, and aberrant modifications have been linked to various diseases like cancer, neurological disorders, cardiovascular diseases, and more recently COVID-19 syndrome (Table 1) (Vodovotz et al., 2020). How epigenetic changes give rise to disease is a broad topic that has been extensively reviewed (Gasser and Li, 2011; Cavalli and Heard, 2019; Lopez-Lopez, 2023; McCulley, 2024; Tollefsbol, 2024). Here we will only touch on a few illustrative examples. In cancers, extensive DNA methylation and histone deacetylation occur within tumor suppressor genes, which results in their silencing and ultimately exacerbates disease. Aberrant DNA methylation and histone demethylation have been shown to promote the epithelial-to-mesenchymal transition in breast cancer cells, causing the cancer to proliferate and metastasize (Serrano-Gomez et al., 2016). Breast tumors defective for ARID1A, a component of the chromatin remodeling complex involved in nucleosome sliding, exhibit altered recruitment of histone deacetylase 2 and uncontrolled cell growth (Xu et al., 2020). Aberrations in epigenetic mechanisms are not limited to breast cancer. In glioblastoma multiforme, a rare and incurable adult brain tumor, hypermethylation of DNA is found in genes that regulate the WNT, Frizzled, and Ras pathways (Horiguchi et al., 2003; Gotze et al., 2010; Lambiv et al., 2011), contributing to cell proliferation. Mutations in ATP-dependent chromatin modifiers, including SMARCB1, BRG1, and BRM, have been identified in primary tumors and tumor-derived cell lines, including rhabdoid tumors, chronic myeloid leukemia, lung cancer, and prostate cancer (Salvatore and Vandenplas, 2003; Cho et al., 2004).
Altered epigenetics are associated with other diseases in addition to cancer. Hutchinson-Gilford progeria syndrome is characterized by abnormal nuclear shape, and increasing heterochromatin can restore normal nuclear morphology in cell lines that model the disease and in patient-derived cells (Scaffidi and Misteli, 2005). Altered DNA methylation patterns are observed in pancreatic islets and adipose and muscle tissues of type 2 diabetic mice and patients (Ling et al., 2008; Barres et al., 2009). In pancreatic islets, hypermethylated DNA can be detected in genes that regulate insulin secretion (Yang et al., 2012; Hall et al., 2013). In Alzheimer’s, altered DNA methylation is observed in disease-associated genes like amyloid precursor protein, Apolipoprotein E, and Ankyrin 1 (Bijarnia-Mahay et al., 2014; Santana et al., 2023). Mutations in the ATRX chromatin remodeling complex cause several X-linked syndromes that manifest with facial dysmorphism, urogenital defects, and α-thalassemia (Gibbons and Higgs, 2000). Mutations in the chromatin remodeler SMARCAL1 are a known cause of Schimke immuno-osseous dysplasia, an autosomal recessive disorder associated with T-cell immunodeficiency, spondyloepiphyseal dysplasia, renal failure, and other symptoms (Boerkoel et al., 2002). Thus, normal cellular physiology depends on precise regulation of the epigenetic state of chromatin and proper function of chromatin-associated molecules including histones and chromatin modifiers. Open questions relate to whether epigenetics influence disease phenotypes through altered nuclear morphology.
4 Emerging techniques and models in the field of nuclear morphology
Over the past 150 years, there have been significant advancements since Sir Lionel Beale first observed alterations in typical nuclear structures in various diseases, including cancer. To this day, the Pap smear, introduced by George Papanicolaou, remains a diagnostic tool for detecting abnormal nuclear structures in cervical tissue samples (Flores et al., 2021). Microscopy serves as an essential means of examining the nuclear condition of cells, offering crucial insights into nuclear shape, nuclear mechanics, protein distribution, and genome arrangement (Kim and Lakadamyali, 2024). Researchers are beginning to elucidate the connections between mechanics and morphology by integrating atomic force microscopy with side-view light sheet microscopy. Offering high spatiotemporal resolution, this approach has been used to observe cells during compression, leading to empirical models for nuclear deformation applicable to atypical nuclear shapes (Hobson et al., 2020; Hobson and Stephens, 2020). RD-SPRITE explores the interactions between chromatin and RNA (Quinodoz et al., 2021). Spatial multi-omics, which combines FISH and immunofluorescence techniques, have been used to chart nuclear architecture, and in situ genome sequencing facilitates the simultaneous visualization and sequencing of the genome at the nucleotide level (Palihati and Saitoh, 2024). Microfluidic devices constructed from polydimethylsiloxane can be used to replicate physiological conditions for the study of intracellular mechanics and dynamics, mimicking conditions relevant to cancer cell invasion and immune cell recruitment (Davidson et al., 2015).
The intricate nature of cells and their organelles poses challenges in studying fundamental molecular processes within living organisms. One approach to addressing this issue is bottom-up synthetic biology, which involves creating life-like systems from molecular components (Blain and Szostak, 2014; Auslander et al., 2017; Gopfrich et al., 2018). This multidisciplinary area incorporates chemistry, biology, and engineering (Sismour and Benner, 2005). One of the challenges in bottom-up synthetic biology is to develop systems that mimic life and sustain physiochemical balance using a minimal set of components (Smigiel et al., 2019). A model system commonly used to study organelle size, morphology, and function is cell-free Xenopus egg extract. Nuclei assembled in Xenopus extract exhibit the typical structures and activities of nuclei found in living cells (Chen and Levy, 2018; Mishra and Levy, 2022b). The open biochemical nature of the system allows for the addition of recombinant proteins and removal of endogenous proteins by immunodepletion, as well as exogenous addition of small molecule inhibitors or activators without pleiotropic effects on transcription, translation, and cell cycle progression (Sengupta et al., 2025). Furthermore, fluorescently labeled proteins allow for live time-lapse microscopy of organelle dynamics (Jevtic and Levy, 2018). Lastly, high-throughput imaging for siRNA screening represents an effective approach to identify regulators of nuclear architecture in an unbiased manner (Schibler et al., 2023). Interestingly, this latter study identified a number of nuclear morphology effectors in two different cell types but there was very little overlap in the hits, highlighting unresolved questions about the complexity of nuclear morphology control.
