- 1Hospital of Stomatology, Guanghua School of Stomatology, Guangdong Provincial Key Laboratory of Stomatology, Sun Yat-sen University, Guangzhou, China
- 2College of Stomatology, Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences, Chongqing Municipal Key Laboratory of Oral Biomedical Engineering of Higher Education, Chongqing Medical University, Chongqing, China
Background: Periodontitis is a highly prevalent chronic inflammatory disease characterized by the progressive destruction of periodontal tissues, which can lead to tooth loss and affect systemic health. This pathological process is driven by both epithelial barrier disruption and a self-perpetuating cycle of dysregulated inflammatory immune responses. Although neurotransmitters, including acetylcholine, are abundant in saliva and gingival crevicular fluid, their role as key mediators of immune homeostasis in the pathogenesis of periodontitis remains poorly understood.
Methods: Utilizing single-cell RNA sequencing (scRNA-seq) data (205,334 cells, 40 human gingival samples) and gingival spatial transcriptomics data (46,230–25 μm2 spots), we revealed that the gingival epithelium exhibits the most significant functional reprogramming of neural signaling pathways in the periodontitis state. Through experiments in vivo and in vitro, we validated the functional role of acetylcholine in periodontitis.
Results: Our findings reveal that cholinergic signals change with the progression of periodontitis and that gingival epithelial cells possess an extensive distribution of non-α7-type nicotinic receptors. The acetylcholine-degrading enzyme, acetylcholinesterase (AChE), is primarily expressed by myeloid immune cells that extensively infiltrate the epithelium, and its expression is significantly upregulated following periodontal treatment. In human oral keratinocytes (HOKs), acetylcholine played a dual role: it promoted epithelial barrier repair by reversing Porphyromonas gingivalis (P. gingivalis)-induced tight junction disruption, yet it also exacerbated inflammation by upregulating key chemokines and inflammasome components. In vivo, mouse models of periodontitis showed that topical application of acetylcholine aggravated periodontal tissue damage.
Conclusion: In conclusion, our results reveal a complex, multifaceted role for acetylcholine in periodontal pathogenesis, highlighting its ability to both protect the epithelial barrier and drive inflammatory tissue destruction. These findings establish a new “neuro-epithelial-immune axis” in the pathogenesis of periodontal disease and reveal potential targets for therapeutic intervention.
1 Introduction
Periodontitis is a prevalent chronic inflammatory disease characterized by the progressive destruction of the tooth-supporting apparatus, including the gingiva, periodontal ligament, and alveolar bone (Slots, 2017). Its pathogenesis is driven by a complex interplay between a dysbiotic microbial community and a dysregulated host immune–inflammatory response (Xu et al., 2020; Li et al., 2025b). At the forefront of this host defense is the gingival epithelium (Takahashi et al., 2019; Easter et al., 2024), which acts as a crucial physical barrier and the first line of defense against invading oral pathogens such as Porphyromonas gingivalis (P. gingivalis) (Wang et al., 2024).
Although the roles of immune cells and microorganisms in periodontitis are well established, the contribution of the nervous system to this intricate disease process remains a frontier of research (Emanuel et al., 2024). The oral mucosa is richly innervated (Moayedi et al., 2022); nerves and neuropeptides influence gingival blood flow and immune cell function (Sakallioglu et al., 2008), thus modulating local inflammation. Acetylcholine is known to regulate inflammation and tissue integrity in various organ systems through the “cholinergic anti-inflammatory pathway,” which typically involves the α7 nicotinic acetylcholine receptor (α7nAChR) and is thought to have a protective effect (Wang et al., 2003; Guzman-Mejia et al., 2018). However, the specific function and regulatory mechanisms of the cholinergic system within the unique inflammatory microenvironment of the gingival tissues are not fully understood.
To comprehensively investigate the cellular and molecular changes underlying periodontitis, we utilized an integrated approach combining single-cell RNA sequencing (scRNA-seq) and our published spatial transcriptomics data (Shen et al., 2024). By analyzing a large cohort of human gingival samples from healthy, periodontitis, and treated periodontitis patients (Caetano et al., 2021; Williams et al., 2021; Chen et al., 2022; Zhu and Huang, 2023), we aimed to determine which neuro-signaling pathways are most affected and map the spatial distribution of their corresponding receptors. In addition, the corresponding functional roles were subsequently validated through in vivo and in vitro experiments.
In this study, we provide a new paradigm for understanding periodontitis pathogenesis by focusing on the neuro-epithelial axis, which reveals a dual, context-dependent role for acetylcholine and provides a theoretical foundation for developing targeted therapeutic strategies.
2 Materials and methods
2.1 Cell culture
The immortalized human oral keratinocyte cell line HOK-16B was seeded at a density of 4 × 105 cells/well in six-well plates. Cells were cultured in defined keratinocyte–SFM basal medium (Gibco, NY, United States) supplemented with 0.2% defined keratinocyte–SFM Growth Supplement (Gibco) and 1% penicillin–streptomycin. Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. Prior to bacterial stimulation, cells were starved for 2 h using growth supplement-free medium.
2.2 Bacterial culture
Porphyromonas gingivalis (P. gingivalis, standard strain ATCC 33277) was cultured anaerobically at 37 °C (80% N2, 10% H2, and 10% CO2) in tryptic soy broth (TSB) supplemented with 0.1% yeast extract, 1 μg/mL menadione, and 5 μg/mL hemin (pH 7.46). Bacteria were inoculated from TSB blood agar plates into liquid medium and grown to the logarithmic phase. Bacterial concentration was measured at 600 nm using spectrophotometry, where OD 1.0 was equivalent to 109/mL of P. gingivalis. After low-speed centrifugation, P. gingivalis was resuspended in growth medium at a concentration of 1010/mL. The multiplicity of infection (MOI) used for bacterial stimulation was 50.
2.3 Cell immunofluorescence
HOK-16B cells were seeded at a density of 2 × 105 cells/well in 12-well plates containing coverslips until reaching 50% confluence, followed by stimulation with P. gingivalis for 24 h. Coverslips were washed three times with PBS and fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature. Cells were then permeabilized with 0.5% Triton X-100 for 10 min and blocked with blocking buffer (1% BSA/5% normal goat serum) for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C: mouse anti-NLRP3 (1:500, Proteintech, Wuhan, China, 68102) and rabbit anti-ASC(PYCARD) (1:500, Proteintech, 10500). Fluorophore-conjugated secondary antibodies were then applied for 1 h: CoraLite Plus 488-Goat Anti-Mouse Recombinant Secondary Antibody (H + L) (1:2000, Proteintech, RGAM002) and CoraLite Plus 647-Goat Anti-Rabbit Recombinant Secondary Antibody (H + L) (1:2000, Proteintech, RGAR005). CoraLite 594-Phalloidin antibody (1:500, Proteintech, PF00003) was incubated for 20 min at room temperature. Coverslips were mounted using Antifade Mounting Medium with DAPI (Beyotime, Shanghai, China, P0131) and imaged using an Olympus FV 3000 Laser Scanning Confocal Microscope. Pseudocolor adjustments were made for the 594 and 647 channels to enhance visual discrimination.
2.4 Enzyme-linked immunosorbent assay
After treating human oral keratinocytes (HOKs) with acetylcholine and P. gingivalis either alone or in combination, the cells were cultured in new medium (for the removal of P. gingivalis) for 2 h, and the supernatant was collected. The levels of CXCL1 and CXCL8 were then measured using an enzyme-linked immunosorbent assay (ELISA) kit (Proteintech, KE00133 and KE00453), according to the manufacturer’s instructions. In brief, samples and standards were added to pre-coated wells, followed by incubation with detection antibodies and HRP-conjugated secondary antibodies. The enzymatic reaction was initiated by adding a TMB substrate and stopped with an acidic solution. The optical density was read at 450 nm using a microplate reader, and protein concentrations were calculated based on a standard curve.
2.5 RNA isolation and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, United States) and reverse-transcribed to first-strand cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Vazyme Biotech, China). Quantitative real-time PCR (qPCR) was performed using SYBR qPCR Master Mix (Yeasen Biotechnology, China) on a Roche LightCycler 480 II System. Human-specific primers were synthesized by Generay Biotech: OCLN (forward 5′-GACTTCAGGCAGCCTCGTTAC-3′; reverse 5′-GCCAGTTGTGTAGTCTGTCTCA-3′), CLDN1 (forward 5′-CCTCCTGGGAGTGATAGCAAT-3′; reverse 5′-GGCAACTAAAATAGCCAGACCT-3′), CDH1 (forward 5′-CGAGAGCTACACGTTCACGG-3′; reverse 5′-GGGTGTCGAGGGAAAAATAGG-3′), and GAPDH: (forward 5′-GGAGCGAGATCCCTCCAAAAT-3′; reverse 5′GGCTGTTGTCATACTTCTCATGG-3′). Gene expression levels were calculated using the 2−ΔΔCT method and normalized to GAPDH as the internal reference gene.
2.6 Patient recruitment and tissue sample collection
Gingival tissue samples from three periodontitis patients were collected from patients undergoing tooth extraction at the Department of Oral and Maxillofacial Surgery, Affiliated Stomatology Hospital of Sun Yat-sen University (Guangzhou, China). All sample collection procedures complied with China’s “Good Clinical Practice for Drug Clinical Trials,” ICH-GCP guidelines, and relevant regulations and were approved by the Medical Ethics Committee of the Affiliated Stomatology Hospital of Sun Yat-sen University (Approval No.: KQEC-2022-14-01). Informed consent was obtained from all participants prior to enrollment. The inclusion criteria for periodontitis patients included the affected tooth having a probing depth (PD) of > 4 mm and a bleeding index (BI) of ≥ 3 upon probing. Exclusion criteria included patients with concurrent systemic diseases.
2.7 Tissue processing and immunofluorescence
Collected gingival tissues were placed in Hank’s balanced salt solution and fixed in 4% PFA for 10 min, followed by further fixation in 4% PFA at 4 °C for 24 h. Tissues were then dehydrated through a graded ethanol series (70%–100%), cleared in xylene, and embedded in paraffin. Continuous 5 μm sections were prepared, baked at 60 °C for 2 h, dewaxed in xylene, and rehydrated through a graded ethanol series for immunofluorescence staining. Antigen retrieval was performed using citrate buffer (pH 6.0) at 95 °C, followed by blocking with blocking buffer (1% BSA/5% normal goat serum) for 1 h at room temperature. Sections were incubated overnight at 4 °C with rabbit anti-acetylcholinesterase polyclonal antibody (1:500, Proteintech, 17975-1-AP), followed by incubation with CoraLite 488-conjugated Goat Anti-Rabbit IgG (H + L) (1:2000, Proteintech, SA00013-2) for 1 h. Sections were mounted using Antifade Mounting Medium with DAPI (Beyotime, P0131) and imaged using an Olympus FV 3000 Laser Scanning Confocal Microscope.
