- 1Department of Basic Health Sciences, College of Applied Medical Sciences, Qassim University, Buraydah, Saudi Arabia
- 2Department of Microbiology the University of Haripur, Haripur, KPK, Pakistan
- 3Islamabad Medical and Dental College, Bharakhau, Islamabad, Pakistan
- 4Microbiology and Biotechnology Research laboratory, Department of Biotechnology, Fatima Jinnah Women University, Rawalpindi, Pakistan
Introduction: Antibiotic resistance is a pressing global challenge, complicating the treatment of infectious diseases caused by multidrug-resistant microorganism. For centuries, medicinal plants have been a cornerstone of natural remedies, offering bioactive compounds with therapeutic potential.
Methods: The study investigate the phytochemical screening and antibacterial efficay of Carica papaya leaf extract, focusing on its impact against three Gram-negative bacterial pathogens i.e., Escherichia coli, Helicobacter pylori, and Salmonella enterica serovar Typhi, are major contributors to gastrointestinal infections worldwide, often leading to severe inflammation and chronic health complications.
Results: The phytochemical screening revealed the presence of phenols and flavonoids, which are key contributors to the extract’s biological activity. GC-MC analysis identified 27 bioactive compounds, with phytol emerging as a prominent constituent, detected at a peak retention time of 18.712 minutes. Antibacterial assays demonstrated significant efficacy, with inhibitory zones ranging from 10 to 20 mm against the tested pathogens. Molecular docking further highlighted phytol’s strong binding affinities to crucial bacterial proteins, including DNA gyrase (E. coli), Vacuolating cytotoxin A (H. pylori) from, and Dihydrofolate reductase (Salmonella enterica serovar Typhi). Notably, phytol exhibited the highest binding energy (-6.64 kcal/mol) with DHFR, indicating a robust interaction that underscores its potential as a targeted antibacterial agent against Salmonella enterica serovar Typhi.
Discussion: These findings position phytol as a promising lead compound for developing novel antibacterial therapies. Its strong activity against multidrug-resistant pathogens suggests potential for further exploration, though additional research is needed to assess its role in resistance modulation or prevention.
1 Introduction
Antimicrobial resistance (AMR) is a critical global health concern affecting several pathogens, including particularly Helicobacter pylori (H. pylori), Escherichia coli (E. coli), and Salmonella enterica serovar Typhi (S. Typhi). In H. pylori, resistance to antibiotics such as metronidazole, fluoroquinolones, and clarithromycin threatens effective eradication therapy, emphasizing the need for alternative treatments and resistance monitoring (Argueta et al., 2022). Similarly, E. coli exhibits resistance to beta-lactams, fluoroquinolones, and carbapenems, with multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains limiting treatment options for sepsis, urinary tract infections, and other infections (Abbas et al., 2023). Factors such as antibiotic overuse, poor infection prevention and control strategies contribute to this issue (Magiorakos et al., 2012). S. enterica serovar Typhi has also shown increasing resistance to commonly used antibiotics, including ampicillin, chloramphenicol, and co-trimoxazole (Sulfamethoxazole and trimethoprim combination), with rising resistance to third-generation cephalosporins and fluoroquinolones (Qamar et al., 2018; Chatham-Stephens, 2019). The appearance of antimicrobial resistance can be ascribed to anthropological, environmental and genetic factors. Human-related factors such as antibiotic overuse and missuse, poor infection prevention and control strategies contribute to antibiotic resistance (Magiorakos et al., 2012). The spread of AMR in S. Typhi is driven by factors like improper antibiotic use, inadequate infection control, poor sanitation and hygiene in public settings (Klemm et al., 2018). AMR is driven by a variety of genetic factors, both intrinsic and acquired, that allow bacteria to survive and thrive in the presence of antibiotics. These include mutations in genes, mobile genetic elements like plasmids, and the spread of resistance genes through horizontal gene transfer (Ahmad et al., 2024).
Plants have long been a source of medicinal agents due to their bioactive compounds, such as alkaloids, flavonoids, and terpenoids, which exhibit anti-inflammatory, antimicrobial, antioxidant, and anticancer properties (Newman and Cragg, 2016). Among these plants, Carica papaya is particularly notable for its rich nutritional and bioactive profile, including vitamins A and C, with trace elements such as potassium, and the enzyme papain (Santana et al., 2019). Papaya’s fruit, leaves, and seeds have been used in traditional medicine for their anti-inflammatory, antioxidant, and antimicrobial properties, with research confirming their potential in treating diseases such as cancer, diabetes, and gastrointestinal disorders (Kong et al., 2021). The seeds exhibit potent antimicrobial effects against Gram-negative bacteria (Ugbogu et al., 2023), while the leaves contain flavonoids that contribute to anti-inflammatory and antioxidant activity (Sharma et al., 2022). The fruit pulp is rich in papain and vitamin C, which are linked to gastrointestinal benefits (Kim et al., 2024). Additionally, papaya holds significant value in the food and pharmaceutical industries, making it a versatile plant for medicinal, nutritional, and industrial applications (Alara et al., 2022).
Molecular docking is a computational technique used to predict the binding affinity and orientation of small molecules within a protein’s active site, playing a crucial role in drug discovery by enabling virtual screening of large compound libraries to identify potential leads for specific protein targets (Meng et al., 2011). This method considers various factors such as hydrogen bonding, hydrophobic interactions, and electrostatic forces to predict the binding energy and conformation of the ligand-protein complex (Morris et al., 2009). Recent advances have improved its accuracy, allowing precise prediction of binding modes and affinities, and have been applied in drug design for diseases like cancer, malaria, tuberculosis, and influenza (Chen et al., 2020; Ejalonibu et al., 2021). Protein-ligand binding, an essential process in biological functions like signal transduction and enzyme inhibition, involves the recognition of molecular features such as shape, charge, and hydrophobicity, with interactions including hydrogen bonding, ionic forces, and van der Waals interactions determining the binding affinity and specificity (Chen et al., 2016). Molecular dynamics simulations and thermodynamic integration have been employed to study binding free energy and kinetics, offering insights into binding mechanisms essential for designing therapeutic agents that modulate protein function (Lazim et al., 2020).
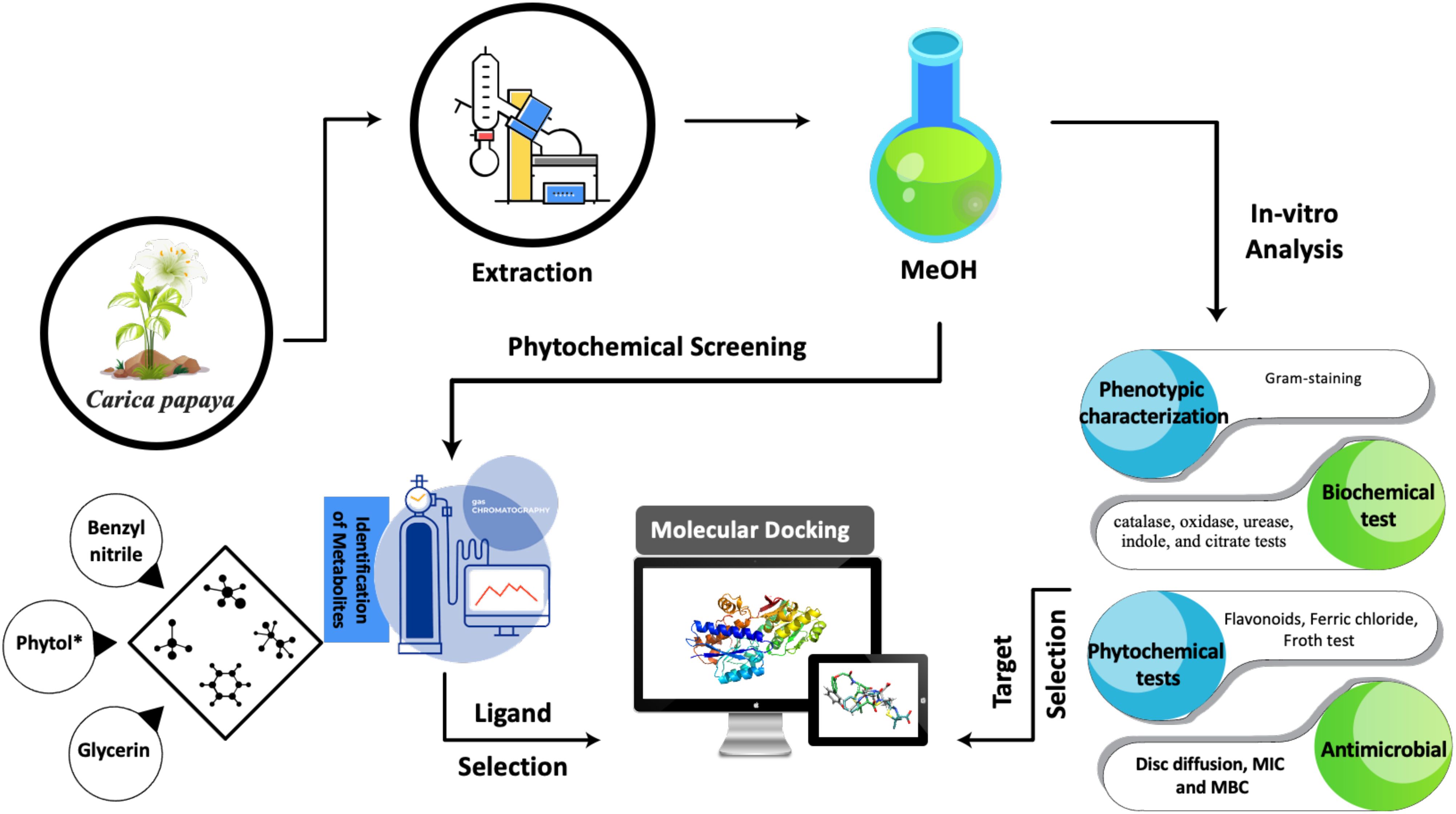
This study investigated the antibacterial activity of Carica papaya leaf extract against three clinically important bacterial pathogens, Helicobacter pylori, Escherichia coli, and Salmonella enterica serovar Typhi. It identified the bioactive compounds responsible for the antibacterial activity and elucidated the molecular interactions between these bioactive compounds and the targeted bacterial proteins using molecular docking simulations, with the ultimate goal of identifying potential lead compounds for developing novel antibacterial therapeutics.
2 Materials and methods
2.1 Inoculation of selected clinical strains
The strains of Helicobacter pylori (ATCC 43504) are commonly used for pathogenicity studies (Kinoshita-Daitoku et al., 2020), Escherichia coli (ATCC 25922) (Jafari et al., 2021; Asghar et al., 2024), and Salmonella enterica serovar Typhi (6539) (Asghar et al., 2024), were obtained from laboratory the department of Microbiology, The University of Haripur Khyber-Pakhtunkhwa Pakistan. H. pylori was cultured in Columbia blood agar supplemented with Dent media, then incubated at 37°C for one week under microaerophilic conditions (Alkharsah et al., 2022). Salmonella enterica serovar Typhi was cultured into the Salmonella-Shigella Agar and incubated for about 24 hours at 37°C (Ruiz et al., 1996). The strain of E. coli was cultured into the MacConkey Agar and incubated for about 24 hours at 37°C (Supriatin et al., 2021).
2.2 Biochemical tests
The biochemical characterization of bacterial isolates included the catalase, oxidase, urease, indole, methyl red, and citrate tests, performed following standard protocols (MacFaddin, 2000). The tests involved the addition of reagents to observe the color shift in the medium, appearance of ring and observing the bubble formation.
2.3 Plant materials
Carica papaya leaves were collected from KTS (Khalabat township) Haripur, Khyber-Pakhtunkhwa. KTS is one of the 30 union councils of Haripur District in the Khyber Pakhtunkhwa of Pakistan with coordinates: 34°13′N 73°02′ECoordinates: 34°13′N 73°02′E
The leaves were thoroughly washed three times with distilled water to remove any impurities and then dried under shade at room temperature. After drying, the leaves were ground into a fine powder using pestle and mortar. The resulting powder was stored in a glass container for further use.
2.4 Leaf extraction
Leaf extracts were prepared following the standard protocol described by Harborne (1998). Twenty grams of dried leaves powder were soaked in 200ml of 80% methanol for one week at room temperature, with the mixture stirred daily using a stirrer. After one week, the mixture was filtered using Whatman filter No.1 filter paper. The filtered extract was then evaporated in a hot air oven for 24 hours at 37°C. The dried leaves extract was stored in sealed bottle at 4°C for further use.
2.5 Phytochemical screening test
Phytochemical screening of Carica papaya leaf extract was conducted to identify secondary metabolites (Sofowora, 1993).
2.6 Gas chromatography-mass spectrometry analysis
The chemical composition of plant extract was assessed using GC-MS on a Perkin Elmer Clarus 600 GC with 600c MS. Two fused silica capillary columns Elite 5-MS (30 m) were used. The oven temperature was initially set to 50°C for 2 minutes and then programmed to 300°C at a rate of 5°C/min. The injector temperature was set between 220 and 250°C. A 1 μl diluted sample(in chloroform) was injected with a 100:1 split ratio. Helium served as the carrier gas (2 mL/min), and analysis utilized the NIST (National Institute of Standards and Technology) library database (Nisa et al., 2020).
