- 1State Key Laboratory of Animal Biotech Breeding, National Engineering Laboratory for Animal Breeding, Breeding and Reproduction of Ministry of Agriculture and Rural Affairs, College of Animal Science and Technology, China Agricultural University, Beijing, China
- 2School of Life Sciences, Westlake University, Hangzhou, Zhejiang, China
- 3College of Animal Science and Technology, Ningxia University, Yinchuan, China
Background: Staphylococcus aureus (S. aureus)-induced bovine mastitis is a major challenge for dairy production, causing significant economic losses. The regulatory mechanisms underlying host cell apoptosis and inflammation during S. aureus infection remain unclear. Therefore, this study investigates the role of N6-methyladenosine (m6A) modification and its reader protein YTHDF2 in regulating mRNA stability, apoptosis, and inflammation in bovine mammary epithelial cells (Mac-T cells) under S. aureus challenge.
Methods: MeRIP-seq, RIP-seq, and RT-qPCR were used to analyze m6A-modified IER3 mRNA and its interaction with YTHDF2. Apoptosis, necrosis, and mitochondrial function were assessed using YO-PRO-1/PI staining and JC-1 assays.
Results: S. aureus infection significantly downregulated YTHDF2 expression in Mac-T cells, leading to destabilization of m6A-modified IER3 mRNA. This resulted in increased reactive oxygen species (ROS) levels, mitochondrial dysfunction, and cell apoptosis. Overexpression of YTHDF2 restored mRNA stability, reduced apoptosis, and preserved mitochondrial function.
Conclusion: YTHDF2 regulates m6A-modified mRNA stability to modulate apoptosis and inflammation during S. aureus infection. These findings provide new insights into understanding the molecular mechanisms of bovine mastitis and provide genetic markers for breeding mastitis-resistant dairy cows.
1 Introduction
Bovine mastitis, particularly caused by Staphylococcus aureus (S. aureus), represents a significant challenge to the dairy industry, resulting in reduced milk production, poor milk quality, substantial economic losses, and compromised animal welfare (Bannerman, 2009; Kang et al., 2016). One of the key pathological events in S. aureus-induced mastitis is the apoptosis of bovine mammary epithelial cells, which are the first line of defense against pathogens, triggering immune recognition and coordinating subsequent immune responses (Aitken et al., 2011). Understanding how S. aureus induces apoptosis in these cells, such as in Mac-T cells, is critical to developing genetic markers for breeding udder health in dairy cows (Wang et al., 2019; Lin et al., 2021; Ogunnaike et al., 2021).
N6-methyladenosine (m6A) is the most abundant RNA modification in eukaryotes and is regulated by methyltransferases, demethylases, and reader proteins (Jiang et al., 2021; Mao et al., 2023; Zhang et al., 2023a). It plays an essential role in regulating key biological processes, including cell differentiation, stress responses, and inflammation (Liu et al., 2020; Su et al., 2023; Zhu et al., 2023). While m6A’s involvement in bacterial infection-induced inflammation has been recognized, its specific mechanisms remain cell type- and pathogen-dependent (Zong et al., 2021; Prall et al., 2023; Liu et al., 2024). Recent studies have shown that m6A levels are significantly altered during S. aureus infection in mammary epithelial cells, suggesting that m6A may be critical in regulating the host cell immune response. However, its precise role in S. aureus-induced inflammation in bovine mammary epithelial cells remains poorly understood.
YTH N6-Methyladenosine RNA Binding Protein F2 (YTHDF2), an m6A reader, promotes mRNA degradation and is implicated in stress responses and inflammation (Guo et al., 2020; Zhang et al., 2022). However, its involvement in regulating immune responses during mastitis has not been fully explored. Given its role in mRNA stability and degradation, YTHDF2 may regulate immune responses in bovine mammary epithelial cells during S. aureus infection, and may play a critical role in modulating the inflammation and apoptosis observed in mastitis.
Immediate early response gene 3 (IER3) (Yoon et al., 2009), also known as IEX-1, is an early response gene induced by various stress stimuli, including cytokines and DNA damage (Pawlikowska et al., 2010; Arlt and Schäfer, 2011). IER3 is involved in apoptosis and cellular stress, interacting with key inflammatory signaling pathways such as NF-κB and PI3K/Akt (Arlt et al., 2003; Sebens Müerköster et al., 2008). Despite its relevance in various pathological conditions (Arlt et al., 2004; You et al., 2007), the precise role of IER3 in S. aureus-induced apoptosis in mammary epithelial cells has not been fully elucidated.
This study investigates how YTHDF2-mediated m6A modification regulates IER3 mRNA stability and its role in S. aureus-induced apoptosis in Mac-T cells. The findings of the current study will be beneficial for understanding the molecular mechanisms underlying mastitis and breeding disease resistance in dairy cows.
2 Materials and methods
2.1 Experimental design and samples information
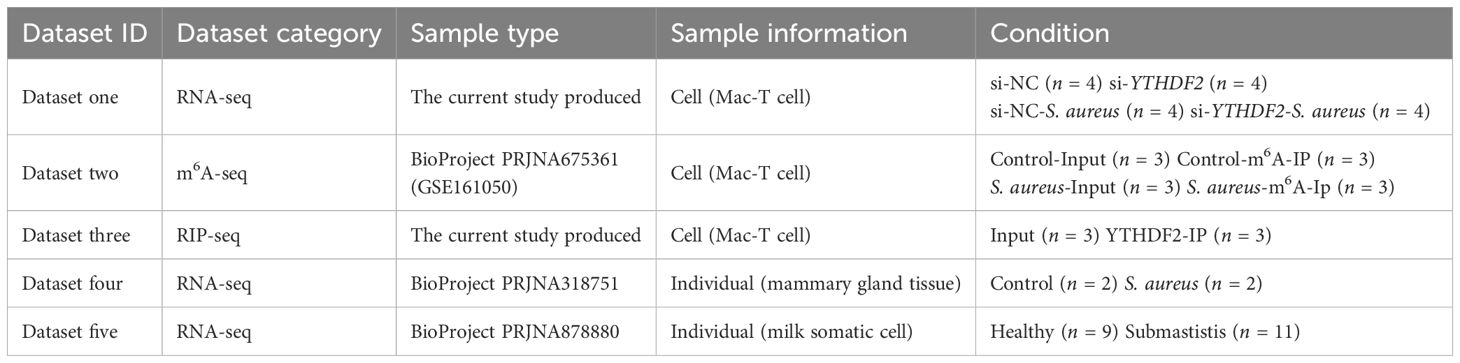
This study aimed to investigate the role of YTHDF2-mediated m6A modification in regulating IER3 mRNA stability during S. aureus-induced apoptosis, using both in vitro and in vivo strategies. The analyses combined data generated by the current study with publicly available datasets (Figure 1, Table 1).
Figure 1. Technical routes of this study. This figure depicts the experimental design to investigate the role of YTHDF2-mediated m6A modification in regulating IER3 mRNA stability during S. aureus-induced apoptosis. The study integrates both in vitro and in vivo approaches. In vitro, RNA-seq (Dataset one) and m6A-seq (Dataset two) were performed to assess gene expression and m6A modifications. RIP-seq (Dataset three) identified YTHDF2-bound mRNAs in Mac-T cells. Functional assays, including detection of apoptosis and oxidative stress, assessed the effects of YTHDF2 knockdown on S. aureus infection. In vivo validation was performed using publicly available RNA-seq datasets from bovine mammary gland tissue (Dataset four) and milk somatic cells from cows with subclinical mastitis (Dataset five).
In vitro, Mac-T cells were divided into four experimental groups, including control (si-NC), S. aureus challenge (si-NC-S. aureus), YTHDF2 knockdown (si-YTHDF2), and YTHDF2 knockdown followed by S. aureus challenge (si-YTHDF2-S. aureus). RNA-seq was performed to assess gene expression differences between these groups (dataset one). To investigate the role of m6A modifications, m6A-seq (dataset two, publicly available) was used to compare S. aureus-infected cells with control cells, focusing on infection-specific m6A modifications. To identify YTHDF2-bound mRNAs, RIP-seq was performed in Mac-T cells (dataset three). This analysis specifically focused on characterizing the direct targets of YTHDF2 in uninfected cells, providing a baseline for understanding its regulatory role in mRNA stability. Functional assays, including YO-PRO-1/PI staining, ROS detection and JC-1 assay, were used to assess apoptosis, oxidative stress, and mitochondrial membrane potential in YTHDF2 knockdown cells during S. aureus infection.
In vivo validation of key gene expression was performed using RNA-seq datasets from bovine mammary gland tissues (dataset four, publicly available) and milk somatic cells from cows with subclinical mastitis (dataset five, publicly available). Regardless of their source, all RNA-seq datasets were processed uniformly to ensure comparability across experiments.
2.2 Cell culture and treatments
2.2.1 Cell culture
Mac-T cells (bovine mammary epithelial cell line) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with GlutaMAX, 10% fetal bovine serum (FBS), and 100 U/mL penicillin-streptomycin (Thermo Fisher Scientific, USA). Cells were maintained at 37°C in a humidified incubator with 5% CO2 and passaged when they reached 80–90% confluence. Initially, cells were seeded in 25-cm² flasks (Corning), and upon reaching logarithmic growth, were transferred to 6-well plates (1 × 106 cells/well), 12-well plates (1 × 105 cells/well), 24-well plates (5 × 104 cells/well), or 96-well plates (5 × 10³ cells/well) for further treatment.
2.2.2 YTHDF2 small interfering RNA transfection
Mac-T cells seeded in 6-well plates at approximately 80% confluence were transfected with 100 pmol of YTHDF2-specific siRNA (Jintuosi, China) or negative control siRNA (si-NC) using 5 μL Lipofectamine 2000 (Thermo Fisher Scientific) in DMEM for 6 hours. Following transfection, the medium was replaced with fresh complete medium, and cells were cultured for an additional 40 hours before further treatment.
2.2.3 Lentivirus packaging and generation of stable cell lines
Lentiviral particles containing YTHDF2 or control vector pNull (Jintuosi, China) were packaged in HEK293T cells by transfecting 3 μg of YTHDF2 plasmid, 3 μg of pMD2.G, and 6 μg of psPAX2 (Lipofectamine 2000, Invitrogen) in Opti-MEM I medium (Gibco). The viral supernatants were collected at 48 and 72 hours post transfection, filtered, and concentrated using Lenti-X Concentrator (Takara Bio).
Mac-T cells were seeded in 6-well plates, and after 24 hours, were infected with lentivirus (25 μL/mL) in the presence of 10 ng/mL polybrene (Sigma-Aldrich). After 48 hours, stable cells were selected using 4 μg/mL puromycin (InvivoGen) for 2 weeks and then maintained in 1 μg/mL puromycin to generate YTHDF2-overexpressing (oe-YTHDF2) cells.
2.3 Cow module sample preparation and treatment
Dataset four was derived from a previously published experiment conducted by our research group, involving two early lactating Holstein cows (Fang et al., 2016). Briefly, mastitis was induced by intramammary infusion of S. aureus suspensions (1 × 106 CFU/mL) into the rear quarters immediately after morning milking, while the front quarters served as controls and received 10 mL sterile saline. Rectal temperature and somatic cell count (SCC, an indicator of leukocyte infiltration and immune response in milk) were measured at baseline (0 hours) and at 6, 12, 18, and 24 hours post infection (Supplementary Table 1). Inflammation onset was defined as SCC > 200,000 cells/mL and rectal temperature above 39.5°C. At 24 hours post infection, mammary gland tissues (udder biopsies) were collected from infected and control quarters, and RNA was extracted for subsequent RNA-seq analysis.
