- 1Center for Reproductive Medicine, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China
- 2Clinical Medicine Research Center of Prenatal Diagnosis and Birth Health in Hubei Province, Wuhan, Hubei, China
- 3Wuhan Clinical Research Center for Reproductive Science and Birth Health, Wuhan, Hubei, China
- 4Second Clinical Hospital of Wuhan University, Wuhan, Hubei, China
Male infertility, accounting for approximately 50% of global infertility cases, is a growing concern in reproductive medicine. A fundamental cause lies in disrupted spermatogenesis—a complex, highly regulated process involving mitotic proliferation, meiotic division, and spermiogenic remodeling. Among the key regulatory pathways, PIWI-interacting RNAs (piRNAs) and their associated PIWI proteins have emerged as essential players in maintaining germline genome integrity and ensuring successful sperm development. However, their clinical relevance remain underexplored. This review provides a comprehensive synthesis of the piRNA pathway’s multifaceted roles across the full spectrum of spermatogenesis. We describe how piRNAs, together with PIWI proteins, silence transposable elements (TEs), guide chromatin remodeling, regulate mRNA translation, and protect sperm from environmental insults. We detail the stage-specific functions of piRNA machinery during spermatocytogenesis, spermatidogenesis, and spermiogenesis, supported by evidence from gene knockout models and cross-species studies. Particular emphasis is placed on piRNA biogenesis, including the primary processing pathway, the ping-pong amplification cycle, and terminal modifications mediated by enzymes such as PNLDC1 and TDRKH. Genetic disruptions in key piRNA pathway genes—including MOV10L1, PNLDC1, SPOCD1, and TDRKH—have been linked to clinical phenotypes such as non-obstructive azoospermia and severe oligozoospermia. We explore how these mutations impair piRNA maturation, compromise TE silencing, and trigger germ cell arrest, highlighting their diagnostic and therapeutic relevance. In addition, we discuss emerging applications of piRNAs as non-invasive biomarkers in seminal plasma, with altered piRNA profiles correlating with reduced sperm count and motility. Beyond pathogenesis, the piRNA pathway presents a promising frontier for reproductive interventions. We examine translational strategies targeting piRNA-associated proteins (e.g., RNF8-MIWI interaction modulators) and the potential for piRNA-guided gene silencing in germ cells. Moreover, we consider the impact of environmental toxins and epigenetic stressors on piRNA dynamics, suggesting new angles for fertility preservation. In summary, this review positions the piRNA pathway as a central regulator of male reproductive health. By integrating molecular biology with clinical genetics, we provide a roadmap for leveraging piRNA biology in the diagnosis, management, and treatment of male infertility.
1 Introduction
Infertility is a growing global health challenge, with the World Health Organization reporting a global infertility rate of 17.5%, with male factor accounting for 50% of cases (Cox et al., 2022). In China, the infertility rate has surged from 12% in 2007 to nearly 18% by 2020 (Qiao et al., 2021). Male infertility is fundamentally linked to spermatogenesis, a highly orchestrated event that produces functional sperm. Spermatogenesis is classically divided into three key stages: spermatocytogenesis, spermatidogenesis, and spermiogenesis. Disruptions in any of these stages—whether caused by genetic mutations, epigenetic alterations, or dysregulated non-coding RNAs—can impair sperm development and lead to infertility (Hosseini et al., 2024).
During spermatogonia formation, mammalian spermatogonial stem cells (SSCs) self-renew through niche signals in mice (Sasaki and Sangrithi, 2023). Subsequently, spermiogenesis involves chromatin condensation mediated by species-specific packaging proteins (protamines P1/P2 in humans, Transition nuclear protein1/2 (Tnp1/2) transition proteins in mice, and sperm nuclear basic proteins (SNBPs) in Drosophila) and morphological specialization, resulting in distinct sperm architectures such as hook-shaped murine heads and elongated Drosophila tails (Tirmarche et al., 2016; Qin et al., 2023; Subash and Kumar 2021). In mice, this process is classically divided into 16 steps based on nuclear and acrosomal morphology (Miyata et al., 2024). Steps 1-8 correspond to the round spermatid phase, characterized by Golgi-derived proacrosomal vesicle formation and flagellar assembly, while steps 9-16 involve elongation, nuclear condensation, and cytoplasmic remodeling to generate mature spermatozoa (Hess and Renato de Franca, 2008). The staging system differs across species; for example, human spermiogenesis is categorized into 6 phases rather than discrete steps, underscoring the necessity of species-specific annotations (Trost et al., 2023; Du et al., 2021). Finally, spermiation entails junction remodeling (e.g., a disintegrin and metalloproteinase 3 (ADAM3) in humans, lactate dehydrogenase A (LDHA) -dependent metabolic regulation in mice) to release mature sperm from Sertoli cells, with residual cytoplasm clearance varying across species (e.g., cytoplasmic droplets in humans versus (vs.) direct detachment in Drosophila) (Ribas-Maynou et al., 2021). Key interspecies divergences span cycle duration (∼74 days in humans vs. ∼10 days in Drosophila), metabolic pathways, and chromatin packaging strategies, as evidenced by recent studies on retinoic acid (RA)-driven SSC differentiation, fragile X-related protein 1 (FXR1) phase separation in translational activation, and protamine-linked infertility (Kang et al., 2022).
A major breakthrough in understanding spermatogenesis came with the discovery of PIWI-interacting RNAs (piRNAs) in 2006, identified in the germ cells of various organisms, including Drosophila, mice, and rats (Aravin et al., 2006; Aravin et al., 2001; Lau et al., 2006; Grivna et al., 2006; Girard et al., 2006; Vagin et al., 2006). piRNAs are a class of non-coding RNAs, ranging from 26 to 31 nucleotides (nt), slightly longer than miRNAs (22–24 nt) and siRNAs (20–25 nt). piRNAs interact with PIWI proteins—a specialized subfamily of the Argonaute (Ago) proteins named after P-element Induced Wimpy testis in Drosophila—forming the piRNA/PIWI complex (Li et al., 2025; Saito et al., 2006). This complex critically safeguards the genomic stability and fidelity of germ cells through targeted silencing of transposable elements (TEs), thereby preventing mutagenic disruptions caused by their aberrant activation (Vandewege et al., 2022; Kalmykova et al., 2005). It is also critical for gametogenesis, particularly in the differentiation and development of germ cells (Ramakrishna et al., 2021). Notably, piRNAs are predominantly derived from TE sequences located in specific genomic regions organized into arrays termed piRNA clusters (Gunawardane et al., 2007; Brennecke et al., 2007). These clusters are categorized as uni-strand or dual-strand clusters based on whether the transcription occurs from one or both DNA strands. Beyond TE silencing, piRNAs are involved in chromatin remodeling, RNA cleavage and stability, and the regulation of apoptosis, all of which are essential for proper germ cell development and spermatogenesis (Shoji et al., 2009; Carmell et al., 2007; Post et al., 2014; Aravin et al., 2006; Brennecke et al., 2007).
The piRNA pathway regulates both transcriptional and post-transcriptional events during spermatogenesis, ensuring the development of functional sperm (Wang et al., 2023b). Its main function is to ensure the suppression of TEs at both transcriptional and post-transcriptional levels, safeguarding genome stability. Transcriptional silencing mechanisms like DNA methylation and histone modifications (e.g., histone H3 lysine 9 di-methylation (H3K9me2)) ensure proper gene regulation and genome stability in spermatogenic cells, while post-transcriptional silencing mechanisms fine-tune mRNA stability in later stages (Legrand and Hobbs, 2018). In addition to protecting the genome, piRNAs are critical for the differentiation and maturation of germ cells (Claro-Linares and Rojas-Rios, 2025). Dysregulation of the piRNA/PIWI pathway has been implicated in various reproductive disorders, most notably male infertility. Studies have shown that mutations or dysfunctions within this pathway can lead to spermatogenic failure, further highlighting its importance in male reproductive health (Liu and Zhang, 2023). piRNA pathway dysregulation may also act as a key driver of oncogenic reprogramming in testicular germ cell tumors (TGCTs) through aberrant activation of proto-oncogenes, positioning this pathway as a potential multilevel biomarker for early detection and precision therapeutics (Wang D. et al., 2023).
This review provides an in-depth exploration of how piRNAs regulate spermatogenesis, focusing on their roles in TEs silencing, maintaining genomic integrity, and guiding germ cell differentiation. We investigate the mutations in piRNA pathway-related genes that lead to male infertility, shedding light on the underlying molecular mechanisms. Insights from knockout models will also be discussed to clarify the phenotypic and mechanistic impacts of piRNA-related gene deficiencies. Beyond understanding these molecular functions, we will explore the therapeutic potential of restoring piRNA function and leveraging gene editing techniques for male infertility treatment, while also highlighting the clinical application of piRNAs as diagnostic biomarkers. By integrating these findings, this review aims to advance the understanding of piRNA pathway and its potential in improving reproductive health outcomes.
2 piRNAs biogenesis and mechanism of action
2.1 piRNA biogenesis
In mammals, piRNAs are classified into two main types based on their expression during spermatogenesis: pre-pachytene piRNAs—first expressed in prospermatogonia, and pachytene piRNAs—expressed after the developing spermatocytes enter the pachytene phase of meiotic prophase I (Deng and Lin, 2002). Pre-pachytene piRNAs are primarily involved in de novo methylation and TEs silencing during embryonic development (Aravin et al., 2008). Pachytene piRNAs are predominantly derived from intergenic regions, 3′UTRs, pseudogenes, and repeat regions, which are thought to regulate mRNAs and lncRNAs via post-transcriptional gene silencing (Ortega et al., 2024).
Most piRNA sequences exhibit rapid evolutionary divergence across species, with minimal sequence conservation particularly between invertebrates and vertebrates (Shi et al., 2013; Chirn et al., 2015). However, mammals possess Eutherian-Conserved piRNA Cluster (ECpiC) loci whose expression demonstrates profound conservation across eutherian evolution. These conserved clusters maintain high expression in diverse mammals (e.g., humans, mice, dogs), suggesting specific adaptation to support eutherian reproductive functions (Chirn et al., 2015). The human piRNA pathway displays distinct evolutionary and functional features compared to other mammals. Despite syntenic conservation of pachytene piRNA loci in eutherians, human pachytene piRNA genes display striking evolutionary dynamics: promoter conservation contrasts sharply with accelerated sequence evolution in transcribed regions, resulting in population diversity indices surpassing most of the other genomic elements (Ozata et al., 2020). This accelerated evolution and hyper-diversity may facilitate differential target gene regulation, potentially influencing parental genome compatibility and thereby constituting a potential driver of reproductive isolation in eutherian lineages.
piRNA biogenesis occurs through two main pathways: the primary pathway and the ping-pong amplification cycle. The primary pathway can be separated into the transcription process and the phased piRNA process. In the transcription process, piRNA precursors are mostly originated primarily from specific genomic regions known as “piRNA clusters” or “piRNA-producing loci”, where they are transcribed by RNA polymerase II (Pol II) as long non-coding RNAs (Brennecke et al., 2007). Then in the phased piRNA process, RNA helicase Armitage activates the transport of Aubergine (Aub)-bound single-stranded piRNA precursors to the outer mitochondrial membrane. Later, piRNA precursors are cleaved by mitochondrial protein Zucchini/Phospholipase D6 (PLD6) into non-overlapping fragments, and polyadenylated at the 3′end. They are subsequently capped at the 5′ end and loaded onto PIWI proteins inside the nucleus, forming mature phased piRNAs with a uridine bias at the 5′ end (Gainetdinov et al., 2018; Brennecke et al., 2007; Mohn et al., 2015; Han et al., 2015). The ping-pong cycle represents a self-amplifying secondary biogenesis pathway majorly taking place in nuage. Cytoplasmic PIWI proteins (e.g., Aub and Ago3) recognize TEs via complementary base pairing and catalyze their endonucleolytic cleavage (Brennecke et al., 2007). The resulting cleavage fragments are subsequently captured by another cytoplasmic PIWI protein, processed into nascent piRNAs, thereby establishing a cyclical “cleavage-reloading” feedback loop (Gunawardane et al., 2007; Brennecke et al., 2007). This cycle plays a critical role particularly during the later stages of spermatogenesis in mice and in Drosophila germ cells (Wang et al., 2023c; Dai et al., 2019). The two pathways of piRNA biogenesis exhibit distinct functional: the primary pathway is responsible for de novo synthesis of the foundational piRNA pool, while the ping-pong cycle employs a self-amplification mechanism to efficiently suppress transposon activity. Their synergistic interaction ensures precise regulation of germ cell development and maintenance of genomic stability (Czech et al., 2018). The mechanism of primary pathway exhibits considerable evolutionary conservation, whereas secondary ping-pong amplification cycle displays marked interspecies divergence. Mammals and Drosophila employ the ping-pong amplification cycle for piRNA biogenesis, whereas Caenorhabditis elegans lacks this machinery. In C. elegans, piRNAs (termed 21U-RNAs) are directly transcribed by RNA polymerase II to form primary piRNAs. These primary piRNAs complex with target RNAs and recruit RNA-dependent RNA polymerases (RdRPs), catalyzing synthesis of secondary 22G-RNAs. These 22G-RNAs subsequently associate with worm-specific Argonaute (WAGO)-clade Argonaute proteins to execute post-transcriptional gene silencing (Pastore et al., 2022; Wang et al., 2023c). “piRNA trimming” is the final step of piRNA biogenesis, which enables the 3′ends of piRNA precursors to achieve their mature lengths. This process is mediated by enzymes such as poly(A)-specific RNase-like domain containing 1 protein (PNLDC1) and Tudor and KH domain-containing protein (TDRKH) (Saxe et al., 2013; Izumi et al., 2016). During piRNA biogenesis, cellular structures such as intermitochondrial cement (IMC), nuage and mitochondria-associated ER membranes (MAMs) play essential roles in TEs silencing and the stabilization of piRNA processing complexes (Aravin et al., 2009; Lim and Kai, 2007). The biogenesis pathway also ensures the protection of piRNAs from degradation through precise terminal modifications, including 3′-end trimming and 2′-O-methylation (Mohn et al., 2014; Zhang et al., 2014). These modifications, mediated by enzymes such as PolyA-specific ribonuclease PARN-1 (a piRNA trimmer) and Small RNA 2′-O-methyltransferase HENN-1 (a 2′-O-methyltransferase), are essential for piRNA stability and proper function. Deficiencies in these enzymes disrupt piRNA maturation, leading to 3′tailing, degradation, and compromised fertility (Pastore et al., 2021).
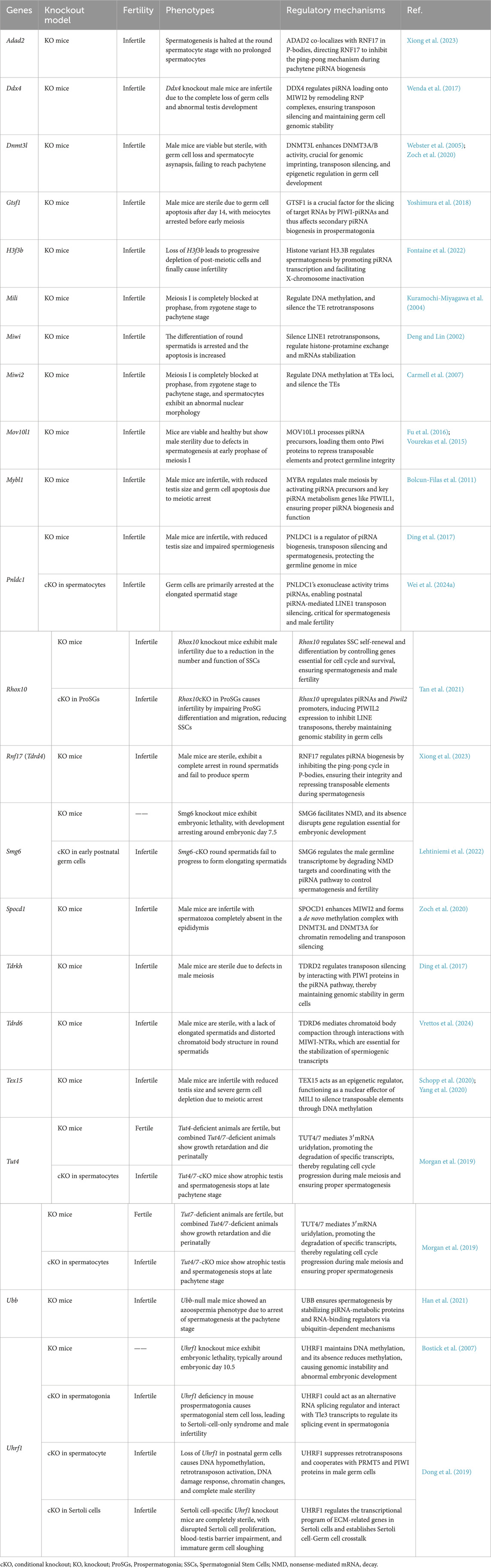
2.2 PIWI proteins and associated factors in the piRNA pathway
In humans, the PIWI protein family comprises four members: PIWIL1 (HIWI), PIWIL2 (HILI), PIWIL3, and PIWIL4 (HIWI2) (Li et al., 2024). In mice, the homologous proteins are designated as MIWI (PIWIL1), MILI (PIWIL2), and MIWI2 (PIWIL4) (Wei H. et al., 2024). The female-specific PIWIL3 is absent in mice and rats but can be identified in golden hamsters and bovine (Tan et al., 2020; Lv et al., 2023). Gene knockout mice of PIWI genes and associated factors in the piRNA pathway exhibit male fertility defects and varying degrees of spermatogenesis abnormalities, as shown in Table 1 (Dong et al., 2019; Bostick et al., 2007; Han et al., 2021; Morgan et al., 2019; Schopp et al., 2020; Yang et al., 2020; Vrettos et al., 2024; Lehtiniemi et al., 2022; Tan et al., 2021; Wei C. et al., 2024; Ding et al., 2017; Bolcun-Filas et al., 2011; Fu et al., 2016; Vourekas et al., 2015; Carmell et al., 2007; Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004; Fontaine et al., 2022; Yoshimura et al., 2018; Webster et al., 2005; Zoch et al., 2020; Wenda et al., 2017; Xiong et al., 2023). Notably, golden hamsters exhibit conserved expression patterns where PIWIL1 and PIWIL2 are predominantly cytoplasmic in prospermatogonia (Lv et al., 2023). PIWIL2 demonstrates dynamic co-localization with RNA-processing proteins mRNA-decapping enzyme 1A (DCP1A), Tudor domain-containing protein 1 (TDRD1), and mitochondrial ATP synthase F (1) complex subunit alpha (ATP5A) within both piP-bodies (a P granule that contains the PIWIL4-TDRD9 module) and IMC (Olotu et al., 2023; Wang X. et al., 2020). In contrast, PIWIL4 displays a dual subcellular distribution: cytoplasmic localization with DCP1A and Probable ATP-dependent RNA helicase DDX6 in piP-bodies coexists with nuclear accumulation, suggesting potential roles in both post-transcriptional regulation and chromatin-level processes (Lv et al., 2023).
During mammalian spermatogenesis, PIWI proteins exhibit stage-specific spatiotemporal dynamics that correlate with distinct regulatory functions. In early meiotic prophase I (leptotene/zygotene stages), PIWIL2 is exclusively expressed and localizes to nuage granules, ribonucleoprotein complexes critical for transposon silencing and mRNA surveillance (Gross, 2024). Upon transition to the pachytene stage, PIWIL1 emerges and forms dynamic co-localization clusters with PIWIL2 within these granules, suggesting synergistic roles in homologous recombination (Wang et al., 2022a).
Disruptions in PIWI proteins can lead to impaired piRNA biogenesis, negatively affecting spermatogenesis. Piwil1-deficient golden hamsters exhibit significantly reduced piRNA levels, causing spermatogenic arrest at the pachytene and zygotene stages, while Piwil2 and Piwil4 deficiencies result in almost complete loss of piRNA loss due to decreased mature gametes, leading to spermatogenic arrest during diplotene for most spermatocytes (Lv et al., 2023). Additionally, PIWI-Ins, a unique module within PIWI proteins, plays a crucial role in piRNA length selection. Deletion of PIWI-Ins in Miwi shifts MIWI(cytoplasmic) to load with shorter piRNAs, causing spermiogenic failure in mice (Wang et al., 2023b).
