Abstract
Persistent infection with high-risk human papillomaviruses (HR-HPVs) is a major etiological factor in the development of cervical cancer, which ranks as the fourth leading cause of cancer-related mortality among women worldwide. HR HPV types 16 and 18 cause more than 70% of all cases. These viruses encode the proteins E1, E2, E1^E4, E4, E5, E6, E7, L1, and L2, through a complex array of polycistronic mRNAs. For decades, research on HPV gene expression has focused predominantly on transcriptional activity and mRNA splicing. In contrast, the mechanisms underlying the translation of mRNAs remain poorly understood. Whereas the translational regulation of E1, E2, E6, and E7 has been elucidated, the translation mechanisms for E5, L1, and, L2 proteins are still unclear. We hypothesized that their translation may occur via internal ribosome entry sites (IRESs). Other critical questions also remain open, including how the viral-cellular chimeric transcripts generated upon virus genome integration into the host DNA are translated, as well as how the translation of polycistronic viral mRNAs is regulated during the differentiation of epithelial cells—a process that is central to HPV-induced carcinogenesis. This review summarizes current knowledge showing that the translation of HPVs mRNAs is subjected to tight regulation, highlights unresolved questions, and discusses potential therapeutic implications of targeting the translational machinery in HPV-related cancers.
Introduction
Cervical cancer ranks fourth in incidence and third in mortality among neoplasias affecting women worldwide. Its burden might reach highest mortality rates in more than 40 low-income countries (National Cancer Institute, 2021; LaVigne et al., 2017; Sung et al., 2021; WHO, 2022). HPV prevalence is markedly higher in emerging regions, reaching 42.2% compared to 22.6% in high-income regions (Pavelescu et al., 2025). Persistent infection of the genital tract with high-risk (HR) human papillomavirus (HPV) is the primary cause of most cervical cancer cases. Among these, HR-HPV types 16 and 18 are responsible for over 70% of cases. Moreover, about 90% of anal cancer, 75% of vaginal cancer, 70% of oropharyngeal and vulvar cancers, and 63% of penile cancers are caused by HR-HPVs across the globe (Alteri et al., 2025). Effective vaccines against HR-HPV have been developed, and HPV vaccination was introduced in 73 countries during 2010–2019 and in 30 additional countries during 2020–2023. Global first-dose HPV coverage among girls increased from 17% in 2019 to 27% in 2023, and full-course coverage rose from 13% in 2019 to 20% in 2023 (Jones et al., 2024). According to World Health Organization, global first-dose coverage further increased to 31% in 2024. However, this is still far below the 2030 target of 90% coverage because their use remains limited or non-existent in many countries (Collaborators, 2025), highlighting cervical cancer as a major public health challenge worldwide.
HR-HPVs infect basal keratinocytes of the mucosal epithelium, leading to the formation of low-grade intraepithelial lesions. Once within the host cells, the viral genome replicates as an extrachromosomal DNA inside the cell nucleus. As epithelial cells divide and differentiate, HPV gene expression couples with cell differentiation, and cells progress to productive viral states, leading to cell proliferation, immortalization, and the development of malignant phenotypes (Nelson and Mirabello, 2023; Obanya et al., 2025; Pavelescu et al., 2025; Rosendo-Chalma et al., 2024). This process has been extensively studied with regard to the transcription and splicing of viral mRNAs (Doomer et al., 2024; Olmedo-Nieva et al., 2018; Wang et al., 2025; Yu et al., 2022) (https://pave.niaid.nih.gov/#explore/transcript_maps). However, the translation of viral mRNAs remains the least studied aspect of viral gene expression (Kajitani and Schwartz, 2022).
Notably, translational control is emerging as a key process of viral proteins synthesis, opening novel avenues to target the translational machinery to combat HVP-induced cervical cancer. We review the old, yet unresolved questions regarding the translation of viral mRNAs, alongside recent advances in our understanding of viral protein synthesis regulation and their implications for the design of new therapeutic strategies.
Translation in eukaryotes
Translation, i.e., the decoding of genetic information of a mRNA into peptide by the ribosome and translation factors using a genetic code, largely determines in all cells protein abundance and the proteome composition in normal physiology, stresses and diseases (Hernández and Tettweiler, 2012). Accordingly, a myriad of mechanisms to regulate mRNA translation exists in cells and virus (Hernández et al., 2020; Hershey et al., 2019; Jan et al., 2016; Kwan and Thompson, 2019). Translation consists of four main stages, namely initiation, elongation, and termination. In some physiological situations a fourth stage may happen for some mRNAs, i.e., ribosome recycling. However, the global process of translation is largely controlled at the initiation step (Hershey et al., 2019). Different mechanism account for the initiation step, namely the canonical mechanism and the non-canonical leaky scanning, ribosome shunting, termination-reinitiation, and IRES-dependent mechanisms.
The canonical mechanism to initiate translation
Most eukaryotic mRNAs initiate translation in the canonic manner, termed cap-dependent mechanism. It begins with the mRNA recruitment via recognition of the cap structure (7-methyl guanosine) at the 5′ of the mRNA by eukaryotic initiation factor (eIF) 4E. During this process, eIF4G binds eIF4E, eIF4A and poly(A)-binding protein (PABP). Subsequently, a free 40S ribosomal subunit in complex with eIF1, eIF1A, eIF3, eIF5 and a ternary complex (TC, consisting of eIF2 bound to an initiator Met-tRNAiMet and GTP) forms a 43S pre-initiation complex (PIC). eIF4G further interacts with the ribosome-bound eIF3 to promote recruitment of the 43S PIC to the mRNA 5′-untranslated region (5′-UTR) (Brito Querido et al., 2024; Pelletier and Sonenberg, 2019). After the mRNA is recruited, the 43S PIC scans the 5′-UTR to reach the translation initiation site (TIS), most frequently an AUG codon (Figure 1A) (Brito Querido et al., 2024; Kwan and Thompson, 2019; Pelletier and Sonenberg, 2019). A context sequence surrounding the TIS, termed the “Kozak motif”, is required for optimal AUG codon recognition by the ribosomal complex. For human mRNAs, this motif consists of the sequence GCCA/GCCAUGG (Hernández et al., 2019). AUG recognition triggers 60S ribosomal subunit joining to assemble an 80S initiation complex which enters in successive rounds of peptide elongation (Brito Querido et al., 2024; Kwan and Thompson, 2019; Pelletier and Sonenberg, 2019). When a stop codon is recognized in the A site of the ribosome eukaryotic releasing factor (eRF) 1 promotes peptide liberation.
FIGURE 1
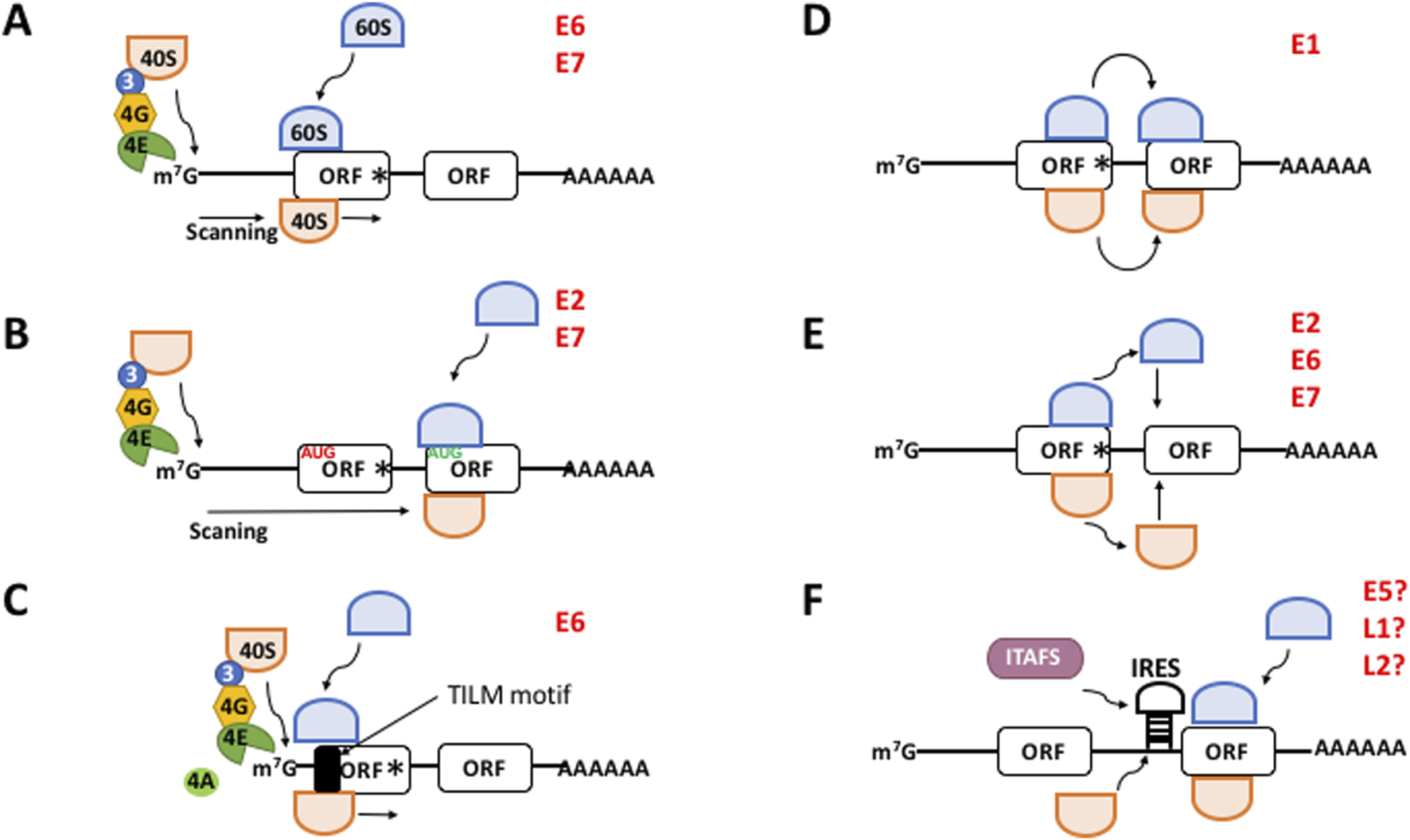
The different mechanisms to initiate translation in eukaryotes. (A) Canonical, cap-, eIF4E-, and scanning dependent mechanism. (B–F) Non-canonical mechanisms. (B) Leaky scanning. mRNAs containing a weak Kozak AUG contexts (in red) allow the ribosomal 40S subunit to skip the first ORF and recognize the downstream AUG (in green) of the next ORF which is in good context. (C) Translation Initiation of Leaderless mRNAs (TILM)-dependent mechanism. Extremely short 5′-UTR and a TILM motif within the E6 ORF promote translation initiation. (D) Ribosome shunting. The 40S ribosome shunts over a transcript segment landing downstream to reach the AUG codon of the next ORF. (E) Termination-reinitiation. After translation has terminated ribosomes quit the mRNA. Then a new ternary complex is loaded onto the 40S ribosome and it resumes scanning to start translation of the next ORF. (F) IRES-dependent mechanism. The 40S ribosome directly binds an IRES next to the AUG start codon to initiate translation. This process may need the help of IRES-trans acting factors (ITAFS). m7G, methyl-7 GTP cap structure of mRNA; 40S, ribosomal subunit 40S; 60S, ribosomal subunit 60S; 3, eIF3; 4G, eIF4G; 4E, eIF4E; Black box, translation initiation of leaderless mRNAs (TILM) motif; ORF, open reading frame; AAAA, poly(A) tail; IRES, internal ribosome entry site. The viral proteins with a proposed mechanism are indicated in red. Hypothetical IRES-driven translation for E5, L1, and L2 is indicated with a?