Dataset five consists of publicly available RNA-seq data from milk somatic cells of cows with subclinical mastitis and healthy control cows (Wang et al., 2024). Cows with subclinical mastitis were selected based on consistently high somatic cell counts (SCC > 350,000 cells/mL) over a period of three or more months, while healthy control cows had SCC < 100,000 cells/mL. The subclinical mastitis group included cows that tested positive for S. aureus in at least one quarter, while the healthy control group consisted of cows with no evidence of mastitis pathogens.
2.4 S. aureus culturing and challenge
The S. aureus strain used in this study is a sequence type 1 (ST1) isolate previously obtained and characterized by our laboratory from milk samples of cows with clinical mastitis (Wang et al., 2014b). Detailed information regarding its virulence factors is summarized in Supplementary Tables 2, 3 and has been described previously (Xing et al., 2025).
S. aureus was cultured overnight in tryptic soy broth (TSB) at 37°C with shaking at 200 rpm. Bacterial cultures were harvested by centrifugation at 5000 rpm for 5 minutes at room temperature, washed twice with sterile phosphate-buffered saline (PBS), and resuspended in PBS at a final concentration of 1 × 109 CFU/mL.
For cell infection, Mac-T cells prepared were challenged with S. aureus at a multiplicity of infection (MOI) of 10:1, reaching a final bacterial concentration of 1 × 107 CFU/mL per well. After incubation at 37°C and 5% CO2 for 6 hours, cells were gently washed with sterile PBS, harvested, and stored at -80°C for subsequent RNA extraction.
2.5 RNA extraction, library preparation, and RNA-seq analysis
2.5.1 RNA extraction and library preparation
Total RNA was extracted from Mac-T cells using TRIzol reagent (Invitrogen). RNA purity and concentration were measured using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, USA). RNA samples were stored at -80°C until use. For RNA-seq, 1 μg of total RNA was reverse-transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Kyoto, Japan). RNA-seq libraries were constructed and sequenced on the Illumina HiSeq2500 platform (Novogene Co., Ltd., Beijing, China), generating 150 bp paired-end reads.
2.5.2 Data preprocessing and mapping
Publicly available RNA-seq datasets (datasets four and five) were downloaded from the SRA database using SRA-Toolkit v2.9.6. All datasets were processed uniformly. Raw read quality was assessed using FastQC v0.11.8, and reads containing adaptors, low quality bases, or undetermined bases were filtered using NGSQCToolkit v2.3.3. Clean reads were then mapped to the bovine reference genome (ARS-UCD1.2) using Hisat2 v2.1.0. SAM files were converted to BAM files with SAMtools v1.9, and read counts were quantified using FeatureCounts (subread v1.6.3).
2.5.3 Normalization and differential expression analysis
Read counts obtained from FeatureCounts were normalized using DESeq2 v1.28.1 to generate normalized read counts across datasets. Differentially expressed genes (DEGs) were identified using DESeq2, with criteria of |log2FC| > 0.58 and p < 0.05. Comparisons included YTHDF2 knockdown vs. control (siYTHDF2 vs. siNc group) and S. aureus challenge vs. control (S. aureus vs. siNc group). Common DEGs between these comparisons were considered as key YTHDF2 target genes in bovine S. aureus mastitis.
2.6 N6-methyladenosine sequencing analysis
m6A sequencing data for Mac-T cells challenged with S. aureus and control cells were downloaded from the publicly available dataset GSE161050 (Li et al., 2021b). In study by Li et al. (2021b), Mac-T cells were co-cultured with heat-inactivated S. aureus (ATCC 29213) for 24 hours, after which RNA was extracted for m6A-seq analysis. Raw data were downloaded from the SRA database and converted to FASTQ format using SRA-Toolkit v2.9.6. 3. Sequence quality was assessed with FastQC v0.11.8, and reads containing adaptors, low quality bases, or undetermined bases were removed using NGSQCToolkit v2.3.3. Clean reads were then mapped to the bovine reference genome (ARS-UCD1.2) using Hisat2 v2.2.1, and SAM files were converted to BAM files using SAMtools v1.9. m6A peaks were called using the R package exomepeak2, with a q-value threshold of 0.05 to filter for significant peaks.
2.7 RNA-binding protein immunoprecipitation (RIP-seq) and analysis
RIP-seq was performed using the RNA Immunoprecipitation Kit (Bes5101, BersinBio, China) according to the manufacturer’s instructions. Briefly, Mac-T cells (1.0 × 107, normal conditions) were lysed in RIP lysis buffer, with 15 μL reserved as input. The remaining 150 μL of lysate was incubated overnight at 4°C with anti-YTHDF2 antibody (Proteintech, Cat No. 24744-1-AP) or rabbit IgG-conjugated protein A/G magnetic beads in IP buffer with RNase inhibitors. Immunoprecipitated RNA was digested with proteinase K, purified using TRIzol reagent, and sequenced on the Illumina HiSeq 2500 platform (Novogene).
For data analysis, RIP-seq peaks were called using the same approach as for m6A-seq analysis. Peak calling was performed using the ChIPseeker package, and the chromosomal distribution of identified peaks was visualized. Motif analysis was then conducted on the common peaks shared between RIP-seq and m6A-seq using the HOMER software to identify potential RNA-binding motifs. These results were visualized using the Integrative Genomics Viewer (IGV, v2.8.0).
2.8 Quantitative real-time PCR and RIP-qPCR
Total RNA was reverse-transcribed into cDNA using the PrimeScript RT reagent kit (Takara, Japan). qRT-PCR was performed on the LightCycler 480 system with SYBR Green I Master Mix (Roche, Switzerland). GAPDH was used as a reference gene, and relative expression levels were calculated using the 2–ΔΔCT method. RIP-qPCR specifically validated enrichment of IER3 mRNA immunoprecipitated by YTHDF2 antibody under both normal and S. aureus challenged conditions, normalized against input RNA. Primer sequences are listed in Supplementary Table 4.
2.9 Western blot
Cell extracts were prepared using RIPA lysis buffer (Beyotime) with 1 mM PMSF and 1× protease inhibitor cocktail (Solarbio). Cells were lysed on ice for 30 minutes, then centrifuged at 12,000 rpm for 5 minutes at 4°C. Equal amounts of protein (20 µg) were loaded onto a 7.5% SDS-PAGE gel, separated, and transferred to PVDF membranes (Millipore, USA). Membranes were blocked with 5% milk in 1× TBST for 1 hour at room temperature, and then incubated with primary antibodies overnight at 4°C: YTHDF2 (1:3000, Proteintech, Cat No. 24744-1-AP) and GAPDH (1:3000, Abcam, ab8245). Membranes were washed three times with TBST (5 minutes each), followed by incubation with secondary antibodies for 1 hour at room temperature. Goat anti-rabbit secondary antibody (1:3000, Thermo Fisher Scientific, Cat No. 32460) was used for YTHDF2, and goat anti-mouse secondary antibody (1:3000, Thermo Fisher Scientific, Cat No. PA1-74421) was used for GAPDH. After three washes with TBST (5 minutes each), band signals were visualized using SuperKine UltraSensitive ECL substrate (Abbkine, BMU102).
2.10 Immunofluorescence
2.10.1 Immunofluorescence staining
Mac-T cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes, and blocked with 5% normal goat serum (Vector) for 1 hour at room temperature. Cells were incubated with anti-YTHDF2 antibody (Proteintech, Cat No. 24744-1-AP) overnight at 4°C, followed by washing and incubation with a fluorescent secondary antibody (Thermo Fisher Scientific, Cat No. 32460) for 1 hour at room temperature. Nuclei were counterstained with DAPI (Sigma-Aldrich) for 5 minutes.
2.10.2 Imaging and analysis
Images were captured using a ZEISS Axio Scope A1 microscope. Fluorescence intensity of YTHDF2 (488 nm) was compared with DAPI (405 nm), and relative expression levels were quantified using image analysis software.
2.11 Cell damage status analysis
Cell damage and viability were assessed after a 6-hour S. aureus challenge using assays for ROS detection, mitochondrial membrane potential (ΔΨm) analysis, and apoptosis (YO-PRO-1/PI staining).
2.11.1 Intracellular reactive oxygen species detection
Intracellular ROS levels were assessed using DCFH-DA and DHE fluorescent dyes (Molecular Probes, Thermo Fisher Scientific) to specifically detect intracellular peroxides and superoxide anions, respectively. Briefly, cells were incubated with 10 μmol/L DCFH-DA for 20 minutes at 37°C, washed twice with PBS, and fluorescence intensity was quantified using a microplate reader at an excitation wavelength of 488 nm and an emission wavelength of 525 nm. ROS levels were expressed as relative fluorescence units (RFU), normalized to control groups.
2.11.2 Mitochondrial membrane potential (ΔΨm) assessment
ΔΨm, an indicator of mitochondrial health and functionality, was assessed using JC-1 dye (Enzo Life Sciences), a fluorescent probe that shifts from red (aggregates, indicating high ΔΨm) to green fluorescence (monomers, indicating low ΔΨm) upon mitochondrial depolarization. After S. aureus challenge, cells were incubated with JC-1 staining solution (5 μg/mL) at 37°C for 20 minutes, washed with PBS, and observed under a fluorescence microscope. Fluorescence intensities were quantified, and mitochondrial depolarization was expressed as the ratio of green fluorescence intensity (monomers) to red fluorescence intensity (aggregates).
2.11.3 Apoptotic cell staining
Apoptosis was assessed using YO-PRO-1/PI staining (Beyotime), a fluorescent method that distinguishes live, early apoptotic, and necrotic cells based on membrane integrity. YO-PRO-1 selectively enters early apoptotic cells and emits green fluorescence (excitation/emission: 491/509 nm), while propidium iodide (PI) penetrates necrotic cells and emits red fluorescence (excitation/emission: 535/617 nm). Cells were incubated with YO-PRO-1/PI staining solution for 20 minutes at 37°C, and fluorescence intensities were measured using confocal laser microscopy and a microplate reader.
2.12 mRNA stability assay
Cells were cultured to 50% confluence and treated with Actinomycin D (5 μg/mL). Samples were collected at 0, 3, and 6 hours after treatment. Total RNA was extracted and analyzed by RT-qPCR, with mRNA levels normalized to GAPDH. mRNA degradation rates were determined according to established protocols (Mukherjee et al., 2011; Wang et al., 2014a).
2.13 Function enrichment analysis
2.13.1 Gene set enrichment analysis
GSEA v4.3.3 was used to identify significantly enriched pathways activated under S. aureus infection, YTHDF2 knockdown (siYTHDF2), and combined siYTHDF2-S. aureus treatment conditions. Genes were ranked by log2 fold-change from normalized expression data. Enrichment significance was assessed by 1,000 permutations, with enriched pathways defined by an FDR q-value < 0.05.
2.13.2 Kyoto encyclopedia of genes and genomes pathway analysis
KEGG pathway analysis of DEGs was conducted using KOBAS 3.0 (http://bioinfo.org/kobas/). Pathways with a p < 0.05 were considered significantly enriched. Visualization of the top 20 pathways (p < 0.05) was performed using ImageGP and ggplot2.