The proper functioning of PIWI proteins and associated factors, is essential for the precise completion of piRNA biogenesis. Mutations or defects in piRNA-related genes have also been shown to disrupt piRNA production and spermatogenesis, leading to male infertility. For a detailed discussion of these mutations in piRNA-related genes, please refer to Section 4.
3 piRNA in spermatogenesis: the regulatory network
The piRNA pathway is an intricate and highly specialized regulatory network that plays a crucial role in the proper development of male germ cells (Masone, 2024). Spermatogenesis, the process by which spermatogonia differentiate into mature spermatozoa, consists of three main stages: spermatocytogenesis, spermatidogenesis, and spermiogenesis. Throughout each stage, piRNAs, in conjunction with their associated PIWI proteins and piRNA pathway-associated proteins, perform vital functions that ensure genomic stability, regulate gene expression, and facilitate chromatin remodeling (Figures 1, 2).
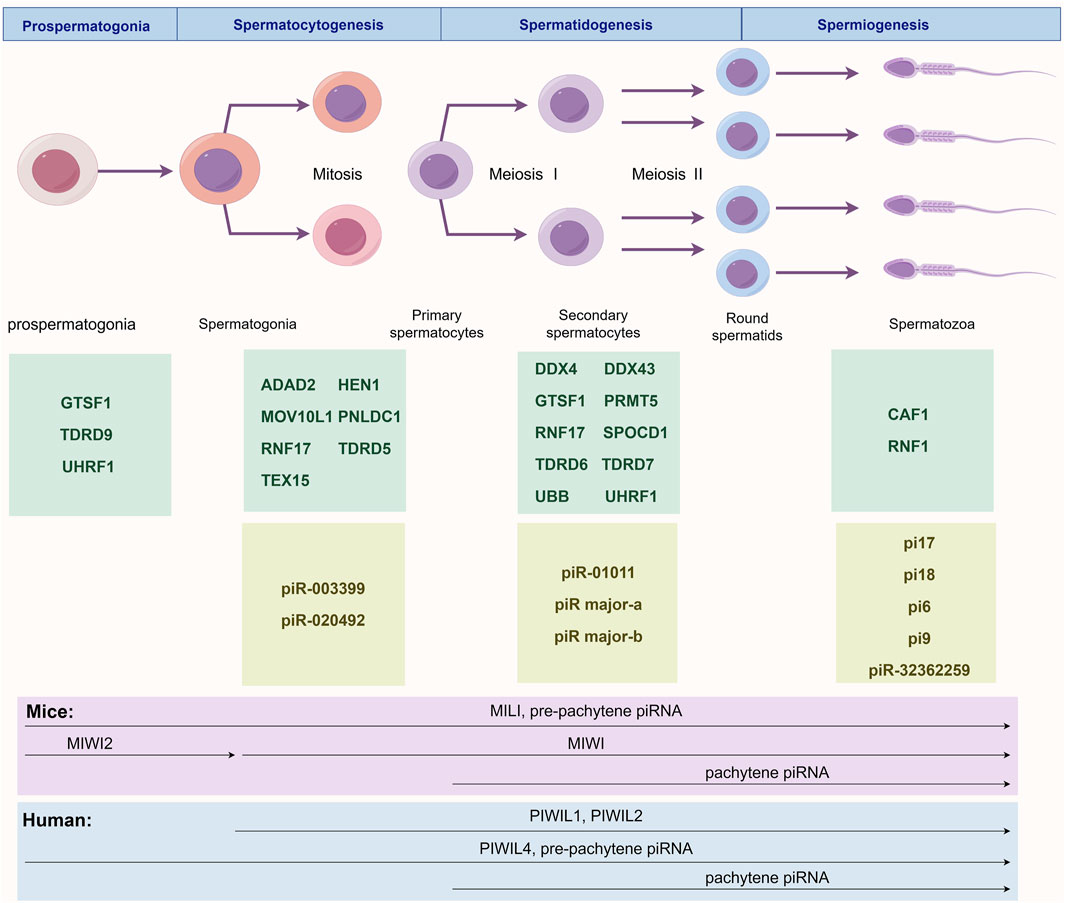
Figure 1. Stage-Specific Regulatory Network of the piRNA/PIWI Pathway During Spermatogenesis. This figure illustrates the major murine piRNAs and pathway proteins involved in prospermatogonia before birth and across three postnatal spermatogenic stages: prospermatogonia (GTSF1, TDRD9 and UHRF1), spermatocytogenesis (ADAD2, HEN1, MOV10L1, PNLDC1, RNF17, TDRD5, TEX15, piR-003399 and piR-020492), spermatidogenesis (DDX4, DDX43, GTSF1, PRMT5, RNF17, SPOCD1, TDRD6, TDRD7, UBB, UHRF1, piR-01011, piR major-a and piR major-b), and spermiogenesis (CAF1, RNF8, pi17, pi18, pi6, pi9 and piR-32362259). The lower panel depicts temporal expression patterns of PIWI proteins and piRNAs in humans and mice. In mice, MILI and pre-pachytene piRNAs appear in prospermatogonia and persist until spermiogenesis; MIWI2 is transiently expressed during prospermatogonia; MIWI expression begins postnatally and continues through spermatogenesis; pachytene piRNAs emerge at meiotic prophase I and remain until sperm maturation. In humans, PIWIL1 and PIWIL2 are expressed throughout postnatal spermatogenesis; PIWIL4 and pre-pachytene piRNAs are active from prospermatogonia through spermiogenesis; pachytene piRNAs follow the same temporal pattern as in mice.
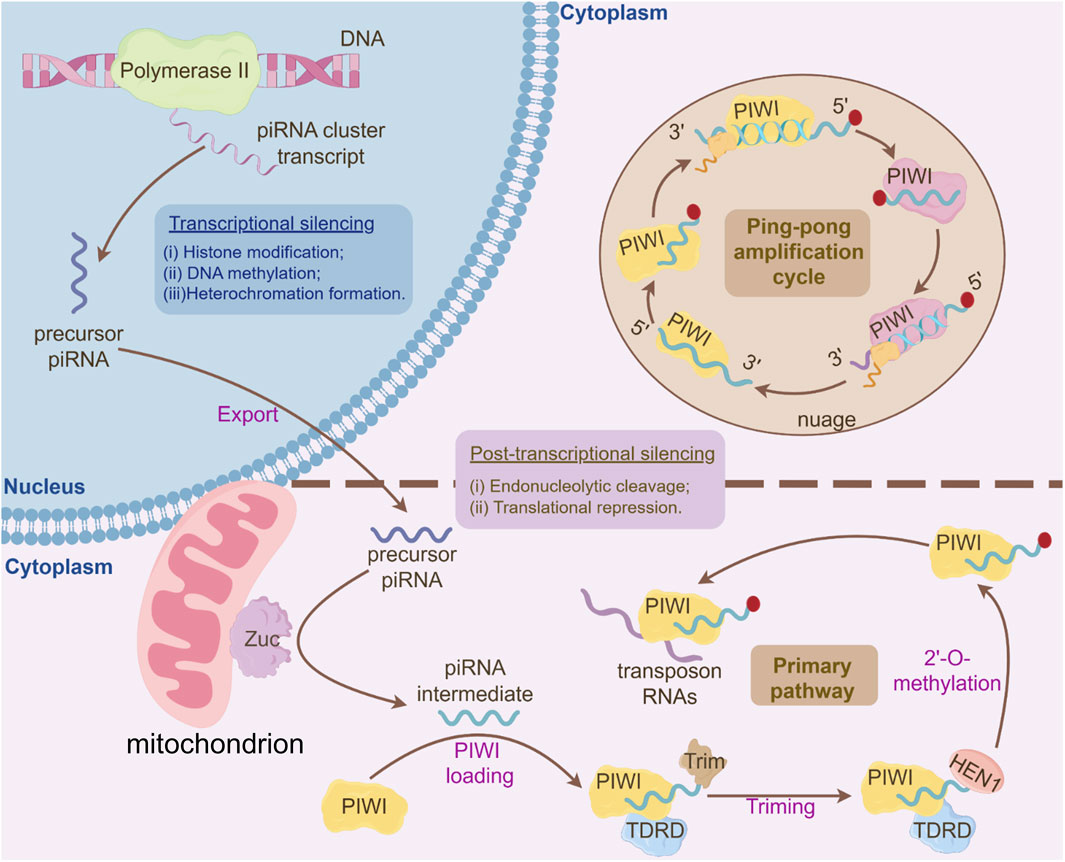
Figure 2. Integrated Mechanisms of piRNA Biogenesis and Gene Silencing Pathways. I. Primary piRNA Processing Pathway: Long single-stranded piRNA precursors, transcribed from piRNA clusters, are exported to the cytoplasm and processed into primary piRNA intermediates. These intermediates undergo 3′-to-5′ trimming by exonucleases to generate mature piRNA lengths. Subsequently, the 3′-ends of piRNAs are 2′-O-methylated by the HEN1 methyltransferase, stabilizing the piRNA molecules. The mature primary piRNAs are then loaded onto PIWI proteins, forming functional piRNA-PIWI complexes. These complexes recognize and bind complementary transposon RNAs in the cytoplasm, initiating downstream silencing mechanisms. II. Ping-Pong Amplification Cycle: The PIWI-piRNA complex binds and cleaves complementary transposon-derived RNAs in the cytoplasm, generating secondary piRNA fragments with a characteristic 10-nt overlap at their 5′ends. These secondary piRNAs are loaded onto PIWI, forming a self-amplifying loop that exponentially enriches piRNA populations targeting active transposons, ensuring robust silencing of transposable elements. III. Transcriptional Silencing in the Nucleus: Nuclear PIWI-piRNA complexes (e.g., PIWI in flies, MIWI2 in mammals) mediate epigenetic silencing through: (1) Histone Modification: Recruitment of histone methyltransferases depositing H3K9me3 marks, promoting heterochromatin assembly; (2) DNA Methylation: Guidance of DNA methyltransferases to target loci, establishing CpG methylation at transposon promoters; (3) Heterochromatin Formation: Cooperative action of H3K9me3-bound HP1 proteins and DNA methylation stabilizes condensed chromatin states, blocking transcriptional machinery access. IV. Post-Transcriptional Silencing in the Cytoplasm: Cytoplasmic PIWI/piRNA complexes bind transposon mRNAs, leading to: Endonucleolytic cleavage (“slicing”) of target RNAs, or Translational repression via recruitment of RNA degradation or inhibition factors.
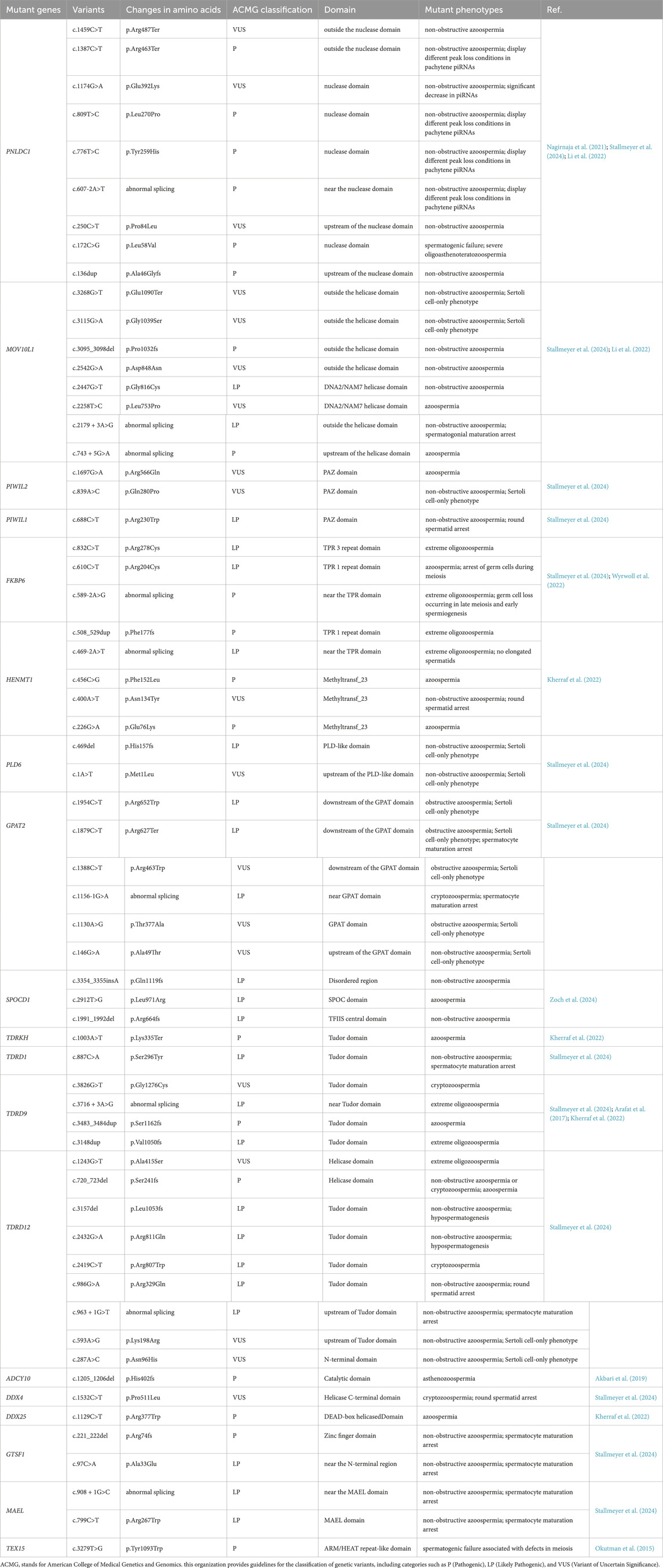
This section explores how mutations in genes associated with the piRNA pathway contribute to male infertility in human, as summarized in Table 2 (Okutman et al., 2015; Akbari et al., 2019; Arafat et al., 2017; Zoch et al., 2024; Kherraf et al., 2022; Wyrwoll et al., 2022; Nagirnaja et al., 2021; Stallmeyer et al., 2024; Li et al., 2022). Additionally, findings from knockout mice models are examined to illustrate the phenotypic consequences and mechanistic insights of piRNA pathway-related gene deficiencies (Table 1). Therefore, we aim to offer a new perspective on elucidating the regulatory network of piRNA in spermatogenesis.
3.1 piRNA pathway in spermatocytogenesis
3.1.1 piRNA pathway protects genomic stability during spermatocytogenesis
Spermatocytogenesis, the mitotic phase of spermatogenesis, involves the differentiation of diploid spermatogonia into primary spermatocytes. Genomic stability is crucial during this stage, as errors in DNA repair or chromosome segregation can lead to infertility or defective offspring (Xie et al., 2021). The piRNA pathway plays an indispensable role in maintaining genomic integrity in mice, primarily by silencing TEs and regulating gene expression.
TEs constitute a substantial portion of the genome and, if left unchecked, can lead to genomic instability. Overactivation of TEs results in DNA damage, apoptosis, sperm abnormalities, and infertility. Specifically, pre-pachytene piRNAs, which are activated during embryogenesis and remain active postnatally, direct the de novo methylation of repetitive elements, ensuring epigenetic silencing. In mice, nuclear PIWI proteins such as MIWI2 mediate transcriptional silencing of TEs, while cytoplasmic PIWI proteins, including MILI and MIWI, regulate post-transcriptional silencing via the ping-pong cycle (Manakov et al., 2015).
3.1.2 PIWI proteins and their multifaceted roles in spermatocytogenesis
Deficiencies in PIWI proteins often lead to early arrest during spermatogenesis. In Atlantic salmon, Almeida et al. reported that the loss of Piwil1 resulted in gametes deletion (Almeida et al., 2022). Some PIWI proteins also exhibit functional redundancy, in golden hamsters, PIWIL2 (cytoplasmic) can partially compensate for the absence of PIWIL4 in silencing specific TEs through the homotypic ping-pong pathway (Lv et al., 2023).
piRNA deficiencies also contribute to spermatogenic failure. In mice, Reddy et al. identified Pirmy and Pirmy-like RNAs as Y chromosome-derived noncoding transcripts that serve as templates for piRNAs, regulating genes critical for male fertility (Reddy et al., 2021). Notably, Y chromosome architecture and sequence content diverge markedly even among closely related species (e.g., Mus musculus vs. Mus caroli); thus, these findings may be context-specific to mice. Disruptions in these piRNAs, particularly due to Y chromosome deletions in the murine system, can cause abnormal sperm protein expression and male infertility (Reddy et al., 2021).
Deficiencies in the piRNA pathway exhibit species-specific phenotypic manifestations. Stallmeyer B et al. performed dual-species phenotypic analysis by characterizing individuals with biallelic high-impact variants in human piRNA pathway genes and generating corresponding knockout mouse models. Their investigation revealed fundamental interspecies divergence: defects in PIWIL2, PLD6, Glycerol-3-phosphate acyltransferase 2 (GPAT2), and TDRD12 produced profound phenotypic consequences in humans, whereas disruptions in TDRD9, Protein maelstrom homolog (MAEL), and PIWIL1 manifested more pronounced abnormalities in murine models. This differential phenotypic penetrance demonstrates that human and mice germlines possess distinct tolerance thresholds to impairments across the piRNA pathway machinery (Stallmeyer et al., 2024). “Trimming” is an important biochemical checkpoint in piRNA biogenesis, yet mutations in associated genes exhibit profound interspecies phenotypic divergence. Patients with PNLDC1 mutations uniformly demonstrate spermatogenic arrest at the pachytene spermatocyte stage, with only rare round spermatid production—representing significantly earlier developmental arrest than the elongated spermatid-stage arrest observed in Pnldc1 knockout mice. While murine models display severe molecular pathologies including profound reduction in MIWI-bound piRNAs and TE derepression, human PNLDC1 mutants exhibit globally extended piRNA lengths without systematically documented TE dysregulation or downregulation in PIWI-bound piRNAs, despite arresting at an earlier spermatogenic stage (Stallmeyer et al., 2024). This species-specific molecular manifestation needs further investigation into the different compensatory pathways in piRNA biogenesis between human and mice.
The piRNA pathway silences TEs through IMC and piP-body complexes (Li et al., 2021; Aravin et al., 2009). Primary piRNAs bind to MILI in prospermatogonia, cleaving TE transcripts to generate secondary piRNAs, which are subsequently loaded onto MILI and MIWI2, enabling nuclear translocation for transcriptional silencing (Aravin et al., 2009). Interspecies divergence characterizes the piRNA pathway’s TE silencing mechanisms. Mice prospermatogonia primarily employ piRNA-guided post-transcriptional degradation for TE suppression, with DNA methylation playing a comparatively limited role (Inoue et al., 2017). Conversely, during meiosis, DNA methylation emerges as the dominant silencing mechanism under the guidance of piRNA pathway proteins including SPOC domain-containing protein 1 (SPOCD1) and MIWI2(nuclear) (Inoue et al., 2017; Kawase and Ichiyanagi, 2022; Zoch et al., 2020). In Drosophila, the piRNA pathway operates through fundamentally distinct regulatory architecture: the Rhino-Deadlock-Cutoff (RDC) complex orchestrates transcription of dual-strand piRNA clusters, enforcing TE silencing at the transcriptional level via histone modification-mediated heterochromatinization (Chen et al., 2021c; Zhang et al., 2021). In humans, piRNAs focus on regulating primate-specific retrotransposable elements, like SINE-VNTR-Alus (SVAs), exhibiting more complex targeting mechanisms than in mice (Fukuda et al., 2022). This phenomenon may be related to humans having a higher proportion of transposons compared to mice (Goodier, 2016; Banuelos-Sanchez et al., 2019).
3.1.3 Key players MOV10L1, GTSF1, TEX15 and TDRD9 in piRNA-mediated regulation
MOV10L1, a testis-specific RNA helicase, serves as a master regulator of piRNA biogenesis. Loss of MOV10L1 function results in pre-pachytene piRNA deficiency, retrotransposon activation, and ultimately, spermatogenic failure and infertility (Fu et al., 2016; Vourekas et al., 2015). Non-obstructive azoospermia is characterized by testicular failure despite patent genital ducts (Agarwal et al., 2021). In non-obstructive azoospermia patients, mutations in MOV10L1 have been linked to reduced piRNA levels and meiotic arrest (Wang et al., 2022b). MOV10L1 also interacts with ribosomes on primary piRNA transcripts, facilitating their translocation beyond stop codons, a process dependent on TDRD5 (Ding et al., 2018). A deficiency in MOV10L1 leads to primary transcripts accumulation and mature piRNA reduction, triggering retrotransposon activation and male infertility (Guan et al., 2021).