5 Conclusion
How nuclear morphology is regulated is a fundamental question of cell biology which remains to be understood completely. This regulation is critical as numerous diseases present with abnormal nuclear phenotypes that are often used by clinicians for diagnostic purposes. Several factors have been identified as contributors to this regulation; however important knowledge gaps still exist. In this review, we have discussed some of the known regulators of nuclear morphology, with a particular focus on how chromatin influences the regulation of nuclear structure and function. Aberrations in epigenetic modifications are the underlying cause for numerous diseases (Farsetti et al., 2023). At the same time, changes in the epigenetic landscape also impact nuclear morphology and function. To what extent aberrant nuclear morphology, altered epigenetic status, and disease phenotype are linked is a critical open question. Simplified model systems like Xenopus egg extract and novel approaches like hydrogel chambers are being increasingly used to mimic physiological cellular conditions and advance our knowledge of fundamental processes like the regulation of nuclear morphology. At the same time, advanced microscopy techniques have improved our ability to visualize component molecules and structures relevant to epigenetics and nuclear morphology. The next decade promises to answer key questions at the intersection of chromatin biology, nuclear morphology, and disease.
Author contributions
SS: Writing – original draft, Conceptualization, Writing – review and editing. HP: Conceptualization, Writing – original draft, Writing – review and editing. DL: Supervision, Writing – review and editing, Conceptualization, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by NIH grants R35GM134885 and P20GM103432.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abuzeid, M. I., Wiebe, R. H., Aksel, S., Shepherd, J., and Yeoman, R. R. (1987). Evidence for a possible cytosol estrogen receptor deficiency in endocervical glands of infertile women with poor cervical mucus. Fertil. Steril. 47 (1), 101–107. doi:10.1016/s0015-0282(16)49943-7
Alber, F., Dokudovskaya, S., Veenhoff, L. M., Zhang, W., Kipper, J., Devos, D., et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450 (7170), 695–701. doi:10.1038/nature06405
Aureille, J., Buffiere-Ribot, V., Harvey, B. E., Boyault, C., Pernet, L., Andersen, T., et al. (2019). Nuclear envelope deformation controls cell cycle progression in response to mechanical force. EMBO Rep. 20 (9), e48084. doi:10.15252/embr.201948084
Auslander, S., Auslander, D., and Fussenegger, M. (2017). Synthetic biology-the synthesis of biology. Angew. Chem. Int. Ed. Engl. 56 (23), 6396–6419. doi:10.1002/anie.201609229
Ayrapetov, M. K., Gursoy-Yuzugullu, O., Xu, C., Xu, Y., and Price, B. D. (2014). DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. U. S. A. 111 (25), 9169–9174. doi:10.1073/pnas.1403565111
Baarlink, C., Plessner, M., Sherrard, A., Morita, K., Misu, S., Virant, D., et al. (2017). A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 19 (12), 1389–1399. doi:10.1038/ncb3641
Barres, R., Osler, M. E., Yan, J., Rune, A., Fritz, T., Caidahl, K., et al. (2009). Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 10 (3), 189–198. doi:10.1016/j.cmet.2009.07.011
Batista, I. A. A., and Helguero, L. A. (2018). Biological processes and signal transduction pathways regulated by the protein methyltransferase SETD7 and their significance in cancer. Signal Transduct. Target Ther. 3, 19. doi:10.1038/s41392-018-0017-6
Bell, E. S., and Lammerding, J. (2016). Causes and consequences of nuclear envelope alterations in tumour progression. Eur. J. Cell Biol. 95 (11), 449–464. doi:10.1016/j.ejcb.2016.06.007
Berg, I. K., Currey, M. L., Gupta, S., Berrada, Y., Nguyen, B. V., Pho, M., et al. (2023). Transcription inhibition suppresses nuclear blebbing and rupture independently of nuclear rigidity. J. Cell Sci. 136 (20), jcs261547. doi:10.1242/jcs.261547
Bijarnia-Mahay, S., Mohan, N., Goyal, D., and Verma, I. C. (2014). Mitochondrial DNA depletion syndrome causing liver failure. Indian Pediatr. 51 (8), 666–668. doi:10.1007/s13312-014-0475-z
Blain, J. C., and Szostak, J. W. (2014). Progress toward synthetic cells. Annu. Rev. Biochem. 83, 615–640. doi:10.1146/annurev-biochem-080411-124036
Boerkoel, C. F., Takashima, H., John, J., Yan, J., Stankiewicz, P., Rosenbarker, L., et al. (2002). Mutant chromatin remodeling protein SMARCAL1 causes schimke immuno-osseous dysplasia. Nat. Genet. 30 (2), 215–220. doi:10.1038/ng821