Non-canonical translation initiation mechanisms
In addition to the canonical pathway, translation initiation may happen via other mechanisms, which mostly account for the translation of internal ORF within polycistronic mRNAs in many viruses.
In the “leaky scanning” (Hinnebusch et al., 2004), mRNAs containing an unfavorable Kozak (i.e., “weak”) motif allow the ribosomal 43S PIC skip the first open reading frame (ORF) and recognize a downstream ORF AUG (Figure 1B). Recently, we demonstrated that E6,E7 mRNAs from HPV-18 possesses extremely short 5′-UTRs (0–5 nucleotides) in cervical tumors which drive the translation of E6 (García et al., 2021), conforming a novel non-canonical mechanism of translation initiation (Figure 1C). In this virus, E6 is translated in a scanning-independent manner but is dependent on the mRNA cap and the poly(A) tail, eIF4E, eIF4A, and on a novel RNA motif within the coding region of the E6 cistron that we termed TILM (for Translation Initiation of Leaderless mRNAs). Thus, in the absence of a 5′-UTR, TILM controls the translation of E6 (García et al., 2021). In the “ribosome shunting” (Kwan and Thompson, 2019), specific cis-elements within the mRNA promote that the 43S PIC bypasses or shunts over a segment of the transcript, lands downstream and resumes scanning to reach the TIS (Figure 1D). In the “termination-reinitiation” (Jackson et al., 2012), after translation has terminated, a new TC is loaded onto a 40S subunit and it resumes scanning to start the translation of a downstream ORF on the same mRNA (Figure 1E). For some other transcripts, translation initiation is driven by an internal ribosome entry site (IRES) (Kwan and Thompson, 2019), a cis-located mRNA region that promotes ribosomal recruitment just nearby the TIS in a cap- and eIF4E-independent manner and without scanning. This process may be promoted by the activity of cellular IRES-trans-acting factors (ITAFs) (Figure 1F).
Function of the viral proteins during infection and carcinogenesis
HPVs encode the early proteins E1, E2, E4, E5, E6, and E7 from the early (E) control region of the viral genome, and the capsid proteins L1 and L2 from the long control region (LCR) (Doorbar et al., 2015; Nelson and Mirabello, 2023; Rosendo-Chalma et al., 2024). E1 is a helicase that, in cooperation with E2, drives viral genome replication. E2 also regulates the viral cycle life and controls the transcription of the E6 and E7 oncogenes. E4 plays a role in virion assembly and facilitates the release of newly formed viral particles. Collectively, E1, E2, and E4 drive integration, replication, and transcription of the viral genome (Bhattacharrjee et al., 2021; Della Fera et al., 2021; Evande et al., 2023). E5 is involved in membrane signaling, cell proliferation, apoptosis, oncogenesis, and angiogenesis (Basukala and Banks, 2021; Estêvão et al., 2019). E6 and E7 are potent oncogenic proteins that interact with a wide array of cellular proteins. Their activity is essential for the development and maintenance of the malignant phenotype. Impairment of these proteins, or the inhibition of their gene expression, induces cellular senescence and growth arrest in HPV-positive cancer cells (Basukala and Banks, 2021; Estêvão et al., 2019). The principal function of E6 is its interaction with the tumor suppressor p53 (Martinez-Zapien et al., 2016). This interaction inhibits apoptosis, thereby allowing cells harboring genomic damage to evade cell death and continue proliferating. In addition, E6 engages with other host proteins, contributing to carcinogenic phenotypes, cellular immortalization, and cell invasion (Panczyszyn et al., 2018). E7 promotes degradation of the tumor suppressor retinoblastoma (pRB), leading abnormal centrosome synthesis and aneuploidy, ultimately resulting in genomic instability (Duensing et al., 2001). In addition, E7 interacts with other cellular targets to promote deregulation of cell proliferation and increased invasive potential. E6 and E7, acting independently or in concert, modulate a wide range of cellular pathways critical for cancer progression. These include immune evasion, dysregulation of cellular energetics, inhibition of growth suppressor pathways, sustained proliferative signaling, epigenetic reprogramming, metastasis, tumor-associated inflammation, and angiogenesis (Basukala and Banks, 2021; Estêvão et al., 2019).
L1 and L2 are structural proteins that constitute the capsomers of the viral capsid. Indeed, current vaccines against HPV are designed to elicit immune responses primarily targeting the strong antigenicity of L1 (Tsakogiiannis et al., 2022). In addition, the L1 gene sequence is used for HPV genotyping, while L2 plays a role in the intracellular transport of viral DNA to the nucleus of infected cells (Doorbar et al., 2015; Graham, 2017; Rosendo-Chalma et al., 2024). In the following sections, we review the mechanisms by which viral proteins are synthesized from a myriad of polycistronic mRNAs.
HR-HPVs proteins are synthesized using different mechanisms to initiate translation
HR-HPV proteins are expressed by polycistronic mRNAs transcribed from the different promoters in the long control region (LCR), the early (E) region and the late (L) region of the HPV-16 and HPV-18 genomes, respectively (Figure 2A) (Doorbar et al., 2015; Graham, 2017; Rosendo-Chalma et al., 2024). Overall, the high complexity of alternative splicing events of viral transcripts produces a plethora of mRNAs with diverse structures. In all mRNAs, the first ORF encodes E6. Full-length E6 is only translated from unspliced transcripts (De la Cruz-Hernández et al., 2005; Mesplede et al., 2012; Moral-Hernández et al., 2010; Schneider-Gädicke and Schwarz, 1986; Smotkin and Wettstein, 1986; Stacey et al., 1995; Toots et al., 2014). In addition, shorter versions of E6 termed E6*I, E6*II, E6*III, E6*IV, E6*V, E6*VI, E6^E7, E6^E7*I, and E6^E7*II are also expressed from alternative splicing events of the same mRNA (Ajiro and Zheng, 2015; Brant et al., 2018; Olmedo-Nieva et al., 2018). E7 is synthesized from several spliced transcripts and from unspliced mRNAs (Doorbar et al., 1990; Durst et al., 1991; Mesplede et al., 2012; Moral-Hernández et al., 2010; Sherman et al., 1992; Smotkin et al., 1989; Tang et al., 2006; Thierry et al., 1987). Moreover, multiple alternatively spliced transcripts and clusters of heterogeneous mRNAs expressed from different transcription start sites have also been identified in cultured cells, raff cultures, and tumorigenic tissues that only contain either E6 or E7 as the sole cistron (Braunstein et al., 1999; Glahder et al., 2003; Grassmann et al., 1996; Rosenstierne et al., 2003; Schneider-Gädicke and Schwarz, 1986; Smotkin and Wettstein, 1986). Figure 2B shows most of the multiple, polycistronic mRNAs transcribed from both the episomal and the integrated HR-HPVs genome (Graham, 2010; Graham, 2017; Wang et al., 2025).