2.13.3 Quantitative trait loci enrichment analysis
QTL enrichment analysis, a genetic approach used to identify genomic regions associated with quantitative phenotypic traits, was conducted to identify cattle QTLs harboring DEGs and 44 YTHDF2-regulated genes involved in S. aureus-induced mastitis. Cattle QTL data were obtained from AnimalQTLdb (https://www.animalgenome.org/cgi-bin/QTLdb/index) (Hu et al., 2022). QTLs within 100 kb upstream or downstream of the target genes were screened.
2.13.4 Transcriptome-wide association studies analysis
Trait associations were predicted using TWAS, an analytical method that integrates gene expression data with genome-wide association studies (GWAS) to identify genetic associations with specific phenotypic traits. The analysis used the cGTEx database (cgtex.roslin.ed.ac.uk) and integrated TWAS results with QTL data for the 44 candidate genes identified in this study.
2.13.5 GWAS enrichment analysis
GWAS enrichment analysis used data from a University of Maryland and USDA collaborative project involving 27,143 dairy cattle (https://figshare.com/s/ea726fa95a5bac158ac1). The analysis focused on 44 complex traits related to production, reproduction, and health. SNPs within a 10 kb flanking region of each candidate gene were screened, and a hypergeometric test was used to assess the overrepresentation of significant SNPs. The Benjamini-Hochberg procedure adjusted for multiple testing, with an adjusted p < 0.05 considered statistically significant.
2.14 Statistical analysis
Statistical analyses were performed using R v4.0.5 and GraphPad Prism v9.0, and quantification of necrotic cell was performed using ImageJ v1.51. Student’s t-test was used to compare differences between S. aureus challenged and control groups. Data are presented as mean ± standard deviation (SD), with significance set at p < 0.05.
3 Results
3.1 S. aureus-induced inflammation in Mac-T cells increases apoptosis and necrosis while significantly reducing YTHDF2 expression
RNA-seq was performed on Mac-T cells from four experimental conditions, including siNC (negative control), siYTHDF2, siNC-S. aureus, and siYTHDF2-S. aureus. Principal component analysis (PCA) revealed a clear separation among these conditions, indicating distinct transcriptional profiles resulting from both S. aureus infection and YTHDF2 knockdown (Figure 2A).
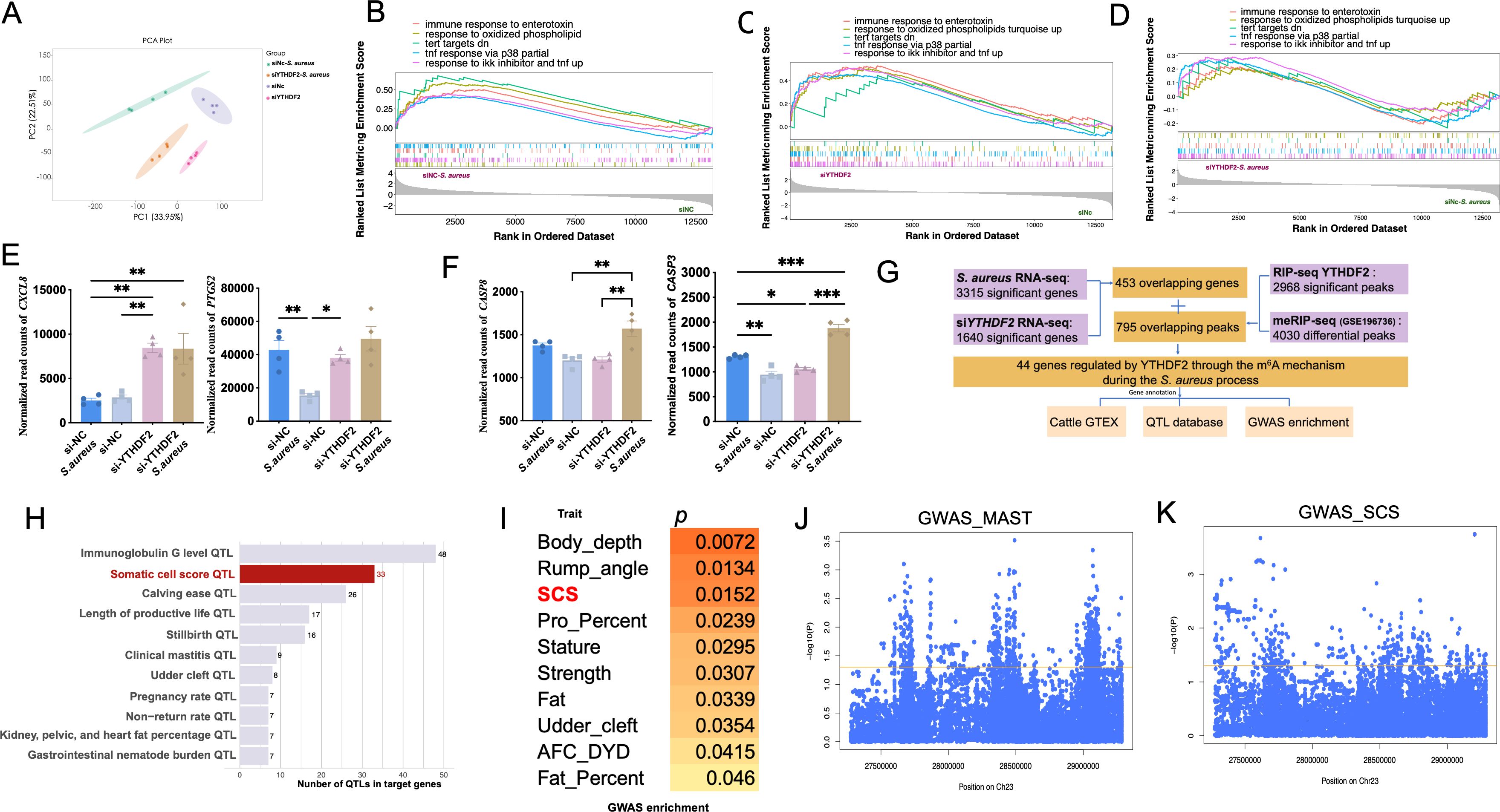
Figure 2. Functional impact of YTHDF2 binding on gene expression in Mac-T cells. (A) PCA of gene expression profiles in different experimental groups: si-NC, si-YTHDF2, si-NC + S. aureus, and si-YTHDF2 + S. aureus. (B) GSEA comparing gene expression in the si-NC + S. aureus group to the si-NC group. (C) GSEA comparing gene expression in the si-YTHDF2 group to the si-NC group. (D) GSEA comparing gene expression in the si-YTHDF2 + S. aureus group to the si-NC + S. aureus group. (E) Expression levels of pro-inflammatory cytokines in different experimental groups, with statistical significance indicated: *p < 0.05, **p < 0.01, ***p < 0.001. (F) Expression levels of apoptosis marker genes in different experimental groups, with statistical significance indicated: *p < 0.05, **p < 0.01, ***p < 0.001. (G) Overlap between RIP-seq and MeRIP-seq data, showing genes regulated by YTHDF2 through m6A modification. (H) QTL analysis of the overlapping differentially expressed genes (DEGs), showing associations with health-related traits in cattle, including immunoglobulin G level, somatic cell score (SCS), and clinical mastitis. (I) GWAS enrichment analysis of the overlapping DEGs, highlighting associations with SCS in cattle. (J, K) Results of SNP analysis within ±1 Mb of IER3, showing associations with mastitis and SCS traits, with the yellow line representing the p = 0.05 threshold.
Gene set enrichment analysis (GSEA) identified key pathways that were activated across the experimental conditions. In siNC-S. aureus cells, pathways associated with inflammation and apoptosis were significantly activated, including immune response to enterotoxin, response to IKK inhibitor and TNF signaling, response to oxidized phospholipids, TNF response via p38, and TERT targets down-regulated genes (Figure 2B). Notably, similar activation of these inflammatory and apoptotic pathways was observed in siYTHDF2 cells, even in the absence of S. aureus infection (Figure 2C), suggesting that YTHDF2 knockdown alone is sufficient to induce inflammation. When comparing siYTHDF2-S. aureus cells to siNC-S. aureus cells, these pathways remained significantly enriched, indicating that YTHDF2 loss exacerbates the inflammatory and apoptotic responses to S. aureus infection (Figure 2D).
Furthermore, pro-inflammatory factors, such as CXCL8 and PTGS2, were significantly upregulated after YTHDF2 knockdown, with the highest expression levels observed in cells with both YTHDF2 knockdown and S. aureus infection (Figure 2E). Similarly, the apoptotic markers CASP3 and CASP8 were significantly upregulated in S. aureus-infected cells, with the highest expression levels detected in the S. aureus-infected YTHDF2 knockdown cells (Figure 2F).
3.2 YTHDF2 regulates m6A-modified genes associated with bovine health traits during S. aureus infection
Differential expression analysis identified 3,315 differentially expressed genes (DEGs) (|log2FC| > 0.58, p < 0.05) between S. aureus-infected cells and control cells (Figure 2F). KEGG enrichment analysis of these DEGs revealed significant activation of apoptosis, TNF signaling, and inflammatory response pathways (Supplementary Figure S1A). A similar RNA-seq analysis comparing YTHDF2 knockdown cells and control cells also identified 3,315 DEGs (|log2FC| > 0.58, p < 0.05), enriched in pathways related to homologous recombination, apoptosis, and DNA repair (Supplementary Figure S1B). Of these, 453 overlapping genes were identified, enriched in pathways such as pyrimidine metabolism, PPAR signaling, and immune response regulation (Supplementary Figure S1C).
To investigate m6A modification during S. aureus infection, m6A-seq was performed using publicly available m6A-seq data (S. aureus infection vs. control). The analysis identified 4,030 distinct m6A methylation sites responsive to S. aureus challenge. To explore the potential regulatory role of YTHDF2 in m6A-modified genes, RIP-seq was performed using control cells. RIP-seq analysis revealed 10,284 peaks with a q-value < 0.05 across the genome and identified 2,968 significantly enriched transcripts (Supplementary Figure S2A).
Integrating the m6A-seq data with the YTHDF2 RNA-binding sites identified in this study, 795 genes with YTHDF2-dependent m6A modifications were obtained. Overlapping results confirmed the DRACH motif (D=A/G/U, R=A/G, H=U/A/C) as the canonical sequence for m6A modification (Supplementary Figure S2B). These findings emphasized YTHDF2’s role in recognizing and binding m6A-modified mRNAs, with significant implications for gene expression regulation in S. aureus-infected cells.