Brennan, K., Shin, J. H., Tay, J. K., Prunello, M., Gentles, A. J., Sunwoo, J. B., et al. (2017). NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci. Rep. 7 (1), 17064. doi:10.1038/s41598-017-17298-x
Bunner, S., Prince, K., Pujadas Liwag, E. M., Eskndir, N., Srikrishna, K., Amonu McCarthy, A., et al. (2025). Decreased DNA density is a better indicator of a nuclear bleb than lamin B loss. J. Cell Sci. 138 (3), jcs262082. doi:10.1242/jcs.262082
Cantwell, H., and Nurse, P. (2019). A homeostatic mechanism rapidly corrects aberrant nucleocytoplasmic ratios maintaining nuclear size in fission yeast. J. Cell Sci. 132 (22), jcs235911. doi:10.1242/jcs.235911
Cavalli, G., and Heard, E. (2019). Advances in epigenetics link genetics to the environment and disease. Nature 571 (7766), 489–499. doi:10.1038/s41586-019-1411-0
Chen, N. Y., Kim, P., Weston, T. A., Edillo, L., Tu, Y., Fong, L. G., et al. (2018). Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. U. S. A. 115 (40), 10100–10105. doi:10.1073/pnas.1812622115
Chen, P., and Levy, D. L. (2018). Nucleus assembly and import in Xenopus laevis egg extract. Cold Spring Harb. Protoc. 2018 (6), pdb prot097196. doi:10.1101/pdb.prot097196
Chen, P., Mishra, S., Prabha, H., Sengupta, S., and Levy, D. L. (2024). Nuclear growth and import can be uncoupled. Mol. Biol. Cell 35 (1), ar1. doi:10.1091/mbc.E23-04-0138
Chen, P., Tomschik, M., Nelson, K. M., Oakey, J., Gatlin, J. C., and Levy, D. L. (2019). Nucleoplasmin is a limiting component in the scaling of nuclear size with cytoplasmic volume. J. Cell Biol. 218 (12), 4063–4078. doi:10.1083/jcb.201902124
Cho, K. S., Elizondo, L. I., and Boerkoel, C. F. (2004). Advances in chromatin remodeling and human disease. Curr. Opin. Genet. Dev. 14 (3), 308–315. doi:10.1016/j.gde.2004.04.015
Chu, C. G., Lang, N., Walsh, E., Zheng, M. D., Manning, G., Shalin, K., et al. (2025). Lamin B loss in nuclear blebs is rupture dependent while increased DNA damage is rupture independent. bioRxiv. doi:10.1101/2025.02.24.639904
Cohen, M., Feinstein, N., Wilson, K. L., and Gruenbaum, Y. (2003). Nuclear pore protein gp210 is essential for viability in HeLa cells and Caenorhabditis elegans. Mol. Biol. Cell 14 (10), 4230–4237. doi:10.1091/mbc.e03-04-0260
Conklin, E. G. (1912). Cell size and nuclear size. J. Exp. Zoology 12 (1), 1–98. doi:10.1002/jez.1400120102
Currey, M. L., Kandula, V., Biggs, R., Marko, J. F., and Stephens, A. D. (2022). A versatile micromanipulation apparatus for biophysical assays of the cell nucleus. Cell Mol. Bioeng. 15 (4), 303–312. doi:10.1007/s12195-022-00734-y
Dahl, K. N., Ribeiro, A. J., and Lammerding, J. (2008). Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102 (11), 1307–1318. doi:10.1161/CIRCRESAHA.108.173989
Damodaran, K., Venkatachalapathy, S., Alisafaei, F., Radhakrishnan, A. V., Sharma Jokhun, D., Shenoy, V. B., et al. (2018). Compressive force induces reversible chromatin condensation and cell geometry-dependent transcriptional response. Mol. Biol. Cell 29 (25), 3039–3051. doi:10.1091/mbc.E18-04-0256
D'Angelo, M. A., and Hetzer, M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18 (10), 456–466. doi:10.1016/j.tcb.2008.07.009
Davidson, P. M., Sliz, J., Isermann, P., Denais, C., and Lammerding, J. (2015). Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. (Camb) 7 (12), 1534–1546. doi:10.1039/c5ib00200a
de Capoa, A., Menendez, F., Poggesi, I., Giancotti, P., Grappelli, C., Marotta, M. R., et al. (1996). Cytological evidence for 5-azacytidine-induced demethylation of the heterochromatic regions of human chromosomes. Chromosome Res. 4 (4), 271–276. doi:10.1007/BF02263676
Devaiah, B. N., Case-Borden, C., Gegonne, A., Hsu, C. H., Chen, Q., Meerzaman, D., et al. (2016). BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 23 (6), 540–548. doi:10.1038/nsmb.3228
Dos Santos, A., Cook, A. W., Gough, R. E., Schilling, M., Olszok, N. A., Brown, I., et al. (2021). DNA damage alters nuclear mechanics through chromatin reorganization. Nucleic Acids Res. 49 (1), 340–353. doi:10.1093/nar/gkaa1202
Doucet, C. M., Talamas, J. A., and Hetzer, M. W. (2010). Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141 (6), 1030–1041. doi:10.1016/j.cell.2010.04.036
Dyer, M. A., Qadeer, Z. A., Valle-Garcia, D., and Bernstein, E. (2017). ATRX and DAXX: mechanisms and mutations. Cold Spring Harb. Perspect. Med. 7 (3), a026567. doi:10.1101/cshperspect.a026567
Edens, L. J., Dilsaver, M. R., and Levy, D. L. (2017). PKC-Mediated phosphorylation of nuclear lamins at a single serine residue regulates interphase nuclear size in xenopus and Mammalian cells. Mol. Biol. Cell 28 (10), 1389–1399. doi:10.1091/mbc.E16-11-0786