FIGURE 2
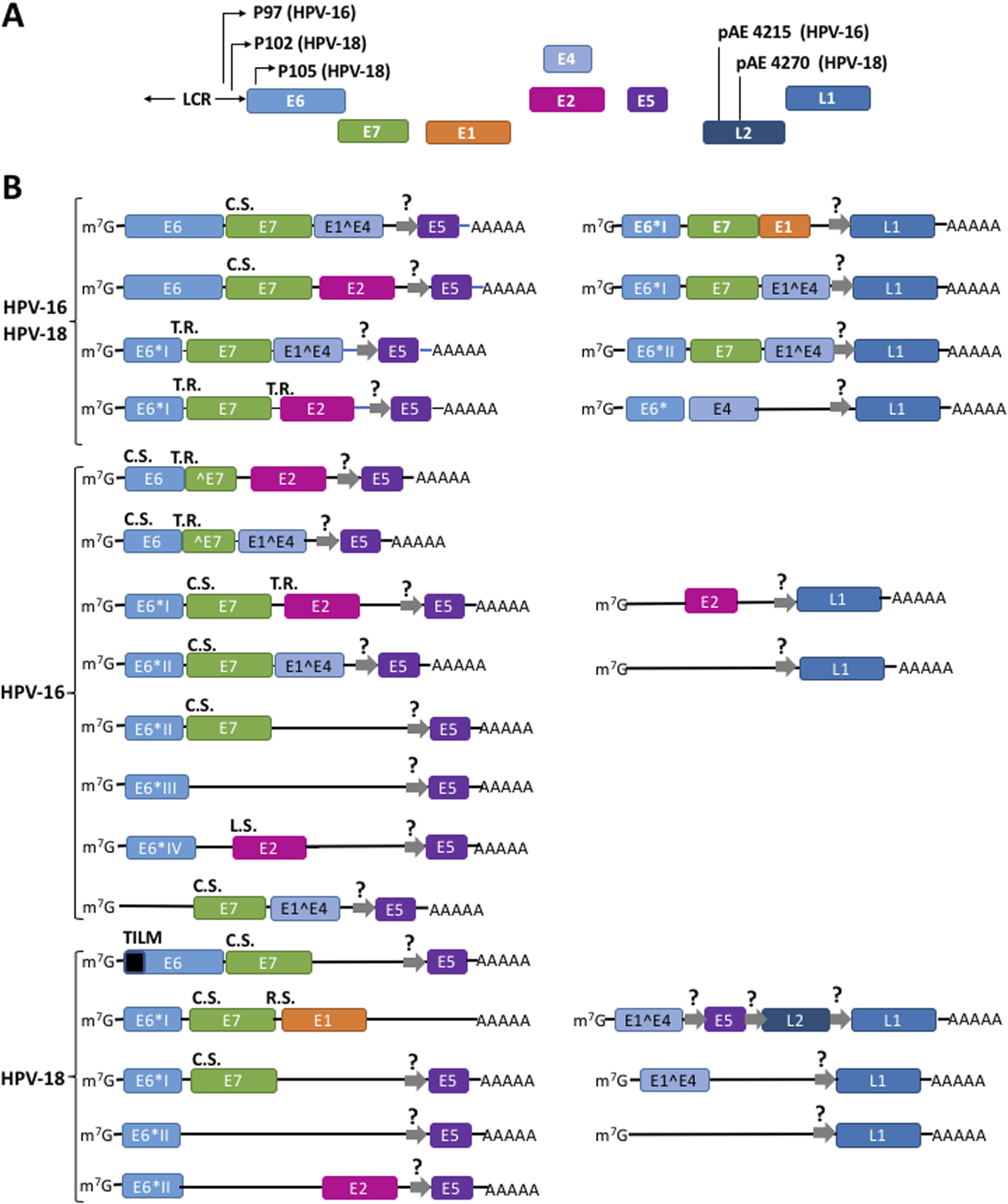
Different mechanisms drive the translation of a plethora of polycistronic mRNAs to synthesize the viral HPV proteins. (A) Structure of HR-HPV 16 and 18 genomic DNA. (B) Some mRNAs originated from episomal genome and from viral DNA integrated into the cellular genome. Thin arrows represent the promoters P97 for HPV-16 and P102 and P105 for HPV-18; pAE, early polyadenylation signals; LCR, long control region; Squares represent exons; lines into mRNAs represent intercistronic regions. R.S., ribosome shunting; T.R., termination-reinitiation; L.S., leaky scanning; C.S., canonical scanning. E5, L1, and L2 cistrons might be translated from hypothetical IRES (thick gray arrows with a question mark). Adapted from Yu et al. (2022), Wang et al. (2025), García et al. (2024), Wang et al. (2025), Yu et al. (2022). A fuller list of the HPVs transcripts is described in the Papilloma Virus Episteme (PaVE) (Doomer et al., 2024; Van Doorslaer et al., 2017) at https://pave.niaid.nih.gov/#explore/transcript_maps and references therein.
Surprisingly, In cultured cells and epithelial rafts the 5′-UTR regions of E6,E7 mRNAs are 7-9 nucleotides in length for HPV-16 (Stacey et al., 2000), and in HPV-18 the most represented transcripts possess either 3 nucleotides long 5′-UTR or completely lack it, i.e., the transcription starts at the A of the AUG codon that initiates translation (García et al., 2021; Romanczuk et al., 1990; Schneider-Gädicke and Schwarz, 1986; Smotkin and Wettstein, 1986; Thierry et al., 1987; Wang et al., 2011). Thus, the structure of viral mRNAs contrasts drastically with that of mRNAs across eukaryotes (both cellular and viral), with a 5′-UTR length average of 53–164 nucleotides (Leppek et al., 2018; Ryczek et al., 2023). In multicellular species, short 5′-UTRs have only been found in the human nuclear-encoded mRNAs harboring the element Translation Initiation of Short 5′-UTR (TISU), consisting of 12–30 nucleotide-long 5′-UTRs (Elfakess and Dikstein, 2008; Elfakess et al., 2011), and some mitochondrial mRNAs with 5′-UTRs shorter than five nucleotides (Gandin et al., 2016). Recently, we demonstrated that in cervical tumors E6,E7 mRNAs from HR HPV-18, HPV-39 and HPV-45 possess 5′-UTRs of 0–5 nucleotides which drive the translation of full-length E6 (García et al., 2021). For HPV-18, E6 is translated in a scanning-independent manner and is dependent on the RNA motif within the E6 coding region termed TILM, that drives the translation of E6 (García et al., 2021). However, the proteins that regulate E6 synthesis via the TILM motif in vivo are unknown. In contrast, the HPV-16 E6 cistron with a 7-9 nucleotides-long 5′-UTR is translated by the canonical mechanism (Stacey et al., 2000).
The synthesis of polycistronic mRNAs raises the fundamental question of how E1, E2, E4, E5, and E7 are translated downstream of E6. Evidence from cervical cancer cell lines suggest that the HPV-16 and HPV-18 E7 cistron is translated from spliced E6*I mRNA via translation termination-reinitiation (Tang et al., 2006). In a cell-free coupled in vitro transcription/translation system using rabbit reticulocyte lysates and a synthetic HPV-16 E6,E7 bicistronic mRNA, Tan et al. showed that full-length E6 translation also promotes termination-reinitiation on the E7 cistron AUG 3 to 6 nucleotides downstream the E6 stop codon (Tan et al., 1994).
The translation HPV-18 E1 has been studied by plasmid transient transfections of the green monkey kidney COS-7 fibroblast-like cultured cells (Remm et al., 1999). The distance between the E7 and E1 ORF consists of 6 nucleotides, and Remm et al. (1999) demonstrated that the translation of the E1 cistron downstream of E7 occurs by ribosome shunting. This group demonstrated that the translation of the E1 ORF is inhibited from monocistronic transcript that might arise from spurious transcription sites or partial degradation, and its translation is strongly favored from polycistronic rather than monocistronic mRNAs as a means to ensure E1 premature synthesis in early stages of the viral infection cycle.
The translation of HPV-16 E2 ORF has been studied in both COS cells and cell-free in vitro translation systems. In the transcripts E6*I,E7,E2,E5, the E7 stop codon and the E2 AUG are separated by 33 nucleotides and the termination-reinitiation mechanism could translate E2 after E7 is translated. On the other hand, in the E6*IV transcript, the E2 AUG is out of frame and upstream of the E6*IV termination codon. Moreover, E2 AUG has a stronger Kozak motif than E6*VI AUG. Thus, avoiding the upstream E6*IV AUG leaky scanning could also be the mechanism to initiate E2 translation (Alloul and Sherman, 1999).
HPV-16 E5 translation has been studied only in the mouse fibroblast NIH3T3 cells (Johnsen et al., 1995). HPV-16 and HPV-18 E1^E4, E5 mRNA is the most abundant transcript found in all cervical neoplasias (Sherman and Alloul, 1992; Stoler et al., 1992). However, HPV-16 E5 is not translated from this mRNA because it is rich in AUG triplets upstream the initiator AUG codon. In contrast, E2 and E5 are well expressed from the E2,E5 transcripts lacking the upstream E^E4 cistron, suggesting a synchronized, positive translational regulation of both E2,E5 ORFs (Johnsen et al., 1995). However, the mechanisms of E5 translation has not been elucidated. We have proposed the possible existence of IRES to drive the expression of E5 ORF (García et al., 2024) (represented in Figure 2B by gray arrows). However, experimental analyses should be performed to prove it.
HPV-18 E7 in COS transfected cells E7 is translated by the canonical mechanism that might result from the translation of a subset of short mRNAs originated from the P105 early promoter lacking the first nucleotides of E6 ORF (Remm et al., 1999). HPV-16 E7 translation was also studied in cell-free rabbit in vitro translation lysates and plasmid transfection of HeLa cells (Stacey et al., 1995; Stacey et al., 2000). In contrast, it was found that E7 ORF is translated by an exceptionally pervasive leaky scanning process in which the ribosomal initiation complex does not recognize the AUG start codon of the upstream E6 cistron, and scans through the mRNA over 13 upstream AUG codons until it reaches the E7 ORF TIS (Stacey et al., 1995; Stacey et al., 2000).