Additionally, MOV10L1 has a role in spermatogonia by silencing TEs through its RNA helicase activity and participates in miRNA-mediated regulation. In cytoplasmic RNA processing bodies (P bodies), MOV10L1 participates in post-transcriptional regulation (Zhu et al., 2015). A study by Fu et al. reported that MOV10 (MOV10L1’s paralog) deficiencies disrupted spermatogonial progenitor cells (SPCs), downregulating essential genes like ETS translocation variant 5 (Etv5), B-cell CLL/lymphoma 6 member B protein (Bcl6b), and Zinc finger and BTB domain containing 16 (Zbtb16), crucial for SPCs proliferation and self-renewal in mice germ cells (Fu et al., 2019). The disruption impairs SPCs’ ability to form seminiferous tubules, further emphasizing the importance of a fully functional piRNA pathway in spermatogenesis (Fu et al., 2019). Notably, in contrast to murine models, Mov10l1 deficiency in golden hamsters results in infertility in both sexes, indicating a critical species-specific divergence in piRNA pathway functionality within female reproductive biology (Hasuwa et al., 2021; Loubalova et al., 2021).
Other proteins involved in the piRNA pathway also play crucial roles in genomic integrity. For instance, in mice, Gametocyte-specific factor 1 (GTSF1) and TDRD9 coordinate transcriptional silencing, with GTSF1 loss leading to depression of long interspersed element 1 (LINE-1)and intracisternal A particle (IAP) retrotransposons in prospermatogonia (Yoshimura et al., 2018). Testis-expressed protein 15 (TEX15), a testis-specific nuclear protein essential for the DNA damage response, works alongside MILI(nuclear) to recruit epigenetic silencing machinery to transposon loci (Yang et al., 2020). Notably, TEX15 is vital for spermatogenesis, but its deficiency does not disrupt piRNA biogenesis (Schopp et al., 2020). Finally, Adenosine deaminase domain-containing protein 2 (ADAD2), a protein highly expressed in male germ cells, regulates piRNA populations in mice. ADAD2 is a key player in the piRNA biogenesis interacting with multiple RNA-binding proteins, including MILI, MIWI, and RING finger protein 17 (RNF17). RNF17 preferentially binds to long precursors of pachytene piRNA clusters, and competitively reducing the opportunity for transposon RNA to interact with PIWI proteins, thereby preventing the over-activation of the ping-pong activity (Wasik et al., 2015). Adad2 deficiency markedly reduces the formation of RNF17 granules, causing the aberrant activation of the piRNA ping-pong cycle, resulting in a higher proportion of secondary piRNAs with a 10th A bias. Deficiency in Adad2 also results in elevated expression of piRNAs derived from transposons, while the cluster-derived pachytene piRNAs significantly decrease, including those associated with MILI and MIWI. This alteration closely resembles the abnormal piRNA patterns observed with RNF17 loss, suggesting a synergistic role of ADAD2 and RNF17 in regulating the genomic source preference of piRNAs (Lu et al., 2023).
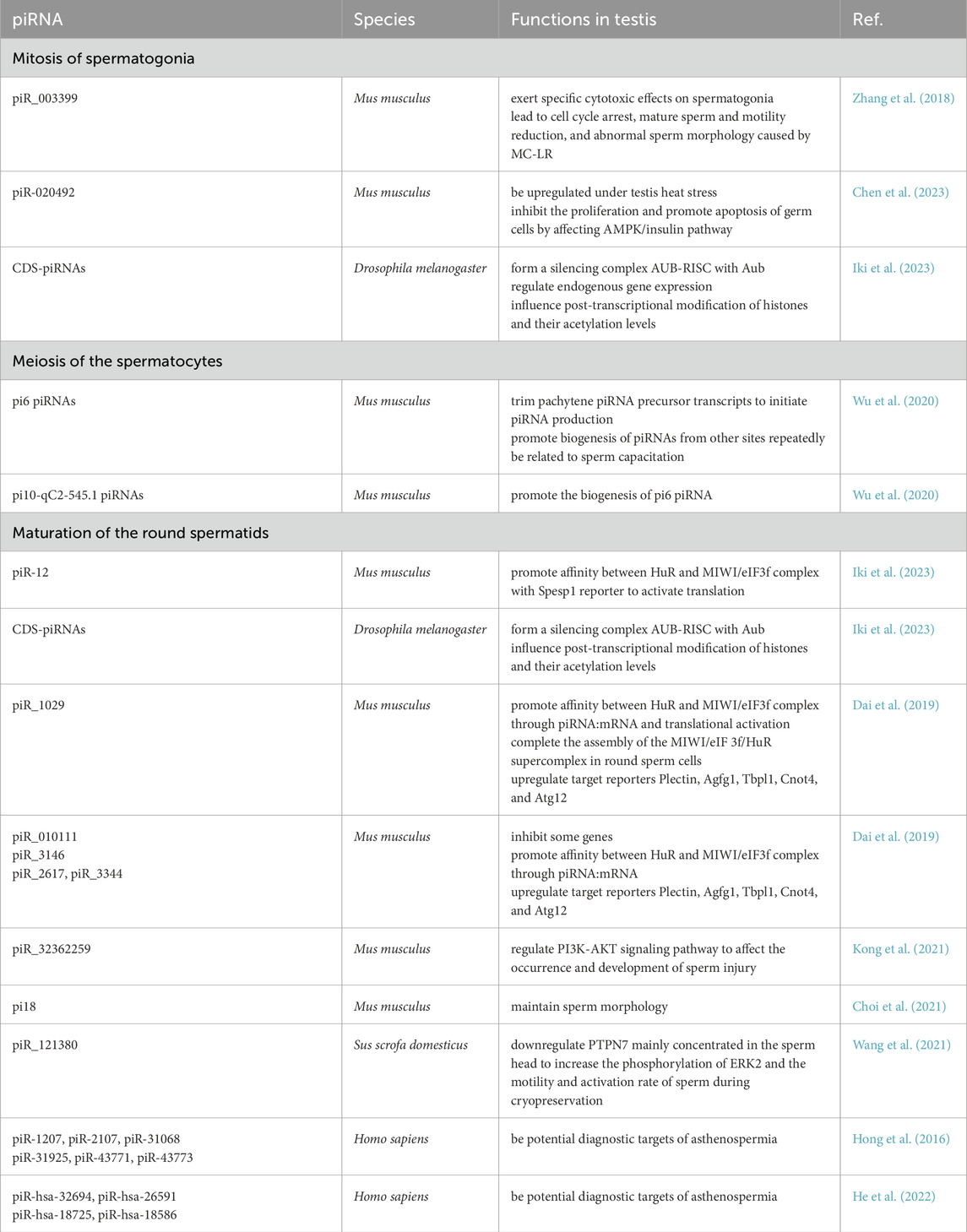
Beyond maintaining genomic integrity, piRNAs also respond to testicular damage from environmental stressors. Chen et al. showed that heat stress in mice upregulates 88 piRNA clusters and downregulates 47 piRNA clusters (Chen et al., 2023). Key piRNA piR-020492 is significantly upregulated, affecting AMP-activated protein kinase (AMPK) and insulin pathways, inhibiting germ cell proliferation, and inducing apoptosis (Chen et al., 2023). Similarly, Zhang et al. found that exposure to Microcystin-LR (MC-LR) elevates piR_003399, causing cytotoxicity in spermatogonia in mice (Zhang et al., 2018). Suppressing piR_003399 increased Cyclin-dependent kinase 6 (CDK6) levels, improving sperm motility and cell cycle progression in mice (Zhang et al., 2018). piR_003399 levels in serum and plasma exhibited dose-correlated variations with MC-LR exposure, paralleling the severity of reproductive impairment in mice in vivo models (Zhang et al., 2018). This supports its utility as a potential circulating biomarker for MC-LR-induced reproductive toxicity. These findings highlight piRNAs’ critical role in protecting testicular health.
In conclusion, the piRNA pathway is indispensable for maintaining genomic stability during spermatocytogenesis by silencing TEs and regulating essential gene expression. Disruptions in this pathway can lead to severe spermatogenic failure and infertility.
3.2 piRNA pathway in spermatidogenesis
3.2.1 piRNAs in maintaining genomic integrity and RNA cleavage
Spermatidogenesis, the second phase of spermatogenesis, involves the transformation of spermatocytes into spermatids through two meiotic divisions. This process begins with Meiosis I, where primary spermatocytes undergo chromosomal recombination and pairing during prophase, then divide into two secondary spermatocytes; Meiosis II follows, producing four haploid spermatids from each primary spermatocyte (Ishiguro, 2024). These spermatids are immature, round or oval-shaped cells lacking motility. The role of piRNAs in this phase is critical for maintaining genomic integrity and ensuring successful meiosis (Newkirk et al., 2017).
PIWI proteins play a central role in RNA cleavage, essential for reproductive function during meiosis. De Fazio et al. found that the RNA-cleaving activity of MILI relies on the conserved disulfide-directed β-hairpin fold (DDH) (Asp-Asp-His) motif in mice (De Fazio et al., 2011). Mutations in this motif (e.g., DAH or ADH) weakened MILI(cytoplasmic)’s cleavage activity, disrupting piRNA production, failing to silence TEs, and leading to spermatogenic arrest during meiosis. Co-factors, such as GTSF1, enhance MIWI(cytoplasmic)’s RNA cleavage function, while DEAD box polypeptide 4 (DDX4) and DDX43 promote the release of cleavage products, accelerating RNA degradation (Arif et al., 2022). Hsieh et al. further demonstrated that piRNAs, such as piR major-a and piR major-b, guide MIWI to cleavage sites, ensuring correct kinetochore assembly and chromosomal separation during meiosis in mice (Hsieh et al., 2020). Similarly, Chen et al. identified sex-specific piRNAs in Drosophila melanogaster that guide transcript cleavage, achieving gene regulation in the testis (Chen et al., 2021b).
Pachytene piRNAs play a central role in RNA cleavage. Cecchini K et al. demonstrated that the core function of murine pachytene piRNAs is to regulate target mRNAs through endonucleolytic cleavage, distinct from miRNA-like mechanisms or transcriptional silencing (Cecchini et al., 2024). Although the majority of these cleavage events do not alter the steady-state abundance of their target mRNAs, the regulation of a limited subset of critical mRNAs is indispensable for male fertility. Furthermore, transposon-derived piRNAs account for a higher proportion of targeting events, yet the high transcriptional activity of most targets buffers the impact of cleavage on their overall abundance. The evolutionary conservation of pachytene piRNAs is likely driven by selective advantages conferred by a minority of functional piRNAs, rather than the activity of the entire population—a feature potentially intrinsic to pachytene piRNA biology.
3.2.2 TE silencing via piRNA-guided methylation and chromatin relaxation
The piRNA pathway enforces TE silencing via species-specific epigenetic mechanisms—including DNA methylation in mice and histone modifications in Drosophila—with disruptions causing germ cell arrest and male infertility. PIWI proteins collaborate with SPOCD1 to establish methylation-dependent silencing of retrotransposons (Zoch et al., 2020). In the murine system, SPOCD1 interacts with MIWI2 and recruits the DNA (cytosine-5)-methyltransferase 3-like protein (DNMT3L)-DNA (cytosine-5)-methyltransferase 3A protein (DNMT3A) methyltransferase complex to nascent transposon loci, facilitating repressive chromatin remodeling. While de novo DNA methylation is largely restricted to embryonic germ cell development, this machinery may primarily enforce maintenance methylation during spermatidogenesis in mice, ensuring persistent TE suppression. Zoch et al. demonstrated that SPOCD1 deficiency disrupts LINE1 and IAP silencing, leading to meiotic arrest and male infertility, despite intact piRNA biogenesis and MIWI2(nuclear) localization (Zoch et al., 2020).
Nuclear protein E3 ubiquitin-protein ligase UHRF1, a five-domain epigenetic regulatory factor, plays a pivotal role in coordinating the piRNA pathway with chromatin remodeling machinery (Dong et al., 2019). UHRF1 interacts with arginine methyltransferase Protein arginine N-methyltransferase 5 (PRMT5), which regulates histone arginine modifications (symmetric dimethylation of histone H4 on Arg 3 (H4R3me2s) and symmetric dimethylation of histone H3 at arginine 2 (H3R2me2s)) (Wang et al., 2015). UHRF1 also controls the localization of key PIWI pathway proteins (MILI, MIWI, and TDRKH), thereby regulating piRNA biogenesis in mice (Li et al., 2021). Dong et al. discovered that UHRF1 forms a complex with PRMT5 and MILI/MIWI in the cytoplasm of mouse spermatocytes, enhancing the cleaving activity of PIWI proteins, which contributes to the post-transcriptional suppression of retrotransposons (Dong et al., 2019). In Uhrf1-deficient mice, piRNA pathway dysfunction results in the loss of pachytene piRNAs, impaired cooperation with PRMT5, global DNA demethylation, retrotransposon upregulation, activation of the DNA damage response, and altered chromatin states, ultimately leading to male infertility (Dong et al., 2019).
The piRNA pathway exhibits interspecies divergence in its mechanisms for TE suppression. FKBP Prolyl Isomerase Family Member 6 (Fkbp6) has been implicated in secondary piRNA biogenesis and synaptonemal complex assembly in mice, with its deficiency leading to meiotic arrest (Xiol et al., 2012; Crackower et al., 2003). In murine models, Fkbp6 loss disrupts synaptonemal complex formation and activates LINE-1 retrotransposons (Wyrwoll et al., 2022). In contrast, human cases of FKBP6 deficiency also cause spermatogenic failure but lack the pronounced LINE-1 derepression observed in mice, suggesting that humans may employ more complex redundant mechanisms and fine-tuning capabilities for TE silencing within the piRNA pathway (Wyrwoll et al., 2022). This phenotypic divergence is similarly observed in TDRD5 deficiency. While loss of Tdrd5 in mice causes retrotransposon derepression and meiotic arrest (Yabuta et al., 2011), human testes harboring TDRD5 mutations exhibit normal LINE-1 open reading frame 1 protein (LINE-1-ORF1p) expression with no observation of TE activation (Guo et al., 2025).
Notably, DNA methylation is not a universal TE silencing mechanism across species. For instance, in Drosophila, DNA methylation associated with the PIWI/piRNA pathway has not been reported. Instead, piRNA-mediated silencing relies predominantly on histone modifications (e.g., trimethylation of histone H3 lysine 9 (H3K9me3)) rather than DNA methylation (Zhang et al., 2021). Thus, the SPOCD1-MIWI2-DNMT3 axis described here reflects a murine-specific adaptation (Zoch et al., 2020). The uncharacterized protein C19ORF84 further bridges SPOCD1 with the methylation machinery, reinforcing piRNA-guided epigenetic silencing in mice. These findings underscore the importance of species context when interpreting piRNA-mediated TE control mechanisms (Zoch et al., 2024).
TE silencing mediated by piRNAs may also be achieved via chromatin modification pathways during spermatidogenesis. Research by Mahadevan et al. highlighted the role of histone H1t in chromatin regions containing TEs, where it interacts with repressive epigenetic markers and proteins, suggesting a mechanism for piRNA-mediated recruitment of repressive chromatin factors in mouse pachytene spermatocytes (Mahadevan et al., 2020). Under the regulation of piRNAs, H1t might facilitate the suppression of TEs through the induction of localized chromatin relaxation, which enables the recruitment of heterochromatin-associated proteins and repeat repressive-associated protein factors, ultimately leading to the formation of closed chromatin repressive structures (Mahadevan et al., 2020). Additionally, piRNA clusters are enriched with the histone variant H3.3B, which is crucial for spermatogenesis. Loss of H3f3b in mice severely impairs piRNA cluster transcription during meiosis, leading to infertility due to defective post-meiotic cells (Fontaine et al., 2022).
3.2.3 piRNAs-mediated mRNA translation and apoptosis regulation
During spermatidogenesis, piRNAs also guide mRNA translation. Dai et al. identified specific MIWI-CLIP clusters on piRNA target sites in key genes like Plectin, Arf-GAP domain and FG repeat-containing protein 1 (Agfg1), TATA box-binding protein-like 1 (Tbpl1), CCR4-NOT transcription complex subunit 4 (Cnot4), and Ubiquitin-like protein ATG12 (Atg12), which are crucial for acrosome formation and spermatid development in mice (Dai et al., 2019). These mRNAs contain AU-rich elements (AREs) in their 3′UTRs, necessary for piRNA-mediated translational activation. This process also inhibits the repressive actions of specific piRNAs, like piR_010111, on some genes. piRNA-mRNA interactions guide MIWI binding to these loci, assembling the MIWI/eIF3f/HuR complex to promote translation. Wang et al. further demonstrated that extended piRNA-mRNA complementarity in mice enhances RNA-binding protein human antigen R (HuR) binding to target mRNA in an ARE-independent manner, expanding the regulatory scope of the piRNA pathway (Wang et al., 2023b).
The interaction between piRNAs and the ubiquitin-proteasome system (UPS) represents an emerging area of research, particularly in the contexts of germline development and cancer. The piRNA pathway has been linked to post-translational protein degradation and apoptosis—the ubiquitin (Ub) pathway. Ub is a highly conserved protein crucial for fertility in both male and female mice (Martin-Villanueva et al., 2021). In mice, the polyubiquitin gene Polyubiquitin-B (Ubb) is composed of a tandem repeat of four ubiquitin-coding units; its primary function is to maintain cellular ubiquitin homeostasis by encoding tandemly repeated ubiquitin units and regulate critical physiological processes (Martin-Villanueva et al., 2021). Further mechanistic studies demonstrated that Ubb deficiency disrupts ubiquitin homeostasis, triggering a cascade of pathological events: (i) depletion of the free ubiquitin pool impairs proteasomal degradation, resulting in abnormal protein accumulation; (ii) dysregulation of germline-specific genes (Ddx4, Tdrd6, Tdrd7, Rnf17) and piRNA pathway effectors (MIWI, MILI) compromises post-transcriptional regulation; and (iii) synergistic dysfunctions culminate in pachytene-stage meiotic arrest, germ cell apoptosis, and complete gametogenesis failure (Han et al., 2021). Charmant O et al. revealed that the ubiquitination of the Paramecium PIWI protein is indispensable in male fertility. Ptiwi09 is predominantly regulated by Gtsf1 (Charmant et al., 2025). This process is initiated when Gtsf1 interacts with the Ptiwi09-maternal polyploid somatic macronucleus (MAC)-scnRNA complex, which is paired with nascent non-coding transcripts from the MAC. This binding triggers Ptiwi09 ubiquitination, facilitating the degradation of both the Ptiwi09 protein and its associated MAC-scnRNAs, compromising genome integrity. In Gtsf1-KD Paramecium, a marked reduction in Ptiwi09 ubiquitination is observed, resulting in abnormal MAC-scnRNA levels and TE activation (Charmant et al., 2025).
Small ubiquitin-like modifier (SUMO) is a highly conserved post-translational modifier that expands the functional diversity of the eukaryotic proteome through dynamic conjugation to target proteins (Celen and Sahin, 2020). Recent studies have revealed an intimate mechanistic link between SUMOylation and the piRNA pathway in female reproductive system. In D. melanogaster, diGly-based proteomic profiling has uncovered widespread SUMOylation of core piRNA pathway components, including nuclear factors such as Piwi and Panoramix (Panx), as well as cytoplasmic nuage constituents like Spindle-E (Spn-E) and Mael (Ninova et al., 2023). Notably, Piwi differentially regulates the SUMOylation status of these substrates, suggesting a hierarchical SUMO-dependent architecture within the piRNA pathway (Ninova et al., 2023). Functional ablation of SUMO disrupts the integrity of nuage, transforming perinuclear Vasa- and Aub-positive granules into dispersed puncta, thereby impairing piRNA biogenesis and post-transcriptional silencing of TEs (Ninova et al., 2023). Parallel work done in hermaphrodite C. elegans has demonstrated that SUMOylation of the germline determinant pharynx and intestine in excess protein 1 (PIE-1) is also essential for both germline fate maintenance and piRNA-mediated TE repression, underscoring the evolutionary conservation of this regulatory axis (Kim et al., 2021).
