Abstract
Tendon injuries represent a significant clinical challenge in the treatment of musculoskeletal disorders due to their restricted intrinsic regenerative capacity and propensity for scar tissue formation. Tendon-derived mesenchymal stem cells (TD-MSCs) are considered a promising therapeutic approach because of their ability to differentiate into tenocytes, modulate inflammation, and secrete trophic factors that facilitate tendon regeneration. However, the molecular mechanisms underlying their effects and therapeutic advantages over other MSC sources remain unclear. This study aimed to elucidate the molecular basis of the therapeutic potential of TD-MSCs through a comprehensive transcriptomic comparison with bone marrow-derived MSCs (BM-MSCs) and evaluate their tenogenic differentiation capacity and regenerative efficacy. We isolated and characterized TD-MSCs and BM-MSCs using flow cytometry, tri-lineage differentiation assays, and proliferation assays (CCK-8) and examined their transcriptomic profiles via RNA sequencing. Subsequently, the tenogenic differentiation potential of TD-MSCs was evaluated in vitro by analyzing the expression of key markers (SCX, COL1, COL3, TN-C, TNMD, DCN, THBS-4, and SOX9) using quantitative reverse transcription-polymerase chain reaction, along with protein levels of SCX, COL1, and COL3 via immunofluorescence. The therapeutic efficacy of TD-MSC treatment was further tested in vivo using a rat model of Achilles tendon injury, with histological and immunohistochemical analyses performed for 6 weeks post-injection. The results showed that TD-MSCs exhibited superior proliferation and a distinct transcriptomic profile, with significantly elevated expression levels of tenogenic genes (COL1 and TN-C) compared to those observed in BM-MSCs. Following tenogenic induction, TD-MSCs showed enhanced differentiation capacity, with increased expression of tenogenic markers and downregulation of chondrogenic markers. In vivo treatment with TD-MSCs improved collagen fiber organization, enhanced structural integrity, and resulted in superior healing outcomes compared to untreated controls. These findings suggest that TD-MSCs possess intrinsic molecular advantages for tendon repair, characterized by enhanced tenogenic gene expression profiles relative to BM-MSCs and superior reparative potential both in vitro and in vivo. This study highlights the therapeutic potential of TD-MSCs for tendon regeneration and provides scientific evidence supporting tissue-specific MSC selection strategies for application in regenerative medicine.
1 Introduction
Tendon injuries represent a significant clinical challenge in musculoskeletal disorders (Leong et al., 2020; He et al., 2024), frequently leading to long-term disability and reduced quality of life. The natural healing process of tendons remains limited, often resulting in inferior mechanical properties that lead to chronic pain and functional impairment (Cottrell et al., 2016; He et al., 2024). Conventional therapeutic methods, including surgical intervention and rehabilitative therapy, typically achieve only partial restoration of native tendon structure and biomechanical function. Although these approaches may facilitate some degree of healing, the regenerated tissue generally lacks an organized extracellular matrix (ECM) and exhibits compromised mechanical strength compared with healthy tendons (Citro et al., 2025). These clinical limitations have driven the interest in regenerative medicine and cell-based therapies as promising options for improving tendon repair
Regenerative medicine approaches, particularly mesenchymal stem cell (MSC)-based therapies, have emerged as promising strategies for treating musculoskeletal disorders and tendinopathy (van den Boom et al., 2020; Mahmoud et al., 2022; Trapana et al., 2024). Many studies involving in vivo models have demonstrated that cell therapy effectively enhances tendon healing (Goldberg et al., 2024; Morya et al., 2024). MSCs demonstrate remarkable potential because of their ability to differentiate into tenocytes, modulate inflammation, and secrete trophic factors that support tissue repair (Costa-Almeida et al., 2019; Pittenger et al., 2019; Zayed et al., 2021; Citro et al., 2025). Importantly, MSCs from different anatomical sources retain a distinct molecular signature reflecting their developmental origins, a phenomenon termed “tissue memory” (Hass et al., 2011). Tissue-specific programming significantly influences regenerative potential via diverse gene expression profiles (Onizuka et al., 2020). Consequently, identifying a population of stem cells in the tendon tissue presents significant therapeutic potential for treating tendon injuries.
Tendon-derived MSCs (TD-MSCs) are clonogenic, multipotent, and capable of expressing both stem cell and tenogenic markers (Lui, 2015). TD-MSCs have been reported to have potential advantages over bone marrow-derived MSCs (BM-MSCs) for tendon repair, with TD-MSCs being more predisposed to tenogenic differentiation (Tan et al., 2012). At a molecular level, TD-MSCs are characterized by elevated expression of key tenogenic marker genes, including scleraxis (SCX), collagen type I (COL1), collagen type III (COL3), tenomodulin (TNMD), and tenascin-C (TN-C) (Tan et al., 2012; He et al., 2024). In addition, TD-MSCs exhibit stronger proliferation and tenogenic differentiation in co-culture with BM-MSCs (Wu et al., 2016). This inherent bias translates into a functionally greater capacity to form a tendon-like matrix in vitro. This superior molecular and functional profile enhances therapeutic outcomes in vivo. For instance, in a rat Achilles tendon rupture model, transplantation of TD-MSCs resulted in superior tendon repair compared to BM-MSCs, characterized by better-organized collagen fiber alignment, higher ultimate failure load, and increased COL1/COL3 expression (Al-Ani et al., 2015). These findings provide direct in vivo evidence that TD-MSCs can enhance tendon structure and biomechanical function, supporting their therapeutic relevance (Tan et al., 2012; Yea et al., 2023; He et al., 2024). However, the underlying molecular mechanisms driving this enhanced regenerative capacity remain poorly defined. Therefore, it is crucial to elucidate the molecular mechanisms via TD-MSCs that enhance their tenogenic potential for advancing clinical translation. Comprehensive transcriptomic analysis using RNA sequencing (RNA-seq) has emerged as a powerful approach for understanding stem cell biology and identifying the genes involved in specific biological processes (Wang et al., 2009). This technology enables the genome-wide characterization of gene expression patterns, providing an unbiased assessment of the molecular signatures that determine the regenerative therapeutic potential of MSCs.
We hypothesized that TD-MSCs would be naturally primed toward tendon regeneration and exhibit distinct gene expression signatures reflecting the enhanced activation of tenogenic regulatory pathways. To validate these predicted characteristics, we aimed to elucidate the molecular basis of the superior tenogenic potential of TD-MSCs by comprehensively comparing their transcriptome with that of BM-MSCs, a widely utilized MSC source for regenerative applications. We evaluated the therapeutic efficacy of TD-MSCs in a rat model of Achilles tendon injury. This integrated approach, which combines transcriptomic analysis with functional characterization, provides crucial mechanistic insights into specific tissue-derived stem cell therapies for tendon regeneration.
2 Materials and methods
2.1 Isolation and culture of MSCs
Sprague–Dawley pathogen-free seven-week-old male rats (n = 6, average weight 245 ± 16 g) were obtained from Samtako Co. (Samtako Bio, Korea) as MSC donors. The animals were maintained in a pathogen-free animal facility under standard laboratory conditions, with unrestricted access to food and water. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Jeonbuk National University, Republic of Korea (NON2024-093).
The animals were euthanized via cervical dislocation. TD-MSCs were isolated and cultured as described previously (Ning et al., 2015). In brief, the Achilles tendon was completely dissected, and the peritendinous connective tissue and bone-tendon junction were removed. Tendons were placed in sterile phosphate-buffered saline (PBS), minced, and digested with collagenase type I (Fujifilm Wako Chemical, Japan) for 2 h at 37 °C. The medium was filtered through a 70-µm strainer and centrifuged (2,000 rpm, 5 min, 4 °C). The final cell pellet was pooled and resuspended in filtered Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% antimycotic antibiotics and cultured at 37 °C in an atmosphere of 5% CO2. The cells were seeded at an initial density of 500 cells/cm2 in a 10 cm culture dish (Wu et al., 2020). Non-adherent cells were removed by changing the medium 48 h post-seeding. Subsequently, adherent cells were cultured to 80% confluence. Cells were trypsinized using 0.25% trypsin–EDTA and subcultured at a standard density of 5,000 cells/cm2 for expansion. BM-MSCs were obtained from the same rats, as described previously (Wu et al., 2023). In brief, the femurs were rinsed with PBS. Using an 18-gauge needle, bone marrow was extracted and transferred to a collection tube with media for flushing. Following centrifugation at 2,000 rpm for 5 min, the cell pellet was pooled, resuspended in media, and filtered twice through a 70-μm strainer. The TD-MSCs and BM-MSCs were cultured under the same conditions. The cells were expanded, cryopreserved in a Cell Banker 1 (Zenogen Pharma Co., Tokyo, Japan), and stored in liquid nitrogen for future use.
2.2 Cell proliferation and viability assay
The proliferative capacity of TD-MSCs and BM-MSCs was evaluated using a cell counting kit-8 colorimetric assay (CCK-8; Sigma-Aldrich, United States). Cells were seeded in 96-well plates at a density of 1 × 104 cells per well (200 μL/well) and incubated for 24 h. Subsequently, 10 µL of CCK-8 reagent was added to each well in the dark, followed by 2 h of incubation at 37 °C with 5% CO2. The absorbance of the viable cells was measured at 450 nm using a microplate spectrometer (Molecular Devices, United States).
2.3 Tri-lineage differentiation of MSCs
The multipotent differentiation potential of TD-MSCs and BM-MSCs was confirmed using adipogenic, osteogenic, and chondrogenic lineage induction under specific in vitro conditions following a previously described protocol (Zayed et al., 2016). For adipogenic differentiation, confluent cells were treated with adipogenic induction medium containing DMEM supplemented with dexamethasone (1 μM), insulin (10 μg/mL), indomethacin (100 μM), and L-ascorbic acid (50 μg/mL) for 7 days. Lipid droplet accumulation was detected using Oil Red O staining. Osteogenic differentiation was induced in an osteogenic medium (DMEM with 50 µM ascorbic acid, 10 mM β-glycerophosphate, and 0.5 µM dexamethasone) for 14 days, and calcium deposition was confirmed using Alizarin Red staining. In chondrogenic differentiation, MSCs were cultured in DMEM supplemented with dexamethasone (100 nM), ascorbic acid (25 μg/mL), and TGF-β1 (10 ng/mL). Glycosaminoglycan deposition was assessed using Alcian Blue staining.
2.4 Immunophenotypic characterization
Flow cytometry analysis was performed according to the MSC criteria proposed by the International Society for Cellular Therapies to characterize the surface marker profile of TD-MSCs and BM-MSCs (CD90, CD45, and CD34). Cells were stained with fluorescein isothiocyanate (FITC)-conjugated antibodies. Mouse anti-rat CD90 (BD PharmingenTM FITC, Cat. #: 561,973, Korea), mouse anti-rat CD45 (BD PharmingenTM FITC, Cat. #: 561,867), and mouse anti-rat CD34 (BD PharmingenTM FITC, Cat. #: 560,238) were used. Isotype-matched IgG1 (BioLegend, Seoul, Korea) was used as a negative control. Fluorescence data were acquired using a NovoCyte® Flow Cytometer (NovoCyte 3000, ACEA Bioscience, Inc., United States), and data analysis was conducted using NovoExpress software (Agilent Technologies, United States).