2.8 Mouse experimental periodontitis model
The Institutional Animal Care and Use Committee (IACUC) at Sun Yat-sen University reviewed and approved the animal use protocol (SYSU-IACUC-2024-000348). This evaluation adhered to the standards of the Animal Welfare Act and the U.S. Public Health Service’s “Guide for the Care and Use of Laboratory Animals.” All animals were housed in a specific pathogen-free (SPF) environment with controlled humidity and temperature on a 12-h light–dark cycle.
Mice were randomly assigned to the HC (healthy control), PD (periodontitis), and PD + ACh treatment groups. Under anesthesia (ketamine/xylazine), periodontitis was induced by placing a 0.15-mm-diameter silk ligature around the maxillary second molars, ligated at both the mesial and distal sides. The ligature was tied securely on the buccal side, and a 1 × 1010 CFU/mL P. gingivalis (Pg) suspension was applied every 3 days under isoflurane anesthesia. Freshly prepared acetylcholine solution was administered daily in the PD + ACh treatment group, while the PD group received a control aqueous solution of the same pH value. After 14 days, the mice were euthanized, and maxillary blocks were harvested and fixed in 4% paraformaldehyde. Mice that experienced premature ligature loss were excluded. Bone loss was assessed using micro-computed tomography (micro-CT). Additionally, a random data-processing method was used to mitigate potential confounding factors.
After decalcification with disodium ethylenediaminetetraacetate (EDTA), the mouse maxillae were embedded in paraffin, sectioned into 5 µm-thick slices, and stained with hematoxylin and eosin (H&E). Serial sections of the buccal and lingual sides of the periodontal area were observed under a microscope, and images were captured using a PANNORAMIC SCAN scanner.
2.9 Statistical analysis
Data were analyzed using GraphPad Prism 10 and expressed as the mean ± standard error of the mean (SEM). Statistical significance was evaluated using t-tests (for two-group comparisons) and one-way ANOVA (three or more groups, followed by Tukey’s HSD post hoc test). Significant differences were denoted as follows: *p < 0.05, **p < 0.01, and ***p < 0.001.
2.10 RNA sequencing
Library preparation: Total RNA was isolated from HOKs using TRIzol reagent. RNA integrity and concentration were evaluated using an Agilent 2100 Bioanalyzer. Polyadenylated mRNA was enriched via oligo(dT)-conjugated magnetic beads and subsequently fragmented in NEB fragmentation buffer containing divalent cations. First-strand cDNA synthesis was performed using the M-MuLV reverse transcriptase system, followed by second-strand synthesis using DNA polymerase I and deoxynucleotide triphosphates (dNTPs). The resulting double-stranded cDNA was purified and subjected to end repair, A-tailing, and adapter ligation. cDNA fragments of approximately 250–300 bp were size-selected using AMPure XP beads, PCR-amplified, and further purified with AMPure XP beads to generate the final library. Initial quantification was conducted using a Qubit 2.0 Fluorometer, followed by dilution of libraries to 1.5 ng/μL. Insert size distribution was assessed using an Agilent 2100 Bioanalyzer, and accurate quantification of library effective concentration (>1.5 nM) was achieved via qRT-PCR to ensure high-quality preparation.
Sequencing and quality control: Libraries were pooled according to effective concentration and subjected to Illumina sequencing, generating 150 bp paired-end reads. During sequencing, four fluorescence-labeled dNTPs, DNA polymerase, and adapter primers were added to the flow cell for amplification. The optical signals were converted to sequencing peaks using computer software to obtain sequence information. Raw image data were base-called using CASAVA into sequence data (reads) in FASTQ format, followed by filtering to remove low-quality data.
Gene expression quantification: Reference genome indices were built using HISAT2 (v2.0.5), and paired-end clean reads were aligned to the human reference genome using HISAT2 (v2.0.5). Read counts mapped to each gene were calculated using featureCounts (v1.5.0-p3).
Data analysis: Differential expression analysis between groups was performed using DESeq2 (v1.46.0). Gene set enrichment analysis (GSEA) for GO functional annotation was conducted using clusterProfiler (v4.14.4). GSEA was performed using fgsea (v1.32.2) and msigdb (v1.14.0).
2.11 Single-cell data analysis
Data acquisition: Previously published human gingival single-cell datasets, including GSE152042, GSE164241, GSE171213, and GSE207502, were integrated. These datasets were carefully selected to represent both healthy and diseased states, encompassing healthy, periodontitis, and treated periodontitis samples, while excluding non-gingival periodontal tissues. The final integrated dataset comprised 40 samples and 205,334 cells.
Integration and clustering: All analyses were performed using the Seurat (v5.1.0) and Harmony (v1.2.3) R packages. Following quality control, 152,073 cells were retained for downstream analysis. After log normalization, highly variable genes were identified, and principal component analysis (PCA) was performed. Batch effects were corrected using Harmony. Cell clusters were identified using Seurat’s Louvain algorithm and visualized with UMAP dimensionality reduction. Cell types were annotated based on canonical markers, and immune cell subtypes were further refined through secondary clustering.
Data analysis: Neural receptor pathway-related gene sets (GO:0099637, GO:0099601, GO:0098962, and GO:0099529; R-HSA-112314) were obtained from the MsigDB website and intersected with highly variable genes (HVGs) from different cell clusters. These were further intersected with significantly differentially expressed genes (DEGs) in epithelial cell populations across groups (healthy and periodontitis), followed by KEGG pathway clustering using clusterProfiler (v4.14.4).
2.12 Spatial transcriptomics data analysis
Stereo-sequencing data were processed as previously described (Shen et al., 2024). Data were captured on a silicon chip covered with 220 nm × 220 nm DNA nanoballs (DNBs). The raw spatial expression matrix was converted into 25 μm2 pseudo-spots (50 × 50 DNBs per spot) and imported into a Seurat object, where it was normalized using the SCTransform function.
Subsequently, we performed PCA for dimensionality reduction. A K-nearest neighbor (KNN) graph was constructed based on the Euclidean distance, and cell clusters were identified using the FindClusters function. The FindAllMarkers function was then used to identify marker genes for each cluster, allowing us to classify the four cell types based on these markers. The gingival spots from Stereo-seq were color-coded to reflect the expression levels of key genes, including cell type identification markers (KRT15, COL1A1, PECAM1, and PTPRC) and acetylcholine receptors.
3 Results
3.1 Single-cell and spatial transcriptomic atlases reveal the cellular landscape of gingival tissues in periodontal health and disease
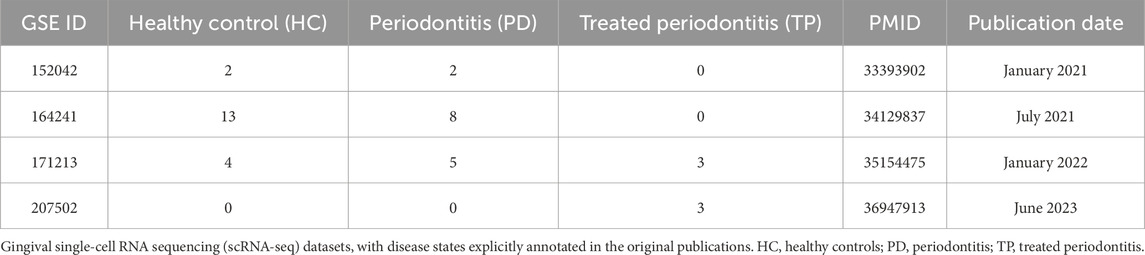
The scRNA-seq datasets comprised 205,334 cells from 40 samples across three clinical categories: 19 healthy controls (HC), 15 periodontitis patients (PD), and 6 treated periodontitis cases (TP) (Table 1; Figure 1A) (Caetano et al., 2021; Williams et al., 2021; Chen et al., 2022; Zhu and Huang, 2023). The spatial transcriptomics data included HC and PD samples with 46,230 spots at a resolution of 25 μm2 (Shen et al., 2024).
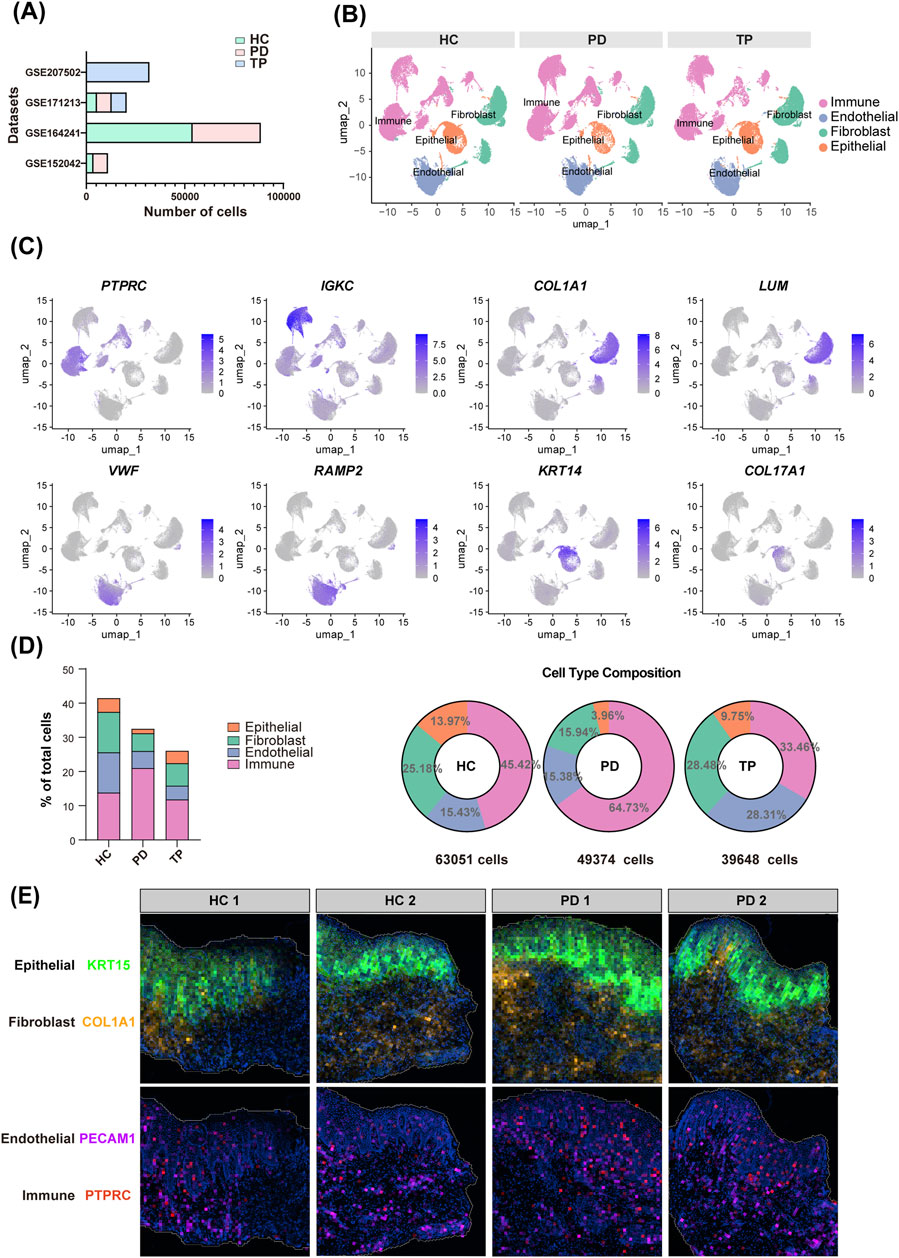
Figure 1. Gingival cellular dynamics landscape during periodontitis progression. (A) Dataset sources (y-axis) and sequenced cell count (x-axis) of gingival single-cell RNA sequencing (scRNA-seq). Groups: healthy control (HC, blue), periodontitis (PD, red), and treated periodontitis (TP, green). (B) Uniform manifold approximation and projection (UMAP) visualization of the integrated single-cell data from HC, PD, and TP groups. (C) Marker gene validation: representative transcripts defining the major cell lineages. (D) The total cell percentage for different cell types across the entire dataset (left) and the proportional composition of cell types within each group (right). (E) Spatial mapping of the gene expressions of KRT15, COL1A1, PECAM1, and PTPRC. Groups: healthy controls (HC) and periodontitis (PD).
