- Laboratory of Developmental Cardiology, Institute of Physiology of the Czech Academy of Sciences, Prague, Czechia
Type 2 diabetes mellitus (T2DM) is a complex metabolic disorder characterized by chronic hyperglycemia, insulin resistance, and progressive β-cell dysfunction. Traditional biomarkers, such as fasting glucose and glycated hemoglobin (HbA1c), offer diagnostic and prognostic value but have limitations in sensitivity and predictive power for disease progression. Recent advances in molecular biology have identified epitranscriptomic modifications as potential biomarkers for T2DM, offering a novel layer of gene expression regulation through reversible RNA modifications. Dysregulation of these modifications has been implicated in insulin resistance, β-cell failure, and diabetes-related complications. Notably, altered levels of N6-methyladenosine (m6A) and its regulatory enzymes, including the eraser fat mass and obesity-associated protein (FTO) and the writer methyltransferase-like 3 (METTL3), have been detected in peripheral blood of T2DM patients, suggesting their potential as promising diagnostic markers. Similarly, circulating levels of pseudouridine (Ψ) have been associated with diabetic complications such as retinopathy and nephropathy. This review highlights the emerging role of epitranscriptomic modifications in T2DM pathophysiology and discusses their translational potential as biomarkers for early detection, disease monitoring, and personalized therapeutic strategies.
1 Introduction
Diabetes mellitus (DM) is a chronic metabolic disorder characterized by persistent hyperglycemia due to defects in insulin secretion, insulin action, or both. Type 2 diabetes mellitus (T2DM), the most prevalent form, is a growing global health challenge, with its incidence driven by increasing obesity rates, sedentary lifestyles, and aging populations. Given its progressive nature and associated microvascular (including nephropathy, retinopathy, and neuropathy) and macrovascular complications (including cardiovascular disease), early and accurate diagnosis is critical for mitigating long-term morbidity and mortality (Benak et al., 2023a). Current diagnostic and monitoring tools, including fasting glucose, oral glucose tolerance tests, fructosamine, glycated hemoglobin (HbA1c), and glycated albumin have limitations in sensitivity, specificity, and predictive power for disease progression (Dorcely et al., 2017; Ahmed et al., 2025). Consequently, there is an urgent need for novel biomarkers that provide more precise risk stratification and early detection of prediabetes and diabetes.
Recent advancements in molecular biology have expanded biomarker research beyond conventional protein and metabolite markers. The study of post-transcriptional modifications in RNA – referred to as epitranscriptomics or RNA epigenetics – has emerged as a promising frontier in diabetes research (Benak et al., 2023a). Like classical epigenetic modifications, epitranscriptomic modifications also regulate gene expression without altering the nucleotide sequence, offering a dynamic and reversible layer of control over cellular function. Aberrations in RNA modifications have been linked to insulin resistance, β-cell dysfunction, and chronic inflammation – hallmarks of T2DM (Benak et al., 2023a). As such, epitranscriptomic biomarkers hold significant potential as diagnostic and prognostic tools (Santos-Pujol et al., 2024), offering novel insights into disease pathophysiology and paving the way for precision medicine in diabetes management. Moreover, their analysis is no longer limited to advanced LC-MS methods but can often be performed using commercial quantification kits, making them more accessible and economically feasible for routine diagnostic testing.
This short review explores the landscape of epitranscriptomic modifications and their regulators, emphasizing their potential role as biomarkers in T2DM. By integrating this emerging knowledge into clinical practice, we may advance early detection strategies and therapeutic interventions for DM and its complications.
2 Epitranscriptomic modifications and their regulators
Epitranscriptomics refers to the study of chemical modifications that occur on RNA molecules, influencing their stability, processing, translation, and degradation (Benak et al., 2024a). Unlike genetic mutations, these modifications are mostly dynamic and reversible, allowing cells to rapidly adapt to physiological and environmental cues. More than 170 distinct RNA modifications have been identified across different RNA species, including messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), and non-coding RNAs (ncRNAs) (Cappannini et al., 2024). These modifications play critical roles in regulating cellular metabolism, differentiation, and stress responses – functions that are particularly relevant in the context of DM.