2.5 Transcriptomic profiling and pathway analysis
RNA-seq was performed to compare the baseline transcriptomic profiles of TD-MSCs and BM-MSCs. Total RNA was isolated from passage 2 (n = 3 per group) using an RNeasy Mini Kit (QIAGEN, Hilden, Germany). Samples with an RNA integrity number (RIN) > 7, confirmed using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States), and with yields of at least 0.5 µg were used for library preparation with the TruSeq Stranded Total RNA Kit (Illumina, San Diego, CA, United States). Paired-end sequencing (2 × 100 bp) was performed on an Illumina platform. Quality-filtered raw reads (Phred Q30) were aligned to the Rattus norvegicus reference genome (rn6) using HISAT2. Transcript abundance was quantified using StringTie software and normalized to the number of transcripts per million. Analysis of differentially expressed genes (DEGs) between TD-MSCs and BM-MSCs was performed using DESeq2, with significance criteria of a fold change ≥2 (|log_2FC| ≥ 1) and p < 0.05. Functional enrichment analysis of DEGs was performed using the g:Profiler for Gene Ontology (GO) terms, including biological process (BP), cellular component (CC), and molecular function (MF), with p-values adjusted using the Benjamini–Hochberg false discovery rate (FDR <0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment (https://www.kegg.jp/kegg/pathway.html) was performed to identify significantly enriched signaling pathways ranked by adjusted p-values and gene ratios (proportion of DEGs within each pathway relative to total pathway genes).
2.6 Assessment of in vitro tenogenic differentiation
TD-MSCs were induced to undergo tenogenic differentiation according to a previously published study (Stanco et al., 2019). Cells were maintained in DMEM (Thermo Fisher Scientific) at 37 °C with 5% CO2 until 80%–90% confluence was reached. Then, the cells were cultured in a complete medium containing CTGF (100 ng/mL; PeproTech®, United States), ascorbic acid (50 μg/mL; Sigma-Aldrich, United States), and BMP-12 (50 ng/mL; BioLegend®, United States). The culture medium was refreshed every 3 days, and the progress in differentiation was monitored. Upon differentiation, quantitative reverse transcription polymerase chain reaction (qRT-PCR) and immunofluorescence staining were used to assess tenogenic marker expression at both the molecular and cellular levels.
2.7 RNA isolation and qRT-PCR
To evaluate the changes in gene expression during tenogenic differentiation, total RNA was isolated from undifferentiated and tenogenic-differentiated TD-MSCs using a GeneAll Biotechnology RNA Extraction Kit (Seoul, Korea). RNA concentration and purity were assessed using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Reverse transcription of mRNA into complementary DNA was performed using the ReverTra Ace® qPCR RT Master Mix with gDNA remover (Toyobo, Japan). qRT-PCR was performed on a CFX Opus 96TM real-time PCR system (Bio-Rad Laboratories, United States) with SYBR Green Master Mix (Toyobo) to assess the expression of key tenogenic marker genes, including SCX, COL1, COL3, TNMD, TN-C, SOX9, thrombospondin 4 (THBS-4), and decorin (DCN), as described in Supplementary Table S1.
2.8 Immunofluorescence
TD-MSCs were cultured at a density of 2 × 104 cells/well in a slide chamber (SPL Life Sciences). Following 7 days of tenogenic induction, the cells were fixed with 4% paraformaldehyde (PFA), washed with PBS, permeabilized using 0.1% Triton X-100 in PBS, and blocked using SuperBlock (ScyTek Laboratories, United States). Immunostaining was performed overnight at 4 °C with primary antibodies against SCX A-7 (mouse, 1:50; Santa Cruz Biotechnology, Dallas, TX, United States), COL1, and COL3 (1:200; Abcam, United Kingdom). After primary antibody binding, the cells were treated with Alexa Fluor 555-conjugated goat anti-mouse IgG for visualizing SCX and Alexa Fluor 488 goat anti-rabbit IgG for visualizing COL1 and COL3 (Invitrogen, United States) for 60 min at room temperature. The cells were mounted with an anti-fade mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, United States). Images were captured using a BX51 fluorescence microscope (Olympus, Tokyo, Japan). Fluorescence intensity was quantified using ImageJ software.
2.9 Rat model of Achilles tendon injury
Sprague Dawley rats (n = 8) were randomly divided into two experimental injection groups (n = 4 each): control (untreated) and TD-MSCs (Figure 1). All rats were anesthetized via intramuscular injection of xylazine (10 mg/kg) and tiletamine/zolazepam (50 mg/kg). The animals were positioned in sternal recumbency, and the surgical area was shaved and disinfected. A small (approximately 1 cm) longitudinal skin incision was made on the right hind limb to expose the Achilles tendon. A standardized transverse transection defect was created at the midpoint of the tendon to preserve the paratenon using a digital micro-caliper with measurements in mm (Supplementary Figure S1; Zhang et al., 2024). The skin was sutured, and then 1 × 106 TD-MSCs in 50 µL PBS were injected into the space between the Achilles tendon and the overlying skin. The control group received a PBS injection. To manage postoperative pain and prevent infection, the animals were administered ketorolac (3 mg/kg IM) and ampicillin (50 mg/kg IM) once daily for 3 days.
FIGURE 1
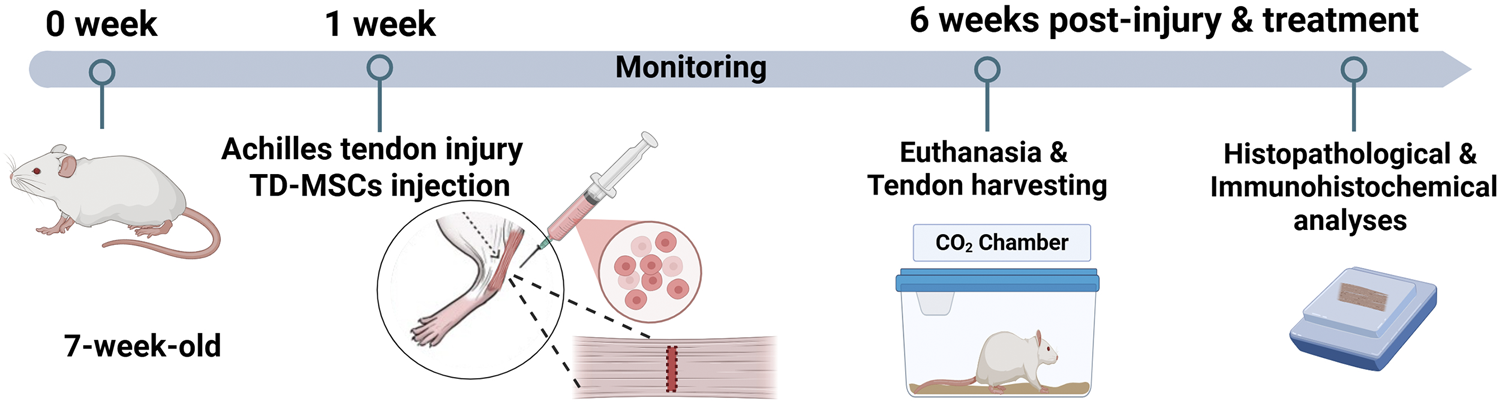
Schematic illustration showing the in vivo experimental design. An accommodation period of 1 week was allowed before the procedure, and TD-MSCs were injected at the same point after tendon defect creation and skin closure (day 0 of surgery). Rats were monitored for 6 weeks and then euthanized for tendon harvests to conduct histopathological and immunohistochemical analyses.
2.10 Histopathological evaluation and immunohistochemical analysis
All rats were euthanized 6 weeks post-injection as previously reported (Freedman et al., 2016; Freedman et al., 2017; Leahy et al., 2022). Achilles tendon tissues were collected from the injury site, immersed in 4% PFA for 48 h, and processed for paraffin embedding according to standard histological procedures. Tissue sections (5-μm thick) were prepared and stained with hematoxylin and eosin (H&E) for morphological examination. A semi-quantitative assessment was performed using a previously established scoring system (Yang et al., 2017). A score of 0 indicates normal tendon structure, while a score of 18 represents the most severe abnormality. Collagen architecture and organization were examined using Masson’s trichrome and picrosirius red staining, according to the manufacturer’s protocols.
Immunohistochemistry (IHC) was performed following the deparaffinization and rehydration of tissue sections. Antigen retrieval was achieved after incubating with proteinase K (120 mg, GeneAll Biotechnology Co., Korea) for 10 min at 37 °C. Nonspecific binding was prevented using SuperBlock (ScyTek Laboratories, United States), followed by overnight incubation at 4 °C with primary antibodies against SCX A-7 (Santa Cruz Biotechnology), COL1, and COL3 (Abcam, United Kingdom), each at a 1:200 dilution. Signal detection was accomplished by incubating for 1 h with a horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) at room temperature. Visualization was performed using a Vectastain DAB Substrate Kit (Vector Laboratories) with hematoxylin counterstaining. Histological examination was conducted using a BX51 light microscope (Olympus Corp., Tokyo, Japan), and IHC staining intensity was quantified using FIJI (ImageJ) software.
2.11 Statistical analysis
All quantitative data were presented as the mean ± standard deviation (SD). Statistical comparisons between groups were performed using an unpaired Student’s t-test. Differences were considered statistically significant at *p < 0.05, **p < 0.01, and ***p < 0.001. Data analysis was conducted using GraphPad Prism software version 8 (GraphPad Software Inc., United States).
3 Results
3.1 Isolation, characterization, and proliferation of TD-MSCs and BM-MSCs
BM-MSCs exhibited a typical spindle-shaped fibroblast-like morphology under an inverted phase-contrast microscope, whereas TD-MSCs were smaller and rounder (Figure 2A). Compared to BM-MSCs, quantitative analysis of cell proliferation on days 1 and 2 revealed a time-dependent increase in the number of TD-MSCs, with a significant increase observed on days 1 (p < 0.01) and 2 (p < 0.001) (Figure 2B). These results suggest that TD-MSCs possess an intrinsically greater in vitro proliferative capacity and regenerative potential than BM-MSCs. Both MSC populations demonstrated robust multipotency, differentiating into adipogenic, osteogenic, and chondrogenic lineages under the appropriate induction conditions (Figure 2C). Adipogenic-differentiated TD-MSCs and BM-MSCs showed Oil Red O-positive lipid droplets, indicating their ability to differentiate into adipocytes. Alizarin Red staining indicated mineralized matrix deposition (calcium nodules) in osteogenic cultures. Notably, TD-MSCs appeared to form more calcium nodules than BM-MSCs. During chondrogenic differentiation, cells were stained blue with Alcian Blue after induction, indicating the presence of a glycosaminoglycan-rich cartilage matrix. TD-MSCs and BM-MSCs were successfully isolated and characterized using flow cytometry (Figure 2D). The level of the MSC marker, CD90, was high in TD-MSCs (98.8%) and BM-MSCs (97.4%), while minimal expression of CD45 and CD34 was observed in TD-MSCs (0.3% and 0.4%, respectively) and BM-MSCs (0.2% and 0.9%, respectively). These findings confirmed the successful isolation of pure TD-MSCs and BM-MSCs.