After quality control, we classified the single-cell populations using classical markers (Williams et al., 2021): immune cells (PTPRC and IGKC), fibroblasts (COL1A1 and LUM), endothelial cells (VWF and RAMP2), and epithelial cells (KRT14 and COL17A1) (Figures 1B,C). Quantitative cell analysis identified epithelial cells as the least abundant population, while immune cells were the most predominant. The PD group showed a pronounced infiltration of immune cells and a concomitant reduction in epithelial cells (Figure 1D), which reflects the pathological inflammatory tissue destruction observed in periodontitis (Kim et al., 2024).
Spatial transcriptomic analysis further revealed cellular distributions within the gingiva (Figure 1E). The expression of KRT15, a basal epithelial marker, clearly delineated the boundary between the epithelium and the lamina propria, while PTPRC, a pan-immune marker, revealed the spatial infiltration of immune cells in periodontitis.
3.2 Epithelial cells show the most pronounced neurotransmitter signaling-related gene reprogramming during periodontitis
A cross-analysis between HVGs and neurotransmitter signaling-related gene sets (Table 2) indicated that endothelial cells exhibited the most extensive potential for neural interactions (Figure 2A). Endothelial cells highly express G proteins (GNG11 and GNAI2), which act as “switches” for neurotransmitter receptor (e.g., GPCR) signal transduction (Liccardo et al., 2022), along with key regulators of signal transduction and vesicle transport, such as RAC1 and RAB11A (Hulsbusch et al., 2015). This observation can be attributed to the dense sympathetic and sensory innervation of microvessels, where endothelial cells rapidly transmit neural signals to regulate local blood flow and inflammatory extravasation (de Juan et al., 2019; Forrester et al., 2024). Epithelial cells characteristically express ADRB2 (β-2 adrenergic receptor)—a classic neurotransmitter receptor (Li et al., 2025a)—and NSG1, which can participate in neurotransmitter receptor endocytosis and transport (Barford et al., 2017), demonstrating the potential of epithelial cells to receive neurotransmitter signals. Fibroblasts express GNAI1 (G protein subunit) and NPTN (neuroplastin, an adhesion molecule) (Ilic et al., 2021), suggesting that fibroblasts might also respond to neural signals. In contrast, immune cells showed the minimal signatures of direct neural interactions, suggesting that their regulation primarily occurs through indirect neuro-stromal-immune crosstalk (Hodo et al., 2020).
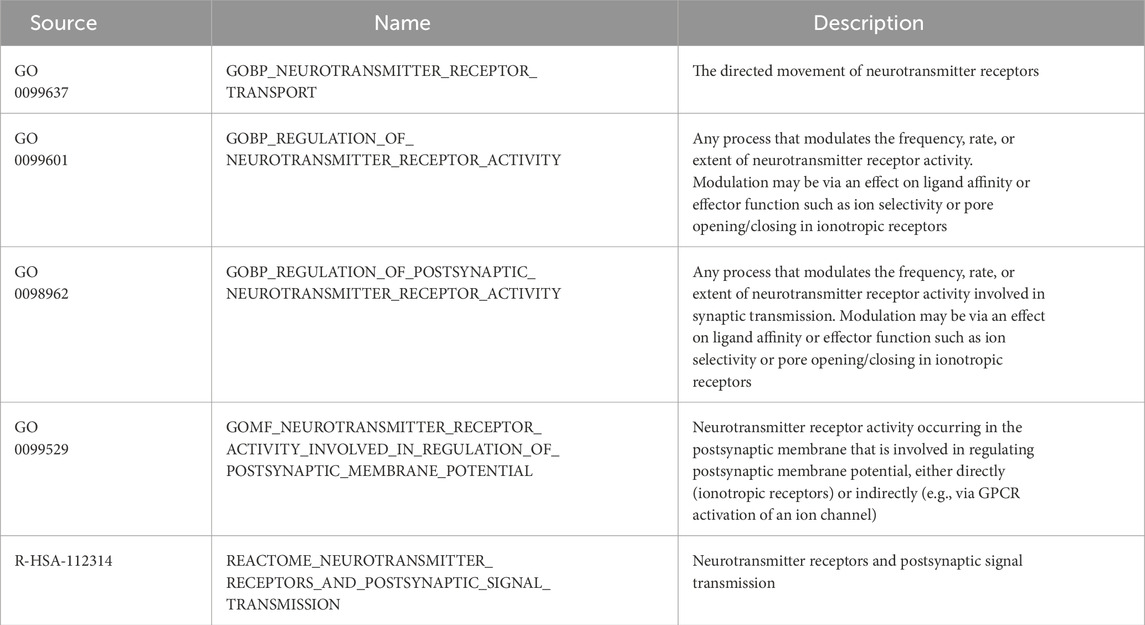
Table 2. Neurotransmitter signaling-related gene sets from the Molecular Signatures Database (MSigDB).
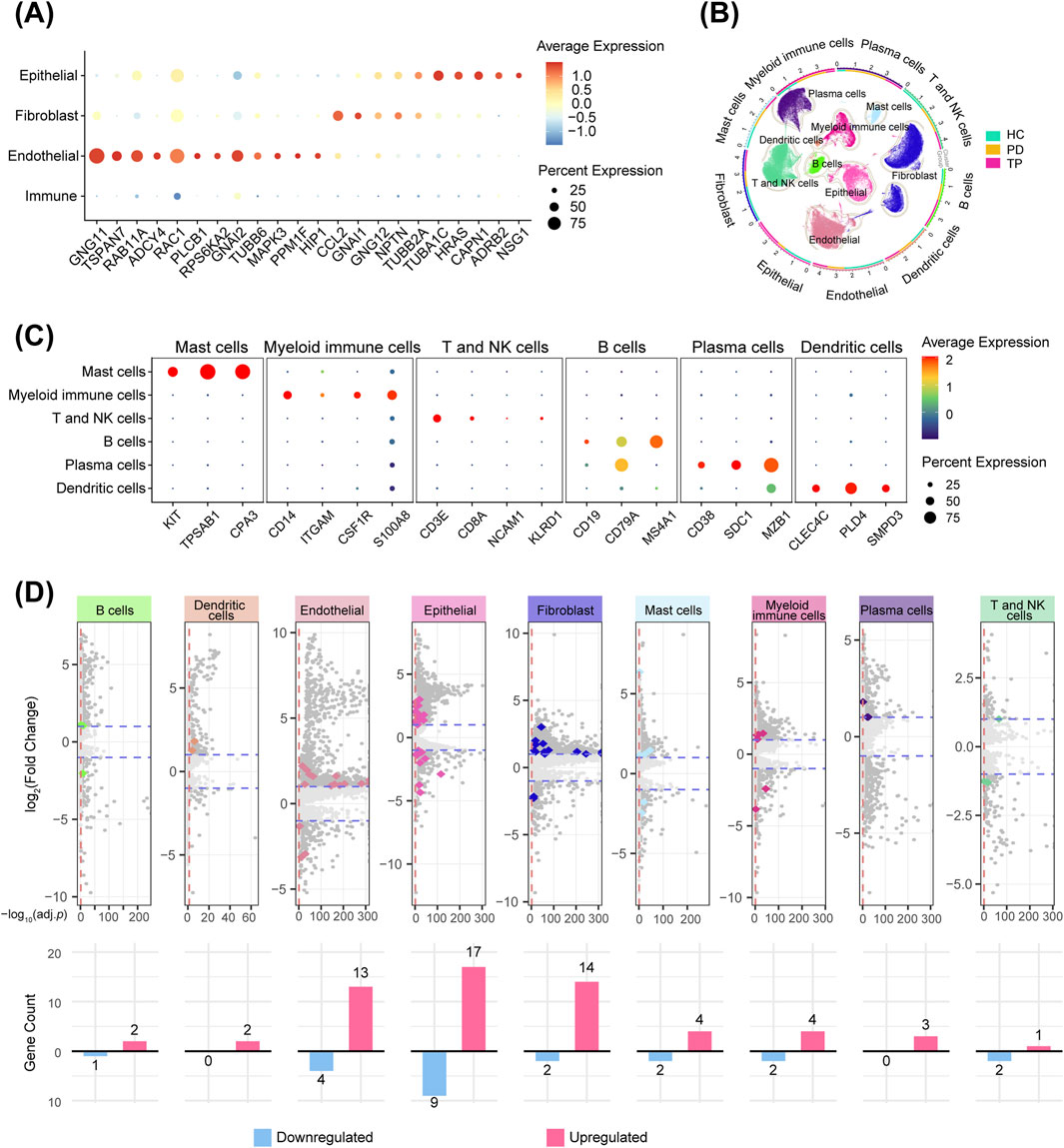
Figure 2. Gingival neurotransmitter signaling-related gene changes during periodontitis progression. (A) Cross-analysis of highly variable genes (HVGs, top 2000) and neurotransmitter signaling-related genes (Table 2) in four gingival cell types. (B) Circular UMAP plots of cell subpopulations. The outer ring and scatter plot colors indicate different cell types, while the inner ring colors represent the cell number ratio among the groups. (C) Markers used for immune cell identification, visualized with a bubble plot. (D) Volcano plots showing DEGs between the PD and HC groups for each cell type. Darker dots represent genes with a |log2-fold change| > 1 and an adjusted p-value <0.05. Colored dots represent genes from the neurotransmitter signaling-related gene sets, and the bar graphs show their counts (upregulated and downregulated in periodontitis).