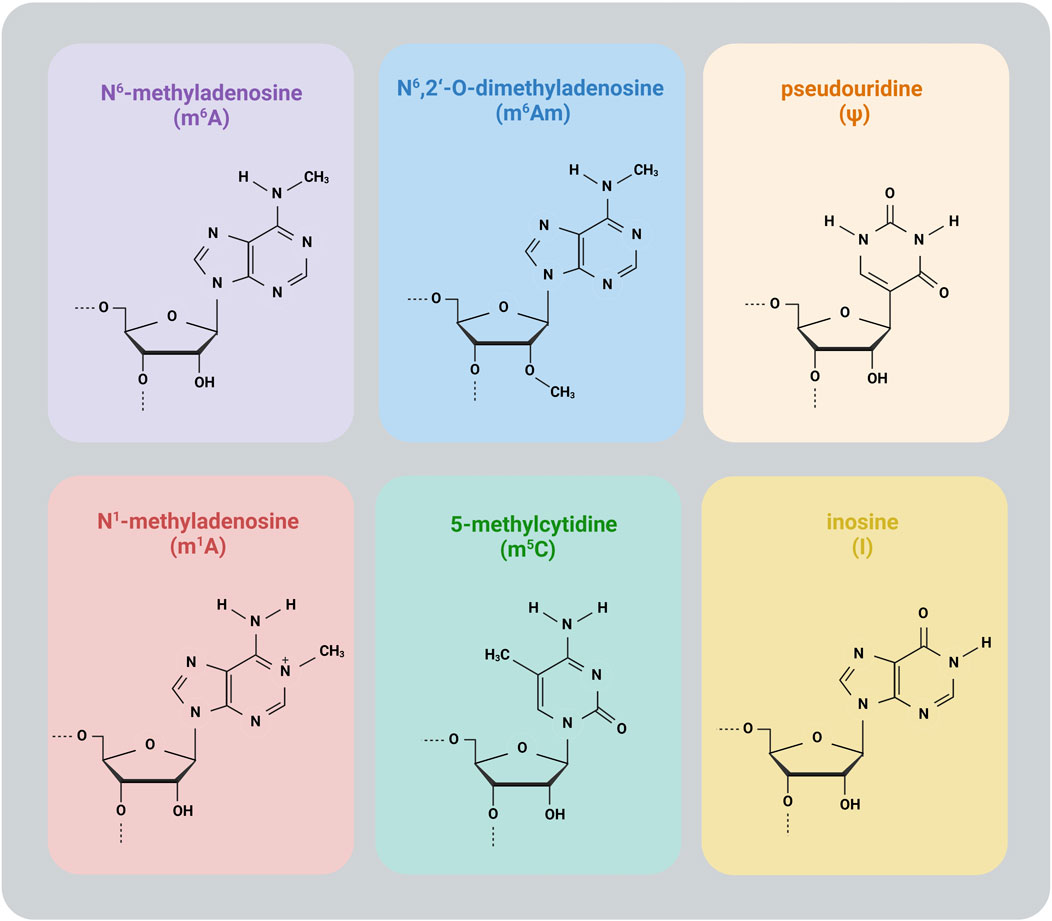
This review covers the following common modifications: N6-methyladenosine (m6A), N6,2′-O-dimethyladenosine (m6Am), N1-methyladenosine (m1A), 5-methylcytidine (m5C), pseudouridine (Ψ) and inosine (I) (Figure 1).
2.1 Reversible RNA modifications
Reversible RNA modifications are primarily regulated by three classes of proteins: writers, readers, and erasers. Writers are enzymes that catalyze the addition of specific modifications to RNA, while readers are proteins that recognize and interpret these modifications, mediating downstream effects. Erasers, in turn, remove modifications, creating a dynamic regulatory system (Benak et al., 2024b). These modifications enable cells to respond rapidly and flexibly to cellular signals and environmental changes.
One of the most prevalent RNA modifications in eukaryotic mRNA – and consequently one of the most extensively studied epitranscriptomic modifications – is N6-methyladenosine (m6A) (Desrosiers et al., 1974; Semenovykh et al., 2022; Benak et al., 2025). This modification plays a crucial role in regulating mRNA stability, splicing, and translation. In addition to mRNA, m6A is also present in various other types of RNA (Desrosiers et al., 1974; Dominissini et al., 2013; Meyer et al., 2012; Oerum et al., 2021). The deposition of m6A is mediated by a multicomponent methyltransferase complex composed of methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms’ tumor 1-associating protein (WTAP) (Wan et al., 2016; Wang et al., 2016). Recognition of m6A is facilitated by a variety of m6A-binding proteins, including YTH domain-containing family proteins (YTHDF1-3) (Zaccara and Jaffrey, 2020; Lasman et al., 2020; Wang et al., 2014; Wang et al., 2015; Shi et al., 2017), YTH domain-containing proteins (YTHDC1-2) (Xiao et al., 2016; Hsu et al., 2017; Ping et al., 2014), insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1-3) (Huang et al., 2018), and heterogeneous nuclear ribonucleoproteins (HNRNPA2B1, HNRNPC, HNRNPD, HNRNPG) (Alarcón et al., 2015; Liu et al., 2015; Song et al., 2019; Liu et al., 2017). The removal of m6A is carried out by demethylases such as fat mass and obesity-associated protein (FTO) (Jia et al., 2011; Benak et al., 2024c) and AlkB homolog 5 (ALKBH5) (Zheng et al., 2013; Wang et al., 2023a). Notably, dysregulation of m6A and its regulators has been observed in various diabetic tissues. This topic has been reviewed in detail (Benak et al., 2023a).
N6,2′-O-dimethyladenosine (m6Am) differs from m6A by the presence of an additional 2′-O-methyl group. In mRNA, m6Am is predominantly found at the mRNA cap, positioned at the transcription start site adjacent to the 7-methylguanosine (m7G) cap structure (Wei et al., 1975; Bokar and Grosjean, 2005). In small nuclear RNA (snRNA), m6Am also occurs at internal sites, where it contributes to pre-mRNA splicing (Mauer et al., 2019). The cap-associated m6Am is introduced by phosphorylated CTD-interacting factor 1 (PCIF1) (Akichika et al., 2019; Sun et al., 2019), whereas methyltransferase-like 4 (METTL4) catalyzes its incorporation at internal snRNA sites (Goh et al., 2020; Chen et al., 2020). Currently, no m6Am-specific readers have been identified, and only a single eraser is known to remove its N6-methylation – the well-characterized m6A demethylase FTO. Studies suggest that FTO predominantly demethylates m6Am in the cytosol, whereas in the nucleus, its primary target is m6A (Wei et al., 2018; Benak et al., 2023b). The relationship between m6Am and diabetes remains unclear. However, since many detection methods do not differentiate between m6A and m6Am (Benak et al., 2023b), and the well-studied FTO enzyme acts on both modifications (Benak et al., 2024c), m6Am is included in this review.