FIGURE 2
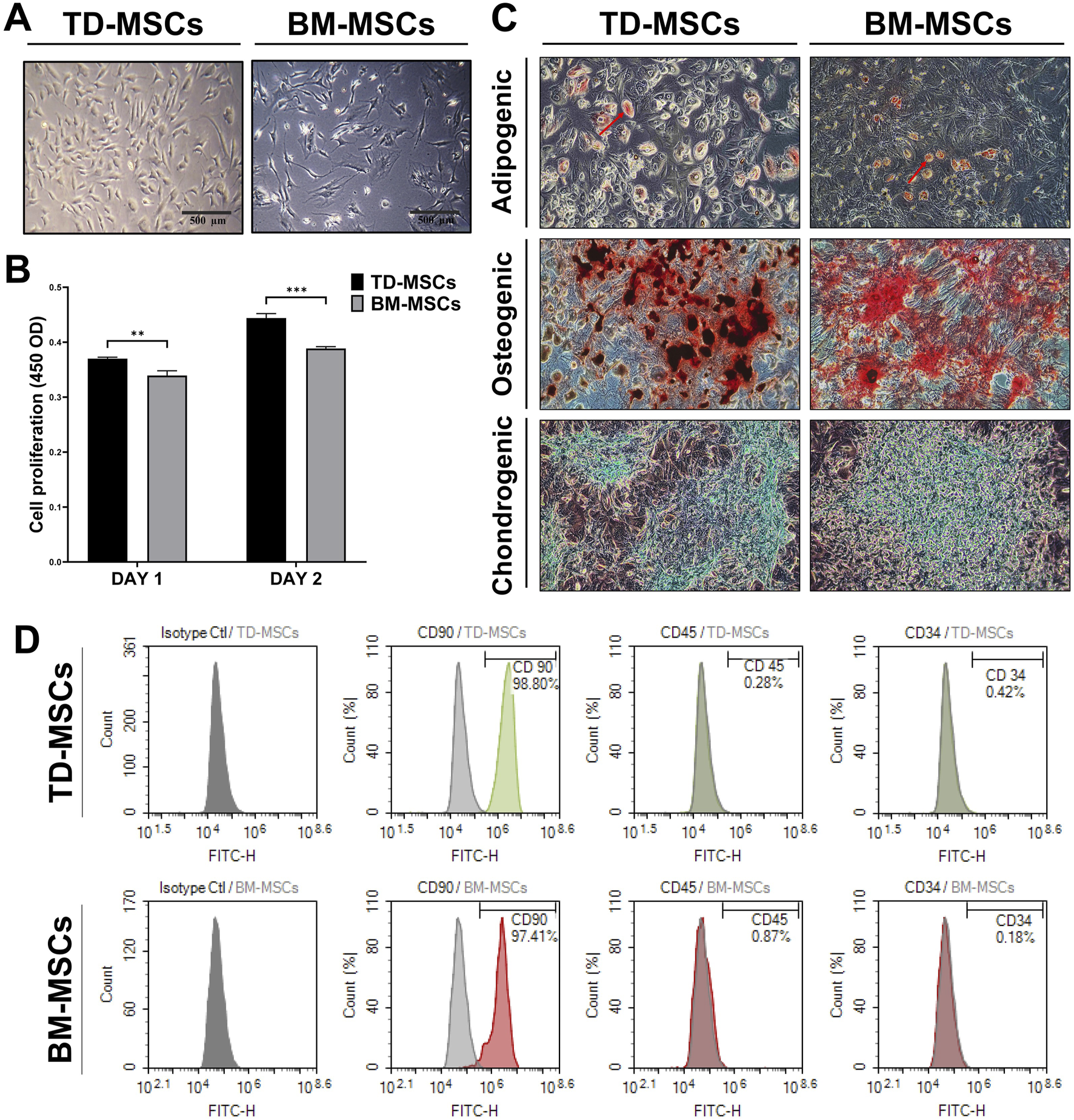
Comparative in vitro characterization of TD-MSCs and BM-MSCs. (A) Representative phase-contrast micrographs of TD-MSCs vs. BM-MSCs highlighting morphological differences (scale bars: 500 μm). (B) Cell proliferation rate on days 1 and 2. Data were presented as the mean ± SD (n = 3); **p < 0.01, and ***p < 0.001. (C) Tri-lineage differentiation potential of TD-MSCs and BM-MSCs (×100), confirmed using Oil Red O staining of lipid droplets for adipogenesis (red arrows), Alizarin Red staining of calcium deposits for osteogenesis, and Alcian Blue staining of glycosaminoglycan-rich matrix for chondrogenesis. (D) Flow cytometric analysis of surface markers. TD-MSCs and BM-MSCs showed >97% positive expression for CD90 and negative for CD45 and CD34 (with less than 1%).
3.2 Comparative transcriptomic profiling of TD-MSCs and BM-MSCs
To compare the baseline transcriptomes of TD-MSCs and BM-MSCs, we performed RNA-seq on undifferentiated cells from each source (n = 3 independent cell isolations; Supplementary Material 1). This analysis identified 2,654 DEGs between TD-MSCs and BM-MSCs (|log2 FC| ≥ 1, FDR <0.05). A clear separation was observed between the MSCs using unsupervised hierarchical clustering and heatmap analysis, suggesting distinct lineage-specific transcriptional profiles (Supplementary Figure S2A). Volcano plot analysis also highlighted the broad transcriptomic divergence between TD-MSCs and BM-MSCs (Supplementary Figure S2B), identifying 1,104 genes that were significantly upregulated and 1,550 genes that were downregulated in TD-MSCs relative to those in BM-MSCs.
DEGs upregulated in TD-MSCs showed strong enrichment for BP of GO terms related to ECM organization and positive regulation of cell migration (Figure 3). Enriched GO terms in the CC category, including ECM and collagen-containing ECM, and the MF category, including integrin and growth factor binding, reflect the ECM adhesion and signaling roles of TD-MSCs in a regenerative environment.
FIGURE 3
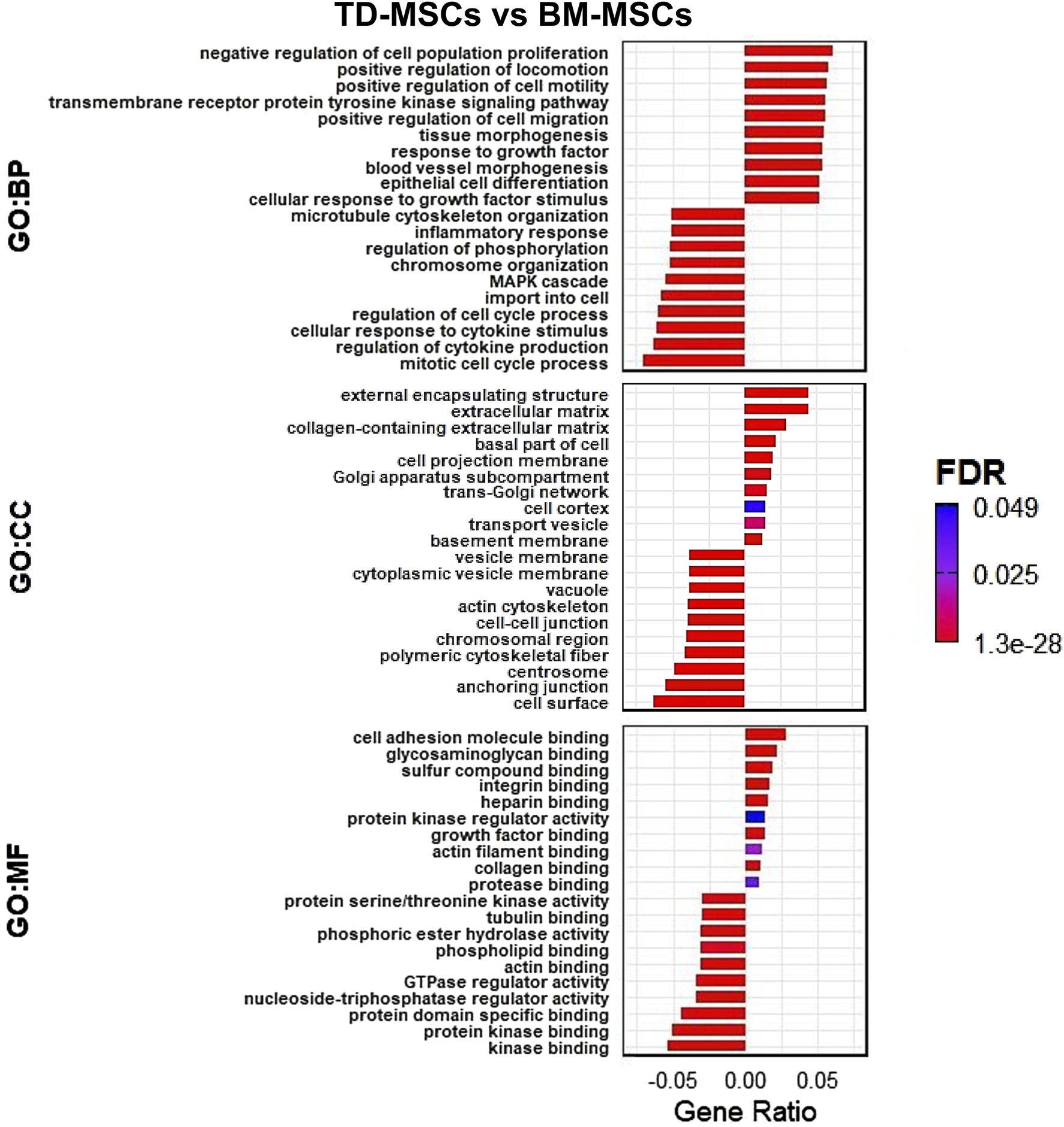
RNA-seq GO transcriptomic comparison of TD-MSCs and BM-MSCs. (A) GO enrichment analysis of DEGs, highlighting top enriched terms in biological processes (BPs), cellular components (CCs), and molecular functions (MFs) for TD-MSC vs. BM-MSC group comparison (bars represent the gene ratio, colored by FDR significance), n = 3 per group.
The schematic workflow (Figure 4A) illustrates the transcriptomic profiling performed to compare TD-MSCs and BM-MSCs. KEGG pathway analysis showed that DEGs highly expressed in TD-MSCs were clustered in key regenerative pathways, such as PI3K–AKT signaling, MAPK signaling, focal adhesion, and ECM–receptor interaction (all FDR <0.001) (Figure 4B). These pathways are vital for cell proliferation, survival, ECM production, and differentiation during tissue repair. In contrast, the upregulated DEGs in BM-MSCs were associated with fewer pro-regenerative pathways; instead, the genes enriched in BM-MSCs included skeletal system development and hematopoietic cell lineage determination, indicating a difference in intrinsic biological predispositions.