Edens, L. J., and Levy, D. L. (2014). cPKC regulates interphase nuclear size during xenopus development. J. Cell Biol. 206 (4), 473–483. doi:10.1083/jcb.201406004
Eskndir, N., Hossain, M., Currey, M. L., Pho, M., Berrada, Y., Lin, K., et al. (2025). DNA damage causes ATM-Dependent heterochromatin loss leading to nuclear softening, blebbing, and rupture. Mol. Biol. Cell 36 (3), br6. doi:10.1091/mbc.E24-05-0232
Esmaeili, M., Blythe, S. A., Tobias, J. W., Zhang, K., Yang, J., and Klein, P. S. (2020). Chromatin accessibility and histone acetylation in the regulation of competence in early development. Dev. Biol. 462 (1), 20–35. doi:10.1016/j.ydbio.2020.02.013
Farhangdoost, N., Horth, C., Hu, B., Bareke, E., Chen, X., Li, Y., et al. (2021). Chromatin dysregulation associated with NSD1 mutation in head and neck squamous cell carcinoma. Cell Rep. 34 (8), 108769. doi:10.1016/j.celrep.2021.108769
Farsetti, A., Illi, B., and Gaetano, C. (2023). How epigenetics impacts on human diseases. Eur. J. Intern Med. 114, 15–22. doi:10.1016/j.ejim.2023.05.036
Fernandez-Martinez, J., and Rout, M. P. (2009). Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 21 (4), 603–612. doi:10.1016/j.ceb.2009.05.001
Flores, L. F., Tader, B. R., Tolosa, E. J., Sigafoos, A. N., Marks, D. L., and Fernandez-Zapico, M. E. (2021). Nuclear dynamics and chromatin structure: implications for pancreatic cancer. Cells 10 (10), 2624. doi:10.3390/cells10102624
Foolchand, A., Mazaleni, S., Ghazi, T., and Chuturgoon, A. A. (2022). A review: highlighting the links between epigenetics, COVID-19 infection, and vitamin D. Int. J. Mol. Sci. 23 (20), 12292. doi:10.3390/ijms232012292
Forman, J., Hine, B., Kaonis, S., and Ghosh, S. (2024). Inhibition of chromatin condensation disrupts planar cell migration. Nucleus 15 (1), 2325961. doi:10.1080/19491034.2024.2325961
Furusawa, T., Rochman, M., Taher, L., Dimitriadis, E. K., Nagashima, K., Anderson, S., et al. (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 6, 6138. doi:10.1038/ncomms7138
Gibbons, R. J., and Higgs, D. R. (2000). Molecular-clinical spectrum of the ATR-X syndrome. Am. J. Med. Genet. 97 (3), 204–212. doi:10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X
Gopfrich, K., Platzman, I., and Spatz, J. P. (2018). Mastering complexity: towards Bottom-up construction of multifunctional eukaryotic synthetic cells. Trends Biotechnol. 36 (9), 938–951. doi:10.1016/j.tibtech.2018.03.008
Gotze, S., Wolter, M., Reifenberger, G., Muller, O., and Sievers, S. (2010). Frequent promoter hypermethylation of wnt pathway inhibitor genes in malignant astrocytic gliomas. Int. J. Cancer 126 (11), 2584–2593. doi:10.1002/ijc.24981
Gregory, T. R. (2005). “Genome size evolution in animals,” in The evolution of the genome. Editor T. R. Gregory (Academic Press).
Hall, E., Dayeh, T., Kirkpatrick, C. L., Wollheim, C. B., Dekker Nitert, M., and Ling, C. (2013). DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Med. Genet. 14, 76. doi:10.1186/1471-2350-14-76
Hara, Y., and Kimura, A. (2009). Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr. Biol. 19 (18), 1549–1554. doi:10.1016/j.cub.2009.07.050
Heijo, H., Shimogama, S., Nakano, S., Miyata, A., Iwao, Y., and Hara, Y. (2020). DNA content contributes to nuclear size control in Xenopus laevis. Mol. Biol. Cell 31 (24), 2703–2717. doi:10.1091/mbc.E20-02-0113
Heo, S. J., Driscoll, T. P., Thorpe, S. D., Nerurkar, N. L., Baker, B. M., Yang, M. T., et al. (2016). Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife 5, e18207. doi:10.7554/eLife.18207
Heo, S. J., Thorpe, S. D., Driscoll, T. P., Duncan, R. L., Lee, D. A., and Mauck, R. L. (2015). Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci. Rep. 5, 16895. doi:10.1038/srep16895
Hobson, C. M., Kern, M., O'Brien, E. T., Stephens, A. D., Falvo, M. R., and Superfine, R. (2020). Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 31 (16), 1788–1801. doi:10.1091/mbc.E20-01-0073
Hobson, C. M., and Stephens, A. D. (2020). Modeling of cell nuclear mechanics: classes, components, and applications. Cells 9 (7), 1623. doi:10.3390/cells9071623
Horiguchi, K., Tomizawa, Y., Tosaka, M., Ishiuchi, S., Kurihara, H., Mori, M., et al. (2003). Epigenetic inactivation of RASSF1A candidate tumor suppressor gene at 3p21.3 in brain tumors. Oncogene 22 (49), 7862–7865. doi:10.1038/sj.onc.1207082
Huang, X., Yao, M., Tian, P., Wong, J. Y. Y., Li, Z., Liu, Z., et al. (2022). Shared genetic etiology and causality between COVID-19 and venous thromboembolism: evidence from genome-wide cross trait analysis and bi-directional Mendelian randomization study. medRxiv. doi:10.1101/2022.05.21.22275413