Mechanistically, SUMOylation acts as a molecular switch linking piRNA-guided silencing to chromatin remodeling. Ninova M et al. discovered that in Drosophila, the SUMO E3 ligase Su(var)2-10 interacts directly with the Piwi–Panx–Arx complex and facilitates the SUMOylation of both itself and associated chromatin factors (Ninova et al., 2020). This modification promotes recruitment of the SetDB1/Windei histone methyltransferase complex, enabling deposition of H3K9me3 marks at transposon loci and transcriptional repression of approximately 60% of TE families (Ninova et al., 2020). These findings establish SUMOylation as an essential scaffold for chromatin-based transposon control, effectively coupling small RNA recognition to epigenetic enforcement.
While the role of SUMOylation in piRNA-mediated silencing is well established in model organisms, its relevance in mammals and its interplay with other post-translational modifications are less clear. Although the role of SUMO in the piRNA pathway has been well studied in the female reproductive system, its function in male germ cells requires further investigation. Additionally, the dynamic regulation of SUMOylation and deSUMOylation, and their impact on piRNA pathway and condensate formation, are in need of further exploration.Taken together, the piRNA pathway is crucial for spermatidogenesis, maintaining genomic integrity, controlling protein degradation, regulating mRNA translation and apoptosis—all essential for the proper development and maturation of spermatids.
3.3 piRNA pathway in spermiogenesis
3.3.1 piRNA’s role in TE silencing during spermiogenesis
Spermiogenesis, the final stage of spermatogenesis, exhibits marked species-specificity in its morphological and molecular progression. This stage contains key molecular events including: proacrosomal vesicle formation, histone-to-protamine transition, acrosome biogenesis flagellar assembly, spermiation and epididymal maturation (Hess and Renato de Franca, 2008). The piRNA pathway and its functional partners regulate critical aspects of murine spermiogenesis, including transposon silencing during nuclear condensation and mRNA surveillance in cytoplasmic droplets.
The primary biological function of piRNAs in spermiogenesis is TEs silencing, particularly LINE1 (L1) retrotransposons, which make up about 20% of the mammalian genome (Doucet et al., 2015). L1 retrotransposition involves RNA polymerase II (RNAPII) synthesizing L1 RNA, which is then cleaved and polyadenylated. The transcript is exported to the cytoplasm and translated into two proteins: ORF1 and ORF2. ORF1 packages L1 RNA for nuclear import, while ORF2 facilitates reverse transcription and integration of L1 into the genome (Miyoshi et al., 2019). piRNA-mediated silencing of L1 is crucial for preventing spermatogenic arrest and infertility in male germ cells (Yang and Wang, 2016). In Pnldc1-deficient mice, piRNA trimming is disrupted, leading to L1 depression and spermatogenic arrest (Ding et al., 2017). TDRKH and PNLDC1 modulate MIWI localization in chromatoid bodies (CBs), promoting L1 silencing in round spermatids (Wei C. et al., 2024). Mutations in the Rhox gene cluster, encoding transcription factors critical for reproductive health, have also been linked to L1 silencing (Tan et al., 2021) (Table 1).
In C. elegans, the cytoplamsic Argonaute protein CSR-1 antagonizes piRNA-mediated silencing by promoting the expression of neo-formed RNAs, protecting germline-expressed genes (Cornes et al., 2022). This regulation facilitates the precise timing of piRNA silencing functions during spermiogenesis. piRNA biogenesis and degradation are tightly controlled, as seen in silkworms, where small RNA 2′-O-methyltransferase (BmHen1) and BmPnldc1 are essential for piRNA stability and TE silencing (Yang et al., 2022).
3.3.2 piRNA-mediated chromatin remodeling, histone-protamine transition and cytoplasmic exclusion
Chromatin remodeling, particularly the histone-to-protamine transition, is another critical process regulated by the piRNA pathway. PIWI proteins, in cooperation with piRNAs, regulate histone modifications during spermiogenesis. In Drosophila, CDS-piRNAs regulate genes involved in histone acetylation, such as newly excysted juveniles (nej), affecting histone acetyltransferase activity (Iki et al., 2023). Loss of aubergine(aub), a PIWI-interacting protein, leads to excessive histone acetylation and disrupted histone-protamine transitions, resulting in defective spermiogenesis (Iki et al., 2023). The THO RNA export complex and Gtsf1 are also involved in piRNA-guided chromatin modifications, such as H3K9me3, to ensure TE silencing during spermatogenesis in Drosophila (Donertas et al., 2013).
The elimination of PIWI proteins is closely linked to histone ubiquitination. Specifically, MIWI and RNF8 regulate H2B ubiquitination and MIWI degradation, both of which are essential for completing spermatogenesis in mice (Gou et al., 2017). MIWI acts as a molecular switch, initiating the histone-to-protamine transition critical for sperm maturation. Disruption of MIWI or RNF8 expression leads to accumulation of MIWI in cytoplasm and male infertility due to abnormal sperm morphology. piRNAs mediate PIWI clearance to ensure successful spermatogenesis (Gou et al., 2017). Zhao et al. found that in late-stage mouse spermatids, MIWI is degraded via the anaphase promoting complex/cyclosome (APC/C)-26S proteasome pathway, and piRNAs facilitate the interaction of MIWI with APC/C substrate subunits. This piRNA-triggered de-ubiquitination and MIWI degradation promote piRNA clearance, aiding sperm maturation (Zhao et al., 2013).
Nuclear condensation and cytoplasmic exclusion represent critical terminal events in spermiogenesis, essential for functional sperm maturation. Nuclear compaction achieves dramatic volumetric reduction of the spermatid nucleus through hyper-condensation of chromatin, forming the streamlined sperm head architecture. Concurrently, cytoplasmic exclusion eliminates superfluous organelles and transcripts via sequestration into residual bodies, purging most of cytoplasmic volume (Hess and Renato de Franca, 2008). This concerted remodeling enhances sperm motility while eliminating redundant mRNAs/proteins that could compromise early embryogenesis. Pachytene piRNAs provide the molecular logic underpinning these transformations. Through pi-RISC mediated large-scale mRNA clearance, they terminate dispensable cellular programs, enabling structural reorganization. Gou LT et al. demonstrated that murine pachytene piRNAs form chromatin assembly factor 1 (CAF1)-containing pi-RISCs in elongating spermatids. These complexes employ imperfect base-pairing to bind 3′UTRs of target mRNAs (e.g., G protein-coupled receptor kinase 4 (Grk4), Transcription factor SOX-6 (Sox6)), where CAF1 catalyzes deadenylation-independent decay. Crucially, this degradation operates independently of MIWI’s slicer activity, but requires MIWI’s piRNA-loading competence (Gou et al., 2014). By orchestrating transcriptome-wide silencing, pachytene piRNAs facilitate nuclear compaction and cytoplasmic exclusion–establishing the molecular framework for spermatid-to-spermatozoon transformation.
3.3.3 piRNA pathway’s impact on sperm motility and morphology
The piRNA pathway is also responsive to environmental factors that affect chromatin remodeling. Liu et al. demonstrated that exposure to silicon dioxide nanoparticles (SiNPs) alters MIWI expression patterns in mice spermatocytes, particularly in round spermatids (Liu et al., 2021). This increased MIWI expression can cause RNF8 retention in the sperm cell cytoplasm, leading to downregulated histone (H2A/H2B) removal and inhibited formation of ubH2A and ubH2B (Liu et al., 2021). The disrupted exchange of histone-to-protamine and chromatin condensation ultimately affecting the round spermatid differentiation and leading to sperm abnormalities (Liu et al., 2021). The study offers insights into how environmental contaminants like SiNPs negatively impact reproductive health. The piRNA pathway is essential for acquiring sperm motility, and deficiencies in piRNAs are often associated with impaired sperm vitality and morphology. Choi et al. found that mice lacking the pachytene piRNA cluster on chromosome 18 (pi18) exhibited severe sperm abnormalities in mice, including malformed heads, excessive acrosome formation, impaired acrosome exocytosis, defects in the axonemal complex of the mitochondrial sheath, and partial loss of outer dense fibers (ODFs) in the tail (Choi et al., 2021). These abnormalities led to poor sperm motility, reduced activity, and infertility. The absence of pi18 primarily disrupted the structural integrity of the trans-Golgi network, which was associated with enlarged proacrosomal vesicles and upregulation of Golgin subfamily A member 2 (GOLGA2) transcripts and protein (Choi et al., 2021).
Furthermore, Kong et al. discovered that piR_32362259 may influence the development of sperm damage by regulating the phosphatidylinositol 3-kinase (PI3K) - protein kinase B (AKT) signaling pathway in mice (Kong et al., 2021). Lower expression levels of this piRNA were found to slow the reduction in sperm vitality, influence the cell cycle, and decrease apoptosis rates. In mice, Wu et al. identified that mutations in pi6 piRNAs led to defective Ca2+ influx during the acrosome reaction, impairing the sperm’s ability to penetrate or bind to the zona pellucida, a critical step in fertilization (Wu et al., 2020).
Pachytene piRNAs are critically involved in maintaining sperm motility. The clusters pi9 and pi17 serve as major sources of murine pachytene piRNAs, collectively accounting for 13.5% of the total pool. Their loss ablates the production of corresponding pachytene piRNAs and damages RNA cleavage activity (Gainetdinov et al., 2023). Cecchini K et al. generated pi9−/− and pi17−/− single mutant male mice, which exhibited only mild reductions in sperm motility and remained fertile—indicating genetic redundancy between these loci. In contrast, pi9−/−pi17−/−double mutants displayed severe phenotypes, exhibiting significantly reduced sperm count and motility, loss of zona pellucida penetration ability, and complete infertility (Cecchini et al., 2024). This study underscores the critical role of pachytene piRNAs in regulating sperm function.
Additionally, Cornes et al. demonstrated that C. elegans spermatocytes lacking PIWI or Argonaute protein HRDE-1 failed to produce pseudopod-like structures observed in wild-type spermatocytes, leading to reduced sperm motility and significantly fewer offspring when crossed with wild-type individuals (Cornes et al., 2022). This highlights the critical role of piRNAs in maintaining sperm motility and ensuring reproductive success. Taken together, specific piRNAs and PIWI/piRNA pathway-associated proteins are involved in different stages of spermatogenesis (Figure 3).
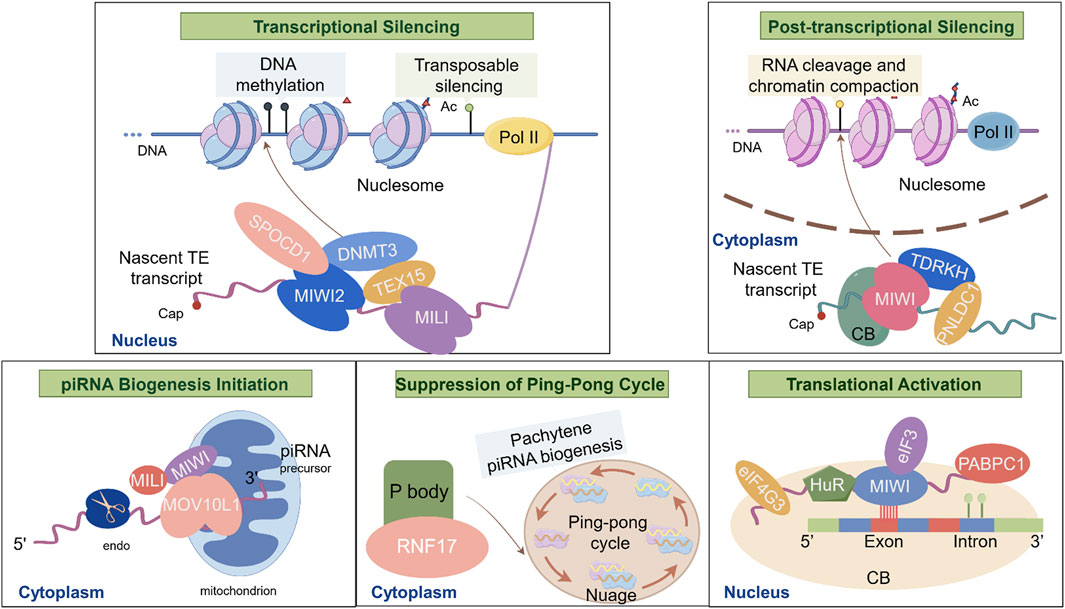
Figure 3. Biological Functions of piRNAs and Major Molecular Mechanisms during spermatogenesis. This schematic illustrates the molecular interactions of piRNAs during spermatogenesis: I. Transcriptional Silencing: TEX15 interacts with MILI to recruit epigenetic silencing complexes (e.g., DNMT3A/3L), targeting LINE1 retrotransposons; SPOCD1 bridges MIWI2 (PIWIL4) with DNMT3L, directing de novo DNA methylation at transposon loci to enforce transcriptional silencing; II. Post-transcriptional Silencing: PNLDC1 and TDRKH localize MIWI to chromatoid bodies, facilitating LINE1 RNA cleavage and chromatin compaction in nucleosome; III. piRNA Biogenesis Initiation: MOV10L1 binds to single-stranded piRNA precursors on mitochondria, recruiting MILI and MIWI to initiate primary piRNA processing; IV. Suppression of the Ping-Pong Cycle by ADAD2: ADAD2 activates RNF17 in cytoplasmic P bodies, inhibiting the amplification of secondary piRNAs via the ping-pong cycle in nuage; V. Translational Activation by MIWI/piRNA Complexes: MIWI/piRNA recruits HuR, eIF3, eIF4G3 and PABPC1 to AREs in target mRNAs (e.g., Plectin, Agfg1), promoting translation essential for acrosome formation in chromtoid bodies.
3.4 Interplay between piRNAs and hormones during spermatogenesis
To date, direct evidence for the impact of piRNAs on hormonal regulation during spermatogenesis remains limited. While piRNAs have been implicated in modulating hormone-related pathways, their capacity to influence systemic hormone levels or directly regulate endocrine signaling has not been fully elucidated. Current findings suggest a potential role for piRNAs in shaping the hormonal environment of the testis, yet mechanistic insights into this axis are still lacking and warrant further investigation.
Most existing data highlight an association between the piRNA/PIWI pathway and testosterone (TT) signaling. In androgen-deficient rat models, Miwi expression is significantly downregulated, whereas follicle-stimulating hormone (FSH) deficiency does not appear to impact Miwi levels, implying that Miwi is predominantly regulated by TT (Gill-Sharma et al., 2012). Supporting this, Kang et al. observed that in normal rats, both PIWI protein and piRNA expression patterns closely follow TT secretion profiles (Kang et al., 2014). Upon exogenous testosterone treatment, piRNA levels significantly increased across treatment groups, with neonatal-to-juvenile testosterone injection leading to elevated PIWI expression and reduced Ago3 expression (Kang et al., 2014). These findings suggest that TT may directly modulate the piRNA pathway to influence testicular function.
In addition to androgens, estrogens have also been shown to impact piRNA pathway components. Pan et al. reported that estradiol administration in mice led to a significant downregulation of both Miwi and Mili at the mRNA and protein levels within the first 3 days of treatment, followed by partial recovery by day 7 (Pan et al., 2012). Notably, this effect was not dose-dependent, and the suppression of Miwi expression was more significant than that of Mili (Pan et al., 2012). This study provided the first in vivo evidence of estrogen-mediated negative regulation of the PIWI pathway in male gonads and suggested a possible antagonistic interaction between estrogen and testosterone in modulating piRNA/PIWI expression.
Corroborating the preclinical findings, clinical data from infertile men further support a link between piRNA dysregulation and hormonal imbalance. Kumar et al., using small RNA sequencing, revealed that men with low serum TT levels (5.9 ± 2.8 nmol/L) displayed distinct piRNA expression profiles, including significant downregulation of piR-hsa-21622 and piR-hsa-28841, compared to other subfertile groups (Kumar et al., 2019). Conversely, in subfertile men with normal TT and FSH levels, hsa-piR-26399 expression was markedly elevated, implying a nuanced relationship between specific piRNA species and hormone status (Kumar et al., 2019).
More recently, Schülke et al. investigated 132 cryptozoospermic patients and 160 men with obstructive azoospermia exhibiting full spermatogenesis (Schulke et al., 2024). Through principal component analysis and hierarchical clustering, they identified that men with Sertoli-cell-only (SCO) phenotype exhibited higher FSH levels, lower TT, reduced testicular volume, and an increased proportion of PIWIL4+ spermatogonia (Schulke et al., 2024). PIWIL4 expression was negatively correlated with the presence of elongated spermatids, suggesting that hormonal dysregulation may influence early spermatogonial PIWI expression and contribute to the SCO phenotype (Schulke et al., 2024). These findings not only underscore the clinical relevance of piRNA-hormone interactions but also hint at novel therapeutic avenues for male infertility.
Together, these studies collectively point toward a functional axis between piRNAs and the hormonal environment in the testis, especially involving TT and estrogen signaling. While the mechanistic underpinnings of how hormones modulate piRNA biogenesis and function remain to be fully elucidated, current evidence suggests that piRNAs may act as both effectors and responders of hormonal cues during spermatogenesis. Unraveling this interplay will be essential for advancing our understanding of male fertility and may offer new biomarkers or targets for intervention in male reproductive disorders.
3.5 Interplay of piRNAs, miRNAs, and siRNAs
piRNAs, miRNAs, and siRNAs represent three major classes of small non-coding RNAs that collectively regulate gene expression and genomic stability. piRNAs primarily function in the germline by silencing TEs and regulating gene expression. miRNAs are ∼22 nt RNAs that suppress gene expression via mRNA decay and translational repression (Bartel, 2004). siRNAs, characterized as 20–24 nt double-stranded RNAs, function as key mediators of RNA interference (RNAi) (Roberts et al., 2016). While they exhibit distinct biogenesis pathways and primary functions, their regulatory networks share numerous molecular components and exhibit significant cross-talk.
3.5.1 Shared molecular machinery
The Ago family proteins serve as central executors across all three pathways. Ago proteins universally bind small RNAs to form the RNA-induced silencing complex (RISC), mediating gene silencing through translational repression and mRNA decay (Azlan et al., 2016). Certain Ago proteins (e.g., human AGO1/2, Drosophila DmAGO2) localize to the nucleus, where they exert additional regulatory functions by promoting DNA methylation and modulating alternative splicing (Tan et al., 2009; Cernilogar et al., 2011; Azlan et al., 2016).
The methyltransferase HEN1 plays a conserved yet pathway-specific role in small RNA stabilization across species. Horwich MD et al. demonstrated that in Drosophila, the methyltransferase activity of DmHen1 majorly functions in the piRNA and siRNA pathways (Horwich et al., 2007). DmHen1 critically maintains piRNA stability and function by catalyzing 3′-terminal 2′-O-methylation in germ cells (Horwich et al., 2007). Within the siRNA pathway, DmHen1 similarly methylates the 3′end of single stranded siRNAs following their loading into Ago2-RISC (Horwich et al., 2007). Intriguingly, a subset of Ago2-bound miRNAs (e.g., miR-277) also undergo DmHen1-mediated methylation (Horwich et al., 2007). This functional conservation extends to the C. elegans HEN1 ortholog henn-1 (Montgomery et al., 2012). Conversely, in N. vectensis, Hen1 primarily methylates miRNAs and piRNAs (Modepalli et al., 2018). Notably, knockdown of PIWI2 in Nematostella vectensis not only reduced piRNA levels but unexpectedly caused a significant decrease in miRNA abundance, suggesting piRNA pathway involvement in miRNA homeostasis, potentially via broad transcriptional regulation (Modepalli et al., 2018).