L1 and L2 are transcribed from the late region of the HR-HPVs genome (Figure 2C) (Doorbar et al., 2015; Graham, 2017; Nelson and Mirabello, 2023; Rosendo-Chalma et al., 2024). So far, no mechanism of mRNA translation has been elucidated for the L1 and L2 mRNAs, but we propose here that they might be translated by IRESs. Further experimental analyses also should be performed to prove it. In Table 1 we summarize the mechanisms used to translate the different HPV-16 and HPV-18 cistrons.
TABLE 1
| Virus | Cistron | Mechanism | Reference |
|---|---|---|---|
| VPH-16 | E1 | Not known | |
| E1^E4 | Not known; possible IRES | This review | |
| E2 | Termination-reinitiation or leaky scanning | Alloul and Sherman (1999) | |
| E5 | Not known; possible IRES | García et al. (2024) | |
| E6 | Canonical | Stacey et al. (2000) | |
| E7 | Termination-reinitiation or leaky scanning | Stacey et al. (1995), Stacey et al. (2000), Tan et al. (1994), Tang et al. (2006) | |
| L1 | Not known; possible IRES | This review | |
| L2 | Not known | ||
| VPH-18 | E1 | Ribosome shunting | Remm et al. (1999) |
| E1^E4 | Not known; possible IRES | This review | |
| E2 | Not known | ||
| E5 | Not known; possible IRES | García et al. (2024) | |
| E6 | Scanning-independent using extremely short (0–3 nucleotides long) 5′-UTRs and an RNA sequence within the ORF; it also requires the mRNA cap and poly(A) tail; it requires eIF4E and eIF4A | García et al. (2021) | |
| E7 | Termination-reinitiation or canonical | Remm et al. (1999), Tang et al. (2006) | |
| L1 | Not known; possible IRES | This review | |
| L2 | Not known; possible IRES | This review |
Mechanism of translation used to translate the viral cistrons.
Codon usage regulates viral mRNA translation
In all organisms, most amino acids are encoded by multiple synonymous codons. However, codon usage is not uniform across the different species. The preference for specific codons over others during mRNA translation is referred to as codon bias or codon preference (Parvathy et al., 2022). Due to the absence of their own translational machinery, viruses have evolved to optimize their codon usage to match that of their host. By aligning their codon usage patterns with the host’s tRNA pool, viruses enhance the translational efficiency of their own proteins (Kaleem et al., 2025; Simón et al., 2021).
Viruses often employ suboptimal codon usage to reduce the translation rate of viral proteins as a strategy to evade host immune surveillance and response. This involves the use of codons that are less favorable for translation (Kaleem et al., 2025; Mordstein et al., 2021). Indeed, in a wide range of HPV types, codon bias shows poor alignment with the average human codon usage preferences (Cladel et al., 2010; Félez-Sánchez et al., 2015; Ren et al., 2025; Zhao and Chen, 2011). Moreover, a study of the codon usage bias across 79 HPVs showed strong differences among the eight cistrons encoding viral proteins toward 18 codons with T at the third position. Among them, L2 ORFs showed the highest codon bias and E4 the lowest (Zhao et al., 2003). The codon bias of E6,E7 mRNAs poorly matches the preferred codons by host cells, which strongly diminishes the rate of protein translation. As expected, changing the rare toward optimal codons of human cells (Müller, 2005) or the co-transfection of different cultured cells with a plasmid expressing tRNASer(CGA) (Gu et al., 2004) enhanced protein synthesis of the viral proteins. Transfection of a codon-optimized HPV-16 E6 gene also increased protein expression in cultured human cell (Lin et al., 2006; Samorski et al., 2006). Regarding E7, supplementation of rat liver tRNAs to the pool of a cell-free in vitro translation system from rabbit reticulocyte (De Pasquale and Kanduc, 1998), or the use of a codon-optimized plasmid transfected into human cells (Steinberg et al., 2005) significantly enhanced E7 synthesis. It was further observed that the codon bias of HPV genes couples viral gene expression with epithelium differentiation throughout virus life cycle and carcinogenesis (Gu et al., 2007; Zhao et al., 2005; Zhou et al., 1999).
The study of codon usage bias has been fundamental to the design of vaccines against HR-HPVs. In the two widely used commercial vaccines, Gardasil and Cervarix, codon optimization was employed to enhance the production of virus-like particles (VLPs) derived from the L1 protein, which serves as the primary immunogenic target (Kaleem et al., 2025; Markowitz and Schiller, 2021).
Integrated HPVs produce chimeric E6, E7 mRNAs fused with cellular genes
The life-cycle of mRNAs is tightly controlled by numerous cis-regulatory elements located at the 5′- and 3′-UTRs that control the translation and transport of the transcripts, are target of nucleases, or bind to a myriad of regulatory molecules (Leppek et al., 2018; Mayr, 2017; Ryczek et al., 2023). Consequently, mutations in the mRNA UTRs may lead to the uncontrolled expression of a wide variety of mRNAs and, often trigger the development of diverse diseases (Chatterjee and Pal, 2009), including cancer (Chan et al., 2023; Huang et al., 2022; Li and Lu, 2013; Schuster and Hsieh, 2019; Silanes et al., 2007; Zhang et al., 2022).
The regulatory elements that control polyadenylation, degradation, localization, and transport of mRNAs are mostly located in the 3′-UTR (Mayr, 2017). In most tumors, HPV DNA is generally integrated into the genome of epithelial cells, unlike productive infections where is found in episomal form (Figure 3). The viral genome integration is a stochastic process over almost all chromosomes. During this, some viral genomes may be integrated into cancer-related genes which may promote tumorigenesis. Moreover, as a consequence of this integration, the E6 and E7 genes lose their natural 3′-UTR, disrupt the viral E2 ORF, and gain a cellular DNA fragment that is transcribed as a viral-cellular chimeric 3′-UTR (Figure 3). The promoter controlling E6 and E7 genes contains binding sites for E2 protein and binding of E2 represses E6, E7 mRNA transcription. Therefore, loss of the E2 cistron leads to an increase in the E6, E7 mRNA transcription and protein synthesis (Jeon et al., 1995; Lace et al., 2011; Vinther et al., 2005; Wentzensen et al., 2002; Wentzensen et al., 2004). Thus, the main consequence of HPV genome integration is the upregulation and stabilization of E6,E7 mRNAs, leading to the E6 and E7 overexpression and the promotion of carcinogenesis (Couturier et al., 1991; Durst et al., 1987; Ehrig et al., 2020; Jeon and Lambert, 1995; Wentzensen et al., 2002; Ziegert et al., 2003).
FIGURE 3
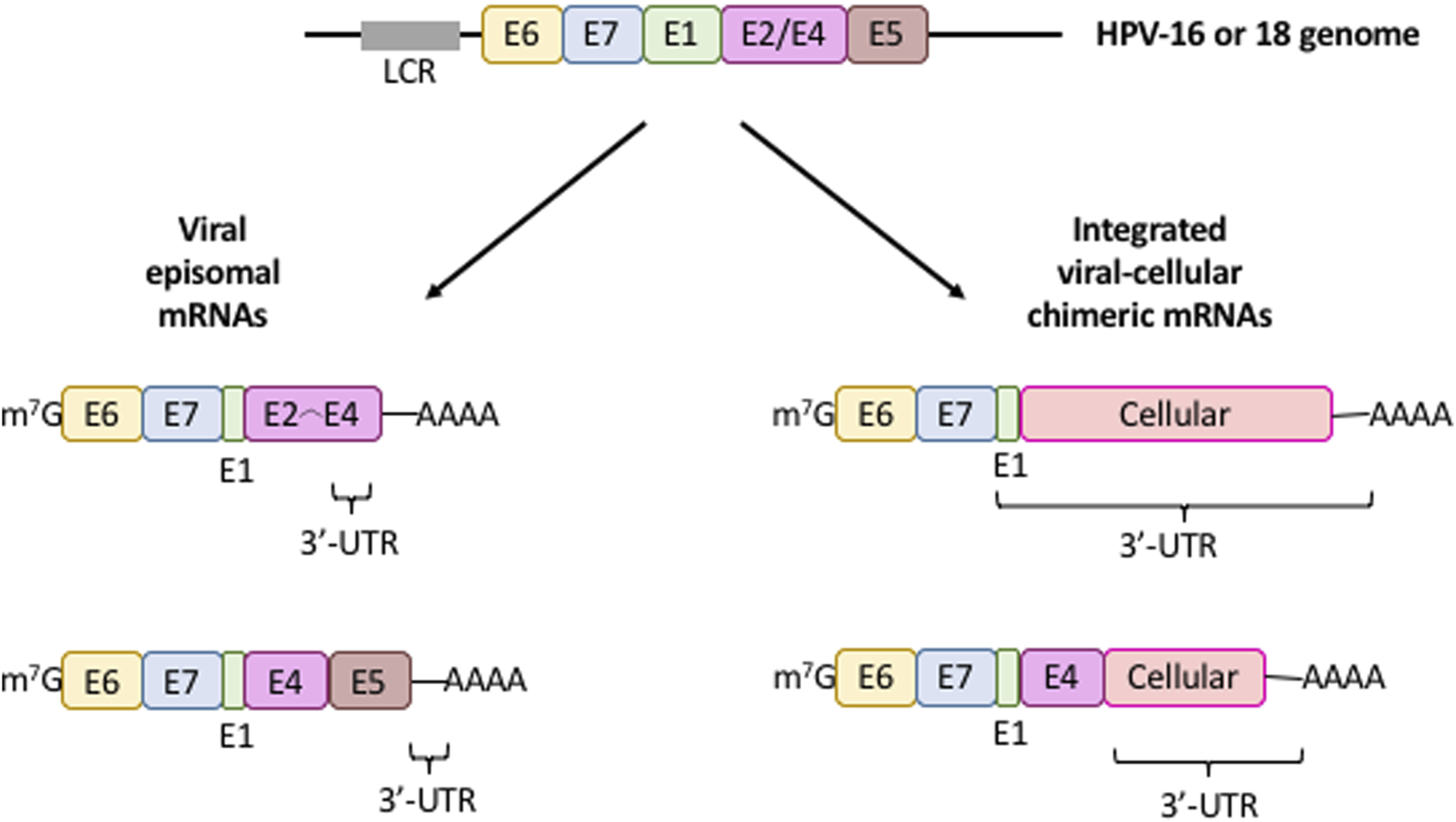
mRNA expression patterns from integrated (fusion) and episomal HPV 16/18 DNA. Diagram illustrating the characteristic structure of viral mRNAs produced from intact episomal genomes (left) or the viral DNA integration into the cellular genome (right). Only the cistrons (colored boxes) controlled by the long control region (LCR, gray box) is depicted. The 3′-UTR of these transcripts is shown. Colored boxes, viral cistrons; m7G, mRNA cap structure; AAAA, mRNa poly(A) tail.