Imbalzano, K. M., Cohet, N., Wu, Q., Underwood, J. M., Imbalzano, A. N., and Nickerson, J. A. (2013). Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One 8 (2), e55628. doi:10.1371/journal.pone.0055628
Jacob, J. T., Nair, R. R., Poll, B. G., Pineda, C. M., Hobbs, R. P., Matunis, M. J., et al. (2020). Keratin 17 regulates nuclear morphology and chromatin organization. J. Cell Sci. 133 (20), jcs254094. doi:10.1242/jcs.254094
Jacobson, E. C., Perry, J. K., Long, D. S., Olins, A. L., Olins, D. E., Wright, B. E., et al. (2018). Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biol. 16 (1), 142. doi:10.1186/s12915-018-0608-2
Janssen, S. M., and Lorincz, M. C. (2022). Interplay between chromatin marks in development and disease. Nat. Rev. Genet. 23 (3), 137–153. doi:10.1038/s41576-021-00416-x
Jevtic, P., Edens, L. J., Li, X., Nguyen, T., Chen, P., and Levy, D. L. (2015). Concentration-dependent effects of nuclear lamins on nuclear size in Xenopus and mammalian cells. J. Biol. Chem. 290 (46), 27557–27571. doi:10.1074/jbc.M115.673798
Jevtic, P., and Levy, D. L. (2015). Nuclear size scaling during xenopus early development contributes to midblastula transition timing. Curr. Biol. 25 (1), 45–52. doi:10.1016/j.cub.2014.10.051
Jevtic, P., and Levy, D. L. (2018). Elucidating nuclear size control in the xenopus model system. Vet. Glas. 72 (1), 1–13. doi:10.2298/VETGL170731012J
Jevtic, P., Schibler, A. C., Wesley, C. C., Pegoraro, G., Misteli, T., and Levy, D. L. (2019). The nucleoporin ELYS regulates nuclear size by controlling NPC number and nuclear import capacity. EMBO Rep. 20 (6), e47283. doi:10.15252/embr.201847283
Jorgensen, P., Edgington, N. P., Schneider, B. L., Rupes, I., Tyers, M., and Futcher, B. (2007). The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell 18 (9), 3523–3532. doi:10.1091/mbc.e06-10-0973
Joseph-Strauss, D., Gorjanacz, M., Santarella-Mellwig, R., Voronina, E., Audhya, A., and Cohen-Fix, O. (2012). Sm protein down-regulation leads to defects in nuclear pore complex disassembly and distribution in C. elegans embryos. Dev. Biol. 365 (2), 445–457. doi:10.1016/j.ydbio.2012.02.036
Kalinin, A. A., Hou, X., Ade, A. S., Fon, G. V., Meixner, W., Higgins, G. A., et al. (2021). Valproic acid-induced changes of 4D nuclear morphology in astrocyte cells. Mol. Biol. Cell 32 (18), 1624–1633. doi:10.1091/mbc.E20-08-0502
Karoutas, A., Szymanski, W., Rausch, T., Guhathakurta, S., Rog-Zielinska, E. A., Peyronnet, R., et al. (2019). The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell Biol. 21 (10), 1248–1260. doi:10.1038/s41556-019-0397-z
Katiyar, A., Zhang, J., Antani, J. D., Yu, Y., Scott, K. L., Lele, P. P., et al. (2022). The Nucleus bypasses obstacles by deforming like a drop with surface tension mediated by lamin A/C. Adv. Sci. (Weinh) 9 (23), e2201248. doi:10.1002/advs.202201248
Kim, H.H.-S., and Lakadamyali, M. (2024). Microscopy methods to visualize nuclear organization in biomechanical studies. Curr. Opin. Biomed. Eng. 30, 100528. doi:10.1016/j.cobme.2024.100528
Kim, T. J., Joo, C., Seong, J., Vafabakhsh, R., Botvinick, E. L., Berns, M. W., et al. (2015). Distinct mechanisms regulating mechanical force-induced Ca²⁺ signals at the plasma membrane and the ER in human MSCs. Elife 4, e04876. doi:10.7554/eLife.04876
Kovacs, M. T., Vallette, M., Wiertsema, P., Dingli, F., Loew, D., Nader, G. P. F., et al. (2023). DNA damage induces nuclear envelope rupture through ATR-Mediated phosphorylation of lamin A/C. Mol. Cell 83 (20), 3659–3668.e10. doi:10.1016/j.molcel.2023.09.023
Kume, K., Cantwell, H., Neumann, F. R., Jones, A. W., Snijders, A. P., and Nurse, P. (2017). A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control. PLoS Genet. 13 (5), e1006767. doi:10.1371/journal.pgen.1006767
Lambiv, W. L., Vassallo, I., Delorenzi, M., Shay, T., Diserens, A. C., Misra, A., et al. (2011). The wnt inhibitory factor 1 (WIF1) is targeted in glioblastoma and has a tumor suppressing function potentially by induction of senescence. Neuro Oncol. 13 (7), 736–747. doi:10.1093/neuonc/nor036
Lanz, M. C., Zatulovskiy, E., Swaffer, M. P., Zhang, L., Ilerten, I., Zhang, S., et al. (2022). Increasing cell size remodels the proteome and promotes senescence. Mol. Cell 82 (17), 3255–3269.e8. doi:10.1016/j.molcel.2022.07.017
Lanz, M. C., Zhang, S., Swaffer, M. P., Ziv, I., Gotz, L. H., Kim, J., et al. (2024). Genome dilution by cell growth drives starvation-like proteome remodeling in Mammalian and yeast cells. Nat. Struct. Mol. Biol. 31 (12), 1859–1871. doi:10.1038/s41594-024-01353-z
Le, H. Q., Ghatak, S., Yeung, C. Y., Tellkamp, F., Gunschmann, C., Dieterich, C., et al. (2016). Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18 (8), 864–875. doi:10.1038/ncb3387