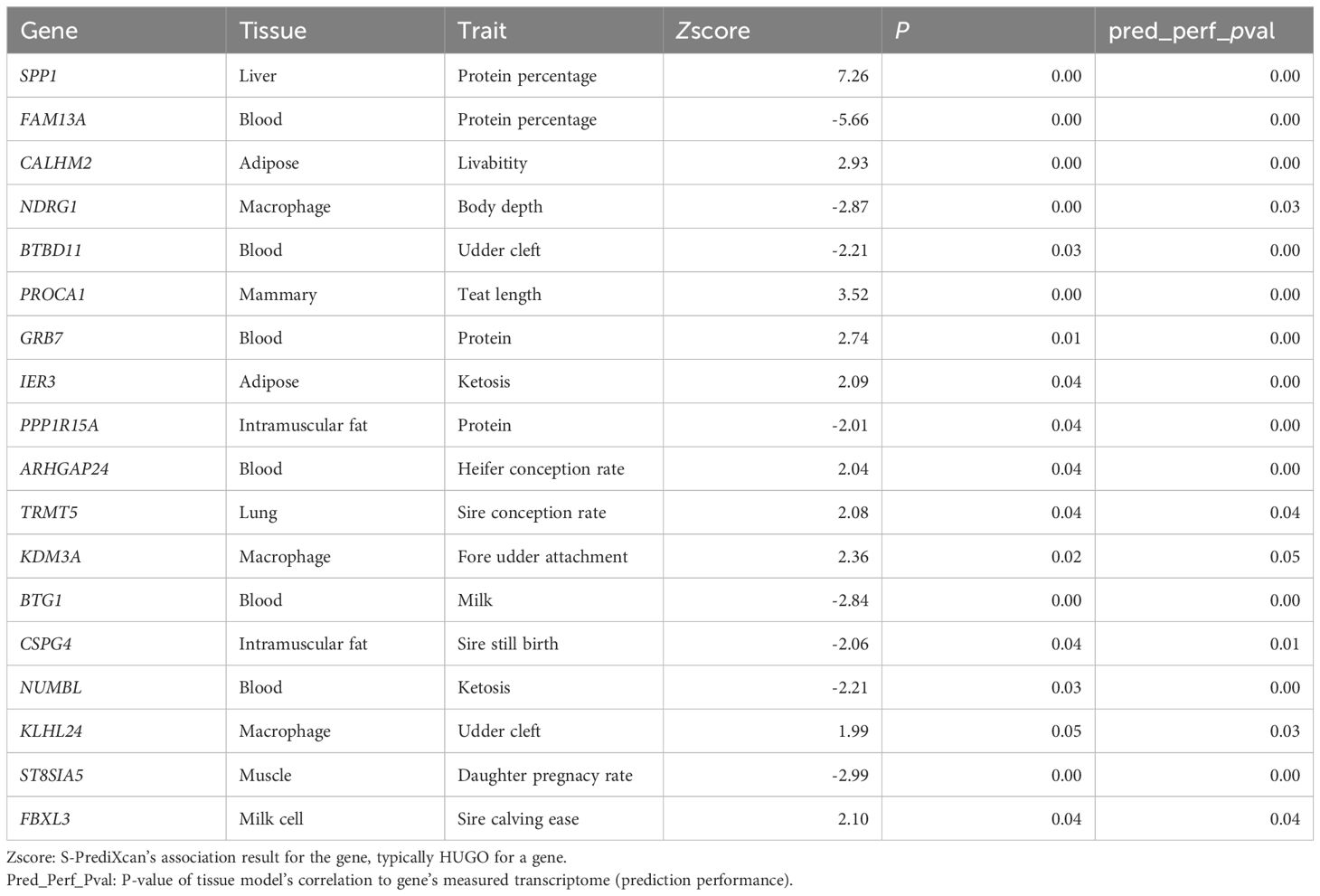
Cross-referencing the identified genes with the 453 overlapping DEGs, 44 candidate genes potentially regulated by YTHDF2-mediated m6A modification during inflammation were identified (Figure 2G, Supplementary Figures S2, D). Gene annotation based on the TWAS database highlighted significant associations between several of these candidate genes (such as SPP1, FAM13A, NDRG1, and IER3) and key bovine health traits, including milk protein percentage, body depth, and ketosis (a metabolic disease of dairy cow) susceptibility (Table 2). Further QTL analysis identified 288 health-related QTLs within ±100 kb of these candidate genes, particularly associated with immunoglobulin G levels, somatic cell score, and clinical mastitis (Figure 2H, Supplementary Figure S2E). GWAS enrichment analysis further confirmed significant enrichment of these candidate gene regions with genomic loci associated with somatic cell score, supporting the role of YTHDF2-mediated m6A modifications in regulating bovine health traits during S. aureus infection (Figure 2I).
Interestingly, IER3 also showed an association with immunoglobulin G levels (Supplementary Figure S2F), suggesting a potential link between this gene and bovine health traits. Key SNPs associated with mastitis and somatic cell score (SCS, a standardized measure of SCC used to assess milk quality and udder health) traits in dairy cows are located within a 1 Mb region of the IER3 gene, and IER3 could be considered as a candidate gene for genetic selection of mastitis resistance (Figures 2J, K).
3.3 Knockdown and overexpression of YTHDF2 reveals its role in S. aureus-induced apoptosis in Mac-T cells
Dataset one RNA-seq analysis showed a significant reduction in YTHDF2 expression in siNC-S. aureus cells compared to siNC controls, with a further reduction observed in siYTHDF2-S. aureus cells, indicating a synergistic effect of S. aureus infection and YTHDF2 knockdown (Figure 3A). This trend was further confirmed by transcriptomic data from milk samples of cows with subclinical mastitis and blood samples from cows infected with S. aureus, providing additional evidence for the downregulation of YTHDF2 during S. aureus infection (Figures 3B, C).
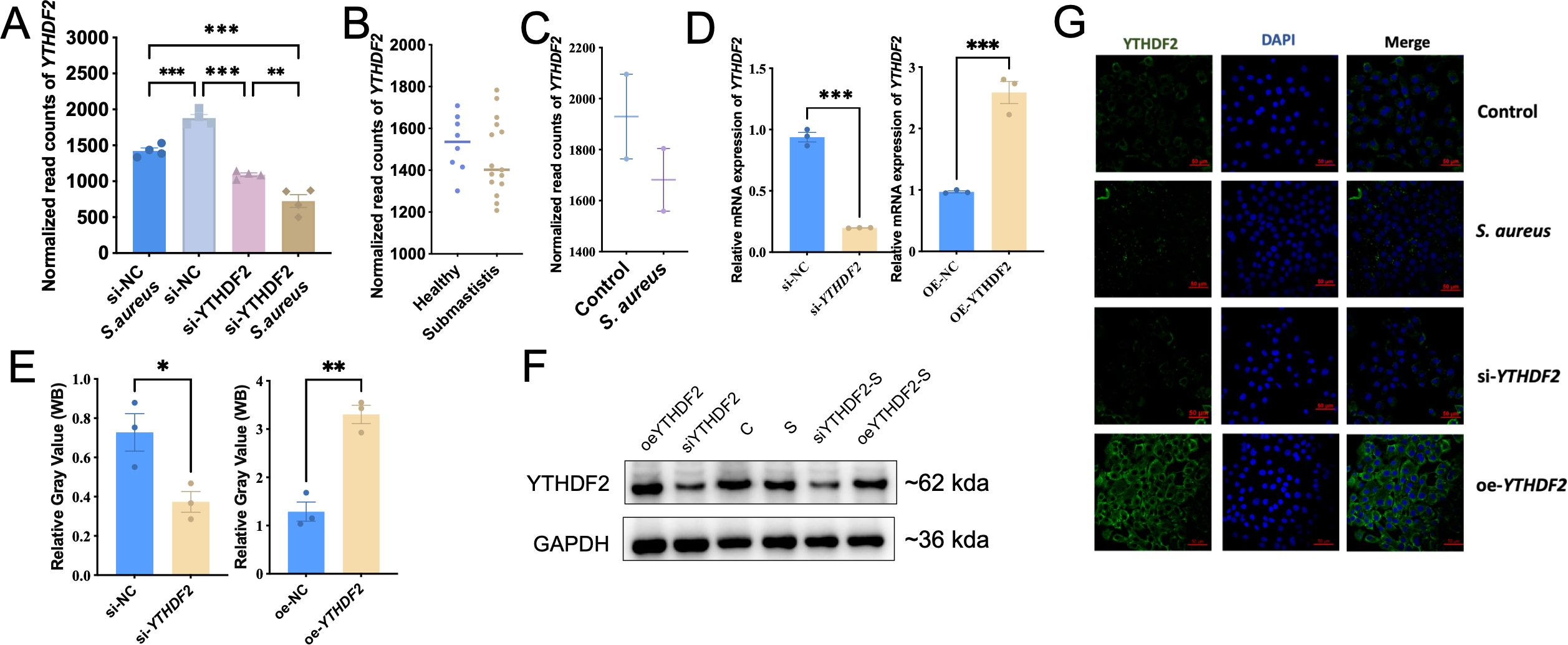
Figure 3. YTHDF2 expression in Mac-T cells under S. aureus challenge. (A) Normalized YTHDF2 expression levels in Mac-T cells from different experimental groups: si-NC, si-YTHDF2, si-NC + S. aureus, and si-YTHDF2 + S. aureus. (B) Normalized YTHDF2 expression in milk samples from subclinical S. aureus mastitis cows and healthy cows. (C) Normalized YTHDF2 expression in mammary gland tissue samples from the bovine model. (D) Relative mRNA expression of YTHDF2 in Mac-T cells transfected with si-NC, si-YTHDF2, oe-NC, and oe-YTHDF2, as determined by RT-qPCR. (E) WB analysis showing YTHDF2 protein levels and relative gray values in Mac-T cells transfected with si-NC, si-YTHDF2, oe-NC and oe-YTHDF2. (F) WB showing YTHDF2 protein levels in different experimental groups. (G) Immunofluorescence images showing YTHDF2 expression (green), DAPI (blue), and merged images in control, S. aureus-challenged, si-YTHDF2 and oe-YTHDF2 Mac-T cells. Statistical significance is indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
Given this observed reduction in YTHDF2 expression during S. aureus-induced mastitis, its potential regulatory role in apoptosis was investigated through both knockdown and overexpression experiments in Mac-T cells. Knockdown of YTHDF2 (si-YTHDF2) significantly reduced its expression at both mRNA and protein levels, as confirmed by RT-qPCR and Western blot analyses, whereas overexpression of YTHDF2 (oe-YTHDF2) resulted in a significant increase in both mRNA and protein levels (Figures 3D, E). Notably, S. aureus infection caused a further decrease in YTHDF2 expression, regardless of the experimental conditions, including both the si-YTHDF2 and oe-YTHDF2 groups (Figure 3F). Immunofluorescence assays confirmed the effective knockdown and overexpression of YTHDF2 protein in the respective experimental conditions (Figure 3G).
Functional assays using YO-PRO-1/PI staining, which detects apoptosis and necrosis based on differential dye uptake, showed that YTHDF2 knockdown significantly enhanced S. aureus-induced apoptosis and necrosis compared to controls (Figure 4A). Conversely, cells overexpressing YTHDF2 showed significantly reduced apoptosis and necrosis following bacterial challenge (Figure 4A). These results were quantitatively validated by microplate reader measurements of YO-PRO-1 and PI fluorescence, where YTHDF2 knockdown resulted in significantly higher fluorescence intensity, whereas YTHDF2 overexpression resulted in lower fluorescence signals compared to controls (Figures 4B–E).