N1-methyladenosine (m1A) is predominantly found in tRNA and rRNA, with a lower abundance in mRNA (Dunn, 1961; Helm et al., 1999; Sharma et al., 2013; Dominissini et al., 2016). Functionally, it influences the structure and stability of tRNA and rRNA, while in mRNA, it plays a role in regulating translation (Dominissini et al., 2016; Oerum et al., 2017; Shima and Igarashi, 2020; Safra et al., 2017; Zhao et al., 2017). Its methylation is catalyzed by tRNA methyltransferase 6 (TRMT6), TRMT61A, TRMT61B, TRMT10C, and ribosomal RNA-processing protein 8 (RRP8, also known as NML) (Safra et al., 2017; Li et al., 2017; Chujo and Suzuki, 2012; Bar-Yaacov et al., 2016; Waku et al., 2016). The demethylation of m1A is carried out by the erasers ALKBH1 and ALKBH3 (Dominissini et al., 2016; Liu et al., 2016; Li et al., 2016a; Chen et al., 2019a). Additionally, FTO, primarily known as an m6A and m6Am eraser also acts as an m1A demethylase in tRNA (Wei et al., 2018). The link between m1A and diabetes remains unclear. However, ALKBH1, an m1A demethylase, was found to be downregulated in pancreatic islet samples from T2DM patients (Wu et al., 2023).
5-methylcytidine (m5C) is a widely distributed RNA modification found across multiple RNA types. It plays a crucial role in regulating RNA export, ribosome biogenesis, translation, and RNA stability (Bohnsack et al., 2019; Squires and Preiss, 2010; Chen et al., 2021). In humans, m5C is deposited by the NOL1/NOP2/SUN domain (NSUN) family proteins (NSUN1-7) as well as DNA methyltransferase homolog DNMT2 (also known as TRDMT1) (Bohnsack et al., 2019; Wang et al., 2023b). Among the m5C-binding proteins, Aly/REF export factor (ALYREF) facilitates nuclear-to-cytoplasmic RNA transport (Yang et al., 2017), whereas Y-box-binding protein 1 (YBX1) stabilizes its target mRNAs by interacting with ELAVL1 (Chen et al., 2019b). The removal of m5C is mediated by ten-eleven translocation (TET) proteins (TET1-3) and ALKBH1. The TET enzymes catalyze the oxidation of m5C to 5-hydroxymethylcytidine (hm5C), while ALKBH1 specifically oxidizes m5C in mitochondrial tRNA, generating 5-formylcytidine (f5C) (Haag et al., 2016; Fu et al., 2014). Notably, 5-methylcytosine also occurs in DNA, where it is often referred to as 5mC. Although the regulatory mechanisms of this modification differ between DNA and RNA, they share certain modifying enzymes, particularly TET proteins, which have been extensively studied in DNA demethylation (Williams et al., 2011). In the context of diabetes, a recent study found that m5C-related genes were significantly differentially expressed in T2DM and showed strong correlations with the majority of T2DM-associated differentially expressed genes in skeletal muscle samples (Song et al., 2022). The m5C reader NSUN2 has been linked to diabetic retinopathy (Wang et al., 2024) and nephropathy (Wang et al., 2025). Additionally, increased expression of Nsun4, Nsun6, and Dnmt2 has been observed in diabetic retinopathy (Wang et al., 2023c). Berberine, a compound known for its protective effects against diabetic nephropathy, has been reported to suppress DNMT2 expression in diabetic nephropathy mouse models (Cai et al., 2024). The m5C eraser TET1 was downregulated in human pancreatic islets from T2DM patients (Bacos et al., 2023) as well as in renal tissues of diabetic nephropathy mouse models (Tan et al., 2021). Another recent study showed that proteins TET1-3 play a critical role in de novo blood vessel formation, aiding the rescue of diabetic ischemic skin (Mohanty et al., 2024). Finally, as previously mentioned, ALKBH1 – a demethylase of both m1A and m5C – was found to be downregulated in pancreatic islet samples from T2DM patients (Li et al., 2016a).
2.2 Irreversible RNA modifications
Unlike reversible RNA modifications, irreversible modifications lack erasers that could dynamically regulate their presence in RNA, thereby limiting their regulation to mRNA turnover.
Pseudouridine (Ψ), a C5-glycoside isomer of uridine (U), was the first RNA modification ever discovered and remains the most abundant, detected across nearly all types of RNA (Cohn, 1951; Xue et al., 2022; Sun et al., 2023). Functionally, Ψ plays a key role in stabilizing RNA structures while simultaneously reducing RNA-binding protein interactions. In mRNA, its most studied role is enhancing stop codon read-through (Sun et al., 2023; Borchardt et al., 2020). The enzymatic conversion of U to Ψ is catalyzed by the pseudouridine synthase (PUS) family, a diverse group of enzymes responsible for this modification (Rintala-Dempsey and Kothe, 2017). To date, 13 PUS enzymes have been identified in eukaryotes (Sun et al., 2023). In humans, this family includes PUS1, PUS3, PUS7, PUS10, PUSL1, PUSL7, TRUB1-2 (TruB pseudouridine synthase 1-2), RPUSD1-4 (RNA pseudouridine synthase D1-4), and DKC1 (dyskerin pseudouridine synthase 1) (Li et al., 2016b). Currently, the only known Ψ-binding protein is the yeast RNA helicase Prp5, which interacts with snRNA (Wu et al., 2016; Levi and Arava, 2021). Diabetic complications, such as diabetic retinopathy and diabetic nephropathy, have been associated with changes in circulating Ψ levels (Sun et al., 2021; Jiang et al., 2024; Mathew et al., 2024; Niewczas et al., 2017); however, the link between Ψ and its regulators in diabetes remains unknown.