Levy, D. L., and Heald, R. (2010). Nuclear size is regulated by importin alpha and Ntf2 in xenopus. Cell 143 (2), 288–298. doi:10.1016/j.cell.2010.09.012
Levy, D. L., and Heald, R. (2012). Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 28, 113–135. doi:10.1146/annurev-cellbio-092910-154158
Lin, D. H., and Hoelz, A. (2019). The structure of the nuclear pore complex (an update). Annu. Rev. Biochem. 88, 725–783. doi:10.1146/annurev-biochem-062917-011901
Ling, C., Del Guerra, S., Lupi, R., Ronn, T., Granhall, C., Luthman, H., et al. (2008). Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 51 (4), 615–622. doi:10.1007/s00125-007-0916-5
Liu, X. Y., Gerges, N., Korshunov, A., Sabha, N., Khuong-Quang, D. A., Fontebasso, A. M., et al. (2012). Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 124 (5), 615–625. doi:10.1007/s00401-012-1031-3
Lu, H., Liu, X., Deng, Y., and Qing, H. (2013). DNA methylation, a hand behind neurodegenerative diseases. Front. Aging Neurosci. 5, 85. doi:10.3389/fnagi.2013.00085
Luger, K., Dechassa, M. L., and Tremethick, D. J. (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13 (7), 436–447. doi:10.1038/nrm3382
Manning, G., Andy, L., Nebiyat, E., Marilena, C., and Stephens, A. D. (2025). Constitutive heterochromatin controls nuclear mechanics, morphology, and integrity through H3K9me3 mediated chromocenter compaction. Nucleus 16(1), 2486816. doi:10.1080/19491034.2025.2486816
Marshall, W. (2002). Size control in dynamic organelles. Trends Cell Biol. 12 (9), 414–419. doi:10.1016/s0962-8924(02)02341-3
Mattei, A. L., Bailly, N., and Meissner, A. (2022). DNA methylation: a historical perspective. Trends Genet. 38 (7), 676–707. doi:10.1016/j.tig.2022.03.010
McCloskey, A., Ibarra, A., and Hetzer, M. W. (2018). Tpr regulates the total number of nuclear pore complexes per cell nucleus. Genes Dev. 32 (19-20), 1321–1331. doi:10.1101/gad.315523.118
Meng, L., Wang, S., Jiang, H., Hua, Y., Yin, B., Huang, X., et al. (2022). Oct4 dependent chromatin activation is required for chicken primordial germ cell migration. Stem Cell Rev. Rep. 18 (7), 2535–2546. doi:10.1007/s12015-022-10371-7
Miranda, T. B., Cortez, C. C., Yoo, C. B., Liang, G., Abe, M., Kelly, T. K., et al. (2009). DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8 (6), 1579–1588. doi:10.1158/1535-7163.MCT-09-0013
Mishra, S., and Levy, D. L. (2022a). Nuclear F-actin and lamin A antagonistically modulate nuclear shape. J. Cell Sci. 135 (13), jcs259692. doi:10.1242/jcs.259692
Mishra, S., and Levy, D. L. (2022b). Visualizing nuclear pore complexes in xenopus egg extracts. Methods Mol. Biol. 2502, 395–405. doi:10.1007/978-1-0716-2337-4_25
Morival, J., Hazelwood, A., and Lammerding, J. (2025). Feeling the force from within - new tools and insights into nuclear mechanotransduction. J. Cell Sci. 138 (5), JCS263615. doi:10.1242/jcs.263615
Neumann, F. R., and Nurse, P. (2007). Nuclear size control in fission yeast. J. Cell Biol. 179 (4), 593–600. doi:10.1083/jcb.200708054
Neurohr, G. E., Terry, R. L., Lengefeld, J., Bonney, M., Brittingham, G. P., Moretto, F., et al. (2019). Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176 (5), 1083–1097. doi:10.1016/j.cell.2019.01.018
Nodelman, I. M., and Bowman, G. D. (2021). Biophysics of chromatin remodeling. Annu. Rev. Biophys. 50, 73–93. doi:10.1146/annurev-biophys-082520-080201
Olins, A. L., Herrmann, H., Lichter, P., Kratzmeier, M., Doenecke, D., and Olins, D. E. (2001). Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp. Cell Res. 268 (2), 115–127. doi:10.1006/excr.2001.5269
Pajerowski, J. D., Dahl, K. N., Zhong, F. L., Sammak, P. J., and Discher, D. E. (2007). Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 104 (40), 15619–15624. doi:10.1073/pnas.0702576104
Palihati, M., and Saitoh, N. (2024). RNA in chromatin organization and nuclear architecture. Curr. Opin. Genet. Dev. 86, 102176. doi:10.1016/j.gde.2024.102176
Papillon-Cavanagh, S., Lu, C., Gayden, T., Mikael, L. G., Bechet, D., Karamboulas, C., et al. (2017). Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 49 (2), 180–185. doi:10.1038/ng.3757
Patta, I., Zand, M., Lee, L., Mishra, S., Bortnick, A., Lu, H., et al. (2024). Nuclear morphology is shaped by loop-extrusion programs. Nature 627 (8002), 196–203. doi:10.1038/s41586-024-07086-9
Pho, M., Berrada, Y., Gunda, A., Lavallee, A., Chiu, K., Padam, A., et al. (2024). Actin contraction controls nuclear blebbing and rupture independent of actin confinement. Mol. Biol. Cell 35 (2), ar19. doi:10.1091/mbc.E23-07-0292
