- 1Department of Infectious Diseases, The First Affiliated Hospital, College of Clinical Medicine, Henan University of Science and Technology, Luoyang, Henan, China
- 2Henan Medical Key Laboratory of Gastrointestinal Microecology and Hepatology, Luoyang, China
- 3Henan Key Laboratory of Cancer Epigenetics, Cancer Institute, The First Affiliated Hospital, College of Clinical Medicine, Medical College of Henan University of Science and Technology, Luoyang, China
Sepsis is an infection-induced syndrome driven primarily by dysregulated host inflammatory responses. This process induces complex physiological changes that provoke systemic inflammation and multi-organ dysfunction, severely threatening survival in advanced cases. N6-methyladenosine (m6A), the most prevalent eukaryotic RNA modification, orchestrates crucial regulatory functions across biological processes and is a focal point in epigenetics. This modification is dynamically controlled by three protein classes: writers that catalyze m6A deposition, erasers that mediate its removal, and readers that decode modification signals. Substantial evidence implicates m6A dysregulation in sepsis-induced multi-organ damage, encompassing cardiovascular dysfunction, acute lung injury, and acute kidney injury. This review synthesizes current mechanistic insights into m6A’s role in sepsis pathogenesis. By delineating how m6A governs inflammatory cascades and organ injury pathways, we evaluate its therapeutic targeting potential, providing translational frameworks for future research.
1 Introduction
Sepsis is currently defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection (Singer et al., 2016). It poses a critical threat to patients due to its high potential to progress to multiple-organ dysfunction syndrome (MODS) and other lethal complications (Wheeler and Bernard, 1999). Globally, sepsis accounts for 20% of annual deaths. (Rudd et al., 2020).Its high mortality rate correlates strongly with heterogeneous manifestations, primarily involving damage to the heart, lungs, kidneys, and other organs. (Wheeler and Bernard, 1999; Borges and Bento, 2024; Gustot et al., 2009; Ricci et al., 2011). The pathogenesis of sepsis is now understood as a dysregulated host response to infection. This response is characterized by a complex and often concurrent interplay between an initial hyperinflammatory phase (frequently manifesting as a “cytokine storm”) (Nedeva, 2021) and a subsequent protracted immunosuppressive state. A critical component of this immunosuppression is the development of immune tolerance, a state of lymphocyte hyporesponsiveness and innate immune paralysis. Key mechanisms include extensive apoptosis of immune cells, T-cell exhaustion, and reprogramming of monocytes/macrophages with diminished antigen-presentation capacity and cytokine production. (Hotchkiss et al., 2013a; Arora et al., 2023). It is precisely this bimodal and dysregulated immune response—the uncontrolled inflammation coupled with compensatory immunosuppression and tolerance—that distinguishes sepsis from an uncomplicated infection, however serious, and underlies the heightened vulnerability to secondary nosocomial infections and later mortality. Consequently, elucidating the pathogenesis of sepsis and identifying targets for early diagnosis and therapeutic intervention have significant clinical implications for improving patient prognoses.
A key clinical indicator of sepsis severity and tissue hypoperfusion is hyperlactatemia, which is particularly prominent in septic shock and strongly correlates with poor outcomes (Garcia-Alvarez et al., 2014). Beyond its role as a metabolic byproduct, lactate is increasingly recognized as a signaling molecule that can influence gene expression through novel epigenetic modifications, such as histone lactylation (Xiong et al., 2022). Gene expression is regulated through heritable, non-DNA-sequence-changing mechanisms across multiple levels. Epigenetic modifications, such as DNA methylation, histone modification, chromatin remodeling, and non-coding RNA (ncRNA)-based regulation, modulate gene activity by altering chromatin accessibility and function (Dai et al., 2024; Bollati and Baccarelli, 2010). In parallel, epitranscriptomic modifications, which refer to post-transcriptional chemical alterations to RNA, represent another critical regulatory layer. Notably, among the >170 identified RNA modifications, m6A methylation regulates all phases of the RNA lifecycle (Zaccara et al., 2019)—such as processing, degradation, nuclear export, and translation—thereby modulating RNA expression and function. This modification is dynamically controlled by three protein classes: “writer” (methyltransferases), “eraser” (demethylases), and “reader” (reader proteins) (An and Duan, 2022).
m6A methylation has been implicated in diverse pathologies, including acute promyelocytic leukemia, (Wu et al., 2025), ischemic brain injury (Xu et al., 2020), and clear-cell renal carcinoma (Strick et al., 2020; Alhammadi et al., 2025). Recent studies suggest its involvement in sepsis pathogenesis. Analysis of gene-expression datasets from 479 sepsis patients by Zhang et al. revealed three sepsis subtypes characterized by heterogeneity in m6A methylation-regulated genes, indicating a link between m6A dysregulation and sepsis heterogeneity (Zhang S. et al., 2020). The link between lactate and m6A adds another layer of complexity. For instance, Xiong et al. demonstrated that in tumor-infiltrating myeloid cells, lactate induces METTL3 expression via H3K18 lactylation (Galle et al., 2022), and this METTL3-mediated m6A modification promotes immunosuppression via JAK/STAT signaling (Xiong et al., 2022) connection suggests a potential mechanism whereby lactate-driven METTL3 induction and subsequent m6A deposition may contribute to the dysregulated immune response and immunosuppression observed in septic patients.
This review synthesizes recent advances in m6A modification within the context of sepsis, outlining its fundamental biology, examining its mechanistic roles in sepsis-induced MODS, and evaluating its potential as a therapeutic target—ultimately aiming to open novel diagnostic or therapeutic avenues for improving sepsis outcomes.
2 m6A methylation: molecular mechanisms and functions
Among over 100 identified RNA chemical modifications, m6A represents the most prevalent and abundant modification in eukaryotic mRNA. This modification occurs at the N6 position of adenosine residues. (Xu Z. et al., 2025; Cappannini et al., 2024). Research confirms its conservation across diverse species—including plants, humans, Drosophila, and other mammals. (Oerum et al., 2021).
Critically, m6A modification levels undergo rapid, reversible reprogramming in response to environmental stimuli (Furci et al., 2024; Dierks et al., 2025; Zhang et al., 2025), developmental stages (Li et al., 2022), and RNA metabolic states (Furci et al., 2024; Dierks et al., 2025). This dynamic regulation enables m6A to participate extensively in RNA-related cellular processes—particularly differentiation and reprogramming—thereby highlighting its broad relevance to disease pathogenesis (Jiang et al., 2021a).
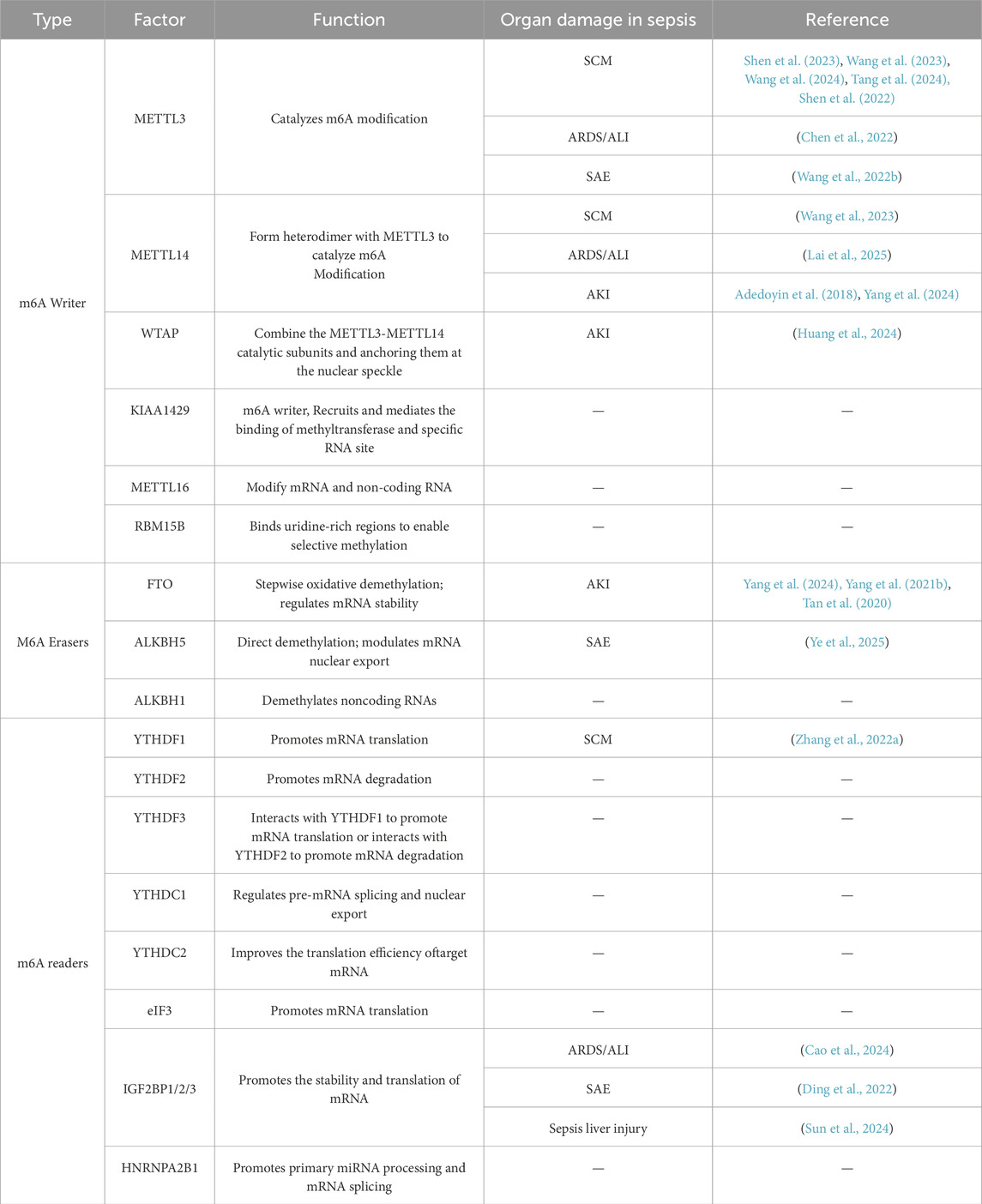
m6A modification is reversible and participates in eukaryotic cell differentiation, proliferation, and apoptosis (Zhang H. et al., 2020). Its regulatory factors fall into three categories: writers, erasers, and readers (as summarized in Table 1).