Inosine is a product of A-to-I editing, a conserved mechanism that contributes to transcriptome diversity as part of the broader RNA editing process, which also encompasses cytosine-to-uridine conversion and nucleotide insertions and deletions (Brennicke et al., 1999; Gott and Emeson, 2000). This modification occurs when the C6-position of adenosine loses a hydrogen-donating amino group, resulting in inosine, which structurally resembles guanosine and can influence various downstream processes. Post-transcriptionally, A-to-I editing can alter codons, create or eliminate splice sites, modify microRNA (miRNA) interactions, and influence RNA base pairing with itself or other RNAs, as well as its binding to RNA-associated proteins. In coding regions, this process can lead to amino acid substitutions, potentially affecting protein function (Nishikura, 2016). Deamination of adenosine to inosine is performed by enzymes belonging to the adenosine deaminase acting on RNA (ADAR) family, which is represented by three ADAR orthologs (ADAR1-3) in mammals. ADAR1 and ADAR2 are widely expressed, while ADAR3 was detected only in the brain (Ganem and Lamm, 2017; Dominis et al., 2011). Both mouse and human β-cells require intact ADAR1 function, as its disruption leads to the accumulation of endogenous double-stranded RNA (dsRNA), activation of an interferon response, islet inflammation, and β-cell failure. These changes closely mimic key aspects of early-stage T1DM (Kneb et al., 2024). Interestingly, inosine supplementation has been reported to protect against T1DM by exerting anti-inflammatory effects and modulating immune responses (Mabley et al., 2003). However, these effects appear to be independent of inosine’s role in RNA editing and are instead linked to its function as a purine metabolite.
3 Epitranscriptomic biomarkers in diabetic patients
Epitranscriptomic modifications have emerged as potential biomarkers for T2DM. Changes in their levels and the expression of its regulatory enzymes in peripheral blood may reflect disease progression and metabolic dysregulation, making them promising candidates for novel diagnostic tools.
Decreased m6A methylation levels have been reported in RNA isolated from the peripheral blood of T2DM patients and diabetic rats (Shen et al., 2015; Onalan et al., 2022). Consistent with this, FTO gene expression – but not ALKBH5 – was found to be significantly upregulated in peripheral blood from T2DM patients (Shen et al., 2015). However, a separate study by Onalan et al. (Onalan et al., 2022) observed increased expression of both demethylases in venous blood samples from T2DM patients. Further supporting the role of FTO, another study confirmed its elevated expression at both gene and protein levels, highlighting a correlation between high FTO levels and T2DM severity (Masoud Abd El Gayed et al., 2021). Similarly, FTO gene expression was upregulated in white blood cells of T2DM patients compared to healthy individuals, with its expression level positively correlated with fasting glucose concentration (Yang et al., 2019). Apart from m6A erasers, METTL3, a key m6A methyltransferase, was found to be downregulated in serum samples from T2DM patients (Zha et al., 2020). Additionally, low serum levels of IGF2BP3, an m6A reader, were associated with a progressively higher risk of developing T2DM (Wu et al., 2023). Collectively, these findings suggest that m6A modifications and their regulatory proteins in peripheral blood could serve as novel epitranscriptomic biomarkers for T2DM (Figure 2). Their potential use in early diagnosis, disease monitoring, and risk assessment warrants further investigation.
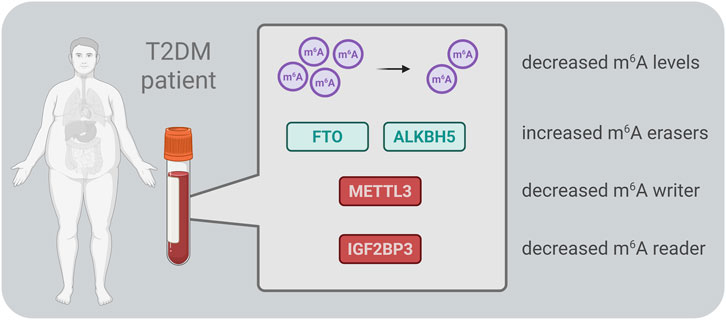
Figure 2. Schematic overview of the main m6A-related enzymes reported in blood samples from patients with type 2 diabetes mellitus (T2DM). ALKBH5 – AlkB homolog 5, FTO – fat mass and obesity-associated protein, IGF2BP3 – insulin-like growth factor 2 mRNA-binding protein 3, METTL3 – methyltransferase-like 3.
Additionally, Ψ has recently been identified as a circulating biomarker related to diabetes complications. Elevated Ψ levels have been associated with the occurrence of diabetic retinopathy (Sun et al., 2021) and have been identified as an early biomarker of diabetic kidney disease in Chinese patients with T2DM (Jiang et al., 2024). Moreover, Ψ levels have been linked to renal function decline and the progression to end-stage renal disease in patients with type 1 diabetes mellitus (T1DM) (Niewczas et al., 2017).
Other RNA modifications and their regulatory enzymes may also play a role in diabetes and its complications, but they remain largely unexplored as potential biomarkers. For example, m1A, m5C, and inosine are among the modifications that have been linked to diabetes-related processes but have yet to be studied in the context of their potential as diagnostic or prognostic biomarkers.