2.7 Antimicrobial activity
The antimicrobial activity was assessed using a modified agar well diffusion technique adapted from the standardized disk diffusion method originally developed by Heatley (1944) and subsequently refined for clinical applications (Balouiri et al., 2016). Mueller-Hinton Agar (MHA) plates were prepared according to CLSI guidelines and sterilized by autoclaving at 121°C for 15 minutes. Following solidification, the plates were inoculated with standardized bacterial suspensions (1.5 × 108 CFU/mL) of the test organisms including H. pylori, S. enterica serovar Typhi, and E. coli, using sterile swabs to create uniform bacterial lawns.
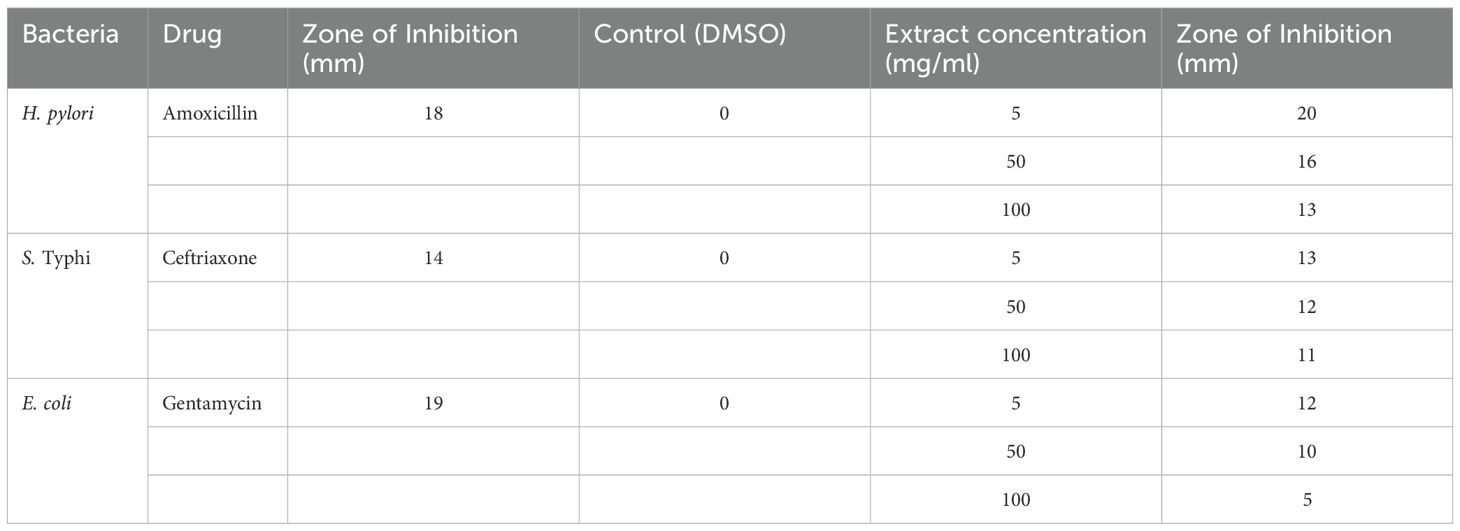
Four equidistant wells (6 mm diameter) were aseptically punched into each agar plate using a sterile cork borer (Nurkhaliza et al., 2024). To prevent lateral diffusion of test compounds while maintaining well integrity, 20 μL of molten MHA was carefully layered at the base of each well and allowed to solidify. The wells were then loaded with three concentrations of the methanolic leaf extract (5 mg/mL, 50 mg/mL, and 100 mg/mL, 100 μL per well) and a negative control (100 μL DMSO) in the fourth well. Antibiotics (amoxicillin, ceftriaxone, and gentamicin) were used as positive control against control for H. pylori, Salmonella enterica serovar Typhi and E. coli respectively. All plates were incubated at 37°C for 24 hours (microaerophilic conditions for H. pylori), after which the diameters of inhibition zones were measured to the nearest millimeter using digital calipers. Each assay was performed in triplicate to ensure reproducibility.
2.8 Determination of MIC and MBC
The minimum inhibitory concentration (MIC) of Carica papaya leaves extract was determined using microdilution method in a 96-well flat-bottom plate (Bauer et al, 1966; Kowalska-Krochmal and Dudek-Wicher, 2021). Wells from A1 to C1 served as negative controls containing 100 µl of nutrient broth. In wells A3 to C12, 100 µl of nutrient broth was dispensed, and serial dilutions of the plant extract were prepared across these wells. A volume of 100 µl of each of the three test pathogens was inoculated into wells A2 to C12. The plate was incubated at 37°C for 24 hours, after which the optical density of each well was measured using spectrophotometry at 520nm to calculate the percentage inhibition.
The minimum bactericidal concentration (MBC) was determined by plating of 20 µl of each dilution from the microdilution assay onto MHA (Mogana et al., 2020). The plates were incubated at 37°C for 24 hours. After incubation, the presence or absence of the bacterial growth was recorded by visual inspection, and zones of inhibition were measured to confirm the bactericidal effect of the extract.
2.9 Selection of ligand and protein
Phytol was selected as the ligand based on its high abundance in the GC-MS analysis of Carica papaya leaf extract. Among the 27 identified compounds, Phytol exhibited the highest peak, indicating its dominance in the extract.
For the target proteins, one representative protein was selected from each of the selected bacterial pathogens. VacA protein from H. pylori, Dihydrofolate Reductase (DHFR) from Salmonella enterica serovar Typhi, and DNA gyrase from E. coli were chosen due to their critical roles in bacterial virulence or survival.
2.10 Protein preparation
The protein structures were carefully prepared for docking simulations to ensure accurate analysis with bioactive compounds (Sastry et al., 2013). Relevant protein sequences of the selected pathogens were identified through the National Centre for Biotechnology Information (NCBI) database. These sequences were then used to retrieve the corresponding two and three-dimensional structures from the Protein Data Bank (PDB), providing reliable templates for the docking studies. Protein preparation involved the removal of non-essential chains and water molecules, followed by the addition of polar hydrogens and the application of Kolman and Gasteiger charges. The modifications were performed using AutoDock tools and Chimera to ensure the proteins were ready for molecular docking analysis (Abbas et al., 2024). This thorough preparation process ensured the structural integrity and compatibility of the proteins for downstream molecular docking simulations, facilitating precise analysis.
2.11 Ligand preparation
To prepare the ligands, PubChem was accessed to retrieve the accurate chemical structures and details of the bioactive compound derived Carica papaya leaves extract. The ligand structure was then converted into suitable format using OpenBabel, ensuring compatibility with the docking software (O’Boyle et al., 2011). Subsequently, Chimera and AutoDock tools were employed to refine both the protein and ligand structures, addressing any structural issues and optimizing the molecular orientations (Butt et al., 2020). This thorough preparation process ensured generation of high-quality inputs for docking simulations, enabling precise analysis of the interactions between the bioactive compounds and bacterial targets.
2.12 Molecular docking
Molecular docking was utilized as a pivotal in-silico technique to investigate the potential mechanisms of action of the bioactive compounds in Carica papaya leaves (Arwansyah et al., 2025). This computational approach simulated the interaction between the identified compounds and key bacterial proteins, providing valuable insights into their binding affinity and specificity. The docking simulations identified potential binding sites and interaction modes, thereby enhancing our understanding of the extract’s efficacy and guiding further experimental validation. Version 4.2.6 of AutoDock was employed to prepare the grid boxes and map files for docking analysis (Abbas et al., 2024). Grid parameter files (GPF) were generated with X, Y, and Z coordinates defining a grid size of 60 x 60 x 60, ensuring sufficient search space for ligand flexibility and rotation. Additional atomic map types, including hydrogen (H), chlorine (Cl), bromine (Br), sulphur (S), phosphorus (P), and fluorine (F), were included to construct the grid box surrounding the active site of the target protein. MGL Tools version 1.5.7 was used to generate the necessary atomic map files by processing the GPF files. Subsequently, grid log files (GLG) containing all atomic maps were created via AUTOGRID. These files served as input for docking calculations, enabling a detailed evaluation of ligand-receptor interactions. The resulting docking conformations were analyzed using Discovery Studio Visualizer, Version 3.1, where 10 docking conformations of the phytol-protein complexes were thoroughly examined to evaluate the potential interactions between the ligand and the bacterial proteins. This systemic approach provided a robust framework for understanding the bioactive potential of the compounds and offered critical insights for subsequent experimental research.
2.13 Pharmacokinetic parameters.
The phytol compound was assessed drug-likeness and pharmacokinetic properties using advanced computational tools. SwissADME (http://www.swissadme.ch) (Daina et al., 2017), was utilized to predict properties such as synthetic accessibility, absorption, and metabolism. Additionally, pkCSM was employed to evaluate key pharmacokinetic parameters, including intestinal absorption, volume of distribution (VDss), CYP metabolism, total clearance (Blundell and Ascher, 2015). Toxicity predictions were performed using ProTox-II (Banerjee et al., 2018), and StopTox (Borba et al., 2022). ProTox-II provided insights into endpoints like carcinogenicity, mutagenicity, acute toxicity, hepatotoxicity, cytotoxicity, immunotoxicity, adverse outcomes (Tox21) pathways, and toxicity targets. Its methodology integrated fragment-based propensities, molecular similarity, frequently observed features, and machine learning algorithms. StopTox applied advanced quantitative structure-activity relationship (QSAR) models to predict toxicity endpoints. StopTox specifically addressed concerns such as inhalation and oral toxicity, skin irritation, and sensitization by using extensive publicly available datasets on toxicity (Borba et al., 2022).
These computational evaluations provided a comprehensive understanding of the pharmacokinetic behavior, drug-likeness, and safety profiles of phytol, supporting its potential as a therapeutic candidate.
3 Results and discussion
3.1 Phenotypic features of selected strains
Distinct colonies of each bacterial strain were observed on specific culture media. H. pylori formed white to greyish colonies on Columbia blood agar, Salmonella enterica serovar Typhi displayed black colonies on Salmonella-Shigella agar, and E. coli produced pink colonies on MacConkey Agar, as illustrated in Figure 1. All pathogens reacted with pink color after Gram-staining confirming them as Gram-negative bacteria (Tripathi and Sapra, 2020). H. pylori appeared as small, curved or spiral-shaped cells (Krzyżek and Gościniak, 2018), Salmonella enterica serovar Typhi as single rods or occasionally in short chains (Arunava Das et al., 2012), and E. coli as single rods, with occasional pairs or short chains (Islam et al., 2016).
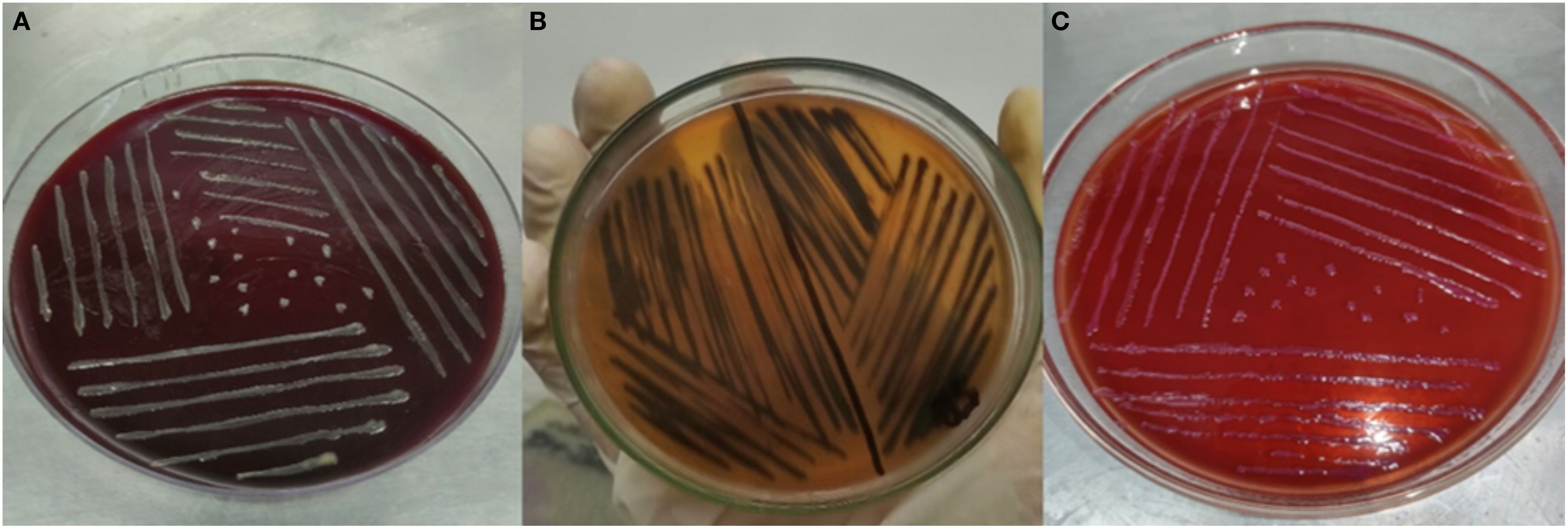
Figure 1. (A) H. pylori on Columbia blood agar. (B) Salmonella enterica serovar Typhi on Salmonella-Shigella agar. (C) E. coli on MacConkey Agar.