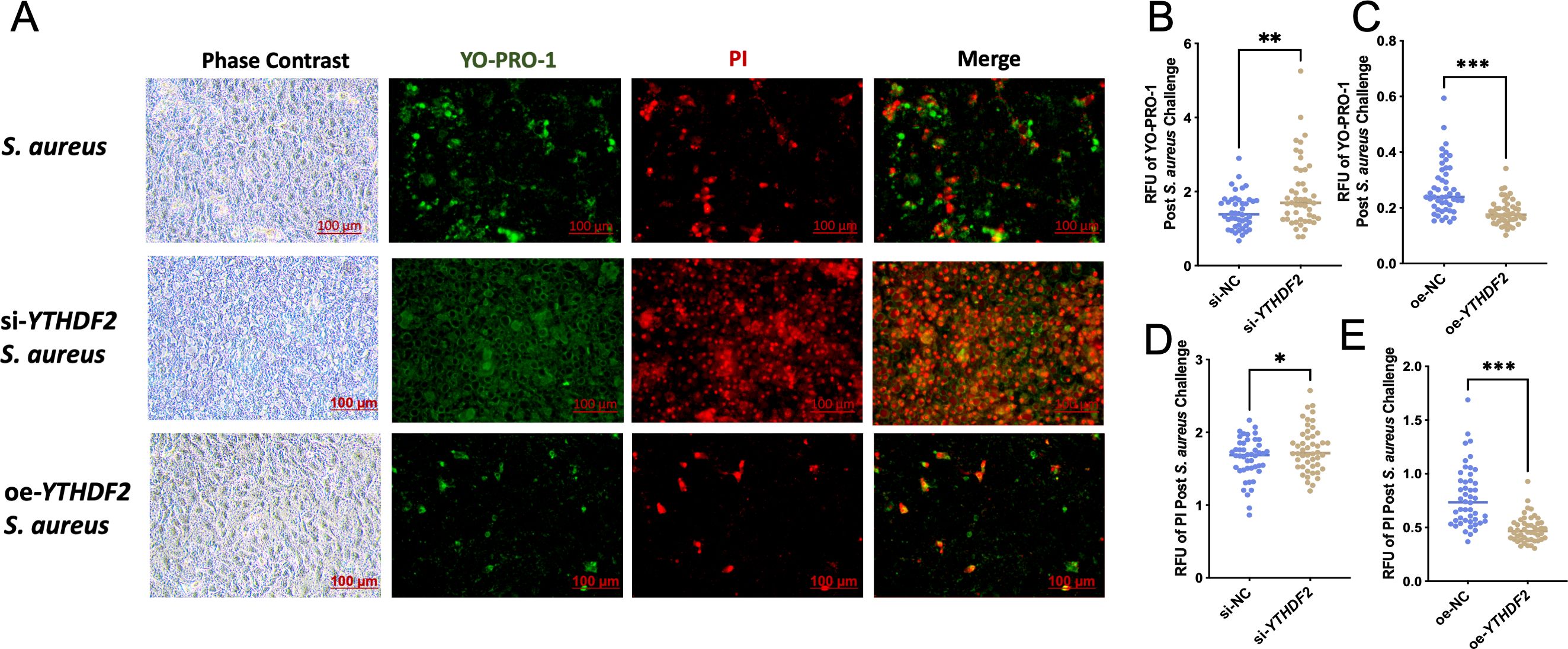
Figure 4. YTHDF2 modulates apoptosis in Mac-T Cells under S. aureus challenge. (A) Phase contrast and YO-PRO-1/PI staining images showing apoptosis and necrosis in S. aureus-challenged, si-YTHDF2 and oe-YTHDF2 Mac-T cells. Green fluorescence indicates apoptotic cells (YO-PRO-1) and red fluorescence indicates necrotic cells (PI). (B-E) Relative fluorescence units (RFU) of YO-PRO-1 and PI in si-NC, si-YTHDF2, oe-NC and oe-YTHDF2Mac-T cells post S. aureus challenge. Statistical significance is indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
3.4 YTHDF2 modulates apoptosis through ROS-mediated mitochondrial dysfunction in Mac-T cells under S. aureus challenge
ROS are known to accumulate during oxidative stress and play a critical role in mediating apoptosis by disrupting mitochondrial function (Maassen et al., 2004; Rizwan et al., 2020). In this study, YTHDF2 knockdown (si-YTHDF2) significantly increased ROS levels in Mac-T cells following S. aureus infection compared to controls (Figure 5A), contributing to mitochondrial dysfunction. Conversely, overexpression of YTHDF2 (oe-YTHDF2) reduced ROS levels and preserved mitochondrial function in S. aureus-challenged cells (Figure 5B). The results of the zymography assay were further supported by fluorescence microscopy, which more clearly demonstrated changes in fluorescence intensity (Figure 5C).
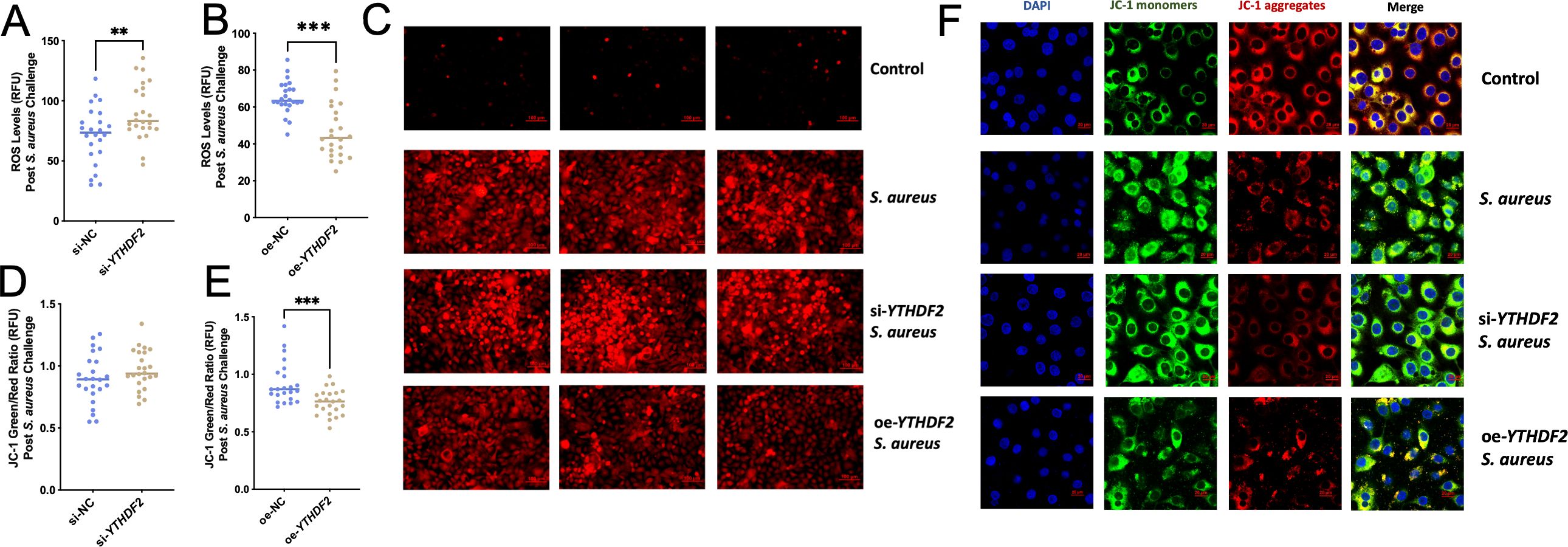
Figure 5. Effects of YTHDF2 on ROS levels and mitochondrial membrane potential in Mac-T Cells under S. aureus challenge. (A, B) RFU of ROS levels in si-NC,si-YTHDF2, oe-NC and oe-YTHDF2 Mac-T cells post S. aureus challenge. Statistical significance is indicated as follows: **p < 0.01, ***p < 0.001. (C) Fluorescent microscopy images showing ROS levels in control, S. aureus-challenged, si-YTHDF2 and oe-YTHDF2 Mac-T cells. (D, E) JC-1 green/red ratio indicating mitochondrial membrane potential (MMP) in si-NC, si-YTHDF2, oe-NC and oe-YTHDF2 Mac-T cells post S. aureus challenge. (F) Confocal microscopy images showing JC-1 monomers (green), JC-1 aggregates (red), and merged images in control, S. aureus-challenged, si-YTHDF2 and oe-YTHDF2 Mac-T cells.
Mitochondrial membrane potential (MMP) was analyzed using the JC-1 assay, as changes in MMP are a hallmark of apoptosis. In si-YTHDF2 cells, the green/red fluorescence ratio was significantly higher, indicating mitochondrial dysfunction (Figure 5D). In contrast, oe-YTHDF2 cells showed a lower green/red ratio, indicating preserved mitochondrial membrane potential (Figure 5E). These findings were further confirmed by confocal microscopy, which showed that si-YTHDF2 cells exhibited increased green fluorescence (MMP loss), whereas oe-YTHDF2 cells showed predominantly red fluorescence (intact MMP) (Figure 5F).
These results establish YTHDF2 as a key regulator of mitochondrial function in S. aureus-infected Mac-T cells. By modulating ROS levels and maintaining mitochondrial membrane potential, YTHDF2 protects cells from oxidative stress and apoptosis, highlighting its essential role in mitigating S. aureus-induced mitochondrial dysfunction and ensuring cellular homeostasis during mastitis.
3.5 IER3 expression is significantly increased in S. aureus-induced apoptosis and necrosis and is regulated by YTHDF2 via m6A modification
IER3, one of the 44 key genes regulated by YTHDF2, was found to be significantly upregulated during S. aureus-induced apoptosis and necrosis. IER3 is a stress-responsive gene associated with apoptosis, as indicated by the GeneCards database (Stelzer et al., 2016). Normalized read counts further supported these findings, with IER3 expression significantly higher in YTHDF2 knockdown cells compared to controls (Figure 6A). Following S. aureus challenge, IER3 expression was further increased in si-YTHDF2 cells, demonstrating that the absence of YTHDF2 amplifies S. aureus-induced upregulation of IER3. This upregulation was also observed in the transcriptomic data of milk samples from cows with subclinical mastitis and in blood transcriptomic data from cows infected with S. aureus, supporting the upregulation of IER3 during infection (Figures 6B, C).

Figure 6. Regulation of IER3 by YTHDF2 in Mac-T cells under S. aureus challenge. (A) Normalized IER3 expression in Mac-T cells from different experimental groups: si-NC, si-YTHDF2, si-NC + S. aureus, and si-YTHDF2 + S. aureus. (B) Normalized IER3 expression in milk samples from subclinical S. aureus mastitis cows and healthy cows. (C) Normalized IER3 expression in mammary gland tissue samples from the bovine model. (D) WB analysis of IER3 expression in si-IER3 Mac-T cells. (E-G) Immunofluorescence images and relative fluorescence unit (RFU) quantification of YO-PRO-1 (green) and PI (red) in si-NC and si-IER3 transfected Mac-T cells after S. aureus challenge. (H) Significant correlation between IER3 gene expression and YTHDF2 levels in Mac-T cells. (I) WB analysis showing the relationship between YTHDF2 and IER3 under different conditions. (J) m6A-MeRIP-seq data showing m6A modification peaks in the IER3 gene in control and S. aureus-challenged Mac-T cells. (K) RIP-qPCR analysis showing m6A peaks and YTHDF2 binding sites in the 3’ UTR region of IER3 under control and S. aureus conditions. (L, M) Half-life (t1/2) of IER3 mRNA, illustrating its stability in Mac-T cells transfected with si-NC, si-YTHDF2, oe-NC, and oe-YTHDF2. Statistical significance is indicated as follows: *p < 0.05,**p < 0.01.