Immunophenotyping using canonical markers identified distinct immune cell subpopulations (Figures 2B,C), enabling a comparative analysis between the periodontitis and healthy groups across all signature subpopulations. The cross-analysis of significant DEGs (p < 0.05), with neurotransmitter signaling-related gene sets, revealed that, among all cell types, gingival epithelial cells exhibited the most distinct rearrangement of neurotransmitter signaling-related genes during the progression of periodontitis (Figure 2D). This finding is particularly significant as the epithelium acts as the primary barrier against periodontal pathogens (Chang et al., 2021). Its unique neurotransmitter-mediated reactivity prompted our further investigation into its potential regulatory mechanisms.
3.3 Acetylcholine signaling and receptors in the periodontal epithelium
Next, we performed enrichment analysis on epithelial DEGs related to neurotransmitter signaling under various periodontitis conditions. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of upregulated genes in PD vs. HC showed that glutamatergic synapse, GABAergic synapse, cholinergic synapse, and chemokine signaling pathways were the most significantly enriched. Among these, the cholinergic synapse pathway showed the most pronounced downregulation in the comparison between TP and PD (Figure 3A), suggesting its dynamic regulation during disease remission. The evolutionarily conserved cholinergic system mediates neuron–peripheral organ communication (Alen, 2022), as exemplified by a 14-fold increase in acetylcholine observed in atopic dermatitis (Wessler et al., 2003). Previous studies have shown that gingival tissues express characteristic acetylcholine receptors (nicotinic/muscarinic) and metabolic enzymes, including acetylcholinesterase (AChE) (Arredondo et al., 2003; Meric et al., 2019). More importantly, acetylcholine levels are significantly elevated in the saliva and gingival crevicular fluid of periodontitis patients (Apatzidou et al., 2018), suggesting that cholinergic signaling may contribute to the progression of periodontitis, although its specific role remains unclear.
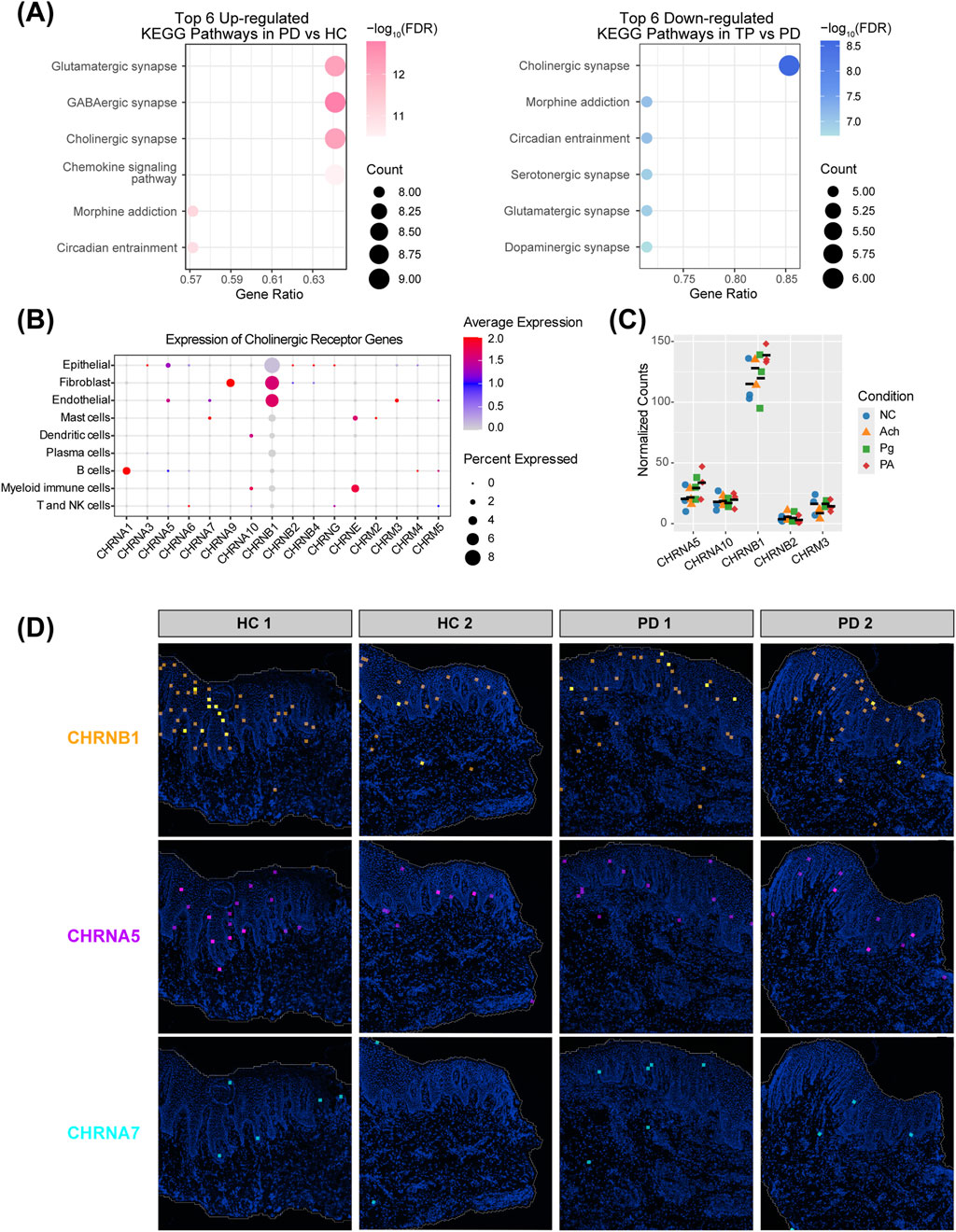
Figure 3. Acetylcholine signaling and receptor distribution in the periodontal epithelium. (A) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of epithelial neurotransmitter signaling-related DEGs. Left: upregulated pathways in PD vs. HC; right: downregulated pathways in TP vs. PD. (B) Expression profiles of acetylcholine receptors across different cell types. Receptors not detected in any cells were excluded. (C) Transcript counts of acetylcholine receptor genes from HOKs RNA-seq data. Five receptors detected in all 12 samples (n = 3 per group), with colored shapes representing groups and black bars indicating the mean. Receptors with low expression (zero counts in some samples) were excluded. (D) Expression of acetylcholine receptors CHRNB1, CHRNA5, and CHRNA7 in gingival spatial transcriptomics data.
Fibroblasts and endothelial cells show a high expression of the cholinergic receptor nicotinic beta 1 subunit (CHRNB1), whereas epithelial cells exhibit a broader but lower level. Given that the epithelium acts as the first barrier against bacterial invasion in periodontitis, is in direct contact with acetylcholine in the gingival crevicular fluid and saliva, and exhibits dynamic changes in the cholinergic synapse pathway across different stages of periodontitis (Figure 3A), we focused our subsequent investigation on epithelial cells. In addition to CHRNB1, epithelial cells also show expression of the cholinergic receptor nicotinic alpha 5 subunit (CHRNA5), with negligible expression of CHRNA7, which is considered an anti-inflammatory “pocket” (Figure 3B) (Wang et al., 2003; Guzman-Mejia et al., 2018). We performed RNA-seq on HOKs, and the captured transcripts showed a similar proportion, with CHRNB1 far exceeding other receptors, followed by CHRNA5, and very low expression of CHRNA7. This expression pattern was further corroborated by the spatial transcriptomics landscape (Figures 3C,D). This unique receptor profile suggests that the role of acetylcholine in the gingival epithelium may differ from the classic α7nAChR-dependent anti-inflammatory pathway.
3.4 Expression patterns of gingival cholinergic metabolic enzymes
The core acetylcholine synthesis gene CHAT was not detected in the whole gingiva scRNA data (data not shown) (Ferreira-Vieira et al., 2016), indicating that resident gingival cells do not produce acetylcholine themselves. Acetylcholine in gingival crevicular fluid and saliva is, therefore, likely to originate mainly from neuronal release. Interestingly, AChE was consistently detected, suggesting active regulation of cholinergic signaling in the gingiva (Korabecny and Soukup, 2021).
Immunofluorescence localized the ACHE protein primarily to immune clusters and vascular endothelium (Figure 4A, circles and arrows). The boxplot analysis shows that the median ACHE expression level in epithelial cells was lower than that in other cell types (Figure 4B). Overall, myeloid immune cells expressed the most ACHE (Figure 4C), even more than endothelial cells and fibroblasts. Further subdivision of immune cells revealed that macrophages, conventional dendritic cells, and monocytes highly expressed ACHE after periodontal treatment (Figures 4D,E). Corroborating this, spatial transcriptomics maps showed the distribution of these cell types with marker genes (Mulder et al., 2021): LYZ (myeloid immune cells), CD68 (macrophages), CD14 (monocytes), and CD1C (dendritic cells) (Figure 4F), revealing that a large number of myeloid immune cells were observed infiltrating the epithelial area. As AChE is a typical secretory hydrolase (Walczak-Nowicka and Herbet, 2021), the significant upregulation of ACHE by myeloid immune cells after periodontal treatment could facilitate the normalization of epithelial cholinergic signaling.