The viral genome integration into the cell genome plays a critical role in triggering carcinogenesis and is one of the molecular features that best defines the transition to carcinogenic transformation (Groves and Coleman, 2015; Groves and Coleman, 2018; Wentzensen et al., 2004). Although the stability of chimeric mRNAs increases (Ehrig et al., 2020; Jeon and Lambert, 1995; Vinther et al., 2005), whether the translation rate of the viral-cellular fusion mRNAs also increases is unknown and deserves further experimental analysis. In Table 2 we summarize the diverse events of viral-cellular fusion transcripts reported in different cells or tissues that generate mRNAs possessing chimeric 3′-UTRs and the consequences in cancer.
TABLE 2
| Virus | Cellular gene fussed with the viral E6,E7 mRNAs | Biological consequence on cancer | Sample sourcea | Integration site | Reference |
|---|---|---|---|---|---|
| HPV-16 | Non-determined | Integration of HPV-16 DNA and expression of virus-cell mRNAs results in the increased stability of E6 and E7 mRNAs and cellular growth | W12 clonal populations and cervical tumor cell lines (SiHa, CaSki) | HPV integration does not occur at any specific locus on the host genome | Jeon and Lambert (1995) |
| HPV-16 | EIF1, MAP4, FAM110B | Viral-cellular fusion transcripts are more stable than episome-derived transcripts, promoting carcinogenesis | Cervical carcinoma tumors | EIF1, MAP4, FAM110B | Ehrig et al. (2020) |
| HPV-16 | MTRF-1, HNRPA3, Rb, SGK1, MYC, Integrin α-2, FAP-1/PTPN13, HMG AT-hook, Fer tyrosine kinase, Plexin D1 | HPV integration is a random process that promotes selection of aggressively expanding cells. Some of those i9ntergation events may hit oncogenes with their concomitant deregulation | HNC tumors, human keratinocytes derived from primary tonsillar or foreskin epithelia immortalized | MTRF-1, HNRPA3, Rb, SGK1, MYC, Integrin α-2, FAP-1/PTPN13, HMG AT-hook, Fer tyrosine kinase, Plexin D1 | Lace et al. (2011) |
| HPV-16, HPV-18 | Non-determined | HPV-16/-18 DNA integration into host cell genome is related to the progression stage of vulvar dysplasia | VIN lesions and cervical carcinoma cell lines (SiHa and Caski) | Non-determined | Hillemanns and Wang (2006) |
| HPV-16, HPV-18, HPV-31, HPV-33, HPV-35 | CYS1, LRP1B, GPD2, LEPREL1, RREB1, MRV11, SSPN, NPM3, KLF7, COG8, RAD51L1, AGBL1, CFTR, EIF1, CEACAM6, MID1 | There is no correlation between HPV type and specific integration loci. HPV integration may occur in transcriptionally active regions and can influence on host gene expression | Ca, CIN3 | CYS1, LRP1B, GPD2, LEPREL1, RREB1, MRV11, SSPN, NPM3, KLF7, COG8, RAD51L1, AGBL1, CFTR, EIF1, CEACAM6, MID1 | Kraus et al. (2008) |
| HPV-2, HPV-6, HPV-11, HPV-13, HPV-16, HPV-18, HPV-30, HPV-31, HPV-35 | Non-determined | The 3′ region HPV-16 mRNA represses the expression at both mRNA and protein level. For other HPV types, except 18, the 3′ region represses mRNA expression by posttranscriptional mechanisms | HaCaT and SiHa cells | Non-determined | Vinther et al. (2005) |
| HPV-16, HPV-18 | N-myc | Integration of HPV genome results in overexpression of the oncogene c-myc and tumor progression | Invasive genital carcinomas | Chromosome band 8q24.1, nearby the protooncogene c-myc; chromosome band 2p24 | Couturier et al. (1991) |
| HPV-16, HPV-18 | Non-determined | The steady-state levels of c-myc mRNA are elevated in HeLa and C4-I cells relative to other Ca cell lines. Thus, This may spurr malignant transformation of cervical cells | Primary Ca and Ca cells (SiHa, SW756, HeLa, and C4-1) | Chromosome regions 20pter->20ql3 and 3p25->3qter, nearby the protooncogenes c-src-1 and c-raf-1; Chromosome region 13ql4-+13q32; Chromosome 12; Chromosome 8, 5′ of the c-myc gene | Durst et al. (1987) |
| HPV-16 | Non-determined | Non-determined | Ca and normal placentas | No cellular sequence specific for the integration is found; some samples showed Alu repetitive sequences in the 5′- or 3′-flanking cellular sequences | Wagatsuma et al. (1990) |
| HPV-16 | Non-determined | Non-determined | Keratinocyte SK-v cells; intraepithelial neoplasms termed bowenoid papulas | Non-determined | Schneider-Maunoury et al. (1987) |
Transcription of chimeric virus-cell 3′-UTR upon the viral DNA integration into the host cells genome.
HNC, head and neck cancer; Ca, Cervical carcinoma; CIN, cervical intraepithelial neoplasia; VaIN, vaginal intraepithelial neoplasia; VIN, vulval intraepithelial neoplasia.
Old questions, new insights
Cervical cancer remains a significant global public health challenge. Despite substantial advances in understanding the translation of HPV mRNAs, several longstanding questions remain unresolved. Among them is the mechanism by which the E5, L1, and L2 cistrons are translated. We speculate that these cistrons may utilize internal ribosome entry sites (IRESs); however, this hypothesis requires experimental validation. For certain viral proteins, such as E7 and E2, alternative translation mechanisms have been proposed. Therefore, further studies are necessary to clarify these observations. Additionally, the impact of cellular fragment fusion on the translation of chimeric virus–host mRNAs remains largely unexplored.
To date, most studies investigating the translation of HPV proteins have been conducted in heterologous systems—such as monkey COS-7 cells, mouse NIH3T3 cells, or rabbit reticulocyte lysates. Therefore, these findings must be validated in human cervical tissues, particularly throughout the process of epithelial differentiation during carcinogenesis.
Understanding the translation mechanisms of viral mRNAs is crucial for the future development of targeted therapies for cervical cancer. This will be instrumental in designing therapeutic strategies that combine translation inhibitors with agents directly targeting viral oncoproteins. Recent findings have highlighted promising pharmacological approaches. Two molecules targeting eIF4E, a key translation initiation factor, are currently being investigated for the treatment of HPV-associated cancers. Cercosporamide has been shown to reduce chemotherapy-induced phosphorylated eIF4E (p-eIF4E) levels, which are implicated in cancer progression, in mouse tumor models (Zhu et al., 2021). Ribavirin, another agent targeting p-eIF4E, has undergone clinical evaluation for HPV-related malignancies. Oral administration of ribavirin resulted in decreased p-eIF4E levels and was well-tolerated (Burman et al., 2022) (clinical trial NCT02308241; ClinicalTrials.gov).
Finally, several drugs are currently undergoing testing in animal models or clinical trials targeting the translation machinery across various cancers (García et al., 2024). These research efforts will be pivotal for the future development of cervical cancer therapies.
Statements
Author contributions
NB-Á: Data curation, Formal Analysis, Investigation, Software, Validation, Visualization, Writing – review and editing. GM: Conceptualization, Formal Analysis, Investigation, Software, Validation, Writing – review and editing. DEV: Conceptualization, Data curation, Formal Analysis, Investigation, Writing – original draft, Writing – review and editing. GH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Project administration, Supervision, Validation, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. GH was supported by the intramural funding program of the Instituto Nacional de Cancerología (INCan). NB-A was supported by INCan. A Secretaría de Ciencia, Humanidades, Tecnología e Innovación (SECIHTI, former CONACyT) fellowship was awarded to GM (Nr. 772862). DEV is a posdoctoral fellow of the SECIHTI Program Estancias posdoctorales por México para la Formación y Consolidación de las y los Investigadores por México, 2022.