Pinto, B. G. G., Oliveira, A. E. R., Singh, Y., Jimenez, L., Goncalves, A. N. A., Ogava, R. L. T., et al. (2020). ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 222 (4), 556–563. doi:10.1093/infdis/jiaa332
Pujadas Liwag, E. M., Acosta, N., Almassalha, L. M., Su, Y. P., Gong, R., Kanemaki, M. T., et al. (2025). Nuclear blebs are associated with destabilized chromatin-packing domains. J. Cell Sci. 138 (3), jcs262161. doi:10.1242/jcs.262161
Quinodoz, S. A., Jachowicz, J. W., Bhat, P., Ollikainen, N., Banerjee, A. K., Goronzy, I. N., et al. (2021). RNA promotes the formation of spatial compartments in the nucleus. Cell 184 (23), 5775–5790.e30. doi:10.1016/j.cell.2021.10.014
Reddy, K. L., Zullo, J. M., Bertolino, E., and Singh, H. (2008). Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452 (7184), 243–247. doi:10.1038/nature06727
Rougier, N., Bourc'his, D., Gomes, D. M., Niveleau, A., Plachot, M., Paldi, A., et al. (1998). Chromosome methylation patterns during Mammalian preimplantation development. Genes Dev. 12 (14), 2108–2113. doi:10.1101/gad.12.14.2108
Ryan, K. J., McCaffery, J. M., and Wente, S. R. (2003). The ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160 (7), 1041–1053. doi:10.1083/jcb.200209116
Salvatore, S., and Vandenplas, Y. (2003). Gastro-oesophageal reflux disease and motility disorders. Best. Pract. Res. Clin. Gastroenterol. 17 (2), 163–179. doi:10.1016/s1521-6918(02)00146-4
Santana, D. A., Smith, M. A. C., and Chen, E. S. (2023). Histone modifications in Alzheimer's disease. Genes (Basel) 14 (2), 347. doi:10.3390/genes14020347
Scaffidi, P., and Misteli, T. (2005). Reversal of the cellular phenotype in the premature aging disease hutchinson-gilford progeria syndrome. Nat. Med. 11 (4), 440–445. doi:10.1038/nm1204
Scaffidi, P., and Misteli, T. (2006). Lamin A-dependent nuclear defects in human aging. Science 312 (5776), 1059–1063. doi:10.1126/science.1127168
Schibler, A. C., Jevtic, P., Pegoraro, G., Levy, D. L., and Misteli, T. (2023). Identification of epigenetic modulators as determinants of nuclear size and shape. Elife 12, e80653. doi:10.7554/eLife.80653
Schreiner, S. M., Koo, P. K., Zhao, Y., Mochrie, S. G., and King, M. C. (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 6, 7159. doi:10.1038/ncomms8159
Schwartzentruber, J., Korshunov, A., Liu, X. Y., Jones, D. T., Pfaff, E., Jacob, K., et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482 (7384), 226–231. doi:10.1038/nature10833
Sengupta, S., Sami, A. B., Gatlin, J. C., and Levy, D. L. (2025). Proteasome inhibition induces microtubule-dependent changes in nuclear morphology. iScience 28 (1), 111550. doi:10.1016/j.isci.2024.111550
Senigagliesi, B., Penzo, C., Severino, L. U., Maraspini, R., Petrosino, S., Morales-Navarrete, H., et al. (2019). The high mobility group A1 (HMGA1) chromatin architectural factor modulates nuclear stiffness in breast cancer cells. Int. J. Mol. Sci. 20 (11), 2733. doi:10.3390/ijms20112733
Serrano-Gomez, S. J., Maziveyi, M., and Alahari, S. K. (2016). Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 15, 18. doi:10.1186/s12943-016-0502-x
Shimi, T., Pfleghaar, K., Kojima, S., Pack, C. G., Solovei, I., Goldman, A. E., et al. (2008). The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22 (24), 3409–3421. doi:10.1101/gad.1735208
Sismour, A. M., and Benner, S. A. (2005). Synthetic biology. Expert Opin. Biol. Ther. 5 (11), 1409–1414. doi:10.1517/14712598.5.11.1409
Skinner, B. M., and Johnson, E. E. (2017). Nuclear morphologies: their diversity and functional relevance. Chromosoma 126 (2), 195–212. doi:10.1007/s00412-016-0614-5
Smigiel, W. M., Lefrancois, P., and Poolman, B. (2019). Physicochemical considerations for bottom-up synthetic biology. Emerg. Top. Life Sci. 3 (5), 445–458. doi:10.1042/ETLS20190017
Stephens, A. D., Banigan, E. J., Adam, S. A., Goldman, R. D., and Marko, J. F. (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 28 (14), 1984–1996. doi:10.1091/mbc.E16-09-0653
Stephens, A. D., Banigan, E. J., and Marko, J. F. (2019a). Chromatin's physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 58, 76–84. doi:10.1016/j.ceb.2019.02.006
Stephens, A. D., Liu, P. Z., Banigan, E. J., Almassalha, L. M., Backman, V., Adam, S. A., et al. (2018). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 29 (2), 220–233. doi:10.1091/mbc.E17-06-0410
Stephens, A. D., Liu, P. Z., Kandula, V., Chen, H., Almassalha, L. M., Herman, C., et al. (2019b). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30 (17), 2320–2330. doi:10.1091/mbc.E19-05-0286
Suganuma, T., and Workman, J. L. (2011). Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80, 473–499. doi:10.1146/annurev-biochem-061809-175347