The writers recognize and bind to m6A-modified RNA, regulating mRNA stability, translation efficiency, splicing, and nuclear export. This group primarily includes the methyltransferase-like proteins methyltransferase-like 3 (METTL3),METTL14 and Wilms’ tumor 1-associating protein (WTAP). Within this complex, METTL3 serves as the catalytic subunit, while METTL14 provides structural support at the active site (Wang et al., 2016). The readers decode the m6A marks and regulate mRNA metabolism through distinct mechanisms. Key examples include YTH domain-containing family proteins (YTHDF1-3 and YTHDC1-2) and eukaryotic translation initiation factor 3 subunit A (eIF3), which recognize m6A sites to modulate target RNA function. The erasers remove m6A modifications from RNA (32), dynamically controlling modification levels and participating in cell development and stress responses. Major erasers include fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5), which mediate m6A demethylation (Kapadia et al., 2025).
These regulatory factors cooperate to determine m6A homeostasis within cells and ensure the precision of m6A methylation, thereby influencing RNA functionality and biological behavior (as illustrated in Figure 1). Research indicates that m6A methylation exhibits dynamic regulatory properties, meaning that its regulatory mechanisms may differ across cell types (Ivanova et al., 2017) and physiological states (Li et al., 2022), thus offering new scientific perspectives (Yang B. et al., 2021).
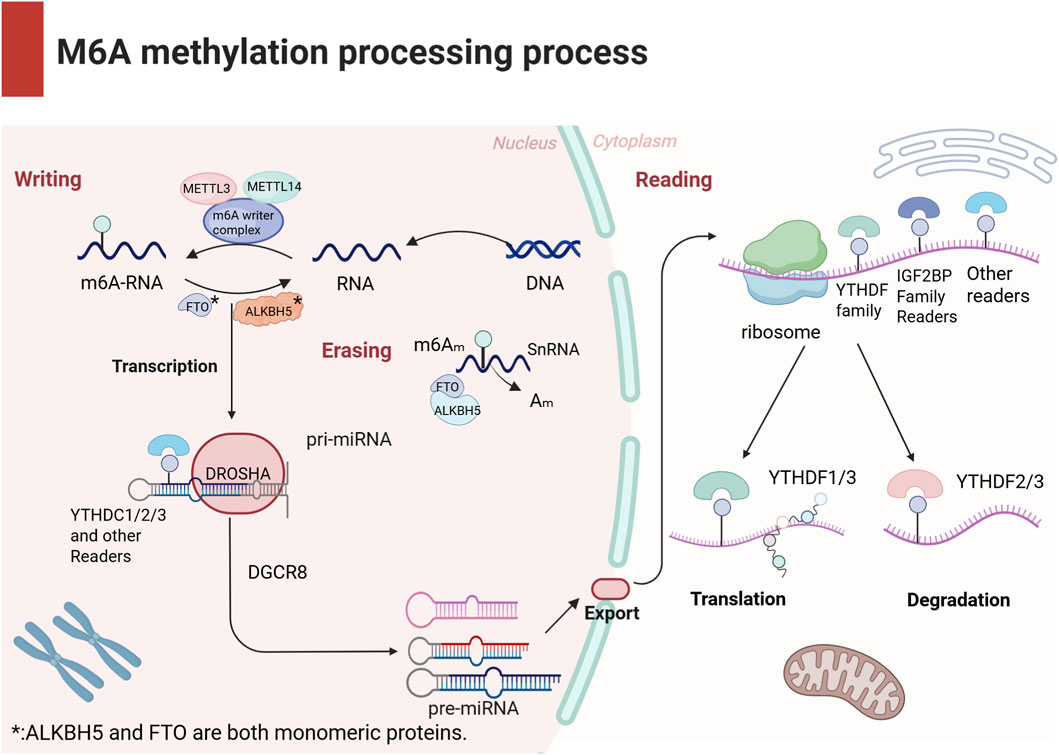
Figure 1. The schematic diagram presents the biological process of m6A modification. m6A methylation is a dynamic and reversible modification regulated by three types of factors: Writers, Readers, and Erasers. Writers are responsible for adding methyl groups; Readers recognize the chemical modification and regulate mRNA metabolism through diverse mechanisms; Erasers remove m6A modifications from RNA, dynamically modulating m6A levels to participate in cellular development and stress responses. These components collectively maintain cellular m6A homeostasis and ensure precision in RNA functional regulation.
m6A methylation critically regulates diverse biological processes (Jiang et al., 2021a). First, it influences gene expression by modulating RNA stability and translation efficiency. For example, m6A methylation can dynamically regulate mRNA stability—either promoting degradation or enhancing stability—in a context-dependent manner (Wei, 2024; Bi et al., 2023). Additionally, m6A governs the RNA lifecycle through its impact on RNA splicing and nuclear export. In immune responses, m6A modifications regulate the effector functions of immune cells, ultimately shaping systemic immunity. Critically, dysregulated m6A methylation is mechanistically linked to multiple pathologies, including cancer, cardiovascular disease, and neurodegenerative disease (An and Duan, 2022).
3 m6A methylation regulates sepsis progression through immune-inflammatory networks
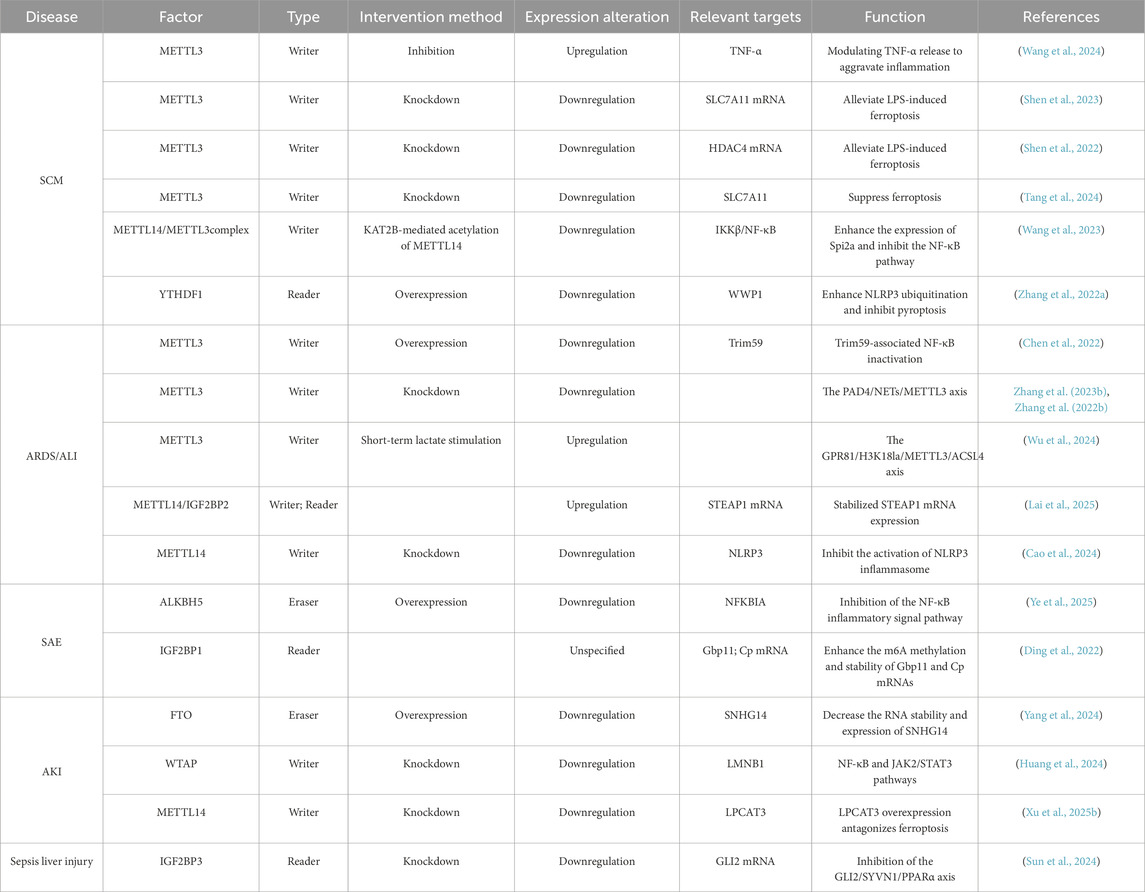
Sepsis is a multisystem disorder characterized by high mortality and complex multidimensional clinical and biological features (Singer et al., 2016). Its heterogeneity stems from diverse factors including host genetics, infection etiology, dysregulated host responses, and multi-organ dysfunction (W et al., 2023). Emerging evidence indicates that m6A methylation plays a critical role in sepsis pathogenesis. (Zhu et al., 2024). Ge et al. demonstrated that elevated WTAP protein and m6A levels correlate strongly with hyperinflammatory responses. Under inflammatory stress, WTAP is upregulated under the regulation of nuclear factor kappa-B(NF-κB) and accelerates the inflammatory response by promoting the expression of numerous pro-inflammatory cytokines in response to various inflammatory stimuli (Ge et al., 2024).