3.2 Biochemical testing
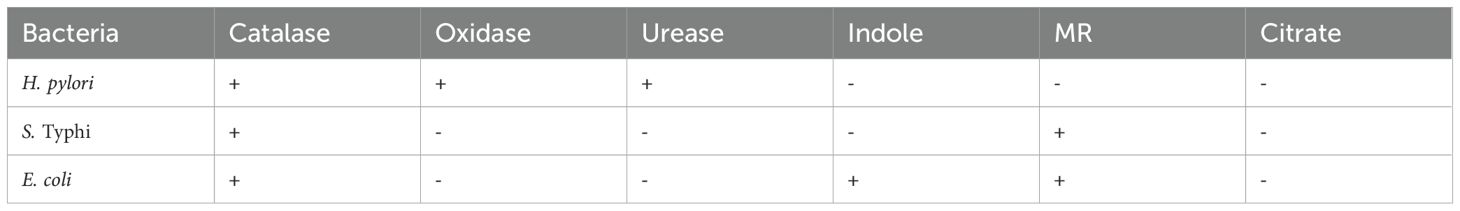
The biochemical test results reveal distinct metabolic characteristics of H. pylori, S. Typhi, and E. coli showcasing their adaptations to specific ecological niches and metabolic pathways (Table 1). The catalase positive results for all three bacteria confirm their ability to degrade hydrogen peroxide, a mechanism essential for combating oxidative stress, particularly in environments exposed to reactive oxygen species. H. pylori, for instance, encounters high levels of oxidative stress in the stomach’s acidic environment, and catalase activity is critical for its survival (Benoit and Maier, 2016). Similarly, S. Typhi, and E. coli, as gut-associated organisms, require catalase for defense against oxidative bursts from host immune response (Supplementary Figure S1) (Chanin et al., 2020; Hahn et al., 2021).
E. coli is oxidase-negative, characteristic of the Enterobacteriaceae family, which relies on facultative anaerobic and can switch between aerobic respiration and fermentation depending on oxygen availability (Zeng et al., 2017). Urease positive activity of H. pylori shows its ability to survive in the acidic gastric environment by neutralizing acid with ammonia, which it produces from urea (Berger, 2000; Kuo et al., 2015). This adaptation is crucial for colonization and pathogenesis in stomach, a unique feature not shared by S. Typhi, and E. coli (Ansari and Yamaoka, 2017; Cheok et al., 2021). The inability of all three bacteria to utilize citrate as the sole carbon source is notable. While some E. coli strains exhibit citrate utilization under aerobic conditions, this variability shows the influence of specific environmental factors on metabolic capabilities. Among the tested strains only E. coli showed the ability to produced indole which was confirmed by the formation of purple ring (Supplementary Figure S1C).
3.3 Phytochemical profile
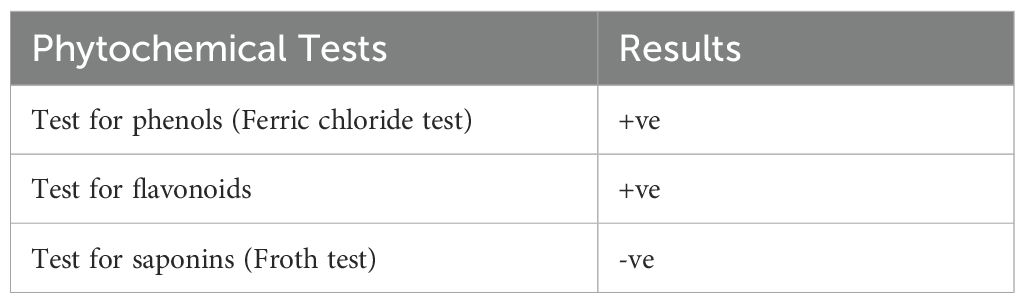
Phytochemical profile of Carica papaya leaves revealed presence of different bioactive compounds, including phenols and flavonoids, while saponins were absent (Table 2). Phenols are known for their antimicrobial, antioxidant, and anti-inflammatory properties, which could contribute to the observed biological activity of the extract (Kauffmann and Castro, 2023). Their presence is indicated by a characteristic brownish-black color reaction with ferric chloride (Supplementary Figure S2). Flavonoids, a class of secondary metabolites, possess well-documented biological activities, including, antimicrobial, antioxidant, and anti-inflammatory effects (Zandavar and Babazad, 2023). The color change observed upon the addition of NaOH solution signifies their presence. Studies have indicated that flavonoids can disrupt bacterial cell walls, inhibit enzymes, or interfere with quorum-sensing mechanisms in bacteria (Nguyen et al., 2024). These findings align with previous studies, such as Prasad et al. (2021), which reported similar results, highlighting the therapeutic potential of the extract.
Phytochemical profile of Carica papaya leaves extract showed that flavonoids and phenols are present in the extract, while saponins are absent, as shown in Table 2.
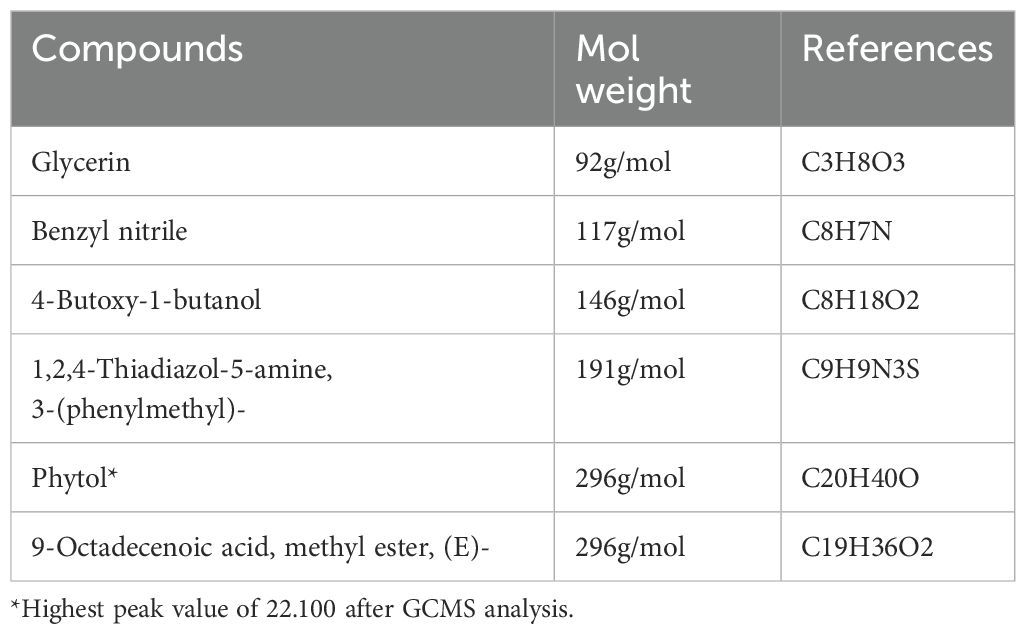
The GC-MS analysis of Carica papaya leaves extract, depicted in the chromatogram (Figure 2), reveals distinct peaks at specific retention times, highlighting the diverse phytochemical composition of the extract, with phytol dominating as the highest peak at a retention time of 18.712 minutes (Figure 2). The analysis identified Phytol, Benzene-(isothiocyanatomethyl), and 9-Octadecenoic acid, methyl ester (E) as the principal bioactive compounds, all known for their antibacterial properties (Al-Seadi et al., 2021). Phytol which was prominent in the identified compounds in the extract is known to be essential in plant defense processes and therefore is a good candidate for possible therapeutic activity (Ghaneian et al., 2015).
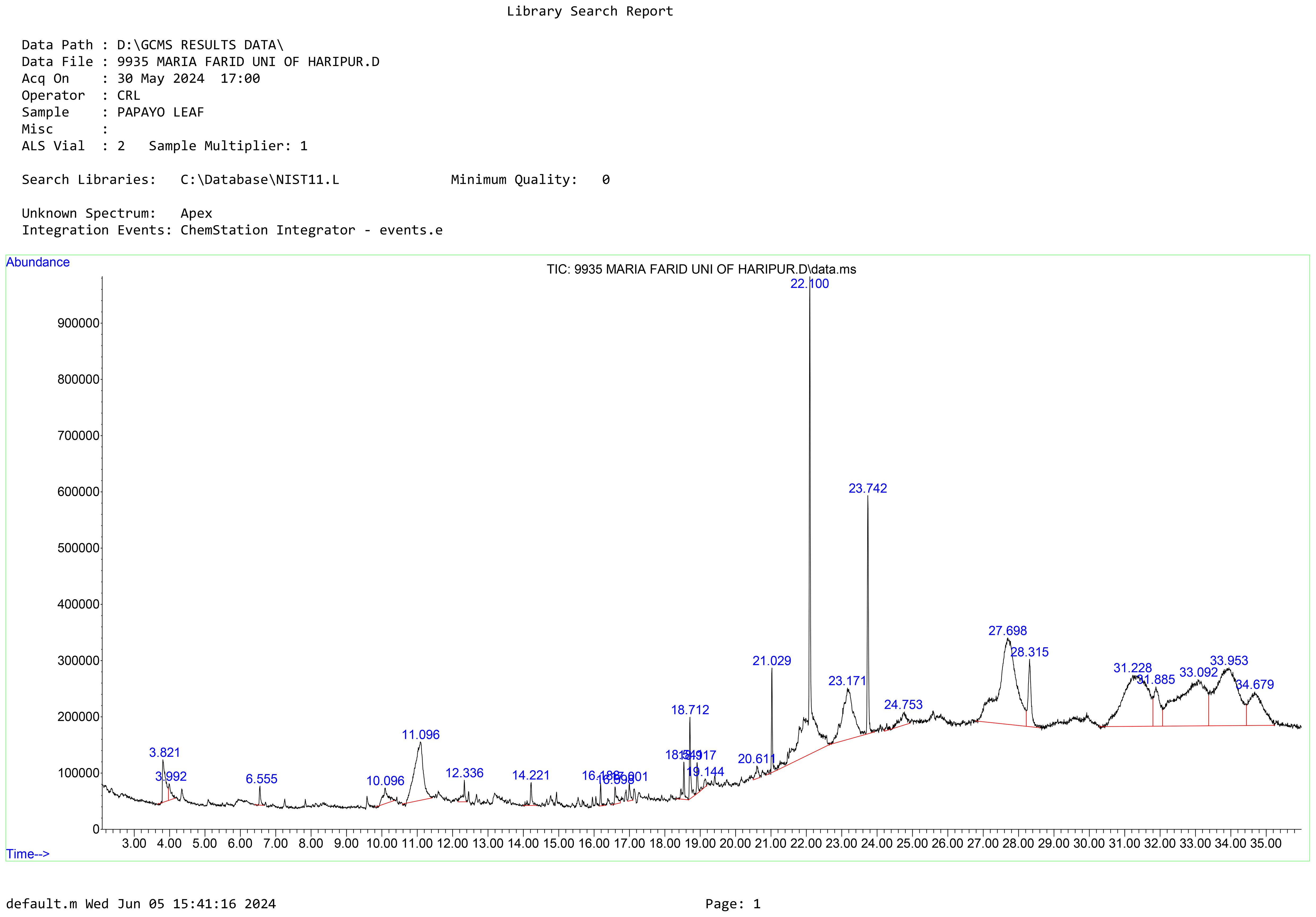
Figure 2. The chromatogram shows peak values of the compounds identified from Carica papaya leaves extract. Peak 22.100 represents phytol which is a compound commonly found in papaya leaves.
Based on these findings, Phytol was selected for further testing to evaluate its antibacterial activity against the three selected bacterial strains. Phytol is a potent bioactive compound known for its antioxidant, antimicrobial, and anti-inflammatory properties. Phytol has also been linked to potential anticancer activity, further emphasizing its pharmacological significance. Other peaks observed in the chromatogram, corresponding to different retention times, likely represent additional phytochemicals such as flavonoids, phenols, saponins, and alkaloids (Table 3). These secondary metabolites are known to contribute Carica papaya’s therapeutic efficacy. For instance, phenols and flavonoids possess string antioxidant properties, which protect against oxidative stress and have been widely documented for their role in disease prevention (Babalola et al., 2024). Moreover, studies by Achukwu et al. (2024) have shown that Carica papaya leaves are rich in phytochemicals that support antimicrobial activity against a range of pathogenic bacteria and fungi. These compounds likely work synergistically, enhancing the plant’s medicinal potential.