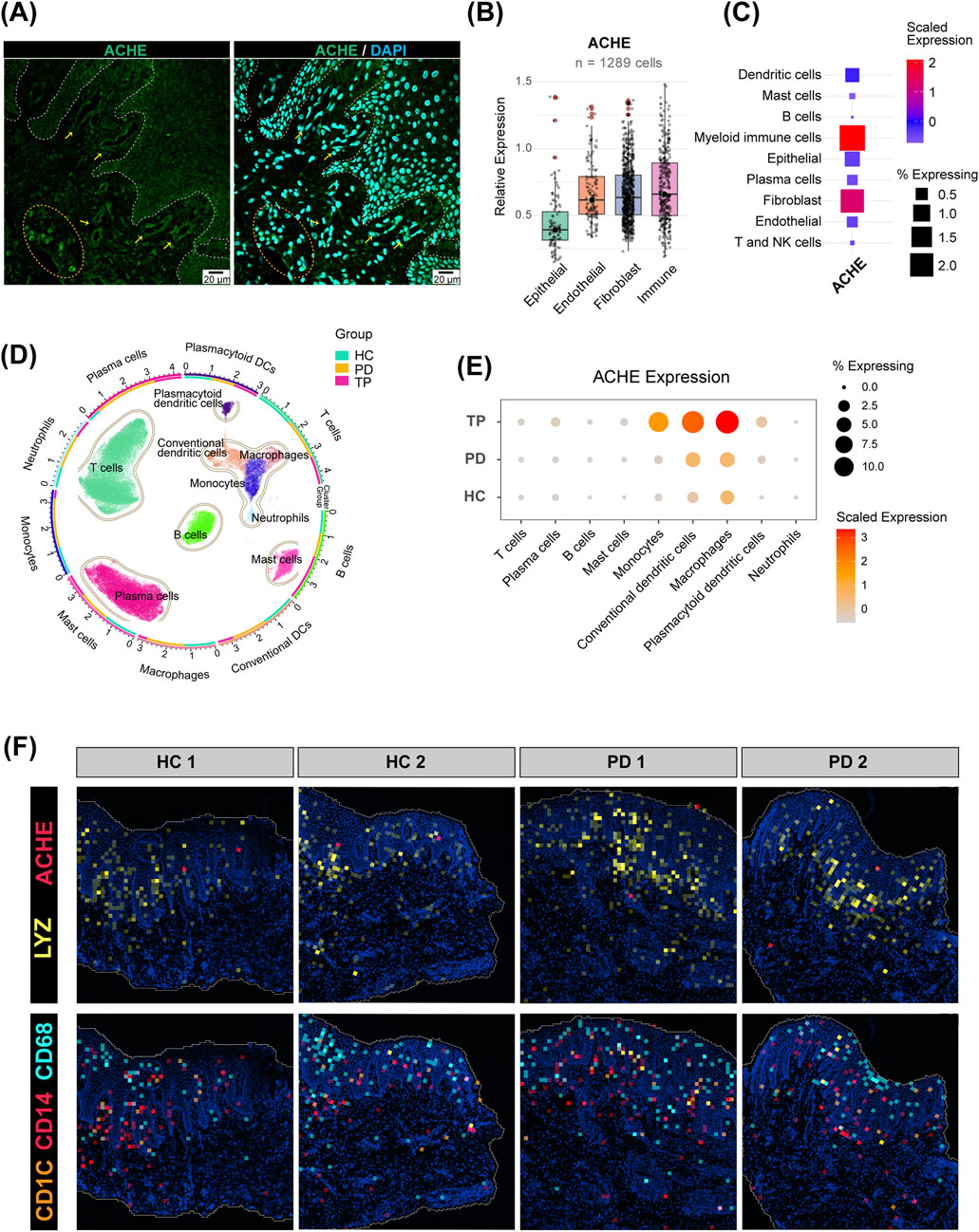
Figure 4. Expression pattern of acetylcholinesterase (AChE) in the gingiva. (A) Immunofluorescence staining of the human gingival epithelium. Cell nuclei are stained with DAPI (blue), and AChE is labeled in green (scale bar: 20 µm). The dashed line delineates the epithelial–connective tissue boundary. (B) Relative expression of AChE in the four major cell types from the scRNA-seq data. Only cells expressing AChE (expression >0) were included (n = 1,289). The center line indicates the median. (C) Expression of AChE in further refined cell subpopulations. (D) UMAP plot of isolated immune cell subpopulations. The outer ring and scatter plot colors indicate different cell types, while the inner ring colors represent the cell number ratio among the groups. (E) Expression of AChE in immune cells from the HC, PD, and TP groups. (F) Spatial distribution of AChE and myeloid immune cell marker genes in the gingiva.
3.5 Acetylcholine promotes periodontal epithelial barrier repair and chemokine expression
The periodontitis-affected gingival epithelium highly expressed tight junction genes claudin-1 (CLDN1) and E-cadherin (CDH1) (p < 0.001), while the average expression of occludin (OCLN) was also elevated, although this was not statistically significant (Figures 5A–C) (Fernandez-Lainez et al., 2023; Wen et al., 2024). Infection of HOKs with P. gingivalis, the key periodontal pathogen (Lamont et al., 2022), significantly downregulated OCLN, CLDN1, and CDH1. However, pretreatment with acetylcholine significantly reversed this epithelial barrier disruption (Figure 5D), indicating that acetylcholine may restore epithelial barrier function by maintaining tight junction integrity during P. gingivalis attack.
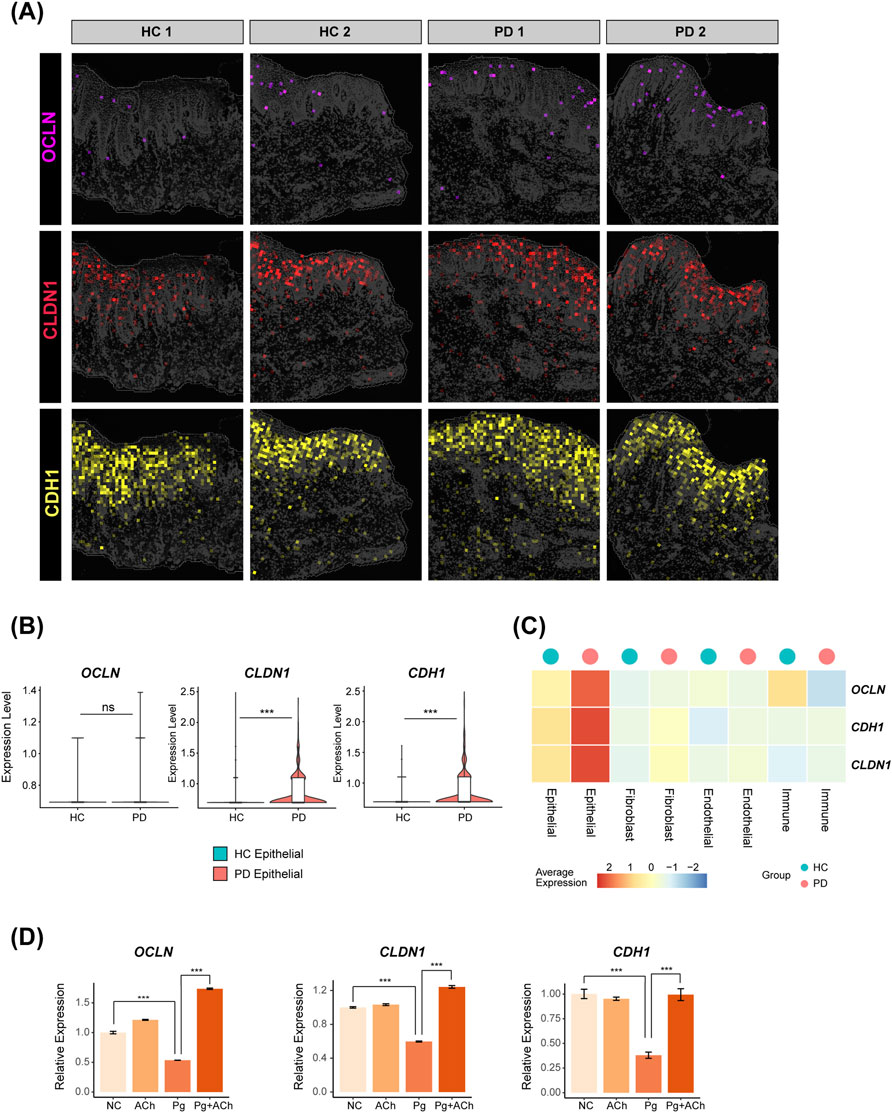
Figure 5. Distribution of tight junction genes in the periodontal gingival epithelium and their regulation by acetylcholine. (A) Expressions of OCLN, CLDN1, and CDH1 in spatial transcriptomics data. (B) Differential comparison of OCLN, CLDN1, and CDH1 in the epithelial subpopulation of HC versus PD groups (***, p < 0.001). (C) Heatmap showing the expressions of OCLN, CLDN1, and CDH1 for each cluster in HC and PD gingiva. (D) Quantitative polymerase chain reaction (qPCR) validation in HOKs. Data are presented as the mean ± standard error of the mean (SEM). ***, p < 0.001.
To further investigate the effects of acetylcholine on the epithelium under periodontitis environment, we performed RNA-seq analysis on HOKs under four experimental conditions: untreated control (NC), acetylcholine (ACh, 100 nM), P. gingivalis infection (Pg, MOI = 50), and combined P. gingivalis infection with acetylcholine treatment (Pg + ACh). Gene set enrichment analysis of Gene Ontology (GSEA-GO) revealed that the P. gingivalis attack induced the upregulation of several biological processes, such as immune regulation (leukocyte/mast cell activation), hydrogen peroxide response, and antibacterial apoptosis triggered by DNA damage (Figure 6A).