To investigate the role of IER3 in apoptosis and necrosis, its expression was silenced in Mac-T cells. RT-qPCR and Western blot analyses confirmed efficient knockdown, with significant reductions in both IER3 mRNA and protein levels (Figure 6D). Functional assays showed that IER3 knockdown significantly reduced S. aureus-induced apoptosis and necrosis. YO-PRO-1/PI staining, visualized by confocal microscopy and quantified by a microplate reader, showed a significant decrease in apoptotic and necrotic cells in IER3 knockdown cells compared to controls (Figure 6E-G). These results suggest that IER3 acts as a pro-apoptotic factor during S. aureus-induced stress.
To further investigate the regulation of IER3 by YTHDF2, a regression analysis was performed to examine the relationship between YTHDF2 and IER3 expression levels. A significant negative correlation was observed between the expression of YTHDF2 and IER3, indicating that increased YTHDF2 expression corresponds to decreased IER3 expression (Figure 6H). Since YTHDF2 is known to promote mRNA degradation (Hou et al., 2021), this negative correlation suggests that YTHDF2 may regulate IER3 through mRNA stability. Further experiments showed that si-YTHDF2 resulted in an increase in IER3 protein levels, while YTHDF2 overexpression resulted in a decrease in IER3 protein expression. Additionally, IER3 protein levels were significantly increased under S. aureus challenge conditions, indicating that YTHDF2 regulates IER3 in response to bacterial stress (Figure 6I).
To explore the mechanism by which YTHDF2 regulates IER3, SRAMP (Structural RNA Modification Annotation and Prediction, a tool for predicting RNA modifications based on sequence and structural features) was used to predict potential m6A modification sites in IER3 mRNA. A high confidence m6A site was identified in the 3’ untranslated region of IER3 (Supplementary Figure S2E). m6A-seq confirmed a significant reduction in the m6A peak of IER3 mRNA following S. aureus challenge (Figure 6J). RIP-seq further validated that YTHDF2 binds to this m6A-modified site, confirming that YTHDF2 regulates IER3 via m6A modification. Since YTHDF2 promotes mRNA degradation, the reduced m6A modification after S. aureus infection likely decreased YTHDF2 binding, slowed IER3 degradation, and resulted in increased expression. RIP-qPCR results showed that reduced YTHDF2 binding after infection resulted in decreased IER3 degradation and increased mRNA stability (Figure 6K). To further support this finding, actinomycin D assays were performed. The half-life (t1/2) of IER3 mRNA increased from 0.36 hours to 0.63 hours in YTHDF2 knockdown cells, indicating increased mRNA stability in the absence of YTHDF2 (Figure 6L). In contrast, the mRNA half-life decreased from 6.59 hours to 1.83 hours in YTHDF2-overexpressing cells, confirming that YTHDF2 destabilizes IER3 mRNA (Figure 6M). These results establish YTHDF2 as a critical regulator of IER3 expression via m6A modification, which affects apoptosis and necrosis in S. aureus-challenged Mac-T cells.
4 Discussion
In previous studies, it has been widely demonstrated that m6A modifications are essential for regulating mRNA stability, degradation, and cellular processes like immune responses and apoptosis (Zhao et al., 2017; Huang et al., 2018; Uzonyi et al., 2023). This study used integrated approaches to investigate the m6A modification in the regulation of gene expression during S. aureus infection. First, RNA-seq analysis (siNC vs. siNC-S. aureus, siNC vs. siYTHDF2) identified infection-responsive transcripts and highlighted those potentially regulated by YTHDF2. This analysis facilitated the identification of genes altered during infection and those affected by YTHDF2. This analysis enabled the identification of genes altered during infection and those affected by YTHDF2. Subsequent analysis by m6A profiling revealed infection-specific differential methylation events, particularly in apoptosis-related genes such as IER3, thus providing insight into the role of m6A modification in the infection process. YTHDF2 RIP-seq was used to define its direct mRNA targets, and the integration of these data with m6A profiling identified infection-specific mRNAs that were directly bound by YTHDF2. These datasets were used to construct the m6A-YTHDF2-IER3 pathway, thereby demonstrating that YTHDF2 regulates IER3 mRNA degradation during S. aureus infection. These findings were further validated in subclinical mastitis and infected samples.
YTHDF2, an m6A “reader” protein, regulates mRNA stability by recognizing m6A modifications and promoting mRNA degradation (He et al., 2019; Chen et al., 2021). This mechanism is essential for controlling gene expression in various biological processes, including immune responses and apoptosis (Guo et al., 2020; Li et al., 2020; Einstein et al., 2021). IER3 is an immediate-early response gene involved in cellular apoptosis and necrosis (Yoon et al., 2009; Ustyugova et al., 2012). During S. aureus infection, IER3 upregulation has been observed to correlates with increased cell death, including apoptosis and necrosis, in Mac-T cells. This suggests that IER3 not only plays a role in cellular stress responses but also contributes to regulating cell survival during infection. Knockdown of YTHDF2 (si-YTHDF2) resulted in a significant increase in IER3 mRNA stability, as indicated by its prolonged half-life. In contrast, overexpression of YTHDF2 (oe-YTHDF2) resulted in a reduction in IER3 mRNA stability. These findings highlight YTHDF2’s role in modulating IER3 stability through m6A modification, which is critical for immune response regulation during bacterial infection.
m6A modification sites, particularly within the 3’ UTR of mRNA, are essential for mRNA stability and are preferentially recognized by YTHDF2 (Guo et al., 2020; Li et al., 2021a). This study confirmed that YTHDF2 targets these specific m6A modification sites within IER3 mRNA, regulating its stability and contributing to the fine-tuning of immune responses. Furthermore, YTHDF2 likely influences key inflammatory pathways, such as NF-κB and PI3K/Akt, which modulate apoptosis and immune responses (Arlt et al., 2003; Sina et al., 2010). By targeting IER3 mRNA, YTHDF2 not only regulates gene expression but also indirectly regulates the immune response and cell death pathways.
Of the 44 YTHDF2-regulated genes identified through m6A modifications, several are involved in immune responses and inflammation. This study confirmed that IER3 is associated with apoptosis regulation, and its m6A-dependent modulation by YTHDF2 underscores the critical role of m6A in inflammatory processes of S. aureus challenge. Previous studies have extensively documented the pivotal role of m6A modifications in regulating immune responses and cellular stress pathways, affecting mRNA stability, translation, and splicing, which modulate key aspects of inflammation and immunity (Jiang et al., 2021; Mao et al., 2023; Zhu et al., 2023). Furthermore, GWAS enrichment analysis demonstrated significant associations between these genes and important bovine health traits, such as somatic cell score (Heringstad et al., 2006; Khan et al., 2023). This suggests that YTHDF2’s regulation of m6A-modified mRNAs not only impacts cellular responses to S. aureus infection but also influences broader health traits in dairy cattle, offering insights into genetic and epigenetic strategies for improving disease resistance.
Oxidative stress and mitochondrial dysfunction are critical mediators of apoptosis in Mac-T cells during S. aureus infection (Malhotra et al., 2019; Zhou et al., 2020). This study shows that knockdown of YTHDF2 significantly increases ROS levels, leading to mitochondrial dysfunction and enhanced apoptosis. These results are consistent with those of studies on bacterial infections, including those caused by E. coli (Jin et al., 2018; Zhang et al., 2023b) and S. aureus (Gao et al., 2017; Xu et al., 2022), where elevated ROS levels contribute to mitochondrial damage and apoptosis (Yang et al., 2016; Pinegin et al., 2018). Specifically, YTHDF2 knockdown exacerbates mitochondrial damage and apoptosis under S. aureus challenge, supporting the notion that YTHDF2 plays a protective role against oxidative damage. In contrast, YTHDF2 overexpression (oe-YTHDF2) mitigates these effects, preserving mitochondrial function and reducing apoptosis. This was evidenced by a lower JC-1 green/red fluorescence ratio in oe-YTHDF2 cells, indicating maintained mitochondrial membrane potential (MMP) and decreased ROS levels. These results suggest that YTHDF2 protects cells from oxidative stress by regulating m6A-modified mRNAs involved in oxidative stress responses, maintaining mitochondrial integrity and cellular health under stress.
Studies have shown that m6A modifications regulate the stability of mRNAs associated with oxidative stress and mitochondrial function (Sun et al., 2021; Yu et al., 2021; Wan et al., 2024). This study, along with ROS and JC-1 assays, further highlights the role of YTHDF2’s role in modulating mitochondrial function and ROS levels. By regulating m6A-modified mRNAs, YTHDF2 contributes to protecting cells from oxidative damage and apoptosis. This highlights how RNA modifications and their reader proteins influence cellular stress responses and mitochondrial function, ultimately impacting cell survival during bacterial infections.
This study provides valuable insights into the role of YTHDF2 in m6A modification, but several limitations should be addressed. First, the research primarily used the Mac-T cell model, which may not fully capture the complexity of cellular interactions in mammary tissue during infection. Future research could incorporate co-culture systems (e.g., Mac-T cells with mammary macrophages) (Yao et al., 2017; Spalinger et al., 2020) or 3D mammary gland tissue models to better simulate immune responses and tissue complexity (Lee et al., 2023). Another limitation is the small sample size, with only two cows tested, which limits the generalizability of the findings. Future studies should include a larger, more diverse sample of cows to confirm the robustness of the results. Additionally, this study primarily focused on the early immune response during S. aureus infection, leaving the chronic phase of mastitis underexplored. Future research should investigate the role of YTHDF2 and IER3 in chronic inflammation, providing a more complete understanding of long-term immune responses.
Mastitis is a major challenge in the dairy industry, with S. aureus being one of the main pathogens responsible for it. This study shows that S. aureus infection decreases YTHDF2 expression while increasing IER3 expression in Mac-T cells, highlighting the role of YTHDF2 in regulating IER3 mRNA stability through m6A modifications (Figure 7). Specifically, YTHDF2 has been shown to promote the degradation of m6A-modified IER3 mRNA, and its downregulation has been observed to increase IER3 levels, which are associated with increased apoptosis and necrosis in infected cells. Furthermore, YTHDF2 plays a critical role in modulating apoptosis, mitochondrial function and ROS levels, thereby supporting cell survival during bacterial infection. In conclusion, this study highlights the essential role of YTHDF2 and IER3 in the inflammatory response of bovine mammary cells to S. aureus infection, providing novel insights into the molecular mechanisms underlying bovine mastitis. The findings of this study provide valuable implications for molecular breeding aimed at improving udder health in dairy cattle.