3.4 Antibacterial activity
The antibiotics used in the study were selected based on their common use in treating selected pathogens in clinical settings of Pakistan. Amoxicillin is a widely prescribed antibiotic for treating H. pylori, particularly in combination therapies for eradicating H. pylori in peptic ulcers and gastritis (Roberts et al., 2022; Vu et al., 2022). Similarly, ceftriaxone, a third-generation cephalosporin is highly effective against Salmonella enterica serovar Typhi (Argimón et al., 2022). Gentamycin, an aminoglycoside, is effective against a wide range of Gram-negative, including E. coli (Karunarathna et al., 2024). The selection of these three antibiotics as positive control was based on their susceptibility against the selected pathogens and their use in clinical practice, allowing for a meaningful comparison with the antimicrobial efficacy of Carica papaya leaves extract. Amoxicillin was very effective against H. pylori as a zone of inhibition of 18 mm was observed. Ceftriaxone and gentamycin were moderately effective against S. Typhi 14 mm, and 19 mm for E. coli respectively. These findings align with research demonstrating the potential of C. papaya against E. coli (Rubaka et al., 2014). This suggest that E. coli might have inherent resistance to gentamycin at low concentrations but could be overcome at higher doses (Gauba and Rahman, 2023). The efficacy of Carica papaya leaves extract has shown great efficacy against H. pylori, Salmonella enterica serovar Typhi and E. coli. The leaf extract exhibited highest activity against H. pylori, with a 20 mm zone of inhibition at 5 mg/ml, which aligns with previous studies highlighting the effectiveness of natural plant extracts against H. pylori due to their bioactive components. For example, Prasad et al. (2021) reported similar findings where papaya leaf extract exhibited potent inhibitory effects against H. pylori, underscoring the plant’s traditional use in treating gastrointestinal infections. This consistency suggests that Carica papaya could be an effective natural alternative for treating H. pylori-related conditions, such as peptic ulcers and gastritis.
However, as the concentration increased, the zone decreased to 13 mm at 100 mg/ml. Similar trends were observed for S. enterica and E. coli, where the zone consistently decreased with increasing extract concentration. Notably, E. coli demonstrated the least susceptibility overall, with the inhibition dropping from 12 mm at 5 mg/ml to 5 mm at 100 mg/ml.
These findings are consistent with previous studies that report antibacterial activity of Carica papaya leaves extract, attributed to phytochemical like alkaloids, flavonoids, and tannins, which may disrupt bacterial cell walls or interfere with metabolic pathways (Lee et al., 2016; Udinyiwe and Omoregie, 2024). However, the observed decrease in efficacy with higher concentrations contrasts with some studies, which suggest a dose-dependent increase in activity, indicating possible differences in extract composition, bacterial susceptibility, or assay conditions. The phenomenon observed, where higher concentrations reduce effectiveness, might be attributed to the “paradoxical effect”, where bioactive compounds may interact more efficiently with bacterial cells at lower concentrations, but at higher levels, they could accumulate and hinder diffusion, reducing efficacy. Additionally, plant extracts may stress the bacteria, triggering defense mechanisms like efflux pumps that expels toxins from the cell, thus reducing the extract’s effectiveness, especially in Gram-negative bacteria, which are known for their robust defense mechanisms (Askarinia et al., 2019; Jubair et al., 2021). Plant extracts often contain large, hydrophobic or poorly water-soluble molecules such as fatty acids or terpenoids (e.g., phytol), which may not effectively diffuse through the agar matrix. This leads to smaller or absent zones of inhibition despite significant antibacterial activity observed in broth dilution assays (CLSI, 2012; Rios et al., 1988). At elevated concentrations, certain phytochemicals present in Carica papaya leaf extract may aggregate, reducing their bioavailability and, consequently, their antimicrobial activity. This aggregation can hinder the diffusion of active compounds into the agar medium, leading to smaller zones of inhibition. Some compounds may exhibit potent activity in liquid broth but may not diffuse well in agar, resulting in underestimation of their activity by zone of inhibition alone (Balouiri et al., 2016).
The antimicrobial activity of Carica papaya leaves extract against all three pathogens is shown in Table 4.
3.5 Antimicrobial activity of Carica papaya leaves extract: MIC and MBC evaluation
Determination of MIC and MBC of H. pylori, S. Typhi, and E. coli in Figure 3 respectively. H. pylori show the highest MIC and MBC values (0.4-0.983), suggesting it is the least sensitive to the extract, while E. coli exhibits the lowest values (0.282-1), indicating higher susceptibility, S. Typhi displayed intermediate sensitivity with MIC ranging from 0.45-0.75 and MBC from 0.5-0.9. The clustering shows that MIC and MBC patterns for H. pylori are more similar, while S. Typhi and E. coli exhibit varied responses. Overall, the results suggest that Carica papaya leaves extract has differential antimicrobial activity.
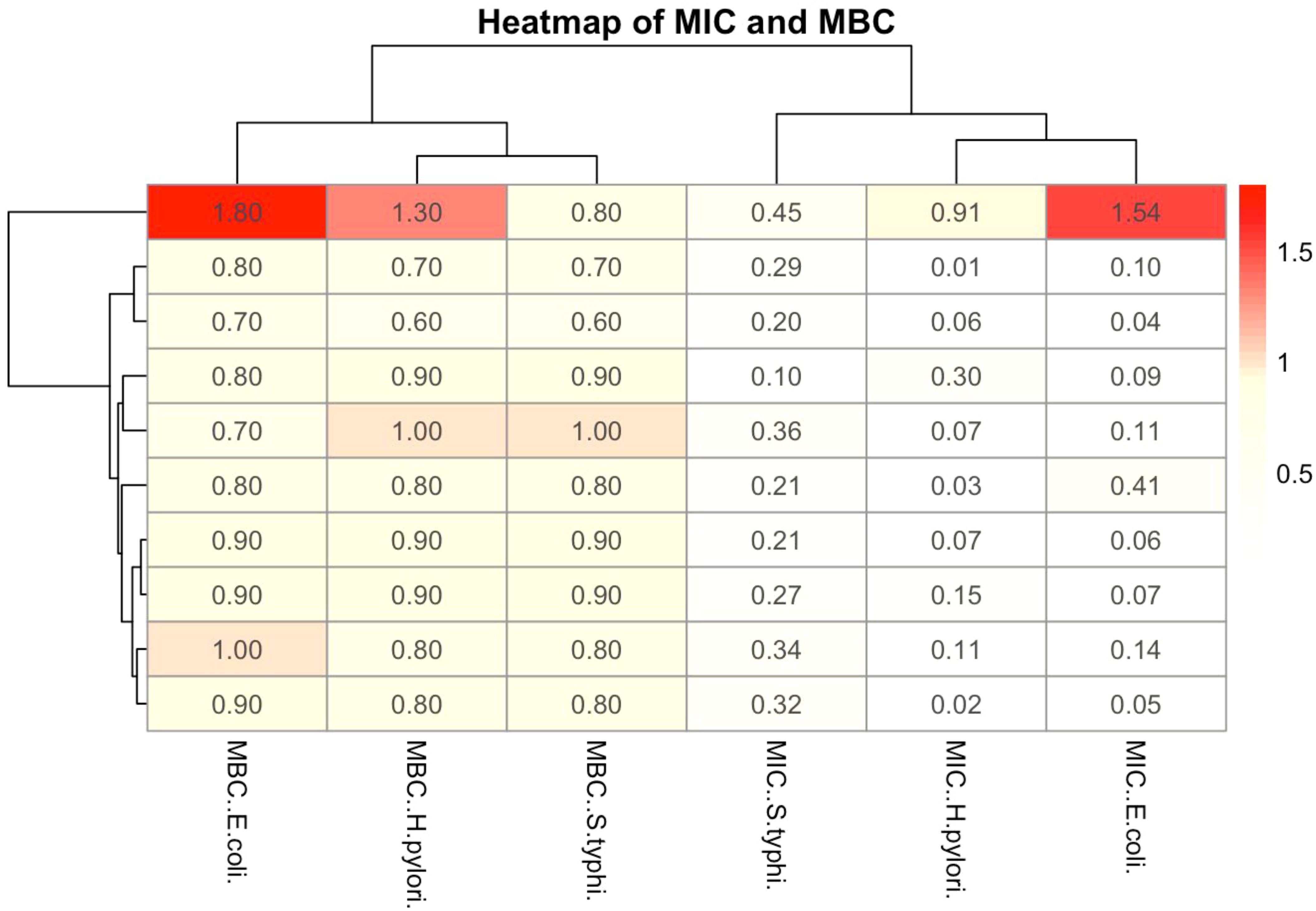
Figure 3. The heatmap illustrates the MIC and MBC of Carica papaya leaves extract against the selected pathogens. The color gradient represents the concentration values, with red indicating higher values and blue indicating lower values.
3.6 Efficacy assessment of Carica papaya extract
The variability in the MIC and MBC values of the leaf extract against the tested strains is illustrated in Figure 4. The MIC and MBC values vary significantly between bacterial strains, with S. Typhi exhibiting the lowest MIC values, suggesting a higher sensitivity to the extract. Conversely, H. pylori and E. coli exhibit slightly higher MIC and MBC values, indicating relatively reduced susceptibility. Notably outliers reflect the variability in response among the bacterial isolates, which may be attributed to intrinsic resistance mechanisms.
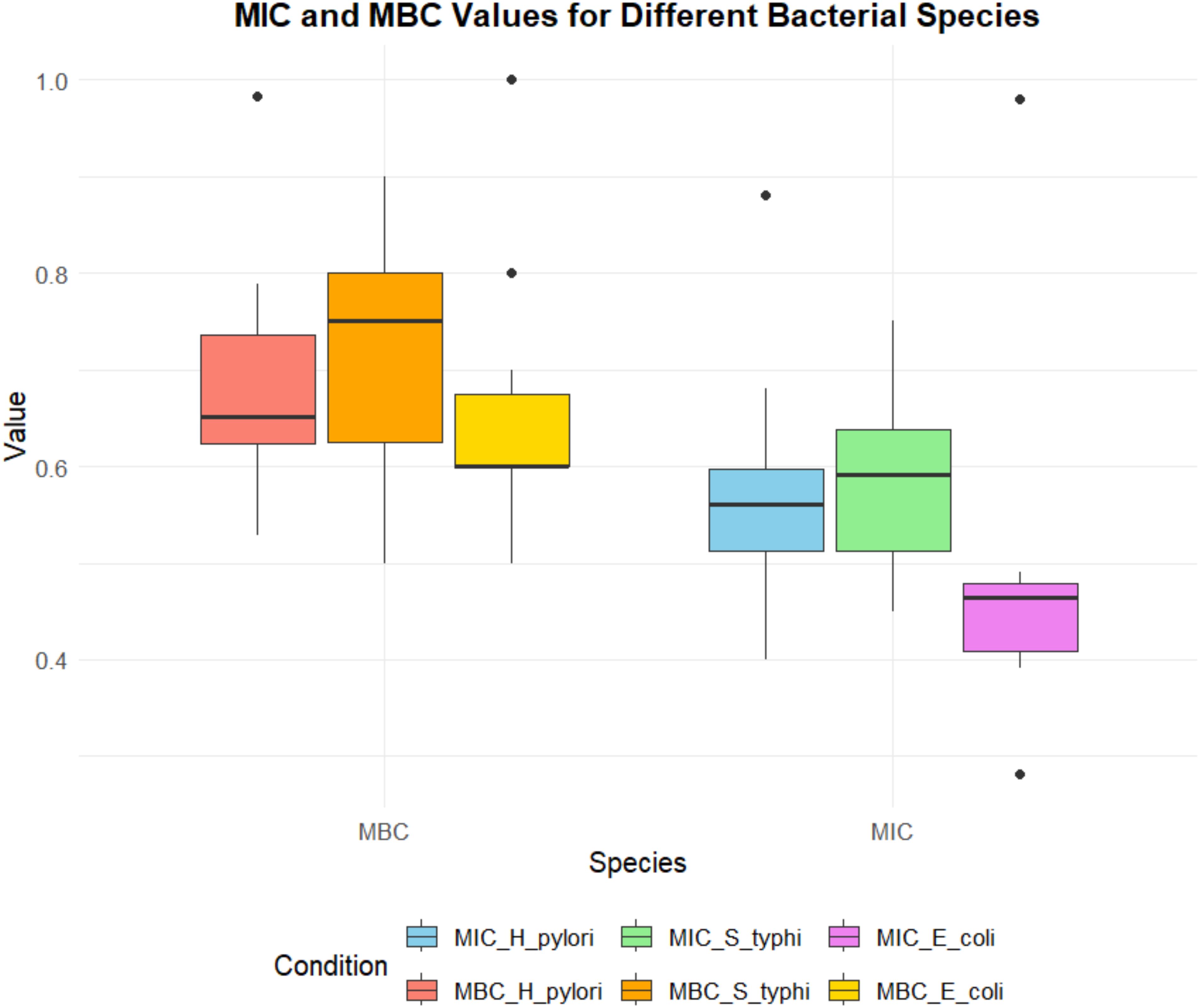
Figure 4. Comparison of MIC (minimum inhibitory concentration) and MBC (minimum bactericidal concentration) values of phytol for different bacterial species, including H. pylori, S. typhi, and E coli. The MIC values are represented in shades of blue, green, and purple, while the MBC values are depicted in shades of red, orange, and yellow. The plot highlights variability across species and conditions, with outliers indicating potential deviations in susceptibility or bactericidal response.