Acknowledgments
This work is part of GM curriculum in the Ph.D. Program on Biochemical Sciences (Doctorado en Ciencias Bioquímicas), Universidad Nacional Autónoma de México. We thank the reviewers’ valuable criticism and suggestions, which significantly improved the manuscript during the revision process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Ajiro M. Zheng Z. M. (2015). E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. mBio6, e02068–e02014. 10.1128/mBio.02068-14
2
Alloul N. Sherman L. (1999). The E2 protein of human papillomavirus type 16 is translated from a variety of differentially spliced polycistroniic mRNAs. J. Gen. Virol.80, 29–37. 10.1099/0022-1317-80-1-29
3
Alteri R. Baptiste D. Bell E. B. (2025). Cancer prevention and early detection. Facts and figures 2025-2026. American Cancer Society. 860025.
4
Basukala O. Banks L. (2021). The not-so-good, the bad and the ugly: HPV E5, E6 and E7 oncoproteins in the orchestration of carcinogenesis. Viruses13, 1892. 10.3390/v13101892
5
Bhattacharrjee R. Das S. S. Biswal S. S. Nath A. Das D. Basu A. et al (2021). Mechanistic role of HPV-associated early proteins in cervical cancer: molecular pathways and targeted therapeutic strategies. Crit. Rev. Oncol. Hematol.174, 103675. 10.1016/j.critrevonc.2022.103675
6
Brant A. C. Menezes A. N. Felix S. P. de Almeida L. M. Sammeth M. Moreira M. A. M. (2018). Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA-Seq: a descriptive study in invasive cervical cancer. Genomics111, 1853–1861. 10.1016/j.ygeno.2018.12.008
7
Braunstein T. H. Madsen B. S. Gavnholt B. Rosenstierne M. W. Koefeld Johnsen C. Norrild B. (1999). Identification of a new promoter in the early region of the human papillomavirus type 16 genome. J. Gen. Virol.80, 3241–3250. 10.1099/0022-1317-80-12-3241
8
Brito Querido J. Días-López I. Ramakrishnan V. (2024). The molecular basis of translation initiation and its regulation in eukaryotes. Nat. Rev. Mol. Cell. Biol.25, 168–186. 10.1038/s41580-023-00624-9
9
Burman B. Drutman S. B. Fury M. G. Wong R. J. Katabi N. Ho A. L. et al (2022). Pharmacodynamic and therapeutic pilot studies of single-agent ribavirin in patients with human papillomavirus-related malignancies. Oral Oncol.128, 105806. 10.1016/j.oraloncology.2022.105806
10
Chan J. J. Tabatabaeian H. Tay Y. (2023). 3' UTR heterogeneity and cancer progression. Trends Cell Biol.33, 568–582. 10.1016/j.tcb.2022.10.001
11
Chatterjee S. Pal J. P. (2009). Role of 5'- and 3'-untranslated regions of mRNAs in human diseases. Biol. Cell101, 251–262. 10.1042/BC20080104
12
Cladel N. M. Bertotto A. Christensen N. D. (2010). Human alpha and beta papillomaviruses use different synonymous codon profiles. Virus Genes40, 329–340. 10.1007/s11262-010-0451-1
13
Collaborators G. V. C. (2025). Global, regional, and national trends in routine childhood vaccination coverage from 1980 to 2023 with forecasts to 2030: a systematic analysis for the global burden of disease study 2023. Lancet406, 235–260. 10.1016/S0140-6736(25)01037-2
14
Couturier J. Sastre-garau X. Schneider-Maunoury S. Labib A. Orth G. (1991). Integration of papillomavirus DNA near myc genes in genital carcinomas and its consequences for proto-oncogene expression. J. Virol.65, 4534–4538. 10.1128/JVI.65.8.4534-4538.1991
15
De la Cruz-Hernández E. D. García-Carrancá A. Mohar-Betancourt A. Dueñas-González A. Contreras-paredes A. Pérez-Cárdenas E. et al (2005). Differential splicing of E6 within human papillomavirus type 18 variants and functional consequences. J. Gen. Virol.86, 2459–2468. 10.1099/vir.0.80945-0
16
De Pasquale C. Kanduc D. (1998). Modulation of HPV16 E7 translation by tRNAs in eukaryotic cell-free translation systems. Biochem. Mol. Biol. Int.45, 1005–1009. 10.1002/iub.7510450518
17
Della Fera A. N. Warburton A. Coursey T. L. Khurana S. Mcbride A. A. (2021). Persistent human papillomavirus infection. Viruses13, 321. 10.3390/v13020321
18
Doomer J. Van Doorslaer K. Afrasiabi C. Browne K. Ezejii S. Kim L. et al (2024). PaVE 2.0: behind the scenes of the papillomavirus episteme. J. Mol. Biol.437, 168925. 10.1016/j.jmb.2024.168925
19
Doorbar J. Parton A. Hartley K. Banks L. Crook T. Stanley M. et al (1990). Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology178, 254–262. 10.1016/0042-6822(90)90401-c
20
Doorbar J. Egawa N. Griffin H. Kranjek C. Mrakami I. (2015). Human papillomavirus molecular biology and disease association. Rev. Med. Virol.25 (Suppl. 1), 2–23. 10.1002/rmv.1822
21
Duensing S. Duensing A. Crum C. P. Munger K. (2001). Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res.61, 2356–2360.
22
Durst M. Croce C. M. Gissmann L. Schwartz E. Huebner K. (1987). Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc. Natl. Acad. Sci. U.S.A.84, 1070–1074. 10.1073/pnas.84.4.1070
23
Durst M. Bosch F. X. Glitz D. Schneider A. zur Hausen H. (1991). Inverse relationship between human papillomavirus (HPV) type 16 early gene expression and cell differentiation in nude mouse epithelial cysts and tumors induced by HPV-positive human cell lines. J. Virol.65, 796–804. 10.1128/JVI.65.2.796-804.1991
24
Ehrig F. Häfner N. Driesch C. Kraus Christiansen I. Beer K. Schmitz M. et al (2020). Differences in stability of viral and viral-cellular fusion transcripts in HPV-induced cervical cancers. Int. J. Mol. Sci.211, 112. 10.3390/ijms21010112
25
Elfakess R. Dikstein R. (2008). A translation initiation element specific to mRNAs with very short 5UTR that also regulates transcription. PLoS One3, e3094. 10.1371/journal.pone.0003094
26
Elfakess R. Sinvani H. Haimov O. Svitkin Y. Sonenberg N. Dikstein R. (2011). Unique translation initiation of mRNAs-containing TISU element. Nucleic Acid. Res.39, 7598–7609. 10.1093/nar/gkr484
27
Estêvão D. Costa N. R. Gil da Costa R. M. Medeiros R. (2019). Hallmarks of HPV carcinogenesis: the role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta Gene Regul. Mech.1862, 153–162. 10.1016/j.bbagrm.2019.01.001
28
Evande R. Rana A. Biswas-Fiss E. E. Biswas S. B. (2023). Protein-DNA interactions regulate human papillomavirus DNA replication, transcription, and oncogenesis. Int. J. Mol. Sci.24, 8493. 10.3390/ijms24108493
29
Félez-Sánchez M. Trosemeier J. H. Bedhomme S. González-Bravo M. I. Kamp C. Bravo I. G. (2015). Cancer, warts, or asymptomatic infections: clinical presentation matches codon usage preferences in human papillomaviruses. Genome Biol. Evol.7, 2117–2135. 10.1093/gbe/evv129
30
Gandin V. Masvidal L. Hulea L. Gravel S. Cargnello M. Mclaughlan S. et al (2016). nanoCAGE reveals 5′UTR features that define specific modes of translation of functionally related mTOR-sensitive mRNAs. Genome Res.26, 636–648. 10.1101/gr.197566.115
31
García A. Maldonado G. González J. L. Svitkin Y. Cantú D. García-Carrancá A. et al (2021). High-risk human papillomavirus-18 uses an mRNA sequence to synthesize oncoprotein E6 in tumors. Proc. Natl. Acad. Sci. U.S.A.118, e2108359118. 10.1073/pnas.2108359118
32
García A. Maldonado G. Hernández G. (2024). Translational control of papillomavirus mRNAs in the spotlight. Trends Cell Biol.34, 703–706. 10.1016/j.tcb.2024.07.001
33
Glahder J. A. Hansen C. N. Vinther J. Madsen B. S. Norrild B. (2003). A promoter within the E6 ORF of human papillomavirus type 16 contributes to the expression of the E7 oncoprotein from a monocistronic mRNA. J. Gen. Virol.84, 3429–3441. 10.1099/vir.0.19250-0
34
Graham S. V. (2010). Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol.5, 1493–1506. 10.2217/fmb.10.107
35
Graham S. V. (2017). Keratinocyte differentiation-dependent human papillomavirus gene regulation. Viruses9, 245. 10.3390/v9090245
36
Grassmann K. Rapp B. Maschek H. Petry K. U. Iftner T. (1996). Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol.70, 2339–2349. 10.1128/JVI.70.4.2339-2349.1996
37
Groves I. J. Coleman N. (2015). Pathogenesis of human papillomavirus-associated mucosal disease. J. Pathol.235, 527–538. 10.1002/path.4496
38
Groves I. J. Coleman N. (2018). Human papillomavirus genome integration in squamous carcinogenesis: what have next-generation sequencing studies taught us?J. Pathol.245, 9–18. 10.1002/path.5058
39
Gu W. Li M. Zhao W. M. Fang N. X. Bu S. Frazzer I. H. et al (2004). tRNASer(CGA) differentially regulates expression of wild-type and codon-modified papillomavirus L1 genes. Nucleic Acid. Res.32, 4448–4461. 10.1093/nar/gkh748
40
Gu W. Ding J. Wang X. de Kluyver R. L. Saunders N. A. Frazer I. H. et al (2007). Generalized substitution of isoencoding codons shortens the duration of papillomavirus L1 protein expression in transiently gene-transfected keratinocytes due to cell differentiation. Nucleic Acid. Res.35, 4820–4832. 10.1093/nar/gkm496
41
Hernández G. Tettweiler G. (2012). “Protein abundance variation,” in Systems biology. Editor MeyerR. A. (Weinheim, Germany: John Wiley), 117–137.