FIGURE 4
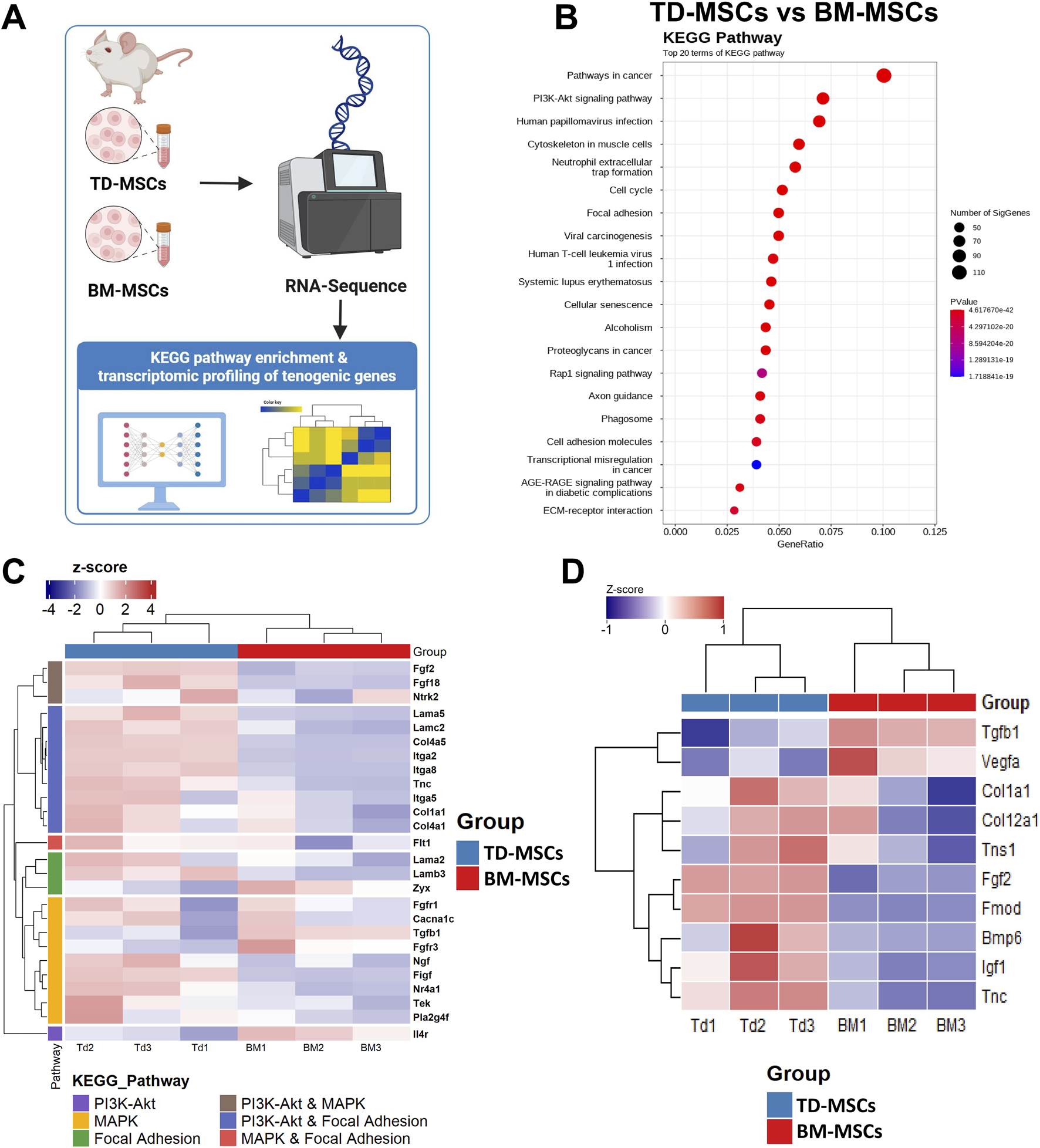
RNA-seq transcriptomic comparison of TD-MSCs and BM-MSCs. (A) Schematic illustration of the experimental workflow, pathway enrichment analysis, and tenogenic gene expression signatures. (B) KEGG pathway enrichment dot plot for DEGs in each pathway, where color indicates significance (FDR), n = 3 per group. (C) Heatmap of selected tenogenic-associated genes (normalized expression Z-scores) in BM-MSCs vs. TD-MSCs, focusing on genes in PI3K–AKT signaling, MAPK signaling, and focal adhesion pathways, n = 3 per group. (D) Heatmap of tenogenic-associated gene expression between BM-MSCs and TD-MSCs. The heatmap presents expression profiles of key tenogenic-associated genes across biological replicates, n = 3 per group.
3.3 Enhanced expression of tendon-specific genes in TD-MSCs compared to that in BM-MSCs
To evaluate the inherent ability of each MSC type for tenogenic differentiation, we performed a hypothesis-driven analysis of the tendon-related genes. The results revealed a significantly higher expression of key tendon ECM genes in TD-MSCs than in BM-MSCs (Figures 4C,D). TD-MSCs showed a marked upregulation of FMOD (507-fold) and COL1A1 (2.3-fold), encoding the primary structural collagens in tendons (Zhang et al., 2019), along with COL11A1 (4.9-fold) and COL1A2 (3.2-fold). Furthermore, TD-MSCs exhibited notable increases in ITGA2 (197.5-fold), ITGA7 (6-fold), ITGA8 (6-fold), and BGN (2.2-fold) expression. Significantly higher expression of genes encoding growth factors and cytokines such as FGF18 (9-fold), FGF2 (5.1-fold), CTGF (3.2-fold), BMP2 (5.7-fold), and BMP6 (5.3-fold) was also detected. Thus, the transcriptional profile of TD-MSCs was indicative of enhanced tendon ECM production and tenogenic signaling compared with BM-MSCs, which exhibited minimal expression of these genes. We further validated the strong tenogenic profile of TD-MSCs versus BM-MSCs based on the increased expression of tendon ECM synthesis and repair genes, such as TN-C (2.2-fold), encoding a glycoprotein critical for cell–ECM interactions during tendon healing (Xu et al., 2021). TD-MSCs also showed significant upregulation of COL4A5 (25.9-fold), contributing to basement membrane formation, and MMP3 (68.4-fold), reflecting active ECM remodeling. The levels of cytokines, such as IL-33 (5.9-fold), were also significantly higher in TD-MSCs than in BM-MSCs. Additionally, TD-MSCs displayed specific upregulation of tenogenic-associated genes (COL1A1, TN-C, FGF2, and FGF18) (Figures 4C,D) within key KEGG pathways, including the PI3K–AKT, MAPK, and focal adhesion pathways. These pathways, which are enriched in TD-MSCs, are known for their essential roles in cell proliferation, differentiation, and ECM interactions necessary for tendon repair relative to BM-MSCs, implying potential differences in the tenogenic differentiation capacity of the MSC types.
3.4 Expression of tenogenic markers in TD-MSCs
Considering the superior baseline tenogenic transcriptomic profile of TD-MSCs compared with that of BM-MSCs, subsequent tenogenic differentiation analyses focused on TD-MSCs. Functional response to tenogenic differentiation was evaluated by determining the expression of key genes associated with tenogenic differentiation (Figure 5A). We hypothesized that baseline differences in RNA-seq might influence the in vitro differentiation potential. TD-MSCs responded strongly to tenogenic induction, with a significant upregulation of SCX (p < 0.05), COL1 (p < 0.01), COL3 (p < 0.05), TN-C (p < 0.05), and THBS-4 (p < 0.05), whereas the expression of DCN and TNMD remained steady (p > 0.05) (Figure 5A), indicating their high tenogenic potential. During differentiation, TD-MSCs downregulated SOX9 (p < 0.05) (Figure 5A), reflecting suppression of the chondrogenic pathway and a tendency to favor the tenogenic lineage. This was supported by the overexpression of tenogenic markers (SCX, COL1, COL3, and TN-C) (Figure 5A), highlighting a strong tenogenic response.
FIGURE 5
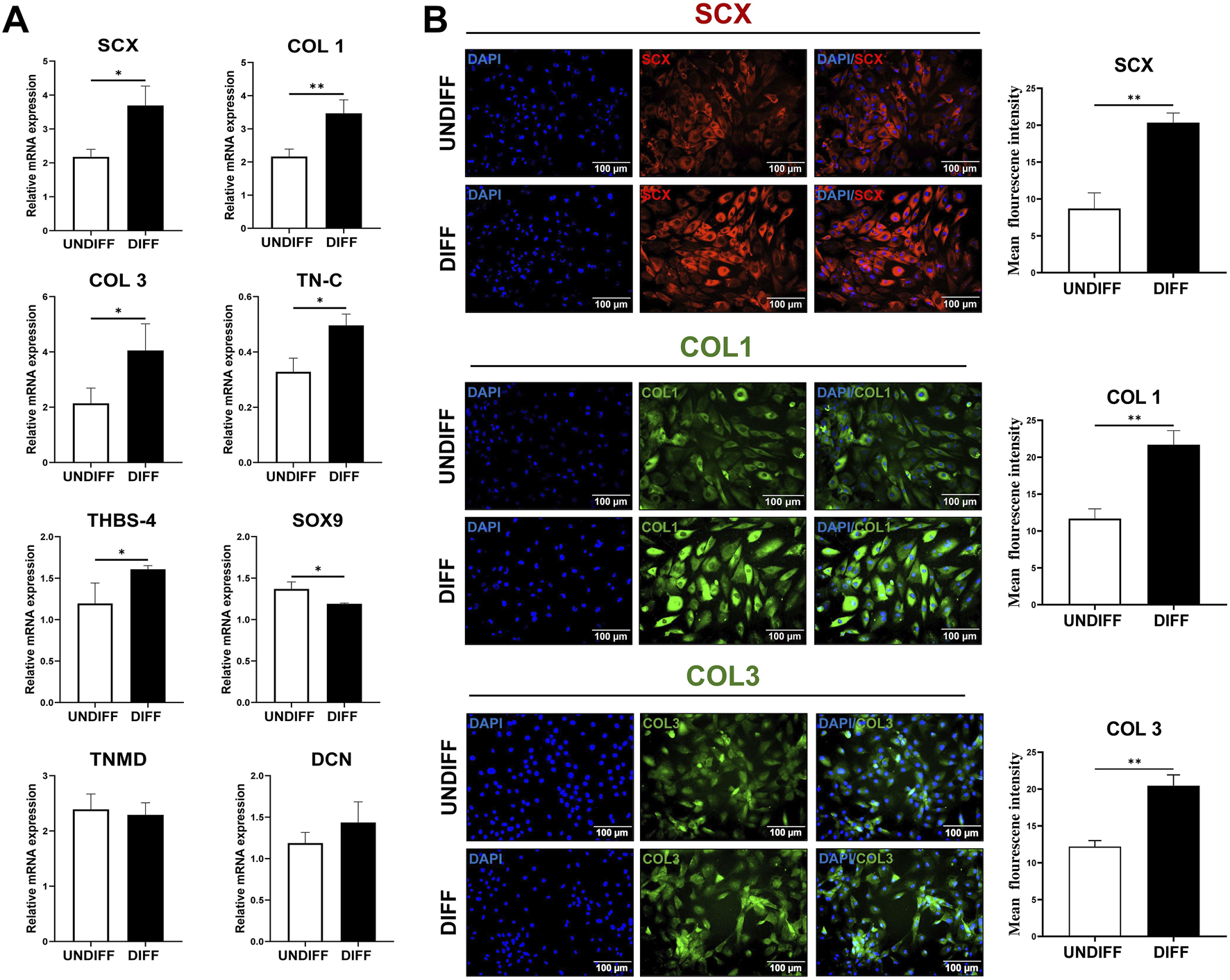
Tenogenic differentiation of TD-MSCs in vitro. (A) Relative mRNA expression of tenogenic marker genes in TD-MSCs under undifferentiated (UNDIFF) vs. differentiated (DIFF) conditions. Target genes included SCX, COL1, COL3, TNMD, TN-C, SOX9, THBS-4, and DCN. Data are presented as the mean ± SD (n = 3 per group); *p < 0.05 and **p < 0.01. (B) Representative immunofluorescence images of TD-MSCs before and after tenogenic induction of tenogenic markers. Cells were stained for SCX (red), COL1 (green), and COL3 (green) and with DAPI (blue) to visualize nuclei (×200, scale bar = 100 µm), n = 3 per group. Tenogenic-differentiated TD-MSCs showed stronger SCX nuclear localization and higher COL1/COL3 expression. Quantification of fluorescence intensity revealed that TD-MSCs exhibited a significant increase in tenogenic marker expression after differentiation (n = 3 per group, **p < 0.01).