3.5.2 Reciprocal regulation of biogenesis and function
Emerging evidence indicates bidirectional crosstalk between these pathways. In Drosophila, Iki T et al. demonstrated that siRNAs can directly initiate piRNA biogenesis: 3′fragments generated by siRNA-directed cleavage of ATPsynbeta mRNA are loaded onto Aub and processed into CDS-piRNAs, evidenced by 5′-end complementarity between the siRNAs and resultant piRNAs (Iki et al., 2023). Furthermore, miRNAs (e.g., miR-316) bound to Ago2 can specify piRNA precursor regions by recognizing antisense sequences within target transcripts (e.g., muc14A), positioning Ago2 to initiate CDS-piRNA biogenesis (Iki et al., 2023).
miRNAs and piRNAs engage in reciprocal regulatory loops critical for developmental processes. Du WW et al. revealed that during murine embryonic development, miR-17-5p disrupts TE silencing by inhibiting the piRNA ping-pong amplification cycle and downregulating Mili and Dnmt3a. Conversely, injection of piR-11 into zygotes competitively attenuated miR-17-5p-mediated suppression of Mili and Dnmt3a, demonstrating mutual regulation essential for normal embryogenesis and TE control (Du et al., 2016).
piRNA-siRNA cooperation is evolutionarily conserved for robust gene silencing. High-throughput sequencing in four nematodes (C. elegans, Caenorhabditis briggsae, Caenorhabditis remanei, Caenorhabditis brenneri) revealed a conserved mechanism where piRNAs guide AGO proteins to cleave target mRNAs, triggering RdRP-dependent amplification of secondary siRNAs (Shi et al., 2013). In C. elegans, Manage KI et al. elucidated that piRNAs bound to the PIWI homolog PRG-1 recognize target mRNAs within P granules (Manage et al., 2020). This initial recognition, facilitated by the Tudor domain protein SIMR-1, nucleates secondary siRNA biogenesis, establishing a synergistic “piRNA-priming, siRNA-amplification” cascade essential for silencing target mRNAs, maintaining genomic integrity, and ensuring proper germ cell development (Manage et al., 2020).
piRNAs, miRNAs, and siRNAs form an interconnected regulatory network through shared biogenesis factors, reciprocal modulation, and overlapping effector mechanisms. These intricate interactions are crucial for fine-tuning gene expression, particularly during germline development and early embryogenesis. Understanding these relationships is fundamental to elucidating how small RNA pathways coordinate to maintain genomic integrity and regulate complex developmental programs.
4 piRNAs and clinical conditions
The piRNA pathway plays a crucial role in male reproductive health, and its dysfunction is linked to infertility. Understanding the dynamic functions of piRNAs at different stages of spermatogenesis is essential for developing novel therapeutic strategies for male infertility (He et al., 2022; Wang et al., 2021) (Table 3).
4.1 piRNAs and their clinical potentials as biomarkers
Due to their germ cell origin, decrease piRNA levels in sperm and seminal plasma may serve as biomarkers of reduced germ cell populations, particularly in azoospermic and oligozoospermic men (Larriba et al., 2023). Hong et al. reported significantly lower expression of piR-1207 and piR-2107 in the seminal plasma and sperm of oligozoospermic men, along with a general reduction in piRNA diversity and abundance (Hong et al., 2016). Moreover, Receiver Operating Characteristic (ROC) curve analysis identified five piRNAs (piR-31068, piR-31925, piR-43771, piR-30198 and piR-43773) as potential biomarkers for non-obstructive azoospermia. Notably, piR-30198 exhibits exclusive downregulation in azoospermia and may serve as a specific diagnostic biomarker distinguishing azoospermia from asthenozoospermia (Hong et al., 2016). Asthenozoospermia is a condition characterized by reduced sperm motility (Shahrokhi et al., 2020). Barceló M et al. compared exosomal microRNAs in seminal plasma exosomes between healthy individuals and patients with obstructive azoospermia to identify a significant downregulation of piR-58527 in the seminal plasma exosomes of obstructive azoospermia patients. This specific piRNA demonstrated precise diagnostic accuracy with high sensitivity and specificity (Barcelo et al., 2018). These findings demonstrate the potential of seminal plasma piRNAs as novel molecular biomarkers for the diagnosis of both non-obstructive azoospermia and obstructive azoospermia.
piRNAs demonstrate diagnostic potential in patients with asthenozoospermia. Hong Y et al. observed no significant alterations in the size or concentration of seminal plasma exosomes between asthenozoospermia patients and normozoospermic controls. However, they identified marked downregulation of piR-1207 and piR-2107 within these exosomes, concomitant with significantly reduced MitoPLD protein expression in spermatozoa in asthenozoospermia patients (Hong et al., 2021). These findings suggest that MitoPLD dysfunction may contribute to asthenozoospermia pathogenesis through impaired piRNA biogenesis. Furthermore, piR-1207 and piR-2107 exhibit promise as diagnostic biomarkers for asthenozoospermia, demonstrating high sensitivity and specificity, though validation in larger cohorts is warranted given the preliminary sample size (Hong et al., 2021). In a further study, He L et al. employed small RNA sequencing and reverse transcription polymerase chain reaction (RT-qPCR) validation to discover significant upregulation of piR-hsa-32694, piR-hsa-26591, piR-hsa-18725, and piR-hsa-18586 in asthenozoospermia patients. ROC analysis revealed exceptional diagnostic performance: piR-hsa-26591 achieved an Area Under the Curve (AUC) of 0.913, while the four-piRNA joint diagnosis model reached an AUC of 0.935 (He et al., 2022).
A clinically significant subset of non-obstructive azoospermia patients exhibits focal spermatogenic activity within the testicular parenchyma. For these individuals, microdissection testicular sperm extraction (micro-TESE) represents a critical surgical intervention, enabling the retrieval of viable sperm from localized regions of preserved spermatogenesis. Contemporary clinical evidence indicates procedure success rates approximating 40% in this patient cohort (Vahidi et al., 2021; Achermann et al., 2021). piRNAs may act as biomarkers in assessment for male infertility treatments. Cao C et al. performed comparative profiling of testicular piRNA expression between successful and unsuccessful micro-TESE groups in non-obstructive azoospermia patients (Cao et al., 2018). Their analysis revealed significant downregulation of 20 piRNAs in the micro-TESE failure group. Notably, hsa-piR-6254 and hsa-piR-17765 exhibited comparable expression levels in seminal plasma and testicular tissue, demonstrating their potential as non-invasive biomarkers for predicting micro-TESE outcomes (Cao et al., 2018). Chen H et al. identified significant downregulation of 8 piRNAs (piR-31704, piR-31843, piR-36659, piR-45048, piR-46102, piR-55522, piR-60351, and piR-61927) in extracellular vesicles (EVs) derived from seminal plasma of non-obstructive azoospermia patients (Chen H. et al., 2021). piR-61927 within seminal plasma EVs also demonstrated efficacy as a biomarker for predicting micro-TESE outcomes (Chen H. et al., 2021). Its non-invasive nature, high diagnostic accuracy (AUC = 0.83), and single-marker predictive capability establish this piRNA as a novel standardized tool for preoperative assessment in non-obstructive azoospermia, potentially reducing unnecessary surgical interventions. However, as a single-center pilot-scale investigation, these findings require validation through multicenter studies with expanded cohorts. Furthermore, developing composite biomarker panels (e.g., integrating piRNAs with hormonal profiles) may enhance predictive performance beyond current capabilities.
Spermatozoal piRNA levels may correlate with intracytoplasmic sperm injection (ICSI) success rates. Cui L et al. compared piRNA expression in spermatozoa between normozoospermic controls and patients with idiopathic male infertility undergoing first ICSI cycles. The infertile cohort exhibited significantly reduced levels of piR-31704 and piR-39888. Conversely, subgroups achieving high 2 pronuclei (2 PN) formation rates demonstrated elevated expression of piR-31704, piR-39888, and piR-40349. However, no significant associations emerged between piRNA levels and conventional semen parameters (motility, morphology), early embryo cleavage rates, top-quality embryo formation, or clinical pregnancy outcomes (Cui et al., 2018). These findings indicate that specific sperm piRNAs may associate with sperm concentration and ICSI fertilization competence, suggesting potential as biomarkers for fertilization potential assessment. Nevertheless, ROC curve analysis revealed suboptimal diagnostic performance for these piRNAs in detecting male infertility (AUC <0.75 for all three), indicating limited immediate clinical utility as standalone diagnostic markers (Cui et al., 2018).
4.2 Genetic and epigenetic alterations in key piRNA biogenesis genes
Mutations in genes involved in the piRNA pathway are frequently linked to male reproductive disorders. Genetic defects in the piRNA biogenesis pathway led to transposon de-repression, which contributes to spermatogenic failure and male infertility. Stallmeyer et al. conducted whole-exome sequencing on 412 idiopathic infertile men identified and identified biallelic pathogenic/likely pathogenic (P/LP) variants in 14 piRNA pathway genes (e.g., PNLDC1, TDRKH, FKBP6); Functional validation in Drosophila models confirmed that these mutations disrupted transposon silencing and triggered germ cell apoptosis, ultimately leading to infertility (Stallmeyer et al., 2024).
Among these genes, PNLDC1 plays a pivotal role in piRNA maturation. Nagirnaja et al. identified mutations in PNLDC1 in four unrelated men with non-obstructive azoospermia, demonstrating reduced expression of PIWIL1, PIWIL4, and other key piRNA pathway proteins in testicular tissue (Nagirnaja et al., 2021). Similarly, Li et al. discovered a homozygous PNLDC1 variant in a Han Chinese patient with severe oligoasthenoteratozoospermia through exome sequencing of 456 infertile patients (Li et al., 2022). Oligoasthenoteratozoospermia is one of the common diseases leading to male infertility, characterized by low sperm concentration, decreased motility and low percent of morphologically normal sperm (Colpi et al., 2018). In the same study, two compound heterozygous variants and a biallelic missense variant in MOV10L1 were identified in patients with non-obstructive azoospermia or severe oligozoospermia (Li et al., 2022). Oligozoospermia is characterized by a sperm concentration below the normattive threshold of 15 million sperm per milliliter of ejaculate observed in healthy males (Choy and Amory, 2020). Sperm concentrations ≤1 million/mL is considered as severe oligozoospermia (Bak et al., 2010). These findings suggest that mutations in key piRNA biogenesis proteins may contribute to dyszoospermia.
Disruptions in the PIWI/piRNA pathway contribute to male infertility through several mechanisms (Figure 4): (i) Reactivation of transposons, causing chromosomal instability and cell death; (ii) Spermatogenic arrest from disrupted gene expression and DNA methylation; (iii) Defective sperm morphology and reduced motility; (iv) Lower sperm count due to deficiencies in key proteins like MOV10L1; and (v) Altered chromatin structure, affecting sperm DNA integrity.
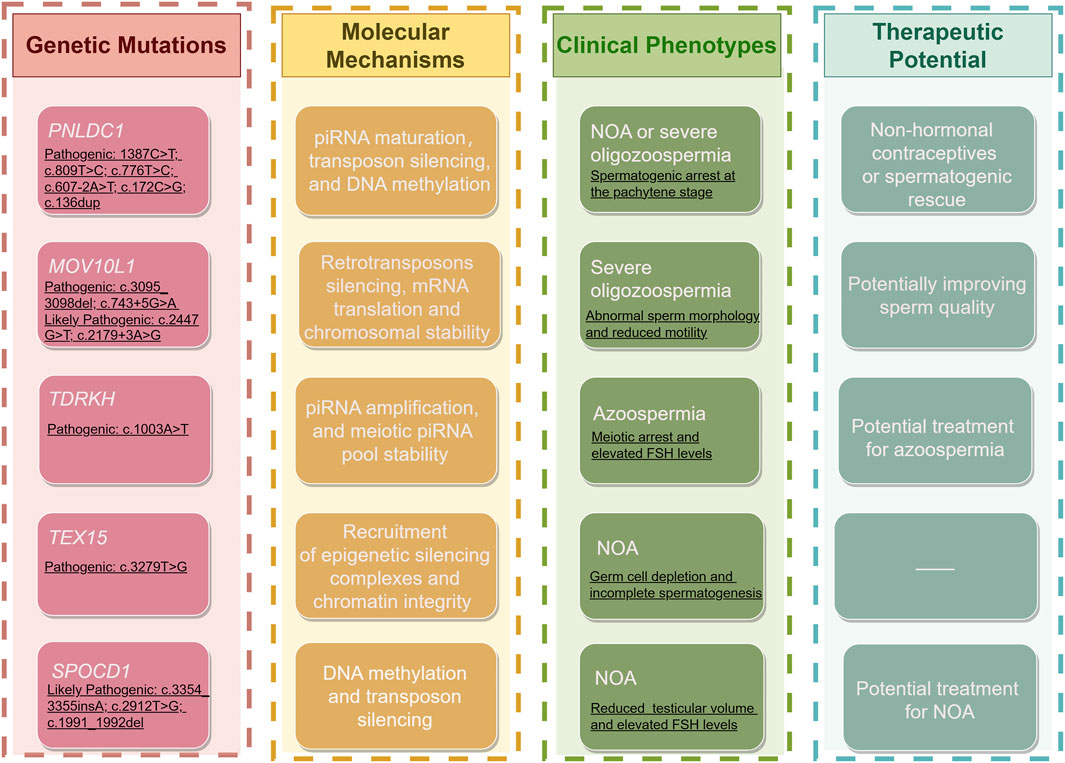
Figure 4. Genetic Defects in piRNA Pathway Genes and Their Implications in Male Infertility. This schematic summarizes the roles of key piRNA biogenesis genes (PNLDC1, MOV10L1, TDRKH, TEX15, and SPOCD1) in male infertility, integrating genetic, molecular, clinical, and therapeutic perspectives. I. Genetic Mutations: Pathogenic and likely pathogenic variants in these five key piRNA biogenesis genes are linked to male infertility. II. Molecular Mechanisms: (1) PNLDC1: Loss of piRNA trimming activity impairs piRNA maturation, leading to LINE1/IAP transposon reactivation and aberrant DNA methylation. (2) MOV10L1: Defective piRNA processing results in mRNA translation errors and chromosomal instability due to unsilenced retrotransposons. (3) TDRKH: Impaired piRNA amplification destabilizes meiotic piRNA pools. (4) TEX15: Failed recruitment of epigenetic silencing complexes (e.g., MILI) permits transposon mobilization, disrupting chromatin integrity. (5) SPOCD1: Compromised piRNA-guided DNA methylation at transposon loci reactivates retroelements. III. Clinical Phenotypes: Shared outcomes include non-obstructive azoospermia (NOA), oligo/azoospermia, spermatogenic arrest, abnormal sperm morphology, reduced motility and elevated FSH levels. IV. Therapeutic Potential: Genetic defects in PNLDC1, MOV10L1, TDRKH, TEX15, and SPOCD1 present actionable targets for male infertility treatment. (1) PNLDC1: Pharmacological modulation may restore piRNA maturation, offering potential for non-hormonal contraceptives or spermatogenic rescue. (2) MOV10L1: Correcting its function could re-establish piRNA biogenesis, improving sperm quality (3) TDRKH: Gene-editing strategies (e.g., CRISPR). to restore transposon silencing may address azoospermia (4) TEX15: Epigenetic regulators (e.g., DNMT inhibitors). could compensate for defective transposon repression. (5) SPOCD1: DNA methyltransferase enhancers might mitigate transposon activity in non-obstructive azoospermia.
4.3 Impact of other piRNA-related genes
Beyond core piRNA biogenesis genes, mutations in piRNA-associated genes have been linked to male infertilitys (Table 2). FKBP6, which participates in secondary piRNA biogenesis, has been found to harbor biallelic loss-of-function (LoF) variants (e.g., p.Arg278Cys, LP) in multiple infertile men, leading to severe oligozoospermia or azoospermia with abnormal sperm morphology (Stallmeyer et al., 2024). Similarly, SPOCD—a key regulator of transposon DNA methylation—has been associated with non-obstructive azoospermia and elevated FSH levels when harboring pathogenic variants such as p.Gln1119fs (Zoch et al., 2024).
Other notable genes include HENMT1, a regulator of piRNA methylation, and the TDRD family, which is essential for arginine methylation and piRNA function. It is noteworthy that the role of HENMT1 in piRNA methylation has thus far been exclusively documented in humans and mice, with no analogous function observed in Drosophila. Homozygous LoF variants in TDRD9 (p.Gly1276Cys, variant of uncertain significance (VUS)) and TDRKH (p.Lys335Ter, P) have been linked to human azoospermia (Kherraf et al., 2022). Moreover, TEX15—which collaborates with MILI to recruit epigenetic silencing machinery to transposon loci—is crucial for transposon suppression in mice. Okutman et al. identified a homozygous premature stop codon in TEX15 in two infertile brothers from a consanguineous Turkish family (Okutman et al., 2015). Likewise, Gou et al. screened 413 men with oligozoospermia and azoospermia and identified D-box heterozygous mutations in HIWI in three individuals. Pedigree analysis suggested that these mutations were either maternally inherited or arose de novo (Gou et al., 2017).
4.4 Treatment protocols and therapeutic potential
To date, treatment protocols for mutations in piRNA-related genes have been largely confined to TESE or micro-TESE, followed by ICSI, with generally poor outcomes. Four PNLDC1 variant carriers with non-obstructive azoospermia underwent underwent TESE/micro-TESE, with no successful pregnancies achieved (Nagirnaja et al., 2021). Stallmeyer et al. performed TESE on 23 of 39 infertile men carrying biallelic variants in 14 distinct piRNA pathway genes. Spermatozoa were retrieved solely in one patient with a TDRD9 variant, yielding extremely few spermatozoa exhibiting abnormal morphology and severely impaired motility. Subsequent ICSI in this TDRD9 variant carrier failed to achieve pregnancy, attributable to compromised sperm morphology, impaired fertilizing capacity, and/or embryonic developmental arrest (Stallmeyer et al., 2024). Wyrwoll et al. reported six individuals with FKBP6 variants exhibiting oligozoospermia or azoospermia. Following fine needle aspiration (FNA), retrieved spermatozoa displayed severe morphological abnormalities (e.g., multiflagellate spermatozoa, head defects, cytoplasmic fragmentation), rendering them unsuitable for assisted reproductive technology (ART) procedures (Wyrwoll et al., 2022). Three infertile men carrying homozygous pathogenic SPOCD1 variants presented with non-obstructive azoospermia. TESE procedures also failed to retrieve usable spermatozoa in all cases (Zoch et al., 2024). piRNA biogenesis protein EXD1 is a protein containing an exonuclease domain that, along with MIWI2 and TDRD12, is involved in secondary piRNA biogenensis and TE silencing in mammalian germ cells (Yang et al., 2016). Hu K et al. reported the first documented LoF variant in the EXD1 gene, identified in a patient presenting with oligoasthenoteratozoospermia characterized by acrosomal hypoplasia. The patient subsequently achieved a healthy live birth following ICSI treatment (Hu et al., 2025).
Notably, Akbari et al. reported a potential therapeutic approach for asthenozoospermia associated with one Adenylate cyclase type 10 (ADCY10) mutation. Supplementation of exogenous cyclic adenosine monophosphate (cAMP) analogue (dibutyryl cAMP sodium salt), dissolved in Human Tubal Fluid (HTF) to prepare a 20 mM stock solution, was employed to elevate cAMP levels, thereby activating the downstream PKA pathway and restoring sperm motility (Akbari et al., 2019). The ultimate pregnancy outcome for this specific patient was not reported. For similar cases of ADCY10 mutation presenting with normal sperm count but impaired motility, a recommended strategy involves in vitro treatment of spermatozoa with cAMP analogue (recommended dose: 0.05 mg/mL, 30-min incubation) followed by ICSI. For patients exhibiting spermatogenic arrest at the round spermatid stage (e.g., some carriers of FKBP6 or PNLDC1 mutations), round spermatid injection (ROSI) represents a theoretical option. However, ROSI currently demonstrates very low clinical success rates and lacks robust evidence supporting its efficacy (Hanson et al., 2021).
ART targeting oocyte activation may represent a promising therapeutic avenue for male infertility caused by piRNA pathway deficiencies. Guo R et al. documented a seminal case involving a TDRD5 mutation in two homozygous variant carriers from China presenting with severe oligoasthenoteratozoospermia (Guo et al., 2025). Sperm from both patients exhibited multiple morphological abnormalities, including multi-headed/multi-flagellated sperm and acrosomal malformations. Testicular analysis revealed significantly reduced expression of key IMC/CB components PIWIL1 and regulator of nonsense transcripts 1 (UPF1), along with a marked decrease in pachytene piRNA abundance. Following failed initial ICSI cycles, both patients underwent a modified protocol combining ICSI with artificial oocyte activation (AOA) using 10 µM calcium ionophore A23187 for 5 min. This intervention elevated fertilization rates to ≥50%, culminating in a live birth in one case (Guo et al., 2025). These outcomes suggest that AOA may offer therapeutic potential for male infertility linked to piRNA-related acrosomal defects.