m6A methylation governs sepsis progression by modulating pro-inflammatory cytokine expression and regulating immune-cell activation and cytokine secretion (Shen et al., 2023). The interdependence between m6A methylation and inflammatory response is well-established (Song et al., 2023; Luo et al., 2021a). The NOD-like receptor family pyrin domain containing 3(NLRP3) inflammasome has been mechanistically linked to septic pathology (Zhang W. et al., 2023). Using Lipopolysaccharide (LPS)-induced septic shock models, Luo et al. showed that FTO inhibition suppresses NLRP3 inflammasome activation through the forkhead box protein O1(FoxO1)/NF-κB signaling pathway in macrophages (Luo et al., 2021b). Correspondingly, Wang et al. revealed that modulating FTO-mediated m6A methylation regulates pyroptosis in sepsis (Wang B. et al., 2022)—a key mechanism driving uncontrolled inflammation. Thus, m6A methylation not only contributes significantly to septic pathogenesis but also represents a promising immunomodulatory target (Figure 2).
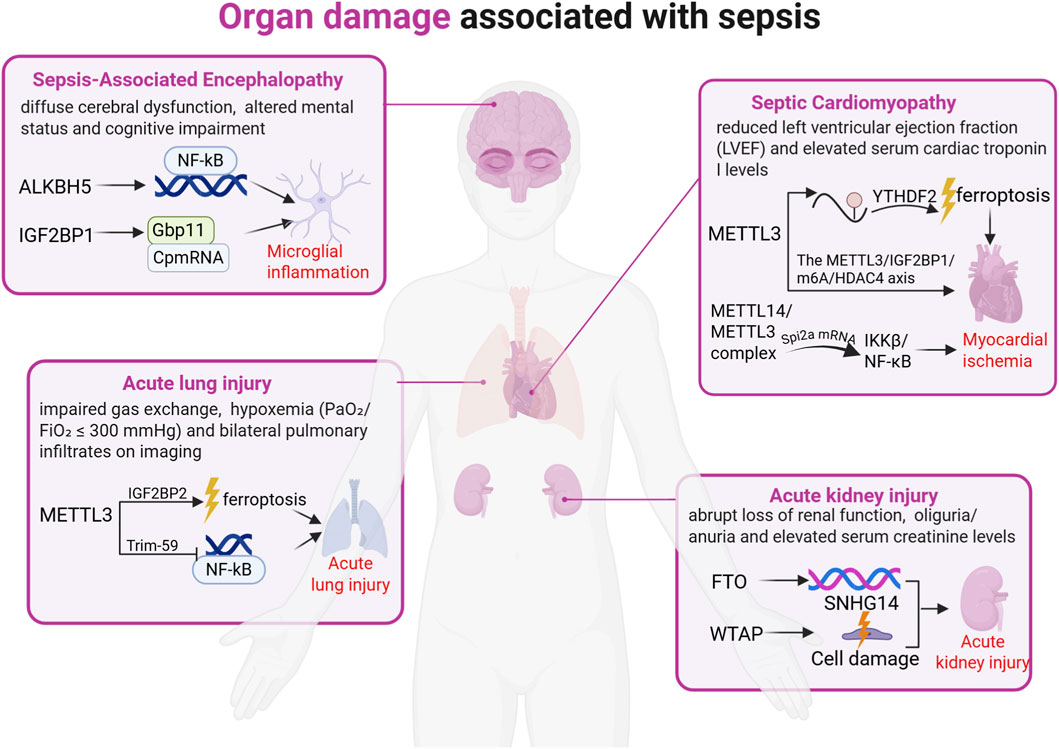
Figure 2. Organ damage associated with sepsis. Severe sepsis is frequently accompanied by organ damage, primarily involving injury to the heart, lungs, kidneys, and brain. Research indicates that sepsis-induced organ damage is driven by aberrant RNA modifications and their regulatory factors, manifested as endothelial cell injury and ferroptosis. The figure above illustrates the role of m6A modification in organ damage during sepsis.
Furthermore, Hotchkiss’ proposition of sepsis as an immunological disorder is supported by autopsy evidence demonstrating immune-cell depletion via apoptosis in deceased patients (Hotchkiss et al., 2013b). Importantly, m6A dysregulation impairs macrophage phagocytic function and disrupts neutrophil chemotaxis, representing critical factors in septic pathology progression (Qian and Cao, 2022). Experimental studies in severe sepsis models indicate that YTHDF1 knockdown alleviates macrophage paralysis and endothelial damage. Mechanistically, YTHDF1 functions as an m6A reader that recognizes m6A modifications on JAK2/STAT3 mRNA and promotes its translation, thereby enhancing JAK-STAT signaling activity. When YTHDF1 is knocked down, its translational enhancement of JAK2/STAT3 mRNA is weakened, resulting in reduced JAK2/STAT3 protein expression (including phosphorylated forms) (Xing et al., 2021). Additionally, m6A methylation mediates negative regulation of serine protease inhibitor 2A (Spi2a) in macrophages, consequently inhibiting the release of pivotal pro-inflammatory cytokines such as tumor necrosis factor-α(TNF-α) and interleukin-6(IL-6), which are central to septic inflammatory cascades (Wang et al., 2023).
4 Role of m6A methylation in sepsis-induced organ dysfunction
MODS represents a severe dysregulated systemic inflammatory state triggered by sepsis, and is characterized by progressive functional deterioration or failure of two or more vital organ systems (Shi et al., 2019) (e.g., heart, lungs, kidneys). As the terminal stage of sepsis, MODS carries a mortality rate of 28%–56% upon diagnosis (Zou et al., 2022). Emerging evidence indicates that m6A methylation modulates sepsis progression through multiple pathways, playing a pivotal role in MODS development (Shen et al., 2023; Zhang S. et al., 2022) (Figure 3). Consequently, elucidating m6A’s functions in sepsis-induced organ dysfunction is crucial for optimizing clinical management and developing novel therapeutics.
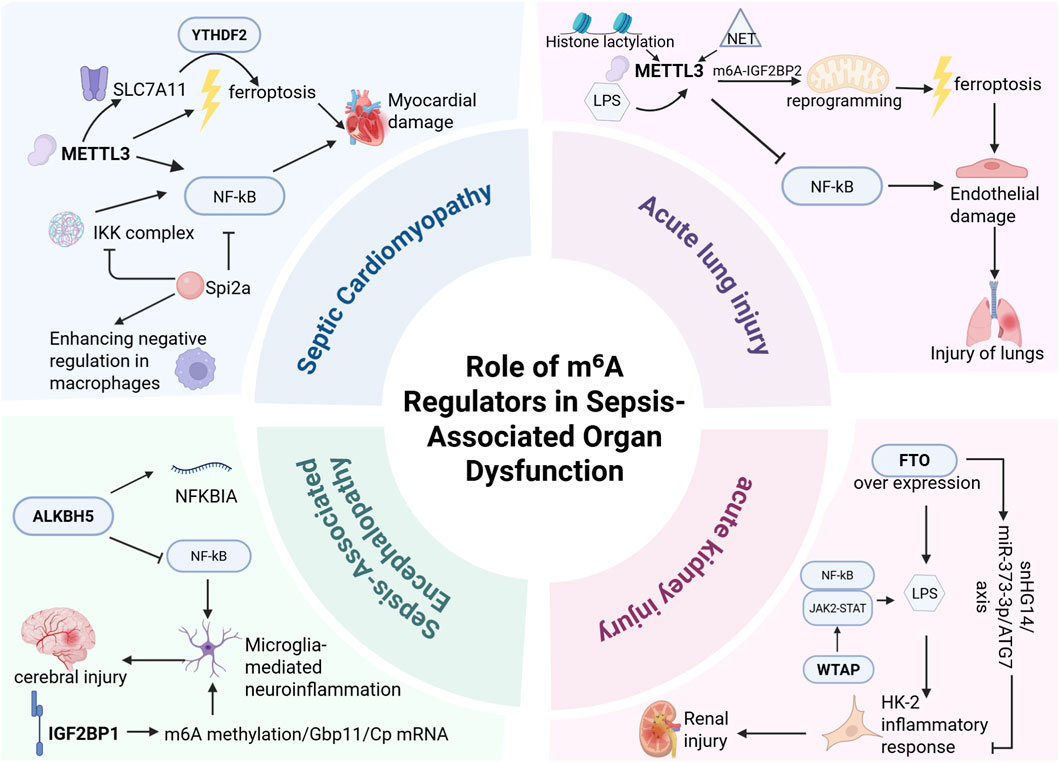
Figure 3. m6A modification plays a critical role in sepsis-associated organ injury, primarily involving sepsis-induced cardiomyopathy (SCM), acute lung injury (ALI), sepsis-associated encephalopathy (SAE), and acute kidney injury (AKI). In SCM, METTL3 exacerbates multi-organ dysfunction by promoting cardiomyocyte ferroptosis and NF-κB activation. In ALI, METTL3 augments m6A-IGF2BP2-dependent mitochondrial metabolic reprogramming to intensify ferroptosis while simultaneously regulating endothelial function through Trim59-mediated NF-κB inactivation, demonstrating high diagnostic and therapeutic value. In SAE, ALKBH5 inhibits NF-κB pathway activation to mitigate microglia-mediated neuroinflammation; IGF2BP1 may regulate microglial inflammatory responses by enhancing m6A methylation and stabilizing Gbp11/Cp mRNAs, emerging as a potential therapeutic target for microglial hyperactivation. In AKI, FTO ameliorates renal injury by suppressing autophagy, reducing RNA stability, and downregulating SNHG14 expression, whereas WTAP promotes LPS-induced inflammation and renal damage in HK-2 cells via NF-κB and JAK2/STAT3 pathway regulation. These findings highlight the significance of m6A regulators as potential therapeutic targets for combating sepsis-induced organ damage.