We measured the mean fluorescence intensity to evaluate the tenogenic capacity of tenogenic differentiation-protein markers. Representative images show immunostaining for SCX (red) and COL1 and COL3 (green) in undifferentiated and differentiated TD-MSCs. TD-MSCs showed a significant increase in SCX-positive nuclei (Figure 5B) and abundant COL1 and COL3 expression (Figure 5B). Quantitative analysis confirmed significantly higher fluorescence intensities in differentiated TD-MSCs, as evidenced by the increased expression of key tenogenic markers (p < 0.01) (Figure 5B).
3.5 In vivo tendon repair efficacy with TD-MSC treatment
Based on the superior profile and tenogenic differentiation capacity of TD-MSCs, we evaluated their therapeutic efficacy in a model of Achilles tendon injury. Rats received an injection of TD-MSCs (surgery day 0) around the Achilles tendon, while the untreated control received PBS only. H&E staining showed that the TD-MSC-treated tendons had a better-organized collagen fiber structure, alignment, and improved cellularity than the untreated control. The average total semi-quantitative histological scores of the TD-MSC treatment group (8.5 ± 2.65) were significantly lower than those of the control group (15 ± 0.82) (p < 0.01) (Figures 6A,B). Masson’s trichrome staining demonstrated increased collagen content and denser connective tissue in the TD-MSC-treated tendons, and picrosirius red staining showed stronger COL1 deposition in the treated tendons (Figure 6A). In IHC analysis, TD-MSC-treated tendons showed significantly higher expression of COL1 and SCX than control tendons (p < 0.01) and significantly lower expression of COL3 (p < 0.05) (Figure 7), reflecting a shift toward a more mature collagen matrix.
FIGURE 6
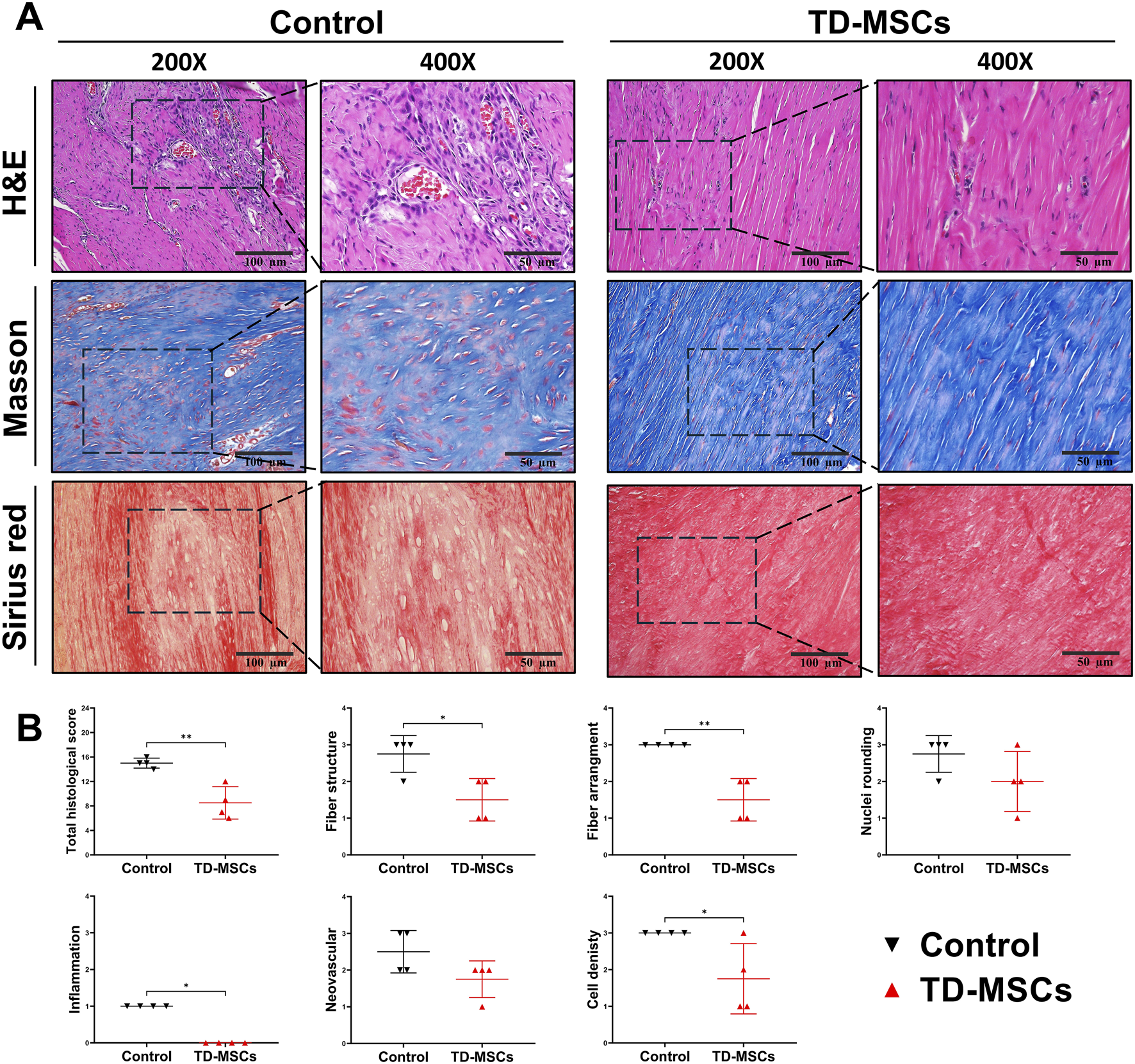
Outcomes of in vivo Achilles tendon healing at 6 weeks post-injury with TD-MSC treatment. (A) Representative histological images from untreated control vs. TD-MSC-treated tendon. Top: H&E staining illustrating overall tissue architecture, cellularity, and fiber alignment. Middle: Masson’s trichrome staining showed collagen deposition and alignment. Bottom: picrosirius red staining showed increased COL1 expression in the TD-MSC-treated group under brightfield (scale bars: 100 μm and 50 µm). (B) Semi-quantitative histological scoring of tendon healing based on H&E-stained sections; TD-MSC-treated tendons showed significantly lower histological scores than controls. Data are presented as the mean ± SD (n = 4 rats per group); *p < 0.05, and **p < 0.01.
FIGURE 7
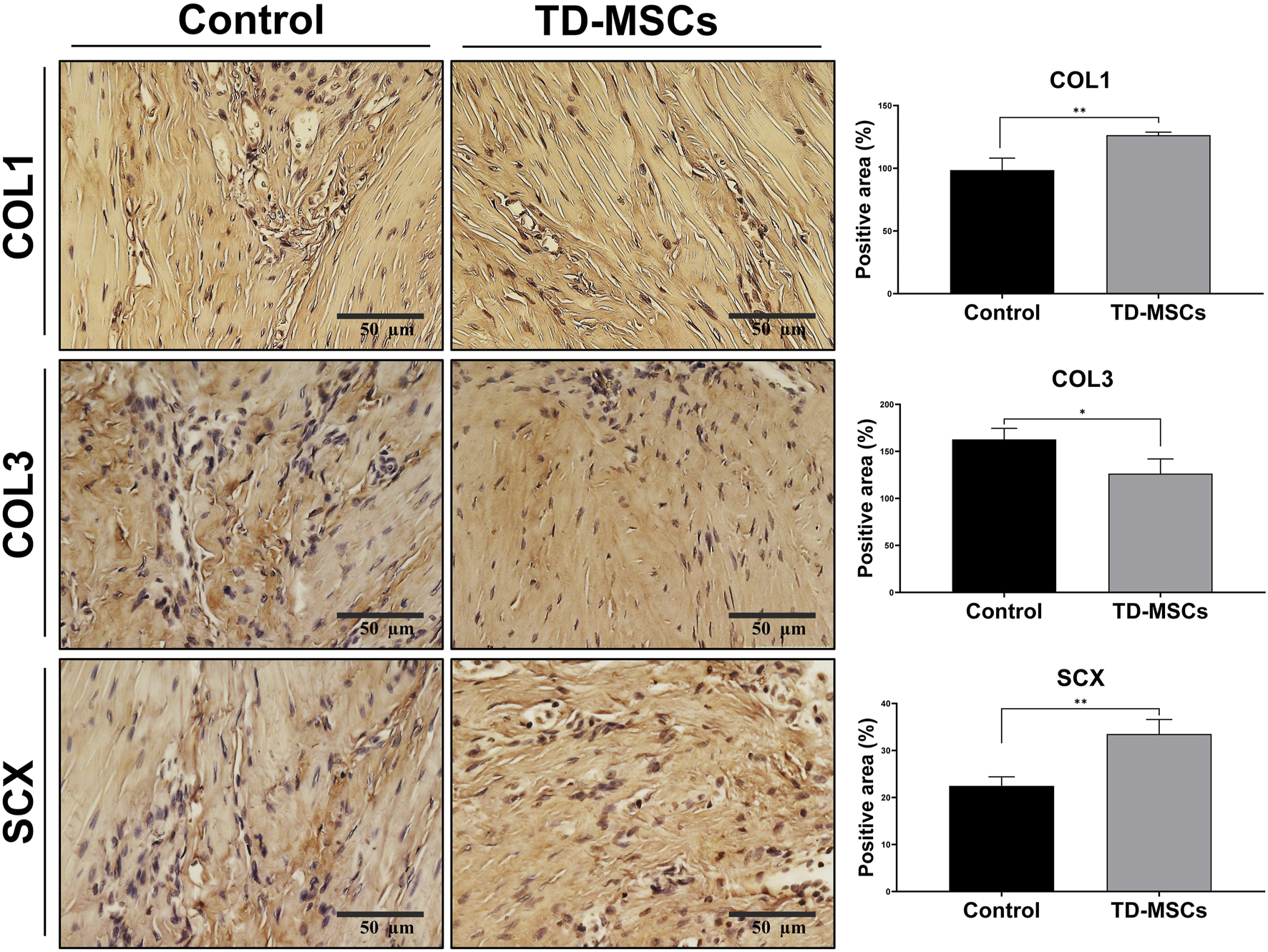
Immunohistochemical evaluation of tendon regeneration markers. Representative IHC images for COL1, COL3, and SCX expression from untreated control and TD-MSC-treated tendons (scale bar: 50 µm), with corresponding quantitative analysis. Data are presented as the mean ± SD (n = 4 animals/group; 3 fields/animal); *p < 0.05, and **p < 0.01.