Importantly, circulating alterations in RNA modifications seem unlikely to exert direct pathogenic effects themselves but rather serve as biomarkers that mirror dysregulated processes in tissues such as pancreatic islets, liver, or kidney. Establishing these tissue–blood relationships will be essential for clarifying underlying mechanisms and for translating biomarker discovery into therapeutic strategies. To this end, integrating blood- and tissue-level epitranscriptomic analyses could refine our understanding of disease pathogenesis, uncover organ-specific vulnerabilities, and guide the development of more precise interventions. Such a dual approach carries translational potential by directly linking biomarker discovery to drug development. Future research should also focus on expanding the scope of epitranscriptomic biomarkers beyond m6A to include m1A, m5C, inosine, and Ψ, as their regulatory mechanisms and clinical significance in DM remain largely unexplored. Elucidating how these modifications influence β-cell function, insulin resistance, and inflammation may open new avenues for early detection, disease monitoring, and therapeutic intervention in DM and its complications.
4 Conclusion
Epitranscriptomic modifications represent a promising frontier in diabetes biomarker research, providing dynamic and often reversible regulation of RNA metabolism. The emerging evidence linking RNA modifications to insulin resistance and β-cell dysfunction underscores their potential as novel diagnostic and prognostic tools. While m6A modifications have been most extensively studied in diabetes, the broader landscape of RNA modifications remains largely unexplored. Future research should focus on validating these biomarkers in large patient cohorts, understanding their mechanistic roles in diabetes pathophysiology, and developing clinically feasible detection methods. Integration of epitranscriptomic signatures into precision medicine approaches may ultimately enhance early diagnosis, risk stratification, and personalized therapeutic interventions in T2DM.
Author contributions
MH: Conceptualization, Funding acquisition, Writing – original draft. DB: Conceptualization, Writing – original draft. KH: Writing – review and editing. PA: Writing – review and editing. JH: Writing – review and editing. MC: Visualization, Writing – review and editing. BaO: Visualization, Writing – review and editing. BvO: Writing – original draft. FK: Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Supported by the project Spa Research Centre (CZ.10.01.01/00/22_001/0000261) under Operational Programme Fair Transformation.
Acknowledgments
Figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. ChatGPT 4.0, a large language model developed by OpenAI, was used for language corrections.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, S., Adnan, H., Khawaja, M. A., and Butler, A. E. (2025). Novel micro-ribonucleic acid biomarkers for early detection of type 2 diabetes mellitus and associated Complications-A literature review. Int. J. Mol. Sci. 26 (2), 753. doi:10.3390/ijms26020753
Akichika, S., Hirano, S., Shichino, Y., Suzuki, T., Nishimasu, H., Ishitani, R., et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363 (6423), eaav0080. doi:10.1126/science.aav0080
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m(6)A-Dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Bacos, K., Perfilyev, A., Karagiannopoulos, A., Cowan, E., Ofori, J. K., Bertonnier-Brouty, L., et al. (2023). Type 2 diabetes candidate genes, including PAX5, cause impaired insulin secretion in human pancreatic islets. J. Clin. Invest 133 (4), e163612. doi:10.1172/JCI163612
Bar-Yaacov, D., Frumkin, I., Yashiro, Y., Chujo, T., Ishigami, Y., Chemla, Y., et al. (2016). Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol. 14 (9), e1002557. doi:10.1371/journal.pbio.1002557
Benak, D., Benakova, S., Plecita-Hlavata, L., and Hlavackova, M. (2023a). The role of m6A and m6Am RNA modifications in the pathogenesis of diabetes mellitus. Front. Endocrinol. (Lausanne) 14, 1223583. doi:10.3389/fendo.2023.1223583
Benak, D., Kolar, F., Zhang, L., Devaux, Y., and Hlavackova, M. (2023b). RNA modification m(6)Am: the role in cardiac biology. Epigenetics 18 (1), 2218771. doi:10.1080/15592294.2023.2218771
Benak, D., Kolar, F., and Hlavackova, M. (2024a). Epitranscriptomic regulations in the heart. Physiol. Res. 73, S185–S198. doi:10.33549/physiolres.935265
Benak, D., Holzerova, K., Hrdlicka, J., Kolar, F., Olsen, M., Karelson, M., et al. (2024b). Epitranscriptomic regulation in fasting hearts: implications for cardiac health. RNA Biol. 21 (1), 1–14. doi:10.1080/15476286.2024.2307732
Benak, D., Sevcikova, A., Holzerova, K., and Hlavackova, M. (2024c). FTO in health and disease. Front. Cell Dev. Biol. 12, 1500394. doi:10.3389/fcell.2024.1500394
Benak, D., Alanova, P., Holzerova, K., Chalupova, M., Opletalova, B., Kolar, F., et al. (2025). Epitranscriptomic regulation of HIF-1: bidirectional regulatory pathways. Mol. Med. 31 (1), 105. doi:10.1186/s10020-025-01149-x
Bohnsack, K. E., Höbartner, C., and Bohnsack, M. T. (2019). Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel) 10 (2), 102. doi:10.3390/genes10020102
Bokar, J. A. (2005). “The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA,” in Fine-tuning of RNA functions by modification and editing. Editor H. Grosjean (Berlin, Heidelberg: Springer Berlin Heidelberg), 141–177.