4.1 m6A methylation and myocardial injury
Septic cardiomyopathy (SCM), a non-ischemic cardiac dysfunction in sepsis, features impaired left/right ventricular systolic or diastolic function, accompanied by cardiomyocyte damage and inflammation-driven pathophysiological alterations (Beesley et al., 2018). Inflammatory cytokines (e.g., IL-6, TNF-α) directly induce cardiomyocyte dysfunction through oxidative stress, calcium mishandling, and mitochondrial damage, leading to hemodynamic instability—manifested as tachycardia, reduced cardiac output, and impaired contractility. These changes exacerbate myocardial ischemia-hypoxia, creating a vicious cycle of injury (Bi et al., 2023).
Recent studies reveal that m6A methylation regulates septic myocardial injury by modulating inflammation and apoptosis. Wang et al. demonstrated METTL3’s protective role in murine sepsis models, where METTL3 inhibition exacerbated multi-organ damage (Wang et al., 2024). Shen et al. further validated METTL3’s interaction with solute carrier family seven member 11 (Slc7a11) via RIP-qPCR and MeRIP-qPCR, showing elevated METTL3 expression and methylation levels in LPS-treated rat cardiomyocytes. METTL3 promotes Slc7a11 mRNA degradation through m6A-dependent mechanisms, intensifying sepsis-induced cardiomyocyte ferroptosis—an iron-dependent, lipid per oxidation-mediated cell death strongly implicated in sepsis pathogenesis. This establishes the METTL3/YTHDF2/Slc7a11 axis as central to septic myocardial injury (Shen et al., 2023).
Supporting this, Tang’s team found that METTL3 silencing suppressed ferroptosis in septic rat cardiomyocytes via Slc7a11 m6A methylation (Tang et al., 2024). Complementarily, Zhang reported that in a mouse model of sepsis, YTHDF1 can inhibit pyroptosis of cells and alleviate the damage caused by sepsis by promoting the ubiquitination of NLRP3 and upregulating the WW domain-containing E3 ubiquitin ligase 1 (Wwp1) (Zhang S. et al., 2022). Wang et al. Identified Spi2a as a novel negative feedback regulator that suppresses cytokine production and myocardial injury in macrophages post LPS challenge by inhibiting inhibitor of kappa B kinase (IKK) complex formation and NF-κB activation. Critically, they proved Spi2a’s m6A methylation sustains macrophage feedback control. Through comprehensive experiments (on cellular, animal, molecular, and clinical specimens), According to Wang et al., the METTL3/METTL14 complex synergistically enhances Spi2a mRNA stability and translation through m6A modification. METTL3 provides the catalytic activity for methylation, while METTL14 stabilizes the complex and enhances substrate recognition. The m6A-modified Spi2a mRNA is then recognized by YTHDF1, which promotes its translational efficiency. This mechanism leads to increased SPI2A expression, subsequently suppressing IKKβ/NF-κB-mediated inflammation (Wang et al., 2023). This indicates that m6A orchestrates SCM pathology at multicellular levels through distinct targets (e.g.,.Spi2a in macrophages), uncovering novel therapeutic avenues that target m6A modifiers (e.g., METTL3, METTL14, or SPI2A). Additionally, Shen et al. implicated METTL3 in septic rat myocardial injury via the METTL3/IGF2BP1/m6A/HDAC4 axis (Shen et al., 2022). Collectively, METTL3 and YTHDF1 emerge as promising diagnostic and therapeutic targets.
4.2 m6A methylation and lung injury
The lungs are highly susceptible to sepsis, and acute respiratory distress syndrome (ARDS) and acute lung injury (ALI) serve as critical prognostic indicators (Wu et al., 2024). ARDS affects 10.4% of ICU patients and 23.4% of mechanically ventilated cases, with overall mortality at 40% (mild: 34.9%; moderate: 40.3%; severe: 46.1%) (Wick et al., 2024). Pathologically, ARDS/ALI features endothelial damage and dysregulated innate immunity. Polymorphonuclear neutrophils (PMNs) and platelets play pivotal roles: recruited PMNs eliminate pathogens via degranulation, phagocytosis, and neutrophil extracellular trap (NET) formation. NETs—extracellular networks of DNA, histones, myeloperoxidase (MPO), cathepsin G, and antimicrobial proteins—neutralize pathogens but paradoxically propagate inflammation and tissue damage when overproduced (Silva et al., 2021; Ma et al., 2017). Studies indicate that enhanced formation of NETs in sepsis-associated ALI/ARDS activates METTL3-mediated m6A modification in alveolar epithelial cells, which regulates the stability of HIF-1α, thereby inducing mitochondrial metabolic reprogramming and ferroptosis, ultimately leading to lung injury.
Mounting evidence implicates METTL3 in sepsis-induced ALI. Ferroptosis (Zhang H. et al., 2023; Lai et al., 2025)—an iron-dependent cell death driven by uncontrolled lipid peroxidation—emerges as a key mechanism (Jiang et al., 2021b). Zhang et al. demonstrated elevated NETs in cecal ligation and puncture (CLP)-induced ALI mice, and showed that NET inhibitors reversed ferroptosis. Integrated RNA-seq and MeRIP-seq revealed that NET-induced METTL3 upregulation exacerbates ferroptosis in alveolar epithelium via m6A-Insulin Like Growth Factor 2 MRNA Binding Protein 2(IGF2BP2)-dependent mitochondrial metabolic reprogramming, thereby offering therapeutic targets to mitigate lung injury and systemic inflammation (Zhang H. et al., 2023). Experiments in Zhang et al.'s laboratory further corroborated this phenomenon (Zhang H. et al., 2022). Chen et al. initially detected reduced global m6A levels in septic patients through colorimetric ELISA assays. Subsequent Western blotting analysis revealed significantly diminished METTL3 expression in the lung tissues of these patients compared to healthy controls, suggesting a potential association between METTL3 dysregulation and sepsis pathogenesis. The team conducted in vivo experiments using METTL3-knockdown murine models versus wild-type counterparts, and demonstrated that METTL3 deficiency exacerbated endothelial barrier disruption, amplified sepsis-induced inflammatory responses, and consequently aggravated pulmonary injury. For in vitro validation, they employed transfection techniques to inhibit METTL3 in LPS-stimulated HULEC-5a cells across multiple time points, and observed impaired endothelial permeability and intensified barrier dysfunction. Furthermore, METTL3 was found to modulate endothelial function in sepsis-induced acute lung injury by inactivating NF-κB through Tripartite Motif Containing 59 (Trim59)-mediated mechanisms (Chen et al., 2022).
Wu et al. validated that histone lactylation induces METTL3-mediated m6A modification to promote ferroptosis (Wu et al., 2024), identifying METTL3 targeting as a viable strategy against septic lung injury. Notably, this regulatory axis exemplifies a broader and highly significant “epigenetic hierarchical network” in sepsis pathogenesis, where upstream histone post-translational modifications (PTMs)orchestrate downstream RNA epigenetic modifications (like m6A) to coordinately amplify the inflammatory response. For instance, metabolic reprogramming during sepsis leads to lactate accumulation, which drives histone lactylation to upregulate METTL3 expression; the increased METTL3 then deposits m6A marks on pro-inflammatory transcripts, enhancing their stability and translation efficiency and further fueling inflammation and lactate production. This creates a positive feed-forward loop that potently exacerbates the cytokine storm and organ damage. Recognizing such multi-layered epigenetic crosstalk not only deepens our mechanistic understanding of septic inflammation but also opens new avenues for therapeutic intervention. Similarly,Lai et al. established an LPS-stimulated human pulmonary microvascular endothelial-cell (HPMEC) model showing that METTL14/IGF2BP2-mediated m6A modification of STEAP1 aggravates ALI (62). Complementarily, Xian et al. reported macrophage NLRP3 inflammasome hyperactivation during ALI/ARDS progression (Xian et al., 2021; Cao et al., 2024). Building on this, Cao’s team identified Nlrp3 as a METTL14 target. They demonstrated that knockdown of IGF2BP2 reduces LPS-induced ALI by downregulating Nlrp3 expression, achieved through a decrease in Nlrp3 transcript stability and inhibition of the Nlrp3 inflammasome activation, thereby highlighting METTL14’s therapeutic potential. Collectively, these findings have transformative potential for advancing diagnostic biomarkers, therapeutic strategies, and prognostic evaluation in sepsis management (Cao et al., 2024).