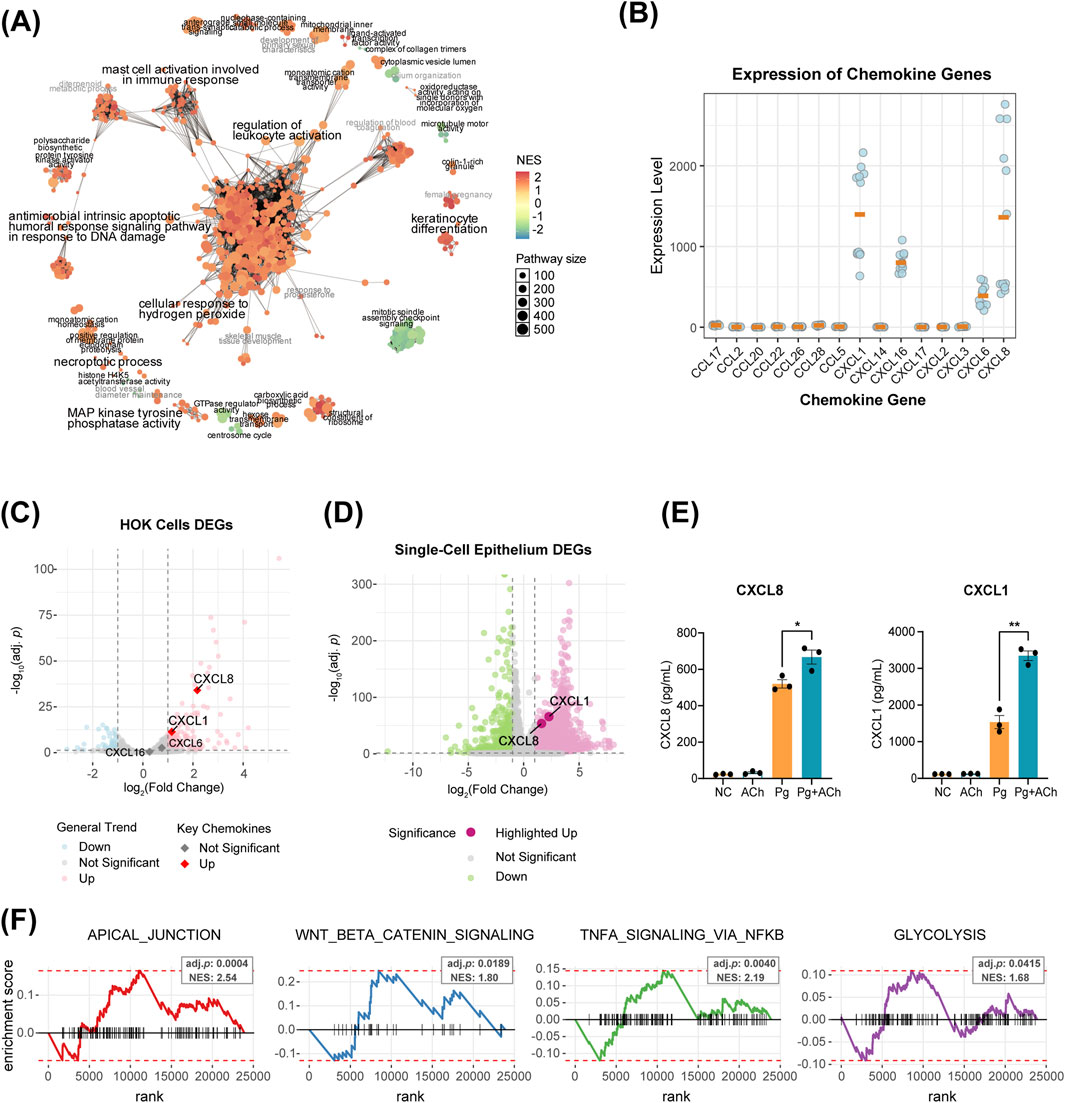
Figure 6. Acetylcholine orchestrates a multi-dimensional defense program in epithelial cells. (A) Network of GSEA-GO enrichment analysis for Pg vs. NC DEGs. Each node represents a GO term, with circle size and color indicating the pathway size and the NES score, respectively. Node-sharing clusters represent functionally related term groups. (B) Transcript counts of CXCL and CCL family genes in HOKs RNA-seq data. Only genes with non-zero counts across all 12 samples are displayed, with orange bars indicating the mean. (C) Volcano plot of DEGs in Pg vs. NC comparison. Colored dots represent significantly differential expression (|log2 fold change| > 1, adjusted p-value <0.05). (D) Volcano plot of PD vs. HC DEGs in epithelial cells from single-cell data. Colored dots represent significantly differential expression (|log2 fold change| > 1, adjusted p-value <0.05). (E) Enzyme-linked immunosorbent assay (ELISA) results of HOK supernatants for CXCL1 and CXCL8. *, p < 0.05 and **, p < 0.01. (F) Enrichment analysis of DEGs from the Pg + Ach vs. Pg groups using the fgsea function, showing four significantly enriched functional categories [adjusted p < 0.05, normalized enrichment score (NES) > 1.5].
Transcriptome analysis showed that the main chemokines expressed by HOKs were CXCL1, CXCL6, CXCL8, and CXCL16 (Figure 6B). Of these, CXCL1 and CXCL8 were significantly upregulated after P. gingivalis infection (Figure 6C) and also upregulated in periodontitis epithelial cells from scRNA-seq data (Figure 6D). ELISA of the supernatant after infection or combined treatment showed that Pg infection promoted the secretion of CXCL1 and CXCL8 proteins by HOKs, and acetylcholine exacerbated this process, further enhancing CXCL1 and CXCL8 secretions (Figure 5E).
Enrichment analysis of DEGs between the Pg + ACh and Pg groups further revealed important regulatory patterns of acetylcholine on the infected epithelium (Figure 6F): upregulation of epithelial repair pathways (APICAL_JUNCTION and WNT/BETA_CATENIN signaling), which corroborates the previous barrier protection findings (Figure 5D) (Mak et al., 2013; Kosumi et al., 2022); and upregulation of TNFA SIGNALING VIA NFKB and GLYCOLYSIS, indicating that acetylcholine exacerbates the inflammatory response and promotes reprogramming of cellular energy metabolism (Chen et al., 2023). In summary, acetylcholine exhibits a dual role in the context of P. gingivalis infection: on one hand, it enhances chemokine-mediated immune recruitment [potentially exacerbating inflammatory damage (Barreiro et al., 2010)], while on the other hand, it promotes epithelial barrier repair and cellular energy supply, demonstrating its complex regulatory function in host–pathogen interactions.
3.6 Acetylcholine exacerbates inflammasome production and periodontal destruction
HOKs were treated with acetylcholine alone, P. gingivalis alone, or P. gingivalis with acetylcholine for 24 h. Acetylcholine enhanced the expression of inflammasome components, including NOD-like receptor family pyrin domain-containing 3 (NLRP3) and apoptosis-associated speck-like protein containing a CARD (ASC, also called PYCARD) (Figures 7A,B). Similar to chemokines, while inflammasome activation helps clear pathogens, it can exacerbate tissue damage (Sharma and Kanneganti, 2021; Fu and Wu, 2023).
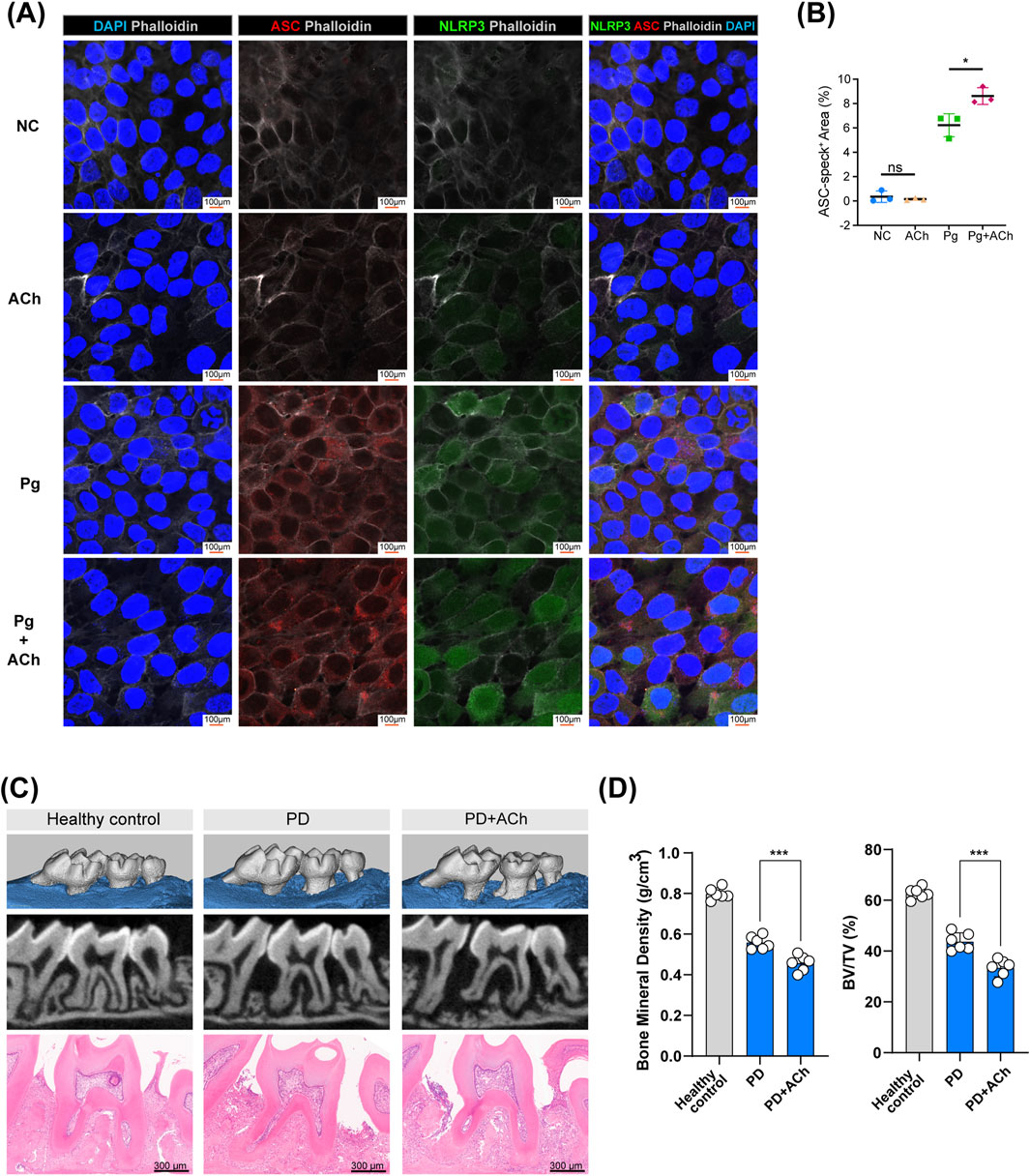
Figure 7. Acetylcholine promotes inflammasome activation and bone loss in periodontitis. (A) Representative confocal images of HOK inflammasome activation after treating with P. gingivalis (Pg), acetylcholine (ACh), or both for 24 h. ASC (red), NLRP3 (green), phalloidin (white), and DAPI (blue); scale bar: 100 µm. (B) Quantification of ASC-positive areas. *, p < 0.05; ns, no difference. (C) Micro-CT scans and H&E staining of maxillary bones from each group (control, PD, and PD + ACh). Scale bar: 300 µm. (n = 6/group) (D) Analysis of bone mineral density (BMD) and the bone volume/tissue volume (BV/TV) ratio in the second molar area from micro-CT scans, represented by individual data points (n = 6/group). ***, p < 0.001.
Mouse experimental periodontitis models, established through silk ligation and P. gingivalis inoculation for 14 days, demonstrated that daily application of acetylcholine exacerbated periodontal tissue destruction (Figure 7C), leading to lower alveolar bone density (Figure 7D). Acetylcholine’s dual effects—barrier protection and pro-inflammatory enhancement—ultimately worsen tissue destruction in the periodontitis environment.