4 Discussion
Tendon tissue engineering has emerged as a promising field offering potential therapeutic strategies for repairing and regenerating damaged tendons and ligaments (Ning et al., 2023; Citro et al., 2025). MSCs derived from various tissues have been studied in tissue regeneration (Mahmoud et al., 2022; Margiana et al., 2022; Zayed et al., 2023; Trapana et al., 2024; Zhidu et al., 2024). Although MSCs share some fundamental characteristics, their tissue-specific characteristics can influence the therapeutic outcomes (Tan et al., 2012; Pizzute et al., 2015). TD-MSCs, which can differentiate into tenocytes, are at the forefront of research as the primary cellular source of tissue-engineered constructs (Zhang et al., 2023). This study evaluated the transcriptomic profiles of TD-MSCs and their potential for tenogenic differentiation and intralesional application in vivo. Considering that the bone marrow is the primary source of MSCs, we initially compared the characteristics of TD-MSCs with those of BM-MSCs according to the guidelines of the International Society for Cellular Therapy. Both cell types adhered to the culture dishes under standard culture conditions and expressed CD90 while lacking the expression of CD45 or CD34 (Rojewski et al., 2008). Although both MSC sources satisfy these standard criteria, BM-MSCs are known to exhibit cellular heterogeneity, and their yield decreases with larger aspiration volumes, which may limit their therapeutic reliability and scalability (Tan et al., 2012; Li et al., 2023). Therefore, TD-MSCs have been proposed as viable alternatives to BM-MSCs (Lui and Chan, 2011). Previous studies have suggested that compared to BM-MSCs, TD-MSCs could display enhanced colony-forming capacity, faster proliferation, and elevated expression of tenogenic markers and tendon-specific ECM components (Bi et al., 2007; Tempfer et al., 2009; Thaker and Sharma, 2012; Al-Ani et al., 2015). These biological characteristics may potentially facilitate tendon tissue repair. Our study supports this hypothesis as our findings indicate that TD-MSCs have a distinct tenogenic transcriptomic signature and exhibit enhanced tendon-specific differentiation and regenerative abilities in vitro and in vivo, supporting their potential utility as a cell source for tendon therapeutic applications.
RNA-seq analysis identified substantial baseline transcriptomic differences between TD-MSCs and BM-MSCs, with DEGs and pathway analyses emphasizing their distinct roles in tendon repair. TD-MSCs exhibited a notable difference, with significant enrichment in the PI3K–AKT, MAPK, and focal adhesion pathways, which are the key pathways involved in ECM remodeling, cell adhesion, proliferation, tenogenesis, collagen production, and tenocyte migration (Han et al., 2019; Zhang et al., 2020; Stańczak et al., 2024). Analysis of tenogenic-associated genes revealed elevated expression of key genes in TD-MSCs, including TN-C, FGF18, and COL1A1, suggesting enhanced ECM remodeling, growth factor signaling, and cell–matrix interactions. These findings indicated the inherent advantage of TD-MSCs in establishing a regenerative microenvironment conducive to tendon healing and enhanced tenogenic differentiation capacity. Notably, the PI3K–AKT pathway was enriched in TD-MSCs, which regulated COL1 production and cell cycle progression during healing (Stańczak et al., 2024). COL1A1 upregulation, crucial for ECM synthesis, has been observed in TD-MSCs (Tan et al., 2012; Yi et al., 2022). The activation of MAPK signaling in TD-MSCs corresponds to previous findings linking this pathway to FGF activity, collagen synthesis, ECM remodeling, and tenogenic differentiation of fibroblasts (Titan et al., 2019; Stańczak et al., 2024). The elevated expression of FGF18 and FGF2 in TD-MSCs further supports the involvement of this pathway in tenogenesis (Titan et al., 2019). Furthermore, this finding is consistent with the results of previous reports, indicating that growth factor pretreatment can promote tenogenic differentiation while minimizing ossification and that TD-MSCs intrinsically express tendon-related genes independent of external stimulation (Citro et al., 2025). Activation of the MAPK pathway also correlated with increased biglycan (BGN) expression, a critical small leucine-rich proteoglycan that regulates collagen fibrillogenesis (Tan et al., 2012; Zhang et al., 2019). In addition, focal adhesion pathway enrichment of integrins and related molecules, such as ITGA2, ITGA8, and TN-C, may improve cell–ECM adhesion and migration, allowing MSCs to respond more effectively to the biomechanical cues essential for tendon remodeling (Xu et al., 2021).
Following the identification of enhanced tenogenic gene expression in TD-MSCs compared to those in BM-MSCs using transcriptomic analysis, we focused our functional experiments on evaluating the differentiation capacity of TD-MSCs. The results confirmed the improved tenogenic differentiation of TD-MSCs in vitro and demonstrated their superior repair ability in vivo compared with untreated controls. Elevated expression of key tenogenic markers, including COL1 and TN-C, suggests that TD-MSCs possess an inherent predisposition toward tendon regeneration. After tenogenic differentiation, TD-MSCs showed upregulation of genes, including SCX, COL1, COL3, TN-C, and THBS-4, compared to that observed in undifferentiated cells, reflecting their strong tendency to differentiate into tenocyte-like cells. SCX, a transcription factor specific to tendon progenitors, regulates COL1 and TNMD and is essential for tendon development and maturation (Shukunami et al., 2006; Gaut and Duprez, 2016). COL1 is the main collagen in mature tenocytes, whereas COL3 supports early wound healing and formation of the epitenon and endotenon (Zhang et al., 2019). TN-C functions as a vital ECM protein that maintains tissue elasticity, especially during tendon injury and healing, when the biomechanical environment is disrupted (Pajala et al., 2009; Zhang et al., 2011; Al-Ani et al., 2015). TN-C is present in mature tendons, where it interacts with integrin receptors and other ECM components, influencing cell–matrix interactions and collagen fiber alignment (Li et al., 2021). Similarly, THBS-4 contributes to ECM organization by binding to various cellular receptors and ligands (Stenina-Adognravi and Plow, 2019). Although these proteins are primarily expressed in tenocytes, their expression also supports the ECM structure across different tissue types (Citro et al., 2025). This enhanced gene expression, validated using qRT-PCR and immunofluorescence, correlated with markedly elevated levels of SCX and collagen-related proteins in TD-MSCs. Moreover, TD-MSCs showed significant downregulation of SOX9, providing evidence that tenogenesis involves the suppression of chondrogenic pathways (Titan et al., 2019; Zhang et al., 2019).
Following the characterization of the TD-MSC population and confirmation of their tenogenic potential, we evaluated the therapeutic efficacy of TD-MSCs in vivo using a tendon injury model. Intratendinous injection of MSCs has become a widely used method for tendon repair in the treatment of musculoskeletal disorders, with promising clinical outcomes (Jo et al., 2018). To accurately assess the effects of TD-MSCs, all rats were euthanized, and their Achilles tendons were analyzed histopathologically and by IHC. H&E staining in the TD-MSC-treated group showed a higher degree of organized collagen fiber arrangement and reduced cellular disarray than in the controls. This improvement was reflected in the significantly lower histological scores, consistent with a previous study demonstrating that TD-MSCs treatment may facilitate tendon healing by promoting the formation of dense, longitudinally aligned collagen fibers and improving tissue organization (Al-Ani et al., 2015). These findings emphasize the importance of regulating collagen composition during tendon healing (Cui et al., 2011; Thankam et al., 2018). In contrast, untreated control tendons show elevated COL3 levels, which are typically associated with disorganized matrix and fibrotic remodeling (Tsai et al., 2006; Xu et al., 2018), whereas an optimal COL1/COL3 ratio is essential for functional tendon regeneration (Matsumoto et al., 2002; Zhang et al., 2020). A previous study has reported that TD-MSCs treatment contributed to connective tissue formation by continuously stimulating COL1 synthesis at the injury site, thereby supporting matrix maturation and mechanical strength (Al-Ani et al., 2015). Early upregulation of COL3 represents a normal part of the initial healing phase, providing temporary stabilization; however, its expression is expected to diminish as healing progresses. Reduced COL3 and increased COL1 levels suggest a shift toward tissue maturation, consistent with the natural remodeling process (Al-Ani et al., 2015). In this study, IHC analysis of the TD-MSC-treated group revealed increased expression of SCX and COL1 and reduced COL3 levels. These results suggest that TD-MSCs support regeneration by modulating the tendon microenvironment and promoting remodeling toward organized repair rather than fibrotic healing. Although previous studies have shown that TD-MSCs can differentiate into tenocytes and produce matrix components (Zhang et al., 2011; Shen et al., 2012; Thaker and Sharma, 2012; Al-Ani et al., 2015), we acknowledge that the present study did not provide direct evidence of transplanted cell tracking. Thus, we cannot assert that TD-MSCs themselves rapidly differentiated and served as the prime source of repair. Instead, our findings may support the therapeutic efficacy of local TD-MSC administration during the acute phase of injury, likely through their paracrine activity and immunomodulatory effects, which stimulated endogenous tendon cells and enhanced remodeling.
While our preclinical data highlight the therapeutic potential of TD-MSCs, translating these cells into clinical settings presents practical challenges. First, deriving MSCs from human tendons, usually obtained through biopsy or routine surgery (Perucca Orfei et al., 2021; Rizzo et al., 2025), is more invasive. However, this process could be made easier with minimally invasive, ultrasound-guided biopsy techniques (Murphy et al., 2013). Second, variability from donors remains a key factor for all cell-based therapies (Zayed and Iohara, 2023). For TD-MSCs, critical factors such as donor age, health status, and the source of the tendon biopsy significantly affect cellular properties (Yan et al., 2020). Future research should establish strict donor selection criteria to ensure consistent clinical outcomes, develop reliable in vitro potency tests to validate cell batches, and consider exploring allogeneic “off-the-shelf” cell banks from well-characterized, healthy donors.