Yahaya et al. (2017) reported in study that the ethanolic leaf extract of Carica papaya exhibited increasing zones of inhibition against E. coli, Shigella spp., P. aeruginosa, and S. Typhi with increasing concentrations up to 200 mg/ml. However, beyond this concentration, a plateau or reduction in activity was noted, suggesting a potential paradoxical effect at higher concentrations. Romasi et al. (2011) research reported that the antibacterial activity of Carica papaya leaf extracts was influenced by pH, with optimal activity observed at acidic pH levels. This indicates that factors such as pH can significantly affect the efficacy of the extract. Evaluation of Antimicrobial Susceptibility of Salmonella Isolated from Household Cockroaches Using Carica Papaya Leaf Extract: The study found that ethanol extracts inhibited Salmonella at lower concentrations (12.5 g and 25 g) but lost effectiveness at higher concentrations (37.5 g and 50 g), indicating ethanol enhances Carica papaya’s antimicrobial activity at specific levels (Abutu and Gbaraba, 2021).
3.7 Protein preparation
The 3D structure of Phytol downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) is shown in Figure 5, and the three proteins were prepared by using Auto Dock tools for docking.
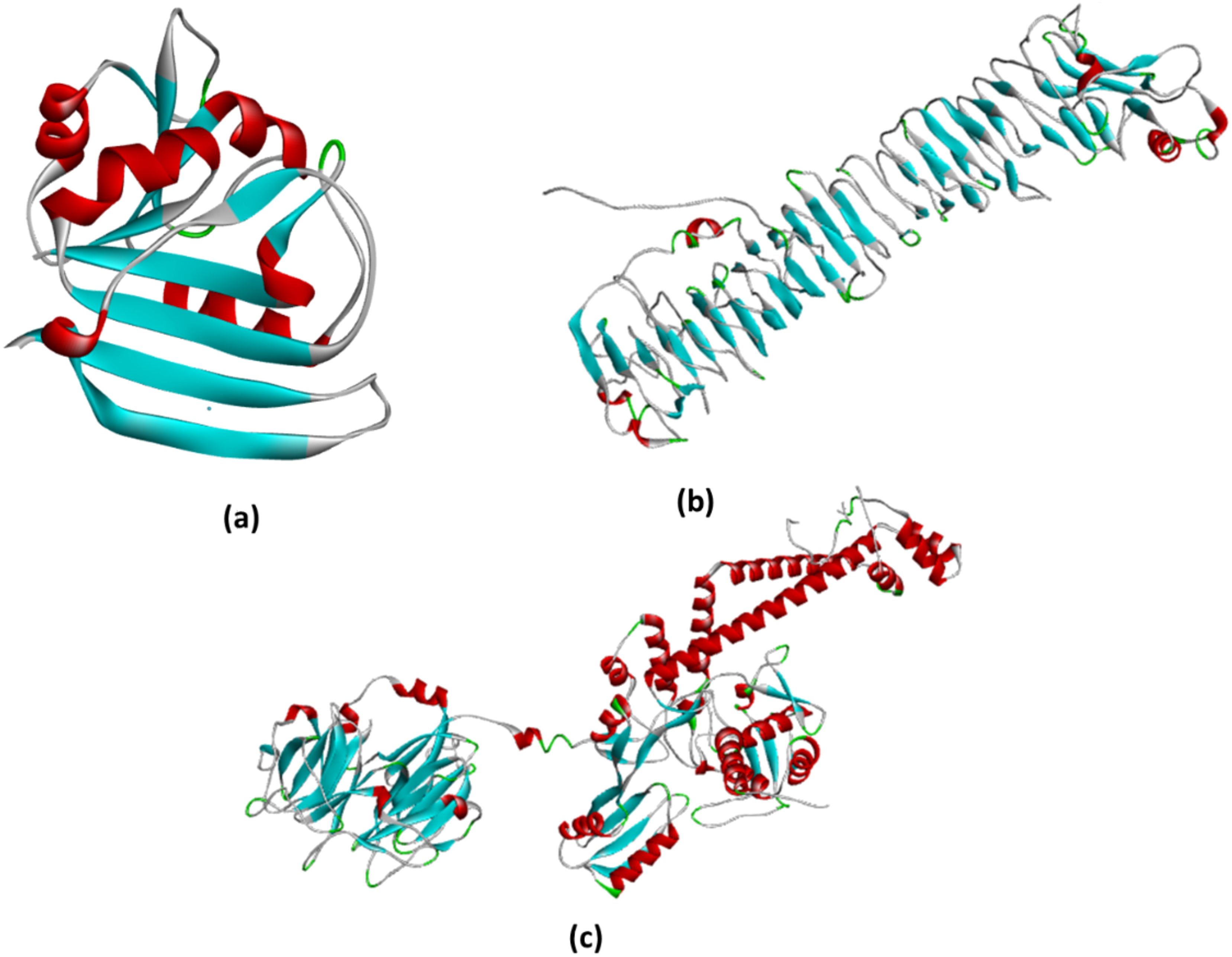
Figure 5. (a) The structure represents the prepared DHFR protein ready to bind with the ligand. All the water molecules were removed, while polar hydrogens and charges were added by using AutoDock tools; (b) The prepared chain from VacA ready to bind with the ligand. All chains except chain A were removed, water molecules removed, while polar hydrogens and charges were added by using AutoDock tools; (c) The DNA gyrase protein was prepared for docking by removing all water molecules and chains, except for chain (a) additionally, polar hydrogens were added, and charges were assigned using AutoDock tools to optimize the structure for docking analysis.
3.8 Molecular docking
Protein-ligand interactions are influenced by various bonded and non-bonded interactions, including hydrogen bonds, electrostatic interactions, and van der Waals forces, all of which contribute to a stable protein-ligand complex. To analyze and compare the binding patterns of docked ligand within target proteins, the ligands with the highest docking scores were extracted and graphically visualized.
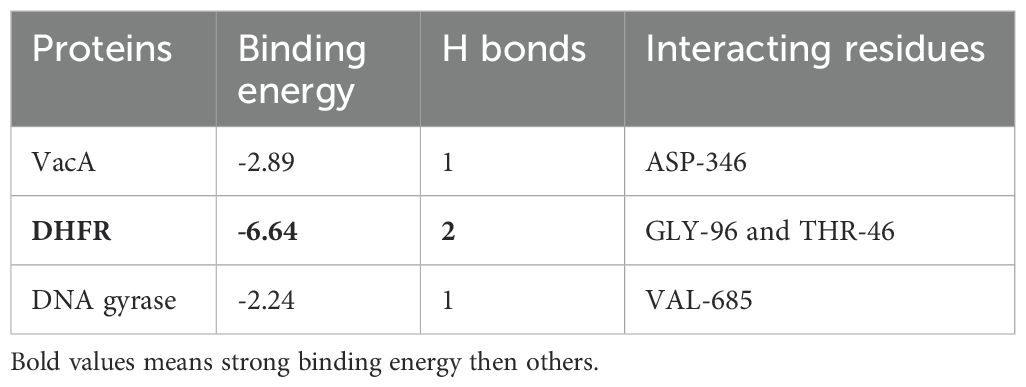
Figure 6 illustrates the optimally docked complexes of target proteins from tested bacterial strains with the phytol. The interaction of phytol with VacA identified ASP-346 as a key interacting residue and with a binding distance of 3.5 Å. For DHFR, two interactions were observed involving the residues GLY-96 and THR-46, with a binding distance of 3.1 Å. Similarly, the interaction of Phytol against DNA gyrase highlighted VAL-685 as the interacting residues.
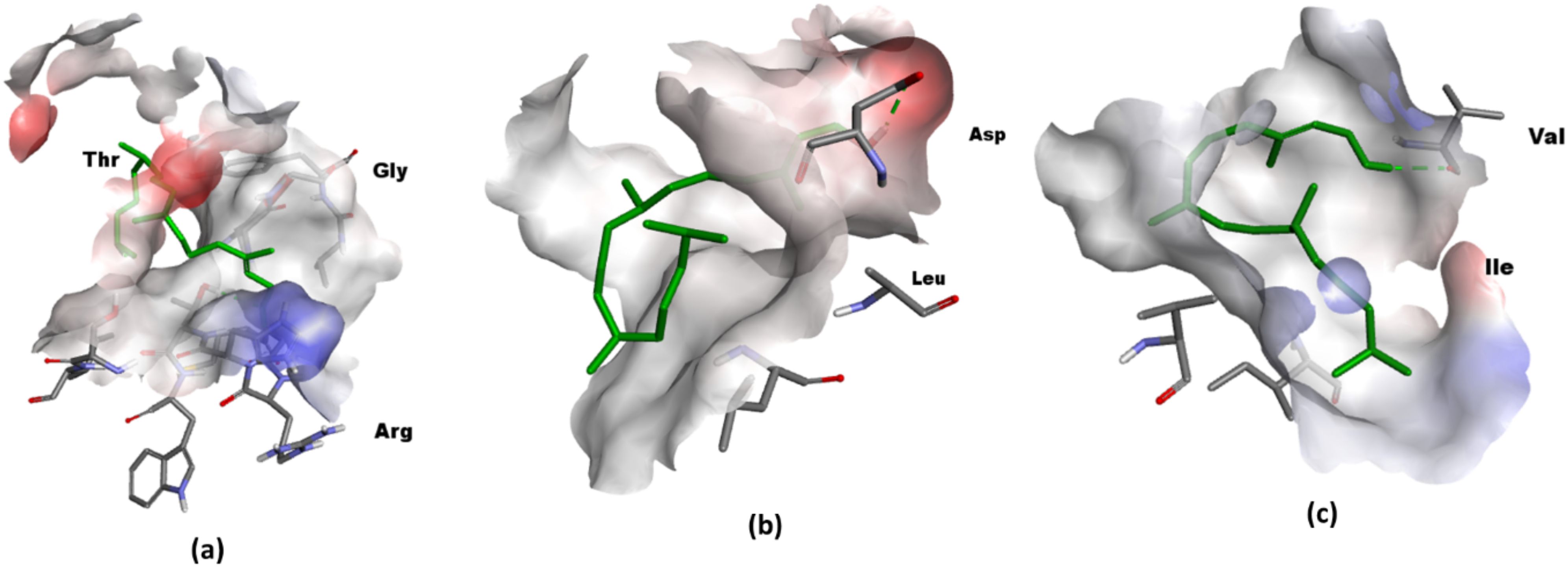
Figure 6. 3D visualization in Discovery Studio (a) Phytol targeting DHFR. The prepared structure of DHFR shows Phytol binding to the protein, with two key interactions identified between the ligand and the residues GLY-96 and THR-46, at a distance of 3.1. (PDB ID: 1rx2); (b) 3D visualization Phytol targeting VacA. The interaction between the protein and ligand highlights ASP-346 as a interacting residue, with a measured distance 3.5 Å (PDB ID: 6nyl) (c) DNA gyrase with the interacting residue VAL-685 ((PDB ID: 6rkw).
Binding energy analyses revealed that phytol exhibited binding energies of -2.89 kcal/mol against VacA, -6.64 kcal/mol against DHFR, and -2.24 kcal/mol against DNA gyrase, as summarized in Table 5. These results underscore phytol’s antibacterial potential, particularly its strong binding affinity to dihydrofolate reductase (DHFR) from Salmonella enterica serovar Typhi (-6.64 kcal/mol). This finding aligns with Asad et al. (2021), who reported phytol’s inhibitory effect on DHFR in similar bacterial strains. In contrast, Phytol’s lower binding energies against VacA from H. pylori (-2.89 kcal/mol) and DNA Gyrase (GyrA) from E. coli (-2.24 kcal/mol) suggest that its efficacy may be more pathogen-specific. This observation is consistent with the study by Sharma et al. (2022), who reported variable binding affinities of phytol across different bacterial targets, indicating potential limitations in its broad-spectrum effectiveness.