4.3 m6A methylation and brain injury
Sepsis-associated encephalopathy (SAE), a frequent neurological complication of sepsis, manifests as brain dysfunction and neuronal damage during systemic inflammation, characterized by delirium, disturbances of consciousness, and cognitive impairment (Bircak-Kuchtova et al., 2023). Emerging evidence implicates m6A methyltransferases in SAE pathogenesis (Ye et al., 2025; Wang H. et al., 2022; Ding et al., 2022; Li et al., 2021; Mittal and Coopersmith, 2014). Wang and colleagues detected serum markers using the enzyme-linked immunosorbent assay method. They found that compared with non-SAE patients, the expression of METTL3 was significantly increased in SAE patients, while the expression of FTO was significantly decreased. (Wang H. et al., 2022).
While microglia in the resting state primarily maintain normal central nervous system function, their excessive activation may contribute to the onset and pathology of SAE. Ye et al. demonstrated through both mechanistic and clinical validation that in a murine model of sepsis, ALKBH5-mediated m6A demethylation stabilizes NF-κB inhibitor alpha (Nfkbia) mRNA, thereby elevating NFKBIA protein levels, suppressing p65 phosphorylation and nuclear translocation, inhibiting the NF-κB signaling pathway, and ultimately alleviating microglia-mediated neuroinflammation; furthermore, in human sepsis patient samples, ALKBH5 expression was found to correlate with disease severity. (Ye et al., 2025). Complementarily, Ding et al. identified IGF2BP1 as a regulator of microglial inflammation in mouse primary microglia through m6A-dependent stabilization of Guanylate Binding Protein 1 (Gbp11) and Cp mRNAs. They proposed IGF2BP1 inhibition as a strategy to mitigate microglial hyperactivation (Ding et al., 2022). Li et al., using primary microglia isolated from newborn (<24 h) Sprague-Dawley (SD) rat brains, further mapped differential m6A modifications in M0-like (resting), M1-like (pro-inflammatory), and M2-like (anti-inflammatory) microglial subtypes, establishing m6A as a key modulator during microglial immune responses. (Li et al., 2021). These collective findings underscore m6A’s role in regulating microglial inflammatory states, and clarify its direct impact on SAE progression and outcomes.
Intriguingly, Wang et al. integrated LC-MS/MS metabolomics and 16S rDNA sequencing to identify gut microbiota dysbiosis in SAE, and detected the expression of serum markers and IL-6 by enzyme-linked immunosorbent assay (ELISA). Comparative analysis of gut microbiota between SAE and non-SAE cohorts revealed a positive correlation between Acinetobacter abundance and METTL3 upregulation. This indicated that targeted METTL3 modulation could restore microbial homeostasis, thereby ameliorating or even therapeutically resolving SAE pathology (Wang H. et al., 2022).
4.4 m6A methylation and kidney injury
Acute kidney injury (AKI) frequently complicates sepsis through pathological mechanisms including microcirculatory dysfunction, dysregulated immune responses, coagulation activation, and renal tubular epithelial damage (Adedoyin et al., 2018). Clinically manifested as abrupt loss of kidney function with oliguria and elevated serum creatinine, sepsis associated-acute kidney injury (SA-AKI) affects >40% of septic patients and represents a major independent risk factor for ICU mortality (Martin et al., 2016; Naime et al., 2018; Alanazi et al., 2024). Current therapeutic strategies—including antimicrobial therapy, fluid resuscitation, vasoactive agents, and renal replacement therapy—demonstrate limited efficacy. Emerging research implicates m6A methylation in regulating ferroptosis during AKI pathogenesis, with METTL14 appearing to be a pivotal regulator of ferroptosis in renal disease progression (Adedoyin et al., 2018; Yang et al., 2024).
Small nucleolar RNA host gene 14(SNHG14) exacerbates renal injury by activating microglia and modulating the miR-373-3p/ATG7 axis in LPS-stimulated HK-2 cells (Yang et al., 2024; Yang N. et al., 2021; Tan et al., 2020). Yang et al. demonstrated that FTO confers nephroprotection in sepsis patients with acute kidney injury (AKI) by suppressing autophagy through RNA destabilization and reduced SNHG14 expression, thus mitigating LPS-induced renal damage. (Yang et al., 2024). Huang et al., using an AKI mouse model established by cecal ligation and puncture (CLP) and an AKI cell model established by treating HK-2 cells with LPS, reported that Wtap knockdown promotes inflammation, ferroptosis, and cellular injury in LPS-treated HK-2 cells by upregulating lamin B1 (Lmnb1) expression while activating NF-κB and JAK2/STAT3 signaling pathways. (Huang et al., 2024). Complementary to these findings, Xu et al., using TCMK-1 cells to establish in vitro AKI models and LPS-treated mice for in vivo AKI models, observed rapid m6A elevation in LPS-challenged murine renal epithelial (TCMK-1) cells. Notably, METTL14 knockdown counteracts LPS-aggravated ferroptosis in these in vivo murine models. (Xu L. et al., 2025).
Collectively, inhibition of METTL14 alleviates both renal injury and ferroptosis in LPS-induced AKI, establishing m6A methylation as a pivotal therapeutic target for future AKI interventions.
4.5 m6A methylation and other organ injuries
The liver critically regulates systemic immune responses during sepsis by means of bacterial clearance, cytokine production, and metabolic adaptations to inflammation (Sun et al., 2020). However, sepsis-induced ischemic hepatic injury, shock-related damage, and secondary sclerosing cholangitis collectively establish the liver as a primary target of sepsis-mediated secondary injury (Strnad et al., 2017). As an independent predictor of ICU outcomes, identifying therapeutic targets for septic liver injury is imperative (Wang et al., 2025). Sun et al. demonstrated that in septic mice,IGF2BP3 interacts with GLI family zinc finger 2 (GLI2) mRNA to stabilize m6A-modified transcripts. Upregulated Gli2 transcriptionally promotes synoviolin 1 (Syvn1) expression, which subsequently enhances degradation of peroxisome proliferator-activated receptor alpha (PPARα). This cascade ultimately exacerbates septic liver injury both in vitro and in vivo by suppressing PPARα-mediated autophagy, establishing the IGF2BP3/GLI2/Syvn1/PPARα axis as a potential therapeutic target (Sun et al., 2024).
In summary, current research demonstrates that m6A RNA methylation—orchestrated through the dynamic interplay of Writers, Erasers, and Readers—precisely regulates key signaling pathways involved in inflammation, apoptosis, and autophagy. This epigenetic mechanism serves as a central driver of inflammatory amplification, tissue-barrier disruption, and cellular dysfunction during sepsis-induced secondary organ injury (Table 2). These findings establish critical targets and pathways for therapeutic intervention while opening novel directions for clinical translation.
5 Conclusion and perspectives
Sepsis-induced multi-organ injury involves complex pathogenic networks. This review has examined mechanisms underlying sepsis-mediated organ damage and delineated the regulatory roles of m6A methylation: Writer, Eraser, and Reader proteins participate dynamically in critical biological processes by post-transcriptionally modulating cellular gene expression, thereby propagating secondary organ injury. These modifications influence RNA fate through splicing, transport, translation, stabilization, and degradation, profoundly impacting sepsis progression.
mRNA methylation and its regulators exhibit broad biological functions. Notably, certain regulators such as METTL3/YTHDF2 exacerbate cellular damage by amplifying inflammatory pathways, while FTO/ALKBH5 confer protective effects by destabilizing pro-inflammatory cytokine mRNAs. Interactions with non-coding RNAs further form regulatory networks influencing sepsis progression. These discoveries provide novel therapeutic insights into organ-specific damage in sepsis. The therapeutic potential of targeting the m6A machinery could be realized through several strategic approaches: 1) Developing small-molecule inhibitors against “Writer” complexes (e.g., METTL3/METTL14) or “Erasers” (e.g., FTO, ALKBH5) to globally reduce or selectively reshape the m6A epitranscriptome; 2) Designing compounds that disrupt the interaction between specific “Reader” proteins (e.g., YTHDF2) and their pro-inflammatory target mRNAs, offering a more precise intervention; 3) Exploiting upstream regulatory cues, such as modulating the lactate-induced histone lactylation that drives METTL3 expression, to indirectly influence m6A deposition; 4) Exploring combination therapies where m6A-targeting agents are used alongside conventional antibiotics or specific pathway inhibitors to achieve synergistic effects and overcome immunosuppression.