This study provided important insights regarding the molecular basis of the transcriptional signature underlying the therapeutic potential of TD-MSCs for tendon regeneration. Our findings support the use of tissue-specific MSC sources in regenerative medicine. However, several limitations must be considered: the number and size of animals used for in vivo experiments were relatively small, healing outcomes were assessed at a single time point (6 weeks), biomechanical testing was not performed, which restricts the ability to correlate histological improvements with functional recovery, and the lack of evaluation of long-term therapeutic durability. In addition, cell tracking was not conducted, so we cannot determine whether injected TD-MSCs engrafted or primarily acted through paracrine mechanisms. Future studies should focus on scaling up TD-MSC production, evaluating their safety and efficacy in larger animal models, extending follow-up periods, and performing biomechanical assessments, including tensile strength and stiffness testing, to directly assess functional recovery.
5 Conclusion
Our findings demonstrate that TD-MSCs possess a distinctive transcriptomic profile characterized by significantly elevated expression of tenogenic genes, including COL1 and TN-C, compared to BM-MSCs. In vitro experiments revealed that TD-MSCs showed robust tenogenic differentiation capacity compared to undifferentiated cells, as evidenced by the increased expression levels of SCX, COL1, COL3, TN-C, and THBS-4. In vivo evaluation showed that TD-MSC treatment improved tendon healing with enhanced collagen organization and structural integrity compared to untreated controls. Overall, our results highlight the tenogenic tendency of TD-MSCs and support their use as a promising cell source for tendon repair applications, warranting further preclinical and clinical investigations.
Statements
Data availability statement
All raw RNA-seq data were deposited in the NCBI Sequence Read Archive (SRA) under accession PRJNA1245624.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of Jeonbuk National University, Republic of Korea (NON2024-093). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HK: Writing – review and editing, Validation, Writing – original draft, Formal analysis, Methodology, Software, Data curation, Visualization, Investigation, Conceptualization. MZ: Conceptualization, Investigation, Writing – review and editing, Methodology, Writing – original draft, Data curation. BK: Supervision, Writing – review and editing. B-HJ: Methodology, Supervision, Writing – original draft, Writing – review and editing. S-IO: Writing – review and editing, Software, Writing – original draft, Resources, Formal analysis, Funding acquisition, Visualization, Methodology, Conceptualization, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National University Development Project of Jeonbuk National University in 2024.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1687816/full#supplementary-material
Abbreviations
TD-MSCs, tendon-derived mesenchymal stem cells; BM-MSCs, bone marrow-derived mesenchymal stem cells; qRT-PCR, quantitative reverse transcription polymerase chain reaction; RNA-seq, RNA sequencing; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
References
1
Al-Ani M. Xu K. Sun Y. Pan L. Xu Z. Yang L. (2015). Study of bone marrow mesenchymal and tendon-derived stem cells transplantation on the regenerating effect of achilles tendon ruptures in rats. Stem Cells Int.2015, 984146. 10.1155/2015/984146
2
Bi Y. Ehirchiou D. Kilts T. M. Inkson C. A. Embree M. C. Sonoyama W. et al (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med.13 (10), 1219–1227. 10.1038/nm1630
3
Citro V. Clerici M. Porta G. D. Maffulli N. Boccaccini A. R. Dale T. P. et al (2025). Tenogenic cues are biochemically and environmentally distinct for tendon stem cells and mesenchymal/stromal stem cells. Stem Cells Int.2025, 9047956. 10.1155/sci/9047956
4
Costa-Almeida R. Calejo I. Gomes M. E. (2019). Mesenchymal stem cells empowering tendon regenerative therapies. Int. J. Mol. Sci.20 (12), 3002. 10.3390/ijms20123002
5
Cottrell J. A. Turner J. C. Arinzeh T. L. O'Connor J. P. (2016). The biology of bone and ligament healing. Foot Ankle Clin.21 (4), 739–761. 10.1016/j.fcl.2016.07.017
6
Cui Q. Wang Z. Jiang D. Qu L. Guo J. Li Z. (2011). HGF inhibits TGF-β1-induced myofibroblast differentiation and ECM deposition via MMP-2 in achilles tendon in rat. Eur. J. Appl. Physiol.111 (7), 1457–1463. 10.1007/s00421-010-1764-4
7
Freedman B. R. Gordon J. A. Bhatt P. R. Pardes A. M. Thomas S. J. Sarver J. J. et al (2016). Nonsurgical treatment and early return to activity leads to improved achilles tendon fatigue mechanics and functional outcomes during early healing in an animal model. J. Orthop. Res.34 (12), 2172–2180. 10.1002/jor.23253
8
Freedman B. R. Salka N. S. Morris T. R. Bhatt P. R. Pardes A. M. Gordon J. A. et al (2017). Temporal healing of achilles tendons after injury in rodents depends on surgical treatment and activity. J. Am. Acad. Orthop. Surg.25 (9), 635–647. 10.5435/jaaos-d-16-00620
9
Gaut L. Duprez D. (2016). Tendon development and diseases. Wiley Interdiscip. Rev. Dev. Biol.5 (1), 5–23. 10.1002/wdev.201
10
Goldberg A. J. Masci L. O’Donnell P. Green R. Brooking D. Bassett P. et al (2024). Autologous bone marrow derived mesenchymal stem cells are safe for the treatment of achilles tendinopathy. Sci. Rep.14 (1), 11421. 10.1038/s41598-024-61399-3
11
Han P. Cui Q. Lu W. Yang S. Shi M. Li Z. et al (2019). Hepatocyte growth factor plays a dual role in tendon-derived stem cell proliferation, migration, and differentiation. J. Cell Physiol.234 (10), 17382–17391. 10.1002/jcp.28360
12
Hass R. Kasper C. Böhm S. Jacobs R. (2011). Different populations and sources of human mesenchymal stem cells (MSC): a comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal9, 12. 10.1186/1478-811x-9-12
13
He W. Jiang C. Zhou P. Hu X. Gu X. Zhang S. (2024). Role of tendon-derived stem cells in tendon and ligament repair: focus on tissue engineer. Front. Bioeng. Biotechnol.12, 1357696. 10.3389/fbioe.2024.1357696
14
Jo C. H. Chai J. W. Jeong E. C. Oh S. Kim P. S. Yoon J. Y. et al (2018). Intratendinous injection of autologous adipose tissue-derived mesenchymal stem cells for the treatment of rotator cuff disease: a first-in-human trial. Stem Cells36 (9), 1441–1450. 10.1002/stem.2855
15
Leahy T. Nuss C. Evans M. K. Fung A. Shetye S. Soslowsky L. J. (2022). Achilles tendon ruptures in middle-aged rats heal poorly compared with those in young and old rats. Am. J. Sports Med.50 (1), 170–181. 10.1177/03635465211055476
16
Leong N. L. Kator J. L. Clemens T. L. James A. Enamoto-Iwamoto M. Jiang J. (2020). Tendon and ligament healing and current approaches to Tendon and ligament regeneration. J. Orthop. Res.38 (1), 7–12. 10.1002/jor.24475
17
Li Y. Wu T. Liu S. (2021). Identification and distinction of tenocytes and tendon-derived stem cells. Front. Cell Dev. Biol.9, 629515. 10.3389/fcell.2021.629515
18
Li J. Wu Z. Zhao L. Liu Y. Su Y. Gong X. et al (2023). The heterogeneity of mesenchymal stem cells: an important issue to be addressed in cell therapy. Stem Cell Res. & Ther.14 (1), 381. 10.1186/s13287-023-03587-y
19
Lui P. P. (2015). Markers for the identification of tendon-derived stem cells in vitro and tendon stem cells in situ - update and future development. Stem Cell Res. Ther.6 (1), 106. 10.1186/s13287-015-0097-y
20
Lui P. P. Chan K. M. (2011). Tendon-derived stem cells (TDSCs): from basic science to potential roles in tendon pathology and tissue engineering applications. Stem Cell Rev. Rep.7 (4), 883–897. 10.1007/s12015-011-9276-0
21
Mahmoud E. E. Mawas A. S. Mohamed A. A. Noby M. A. Abdel-Hady A. A. Zayed M. (2022). Treatment strategies for meniscal lesions: from past to prospective therapeutics. Regen. Med.17 (8), 547–560. 10.2217/rme-2021-0080
22
Margiana R. Markov A. Zekiy A. O. Hamza M. U. Al-Dabbagh K. A. Al-Zubaidi S. H. et al (2022). Clinical application of mesenchymal stem cell in regenerative medicine: a narrative review. Stem Cell Res. & Ther.13 (1), 366. 10.1186/s13287-022-03054-0
23
Matsumoto F. Trudel G. Uhthoff H. K. (2002). High collagen type I and low collagen type III levels in knee joint contracture: an immunohistochemical study with histological correlate. Acta Orthop. Scand.73 (3), 335–343. 10.1080/000164702320155365
24
Morya V. K. Shahid H. Lang J. Kwak M. K. Park S. H. Noh K. C. (2024). Advancements in therapeutic approaches for degenerative tendinopathy: evaluating efficacy and challenges. Int. J. Mol. Sci.25 (21), 11846. 10.3390/ijms252111846
25
Murphy R. J. Floyd Dean B. J. Wheway K. Watkins B. Morrey M. E. Carr A. J. (2013). A novel minimally invasive ultrasound-guided technique to biopsy supraspinatus tendon. Operative Tech. Orthop.23 (2), 56–62. 10.1053/j.oto.2013.05.003
26
Ning L. J. Zhang Y. J. Zhang Y. Qing Q. Jiang Y. L. Yang J. L. et al (2015). The utilization of decellularized tendon slices to provide an inductive microenvironment for the proliferation and tenogenic differentiation of stem cells. Biomaterials52, 539–550. 10.1016/j.biomaterials.2015.02.061
27
Ning C. Li P. Gao C. Fu L. Liao Z. Tian G. et al (2023). Recent advances in tendon tissue engineering strategy. Front. Bioeng. Biotechnol.11, 1115312–2023. 10.3389/fbioe.2023.1115312
28
Onizuka S. Yamazaki Y. Park S. J. Sugimoto T. Sone Y. Sjöqvist S. et al (2020). RNA-sequencing reveals positional memory of multipotent mesenchymal stromal cells from oral and maxillofacial tissue transcriptomes. BMC Genomics21 (1), 417. 10.1186/s12864-020-06825-2