3.9 Pharmacokinetic parameters
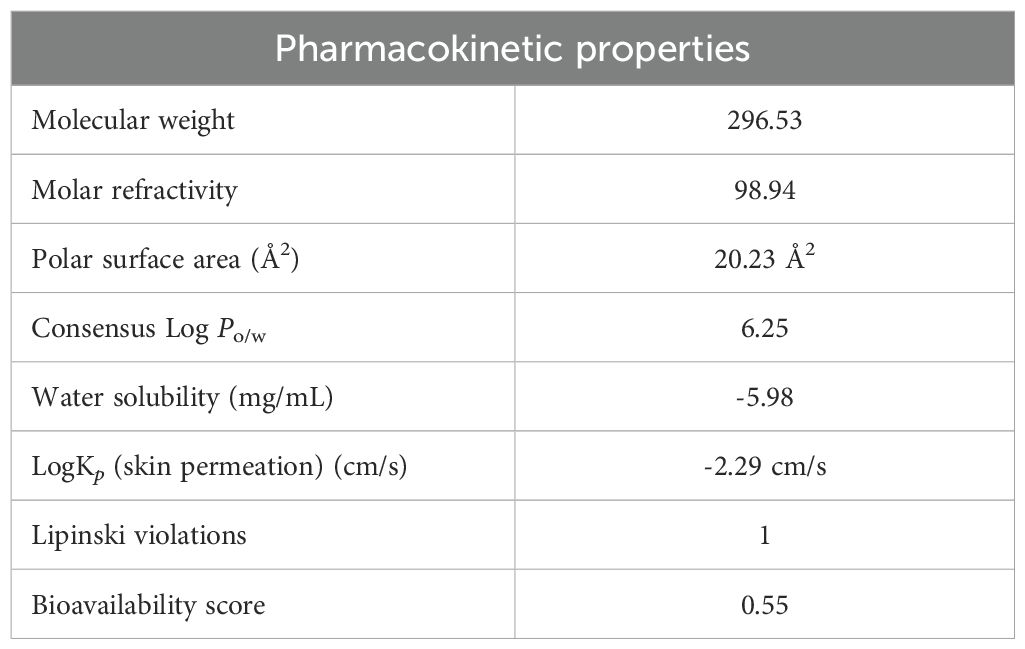
Phytol’s pharmacokinetics properties, evaluated through in-silico ADME tools, pkCSM, Stoptox, Pro-Tox II, underscore its potential as a drug candidate. With a molecular weight of 296.53, it falls within the range suitable for drug-likeness and its polar surface area of 20.23 Å2 supports good membrane permeability and absorption (Almeida-Bezerra et al., 2024). Phytol’s high lipophilicity (Log Po/w 6.25) enhances its ability to penetrate lipid membrane, while a single Lipinski rule violation suggests acceptable drug-likeness (Table 6).
The radar chart (Figure 7) illustrates the physiochemical attribute of phytol, emphasizing its high lipophilicity, moderate molecular size, low polarity, and significant molecular flexibility, which collectively enhance its hydrophobic interactions and adaptability to diverse molecular targets. Additional evaluations indicate moderate intestinal absorption (90.64%), low skin permeability (-2.631) and minimal blood-brain barrier penetration (0.793). As a substrate for CYP3A4 and an inhibitor of CYP1A2, phytol engages in specific metabolic pathways. Furthermore, its non-hepatotoxic, non-mutagenic and is non-carcinogenic profile reinforce its therapeutic potential while warranting further exploration for drug development (Supplementary Tables S1–S3).
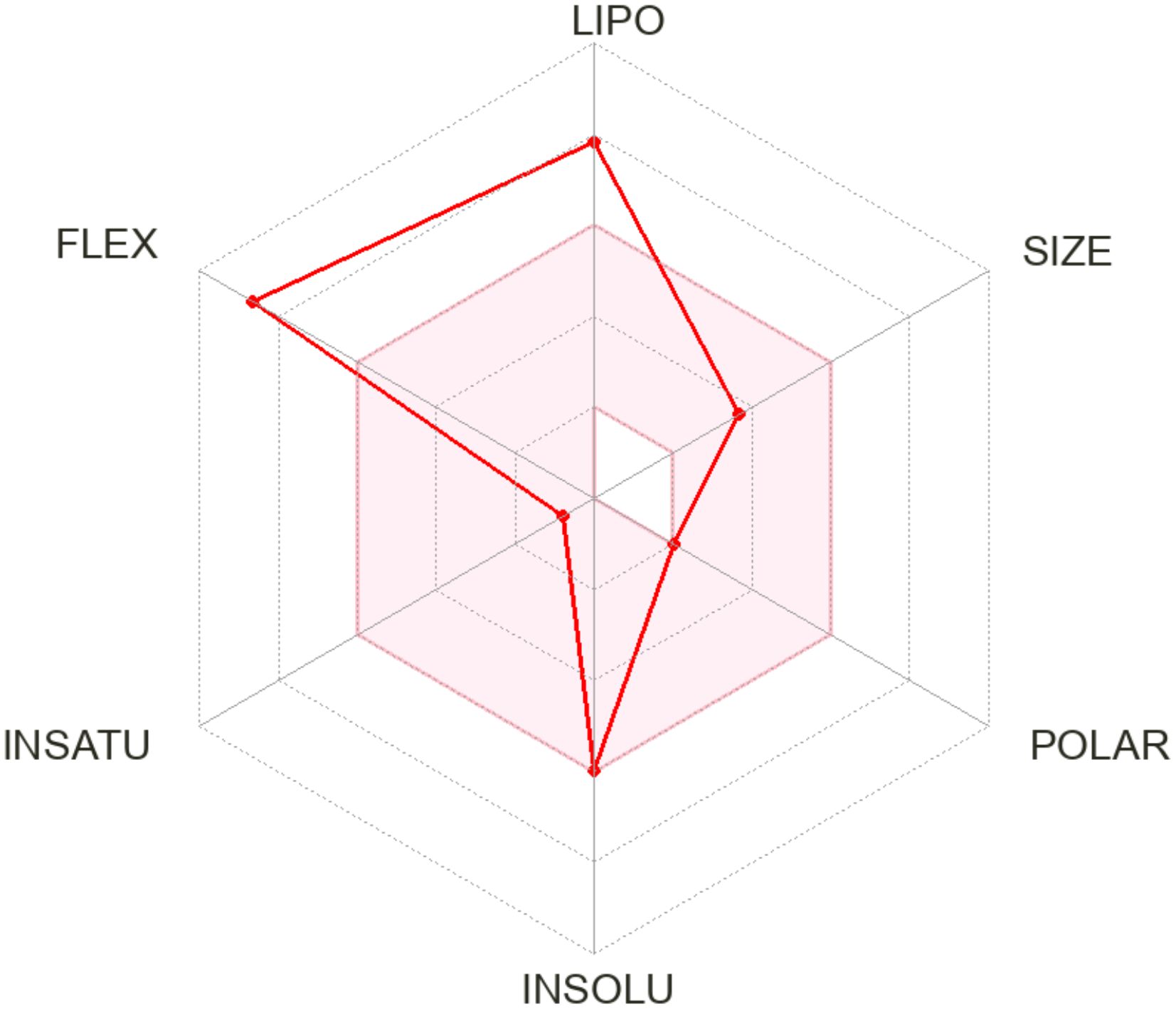
Figure 7. The radar chart illustrates the physiochemical properties of the phytol across six parameters: lipophilicity (LIPO), size (SIZE), polarity (POLAR), water insolubility (INSOLU), degree of unsaturation (UNSATU), and molecular flexibility (FLEX). Each axis represents a distinct property, with higher values indicating greater intensity.
3.10 Protein-protein interaction network analysis
The Figure 8 illustrates three distinct protein-protein interaction (PPI) networks that reveal critical pathway disruptions in bacterial pathogens. In H. pylori, the network centers on the virulence factor VacA, which interacts with UreB (urease subunit) and HpaA (involved in nickel cation binding), demonstrating coordinated roles in host cell damage, acid neutralization. Nickel acts as a virulence determinant in H. pylori by serving as an essential cofactor for urease, an enzyme critical for gastric colonization through acid resistance. Notably, studies have shown that H. pylori accumulate nickel at concentrations 50 times higher than those found in E. coli (Manente et al., 2008; Shah et al., 2022). Therefore, a supply of nickel is crucial for its survival in the stomach. The interactions imply synergy—UreB may enhance VacA’s cytotoxic effects by altering local pH, while HpaA could localize VacA to host cell surfaces for targeted toxin delivery (Vitoriano et al., 2011). Together, these interactions likely disrupt host pathways (e.g., mitochondrial apoptosis, immune signaling) to sustain infection, offering potential targets for anti-H. pylori therapies (Shah et al., 2022).
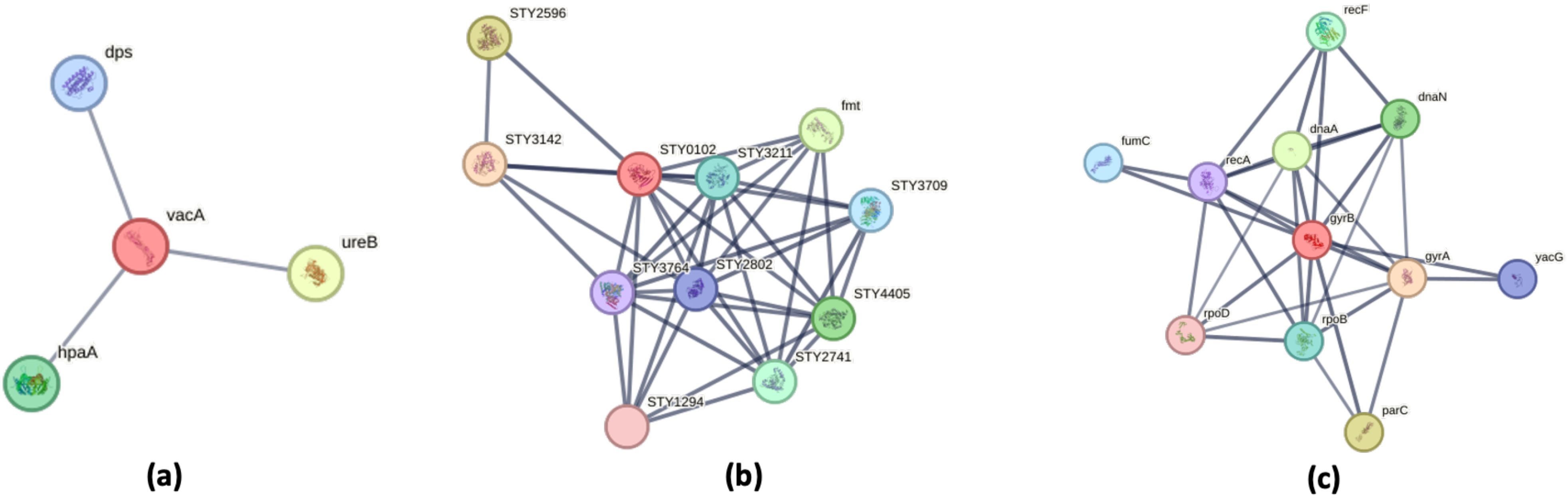
Figure 8. Protein–Protein Interaction Networks of Bacterial Virulence and Adaptation Factors. Protein–protein interaction (PPI) networks were generated using the STRING database, with PPI enrichment p-value: 2.04e-07. Proteins involved in localization are highlighted in red. (a) Helicobacter pylori: The PPI network is centered around the vacuolating cytotoxin VacA, a key virulence factor involved in host cell interaction. (b) Salmonella Typhi: The network centers on dihydrofolate reductase (DHFR, STY0102), a pivotal metabolic enzyme that acts as a hub connecting multiple biosynthetic pathways. (c) Escherichia coli: The DNA gyrase subunit B (GyrB) occupies a central position in the interaction network, underscoring its role as a master regulator of genomic processes including DNA replication, repair, and transcription.
In S. Typhi, dihydrofolate reductase (DHFR, STY0102) serves as a metabolic nexus (He et al., 2020), connecting folate metabolism to nucleotide synthesis through interactions with thymidylate synthase (STY3142) and folylpolyglutamate synthase (STY2596) (Joshi et al., 2022; Shen and Downs, 2024). This network not only supports bacterial proliferation but also reveals compensatory pathways that could be co-targeted with DHFR inhibitors like trimethoprim to overcome resistance.
The protein-protein interaction (PPI) network of DNA gyrase subunit B (GyrB) in E. coli reveals its central role as a master regulator of genomic processes (Menon and Piramanayakam, 2021). As the catalytic core of DNA gyrase, GyrB partners with GyrA to control DNA supercoiling (Nöllmann et al., 2007). The network demonstrates GyrB’s multifaceted interactions with key cellular components, it coordinates with DnaA to initiate replication and DnaN to maintain processivity (Katayama et al., 2017), interfaces with RpoB/RpoD to regulate transcription, and connects to RecF/RecA for DNA repair. Targeting GyrB alongside topoisomerase IV (ParC) could amplify the effect of quinolone antibiotics by simultaneously disrupting chromosome topology and associated pathways (Bansal and Tandon, 2011; Gurram and Azam, 2021).
Collectively, these networks expose vulnerable points in bacterial physiology—virulence factor coordination (H. pylori), metabolic resilience (S. Typhi), and genome-metabolism crosstalk (E. coli)—providing a roadmap for multitarget therapies to disrupt pathogenesis and combat antibiotic resistance.