We recognize that targeting ubiquitously expressed enzymes like METTL3 presents a specificity challenge, which is reflected in their context-dependent roles across different organs. For instance, METTL3 exacerbates injury in cardiomyocytes and alveolar epithelial cells by promoting ferroptosis, whereas in pulmonary endothelial cells and the gut, it exhibits protective effects by maintaining barrier integrity and modulating inflammatory responses. This apparent contradiction is not a paradox but can be explained by an emerging paradigm: m6A regulates sepsis through several evolutionarily conserved, cross-organ pathways—primarily by amplifying inflammatory signaling, programmed cell death, and metabolic reprogramming, which collectively drive the pathological process. The key insight is that the same pathway (e.g., NF-κB or ferroptosis) may produce opposing outcomes in different tissues due to cell-type-specific molecular targets. For example, METTL3-mediated m6A modification promotes NF-κB activation in macrophages (Wang et al., 2023), yet suppresses it in pulmonary endothelial cells via Trim59 (65). Similarly, while ferroptosis is universally pathogenic, its triggering mechanisms vary significantly. This paradigm reveals that the core pathways are shared, but the cellular context determines the final, organ-specific effects.
Our understanding of METTL3 and METTL14 in sepsis is currently confined to their m6A-dependent functions, this emerging paradigm from other fields highlights a critical, non-canonical dimension of their functionality. The findings by Dou et al. and Liu et al. provide a foundational rationale for hypothesizing that METTL3 may act as a transcriptional co-activator on inflammatory gene promoters, (Liu et al., 2021), while METTL14 may engage in direct chromatin regulation, as exemplified by its interaction with H3K27me3 and recruitment of KDM6B (Dou et al., 2023), provides a mechanistic precedent for METTL14 acting beyond the Methyltransferase Complex (MTC).
As a pivotal RNA modification, m6A methylation has garnered substantial research interest in sepsis-related organ injury in recent years. Despite extensive investigations into its roles in sepsis, the precise regulatory mechanisms remain incompletely elucidated, which presents ongoing challenges. Key knowledge gaps include: undefined interactions among m6A regulatory factors during sepsis; potential organ-specific regulatory factors within m6A networks that may explain injury heterogeneity (with such factors potentially serving as novel biomarkers for sepsis severity, organ-injury risk, and treatment prognosis); Translating these findings into clinical applications faces significant hurdles. Currently, no clinical trials are specifically investigating m6A-targeted therapies for sepsis or infectious diseases, underscoring the nascent stage of this field. The path to clinical translation is fraught with challenges, primarily due to the context-dependent nature of m6A function, which varies by cell type, pathological phase, and target gene, raising concerns about therapeutic specificity and potential off-target effects. Furthermore, achieving organ- or cell-selective drug delivery remains a major pharmacological bottleneck. Lastly, the essential physiological roles of m6A regulators necessitate a careful assessment of the safety profile and a narrow therapeutic window in critically ill septic patients. Crucially, most current conclusions are derived from murine and in vitro models, which means that clinical studies in sepsis patients are needed to validate the relationships between m6A dysregulation and secondary organ damage.
In summary, targeting m6A regulators holds great potential for sepsis diagnosis, treatment, and prognosis, yet comprehensive research remains essential to fully harness their therapeutic capabilities.
Author contributions
LZ: Investigation, Writing – review and editing, Visualization, Conceptualization, Formal Analysis, Writing – original draft. WC: Conceptualization, Writing – review and editing, Formal Analysis, Writing – original draft, Visualization, Investigation. YL: Conceptualization, Writing – review and editing, Investigation, Methodology. SZ: Investigation, Conceptualization, Writing – review and editing, Methodology. BY: Formal Analysis, Writing – review and editing, Methodology, Conceptualization. KL: Methodology, Investigation, Formal Analysis, Writing – review and editing. XG: Conceptualization, Validation, Supervision, Funding acquisition, Writing – review and editing. XH: Validation, Conceptualization, Supervision, Funding acquisition, Writing – review and editing.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This work was supported by the Science and technology Research program of Henan Province (NO 252102311125) and The Third HeLuo Youth Talent Support Project (2024HLTJ15).
Acknowledgments
AcknowledgementsThe figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adedoyin, O., Boddu, R., Traylor, A., Lever, J. M., Bolisetty, S., George, J. F., et al. (2018). Heme oxygenase-1 mitigates ferroptosis in renal proximal tubule cells. Am. J. Physiol. Ren. Physiol. 314 (5), F702-F714–f14. doi:10.1152/ajprenal.00044.2017
Alanazi, W. A., Alharbi, T., Bin Anzan, K. M., Alyahiya, M. K., El-Nagar, D. M., Alanazi, M. M., et al. (2024). The role of dapagliflozin in the modulation of hypothermia and renal injury caused by septic shock in euglycemic and hyperglycemic rat models. Curr. Mol. Pharmacol. 17, e18761429329635. doi:10.2174/0118761429329635241016054513
Alhammadi, M. A., Ilce, B. Y., Bhamidimarri, P. M., Bouzid, A., Ali, N., Alhamidi, R. S., et al. (2025). Analysis of genotype and expression of FTO and ALKBH5 in a MENA-region renal cell carcinoma cohort. Cancers (Basel) 17 (9), 1395. doi:10.3390/cancers17091395
An, Y., and Duan, H. (2022). The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21 (1), 14. doi:10.1186/s12943-022-01500-4
Arora, J., Mendelson, A. A., and Fox-Robichaud, A. (2023). Sepsis: network pathophysiology and implications for early diagnosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 324 (5), R613–R624. doi:10.1152/ajpregu.00003.2023
Beesley, S. J., Weber, G., Sarge, T., Nikravan, S., Grissom, C. K., Lanspa, M. J., et al. (2018). Septic cardiomyopathy. Crit. Care Med. 46 (4), 625–634. doi:10.1097/CCM.0000000000002851
Bi, C. F., Liu, J., Hu, X. D., Yang, L. S., and Zhang, J. F. (2023). Novel insights into the regulatory role of N6-methyladenosine methylation modified autophagy in sepsis. Aging (Albany NY) 15 (24), 15676–15700. doi:10.18632/aging.205312
Bircak-Kuchtova, B., Chung, H. Y., Wickel, J., Ehler, J., and Geis, C. (2023). Neurofilament light chains to assess sepsis-associated encephalopathy: are we on the track toward clinical implementation? Crit. Care 27 (1), 214. doi:10.1186/s13054-023-04497-4
Bollati, V., and Baccarelli, A. (2010). Environmental epigenetics. Hered. (Edinb) 105 (1), 105–112. doi:10.1038/hdy.2010.2
Borges, A., and Bento, L. (2024). Organ crosstalk and dysfunction in sepsis. Ann. Intensive Care 14 (1), 147. doi:10.1186/s13613-024-01377-0
Cao, F., Chen, G., Xu, Y., Wang, X., Tang, X., Zhang, W., et al. (2024). METTL14 contributes to acute lung injury by stabilizing NLRP3 expression in an IGF2BP2-dependent manner. Cell Death Dis. 15 (1), 43. doi:10.1038/s41419-023-06407-6
Cappannini, A., Ray, A., Purta, E., Mukherjee, S., Boccaletto, P., Moafinejad, S. N., et al. (2024). MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 52 (D1), D239–D244. doi:10.1093/nar/gkad1083
Chen, Y., Wu, Y., Zhu, L., Chen, C., Xu, S., Tang, D., et al. (2022). METTL3-Mediated N6-Methyladenosine modification of Trim59 mRNA protects against sepsis-induced acute respiratory distress syndrome. Front. Immunol. 13, 897487. doi:10.3389/fimmu.2022.897487
Dai, W., Qiao, X., Fang, Y., Guo, R., Bai, P., Liu, S., et al. (2024). Epigenetics-targeted drugs: current paradigms and future challenges. Signal Transduct. Target Ther. 9 (1), 332. doi:10.1038/s41392-024-02039-0