Environmental stressors, such as oxidative stress and endocrine disruptors, further exacerbate infertility by impairing piRNA-mediated transposon silencing in gonad cells and germ cells. This underscores the urgent need for research into infertility treatments targeting this pathway (Munzel and Daiber, 2018). piRNAs may function as natural tumor suppressors in humans. Yuan J et al. discovered that piR-26441 directly binds to and upregulates YTH domain-containing protein 1 (Ythdc1) expression, which increases m6A modification to destabilize elongation factor Ts (Tsfm) mRNA (Yuan et al., 2025). This leads to reduced Tsfm expression, thereby suppressing mitochondrial oxidative phosphorylation (OXPHOS) and decreasing Complex I activity in human mitochondrion. Consequently, mitochondrial dysfunction and reactive oxygen species (ROS) accumulation occur, resulting in DNA damage and apoptosis. This study links piRNAs with oxidative stress, indicating that piRNAs may serve as a promising diagnostic biomarker for OXPHOS-associated ovarian cancer (Yuan et al., 2025).
Notably, piRNA-mediated interference (piRNAi) offers a precise and scalable gene-silencing strategy. Priyadarshini et al. demonstrated that in C. elegans, piRNAi enables targeted gene regulation via auxin-mediated degradation of PRG-1, leading to transgenerational epigenetic silencing (Priyadarshini et al., 2022). Similarly, Fabry et al. engineered a rapidly degrading PIWI protein after embryo formation in Drosophila, providing insights into the role of maternally deposited PIWI proteins in transposon regulation (Fabry et al., 2021). This strategy could be adapted to study the effects of sperm-derived PIWI proteins on embryogenesis.
As previously established, piRNAs demonstrate diagnostic potential for male infertility and micro-TESE outcome prediction. However, current biomarker researches on piRNAs remain predominantly focused on oncological applications. Multiple piRNAs, including piR-9491, piR-018569, piR-5937, piR-28876, piR-020619, and piR-020450, have been validated as high-performance diagnostic biomarkers across gastrointestinal and renal malignancies. These molecules consistently demonstrate superior sensitivity and robust AUC values, frequently outperforming established biomarkers in both early detection accuracy and diagnostic specificity (Wang Z. et al., 2020; Vychytilova-Faltejskova et al., 2018; Sohn et al., 2023). In stark contrast, the clinical translation of piRNA as biomarkers for male infertility diagnostics and treatments demands expanded mechanistic investigation and multi-center validation studies.
Overall, therapeutic interventions targeting the piRNA pathway hold significant promise for treating male infertility. Strategies such as modulating RNF8-MIWI interactions or targeting PNLDC1 could lead to novel fertility treatments. Additionally, the piRNA pathway may offer innovative approaches to non-hormonal contraception, broadening reproductive health options. As research advances, a deeper understanding of piRNA dynamics in testicular tissues and spermatozoa could pave the way for improved fertility treatments and reproductive health strategies.
5 Discussion, limitations and further perspectives
The piRNA pathway emerges as a central regulatory axis in spermatogenesis, with its dysfunction intricately linked to male infertility. This review consolidates evidence from animal models and human genetic studies, demonstrating that inherited defects in piRNA biogenesis genes (e.g., PNLDC1, MOV10L1, TDRD9) disrupt transposon silencing, induce genomic instability, and impair germ cell differentiation. Notably, mutations in these genes are strongly associated with clinical phenotypes such as non-obstructive azoospermia and oligoasthenoteratozoospermia, underscoring the essential role of piRNA pathway in male fertility.
A key advancement highlighted here is the identification of piRNAs as diagnostic biomarkers in seminal plasma (e.g., piR-hsa-26591 for asthenozoospermia) and spermatozoa, offering non-invasive tools for male infertility evaluation. Furthermore, the stage-specific dynamics of PIWI proteins (such as MIWI’s role in chromatin compaction during spermiogenesis and MIWI2’s nuclear localization for TE methylation) reveal spatiotemporal precision in piRNA-mediated regulation. These findings bridge gaps between animal studies and human pathology, particularly in explaining how piRNA defects lead to meiotic arrest or sperm structural abnormalities.
However, limitations persist. While knockout models (e.g., Piwil1−/− mice) robustly recapitulate human infertility phenotypes, translational challenges remain. For instance, the functional validation of rare human variants (e.g., HIWI mutations) is often constrained by limited patient-derived germline samples. Additionally, the interplay between piRNAs and environmental stressors (e.g., oxidative stress, endocrine disruptors) warrants deeper exploration, as these factors may exacerbate piRNA pathway dysregulation in clinical settings.
Current evidence indicates that therapeutic options for individuals carrying pathogenic variants in piRNA-related genes remain limited, often yielding suboptimal outcomes. A critical gap exists in the widespread lack of precisely targeted therapies directed against the piRNA pathway. While exogenous administration of cAMP analogues represents a potential targeted strategy for infertility caused by ADCY10 mutations, this approach has not advanced to clinical trials. Strategies such as modulating PIWI protein interactions (e.g., targeting the RNF8-MIWI complex), targeting key piRNA pathway components (e.g., PNLDC1, MOV10L1), or utilizing piRNAi hold promise for novel fertility treatments. However, their development is currently only confined to preclinical animal models. Although the piRNA pathway exhibits a high degree of evolutionary conservation, rigorous experimental validation is essential to establish the efficacy and feasibility of these therapeutic concepts. Furthermore, while emerging clinical data suggest piRNA expression levels may serve as a biomarker for the extent of environmentally induced male fertility impairment, comprehensive clinical data collection and further investigation are imperative. Additionally, the piRNA pathway may offer innovative approaches to non-hormonal contraception, broadening reproductive health options. As research advances, a deeper understanding of piRNA dynamics in testicular tissues and spermatozoa could pave the way for improved fertility treatments and reproductive health strategies.
The therapeutic potential of piRNA pathway has taken initial steps towards clinical translation from foundational research. However, significant practical hurdles remain. First, interspecies divergences between animal models and humans (e.g., species-specific TE silencing mechanisms) pose substantial challenges for extrapolating preclinical findings. Most interventional strategies remain confined to in vitro or preclinical animal models. Future progress will necessitate large-scale clinical validation and the refinement of targeted delivery systems (e.g., spermatogonia-specific vectors) to advance piRNA-based therapies. Second, delivery obstacles impede the development of piRNA therapeutics, including in vivo instability, off-target effects, and inadequate tissue-specific targeting. A primary physiological barrier is the blood-testis barrier (BTB), which rigorously regulates molecular transit into the seminiferous tubules. Composed of tight junctions, gap junctions, and basement membranes between adjacent Sertoli cells, the BTB maintains the spermatogenic microenvironment and protects germ cells from immune attack (Du et al., 2023). This highly selective barrier excludes most exogenous agents, including nucleic acid-based therapeutics and their delivery vehicles, from reaching target cells (e.g., spermatogonia, spermatocytes). Nanostructured lipid carriers (NLCs) represent a promising delivery vehicle candidate due to their demonstrated capacity to BTB, yet their therapeutic applicability requires further investigation (Tanyapanyachon et al., 2023). Direct supplementation of piRNAs or piRNA pathway components may disrupt epigenetic regulation in both germ and non-germ cells (e.g., Leydig cells, Sertoli cells), potentially leading to secondary spermatogenic defects or even testicular germ cell tumors. Moreover, piRNA functionality depends critically on intact secondary structure (Balaratnam et al., 2019). Certain delivery vehicles (e.g., cationic polymers) may compromise piRNA integrity, preventing formation of functional pi-RISCs or accelerating degradation, thereby abolishing in vivo activity. Third, safety concerns regarding germline gene editing technologies warrant careful consideration, particularly given ongoing ethical debates. Finally, large-scale, validated clinical cohorts are urgently needed to establish piRNA-based biomarkers with robust sensitivity and specificity for male infertility.
Future directions should prioritize:
(i) Mechanistic studies: Elucidating post-transcriptional roles of pachytene piRNAs in mRNA translation and their crosstalk with ubiquitin-proteasome systems.
(ii) Therapeutic innovation: Developing small-molecule inhibitors (e.g., targeting RNF8-MIWI interactions) or gene-editing strategies to restore piRNA function in defective germ cells.
(iii) Clinical translation: Validating piRNA biomarkers in large cohorts and exploring non-hormonal contraceptives via PNLDC1 modulation.
(iv) Environmental interactions: Investigating how pollutants or metabolic stressors impair piRNA networks, informing preventive strategies.
Author contributions
ZH: Project administration, Validation, Formal Analysis, Data curation, Resources, Methodology, Writing – review and editing, Investigation, Writing – original draft, Conceptualization. SH: Data curation, Investigation, Writing – original draft, Writing – review and editing, Formal Analysis. LL: Methodology, Investigation, Data curation, Formal Analysis, Visualization, Resources, Writing – review and editing, Writing – original draft, Software. YG: Writing – original draft, Software, Writing – review and editing, Resources, Investigation, Methodology, Visualization, Data curation. BM: Validation, Methodology, Data curation, Writing – original draft, Resources, Writing – review and editing. QF: Resources, Writing – original draft, Validation, Writing – review and editing, Data curation, Investigation, Methodology. YZ: Writing – review and editing, Methodology, Supervision, Writing – original draft, Investigation, Funding acquisition, Formal Analysis, Validation, Conceptualization, Project administration. MW: Funding acquisition, Software, Validation, Formal Analysis, Writing – review and editing, Resources, Investigation, Data curation, Methodology, Conceptualization, Supervision, Writing – original draft, Visualization, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project has received funding from National Natural Science Foundation of China (NSFC) (No. 82301817 to MW), Discipline construction project of Zhongnan Hospital of Wuhan University (No. XKJS202001 to YZ), the Health Commission of Hubei Provincial General Program (No. WJ2023M049 to MW), and the Fundamental Research Funds for the Central Universities (No. 2042025YXB003 to MW).
Acknowledgments
The authors thank to all the editors and reviewers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fcell.2025.1710695.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achermann, A. P. P., Pereira, T. A., and Esteves, S. C. (2021). Microdissection testicular sperm extraction (micro-TESE) in men with infertility due to nonobstructive azoospermia: summary of current literature. Int. Urol. Nephrol. 53 (11), 2193–2210. doi:10.1007/s11255-021-02979-4
Agarwal, A., Baskaran, S., Parekh, N., Cho, C. L., Henkel, R., Vij, S., et al. (2021). Male infertility. Lancet 397 (10271), 319–333. doi:10.1016/S0140-6736(20)32667-2
Akbari, A., Pipitone, G. B., Anvar, Z., Jaafarinia, M., Ferrari, M., Carrera, P., et al. (2019). ADCY10 frameshift variant leading to severe recessive asthenozoospermia and segregating with absorptive hypercalciuria. Hum. Reprod. 34 (6), 1155–1164. doi:10.1093/humrep/dez048
Almeida, F. L., Skaftnesmo, K. O., Andersson, E., L, K., R B, E., B, N., et al. (2022). The Piwil1 N domain is required for germ cell survival in Atlantic salmon. Front. Cell Dev. Biol. 10, 977779. doi:10.3389/fcell.2022.977779
Arafat, M., Har-Vardi, I., Harlev, A., Levitas, E., Zeadna, A., Abofoul-Azab, M., et al. (2017). Mutation in TDRD9 causes non-obstructive azoospermia in infertile men. J. Med. Genet. 54 (9), 633–639. doi:10.1136/jmedgenet-2017-104514
Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M., and Gvozdev, V. A. (2001). Double-stranded RNA-Mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11 (13), 1017–1027. doi:10.1016/s0960-9822(01)00299-8
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 (7099), 203–207. doi:10.1038/nature04916
Aravin, A. A., Sachidanandam, R., Bourc'his, D., Schaefer, C., Pezic, D., Toth, K. F., et al. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31 (6), 785–799. doi:10.1016/j.molcel.2008.09.003
Aravin, A. A., van der Heijden, G. W., Castaneda, J., Vagin, V. V., Hannon, G. J., and Bortvin, A. (2009). Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 5 (12), e1000764. doi:10.1371/journal.pgen.1000764
Arif, A., Bailey, S., Izumi, N., Anzelon, T. A., Ozata, D. M., Andersson, C., et al. (2022). GTSF1 accelerates target RNA cleavage by PIWI-clade Argonaute proteins. Nature 608 (7923), 618–625. doi:10.1038/s41586-022-05009-0
Azlan, A., Dzaki, N., and Azzam, G. (2016). Argonaute: the executor of small RNA function. J. Genet. Genomics 43 (8), 481–494. doi:10.1016/j.jgg.2016.06.002
Bak, C. W., Song, S. H., Yoon, T. K., Lim, J. J., Shin, T. E., and Sung, S. (2010). Natural course of idiopathic oligozoospermia: comparison of mild, moderate and severe forms. Int. J. Urol. 17 (11), 937–943. doi:10.1111/j.1442-2042.2010.02628.x
Balaratnam, S., Hettiarachchilage, M., West, N., Piontkivska, H., and Basu, S. (2019). A secondary structure within a human piRNA modulates its functionality. Biochimie 157, 72–80. doi:10.1016/j.biochi.2018.11.002
Banuelos-Sanchez, G., Sanchez, L., Benitez-Guijarro, M., Sanchez-Carnerero, V., Salvador-Palomeque, C., Tristan-Ramos, P., et al. (2019). Synthesis and characterization of specific reverse transcriptase inhibitors for Mammalian LINE-1 retrotransposons. Cell Chem. Biol. 26 (8), 1095–1109. doi:10.1016/j.chembiol.2019.04.010
Barcelo, M., Mata, A., Bassas, L., and Larriba, S. (2018). Exosomal microRNAs in seminal plasma are markers of the origin of azoospermia and can predict the presence of sperm in testicular tissue. Hum. Reprod. 33 (6), 1087–1098. doi:10.1093/humrep/dey072
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Bolcun-Filas, E., Bannister, L. A., Barash, A., Schimenti, K. J., Hartford, S. A., Eppig, J. J., et al. (2011). A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138 (15), 3319–3330. doi:10.1242/dev.067645
Bostick, M., Kim, J. K., Esteve, P. O., Clark, A., Pradhan, S., and Jacobsen, S. E. (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317 (5845), 1760–1764. doi:10.1126/science.1147939
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128 (6), 1089–1103. doi:10.1016/j.cell.2007.01.043
Cao, C., Wen, Y., Wang, X., Fang, N., Yuan, S., and Huang, X. (2018). Testicular piRNA profile comparison between successful and unsuccessful micro-TESE retrieval in NOA patients. J. Assist. Reprod. Genet. 35 (5), 801–808. doi:10.1007/s10815-018-1134-4
Carmell, M. A., Girard, A., van de Kant, H. J., Bourc'his, D., Bestor, T. H., de Rooij, D. G., et al. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12 (4), 503–514. doi:10.1016/j.devcel.2007.03.001
Cecchini, K., Ajaykumar, N., Bagci, A., Zamore, P. D., and Gainetdinov, I. (2024). Mouse pachytene piRNAs cleave hundreds of transcripts, but alter the steady-state abundance of only a minority of targets. bioRxiv, 2024.11.02.621675. doi:10.1101/2024.11.02.621675
Celen, A. B., and Sahin, U. (2020). Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts. FEBS J. 287 (15), 3110–3140. doi:10.1111/febs.15319
Cernilogar, F. M., Onorati, M. C., Kothe, G. O., Burroughs, A. M., Parsi, K. M., Breiling, A., et al. (2011). Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480 (7377), 391–395. doi:10.1038/nature10492
Charmant, O., Gruchota, J., Arnaiz, O., Nowak, K. P., Moisan, N., Zangarelli, C., et al. (2025). The PIWI-interacting protein Gtsf1 controls the selective degradation of small RNAs in Paramecium. Nucleic Acids Res. 53 (1), gkae1055. doi:10.1093/nar/gkae1055
Chen, H., Xie, Y., Li, Y., Zhang, C., Lv, L., Yao, J., et al. (2021a). Outcome prediction of microdissection testicular sperm extraction based on extracellular vesicles piRNAs. J. Assist. Reprod. Genet. 38 (6), 1429–1439. doi:10.1007/s10815-021-02101-8
Chen, P., Kotov, A. A., Godneeva, B. K., Bazylev, S. S., Olenina, L. V., and Aravin, A. A. (2021b). piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes Dev. 35 (11-12), 914–935. doi:10.1101/gad.345041.120
Chen, P., Luo, Y., and Aravin, A. A. (2021c). RDC complex executes a dynamic piRNA program during Drosophila spermatogenesis to safeguard male fertility. PLoS Genet. 17 (9), e1009591. doi:10.1371/journal.pgen.1009591
Chen, W., Zhao, Z., Cen, S., Lv, D., Wu, J., Zhou, X., et al. (2023). Exposure to elevated temperature affects the expression of PIWI-interacting RNAs and associated transcripts in mouse testes. Andrology 11 (4), 724–737. doi:10.1111/andr.13381
Chirn, G. W., Rahman, R., Sytnikova, Y. A., Matts, J. A., Zeng, M., Gerlach, D., et al. (2015). Conserved piRNA expression from a distinct set of piRNA cluster loci in Eutherian mammals. PLoS Genet. 11 (11), e1005652. doi:10.1371/journal.pgen.1005652
Choi, H., Wang, Z., and Dean, J. (2021). Sperm acrosome overgrowth and infertility in mice lacking chromosome 18 pachytene piRNA. PLoS Genet. 17 (4), e1009485. doi:10.1371/journal.pgen.1009485