4 Conclusion
Phytol demonstrated significant potential as a bioactive compound with antibacterial activities. Our findings suggest that phytol effectively inhibits bacterial growth, bactericidal effect, highlighting its potential as both an inhibitors and bactericidal agent. The variation in MIC and MBC values across different species also suggest that phytol’s antibacterial efficacy may be more pronounced against certain pathogens. The pharmacokinetic profile, as assessed through various bioinformatics tools shows a favorable molecular weight and molar refractivity, positioning it as a viable candidate for drug development. The molecular docking analysis exhibits varying binding affinities against the target proteins from different bacteria, with notable implications for its antibacterial activity. The strongest binding energy was observed with Dihydrofolate Reductase (DHFR) from Salmonella enterica serovar Typhi, suggesting a robust interaction that may enhance its potential as an antibacterial agent against this pathogen. These findings support the hypothesis that Phytol could serve as a promising lead compound for further investigation into its antibacterial properties, particularly against Salmonella enterica serovar Typhi.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
FSA: Project Administration, Writing – original draft, Writing – review & editing, Formal analysis. MF: Investigation, Methodology, Software, Writing – original draft. MS: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. SN: Investigation, Methodology, Software, Writing – review & editing. SS: Writing – review & editing. SA: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The Researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University, Buraydah, Saudi Arabia for financial support (QU-APC-2025). Authors would also like to acknowledge the Department of Microbiology, The University of Haripur, Pakistan for providing research facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1564787/full#supplementary-material
References
Abbas, S., Yasmin, A., Maqbool, N., Shah, A. A., and Fariq, A. (2023). Insights into the microbiological and virulence characteristics of bacteria in orthopedic implant infections: A study from Pakistan. PloS One 18, e0292956. doi: 10.1371/journal.pone.0292956
Abbas, S., Yasmin, A., Mujawar, S., Bibi, M., Kazmi, A., and Rehman, S. U. (2024). Identification of the plausible drug target via network/genome analysis and its molecular interaction studies against multidrug resistance bacterial pathogens. Cell. Microbiol. 2024, 6635476. doi: 10.1155/2024/6635476
Abutu, A. O. and Gbaraba, P. V. (2021). Evaluation of antimicrobial susceptibility of Salmonella isolated from household cockroaches using Carica papaya leaf extract. Int. J. Res. Sci. Innovation (IJRSI) 8, 23–28.
Achukwu, N. O., Aniobi, C. C., Isiofia, P. O., Ajare, C. C., and Agu, A. N. (2024). Phytochemical constituents and antimicrobial activity of Carica papaya leaf extract on some clinical isolates. Sokoto J. Med. Lab. Sci. 9, 333–339. doi: 10.4314/sokjmls.v9i2.39
Ahmed, S. K., Hussein, S., Qurbani, K., Ibrahim, R. H., Fareeq, A., Mahmood, K. A., et al. (2024). Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health. 2, 100081. doi: 10.1016/j.glmedi.2024.100081
Alara, O. R., Abdurahman, N. H., and Alara, J. A. (2022). Carica papaya: comprehensive overview of the nutritional values, phytochemicals and pharmacological activities. Adv. traditional Med. 22, 17–47.
Alkharsah, K. R., Aljindan, R. Y., Alamri, A. M., Alomar, A. I., and Al-Quorain, A. A. (2022). Molecular characterization of Helicobacter pylori clinical isolates from Eastern Saudi Arabia. Saudi Med. J. 43, 1128. doi: 10.15537/smj.2022.43.10.20220355
Almeida-Bezerra, J. W., Menezes, S. A., Silva, J. T. D. C., de Sousa, S. G., Alves, D. S., Alencar, G. G., et al. (2024). Analysis of the antibiotic-potentiating activity, absorption, distribution, metabolism, and excretion (ADME) and the molecular docking properties of phytol against multi-drug-resistant (MDR) strains. Antibiotics 13, 1171. doi: 10.3390/antibiotics13121171
Al-Seadi, H. L., Sabti, M. Z., and Taain, D. A. (2021). “GC-MS analysis of papaya leaf extract (Carica papaya L.),” in IOP Conference Series: Earth and Environmental Science (IOP Publishing), vol. 910 (1), 012011.
Ansari, S. and Yamaoka, Y. (2017). Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 22, p.e12386. doi: 10.1111/hel.2017.22.issue-4
Argimón, S., Nagaraj, G., Shamanna, V., Sravani, D., Vasanth, A. K., Prasanna, A., et al. (2022). Circulation of third-generation cephalosporin resistant Salmonella Typhi in Mumbai, India. Clin. Infect. Dis. 74, 2234–2237. doi: 10.1093/cid/ciab897
Argueta, E. A., Ho, J. J. C., Elfanagely, Y., D’Agata, E., and Moss, S. F. (2022). Clinical implication of drug resistance for H. pylori management. Antibiotics 11, 1684.
Arunava Das, A. D., Hari, S. S., Umachandran Shalini, U. S., Arumugam Ganeshkumar, A. G., and Magudeshwaran Karthikeyan, M. K. (2012). Molecular characterization of Salmonella enterica serovar Typhi isolated from typhoidial humans. Malaysian Journal of Microbiology 8 (3), 148–155. doi: 10.5555/20133057461
Arwansyah, A., Rahmawati, S., Nuryanti, S., Yusuf, Y., and Arif, A. R. (2025). Molecular investigation on active compounds in papaya leaves (Carica papaya linn) as anti-malaria using network pharmacology, molecular docking, clustering-based analysis and molecular dynamics simulation. Phytomedicine Plus 1 (5), 100713. doi: 10.2139/ssrn.5007313
Asad, M. H., Zubair, H. M., and Khan, M. I. (2021). Molecular docking and simulation studies of phytol as a potential inhibitor of DNA gyrase. Journal of Molecular Graphics and Modelling 101, 107713.
Asghar, M., Khan, T. A., Séraphin, M. N., Schimke, L. F., Cabral-Marques, O., Haq, I. U., et al. (2024). Exploring the antimicrobial resistance profile of Salmonella Typhi and its clinical burden. Antibiotics 13, 765. doi: 10.3390/antibiotics13080765
Askarinia, M., Ganji, A., Jadidi-Niaragh, F., Hasanzadeh, S., Mohammadi, B., Ghalamfarsa, F., et al. (2019). A review on medicinal plant extracts and their active ingredients against methicillin-resistant and methicillin-sensitive Staphylococcus aureus. J. Herbmed Pharmacol. 8, 173–184. doi: 10.15171/jhp.2019.27
Babalola, B. A., Akinwande, A. I., Otunba, A. A., Adebami, G. E., Babalola, O., and Nwufo, C. (2024). Therapeutic benefits of Carica papaya: A review on its pharmacological activities and characterization of papain. Arabian J. Chem. 17, 105369. doi: 10.1016/j.arabjc.2023.105369
Balouiri, M., Sadiki, M., and Ibnsouda, S. K. (2016). Methods for in vitro evaluating antimicrobial activity: A review. J. pharmaceutical Anal. 6, pp.71–pp.79. doi: 10.1016/j.jpha.2015.11.005
Banerjee, P., Eckert, A. O., Schrey, A. K., and Preissner, R. (2018). ProTox-II: a web server for the prediction of toxicity of chemicals. Nucleic Acid Research 46, W257–WW63. doi: 10.1093/nar/gky318
Bansal, S. and Tandon, V. (2011). Contribution of mutations in DNA gyrase and topoisomerase IV genes to ciprofloxacin resistance in Escherichia coli clinical isolates. Int. J. antimicrobial Agents 37, 253–255. doi: 10.1016/j.ijantimicag.2010.11.022
Bauer, A. W., Kirby, W. M. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Benoit, S. L. and Maier, R. J. (2016). Helicobacter catalase devoid of catalytic activity protects the bacterium against oxidative stress. Journal of Biological Chemistry 291 (45), 23366-23373.
Berger, A. (2000). Scientists discover how helicobacter survives gastric acid. BMJ. 320, 268. doi: 10.1136/bmj.320.7230.268
Blundell, T. L. and Ascher, D. B. (2015). pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. medicinal Chem. 58, 4066–4072. doi: 10.1021/acs.jmedchem.5b00104
Borba, J. V., Alves, V. M., Braga, R. C., Korn, D. R., Overdahl, K., Silva, A. C., et al. (2022). STopTox: An in-silico alternative to animal testing for acute systemic and topical toxicity. Environ. Health Perspect. 130 (2), 027012. doi: 10.1289/EHP9341
Butt, S. S., Badshah, Y., Shabbir, M., and Rafiq, M. (2020). Molecular docking using chimera and autodock vina software for nonbioinformaticians. JMIR Bioinf. Biotechnol. 1, e14232. doi: 10.2196/14232
Chanin, R. B., Winter, M. G., Spiga, L., Hughes, E. R., Zhu, W., Taylor, S. J., et al. (2020). Epithelial-derived reactive oxygen species enable AppBCX-mediated aerobic respiration of Escherichia coli during intestinal inflammation. Cell host Microbe 28, 780–788. doi: 10.1016/j.chom.2020.09.005
Chatham-Stephens, K. (2019). Emergence of extensively drug-resistant Salmonella Typhi infections among travelers to or from Pakistan—United States 2016–2018. MMWR. Morbidity Mortality Weekly Rep. 68, 11-13. doi: 10.15585/mmwr.mm6801a3
Chen, D., Oezguen, N., Urvil, P., Ferguson, C., Dann, S. M., and Savidge, T. C. (2016). Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2, e1501240. doi: 10.1126/sciadv.1501240
Chen, G., Seukep, A.J., and Guo, M. (2020). Recent advances in molecular docking for the research and discovery of potential marine drugs. Marine drugs 18 (11), 545. doi: 10.3390/md18110545
Cheok, Y. Y., Lee, C. Y. Q., Cheong, H. C., Vadivelu, J., Looi, C. Y., Abdullah, S., et al. (2021). An overview of Helicobacter pylori survival tactics in the hostile human stomach environment. Microorganisms 9, p.2502. doi: 10.3390/microorganisms9122502
Clinical and Laboratory Standards Institute (2012). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard (9th ed.) (CLSI document M07-A9) (Clinical and Laboratory Standards Institute).
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. reports. 7, 42717. doi: 10.1038/srep42717
Ejalonibu, M. A., Ogundare, S. A., Elrashedy, A. A., Ejalonibu, M. A., Lawal, M. M., Mhlongo, N. N., et al. (2021). Drug discovery for Mycobacterium tuberculosis using structure-based computer-aided drug design approach. Int. J. Mol. Sci. 22, 13259. doi: 10.3390/ijms222413259
Gauba, A. and Rahman, K. M. (2023). Evaluation of antibiotic resistance mechanisms in gram-negative bacteria. Antibiotics 12, p.1590. doi: 10.3390/antibiotics12111590
Ghaneian, M. T., Ehrampoush, M. H., Jebali, A., Hekmatimoghaddam, S., and Mahmoudi, M. (2015). Antimicrobial activity, toxicity and stability of phytol as a novel surface disinfectant. Environ. Health Eng. Manage. J. 2, pp.13–pp.16,
Gurram, S. R. and Azam, M. A. (2021). GyrB inhibitors as potential antibacterial agents: A review. Monatshefte für Chemie-Chemical Monthly 152, 725–744. doi: 10.1007/s00706-021-02800-z
Hahn, M. M., González, J. F., and Gunn, J. S. (2021). Salmonella biofilms tolerate hydrogen peroxide by a combination of extracellular polymeric substance barrier function and catalase enzymes. Front. Cell. Infection Microbiol. 11, 683081. doi: 10.3389/fcimb.2021.683081
Harborne, J. B. (1998). Phytochemical methods a guide to modern techniques of plant analysis, springer science & business media, 3rd ed.
He, J., Qiao, W., An, Q., Yang, T., and Luo, Y. (2020). Dihydrofolate reductase inhibitors for use as antimicrobial agents. Eur. J. Medicinal Chem. 195, 112268. doi: 10.1016/j.ejmech.2020.112268
Islam, M. A., Kabir, S. M. L., and Seel, S. K. (2016). Molecular detection and characterization of Escherichia coli isolated from raw milk sold in different markets of Bangladesh. Bangladesh J. Veterinary Med. 14, pp.271–pp.275. doi: 10.3329/bjvm.v14i2.31408
Jafari, E., Oloomi, M., and Bouzari, S. (2021). Characterization of antimicrobial susceptibility, extended-spectrum β-lactamase genes and phylogenetic groups of Shigatoxin producing Escherichia coli isolated from patients with diarrhea in Iran. Ann. Clin. Microbiol. Antimicrobials 20, 1–9. doi: 10.1186/s12941-021-00430-1
Joshi, T., Sharma, P., Joshi, T., Mathpal, S., Pande, V., and Chandra, S. (2022). Repurposing of FDA approved drugs against Salmonella enteric serovar Typhi by targeting dihydrofolate reductase: an in-silico study. J. Biomolecular Structure Dynamics 40, 3731–3744. doi: 10.1080/07391102.2020.1850356
Jubair, N., Rajagopal, M., Chinnappan, S., Abdullah, N. B., and Fatima, A. (2021). Review on the antibacterial mechanism of plant-derived compounds against multidrug-resistant bacteria (MDR). Evidence-Based Complementary Altern. Med. 2021, 3663315. doi: 10.1155/2021/3663315
Karunarathna, I., Gunasena, P., Gunathilake, S., Hapuarachchi, T., Ekanayake, U., Rajapaksha, S., et al. (2024). Clinical applications of gentamicin in treating gram-negative infections.