Dierks, D., Shachar, R., Nir, R., Garcia-Campos, M. A., Uzonyi, A., Wiener, D., et al. (2025). Passive shaping of intra- and intercellular m6A dynamics via mRNA metabolism. Elife 13. doi:10.7554/eLife.100448
Ding, L., Wu, H., Wang, Y., Li, Y., Liang, Z., Xia, X., et al. (2022). m6A Reader Igf2bp1 regulates the inflammatory responses of microglia by stabilizing Gbp11 and Cp mRNAs. Front. Immunol. 13, 872252. doi:10.3389/fimmu.2022.872252
Dou, X., Huang, L., Xiao, Y., Liu, C., Li, Y., Zhang, X., et al. (2023). METTL14 is a chromatin regulator independent of its RNA N6-methyladenosine methyltransferase activity. Protein Cell 14 (9), 683–697. doi:10.1093/procel/pwad009
Furci, L., Berthelier, J., and Saze, H. (2024). RNA N6-adenine methylation dynamics impact Hyaloperonospora arabidopsidis resistance in Arabidopsis. Plant Physiol. 196 (2), 745–753. doi:10.1093/plphys/kiae373
Galle, E., Wong, C. W., Ghosh, A., Desgeorges, T., Melrose, K., Hinte, L. C., et al. (2022). H3K18 lactylation marks tissue-specific active enhancers. Genome Biol. 23 (1), 207. doi:10.1186/s13059-022-02775-y
Garcia-Alvarez, M., Marik, P., and Bellomo, R. (2014). Sepsis-associated hyperlactatemia. Crit. Care 18 (5), 503. doi:10.1186/s13054-014-0503-3
Ge, Y., Chen, R., Ling, T., Liu, B., Huang, J., Cheng, Y., et al. (2024). Elevated WTAP promotes hyperinflammation by increasing m6A modification in inflammatory disease models. J. Clin. Invest 134 (14), e177932. doi:10.1172/JCI177932
Gustot, T., Durand, F., Lebrec, D., Vincent, J. L., and Moreau, R. (2009). Severe sepsis in cirrhosis. Hepatology 50 (6), 2022–2033. doi:10.1002/hep.23264
Hotchkiss, R. S., Monneret, G., and Payen, D. (2013a). Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 13 (12), 862–874. doi:10.1038/nri3552
Hotchkiss, R. S., Monneret, G., and Payen, D. (2013b). Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect. Dis. 13 (3), 260–268. doi:10.1016/S1473-3099(13)70001-X
Huang, F., Wang, Y., Lv, X., and Huang, C. (2024). WTAP-mediated N6-methyladenosine modification promotes the inflammation, mitochondrial damage and ferroptosis of kidney tubular epithelial cells in acute kidney injury by regulating LMNB1 expression and activating NF-κB and JAK2/STAT3 pathways. J. Bioenerg. Biomembr. 56 (3), 285–296. doi:10.1007/s10863-024-10015-0
Ivanova, I., Much, C., Di Giacomo, M., Azzi, C., Morgan, M., Moreira, P. N., et al. (2017). The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67 (6), 1059–1067. doi:10.1016/j.molcel.2017.08.003
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021a). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6 (1), 74. doi:10.1038/s41392-020-00450-x
Jiang, X., Stockwell, B. R., and Conrad, M. (2021b). Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22 (4), 266–282. doi:10.1038/s41580-020-00324-8
Kapadia, B., Roychowdhury, A., Kayastha, F., Lee, W. S., Nanaji, N., Windle, J., et al. (2025). m6A eraser ALKBH5/treRNA1/DDX46 axis regulates BCR expression. Neoplasia 62, 101144. doi:10.1016/j.neo.2025.101144
Lai, J., Yu, S., Li, X., Wei, Q., and Qin, J. (2025). mettl14/igf2bp2-mediated m6a modification of steap1 aggravates acute lung injury induced by sepsis. Shock 63 (2), 217–225. doi:10.1097/SHK.0000000000002456
Li, Q., Wen, S., Ye, W., Zhao, S., and Liu, X. (2021). The potential roles of m(6)A modification in regulating the inflammatory response in microglia. J. Neuroinflammation 18 (1), 149. doi:10.1186/s12974-021-02205-z
Li, S., Yang, Q., Jiao, R., Xu, P., Sun, Y., and Li, X. (2022). m6A topological transition coupled to developmental regulation of gene expression during Mammalian tissue development. Front. Cell Dev. Biol. 10, 916423. doi:10.3389/fcell.2022.916423
Liu, P., Li, F., Lin, J., Fukumoto, T., Nacarelli, T., Hao, X., et al. (2021). m(6)A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nat. Cell Biol. 23 (4), 355–365. doi:10.1038/s41556-021-00656-3
Luo, J., Xu, T., and Sun, K. (2021a). N6-Methyladenosine RNA modification in inflammation: roles, mechanisms, and applications. Front. Cell Dev. Biol. 9, 670711. doi:10.3389/fcell.2021.670711
Luo, J., Wang, F., Sun, F., Yue, T., Zhou, Q., Yang, C., et al. (2021b). Targeted inhibition of FTO demethylase protects mice against LPS-Induced septic shock by suppressing NLRP3 inflammasome. Front. Immunol. 12, 663295. doi:10.3389/fimmu.2021.663295
Ma, X. L., Song, F. F., Zhang, H., Huan, X., and Li, S. Y. (2017). Compositional monosaccharide analysis of Morus nigra Linn by HPLC and HPCE quantitative determination and comparison of polysaccharide from Morus nigra Linn by HPCE and HPLC. Curr. Pharm. Anal. 13 (5), 433–437. doi:10.2174/1573412913666170330150807
Martin, L., Koczera, P., Zechendorf, E., and Schuerholz, T. (2016). The endothelial glycocalyx: new diagnostic and therapeutic approaches in sepsis. Biomed. Res. Int. 2016, 3758278. doi:10.1155/2016/3758278
Mittal, R., and Coopersmith, C. M. (2014). Redefining the gut as the motor of critical illness. Trends Mol. Med. 20 (4), 214–223. doi:10.1016/j.molmed.2013.08.004
Naime, A. C. A., Ganaes, J. O. F., and Lopes-Pires, M. E. (2018). Sepsis: the involvement of platelets and the Current treatments. Curr. Mol. Pharmacol. 11 (4), 261–269. doi:10.2174/1874467211666180619124531
Nedeva, C. (2021). Inflammation and cell death of the innate and adaptive immune System during sepsis. Biomolecules 11 (7), 1011. doi:10.3390/biom11071011
Oerum, S., Meynier, V., Catala, M., and Tisné, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49 (13), 7239–7255. doi:10.1093/nar/gkab378
Qian, W., and Cao, Y. (2022). An overview of the effects and mechanisms of m6 A methylation on innate immune cells in sepsis. Front. Immunol. 13, 1041990. doi:10.3389/fimmu.2022.1041990
Ricci, Z., Polito, A., Polito, A., and Ronco, C. (2011). The implications and management of septic acute kidney injury. Nat. Rev. Nephrol. 7 (4), 218–225. doi:10.1038/nrneph.2011.15
Rudd, K. E., Johnson, S. C., Agesa, K. M., Shackelford, K. A., Tsoi, D., Kievlan, D. R., et al. (2020). Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the global Burden of Disease Study. Lancet 395 (10219), 200–211. doi:10.1016/S0140-6736(19)32989-7
Shen, H., Xie, K., Li, M., Yang, Q., and Wang, X. (2022). N(6)-methyladenosine (m(6)A) methyltransferase METTL3 regulates sepsis-induced myocardial injury through IGF2BP1/HDAC4 dependent manner. Cell Death Discov. 8 (1), 322. doi:10.1038/s41420-022-01099-x
Shen, H., Xie, K., Tian, Y., and Wang, X. (2023). N6-methyladenosine writer METTL3 accelerates the sepsis-induced myocardial injury by regulating m6A-dependent ferroptosis. Apoptosis 28 (3-4), 514–524. doi:10.1007/s10495-022-01808-y
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: Context-Dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Silva, C. M. S., Wanderley, C. W. S., Veras, F. P., Sonego, F., Nascimento, D. C., Gonçalves, A. V., et al. (2021). Gasdermin D inhibition prevents multiple organ dysfunction during sepsis by blocking NET formation. Blood 138 (25), 2702–2713. doi:10.1182/blood.2021011525
Singer, M., Deutschman, C. S., Seymour, C. W., Shankar-Hari, M., Annane, D., Bauer, M., et al. (2016). The third international consensus definitions for sepsis and septic shock (Sepsis-3). Jama 315 (8), 801–810. doi:10.1001/jama.2016.0287
Song, B., Zeng, Y., Cao, Y., Zhang, J., Xu, C., Pan, Y., et al. (2023). Emerging role of METTL3 in inflammatory diseases: mechanisms and therapeutic applications. Front. Immunol. 14, 1221609. doi:10.3389/fimmu.2023.1221609
Strick, A., von Hagen, F., Gundert, L., Klümper, N., Tolkach, Y., Schmidt, D., et al. (2020). The N(6) -methyladenosine (m(6) A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 125 (4), 617–624. doi:10.1111/bju.15019
Strnad, P., Tacke, F., Koch, A., and Trautwein, C. (2017). Liver - guardian, modifier and target of sepsis. Nat. Rev. Gastroenterol. Hepatol. 14 (1), 55–66. doi:10.1038/nrgastro.2016.168
Sun, J., Zhang, J., Wang, X., Ji, F., Ronco, C., Tian, J., et al. (2020). Gut-liver crosstalk in sepsis-induced liver injury. Crit. Care 24 (1), 614. doi:10.1186/s13054-020-03327-1