42
Hernández G. Osnaya V. G. Pérez-Martínez X. (2019). Conservation and variability of the AUG initiation codon context in eukaryotes. Trends Biochem. Sci.44, 1009–1021. 10.1016/j.tibs.2019.07.001
43
Hernández G. García A. Lasko P. Sonenberg N. (2020). Unorthodox mechanisms to initiate translation open novel paths for gene expression. J. Mol. Biol.432, 166702. 10.1016/j.jmb.2020.10.035
44
Hershey J. W. B. Sonenberg N. Mathews M. B. (2019). Principles of translational control. Cold Spring Harb. Perspect. Biol.11, a032607. 10.1101/cshperspect.a032607
45
Hillemanns P. Wang X. (2006). Integration of HPV-16 and HPV-18 DNA in vulvar intraepithelial neoplasia. Gynecol. Oncol.100, 276–282. 10.1016/j.ygyno.2005.10.003
46
Hinnebusch A. G. Dever T. E. Sonenberg N. (2004). “Mechanisms and regulation of protein synthesis initiation in eukaryotes,” in Protein synthesis and ribosome structure. Editors NierhausK. H.WilsonD. N. (Weinheim: Wiley-VCH Verlag GmbH and Co. KGaA), 241–322.
47
Huang D. Wang X. Huang Z. Liu Y. Liu X. Gin T. et al (2022). 3' untranslated regions of tumor suppressor genes evolved specific features to favor cancer resistance. Oncogene41, 3278–3288. 10.1038/s41388-022-02343-5
48
Jackson R. J. Hellen C. U. Pestova T. V. (2012). Termination and post-terminationevents in eukaryotic translation. Adv. Protein Chem. Struct. Biol.86, 45–93. 10.1016/B978-0-12-386497-0.00002-5
49
Jan E. Mohr I. Walsh D. (2016). A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol.3, 283–307. 10.1146/annurev-virology-100114-055014
50
Jeon S. Lambert P. F. (1995). Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. U. S. A.92, 1654–1658. 10.1073/pnas.92.5.1654
51
Jeon S. Allen-Hoffmann B. L. Lambert P. F. (1995). Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol.69, 2989–2997. 10.1128/JVI.69.5.2989-2997.1995
52
Johnsen C. K. Stanley M. Norrild B. (1995). Analysis of human papillomavirus type 16 E5 oncogene expression in vitro and from bicistronic messenger RNAs. Intervirology38, 339–345. 10.1159/000150461
53
Jones C. E. Danovaro-Holliday M. C. Mwinnyaa G. Gacic-Dobo M. Francis L. Grevendonk J. et al (2024). Routine vaccination coverage - Worldwide, 2023. MMWR Morb. Mortal. Wkly. Rep.73, 978–984. 10.15585/mmwr.mm7343a4
54
Kajitani N. Schwartz S. (2022). The role of RNA-binding proteins in the processing of mRNAs produced by carcinogenic papillomaviruses. Semin. Cancer Biol.86, 482–496. 10.1016/j.semcancer.2022.02.014
55
Kaleem S. Dahal U. D’evi S. Kour B. Kour S. A. (2025). Codon usage evolution in viruses: implications for survival and pathogenicity. J. Mol. Evol. 10.1007/s00239-025-10263-7
56
Kraus I. Driesch C. Vinokurova S. Hovig E. A. S. von Knebe M. Doeberitz M. et al (2008). The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res.68, 2514–2522. 10.1158/0008-5472.CAN-07-2776
57
Kwan T. Thompson S. R. (2019). Noncanonical translation initiation in eukaryotes. Cold Spring Harb. Perspect. Biol.11, a032672. 10.1101/cshperspect.a032672
58
Lace M. J. Anson J. R. Klussmann J. P. Wang D. H. Smith E. M. Haugen T. H. et al (2011). Human papillomavirus type 16 (HPV-16) genomes integrated in head and neck cancers and in HPV-16-immortalized human keratinocyte clones express chimeric virus-cell mRNAs similar to those found in cervical cancers. J. Virol.85, 1645–1654. 10.1128/JVI.02093-10
59
LaVigne A. W. Triedman S. A. Randall T. C. Trimble E. L. Viswanathan A. N. (2017). Cervical cancer in low and middle income countries: addressing barriers to radiotherapy delivery. Gynecol. Oncol. Rep.22, 16–20. 10.1016/j.gore.2017.08.004
60
Leppek K. Das R. Barna M. (2018). Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol.19, 158–174. 10.1038/nrm.2017.103
61
Li J. Lu X. (2013). The emerging roles of 3' untranslated regions in cancer. Cancer Lett.337, 22–25. 10.1016/j.canlet.2013.05.034
62
Lin C. T. Tsai Y. C. He L. Calizo R. Chou H. H. Chang T. C. et al (2006). A DNA vaccine encoding a codon-optimized human papillomavirus type 16 E6 gene enhances CTL response and anti-tumor activity. J. Biomed. Sci.13, 481–488. 10.1007/s11373-006-9086-6
63
Markowitz L. Schiller J. T. (2021). Human papillomavirus vaccines. J. Infect. Dis.224, S367–S378. 10.1093/infdis/jiaa621
64
Martinez-Zapien D. Ruiz F. X. Poirson J. Mitschler A. Ramirez J. Forster A. et al (2016). Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature539, 541–545. 10.1038/nature16481
65
Mayr C. (2017). Regulation by 3′-untranslated regions. Annu. Rev. Genet.51, 171–194. 10.1146/annurev-genet-120116-024704
66
Mesplede T. Gagnon D. Bergeron-Labrecque F. Azar I. Senechal H. Coutlée F. et al (2012). p53 degradation activity, expression, and subcellular localization of E6 proteins from 29 human papillomavirus genotypes. J. Virol.86, 94–107. 10.1128/JVI.00751-11
67
Moral-Hernández O. López-Urrutia E. Bonilla-Moreno R. Martínez-Salazar M. Arechaga-Ocampo E. Berumen J. et al (2010). The HPV-16 E7 oncoprotein is expressed mainly from the unspliced E6/E7 transcript in cervical carcinoma C33-A cells. Arch. Virol.155, 1959–1970. 10.1007/s00705-010-0787-9
68
Mordstein C. Cano L. Morales A. C. Young B. Ho A. T. Rice A. M. et al (2021). Transcription, mRNA export, and immune evasion shape the codon usage of viruses. Genome Biol. Evol.13, evab106. 10.1093/gbe/evab106
69
Müller M. (2005). Codon optimization of papillomavirus genes. Methods Mol. Med.119, 433–444. 10.1385/1-59259-982-6:433
70
National Cancer Institute (2021). Cervical cancer causes, risk factors, and prevention. Washington: National Cancer Institute.