Katayama, T., Kasho, K., and Kawakami, H. (2017). The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 8, 2496. doi: 10.3389/fmicb.2017.02496
Kauffmann, A. C. and Castro, V. S. (2023). Phenolic compounds in bacterial inactivation: a perspective from Brazil. Antibiotics 12, p.645. doi: 10.3390/antibiotics12040645
Kim, J. H., Kim, S. L., Kim, K. C., Perera, R. M. T. D., Kim, S. C., and Lee, D. S. (2024). Bioactive constituents from Carica papaya fruit: implications for drug discovery and pharmacological applications. Appl. Biol. Chem. 67, 1–23. doi: 10.1186/s13765-023-00849-4
Kinoshita-Daitoku, R., Ogura, Y., Kiga, K., Maruyama, F., Kondo, T., Nakagawa, I., et al. (2020). Complete genome sequence of Helicobacter pylori strain ATCC 43504, a type of strain that can infect gerbils. Microbiol. Resource Announcements 9, 10–1128. doi: 10.1128/MRA.00105-20
Klemm, E. J., Shakoor, S., Page, A. J., Qamar, F. N., Judge, K., Saeed, D. K., et al. (2018). Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9, e00105–e00118. doi: 10.1128/mBio.00105-18
Kong, Y. R., Jong, Y. X., Balakrishnan, M., Bok, Z. K., Weng, J. K. K., Tay, K. C., et al. (2021). Beneficial role of Carica papaya extracts and phytochemicals on oxidative stress and related diseases: A mini review. Biol. (Basel) 10, 287. doi: 10.3390/biology10040287
Kowalska-Krochmal, B. and Dudek-Wicher, R. (2021). The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens 10, 165. doi: 10.3390/pathogens10020165
Krzyżek, P. and Gościniak, G. (2018). Morphology of Helicobacter pylori as a result of peptidoglycan and cytoskeleton rearrangements. Gastroenterology Review/Przegląd Gastroenterologiczny 13, pp.182–pp.195. doi: 10.5114/pg.2018.78284
Kuo, C. J., Guillemin, K., and Amieva, M. R. (2015). Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell host Microbe 18, 147–156. doi: 10.1016/j.chom.2015.07.002
Lazim, R., Suh, D., and Choi, S. (2020). Advances in molecular dynamics simulations and enhanced sampling methods for the study of protein systems. Int. J. Mol. Sci. 21, 6339. doi: 10.3390/ijms21176339
Lee, W., Woo, E. R., and Lee, D. G. (2016). Phytol has antibacterial property by inducing oxidative stress response in Pseudomonas aeruginosa. Free Radical Res. 50, 1309–1318. doi: 10.1080/10715762.2016.1241395
MacFaddin, J. F. (2000). Biochemical tests for identification of medical bacteria. 3rd ed (Lippincott Williams & Wilkins).
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. infection 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Manente, L., Perna, A., Buommino, E., Altucci, L., Lucariello, A., Citro, G., et al. (2008). The Helicobacter pylori’s protein VacA has direct effects on the regulation of cell cycle and apoptosis in gastric epithelial cells. J. Cell. Physiol. 214, pp.582–pp.587. doi: 10.1002/jcp.v214:3
Meng, X. Y., Zhang, H.X., Mezei, M., and Cui, M. (2011). Molecular docking: A powerful tool for drug discovery. J. Computer-Aided Mol. Design 7 (2), 146–57. doi: 10.2174/157340911795677602
Menon, S. S. and Piramanayakam, S. (2021). Insilico prediction of gyr A and gyr B in Escherichia coli insights the DNA-Protein interaction in prokaryotes. Int. J. Multidiscip. Res. Growth Evaluation,(IJMRD) 2, 709–714.
Mogana, R., Adhikari, A., Tzar, M. N., Ramliza, R., and Wiart, C.J.B.C.M. (2020). Antibacterial activities of the extracts, fractions and isolated compounds from Canarium patentinervium Miq. against bacterial clinical isolates. BMC complementary Med. therapies 20, 1–11. doi: 10.1186/s12906-020-2837-5
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi: 10.1002/jcc.v30:16
Newman, D. J. and Cragg, G. M. (2016). Natural products as sources of new drugs over the last 25 years. J. Natural Products 79, 629–661. doi: 10.1021/acs.jnatprod.5b01055
Nguyen, A. N. X., Thirapanmethee, K., Audshasai, T., Khuntayaporn, P., and Chomnawang, M. T. (2024). Insights into molecular mechanisms of phytochemicals in quorum sensing modulation for bacterial biofilm control. Arch. Microbiol. 206, p.459. doi: 10.1007/s00203-024-04171-5
Nisa, S., Khan, N., Shah, W., Sabir, M., Khan, W., Bibi, Y., et al. (2020). Identification and bioactivities of two endophytic fungi Fusarium fujikuroi and Aspergillus tubingensis from foliar parts of Debregeasia salicifolia. Arabian J. Sci. Eng. 45, 4477–4487. doi: 10.1007/s13369-020-04454-1
Nöllmann, M., Stone, M. D., Bryant, Z., Gore, J., Crisona, N. J., Hong, S. C., et al. (2007). Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 14, 264–271. doi: 10.1038/nsmb1213
Nurkhaliza, F., Risana, M. Z., Pubasari, A., Priatmoko, S., Prastya, M. E., and Andreani, A. S. (2024). “Comparative study of well diffusion and disc diffusion method to investigate the antibacterial properties of silver nanoparticles synthesized from Curcuma longa extracts,” in E3S Web of Conferences, vol. 503. (EDP Sciences), 09003.
O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., and Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. J. cheminformatics 3, 1–14. doi: 10.1186/1758-2946-3-33
Prasad, S. K., Bhatia, S., and Singh, R. (2021). Inhibition of Helicobacter pylori by Carica papaya leaf extract: A potential therapy for gastric diseases. J. Ethnopharmacology 267, 113686.
Qamar, F. N., Yousafzai, M. T., Khalid, M., Kazi, A. M., Lohana, H., Karim, S., et al. (2018). Outbreak investigation of ceftriaxone-resistant Salmonella enterica serotype Typhi and its risk factors among the general population in Hyderabad, Pakistan: a matched case-control study. Lancet Infect. Dis. 18, pp.1368–1376. doi: 10.1016/S1473-3099(18)30483-3
Rios, J. L., Recio, M. C., and Villar, A. (1988). Screening methods for natural products with antimicrobial activity: a review of the literature. J. ethnopharmacology 23, 127–149. doi: 10.1016/0378-8741(88)90001-3
Roberts, L. T., Issa, P. P., Sinnathamby, E. S., Granier, M., Mayeux, H., Eubanks, T. N., et al. (2022). Helicobacter pylori: A review of current treatment options in clinical practice. Life 12, 2038. doi: 10.3390/life12122038
Romasi, E. F., Karina, R., and Tjandrawinata, R. R. (2011). Antibacterial activity of papaya (Carica papaya L.) leaf extracts against pathogenic bacteria. Makara Journal of Technology, 15 (2), p.12.
Rubaka, C., Ndakidemi, P., and Shahada, H. M. M. F. (2014). Individual and combined antibacterial activity of crude extracts from medicinal plants Carissa spinarum Linn and Carica papaya Linn. European Journal of Medicinal Plants 4 (12), 1513–1523. doi: 10.9734/EJMP/2014/10599
Ruiz, J., Nunez, M. L., Diaz, J., Lorente, I., Perez, J., and Gomez, J. (1996). Comparison of five plating media for isolation of Salmonella species from human stools. J. Clin. Microbiol. 34, 686–688. doi: 10.1128/jcm.34.3.686-688.1996
Santana, L. F., Inada, A. C., Espírito Santo, B. L. S. D., Filiú, W. F. O., Pott, A., Alves, F. M., et al. (2019). Nutraceutical potential of Carica papaya in metabolic syndrome. Nutrients 11, 1608. doi: 10.3390/nu11071608
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., and Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Computer-Aided Mol. Design 27, 221–234. doi: 10.1007/s10822-013-9644-8
Shah, S. A. R., Rahman, H., Qasim, M., Akram, M. S., Saygideger, Y., Puspita, N., et al. (2022). Differential proteomics of helicobacter pylori isolates from gastritis, ulcer, and cancer patients: first study from northwest Pakistan. Medicina 58, 1168. doi: 10.3390/medicina58091168
Sharma, A., Sharma, R., Sharma, M., Kumar, M., Barbhai, M. D., Lorenzo, J. M., et al. (2022). Carica papaya L. leaves: Deciphering its antioxidant bioactives, biological activities, innovative products, and safety aspects. Oxid. Med. Cell. Longevity 2022, 2451733.
Sharma, P. R., Srivastava, S., and Kumar, A. (2022). Evaluation of phytol as a potential dihydrofolate reductase inhibitor: A molecular docking study. ChemistrySelect 7, e202201146.
Shen, W. and Downs, D. M. (2024). Tetrahydrofolate levels influence 2-aminoacrylate stress in Salmonella enterica. J. Bacteriology 206, pp.e00042–24. doi: 10.1128/jb.00042-24
Sofowora, A. (1993). Medicinal Plants and Traditional Medicine in Africa, 2nd ed. Sunshine House, Ibadan. 134-156.
Supriatin, Y., Sumirat, V. A., and Herdiani, M. (2021). “Growth Analysis of escherichia coli and Salmonella typhi on MacConkey Agar Modification,” in Journal of Physics: Conference Series (IOP Publishing), vol. 1764 (1), 012207.
Tripathi, N., Zubair, M., and Sapra, A. (2025). Gram Staining. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing.
Udinyiwe, O. C. and Omoregie, A. E. (2024). Determination of the Mineral Content, Phytochemical Properties, and the Antimicrobial Properties of the Seed Extracts of Carica papaya on some Clinical Isolates. Matrix Sci. Pharma 8, 37–44. doi: 10.4103/mtsp.mtsp_10_24
Ugbogu, E. A., Dike, E. D., Uche, M. E., Etumnu, L. R., Okoro, B. C., Ugbogu, O. C., et al. (2023). Ethnomedicinal uses, nutritional composition, phytochemistry and potential health benefits of Carica papaya. Pharmacol. Research-Modern Chin. Med. 7, 100266. doi: 10.1016/j.prmcm.2023.100266
Vitoriano, I., Rocha-Gonçalves, A., Carvalho, T., Oleastro, M., Calado, C. R., and Roxo-Rosa, M. (2011). Antigenic diversity among Portuguese clinical isolates of Helicobacter pylori. Helicobacter 16, 153–168. doi: 10.1111/j.1523-5378.2011.00825.x
Vu, T. B., Tran, T. N. Q., Tran, T. Q. A., Vu, D. L., and Hoang, V. T. (2022). Antibiotic resistance of Helicobacter pylori in patients with peptic ulcer. Medicina 59, p.6. doi: 10.3390/medicina59010006
Yahaya, M. S., Haruna, A., and Abubakar, S. (2017). Antibacterial activity of Carica papaya leaf extract on some bacterial pathogens. Greener J. Biol. Sci. 7, 1–5. doi: 10.15580/GJBS.2017.1.122916224
Zandavar, H. and Babazad, M. A. (2023). “Secondary metabolites: Alkaloids and flavonoids in medicinal plants,” in Herbs and Spices-new Advances (IntechOpen). doi: 10.5772/intechopen.108030
Keywords: antibiotic resistance of bacterial strains, inflammation, gastrointestinal infections, Carica papaya, antimicrobial compounds, molecular docking
Citation: Alhodieb FS, Farid M, Sabir M, Nisa S, Sarwar S and Abbas S (2025) Exploring the bioactive compounds of Carica papaya leaves: phytol’s role in combatting antibiotic-resistant bacteria. Front. Cell. Infect. Microbiol. 15:1564787. doi: 10.3389/fcimb.2025.1564787
Received: 22 January 2025; Accepted: 21 May 2025;
Published: 07 July 2025.
Edited by:
Cynthia A. Danquah, Kwame Nkrumah University of Science and Technology, GhanaReviewed by:
Sudharshan S. J., University of Tennessee, United StatesNeha Kapoor, Suresh Gyan Vihar University, India
Copyright © 2025 Alhodieb, Farid, Sabir, Nisa, Sarwar and Abbas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sidra Abbas, c2lkcmFhc0BnbWFpbC5jb20=
 Fahad Saad Alhodieb
Fahad Saad Alhodieb Maira Farid2
Maira Farid2 Maimoona Sabir
Maimoona Sabir Sobia Nisa
Sobia Nisa Sumaira Sarwar
Sumaira Sarwar Sidra Abbas
Sidra Abbas