29
Pajala A. Melkko J. Leppilahti J. Ohtonen P. Soini Y. Risteli J. (2009). Tenascin-C and type I and III collagen expression in total achilles tendon rupture. An immunohistochemical study. Histol. Histopathol.24 (10), 1207–1211. 10.14670/hh-24.1207
30
Perucca Orfei C. Bowles A. C. Kouroupis D. Willman M. A. Ragni E. Kaplan L. D. et al (2021). Human tendon stem/progenitor cell features and functionality are highly influenced by in vitro culture conditions. Front. Bioeng. Biotechnol.9, 711964. 10.3389/fbioe.2021.711964
31
Pittenger M. F. Discher D. E. Péault B. M. Phinney D. G. Hare J. M. Caplan A. I. (2019). Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen. Med.4, 22. 10.1038/s41536-019-0083-6
32
Pizzute T. Lynch K. Pei M. (2015). Impact of tissue-specific stem cells on lineage-specific differentiation: a focus on the musculoskeletal system. Stem Cell Rev. Rep.11 (1), 119–132. 10.1007/s12015-014-9546-8
33
Rizzo C. Coronel L. De Lorenzis E. Rubortone P. Möller I. Miguel Perez M. et al (2025). Minimally invasive ultrasound-guided biopsy of the common extensor tendon enthesis: a cadaveric study to standardise the technique. RMD Open11 (2), e005328. 10.1136/rmdopen-2024-005328
34
Rojewski M. T. Weber B. M. Schrezenmeier H. (2008). Phenotypic characterization of mesenchymal stem cells from various tissues. Transfus. Med. Hemother35 (3), 168–184. 10.1159/000129013
35
Shen W. Chen J. Yin Z. Chen X. Liu H. Heng B. C. et al (2012). Allogenous tendon stem/progenitor cells in silk scaffold for functional shoulder repair. Cell Transpl.21 (5), 943–958. 10.3727/096368911x627453
36
Shukunami C. Takimoto A. Oro M. Hiraki Y. (2006). Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev. Biol.298 (1), 234–247. 10.1016/j.ydbio.2006.06.036
37
Stanco D. Caprara C. Ciardelli G. Mariotta L. Gola M. Minonzio G. et al (2019). Tenogenic differentiation protocol in xenogenic-free media enhances tendon-related marker expression in ASCs. PLoS One14 (2), e0212192. 10.1371/journal.pone.0212192
38
Stańczak M. Kacprzak B. Gawda P. (2024). Tendon cell biology: effect of mechanical loading. Cell Physiol. Biochem.58 (6), 677–701. 10.33594/000000743
39
Stenina-Adognravi O. Plow E. F. (2019). Thrombospondin-4 in tissue remodeling. Matrix Biol.75-76, 300–313. 10.1016/j.matbio.2017.11.006
40
Tan Q. Lui P. P. Rui Y. F. Wong Y. M. (2012). Comparison of potentials of stem cells isolated from tendon and bone marrow for musculoskeletal tissue engineering. Tissue Eng. Part A18 (7-8), 840–851. 10.1089/ten.TEA.2011.0362
41
Tempfer H. Wagner A. Gehwolf R. Lehner C. Tauber M. Resch H. et al (2009). Perivascular cells of the supraspinatus tendon express both tendon- and stem cell-related markers. Histochem Cell Biol.131 (6), 733–741. 10.1007/s00418-009-0581-5
42
Thaker H. Sharma A. K. (2012). Engaging stem cells for customized tendon regeneration. Stem Cells Int.2012, 309187. 10.1155/2012/309187
43
Thankam F. G. Roesch Z. K. Dilisio M. F. Radwan M. M. Kovilam A. Gross R. M. et al (2018). Association of inflammatory responses and ECM disorganization with HMGB1 upregulation and NLRP3 inflammasome activation in the injured rotator cuff tendon. Sci. Rep.8 (1), 8918. 10.1038/s41598-018-27250-2
44
Titan A. L. Foster D. S. Chang J. Longaker M. T. (2019). Flexor tendon: development, healing, adhesion formation, and contributing growth factors. Plast. Reconstr. Surg.144 (4), 639e–647e. 10.1097/prs.0000000000006048
45
Trapana J. Weinerman J. Lee D. Sedani A. Constantinescu D. Best T. M. et al (2024). Cell-based therapy in the treatment of musculoskeletal diseases. Stem Cells Transl. Med.13 (10), 959–978. 10.1093/stcltm/szae049
46
Tsai W. C. Pang J. H. Hsu C. C. Chu N. K. Lin M. S. Hu C. F. (2006). Ultrasound stimulation of types I and III collagen expression of tendon cell and upregulation of transforming growth factor beta. J. Orthop. Res.24 (6), 1310–1316. 10.1002/jor.20130
47
van den Boom N. A. C. Winters M. Haisma H. J. Moen M. H. (2020). Efficacy of stem cell therapy for tendon disorders: a systematic review. Orthop. J. Sports Med.8 (4), 2325967120915857. 10.1177/2325967120915857
48
Wang Z. Gerstein M. Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet.10 (1), 57–63. 10.1038/nrg2484
49
Wu T. Liu Y. Wang B. Sun Y. Xu J. Yuk-Wai L. W. et al (2016). The use of cocultured mesenchymal stem cells with tendon-derived stem cells as a better cell source for tendon repair. Tissue Eng. Part A22 (19-20), 1229–1240. 10.1089/ten.TEA.2016.0248
50
Wu Y. F. Chen C. Tang J. B. Mao W. F. (2020). Growth and stem cell characteristics of tendon-derived cells with different initial seeding densities: an in vitro study in Mouse flexor tendon cells. Stem Cells Dev.29 (15), 1016–1025. 10.1089/scd.2020.0036
51
Wu S. Zhang L. Zhang R. Yang K. Wei Q. Jia Q. et al (2023). Rat bone marrow mesenchymal stem cells induced by rrPDGF-BB promotes bone regeneration during distraction osteogenesis. Front. Bioeng. Biotechnol.11, 1110703. 10.3389/fbioe.2023.1110703
52
Xu K. Sun Y. Kh Al-Ani M. Wang C. Sha Y. Sung K. P. et al (2018). Synergistic promoting effects of bone morphogenetic protein 12/connective tissue growth factor on functional differentiation of tendon derived stem cells and patellar tendon window defect regeneration. J. Biomech.66, 95–102. 10.1016/j.jbiomech.2017.11.004
53
Xu K. Shao Y. Xia Y. Qian Y. Jiang N. Liu X. et al (2021). Tenascin-C regulates migration of SOX10 tendon stem cells via integrin-α9 for promoting patellar tendon remodeling. Biofactors47 (5), 768–777. 10.1002/biof.1759
54
Yan Z. Yin H. Brochhausen C. Pfeifer C. G. Alt V. Docheva D. (2020). Aged tendon stem/progenitor cells are less competent to form 3D tendon organoids due to cell autonomous and matrix production deficits. Front. Bioeng. Biotechnol.8, 406. 10.3389/fbioe.2020.00406
55
Yang Z. Cao H. Gao S. Yang M. Lyu J. Tang K. (2017). Effect of Tendon stem cells in Chitosan/β-Glycerophosphate/Collagen hydrogel on achilles tendon healing in a rat model. Med. Sci. Monit.23, 4633–4643. 10.12659/msm.906747
56
Yea J. H. Kim Y. Jo C. H. (2023). Comparison of mesenchymal stem cells from bone marrow, umbilical cord blood, and umbilical cord tissue in regeneration of a full-thickness tendon defect in vitro and in vivo. Biochem. Biophysics Rep.34, 101486. 10.1016/j.bbrep.2023.101486
57
Yi X. M. Lian H. Li S. (2022). Signaling and functions of interleukin-33 in immune regulation and diseases. Cell Insight1 (4), 100042. 10.1016/j.cellin.2022.100042
58
Zayed M. Iohara K. (2023). Age related senescence, apoptosis, and inflammation profiles in periodontal ligament cells from canine teeth. Curr. Mol. Med.23 (8), 808–814. 10.2174/1566524022666220520124630
59
Zayed M. N. Schumacher J. Misk N. Dhar M. S. (2016). Effects of pro-inflammatory cytokines on chondrogenesis of equine mesenchymal stromal cells derived from bone marrow or synovial fluid. Vet. J.217, 26–32. 10.1016/j.tvjl.2016.05.014
60
Zayed M. Adair S. Dhar M. (2021). Effects of normal synovial fluid and interferon gamma on chondrogenic capability and immunomodulatory potential respectively on equine mesenchymal stem cells. Int. J. Mol. Sci.22 (12), 6391. 10.3390/ijms22126391
61
Zayed M. Kook S. H. Jeong B. H. (2023). Potential therapeutic use of stem cells for prion diseases. Cells12 (19), 2413. 10.3390/cells12192413
62
Zhang J. Li B. Wang J. H. (2011). The role of engineered tendon matrix in the stemness of tendon stem cells in vitro and the promotion of tendon-like tissue formation in vivo. Biomaterials32 (29), 6972–6981. 10.1016/j.biomaterials.2011.05.088
63
Zhang Y. J. Qing Q. Zhang Y. J. Ning L. J. Cui J. Yao X. et al (2019). Enhancement of tenogenic differentiation of rat tendon-derived stem cells by biglycan. J. Cell Physiol.234 (9), 15898–15910. 10.1002/jcp.28247
64
Zhang M. Liu H. Cui Q. Han P. Yang S. Shi M. et al (2020). Tendon stem cell-derived exosomes regulate inflammation and promote the high-quality healing of injured tendon. Stem Cell Res. Ther.11 (1), 402. 10.1186/s13287-020-01918-x
65
Zhang H. Dai Y. Long H. Cao R. Shi L. Zhao J. et al (2023). Tendon stem/progenitor Cell-laden nanofiber hydrogel enhanced functional repair of patellar tendon. Tissue Eng. Part A29 (5-6), 150–160. 10.1089/ten.TEA.2022.0183
66
Zhang K. Zhang P. Shi G. Wang L. Sun C. Xiang W. (2024). Tendon extracellular-matrix-derived tissue engineering micro-tissue for achilles tendon injury regeneration in rats. J. Orthop. Surg. Res.19 (1), 377. 10.1186/s13018-024-04863-0
67
Zhidu S. Ying T. Rui J. Chao Z. (2024). Translational potential of mesenchymal stem cells in regenerative therapies for human diseases: challenges and opportunities. Stem Cell Res. & Ther.15 (1), 266. 10.1186/s13287-024-03885-z
Summary
Keywords
mesenchymal stem cells, Achilles tendon, tendon-derived mesenchymal stem cells, bone marrow, RNA-seq, tendon repair
Citation
Khaled H, Zayed M, Kim B, Jeong B-H and Oh S-I (2025) Enhanced tenogenic potential of tendon-derived mesenchymal stem cells: transcriptomic profiling and in vivo validation. Front. Cell Dev. Biol. 13:1687816. doi: 10.3389/fcell.2025.1687816
Received
18 August 2025
Accepted
22 September 2025
Published
16 October 2025
Volume
13 - 2025
Edited by
Diana Hernandez, Anthony Nolan, United Kingdom
Reviewed by
Kristin Bowers, Medtronic, United States
Ya Fang Wu, Affiliated Hospital 2 of Nantong Univeristy, China
Updates
Copyright
© 2025 Khaled, Zayed, Kim, Jeong and Oh.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Hoon Jeong, bhjeong@jbnu.ac.kr; Sang-Ik Oh, sioh@jbnu.ac.kr
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.