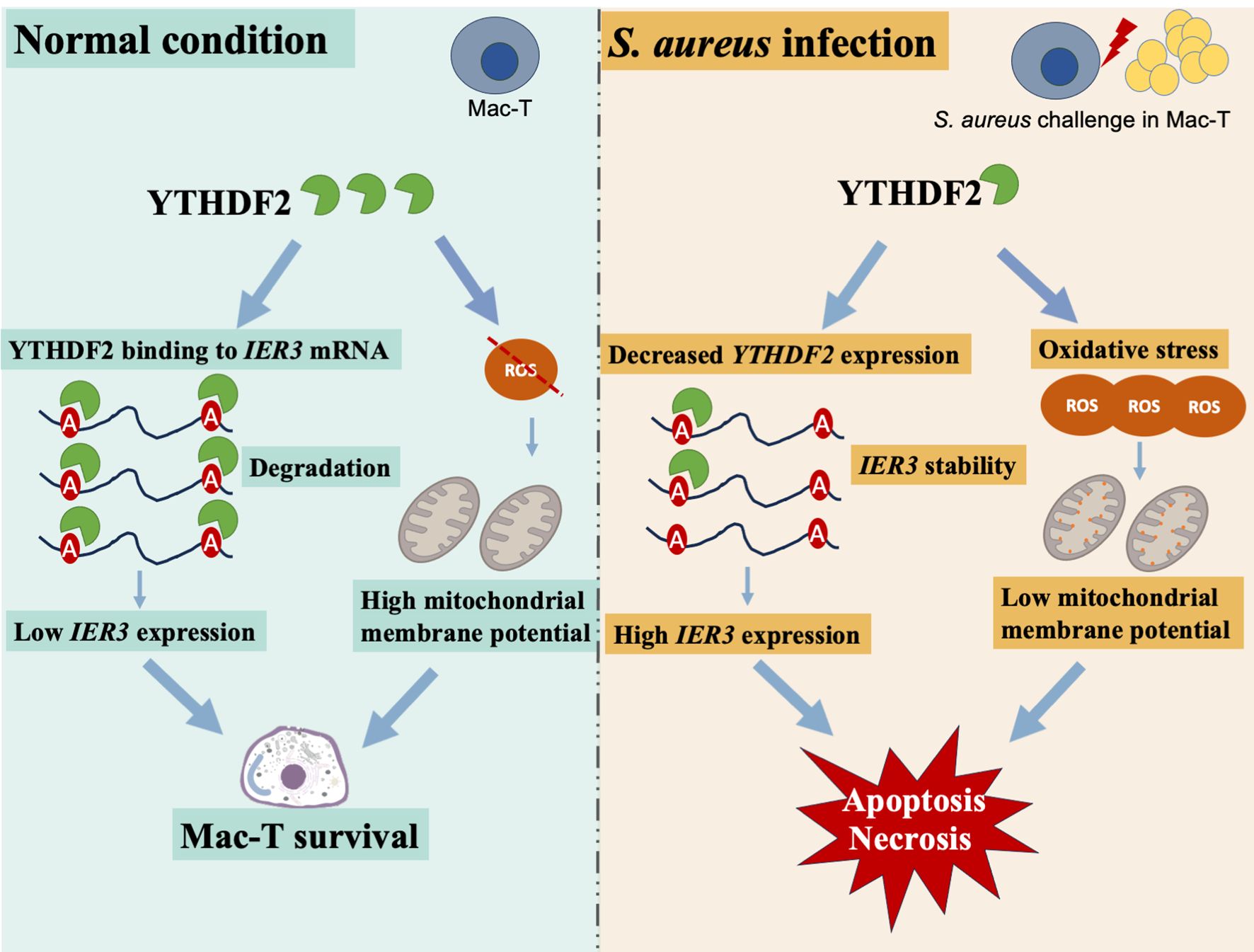
Figure 7. Mechanistic insights into YTHDF2 regulation of IER3 mRNA stability and cell death in S. aureus-induced Mac-T cells.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by the Animal Welfare Committee of China Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YX: Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. SM: Funding acquisition, Investigation, Validation, Writing – review & editing. SC: Validation, Writing – review & editing. XT: Validation, Writing – original draft. ZZ: Validation, Writing – original draft. YS: Investigation, Writing – original draft. XW: Methodology, Writing – review & editing. YY: Conceptualization, Funding acquisition, Methodology, Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Key Research and Development Program of China (2023YFF1000902 and 2021YFD1200903), the Young Scientists Fund of the National Natural Science Foundation of China (Grant 32302706), the earmarked fund for CARS-36, the Program for Changjiang Scholars and the Innovative Research Team in University (Grant IRT-15R62), and the Seed Fund from China Agricultural University (CAU). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1542647/full#supplementary-material
Supplementary Figure 1 | KEGG pathway enrichment. (A) KEGG pathway enrichment analysis of DEGs post S. aureus challenge. (B) KEGG pathway enrichment of differentially expressed genes (DEGs) between treatment groups. (C) KEGG pathway enrichment of common DEGs across conditions.
Supplementary Figure 2 | Effects of YTHDF2 knockdown on necrosis and m6A-modified gene regulation in Mac-T cells under S. aureus challenge. (A) Chromosomal distribution of transcription factor binding sites across the bovine genome. (B) Binding motif identified by HOMER showing the overlap between YTHDF2 RIP-seq and m6A-MeRIP-seq data. (C) Intersection analysis of gene sets from RIP-seq, JN-seq, and DE-m6A-seq, highlighting shared gene overlaps. (D) Venn diagram showing the overlap of DEGs among different experimental comparisons (N: si-NC, Y: si-YTHDF2, and JN: si-NC + S. aureus). (E) QTL associations with bovine traits, including disease susceptibility and production traits. (F) QTL enrichment analysis of the IER3 gene, showing associations with immunoglobulin G levels and disease resistance.
Supplementary Figure 3 | Effects of YTHDF2 knockdown on necrosis and m6A-modified gene regulation in Mac-T cells under S. aureus challenge. (A, B) WB analysis showing YTHDF2 protein levels and relative gray values in Mac-T cells transfected with si-NC, si-YTHDF2, oe-NC and oe-YTHDF2. (C) Propidium iodide (PI) staining showing necrotic cells (red fluorescence) in si-NC and si-YTHDF2-transfected Mac-T cells post-S. aureus challenge. Dark field and bright field images are shown for comparison. (D) Correlation heatmap displaying expression relationships between m6A-modified DEGs regulated by YTHDF2. Significant correlations are indicated by P < 0.05. (E) Predicted m6A modification site on target mRNA, with the highlighted position located within the sequencing data obtained in this study. (F) Confidence score plot for predicted m6A sites across the transcript, showing regions of very high, high, moderate, and low confidence, with marked positions falling within the sequencing results from this study.
References
Aitken, S. L., Corl, C. M., and Sordillo, L. M. (2011). Immunopathology of mastitis: insights into disease recognition and resolution. J. Mammary Gland Biol. Neoplasia 16, 291–304. doi: 10.1007/s10911-011-9230-4
Arlt, A., Kruse, M. L., Breitenbroich, M., Gehrz, A., Koc, B., Minkenberg, J., et al. (2003). The early response gene IEX-1 attenuates NF-kappaB activation in 293 cells, a possible counter-regulatory process leading to enhanced cell death. Oncogene 22, 3343–3351. doi: 10.1038/sj.onc.1206524
Arlt, A., Minkenberg, J., Kocs, B., Grossmann, M., Kruse, M. L., Fölsch, U. R., et al. (2004). The expression of immediate early gene X-1 (IEX-1) is differentially induced by retinoic acids in NB4 and KG1 cells: possible implication in the distinct phenotype of retinoic acid-responsive and -resistant leukemic cells. Leukemia 18, 1646–1655. doi: 10.1038/sj.leu.2403481
Arlt, A. and Schäfer, H. (2011). Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur. J. Cell Biol. 90, 545–552. doi: 10.1016/j.ejcb.2010.10.002
Bannerman, D. D. (2009). Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows1. J. Anim. Sci. 87, 10–25. doi: 10.2527/jas.2008-1187
Chen, G., Zhao, Q., Yuan, B., Wang, B., Zhang, Y., Li, Z., et al. (2021). ALKBH5-modified HMGB1-STING activation contributes to radiation induced liver disease via innate immune response. Int. J. Radiat. Oncol. Biol. Phys. 111, 491–501. doi: 10.1016/j.ijrobp.2021.05.115
Einstein, J. M., Perelis, M., Chaim, I. A., Meena, J. K., Nussbacher, J. K., Tankka, A. T., et al. (2021). Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell 81, 3048–3064.e3049. doi: 10.1016/j.molcel.2021.06.014
Fang, L., Hou, Y., An, J., Li, B., Song, M., Wang, X., et al. (2016). Genome-wide transcriptional and post-transcriptional regulation of innate immune and defense responses of bovine mammary gland to Staphylococcus aureus. Front. Cell Infect. Microbiol. 6. doi: 10.3389/fcimb.2016.00193
Gao, P., Davies, J., and Kao, R. Y. T. (2017). Dehydrosqualene Desaturase as a Novel Target for Anti-Virulence Therapy against Staphylococcus aureus. mBio 8 (5), e01224-17. doi: 10.1128/mBio.01224-17
Guo, X., Li, K., Jiang, W., Hu, Y., Xiao, W., Huang, Y., et al. (2020). RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol. Cancer 19, 91. doi: 10.1186/s12943-020-01158-w
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18, 176. doi: 10.1186/s12943-019-1109-9
Heringstad, B., Gianola, D., Chang, Y. M., Odegård, J., and Klemetsdal, G. (2006). Genetic associations between clinical mastitis and somatic cell score in early first-lactation cows. J. Dairy Sci. 89, 2236–2244. doi: 10.3168/jds.S0022-0302(06)72295-0
Hou, G., Zhao, X., Li, L., Yang, Q., Liu, X., Huang, C., et al. (2021). SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 49, 2859–2877. doi: 10.1093/nar/gkab065
Hu, Z. L., Park, C. A., and Reecy, J. M. (2022). Bringing the Animal QTLdb and CorrDB into the future: meeting new challenges and providing updated services. Nucleic Acids Res. 50, D956–d961. doi: 10.1093/nar/gkab1116
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 6, 74. doi: 10.1038/s41392-020-00450-x
Jin, M., Lu, J., Chen, Z., Nguyen, S. H., Mao, L., Li, J., et al. (2018). Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ. Int. 120, 421–430. doi: 10.1016/j.envint.2018.07.046
Kang, S. J., Cho, Y. I., Kim, K. H., and Cho, E. S. (2016). Proteomic analysis to elucidate the antibacterial action of silver ions against bovine mastitis pathogens. Biol. Trace Element Res. 171, 101–106. doi: 10.1007/s12011-015-0510-5
Khan, M. Z., Wang, J., Ma, Y., Chen, T., Ma, M., Ullah, Q., et al. (2023). Genetic polymorphisms in immune- and inflammation-associated genes and their association with bovine mastitis resistance/susceptibility. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1082144
Lee, J., Liu, Y., Ray, E., Giuliano, A. E., and Cui, X. (2023). Human breast organoid models for lactation research. Reprod. Breed. 3, 125–130. doi: 10.1016/j.repbre.2023.08.003
Li, T., Lin, C., Zhu, Y., Xu, H., Yin, Y., Wang, C., et al. (2021b). Transcriptome Profiling of m(6)A mRNA Modification in Bovine Mammary Epithelial Cells Treated with Escherichia coli. Int. J. Mol. Sci. 22 (12), 6254. doi: 10.3390/ijms22126254
Li, J., Xie, H., Ying, Y., Chen, H., Yan, H., He, L., et al. (2020). YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer 19, 152. doi: 10.1186/s12943-020-01267-6
Li, H., Zhang, N., Jiao, X., Wang, C., Sun, W., He, Y., et al. (2021a). Downregulation of microRNA-6125 promotes colorectal cancer growth through YTHDF2-dependent recognition of N6-methyladenosine-modified GSK3β. Clin. Transl. Med. 11, e602. doi: 10.1002/ctm2.602
Lin, C., Zhu, Y., Hao, Z., Xu, H., Li, T., Yang, J., et al. (2021). Genome-wide analysis of LncRNA in bovine mammary epithelial cell injuries induced by Escherichia coli and Staphylococcus aureus. Int. J. Mol. Sci. 22 (18), 9719. doi: 10.3390/ijms22189719
Liu, J., Li, K., Cai, J., Zhang, M., Zhang, X., Xiong, X., et al. (2020). Landscape and Regulation of m(6)A and m(6)Am Methylome across Human and Mouse Tissues. Mol. Cell 77, 426–440.e426. doi: 10.1016/j.molcel.2019.09.032
Liu, Y., Song, R., Lu, Z., Zhao, L., Zhan, X., Li, Y., et al. (2024). The RNA m(6)A demethylase ALKBH5 drives emergency granulopoiesis and neutrophil mobilization by upregulating G-CSFR expression. Cell Mol. Immunol. 21, 6–18. doi: 10.1038/s41423-023-01115-9