Choy, J. T., and Amory, J. K. (2020). Nonsurgical management of oligozoospermia. J. Clin. Endocrinol. Metab. 105 (12), e4194–e4207. doi:10.1210/clinem/dgaa390
Claro-Linares, F., and Rojas-Rios, P. (2025). PIWI proteins and piRNAs: key regulators of stem cell biology. Front. Cell Dev. Biol. 13, 1540313. doi:10.3389/fcell.2025.1540313
Colpi, G. M., Francavilla, S., Haidl, G., Link, K., Behre, H. M., Goulis, D. G., et al. (2018). European academy of andrology guideline management of oligo-astheno-teratozoospermia. Andrology 6 (4), 513–524. doi:10.1111/andr.12502
Cornes, E., Bourdon, L., Singh, M., Mueller, F., Quarato, P., Wernersson, E., et al. (2022). piRNAs initiate transcriptional silencing of spermatogenic genes during C. elegans germline development. Dev. Cell 57 (2), 180–196.e7. doi:10.1016/j.devcel.2021.11.025
Cox, C. M., Thoma, M. E., Tchangalova, N., Mburu, G., Bornstein, M. J., Johnson, C. L., et al. (2022). Infertility prevalence and the methods of estimation from 1990 to 2021: a systematic review and meta-analysis. Hum. Reprod. Open 2022 (4), hoac051. doi:10.1093/hropen/hoac051
Crackower, M. A., Kolas, N. K., Noguchi, J., Sarao, R., Kikuchi, K., Kaneko, H., et al. (2003). Essential role of Fkbp6 in Male fertility and homologous chromosome pairing in meiosis. Science 300 (5623), 1291–1295. doi:10.1126/science.1083022
Cui, L., Fang, L., Shi, B., Qiu, S., and Ye, Y. (2018). Spermatozoa expression of piR-31704, piR-39888, and piR-40349 and their correlation to sperm concentration and fertilization rate after ICSI. Reprod. Sci. 25 (5), 733–739. doi:10.1177/1933719117725822
Czech, B., Munafo, M., Ciabrelli, F., Eastwood, E. L., Fabry, M. H., Kneuss, E., et al. (2018). piRNA-Guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52, 131–157. doi:10.1146/annurev-genet-120417-031441
Dai, P., Wang, X., Gou, L. T., Li, Z. T., Wen, Z., Chen, Z. G., et al. (2019). A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell 179 (7), 1566–1581. doi:10.1016/j.cell.2019.11.022
De Fazio, S., Bartonicek, N., Di Giacomo, M., Abreu-Goodger, C., Sankar, A., Funaya, C., et al. (2011). The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480 (7376), 259–263. doi:10.1038/nature10547
Deng, W., and Lin, H. (2002). Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2 (6), 819–830. doi:10.1016/s1534-5807(02)00165-x
Ding, D., Liu, J., Dong, K., Midic, U., Hess, R. A., Xie, H., et al. (2017). PNLDC1 is essential for piRNA 3' end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun. 8 (1), 819. doi:10.1038/s41467-017-00854-4
Ding, D., Liu, J., Midic, U., Wu, Y., Dong, K., Melnick, A., et al. (2018). TDRD5 binds piRNA precursors and selectively enhances pachytene piRNA processing in mice. Nat. Commun. 9 (1), 127. doi:10.1038/s41467-017-02622-w
Donertas, D., Sienski, G., and Brennecke, J. (2013). Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 27 (15), 1693–1705. doi:10.1101/gad.221150.113
Dong, J., Wang, X., Cao, C., Wen, Y., Sakashita, A., Chen, S., et al. (2019). UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in Male germ cells. Nat. Commun. 10 (1), 4705. doi:10.1038/s41467-019-12455-4
Doucet, A. J., Wilusz, J. E., Miyoshi, T., Liu, Y., and Moran, J. V. (2015). A 3' Poly(A) tract is required for LINE-1 retrotransposition. Mol. Cell 60 (5), 728–741. doi:10.1016/j.molcel.2015.10.012
Du, W. W., Yang, W., Xuan, J., Gupta, S., Krylov, S. N., Ma, X., et al. (2016). Reciprocal regulation of miRNAs and piRNAs in embryonic development. Cell Death Differ. 23 (9), 1458–1470. doi:10.1038/cdd.2016.27
Du, L., Chen, W., Cheng, Z., Wu, S., He, J., Han, L., et al. (2021). Novel gene regulation in normal and abnormal spermatogenesis. Cells 10 (3), 666. doi:10.3390/cells10030666
Du, S., Li, W., Zhang, Y., Xue, Y., Hou, X., Yan, J., et al. (2023). Cholesterol-amino-phosphate (CAP) derived lipid nanoparticles for delivery of self-amplifying RNA and restoration of spermatogenesis in infertile mice. Adv. Sci. (Weinh) 10 (11), e2300188. doi:10.1002/advs.202300188
Fabry, M. H., Falconio, F. A., Joud, F., Lythgoe, E. K., Czech, B., and Hannon, G. J. (2021). Maternally inherited piRNAs direct transient heterochromatin formation at active transposons during early drosophila embryogenesis. Elife 10, e68573. doi:10.7554/eLife.68573
Fontaine, E., Papin, C., Martinez, G., Le Gras, S., Nahed, R. A., Héry, P., et al. (2022). Dual role of histone variant H3.3B in spermatogenesis: positive regulation of piRNA transcription and implication in X-chromosome inactivation. Nucleic Acids Res. 50 (13), 7350–7366. doi:10.1093/nar/gkac541
Fu, Q., Pandey, R. R., Leu, N. A., Pillai, R. S., and Wang, P. J. (2016). Mutations in the MOV10L1 ATP hydrolysis motif cause piRNA biogenesis failure and Male sterility in mice. Biol. Reprod. 95 (5), 103. doi:10.1095/biolreprod.116.142430
Fu, K., Tian, S., Tan, H., Wang, C., Wang, H., Wang, M., et al. (2019). Biological and RNA regulatory function of MOV10 in Mammalian germ cells. BMC Biol. 17 (1), 39. doi:10.1186/s12915-019-0659-z
Fukuda, K., Makino, Y., Kaneko, S., Shimura, C., Okada, Y., Ichiyanagi, K., et al. (2022). Potential role of KRAB-ZFP binding and transcriptional states on DNA methylation of retroelements in human Male germ cells. Elife 11, e76822. doi:10.7554/eLife.76822
Gainetdinov, I., Colpan, C., Arif, A., Cecchini, K., and Zamore, P. D. (2018). A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in Most animals. Mol. Cell 71 (5), 775–790. doi:10.1016/j.molcel.2018.08.007
Gainetdinov, I., Vega-Badillo, J., Cecchini, K., Bagci, A., Colpan, C., De, D., et al. (2023). Relaxed targeting rules help PIWI proteins silence transposons. Nature 619 (7969), 394–402. doi:10.1038/s41586-023-06257-4
Gill-Sharma, M. K., Choudhuri, J., Ansari, M. A., and D'Souza, S. (2012). Putative molecular mechanism underlying sperm chromatin remodelling is regulated by reproductive hormones. Clin. Epigenetics 4 (1), 23. doi:10.1186/1868-7083-4-23
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds Mammalian piwi proteins. Nature 442 (7099), 199–202. doi:10.1038/nature04917
Goodier, J. L. (2016). Restricting retrotransposons: a review. Mob. DNA 7, 16. doi:10.1186/s13100-016-0070-z
Gou, L. T., Dai, P., Yang, J. H., Xue, Y., Hu, Y. P., Zhou, Y., et al. (2014). Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24 (6), 680–700. doi:10.1038/cr.2014.41
Gou, L. T., Kang, J. Y., Dai, P., Wang, X., Li, F., Zhao, S., et al. (2017). Ubiquitination-deficient mutations in human piwi cause Male infertility by impairing histone-to-protamine exchange during spermiogenesis. Cell 169 (6), 1090–1104. doi:10.1016/j.cell.2017.04.034
Grivna, S. T., Beyret, E., Wang, Z., and Lin, H. (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20 (13), 1709–1714. doi:10.1101/gad.1434406
Gross, P. (2024). Spatiotemporal regulation of germline genes by piRNAs. Nat. Genet. 56 (11), 2295. doi:10.1038/s41588-024-02004-1
Guan, Y., Keeney, S., Jain, D., and Wang, P. J. (2021). Yama, a mutant allele of Mov10l1, disrupts retrotransposon silencing and piRNA biogenesis. PLoS Genet. 17 (2), e1009265. doi:10.1371/journal.pgen.1009265
Gunawardane, L. S., Saito, K., Nishida, K. M., Miyoshi, K., Kawamura, Y., Nagami, T., et al. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in drosophila. Science 315 (5818), 1587–1590. doi:10.1126/science.1140494
Guo, R., Ding, B., Hu, K., Wang, K., Tang, D., Wang, X., et al. (2025). Homozygous deleterious variants in the C-terminal of TDRD5 impair spermiogenesis, causing severe oligoasthenoteratozoospermia in humans. Andrology 13 (5), 1270–1284. doi:10.1111/andr.13676
Han, B. W., Wang, W., Li, C., Weng, Z., and Zamore, P. D. (2015). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348 (6236), 817–821. doi:10.1126/science.aaa1264
Han, B., Jung, B. K., Park, S. H., Song, K. J., Anwar, M. A., Ryu, K. Y., et al. (2021). Polyubiquitin gene ubb is required for upregulation of piwi protein level during mouse testis development. Cell Death Discov. 7 (1), 194. doi:10.1038/s41420-021-00581-2
Hanson, B. M., Kohn, T. P., Pastuszak, A. W., Scott, R. T., Cheng, P. J., and Hotaling, J. M. (2021). Round spermatid injection into human oocytes: a systematic review and meta-analysis. Asian J. Androl. 23 (4), 363–369. doi:10.4103/aja.aja_85_20
Hasuwa, H., Iwasaki, Y. W., Au Yeung, W. K., Ishino, K., Masuda, H., Sasaki, H., et al. (2021). Production of functional oocytes requires maternally expressed PIWI genes and piRNAs in golden hamsters. Nat. Cell Biol. 23 (9), 1002–1012. doi:10.1038/s41556-021-00745-3
He, L., Wu, X., Wu, R., Guo, P., He, W., Sun, W., et al. (2022). Seminal plasma piRNA array analysis and identification of possible biomarker piRNAs for the diagnosis of asthenozoospermia. Exp. Ther. Med. 23 (5), 347. doi:10.3892/etm.2022.11275
Hess, R. A., and Renato de Franca, L. (2008). Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 636, 1–15. doi:10.1007/978-0-387-09597-4_1
Hong, Y., Wang, C., Fu, Z., Liang, H., Zhang, S., Lu, M., et al. (2016). Systematic characterization of seminal plasma piRNAs as molecular biomarkers for Male infertility. Sci. Rep. 6, 24229. doi:10.1038/srep24229
Hong, Y., Wu, Y., Zhang, J., Yu, C., Shen, L., Chen, H., et al. (2021). Decreased piRNAs in infertile semen are related to downregulation of sperm MitoPLD expression. Front. Endocrinol. (Lausanne) 12, 696121. doi:10.3389/fendo.2021.696121
Horwich, M. D., Li, C., Matranga, C., Vagin, V., Farley, G., Wang, P., et al. (2007). The drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17 (14), 1265–1272. doi:10.1016/j.cub.2007.06.030
Hosseini, M., Khalafiyan, A., Zare, M., Karimzadeh, H., Bahrami, B., Hammami, B., et al. (2024). Sperm epigenetics and Male infertility: unraveling the molecular puzzle. Hum. Genomics 18 (1), 57. doi:10.1186/s40246-024-00626-4
Hsieh, C. L., Xia, J., and Lin, H. (2020). MIWI prevents aneuploidy during meiosis by cleaving excess satellite RNA. EMBO J. 39 (16), e103614. doi:10.15252/embj.2019103614
Hu, K., Sha, X., Zu, C., Yang, F., Wang, G., Cheng, H., et al. (2025). Novel homozygous variants in piRNA pathway factors lead to Male infertility in humans. Reprod. Biomed. Online 51 (3), 104974. doi:10.1016/j.rbmo.2025.104974
Iki, T., Kawaguchi, S., and Kai, T. (2023). miRNA/SiRNA-Directed pathway to produce noncoding piRNAs from endogenous protein-coding regions ensures drosophila spermatogenesis. Sci. Adv. 9 (29), eadh0397. doi:10.1126/sciadv.adh0397
Inoue, K., Ichiyanagi, K., Fukuda, K., Glinka, M., and Sasaki, H. (2017). Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during Male germ-cell development in mice. PLoS Genet. 13 (7), e1006926. doi:10.1371/journal.pgen.1006926
Ishiguro, K. I. (2024). Mechanisms of meiosis initiation and meiotic prophase progression during spermatogenesis. Mol. Asp. Med. 97, 101282. doi:10.1016/j.mam.2024.101282
Izumi, N., Shoji, K., Sakaguchi, Y., Honda, S., Kirino, Y., Suzuki, T., et al. (2016). Identification and functional analysis of the Pre-piRNA 3' trimmer in silkworms. Cell 164 (5), 962–973. doi:10.1016/j.cell.2016.01.008
Kalmykova, A. I., Klenov, M. S., and Gvozdev, V. A. (2005). Argonaute protein PIWI controls mobilization of retrotransposons in the drosophila Male germline. Nucleic Acids Res. 33 (6), 2052–2059. doi:10.1093/nar/gki323
Kang, H. J., Moon, M. J., Lee, H. Y., and Han, S. W. (2014). Testosterone alters testis function through regulation of piRNA expression in rats. Mol. Biol. Rep. 41 (10), 6729–6735. doi:10.1007/s11033-014-3558-y
Kang, J. Y., Wen, Z., Pan, D., Zhang, Y., Li, Q., Zhong, A., et al. (2022). LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs. Science 377 (6607), eabj6647. doi:10.1126/science.abj6647
Kawase, M., and Ichiyanagi, K. (2022). The expression dynamics of piRNAs derived from Male germline piRNA clusters and retrotransposons. Front. Cell Dev. Biol. 10, 868746. doi:10.3389/fcell.2022.868746
Kherraf, Z. E., Cazin, C., Bouker, A., Fourati Ben Mustapha, S., Hennebicq, S., Septier, A., et al. (2022). Whole-exome sequencing improves the diagnosis and care of men with non-obstructive azoospermia. Am. J. Hum. Genet. 109 (3), 508–517. doi:10.1016/j.ajhg.2022.01.011
Kim, H., Ding, Y. H., Lu, S., Zuo, M. Q., Tan, W., Conte, D., et al. (2021). PIE-1 SUMOylation promotes germline fates and piRNA-dependent silencing in C. elegans. Elife 10, e63300. doi:10.7554/eLife.63300
Kong, L., Wu, Y., Hu, W., Liu, L., Xue, Y., and Liang, G. (2021). Mechanisms underlying reproductive toxicity induced by nickel nanoparticles identified by comprehensive gene expression analysis in GC-1 spg cells. Environ. Pollut. 275, 116556. doi:10.1016/j.envpol.2021.116556
Kumar, K., Trzybulska, D., Tsatsanis, C., Giwercman, A., and Almstrup, K. (2019). Identification of circulating small non-coding RNAs in relation to male subfertility and reproductive hormones. Mol. Cell Endocrinol. 492, 110443. doi:10.1016/j.mce.2019.05.002
Kuramochi-Miyagawa, S., Kimura, T., Ijiri, T. W., Isobe, T., Asada, N., Fujita, Y., et al. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131 (4), 839–849. doi:10.1242/dev.00973
Larriba, S., Vigues, F., and Bassas, L. (2023). Using small non-coding RNAs in extracellular vesicles of semen as biomarkers of Male reproductive System health: opportunities and challenges. Int. J. Mol. Sci. 24 (6), 5447. doi:10.3390/ijms24065447
Lau, N. C., Seto, A. G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D. P., et al. (2006). Characterization of the piRNA complex from rat testes. Science 313 (5785), 363–367. doi:10.1126/science.1130164
Legrand, J. M. D., and Hobbs, R. M. (2018). RNA processing in the male germline: mechanisms and implications for fertility. Semin. Cell Dev. Biol. 79, 80–91. doi:10.1016/j.semcdb.2017.10.006
Lehtiniemi, T., Bourgery, M., Ma, L., Ahmedani, A., Makela, M., Asteljoki, J., et al. (2022). SMG6 localizes to the chromatoid body and shapes the male germ cell transcriptome to drive spermatogenesis. Nucleic Acids Res. 50 (20), 11470–11491. doi:10.1093/nar/gkac900
Li, Y., Zhang, Y., and Liu, M. (2021). Knockout gene-based evidence for PIWI-Interacting RNA pathway in mammals. Front. Cell Dev. Biol. 9, 681188. doi:10.3389/fcell.2021.681188
Li, L., Tan, Y. Q., and Lu, L. Y. (2022). Defective piRNA processing and azoospermia. N. Engl. J. Med. 386 (17), 1675–1676. doi:10.1056/NEJMc2116008
Li, Z., Li, Z., Zhang, Y., Zhou, L., Xu, Q., Li, L., et al. (2024). Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing. Nat. Struct. Mol. Biol. 31 (8), 1222–1231. doi:10.1038/s41594-024-01287-6
Li, Z., Xu, Q., Zhong, J., Zhang, Y., Zhang, T., Ying, X., et al. (2025). Structural insights into RNA cleavage by PIWI Argonaute. Nature 639 (8053), 250–259. doi:10.1038/s41586-024-08438-1
Lim, A. K., and Kai, T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 104 (16), 6714–6719. doi:10.1073/pnas.0701920104
Liu, C., and Zhang, F. (2023). PIWI-specific insertion module: a newly identified regulatory element essential for longer piRNAs loading and male fertility. Sci. China Life Sci. 66 (7), 1708–1710. doi:10.1007/s11427-023-2402-x
Liu, J., Li, X., Zhou, G., Zhang, Y., Sang, Y., Wang, J., et al. (2021). Silica nanoparticles inhibiting the differentiation of round spermatid and chromatin remodeling of haploid period via MIWI in mice. Environ. Pollut. 284, 117446. doi:10.1016/j.envpol.2021.117446
Loubalova, Z., Fulka, H., Horvat, F., Pasulka, J., Malik, R., Hirose, M., et al. (2021). Formation of spermatogonia and fertile oocytes in golden hamsters requires piRNAs. Nat. Cell Biol. 23 (9), 992–1001. doi:10.1038/s41556-021-00746-2
Lu, Y., Nagamori, I., Kobayashi, H., Kojima-Kita, K., Shirane, K., Chang, H. Y., et al. (2023). ADAD2 functions in spermiogenesis and piRNA biogenesis in mice. Andrology 11 (4), 698–709. doi:10.1111/andr.13400
Lv, X., Xiao, W., Lai, Y., Zhang, Z., Zhang, H., Qiu, C., et al. (2023). The non-redundant functions of PIWI family proteins in gametogenesis in golden hamsters. Nat. Commun. 14 (1), 5267. doi:10.1038/s41467-023-40650-x
Mahadevan, I. A., Kumar, S., and Rao, M. R. S. (2020). Linker histone variant H1t is closely associated with repressed repeat-element chromatin domains in pachytene spermatocytes. Epigenetics Chromatin 13 (1), 9. doi:10.1186/s13072-020-00335-x
Manage, K. I., Rogers, A. K., Wallis, D. C., Uebel, C. J., Anderson, D. C., Nguyen, D. A. H., et al. (2020). A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans. Elife 9, e56731. doi:10.7554/eLife.56731
Manakov, S. A., Pezic, D., Marinov, G. K., Pastor, W. A., Sachidanandam, R., and Aravin, A. A. (2015). MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 12 (8), 1234–1243. doi:10.1016/j.celrep.2015.07.036
Martin-Villanueva, S., Gutierrez, G., Kressler, D., and de la Cruz, J. (2021). Ubiquitin and ubiquitin-like proteins and domains in ribosome production and function: chance or necessity? Int. J. Mol. Sci. 22 (9), 4359. doi:10.3390/ijms22094359
Masone, M. C. (2024). piRNA pathway disruption in human infertility. Nat. Rev. Urol. 21 (10), 577. doi:10.1038/s41585-024-00946-z
Miyata, H., Shimada, K., Kaneda, Y., and Ikawa, M. (2024). Development of functional spermatozoa in Mammalian spermiogenesis. Development 151 (14), dev202838. doi:10.1242/dev.202838
Miyoshi, T., Makino, T., and Moran, J. V. (2019). Poly(ADP-Ribose) polymerase 2 recruits replication protein A to sites of LINE-1 integration to facilitate retrotransposition. Mol. Cell 75 (6), 1286–1298. doi:10.1016/j.molcel.2019.07.018
Modepalli, V., Fridrich, A., Agron, M., and Moran, Y. (2018). The methyltransferase HEN1 is required in Nematostella vectensis for microRNA and piRNA stability as well as larval metamorphosis. PLoS Genet. 14 (8), e1007590. doi:10.1371/journal.pgen.1007590
Mohn, F., Sienski, G., Handler, D., and Brennecke, J. (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in drosophila. Cell 157 (6), 1364–1379. doi:10.1016/j.cell.2014.04.031
Mohn, F., Handler, D., and Brennecke, J. (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for zucchini-dependent, phased piRNA biogenesis. Science 348 (6236), 812–817. doi:10.1126/science.aaa1039