Borchardt, E. K., Martinez, N. M., and Gilbert, W. V. (2020). Regulation and function of RNA pseudouridylation in human cells. Annu. Rev. Genet. 54, 309–336. doi:10.1146/annurev-genet-112618-043830
Brennicke, A., Marchfelder, A., and Binder, S. (1999). RNA editing. FEMS Microbiol. Rev. 23 (3), 297–316. doi:10.1111/j.1574-6976.1999.tb00401.x
Cai, S., Zhu, H., Chen, L., Yu, C., Su, L., Chen, K., et al. (2024). Berberine inhibits KLF4 promoter methylation and ferroptosis to ameliorate diabetic nephropathy in mice. Chem. Res. Toxicol. 37 (10), 1728–1737. doi:10.1021/acs.chemrestox.4c00263
Cappannini, A., Ray, A., Purta, E., Mukherjee, S., Boccaletto, P., Moafinejad, S. N., et al. (2024). MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 52 (D1), D239–d244. doi:10.1093/nar/gkad1083
Chen, Z., Qi, M., Shen, B., Luo, G., Wu, Y., Li, J., et al. (2019a). Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47 (5), 2533–2545. doi:10.1093/nar/gky1250
Chen, X., Sun, B. F., Yang, Y., Han, Y. N., Yuan, X., Chen, R. X., et al. (2019b). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21 (8), 978–990. doi:10.1038/s41556-019-0361-y
Chen, H., Gu, L., Orellana, E. A., Wang, Y., Guo, J., Liu, Q., et al. (2020). METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 30 (6), 544–547. doi:10.1038/s41422-019-0270-4
Chen, Y. S., Yang, W. L., Zhao, Y. L., and Yang, Y. G. (2021). Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 12 (4), e1639. doi:10.1002/wrna.1639
Chujo, T., and Suzuki, T. (2012). Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. Rna 18 (12), 2269–2276. doi:10.1261/rna.035600.112
Cohn, W. E. (1951). Some results of the applications of ion-exchange chromatography to nucleic acid chemistry. J. Cell Physiol. Suppl. 38 (Suppl. 1), 21–40. doi:10.1002/jcp.1030380405
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Amariglio, N., and Rechavi, G. (2011). Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis 32 (11), 1569–1577. doi:10.1093/carcin/bgr124
Dominissini, D., Moshitch-Moshkovitz, S., Salmon-Divon, M., Amariglio, N., and Rechavi, G. (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 8 (1), 176–189. doi:10.1038/nprot.2012.148
Dominissini, D., Nachtergaele, S., Moshitch-Moshkovitz, S., Peer, E., Kol, N., Ben-Haim, M. S., et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530 (7591), 441–446. doi:10.1038/nature16998
Dorcely, B., Katz, K., Jagannathan, R., Chiang, S. S., Oluwadare, B., Goldberg, I. J., et al. (2017). Novel biomarkers for prediabetes, diabetes, and associated complications. Diabetes Metab. Syndr. Obes. 10, 345–361. doi:10.2147/DMSO.S100074
Dunn, D. B. (1961). The occurrence of 1-methyladenine in ribonucleic acid. Biochim. Biophys. Acta 46, 198–200. doi:10.1016/0006-3002(61)90668-0
Fu, L., Guerrero, C. R., Zhong, N., Amato, N. J., Liu, Y., Liu, S., et al. (2014). Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 136 (33), 11582–11585. doi:10.1021/ja505305z
Ganem, N. S., and Lamm, A. T. (2017). A-to-I RNA editing - thinking beyond the single nucleotide. RNA Biol. 14 (12), 1690–1694. doi:10.1080/15476286.2017.1364830
Goh, Y. T., Koh, C. W. Q., Sim, D. Y., Roca, X., and Goh, W. S. S. (2020). METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 48 (16), 9250–9261. doi:10.1093/nar/gkaa684
Gott, J. M., and Emeson, R. B. (2000). Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34, 499–531. doi:10.1146/annurev.genet.34.1.499
Haag, S., Sloan, K. E., Ranjan, N., Warda, A. S., Kretschmer, J., Blessing, C., et al. (2016). NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. Embo J. 35 (19), 2104–2119. doi:10.15252/embj.201694885
Helm, M., Giegé, R., and Florentz, C. (1999). A watson-crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38 (40), 13338–13346. doi:10.1021/bi991061g
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27 (9), 1115–1127. doi:10.1038/cr.2017.99
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jiang, J. J., Sham, T. T., Gu, X. F., Chan, C. O., Dong, N. P., Lim, W. H., et al. (2024). Insights into serum metabolic biomarkers for early detection of incident diabetic kidney disease in Chinese patients with type 2 diabetes by random forest. Aging (Albany NY) 16 (4), 3420–3530. doi:10.18632/aging.205542
Knebel, U. E., Peleg, S., Dai, C., Cohen-Fultheim, R., Jonsson, S., Poznyak, K., et al. (2024). Disrupted RNA editing in beta cells mimics early-stage type 1 diabetes. Cell Metab. 36 (1), 48–61.e6. doi:10.1016/j.cmet.2023.11.011
Lasman, L., Krupalnik, V., Viukov, S., Mor, N., Aguilera-Castrejon, A., Schneir, D., et al. (2020). Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 34 (19-20), 1373–1391. doi:10.1101/gad.340695.120
Levi, O., and Arava, Y. S. (2021). Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res. 49 (1), 432–443. doi:10.1093/nar/gkaa1178
Li, X., Xiong, X., Wang, K., Wang, L., Shu, X., Ma, S., et al. (2016a). Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 12 (5), 311–316. doi:10.1038/nchembio.2040
Li, X., Ma, S., and Yi, C. (2016b). Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 33, 108–116. doi:10.1016/j.cbpa.2016.06.014