Maassen, J. A., LM, T. H., Van Essen, E., Heine, R. J., Nijpels, G., Jahangir Tafrechi, R. S., et al. (2004). Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes 53 Suppl 1, S103–S109. doi: 10.2337/diabetes.53.2007.s103
Malhotra, S., Hayes, D., Jr., and Wozniak, D. J. (2019). Cystic fibrosis and pseudomonas aeruginosa: the host-microbe interface. Clin. Microbiol. Rev. 32 (3), e00138-18. doi: 10.1128/cmr.00138-18
Mao, Y., Jiang, F., Xu, X. J., Zhou, L. B., Jin, R., Zhuang, L. L., et al. (2023). Inhibition of IGF2BP1 attenuates renal injury and inflammation by alleviating m6A modifications and E2F1/MIF pathway. Int. J. Biol. Sci. 19, 593–609. doi: 10.7150/ijbs.78348
Mukherjee, N., Corcoran, D. L., Nusbaum, J. D., Reid, D. W., Georgiev, S., Hafner, M., et al. (2011). Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339. doi: 10.1016/j.molcel.2011.06.007
Ogunnaike, M., Wang, H., and Zempleni, J. (2021). Bovine mammary alveolar MAC-T cells afford a tool for studies of bovine milk exosomes in drug delivery. Int. J. Pharm. 610, 121263. doi: 10.1016/j.ijpharm.2021.121263
Pawlikowska, P., Leray, I., de Laval, B., Guihard, S., Kumar, R., Rosselli, F., et al. (2010). ATM-dependent expression of IEX-1 controls nuclear accumulation of Mcl-1 and the DNA damage response. Cell Death Differentiation 17, 1739–1750. doi: 10.1038/cdd.2010.56
Pinegin, B., Vorobjeva, N., Pashenkov, M., and Chernyak, B. (2018). The role of mitochondrial ROS in antibacterial immunity. J. Cell Physiol. 233, 3745–3754. doi: 10.1002/jcp.26117
Prall, W., Sheikh, A. H., Bazin, J., Bigeard, J., Almeida-Trapp, M., Crespi, M., et al. (2023). Pathogen-induced m6A dynamics affect plant immunity. Plant Cell 35, 4155–4172. doi: 10.1093/plcell/koad224
Rizwan, H., Pal, S., Sabnam, S., and Pal, A. (2020). High glucose augments ROS generation regulates mitochondrial dysfunction and apoptosis via stress signalling cascades in keratinocytes. Life Sci. 241, 117148. doi: 10.1016/j.lfs.2019.117148
Sebens Müerköster, S., Rausch, A. V., Isberner, A., Minkenberg, J., Blaszczuk, E., Witt, M., et al. (2008). The apoptosis-inducing effect of gastrin on colorectal cancer cells relates to an increased IEX-1 expression mediating NF-κB inhibition. Oncogene 27, 1122–1134. doi: 10.1038/sj.onc.1210728
Sina, C., Arlt, A., Gavrilova, O., Midtling, E., Kruse, M. L., Müerköster, S. S., et al. (2010). Ablation of gly96/immediate early gene-X1 (gly96/iex-1) aggravates DSS-induced colitis in mice: role for gly96/iex-1 in the regulation of NF-kappaB. Inflammation Bowel Dis. 16, 320–331. doi: 10.1002/ibd.21066
Spalinger, M. R., Sayoc-Becerra, A., Santos, A. N., Shawki, A., Canale, V., Krishnan, M., et al. (2020). PTPN2 regulates interactions between macrophages and intestinal epithelial cells to promote intestinal barrier function. Gastroenterology 159, 1763–1777.e1714. doi: 10.1053/j.gastro.2020.07.004
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The geneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinf. 54, 1.30.31–31.30.33. doi: 10.1002/cpbi.5
Su, T., Zhang, N., Wang, T., Zeng, J., Li, W., Han, L., et al. (2023). Super enhancer-regulated LncRNA LINC01089 induces alternative splicing of DIAPH3 to drive hepatocellular carcinoma metastasis. Cancer Res. 83, 4080–4094. doi: 10.1158/0008-5472.Can-23-0544
Sun, L., Wan, A., Zhou, Z., Chen, D., Liang, H., Liu, C., et al. (2021). RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut 70, 1698–1712. doi: 10.1136/gutjnl-2020-320652
Ustyugova, I. V., Zhi, L., Abramowitz, J., Birnbaumer, L., and Wu, M. X. (2012). IEX-1 deficiency protects against colonic cancer. Mol. Cancer Res. 10, 760–767. doi: 10.1158/1541-7786.Mcr-11-0556
Uzonyi, A., Dierks, D., Nir, R., Kwon, O. S., Toth, U., Barbosa, I., et al. (2023). Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol. Cell 83, 237–251.e237. doi: 10.1016/j.molcel.2022.12.026
Wan, F., Qiu, F., Deng, Y., Hu, H., Zhang, Y., Zhang, J. Y., et al. (2024). Knockdown of YTHDF2 initiates ERS-induced apoptosis and cancer stemness suppression by sustaining GLI2 stability in cervical cancer. Transl. Oncol. 46, 101994. doi: 10.1016/j.tranon.2024.101994
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, H., Wang, X., Li, X., Wang, Q., Qing, S., Zhang, Y., et al. (2019). A novel long non-coding RNA regulates the immune response in MAC-T cells and contributes to bovine mastitis. FEBS J. 286, 1780–1795. doi: 10.1111/febs.14783
Wang, X., Wang, X., Wang, Y., Guo, G., Usman, T., Hao, D., et al. (2014b). Antimicrobial resistance and toxin gene profiles of Staphylococcus aureus strains from Holstein milk. Lett. Appl. Microbiol. 58, 527–534. doi: 10.1111/lam.12221
Wang, M., Yang, N., Laterrière, M., Gagné, D., Omonijo, F., and Ibeagha-Awemu, E. M. (2024). Multi-omics integration identifies regulatory factors underlying bovine subclinical mastitis. J. Anim. Sci. Biotechnol. 15, 46. doi: 10.1186/s40104-024-00996-8
Xing, Y., Mi, S., Dari, G., Zhang, Z., Chen, S., and Yu, Y. (2025). Ferroptosis-related genes as molecular markers in bovine mammary epithelial cells challenged with Staphylococcus aureus. Int. J. Mol. Sci. 26 (6), 2506. doi: 10.3390/ijms26062506
Xu, L., Zhan, W., Deng, Y., Liu, X., Gao, G., Sun, X., et al. (2022). ROS turn nanoparticle fluorescence on for imaging Staphylococcus aureus infection in vivo. Adv. Healthc Mater 11, e2200453. doi: 10.1002/adhm.202200453
Yang, Y., Karakhanova, S., Hartwig, W., D’Haese, J. G., Philippov, P. P., Werner, J., et al. (2016). Mitochondria and mitochondrial ROS in cancer: novel targets for anticancer therapy. J. Cell Physiol. 231, 2570–2581. doi: 10.1002/jcp.25349
Yao, Y., Wang, Z. C., Liu, J. X., Ma, J., Chen, C. L., Deng, Y. K., et al. (2017). Increased expression of TIPE2 in alternatively activated macrophages is associated with eosinophilic inflammation and disease severity in chronic rhinosinusitis with nasal polyps. Int. Forum Allergy Rhinol. 7, 963–972. doi: 10.1002/alr.21984
Yoon, S., Ha, H.-J., Kim, Y.-H., Won, M., Park, M., Ko, J.-J., et al. (2009). IEX-1-induced cell death requires BIM and is modulated by MCL-1. Biochem. Biophys. Res. Commun. 382, 400–404. doi: 10.1016/j.bbrc.2009.03.037
You, F., Osawa, Y., Hayashi, S., and Nakashima, S. (2007). Immediate early gene IEX-1 induces astrocytic differentiation of U87-MG human glioma cells. J. Cell Biochem. 100, 256–265. doi: 10.1002/jcb.21082
Yu, F., Wei, J., Cui, X., Yu, C., Ni, W., Bungert, J., et al. (2021). Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 49, 5779–5797. doi: 10.1093/nar/gkab415
Zhang, X. W., An, M. X., Huang, Z. K., Ma, L., Zhao, D., Yang, Z., et al. (2023b). Lpp of Escherichia coli K1 inhibits host ROS production to counteract neutrophil-mediated elimination. Redox Biol. 59, 102588. doi: 10.1016/j.redox.2022.102588
Zhang, M., Wang, J., Jin, Y., Zheng, Q., Xing, M., Tang, Y., et al. (2022). YTHDF2-mediated FGF14-AS2 decay promotes osteolytic metastasis of breast cancer by enhancing RUNX2 mRNA translation. Br. J. Cancer 127, 2141–2153. doi: 10.1038/s41416-022-02006-y
Zhang, H., Wu, D., Wang, Y., Guo, K., Spencer, C. B., Ortoga, L., et al. (2023a). METTL3-mediated N6-methyladenosine exacerbates ferroptosis via m6A-IGF2BP2-dependent mitochondrial metabolic reprogramming in sepsis-induced acute lung injury. Clin. Transl. Med. 13, e1389. doi: 10.1002/ctm2.1389
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi: 10.1038/nrm.2016.132
Zhou, T., Hu, R., Wang, L., Qiu, Y., Zhang, G., Deng, Q., et al. (2020). An AIE-active conjugated polymer with high ROS-generation ability and biocompatibility for efficient photodynamic therapy of bacterial infections. Angew Chem. Int. Ed Engl. 59, 9952–9956. doi: 10.1002/anie.201916704
Zhu, Z. M., Huo, F. C., Zhang, J., Shan, H. J., and Pei, D. S. (2023). Crosstalk between m6A modification and alternative splicing during cancer progression. Clin. Transl. Med. 13, e1460. doi: 10.1002/ctm2.1460
Keywords: bovine mastitis, Staphylococcus aureus, YTHDF2, m6A modification, apoptosis and necrosis
Citation: Xing Y, Mi S, Chen S, Tao X, Zhang Z, Shi Y, Wang X and Yu Y (2025) YTHDF2-mediated m6A modification regulates mRNA stability of Immediate early response gene 3 to modulate cell death in Staphylococcus aureus-induced bovine mastitis. Front. Cell. Infect. Microbiol. 15:1542647. doi: 10.3389/fcimb.2025.1542647
Received: 10 December 2024; Accepted: 06 May 2025;
Published: 30 May 2025.
Edited by:
Mathieu Coureuil, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Bruno Sargueil, UMR8015 Laboratoire de cristallographie et RMN biologiques, FranceHan Sang Yoo, Seoul National University, Republic of Korea
Copyright © 2025 Xing, Mi, Chen, Tao, Zhang, Shi, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Yu, eXV5aW5nQGNhdS5lZHUuY24=
 Yue Xing1
Yue Xing1 Siyuan Mi
Siyuan Mi Xingping Wang
Xingping Wang Ying Yu
Ying Yu