4 Discussion
In this study, scRNA-seq and spatial transcriptomics analyses, combined with in vivo and in vitro experiments, reveal a novel dual role for acetylcholine signaling in periodontitis. We first identified that although multiple cell types interact with neural signals, gingival epithelial cells undergo the most significant reprogramming of neurotransmitter-related gene expression, particularly involving the cholinergic synapse pathway, during the progression from health to periodontitis. Investigating this further, we found that epithelial cells express a distinct cholinergic receptor profile, characterized by high levels of non-α7 nicotinic receptors CHRNB1 and CHRNA5, with minimal expression of the canonical anti-inflammatory receptor CHRNA7. Regarding the source and regulation of acetylcholine, we observed negligible expression of the synthesis enzyme CHAT in resident gingival cells, suggesting that acetylcholine likely originates from neural release or fluids such as saliva. Conversely, the degrading enzyme AChE is prominently expressed, primarily by infiltrating myeloid immune cells, and its levels increase significantly following periodontal treatment, indicating active regulation of local acetylcholine concentrations. Functionally, in vitro experiments on HOKs demonstrated a dual role for acetylcholine in the context of P. gingivalis infection: it promoted epithelial barrier repair by counteracting pathogen-induced tight junction disruption and simultaneously exacerbated the inflammatory response by increasing the expression and secretion of pro-inflammatory chemokines (CXCL1 and CXCL8) and inflammasome components (NLRP3 and ASC). This observation is particularly significant, given that epithelial cells serve as the first line of defense against periodontal pathogenic bacteria and are in direct contact with a large amount of neurotransmitters in saliva and gingival crevicular fluid (Sakallioglu et al., 2008; Takahashi et al., 2019). Crucially, in vivo validation in a mouse model showed that the net effect of topical acetylcholine application was an aggravation of periodontal tissue destruction and bone loss. Collectively, these findings establish a previously unappreciated “neuro-epithelial-immune axis” driven by acetylcholine in periodontitis.
A key finding of our study is the expression profile of acetylcholine receptors in gingival epithelial cells, with CHRNB1 and CHRNA5 being highly expressed, while the classic anti-inflammatory receptor CHRNA7 shows minimal expression (Burke et al., 2024). This pattern, also confirmed in HOKs, strongly suggests that acetylcholine does not exert its classic α7nAChR-mediated anti-inflammatory effects in the gingival epithelium (Hone and McIntosh, 2023).
Furthermore, our analysis of cholinergic metabolic enzymes provides an insight into the gingival acetylcholine microenvironment. The absence of CHAT transcription in resident cells suggests that acetylcholine may originate from the dense local neural network (Ferreira-Vieira et al., 2016) and from saliva and gingival crevicular fluid. In contrast, the prominent expression of AChE in myeloid immune cells and the vascular endothelium highlights the active capacity of the gingiva to regulate acetylcholine levels (Petsakou et al., 2023). The spatial transcriptomics landscape demonstrates a substantial infiltration of myeloid immune cells into the periodontal epithelium, and the significant upregulation of AChE in these myeloid cells after periodontal treatment suggests a potential feedback loop that modulates the local acetylcholine concentration, thereby facilitating the restoration of epithelial homeostasis.
Our in vitro and in vivo experiments demonstrate the complex and seemingly contradictory effects of acetylcholine. On the one hand, it acts as a protective agent by restoring the integrity of tight junctions compromised by P. gingivalis infection. This is consistent with the spatial transcriptomics finding of a general upregulation of tight junction proteins in the periodontitis-affected epithelium and highlights its potential in promoting tissue repair and maintaining mucosal defense (France and Turner, 2017). On the other hand, acetylcholine exacerbates inflammation by upregulating pro-inflammatory chemokines (CXCL1 and CXCL8) and inflammasome components (NLRP3 and ASC) (Plemmenos et al., 2021; Fu and Wu, 2023). The fact that the topical application of exogenous acetylcholine ultimately aggravated periodontal destruction in a mouse model confirms that its pro-inflammatory effects outweigh its barrier-protective role in the context of active disease, leading to the heightened tissue damage and reduced alveolar bone density (Zhang et al., 2024).
Despite these compelling findings, our study has certain limitations. The causal relationships between specific cell types and neural signaling changes require further functional validation. Furthermore, our in vitro experiments utilized acetylcholine without the addition of AChE inhibitors. Given that keratinocytes may endogenously produce cholinesterases, the stability and effective concentration of acetylcholine in the culture medium could be impacted, which represents a limitation of this study. Additionally, although the mouse model is valuable, it may not fully recapitulate the complexity of human periodontal disease. Future studies should aim to elucidate the specific molecular pathways by which CHRNB1 and CHRNA5 mediate their pro-inflammatory and barrier-protective effects. Investigating whether pharmacological targeting of these receptors could be a viable therapeutic strategy for modulating periodontal inflammation and promoting tissue regeneration is a promising direction for future research.
Data availability statement
The RNA-seq data are listed under GSE297378 (GEO database), and the scRNA-seq data of gingiva are listed under GSE152042, 164241, 171213 and 207502 (GEO database). The spatial transcriptomics data are listed under HRA003217 in the Genome Sequence Archive (GSA) database.
Ethics statement
The studies involving humans were approved by the Medical Ethics Committee of the Affiliated Stomatology Hospital of Sun Yat-sen University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) at Sun Yat-sen University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SX: Conceptualization, Formal analysis, Funding acquisition, Methodology, Validation, Visualization, Writing – original draft. JG: Conceptualization, Formal analysis, Funding acquisition, Methodology, Validation, Visualization, Writing – original draft. SY: Investigation, Methodology, Resources, Writing – original draft. BC: Conceptualization, Data curation, Formal analysis, Writing – review and editing. JX: Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – review and editing.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (grant no. U23A20445 and 82201082), the Medical Scientific Research Foundation of Guangdong Province (grant no. B2025282), the Natural Science Foundation of Chongqing (grant no. cstc2021jcyj-bshX0086), and the China Postdoctoral Science Foundation (grant no. 2024M753791).
Acknowledgments
AcknowledgementsThe authors would like to thank Jianming Zeng (University of Macau), all the members of his bioinformatics team, and bio-trainees for generously sharing their experience and codes.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alen, N. V. (2022). The cholinergic anti-inflammatory pathway in humans: state-of-the-art review and future directions. Neurosci. Biobehav Rev. 136, 104622. doi:10.1016/j.neubiorev.2022.104622
Apatzidou, D. A., Iskas, A., Konstantinidis, A., Alghamdi, A. M., Tumelty, M., Lappin, D. F., et al. (2018). Clinical associations between acetylcholine levels and cholinesterase activity in saliva and gingival crevicular fluid and periodontal diseases. J. Clin. Periodontol. 45 (10), 1173–1183. doi:10.1111/jcpe.12989
Arredondo, J., Hall, L. L., Ndoye, A., Chernyavsky, A. I., Jolkovsky, D. L., and Grando, S. A. (2003). Muscarinic acetylcholine receptors regulating cell cycle progression are expressed in human gingival keratinocytes. J. Periodontal Res. 38 (1), 79–89. doi:10.1034/j.1600-0765.2003.01006.x
Barford, K., Yap, C. C., Dwyer, N. D., and Winckler, B. (2017). The related neuronal endosomal proteins NEEP21 (Nsg1) and P19 (Nsg2) have divergent expression profiles in vivo. J. Comp. Neurol. 525 (8), 1861–1878. doi:10.1002/cne.24168
Barreiro, O., Martin, P., Gonzalez-Amaro, R., and Sanchez-Madrid, F. (2010). Molecular cues guiding inflammatory responses. Cardiovasc Res. 86 (2), 174–182. doi:10.1093/cvr/cvq001
Burke, S. M., Avstrikova, M., Noviello, C. M., Mukhtasimova, N., Changeux, J. P., Thakur, G. A., et al. (2024). Structural mechanisms of alpha7 nicotinic receptor allosteric modulation and activation. Cell 187 (5), 1160–1176 e1121. doi:10.1016/j.cell.2024.01.032
Caetano, A. J., Yianni, V., Volponi, A., Booth, V., D'Agostino, E. M., and Sharpe, P. (2021). Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease. Elife 10, e62810. doi:10.7554/eLife.62810
Chang, A. M., Kantrong, N., and Darveau, R. P. (2021). Maintaining homeostatic control of periodontal epithelial tissue. Periodontol 86 (1), 188–200. doi:10.1111/prd.12369
Chen, Y., Wang, H., Yang, Q., Zhao, W., Chen, Y., Ni, Q., et al. (2022). Single-cell RNA landscape of the osteoimmunology microenvironment in periodontitis. Theranostics 12 (3), 1074–1096. doi:10.7150/thno.65694
Chen, Y., Fang, Z. M., Yi, X., Wei, X., and Jiang, D. S. (2023). The interaction between ferroptosis and inflammatory signaling pathways. Cell Death Dis. 14 (3), 205. doi:10.1038/s41419-023-05716-0