Montgomery, T. A., Rim, Y. S., Zhang, C., Dowen, R. H., Phillips, C. M., Fischer, S. E., et al. (2012). PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8 (4), e1002616. doi:10.1371/journal.pgen.1002616
Morgan, M., Kabayama, Y., Much, C., Ivanova, I., Di Giacomo, M., Auchynnikava, T., et al. (2019). A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and Mammalian spermatogenesis. Cell Res. 29 (3), 221–232. doi:10.1038/s41422-018-0128-1
Munzel, T., and Daiber, A. (2018). Environmental stressors and their impact on health and disease with focus on oxidative stress. Antioxid. Redox Signal 28 (9), 735–740. doi:10.1089/ars.2017.7488
Nagirnaja, L., Morup, N., Nielsen, J. E., Stakaitis, R., Golubickaite, I., Oud, M. S., et al. (2021). Variant PNLDC1, defective piRNA processing, and azoospermia. N. Engl. J. Med. 385 (8), 707–719. doi:10.1056/NEJMoa2028973
Newkirk, S. J., Lee, S., Grandi, F. C., Gaysinskaya, V., Rosser, J. M., Vanden Berg, N., et al. (2017). Intact piRNA pathway prevents L1 mobilization in Male meiosis. Proc. Natl. Acad. Sci. U. S. A. 114 (28), E5635-E5644–E5644. doi:10.1073/pnas.1701069114
Ninova, M., Chen, Y. A., Godneeva, B., Rogers, A. K., Luo, Y., Fejes Toth, K., et al. (2020). Su(var)2-10 and the SUMO pathway link piRNA-Guided target recognition to chromatin silencing. Mol. Cell 77 (3), 556–570. doi:10.1016/j.molcel.2019.11.012
Ninova, M., Holmes, H., Lomenick, B., Fejes Toth, K., and Aravin, A. A. (2023). Pervasive SUMOylation of heterochromatin and piRNA pathway proteins. Cell Genom 3 (7), 100329. doi:10.1016/j.xgen.2023.100329
Okutman, O., Muller, J., Baert, Y., Serdarogullari, M., Gultomruk, M., Piton, A., et al. (2015). Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum. Mol. Genet. 24 (19), 5581–5588. doi:10.1093/hmg/ddv290
Olotu, O., Dowling, M., Homolka, D., Wojtas, M. N., Tran, P., Lehtiniemi, T., et al. (2023). Intermitochondrial cement (IMC) harbors piRNA biogenesis machinery and exonuclease domain-containing proteins EXD1 and EXD2 in mouse spermatocytes. Andrology 11 (4), 710–723. doi:10.1111/andr.13361
Ortega, J., Wahba, L., Seemann, J., Chen, S. Y., Fire, A. Z., and Arur, S. (2024). Pachytene piRNAs control discrete meiotic events during spermatogenesis and restrict gene expression in space and time. Sci. Adv. 10 (40), eadp0466. doi:10.1126/sciadv.adp0466
Ozata, D. M., Yu, T., Mou, H., Gainetdinov, I., Colpan, C., Cecchini, K., et al. (2020). Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 4 (1), 156–168. doi:10.1038/s41559-019-1065-1
Pan, Y., Hu, M., Liang, H., Wang, J. J., and Tang, L. J. (2012). The expression of the PIWI family members miwi and Mili in mice testis is negatively affected by estrogen. Cell Tissue Res. 350 (1), 177–181. doi:10.1007/s00441-012-1447-z
Pastore, B., Hertz, H. L., Price, I. F., and Tang, W. (2021). pre-piRNA trimming and 2'-O-methylation protect piRNAs from 3' tailing and degradation in C. elegans. Cell Rep. 36 (9), 109640. doi:10.1016/j.celrep.2021.109640
Pastore, B., Hertz, H. L., and Tang, W. (2022). Comparative analysis of piRNA sequences, targets and functions in nematodes. RNA Biol. 19 (1), 1276–1292. doi:10.1080/15476286.2022.2149170
Post, C., Clark, J. P., Sytnikova, Y. A., Chirn, G. W., and Lau, N. C. (2014). The capacity of target silencing by drosophila PIWI and piRNAs. Rna 20 (12), 1977–1986. doi:10.1261/rna.046300.114
Priyadarshini, M., Ni, J. Z., Vargas-Velazquez, A. M., Gu, S. G., and Frokjaer-Jensen, C. (2022). Reprogramming the piRNA pathway for multiplexed and transgenerational gene silencing in C. elegans. Nat. Methods 19 (2), 187–194. doi:10.1038/s41592-021-01369-z
Qiao, J., Wang, Y., Li, X., Jiang, F., Zhang, Y., Ma, J., et al. (2021). A lancet commission on 70 years of women's reproductive, maternal, newborn, child, and adolescent health in China. Lancet 397 (10293), 2497–2536. doi:10.1016/S0140-6736(20)32708-2
Qin, D., Gu, Y., Zhang, Y., Wang, S., Jiang, T., Wang, Y., et al. (2023). Phase-separated CCER1 coordinates the histone-to-protamine transition and Male fertility. Nat. Commun. 14 (1), 8209. doi:10.1038/s41467-023-43480-z
Ramakrishna, N. B., Murison, K., Miska, E. A., and Leitch, H. G. (2021). Epigenetic regulation during primordial germ cell development and differentiation. Sex Dev. 15 (5-6), 411–431. doi:10.1159/000520412
Reddy, H. M., Bhattacharya, R., Tiwari, S., Mishra, K., Annapurna, P., Jehan, Z., et al. (2021). Y chromosomal noncoding RNAs regulate autosomal gene expression via piRNAs in mouse testis. BMC Biol. 19 (1), 198. doi:10.1186/s12915-021-01125-x
Ribas-Maynou, J., Garcia-Bonavila, E., Hidalgo, C. O., Catalan, J., Miro, J., and Yeste, M. (2021). Species-specific differences in sperm chromatin decondensation between Eutherian mammals underlie distinct lysis requirements. Front. Cell Dev. Biol. 9, 669182. doi:10.3389/fcell.2021.669182
Roberts, T. C., Ezzat, K., El Andaloussi, S., and Weinberg, M. S. (2016). Synthetic SiRNA delivery: progress and prospects. Methods Mol. Biol. 1364, 291–310. doi:10.1007/978-1-4939-3112-5_23
Saito, K., Nishida, K. M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., et al. (2006). Specific association of piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the drosophila genome. Genes Dev. 20 (16), 2214–2222. doi:10.1101/gad.1454806
Sasaki, K., and Sangrithi, M. (2023). Developmental origins of Mammalian spermatogonial stem cells: new perspectives on epigenetic regulation and sex chromosome function. Mol. Cell Endocrinol. 573, 111949. doi:10.1016/j.mce.2023.111949
Saxe, J. P., Chen, M., Zhao, H., and Lin, H. (2013). Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 32 (13), 1869–1885. doi:10.1038/emboj.2013.121
Schopp, T., Zoch, A., Berrens, R. V., Auchynnikava, T., Kabayama, Y., Vasiliauskaite, L., et al. (2020). TEX15 is an essential executor of MIWI2-directed transposon DNA methylation and silencing. Nat. Commun. 11 (1), 3739. doi:10.1038/s41467-020-17372-5
Schulke, L. C., Wistuba, J., Nordhoff, V., Behre, H. M., Cremers, J. F., Kliesch, S., et al. (2024). Identification of two hidden clinical subgroups among men with idiopathic cryptozoospermia. Hum. Reprod. 39 (5), 892–901. doi:10.1093/humrep/deae013
Shahrokhi, S. Z., Salehi, P., Alyasin, A., Taghiyar, S., and Deemeh, M. R. (2020). Asthenozoospermia: cellular and molecular contributing factors and treatment strategies. Andrologia 52 (2), e13463. doi:10.1111/and.13463
Shi, Z., Montgomery, T. A., Qi, Y., and Ruvkun, G. (2013). High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 23 (3), 497–508. doi:10.1101/gr.149112.112
Shoji, M., Tanaka, T., Hosokawa, M., Reuter, M., Stark, A., Kato, Y., et al. (2009). The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse Male germline. Dev. Cell 17 (6), 775–787. doi:10.1016/j.devcel.2009.10.012
Sohn, E. J., Han, M. E., Park, Y. M., Kim, Y. H., and Oh, S. O. (2023). The potential of piR-823 as a diagnostic biomarker in oncology: a systematic review. PLoS One 18 (12), e0294685. doi:10.1371/journal.pone.0294685
Stallmeyer, B., Buhlmann, C., Stakaitis, R., Dicke, A. K., Ghieh, F., Meier, L., et al. (2024). Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human Male infertility. Nat. Commun. 15 (1), 6637. doi:10.1038/s41467-024-50930-9
Subash, S. K., and Kumar, P. G. (2021). Spermatogonial stem cells: a story of self-renewal and differentiation. Front. Biosci. Landmark Ed. 26 (1), 163–205. doi:10.2741/4891
Tan, G. S., Garchow, B. G., Liu, X., Yeung, J., Morris, J. P. t., Cuellar, T. L., et al. (2009). Expanded RNA-Binding activities of Mammalian argonaute 2. Nucleic Acids Res. 37 (22), 7533–7545. doi:10.1093/nar/gkp812
Tan, M., Tol, H., Rosenkranz, D., Roovers, E. F., Damen, M. J., Stout, T. A. E., et al. (2020). PIWIL3 forms a complex with TDRKH in mammalian oocytes. Cells 9 (6), 1356. doi:10.3390/cells9061356
Tan, K., Kim, M. E., Song, H. W., Skarbrevik, D., Babajanian, E., Bedrosian, T. A., et al. (2021). The rhox gene cluster suppresses germline LINE1 transposition. Proc. Natl. Acad. Sci. U. S. A. 118 (23), e2024785118. doi:10.1073/pnas.2024785118
Tanyapanyachon, P., Dana, P., Thumsongsiri, N., Chonniyom, W., and Saengkrit, N. (2023). Interrupting the blood-testis barrier with a flutamide-loaded nanostructured lipid carrier: a novel nonsurgical contraceptive approach for Male animals. Theriogenology 206, 96–105. doi:10.1016/j.theriogenology.2023.04.023
Tirmarche, S., Kimura, S., Dubruille, R., Horard, B., and Loppin, B. (2016). Unlocking sperm chromatin at fertilization requires a dedicated egg thioredoxin in drosophila. Nat. Commun. 7, 13539. doi:10.1038/ncomms13539
Trost, N., Mbengue, N., and Kaessmann, H. (2023). The molecular evolution of Mammalian spermatogenesis. Cells Dev. 175, 203865. doi:10.1016/j.cdev.2023.203865
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 (5785), 320–324. doi:10.1126/science.1129333
Vahidi, S., Horoki, A. Z., Talkhooncheh, M. H., Jambarsang, S., Marvast, L. D., Sadeghi, A., et al. (2021). Success rate and ART outcome of microsurgical sperm extraction in non obstructive azoospermia: a retrospective study. Int. J. Reprod. Biomed. 19 (9), 781–788. doi:10.18502/ijrm.v19i9.9710
Vandewege, M. W., Patt, R. N., Merriman, D. K., Ray, D. A., and Hoffmann, F. G. (2022). The PIWI/piRNA response is relaxed in a rodent that lacks mobilizing transposable elements. Rna 28 (4), 609–621. doi:10.1261/rna.078862.121
Vourekas, A., Zheng, K., Fu, Q., Maragkakis, M., Alexiou, P., Ma, J., et al. (2015). The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 29 (6), 617–629. doi:10.1101/gad.254631.114
Vrettos, N., Oppelt, J., Zoch, A., Sgourdou, P., Yoshida, H., Song, B., et al. (2024). MIWI N-terminal arginines orchestrate generation of functional pachytene piRNAs and spermiogenesis. Nucleic Acids Res. 52 (11), 6558–6570. doi:10.1093/nar/gkae193
Vychytilova-Faltejskova, P., Stitkovcova, K., Radova, L., Sachlova, M., Kosarova, Z., Slaba, K., et al. (2018). Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of Colon cancer. Cancer Epidemiol. Biomarkers Prev. 27 (9), 1019–1028. doi:10.1158/1055-9965.EPI-18-0318
Wang, Y., Zhu, T., Li, Q., Liu, C., Han, F., Chen, M., et al. (2015). Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 5, 11031. doi:10.1038/srep11031
Wang, X., Lv, C., Guo, Y., and Yuan, S. (2020a). Mitochondria associated germinal structures in spermatogenesis: piRNA pathway regulation and beyond. Cells 9 (2), 399. doi:10.3390/cells9020399
Wang, Z., Yang, H., Ma, D., Mu, Y., Tan, X., Hao, Q., et al. (2020b). Serum PIWI-interacting RNAs piR-020619 and piR-020450 are promising novel biomarkers for early detection of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 29 (5), 990–998. doi:10.1158/1055-9965.EPI-19-1148
Wang, Y., Yuan, X., Ali, M. A., Qin, Z., Zhang, Y., and Zeng, C. (2021). piR-121380 is involved in cryo-capacitation and regulates post-thawed boar sperm quality through phosphorylation of ERK2 via targeting PTPN7. Front. Cell Dev. Biol. 9, 792994. doi:10.3389/fcell.2021.792994
Wang, X., Gou, L. T., and Liu, M. F. (2022a). Noncanonical functions of PIWIL1/piRNAs in animal Male germ cells and human diseases†. Biol. Reprod. 107 (1), 101–108. doi:10.1093/biolre/ioac073
Wang, X., Tan, Y. Q., and Liu, M. F. (2022b). Defective piRNA processing and azoospermia. N. Engl. J. Med. 386 (17), 1674–1675.
Wang, D., Liu, B., and Zhang, Z. (2023a). Accelerating the understanding of cancer biology through the lens of genomics. Cell 186 (8), 1755–1771. doi:10.1016/j.cell.2023.02.015
Wang, X., Lin, D. H., Yan, Y., Wang, A. H., Liao, J., Meng, Q., et al. (2023b). The PIWI-Specific insertion module helps load longer piRNAs for translational activation essential for Male fertility. Sci. China Life Sci. 66 (7), 1459–1481. doi:10.1007/s11427-023-2390-5
Wang, X., Ramat, A., Simonelig, M., and Liu, M. F. (2023c). Emerging roles and functional mechanisms of PIWI-Interacting RNAs. Nat. Rev. Mol. Cell Biol. 24 (2), 123–141. doi:10.1038/s41580-022-00528-0
Wasik, K. A., Tam, O. H., Knott, S. R., Falciatori, I., Hammell, M., Vagin, V. V., et al. (2015). RNF17 blocks promiscuous activity of PIWI proteins in mouse testes. Genes Dev. 29 (13), 1403–1415. doi:10.1101/gad.265215.115
Webster, K. E., O'Bryan, M. K., Fletcher, S., Crewther, P. E., Aapola, U., Craig, J., et al. (2005). Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 102 (11), 4068–4073. doi:10.1073/pnas.0500702102
Wei, C., Yan, X., Mann, J. M., Geng, R., Wang, Q., Xie, H., et al. (2024a). PNLDC1 catalysis and postnatal germline function are required for piRNA trimming, LINE1 silencing, and spermatogenesis in mice. PLoS Genet. 20 (9), e1011429. doi:10.1371/journal.pgen.1011429
Wei, H., Gao, J., Lin, D. H., Geng, R., Liao, J., Huang, T. Y., et al. (2024b). piRNA loading triggers MIWI translocation from the intermitochondrial cement to chromatoid body during mouse spermatogenesis. Nat. Commun. 15 (1), 2343. doi:10.1038/s41467-024-46664-3
Wenda, J. M., Homolka, D., Yang, Z., Spinelli, P., Sachidanandam, R., Pandey, R. R., et al. (2017). Distinct roles of RNA helicases MVH and TDRD9 in PIWI slicing-triggered mammalian piRNA biogenesis and function. Dev. Cell 41 (6), 623–637. doi:10.1016/j.devcel.2017.05.021
Wu, P. H., Fu, Y., Cecchini, K., Ozata, D. M., Arif, A., Yu, T., et al. (2020). The evolutionarily conserved piRNA-producing locus pi6 is required for Male mouse fertility. Nat. Genet. 52 (7), 728–739. doi:10.1038/s41588-020-0657-7
Wyrwoll, M. J., Gaasbeek, C. M., Golubickaite, I., Stakaitis, R., Oud, M. S., Nagirnaja, L., et al. (2022). The piRNA-pathway factor FKBP6 is essential for spermatogenesis but dispensable for control of meiotic LINE-1 expression in humans. Am. J. Hum. Genet. 109 (10), 1850–1866. doi:10.1016/j.ajhg.2022.09.002
Xie, Y., Wei, B. H., Ni, F. D., and Yang, W. X. (2021). Conversion from spermatogonia to spermatocytes: extracellular cues and downstream transcription network. Gene 764, 145080. doi:10.1016/j.gene.2020.145080
Xiol, J., Cora, E., Koglgruber, R., Chuma, S., Subramanian, S., Hosokawa, M., et al. (2012). A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 47 (6), 970–979. doi:10.1016/j.molcel.2012.07.019
Xiong, M., Yin, L., Gui, Y., Lv, C., Ma, X., Guo, S., et al. (2023). ADAD2 interacts with RNF17 in P-bodies to repress the Ping-pong cycle in pachytene piRNA biogenesis. J. Cell Biol. 222 (5), e202206067. doi:10.1083/jcb.202206067
Yabuta, Y., Ohta, H., Abe, T., Kurimoto, K., Chuma, S., and Saitou, M. (2011). TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 192 (5), 781–795. doi:10.1083/jcb.201009043
Yang, F., and Wang, P. J. (2016). Multiple LINEs of retrotransposon silencing mechanisms in the Mammalian germline. Semin. Cell Dev. Biol. 59, 118–125. doi:10.1016/j.semcdb.2016.03.001
Yang, Z., Chen, K. M., Pandey, R. R., Homolka, D., Reuter, M., Janeiro, B. K., et al. (2016). PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol. Cell 61 (1), 138–152. doi:10.1016/j.molcel.2015.11.009
Yang, F., Lan, Y., Pandey, R. R., Homolka, D., Berger, S. L., Pillai, R. S., et al. (2020). TEX15 associates with MILI and silences transposable elements in Male germ cells. Genes Dev. 34 (11-12), 745–750. doi:10.1101/gad.335489.119
Yang, X., Chen, D., Zheng, S., Yi, M., Liu, Z., Liu, Y., et al. (2022). BmHen1 is essential for eupyrene sperm development in Bombyx mori but PIWI proteins are not. Insect Biochem. Mol. Biol. 151, 103874. doi:10.1016/j.ibmb.2022.103874
Yoshimura, T., Watanabe, T., Kuramochi-Miyagawa, S., Takemoto, N., Shiromoto, Y., Kudo, A., et al. (2018). Mouse GTSF1 is an essential factor for secondary piRNA biogenesis. EMBO Rep. 19 (4), e42054. doi:10.15252/embr.201642054
Yuan, J., Xie, B. M., Ji, Y. M., Bao, H. J., Wang, J. L., Cheng, J. C., et al. (2025). piR-26441 inhibits mitochondrial oxidative phosphorylation and tumorigenesis in ovarian cancer through m6A modification by interacting with YTHDC1. Cell Death Dis. 16 (1), 25. doi:10.1038/s41419-025-07340-6
Zhang, Z., Wang, J., Schultz, N., Zhang, F., Parhad, S. S., Tu, S., et al. (2014). The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157 (6), 1353–1363. doi:10.1016/j.cell.2014.04.030
Zhang, L., Meng, X., Xiang, Z., Li, D., and Han, X. (2018). From the cover: roles of mmu_piR_003399 in microcystin-leucine arginine-induced reproductive toxicity in the spermatogonial cells and testis. Toxicol. Sci. 161 (1), 159–170. doi:10.1093/toxsci/kfx209
Zhang, G., Yu, T., Parhad, S. S., Ho, S., Weng, Z., and Theurkauf, W. E. (2021). piRNA-independent transposon silencing by the drosophila THO complex. Dev. Cell 56 (18), 2623–2635.e5. doi:10.1016/j.devcel.2021.08.021
Zhao, S., Gou, L. T., Zhang, M., Zu, L. D., Hua, M. M., Hua, Y., et al. (2013). piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev. Cell 24 (1), 13–25. doi:10.1016/j.devcel.2012.12.006
Zhu, X., Zhi, E., and Li, Z. (2015). MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis. Reproduction 149 (5), R229–R235. doi:10.1530/REP-14-0569
Zoch, A., Auchynnikava, T., Berrens, R. V., Kabayama, Y., Schopp, T., Heep, M., et al. (2020). SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 584 (7822), 635–639. doi:10.1038/s41586-020-2557-5
Keywords: PIWI proteins, spermatogenesis, male infertility, transposable elements, epigenetic regulation, piRNA biomarkers
Citation: Hong Z, Huang S, Li L, Gao Y, Ma B, Fan Q, Zhang Y and Wang M (2025) Cracking the code: how piRNA pathway shapes spermatogenesis and combats male infertility. Front. Cell Dev. Biol. 13:1657744. doi: 10.3389/fcell.2025.1657744
Received: 01 July 2025; Accepted: 04 September 2025;
Published: 22 September 2025; Corrected: 06 October 2025.
Edited by:
Anthony Valverde, Costa Rica Institute of Technology, Costa RicaReviewed by:
Hui Li, Guangxi University, ChinaSoumya Ranjan Jena, Ravenshaw University, India
Patricia Rojas Ríos, University of Seville, Spain
Onur Eroglu, Bilecik Şeyh Edebali University, Türkiye
Copyright © 2025 Hong, Huang, Li, Gao, Ma, Fan, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Wang, d2FuZ21laTE5OTBAd2h1LmVkdS5jbg==; Yuanzhen Zhang, emhhbmd5dWFuemhlbkB3aHUuZWR1LmNu
†These authors have contributed equally to this work
 Zhidan Hong
Zhidan Hong Sihan Huang
Sihan Huang Li Li1†
Li Li1† Yuanzhen Zhang
Yuanzhen Zhang Mei Wang
Mei Wang