Li, X., Xiong, X., Zhang, M., Wang, K., Chen, Y., Zhou, J., et al. (2017). Base-resolution mapping reveals distinct m(1)A methylome in Nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68 (5), 993–1005. doi:10.1016/j.molcel.2017.10.019
Liu, N., Dai, Q., Zheng, G., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, F., Clark, W., Luo, G., Wang, X., Fu, Y., Wei, J., et al. (2016). ALKBH1-Mediated tRNA demethylation regulates translation. Cell 167 (3), 816–828. doi:10.1016/j.cell.2016.09.038
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi:10.1093/nar/gkx141
Mabley, J. G., Rabinovitch, A., Suarez-Pinzon, W., Haskó, G., Pacher, P., Power, R., et al. (2003). Inosine protects against the development of diabetes in multiple-low-dose streptozotocin and nonobese diabetic mouse models of type 1 diabetes. Mol. Med. 9 (3-4), 96–104. doi:10.2119/2003-00016.mabley
Masoud Abd El Gayed, E., Kamal El Din Zewain, S., Ragheb, A., and ElNaidany, S. S. (2021). Fat mass and obesity-associated gene expression and disease severity in type 2 diabetes mellitus. Steroids 174, 108897. doi:10.1016/j.steroids.2021.108897
Mathew, A. V., Kayampilly, P., Byun, J., Nair, V., Afshinnia, F., Chai, B., et al. (2024). Tubular dysfunction impairs renal excretion of pseudouridine in diabetic kidney disease. Am. J. Physiol. Ren. Physiol. 326 (1), F30–f38. doi:10.1152/ajprenal.00252.2022
Mauer, J., Sindelar, M., Despic, V., Guez, T., Hawley, B. R., Vasseur, J. J., et al. (2019). FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15 (4), 340–347. doi:10.1038/s41589-019-0231-8
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mohanty, S. K., Singh, K., Kumar, M., Verma, S. S., Srivastava, R., Gnyawali, S. C., et al. (2024). Vasculogenic skin reprogramming requires TET-mediated gene demethylation in fibroblasts for rescuing impaired perfusion in diabetes. Nat. Commun. 15 (1), 10277. doi:10.1038/s41467-024-54385-w
Niewczas, M. A., Mathew, A. V., Croall, S., Byun, J., Major, M., Sabisetti, V. S., et al. (2017). Circulating modified metabolites and a risk of ESRD in patients with type 1 diabetes and chronic kidney disease. Diabetes Care 40 (3), 383–390. doi:10.2337/dc16-0173
Nishikura, K. (2016). A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17 (2), 83–96. doi:10.1038/nrm.2015.4
Oerum, S., Dégut, C., Barraud, P., and Tisné, C. (2017). m1A post-transcriptional modification in tRNAs. Biomolecules 7 (1), 20. doi:10.3390/biom7010020
Oerum, S., Meynier, V., Catala, M., and Tisné, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49 (13), 7239–7255. doi:10.1093/nar/gkab378
Onalan, E., Yakar, B., Karakulak, K., Kaymaz, T., and Donder, E. (2022). m(6)A RNA, FTO, ALKBH5 expression in type 2 diabetic and obesity patients. J. Coll. Physicians Surg. Pak 32 (9), 1143–1148. doi:10.29271/jcpsp.2022.09.1143
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Rintala-Dempsey, A. C., and Kothe, U. (2017). Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14 (9), 1185–1196. doi:10.1080/15476286.2016.1276150
Safra, M., Sas-Chen, A., Nir, R., Winkler, R., Nachshon, A., Bar-Yaacov, D., et al. (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551 (7679), 251–255. doi:10.1038/nature24456
Santos-Pujol, E., Quero-Dotor, C., and Esteller, M. (2024). Clinical perspectives in epitranscriptomics. Curr. Opin. Genet. Dev. 87, 102209. doi:10.1016/j.gde.2024.102209
Semenovykh, D., Benak, D., Holzerova, K., Cerna, B., Telensky, P., Vavrikova, T., et al. (2022). Myocardial m6A regulators in postnatal development: effect of sex. Physiol. Res. 71 (6), 877–882. doi:10.33549/physiolres.934970
Sharma, S., Watzinger, P., Kötter, P., and Entian, K. D. (2013). Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 41 (10), 5428–5443. doi:10.1093/nar/gkt195
Shen, F., Huang, W., Huang, J. T., Xiong, J., Yang, Y., Wu, K., et al. (2015). Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 100 (1), E148–E154. doi:10.1210/jc.2014-1893
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Shima, H., and Igarashi, K. (2020). N 1-methyladenosine (m1A) RNA modification: the key to ribosome control. J. Biochem. 167 (6), 535–539. doi:10.1093/jb/mvaa026
Song, H., Feng, X., Zhang, H., Luo, Y., Huang, J., Lin, M., et al. (2019). METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 15 (8), 1419–1437. doi:10.1080/15548627.2019.1586246
Song, Y., Jiang, Y., Shi, L., He, C., Zhang, W., Xu, Z., et al. (2022). Comprehensive analysis of key m5C modification-related genes in type 2 diabetes. Front. Genet. 13, 1015879. doi:10.3389/fgene.2022.1015879
Squires, J. E., and Preiss, T. (2010). Function and detection of 5-methylcytosine in eukaryotic RNA. Epigenomics 2 (5), 709–715. doi:10.2217/epi.10.47
Sun, H., Zhang, M., Li, K., Bai, D., and Yi, C. (2019). Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 29 (1), 80–82. doi:10.1038/s41422-018-0117-4
Sun, Y., Zou, H., Li, X., Xu, S., and Liu, C. (2021). Plasma Metabolomics reveals Metabolic profiling for diabetic retinopathy and disease progression. Front. Endocrinol. (Lausanne) 12, 757088. doi:10.3389/fendo.2021.757088