de Juan, A., Ince, L. M., Pick, R., Chen, C. S., Molica, F., Zuchtriegel, G., et al. (2019). Artery-associated sympathetic innervation drives rhythmic vascular inflammation of arteries and veins. Circulation 140 (13), 1100–1114. doi:10.1161/CIRCULATIONAHA.119.040232
Easter, Q. T., Fernandes Matuck, B., Beldorati Stark, G., Worth, C. L., Predeus, A. V., Fremin, B., et al. (2024). Single-cell and spatially resolved interactomics of tooth-associated keratinocytes in periodontitis. Nat. Commun. 15 (1), 5016. doi:10.1038/s41467-024-49037-y
Emanuel, E., Arifuzzaman, M., and Artis, D. (2024). Epithelial-neuronal-immune cell interactions: implications for immunity, inflammation, and tissue homeostasis at mucosal sites. J. Allergy Clin. Immunol. 153 (5), 1169–1180. doi:10.1016/j.jaci.2024.02.004
Fernandez-Lainez, C., Aan de Stegge, M., Silva-Lagos, L. A., Lopez-Velazquez, G., and de Vos, P. (2023). β(2 → 1)-β(2 → 6) and β(2 → 1) fructans protect from impairment of intestinal tight junction's gene expression and attenuate human dendritic cell responses in a fructan-dependent fashion. Carbohydr. Polym. 320, 121259. doi:10.1016/j.carbpol.2023.121259
Ferreira-Vieira, T. H., Guimaraes, I. M., Silva, F. R., and Ribeiro, F. M. (2016). Alzheimer's disease: targeting the cholinergic system. Curr. Neuropharmacol. 14 (1), 101–115. doi:10.2174/1570159x13666150716165726
Forrester, E. A., Benitez-Angeles, M., Redford, K. E., Rosenbaum, T., Abbott, G. W., Barrese, V., et al. (2024). Crucial role for sensory nerves and Na/H exchanger inhibition in dapagliflozin- and empagliflozin-induced arterial relaxation. Cardiovasc Res. 120 (14), 1811–1824. doi:10.1093/cvr/cvae156
France, M. M., and Turner, J. R. (2017). The mucosal barrier at a glance. J. Cell Sci. 130 (2), 307–314. doi:10.1242/jcs.193482
Fu, J., and Wu, H. (2023). Structural mechanisms of NLRP3 inflammasome assembly and activation. Annu. Rev. Immunol. 41, 301–316. doi:10.1146/annurev-immunol-081022-021207
Guzman-Mejia, F., Lopez-Rubalcava, C., and Gonzalez-Espinosa, C. (2018). Stimulation of nAchRα7 receptor inhibits TNF synthesis and secretion in response to LPS treatment of mast cells by targeting ERK1/2 and TACE activation. J. Neuroimmune Pharmacol. 13 (1), 39–52. doi:10.1007/s11481-017-9760-7
Hodo, T. W., de Aquino, M. T. P., Shimamoto, A., and Shanker, A. (2020). Critical neurotransmitters in the neuroimmune network. Front. Immunol. 11, 1869. doi:10.3389/fimmu.2020.01869
Hone, A. J., and McIntosh, J. M. (2023). Nicotinic acetylcholine receptors: therapeutic targets for novel ligands to treat pain and inflammation. Pharmacol. Res. 190, 106715. doi:10.1016/j.phrs.2023.106715
Hulsbusch, N., Solis, G. P., Katanaev, V. L., and Stuermer, C. A. (2015). Reggie-1/Flotillin-2 regulates integrin trafficking and focal adhesion turnover via Rab11a. Eur. J. Cell Biol. 94 (11), 531–545. doi:10.1016/j.ejcb.2015.07.003
Ilic, K., Mlinac-Jerkovic, K., Sedmak, G., Rosenzweig, I., and Kalanj-Bognar, S. (2021). Neuroplastin in human cognition: review of literature and future perspectives. Transl. Psychiatry 11 (1), 394. doi:10.1038/s41398-021-01509-1
Kim, T. S., Ikeuchi, T., Theofilou, V. I., Williams, D. W., Greenwell-Wild, T., June, A., et al. (2024). Epithelial-derived interleukin-23 promotes oral mucosal immunopathology. Immunity 57 (4), 859–875.e11. doi:10.1016/j.immuni.2024.02.020
Korabecny, J., and Soukup, O. (2021). Cholinesterase research. Biomolecules 11 (8), 1121. doi:10.3390/biom11081121
Kosumi, H., Watanabe, M., Shinkuma, S., Nohara, T., Fujimura, Y., Tsukiyama, T., et al. (2022). Wnt/beta-catenin signaling stabilizes hemidesmosomes in keratinocytes. J. Invest Dermatol 142 (6), 1576–1586 e1572. doi:10.1016/j.jid.2021.10.018
Lamont, R. J., Fitzsimonds, Z. R., Wang, H., and Gao, S. (2022). Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontol. 2000 89 (1), 154–165. doi:10.1111/prd.12425
Li, W., Zhu, H., Zou, X., Ye, H., Zhong, J., Xiang, S., et al. (2025a). A brain-to-lung signal from GABAergic neurons to ADRB2(+) interstitial macrophages promotes pulmonary inflammatory responses. Immunity 58 (8), 2069–2085.e9. doi:10.1016/j.immuni.2025.05.005
Li, Y., He, X., Luo, G., Zhao, J., Bai, G., and Xu, D. (2025b). Innovative strategies targeting oral microbial dysbiosis: unraveling mechanisms and advancing therapies for periodontitis. Front. Cell Infect. Microbiol. 15, 1556688. doi:10.3389/fcimb.2025.1556688
Liccardo, F., Luini, A., and Di Martino, R. (2022). Endomembrane-based signaling by GPCRs and G-Proteins. Cells 11 (3), 528. doi:10.3390/cells11030528
Mak, P., Chang, C., Pursell, B., and Mercurio, A. M. (2013). Estrogen receptor beta sustains epithelial differentiation by regulating prolyl hydroxylase 2 transcription. Proc. Natl. Acad. Sci. U. S. A. 110 (12), 4708–4713. doi:10.1073/pnas.1221654110
Meric, P., Buduneli, N., Kanmaz, B., Gurlek, O., Comlekoglu, E., Calvert, G., et al. (2019). Cholinergic signalling mechanisms and early implant healing phases in healthy versus generalized aggressive periodontitis patients: a prospective, case-control study. J. Clin. Periodontol. 46 (11), 1155–1163. doi:10.1111/jcpe.13185
Moayedi, Y., Michlig, S., Park, M., Koch, A., and Lumpkin, E. A. (2022). Localization of TRP channels in healthy oral mucosa from human donors. eNeuro 9 (6), ENEURO.0328–21.2022. doi:10.1523/ENEURO.0328-21.2022
Mulder, K., Patel, A. A., Kong, W. T., Piot, C., Halitzki, E., Dunsmore, G., et al. (2021). Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54 (8), 1883–1900.e5. doi:10.1016/j.immuni.2021.07.007
Petsakou, A., Liu, Y., Liu, Y., Comjean, A., Hu, Y., and Perrimon, N. (2023). Cholinergic neurons trigger epithelial Ca(2+) currents to heal the gut. Nature 623 (7985), 122–131. doi:10.1038/s41586-023-06627-y
Plemmenos, G., Evangeliou, E., Polizogopoulos, N., Chalazias, A., Deligianni, M., and Piperi, C. (2021). Central regulatory role of cytokines in periodontitis and targeting options. Curr. Med. Chem. 28 (15), 3032–3058. doi:10.2174/0929867327666200824112732
Sakallioglu, E. E., Lutfioglu, M., Sakallioglu, U., Diraman, E., Pamuk, F., and Odyakmaz, S. (2008). Local peptidergic innervation of gingiva in smoking and non-smoking periodontitis patients. J. Periodontol. 79 (8), 1451–1456. doi:10.1902/jop.2008.070667
Sharma, B. R., and Kanneganti, T. D. (2021). NLRP3 inflammasome in cancer and metabolic diseases. Nat. Immunol. 22 (5), 550–559. doi:10.1038/s41590-021-00886-5
Shen, Z., Zhang, R., Huang, Y., Chen, J., Yu, M., Li, C., et al. (2024). The spatial transcriptomic landscape of human gingiva in health and periodontitis. Sci. China Life Sci. 67 (4), 720–732. doi:10.1007/s11427-023-2467-1
Slots, J. (2017). Periodontitis: facts, fallacies and the future. Periodontol 75 (1), 7–23. doi:10.1111/prd.12221
Takahashi, N., Sulijaya, B., Yamada-Hara, M., Tsuzuno, T., Tabeta, K., and Yamazaki, K. (2019). Gingival epithelial barrier: regulation by beneficial and harmful microbes. Tissue Barriers 7 (3), e1651158. doi:10.1080/21688370.2019.1651158
Walczak-Nowicka, L. J., and Herbet, M. (2021). Acetylcholinesterase inhibitors in the treatment of neurodegenerative diseases and the role of acetylcholinesterase in their pathogenesis. Int. J. Mol. Sci. 22 (17), 9290. doi:10.3390/ijms22179290
Wang, H., Yu, M., Ochani, M., Amella, C. A., Tanovic, M., Susarla, S., et al. (2003). Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 421 (6921), 384–388. doi:10.1038/nature01339
Wang, S. Y., Cai, Y., Hu, X., Li, F., Qian, X. H., Xia, L. Y., et al. (2024). P. gingivalis in oral-prostate axis exacerbates benign prostatic hyperplasia via IL-6/IL-6R pathway. Mil. Med. Res. 11 (1), 30. doi:10.1186/s40779-024-00533-8
Wen, G., Eder, K., and Ringseis, R. (2024). Comparative evaluation of the modulatory role of 1,25-dihydroxy-vitamin D(3) on endoplasmic reticulum stress-induced effects in 2D and 3D cultures of the intestinal porcine epithelial cell line IPEC-J2. J. Anim. Sci. Biotechnol. 15 (1), 153. doi:10.1186/s40104-024-01112-6
Wessler, I., Reinheimer, T., Kilbinger, H., Bittinger, F., Kirkpatrick, C. J., Saloga, J., et al. (2003). Increased acetylcholine levels in skin biopsies of patients with atopic dermatitis. Life Sci. 72 (18-19), 2169–2172. doi:10.1016/s0024-3205(03)00079-1
Williams, D. W., Greenwell-Wild, T., Brenchley, L., Dutzan, N., Overmiller, A., Sawaya, A. P., et al. (2021). Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 184 (15), 4090–4104.e15. doi:10.1016/j.cell.2021.05.013
Xu, S., Jiang, C., Liu, H., Zhang, H., Liao, H., Wang, X., et al. (2020). Integrin-α9 and its corresponding ligands play regulatory roles in chronic periodontitis. Inflammation 43 (4), 1488–1497. doi:10.1007/s10753-020-01226-9
Zhang, M., Liu, Y., Afzali, H., and Graves, D. T. (2024). An update on periodontal inflammation and bone loss. Front. Immunol. 15, 1385436. doi:10.3389/fimmu.2024.1385436
Keywords: single-cell RNA sequencing, spatial transcriptomics, gingival epithelium, periodontitis, acetylcholine, cholinergic signaling
Citation: Xu S, Guo J, Yang S, Cheng B and Xia J (2025) Acetylcholine in the gingival epithelium drives the pathogenesis of periodontitis. Front. Cell Dev. Biol. 13:1701252. doi: 10.3389/fcell.2025.1701252
Received: 08 September 2025; Accepted: 29 October 2025;
Published: 20 November 2025.
Edited by:
Jihong Yang, BoYu Intelligent Health Innovation Laboratory, ChinaCopyright © 2025 Xu, Guo, Yang, Cheng and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Cheng, Y2hlbmdiaW5AbWFpbC5zeXN1LmVkdS5jbg==; Juan Xia, eGlhanVhbkBtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Shihan Xu
Shihan Xu Jiaxin Guo1†
Jiaxin Guo1† Bin Cheng
Bin Cheng Juan Xia
Juan Xia