Swift, J., and Discher, D. E. (2014). The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 127 (Pt 14), 3005–3015. doi:10.1242/jcs.149203
Swift, J., Ivanovska, I. L., Buxboim, A., Harada, T., Dingal, P. C., Pinter, J., et al. (2013). Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341 (6149), 1240104. doi:10.1126/science.1240104
Tamashunas, A. C., Tocco, V. J., Matthews, J., Zhang, Q., Atanasova, K. R., Paschall, L., et al. (2020). High-throughput gene screen reveals modulators of nuclear shape. Mol. Biol. Cell 31 (13), 1392–1402. doi:10.1091/mbc.E19-09-0520
Tamura, K., and Hara-Nishimura, I. (2011). Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2 (3), 168–172. doi:10.4161/nucl.2.3.16175
Titus, L. C., Dawson, T. R., Rexer, D. J., Ryan, K. J., and Wente, S. R. (2010). Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol. Biol. Cell 21 (6), 1072–1087. doi:10.1091/mbc.e09-07-0615
Toth, K. F., Knoch, T. A., Wachsmuth, M., Frank-Stohr, M., Stohr, M., Bacher, C. P., et al. (2004). Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 117 (Pt 18), 4277–4287. doi:10.1242/jcs.01293
Varambally, S., Dhanasekaran, S. M., Zhou, M., Barrette, T. R., Kumar-Sinha, C., Sanda, M. G., et al. (2002). The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419 (6907), 624–629. doi:10.1038/nature01075
Vodovotz, Y., Barnard, N., Hu, F. B., Jakicic, J., Lianov, L., Loveland, D., et al. (2020). Prioritized research for the prevention, treatment, and reversal of chronic disease: recommendations from the lifestyle medicine research summit. Front. Med. (Lausanne) 7, 585744. doi:10.3389/fmed.2020.585744
Vukovic, L. D., Chen, P., Mishra, S., White, K. H., Gigley, J. P., and Levy, D. L. (2021). Nuclear transport factor 2 (NTF2) suppresses WM983B metastatic melanoma by modifying cell migration, metastasis, and gene expression. Sci. Rep. 11 (1), 23586. doi:10.1038/s41598-021-02803-0
Vukovic, L. D., Jevtic, P., Edens, L. J., and Levy, D. L. (2016). New insights into mechanisms and functions of nuclear size regulation. Int. Rev. Cell Mol. Biol. 322, 1–59. doi:10.1016/bs.ircmb.2015.11.001
Woulfe, J. (2008). Nuclear bodies in neurodegenerative disease. Biochim. Biophys. Acta 1783 (11), 2195–2206. doi:10.1016/j.bbamcr.2008.05.005
Xia, Y., Ivanovska, I. L., Zhu, K., Smith, L., Irianto, J., Pfeifer, C. R., et al. (2018). Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol. 217 (11), 3796–3808. doi:10.1083/jcb.201711161
Xu, G., Chhangawala, S., Cocco, E., Razavi, P., Cai, Y., Otto, J. E., et al. (2020). ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat. Genet. 52 (2), 198–207. doi:10.1038/s41588-019-0554-0
Xu, P., Zhang, B., Luo, Q., and Song, G. (2023). Mechanical stretch promotes tenocyte migration via chromatin remodelling-mediated nuclear morphology changes. Wound Repair Regen. 31 (3), 410–414. doi:10.1111/wrr.13080
Yang, B. T., Dayeh, T. A., Volkov, P. A., Kirkpatrick, C. L., Malmgren, S., Jing, X., et al. (2012). Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol. Endocrinol. 26 (7), 1203–1212. doi:10.1210/me.2012-1004
Yang, Z., Maciejowski, J., and de Lange, T. (2017). Nuclear envelope rupture is enhanced by loss of p53 or Rb. Mol. Cancer Res. 15 (11), 1579–1586. doi:10.1158/1541-7786.MCR-17-0084
Yoo, C. B., Cheng, J. C., and Jones, P. A. (2004). Zebularine: a new drug for epigenetic therapy. Biochem. Soc. Trans. 32 (Pt 6), 910–912. doi:10.1042/BST0320910
Zink, D., Fischer, A. H., and Nickerson, J. A. (2004). Nuclear structure in cancer cells. Nat. Rev. Cancer 4 (9), 677–687. doi:10.1038/nrc1430
Keywords: nuclear size, nuclear shape, chromatin structure, chromatin modifications, cancer, epigenetics, Xenopus egg extract, microscopy
Citation: Sengupta S, Prabha H and Levy DL (2025) How the chromatin landscape influences nuclear morphology. Front. Cell Dev. Biol. 13:1634252. doi: 10.3389/fcell.2025.1634252
Received: 23 May 2025; Accepted: 24 June 2025;
Published: 03 July 2025.
Edited by:
Coral Zhou, University of Kansas, United StatesReviewed by:
Abigail Buchwalter, University of California San Francisco, United StatesHelena Cantwell, University of California, Berkeley, United States
Andrew Stephens, University of Massachusetts Amherst, United States
Copyright © 2025 Sengupta, Prabha and Levy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sourabh Sengupta, c291cmFiaC5zZW5ndXB0YUB1dHNvdXRod2VzdGVybi5lZHU=; Daniel L. Levy, ZGxldnkxQHV3eW8uZWR1
†These authors have contributed equally to this work and share first authorship
 Sourabh Sengupta
Sourabh Sengupta Haritha Prabha
Haritha Prabha Daniel L. Levy
Daniel L. Levy