71
Nelson C. W. Mirabello L. (2023). Human papillomavirus genomics: understanding carcinogenicity. Tumour Virus Res.15, 200258. 10.1016/j.tvr.2023.200258
72
Obanya D. I. Wootton L. M. Morgan E. L. (2025). Advances in understanding the mechanisms of the human papillomavirus oncoproteins. Biochem. Soc. Trans.53, 565–577. 10.1042/BST20253041
73
Olmedo-Nieva L. Munoz-Bello J. O. Contreras-Paredes A. Lizano M. (2018). The role of E6 spliced isoforms (E6*) in human papillomavirus-induced carcinogenesis. Viruses10, 45. 10.3390/v10010045
74
Panczyszyn A. Boniewska-Bernacka E. Glab G. (2018). Telomers and telomerase during human papillomavirus-induced papillomavirus. Mol. Diagn. Ther.22, 421–430. 10.1007/s40291-018-0336-x
75
Parvathy S. T. Udayasuriyan V. Bhadana V. (2022). Codon usage bias. Mol. Biol. Rep.49, 539–565. 10.1007/s11033-021-06749-4
76
Pavelescu L. A. Mititelu-Zafiu N. L. Mindru D. E. Vladareanu R. Curici A. (2025). Molecular insights into HPV-driven cervical cancer: oncoproteins, immune evasion, and epigenetic modifications. Microorganisms13, 1000. 10.3390/microorganisms13051000
77
Pelletier J. Sonenberg N. (2019). The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem.88, 307–335. 10.1146/annurev-biochem-013118-111042
78
Remm M. Remm A. Ustav M. (1999). Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol.73, 3062–3070. 10.1128/JVI.73.4.3062-3070.1999
79
Ren J. Lii Q. Shen W. Tan X. (2025). Decoding codon usage patterns in high-risk human papillomavirus genomes: a comprehensive analysis. Curr. Microbiol.82, 148. 10.1007/s00284-025-04131-2
80
Romanczuk H. Thierry F. Howley P. M. (1990). Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol.64, 2849–2859. 10.1128/JVI.64.6.2849-2859.1990
81
Rosendo-Chalma P. Antonio-Vejar V. Tejedor J. G. O. Segarra J. O. Crespo B. V. Bigoni-Ordoñez G. D. (2024). The hallmarks of cervical cancer: molecular mechanisms induced by human papillomavirus. Biology13, 77. 10.3390/biology13020077
82
Rosenstierne M. W. Vinther J. Hansen C. N. Prydsoe M. Norrild B. (2003). Identification and characterization of a cluster of transcription start sites located in the E6 ORF of human papillomavirus type 16. J. Gen. Virol.84, 2909–2920. 10.1099/vir.0.19332-0
83
Ryczek N. Łyś A. Makałowska I. (2023). The functional meaning of 5'UTR in protein-coding genes. Int. J. Mol. Sci.24, 2976. 10.3390/ijms24032976
84
Samorski R. Gissmann L. Osen W. (2006). Codon optimized expression of HPV 16 E6 renders target cells susceptible to E6-specific CTL recognition. Immunol. Lett.107, 41–49. 10.1016/j.imlet.2006.07.003
85
Schneider-Gädicke A. Schwarz E. (1986). Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J.5, 2285–2292. 10.1002/j.1460-2075.1986.tb04496.x
86
Schneider-Maunoury S. Croissant O. Orth G. (1987). Integration of human papillomavirus type 16 DNA sequences: a possible early event in the progression of genital tumors. J. Virol.61, 3295–3298. 10.1128/JVI.61.10.3295-3298.1987
87
Schuster S. L. Hsieh A. C. (2019). The untranslated regions of mRNAs in cancer. Trends Cancer5, 245–262. 10.1016/j.trecan.2019.02.011
88
Sherman L. Alloul N. (1992). Human papillomavirus type 16 expresses a variety of alternatively spliced mRNAs putatively encoding the E2 protein. Virology191, 953–959. 10.1016/0042-6822(92)90271-p
89
Sherman L. Alloul N. Golan I. Durst M. Baram A. (1992). Expression and splicing patterns of human papillomavirus type-16 mRNAs in pre-cancerous lesions and carcinomas of the cervix, in human keratinocytes immortalized by HPV 16, and in cell lines established from cervical cancers. Int. J. Cancer50, 356–364. 10.1002/ijc.2910500305
90
Silanes I. L. Quezada M. P. Esteller M. (2007). Aberrant regulation of messenger RNA 3'-untranslated region in human cancer. Cell Oncol.29, 1–17. 10.1155/2007/586139
91
Simón D. Cristina J. Musto H. (2021). Nucleotide composition and codon usage across viruses and their respective hosts. Front. Microbiol.12, 646300. 10.3389/fmicb.2021.646300
92
Smotkin D. Wettstein F. O. (1986). Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. U. S. A.83, 4680–4684. 10.1073/pnas.83.13.4680
93
Smotkin D. Prokoph H. Wettstein F. O. (1989). Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J. Virol.63, 1441–1447. 10.1128/JVI.63.3.1441-1447.1989
94
Stacey S. N. Jordan D. Snijders P. J. Mackett M. Walboomers J. M. Arrand J. R. (1995). Translation of the human papillomavirus type 16 E7 oncoprotein from bicistronic mRNA is independent of splicing events within the E6 open reading frame. J. Virol.69, 7023–7031. 10.1128/JVI.69.11.7023-7031.1995
95
Stacey S. N. Jordan D. Williamson A. J. K. Brown M. Coote J. H. Arrand J. R. (2000). Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol.74, 7284–7297. 10.1128/jvi.74.16.7284-7297.2000
96
Steinberg T. Ohlschlager P. Sehr P. Osen W. Gissmann L. (2005). Modification of HPV 16 E7 genes: correlation between the level of protein expression and CTL response after immunization of C57BL/6 mice. Vaccine23, 1149–1157. 10.1016/j.vaccine.2004.08.027
97
Stoler M. H. Rhodes C. R. Whitbeck A. Wolinsky S. M. Chow I. T. Broker T. R. (1992). Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol.23, 117–128. 10.1016/0046-8177(92)90232-r
98
Sung H. Ferlay J. Siegel R. L. Laversanne M. Soerjomataram I. Jemal A. et al (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249. 10.3322/caac.21660
99
Tan T. M. C. Gloss B. Bernard H. U. Ting R. C. Y. (1994). Mechanism of translation of the bicistronic mRNA encoding human papillomavirus type 16 E6-E7 genes. J. Gen. Virol.75, 2663–2670. 10.1099/0022-1317-75-10-2663
100
Tang S. Tao M. McCoy J. P. Zheng Z. M. (2006). The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol.80, 4249–4263. 10.1128/JVI.80.9.4249-4263.2006
101
Thierry F. Heard J. M. Dartmann K. Yaniv M. (1987). Characterization of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J. Virol.61, 134–142. 10.1128/JVI.61.1.134-142.1987
102
Toots M. Mannik A. Kivi G. Ustav M. Ustav E. Ustav M. (2014). The transcription map of human papillomavirus type 18 during genome replication in U2OS cells. PLoS One9, e116151. 10.1371/journal.pone.0116151
103
Tsakogiiannis D. Nikolaidis M. Zagourii F. Zografos E. Kottaridi C. al e. et al (2022). Mutation profile of HPV16 L1 and L2 genes in different geographic areas. Viruses15, 141. 10.3390/v15010141
104
Van Doorslaer K. Li Z. Xirasagar S. Maes P. Kaminsky D. Liiou D. et al (2017). The papillomavirus episteme: a major update to the papillomavirus sequence database. Nucleic Acid. Res.45, D499–D506. 10.1093/nar/gkw879
105
Vinther J. Rosenstierne M. W. Kristiansen K. Norrild B. (2005). The 3' region of human papillomavirus type 16 early mRNAs decrease expression. BMC Infect. Dis.5, 83. 10.1186/1471-2334-5-83
106
Wagatsuma M. Hashimoto K. Matsukura T. (1990). Analysis of integrated human papillomavirus type 16 DNA in cervical cancers: amplification of viral sequences together with cellular flanking sequences. J. Virol.64, 813–821. 10.1128/JVI.64.2.813-821.1990
107
Wang X. Meyers C. Wang H. K. Chow L. T. Zheng Z.M. (2011). Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J. Virol.85, 8080–8092. 10.1128/JVI.00670-11
108
Wang Y. Chen F. Qu W. Gong Y. Wang Y. Chen L. et al (2025). Alternative splicing in the genome of HPV and its regulation. Front. Cell. Infect. Microbiol. 10.3389/fcimb.2024.1443868
109
Wentzensen N. Ridder R. Klaes R. Vinukurova S. Schaefer U. Doeberitz M. v. K. (2002). Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene21, 419–426. 10.1038/sj.onc.1205104
110
Wentzensen N. Vinokurova S. von Knebel Doeberitz M. (2004). Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res.64, 3878–3884. 10.1158/0008-5472.CAN-04-0009
111
WHO (2022). International agency for research on cancer. Geneva: WHO. Available online at: https://www.iarc.who.int/cancer-type/cervical-cancer/900-world-fact-sheet.pdf (Accessed on September 15th, 2025).
112
Yu L. Majerciack V. Zheng Z.-M. (2022). HPV16 and HPV18 genome structure, expression, and post-transcriptional regulation. Int. J. Mol. Sci.23, 4943. 10.3390/ijms23094943
113
Zhang Y. Yang M. Yang S. Hong S. (2022). Role of noncoding RNAs and untranslated regions in cancer: a review. Medicine101, e30045. 10.1097/MD.0000000000030045
114
Zhao K. N. Chen J. (2011). Codon usage roles in human papillomavirus. Rev. Med. Virol.21, 397–411. 10.1002/rmv.707
115
Zhao K. N. Liu W. J. Frazer I. H. (2003). Codon usage bias and A + T content variation in human papillomavirus genomes. Virus Res.98, 95–104. 10.1016/j.virusres.2003.08.019
116
Zhao K. N. Gu W. Fang N. X. Saunders N. A. Frazer I. H. (2005). Gene codon composition determines differentiation-dependent expression of a viral capsid gene in keratinocytes in vitro and in vivo. Mol. Cell Biol.25, 8643–8655. 10.1128/MCB.25.19.8643-8655.2005
117
Zhou J. Liu W. J. Peng S. W. Sun X. Y. Frazer I. (1999). Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol.73, 4972–4982. 10.1128/JVI.73.6.4972-4982.1999
118
Zhu Y. Wang C. Li M. Yang X. (2021). Targeting of MNK/eIF4E overcomes chemoresistance in cervical cancer. J. Pharm. Pharmacol.73, 1418–1426. 10.1093/jpp/rgab094
119
Ziegert C. Wentzensen N. Vinokurova S. Kisseljov F. Einekel J. Hoeckel M. et al (2003). Comprehensive analysis of HPV ntegration locii in anogenital lessions conbining transcript and genome-based amplification techniques. Oncogene22, 3984. 10.1038/sj.onc.1206629
Summary
Keywords
papillomavirus, cervical cancer, translational control, eIF4E, codon usage, untranslated region
Citation
Baranda-Ávila N, Maldonado G, Vélez DE and Hernández G (2025) Translational regulation of human papillomavirus mRNAs in carcinogenesis: old questions and new insights. Front. Cell Dev. Biol. 13:1676001. doi: 10.3389/fcell.2025.1676001
Received
29 July 2025
Accepted
23 September 2025
Published
07 October 2025
Volume
13 - 2025
Edited by
Jaime Villegas, Andrés Bello University, Chile
Reviewed by
Omid Vakili, Isfahan University of Medical Sciences, Iran
Adán Arizmendi Izazaga, Autonomous University of Guerrero, Mexico
Updates
Copyright
© 2025 Baranda-Ávila, Maldonado, Vélez and Hernández.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Greco Hernández, ghernandezr@incan.edu.mx, greco.hernandez@tec.mx
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.