Sun, H., Li, K., Liu, C., and Yi, C. (2023). Regulation and functions of non-m(6)A mRNA modifications. Nat. Rev. Mol. Cell Biol. 24, 714–731. doi:10.1038/s41580-023-00622-x
Tan, Y., Cao, H., Li, Q., and Sun, J. (2021). The role of transcription factor Ap1 in the activation of the Nrf2/ARE pathway through TET1 in diabetic nephropathy. Cell Biol. Int. 45 (8), 1654–1665. doi:10.1002/cbin.11599
Waku, T., Nakajima, Y., Yokoyama, W., Nomura, N., Kako, K., Kobayashi, A., et al. (2016). NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci. 129 (12), 2382–2393. doi:10.1242/jcs.183723
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. doi:10.1038/nature18298
Wang, Y., Traugot, C. M., Bubenik, J. L., Li, T., Sheng, P., Hiers, N. M., et al. (2023a). N(6)-methyladenosine in 7SK small nuclear RNA underlies RNA polymerase II transcription regulation. Mol. Cell 83 (21), 3818–3834.e7. doi:10.1016/j.molcel.2023.09.020
Wang, Y. Y., Tian, Y., Li, Y. Z., Liu, Y. F., Zhao, Y. Y., Chen, L. H., et al. (2023b). The role of m5C methyltransferases in cardiovascular diseases. Front. Cardiovasc Med. 10, 1225014. doi:10.3389/fcvm.2023.1225014
Wang, X., Li, X., Zong, Y., Yu, J., Chen, Y., Zhao, M., et al. (2023c). Identification and validation of genes related to RNA methylation modification in diabetic retinopathy. Curr. Eye Res. 48 (11), 1034–1049. doi:10.1080/02713683.2023.2238144
Wang, R., Xue, W., Kan, F., Zhang, H., Wang, D., Wang, L., et al. (2024). NSUN2 affects diabetic retinopathy progression by regulating MUC1 expression through RNA m(5)C methylation. J. Transl. Med. 22 (1), 476. doi:10.1186/s12967-024-05287-4
Wang, R., Qu, J., Chen, M., Han, T., Liu, Z., and Wang, H. (2025). NSUN2 knockdown inhibits macrophage infiltration in diabetic nephropathy via reducing N5-methylcytosine methylation of SOCS1. Int. Urol. Nephrol. 57 (2), 643–653. doi:10.1007/s11255-024-04214-2
Wei, C., Gershowitz, A., and Moss, B. (1975). N6, O2'-dimethyladenosine a novel methylated ribonucleoside next to the 5' terminal of animal cell and virus mRNAs. Nature 257 (5523), 251–253. doi:10.1038/257251a0
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71 (6), 973–985. doi:10.1016/j.molcel.2018.08.011
Williams, K., Christensen, J., and Helin, K. (2011). DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 13 (1), 28–35. doi:10.1038/embor.2011.233
Wu, G., Adachi, H., Ge, J., Stephenson, D., Query, C. C., and Yu, Y. T. (2016). Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. Embo J. 35 (6), 654–667. doi:10.15252/embj.201593113
Wu, X., Wang, W., Fan, S., You, L., Li, F., Zhang, X., et al. (2023). U-shaped association between serum IGF2BP3 and T2DM: a cross-sectional study in Chinese population. J. Diabetes 15 (4), 349–361. doi:10.1111/1753-0407.13378
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xue, C., Chu, Q., Zheng, Q., Jiang, S., Bao, Z., Su, Y., et al. (2022). Role of main RNA modifications in cancer: N(6)-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct. Target Ther. 7 (1), 142. doi:10.1038/s41392-022-01003-0
Yang, X., Yang, Y., Sun, B. F., Chen, Y. S., Xu, J. W., Lai, W. Y., et al. (2017). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 27 (5), 606–625. doi:10.1038/cr.2017.55
Yang, Y., Shen, F., Huang, W., Qin, S., Huang, J. T., Sergi, C., et al. (2019). Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 104 (3), 665–673. doi:10.1210/jc.2018-00619
Zaccara, S., and Jaffrey, S. R. (2020). A unified model for the function of YTHDF proteins in regulating m(6)A-Modified mRNA. Cell 181 (7), 1582–1595. doi:10.1016/j.cell.2020.05.012
Zha, X., Xi, X., Fan, X., Ma, M., Zhang, Y., and Yang, Y. (2020). Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. Aging (Albany NY) 12 (9), 8137–8150. doi:10.18632/aging.103130
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18 (1), 31–42. doi:10.1038/nrm.2016.132
Keywords: diabetes, biomarkers, epitranscriptomics, RNA modification, m6A, pseudouridine
Citation: Hlavackova M, Benak D, Holzerova K, Alanova P, Hrdlicka J, Chalupova M, Opletalova B, Ostadal B and Kolar F (2025) Epitranscriptomic signatures in blood: emerging biomarkers for diagnosis of diabetes and its complications. Front. Cell Dev. Biol. 13:1656769. doi: 10.3389/fcell.2025.1656769
Received: 30 June 2025; Accepted: 09 October 2025;
Published: 21 October 2025.
Edited by:
Liliana Burlibasa, University of Bucharest, RomaniaReviewed by:
Shafeeq Ahmed Mohammed, University Hospital Zürich, SwitzerlandCopyright © 2025 Hlavackova, Benak, Holzerova, Alanova, Hrdlicka, Chalupova, Opletalova, Ostadal and Kolar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marketa Hlavackova, bWFya2V0YS5obGF2YWNrb3ZhQGZndS5jYXMuY3o=
†These authors have contributed equally to this work and share first authorship
 Marketa Hlavackova
Marketa Hlavackova Daniel Benak
Daniel Benak Kristyna Holzerova
Kristyna Holzerova Petra Alanova
Petra Alanova Jaroslav Hrdlicka
Jaroslav Hrdlicka Miloslava Chalupova
Miloslava Chalupova