Sun, C., Gao, M., Hu, H., Qi, J., Tang, Y., Cao, X., et al. (2024). IGF2BP3 modified GLI2 transcriptionally regulates SYVN1 and facilitates sepsis liver injury through autophagy. iScience 27 (6), 109870. doi:10.1016/j.isci.2024.109870
Tan, J., Fan, J., He, J., Zhao, L., and Tang, H. (2020). Knockdown of LncRNA DLX6-AS1 inhibits HK-2 cell pyroptosis via regulating miR-223-3p/NLRP3 pathway in lipopolysaccharide-induced acute kidney injury. J. Bioenerg. Biomembr. 52 (5), 367–376. doi:10.1007/s10863-020-09845-5
Tang, Z., Huang, X., Mei, H., and Zheng, Z. (2024). Silencing of METTL3 suppressed ferroptosis of myocardial cells by m6A modification of SLC7A11 in a YTHDF2 manner. J. Bioenerg. Biomembr. 56 (2), 149–157. doi:10.1007/s10863-024-10006-1
Wang, W., and Liu, C. F. (2023). Sepsis heterogeneity. World J. Pediatr. 19 (10), 919–927. doi:10.1007/s12519-023-00689-8
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, B., Liu, Y., Jiang, R., Liu, Z., Gao, H., Chen, F., et al. (2022a). Emodin relieves the inflammation and pyroptosis of lipopolysaccharide-treated 1321N1 cells by regulating methyltransferase-like 3 -mediated NLR family pyrin domain containing 3 expression. Bioengineered 13 (3), 6740–6749. doi:10.1080/21655979.2022.2045836
Wang, H., Wang, Q., Chen, J., and Chen, C. (2022b). Association among the gut microbiome, the serum metabolomic profile and RNA m(6)A methylation in sepsis-associated encephalopathy. Front. Genet. 13, 859727. doi:10.3389/fgene.2022.859727
Wang, X., Ding, Y., Li, R., Zhang, R., Ge, X., Gao, R., et al. (2023). N(6)-methyladenosine of Spi2a attenuates inflammation and sepsis-associated myocardial dysfunction in mice. Nat. Commun. 14 (1), 1185. doi:10.1038/s41467-023-36865-7
Wang, S., Shen, S., Cheng, N., Zhou, W., Yu, W., Liang, D., et al. (2024). The role of m6A methylation genes in predicting poor prognosis in sepsis: identifying key biomarkers and therapeutic targets. Eur. J. Med. Res. 29 (1), 608. doi:10.1186/s40001-024-02194-8
Wang, B., Wu, X., Cheng, J., Ye, J., Zhu, H., and Liu, X. (2025). Regulatory role of S1P and its receptors in sepsis-induced liver injury. Front. Immunol. 16, 1489015. doi:10.3389/fimmu.2025.1489015
Wei, G. (2024). RNA m6A modification, signals for degradation or stabilisation? Biochem. Soc. Trans. 52 (2), 707–717. doi:10.1042/BST20230574
Wheeler, A. P., and Bernard, G. R. (1999). Treating patients with severe sepsis. N. Engl. J. Med. 340 (3), 207–214. doi:10.1056/NEJM199901213400307
Wick, K. D., Ware, L. B., and Matthay, M. A. (2024). Acute respiratory distress syndrome. Bmj 387, e076612. doi:10.1136/bmj-2023-076612
Wu, D., Spencer, C. B., Ortoga, L., Zhang, H., and Miao, C. (2024). Histone lactylation-regulated METTL3 promotes ferroptosis via m6A-modification on ACSL4 in sepsis-associated lung injury. Redox Biol. 74, 103194. doi:10.1016/j.redox.2024.103194
Wu, H., Lai, G. Q., Cheng, R., Huang, H., Wang, J., Liu, Z., et al. (2025). Discovery of covalent and cell-active ALKBH5 inhibitors with potent antileukemia effects in vivo. Angew. Chem. Int. Ed. Engl. 64 (18), e202424928. doi:10.1002/anie.202424928
Xian, H., Liu, Y., Rundberg Nilsson, A., Gatchalian, R., Crother, T. R., Tourtellotte, W. G., et al. (2021). Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation. Immunity 54 (7), 1463–77.e11. doi:10.1016/j.immuni.2021.05.004
Xing, Y., Cheng, D., Shi, C., and Shen, Z. (2021). The protective role of YTHDF1-knock down macrophages on the immune paralysis of severe sepsis rats with ECMO. Microvasc. Res. 137, 104178. doi:10.1016/j.mvr.2021.104178
Xiong, J., He, J., Zhu, J., Pan, J., Liao, W., Ye, H., et al. (2022). Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 82 (9), 1660–77.e10. doi:10.1016/j.molcel.2022.02.033
Xu, K., Mo, Y., Li, D., Yu, Q., Wang, L., Lin, F., et al. (2020). N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic Dis. 11, 2040622320916024. doi:10.1177/2040622320916024
Xu, Z., Sun, B., Wang, W., Fan, Y., Su, J., Sun, J., et al. (2025a). Research progress on m6A and drug resistance in gastrointestinal tumors. Front. Pharmacol. 16, 1565738. doi:10.3389/fphar.2025.1565738
Xu, L., Wang, Q. J., Nie, M. X., and Chen, Z. F. (2025b). Methyltransferase-like 14 promotes ferroptosis in sepsis-induced acute kidney injury via increasing the m6A methylation modification of LPCAT3. Mol. Genet. Genomics 300 (1), 16. doi:10.1007/s00438-024-02219-1
Yang, B., Wang, J. Q., Tan, Y., Yuan, R., Chen, Z. S., and Zou, C. (2021a). RNA methylation and cancer treatment. Pharmacol. Res. 174, 105937. doi:10.1016/j.phrs.2021.105937
Yang, N., Wang, H., Zhang, L., Lv, J., Niu, Z., Liu, J., et al. (2021b). Long non-coding RNA SNHG14 aggravates LPS-induced acute kidney injury through regulating miR-495-3p/HIPK1. Acta Biochim. Biophys. Sin. (Shanghai) 53 (6), 719–728. doi:10.1093/abbs/gmab034
Yang, N., Yan, N., Bai, Z., Du, S., Zhang, J., Zhang, L., et al. (2024). FTO attenuates LPS-induced acute kidney injury by inhibiting autophagy via regulating SNHG14/miR-373-3p/ATG7 axis. Int. Immunopharmacol. 128, 111483. doi:10.1016/j.intimp.2023.111483
Ye, C., Huang, X., Tong, Y., Chen, Y., Zhao, X., Xie, W., et al. (2025). Overexpression of ALKBH5 alleviates LPS induced neuroinflammation via increasing NFKBIA. Int. Immunopharmacol. 144, 113598. doi:10.1016/j.intimp.2024.113598
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20 (10), 608–624. doi:10.1038/s41580-019-0168-5
Zhang, S., Liu, F., Wu, Z., Xie, J., Yang, Y., and Qiu, H. (2020a). Contribution of m6A subtype classification on heterogeneity of sepsis. Ann. Transl. Med. 8 (6), 306. doi:10.21037/atm.2020.03.07
Zhang, H., Shi, X., Huang, T., Zhao, X., Chen, W., Gu, N., et al. (2020b). Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 48 (11), 6251–6264. doi:10.1093/nar/gkaa347
Zhang, S., Guan, X., Liu, W., Zhu, Z., Jin, H., Zhu, Y., et al. (2022a). YTHDF1 alleviates sepsis by upregulating WWP1 to induce NLRP3 ubiquitination and inhibit caspase-1-dependent pyroptosis. Cell Death Discov. 8 (1), 244. doi:10.1038/s41420-022-00872-2
Zhang, H., Liu, J., Zhou, Y., Qu, M., Wang, Y., Guo, K., et al. (2022b). Neutrophil extracellular traps mediate m(6)A modification and regulates sepsis-associated acute lung injury by activating ferroptosis in alveolar epithelial cells. Int. J. Biol. Sci. 18 (8), 3337–3357. doi:10.7150/ijbs.69141
Zhang, W., Jiang, H., Huang, P., Wu, G., Wang, Q., Luan, X., et al. (2023a). Dracorhodin targeting CMPK2 attenuates inflammation: a novel approach to sepsis therapy. Clin. Transl. Med. 13 (10), e1449. doi:10.1002/ctm2.1449
Zhang, H., Wu, D., Wang, Y., Guo, K., Spencer, C. B., Ortoga, L., et al. (2023b). METTL3-mediated N6-methyladenosine exacerbates ferroptosis via m6A-IGF2BP2-dependent mitochondrial metabolic reprogramming in sepsis-induced acute lung injury. Clin. Transl. Med. 13 (9), e1389. doi:10.1002/ctm2.1389
Zhang, T., Yang, H., Yu, Q., Zhang, Y., Zhang, Y., Zhu, X., et al. (2025). Dynamic, single-cell monitoring of RNA modifications response to viral infection using a genetically encoded live-cell RNA methylation sensor. Angew. Chem. Int. Ed. Engl. 64 (9), e202418003. doi:10.1002/anie.202418003
Zhu, L., Zhang, H., Zhang, X., and Xia, L. (2024). RNA m6A methylation regulators in sepsis. Mol. Cell Biochem. 479 (9), 2165–2180. doi:10.1007/s11010-023-04841-w
Keywords: m6A methylation, sepsis, organ injury, inflammatory response, RNA methylation
Citation: Zhang L, Chen W, Liu Y, Zhang S, Yin B, Liu K, Gu X and Hu X (2025) Research advances in m6A methylation and sepsis. Front. Cell Dev. Biol. 13:1682283. doi: 10.3389/fcell.2025.1682283
Received: 08 August 2025; Accepted: 04 November 2025;
Published: 18 November 2025.
Edited by:
Douglas Mark Ruden, Wayne State University, United StatesReviewed by:
Hailin Wang, Chinese Academy of Sciences (CAS), ChinaMilena Leseva, Stephan Angeloff Institute of Microbiology, Bulgaria
Copyright © 2025 Zhang, Chen, Liu, Zhang, Yin, Liu, Gu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinjun Hu, aHhqNTEyOUAxNjMuY29t; Xinyu Gu, aGtkZ3V4eUAxNjMuY29t
†These authors share first authorship
 Lifan Zhang
Lifan Zhang Wenjuan Chen1,2†
Wenjuan Chen1,2† Xinyu Gu
Xinyu Gu Xinjun Hu
Xinjun Hu